
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 05 September 2016
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 7 - 2016 | https://doi.org/10.3389/fmicb.2016.01374
β-lactamases are the primary cause of resistance to β-lactams among members of the family Enterobacteriaceae. SHV enzymes have emerged in Enterobacteriaceae causing infections in health care in the last decades of the Twentieth century, and they are now observed in isolates in different epidemiological settings both in human, animal and the environment. Likely originated from a chromosomal penicillinase of Klebsiella pneumoniae, SHV β-lactamases currently encompass a large number of allelic variants including extended-spectrum β-lactamases (ESBL), non-ESBL and several not classified variants. SHV enzymes have evolved from a narrow- to an extended-spectrum of hydrolyzing activity, including monobactams and carbapenems, as a result of amino acid changes that altered the configuration around the active site of the β -lactamases. SHV-ESBLs are usually encoded by self-transmissible plasmids that frequently carry resistance genes to other drug classes and have become widespread throughout the world in several Enterobacteriaceae, emphasizing their clinical significance.
Thanks to their ability to inhibit cell wall biosynthesis, β-lactams remained the first-line defense against bacterial infections for over 20 years, before resistant bacteria appeared in clinical practice.
Resistance to this class of drugs can be the result of antibiotic target site alteration, prevention of antibiotic access by altered permeability or forced efflux, or antibiotic degradation (Wilke et al., 2005). The latter, represents the primary resistance mechanism in Gram-negative bacteria producing β-lactamase enzymes able to covalently bind the carbonyl moiety of the β-lactam ring and hydrolyze its amide bond (Fisher et al., 2005). Naturally occurring chromosomally located β-lactamases are quite common in Gram-negative bacteria; likely evolved from penicillin-binding proteins, when produced in small quantity they do not significantly contribute to antibiotic resistance. It was the appearance of the first plasmid-mediated β-lactamase TEM-1 (Datta and Kontomichalou, 1965) to designate the beginning of an unstoppable phenomenon in the 1960s. Ever since, the introduction of new natural or synthetic drugs to replace old ones in an attempt to limit the insurgence of antibiotic resistant bacteria triggered a chain reaction providing bacteria with a constant selective pressure driving the expansion of different resistance mechanisms (Medeiros, 1997).
In recent years β-lactamases have extensively diversified in response to the clinical use of new generations of β-lactams (penicillin, cephalosporins, carbapenems, and monobactams) leading to the need of classification schemes. Based on primary structure (Ambler, 1980), enzymatic properties and biochemical attributes (Bush et al., 1995), and the increasingly available amino acid sequences (Bush and Jacoby, 2010) four major classes (A, B, C, D) can be acknowledged. Serine β-lactamases belonging to class A are the most abundant (Philippon et al., 2016), with more than 500 enzymes, including the most clinically significant extended spectrum β-lactamases (ESBL) variants, i.e., CTX-M-, TEM-, and SHV-type enzymes (Bush and Fisher, 2011).
Although, SHV enzymes did not undergo the explosive dissemination observed for CTX-M-type variants (Canton et al., 2012), in recent years they have been found in several Enterobacteriaceae outside of the typical clinical hosts Klebsiella pneumoniae and Escherichia coli, with a rising allele variability (http://www.lahey.org/studies), and in different environmental niches. Many admirable works describing the biochemistry, the genetics and the evolution of SHV β-lactamases have appeared over the last years. The aim of this review is to provide the readers with an updated overview on SHV β-lactamases, their amino acid variants and spectrum of activity, and to describe the occurrence of plasmid-associated SHV enzymes in Enterobacteriaceae and their epidemiological significance.
The first blaSHV-1 gene was identified in the 1970s in E. coli (Pitton, 1972). The encoded enzyme SHV−1 (sulfhydryl reagent variable) proved its activity against penicillins and first generation cephalosporins (Matthew et al., 1979) and was confirmed part of the conjugative plasmid p453 (Barthélémy et al., 1988; Table 1). The most likely ancestor of the plasmid-mediated SHV−1 is a chromosomal species-specific penicillinase detected in fecal K. pneumoniae isolates from neonates (Haeggman et al., 1997). The enzyme showed a typical antibiogram with penicillin rather than cephalosporin resistance and a marked inhibition by clavulanic acid. How blaSHV-1 moved from the chromosome to the plasmid does not have a conclusive explanation since the proposed association with a transposable element (Nugent and Hedges, 1979) has not been confirmed.
As of today, 1891 SHV allelic variants have been described, having developed resistance to 3rd generation cephalosporin (Tzouvelekis and Bonomo, 1999), monobactam and carbapenems (Poirel et al., 2003). Only a small proportion is biochemically and/or genetically characterized (http://www.lahey.org/studies). SHV β-lactamases can be divided into three subgroups on the basis of molecular characteristics or functional properties: (i) subgroup 2b (n = 37), able to hydrolyze penicillins and early cephalosporins (cephaloridine and cephalothin) and strongly inhibited by clavulanic acid and tazobactam; (ii) subgroup 2br (n = 7), broad-spectrum β-lactamases that acquired resistance to clavulanic acid; and (iii) subgroup 2be (n = 46), comprises ESBLs that can also hydrolyze one or more oxyimino β-lactams (cefotaxime, ceftazidime, and aztreonam). More than half of these variants (n = 99) has not been classified yet due to absence of biochemical characterization.
Figure 1 illustrates a phylogenetic analysis of 149 out of the 189 SHV β-lactamase variants whose amino acid sequences were available online (http://www.lahey.org/studies), as of July 2016. Unlike other β-lactamase families (D'andrea et al., 2013; Evans and Amyes, 2014), there is no clear clustering of the different subgroups, as also mirrored by gene based analysis (Supplementary Figure S1). Among the majority of unclassified variants, subgroup 2b and the few 2br variants are scattered all over the tree. Subgroup 2be showed clustering of most of the ESBL variants (including SHV-2a, SHV-5, and SHV-12), together with few non-classified enzymes (SHV−29, SHV−152, SHV−153, SHV−160, and SHV−165). It has been proposed that SHV β-lactamases descended from an unidentified ancestor holding an extended spectrum phenotype (2be) and that subgroup 2b derived from it (Hall and Barlow, 2004). Our analysis showed that several of SHV ESBL variants were scattered along the tree with short branch lengths with neighboring 2b or unknown variants within the SHV phylogeny (i.e., SHV−40, SHV−11, and SHV−35; Figure 1), supporting the hypothesis that they evolved from multiple variants, probably within the antibiotic era. Among the non-ESBL variants, blaSHV-11 represents one of the most successful and, together with blaSHV-1, the likely source of evolution for the existing SHV ESBL variants. blaSHV-11 was first identified as plasmid-encoded in clinical K. pneumoniae from Switzerland (Nüesch-Inderbinen et al., 1997) and ever since has been isolated worldwide.
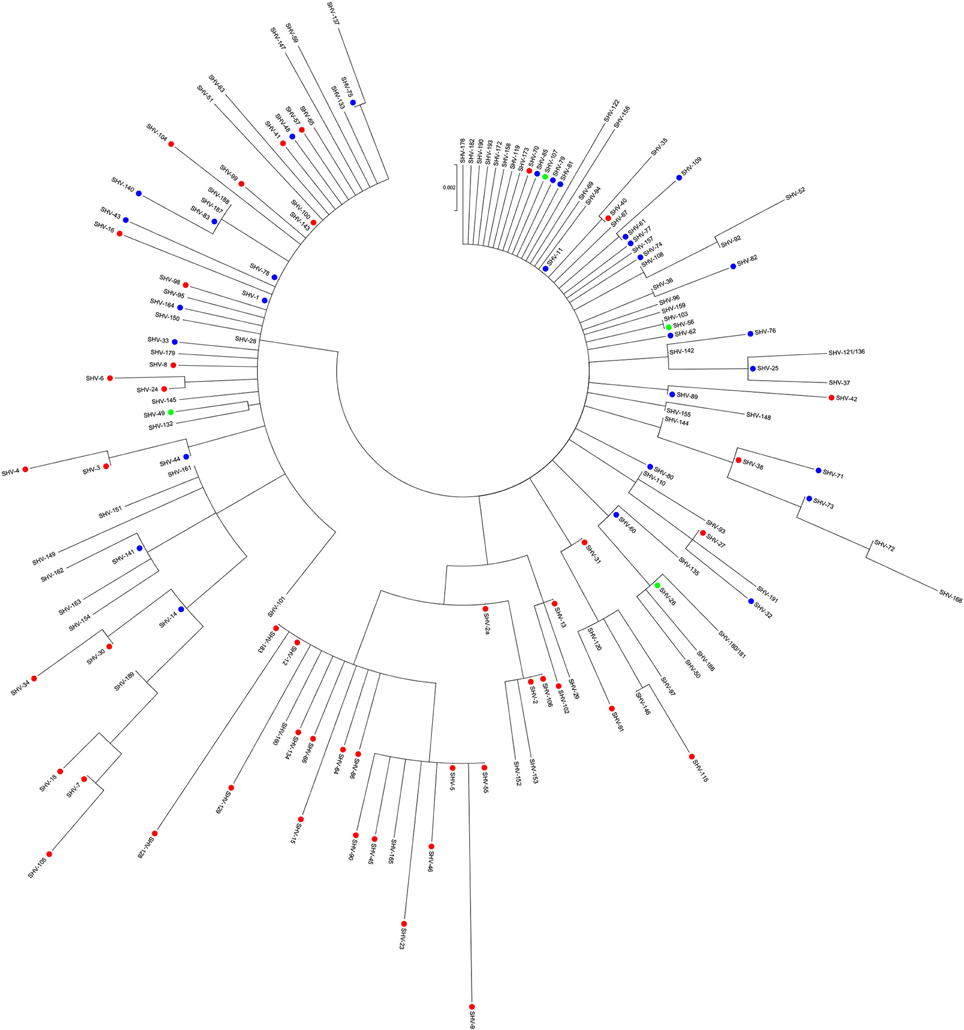
Figure 1. Maximum likelihood amino acid tree of 149 SHV-type β-lactamases. Variants whose sequence has not been released in GenBank as of July 2016, that show partial sequence or are identical to others (http://www.lahey.org/studies/) were not included in the analysis. SHV-180 and SHV-181 share the same sequence as well as SHV-121 and SHV-136. The tree was implemented in Mega version 6.06 (Tamura et al., 2013). Solid circles represent: red, extended-spectrum β-lactamases (2be; n = 46); green, broad-spectrum β-lactamases (2br, n = 5); and blue, penicillinases (2b, n = 30). Unclassified alleles are reported in black (n = 68).
Although, nearly displaced, together with TEM, by CTX-M enzymes over the years (Canton et al., 2012), 46 ESBL blaSHV genes have been described so far (Table 1). The first report of SHV-mediated resistance to third-generation cephalosporins was in 1983 with the isolation and characterization of blaSHV-2, encoded by plasmid pBP60 in a German clinical isolate of Klebsiella ozaenae and showing only a few nucleotide mismatches with blaSHV-1 (Kliebe et al., 1985). In a few years four other ESBL variants were identified as plasmid-encoded in clinical K. pneumoniae, showing variable gene homologies with the blaSHV-1 and blaSHV-2 sequences (50–90%): blaSHV-2a encoded by conjugative plasmid pZMP1 (Podbielski et al., 1991); blaSHV-3 on pUD18 (Nicolas et al., 1989); blaSHV-4, widely disseminated from France as a result of a single K. pneumoniae clone diffusion (Arlet et al., 1990, 1994); and blaSHV-5 able to hydrolyze broad-spectrum cephalosporins and monobactams (Gutmann et al., 1989). Of these first variants, the most epidemiologically successful were blaSHV-2a and blaSHV-5, which will be further discussed, together with blaSHV-2 and blaSHV-12, in a dedicated paragraph (Section Expansion toward New Ecological Niches). Interestingly, blaSHV-3 and blaSHV-4 have been only sporadically detected since their first description. blaSHV-3 seems to be geographically restricted to the USA where it was detected in E. coli of animal origin, associated with other antibiotic resistance genes such as blaCTX-M-15, blaCTX-M-24, blaCMY-2, and/or blaTEM-1 (Shaheen et al., 2011). blaSHV-4 was identified also in Enterobacter aerogenes and Citrobacter diversus in different countries (Arpin et al., 1996; El Harrif-Heraud et al., 1997; Pitout et al., 1998).
The last two decades witnessed the appearance of several new variants (blaSHV-7, blaSHV-8, blaSHV-9, blaSHV-31, blaSHV-38, blaSHV-40, blaSHV-41, and blaSHV-42) whose dissemination was restricted to limited cases (Supplementary Table S1). A few variants seem to be geographically constrained: (i) blaSHV-106, only described in Portuguese isolates of K. pneumoniae together with blaTEM-1, and/or blaCTX-M-32 (Mendonça et al., 2009); (ii) blaSHV-55, in Portugal (Mendonça et al., 2006; Machado et al., 2007) and recently in Brazil (Dropa et al., 2015); and (iii) blaSHV-57, in E. coli isolates from Taiwan and China (Ma et al., 2005; Tian et al., 2012). A variant worth to mention is blaSHV-27 (Corkill et al., 2001), that has been detected on different plasmids in E. coli, K. pneumoniae and Enterobacter cloacae, associated with a vast array of antibiotic resistance genes (blaDHA−1, blaTEM-1, blaTEM−1b, blaCMY-2, blaIMP, blaCTX-M-14, blaCTX-M-15, blaSHV-12, blaSHV-45, blaOXA-1, dfrA5, ereA2; Muratani et al., 2006; Abbassi et al., 2008; Kiratisin et al., 2008; Duval et al., 2009; Hammami et al., 2011).
Most of SHV ESBLs (25 out of 46) are unique cases, with only one report so far. Seventeen variants are exclusively found in clinical K. pneumoniae: blaSHV-6, blaSHV-13, blaSHV-16, blaSHV-18, blaSHV-23, blaSHV-45, blaSHV-64,blaSHV-66, blaSHV-86, blaSHV-90, blaSHV-91, blaSHV-98, blaSHV-99, blaSHV-100,blaSHV−104,blaSHV-105, and blaSHV-134. These variants have been described worldwide (Brazil, Portugal, Algeria, USA, Tunisia, Netherlands, France, South Africa, Colombia, and China) and are mostly associated to plasmids (Table 1). Some of these variants are sporadically accompanied by other antibiotic resistance genes like in the case of: (i) blaSHV-45 encoded by an IncA/C plasmid together with blaCTX-M-2 and blaSHV-27 (Dropa et al., 2015); (ii) blaSHV-134 encoded by an IncFII plasmid accompanied by a second plasmid carrying blaVIM−1 (Sánchez-Romero et al., 2012); (iii) and blaSHV-105, conferring reduced susceptibility to ceftazidime, ceftriaxone, and aztreonam together with blaSHV-1, and blaSHV-5 (Jones et al., 2009). One of the oldest variants, blaSHV-6, was only described in France in 1991 in a K. pneumoniae clinical case (Arlet et al., 1991). It might be speculated that the 180 kb plasmid encoding blaSHV-6 and conferring decreased susceptibility to ceftazidime and aztreonam was not stable or it reduced bacterial strain fitness preventing a successful dissemination.
Four variants have been described only in clinical E. coli: (i) blaSHV-15, described together with blaCMY-2 in a strain imported from India into the United Kingdom (http://www.lahey.org/studies/); (ii) blaSHV-24, identified in Japan on a transferable 150 Kb plasmid conferring high-level resistance to ceftazidime but not cefotaxime and cefazolin (Kurokawa et al., 2000); emergence of SHV-24 might have been driven by the extensive use of ceftazidime in Japan, enabling bacterial survival in high concentrations of this drug; (iii) blaSHV-102, recovered in a Spanish hospital and hydrolyzing cefotaxime and ceftazidime (Vinué et al., 2008); (iv) and blaSHV-129, detected in an abscess specimen from a patient hospitalized in Italy in 2008 (Lascols et al., 2012).
blaSHV-46 was only described on a 70 Kb conjugative plasmid also carrying blaTEM-1 and blaKPC−2 in a carbapenem-resistant strain of Klebsiella oxytoca from the urine of a hospitalized patient in New York (USA) in 1998 (Yigit et al., 2003). Finally, blaSHV-34 is an interesting example of extended-spectrum β-lactamase encoded by an epidemic plasmid circulating among Citrobacter koseri, E. coli, and K. pneumoniae in the same US hospital between 1998 and 2000 (Heritage et al., 2003).
Majority of SHV ESBLs have been detected in K. pneumoniae or E. coli (Table 1). blaSHV-30 was the first variant to be detected in an E. cloacae isolate from a blood culture from a solid-organ transplant recipient in the USA in 2003 (Szabó et al., 2005). The gene, previously described in K. pneumoniae and Salmonella (Mulvey et al., 2004; Whichard et al., 2007), was located on a 9.4 Kb plasmid and contributed together with chromosomal ampC, blaSHV-7, and blaTEM-1 to the antibiotic resistance profile of the E. cloacae isolate, the first of its kind producing two different SHV enzymes. Three other novel ESBL variants have been solely identified as plasmid-encoded in clinical E. cloacae: (i) blaSHV-70, from a Chinese patient with history of ceftazidime treatment (Ling et al., 2006) and observed in other clinical Chinese settings (Liu et al., 2008); (ii) blaSHV-128, isolated in Tunisia in 2009, located on an IncFII conjugative plasmid, and conferring resistance to all β-lactams except imipenem (Bourouis et al., 2015); (iii) and blaSHV-183, for which additional description is not available (http://www.lahey.org/studies/).
Extended-spectrum SHV β-lactamases belong to functional group 2be, while very recently they were assigned to subclass A1 of serine β-lactamases, clustering with TEM and CTX-M enzymes among other clinically relevant β-lactamases (Bush, 2013; Philippon et al., 2016). SHV ESBLs consist of two subdomains: an α/β that includes an antiparallel five-stranded β-sheet flanked by α-helices, and an all-α-helical subdomain (Matagne et al., 1998). Similar to TEM β-lactamases (Jelsch et al., 1993), the active site is located within the cleft created by the subdomains and it contains the Ser70 residue that mediates the nucleophilic attack on the carbonyl group of the β-lactam ring. In the vicinity of this serine residue, several conserved structural and functional amino acid motifs have been identified. These include the Ser70XXLys (“SXXK” motif, with X representing variable amino acids), the Ser130AspAsn (“SDN” motif), the Glu166XXLysAsn (“EXXLN” motif), and the Lys234Thr/SerGly (“KTG” motif) (Bush, 2013).
Each SHV ESBL has one (SHV-2, SHV-6, SHV-8, SHV-24, SHV-27, SHV-38, SHV-41, SHV-57, SHV-98, SHV-99, SHV-102, and SHV-104) to six (SHV-128) amino acid substitutions when compared to SHV-1 (Table 2), indicating that even a single amino acid substitution is enough to convey an extended-spectrum phenotype. Therefore, we can speculate that other SHV ESBLs may still evolve from a parental SHV β-lactamase due to single spectrum-extending substitutions, although the majority of them have possibly emerged through a stepwise acquisition of several mutations (substitutions, deletions and/or insertions) from pre-existing extended-spectrum SHV variants.
Among SHV ESBLs, amino acid substitutions are predominantly located at positions Leu35, Gly238, and Glu240, while other less frequent but critical substitutions for the extended-spectrum phenotype occur on several amino acids including Ile8, Arg43, Glu64, Gly156, Asp179, and Arg205 (Table 2). Although, most of these residues are not involved directly in β-lactams hydrolysis, they result in the enhancement or relaxation of the active site, enabling it to accommodate and to efficiently react with oxyimino-β-lactams (Tzouvelekis and Bonomo, 1999). Amino acid substitutions on some of these positions (Arg43, Asp179, Arg205, Gly238, and Glu240) have been also associated with the expansion toward an ESBL phenotype among TEM enzymes (Knox, 1995).
Residue Leu35 is located further away from the active site of class A β-lactamases and its substitution to Gln (e.g., SHV-2a, SHV-12) has been suggested to have an indirect role in enhancing the extended-spectrum capability of SHV β-lactamases (Nüesch-Inderbinen et al., 1997). In contrast, Gly238 and Glu240 amino acids are part of the active site lying near the R1 side chain of the β-lactam (Huletsky et al., 1993). Substitutions in Gly238 either to Ser (e.g., SHV-2, SHV-2a) or Ala (e.g., SHV-13, SHV-18) displace the β3-strand from the reactive Ser70, resulting in a slightly expanded active site. This conformational change improves the binding to and the accommodation of newer cephalosporins with large C7 substituents, thereby expanding the substrate spectrum of these SHV ESBLs to include cefotaxime and to a lesser extent to ceftazidime (Huletsky et al., 1993; Matagne et al., 1998; Nukaga et al., 2003). It has been suggested that Glu240 substitutions to Arg (SHV-86) or Lys (e.g., SHV-4, SHV-5) cause the ammonium group of the long side-chains of these residues to form an electrostatic bond with the carboxylic acid group on the oxyimino-substituents of ceftazidime and aztreonam (Knox, 1995). This interaction has a dual effect on the hydrolysis of ceftazidime by improving initial binding and facilitating proper positioning within the SHV β-lactamase, whereas the hydrolysis of other β-lactams is less affected (Huletsky et al., 1993). Gly238Ser and Glu240Lys amino acid substitutions characterize the majority of SHV ESBLs (Table 2) and mirror those seen in extended-spectrum TEM β-lactamases. Interestingly, a plethora of extended-spectrum SHV and TEM β-lactamases exhibit higher levels of hydrolytic activity against ceftazidime than against cefotaxime (ceftazidimases) (Table 3). This phenotype was attributed to the Glu240Lys substitution, in contrast with most CTX-M β-lactamases lacking this critical substitution and only showing a cefotaximase activity, (Bonnet, 2004).
Among the less frequent but critical substitutions, Ile8Phe in the signal sequence of the precursor of SHV ESBLs (e.g., SHV-7, SHV-18) has been associated with a more efficient β-lactamase transfer into the periplasm (Randegger et al., 2000), a proof that, beside enzymatic structure and gene expression, also the rate of transfer plays a role in resistance phenotype. On the contrary, Arg43Ser (e.g., SHV-7, SHV-18) and Gly156Asp (SHV-27, SHV-45, SHV-105) substitutions affect the structural arrangement of the conserved residues 64–69 and 166–170, respectively. These changes, opposite to the active site cavity (Ser70) for the hydrolysis of the β-lactam molecules, expand the active site to accommodate bulkier cephalosporins (Knox, 1995; Corkill et al., 2001). Asp179 amino acid is highly conserved among subclass A1 of serine β-lactamases and together with Arg164 form a salt bridge that links the two ends of the Ω loop. Substitutions Asp179Ala (SHV-6), Asp179Asn (SHV-8) and Asp179Gly (SHV-24) result in the elimination of the salt bridge with subsequent increase in ceftazidime resistance (Sowek et al., 1991). Several other amino acid substitutions (Table 2) have been described as either responsible for or possibly contributing to the ESBL phenotype, the detailed description of which exceeds the scope of this review.
Apart from point mutations leading to amino acid substitutions, frame shift mutations have been observed with very low occurrence among SHV ESBLs resulting in amino acid insertions (Arpin et al., 2001; Ramdani-Bouguessa et al., 2011) or deletions (Prinarakis et al., 1996). However, their role in the rising of the extended-spectrum phenotype remains unclear. SHV ESBL variants falling in this category are: (i) SHV-9, with the deletion of Gly54 (Prinarakis et al., 1996); (ii) SHV-16, with a 5-amino acid sequence duplication (Asp163aArgGluTrpGluThr-Asp163bArgGluTrpGluThr) of the amino acids between 163 and 167, including Glu166 in the Ω loop (Arpin et al., 2001); (iii) SHV-100, with a 13-amino acid insertion (SerGluSerGlnLeuSerGlyArgValGlyMetIleGlu) between amino acids 35 and 36 (Ramdani-Bouguessa et al., 2011); and (iv) SHV-183, with an Ala insertion between amino acids 186 and 187 (http://www.lahey.org/studies). Of note, the duplication observed in SHV-16 was shown to increase the conformational flexibility of the catalytic region facilitating the access of bulkier cephalosporins, such as ceftazidime, but resulted in enzymatic instability (Arpin et al., 2001). This finding could explain the low incidence of frame shift mutations among extended-spectrum SHV β-lactamases, due to a deleterious effect on the enzymes.
Overall, the available SHV ESBL kinetic parameters show that most of the substitutions lead to more efficient hydrolysis of oxyimino-β-lactams than penicillins, as depicted by the low Kcat values for penicillins (Table 3). While they retain their ability to hydrolyze penicillins, they are not catalytically so efficient compared to SHV-1 (Bush and Singer, 1989) and this is due to the decreased strength of the crucial hydrogen-bonding network needed for penicillin catalysis (turnover). As a consequence, since β-lactam inhibitors (clavulanic acid, sulbactam, and tazobactam) are structurally very similar to penicillin substrates, SHV ESBLs also exhibit increased susceptibility to β-lactam inhibitors compared to SHV-1 (Table 3) leading to less inhibitor required for inactivation (lower Ki and IC50s; Tzouvelekis and Bonomo, 1999).
There are at least 46 known SHV-ESBL genes together with more than 150 non-ESBL or unclassified alleles to date (http://www.lahey.org/studies/). Accurate identification of these variants is essential for surveillance and for epidemiological studies of transmission mode, particularly in clinical setting, where appropriate antimicrobial therapy is critical.
A panel of different phenotypic confirmatory tests is available to determine the presence of extended-spectrum β-lactamases, including SHV-variants: minimum inhibitory concentration (MIC) determination of β-lactam with and without clavulanic acid, double disk synergy test (DDST), inhibitor potentiated disk diffusion test (IPDDT), three-dimensional test (TDT) and commercially available methods (Etest for ESBLs, Vitek ESBL cards, MicroScan panels, and BD Phoenix Automated Microbiology System (Bradford, 2001; Paterson and Bonomo, 2005). Standard microbiological procedures can take up to several days for culture, isolation and characterization and many comparative studies have shown that PCR-based methods have higher sensitivity (Bedenic et al., 2001, 2007; Singh et al., 2012), mostly due to variable levels of gene expression. Therefore, PCR and nucleotide sequence analysis (Stürenburg et al., 2003), together with various PCR-based methods, remain the gold standard for extended-spectrum β-lactamase SHV-variants identification.
Chanawong and colleagues developed a PCR-restriction fragment length polymorphism (PCR-RFLP) method to allow the identification of new SHV β-lactamases variants through detection of known mutations that alter recognition sites of restriction endonucleases (Chanawong et al., 2000). PCR-RFLP complements pre-existing PCR-single strand conformational polymorphism (PCR-SSCP) limited by partial gene amplification, thus missing potential mutation sites (M'Zali et al., 1998). PCR-RFLP can also be used in combination with restriction site insertion-PCR (RSI-PCR), a method based on primers mismatches, allowing the unambiguous identification of up to 27 SHV variants by point mutation (Chanawong et al., 2001a). Fluorescently labeled hybridization probes followed by melting curve analysis can also be used to discriminate between ESBL and non-ESBL blaSHV genes (Randegger and Hächler, 2001). This method, termed the SHV melting curve mutation detection method, is also able to categorize SHV ESBL producers into phenotypically relevant subgroups: (i) weak ceftazidime resistance (SHV-6 and SHV-8); (ii) significant resistance to cefotaxime and ceftriaxone and moderate resistance to ceftazidime (SHV-2, SHV-2a, and SHV-3); and (iii), most effective against all expanded-spectrum cephalosporins (SHV-4, SHV-5, SHV-9, and SHV-12). Combined systems can also be developed ad hoc to rapidly screen local epidemiological settings (Chia et al., 2005). A modified SHV melting-curve mutation detection method able to distinguish between prevalent Taiwanese blaSHV genes (SHV-1, SHV-2, SHV-2a, SHV-5, SHV-11, and SHV-12) was combined with a multiplex PCR to identify different β-lactamases genes (blaSHV, blaCTX-M-3-like, and blaCTX-M-14). The design of this method can be easily adapted to other geographic areas where different ESBLs are prevalent. Multiplex real-time PCR assays for the fast detection of extended-spectrum β-lactamase and carbapenemase genes were developed with differential melting curves able to recognize up to 120 different SHV allelic variants (Singh et al., 2016).
New techniques for ESBL detection are employed alongside PCR-based methods these days. Loop-mediated isothermal amplification (LAMP) was applied to detect SHV- and other ESBL-producing bacteria in meat and proved to be more specific and sensitive than MacConkey agar or cefpodoxime disc methods (Anjum et al., 2013). Commercial DNA microarrays are also proving themselves to be accurate, with sensitivity and specificity values for ESBL detection being high. Up to 53 SHV-variants can be covered on a same array (Leinberger et al., 2010), but on the other hand some alleles may fail to be detected (i.e., SHV-12), as previously reported (Stuart et al., 2012). Because arrays have major limitations to detect novel genes or variants, PCR and sequencing remains essential. Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (MS) is routinely used for bacteria identification and has been recently applied to detect ESBL-producing Enterobacteriaceae from positive blood cultures in clinical practice (Jung et al., 2014; Oviaño et al., 2014). Although, this methodology has yet to be fully validated, preliminary results show 99% sensitivity and 100% specificity, and denote a novel approach to categorize bacteria as ESBL producers.
Pyrosequencing combines standard PCR and sequencing by synthesis to rapidly determine the sequence of a target DNA region; it has been extensively used for the detection of bacterial resistance genes and bacterial community composition (Tang et al., 2016; Tian et al., 2016). This technique has been used to perform mutation analysis of blaSHV to resolve heterogeneous sequences in clinical isolates of K. pneumoniae containing more than one SHV variant (Haanperä et al., 2008). An alternative protocol for pyrosequencing is the single-nucleotide polymorphism (SNP), ideal for the sequencing of mixed templates and determination of SNPs at the position of interest. This protocol has been applied to discriminate between eight blaSHV variants from clinical isolates of E. coli and K. pneumoniae, reporting great reproducibility and ability to discriminate between sequences (Jones et al., 2009). A multiplex pyrosequencing assay coupled with qPCR amplification has also been recently developed to enable rapid and accurate detection of blaSHV and blaTEM -producing Enterobacteriaceae (Deccache et al., 2015). Overall, pyrosequencing can be a useful epidemiological tool for the exact identification of blaSHV as a prerequisite for analyzing the spread of certain SHV variants.
Finally, the advent of whole genome sequencing (WGS) has taken differentiation of bacterial strains and identification of the associated antibiotic resistance gene cargo to another level. Aside from the phylogenetic analysis that WGS provides, the complete resistome of a strain can be unraveled as well as its mobilome, i.e., the mobile genetic elements that are associated with antibiotic resistance diffusion. Only this information can provide us with full understanding of complex genomic structures as observed, for example, in clinical K. pneumoniae genomes carrying (i) nineteen antibiotic resistance genes including blaOXA-1 and blaSHV-28 in the chromosome, blaNDM-1 in a plasmid, and blaOXA-232 in a second plasmid (Kwon et al., 2016); (ii) β-lactamase genes blaKPC−2, blaSHV-11, blaTEM−169, and blaOXA-9, together with aac(6′-)Ib, aadA2, and aph(3′-)Ia as aminoglycoside resistance encoding genes, mph(A) for macrolides, oqxA and oqxB for quinolone, catA1 for phenicol, sul1 for sulfonamide, and dfrA12 for trimethoprim (Lee et al., 2014); or (iii) six different plasmids, adding up to 0.43 Mbp, coding for six β-lactamases (blaSHV-12, blaOXA-9, blaTEM-1, blaCTX-M-2, and blaKPC−2), together with blaSHV-110 and adhesin-related gene clusters on the chromosome (Perreira Ramos et al., 2014).
Over the last years the presence of antibiotics as well as antibiotic resistant bacteria has been shown outside the clinical environment, including water, soil and, most notably, food producing animals. When looking at SHV-variants distribution it is evident that in recent years, as for most extended-spectrum β-lactamases (Canton et al., 2012), their presence has been confirmed in virtually all ecological niches (Supplementary Table S1), making it more challenging to restrain antibiotic resistance diffusion. The most representative cases and variants will be discussed.
In an effort to control the release of antibiotics and antibiotic resistant bacteria in the environment, aquatic environments are being investigated worldwide, whether they be natural, drinking or wastewaters. The latter are particularly worrisome given the high prevalence of blaSHV alleles, as observed in untreated hospital wastewater in Australia (Gündogdu et al., 2013), their possible association with determinants of quinolone and other β-lactamase resistance (Calhau et al., 2015; Osinska et al., 2016), and their relatively easy transmission to surface water through waste water treatment plant discharges (Marti et al., 2013). Studies showed that SHV types, together with CTX-M and OXA genes can be significantly decreased by biological treatments such as activated sludge processing and anaerobic digestion, although not all can be effectively eliminated (Yi et al., 2015).
Urban waters are also exposed to relatively high population densities and therefore are often unprotected from biological contaminants, with people playing a crucial role in antibiotic resistance dissemination in the environment. Unusual finding of SHV-producing Stenotrophomonas maltophilia in a swimming recreational Serbian lake and its transient presence during summer months can be considered as a proof of its anthropogenic origin, given its nature of emerging nosocomial pathogen (Novovic et al., 2015). Similar conclusions can be drawn for SHV-producing K. pneumoniae and E. cloacae isolated from a Bangladeshi lake, which receives waste water from surrounding residents, commercial buildings and clinics in Dhaka city (Haque et al., 2014), as well as for artificial water reservoirs in Poland (Wolny-Koladka and Lenart-Boron, 2016), or urban surface waters in Malaysia (Tissera and Lee, 2013). In recent surveillance studies of different rivers and lakes in Switzerland, blaSHV-12-producing Enterobacteriaceae were isolated only in 4% of the cases (Zurfluh et al., 2013), although this variant is predominant in clinical Swiss isolates (Nüesch-Inderbinen et al., 1997). blaSHV-12 was also detected in Enterobacteriaceae from seawater, together with tet(A) and sul2 in Portugal (Alves et al., 2014), and plasmid-encoded together with blaTEM-1 and/or blaCTX-M-1 in Croatia (Maravic et al., 2015). Finally, data on ESBL-producing Enterobacteriaceae isolated from drinking water is also increasing, reporting SHV alleles in rural water reservoirs in China (Zhang et al., 2015), or drinking water sources for First Nation communities in Canada (Fernando et al., 2016).
Food producing animals have become subject of increasing interest after several studies demonstrated that resistant strains of animal origin can be associated to human infections, possibly through the food chain (Hasman et al., 2005). Majority of SHV variants in this reservoir belong to blaSHV-2, blaSHV-2a, blaSHV-5, and blaSHV-12 (Supplementary Table S1) owing to their successful association with conjugative plasmids (see Section Plasmid epidemiology of blaSHV-2, blaSHV-2a, blaSHV-5, and blaSHV-12).
Surveillance activities in healthy animals worldwide are generating a tremendous amount of data on ESBL distribution. Most SHV β-lactamase producers are E. coli from swine and broiler fecal samples as observed in China (Tian et al., 2012); in Spain, with blaSHV-2 associated with blaCTX-M-9 and blaSHV-12 with blaCTX-M-1, in pigs and broilers respectively (Blanc et al., 2006); in layers, cattle, and broilers but not in swine in Japan (Hiki et al., 2013; Kameyama et al., 2013); and in the Netherlands, where healthy broilers carried blaSHV-2 in combination with blaTEM-1 or blaTEM−135 (Dierikx et al., 2010). Other Enterobacteriaceae like K. pneumoniae and Citrobacter freundii were positive for blaSHV-2 or blaSHV-12 from poultry and swine, respectively (Machado et al., 2008).
Finding ESBL producers in food producing animals is also mirrored by positive food samples worldwide, mostly retail chicken meat, as reported in Tunisia, with E. coli carrying blaSHV-5 isolated from different butcheries, supermarkets, and local markets (Jouini et al., 2013), or Salmonella enterica carrying blaSHV-12 in Japan (Noda et al., 2015). The cross-contamination between food producing animals and retail meat has been internationally demonstrated due to the detection of plasmid-borne SHV variants, such as blaSHV-2 and blaSHV-2a from Canadian chicken meat and abattoir chicken cecum (Pouget et al., 2013) or blaSHV-2 and blaTEM-1 in Japan (Hiroi et al., 2011), presenting the potential for horizontal transfer between Enterobacteriaceae as a high public health concern.
SHV β-lactamase producing Enterobacteriaceae have been detected also in diseased animals, as reported for septicemic broilers due to avian pathogenic E. coli encoding a remarkable array of antibiotic resistance genes (dfrA17-aadA5, blaTEM-1, blaCTX-M-15, blaOXA-1, blaSHV-2, tet(A), tet(E), qnrB2, aac(6)-Ib-cr) (Ahmed et al., 2013); for K. pneumoniae isolated from bovine mastitis in the United Kingdom (Timofte et al., 2014) and Egypt (Ahmed and Shimamoto, 2011); and for multidrug resistant S. enterica serotypes Enteritidis and Typhimurium isolated from diarrheic calves (Ahmed et al., 2009).
Finally, blaSHV-27 is the only other SHV variant frequently reported as chromosomally located in K. pneumoniae from swine, in association with blaSHV-11 and blaCTX-M-1 in China (Zou et al., 2011); in E. coli isolated from farmed fish together with non ESBLs blaSHV-1, blaSHV-11, blaSHV-25, and blaSHV-26 (Jiang et al., 2012); and in opportunistic pathogens asymptomatically colonizing healthy milk cows (Hammad and Shimamoto, 2011).
ESBL diffusion has been studied extensively in Enterobacteriaceae from humans and livestock, whereas information on antibiotic resistance in the environment is still limited. Yet, the dissemination success of blaSHV-12 is confirmed by its introduction into the wildlife, notably in birds, as reported in Spain (Alcalá et al., 2015), the Netherlands (Veldman et al., 2013), Poland (Literak et al., 2010), and the Czech Republic (Dolejská et al., 2009). This success is likely associated to predominant avian clones and to efficient plasmids (Table 4, Figure 3) of the IncN incompatibility group, described to be more frequent in pathogenic than in commensal avian and human E. coli strains (Johnson et al., 2007). blaSHV-5 was also detected in E. coli from several birds of prey in Portugal, alone or in associations with blaTEM−1b (Pinto et al., 2010).
Emergence of Enterobacteriaceae producing β-lactamases in companion animals have been gradually reported, with CTX-M enzymes being prevalent as observed in the human scenario (Rubin and Pitout, 2014). Few studies, on both healthy and diagnostic clinical canine and feline samples, report finding other ESBL variants including blaSHV-3 in the USA (Shaheen et al., 2011), blaSHV-2 in Mexico (Rocha-Gracia et al., 2015), blaSHV-12 in Italy and Poland (Carattoli et al., 2005b; Rzewuska et al., 2015) and blaSHV-12 in association with blaOXA-48, blaCMY-2, blaTEM-1, aac(6′)-Ib-cr, and qnrB2 in Germany (Stolle et al., 2013).
Lastly, SHV variants have been detected in imported vegetables in Switzerland together with blaSHV-12 for the first time in the opportunistic foodborne pathogen Cronobacter sakazakii whose potential to cause bacteremia and meningitis is an actual concern (Zurfluh et al., 2015). Similar results were observed in vegetables collected in South Korea (Kim et al., 2015), salads in the Netherlands (Reuland et al., 2014), and Spain (Egea et al., 2011), displaying a new route of introduction for ESBLs and pathogenic Enterobacteriaceae.
The role of plasmids in the successful spread of β–lactamase genes has been extensively described (Carattoli, 2009, 2013) and, among the SHV family, it finds its best examples in blaSHV-2, blaSHV-2a, blaSHV-5, and blaSHV-12. Combination of these alleles with different dissemination machineries has brought the enzymes to reach diverse niches worldwide (Figure 2). Plasmids belonging to seven replicon types (A/C, F, HI2, I1, L/M, N, X3) have been shown to drive the epidemiology of these four predominant SHV ESBLs, although their distribution varies on the plasmid families (Table 4). Other rep families that have been only incidentally associated with extended-spectrum SHV β–lactamases include the ColE, K, P, and R (Table 4).
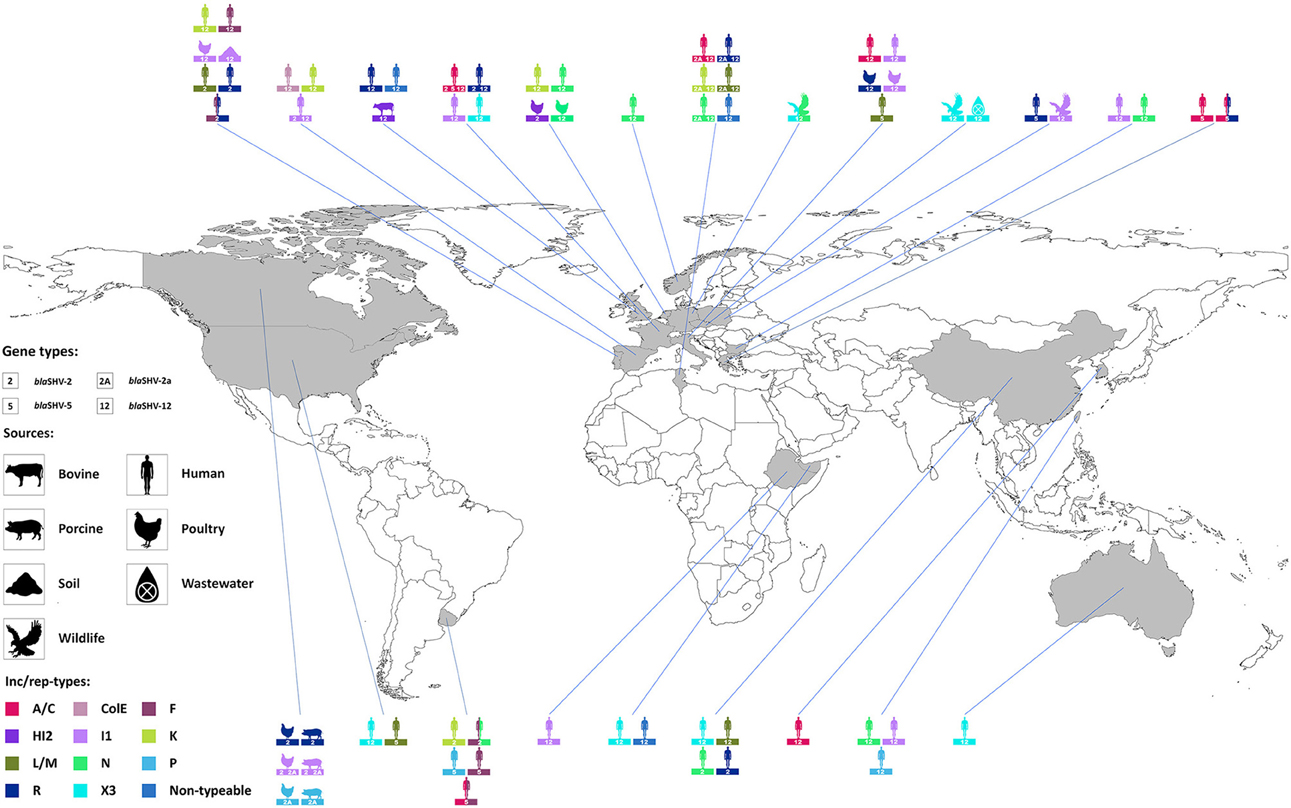
Figure 2. Worldwide distribution of plasmid families encoding blaSHV-2, blaSHV-2a, blaSHV-5, and blaSHV-12. Inc/rep types are represented in different colors; diverse symbols depict human, bovine, porcine, poultry, wildlife, soil, or wastewater sources. For a complete full reference list see Table 4.
blaSHV-12 has been identified on mostly conjugative broad-host range IncA/C plasmids in a variety of bacterial species, including E. coli, Proteus mirabilis and Aeromonas caviae, isolated from clinical samples in Tunisia, France, Korea and Italy (Marcadé et al., 2009; Song et al., 2011; Mnif et al., 2013; Antonelli et al., 2016). E. coli isolates recovered from clinical specimens encoding blaSHV-2, blaSHV-2a, and blaSHV-5 have been also identified in Tunisia and France (Marcadé et al., 2009; Mnif et al., 2013), whereas Providencia stuartii isolates encoding blaSHV-5 on either IncA/C or multireplicon IncA/C-R plasmids have been reported from different outbreaks in Greece (Giakkoupi et al., 2015; Oikonomou et al., 2016). Interestingly, these IncA/C plasmids (130–220 Kb) often carried multiple resistance genes, conferring multidrug resistant phenotypes (Giakkoupi et al., 2015; Antonelli et al., 2016; Oikonomou et al., 2016), resulting in the proliferation of the SHV ESBLs by co-selection.
Plasmids belonging to the narrow-host range IncF group, including plasmids with fused replicons, have been reported to accommodate blaSHV-12 among clinical E. coli isolates from France (IncFII), Tunisia (IncFIA-FIB, IncFII-FIA, IncFII-FIA-FIB) and United Kingdom (IncFIB), but also among food-producing animals from Italy (IncFIB) (Marcadé et al., 2009; Bortolaia et al., 2010; Doumith et al., 2012; Mnif et al., 2013). IncF plasmids account for the dissemination of blaSHV-2 gene among E. coli from both clinical specimens in France (IncFIB, IncFII, IncFII-FIB) and food-producing animals (avian and porcine sources) in Canada (IncFIB), as well as in clinical K. pneumoniae isolates belonging to ST654 and ST15 from China (IncFIB) and Portugal (IncFII), respectively (Marcadé et al., 2009; Wang et al., 2012; Pouget et al., 2013; Rodrigues et al., 2014). Finally, clinical E. coli and K. pneumoniae from Tunisia were found to encode blaSHV-2a (Elhani et al., 2010; Mnif et al., 2013), clinical E. coli from Poland encoded blaSHV-5 on IncF plasmids, as well as clinical K. pneumoniae and Serratia marcescens from Uruguay (García-Fulgueiras et al., 2011; Zienkiewicz et al., 2013), whereas the same plasmids have been associated with less prevalent SHV ESBLs (blaSHV-55 and blaSHV-106) in clinical K. pneumoniae isolates from Portugal (Rodrigues et al., 2014).
In contrast with the IncA/C and IncF plasmids, the broad-host range IncHI2 group is responsible mainly for the dissemination of blaSHV-12, although this group has been found incidentally to also accommodate blaSHV-2a (Mnif et al., 2013). Plasmids of this group varying in sizes (95–400 Kb) have been reported to encode blaSHV-12 in various bacterial species, such as E. coli, K. pneumoniae, E. cloacae, and at least three S. enterica serotypes (Bredeney, Concord, and Senftenberg) from human specimens with diverse geographical origin (Netherlands, Portugal, Spain, Taiwan, Tunisia; Elhani et al., 2010; Veldman et al., 2010; Herrera-Leon et al., 2011; Mnif et al., 2013; Rodrigues et al., 2014; Chen C. M. et al., 2015). Apart from blaSHV-12, some of these conjugative plasmids have been reported to co-encode for other resistance genes, including additional SHV ESBLs (blaCTX-M-3, blaCTX-M-14; Veldman et al., 2010; Chen C. M. et al., 2015).
The IncI1 group, consisting of narrow-host range mostly conjugative plasmids, ranks amongst the top facilitators of blaSHV-2, blaSHV-2a, and blaSHV-12 genes. The range of bacterial species they have encountered is limited to E. coli, K. pneumoniae, E. cloacae, and the S. enterica serotypes Concord, Enteritidis, Heidelberg and Kiambu. Nevertheless, IncI1 plasmids (19–340 Kb) occur in very diverse settings: blaSHV-2- and blaSHV-12-encoding isolates from human infections (Bulgaria, France, Italy, Spain, Taiwan; Tato et al., 2007; Marcadé et al., 2009; Accogli et al., 2013; de Toro et al., 2013; Markovska et al., 2014; Chen C. M. et al., 2015) and colonization (Ethiopia) (Fabre et al., 2009); blaSHV-2-, blaSHV-2a–, and blaSHV-12-encoding isolates from poultry (Canada, Italy, Portugal; Bortolaia et al., 2010, 2011; Accogli et al., 2013; Pouget et al., 2013; Jones-Dias et al., 2015), blaSHV-2- and blaSHV-2a-encoding isolates from pigs (Canada) (Pouget et al., 2013); blaSHV-12-encoding isolates from aquatic birds (Poland) (Literak et al., 2010); and blaSHV-12-encoding isolates from farming soil (Portugal) (Jones-Dias et al., 2016). Remarkably, blaSHV-12 on IncI1 plasmids belonging to pST26 have been identified among E. coli isolates of human and animal origin (Accogli et al., 2013; Jones-Dias et al., 2015), indicating the potential transmission of these blaSHV-12-encoding vehicles from human to animals and/or vice versa.
The broad-host range IncL/M and IncN plasmids contribute to a lesser extent to the epidemiology of blaSHV-2, blaSHV-2a, blaSHV-5, and blaSHV-12 than the above-mentioned families. IncL/M plasmids (60–90 Kb) carrying SHV ESBL genes have been reported only among E. coli, K. pneumoniae, K. oxytoca, and S. enterica serotype Typhimurium of human origin in Portugal (blaSHV-2), Tunisia (blaSHV-2a, blaSHV-12), Italy (blaSHV-5), USA (blaSHV-5), and recently in China (blaSHV-12) (Villa et al., 2000; Elhani et al., 2010; Mnif et al., 2013; Preston et al., 2014; Rodrigues et al., 2014; Liu et al., 2015). The same bacterial species mostly from human sources carry IncN plasmids (~50 Kb) encoding blaSHV-2 (China, Senegal), blaSHV-2a (Tunisia) or blaSHV-12 (Bulgaria, Netherlands, Norway, Tunisia; Samuelsen et al., 2011; Naseer et al., 2012; Wang et al., 2012; Dierikx et al., 2013; Mnif et al., 2013; Harrois et al., 2014; Markovska et al., 2014). Interestingly, the presence of IncN (pST1) plasmids encoding blaSHV-12 has been reported among E. coli from human and animal sources (Dierikx et al., 2013), mirroring the situation for IncI1 plasmids and underscoring the contribution of this plasmid family in the transmission of blaSHV-12 within or between these niches.
The IncX3 plasmid subgroup consists of narrow-host range plasmids and plays an important role in the exclusive dissemination of blaSHV-12. Conjugative plasmids (40–60 Kb) of this subgroup have been identified in diverse bacterial species (E. coli, K. pneumoniae, C. freundii, E. aerogenes, E. cloacae), sources (human, animal, environment) and geographical areas (Australia, China, Czech Republic, France, United Arab Emirates, US; Ho et al., 2012; Du et al., 2013; Kassis-Chikhani et al., 2013; Sonnevend et al., 2013; Wang et al., 2014; Feng et al., 2015; Hargreaves et al., 2015; Partridge et al., 2015; Dobiasova and Dolejska, 2016; Huang et al., 2016). Interestingly, the majority of these plasmids appear to co-harbor carbapenemase genes (blaKPC-2, blaNDM-1), whereas the co-localization of SHV ESBL and carbapenemase genes was reported only on IncA/C or IncA/C-R (blaVIM−1), IncL/M (blaKPC−2), and IncN (blaVIM−1) plasmids (Samuelsen et al., 2011; Naseer et al., 2012; Giakkoupi et al., 2015; Oikonomou et al., 2016), enhancing the plasmid potential maintenance among bacterial populations and the subsequent preservation and dissemination of the SHV ESBL genes.
blaSHV-12 has been incidentally found on: (i) a ColE plasmid from S. enterica serotype Typhimurium DT104b in Spain (Herrera-Leon et al., 2011); (ii) an IncK plasmid from K. pneumoniae in the United Kingdom (Timofte et al., 2014); (iii) an IncP plasmid from E. cloacae in Taiwan (Chen C. M. et al., 2015); and (iv) a plasmid assigned to the R replicon type from K. pneumoniae in Portugal (Rodrigues et al., 2014). E. coli and K. pneumoniae encoding blaSHV-2 on IncK plasmids were recovered from animal and human sources in the Netherlands and in Uruguay, respectively (Dierikx et al., 2010; García-Fulgueiras et al., 2011). IncP plasmids encoding blaSHV-2a from animals in Canada and blaSHV-5 from human in Uruguay have also been reported (García-Fulgueiras et al., 2011; Pouget et al., 2013). Finally, a number of reports highlight the presence of blaSHV-12 on mostly conjugative non-typeable plasmids, according to the PCR-based replicon-typing scheme (Carattoli et al., 2005a). These plasmids of human origin, varying between 50 and 140 Kb in size, were mostly detected among E. coli from the United Kingdom (Doumith et al., 2012) and K. pneumoniae from Tunisia (Elhani et al., 2010) and United Arab Emirates (Sonnevend et al., 2013), underscoring that their dissemination is wider than we know.
Analysis of the sequences bracketing several SHV ESBL genes (blaSHV-2, blaSHV-2a, blaSHV-5, blaSHV-12, blaSHV-106, and blaSHV-134) among Gram-negative bacteria, including Enterobacteriaceae and non-fermenters, revealed that these β-lactamase genes are mostly associated with the IS26 element (Table 4, Supplementary Table S1). Beside SHV ESBLs, this member of the IS6 insertion sequence family (Mahillon and Chandler, 1998), has been associated with a plethora of resistance genes (Allard et al., 1993; Miriagou et al., 2005; Post and Hall, 2009; Cain et al., 2010; Hordijk et al., 2011) and has been found to contribute to their expression by supplying a promoter -35 box that can be coupled with a -10 box in the adjacent DNA (Lee et al., 1990; Cain and Hall, 2011). In contrast to other insertions sequences, it has been suggested that IS26 transposes preferentially within plasmids rather than into the chromosome (He et al., 2015), possibly explaining the linkage of IS26 and the four predominant SHV ESBL genes with IncA/C (blaSHV-5, blaSHV-12), IncF (blaSHV-2a, blaSHV-5, blaSHV-12), IncHI2 (blaSHV-12), IncI1 (blaSHV-12), IncL/M (blaSHV-2a, blaSHV-5), IncN (blaSHV-2), IncP (blaSHV-12), IncX3 (blaSHV-12), and non-typeable (blaSHV-12) plasmids (Table 4). Similarly to most antibiotic resistance genes, IS26-mediated mobilization of SHV ESBL genes on conjugative plasmids facilitated their subsequent intra- and inter-species dissemination (Table 4). Available sequences of transposons flanked by copies of intact and/or truncated IS26 elements (Figure 3) and coding for SHV ESBL genes show the presence of other co-linear genes originating from the chromosome of K. pneumoniae (i.e., fucA, ygbI, ygbK, ygbJ, ygbM, deoR; Ho et al., 2012; Wang et al., 2012; Du et al., 2013; Kassis-Chikhani et al., 2013; Preston et al., 2014; Chen C. M. et al., 2015; Feng et al., 2015; Giakkoupi et al., 2015), likely underscoring the involvement of IS26 in the mobilization of blaSHV from the chromosome of K. pneumoniae, as previously suggested (Haeggman et al., 1997).
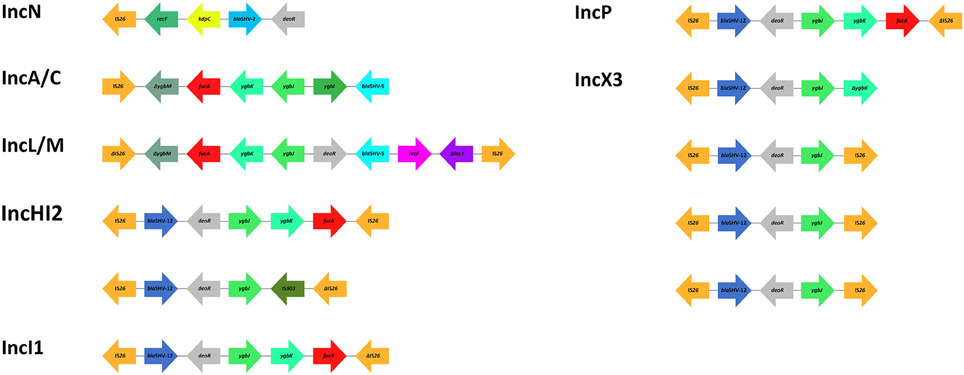
Figure 3. Schematic representation of blaSHV genetic surroundings and IS26 association. Common occurring features are color coded. Map is not to scale. References: IncN (Wang et al., 2012); IncA/C (Giakkoupi et al., 2015); IncL/M (Preston et al., 2014); IncHI2, IncI1, IncP (Chen C. M. et al., 2015); IncX3 (Ho et al., 2012; Du et al., 2013; Kassis-Chikhani et al., 2013; Feng et al., 2015).
SHV β–lactamases have virtually invaded all human, environmental and animal sceneries, mostly associated to Enterobacteriaceae. In recent years, the first reports of alternative bacterial hosts have been described, notably in Aeromonads, ubiquitous in aquatic habitats and occasionally able to cause human infections. blaSHV-12 was detected, in association with blaFOX-2 and blaCTX-M-15, on the chromosome of the foodborne pathogens A. caviae and Aeromonas hydrophila from wild-growing mussels from Croatia (Maravić et al., 2013). The first identification of plasmid-encoded SHV-12, together with VIM-1, occurred in clinical A. caviae accountable for a newborn bloodstream infection (Antonelli et al., 2016). The coproduction of these enzymes highlights the potential risks for public health and the role of Aeromonads as reservoirs and dissemination tools of resistance determinants in both environmental and clinical settings.
Occasionally, SHV ESBL-producing Pseudomonas aeruginosa can be detected in nosocomial settings and can pose a serious threat as healthcare-associated infection in many regions of the world. blaSHV-2a was first identified on the chromosome of a 1995 clinical P. aeruginosa strain, with high sequence homology to plasmid pMPA2a from K. pneumoniae indicating a likely enterobacterial gene origin (Naas et al., 1999). Subsequent studies demonstrated the insertion of blaSHV alleles into P. aeruginosa chromosome: blaSHV in China (Chen Z. et al., 2015) and Iran (Shahcheraghi et al., 2009); blaSHV-5 (Poirel et al., 2004) and blaSHV-12 (Neonakis et al., 2003) in Greece; and blaSHV-2a in Tunisia (Mansour et al., 2009) and France (Hocquet et al., 2010; Jeannot et al., 2013). The role of IS26 in the mobilization of blaSHV-12 was demonstrated by the chromosomal insertion of an IS26 composite transposon (>24 kb) thanks to the co-mobilization of antibiotic resistance aac(6′)-Ib, which confers amikacin resistance, likely occurred during the clinical course of a burn infection, immediately after amikacin administration (Uemura et al., 2010). blaSHV-5, blaSHV-11, blaSHV-12 were also detected in different combinations, together with blaTEM-1b, on various plasmids in Thailand (Chanawong et al., 2001b).
Finally, one of the most effective associations outside of the Enterobacteriaceae is with Acinetobacter baumannii, contributing to the worrisome spread of ESBL-producing strains especially in clinical outbreaks (Blackwell et al., 2016). Plasmid transfer from nosocomial SHV-encoding Enterobacteriaceae seems to be responsible for this phenomenon, as observed for SHV-12 in the Netherlands (Naiemi et al., 2005), or SHV-5 in the USA, a country where this variant is the most prevalent ESBL gene in Enterobacteriaceae (Naas et al., 2007).
Among all ESBL SHV β-lactamases, few enzymes deserve special consideration because of their unique enzymatic features.
SHV-38 is a unique allelic variant of the SHV family to have an expanded-spectrum to carbapenems. It was first described in K. pneumoniae from France (Poirel et al., 2003) and it holds a point mutation (Ala146Val) compared to the chromosome-encoded SHV-1. Among all 46 available SHV ESBL variants, only SHV-38 possess the Ala146Val substitution (Table 2), likely inducing subtle structural conformational changes favoring imipenem but not meropenem hydrolysis (Walther-Rasmussen and Høiby, 2007).
SHV-129 is a novel clinically acquired variant identified in 2012 from an Italian E. coli isolate (Table 1; Lascols et al., 2012) and it represents an interesting example of enzyme evolution due to antibiotic pressure. Alongside two well-known amino acid substitutions (Gly238Ser, Glu240Lys), SHV-129 contains new substitutions, Arg275Leu and Asn146Asp). The latter was recently demonstrated to be the first global suppressor substitution identified in the SHV β-lactamase family (Winkler and Bonomo, 2016), likely helping in protein stabilization and functionality, as well as in the ability of the enzyme to acquire additional substitutions. It is also proposed that due to the increasing clinical use of cefepime, SHV-129 might have evolved from SHV-2 or SHV-5 in an alternative conformation to expand its spectrum to hydrolyze cefepime, as mirrored by the kinetic parameters of the three enzymes (Table 3).
Finally, SHV-2 can be located on both chromosome and self-transmissible plasmids (Table 4; Supplementary Table S1). Association of blaSHV-2 with RCS47, a P1-like bacteriophage that infects and lysogenizes E. coli and several other enteric bacteria, was recently reported (Billard-Pomares et al., 2014). blaSHV-2 is flanked by two IS26 elements that likely drove the insertion in the phage backbone. The P1-like prophages were found with high prevalence in natural E. coli of both animal and human origin, including ESBL-producing isolates. This kind of association was already reported for other β-lactamases (blaTEM, blaCTX-M, and mecA) from river and urban sewage water (von Wintersdorff et al., 2016), suggesting that bacteriophages might play a wider role in favoring horizontal transfer of antibiotic resistance determinants than initially thought (Muniesa et al., 2013).
Tzouvelekis and Bonomo suggested than “it will not be surprising if (SHV) enzymes will continue to expand their substrate spectrum as long as the current antibiotics, or novel ones derived from the basic β-lactam structure, are used” (Tzouvelekis and Bonomo, 1999). In the last two decades we observed the appearance of multiple SHV-type variants, with few ones significantly expanding their substrate. One exception is represented by SHV-38, the only known SHV allelic variant able to hydrolyze carbapenems (Poirel et al., 2003), a feature that has not been associated with any TEM or CTX-M enzyme. In this image resides the fate of SHV extended β-lactamases, unable to undergo the dominant propagation observed, for instance, for the CTX-M family but yet contributing to β-lactam resistance in a not negligible way.
The persistence of SHV enzymes in the bacterial community might also be secured by co-selection with emerging resistance genes. Association of blaSHV-12 with IncX3 plasmids carrying carbapenemase genes blaKPC−2 and blaNDM has been observed in recent years (Table 4) and it seems to be a phenomenon occurring in clinical carbapenem-resistant Enterobacteriaceae worldwide (Kassis-Chikhani et al., 2013; Sonnevend et al., 2013; Partridge et al., 2015; Huang et al., 2016). As highlighted in this review, the association of successful variants blaSHV-2, blaSHV-2a, blaSHV-5, and blaSHV-12 with different families of conjugative plasmids (IncA/C, IncF, IncHI2) might also underlie the colonization of virtually all ecological niches encompassing food producing animals, aquatic environment, wildlife, companion animals, and vegetables. Plasmid mediated transfer from nosocomial Enterobacteriaceae enabled SHV dispersion toward alternative bacterial hosts such as the emerging nosocomial pathogens of aquatic origin S. maltophilia and A. caviae, or contributed to the worrisome spread of ESBL-producing strains of A. baumannii and P. aeruginosa. Most interestingly, the ubiquitous presence of SHV ESBL genes and plasmids is suggestive for transmission in human, animals, and the environment, most likely through the food chain, highlighting the potential risks for public health and endorsing a one health research approach.
Overall, SHV ESBL enzymes have kept a stable role in antibiotic resistance over the years. Allele diversification is still occurring, the latest variant being identified in E. cloacae in 2014 (blaSHV-183), and effective associations with new genetic platforms are taking place helping expansion toward novel bacterial hosts and reservoirs.
The paper was written by AL and DC, and reviewed by DM. All authors discussed, read, contributed to and approved the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to thank Mr. Bo Derks for valuable technical help in Figure 2 production. AL was partially supported by the ESBLAT project (BO-22.04-008-001). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01374
1. ^Of the 194 variants available on line (http://www.lahey.org/studies), 5 were withdrawn or invalidated as only partial sequence.
Abbassi, M. S., Torres, C., Achour, W., Vinué, L., Sáenz, Y., Costa, D., et al. (2008). Genetic characterisation of CTX-M-15-producing Klebsiella pneumoniae and Escherichia coli strains isolated from stem cell transplant patients in Tunisia. Int. J. Antimicrob. Agents 32, 308–314. doi: 10.1016/j.ijantimicag.2008.04.009
Accogli, M., Fortini, D., Giufrè, M., Graziani, C., Dolejska, M., Carattoli, A., et al. (2013). IncI1 plasmids associated with the spread of CMY-2, CTX-M-1 and SHV-12 in Escherichia coli of animal and human origin. Clin. Microbiol. Infect. 19, E238–E240. doi: 10.1111/1469-0691.12128
Ahmed, A. M., and Shimamoto, T. (2011). Molecular characterization of antimicrobial resistance in Gram-negative bacteria isolated from bovine mastitis in Egypt. Microbiol. Immunol. 55, 318–327. doi: 10.1111/j.1348-0421.2011.00323.x
Ahmed, A. M., Shimamoto, T., and Shimamoto, T. (2013). Molecular characterization of multidrug-resistant avian pathogenic Escherichia coli isolated from septicemic broilers. Int. J. Med. Microbiol. 303, 475–483. doi: 10.1016/j.ijmm.2013.06.009
Ahmed, A. M., Younis, E. E., Ishida, Y., and Shimamoto, T. (2009). Genetic basis of multidrug resistance in Salmonella enterica serovars Enteritidis and Typhimurium isolated from diarrheic calves in Egypt. Acta Trop. 111, 144–149. doi: 10.1016/j.actatropica.2009.04.004
Alcalá, L., Alonso, C. A., Simón, C., González-Esteban, C., Orós, J., Rezusta, A., et al. (2015). Wild birds, frequent carriers of Extended-Spectrum β-Lactamase (ESBL) producing Escherichia coli of CTX-M and SHV-12 types. Microb. Ecol. doi: 10.1007/s00248-015-0718-0. [Epub ahead of print].
Allard, J. D., Gibson, M. L., Vu, L. H., Nguyen, T. T., and Bertrand, K. P. (1993). Nucleotide sequence of class D tetracycline resistance genes from Salmonella ordonez. Mol. Gen. Genet. 237, 301–305. doi: 10.1007/bf00282811
Alves, M. S., Pereira, A., Araújo, S. M., Castro, B. B., Correia, A. C., and Henriques, I. (2014). Seawater is a reservoir of multi-resistant Escherichia coli, including strains hosting plasmid-mediated quinolones resistance and extended-spectrum β-lactamases genes. Front. Microbiol. 5:426. doi: 10.3389/fmicb.2014.00426
Ambler, R. P. (1980). The structure of β-lactamases. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 289, 321–331. doi: 10.1098/rstb.1980.0049
Anjum, M. F., Lemma, F., Cork, D. J., Meunier, D., Murphy, N., North, S. E., et al. (2013). Isolation and detection of extended spectrum β-lactamase (ESBL)-producing Enterobacteriaceae from meat using chromogenic agars and isothermal loop-mediated amplification (LAMP) assays. J. Food Sci. 78, M1892–M1898. doi: 10.1111/1750-3841.12297
Antonelli, A., D'andrea, M. M., Montagnani, C., Bartolesi, A. M., Di Pilato, V., Fiorini, P., et al. (2016). Newborn bacteraemia caused by an Aeromonas caviae producing the VIM-1 and SHV-12 β-lactamases, encoded by a transferable plasmid. J. Antimicrob. Chemother. 71, 272–274. doi: 10.1093/jac/dkv304
Arlet, G., Rouveau, M., Bengoufa, D., Nicolas, M. H., and Philippon, A. (1991). Novel transferable extended-spectrum β-lactamase (SHV-6) from Klebsiella pneumoniae conferring selective resistance to ceftazidime. FEMS Microbiol. Lett. 81, 57–62. doi: 10.1016/0378-1097(91)90471-l
Arlet, G., Rouveau, M., Casin, I., Bouvet, P. J., Lagrange, P. H., and Philippon, A. (1994). Molecular epidemiology of Klebsiella pneumoniae strains that produce SHV-4 β-lactamase and which were isolated in 14 French hospitals. J. Clin. Microbiol. 32, 2553–2558.
Arlet, G., Sanson-le Pors, M. J., Rouveau, M., Fournier, G., Marie, O., Schlemmer, B., et al. (1990). Outbreak of nosocomial infections due to Klebsiella pneumoniae producing SHV-4 β-lactamase. Eur. J. Clin. Microbiol. Infect. Dis. 9, 797–803. doi: 10.1007/BF01967377
Arpin, C., Coze, C., Rogues, A. M., Gachie, J. P., Bebear, C., and Quentin, C. (1996). Epidemiological study of an outbreak due to multidrug-resistant Enterobacter aerogenes in a medical intensive care unit. J. Clin. Microbiol. 34, 2163–2169.
Arpin, C., Labia, R., Andre, C., Frigo, C., El Harrif, Z., and Quentin, C. (2001). SHV-16, a β-lactamase with a pentapeptide duplication in the omega loop. Antimicrob. Agents Chemother. 45, 2480–2485. doi: 10.1128/AAC.45.9.2480-2485.2001
Barthélémy, M., Peduzzi, J., and Labia, R. (1988). Complete amino acid sequence of p453-plasmid-mediated PIT-2 β-lactamase (SHV-1). Biochem. J. 251, 73–79. doi: 10.1042/bj2510073
Bedenic, B., Randegger, C., Boras, A., and Haechler, H. (2001). Comparison of five different methods for detection of SHV extended-spectrum β-lactamases. J. Chemother. 13, 24–33. doi: 10.1179/joc.2001.13.1.24
Bedenic, B., Vranes, J., Mihaljevic, L. J., Tonkic, M., Sviben, M., Plecko, V., et al. (2007). Sensitivity and specificity of various β-lactam antibiotics and phenotypical methods for detection of TEM, SHV and CTX-M extended-spectrum β-lactamases. J. Chemother. 19, 127–139. doi: 10.1179/joc.2007.19.2.127
Ben Achour, N., Belhadj, O., Galleni, M., Ben Moussa, M., and Mercuri, P. S. (2014). Study of a natural mutant SHV-type b-lactamase, SHV-104, from Klebsiella pneumoniae. Int. J. Microbiol. 2014, 6. doi: 10.1155/2014/548656
Billard-Pomares, T., Fouteau, S., Jacquet, M. E., Roche, D., Barbe, V., Castellanos, M., et al. (2014). Characterization of a P1-like bacteriophage carrying an SHV-2 extended-spectrum β-lactamase from an Escherichia coli strain. Antimicrob. Agents Chemother. 58, 6550–6557. doi: 10.1128/AAC.03183-14
Blackwell, G. A., Hamidian, M., and Hall, R. M. (2016). IncM plasmid R1215 is the source of chromosomally located regions containing multiple antibiotic resistance genes in the globally disseminated Acinetobacter baumannii GC1 and GC2 clones. mSphere 1:e00117-16. doi: 10.1128/mSphere.00117-16
Blanc, V., Mesa, R., Saco, M., Lavilla, S., Prats, G., Miró, E., et al. (2006). ESBL- and plasmidic class C β-lactamase-producing E. coli strains isolated from poultry, pig and rabbit farms. Vet. Microbiol. 118, 299–304. doi: 10.1016/j.vetmic.2006.08.002
Bonnet, R. (2004). Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48, 1–14. doi: 10.1128/AAC.48.1.1-14.2004
Bortolaia, V., Guardabassi, L., Trevisani, M., Bisgaard, M., Venturi, L., and Bojesen, A. M. (2010). High diversity of Extended-Spectrum β-Lactamases in Escherichia coli isolates from Italian broiler flocks. Antimicrob. Agents Chemother. 54, 1623–1626. doi: 10.1128/AAC.01361-09
Bortolaia, V., Larsen, J., Damborg, P., and Guardabassi, L. (2011). Potential pathogenicity and host range of Extended-Spectrum β-Lactamase-producing Escherichia coli isolates from healthy poultry. Appl. Environ. Microbiol. 77, 5830–5833. doi: 10.1128/AEM.02890-10
Bourouis, A., Ben Moussa, M., and Belhadj, O. (2015). Multidrug-resistant phenotype and isolation of a novel SHV- β-lactamase variant in a clinical isolate of Enterobacter cloacae. J. Biomed. Sci. 22, 1–7. doi: 10.1186/s12929-015-0131-5
Bradford, P. A. (2001). Extended-spectrum β-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14, 933–951. doi: 10.1128/CMR.14.4.933-951.2001
Bradford, P. A., Urban, C., Jaiswal, A., Mariano, N., Rasmussen, B. A., Projan, S. J., et al. (1995). SHV-7, a novel cefotaxime-hydrolyzing β-lactamase, identified in Escherichia coli isolates from hospitalized nursing home patients. Antimicrob. Agents Chemother. 39, 899–905. doi: 10.1128/AAC.39.4.899
Bush, K. (2013). The ABCD's of β-lactamase nomenclature. J. Infect. Chemother. 19, 549–559. doi: 10.1007/s10156-013-0640-7
Bush, K., and Fisher, J. F. (2011). Epidemiological expansion, structural studies, and clinical challenges of new β-lactamases from Gram-negative bacteria. Annu. Rev. Microbiol. 65, 455–478. doi: 10.1146/annurev-micro-090110-102911
Bush, K., and Jacoby, G. A. (2010). Updated functional classification of β-lactamases. Antimicrob. Agents Chemother. 54, 969–976. doi: 10.1128/AAC.01009-09
Bush, K., Jacoby, G. A., and Medeiros, A. A. (1995). A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39, 1211–1233. doi: 10.1128/AAC.39.6.1211
Bush, K., and Singer, S. B. (1989). New plasmid borne β-lactamases - biochemical characteristics of extended broad-spectrum β-lactamases. Infection 17, 429–433. doi: 10.1007/BF01645566
Cain, A. K., and Hall, R. M. (2011). Transposon Tn5393e Carrying the aphA1-Containing Transposon Tn6023 upstream of strAB does not confer resistance to Streptomycin. Microb. Drug Resist. 17, 389–394. doi: 10.1089/mdr.2011.0037
Cain, A. K., Liu, X., Djordjevic, S. P., and Hall, R. M. (2010). Transposons related to Tn1696 in IncHI2 Plasmids in multiply Antibiotic Resistant Salmonella enterica Serovar Typhimurium from Australian Animals. Microb. Drug Resist. 16, 197–202. doi: 10.1089/mdr.2010.0042
Calhau, V., Mendes, C., Pena, A., Mendonça, N., and Da Silva, G. J. (2015). Virulence and plasmidic resistance determinants of Escherichia coli isolated from municipal and hospital wastewater treatment plants. J. Water Health 13, 311–318. doi: 10.2166/wh.2014.327
Cantón, R., González-Alba, J. M., and Galán, J. C. (2012). CTX-M enzymes: origin and diffusion. Front. Microbiol. 3:110. doi: 10.3389/fmicb.2012.00110
Carattoli, A. (2009). Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53, 2227–2238. doi: 10.1128/AAC.01707-08
Carattoli, A. (2013). Plasmids and the spread of resistance. Int. J. Med. Microbiol. 303, 298–304. doi: 10.1016/j.ijmm.2013.02.001
Carattoli, A., Bertini, A., Villa, L., Falbo, V., Hopkins, K. L., and Threlfall, E. J. (2005a). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63, 219–228. doi: 10.1016/j.mimet.2005.03.018
Carattoli, A., Lovari, S., Franco, A., Cordaro, G., Di Matteo, P., and Battisti, A. (2005b). Extended-spectrum β-lactamases in Escherichia coli isolated from dogs and cats in Rome, Italy, from 2001 to 2003. Antimicrob. Agents Chemother. 49, 833–835. doi: 10.1128/AAC.49.2.833-835.2005
Chanawong, A., M'Zali, F. H., Heritage, J., Lulitanond, A., and Hawkey, P. M. (2000). Characterisation of extended-spectrum β-lactamases of the SHV family using a combination of PCR-single strand conformational polymorphism (PCR-SSCP) and PCR-restriction fragment length polymorphism (PCR-RFLP). FEMS Microbiol. Lett. 184, 85–89. doi: 10.1111/j.1574-6968.2000.tb08995.x
Chanawong, A., M'Zali, F. H., Heritage, J., Lulitanond, A., and Hawkey, P. M. (2001a). Discrimination of SHV β-lactamase genes by restriction site insertion-PCR. Antimicrob. Agents Chemother. 45, 2110–2114. doi: 10.1128/AAC.45.7.2110-2114.2001
Chanawong, A., M'Zali, F. H., Heritage, J., Lulitanond, A., and Hawkey, P. M. (2001b). SHV-12, SHV-5, SHV-2a and VEB-1 extended-spectrum β-lactamases in Gram-negative bacteria isolated in a university hospital in Thailand. J. Antimicrob. Chemother. 48, 839–852. doi: 10.1093/jac/48.6.839
Chen, C. M., Yu, W. L., Huang, M., Liu, J. J., Chen, I. C., Chen, H. F., et al. (2015). Characterization of IS26-composite transposons and multidrug resistance in conjugative plasmids from Enterobacter cloacae. Microbiol. Immunol. 59, 516–525. doi: 10.1111/1348-0421.12289
Chen, Z., Niu, H., Chen, G., Li, M., Li, M., and Zhou, Y. (2015). Prevalence of ESBLs-producing Pseudomonas aeruginosa isolates from different wards in a Chinese teaching hospital. Int. J. Clin. Exp. Med. 8, 19400–19405.
Chia, J. H., Chu, C., Su, L. H., Chiu, C. H., Kuo, A. J., Sun, C. F., et al. (2005). Development of a multiplex PCR and SHV melting-curve mutation detection system for detection of some SHV and CTX-M β-lactamases of Escherichia coli, Klebsiella pneumoniae, and Enterobacter cloacae in Taiwan. J. Clin. Microbiol. 43, 4486–4491. doi: 10.1128/JCM.43.9.4486-4491.2005
Corkill, J. E., Cuevas, L. E., Gurgel, R. Q., Greensill, J., and Hart, C. A. (2001). SHV-27, a novel cefotaxime-hydrolysing β-lactamase, identified in Klebsiella pneumoniae isolates from a Brazilian hospital. J. Antimicrob. Chemother. 47, 463–465. doi: 10.1093/jac/47.4.463
D'Andrea, M. M., Arena, F., Pallecchi, L., and Rossolini, G. M. (2013). CTX-M-type β-lactamases: a successful story of antibiotic resistance. Int. J. Med. Microbiol. 303, 305–317. doi: 10.1016/j.ijmm.2013.02.008
Datta, N., and Kontomichalou, P. (1965). Penicillinase synthesis controlled by infectious R factors in Enterobacteriaceae. Nature 208, 239–241. doi: 10.1038/208239a0
Deccache, Y., Irenge, L. M., Ambroise, J., Savov, E., Marinescu, D., Chirimwami, R. B., et al. (2015). A qPCR and multiplex pyrosequencing assay combined with automated data processing for rapid and unambiguous detection of ESBL-producers Enterobacteriaceae. AMB Express 5:136. doi: 10.1186/s13568-015-0136-1
de Toro, M., García, P., Rodríguez, I., Rojo-Bezares, B., Helmuth, R., Sáenz, Y., et al. (2013). Characterisation of plasmids implicated in the mobilisation of extended-spectrum and AmpC β-lactamase genes in clinical Salmonella enterica isolates and temporal stability of the resistance genotype. Int. J. Antimicrob. Agents 42, 167–172. doi: 10.1016/j.ijantimicag.2013.04.016
Dierikx, C., Van Der Goot, J., Fabri, T., Van Essen-Zandbergen, A., Smith, H., and Mevius, D. (2013). Extended-spectrum-β-lactamase- and AmpC-β-lactamase-producing Escherichia coli in Dutch broilers and broiler farmers. J. Antimicrob. Chemother. 68, 60–67. doi: 10.1093/jac/dks349
Dierikx, C., van Essen-Zandbergen, A., Veldman, K., Smith, H., and Mevius, D. (2010). Increased detection of extended spectrum β-lactamase producing Salmonella enterica and Escherichia coli isolates from poultry. Vet. Microbiol. 145, 273–278. doi: 10.1016/j.vetmic.2010.03.019
Dobiasova, H., and Dolejska, M. (2016). Prevalence and diversity of IncX plasmids carrying fluoroquinolone and β-lactam resistance genes in Escherichia coli originating from diverse sources and geographical areas. J. Antimicrob. Chemother. 71, 2118–2124. doi: 10.1093/jac/dkw144
Dolejská, M., Bierošová, B., Kohoutová, L., Literák, I., and Čížek, A. (2009). Antibiotic-resistant Salmonella and Escherichia coli isolates with integrons and extended-spectrum β-lactamases in surface water and sympatric black-headed gulls. J. Appl. Microbiol. 106, 1941–1950. doi: 10.1111/j.1365-2672.2009.04155.x
Doumith, M., Dhanji, H., Ellington, M. J., Hawkey, P., and Woodford, N. (2012). Characterization of plasmids encoding extended-spectrum -lactamases and their addiction systems circulating among Escherichia coli clinical isolates in the UK. J. Antimicrob. Chemother. 67, 878–885. doi: 10.1093/jac/dkr553
Dropa, M., Balsalobre, L. C., Lincopan, N., Matté, G. R., and Matté, M. H. (2015). Complex class 1 integrons harboring CTX-M-2-encoding genes in clinical Enterobacteriaceae from a hospital in Brazil. J. Infect. Dev. Ctries 9, 890–897. doi: 10.3855/jidc.6241
Du, X. X., Wang, J. F., Fu, Y., Zhao, F., Chen, Y., Wang, H. P., et al. (2013). Genetic characteristics of blaNDM-1-positive plasmid in Citrobacter freundii isolate separated from a clinical infectious patient. J. Med. Microbiol. 62, 1332–1337. doi: 10.1099/jmm.0.057091-0
Duval, V., Maiga, I., Maiga, A., Guillard, T., Brasme, L., Forte, D., et al. (2009). High prevalence of CTX-M-type β-lactamases among clinical isolates of Enterobacteriaceae in Bamako, Mali. Antimicrob. Agents Chemother. 53, 4957–4958. doi: 10.1128/AAC.00675-09
Egea, P., López-Cerero, L., Navarro, M. D., Rodríguez-Baño, J., and Pascual, A. (2011). Assessment of the presence of extended-spectrum β-lactamase-producing Escherichia coli in eggshells and ready-to-eat products. Eur. J. Clin. Microbiol. Infect. Dis. 30, 1045–1047. doi: 10.1007/s10096-011-1168-3
Elhani, D., Bakir, L., Aouni, M., Passet, V., Arlet, G., Brisse, S., et al. (2010). Molecular epidemiology of extended-spectrum β-lactamase-producing Klebsiella pneumoniae strains in a university hospital in Tunis, Tunisia, 1999-2005. Clin. Microbiol. Infect. 16, 157–164. doi: 10.1111/j.1469-0691.2009.03057.x
El Harrif-Heraud, Z., Arpin, C., Benliman, S., and Quentin, C. (1997). Molecular epidemiology of a nosocomial outbreak due to SHV-4-producing strains of Citrobacter diversus. J. Clin. Microbiol. 35, 2561–2567.
Espinal, P., Garza-Ramos, U., Reyna, F., Rojas-Moreno, T., Sanchez-Perez, A., Carrillo, B., et al. (2010). Identification of SHV-type and CTX-M-12 extended-spectrum β-lactamases (ESBLs) in multiresistant Enterobacteriaceae from Colombian Caribbean hospitals. J. Chemother. 22, 160–164. doi: 10.1179/joc.2010.22.3.160
Essack, S. Y., Hall, L. M. C., and Livermore, D. M. (2004). Klebsiella pneumoniae isolate from South Africa with multiple TEM, SHV and AmpC β-lactamases. Int. J. Antimicrob. Agents 23, 398–400. doi: 10.1016/j.ijantimicag.2003.08.010
Evans, B. A., and Amyes, S. G. (2014). OXA β-lactamases. Clin. Microbiol. Rev. 27, 241–263. doi: 10.1128/CMR.00117-13
Fabre, L., Delauné, A., Espié, E., Nygard, K., Pardos, M., Polomack, L., et al. (2009). Chromosomal integration of the extended-spectrum β-lactamase gene blaCTX-M-15 in Salmonella enterica serotype Concord isolates from internationally adopted children. Antimicrob. Agents Chemother. 53, 1808–1816. doi: 10.1128/AAC.00451-08
Feng, Y., Yang, P., Xie, Y., Wang, X., Mcnally, A., and Zong, Z. (2015). Escherichia coli of sequence type 3835 carrying blaNDM-1, blaCTX-M-15, blaCMY-42 and blaSHV-12. Sci. Rep. 5:12275. doi: 10.1038/srep12275
Fernando, D. M., Tun, H. M., Poole, J., Patidar, R., Li, R., Mi, R., et al. (2016). Detection of antibiotic resistance genes in source and drinking water samples from a first nation community in Canada. Appl. Environ. Microbiol. 82, 4767–4775. doi: 10.1128/aem.00798-16
Fisher, J. F., Meroueh, S. O., and Mobashery, S. (2005). Bacterial resistance to β-lactam antibiotics: compelling opportunism, compelling opportunity. Chem. Rev. 105, 395–424. doi: 10.1021/cr030102i
García-Fulgueiras, V., Bado, I., Mota, M. I., Robino, L., Cordeiro, N. F., Varela, A., et al. (2011). Extended-spectrum β-lactamases and plasmid-mediated quinolone resistance in enterobacterial clinical isolates in the paediatric hospital of Uruguay. J. Antimicrob. Chemother. 66, 1725–1729. doi: 10.1093/jac/dkr222
Giakkoupi, P., Tryfinopoulou, K., Polemis, M., Pappa, O., Miriagou, V., and Vatopoulos, A. (2015). Circulation of a multiresistant, conjugative, IncA/C plasmid within the nosocomial Providencia stuartii population in the Athens area. Diagn. Microbiol. Infect. Dis. 82, 62–64. doi: 10.1016/j.diagmicrobio.2015.02.009
Gündogdu, A., Jennison, A. V., Smith, H. V., Stratton, H., and Katouli, M. (2013). Extended-spectrum β-lactamase producing Escherichia coli in hospital wastewaters and sewage treatment plants in Queensland, Australia. Can. J. Microbiol. 59, 737–745. doi: 10.1139/cjm-2013-0515
Gutmann, L., Ferré, B., Goldstein, F. W., Rizk, N., Pinto-Schuster, E., Acar, J. F., et al. (1989). SHV-5, a novel SHV-type β-lactamase that hydrolyzes broad-spectrum cephalosporins and monobactams. Antimicrob. Agents Chemother. 33, 951–956. doi: 10.1128/AAC.33.6.951
Haanperä, M., Forssten, S. D., Huovinen, P., and Jalava, J. (2008). Typing of SHV extended-spectrum β-lactamases by pyrosequencing in Klebsiella pneumoniae strains with chromosomal SHV β-lactamase. Antimicrob. Agents Chemother. 52, 2632–2635. doi: 10.1128/AAC.01259-07
Haeggman, S., Löfdahl, S., and Burman, L. G. (1997). An allelic variant of the chromosomal gene for class A β-lactamase K2, specific for Klebsiella pneumoniae, is the ancestor of SHV-1. Antimicrob. Agents Chemother. 41, 2705–2709.
Hall, B. G., and Barlow, M. (2004). Evolution of the serine β-lactamases: past, present and future. Drug Resist. Updat. 7, 111–123. doi: 10.1016/j.drup.2004.02.003
Hammad, A. M., and Shimamoto, T. (2011). Asymptomatic intramammary infection with multidrug-resistant Gram-negative bacteria in a research dairy farm: incidence and genetic basis of resistance. J. Vet. Med. Sci. 73, 1089–1092. doi: 10.1292/jvms.10-0361
Hammami, S., Boubaker, I. B.-B., Saidani, M., Lakhal, E., Hassen, A. B., Kamoun, A., et al. (2011). Characterization and molecular epidemiology of extended spectrum β-lactamase producing Enterobacter cloacae isolated from a Tunisian hospital. Microb. Drug Resist. 18, 59–65. doi: 10.1089/mdr.2011.0074
Haque, A., Yoshizumi, A., Saga, T., Ishii, Y., and Tateda, K. (2014). ESBL-producing Enterobacteriaceae in environmental water in Dhaka, Bangladesh. J. Infect. Chemother. 20, 735–737. doi: 10.1016/j.jiac.2014.07.003
Hargreaves, M. L., Shaw, K. M., Dobbins, G., Snippes Vagnone, P. M., Harper, J. E., Boxrud, D., et al. (2015). Clonal dissemination of Enterobacter cloacae harboring blaKPC-3 in the upper midwestern United States. Antimicrob. Agents Chemother. 59, 7723–7734. doi: 10.1128/AAC.01291-15
Harrois, D., Breurec, S., Seck, A., Delauné, A., Hello, S. L., Gándara, M. P. D. L., et al. (2014). Prevalence and characterization of extended-spectrum β-lactamase-producing clinical Salmonella enterica isolates in Dakar, Senegal, from 1999 to 2009. Clin. Microbiol. Infect. 20, O109–O116. doi: 10.1111/1469-0691.12339
Hasman, H., Mevius, D., Veldman, K., Olesen, I., and Aarestrup, F. M. (2005). β-Lactamases among extended-spectrum β-lactamase (ESBL)-resistant Salmonella from poultry, poultry products and human patients in The Netherlands. J. Antimicrob. Chemother. 56, 115–121. doi: 10.1093/jac/dki190
He, S., Hickman, A. B., Varani, A. M., Siguier, P., Chandler, M., Dekker, J. P., et al. (2015). Insertion sequence IS26 reorganizes Plasmids in clinically isolated multidrug-resistant Bacteria by replicative transposition. mBio 6:e00762-15. doi: 10.1128/mBio.00762-15
Heritage, J., Chambers, P. A., Tyndall, C., and Buescher, E. S. (2003). SHV-34: an extended-spectrum β-lactamase encoded by an epidemic plasmid. J. Antimicrob. Chemother. 52, 1015–1017. doi: 10.1093/jac/dkh017
Herrera-Leon, S., Gonzalez-Sanz, R., Herrera-Leon, L., and Echeita, M. A. (2011). Characterization of multidrug-resistant Enterobacteriaceae carrying plasmid-mediated quinolone resistance mechanisms in Spain. J. Antimicrob. Chemother. 66, 287–290. doi: 10.1093/jac/dkq423
Hiki, M., Usui, M., Kojima, A., Ozawa, M., Ishii, Y., and Asai, T. (2013). Diversity of plasmid replicons encoding the blaCMY-2 gene in broad-spectrum cephalosporin-resistant Escherichia coli from livestock animals in Japan. Foodborne Pathog. Dis. 10, 243–249. doi: 10.1089/fpd.2012.1306
Hiroi, M., Harada, T., Kawamori, F., Takahashi, N., Kanda, T., Sugiyama, K., et al. (2011). A survey of β-lactamase-producing Escherichia coli in farm animals and raw retail meat in Shizuoka Prefecture, Japan. Jpn. J. Infect. Dis. 64, 153–155.
Ho, P. L., Li, Z., Lo, W. U., Cheung, Y. Y., Lin, C. H., Sham, P. C., et al. (2012). Identification and characterization of a novel incompatibility group X3 plasmid carrying blaNDM-1 in Enterobacteriaceae isolates with epidemiological links to multiple geographical areas in China. Emerg. Microbes Infect. 1, e39. doi: 10.1038/emi.2012.37
Hocquet, D., Plesiat, P., Dehecq, B., Mariotte, P., Talon, D., and Bertrand, X. (2010). Nationwide investigation of extended-spectrum β-lactamases, metallo-β-lactamases, and extended-spectrum oxacillinases produced by ceftazidime-resistant Pseudomonas aeruginosa strains in France. Antimicrob. Agents Chemother. 54, 3512–3515. doi: 10.1128/AAC.01646-09
Hordijk, J., Bosman, A. B., van Essen-Zandbergen, A., Veldman, K., Dierikx, C., Wagenaar, J. A., et al. (2011). qnrB19 Gene Bracketed by IS26 on a 40-Kilobase IncR Plasmid from an Escherichia coli Isolate from a Veal Calf. Antimicrobial. Agents Chemother. 55, 453–454. doi: 10.1128/AAC.00866-10
Huang, Y., Yu, X., Xie, M., Wang, X., Liao, K., Xue, W., et al. (2016). Widespread dissemination of carbapenem-resistant Escherichia coli sequence tpe 167 strains harboring blaNDM-5 in clinical settings in China. Antimicrob. Agents Chemother. 60, 4364–4368. doi: 10.1128/AAC.00859-16
Huletsky, A., Knox, J. R., and Levesque, R. C. (1993). Role of Ser-238 and Lys-240 in the hydrolysis of third-generation cephalosporins by SHV-type β-lactamases probed by site-directed mutagenesis and three-dimensional modeling. J. Biol. Chem. 268, 3690–3697.
Jeannot, K., Fournier, D., Müller, E., Cholley, P., and Plesiat, P. (2013). Clonal dissemination of Pseudomonas aeruginosa isolates producing extended-spectrum β-lactamase SHV-2a. J. Clin. Microbiol. 51, 673–675. doi: 10.1128/JCM.02313-12
Jelsch, C., Mourey, L., Masson, J. M., and Samama, J. P. (1993). Crystal structure of Escherichia coli TEM1 β-lactamase at 1.8A resolution. Proteins 16, 364–383. doi: 10.1002/prot.340160406
Jiang, H.-X., Tang, D., Liu, Y.-H., Zhang, X.-H., Zeng, Z.-L., Xu, L., et al. (2012). Prevalence and characteristics of β-lactamase and plasmid-mediated quinolone resistance genes in Escherichia coli isolated from farmed fish in China. J. Antimicrob. Chemother. 67, 2350–2353. doi: 10.1093/jac/dks250
Johnson, T. J., Wannemuehler, Y. M., Johnson, S. J., Logue, C. M., White, D. G., Doetkott, C., et al. (2007). Plasmid replicon typing of commensal and pathogenic Escherichia coli isolates. Appl. Environ. Microbiol. 73, 1976–1983. doi: 10.1128/AEM.02171-06
Jones, C. H., Ruzin, A., Tuckman, M., Visalli, M. A., Petersen, P. J., and Bradford, P. A. (2009). Pyrosequencing using the single-nucleotide polymorphism protocol for rapid determination of TEM- and SHV-type extended-spectrum β-lactamases in clinical isolates and identification of the novel β-lactamase genes blaSHV-48, blaSHV-105, and blaTEM-155. Antimicrob. Agents Chemother. 53, 977–986. doi: 10.1128/AAC.01155-08
Jones-Dias, D., Manageiro, V., and Caniça, M. (2016). Influence of agricultural practice on mobile bla genes: IncI1-bearing CTX-M, SHV, CMY and TEM in Escherichia coli from intensive farming soils. Environ. Microbiol. 18, 260–272. doi: 10.1111/1462-2920.13021
Jones-Dias, D., Manageiro, V., Martins, A. P., Ferreira, E., and Caniça, M. (2015). New class 2 integron In2-4 among IncI1-positive Escherichia coli isolates carrying ESBL and PMAβ genes from food animals in Portugal. Foodborne Pathog. Dis. 13, 36–39. doi: 10.1089/fpd.2015.1972
Jouini, A., Slama, K. B., Klibi, N., Sallem, R. B., Estepa, V., Vinué, L., et al. (2013). Lineages and virulence gene content among extended-spectrum β-Lactamase producing Escherichia coli strains of food origin in Tunisia. J. Food Prot. 76, 323–327. doi: 10.4315/0362-028X.JFP-12-251
Jung, J. S., Popp, C., Sparbier, K., Lange, C., Kostrzewa, M., and Schubert, S. (2014). Evaluation of matrix-assisted laser desorption ionization–time of flight mass spectrometry for rapid detection of β-lactam resistance in Enterobacteriaceae derived from blood cultures. J. Clin. Microbiol. 52, 924–930. doi: 10.1128/JCM.02691-13
Kameyama, M., Chuma, T., Yabata, J., Tominaga, K., Iwata, H., and Okamoto, K. (2013). Prevalence and epidemiological relationship of CMY-2 AmpC β-cactamase and CTX-M extended-spectrum β-lactamase-producing Escherichia coli isolates from broiler farms in Japan. J. Vet. Med. Sci. 75, 1009–1015. doi: 10.1292/jvms.12-0453
Kassis-Chikhani, N., Frangeul, L., Drieux, L., Sengelin, C., Jarlier, V., Brisse, S., et al. (2013). Complete nucleotide sequence of the first KPC-2- and SHV-12-encoding IncX plasmid, pKpS90, from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 57, 618–620. doi: 10.1128/AAC.01712-12
Kim, H.-S., Chon, J.-W., Kim, Y.-J., Kim, D.-H., Kim, M.-S., and Seo, K.-H. (2015). Prevalence and characterization of extended-spectrum-β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in ready-to-eat vegetables. Int. J. Food Microbiol. 207, 83–86. doi: 10.1016/j.ijfoodmicro.2015.04.049
Kiratisin, P., Apisarnthanarak, A., Laesripa, C., and Saifon, P. (2008). Molecular characterization and epidemiology of extended-spectrum- β-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolates causing health care-associated infection in Thailand, where the CTX-M family is endemic. Antimicrob. Agents Chemother. 52, 2818–2824. doi: 10.1128/AAC.00171-08
Kliebe, C., Nies, B. A., Meyer, J. F., Tolxdorff-Neutzling, R. M., and Wiedemann, B. (1985). Evolution of plasmid-coded resistance to broad-spectrum cephalosporins. Antimicrob. Agents Chemother. 28, 302–307. doi: 10.1128/AAC.28.2.302
Knox, J. R. (1995). Extended-spectrum and inhibitor-resistant TEM-type β-lactamases: mutations, specificity, and three-dimensional structure. Antimicrob. Agents Chemother. 39, 2593–2601. doi: 10.1128/AAC.39.12.2593
Kurokawa, H., Yagi, T., Shibata, N., Shibayama, K., Kamachi, K., and Arakawa, Y. (2000). A new SHV-derived extended-spectrum β-Lactamase (SHV-24) that hydrolyzes ceftazidime through a single-amino-acid substitution (D179G) in the Ω-Loop. Antimicrob. Agents Chemother. 44, 1725–1727. doi: 10.1128/AAC.44.6.1725-1727.2000
Kwon, T., Jung, Y. H., Lee, S., Yun, M. R., Kim, W., and Kim, D. W. (2016). Comparative genomic analysis of Klebsiella pneumoniae subsp. pneumoniae KP617 and PittNDM01, NUHL24835, and ATCC BAA-2146 reveals unique evolutionary history of this strain. Gut Pathog. 8, 34. doi: 10.1186/s13099-016-0117-1
Lascols, C., Hackel, M., Hujer, A. M., Marshall, S. H., Bouchillon, S. K., Hoban, D. J., et al. (2012). Using nucleic acid microarrays to perform molecular epidemiology and detect novel β-lactamases: a snapshot of extended-spectrum β-lactamases throughout the world. J. Clin. Microbiol. 50, 1632–1639. doi: 10.1128/JCM.06115-11
Lee, K. Y., Hopkins, J. D., and Syvanen, M. (1990). Direct involvement of Is26 in an Antibiotic-Resistance Operon. J. Bacteriol. 172, 3229–3236.
Lee, Y., Kim, B. S., Chun, J., Yong, J. H., Lee, Y. S., Yoo, J. S., et al. (2014). Clonality and resistome analysis of KPC-producing Klebsiella pneumoniae strain isolated in Korea using whole genome sequencing. Biomed. Res. Int. 2014:352862. doi: 10.1155/2014/352862
Leinberger, D. M., Grimm, V., Rubtsova, M., Weile, J., Schroppel, K., Wichelhaus, T. A., et al. (2010). Integrated detection of extended-spectrum-β-lactam resistance by DNA microarray-based genotyping of TEM, SHV, and CTX-M genes. J. Clin. Microbiol. 48, 460–471. doi: 10.1128/JCM.00765-09
Ling, B.-D., Liu, G., Xie, Y.-E., Zhou, Q.-X., Zhao, T.-K., Li, C.-Q., et al. (2006). Characterisation of a novel extended-spectrum b-lactamase, SHV-70, from a clinical isolate of Enterobacter cloacae in China. Int. J. Antimicrob. Agents 27, 355–356. doi: 10.1016/j.ijantimicag.2006.02.003
Literak, I., Dolejska, M., Janoszowska, D., Hrusakova, J., Meissner, W., Rzyska, H., et al. (2010). Antibiotic resistant Escherichia coli bacteria, including strains with genes encoding the extended-spectrum β-lactamase and qnrS, in waterbirds on the Baltic Sea coast of Poland. Appl. Environ. Microbiol. 76, 8126–8134. doi: 10.1128/AEM.01446-10
Liu, G., Ling, B. D., Zeng, Y., Lin, L., Xie, Y. E., and and, Lei, J. (2008). Molecular characterization of extended-spectrum β-lactamases produced by clinical isolates of Enterobacter cloacae from a teaching hospital in China. Jpn. J. Infect. Dis. 61, 286–289.
Liu, Y., Wan, L. G., Deng, Q., Cao, X. W., Yu, Y., and Xu, Q. F. (2015). First description of NDM-1-, KPC-2-, VIM-2- and IMP-4-producing Klebsiella pneumoniae strains in a single Chinese teaching hospital. Epidemiol. Infect. 143, 376–384. doi: 10.1017/S0950268814000995
Ma, L., Alba, J., Chang, F.-Y., Ishiguro, M., Yamaguchi, K., Siu, L. K., et al. (2005). Novel SHV-derived extended-spectrum β-lactamase, SHV-57, that confers resistance to ceftazidime but not cefazolin. Antimicrob. Agents Chemother. 49, 600–605. doi: 10.1128/AAC.49.2.600-605.2005
Machado, E., Coque, T. M., Cantón, R., Novais, A., Sousa, J. C., Baquero, F., et al. (2007). High diversity of extended-spectrum β-lactamases among clinical isolates of Enterobacteriaceae from Portugal. J. Antimicrob. Chemother. 60, 1370–1374. doi: 10.1093/jac/dkm381
Machado, E., Coque, T. M., Cantón, R., Sousa, J. C., and Peixe, L. (2008). Antibiotic resistance integrons and extended-spectrum β-lactamases among Enterobacteriaceae isolates recovered from chickens and swine in Portugal. J. Antimicrob. Chemother. 62, 296–302. doi: 10.1093/jac/dkn179
Mansour, W., Dahmen, S., Poirel, L., Charfi, K., Bettaieb, D., Boujaafar, N., et al. (2009). Emergence of SHV-2a extended-spectrum β-lactamases in clinical isolates of Pseudomonas aeruginosa in a university hospital in Tunisia. Microb. Drug. Resist. 15, 295–301. doi: 10.1089/mdr.2009.0012
Maravic, A., Skocibusic, M., Cvjetan, S., Samanic, I., Fredotovic, Z., and Puizina, J. (2015). Prevalence and diversity of extended-spectrum-β-lactamase-producing Enterobacteriaceae from marine beach waters. Mar. Pollut. Bull. 90, 60–67. doi: 10.1016/j.marpolbul.2014.11.021
Maravić, A., Skočibušič, M., Samanić, I., Fredotovič, Z., Cvjetan, S., Jutronić, M., et al. (2013). Aeromonas spp. simultaneously harbouring blaCTX-M-15, blaSHV-12, blaPER-1 and blaFOX-2, in wild-growing Mediterranean mussel (Mytilus galloprovincialis) from Adriatic Sea, Croatia. Int. J. Food Microbiol. 166, 301–308. doi: 10.1016/j.ijfoodmicro.2013.07.010
Marcadé, G., Deschamps, C., Boyd, A., Gautier, V., Picard, B., Branger, C., et al. (2009). Replicon typing of plasmids in Escherichia coli producing extended-spectrum β-lactamases. J. Antimicrob. Chemother. 63, 67–71. doi: 10.1093/jac/dkn428
Markovska, R., Schneider, I., Ivanova, D., Mitov, I., and Bauernfeind, A. (2014). Predominance of IncL/M and IncF plasmid types among CTX-M-ESBL-producing Escherichia coli and Klebsiella pneumoniae in Bulgarian hospitals. APMIS 122, 608–615. doi: 10.1111/apm.12204
Marti, E., Jofre, J., and Balcazar, J. L. (2013). Prevalence of antibiotic resistance genes and bacterial community composition in a river influenced by a wastewater treatment plant. PLoS ONE 8:e78906. doi: 10.1371/journal.pone.0078906
Matagne, A., Lamotte-Brasseur, J., and Frère, J. M. (1998). Catalytic properties of class A β-lactamases: efficiency and diversity. Biochem. J. 330(Pt 2), 581–598.
Matthew, M., Hedges, R. W., and Smith, J. T. (1979). Types of β-lactamase determined by plasmids in gram-negative bacteria. J. Bacteriol. 138, 657–662.
Mazzariol, A., Roelofsen, E., Koncan, R., Voss, A., and Cornaglia, G. (2007). Detection of a new SHV-type Extended-spectrum β-Lactamase, SHV-31, in a Klebsiella pneumoniae strain causing a large nosocomial outbreak in The Netherlands. Antimicrob. Agents Chemother. 51, 1082–1084. doi: 10.1128/AAC.00909-06
Medeiros, A. A. (1997). Evolution and dissemination of β-lactamases accelerated by generations of β-lactam antibiotics. Clin. Infect. Dis. 24 (Suppl. 1), S19–S45.
Mendonça, N., Ferreira, E., and Caniça, M. (2006). Occurrence of a novel SHV-type enzyme (SHV-55) among isolates of Klebsiella pneumoniae from Portuguese origin in a comparison study for extended-spectrum β-lactamase–producing evaluation. Diagn. Microbiol. Infect. Dis. 56, 415–420. doi: 10.1016/j.diagmicrobio.2006.06.023
Mendonça, N., Ferreira, E., Louro, D., and Caniça, M. (2009). Molecular epidemiology and antimicrobial susceptibility of extended- and broad-spectrum β-lactamase-producing Klebsiella pneumoniae isolated in Portugal. Int. J. Antimicrob. Agents 34, 29–37. doi: 10.1016/j.ijantimicag.2008.11.014
Miriagou, V., Carattoli, A., Tzelepi, E., Villa, L., and Tzouvelekis, L. S. (2005). IS26-Associated In4-type integrons forming multiresistance loci in enterobacterial plasmids. Antimicrobial. Agents Chemother. 49, 3541–3543. doi: 10.1128/AAC.49.8.3541-3543.2005
Mnif, B., Harhour, H., Jdidi, J., Mahjoubi, F., Genel, N., Arlet, G., et al. (2013). Molecular epidemiology of extended-spectrum β-lactamase-producing Escherichia coli in Tunisia and characterization of their virulence factors and plasmid addiction systems. BMC Microbiol. 13:147. doi: 10.1186/1471-2180-13-147
Mulvey, M. R., Bryce, E., Boyd, D., Ofner-Agostini, M., Christianson, S., Simor, A. E., et al. (2004). Ambler class A extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella spp. in Canadian hospitals. Antimicrob. Agents Chemother. 48, 1204–1214. doi: 10.1128/AAC.48.4.1204-1214.2004
Muniesa, M., Colomer-Lluch, M., and Jofre, J. (2013). Potential impact of environmental bacteriophages in spreading antibiotic resistance genes. Future Microbiol. 8, 739–751. doi: 10.2217/fmb.13.32
Muratani, T., Kobayashi, T., and Matsumoto, T. (2006). Emergence and prevalence of β-lactamase-producing Klebsiella pneumoniae resistant to cephems in Japan. Int. J. Antimicrob. Agents 27, 491–499. doi: 10.1016/j.ijantimicag.2006.03.007
M'Zali, F. H., Heritage, J., Gascoyne-Binzi, D. M., Snelling, A. M., and Hawkey, P. M. (1998). PCR single strand conformational polymorphism can be used to detect the gene encoding SHV-7 extended-spectrum β-lactamase and to identify different SHV genes within the same strain. J. Antimicrob. Chemother. 41, 123–125. doi: 10.1093/jac/41.1.123
Naas, T., Namdari, F., Réglier-Poupet, H., Poyart, C., and Nordmann, P. (2007). Panresistant extended-spectrum β-lactamase SHV-5-producing Acinetobacter baumannii from New York City. J. Antimicrob. Chemother. 60, 1174–1176. doi: 10.1093/jac/dkm366
Naas, T., Philippon, L., Poirel, L., Ronco, E., and Nordmann, P. (1999). An SHV-derived extended-spectrum β-lactamase in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43, 1281–1284.
Naiemi, N. A., Duim, B., Savelkoul, P. H. M., Spanjaard, L., de Jonge, E., Bart, A., et al. (2005). Widespread transfer of resistance genes between bacterial species in an intensive care unit: implications for hospital epidemiology. J. Clin. Microbiol. 43, 4862–4864. doi: 10.1128/JCM.43.9.4862-4864.2005
Naseer, U., Eriksen, B. O., Sundsfjord, A., and Samuelsen, Ø. (2012). Fecal colonization of VIM-1-producing Klebsiella pneumoniae and in vivo transfer of multidrug-resistant IncN plasmid in a renal transplant patient. Diagn. Microbiol. Infect. Dis. 72, 363–366. doi: 10.1016/j.diagmicrobio.2011.12.010
Neonakis, I. K., Scoulica, E. V., Dimitriou, S. K., Gikas, A. I., and Tselentis, Y. J. (2003). Molecular epidemiology of extended-spectrum β-lactamases produced by clinical isolates in a university hospital in Greece: detection of SHV-5 in Pseudomonas aeruginosa and prevalence of SHV-12. Microb. Drug Resist. 9, 161–165. doi: 10.1089/107662903765826750
Nicolas, M. H., Jarlier, V., Honore, N., Philippon, A., and Cole, S. T. (1989). Molecular characterization of the gene encoding SHV-3 β-lactamase responsible for transferable cefotaxime resistance in clinical isolates of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 33, 2096–2100. doi: 10.1128/AAC.33.12.2096
Noda, T., Murakami, K., Etoh, Y., Okamoto, F., Yatsuyanagi, J., Sera, N., et al. (2015). Increase in resistance to extended-spectrum cephalosporins in Salmonella isolated from retail chicken products in Japan. PLoS ONE 10:e0116927. doi: 10.1371/journal.pone.0133094
Novovic, K., Filipic, B., Veljovic, K., Begovic, J., Mirkovic, N., and Jovcic, B. (2015). Environmental waters and blaNDM-1 in Belgrade, Serbia: endemicity questioned. Sci. Total. Environ. 511, 393–398. doi: 10.1016/j.scitotenv.2014.12.072
Nüesch-Inderbinen, M. T., Kayser, F. H., and Hächler, H. (1997). Survey and molecular genetics of SHV β-lactamases in Enterobacteriaceae in Switzerland: two novel enzymes, SHV-11 and SHV-12. Antimicrob. Agents Chemother. 41, 943–949.
Nugent, M. E., and Hedges, R. W. (1979). The nature of genetic determinants for the SHV-1 β-lactamase. Mol. Gen. Genet. 175, 239–243. doi: 10.1007/BF00397222
Nukaga, M., Mayama, K., Hujer, A. M., Bonomo, R. A., and Knox, J. R. (2003). Ultrahigh resolution structure of a class A β-lactamase: on the mechanism and specificity of the extended-spectrum SHV-2 enzyme. J. Mol. Biol. 328, 289–301. doi: 10.1016/S0022-2836(03)00210-9
Oikonomou, O., Liakopoulos, A., Phee, L. M., Betts, J., Mevius, D., and Wareham, D. W. (2016). Providencia stuartii isolates from Greece: co-carriage of cephalosporin (blaSHV-5, blaVEB-1), carbapenem (blaVIM-1), and aminoglycoside (rmtB) resistance determinants by a multidrug-resistant outbreak clone. Microb. Drug Resist. 22, 379–386. doi: 10.1089/mdr.2015.0215
Osinska, A., Harnisz, M., and Korzeniewska, E. (2016). Prevalence of plasmid-mediated multidrug resistance determinants in fluoroquinolone-resistant bacteria isolated from sewage and surface water. Environ. Sci. Pollut. Res. Int. 23, 10818–10831. doi: 10.1007/s11356-016-6221-4
Oviaño, M., Fernández, B., Fernández, A., Barba, M. J., Mouriño, C., and Bou, G. (2014). Rapid detection of Enterobacteriaceae producing extended spectrum β-lactamases directly from positive blood cultures by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Clin. Microbiol. Infect. 20, 1146–1157. doi: 10.1111/1469-0691.12729
Partridge, S. R., Ginn, A. N., Wiklendt, A. M., Ellem, J., Wong, J. S., Ingram, P., et al. (2015). Emergence of blaKPC carbapenemase genes in Australia. Int. J. Antimicrob. Agents 45, 130–136. doi: 10.1016/j.ijantimicag.2014.10.006
Paterson, D. L., and Bonomo, R. A. (2005). Extended-spectrum β-lactamases: a clinical update. Clin. Microbiol. Rev. 18, 657–686. doi: 10.1128/CMR.18.4.657-686.2005
Péduzzi, J., Barthélémy, M., Tiwari, K., Mattioni, D., and Labia, R. (1989). Structural features related to hydrolytic activity against ceftazidime of plasmid-mediated SHV-type CAZ-5 β-lactamase. Antimicrob. Agents Chemother. 33, 2160–2163. doi: 10.1128/AAC.33.12.2160
Perreira Ramos, P. I., Picão, R. C., Almeida, L. G., Lima, N. C., Girardello, R., Vivan, A. C., et al. (2014). Comparative analysis of the complete genome of KPC-2-producing Klebsiella pneumoniae Kp13 reveals remarkable genome plasticity and a wide repertoire of virulence and resistance mechanisms. BMC Genomics 15:54. doi: 10.1186/1471-2164-15-54
Philippon, A., Slama, P., Dény, P., and Labia, R. (2016). A structure-based classification of class A β-lactamases, a broadly diverse family of enzymes. Clin. Microbiol. Rev. 29, 29–57. doi: 10.1128/CMR.00019-15
Pinto, L., Radhouani, H., Coelho, C., Martins Da Costa, P., Simões, R., Brandão, R. M. L., et al. (2010). Genetic detection of extended-spectrum β-lactamase-containing Escherichia coli isolates from birds of prey from Serra da Estrela Natural Reserve in Portugal. Appl. Environ. Microbiol. 76, 4118–4120. doi: 10.1128/AEM.02761-09
Pitout, J. D. D., Thomson, K. S., Hanson, N. D., Ehrhardt, A. F., Coudron, P., and Sanders, C. C. (1998). Plasmid-mediated resistance to expanded-spectrum cephalosporins among Enterobacter aerogenes strains. Antimicrob. Agents Chemother. 42, 596–600.
Pitton, J. (1972). “Mechanisms of bacterial resistance to antibiotics,” in Review of Physiology, ed R. Adirna (Berlin: Springer-Verlag), 15–93.
Podbielski, A., Schönling, J., Melzer, B., Warnatz, K., and Leusch, H.-G. (1991). Molecular characterization of a new plasmid-encoded SHV-type β-lactamase (SHV-2 variant) conferring high-level cefotaxime resistance upon Klebsiella pneumoniae. J. Gen. Microbiol. 137, 569–578. doi: 10.1099/00221287-137-3-569
Poirel, L., Héritier, C., Podglajen, I., Sougakoff, W., Gutmann, L., and Nordmann, P. (2003). Emergence in Klebsiella pneumoniae of a chromosome-encoded SHV β-Lactamase that compromises the efficacy of imipenem. Antimicrob. Agents Chemother. 47, 755–758. doi: 10.1128/AAC.47.2.755-758.2003
Poirel, L., Lebessi, E., Castro, M., Fèvre, C., Foustoukou, M., and Nordmann, P. (2004). Nosocomial outbreak of extended-spectrum β-lactamase SHV-5-producing isolates of Pseudomonas aeruginosa in Athens, Greece. Antimicrob. Agents Chemother. 48, 2277–2279. doi: 10.1128/AAC.48.6.2277-2279.2004
Post, V., and Hall, R. M. (2009). AbaR5, a large multiple-antibiotic resistance region found in Acinetobacter baumannii. Antimicrobial. Agents Chemother. 53, 2667–2671. doi: 10.1128/AAC.01407-08
Pouget, J. G., Coutinho, F. J., Reid-Smith, R. J., and Boerlin, P. (2013). Characterization of blaSHV genes on plasmids from Escherichia coli and Salmonella enterica isolates from Canadian food animals (2006-2007). Appl. Environ. Microbiol. 79, 3864–3866. doi: 10.1128/AEM.00355-13
Preston, K. E., Hitchcock, S. A., Aziz, A. Y., and Tine, J. A. (2014). The complete nucleotide sequence of the multi-drug resistance-encoding IncL/M plasmid pACM1. Plasmid 76, 54–65. doi: 10.1016/j.plasmid.2014.08.005
Prinarakis, E. E., Tzelepi, E., Gazouli, M., Mentis, A. F., and Tzouvelekis, L. S. (1996). Characterization of a novel SHV α-lactamase variant that resembles the SHV-5 enzyme. FEMS Microbiol. Lett. 139, 229–234.
Ramdani-Bouguessa, N., Manageiro, V., Jones-Dias, D., Ferreira, E., Tazir, M., and Caniça, M. (2011). Role of SHV β-lactamase variants in resistance of clinical Klebsiella pneumoniae strains to β-lactams in an Algerian hospital. J. Med. Microbiol. 60, 983–987. doi: 10.1099/jmm.0.030577-0
Randegger, C. C., and Hächler, H. (2001). Real-time PCR and melting curve analysis for reliable and rapid detection of SHV extended-spectrum β-lactamases. Antimicrob. Agents Chemother. 45, 1730–1736. doi: 10.1128/AAC.45.6.1730-1736.2001
Randegger, C. C., Keller, A., Irla, M., Wada, A., and Hächler, H. (2000). Contribution of natural amino acid substitutions in SHV extended-spectrum β-lactamases to resistance against various β-lactams. Antimicrob. Agents Chemother. 44, 2759–2763. doi: 10.1128/AAC.44.10.2759-2763.2000
Rasheed, J. K., Anderson, G. J., Yigit, H., Queenan, A. M., Doménech-Sánchez, A., Swenson, J. M., et al. (2000). Characterization of the extended-spectrum β-lactamase reference strain, Klebsiella pneumoniae K6 (ATCC 700603), which produces the Novel enzyme SHV-18. Antimicrob. Agents Chemother. 44, 2382–2388. doi: 10.1128/AAC.44.9.2382-2388.2000
Rasheed, J. K., Jay, C., Metchock, B., Berkowitz, F., Weigel, L., Crellin, J., et al. (1997). Evolution of extended-spectrum β-lactam resistance (SHV-8) in a strain of Escherichia coli during multiple episodes of bacteremia. Antimicrob. Agents Chemother. 41, 647–653.
Reuland, E. A., Al Naiemi, N., Raadsen, S. A., Savelkoul, P. H., Kluytmans, J. A., and Vandenbroucke-Grauls, C. M. (2014). Prevalence of ESBL-producing Enterobacteriaceae in raw vegetables. Eur. J. Clin. Microbiol. Infect. Dis. 33, 1843–1846. doi: 10.1007/s10096-014-2142-7
Rocha-Gracia, R. C., Cortés-Cortés, G., Lozano-Zarain, P., Bello, F., Martínez-Laguna, Y., and Torres, C. (2015). Faecal Escherichia coli isolates from healthy dogs harbour CTX-M-15 and CMY-2 β-lactamases. Vet. J. 203, 315–319. doi: 10.1016/j.tvjl.2014.12.026
Rodrigues, C., Machado, E., Ramos, H., Peixe, L., and Novais, Â. (2014). Expansion of ESBL-producing Klebsiella pneumoniae in hospitalized patients: a successful story of international clones (ST15, ST147, ST336) and epidemic plasmids (IncR, IncFIIK). Int. J. Antimicrob. Agents 304, 1100–1108. doi: 10.1016/j.ijmm.2014.08.003
Rubin, J. E., and Pitout, J. D. D. (2014). Extended-spectrum β-lactamase, carbapenemase and AmpC producing Enterobacteriaceae in companion animals. Vet. Microbiol. 170, 10–18. doi: 10.1016/j.vetmic.2014.01.017
Rzewuska, M., Stefanska, I., Kizerwetter-Swida, M., Chrobak-Cmiel, D., Szczygielska, P., Lesniak, M., et al. (2015). Characterization of extended-spectrum-β-lactamases produced by Escherichia coli strains isolated from dogs in Poland. Pol. J. Microbiol. 64, 285–288.
Samuelsen, Ø., Toleman, M. A., Hasseltvedt, V., Fuursted, K., Leegaard, T. M., Walsh, T. R., et al. (2011). Molecular characterization of VIM-producing Klebsiella pneumoniae from Scandinavia reveals genetic relatedness with international clonal complexes encoding transferable multidrug resistance. Clin. Microbiol. Infect. 17, 1811–1816. doi: 10.1111/j.1469-0691.2011.03532.x
Sánchez-Romero, I., Asensio, A., Oteo, J., Muñoz-Algarra, M., Isidoro, B., Vindel, A., et al. (2012). Nosocomial outbreak of VIM-1-producing Klebsiella pneumoniae isolates of multilocus sequence type 15: molecular basis, clinical risk factors, and outcome. Antimicrob. Agents Chemother. 56, 420–427. doi: 10.1128/AAC.05036-11
Shahcheraghi, F., Nikbin, V. S., and Feizabadi, M. M. (2009). Prevalence of ESBLs genes among multidrug-resistant isolates of Pseudomonas aeruginosa isolated from patients in Tehran. Microb. Drug Resist. 15, 37–39. doi: 10.1089/mdr.2009.0880
Shaheen, B. W., Nayak, R., Foley, S. L., Kweon, O., Deck, J., Park, M., et al. (2011). Molecular characterization of resistance to extended-spectrum cephalosporins in clinical Escherichia coli isolates from companion animals in the United States. Antimicrob. Agents Chemother. 55, 5666–5675. doi: 10.1128/AAC.00656-11
Singh, K., Mangold, K. A., Wyant, K., Schora, D. M., Voss, B., Kaul, K. L., et al. (2012). Rectal screening for Klebsiella pneumoniae carbapenemases: comparison of real-time PCR and culture using two selective screening agar plates. J. Clin. Microbiol. 50, 2596–2600. doi: 10.1128/JCM.00654-12
Singh, P., Pfeifer, Y., and Mustapha, A. (2016). Multiplex real-time PCR assay for the detection of extended-spectrum β-lactamase and carbapenemase genes using melting curve analysis. J. Microbiol. Methods 124, 72–78. doi: 10.1016/j.mimet.2016.03.014
Song, W., Kim, J., Bae, I. K., Jeong, S. H., Seo, Y. H., Shin, J. H., et al. (2011). Chromosome-encoded AmpC and CTX-M extended-spectrum β-lactamases in clinical isolates of Proteus mirabilis from Korea. Antimicrob. Agents Chemother. 55, 1414–1419. doi: 10.1128/AAC.01835-09
Sonnevend, A., Al Baloushi, A., Ghazawi, A., Hashmey, R., Girgis, S., Hamadeh, M. B., et al. (2013). Emergence and spread of NDM-1 producer Enterobacteriaceae with contribution of IncX3 plasmids in the United Arab Emirates. J. Med. Microbiol. 62, 1044–1050. doi: 10.1099/jmm.0.059014-0
Sowek, J. A., Singer, S. B., Ohringer, S., Malley, M. F., Dougherty, T. J., Gougoutas, J. Z., et al. (1991). Substitution of lysine at position 104 or 240 of TEM-1pTZ18R β-lactamase enhances the effect of serine-164 substitution on hydrolysis or affinity for cephalosporins and the monobactam aztreonam. Biochemistry 30, 3179–3188. doi: 10.1021/bi00227a004
Stolle, I., Prenger-Berninghoff, E., Stamm, I., Scheufen, S., Hassdenteufel, E., Guenther, S., et al. (2013). Emergence of OXA-48 carbapenemase-producing Escherichia coli and Klebsiella pneumoniae in dogs. J. Antimicrob. Chemother. 68, 2802–2808. doi: 10.1093/jac/dkt259
Stuart, J. C., Voets, G., Scharringa, J., Fluit, A. C., and Leverstein-Van Hall, M. A. (2012). Detection of carbapenemase-producing Enterobacteriaceae with a commercial DNA microarray. J. Med. Microbiol. 61, 809–812. doi: 10.1099/jmm.0.041673-0
Stürenburg, E., Sobottka, I., Feucht, H.-H., Mack, D., and Laufs, R. (2003). Comparison of BDPhoenix and VITEK2 automated antimicrobial susceptibility test systems for extended-spectrum β-lactamase detection in Escherichia coli and Klebsiella species clinical isolates. Diagn. Microbiol. Infect. Dis. 45, 29–34. doi: 10.1016/S0732-8893(02)00481-9
Szabó, D., Melan, M. A., Hujer, A. M., Bonomo, R. A., Hujer, K. M., Bethel, C. R., et al. (2005). Molecular analysis of the simultaneous production of two SHV-type extended-spectrum β-lactamases in a clinical isolate of Enterobacter cloacae by using single-nucleotide polymorphism genotyping. Antimicrob. Agents Chemother. 49, 4716–4720. doi: 10.1128/AAC.49.11.4716-4720.2005
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tang, J., Bu, Y., Zhang, X. X., Huang, K., He, X., Ye, L., et al. (2016). Metagenomic analysis of bacterial community composition and antibiotic resistance genes in a wastewater treatment plant and its receiving surface water. Ecotoxicol. Environ. Saf. 132, 260–269. doi: 10.1016/j.ecoenv.2016.06.016
Tato, M., Coque, T. M., Ruiz-Garbajosa, P., Pintado, V., Cobo, J., Sader, H. S., et al. (2007). Complex clonal and plasmid epidemiology in the first outbreak of Enterobacteriaceae infection involving VIM-1 metallo-β-Lactamase in Spain: toward endemicity? Clin. Infect. Dis. 45, 1171–1178. doi: 10.1086/522288
Tian, G.-B., Wang, H.-N., Zhang, A.-Y., Zhang, Y., Fan, W.-Q., Xu, C.-W., et al. (2012). Detection of clinically important β-lactamases in commensal Escherichia coli of human and swine origin in western China. J. Med. Microbiol. 61, 233–238. doi: 10.1099/jmm.0.036806-0
Tian, Z., Zhang, Y., Yu, B., and Yang, M. (2016). Changes of resistome, mobilome and potential hosts of antibiotic resistance genes during the transformation of anaerobic digestion from mesophilic to thermophilic. Water Res. 98, 261–269. doi: 10.1016/j.watres.2016.04.031
Timofte, D., Maciuca, I. E., Evans, N. J., Williams, H., Wattret, A., Fick, J. C., et al. (2014). Detection and molecular characterization of Escherichia coli CTX-M-15 and Klebsiella pneumoniae SHV-12 β-lactamases from bovine mastitis isolates in the United Kingdom. Antimicrob. Agents Chemother. 58, 789–794. doi: 10.1128/AAC.00752-13
Tissera, S., and Lee, S. M. (2013). Isolation of Extended Spectrum β-lactamase (ESBL) producing bacteria from urban surface waters in Malaysia. Malays. J. Med. Sci. 20, 14–22.
Uemura, S., Yokota, S., Mizuno, H., Sakawaki, E., Sawamoto, K., Maekawa, K., et al. (2010). Acquisition of a transposon encoding extended-spectrum β-lactamase SHV-12 by Pseudomonas aeruginosa isolates during the clinical course of a burn patient. Antimicrob. Agents Chemother. 54, 3956–3959. doi: 10.1128/AAC.00110-10
Veldman, K., Dierikx, C., Van Essen-Zandbergen, A., Van Pelt, W., and Mevius, D. (2010). Characterization of multidrug-resistant, qnrB2-positive and extended-spectrum-β-lactamase-producing Salmonella Concord and Salmonella Senftenberg isolates. J. Antimicrob. Chemother. 65, 872–875. doi: 10.1093/jac/dkq049
Veldman, K., van Tulden, P., Kant, A., Testerink, J., and Mevius, D. (2013). Characteristics of cefotaxime-resistant Escherichia coli from wild birds in The Netherlands. Appl. Environ. Microbiol. 79, 7556–7561. doi: 10.1128/AEM.01880-13
Villa, L., Pezzella, C., Tosini, F., Visca, P., Petrucca, A., and Carattoli, A. (2000). Multiple-antibiotic resistance mediated by structurally related IncL/M plasmids carrying an extended-spectrum β-lactamase gene and a class 1 integron. Antimicrob. Agents Chemother. 44, 2911–2914. doi: 10.1128/AAC.44.10.2911-2914.2000
Vinué, L., Lantero, M., Sáenz, Y., Somalo, S., de Diego, I., Pérez, F., et al. (2008). Characterization of extended-spectrum β-lactamases and integrons in Escherichia coli isolates in a Spanish hospital. J. Med. Microbiol. 57, 916–920. doi: 10.1099/jmm.0.47723-0
von Wintersdorff, C. J. H., Penders, J., van Niekerk, J. M., Mills, N. D., Majumder, S., van Alphen, L. B., et al. (2016). Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 7:173. doi: 10.3389/fmicb.2016.00173
Walther-Rasmussen, J., and Høiby, N. (2007). Class A carbapenemases. J. Antimicrob. Chemother. 60, 470–482. doi: 10.1093/jac/dkm226
Wang, X. J., Xu, X. L., Li, Z. W., Chen, H. B., Wang, Q., Yang, P. H., et al. (2014). An outbreak of a nosocomial NDM-1-producing Klebsiella pneumoniae ST147 at a teaching hospital in mainland China. Microb. Drug Resist. 20, 144–149. doi: 10.1089/mdr.2013.0100
Wang, X. R., Chen, J. C., Kang, Y., Jiang, N., An, S. C., and Gao, Z. C. (2012). Prevalence and characterization of plasmid-mediated blaESBL with their genetic environment in Escherichia coli and Klebsiella pneumoniae in patients with pneumonia. Chin. Med. J. (Engl.) 125, 894–900.
Whichard, J., Gay, K., Stevenson, J., Joyce, K., Cooper, K., Omondi, M., et al. (2007). Human Salmonella and concurrent decreased susceptibility to quinolones and extended-spectrum cephalosporins. Emerg. Infect. Dis. 13, 1681. doi: 10.3201/eid1311.061438
Wilke, M. S., Lovering, A. L., and Strynadka, N. C. (2005). β-lactam antibiotic resistance: a current structural perspective. Curr. Opin. Microbiol. 8, 525–533. doi: 10.1016/j.mib.2005.08.016
Winkler, M. L., and Bonomo, R. A. (2016). SHV-129: a gateway to global suppressors in the SHV β-lactamase family? Mol. Biol. Evol. 33, 429–441. doi: 10.1093/molbev/msv235
Wolny-Koladka, K., and Lenart-Boron, A. (2016). Phenotypic and molecular assessment of drug resistance profile and genetic diversity of waterborne. Water Air Soil Pollut. 227, 146. doi: 10.1007/s11270-016-2833-z
Yi, T., Kim, T. G., and Cho, K. S. (2015). Fate and behavior of extended-spectrum β-lactamase-producing genes in municipal sewage treatment plants. J. Environ. Sci. Health A Tox. Hazard Subst. Environ. Eng. 50, 1160–1168. doi: 10.1080/10934529.2015.1047673
Yigit, H., Queenan, A. M., Rasheed, J. K., Biddle, J. W., Domenech-Sanchez, A., Alberti, S., et al. (2003). Carbapenem-resistant strain of Klebsiella oxytoca harboring carbapenem-hydrolyzing β-lactamase KPC-2. Antimicrob. Agents Chemother. 47, 3881–3889. doi: 10.1128/AAC.47.12.3881-3889.2003
Yuan, M., Hall, L. M. C., Savelkoul, P. H. M., Vandenbroucke-Grauls, C. M. J. E., and Livermore, D. M. (2000). SHV-13, a novel extended-spectrum β-lactamase, in Klebsiella pneumoniae isolates from patients in an intensive care unit in Amsterdam. Antimicrob. Agents Chemother. 44, 1081–1084. doi: 10.1128/AAC.44.4.1081-1084.2000
Zhang, H., Zhou, Y., Guo, S., and Chang, W. (2015). Multidrug resistance found in extended-spectrum β-lactamase-producing Enterobacteriaceae from rural water reservoirs in Guantao, China. Front. Microbiol. 6:267. doi: 10.3389/fmicb.2015.00267
Zienkiewicz, M., Kern-Zdanowicz, I., Carattoli, A., Gniadkowski, M., and Ceglowski, P. (2013). Tandem multiplication of the IS26-flanked amplicon with the blaSHV-5 gene within plasmid p1658/97. FEMS Microbiol. Lett. 341, 27–36. doi: 10.1111/1574-6968.12084
Zou, L.-K., Wang, H.-N., Zeng, B., Zhang, A.-Y., Li, J.-N., Li, X.-T., et al. (2011). Phenotypic and genotypic characterization of β-lactam resistance in Klebsiella pneumoniae isolated from swine. Vet. Microbiol. 149, 139–146. doi: 10.1016/j.vetmic.2010.09.030
Zuo, B., Liu, Z., Wang, H., Yang, Y., Chen, J., and Ye, H. (2006). Genotype of TEM- and SHV-type β-lactamase producing Klebsiella pneumoniae in Guangzhou area [Article in Chinese]. Zhonghua Yi Xue Za Zhi 86, 2928–2932.
Zurfluh, K., Hächler, H., Nüesch-Inderbinen, M., and Stephan, R. (2013). Characteristics of extended-spectrum β-lactamase- and carbapenemase-producing Enterobacteriaceae isolates from rivers and lakes in Switzerland. Appl. Environ. Microbiol. 79, 3021–3026. doi: 10.1128/AEM.00054-13
Zurfluh, K., Nüesch-Inderbinen, M., Morach, M., Zihler Berner, A., Hächler, H., and Stephan, R. (2015). Extended-spectrum-β-lactamase-producing Enterobacteriaceae isolated from vegetables imported from the Dominican Republic, India, Thailand, and Vietnam. Appl. Environ. Microbiol. 81, 3115–3120. doi: 10.1128/AEM.00258-15
Keywords: β-lactamase, ESBL, blaSHV, SHV-2, SHV-5, SHV-12, plasmid, Enterobacteriaceae
Citation: Liakopoulos A, Mevius D and Ceccarelli D (2016) A Review of SHV Extended-Spectrum β-Lactamases: Neglected Yet Ubiquitous. Front. Microbiol. 7:1374. doi: 10.3389/fmicb.2016.01374
Received: 20 July 2016; Accepted: 19 August 2016;
Published: 05 September 2016.
Edited by:
Axel Cloeckaert, French National Institute for Agricultural Research (INRA), FranceReviewed by:
Sebastian Guenther, Free University of Berlin, GermanyCopyright © 2016 Liakopoulos, Mevius and Ceccarelli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniela Ceccarelli, ZGFuaWVsYS5jZWNjYXJlbGxpQHd1ci5ubA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.