- 1Department of Molecular Genetics and Microbiology, Stony Brook University, Stony Brook, NY, USA
- 2Veterans Administration Medical Center, Northport, NY, USA
Sphingolipids form of a unique and complex group of bioactive lipids in fungi. Structurally, sphingolipids of fungi are quite diverse with unique differences in the sphingoid backbone, amide linked fatty acyl chain and the polar head group. Two of the most studied and conserved sphingolipid classes in fungi are the glucosyl- or galactosyl-ceramides and the phosphorylinositol containing phytoceramides. Comprehensive structural characterization and quantification of these lipids is largely based on advanced analytical mass spectrometry based lipidomic methods. While separation of complex lipid mixtures is achieved through high performance liquid chromatography, the soft – electrospray ionization tandem mass spectrometry allows a high sensitivity and selectivity of detection. Herein, we present an overview of lipid extraction, chromatographic separation and mass spectrometry employed in qualitative and quantitative sphingolipidomics in fungi.
Introduction
As a group, sphingolipids are essential components of all eukaryotic cell membranes (Dickson and Lester, 2002). In fungi, sphingolipids play an important role in a variety of biological processes like cell division (Epstein et al., 2012), heat stress response (Jenkins et al., 1997), acid/alkaline tolerance (Luberto et al., 2001; Rittershaus et al., 2006), hyphae formation (Oura and Kajiwara, 2010), domain formation (Marquês et al., 2015) spore germination (Levery et al., 2002), endocytosis (Zanolari et al., 2000), signal transduction (Obeid et al., 2002), apoptosis (Cheng et al., 2003), pathogenesis and virulence (Luberto et al., 2001; Rittershaus et al., 2006). Endowed with unique chemical structure and synthesized by fungal specific enzymes, these sphingolipids are ideal drug targets (Mor et al., 2015). In this context, it is important to characterize sphingolipids in greater detail.
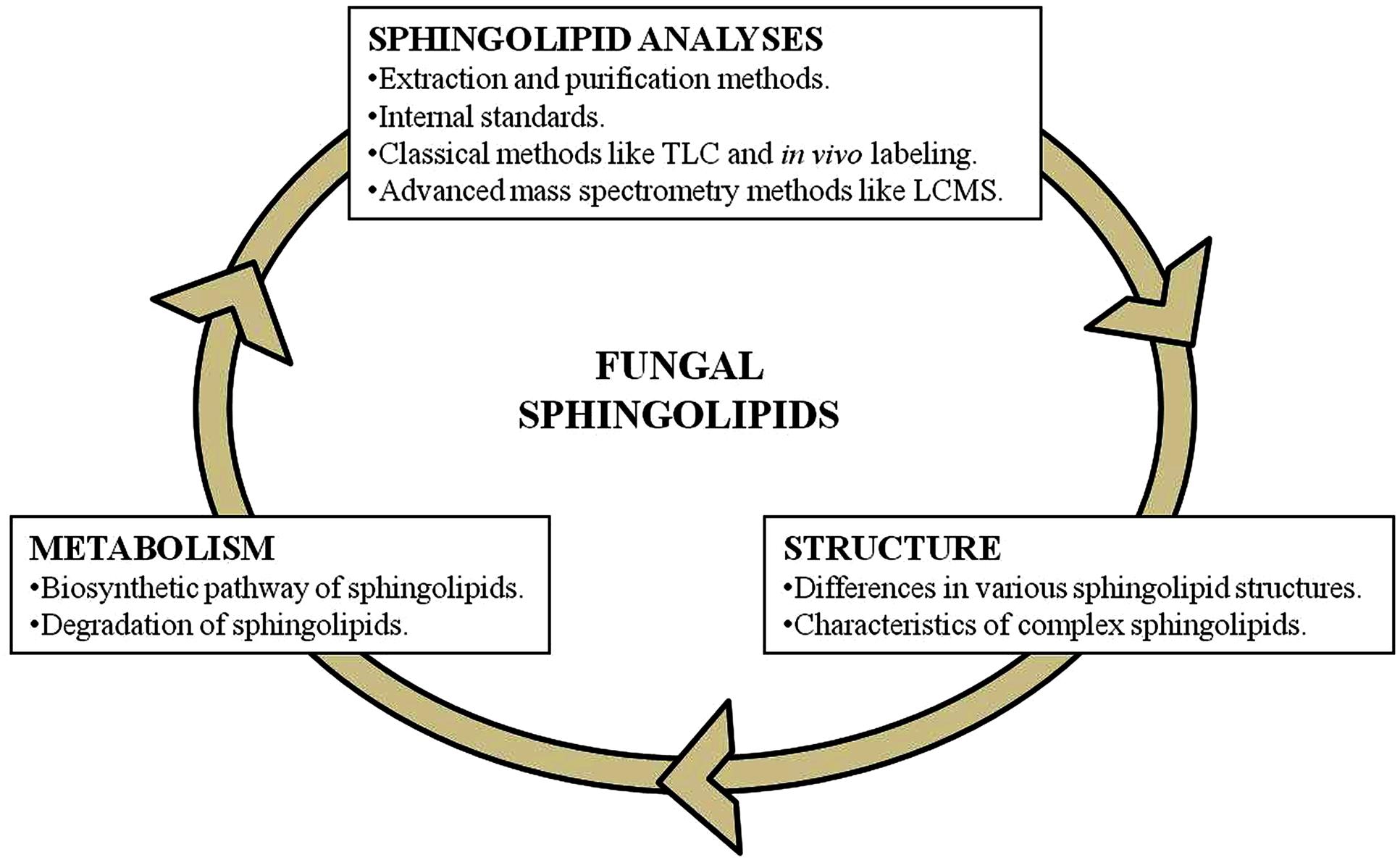
Characterization of various fungal sphingolipids requires the understanding of three key components (Figure 1). These are: the pathways of biosynthesis and degradation; structures of various sphingolipids being synthesized; understanding the ways for efficient extraction of sphingolipids from the cell and the precise methods for their analysis. A tremendous amount of literature in the field of fungal lipid metabolism allows us to categorically understand these components. In the sections below, we have described a literature based review of these key components of sphingolipid characterization, with an emphasis on mass spectrometry based structural and functional characterization of sphingolipids from pathogenic fungi.
In general, the structure of sphingolipids comprise a LCB backbone amide- linked to a fatty acid at C2 position and ester linked to a polar head group at C1 position (Del Poeta et al., 2014). There is a large diversity in sphingoid bases of mammals (Kendall et al., 2015), plants (Luttgeharm et al., 2015), and fungi (Del Poeta et al., 2014). In fungal cells, de novo sphingolipid biosynthesis begins with condensation of L-serine and palmitoyl-CoA to form 3-ketodihydrosphingosine which is then reduced to dihydrosphingosine (d18:0 backbone; sphinganine; Figure 2A; Buede et al., 1991; Nagiec et al., 1994). This step leads to the formation of an 18 C containing LCB. Dihydrosphingosine is then amide linked to a fatty acid (usually α-hydroxylated, 18 or 16 C containing) by ceramide synthases to form dihydroceramide (Lahiri and Futerman, 2005). Further, a Δ4-desaturation of the sphingoid backbone of dihydroceramide forms ceramide (d18:1 backbone; 4-sphingenine; Ternes et al., 2011; Rodriguez-Cuenca et al., 2015). Next, a Δ8-desaturation of the sphingoid backbone of Δ4-ceramide leads to formation of Δ4, Δ8-ceramide (d18:2 backbone; 4,8-sphingadienine; Sperling et al., 2000; Figure 2A). A sphingolipid C9-methyl transferase catalyzes the addition of a methyl group at C9 position of the 4,8-sphingadienine base of Δ4, Δ8-ceramide to form 9-methyl-Δ4, Δ8-ceramide (d19:2 backbone; 9-methyl-4,8-sphingadienine; Ternes et al., 2006; Singh et al., 2012; Figure 2A). Finally, a Gcs1 catalyzes the transfer of glucose moiety from the UDP-glucose onto the C1 hydroxyl group of the ceramide forming glucosylceramide (Rittershaus et al., 2006). Although all the enzymatic steps are not well characterized, some have been studied in fungi like Cryptococcus neoformans (Rittershaus et al., 2006; Singh et al., 2012), Aspergillus nidulans (Li et al., 2006), A. fumigatus (Levery et al., 2002; Kotz et al., 2010), Fusarium graminearum (Rittenour et al., 2011) and Candida albicans (Cheon et al., 2012). Notably, glucosylceramide biosynthesis is absent in Saccharomyces cerevisiae and C. glabrata (Leipelt et al., 2001).
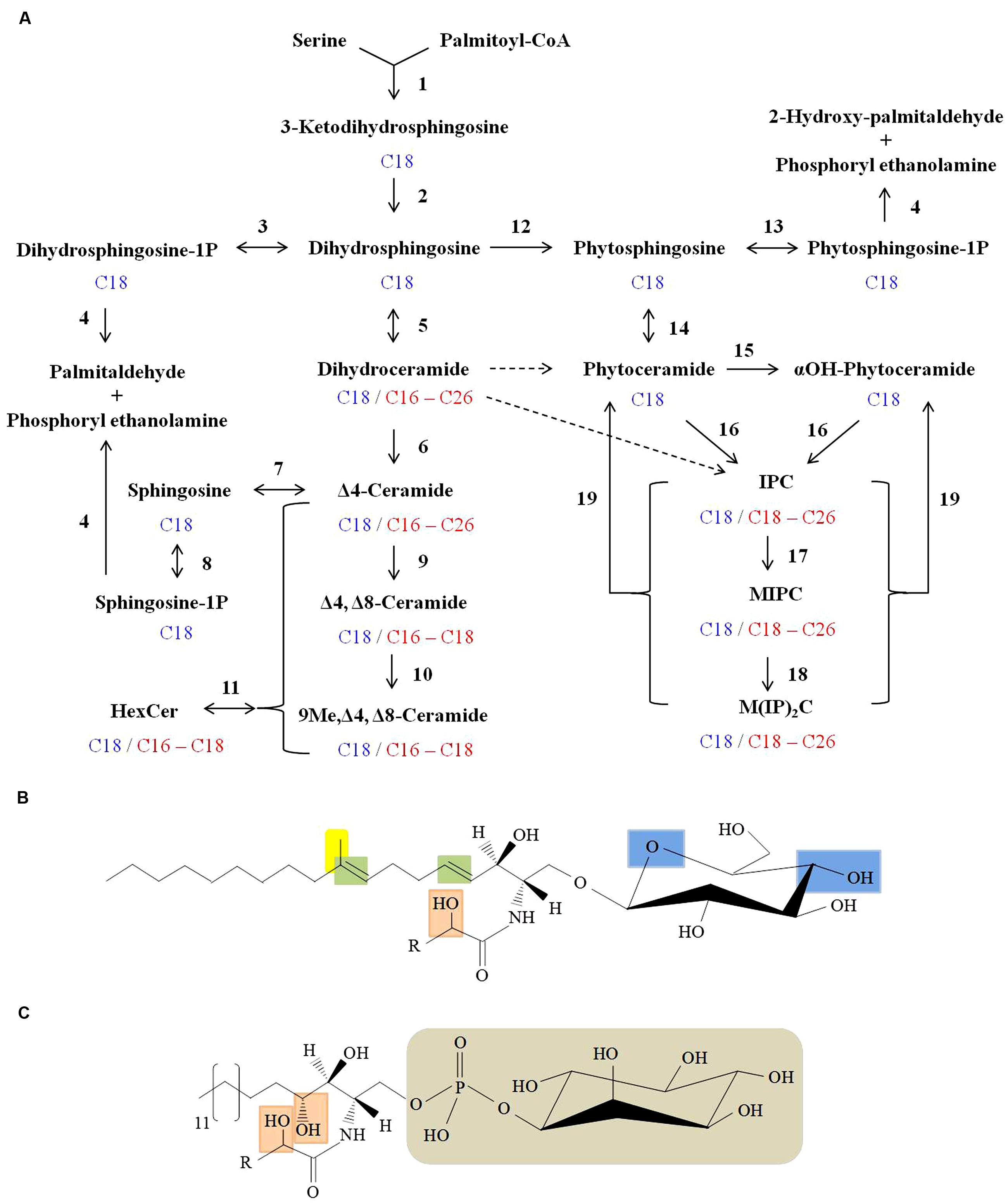
FIGURE 2. Sphingolipid biosynthesis in fungi. (A) Scheme of fungal sphingolipid biosynthetic pathway. Enzymatic steps with reverse reaction shown in brackets are as follows: (1) Serine C-palmitoyl transferase; (2) 3-ketosphinganine reductase; (3) Dihydrosphingosine kinase [Dihydrosphingosine-1P-phosphatase]; (4) Long-chain base (LCB) phosphate lyase; (5) Dihydroceramide synthase; (6) Dihydroceramide C4 desaturase; (7) Ceramidase [Ceramide synthase]; (8) Sphingosine kinase [Sphingosine-1P-phosphatase]; (9) Ceramide C8 desaturase; (10) Sphingolipid C9 methyltransferase (Smt1); (11) Hexosyl ceramide synthase [Hexosyl ceramidase]; (12) Dihydrosphingosine C4-hydroxylase (13) Phytosphingosine kinase [Phytosphingosine -1P-phosphatase]; (14) Ceramide synthase [Ceramidase]; (15) Sphingolipid α-hydroxylase; (16) IPC synthase; (17) IPC mannosyl transferase; (18) Inositolphosphotransferase; (19) Inositol phosphosphingolipid phospholipase C. Major sphingoid backbone in fungi is composed of 18 C-atoms (shown in blue) and the fatty acyls attached to various sphingolipids comprise of 16–26 C-atoms (shown in red). Putative conversion steps of dihydroceramide to phytoceramide and to IPC are represented by dashed arrows. (B) Structure of fungal glucosylceramide. Long chain sphingoid backbone amide linked to a fatty acyl and linked by β-glycosidic bond to a polar head group (glucose) at C1 position. Unique features of fungal glucosylceramide are: Δ4 and Δ8 double bonds in the sphingoid backbone (shown in green), 9-methylation in the sphingoid backbone (shown in yellow), α-hydroxyl fatty acyl (shown in orange) and hydroxyl groups in the hexose moiety (shown in blue). (C) Structure of fungal IPC. The distinguished features of IPC’s in fungi are: C3-hydroxylation of sphingoid backbone and α-hydroxyl fatty acyl (both shown in orange), phosphate linked inositol group (shown in grey). ‘R’ represents 16–24 carbons in the fatty acyl chain.
In plants, 8-sphingenine (d18:1) and 4,8-sphingenine (d18:2) represent the major sphingoid bases, however, presence of the cis- and trans- isomeric forms result in nine different C18 sphingoid bases (Ohnishi et al., 1983). This results in a complex mixture of glucosylceramide pool in plants (Imai et al., 2012). In mammals, the major sphingoid base is 4-sphingenine (d18:1) which is linked to 16 C fatty acid in the glucosylceramide structure (Leipelt et al., 2001). In contrast, the fungal glucosylceramide structure is rather unique. The sphingoid backbone is composed of 9-methyl-4,8-sphingadienine which is amide linked to α-hydroxylated C18:0 fatty acid (Figure 2B; Del Poeta et al., 2014). Certain fungal species contain galactose instead of glucose in this cerebroside structure (Warnecke and Heinz, 2003). The α-hydroxylated C16:0 and α-hydroxylated C18:1 are the two other major fatty acyls reported in the fungal cerebroside structure (Barreto-Bergter et al., 2004). The structures of different cerebrosides produced by fungi have been extensively reviewed by Barreto-Bergter et al. (2011).
Fungi also produce phytoceramide (Heung et al., 2006). Dihydrosphingosine C4-hydroxylase catalyzes the addition of a hydroxyl group at C4 position of dihydrosphingosine backbone to produce phytosphingosine (t18:0 backbone; 4-hydroxysphinganine; Li et al., 2007; Figure 2A). Ceramide synthases then transfer a non-hydroxylated fatty acid (18, 24 or 26 C containing) to amide group at C2 position to form phytoceramide (Cheon et al., 2012). The fatty acyl moiety of the phytoceramides is α-hydroxylated by a sphingolipid α-hydroxylase to form αOH-phytoceramides (Haak et al., 1997). Both phytoceramides and αOH-phytoceramides are further converted to complex phosphosphingolipids such as IPC (transfer of phosphorylinositol group to phytoceramide), MIPC, (transfer of mannosyl group to IPC) and M(IP)2C, (transfer of a second phosphorylinositol group to MIPC; Guan and Wenk, 2008). Although IPC derivatives are not reported in mammals, certain plants and kinetoplastid protozoa do produce IPC derivatives (Heung et al., 2006).
The t18:0 phytosphingosine backbone, C2 hygroxylated fatty acyls and phosphorylinositol containing polar head group are the unique feature of IPC derivatives (Figure 2C). IPC structures and pathway of synthesis have been characterized in several fungi like S. cereviseae (Ejsing et al., 2009; Voynova et al., 2014), C. neoformans (Luberto et al., 2001), A. fumigatus (Kuroda et al., 1999; Heidler and Radding, 2000), A. nidulans (Bennion et al., 2003), Histoplasma capsulatum (Guimarães et al., 2014), and C. albicans (Singh et al., 2010; Cheon et al., 2012).
The re-cycling of fungal sphingolipids involves the production of sphingosine, sphingosine-1-phosphate, dihydrosphingosine-1-phosphate, and phytosphingosine-1-phosphate (Figure 2A). The ultimate degradation steps of sphingolipids are constituted by the catabolism of dihydrosphingosine-1-phosphate and sphingosine-1-phosphate into palmitaldehyde and phosphoryl ethanolamine (Kim et al., 2000; Serra and Saba, 2010) and the phytosphingosine-1-phosphate into 2-hydroxy-palmitaldehyde and phosphoryl ethanolamine (Kondo et al., 2014), by the long-chain basse-1-phosphate lyases (Figure 2A).
The ability of fungal cells to produce sphingolipids with different sphingoid backbone structures adds to the complexity of the lipid mixtures (Guimarães et al., 2014). In addition to the backbone structure, the degree of unsaturation, fatty acyl chain lengths, methylation and hydroxylation modifications, all make it quite difficult to analyze these lipids using classical techniques like in vivo labeling (Chigorno et al., 2006), TLC (Urban et al., 1980) and GC (Cacas et al., 2012). Therefore, advanced analytical methods for the analyses of these lipids are employed to characterize their structures.
In last two decades, several mass spectrometry techniques have been used to identify and characterize the total sphingolipid pool or the “sphingolipidome.” A wide variety of mass spectrometry based methods are available in literature that allows accurate analyses of these complex sphingolipid mixtures (Merrill et al., 2005, 2009; Haynes et al., 2009; Wenk, 2010; Köfeler et al., 2012). Below we describe methods of extraction, chromatographic separation and the mass spectrometry based strategies to analyze sphingolipids.
Lipid Extraction
Extraction of lipids is the most crucial step for lipid analysis by both classical and high throughput techniques. Currently several modified adaptations of Folch method (Folch et al., 1957) and Bligh and Dyer method (Bligh and Dyer, 1959) are being employed to extract lipids from fungal cells (Prasad and Ghannoum, 1996; Schneiter and Daum, 2006; Ejsing et al., 2009; Haynes et al., 2009). In our laboratory and others, we use the method described by Mandala et al. (1995) for lipid extraction. This method has shown a good extraction efficiency and reproducibility for lipid analyses. The scheme of lipid extraction from fungal cells is shown in Figure 3. Fungal cells are extracted in ethanol: dH2O:diethylether:pyridine:NH4OH (15:15:5:1:0.018; v/v) at 60°C for 1 h as described previously (Hanson and Lester, 1980). Lipid extract is then subjected to a solution of methanol:chloroform (2:1; v/v) followed by addition of 1/3rd volume chloroform and 1/3rd volume dH2O and the lower organic phase reserved (Bligh and Dyer, 1959). Organic phase is dried in SpeedVac, flushed with N2 and stored in -20°C. At this stage, these lipid extracts can be used for the estimation of inorganic phosphate (Pi) content, dry lipid weight or for the isolation and purification specific lipid classes (Merrill et al., 2009; Rana et al., 2015). Both Pi content and dry lipid weight have been used to normalize the lipid amounts. Several groups have employed SPE techniques to further purify specific sphingolipid groups (Bodennec et al., 2000; Barreto-Bergter et al., 2004). Extensive lipid purification by SPE is cumbersome and time consuming, and is usually not required for routine lipid analysis; however, is preferred for a thorough structural characterization of complex glycosphingolipids (Barreto-Bergter et al., 2004, 2011).
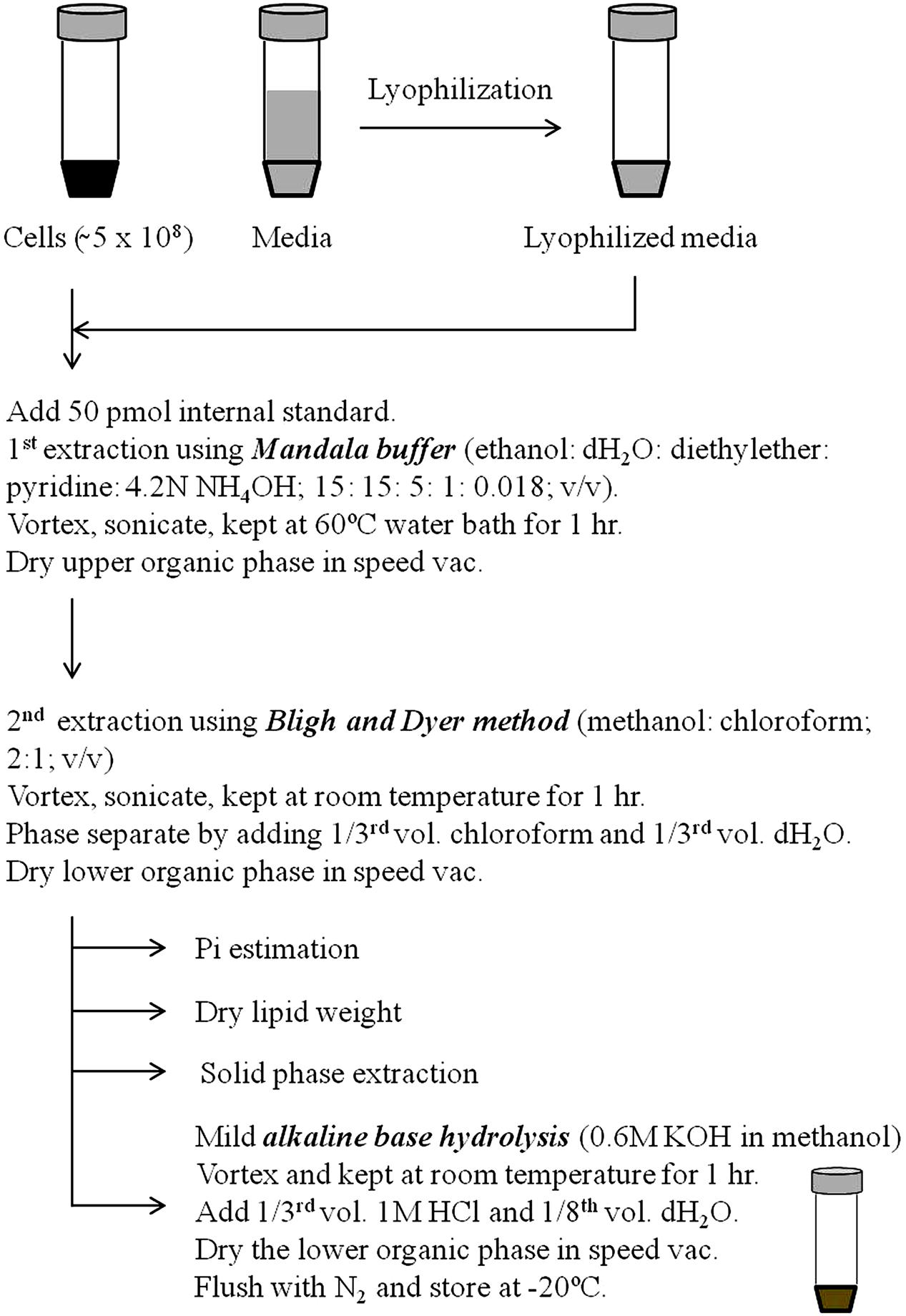
FIGURE 3. Procedure of sphingolipid extraction. Prior to extraction, fungal cell extracts or lyophilized media are spiked with suitable internal standards. First extraction is performed using Mandala extraction buffer (ethanol: dH2O: diethyl ether: pyridine: 4.2N NH4OH; 15: 15: 5: 1: 0.018; v/v). Subsequently, a second Bligh and Dyer extraction is performed using methanol and chloroform. Samples at this stage can be used to determine the Pi content, lipid dry weight or to purify specific sphingolipid components using solid phase extraction. Finally, samples are base hydrolyzed using mild alkaline base hydrolysis (0.6 M KOH in methanol) to remove the glycerol backbone containing lipid contaminants. For certain fungal masses it is required to first crush the sample using glass beads in the lipid extraction buffer itself or to make the sample into powder using pestle and mortar in liquid N2 prior to extraction.
Glycerolipids are the major lipid contaminants that are co-extracted with sphingolipids (Bligh and Dyer, 1959). A mild alkaline methanolysis (Clarke and Dawson, 1981), usually for 60 min at room temperature is sufficient to hydrolyze the ester linkages of fatty acyls of glycerolipids. This allows a clean extraction of alkaline-stable components, which are highly enriched in sphingolipids.
For accurate quantification of lipids by mass spectrometry, internal standards must be added prior to lipid extraction (Rana et al., 2015). Although, absolute quantification of each lipid species by mass spectrometry requires the use of isotope-labeled standard for that species (Ecker and Liebisch, 2014); however, presently this is not possible due to high synthesis costs and the large number of lipids being analyzed. Addition of one internal standard per lipid class being analyzed is widely accepted (LIPID MAPS Consortium). This is primarily because the ionization of lipid species is largely dependent upon the specific head group rather than the attached fatty acyls (Köfeler et al., 2012). The C17-sphingoid backbone lipids (Avanti Polar Lipids Inc., Alabaster, AL, USA; Matreya Inc., Pleasant Gap, PA, USA) are routinely used as internal standards as these closely resemble C18 lipids in physicochemical properties and ionization efficiencies (Rana et al., 2015). Quantification of sphingolipid species with different chain lengths can be achieved using the calibration curves of closest chain length standards (Rana et al., 2015).
Methods of Lipid Analysis
In Vivo Labeling, Thin-layer Chromatography and Gas Chromatography Mass Spectrometry
For several decades, in vivo labeling has been used to characterize the sphingolipid metabolic pathways in fungi (Chigorno et al., 2006). The technique involves the uptake of a radiolabeled precursor by cells. The radiolabeled precursor then gets incorporated into the complex lipid structures. Heavy labeled [14C]-palmitate and [14C]-serine are the most commonly used labeled precursors to follow sphingolipid synthesis (Chigorno et al., 2006). However, these labels get incorporated into other untargeted lipids and give high background signals. [3H]-dihydrosphingosine provides a more sphingolipid specific labeling (Karashima et al., 2013). [3H]-inositol has been used to focus on IPC derivatives (Haak et al., 1997) and [3H]- or [14C]- glucose or galactose have been used to focus on glycosphingolipids (Sasaki, 1981). Unfortunately, the very long half-life of these radiolabeled precursors presents a serious risk to health if exposed and is a challenge during disposal (Ecker and Liebisch, 2014).
Lipid extracts from labeled or unlabeled cells can be resolved using the TLC technique (Figure 4A). It is a simple, cost effective, fast and sensitive method to qualitatively characterize lipids. Identification of broad lipid classes by TLC is simple and achieved by comparing their mobility to the standard (Urban et al., 1980). The most commonly used solvent systems to resolve sphingolipids on a TLC include: chloroform:methanol:dH2O (65:25:4; v/v), chloroform:methanol:4.2N NH4OH (9:7:2 and 40:10:1; v/v) and chloroform:methanol:acetic acid (90:2:8; v/v Mandala et al., 1995; Bodennec et al., 2000; Barreto-Bergter et al., 2004). Unlabeled sphingolipids are visualized using iodine (Palumbo and Zullo, 1987) or orcinol/resorcinol staining (Tadano and Ishizuka, 1982) and the labeled sphingolipid are visualized by autoradiography on an X-ray film (Kondo et al., 2014).
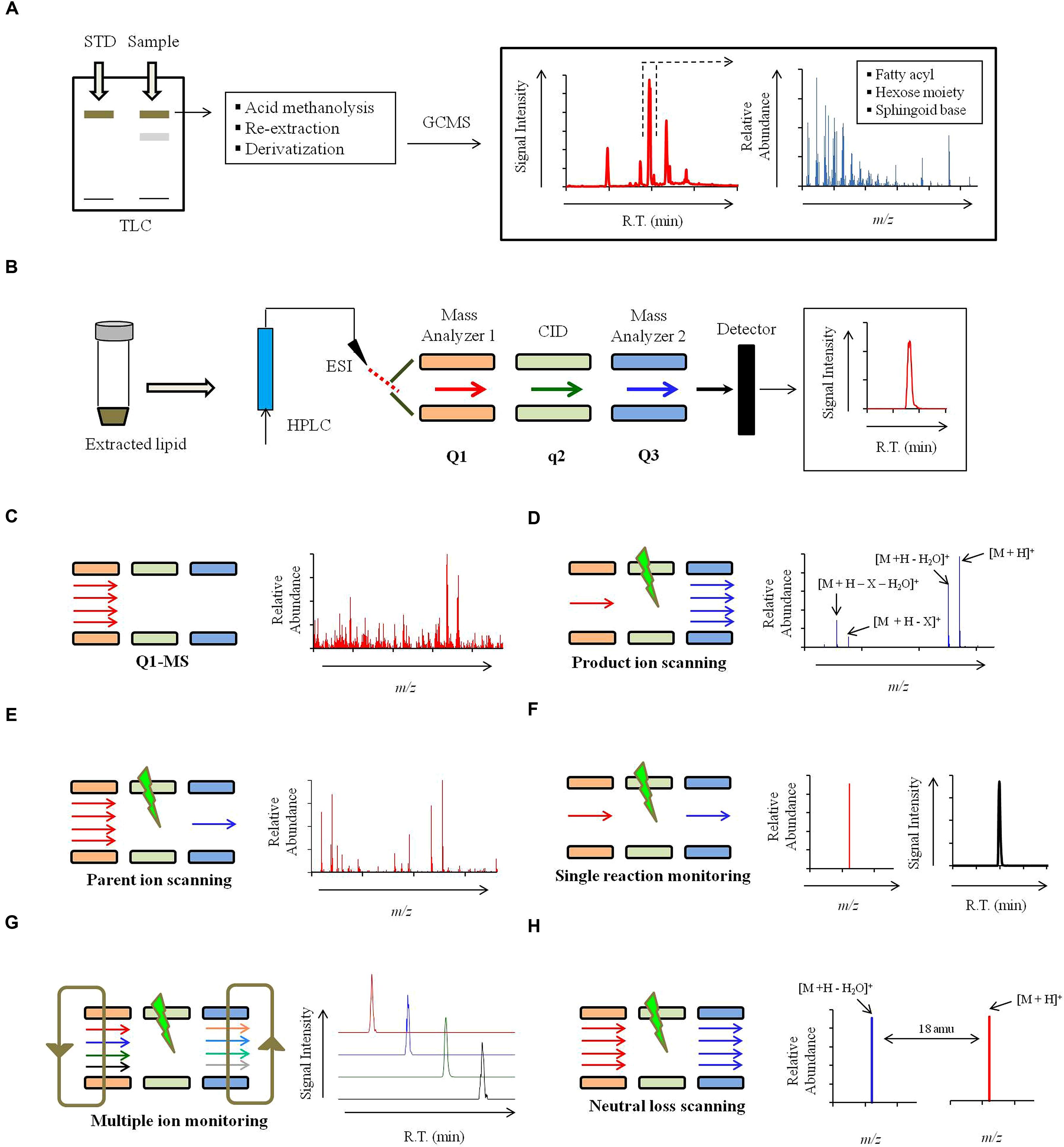
FIGURE 4. Strategy for mass spectrometry based identification of sphinglipids. (A) Alternate strategies to confirm the sphingolipid structure. Qualitative analysis of sphingolipids can be performed using commercially available standards by High performance thin-layer chromatography (HP-TLC) or TLC. Sphingolipid components like sphingoid backbone, sugar and fatty can be analyzed using Gas chromatography mass spectrometry (GCMS). For sphingoid base and sugar analysis, samples are prepared using following steps: acid methanolysis (1 N HCl in methanol), N-acetylation (pyridine and acetic anhydride) and per-O-trimethylsilylation [TriSil reagent; 1-(Trimethylsilyl)imidazole – Pyridine mixture]. Sphingoid bases and sugars are analyzed as N-acetyl-per-O-trimethylsilyl derivatives and monosaccharide methyl glycosides, respectively. For fatty acyl analysis, samples are prepared using acid methanolysis, re-extraction in n-hexane and per-O-trimethylsilylated (TriSil) to form fatty acid methyl esters (FAMEs). (B) A scheme of HPLC-ESI-MS/MS. Lipids are resolved by HPLC and thereafter analyzed by tandem mass spectrometry (MS/MS). ‘Q1’ and ‘Q3’ quadruples are mass analyzers and the quadrupole ‘q’ operates in radio frequency leading to collision-induced dissociation (CID). Electrospray ionization (ESI) or Atmospheric pressure chemical ionization (APCI, not shown) is used as ionization methods. (C) Scheme for Q1-MS or full scan. The molecular ions or m/z ratio are recorded in Q1. (D) Scheme for product ion scanning. Only a select ion with specific m/z is recorded in Q1 and undergoes CID. Resultant ions are recorded in Q3. ‘M’ represents the molecular ion; ‘X’ represents a fragment loss. (E) Scheme for parent ion scanning. A full scan of ions recorded in Q1 and following CID, only a select fragment ion is recorded in Q3. (F) Scheme for single-reaction monitoring (SRM). A preselected ion is recorded in Q1, a select parent ion undergoes CID and a select fragment ion is recorded in Q3. (G) Scheme for multiple-reaction monitoring (MRM). Multiple SRM’s can be set up simultaneously depending upon the scanning efficiency of the instrument. (H) Scheme for neutral loss (NL) scanning. Ions are recorded in both Q1 and Q3 with a set mass difference between the two.
Structural composition of the purified sphingolipids can be determined using GC-EI-MS (Cacas et al., 2012). Only the volatile components that can be carried by the carrier gas (He) can be analyzed by GCMS. Most sphingolipid structures are non-volatile. For this reason, sphingolipids are hydrolyzed to release the fatty acid, sugar, and LCB components, which are then derivatized into methyl esters or TMS derivatives and analyzed by GC-EI-MS (Figure 4A; Christie, 1989; Matsubara and Hayashi, 1991; Johnson and Brown, 1992; Merkle and Poppe, 1994). GC-EI-MS is not a preferred technique to analyze sphingolipids because fragmentation obtained in EI-MS is not very consistent as high energy electrons are used for fragmentation, strong ionization may completely destroy the molecular ion and extensive derivatization may generate complicated spectral patterns with poor resolution (Cacas et al., 2012).
High-performance Liquid Chromatography (HPLC)
Various sphingolipid classes can readily be separated by HPLC on both the normal phase C18 and reverse phase C8 columns. Normal phase HPLC utilizes the head group properties to obtain separation (for example, ceramides and hexosylceramides; Merrill et al., 2005). However, the reverse phase HPLC using C8 column is capable of resolving sphingolipid classes based on their hydrophobicity of carbon backbone and degree of unsaturation (for example, sphingosine and dihydrosphingosine; Rana et al., 2015). During most of the routine sphingolipid analysis the reverse phase HPLC when coupled with mass spectrometry is a powerful tool for simultaneous separation and detection of sphingolipid species. The main goal of HPLC is to chromatographically separate the sphingolipid species that cannot be resolved based on m/z (mass-to-charge) by the mass spectrometers, like isomeric and isobaric species, and to improve the sensitivity of detection. Several different buffer systems have been described as mobile phases (Merrill et al., 2005). The binary buffer system most frequently used as the mobile phase in sphingolipid analysis is: 2 mM ammonium formate + 0.2% formic acid in H2O (Buffer 1) and 1 mM ammonium formate + 0.2% formic acid in methanol (Buffer 2). A gradient elution of analytes using buffers 1 and 2 allows complete separation of sphingolipid species. Additional separation on HPLC may be achieved by lowering the flow rate of the mobile phase and changing the gradient conditions (Rana et al., 2015).
Fungal lipid extracts may contain a mixture of glucosyl- and galactosyl- ceramides (Barreto-Bergter et al., 2004). Due to a remarkable similarity in their backbone structures these lipid species are difficult to separate. These lipid species can be chromatographically separated using HILIC on silica based columns using isocratic elution (Zama et al., 2009). The elution buffer contains acetonitrile:methanol:acetic acid (97:2:1; v/v) + 5 mM ammonium acetate (Merrill et al., 2005).
Advanced Mass Spectrometry
A wide variety of mass spectrometry platforms are now available that can be used to analyze the molecular composition of complex lipid mixtures (Köfeler et al., 2012). Mass spectrometry is a structure-based analysis of biological molecules (Wenk, 2010), and has been extensively used to characterize various sphingolipid species (Sugawara et al., 2010; Brügger, 2014; Li et al., 2014; Ogiso et al., 2014).
Several different ionization methods have been used in sphingolipid analysis, for example, the ESI (Han and Gross, 1994), MALDI (Jones et al., 2014), and FAB (Ahn et al., 2009). These ionization techniques are coupled with mass analyzers like MS/MS (Shaner et al., 2009), MSn (Ito et al., 2013), TOF (Jia et al., 2015), FT-ICR (Guo et al., 2012), and QqTOF MS (Ejsing et al., 2006). The advantages and disadvantages of these techniques have been described in detail previously (Köfeler et al., 2012).
For sphingolipidomic purposes, ESI-MS/MS approach is most extensively employed. Coupled with reverse phase HPLC, ESI-MS/MS provides a simple, sensitive, and structure specific quantification of sphingolipids. Although the setup of other lipidomic platforms may be different but the basic concepts of mass spectrometry remain the same.
The primary requisition of any analytes to be analyzed by mass spectrometry depends on the fact that whether they can be readily ionized in gaseous phase without in source fragmentation (Han and Gross, 1994). ESI is a soft-ionization technique that allows the formation of intact positively or negatively charged ions that are then transmitted to the mass analyzers. In a typical ESI source, analytes in solvents are infused into the ion source via a narrow capillary needle. A high voltage, positive or negative depending upon the lipids being targeted, is applied between the tip of the needle and the inlet of mass analyzer. This results in the formation of a charged droplet which gets desolvated under high voltage, vacuum and a drying N2 gas, leading to the production of charged molecular ions in gaseous phase (Brügger, 2014). Sphingolipids are readily ionized in ESI source to form [M + H]+ molecular ions for long chain sphingoid bases, ceramides, and glycosphingolipids, or to from [M-H]- molecular ions for IPC derivatives (Shaner et al., 2009).
A scheme for HPLC-ESI-MS/MS is summarized in Figure 4B. Different scanning approaches have developed around this triple quadrupole set up (Han and Gross, 1994; Merrill et al., 2005; Bielawski et al., 2006; Guan and Wenk, 2006; Brügger, 2014; Rana et al., 2015), and these are:
1) Full scan (Figure 4C): In a full scan, the m/z of molecular ions generated in ESI are recorded in the first quadrupole or the first mass analyzer. Full scan approach is widely used in non-targeted lipidomics. However, no structural information can be deduced from these scanning.
2) Product ion scanning (Figure 4D): In this mode, the first mass analyzer allows a molecular ion of specific m/z to pass to the collision cell. Based on the applied CE the molecular ion is fragmented into product ions using an inert collision gas (N2 or Ar) via RF; this process is referred as CID. The m/z of these fragments is recorded in the third quadrupole or the second mass analyzer. For sphingolipids, the product ion analysis provides valuable structural information about the sphingoid backbone, polar head groups and the fatty acyl attached. For example, in positive ion mode, at high CE, glucosylceramide is fragmented into a characteristic ion representing the loss of double dehydration product of sphingoid backbone (Figure 4D, Table 1). Similar ions characteristic to specific sphingolipid structures are identified and further used for lipid detection. Optimization of CE is important to obtain correct fragmentation of the molecular ions into desired daughter ions. CE required for achieving CID of different sphingolipid molecular species may vary depending upon hydroxylations, unsaturations, and carbon chain length.
3) Parent- or precursor-ion scanning (PREIS; Figure 4E): Here, the mass analyzer records molecular ions based on their m/z, these ions undergo CID, and only a select daughter ion is passed into the third quadrupole. This mode is very useful in the analysis of sphingolipid species which posses a common backbone or headgroup fragment.
4) Single-reaction monitoring (SRM; Figure 4F): Only a select parent ion is recorded in the first quadrupole and undergoes CID and m/z of a select daughter ion is recorded in the third quadrupole. SRM’s are highly selective and very sensitive methods.
5) Multiple-reaction monitoring (MRM; Figure 4G): Several SRM reactions can be simultaneously recorded by the mass spectrometer, however, this largely depends upon the scanning speed of the instrument. MRM scanning represents a targeted lipidomics approach and is quite successfully used for the analysis of sphingolipids.
6) Neutral loss (NL) scanning (Figure 4H): Both first and third quadrupole records the m/z, with a constant mass offset between them. For example, a NL of 18 amu is set, which represents a loss of water molecule from the molecular ion and is characteristic of sphingolipid structures (Rana et al., 2015).
A large amount of literature containing the fragmentation pattern of different sphingolipid classes is now available. Most of these fragmentation data (product ions) were acquired using direct infusion ESI-MS/MS approaches. This, however, requires extensive offline purification of sphingolipid of interest, usually using SPE (Barreto-Bergter et al., 2004). Fortunately a large amount of literature containing the fragmentation patterns of various fungal sphingolipids is available and sphingolipid class-specific fragment and precursor ions are known for most classes (Guan and Wenk, 2006). In targeted lipidomics, the unique precursor and parent ions are selected for each sphingolipid species and a MRM based method is employed for analyses (Bielawski et al., 2006).
Analysis of Fungal Sphingolipids
By using a precursor m/z and product m/z ion pairs of molecular species of interest, an accurate detection and quantification can be done. Although the complete sphingolipidome for most fungi is yet to be determined, many sphingolipid specific backbones and polar head groups have been characterized (summarized in Table 1). Targeted methods using PREIS and MRM scanning methods have been developed using these fragments to trace the sphingolipid metabolism of many fungi (Bielawski et al., 2006; Guan and Wenk, 2006; Ejsing et al., 2009; Sugawara et al., 2010; Brügger, 2014; Li et al., 2014; Ogiso et al., 2014).
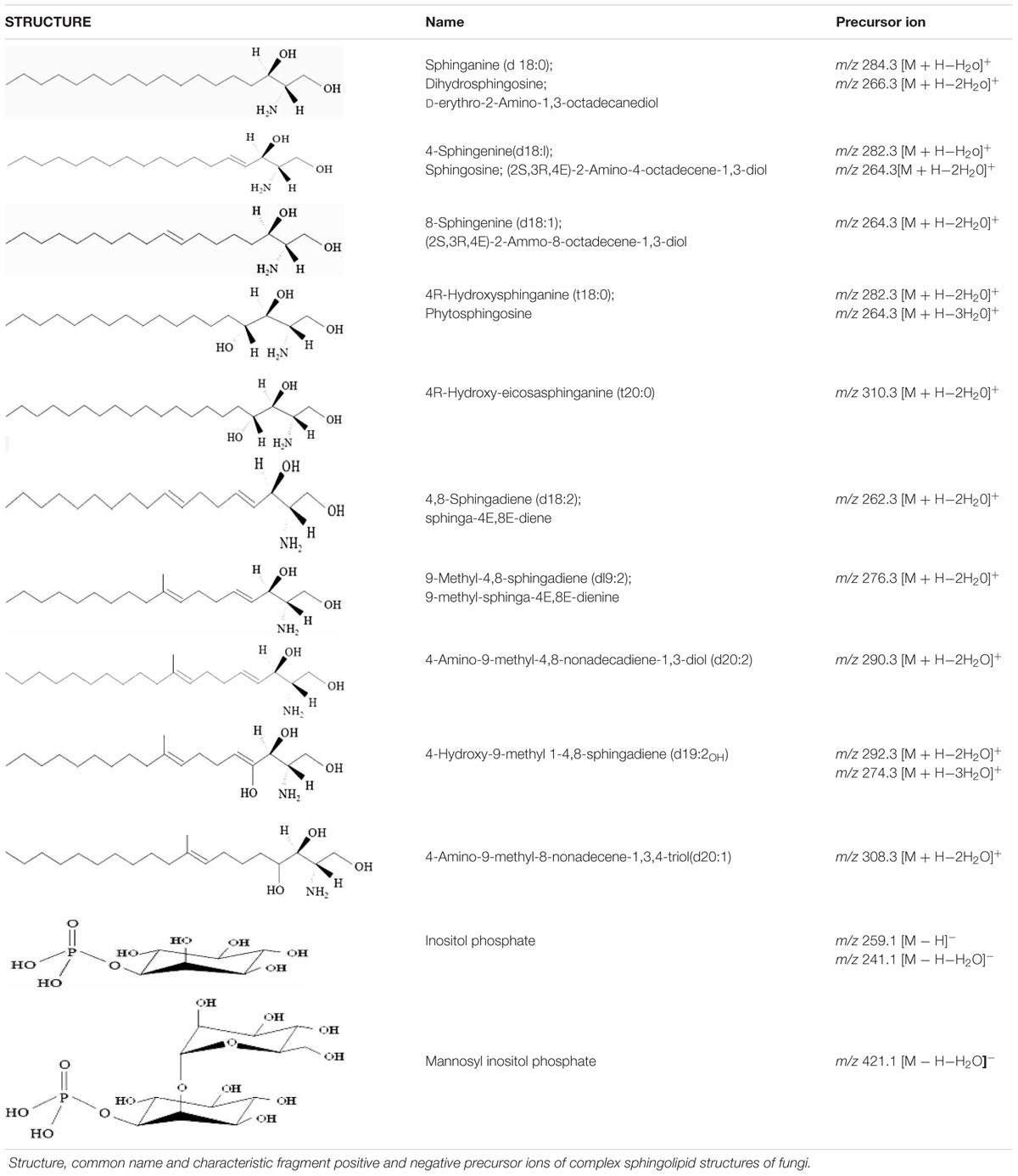
For example, dihydrosphingosine (d18:0) and sphingosine (d18:1) can be identified using precursor ions of m/z 302 and 300, and product ions of m/z 266 and 264, respectively, in the positive ion mode. Ions of m/z 266 and 264 are a result of loss of two water molecules from the sphingoid backbone. Loss of one water molecule from the backbone results ions of m/z 284 and 282. However, the lipid extracts may contain a mixture of d18:1 backbone with a difference in the position of double bonds, like 4-Sphingenine (major species) and 8-Sphingenine. In this situation, a prior chromatographic separation of these species is important and can be achieved by optimizing the mobile phase. The d18:0 backbone fragment ion m/z 266 is used to analyze dihydroceramide species while the d18:1 backbone fragment ion m/z 264 is used to analyze ceramide species. The phytosphingosine backbone (t18:0) is abundant in fungal lipid extract, and is constituent of phytoceramides and IPC derivatives. A precursor ion m/z 282 is characteristic to t18:0 backbone and results from the loss of two water molecules. Similarly, a rather less abundant t20:0 (4R-Hydroxy-eicosasphinganine) backbone is also detectable in phytoceramide and IPC structures using the positive ion precursor m/z 310. The presence of phosphorylinositol group/s in IPC, MIPC, and M(IP)2C structure often results in poor ionization efficiencies in positive ion mode. However, these lipids are readily ionized using negative ion mode and can be detected as [M–H]-. The precursor ion with m/z 241 which represents a dehydration product of the ion m/z 259 (the phosphorylinositol group) and can be used to analyze IPC and M(IP)2C species. Similarly, the precursor ion m/z 421 is used to analyze MIPC species (Table 1; Guan and Wenk, 2006; Ejsing et al., 2009; Sugawara et al., 2010).
The mass spectrometric analyses of purified lipids, especially cerebrosides, from different fungal sources have revealed several other complex backbone structures (Barreto-Bergter et al., 2011). Among these 9-methyl-4,8-sphingadiene (d19:2) is predominant in most fungal species like Cryptococcus, Aspergillus, Candida, and several others (Del Poeta et al., 2014). The d19:2 backbone has a characteristic fragment of m/z 276 that results from the loss of two water molecules. The corresponding hexosylceramide can be identified in positive ion mode using [M + H]+ as the parent ion. The [M + H]+ ions 756, 754, and 728 represent three most abundant hexosylceramides in fungi. The 2-hydroxy fatty acyl group in these structures is C18:0, C18:1, and C16:0. It is important to mention here that often the ions for these lipid species present themselves as Na+ or K+ adducts in positive ion or Cl- adduct in the negative ion mode. Analysis of such samples requires optimization of the CE and accounting for the altered mass during the analysis. Interestingly, the differential fragmentation pattern of Cl- adduct of hexosylceramide in negative ESI-MS/MS mode can be used to identify the nature of the hexose moiety (glucose or galactose). This achievable by monitoring the peak intensity ratios of two characteristic product ions: m/z 179 and 89. The ion intensity patterns of 179 < 89 and 179 > 89 represent galactose and glucose moiety in the hexosylceramide structure, respectively (Han and Cheng, 2005).
The 4,8-Sphingadiene (d18:2) backbone containing sphingolipid species are also common in fungi. These can be analyzed using a precursor ion m/z 262. Although, the cis- or trans- isomers of d19:2 and d18:2 are not reported in fungi, however, an HPLC-ESI-MS/MS based method for analysis of these was recently described using plant lipid extracts (Imai et al., 2012). Several unique sphingoid backbones in cerebroside structures have also been identified in fungi like 4-Hydroxy-9-methyl-4,8-sphingadiene (d19:2OH) in Scedosporium apiospermum and Pseudallescheria boydii (Pinto et al., 2002; Rollin-Pinheiro et al., 2014), 4-Amino-9-methyl-8-nonadecene-1,3,4-triol (d20:1) and 4-Amino-9-methyl-4,8-nonadecadiene-1,3-diol (d20:2) in Cladosporium resinae (Barreto-Bergter et al., 2011).
Our ability to accurately quantify different sphingolipid structures has been efficiently used to establish the sphingolipid biosynthetic pathways in fungi. Considering that differences in the biosynthesis of various sphingolipid structures can have drastic physiological affect on fungi, it becomes necessary to understand the enzymatic steps involved therein. This has been successfully achieved for several fungal species by comparing the sphingolipid profiles of the wild type and various sphingolipid pathway mutants (Kuroda et al., 1999; Heidler and Radding, 2000; Luberto et al., 2001; Bennion et al., 2003; Ejsing et al., 2009; Cheon et al., 2012; Guimarães et al., 2014; Voynova et al., 2014). In a similar scenario, our lab worked out the glucosylceramide biosynthetic pathway of C. neofomans (shown in Figure 5A; Rittershaus et al., 2006; Singh et al., 2012). Lipid analysis of wild type C. neofomans strain (CnWT) showed that 1′-β-D-glucosyl-2′-N-hydroxyoctadecanoyl-9-methyl-4,8-sphingadiene (or Δ4–Δ8,9Me-GlcCer, detected using the SRM transition of m/z 756 to 276; loss of double dehydrated sphingoid base) was the most abundant cerebroside structure (Figure 5B, panel 1). The corresponding ceramide species 2′-N-hydroxyoctadecanoyl-9-methyl-4,8-sphingadiene (or Δ4–Δ8,9Me-Cer, detected using the SRM transition of m/z 594 to 576; water loss) was also detectable. The deletion of SMT1 (CnΔsmt1) abolishes the Smt1 activity, leading to an accumulation of un-methylated ceramide and glucosylceramide structures (Figure 5B, panel 2). These are 2′-N-hydroxyoctadecanoyl-4,8-sphingadiene (or Δ4–Δ8-Cer, detected using the SRM transition of m/z 580 to 562; water loss) and 1′-β-D-glucosyl-2′-N-hydroxyoctadecanoyl-4,8-sphingadiene (or Δ4–Δ8-GlcCer, detected using the SRM transition of m/z 742 to 262; loss of double dehydrated sphingoid base), respectively. The Δ4–Δ8,9Me-Cer and Δ4–Δ8,9Me-GlcCer structures are absent in CnΔsmt1 mutant. These data confirmed that methylation of ceramide by Smt1 is a pre-requisite to the formation of methylated glucosylceramide. A deletion of glucosylceramide synthase GCS1 (CnΔsmt1) leads to an accumulation of Δ4–Δ8,9Me-Cer; however, all glucosylceramide structures are completely absent, suggesting that Gcs1 is the key Gcs1 in C. neofomans. A similar approach can also be used to study the function of Sld8, (uncharacterized in C. neoformans), where 2′-N-hydroxyoctadecanoyl-4-sphingenine (Δ4-Cer) can be detected using the SRM transition of m/z 582 to 564 (water loss) and 1′-β-D-glucosyl-2′-N-hydroxyoctadecanoyl-4-sphingenine (Δ4- GlcCer) can be detected using the SRM transition of m/z 744 to 264 (loss of double dehydrated sphingoid base). Thus, combined with genetic approaches, mass spectrometry becomes extremely useful in understanding the lipid metabolism.
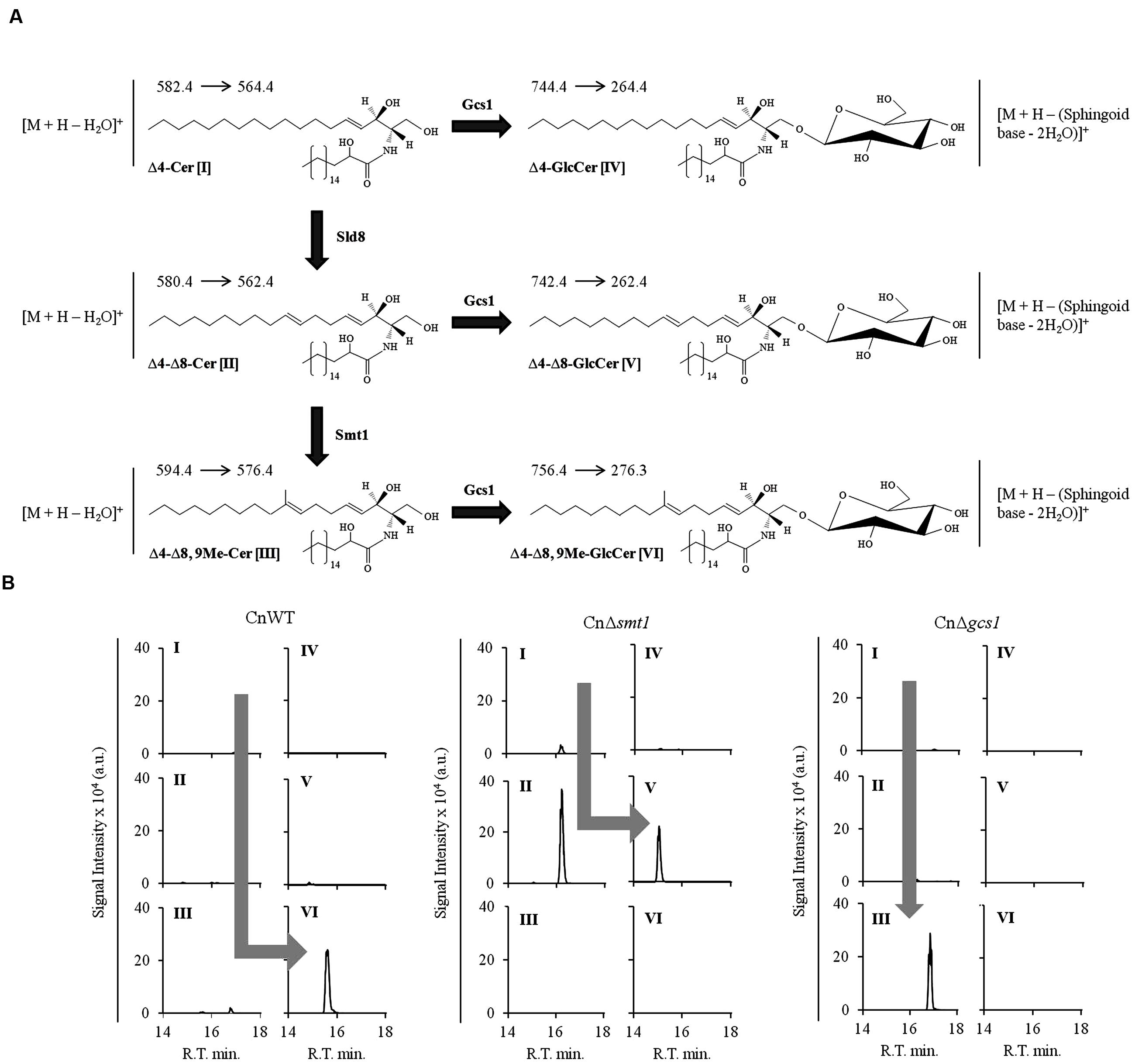
FIGURE 5. Analysis of ceramides and glucosylceramides of Cryptococcus neoformans using MRM approach. (A) Partial pathway and structures of glucosylceramide biosynthetic pathway of C. neoformans. Δ4-Cer [I] is converted to Δ4, Δ8-Cer [II] by Δ8-desaturase (Sld8). Δ4–8-Cer is then methylated at C9 position of sphingoid backbone to from Δ4–Δ8,9Me-Cer [III] by Smt1. A Gcs1 transfers the glucose group to sphingoid backbone of ceramide structures at C1 position forming a β1-3-glycosidic bond. This results in formation of Δ4-GlcCer (IV), Δ4, Δ8-GlcCer (V) and Δ4–Δ8,9Me-GlcCer (VI) structures. The amide-linked fatty acyl in these structures is α-hydroxylated, a key feature. The respective SRM transitions used for the identification and quantification of these structures are shown as (Parent ion→Daughter ion). (B) Analysis of ceramide and glucosylcamides in sphingolipid pathway mutants of C. neoformans. The wild type strain CnWT shows a major pool of Δ4–Δ8,9Me-GlcCer (VI), where all other structures are present in negligible amounts. The CnΔsmt1 strain does not produce Δ4–Δ8,9Me-GlcCer (VI) structure and accumulates Δ4, Δ8-Cer [II] which is converted to Δ4, Δ8-GlcCer [IV] by Gcs1. The CnΔgcs1 shows an accumulation of Δ4–Δ8,9Me-Cer (III). The pathway trends are represented by gray arrows in the wild type and mutant strains.
Although, HPLC-ESI-MS/MS presents as a powerful tool for qualitative and quantitative analysis of sphingolipids in fungi as well as other biological systems. It is important to note that ESI-MS/MS only gives limited information about the structure, especially if we are looking at an uncharacterized structure. For example, it is not possible to assign positions of double bonds, hydroxyl groups, and methylation based on limited fragmentation information. Extensive MSn is necessary to provide some insights into the possible arrangement of the structure (Serb et al., 2009). However, such studies require more sophisticated instrumentation with high mass resolution, high sensitivity and sub part per-million accuracy are required. Confirmation of the exact structure is only possible using NMR spectroscopy of the purified lipid species (Sarkar et al., 2015). A routine sphingolipidome profiling of fungal lipids provides limited information regarding the rate at which these structures are synthesized or the activity of the enzymes that synthesize them. More detailed information of the metabolic flux of sphingolipid species may be achievable by using the labeling approaches like stable isotope (Ecker and Liebisch) or isotopomer analysis (Hellerstein and Neese, 1999). Regardless of its limitations HPLC-ESI-MS/MS remains a method of choice for quantitative sphingolipidome profiling.
Author Contributions
All authors listed, have made substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work was supported by NIH Grants AI56168, AI71142, AI116420, AI100631, and T32AI007539 and by a Merit Review Grant I01BX002624 from the Veterans Affairs Program in Biomedical Laboratory Research and Development to MD.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
We thank the valuable inputs from Arielle Bryan.
Abbreviations
CE, collision energy; CID, collision-induced dissociation; ESI, electrospray ionization; FAB, fast atom bombardment; FT-ICR, Fourier-transform ion cyclotron resonance; GC, gas chromatography; GC-EI-MS, gas chromatography-electron impact-mass spectrometry; Gcs1, glucosylceramide synthase; HILIC, hydrophilic-interaction liquid chromatography; HPLC, high-performance liquid chromatography; IPC, inositolphosphoryl ceramide; LCB, long-chain base; MALDI, matrix-assisted laser desorption/ionization; MIPC, mannosyl-inositol phosphorylceramide; MS/MS, tandem mass spectrometry; MSn, multiple-stage mass spectrometry; (M(IP)2C, mannosyldiinositolphosphoryl ceramide; MRM, multiple-reaction monitoring; NL, neutral loss; NMR, nuclear magnetic resonance spectroscopy; PREIS, precursor-ion scanning; QqTOF MS, hybrid quadrupole time-of–flight mass spectrometer; RF, radiofrequency; SRM, single-reaction monitoring; Smt1, sphingolipid C9 methyltransferase; Sld8, Δ8-desaturase; SPE, solid-phase extraction; TLC, thin-layer chromatography; TMS, trimethylsilyl; TOF, time-of-flight; UDP, uridine 5-diphosphate.
References
Ahn, Y. M., Lee, W. W., Jung, J. H., Lee, S. G., and Hong, J. (2009). Structural determination of glucosylceramides isolated from marine sponge by fast atom bombardment collision-induced dissociation linked scan at constant B/E. J. Mass Spectrom. 44, 1698–708. doi: 10.1002/jms.1678
Barreto-Bergter, E., Pinto, M. R., and Rodrigues, M. L. (2004). Structure and biological functions of fungal cerebrosides. An. Acad. Bras. Cienc. 76, 67–84. doi: 10.1590/S0001-37652004000100007
Barreto-Bergter, E., Sassaki, G. L., and de Souza, L. M. (2011). Structural analysis of fungal cerebrosides. Front. Microbiol. 2:239. doi: 10.3389/fmicb.2011.00239
Bennion, B., Park, C., Fuller, M., Lindsey, R., Momany, M., Jennemann, R., et al. (2003). Glycosphingolipids of the model fungus Aspergillus nidulans: characterization of GIPCs with oligo-alpha-mannose-type glycans. J. Lipid Res. 44, 2073–2088. doi: 10.1194/jlr.M300184-JLR200
Bielawski, J., Szulc, Z. M., Hannun, Y. A., and Bielawska, A. (2006). Simultaneous quantitative analysis of bioactive sphingolipids by high-performance liquid chromatography-tandem mass spectrometry. Methods 39, 82–91. doi: 10.1016/j.ymeth.2006.05.004
Bligh, E. G., and Dyer, W. J. (1959). A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917. doi: 10.1139/o59-099
Bodennec, J., Koul, O., Aguado, I., Brichon, G., Zwingelstein, G., and Portoukalian, J. (2000). A procedure for fractionation of sphingolipid classes by solid-phase extraction on aminopropyl cartridges. J. Lipid Res. 41, 1524–1531.
Brügger, B. (2014). Lipidomics: analysis of the lipid composition of cells and subcellular organelles by electrospray ionization mass spectrometry. Annu. Rev. Biochem. 83, 79–98. doi: 10.1146/annurev-biochem-060713-035324
Buede, R., Rinker-Schaffer, C., Pinto, W. J., Lester, R. L., and Dickson, R. C. (1991). Cloning and characterization of LCB1, a Saccharomyces gene required for biosynthesis of the long-chain base component of sphingolipids. J. Bacteriol. 173, 4325–4332.
Cacas, J. L., Melser, S., Domergue, F., Joubès, J., Bourdenx, B., Schmitter, J. M., et al. (2012). Rapid nanoscale quantitative analysis of plant sphingolipid long-chain bases by GC-MS. Anal. Bioanal. Chem. 403, 2745–2755. doi: 10.1007/s00216-012-6060-1
Cheng, J., Park, T. S., Chio, L. C., Fischl, A. S., and Ye, X. S. (2003). Induction of apoptosis by sphingoid long-chain bases in Aspergillus nidulans. Mol. Cell. Biol. 23, 163–177. doi: 10.1128/MCB.23.1.163-177.2003
Cheon, S. A., Bal, J., Song, Y., Hwang, H. M., Kim, A. R., Kang, W. K., et al. (2012). Distinct roles of two ceramide synthases, CaLag1p and CaLac1p, in the morphogenesis of Candida albicans. Mol. Microbiol. 83, 728–745. doi: 10.1111/j.1365-2958.2011.07961.x
Chigorno, V., Sciannamblo, M., Mikulak, J., Prinetti, A., and Sonnino, S. (2006). Efflux of sphingolipids metabolically labeled with [1-3H]sphingosine, L-[3- 3H]serine and [9,10- 3H]palmitic acid from normal cells in culture. Glycoconj. J. 23, 159–165. doi: 10.1007/s10719-006-7921-7
Clarke, N. G., and Dawson, R. M. (1981). Alkaline O leads to N-transacylation. A new method for the quantitative deacylation of phospholipids. Biochem. J. 195, 301–306. doi: 10.1042/bj1950301
Del Poeta, M., Nimrichter, L., Rodrigues, M. L., and Luberto, C. (2014). Synthesis and biological properties of fungal glucosylceramide. PLoS Pathog. 10:e1003832. doi: 10.1371/journal.ppat.1003832
Dickson, R. C., and Lester, R. L. (2002). Sphingolipid functions in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1583, 13–25. doi: 10.1016/S1388-1981(02)00210-X
Ecker, J., and Liebisch, G. (2014). Application of stable isotopes to investigate the metabolism of fatty acids, glycerophospholipid and sphingolipid species. Prog. Lipid Res. 54, 14–31. doi: 10.1016/j.plipres.2014.01.002
Ejsing, C. S., Moehring, T., Bahr, U., Duchoslav, E., Karas, M., Simons, K., et al. (2006). Collision-induced dissociation pathways of yeast sphingolipids and their molecular profiling in total lipid extracts: a study by quadrupole TOF and linear ion trap-orbitrap mass spectrometry. J. Mass Spectrom. 41, 372–389. doi: 10.1002/jms.997
Ejsing, C. S., Sampaio, J. L., Surendranath, V., Duchoslav, E., Ekroos, K., Klemm, R. W., et al. (2009). Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 106, 2136–2141. doi: 10.1073/pnas.0811700106
Epstein, S., Castillon, G. A., Qin, Y., and Riezman, H. (2012). An essential function of sphingolipids in yeast cell division. Mol. Microbiol. 84, 1018–1032. doi: 10.1111/j.1365-2958.2012.08087.x
Folch, J., Lees, M., and Sloane Stanley, G. H. (1957). A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226, 497–509.
Guan, X., and Wenk, M. R. (2008). Biochemistry of inositol lipids. Front. Biosci. 13, 3239–3251. doi: 10.2741/2923
Guan, X. L., and Wenk, M. R. (2006). Mass spectrometry-based profiling of phospholipids and sphingolipids in extracts from Saccharomyces cerevisiae. Yeast 23, 465–477. doi: 10.1002/yea.1362
Guimarães, L. L., Toledo, M. S., Ferreira, F. A., Straus, A. H., and Takahashi, H. K. (2014). Structural diversity and biological significance of glycosphingolipids in pathogenic and opportunistic fungi. Front. Cell. Infect. Microbiol. 4:138. doi: 10.3389/fcimb.2014.00138
Guo, Y., Wang, X., Qiu, L., Qin, X., Liu, H., Wang, Y., et al. (2012). Probing gender-specific lipid metabolites and diagnostic biomarkers for lung cancer using Fourier transform ion cyclotron resonance mass spectrometry. Clin. Chim. Acta 414, 135–141. doi: 10.1016/j.cca.2012.08.010
Haak, D., Gable, K., Beeler, T., and Dunn, T. (1997). Hydroxylation of Saccharomyces cerevisiae ceramides requires Sur2p and Scs7p. J. Biol. Chem. 272, 29704–29710. doi: 10.1074/jbc.272.47.29704
Han, X., and Cheng, H. (2005). Characterization and direct quantitation of cerebroside molecular species from lipid extracts by shotgun lipidomics. J. Lipid Res. 46, 163–175. doi: 10.1194/jlr.D400022-JLR200
Han, X., and Gross, R. W. (1994). Electrospray ionization mass spectroscopic analysis of human erythrocyte plasma membrane phospholipids. Proc. Natl. Acad. Sci. U.S.A. 91, 10635–10639. doi: 10.1073/pnas.91.22.10635
Hanson, B. A., and Lester, R. L. (1980). The extraction of inositol-containing phospholipids and phosphatidylcholine from Saccharomyces cerevisiae and Neurospora crassa. J. Lipid Res. 21, 309–315.
Haynes, C. A., Allegood, J. C., Park, H., and Sullards, M. C. (2009). Sphingolipidomics: methods for the comprehensive analysis of sphingolipids. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877, 2696–2708. doi: 10.1016/j.jchromb.2008.12.057
Heidler, S. A., and Radding, J. A. (2000). Inositol phosphoryl transferases from human pathogenic fungi. Biochim. Biophys. Acta 1500, 147–152. doi: 10.1016/S0925-4439(99)00097-6
Hellerstein, M. K., and Neese, R. A. (1999). Mass isotopomer distribution analysis at eight years: theoretical, analytic, and experimental considerations. Am. J. Physiol. 276(6 Pt 1), E1146–E1170.
Heung, L. J., Luberto, C., and Del Poeta, M. (2006). Role of sphingolipids in microbial pathogenesis. Infect. Immun. 74, 28–39. doi: 10.1128/IAI.74.1.28-39.2006
Imai, H., Hattori, H., and Watanabe, M. (2012). An improved method for analysis of glucosylceramide species having cis-8 and trans-8 isomers of sphingoid bases by LC-MS/MS. Lipids 47, 1221–1229. doi: 10.1007/s11745-012-3725-7
Ito, E., Waki, H., Miseki, K., Shimada, T., Sato, T. A., Kakehi, K., et al. (2013). Structural characterization of neutral glycosphingolipids using high-performance liquid chromatography-electrospray ionization mass spectrometry with a repeated high-speed polarity and MSn switching system. Glycoconj. J. 30, 881–888. doi: 10.1007/s10719-013-9492-8
Jenkins, G. M., Richards, A., Wahl, T., Mao, C., Obeid, L., and Hannun, Y. (1997). Involvement of yeast sphingolipids in the heat stress response of Saccharomyces cerevisiae. J. Biol. Chem. 272, 32566–32572. doi: 10.1074/jbc.272.51.32566
Jia, Z., Li, S., Cong, P., Wang, Y., Sugawara, T., Xue, C., et al. (2015). High throughput analysis of cerebrosides from the sea cucumber Pearsonothria graeffei by liquid chromatography-quadrupole-time-of-flight mass spectrometry. J. Oleo Sci. 64, 51–60. doi: 10.5650/jos.ess14136
Johnson, S. B., and Brown, R. E. (1992). Simplified derivatization for determining sphingolipid fatty acyl composition by gas chromatography-mass spectrometry. J. Chromatogr. 605, 281–286. doi: 10.1016/0021-9673(92)85248-R
Jones, E. E., Dworski, S., Canals, D., Casas, J., Fabrias, G., Schoenling, D., et al. (2014). On-tissue localization of ceramides and other sphingolipids by MALDI mass spectrometry imaging. Anal. Chem. 86, 8303–8311. doi: 10.1021/ac501937d
Karashima, T., Kajiwara, K., and Funato, K. (2013). Metabolic labeling of yeast sphingolipids with radioactive D-erythro-[4,5-3H]dihydrosphingosine. Bio Protoc. 3:e862.
Kendall, A. C., Pilkington, S. M., Massey, K. A., Sassano, G., Rhodes, L. E., and Nicolaou, A. (2015). Distribution of bioactive lipid mediators in human skin. J. Invest. Dermatol. 135, 1510–1520. doi: 10.1038/jid.2015.41
Kim, S., Fyrst, H., and Saba, J. (2000). Accumulation of phosphorylated sphingoid long chain bases results in cell growth inhibition in Saccharomyces cerevisiae. Genetics 156, 1519–1529.
Köfeler, H. C., Fauland, A., Rechberger, G. N., and Trötzmüller, M. (2012). Mass spectrometry based lipidomics: an overview of technological platforms. Metabolites 2, 19–38. doi: 10.3390/metabo2010019
Kondo, N., Ohno, Y., Yamagata, M., Obara, T., Seki, N., Kitamura, T., et al. (2014). Identification of the phytosphingosine metabolic pathway leading to odd-numbered fatty acids. Nat. Commun. 5:5338. doi: 10.1038/ncomms6338
Kotz, A., Wagener, J., Engel, J., Routier, F., Echtenacher, B., Pich, A., et al. (2010). The mitA gene of Aspergillus fumigatus is required for mannosylation of inositol-phosphorylceramide, but is dispensable for pathogenicity. Fungal Genet. Biol. 47, 169–178. doi: 10.1016/j.fgb.2009.10.001
Kuroda, M., Hashida-Okado, T., Yasumoto, R., Gomi, K., Kato, I., and Takesako, K. (1999). An aureobasidin A resistance gene isolated from Aspergillus is a homolog of yeast AUR1, a gene responsible for inositol phosphorylceramide (IPC) synthase activity. Mol. Gen. Genet. 261, 290–296. doi: 10.1007/s004380050969
Lahiri, S., and Futerman, A. H. (2005). LASS5 is a bona fide dihydroceramide synthase that selectively utilizes palmitoyl-CoA as acyl donor. J. Biol. Chem. 280, 33735–33738. doi: 10.1074/jbc.M506485200
Leipelt, M., Warnecke, D., Zahringer, U., Ott, C., Muller, F., Hube, B., et al. (2001). Glucosylceramide synthases, a gene family responsible for the biosynthesis of glucosphingolipids in animals, plants, and fungi. J. Biol. Chem. 276, 33621–33629. doi: 10.1074/jbc.m104952200
Levery, S. B., Momany, M., Lindsey, R., Toledo, M. S., Shayman, J. A., Fuller, M., et al. (2002). Disruption of the glucosylceramide biosynthetic pathway in Aspergillus nidulans and Aspergillus fumigatus by inhibitors of UDP-Glc:ceramide glucosyltransferase strongly affects spore germination, cell cycle, and hyphal growth. FEBS Lett. 525, 59–64. doi: 10.1016/S0014-5793(02)03067-3
Li, M., Yang, L., Bai, Y., and Liu, H. (2014). Analytical methods in lipidomics and their applications. Anal. Chem. 86, 161–75. doi: 10.1021/ac403554h
Li, S., Bao, D., Yuen, G., Harris, S. D., and Calvo, A. M. (2007). basA regulates cell wall organization and asexual/sexual sporulation ratio in Aspergillus nidulans. Genetics 176, 243–253. doi: 10.1534/genetics.106.068239
Li, S., Du, L., Yuen, G., and Harris, S. D. (2006). Distinct ceramide synthases regulate polarized growth in the filamentous fungus Aspergillus nidulans. Mol. Biol. Cell 17, 1218–1227. doi: 10.1091/mbc.E05-06-0533
Luberto, C., Toffaletti, D. L., Wills, E. A., Tucker, S. C., Casadevall, A., Perfect, J. R., et al. (2001). Roles for inositol-phosphoryl ceramide synthase 1 (IPC1) in pathogenesis of C. neoformans. Genes Dev. 15, 201–212. doi: 10.1101/gad.856001
Luttgeharm, K. D., Chen, M., Mehra, A., Cahoon, R. E., Markham, J. E., and Cahoon, E. B. (2015). Overexpression of Arabidopsis ceramide synthases differentially affects growth, sphingolipid metabolism, programmed cell death, and mycotoxin resistance. Plant Physiol. 169, 1108–1117. doi: 10.1104/pp.15.00987
Mandala, S. M., Thornton, R. A., Frommer, B. R., Curotto, J. E., Rozdilsky, W., Kurtz, M. B., et al. (1995). The discovery of australifungin, a novel inhibitor of sphinganine N-acyltransferase from Sporormiella australis. Producing organism, fermentation, isolation, and biological activity. J. Antibiot. (Tokyo) 48, 349–356. doi: 10.7164/antibiotics.48.349
Marquês, J. T., Cordeiro, A. M., Viana, A. S., Herrmann, A., Marinho, H. S., and de Almeida, R. F. (2015). Formation and properties of membrane-ordered domains by phytoceramide: role of sphingoid base hydroxylation. Langmuir 31, 9410–9421. doi: 10.1021/acs.langmuir.5b02550
Matsubara, T., and Hayashi, A. (1991). Fragmentation pathways of O-trimethylsilyl ethers of dihydroxy long-chain bases analysed by linked-scan mass spectrometry. J. Chromatogr. 562, 119–124. doi: 10.1016/0378-4347(91)80570-3
Merkle, R. K., and Poppe, I. (1994). Carbohydrate composition analysis of glycoconjugates by gas-liquid chromatography/mass spectrometry. Methods Enzymol. 230, 1–15. doi: 10.1016/0076-6879(94)30003-8
Merrill, A. H. Jr., Stokes, T. H., Momin, A., Park, H., Portz, B. J., Kelly, S., et al. (2009). Sphingolipidomics: a valuable tool for understanding the roles of sphingolipids in biology and disease. J. Lipid Res. 50(Suppl.), S97–S102. doi: 10.1194/jlr.R800073-JLR200
Merrill, A. H. Jr., Sullards, M. C., Allegood, J. C., Kelly, S., and Wang, E. (2005). Sphingolipidomics: high-throughput, structure-specific, and quantitative analysis of sphingolipids by liquid chromatography tandem mass spectrometry. Methods 36, 207–224. doi: 10.1016/j.ymeth.2005.01.009
Mor, V., Rella, A., Farnoud, A. M., Singh, A., Munshi, M., Bryan, A., et al. (2015). Identification of a new class of antifungals targeting the synthesis of fungal sphingolipids. MBio 6:e00647. doi: 10.1128/mBio.00647-15
Nagiec, M. M., Baltisberger, J. A., Wells, G. B., Lester, R. L., and Dickson, R. C. (1994). The LCB2 gene of Saccharomyces and the related LCB1 gene encode subunits of serine palmitoyltransferase, the initial enzyme in sphingolipid synthesis. Proc. Natl. Acad. Sci. U.S.A. 91, 7899–7902. doi: 10.1073/pnas.91.17.7899
Obeid, L. M., Okamoto, Y., and Mao, C. (2002). Yeast sphingolipids: metabolism and biology. Biochim. Biophys. Acta 1585, 163–171. doi: 10.1016/S1388-1981(02)00337-2
Ogiso, H., Taniguchi, M., Araya, S., Aoki, S., Wardhani, L. O., Yamashita, Y., et al. (2014). Comparative analysis of biological sphingolipids with glycerophospholipids and diacylglycerol by LC-MS/MS. Metabolites 4, 98–114. doi: 10.3390/metabo4010098
Ohnishi, M., Ito, S., and Fujino, Y. (1983). Characterization of sphingolipids in spinach leaves. Biochim. Biophys. Acta 752, 416–422. doi: 10.1016/0005-2760(83)90271-0
Oura, T., and Kajiwara, S. (2010). Candida albicans sphingolipid C9-methyltransferase is involved in hyphal elongation. Microbiology 156(Pt 4), 1234–1243. doi: 10.1099/mic.0.033985-0
Palumbo, G., and Zullo, F. (1987). The use of iodine staining for the quantitative analysis of lipids separated by thin layer chromatography. Lipids 22, 201–205. doi: 10.1007/BF02537303
Pinto, M. R., Rodrigues, M. L., Travassos, L. R., Haido, R. M., Wait, R., and Barreto-Bergter, E. (2002). Characterization of glucosylceramides in Pseudallescheria boydii and their involvement in fungal differentiation. Glycobiology 12, 251–260. doi: 10.1093/glycob/12.4.251
Rana, N. A., Singh, A., Del Poeta, M., and Hannun, Y. A. (2015). “Qualitative and quantitative measurements of sphingolipids by mass spectrometry,” in Bioactive Sphingolipids in Cancer Biology and Therapy, eds Y. A. Hannun, C. Luberto, C. Mao, and L. M. Obeid (Cham: Springer International Publishing), 313–338.
Rittenour, W. R., Chen, M., Cahoon, E. B., and Harris, S. D. (2011). Control of glucosylceramide production and morphogenesis by the Bar1 ceramide synthase in Fusarium graminearum. PLoS ONE 6:e19385. doi: 10.1371/journal.pone.0019385
Rittershaus, P. C., Kechichian, T. B., Allegood, J., Merrill, A. H. J., Hennig, M., Luberto, C., et al. (2006). Glucosylceramide is an essential regulator of pathogenicity of Cryptococcus neoformans. J. Clin. Invest. 116, 1651–1659. doi: 10.1172/jci27890
Rodriguez-Cuenca, S., Barbarroja, N., and Vidal-Puig, A. (2015). Dihydroceramide desaturase 1, the gatekeeper of ceramide induced lipotoxicity. Biochim. Biophys. Acta. 1851, 40–50. doi: 10.1016/j.bbalip.2014.09.021
Rollin-Pinheiro, R., Liporagi-Lopes, L. C., de Meirelles, J. V., Souza, L. M., and Barreto-Bergter, E. (2014). Characterization of Scedosporium apiospermum glucosylceramides and their involvement in fungal development and macrophage functions. PLoS ONE 9:e98149. doi: 10.1371/journal.pone.0098149
Sarkar, A., Drouillard, S., Rivet, A., and Perez, S. (2015). Databases of conformations and NMR structures of glycan determinants. Glycobiology 25, 1480–1490. doi: 10.1093/glycob/cwv054
Sasaki, T. (1981). Extensive radioactive labeling of glycolipids in cultured hamster fibroblasts by incubation of whole cells with UDP-[14C]glucose and UDP-[14C]galactose. Biochim. Biophys. Acta 666, 418–425. doi: 10.1016/0005-2760(81)90301-5
Serb, A., Schiopu, C., Flangea, C., Sisu, E., and Zamfir, A. D. (2009). Top-down glycolipidomics: fragmentation analysis of ganglioside oligosaccharide core and ceramide moiety by chip-nanoelectrospray collision-induced dissociation MS2-MS6. J. Mass Spectrom. 44, 1434–1442. doi: 10.1002/jms.1625
Serra, M., and Saba, J. D. (2010). Sphingosine 1-phosphate lyase, a key regulator of sphingosine 1-phosphate signaling and function. Adv. Enzyme Regul. 50, 349–362. doi: 10.1016/j.advenzreg.2009.10.024
Shaner, R. L., Allegood, J. C., Park, H., Wang, E., Kelly, S., Haynes, C. A., et al. (2009). Quantitative analysis of sphingolipids for lipidomics using triple quadrupole and quadrupole linear ion trap mass spectrometers. J. Lipid Res. 50, 1692–1707. doi: 10.1194/jlr.D800051-JLR200
Singh, A., Prasad, T., Kapoor, K., Mandal, A., Roth, M., Welti, R., et al. (2010). Phospholipidome of Candida: each species of Candida has distinctive phospholipid molecular species. OMICS 14, 665–677. doi: 10.1089/omi.2010.0041
Singh, A., Wang, H., Silva, L. C., Na, C., Prieto, M., Futerman, A. H., et al. (2012). Methylation of glycosylated sphingolipid modulates membrane lipid topography and pathogenicity of Cryptococcus neoformans. Cell. Microbiol. 14, 500–516. doi: 10.1111/j.1462-5822.2011.01735.x
Sperling, P., Lee, M., Girke, T., Zähringer, U., Stymne, S., and Heinz, E. (2000). A bifunctional delta-fatty acyl acetylenase/desaturase from the moss Ceratodon purpureus. A new member of the cytochrome b5 superfamily. Eur. J. Biochem. 267, 3801–3811. doi: 10.1046/j.1432-1327.2000.01418.x
Sugawara, T., Aida, K., Duan, J., and Hirata, T. (2010). Analysis of glucosylceramides from various sources by liquid chromatography-ion trap mass spectrometry. J. Oleo Sci. 59, 387–394. doi: 10.5650/jos.59.387
Tadano, K., and Ishizuka, I. (1982). Isolation and characterization of the sulfated gangliotriaosylceramide from rat kidney. J. Biol. Chem. 257, 1482–1490.
Ternes, P., Sperling, P., Albrecht, S., Franke, S., Cregg, J. M., Warnecke, D., et al. (2006). Identification of fungal sphingolipid C9-methyltransferases by phylogenetic profiling. J. Biol. Chem. 281, 5582–5592. doi: 10.1074/jbc.M512864200
Ternes, P., Wobbe, T., Schwarz, M., Albrecht, S., Feussner, K., Riezman, I., et al. (2011). Two pathways of sphingolipid biosynthesis are separated in the yeast Pichia pastoris. J. Biol. Chem. 86, 11401–1114. doi: 10.1074/jbc.M110.193094
Urban, P. F., Harth, S., Freysz, L., and Dreyfus, H. (1980). Brain and retinal ganglioside composition from different species determined by TLC and HPTLC. Adv. Exp. Med. Biol. 125, 149–157. doi: 10.1007/978-1-4684-7844-0-14
Voynova, N. S., Mallela, S. K., Vazquez, H. M., Cerantola, V., Sonderegger, M., Knudsen, J., et al. (2014). Characterization of yeast mutants lacking alkaline ceramidases YPC1 and YDC1. FEMS Yeast Res. 14, 776–788. doi: 10.1111/1567-1364.12169
Warnecke, D., and Heinz, E. (2003). Recently discovered functions of glucosylceramides in plants and fungi. Cell. Mol. Life Sci. 60, 919–941.
Wenk, M. R. (2010). Lipidomics: new tools and applications. Cell 143, 888–895. doi: 10.1016/j.cell.2010.11.033
Zama, K., Hayashi, Y., Ito, S., Hirabayashi, Y., Inoue, T., Ohno, K., et al. (2009). Simultaneous quantification of glucosylceramide and galactosylceramide by normal-phase HPLC using O-phtalaldehyde derivatives prepared with sphingolipid ceramide N-deacylase. Glycobiology 19, 767–775. doi: 10.1093/glycob/cwp047
Keywords: sphingolipids, high performance liquid chromatography, electrospray ionization tandem mass spectrometry, fungi, fungal infections, ceramide
Citation: Singh A and Del Poeta M (2016) Sphingolipidomics: An Important Mechanistic Tool for Studying Fungal Pathogens. Front. Microbiol. 7:501. doi: 10.3389/fmicb.2016.00501
Received: 27 December 2015; Accepted: 28 March 2016;
Published: 14 April 2016.
Edited by:
Agostinho Carvalho, University of Minho, PortugalReviewed by:
Igor C. Almeida, University of Texas at El Paso, USALauren Ashley Cowart, Medical University of South Carolina, USA
Copyright © 2016 Singh and Del Poeta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maurizio Del Poeta, bWF1cml6aW8uZGVscG9ldGFAc3Rvbnlicm9vay5lZHU=
 Ashutosh Singh
Ashutosh Singh Maurizio Del Poeta
Maurizio Del Poeta