
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 30 March 2016
Sec. Virology
Volume 7 - 2016 | https://doi.org/10.3389/fmicb.2016.00334
This article is part of the Research Topic New Insights in Oncogenic Human Herpesvirus Biology View all 12 articles
Manipulation of cell cycle is a commonly employed strategy of viruses for achieving a favorable cellular environment during infection. Kaposi’s sarcoma-associated herpesvirus (KSHV), the primary etiological agent of several human malignancies including Kaposi’s sarcoma, and primary effusion lymphoma, encodes several oncoproteins that deregulate normal physiology of cell cycle machinery to persist with endothelial cells and B cells and subsequently establish a latent infection. During latency, only a small subset of viral proteins is expressed. Latency-associated nuclear antigen (LANA) is one of the latent antigens shown to be essential for transformation of endothelial cells in vitro. It has been well demonstrated that LANA is critical for the maintenance of latency, episome DNA replication, segregation and gene transcription. In this review, we summarize recent studies and address how LANA functions as an oncoprotein to steer host cell cycle-related events including proliferation and apoptosis by interacting with various cellular and viral factors, and highlight the potential therapeutic strategy of disrupting LANA-dependent signaling as targets in KSHV-associated cancers.
The eukaryotic cell cycle is divided into four phases: G1, S, G2, and M. The G1 phase is the first gap for cells to organize themselves prior to DNA replication. Any decisive events during G1 phase will determine whether the cell continues to proceed for division, pauses, or exits the cell cycle and enters the cell apoptosis pathway. The S phase is the stage for DNA synthesis, and hence genome duplication. The G2 phase is the second gap for cells to prepare the process of mitosis, and the associated cell division of two daughter cells, when the duplicated chromosomes are segregated into separated nuclei and cytokinesis. In addition, G2 phase also provides an opportunity for recognition and repair of damaged DNA. Thus, the G1 and G2 phase are called checkpoints for DNA replication and mitosis during cell cycle, respectively (Gabrielli et al., 2012). Strict regulation of cell division is critical for the normal development and maintenance of multicellular organisms. Loss of control of cell division will ultimately lead to cancer (Kastan and Bartek, 2004). In the past three decades, the studies of basic mechanism of cell cycle have led to a better understanding of how the molecular events required for cell division are controlled and coordinated (Gabrielli et al., 2012). The key elements in the basis of cell cycle regulation are the periodic synthesis and destruction of cyclins, which associate with or activate cyclin-dependent kinases (Cdks) (Dai and Grant, 2011). Although at least 16 cyclins and 9 Cdks have been identified in mammalian cells, not all cyclins and Cdks is necessary to regulate the cell cycle, some have been shown act as regulators of transcription, DNA repair or apoptosis (Johnson and Walker, 1999). In addition to the interaction between cyclin and Cdks, there are several levels of regulation including cyclin-dependent kinase inhibitors (CdkIs) and ubiquitin-mediated proteolysis which are also involved in controlling the activity of Cdks during the cell cycle (Kastan and Bartek, 2004).
Manipulation of the host cell cycle is a frequent strategy for viruses to evade host cells, presumably in order to achieve a cellular environment favorable for their replication (Nascimento et al., 2012). Due to the complex and interactive nature of intracellular signaling pathways in controlling cell division, which could provide many opportunities for viral manipulation, the important effect of viral regulation on cell cycle dynamics are the consequences for driving neoplastic transformation. This offers a rational approach to the control of virus causing cancers (Gabrielli et al., 2012). The study of host evasion strategies for cell cycle manipulation evolved by viruses will undoubtedly reveal new control mechanisms and their corresponding cellular signaling pathways.
Kaposi’s sarcoma-associated herpesvirus (KSHV), also known as human herpesvirus type 8 (HHV-8), is a gamma-herpesvirus associated with several human malignancies including Kaposi’s sarcoma (KS), primary effusion lymphomas (PEL), and multicentric Castleman’s disease (MCD) (Dupin et al., 1999). As shown in Figure 1A, the KSHV genome is an approximately 140 kb long unique coding region (LUR) that is flanked by multiple, non-coding terminal repeat (TR) units with high GC content (Russo et al., 1996; Ohsaki and Ueda, 2012). The LUR encodes about 90 open reading frames (ORFs), 12 microRNAs and several ncRNAs (Russo et al., 1996; Toth et al., 2013). Like all herpesviruses, KSHV exhibits two distinct phases of infection: latency and lytic replication. In primary infection, KSHV enters a latency whereby the viral genome circularizes and exists as nuclear episome through multiple host cell divisions. During latent infection, only a subset of viral genes including latency-associated nuclear antigen (LANA, ORF73), v-Cyclin (ORF72), v-FLIP (ORF71), and Kaposin (K12) are expressed (Figure 1A). Upon stimulation such as chemical agents or environmental stress, KSHV could be reactivated from latency to lytic replication and in turn produce infectious virion progeny (Yu et al., 1999; Davis et al., 2001).
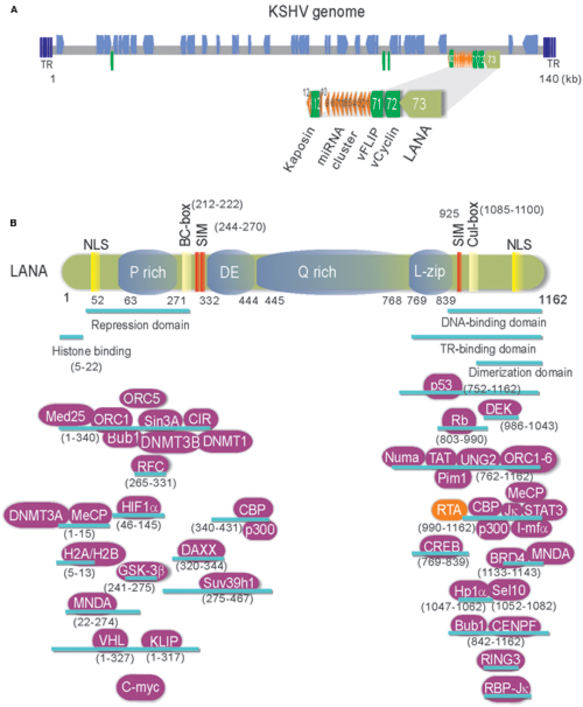
FIGURE 1. (A) Schematic and location of the KSHV latent genes including miRNA cluster. Bottom: The major latency locus (ORF73/LANA, ORF72/v-Cyclin, ORF71/vFLIP, and K12/Kaposin) of KSHV is shown in an enlarged view. Position of 12 pre-miRNA cluster is shown in red triangle. (B) The structure and functional motifs of Latency-associated nuclear antigen (LANA). LANA consists of 1162 amino acids. Numbers indicate the amino acids (aa). Repetitive regions and key motif of LANA are noted. P, Proline; DE, Aspartic acid and Glutamic acid; Q, Glutamine; L, Leucine; NLS, nuclear localized sequence; BC, Elongin B and C; Cul: Cullin5; SIM, SUMO-interacting motif. The binding regions of LANA-associated cellular and viral proteins are listed at the bottom panel.
Many evidences have shown that KSHV establishes a stable latent infection which plays an essential role in KSHV-induced malignancies and pathogenesis (Dupin et al., 1999; Katano et al., 2000; Parravicini et al., 2000). Serological analysis of the infected cells by immunofluorescence and immunohistochemistry indicated that LANA is one of only a few latent proteins consistently present in all KSHV-infected tumor cells of KS, PEL, and MCD (Dupin et al., 1999; Katano et al., 2000; Parravicini et al., 2000). It has been well demonstrated that LANA is a multi-functional protein with approximately 1162 amino acids in length, through directly or indirectly interacting with many other molecules in different signaling pathways including apoptosis, cell proliferation and gene transcription (Figure 1B). Many studies have shown that LANA contains unique nuclear localization signal (NLS) and is localized in the nuclei and interacts with host cellular DNA and KSHV genome in a punctate pattern during interphase and mitosis (Ballestas et al., 1999; Cotter and Robertson, 1999). This indicates that LANA is a cell-cycle related protein. LANA contains a large internal region of acidic and glutamine-rich repeat, and separating the amino and carboxyl terminal region of LANA, although it has been noted that the length of the internal repeat region varies between different KSHV isolates (Gao et al., 1999). LANA encodes various functional motifs to specifically recruit different target molecules. For examples, LANA has a chromosome-binding motif within 5–13 amino acids to bind with histones H2A/H2B (Barbera et al., 2006), a motif to recognize DNA sequence within the TR region of KSHV (Srinivasan et al., 2004; Kelley-Clarke et al., 2009), a region for LANA oligomerization (Komatsu et al., 2004), a SOCS-like box motif to recruit EC5S (Elongin BC-Cullin5) ubiquitin complex (Cai Q.-L. et al., 2006), a SIM motif to bind with SUMO molecules (Cai et al., 2013), and so on. Interestingly, the phenomenon that LANA drives transgenic mice developing splenic follicular hyperplasia (Fakhari et al., 2006), and transforms primary REF cells when conjunction with h-Ras (Radkov et al., 2000), as well as LANA upregulates the transcriptional activity of human telomerase promoter through interaction with Sp1 (Knight et al., 2001; Verma et al., 2004), supporting the notion that LANA acts as an oncoprotein to contribute to the pathogenesis of KSHV infection. In regard to how LANA plays a critical role in KSHV episome persistence, DNA replication and gene transcription, as well as program switch of latency and lytic replication, two reviews have been summarized recently (Ballestas and Kaye, 2011; Uppal et al., 2014). However, although it is clear that LANA binds to nucleosomal proteins throughout the cell cycle, how LANA associates with the regulators of cell cycle to driving cell proliferation remains elusive. Here, we summarize and highlight the recent progression of cell cycle regulatory functions of LANA.
Both control of cell cycle checkpoints and inhibition of apoptosis are hallmarks of tumor cell proliferation including KSHV-infected KS and PEL cells. Many studies have shown that LANA not only blocks tumor suppressor pathways, but also enhances expression of oncogenes which involve in cell cycle regulation, supporting the role of LANA in cell transformation and growth. For examples, the inhibitors of DNA binding (Id) have been demonstrated to involve in cell cycle regulation through deregulating expression of p21 (Nickoloff et al., 2000). Moreover, Id-1 could directly interact with the p16 promoter to drive cell proliferation (Alani et al., 2001). The introduction of LANA in endothelial cells dramatically increased Id-1 expression, suggesting that LANA may drive cell proliferation by targeting Id-1 expression (Tang et al., 2003). Many other oncoproteins associated with prolonged cellular survival are also affected by LANA. For examples, LANA activates telomerase expression (Verma et al., 2004), stabilizes c-myc and HIF1α oncoproteins and promotes its transcriptional activity (Cai Q. et al., 2006; Bubman et al., 2007; Cai et al., 2007; Liu et al., 2007b), increases level of ICN in Notch signaling pathway (Lan et al., 2007), upregulates expression of Aurora Kinase (Cai et al., 2012), as well as associates with and upregulates Pim1 kinase activity (Bajaj et al., 2006; Cheng et al., 2009). On the other hand, LANA also functions as a component of E3 ubiquitin ligase to target tumor suppressors like p53 and VHL for degradation, which creates a favorable environment for cell growth. LANA also affects the tumor suppressor p73 stability and subnuclear localization to contribute to the survival of PEL cells (Santag et al., 2013). Due to activation of TGF-β signaling pathway in many cell types results in inhibition of cell growth and induction of apoptosis, LANA suppresses the promoter of TGF-β type II receptor (TβRII) through epigenetic silencing (Di Bartolo et al., 2008), which may lead to cell proliferation by activation of c-myc, p15, and Cdc25A. To further explore the role of LANA on cell cycle regulation, our recent studies revealed that LANA upregulates transcription of 59 of 116 cell-cycle genes (including CDK4, Cdc25A/C, BAX, and BCL2), and only downregulates eight genes (Figure 2) (Cai et al., 2010; Gan et al., 2015). This indicates that LANA plays a positive instead of negative regulation on process of cell progression.
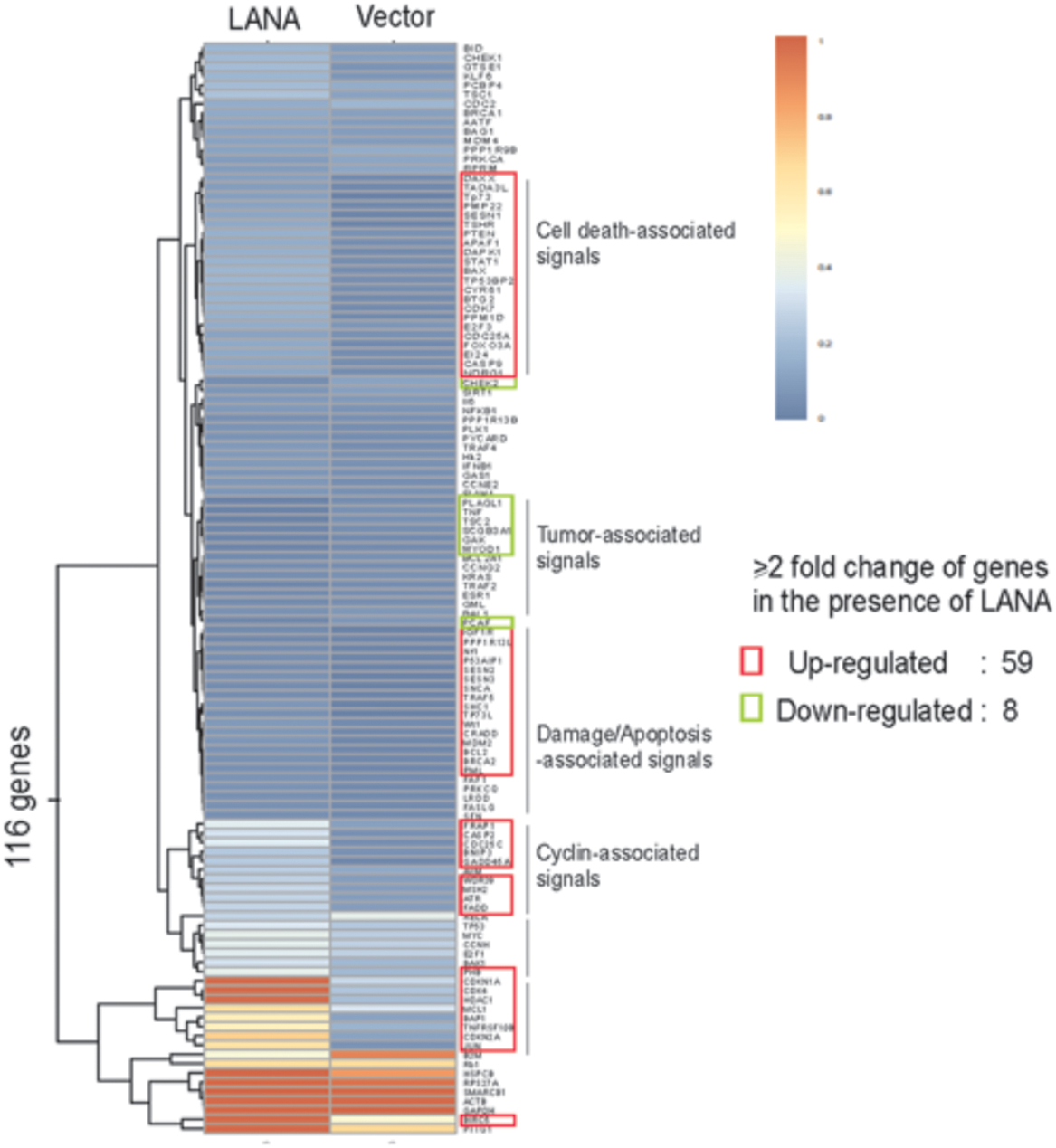
FIGURE 2. Hierarchical clustering of genes profiling related to cell cycle in BJAB cells with LANA-RFP stable expression or RFP vector alone. Data analysis revealed that 67 out of 116 genes are changed by over twofold in their expression in the presence of LANA.
Upon growth factors such as epidermal growth factor (EGF) and insulin-like growth factor (IGF), the sequential activation of the two kinase complexes, Cdk4/6-cyclin D, and Cdk2-cyclin E, is the key event that leads to cell cycle progression (Figure 3). These activated complexes could phosphorylate tumor suppressor retinoblastoma (RB), and in turn dissociate E2F from RB and accumulate E2F. In the LANA-expressing cells, LANA interacts with RB and enhances the transcriptional activation of E2F-responsive genes (Hume and Kalejta, 2009). In addition, LANA has also been shown to interact with glycogen synthase kinase (GSK-3β), a kinase involved in phosphorylation and subsequent degradation of many cell-cycle regulators including c-Myc and Cyclin D (Fujimuro et al., 2003; Bubman et al., 2007; Liu et al., 2007a,b). Recent studies showed that the interaction of GSK-3 with LANA could lead to the accumulated expression of iASPP and in turn degradation of p53 for cell growth (Woodard et al., 2015). Our previous studies further demonstrated that p53 can be degraded by LANA-mediated recruitment of the cellular EC5S (Elongin BC-Cullin 5-Rbx1) ubiquitin complex (Cai Q.-L. et al., 2006), as well as involvement of the Serine/Threonine oncogenic kinase Aurora A (Cai et al., 2012).
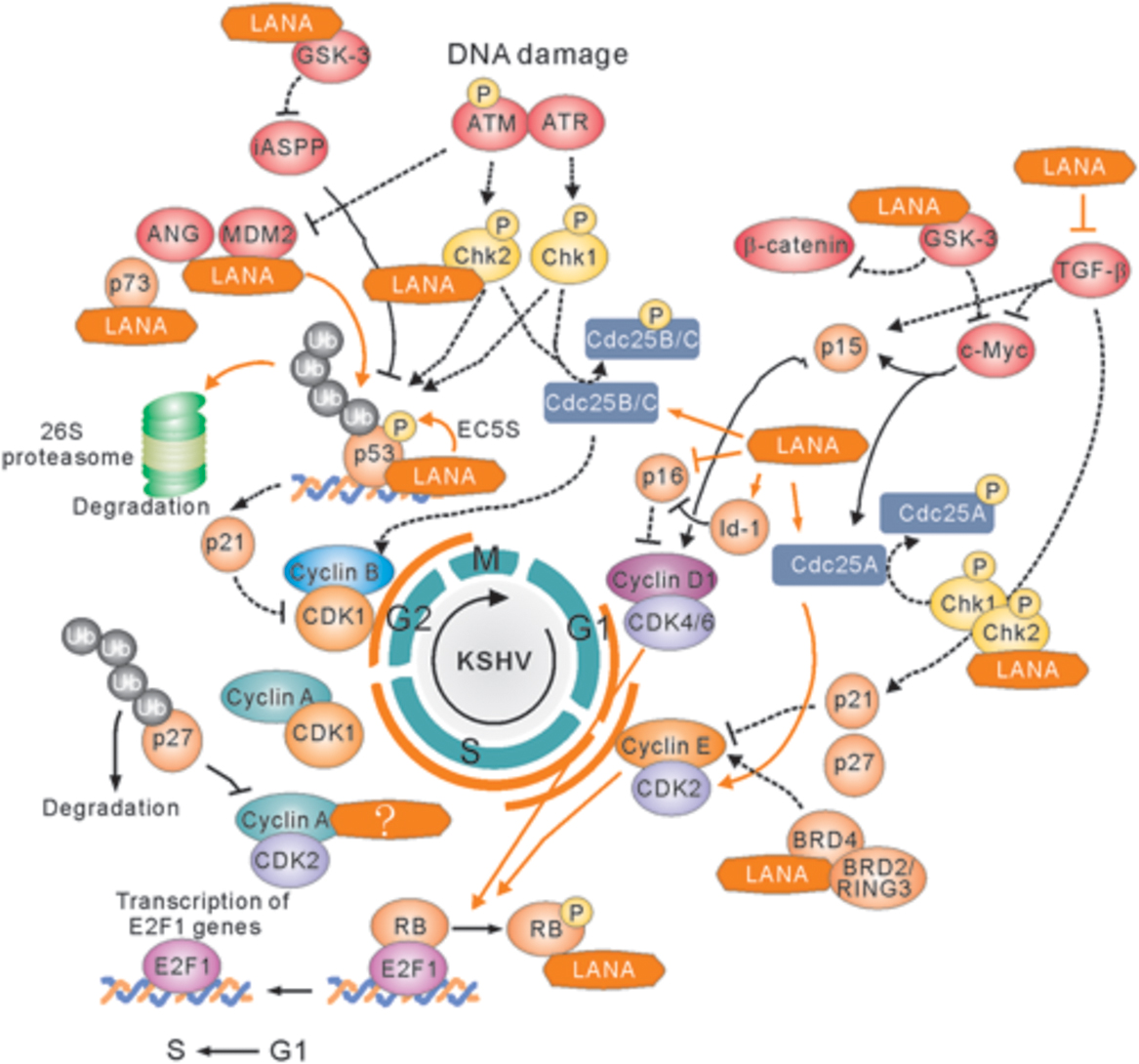
FIGURE 3. Latency-associated nuclear antigen-mediated deregulation of cell cycle. LANA associates with many cellular proteins including GSK-3, c-Myc, Chk2, BRD4, and BRD2/RING3, to activate Cyclin D1-CDK4/6 and Cyclin E-CDK2 complex, which results in the hyperphosphorylation form of retinoblastoma protein (RB). Hypersphophorylation of RB prevents its interaction with E2F, and releases E2F to activate expression of genes required for entry into the S phase. LANA also interacts with RB to facilitate G1/S phase transition. On the other hand, LANA not only recruits E3-ligase complex (including Elongin BC-Cullin 5, EC5S) or MDM2 to target p53 for ubiquitin-mediated degradation, but also associates with many p53-associated pathway regulators including p73, iASPP, and Chk2 to induce Cyclin B-CDK1 complex for driving G2/M phase transition. The positive and negative regulation by LANA is shown by solid orange line. The cellular signaling pathway blocked by LANA is indicated by dot line.
In response to DNA damage, entry of cells into S phase is prevented by activation of the two transducing kinases, ATM/ATR and Chk1/Chk2, and followed by Cdc25A phosphorylation and p53 activation. Phosphorylated Cdc25A pathway has a faster inhibitory impact on the cell cycle progression (Lukas et al., 2004). In contrast, both Chk1/Chk2 and ATM/ATR-mediated phosphorylation on Serine 15 of p53 signaling can further prolong G1 arrest. Recent studies showed that LANA directly interacts with Chk2 to block ATM/ATR-mediated apoptosis and potential activation of Cdc25A (Kumar et al., 2014). Meanwhile, Mdm2 (the negative regulator of p53) is inactivated by ATM/ATR, and association of p53 with p300 results in increased transcriptional activity of p300. The p53-mediated induction of downstream genes including p21 blocks the G1/S progression which promoted by Cdk2-cyclin E kinase expression. The inhibitors of p53-MdM2 interactions including nutlin-3 interfere with the formation of Mdm2-p53-LANA complex and cause G1 arrest and apoptosis of PEL cells (Petre et al., 2007; Sarek et al., 2007; Ye et al., 2012), further supporting the notion that modulation of p53-dependent pathways by LANA is critical for KSHV to prevent cell cycle arrest and apoptosis.
In addition, LANA has been shown to bind and block p53-mediated transcriptional activity and in turn inhibit p53-induced cell apoptosis (Friborg et al., 1999; Si and Robertson, 2006; Lu et al., 2009). It has been demonstrated that LANA can also positively regulate cell cycle-dependent promoters, and promote the G1/S transition of cells by overcoming the serum starvation, overexpression of cyclin-dependent kinase inhibitor p16, or BRD4 and BRD2/RING3-induced G1 cell cycle arrest (Platt et al., 1999; Direkze and Laman, 2004; An et al., 2005; Viejo-Borbolla et al., 2005; Ottinger et al., 2006). Moreover, Disruption of Annexin A2 with LANA results in downregulation of cell cycle-asociated CDK6, cyclin D, E, and A protein, indicating that LANA upregulates cyclin D, E, and A proteins during the G1/S phase of cell cycle (Paudel et al., 2012a). In consistent, to maintain the viral genome in each cell cycle, LANA recruits proliferating cell nuclear antigen (PCNA) via Bub1 and replication factor C (RFC) to initiate DNA replication of viral genome during S phase, and enhances survival of KSHV-infected cells in response to UV-induced DNA damage (Sun et al., 2014, 2015). In addition, to enhance the survival of KSHV-infected cells, LANA upregulates the angiogenic multifunctional protein angiongenin (ANG) (Sadagopan et al., 2009). Further studies showed that LANA interacts with ANG/annexin A2 to contribute to the binding of LANA with p53 (Paudel et al., 2012a,b).
DNA damage is a common phenomenon through exposure to a variety of environmental stresses, including abnormally low oxygen, or nutrients. Before cells enter mitosis, the G2/M checkpoint responds directly to DNA damage by repairing DNA breaks or alternately by holding cell cycle progression and/or by undergoing programmed cell death. The key target of G2 arrest is the mitosis promoting complex Cdk1-cyclin B. In response to different type of DNA damage, the ATM-Chk2 or the ATR-Chk1 signal pathway is activated to arrest the cell in G2 phase by blocking activation of Cdk1-Cyclin B or inhibition of Cdc25B/C phosphatase (an enzyme normally activates Cdk1 at G2/M transition). The increased phosphorylation of Cdc25B/C is observed during G2 arrest. Both Chk1 and Chk2 kinase are able to phosphorylate Cdc25B/C in response to DNA damage. The ATM/ATR kinase not only activates Chk1 and Chk2 but also phosphorylates p53 on its serine 15. Phosphorylation of p53 prevents p53 binding with MdM2, thereby stabilizing p53. The stabilized p53 in turn up-regulates the Cdk inhibitor p21 at the G1 checkpoint, which eventually leads to cell arrest or apoptosis. Although no literatures reported that LANA could directly target Cdc25B/C or Cdk1-cyclin B complex, many molecules [including p73 (Santag et al., 2013), ANG (Sadagopan et al., 2009), Mdm2 (Sarek et al., 2007), and GSK-3 (Woodard et al., 2015)] have been shown to interaction with LANA for blocking p53-mediated apoptosis during G2/M checkpoint. Recent studies showed that LANA upregulates the expression of survivin, and recruits Aurora kinase B to induce phosphorylation of Survivin at Tyrosine 34 (Lu et al., 2009, 2014), which potentially drive host cell proliferation during mitosis. Moreover, in human B cells, it has been demonstrated that LANA directly interacts with Chk2 (the ATM/ATR signaling effector) to relieve the nocodazole-induced G2/M checkpoint arrest (Kumar et al., 2014). This indicates that LANA also involve in the deregulation of G2/M checkpoint during cell progression (Figure 3).
In the view of the facts that in addition to promoting cell cycle arrest, most members of herpesviruses family also activate several factors to induce cell cycle progression. It is of importance and complex to elucidate how KSHV-encoded LANA balances the interaction between herpesvirus and cell cycle regulatory mechanisms. In the past two decades, numerous studies have shown that LANA is a multifunctional oncoprotein ubiquitously expressed in the KSHV-infected cells, and modulates various cellular pathways to drive cell proliferation. To seek the chemical compounds to efficiently block the KSHV-driven cell proliferation and its associated cancers, Gao and Schulz groups have shown that the small-molecule inhibitors Nutlin-3 and RETRA, which disrupt the interaction of p53 and p73 with MDM2, are efficient to individually induce apoptotic cell death in a p53 and p73-dependent manner (Ye et al., 2012; Santag et al., 2013). Glycyrrhizic acid (GA), a triterpenoid compound shown to inhibit the lytic replication of herpesviruses, is able to reduce the expression of LANA and leads to G1 cell cycle arrest and p53-mediated apoptosis of PEL cells (Curreli et al., 2005). Gamma-secretase inhibitor targeting LANA-mediated Notch pathway also inhibits cell growth and death (Liu et al., 2010). Taken together, these studies now clearly show that in addition to regulating transcription, chromatin remodeling, episome maintenance, DNA replication, and control of latency and lytic reactivation, LANA also plays a critical role in KSHV-mediated tumorigenesis by regulating cell cycle machinery. Understanding the role of LANA in regulation of cell proliferation, particularly on regulation of episome replication and segregation during cell cycle, will lead to a better understanding of cellular growth control processes, which will open an opportunity target to prevent and treat KSHV-associated malignancies.
FW, QC wrote, and reviewed the manuscript, JG, CW, CZ analyzed data.
This work is supported by the Research and Innovation Program of the Shanghai Municipal Education (13zz011 to QC), Shanghai Sailing Program (15YF1400900 to CZ), the National Natural Science Foundation of China (81471930 to QC, 81402542 to FW, 81501739 to CZ), and the National Key Basic Research “973” program of China (2012CB519001 to QC). FW is a scholar of Pujiang Talents in Shanghai. QC is a scholar of New Century Excellent Talents in University of China.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to apologize to the many researchers who have contributed to this area of research but have not been cited in this review due to space limitations.
Alani, R. M., Young, A. Z., and Shifflett, C. B. (2001). Id1 regulation of cellular senescence through transcriptional repression of p16/Ink4a. Proc. Natl. Acad. Sci. U.S.A. 98, 7812–7816. doi: 10.1073/pnas.141235398
An, F. Q., Compitello, N., Horwitz, E., Sramkoski, M., Knudsen, E. S., and Renne, R. (2005). The latency-associated nuclear antigen of Kaposi’s sarcoma-associated herpesvirus modulates cellular gene expression and protects lymphoid cells from p16 INK4A-induced cell cycle arrest. J. Biol. Chem. 280, 3862–3874. doi: 10.1074/jbc.M407435200
Bajaj, B. G., Verma, S. C., Lan, K., Cotter, M. A., Woodman, Z. L., and Robertson, E. S. (2006). KSHV encoded LANA upregulates Pim-1 and is a substrate for its kinase activity. Virology 351, 18–28. doi: 10.1016/j.virol.2006.03.037
Ballestas, M. E., Chatis, P. A., and Kaye, K. M. (1999). Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284, 641–644. doi: 10.1126/science.284.5414.641
Ballestas, M. E., and Kaye, K. M. (2011). The latency-associated nuclear antigen, a multifunctional protein central to Kaposi’s sarcoma-associated herpesvirus latency. Future Microbiol. 6, 1399–1413. doi: 10.2217/fmb.11.137
Barbera, A. J., Chodaparambil, J. V., Kelley-Clarke, B., Joukov, V., Walter, J. C., Luger, K., et al. (2006). The nucleosomal surface as a docking station for Kaposi’s sarcoma herpesvirus LANA. Science 311, 856–861. doi: 10.1126/science.1120541
Bubman, D., Guasparri, I., and Cesarman, E. (2007). Deregulation of c-Myc in primary effusion lymphoma by Kaposi’s sarcoma herpesvirus latency-associated nuclear antigen. Oncogene 26, 4979–4986. doi: 10.1038/sj.onc.1210299
Cai, Q., Cai, S., Zhu, C., Verma, S. C., Choi, J. Y., and Robertson, E. S. (2013). A unique SUMO-2-interacting motif within LANA is essential for KSHV latency. PLoS Pathog. 9:e1003750. doi: 10.1371/journal.ppat.1003750
Cai, Q., Lan, K., Verma, S. C., Si, H., Lin, D., and Robertson, E. S. (2006). Kaposi’s sarcoma-associated herpesvirus latent protein LANA interacts with HIF-1 alpha to upregulate RTA expression during hypoxia: latency control under low oxygen conditions. J. Virol. 80, 7965–7975. doi: 10.1128/JVI.00689-06
Cai, Q., Murakami, M., Si, H., and Robertson, E. S. (2007). A potential alpha-helix motif in the amino terminus of LANA encoded by Kaposi’s sarcoma-associated herpesvirus is critical for nuclear accumulation of HIF-1alpha in normoxia. J. Virol. 81, 10413–10423. doi: 10.1128/JVI.00611-07
Cai, Q., Verma, S. C., Choi, J. Y., Ma, M., and Robertson, E. S. (2010). Kaposi’s sarcoma-associated herpesvirus inhibits interleukin-4-mediated STAT6 phosphorylation to regulate apoptosis and maintain latency. J. Virol. 84, 11134–11144. doi: 10.1128/JVI.01293-10
Cai, Q., Xiao, B., Si, H., Cervini, A., Gao, J., Lu, J., et al. (2012). Kaposi’s sarcoma herpesvirus upregulates Aurora A expression to promote p53 phosphorylation and ubiquitylation. PLoS Pathog 8:e1002566. doi: 10.1371/journal.ppat.1002566
Cai, Q.-L., Knight, J. S., Verma, S. C., Zald, P., and Robertson, E. S. (2006). EC5S ubiquitin complex is recruited by KSHV latent antigen LANA for degradation of the VHL and p53 tumor suppressors. PLoS Pathog. 2:e116. doi: 10.1371/journal.ppat.0020116
Cheng, F., Weidner-Glunde, M., Varjosalo, M., Rainio, E. M., Lehtonen, A., Schulz, T. F., et al. (2009). KSHV reactivation from latency requires Pim-1 and Pim-3 kinases to inactivate the latency-associated nuclear antigen LANA. PLoS Pathog. 5:e1000324. doi: 10.1371/journal.ppat.1000324
Cotter, M. A. II, and Robertson, E. S. (1999). The latency-associated nuclear antigen tethers the Kaposi’s sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology 264, 254–264. doi: 10.1006/viro.1999.9999
Curreli, F., Friedman-Kien, A. E., and Flore, O. (2005). Glycyrrhizic acid alters Kaposi sarcoma-associated herpesvirus latency, triggering p53-mediated apoptosis in transformed B lymphocytes. J. Clin. Invest. 115, 642–652. doi: 10.1172/JCI23334
Dai, Y., and Grant, S. (2011). Methods to study cancer therapeutic drugs that target cell cycle checkpoints. Methods Mol. Biol. 782, 257–304. doi: 10.1007/978-1-61779-273-1_19
Davis, D. A., Rinderknecht, A. S., Zoeteweij, J. P., Aoki, Y., Read-Connole, E. L., Tosato, G., et al. (2001). Hypoxia induces lytic replication of Kaposi sarcoma-associated herpesvirus. Blood 97, 3244–3250. doi: 10.1182/blood.V97.10.3244
Di Bartolo, D. L., Cannon, M., Liu, Y. F., Renne, R., Chadburn, A., Boshoff, C., et al. (2008). KSHV LANA inhibits TGF-beta signaling through epigenetic silencing of the TGF-beta type II receptor. Blood 111, 4731–4740. doi: 10.1182/blood-2007-09-110544
Direkze, S., and Laman, H. (2004). Regulation of growth signalling and cell cycle by Kaposi’s sarcoma-associated herpesvirus genes. Int. J. Exp. Pathol. 85, 305–319. doi: 10.1111/j.0959-9673.2004.00407.x
Dupin, N., Fisher, C., Kellam, P., Ariad, S., Tulliez, M., Franck, N., et al. (1999). Distribution of human herpesvirus-8 latently infected cells in Kaposi’s sarcoma, multicentric Castleman’s disease, and primary effusion lymphoma. Proc. Natl. Acad. Sci. U.S.A. 96, 4546–4551. doi: 10.1073/pnas.96.8.4546
Fakhari, F. D., Jeong, J. H., Kanan, Y., and Dittmer, D. P. (2006). The latency-associated nuclear antigen of Kaposi sarcoma-associated herpesvirus induces B cell hyperplasia and lymphoma. J. Clin. Invest. 116, 735–742. doi: 10.1172/JCI26190
Friborg, J. Jr., Kong, W., Hottiger, M. O., and Nabel, G. J. (1999). p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 402, 889–894. doi: 10.1038/47266
Fujimuro, M., Wu, F. Y., ApRhys, C., Kajumbula, H., Young, D. B., Hayward, G. S., et al. (2003). A novel viral mechanism for dysregulation of beta-catenin in Kaposi’s sarcoma-associated herpesvirus latency. Nat. Med. 9, 300–306. doi: 10.1038/nm829
Gabrielli, B., Brooks, K., and Pavey, S. (2012). Defective cell cycle checkpoints as targets for anti-cancer therapies. Front. Pharmacol. 3:9. doi: 10.3389/fphar.2012.00009
Gan, J., Wang, C., Jin, Y., Guo, Y., Xu, F., Zhu, Q., et al. (2015). Proteomic profiling identifies the SIM-associated complex of KSHV-encoded LANA. Proteomics 15, 2023–2037. doi: 10.1002/pmic.201400624
Gao, S. J., Zhang, Y. J., Deng, J. H., Rabkin, C. S., Flore, O., and Jenson, H. B. (1999). Molecular polymorphism of Kaposi’s sarcoma-associated herpesvirus (Human herpesvirus 8) latent nuclear antigen: evidence for a large repertoire of viral genotypes and dual infection with different viral genotypes. J. Infect. Dis. 180, 1466–1476. doi: 10.1086/315098
Hume, A. J., and Kalejta, R. F. (2009). Regulation of the retinoblastoma proteins by the human herpesviruses. Cell Div. 4:1. doi: 10.1186/1747-1028-4-1
Johnson, D. G., and Walker, C. L. (1999). Cyclins and cell cycle checkpoints. Annu. Rev. Pharmacol. Toxicol. 39, 295–312. doi: 10.1146/annurev.pharmtox.39.1.295
Kastan, M. B., and Bartek, J. (2004). Cell-cycle checkpoints and cancer. Nature 432, 316–323. doi: 10.1038/nature03097
Katano, H., Sato, Y., Kurata, T., Mori, S., and Sata, T. (2000). Expression and localization of human herpesvirus 8-encoded proteins in primary effusion lymphoma, Kaposi’s sarcoma, and multicentric Castleman’s disease. Virology 269, 335–344. doi: 10.1006/viro.2000.0196
Kelley-Clarke, B., De Leon-Vazquez, E., Slain, K., Barbera, A. J., and Kaye, K. M. (2009). Role of Kaposi’s sarcoma-associated herpesvirus C-terminal LANA chromosome binding in episome persistence. J. Virol. 83, 4326–4337. doi: 10.1128/JVI.02395-08
Knight, J. S., Cotter, M. A. II, and Robertson, E. S. (2001). The latency-associated nuclear antigen of Kaposi’s sarcoma-associated herpesvirus transactivates the telomerase reverse transcriptase promoter. J. Biol. Chem. 276, 22971–22978. doi: 10.1074/jbc.M101890200
Komatsu, T., Ballestas, M. E., Barbera, A. J., Kelley-Clarke, B., and Kaye, K. M. (2004). KSHV LANA1 binds DNA as an oligomer and residues N-terminal to the oligomerization domain are essential for DNA binding, replication, and episome persistence. Virology 319, 225–236. doi: 10.1016/j.virol.2003.11.002
Kumar, A., Sahu, S. K., Mohanty, S., Chakrabarti, S., Maji, S., Reddy, R. R., et al. (2014). Kaposi sarcoma herpes virus latency associated nuclear antigen protein release the G2/M cell cycle blocks by modulating ATM/ATR mediated checkpoint pathway. PLoS ONE 9:e100228. doi: 10.1371/journal.pone.0100228
Lan, K., Verma, S. C., Murakami, M., Bajaj, B., Kaul, R., and Robertson, E. S. (2007). Kaposi’s sarcoma herpesvirus-encoded latency-associated nuclear antigen stabilizes intracellular activated Notch by targeting the Sel10 protein. Proc. Natl. Acad. Sci. U.S.A. 104, 16287–16292. doi: 10.1073/pnas.0703508104
Liu, J., Martin, H., Shamay, M., Woodard, C., Tang, Q. Q., and Hayward, S. D. (2007a). Kaposi’s sarcoma-associated herpesvirus LANA protein downregulates nuclear glycogen synthase kinase 3 activity and consequently blocks differentiation. J. Virol. 81, 4722–4731. doi: 10.1128/JVI.02548-06
Liu, J., Martin, H. J., Liao, G., and Hayward, S. D. (2007b). The Kaposi’s sarcoma-associated herpesvirus LANA protein stabilizes and activates c-Myc. J. Virol. 81, 10451–10459. doi: 10.1128/JVI.00804-07
Liu, R., Li, X., Tulpule, A., Zhou, Y., Scehnet, J. S., Zhang, S., et al. (2010). KSHV-induced notch components render endothelial and mural cell characteristics and cell survival. Blood 115, 887–895. doi: 10.1182/blood-2009-08-236745
Lu, J., Jha, H. C., Verma, S. C., Sun, Z., Banerjee, S., Dzeng, R., et al. (2014). Kaposi’s sarcoma-associated herpesvirus-encoded LANA contributes to viral latent replication by activating phosphorylation of survivin. J. Virol. 88, 4204–4217. doi: 10.1128/JVI.03855-13
Lu, J., Verma, S. C., Murakami, M., Cai, Q., Kumar, P., Xiao, B., et al. (2009). Latency-associated nuclear antigen of Kaposi’s sarcoma-associated herpesvirus (KSHV) upregulates survivin expression in KSHV-Associated B-lymphoma cells and contributes to their proliferation. J. Virol. 83, 7129–7141. doi: 10.1128/JVI.00397-09
Lukas, J., Lukas, C., and Bartek, J. (2004). Mammalian cell cycle checkpoints: signalling pathways and their organization in space and time. DNA Repair (Amst.) 3, 997–1007. doi: 10.1016/j.dnarep.2004.03.006
Nascimento, R., Costa, H., and Parkhouse, R. M. (2012). Virus manipulation of cell cycle. Protoplasma 249, 519–528. doi: 10.1007/s00709-011-0327-9
Nickoloff, B. J., Chaturvedi, V., Bacon, P., Qin, J. Z., Denning, M. F., and Diaz, M. O. (2000). Id-1 delays senescence but does not immortalize keratinocytes. J. Biol. Chem. 275, 27501–27504. doi: 10.1074/jbc.C000311200
Ohsaki, E., and Ueda, K. (2012). Kaposi’s sarcoma-associated herpesvirus genome replication, partitioning, and maintenance in latency. Front. Microbiol. 3:7. doi: 10.3389/fmicb.2012.00007
Ottinger, M., Christalla, T., Nathan, K., Brinkmann, M. M., Viejo-Borbolla, A., and Schulz, T. F. (2006). Kaposi’s sarcoma-associated herpesvirus LANA-1 interacts with the short variant of BRD4 and releases cells from a BRD4- and BRD2/RING3-induced G1 cell cycle arrest. J. Virol. 80, 10772–10786. doi: 10.1128/JVI.00804-06
Parravicini, C., Chandran, B., Corbellino, M., Berti, E., Paulli, M., Moore, P. S., et al. (2000). Differential viral protein expression in Kaposi’s sarcoma-associated herpesvirus-infected diseases: Kaposi’s sarcoma, primary effusion lymphoma, and multicentric Castleman’s disease. Am. J. Pathol. 156, 743–749. doi: 10.1016/S0002-9440(10)64940-1
Paudel, N., Sadagopan, S., Balasubramanian, S., and Chandran, B. (2012a). Kaposi’s sarcoma-associated herpesvirus latency-associated nuclear antigen and angiogenin interact with common host proteins, including annexin A2, which is essential for survival of latently infected cells. J. Virol. 86, 1589–1607. doi: 10.1128/JVI.05754-11
Paudel, N., Sadagopan, S., Chakraborty, S., Sarek, G., Ojala, P. M., and Chandran, B. (2012b). Kaposi’s sarcoma-associated herpesvirus latency-associated nuclear antigen interacts with multifunctional angiogenin to utilize its antiapoptotic functions. J. Virol. 86, 5974–5991. doi: 10.1128/JVI.00070-12
Petre, C. E., Sin, S. H., and Dittmer, D. P. (2007). Functional p53 signaling in Kaposi’s sarcoma-associated herpesvirus lymphomas: implications for therapy. J. Virol. 81, 1912–1922. doi: 10.1128/JVI.01757-06
Platt, G. M., Simpson, G. R., Mittnacht, S., and Schulz, T. F. (1999). Latent nuclear antigen of Kaposi’s sarcoma-associated herpesvirus interacts with RING3, a homolog of the Drosophila female sterile homeotic (fsh) gene. J. Virol. 73, 9789–9795.
Radkov, S. A., Kellam, P., and Boshoff, C. (2000). The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nat. Med. 6, 1121–1127. doi: 10.1038/80459
Russo, J. J., Bohenzky, R. A., Chien, M. C., Chen, J., Yan, M., Maddalena, D., et al. (1996). Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. U.S.A. 93, 14862–14867. doi: 10.1073/pnas.93.25.14862
Sadagopan, S., Sharma-Walia, N., Veettil, M. V., Bottero, V., Levine, R., Vart, R. J., et al. (2009). Kaposi’s sarcoma-associated herpesvirus upregulates angiogenin during infection of human dermal microvascular endothelial cells, which induces 45S rRNA synthesis, antiapoptosis, cell proliferation, migration, and angiogenesis. J. Virol. 83, 3342–3364. doi: 10.1128/JVI.02052-08
Santag, S., Jager, W., Karsten, C. B., Kati, S., Pietrek, M., Steinemann, D., et al. (2013). Recruitment of the tumour suppressor protein p73 by Kaposi’s Sarcoma Herpesvirus latent nuclear antigen contributes to the survival of primary effusion lymphoma cells. Oncogene 32, 3676–3685. doi: 10.1038/onc.2012.385
Sarek, G., Kurki, S., Enback, J., Iotzova, G., Haas, J., Laakkonen, P., et al. (2007). Reactivation of the p53 pathway as a treatment modality for KSHV-induced lymphomas. J. Clin. Invest. 117, 1019–1028. doi: 10.1172/JCI30945
Si, H., and Robertson, E. S. (2006). Kaposi’s sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen induces chromosomal instability through inhibition of p53 function. J. Virol. 80, 697–709. doi: 10.1128/JVI.80.2.697-709.2006
Srinivasan, V., Komatsu, T., Ballestas, M. E., and Kaye, K. M. (2004). Definition of sequence requirements for latency-associated nuclear antigen 1 binding to Kaposi’s sarcoma-associated herpesvirus DNA. J. Virol. 78, 14033–14038. doi: 10.1128/JVI.78.24.14033-14038.2004
Sun, Q., Tsurimoto, T., Juillard, F., Li, L., Li, S., De Leon Vazquez, E., et al. (2014). Kaposi’s sarcoma-associated herpesvirus LANA recruits the DNA polymerase clamp loader to mediate efficient replication and virus persistence. Proc. Natl. Acad. Sci. U.S.A. 111, 11816–11821. doi: 10.1073/pnas.1404219111
Sun, Z., Jha, H. C., and Robertson, E. S. (2015). Bub1 in complex with LANA recruits PCNA to regulate kaposi’s sarcoma-associated herpesvirus latent replication and DNA translesion synthesis. J. Virol. 89, 10206–10218. doi: 10.1128/JVI.01524-15
Tang, J., Gordon, G. M., Muller, M. G., Dahiya, M., and Foreman, K. E. (2003). Kaposi’s sarcoma-associated herpesvirus latency-associated nuclear antigen induces expression of the helix-loop-helix protein Id-1 in human endothelial cells. J. Virol. 77, 5975–5984. doi: 10.1128/JVI.77.10.5975-5984.2003
Toth, Z., Brulois, K., and Jung, J. U. (2013). The chromatin landscape of Kaposi’s sarcoma-associated herpesvirus. Viruses 5, 1346–1373. doi: 10.3390/v5051346
Uppal, T., Banerjee, S., Sun, Z., Verma, S. C., and Robertson, E. S. (2014). KSHV LANA–the master regulator of KSHV latency. Viruses 6, 4961–4998. doi: 10.3390/v6124961
Verma, S. C., Borah, S., and Robertson, E. S. (2004). Latency-associated nuclear antigen of Kaposi’s sarcoma-associated herpesvirus up-regulates transcription of human telomerase reverse transcriptase promoter through interaction with transcription factor Sp1. J. Virol. 78, 10348–10359. doi: 10.1128/JVI.78.19.10348-10359.2004
Viejo-Borbolla, A., Ottinger, M., Bruning, E., Burger, A., Konig, R., Kati, E., et al. (2005). Brd2/RING3 interacts with a chromatin-binding domain in the Kaposi’s Sarcoma-associated herpesvirus latency-associated nuclear antigen 1 (LANA-1) that is required for multiple functions of LANA-1. J. Virol. 79, 13618–13629. doi: 10.1128/JVI.79.21.13618-13629.2005
Woodard, C., Liao, G., Goodwin, C. R., Hu, J., Xie, Z., Dos Reis, T. F., et al. (2015). A screen for extracellular signal-regulated kinase-primed glycogen synthase kinase 3 substrates identifies the p53 inhibitor iASPP. J. Virol. 89, 9232–9241. doi: 10.1128/JVI.01072-15
Ye, F., Lattif, A. A., Xie, J., Weinberg, A., and Gao, S. (2012). Nutlin-3 induces apoptosis, disrupts viral latency and inhibits expression of angiopoietin-2 in Kaposi sarcoma tumor cells. Cell Cycle 11, 1393–1399. doi: 10.4161/cc.19756
Keywords: cell cycle, LANA, KSHV
Citation: Wei F, Gan J, Wang C, Zhu C and Cai Q (2016) Cell Cycle Regulatory Functions of the KSHV Oncoprotein LANA. Front. Microbiol. 7:334. doi: 10.3389/fmicb.2016.00334
Received: 12 November 2015; Accepted: 03 March 2016;
Published: 30 March 2016.
Edited by:
Erle S. Robertson, University of Pennsylvania, USAReviewed by:
Masahiro Fujimuro, Kyoto Pharmaceutical University, JapanCopyright © 2016 Wei, Gan, Wang, Zhu and Cai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiliang Cai, cWlsaWFuZ0BmdWRhbi5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.