- 1Department of Environment and Agriculture, Centre for Crop and Disease Management, Curtin University, Bentley, WA, Australia
- 2Institute of Life Science, School of Medicine, Swansea University, Swansea, UK
- 3School of Veterinary and Life Sciences, Murdoch University, Murdoch, WA, Australia
Pyrenophora teres f. sp. teres is the cause of net form of net blotch (NFNB), an economically important foliar disease in barley (Hordeum vulgare). Net and spot forms of net blotch are widely controlled using site-specific systemic fungicides. Although resistance to succinate dehydrogenase inhibitors and quinone outside inhibitors has been addressed before in net blotches, mechanisms controlling demethylation inhibitor resistance have not yet been reported at the molecular level. Here we report the isolation of strains of NFNB in Australia since 2013 resistant to a range of demethylase inhibitor fungicides. Cyp51A:KO103-A1, an allele with the mutation F489L, corresponding to the archetype F495I in Aspergillus fumigatus, was only present in resistant strains and was correlated with resistance factors to various demethylase inhibitors ranging from 1.1 for epoxiconazole to 31.7 for prochloraz. Structural in silico modeling of the sensitive and resistant CYP51A proteins docked with different demethylase inhibitor fungicides showed how the interaction of F489L within the heme cavity produced a localized constriction of the region adjacent to the docking site that is predicted to result in lower binding affinities. Resistant strains also displayed enhanced induced expression of the two Cyp51A paralogs and of Cyp51B genes. While Cyp51B was found to be constitutively expressed in the absence of fungicide, Cyp51A was only detected at extremely low levels. Under fungicide induction, expression of Cyp51B, Cyp51A2, and Cyp51A1 was shown to be 1.6-, 3,- and 5.3-fold higher, respectively in the resistant isolate compared to the wild type. These increased levels of expression were not supported by changes in the promoters of any of the three genes. The implications of these findings on demethylase inhibitor activity will require current net blotch management strategies to be reconsidered in order to avoid the development of further resistance and preserve the lifespan of fungicides in use.
Introduction
Pyrenophora teres f. sp. teres (Ptt) (P. teres Drechsler; anamorph Drechslera teres [Sacc.] Shoem.) is a necrotrophic fungal pathogen and the cause of net form of net blotch (NFNB) that, together with spot form of net blotch (SFNB), is one of the most important diseases of barley (Hordeum vulgare). Worldwide, net blotches are commonly responsible for barley yield losses of 10–40%, and in some cases losses of up to 100% can occur (Liu et al., 2011). NFNB is a major disease of most barley growing regions including Europe (Leišova et al., 2005; Serenius et al., 2007; Ficsor et al., 2014), North America (Tekauz, 1990; Steffenson et al., 1991), and North Africa (Boungab et al., 2012). In Australia, the value of disease control for NFNB is estimated at $98 million annually with average direct costs of $19 million annually, making it the most significant necrotrophic barley disease after SFNB (Murray and Brennan, 2010). Along with cultural practices, the main control measures are the application of effective fungicides and the use of cultivars with genetic resistance. However, due to the lack of highly resistant cultivars, net blotch diseases are mainly controlled using fungicides (Sierotzki et al., 2007). Fungicides of the quinone outside inhibitor (QoI) and the succinate dehydrogenase inhibitor (SDHI) classes are used in the control of Ptt, especially in Europe, where mutations in the respective target genes cytochrome b and the succinate dehydrogenase complex (SdhB, SdhC, and SdhD) associated with resistance in Ptt have now been widely reported (Semar et al., 2007; Sierotzki et al., 2007; Rehfus et al., 2016). In Australia, the fungicides used against Ptt are predominantly of the azole or demethylase inhibitor (DMI) group (APVMA, 2016).
The DMIs are a group of site-specific systemic fungicides and are the most important compounds used for the control of fungal pathogens in both medicine and agriculture (Becher and Wirsel, 2012; Ishii and Hollomon, 2015). This fungicide class is comprised of a large number of structurally diverse compounds, all having in common the presence of a N-substituted five or six-membered heterocyclic ring (Hof, 2001). The target site of all DMIs is the enzyme CYP51, a cytochrome P450 sterol 14α-demethylase essential to the biosynthesis of fungal sterols (López-Ruiz et al., 2010). Ergosterol is the most common sterol in fungi, being a vital component of the fungal cell membrane and essential for fungal growth (Köllerm, 2003).
Many cases of resistance to DMI fungicides have been documented in phytopathogenic fungi (Délye et al., 1997; Erickson and Wilcox, 1997; Fraaije et al., 2007; Ghosoph et al., 2007; Omrane et al., 2015). Acquired resistance to DMIs has also been widely reported in fungal pathogens of humans (Kanafani and Perfect, 2008; Morio et al., 2010; Howard and Arendrup, 2011; Becher and Wirsel, 2012). In filamentous fungi there are three principle known mechanisms of resistance; (1) target site modification, where point mutations in the Cyp51 gene result in amino acid substitutions altering the structure of the CYP51 protein, and thus reducing the binding affinity of the fungicide to the enzyme. Point mutations have been observed to cause varying levels of cross resistance to different DMIs (Cools and Fraaije, 2013); (2) overexpression of the target gene(s) Cyp51. Increased production of the CYP51 enzyme results in a general reduction of sensitivity across all the DMIs (Ishii and Hollomon, 2015); (3) increased efflux by overexpression of membrane-bound drug transporters, reducing the accumulation of fungicide at the target site. Enhanced efflux tends to result in a phenotype of broad-spectrum resistance across the DMIs and unrelated fungicide classes (Ishii and Hollomon, 2015).
Several species of Ascomycete fungi have been shown to carry multiple Cyp51 paralogs, including Aspergillus spp., Fusarium spp., Penicillium digitatum, and Rhynchosporium commune (Becher et al., 2011; Hawkins et al., 2014). Cyp51B is carried by all ascomycetes. Some species also carry a paralog termed Cyp51A (Becher et al., 2011). In species with three Cyp51 genes, the third Cyp51 is either a duplicated copy of Cyp51A (as in A. flavus and A. oryzae) or of Cyp51B (as in A. terreus) or a unique paralog termed Cyp51C (in Fusarium spp.) (Becher et al., 2011). In R. commune one of the Cyp51 paralogs is a pseudogenized duplication of Cyp51A termed Cyp51A-p (Hawkins et al., 2014).
In fungi with multiple Cyp51 paralogs, Cyp51-mediated azole resistance is often associated with mutations in or overexpression of the Cyp51A gene (Fan et al., 2013; Brunner et al., 2015). For example, in A. fumigatus mutations correlated with DMI resistance have been found only in the Cyp51A paralog and not in Cyp51B (Becher et al., 2011). Resistance to DMIs in A. fumigatus is also mediated by overexpression of the Cyp51A paralog, which in combination with mutations in Cyp51A results in cross-resistance (Mellado et al., 2007). Similarly, overexpression of the Cyp51A gene but not Cyp51B has been demonstrated as a mechanism for azole resistance in A. flavus, A. niger, A. parasiticus, and Magnaporthe oryzae (Yan et al., 2011; Fan et al., 2013). In P. digitatum, azole resistance has been shown to result from the overexpression of both Cyp51A and Cyp51B (Sun et al., 2013). In R. commune, azole resistance is associated with the presence of Cyp51A (Hawkins et al., 2014). An analysis of R. commune isolates showed no correlation between Cyp51B sequence and azole sensitivity, so the authors hypothesized that the selection pressure of azoles is driving the observed accumulation of polymorphisms in the sequence of Cyp51A (Brunner et al., 2015).
Resistance to the DMI fungicide triadimenol was first reported in P. teres isolates from New Zealand (Sheridan et al., 1985). Subsequent studies showed this phenotype emerged in both New Zealand and the United Kingdom from the early 1980s onwards (Sheridan et al., 1987). Crossing studies determined that in P. teres this resistance segregated to a single major genetic locus (Peever and Milgroom, 1992), and that resistance to other DMIs, including propiconazole, imazalil and fenarimol, was also genetically correlated (Peever and Milgroom, 1993). These studies did not distinguish the net and spot types of P. teres, although subsequent phylogenetic analysis has shown that the two formae speciales are genetically isolated and should be considered as distinct species (Rau et al., 2007). In Ptt specifically, an increase in tolerance to the DMI prochloraz has also been observed in isolates from Finland (Serenius and Manninen, 2008).
Here we report the discovery in P. teres f. sp. teres of resistance to multiple DMIs. Resistance was correlated with two genetic changes. Resistant isolates carried a novel mutation in a copy of the Cyp51A gene. Structural analysis of the CYP51A target revealed a strong correlation between this mutation and the resistance levels detected. These isolates also displayed overexpression of both copies of the Cyp51A gene as well as overexpression of the Cyp51B.
Materials and Methods
Fungal Isolates
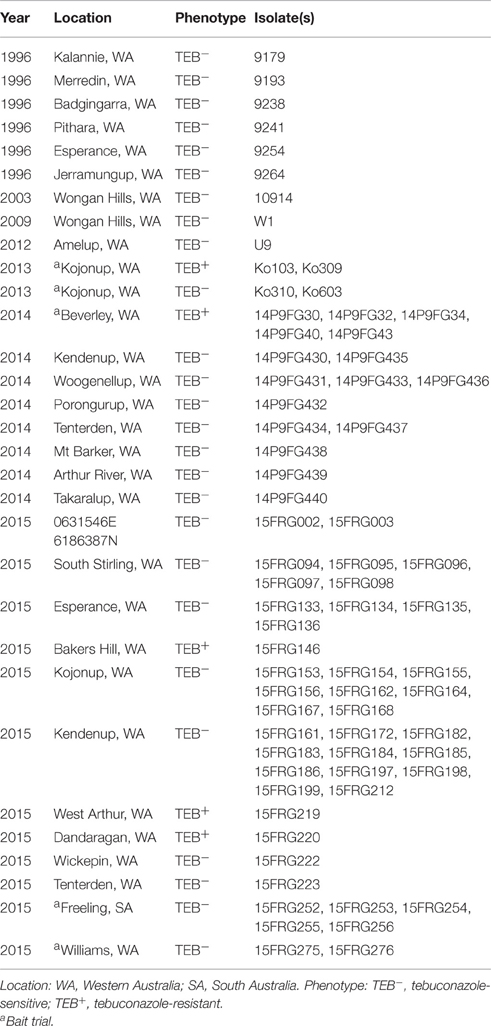
The Ptt isolates used in this study are listed in Table 1. Isolates from 1996 to 2003 were collected by the Department of Agriculture and Food, Western Australia (South Perth, Western Australia). Isolates from 2009 onwards were collected and isolated by the authors from a combination of especially designed bait trials (years 2013, 2014, and 2015) and sampling trips. Bait trials sown with the NFNB susceptible variety Hindmarsh were designed to work as a fungicide resistance early warning system. 2 × 4 m plots were sprayed with either 1 × or 2 × the maximum registered dose of the fungicides epoxiconazole, tebuconazole, prothioconazole, and azoxystrobin, at growth stages GS31 and GS39. Treatments were replicated three times. Leaf samples from bait trials were collected 7 days following to the second spray application.
Lesions were excised from dried leaf samples and surface sterilized for 30 s in 70% (v/v) ethanol, 60 s in 10% (v/v) NaOCl, and rinsed for 60 s in sterile deionized H2O. Surface-sterilized lesions were transferred to 1.5% (w/v) tap water agar plates amended with ampicillin (100 μg mL−1), streptomycin (100 μg mL−1), and neomycin (50 μg mL−1), and grown at room temperature for 7 days. Subsequent hyphal growth was sub-cultured to V8-potato-dextrose agar (10 g potato-dextrose agar, 3 g CaCO3, 15 g agar, 150 mL V8 juice in 850 mL sterile deionized H2O; V8PDA) plates and incubated at room temperature under white light for 5 days. To induce sporulation, sterile deionized H2O was applied to hyphae and flattened gently with a glass rod; the plate was then incubated at room temperature under near-UV for 24 h, followed by 15°C in darkness for 24 h. For each isolate, a single conidium was transferred to a new V8PDA plate. Mycelial plugs of this culture were stored at −80°C and these were used for all subsequent testing.
In vitro Fungicide Sensitivity Testing
Mycelial plugs of monoconidial Ptt isolates were inoculated on V8PDA plates and incubated at room temperature under white light for 5 days. Sporulation was induced as described above. Spores were harvested by flooding plates with 10 mL sterile deionized H2O and scraping the mycelia with glass rod. The suspension was then filtered through two layers of sterile cheesecloth and centrifuged at 3220 g for 10 min at 4°C. The pellet was resuspended in 1 mL sterile deionized H2O and spore density estimated using a haemocytometer. The suspension was adjusted to produce a stock with a spore concentration of 1 × 104 mL−1.
Pure technical grade fungicide compounds were dissolved in absolute ethanol, and serial dilutions were prepared so that the total volume of fungicide/solvent added to each well was kept constant. A master mix was prepared consisting of 0.5% (v/v) fungicide stock, or an equivalent volume of ethanol (for the zero fungicide control), in Yeast Bacto Acetate liquid medium (10 g yeast extract, 10 g Bacto peptone, 10 g sodium acetate in 1 L sterile deionized H2O; YBA). A total volume of 95 μL was added to each well of a 96-well microtiter plate. The range of concentrations used for each fungicide was adapted to the response of each isolate to the fungicide tested in order to obtain a dose-response curve. In order to exclude the activity of alternative oxidase, salicylhydroxamic acid was included in the medium at a final concentration 50 μM for tests involving azoxystrobin.
A 5 μL volume of spore stock was inoculated to 95 μL of media with fungicide to a final concentration of 500 spores mL−1; at least two biological replicates with up to three technical replicates were inoculated for each isolate. Immediately following inoculation, optical density (OD) was measured at 405 nm wavelength in a Synergy HT microplate reader (BioTek). Plates were incubated for 96 h at room temperature in darkness, after which OD was again measured. Final OD values were normalized with readings taken immediately following inoculation. The concentration of fungicide resulting in 50% reduction in growth (EC50) was calculated in Microsoft Excel by linear regression of log10-transformed percentage reduction in OD compared to zero fungicide control, against log10-transformed concentration of fungicide. Estimate of EC50 was made with the linear portion of the dose response, using the formula , from the regression y = m.log(concentration)+b. The resistance factor (RF) of each resistant isolate was calculated as a ratio of the EC50 to the mean EC50 of all sensitive isolates collected in the years 1996 to 2012. All EC50 values were log10-transformed prior to statistical analysis (Liang et al., 2015), statistical analysis was performed with IBM SPSS (Statistical Product and Service Solutions, version 24.0, Armonk, NY, USA). EC50 values of populations were compared using Student's t-test with the significance threshold set at 5%; correlations between EC50 values were determined with Spearman's rank order correlation with the significance threshold set at 1%.
Discriminatory Dose Screening
A 4 mm-diameter mycelial plug taken from the colony edge of a 5 day culture grown on V8PDA was inoculated to the center of a YBA agar plate amended with tebuconazole to a final concentration 10 μg mL−1, or equivalent volume of ethanol solvent for the zero fungicide control. Two biological replicates were inoculated for each isolate and plates were incubated for 96 h at room temperature in darkness, after which time the presence or absence of growth on the plate was visually assessed. Isolates able to grow at 10 μg mL−1 were considered to be resistant.
Cloning and Sequencing of Cyp51A and Cyp51B Genes
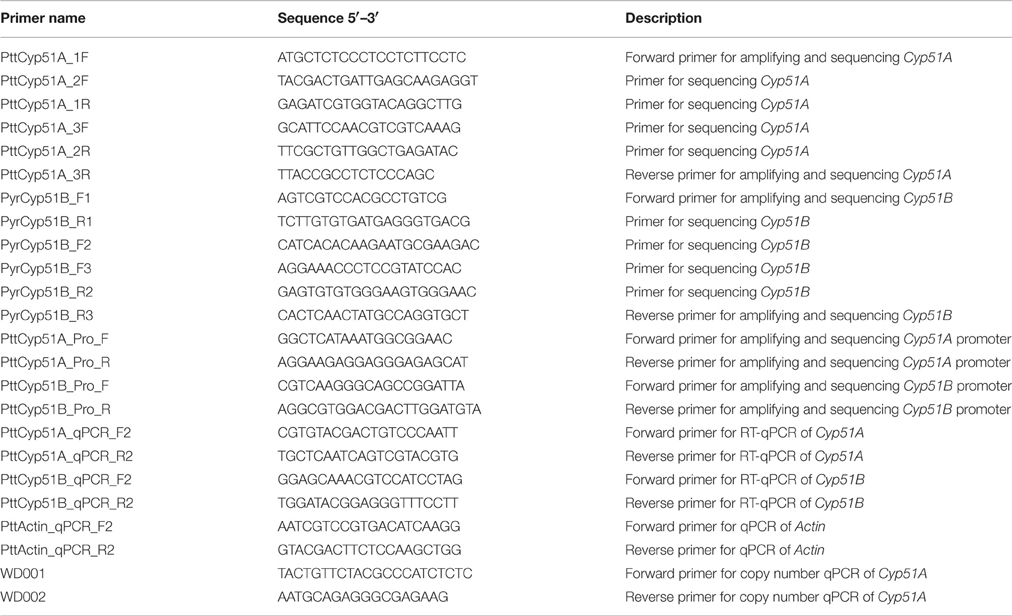
Mycelia of Ptt was snap-frozen in liquid nitrogen and ground with tungsten beads in a Retsch MM301 Mixer-Mill (Retsch GmbH). Genomic DNA was isolated using a Biosprint 15 instrument and Biosprint DNA Plant Kit (QIAGEN) as per manufacturer's instructions. For sequencing of Cyp51A promoter, Cyp51B coding sequence and Cyp51B promoter, PCR amplification was carried out in 50 μL reaction volume containing 0.25 μL MyTaq DNA Polymerase (5 U μL−1, Bioline), 10.0 μL 5 × reaction buffer, 2.5 μL each forward and reverse primer (5 μM; Table 2), 2.5 μL DNA template (100 ng μL−1), and 32.25 μL H2O. PCR parameters were initial denaturation at 94°C for 2 min, followed by 35 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 2 min, followed by a final extension at 72°C for 5 min. PCR products were purified with a GenElute PCR Clean-Up Kit (Sigma). Purified PCR products were sequenced by Macrogen (Seoul, South Korea). Sequences were assembled and analyzed using Geneious 6 software (Biomatters), alignments performed using ClustalW algorithm (Thompson et al., 1994) with IUB scoring matrix, gap opening penalty 15, gap extension penalty 6.66.
For cloning and sequencing of Cyp51A gene, PCR amplification was carried out in a 50 μL reaction volume containing 0.5 μL Phusion High Fidelity DNA Polymerase (2 U μL−1, Thermo Scientific), 10 μL 5 × reaction buffer, 1.5 μL DMSO, 5 μL dNTPs (2 mM), 2.5 μL each forward and reverse primer (10 μM; Table 2), 1 μL genomic DNA template (100 ng μL−1), 27 μL H2O. PCR parameters were initial denaturation at 98°C for 30 s, followed by 35 cycles of 98°C for 10 s, 69°C for 30 s, and 72°C for 1 min, followed by a final extension at 72°C for 10 min. PCR products were purified as described above. Purified PCR products were A-tailed with DyNAzyme II DNA Polymerase (2 U μL−1, Thermo Scientific), ligated into pGEM-T Easy Vector (Promega) as per manufacturer's instructions and transformed to XL10 Ultracompetent cells (Agilent). Plasmid DNA was prepared with a GenElute Plasmid Miniprep Kit (Sigma) and cloned inserts sequenced by Macrogen (Seoul, South Korea). Sequences were assembled and analyzed using Geneious 6 software (Biomatters), alignments performed using ClustalW algorithm (Thompson et al., 1994) with Blosum scoring matrix, gap opening penalty 10, gap extension penalty 0.5, free end gaps. Translated amino acid sequences were aligned to the archetypal CYP51A (Mair et al., 2016) amino acid sequence from A. fumigatus (GenBank accession no. AAK73659), and the CYP51B sequence from P. digitatum (GenBank accession no. ADO85402). Amino acid substitutions associated with resistance are described with reference to the archetype “mutation label” (italicized) throughout text as previously described (Mair et al., 2016). Sequences have been deposited to GenBank (accession numbers KX578217-KX578221).
Growth of Fungal Cultures and Nucleic Acid Extraction for Quantitative PCR Analysis and Sequencing
Isolates Ko103 and 9193 were inoculated in 60 mL Fries liquid medium Number 2 (Fries, 1938) to a concentration of 500 spores mL−1 and grown at room temperature, 150 rpm in darkness with three biological replicates per isolate of both treatment and control cultures. Growth curve analysis of Ko103 and 9193 showed they had similar rates of growth (data not shown). At 64 h post-inoculation, when cultures were in the exponential phase of growth, tebuconazole in ethanol solution was added to the media to a final concentration equivalent to the EC50 of each respective isolate (EC50 = 3.9 μg mL−1 for Ko103; 0.26 μg mL−1 for 9193) as described previously (Cools et al., 2012). For the control cultures an equivalent volume of ethanol solvent was added with no tebuconazole. At 112 h post-inoculation, cultures were harvested and freeze-dried. RNA was extracted with Trizol reagent (Invitrogen), treated with RNase-Free DNase (QIAGEN), and purified with RNeasy Plant Mini Kit (QIAGEN). cDNA synthesis was performed with iScript cDNA Synthesis Kit (BioRad). Genomic DNA was extracted using the CTAB method (Saghai-Maroof et al., 1984). Concentration of nucleic acid was quantitated using a Quantus Fluorometer (Promega).
Quantitative PCR Analysis of Gene Expression and Gene Copy Number
Quantitative RT-PCR (qPCR) analysis of the Cyp51A and Cyp51B genes was conducted with QuantiTech SYBR Green Mastermix (QIAGEN) on the BioRad CFX96 qPCR system using Actin as the endogenous control (NCBI accession no. XM_003298028) and primers listed in Table 2. All reactions were carried out with three biological replicates and three technical replicates each. Relative transcript abundances were calculated using the 2−ΔCT method (Livak and Schmittgen, 2001). Statistical analysis was performed with IBM SPSS, relative expression ratios were compared using Mann–Whitney U-test with the significance threshold set at 5%. Gene copy number estimation by qPCR was performed as previously described (Solomon et al., 2008), using four biological replicates of genomic DNA and with Actin as the single copy control.
Genome and RNA Sequencing
All RNA sequencing and cDNA library construction was conducted by Novogene Bioinformatics Technology (Beijing, China). RNA was sequenced using Illumina HiSeq platform. Differential expression analysis was performed with edgeR (Robinson et al., 2010). Statistical analysis was performed with IBM SPSS, normalized read counts were compared using Student's t-test with the significance threshold set at 5%. Whole genome sequencing and library construction was performed by the Australian Genome Research Facility (Nedlands, Western Australia) using Illumina MiSeq platform. Raw Illumina reads were analyzed using FastQC (version 0.10.1; Andrews, 2010) for quality control. The filtered sequencing reads were assembled using SPAdes assembler (version 3.6.2; Nurk et al., 2013). Coverage cut-off was disabled and the number of mismatches and short indels was reduced by incurring SPAdes' build-in post processing module MismatchCorrector, which utilizes BWA tool (Li and Durbin, 2009). The obtained assembly was scaffolded using SSPACE (version 3.0; Boetzer et al., 2011). Annotation of the scaffolded assemblies was achieved by using Prokka tool (version 1.11; Seemann, 2014).
In silico Structural Modeling
Structural modeling of CYP51A sensitive allele 9193-A1 and KO103-A1 allele carrying the F489L mutation was undertaken using an automated homology modeling platform as previously described for Mycosphaerella graminicola CYP51 and mutants (Mullins et al., 2011). Ligand docking of azoles was also carried out as previously described, and with AutoDock Vina (Trott and Olson, 2010), cross-referenced and corroborated using the heme-azole positions of the CYP51-azole co-crystallized structures of PDB:5EAB (Saccharomyces cerevisiae CYP51 complexed with S-tebuconazole) and PDB:5EAF (Saccharomyces cerevisiae CYP51 complexed with fluquinconazole).
Results
Identification of DMI Fungicide Resistance in Ptt
The fungicide sensitivities of monoconidial Ptt strains collected from 1996 to 2013 were determined in vitro via microtiter assay in order to establish EC50 levels to several DMIs as well as to fungicides from different mode of action groups. Isolates were screened initially with tebuconazole, epoxiconazole, and prothioconazole (Table 3), and a subset of these isolates tested against QoI fungicide azoxystrobin and the SDHI fungicide boscalid (Table S1). Two isolates, Ko103 and Ko309, were identified with reduced sensitivity to tebuconazole (EC50 of 3.9 and 2.4 μg mL−1, RF of 16.4 and 10.1, respectively) compared to the isolates collected during 1996–2012 (EC50 range 0.06–0.34 μg mL−1, mean EC50 = 0.24 μg mL−1). These two isolates also displayed a level of reduced sensitivity to other triazole fungicides, including epoxiconazole (EC50 of 0.23 and 0.16 μg mL−1, RF of 2.0 and 1.4, respectively) and prothioconazole (EC50 of 0.18 and 0.12 μg mL−1, RF of 2.7 and 1.6, respectively). By comparison, the EC50 values of the isolates collected during 1996–2012 were for epoxiconazole 0.02–0.23 μg mL−1 (mean EC50 = 0.11 μg mL−1) and for prothioconazole 0.03–0.21 μg mL−1 (mean EC50 = 0.07 μg mL−1). The analysis showed no significant differences in sensitivities of the DMI-resistant isolates to the non-DMI fungicides azoxystrobin [t(10) = −0.62, p = 0.548] and boscalid [t(9) = −0.65, p = 0.530], compared to the wild-type.
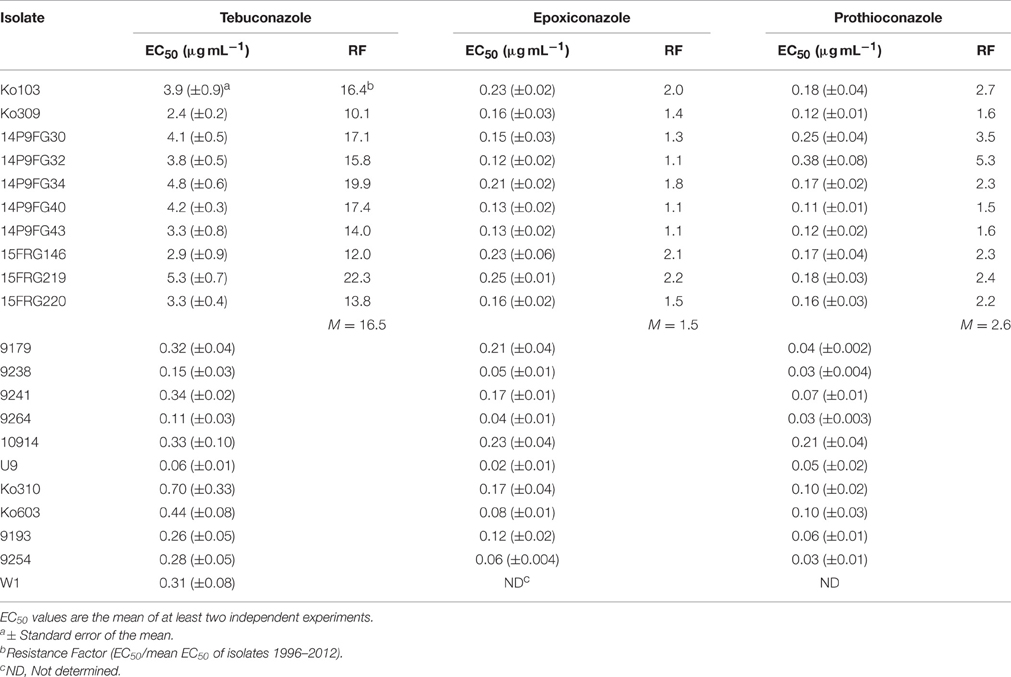
Table 3. EC50 and RF values of Pyrenophora teres f. sp. teres isolates to tebuconazole, epoxiconazole, and prothioconazole.
The EC50 results from microtiter assays were used to define a cut-off concentration in order to distinguish the two tebuconazole sensitivity groups identified in the above screening. A total of 58 isolates collected in 2014 and 2015 were pre-screened on a discriminatory dose of 10 μg mL−1 tebuconazole (Table 1). Pre-screening identified a further eight isolates showing a reduced sensitivity to tebuconazole (Table 1). These eight new isolates were selected for more detailed analysis by EC50 screening on the previously tested DMI fungicides tebuconazole, epoxiconazole, and prothioconazole (Table 3). These isolates identified in pre-screening showed similar patterns of sensitivity to the original two resistant isolates Ko103 and Ko309, with RFs to tebuconazole of 12–19.9 (mean RF 16.5), to epoxiconazole 1.1–2.2 (mean RF 1.5), and to prothioconazole 1.5–5.3 (mean RF 2.6). The reduced sensitivity of the 10 isolates to DMIs was shown to be significant for tebuconazole [t(12.56) = −12.32, p < 0.001], prothioconazole [t(18) = −4.57, p < 0.001], and epoxiconazole [t(10.99) = −2.41, p = 0.035], compared to the wild-types. There were significant positive correlations between the EC50 values of tebuconazole-prothioconazole [rS(18) = 0.777, p < 0.001], tebuconazole-epoxiconazole [rS(18) = 0.570, p = 0.009], and prothioconazole-epoxiconazole [rS(18) = 0.593, p = 0.006].
These 10 tebuconazole-resistant isolates, and two tebuconazole-sensitive isolates, 9193 and 9254, were also tested for their sensitivity to two additional DMI compounds, propiconazole and metconazole (Table 4). The 10 tebuconazole-resistant isolates also showed reduced sensitivity to these DMIs compared to the two tebuconazole-sensitive isolates, with RFs to propiconazole of 5.7–9.7 (mean RF 7.7), and to metconazole 8.9–25.3 (mean RF 13.8). The two tebuconazole-sensitive isolates, 9193 and 9254, and six tebuconazole-resistant isolates (Ko103, Ko309, 14P9FG40, 14P9FG43, 15FRG146, and 15FRG220) were also tested for their sensitivity to a further four additional DMI compounds: triadimenol, triticonazole, difenoconazole, and prochloraz (Table 5). The six tebuconazole-resistant isolates also showed reduced sensitivity to these DMIs compared to the two tebuconazole-sensitive isolates, with RFs to triadimenol of 3.1–4.2 (mean RF 3.4), to triticonazole 10.8–19.1 (mean RF 15.6), to difenoconazole 10.2–15.2 (mean RF 12.4), and to prochloraz 17.8–31.7 (mean RF 27.7).
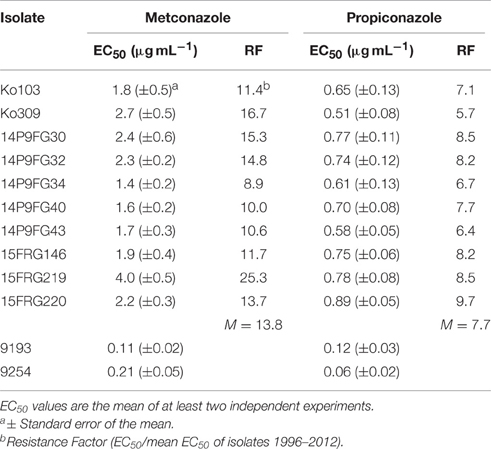
Table 4. EC50 and RF values of Pyrenophora teres f. sp. teres isolates to metconazole and propiconazole.
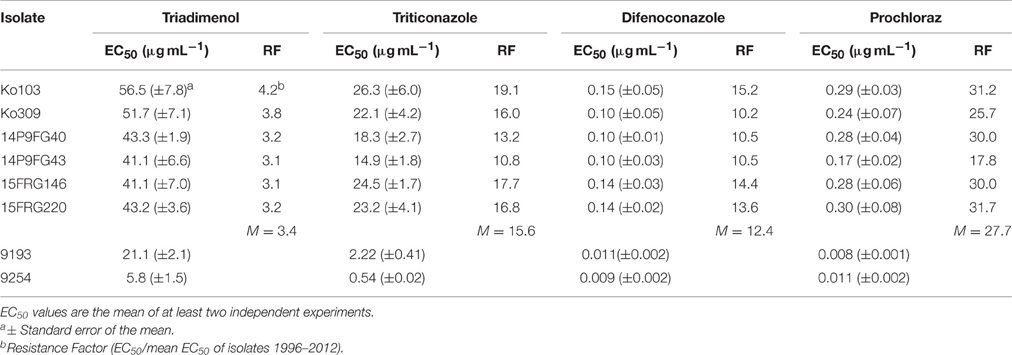
Table 5. EC50 and RF values of Pyrenophora teres f. sp. teres isolates to triadimenol, triticonazole, difenoconazole, and prochloraz.
DMI Resistance in Ptt Is Correlated with Mutation F489L in One of the Cyp51A Alleles
Sequencing of the 1.68 kb Cyp51B gene in all the tebuconazole-resistant isolates and in seven tebuconazole-sensitive isolates (W1, 9193, 9254, 9179, 9238, 9264, and 10914) showed the existence of a single allele of Cyp51B in all cases. There were no changes observed when the sequences were aligned to the Cyp51B reference of Ptt isolate 0–1 deposited in GenBank (accession no. NW_003523339).
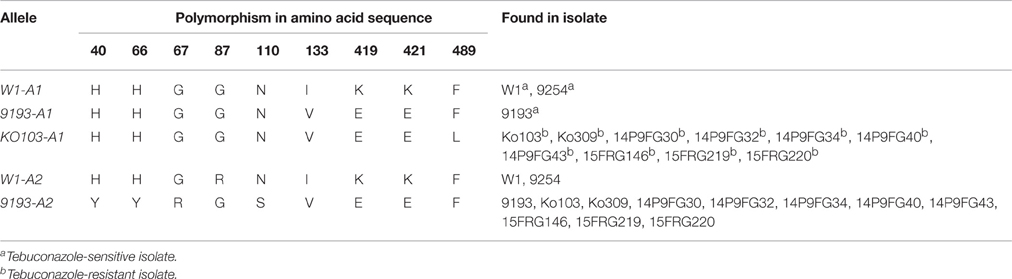
Genome sequencing of the isolates Ko103 and 9193 showed they both carried two copies of the Cyp51A gene (data not shown). This observation was later confirmed by qPCR (Figure S2). Subsequently, both copies of the Cyp51A gene were cloned and sequenced in 10 tebuconazole-resistant isolates and in three tebuconazole-sensitive isolates (W1, 9193, 9254), revealing the existence of five alleles of Cyp51A, assorted into three genotypes (Table 6). Sensitive isolates W1 and 9254 both carry the two alleles termed W1-A1 and W1-A2, which differ by only one nucleotide at base 259, resulting in a substitution of arginine for glycine at position 87 of the translated amino acid sequence. The sensitive isolate 9193 carries the alleles termed 9193-A1 and 9193-A2, with both of these alleles differing from the W1-A1 and W1-A2 alleles at four nucleotides including three non-synonymous changes, resulting in the point mutations V133I, K419E, and K421E in the amino acid sequences. The two 9193 alleles differ between each other by seven nucleotides including four non-synonymous changes, resulting in the point mutations H40Y, H66Y, G67R, and N110S in the amino acid sequence. The amino acid sequence of allele 9193-A1 is identical to that of the reference sequence of isolate 0–1 from GenBank (accession no. XP_003303644; Figure 1). All 10 resistant isolates also carry the 9193-A2 allele as well as a unique allele labeled KO103-A1. The KO103-A1 allele is identical to the 9193-A1 allele except for a C to A transversion at base 1467 resulting in a substitution of leucine for phenylalanine at position 489 of the amino acid sequence. Thus, the F489L amino acid substitution represents the only polymorphism of Cyp51A segregating the resistant from the sensitive isolates. An alignment of the amino acid sequences of the CYP51A variants to the CYP51A amino acid sequence from A. fumigatus (Figure 1) revealed the F489L substitution to be orthologous to the archetype F495I mutation associated with DMI resistance in A. fumigatus and is therefore given the mutation label F495L. The F495L amino acid substitution is also homologous to the DMI resistance mutation F506I in P. digitatum CYP51B (Figure 1).
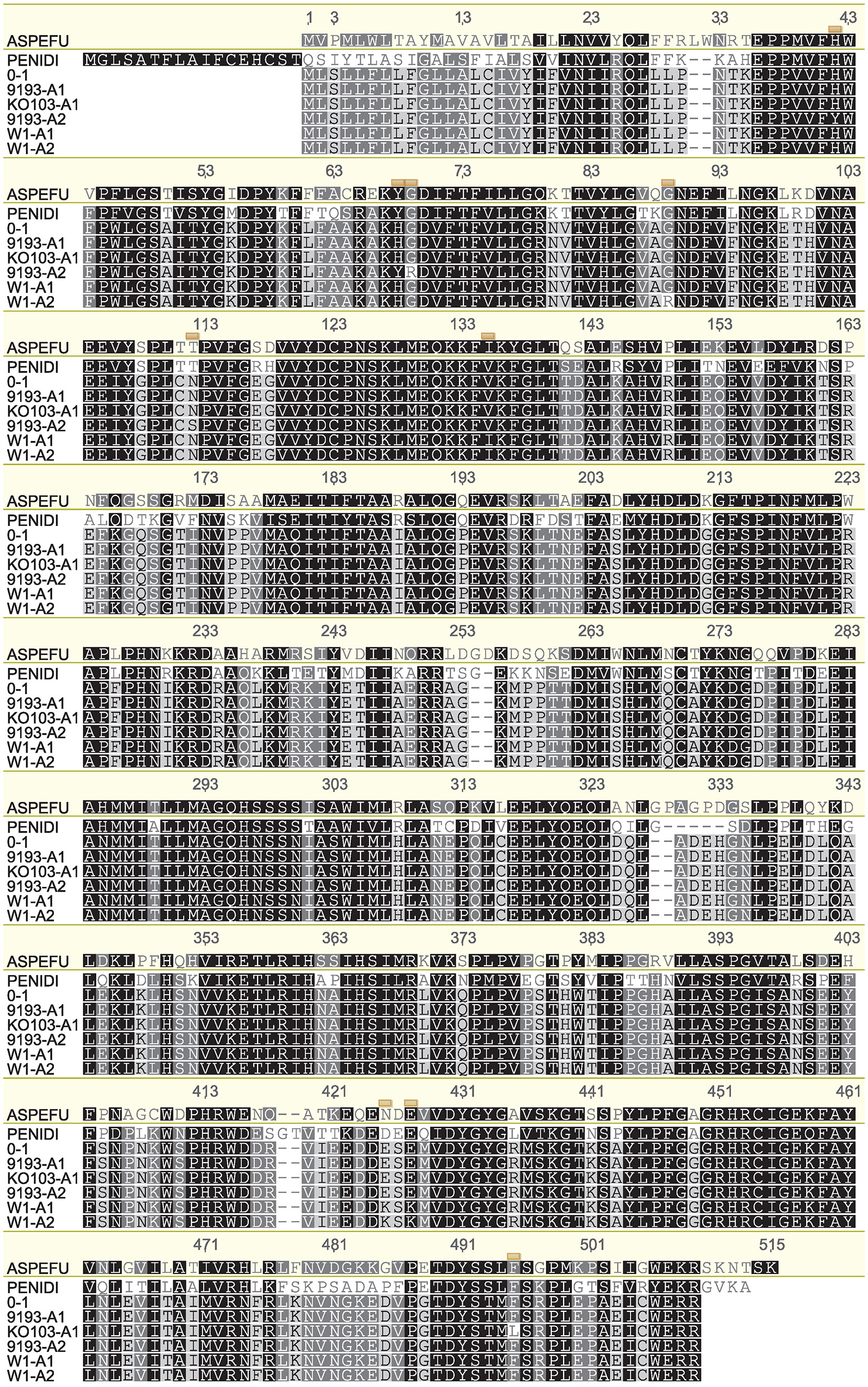
Figure 1. Alignment of amino acid sequences corresponding to the different Cyp51A alleles. Translated amino acid sequences of the five P. teres f. sp. teres Cyp51A alleles detected in this study (KO103-A1, 9193-A1, 9193-A2, W1-A1, W1-A2), aligned with CYP51A from the reference isolate 0–1 (GenBank accession no. XP_003303644), Aspergillus fumigatus CYP51A (ASPEFU, GenBank accession no. AAK73659), and Penicillium digitatum CYP51B (PENIDI, GenBank accession no. ADO85402). Alignment is numbered according to A. fumigatus CYP51A. The positions of polymorphisms in the P. teres f. sp. teres CYP51A sequences are indicated by yellow boxes. The amino acid sequence of 9193-A1 is identical to the 0–1 reference sequence. KO103-A1 differs from 9193-A1 by the amino acid substitution F489L, orthologous to F495I in A. fumigatus and F506I in P. digitatum. Alignment generated in Geneious version 6.1 software (Biomatters) using ClustalW algorithm with Blosum scoring matrix, gap opening penalty 10, gap extension penalty 0.5, free end gaps.
The F495L Mutation in Cyp51A Results in Reduced Binding Affinity to Azole Fungicides
Structural modeling was performed on the CYP51A variants 9193-A1 and KO103-A1 docked with the DMIs difenoconazole, prochloraz and tebuconazole (Table 7, Figure 2). The KO103-A1 allele with the F495L substitution had a lower predicted binding affinity for all three DMIs tested when compared to the 9193-A1 allele carrying F495. There were consistently more residues in close proximity to the docked azole compounds in KO103-A1 than in 9193-A1, suggesting a general diminishing of the binding cavity, but particularly markedly around the M288-H292 helical region. Residues A289 and H292 are consistently implicated to be in close proximity to the bound azole compounds in both 9193-A1 and KO103-A1, but in KO103-1 the close proximity is extended to M288 and G290.
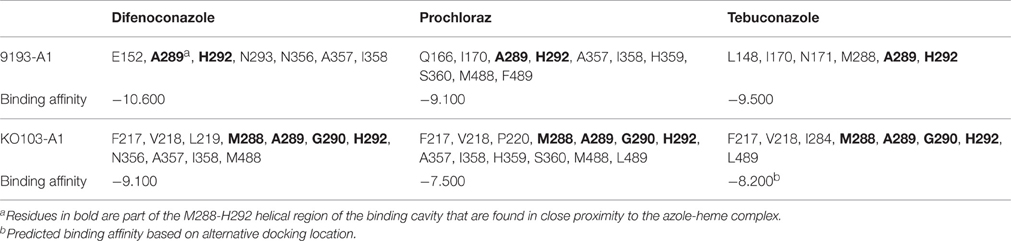
Table 7. Residues within 3 Å of the docked azoles and predicted binding affinities (kcal mol−1) of difenoconazole, prochloraz, and tebuconazole docked in 9193-A1 and KO103-A1.
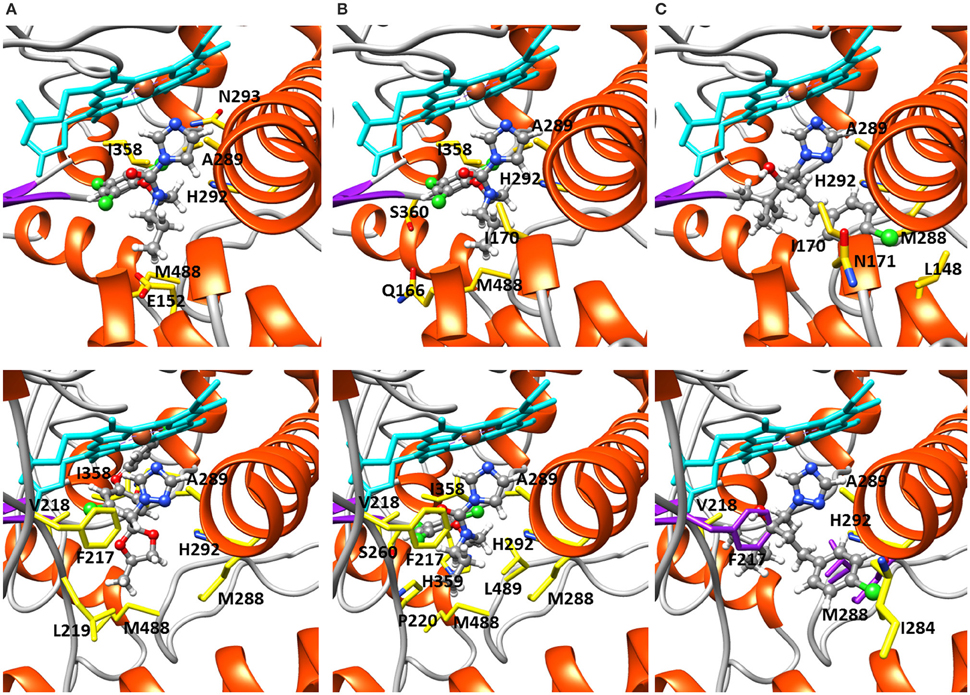
Figure 2. Structural modeling of 9193-A1 and KO103-A1 docked with difenoconazole, prochloraz, and tebuconazole. 9193-A1 (top row), KO103-A1 (bottom row). Fungicides in ball and stick, heme groups in cyan, showing the location of each azole and interacting residues shown in yellow and labeled. (A) Difenoconazole and (B) prochloraz occupy the binding cavities of 9193-1 and KO103-1 unhindered. (C) Tebuconazole binding is unhindered in 9193-1, but blocked by F217, M288, and L489 (in purple) in the KO103-1 mutant, as indicated by the spatial conflict. F217 is located away from the binding cavity in 9193-1.
The changes associated with CYP51A 9193-A1 do not prevent heme iron-coordinated binding of difenoconazole, prochloraz, or tebuconazole. In KO103-A1, however, there is substantial conformational change compared with 9193-A1, the foremost being movement of the long I helix at the amino terminal end around M288, leading to a constriction of the binding cavity; and the insertion of a section of beta turn, including F217 and V218, into the border of the heme cavity. In that regard, it is interesting to note that tebuconazole was predicted to dock adjacent to M288 in 9193-A1. The site of the primary mutation, F489, is itself within a 3 Å range of prochloraz in 9193-A1, and L489 (F495L) is again adjacent to prochloraz in KO103-A1 and is one of the three amino acids, along with F217 and M288 that prevents normal docking of tebuconazole.
DMI Resistance in Ptt Is Associated with Overexpression of the Cyp51A and Cyp51B Genes
To investigate the role of Cyp51A and Cyp51B expression on resistance, qPCR analysis and RNA sequencing of sensitive and resistant isolates induced with their respective EC50 tebuconazole concentrations were undertaken. Both experiments confirmed constitutive expression of the Cyp51B gene in the ethanol control samples (Figures 3A, 4A). By contrast, Cyp51A1/A2 alleles were not detectable (RNA sequencing) or detected at extremely low levels (qPCR, no distinction between alleles A1 and A2) in the control samples of either isolate (Figures 3B, 4B). Conditions that imposed a similar growth constraint on both isolates were used for studying the expression of Cyp51A and Cyp51B under fungicide induction (Cools et al., 2012). qPCR analysis comparing the tebuconazole-sensitive isolate 9193 and the resistant isolate Ko103 grown at their respective EC50 revealed the expression of Cyp51B to be 1.6-fold higher in the resistant compared to the sensitive isolate, although this difference was not considered significant (Mann–Whitney U = 33, p = 0.546; Figure 3A). The expression of Cyp51A was significantly higher in Ko103 than in 9193 (Mann–Whitney U = 0, p < 0.001), with a 5.2-fold increase in the resistant compared to the sensitive isolate (Figure 3B). These observations were validated by read density analysis of RNA sequencing data of the two isolates showing the expression of Cyp51B to be 1.6-fold higher in the resistant compared to the sensitive isolate at EC50 tebuconazole, which was considered significant {t(4) = 3.44, p = 0.026, 95% CI [794, 7436]; Figure 4A}. The expression of the Cyp51A genes were on average 3.6-fold higher in the resistant compared to the sensitive isolate when grown at their respective tebuconazole EC50; RNA sequencing analysis also confirmed that both copies of Cyp51A were expressed in both isolates and further dissected the relative expression of the two Cyp51A copies (Figure 4B). The expression levels of the Cyp51A1 and Cyp51A2 alleles were shown to be 5.3-fold higher {t(4) = 8.46, p = 0.001, 95% CI [275, 545]} and three-fold higher {t(4) = 6.78, p = 0.002, 95% CI [299, 714]} in Ko103 than in 9193, respectively. The Cyp51A2 gene was revealed to be expressed at a significantly higher level than the Cyp51A1 gene in both isolate 9193 {2.6-fold higher, t(4) = −6.84, p = 0.002, 95% CI [−219, −92.4]} and in Ko103 {1.5-fold higher, t(4) = −2.92, p = 0.043, 95% CI [−491, −12.7]}. The results support the conclusion that Cyp51A expression appears to be induced by the presence of tebuconazole, especially in the resistant isolate.
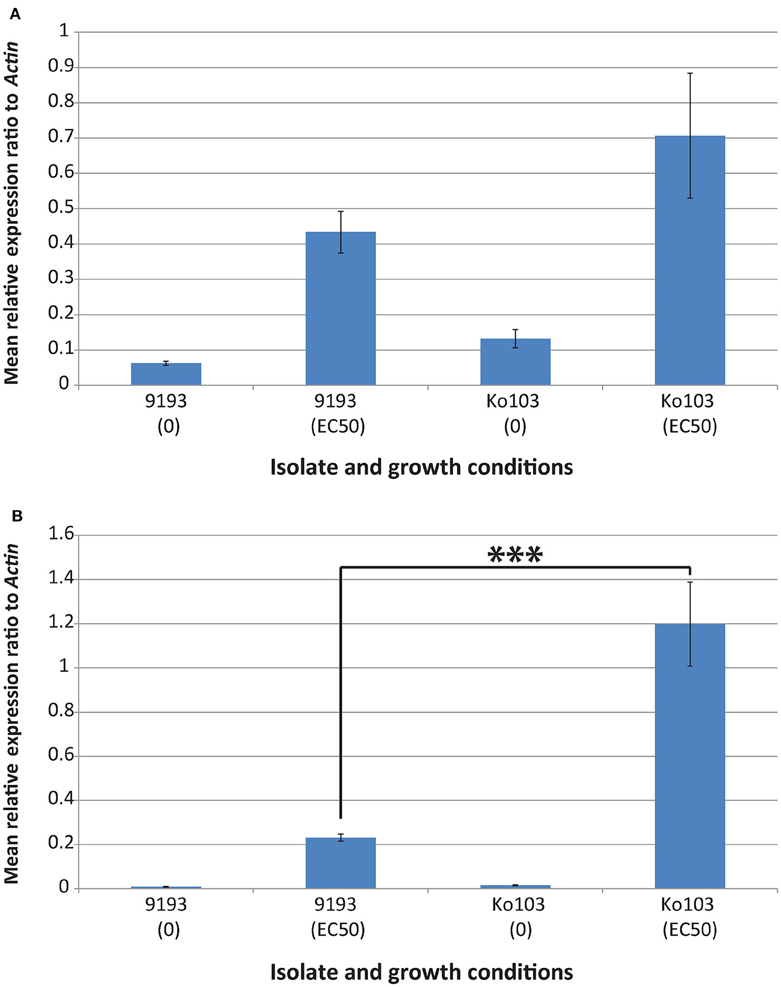
Figure 3. Effect of tebuconazole on Cyp51A and Cyp51B gene expression in isolates 9193 and Ko103. (A) qPCR confirmed constitutive expression of the Cyp51B gene in the ethanol control samples of both isolates. When grown under conditions that imposed a similar constraint on both isolates (EC50 tebuconazole), the expression of Cyp51B was found to be 1.6-fold higher in the resistant compared to the sensitive isolate. (B) qPCR showed Cyp51A (no distinction between alleles A1 and A2) to be expressed at extremely low levels in the control samples of either isolate. When grown under conditions that imposed a similar constraint on both isolates (EC50 tebuconazole), the expression of Cyp51A was 5.2-fold higher in the resistant compared to the sensitive isolate. Cyp51A expression appears to be induced by the presence of tebuconazole. Mean relative expression of genes of interest calculated by 2−ΔCT, normalized to Actin as the endogenous control (±Standard error of the mean, n = 3 biological replicates, three technical replicates per biological replicate). ***p < 0.001 (Mann–Whitney U-test).
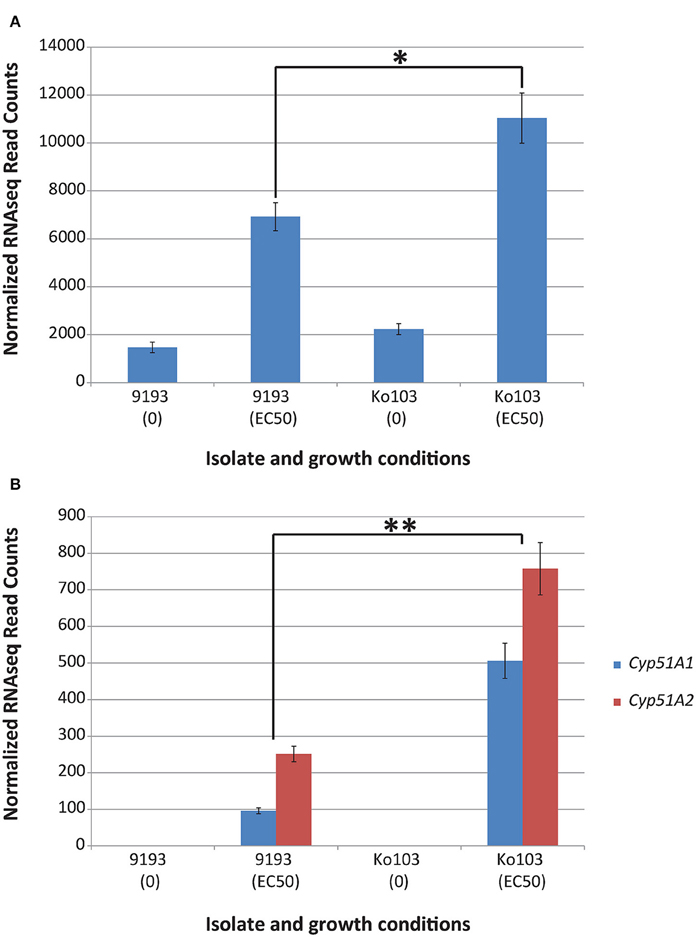
Figure 4. RNAseq read counts of Cyp51A1, Cyp51A2, and Cyp51B in isolates 9193 and Ko103 under tebuconazole treatment. (A) Mean RNAseq read counts of Cyp51B. Analysis of isolates Ko103 and 9193 showed the expression of Cyp51B to be 1.6-fold higher in the resistant compared to the sensitive isolate when grown under their corresponding EC50 tebuconazole. (B) Mean RNAseq read counts of Cyp51A1 and Cyp51A2. Expression of Cyp51A1 and Cyp51A2 was not detectable (zero reads) in the control samples of either isolate. Read density analysis of RNA sequencing data of the two isolates confirmed that both copies of Cyp51A were expressed in both isolates. The expression levels of the Cyp51A1 and Cyp51A2 alleles in Ko103 and 9193 cultures amended with their corresponding EC50 tebuconazole, were shown to be 5.3- and 3-fold higher in Ko103 than in 9193, respectively. Differential expression analysis and read count normalization achieved by edgeR (±Standard error of the mean, n = 3 biological replicates). *p < 0.05; **p < 0.01 (Student's t-test).
Cyp51 Gene Overexpression in Ptt Is Not Correlated with Changes in the Nucleotide Sequences of the Cyp51A or Cyp51B Promoters
Initially primers were used to amplify and sequence a 571 bp region upstream of the start codon of the Cyp51B gene in isolates 9193 and Ko103, revealing no changes at the nucleotide level between the two isolates. Subsequent genome sequencing of the two isolates allowed the investigation to be further extended to 1000 bp upstream of the start codon of the Cyp51B gene in isolates 9193 and Ko103, and also revealed no changes at the nucleotide level between the two isolates. The sequence data generated allowed the analysis of a region extending over 800 bp upstream of the start codons of both of the Cyp51A1 and A2 alleles, revealing that the sequence of the Cyp51A1 and A2 putative promoters are identical between isolates 9193 and Ko103 (Figure S3). However, comparing the promoter sequence of the Cyp51A1 and Cyp51A2 paralogs revealed several differences between them, including an 8 bp insert located 471 bp upstream of the start codon in the Cyp51A2 promoter.
Discussion
In Australia, emergence of multi-DMI resistance in Ptt has been observed in isolates collected from the years 2013 onwards. Resistant isolates were geographically widely dispersed across Western Australia, originating in Kojonup, Beverley, Bakers Hill, West Arthur, and Dandaragan (Figure 5). We used bait trials in these studies so as to increase the likelihood of finding resistant isolates even when their frequency is still low. We therefore cannot reliably estimate the frequency of the resistant isolates, but it is clearly at a significant level in Western Australian populations of Ptt (Table 1).
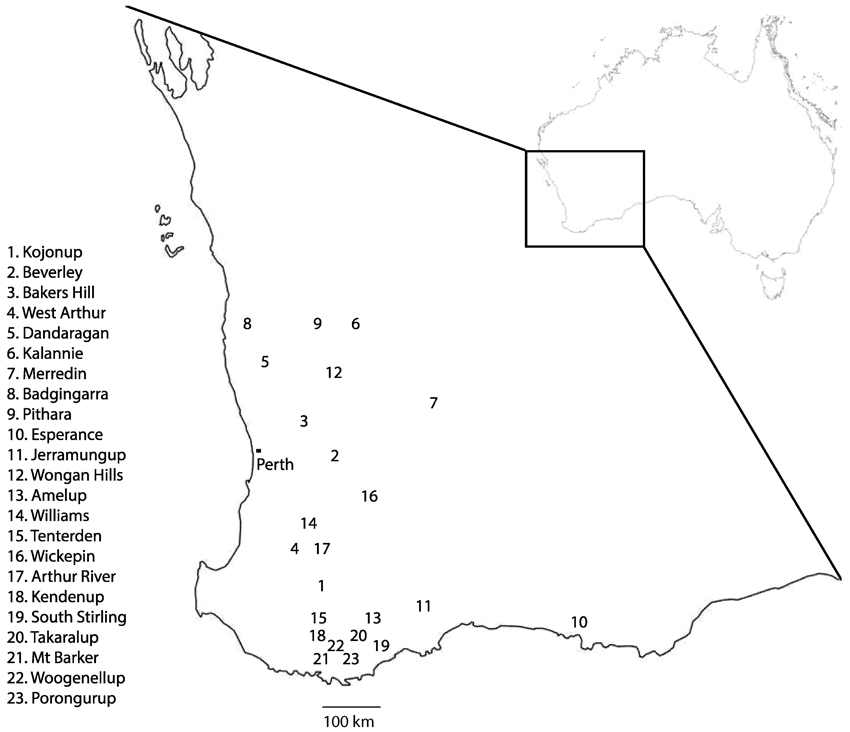
Figure 5. Map of Western Australia showing geographic origin of Pyrenophora teres f. sp. teres isolates. Leaf samples were collected from 23 separate locations in Western Australia from a combination of field trips and bait trials. Locations 1, 2, and 14 were specially designed bait trials. Resistant strains of P. teres f. sp. teres were isolated from samples taken in locations 1–5. Adapted from “Outline map of Australia” (http://www.ga.gov.au/metadata-gateway/metadata/record/gcat_61754) by Commonwealth of Australia (Geoscience Australia), used under CC BY 4.0.
Resistant isolates displayed high resistance factors (mean RF > 10) to tebuconazole, metconazole, triticonazole, difenoconazole, and prochloraz, and lower RFs (mean RF < 10) to epoxiconazole, prothioconazole, propiconazole, and triadimenol (Table 3). Isolates of this phenotype showed no observable reduction in sensitivity to QoI or SDHI fungicides. Although Ptt is a sexually recombining organism its reproduction occurs mostly asexually (Lehmensiek et al., 2010); therefore analysis of simple-sequence repeat markers conducted shortly after the emergence of a mutation should reveal whether the mutants are clonally related and derive from a single event, or whether this resistance has emerged multiple times independently as a response to the common selection pressure of azole fungicide use.
Two potential mechanisms of resistance have been observed in our data and may both contribute to the resistant phenotype. The observation that all DMI-resistant strains, and none of the DMI-sensitive strains, carry an identical point mutation resulting in the amino acid substitution F495L of the DMI target CYP51A, strongly supports the relationship of this mutation with azole fungicide resistance. The structural modeling and analysis of the CYP51A sensitive allele 9193-A1 and resistant allele KO103-A1 suggests that the F495L mutation has led to notable rearrangements of particular structural elements of the heme cavity, leading to localized constrictions, bringing more residues into proximity with all three of the azoles tested, most acutely in the region of the cavity adjacent to the docking site for the bulky less elongated tebuconazole, which provides a structural mechanism for the observed lack of sensitivity to azoles in isolates carrying the allele KO103-A1.
This conclusion is reinforced by an alignment of the Ptt CYP51A with its ortholog from A. fumigatus, showing that the F489L mutation is orthologous to F495I in A. fumigatus CYP51A. This mutation is associated with resistance to the DMI fungicides itraconazole and posaconazole in A. fumigatus (Mellado et al., 2007; Liu et al., 2015). Structural modeling of A. fumigatus CYP51A has shown that the F495I residue is close to the substrate binding pocket (Liu et al., 2016). The F495L mutation in Ptt and A. fumigatus CYP51A is also homologous to the F506I substitution in P. digitatum CYP51B, which is correlated with resistance to prochloraz (Wang et al., 2014). In A. fumigatus the F495I mutation has been found only in combination with other changes in the CYP51A amino acid sequence such as L98H or S297T (Mellado et al., 2007; Lockhart et al., 2011). In P. digitatum, the F506I substitution is found only in combination with G459S in CYP51B (Wang et al., 2014). By contrast, in Ptt the F495L mutation is the sole polymorphism of CYP51A to be found only in resistant and not in sensitive isolates.
Point mutations in the amino acid sequence of CYP51 targets occurring in conjunction with overexpression of the corresponding genes have been described in DMI-resistant strains of several pathogens. The predominant mechanisms of azole resistance in A. fumigatus is termed CYP51A TR/L98H, a combination of the amino acid substitution L98H with a 34 bp tandem repeat in the Cyp51A promoter that results in an up to eight-fold increase in expression (Mellado et al., 2007; Snelders et al., 2008; Lockhart et al., 2011). In Pyrenopeziza brassicae, the isolates with the highest levels of azole resistance have a combination of inducible Cyp51B overexpression and the amino acid substitution S508T (S524T) (Carter et al., 2014). In azole-resistant isolates of Erysiphe necator, the CYP51B Y136F (Y137F) amino acid substitution co-occurs with a 1.4 to 19-fold increase in expression (Rallos and Baudoin, 2016). In our study, induced overexpression of both the Cyp51A genes (A1 and A2) and the Cyp51B gene has been demonstrated in a DMI-resistant strain compared to a DMI-sensitive wild-type and this may contribute to the observed phenotype of DMI cross-resistance. Alternatively, the observed increased expression of the Cyp51 genes may be a compensatory mechanism to maintain sterol composition if the F495L mutation results in reduced CYP51A enzyme efficiency. Subsequent heterologous expression and enzymatic activity studies should determine the effect of the observed mutation on the function of CYP51A, and may clarify the role of target overexpression and dissect its contribution to DMI resistance.
Overexpression of Cyp51 is typically caused by insertions of tandem repeats or transposable elements in the promoter region (Price et al., 2015). As described above, in A. fumigatus overexpression of Cyp51A is caused by a 34 bp tandem duplication in the promoter (Mellado et al., 2007). Similarly in P. digitatum a 126 bp tandem repeat acts as a transcriptional enhancer for Cyp51A expression in DMI-resistant isolates (Hamamoto et al., 2000). In Pyrenopeziza brassicae insertions of 46, 151, and 232 bp in the predicted promoter region of Cyp51B are correlated with overexpression (Carter et al., 2014). In azole-resistant isolates of Zymoseptoria tritici, overexpression of the Cyp51B gene is correlated with a 120 bp insertion in the predicted promoter region (Cools et al., 2012). In Ptt we were unable to find any changes at the nucleotide level in the promoter regions of Cyp51A1, Cyp51A2, and Cyp51B of a strain overexpressing these genes, when compared to a wild-type. An overexpressing Cyp51 phenotype that cannot be linked to promoter changes has previously been reported in azole-resistant isolates of some fungal pathogens including A. fumigatus and Villosiclava virens (Arendrup et al., 2010; Wang et al., 2015). Future studies of possible trans-regulatory changes due to mutations in transcription factors, and analyses of the DNA methylation and histone modification of the promoters may elucidate whether such regulatory elements could be involved in the overexpression of these genes in Ptt.
Interestingly, although Ptt shares the same host as P. teres f. sp. maculata (Ptm), the other forma specialis responsible for SFNB and the predominant form of net blotch disease in Australia (McLean et al., 2010; Murray and Brennan, 2010), azole resistance in this pathogen has not been reported yet. This can probably be explained by the comparatively lower exposure of Ptm to DMI fungicides through time. Until comparatively recently, Ptt was the predominant net blotch form in Western Australia. Ptm was first reported in southern Western Australia in 1995, having previously only been found in the northern wheatbelt. Ptm appears to have spread in south-west WA from that time aided by the introduction of stubble retention practices and SFNB sensitive varieties including Franklin (1989), Gairdner (1998), and Baudin (2002) (Paynter et al., 1999; Paynter and Fettell, 2011; Gupta et al., 2012). The prevalence of these varieties in high rainfall areas together with treatment with significant levels of DMI fungicides (Geoff Thomas, personal communication) may underlie the emergence of resistance in Ptt first. However, current coexistence of Ptt and Ptm raises the possibility of fungicide resistance transfer to Ptm by rare hybridization events (McLean et al., 2014). A more likely scenario, however, is independent acquisition of resistance by Ptm under the same selection pressures. In view of the results presented here, there is an urgent need for adequate anti-resistance management strategies in both types of net blotch disease.
The evolution of new DMI resistant strains in Ptt would provide additional challenges to the barley industry. Improved knowledge of fungicide resistance evolution and of the molecular mechanisms by which this occurs will be necessary to implement suitable control strategies that will reduce the likelihood of fungicide resistance outbreaks.
Author Contributions
FL designed the project. WM, RO, WD, and SE contributed to the conception of the work. WM and NB performed the experiments. WM, WD, and PW performed all the bioinformatics analyses. JM and SW performed all the analysis and interpretation of structural modeling data. WM, WD, PW, RO, JM, SW, NB, SE, and FL wrote and edited the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Authors wish to thank James Hane and Alison Testa for bioinformatics support. The authors wish to thank Kejal Dodhia and Steven Chang for the technical support. The authors wish to thank Geoff Thomas for his advice in drafting the discussion. This study was supported by the Centre for Crop and Disease Management, a joint initiative of Curtin University and the Grains Research and Development Corporation—research grant CUR00023 (programme 9).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01279
References
Andrews, S. (2010). FastQC A Quality Control Tool for High Throughput Sequence Data. Babraham Bioinformatics. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (Accessed May 25, 2016).
APVMA (2016). PubCRIS: Public Chemical Registration Information System Search. Australian Pesticides and Veterinary Medicines Authority. Available online at: https://portal.apvma.gov.au/pubcris
Arendrup, M. C., Mavridou, E., Mortensen, K. L., Snelders, E., Frimodt-Møller, N., Khan, H., et al. (2010). Development of azole resistance in Aspergillus fumigatus during azole therapy associated with change in virulence. PLoS ONE 5:e10080. doi: 10.1371/journal.pone.0010080
Becher, R., Weihmann, F., Deising, H. B., and Wirsel, S. G. (2011). Development of a novel multiplex DNA microarray for Fusarium graminearum and analysis of azole fungicide responses. BMC Genomics 12:52. doi: 10.1186/1471-2164-12-52
Becher, R., and Wirsel, S. G. (2012). Fungal cytochrome P450 sterol 14α-demethylase (CYP51) and azole resistance in plant and human pathogens. Appl. Microbiol. Biotechnol. 95, 825–840. doi: 10.1007/s00253-012-4195-9
Boetzer, M., Henkel, C. V., Jansen, H. J., Butler, D., and Pirovano, W. (2011). Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27, 578–579. doi: 10.1093/bioinformatics/btq683
Boungab, K., Belabid, L., Fortas, Z., and Bayaa, B. (2012). Pathotype diversity among Algerian isolates of Pyrenophora teres f. teres. Phytopathol. Mediterr. 51, 577–586. doi: 10.14601/Phytopathol_Mediterr-10561
Brunner, P. C., Stefansson, T. S., Fountaine, J., Richina, V., and McDonald, B. A. (2015). A global analysis of CYP51 diversity and azole sensitivity in Rhynchosporium commune. Phytopathology 106, 355–361. doi: 10.1094/PHYTO-07-15-0158-R
Carter, H. E., Fraaije, B. A., West, J. S., Kelly, S. L., Mehl, A., Shaw, M. W., et al. (2014). Alterations in the predicted regulatory and coding regions of the sterol 14α-demethylase gene (CYP51) confer decreased azole sensitivity in the oilseed rape pathogen Pyrenopeziza brassicae. Mol. Plant Pathol. 15, 513–522. doi: 10.1111/mpp.12106
Cools, H. J., Bayon, C., Atkins, S., Lucas, J. A., and Fraaije, B. A. (2012). Overexpression of the sterol 14α-demethylase gene (MgCYP51) in Mycosphaerella graminicola isolates confers a novel azole fungicide sensitivity phenotype. Pest Manag. Sci. 68, 1034–1040. doi: 10.1002/ps.3263
Cools, H. J., and Fraaije, B. A. (2013). Update on mechanisms of azole resistance in Mycosphaerella graminicola and implications for future control. Pest Manag. Sci. 69, 150–155. doi: 10.1002/ps.3348
Délye, C., Laigret, F., and Corio-Costet, M. F. (1997). A mutation in the 14 alpha-demethylase gene of Uncinula necator that correlates with resistance to a sterol biosynthesis inhibitor. Appl. Environ. Microbiol. 63, 2966–2970.
Erickson, E. O., and Wilcox, W. F. (1997). Distributions of sensitivities to three sterol demethylation inhibitor fungicides among populations of uncinula necator sensitive and resistant to triadimefon. Phytopathology 87, 784–791. doi: 10.1094/PHYTO.1997.87.8.784
Fan, J., Urban, M., Parker, J. E., Brewer, H. C., Kelly, S. L., Hammond-Kosack, K. E., et al. (2013). Characterization of the sterol 14α-demethylases of Fusarium graminearum identifies a novel genus-specific CYP51 function. New Phytol. 198, 821–835. doi: 10.1111/nph.12193
Ficsor, A., Tóth, B., Varga, J., Tomcsányi, A., Mészáros, K., Kótai, É., et al. (2014). Variability of Pyrenophora teres f. teres in Hungary as revealed by mating type and RAPD analyses. J. Plant Pathol. 96, 515–523. doi: 10.4454/JPP.V96I3.020
Fraaije, B. A., Cools, H. J., Kim, S. H., Motteram, J., Clark, W. S., and Lucas, J. A. (2007). A novel substitution I381V in the sterol 14α-demethylase (CYP51) of Mycosphaerella graminicola is differentially selected by azole fungicides. Mol. Plant Pathol. 8, 245–254. doi: 10.1111/j.1364-3703.2007.00388.x
Fries, N. (1938). Über die bedeutung von wuchsstoffen für das wachstum verschiedener Pilze. Symbol. Botan. Upsalienses 3, 188.
Ghosoph, J. M., Schmidt, L. S., Margosan, D. A., and Smilanick, J. L. (2007). Imazalil resistance linked to a unique insertion sequence in the PdCYP51 promoter region of Penicillium digitatum. Postharvest Biol. Technol. 44, 9–18. doi: 10.1016/j.postharvbio.2006.11.008
Gupta, S., Loughman, R., D'antuono, M., and Bradley, J. (2012). Characterisation and diversity of Pyrenophora teres f. maculata isolates in Western Australia. Aust. Plant Pathol. 41, 31–40. doi: 10.1007/s13313-011-0083-5
Hamamoto, H., Hasegawa, K., Nakaune, R., Lee, Y. J., Makizumi, Y., Akutsu, K., et al. (2000). Tandem repeat of a transcriptional enhancer upstream of the sterol 14α-demethylase gene (CYP51) in Penicillium digitatum. Appl. Environ. Microbiol. 66, 3421–3426. doi: 10.1128/AEM.66.8.3421-3426.2000
Hawkins, N. J., Cools, H. J., Sierotzki, H., Shaw, M. W., Knogge, W., Kelly, S. L., et al. (2014). Paralog re-emergence: a novel, historically contingent mechanism in the evolution of antimicrobial resistance. Mol. Biol. Evol. 31, 1793–1802. doi: 10.1093/molbev/msu134
Hof, H. (2001). Critical annotations to the use of azole antifungals for plant protection. Antimicrob. Agents Chemother. 45, 2987–2990. doi: 10.1128/AAC.45.11.2987-2990.2001
Howard, S. J., and Arendrup, M. C. (2011). Acquired antifungal drug resistance in Aspergillus fumigatus: epidemiology and detection. Med. Mycol. 49, S90–S95. doi: 10.3109/13693786.2010.508469
Ishii, H., and Hollomon, D. W. (2015). Fungicide Resistance in Plant Pathogens: Principles and a Guide to Practical Management. Tokyo: Springer.
Kanafani, Z. A., and Perfect, J. R. (2008). Resistance to antifungal agents: mechanisms and clinical impact. Clin. Infect. Dis. 46, 120–128. doi: 10.1086/524071
Köllerm, W. (2003). “Fungicides, sterol biosynthesis inhibitors,” in Encyclopedia of Agrochemicals, ed J. R. Plimmer (Hoboken, NJ: John Wiley & Sons, Inc.), 628–640.
Lehmensiek, A., Bester-van der Merwe, A. E., Sutherland, M. W., Platz, G., Kriel, W. M., Potgieter, G. F., et al. (2010). Population structure of South African and Australian Pyrenophora teres isolates. Plant Pathol. 59, 504–515. doi: 10.1111/j.1365-3059.2009.02231.x
Leišova, L., MinariˇḰova, V., Kučera, L., and Ovesna, J. (2005). Genetic diversity of Pyrenophora teres isolates as detected by AFLP analysis. J. Phytopathol. 153, 569–578. doi: 10.1111/j.1439-0434.2005.01019.x
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Liang, H. J., Li, J. L., Di, Y. L., Zhang, A. S., and Zhu, F. X. (2015). Logarithmic transformation is essential for statistical analysis of fungicide EC50 values. J. Phytopathol. 163, 456–464. doi: 10.1111/jph.12342
Liu, M., Zeng, R., Zhang, L., Li, D., Lv, G., Shen, Y., et al. (2015). Multiple cyp51A-based mechanisms identified in azole-resistant isolates of Aspergillus fumigatus from China. Antimicrob. Agents Chemother. 59, 4321–4325. doi: 10.1128/AAC.00003-15
Liu, M., Zheng, N., Li, D., Zheng, H., Zhang, L., Ge, H., et al. (2016). cyp51A-based mechanism of azole resistance in Aspergillus fumigatus: illustration by a new 3D structural model of Aspergillus fumigatus CYP51A protein. Med. Mycol. 54, 400–408. doi: 10.1093/mmy/myv102
Liu, Z., Ellwood, S. R., Oliver, R. P., and Friesen, T. L. (2011). Pyrenophora teres: profile of an increasingly damaging barley pathogen. Mol. Plant Pathol. 12, 1–19. doi: 10.1111/j.1364-3703.2010.00649.x
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lockhart, S. R., Frade, J. P., Etienne, K. A., Pfaller, M. A., Diekema, D. J., and Balajee, S. A. (2011). Azole resistance in Aspergillus fumigatus isolates from the ARTEMIS global surveillance is primarily due to the TR/L98H mutation in the cyp51A gene. Antimicrob. Agents Chemother. 55, 4465–4468. doi: 10.1128/AAC.00185-11
López-Ruiz, F. J., Pérez-García, A., Fernández-Ortuño, D., Romero, D., García, E., de Vicente, A., et al. (2010). Sensitivities to DMI fungicides in populations of Podosphaera fusca in south central Spain. Pest Manag. Sci. 66, 801–808. doi: 10.1002/ps.1948
Mair, W., Lopez-Ruiz, F, Stammler, G., Clark, W., Burnett, F., Hollomon, D., et al. (2016). Proposal for a unified nomenclature for target site mutations associated with resistance to fungicides. Pest Manag. Sci. 72, 1449–1459. doi: 10.1002/ps.4301
McLean, M., Martin, A., Gupta, S., Sutherland, M., Hollaway, G., and Platz, G. (2014). Validation of a new spot form of net blotch differential set and evidence for hybridisation between the spot and net forms of net blotch in Australia. Aust. Plant Pathol. 43, 223–233. doi: 10.1007/s13313-014-0285-8
McLean, M. S., Howlett, B. J., and Hollaway, G. J. (2010). Spot form of net blotch, caused by Pyrenophora teres f. maculata, is the most prevalent foliar disease of barley in Victoria, Australia. Aust. Plant Pathol. 39, 46–49. doi: 10.1071/AP09054
Mellado, E., Garcia-Effron, G., Alcázar-Fuoli, L., Melchers, W. J., Verweij, P. E., Cuenca-Estrella, M., et al. (2007). A new Aspergillus fumigatus resistance mechanism conferring in vitro cross-resistance to azole antifungals involves a combination of cyp51A alterations. Antimicrob. Agents Chemother. 51, 1897–1904. doi: 10.1128/AAC.01092-06
Morio, F., Loge, C., Besse, B., Hennequin, C., and Le Pape, P. (2010). Screening for amino acid substitutions in the Candida albicans Erg11 protein of azole-susceptible and azole-resistant clinical isolates: new substitutions and a review of the literature. Diagn. Microbiol. Infect. Dis. 66, 373–384. doi: 10.1016/j.diagmicrobio.2009.11.006
Mullins, J. G., Parker, J. E., Cools, H. J., Togawa, R. C., Lucas, J. A., Fraaije, B. A., et al. (2011). Molecular modelling of the emergence of azole resistance in Mycosphaerella graminicola. PLoS ONE 6:e20973. doi: 10.1371/journal.pone.0020973
Murray, G. M., and Brennan, J. P. (2010). Estimating disease losses to the Australian barley industry. Aust. Plant Pathol. 39, 85–96. doi: 10.1071/AP09064
Nurk, S., Bankevich, A., Antipov, D., Gurevich, A. A., Korobeynikov, A., Lapidus, A., et al. (2013). Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J. Comput. Biol. 20, 714–737. doi: 10.1089/cmb.2013.0084
Omrane, S., Sghyer, H., Audéon, C., Lanen, C., Duplaix, C., Walker, A.-S., et al. (2015). Fungicide efflux and the MgMFS1 transporter contribute to the multidrug resistance phenotype in Zymoseptoria tritici field isolates. Environ. Microbiol. 17, 2805–2823. doi: 10.1111/1462-2920.12781
Paynter, B., and Fettell, N. (2011). “Chapter 9: cultural practices: focus on major barleyproducing regions,” in Barley Production, Improvement, and Uses, ed S. E. Ullrich (Ames, IA: Blackwell Publishing Ltd.), 258–281.
Paynter, B., Young, K., Shackley, B., and Jettner, R. (1999). “Growing gairdner barley for malting in Western Australia,” in Proceedings of the 9th Australian Barley Technical Symposium (Canberra, ACT).
Peever, T. L., and Milgroom, M. G. (1992). Inheritance of triadimenol resistance in Pyrenophora teres. Phytopathology 82, 821–828. doi: 10.1094/Phyto-82-821
Peever, T. L., and Milgroom, M. G. (1993). Genetic correlations in resistance to sterol biosynthesis-inhibiting fungicides in Pyrenophora teres. Phytopathology 83, 1076–1082. doi: 10.1094/Phyto-83-1076
Price, C. L., Parker, J. E., Warrilow, A. G. S., Kelly, D. E., and Kelly, S. L. (2015). Azole fungicides – understanding resistance mechanisms in agricultural fungal pathogens. Pest Manag. Sci. 71, 1054–1058. doi: 10.1002/ps.4029
Rallos, L. E. E., and Baudoin, A. B. (2016). Co-occurrence of two allelic variants of CYP51 in Erysiphe necator and their correlation with over-expression for DMI resistance. PLoS ONE 11:e0148025. doi: 10.1371/journal.pone.0148025
Rau, D., Attene, G., Brown, A. H. D., Nanni, L., Maier, F. J., Balmas, V., et al. (2007). Phylogeny and evolution of mating-type genes from Pyrenophora teres, the causal agent of barley “net blotch” disease. Curr. Genet. 51, 377–392. doi: 10.1007/s00294-007-0126-1
Rehfus, A., Miessner, S., Achenbach, J., Strobel, D., Bryson, R., and Stammler, G. (2016). Emergence of succinate dehydrogenase inhibitor resistance of Pyrenophora teres in Europe. Pest Manag. Sci. 72, 1977–1988. doi: 10.1002/ps.4244
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Saghai-Maroof, M. A., Soliman, K. M., Jorgensen, R. A., and Allard, R. W. (1984). Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. U.S.A. 81, 8014–8018. doi: 10.1073/pnas.81.24.8014
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Semar, M., Strobel, D., Koch, A., Klappach, K., and Stammler, G. (2007). Field efficacy of pyraclostrobin against populations of Pyrenophora teres containing the F129L mutation in the cytochrome b gene. J. Plant Dis. Protect. 114, 117–119. doi: 10.1007/BF03356718
Serenius, M., and Manninen, O. (2008). Prochloraz tolerance of Pyrenophora teres population in Finland. Agric. Food Sci. 15, 35–42. doi: 10.2137/145960606777245588
Serenius, M., Manninen, O., Wallwork, H., and Williams, K. (2007). Genetic differentiation in Pyrenophora teres populations measured with AFLP markers. Mycol. Res. 111, 213–223. doi: 10.1016/j.mycres.2006.11.009
Sheridan, J. E., Grbavac, N., and Sheridan, M. H. (1985). Triadimenol insensitivity in Pyrenophora teres. Trans.Br. Mycol. Soc. 85, 338–341. doi: 10.1016/S0007-1536(85)80198-4
Sheridan, J., Grbavac, Sheridan, M. H., and Soteros, J. (1987). Further studies on triadimenol resistance in the barley net blotch pathogen Pyrenophora teres. N. Z. J. Agric. Res. 30, 101–105. doi: 10.1080/00288233.1987.10430483
Sierotzki, H., Frey, R., Wullschleger, J., Palermo, S., Karlin, S., Godwin, J., et al. (2007). Cytochrome b gene sequence and structure of Pyrenophora teres and P. tritici-repentis and implications for QoI resistance. Pest Manag. Sci. 63, 225–233. doi: 10.1002/ps.1330
Snelders, E., van der Lee, H. A., Kuijpers, J., Rijs, A. J. M., Varga, J., Samson, R. A., et al. (2008). Emergence of azole resistance in Aspergillus fumigatus and spread of a single resistance mechanism. PLoS Med. 5:e219. doi: 10.1371/journal.pmed.0050219
Solomon, P. S., Ipcho, S. V., Hane, J. K., Tan, K.-C., and Oliver, R. P. (2008). A quantitative PCR approach to determine gene copy number. Fungal Genet. Rep. 55, 5–8. Available online at: http://www.fgsc.net/FGR/FGR55/FGR55Solomon.htm (Accessed May 27, 2016).
Steffenson, B., Webster, R., and Jackson, L. (1991). Reduction in yield loss using incomplete resistance to Pyrenophora teres f. teres in barley. Plant Dis. 75, 96–100. doi: 10.1094/PD-75-0096
Sun, X., Xu, Q., Ruan, R., Zhang, T., Zhu, C., and Li, H. (2013). PdMLE1, a specific and active transposon acts as a promoter and confers Penicillium digitatum with DMI resistance. Environ. Microbiol. Rep. 5, 135–142. doi: 10.1111/1758-2229.12012
Tekauz, A. (1990). Characterization and distribution of pathogenic variation in Pyrenophora teres f. teres and P. teres f. maculata from western Canada. Can. J. Plant Pathol. 12, 141–148. doi: 10.1080/07060669009501017
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Trott, O., and Olson, A. J. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461. doi: 10.1002/jcc.21334
Wang, F., Lin, Y., Yin, W. X., Peng, Y. L., Schnabel, G., Huang, J. B., et al. (2015). The Y137H mutation of VvCYP51 gene confers the reduced sensitivity to tebuconazole in Villosiclava virens. Sci. Rep. 5:17575. doi: 10.1038/srep17575
Wang, J., Yu, J., Liu, J., Yuan, Y., Li, N., He, M., et al. (2014). Novel mutations in CYP51B from Penicillium digitatum involved in prochloraz resistance. J. Microbiol. 52, 762–770. doi: 10.1007/s12275-014-4112-2
Yan, X., Ma, W. B., Li, Y., Wang, H., Que, Y. W., Ma, Z. H., et al. (2011). A sterol 14alpha-demethylase is required for conidiation, virulence and for mediating sensitivity to sterol demethylation inhibitors by the rice blast fungus Magnaporthe oryzae. Fungal Genet. Biol. 48, 144–153. doi: 10.1016/j.fgb.2010.09.005
Keywords: Cyp51, azole, DMI, resistance, net blotch, Pyrenophora teres, overexpression, mutation
Citation: Mair WJ, Deng W, Mullins JGL, West S, Wang P, Besharat N, Ellwood SR, Oliver RP and Lopez-Ruiz FJ (2016) Demethylase Inhibitor Fungicide Resistance in Pyrenophora teres f. sp. teres Associated with Target Site Modification and Inducible Overexpression of Cyp51. Front. Microbiol. 7:1279. doi: 10.3389/fmicb.2016.01279
Received: 03 June 2016; Accepted: 03 August 2016;
Published: 19 August 2016.
Edited by:
Sabine Fillinger, Institut National de la Recherche Agronomique, FranceReviewed by:
Nichola Jane Hawkins, Rothamsted Research (BBSRC), UKPatrick C. Brunner, Eidgenössische Technische Hochschule Zürich, Switzerland
Copyright © 2016 Mair, Deng, Mullins, West, Wang, Besharat, Ellwood, Oliver and Lopez-Ruiz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Francisco J. Lopez-Ruiz, ZnJhbi5sb3BlenJ1aXpAY3VydGluLmVkdS5hdQ==
 Wesley J. Mair
Wesley J. Mair Weiwei Deng
Weiwei Deng Jonathan G. L. Mullins
Jonathan G. L. Mullins Samuel West2
Samuel West2 Penghao Wang
Penghao Wang Richard P. Oliver
Richard P. Oliver Francisco J. Lopez-Ruiz
Francisco J. Lopez-Ruiz