- 1State Key Laboratory of Oral Diseases, West China Hospital of Stomatology, Sichuan University, Chengdu, China
- 2Department of Dentistry, Yan'an Hospital Affiliated to Kunming Medical University, Kunming, China
- 3Chengdu Institute of Biology, Chinese Academy of Sciences, Chengdu, China
- 4Department of Endodontics and Operative Dentistry School of Stomatology, Capital Medical University, Beijing, China
Dental caries is a biofilm-dependent disease that largely relies on the ability of Streptococcus mutans to synthesize exopolysaccharides. Although the rnc gene is suggested to be involved in virulence mechanisms in many other bacteria, the information regarding it in S. mutans is very limited. Here, using deletion or overexpression mutant assay, we demonstrated that rnc in S. mutans significantly positively regulated exopolysaccharide synthesis and further altered biofilm formation. Meanwhile, the cariogenecity of S. mutans was decreased by deletion of rnc in a specific pathogen-free (SPF) rat model. Interestingly, analyzing the expression at mRNA level, we found the downstream vic locus was repressed by rnc in S. mutans. Using deep sequencing and bioinformatics analysis, for the first time, three putative microRNA-size small RNAs (msRNAs) targeting vicRKX were predicted in S. mutans. The expression levels of these msRNAs were negatively correlated with vicRKX but positively correlated with rnc, indicating rnc probably repressed vicRKX expression through msRNAs at the post-transcriptional level. In all, the results present that rnc has a potential role in the regulation of exopolysaccharide synthesis and can affect vicRKX expressions via post-transcriptional repression in S. mutans. This study provides an alternative avenue for further research aimed at preventing caries.
Introduction
Dental caries is one of the most prevalent and costly biofilm-dependent oral infectious diseases, affecting the majority of the world's population (Selwitz et al., 2007). Streptococcus mutans is considered to be the principal pathogen responsible for dental caries (Takahashi and Nyvad, 2008). The ability to synthesize exopolysaccharides, which contribute to the adhesion and formation of tenacious biofilms on tooth surfaces, is recognized as one of the most important virulence factors related to dental caries formation (Nobbs et al., 2009; Bowen and Koo, 2011). As exopolysaccharide is an important virulence factor, exopolysaccharide synthesis-related genes are optimal targets for the development of anti-caries compounds (Chau et al., 2015; Zhang et al., 2015). Among these genes, vicRKX, which are known to encode the VicRK signal transduction system (TCS), are crucial to the pathogenicity of S. mutans, especially for exopolysaccharide formation (Senadheera et al., 2005, 2007). Basically, the vicRKX positively regulate exopolysaccharide production by VicR specifically binding to the promoter regions of genes responsible for the synthesis of glucans and fructans (Senadheera et al., 2005).
The rnc gene is recognized as the encoding gene of ribonuclease III (RNase III) (March et al., 1985; Watson and Apirion, 1985). The double-strand-specific endonuclease RNase III is highly conserved in bacteria and eukaryotes and has a critical role in physiology regulation, where it can promote mRNA maturation (Viegas et al., 2011). Much more studies have showed that rnc can be instrumental in virulence mechanisms in bacteria (Haddad et al., 2013; Hotto et al., 2015). However, the information regarding rnc in S. mutans is very limited. Previously, we investigated the genome of S. mutans (accession no. AE014133) (Ajdic et al., 2002) and identified an interesting phenomenon: the rnc gene is located on upstream of vicRKX tricistronic operon. It is suggested that genes located nearby are functionally related and make up a part of a metabolic pathway (Okuda et al., 2007). Thus, we hypothesize that rnc may be involved in the regulation of exopolysaccharide synthesis.
Over the last decade, studies of RNA-mediated regulation by small noncoding RNA molecules in eukaryotes and prokaryotes have demonstrated the major roles these molecules play in post-transcriptional regulation in various biological processes (Bartel, 2004; Archambaud et al., 2013). MicroRNAs (miRNAs), known as a special kind of small noncoding RNAs, are endogenous ~23-nt RNAs that can play important gene-regulatory roles in animals and plants by pairing to the mRNAs of protein-coding genes to direct their post-transcriptional repression (Bartel, 2009). In vitro, bacterial RNase III can be used to produce small noncoding RNA molecules as Dicer in eukaryotes, which is the essential enzyme to process miRNAs (Yang et al., 2002). It is suggested that they all have RIIID and dsRNA-binding domain that allows RNase III functions like Dicer, though more complex RIIID-containing proteins are found in Dicer (Bernstein et al., 2001; Calin-Jageman and Nicholson, 2003). Meanwhile, recent parallel studies have provided evidence for the high abundance of discrete microRNA-size small RNAs (msRNAs) in S. mutans and Escherichia coli (Lee and Hong, 2012; Kang et al., 2013). Overall, these findings suggest that msRNAs may participate in rnc-related regulation as post-transcriptional regulators in bacteria.
In this work, we identified the role of the rnc in exopolysaccharide synthesis, cariogenecity and its influence on the downstream vicRKX expression in S. mutans by mutant analyses and animal study. Furthermore, we detected the expressions of the candidate msRNAs targeting vicRKX by deep sequencing for analyzing the possible molecular mechanism. Here, we describe an additional level of control of S. mutans cariogenicity, msRNA-mediated post-transcriptional repression, which enriches the knowledge of virulence regulation in S. mutans. This paper provides an alternative avenue for further caries prevention research.
Materials and Methods
Bacterial Strains
The parent and rnc mutant strains of S. mutans, plasmid pDL278 and amplicons PcErm and aRnc used in this study are detailed in the Table S1. Synthetic oligonucleotides (Sangon Biotech, Shanghai, China) used and generated are listed in Table S2. The rnc gene sequence was obtained from the S. mutans UA159 genome dataset (http://www.ncbi.nlm.nih.gov/nuccore/AE014133.2) (Ajdic et al., 2002). To delete the rnc gene from S. mutans UA159, we used a ligation-PCR mutagenesis strategy, as previously described (Lau et al., 2002). The resulting rnc insertion–deletion mutant was named Smurnc. The rnc gene overexpression strain was constructed using the pDL278 shuttle vector with insertion of aRnc and was designated as Smurnc+. Ligation constructs and the shuttle vector were separately introduced into S. mutans UA159 using competence-simulating peptide (CSP)-induced natural transformation, and transformants that were independently resistant to erythromycin and spectinomycin were selected for recombination into the chromosome using PCR, followed by nucleotide sequence analysis. The level of rnc expression in the resulting mutants was monitored and compared to its level of expression in UA159 by quantitative real-time PCR (qRT-RCR) (Supplementary Material).
Bacterial Growth Conditions
Unless stated otherwise, strains were grown in brain heart infusion (BHI) medium (Oxoid, Basingstoke, England) in an atmosphere of 80% N2 and 20% CO2 at 37°C. Appropriate antibiotics were added when culturing the mutant strains, i.e., erythromycin (10 μg/mL) for Smurnc, and spectinomycin (1200 μg/mL) for Smurnc+. Overnight cultures of UA159, Smurnc, and Smurnc+ were subcultured at 1:20 in fresh BHI for 2–3 h and the optical density (OD) at 600 nm (0.3) was then determined using an ultraviolet spectrophotometer system (Helsinki, Finland).
Exopolysaccharide Production Assay
Modified biofilm growth was achieved as previously described (Yang et al., 2013). Each well of a 24-well plate, containing 2.0 ml sterile BHI with 1% sucrose, was inoculated with 0.5 ml of an exponential culture (OD600nm = 0.3) of S. mutans and incubated anaerobically for 24 h. The established mature biofilms were used to measure the amounts of water-insoluble exopolysaccharides (WIGs) and water-soluble exopolysaccharides (WSGs) by anthrone–sulfuric acid colorimetric assay (Cury et al., 1997). All assays were performed in triplicate from at least three different experiments.
Biofilm Formation Assays
Samples were obtained after 24 h of in vitro biofilm synthesis and from the in vivo SPF rat model. To determine the production and distribution of exopolysaccharides in the parent and rnc mutant S. mutans biofilms, we used scanning electron microscopy (SEM; FEI, Hillsboro, OR) (Li et al., 2001) and confocal laser scanning microscopy (CLSM; TSP SP2; Leica, Germany) based on in situ labeling of the exopolysaccharides with 1 μM Alexa Fluor 647 (Invitrogen, Eugene, OR, USA) and bacterial cells with 2.5 μM SYTO 9 (Invitrogen, Carlsbad, CA, USA) (Koo et al., 2010). The biofilms were washed three times with physiological saline and observed under confocal microscopy using Ar (514/488 nm) and He-Ne (543 nm) lasers. Under CLSM, the bacteria were stained green, while exopolysaccharides were stained red. A three-dimensional reconstruction of the biofilms from CLSM was analyzed using Imaris 7.0.0 software (Bitplane, Zurich, Switzerland). Three independent biofilm experiments were performed and images of three random fields of each group were captured.
Animal Study for Detecting Cariogenecity
All experiments involving rats were performed in accordance with the Chinese State Key Laboratory of Oral Diseases guidelines for animal welfare (NO.SCXK (111) 2009-09). Caries-susceptible, specific pathogen-free (SPF) Osborne-Mendel rats (IVC Experimental Animal Center of Public Health, Sichuan University, Chengdu, China) were used to investigate the in vivo effects of rnc mutations on the formation of dental caries. Four experimental groups were used: a blank was used as the negative control, UA159 was used as the positive control, Smurnc, and Smurnc+, where each group comprised 10 test animals. On days 23–28, each rat was infected orally once daily using 200 μl of a dense bacterial suspension that contained the UA159 strain, or the rnc deletion or expression mutants. The animals were sacrificed on day 50. The lower jaws were divided into right and left pieces and after three washes in phosphate-buffered saline, two pieces were selected for SEM and CLSM. The remaining section of the jaw was subjected to sonication using a Branson Sonifier 450 (Branson, Danbury, CT, USA) set for 10 s pulses at 70 W. The homogenized suspensions were stored at −20°C until further use. The lower jaws were dissected and immersed in fixative (10% buffered formalin phosphate) for a minimum of 72 h. The mandibular molars were sectioned and scored to determine fissure caries according to a modified Keyes system in this study. Detailed information about the modified Keyes system is provided in the Supplementary Material.
Gene Expression Assay by qRT-PCR
qRT-PCR was performed to determine the transcript levels in samples of UA159 and rnc mutants in vitro and in vivo. RNA was extracted and purified using the classical TRIzol–chloroform protocol (Invitrogen, Carlsbad, CA). RNA purity (A260/A280) and concentration was assessed by a NanoDrop2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA). Total RNA reverse transcription was performed with the RNeasy purification kit (QIAGEN, Valencia, CA, USA). qPCR was conducted as described by the manufacturer using a Bio-Rad CFX96 TM Real-time System (Bio-Rad, Hercules, CA, USA) and the Quantitect SYBR-Green PCR kit (QIAGEN, Valencia, CA, USA). qPCR was performed using specific primers for the vicRKX genes, and gyrA as a reference gene (Senadheera et al., 2005). All primers for RT-qPCR were obtained commercially (Sangon Biotech, Shanghai, China) and are listed in Table S2. Relative fold changes were determined using the 2−ΔΔCT method. We used technical replicates for each gene tested and we used at least three biological replicates in each experiment.
Genatic Analyses of rnc Promoter Structure
In the S. mutans UA159 dataset, we detected 108 intergenic base pairs between the vicX and rnc genes (gb|AE014133.2|:1443948–1444056). We first analyzed these intergenic noncoding sequences by FGENESB and BPROM programs for operon and promoter prediction, respectively (http://linux1.softberry.com/berry.phtml) (Solovyev and Tatarinova, 2011). Co-transcription assay were conducted with templates of DNA and reverse transcribed cDNA extracted from UA159. Genomic DNA was extracted and purified by using QIAamp DNA micro Kit (QIAGEN Sciences, MD, USA) and cDNA was conducted as previously described. qPCR was performed by using the specific primes for this assay (Table S2), and rnc as a reference gene.
Identification of msRNAs by Deep Sequencing
Samples of UA159, Smurnc, and Smurnc+ were grown in fresh BHI until mid-exponential phase. Total RNA was isolated as described above. Briefly, cDNA libraries were generated according to the Illumina TruSeq™ SmallRNA sample preparation protocol (online Figure S2). Size-fractionated RNA fragments of ca 18–150 nt in length were isolated using gel extraction and ethanol precipitation. After ligation with a pair of adaptors 5′ (5′-GUUCAGAGUUCUACAGUCCGACGAUC-3′) and 3′ (5′-UGGAAUUCUCGGGUGCCAAGG-3′), small RNA molecules were subjected to reverse transcription-PCR to obtain single-stranded cDNAs and then further amplified by PCR. After purification, the PCR products were used for sequencing by Illumina technology on HiSeq™ 2000 (Illumina, San Diego, CA, USA) by BGI, Shenzhen China.
Bioinformatic Analysis for Selecting msRNAs
Raw sequences were processed using Illumina PIPELINE software and then subjected to a series of data filtration steps for analyses. After filtering out low quality reads and trimming the adaptor sequences, high quality clean sequences mapping to intergenic region (IGR) and antisense mRNA (AM) were selected. The annotated sequences were further screened to remove rRNA and tRNA by searching against the NCBI Genbank database (http://www.ncbi.nlm.nih.gov/nuccore/AE014133) and the Rfam database (http://www.sanger.ac.uk/software/Rfam). RNAfold software (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi) was employed to predict msRNA candidates by exploring the secondary structure and the minimum free energy. Sequences and structures of the putative msRNAs satisfied the criteria of forming hairpin miRNAs, and secondary structures of hairpins have free energy of hybridization ≤ 0 kcal/mol. Differential-expression analysis was performed using DEseq (Anders and Huber, 2010). Differential expression of msRNAs was performed based on the reads from the UA159, Smurnc, and Smurnc+ libraries. Fold-change was calculated according the following equation: log2 Ratio = log2 (Smurnc reads/UA159 reads). P-value was calculated as previously described (Audic and Claverie, 1997). If the standard expression of a given msRNA was zero, its expression value was modified to 0.01. Significant difference in msRNA expression was assigned to sequences with a P < 0.05 and |log2Ratio| ≥ 6.64. Target gene predictions were obtained from IntaRNA (http://rna.informatik.uni-freiburg.de:8080/IntaRNA.jsp). GO enrichment analysis of target gene candidates was carried out using the GO terms in the database (http://www.geneontology.org/) and we set the candidates as vicRKX. Mis-matching analyses were conducted based the base-pairing between candidate msRNAs and target gene mRNA.
Stem-Loop qRT-PCR for Verifying msRNA Expression
Stem-loop qRT-PCR with SYBR Green was performed to verify the expression patterns revealed by RNA-seq (Chen et al., 2005). Total RNA was isolated from UA159 and rnc mutant strains as described above, and first strand cDNA was synthesized using specific stem-loop primers listed in Table S3. The reverse transcripts were used with the RevertAid First Strand cDNA Synthesis kit (RevertAid, Thermo scientific) following the manufacturer's protocol. qPCR was performed using SYBR® Green Realtime PCR Master Mix (QIAGEN, Valencia, CA, USA) and carried out in a Bio-Rad CFX96 TM Real-time System (Bio-Rad, Hercules, CA, USA). The reaction conditions were 95°C for 2 min, followed by 45 cycles of 95°C for 15 s, 45–50°C (according to the Tm of each pair of primers) for 40 s, 60°C for 30 s, and then fluorescence levels were measured at 60°C. The expression level of each msRNA gene was determined based on three replicates.
Statistical Analyses
Statistical analyses of the data were performed using SPSS 16.0 (SPSS Inc., Chicago, IL, U.S.). The Shapiro–Wilk test and Bartlett's test were first used to assess whether the data were parametric or not. For parametric testing, Fisher's tests and one-way ANOVA were used to detect the significant effects of variables. For nonparametric testing, the Kruskal–Wallis test and least significant difference (LSD) multiple comparisons were used. The differences of the means of data were considered significant if the P < 0.05.
Results
rnc Promotes Exopolysaccharide Synthesis and Alters Biofilm Morphology
To explore the biological function of rnc, we successfully constructed an rnc deletion mutant Smurnc and rnc overexpression mutant Smurnc+. We found Smurnc and Smurnc+ possessed similar growth rates compared to UA159 in planktonic and biofilm forms (Figure S1). Detected by the anthrone assay, it showed that both WSGs and WIGs decreased significantly in Smurnc but increased in Smurnc+ (P < 0.05, Figures 1A,B). However, the real production of WIGs in Smurnc+ (0.0036 ± 0.0001 mg/mL) was slightly elevated comparing to UA159 (0.0032 ± 0.0001 mg/mL). CLSM was used to observe the biofilms in three-dimensions in both parent UA159 or rnc mutant strains (Figure 1C). In UA159, the exopolysaccharides and bacteria formed a compact lattice-like structure with large “mushroom-like” clumps of colonies and conspicuous channels. In contrast, Smurnc exhibited a reduced capacity to synthesize an exopolysaccharides matrix similar with anthrone assay. For Smurnc+, exopolysaccharides was slightly less than UA159. The microcolonies of the rnc mutants, including Smurnc and Smurnc+, were decreased in terms of both size and number, and their biofilms lost their typical branch-like structure. The decreased bacteria colony may be caused by the abnormal cell arrangement shown by SEM (Figure 1D). According to the results of SEM, in Smurnc+ the bacteria preferred to gather into dense clusters covered by floating exopolysaccharides and in Smurnc bacteria appeared to scatter to form larger tunnels among the clumps. The biofilm of them both were less condensed than the biofilm in UA159.
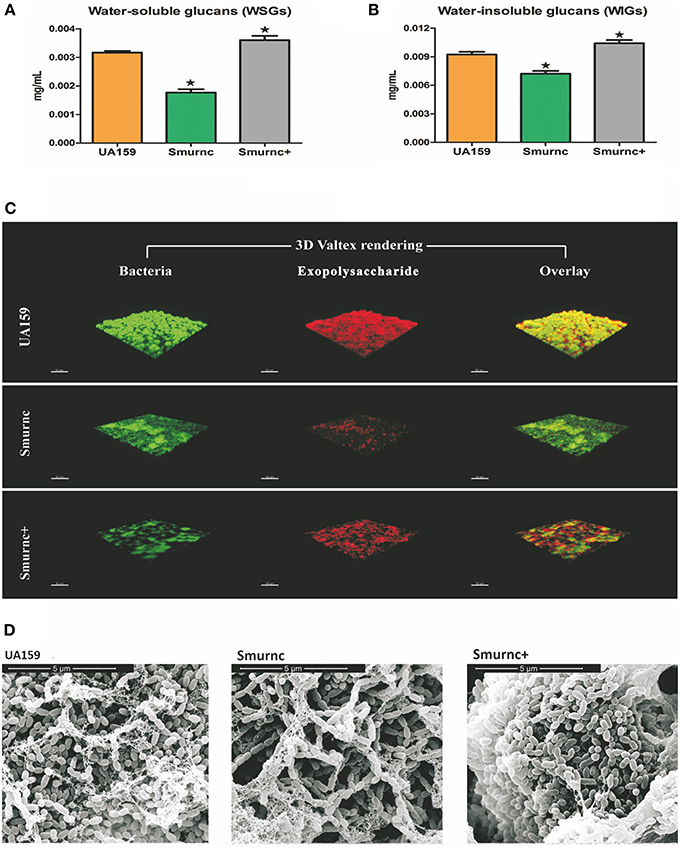
Figure 1. The effects of rnc gene on exopolysaccharide synthesis and biofilm morphology in S. mutans. The anthrone assay was used to determine glucan productions in UA159, Smurnc, and Smurnc+ biofilms at 24 h. WSGs (A) and WIGs (B) were both calculated. The Smurnc strain decreased the amount of glucans, whereas the Smurnc+ strain increased the amount. The error bars represent standard deviation values. Asterisks indicate significant differences (P < 0.05). (C) Representative three-dimensional images of production and distribution of exopolysaccharide matrix in the biofilm architecture. Mature biofilms (24 h) formed by S. mutans UA159 and rnc mutant strains in the presence of 1% (wt/vol) sucrose by CLSM. Images were taken at 63× magnification. (D) SEM observation of the architecture of S. mutans mature biofilms (24 h). Images of biofilm structures formed by UA159 and rnc mutant strains were taken at 20,000× magnification.
rnc Affects the Cariogenicity of S. mutans in a SPF Rat Model
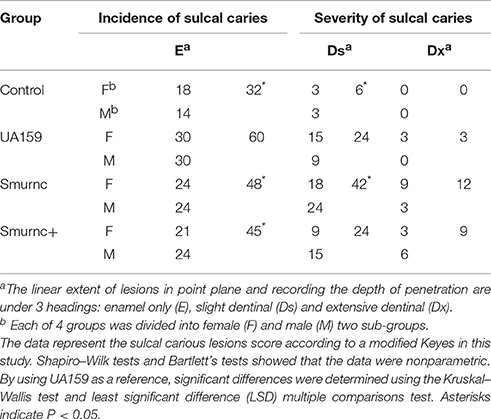
We next examined the role of rnc in the pathology of dental caries in a SPF rat model. Parent and rnc mutant strains were inoculated in the rat oral cavity. The lower dentition in rats was used to investigate the effects of the rnc gene on cariogenic characteristics. We verified that a knockout of rnc reduced the cariogenicity of S. mutans (Table 1). Smurnc and Smurnc+ exhibited a significantly decreased number of sulcal caries compared with those in the UA159 group after three weeks in vivo (P < 0.05, E). Interestingly, the severity of limited sulcal caries, designated as slight dentinal (Ds), was increased in the Smurnc groups, while the Smurnc+ mutants did not show any differences in comparison to UA159.
Furthermore, the rnc mutants had changes in exopolysaccharide production in the in vivo study. In a representative three-dimensional view obtained by CLSM, the mutants exhibited a similar inclination toward exopolysaccharide synthesis compared with that observed in vitro. In particular, compared to the parent strain UA159, Smurnc exhibited a significantly decreased level of exopolysaccharides whereas Smurnc+ showed a similar level of exopolysaccharide production (Figure 2A). To obtain a better view of the exopolysaccharide distribution in the biofilms, SEM images of rat molars were also captured. At 20,000× magnification, we observed that the exopolysaccharides produced by the rnc mutants was much sparser and thinner than that produced by UA159 (Figure 2B).
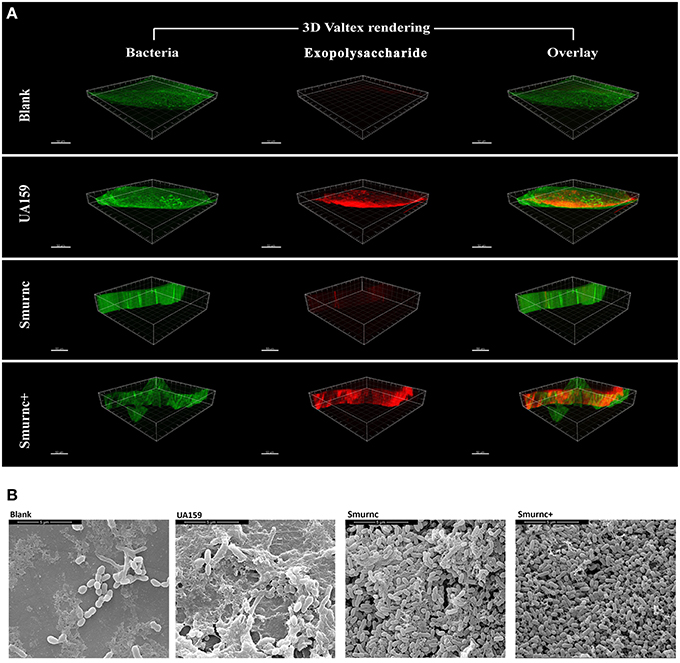
Figure 2. The influences of rnc gene on S. mutans cariogenicity in the SPF rat model. (A) Representative three-dimensional images of biofilms that formed on occlusal plane of rat mandibular molars in four different groups by CLSM. Images were taken at 63× magnification. (B) SEM observation of the architecture of biofilms that formed on occlusal plane of rat mandibular molars. Images were taken at 20,000× magnification.
rnc Represses vicRKX Expression In vitro and In vivo
Our previous studies identified that rnc was involved in exopolysaccharide synthesis, biofilm formation and cariogenicity in S. mutans. We analyzed the expressions of several exopolysaccheride synthesis-related genes in Smurnc and found these genes all up-regulated (Figure S3). For exploring the promising repression mechanism of rnc, we first analyzed the specific expression of vicRKX at the transcriptional level in parent UA159 and rnc mutant strains both in vitro biofilm study and in vivo animal study. The trend of vicRKX expressions was similar both in vitro and in vivo. Specifically, when compared to parent strain UA159, the transcription levels of all of the vicR/K/X genes increased remarkably in Smurnc (P < 0.05, Figure 3), whereas vicR transcription was reduced in Smurnc+ (P < 0.05, Figures 3A,D) and vicK/X transcription appeared stable level. Based on these results, we further investigated the molecular mechanism underlying this peculiar phenomenon.
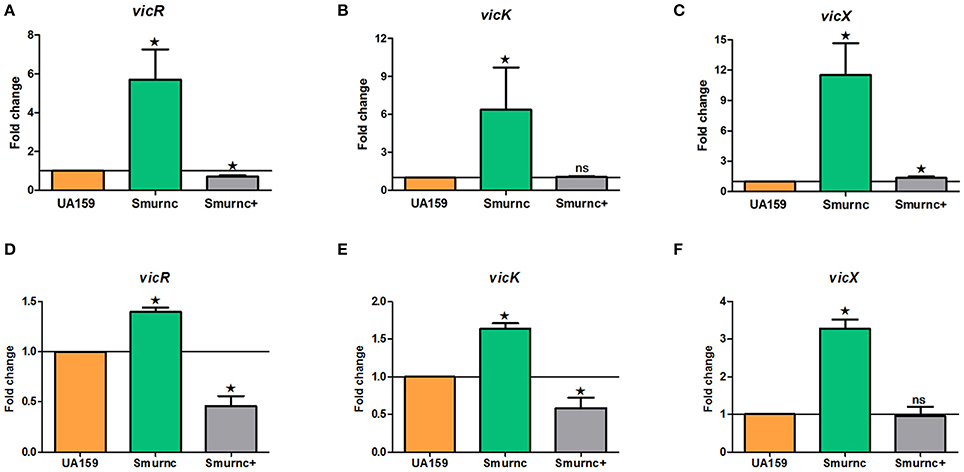
Figure 3. Dynamics of the expression of vicR/K/X genes in interaction with rnc gene among three types of strains. (A–C) Relative expression levels of vicR/K/X were calculated during the mid-exponential phase in vitro. (D–F) Relative expression levels of vicR/K/X were calculated in SPF rat model in vivo. The fold changes were shown after standardization relative to gyrA using UA159 as a reference. The data were represented as the means and standard deviation values. Asterisks indicate significant differences (P < 0.05); ns, no significant difference.
Genetic Analysis Indicates rnc Is Not a Part of vicRKX Tricistronic Operon
Knowledge of the co-transcription relationship between a regulator gene and structural genes is helpful to illustrate its potential regulation mechanism. Thus, we investigated the transcriptional relationship between rnc and the vic locus.
We analyzed the NCBI database and found that rnc and the vicRKX tricistronic operon were transcribed in the same direction and that they were separated by 108 bp of intergenic noncoding DNA. In most organisms, knowledge of operon structure is based on computational methods. We first used FGENESB to predict and it showed that these genes were not located in one operon. Then, we employed BPROM to analyze the existence of a promoter within the 108 intergenic base pairs. Similar to FGENESB, it identified a promoter that could initiate rnc transcription. The predicted genetic structure of rnc and the vicRKX locus are shown in Figure 4A. Furthermore, the co-transcription assay directly demonstrated that rnc and vicX were not transcribed as a single mRNA (Figure 4B). The results of these experiments showed that rnc and vicRKX were not located in the same operon and that they are expressed separately.
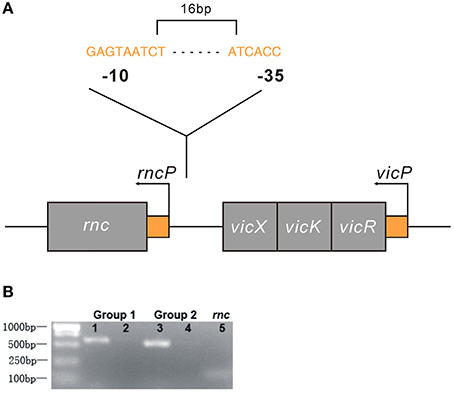
Figure 4. Genetic analyses of the rnc and vicRKX gene loci. (A) Predicted genetic structure of rnc and vicRKX loci by FGENESB and BPROM. The fragment shown was from the S. mutans chromosome, which contained the intact rnc, vicX, vicK, and vicR genes, shown from left to right. The entire sequence was known for this region. Arrows indicated the initiation sites for the vicP and rncP promoters. The supposed sequences of -10 and -35 boxes in rncP were GAGTAATCT and ATCACC, respectively. (B) PCR of rnc and vicX co-transcription assays. Two sets of specific PCR primers were used for the co-transcription assay. Lanes 1 and 3 were used for UA159 DNA, and lanes 2 and 4 were used for UA159 cDNA, which was reverse transcribed from RNA. Line 5 was used as a reference to verify the feasibility of the cDNA.
rnc Represses vicRKX Expressions at the Post-Transcriptional Level via msRNAs
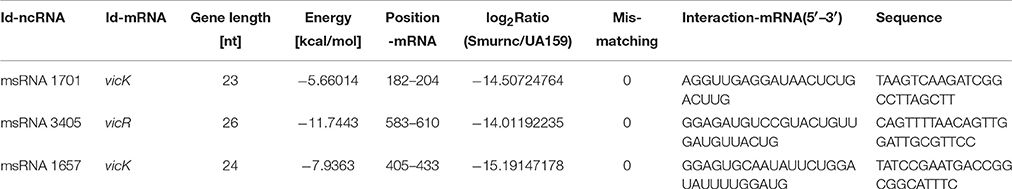
Deep sequencing allows the identification and quantification of small noncoding RNA molecules. Using this technique, we analyzed msRNAs targeting vicRKX in UA159 and the rnc mutants. The detailed filtering steps are shown in Figure S4. Briefly, we removed low-quality reads and chose clean reads in cultures of UA159 and the rnc mutants. Taking several bioinformatic analyses into consideration, we finally extracted three putative molecules as validated msRNAs, which correspond to msRNA 1701, 3405, and 1657 as detailed in Table 2. The ability of miRNAs to fold back on themselves to form distinctive hairpin structures is known to be a critical factor that differentiates these from other classes of small RNAs (Bartel, 2009). Thus, we predicted the potential secondary structures of these msRNAs (Figure 5A), which showed they all could form hairpins structures.
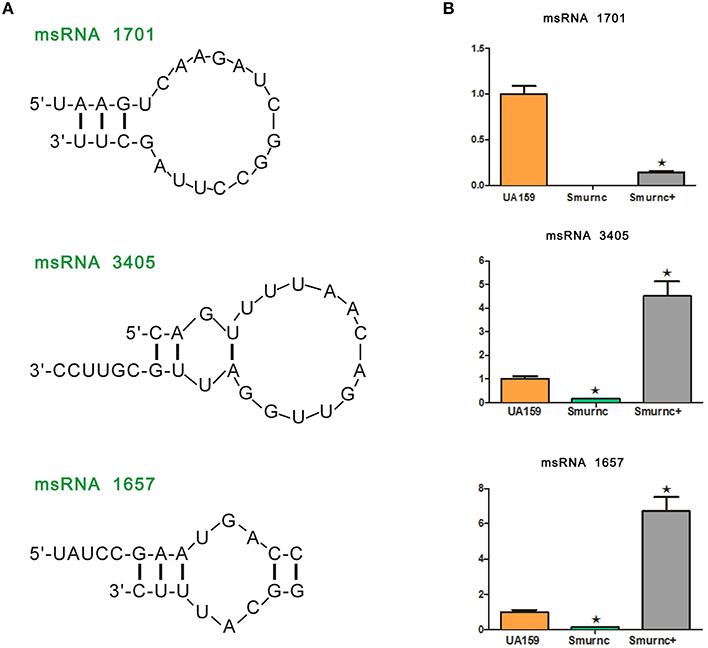
Figure 5. Presentation and validation of the deep sequencing data by secondary structure prediction and expression analyses. (A) Predicted secondary RNA structures formed by three validated msRNAs, registered by deep sequencing. (B) Comparison of expression levels of three validated msRNAs in UA159 and rnc mutations by stem loop RT-PCR. The fold changes were shown after standardization relative to 16s RNA using UA159 as a reference. The data were represented as the means and standard deviation values. Asterisks indicate significant differences (P < 0.05); ns, no significant difference.
Bioinformatic analysis predicted the presence of these particular msRNAs and stem-loop qRT-PCR was applied to validate the deep sequencing data. Expression levels of the three selected msRNAs were calculated using qPCR, and it revealed a rough correlation between the number of msRNAs and their cellular level. More importantly, expression of the three msRNAs corresponded to that seen with deep sequencing, suggesting that deep sequencing data were reliable. We found that there were significant differences in the expression of selected msRNAs between UA159 and the rnc mutants (P < 0.05, Figure 5B). That is, compared with the UA159 strain, the expression of msRNA 1701, 3405, and 1657 in the Smurnc was significantly down-regulated by 0.01-, 0.15- and 0.13-fold, respectively. Meanwhile, we found their expression levels were negatively correlated with vicRKX but positively correlated with rnc by combining mRNA and msRNA qPCR analyses, indicating rnc probably regulates vicRKX expression through msRNAs by post-transcriptional repression.
Discussion
The rnc gene has long been considered as a regulator that is involved in bacterial physiology. Previous investigations showed in Streptomyces coelicolor rnc was essential for the production of antibiotics actinorhodin and undecylprodigiosin (Sello and Buttner, 2008; Xu et al., 2010). In Streptomyces antibioticus, it has been suggested as a global regulator of actinomycin production (Lee et al., 2013). In the present study, we found that the expression of rnc was positively correlated with exopolysaccharide synthesis. Here, we employed the anthrone assay to detect the production of WSGs and WIGs. Comparing to UA159, the amounts of WSGs and WIGs in Smurnc was significantly declined, which could be used to explain the less condensed biofilm structure. It is suggested that reductions in the exopolysaccharide matrix might have resulted in weakened connections among microcolonies and, therefore, the presence of more scattered microcolonies. The disorder of biofilm formation, which was mainly caused by altered exopolysaccharide production, could be vividly detected by CLSM and SEM in vitro and in vivo. Similar with our study, exopolysaccharide is considered to provide mechanical integrity/stability for biofilm formation, and provide supporting frame for continuous growth of the microcolonies (Koo et al., 2009, 2013). Interestingly, the production of exopolysaccharides was not representative in Smurnc+ (Figures 1C, 2A). In other words, the Smurnc+ strain exhibited different amounts of exopolysaccharide matrix in different conditions. It could be speculated that the introduction of an exogenous plasmid vector carrying rnc gene may interfere with the role of the whole genome in regulating intracellular homeostasis in S. mutans. All the available evidences showed that exopolysaccharide is a critical virulence factor of dental caries (Bowen, 2016). It provides an abundance of primary binding sites and forms the core of the matrix-scaffold in cariogenic biofilms (Nobbs et al., 2009; Bowen and Koo, 2011; Xiao et al., 2012). Collectively, our analysis of the rnc demonstrate that rnc has a potential biological function where by it can regulate exopolysaccharide synthesis and further alter the biofilm morphology and the cariogenicity of S. mutans. Therefore, we propose that rnc is a new anti-virulence gene locus and it will provide important insights into the genetic pathways that control exopolysaccharide synthesis in S. mutans.
Herein, we firstly studied the expression of several exopolysaccharide synthesis-related genes in Smurnc. However, the results showed that all these genes were typically repressed by rnc (Figure S3). It indicated that the regulation of rnc on exopolysaccharide synthesis was conducted by a genetic network involving not only formation-related but also disintegration-related genes. Interestingly, the results of this study showed that the expressions of vicRKX and gtfB both were markedly increased compared to UA159, which was consistent with VicR being a positive regulator of the gtf genes of previous studies (Senadheera et al., 2005, 2007). The rnc-mediated regulation of exopolysaccharides needs more exploration and our further research will focus on this proposal. TCS is one of the most prevalent means to mediate the response of bacteria to a wide range of signals and stimuli (Laub and Goulian, 2007). Among these genes, the vic locus has been reported to encode VicRK TCS and it underscores its tremendous versatility and utility to S. mutans (Senadheera et al., 2009, 2012; Ayala et al., 2014). Indeed, the localization of vicRKX and rnc is conjoint in S. mutans genome. Thus, we chose the vicRKX genes for an in-depth study of the possible regulatory mechanism of rnc-mediated repression.
Determining whether the regulatory effects of rnc on vicRKX are limited to one operon would help us better understand rnc-mediated virulence expression in S. mutans genome. Here, all the genetic location analyses showed that rnc was not a part of vicRKX tricistronic operon. Each operon is a series of genes transcribed in a single mRNA, often identified by the presence of promoters and terminators. In bacteria, regulator genes can be located within an operon, adjacent to it, or far away from it. It has been suggested that bacterial genes with similar functions may be located near each other or even in a single operon, which are major structural and regulatory features of prokaryotic genomes (Okuda et al., 2007). However, there are several global regulators that can regulate distal genes and these seem to have more powerful effects (Bratlie et al., 2010). Considering the global regulation of rnc in S. antibioticus, rnc-mediated regulation in S. mutans appears to operate in a similar fashion (Lee et al., 2013).
For the first time, we identified three putative msRNAs that target vicRKX in S. mutans, and their expression levels were negatively correlated with vicRKX but positively correlated with rnc. In S. mutans, it appears that rnc participates in the post-transcriptional regulation of vicRKX. The rnc gene is believed to encode RNase III in S. mutans (Fonfara et al., 2014). Members of the RNase III family include bacterial RNase III and the eukaryotic proteins Drosha and Dicer (Court et al., 2013). All of these proteins have similar core domains, RIIID and dsRBD (Calin-Jageman and Nicholson, 2003). Drosha and Dicer are critical in miRNA-related processes. In eukaryotes, Drosha catalyzes the initial processing of the stem loop (hairpins) of primary miRNAs (pri-miRNAs) into short, hairpin, precursor miRNAs (pre-miRNAs), and Dicer further processes pre-miRNAs into miRNAs, which regulate more than half of all mammalian coding sequences via RNA interference (RNAi) (Tomari and Zamore, 2005; Friedman et al., 2009). A recent study reported a novel RNA cleavage event in eukaryotes and it was suggested that some of the miRNAs generated by Dicer might be obtained via this cleavage process, which is the same as that mediated by RNase III in E. coli (Gu et al., 2012). The existence of msRNAs in E. coli has been predicted and verified (Kang et al., 2013). This newly identified cleavage event may therefore represent a possible mechanism for msRNA production in bacteria. Here three msRNAs were predicted to target the downstream vicRKX and their expression levels were detected. The results showed that the expression of rnc directly and positively affected the levels of the three msRNAs. The evidence here implies that rnc regulates msRNA expression or is involved in their synthesis. Further investigation of the formation of msRNAs and the specific functions of msRNA-related mechanisms should provide new insights into post-transcriptional regulation in prokaryotes and, more importantly, their evolution.
Author Contributions
TH and X-RM designed research; M-YM, Y-MY, K-ZL, LL, ML, YY, J-XY, and RZ acquired the data; M-YM and Y-MY analyzed and interpreted the data; and M-YM wrote the main manuscript text. All authors discussed the results and commented on the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors declare no competing financial interests. We thank Dr Yaling Liu for critical reading the manuscript and providing useful discussions. This work was supported by grant from National Nature Science Foundation of China (grant number: 81400507 to Y-MY) and Science and Technology Support Projects of Sichuan Province (grant number: 2014SZ0024 to Y-MY).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00687
Table S1. Bacterial strains, plasmid, and amplicons used in this study.
Table S2. List of oligonucleotide primers used in this study.
Table S3. Sequences of primers used for stem-loop qRT-PCR analysis.
Figure S1. Growth curves of S. mutans UA159, Smurnc, and Smurnc+. Each datum point is the average of nine independent OD values per sample. The results shown are representative of three independent experiments conducted with the mutants and UA159 parent strains.
Figure S2. Workflow to generate cDNA libraries. The directions of the arrow marks denote the flow of total RNA processing and sequential.
Figure S3. rnc affected the expression of several exopolysaccheride- related genes at the transcriptional level. The relative levels of target gene expression in UA159 and Smurnc were determined at mid-exponential phase in vitro. The fold changes are shown after standardization relative to gyrA using UA159 as a reference.
Figure S4. Workflow to analyze and select the validated msRNAs. The directions of the arrows denote the flow of data processing and sequences. The shapes of the boxes have no particular significance, while the descriptions within the boxes represent the steps corresponding to data filtering.
References
Ajdic, D., McShan, W. M., McLaughlin, R. E., Savic, G., Chang, J., Carson, M. B., et al. (2002). Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. U.S.A. 99, 14434–14439. doi: 10.1073/pnas.172501299
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. doi: 10.1186/gb-2010-11-10-r106
Archambaud, C., Sismeiro, O., Toedling, J., Soubigou, G., Becavin, C., Lechat, P., et al. (2013). The intestinal microbiota interferes with the microRNA response upon oral Listeria infection. MBio 4, e00707–00713. doi: 10.1128/mBio.00707-13
Audic, S., and Claverie, J. M. (1997). The significance of digital gene expression profiles. Genome Res. 7, 986–995.
Ayala, E., Downey, J. S., Mashburn-Warren, L., Senadheera, D. B., Cvitkovitch, D. G., and Goodman, S. D. (2014). A biochemical characterization of the DNA binding activity of the response regulator VicR from Streptococcus mutans. PLoS ONE 9:e108027. doi: 10.1371/journal.pone.0108027
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. doi: 10.1016/S0092-8674(04)00045-5
Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. doi: 10.1016/j.cell.2009.01.002
Bernstein, E., Caudy, A. A., Hammond, S. M., and Hannon, G. J. (2001). Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366. doi: 10.1038/35053110
Bowen, W. H. (2016). Dental caries - not just holes in teeth! A perspective. Mol. Oral Microbiol. 31, 228–233. doi: 10.1111/omi.12132
Bowen, W. H., and Koo, H. (2011). Biology of Streptococcus mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res. 45, 69–86. doi: 10.1159/00032459
Bratlie, M. S., Johansen, J., and Drablos, F. (2010). Relationship between operon preference and functional properties of persistent genes in bacterial genomes. BMC Genomics 11:71. doi: 10.1186/1471-2164-11-71
Calin-Jageman, I., and Nicholson, A. W. (2003). RNA structure-dependent uncoupling of substrate recognition and cleavage by Escherichia coli ribonuclease III. Nucleic Acids Res. 31, 2381–2392. doi: 10.1093/Nar/Gkg329
Chau, N. P., Pandit, S., Cai, J. N., Lee, M. H., and Jeon, J. G. (2015). Relationship between fluoride release rate and anti-cariogenic biofilm activity of glass ionomer cements. Dent. Mater. 31, e100–108. doi: 10.1016/j.dental.2014.12.016
Chen, C., Ridzon, D. A., Broomer, A. J., Zhou, Z., Lee, D. H., Nguyen, J. T., et al. (2005). Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 33, e179. doi: 10.1093/nar/gni178
Court, D. L., Gan, J. H., Liang, Y. H., Shaw, G. X., Tropea, J. E., Costantino, N., et al. (2013). RNase III: genetics and function; structure and mechanism. Annu. Rev. Genet. 47, 405. doi: 10.1146/annurev-genet-110711-155618
Cury, J. A., Rebello, M. A. B., and Del Bel Cury, A. A. (1997). In situ relationship between sucrose exposure and the composition of dental plaque. Caries Res. 31, 356–360.
Fonfara, I., Le Rhun, A., Chylinski, K., Makarova, K. S., Lecrivain, A. L., Bzdrenga, J., et al. (2014). Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic Acids Res. 42, 2577–2590. doi: 10.1093/nar/gkt1074
Friedman, R. C., Farh, K. K., Burge, C. B., and Bartel, D. P. (2009). Most Mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105. doi: 10.1101/gr.082701.108
Gu, S., Jin, L., Zhang, Y., Huang, Y., Zhang, F., Valdmanis, P. N., et al. (2012). The loop position of shRNAs and pre-miRNAs is critical for the accuracy of dicer processing in vivo. Cell 151, 900–911. doi: 10.1016/j.cell.2012.09.042.
Haddad, N., Saramago, M., Matos, R. G., Prevost, H., and Arraiano, C. M. (2013). Characterization of the biochemical properties of Campylobacter jejuni RNase III. Biosci. Rep. 33:e00082. doi: 10.1042/BSR20130090
Hotto, A. M., Castandet, B., Gilet, L., Higdon, A., Condon, C., and Stern, D. B. (2015). Arabidopsis chloroplast mini-ribonuclease III participates in rRNA maturation and intron recycling. Plant Cell 27, 724–740. doi: 10.1105/tpc.114.134452
Kang, S. M., Choi, J. W., Lee, Y., Hong, S. H., and Lee, H. J. (2013). Identification of microRNA-size, small RNAs in Escherichia coli. Curr. Microbiol. 67, 609–613. doi: 10.1007/s00284-013-0411-9
Koo, H., Falsetta, M. L., and Klein, M. I. (2013). The exopolysaccharide matrix: a virulence determinant of cariogenic biofilm. J. Dent. Res. 92, 1065–1073. doi: 10.1177/0022034513504218
Koo, H., Xiao, J., and Klein, M. I. (2009). Extracellular polysaccharides matrix–an often forgotten virulence factor in oral biofilm research. Int. J. Oral. Sci. 1, 229–234. doi: 10.4248/IJOS.09086
Koo, H., Xiao, J., Klein, M. I., and Jeon, J. G. (2010). Exopolysaccharides produced by Streptococcus mutans glucosyltransferases modulate the establishment of microcolonies within multispecies biofilms. J. Bacteriol. 192, 3024–3032. doi: 10.1128/Jb.01649-09
Lau, P. C. Y., Sung, C. K., Lee, J. H., Morrison, D. A., and Cvitkovitch, D. G. (2002). PCR ligation mutagenesis in transformable Streptococci: application and efficiency. J. Microbiol. Methods 49, 193–205. doi: 10.1016/S0167-7012(01)00369-4
Laub, M. T., and Goulian, M. (2007). Specificity in two-component signal transduction pathways. Annu. Rev. Genetics 41, 121–145. doi: 10.1146/annurev.genet.41.042007.170548
Lee, H. J., and Hong, S. H. (2012). Analysis of microRNA-size, small RNAs in Streptococcus mutans by deep sequencing. FEMS Microbiol. Lett. 326, 131–136. doi: 10.1111/j.1574-6968.2011.02441.x
Lee, J. H., Gatewood, M. L., and Jones, G. H. (2013). RNase III is required for actinomycin production in Streptomyces antibioticus. Appl. Environ. Microbiol. 79, 6447–6451. doi: 10.1128/AEM.02272-13
Li, Y. H., Hanna, M. N., Svensater, G., Ellen, R. P., and Cvitkovitch, D. G. (2001). Cell density modulates acid adaptation in Streptococcus mutans: implications for survival in biofilms. J. Bacteriol. 183, 6875–6884. doi: 10.1128/JB.183.23.6875-6884.2001
March, P. E., Ahnn, J., and Inouye, M. (1985). The DNA-Sequence of the Gene (Rnc) Encoding Ribonuclease-Iii of Escherichia coli. Nucleic Acids Res. 13, 4677–4685. doi: 10.1093/nar/13.13.4677
Nobbs, A. H., Lamont, R. J., and Jenkinson, H. F. (2009). Streptococcus adherence and colonization. Microbiol. Mol. Biol. Rev. 73, 407–450. doi: 10.1128/MMBR.00014-09
Okuda, S., Kawashima, S., Kobayashi, K., Ogasawara, N., Kanehisa, M., and Goto, S. (2007). Characterization of relationships between transcriptional units and operon structures in Bacillus subtilis and Escherichia coli. BMC Genomics 8:38. doi: 10.1186/1471-2164-8-48
Sello, J. K., and Buttner, M. J. (2008). The gene encoding RNase III in Streptomyces coelicolor is transcribed during exponential phase and is required for antibiotic production and for proper sporulation. J. Bacteriol. 190, 4079–4083. doi: 10.1128/JB.01889-07
Selwitz, R. H., Ismail, A. I., and Pitts, N. B. (2007). Dental caries. Lancet 369, 51–59. doi: 10.1016/S0140-6736(07)60031-2
Senadheera, D. B., Cordova, M., Ayala, E. A., Chavez de Paz, L. E., Singh, K., Downey, J. S., et al. (2012). Regulation of bacteriocin production and cell death by the VicRK signaling system in Streptococcus mutans. J. Bacteriol. 194, 1307–1316. doi: 10.1128/JB.06071-11
Senadheera, D., Krastel, K., Mair, R., Persadmehr, A., Abranches, J., Burne, R. A., et al. (2009). Inactivation of VicK affects acid production and acid survival of Streptococcus mutans. J. Bacteriol. 191, 6415–6424. doi: 10.1128/JB.00793-09.
Senadheera, M. D., Guggenheim, B., Spatafora, G. A., Huang Y. C., Choi, J., Hung, D. C., et al. (2005). A VicRK signal transduction system in Streptococcus mutans affects gtfBCD, gbpB, and ftf expression, biofilm formation, and genetic competence development. J. Bacteriol. 187, 4064–4076. doi: 10.1128/Jb.187.12.4064-4076.2005
Senadheera, M. D., Lee, A. W., Hung, D. C., Spatafora, G. A., Goodman, S. D., and Cvitkovitch, D. G. (2007). The Streptococcus mutans vicX gene product modulates gtfB/C expression, biofilm formation, genetic competence, and oxidative stress tolerance. J. Bacteriol. 189, 1451–1458. doi: 10.1128/JB.01161-06
Solovyev, V. V., and Tatarinova, T. V. (2011). Towards the integration of genomics, epidemiological and clinical data. Genome Med. 3:48. doi: 10.1186/Gm264
Takahashi, N., and Nyvad, B. (2008). Caries ecology revisited: microbial dynamics and the caries process. Caries Res. 42, 409–418. doi: 10.1159/000159604
Tomari, Y., and Zamore, P. D. (2005). Perspective: machines for RNAi. Genes Dev. 19, 517–529. doi: 10.1101/gad.1284105
Viegas, S. C., Silva, I. J., Saramago, M., Domingues, S., and Arraiano, C. M. (2011). Regulation of the small regulatory RNA MicA by ribonuclease III: a target-dependent pathway. Nucleic Acids Res. 39, 2918–2930. doi: 10.1093/nar/gkq1239
Watson, N., and Apirion, D. (1985). Molecular cloning of the gene for the RNA-processing enzyme RNase III of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 82, 849–853.
Xiao, J., Klein, M. I., Falsetta, M. L., Lu, B., Delahunty, C. M., Yates, J. R. III, et al. (2012). The exopolysaccharide matrix modulates the interaction between 3D architecture and virulence of a mixed-species oral biofilm. PLoS Pathog 8:e1002623. doi: 10.1371/journal.ppat.1002623
Xu, W., Huang, J., Lin, R., Shi, J., and Cohen, S. N. (2010). Regulation of morphological differentiation in S. coelicolor by RNase III (AbsB) cleavage of mRNA encoding the AdpA transcription factor. Mol. Microbiol. 75, 781–791. doi: 10.1111/j.1365-2958.2009.07023.x
Yang, D., Buchholz, F., Huang, Z., Goga, A., Chen, C. Y., Brodsky, F. M., et al. (2002). Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 99, 9942–9947. doi: 10.1073/pnas.152327299
Yang, Y. M., Jiang, D., Qiu, Y. X., Fan, R., Zhang, R., Ning, M. Z., et al. (2013). Effects of combined exogenous dextranase and sodium fluoride on Streptococcus mutans 25175 monospecies biofilms. Am. J. Dent. 26, 239–243.
Keywords: rnc gene, vicRKX, post-transcriptional regulation, microRNA-size small RNAs, exopolysacchrides, Streptococcus mutans, dental caries
Citation: Mao M-Y, Yang Y-M, Li K-Z, Lei L, Li M, Yang Y, Tao X, Yin J-X, Zhang R, Ma X-R and Hu T (2016) The rnc Gene Promotes Exopolysaccharide Synthesis and Represses the vicRKX Gene Expressions via MicroRNA-Size Small RNAs in Streptococcus mutans. Front. Microbiol. 7:687. doi: 10.3389/fmicb.2016.00687
Received: 30 January 2016; Accepted: 26 April 2016;
Published: 10 May 2016.
Edited by:
Yuji Morita, Aichi Gakuin University, JapanReviewed by:
Shivangi Agarwal, Northwestern University, USALin Zeng, University of Florida, USA
Grace Ann Spatafora, Middlebury College, USA
Copyright © 2016 Mao, Yang, Li, Lei, Li, Yang, Tao, Yin, Zhang, Ma and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin-Rong Ma, bWF4ckBjaWIuYWMuY24=;
Tao Hu, aHV0YW9Ac2N1LmVkdS5jbg==
†Co-first author
 Meng-Ying Mao
Meng-Ying Mao Ying-Ming Yang
Ying-Ming Yang Ke-Zeng Li
Ke-Zeng Li Lei Lei
Lei Lei Meng Li
Meng Li Yan Yang
Yan Yang Xiang Tao
Xiang Tao Jia-Xin Yin
Jia-Xin Yin Ru Zhang
Ru Zhang Xin-Rong Ma
Xin-Rong Ma Tao Hu
Tao Hu