
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 26 April 2016
Sec. Plant Pathogen Interactions
Volume 7 - 2016 | https://doi.org/10.3389/fmicb.2016.00493
This article is part of the Research Topic Harnessing useful rhizosphere microorganisms for pathogen and pest biocontrol View all 26 articles
Effective translation of research findings from laboratory to agricultural fields is essential for the success of biocontrol or growth promotion trials employing beneficial microorganisms. The rhizosphere is to be viewed holistically as a dynamic ecological niche comprising of diverse microorganisms including competitors and noxious antagonists to the bio-inoculant. This study was undertaken to assess the effects due to the soil application of an endophytic bacterium with multiple pathogen antagonistic potential on native bacterial community and its sustenance in agricultural soil. Pseudomonas aeruginosa was employed as a model system considering its frequent isolation as an endophyte, wide antagonistic effects reported against different phytopathogens and soil pests, and that the species is a known human pathogen which makes its usage in agriculture precarious. Employing the strain ‘GNS.13.2a’ from banana, its survival in field soil and the effects upon soil inoculation were investigated by monitoring total culturable bacterial fraction as the representative indicator of soil microbial community. Serial dilution plating of uninoculated control versus P. aeruginosa inoculated soil from banana rhizosphere indicated a significant reduction in native bacterial cfu soon after inoculation compared with control soil as assessed on cetrimide- nalidixic acid selective medium against nutrient agar. Sampling on day-4 showed a significant reduction in P. aeruginosa cfu in inoculated soil and a continuous dip thereafter registering >99% reduction within 1 week while the native bacterial population resurged with cfu restoration on par with control. This was validated in contained trials with banana plants. Conversely, P. aeruginosa showed static cfu or proliferation in axenic-soil. Lateral introduction of soil microbiome in P. aeruginosa established soil under axenic conditions or its co-incubation with soil microbiota in suspension indicated significant adverse effects by native microbial community. Direct agar-plate challenge assays with individual environmental bacterial isolates displayed varying interactive or antagonistic effects. In effect, the application of P. aeruginosa in rhizospheric soil did not serve any net benefit in terms of sustained survival. Conversely, it caused a disturbance to the native soil bacterial community. The findings highlight the need for monitoring the bio-inoculant(s) in field-soil and assessing the interactive effects with native microbial community before commercial recommendation.
With increasing awareness about the hazardous effects of agrochemicals employed in crop husbandry, there is an impetus on the usage of safe and effective microorganisms in agriculture toward protection against biotic and abiotic stresses and in crop production (Zarb et al., 2005; Thomas and Upreti, 2015). It is often observed that the growth promotion effects or the antagonistic potential shown by the bacterial strains in laboratory assays are not translated to effective biocontrol strategies in the field. The conditions in the field are different influenced by soil, water, and edaphic factors as well as the native microbial community (van Veen et al., 1997; Bakker et al., 2013; Tyc et al., 2014). The successful performance of a bio-inoculant in the agricultural field is governed by its ability to survive in field soil and the interactive effects with the native microbiome. The introduced organisms in soil are vulnerable to physical stresses and antagonistic effects by other microorganisms (Acea et al., 1988; van Veen et al., 1997). It is also important that the candidate bio-inoculant shall not cause undue biological perturbation in the native soil microbial community. Effects due to the introduced organisms on resident soil microcosm is a topic of much interest to the microbiologists (Trabelsi and Mhamdi, 2013; Tyc et al., 2014).
A gradual reduction in the population of the introduced organism in soil/field has been documented in several instances (van Veen et al., 1997; Matos et al., 2005; Kröber et al., 2014). As for the interaction effects, studies employing Azospirillum brasilense in maize rhizospehere (Herschkovitz et al., 2005) or Bacillus amyloliquefaciens in lettuce rhizosphere through molecular tools (Chowdhury et al., 2013; Kröber et al., 2014) have indicated only little or marginal changes in the rhizosphere bacterial community. The bioinoculant consortium of Bacillus megaterium, Pseudomonas fluorescens, and Trichoderma harzianum also did not impart any negative effects on rhizospheric microbial community (Gupta R. et al., 2015). On the other hand, significant modifying effects due to the introduced biocontrol agent Pseudomonas jessenii on the soil bacterial community composition of lettuce rhizosphere influenced by soil type and time span after inoculation have been documented (Schreiter et al., 2014). The microbial community varies from soil to soil and location to location. This is largely constituted by non-cultivable organisms whose community profiles can be studied deploying molecular tools (Ogram et al., 2007; Bakker et al., 2013). Cultivation based methods may not provide a full account of metabolically active cells, yet it can be a powerful tool in different spheres of microbiology and in the modern era of genomics towards exploitation of beneficial organisms or for further physiological, molecular and application studies (Steward and Rappe, 2007; Epstein, 2013; Prakash et al., 2013). Here, we consider that monitoring the cultivable bacterial community in a confined environment could serve as a representative of soil microbiota to assess the effects due the introduced organism.
Endophytic microorganisms are plant internal inhabitants and are often isolated from surface sterilized tissue or through vacuum extraction (Hallmann et al., 1997; Hardoim et al., 2015). Endophytes are known to benefit plants through growth promotion and antagonistic effects on phytopathogens and pests besides facilitating phytoremediation (Ryan et al., 2008; Gaiero et al., 2013; Kumar et al., 2014). The potential for intra-tissue colonization and their intimate association with the host (Thomas and Reddy, 2013; Thomas and Sekhar, 2014) make endophytes more valuable agents in biocontrol applications over the rhizospheric organisms (Turner et al., 2013; Upreti and Thomas, 2015). Endophytes are normally considered to be recruited by the host plant primarily from the soil community through roots (Hallmann et al., 1997; Hardoim et al., 2015). The methods for the delivery of endophytes in agriculture range from inoculation of seed, seedling or other planting propagules to soil drenching, stem injection, and foliar sprays (Hallmann et al., 1997; Puri et al., 2015). When an antagonistic microorganism is to be employed as a biocontrol agent against soil-borne pathogens or pests, it warrants that the organism be applied through soil-drenching to neutralize the pathogen/pest in the root-zone from where the endophytes should migrate to the host. In this respect, the survival of endophytic microorganisms under field conditions for a longer duration assumes significance. Endophytes invariably return to the soil at the end of the life span of the host/organ and are thus having a phase in the soil environment before re-colonizing the host (Thomas and Soly, 2009).
Recently, we cultured several bacterial endophytes from the sucker-derived shoot-tips of banana (Musa sp.) cv. Grand Naine (Sekhar and Thomas, 2015). The prime target was to explore by testing these isolates for potential biocontrol of banana wilt disease caused by the soil-borne vascular pathogen Fusarium oxysporum f. sp. cubense (Foc), a serious limiting factor in major banana growing areas world over (Stover, 1962; Ploetz et al., 2003). Evaluating the 47 endophytic banana strains against Foc, one isolate (GNS.13.2a) exhibited significant antagonistic activity against the pathogen in direct in vitro challenge assays and this isolate was identified as Pseudomonas aeruginosa (Sekhar and Thomas, 2015). This isolate also displayed antagonistic effects against the soil-borne pathogens, Ralstonia solanacearum from tomato and Erwinia carotovora from banana and thus a potential biocontrol agent against multiple diseases. P. aeruginosa is also known to be a human pathogen and there are concerns about its usage in agriculture (Matos et al., 2005; Kumar et al., 2013). Therefore, we did not consider our endophytic strain from banana as a biocontrol candidate for agricultural applications.
Pseudomonas aeruginosa has been frequently isolated as endophytes from different crop plants and most of such strains have been reported to show inhibitory activity against various phyto-pathogens. These include antagonistic activity against Phytophthora capsici in black pepper (Aravind et al., 2009), Sclerotium rolfsii in cucumber (Pandey et al., 2012), Colletotrichum gloeosporioides in chili (Allu et al., 2014), Pythium myriotylum in ginger (Jasim et al., 2014), Fusarium oxysporum in cotton (Yasmin et al., 2014) and wheat (Gupta G. et al., 2015), Ralstonia solanacearum in tomato (Maji and Chakrabartty, 2014) and Xanthomonas sp. infecting different crops (Spago et al., 2014). Endophytic P. aeruginosa is also known to be an effective biocontrol agent against nematodes (Ali et al., 2002; Kumar et al., 2013). Further, some strains of P. aeruginosa have been reported as plant growth promoters (Pandey et al., 2012; Gupta et al., 2013; Gupta G. et al., 2015) or useful in weed management (Lakshmi et al., 2015). Thus the endophytic strain of P. aeruginosa from banana (Sekhar and Thomas, 2015) appeared to form a good model system for studying the twin aspects of (i), the ability of an endophytic strain to survive in soil and (ii), effects due to the introduced microorganism with pathogen-antagonistic potential on native soil microbial community. Further, Pseudomonas sp. represents one of the most abundant genera of the root microbiome (Zamioudis et al., 2013). The present investigations were undertaken to assess the survivability of the endophytic P. aeruginosa strain from banana under non-axenic conditions in agricultural soil and rhizosphere and to gauge the effects due to the introduced organism on native microbiome by monitoring the gross cultivable bacterial community as the representative indicator of soil microbiota.
Endophytic P. aeruginosa strain ‘GNS13.2a’ (NCBI 16S rRNA gene accession number KP798813) isolated from the deep-seated shoot-tip tissue of banana sucker cv. Grand Naine (Musa sp.; AAA genome) in a previous study (Sekhar and Thomas, 2015), referred to as Pau hereafter, was employed as the test organism in this study. The isolate was maintained as glycerol (30%) stock at -80°C and revived on nutrient agar (NA) followed by single colony perpetuation at each culturing. This strain was not specifically tested for human or animal pathogenicity. The experiments were conducted under containment followed by destruction of all biosamples through autoclaving or formaldehyde drenching. A Class-II vertical airflow cabinet with ULPA filter (Esco Biotech, Pvt. Ltd., Singapore) was employed during axenic works.
To select the appropriate media for capturing the culturable bacterial community and for monitoring Pau specifically, NA was tested compared with two known Pau selective media. These included (i), NA containing 60 μg ml-1 kanamycin and 50 μg ml-1 2,3,5-triphenyl tetrazolium chloride (Kan+TTC:NA) as used by Kumar et al. (2013), and (ii), cetrimide agar base added with 15 μg ml-1 nalidixic acid (Goto and Enomoto, 1970) as employed by Deredjian et al. (2014). The above media were tested using pure culture of Pau. For this, six decimal dilutions (101 to 106) in sterile distilled water (SDW) considering 0.1 OD600 nm overnight NA colony-derived culture as 100 ‘anchored stock’ (Thomas et al., 2015) were applied through SATS approach (Thomas et al., 2012). The plates were observed at 30°C for 1–4 days for Pau cfu. The dilution level that yielded cfu in the 30–300 range was selected based on which cfu ml-1 of 0.1 OD stock in different media was worked out (Thomas et al., 2015).
Further, pure culture of Pau, irrigation grade water and rhizospheric field soil were tested on the above three media to ascertain the suitability for harnessing Pau cfu specifically. Here, the six serial dilutions from Pau anchored stock as above were tested by spotting 20 μl aliquots side by side in six sectors in a 9-cm Petri dish, a method designated as single plate-serial dilution spotting (SP-SDS) (Thomas et al., 2015). The anchored stocks (100) constituted 0.1 OD suspension for pure bacterial cultures, original sample for irrigation water and 1 g per 10 ml SDW for soil samples. Irrigation water was tested directly and after mixing with Pau stock. Soil sample suspensions were tested similarly after mixing with Pau culture employing six serial dilutions. The plates after surface drying were observed for bacterial cfu and specificity for Pau detection over 7 days.
Based on the results, NA was selected as the standard medium for growing pure cultures of Pau and for total culturable bacterial monitoring. Cetrimide-nalidixic acid-agar (CNA) was identified as the selective medium for the specific monitoring of Pau with obvious colony development within 24 h similar to NA. The GNS13.2a isolate had kanamycin and rifampicin resistance as genetic markers besides nalidixic acid. Modification of CNA with 60 μg ml-1 kanamycin was tried which delayed the colony growth and enumeration by 1 day with no extra benefit on cfu or selection specificity. Cetrimide agar with 50 μg ml-1 rifampcin (Matos et al., 2005) also did not offer any advantage over CNA while testing field soil or irrigation water for Pau. Further, 1–2 days-old NA plate cultures of Pau were tested directly and after refrigeration for cfu ml-1 on NA and CNA (six replications) to ensure that the inocula employed had good viability. All media formulations and supplements were sourced from M/s Hi Media Biosciences, Mumbai, India.
As a prerequisite to ascertain whether the water used for irrigation, or the rhizospheric soil-mix employed in pot-culture trials harbored any Pau, both were checked 4 days prior to the start of the soil-monitoring experiment. Piped irrigation-grade water collected and stored in an open plastic tank in the glasshouse 1 day before was generally used for watering the soil. Aliquots of 100 μl were applied directly on CNA plates (100 nos) which altogether offered a gross detection sensitivity of 0.1 cfu ml-1. The soil stock was comprised of a 1:1:1 blend of rhizospheric soil where banana was being gown, river sand and well-decayed farm yard manure (pH 7.35 ± 0.44) without any chemical or other sterilization treatments. The soil mix (50 g in 500 ml autoclaved water; 100 stock) was shake incubated for 1 h at 200 rpm and plated at 101 dilution on CNA plates (100 nos) offering a detection sensitivity of 1 cfu g-1 soil. The plates were monitored for microbial cfu and distinct colony types for 4 days in comparison with pure culture of Pau. Two colony morphotypes that emerged on CNA from water (2–4 × 101 cfu ml-1) and soil (1–4 × 102 cfu g-1) and distinct from Pau reference colonies in appearance were taken for identification through 16S rRNA gene sequence analysis as described elsewhere (Sekhar and Thomas, 2015). Mix culture of these organisms was further tested on NA and CNA along with pure Pau culture through SATS to ensure their distinction from Pau colonies.
To monitor the survival of Pau in inoculated versus control soils, the rhizospheric soil mix from the same lot as above in pots was employed as it was not possible to get a clear estimate under field conditions. Plastic pots (7′′ height and 6.5′′ diameter) were provided with 2.5 kg of dry soil-mix per pot and watered to field capacity. After 24 h under glasshouse conditions, eight replicate pots were drenched with 250 ml of 0.1 OD Pau suspension (about 108 cfu ml-1) from day-2 NA plate colonies (Pau+ soil; approximately Pau cfu of about 107 g-1) and the control pots (Pau- soil) were applied with equal volume of SDW. First soil sampling was done on the same day (day-0) within 30–60 min. This involved inserting a 15 ml sterile Falcon tube to the soil to its full length (to a depth of approximately 10 cm) during which approximately 2 cm of compacted soil (approximately 2–2.5 g) was collected inside the tube. The sample from eight such pots was pooled and mixed thoroughly after removing the stony particles and breaking the lumps. 10 g soil was dispersed in 100 ml of SDW (100 stock) in a sterilized container and Pau and non-Pau cfu were assessed through SATS employing four replications per dilution. The Pau- soil samples were also processed similarly and monitored on NA and CNA. Pau colonies were located on NA based on the bluish green tinge while on CNA they appeared as shining/fluorescent cream-yellow colonies. The cumulative cfu on day-4 was used for estimating the Pau versus non-Pau cfu which was expressed as cfu g-1 soil. A simple assessment of the extent of bacterial variability was made on day-4 by counting the different colony types formed on NA at dilutions (103 or 104) that yielded well-delineated colonies.
The pots were left open under glasshouse conditions (day temperature range of 25–30°C; irradiance of 500–600 μE m-1 s-1 for 8–10 h) using 250 ml of irrigation-grade piped water per pot daily in the afternoon. Sampling of Pau+ and Pau- soils was repeated on days-4, 7, 14, 21, and 28 with sample collection during the 30–60 min time span after watering. The irrigation water sample was routinely monitored for bacterial cfu by plating on NA and CNA. The pots thereafter were left without watering allowing the soil-mix to dry completely. The survival of Pau under dry soil conditions was monitored as above after another 4 weeks preceded by watering 4 h before sampling. This soil was monitored for Pau again after 48 h.
To assess the effect due to the moistening of dry soil in altering the total culturable bacterial cfu and the contribution of irrigating water to it, a further trial was set up. The dry soil-mix (2.5 kg in plastic pots) was applied with 500 ml of (i) sterile water or (ii) irrigation-grade water (3.2 × 105 cfu ml-1). The initial soil bacterial cfu was monitored within 30–60 min on NA. Thereafter, the pots were kept covered with polythene sheet to avoid the lateral introduction of organisms, and were monitored for bacterial cfu again after 24 h. A third treatment involved rhizospheric soil from pots planted with tomato (6 weeks post-planting). These pots were maintained under glasshouse conditions, watered daily with irrigation-grade water (250 ml) and cfu estimations were undertaken as above.
A validation trial was undertaken employing a new batch of rhizospheric soil-mix watered to field capacity one day prior to Pau inoculation as discussed above (Pau+ and Pau- soils) employing eight replicate pots. Another experiment was set up employing pots which were planted with tissue-cultured banana ‘Grand Naine’ saplings for the preceding 2 months (pH 7.13 ± 0.37) with daily inundation using irrigation-grade water. The Pau+ and Pau- soils were monitored for Pau and gross cultivable bacterial flora on days-0, 4, and 7 employing CNA and NA after clearing the root tissues. Irrigation was practiced daily as above with periodic monitoring of water employing NA and CNA.
For this, the soil-mix from the same lot described above was employed. Dry rhizospheric soil-mix (200 g) was wetted to field capacity employing 50 ml distilled water in 300 ml volume glass bottles with wide mouth (50 mm diameter). The screw capped bottles were subjected to autoclaving for 20 min at 121°C (1.1 kg cm-2) on three consecutive days. On the third day, the bottles were kept open in the vertical airflow cabinet for 1 h followed by the addition of 10 ml of 1.0 OD Pau culture (about 109 cfu ml-1) per bottle employing eight replications. The control bottles were provided with SDW. The soil was monitored on the same day (day-0) and thereafter on days 1, 4, 7, 14, 21, and 28 on CNA for the extent of Pau cfu and on NA to check for any lateral introduction of non-Pau cfu during samplings. The soil from eight replicate Pau+ and Pau- bottles was collected separately in 50 ml tubes, weighed aseptically and dispersed in sterile water (1 g per 10 ml) followed by SATS of decimal dilutions on NA and CNA.
With a view to assess the response of established Pau flora in axenic soil to the exposure to native soil microbiota, Pau population was initiated in 3× autoclaved soil-mix in glass bottles as above followed by the introduction of soil microbiome. For this, the baseline Pau cfu was assessed by pooling the soil from two sets of Pau inoculated bottles 2 weeks post-inoculation (Pau+ axenic soil sets-I and II) employing four replications. The set-I was applied with 5 ml of SDW while the set-II was added with 5 ml of the supernatant from the soil-mix suspended in SDW (1 g per 10 ml). Pau and non-Pau cfu were assessed on NA after 48 h. Thereafter, the set-I was applied with 5 ml SDW while the set-II was applied with 5 ml of 1.0 OD suspension of pooled bacterial inoculum prepared from NA colonies of soil bacteria derived from the plating of soil-mix 2 days before. The bottles were kept open in the vertical airflow cabinet for 4 h to evaporate away the excess moisture, and the Pau versus non-Pau cfu was assessed on NA and CNA soon after.
This trial was undertaken to ascertain whether the low cfu of non-Pau isolates on NA during the day-0 monitoring of Pau+ soil arose from the antagonistic effects by Pau on the agar plate or due to the interactive effects in the soil itself. The soil suspensions of Pau+ soil samples from glasshouse pots prepared on days-0, 4, 7, or 14 in SDW and left under sealed conditions in 50 ml falcon tubes (to avoid the lateral introduction of microorganisms) were monitored on CNA and NA after 7 days of stationary incubation. The cfu on the date of original sampling (days-0, 4, 7, or 14) was adopted as the base reference point. As a control to test the ability of the organism to survive under static non-aerated conditions, the axenic culture of Pau in SDW under identical conditions was employed.
With a view to assess that the cfu reduction of Pau was arising from the interactive effects between Pau and soil bacteria, 10 random representative colony types that developed on NA from Pau+ soil during the day-0 sampling of soil in the first trial were selected. A bacterial lawn of individual soil isolates was prepared on NA by applying 100 μl of 0.1 OD inoculum in peptone-salt (Thomas et al., 2012) in 9-cm diameter plates. After allowing 1 h for the organism to establish, 25 μl of 0.1 OD Pau inoculum prepared in SDW was applied at the center of the agar plate (6–7 mm diameter well) followed by air-drying in the vertical airflow cabinet for 25–30 min. The reaction of Pau to the test isolate and vice versa was assessed based on (i), the diameter of spreading Pau colony growth from the center of the well, (ii), the extent of clear zone, if any, between Pau and the isolate in the lawn, and (iii), the extent to which the lawn of the test isolate was pushed to the outer edge of NA plate by Pau. The test plates were scored on a – to ++++ scale for the above characteristics representing none, low, medium, high or significant. Based on the pooled information, four categories of interactive effects were documented: (i), no mutual antagonism (with or without dominance by Pau), (ii), aggressive antagonistic effect by Pau on the soil isolate, (iii), significant anti-Pau effect by the soil isolate, and (iv), mutual antagonism. The experiment was repeated with additional 20 distinct colony types selected from NA plates that were employed for the monitoring of Pau+ pot-soil on day-7 (4th day after SATS) and 20 colony types from the control soil.
A direct confrontation assay was set up where the lawn of Pau on NA prepared using 100 μl of 0.1 OD culture 4 h after plating was applied with 5 μl of 0.1 OD inoculums of the test organisms. Based on the outcome, preparation of Pau lawn on NA using 0.001 OD inoculum (based on pre-trials employing 100 μl of 0.01, 0.001, or 0.0001 OD inoculum) followed by spotting with the challenge isolate as above (5 μl of 0.1 or 1.0 OD inocula) was tried. The ability of different isolates to grow or establish on Pau lawn at different strengths or any clear-zone development between Pau and the test isolate was recorded 1–4 days from the start of the experiment.
The ability of Pau strain to survive in soil vis-à-vis the interactive effects on soil bacterial community were assessed primarily based on the cumulative cfu data recorded as on day-4 from the date of plating on CNA and NA. The survival of Pau was assessed based on the percent difference in cfu keeping the day-0 cfu in soil as the reference point. The extent of non-Pau cfu in Pau+ soils was assessed with reference to the native bacterial cfu in Pau- control soil samples on NA. During soil samplings, eight replications were employed for Pau+ and Pau- treatments. For all cfu comparisons, unless mentioned differently, two independent serial dilutions were prepared from pooled soil/water samples and applied as per SATS on two plates per dilution, thus constituting four replications. For statistical analysis, the cfu data were subjected to logarithmic transformation and Single Factor ANOVA or t-test (assuming equal variance) using the Data Analysis package of Microsoft Excel 2010. The mean (logarithmic scale) ± standard deviation values are presented.
Testing the Pau serial dilutions on three media, NA and CNA showed colonies in the 30–300 range at 105 dilution of the original 0.1 OD stock within 18–24 h while it took an extra day for colony emergence on Kan+TTC:NA with obvious cfu at 101 to 103 dilutions of the stock culture only. CNA gave similar cfu estimates as for NA within 24 h while Kan+TTC:NA showed significantly low estimate (P < 0.0001; Figure 1A). Pau colonies on NA showed a blue-green tinge and they tended to fade away after 4–5 days whereas on CNA they remained delineated with a fluorescing yellow shade for several days. A comparison of Pau day-1, day-2 NA source cultures versus day-1 NA plate culture refrigerated for two nights showed similar cfu estimates for different age groups on both NA and CAN (P = 0.104; Figure 1B). This endorsed the usage of day-1 or day-2 NA cultures directly or after 1–2 days of refrigeration as start culture without loss of viability.
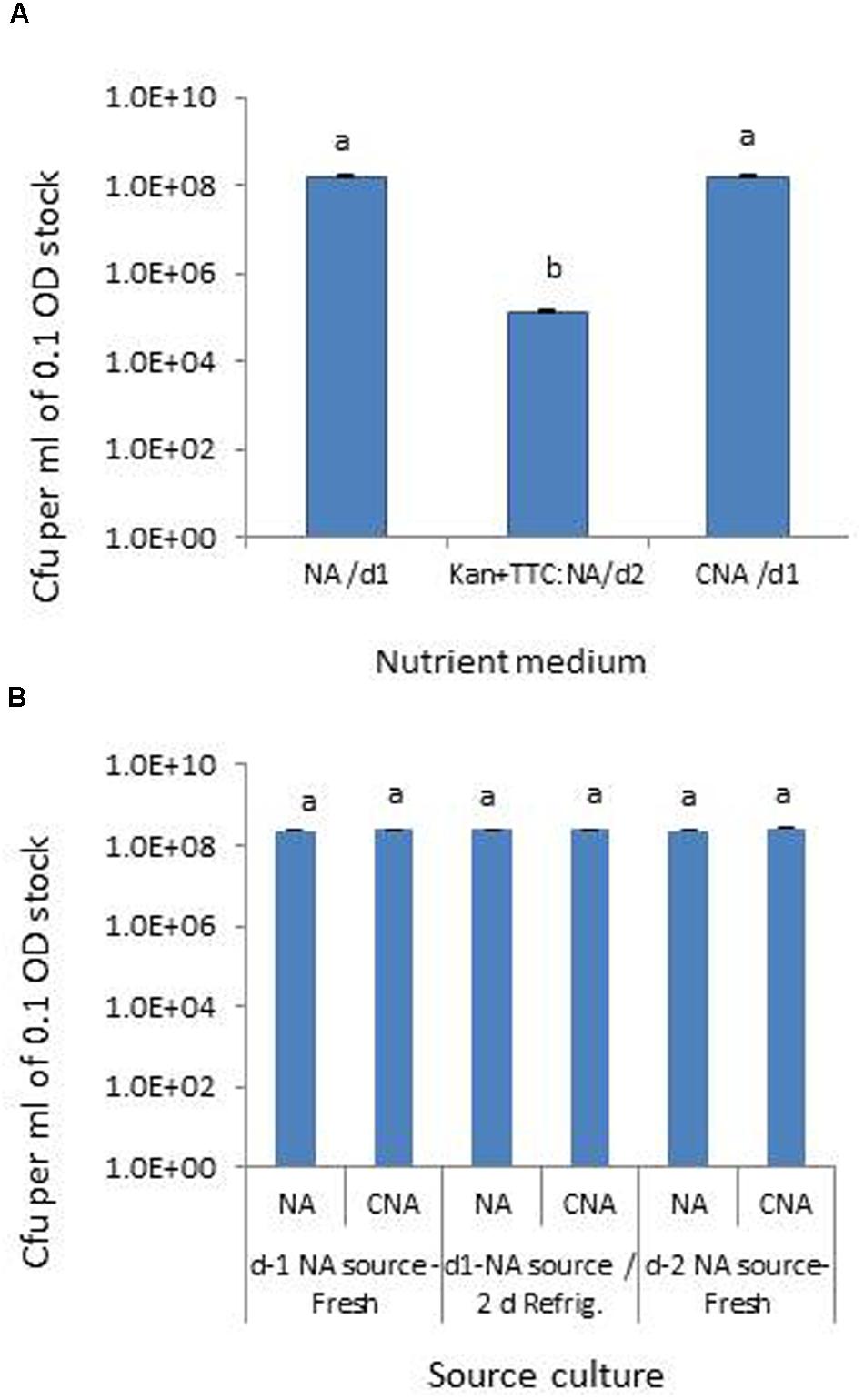
FIGURE 1. Assessing different media for the selective monitoring of Pseudomonas aeruginosa (Pau) based on cfu ml-1 of 0.1 OD anchored stock (A) and testing of source Pau cultures of different ages for cell viability based on cfu (B). NA/d1 and CNA/d1 in (A) indicate cfu recorded on respective media on day-1 and Kan+TTC:NA/d2 cfu observed on day-2. Vertical bars indicate standard deviation. Bars with same letters are not significantly different at P = 0.05.
Now, testing the different dilutions of Pau pure culture, irrigation-grade water and soil samples (with or without Pau addition) in SP-SDS on the above mentioned three media (Supplementary Figures S1A–E) indicated (i), similar cfu at 105 dilution in NA and CNA on day-1 itself, (ii), Kan+TTC:NA required an extra day for colony growth displaying countable cfu at 102 dilution, (iii), Kan+TTC:NA supported the growth of several non-Pau organisms from irrigation water and soil, (iv), NA displayed considerable bacterial variability considering the diverse colony types, (v), CNA did not support any water or soil isolates, (vi), Pau-mixed irrigation water and soil showed characteristic Pau type colonies on NA distinguishable from the native bacterial flora based on colony features, and (vii), CNA showed pure Pau colonies with cfu on par with Pau colony counts on NA. It was quite striking that Kan+TTC:NA supported the growth of diverse microorganisms including fungi from soil while NA normally did not support fungal growth. This was validated in a repeat trial where the irrigating water and soil suspensions were mixed with 102 serial dilution of Pau 0.1 OD stock (data not shown). Pau colonies from Pau-added soil and water were clearly distinguishable on NA when they were not entirely masked by the native bacterial colonies at higher cfu.
Testing up to 10 ml water sample directly for the presence of Pau (100 μl in 100 CNA plates) indicated some Pau-unlike cfu in the range of 1–8 per 100 μl (1.0–8.0 × 101 cfu ml-1) which was too low considering the cfu ml-1 of about 108 for 0.1 OD Pau stock. The soil sample used at 101 dilution showed a meager 1–4 cfu per plate (1.0–4.0 × 102 cfu g-1 soil) which did not include any Pau as per the colony characteristics. There were two colony types other than Pau that were supported on CNA. These were identified as Pseudomonas plecoglossicida (predominant) and Pantoea ananatis (occasional). Testing the mix culture of Pau with the above two organisms indicated that Pau colonies were clearly distinguishable on NA and CNA based on the bluish green/yellow tinge and colony size by day-2 (Figure 2).
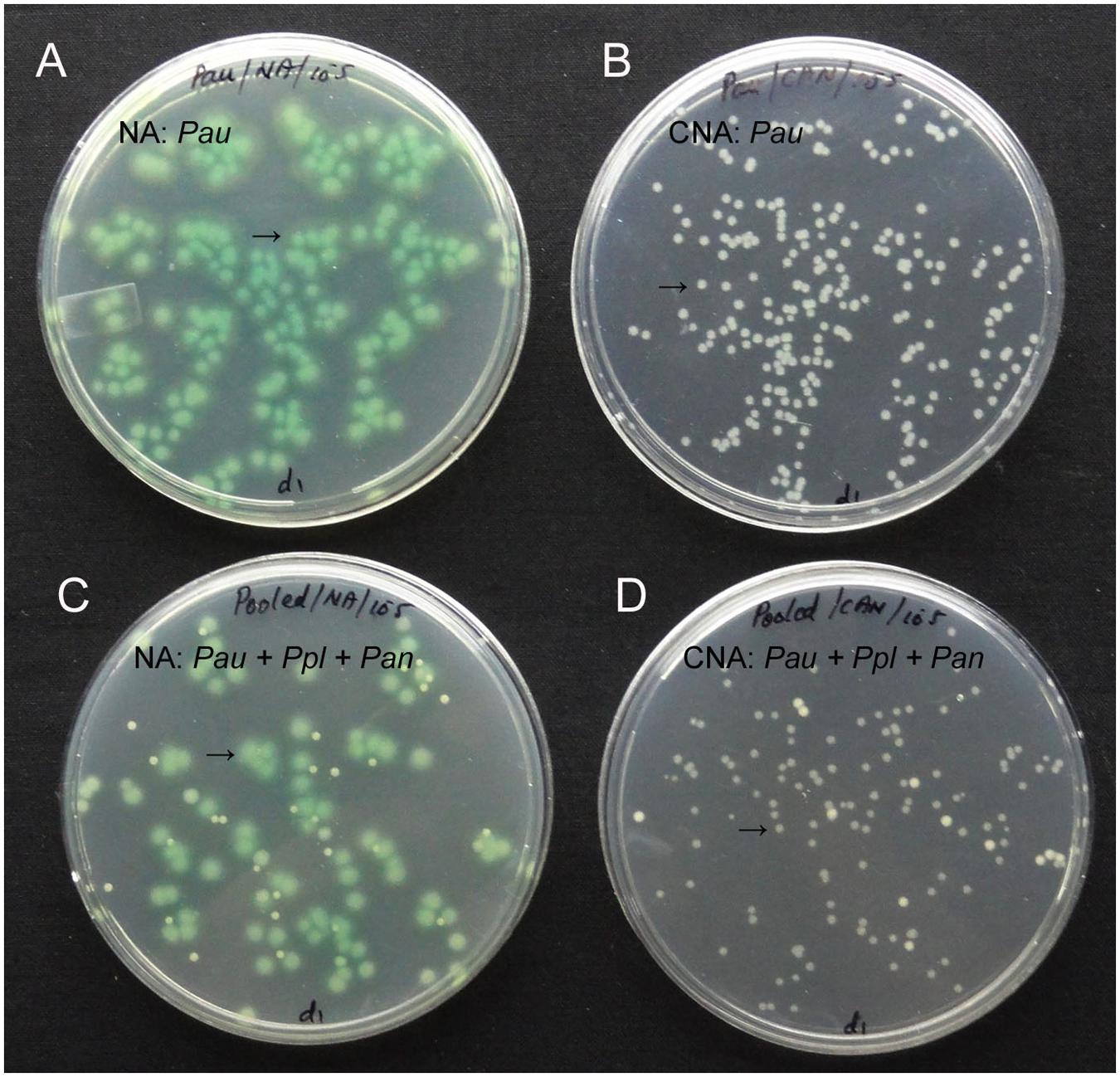
FIGURE 2. Pure culture of P. aeruginosa (Pau) showing cfu at 105 of the 0.1 OD anchored stock as bluish-green tinged larger colonies on nutrient agar (NA, A) or as white distinct colonies on CNA (B), or a mix culture with P. plecoglossicida (Ppl) and Pantoea ananatis (Pan) showing clearly identifiable Pau colonies on NA (C) and CNA (D; Pau indicated by arrow).
The salient observations from the monitoring of Pau in potted Pau+ soils on the date of inoculum application (Figures 3 and 4) included: (i) the Pau+ soil yielded only Pau colonies on CNA to the tune of 7.1 × 106 g-1, (ii), both Pau and non-Pau colonies were easily identified on NA registering a slightly higher Pau cfu (9.7 × 106 g-1) than on CNA, (iii), no fresh Pau colonies emerged after day-1 on NA from Pau+ soil while non-Pau colonies continued to emerge for 2–4 days, and (iv), the non-Pau cfu in Pau+ soil was significantly lower (4.6 × 106 g-1) than the corresponding value for un-inoculated control (1.8 × 107 g-1; P = 0.0071). Pau- soil did not exhibit any colony growth on CNA nor yielded any Pau-like colonies on NA (Figures 3 and 4). Beyond 4–5 days, Pau colonies tended to fade away or vanish from NA plates, also causing the waning of a major share of non-Pau colonies. Thus, the gross cfu estimate on day-4 with colony enumerations on days-1, 2, and 4 was adopted for monitoring Pau versus non-Pau cfu on NA. Marking the initially formed colonies on the reverse of the plate facilitated clearer cfu enumeration.
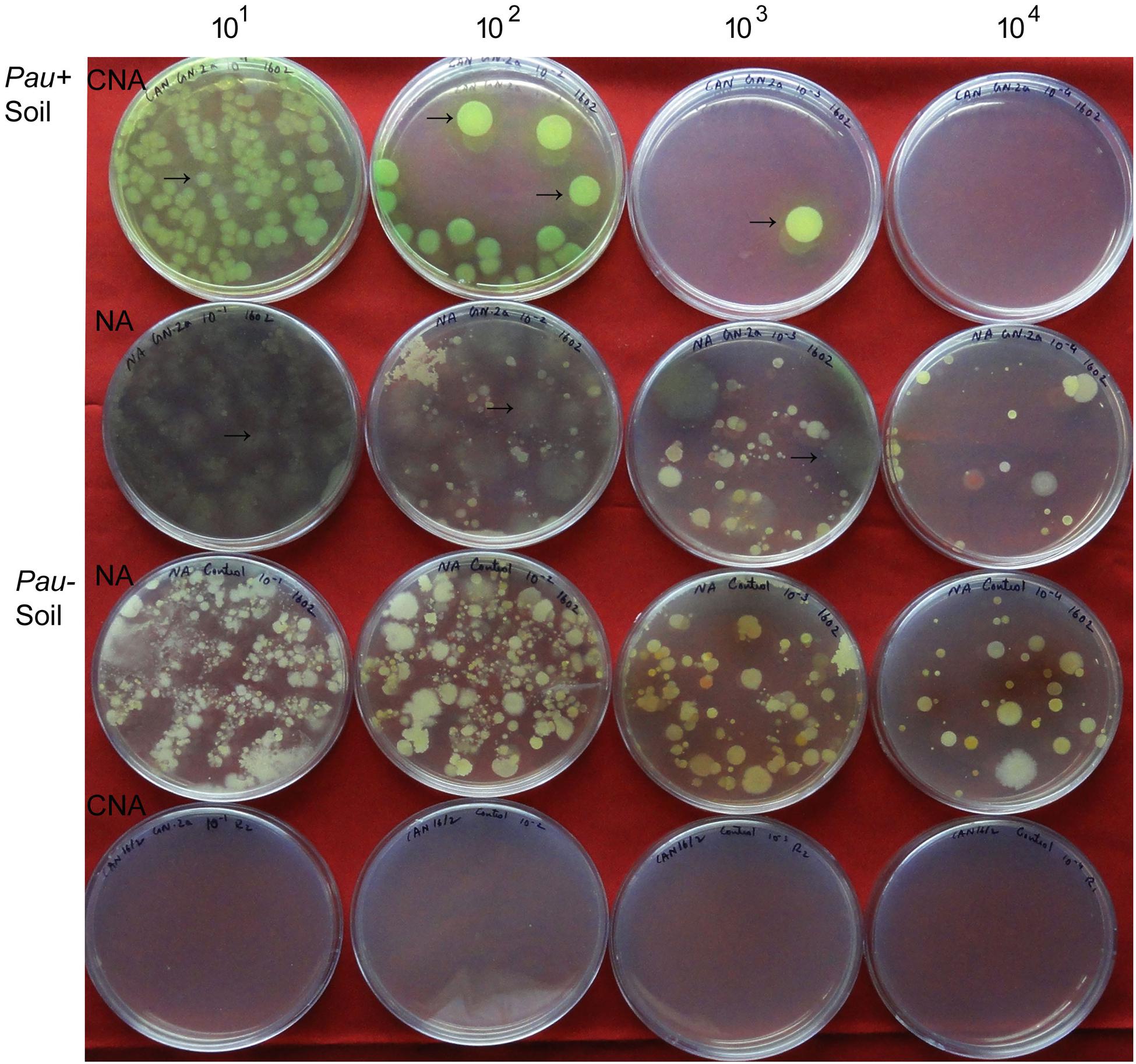
FIGURE 3. Plating of serial dilutions (101 to 104) of Pau+ and Pau- rhizospheric soil mix on CNA and nutrient agar (NA) at the start of the experiment (day-0) and documentation of Pau (→) and non-Pau cfu as on day-4 after plating (row 1: Pau colonies from Pau+ soil on CNA; row 2: Pau cfu on NA appearing brownish and vanishing gradually with evident non-Pau bacterial cfu; row 3: Pau- soil showing non-Pau colonies on NA; row 4: no colony growth on CNA from Pau- soil; Colony counts were taken on days 1, 2, and 4 with the marking of initially formed ones on the reverse of the plate).
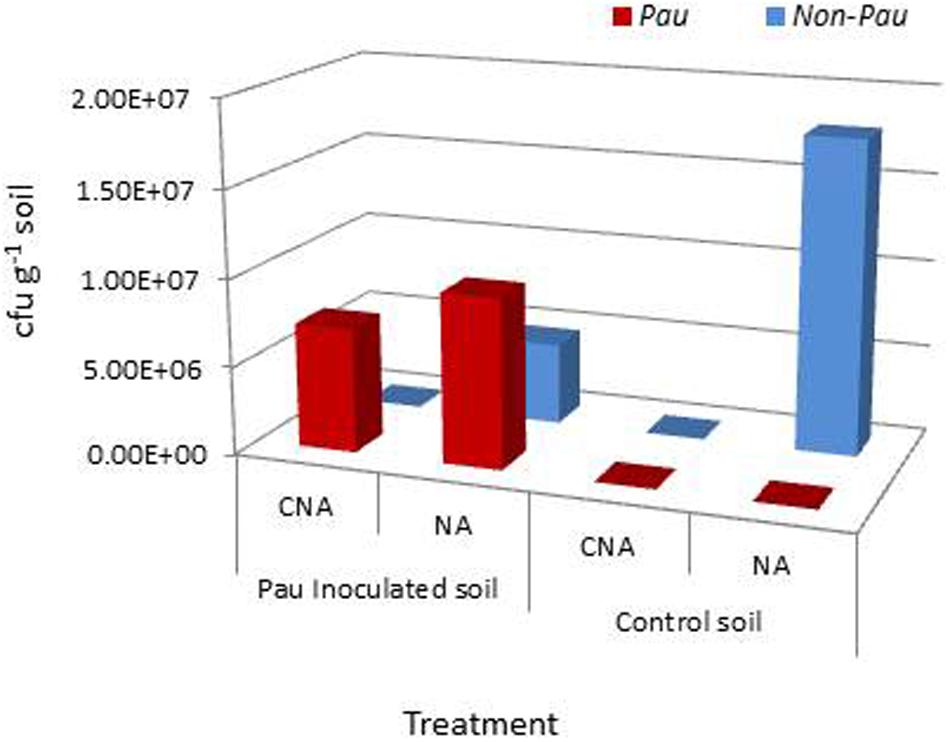
FIGURE 4. Extent of P. aeruginosa (Pau) and non-Pau bacterial cfu in potted rhizospheric soil mix within 30–60 min of Pau application as per the cfu estimations on the selective CNA medium and NA; (cumulative of cfu recorded on day-1 to day-4).
Monitoring the Pau+ and Pau- soils in the pots kept under glasshouse conditions on day-4 indicated a significant reduction in Pau cfu in Pau+ soil compared with the day-0 cfu registering 2.2 × 105 (-97%; P < 0.0001) and 2.5 × 105 cfu g-1 (-97.4%; P < 0.0001) on CNA and NA, respectively (Figure 5). On the other hand, the non-Pau cfu in Pau+ soil showed a rise to 1.3 × 107 g-1 (from day-0 cfu of 4.6 × 106 g-1; 182%; P < 0.0001). This indicated a gradual build-up of native bacterial community following the initial counter effect by Pau. The cfu of non-Pau organisms on NA from control soil (3.4 × 107 g-1) also showed a significant increase from day-0 (1.8 × 107 g-1; 93.3%; P = 0.0016) to day-4 indicating a surge in native bacterial community following the watering of dry soil which was commenced 1 day prior to the start of the trial.
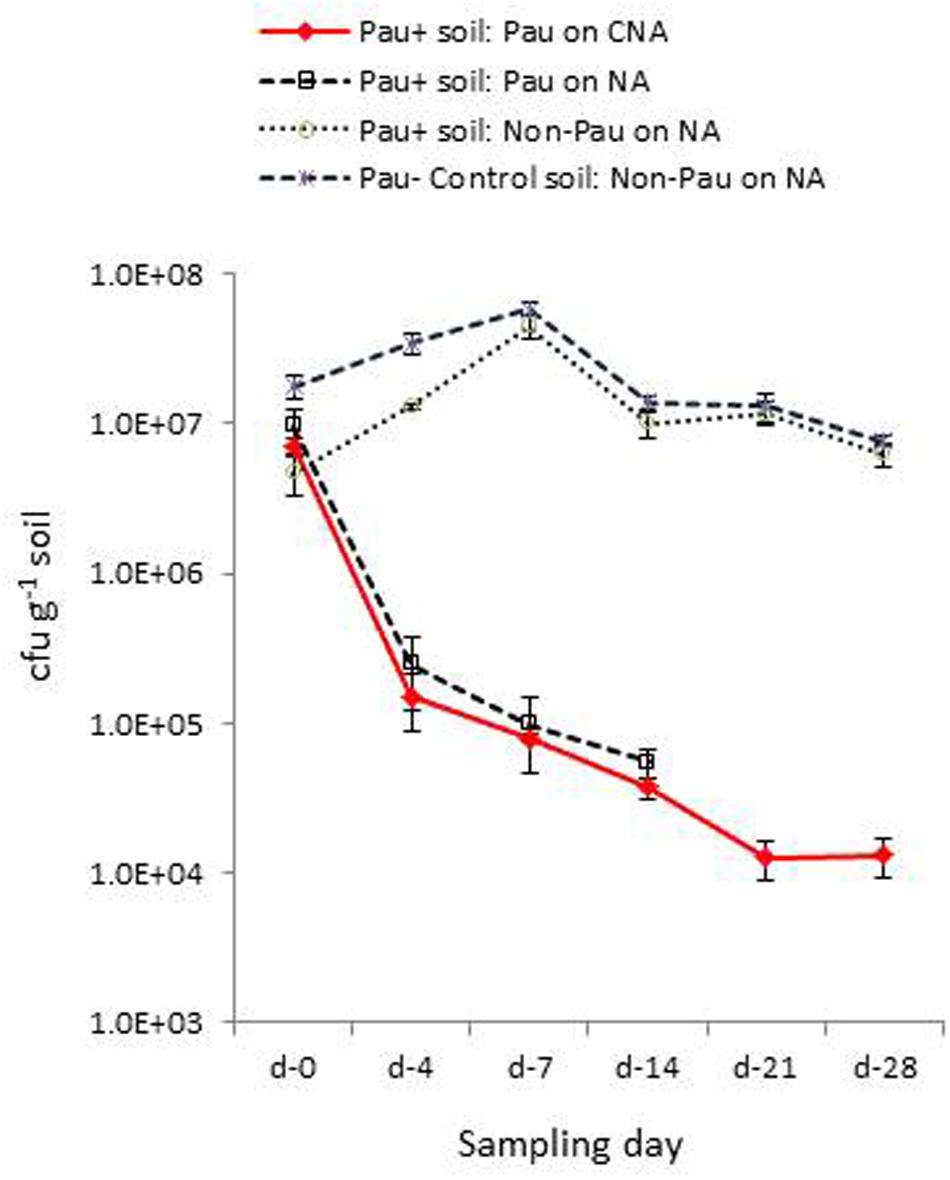
FIGURE 5. Extent of P. aeruginosa (Pau) and non-Pau bacterial cfu in potted Pau-inoculated (Pau+) and control (Pau-) rhizospheric soil mix on different sampling dates commencing from the date of Pau application (day-0) as per the cfu estimations on selective CNA and NA (cumulative of cfu recorded on day-1 to day-4; vertical bars indicate standard deviation).
The day-7 sampling indicated a further reduction in Pau cfu from day-4 cfu in Pau+ soil (7.9 × 104; P < 0.001, and 9.8 × 104 g-1; P = 0.002, respectively, on CNA and NA). The non-Pau cfu here showed a significant increase (4.5 × 107 g-1) over the day-0 (4.7 × 106 g-1; P < 0.001) and day-4 cfu (1.3 × 107 g-1; P < 0.001) indicating native bacterial community build up after the initial attack by Pau. The level attained here by day-7 (4.5 × 107 g-1) reached nearly close to that recorded for the un-inoculated soil (5.8 × 107 g-1; P = 0.05). The sampling done on day-14 indicated a further drop in Pau population which continued through day-21 and almost stabilized by day-28 (Figure 5). It was not feasible to document Pau cfu of Pau+ soil on NA at this stage due to the too few colonies which were masked by high non-Pau cfu. During this phase both the Pau+ and Pau- sets also displayed a gradual reduction in non-Pau cfu indicating stabilization of native bacterial community to similar cfu levels in both cases.
Monitoring the Pau+ soil after another 4 weeks with an intervening 4-weeks dry spell showed barely any Pau (1.0 × 102 to 2.0 × 102 cfu g-1) indicating poor survival of Pau. The native non-Pau population in both Pau+ and Pau- soils appeared comparable which was similar to the levels documented before the onset of 1 month dry spell (7.4 × 106 and 7.9 × 106, respectively; P = 0.66). The sampling 48 h after the rehydration of above soils showed a slightly more but no significant increase in Pau colonies in Pau+ soil (1.4 × 103 cfu g-1). It was striking to note that the cfu of non-Pau organisms capable of growing on CNA showed an increase in Pau+ soil (4.0 × 103 cfu g-1) while the Pau- soil did not show such colonies. In other words, the non-Pau organisms supported on CNA appeared more rampant in Pau+ soil than in un-inoculated soil.
The irrigating water did not show any Pau colonies on CNA during the periodic monitoring. The control soil sample also did not yield any Pau-like colonies on CNA but the Pau+ soil showed a few Pau-unlike colonies capable of growing on CNA during the 8 weeks monitoring. Identification of these distinct colony morphotypes confirmed them to be not Pau; these included P. plecoglossicida, P. monteilii, P. taiwanensis, and Achromobacter sp. detected in very few counts on CNA (0–4 cfu) applied with the 101 dilution of soil suspension. These were distinguishable from Pau colonies when grown together based on color and size on CNA and NA. Randomly selected supposedly Pau-colonies were confirmed to be so with 16S rRNA typing.
The trial assessing the contribution of soil wetting to the build-up of native bacterial cfu in dry soil and the relative contribution from the irrigation water indicated that the cfu hike in dry soil 24 h after the application of sterile water was mainly contributed by the activation of bacterial cells to cultivation and/or their multiplication than the direct contribution from irrigating water. This was evident from the cfu levels in the treatment inundated with irrigation-grade water (3 × 105 cfu ml-1) which showed a relatively lower amount of cfu increase (Figure 6). The rhizospheric soil of tomato showed identical cfu during day-0 and day-1 samplings indicating a microbially buffered condition therein.
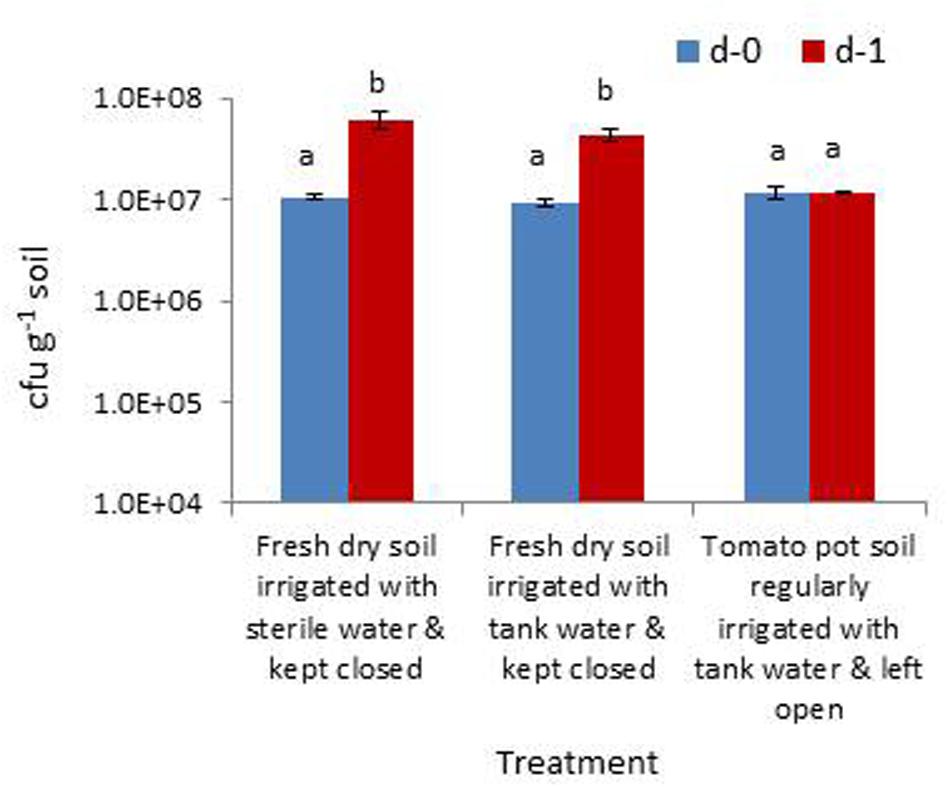
FIGURE 6. Assessing the extent of increase in gross bacterial cfu due to the moistening of dry rhizospheric soil mix with sterile water, the contribution from irrigating water and the monitoring of rhizospheric soil of tomato (Bars with same letters are not significantly different at P = 0.05).
The low survival of Pau in field soil and the adverse effect on soil bacterial community were verified in this repeat trial where the dry soil was watered to field capacity 1 day prior and applied with the Pau inoculum/sterile water as discussed above (Figure 7A). Monitoring of soil on day-0, day-4, and day-7 for Pau and non-Pau bacterial cfu on CNA and NA endorsed that (i), Pau was less fit to survive in soil, (ii), Pau application disturbed the native bacterial community instantly and (iii), the soil bacterial community showed a revival following the reduction in Pau population.
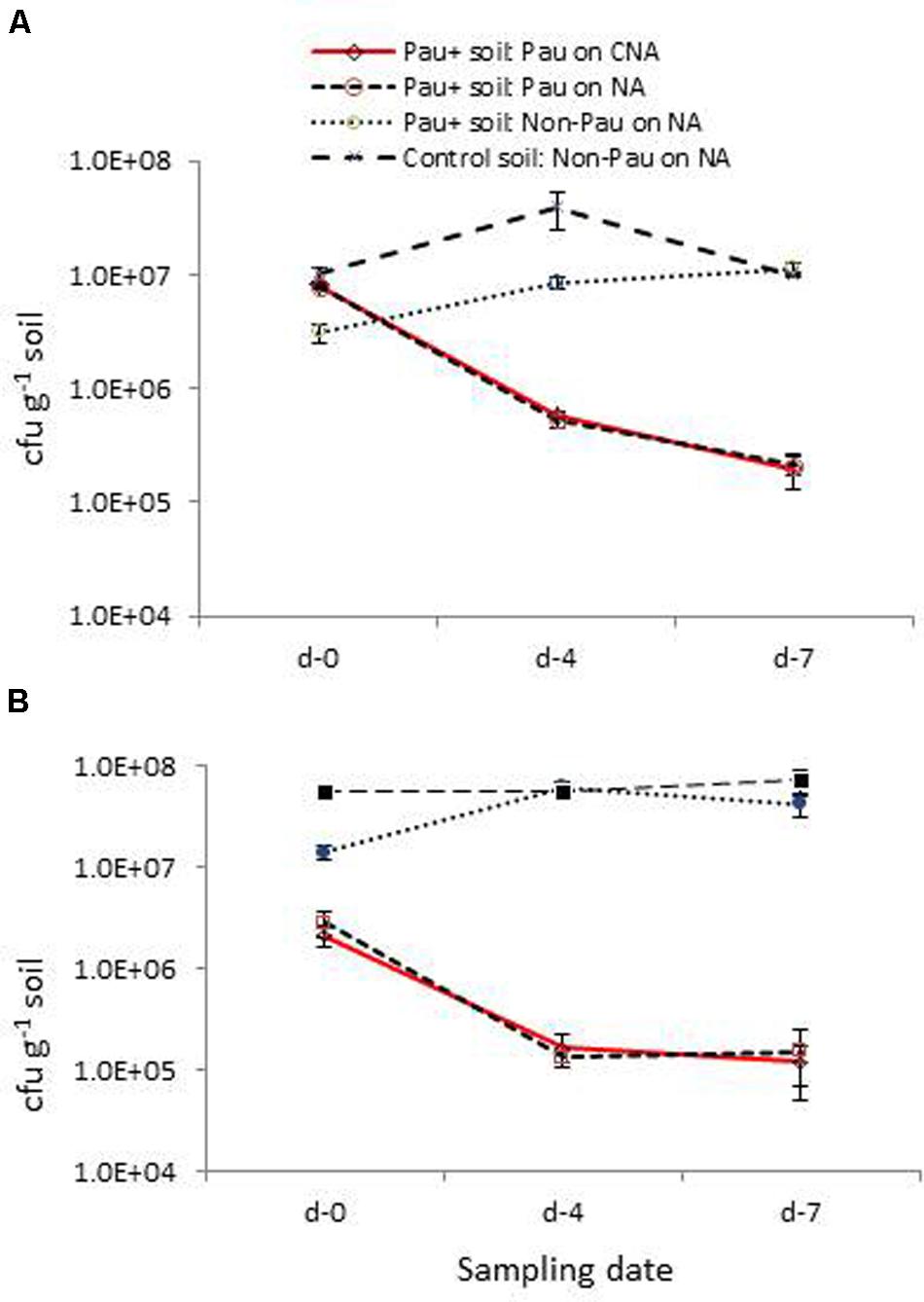
FIGURE 7. Validation trials employing fresh field soil (A) and micro-flora stabilized banana rhizospheric soil (B): extent of P. aeruginosa (Pau) and non-Pau organisms in potted Pau-inoculated (Pau+) and control (Pau-) field soils on different sampling dates commencing from the date of Pau application (day-0) as per the cfu estimations on CNA and NA (cumulative of cfu recorded on day-1 to day-4; vertical bars indicate standard deviation).
The parallel trial employing pots which were planted with banana ‘Grand Naine’ plants showed a similar pattern of significant reduction in Pau cfu within a weeks’ time as in the trial employing dry soils (Figure 7B). The native bacterial cfu in Pau+ soil also appeared significantly low compared with the respective figure for Pau- soil initially and then showed a gradual cfu build up as observed earlier. One striking observation in this trial was a stable gross bacterial cfu for Pau- rhizospheric soil from day-0 to day-7 in line with the observations on tomato rhizosphere soil indicating a stabilized condition when watering was practiced on a regular basis.
Monitoring of irrigation water confirmed that there was no lateral introduction of Pau. On the other hand, a significant share of non-Pau cfu and considerable diversity (25–30 distinct colony types) were observed in irrigation water. This explained the inoculum and diversity build-up in Pau+ soil after the initial adverse effect on native bacterial community by Pau. While the direct piped water from the irrigation source tank showed 4.3 × 104 to 7.6 × 104 cfu ml-1 with ≥15–20 diverse colony types, the stored water in the glasshouse in the open tank showed 1.3 × 105 to 3.2 × 105 cfu ml-1 with ≥25–30 colony types. This explained to some extent the variations in the non-Pau cfu and diversity detected in the Pau+ and Pau- soils on some sampling dates.
On the date of inoculation of axenic soil with Pau, CNA and NA registered 1.30 × 108 and 1.33 × 108 Pau-cfu g-1, respectively, (P > 0.05). During the monitoring over the next 1 week, a significant increase in Pau cfu was observed under the axenic conditions as monitored on CNA and NA (Figure 8). This amounted to 2.5×, 4.0×, 5.5× increase over the day-0 base cfu as on day-1, day-4, and day-7, respectively, on CNA (P < 0.001 in all instances on both CNA and NA). Thereafter, Pau cfu showed a gradual dip but the population was still higher than that of the base level (2.8×, 2.6×, and 2.4× on day-14, day-21, and day-28, respectively). The corresponding figures while monitoring the samples on NA were 2.3×, 4.5×, 6.0×, 3.4×, 3.3×, and 2.7×, respectively, over the day-4 cfu. The un-inoculated control soil did not show any colony growth ensuring the aseptic conditions following 3× autoclaving.
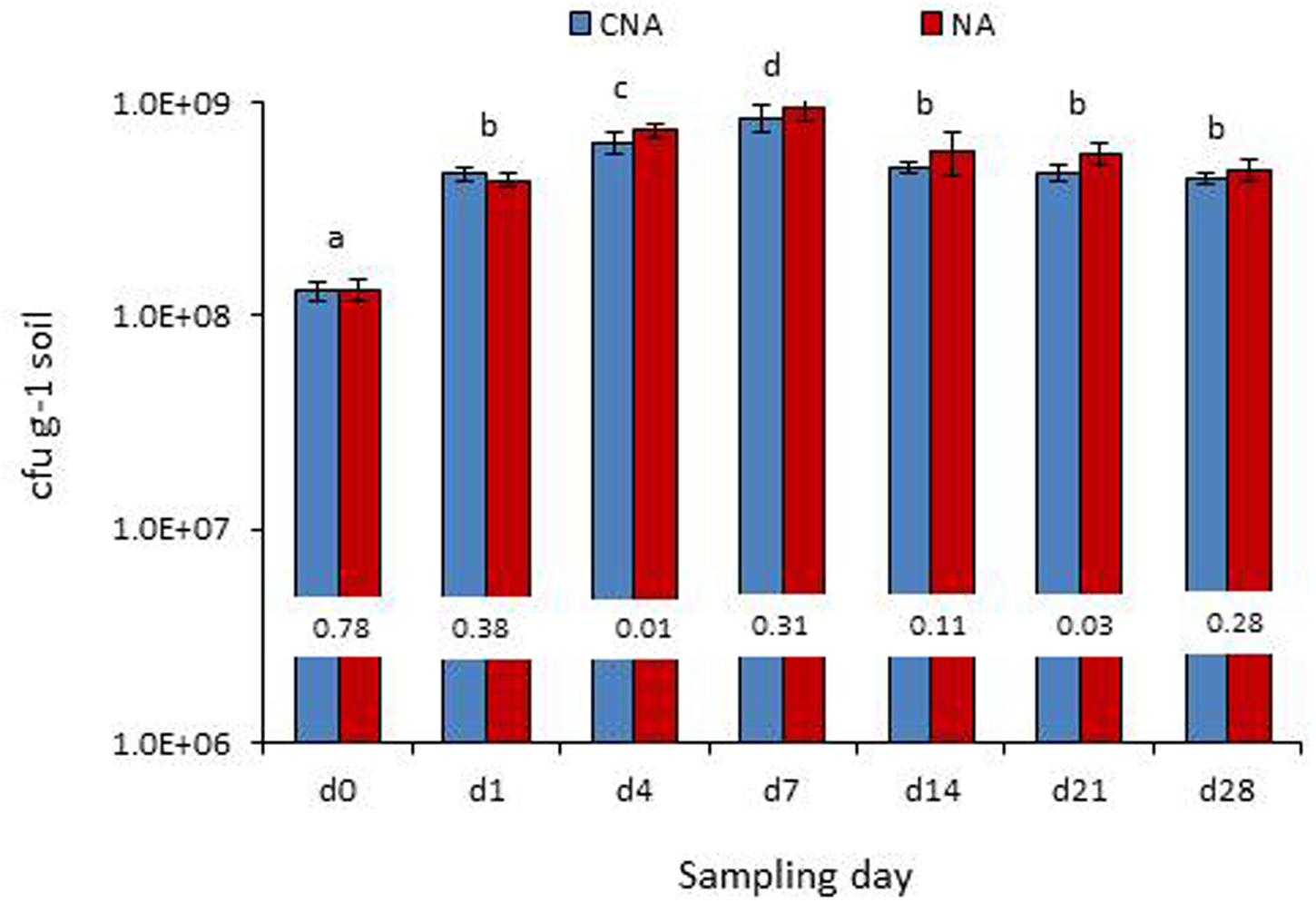
FIGURE 8. Monitoring of the P. aeruginosa (Pau) inoculated soil under axenic conditions (bottles which were maintained under axenic conditions) employing the selective CNA and NA on different dates from the initial inoculation (Bars with same letters are not significantly different at P = 0.05; Values on the bar indicate P-value at 0.05 between the Pau cfu on CNA and NA).
Both the sets of bottles (axenic-I and II) displayed similar Pau cfu at the start of the experiment (Table 1). The monitoring 2 days after the addition of SDW to set-1 showed an increase in Pau cfu by 4.6× over the cfu at the start of the experiment while set-II applied with soil suspension showed only 3.2× hike over control which was significantly low compared with the Pau+ soil. The non-Pau cfu was not assessable as the incorporation of soil suspension added only an estimated 2 × 104 cfu g-1 which was masked by Pau during the monitoring on NA plates. The monitoring of soil 4 h after the addition of NA-grown soil inoculum showed a 50% reduction in Pau cfu over the pre-application cfu (with the detection of a high non-Pau cfu). On the other hand, SDW-applied control set under axenic conditions showed a 43% increase in Pau cfu over the weighted average. The results overall indicated an adverse effect on established Pau cfu with the laterally introduced soil microbiome.
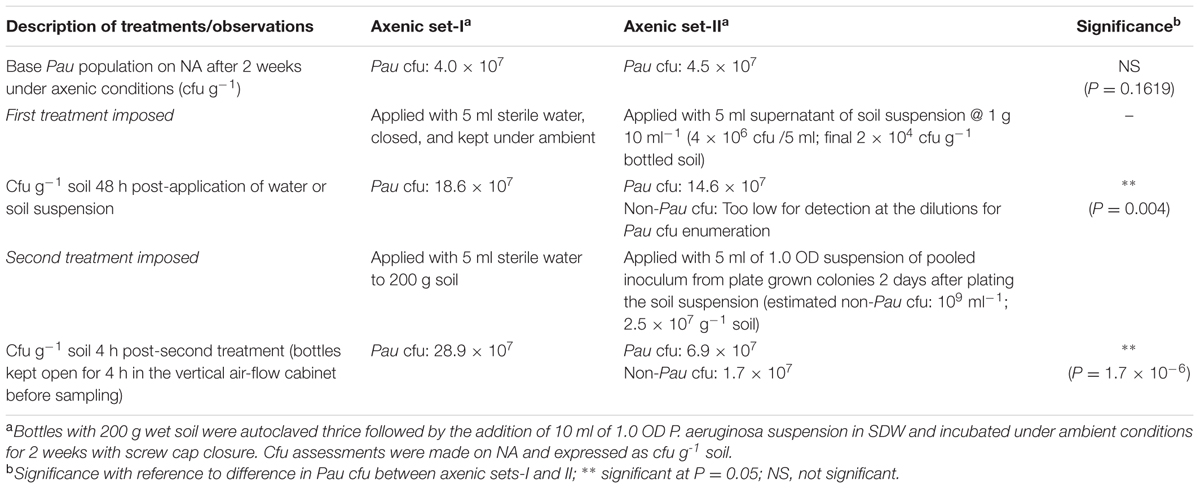
TABLE 1. Effect due to the lateral introduction of soil microbial suspension or soil derived bacterial inoculum on the population of Pseudomonas aeruginosa established in axenic soil for 2 weeksa.
The monitoring of the soil suspensions prepared on days-0, 4, 7 or 14, a week later indicated that Pau was counteracted by other organisms in the suspension significantly affecting its population levels (Figure 9). Soil suspension also offered the entire soil microbiome including bacterial, fungal, and protozoan mircocosms in cultivable and non-cultivable forms. The monitoring of pure Pau suspension under identical axenic conditions indicated that the organism was capable of surviving stationary incubation with no change in cfu. The non-Pau community showed a revival after the initial attack by Pau in the sample prepared on day-0. The suspensions from days-4, 7, and 14 sampling showed a less fitness for the survival of both Pau and non-Pau sets.
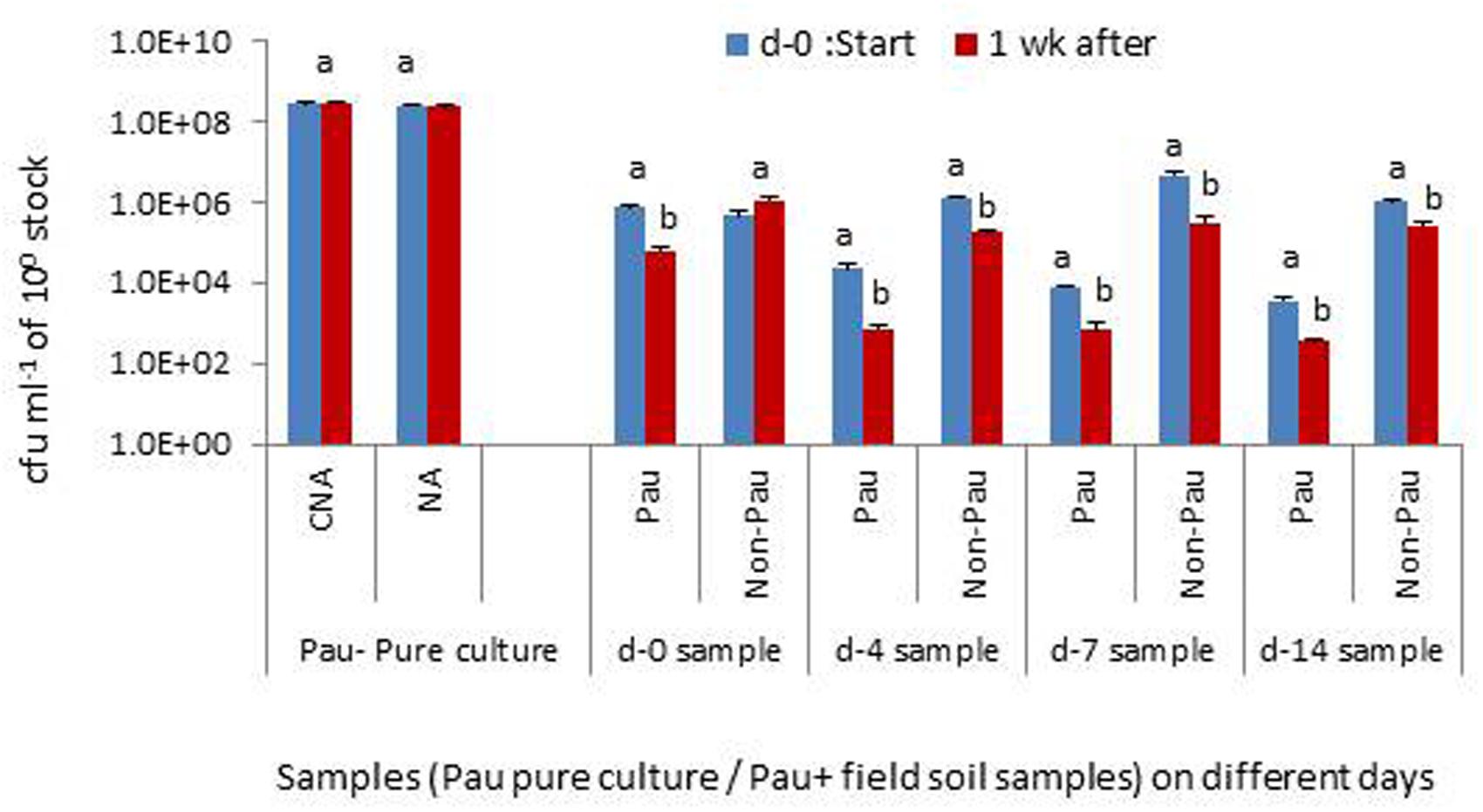
FIGURE 9. Testing the interactive effect of P. aeruginosa (Pau) with soil microbiota in suspension employing the soil suspensions prepared on days 0, 4, 7 or 14, with static incubation for 1 week after the preparation, in comparison with the pure culture employing CNA for Pau and NA for non-Pau estimations (100 stock refers to 0.1 OD anchored stock for the pure culture and 1 g per 10 ml for the soil suspension; Bars with different letters in a treatment set are significantly different at P = 0.05).
The control NA applied with Pau in the agar-well at the center of the plate showed growth covering almost 30–35 mm with a plate radius of 45 mm over 3–4 days. In dual culture plates, the growth of Pau varied from nil or negligible around the pit to quite active growth to the extent of 15–20 mm radius as documented after 3–4 days of application. A clear-zone was evident between Pau and the soil isolates in most instances. The extent of lawn formed by the soil strains in dual culture plates also varied significantly depending on the isolate ranging from full growth reaching the pit domineering over Pau to the extent of being pushed to the outer periphery. In extreme cases, there was no obvious lawn development at all. Based on the above observations, four different categories of responses were documented: (i), null effect, (ii), severe antagonistic effect by Pau on the soil isolate, (iii), severe anti-Pau effect displayed by the soil isolate, and (iv), mutual antagonistic effects (Supplementary Table S1).
The spot application of soil isolates on Pau lawn formed on NA after plating the 0.1 OD culture and allowing 1 h for Pau to establish wholly inhibited the growth of test isolates. No obvious colony growth was observed from the applied spots even after 3–4 days in all the 50 test isolates. Reducing the Pau cell population employing 0.1, 0.01, or 0.001 OD inoculums also showed proper lawn development. Use of 0.001 OD Pau-inoculum for lawn plating followed by immediate spotting of soil isolate using 0.1 or 1.0 OD inoculum showed the growth of the test organism at the applied spots at 1.0 OD in a few instances but not at 0.1 OD (Supplementary Table S1). Thus, the presence of Pau even at lower cell densities before the addition of challenge isolate appeared to inhibit their establishment under the nutrient rich conditions in the plates.
The present investigations have brought out valuable information on the effects due to the introduction of an agriculturally significant bacterium, P. aeruginosa, possessing multiple pathogen-antagonism potential on the native soil microbial community and the response of the soil microbiome to the introduced microorganism. Besides, it has added the much desired information on the survivability of an endophytic strain artificially inoculated into the field soil which is generally lacking. For an organism to be effective as a biocontrol or growth promoting agent in agriculture, it should be able to sustain under field soil-conditions in sufficient populations for a minimum duration (van Veen et al., 1997). It should not disturb the native microbial community drastically which otherwise be playing diverse significant roles in the rhizosphere (Hartmann et al., 2009; Trabelsi and Mhamdi, 2013). Contrary to this, P. aeruginosa proved to be not a competitive inoculant for soil and its application in agricultural soil instantly affected the non-target organisms, thereby disturbing the soil microbial community dynamics. The effect, however, was transient as the non- P. aeruginosa cfu showed restoration on par with control soil within 1 week. This perhaps was contributed by the multiplication of survivors or the addition of new organisms through water/air, with the reduction in the population of the inoculant. The present trials were carried out in pot cultures considering the feasibility of monitoring the inoculant and the soil microbiome unlike the open field but employing the rhizospheric soil with the natural soil associates.
Studies exploring the effects due to the introduced organism on soil bacterial or microbial community depict varying conclusions depending on the organism or the soil type (Herschkovitz et al., 2005; Chowdhury et al., 2013; Kröber et al., 2014; Schreiter et al., 2014; Gupta R. et al., 2015). Most of the above studies have relied on molecular approaches/metagenomics rather than cultivation which on one hand gave a wider community impact effect but did not involve verification with direct challenge assays. Both cultivation-based and cultivation-independent approaches have inherent advantages and disadvantages. In this report, the cultivable fraction was analyzed as a whole representative of the soil microcosm and it facilitated the documentation of four categories of interaction effects varying from severe antagonistic effect by P. aeruginosa on the soil isolate, severe anti- P. aeruginosa effect displayed by the soil isolate or mutual antagonistic effects to nil effects. We did not target the identification of different isolates as the soil and irrigation water contained multitude of organisms, the cultivable fraction constitutes a minor fraction of the microbial community and that the soil microbial communities vary with location and time.
Pseudomonas aeruginosa was employed in this study as a model system considering the wide range of antagonistic activity reported against various pathogens including bacteria (Maji and Chakrabartty, 2014; Spago et al., 2014) and fungi (Aravind et al., 2009; Allu et al., 2014) and pests like nematodes (Ali et al., 2002; Kumar et al., 2013). The bacterium is known to produce different antimicrobial compounds which include mainly phenazines (Anjaiah et al., 1998; Mavrodi et al., 2006; Jasim et al., 2014). The putative biocontrol agent exerted significant adverse effects on non-target native microbiome at variance from the conventional wisdom of nurturing the soil microbiome. It is likely that such responses vary with the aggressiveness of the inoculants or the extent of antimicrobial activity. Considering the spectrum of antagonistic effects by P. aeruginosa cited above, it could be viewed as an aggressive species to which the soil community perhaps needed to respond likewise. This proposal is hypothetical at this stage but forms an aspect for future research. The observations in this study emphasize the need for pre-testing the bio-inoculant for possible adverse effects on native microbial community before recommending it as a potential biocontrol agent, particularly for organisms with broad anti-microbial activity.
Pseudomonas aeruginosa, in this study, proved to be a poor survivor in agricultural field soil. The sustenance and proliferation of the strain under gnotobiotic conditions but poor survival in field soil suggested that this was not a mere fitness issue but mostly due to the antagonistic effects by the native microbial community. This was endorsed by the observations from the lateral introduction of soil-microbiome in Pau-established axenic soil and the interactive effects of Pau in soil suspension both of which involved the gross microbial community including cultivable and non-cultivable microorganisms. The direct confrontation assays in agar plates also endorsed the significant role of microbe–microbe interactions though it involved a representative group of cultivable environmental bacteria. The soil microbial community includes bacteria, fungi, and viruses. The introduced organism faces challenge due to the antagonistic effects or competition from native microorganisms and is vulnerable to predation by protozoa which could affect its survivability (England et al., 1993; van Veen et al., 1997; Trabelsi and Mhamdi, 2013; Tyc et al., 2014, 2015).
Besides the above biotic factors, the environmental fitness of the introduced organisms in soil is also influenced by several abiotic factors such as soil type, pH, temperature, water content, periodicity of wetting and drying, and soil constituents including organic carbon and mineral nutrition (Acea et al., 1988; van Veen et al., 1997; Bakker et al., 2013; Trabelsi and Mhamdi, 2013; Tyc et al., 2014, 2015). The inoculant is also vulnerable to desiccation. Considering that the same soil-mix was employed for axenic studies and that P. aeruginosa monitoring in field soil was carried out in pot trials under glasshouse conditions where there was a better control over watering and edaphic factors, the observations suggest that the poor survival of P. aeruginosa under field conditions was governed by interactions with the soil microbiome rather than due to environmental factors. The situation in the field may be still harsher with alternating spells of wetting and drying or prolonged dry/wet spells compared to the controlled conditions in the glasshouse.
The low fitness of P. aeruginosa strain employed in this study did not appear to arise from its endophytic origin as the same has been reported with environmental strains too. Weir et al. (1996) observed a 100-fold reduction in the cfu of P. aeruginosa in dry soil inoculated with rifampicin-resistant environmental strain UG2Lr over 3 weeks in comparison with cells encapsulated in dry alginate beads before application. A significant interaction effect between P. aeruginosa and the soil community was documented in wheat rhizosphere that the invasibility was inversely related to the level of native microbial diversity (Matos et al., 2005). Adopting cultivation-based as well as molecular monitoring, P. aeruginosa was not detected in most native agricultural soils but sparsely observed in manure-amended soils although the bacterium showed good survival in organic-manures (Deredjian et al., 2014). Further, employing clinical and environmental strains of P. aeruginosa, the introduced organism showed a decline to below detectable levels after 3–5 weeks under non-sterile microcosms while the population was maintained high under sterilized microcosms (Deredjian et al., 2014). It may be argued that an endophytic strain is more suitable for plant colonization. The ‘GNS.13.2a’ strain showed colonization of banana roots. However, it is imperative that endophytic microorganisms go through a phase in the soil or environment at the end of the life of the plant or the organ. Besides, soil drenching of inoculum constitutes the best form of application toward biocontrol of soil-borne pathogens. Decline in the populations of introduced bacteria in soils have been documented for a host of organisms irrespective of the source of their original isolation (van Veen et al., 1997). The form in which endophytes survive intra-plant or in soil may be different from the nutrient rich monoculture during in vitro growing.
There is a general criticism that the significant pathogen and pest antagonistic effects displayed by the test organisms in the laboratory trials are not always translated to successful biocontrol strategies in the field (Acea et al., 1988; van Veen et al., 1997; Upreti and Thomas, 2015). In effect, the application of Pau in field soil did not serve any net beneficial effect in terms of its sustained survival. It rather caused an unwarranted transient disturbance in the soil community dynamics which otherwise was at harmony as observed with the undisturbed banana and tomato control rhizospheric soils. The population of surviving Pau in field soil applied with nearly 107 cfu g-1 reached barely 0.01–0.025% of the initial population within 1 week in the instance of immediately wetted soil, and about 0.05% level for the microbially buffered banana rhizospheric soil. Such low population level in soil would perhaps be insufficient to offer a formidable and sustained protection against pathogens which survive in the field even under harsh conditions.
It is pertinent to mention that a vast majority of trials recommending the use of pest/pathogen antagonistic organisms are based on mere laboratory assays with no systematic monitoring of the survival of the organism or the effect due to the introduced organism on the native microorganisms. The observations here suggest the need for investigations on soil survival and microbe–microbe interaction effects before recommending the commercial adoption of bio-inoculants as the preservation and nurturing of native flora is also important in sustainable agriculture. The significance of microbe–microbe interactions under field conditions has not received much attention which is being gradually recognized now (Trabelsi and Mhamdi, 2013; Tyc et al., 2014, 2015). This also calls for the detailed analysis of microbe–microbe interactions while formulating microbial consortia rather than mere compatibility testing in nutrient plates and also testing the combined effect of the microbial consortium on soil microbiome.
Pseudomonas aeruginosa strains are known to be a human pathogen (Alonso et al., 1999; Morita et al., 2014). Therefore, it does not form a choice candidate toward biocontrol or plant growth promotion applications unless it is vividly established that the isolates infecting humans or animals and the plant/soil associated isolates are different and the possibility of horizontal gene transfers are low. The study by Kumar et al. (2013) showed the uniqueness of the endophytic isolate from pepper from other clinical isolates but the former also possessed the virulent genes as the clinical strains. A high degree of genomic conservation between P. aeruginosa isolates from diverse environments including clinical strains has been documented which raises concerns about the usage of this organism in agriculture (Grosso-Becerra et al., 2014). Therefore, authentic information on the relatedness between the plant isolates and human opportunistic pathogenic isolates, effect on native endophytes and the possible transmission to different plant parts all need to be considered before recommending Pau in agricultural applications. P. aeruginosa is also reported as a plant pathogen (Walker et al., 2004).
Reports highlighting the benefits of P. aeruginosa in terms of plant growth promotion and antagonistic effects on phytopathogens and pests and recommending the organism in agriculture are continuing to emerge as per the recent research publications cited. These studies do not mention about the monitoring of the survival and the effects on native microflora nor do they consider the potential hazards to human or animal systems. Whether the beneficial effects arise from the endophytic colonization by P. aeruginosa and any consequential effects on the endophytic microbiome need further investigations. The potential of transmission of the inoculant to banana fruits which is consumed in fresh form is another aspect for future research. Such investigations would be strengthened with the adoption of metagenome based analyses (Knief, 2014) facilitating interaction studies between the introduced strain and native microbiome covering cultivable and non-cultivable communities.
Pseudomonas aeruginosa proved to be a poor survivor in agricultural soil with a quick decline in the cfu of the inoculant within a week while it showed survival and proliferation under axenic conditions. Its application in the rhizospheric soil caused an unwarranted disturbance to the native soil bacterial community which in turn fought back and showed restoration of population on par with the control soil within this time span. The observations explain the cause of poor translation of some of the laboratory results ascribable to the poor survival of the inoculant in agricultural soil and highlight the need for monitoring the sustenance and performance of the introduced organism under field soil conditions which would be different from the nutrient-rich conditions during in vitro assays. It also pinpoints the essentiality to assess the interactive effects of the bio-inoculant with native microbial community and the plausible adverse effects on resident soil flora before commercial recommendation.
The experiments were planned and executed together by PT and AS. PT undertook the data analysis, interpretation, and manuscript preparation. This publication bears the Institute contribution No. 38/2015.
The study was funded partly under the ICAR-AMAAS Net-work project “Exploration of the endophytic microbial diversity in horticultural crops through metagenomics and cultivation” funded through the ICAR-National Bureau of Agriculturally Important Microorganisms, Mau Nath Bhanjan, Uttar Pradesh, India and partly by ICAR- Indian Institute of Horticultural Research, Bengaluru, India.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The help by Shaik Sadiq Pasha during the conduct of some of the trials is gratefully acknowledged. The authors thank Dr. T. P. Rajendran, former Officer on Special Duty/Acting Director, ICAR-National Institute of Biotic Stress Management, Raipur, India for the critical reading of the manuscript and the suggestions.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00493
FIGURE S1 | Testing of media formulations for the selective monitoring of Pseudomonas aeruginosa (Pau) by spotting six serial dilutions of the socks in a SP-SDS format employing pure culture of Pau (A), irrigation water (B), field soil (C), irrigation water mixed with Pau (D) and field soil sample mixed with Pau (E). NA, Kan+TTC NA and CNA represent nutrient agar, formulation as per Kumar et al. (2013) and CNA as per Goto and Enomoto (1970), respectively; f > indicates fungal colony growth.
TABLE S1 | Testing the interactive effects between banana endophytic strain of Pseudomonas aeruginosa with soil bacterial isolates in agar plate assays.
CNA, Cetrimide- nalidixic acid- agar medium; Kan+TTC:NA medium, NA containing kanamycin and 2,3,5-triphenyl tetrazolium chloride; NA, nutrient agar; Pau, Pseudomonas aeruginosa banana strain ‘GNS13.2a’; SATS, spotting- and- tilt- spreading; SDW, sterile distilled water; SP-SDS, single plate – serial dilution spotting.
Acea, M. J., Moore, C. R., and Alexander, M. (1988). Survival and growth of bacteria introduced into soil. Soil. Biol. Biochem. 20, 509–515. doi: 10.1016/0038-0717(88)90066-1
Ali, N. I., Siddiqui, I. A., Shaukat, S. S., and Zaki, M. J. (2002). Nematicidal activity of some strains of Pseudomonas spp. Soil. Biol. Biochem. 34, 1051–1058. doi: 10.1016/S0038-0717(02)00029-9
Allu, S., Kumar, N. P., and Audipudi, A. V. (2014). Isolation, biochemical and PGP characterization of endophytic Pseudomonas aeruginosa isolated from chilli red fruit antagonistic against chilli anthracnose disease. Int. J. Curr. Microbiol. Appl. Sci. 3, 318–329.
Alonso, A., Rojo, F., and Martínez, J. L. (1999). Environmental and clinical isolates of Pseudomonas aeruginosa show pathogenic and biodegradative properties irrespective of their origin. Environ. Microbiol. 1, 421–430. doi: 10.1046/j.1462-2920.1999.00052.x
Anjaiah, V., Koedam, N., Nowark-Thompson, B., Loper, J. E., Hofte, M., Tambong, J. T., et al. (1998). Involvement of phenazines and anthranilate in the antagonism of Pseudomonas aeruginosa PNA1 and Tn5 derivatives toward Fusarium spp. and Pythium spp. Mol. Plant Microb. Interact. 11, 847–854. doi: 10.1094/MPMI.1998.11.9.847
Aravind, R., Kumar, A., Eapen, S. J., and Ramana, K. V. (2009). Endophytic bacterial flora in root and stem tissues of black pepper (Piper nigrum L.) genotype: isolation, identification and evaluation against Phytophthora capsici. Lett. Appl. Microbiol. 48, 58–64. doi: 10.1111/j.1472-765X.2008.02486.x
Bakker, P. A. H. M., Berendsen, R. L., Doornbos, R. F., Wintermans, P. C. A., and Pieterse, C. M. J. (2013). The rhizosphere revisited: root microbiomics. Front. Plant. Sci. 4:165. doi: 10.3389/fpls.2013.00165
Chowdhury, S. P., Dietel, K., Rändler, M., Schmid, M., Junge, H., Borriss, R., et al. (2013). Effects of Bacillus amyloliquefaciens fzb42 on lettuce growth and health under pathogen pressure and its impact on the rhizosphere bacterial community. PLoS ONE 8:e68818. doi: 10.1371/journal.pone.0068818
Deredjian, A., Colinon. C., Hien, E., Brothier, E., Youenou, B., Cournoyer, B., et al. (2014). Low occurrence of Pseudomonas aeruginosa in agricultural soils with and without organic amendment. Front. Cell. Infect. Microbiol. 4:53. doi: 10.3389/fcimb.2014.00053
England, L. S., Lee, H., and Trevors, J. T. (1993). Bacterial survival in soil: effect of clays and protozoa. Soil. Biol. Biochem. 25, 525–531. doi: 10.1016/0038-0717(93)90189-I
Epstein, S. S. (2013). The phenomenon of microbial uncultivability. Curr. Opin. Microbiol. 16, 636–642. doi: 10.1016/j.mib.2013.08.003
Gaiero, J. R., McCall, C. A., Thompson, K. A., Day, N. J., Best, A. S., and Dunfield, K. E. (2013). Inside the root microbiome: bacterial root endophytes and plant growth promotion. Am. J. Bot. 100, 1738–1750. doi: 10.3732/ajb.1200572
Goto, S., and Enomoto, S. (1970). Nalidixic acid cetrimide agar: a new selective plating medium for the selective isolation of Pseudomonas aeruginosa. Jpn. J. Microbiol. 14, 65–72. doi: 10.1111/j.1348-0421.1970.tb00492.x
Grosso-Becerra, M. V., Santos-Medellín, C., González-valdez, A., Méndez, J. L., Delgado, G., Morales-Espinosa, R., et al. (2014). Pseudomonas aeruginosa clinical and environmental isolates constitute a single population with high phenotypic diversity. BMC Genomics 15:318. doi: 10.1186/1471-2164-15-318
Gupta, G., Gupta, A. G., and Jha, P. N. (2015). Biocontrolling and colonization potential of Pseudomonas aeruginosa PM389 Isolated from Pennisetum glaucum under field conditions. Int. J Res. Emerg. Sci. Technol. 2, 104–116.
Gupta, G., Panwar, J., and Jha, P. N. (2013). Natural occurrence of Pseudomonas aeruginosa, a dominant cultivable diazotrophic endophytic bacterium colonizing Pennisetum glaucum (L.) R. Br. Appl. Soil. Ecol. 64, 252–261. doi: 10.1016/j.apsoil.2012.12.016
Gupta, R., Bisaria, V. S., and Sharma, S. (2015). Effect of agricultural amendments on Cajanus cajan (pigeon pea) and its rhizospheric microbial communities – a comparison between chemical fertilizers and bioinoculants. PLoS ONE 10:e0132770. doi: 10.1371/journal.pone.0132770
Hallmann, J., Quadt-Hallmann, A., Mahaffee, W. F., and Kloepper, J. W. (1997). Bacterial endophytes in agricultural crops. Can. J. Microbiol. 43, 895–914. doi: 10.1139/m97-131
Hardoim, P. R., van Overbeek, L. S., Berg, G., Pirttilä, A. M., Compant, S., Campisano, A., et al. (2015). The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 79, 293–320. doi: 10.1128/MMBR.00050-14
Hartmann, A., Schmid, M., van Tuinen, D., and Berg, G. (2009). Plant-driven selection of microbes. Plant Soil. 321, 235–257. doi: 10.1007/s11104-008-9814-y
Herschkovitz, Y., Lerner, A., Davidov, Y., Rothballer, M., Hartmann, A., Okon, Y., et al. (2005). Inoculation with the plant-growth-promoting rhizobacterium Azospirillum brasilense causes little disturbance in the rhizosphere and rhizoplane of maize (Zea mays). Microb. Ecol. 50, 277–288. doi: 10.1007/s00248-004-0148-x
Jasim, B., Anisha, C., Rohini, S., Kurian, J. M., Jyothis, M., and Radhakrishnan, E. K. (2014). Phenazine carboxylic acid production and rhizome protective effect of endophytic Pseudomonas aeruginosa isolated from Zingiber officinale. World J. Microbiol. Biotechnol. 30, 1649–1654. doi: 10.1007/s11274-013-1582-z
Knief, C. (2014). Analysis of plant microbe interactions in the era of next generation sequencing technologies. Front. Plant Sci. 5:216. doi: 10.3389/fpls.2014.00216
Kröber, M., Wibberg, D., Grosch, R., Eikmeyer, F. G., Verwaijen, B., Chowdhury, S. P., et al. (2014). Effect of the biocontrol strain Bacillus amyloliquefaciens FZB42 on the microbial community in the rhizosphere of lettuce under field conditions analyzed by whole metagenome sequencing. Front. Microbiol. 5:252. doi: 10.3389/fmicb.2014.00252
Kumar, A., Munder, A., Aravind, R., Eapen, S. J., Tummler, B., and Raaijmakers, J. M. (2013). Friend or foe: genetic and functional characterization of plant endophytic Pseudomonas aeruginosa. Environ. Microbiol. 15, 764–779. doi: 10.1111/1462-2920.12031
Kumar, U., Aggarwal, C., Paul, S., and Annapurna, K. (2014). Endophytes as biocontrol agents of plant pathogens and insects. Kavaka 41, 92–95.
Lakshmi, V., Kumari, S., Singh, A., and Prabha, C. (2015). Isolation and characterization of deleterious Pseudomonas aeruginosa KC1 from rhizospheric soils and its interaction with weed seedlings. J. King Saud Univ. Sci. 27, 113–119. doi: 10.1016/j.jksus.2014.04.007
Maji, S., and Chakrabartty, P. K. (2014). Biocontrol of bacterial wilt of tomato caused by Ralstonia solanacearum by isolates of plant growth promoting rhizobacteria. Aust. J. Crop Sci. 8, 208–214.
Matos, A., Kerkhof, L., and Garland, J. L. (2005). Effects of microbial community diversity on the survival of Pseudomonas aeruginosa in the wheat rhizosphere. Microb. Ecol. 49, 257–264. doi: 10.1007/s00248-004-0179-3
Mavrodi, D. V., Blankenfeldt, W., and Thomashow, L. S. (2006). Phenazine compounds in fluorescent Pseudomonas spp. biosynthesis and regulation. Annu. Rev. Phytopathol. 44, 417–445. doi: 10.1146/annurev.phyto.44.013106.145710
Morita, Y., Tomida, J., and Kawamura, Y. (2014). Responses of Pseudomonas aeruginosa to antimicrobials. Front. Microbiol. 4:22. doi: 10.3389/fmicb.2013.00422
Ogram, A., Castro, H., and Chauhan, A. (2007). “Methods of soil microbial community analysis” in Manual of Environmental Microbiology, eds C. J. Hurst, R. L. Crawford, J. L. Garland, D. A. Lipson, A. L. Mills, L. D. Stetzenbach (Washington, DC: ASM Press), 652–662.
Pandey, P. K., Yadav, S. K., Singh, A., Sarma, B. K., Mishra, A., and Singh, H. B. (2012). Cross-species alleviation of biotic and abiotic stresses by the endophyte Pseudomonas aeruginosa PW09. J. Phytopath. 160, 532–539. doi: 10.1111/j.1439-0434.2012.01941.x
Ploetz, R., Thomas, J. E., and Slabaugh, W. R. (2003). “Diseases of banana and plantains,” in Diseases of Tropical Fruit Crops, ed. R. C. Ploetz (Wallingford: CABI Publishing, CABI International), 73–134. doi: 10.1079/9780851993904.0073
Prakash, O., Shouche, Y., Jangid, K., and Kostka, J. E. (2013). Microbial cultivation and the role of microbial resource centers in the omics era. Appl. Microbiol. Biotechnol. 97, 51–62. doi: 10.1007/s00253-012-4533-y
Puri, A., Padda, K. P., and Chanway, C. P. (2015). Can a diazotrophic endophyte originally isolated from lodgepole pine colonize an agricultural crop (corn) and promote its growth? Soil Biol. Biochem. 89, 210–216. doi: 10.1016/j.soilbio.2015.07.012
Ryan, R. P., Germaine, K., Franks, A., Ryan, D. J., and Dowling, D. N. (2008). Bacterial endophytes: recent development and applications. FEMS Microbiol. Lett. 278, 1–9. doi: 10.1111/j.1574-6968.2007.00918.x
Schreiter, S., Ding, G. C., Grosch, R., Kropf, S., Antweiler, K., and Smalla, K. (2014). Soil type-dependent effects of a potential biocontrol inoculant on indigenous bacterial communities in the rhizosphere of field-grown lettuce. FEMS Microbiol. Ecol. 90, 718–730. doi: 10.1111/1574-6941.12430
Sekhar, A. C., and Thomas, P. (2015). Isolation and identification of shoot-tip associated endophytic bacteria from banana cv. Grand Naine and testing for antagonistic activity against Fusarium oxysporum f. sp. cubense. Am. J. Plant Sci. 6, 943–954. doi: 10.4236/ajps.2015.67101
Spago, F. R., Ishii Mauro, C. S., Oliveira, A. G., Berangera, J. P. O., Celya, M. V. T., Stanganellia, M. M., et al. (2014). Pseudomonas aeruginosa produces secondary metabolites that have biological activity against plant pathogenic Xanthomonas species. Crop Prot. 62, 46–54. doi: 10.1016/j.cropro.2014.04.011
Steward, G. F., and Rappe, M. S. (2007). What’s the ‘meta’ with metagenomics? ISME J. 1, 100–102. doi: 10.1038/ismej.2007.25
Stover, R. H. (1962). Fusarial Wilt (Panama disease) of Bananas and Other Musa species. Kew: Commonwealth Mycological Institute.
Thomas, P., and Reddy, M. K. (2013). Microscopic elucidation of abundant endophytic bacteria colonizing the cell wall – plasma membrane peri-space in the shoot-tip tissue of banana. AoB Plants 5:plt011. doi: 10.1093/aobpla/plt011
Thomas, P., and Sekhar, A. C. (2014). Live cell imaging reveals extensive intracellular cytoplasmic colonization of banana genotypes by normally non-cultivable endophytic bacteria. AoB Plants 6:plu002. doi: 10.1093/aobpla/plu002
Thomas, P., Sekhar, A. C., and Mujawar, M. M. (2012). Non-recovery of varying proportions of viable bacteria during spread-plating governed by the extent of spreader usage and proposal for an alternate spotting-spreading approach to maximize the CFU. J. Appl. Microbiol. 113, 339–350. doi: 10.1111/j.1365-2672.2012.05327.x
Thomas, P., Sekhar, A. C., Upreti, R., Mujawar, M. M., and Pasha, S. S. (2015). Optimization of ‘single plate - serial dilution spotting’ (SP-SDS) with sample anchoring as an assured method for bacterial and yeast cfu enumeration and single colony isolation from diverse samples. Biotechnol. Rep. 8, 45–55. doi: 10.1016/j.btre.2015.08.003
Thomas, P., and Soly, T. A. (2009). Endophytic bacteria associated with growing shoot tips of banana (Musa sp.) cv. Grand Naine and the affinity of endophytes to the host. Microb. Ecol. 58, 952–964. doi: 10.1007/s00248-009-9559-z
Thomas, P., and Upreti, R. (2015). Evaluation of tomato seedling root-associated bacterial endophytes towards organic seedling production. Org. Agric. 1–10. doi: 10.1007/s13165-015-0111-9
Trabelsi, D., and Mhamdi, R. (2013). Microbial inoculants and their impact on soil microbial communities: a review. BioMed. Res. Int. 2013:863240. doi: 10.1155/2013/863240
Turner, T. R., James, E. K., and Poole, P. S. (2013). The plant microbiome. Gen. Biol. 14, 209. doi: 10.1186/gb-2013-14-6-209
Tyc, O., van den Berg, M., Gerards, S., van Veen, J. A., Raaijmakers, J. M., de Boer, W., et al. (2014). Impact of interspecific interactions on antimicrobial activity among soil bacteria. Front. Microbiol. 5:567. doi: 10.3389/fmicb.2014.00567
Tyc, O., Wolf, A. B., and Garbeva, P. (2015). The effect of phylogenetically different bacteria on the fitness of Pseudomonas fluorescens in sand microcosms. PLoS ONE 10:e0119838. doi: 10.1371/journal.pone.0119838
Upreti, R., and Thomas, P. (2015). Root-associated bacterial endophytes from Ralstonia solanacearum resistant and susceptible tomato cultivars and their pathogen antagonistic effects. Front. Microbiol. 6:255. doi: 10.3389/fmicb.2015.00255
van Veen, J. A., van Overbeek, L. S., and van Elsas, J. D. (1997). Fate and activity of microorganisms introduced into soil. Microbiol. Mol. Biol. Rev. 61, 121–135.
Walker, T. S., Bais, H. P., Déziel, E., Schweizer, H. P., Rahme, L. G., Fall, R., et al. (2004). Pseudomonas aeruginosa-plant root interactions. Pathogenicity, biofilm formation and root exudation. Plant Physiol. 134, 320–331. doi: 10.1104/pp.103.027888
Weir, S. C., Lee, H., and Trevors, J. T. (1996). Survival of free and alginate-encapsulated Pseudomonas aeruginosa UG2Lr in soil treated with disinfectants. J. Appl. Bacteriol. 80, 19–25. doi: 10.1111/j.1365-2672.1996.tb03184.x
Yasmin, S., Hafeeza, F. Y., and Rasul, G. (2014). Evaluation of Pseudomonas aeruginosa Z5 for biocontrol of cotton seedling disease caused by Fusarium oxysporum. Biocontrol. Sci. Technol. 24, 1227–1242. doi: 10.1080/09583157.2014.932754
Zamioudis, C., Mastranesti, P., Dhonukshe, P., Blilou, I., and Pieterse, C. M. J. (2013). Unraveling root developmental programs initiated by beneficial Pseudomonas bacteria. Plant Physiol. 162, 304–318. doi: 10.1104/pp.112.212597
Keywords: antagonistic effect, bacterial endophytes, banana, biological control, microbe–microbe interactions, Musa sp., Pseudomonas aeruginosa, soil microbial community
Citation: Thomas P and Sekhar AC (2016) Effects Due to Rhizospheric Soil Application of an Antagonistic Bacterial Endophyte on Native Bacterial Community and Its Survival in Soil: A Case Study with Pseudomonas aeruginosa from Banana. Front. Microbiol. 7:493. doi: 10.3389/fmicb.2016.00493
Received: 28 October 2015; Accepted: 27 March 2016;
Published: 26 April 2016.
Edited by:
Corné M. J. Pieterse, Utrecht University, NetherlandsReviewed by:
Anton Hartmann, Helmholtz Zentrum München, GermanyCopyright © 2016 Thomas and Sekhar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pious Thomas, cGlvdXN0QGdtYWlsLmNvbQ==; cGlvdXN0QGlpaHIucmVzLmlu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.