- Department of Biología Molecular, Facultad de Ciencias Exactas, Químicas y Naturales, Universidad Nacional de Río Cuarto, Río Cuarto, Argentina
Many species or strains of the genus Pseudomonas have been characterized as plant growth promoting rhizobacteria (PGPR). We used a combination of phenotypic and genotypic techniques to analyze the community of fluorescent Pseudomonas strains in the rhizosphere of commercially grown Mentha piperita (peppermint). Biochemical techniques, Amplified rDNA Restriction Analysis (ARDRA), and 16S rRNA gene sequence analysis revealed that the majority of the isolated native fluorescent strains were P. putida. Use of two Repetitive Sequence-based PCR (rep-PCR) techniques, BOX-PCR and ERIC-PCR, allowed us to evaluate diversity among the native strains and to more effectively distinguish among them. PGPR activity was tested for the native strains and reference strain P. fluorescens WCS417r. Micropropagated M. piperita plantlets were exposed to microbial volatile organic compounds (mVOCs) emitted by the bacterial strains, and plant biomass parameters and production of essential oils (EOs) were measured. mVOCs from 11 of the native strains caused an increase in shoot fresh weight. mVOCs from three native strains (SJ04, SJ25, SJ48) induced changes in M. pierita EO composition. The mVOCs caused a reduction of metabolites in the monoterpene pathway, for example menthofuran, and an increase in menthol production. Menthol production is the primary indicator of EO quality. The mVOCs produced by native strains SJ04, SJ25, SJ48, and strain WCS417r were analyzed. The obtained mVOC chromatographic profiles were unique for each of the three native strains analyzed, containing varying hydrocarbon, aromatic, and alogenic compounds. The differential effects of the strains were most likely due to the specific mixtures of mVOCs emitted by each strain, suggesting a synergistic effect occurs among the compounds present.
Introduction
Peppermint (Mentha piperita L.; family Labiatae) is a popular, commonly-used flavoring agent worldwide. Fresh or dried leaves of Mentha species are used primarily as condiments. Essential oils (EOs) of these plants, which are produced and stored in glandular hairs, are used as food and beverage flavorings, as fragrances, and as fungicides or insecticides in a variety of pharmaceutical and industrial products (Harrewijn et al., 2001; Ram et al., 2006). EOs are secondary metabolites whose production is associated with primary metabolism and with availability of soil nutrients (Shulka et al., 1992).
Among the many known PGPR genera, Pseudomonas has received the most research attention because it is widely distributed in various environments and is easy to culture under laboratory conditions (Palleroni, 2005). Members of the “fluorescent Pseudomonas group” produce a water-soluble fluorescent green-yellow pigment termed pseudobactin or pyoverdine (Stanier et al., 1966). This characteristic peptide displays iron-binding activity and is associated with pathogenic or growth-promoting properties (Fuchs et al., 2001; Mehri et al., 2011). Fluorescent Pseudomonas strains have been isolated from rhizospheric soils of numerouscropplants, including cotton (Hu et al., 2005), rice (Loaces et al., 2011), banana (Naik et al., 2008), rape (Patten and Glick, 2002), sugar cane (Rameshkumar et al., 2012), wheat, and barley (Mavrodi et al., 2001; Parejko et al., 2012). The two major fluorescent Pseudomonas species that display PGPR activity are P. fluorescens and P. putida (Kloepper and Schroth, 1978). Few studies to date have focused on PGPR isolated from rhizospheres of aromatic plant species because of the presumption that EOs may be released in root exudates and display antimicrobial activity (Chen et al., 2004). We used phenotyping and genotyping techniques to isolate, identify, and characterize fluorescent Pseudomonas strains from M. piperita rhizosphere of M. piperita. We then evaluated PGPR activity of these strains on micropropagated M. piperita plants, including mVOC-mediated effects on plant biomass and EO production.
Materials and Methods
Sample Collection and Strain Isolation
A total of 20 M. piperita plants were collected randomly from a commercial crop (San José) grown in the San Javier department, Córdoba province, Argentina (30°30′00.0″S, 64°30′00.0″W). Samples were placed individually in plastic bags, stored at 4°C, transported and processed within 24 h. Five roots were washed in sterile distilled water to remove loosely adherent soil, and then rotary shaken 15 min in 100 ml sterile phosphate buffer, pH 7.5, to obtain a rhizospheric soil suspension. Serial dilutions were prepared from the suspension, and 0.1 ml of each dilution was spread on Nutrient Agar medium (Laboratorios Britania, Buenos Aires, Argentina) and on King B medium (King et al., 1954) and incubated at 28°C for 48 h. After complete evaporation of buffer from soil, total bacteria and fluorescent bacteria under UV light were counted as colony forming units (CFU) per g rhizospheric soil (Mehnaz et al., 2009). Bacterial strains were purified on King B medium, identified, and characterized. Fifty fluorescent strains were isolated (Table 1), and cryopreserved at −80°C in 20% glycerol (v/v). A single colony from each strain was grown aerobically on LB medium (Luria and Burrous, 1955) and incubated on a rotary shaker (200 rpm) for 24 h at 28°C prior to testing. This isolation from roots was performed four times.
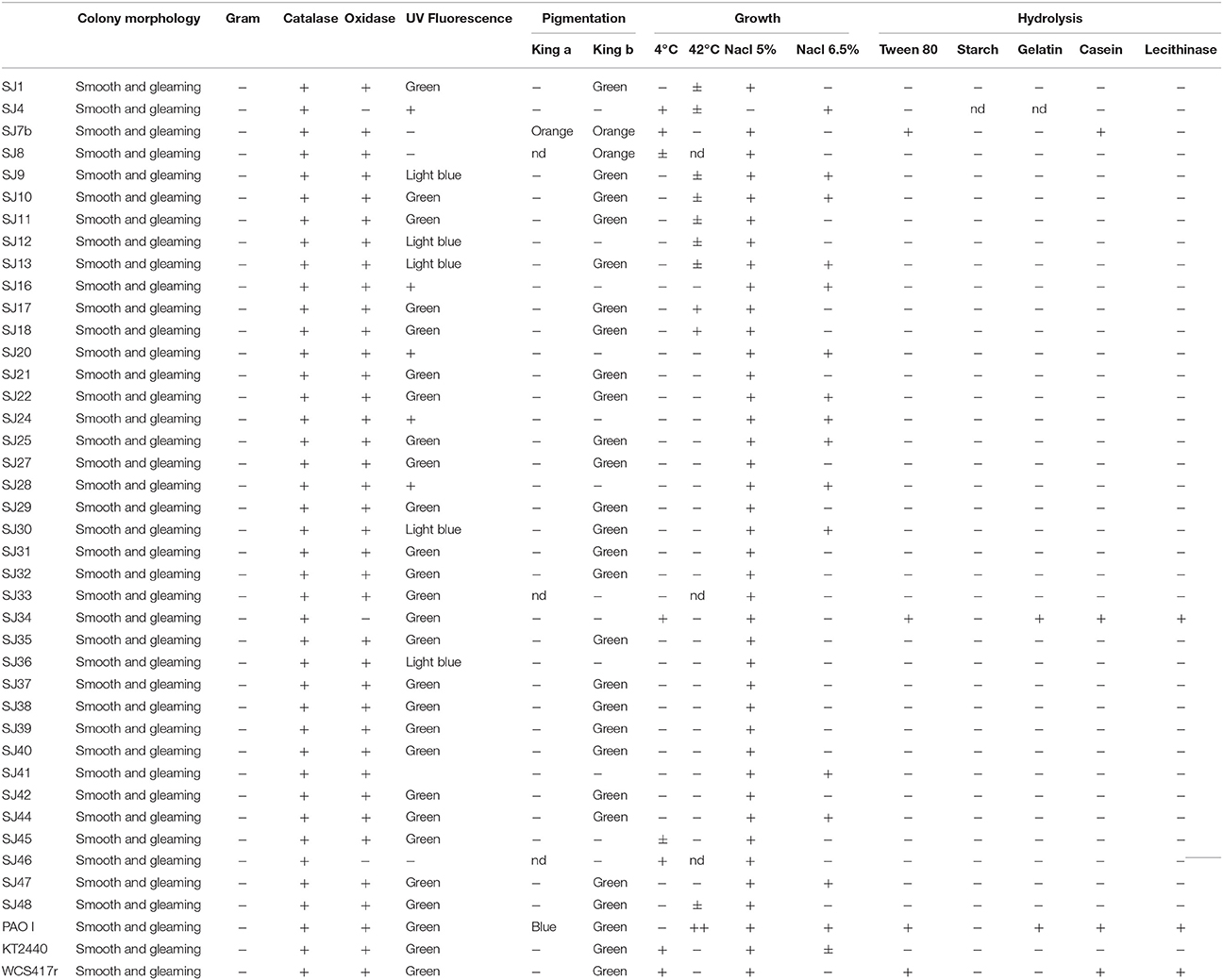
Table 1. Phenotypic characterization of fluorescent Pseudomonas strains, based on morphological, physiological, and biochemical evaluation of native strains isolated from the M. piperita rhizosphere.
Analysis of EO Components in Root Exudates
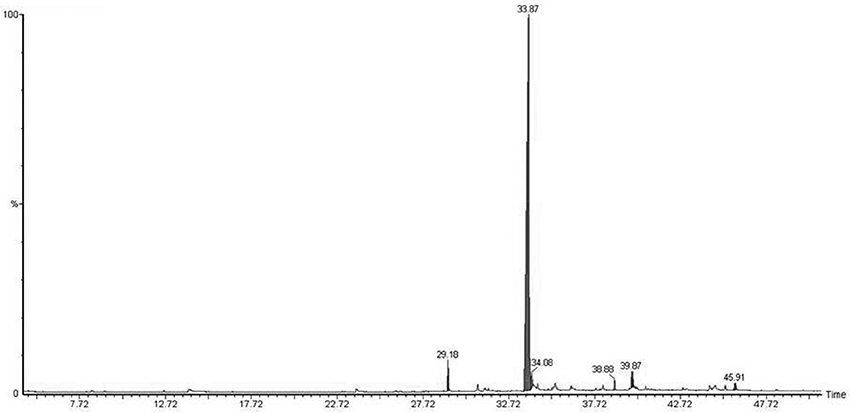
Micropropagated M. piperita plantlets were grown on MS liquid medium (Santoro et al., 2011) for 30 days. The medium was then filtered (pore diameter 0.46 μm), and the volatile fraction was extracted with dichloromethane. EOs were analyzed and detected by gas chromatography/mass spectrometry (GC-MS; Figure 1; Table 2). Flame ionization detector (FID) response factors for each compound generated essentially equal areas (differences<5%). Chemical analyses were performed by GC (model Clarus 600, PerkinElmer; Waltham, MA, USA) with a CBP-1 capillary column (30 m × 0.25 mm, film thickness 0.25 mm) and mass-selective detector. Analytical conditions: injector temperature 250°C, detector temperature 270°C; oven temperature programmed from 60°C (3 min) to 240°C at 4°C min−1; carrier gas ¼ helium at constant flow 0.9 ml min−1; source 70 eV. EO components were identified based on mass spectra and retention times, in comparison with standards.
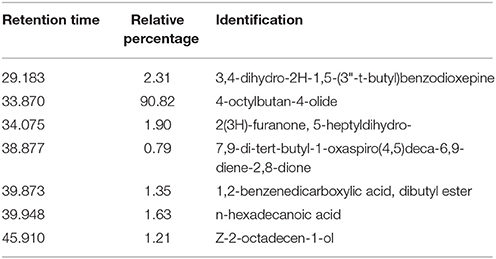
Table 2. Identification and relative percentages of major chemical signals detected in M. piperita root exudates, based on GC-MS analysis and comparison with the NIST database, with acceptance of SI ≥ 800.
Phenotypic Characterization
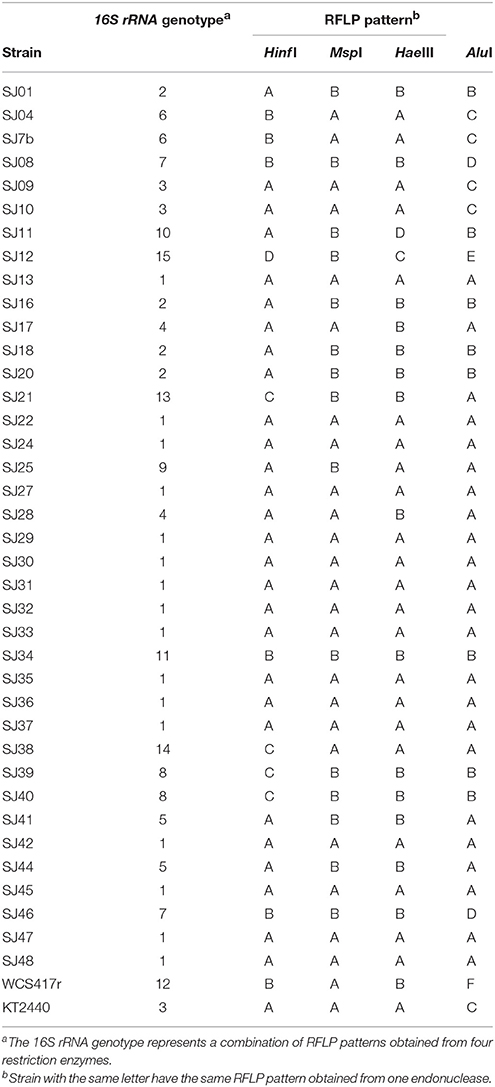
Fluorescent bacteria were phenotypically characterized based on morphological, physiological, and biochemical criteria (Lysenko, 1961; Stanier et al., 1966; Palleroni, 2005; Table 1). The tests and parameters included colony morphology, Gram determination, catalase determination (Urzí et al., 2001), oxidase determination (kit fromLaboratorios Britania), fluorescence color under UV illumination, pigment production (King et al., 1954), temperature tolerance, growth at 5 and 6.5% salt concentration, gelatin hydrolysis (Medina and Baresi, 2007), Tween 80 hydrolysis (Sierra, 1957), starch hydrolysis (Gordon and Mihm, 1956), casein hydrolysis (Salisbury and Lykos, 1972), and lecithinase production (Sneath, 1956). Each test was performed twice.
DNA Extraction
A single colony was inoculated on 3 ml LB medium and incubated at 28°C for 24 h. A 2-ml culture sample taken at late exponential growth phase was subjected to DNA extraction using a Genomic DNA Purification Kit (Fermentas, Thermo Fisher Scientific; Pittsburgh, PA, USA) according to the manufacturer's instructions.
Genotypic Testing
Amplified Ribosomal DNA Restriction Analysis (ARDRA)
16S rRNA gene amplification was performed, primers and methodology are specified in Table 3. A single specific band (~1400 bp) was observed by UV transillumination, and endonuclease digestion analysis of each amplicon was performed separately with MspI, Hinf I, HaeIII, and AluI (Achouak et al., 2000; Wu et al., 2009; Mehri et al., 2011; Nievas et al., 2012) according to the manufacturer's (Fermentas') instructions. Reaction mixtures were incubated overnight at 37°C. Total volume of each restriction-digested product was evaluated by 3% (w/v) agarose-ethidium bromide stained gel electrophoresis and photographed for subsequent analysis.
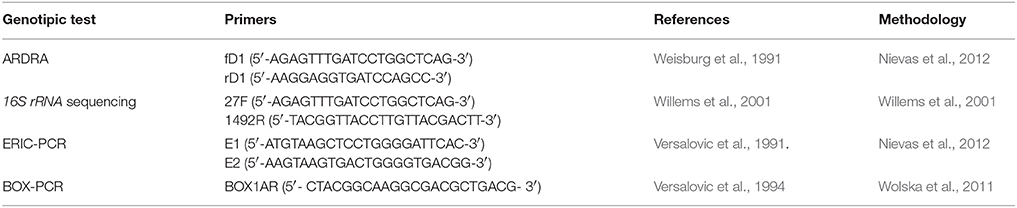
Table 3. Primers sequences used for genotypic analysis of native fluorescent Pseudomonas and reference strains.
16S rRNA Gene Nucleotide Sequence Analysis
Nucleotide sequence analysis of the 16S rRNA gene was performed for 17 native strains (Table 4). Direct PCR was performed with 1 μl DNA template in a 20-μl PCR reaction mixture using primer and methodology specified in Table 3. Purified PCR products (~1400 bp) were sequenced by Macrogen, Inc. (Seoul, Korea) using an automated DNA sequencing system (model 3730XL, Applied Biosystems; Foster City, CA, USA). Sequence identities were determined by BLAST search program (National Center for Biotechnology Information [NCBI]; Bethesda, MD, USA; Altschul et al., 1997). Nucleotide sequences of the 16S rRNA gene for each analyzed strain were deposited in GenBank with respective accession numbers (Table 5).
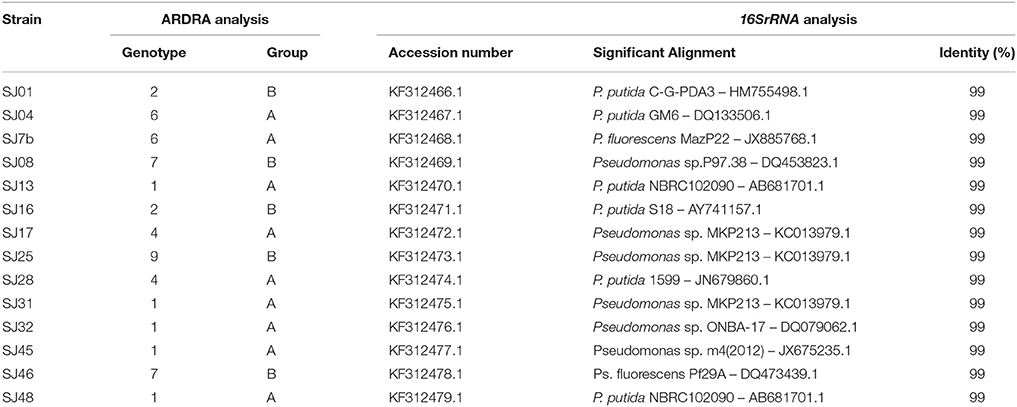
Table 5. BLAST analysis of 16SrRNA gene sequences of native strains isolated from M. piperita rhizosphere.
To generate a phylogenetic tree reflecting evolutionary relationships among fluorescent Pseudomonas strains, phylogenetic analysis was performed using the MEGA 4.0.2 software program (Tamura et al., 2007). Multiple alignments were made with the Clustal W software program (Higgins et al., 1994). Aligned sequences were used to construct a phylogenetic tree based on the neighbor-joining method (Saitou and Nei, 1987), using the Kimura two-parameter model (Kimura, 1980; Figure 2).
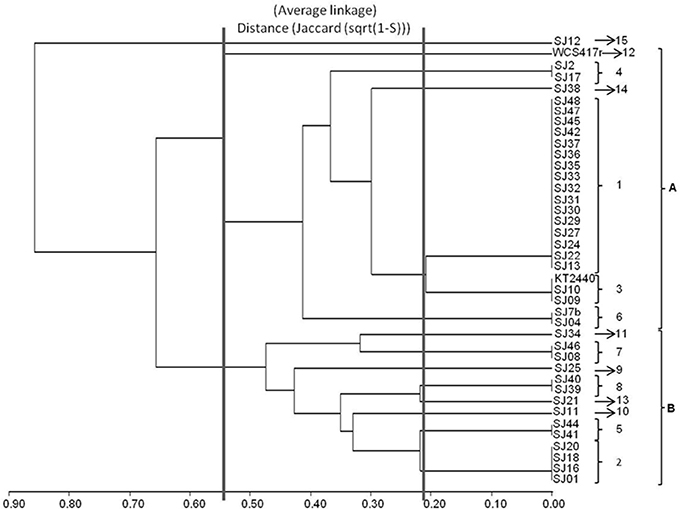
Figure 2. Dendrogram generated from RFLP patterns of 16S rRNA genes of native and reference strains of fluorescent Pseudomonas, using the UPGMA algorithm and Jaccard coefficient.
ERIC-PCR Fingerprinting
Enterobacterial Repetitive Intergenic Consensus (ERIC) fragments were analyzed for 40 native fluorescent strains. Amplification was performed using primers and methodology specified in Table 3. PCR products were separated by 1.2% (w/v) agarose gel electrophoresis in TAE 1X buffer, stained with ethidium bromide, and photographed for subsequent analysis.
BOX-PCR Fingerprinting
Repetitive extragenic palindromic sequences (BOX-PCR) were evaluated for the 40 strains. Amplification was performed with primer and methodology specified in Table 3. Amplified products were loaded on 2% (w/v) agarose gel electrophoresis, stained with ethidium bromide, and run at 60 V for 30 min and90 V for 120 min. The obtained gel was photographed for latter analysis.
Analysis of Genotypic Data
Fingerprints generated by ARDRA and by Repetitive Sequence-based PCR (rep-PCR) techniques (ERIC-PCR, BOX-PCR) as above were analyzed manually. Presence or absence of each band in the PCR profile was coded as a binary matrix. Genetic similarity and clustering analysis for each generated matrix were performed using the Infogen software program, v. 2011 (Universidad Nacional de Córdoba, Argentina). Genetic distances among native fluorescent strains were evaluated using a Jaccard coefficient. Clustering was visualized by construction of dendrograms using the Unweighted Pair-Group Method with Arithmetic Average (UPGMA) algorithm (Di Renzo et al., 2001; Balzarini, 2003).
Evaluation of Growth-Promoting Effects
Bacterial Suspension
Strains were grown aerobically on LB liquid medium for 24 h at 30°C with rotary shaking. Saturated cultures were diluted in sterile distilled water to final concentration 109 CFU ml−1.
Exposure to mVOCs
Plastic Petri dishes (90 × 15 mm) with a center partition (I-plate, Fisher Scientific) were used. One side of the dish contained MS solid medium, and the other contained Hoagland medium (Hoagland and Arnon, 1938) added with 3% sucrose (w/v) and 1.5% agar (w/v). Bacterial suspension as above (one drop; 20 μl) was placed on the Hoagland side. Distilled water (one drop) was used as negative control. A single node from a young shoot of aseptically cultured M. piperita (Santoro et al., 2011) was placed on the opposite side of the partition. In this way, plants were exposed to mVOCs without physical contact. Dishes were parafilm-sealed, arranged in a completely randomized design, and placed in a growth chamber under controlled conditions of light (16/8-h light/dark cycle), temperature (22 ± 2°C), and relative humidity (~70%). Five plants were used for each treatment, and experiments were replicated four times.
Plant Biomass and Morphology
Plants exposed to mVOCs were harvested after 30 days and evaluated for shoot length, root length, shoot fresh weight, root dry weight, node number, leaf number, and shoot ramification number (Santoro et al., 2011).
Extraction and Analysis of EOs
Each shoot sample was weighed, subjected to hydrodistillation in a micro Clevenger-like apparatus for 14 min, and the volatile fraction was extracted with dichloromethane. An internal standard (1 μl γ-dodecalactone in 100 μl ethanol) was added during the distillation procedure. Oil components were initially identified based on mass spectral and retention time data and confirmed by direct comparison with commercial standards (Sigma-Aldrich). FID response factors for each compound generated essentially equal areas (differences <5%). For comparison of results under different treatments, response factors for individual compounds were assumed to be equal (Shukui et al., 2005). Chemical analyses of EOs were performed as described in Section Analysis of EO Components in Root Exudates. For each plant, areas corresponding to analyzed compounds were added, and total weight of EOs from each sample was expressed relative to area of the internal standard. Shoots were weighed, and yield was expressed as μg g−1 fresh weight.
Statistical Analysis
Data were subjected to analysis of variance (ANOVA) followed by comparison of multiple treatment levels with control, using post hoc Fisher's Least Significant Difference (LSD) test. Infostat software program, v. 2.0 (Group Infostat, Universidad Nacional de Córdoba, Argentina) was used for all statistical analyses.
Chemical Analysis of mVOCs
mVOC Collection
The three bacterial strains that had the greatest effect on plant biomass and EO production (SJ04, SJ25, SJ48) were cultured for mVOC analysis, and WCS417r was included as reference strain. Bacteria were cultured in sealed 100-ml Kitasato flasksin Hoagland medium plus 0.4% TSB for 48 h at 30°C. The flasks were then connected to a vacuum pump to guarantee air flow, and the flaskinlet was covered with a cap that had been pre-drilled and modified with a cotton-filled glass tube to control air flux to the collection trap. Air exited the chamber through a reusable glass collection trap packed with 30 mg Super-Q absorbent (80–100 mesh; Alltech Assoc., Deerfield, IL, USA) to capture mVOCs. Prior to mVOC collection, eachfilter was rinsed with 2 ml dichloromethane and 2 ml hexaneto remove impurities. Flasks were then placed on a heating block at 50°C with magnetic stirring for 30 min. The filters were immediately eluted from absorbent traps with 200 μl dichloromethane, and collected mVOCs were analyzed by GC-MS as described below.
GC-MS Analysis and Conditions
Filter samples were concentrated with nitrogen gas, resuspended in ethanol, and injected into a Perkin-Elmer Clarus600 gas chromatograph equipped with DB5 column (60 m, i.d. 0.25 mm, film thickness 0.25 μm; J&W Scientific, Folsom, CA, USA) and mass-selective detector. GC-MS conditions: 25-minruns; injection port operated in splitless mode with constant He flow 1.0 ml/min; initial oven temperature 33°C, held for 3 min, ramped at 10°C min−1 to 180°C, ramped at40 min−1 to 220°C, and held for 5 min; HP quadrupole mass spectrometer operated in electron ionization mode at 70 eV; source temperature 200°C; quadrupole temperature 150°C; continuous scan from m/z 40 to 500. Volatile components were identified by comparison of retention times and recorded mass spectra against the NIST database, with acceptance of similarity index (SI) ≥ 800.
Results and Discussion
Sample and Strain Isolation
Pseudomonas is the most extensively studied of the known PGPR genera because the species are widely distributed in many different environments and are easy to culture under laboratory conditions. For isolation of fluorescent Pseudomonas strains, rhizospheric soil from a commercial M. piperita crop as described in Section Sample Collection and Strain Isolation was collected, with total bacteria count 3.05 × 107 CFU g−1. Similar results were obtained in previous studies of rhizospheric soil from other crops (Torsvik et al., 1996; Houlden et al., 2008; Mehnaz et al., 2009). In total 4.7 × 105 CFU g−1 fluorescent bacteria were counted, corresponding to 1.54% of total bacteria. Fifty colony isolates were collected based on observation of fluorescence under UV light (Table 1).
Various types of soil used for crop production support highly diverse microbial communities, and introduction of crop plants affects those communities by promoting or suppressing growth of indigenous bacteria (Houlden et al., 2008; Agaras et al., 2012). Whether the primary determinant of microbial community structure is soil properties or introduced plants remains controversial. Plants clearly have a substantial effect on microbial communities based on the composition of their root exudates, which is closely associated with nutritional status and growth stage (Houlden et al., 2008). The proportion of fluorescent bacteria relative to total count of culturable bacteria in rhizospheric soil or bulk soil has not been addressed in previous studies. Agaras et al. (2012) reported that fluorescent strains accounted for 10–94% of total Pseudomonas in various crop soils and bulk soils. Results of the present study suggest that selection by M. piperita root exudates may explain the much lower count (~107 CFU g−1) of culturable bacteria for this species than for other crop plants (~1010 CFU g−1; Agaras et al., 2012; Bogino et al., 2013).
Analysis of Root Exudates
Native bacterial strains have not been isolated from the rhizosphere of aromatic crop plants in previous studies, because EOs present in root exudates are generally presumed to have a toxic effect on microorganisms (Chen et al., 2004). We analyzed root exudates from in vitro cultured M. piperita by GC-MS (Figure 1), and evaluated fragmentation patterns by comparison with the NIST database, with acceptance of SI ≥ 800 (Table 2). No monoterpenes were detected in the root exudates, indicating that bacterial communities can be established in the M. piperita rhizosphere. The absence of EOs in root exudates suggests that the low total bacterial count did not reflect selection based on plant antimicrobial properties, e.g., inhibition or suppression of bacterial growth. Rather, the low count may result from competition or inhibition of established communities through direct growth inhibition by microbial compounds, or from adaptation by bacteria to the soil environment, edaphic conditions, and/or altitude.
Phenotypic Testing
All 50 isolated strains were subjected to phenotypic testing. Three strains representing the main groups of fluorescent Pseudomonas were used as references: P. fluorescens WCS417r, P. putida KT2440, and P. aeruginosa PAO I (Lysenko, 1961; Stanier et al., 1966; Palleroni, 2005). Biochemical characteristics of the isolated strains are summarized in Table 1. In terms of biochemical responses, 33 strains (66% of total) were most similar to P. putida KT2440, one (SJ34) was similar to P. fluorescens WCS417R, and none were similar to P. aeruginosa PAO I. The 50 strains were subjected to genotypic testing (Sec. 3.4) for identification of Pseudomonas species, and screened for PGPR activity mediated by production of mVOCs (Section Plant Growth-Promoting Effects of mVOCs). Strains SJ04, SJ7b, and SJ46 were included in the genotypic testing because they showed the strongest promoting effects on biomass and EO production of micropropagated M. piperita plants.
Genotypic Testing
Three molecular techniques (ARDRA, ERIC-PCR fingerprinting, BOX-PCR fingerprinting) were applied to evaluate similarities through clustering analysis of the isolated fluorescent Pseudomonas strains. Forty native strains and two reference strains (P. fluorescens WCS417r, P. putida KT2440) were evaluated. P. aeruginosa PAO I was excluded because none of the native strains showed biochemical similarity to it.
ARDRA
The ARDRA technique (analysis of16S rRNA gene fragments produced by digestion with restriction enzymes) has been widely applied in genetic diversity studies of Pseudomonas and other rhizobacteria. It has been used successfully for analysis of fluorescent Pseudomonas communities isolated from rhizospheres of various crop plants under a variety of environmental and climatic conditions (Prosser, 2002; Mehri et al., 2011; Nievas et al., 2012). It has also been used to study variation of Pseudomonas communities as a function of crop management strategies (Achouak et al., 2000) and application of pesticides or other chemical compounds (Viti and Giovannetti, 2005; Braun et al., 2006; Wang et al., 2008; Wu et al., 2009). The present study is the first to apply ARDRA to PGPR isolates from the M. piperita rhizosphere.
We used restriction endonucleases Alu I, Msp I, Hinf I, and Hae III because of their ability to discriminate among Pseudomonas strains (Achouak et al., 2000; Wu et al., 2009; Mehri et al., 2011). For each analyzed strain, a single amplicon (~1400 bp) was obtained using the primers rD1 and fD1. Only seven of the 40 markers gave monomorphic bands. AluI had the highest polymorphism, generating six different Restriction Fragment Length Polymorphism (RFLP) patterns. Hinf I and Hae III each generated four RFLP patterns, and MspI generated two RFLP patterns, resulting in less informative restriction (Table 4). 16S rRNA RFLP patterns obtained using each of the endonucleases were combined to obtain a general genotype for each strain. Genotype 1, the most frequent, was present in 16 native fluorescent strains. The genotype of P. putida KT2440 was also found in two other strains, while the genotype of P. fluorescensWCS417r was unique (Table 4).
A dendrogram was constructed as described in Section Analysis of Genotypic Data to determine genetic distances among native Pseudomonas strains (Figure 2). At 35% similarity, the dendrogram showed a division into two major clusters (A and B). At 45% similarity, cluster A grouped 25 strains having genotypes 1, 3 (including P. putida KT2440), 4, 6, 12 (including P. fluorescens WCS417r), and 14. Within cluster A, genotypes 1 and 3 were highly related (80%similarity). Cluster B grouped the remaining strains at 35% similarity. These strains had genotypes 2, 5, 7, 8, 9, 10, and 13. Within cluster B, genotypes 8 and 13, and 5 and 2 were highly related (80% similarity).
The genotypic analysis revealed high correlations between P. putida KT2440 and ≥ 50% of total fluorescent strains isolated from the M. piperita rhizosphere, consistent with the results of biochemical analysis. Strains with genotype 7 differed from P. putida KT2440 in both genotypic and phenotypic analyses. Strains with genotype 10, 2, 13, 9, 8, and 5 displayed genotypic divergence from P. putida KT2440 in spite of phenotypic similarities. Similar divergence was observed between strains with genotype 11 and P. fluorescens WCS417r (Table 4; Figure 2).
In a comparison of the phenotypic and genotypic results, clustering discrepancies may result from the differing characteristics evaluated by the methodologies. Phenotypic testing detects metabolic activities that may be shared among various species within a particular genus, whereas genotypic testing detects similarities or distances among genetic markers generated from a defined gene. Regardless of these considerations, 60% of native fluorescent rhizobacterial strains displayed high biochemical and genetic similarity with P. putida KT2440, and 50% of strains showed 80% genotypic similarity with KT2440. P. putida is clearly the major fluorescent strain present in the M. piperita rhizosphere.
Results have shown that the16SrRNA gene is highly conserved, causing difficulty in differentiating among closely related species. It is therefore preferable to apply two or more molecular techniques. Inclusion of techniques that identify sequences repeated along the whole genome permits analysis of more data, and of differences in sequences of more than one gene (Grimont, 2002; Rangel-Castro et al., 2002; Gomila et al., 2007; Naik et al., 2008; Mehri et al., 2011).
16S rRNA Gene Nucleotide Sequence Analysis
The analyzed strains were selected based on their abundance of genotypes and distribution in clusters A and B (Table 4). PGPR activity of strains on micropropagated M. piperita plantlets mediated by mVOC production (Section Plant Growth-Promoting Effects of mVOCs) was also taken into consideration.
The BLAST software program was used to analyze the obtained 16S rRNA gene sequences and to establish their alignment and identity with reference strains (Altschul et al., 1997). All of the analyzed strains had identities within the genus Pseudomonas. The native strains had high percentage identity with P. putida and P. fluorescens. Strains SJ13 and SJ48, and SJ17 and SJ25,had the same identity (Table 5). They should be considered different strains of the same species, in view of their very low similarity in ERIC-PCR and BOX-PCR analyses (Sections ERIC-PCR, BOX-PCR), which deeply probe strain diversity.
A phylogenetic tree was constructed to determine relationships and evolutionary distances among the fluorescent Pseudomonas strains isolated in this study, and the major, well-known species of this genus (Figure 3). We utilized the GenBank database because of its size and the ability to download sequences closely related to the fluorescent group (Altschul et al., 1997). Two major clusters appear in the phylogenetic tree: the first contains P. putida, P. fluorescens, P. protegens, P. syringae, P. chlororaphis, and P. corrugate; the second contains P. fulva, P. mendocina, P. nitroreducens, P. stutzeri, P. aeruginosa, and P. alcaligenes. The first cluster includes all the native fluorescent Pseudomonas strains isolated from the M. piperita rhizosphere, and can be divided into two subclusters centered on P. putida and P. fluorescens.
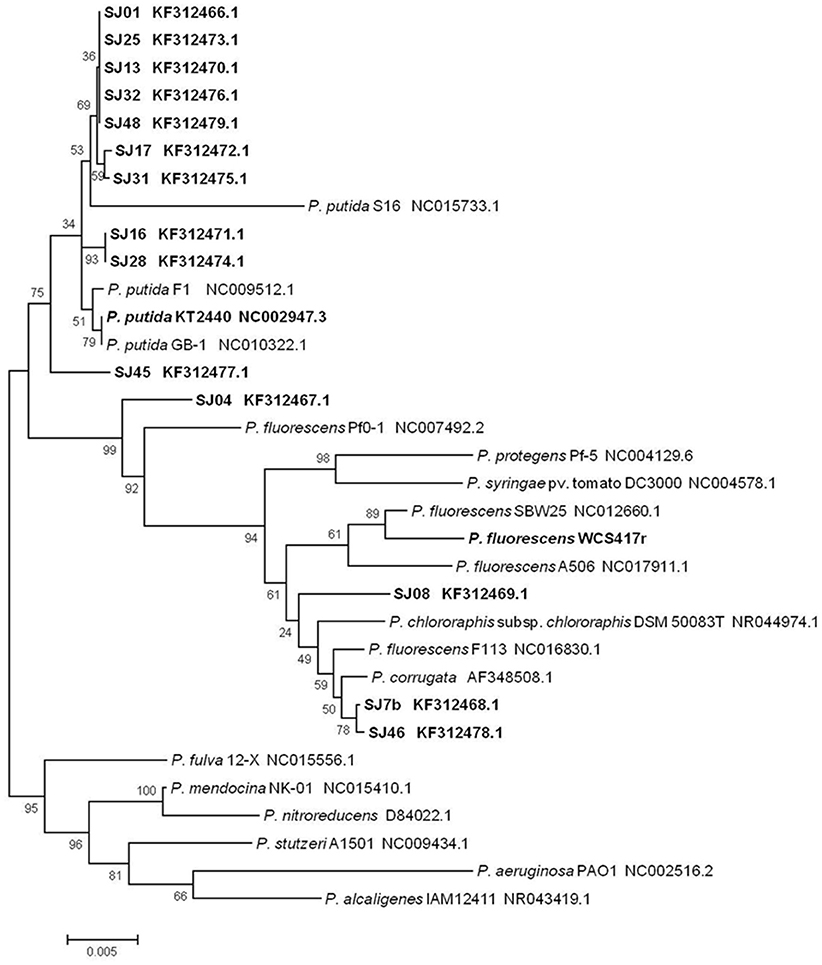
Figure 3. Phylogenetic tree of native and reference strains of fluorescent Pseudomonas based on aligned 16S rRNA sequences. Native strains isolated from the M. piperita rhizosphere are indicated by boldface. The tree was constructed based on multiple alignments and the neighbor-joining method, using the MEGA 4.0.2 software program. Bootstrap values on branches are shown as percentages from 1000 replications. Scale bar at bottom represents 5 nt substitution per 1000 nt sequences. GenBank accession numbers are shown.
Most of the native strains analyzed had high similarity with P. putida. The strains most strongly related to P. putida were SJ16 and SJ28, followed by SJ01, SJ13, SJ17, SJ25, SJ31, SJ32, and SJ48. Strain SJ45 was distant from this group. These findings are consistent with genotypic and phenotypic analyses, and confirm that P. putida is the major fluorescent species present in the M. piperita rhizosphere.
The remaining four strains were part of the P. fluorescens subcluster and were notably distant from the P. putida subcluster, again consistent with results of genotypic and phenotypic analyses. Strain SJ04was intermediate in phylogenetic position between the two subclusters, and showed a phylogenetic relationship with P. fluorescens in spite of its identity with P. putida (Table 4). Strains SJ7b and SJ46 had a stronger phylogenetic relationship with P. corrugate than with P. fluorescens, consistent with results of BLAST analysis. Strain SJ08 had a phylogenetic relationship with P. fluorescens and P. chlororaphis.
ERIC-PCR
Of the 40 native fluorescent strains analyzed, only 38 generated an appropriate profile; SJ32 and SJ35 did not show amplification under the conditions applied. The primers used generated 67 markers with sizes ranging from 100 to 1700 bp, represented only by polymorphic bands. The pairs SJ21/SJ22, SJ17/SJ18, and SJ38/SJ39 showed identical fingerprinting (100% similarity); i.e., each of these pairs was likely the same strain.
A dendrogram was constructed to determine genetic distance or proximity among the isolated strains (Figure 4). Clusters were generally observed at greater genetic distances than in ARDRA analysis, most likely because of the higher number of markers generated and their distribution along the whole genome. At 50% similarity, four groups (I-IV) were formed. The main group (II) included 19 strains and was divided into three subgroups (A-C). Within these subgroups, the most closely related strains were SJ16 and SJ20 (70% similarity), SJ29 and SJ47 (70% similarity), and strains SJ45 and SJ46 (65% similarity). Group I included reference strains P. fluorescensWCS417r and P. Putida KT2440 with 55% similarity. Groups III and IV were smaller than groups I and II.
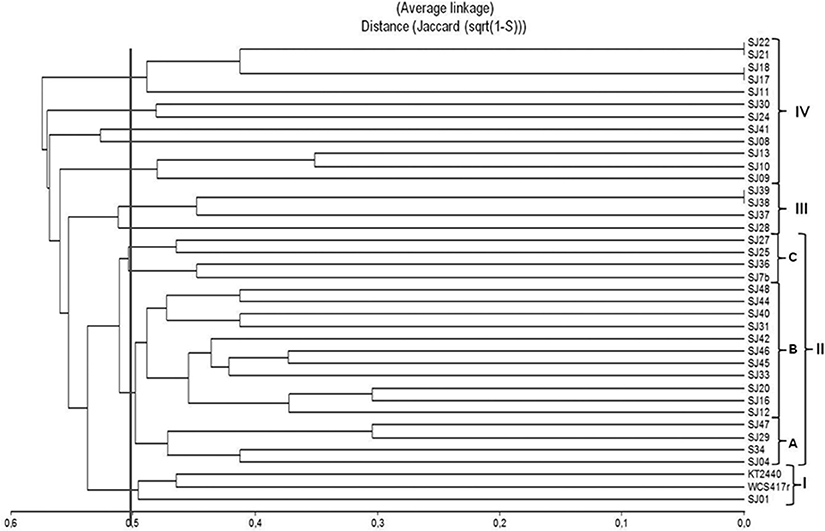
Figure 4. Dendrogram based on ERIC-PCR profiles, generated from native, and reference Pseudomonas strains using the UPGMA algorithm and Jaccard coefficient.
ERIC-PCR analysis did not allow us to distinguish between the two fluorescent reference strains because they presented 50% similarity, the same as observed for various native strains (Figure 4). In previous studies, this technique was used to evaluate diversity/similarity within groups of native Pseudomonas strains, and gave comparable degrees of similarity (Achouak et al., 2000; Wolska and Szweda, 2008; Selvakumar et al., 2009; Vyas et al., 2009). In contrast, it did not give high degrees of similarity for native fluorescent Pseudomonas isolated from the M. piperita rhizosphere, suggesting that these populations have considerable genetic diversity in spite of their phylogenetic affiliation with P. putida.
BOX-PCR
Each of the 40 analyzed strains generated an appropriate profile using the primer BOXA1R. This technique allowed a high degree of discrimination among native fluorescent strains isolated from the M. piperita rhizosphere. It is frequently applied for grouping of Pseudomonas strains isolated from soil samples (Naik et al., 2008; Vyas et al., 2009; Mehri et al., 2011; Parejko et al., 2012). Of the 40 strains, 39 generated an appropriate profile, whereas strain SJ25 did not undergo amplification under the conditions applied. In comparison with ERIC-PCR, BOX-PCR analysis generated a larger number of markers. A total of 104 discrete polymorphic bands were scored, ranging in size from 250 to 2000 bp, with 8 to 20 bands for each strain. Six strains presented the same profile, and were presumably identical.
A dendrogram was constructed to determine genetic distance or proximity among the isolated native strains (Figure 5). The BOX-PCR dendrogram was highly complex, and showed formation of a larger number of clusters at higher similarities. All strains grouped at 48% similarity, except for SJ01 which separated from the other strains at lower similarity. A large cluster (1) was formed at 50% similarity. It included the majority of strains (29), and was divided into five subclusters (A–E). The main subcluster (A), formed at ~55% similarity, included 15 of the native strains and reference strain P. fluorescens WCS417r. Within subcluster A, 100% similarity was obtained for three groups of strains: SJ09/SJ10/SJ13, SJ38/SJ39, and SJ31/SJ32. A smaller cluster (2) included high-similarity groups; e.g., SJ21/SJ22/SJ27 showed 100% similarity among themselves and 85% similarity with SJ29.SJ17/SJ18 grouped at 85% similarity. Reference strain P. putida KT2440 was not part of either cluster 1 or 2, and showed only low similarity (~55%) with cluster 1.
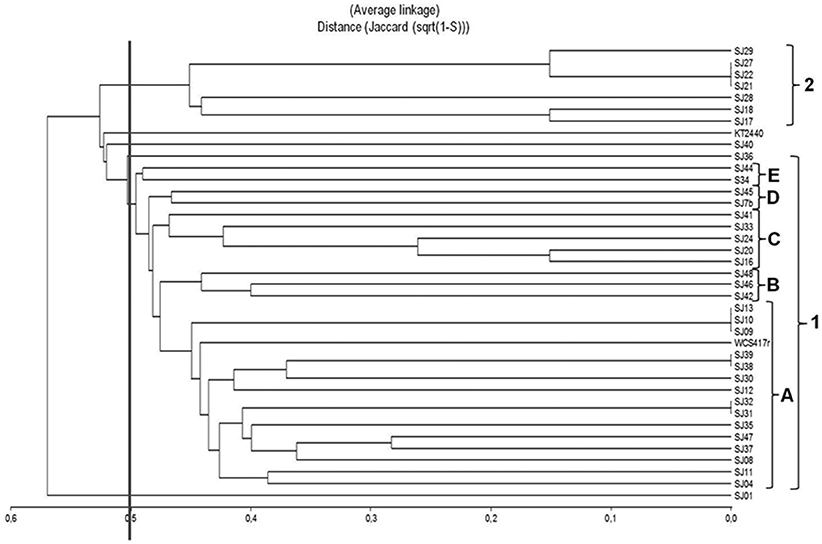
Figure 5. Dendrogram based on BOX-PCR profiles, generated from native, and reference Pseudomonas strains using the UPGMA algorithm and Jaccard coefficient.
In contrast to ERIC-PCR, BOX-PCR analysis did not give high similarity levels among reference strain P. putida KT2440 and the native strains, but did give high similarity levels among reference strain P. fluorescens WCS417r and the native strains. These rep-PCR techniques, applied together, thus provide a valuable tool for evaluating diversity of native fluorescent Pseudomonas strains isolated from the M. piperita rhizosphere, and most likely from rhizospheres of other aromatic species as well.
Comparison of Molecular Techniques Applied
In a comparison of the two rep-PCR techniques, ERIC-PCR showed better discrimination among native strains. The number of markers obtained was lower, resulting in relatively easy analysis and a high degree of clustering, i.e., lower similarity among native fluorescent Pseudomonas strains isolated from the M. piperita rhizosphere. At 80% similarity, 35 groups were obtained from ERIC-PCR analysis, vs. 30 groups obtained from BOX-PCR analysis. ERIC-PCR was useful for detailed study of differences in genomic DNA among closely related strains. BOX-PCR was useful for grouping strains while eliminating small differences among them, resulting in larger clusters. BOX-PCR produced a larger number (6) of duplicate profiles (Figure 4), i.e., a larger number of identical strains, than the number (3) obtained from ERIC-PCR (Figure 3). Previous studies gave similar degrees of heterogeneity by application of rep-PCR analysis in groups of isolated Pseudomonas strains (Selvakumar et al., 2009; Vyas et al., 2009; Wolska et al., 2011).
Simultaneous application of ARDRA and rep-PCR techniques could not distinguish among P. fluorescens and P. putida strains because of the 50% similarity with reference strains. However, both types of analysis revealed close relationships within the SJ09/SJ10/SJ13, SJ16/SJ20, and SJ31/SJ32 groups. The SJ17/SJ18, SJ21/SJ22, and SJ38/SJ39 groups showed high similarity in rep-PCR but lower similarity in ARDRA. Although BOX-PCR and ERIC-PCR sequences allow us to deeply probe genomic differences, they cannot be used as a tool to identify or determine phylogenetic positions of native fluorescent strains. Phylogenetic positioning is based on comparison of 16S rRNA gene sequences. ARDRA identifies small differences among native strains by evaluating this particular gene of interest. The combination of ARDRA and rDNA 16S sequencing clearly identified P. putida as the major fluorescent species in M. piperita rhizospheric soil.
Plant Growth-Promoting Effects of mVOCs
Biomass
Plants were analyzed after 30 days incubation with mVOCs. A significant (p ≤ 0.05) effect by mVOCs from native fluorescent Pseudomonas was observed for shoot fresh weight, which was 3.0 to 4.5-fold higher than control value. Values for shoot ramification number, node number, leaf number, root length, and dry weight did not differ significantly from control values (p > 0.05; Table 6). In one of our previous studies (Santoro et al., 2011), mVOCs from reference strain P. fluorescensWCS417r similarly increased shoot fresh weight of micropropagated M. piperita, but had no effect on leaf number or ramification number. The increase of shoot fresh weight can be attributed to greater leaf and stem size, leading to increased aerial biomass. We also observed a similar effect in Ocimum basilicum (basil) plants exposed to mVOCs from B. subtilis (Banchio et al., 2009). Zhang et al. (2007) explained the effect of mVOC exposure on plant biomass by demonstrating that stimulated genes were associated with cell wall structure, allowing expansion and reduced rigidity. They also observed reduced mVOC exposure in leaves, leading to enhanced shoot development through stimulation of elongation, and cellular differentiation (Zhang et al., 2008).
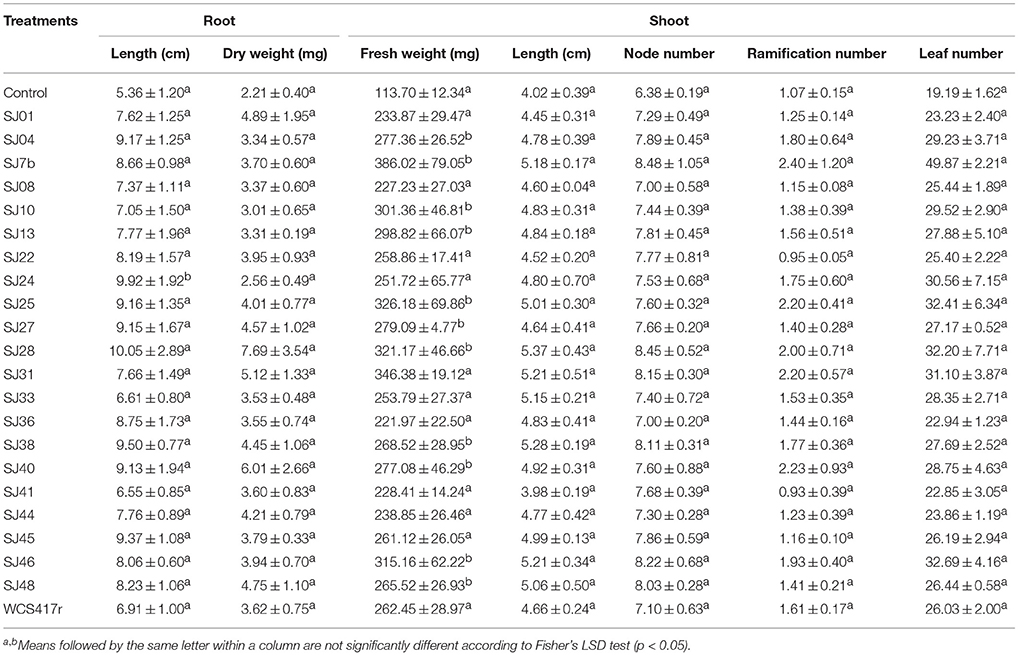
Table 6. Effects of mVOCs emitted by native fluorescent Pseudomonas strains on biomass parameters of micropropagated M. piperita plantlets.
EO Production
We examined the effect of mVOCs on EO production in micropropagated M. piperita plantlets. The major components of extracted EOs were (−) limonene, terpineol, (−) menthone, (+) menthofuran, (−) menthol, (+) pulegone, and (−) menthyl acetate.
Exposure to mVOCs from strains SJ25 and SJ27 resulted in 86 and 57% increases, respectively, in total EO production relative to control (Figure 6). Limonene content was increased 2-fold by exposure to SJ04, SJ28, or SJ7b mVOCs, and 4-fold by SJ48mVOCs (Figure 7). Menthol content was increased three-fold by SJ04 mVOCs (Figure 8). Menthofuran Limonene content was reduced by exposure to mVOCs from all strains, most notably (20% reduction) by SJ48 mVOCs (Figure 7). Menthyl acetate content was increased four-fold by SJ7b mVOCs (Figure 9). The changes mentioned above were statistically significant at p ≤ 0.05. Changes in menthone, terpineol, and pulegone content produced by exposure to mVOCs were not significant (p > 0.05; data not shown).
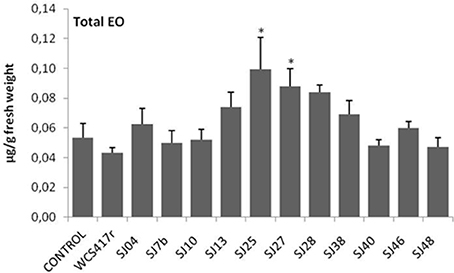
Figure 6. Effects of exposure to mVOCs of native and reference strains of fluorescent Pseudomonason EO content of micropropagated M. piperita plantlets. *Significant difference (p ≤ 0.05) from control value.
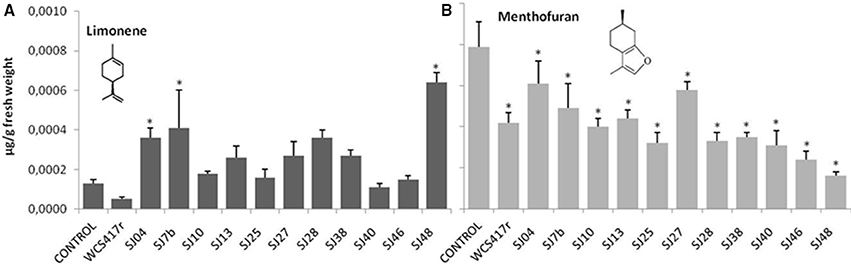
Figure 7. Effects of exposure to mVOCs of native and reference strains of fluorescent Pseudomonason limonene and menthofuran content of micropropagated M. piperita. *Significant difference (p ≤ 0.05) from control value.
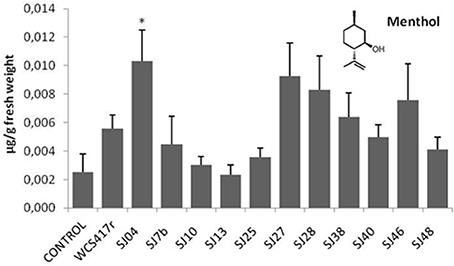
Figure 8. Effects of exposure to mVOCs of native and reference strains of fluorescent Pseudomonason menthol content of micropropagated M. piperita. *Significant difference (p ≤ 0.05) from control value.
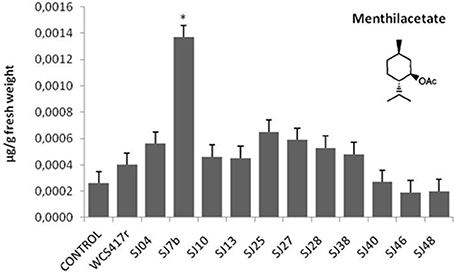
Figure 9. Effects of exposure to mVOCs of native and reference strains of fluorescent Pseudomonason menthylacetate content of micropropagated M. piperita. *Significant difference (p ≤ 0.05) from control value.
Content of menthol, the major monoterpene, was significantly elevated only by SJ04 mVOCs. Monoterpenes are the commercially valuable components among EOs (Gupta et al., 2002). The observed reduction of menthofuran content is related to the increase in menthol, because menthofuran represents a branch in the flux of terpene precursors to final production of menthol (Lange and Croteau, 1999; Turner and Croteau, 2004). The increased levels caused by SJ04, SJ7b, and SJ48 mVOCs may reflect a lack of maturation of EOs, considering that enzymes involved in later reactions are activated at ~30 days of foliar growth (McConkey et al., 2000). Thus, our findings suggest that mVOCs produced by native fluorescent strains stimulate monoterpene synthesis in micropropagated M. piperita.
Enhanced production of secondary metabolites may depend directly on improved nutritional status and on primary metabolism of plants following PGPR inoculation (Shulka et al., 1992; Giri et al., 2003; Farag et al., 2006). Studies by Zhang et al. (2007, 2008, 2009) suggest that plants exposed to mVOCs experience high availability of nutrients for growth and increased level of photosynthesis. These phenomena could lead to improved distribution of energy to support synthesis of secondary metabolites such as monoterpenes. Alternatively, they could reflect defensive responses of plants to bacterial colonization following PGPR inoculation, in view of the known antimicrobial properties of EOs (Hammerschmidt and Nicholson, 1999; Sangwan et al., 2001; Wittstock and Gershenzon, 2002). Studies by Radruppa et al. (2010) demonstrated induced systemic resistance (ISR) mediated by ethylene and salicylic acid in A. thaliana exposed to acetoin, a component of mVOCs produced by B. subtilis (Farag et al., 2006). Even in the absence of contact between a plant and bacterial mVOCs, enhanced EO production resulting from improved nutritional status could be mediated by improved defensive ability of the plant following mVOC exposure.
Chemical Analysis of mVOCs
mVOCs of native strains SJ04, SJ25, and SJ48, and reference strain P. fluorescens WCS417r, were analyzed using Super Q® filters. Only compounds not present in control samples (Table 7) were considered. Strain SJ48 showed the greatest mVOC production. Strains SJ25 and WCS417r had the same chromatographic profile. A total of 11 components were identified, belonging to 3 classes: hydrocarbons (cyclohexane, decane, dodecane, 2,6,10-trimethyl-tetradecane, dotriacontane, 11-decyl-docosane), aromatic compounds (methyl-benzene, 2-benzyloxy-ethanamine, 1-methylnonadecyl-benzene, 1-(N-phenylcarbamyl)-2-morpholinocyclohexene), and halogenated compounds (1-chlorooctadecane). Hydrocarbons such as cyclohexane and dodecane were detected in strains of the genera Bacillus (Farag et al., 2006) and Paenibacillus (Lee et al., 2012), and were found to increase plant biomass in A. thaliana. They also induced ISR status, even though they were not the major mVOC components. The hydrocarbons decane and dodecane were detected in Pseudomonas strains, but did not display specific biocontrol activity against the plant pathogenic fungus Sclerotinia sclerotiorum (Fernando et al., 2005).
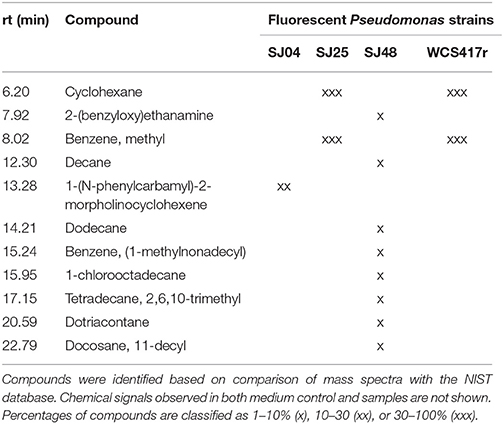
Table 7. mVOCs identified in headspace extracts of native and reference strains of fluorescent Pseudomonas.
We conclude that the obtained chromatographic profiles are characteristic of the three native strains analyzed. This is interesting considering that the strains were isolated from the same location and presumably subjected to similar selective pressures.
It should be noted that low chemical diversity in a pool of mVOCs does not necessarily relfect low activity of the producer strain. Previous studies, even when focused on the major compound in the mVOC chemical profile, have consistently concluded that the degree of the observed effect reflects synergism among the compounds present. Even when investigators evaluated promoting activity using standards, they did not observe the same level of response in the parameters analyzed in comparison with the entire mVOC pool (Fernando et al., 2005; Farag et al., 2006; Radruppa et al., 2010; Groenhagen et al., 2013).
From a functional point of view, the various mVOC pools did not generate the same effects. Each of the native fluorescent bacterial strains analyzed had different PGPR activity. Although the strains all produced an increase in total fresh weight, their enhancing effect on EO content was variable, be causing of differing mixtures of emitted mVOCs. For example, mVOCs produced by strain SJ25 increased total EO content without improving the composition. In contrast, SJ04 and SJ48mVOCs improved the composition of EOs produced by the plants. Exposure of plants to SJ04 mVOCs resulted in decreased menthofuran and increased limonene and menthol contents. Exposure to SJ48 mVOCs similarly resulted in decreased menthofuran and increased limonene. These strains improved the quality of extracted EOs because of their ability to reduce menthofuran production, causing a diversion to the menthol production pathway, the primary indicator of EO quality (Lange and Croteau, 1999; Turner and Croteau, 2004). Clearly, differing mVOC pools emitted by bacteria induce differential responses in micropropagated M. piperita, and allow us to improve both quantity and quality of EO production.
Conclusion
We identified Pseudomonas putida as the predominant fluorescent PGPR species present in rhizospheric soil of Mentha piperita (peppermint), using a combination of phenotypic testing and molecular techniques (ARDRA, rDNA 16S sequence analysis). This species accounted for ~60% of native strains analyzed. Other Pseudomonas strains present in the rhizospheric soil were also identified. The whole genome of the fluorescent bacterial population isolated was evaluated using rep-PCR techniques (ERIC-PCR, BOX-PCR), which revealed the overall complexity of the population and the degree of dissimilarity among strains.
This is the first study focused on fluorescent bacterial populations in rhizospheric soil of aromatic plants in general and M. piperita in particular. The isolated native bacterial strains promoted M. piperita growth, through mVOC production. Exposure of plants to mVOCs results in increased shoot fresh weight and EO production, through improved quality of EO composition. The obtained chromatographic profiles were characteristic of the three native strains analyzed, and each mVOC pool generated a different level of PGPR activity in micropropagated M. piperita plantlets, which is interesting considering that the strains were isolated from the same location and presumably subjected to similar selective pressures. Our findings provide a basis for extensive further studies, and demonstrate the importance of PGPR properties in analysis of isolated native strains, in terms of phylogenetic position and adaptation to environmental and edaphic conditions. The combined analytical approach used represents a significant biotechnological advance in understanding of growth promotion of aromatic plants by rhizobacteria.
Author Contributions
Conceived and designed the experiments: MS, EB. Performed the experiments: MS, PB, NN, LC. Analyzed the data: MS, PB, NN, LC, WG, EB. Contributed reagents/materials/analysis tools: MS, PB, NN, LC, WG, EB. Wrote the manuscript: MS, PB, EB.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by grants from the Secretaría de Ciencia y Técnica de la Universidad Nacional de Río Cuarto, the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), and the Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT), Argentina. EB, WG, and PB are Career Members of CONICET. NN, LC, and MS have fellowships from CONICET. The authors are grateful to Dr. S. Anderson for English editing of the manuscript.
References
Achouak, W., Thiery, J. M., Rouband, P., and Heulin, T. (2000). Impact of crop management on intraspecific diversity of Pseudomonas corrugata in bulk soil. FEMS Microbiol. Ecol. 31, 11–19. doi: 10.1111/j.1574-6941.2000.tb00666.x
Agaras, B., Wall, L. G., and Valverde, C. (2012). Specific enumeration and analysis of the community structure of culturable pseudomonads in agricultural soils under no-till management in Argentina. Appl. Soil Ecol. 61, 305–319. doi: 10.1016/j.apsoil.2011.11.016
Altschul, S., Madden, T., Schäffer, A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Balzarini, M. (2003). Aplicaciones del Análisis Estadístico Multivariado. Material teórico. Maestría en estadística aplicada; Universidad Nacional de Córdoba, Córdoba.
Banchio, E., Xie, X., Zhang, H., and Paré, P. W. (2009). Soil bacteria elevate essential oil accumulation and emissions in sweet basil. J. Agric. Food Chem. 57, 653–657. doi: 10.1021/jf8020305
Bogino, P., Abod, A., Nievas, F., and Giordano, W. (2013). Water-limiting conditions alter the structure and biofilm-forming ability of bacterial multispecies communities in the Alfalfarhizosphere. PLoS ONE 8:e79614. doi: 10.1371/journal.pone.0079614
Braun, B., Böckelmann, U., Grohmann, E., and Szewzyk, U. (2006). Polyphasic characterization of the bacterial community in an urban soil profile with in situ and culture-dependent methods. Appl. Soil Ecol. 31, 267–279. doi: 10.1016/j.apsoil.2005.05.003
Chen, F., Ro, D.-K., Petri, J., Gershenzon, J., Bohlmann, J., Pichersky, E., et al. (2004). Characterization of a root-specific Arabidopsis terpene synthase responsible for the formation of the volatile monoterpene 1,8-Cineole. Plant Physiol. 135, 1956–1966. doi: 10.1104/pp.104.044388
Di Renzo, J. A., Cas anoves, F., Gonzalez, L. A., Tablada, E. M., Díaz, M. P., Robledo, C. W., et al. (2001). Estadística para las Ciencias Agropecuarias, Cuarta Edición. Córdoba: Editorial Triunfar.
Farag, M. A., Ryu, C.-M., Sumner, L. W., and Paré, P. W. (2006). GC–MS SPME profiling of rhizobacterial volatiles reveals prospective inducers of growth promotion and induced systemic resistance in plants. Phytochemistry 67, 2262–2268. doi: 10.1016/j.phytochem.2006.07.021
Fernando, D. W., Ramarathnam, R., Krishnamoorthy, A., and Savchuk, S. (2005). Identification and use of potential bacterial organic antifungal volatiles in biocontrol. Soil Biol. Biochem. 37, 955–964. doi: 10.1016/j.soilbio.2004.10.021
Fuchs, R., Schäfer, M., Geoffroy, V., and Meyer, J.-M. (2001). Siderotyping – A powerful tool for the characterization of pyoverdines. Curr. Top. Med. Chem. 1, 31–57. doi: 10.2174/1568026013395542
Giri, B., Kapoor, R., and Mukerji, K. G. (2003). Influence of arbuscular mycorrhizal fungi and salinity on growth, biomass and mineral nutrition of Acacia auriculiformis. Biol. Fertil. Soils 38, 170–175. doi: 10.1007/s00374-003-0636-z
Gomila, M., Ramirez, A., and Lalucat, J. (2007). Diversity of environmental Mycobacterium isolates from hemodialysis water as shown by a multigene sequencing approach. Appl. Environ. Microbiol. 73, 3787–3797. doi: 10.1128/AEM.02934-06
Gordon, R. E., and Mihm, J. M. (1956). A comparative study of some strains received as Nocardia. J. Bacteriol. 73, 15–27.
Grimont, P. A. D. (2002). Risque bactériologique: repérage des souches. C. R. Biol. 325, 901–906. doi: 10.1016/S1631-0691(02)01499-3
Groenhagen, U., Baumgartner, R., Bailly, A., Gardiner, A., Eberl, L., Schulz, S., et al. (2013). Production of bioactive volatiles by different Burkholderia ambifaria strains. J. Chem. Ecol. 39, 892–906. doi: 10.1007/s10886-013-0315-y
Gupta, M. L., Prasad, A., Ram, M., and Kumar, S. (2002). Effect of vesicular-arbuscular mycorrhizal (VAM) fungus Glomus fasciculatum on the essential oil yield related characters and nutrient acquisition in the crops of different cultivars of menthol mint (Mentha arvensis) under field conditions. Bioresour. Technol. 81, 77–79. doi: 10.1016/S0960-8524(01)00109-2
Hammerschmidt, R., and Nicholson, R. (1999). “A survey of plant defense responses to pathogens,” in Induced Plant Defenses against Pathogens and Herbivores: Biochemistry, Ecology, and Agriculture, eds A. Agrawal, S. Tuzun, and E. Bent (Minnesota, SD: APS Press, St. Paul), 55–57.
Harrewijn, P., van Oosten, A. M., and Piron, P. G. M. (2001). Natural terpenoids as messengers. A multidisciplinary study of their production, biological functions and practical applications. Ann. Bot. 90, 299–300. doi: 10.1093/aob/mcf187
Higgins, D., Thompson, J., Gibson, T., Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Hoagland, D. R., and Arnon, D. I. (1938). The Water-Culture Method for Growing Plants without Soil. Univ. Calif. Coll. Agric. Exp. Sta. Circ., Berkeley.
Houlden, A., Timms-Wilson, T. M., Day, M. J., and Bailey, M. J. (2008). Influence of plant developmental stage on microbial community structure and activity in the rhizosphere of three field crops. FEMS Microbiol. Ecol. 65, 193–201. doi: 10.1111/j.1574-6941.2008.00535.x
Hu, H. B., Xu, Y. Q., Chen, F., Zhang, X. H., and Hur, B. K. (2005). Isolation and characterization of a new fluorescent Pseudomonas strain that produces both phenazine-1-carboxylic acid and pyoluteorin. J. Microbiol. Biotechnol. 15, 86–90.
Kimura, M. (1980). A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
King, E. O., Ward, M. K., and Randey, D. E. (1954). Two simple media for the demonstration of pyocyanin and fluorescein. J. Lab. Clin. Med. 44, 301–307.
Kloepper, J. W., and Schroth, M. N. (1978). “Plant growth-promoting rhizobacteria in radish,” in Proceeding of the 4th International Conference on Plant Pathogenic Bacteria, Vol. 2. Angers: Station de Pathologic Vegetal et Phytobacteriologic, 879–882.
Lange, B. M., and Croteau, R. (1999). Genetic engineering of essential oil production in mint. Curr. Opin. Plant Biol. 2, 139–144. doi: 10.1016/S1369-5266(99)80028-4
Lee, B., Farag, M. A., Park, H. B., Kloepper, J. W., Lee, S. H., and Ryu, C.-M. (2012). Induced resistance by a long-chain bacterial volatile: elicitation of plant systemic defense by a C13 volatile produced by Paenibacillus polymyxa. PLoS ONE 7:e48744. doi: 10.1371/journal.pone.0048744
Loaces, I., Ferrando, L., and Scavino, A. F. (2011). Dynamics, diversity and function of endophytic siderophore-producing bacteria in rice. Microb. Ecol. 61, 606–618. doi: 10.1007/s00248-010-9780-9
Luria, S. E., and Burrous, J. W. (1955). Hybridization between Escherichia coli and Shigella. J. Bacteriol. 74, 461–476.
Lysenko, O. (1961). Pseudomonas - an attempt at a general classification. J. Gen. Microbiol. 25, 379–408. doi: 10.1099/00221287-25-3-379
Mavrodi, D. V., Bonsall, R. F., Delaney, S. M., Soule, M. J., Phillips, G., and Thomashow, L. S. (2001). Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J. Bacteriol. 183, 6454–6465. doi: 10.1128/JB.183.21.6454-6465.2001
McConkey, M. E., Gershenzon, J., and Croteau, R. B. (2000). Developmental regulation of monoterpene biosynthesis in the glandular trichomes of peppermint. Plant Physiol. 122, 215–224. doi: 10.1104/pp.122.1.215
Medina, P., and Baresi, L. (2007). Rapid identification of gelatin and casein hydrolysis using TCA. J. Microbiol. Meth. 69, 391–393. doi: 10.1016/j.mimet.2007.01.005
Mehnaz, S., Weselowski, B., Aftab, F., Zahid, S., Lazarovits, G., and Iqbal, J. (2009). Isolation, characterization and effect of fluorescent pseudomonads on micropropagated sugarcane. Can. J. Microbiol. 55, 1007–1011. doi: 10.1139/W09-050
Mehri, I., Turki, Y., Chair, M., Chérif, H., Hassen, A., Meyer, J. M., et al. (2011). Genetic and functional heterogeneities among fluorescent Pseudomonas isolated from environmental samples. J. Gen. Appl. Microbiol. 57, 101–114. doi: 10.2323/jgam.57.101
Naik, P. R., Raman, G., Narayanan, K. B., and Sakthivel, N. (2008). Assessment of genetic and functional diversity of phosphate solubilizing fluorescent pseudomonads isolated from rhizospheric soil. BMC Microbiol. 8:230. doi: 10.1186/1471-2180-8-230
Nievas, F., Bogino, P., Nocelli, N., and Giordano, W. (2012). Genotypic analysis of isolated penaut-nodulating rhizobial strains reveals differences among populations obtained from soils with different cropping histories. Appl. Soil Ecol. 53, 74–82. doi: 10.1016/j.apsoil.2011.11.010
Palleroni, N. J. (2005). “Genus Pseudomonas,” in Bergey's Manual of Systematic Bacteriology, 2nd Edn, Vol. 2., eds D. J Brenner, N. R. Krieg, and J. T. Staley, (New York, NY: Springer), 323–379.
Parejko, J. A., Mavrodi, D. V., Mavrodi, O. V., Weller, D. M., and Thomashow, L. S. (2012). Population structure and diversity of phenazine-1-carboxylic acid producing fluorescent Pseudomonas spp. from dryland cereal fields of central Washington State (USA). Microb. Ecol. 64, 226–241. doi: 10.1007/s00248-012-0015-0
Patten, C. L., and Glick, B. R. (2002). Role of Pseudomonas putida indole acetic acid in development of the host plant root system. Appl. Environ. Microbiol. 68, 3795–3801. doi: 10.1128/AEM.68.8.3795-3801.2002
Prosser, J. I. (2002). Molecular and functional diversity in soil microorganisms. Plant Soil 244, 9–17. doi: 10.1023/A:1020208100281
Radruppa, T., Biedrzycki, M. L., Kunjeti, S. G., Donofrio, N. M., Czymmek, K. J., Paré, P. W., et al. (2010). The rhizobacterial elicitor acetoin induces systemic resistance in Arabidopsis thaliana. Commun. Integr. Biol. 3, 130–138. doi: 10.4161/cib.3.2.10584
Ram, D., Ram, M., and Singh, R. (2006). Optimization of water and nitrogen application to menthol mint (Mentha arvensis L.) through sugarcane trash mulch in a sandy loam soil of semi-arid subtropical climate. Bioresour. Technol. 97, 886–893. doi: 10.1016/j.biortech.2005.04.047
Rameshkumar, N., Ayyadurai, N., Kayalvizhi, N., and Gunasekaran, P. (2012). Genotypic and phenotypic diversity of PGPR fluorescent pseudomonads isolated from the rhizosphere of sugarcane (Saccharum officinarum L.). J. Microbiol. Biotechnol. 22, 13–24. doi: 10.4014/jmb.1107.07025
Rangel-Castro, J. I., Levenfors, J. J., and Danell, E. (2002). Physiological and genetic characterization of fluorescent Pseudomonas associated with Cantharellus cibarius. Can. J. Microbiol. 48, 739–748. doi: 10.1139/w02-062
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Salisbury, W. A., and Lykos, J. J. (1972). Hydrolysis of casein: a differential aid for the indentification of Serratia marcescens. J. Clin. Path. 25, 1083–1085. doi: 10.1136/jcp.25.12.1083
Sangwan, V., Foulds, I., Singh, J., and Dhindsa, R. S. (2001). Cold–activation of Brassica napus BN115 promoter is mediated by structural changes in membranes and cytoskeleton, and requires Ca2+ influx. Plant J. 27, 1–12. doi: 10.1046/j.1365-313x.2001.01052.x
Santoro, M. V., Zygadlo, J., Giordano, W., and Banchio, E. (2011). Volatile organic compounds from rhizobacteria increase biosynthesis of essential oils and growth parameters in peppermint (Mentha piperita). Plant Physiol. Biochem. 49, 177–1082. doi: 10.1016/j.plaphy.2011.07.016
Selvakumar, G., Piyush, J., Mishra, P. K., Bisht, J. K., and Gupta, H. S. (2009). Mountain aspect influences the genetic clustering of rhizospheric psychrotolerant phosphate solubilizing Pseudomonads in the Uttarakhand Himalayas. Curr. Microbiol. 50, 432–438. doi: 10.1007/s00284-009-9456-1
Shukui, Z., Lu, X., Dong, L., Xing, J., Su, X., Kong, H., et al. (2005). Quantitative determination of compounds in tobacco essential oils by comprehensive two-dimensional gas chromatography coupled to time-of-flight mass spectrometry. J. Chromatogr. 1086, 107–114. doi: 10.1016/j.chroma.2005.04.007
Shulka, A., Abad Farooqi, A. H., Shukla, Y. N., and Sharma, S. (1992). Effect of triacontanol and chlormequat on growth, plant hormones and artemisinin yield in Artemisia annuaL. Plant Growth Regul. 11, 165–171. doi: 10.1007/BF00024071
Sierra, G. (1957). A simple method for the detection of lipolytic activity of microorganisms and some observations on the influence of the contact between cells and fatty substrates. Antonie Van Leeuwenhoek 23, 15–22. doi: 10.1007/BF02545855
Sneath, P. H. A. (1956). Cultural and biochemical characteristics of the genus Chromobacterium. J. Gen. Microb. 15, 70–98. doi: 10.1099/00221287-15-1-70
Stanier, R. Y., Palleroni, N. J., and Doudoroff, M. (1966). The aerobic pseudomonads: a taxonomic study. J. Gen. Microbiol. 43, 159–271. doi: 10.1099/00221287-43-2-159
Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599. doi: 10.1093/molbev/msm092
Torsvik, V., Sorheim, R., and Goksoyr, J. (1996). Total bacterial diversity in soil and sediment communities-a review. J. Ind. Microbiol. 17, 170–178. doi: 10.1007/BF01574690
Turner, G. W., and Croteau, R. (2004). Organization of monoterpene biosynthesis in Mentha. Immunocytochemical localizations of geranyl diphosphate synthase, limonene-6-hydroxylase, isopiperitenol dehydrogenase and pulegone reductase. Plant Physiol. 136, 4215–4227. doi: 10.1104/pp.104.050229
Urzí, C., Brusetti, L., Salamone, P., Sorlini, C., Stackebrandt, E., and Daffonchio, D. (2001). Biodiversity of Geodermatophilaceae isolated from altered stones and monuments in the Mediterranean basin. Environ. Microb. 3, 471–479. doi: 10.1046/j.1462-2920.2001.00217.x
Versalovic, J., Koeuth, T., and Lupski, J. R. (1991). Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19, 6823–6831. doi: 10.1093/nar/19.24.6823
Versalovic, J. S. M., de Bruijn, F. J., and Lupski, J. R. (1994). Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol. Cell Biol. 5, 25–40.
Viti, C., and Giovannetti, L. (2005). Characterization of cultivable heterotrophic bacterial communities in Cr-polluted and unpolluted soils using Biolog and ARDRA approaches. Appl. Soil Ecol. 28, 101–112. doi: 10.1016/j.apsoil.2004.07.008
Vyas, P., Rahi, P., and Gulati, A. (2009). Stress tolerance and genetic variability of phosphate-solubilizing fluorescent Pseudomonas from the cold deserts of the trans-Himalayas. Microb. Ecol. 58, 425–434. doi: 10.1007/s00248-009-9511-2
Wang, M. C., Liu, Y. H., Wang, Q., Gong, M., Hua, X. M., Pang, Y. J., et al. (2008). Impacts of methamidophos on the biochemical, catabolic, and genetic characteristics of soil microbial communities. Soil Biol. Biochem. 40, 778–788. doi: 10.1016/j.soilbio.2007.10.012
Weisburg, W. G., Barns, S. M., Pelletier, D. A., and Lane, D. J. (1991). 16S Ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703.
Willems, A., Coopman, R., and Gillis, M. (2001). Phylogenetic and DNA–DNA hybridization analyses of Bradyrhizobium species. Int. J. Syst. Evol. Microbiol. 51, 111–117. doi: 10.1099/00207713-51-1-111
Wittstock, U., and Gershenzon, J. (2002). Constitutive plant toxins and their role in plant defense. Curr. Opin. Plant Biol. 5, 300–307. doi: 10.1016/S1369-5266(02)00264-9
Wolska, K., Kot, B., Jakubczak, A., and Rymuza, K. (2011). BOX-PCR is an adequate tool for typing of clinical Pseudomonas aeruginosa isolates. Folia Histochem. Cytobiol. 49, 734–738. doi: 10.5603/FHC.2011.0098
Wolska, K., and Szweda, P. (2008). A Comparative evaluation of PCR ribotyping and ERIC PCR for determining the diversity of clinical Pseudomonas aeruginosa isolates. Pol. J. Microbiol. 57, 157–163.
Wu, M., Zhang, X., Zhang, H., Zhang, Y., Li, X., Zhou, Q., et al. (2009). Soil Pseudomonas community structure and its antagonism towards Rhizoctonia solani under the stress of acetochlor. Bull. Environ. Contam. Toxicol. 83, 313–317. doi: 10.1007/s00128-009-9731-7
Zhang, H., Kim, M. S., Krishnamachari, V., Payton, P., Sun, Y., and Crimson, M. (2007). Rhizobacterial volatile emissions regulate auxin homeostasis and cell expansion in Arabidopsis. Planta 226, 839–851. doi: 10.1007/s00425-007-0530-2
Zhang, H., Sun, Y., Xie, X., Kim, M. S., Dowd, S. E., and Paré, P. W. (2009). A soil bacterium regulates plant acquisition of iron via deficiency-inducible mechanisms. Plant J. 58, 568–577. doi: 10.1111/j.1365-313X.2009.03803.x
Keywords: Mentha, fluorescent Pseudomonas, genotyping, ARDRA, essential oils, microbial volatile organic compounds
Citation: Santoro MV, Bogino PC, Nocelli N, Cappellari LdR, Giordano WF and Banchio E (2016) Analysis of Plant Growth-Promoting Effects of Fluorescent Pseudomonas Strains Isolated from Mentha piperita Rhizosphere and Effects of Their Volatile Organic Compounds on Essential Oil Composition. Front. Microbiol. 7:1085. doi: 10.3389/fmicb.2016.01085
Received: 25 March 2016; Accepted: 28 June 2016;
Published: 19 July 2016.
Edited by:
Dirk Balmer, Syngenta Crop Protection, SwitzerlandReviewed by:
Bryan Bailey, United States Department of Agriculture, USALei Zhang, North Carolina State University, USA
Copyright © 2016 Santoro, Bogino, Nocelli, Cappellari, Giordano and Banchio. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Erika Banchio, ZWJhbmNoaW9AZXhhLnVucmMuZWR1LmFy
 Maricel V. Santoro
Maricel V. Santoro Erika Banchio
Erika Banchio