
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
OPINION article
Front. Microbiol., 31 May 2016
Sec. Microbial Symbioses
Volume 7 - 2016 | https://doi.org/10.3389/fmicb.2016.00834
This article is part of the Research TopicBifidobacteria and their role in the human gut microbiotaView all 23 articles
Members of the genus Bifidobacterium are abundant in the stool of most human infants during the initial exclusively milk-fed period of life, especially at an age of 2–3 months (Harmsen et al., 2000; Favier et al., 2002; Mariat et al., 2009; Coppa et al., 2011; Turroni et al., 2012; Yatsunenko et al., 2012; Tannock et al., 2013; Barrett et al., 2015). Bifidobacteria dominate the stool microbiota regardless of whether the infants are fed human milk or formula based on ruminant milk (cow or goat). However, bifidobacteria have about 20% higher relative abundances in human milk-fed compared to formula-fed babies (Tannock et al., 2013). The greater abundance of bifidobacteria in human-milk-fed infants can, at least in part, be explained by the fact that bifidobacterial species that are enriched in the infant bowel can utilize Human Milk Oligosaccharides (HMO) or their components as growth substrates (Sela et al., 2008; LoCascio et al., 2010; Garrido et al., 2013). It could be anticipated, therefore, that bifidobacteria would be detectable in the stool microbiota of every child nourished at the breast because of the supply of appropriate growth substrates. This expectation is not borne out completely because a proportion of infants have very low abundance or undetectable bifidobacteria as members of the fecal microbiota regardless of breast milk or formula feeding (Young et al., 2004; Gore et al., 2008; Tannock et al., 2013). Antibiotics had not been administered to these infants. How then can the absence of bifidobacteria be explained?
The “bifidobacteria-negative” babies have been detected in both human milk and formula-fed infants. Therefore, a bacterial growth substrate effect seems unlikely. While human milk is rich in HMO and ruminant milk lacks these complex molecules (although simpler forms such as sialylated-lactose are present in very small amounts), bifidobacteria are still the most abundant taxon in the feces of infants fed formula un-supplemented with galacto- or fructo-oligosaccharides (Tannock et al., 2013). In this case, lactose and/or glycoproteins and glycolipids are probable growth substrates for bifidobacteria (Turroni et al., 2010; Bottacini et al., 2014; O'Callaghan et al., 2015) in the bowel of exclusively milk fed infants. There is, however, a need to support genomic analysis of bifidobacteria with culture-based investigations of bifidobacterial nutrition based on substrates present in the bowel of exclusively milk-fed babies (other than HMO).
An obvious reason for bifidobacteria-negative feces is that the detection methods lack sufficient sensitivity. Culture-based methods usually have a lower detection limit of 1 × 103 per gram, fluorescent in situ hybridization (FISH) 1 × 106 − 107 per gram (manual or digital counts respectively) or ~4 × 104 by flow cytometry, and denaturing gradient gel electrophoresis of PCR amplicons ~1 × 105 – 106 cells (Welling et al., 1997; Jansen et al., 1999; Zoetendal et al., 2001, 2002) or 1 × 104 using internal transcribed spacer targets (Milani et al., 2014). High throughput DNA sequencing methods, such as Illumina, generate tens of thousands of 16S rRNA gene sequences per DNA sample, but there may be several hundred OTU per sample. Thus, taxa present in very low abundance could be missed. However, reference to rarefaction curves (alpha diversity) during sequence analysis will show whether coverage of the microbiota is near complete or not. Therefore, while lack of sufficient sensitivity of detection methods remains a possibility, it probably does not provide the total explanation.
Most fecal microbiota studies examine a single fecal sample from each participating individual. Comprehensive temporal studies of the fecal microbiota to determine day-to-day variations in composition have not been reported. It is possible that bifidobacteria are present in the feces of all children during early life but that, on some days, the bifidobacterial population falls to undetectable levels. Populations of bifidobacteria in the feces of some adults without diseases are dynamic in terms of strain composition, so there is some support for a concept of temporal instability in the bifidobacterial population of the microbiota (McCartney et al., 1996). Figure 1A shows data from feces collected at intervals from infants during the first 12 weeks of life. In the example, fluctuations in the abundances of bifidobacteria were seen, varying from very low abundance to absence, in feces of individual children. Strikingly, bifidobacteria were not detected in any of the fecal samples of one child. Therefore, bifidobacteria-free infants do seem to be a real phenomenon.
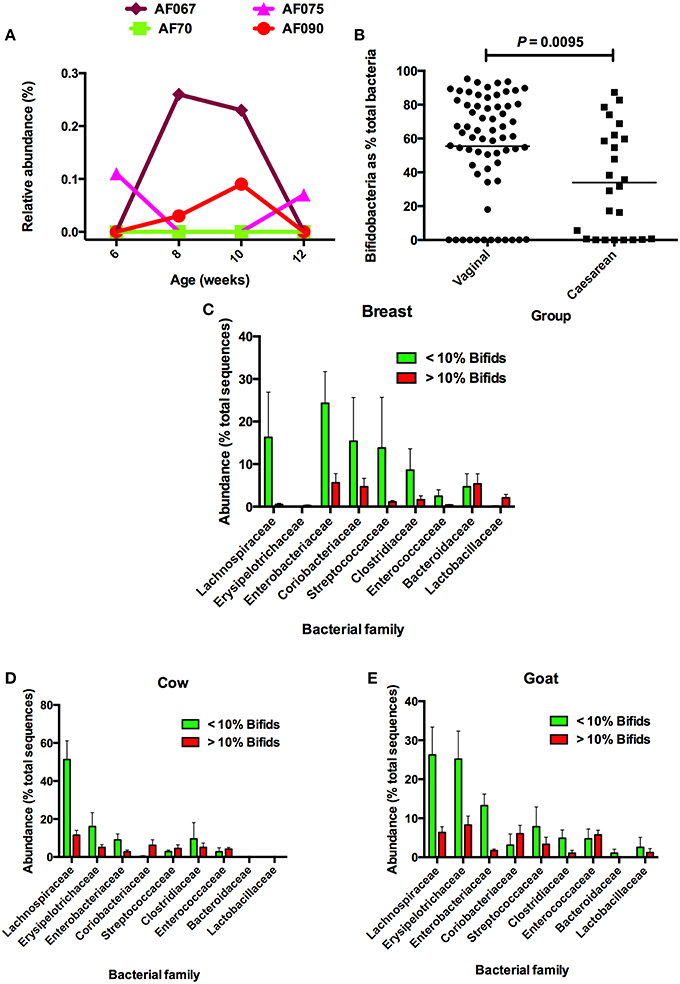
Figure 1. (A) Examples of babies without bifidobacteria, or very low abundances of bifidobacteria, in feces during the first 12 weeks of life. Bifidobacteria were undetectable by DNA sequencing of 16S rRNA gene amplicons at 6 and 12 weeks in the feces of AF067; 6, 8, 10, and 12 weeks in AF70; 8 and 10 weeks in AF075; and 6 and 12 weeks in AF090. (B) Comparison of bifidobacerial abundances in the feces of infants delivered vaginally or by cesarean. Note that in both groups, bifidobacteria were not detected in some infants. (C) Comparison of abundances of bacterial families in microbiotas of breast milk-fed infants in relation to abundances of bifidobacteria. (D) Comparison of abundances of bacterial families in microbiotas of cow milk formula-fed infants in relation to abundances of bifidobacteria. (E) Comparison of abundances of bacterial families in microbiotas of goat milk formula-fed infants in relation to abundances of bifidobacteria. Note that Lachnospiraceae have increased abundances when bifidobacteria have low relative abundance. Figures after Tannock et al. (2013), reproduced with permission.
A window of opportunity is a short time period during which an otherwise unattainable opportunity exists. After the window of opportunity closes, the opportunity ceases to exist. Caufield was the first to describe the “window of infectivity” in the acquisition of commensal bacteria. His example was Streptococcus mutans in the oral cavity of children (Caufield et al., 1993; Li and Caufield, 1995). This bacterial species is associated with dental plaque, thus the window of infectivity coincided with the eruption of the first molars. Prior to this, a habitat for S. mutans is not available in the oral cavity of children for this species. The Caufield hypothesis reminds us that many factors have to coincide to favor the establishment of a commensal in a body site. Cesarean-delivered babies have lower prevalences of bifidobacteria in their feces in early life (Figure 1B). By analogy to Caufield's studies, this probably relates to a lack of favorable opportunities for bifidobacteria to colonize the bowel relative to the vaginal birth process. Notably, we found that 36% of cesarean-derived babies lacked bifidobacteria, whereas 18% of vaginally delivered infants were bifidobacteria-free at 2 months of age (Tannock et al., 2013).
If bifidobacteria have not colonized the bowel of certain infants, they are likely to be replaced by other taxa, which may have the requisite metabolic properties to fill the vacant ecological niche. In a study of the fecal microbiotas of Australian babies that were breast milk- or formula-fed, we compared the relative abundances of bacterial taxa in infants that had very low (< 10%) or higher (>10%) bifidobacterial content (Tannock et al., 2013). Analysis of the compositions of these microbiotas showed that when Bifidobacteriaceae abundance was low, Lachnospiraceae abundances tended to be greater in babies in all dietary groups (Figures 1C–E). There was also a tendency for Erysipelotrichaceae abundances to be greater in formula-fed babies with low bifidobacterial abundances, being much more evident in the case of goat milk-fed infants. These observations suggest that, yes, other taxa might replace bifidobacteria in the fecal microbiota of some children.
The absence of bifidobacteria in the bowel may be detrimental for infant development. The curious phenomenon whereby mother's milk contains substances not used in the nutrition of the offspring, but which fertilize bifidobacterial growth, is unique to humans. There must be a good reason for this. Enriching bifidobacterial populations in the bowel tends to minimize the abundance of other bacterial species, so a competitive exclusion function could be ascribed to HMO. Additionally, HMO may act as “decoys” in the bowel by binding to pathogens (bacteria and viruses) and their toxins and thus limiting contact with mucosal surfaces (Kunz et al., 2000). The large diversity of HMO structures that is known to occur in human milk suggests a large diversity of decoy functions (Pacheco et al., 2015). Irrespective of where in the World babies live, their gut microbiomes are enriched in genes involved in the de novo biosynthesis of folate (Yatsunenko et al., 2012). In contrast, the microbiome of adults favors synthesis of another B vitamin, cobalamin. Folate synthesis is an attribute of bifidobacteria and folate can be absorbed from the large bowel, so enrichment of bifidobacteria in the infant bowel may provide an important contribution to infant nutrition (Aufreiter et al., 2009; D'Aimmo et al., 2012; Lakoff et al., 2014). Folate functions as a coenzyme or co-substrate in single-carbon transfers in the synthesis of nucleic acids and metabolism of amino acids. One of the most important folate-dependent reactions is the conversion of homocysteine to methionine in the synthesis of S-adenosyl-methionine, an important methyl donor. Another folate-dependent reaction, the methylation of deoxyuridylate to thymidylate in the formation of DNA, is required for proper cell division (Crider et al., 2012). Neonatal nutrition could, indeed, be the very important reason for the HMO-bifidobacteria-infant paradigm. The foundation of brain structure and function is set early in life through genetic, biological and psychosocial influences. The rate of neonatal brain growth exceeds that of any other organ or body tissue (Wang, 2012). The infant is born with neurons already formed but the synaptic connections between these cells are mostly established and elaborated after birth causing a large nutritional demand for biosynthesis of gangliosides (Svennerholm et al., 1989). Nutrition of the infant in early life affects brain developmental processes including cognition (Uauy and Peirano, 1999; Uauy et al., 2001). While long-chain fatty acids (such as docosahexaenoic acid) have been the focus of much of the research in this field, tantalizing research evidence now indicates that sialic acid (N-acetyl-neuraminic acid), a 9-carbon carbohydrate, is also an essential nutrient for optimal brain development and cognition (Gibson, 1999; Meldrum et al., 2012; Wang, 2012). Strikingly, cortical tissue from human brain contains up to 4 times more sialic acid than that of other mammals tested (Wang et al., 1998). Moreover, the sialic acid concentration in the brain of breast milk-fed babies is higher than in that of formula-fed infants (Wang et al., 2003). These facts correlate with the unique biochemistry of human milk and the unique bacteriology of the infant bowel. Intriguingly, Ruhaak et al. (2014) have reported the detection of sialylated oligosaccharides (3′ sialyl-lactose, 6′ sialyl-lactose, 3′ sialyl-lactosamine, 6′ sialyl-lactosamine) that might result from the hydrolysis of HMO, in the blood of human infants. Thus, bifidobacterial biochemistry in the bowel may have extra-intestinal, nutritional influences important in brain development. However, perhaps the taxa that are abundant in the bowel of infants in the absence of bifidobacteria can carry out these same functions? This interesting possibility remains to be investigated.
Rene Dubos explored in a number of books the interplay between environmental forces and the physical, mental, and spiritual development of humankind. His article published in the journal Pediatrics entitled “Biological Freudianism: lasting effects of early environmental influences” encapsulated this theme (Dubos et al., 1966). Drawing on the results of experiments conducted with specific-pathogen-free mice, the authors concluded that “From all points of view, the child is truly the father of the man, and for this reason we need to develop an experimental science that might be called biological Freudianism. Socially and individually the response of human beings to the conditions of the present is always conditioned by the biological remembrance of things past.”
Biological Freudianism is clearly of relevance to the concept that the first 1000 days, between conception and the child's second birthday, offer a unique window of opportunity to shape healthier and more prosperous futures. Nutrition during this 1000 day window can have a profound impact on a child's ability to grow, and learn. The influences of the microbiota on the development of the child during early life are potentially very important, and much longitudinal research is required to clarify whether there are continuing, medically important impacts of the microbiota, inlcuding the bifidobacteria, that last throughout the lifetime of humans. Comparisons of the cognitive development and general health status of children that had been bifidobacteria-free, and children that were ex-bifidobacteria-free then intentionally exposed to bifidobacteria, in a longitudinal study extending perhaps 10 or 20 years, would tell us whether these bacteria optimize short and/or long term human development and health.
GT wrote the article. BL, PL, and KW provided data described in the article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Aufreiter, S., Gregory, J. F., Pfeiffer, C. M., Fazil, Z., Kim, Y.-I., Marcon, N., et al. (2009). Folate is absorbed across the colon of adults: evidence from cecal infusion of 13C-labelled [6S]-5-formaltetrahydrofolic acid. Am. J. Clin. Nutr. 90, 116–123. doi: 10.3945/ajcn.2008.27345
Barrett, E., Deshpandey, A. K., Ryan, C. A., Dempsey, E. M., Murphy, B., O'sullivan, L., et al. (2015). The neonatal gut harbours distinct bifidobacterial strains. Arch. Dis. Child. Fetal Neonatal Ed. 100, F405–F410. doi: 10.1136/archdischild-2014-306110
Bottacini, F., Motherway, M. O., Kucynski, J., O'Connell, K. J., Serafini, F., Duranti, S., et al. (2014). Comparative genomics of the Bifidobacterium breve taxon. BMC Genom. 15:170. doi: 10.1186/1471-2164-15-170
Caufield, P. W., Cutter, G. R., and Dasanayake, A. P. (1993). Initial acquisition of mutans streptococci by infants: evidence for a discrete window of infectivity. J. Dent. Res. 72, 37–45. doi: 10.1177/00220345930720010501
Coppa, G. V., Gabrielli, O., Zampini, L., Galeazzi, T., Ficcadenti, A., Padella, L., et al. (2011). Oligosaccharides in 4 different milk groups, Bifidobacteria and Ruminococcus obeum. J. Pediatr. Gastroenterol. Nutr. 53, 80–87. doi: 10.1097/MPG.0b013e3182073103
Crider, K. S., Young, T. P., Berry, R. J., and Bailey, L. B. (2012). Folate and DNA methylation: a review of molecular mechanisms and the evidence for folate's role. Adv. Nutr. 3, 21–38. doi: 10.3945/an.111.000992
D'Aimmo, M. R., Mattarelli, P., Biavati, B., Carlsson, N. G., and Andlid, T. (2012). The potential of bifidobacteria as a source of natural folate. J. Appl. Microbiol. 112, 975–984. doi: 10.1111/j.1365-2672.2012.05261
Dubos, R., Savage, D., Schaedler, R., and Biological Freudianism (1966). Lasting effects of early environmental influences. Pediatric 38, 789–800.
Favier, C. F., Vaughan, E. E., De Vos, W. M., and Akkermans, A. D. L. (2002). Molecular monitoring of succession of bacterial communities in human neonates. Appl. Environ. Microbiol. 68, 219–226. doi: 10.1128/AEM.68.1.219-226.2002
Garrido, D., Dallas, D. C., and Mills, D. A. (2013). Consumption of human milk glycoconjugates by infant-associated bifidobacteria: mechanisms and implications. Microbiology 159, 649–664. doi: 10.1099/mic.0.064113-0
Gibson, R. A. (1999). Long-chain polyunsaturated fatty acids and infant development. Lancet 354, 1919–1920. doi: 10.1016/S0140-6736(99)00300-1
Gore, C., Munro, K., Lay, C., Bibiloni, R., Morris, J., Woodcock, A., et al. (2008). Bifidobacterium pseudocatenulatum is associated with atopic eczema: a nested case-control study investigating the fecal microbiota of infants. J. Allergy. Clin. Immunol. 121, 135–140. doi: 10.1016/j.jaci.2007.07.061
Harmsen, H. J., Wildeboer-Veloo, A. C., Raangs, G. C., Wagendorp, A. A., Klijn, N., Bindels, J. G., et al. (2000). Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J. Pediatr. Gastroenterol. Nutr. 30, 61–67. doi: 10.1097/00005176-200001000-00019
Jansen, G. J., Wildeboer-Veloo, A. C., Tonk, R. H., Franks, A. H., and Welling, G. W. (1999). Development and validation of an automated, microscopy-based method for enumeration of groups of intestinal bacteria. J. Microbiol. Methods 37, 215–221. doi: 10.1016/S0167-7012(99)00049-4
Kunz, C., Rudloff, S., Baier, W., Klein, N., and Strobel, S. (2000). Oligosaccharides in human milk: structural, functional, and metabolic aspects. Annu. Rev. Microbiol. 20, 699–722. doi: 10.1146/annurev.nutr.20.1.699
Lakoff, A., Fazili, Z., Aufreiter, S., Pfeiffer, C. M., Connolly, B., Gregory, J. F., et al. (2014). Folate is absorbed across the human colon: evidence using enteric-coated caplets containing 13C-labeled [6S]-5-formyltetrahydrofolate. Am. J. Clin. Nutr. 100, 1278–1286. doi: 10.3945/ajcn.114.091785
Li, Y., and Caufield, P. W. (1995). The fidelity of initial acquisition of mutans streptococci by infants from their mothers. J. Dent. Res. 74, 681–685. doi: 10.1177/00220345950740020901
LoCascio, R. G., Desai, P., Sela, D. A., Weimer, B., and Mills, D. A. (2010). Broad conservation of milk utilization genes in Bifidobacterium longum subsp. infantis as revealed by comparative genomic hybridization. Appl. Environ. Microbiol. 76, 7373–7381. doi: 10.1128/AEM.00675-10
Mariat, D., Firmesse, O., Levenez, F., Guimaraes, V. D., Sokol, H., Doré, J., et al. (2009). The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 9:123. doi: 10.1186/1471-2180-9-123
McCartney, A. L., Wenzhi, W., and Tannock, G. W. (1996). Molecular analysis of the composition of the bifidobacterial and lactobacillus microflora of humans. Appl. Environ. Microbiol. 62, 4608–4613.
Meldrum, S. J., D'Vaz, N., Simmer, K., Dunstan, J. A., Hird, K., and Prescott, S. L. (2012). Effects of high-dose fish oil supplementation during early infancy on neurodevelopment and language: a randomized controlled trial. Br. J. Nutr. 108, 1443–1454. doi: 10.1017/S0007114511006878
Milani, C., Lugli, G. A., Turroni, F., Mancabelli, L., Duranti, S., Viappiani, A., et al. (2014). Evaluation of bifidobacterial community composition in the human gut by means of a targeted amplicon sequencing (ITS)protocol. FEMS Microbiol. Ecol. 90, 493–503. doi: 10.1111/1574-6941.12410
O'Callaghan, A., Bottacini, F., Motherway, M. O., and van Sinderen, D. (2015). Pangenome analysis of Bifidobacterium longum and site-directed mutagenesis through by-pass of restriction-modification systems. BMC Genom. 16:832. doi: 10.1186/s12864-015-1968-4
Pacheco, A. R., Barile, D., Underwood, M. A., and Mills, D. A. (2015). The impact of the milk glycobiome on the neonate gut microbiota. Ann. Rev. Anim. Sci. 3, 419–445. doi: 10.1146/annurev-animal-022114-111112
Ruhaak, L. R., Stroble, C., Underwood, M. A., and Lebrilla, C. B. (2014). Detection of milk oligosaccharides in plasma of infants. Anal. Bioanal. Chem. 406, 5775–5784. doi: 10.1007/s00216-014-8025-z
Sela, D. A., Chapman, J., Adeuya, A., Kim, J. H., Chen, F., Whitehead, T. R., et al. (2008). The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl. Acad. Sci.U.S.A. 2, 18964–18969. doi: 10.1073/pnas.0809584105
Svennerholm, L., Boström, K., Fredman, P., Mansson, J. E., Rosengren, B., and Rynmark, B. M. (1989). Human brain gangliosides: developmental changes from early fetal stage to advanced age. Biochim. Biophys. Acta 1005, 109–117. doi: 10.1016/0005-2760(89)90175-6
Tannock, G. W., Lawley, B., Munro, K., Pathmanathan, S. G., Zhou, S. J., Makrides, M., et al. (2013). Comparison of the compositions of the stool microbiotas of infants fed goat milk formula, cow milk-based formula, or breast milk. Appl. Environ. Microbiol. 79, 3040–3048. doi: 10.1128/AEM.03910-12
Turroni, F., Bottacini, F., Foroni, E., Mulder, I., Kim, J.-H., Zomer, A., et al. (2010). Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc. Natl. Acad. Sci. U.S.A. 107, 19514–19519. doi: 10.1073/pnas.1011100107
Turroni, F., Peano, C., Pass, D. A., Foroni, E., Severgnini, M., Claesson, M. J., et al. (2012). Diversity of bifidobacteria within the infant gut microbiota. PLoS ONE 7:e36957. doi: 10.1371/journal.pone.0036957
Uauy, R., Mena, P., and Peirano, P. (2001). Mechanisms for nutrient effects on brain development and cognition. Nestle Nutr. Workshop Ser. Clin. Perform Programme 5, 41–70. doi: 10.1159/000061845
Uauy, R., and Peirano, P. (1999). Breast is best: human milk is the optimal food for brain development. Amer. J. Clin. Nutr. 70, 433–434.
Wang, B. (2012). Molecular mechanism underlying sialic acid as an essential nutrient for brain development and cognition. Adv. Nutr. 3, 465S–472S. doi: 10.3945/an.112.001875
Wang, B., McVeagh, P., Petocz, P., and Brand-Miller, J. (2003). Brain ganglioside and glycoprotein sialic acid in breast-fed compared with formula-fed infants. Am. J. Clin. Nutr. 78, 1024–1029.
Wang, B., Miller, J. B., McNeil, Y., and McVeagh, P. (1998). Sialic acid concentration of brain gangliosides: variation among eight mammalian species. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 19, 435–439. doi: 10.1016/S1095-6433(97)00445-5
Welling, G. W., Elfferich, P., Raangs, G. C., Wildeboer-Veloo, A. C., Jansen, G. J., and Degener, J. E. (1997). 16S ribosomal RNA-taggd oligonucleotide probes for monitoring of intestinal tract bacteria. Scand. J. Gastroenterol. 222, 17–19. doi: 10.1080/00365521.1997.11720711
Yatsunenko, T., Rey, F. E., Manary, M. J., Trehan, I., Dominguez-Bello, M. G., Contreras, M., et al. (2012). Human gut microbiome viewed across age and geography. Nature 486, 222–227. doi: 10.1038/nature11053
Young, S. L., Simon, M. A., Baird, M. A., Tannock, G. W., Bibiloni, R., Spencely, K., et al. (2004). Bifidobacterial species differentially affect expression of cell surface markers and cytokines of dendritic cells harvested from cord blood. Clin. Diagn. Lab. Immunol. 11, 686–690. doi: 10.1128/cdli.11.4.686-690.2004
Zoetendal, E. G., Ben-Amor, K., Akkermans, A. D., Abee, T., and de Vos, W. M. (2001). DNA isolation protocols affect the detection limit of PCR approaches of bacteria in samples from the human gastrointestinal tract. Syst. Appl. Microbiol. 24, 405–410. doi: 10.1078/0723-2020-00060
Zoetendal, E. G., von Wright, A., Vilponen-Samela, T., Ben-Amor, K., Akkermans, A. D., and de Vos, W. M. (2002). Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl. Environ. Microbiol. 68, 3401–3407. doi: 10.1128/AEM.68.7.3401-3407.2002
Keywords: bifidobacteria, infants, fecal microbiota, Lachnospiraceae, child development
Citation: Tannock GW, Lee PS, Wong KH and Lawley B (2016) Why Don't All Infants Have Bifidobacteria in Their Stool? Front. Microbiol. 7:834. doi: 10.3389/fmicb.2016.00834
Received: 24 February 2016; Accepted: 17 May 2016;
Published: 31 May 2016.
Edited by:
Francesca Turroni, University College Cork, IrelandReviewed by:
Sabrina Duranti, University of Parma, ItalyCopyright © 2016 Tannock, Lee, Wong and Lawley. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gerald W. Tannock, Z2VyYWxkLnRhbm5vY2tAb3RhZ28uYWMubno=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.