
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol. , 31 May 2016
Sec. Evolutionary and Genomic Microbiology
Volume 7 - 2016 | https://doi.org/10.3389/fmicb.2016.00829
Integrative conjugative elements (ICEs) are a class of bacterial mobile elements that have the ability to mediate their own integration, excision, and transfer from one host genome to another by a mechanism of site-specific recombination, self-circularisation, and conjugative transfer. Members of the SXT/R391 ICE family of enterobacterial mobile genetic elements display an unusual UV-inducible sensitization function which results in stress induced killing of bacterial cells harboring the ICE. This sensitization has been shown to be associated with a stress induced overexpression of a mobile element encoded conjugative transfer gene, orf43, a traV homolog. This results in cell lysis and release of a circular form of the ICE. Induction of this novel system may allow transfer of an ICE, enhancing its survival potential under conditions not conducive to conjugative transfer.
Integrative conjugative elements (ICEs) are a class of diverse bacterial mobile elements that are characterized by their ability to mediate and encode all determinants for their own integration, excision, and transfer from one host genome to another by a mechanism of site-specific recombination, self-circularisation, and conjugative transfer (Taviani et al., 2009; Michael et al., 2012). In general the elements are mosaic in structure containing Phage-like integration systems, plasmid like transfer and maintenance systems and accumulated accessory genes with homology to a variety of organisms suggesting evolutionary accumulation via passage and residency in a variety of host organisms (Boltner et al., 2002; Pembroke et al., 2002). They are a major factor in the evolution of bacterial genomes allowing bacteria to rapidly acquire new phenotypic traits and adaptive functions such as resistance to antimicrobial compounds and heavy metals, virulence mechanisms, metabolic pathways (such as pathways for the degradation of xenobiotic pollutants) and the ability to resist bacteriophage infection (Ryan et al., 2009; Wozniak and Waldor, 2010; Guglielmini et al., 2011; Van Houdt et al., 2012). There are a growing number of ICEs being reported, many associated with multiple drug resistance and adaptive traits and archived in the ICE Berg Database1 (Bi et al., 2012).
The core set of genes required for a functionally active ICE are divided into three distinct modules known as the maintenance, conjugation, and regulation module (Wozniak and Waldor, 2009). The maintenance module contains the genes responsible for the stable integration of an ICE into a host genome and is defined by the presence of two genes int and xis (Storrs et al., 1991). The ICE integrase (int) is a recombinase which catalyzes the recombination reaction between specific recognition sites in a host genome (attB) and the circular ICE element usually called attP (located on the ICE; McGrath and Pembroke, 2004). In addition to the integrase most ICEs encode a recombination directionality factor (RDF) termed Xis which stimulates excision of ICE elements for transfer (Lewis and Hatfull, 2001; Abbani et al., 2005; O’Halloran et al., 2007; Wozniak and Waldor, 2010). ICEs transfer via the conjugation module and utilize conjugation mechanisms that are highly similar to those of conjugative plasmids. Gram negative ICEs typically use type IV secretion systems for conjugative transfer (Burrus and Waldor, 2004; Wozniak and Waldor, 2010; Guglielmini et al., 2011; Bellanger et al., 2014). ICE elements generally contain a regulatory module to control various aspects of ICE metabolism. ICESt1 and the SXT/R391 family have been found to have regulatory systems containing “phage-like” regulatory genes that are induced by DNA damaging agents (Beaber and Waldor, 2004; O’Halloran et al., 2007; Bellanger et al., 2008).
The SXT/R391 family of ICEs is one of the largest of the ICE families with >100 elements being identified experimentally or bioinformatically from this group to date (Bi et al., 2012). This group was originally recognized following the sequencing of R391 (Boltner et al., 2002). Since then a large number of SXT/R391 elements have been identified based on similar mosaic structure while earlier elements dating back to the 1970s such as R997 and pMERPH have also been shown to be members of this group (McGrath et al., 2006). ICEs of the SXT/R391 family have been identified in both clinical (human and veterinary) and environmental isolates of Gammaproteobacteria in the main (Burrus and Waldor, 2004; Wozniak et al., 2009) which is significant as the integration site attB within the prfC gene (a 17 bp site at the 5′ end of the gene; Hochhut and Waldor, 1999; McGrath and Pembroke, 2004; Wozniak and Waldor, 2010) is highly conserved within this bacterial group (Armshaw and Pembroke, 2013c).
Since the start of the 1990s, SXT/R391 ICEs have been found to be widespread in both environmental and clinical Vibrio cholerae isolates from Asia and Africa (Burrus and Waldor, 2004; Spagnoletti et al., 2014). SXT/R391 ICEs have been found in all isolates recovered from cholera patients in Haiti (Ceccarelli et al., 2013) and are naturally occurring in many other enterobacteriaceae (Juiz-Rio et al., 2005; Pembroke and Piterina, 2006).
The ICE SXT (99 kb) is one of two archetypal members of the SXT/R391 family that was initially isolated from a multidrug resistant clinical isolate of V. cholerae O139 in India in 1992 (Beaber et al., 2002b). The second archetypal member of the SXT/R391 family is ICE R391 (89 kb), which was first discovered in 1967 in an isolate of Providencia rettgeri from South Africa (Boltner et al., 2002).
All elements contain a conserved core set of genes (49 genes- 29 of known function and 20 hypothetical genes, see Figure 1) and sequences that facilitate regulation of element functions, their integration/excision and their conjugative transfer (Wozniak et al., 2009). Studies by Beaber et al. (2002a), Wozniak et al. (2009), and Lei et al. (2016) proposed that SXT/R391 family members contained five “hotspots” and five variable regions into which accessory (non-core) genes integrated at specific locations within the core genome. These genes code for proteins involved in antibiotic and heavy metal resistance, restriction modification systems, DNA repair systems, and many other functions.
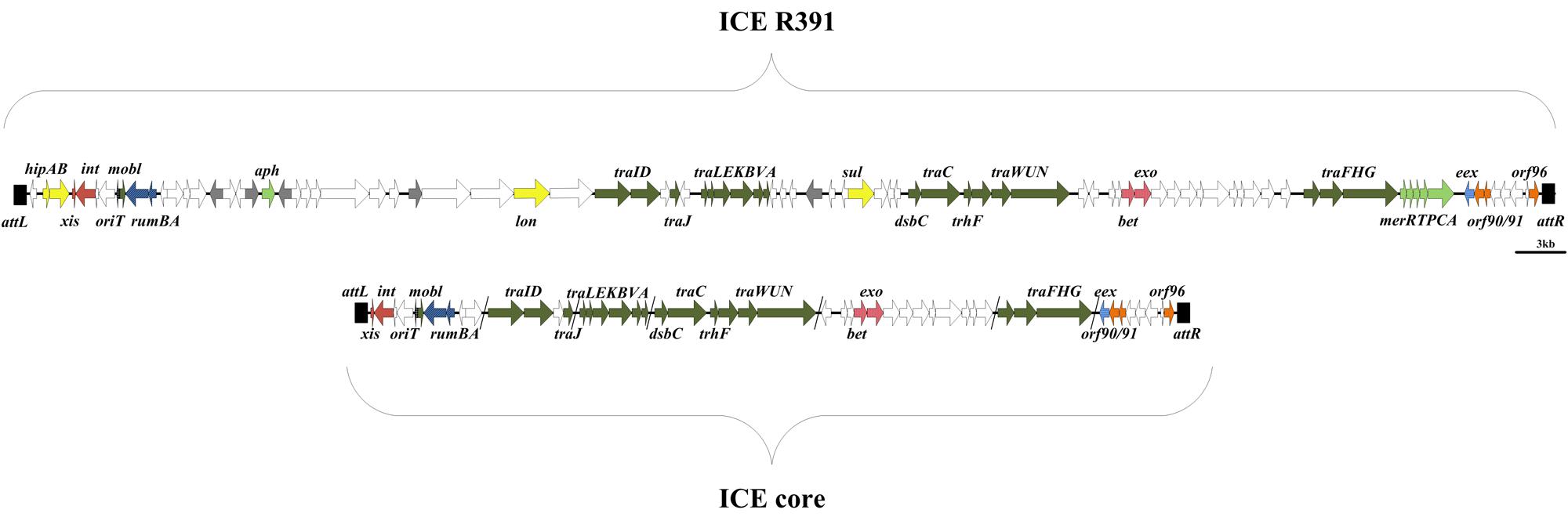
FIGURE 1. Molecular map of the ICE R391 showing the location of the genes associated with the 89 kb element (Boltner et al., 2002; Armshaw and Pembroke, 2013c). orf4 encoding the excisionase Jef/xis (O’Halloran et al., 2007) is located at the left end of the element in the integrated form. Orf43 here designated traV is located in the center of the element within a cluster of ICE transfer genes (Armshaw and Pembroke, 2013a,b, 2015). Underneath the ICE core is illustrated containing genes that are common to all ICE elements (Wozniak et al., 2009).
The SOS response is a global response to DNA damage in which the cell cycle is arrested and DNA repair and mutagenesis are induced. The system involves the RecA protein (Michel, 2005). In 2004 it was discovered that repression of setC and setD in ICE SXT (orf90 and 91 in ICE R391), which are homologous to transcriptional enhancers, resulted in induction of the SOS response, which promotes autoproteolysis of SetR (Orf96) (Beaber et al., 2004). SetR/Orf96 encodes a cI-like repressor protein with homology to the phage λ434 cI repressor. (Boltner et al., 2002) This in turn was found to upregulate the transfer of the ICE SXT element. Poulin-Laprade et al. (2015) demonstrated that expression of genes associated with the conjugative function of the SXT/R391 ICE family are tightly regulated by SetR/Orf96. This represses the expression of the transcriptional enhancer genes setC/orf90 and setD/orf91. The products of these genes activate transcription of ICE encoded int, jef (xis) and conjugation-associated operons (O’Halloran et al., 2007; Poulin-Laprade et al., 2015).
Members of the ICE SXT/R391 family have been shown to induce an unusual UV-inducible sensitization function following DNA damage (Pembroke and Stevens, 1984; McGrath et al., 2006). In the early 1980s it was noted that the presence of R391 and R997 sensitized a strain of Escherichia coli AB1157 to UV irradiation, significantly decreasing post-irradiation cell survival rates. It was thought that this sensitization effect was most likely due to interference with one of the RecA∗-induced DNA damage repair pathways of E. coli as the effect was shown to be recA-dependent but lexA-independent (Pembroke and Stevens, 1984). The mechanism could not be elucidated at the time and no rationale as to why the R391 element (ICE R391) should possess this effect on host cells was proposed.
Analysis of the nature of UV sensitization associated with ICE R391 was then investigated again using a structured deletion library of the entire ICE (Armshaw and Pembroke, 2013a,b). It appeared that deletion of ICE encoded orf90 and 91 (the R391 transcriptional enhancers) and a conjugative transfer gene orf43 (encoding a TraV homolog) abolished the induced sensitization. This was interesting as orf43 was found to be one of the most conserved of the “core genes” of the SXT/R391 family with all homologs (found to date) being ≥98% similar, indicating that it may be under evolutionary pressure to stay conserved. Orf43 is a TraV homolog which forms part of the mating pore during conjugation (Alvarez-Martinez and Christie, 2009). Characterization of how this gene is expressed revealed that UV and other DNA damaging agents induced the hosts recA gene who’s product RecA∗ cleaved the ICE R391 repressor orf96. Cleavage of orf96 resulted in induction of orf90 and 91 (Armshaw and Pembroke, 2013c; Poulin-Laprade et al., 2015).
It had previously been demonstrated that induction of orf90/91 resulted in the upregulation of expression of orf4 which encodes for the ICE R391 excisionase, Jef (J excisionase). Induction of Jef, which is also known as Xis (O’Halloran et al., 2007), resulted in an increased copy number of the circular transfer intermediate of ICE R391 and hence an increased conjugation rate to recipients cells (O’Halloran et al., 2007). This was also shown by Poulin-Laprade et al. (2015).
It was of interest to then examine if this expression hierarchy, which resulted in induction of orf43 (Armshaw and Pembroke, 2013a, 2015), was associated with Jef/Xis and indeed what was the mechanism of sensitization resulting from orf43 induction.
Cloning and controlled expression of orf43 was utilized initially to probe the nature of the ‘UV sensitization’ (Armshaw and Pembroke, 2015). Upon overexpression, as occurs upon UV induction controlled by orf90/91, increased cell permeability and cell lysis was observed (Armshaw and Pembroke, 2015) consistent with increased pore formation within the ICE R391 cell. Transmission electronic microscopy (TEM) analysis revealed significant cell lysis (Armshaw and Pembroke, 2015) consistent with cell death and the sensitization observed following native orf43 induction.
Although, the significance of such a detrimental effect was not immediately obvious when the two phenomenon (increased circular form of the mobile element and cell bursting) were linked, a testable hypothesis emerged. On the one hand DNA damage induces excision of the element (O’Halloran et al., 2007; Poulin-Laprade et al., 2015) while at the same time causing cell lysis. It was reasoned that this lysis might allow for a ‘trap-door’ effect whereby the circular intermediate (the excised ICE element), might escape in the absence of a functional conjugative mechanism such as might be the case in severely damaged cells. This hypothesis was tested using specific deletion mutants of ICE R391 that were deficient in conjugative transfer (Armshaw and Pembroke, 2013a). These strains were unable to undergo conjugative transfer at any detectable levels. However, upon orf43 induction as occurs in cells with DNA damage that contained conjugative defective ICE R391, it was determined that low levels of detectable transfer could be restored (Armshaw and Pembroke, 2015). Effectively, the apparent UV sensitization/DNA damage allows the formation of a ‘trap-door’ to allow ICE survival upon significant DNA damage to the host cell. This mechanism could allow SXT/R391 ICEs, which are missing necessary conjugative genes (possible examples of these were identified by Spagnoletti et al., 2014, however, alternative explanations for selective loss of conjugation functions post-transfer may apply) to transfer via the ‘trap-door’ mechanism at low levels. The elements are then taken up by other cells in “apparent transformation” In Armshaw and Pembroke (2015); experiments were carried out with gene knockouts. The conjugation apparatus genes orf40–orf44 were knocked out. Transfer rates of <10-10 were found. When an expression plasmid with orf43 was added to this knockout transfer was restored at rates of 1.54 × 10-7 (without UV) and 1.01 × 10-6 (with UV). Addition of DNase abolished transfer (Transfer rates of <10-10). This indicated that some form of transformation must have occurred.
This mechanism (Figure 2) could also provide a back-up escape route to allow conjugation levels discussed above, so that if the cell were so damaged that conjugation cannot take place then a secondary escape mechanism would be available to the element. This appears to be a novel mechanism, which we term a ‘trap-door,’ of mobile element survival and presumably occurs in tandem with formation of circular transfer intermediate, which is subsequently released as a result of pore formation and cell lysis making the element available for transformation allowing for some ICE survival in extreme environments. This proposed mechanism is so far unique to the SXT/R391 family and no comparable escape mechanism has been found in other mobile genetic elements thus far.
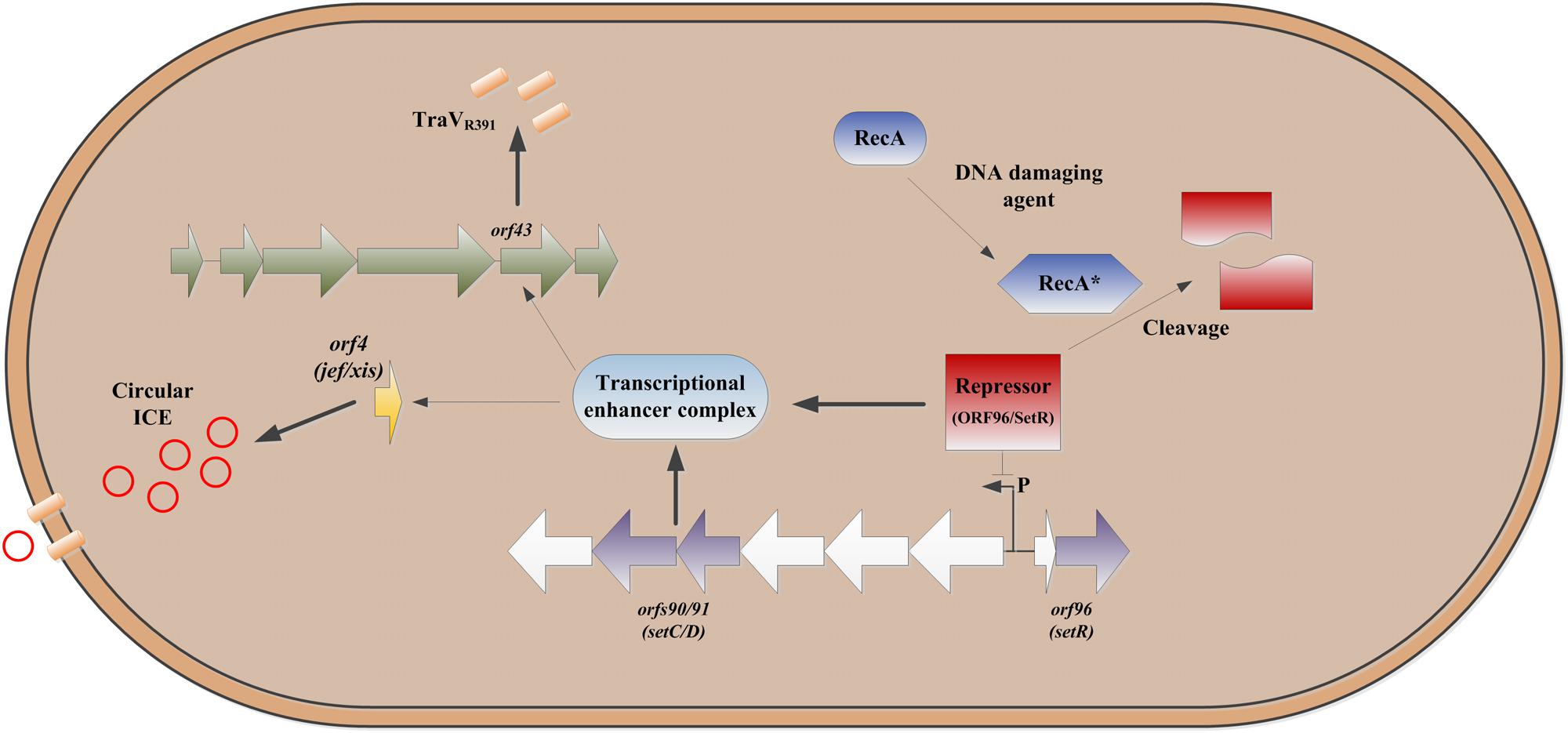
FIGURE 2. Outline of the escape mechanism proposed for ICE’s of the SXT/R391 group. DNA damage causes cleavage of orf96 leading to upregulation of expression of orf90/91 (Beaber et al., 2004), which in turns leads to induction of orf4 and production of the ICE excisionase (Jef/Xis) which results in increased levels of the circular form of the ICE (O’Halloran et al., 2007). UV irradiation also results in increased expression of orf43 which results in overproduction of the ICE TraV homolog (Armshaw and Pembroke, 2013a,b, 2015) which results in increased cell porosity and cell bursting providing an escape or ‘trap-door’ for the ICE.
1. DNA of the bacterial cell is damaged (by UV, chemical DNA damaging agent such as mitomycin C, etc.) creating single stranded DNA which stimulates RecA to become its active form RecA∗ (O’Reilly and Kreuzer, 2004).
2. RecA∗ cleaves the putative ICE SXT/R391 setCD/orf90-91 repressor protein setR/orf96 (Pembroke and Stevens, 1984; Beaber et al., 2004; McGrath et al., 2005).
3. Cleavage of the putative repressor protein setR/orf96 causes up-regulated expression of the putative transcriptional activator complex encoded by setCD/orf90-91 (Beaber et al., 2004).
4. The putative transcriptional activator complex encoded by setCD/orf90-91 binds a regulatory region upstream of orf43, upregulating the expression of TraVR391 (orf43) in a manner that is cytotoxic to the host cell (Armshaw and Pembroke, 2013b, 2015; Poulin-Laprade et al., 2015).
5. The putative transcriptional activator complex encoded by setCD/orf90-91 also binds a regulatory region upstream of orf4 which encodes the Jef protein, a protein that mediates the excision of the integrated SXT/R391 ICE from the bacterial chromosome (O’Halloran et al., 2007; Poulin-Laprade et al., 2015).
6. The cytotoxic effect is the result of damage to the cells outer membrane caused via the action of TraVR391 which causes the formation of pores in the membrane which allow escape of the excised SXT/R391 ICE element (the ‘trap–door’) and the death of the cell (Armshaw and Pembroke, 2013b, 2015) (Figure 2).
7. The SXT/R391 ICE element is then available to be taken (via ‘apparent transformation’) into other bacterial cells (Armshaw and Pembroke, 2015)
All authors listed, have made substantial, direct and intellectual contribution to the work, and approved it for publication.
This work was funded by the Irish Research Council for Science, Engineering and Technology (IRSCET) to PA.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to thank Dr. John O’Halloran for helpful discussion.
Abbani, M., Iwahara, M., and Clubb, R. T. (2005). The structure of the excisionase (Xis) protein from conjugative transposon Tn916 provides insights into the regulation of heterobivalent tyrosine recombinases. J. Mol. Biol. 347, 11–25. doi: 10.1016/j.jmb.2005.01.019
Alvarez-Martinez, C. E., and Christie, P. J. (2009). Biological diversity of prokaryotic type IV secretion systems. Microbiol. Mol. Biol. Rev. 73, 775–808. doi: 10.1128/MMBR.00023-09
Armshaw, P., and Pembroke, J. T. (2013a). Generation and analysis of an ICE R391 deletion library identifies genes involved in the element encoded UV-inducible cell-sensitising function. FEMS Microbiol. Lett. 342, 45–53. doi: 10.1111/1574-6968.12107
Armshaw, P., and Pembroke, J. T. (2013b). Control of expression of the ICE R391 encoded UV-inducible cell-sensitising function. BMC Microbiol. 13:195. doi: 10.1186/1471-2180-13-195
Armshaw, P., and Pembroke, J. (2013c). “Integrative conjugative elements (ICEs) of the SXT/R391 group as vehicles for acquisition of resistance determinants, stable maintenance and transfer to a wide range of enterobacterial pathogens,” in Microbial Pathogens and Strategies for Combating Them: Science, Technology and Education, ed. A. Méndez-Vilas (Badajoz: Formatex Research Center), 439–446.
Armshaw, P., and Pembroke, J. T. (2015). Examination of the cell sensitizing gene orf43 of ICE R391 suggests a role in ICE transfer enhancement to recipient cells. FEMS Microbiol. Lett. 362:fnu057 doi: 10.1093/femsle/fnu057
Beaber, J. W., Burrus, V., Hochhut, B., and Waldor, M. K. (2002a). Comparison of SXT and R391, two conjugative integrating elements: definition of a genetic backbone for the mobilization of resistance determinants. Cell. Mol. Life Sci. 59, 2065–2070. doi: 10.1007/s000180200006
Beaber, J. W., Hochhut, B., and Waldor, M. K. (2002b). Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J. Bacteriol. 184, 4259–4269. doi: 10.1128/JB.184.15.4259-4269.2002
Beaber, J. W., Hochhut, B., and Waldor, M. K. (2004). SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427, 72–74. doi: 10.1038/nature02241
Beaber, J. W., and Waldor, M. K. (2004). Identification of operators and promoters that control SXT conjugative transfer. J. Bacteriol. 186, 5945–5949. doi: 10.1128/JB.186.17.5945-5949.2004
Bellanger, X., Morel, C., Decaris, B., and Guédon, G. (2008). Regulation of excision of integrative and potentially conjugative elements from Streptococcus Thermophilus: role of the arp1 repressor. J. Mol. Microbiol. Biotechnol. 14, 16–21. doi: 10.1159/000106078
Bellanger, X., Payot, S., Leblond-Bourget, N., and Guédon, G. (2014). Conjugative and mobilizable genomic islands in bacteria: evolution and diversity. FEMS Microbiol. Rev. 38, 720–760. doi: 10.1111/1574-6976.12058
Bi, D., Xu, Z., Harrison, E. M., Tai, C., Wei, Y. Q., He, X. Y., et al. (2012). ICEberg: a web-based resource for integrative and conjugative elements found in Bacteria. Nucleic Acids Res. 40:D621–D626. doi: 10.1093/nar/gkr846
Boltner, D., MacMahon, C., Pembroke, J. T., Strike, P., and Osborn, A. M. (2002). R391: a conjugative integrating mosaic comprised of phage, plasmid, and transposon elements. J. Bacteriol. 184, 5158–5169. doi: 10.1128/JB.184.18.5158-5169.2002
Burrus, V., and Waldor, M. K. (2004). Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 155, 376–386. doi: 10.1016/j.resmic.2004.01.012
Ceccarelli, D., Spagnoletti, M., Hasan, N. A., Lansing, S., Huq, A., and Colwell, R. R. (2013). A new integrative conjugative element detected in Haitian isolates of Vibrio cholerae non-O1/non-O139. Res. Microbiol. 164, 891–893. doi: 10.1016/j.resmic.2013.08.004
Guglielmini, J., Quintais, L., Garcillan-Barcia, M. P., de la Cruz, F., and Rocha, E. P. (2011). The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet. 7:e1002222. doi: 10.1371/journal.pgen.1002222
Hochhut, B., and Waldor, M. K. (1999). Site-specific integration of the conjugal Vibrio cholerae SXT element into prfC. Mol. Microbiol. 32, 99–110. doi: 10.1046/j.1365-2958.1999.01330.x
Juiz-Rio, S., Osorio, C. R., de Lorenzo, V., and Lemos, M. L. (2005). Subtractive hybridization reveals a high genetic diversity in the fish pathogen Photobacterium damselae subsp. Piscicida: evidence Of A Sxt-Like Element. Microbiology 151, 2659–2669. doi: 10.1099/mic.0.27891-0
Lei, C.-W., Zhang, A.-Y., Wang, H.-N., Liu, B.-H., Yang, L.-Q., and Yang, Y.-Q. (2016). Characterization of SXT/R391 integrative and conjugative elements in Proteus mirabilis isolates from food-producing animals in China. Antimicrob. Agents Chemother. 60, 1935–1938. doi: 10.1128/AAC.02852-15
Lewis, J. A., and Hatfull, G. F. (2001). Control of directionality in integrase-mediated recombination: examination of recombination directionality factors (RDFs) including Xis and Cox proteins. Nucleic Acids Res. 29, 2205–2216. doi: 10.1093/nar/29.11.2205
McGrath, B. M., O’Halloran, J. A., and Pembroke, J. T. (2005). Pre-exposure to UV irradiation increases the transfer frequency of the IncJ conjugative transposon-like elements R391, R392, R705, R706, R997 and pMERPH and is recA+ dependent. FEMS Microbiol. Lett. 243, 461–465. doi: 10.1016/j.femsle.2005.01.013
McGrath, B. M., O’Halloran, J. A., Piterina, A., and Pembroke, J. T. (2006). Molecular probes to detect IncJ elements: a family of integrating, antibiotic resistance mobile genetic elements. J. Microbiol. Methods 66, 32–42 doi: 10.1016/j.mimet.2005.10.004
McGrath, B. M., and Pembroke, J. T. (2004). Detailed analysis of the insertion site of the mobile elements R997, pMERPH, R392, R705 and R391 in E. coli K12. FEMS Microbiol. Lett. 237, 19–26. doi: 10.1111/j.1574-6968.2004.tb09673.x
Michel, B. (2005). After 30 years of study, the bacterial SOS response still surprises us. PLoS Biol. 3:e255. doi: 10.1371/journal.pbio.0030255
Michael, G. B., Kadlec, K., Sweeney, M. T., Brzuszkiewicz, E., Liesegang, H., Daniel, R., et al. (2012). ICEPmu1, an integrative conjugative element (ICE) of Pasteurella multocida: structure and transfer. J. Antimicrob. Chemother. 67, 91–100. doi: 10.1093/jac/dkr411
O’Halloran, J. A., McGrath, B. M., and Pembroke, J. T. (2007). The orf4 gene of the enterobacterial ICE, R391, encodes a novel UV-inducible recombination directionality factor, Jef, involved in excision and transfer of the ICE. FEMS Microbiol. Lett. 272, 99–105. doi: 10.1111/j.1574-6968.2007.00747.x
O’Reilly, E. K., and Kreuzer, K. N. (2004). Isolation of SOS constitutive mutants of Escherichia coli. J. Bacteriol. 186, 7149–7160. doi: 10.1128/JB.186.21.7149-7160.2004
Pembroke, J. T., MacMahon, C., and McGrath, B. (2002). The role of conjugative transposons in the Enterobacteriaceae. Cell. Mol. Life Sci. 59, 2055–2064. doi: 10.1007/s000180200005
Pembroke, J. T., and Piterina, A. V. (2006). A novel ICE in the genome of Shewanella putrefaciens W3-18-1: comparison with the SXT/R391 ICE-like elements. FEMS Microbiol. Lett. 264, 80–88. doi: 10.1111/j.1574-6968.2006.00452.x
Pembroke, J. T., and Stevens, E. (1984). The effect of plasmid R391 and Other IncJ plasmids on the survival of Escherichia coli after UV Irradiation. J. Gen. Microbiol. 130, 1839–1844. doi: 10.1099/00221287-130-7-1839
Poulin-Laprade, D., Matteau, D., Jacques, P. E., Rodrigue, S., and Burrus, V. (2015). Transfer activation of SXT/R391 integrative and conjugative elements: unraveling the SetCD regulon. Nucleic. Acids Res. 43, 2045–2056. doi: 10.1093/nar/gkv071
Ryan, M. P., Pembroke, J. T., and Adley, C. C. (2009). Novel Tn4371-ICE like element in Ralstonia pickettii and Genome mining for comparative elements. BMC Microbiol. 9:242. doi: 10.1186/1471-2180-9-242
Spagnoletti, M., Ceccarelli, D., Rieux, A., Fondi, M., Taviani, E., Fani, R., et al. (2014). Acquisition and evolution of SXT-R391 integrative conjugative elements in the seventh-pandemic Vibrio cholerae lineage. MBio 5:e01356-14. doi: 10.1128/mBio.01356-14
Storrs, M. J., Poyart-Salmeron, C., Trieu-Cuot, P., and Courvalin, P. (1991). Conjugative transposition of Tn916 requires the excisive and integrative activities of the transposon-encoded integrase. J. Bacteriol. 173, 4347–4352.
Taviani, E., Grim, C. J., Chun, J., Huq, A., and Colwell, R. R. (2009). Genomic analysis of a novel integrative conjugative element in Vibrio cholerae. FEBS Lett. 583, 3630–3636 doi: 10.1016/j.febslet.2009.10.041
Van Houdt, R., Toussaint, A., Ryan, M. P., Pembroke, J. T., Mergeay, M., and Adley, C. C. (2012). “The Tn4371 ICE family of bacterial mobile genetic elements,” in Bacterial Integrative Mobile Genetic Elements, eds A. P. Roberts and P. Mullany (Austin, TX: Landes Bioscience).
Wozniak, R. A., Fouts, D. E., Spagnoletti, M., Colombo, M. M., Ceccarelli, D., Garriss, G., et al. (2009). Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. PLoS Genet. 5:e1000786. doi: 10.1371/journal.pgen.1000786
Wozniak, R. A., and Waldor, M. K. (2009). A toxin-antitoxin system promotes the maintenance of an integrative conjugative element. PLoS Genet. 5:e1000439. doi: 10.1371/journal.pgen.1000439
Keywords: integrative conjugative elements (ICEs), UV sensitization, ‘Trap-Door’ escape
Citation: Ryan MP, Armshaw P and Pembroke JT (2016) SXT/R391 Integrative and Conjugative Elements (ICEs) Encode a Novel ‘Trap-Door’ Strategy for Mobile Element Escape. Front. Microbiol. 7:829. doi: 10.3389/fmicb.2016.00829
Received: 19 January 2016; Accepted: 17 May 2016;
Published: 31 May 2016.
Edited by:
John R. Battista, Louisiana State University and A&M College, USAReviewed by:
Lanming Chen, Shanghai Ocean University, ChinaCopyright © 2016 Ryan, Armshaw and Pembroke. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: J. Tony Pembroke, dG9ueS5wZW1icm9rZUB1bC5pZQ==; Michael P. Ryan, bWljaGFlbC5wLnJ5YW5AdWwuaWU=; Patricia Armshaw, cGF0cmljaWEuYXJtc2hhd0B1bC5pZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.