- 1Rowland Institute at Harvard, Harvard University, Cambridge, MA, USA
- 2Department of Biological Sciences, Vanderbilt University, Nashville, TN, USA
- 3Department of Pathology, Microbiology, and Immunology, Vanderbilt University, Nashville, TN, USA
- 4Institute of Integrative Biology, University of Liverpool, Liverpool, UK
The parasitoid wasp genus Nasonia (Hymenoptera: Chalcidoidea) is a well-established model organism for insect development, evolutionary genetics, speciation, and symbiosis. The host-microbiota assemblage which constitutes the Nasonia holobiont (a host together with all of its associated microbes) consists of viruses, two heritable bacterial symbionts and a bacterial community dominated in abundance by a few taxa in the gut. In the wild, all four Nasonia species are systematically infected with the obligate intracellular bacterium Wolbachia and can additionally be co-infected with Arsenophonus nasoniae. These two reproductive parasites have different transmission modes and host manipulations (cytoplasmic incompatibility vs. male-killing, respectively). Pioneering studies on Wolbachia in Nasonia demonstrated that closely related Nasonia species harbor multiple and mutually incompatible Wolbachia strains, resulting in strong symbiont-mediated reproductive barriers that evolved early in the speciation process. Moreover, research on host-symbiont interactions and speciation has recently broadened from its historical focus on heritable symbionts to the entire microbial community. In this context, each Nasonia species hosts a distinguishable community of gut bacteria that experiences a temporal succession during host development and members of this bacterial community cause strong hybrid lethality during larval development. In this review, we present the Nasonia species complex as a model system to experimentally investigate questions regarding: (i) the impact of different microbes, including (but not limited to) heritable endosymbionts, on the extended phenotype of the holobiont, (ii) the establishment and regulation of a species-specific microbiota, (iii) the role of the microbiota in speciation, and (iv) the resilience and adaptability of the microbiota in wild populations subjected to different environmental pressures. We discuss the potential for easy microbiota manipulations in Nasonia as a promising experimental approach to address these fundamental aspects.
Introduction
Bacterial symbionts are widely recognized as important drivers of insect physiology, development, behavior, reproduction, nutrition, and evolution (Buchner, 1965; Moran, 2007; Douglas, 2010, 2015). Historically, symbiosis research focused primarily on binary interactions between insect hosts and particular symbionts, whether they are harmful or helpful (Duron et al., 2008; Moran et al., 2008; Moya et al., 2008; Werren et al., 2008). The advent of new DNA sequencing technologies over the last 10 years resulted in what has recently been termed ‘the microbiome revolution’ (Blaser, 2014), providing an unprecedented wealth of information on insect microbiotas from various species. Biologists now recognize that symbioses are shaped by complex multipartite interactions, not only between the host and its associated microbes, but also between different members of the microbial community and the environment. This understanding has led to the view of hosts as complex ecosystems (McFall-Ngai et al., 2013; Sicard et al., 2014), and to the recognition that a more holistic approach is needed to understand the role of the microbiota in major facets of host biology (Gilbert et al., 2012). In insects, the microbiota can modulate numerous host phenotypes spanning development (Shin et al., 2011), nutrition (Chandler et al., 2011; He et al., 2013; Wong et al., 2014), immunity (Chu and Mazmanian, 2013), vector competence and susceptibility to pathogen infection (Dong et al., 2009; Koch and Schmid-Hempel, 2012), among others. The microbiota can also mediate reproductive isolation and thus the mechanisms that drive speciation (Brucker and Bordenstein, 2012c, 2013; Shropshire and Bordenstein, 2016), underscoring the need to understand host-microbiota dynamics over evolutionary timescales.
The recognition of the significance and complexity of host-microbiota interactions has led to the revival of old terms and the establishment of new ones to describe host-microbiota assemblages: As such, the term “holobiont”, originally coined by Margulis (1991), is now frequently used to refer to a host together with its entire microbial consortium, while the “hologenome” encompasses the genomes of all members of the holobiont (Rosenberg et al., 2007; Zilber-Rosenberg and Rosenberg, 2008). These terms provide structural definitions that can be universally applied to any host-microbiota assemblage. Moreover, they are pluralistic in that they encompass constant or inconstant, intracellular or extracellular, horizontally or vertically transmitted, harmful or helpful microbial symbionts (Rosenberg et al., 2007; Bordenstein and Theis, 2015; Theis et al., 2016). This perception of a holobiont therefore embraces both competition and cooperation between a host and its associated microbes. This is particularly obvious in the case of symbionts like Wolbachia, which override host reproduction to increase their own transmission (Duron et al., 2008; Werren et al., 2008). More generally, the microbial partners present in a host organism contribute to the “extended phenotype” of this particular host-symbiont assemblage, i.e., the holobiont. However, many aspects regarding holobionts need to be elucidated: For instance, one may ask whether phenotypic variation in traits, caused by different holobiont assemblies, could drive a multigenerational response to selection, as originally proposed as part of the hologenome concept of evolution (Rosenberg et al., 2007; Zilber-Rosenberg and Rosenberg, 2008). Moreover, if there is a response to selection, does it occur at the host, microbe, or microbial community level? While the broad utility of the hologenome concept remains debated (Bordenstein and Theis, 2015; Moran and Sloan, 2015; Douglas and Werren, 2016; Theis et al., 2016), it is clear that the microbiome represents an important component of insect biology as well as a source of phenotypic and evolutionary novelty.
With tools available to investigate the diversity and complexity of host-microbe associations, the next challenge will be to disentangle the holobiont in a functional context to understand (i) how different microbes, alone or in synergy, contribute to host phenotype and fitness; (ii) the role of the host, the symbionts and the environment in establishing and regulating the microbiota with each generation; (iii) the role of the microbiota in evolutionary processes such as speciation; and (iv) the resilience and adaptability of the microbiota in wild populations subjected to different environmental pressures.
In this review, we present the parasitoid wasp genus Nasonia (Hymenoptera: Chalcidoidea) as an excellent model to experimentally investigate fundamental aspects and evolutionary dynamics of host-microbiota interactions. In particular, we focus on how symbiotic bacteria – both intracellular and the extracellular microbiota – influence Nasonia biology, reproduction, and speciation.
Nasonia As A Model Organism
The parasitoid wasp genus Nasonia (also referred to as “jewel wasp”) is a species complex comprised of four interfertile species: N. vitripennis, N. longicornis, N. giraulti, and N. oneida (Figure 1A) (Darling and Werren, 1990; Raychoudhury et al., 2010a). The older species N. vitripennis is estimated to have diverged from the three younger species 1 million years ago (mya). The other species were discovered only in the last 26 years and have diverged 0.4 mya in the case of N. longicornis and N. giraulti and 0.3 mya in the case of N. giraulti and N. oneida (Werren and Loehlin, 2009; Werren et al., 2010). While N. vitripennis is cosmopolitan (Figure 1B), the three younger species have only been observed in North America, where they show species-specific distributions: N. longicornis is restricted to the west, N. giraulti to the northeast and the most recently discovered species N. oneida has so far only been observed in New York state (Figure 1C) (Darling and Werren, 1990; Raychoudhury et al., 2010a). All species are parasitoids of fly pupae (Werren and Loehlin, 2009; Desjardins et al., 2010). Adult Nasonia females lay their eggs within the fly puparium (Figure 1D) and inject a venom that prevents the fly from mounting an immune response against the intruders (Danneels et al., 2014). A single fly pupa may be parasitized by multiple females of the same or different species (superparasitism and multiparasitism, respectively) (Darling and Werren, 1990). Under laboratory conditions (constant temperature of 25°C), Nasonia has a short generation time of only 14 days: Larvae emerge 24-36 h after egg laying and undergo four larval instars (during which they feed on the fly host), followed by pupation after 7-8 days and emergence from the fly as adults (Figure 1D).
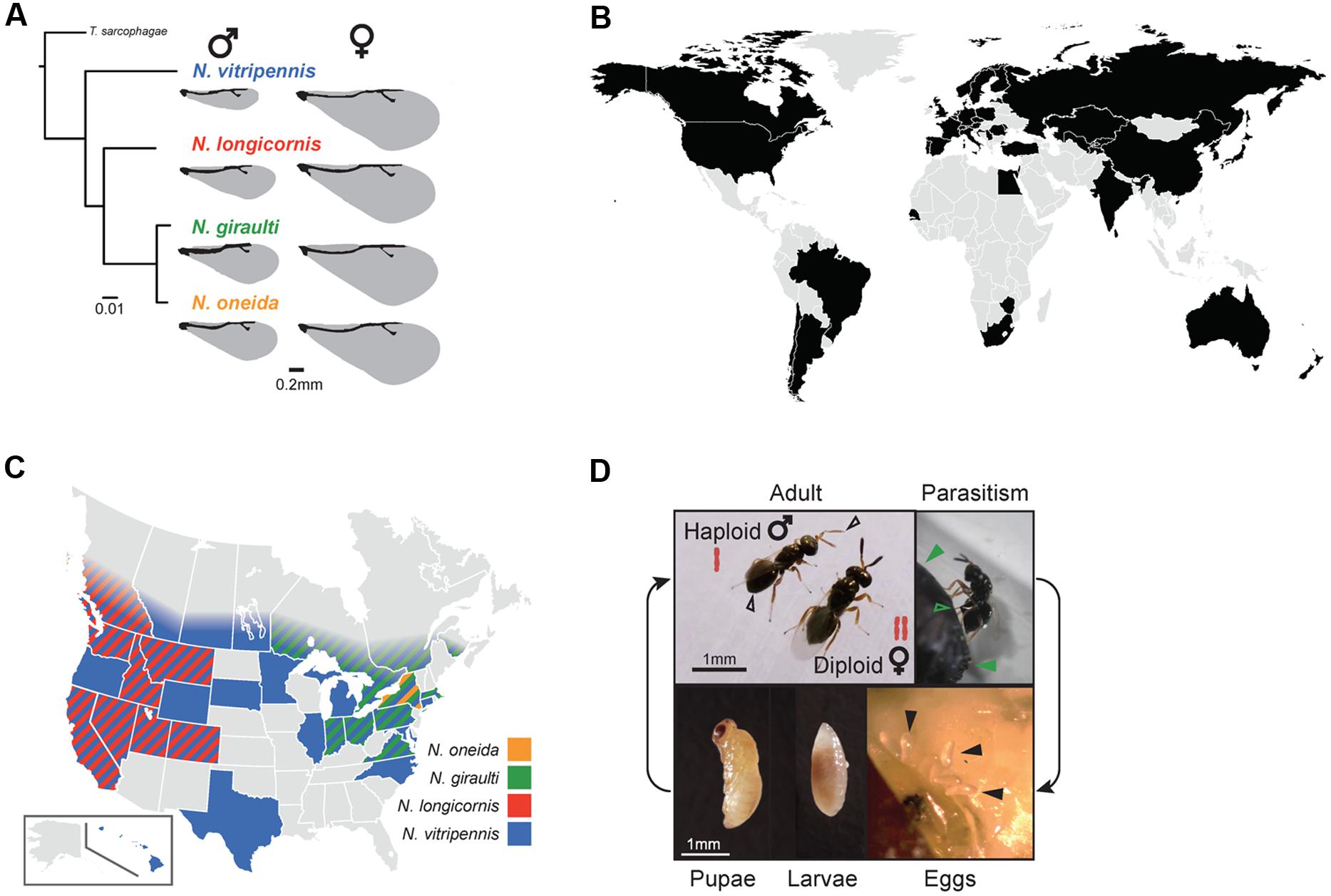
FIGURE 1. (A) Phylogenetic relationships within the Nasonia species complex based on the CO1 gene. The parasitoid wasp Trichomalopsis sarcophagae was used as outgroup. The scale bar indicates substitutions per site. Note the morphological differences in wing size between species and genders (drawing based on Loehlin et al., 2010). (B) Countries in which Nasonia has been observed, based on published records of N. vitripennis [Universal Chalcidoidea Database (Noyes, 2016) and (Raychoudhury et al., 2009, 2010b; Paolucci et al., 2013)]. The gray color indicates countries for which no observations are documented, in most cases due to missing sampling information. For all countries except the US, records state that the observed species was Nasonia vitripennis – however, many of these observations were made before the discovery of the three younger species in the US. Therefore, this global map shows observations of Nasonia without distinguishing between species. (C) Observations of all four Nasonia species in the US and Canada, based on published information (Darling and Werren, 1990; Raychoudhury et al., 2009, 2010a,b). (D) Nasonia life cycle from oviposition to adulthood. Sex-specific differences in ploidy are indicated for adult males and females. Additional sexual dimorphisms include smaller wing size, less pigmented antennae and rounded abdomen in males (open black arrows). Parasitism is representing by a female wasp ovipositing into a fly pupa (closed green arrows: Fly pupa; open green arrows: Ovipositor of the wasp). Embryos are approximately 100 μm by 500 μm in size (closed black arrows). The Nasonia larva and pupa were photographed outside of their fly host. Photo credit: Matthew C. Johnson © 2016
Nasonia is a well-established model for insect development (Rosenberg et al., 2014), behavior (Bertossa et al., 2013), sex determination (Beukeboom and van de Zande, 2010; Verhulst et al., 2010), evolutionary genetics (Desjardins et al., 2010, 2013; Loehlin et al., 2010), immunity (Tian et al., 2010; Brucker et al., 2012; Sackton et al., 2013), speciation (Breeuwer and Werren, 1995; Ellison et al., 2008; Buellesbach et al., 2013; Gibson et al., 2013) and symbiosis with the reproductive parasites Wolbachia (Breeuwer and Werren, 1990; Bordenstein et al., 2001; Raychoudhury et al., 2009) and Arsenophonus (Huger et al., 1985; Gherna et al., 1991; Ferree et al., 2008). In addition, Nasonia is emerging as a model for insect-gut microbiota symbioses across recent evolutionary time periods and speciation events (Brucker and Bordenstein, 2012b, 2013). Major reasons for this versatility are its ease of rearing in the laboratory, the ability to establish interspecies hybrids after curing of Wolbachia, and the advantages of haplodiploid sex determination, wherein males and females develop from unfertilized (haploid) and fertilized (diploid) eggs, respectively (Figure 1D). Haplodiploidy is particularly useful for quantitative genetics as all recessive alleles are expressed in the male haploid state (Werren and Loehlin, 2009).
Perhaps the most recognizable aspect of Nasonia biology is the body of literature on speciation. Several types of hybrid maladies have been studied in Nasonia, including cytonuclear, cytoplasmic and microbiota-nuclear incompatibilities (Figure 2). Specifically, cytonuclear incompatibilities exist between mitochondrial and nuclear genes presumably involved in the oxidative phosphorylation pathway, leading to reduced energy production and hybrid fitness (Ellison et al., 2008; Niehuis et al., 2008; Koevoets et al., 2012; Gibson et al., 2013). On the other hand, cytoplasmic and microbiota-nuclear incompatibilities result from the influence of bacterial endosymbionts (e.g., Wolbachia) or the extracellular microbiota on reproductive fitness, either by affecting offspring viability at the embryonic stage or by altering the immune response of developing larvae, respectively (Bordenstein et al., 2001; Brucker and Bordenstein, 2013). These incompatibilities influence Nasonia reproductive isolation in an additive fashion, since removal of one incompatibility does not necessarily remove the others (Brucker and Bordenstein, 2013).
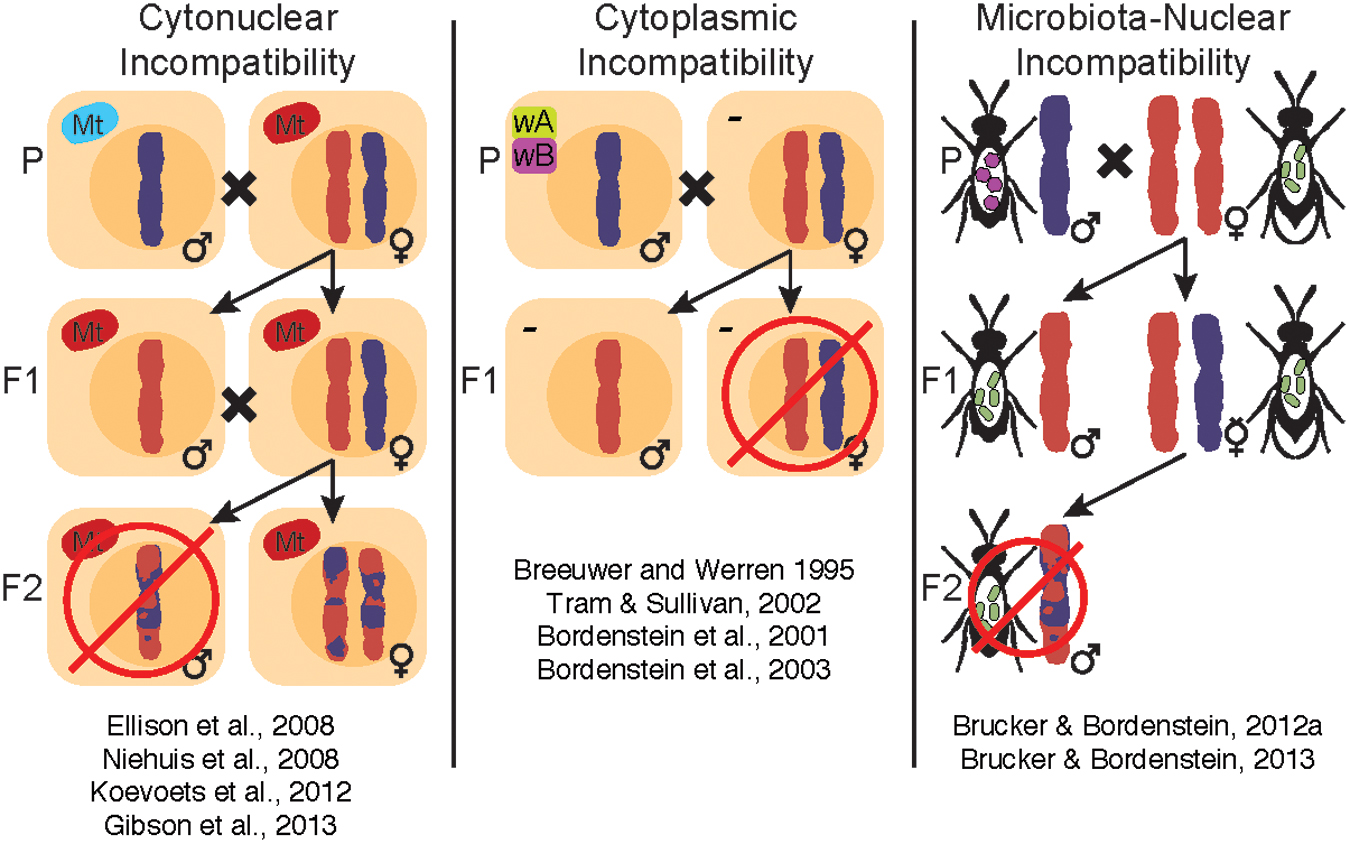
FIGURE 2. Within the Nasonia clade, there are three published sources of hybrid incompatibilities: cytonuclear, cytoplasmic, and microbiota-nuclear incompatibilities. Cytonuclear incompatibilities, or negative interactions between mitochondria and the nuclear genome, are associated with lethality in F2 males from younger interspecific crosses (N. giraulti and N. longicornis) and near complete lethality in older interspecific crosses (N. vitripennis and N. giraulti or N. longicornis). Hybrid lethality has some plasticity due to environmental factors (Koevoets et al., 2012), but clear cytonuclear incompatibilities that complicate development and gene regulation (Ellison et al., 2008; Niehuis et al., 2008; Koevoets et al., 2012; Gibson et al., 2013) have been genetically mapped across the Nasonia genomes. Cytoplasmic incompatibilities are a consequence of infection with different Wolbachia strains (wA and wB), which causes post-fertilization chromatin defects that result in inviable fertilized eggs (Bordenstein et al., 2001, 2003; Tram and Sullivan, 2002). Finally, microbiota-nuclear incompatibilities result from negative interactions between the microbiota and host genome and lead to hybrid lethality, altered microbial communities and innate immune regulation (Brucker and Bordenstein, 2013). The collective influences of these incompatibilities on Nasonia make it a powerful model for evolutionary and symbiotic studies of speciation and reproductive isolation. How these incompatibilities have evolved relative to each other is an important avenue for future research.
The growing interest in Nasonia has also resulted in a wealth of available resources, many of which are advantageous for the study of host-microbe interactions: Annotated genomes are available for all species except N. oneida (Werren et al., 2010), together with an extensive genetic toolbox (reviewed in Werren and Loehlin, 2009; Lynch, 2015), transcriptome and methylome for N. vitripennis (Sackton et al., 2013; Wang et al., 2013; Beeler et al., 2014), a well-characterized complex innate immune system (Tian et al., 2010; Brucker et al., 2012; Sackton et al., 2013) and a procedure for host genetic manipulation via RNAi (Lynch and Desplan, 2006; Werren et al., 2009). The most promising technique for the purpose of this review is the recently developed in vitro rearing technique, allowing the successful rearing of Nasonia from embryos to adults outside of its fly host (Brucker and Bordenstein, 2012a; Shropshire et al., 2016), thereby providing the means to establish axenic and gnotobiotic lineages.
Two Reproductive Parasites With Different Lifestyles
A prominent feature of insect symbioses is that many species entertain long-lasting associations with heritable obligate mutualistic endosymbionts (primary symbionts) (Moran et al., 2008; Moya et al., 2008; Koga et al., 2013). While this type of symbiosis is absent in Nasonia, all Nasonia species harbor a different type of heritable endosymbiont - reproductive parasites. Instead of conferring an obvious benefit to their host, these bacteria are facultative symbionts that have evolved different strategies to manipulate host reproduction in order to promote their own vertical transmission from mother to offspring (Hurst and Frost, 2015). The most common phenotype in insects is cytoplasmic incompatibility (CI), a reproductive incompatibility between sperm and egg preventing normal mitosis (Serbus et al., 2008). Other reproductive manipulations result in female-biased sex-ratios caused by parthenogenesis, male-killing or the feminization of genetic males (Stouthamer et al., 1993; Hurst et al., 2003; Narita et al., 2007; Bouchon et al., 2008). The common theme of these reproductive manipulations is that they increase the number of infected females in host populations, thereby enhancing maternal symbiont transmission. Bacteria of the genus Wolbachia (Alphaproteobacteria) are by far the best-studied and the most frequently encountered reproductive parasites (Hilgenboecker et al., 2008; Werren et al., 2008; Sicard et al., 2014), but other bacteria (Rickettsia (Alphaproteobacteria), Arsenophonus (Gammaproteobacteria), Cardinium and Flavobacterium (Bacteroidetes) and Spiroplasma (Mollicutes)] are also able to induce at least one reproductive manipulation (Duron et al., 2008; Hurst and Frost, 2015).
In the wild, all four Nasonia species are ubiquitously infected (100%) with Wolbachia in North America and Eurasia (Bordenstein et al., 2001; van Opijnen et al., 2005; Raychoudhury et al., 2009; Raychoudhury et al., 2010b), while a fraction of N. vitripennis and N. longicornis females (approximately 5%) additionally carry Arsenophonus nasoniae (Gherna et al., 1991; Balas et al., 1996). These symbionts are highly different in terms of reproductive manipulation -Wolbachia induces CI, while Arsenophonus is a male-killer (historically referred to as the ‘Son-Killer’ in Nasonia (Skinner, 1985; Gherna et al., 1991)). The two symbionts also differ in their vertical transmission mechanisms: Wolbachia are transmitted transovarially (Breeuwer et al., 1992), while Arsenophonus depends on horizontal or environmental transmission and establishes new infections after ingestion (Werren et al., 1986; Gherna et al., 1991; Parratt et al., 2016). Therefore, these associations provide ample opportunities for studying the evolution of different symbiotic lifestyles in bacteria and the role of bacterial endosymbionts on host physiology and evolution, including reproductive processes and speciation.
Wolbachia, An Influential Partner Over Evolutionary Time-Scales
Wolbachia are widespread obligate intracellular Alphapro-teobacteria, estimated to infect 40-65% of insect species (Hilgenboecker et al., 2008; Werren et al., 2008; Zug and Hammerstein, 2012). While being primarily maternally transmitted, horizontal transfers have frequently occurred over evolutionary time-scales (O’Neill et al., 1992; Rousset et al., 1992; Werren et al., 1995). To date, Wolbachia strains are divided into 16 clades, referred to as “supergroups” A-Q (Lo et al., 2007; Ramírez-Puebla et al., 2015).
The Nasonia species complex has been a major model system for Wolbachia-insect symbioses for more than 25 years (Breeuwer and Werren, 1990). This young species complex has enabled scientists to reconstruct the history of Wolbachia acquisitions and transmission routes across the Nasonia clade (van Opijnen et al., 2005; Raychoudhury et al., 2009). Moreover, the ability to produce interspecies hybrids has been exploited to introgress the cytotype (including the Wolbachia) of a given species into the nuclear genotype of another, thereby providing insights into Wolbachia-host genotype interactions, different modes of CI and the role of Wolbachia in speciation (Breeuwer and Werren, 1990, 1993b, Bordenstein and Werren, 1998; Bordenstein et al., 2001, 2003; Chafee et al., 2011; Raychoudhury and Werren, 2012).
The emerging picture of the Nasonia-Wolbachia association is as follows: The four Nasonia species together harbor 11 different Wolbachia strains from the A and B supergroups (Figure 3A) (Raychoudhury et al., 2009), two major arthropod-Wolbachia clades that diverged about 60 million years ago (Werren et al., 1995). N. vitripennis carries two Wolbachia strains (one from each supergroup), while the three younger species are all triple infected: N. giraulti and N. oneida both harbor two supergroup A strains and one supergroup B strain, whereas N. longicornis harbors one supergroup A strain and two supergroup B strains (Figure 3A) (Raychoudhury et al., 2009). Comparing the phylogenetic relationships between these Wolbachia strains with host phylogenies based on nuclear and mitochondrial genes revealed that several Wolbachia strains were most likely acquired independently via horizontal transfers from other insects, including Drosophila spp. (supergroup A strains of N. giraulti and N. longicornis), the parasitoid wasp Muscidifurax uniraptor (supergroup A strain of N. vitripennis) as well as the blowfly Protocalliphora sialia (supergroup B strain of N. vitripennis) (van Opijnen et al., 2005; Raychoudhury et al., 2009). The latter cases point towards an ecological interaction as the source of the Wolbachia transfers to N. vitripennis, since both Nasonia and M. uniraptor parasitize blowflies. Indeed, horizontal transfers between parasitoids and their fly hosts as well as between different parasitoid species infecting the same host are known to occur occasionally (Heath et al., 1999; Vavre et al., 1999; Huigens et al., 2004).
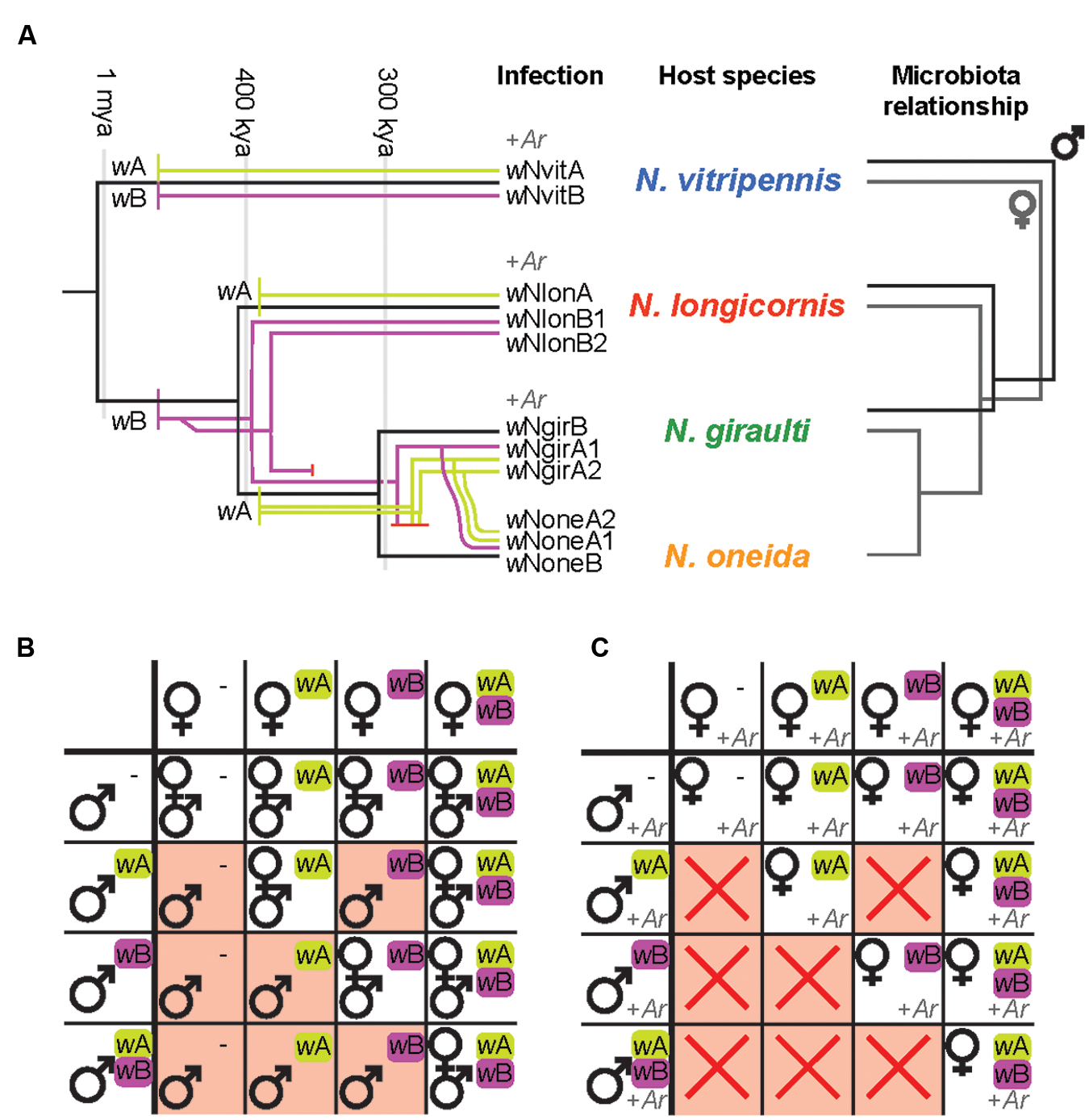
FIGURE 3. (A) Wolbachia-Nasonia associations and phylosymbiosis (modified from Raychoudhury et al., 2009; Brucker and Bordenstein, 2012b,c). Wolbachia acquisitions and subsequent divergence are overlaid on the Nasonia phylogeny. Strains from Wolbachia supergroup A are represented in green, strains from supergroup B in purple. Arsenophonus (+Ar) has been found to infect three species of Nasonia. The microbial community relationships parallel the host phylogeny, indicating species-specific microbiota assemblies that establish phylosymbiosis. This pattern has been observed in males for three species (Brucker and Bordenstein, 2012b, 2013) as well as in females for all four species (R. M. Brucker and S. R. Bordenstein, personal communication). (B) Impact of Wolbachia-induced CI on offspring production. Wolbachia present in males induce a sperm modification that needs to be rescued by the same Wolbachia strain in the fertilized egg for normal offspring production. CI (red background) occurs if the female is uninfected (-) (unidirectional CI) or harbors a different Wolbachia strain (bidirectional CI) and results in male-only (or male-biased) broods due to loss of the paternal chromosomes. Note that although Wolbachia modify male sperm, the symbiont is only maternally transmitted. Offspring will therefore harbor the same Wolbachia strain(s) as their mothers. wA/wB indicate different Wolbachia strains. (C) Impact of Arsenophonus-induced male-killing on offspring production, in combination with Wolbachia-mediated CI. Male-killing results in all-female broods in the absence of CI (white background) and no offspring production in combination with CI (red crosses), since the males that are not affected by CI would be killed by male-killing.
The supergroup B Wolbachia of N. longicornis and N. giraulti were acquired prior to the speciation of the two species and subsequently co-diverged with their hosts (Figure 3A) (van Opijnen et al., 2005; Raychoudhury et al., 2009). Hence, the supergroup B strain from N. giraulti is nearly identical to one of the B strains from N. longicornis and the estimated divergence time of the two Wolbachia strains coincides with the divergence of their host species, i.e., about 0.4-0.5 mya (van Opijnen et al., 2005; Raychoudhury et al., 2009). In addition, the second B strain of N. longicornis is estimated to have diverged from the other B strains about 1.5 mya. Considering that this time point was long before the speciation of the two host species, it is likely that the common ancestor of N. longicornis and N. giraulti harbored two B strains, one of which was lost in N. giraulti after the speciation event (Raychoudhury et al., 2009). A similar co-divergence event between the A strains of N. longicornis and N. giraulti is possible, but the similarity of these strains with Wolbachia from several Drosophila species currently makes it impossible to rule out independent horizontal transfer events (Raychoudhury et al., 2009). The three Wolbachia strains in the recently discovered species N. oneida are identical (for 5 house-keeping genes and the wsp gene) to those of the closely related N. giraulti and have likely been acquired via hybridisation between the two species, resulting in a mitochondrial-Wolbachia sweep from N. giraulti to N. oneida (Figure 3A) (Raychoudhury et al., 2009). Future phylogenomic comparisons based on the entire genomes of the different Wolbachia strains will be needed to obtain a higher resolution. Nonetheless, these findings illustrate a high Wolbachia diversity in the Nasonia species complex, along with various patterns of Wolbachia transfers within a single insect genus.
One of the most prominent aspects of Wolbachia is undoubtedly its ability to manipulate host reproduction in various ways (Werren et al., 2008). All Nasonia-associated Wolbachia induce CI, a reproductive incompatibility consisting of two components: A symbiont-induced modification of the paternal chromosomes during spermatogenesis that needs to be ‘rescued’ by the same symbiont being present in the fertilized egg (Tram and Sullivan, 2002; Serbus et al., 2008). If the female is uninfected (unidirectional CI) or harbors a different bacterial strain (bidirectional CI), the modification may not be rescued, resulting in CI (Figure 3B). Consequently, infected females have a fitness advantage over uninfected females, since they can reproduce successfully with all available males, regardless of male infection status. Similarly, bidirectional CI results in a fitness benefit for multiply infected females since they are at lower risk to suffer from CI (Mouton et al., 2003, 2004). The fact that all Nasonia species generally harbor double or triple infections that are mutually incompatible reinforces bidirectional incompatibility between all species pairs, with the notable exception of N. giraulti and N. oneida, whose Wolbachia strains are identical (Breeuwer and Werren, 1990; Breeuwer et al., 1992; Bordenstein and Werren, 1998, 2007; Bordenstein et al., 2001, 2003; Raychoudhury et al., 2010a). While the exact molecular mechanisms of CI are still not understood, it is evident that the paternal chromosomes fail to condense correctly and may be lost during the first mitotic division, causing embryo mortality in diploid organisms (Tram and Sullivan, 2002). In contrast, in haplodiploid insects such as Nasonia, CI can be manifested in different ways. The most extreme phenotypes are Male Development and Female Mortality (Vavre et al., 2000; Bordenstein et al., 2003; Vavre et al., 2009). In the first case, the paternal genome is completely lost, which restores haploidy in fertilized eggs and results in the conversion of female into male offspring. Hence, this type of CI is characterized by all-male broods (Figure 3B) with little or no embryonic mortality compared to compatible crosses. In contrast, an incomplete loss of the paternal chromosomes would instead cause aneuploidy and a high mortality of fertilized eggs (i.e., female offspring), resulting in smaller broods with male-biased sex-ratios (Bordenstein et al., 2003). It has been hypothesized that paternal chromosome loss was mediated by the intensity of Wolbachia-induced sperm modification, with less efficient modifications leading to only partial chromosome loss and aneuploidy in fertilized eggs (Vavre et al., 2009). While CI-induced mortality seems to be common in haplodiploid species, including the younger Nasonia species N. longicornis and N. giraulti (Bordenstein et al., 2003; Dedeine et al., 2004; Vavre et al., 2009), N. vitripennis is an exception from the rule in that the Male Development type is the predominant CI phenotype in this species (Breeuwer and Werren, 1990; Bordenstein et al., 2003). Interestingly, interspecies crosses and introgression experiments between N. vitripennis and N. giraulti revealed that this rare CI phenotype is determined by the N. vitripennis nuclear genotype rather than Wolbachia-related effects, since it is even observed in incompatible crosses between infected N. giraulti males and uninfected N. vitripennis females (Bordenstein et al., 2003). These results show that the host genotype may determine the fate of the paternal chromosomes in fertilized eggs, independent of Wolbachia (Bordenstein et al., 2003). On an evolutionary scale, the conversion of incompatible fertilized eggs into viable haploid males may represent a selective advantage for the more widely distributed N. vitripennis, in that it prevents embryo mortality as a consequence of incompatible matings with its microsympatric sister species (Bordenstein et al., 2003). Taken together, the above illustrates the preeminent role of Nasonia as a model to understand host-Wolbachia interactions, notably in terms of diverse acquisition routes, Wolbachia-host genotype interactions and modes of CI.
Arsenophonus, the Son-Killer
The genus Arsenophonus lies in the Gammaproteobacteria and is most closely related to the genera Proteus, Providencia, and Photorhabdus. Members of this genus establish diverse symbiotic interactions with insect hosts, with around 5% of insect species estimated to carry the symbiont (Duron et al., 2008). Many, but not all interactions are based on heritable symbiosis – passage from female to her progeny. However, unlike many heritable symbionts (e.g., Wolbachia), there is substantial diversity in the transmission biology of this microbe. Some strains maintain substantial ‘infectious’ transmissions through the environment, others show vertical transmission via maternal transfer of the microbe to the egg surface followed by ingestion of the symbiont by the hatching larva, and others show classical maternal inheritance through the egg cytoplasm (see Table 1 for the diversity of Arsenophonus-host interactions).
Arsenophonus nasoniae, the symbiont of Nasonia wasps, represents the type species of the genus (Gherna et al., 1991). The discovery of the symbiont followed observations of maternally inherited sex ratio biases in certain N. vitripennis isofemale lines. These lineages were typified by the presence of all (or near all) female broods associated with the death of male progeny, referred to as the ‘son-killer’ trait (Figure 3C) (Skinner, 1985). The trait was heritable through the female line. Subsequent microscopical examination recorded diffuse microbial infections throughout the soma of affected females (Huger et al., 1985). Unusual for heritable microbes, the infection was relatively easily isolated into pure culture, and reinjection into fly pupae alongside ovipositing Nasonia led to establishment of the son-killer trait, fulfilling Koch’s postulates for this microbe (Werren et al., 1986). Later, male-killing was observed to be confined to unfertilized (rather than simply haploid) eggs, and associated with destruction of the maternal centrosome upon which unfertilized eggs depend for development, while diploids, which also inherit a paternal centrosome, do not (Ferree et al., 2008).
The early experiments of Skinner (1985) demonstrated that unlike other heritable microbes known at the time, A. nasoniae possessed the capacity for infectious transmission in addition to maternal inheritance. He observed that when an infected and uninfected female used the same host fly pupa (superparasitism), the progeny from the uninfected female acquired the infection, which would then be passed on in turn by them to their progeny. The infectious transmission is a result of the microbe not being inherited within eggs, but passed through calyx fluid deposited in the fly pupa during oviposition. The microbe grows saprophytically inside the deceased fly puparium, and is ingested by the developing Nasonia larvae, whether they are the progeny of the infected female or are derived from co-parasitising individuals. Following ingestion, the microbe enters the wasp through the gut epithelium to produce the diffuse infection seen in the adult soma. This infection includes presence in the calyx gland, thus ensuring A. nasoniae onward transmission. Arsenophonus thus has a saprophytic stage, a stage where it is a component of the gut microbiota, and a systemic infection stage.
This unusual life cycle is distinct from many other heritable microbe-host interactions and has important biological consequences in terms of the population dynamics of A. nasoniae infections in natural populations. First, superparasitism (and the infectious transmission that follows from it) apparently represents a necessary condition for the maintenance of the microbe (Parratt et al., 2016). In laboratory emulation, A. nasoniae was lost from wasp populations where females were forced to oviposit alone, because vertical transmission is leaky (10% of progeny do not inherit the microbe). In contrast, where females are forced to oviposit in patches with other females and superparasitism rates are high, the infection spreads to fixation in just 3-5 generations. In experiments where the opportunity for superparasitism varies between these global absence/presence extremes, the prevalence achieved is a function of the superparasitism opportunity. Thus, in contrast to other heritable reproductive parasitic microbes, infectious transmission is necessary for A. nasoniae maintenance, and son-killing appears to represent a secondary benefit to the microbe.
A likely consequence of this transmission biology is also the presence of multiple A. nasoniae strains within an individual. The presence of co-infections is likely to erode the correlation between host and microbe fitness that selects for benign and beneficial infections, as the ability to compete (and outcompete) other strains is also a fitness-related trait for the microbe. In this context, it is notable that the genome carries colicin elements whose canonical function is to disable cells that do not carry the element (Wilkes et al., 2010). Further, A. nasoniae represents an antagonist of Nasonia (male-killing makes the element parasitic) that may drive host evolution to prevent its acquisition; as such it may be a driver of immune system evolution that may impact upon other microbiota members.
The transmission pathway also has an important consequence for the movement of the symbiont within chalcid wasp communities. (Duron et al., 2010) noted that uninfected individuals of a different species acquired infection during multiparasitism with an A. nasoniae infected N. vitripennis. This exposure resulted in the efficient transfer of the symbiont into the second species with efficiency comparable to that seen during superparasitism. Using ecological data on multiparasitism rates, the authors concluded that A. nasoniae in a female N. vitripennis had a 12% chance of transfer to N. giraulti in nature. Further to this, the ability of A. nasoniae to be maintained in different species of chalcid wasps in the laboratory was (as above) related to their tendency to superparasitize, with A. nasoniae being lost in species where females were reluctant to utilize already parasitized pupae (Parratt et al., 2016). These results collectively indicate this heritable microbe passes readily between species through multiparasitism, and may pass freely amongst sympatric Nasonia species. Closely related A. nasoniae strains have been retrieved from other chalcid parasitoids (Taylor et al., 2011; Bohacsova et al., 2016), indicating this symbiont infects a wide range of parasitic wasps.
Aside from these biological consequences, the unusual biology of A. nasoniae makes the system more manipulable than most heritable microbe-host interactions. The saprophytic life style stage is almost certainly the reason this microbe grows readily in cell free culture, a property distinct from other heritable microbes (Werren et al., 1986). Further, the strains can easily be reintroduced into the wasp by injection into the host pupa allowing targeted and controlled study of the interaction of the microbe with different wasp genotypes and other elements of the microbiome. Finally, the microbe has intact systems for recombination, and is likely to be genetically manipulable, which would make this one of the few heritable microbes in which functional genetics, both forward and reverse, are possible. However, this ability comes with the caveat that the microbe is not necessarily a reflective model for heritable symbioses in general – it is genetically and biologically more similar to pathogens and gut ‘commensals’ than the classically considered heritable microbiota.
Meeting in the Same Host: Multipartite Interactions between Reproductive Parasites
The previous sections have illustrated our growing knowledge regarding binary interactions of Nasonia with either Wolbachia or Arsenophonus. This leads us to an as yet unexplored question: Do the two reproductive parasites also interact with each other in the same host? And if so, what is the outcome? The effects of co-infections studied to date vary immensely. For instance, Wolbachia and Asaia occupy different niches when co-infecting mosquitoes (Hughes et al., 2014; Rossi et al., 2015). In contrast, male-killing Spiroplasma (Mollicutes) have been observed to negatively affect Wolbachia densities when co-infecting D. melanogaster, while Wolbachia had no impact on Spiroplasma (Goto et al., 2006). Phenotypically, Wolbachia has been shown to interfere with Cardinium-induced CI in the spider mite Bryobia sarothamni, although little is known about the underlying mechanisms (Ros and Breeuwer, 2009). If Arsenophonus also had a negative impact on Wolbachia, co-infection could directly affect CI (and thereby population dynamics) in Nasonia, since the strength of CI is dependent on Wolbachia density (Breeuwer and Werren, 1993a). How Arsenophonus and Wolbachia interact remains to be determined, but the symbionts may well represent important drivers of each other’s biology.
The co-existence of different symbionts in the same host environment also creates conditions that are permissive for the exchange of genetic information between symbionts. Hence, the detection of a lateral gene transfer from Wolbachia to the genome of the A. nasoniae strain infecting N. vitripennis (Darby et al., 2010) is interesting in several ways: First, its presence indicates that cross-talk between the two symbionts can occur. Second, the transferred gene is highly similar to a Wolbachia surface protein and likely has a functional role at the host-symbiont interface (Darby et al., 2010). Bacteriophages associated with either bacterium could be potential agents for lateral gene transfers between these two symbionts (and potentially other members of the Nasonia microbiome). Indeed, both Wolbachia and Arsenophonus are known to have associated phages, a rare feature in bacterial endosymbionts and a potential source of genomic innovation mediating the adaptive plasticity of both symbionts (Kent and Bordenstein, 2010; Duron, 2014). Bacteriophage WO is extremely widespread in arthropod-Wolbachia (Masui et al., 2000; Bordenstein and Wernegreen, 2004; Wu et al., 2004; Braquart-Varnier et al., 2005; Gavotte et al., 2007) and renowned for its ability to jump between different Wolbachia strains, especially when infecting the same host (Gavotte et al., 2004; Gavotte et al., 2007; Chafee et al., 2010; Kent et al., 2011). This has led to the “intracellular arena hypothesis” whereby genetic material can be exchanged between different bacterial endosymbionts co-occurring in the same intracellular environment (Bordenstein and Wernegreen, 2004). Indeed, the discovery of a Wolbachia gene in the genome of Arsenophonus (Darby et al., 2010), a prophage-flanked region of the wMel genome on a plasmid of a Rickettsia strain infecting the tick Ixodes scapularis (Ishmael et al., 2009), and numerous lateral gene transfers from other bacteria associated with phage regions in the wBol1 genome (Duplouy et al., 2013) strongly indicate that WO phages may also vehicle genetic material between Wolbachia and other bacterial taxa. Moreover, sequence similarities between phage WO and eukaryotic genes suggest a history of lateral genetic transfers between the two entities (Bordenstein and Bordenstein, 2016). Interestingly, active lytic phages might also be implicated in Wolbachia regulation and reduce the strength of CI via a phage-mediated reduction of Wolbachia titers (Bordenstein et al., 2006; Bordenstein and Bordenstein, 2011). Regarding Arsenophonus, it has recently been shown that the majority of strains harbor the phage APSE (Duron, 2014), previously known from the protective aphid secondary symbiont Hamiltonella defensa (Oliver et al., 2003, 2009), suggesting a transfer of phage elements between Arsenophonus and Hamiltonella. Such an exchange would be all the more relevant as this phage encodes toxicity genes mediating defense against natural enemies of aphids, which happen to be – parasitoid wasps (Oliver et al., 2009). The role of these genes in Arsenophonus, a symbiont of parasitoid wasps, remains to be elucidated. Taken together, this section illustrates the complexity and diversity of potential interactions between only two bacterial symbionts infecting the same host organism, without even considering the complexity of the microbiome as a whole.
Gaining in Complexity: The Nasonia Microbiota
Tremendous progress has been made regarding our understanding of the intricate relationships between diverse insects and their co-evolved primary symbionts, particularly regarding metabolic complementarity and metabolite exchange between different partners (McCutcheon et al., 2009; McCutcheon and Moran, 2010; Wilson et al., 2010; Hansen and Moran, 2011; McCutcheon and von Dohlen, 2011; Husnik et al., 2013; Luan et al., 2015) and adaptations of the host immune system to recognize and regulate resident symbionts (Wang et al., 2009; Login et al., 2011; Futahashi et al., 2013; Kim et al., 2013; Shigenobu and Stern, 2013; Masson et al., 2015). However, achieving the same level of insight into host-symbiont cross-talk for highly complex insect microbiotas remains challenging. Many host-associated microbes may not be culturable and therefore impossible to manipulate outside of the host’s body. Hence, we need a study system where the host (i) is easy to rear in the lab; (ii) genetically tractable with resources available for genomic/transcriptomic or immunity-related investigations; (iii) has a complex but well-characterized microbiota; and (iv) this microbiota can be relatively easily manipulated in the host organism, which can be an asset for testing the influence of the microbiome on host traits. Previous work demonstrates that Nasonia maintains a relatively high level of microbial diversity, microbiome functionality, and experimental tractability, even while kept under laboratory conditions.
Microbiota Diversity
The bacterial diversity of Nasonia has been described in lab-reared larvae, pupae and adult males for the three Nasonia species N. vitripennis, N. giraulti, and N. longicornis (Figure 3) (Brucker and Bordenstein, 2012b, 2013). Microbial diversity in these strains ranged from 44 to 83 OTUs at a 97% identity cutoff and varies between host species and developmental stages (Figure 4). Overall bacterial diversity in Nasonia is similar to other Hymenoptera, such as honey bees (Apis mellifera, 82-116 OTUs), bumblebees (Bombus sp., 33-47 OTUs), and fungus farming ants (Mycocepurus smithii, an average of 52 OTUs) (Martinson et al., 2011; Cariveau et al., 2014; Corby-Harris et al., 2014; Kellner et al., 2015). Like most insects, the Nasonia microbiota is dominated by members of the Proteobacteria phylum. The average Nasonia microbiota in adult males is composed of 74.4% Proteobacteria, 15.7% Actinobacteria, and 9.5% Firmicutes (Figure 4) (Brucker and Bordenstein, 2013). Interestingly, at the bacterial genus level, there are three major taxa (Gammaproteobacteria) that account for up to 75% of the male microbiota: Providencia, Proteus, and Morganella (Figure 4). Alone, Providencia sp. compose 59, 68, and 41% of the microbiota in N. vitripennis, N. giraulti, and N. longicornis, respectively. Comparatively, Nasonia is a more tractable laboratory model for controlled experiments and is consistently comprised of 4-5 OTUs that make up the majority of all bacterial sequences.
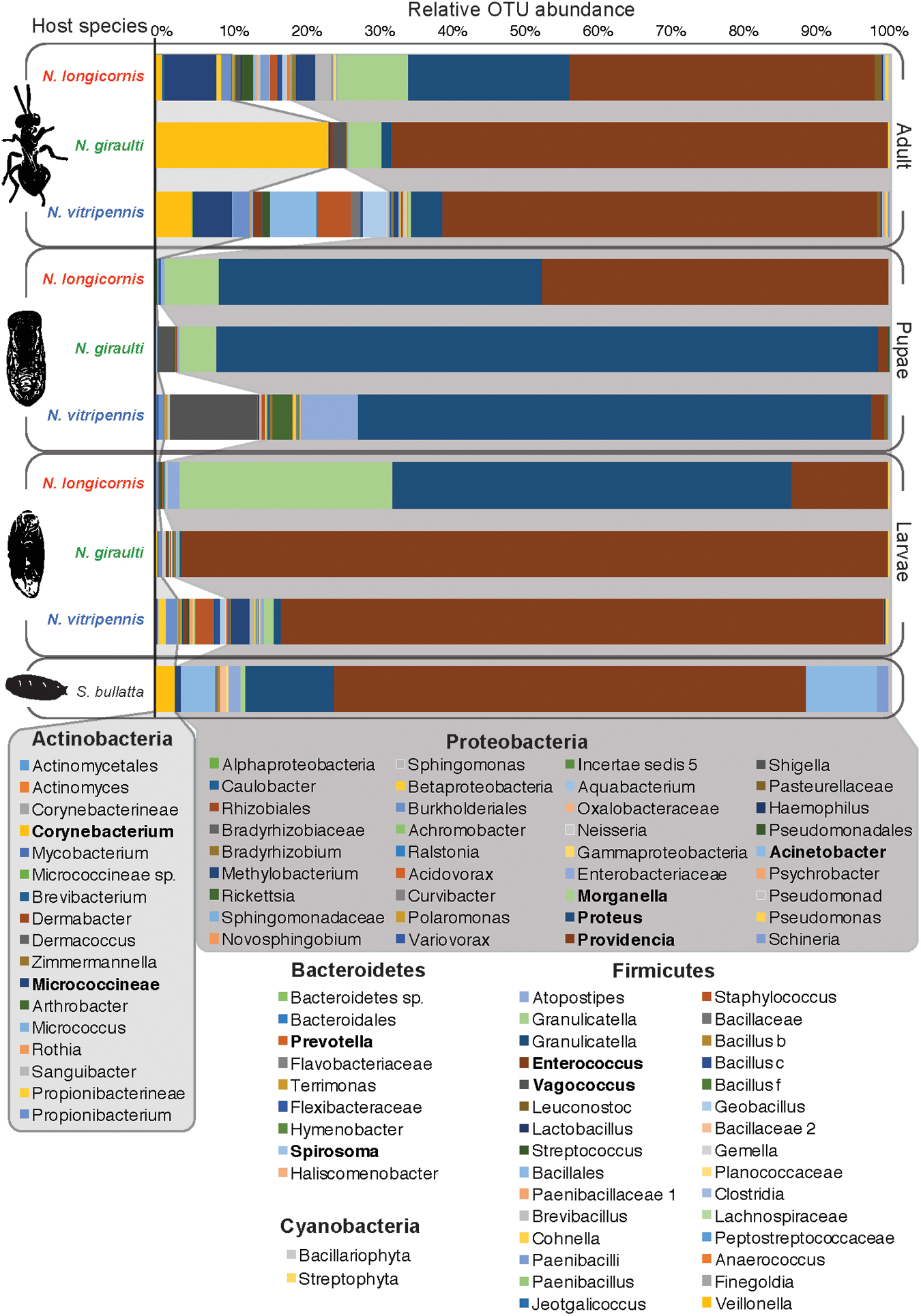
FIGURE 4. The relative abundance of bacterial OTUs observed in male Nasonia throughout development (Brucker and Bordenstein, 2013). The OTUs represented across the three Nasonia species and their S. bullata fly host are dominated by Actinobacteria, Firmicutes, and especially Proteobacteria. Three genera, Providencia, Proteus, and Morganella, are particularly dominant across all samples. However, their relative abundances differ according to host species and developmental stage. The unparasitized S. bullata fly host is similarly dominated by Proteobacteria, specifically the genus Providencia. Emboldened OTUs are observed at higher frequencies in one or more samples. It is important to note that many of the rarer OTUs have also been observed in other studies (Brucker and Bordenstein, 2012b and personal communication).
The two genera Providencia and Proteus are often the most dominant OTUs observed in the three wasp species throughout their development. These same two OTUs are frequently found in the fly host as well, which could represent a natural reservoir of the bacteria for Nasonia. Notably, Nasonia undergoes bacterial community successions throughout its development: The microbial community remains relatively simple when the developing larvae are feeding on the likewise relatively simple microbiota of the fly pupa. Then, microbiota composition shifts during pupation, a time when the wasp is no longer feeding, and again before emergence as adult wasps (Figure 4). As such, Providencia and Proteus represent 95-100% of the microbiota in larvae (Brucker and Bordenstein, 2012b, 2013). Although the microbiota of pupae is less diverse than the microbiota of adults, both tend to exhibit a reduction in the dominance of Proteobacteria (Brucker and Bordenstein, 2012b, 2013).
While little is known about the specific functional roles of the microbiota in Nasonia, several of the major bacterial genera have been previously studied in other insect models. For instance, Proteus has been shown to control the gut microenvironment in blowflies from overgrowth by other bacteria (Erdmann et al., 1984). This colonization resistance could be important for Nasonia, which feed on a decaying pupal fly host. Another major taxon, Providencia, has been implicated in two symbiotic roles: (i) Providing vitamin B to the blood-feeding leech Haementeria officinalis (Manzano-Marin et al., 2015) and (ii) acting as a natural control against the insect pathogen Paenibacillus in the Japanese honeybee Apis cerana japonica (Yoshiyama and Kimura, 2009).
Ongoing studies are now testing the functional significance of the microbiota in different species of Nasonia to determine their role in host development, e.g., through immune regulation, nutrition, and other mechanisms. In addition, transplantations of Nasonia microbiotas between host species will elucidate whether interspecific microbiotas alter host development traits such as larval size, larval and pupal development time or adult viability in comparison to intraspecific microbiotas. In this context, studies in D. melanogaster have demonstrated that axenic individuals suffer from developmental defects along with smaller body size and an altered nutrient metabolism (Shin et al., 2011; Newell and Douglas, 2014). These defects can be rescued by the acetic acid bacterium Acetobacter pomorum, which promotes larval growth and reduces lipid and sugar levels by modulating insulin signaling (Shin et al., 2011). In addition, Lactobacillus plantarum promotes larval growth in conditions of nutrient scarcity by enhancing protein assimilation and TOR-dependent hormonal growth signals (Storelli et al., 2011). In turn, the Drosophila innate immune response is fine-tuned to maintain gut microbiota homeostasis and responds to bacterial pathogens via ROS-production triggered by bacteria-derived uracil, which is released by various opportunistic pathogens but not autochthonous gut microbes (Ryu et al., 2008; Lee et al., 2013).
The microbial community is not limited to bacteria. Nasonia also harbors a diverse set of viruses (Bordenstein and Bordenstein, personal communication) and fungi, and their functional effects on the holobiont await further investigation. While no studies to date have investigated Nasonia’s fungal microbiota, the original draft of the Nasonia transcriptome revealed three novel single-stranded RNA viruses: NvitV-1, -2, -3 (Oliveira et al., 2010). These viruses were not previously found in other insect hosts, though they are related to the Picornavirales—a known order of insect pathogens. The observation of novel viruses in the system is interesting from the perspective that viruses are known to influence the biology of other parasitoid wasps. For instance, the virus Leptopilina boulardi Filamentous Virus (LbFV) manipulates the foraging behavior of its solitary parasitoid wasp host, Leptopilina boulardi, by inducing superparasitism (Varaldi et al., 2003, 2005, 2006). The virus is injected into the fly host together with the parasitoid eggs, allowing it to spread horizontally to uninfected individuals. In contrast to this infectious virus, polydnavirus-like particles have been integrated into the genomes of braconid and ichneumonid wasps and encode particles that contain wasp DNA and proteins which, when injected into the host with the parasitoids’ eggs, enable evasion or direct suppression of the host’s immune response against the parasitoid, thereby contributing significantly to parasitoid fitness (reviewed in Federici and Bigot, 2003).
Establishment and Transmission of the Microbiota
The changes in the bacterial community throughout development raise questions as to how the Nasonia microbial community assembles through metamorphosis. The answer is not yet clear in any animal system but the patterns exhibited by Nasonia offer an opportunity to better understand how animals change developmentally and anatomically with their microbiota. Since Nasonia embryos are directly deposited within fly host pupae via oviposition, both maternal and fly host microbes could contribute to the initial microbiota assembly of Nasonia larvae. Based on the transmission of microbes in Drosophila (Bakula, 1969), it is possible that Nasonia acquire their first non-endosymbiotic bacteria through ingestion of the chorion during hatching. Alternatively, the microbial community could be passaged via maternal deposition of calyx fluid and venom – using the same process of transmission as Arsenophonus (Huger et al., 1985; Werren et al., 1986). During this event, rare microbes could be introduced into the Nasonia microbiota. Subsequent colonization of the microbiota would then occur through feeding on the fly host. However, the excretion of the larval gut content prior to pupation presents a marked bottleneck for the microbiota. As larvae and pupae develop, it is possible that Nasonia species-specific innate immune genes regulate this community, which would parallel species-specific antimicrobial regulation of the microbiota in Hydra (Franzenburg et al., 2013). On the other hand, the innate immune response of honey bees has been shown to be strongly reduced during the pupal stage compared to larvae and adults (Gatschenberger et al., 2013). If this pattern is consistent across the order Hymenoptera, then a weaker immune regulation during the pupal stage could be influential in the mechanisms that establish the new host species-specific microbiota.
An important aspect that is often overlooked is that microbiota composition may not be regulated solely by host mechanisms, but also through interactions between the microbes themselves. From the microbial perspective, a host organism represents an ecosystem consisting of different microhabitats (i.e., niches) (Sicard et al., 2014), and microbes can be expected to differ in their preference for particular niches. Given that the Nasonia microbiota consists at least partly of bacteria acquired from its fly host during larval development, one might ask whether the transfer to Nasonia as a new host results in fitness consequences for the microbes. While some might be opportunistic and able to find suitable niches, Nasonia may represent a dead-end for other microbes, either due to host factors or competition with other bacteria. The latter may be due to competition for a shared resource/niche and/or by direct interference, for instance via the production of bacteriocidal toxins. Moreover, there may be indirect interactions, mediated through host mechanisms. A particular bacterium may, for instance, activate or suppress the host innate immune system, which then affects the proliferation of other bacteria.
An as yet unexplored but highly relevant aspect is the role of heritable symbionts in the establishment and composition of the Nasonia microbiota. Wolbachia, for instance, are generally highly abundant in various host tissues, thereby limiting available niches and resources for other bacteria (Dittmer et al., 2014, 2016). It is also known to influence other aspects of its host environment, such as immunity, apoptosis and iron homeostasis (Braquart-Varnier et al., 2008; Brownlie et al., 2009; Kremer et al., 2009, 2012; Kambris et al., 2010; Pan et al., 2012). Considering that Wolbachia is ubiquitous in Nasonia under natural conditions, Wolbachia infection represents the natural infection status of Nasonia. Arsenophonus, on the other hand, is a more variable heritable symbiont in this system and has the ability to efficiently infect uninfected individuals via horizontal/environmental transmission within the same fly host (Duron et al., 2010; Parratt et al., 2016). Investigating the impact of Arsenophonus infection on establishment and composition of the wider Nasonia microbiota therefore constitutes a promising line of future research.
Phylosymbiosis
Microbiota composition is shaped by both host and environmental factors (e.g., immunity and diet, respectively (Ley et al., 2008; Ochman et al., 2010; Chandler et al., 2011; Colman et al., 2012; Ridley et al., 2012). While the first can be considered deterministic, the latter would be rather stochastic. However, it can be challenging to disentangle these two components and to precisely determine the relative roles of the host versus the environment on the establishment of a species’ microbial community. Controlled conditions can provide evidence for host-microbiota interactions by removing confounding variations in diet, age and gender, for instance. Indeed, under a controlled experimental design, three Nasonia species were found to harbor distinguishable microbiotas whose beta-diversity relationships parallel host phylogeny at all developmental stages (Figure 3A) (Brucker and Bordenstein, 2012b,c). The congruence of host phylogeny and dendrograms reflecting relationships in microbiota composition has since been dubbed “phylosymbiosis” (Brucker and Bordenstein, 2013). For a particular set of closely related species, phylosymbiosis predicts that intraspecific microbial communities are more similar than interspecific communities (Bordenstein and Theis, 2015). Based on that, one could hypothesize that (i) microbiota-based models should predict host species origin with high accuracy, and (ii) various topological congruence analyses of host phylogeny and microbiota dendrograms will reveal significant degrees of phylosymbiosis. Furthermore, if phylosymbiosis were driven by both evolutionary and ecological forces, we might also observe that experimental transplants of autochthonous (intraspecific) versus allochthonous (interspecific) microbiota will drive reductions in host survival and fitness. In addition to Nasonia, the pattern of phylosymbiosis is evident in Hominidae (Ochman et al., 2010; Moeller et al., 2016), Hydra (Fraune and Bosch, 2007; Franzenburg et al., 2013), sponges (Easson and Thacker, 2014), ants (Sanders et al., 2014), and bats (Phillips et al., 2012). One future area of investigation will be to understand the factors influencing phylosymbiosis, e.g., fine-tuned host immune mechanisms and/or different transmission modes (i.e., through maternal transmission or environmental acquisition).
The Microbiota and Reproductive Isolation
Our growing knowledge of the many ways in which microbial symbionts can induce changes in host phenotypic traits raises the question - to what extent do the microbiota contribute to host diversification, reproductive isolation barriers, and speciation (see Brucker and Bordenstein, 2012c; Vavre and Kremer, 2014; Shropshire and Bordenstein, 2016 for recent reviews)? Isolation barriers can be either pre-mating or post-mating. Pre-mating reproductive barriers may be driven by ecological or behavioral isolation. For instance, particular bacterial symbionts can confer novel traits (e.g., increased thermal tolerance or adaptation to new host plants), allowing their insect host to exploit new ecological niches (Montllor et al., 2002; Ferrari et al., 2004; Tsuchida et al., 2004). Niche expansions such as these can result in geographically or sympatrically isolated populations and, given enough time, lead to speciation. In addition, the microbiota has been implicated in behavioral changes related to mate choice, which may result in symbiont-driven behavioral isolation due to differences in courtship or mate discrimination (reviewed in Shropshire and Bordenstein, 2016). For example, Wolbachia plays a crucial role in driving pre-mating isolation between semispecies of the Drosophila paulistorum species complex (Miller et al., 2010). In addition, the gut microbiota influences kin recognition and mating investment in several Drosophila species (Lize et al., 2014). Specifically, both D. bifasciata and D. melanogaster are able to distinguish between mates that have a more similar or dissimilar microbiota to themselves (Sharon et al., 2010; Lize et al., 2014). This results in a tendency for assortative mating in D. melanogaster after feeding on different food sources (Sharon et al., 2010), although this behavior was replicated only in inbred laboratory lines (Najarro et al., 2015). Similarly, mate selection in scarab beetles is dependent upon immune competence that the females sense in the bacterial-derived male pheromones secretions (Leal, 1998; Vasanthakumar et al., 2008; Andert et al., 2010).
In contrast, post-mating reproductive barriers may be driven by genetic conflicts between host and microbes (i.e., Wolbachia) or a breakdown in holobiont complexes. In Nasonia, both types occur. Wolbachia-induced CI in this system is a pre-eminent case of symbiont-assisted isolation in which nearly complete CI levels (Figure 2) between the Nasonia species cause F1 hybrid lethality that is reversible by curing the Wolbachia infections. In other words, the interspecific F1 isolation is essentially undone with antibiotics that restore production of viable F1 hybrids (Breeuwer and Werren, 1990, 1995; Bordenstein et al., 2001; Raychoudhury et al., 2010a). The study system is notable in that it provided the opportunity to investigate whether Wolbachia-induced CI evolved early or late in the speciation process, i.e., before or after other interspecific pre- or post-mating barriers. While the “older” species pair, N. vitripennis and N. giraulti, diverged ∼1 million years ago and evolved other post-mating barriers such as high F2 hybrid mortality and abnormal courtship behavior (Breeuwer and Werren, 1995; Bordenstein et al., 2001), the very young species pair, N. giraulti and N. longicornis, diverged only ∼400,000 years ago and produce viable and fertile hybrids (Bordenstein et al., 2001). This observation indicates that Wolbachia-induced reproductive isolation via CI preceded the evolution of other post-mating barriers in the younger species pair, and therefore is the first major step in the speciation process (Bordenstein et al., 2001). The even younger species pair, N. giraulti and N. oneida, represents an interesting case in this context: N. oneida females show strong mate discrimination against N. giraulti males, but not vice versa, resulting in strong but incomplete and asymmetrical pre-mating isolation (Raychoudhury et al., 2010a). Moreover, the mate discrimination phenotype in N. oneida is recessive and lost in F1 hybrid females (Raychoudhury et al., 2010a). The impact of Wolbachia on this speciation event is unfortunately blurred by the recent Wolbachia-mitochondrial sweep from N. giraulti into N. oneida (Raychoudhury et al., 2009), which eliminated any Wolbachia-induced incompatibilities that may have existed previously. Therefore, pre-mating isolation is the only barrier currently preventing hybridisation between the two species.
An additional microbiota-mediated reproductive barrier has recently been uncovered in Nasonia, manifested as strong F2 hybrid lethality in interspecies crosses after curing of Wolbachia (Brucker and Bordenstein, 2013). Specifically, hybrid lethality between Nasonia vitripennis and Nasonia giraulti is reversed through removal of the Nasonia gut microbiota, and can be reinstated by inoculating germ-free hybrids with Nasonia-derived bacterial cultures (Brucker and Bordenstein, 2013). Nasonia hybrid lethality was also characterized by an altered gut microbiota in which a rare microbial genus became abundant in hybrids. The change in bacterial community structure was coupled with aberrant host immune gene expression (specifically differential regulation of serine proteases, antimicrobial peptides, and several signaling molecules from the IMD and Toll pathways) compared to the parental species (Brucker and Bordenstein, 2013). In this case, hybrid breakdown at the holobiont level led to severe mortality. This is the first study, to our knowledge, in which the microbial community contributes to hybrid mortality (Figure 2).
Changes in the microbiota could also result in other microbe-dependent reproductive barriers, similar to phenotypes observed in various animal systems, e.g., in terms of development time, behavior, and fecundity (Brucker and Bordenstein, 2012c). For example, species-specific cuticular hydrocarbons help in mate discrimination in Nasonia (Buellesbach et al., 2013), but the impact of the microbiota on mate-choice is unknown. The behavioral issues that arise in hybrids (Clark et al., 2010) may therefore have microbial underpinnings.
Germ-Free and Gnotobiotic Rearing
A powerful aspect of the Nasonia model lies in the ability to selectively rear (or co-rear) Nasonia hosts in germ-free (Brucker and Bordenstein, 2012a; Shropshire et al., 2016), gnotobiotic (harboring a known, controlled microbiota) and transbiotic (harboring the microbiota of a different species) conditions. The ability to inoculate germ-free Nasonia larvae with monocultures or whole microbial communities will enable high precision studies that deconstruct the effects of specific microbial functions in Nasonia. These studies have the benefit of being implemented at any stage throughout the Nasonia developmental process, which is important to understand the assembly and regulation of the Nasonia microbiota. Specific host genes could also be knocked down to observe their direct effects on microbiota assembly and host-microbe interactions in the four different Nasonia species. With rising interest in utilizing CRISPR genome editing in Nasonia (Lynch, 2015), host gene addition and removal could soon be incorporated into the toolbox available for deciphering hologenomic interactions. With the unique environmental controls afforded by the Nasonia rearing system, there is ample opportunity to study microbiota influences on Nasonia development and fitness. With these tools at hand, the Nasonia model could also be used to experimentally test hologenomic evolution, for instance by exposing a wasp population to a selective pressure (e.g., an environmental stressor) and subsequently monitor (i) whether changes in the microbiota correlate with changes in host life history traits or behavior and (ii) whether this shift in the microbial community persists over multiple generations, as long as the selective pressure persists.
Perspectives and Concluding Remarks
This review highlights the important contribution Nasonia has made regarding our understanding of the manifold roles of both heritable symbionts and the general microbiota on host fitness and evolution. By conducting full microbial community transplantation experiments using the established Nasonia in vitro rearing system, further studies can elucidate the genome-by-microbiome interactions that cause reproductive barriers in Nasonia. This, among many other advantages highlighted in this review, reinforces the Nasonia holobiont as a highly versatile and tractable model, allowing for a controlled approach to host-microbe analyses. An important direction for future research will be to investigate Nasonia-microbiota dynamics in more ecologically relevant settings and to test whether the patterns observed thus far in a small number of inbred laboratory lineages hold true under natural conditions. For instance, is phylosymbiosis evident in wild populations? Does the microbiota of a given Nasonia species change when parasitizing different fly species? Is there an exchange of microbes between different Nasonia species parasitizing the same fly host or does each species still ‘filter’ its characteristic microbiota from the common pool? Does the microbiota remain stable under different environmental conditions (e.g., when exposed to environmental stressors) or does it evolve in structure and function in a way that is malleable under given circumstances? It is beyond doubt that Nasonia will continue to provide valuable insights regarding the evolution of host-microbe interactions, especially as new tools for microbiome manipulation and functional interactions become available.
Author Contributions
JD, EO, JS, SB, GH, and RB contributed ideas and concepts. JD, EO, GH, and RB wrote the paper. JD, EO, and RB made the figures. JD, EO, JS, SB, GH, and RB finalized the paper and figures.
Funding
Support for this work was provided by the Rowland Institute at Harvard University to RB and National Science Foundation Awards DEB 1046149 and IOS 1456778 to SB.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
We would like to thank Matthew C. Johnson for contributing pictures of the different Nasonia life stages.
References
Andert, J., Marten, A., Brandl, R., and Brune, A. (2010). Inter- and intraspecific comparison of the bacterial assemblages in the hindgut of humivorous scarab beetle larvae (Pachnoda spp.). FEMS Microbiol. Ecol. 74, 439–449. doi: 10.1111/j.1574-6941.2010.00950.x
Bakula, M. (1969). The persistence of a microbial flora during postembryogenesis of Drosophila melanogaster. J. Invertebr. Pathol. 14, 365–374. doi: 10.1016/0022-2011(69)90163-3
Balas, M. T., Lee, M. H., and Werren, J. H. (1996). Distribution and fitness effects of the son-killer bacterium in Nasonia. Evol. Ecol. 10, 593–607. doi: 10.1007/BF01237709
Beeler, S. M., Wong, G. T., Zheng, J. M., Bush, E. C., Remnant, E. J., Oldroyd, B. P., et al. (2014). Whole-genome DNA methylation profile of the jewel wasp (Nasonia vitripennis). G3 (Bethesda) 4, 383–388. doi: 10.1534/g3.113.008953
Bertossa, R. C., van Dijk, J., Diao, W., Saunders, D., Beukeboom, L. W., and Beersma, D. G. (2013). Circadian rhythms differ between sexes and closely related species of Nasonia wasps. PLoS ONE 8:e60167. doi: 10.1371/journal.pone.0060167
Beukeboom, L. W., and van de Zande, L. (2010). Genetics of sex determination in the haplodiploid wasp Nasonia vitripennis (Hymenoptera: Chalcidoidea). J. Genet. 89, 333–339. doi: 10.1007/s12041-010-0045-7
Blaser, M. J. (2014). The microbiome revolution. J. Clin. Invest. 124, 4162–4165. doi: 10.1172/JCI78366
Bohacsova, M., Mediannikov, O., Kazimirova, M., Raoult, D., and Sekeyova, Z. (2016). Arsenophonus nasoniae and Rickettsiae infection of Ixodes ricinus due to parasitic wasp Ixodiphagus hookeri. PLoS ONE 11:e0149950. doi: 10.1371/journal.pone.0149950
Bordenstein, S. R., and Bordenstein, S. R. (2011). Temperature affects the tripartite interactions between bacteriophage WO, Wolbachia, and cytoplasmic incompatibility. PLoS ONE 6:e29106. doi: 10.1371/journal.pone.0029106
Bordenstein, S. R., Marshall, M. L., Fry, A. J., Kim, U., and Wernegreen, J. J. (2006). The tripartite associations between bacteriophage, Wolbachia, and arthropods. PLoS Pathog. 2:e43. doi: 10.1371/journal.ppat.0020043
Bordenstein, S. R., O’Hara, F. P., and Werren, J. H. (2001). Wolbachia-induced incompatibility precedes other hybrid incompatibilities in Nasonia. Nature 409, 707–710. doi: 10.1038/35055543
Bordenstein, S. R., and Theis, K. R. (2015). Host biology in light of the microbiome: ten principles of holobionts and hologenomes. PLoS Biol. 13:e1002226. doi: 10.1371/journal.pbio.1002226
Bordenstein, S. R., Uy, J. J., and Werren, J. H. (2003). Host genotype determines cytoplasmic incompatibility type in the haplodiploid genus Nasonia. Genetics 164, 223–233.
Bordenstein, S. R., and Wernegreen, J. J. (2004). Bacteriophage flux in endosymbionts (Wolbachia): infection frequency, lateral transfer, and recombination rates. Mol. Biol. Evol. 21, 1981–1991. doi: 10.1093/molbev/msh211
Bordenstein, S. R., and Werren, J. H. (1998). Effects of A and B Wolbachia and host genotype on interspecies cytoplasmic incompatibility in Nasonia. Genetics 148, 1833–1844.
Bordenstein, S. R., and Werren, J. H. (2007). Bidirectional incompatibility among divergent Wolbachia and incompatibility level differences among closely related Wolbachia in Nasonia. Heredity (Edinb) 99, 278–287. doi: 10.1038/sj.hdy.6800994
Bordenstein, S. R., and Bordenstein, S. R. (2016). Eukaryotic association module in phage WO genomes from Wolbachia. bioRxiv. doi: 10.1101/049049
Bouchon, D., Cordaux, R., and Grève, P. (2008). “Feminizing Wolbachia and the evolution of sex determination in isopods,” in Insect Symbiosis, eds K. Bourtzis and T. A. Miller (Boca Raton, FL: CRC Press), 273–296.
Braquart-Varnier, C., Greve, P., Felix, C., and Martin, G. (2005). Bacteriophage WO in Wolbachia infecting terrestrial isopods. Biochem. Biophys. Res. Commun. 337, 580–585. doi: 10.1016/j.bbrc.2005.09.091
Braquart-Varnier, C., Lachat, M., Herbinière, J., Johnson, M., Caubet, Y., Bouchon, D., et al. (2008). Wolbachia mediate variation of host immunocompetence. PLoS ONE 3:e3286. doi: 10.1371/journal.pone.0003286
Breeuwer, J. A., Stouthamer, R., Barns, S. M., Pelletier, D. A., Weisburg, W. G., and Werren, J. H. (1992). Phylogeny of cytoplasmic incompatibility microorganisms in the parasitoid wasp genus Nasonia (Hymenoptera: Pteromalidae) based on 16S ribosomal DNA sequences. Insect Mol. Biol. 1, 25–36. doi: 10.1111/j.1365-2583.1993.tb00074.x
Breeuwer, J. A., and Werren, J. H. (1990). Microorganisms associated with chromosome destruction and reproductive isolation between two insect species. Nature 346, 558–560. doi: 10.1038/346558a0
Breeuwer, J. A., and Werren, J. H. (1993a). Cytoplasmic incompatibility and bacterial density in Nasonia vitripennis. Genetics 135, 565–574.
Breeuwer, J. A., and Werren, J. H. (1993b). Effect of genotype on cytoplasmic incompatibility in two species of Nasonia. Heredity 70, 428–436. doi: 10.1038/hdy.1993.60
Breeuwer, J. A., and Werren, J. H. (1995). Hybrid breakdown between two haplodiploid species: the role of nuclear and cytoplasmic genes. Evolution 49, 705–717. doi: 10.2307/2410324
Bressan, A. (2014). Emergence and evolution of Arsenophonus bacteria as insect-vectored plant pathogens. Infect. Genet. Evol. 22, 81–90. doi: 10.1016/j.meegid.2014.01.004
Brownlie, J. C., Cass, B. N., Riegler, M., Witsenburg, J. J., Iturbe-Ormaetxe, I., McGraw, E. A., et al. (2009). Evidence for metabolic provisioning by a common invertebrate endosymbiont, Wolbachia pipientis, during periods of nutritional stress. PLoS Pathog. 5:e1000368. doi: 10.1371/journal.ppat.1000368
Brucker, R. M., and Bordenstein, S. R. (2012a). In vitro cultivation of the hymenoptera genetic model, Nasonia. PLoS ONE 7:e51269. doi: 10.1371/journal.pone.0051269
Brucker, R. M., and Bordenstein, S. R. (2012b). The roles of host evolutionary relationships (genus: Nasonia) and development in structuring microbial communities. Evolution 66, 349–362. doi: 10.1111/j.1558-5646.2011.01454.x
Brucker, R. M., and Bordenstein, S. R. (2012c). Speciation by symbiosis. Trends Ecol. Evol. 27, 443–451. doi: 10.1016/j.tree.2012.03.011
Brucker, R. M., and Bordenstein, S. R. (2013). The hologenomic basis of speciation: gut bacteria cause hybrid lethality in the genus Nasonia. Science 341, 667–669. doi: 10.1126/science.1240659
Brucker, R. M., Funkhouser, L. J., Setia, S., Pauly, R., and Bordenstein, S. R. (2012). Insect innate immunity database (IIID): an annotation tool for identifying immune genes in insect genomes. PLoS ONE 7:e45125. doi: 10.1371/journal.pone.0045125
Buchner, P. (1965). Endosymbiosis of Animals with Plant Microorganisms. New York, NY: Interscience Publishers.
Buellesbach, J., Gadau, J., Beukeboom, L. W., Echinger, F., Raychoudhury, R., Werren, J. H., et al. (2013). Cuticular hydrocarbon divergence in the jewel wasp Nasonia: evolutionary shifts in chemical communication channels? J. Evol. Biol. 26, 2467–2478. doi: 10.1111/jeb.12242
Cariveau, D. P., Elijah Powell, J., Koch, H., Winfree, R., and Moran, N. A. (2014). Variation in gut microbial communities and its association with pathogen infection in wild bumble bees (Bombus). ISME J. 8, 2369–2379. doi: 10.1038/ismej.2014.68
Chafee, M. E., Funk, D. J., Harrison, R. G., and Bordenstein, S. R. (2010). Lateral phage transfer in obligate intracellular bacteria (Wolbachia): verification from natural populations. Mol. Biol. Evol. 27, 501–505. doi: 10.1093/molbev/msp275
Chafee, M. E., Zecher, C. N., Gourley, M. L., Schmidt, V. T., Chen, J. H., Bordenstein, S. R., et al. (2011). Decoupling of host-symbiont-phage coadaptations following transfer between insect species. Genetics 187, 203–215. doi: 10.1534/genetics.110.120675
Chandler, J. A., Lang, J. M., Bhatnagar, S., Eisen, J. A., and Kopp, A. (2011). Bacterial communities of diverse Drosophila species: ecological context of a host-microbe model system. PLoS Genet. 7:e1002272. doi: 10.1371/journal.pgen.1002272
Chu, H., and Mazmanian, S. K. (2013). Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat. Immunol. 14, 668–675. doi: 10.1038/ni.2635
Clark, M. E., O’Hara, F. P., Chawla, A., and Werren, J. H. (2010). Behavioral and spermatogenic hybrid male breakdown in Nasonia. Heredity (Edinb.) 104, 289–301. doi: 10.1038/hdy.2009.152
Colman, D. R., Toolson, E. C., and Takacs-Vesbach, C. D. (2012). Do diet and taxonomy influence insect gut bacterial communities? Mol. Ecol. 21, 5124–5137. doi: 10.1111/j.1365-294X.2012.05752.x
Corby-Harris, V., Maes, P., and Anderson, K. E. (2014). The bacterial communities associated with honey bee (Apis mellifera) foragers. PLoS ONE 9:e95056. doi: 10.1371/journal.pone.0095056
Danneels, E. L., Gerlo, S., Heyninck, K., Van Craenenbroeck, K., De Bosscher, K., Haegeman, G., et al. (2014). How the venom from the ectoparasitoid Wasp Nasonia vitripennis exhibits anti-inflammatory properties on mammalian cell lines. PLoS ONE 9:e96825. doi: 10.1371/journal.pone.0096825
Darby, A. C., Choi, J. H., Wilkes, T., Hughes, M. A., Werren, J. H., Hurst, G. D., et al. (2010). Characteristics of the genome of Arsenophonus nasoniae, son-killer bacterium of the wasp Nasonia. Insect Mol. Biol. 19(Suppl. 1), 75–89. doi: 10.1111/j.1365-2583.2009.00950.x
Darling, D. C., and Werren, J. H. (1990). Biosystematics of Nasonia (Hymenoptera: Pteromalidae): two new species reared from birds’ nests in North America. Ann. Entomol. Soc. Am. 83, 352–370. doi: 10.1093/aesa/83.3.352
Dedeine, F., Vavre, F., Shoemaker, D. D., and Bouletreau, M. (2004). Intra-individual coexistence of a Wolbachia strain required for host oogenesis with two strains inducing cytoplasmic incompatibility in the wasp Asobara tabida. Evolution 58, 2167–2174. doi: 10.1111/j.0014-3820.2004.tb01595.x
Desjardins, C. A., Gadau, J., Lopez, J. A., Niehuis, O., Avery, A. R., Loehlin, D. W., et al. (2013). Fine-scale mapping of the Nasonia genome to chromosomes using a high-density genotyping microarray. G3 (Bethesda) 3, 205–215. doi: 10.1534/g3.112.004739
Desjardins, C. A., Perfectti, F., Bartos, J. D., Enders, L. S., and Werren, J. H. (2010). The genetic basis of interspecies host preference differences in the model parasitoid Nasonia. Heredity (Edinb.) 104, 270–277. doi: 10.1038/hdy.2009.145
Dittmer, J., Beltran-Bech, S., Lesobre, J., Raimond, M., Johnson, M., and Bouchon, D. (2014). Host tissues as microhabitats for Wolbachia and quantitative insights into the bacterial community in terrestrial isopods. Mol. Ecol. 23, 2619–2635. doi: 10.1111/mec.12760
Dittmer, J., Lesobre, J., Moumen, B., and Bouchon, D. (2016) Host origin and tissue microhabitat shaping the microbiota of the terrestrial isopod Armadillidium vulgare. FEMS Microbiol. Ecol. 92:fiw063.
Dong, Y., Manfredini, F., and Dimopoulos, G. (2009). Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog. 5:e1000423. doi: 10.1371/journal.ppat.1000423
Douglas, A. E. (2015). Multiorganismal insects: diversity and function of resident microorganisms. Annu. Rev. Entomol. 60, 17–34. doi: 10.1146/annurev-ento-010814-020822
Douglas, A. E, and Werren, J. H. (2016). Holes in the hologenome: why host-microbe symbioses are not holobionts. MBio 7:e02099.
Duplouy, A., Iturbe-Ormaetxe, I., Beatson, S. A., Szubert, J. M., Brownlie, J. C., McMeniman, C. J., et al. (2013). Draft genome sequence of the male-killing Wolbachia strain wBol1 reveals recent horizontal gene transfers from diverse sources. BMC Genomics 14:20. doi: 10.1186/1471-2164-14-20
Duron, O. (2014). Arsenophonus insect symbionts are commonly infected with APSE, a bacteriophage involved in protective symbiosis. FEMS Microbiol. Ecol. 90, 184–194. doi: 10.1111/1574-6941.12381
Duron, O., Bouchon, D., Boutin, S., Bellamy, L., Zhou, L., Engelstädter, J., et al. (2008). The diversity of reproductive parasites among arthropods: Wolbachia do not walk alone. BMC Biol. 6:27. doi: 10.1186/1741-7007-6-27
Duron, O., Wilkes, T. E., and Hurst, G. D. (2010). Interspecific transmission of a male-killing bacterium on an ecological timescale. Ecol. Lett. 13, 1139–1148. doi: 10.1111/j.1461-0248.2010.01502.x
Easson, C. G., and Thacker, R. W. (2014). Phylogenetic signal in the community structure of host-specific microbiomes of tropical marine sponges. Front. Microbiol. 5:532. doi: 10.3389/fmicb.2014.00532
Ellison, C. K., Niehuis, O., and Gadau, J. (2008). Hybrid breakdown and mitochondrial dysfunction in hybrids of Nasonia parasitoid wasps. J. Evol. Biol. 21, 1844–1851. doi: 10.1111/j.1420-9101.2008.01608.x
Erdmann, G. R., Bromel, M., Gassner, G., Freeman, T. P., and Fischer, A. (1984). Antibacterial activity demonstrated by culture filtrates of Proteus mirabilis isolated from screwworm (Cochliomyia hominivorax) (Diptera: Calliphoridae) larvae. J. Med. Entomol. 21, 159–164. doi: 10.1093/jmedent/21.2.159
Federici, B. A., and Bigot, Y. (2003). Origin and evolution of polydnaviruses by symbiogenesis of insect DNA viruses in endoparasitic wasps. J. Insect Physiol. 49, 419–432. doi: 10.1016/S0022-1910(03)00059-3
Ferrari, J., Darby, A. C., Daniell, T. J., Godfray, H. C. J., and Douglas, A. E. (2004). Linking the bacterial community in pea aphids with host-plant use and natural enemy resistance. Ecol. Entomol. 29, 60–65. doi: 10.1111/j.1365-2311.2004.00574.x
Ferree, P. M., Avery, A., Azpurua, J., Wilkes, T., and Werren, J. H. (2008). A bacterium targets maternally inherited centrosomes to kill males in Nasonia. Curr. Biol. 18, 1409–1414. doi: 10.1016/j.cub.2008.07.093
Franzenburg, S., Walter, J., Künzel, S., Wang, J., Baines, J. F., Bosch, T. C., et al. (2013). Distinct antimicrobial peptide expression determines host species-specific bacterial associations. Proc. Natl. Acad. Sci. U.S.A. 110, E3730–E3738. doi: 10.1073/pnas.1304960110
Fraune, S., and Bosch, T. C. (2007). Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc. Natl. Acad. Sci. U.S.A. 104, 13146–13151. doi: 10.1073/pnas.0703375104
Futahashi, R., Tanaka, K., Tanahashi, M., Nikoh, N., Kikuchi, Y., Lee, B. L., et al. (2013). Gene expression in gut symbiotic organ of stinkbug affected by extracellular bacterial symbiont. PLoS ONE 8:e64557. doi: 10.1371/journal.pone.0064557
Gatschenberger, H., Azzami, K., Tautz, J., and Beier, H. (2013). Antibacterial immune competence of honey bees (Apis mellifera) is adapted to different life stages and environmental risks. PLoS ONE 8:e66415. doi: 10.1371/journal.pone.0066415
Gavotte, L., Henri, H., Stouthamer, R., Charif, D., Charlat, S., Boulétreau, M., et al. (2007). A Survey of the bacteriophage WO in the endosymbiotic bacteria Wolbachia. Mol. Biol. Evol. 24, 427–435. doi: 10.1093/molbev/msl171
Gavotte, L., Vavre, F., Henri, H., Ravallec, M., Stouthamer, R., and Boulétreau, M. (2004). Diversity, distribution and specificity of WO phage infection in Wolbachia of four insect species. Insect Mol. Biol. 13, 147–153. doi: 10.1111/j.0962-1075.2004.00471.x
Gherna, R. L., Werren, J. H., Weisburg, W., Cote, R., Woese, C. R., Mandelc, L., et al. (1991). Arsenophonus nasoniae gen. nov., sp. nov. the causative agent of the son-killer trait in the parasitic wasp Nasonia vitripennis. Int. J. Syst. Bacteriol. 41, 563–565. doi: 10.1099/00207713-41-4-563
Gibson, J. D., Niehuis, O., Peirson, B. R., Cash, E. I., and Gadau, J. (2013). Genetic and developmental basis of F2 hybrid breakdown in Nasonia parasitoid wasps. Evolution 67, 2124–2132. doi: 10.1111/evo.12080
Gilbert, S. F., Sapp, J., and Tauber, A. I. (2012). A symbiotic view of life: we have never been individuals. Q. Rev. Biol. 87, 325–341. doi: 10.1086/668166
Goto, S., Anbutsu, H., and Fukatsu, T. (2006). Asymmetrical interactions between Wolbachia and Spiroplasma endosymbionts coexisting in the same insect host. Appl. Environ. Microbiol. 72, 4805–4810. doi: 10.1128/AEM.00416-06
Hansen, A. K., and Moran, N. A. (2011). Aphid genome expression reveals host-symbiont cooperation in the production of amino acids. Proc. Natl. Acad. Sci. U.S.A. 108, 2849–2854. doi: 10.1073/pnas.1013465108
He, S., Ivanova, N., Kirton, E., Allgaier, M., Bergin, C., Scheffrahn, R. H., et al. (2013). Comparative metagenomic and metatranscriptomic analysis of hindgut paunch microbiota in wood- and dung-feeding higher termites. PLoS ONE 8:e61126. doi: 10.1371/journal.pone.0061126
Heath, B. D., Butcher, R. D., Whitfield, W. G., and Hubbard, S. F. (1999). Horizontal transfer of Wolbachia between phylogenetically distant insect species by a naturally occurring mechanism. Curr. Biol. 9, 313–316. doi: 10.1016/S0960-9822(99)80139-0
Hilgenboecker, K., Hammerstein, P., Schlattmann, P., Telschow, A., and Werren, J. H. (2008). How many species are infected with Wolbachia? - A statistical analysis of current data. FEMS Microbiol. Lett. 281, 215–220. doi: 10.1111/j.1574-6968.2008.01110.x
Huger, A. M., Skinner, S. W., and Werren, J. H. (1985). Bacterial infections associated with the son-killer trait in the parasitoid wasp Nasonia (=Mormoniella) vitripennis (Hymenoptera: Pteromalidae). J. Invertebr. Pathol. 46, 272–280. doi: 10.1016/0022-2011(85)90069-2
Hughes, G. L., Dodson, B. L., Johnson, R. M., Murdock, C. C., Tsujimoto, H., Suzuki, Y., et al. (2014). Native microbiome impedes vertical transmission of Wolbachia in Anopheles mosquitoes. Proc. Natl. Acad. Sci. U.S.A. 111, 12498–12503. doi: 10.1073/pnas.1408888111
Huigens, M. E., de Almeida, R. P., Boons, P. A., Luck, R. F., and Stouthamer, R. (2004). Natural interspecific and intraspecific horizontal transfer of parthenogenesis-inducing Wolbachia in Trichogramma wasps. Proc. Biol. Sci. 271, 509–515. doi: 10.1098/rspb.2003.2640
Hurst, G. D., and Frost, C. L. (2015). Reproductive parasitism: maternally inherited symbionts in a biparental world. Cold Spring Harb. Perspect. Biol. 7:a017699. doi: 10.1101/cshperspect.a017699
Hurst, G. D., Jiggins, F. M., and Majerus, M. E. (2003). “Inherited microorganisms that selectively kill male hosts: the hidden players of insect evolution?,” in Insect Symbiosis, eds K. Bourtzis and T. A. Miller (Boca Raton, FL: CRC Press), 177–197.
Husnik, F., Nikoh, N., Koga, R., Ross, L., Duncan, R. P., Fujie, M., et al. (2013). Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis. Cell 153, 1567–1578. doi: 10.1016/j.cell.2013.05.040
Hypsa, V., and Dale, C. (1997). In vitro culture and phylogenetic analysis of “Candidatus Arsenophonus triatominarum,” an intracellular bacterium from the triatomine bug, Triatoma infestans. Int. J. Syst. Bacteriol. 47, 1140–1144. doi: 10.1099/00207713-47-4-1140
Ishmael, N., Dunning Hotopp, J. C., Ioannidis, P., Biber, S., Sakamoto, J., Siozios, S., et al. (2009). Extensive genomic diversity of closely related Wolbachia strains. Microbiology 155, 2211–2222. doi: 10.1099/mic.0.027581-0
Kambris, Z., Blagborough, A. M., Pinto, S. B., Blagrove, M. S., Godfray, H. C., Sinden, R. E., et al. (2010). Wolbachia stimulates immune gene expression and inhibits Plasmodium development in Anopheles gambiae. PLoS Pathog. 6:e1001143. doi: 10.1371/journal.ppat.1001143
Kellner, K., Ishak, H. D., Linksvayer, T. A., and Mueller, U. G. (2015). Bacterial community composition and diversity in an ancestral ant fungus symbiosis. FEMS Microbiol. Ecol. 91:fiv073.
Kent, B. N., and Bordenstein, S. R. (2010). Phage WO of Wolbachia: lambda of the endosymbiont world. Trends Microbiol. 18, 173–181. doi: 10.1016/j.tim.2009.12.011
Kent, B. N., Salichos, L., Gibbons, J. G., Rokas, A., Newton, I. L., Clark, M. E., et al. (2011). Complete bacteriophage transfer in a bacterial endosymbiont (Wolbachia) determined by targeted genome capture. Genome Biol. Evol. 3, 209–218. doi: 10.1093/gbe/evr007
Kim, J. K., Kim, N. H., Jang, H. A., Kikuchi, Y., Kim, C. H., Fukatsu, T., et al. (2013). Specific midgut region controlling the symbiont population in an insect-microbe gut symbiotic association. Appl. Environ. Microbiol. 79, 7229–7233. doi: 10.1128/AEM.02152-13
Kirkness, E. F., Haas, B. J., Sun, W., Braig, H. R., Perotti, M. A., Clark, J. M., et al. (2010). Genome sequences of the human body louse and its primary endosymbiont provide insights into the permanent parasitic lifestyle. Proc. Natl. Acad. Sci. U.S.A. 107, 12168–12173. doi: 10.1073/pnas.1003379107
Koch, H., and Schmid-Hempel, P. (2012). Gut microbiota instead of host genotype drive the specificity in the interaction of a natural host-parasite system. Ecol. Lett. 15, 1095–1103. doi: 10.1111/j.1461-0248.2012.01831.x
Koevoets, T., Niehuis, O., van de Zande, L., and Beukeboom, L. W. (2012). Hybrid incompatibilities in the parasitic wasp genus Nasonia: negative effects of hemizygosity and the identification of transmission ratio distortion loci. Heredity (Edinb.) 108, 302–311. doi: 10.1038/hdy.2011.75
Koga, R., Bennett, G. M., Cryan, J. R., and Moran, N. A. (2013). Evolutionary replacement of obligate symbionts in an ancient and diverse insect lineage. Environ. Microbiol. 15, 2073–2081. doi: 10.1111/1462-2920.12121
Kremer, N., Charif, D., Henri, H., Gavory, F., Wincker, P., Mavingui, P., et al. (2012). Influence of Wolbachia on host gene expression in an obligatory symbiosis. BMC Microbiol. 12(Suppl. 1):S7. doi: 10.1186/1471-2180-12-S1-S7
Kremer, N., Voronin, D., Charif, D., Mavingui, P., Mollereau, B., and Vavre, F. (2009). Wolbachia interferes with ferritin expression and iron metabolism in insects. PLoS Pathog. 5:e1000630. doi: 10.1371/journal.ppat.1000630
Leal, W. S. (1998). Chemical ecology of phytophagous scarab beetles. Annu. Rev. Entomol. 43, 39–61. doi: 10.1146/annurev.ento.43.1.39
Lee, K. A., Kim, S. H., Kim, E. K., Ha, E. M., You, H., Kim, B., et al. (2013). Bacterial-derived uracil as a modulator of mucosal immunity and gut-microbe homeostasis in Drosophila. Cell 153, 797–811. doi: 10.1016/j.cell.2013.04.009
Ley, R. E., Hamady, M., Lozupone, C., Turnbaugh, P. J., Ramey, R. R., Bircher, J. S., et al. (2008). Evolution of mammals and their gut microbes. Science 320, 1647–1651. doi: 10.1126/science.1155725
Lize, A., McKay, R., and Lewis, Z. (2014). Kin recognition in Drosophila: the importance of ecology and gut microbiota. ISME J. 8, 469–477. doi: 10.1038/ismej.2013.157
Lo, N., Paraskevopoulos, C., Bourtzis, K., O’Neill, S. L., Werren, J. H., Bordenstein, S. R., et al. (2007). Taxonomic status of the intracellular bacterium Wolbachia pipientis. Int. J. Syst. Evol. Microbiol. 57, 654–657. doi: 10.1099/ijs.0.64515-0
Loehlin, D. W., Oliveira, D. C., Edwards, R., Giebel, J. D., Clark, M. E., Cattani, M. V., et al. (2010). Non-coding changes cause sex-specific wing size differences between closely related species of Nasonia. PLoS Genet. 6:e1000821. doi: 10.1371/journal.pgen.1000821
Login, F. H., Balmand, S., Vallier, A., Vincent-Monégat, C., Vigneron, A., Weiss-Gayet, M., et al. (2011). Antimicrobial peptides keep insect endosymbionts under control. Science 334, 362–365. doi: 10.1126/science.1209728
Luan, J. B., Chen, W., Hasegawa, D. K., Simmons, A. M., Wintermantel, W. M., Ling, K. S., et al. (2015). Metabolic coevolution in the bacterial symbiosis of whiteflies and related plant sap-feeding insects. Genome Biol. Evol. 7, 2635–2647. doi: 10.1093/gbe/evv170
Lynch, J. A. (2015). The expanding genetic toolbox of the wasp Nasonia vitripennis and its relatives. Genetics 199, 897–904. doi: 10.1534/genetics.112.147512
Lynch, J. A., and Desplan, C. (2006). A method for parental RNA interference in the wasp Nasonia vitripennis. Nat. Protoc. 1, 486–494. doi: 10.1038/nprot.2006.70
Manzano-Marin, A., Oceguera-Figueroa, A., Latorre, A., Jimenez-Garcia, L. F., and Moya, A. (2015). Solving a bloody mess: B-Vitamin independent metabolic convergence among gammaproteobacterial obligate endosymbionts from blood-feeding arthropods and the leech Haementeria officinalis. Genome Biol. Evol. 7, 2871–2884. doi: 10.1093/gbe/evv188
Margulis, L. (1991). “Symbiogenesis and symbionticism,” in Symbiosis as a Source of Evolutionary Innovation: Speciation and Morphogenesis, eds L. Margulis and R. Fester (Cambridge, MA: MIT Press), 1–14.
Martinson, V. G., Danforth, B. N., Minckley, R. L., Rueppell, O., Tingek, S., and Moran, N. A. (2011). A simple and distinctive microbiota associated with honey bees and bumble bees. Mol. Ecol. 20, 619–628. doi: 10.1111/j.1365-294X.2010.04959.x
Masson, F., Vallier, A., Vigneron, A., Balmand, S., Vincent-Monégat, C., Zaidman-Rémy, A., et al. (2015). Systemic infection generates a local-like immune response of the bacteriome organ in insect symbiosis. J. Innate Immun. 7, 290–301. doi: 10.1159/000368928
Masui, S., Kamoda, S., Sasaki, T., and Ishikawa, H. (2000). Distribution and evolution of bacteriophage WO in Wolbachia, the endosymbiont causing sexual alterations in arthropods. J. Mol. Evol. 51, 491–497.
McCutcheon, J. P., McDonald, B. R., and Moran, N. A. (2009). Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc. Natl. Acad. Sci. U.S.A. 106, 15394–15399. doi: 10.1073/pnas.0906424106
McCutcheon, J. P., and Moran, N. A. (2010). Functional convergence in reduced genomes of bacterial symbionts spanning 200 My of evolution. Genome Biol. Evol. 2, 708–718. doi: 10.1093/gbe/evq055
McCutcheon, J. P., and von Dohlen, C. D. (2011). An interdependent metabolic patchwork in the nested symbiosis of mealybugs. Curr. Biol. 21, 1366–1372. doi: 10.1016/j.cub.2011.06.051
McFall-Ngai, M., Hadfield, M. G., Bosch, T. C., Carey, H. V., Domazet-Lošo, T., Douglas, A. E., et al. (2013). Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. U.S.A. 110, 3229–3236. doi: 10.1073/pnas.1218525110
Miller, W. J., Ehrman, L., and Schneider, D. (2010). Infectious speciation revisited: impact of symbiont-depletion on female fitness and mating behavior of Drosophila paulistorum. PLoS Pathog. 6:e1001214. doi: 10.1371/journal.ppat.1001214
Moeller, A. H., Caro-Quintero, A., Mjungu, D., Georgiev, A. V., Lonsdorf, E. V., Muller, M. N., et al. (2016). Cospeciation of gut microbiota with hominids. Science 353, 380–382. doi: 10.1126/science.aaf3951
Montllor, C. B., Maxmen, A., and Purcell, A. H. (2002). Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol. Entomol. 27, 189–195. doi: 10.1046/j.1365-2311.2002.00393.x
Moran, N. A. (2007). Symbiosis as an adaptive process and source of phenotypic complexity. Proc. Natl. Acad. Sci. U.S.A. 104(Suppl. 1), 8627–8633. doi: 10.1073/pnas.0611659104
Moran, N. A., McCutcheon, J. P., and Nakabachi, A. (2008). Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 42, 165–190. doi: 10.1146/annurev.genet.41.110306.130119
Moran, N. A., and Sloan, D. B. (2015). The hologenome concept: helpful or hollow? PLoS Biol. 13:e1002311. doi: 10.1371/journal.pbio.1002311
Mouton, L., Dedeine, F., Henri, H., Boulétreau, M., Profizi, N., and Vavre, F. (2004). Virulence, multiple infections and regulation of symbiotic population in the Wolbachia-Asobara tabida symbiosis. Genetics 168, 181–189. doi: 10.1534/genetics.104.026716
Mouton, L., Henri, H., Bouletreau, M., and Vavre, F. (2003). Strain-specific regulation of intracellular Wolbachia density in multiply infected insects. Mol. Ecol. 12, 3459–3465. doi: 10.1046/j.1365-294X.2003.02015.x
Moya, A., Pereto, J., Gil, R., and Latorre, A. (2008). Learning how to live together: genomic insights into prokaryote-animal symbioses. Nat. Rev. Genet. 9, 218–229. doi: 10.1038/nrg2319
Najarro, M. A., Sumethasorn, M., Lamoureux, A., and Turner, T. L. (2015). Choosing mates based on the diet of your ancestors: replication of non-genetic assortative mating in Drosophila melanogaster. PeerJ 3:e1173. doi: 10.7717/peerj.1173
Narita, S., Kageyama, D., Nomura, M., and Fukatsu, T. (2007). Unexpected mechanism of symbiont-induced reversal of insect sex: feminizing Wolbachia continuously acts on the butterfly Eurema hecabe during larval development. Appl. Environ. Microbiol. 73, 4332–4341. doi: 10.1128/AEM.00145-07
Newell, P. D., and Douglas, A. E. (2014). Interspecies interactions determine the impact of the gut microbiota on nutrient allocation in Drosophila melanogaster. Appl. Environ. Microbiol. 80, 788–796. doi: 10.1128/AEM.02742-13
Niehuis, O., Judson, A. K., and Gadau, J. (2008). Cytonuclear genic incompatibilities cause increased mortality in male F2 hybrids of Nasonia giraulti and N. vitripennis. Genetics 178, 413–426. doi: 10.1534/genetics.107.080523
Novakova, E., Husnik, F., Sochova, E., and Hypsa, V. (2015). Arsenophonus and sodalis symbionts in louse flies: an analogy to the wigglesworthia and sodalis system in tsetse flies. Appl. Environ. Microbiol. 81, 6189–6199. doi: 10.1128/AEM.01487-15
Noyes, J. S. (2016) Universal Chalcidoidea Database. World Wide Web Electronic Publication. Available at: http://www.nhm.ac.uk/chalcidoids
Ochman, H., Worobey, M., Kuo, C. H., Ndjango, J. B., Peeters, M., Hahn, B. H., et al. (2010). Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLoS Biol. 8:e1000546. doi: 10.1371/journal.pbio.1000546
Oliveira, D. C., Hunter, W. B., Ng, J., Desjardins, C. A., Dang, P. M., and Werren, J. H. (2010). Data mining cDNAs reveals three new single stranded RNA viruses in Nasonia (Hymenoptera: Pteromalidae). Insect Mol. Biol. 19(Suppl. 1), 99–107. doi: 10.1111/j.1365-2583.2009.00934.x
Oliver, K. M., Degnan, P. H., Hunter, M. S., and Moran, N. A. (2009). Bacteriophages encode factors required for protection in a symbiotic mutualism. Science 325, 992–994. doi: 10.1126/science.1174463
Oliver, K. M., Russell, J. A., Moran, N. A., and Hunter, M. S. (2003). Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc. Natl. Acad. Sci. U.S.A. 100, 1803–1807. doi: 10.1073/pnas.0335320100
O’Neill, S. L., Giordano, R., Colbert, A. M., Karr, T. L., and Robertson, H. M. (1992). 16S rRNA phylogenetic analysis of the bacterial endosymbionts associated with cytoplasmic incompatibility in insects. Proc. Natl. Acad. Sci. U.S.A. 89, 2699–2702. doi: 10.1073/pnas.89.7.2699
Pan, X., Zhou, G., Wu, J., Bian, G., Lu, P., Raikhel, A. S., et al. (2012). Wolbachia induces reactive oxygen species (ROS)-dependent activation of the Toll pathway to control dengue virus in the mosquito Aedes aegypti. Proc. Natl. Acad. Sci. U.S.A. 109, E23–E31. doi: 10.1073/pnas.1116932108
Paolucci, S., van de Zande, L., and Beukeboom, L. W. (2013). Adaptive latitudinal cline of photoperiodic diapause induction in the parasitoid Nasonia vitripennis in Europe. J. Evol. Biol. 26, 705–718. doi: 10.1111/jeb.12113
Parratt, S. R., Frost, C. L., Schenkel, M. A., Rice, A., Hurst, G. D., and King, K. C. (2016). Superparasitism drives heritable symbiont epidemiology and host sex ratio in a wasp. PLoS Pathog. 12:e1005629. doi: 10.1371/journal.ppat.1005629
Phillips, C. D., Phelan, G., Dowd, S. E., McDonough, M. M., Ferguson, A. W., Delton Hanson, J., et al. (2012). Microbiome analysis among bats describes influences of host phylogeny, life history, physiology, and geography. Mol. Ecol. 21, 2617–2627. doi: 10.1111/j.1365-294X.2012.05568.x
Ramírez-Puebla, S. T., Servín-Garcidueñas, L. E., Ormeño-Orrillo, E., Vera-Ponce de León, A., Rosenblueth, M., Delaye, L., et al. (2015). Species in Wolbachia? Proposal for the designation of ‘Candidatus Wolbachia bourtzisii’, ‘Candidatus Wolbachia onchocercicola’, ‘Candidatus Wolbachia blaxteri’, ‘Candidatus Wolbachia brugii’, ‘Candidatus Wolbachia taylori’, ‘Candidatus Wolbachia collembolicola’ and ‘Candidatus Wolbachia multihospitum’ for the different species within Wolbachia supergroups. Syst. Appl. Microbiol. 38, 390–399. doi: 10.1016/j.syapm.2015.05.005
Raychoudhury, R., Baldo, L., Oliveira, D. C., and Werren, J. H. (2009). Modes of acquisition of Wolbachia: horizontal transfer, hybrid introgression, and codivergence in the Nasonia species complex. Evolution 63, 165–183. doi: 10.1111/j.1558-5646.2008.00533.x
Raychoudhury, R., Desjardins, C. A., Buellesbach, J., Loehlin, D. W., Grillenberger, B. K., Beukeboom, L., et al. (2010a). Behavioral and genetic characteristics of a new species of Nasonia. Heredity (Edinb) 104, 278–288. doi: 10.1038/hdy.2009.147
Raychoudhury, R., Grillenberger, B. K., Gadau, J., Bijlsma, R., van de Zande, L., Werren, J. H., et al. (2010b). Phylogeography of Nasonia vitripennis (Hymenoptera) indicates a mitochondrial-Wolbachia sweep in North America. Heredity (Edinb.) 104, 318–326. doi: 10.1038/hdy.2009.160
Raychoudhury, R., and Werren, J. H. (2012). Host genotype changes bidirectional to unidirectional cytoplasmic incompatibility in Nasonia longicornis. Heredity 108, 105–114. doi: 10.1038/hdy.2011.53
Ridley, E. V., Wong, A. C., Westmiller, S., and Douglas, A. E. (2012). Impact of the resident microbiota on the nutritional phenotype of Drosophila melanogaster. PLoS ONE 7:e36765. doi: 10.1371/journal.pone.0036765
Ros, V. I., and Breeuwer, J. A. (2009). The effects of, and interactions between, Cardinium and Wolbachia in the doubly infected spider mite Bryobia sarothamni. Heredity (Edinb.) 102, 413–422. doi: 10.1038/hdy.2009.4
Rosenberg, E., Koren, O., Reshef, L., Efrony, R., and Zilber-Rosenberg, I. (2007). The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 5, 355–362. doi: 10.1038/nrmicro1635
Rosenberg, M. I., Brent, A. E., Payre, F., and Desplan, C. (2014). Dual mode of embryonic development is highlighted by expression and function of Nasonia pair-rule genes. Elife 3:e01440. doi: 10.7554/eLife.01440
Rossi, P., Ricci, I., Cappelli, A., Damiani, C., Ulissi, U., Mancini, M. V., et al. (2015). Mutual exclusion of Asaia and Wolbachia in the reproductive organs of mosquito vectors. Parasit Vectors 8:278. doi: 10.1186/s13071-015-0888-0
Rousset, F., Bouchon, D., Pintureau, B., Juchault, P., and Solignac, M. (1992). Wolbachia endosymbionts responsible for various alterations of sexuality in arthropods. Proc. Biol. Sci. 250, 91–98. doi: 10.1098/rspb.1992.0135
Ryu, J. H., Kim, S. H., Lee, H. Y., Bai, J. Y., Nam, Y. D., Bae, J. W., et al. (2008). Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science 319, 777–782. doi: 10.1126/science.1149357
Sackton, T. B., Werren, J. H., and Clark, A. G. (2013). Characterizing the infection-induced transcriptome of Nasonia vitripennis reveals a preponderance of taxonomically-restricted immune genes. PLoS ONE 8:e83984. doi: 10.1371/journal.pone.0083984
Sanders, J. G., Powell, S., Kronauer, D. J., Vasconcelos, H. L., Frederickson, M. E., and Pierce, N. E. (2014). Stability and phylogenetic correlation in gut microbiota: lessons from ants and apes. Mol. Ecol. 23, 1268–1283. doi: 10.1111/mec.12611
Serbus, L. R., Casper-Lindley, C., Landmann, F., and Sullivan, W. (2008). The genetics and cell biology of Wolbachia-host interactions. Annu. Rev. Genet. 42, 683–707. doi: 10.1146/annurev.genet.41.110306.130354
Sharon, G., Segal, D., Ringo, J. M., Hefetz, A., Zilber-Rosenberg, I., and Rosenberg, E. (2010). Commensal bacteria play a role in mating preference of Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 107, 20051–20056. doi: 10.1073/pnas.1009906107
Shigenobu, S., and Stern, D. L. (2013). Aphids evolved novel secreted proteins for symbiosis with bacterial endosymbiont. Proc. Biol. Sci. 280:20121952. doi: 10.1098/rspb.2012.1952
Shin, S. C., Kim, S. H., You, H., Kim, B., Kim, A. C., Lee, K. A., et al. (2011). Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 334, 670–674. doi: 10.1126/science.1212782
Shropshire, J. D., and Bordenstein, S. R. (2016). Speciation by symbiosis: the microbiome and behavior. MBio 7:e01785-15.
Shropshire, J. D., van Opstal, E. J., and Bordenstein, S. R. (2016). An optimized approach to germ-free rearing in the jewel wasp Nasonia. PeerJ Preprints 4:e2088v1. doi: 10.7287/peerj.preprints.2088v1
Sicard, M., Dittmer, J., Greve, P., Bouchon, D., and Braquart-Varnier, C. (2014). A host as an ecosystem: Wolbachia coping with environmental constraints. Environ. Microbiol. 16, 3583–3607. doi: 10.1111/1462-2920.12573
Skinner, S. W. (1985). Son-killer: a third extrachromosomal factor affecting the sex ratio in the parasitoid wasp, Nasonia (=Mormoniella) vitripennis. Genetics 109, 745–759.
Storelli, G., Defaye, A., Erkosar, B., Hols, P., Royet, J., and Leulier, F. (2011). Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab. 14, 403–414. doi: 10.1016/j.cmet.2011.07.012
Stouthamer, R., Breeuwert, J. A., Luck, R. F., and Werren, J. H. (1993). Molecular identification of microorganisms associated with parthenogenesis. Nature 361, 66–68. doi: 10.1038/361066a0
Taylor, G. P., Coghlin, P. C., Floate, K. D., and Perlman, S. J. (2011). The host range of the male-killing symbiont Arsenophonus nasoniae in filth fly parasitioids. J. Invertebr. Pathol. 106, 371–379. doi: 10.1016/j.jip.2010.12.004
Theis, K. R., Dheilly, N. M., Klassen, J. L., Brucker, R. M., Baines, J. F., Bosch, T. C. G., et al. (2016). Getting the hologenome concept right: an eco-evolutionary framework for hosts and their microbiomes. mSystems 1, e00028-16. doi: 10.1128/mSystems.00028-16
Tian, C., Gao, B., Fang, Q., Ye, G., and Zhu, S. (2010). Antimicrobial peptide-like genes in Nasonia vitripennis: a genomic perspective. BMC Genomics 11:187. doi: 10.1186/1471-2164-11-187
Tram, U., and Sullivan, W. (2002). Role of delayed nuclear envelope breakdown and mitosis in Wolbachia-induced cytoplasmic incompatibility. Science 296, 1124–1126. doi: 10.1126/science.1070536
Tsuchida, T., Koga, R., and Fukatsu, T. (2004). Host plant specialization governed by facultative symbiont. Science 303:1989. doi: 10.1126/science.1094611
van Opijnen, T., Baudry, E., Baldo, L., Bartos, J., and Werren, J. H. (2005). Genetic variability in the three genomes of Nasonia: nuclear, mitochondrial and Wolbachia. Insect Mol. Biol. 14, 653–663. doi: 10.1111/j.1365-2583.2005.00595.x
Varaldi, J., Bouletreau, M., and Fleury, F. (2005). Cost induced by viral particles manipulating superparasitism behaviour in the parasitoid Leptopilina boulardi. Parasitology 131, 161–168. doi: 10.1017/S0031182005007602
Varaldi, J., Fouillet, P., Ravallec, M., López-Ferber, M., Boulétreau, M., Fleury, F., et al. (2003). Infectious behavior in a parasitoid. Science 302:1930. doi: 10.1126/science.1088798
Varaldi, J., Ravallec, M., Labrosse, C., Lopez-Ferber, M., Boulétreau, M., and Fleury, F. (2006). Artifical transfer and morphological description of virus particles associated with superparasitism behaviour in a parasitoid wasp. J. Insect Physiol. 52, 1202–1212. doi: 10.1016/j.jinsphys.2006.09.002
Vasanthakumar, A., Handelsman, J., Schloss, P. D., Bauer, L. S., and Raffa, K. F. (2008). Gut microbiota of an invasive subcortical beetle, Agrilus planipennis Fairmaire, across various life stages. Environ. Entomol. 37, 1344–1353.
Vavre, F., Fleury, F., Lepetit, D., Fouillet, P., and Bouletreau, M. (1999). Phylogenetic evidence for horizontal transmission of Wolbachia in host-parasitoid associations. Mol. Biol. Evol. 16, 1711–1723. doi: 10.1093/oxfordjournals.molbev.a026084
Vavre, F., Fleury, F., Varaldi, J., Fouillet, P., and Bouletreau, M. (2000). Evidence for female mortality in Wolbachia-mediated cytoplasmic incompatibility in haplodiploid insects: epidemiologic and evolutionary consequences. Evolution 54, 191–200. doi: 10.1554/0014-3820(2000)054[0191:EFFMIW]2.0.CO;2
Vavre, F., and Kremer, N. (2014). Microbial impacts on insect evolutionary diversification: from patterns to mechanisms. Curr Opin Insect Sci. 4, 29–34. doi: 10.1016/j.cois.2014.08.003
Vavre, F., Mouton, L., and Pannebakker, B. A. (2009). Drosophila-parasitoid communities as model systems for host-Wolbachia interactions. Adv. Parasitol. 70, 299–331. doi: 10.1016/S0065-308X(09)70012-0
Verhulst, E. C., Beukeboom, L. W., and van de Zande, L. (2010). Maternal control of haplodiploid sex determination in the wasp Nasonia. Science 328, 620–623. doi: 10.1126/science.1185805
Wang, J., Wu, Y., Yang, G., and Aksoy, S. (2009). Interactions between mutualist Wigglesworthia and tsetse peptidoglycan recognition protein (PGRP-LB) influence trypanosome transmission. Proc. Natl. Acad. Sci. U.S.A. 106, 12133–12138. doi: 10.1073/pnas.0901226106
Wang, X., Wheeler, D., Avery, A., Rago, A., Choi, J. H., Colbourne, J. K., et al. (2013). Function and evolution of DNA methylation in Nasonia vitripennis. PLoS Genet. 9:e1003872. doi: 10.1371/journal.pgen.1003872
Werren, J. H., Baldo, L., and Clark, M. E. (2008). Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6, 741–751. doi: 10.1038/nrmicro1969
Werren, J. H., and Loehlin, D. W. (2009). The parasitoid wasp Nasonia: an emerging model system with haploid male genetics. Cold Spring Harb Protoc. 2009:pdb emo134.
Werren, J. H., Loehlin, D. W., and Giebel, J. D. (2009). Larval RNAi in Nasonia (parasitoid wasp). Cold Spring Harb. Protoc. 2009:pdb.prot5311.
Werren, J. H., Richards, S., Desjardins, C. A., Niehuis, O., Gadau, J., Colbourne, J. K., et al. (2010). Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science 327, 343–348. doi: 10.1126/science.1178028
Werren, J. H., Skinner, S. W., and Huger, A. M. (1986). Male-killing bacteria in a parasitic wasp. Science 231, 990–992. doi: 10.1126/science.3945814
Werren, J. H., Zhang, W., and Guo, L. R. (1995). Evolution and phylogeny of Wolbachia: reproductive parasites of arthropods. Proc. Biol. Sci. 261, 55–63. doi: 10.1098/rspb.1995.0117
Wilkes, T. E., Darby, A. C., Choi, J. H., Colbourne, J. K., Werren, J. H., and Hurst, G. D. (2010). The draft genome sequence of Arsenophonus nasoniae, son-killer bacterium of Nasonia vitripennis, reveals genes associated with virulence and symbiosis. Insect Mol. Biol. 19(Suppl. 1), 59–73. doi: 10.1111/j.1365-2583.2009.00963.x
Wilson, A. C., Ashton, P. D., Calevro, F., Charles, H., Colella, S., Febvay, G., et al. (2010). Genomic insight into the amino acid relations of the pea aphid, Acyrthosiphon pisum, with its symbiotic bacterium Buchnera aphidicola. Insect Mol. Biol. 19(Suppl. 2), 249–258. doi: 10.1111/j.1365-2583.2009.00942.x
Wong, A. C., Dobson, A. J., and Douglas, A. E. (2014). Gut microbiota dictates the metabolic response of Drosophila to diet. J. Exp. Biol. 217, 1894–1901. doi: 10.1242/jeb.101725
Wu, M., Sun, L. V., Vamathevan, J., Riegler, M., Deboy, R., Brownlie, J. C., et al. (2004). Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2:E69. doi: 10.1371/journal.pbio.0020069
Wulff, J. A., and White, J. A. (2015). The endosymbiont arsenophonus provides a general benefit to soybean aphid (hemiptera: aphididae) regardless of host plant resistance (Rag). Environ. Entomol. 44, 574–581. doi: 10.1093/ee/nvv031
Yoshiyama, M., and Kimura, K. (2009). Bacteria in the gut of Japanese honeybee, Apis cerana japonica, and their antagonistic effect against Paenibacillus larvae, the causal agent of American foulbrood. J. Invertebr. Pathol. 102, 91–96. doi: 10.1016/j.jip.2009.07.005
Zilber-Rosenberg, I., and Rosenberg, E. (2008). Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol. Rev. 32, 723–735. doi: 10.1111/j.1574-6976.2008.00123.x
Keywords: host-symbiont interactions, microbiome, Nasonia, symbiosis, Wolbachia, Arsenophonus
Citation: Dittmer J, van Opstal EJ, Shropshire JD, Bordenstein SR, Hurst GDD and Brucker RM (2016) Disentangling a Holobiont – Recent Advances and Perspectives in Nasonia Wasps. Front. Microbiol. 7:1478. doi: 10.3389/fmicb.2016.01478
Received: 30 June 2016; Accepted: 05 September 2016;
Published: 23 September 2016.
Edited by:
Sebastian Fraune, University of Kiel, GermanyReviewed by:
Natacha Kremer, Claude Bernard University Lyon 1, FranceFranck Dedeine, François Rabelais University, France
Copyright © 2016 Dittmer, van Opstal, Shropshire, Bordenstein, Hurst and Brucker. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robert M. Brucker, YnJ1Y2tlckByb3dsYW5kLmhhcnZhcmQuZWR1
 Jessica Dittmer
Jessica Dittmer Edward J. van Opstal
Edward J. van Opstal J. Dylan Shropshire
J. Dylan Shropshire Seth R. Bordenstein
Seth R. Bordenstein Gregory D. D. Hurst
Gregory D. D. Hurst Robert M. Brucker
Robert M. Brucker