- Lantibiotic Immunity and Resistance, Institute of Biochemistry, Heinrich-Heine-University, Duesseldorf, Germany
Lantibiotics are potential alternatives to antibiotics because of their broad-range killing spectrum. The producer strain is immune against its own synthesized lantibiotic via the expression of two proteins LanI and LanFEG. Recently, gene operons are found in mainly human pathogenic strains, which confer resistance against lantibiotics. Of all the lantibiotics discovered till date, nisin produced by some Lactococcus lactis strains is the most prominent member. Nisin has multiple mode of actions of which binding to the cell wall precursor lipid II and subsequent insertion into the bacterial membrane to form pores are the most effective. The nisin producing strains express the lipoprotein NisI to prevent a suicidal effect. NisI binds nisin, inducing a reversible cell clustering to prevent nisin from reaching the membrane. Importantly NisI does not modify nisin and releases it as soon as the concentration in the media drops below a certain level. The human pathogen Streptococcus agalactiae is naturally resistant against nisin by expressing a resistance protein called SaNSR, which is a nisin degrading enzyme. By cleaving off the last six amino acids of nisin, its effectiveness is 100-fold reduced. This cleavage reaction appears to be specific for nisin since SaNSR recognizes the C-terminal located lanthionine rings. Recently, the structures of both NisI and SaNSR were determined by NMR and X-ray crystallography, respectively. Furthermore, for both proteins the binding site for nisin was determined. Within this review, the structures of both proteins and their different defense mechanisms are described.
Introduction
Since the beginning of the last century, the heterogeneous group of bacteriocins have become an interesting research topic for various applications like food preservatives or pharmaceutical purposes as antibiotic alternatives (Cleveland et al., 2001; Cotter et al., 2012). Bacteriocins are small, ribosomally synthesized peptides of which some possess high antimicrobial activity (Tagg et al., 1976; Cotter et al., 2005b).
Within the group of bacteriocins a large family exists, which are called lantibiotics (Jung, 1991; Willey and van der Donk, 2007; Bierbaum and Sahl, 2009; Alvarez-Sieiro et al., 2016). These are small antimicrobial peptides, which are post-translationally modified and contain uncommon amino acids (Deyhydrobutyrine or Dehydroalanine). The linkage of these with cysteine residues results in the formation of characteristic thioether bridges, called lanthionine rings. These lanthionine rings ensure the high antimicrobial activity against various bacteria. Lantibiotics are highly potent as depicted by the observation that already nanomolar concentrations are sufficient to fulfill their antimicrobial activity (Delves-Broughton et al., 1996; Chatterjee et al., 2005). Furthermore, the lanthionine rings ensure that lantibiotics are intrinsically resistant against proteolysis by unspecific degrading enzymes.
Active lantibiotics are able to inhibit the growth of Gram-positive as well as Gram-negative bacteria. Some exhibit multiple modes of action, of which binding to lipid II, thereby inhibiting cell wall synthesis, and pore formation are the most prominent ones (Héchard and Sahl, 2002; Bierbaum and Sahl, 2009). Due to the high activitiy in combination with the high stability lantibiotics are considered as usefull compounds for medical treatment and food preservatives. One example is nisin produced by some Lactococcus lactis strains which is linked to the potential biomedical application against bacterial mastitis, treatment of methicillin-resistant Staphylococcus aureus (MRSA) and enterococcal infections. Other examples are the lantibiotics Gallidermin/Epidermin, which are associated with acne, eczema, follicultis, impetigo as possible compound for treatment (Cotter et al., 2005a).
Within the lantibiotic producer strains, the structural genes for the lantibiotic itself (lanA) as well as for its biosynthesis and modification (lanBC or lanM), transport (lanT, lanT+C39 peptidase or lanT+C39 peptidase+lanM) across the cellular membrane are with a few exceptions encoded on a single gene cluster (Chatterjee et al., 2005; Willey and van der Donk, 2007; Alkhatib et al., 2012; Singh and Sareen, 2014). Furthermore, a two-component system (lanK and lanR) is present on this operon, which up-regulates the expression of these genes (Kuipers et al., 1995; Qiao et al., 1996).
In order to prevent the harmful effect of the secreted and activated lantibiotic on their own membrane, additional genes (lanI and lanFEG) encode a lantibiotic specific (auto-)immunity system. Interestingly, although lantibiotics are grouped in different classes based on their size and specific activities (Willey and van der Donk, 2007; Arnison et al., 2013), the lantibiotic specific (auto-)immunity system genes seem to be conserved throughout the species (Alkhatib et al., 2012). The genes of the (auto-)immunity system encode for the following proteins: LanI, a membrane associated lipoprotein, and LanFEG, an ABC transporter localized in the cellular membrane (Draper et al., 2008, 2015). Some lantibiotic strains contain only one of the (auto-)immunity proteins, which correlates with the activity of the produced lantibiotic, which can be membrane binding and/or pore formation activity.
Nisin secreted by some L. lactis strains is the most prominent lantibiotic. Due to its high bactericidal activity in combination with low toxicity in humans, nisin has already been used for decades as a natural preservative in the food industry (Delves-Broughton et al., 1996). Active nisin consists of 34 amino acids and contains five stereo-specifically installed lanthionine-based rings. The first three rings (rings A–C) are separated from the last two intertwined rings (rings D–E) by a flexible hinge region (Van de Ven et al., 1991; Zhou et al., 2015). Rings A and B are able to bind lipid II, thereby inhibiting cell growth (Hsu et al., 2004), whereas the hinge region and rings D and E, are able to flip into the membrane (Wiedemann et al., 2001; van Heusden et al., 2002; Hasper et al., 2004). This creates pores, which leads to an immediate efflux of nutrients and small compounds, resulting in cell death (Ruhr and Sahl, 1985; Breukink et al., 1999).
The nisin producing L. lactis strains are immune against the high antimicrobial activity of nisin via the expression of an distinct immunity system consisting of the lipoprotein NisI and ABC transporter NisFEG (Figure 1). When these proteins are expressed, the producing strains survives a high level of immunity against nisin, of up to ~750 nM nisin (1000 IU/ml; Ra et al., 1996). This concentration is a >100-fold higher than the 4–6 nM observed in L. lactis strain lacking this immunity system. Interestingly, both lipoprotein and ABC transporter act cooperatively and each of them displays only 10–30% of the full immunity levels when expressed alone (Ra et al., 1999; Stein et al., 2003; AlKhatib et al., 2014a,b).
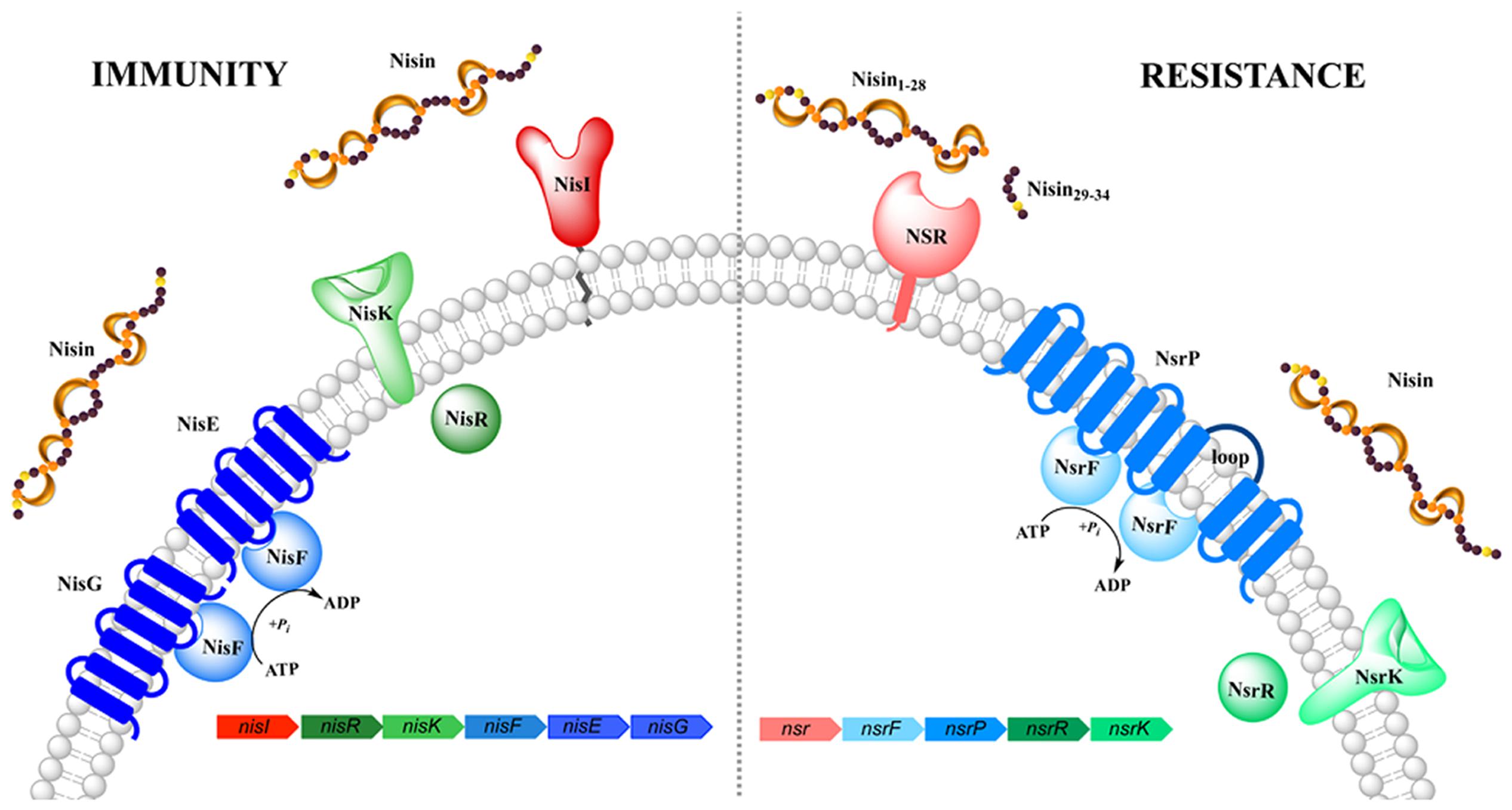
FIGURE 1. Schematic representation of the nisin (auto-)immunity and resistance system. Both systems comprise of a lipoprotein/membrane-associated protein (colored in red/pink, respectively), an ABC transporter (depicted in blue) and a two-component system (shown in green). Functionally similar genes are color coded identically with the exception of NisI and NSR.
Such an (auto)-immunity system is conserved and present in al most all lantibiotic producing strain. For example, the subtilin producing Bacillus subtilis strain expresses SpaI and SpaFEG (Klein and Entian, 1994); the epidermin producer Staphylococcus epidermidis expresses EpiH and EpiFEG (Peschel and Götz, 1996; Otto et al., 1998); the gallidermin producer Staphylococcus gallinarium expresses GdmH and GdmFEG and the Pep5 producer S. epidermidis expresses PepI (Reis et al., 1994). More examples as well as their operon structure are highlighted in (Alkhatib et al., 2012).
Recently, however, some operons were described encoding a protein defense system against lantibiotics although the host strain is not producing these lantibiotics itself. Interestingly, these operon are mainly found in human pathogenic strains. The expression of the genes localized on these operon results in a resistance against the lantibiotic. For example: lctGEFlcrXRS in Streptococcus mutans conferring resistance against nisin, nukacin ISK-I and lacticin 481 (Kawada-Matsuo et al., 2013), and graXSR/vraFG in S. aureus provides resistance against various lantibiotics including nisin and nukacin ISK-I (Meehl et al., 2007; Falord et al., 2011, 2012). The cprABCK-R operon from Clostridium difficile confers even resistance against multiple lantibiotics of which nisin, gallidermin, subtilin, and mutacin 1140 (McBride and Sonenshein, 2011; Suárez et al., 2013).
In Streptococcus agalactiae, the operon identified confers resistance against the lantibiotic nisin and resembles the genetic architecture of the nisI and nisFEG immunity genes found in the producing L. lactis strain (Khosa et al., 2013; Figure 1). The genes encoded on this operon are called nsr (encoding for nisin resistance protein) and nsrFP (encoding for an ABC transporter), which are probably auto-regulated by a two-component system formed by nsrR and nsrK (Khosa et al., 2013). The solely expressed membrane-associated nisin resistance protein (also known as NSR) has been shown to confer 20-fold resistance against nisin, when expressed in a sensitive L. lactis strain (Khosa et al., 2013). Due to the high sequence homology of NSR, especially the specific TASSAEM region, homologs of this resistance protein have been identified in several other strains such as S. epidermidis among others (Khosa et al., 2013).
Recently, the structures of both nisin (auto-)immunity protein NisI from L. lactis (Hacker et al., 2015) as well as the resistance protein NSR from S. agalactiae (SaNSR; Khosa et al., 2016) have been revealed. They both, although sharing a low sequence similarity of only 23%, protect the cell membrane against the presence of high concentrations of nisin albeit by a different mechanism. Both proteins can be considered as role models for the (auto-)immunity and resistance system of other lantibiotics. Within this review, the function and structure of both proteins will be compared and described.
The Nisin Immunity Protein NisI
In nisin producing L. lactis strains, the LanI immunity protein is called NisI, which is a 27.8 kDa in size comprising of 245 amino acid residues (Kuipers et al., 1993; Table 1). The sequence contains a signal peptide which is cleaved off after secretion followed by a site for lipidation (Cys-1 in mature NisI; Kuipers et al., 1993; Sutcliffe and Russell, 1995). The resulting mature NisI (226 amino acid residues and 25.8 kDa in size) is lipid-anchored at the outside of the cytoplasmic membrane (Ra et al., 1996). Approximately, one-third of NisI escapes this lipid modification and is thereby released into the extracellular environment in a lipid-free form that forms an additional mechanism of immunity (Koponen et al., 2004; Takala et al., 2004).
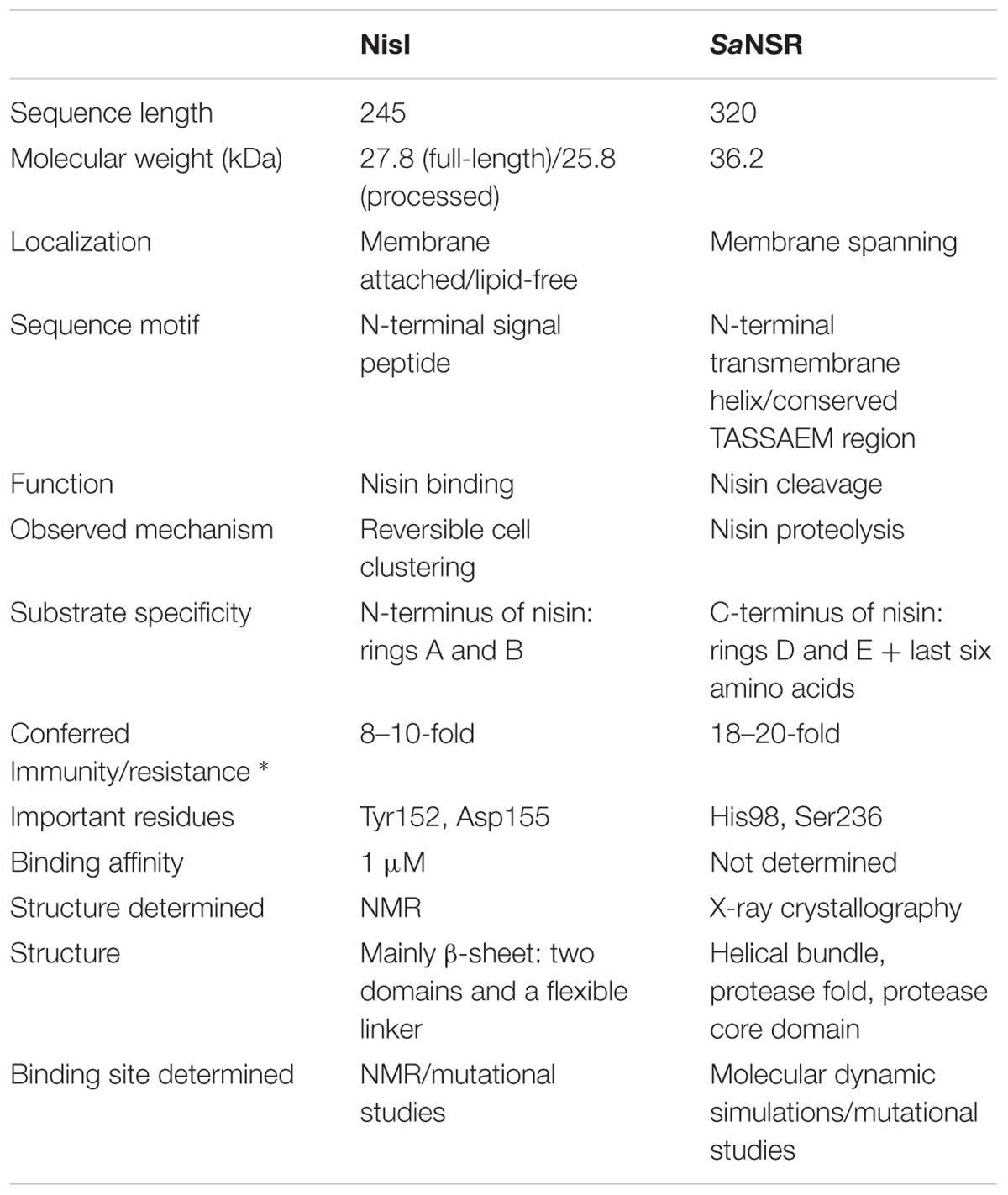
TABLE 1. Characteristics of the nisin immunity protein NisI and nisin resistance protein NSR (* IC50 compared against sensitive Lactococcus lactis strain NZ9000; Sun et al., 2009; AlKhatib et al., 2014a; Hacker et al., 2015; Khosa et al., 2016).
NisI exhibits two functions preventing nisin to reach the cellular membrane. First NisI binds nisin, thereby protecting the nisin-producing bacteria. Important, here is the fact that NisI does not modify or degrade nisin as shown by several independent studies (Qiao et al., 1995; Ra et al., 1996, 1999; Stein et al., 2003; Koponen et al., 2004; AlKhatib et al., 2014a). Initially, the involvement of NisI in immunity was identified via the expression of solely NisI which increased the nisin resistance of both E. coli and L. lactis (Kuipers et al., 1993). Furthermore, the significance of NisI in the overall nisin immunity of L. lactis cells was observed via deletion of the nisI gene within the whole nisin operon. The resulting nisI knockout strain was more sensitive to nisin than the fully equipped L. lactis strain (Siegers and Entian, 1995). Interestingly, the knockout strain of the nisI gene had more adverse effects than the knockout of the nisFEG genes, leading to the hypothesis that the NisI protein plays a more effective role in the immunity against nisin.
In order to obtain further information on the functioning of the nisI gene, it was expressed in a nisin-sensitive L. lactis NZ900 strain that contains the nisRK two-component system but lacks the rest of the nisin operon and especially the immunity system encoding genes. When NisI was expressed in this strain, 8–10-fold more nisin was needed to inhibit the cell growth by 50% (IC50; AlKhatib et al., 2014a; Figure 2A). Various studies have shown the importance of the C-terminus of NisI for its activity, especially the last 22 amino acids (Takala and Saris, 2006; AlKhatib et al., 2014a). A deletion of the last 22 residues, reduces the activity of NisI to 30–34% (AlKhatib et al., 2014a). Furthermore, a deletion of only the last five residues decreases the immunity conferred by NisI to approximately 78% (Takala and Saris, 2006). It appears that at low nisin concentration in the environment, the binding capacity of NisI is enough to provide immunity up to a concentration of 60 nM, where the cells are still able to grow. Since the immunity conferred by NisI is a result of its binding to nisin, it is somewhat surprising that NisI in vitro displayed a rather weak affinity for nisin as seen by a KD of approximately 0.6–2 μM (Takala et al., 2004; Hacker et al., 2015). However, this is likely due to the fact that the in vitro experimental setup lacks the membrane environment.
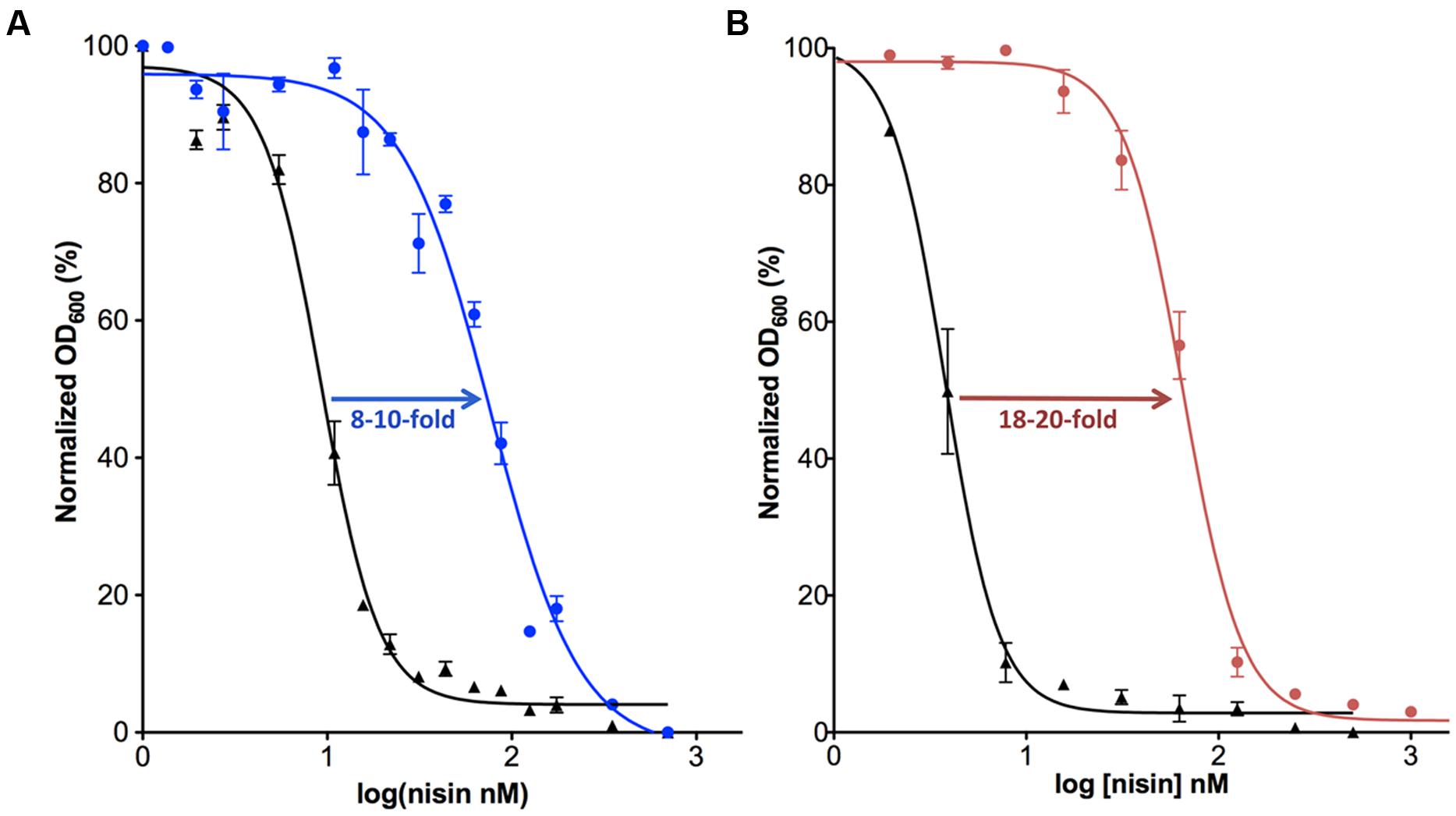
FIGURE 2. Activity determination of NisI and SaNSR. The IC50 value was determined for the nisin sensitive Lactococcus lactis NZ9000 strain (black) in comparison with the same strain harboring a plasmid containing the nisI (A; shown in blue) or nsr (B; shown in pink) gene.
Recently, in addition to the binding capability of NisI, a second mechanism for conferring immunity by NisI was also described. Upon addition of nisin to nisI expressing L. lactis cells, the cells start to cluster. Especially at nisin concentrations above the determined IC50 value, the cells cluster to form large chains up to a number of 30 cells (AlKhatib et al., 2014a). Importantly, this clustering is only observed when both NisI and nisin are present. Due to this clustering, nisin is unable to reach lipid II. Thereby the activity of nisin, especially the pore forming ability is inhibited as observed via a so-called Sytox assay (AlKhatib et al., 2014a). This clustering mechanism is reversible and as soon as the the external nisin concentration drops below the IC50 value, the L. lactis cells start growing normally again (AlKhatib et al., 2014a). Thus, when the concentration of nisin increases above a certain threshold (in the reported study around 60–70 nM), which coincides with the measured IC50 values, the presence of both NisI and nisin induces a clustering of the L. lactis cells. In the nisin sensitive strains lacking nisI or upon expressing a nisI variant lacking the C-terminally located 22 amino acids, this clustering is not observed, leading to the assumption that the C-terminus of NisI is responsible for this clustering phenomenon (AlKhatib et al., 2014a). This is inline with studies describing that the C-terminus of NisI (last 21 amino acids) interacts with nisin and provides specificity to NisI (Takala and Saris, 2006). Although, this clustering is an interesting observation it has not been unraveled how NisI mediates this.
Since the first two rings of nisin (N-terminal region) are crucial for its binding to lipid II (Wiedemann et al., 2001), which is an initial step in the activity of nisin and also essential for pore formation by nisin, it is a reasonable assumption that NisI recognizes the N-terminus of nisin, presumably the first two rings. Thereby, NisI would directly interfere with the initial and crucial binding event of nisin with lipid II.
The Nisin Resistance Protein SaNSR
The counter part of NisI in the nisin resistance system of S. agalactiae is the nisin resistance protein (SaNSR), which contains 320 amino acids and has a molecular weight of 36.2 kDa (Froseth and McKAY, 1991; Khosa et al., 2013; Table 1). SaNSR has a N-terminus hydrophobic region predicted to encode a transmembrane helix (Khosa et al., 2013), thereby SaNSR would be localized at the bacterial membrane (Sun et al., 2009). SaNSR is a nisin degrading enzyme (Sun et al., 2009). Since the lanthionine rings usually cause steric hindrance, thereby inhibiting any protease cleavage a nisin degradation mechanism is quite unique (Wiedemann et al., 2001). SaNSR cleaves off the last six amino acids of nisin, yielding two fragments: nisin1-28 and nisin29-34. This nisin1-28 variant when purified displays 100-fold less bactericidal activity and significantly less affinity toward the bacterial membrane (Sun et al., 2009; AlKhatib et al., 2014a). Thus, the non-producing strains become resistant against nisin by reducing its effectiveness. SaNSR confers 18–20-fold resistance in a nisin sensitive L. lactis strain as determined by IC50 assays (Figure 2B). SaNSR belongs to the S41 family of peptidases and contains a highly conserved TASSAEM sequence motif, which harbors the catalytically active serine residue at position 236 (Table 1; Khosa et al., 2013, 2016). This 18–20-fold resistance mediated by SaNSR when expressed in nisin sensitive L. lactis cells is almost lost when mutating this Ser236 residue (Khosa et al., 2013).
Furthermore, the resistance mediated by SaNSR in nisin sensitive L. lactis strain dropped to mere 1.4–1.7-fold for the nisin variants lacking the rings D and E or only E (Khosa et al., 2016). Additionally, removing the last 6 or 12 amino acids of nisin (nisin1-28 and nisin1-22, respectively) completely abolished the resistance, clearly indicating the importance of the last ring as well as the C-terminal tail of nisin for recognition by SaNSR. This is in contrast to the specificity of NisI. Nisin shares a high sequence similarity of around 63% with subtilin from B. subtilis, and like nisin the lantibiotic subtilomycin also harbors five lanthionine-based rings (Barbosa et al., 2015). Therefore, it might be possible that SaNSR exhibit a broader substrate specificity including resistance toward other lantibiotics as well. Such a multiple lantibiotic resistance specificity has been previously described for an other lantibiotic resistance operon system of C. difficile (McBride and Sonenshein, 2011; Suárez et al., 2013).
Structures of the Immunity Protein NisI and the Resistance Protein NSR
The structure of NisI from L. lactis was solved using NMR spectroscopy (Hacker et al., 2015). NisI is a two-domain and predominantly a β-sheet containing protein (Figure 3A). The N-terminal part (1–111 residues) is connected to the C-terminal domain (120–226) via a flexible linker (112–119). Interestingly and rather unusual is that both domains adopt a similar unique fold, which has been previously observed for SpaI from B. subtilis, the immunity protein against subtilin (Christ et al., 2012). The core of the N- and C- terminal domains of NisI are formed by a seven-stranded antiparallel twisted β-sheet in the strand order β1-β2-β3-β8-β7-β6b-β4a. An extended β-hairpin is formed by strands β4b and β6a that is stabilized by hydrophobic packing interactions with residues from β1 and β2. In addition, the β-hairpin is flanked by a short 310 helix.
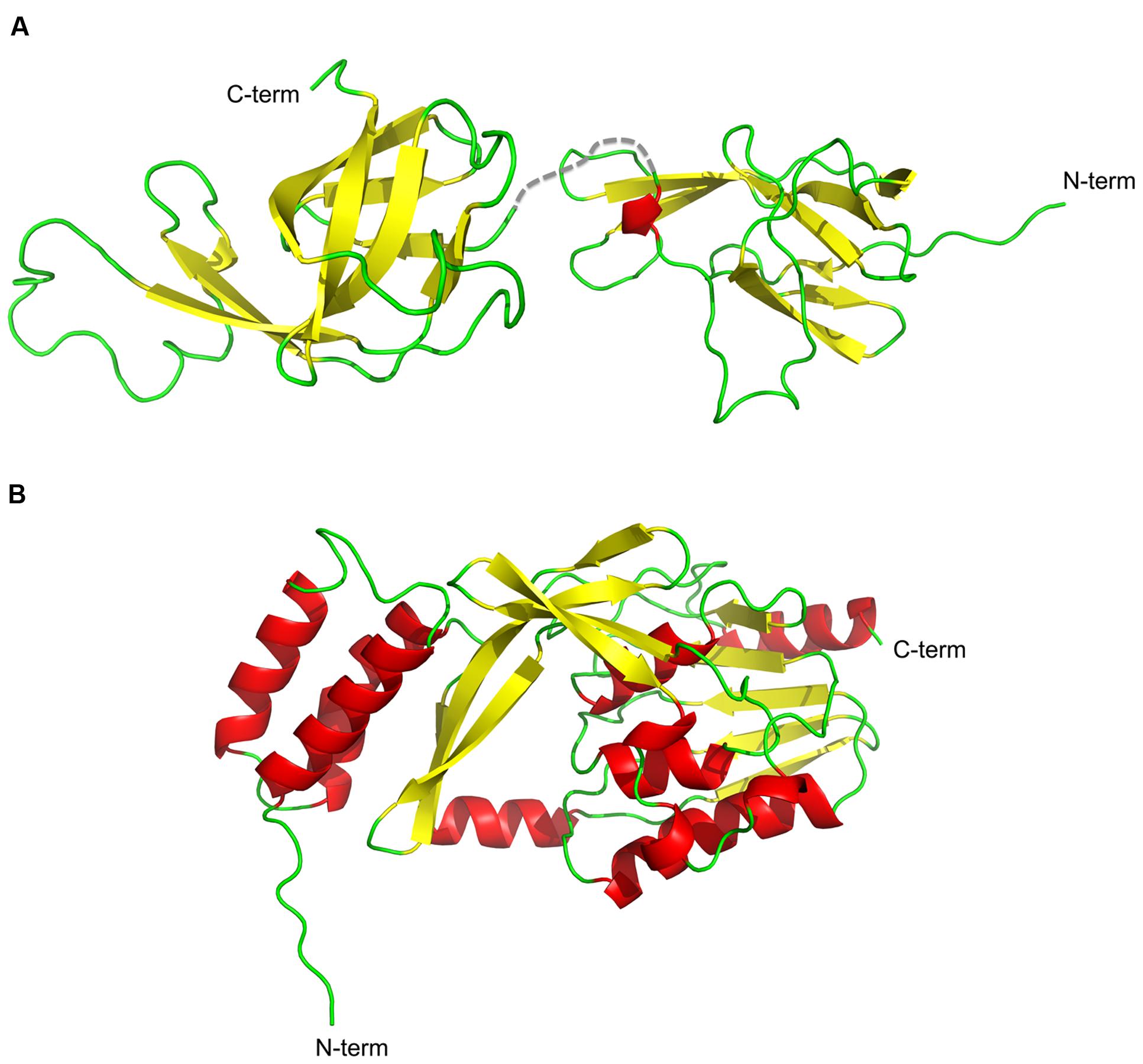
FIGURE 3. Cartoon representation of the structures of NisI and SaNSR. The structures of (A) NisI (PDB codes: 2N32 and 2N2E) and (B) SaNSR (PDB code: 4Y68) are shown with the secondary elements color coded as red for helices, yellow for sheets and green for the loops. The figure was created with Pymol.
NisI lacks the N-terminus unstructured region, which is present in SpaI and supposedly allows interaction with the host membrane (Christ et al., 2012; Hacker et al., 2015). Since it was proposed that NisI interacts with the membrane, an elegant set of experiments were performed with the single expressed domains, which revealed that only the N-terminal domain possesses affinity toward the membrane environment (Hacker et al., 2015; Figure 4). Therefore, this domain is thought to be localized at the membrane surface. On the contrary, the C-terminal domain of NisI does not bind to lipids. Instead this C-terminal domain binds nisin as observed in NMR titration experiments. This is inline with the previous observations that the C-terminally located last 22 amino acids are important for the functioning of NisI in vivo. Although structurally similar, both domains of NisI differ in their surface properties. While the surface of the N-terminal domain of NisI is highly positively charged and interacts with membranes, the C-terminal domain has negatively charged surface with hydrophobic patches and is able to bind nisin (Figure 4A), thereby modulating the membrane affinity of the N-terminal domain of NisI by shielding its membrane binding surface (Hacker et al., 2015; Figure 4A). NisI has not been shown to interact with other lantibiotics and displays rather high substrate specificity. A protein consisting of the N-terminal SpaI domain fused to the C-terminal domain of NisI, however, displays immunity against nisin (Takala and Saris, 2006). This highlights that the nisin binding site is localized within the C-terminus of NisI. Since subtilin and nisin share high homology in terms of sequence as well as lanthioinine ring positions it is not surprising that the domain structure of NisI and SpaI are structurally similar. This in contrast to the structure MlbQ from the actinomycete Microbispora ATCC PTA-5024 conferring resistance against NAI-107 which was solved by NMR (Pozzi et al., 2015). No significant sequence identity as well as structural similarity was observed between NisI and MlbQ. This likely is due to the larger difference between nisin and NAI-107 in sequence as well as lanthionine ring composition. It seems that the LanI immunity proteins evolved differently and are highly specific for their cognate lantibiotic.
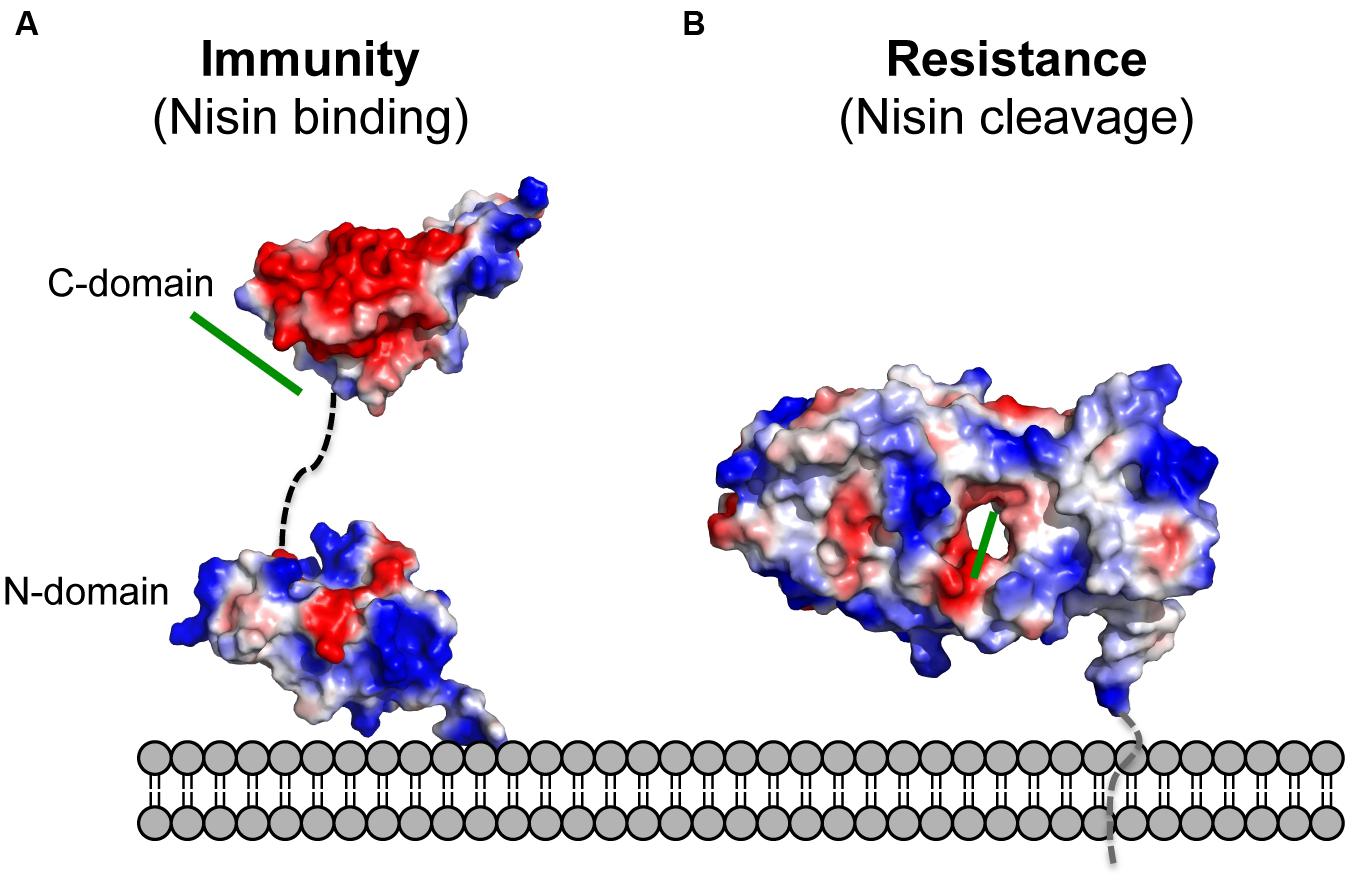
FIGURE 4. Electrostatic surface potential of the structures of NisI and SaNSR. The electrostatic surface potential of (A) NisI (PDB codes: 2N32 and 2N2E) and (B) SaNSR (PDB code: 4Y68) structures is shown. Negatively charged surface areas are colored in red, while the positively charged areas colored in blue and white areas correspond to hydrophobic surfaces. The determined nisin binding site for both proteins are highlighted with a green line. The figure was created with Pymol.
The crystal structure of nisin resistance protein from S. agalactiae (SaNSR; without the N-terminal transmembrane helix) was solved using X-ray crystallography and consists of eleven α-helices and eleven β-strands (Figure 3B; Khosa et al., 2016). SaNSR is composed of three domains: an N-terminal helical bundle comprising of 65 amino acid residues (Lys31-Gly96), form helices α1-α3. This domain ends in a triple glycine motif before entering the protease cap domain. This protease cap domain consists of helix α4 and a β-hairpin structure formed by two strands and forms a lid-like structure above the tunnel. The last domain called the protease core is formed by six strands βb-βg and five helices α5-α9 and adopts a ‘protease fold’ domain as observed in other S41 peptidases (Liao et al., 2000; Kim et al., 2002; Mastny et al., 2013). Together they form a hydrophobic tunnel of ~10 Å width. This hydrophobic, negatively charged tunnel is responsible for binding to nisin by ‘roping in’ the peptide (Figure 4B). The protease core domain also contains the highly conserved TASSAEM region that harbors the previously identified catalytically active serine at position 236 (Khosa et al., 2013).
The active center of SaNSR consists of a catalytic dyad formed by residues Ser236 (Khosa et al., 2013), which is part of the TASSAEM motif, and His98 as determined by mutational analysis (Khosa et al., 2016) and also described for some other proteases (Page and Di Cera, 2008).
Unfortunately, the crystal structure lacks the substrate nisin. Instead a peptide called N-pep, belonging to symmetry-related molecule was bound within the tunnel. This information was used for molecular dynamic simulation studies to determine the nisin-binding site. It was observed that nisin is stably bound in the tunnel formed in between the domains of SaNSR. Additionally, the residues forming the hydrophobic interactions for proper orientation of rings D and E of nisin are embedded in the protease core domain within or on the outside of the tunnel that is situated in the middle of SaNSR protein.
The model of SaNSR/nisin complex demonstrates the significance of C-terminally located lanthionine rings D and E of nisin for substrate specificity (Wiedemann et al., 2001). The importance of these rings was also highlighted by mutational analyses of nisin. Here, SaNSR did not recognize nisin variants lacking the last or the last two rings.
Conclusion
The expression of NisI and NSR reduce the activity of nisin. In the nisin producing strains, the immunity protein NisI solely binds nisin thereby reducing the amount of nisin reaching the lipid II target molecule in the membrane. To achieve this NisI not only binds to nisin but also induces a “shielding mechanism” of the L. lactis cells. Immunity is thereby provided without harming or modifying the own lantibiotic produced.
In contrast, NSR present in nisin non-producing strains cleaves nisin in two parts, thus, reducing the ability of nisin to form pores in the membrane.
NisI and NSR, however, represent the first line of defense and within both immunity and the resistance operons, an additional protein system is also encoded. In case of the immunity system, it is the ABC transporter NisFEG and for the resistance system, it is NsrFP. It has been shown that NisFEG and NisI act cooperatively. However, for the nsr system such a cooperative mechanism has not been detected so far and it has to be further investigated whether these resistance proteins are also able to act together in their battle against the lantibiotic nisin. Although, lantibiotics are considered to be powerfull biological antimicrobial compounds already resistance mechanisms are present in some human pathogenic strains. This hampers the usage of lantibiotics for medical purposes. A detailed knowledge about these resistance mechanisms would pave a way to bypass these inherently present systems.
Author Contributions
SK, ML, and SS wrote the manuscript. SK, ML prepared the figures. SK, ML, and SS finalized the manuscript.
Funding
We are also grateful to Heinrich Heine International Graduate School of Protein Science and Technology (iGRASPseed) for providing a scholarship to SK and all the group members for the stimulating discussions.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Lutz Schmitt for fruitful discussions, encouragement, support and invaluable advice. We would like to thank Jens Wöhnert (Frankfurt University, Germany) for providing the coordinates of the NisI structure.
References
Alkhatib, Z., Abts, A., Mavaro, A., Schmitt, L., and Smits, S. H. (2012). Lantibiotics: how do producers become self-protected? J. Biotechnol. 159, 145–154. doi: 10.1016/j.jbiotec.2012.01.032
AlKhatib, Z., Lagedroste, M., Fey, I., Kleinschrodt, D., Abts, A., and Smits, S. H. (2014a). Lantibiotic immunity: inhibition of nisin mediated pore formation by NisI. PLoS ONE 9:e102246. doi: 10.1371/journal.pone.0102246
AlKhatib, Z., Lagedroste, M., Zaschke, J., Wagner, M., Abts, A., Fey, I., et al. (2014b). The C-terminus of nisin is important for the ABC transporter NisFEG to confer immunity in Lactococcus lactis. Microbiol. Open 3, 752–763. doi: 10.1002/mbo3.205
Alvarez-Sieiro, P., Montalban-Lopez, M., Mu, D., and Kuipers, O. P. (2016). Bacteriocins of lactic acid bacteria: extending the family. Appl. Microbiol. Biotechnol. 100, 2939–2951. doi: 10.1007/s00253-016-7343-9
Arnison, P. G., Bibb, M. J., Bierbaum, G., Bowers, A. A., Bugni, T. S., Bulaj, G., et al. (2013). Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30, 108–160. doi: 10.1039/c2np20085f
Barbosa, J., Caetano, T. N., and Mendo, S. N. (2015). Class I and Class II Lanthipeptides Produced by Bacillus spp. J. Nat. Prod. 11, 2850–2866. doi: 10.1021/np500424y
Bierbaum, G., and Sahl, H. G. (2009). Lantibiotics: mode of action, biosynthesis and bioengineering. Curr. Pharm. Biotechnol. 10, 2–18. doi: 10.2174/138920109787048616
Breukink, E., Wiedemann, I., Van Kraaij, C., Kuipers, O., Sahl, H.-G., and De Kruijff, B. (1999). Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 286, 2361–2364. doi: 10.1126/science.286.5448.2361
Chatterjee, C., Paul, M., Xie, L., and van der Donk, W. A. (2005). Biosynthesis and mode of action of lantibiotics. Chem. Rev. 105, 633–684. doi: 10.1021/cr030105v
Christ, N. A., Bochmann, S., Gottstein, D., Duchardt-Ferner, E., Hellmich, U. A., Düsterhus, S., et al. (2012). The First structure of a lantibiotic immunity protein, SpaI from Bacillus subtilis, reveals a novel fold. J. Biol. Chem. 287, 35286–35298. doi: 10.1074/jbc.M112.401620
Cleveland, J., Montville, T. J., Nes, I. F., and Chikindas, M. L. (2001). Bacteriocins: safe, natural antimicrobials for food preservation. Int. J. Food Microbiol. 71, 1–20. doi: 10.1016/S0168-1605(01)00560-8
Cotter, P. D., Hill, C., and Ross, R. P. (2005a). Bacterial lantibiotics: strategies to improve therapeutic potential. Curr. Protein Pept. Sci. 6, 61–75. doi: 10.2174/1389203053027584
Cotter, P. D., Hill, C., and Ross, R. P. (2005b). Bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 3, 777–788. doi: 10.1038/nrmicro1273
Cotter, P. D., Ross, R. P., and Hill, C. (2012). Bacteriocins—a viable alternative to antibiotics? Nat. Rev. Microbiol. 11, 95–105. doi: 10.1038/nrmicro2937
Delves-Broughton, J., Blackburn, P., Evans, R., and Hugenholtz, J. (1996). Applications of the bacteriocin, nisin. Antonie Van Leeuwenhoek 69, 193–202. doi: 10.1007/BF00399424
Draper, L. A., Cotter, P. D., Hill, C., and Ross, R. P. (2015). Lantibiotic Resistance. Microbiol. Mol. Biol. Rev. 79, 171–191. doi: 10.1128/MMBR.00051-14
Draper, L. A., Ross, R. P., Hill, C., and Cotter, P. D. (2008). Lantibiotic immunity. Curr. Protein Pept. Sci. 9, 39–49. doi: 10.2174/138920308783565750
Falord, M., Karimova, G., Hiron, A., and Msadek, T. (2012). GraXSR proteins interact with the VraFG ABC transporter to form a five-component system required for cationic antimicrobial peptide sensing and resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 56, 1047–1058. doi: 10.1128/AAC.05054-11
Falord, M., Mäder, U., Hiron, A., Debarbouille, M., and Msadek, T. (2011). Investigation of the Staphylococcus aureus GraSR regulon reveals novel links to virulence, stress response and cell wall signal transduction pathways. PLoS ONE 6:e21323. doi: 10.1371/journal.pone.0021323
Froseth, B. R., and McKAY, L. L. (1991). Molecular characterization of the nisin resistance region of Lactococcus lactis subsp. lactis biovar diacetylactis DRC3. Appl. Environ. Microbiol. 57, 804–811.
Hacker, C., Christ, N. A., Duchardt-Ferner, E., Korn, S., Göbl, C., Berninger, L., et al. (2015). The solution structure of the lantibiotic immunity protein NisI and its interactions with nisin. J. Biol. Chem. 48, 28869–28886. doi: 10.1074/jbc.M115.679969
Hasper, H. E., de Kruijff, B., and Breukink, E. (2004). Assembly and stability of nisin-lipid II pores. Biochemistry 43, 11567–11575. doi: 10.1021/bi049476b
Héchard, Y., and Sahl, H.-G. (2002). Mode of action of modified and unmodified bacteriocins from Gram-positive bacteria. Biochimie 84, 545–557. doi: 10.1016/S0300-9084(02)01417-7
Hsu, S.-T. D., Breukink, E., Tischenko, E., Lutters, M. A. G., de Kruijff, B., Kaptein, R., et al. (2004). The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics. Nat. Struct. Mol. Biol. 11, 963–967. doi: 10.1038/nsmb830
Jung, G. (1991). Lantibiotics-ribosomally sythesized biologically active polypeptides containing sulfide bridges and α,β-Didehyroamino acids. Angew. Chem. Int. Ed. Engl. 30, 1051–1068. doi: 10.1002/anie.199110513
Kawada-Matsuo, M., Oogai, Y., Zendo, T., Nagao, J., Shibata, Y., Yamashita, Y., et al. (2013). Involvement of the novel two-component NsrRS and LcrRS systems in distinct resistance pathways against Nisin A and Nukacin ISK-1 in Streptococcus mutans. Appl. Environ. Microbiol. 79, 4751–4755. doi: 10.1128/AEM.00780-13
Khosa, S., AlKhatib, Z., and Smits, S. H. (2013). NSR from Streptococcus agalactiae confers resistance against nisin and is encoded by a conserved nsr operon. Biol. Chem. 394, 1543–1549. doi: 10.1515/hsz-2013-0167
Khosa, S., Frieg, B., Mulnaes, D., Kleinschrodt, D., Hoeppner, A., Gohlke, H., et al. (2016). Structural basis of lantibiotic recognition by the nisin resistance protein from Streptococcus agalactiae. Sci. Rep. 6:18679. doi: 10.1038/srep18679
Kim, J.-S., Groll, M., Musiol, H.-J., Behrendt, R., Kaiser, M., Moroder, L., et al. (2002). Navigation inside a protease: substrate selection and product exit in the tricorn protease from Thermoplasma acidophilum. J. Mol. Biol. 324, 1041–1050. doi: 10.1016/S0022-2836(02)01153-1
Klein, C., and Entian, K. (1994). Genes involved in self-protection against the lantibiotic subtilin produced by Bacillus subtilis ATCC 6633. Appl. Environ. Microbiol. 60, 2793–2801.
Koponen, O., Takala, T. M., Saarela, U., Qiao, M., and Saris, P. E. (2004). Distribution of the NisI immunity protein and enhancement of nisin activity by the lipid-free NisI. FEMS Microbiol. Lett. 231, 85–90. doi: 10.1016/S0378-1097(03)00934-0
Kuipers, O. P., Beerthuyzen, M. M., de Ruyter, P. G., Luesink, E. J., and de Vos, W. M. (1995). Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J. Biol. Chem. 270, 27299–27304. doi: 10.1074/jbc.270.45.27299
Kuipers, O. P., Beerthuyzen, M. M., Siezen, R. J., and Vos, W. M. (1993). Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Eur. J. Biochem. 216, 281–291. doi: 10.1111/j.1432-1033.1993.tb18143.x
Liao, D.-I., Qian, J., Chisholm, D. A., Jordan, D. B., and Diner, B. A. (2000). Crystal structures of the photosystem II D1 C-terminal processing protease. Nat. Struct. Mol. Biol. 7, 749–753. doi: 10.1038/78973
Mastny, M., Heuck, A., Kurzbauer, R., Heiduk, A., Boisguerin, P., Volkmer, R., et al. (2013). CtpB assembles a gated protease tunnel regulating cell-cell signaling during spore formation in Bacillus subtilis. Cell 155, 647–658. doi: 10.1016/j.cell.2013.09.050
McBride, S. M., and Sonenshein, A. L. (2011). Identification of a genetic locus responsible for antimicrobial peptide resistance in Clostridium difficile. Infect. Immun. 79, 167–176. doi: 10.1128/IAI.00731-10
Meehl, M., Herbert, S., Götz, F., and Cheung, A. (2007). Interaction of the GraRS two-component system with the VraFG ABC transporter to support vancomycin-intermediate resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 51, 2679–2689. doi: 10.1128/AAC.00209-07
Otto, M., Peschel, A., and Götz, F. (1998). Producer self-protection against the lantibiotic epidermin by the ABC transporter EpiFEG of Staphylococcus epidermidis Tü3298. FEMS Microbiol. Lett. 166, 203–211. doi: 10.1111/j.1574-6968.1998.tb13891.x
Page, M., and Di Cera, E. (2008). Serine peptidases: classification, structure and function. Cell. Mol. Life Sci. 65, 1220–1236. doi: 10.1007/s00018-008-7565-9
Peschel, A., and Götz, F. (1996). Analysis of the Staphylococcus epidermidis genes epiF,-E, and-G involved in epidermin immunity. J. Bacteriol. 178, 531–536.
Pozzi, R., Coles, M., Linke, D., Kulik, A., Nega, M., Wohlleben, W., et al. (2015). Distinct mechanisms contribute to immunity in the lantibiotic NAI-107 producer strain Microbispora ATCC PTA-5024. Environ. Microbiol. 18, 118–132. doi: 10.1111/1462-2920.12892
Qiao, M., Immonen, T., Koponen, O., and Saris, P. E. (1995). The cellular location and effect on nisin immunity of the NisI protein from Lactococcus lactis N8 expressed in Escherichia coli and L. lactis. FEMS Microbiol. Lett. 131, 75–80. doi: 10.1111/j.1574-6968.1995.tb07757.x
Qiao, M., Ye, S., Koponen, O., Ra, R., Usabiaga, M., Immonen, T., et al. (1996). Regulation of the nisin operons in Lactococcus lactis N8. J. Appl. Bacteriol. 80, 626–634. doi: 10.1111/j.1365-2672.1996.tb03267.x
Ra, R., Beerthuyzen, M. M., de Vos, W. M., Saris, P. E., and Kuipers, O. P. (1999). Effects of gene disruptions in the nisin gene cluster of Lactococcus lactis on nisin production and producer immunity. Microbiology 145, 1227–1233. doi: 10.1099/13500872-145-5-1227
Ra, S. R., Qiao, M., Immonen, T., Pujana, I., and Saris, P. E. (1996). Genes responsible for nisin synthesis, regulation and immunity form a regulon of two operons and are induced by nisin in Lactoccocus lactis N8. Microbiology 142, 1281–1288. doi: 10.1099/13500872-142-5-1281
Reis, M., Eschbach-Bludau, M., Iglesias-Wind, M. I., Kupke, T., and Sahl, H.-G. (1994). Producer immunity towards the lantibiotic Pep5: identification of the immunity gene pepI and localization and functional analysis of its gene product. Appl. Environ. Microbiol. 60, 2876–2883.
Ruhr, E., and Sahl, H.-G. (1985). Mode of action of the peptide antibiotic nisin and influence on the membrane potential of whole cells and on cytoplasmic and artificial membrane vesicles. Antimicrob. Agents Chemother. 27, 841–845. doi: 10.1128/AAC.27.5.841
Siegers, K., and Entian, K. (1995). Genes involved in immunity to the lantibiotic nisin produced by Lactococcus lactis 6F3. Appl. Environ. Microbiol. 61, 1082–1089.
Singh, M., and Sareen, D. (2014). Novel LanT associated lantibiotic clusters identified by genome database mining. PLoS ONE 9:e91352. doi: 10.1371/journal.pone.0091352
Stein, T., Heinzmann, S., Solovieva, I., and Entian, K.-D. (2003). Function of Lactococcus lactis nisin immunity genes nisI and nisFEG after coordinated expression in the surrogate host Bacillus subtilis. J. Biol. Chem. 278, 89–94. doi: 10.1074/jbc.M207237200
Suárez, J. M., Edwards, A. N., and McBride, S. M. (2013). The Clostridium difficile cpr locus is regulated by a noncontiguous two-component system in response to type A and B lantibiotics. J. Bacteriol. 195, 2621–2631. doi: 10.1128/JB.00166-13
Sun, Z., Zhong, J., Liang, X., Liu, J., Chen, X., and Huan, L. (2009). Novel mechanism for nisin resistance via proteolytic degradation of nisin by the nisin resistance protein NSR. Antimicrob. Agents Chemother. 53, 1964–1973. doi: 10.1128/AAC.01382-08
Sutcliffe, I. C., and Russell, R. (1995). Lipoproteins of Gram-positive bacteria. J. Bacteriol. 177:1123.
Tagg, J. R., Dajani, A. S., and Wannamaker, L. W. (1976). Bacteriocins of Gram-positive bacteria. Bacteriol. Rev. 40, 722–756.
Takala, T. M., Koponen, O., Qiao, M., and Saris, P. E. (2004). Lipid-free NisI: interaction with nisin and contribution to nisin immunity via secretion. FEMS Microbiol. Lett. 237, 171–177. doi: 10.1111/j.1574-6968.2004.tb09693.x
Takala, T. M., and Saris, P. E. (2006). C terminus of NisI provides specificity to nisin. Microbiology 152, 3543–3549. doi: 10.1099/mic.0.29083-0
Van de Ven, F., Van den Hooven, H., Konings, R., and Hilbers, C. (1991). “The spatial structure of nisin in aqueous solution,” in Nisin and Novel Lantibiotics, eds G. Jung and H.-G. Sahl (Leiden: Escom Publishers), 35–42.
van Heusden, H. E., de Kruijff, B., and Breukink, E. (2002). Lipid II induces a transmembrane orientation of the pore-forming peptide lantibiotic nisin. Biochemistry 41, 12171–12178. doi: 10.1021/bi026090x
Wiedemann, I., Breukink, E., van Kraaij, C., Kuipers, O. P., Bierbaum, G., de Kruijff, B., et al. (2001). Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J. Biol. Chem. 276, 1772–1779. doi: 10.1074/jbc.M006770200
Willey, J. M., and van der Donk, W. A. (2007). Lantibiotics: peptides of diverse structure and function. Annu. Rev. Microbiol. 61, 477–501. doi: 10.1146/annurev.micro.61.080706.093501
Keywords: lantibiotics, nisin, resistance, immunity, human pathogen, antimicrobial peptides
Citation: Khosa S, Lagedroste M and Smits SHJ (2016) Protein Defense Systems against the Lantibiotic Nisin: Function of the Immunity Protein NisI and the Resistance Protein NSR. Front. Microbiol. 7:504. doi: 10.3389/fmicb.2016.00504
Received: 25 January 2016; Accepted: 29 March 2016;
Published: 12 April 2016.
Edited by:
Yuji Morita, Aichi Gakuin University, JapanReviewed by:
Atte Von Wright, University of Eastern Finland, FinlandOscar P. Kuipers, University of Groningen, Netherlands
Copyright © 2016 Khosa, Lagedroste and Smits. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sander H. J. Smits, c2FuZGVyLnNtaXRzQGhodS5kZQ==
 Sakshi Khosa
Sakshi Khosa Marcel Lagedroste
Marcel Lagedroste Sander H. J. Smits
Sander H. J. Smits