
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 08 April 2016
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 7 - 2016 | https://doi.org/10.3389/fmicb.2016.00477
This article is part of the Research Topic Surveying Antimicrobial Resistance, Approaches, Issues, and Challenges to overcome View all 41 articles
 Haihong Hao1,2†
Haihong Hao1,2† Shengxi Zhou2†
Shengxi Zhou2† Guyue Cheng2
Guyue Cheng2 Menghong Dai2
Menghong Dai2 Xu Wang1
Xu Wang1 Zhenli Liu1
Zhenli Liu1 Yulian Wang1,2*
Yulian Wang1,2* Zonghui Yuan1,2*
Zonghui Yuan1,2*To evaluate microbiological safety of tulathromycin on human intestinal bacteria, tulathromycin (0, 0.1, 1, 10, and 100 μg/mL) was added into Chemostats. Before and after drug exposure, we monitored (1) population, SCFA products, antimicrobial resistance, and colonization resistance of gut microbiota, and (2) the antimicrobial resistance genes, transferability, virulent genes, pathogenicity of Enterococus faecalis. Results showed that low level of tulathromycin did not exhibit microbiological hazard on resistance selection and colonization resistance. However, high level of tulathromycin (10 and 100 μg/mL) may disturb colonization resistance of human gut microbiota and select antimicrobial resistant E. faecalis. Most of the selected resistant E. faecalis carried resistant gene of ermB, transferable element of Tn1545 and three virulence genes (esp, cylA, and ace). One of them (E. faecalis 143) was confirmed to have higher horizontal transfer risk and higher pathogenicity. The calculated no observable adverse effect concentration (NOAEC) and microbiological acceptable daily intake (mADI) in our study was 1 μg/mL and 14.66 μg/kg.bw/day, respectively.
Tulathromycin is the first and only member of the triamilide sub-class of macrolide. Tulathromycin is used therapeutically in treatment of respiratory disease in swine and cattle at a single dose of 2.5 mg/kg.bw. It has been registered in more than 30 countries across America, Europe, Oceania, and Asia and played important role in veterinary medicine (FDA, 2004). This agent is characterized by rapid absorption from the injection site, extensive distribution to tissue, and slow elimination. The withdrawal period for cattle and swine is 22 and 5 days, respectively. This drug is excreted primarily unchanged (90%) in feces (2/3) and in urine (1/3), suggesting that it may reach to human colon and remain its antimicrobial activity (Benchaoui et al., 2004; EMEA, 2004; Nowakowski et al., 2004).
The residue of tulathromycin in animal food may have unintended microbiological effects on human gut microbiota. The possible harmful effects may be: (1) shifts in bacterial counts and biochemistry, (2) destruction of colonization resistance, (3) emergence of antimicrobial resistant bacteria (Cerniglia and Kotarski, 1999; Nutsch et al., 2005). Additionally, antibiotic resistant bacteria may have enhanced fitness or high virulence (Mundy et al., 2000). Human intestinal strains may also serve as reservoirs for antimicrobial resistant determinations and mediates the gene transfer (Salyers et al., 2004).
Based on in vitro antibiotic susceptibility tests, microbiological ADIs recommended by Food and Drug Administration (FDA), European Medicine Agency (EMA), and Australian Pesticides and Veterinary Medicines Authority (APVMA) were 50, 10.99, and 5 μg/kg.bw/d, respectively. The large difference in these mADI may due to the disadvantage and limitation of short-term in vitro antibiotic susceptibility tests in evaluation of long-term effects of tulathromycin residues posed on human intestinal bacteria and in analyze of impact of tulathromycin on colonization resistance and emergence of antimicrobial resistance of human colonic microbiota (Cerniglia and Kotarski, 2005). Therefore, microbiological safety assessment measures of tulathromycin residues is largely inadequate. It is largely unknown that what is the effect of long-term exposure to tulathromycin on colonization resistance of human intestinal microbiota and on antimicrobial resistance development, virulence change, and gene transfer in some specific intestinal bacteria.
In the present article, the chemostat model of human colonic gut, which is an approach recommended by VICH (VICH GL-36) and some previous studies (Carman and Woodburn, 2001; Carman et al., 2004, 2005), is used to evaluate microbiological safety of tulathromycin on human intestinal microbiota.
The tulathromycin (TUL), erythromycin (ERY), and lincomycin (LIN) were dissolved in methanol and then diluted with deionized water. The ciprofloxacin (CIP), tetracycline (TET), and rifampicin (RIF) were dissolved in deionized water. Tulathromycin was purchased from Liu He animal Pharmaceutical Co., Ltd. (Qingdao, China). The erythromycin, lincomycin, ciprofloxacin, tetracycline, and rifampicin were purchased from SIGMA.
The E. coli ATCC 25922, Enterococcus faecalis ATCC29212, and Micrococcus luteus ATCC 9341 were purchased from American Type Culture Collection (ATCC). The E. coli ATCC 25922 and E. faecalis ATCC29212 were used as quality controls for species identification and minimum inhibitory concentration (MIC) determination. The M. luteus ATCC 9341 were used as an indicator for determination of the concentration of tulathromycin. E. faecalis JH2-2 (Rifampicin MIC > 50 μg/mL) and E. coli NK5449 (Rifampicin MIC > 100 μg/mL), purchased from Belgian coordinated collections of microorganisms and Institute of microbiology in Chinese academy of sciences, respectively, were used as receptors for conjugation test. The Salmonella Typhimurium SI3 was a ciprofloxacin resistant strain which was selected in our previous study (Sun et al., 2011). This strain was used as the challenge strain for evaluating the colonization resistance of chemostat.
Fresh fecal samples from six adult volunteers who had no history of antibiotic usage and gastrointestinal disturbance within the preceding 3 months were prepared following previous method (Carman and Woodburn, 2001; Hao et al., 2013). The use of human fecal samples was approved by the ethical Committee of Huazhong Agricultural University (approval number hzauch 2013-002).
The four predominant intestinal bacteria (E. coli, Enterococcus, Bifidobacterium, and Bacteroides fragilis) were isolated from the fresh fecal samples of six adult volunteers by selective agars (Eosin methylene blue agar, bile esculin azide agar, BBL media, and bacteroides bile esculin agar). Ten isolates of each species were obtained from fresh fecal sample of each volunteer. After species identification by classic biochemical tests and ABI 3130 system (Applied Biosystem, USA), 32 isolated strains of each species were subjected to MIC determination by agar dilution method provide by CLSI (document M7-A5 for aerobic bacterial and document M11-A5 for anaerobic bacterial). The E. coli ATCC25922 and E. faecalis ATCC 29212 were used as quality control for MIC determination. The minimum concentration to inhibit 50% of isolates (MIC50) and the minimum concentration to inhibit 90% of isolates (MIC90) was calculated based on the MIC distribution and estimated by probability analysis using SPSS statistical package (Kays and Graff, 2002; Xu et al., 2013).
The preliminary test for selection of tulathromycin concentration was carried out following the method in previous study (Hao et al., 2013). Briefly, different concentrations of tulathromycin (0, 0.5, 10, 20, 40, 60, 80, 100 μg/mL) were mixed with 50 mL prepared fecal samples at the final concentration of 20% (w/v). The activity of tulathromycin on populations of four different bacteria (E. coli, Enterococcus, Bifidobacterium, and B. fragilis) were determined by bacterial colony count using selective agars. The highest concentration of tulathromycin was selected for further experiments as it was able to significantly change the population of four predominant intestinal bacteria.
The low to intermediate concentration of tulathromycin (0.1, 1, and 10 μg/mL) were designed based on the acceptable daily intake (ADI) recommended by FDA (50 μg/kg.bw/d), EMA (10.99 μg/kg.bw/d), and APVMA (5 μg/kg.bw/d). In those designed concentration, 1 μg/mL tulathromycin was the mean MIC50-value to the most sensitive Bifidobacterium strains. One group was drug free methanol control (0 μg/mL tulathromycin).
The establishment of chemostat models and the time schedule were carried out following the method in previous studies (Carman and Woodburn, 2001; Hao et al., 2013). Briefly, 50 mL prepared fecal suspension at the final concentration of 20% (w/v) was inoculated into each culture vessel. After 7 h incubation, culture medium was pumped into and out of the culture vessel at a uniform rate of 35 mL/h to maintain 500 mL of culture medium.
After 7 days running, the chemostats reached to a steady state. From 7th to 13th day, the chemostats were kept on running on steady state without administration of tulathromycin(Chen et al., 2011, 2014). From 14th to 20th day, five designed concentrations of tulathromycin (0, 0.1, 1, 10, and 100 μg/mL) were, respectively, infused into the culture medium. During 20th–22nd day, the tulathromycin was withdrawn from chemostat models and 1 mL of 6 × 108 CFU/mL (2X McFarland) Salmonella Typhimurium SI3 were daily inoculated into each chemostat. Then the chemostats continued to run for another 7 days (to 29th day) without challenge of Salmonella Typhimurium (Moffatt, 2007).
During day 7–20, samples were daily taken from each chemostat. The changes of four predominant bacteria (E. coli, Enterococcus, B. fragilis, and Bifidobacterium) were measured by viable cell counting (Hao et al., 2013, 2015).
During day 7–20, samples were daily taken from each chemostat. The concentration of three primary SCFAs (acetic acid, butryric acid, and propionic acid) were monitored using a developed gas chromatographic (GC) method (Hao et al., 2013, 2015).
From day 20th to 29th, samples were taken from each chemostat to monitor the population of Salmonella Typhimurium SI3 using bismuth sulfite (BS) agar containing 4 μg/mL ciprofloxacin(Sun et al., 2011; Hao et al., 2013, 2015). The colonization resistance was disrupted if Salmonella Typhimurium SI3 was successfully colonized into chemostat models.
During day 8–20, samples were daily taken from each chemostat. The change of resistance rate was also monitored following the method in previous study (Hao et al., 2013, 2015). The resistance rates of E. coli and Enterococcus were equal to the number of resistant colonies on the selective agars containing 4-fold MIC90 of tulathromycin divided by the number of total colonies on the selective agars without tulathromycin. The 4-fold MIC90 was used for selection of tulathromycin resistance on basis of VICH GL-36 document and some previous reports of resistance selection (Allen and Bierman, 2009; Kadlec et al., 2011).
During day 8–20, a total of 70 Enterococcus isolates were random selected from samples in the chemostat containing 100 μg/mL tulathromycin. After species identification by biochemical tests and PCR amplification, 64 isolates were identified as E. faecalis. Among these E. faecalis, 34 isolates were obtained before the administration of tulathromycin, while 30 isolates were selected after the treatment with tulathromycin. The susceptibility of the 64 E. faecalis to macrolide, lincosamides, and tetracycline were determined by agar dilution method.
The genes involved in macrolide-lincosamides-streptogramins (MLS) resistance with a methylation mechanism were determined by PCR amplification of known erm genes using specific primers for ermA, ermB, and ermC (see Table 1). The genes involved in antibiotic efflux systems were determined using specific primers for the mefA/E gene (see Table 1). The PCR procedure was followed the method in previous published paper (Portillo et al., 2000).
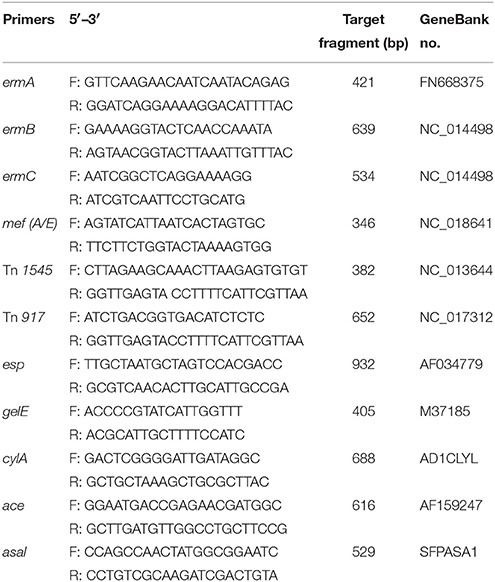
Table 1. PCR primers and products for detection of macrolides resistance gene and transposons and virulence determinants in E. faecalis.
The macrolide-resistant isolates containing ermB gene were selected to determine the presence of transposons Tn1545 and Tn917 using the primers (see Table 1) and method established in previous study (Okitsu et al., 2005). Filter mating method was used to investigate the conjugative transfer of the selected macrolide resistant E. faecalis. A representative macrolide-resistant isolate containing ermB and composite transposon was selected as donor strain. The recipients were two rifampicin-resistant strains, E. faecalis JH2-2 (Rif MIC > 50 μg/mL) and E. coli NK5449 (Rif MIC > 100 μg/mL). The donor and recipient strains were mixed into the conjugation system at the ratio of 1:9 and incubated on the serum agar for 48 h at 37°C. The number of donor, recipient and transconjugants were counted by agar plate containing rifampicin and/or erythromycin.
The presence of virulence factors, including the surface protein (esp), haemolysin activator (cylA), gelatinase (gelE), collagen binding protein (ace), and aggregation substances (asal), were determined by PCR in the 64 E. faecalis isolates obtained in the pre- and post-treatment of tulathromycin in the chemostat. PCR primers used in this study were listed in Table 1. The annealing temperatures of five virulence factors were 65°C for esp, 51°C for gelE, 61°C for CylA, 61°C for ace and 63°C for asal gene, respectively.
In order to compare the changes of pathogenicity of E. faecalis, two representative E. faecalis isolates (E. faecalis 143 and E. faecalis 174) were subjected to determine the median lethal dose (LD50). Briefly, the 72 SPF Kunming mice with body weight of 25–28 g were randomly divided into nine groups with half males and half females. The groups were, respectively, challenged with E. faecalis isolate by intraperitoneal injection of 0.5 mL diluted concentration of bacteria suspension (107–1010 CFU/mouse). The dead mice were carefully dissected in a bacteria-free operating environment. The LD50 was calculated by improved Karber method. All experimental procedures were performed according to the guidelines of the committee on the use and care of the laboratory animals in Hubei province China. The study was approved by the Animal Care Center, Hubei Science and Technology Agency in China. All the animals were monitored throughout the study for any sign of adverse effect.
All experiments were performed in triplicate. The statistical analysis was performed following the previous study (Hao et al., 2013, 2015). Briefly, a “mean pre-treatment” level and 95% prediction interval (PI95) for each parameter was set up based on the data obtained from each culture vessel during steady state (day 7–13) and used as reference boundaries for assessing the effect of tulathromycin on each parameter in the drug exposure period (day 14–20).
The NOAEC was the no observable adverse effect concentration in this study. The microbiological acceptable daily intake (mADI) for tulathromycin was calculated following the guideline of VICH GL-36. The formula used for mADI calculation was that mADI = NOAEC × daily fecal bolus/(fraction of oral dose available for micro-organisms × weight of human).
The 32 strains of each species (E. coli, Enterococcus, Bifidobacterium, and B. fragilis) were isolated from fresh fecal samples of six volunteers. The MIC for 90% of isolates (MIC90) of tulathromycin for E. coli, Enterococcus, Bifidobacterium, and B. fragilis were 5.53, 7.87, 1.35, and 2.30 μg/mL, respectively. After dilution, the methanol used for dissolving tulathromycin did not exert some antimicrobial effect.
From the preliminary test, 100 μg/mL was the highest concentration of tulathromycin which could significantly change the population of four predominant intestinal bacteria. Based on the ADI-values recommended by FDA, EMA, and APVMA, the lower concentration of tulathromycin were designed as 0.1, 1, and 10 μg/mL.
The population of E. coli was kept between 3.667 × 106 and 5.482 × 106 CFU/mL in steady state based on statistical data among five chemostats (Figure 1A). Three days after administration of 100 μg/mL tulathromycin, the population of E. coli was decreased to 6.556 × 105 CFU/mL which was ~6-fold lower than 95% lower confidence limit. However, the three low concentration of tulathromycin (0.1, 1, and 10 μg/mL) did not significantly change the number of E. coli.
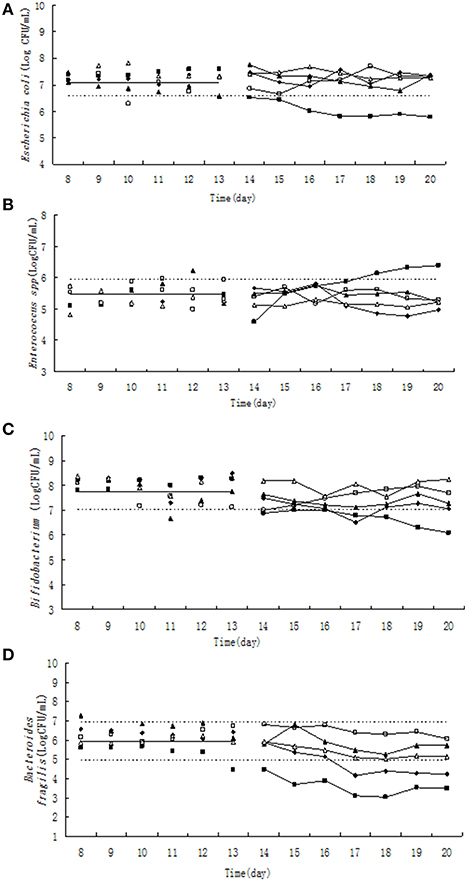
Figure 1. Effect of tulathromycin on Escherichia coli (A), Enterococcus (B), Bifidobacterium (C), Bacteroides fragilis (D) in chemostat model. □, 0 μg/ mL; Δ, 0.1 μg/ mL; ▴, 1 μg/mL; ♦, 10 μg/mL; ■, 100 μg/mL; —————, pre-treatment mean; - - - - - - - -, 95% predicated interval.
Pre-treatment of tulathromycin (day 8–13), the Enterococcus counts was in the range of 9.526 × 104–8.674 × 105 CFU/mL (Figure 1B). Under exposure to 10 μg/mL tulathromycin, Enterococcus count was decreased ~1.6-fold in the 5th day of post-treatment. On the contrary, the total number of Enterococcus was increased almost 3-fold from 18th day after administration of 100 μg/mL tulathromycin.
The number of Bifidobacterium varied from 1.105 × 107 to 2.70 × 108 CFU/mL (Figure 1C) during day 8–13. The administration of 100 μg/mL tulathromycin significantly decreased the number of Bifidobacterium in the chemostat. It was shown that Bifidobacterium count was decreased ~10-fold lower than 95% lower confidence limit.
Significant and dose-dependent effects of tulathromycin on B. fragilis counts were observed in all tested concentration of tulathromycin. After treatment by 10 and 100 μg/mL tulathromycin, B. fragilis count was decreased 6- and 90-fold lower than 95% lower confidence limit, respectively (Figure 1D).
The relative low concentration of tulathromycin (0, 0.1, and 1 μg/mL) did not significantly change the molar concentration of acetic acid, propionc acid and butyric acid (Figure 2). Under exposure to 10 μg/mL tulathromycin, the concentration of acetic acid and propionc acid was slightly decreased. Upon the administration of 100 μg/mL tulathromycin, the concentration of acetic acid, propionc acid, and butyric acid were decreased ~1.3, 1.3, and 2.68-fold lower than 95% lower confidence limit, respectively. Among the three dominant SCFAs, the concentration of propionc acid was changed remarkably under exposure of tulathromycin and present significant dose-dependent effect of tulathromycin.
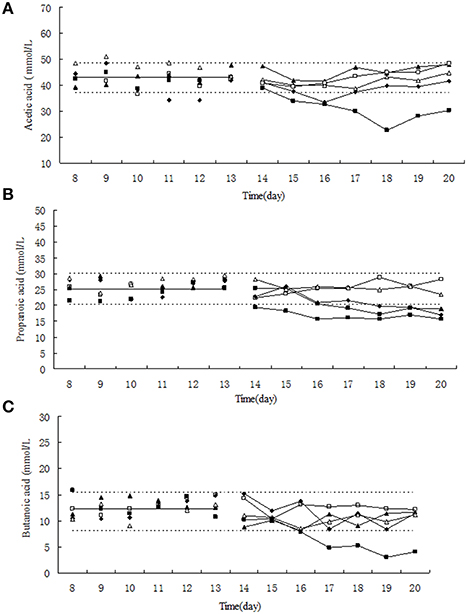
Figure 2. Effect of tulathromycin on acetic acid (A), propionic acid (B), butyric acid (C) in chemotat models. □, 0 μg/mL; Δ, 0.1 μg/mL; ▴, 1 μg/mL; ♦, 10 μg/mL; ■, 100 μg/mL; —————, pre-treatment mean; - - - - - - - -, 95% predicated interval.
In the single growth control group, the growth of Salmonella Typhimurium was stable at the level of 3.75 × 107–1.54 × 108 CFU/mL, indicating that Salmonella Typhimurium grew well in the chemostat model containing no drug and gut microbiome (Figure 3). However, the colony number of Salmonella Typhimurium tapered to 102 CFU/mL on day 31 in the chemostat containing stable gut microbiome, suggesting that the gut microbiome may establish colonization resistance to exogenous pathogens. After treatment of lower concentration of tulathromycin (10, 1, and 0.1 μg/mL), the number of Salmonella Typhimurium was also decreased to the level of 102 CFU/mL. In contrast, the colonization number of Salmonella Typhimurium was in the range of 2.08 × 105–7.15 × 106 CFU/mL in the chemostat administrated by highest concentration of tulathromycin (100 μg/mL), suggesting that the high concentration of drug may disturb the colonization resistance of gut microbiome and induce the proliferation of pathogen (Figure 3).
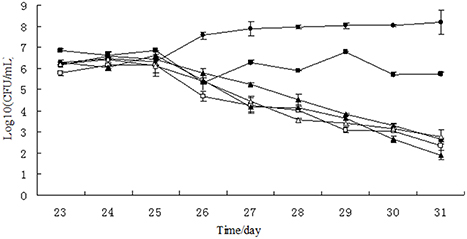
Figure 3. Growth curves of Salmonella Typhimurium in chemostat models. □, 0 μg/mL; Δ, 0.1 μg/mL; ▴, 1 μg/mL; ♦, 10 μg/mL; ■, 100 μg/ mL; ∙, growth control.
Before the treatment of tulathromycin, 10–30% of tulathromycin resistance was observed in Escherichia coli and in Enterococcus strains (Figures 4A,B). Under exposure to 10 μg/mL tulathromycin, the percentage of resistant Enterococcus was slight increased (Figure 4B). After treatment with 100 μg/mL tulathromycin, the resistant rates in E. coli and Enterococcus were obviously higher than control group, especially in Enterococcus, it reach up to 80% (Figures 4A,B). After administration of different concentration of tulathromycin, regular changes were not observed in Bifidobacterium and Bacteroides fragilis (Figures 4C,D).
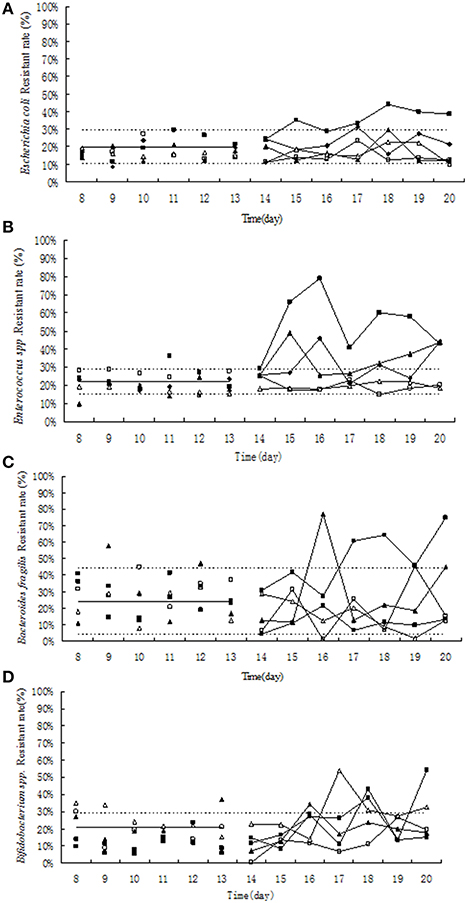
Figure 4. Effect of tulathromycin on resistance in Escherichia coli (A), Enterococcus (B), Bifidobacterium (C), Bacteroides fragilis (D) in chemostat model. □, 0 μg/mL; Δ, 0.1 μg/mL; ▴, 1 μg/mL; ♦, 10 μg/ mL; ■, 100 μg/ mL; —————, pre-treatment mean; - - - - - - - -, 95%predicated interval.
After species confirmation, 34 and 30 E. faecalis isolates were selected from chemostats pre- and post-treatment, respectively. The MIC50 and MIC90 of these 64 E. faecalis strains to erythromycin, tulathromycin, lincomycin, and tetracycline were showed in Table 2. The 34 E. faecalis strains isolated from chemostats before administration of tulathromycin exhibited high susceptibility to erythromycin, tulathromycin and tetracycline (MIC ≤ 4 μg/mL), and low resistance to lincomycin (MIC = 16, 32 μg/mL). However, after treatment with tulathromycin, 30 E. faecalis isolates (except for the strains of 141, 142, 144, and 164) showed high-level resistance to the four tested antibiotics (MIC ≥ 4 μg/mL). The MIC50 of erythromycin, tulathromycin, lincomycin, and tetracycline to E. faecalis were considerably increased for 88.9, 49.8, 5.1, and 45.4-fold, respectively.
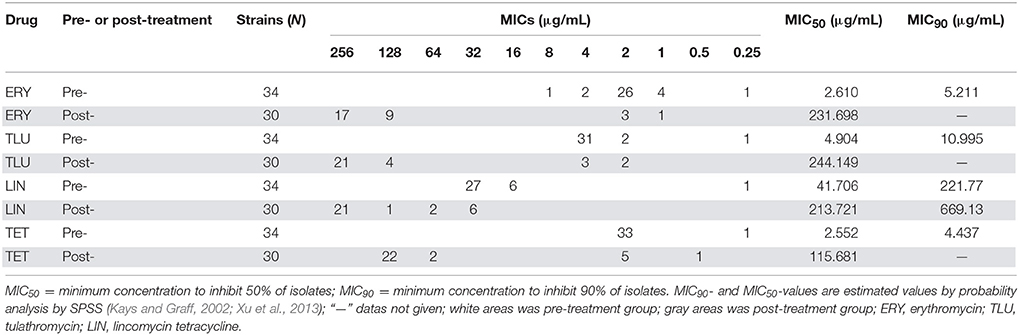
Table 2. Comparison of MICs of separated E. faecalis strains, before and after treated with four drugs.
About 88.46% of the tulathromycin resistant E. faecalis isolated from chemostats harbored macrolide resistance associated gene of ermB (Table 3). However, the tested macrolide resistance associated genes (ermABC and mefA/E) were free in the tulathromycin resistant E. faecalis strains (Table 3).
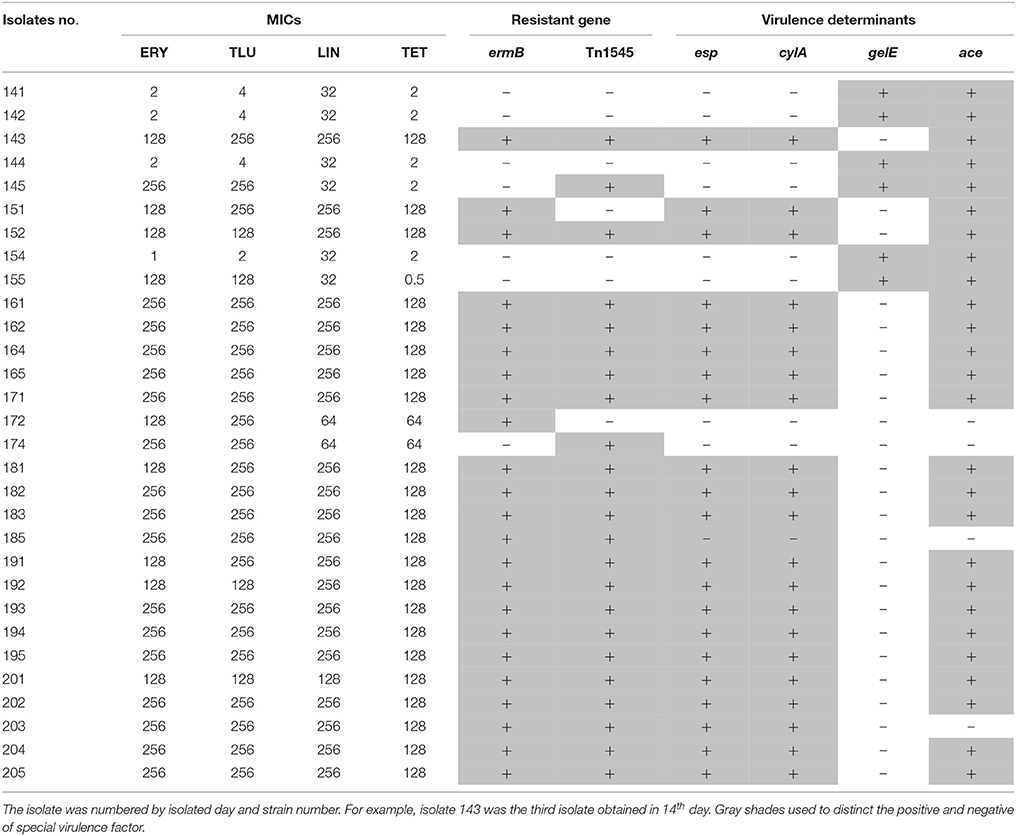
Table 3. The relationship of MIC, resistant gene and virulence gene in E. faecalis isolated during drug administration.
All of ermB containing E. faecalis strains also harbored composite transposon of Tn1545 (Table 3). When the E. faecalis 143 strain containing both ermB and Tn1545 was selected as donor strain and subjected to conjugation transfer test, the result showed that macrolide resistant determinants could transfer to E. faecalis JH2-2 and E. coli NK5449 at the frequencies of 10−6–10−7 (Table 4).
In the 34 E. faecalis stains selected in the pre-treatment, positive ratios of esp, gelE, clyA, ace, and asal were 0, 61.76, 0, 58.8% and 0, respectively. However, these changed into 70, 20, 70, 83.3% and 0, respectively, in the 30 E. faecalis stains selected in the post-treatment (Figure 5). The positive rates of surface protein (esp) and hemolysin activator (cylA) were significantly increased (0–70%). To the contrary, the carriage ratios of gelatinase (gelE) had a downside (61.76–20%). Aggregation substances (asal) were not found in all of the 64 strains E. faecalis. As shown in Table 3, most of the antimicrobial resistance in E. faecalis selected after treating with tulathromycin carried resistance gene of ermB, transferable element of Tn1545 and three virulence gens (esp, cylA, and ace).
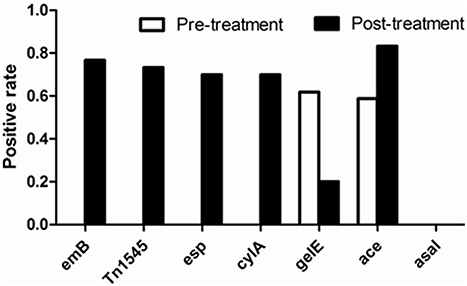
Figure 5. Comparison of positive rate of resistant determinants and virulence genes of E. faecalis before and after experiment with drug.
After infection with E. faecalis 143 (harboring ermB, Tn1545, esp, cylA, and ace), mice died within 8 h which was earlier than the death of mice infected with E. faecalis 174 (free of ermB, esp, cylA, and ace). The LD50 of the two E. faecalis strains was calculated by improved Karber method. The detailed results were shown in Table 5. The LD50 E. faecalis 143 (5.970 × 109 μg/mL) was three-times higher than that of E. faecalis 174(1.603 × 109 μg/mL), suggesting that E. faecalis 143 strain harboring virulence factors (esp, cylA, and ace) was more virulent than the strain without these two virulence genes.
Based on our results, the NOAEC was set up as 1 μg/mL. The daily fecal bolus was known as 220 mL. The fraction of oral dose available for microorganisms in the intestinal tract was 0.25 which can be split into two factors of 0.5 each. One factor of 0.5 was based on results for reduced availability of the substance due to interaction with fecal matter (EMEA, 2004) and the other factor of 0.5 was based on the impact of acidic colonic pH on tulathromycin availability for gut bacteria(USFDA, 2004). The weight of human was 60 kg/person. Therefore, our established mADI was 14.66 μg/kg.bw/d.
This study is the first of its kind to assess the effect of tulathromycin on human microbiota using the chemostat model. In addition, this study concentrated on the antimicrobial resistance development and virulence change of E. faecalis isolates in chemostats under long term exposure to a range of tulathromycin concentrations.
The present study showed that the intermediate concentration (10 μg/mL) slightly change the population of Enterococci and Bacteroides fragilis, while the highest concentration (100 μg/mL) inhibited the growth of three type of bacteria except for Enterococcus. This result was similar with our previous results that the numbers of some intestinal bacteria (e.g., B. fragilis) underwent significant changes during exposure to tilmicosin (Hao et al., 2015). As one of the most predominant bacteria in the intestinal microbiota, B. fragilis may be a most sensitive indicator of gut microbiome under exposure to macrolide drugs including erythromycin, tilmicosin, and tulathromycin (Carman et al., 2005; Merck sharp and D. Corp., 2011; Hao et al., 2015).
The present study and our previous investigation suggested that in the chemostat model high concentration of macrolide drugs (tilmicosin and tulathromycin) could significantly reduce the population of B. fragilis and subsequently resulted into the decline of the three main SCFAs especially propionic acid (Hao et al., 2015). The related change of propionic acid with B. fragilis population that we found was also observed in human flora associated (HFA) mice model (MacNeil, 2005).
Similar to previous studies, the colonization resistance of human intestinal microbiota in the chemostat was disrupted under 7 days exposure to high concentration of antibiotics (Carman et al., 2004; Hao et al., 2013). Coincidently, in our chemostat model, there were significant alterations in the populations of E. coli, B. fragilis, Enterococcus, and Bifidobacterium which appeared to be indicator of intestinal microbiota and contributed a lot to the colonization resistance (Corpet, 1993; Nuding et al., 2013). As previously reported, 105 CFU/mL of E. coli had a complete inhibitory effect on the growth of Salmonella (Carman et al., 2004). However, in our previous result, the treatment of tilmicosin did change the colonization resistance (Hao et al., 2015). This difference suggested that disruption of colonization resistance was largely depended on antimicrobial agents and their concentrations (Carman et al., 2004; Ferreira et al., 2011).
Similar to previous results, resistance rate in E. coli did not change in the chemostat and HFA rodent models treated by tilmicosin (EMEA, 1997; Cerniglia and Kotarski, 2005; Hao et al., 2015). This may be due to the inherent resistance to macrolide drugs in E. coli (Phuc Nguyen et al., 2009). However, the resistance rate of Enterococcus, Bifidobacterium, and B. fragilis was significantly increased under the selective pressure of 100 and 10 μg/mL tulathromycin in our study. Consistently, the subtherapeutic and therapeutic administration of tulathromycin also significantly increased the proportion of erythromycin resistant enterococci in beef cattle (Zaheer et al., 2013).
Our study showed that most of the isolated tulathromycin resistant enterococcicontained ermB. The ermB gene in different gram-positive bacteria has been well-documented (Schmitz et al., 2000; Perreten et al., 2005; Littauer et al., 2006; Tremblay et al., 2011; Zmantar et al., 2011). The occurrence of macrolide resistance mediated by ermB was also found in enterococci originating from swine and cattle due to the subtherapeutic use of tylosin and tulathromycin (Jackson et al., 2004; Chen et al., 2008; Zaheer et al., 2013).
Tn1545 was found in almost of the high-level macrolides resistant Enterococcus harboring ermB gene. Our conjugation test also showed that the Enterococcus isolate containing Tn1545 and ermB can transfer its resistance to E. faecalis JH2-2 and E. coli NK5449. The transferability of ermB located in transposon may play an important role on the increase of resistance rate in Enterococcus, Bifidobacterium, and B. fragilis (Okitsu et al., 2005; Ciric et al., 2013).
Both the previous study and our study found that gelE gene was one of the predominant virulence genes in E. faecalis (Di Rosa et al., 2006). A positive correlation between macrolide resistance and gelE virulence gene was observed in an epidemiological investigation (Zou et al., 2011; Lins et al., 2013). High level expression of gelE has also been observed in multi-drug resistant Enterococci and in macrolide resistant E. feacalis containing ermB gene (Arciola et al., 2008; Hao et al., 2015). However, the E. feacalis containing both ermB gene and gelatinase (gelE) did not occur in our study.
Most of E. faecalis harboring both virulence factors (esp and cylA) and resistance gene of ermB were selected after treatment with tulathromycin, however, no E. faecalis isolates containing these two virulence genes was found before tulathromycin treatment. Coincidently, recent studies also found the existence of a large pool of potentially virulent and multidrug resistant E. faecalis in diseased farm animals (Seputiene et al., 2012). The presence and expression of some virulence determinants (e.g., esp and cylA) may enhance the colonization and invasion of E. faecalis to the epithelial cell of host (Trieu-Cuot et al., 1990; Littauer et al., 2006; Heikens et al., 2009; Johanson et al., 2012; Cafini et al., 2015; Kafil and Mobarez, 2015). The acute toxicity test with SPF mice indicated that E. faecalis harboring the macrolide resistant gene of ermB and two virulence genes (esp and cylA) had higher toxicity and pathogenicity.
Our established mADI (14.6 μg/kg.bw/d) was slightly higher than the mADI (10.97 μg/kg.bw/d) recommended by EMA-CVMP and the mADI (5 μg/kg.bw/d) recommended APVMA. Recently, EMA-CVMP revised the mADI of tulathromycin to 55 μg/kg.bw/d (EMA/CVMP, 2015). As a new approved drug, more work of safety assessment needs to be done to establish the final mADI of tulathromycin.
In conclusion, the established NOAEC and mADI in our study was 1 μg/mL and 14.66 μg/kg.bw/day, respectively. The low concentration of tulathromycin had no significant microbiological hazard with regard to colonization resistance and antimicrobial resistance. However, the long-term exposure to high concentration of tulathromycin (100 μg/mL) may damage the colonization resistance of human gut microbiota and induce the development of antimicrobial resistance in Enterococcus. The E. feacalis containing both transferable resistance determinant (ermB in Tn1545) and virulence genes (esp and cylA) were selected after administration of high level tulathromycin. More work need to be done to systematically assessment the safety of tulathromycin on human public health.
Experiment designation: HH, SZ, YW, and ZY; Experiment implement: SZ, HH, and ZL; Data analysis: SZ, HH, XW, YW, and ZY; Manuscript writing: SZ and HH; Manuscript modification: HH, GC, MD, YW, and ZY.
This work was supported by Grants from National Basic Research program of China (2013CB127200), project supported by the morning program of Wuhan in China (2015070404010191), Fundamental Research Funds for the Central Universities (2662015PY035), National Natural Science Foundation of China (31101856 and 31272614), and National Program for Risk Assessment of Quality and Safety of Livestock and Poultry Products (GJFP2016008).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We give our sincere appreciation to Carl E. Cerniglia and Sangeeta Khare from National Center of Toxicological Research, USFDA due to their help on modification of this manuscript.
Allen, G. P., and Bierman, B. C. (2009). In vitro analysis of resistance selection by linezolid in vancomycin-susceptible and -resistant Enterococcus faecalis and Enterococcus faecium. Int. J. Antimicrob. Agents 34, 21–24. doi: 10.1016/j.ijantimicag.2008.12.011
Arciola, C. R., Baldassarri, L., Campoccia, D., Creti, R., Pirini, V., Huebner, J., et al. (2008). Strong biofilm production, antibiotic multi-resistance and high gelE expression in epidemic clones of Enterococcus faecalis from orthopaedic implant infections. Biomaterials 29, 580–586. doi: 10.1016/j.biomaterials.2007.10.008
Benchaoui, H. A., Nowakowski, M., Sherington, J., Rowan, T. G., and Sunderland, S. J. (2004). Pharmacokinetics and lung tissue concentrations of tulathromycin in swine. J. Vet. Pharmacol. Ther. 27, 203–210. doi: 10.1111/j.1365-2885.2004.00586.x
Cafini, F., Gómez-Aguado, F., Corcuera, M. T., Ramos, C., Bas, P., Collado, L., et al. (2015). Genotypic and phenotypic diversity in Enterococcus faecalis: is agar invasion a pathogenicity score? Rev. Esp. Quimioter. 28, 101–108. doi: 10.1016/j.micpath.2015.11.017
Carman, R. J., Simon, M. A., Fernandez, H., Miller, M. A., and Bartholomew, M. J. (2004). Ciprofloxacin at low levels disrupts colonization resistance of human fecal microflora growing in chemostats. Regul. Toxicol. Pharmacol. 40, 319–326. doi: 10.1016/j.yrtph.2004.08.005
Carman, R. J., Simon, M. A., Petzold, H. E. III, Wimmer, R. F., Batra, M. R., Fernandez, A. H., et al. (2005). Antibiotics in the human food chain: establishing no effect levels of tetracycline, neomycin, and erythromycin using a chemostat model of the human colonic microflora. Regul. Toxicol. Pharmacol. 43, 168–180. doi: 10.1016/j.yrtph.2005.06.005
Carman, R. J., and Woodburn, M. A. (2001). Effects of low levels of ciprofloxacin on a chemostat model of the human colonic microflora. Regul. Toxicol. Pharmacol. 33, 276–284. doi: 10.1006/rtph.2001.1473
Cerniglia, C. E., and Kotarski, S. (1999). Evaluation of veterinary drug residues in food for their potential to affect human intestinal microflora. Regul. Toxicol. Pharmacol. 29, 238–261. doi: 10.1006/rtph.1999.1300
Cerniglia, C. E., and Kotarski, S. (2005). Approaches in the safety evaluations of veterinary antimicrobial agents in food to determine the effects on the human intestinal microflora. J. Vet. Pharmacol. Ther. 28, 3–20. doi: 10.1111/j.1365-2885.2004.00595.x
Chen, J., Fluharty, F. L., St-Pierre, N., Morrison, M., and Yu, Z. (2008). Technical note: occurrence in fecal microbiota of genes conferring resistance to both macrolide-lincosamide-streptogramin B and tetracyclines concomitant with feeding of beef cattle with tylosin. J. Anim. Sci. 86, 2385–2391. doi: 10.2527/jas.2007-0705
Chen, T., Li, S., and Wei, H. (2014). Antibiotic resistance capability of cultured human colonic microbiota growing in a chemostat model. Appl. Biochem. Biotechnol. 173, 765–774. doi: 10.1007/s12010-014-0882-6
Chen, T., Yuan, J., Feng, X., Wei, H., and Hua, W. (2011). Effects of enrofloxacin on the human intestinal microbiota in vitro. Int. J. Antimicrob. Agents 37, 567–571. doi: 10.1016/j.ijantimicag.2011.01.013
Ciric, L., Jasni, A., de Vries, L. E., Agersø, Y., Mullany, P., and Roberts, A. P. (2013). “Landes bioscience,” in Madame Curie Bioscience Database [Internet], Bacterial Integrative Mobile Genetic Elements, eds A. P. Roberts and P. Mullany (Austin, TX: Landes Bioscience), 2000–2013.
Corpet, D. E. (1993). An evaluation of methods to assess the effect of antimicrobial residues on the human gut flora. Vet. Microbiol. 35, 199–212. doi: 10.1016/0378-1135(93)90145-W
Di Rosa, R., Creti, R., Venditti, M., D'Amelio, R., Arciola, C. R., Montanaro, L., et al. (2006). Relationship between biofilm formation, the enterococcal surface protein (Esp) and gelatinase in clinical isolates of Enterococcus faecalis and Enterococcus faecium. FEMS Microbiol. Lett. 256, 145–150. doi: 10.1111/j.1574-6968.2006.00112.x
EMA/CVMP (2015). Tulathromycin (Modification of the Microbiological ADI and MRLs in Bovine and Porcine Species). EMA/CVMP. Available online at: http://www.ema.europa.eu/docs/en_GB/document_library/Maximum_Residue_Limits_-_Report/2015/02/WC500182166.pdf
EMEA (1997). Committee for Veterinary Medicinal Products, Tilmicosin, Summary Report (1). E. CVMP. EMEA/MRL/318/97-FINAL.
EMEA (2004). Committee for Veterinary Medicinal Products, Tulathromycin, Summary Report (2). EMEA-CVMP. EMEA/MRL/894/04-Final.
FDA (2004) Tulathromycin Microbiological Effects on Bacteria of Humanhealth Concern A Qualitative Risk Estimation, Vol. 9, 1–58.
Ferreira, R. B., Gill, N., Willing, B. P., Antunes, L. C., Russell, S. L., Croxen, M. A., et al. (2011). The intestinal microbiota plays a role in Salmonella-induced colitis independent of pathogen colonization. PLoS ONE 6:e20338. doi: 10.1371/journal.pone.0020338
Hao, H., Guo, W., Iqbal, Z., Cheng, G., Wang, X., Dai, M., et al. (2013). Impact of cyadox on human colonic microflora in chemostat models. Regul. Toxicol. Pharmacol. 67, 335–343. doi: 10.1016/j.yrtph.2013.08.011
Hao, H., Yao, J., Wu, Q., Wei, Y., Dai, M., Iqbal, Z., et al. (2015). Microbiological toxicity of tilmicosin on human colonic microflora in chemostats. Regul. Toxicol. Pharmacol. 73, 201–208. doi: 10.1016/j.yrtph.2015.07.008
Heikens, E., Leendertse, M., Wijnands, L. M., van Luit-Asbroek, M., Bonten, M. J., van der Poll, T., et al. (2009). Enterococcal surface protein Esp is not essential for cell adhesion and intestinal colonization of Enterococcus faecium in mice. BMC Microbiol. 9:19. doi: 10.1186/1471-2180-9-19
Jackson, C. R., Fedorka-Cray, P. J., Barrett, J. B., and Ladely, S. R. (2004). Effects of tylosin use on erythromycin resistance in enterococci isolated from swine. Appl. Environ. Microbiol. 70, 4205–4210. doi: 10.1128/AEM.70.7.4205-4210.2004
Johanson, J. J., Feriancikova, L., and Xu, S. (2012). Influence of enterococcal surface protein (esp) on the transport of Enterococcus faecium within saturated quartz sands. Environ. Sci. Technol. 46, 1511–1518. doi: 10.1021/es203265m
Kadlec, K., Brenner Michael, G., Sweeney, M. T., Brzuszkiewicz, E., Liesegang, H., Daniel, R., et al. (2011). Molecular basis of macrolide, triamilide, and lincosamide resistance in Pasteurella multocida from bovine respiratory disease. Antimicrob. Agents Chemother. 55, 2475–2477. doi: 10.1128/AAC.00092-11
Kafil, H. S., and Mobarez, A. M. (2015). Spread of enterococcal surface protein in antibiotic resistant Enterococcus faecium and Enterococcus faecalis isolates from urinary tract infections. Open Microbiol. J. 26, 14–17. doi: 10.2174/1874285801509010014
Kays, M. B., and Graff, M. A. (2002). Broth microdilution and E-test for determining fluoroquinolone activity against Streptococcus pneumoniae. Ann. Pharmacother. 36, 416–422. doi: 10.1345/aph.1A119
Lins, R. X., de Oliveira Andrade, A., Hirata, R. Jr., Wilson, M. J., Lewis, M. A., Williams, D. W., et al. (2013). Antimicrobial resistance and virulence traits of Enterococcus faecalis from primary endodontic infections. J. Dent. 41, 779–786. doi: 10.1016/j.jdent.2013.07.004
Littauer, P., Caugant, D., Sangvik, M., Høiby, E., Sundsfjord, A., and Simonsen, G. (2006). Macrolide-resistant Streptococcus pyogenes in Norway: population structure and resistance determinants. Antimicrob. Agents Chemother. 50, 1896–1899. doi: 10.1128/AAC.50.5.1896-1899.2006
MacNeil, J. D. (2005). The joint food and agriculture organization of the United Nations/World Health Organization Expert Committee on Food Additives and its role in the evaluation of the safety of veterinary drug residues in foods. AAPS J. 7, E274–E280. doi: 10.1208/aapsj070228
Merck sharp D. Corp. (2011). Antimicrobial Activity-Merck Veterianry Manual. London: Merck sharp and D. Corp.
Moffatt, L. (2007). Draxxin Injectable Solution-APVMA Product Number 5930. Australian Pesticides and Veterinary Medicines Authority. Kingston, ACT. Available online at: http://apvma.gov.au/sites/default/files/publication/14156-prs-draxxin.pdf
Mundy, L., Sahm, D., and Gilmore, M. (2000). Relationships between enterococcal virulence and antimicrobial resistance. Clin. Microbiol. Rev. 13, 513–522. doi: 10.1128/CMR.13.4.513-522.2000
Nowakowski, M. A., Inskeep, P. B., Risk, J. E., Skogerboe, T. L., Benchaoui, H. A., Meinert, T. R., et al. (2004). Pharmacokinetics and lung tissue concentrations of tulathromycin, a new triamilide antibiotic, in cattle. Vet. Ther. 5, 60–74.
Nuding, S., Antoni, L., and Stange, E. F. (2013). The host and the flora. Dig. Dis. 31, 286–292. doi: 10.1159/000354680
Nutsch, R. G., Hart, F. J., Rooney, K. A., Weigel, D. J., Kilgore, W. R., and Skogerboe, T. L. (2005). Efficacy of tulathromycin injectable solution for the treatment of naturally occurring swine respiratory disease. Vet. Ther. 6, 214.
Okitsu, N., Kaieda, S., Yano, H., Nakano, R., Hosaka, Y., Okamoto, R., et al. (2005). Characterization of ermB gene transposition by Tn1545 and Tn917 in macrolide-resistant Streptococcus pneumoniae isolates. J. Clin. Microbiol. 43, 168–173. doi: 10.1128/JCM.43.1.168-173.2005
Perreten, V., Vorlet-Fawer, L., Slickers, P., Ehricht, R., Kuhnert, P., and Frey, J. (2005). Microarray-based detection of 90 antibiotic resistance genes of gram-positive bacteria. J. Clin. Microbiol. 43, 2291–2302. doi: 10.1128/JCM.43.5.2291-2302.2005
Phuc Nguyen, M. C., Woerther, P. L., Bouvet, M., Andremont, A., Leclercq, R., and Canu, A. (2009). Escherichia coli as reservoir for macrolide resistance genes. Emerging Infect. Dis. 15, 1648–1650. doi: 10.3201/eid1510.090696
Portillo, A., Ruiz-Larrea, F., Zarazaga, M., Alonso, A., Martinez, J. L., and Torres, C. (2000). Macrolide resistance genes in Enterococcus spp. Antimicrob. Agents Chemother. 44, 967–971. doi: 10.1128/AAC.44.4.967-971.2000
Salyers, A. A., Gupta, A., and Wang, Y. (2004). Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 12, 412–416. doi: 10.1016/j.tim.2004.07.004
Schmitz, F.-J., Sadurski, R., Kray, A., Boos, M., Geisel, R., Köhrer, K., et al. (2000). Prevalence of macrolide-resistance genes in Staphylococcus aureus and Enterococcus faecium isolates from 24 European university hospitals. J. Antimicrob. Chemother. 45, 891–894. doi: 10.1093/jac/45.6.891
Seputiene, V., Bogdaite, A., Ruzauskas, M., and Suziedeliene, E. (2012). Antibiotic resistance genes and virulence factors in Enterococcus faecium and Enterococcus faecalis from diseased farm animals: pigs, cattle and poultry. Pol. J. Vet. Sci. 15, 431–438.
Sun, Y., Dai, M., Hao, H., Wang, Y., Huang, L., Almofti, Y. A., et al. (2011). The role of RamA on the development of ciprofloxacin resistance in Salmonella enterica serovar Typhimurium. PLoS ONE 6:e23471. doi: 10.1371/journal.pone.0023471
Tremblay, C.-L., Letellier, A., Quessy, S., Boulianne, M., Daignault, D., and Archambault, M. (2011). Multiple-antibiotic resistance of Enterococcus faecalis and Enterococcus faecium from cecal contents in broiler chicken and turkey flocks slaughtered in Canada and plasmid colocalization of tetO and ermB genes. J. Food Prot. 74, 1639–1648. doi: 10.4315/0362-028X.JFP-10-451
Trieu-Cuot, P., Poyart-Salmeron, C., Carlier, C., and Courvalin, P. (1990). Nucleotide sequence of the erythromycin resistance gene of the conjugative transposon Tn1545. Nucleic Acids Res. 18, 3660–3660. doi: 10.1093/nar/18.12.3660
USFDA (2004). Tulathromycin Microbiological Effect. USFDA. Available online at: http://www.fda.gov/downloads/AdvisoryCommittees/CommitteesMeetingMaterials/VeterinaryMedicineAdvisoryCommittee/UCM127196.pdf
Xu, Y., Gao, C., Li, X., He, Y., Zhou, L., Pang, G., et al. (2013). In vitro antifungal activity of silver nanoparticles against ocular pathogenic filamentous fungi. J. Ocul. Pharmacol. Ther., 29, 270–274. doi: 10.1089/jop.2012.0155
Zaheer, R., Cook, S. R., Klima, C. L., Stanford, K., Alexander, T., Topp, E., et al. (2013). Effect of subtherapeutic vs. therapeutic administration of macrolides on antimicrobial resistance in Mannheimia haemolytica and Enterococci isolated from beef cattle. Front. Microbiol. 4:133. doi: 10.3389/fmicb.2013.00133
Zmantar, T., Kouidhi, B., Miladi, H., and Bakhrouf, A. (2011). Detection of macrolide and disinfectant resistance genes in clinical Staphylococcus aureus and coagulase-negative Staphylococci. BMC Res. Notes 4:453. doi: 10.1186/1756-0500-4-453
Keywords: tulathromycin, gut microbiota, colonization resistance, antimicrobial resistance, chemostat
Citation: Hao H, Zhou S, Cheng G, Dai M, Wang X, Liu Z, Wang Y and Yuan Z (2016) Effect of Tulathromycin on Colonization Resistance, Antimicrobial Resistance, and Virulence of Human Gut Microbiota in Chemostats. Front. Microbiol. 7:477. doi: 10.3389/fmicb.2016.00477
Received: 07 January 2016; Accepted: 22 March 2016;
Published: 08 April 2016.
Edited by:
Alexandre Gonçalves, University of Trás-os-Montes e Alto Douro, PortugalReviewed by:
Atte Von Wright, University of Eastern Finland, FinlandCopyright © 2016 Hao, Zhou, Cheng, Dai, Wang, Liu, Wang and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yulian Wang, d2FuZ3l1bGlhbkBtYWlsLmh6YXUuZWR1LmNu;
Zonghui Yuan, eXVhbjU4MDJAbWFpbC5oemF1LmVkdS5jbg==
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.