- 1The ithree Institute, University of Technology Sydney, Sydney, NSW, Australia
- 2NSW Department of Primary Industries, Elizabeth Macarthur Agricultural Institute, Sydney, NSW, Australia
- 3Institute of Food Research, Norwich Research Park, Norwich, UK
Contamination of waste effluent from hospitals and intensive food animal production with antimicrobial residues is an immense global problem. Antimicrobial residues exert selection pressures that influence the acquisition of antimicrobial resistance and virulence genes in diverse microbial populations. Despite these concerns there is only a limited understanding of how antimicrobial residues contribute to the global problem of antimicrobial resistance. Furthermore, rapid detection of emerging bacterial pathogens and strains with resistance to more than one antibiotic class remains a challenge. A comprehensive, sequence-based genomic epidemiological surveillance model that captures essential microbial metadata is needed, both to improve surveillance for antimicrobial resistance and to monitor pathogen evolution. Escherichia coli is an important pathogen causing both intestinal [intestinal pathogenic E. coli (IPEC)] and extraintestinal [extraintestinal pathogenic E. coli (ExPEC)] disease in humans and food animals. ExPEC are the most frequently isolated Gram negative pathogen affecting human health, linked to food production practices and are often resistant to multiple antibiotics. Cattle are a known reservoir of IPEC but they are not recognized as a source of ExPEC that impact human or animal health. In contrast, poultry are a recognized source of multiple antibiotic resistant ExPEC, while swine have received comparatively less attention in this regard. Here, we review what is known about ExPEC in swine and how pig production contributes to the problem of antibiotic resistance.
Antimicrobial Resistance in Agricultural and Clinical Settings
Meat production farming practices have increased in scale over the past 30 years (Landers et al., 2012) and are predicted to continue to increase in line with rising incomes in low- and middle-income countries (Van Boeckel et al., 2015). This rationalization of farming has led to intensification of animal production and greater reliance on antimicrobials to control infectious disease, improve feed conversion efficiency, and promote animal growth (Levy, 1992; Sarmah et al., 2006; FAO, 2009; Krishnasamy et al., 2015; Van Boeckel et al., 2015). Modeling studies estimate that global antimicrobial consumption will increase by 67% from an estimated 63,151 tons in 2010 to 105,596 tons in 2030 (Van Boeckel et al., 2015). Much of this rise in consumption is expected to come from China, India, Russia, South Africa, and Brazil; countries where meat consumption is growing due to rising incomes (Van Boeckel et al., 2015). Large scale animal production facilities generate huge volumes of animal waste that is contaminated with veterinary antibiotics. While estimates vary, it is thought that in the USA, 11–14 million kilograms of antibiotics are used annually in the production of animals for food (i.e., livestock; Institute of Medicine; National Research Council; Panel on Animal Health, 1999; Landers et al., 2012). Almost half of these are used for non-therapeutic purposes, which is significantly more than the estimated 1.4 million kilograms of antibiotics used in human medicine (Institute of Medicine; National Research Council; Panel on Animal Health, 1999). Many classes of antibiotics are used in animal production including β-lactams, sulfonamides, tetracyclines, streptogramins, macrolides, lincosamides, polyethers, quinoxalines, elfamycins, glycolipids, arsenicals, and polypeptides (Sarmah et al., 2006). How these compounds are modified within the animal, how they persist in soils and waterways after they are excreted, and their fate when animal slurries are used to fertilize pastures are complex and poorly understood (Sarmah et al., 2006; Zhu et al., 2013). Antimicrobial residues that contaminate groundwater tables, aquatic environments, and land used for crop production exert selection pressure on microbial populations that may promote the lateral transfer of resistance and virulence genes between populations, and contributes to the emergence of novel pathogenic profiles (Venturini et al., 2010, 2013; Baquero and Tobes, 2013; Baquero et al., 2013; Roy Chowdhury et al., 2015). It is perhaps unsurprising that commensal gastrointestinal bacterial populations are commonly resistant to more than one antimicrobial (Bettelheim et al., 2003; Diarrassouba et al., 2007; Bailey et al., 2010) and consequently the frequency of community-acquired infections with resistance to multiple antimicrobials are also increasing (Onken et al., 2015; Paniagua-Contreras et al., 2015; Yahiaoui et al., 2015). Furthermore, surveys have shown the presence of antibiotic resistance genes in the gastrointestinal tracts of humans and animals that have never received antibiotic interventions, specifically in: human feces from remote communities (Bartoloni et al., 2009; Ruppe et al., 2009; Eisenberg et al., 2012), migratory bird populations (Foti et al., 2011; Carroll et al., 2015; Jamborova et al., 2015; Stedt et al., 2015) and various wildlife species (Allen et al., 2010; Baez et al., 2015; Jobbins and Alexander, 2015; Katakweba et al., 2015). This demonstrates that resistance genes in host bacteria can readily spread to naïve microbial populations via waterways, air currents, road incursions into remote areas, animal and bird migration, and land clearance (Tsubokura et al., 1995; Middleton and Ambrose, 2005; Ewers et al., 2009). The application of manure onto pasture from ruminants that have not received recent antibiotic therapy is known to stimulate the expansion of resident flora encoding resistance to β-lactam antibiotics (Udikovic-Kolic et al., 2014). These observations demonstrate our limited understanding of the impact of the application animal waste to agriculture. The mechanisms by which mobile elements flux through microbial populations are not fully understood, but it is clear that efforts must be made to reduce environmental contamination sourced from human and animal effluent.
Antimicrobial resistance is largely an ecological problem (Summers, 2002, 2006; Stokes and Gillings, 2011; Landers et al., 2012; Berendonk et al., 2015; Gillings et al., 2015). In general, studies have focused on the emergence and carriage of resistance genes in certain key microbial populations, without an appreciation for the wider complexity of the problem, or its origins. Hence, there are numerous under-reported microbial sources of resistance genes. Antibiotic resistance genes often traffic in association with transposons that carry mercury resistance operons; these play an important role in transforming mercury to less biologically toxic forms, thereby regulating the availability of toxic mercury compounds in the environment. Mercury-resistance gene operons have evolved as key components of Tn3-family transposons and these transposons are globally disseminated. Mercury has been released into the environment via geological processes over millions of years and, although anthropogenic activity over the past two centuries has contributed significantly to the release of mercury into waterways and the atmosphere, it is likely that mercury levels were higher prior to the industrial revolution than they are now as a result of extensive volcanic activity (Osborn et al., 1997; Barkay et al., 2003). Despite this, large scale surveillance studies of antimicrobial resistance fail to screen microbial populations for the presence of mercury-resistance genes (Djordjevic et al., 2013). Resistance genes can also accumulate in microbial populations in soils that have been fertilized with animal waste and treated effluent (Heuer and Smalla, 2007; Chee-Sanford et al., 2009; Heuer et al., 2011a,b; Kristiansson et al., 2011; Baquero et al., 2013; Berendonk et al., 2015). Meat, seafood, and vegetables are also important and under-recognized sources of antimicrobial resistance genes in bacteria (Johnson et al., 2005a; Agerso et al., 2014; Kim and Woo, 2014; Ahmed and Shimamoto, 2015a,b; Ben Said et al., 2015; Le et al., 2015; van Hoek et al., 2015; Zhang et al., 2015; Zurfluh et al., 2015).
Anthropogenic industrial processes generate heavy metals including cadmium, lead, arsenic, copper, silver, and a wide range of synthetic organic compounds that pollute natural ecosystems. As such, heavy metal contamination represents yet another selective pressure for the development of antimicrobial resistance (Baker-Austin et al., 2006; Castillo et al., 2008; Li et al., 2010; Zhu et al., 2013). Genes coding for resistance to silver, copper, arsenic and antimony are associated with complex resistance gene loci in bacteria resistant to more than one antimicrobial and have great significance to human, livestock, and plant health (Berg et al., 2005; Seiler and Berendonk, 2012; Hobman and Crossman, 2015; Bondarczuk et al., 2016). The widespread occurrence of genes coding for resistance to heavy metals has, in part, been demonstrated in studies using information mined from publicly available genome sequences highlighting the value of genomic surveillance studies (Hobman and Crossman, 2015).
Genome-wide studies of genes coding for resistance to antibiotics, biocides and heavy metals have shed light on how these genes co-assemble on mobile genetic elements. In an analysis of 2522 fully sequenced bacterial genomes and 4582 plasmid sequences, Pal et al. (2015) were able to identify examples where antimicrobial resistance genes co-occurred. The authors found that the most likely co-selection scenario occurred in bacterial strains that carried plasmid-borne antibiotic resistance genes and biocide/heavy metal resistance genes on the same chromosome. A plasmid localized gene cluster coding for a cadmium/zinc resistance gene (cadD) with aminoglycoside and macrolide resistance genes was also identified (Pal et al., 2015). Resistance to these combinations of antimicrobials would benefit bacterial populations found in intensive animal production systems where cadmium and zinc are both used to promote growth and aminoglycosides and macrolides are frequently administered for the treatment of Gram-negative and Gram-positive infections, respectively (Castillo et al., 2008; Li et al., 2010). It is clear that plasmids can carry combinations of genes coding for resistance to more than one microbial agent alongside genes coding for resistance to biocides and heavy metals. Invariably these plasmids are large, conjugative and commonly found in pathogens from hospital and intensive farming environments (Chen et al., 2007; Woodford et al., 2009; Venturini et al., 2010, 2013; Pal et al., 2015). Sub-lethal levels of antibacterial biocides and heavy metals found in mildly polluted aquatic and land-based ecosystems may be sufficient to maintain these mobile elements carrying multiple antibiotic, biocide and heavy metal resistance genes (Stepanauskas et al., 2006; Gullberg et al., 2014).
Although plasmids can vary in size and coding ability, their size is generally positively correlated with the size of the host chromosome (Smillie et al., 2010). Plasmids conferring multiple-drug resistance are usually large, as they code for a suite of resistance genes, alongside associated integrons, transposition factors, and genes for heavy metal resistance, which can be in excess of 20,000 bp or more in size. Small plasmids that acquire resistance to more than one antimicrobial are, however, not without precedent. Labar et al. (2012) described the sequence of a small plasmid (pASL01A; 27,072 bp) that codes for resistance to ampicillin, streptomycin, sulfonamides, trimethoprim, and mercury (Labar et al., 2012). Most of the plasmid (21,904 bp) is comprised of a Tn21 derivate mercury resistance transposon that carries a class 1 integron with a trimethoprim resistance dfrA7 gene cassette and Tn6029C (Roy Chowdhury et al., 2015). Tn6029C comprises a blaTEM-1-IS26-repA/C-sul2-strA-strB gene cluster flanked by direct copies of IS26 (Labar et al., 2012; Roy Chowdhury et al., 2015). The plasmid backbone (5,168 bp) and related plasmids are widespread in enterobacterial populations in Africa (Labar et al., 2012).
Antimicrobials, including heavy metals, are used widely in animal production to treat clinical disease, to prevent disease outbreaks during critical or vulnerable periods, and to promote growth (McEwen and Fedorka-Cray, 2002; Landers et al., 2012). The use of antibiotics to promote growth is a phenomenon whereby animals receiving very low doses of antimicrobials appear healthier and grow larger (Roy Chowdhury et al., 2014). Although, the mechanism for this growth promoting effect is still poorly understood, the majority of pigs produced in the USA are exposed to tetracyclines and tylosin to prevent disease and promote growth (Apley et al., 2012). While these approaches have proved effective in controlling disease and aiding farms globally, the uninhibited use of antimicrobials also provides the selection pressure that drives antimicrobial resistance (Pulcini and Gyssens, 2013). As such, concerted efforts to reduce the use of antimicrobials in food production should be a priority.
Given that the persistence and spread of resistance is intrinsically linked with the presence and activity of antimicrobials, it is significant that 30–90% of ingested antibiotics are excreted in an un-metabolized or only partially metabolized form (Sarmah et al., 2006) into waste treatment plants and animal waste-holding facilities. Antibiotic residues are known to persist in secondary effluent despite the treatment process (Sarmah et al., 2006; Watkinson et al., 2007; Martinez, 2008; Le-Minh et al., 2010; Bondarczuk et al., 2016). Furthermore, the pH, temperature, nutrient concentration, and bacterial loads in waste treatment plants are conducive to the evolution and spread of antimicrobial resistance (and virulence) genes (Andersen, 1993; Dolejska et al., 2011). This is in part evidenced by an increase in the presence of the class 1 integrase gene, a reliable proxy for resistance to more than one antimicrobial in bacterial populations sampled in wastewater treatment plants (LaPara et al., 2011; Ma et al., 2013; Gillings et al., 2015), and an increase in the number of resistance genes in wastewater treatment plants and rural domestic wastewater treatment systems (Gao et al., 2012; Chen and Zhang, 2013; Ju et al., 2015; Mao et al., 2015). This indicates that controlling the use of antimicrobials is a prerequisite for slowing the development and spread of pathogens with multiple antimicrobial resistance mechanisms.
Antimicrobial Resistance in Escherichia coli
Escherichia coli is a widespread and abundant organism capable of causing a wide range of gastrointestinal and extra-intestinal diseases. It can survive and proliferate in a diversity of terrestrial and aquatic environments (van Elsas et al., 2011). As such, its genetic profile is shaped profoundly by horizontal gene transfer, and it is a useful marker species for understanding how antimicrobial resistance and virulence genes accumulate over evolutionary time. Horizontal gene transfer events are known to generate novel combinations of putative and established virulence factors, and generate novel pathotypes of E. coli (Wu et al., 2007; Bielaszewska et al., 2014). Although, E. coli is a commensal inhabitant of the gastrointestinal tract of warm-blooded animals, pathotypes have evolved that cause intestinal [intestinal pathogenic E. coli (IPEC)] and extra-intestinal [extraintestinal pathogenic E. coli (ExPEC)] diseases. Different pathotypes of E. coli cause diarrheal disease, hemolytic uremic syndrome (HUS), urinary tract infection (UTI), pyelonephritis, septicemia, meningitis, and respiratory disease (pneumonia) in humans and animals. Known IPEC pathotypes include the attaching and effacing E. coli (AEEC), Shiga-toxigenic E. coli (STEC), enteroaggregative E. coli (EAEC), enterotoxigenic E. coli (ETEC), enteroinvasive E. coli (EIEC), diffuse adhering E. coli (DAEC) and the recently described enteroaggregative hemorrhagic E. coli (EAHEC; Kaper et al., 2004; Brzuszkiewicz et al., 2011). EHEC are a subgroup of STEC, which colonizes the terminal regions of the gastrointestinal tract of ruminants. It is notable that different serotypes of E. coli preferentially colonize different ruminant hosts (Djordjevic et al., 2001, 2004; Hornitzky et al., 2002) and carry different Shiga toxin gene variants (Ramachandran et al., 2001; Brett et al., 2003a,b). These studies underscore the importance of understanding the ecology of disease-causing organisms and how niche adaptation influences lateral gene transfer. Notably, however, ruminants are not a recognized source of ExPEC (Manges and Johnson, 2015).
Many recent molecular surveillance studies on E. coli report the frequency of genes that code for extended-spectrum beta lactamase enzymes (ESBL’s), which are indicative of resistance to penicillin, cephalosporin, and other clinically important antibiotics, but fail to screen for resistance to the older, first generation antibiotics (Bean et al., 2009; Ewers et al., 2012; Glasner et al., 2013; Roschanski et al., 2015). Genes that code for resistance to first generation antibiotics are often carried in bacteria containing class 1 integrons (Johnson et al., 2005b; Jakobsen et al., 2011; Hansen et al., 2014). From a diagnostic perspective, recent genomic studies on an outbreak of EAHEC and EAEC strain O104:H4, highlighted this issue (Ahmed et al., 2012). EAHEC is a recently emerged E. coli pathotype and EAEC strain O104:H4 has acquired the Shiga toxin gene stx2a, a diagnostic marker of EHEC. Outbreak strains were found to be resistant to ampicillin, streptomycin, sulfamethoxazole, trimethoprim, and tetracycline but little effort was given to determining how the genes that coded for these antibiotic resistance phenotypes were assembled (Ahmed et al., 2012). We were subsequently able to show that the genes coding for resistance to these antibiotics were assembled in Tn6029D, a transposon that abuts a class 1 integron in a Tn21/Tn1721 hybrid backbone (Roy Chowdhury et al., 2015). The resistance region resided in genomic island 3 which also carried the virulence gene ag43 encoding a self-associating serine protease auto-transporter, important for biofilm formation. This is an example of how antimicrobial resistance genes and virulence genes co-localize on the same mobile genetic element (Roy Chowdhury et al., 2015). Chromosomal islands are increasingly associated with carriage of class 1 integrons encoding resistance to multiple antibiotics of clinical significance (Roy Chowdhury et al., 2016).
Salmonella genomic island 1 (Boyd et al., 2001; Levings et al., 2005), Salmonella genomic island 2 (Levings et al., 2008) and several large plasmids in EHEC strain O26:H (Venturini et al., 2010), and an atypical EPEC, strain O111 (Venturini et al., 2013) are also examples of virulence and resistance genes being carried on the same mobile element. Strains of E. coli that carry genes that are markers for more than one E. coli pathotype pose a dilemma for diagnostic laboratories. There are numerous reports describing novel E. coli strains with combinations of virulence genes that are diagnostic of more than one E. coli pathotype; this suggests that newly emerging strains are increasingly likely to have complex resistance loci facilitated by the clustering of mobile element-associated resistance and virulence genes. In a study of 265 cases of E. coli causing UTI, 28 carried virulence genes typical of IPEC (Bielaszewska et al., 2014). Several other examples of hybrid E. coli strains have been described and their presence demonstrates the need to have genomic surveillance strategies in place to determine how antibiotic resistance and virulence genes assemble in emerging pathogens (Wallace-Gadsden et al., 2007; Abe et al., 2008; Bielaszewska et al., 2014; Mariani-Kurkdjian et al., 2014).
Extraintestinal pathogenic E. coli is a phylogenetically diverse group comprised of a broad range of E. coli sequence types (STs). ExPEC are fecal E. coli that rarely cause intestinal disease but have acquired a wide variety of virulence gene cargo that facilities their ability to cause disease at extraintestinal sites, particularly in the urinary tract. ExPEC are the most frequently isolated Gram negative bacterial pathogen infecting humans incurring significant mortality and morbidity. Notably, ExPEC are a leading cause of sepsis and are increasingly resistant to multiple antibiotics posing a serious health concern (Manges and Johnson, 2015; Poolman and Wacker, 2016). ExPEC have a fecal origin but rarely cause gastrointestinal disease. The acquisition of a diverse range of virulence factors by lateral gene transfer has armed ExPEC with an ability to cause life-threatening blood-borne, urinary tract, respiratory, skin and soft tissue infections, and meningitis in humans of all ages (Russo and Johnson, 2003; Poolman and Wacker, 2016). ExPEC vary widely in the combinations of putative virulence genes that they carry. Many genes encode for adhesins, iron acquisition systems and other putative virulence attributes and this genetic redundancy complicates efforts to develop broadly applicable molecular diagnostic tests to detect ExPEC (Johnson et al., 2001; Spurbeck et al., 2012). Importantly, livestock are important reservoirs for ExPEC (Ewers et al., 2007, 2009; Manges et al., 2007; Vincent et al., 2010; Nordstrom et al., 2013; Manges and Johnson, 2015). Virulence genes are often shared among E. coli isolates from samples collected from humans, and the meat and feces of intensively reared livestock, particularly poultry (Rodriguez-Siek et al., 2005; Vandekerchove et al., 2005; Jakobsen et al., 2010a,b). Consistent with this view, ExPEC carry genes coding for resistance to antimicrobials frequently used in veterinary medicine. Hospital diagnostics laboratories do not screen for most of the antibiotics used in veterinary medicine and so resistance to these is under-reported. In a Chinese study of 315 ExPEC isolates from various pig tissues, more than 63% exhibited resistance to 10 antibiotics including ampicillin, trimethoprim, sulfadimidine, tetracycline, neomycin, streptomycin, and kanamycin (Tan et al., 2012). The genes blaTEM-1 (ampicillin resistance); strAB (streptomycin resistance); sul1 and, sul2 (sulfonamide resistance); aphA1 (kanamycin and neomycin resistance); tetA, tetB, and tetG (tetracycline resistance); as well as various dfrA genes (trimethoprim resistance) coded for resistance to these antibiotics.
The class 1 integron is widespread in clinical settings. Bacteria that are resistant to more than one antimicrobial often carry class 1 integrons and their presence is a reliable indicator of multiple antimicrobial resistance (Leverstein-van Hall et al., 2003). The class 1 integron structures identified in most clinical samples are thought to be derived from the capture of a chromosomally located class 1 integron, prevalent within environmentally abundant bacterial species belonging to the broad β-proteobacterium group, by a Tn402 family transposon (Gillings et al., 2008; Gillings, 2014). Global spread of the class 1 integron is in part a result of the propensity for the Tn402 family of transposons to target res sites found on a wide variety of conjugative and mobile plasmids, including the widespread mercury-resistance transposon, Tn21, and the broader Tn3 family (Liebert et al., 1999; Gillings et al., 2008, 2015). For this and a variety of other reasons described below, resistance genes cluster on mobile elements. Clinical class 1 integrons have subsequently evolved, typically by the loss of most of the Tn402 tniB transposition gene, and partial fusion of the qacE gene (coding for an efflux pump for disinfectants, Hall et al., 1994; Partridge et al., 2001) with the sulfonamide resistance gene sul1. The most commonly observed structure observed in clinical isolates (Figure 1A) is comprised of a 5′ conserved segment (5′-CS), a 3′-conserved segment (3′CS) and a variable region inserted in between them. The 5′-CS contains the intI1 gene encoding the class 1 integrase; the Tn402 inverted repeat, the two promoters, one required for the transcription of the integrase and the other for the gene cassettes and the attI1 recombination site. The variable region contains a diverse array of resistance gene cassettes (Partridge et al., 2009) and the associated attC sites. The 3′ conserved segment (3′-CS) most frequently contains qacEΔ1 and sul1 genes, remnants of the transposition module and a terminal Tn402 inverted repeat. Most gene cassettes found in clinical class 1 integrons from nosocomial and livestock settings code for resistance to trimethoprim (dfrA) and aminoglycosides (aadA); despite this over 130 different gene cassettes have been described (Partridge et al., 2009) and, to date, 410 annotated variants are recognized (Tsafnat et al., 2011).
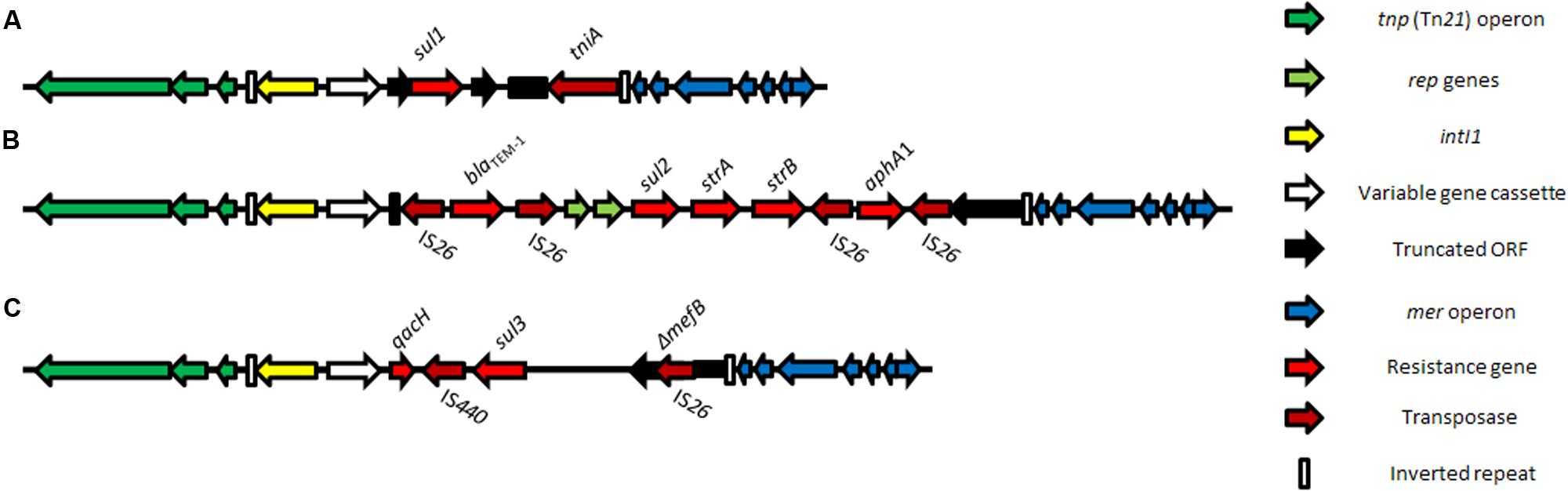
FIGURE 1. Class 1 integron structures associated with the three sulfonamide resistance gene alleles sul1 (A), sul2 (B), and sul3 (C), shown in a Tn21 backbone.
Further variation in the structure of the 3′-CS has also been described (Pan et al., 2006; Cain et al., 2010; Dawes et al., 2010; Saenz et al., 2010; Venturini et al., 2010, 2013; Mattiello et al., 2015). In one instance, the 3′-CS was almost completely lost and had been replaced with the compound transposon, Tn6026 (Venturini et al., 2013). The Tn6026 structure contains the beta-lactamase blaTEM-1 gene; the sulfonamide resistance gene, sul2; the streptomycin resistance gene strAB; the neomycin/kanamycin resistance gene, aphA1 and it is flanked by direct copies of IS26 (Figure 1B). Tn6026 is located between attI, the insertion point of the gene cassette, and the defective tni module of a clinical class 1 integron, replacing all but the first 24 bp of the 3′-CS (Cain et al., 2010; Dawes et al., 2010; Venturini et al., 2010, 2013). A sul3 variant structure associated with IS26 has also emerged over the last decade from human- and animal-sourced isolates (Figure 1C). The first report of sul3 was from Swiss pigs and humans in Grape et al. (2003) and Perreten and Boerlin (2003). Since then, sul3 has been reported in samples from a number of countries (Pan et al., 2006; Saenz et al., 2010; Tang et al., 2011; Mattiello et al., 2015; Wyrsch et al., 2015; Moran et al., 2016). These atypical class 1 integrons (Figure 1) are often components of larger complex resistance regions, hosted on plasmids that also carry virulence genes (Venturini et al., 2010, 2013).
Small mobile elements, including insertion sequences (IS1, IS26, ISEcp1c, and IS6100 are of note), Insertion Sequence Common Regions (ISCRs) and composite transposons formed from two or more IS elements, play an important role in reshaping the 3′-CS of clinical class 1 integrons (Daly et al., 2005; Dionisi et al., 2009; Venturini et al., 2010). The genes blaSHV-11, and blaSHV-12 (coding for β-lactam resistance; Ford and Avison, 2004; Dionisi et al., 2009); qnrB19 (coding for quinolone resistance; Dionisi et al., 2009); and aphA1 (coding for kanamycin/neomycin resistance; Cain et al., 2010) have been shown to be flanked by direct copies of IS26. The mechanism of action of IS26 has been the subject of several recent studies (Harmer et al., 2014; Harmer and Hall, 2015; He et al., 2015). These studies show that resistance loci containing IS26 can be hotspots for the capture of further resistance genes and other genetic cargo flanked by IS26 and demonstrates the important role of IS26 and other mobile elements in the assembly and re-assembly of novel complex resistance loci.
Currently, genomic epidemiological surveillance relies heavily on whole genome and metagenomic sequence information generated using short-read (<400 bp) sequencing platforms. A fall in the cost of high throughput, next generation sequencing with platforms that generate short-read lengths means they have become the leading technology for characterizing microbial evolution and uncultured microbial consortia, and to catalog antimicrobial resistance and virulence gene carriage. However, short-read sequencing technologies are unable to assemble complex resistance gene loci, or generate complete genome sequences, without which it is difficult to characterize mobile genetic elements rapidly and reliably. These limitations severely restrict efforts to comprehend the role of lateral gene transfer in the evolution of microbial pathogens and antimicrobial resistance, particularly in regards to the microevolutionary events that lead to the generation of novel complex resistance loci. Long-read sequencing technologies have improved significantly over the past 5 years and are beginning to address this shortfall, but are currently either cost prohibitive for large-scale genomic epidemiological surveillance or require further improvements in base call accuracy (Loman and Pallen, 2015).
Genes that code for resistance to older-generation antibiotics are often incorporated within complex resistance regions. In part, this provides a plausible explanation for why these genes persist despite enforced restrictions on prescribing of older-generation antibiotics. In the UK, strict limitations on the prescription of sulfonamide drugs were enforced in Anonymous (1995). However, functional sul genes persist in clinically important Gram negative bacteria and their frequency has not declined since the restriction was imposed (Enne et al., 2001; Bean et al., 2005, 2009). A similar scenario unfolded in Finland when trimethoprim was withdrawn from use for the treatment of UTI (Heikkila et al., 1990). In the Enterobacteriaceae, resistance to sulfonamides, trimethoprim, ampicillin, tetracycline, and fluoroquinolones remains high (Enne et al., 2001; Nozarian and Abdollahi, 2015; Paniagua-Contreras et al., 2015; Uzun et al., 2015; Yahiaoui et al., 2015). Carriage rates for sul1, sul2, or sul3 genes in E. coli isolated from retail meat, livestock, healthy humans, and humans with clinical disease also remains a problem (Enne et al., 2001; Blahna et al., 2006; Frank et al., 2007; Altalhi et al., 2010; Bailey et al., 2010; Saenz et al., 2010; Vogt et al., 2014; Yahiaoui et al., 2015). Carriage of sul genes is linked with the presence of both typical and atypical class 1 integrons, including genes that code for resistance to ampicillin, streptomycin, and trimethoprim (Bettelheim et al., 2003; Bean et al., 2009; Bailey et al., 2010; Soufi et al., 2011; Ewers et al., 2014; Yahiaoui et al., 2015). Combinations of genes coding for resistance to first generation antibiotics contribute significantly to the multiple antimicrobial resistance phenotypes observed in commensal E. coli (Bettelheim et al., 2003; Vinue et al., 2008, 2010; Bailey et al., 2010). Genetic studies that have examined resistance to a broad spectrum of antibiotics frequently report the presence of blaTEM-1, sul1, sul2, strAB, tetA(A), and a range of aadA and dfrA genes (Soufi et al., 2011; Labar et al., 2012; Ewers et al., 2014; Yamamoto et al., 2014; Yahiaoui et al., 2015). The streptomycin resistance gene, strAB, and the sul2 sulfonamide resistance gene are also spread via small plasmids such as RSF1010 (Yau et al., 2010), and IncQ plasmids that are related to RSF1010 (Tietze et al., 1989). The strAB gene and the adjacent inverted repeat IRR, as seen in RSF1010, probably had their origins in Tn5393c, while sul2 was recruited from a CR2-containing element (Yau et al., 2010). These molecular events describing the evolution of the sul2-strAB configuration observed in RSF1010 have been described previously (Yau et al., 2010). The sul2-strAB gene cluster and flanking sequences from RSF1010 have been captured onto Tn21 derivative transposons, on large and small plasmid backbones, and in chromosomal islands; they are often in association with IS26 and are distributed widely (Daly et al., 2005; Cain et al., 2010; Venturini et al., 2010, 2013; Labar et al., 2012; Reid et al., 2015; Roy Chowdhury et al., 2015).
E. coli in Intensive Production Animals
Porcine Enterotoxigenic Escherichia coli (ETEC)
Enterotoxigenic Escherichia coli (ETEC) are the causative agents of three distinct diseases in young pigs: neonatal, pre-weaning, and post-weaning diarrhea (Fairbrother et al., 1988; Noamani et al., 2003). These diseases vary in severity and are categorized by the age at which the pig develops symptoms. Diarrhea caused by ETEC strains, generally known as enteric colibacillosis, is a problem in all countries involved in pig production, and various methods have been employed to protect pigs from ETEC infection and any subsequent diarrheal disease. These methods include, but are not limited to, treatment with antibiotics, vaccination, and feed modulation (Fairbrother et al., 2005; Virdi et al., 2013). While these protective steps have helped reduce levels of ETEC infection, particularly in neonatal and pre-weaning colibacillosis, antimicrobial resistance has become a global challenge that is severely limiting antimicrobials as a treatment option.
Pathogenic E. coli express a wide variety of functional molecules including toxins, adhesins, autotransporters, and invasins (Nataro and Kaper, 1998; Kaper et al., 2004), which enable the E. coli cells to damage host tissues. In porcine ETEC, these molecules, referred to as virulence factors, include heat-stable (ST) and heat-labile (LT) enterotoxins, both of which can cause diarrhea via different molecular pathways within the pig’s gastrointestinal tract (Nataro and Kaper, 1998). Various adhesins have evolved in ETEC and are expressed on the bacterial cell surface, allowing adhesion to the host’s gastrointestinal tract. In pigs, the K88 (F4), K99 (F5), 987P (F6), F18 and F41 fimbrial adhesins all provide host intestinal cell binding, with K88, K99 and F18 fimbriae being particularly prevalent (Nataro and Kaper, 1998; Nagy and Fekete, 1999; Koh et al., 2008). Interestingly, the K88 and F18 fimbrial adhesins are often associated with pre- and post-weaning diarrhea, whilst the K99 fimbria has been associated with neonatal diarrhea in pigs (DebRoy and Maddox, 2001). These binding effects are thought to be supplemented by other common adhesins coded by genes such as fimH (Bouckaert et al., 2006) and eaeH (Sheikh et al., 2014), which are known to provide additional post-attachment binding. Chapman et al. (2006) employed multiplex PCR assays targeting 58 virulence genes to screen 52 clinical E. coli isolates from episodes of porcine neonatal and post-weaning diarrhea, and 23 commensal E. coli isolates from healthy pigs. Of the 58 genes, 17 were useful in distinguishing commensal from clinical E. coli, nine of which (iha, hlyA, aidA, east1, aah, fimH, iroNE.coli, traT, and saa) were identified for the first time in clinical porcine isolates in Australia (Chapman et al., 2006).
In an attempt to combat E. coli expressing these and other virulence genes, antimicrobials have been employed as both curative and prophylactic medicines on farms worldwide. This has placed a selection pressure on both targeted and surrounding microbial populations, and as such resistance to these antimicrobials has become extremely prevalent. For pathogenic E. coli in general, the definition and interpretation of virulence has been tempered based on results from large-scale microbial genome sequencing projects (Falkow, 1988; Pallen and Wren, 2007). ExPEC reside in the gut of their host, often in a commensal state, but have the genetic repertoire of virulence genes that enables them to cause life-threatening extraintestinal infections. Due to the degree of redundancy inherent in many ExPEC virulence genes, it remains a challenge to define virulence in ExPEC. These ExPEC-associated virulence genes have also been found in isolates sourced from humans, poultry and swine but rarely in cattle and other ruminants (Johnson et al., 2001; Tan et al., 2012; Manges and Johnson, 2015). Poultry are a well-recognized reservoir of ExPEC that have the potential to cause human disease but the role of swine in this regard has not been investigated sufficiently.
Current Status of Global Antimicrobial Use and Its Influence on the Porcine E. coli Resistome
Rationalization of farming practices has had profound effects on the spread of resistance to antimicrobials, and consequently both human and animal health – and the problem continues to grow (FAO, 2009). An estimated 210 million kilograms of antibiotics are produced annually in China, making it the largest producer and consumer of antibiotics globally. Approximately half of the antibiotics produced are used in livestock production (Hvistendahl, 2012; Zhu et al., 2013). China is also the largest producer and consumer of pork; 690 million pigs were produced in 2011 and 38.1 kg of pork are consumed per person per year (Krishnasamy et al., 2015), now reportedly almost double the pork consumption of the USA population. China is said to contain half the world’s pig population, with five times the production rate of the USA (the former largest production center) and a higher per capita meat consumption than any developing nation other than Brazil and Latin America (FAO, 2009). Trends are similar for poultry production in China (Krishnasamy et al., 2015). An estimated 34 million kilograms of antibiotics, particularly tetracyclines, sulfonamides, macrolides, and penicillins were used in Chinese pig production during 2012 (Krishnasamy et al., 2015). Combination therapies of chlortetracycline with sulfathiazole and penicillin, and chlortetracycline with sulfamethazine and penicillin, are used widely. Other individual antibiotics that are used less extensively include tylosin, chlortetracycline, oxytetracycline, bacitracin, and bambermycin amongst others (Krishnasamy et al., 2015). Furthermore, pig production in China is responsible for the production of approximately 618 billion kilograms of manure per annum (Wang et al., 2006), much of which is contaminated with antibiotic residues and heavy metals, particularly tetracyclines, sulfonamides and copper; some residues are present at levels in the order of 100s of milligrams per kilogram (Pan et al., 2011; Qiao et al., 2012; Zhu et al., 2013). Elevated levels of zinc and arsenic have also been detected in these studies. Of 149 unique antimicrobial resistance genes 63 were detected significantly more frequently in microbes from manure at different stages of processing on three large pig production farms in China, compared with microbes in manure from control animals that received no antimicrobials; this included genes coding for resistance to antibiotics used to treat humans (Zhu et al., 2013). Samples of pig manure from China were also rich in microbes carrying transposase genes, notably those belonging to the IS6-family, predominantly IS26 transposase (Zhu et al., 2013).
In the USA, estimates suggest that the usage of tetracyclines is as high as in China, both in the combinations described above, and as stand-alone treatments. Additionally, bacitracin, carbadox, lincomycin, neomycin, penicillin, tiamulin, tilmicosin, tylosin, and viginiamycin have all been used in the USA pig production industry (Apley et al., 2012). The USA and China combined produce the majority of the world’s pigs through intensive farming practices (FAO, 2009), with China producing up to five times more than the USA (Krishnasamy et al., 2015). However, other countries also have pig production systems that rely heavily on antibiotic usage. In Alberta, Canada, the same suite of antibiotics is used as in the USA, with the addition of dimetridazole (Rajic et al., 2006). A similar pattern of use has also emerged in an Australian survey of pig production, with the use of apramycin and neomycin, penicillins, macrolides, sulfonamides, tetracyclines, lincomycin and spectinomycin, tiamulin, olaquindox, ceftiofur, and dimetridazole reported (Jordan et al., 2009). A number of these antibiotics, including lincomycin, spectinomycin, and dimetridazole are also used in the treatment of infectious human diseases. In an Australia-wide study of 114 porcine-derived E. coli, resistance to numerous antibiotics was common: tetracycline (88.6%), ampicillin (71.05%), trimethoprim/sulfamethoxazole (67.5%), streptomycin (69.3%), chloramphenicol (44.74%), neomycin (35.96%), apramycin (34.21%), gentamicin (28.95%), florphenicol (26.32), cefalotin (24.56%), and spectinomycin (21.93%). Resistance to imipenem and amikacin was not detected (Abraham et al., 2015). Of the 114 isolates evaluated 79% were classified as resistant to antibiotics from three or more different classes. Resistance to extended-spectrum cephalosporins (3%) and fluoroquinolones (1%) was detected in isolates belonging to some E. coli lineages (ST117, ST744, ST10, and ST1) albeit infrequently (Abraham et al., 2015).
Colistin is a polymyxin antimicrobial used to treat extensively drug-resistant Gram negative infections. It was first used in the 1950s but its use in humans has declined due to concerns about nephrotoxicity and neurotoxicity (Nation and Li, 2009). China, India, and Europe still use colistin extensively for agricultural purposes. In Europe, colistin is used to treat enterobacterial infections in pigs (neonatal diarrhea), poultry (colibacillosis), cattle (neonatal diarrhea in veal calves), sheep, and goats (Timmerman et al., 2006; Pardon et al., 2012); in 2010 it was the fifth most commonly sold antimicrobial after tetracycline, penicillins, sulfonamides, and macrolides (European Medicines Agency [EMA], 2013). Colistin is also used in aquaculture (Xu et al., 2012). The recent emergence of colistin resistance in commensal porcine E. coli isolates in China via the acquisition of plasmid-mediated mcr-1 gene has serious implications for the treatment of pan-drug-resistant Gram-negative bacteria, particularly isolates of the extremely drug resistant Acinetobacter baumannii and carbapenemase-resistant Klebsiella pneumoniae (Liu et al., 2015). The seriousness of this finding is clear due to: (i) high in vitro transfer rates among E. coli by conjugation and the ability of the mcr-1 plasmid to transfer and be maintained in globally dispersed pathogenic clones such as E. coli ST131, K. pneumoniae ST11, and Pseudomonas aeruginosa; (ii) the high frequency of carriage of mcr-1 in E. coli from livestock and in retail meats sourced from southern China; (iii) detection of mcr-1-like genes in Malaysia (Liu et al., 2016). The apparent high rates of carriage of mcr-1 in isolates of E. coli from animals and the apparent low incidence in human-derived E. coli populations has led to speculation that the heavy use of colistin in agriculture in China has been the main driver for the emergence of plasmid-mediated colistin resistance (European Medicines Agency [EMA], 2013; Liu et al., 2016).
Genes conferring antimicrobial resistance have been identified in E. coli from diverse pig-related sources (Table 1). The genes blaTEM, sul1/2/3, aadA, dfrA, and tetA/B/C feature prominently. In order, these genes confer resistance to penicillin, sulfonamides (such as sulphamethoxazole), streptomycin and spectinomycin, trimethoprim, and tetracyclines; all of these are used in the treatment of animals. Additional clinically important cases of resistance are also identified sporadically in the literature, specifically to chloramphenicol (cmlA, cat), streptothricin (estX/sat), florfenicol (flo), quinolones (qnr), streptomycin (strAB), and fosfomycin (fosA). There are also a number of known resistance mutations in the gyr and pmr genes that confer elevated resistance to quinolones and colistin respectively. Already the presence of these genes together in the same isolate and from disparate countries indicates they are both genetically linked in antimicrobial resistance-encoding loci, and globally disseminated. Detection of the genes blaOXA, blaSHV, and blaCTX-M coding for the extended-spectrum beta-lactamases, which are relevant to human medicine, is also of concern (Randall et al., 2014). Of note, these data were collected from a variety of publications for each country, and show some selection bias concerning the genes detected. Whole genome sequencing has become a tool to rectify this bias, and will continue to feature in the detection and characterisation of antimicrobial resistance, its evolution and dissemination (Salipante et al., 2015).
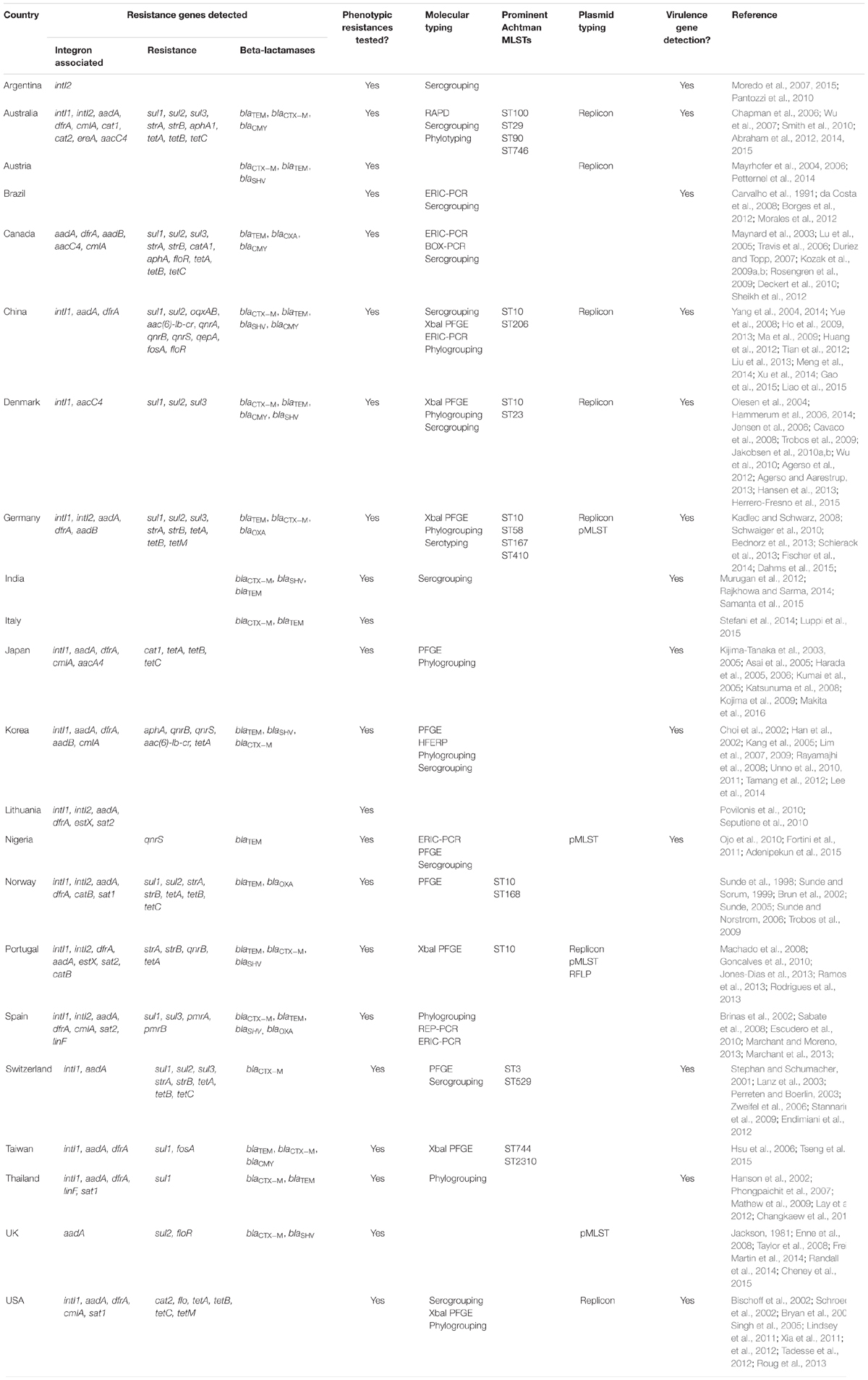
TABLE 1. Molecular characterisation of published antimicrobial resistant, porcine-derived Escherichia coli isolates.
Much needs to be done to reduce the reliance on antimicrobials in food production. There is a growing body of evidence suggesting that food production may be a key driver in the evolution of multiple drug resistance in a number of bacterial species of clinical significance to human health. Livestock represent a reservoir of E. coli pathotypes with resistance to more than one antimicrobial. While pigs and poultry are significant sources of ExPEC, cattle are the major reservoirs for Shiga toxin producing E. coli (EHEC; Manges and Johnson, 2015) and atypical EPEC (Hornitzky et al., 2005; Dawes et al., 2010). The consequences for the environment of large-scale intensive animal production and a growing human population, estimated to be 9 billion by 2050 (United Nations, 2015) are not fully understood. Much remains to be done to determine how antimicrobial resistance genes are assembled on mobile genetic elements and to identify their unique genetic features. Diagnostics tests that target unique features will be useful in tracking the movement of mobile elements that are carrying complex resistance loci and to identify hotspots where they reside. Metagenomic approaches will have a major role to play in understanding the scale of the antimicrobial resistance problem, detecting and tracking emerging pathogens, and the role of gut flora in human and animal health. Studies on microbial populations from the gastrointestinal tracts of intensively reared livestock are needed to identify and characterize novel pathotypes that will undoubtedly emerge in response to the selection pressures that are commonplace in large-scale animal production facilities. Genomic surveillance studies will have an important role to play in identifying complex resistance loci, particularly how they move through different production systems and natural ecosystems (aquatic and terrestrial), and the impact they have on the carriage of multiple antimicrobial resistance genes in bacteria that colonize and infect humans and animals. Currently, there are just three complete porcine-derived E. coli chromosomal sequences published, as well as a variety of complete plasmid sequences, the majority of which have been derived from the study of isolates with closed genome sequences (Table 2). This is at odds with the significant role that intensive pig production plays in the release of antibiotic residues into the environment, and we expect more focus to be placed on the sequencing of pig-associated E. coli isolates in the near future.
Author Contributions
EW drafted an early version of the manuscript and produced the figure and tables. PR and IC contributed to several sections in the manuscript. TC and JH edited drafts of the manuscript. SD wrote most the manuscript.
Funding
The research in this article is part of the Ausgem partnership and supported by an Australian Research Council Linkage Project grant LP150100912.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
We wish to acknowledge J. K. Pell Consulting for copyediting the manuscript.
References
Abe, C. M., Salvador, F. A., Falsetti, I. N., Vieira, M. A., Blanco, J., Blanco, J. E., et al. (2008). Uropathogenic Escherichia coli (UPEC) strains may carry virulence properties of diarrhoeagenic E. coli. FEMS Immunol. Med. Microbiol. 52, 397–406. doi: 10.1111/j.1574-695X.2008.00388.x
Abraham, S., Chin, J., Brouwers, H. J., Zhang, R., and Chapman, T. A. (2012). Molecular serogrouping of porcine enterotoxigenic Escherichia coli from Australia. J. Microbiol. Methods 88, 73–76. doi: 10.1016/j.mimet.2011.10.016
Abraham, S., Jordan, D., Wong, H. S., Johnson, J. R., Toleman, M. A., Wakeham, D. L., et al. (2015). First detection of extended-spectrum cephalosporin- and fluoroquinolone-resistant Escherichia coli in Australian food-producing animals. J. Glob. Antimicrob. Resist. 3, 273–277. doi: 10.1016/j.jgar.2015.08.002
Abraham, S., Trott, D. J., Jordan, D., Gordon, D. M., Groves, M. D., Fairbrother, J. M., et al. (2014). Phylogenetic and molecular insights into the evolution of multidrug-resistant porcine enterotoxigenic Escherichia coli in Australia. Int. J. Antimicrob. Agents 44, 105–111. doi: 10.1016/j.ijantimicag.2014.04.011
Adenipekun, E. O., Jackson, C. R., Oluwadun, A., Iwalokun, B. A., Frye, J. G., Barrett, J. B., et al. (2015). Prevalence and antimicrobial resistance in Escherichia coli from food animals in Lagos, Nigeria. Microb. Drug Resist. 21, 358–365. doi: 10.1089/mdr.2014.0222
Agerso, Y., and Aarestrup, F. M. (2013). Voluntary ban on cephalosporin use in Danish pig production has effectively reduced extended-spectrum cephalosporinase-producing Escherichia coli in slaughter pigs. J. Antimicrob. Chemother. 68, 569–572. doi: 10.1093/jac/dks427
Agerso, Y., Aarestrup, F. M., Pedersen, K., Seyfarth, A. M., Struve, T., and Hasman, H. (2012). Prevalence of extended-spectrum cephalosporinase (ESC)-producing Escherichia coli in Danish slaughter pigs and retail meat identified by selective enrichment and association with cephalosporin usage. J. Antimicrob. Chemother. 67, 582–588. doi: 10.1093/jac/dkr507
Agerso, Y., Jensen, J. D., Hasman, H., and Pedersen, K. (2014). Spread of extended spectrum cephalosporinase-producing Escherichia coli clones and plasmids from parent animals to broilers and to broiler meat in a production without use of cephalosporins. Foodborne Pathog. Dis. 11, 740–746. doi: 10.1089/fpd.2014.1742
Ahmed, A. M., and Shimamoto, T. (2015a). Molecular analysis of multidrug resistance in Shiga toxin-producing Escherichia coli O157:H7 isolated from meat and dairy products. Int. J. Food Microbiol. 193, 68–73. doi: 10.1016/j.ijfoodmicro.2014.10.014
Ahmed, A. M., and Shimamoto, T. (2015b). Molecular characterization of multidrug-resistant Shigella spp. of food origin. Int. J. Food Microbiol. 194, 78–82. doi: 10.1016/j.ijfoodmicro.2014.11.013
Ahmed, S. A., Awosika, J., Baldwin, C., Bishop-Lilly, K. A., Biswas, B., Broomall, S., et al. (2012). Genomic comparison of Escherichia coli O104:H4 isolates from 2009 and 2011 reveals plasmid, and prophage heterogeneity, including shiga toxin encoding phage stx2. PLoS ONE 7:e48228. doi: 10.1371/journal.pone.0048228
Allen, H. K., Donato, J., Wang, H. H., Cloud-Hansen, K. A., Davies, J., and Handelsman, J. (2010). Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 8, 251–259. doi: 10.1038/nrmicro2312
Altalhi, A. D., Gherbawy, Y. A., and Hassan, S. A. (2010). Antibiotic resistance in Escherichia coli isolated from retail raw chicken meat in Taif, Saudi Arabia. Foodborne Pathog. Dis. 7, 281–285. doi: 10.1089/fpd.2009.0365
Andersen, S. R. (1993). Effects of waste water treatment on the species composition and antibiotic resistance of coliform bacteria. Curr. Microbiol. 26, 97–103. doi: 10.1007/bf01577343
Anonymous. (1995). Co-trimoxazole use restricted. Drug Ther. Bull. 33, 92–93. doi: 10.1136/dtb.1995.331292
Apley, M. D., Bush, E. J., Morrison, R. B., Singer, R. S., and Snelson, H. (2012). Use estimates of in-feed antimicrobials in swine production in the United States. Foodborne Pathog. Dis. 9, 272–279. doi: 10.1089/fpd.2011.0983
Asai, T., Kojima, A., Harada, K., Ishihara, K., Takahashi, T., and Tamura, Y. (2005). Correlation between the usage volume of veterinary therapeutic antimicrobials and resistance in Escherichia coli isolated from the feces of food-producing animals in Japan. Jpn. J. Infect. Dis. 58, 369–372.
Baez, J., Hernandez-Garcia, M., Guamparito, C., Diaz, S., Olave, A., Guerrero, K., et al. (2015). Molecular characterization and genetic diversity of ESBL-producing Escherichia coli colonizing the migratory Franklin’s gulls (Leucophaeus pipixcan) in Antofagasta, North of Chile. Microb. Drug Resist. 21, 111–116. doi: 10.1089/mdr.2014.0158
Bailey, J. K., Pinyon, J. L., Anantham, S., and Hall, R. M. (2010). Commensal Escherichia coli of healthy humans: a reservoir for antibiotic-resistance determinants. J. Med. Microbiol. 59, 1331–1339. doi: 10.1099/jmm.0.022475-0
Baker-Austin, C., Wright, M. S., Stepanauskas, R., and McArthur, J. V. (2006). Co-selection of antibiotic and metal resistance. Trends Microbiol. 14, 176–182. doi: 10.1016/j.tim.2006.02.006
Baquero, F., Tedim, A. P., and Coque, T. M. (2013). Antibiotic resistance shaping multi-level population biology of bacteria. Front. Microbiol. 4:15. doi: 10.3389/fmicb.2013.00015
Baquero, F., and Tobes, R. (2013). Bloody coli: a gene cocktail in Escherichia coli O104:H4. MBio 4:e00066-13. doi: 10.1128/mBio.00066-13
Barkay, T., Miller, S. M., and Summers, A. O. (2003). Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol. Rev. 27, 355–384. doi: 10.1016/S0168-6445(03)00046-9
Bartoloni, A., Pallecchi, L., Rodriguez, H., Fernandez, C., Mantella, A., Bartalesi, F., et al. (2009). Antibiotic resistance in a very remote Amazonas community. Int. J. Antimicrob. Agents 33, 125–129. doi: 10.1016/j.ijantimicag.2008.07.029
Bean, D. C., Livermore, D. M., and Hall, L. M. (2009). Plasmids imparting sulfonamide resistance in Escherichia coli: implications for persistence. Antimicrob. Agents Chemother. 53, 1088–1093. doi: 10.1128/AAC.00800-08
Bean, D. C., Livermore, D. M., Papa, I., and Hall, L. M. (2005). Resistance among Escherichia coli to sulphonamides and other antimicrobials now little used in man. J. Antimicrob. Chemother. 56, 962–964. doi: 10.1093/jac/dki332
Bednorz, C., Guenther, S., Oelgeschlager, K., Kinnemann, B., Pieper, R., Hartmann, S., et al. (2013). Feeding the probiotic Enterococcus faecium strain NCIMB 10415 to piglets specifically reduces the number of Escherichia coli pathotypes that adhere to the gut mucosa. Appl. Environ. Microbiol. 79, 7896–7904. doi: 10.1128/AEM.03138-13
Ben Said, L., Jouini, A., Klibi, N., Dziri, R., Alonso, C. A., Boudabous, A., et al. (2015). Detection of extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae in vegetables, soil and water of the farm environment in Tunisia. Int. J. Food Microbiol. 203, 86–92. doi: 10.1016/j.ijfoodmicro.2015.02.023
Berendonk, T. U., Manaia, C. M., Merlin, C., Fatta-Kassinos, D., Cytryn, E., Walsh, F., et al. (2015). Tackling antibiotic resistance: the environmental framework. Nat. Rev. Microbiol. 13, 310–317. doi: 10.1038/nrmicro3439
Berg, J., Tom-Petersen, A., and Nybroe, O. (2005). Copper amendment of agricultural soil selects for bacterial antibiotic resistance in the field. Lett. Appl. Microbiol. 40, 146–151. doi: 10.1111/j.1472-765X.2004.01650.x
Bettelheim, K. A., Hornitzky, M. A., Djordjevic, S. P., and Kuzevski, A. (2003). Antibiotic resistance among verocytotoxigenic Escherichia coli (VTEC) and non-VTEC isolated from domestic animals and humans. J. Med. Microbiol. 52, 155–162. doi: 10.1099/jmm.0.04903-0
Bielaszewska, M., Schiller, R., Lammers, L., Bauwens, A., Fruth, A., Middendorf, B., et al. (2014). Heteropathogenic virulence and phylogeny reveal phased pathogenic metamorphosis in Escherichia coli O2:H6. EMBO Mol. Med. 6, 347–357. doi: 10.1002/emmm.201303133
Bischoff, K. M., White, D. G., McDermott, P. F., Zhao, S., Gaines, S., Maurer, J. J., et al. (2002). Characterization of chloramphenicol resistance in beta-hemolytic Escherichia coli associated with diarrhea in neonatal swine. J. Clin. Microbiol. 40, 389–394. doi: 10.1128/JCM.40.2.389-394.2002
Blahna, M. T., Zalewski, C. A., Reuer, J., Kahlmeter, G., Foxman, B., and Marrs, C. F. (2006). The role of horizontal gene transfer in the spread oftrimethoprim–sulfamethoxazole resistance among uropathogenic Escherichia coli in Europe and Canada. J. Antimicrob. Chemother. 57, 666–672. doi: 10.1093/jac/dkl020
Bondarczuk, K., Markowicz, A., and Piotrowska-Seget, Z. (2016). The urgent need for risk assessment on the antibiotic resistance spread via sewage sludge land application. Environ. Int. 87, 49–55. doi: 10.1016/j.envint.2015.11.011
Borges, C. A., Beraldo, L. G., Maluta, R. P., Cardozo, M. V., Guth, B. E., Rigobelo, E. C., et al. (2012). Shiga toxigenic and atypical enteropathogenic Escherichia coli in the feces and carcasses of slaughtered pigs. Foodborne Pathog. Dis. 9, 1119–1125. doi: 10.1089/fpd.2012.1206
Bouckaert, J., Mackenzie, J., de Paz, J. L., Chipwaza, B., Choudhury, D., Zavialov, A., et al. (2006). The affinity of the FimH fimbrial adhesin is receptor-driven and quasi-independent of Escherichia coli pathotypes. Mol. Microbiol. 61, 1556–1568. doi: 10.1111/j.1365-2958.2006.05352.x
Boyd, D., Peters, G. A., Cloeckaert, A., Boumedine, K. S., Chaslus-Dancla, E., Imberechts, H., et al. (2001). Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183, 5725–5732. doi: 10.1128/jb.183.19.5725-5732.2001
Brett, K. N., Hornitzky, M. A., Bettelheim, K. A., Walker, M. J., and Djordjevic, S. P. (2003a). Bovine non-O157 Shiga toxin 2-containing Escherichia coli isolates commonly possess stx2-EDL933 and/or stx2vhb Subtypes. J. Clin. Microbiol. 41, 2716–2722. doi: 10.1128/jcm.41.6.2716-2722.2003
Brett, K. N., Ramachandran, V., Hornitzky, M. A., Bettelheim, K. A., Walker, M. J., and Djordjevic, S. P. (2003b). stx1c Is the most common Shiga toxin 1 subtype among Shiga toxin-producing Escherichia coli isolates from sheep but not among isolates from cattle. J. Clin. Microbiol. 41, 926–936. doi: 10.1128/JCM.41.3.926-936.2003
Brinas, L., Zarazaga, M., Saenz, Y., Ruiz-Larrea, F., and Torres, C. (2002). Beta-lactamases in ampicillin-resistant Escherichia coli isolates from foods, humans, and healthy animals. Antimicrob. Agents Chemother. 46, 3156–3163. doi: 10.1128/AAC.46.10.3156-3163.2002
Brun, E., Holstad, G., Kruse, H., and Jarp, J. (2002). Within-sample and between-sample variation of antimicrobial resistance in fecal Escherichia coli isolates from pigs. Microb. Drug Resist. 8, 385–391. doi: 10.1089/10766290260469660
Bryan, A., Shapir, N., and Sadowsky, M. J. (2004). Frequency and distribution of tetracycline resistance genes in genetically diverse, nonselected, and nonclinical Escherichia coli strains isolated from diverse human and animal sources. Appl. Environ. Microbiol. 70, 2503–2507. doi: 10.1128/AEM.70.4.2503-2507.2004
Brzuszkiewicz, E., Thurmer, A., Schuldes, J., Leimbach, A., Liesegang, H., Meyer, F. D., et al. (2011). Genome sequence analyses of two isolates from the recent Escherichia coli outbreak in Germany reveal the emergence of a new pathotype: entero-aggregative-haemorrhagic Escherichia coli (EAHEC). Arch. Microbiol. 193, 883–891. doi: 10.1007/s00203-011-0725-6
Cain, A. K., Liu, X., Djordjevic, S. P., and Hall, R. M. (2010). Transposons related to Tn1696 in IncHI2 plasmids in multiply antibiotic resistant Salmonella enterica serovar Typhimurium from Australian animals. Microb. Drug Resist. 16, 197–202. doi: 10.1089/mdr.2010.0042
Carroll, D., Wang, J., Fanning, S., and McMahon, B. J. (2015). Antimicrobial resistance in wildlife: implications for public health. Zoonoses Public Health 62, 534–542. doi: 10.1111/zph.12182
Carvalho, A. C., Avila, F. A., Schocken-Iturrino, R. P., Quintana, J. L., and Albertini, P. E. (1991). Virulence factors in Escherichia coli strains isolated from pigs in the Ribeirao Preto region, State of Sao Paulo, Brazil. Rev. Elev. Med. Vet. Pays Trop. 44, 49–52.
Castillo, M., Martin-Orue, S. M., Taylor-Pickard, J. A., Perez, J. F., and Gasa, J. (2008). Use of mannanoligosaccharides and zinc chelate as growth promoters and diarrhea preventative in weaning pigs: effects on microbiota and gut function. J. Anim. Sci. 86, 94–101. doi: 10.2527/jas.2005-686
Cavaco, L. M., Frimodt-Moller, N., Hasman, H., Guardabassi, L., Nielsen, L., and Aarestrup, F. M. (2008). Prevalence of quinolone resistance mechanisms and associations to minimum inhibitory concentrations in quinolone-resistant Escherichia coli isolated from humans and swine in Denmark. Microb. Drug Resist. 14, 163–169. doi: 10.1089/mdr.2008.0821
Changkaew, K., Intarapuk, A., Utrarachkij, F., Nakajima, C., Suthienkul, O., and Suzuki, Y. (2015). Antimicrobial resistance, extended-spectrum beta-lactamase productivity, and class 1 integrons in Escherichia coli from healthy swine. J. Food Prot. 78, 1442–1450. doi: 10.4315/0362-028X.JFP-14-445
Chapman, T. A., Wu, X. Y., Barchia, I., Bettelheim, K. A., Driesen, S., Trott, D., et al. (2006). Comparison of virulence gene profiles of Escherichia coli strains isolated from healthy and diarrheic swine. Appl. Environ. Microbiol. 72, 4782–4795. doi: 10.1128/AEM.02885-05
Chee-Sanford, J. C., Mackie, R. I., Koike, S., Krapac, I. G., Lin, Y. F., Yannarell, A. C., et al. (2009). Fate and transport of antibiotic residues and antibiotic resistance genes following land application of manure waste. J. Environ. Qual. 38, 1086–1108. doi: 10.2134/jeq2008.0128
Chen, H., and Zhang, M. (2013). Occurrence and removal of antibiotic resistance genes in municipal wastewater and rural domestic sewage treatment systems in eastern China. Environ. Int. 55, 9–14. doi: 10.1016/j.envint.2013.01.019
Chen, Y. T., Lauderdale, T. L., Liao, T. L., Shiau, Y. R., Shu, H. Y., Wu, K. M., et al. (2007). Sequencing and comparative genomic analysis of pK29, a 269-kilobase conjugative plasmid encoding CMY-8 and CTX-M-3 beta-lactamases in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 51, 3004–3007. doi: 10.1128/AAC.00167-07
Cheney, T. E., Smith, R. P., Hutchinson, J. P., Brunton, L. A., Pritchard, G., and Teale, C. J. (2015). Cross-sectional survey of antibiotic resistance in Escherichia coli isolated from diseased farm livestock in England and Wales. Epidemiol. Infect. 143, 2653–2659. doi: 10.1017/s0950268814003963
Choi, C., Ham, H. J., Kwon, D., Kim, J., Cheon, D. S., Min, K., et al. (2002). Antimicrobial susceptibility of pathogenic Escherichia coli isolated from pigs in Korea. J. Vet. Med. Sci. 64, 71–73. doi: 10.1292/jvms.64.71
da Costa, M. M., Drescher, G., Maboni, F., Weber, S., de Avila Botton, S., Vainstein, M. H., et al. (2008). Virulence factors and antimicrobial resistance of Escherichia coli isolated from urinary tract of swine in southern of Brazil. Braz. J. Microbiol. 39, 741–743. doi: 10.1590/s1517-838220080004000027
Dahms, C., Hubner, N. O., Kossow, A., Mellmann, A., Dittmann, K., and Kramer, A. (2015). Occurrence of ESBL-producing Escherichia coli in livestock and farm workers in mecklenburg-western pomerania, Germany. PLoS ONE 10:e0143326. doi: 10.1371/journal.pone.0143326
Daly, M., Villa, L., Pezzella, C., Fanning, S., and Carattoli, A. (2005). Comparison of multidrug resistance gene regions between two geographically unrelated Salmonella serotypes. J. Antimicrob. Chemother. 55, 558–561. doi: 10.1093/jac/dki015
Dawes, F. E., Kuzevski, A., Bettelheim, K. A., Hornitzky, M. A., Djordjevic, S. P., and Walker, M. J. (2010). Distribution of class 1 integrons with IS26-mediated deletions in their 3’-conserved segments in Escherichia coli of human and animal origin. PLoS ONE 5:e12754. doi: 10.1371/journal.pone.0012754
DebRoy, C., and Maddox, C. W. (2001). Identification of virulence attributes of gastrointestinal Escherichia coli isolates of veterinary significance. Anim. Health Res. Rev. 2, 129–140.
Deckert, A., Gow, S., Rosengren, L., Leger, D., Avery, B., Daignault, D., et al. (2010). Canadian integrated program for antimicrobial resistance surveillance (CIPARS) farm program: results from finisher pig surveillance. Zoonoses Public Health 57(Suppl. 1), 71–84. doi: 10.1111/j.1863-2378.2010.01356.x
Diarrassouba, F., Diarra, M. S., Bach, S., Delaquis, P., Pritchard, J., Topp, E., et al. (2007). Antibiotic resistance and virulence genes in commensal Escherichia coli and Salmonella isolates from commercial broiler chicken farms. J. Food Prot. 70, 1316–1327.
Dionisi, A. M., Lucarelli, C., Owczarek, S., Luzzi, I., and Villa, L. (2009). Characterization of the plasmid-borne quinolone resistance gene qnrB19 in Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 53, 4019–4021. doi: 10.1128/AAC.00294-09
Djordjevic, S. P., Hornitzky, M. A., Bailey, G., Gill, P., Vanselow, B., Walker, K., et al. (2001). Virulence properties and serotypes of Shiga toxin-producing Escherichia coli from healthy Australian slaughter-age sheep. J. Clin. Microbiol. 39, 2017–2021. doi: 10.1128/JCM.39.5.2017-2021.2001
Djordjevic, S. P., Ramachandran, V., Bettelheim, K. A., Vanselow, B. A., Holst, P., Bailey, G., et al. (2004). Serotypes and virulence gene profiles of shiga toxin-producing Escherichia coli strains isolated from feces of pasture-fed and lot-fed sheep. Appl. Environ. Microbiol. 70, 3910–3917. doi: 10.1128/AEM.70.7.3910-3917.2004
Djordjevic, S. P., Stokes, H. W., and Roy Chowdhury, P. (2013). Mobile elements, zoonotic pathogens and commensal bacteria: conduits for the delivery of resistance genes into humans, production animals and soil microbiota. Front. Microbiol. 4:86. doi: 10.3389/fmicb.2013.00086
Dolejska, M., Frolkova, P., Florek, M., Jamborova, I., Purgertova, M., Kutilova, I., et al. (2011). CTX-M-15-producing Escherichia coli clone B2-O25b-ST131 and Klebsiella spp. isolates in municipal wastewater treatment plant effluents. J. Antimicrob. Chemother. 66, 2784–2790. doi: 10.1093/jac/dkr363
Duriez, P., and Topp, E. (2007). Temporal dynamics and impact of manure storage on antibiotic resistance patterns and population structure of Escherichia coli isolates from a commercial swine farm. Appl. Environ. Microbiol. 73, 5486–5493. doi: 10.1128/AEM.00218-07
Eisenberg, J. N., Goldstick, J., Cevallos, W., Trueba, G., Levy, K., Scott, J., et al. (2012). In-roads to the spread of antibiotic resistance: regional patterns of microbial transmission in northern coastal Ecuador. J. R. Soc. Interface 9, 1029–1039. doi: 10.1098/rsif.2011.0499
Endimiani, A., Rossano, A., Kunz, D., Overesch, G., and Perreten, V. (2012). First countrywide survey of third-generation cephalosporin-resistant Escherichia coli from broilers, swine, and cattle in Switzerland. Diagn. Microbiol. Infect. Dis. 73, 31–38. doi: 10.1016/j.diagmicrobio.2012.01.004
Enne, V. I., Cassar, C., Sprigings, K., Woodward, M. J., and Bennett, P. M. (2008). A high prevalence of antimicrobial resistant Escherichia coli isolated from pigs and a low prevalence of antimicrobial resistant E. coli from cattle and sheep in Great Britain at slaughter. FEMS Microbiol. Lett. 278, 193–199. doi: 10.1111/j.1574-6968.2007.00991.x
Enne, V. I., Livermore, D. M., Stephens, P., and Hall, L. M. (2001). Persistence of sulphonamide resistance in Escherichia coli in the UK despite national prescribing restriction. Lancet 357, 1325–1328. doi: 10.1016/S0140-6736(00)04519-0
Escudero, E., Vinue, L., Teshager, T., Torres, C., and Moreno, M. A. (2010). Resistance mechanisms and farm-level distribution of fecal Escherichia coli isolates resistant to extended-spectrum cephalosporins in pigs in Spain. Res. Vet. Sci. 88, 83–87. doi: 10.1016/j.rvsc.2009.05.021
European Medicines Agency [EMA] (2013). Use of Colistin Products in Animals within the European Union: Development of Resistance and Possible Impact on Human and Animal Health. London: European Medicines Agency.
Ewers, C., Bethe, A., Semmler, T., Guenther, S., and Wieler, L. H. (2012). Extended-spectrum beta-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: a global perspective. Clin. Microbiol. Infect. 18, 646–655. doi: 10.1111/j.1469-0691.2012.03850.x
Ewers, C., Bethe, A., Stamm, I., Grobbel, M., Kopp, P. A., Guerra, B., et al. (2014). CTX-M-15-D-ST648 Escherichia coli from companion animals and horses: another pandemic clone combining multiresistance and extraintestinal virulence? J. Antimicrob. Chemother. 69, 1224–1230. doi: 10.1093/jac/dkt516
Ewers, C., Guenther, S., Wieler, L. H., and Schierack, P. (2009). Mallard ducks - a waterfowl species with high risk of distributing Escherichia coli pathogenic for humans. Environ. Microbiol. Rep. 1, 510–517. doi: 10.1111/j.1758-2229.2009.00058.x
Ewers, C., Li, G., Wilking, H., Kiessling, S., Alt, K., Antao, E. M., et al. (2007). Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: how closely related are they? Int. J. Med. Microbiol. 297, 163–176. doi: 10.1016/j.ijmm.2007.01.003
Fairbrother, J. M., Lariviere, S., and Johnson, W. M. (1988). Prevalence of fimbrial antigens and enterotoxins in nonclassical serogroups of Escherichia coli isolated from newborn pigs with diarrhea. Am. J. Vet. Res. 49, 1325–1328.
Fairbrother, J. M., Nadeau, E., and Gyles, C. L. (2005). Escherichia coli in postweaning diarrhea in pigs: an update on bacterial types, pathogenesis, and prevention strategies. Anim. Health Res. Rev. 6, 17–39. doi: 10.1079/AHR2005105
Falkow, S. (1988). Molecular Koch’s postulates applied to microbial pathogenicity. Rev. Infect. Dis. 10(Suppl. 2), S274–S276. doi: 10.1093/cid/10.Supplement_2.S274
Fekete, P. Z., Brzuszkiewicz, E., Blum-Oehler, G., Olasz, F., Szabo, M., Gottschalk, G., et al. (2012). DNA sequence analysis of the composite plasmid pTC conferring virulence and antimicrobial resistance for porcine enterotoxigenic Escherichia coli. Int. J. Med. Microbiol. 302, 4–9. doi: 10.1016/j.ijmm.2011.07.003
Fischer, J., Rodriguez, I., Baumann, B., Guiral, E., Beutin, L., Schroeter, A., et al. (2014). blaCTX-M-(1)(5)-carrying Escherichia coli and Salmonella isolates from livestock and food in Germany. J. Antimicrob. Chemother. 69, 2951–2958. doi: 10.1093/jac/dku270
Ford, P. J., and Avison, M. B. (2004). Evolutionary mapping of the SHV beta-lactamase and evidence for two separate IS26-dependent blaSHV mobilization events from the Klebsiella pneumoniae chromosome. J. Antimicrob. Chemother. 54, 69–75. doi: 10.1093/jac/dkh251
Fortini, D., Fashae, K., Garcia-Fernandez, A., Villa, L., and Carattoli, A. (2011). Plasmid-mediated quinolone resistance and beta-lactamases in Escherichia coli from healthy animals from Nigeria. J. Antimicrob. Chemother. 66, 1269–1272. doi: 10.1093/jac/dkr085
Foti, M., Rinaldo, D., Guercio, A., Giacopello, C., Aleo, A., De Leo, F., et al. (2011). Pathogenic microorganisms carried by migratory birds passing through the territory of the island of Ustica, Sicily (Italy). Avian. Pathol. 40, 405–409. doi: 10.1080/03079457.2011.588940
Frank, T., Gautier, V., Talarmin, A., Bercion, R., and Arlet, G. (2007). Characterization of sulphonamide resistance genes and class 1 integron gene cassettes in Enterobacteriaceae, Central African Republic (CAR). J. Antimicrob. Chemother. 59, 742–745. doi: 10.1093/jac/dkl538
Freire Martin, I., AbuOun, M., Reichel, R., La Ragione, R. M., and Woodward, M. J. (2014). Sequence analysis of a CTX-M-1 IncI1 plasmid found in Salmonella 4,5,12:i:-, Escherichia coli and Klebsiella pneumoniae on a UK pig farm. J. Antimicrob. Chemother. 69, 2098–2101. doi: 10.1093/jac/dku098
Gao, L., Hu, J., Zhang, X., Wei, L., Li, S., Miao, Z., et al. (2015). Application of swine manure on agricultural fields contributes to extended-spectrum beta-lactamase-producing Escherichia coli spread in Tai’an, China. Front. Microbiol. 6:313. doi: 10.3389/fmicb.2015.00313
Gao, P., Munir, M., and Xagoraraki, I. (2012). Correlation of tetracycline and sulfonamide antibiotics with corresponding resistance genes and resistant bacteria in a conventional municipal wastewater treatment plant. Sci. Total Environ. 42, 173–183. doi: 10.1016/j.scitotenv.2012.01.061
Garcia-Cobos, S., Kock, R., Mellmann, A., Frenzel, J., Friedrich, A. W., and Rossen, J. W. (2015). Molecular typing of Enterobacteriaceae from pig holdings in North-Western Germany reveals extended-spectrum and AmpC beta-lactamases producing but no carbapenem resistant ones. PLoS ONE 10:e0134533. doi: 10.1371/journal.pone.0134533
Gillings, M. R. (2014). Integrons: past, present, and future. Microbiol. Mol. Biol. Rev. 78, 257–277. doi: 10.1128/MMBR.00056-13
Gillings, M. R., Boucher, Y., Labbate, M., Holmes, A., Krishnan, S., Holley, M., et al. (2008). The evolution of class 1 integrons and the rise of antibiotic resistance. J. Bacteriol. 190, 5095–5100. doi: 10.1128/JB.00152-08
Gillings, M. R., Gaze, W. H., Pruden, A., Smalla, K., Tiedje, J. M., and Zhu, Y. G. (2015). Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J. 9, 1269–1279. doi: 10.1038/ismej.2014.226
Glasner, C., Albiger, B., Buist, G., Tambic Andrasevic, A., Canton, R., Carmeli, Y., et al. (2013). Carbapenemase-producing Enterobacteriaceae in europe: a survey among national experts from 39 countries, February 2013. Euro. Surveill. 18:3. doi: 10.2807/1560-7917.ES2013.18.28.20525
Goncalves, A., Torres, C., Silva, N., Carneiro, C., Radhouani, H., Coelho, C., et al. (2010). Genetic characterization of extended-spectrum beta-lactamases in Escherichia coli isolates of pigs from a Portuguese intensive swine farm. Foodborne Pathog. Dis. 7, 1569–1573. doi: 10.1089/fpd.2010.0598
Grape, M., Sundstrom, L., and Kronvall, G. (2003). Sulphonamide resistance gene sul3 found in Escherichia coli isolates from human sources. J. Antimicrob. Chemother. 52, 1022–1024. doi: 10.1093/jac/dkg473
Gullberg, E., Albrecht, L. M., Karlsson, C., Sandegren, L., and Andersson, D. I. (2014). Selection of a multidrug resistance plasmid by sublethal levels of antibiotics and heavy metals. MBio 5:e01918-14. doi: 10.1128/mBio.01918-14
Hall, R. M., Brown, H. J., Brookes, D. E., and Stokes, H. W. (1994). Integrons found in different locations have identical 5’ ends but variable 3’ ends. J. Bacteriol. 176, 6286–6294.
Hammerum, A. M., Larsen, J., Andersen, V. D., Lester, C. H., Skovgaard Skytte, T. S., Hansen, F., et al. (2014). Characterization of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli obtained from Danish pigs, pig farmers and their families from farms with high or no consumption of third- or fourth-generation cephalosporins. J. Antimicrob. Chemother. 69, 2650–2657. doi: 10.1093/jac/dku180
Hammerum, A. M., Sandvang, D., Andersen, S. R., Seyfarth, A. M., Porsbo, L. J., Frimodt-Moller, N., et al. (2006). Detection of sul1, sul2 and sul3 in sulphonamide resistant Escherichia coli isolates obtained from healthy humans, pork and pigs in Denmark. Int. J. Food Microbiol. 106, 235–237. doi: 10.1016/j.ijfoodmicro.2005.06.023
Han, D. U., Choi, C., Kim, J., Cho, W. S., Chung, H. K., Ha, S. K., et al. (2002). Anti-microbial susceptibility for east1 + Escherichia coli isolated from diarrheic pigs in Korea. J. Vet. Med. B Infect. Dis. Vet. Public Health 49, 346–348. doi: 10.1046/j.1439-0450.2002.00577.x
Hansen, F., Olsen, S. S., Heltberg, O., Justesen, U. S., Fuglsang-Damgaard, D., Knudsen, J. D., et al. (2014). Characterization of third-generation cephalosporin-resistant Escherichia coli from bloodstream infections in Denmark. Microb. Drug Resist. 20, 316–324. doi: 10.1089/mdr.2013.0157
Hansen, K. H., Damborg, P., Andreasen, M., Nielsen, S. S., and Guardabassi, L. (2013). Carriage and fecal counts of cefotaxime M-producing Escherichia coli in pigs: a longitudinal study. Appl. Environ. Microbiol. 79, 794–798. doi: 10.1128/AEM.02399-12
Hanson, R., Kaneene, J. B., Padungtod, P., Hirokawa, K., and Zeno, C. (2002). Prevalence of Salmonella and E. coli, and their resistance to antimicrobial agents, in farming communities in northern Thailand. S. Asian J. Trop. Med. Public Health 33(Suppl. 3), 120–126.
Harada, K., Asai, T., Kojima, A., Ishihara, K., and Takahashi, T. (2006). Role of coresistance in the development of resistance to chloramphenicol in Escherichia coli isolated from sick cattle and pigs. Am. J. Vet. Res. 67, 230–235. doi: 10.2460/ajvr.67.2.230
Harada, K., Asai, T., Kojima, A., Oda, C., Ishihara, K., and Takahashi, T. (2005). Antimicrobial susceptibility of pathogenic Escherichia coli isolated from sick cattle and pigs in Japan. J. Vet. Med. Sci. 67, 999–1003. doi: 10.1292/jvms.67.999
Harmer, C. J., and Hall, R. M. (2015). IS26-mediated precise excision of the IS26-aphA1a translocatable unit. MBio 6, e1866–e1815. doi: 10.1128/mBio.01866-15
Harmer, C. J., Moran, R. A., and Hall, R. M. (2014). Movement of IS26-associated antibiotic resistance genes occurs via a translocatable unit that includes a single IS26 and preferentially inserts adjacent to another IS26. mBio 5:e01801-14. doi: 10.1128/mBio.01801-14
He, S., Hickman, A. B., Varani, A. M., Siguier, P., Chandler, M., Dekker, J. P., et al. (2015). Insertion Sequence IS26 Reorganises Plasmids in Clinically Isolated Multidrug-Resistant Bacteria by Replicative Transposition. mBio 6:e00762. doi: 10.1128/mBio.00762-15
Heikkila, E., Sundstrom, L., and Huovinen, P. (1990). Trimethoprim resistance in Escherichia coli isolates from a geriatric unit. Antimicrob. Agents Chemother. 34, 2013–2015. doi: 10.1128/AAC.34.10.2013
Herrero-Fresno, A., Larsen, I., and Olsen, J. E. (2015). Genetic relatedness of commensal Escherichia coli from nursery pigs in intensive pig production in Denmark and molecular characterization of genetically different strains. J. Appl. Microbiol. 119, 342–353. doi: 10.1111/jam.12840
Heuer, H., Schmitt, H., and Smalla, K. (2011a). Antibiotic resistance gene spread due to manure application on agricultural fields. Curr. Opin. Microbiol. 14, 236–243. doi: 10.1016/j.mib.2011.04.009
Heuer, H., Solehati, Q., Zimmerling, U., Kleineidam, K., Schloter, M., Muller, T., et al. (2011b). Accumulation of sulfonamide resistance genes in arable soils due to repeated application of manure containing sulfadiazine. Appl. Environ. Microbiol. 77, 2527–2530. doi: 10.1128/AEM.02577-10
Heuer, H., and Smalla, K. (2007). Manure and sulfadiazine synergistically increased bacterial antibiotic resistance in soil over at least two months. Environ. Microbiol. 9, 657–666. doi: 10.1111/j.1462-2920.2006.01185.x
Ho, P. L., Chan, J., Lo, W. U., Law, P. Y., and Chow, K. H. (2013). Plasmid-mediated fosfomycin resistance in Escherichia coli isolated from pig. Vet. Microbiol. 162, 964–967. doi: 10.1016/j.vetmic.2012.09.023
Ho, P. L., Wong, R. C., Chow, K. H., and Que, T. L. (2009). Distribution of integron-associated trimethoprim-sulfamethoxazole resistance determinants among Escherichia coli from humans and food-producing animals. Lett. Appl. Microbiol. 49, 627–634. doi: 10.1111/j.1472-765X.2009.02717.x
Hobman, J. L., and Crossman, L. C. (2015). Bacterial antimicrobial metal ion resistance. J. Med. Microbiol. 64, 471–497. doi: 10.1099/jmm.0.023036-0
Hornitzky, M. A., Mercieca, K., Bettelheim, K. A., and Djordjevic, S. P. (2005). Bovine feces from animals with gastrointestinal infections are a source of serologically diverse atypical enteropathogenic Escherichia coli and Shiga toxin-producing E. coli strains that commonly possess intimin. Appl. Environ. Microbiol. 71, 3405–3412. doi: 10.1128/aem.71.7.3405-3412.2005
Hornitzky, M. A., Vanselow, B. A., Walker, K., Bettelheim, K. A., Corney, B., Gill, P., et al. (2002). Virulence properties and serotypes of Shiga toxin-producing Escherichia coli from healthy Australian cattle. Appl. Environ. Microbiol. 68, 6439–6445. doi: 10.1128/aem.68.12.6439-6445.2002
Hsu, S. C., Chiu, T. H., Pang, J. C., Hsuan-Yuan, C. H., Chang, G. N., and Tsen, H. Y. (2006). Characterisation of antimicrobial resistance patterns and class 1 integrons among Escherichia coli and Salmonella enterica serovar Choleraesuis strains isolated from humans and swine in Taiwan. Int. J. Antimicrob. Agents 27, 383–391. doi: 10.1016/j.ijantimicag.2005.11.020
Huang, S. Y., Zhu, X. Q., Wang, Y., Liu, H. B., Dai, L., He, J. K., et al. (2012). Co-carriage of qnrS1, floR, and bla(CTX-M-14) on a multidrug-resistant plasmid in Escherichia coli isolated from pigs. Foodborne Pathog. Dis. 9, 896–901. doi: 10.1089/fpd.2012.1131
Hvistendahl, M. (2012). Public health. China takes aim at rampant antibiotic resistance. Science 336:795. doi: 10.1126/science.336.6083.795
Institute of Medicine; National Research Council; Panel on Animal Health (1999). The Use of Drugs in Food Animals: Benefits and Risks. Washington, DC: National Academy Press.
Jackson, G. (1981). A survey of antibiotic resistance of Escherichia coli isolated from farm animals in Great Britain from 1971 to 1977. Vet. Rec. 108, 325–328. doi: 10.1136/vr.108.15.325
Jakobsen, L., Garneau, P., Kurbasic, A., Bruant, G., Stegger, M., Harel, J., et al. (2011). Microarray-based detection of extended virulence and antimicrobial resistance gene profiles in phylogroup B2 Escherichia coli of human, meat and animal origin. J. Med. Microbiol. 60, 1502–1511. doi: 10.1099/jmm.0.033993-0
Jakobsen, L., Kurbasic, A., Skjot-Rasmussen, L., Ejrnaes, K., Porsbo, L. J., Pedersen, K., et al. (2010a). Escherichia coli isolates from broiler chicken meat, broiler chickens, pork, and pigs share phylogroups and antimicrobial resistance with community-dwelling humans and patients with urinary tract infection. Foodborne Pathog. Dis. 7, 537–547. doi: 10.1089/fpd.2009.0409
Jakobsen, L., Spangholm, D. J., Pedersen, K., Jensen, L. B., Emborg, H. D., Agerso, Y., et al. (2010b). Broiler chickens, broiler chicken meat, pigs and pork as sources of ExPEC related virulence genes and resistance in Escherichia coli isolates from community-dwelling humans and UTI patients. Int. J. Food Microbiol. 142, 264–272. doi: 10.1016/j.ijfoodmicro.2010.06.025
Jamborova, I., Dolejska, M., Vojtech, J., Guenther, S., Uricariu, R., Drozdowska, J., et al. (2015). Plasmid-mediated resistance to cephalosporins and fluoroquinolones in various Escherichia coli sequence types isolated from rooks wintering in Europe. Appl. Environ. Microbiol. 81, 648–657. doi: 10.1128/AEM.02459-14
Jensen, V. F., Jakobsen, L., Emborg, H. D., Seyfarth, A. M., and Hammerum, A. M. (2006). Correlation between apramycin and gentamicin use in pigs and an increasing reservoir of gentamicin-resistant Escherichia coli. J. Antimicrob. Chemother. 58, 101–107. doi: 10.1093/jac/dkl201
Jobbins, S. E., and Alexander, K. A. (2015). From whence they came – antibiotic-resistant Escherischia coli in African wildlife. J. Wildl. Dis. 51, 811–820. doi: 10.7589/2014-11-257
Johnson, J. R., Delavari, P., Kuskowski, M., and Stell, A. L. (2001). Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J. Infect. Dis. 183, 78–88. doi: 10.1086/317656
Johnson, J. R., Kuskowski, M. A., Smith, K., O’Bryan, T. T., and Tatini, S. (2005a). Antimicrobial-resistant and extraintestinal pathogenic Escherichia coli in retail foods. J. Infect. Dis. 191, 1040–1049. doi: 10.1086/428451
Johnson, J. R., Murray, A. C., Kuskowski, M. A., Schubert, S., Prere, M. F., Picard, B., et al. (2005b). Distribution and characteristics of Escherichia coli clonal group A. Emerg. Infect. Dis. 11, 141–145. doi: 10.3201/eid1101.040418
Johnson, T. J., Shepard, S. M., Rivet, B., Danzeisen, J. L., and Carattoli, A. (2011). Comparative genomics and phylogeny of the IncI1 plasmids: a common plasmid type among porcine enterotoxigenic Escherichia coli. Plasmid 66, 144–151. doi: 10.1016/j.plasmid.2011.07.003
Jones-Dias, D., Manageiro, V., Francisco, A. P., Martins, A. P., Domingues, G., Louro, D., et al. (2013). Assessing the molecular basis of transferable quinolone resistance in Escherichia coli and Salmonella spp. from food-producing animals and food products. Vet. Microbiol. 167, 523–531. doi: 10.1016/j.vetmic.2013.08.010
Jordan, D., Chin, J. J., Fahy, V. A., Barton, M. D., Smith, M. G., and Trott, D. J. (2009). Antimicrobial use in the Australian pig industry: results of a national survey. Aust. Vet. J. 87, 222–229. doi: 10.1111/j.1751-0813.2009.00430.x
Ju, F., Li, B., Ma, L., Wang, Y., Huang, D., and Zhang, T. (2015). Antibiotic resistance genes and human bacterial pathogens: co-occurrence, removal, and enrichment in municipal sewage sludge digesters. Water Res. 91, 1–10. doi: 10.1016/j.watres.2015.11.071
Ju, W., Shen, J., Li, Y., Toro, M. A., Zhao, S., Ayers, S., et al. (2012). Non-O157 Shiga toxin-producing Escherichia coli in retail ground beef and pork in the Washington D.C. area. Food Microbiol. 32, 371–377. doi: 10.1016/j.fm.2012.07.017
Kadlec, K., and Schwarz, S. (2008). Analysis and distribution of class 1 and class 2 integrons and associated gene cassettes among Escherichia coli isolates from swine, horses, cats and dogs collected in the BfT-GermVet monitoring study. J. Antimicrob. Chemother. 62, 469–473. doi: 10.1093/jac/dkn233
Kang, S. G., Lee, D. Y., Shin, S. J., Ahn, J. M., and Yoo, H. S. (2005). Changes in patterns of antimicrobial susceptibility and class 1 integron carriage among Escherichia coli isolates. J. Vet. Sci. 6, 201–205.
Kaper, J. B., Nataro, J. P., and Mobley, H. L. (2004). Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2, 123–140. doi: 10.1038/nrmicro818
Katakweba, A. A., Moller, K. S., Muumba, J., Muhairwa, A. P., Damborg, P., Rosenkrantz, J. T., et al. (2015). Antimicrobial resistance in faecal samples from buffalo, wildebeest and zebra grazing together with and without cattle in Tanzania. J. Appl. Microbiol. 118, 966–975. doi: 10.1111/jam.12738
Katsunuma, Y., Hanazumi, M., Fujisaki, H., Minato, H., Kataoka, Y., Sawada, T., et al. (2008). Comparison of pulsed-field gel electrophoresis patterns of antimicrobial-resistant Escherichia coli and enterococci isolates from the feces of livestock and livestock farmers in Japan. J. Gen. Appl. Microbiol. 54, 39–50. doi: 10.2323/jgam.54.39
Kijima-Tanaka, M., Ishihara, K., Kojima, A., Morioka, A., Nagata, R., Kawanishi, M., et al. (2005). A national surveillance of Shiga toxin-producing Escherichia coli in food-producing animals in Japan. J. Vet. Med. B Infect. Dis. Vet. Public Health 52, 230–237. doi: 10.1111/j.1439-0450.2005.00852.x
Kijima-Tanaka, M., Ishihara, K., Morioka, A., Kojima, A., Ohzono, T., Ogikubo, K., et al. (2003). A national surveillance of antimicrobial resistance in Escherichia coli isolated from food-producing animals in Japan. J. Antimicrob. Chemother. 51, 447–451. doi: 10.1093/jac/dkg014
Kim, S., and Woo, G. J. (2014). Prevalence and characterization of antimicrobial-resistant Escherichia coli isolated from conventional and organic vegetables. Foodborne Pathog. Dis. 11, 815–821. doi: 10.1089/fpd.2014.1771
Koh, S. Y., George, S., Brozel, V., Moxley, R., Francis, D., and Kaushik, R. S. (2008). Porcine intestinal epithelial cell lines as a new in vitro model for studying adherence and pathogenesis of enterotoxigenic Escherichia coli. Vet. Microbiol. 130, 191–197. doi: 10.1016/j.vetmic.2007.12.018
Kojima, A., Asai, T., Ishihara, K., Morioka, A., Akimoto, K., Sugimoto, Y., et al. (2009). National monitoring for antimicrobial resistance among indicator bacteria isolated from food-producing animals in Japan. J. Vet. Med. Sci. 71, 1301–1308. doi: 10.1292/jvms.001301
Kozak, G. K., Boerlin, P., Janecko, N., Reid-Smith, R. J., and Jardine, C. (2009a). Antimicrobial resistance in Escherichia coli isolates from swine and wild small mammals in the proximity of swine farms and in natural environments in Ontario, Canada. Appl. Environ. Microbiol. 75, 559–566. doi: 10.1128/AEM.01821-08
Kozak, G. K., Pearl, D. L., Parkman, J., Reid-Smith, R. J., Deckert, A., and Boerlin, P. (2009b). Distribution of sulfonamide resistance genes in Escherichia coli and Salmonella isolates from swine and chickens at abattoirs in Ontario and Quebec, Canada. Appl. Environ. Microbiol. 75, 5999–6001. doi: 10.1128/AEM.02844-08
Krishnasamy, V., Otte, J., and Silbergeld, E. (2015). Antimicrobial use in Chinese swine and broiler poultry production. Antimicrob. Resist. Infect. Control 4:17. doi: 10.1186/s13756-015-0050-y
Kristiansson, E., Fick, J., Janzon, A., Grabic, R., Rutgersson, C., Weijdegard, B., et al. (2011). Pyrosequencing of antibiotic-contaminated river sediments reveals high levels of resistance and gene transfer elements. PLoS ONE 6:e17038. doi: 10.1371/journal.pone.0017038
Kumai, Y., Suzuki, Y., Tanaka, Y., Shima, K., Bhadra, R. K., Yamasaki, S., et al. (2005). Characterization of multidrug-resistance phenotypes and genotypes of Escherichia coli strains isolated from swine from an abattoir in Osaka, Japan. Epidemiol. Infect. 133, 59–70. doi: 10.1017/S0950268804003280
Labar, A. S., Millman, J. S., Ruebush, E., Opintan, J. A., Bishar, R. A., Aboderin, A. O., et al. (2012). Regional dissemination of a trimethoprim-resistance gene cassette via a successful transposable element. PLoS ONE 7:e38142. doi: 10.1371/journal.pone.0038142
Landers, T. F., Cohen, B., Wittum, T. E., and Larson, E. L. (2012). A review of antibiotic use in food animals: perspective, policy, and potential. Public Health Rep. 127, 4–22.
Lanz, R., Kuhnert, P., and Boerlin, P. (2003). Antimicrobial resistance and resistance gene determinants in clinical Escherichia coli from different animal species in Switzerland. Vet. Microbiol. 91, 73–84. doi: 10.1016/S0378-1135(02)00263-8
LaPara, T. M., Burch, T. R., McNamara, P. J., Tan, D. T., Yan, M., and Eichmiller, J. J. (2011). Tertiary-treated municipal wastewater is a significant point source of antibiotic resistance genes into Duluth-Superior Harbor. Environ. Sci. Technol. 45, 9543–9549. doi: 10.1021/es202775r
Lay, K. K., Koowattananukul, C., Chansong, N., and Chuanchuen, R. (2012). Antimicrobial resistance, virulence, and phylogenetic characteristics of Escherichia coli isolates from clinically healthy swine. Foodborne Pathog. Dis. 9, 992–1001. doi: 10.1089/fpd.2012.1175
Le, Q. P., Ueda, S., Nguyen, T. N., Dao, T. V., Van Hoang, T. A., Tran, T. T., et al. (2015). Characteristics of extended-spectrum beta-lactamase-producing Escherichia coli in retail meats and shrimp at a local market in Vietnam. Foodborne Pathog. Dis. 12, 719–725. doi: 10.1089/fpd.2015.1954
Lee, M., Shin, E., and Lee, Y. (2014). Antimicrobial resistance and integron profiles in multidrug-resistant Escherichia coli isolates from pigs. Foodborne Pathog. Dis. 11, 988–997. doi: 10.1089/fpd.2014.1795
Le-Minh, N., Khan, S. J., Drewes, J. E., and Stuetz, R. M. (2010). Fate of antibiotics during municipal water recycling treatment processes. Water Res. 44, 4295–4323. doi: 10.1016/j.watres.2010.06.020
Leverstein-van Hall, M. A., He, M. B., Ar, T. D., Paauw, A., Fluit, A. C., and Verhoef, J. (2003). Multidrug resistance among Enterobacteriaceae is strongly associated with the presence of integrons and is independent of species or isolate origin. J. Infect. Dis. 187, 251–259. doi: 10.1086/345880
Levings, R. S., Djordjevic, S. P., and Hall, R. M. (2008). SGI2, a relative of Salmonella genomic island SGI1 with an independent origin. Antimicrob. Agents Chemother. 52, 2529–2537. doi: 10.1128/AAC.00189-08
Levings, R. S., Lightfoot, D., Partridge, S. R., Hall, R. M., and Djordjevic, S. P. (2005). The genomic island SGI1, containing the multiple antibiotic resistance region of Salmonella enterica serovar Typhimurium DT104 or variants of it, is widely distributed in other S. enterica serovars. J. Bacteriol. 187, 4401–4409. doi: 10.1128/jb.187.13.4401-4409.2005
Levy, S. B. (1992). The Antibiotic Paradox: How Miracle Drugs Are Destroying the Miracle. New York, NY: Plenum.
Li, Y. X., Xiong, X., Lin, C. Y., Zhang, F. S., Wei, L., and Wei, H. (2010). Cadmium in animal production and its potential hazard on Beijing and Fuxin farmlands. J. Hazard. Mater. 177, 475–480. doi: 10.1016/j.jhazmat.2009.12.057
Liao, X. P., Xia, J., Yang, L., Li, L., Sun, J., Liu, Y. H., et al. (2015). Characterization of CTX-M-14-producing Escherichia coli from food-producing animals. Front. Microbiol. 6:1136. doi: 10.3389/fmicb.2015.01136
Liebert, C. A., Hall, R. M., and Summers, A. O. (1999). Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63, 507–522.
Lim, S. K., Lee, H. S., Nam, H. M., Cho, Y. S., Kim, J. M., Song, S. W., et al. (2007). Antimicrobial resistance observed in Escherichia coli strains isolated from fecal samples of cattle and pigs in Korea during 2003-2004. Int. J. Food Microbiol. 116, 283–286. doi: 10.1016/j.ijfoodmicro.2006.12.014
Lim, S. K., Lee, H. S., Nam, H. M., Jung, S. C., and Bae, Y. C. (2009). CTX-M-type beta-lactamase in Escherichia coli isolated from sick animals in Korea. Microb. Drug Resist. 15, 139–142. doi: 10.1089/mdr.2009.0901
Lindsey, R. L., Frye, J. G., Thitaram, S. N., Meinersmann, R. J., Fedorka-Cray, P. J., and Englen, M. D. (2011). Characterization of multidrug-resistant Escherichia coli by antimicrobial resistance profiles, plasmid replicon typing, and pulsed-field gel electrophoresis. Microb. Drug Resist. 17, 157–163. doi: 10.1089/mdr.2010.0148
Liu, B. T., Yang, Q. E., Li, L., Sun, J., Liao, X. P., Fang, L. X., et al. (2013). Dissemination and characterization of plasmids carrying oqxAB-bla CTX-M genes in Escherichia coli isolates from food-producing animals. PLoS ONE 8:e73947. doi: 10.1371/journal.pone.0073947
Liu, C., Zheng, H., Yang, M., Xu, Z., Wang, X., Wei, L., et al. (2015). Genome analysis and in vivo virulence of porcine extraintestinal pathogenic Escherichia coli strain PCN033. BMC Genomics 16:717. doi: 10.1186/s12864-015-1890-9
Liu, Y.-Y., Wang, Y., Walsh, T. R., Yi, L.-X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Loman, N. J., and Pallen, M. J. (2015). Twenty years of bacterial genome sequencing. Nat. Rev. Microbiol. 13, 787–794. doi: 10.1038/nrmicro3565
Lu, Z., Lapen, D., Scott, A., Dang, A., and Topp, E. (2005). Identifying host sources of fecal pollution: diversity of Escherichia coli in confined dairy and swine production systems. Appl. Environ. Microbiol. 71, 5992–5998. doi: 10.1128/aem.71.10.5992-5998.2005
Luppi, A., Bonilauri, P., Dottori, M., Gherpelli, Y., Biasi, G., Merialdi, G., et al. (2015). Antimicrobial resistance of F4+ Escherichia coli isolated from Swine in Italy. Transbound. Emerg. Dis. 62, 67–71. doi: 10.1111/tbed.12081
Ma, J., Zeng, Z., Chen, Z., Xu, X., Wang, X., Deng, Y., et al. (2009). High prevalence of plasmid-mediated quinolone resistance determinants qnr, aac(6’)-Ib-cr, and qepA among ceftiofur-resistant Enterobacteriaceae isolates from companion and food-producing animals. Antimicrob. Agents Chemother. 53, 519–524. doi: 10.1128/AAC.00886-08
Ma, L., Zhang, X. X., Zhao, F., Wu, B., Cheng, S., and Yang, L. (2013). Sewage treatment plant serves as a hot-spot reservoir of integrons and gene cassettes. J. Environ. Biol. 34, 391–399.
Machado, E., Coque, T. M., Canton, R., Sousa, J. C., and Peixe, L. (2008). Antibiotic resistance integrons and extended-spectrum {beta}-lactamases among Enterobacteriaceae isolates recovered from chickens and swine in Portugal. J. Antimicrob. Chemother. 62, 296–302. doi: 10.1093/jac/dkn179
Makita, K., Goto, M., Ozawa, M., Kawanishi, M., Koike, R., Asai, T., et al. (2016). Multivariable analysis of the association between antimicrobial use and antimicrobial resistance in Escherichia coli isolated from apparently healthy pigs in Japan. Microb. Drug Resist. 22, 28–39. doi: 10.1089/mdr.2014.0311
Manges, A. R., and Johnson, J. R. (2015). Reservoirs of extraintestinal pathogenic Escherichia coli. Microbiol. Spectr. 3. doi: 10.1128/microbiolspec.UTI-0006-2012
Manges, A. R., Smith, S. P., Lau, B. J., Nuval, C. J., Eisenberg, J. N., Dietrich, P. S., et al. (2007). Retail meat consumption and the acquisition of antimicrobial resistant Escherichia coli causing urinary tract infections: a case-control study. Foodborne Pathog. Dis. 4, 419–431. doi: 10.1089/fpd.2007.0026
Mao, D., Yu, S., Rysz, M., Luo, Y., Yang, F., Li, F., et al. (2015). Prevalence and proliferation of antibiotic resistance genes in two municipal wastewater treatment plants. Water Res. 85, 458–466. doi: 10.1016/j.watres.2015.09.010
Marchant, M., and Moreno, M. A. (2013). Dynamics and diversity of Escherichia coli in animals and system management of the manure on a commercial farrow-to-finish pig farm. Appl. Environ. Microbiol. 79, 853–859. doi: 10.1128/AEM.02866-12
Marchant, M., Vinue, L., Torres, C., and Moreno, M. A. (2013). Change of integrons over time in Escherichia coli isolates recovered from healthy pigs and chickens. Vet. Microbiol. 163, 124–132. doi: 10.1016/j.vetmic.2012.12.011
Mariani-Kurkdjian, P., Lemaitre, C., Bidet, P., Perez, D., Boggini, L., Kwon, T., et al. (2014). Haemolytic-uraemic syndrome with bacteraemia caused by a new hybrid Escherichia coli pathotype. New Microbes New Infect 2, 127–131. doi: 10.1002/nmi2.49
Martinez, J. L. (2008). Antibiotics and antibiotic resistance genes in natural environments. Science 321, 365–367. doi: 10.1126/science.1159483
Mathew, A. G., Liamthong, S., Lin, J., and Hong, Y. (2009). Evidence of class 1 integron transfer between Escherichia coli and Salmonella spp. on livestock farms. Foodborne Pathog. Dis. 6, 959–964. doi: 10.1089/fpd.2009.0263
Mattiello, S. P., Drescher, G., Barth, V. C. Jr., Ferreira, C. A., and Oliveira, S. D. (2015). Characterization of antimicrobial resistance in Salmonella enterica strains isolated from Brazilian poultry production. Antonie Van Leeuwenhoek 108, 1227–1238. doi: 10.1007/s10482-015-0577-1
Maynard, C., Fairbrother, J. M., Bekal, S., Sanschagrin, F., Levesque, R. C., Brousseau, R., et al. (2003). Antimicrobial resistance genes in enterotoxigenic Escherichia coli O149:K91 isolates obtained over a 23-year period from pigs. Antimicrob. Agents Chemother. 47, 3214–3221. doi: 10.1128/AAC.47.10.3214-3221.2003
Mayrhofer, S., Paulsen, P., Smulders, F. J., and Hilbert, F. (2004). Antimicrobial resistance profile of five major food-borne pathogens isolated from beef, pork and poultry. Int. J. Food Microbiol. 97, 23–29. doi: 10.1016/j.ijfoodmicro.2004.04.006
Mayrhofer, S., Paulsen, P., Smulders, F. J., and Hilbert, F. (2006). Antimicrobial resistance in commensal Escherichia coli isolated from muscle foods as related to the veterinary use of antimicrobial agents in food-producing animals in Austria. Microb. Drug Resist. 12, 278–283. doi: 10.1089/mdr.2006.12.278
McEwen, S. A., and Fedorka-Cray, P. J. (2002). Antimicrobial use and resistance in animals. Clin. Infect. Dis. 34(Suppl. 3), S93–S106. doi: 10.1086/340246
Meng, Q., Bai, X., Zhao, A., Lan, R., Du, H., Wang, T., et al. (2014). Characterization of Shiga toxin-producing Escherichia coli isolated from healthy pigs in China. BMC Microbiol. 14:5. doi: 10.1186/1471-2180-14-5
Middleton, J. H., and Ambrose, A. (2005). Enumeration and antibiotic resistance patterns of fecal indicator organisms isolated from migratory Canada geese (Branta canadensis). J. Wildl. Dis. 41, 334–341. doi: 10.7589/0090-3558-41.2.334
Morales, A. S., Fragoso de Araujo, J., de Moura Gomes, V. T., Reis Costa, A. T., dos Prazeres Rodrigues, D., Porfida Ferreira, T. S., et al. (2012). Colistin resistance in Escherichia coli and Salmonella enterica strains isolated from swine in Brazil. Sci. World J. 2012:109795. doi: 10.1100/2012/109795
Moran, R. A., Holt, K. E., and Hall, R. M. (2016). pCERC3 from a commensal ST95 Escherichia coli: a ColV virulence-multiresistance plasmid carrying a sul3-associated class 1 integron. Plasmid 84–84, 11–19. doi: 10.1016/j.plasmid.2016.02.002
Moredo, F. A., Pineyro, P. E., Marquez, G. C., Sanz, M., Colello, R., Etcheverria, A., et al. (2015). Enterotoxigenic Escherichia coli subclinical infection in pigs: bacteriological and genotypic characterization and antimicrobial resistance profiles. Foodborne Pathog. Dis. 12, 704–711. doi: 10.1089/fpd.2015.1959
Moredo, F. A., Vigo, G. B., Cappuccio, J. A., Pineyro, P., Perfumo, C. J., and Giacoboni, G. I. (2007). Antimicrobial resistance of Escherichia coli isolated from pigs in Argentina. Rev. Argent Microbiol. 39, 227–229.
Murugan, K., Prabhakaran, P., Al-Sohaibani, S., and Sekar, K. (2012). Identification of source of faecal pollution of Tirumanimuttar River, Tamilnadu, India using microbial source tracking. Environ. Monit. Assess. 184, 6001–6012. doi: 10.1007/s10661-011-2398-7
Nagy, B., and Fekete, P. Z. (1999). Enterotoxigenic Escherichia coli (ETEC) in farm animals. Vet. Res. 30, 259–284.
Nataro, J. P., and Kaper, J. B. (1998). Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11, 142–201.
Nation, R. L., and Li, J. (2009). Colistin in the 21st century. Curr. Opin. Infect. Dis. 22, 535–543. doi: 10.1097/QCO.0b013e328332e672
Noamani, B. N., Fairbrother, J. M., and Gyles, C. L. (2003). Virulence genes of O149 enterotoxigenic Escherichia coli from outbreaks of postweaning diarrhea in pigs. Vet. Microbiol. 97, 87–101. doi: 10.1016/j.vetmic.2003.08.006
Nordstrom, L., Liu, C. M., and Price, L. B. (2013). Foodborne urinary tract infections: a new paradigm for antimicrobial-resistant foodborne illness. Front. Microbiol. 4:29. doi: 10.3389/fmicb.2013.00029
Nozarian, Z., and Abdollahi, A. (2015). Microbial etiology and antimicrobial susceptibility of bacteria implicated in urinary tract infection in Tehran, Iran. Iran J. Pathol. 10, 54–60.
Ojer-Usoz, E., Gonzalez, D., Vitas, A. I., Leiva, J., Garcia-Jalon, I., Febles-Casquero, A., et al. (2013). Prevalence of extended-spectrum beta-lactamase-producing Enterobacteriaceae in meat products sold in Navarra, Spain. Meat Sci. 93, 316–321. doi: 10.1016/j.meatsci.2012.09.009
Ojo, O. E., Ajuwape, A. T., Otesile, E. B., Owoade, A. A., Oyekunle, M. A., and Adetosoye, A. I. (2010). Potentially zoonotic shiga toxin-producing Escherichia coli serogroups in the faeces and meat of food-producing animals in Ibadan, Nigeria. Int. J. Food Microbiol. 142, 214–221. doi: 10.1016/j.ijfoodmicro.2010.06.030
Olesen, I., Hasman, H., and Aarestrup, F. M. (2004). Prevalence of beta-lactamases among ampicillin-resistant Escherichia coli and Salmonella isolated from food animals in Denmark. Microb. Drug Resist. 10, 334–340. doi: 10.1089/mdr.2004.10.334
Onken, A., Said, A. K., Jorstad, M., Jenum, P. A., and Blomberg, B. (2015). Prevalence and antimicrobial resistance of microbes causing bloodstream infections in Unguja, Zanzibar. PLoS ONE 10:e0145632. doi: 10.1371/journal.pone.0145632
Osborn, A. M., Bruce, K. D., Strike, P., and Ritchie, D. A. (1997). Distribution, diversity and evolution of the bacterial mercury resistance (mer) operon. FEMS Microbiol. Rev. 19, 239–262. doi: 10.1111/j.1574-6976.1997.tb00300.x
Pal, C., Bengtsson-Palme, J., Kristiansson, E., and Larsson, D. G. (2015). Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genomics 16:964. doi: 10.1186/s12864-015-2153-5
Pallen, M. J., and Wren, B. W. (2007). Bacterial pathogenomics. Nature 449, 835–842. doi: 10.1038/nature06248
Pan, J. C., Ye, R., Meng, D. M., Zhang, W., Wang, H. Q., and Liu, K. Z. (2006). Molecular characteristics of class 1 and class 2 integrons and their relationships to antibiotic resistance in clinical isolates of Shigella sonnei and Shigella flexneri. J. Antimicrob. Chemother. 58, 288–296. doi: 10.1093/jac/dkl228
Pan, X., Qiang, Z., Ben, W., and Chen, M. (2011). Residual veterinary antibiotics in swine manure from concentrated animal feeding operations in Shandong Province, China. Chemosphere 84, 695–700. doi: 10.1016/j.chemosphere.2011.03.022
Paniagua-Contreras, G. L., Monroy-Perez, E., Rodriguez-Moctezuma, J. R., Dominguez-Trejo, P., Vaca-Paniagua, F., and Vaca, S. (2015). Virulence factors, antibiotic resistance phenotypes and O-serogroups of Escherichia coli strains isolated from community-acquired urinary tract infection patients in Mexico. J. Microbiol. Immunol. Infect. doi: 10.1016/j.jmii.2015.08.005 [Epub ahead of print].
Pantozzi, F. L., Moredo, F. A., Vigo, G. B., and Giacoboni, G. I. (2010). Antimicrobial resistance in indicator and zoonotic bacteria isolated from domestic animals in Argentina. Rev. Argent Microbiol. 42, 49–52. doi: 10.1590/s0325-75412010000100011
Pardon, B., Catry, B., Dewulf, J., Persoons, D., Hostens, M., De Bleecker, K., et al. (2012). Prospective study on quantitative and qualitative antimicrobial and anti-inflammatory drug use in white veal calves. J. Antimicrob. Chemother. 67, 1027–1038. doi: 10.1093/jac/dkr570
Partridge, S. R., Brown, H. J., Stokes, H. W., and Hall, R. M. (2001). Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob. Agents Chemother. 45, 1263–1270. doi: 10.1128/aac.45.4.1263-1270.2001
Partridge, S. R., Tsafnat, G., Coiera, E., and Iredell, J. R. (2009). Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol. Rev. 33, 757–784. doi: 10.1111/j.1574-6976.2009.00175.x
Perreten, V., and Boerlin, P. (2003). A new sulfonamide resistance gene (sul3) in Escherichia coli is widespread in the pig population of Switzerland. Antimicrob. Agents Chemother. 47, 1169–1172. doi: 10.1128/AAC.47.3.1169-1172.2003
Petternel, C., Galler, H., Zarfel, G., Luxner, J., Haas, D., Grisold, A. J., et al. (2014). Isolation and characterization of multidrug-resistant bacteria from minced meat in Austria. Food Microbiol. 44, 41–46. doi: 10.1016/j.fm.2014.04.013
Phongpaichit, S., Liamthong, S., Mathew, A. G., and Chethanond, U. (2007). Prevalence of class 1 integrons in commensal Escherichia coli from pigs and pig farmers in Thailand. J. Food Prot. 70, 292–299.
Poolman, J. T., and Wacker, M. (2016). Extraintestinal pathogenic Escherichia coli, a common human pathogen: challenges for vaccine development and progress in the field. J. Infect. Dis. 213, 6–13. doi: 10.1093/infdis/jiv429
Povilonis, J., Seputiene, V., Ruzauskas, M., Siugzdiniene, R., Virgailis, M., Pavilonis, A., et al. (2010). Transferable class 1 and 2 integrons in Escherichia coli and Salmonella enterica isolates of human and animal origin in Lithuania. Foodborne Pathog. Dis. 7, 1185–1192. doi: 10.1089/fpd.2010.0536
Pulcini, C., and Gyssens, I. C. (2013). How to educate prescribers in antimicrobial stewardship practices. Virulence 4, 192–202. doi: 10.4161/viru.23706
Qiao, M., Chen, W., Su, J., Zhang, B., and Zhang, C. (2012). Fate of tetracyclines in swine manure of three selected swine farms in China. J. Environ. Sci. (China) 24, 1047–1052. doi: 10.1016/S1001-0742(11)60890-5
Quesada, A., Porrero, M. C., Tellez, S., Palomo, G., Garcia, M., and Dominguez, L. (2015). Polymorphism of genes encoding PmrAB in colistin-resistant strains of Escherichia coli and Salmonella enterica isolated from poultry and swine. J. Antimicrob. Chemother. 70, 71–74. doi: 10.1093/jac/dku320
Rajic, A., Reid-Smith, R., Deckert, A. E., Dewey, C. E., and McEwen, S. A. (2006). Reported antibiotic use in 90 swine farms in Alberta. Can. Vet. J. 47, 446–452.
Rajkhowa, S., and Sarma, D. K. (2014). Prevalence and antimicrobial resistance of porcine O157 and non-O157 Shiga toxin-producing Escherichia coli from India. Trop. Anim. Health Prod. 46, 931–937. doi: 10.1007/s11250-014-0587-4
Ramachandran, V., Hornitzky, M. A., Bettelheim, K. A., Walker, M. J., and Djordjevic, S. P. (2001). The common ovine Shiga toxin 2-containing Escherichia coli serotypes and human isolates of the same serotypes possess a Stx2d toxin type. J. Clin. Microbiol. 39, 1932–1937. doi: 10.1128/JCM.39.5.1932-1937.2001
Ramos, S., Silva, N., Dias, D., Sousa, M., Capelo-Martinez, J. L., Brito, F., et al. (2013). Clonal diversity of ESBL-producing Escherichia coli in pigs at slaughter level in Portugal. Foodborne Pathog. Dis. 10, 74–79. doi: 10.1089/fpd.2012.1173
Randall, L. P., Lemma, F., Rogers, J. P., Cheney, T. E., Powell, L. F., and Teale, C. J. (2014). Prevalence of extended-spectrum-beta-lactamase-producing Escherichia coli from pigs at slaughter in the UK in 2013. J. Antimicrob. Chemother. 69, 2947–2950. doi: 10.1093/jac/dku258
Rayamajhi, N., Kang, S. G., Lee, D. Y., Kang, M. L., Lee, S. I., Park, K. Y., et al. (2008). Characterization of TEM-, SHV- and AmpC-type beta-lactamases from cephalosporin-resistant Enterobacteriaceae isolated from swine. Int. J. Food Microbiol. 124, 183–187. doi: 10.1016/j.ijfoodmicro.2008.03.009
Reid, C. J., Roy Chowdhury, P., and Djordjevic, S. P. (2015). Tn6026 and Tn6029 are found in complex resistance regions mobilised by diverse plasmids and chromosomal islands in multiple antibiotic resistant Enterobacteriaceae. Plasmid 80, 127–137. doi: 10.1016/j.plasmid.2015.04.005
Rodrigues, C., Machado, E., Peixe, L., and Novais, A. (2013). IncI1/ST3 and IncN/ST1 plasmids drive the spread of blaTEM-52 and blaCTX-M-1/-32 in diverse Escherichia coli clones from different piggeries. J. Antimicrob. Chemother. 68, 2245–2248. doi: 10.1093/jac/dkt187
Rodriguez-Siek, K. E., Giddings, C. W., Doetkott, C., Johnson, T. J., Fakhr, M. K., and Nolan, L. K. (2005). Comparison of Escherichia coli isolates implicated in human urinary tract infection and avian colibacillosis. Microbiology 151, 2097–2110. doi: 10.1099/mic.0.27499-0
Roschanski, N., Friese, A., von Salviati-Claudius, C., Hering, J., Kaesbohrer, A., Kreienbrock, L., et al. (2015). Prevalence of carbapenemase producing Enterobacteriaceae isolated from German pig-fattening farms during the years 2011-2013. Vet. Microbiol. doi: 10.1016/j.vetmic.2015.11.030 [Epub ahead of print].
Rosengren, L. B., Waldner, C. L., and Reid-Smith, R. J. (2009). Associations between antimicrobial resistance phenotypes, antimicrobial resistance genes, and virulence genes of fecal Escherichia coli isolates from healthy grow-finish pigs. Appl. Environ. Microbiol. 75, 1373–1380. doi: 10.1128/AEM.01253-08
Roug, A., Byrne, B. A., Conrad, P. A., and Miller, W. A. (2013). Zoonotic fecal pathogens and antimicrobial resistance in county fair animals. Comp. Immunol. Microbiol. Infect. Dis. 36, 303–308. doi: 10.1016/j.cimid.2012.11.006
Roy Chowdhury, P., Charles, I. G., and Djordjevic, S. P. (2015). A role for Tn6029 in the evolution of the complex antibiotic resistance gene loci in genomic island 3 in enteroaggregative hemorrhagic Escherichia coli O104:H4. PLoS ONE 10:e0115781. doi: 10.1371/journal.pone.0115781
Roy Chowdhury, P., McKinnon, J., Wyrsch, E., Hammond, J. M., Charles, I. G., and Djordjevic, S. P. (2014). Genomic interplay in bacterial communities: implications for growth promoting practices in animal husbandry. Front. Microbiol. 5:394. doi: 10.3389/fmicb.2014.00394
Roy Chowdhury, P., Scott, M., Worden, P., Huntington, P., Hudson, B., Karagiannis, T., et al. (2016). Genomic islands 1 and 2 play key roles in the evolution of extensively drug-resistant ST235 isolates of Pseudomonas aeruginosa. Open Biol. 6:150175. doi: 10.1098/rsob.150175
Ruppe, E., Woerther, P. L., Diop, A., Sene, A. M., Da Costa, A., Arlet, G., et al. (2009). Carriage of CTX-M-15-producing Escherichia coli isolates among children living in a remote village in Senegal. Antimicrob. Agents Chemother. 53, 3135–3137. doi: 10.1128/AAC.00139-09
Russo, T. A., and Johnson, J. R. (2003). Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect. 5, 449–456. doi: 10.1016/S1286-4579(03)00049-2
Sabate, M., Prats, G., Moreno, E., Balleste, E., Blanch, A. R., and Andreu, A. (2008). Virulence and antimicrobial resistance profiles among Escherichia coli strains isolated from human and animal wastewater. Res Microbiol 159, 288–293. doi: 10.1016/j.resmic.2008.02.001
Saenz, Y., Vinue, L., Ruiz, E., Somalo, S., Martinez, S., Rojo-Bezares, B., et al. (2010). Class 1 integrons lacking qacEDelta1 and sul1 genes in Escherichia coli isolates of food, animal and human origins. Vet. Microbiol. 144, 493–497. doi: 10.1016/j.vetmic.2010.01.026
Salipante, S. J., Roach, D. J., Kitzman, J. O., Snyder, M. W., Stackhouse, B., Butler-Wu, S. M., et al. (2015). Large-scale genomic sequencing of extraintestinal pathogenic Escherichia coli strains. Genome Res. 25, 119–128. doi: 10.1101/gr.180190.114
Samanta, I., Joardar, S. N., Mahanti, A., Bandyopadhyay, S., Sar, T. K., and Dutta, T. K. (2015). Approaches to characterize extended spectrum beta-lactamase/beta-lactamase producing Escherichia coli in healthy organized vis-a-vis backyard farmed pigs in India. Infect. Genet. Evol. 36, 224–230. doi: 10.1016/j.meegid.2015.09.021
Sarmah, A. K., Meyer, M. T., and Boxall, A. B. (2006). A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment. Chemosphere 65, 725–759. doi: 10.1016/j.chemosphere.2006.03.026
Schierack, P., Rodiger, S., Kuhl, C., Hiemann, R., Roggenbuck, D., Li, G., et al. (2013). Porcine E. coli: virulence-associated genes, resistance genes and adhesion and probiotic activity tested by a new screening method. PLoS ONE 8:e59242. doi: 10.1371/journal.pone.0059242
Schroeder, C. M., Zhao, C., DebRoy, C., Torcolini, J., Zhao, S., White, D. G., et al. (2002). Antimicrobial resistance of Escherichia coli O157 isolated from humans, cattle, swine, and food. Appl. Environ. Microbiol. 68, 576–581. doi: 10.1128/AEM.68.2.576-581.2002
Schwaiger, K., Holzel, C., and Bauer, J. (2010). Resistance gene patterns of tetracycline resistant Escherichia coli of human and porcine origin. Vet. Microbiol. 142, 329–336. doi: 10.1016/j.vetmic.2009.09.066
Seiler, C., and Berendonk, T. U. (2012). Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front. Microbiol. 3:399. doi: 10.3389/fmicb.2012.00399
Seputiene, V., Povilonis, J., Ruzauskas, M., Pavilonis, A., and Suziedeliene, E. (2010). Prevalence of trimethoprim resistance genes in Escherichia coli isolates of human and animal origin in Lithuania. J. Med. Microbiol. 59, 315–322. doi: 10.1099/jmm.0.015008-0
Sheikh, A., Luo, Q., Roy, K., Shabaan, S., Kumar, P., Qadri, F., et al. (2014). Contribution of the highly conserved EaeH surface protein to enterotoxigenic Escherichia coli pathogenesis. Infect. Immun. 82, 3657–3666. doi: 10.1128/IAI.01890-14
Sheikh, A. A., Checkley, S., Avery, B., Chalmers, G., Bohaychuk, V., Boerlin, P., et al. (2012). Antimicrobial resistance and resistance genes in Escherichia coli isolated from retail meat purchased in Alberta, Canada. Foodborne Pathog. Dis. 9, 625–631. doi: 10.1089/fpd.2011.1078
Shepard, S. M., Danzeisen, J. L., Isaacson, R. E., Seemann, T., Achtman, M., and Johnson, T. J. (2012). Genome sequences and phylogenetic analysis of K88- and F18-positive porcine enterotoxigenic Escherichia coli. J. Bacteriol. 194, 395–405. doi: 10.1128/JB.06225-11
Singh, R., Schroeder, C. M., Meng, J., White, D. G., McDermott, P. F., Wagner, D. D., et al. (2005). Identification of antimicrobial resistance and class 1 integrons in Shiga toxin-producing Escherichia coli recovered from humans and food animals. J. Antimicrob. Chemother. 56, 216–219. doi: 10.1093/jac/dki161
Smillie, C., Garcillan-Barcia, M. P., Francia, M. V., Rocha, E. P., and de la Cruz, F. (2010). Mobility of plasmids. Microbiol. Mol. Biol. Rev. 74, 434–452. doi: 10.1128/MMBR.00020-10
Smith, M. G., Jordan, D., Chapman, T. A., Chin, J. J., Barton, M. D., Do, T. N., et al. (2010). Antimicrobial resistance and virulence gene profiles in multi-drug resistant enterotoxigenic Escherichia coli isolated from pigs with post-weaning diarrhoea. Vet. Microbiol. 145, 299–307. doi: 10.1016/j.vetmic.2010.04.004
Soufi, L., Saenz, Y., Vinue, L., Abbassi, M. S., Ruiz, E., Zarazaga, M., et al. (2011). Escherichia coli of poultry food origin as reservoir of sulphonamide resistance genes and integrons. Int. J. Food Microbiol. 144, 497–502. doi: 10.1016/j.ijfoodmicro.2010.11.008
Spurbeck, R. R., Dinh, P. C. Jr., Walk, S. T., Stapleton, A. E., Hooton, T. M., Nolan, L. K., et al. (2012). Escherichia coli isolates that carry vat, fyuA, chuA, and yfcV efficiently colonize the urinary tract. Infect. Immun. 80, 4115–4122. doi: 10.1128/IAI.00752-12
Stannarius, C., Burgi, E., Regula, G., Zychowska, M. A., Zweifel, C., and Stephan, R. (2009). Antimicrobial resistance in Escherichia coli strains isolated from Swiss weaned pigs and sows. Schweiz Arch Tierheilkd. 151, 119–125. doi: 10.1024/0036-7281.151.3.119
Stedt, J., Bonnedahl, J., Hernandez, J., Waldenstrom, J., McMahon, B. J., Tolf, C., et al. (2015). Carriage of CTX-M type extended spectrum beta-lactamases (ESBLs) in gulls across Europe. Acta Vet. Scand. 57:74. doi: 10.1186/s13028-015-0166-3
Stefani, S., Giovanelli, I., Anacarso, I., Condo, C., Messi, P., de Niederhausern, S., et al. (2014). Prevalence and characterization of extended-spectrum beta-lactamase-producing Enterobacteriaceae in food-producing animals in Northern Italy. New Microbiol. 37, 551–555.
Stepanauskas, R., Glenn, T. C., Jagoe, C. H., Tuckfield, R. C., Lindell, A. H., King, C. J., et al. (2006). Coselection for microbial resistance to metals and antibiotics in freshwater microcosms. Environ. Microbiol. 8, 1510–1514. doi: 10.1111/j.1462-2920.2006.01091.x
Stephan, R., and Schumacher, S. (2001). Resistance patterns of non-O157 Shiga toxin-producing Escherichia coli (STEC) strains isolated from animals, food and asymptomatic human carriers in Switzerland. Lett. Appl. Microbiol. 32, 114–117. doi: 10.1046/j.1472-765x.2001.00867.x
Stokes, H. W., and Gillings, M. R. (2011). Gene flow, mobile genetic elements and the recruitment of antibiotic resistance genes into Gram-negative pathogens. FEMS Microbiol. Rev. 35, 790–819. doi: 10.1111/j.1574-6976.2011.00273.x
Summers, A. O. (2002). Generally overlooked fundamentals of bacterial genetics and ecology. Clin. Infect. Dis. 34(Suppl. 3), S85–S92. doi: 10.1086/340245
Summers, A. O. (2006). Genetic linkage and horizontal gene transfer, the roots of the antibiotic multi-resistance problem. Anim. Biotechnol. 17, 125–135. doi: 10.1080/10495390600957217
Sun, J., Deng, H., Li, L., Chen, M. Y., Fang, L. X., Yang, Q. E., et al. (2015). Complete nucleotide sequence of cfr-carrying IncX4 plasmid pSD11 from Escherichia coli. Antimicrob. Agents Chemother. 59, 738–741. doi: 10.1128/AAC.04388-14
Sunde, M. (2005). Prevalence and characterization of class 1 and class 2 integrons in Escherichia coli isolated from meat and meat products of Norwegian origin. J. Antimicrob. Chemother. 56, 1019–1024. doi: 10.1093/jac/dki377
Sunde, M., Fossum, K., Solberg, A., and Sorum, H. (1998). Antibiotic resistance in Escherichia coli of the normal intestinal flora of swine. Microb. Drug Resist. 4, 289–299. doi: 10.1089/mdr.1998.4.289
Sunde, M., and Norstrom, M. (2006). The prevalence of, associations between and conjugal transfer of antibiotic resistance genes in Escherichia coli isolated from Norwegian meat and meat products. J. Antimicrob. Chemother. 58, 741–747. doi: 10.1093/jac/dkl294
Sunde, M., and Sorum, H. (1999). Characterization of integrons in Escherichia coli of the normal intestinal flora of swine. Microb. Drug Resist. 5, 279–287. doi: 10.1089/mdr.1999.5.279
Tadesse, D. A., Zhao, S., Tong, E., Ayers, S., Singh, A., Bartholomew, M. J., et al. (2012). Antimicrobial drug resistance in Escherichia coli from humans and food animals, United States, 1950-2002. Emerg. Infect. Dis. 18, 741–749. doi: 10.3201/eid1805.111153
Tamang, M. D., Nam, H. M., Chae, M. H., Kim, S. R., Gurung, M., Jang, G. C., et al. (2012). Prevalence of plasmid-mediated quinolone resistance determinants among Escherichia coli isolated from food animals in Korea. Foodborne Pathog. Dis. 9, 1057–1063. doi: 10.1089/fpd.2012.1225
Tan, C., Tang, X., Zhang, X., Ding, Y., Zhao, Z., Wu, B., et al. (2012). Serotypes and virulence genes of extraintestinal pathogenic Escherichia coli isolates from diseased pigs in China. Vet. J. 192, 483–488. doi: 10.1016/j.tvjl.2011.06.038
Tang, X., Tan, C., Zhang, X., Zhao, Z., Xia, X., Wu, B., et al. (2011). Antimicrobial resistances of extraintestinal pathogenic Escherichia coli isolates from swine in China. Microb. Pathog. 50, 207–212. doi: 10.1016/j.micpath.2011.01.004
Taylor, N. M., Davies, R. H., Ridley, A., Clouting, C., Wales, A. D., and Clifton-Hadley, F. A. (2008). A survey of fluoroquinolone resistance in Escherichia coli and thermophilic Campylobacter spp. on poultry and pig farms in Great Britain. J. Appl. Microbiol. 105, 1421–1431. doi: 10.1111/j.1365-2672.2008.03877.x
Tian, G. B., Wang, H. N., Zhang, A. Y., Zhang, Y., Fan, W. Q., Xu, C. W., et al. (2012). Detection of clinically important beta-lactamases in commensal Escherichia coli of human and swine origin in western China. J. Med. Microbiol. 61, 233–238. doi: 10.1099/jmm.0.036806-0
Tietze, E., Tschape, H., and Voigt, W. (1989). Characterization of new resistance plasmids belonging to incompatibility group IncQ. J. Basic Microbiol. 29, 695–706. doi: 10.1002/jobm.3620291013
Timmerman, T., Dewulf, J., Catry, B., Feyen, B., Opsomer, G., de Kruif, A., et al. (2006). Quantification and evaluation of antimicrobial drug use in group treatments for fattening pigs in Belgium. Prev. Vet. Med. 74, 251–263. doi: 10.1016/j.prevetmed.2005.10.003
Travis, R. M., Gyles, C. L., Reid-Smith, R., Poppe, C., McEwen, S. A., Friendship, R., et al. (2006). Chloramphenicol and kanamycin resistance among porcine Escherichia coli in Ontario. J. Antimicrob. Chemother. 58, 173–177. doi: 10.1093/jac/dkl207
Trobos, M., Christensen, H., Sunde, M., Nordentoft, S., Agerso, Y., Simonsen, G. S., et al. (2009). Characterization of sulphonamide-resistant Escherichia coli using comparison of sul2 gene sequences and multilocus sequence typing. Microbiology 155, 831–836. doi: 10.1099/mic.0.024190-0
Tsafnat, G., Copty, J., and Partridge, S. R. (2011). RAC: repository of antibiotic resistance cassettes. Database (Oxford) 2011:bar054. doi: 10.1093/database/bar054
Tseng, S. P., Wang, S. F., Kuo, C. Y., Huang, J. W., Hung, W. C., Ke, G. M., et al. (2015). Characterization of fosfomycin resistant extended-spectrum beta-lactamase-producing Escherichia coli isolates from human and pig in Taiwan. PLoS ONE 10:e0135864. doi: 10.1371/journal.pone.0135864
Tsubokura, M., Matsumoto, A., Otsuki, K., Animas, S. B., and Sanekata, T. (1995). Drug resistance and conjugative R plasmids in Escherichia coli strains isolated from migratory waterfowl. J. Wildl. Dis. 31, 352–357. doi: 10.7589/0090-3558-31.3.352
Udikovic-Kolic, N., Wichmann, F., Broderick, N. A., and Handelsman, J. (2014). Bloom of resident antibiotic-resistant bacteria in soil following manure fertilisation. Proc. Natl. Acad. Sci. U.S.A. 111, 15202–15207. doi: 10.1073/pnas.1409836111
Unno, T., Han, D., Jang, J., Lee, S. N., Kim, J. H., Ko, G., et al. (2010). High diversity and abundance of antibiotic-resistant Escherichia coli isolated from humans and farm animal hosts in Jeonnam Province, South Korea. Sci. Total Environ. 408, 3499–3506. doi: 10.1016/j.scitotenv.2010.04.046
Unno, T., Han, D., Jang, J., Widmer, K., Ko, G., Sadowsky, M. J., et al. (2011). Genotypic and phenotypic trends in antibiotic resistant pathogenic Escherichia coli isolated from humans and farm animals in South Korea. Microbes Environ. 26, 198–204. doi: 10.1264/jsme2.ME10194
Uzun, C., Oncul, O., Gumus, D., Alan, S., Dayioglu, N., and Kucuker, M. A. (2015). Virulence genes and antibiotic susceptibilities of uropathogenic E. coli strains. Clin. Lab. 61, 941–950.
Van Boeckel, T. P., Brower, C., Gilbert, M., Grenfell, B. T., Levin, S. A., Robinson, T. P., et al. (2015). Global trends in antimicbial use in food animals. Proc. Natl. Acad. Sci. U.S.A. 112, 5649–5654. doi: 10.1073/pnas.1503141112
van Elsas, J. D., Semenov, A. V., Costa, R., and Trevors, J. T. (2011). Survival of Escherichia coli in the environment: fundamental and public health aspects. ISME J. 5, 173–183. doi: 10.1038/ismej.2010.80
van Hoek, A. H., Veenman, C., van Overbeek, W. M., Lynch, G., de Roda Husman, A. M., and Blaak, H. (2015). Prevalence and characterization of ESBL- and AmpC-producing Enterobacteriaceae on retail vegetables. Int. J. Food Microbiol. 204, 1–8. doi: 10.1016/j.ijfoodmicro.2015.03.014
Vandekerchove, D., Vandemaele, F., Adriaensen, C., Zaleska, M., Hernalsteens, J. P., De Baets, L., et al. (2005). Virulence-associated traits in avian Escherichia coli: comparison between isolates from colibacillosis-affected and clinically healthy layer flocks. Vet. Microbiol. 108, 75–87. doi: 10.1016/j.vetmic.2005.02.009
Venturini, C., Beatson, S. A., Djordjevic, S. P., and Walker, M. J. (2010). Multiple antibiotic resistance gene recruitment onto the enterohemorrhagic Escherichia coli virulence plasmid. FASEB J. 24, 1160–1166. doi: 10.1096/fj.09-144972
Venturini, C., Hassan, K. A., Roy Chowdhury, P., Paulsen, I. T., Walker, M. J., and Djordjevic, S. P. (2013). Sequences of two related multiple antibiotic resistance virulence plasmids sharing a unique IS26-related molecular signature isolated from different Escherichia coli pathotypes from different hosts. PLoS ONE 8:e78862. doi: 10.1371/journal.pone.0078862
Vincent, C., Boerlin, P., Daignault, D., Dozois, C. M., Dutil, L., Galanakis, C., et al. (2010). Food reservoir for Escherichia coli causing urinary tract infections. Emerg. Infect. Dis. 16, 88–95. doi: 10.3201/eid1601.091118
Vinue, L., Saenz, Y., Rojo-Bezares, B., Olarte, I., Undabeitia, E., Somalo, S., et al. (2010). Genetic environment of sul genes and characterisation of integrons in Escherichia coli isolates of blood origin in a Spanish hospital. Int. J. Antimicrob. Agents 35, 492–496. doi: 10.1016/j.ijantimicag.2010.01.012
Vinue, L., Saenz, Y., Somalo, S., Escudero, E., Moreno, M. A., Ruiz-Larrea, F., et al. (2008). Prevalence and diversity of integrons and associated resistance genes in faecal Escherichia coli isolates of healthy humans in Spain. J. Antimicrob. Chemother. 62, 934–937. doi: 10.1093/jac/dkn331
Virdi, V., Coddens, A., De Buck, S., Millet, S., Goddeeris, B. M., Cox, E., et al. (2013). Orally fed seeds producing designer IgAs protect weaned piglets against enterotoxigenic Escherichia coli infection. Proc. Natl. Acad. Sci. U.S.A. 110, 11809–11814. doi: 10.1073/pnas.1301975110
Vogt, D., Overesch, G., Endimiani, A., Collaud, A., Thomann, A., and Perreten, V. (2014). Occurrence and genetic characteristics of third-generation cephalosporin-resistant Escherichia coli in Swiss retail meat. Microb. Drug Resist. 20, 485–494. doi: 10.1089/mdr.2013.0210
Wallace-Gadsden, F., Johnson, J. R., Wain, J., and Okeke, I. N. (2007). Enteroaggregative Escherichia coli related to uropathogenic clonal group A. Emerg. Infect. Dis 13, 757–760. doi: 10.3201/eid1305.061057
Wang, F., Ma, W., Dou, Z., Ma, L., Liu, X., Xu, J., et al. (2006). The estimation of the production amount of animal manure and its environmental effect in China. China Environ. Sci. 26, 614–617.
Watkinson, A. J., Murby, E. J., and Costanzo, S. D. (2007). Removal of antibiotics in conventional and advanced wastewater treatment: implications for environmental discharge and wastewater recycling. Water Res. 41, 4164–4176. doi: 10.1016/j.watres.2007.04.005
Woodford, N., Carattoli, A., Karisik, E., Underwood, A., Ellington, M. J., and Livermore, D. M. (2009). Complete nucleotide sequences of plasmids pEK204, pEK499, and pEK516, encoding CTX-M enzymes in three major Escherichia coli lineages from the United Kingdom, all belonging to the international O25:H4-ST131 clone. Antimicrob. Agents Chemother. 53, 4472–4482. doi: 10.1128/AAC.00688-09
Wu, S., Dalsgaard, A., Hammerum, A. M., Porsbo, L. J., and Jensen, L. B. (2010). Prevalence and characterization of plasmids carrying sulfonamide resistance genes among Escherichia coli from pigs, pig carcasses and human. Acta Vet. Scand. 52:47. doi: 10.1186/1751-0147-52-47
Wu, X. Y., Chapman, T., Trott, D. J., Bettelheim, K., Do, T. N., Driesen, S., et al. (2007). Comparative analysis of virulence genes, genetic diversity, and phylogeny of commensal and enterotoxigenic Escherichia coli isolates from weaned pigs. Appl. Environ. Microbiol. 73, 83–91. doi: 10.1128/AEM.00990-06
Wyrsch, E., Roy Chowdhury, P., Abraham, S., Santos, J., Darling, A. E., Charles, I. G., et al. (2015). Comparative genomic analysis of a multiple antimicrobial resistant enterotoxigenic E. coli O157 lineage from Australian pigs. BMC Genomics 16:165. doi: 10.1186/s12864-015-1382-y
Xia, X., Meng, J., Zhao, S., Bodeis-Jones, S., Gaines, S. A., Ayers, S. L., et al. (2011). Identification and antimicrobial resistance of extraintestinal pathogenic Escherichia coli from retail meats. J. Food Prot. 74, 38–44. doi: 10.4315/0362-028X.JFP-10-251
Xu, X., Cui, S., Zhang, F., Luo, Y., Gu, Y., Yang, B., et al. (2014). Prevalence and characterization of cefotaxime and ciprofloxacin co-resistant Escherichia coli isolates in retail chicken carcasses and Ground Pork, China. Microb. Drug Resist. 20, 73–81. doi: 10.1089/mdr.2012.0224
Xu, Y., Tian, X., Ren, C., Huang, H., Zhang, X., Gong, X., et al. (2012). Analysis of colistin A and B in fishery products by ultra performance liquid chromatography with positive electrospray ionization tandem mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 899, 14–20. doi: 10.1016/j.jchromb.2012.04.028
Yahiaoui, M., Robin, F., Bakour, R., Hamidi, M., Bonnet, R., and Messai, Y. (2015). Antibiotic resistance, virulence, and genetic background of community-acquired uropathogenic Escherichia coli from Algeria. Microb. Drug Resist. 21, 516–526. doi: 10.1089/mdr.2015.0045
Yamamoto, S., Nakano, M., Kitagawa, W., Tanaka, M., Sone, T., Hirai, K., et al. (2014). Characterization of multi-antibiotic-resistant Escherichia coli isolated from beef cattle in Japan. Microbes Environ. 29, 136–144. doi: 10.1264/jsme2.ME13173
Yang, H., Chen, S., White, D. G., Zhao, S., McDermott, P., Walker, R., et al. (2004). Characterization of multiple-antimicrobial-resistant Escherichia coli isolates from diseased chickens and swine in China. J. Clin. Microbiol. 42, 3483–3489. doi: 10.1128/jcm.42.8.3483-3489.2004
Yang, T., Zeng, Z., Rao, L., Chen, X., He, D., Lv, L., et al. (2014). The association between occurrence of plasmid-mediated quinolone resistance and ciprofloxacin resistance in Escherichia coli isolates of different origins. Vet. Microbiol. 170, 89–96. doi: 10.1016/j.vetmic.2014.01.019
Yau, S., Liu, X., Djordjevic, S. P., and Hall, R. M. (2010). RSF1010-like plasmids in Australian Salmonella enterica serovar Typhimurium and origin of their sul2-strA-strB antibiotic resistance gene cluster. Microb. Drug Resist. 16, 249–252. doi: 10.1089/mdr.2010.0033
Yue, L., Jiang, H. X., Liao, X. P., Liu, J. H., Li, S. J., Chen, X. Y., et al. (2008). Prevalence of plasmid-mediated quinolone resistance qnr genes in poultry and swine clinical isolates of Escherichia coli. Vet. Microbiol. 132, 414–420. doi: 10.1016/j.vetmic.2008.05.009
Zhang, R., Sun, B., Wang, Y., Lei, L., Schwarz, S., and Wu, C. (2015). Characterization of a cfr-carrying plasmid from porcine Escherichia coli that closely resembles plasmid pEA3 from the plant pathogen Erwinia amylovora. Antimicrob Agents Chemother. 60, 658–661. doi: 10.1128/AAC.02114-15
Zhang, W. J., Xu, X. R., Schwarz, S., Wang, X. M., Dai, L., Zheng, H. J., et al. (2014). Characterization of the IncA/C plasmid pSCEC2 from Escherichia coli of swine origin that harbours the multiresistance gene cfr. J. Antimicrob. Chemother. 69, 385–389. doi: 10.1093/jac/dkt355
Zhu, Y. G., Johnson, T. A., Su, J. Q., Qiao, M., Guo, G. X., Stedtfeld, R. D., et al. (2013). Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. U.S.A. 110, 3435–3440. doi: 10.1073/pnas.1222743110
Zurfluh, K., Poirel, L., Nordmann, P., Klumpp, J., and Stephan, R. (2015). First detection of Klebsiella variicola producing OXA-181 carbapenemase in fresh vegetable imported from Asia to Switzerland. Antimicrob. Resist. Infect. Control 4:38. doi: 10.1186/s13756-015-0080-5
Keywords: multiple antibiotic resistance, animal production, environmental pollutants, genomic epidemiology, agriculture, Escherichia coli
Citation: Wyrsch ER, Roy Chowdhury P, Chapman TA, Charles IG, Hammond JM and Djordjevic SP (2016) Genomic Microbial Epidemiology Is Needed to Comprehend the Global Problem of Antibiotic Resistance and to Improve Pathogen Diagnosis. Front. Microbiol. 7:843. doi: 10.3389/fmicb.2016.00843
Received: 16 February 2016; Accepted: 22 May 2016;
Published: 15 June 2016.
Edited by:
Patrícia Poeta, University of Trás-os-Montes and Alto Douro, PortugalReviewed by:
Francisco Dionisio, University of Lisbon, PortugalManuela Caniça, National Health Institute Doutor Ricardo Jorge, Portugal
Copyright © 2016 Wyrsch, Roy Chowdhury, Chapman, Charles, Hammond and Djordjevic. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Steven P. Djordjevic, c3RldmVuLmRqb3JkamV2aWNAdXRzLmVkdS5hdQ==
 Ethan R. Wyrsch
Ethan R. Wyrsch Piklu Roy Chowdhury
Piklu Roy Chowdhury Toni A. Chapman
Toni A. Chapman Ian G. Charles
Ian G. Charles Jeffrey M. Hammond
Jeffrey M. Hammond Steven P. Djordjevic
Steven P. Djordjevic