
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 01 March 2016
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 7 - 2016 | https://doi.org/10.3389/fmicb.2016.00219
Emergence of antimicrobial resistance coupled with the slowdown in discovery of new antimicrobial compounds points to serious consequences for human health. Therefore, scientists are looking for new antimicrobial compounds from unique and understudied ecosystems such as tropical peat swamp forests. Over the course of isolating antimicrobial producing bacteria from North Selangor tropical peat swamp forest, Malaysia, a Gram variable, rod shaped, endospore forming, facultative anaerobic novel strain MSt1T that exerts potent and broad spectrum antimicrobial activity was isolated. Phylogenetic analysis using 16S rRNA gene sequences showed that strain MSt1T belonged to the genus Paenibacillus with the highest similarity to Paenibacillus elgii SD17T (99.5%). Whole genome comparison between strain MSt1T with its closely related species using average nucleotide identity (ANI) revealed that similarity between strain MSt1T with P. elgii B69 (93.45%) and Paenibacillus ehimensis A2 (90.42%) was below the recommended threshold of 95%. Further analysis using in silico pairwise DDH also showed that similarity between strain MSt1T with P. elgii B69 (55.4%) and P. ehimensis A2 (43.7%) was below the recommended threshold of 70%. Strain MSt1T contained meso-diaminopilemic acid in the cell wall and MK-7 as the major menaquinone. The major fatty acids of strain MSt1T were anteiso-C15:0 (48.2%) and C16:0 (29.0%) whereas the polar lipid profile consisted of phosphatidylglycerol, phosphatidylethanolamine, diphosphatidylglycerol, one unknown lipid, two unknown glycolipids, and one unknown phospholipid. Total DNA G+C content of strain MSt1T was 51.5 mol%. The extract from strain MSt1T exerted strong antimicrobial activity against Escherichia coli ATCC 25922 (MIC = 1.5 μg/mL), MRSA ATCC 700699 (MIC = 25 μg/mL) and Candida albicans IMR (MIC = 12.5 μg/mL). Partially purified active fraction exerted a strong effect against E. coli ATCC 25922 resulting in cell rupture when viewed with SEM. Based on distinctive taxonomic differences between strain MSt1T when compared to its closely related type species, we propose that strain MSt1T represents a novel species within the genus of Paenibacillus, for which the name Paenibacillus tyrfis sp. nov. (= DSM 100708T = MCCC 1K01247T) is proposed.
Antimicrobial resistant bacteria (ARB) are a major concern because infections by ARB are directly responsible for not only increased healthcare costs but also high mortality rates (Boucher et al., 2009; Davies and Davies, 2010). Infections by ARB cost approximately $55 billion annually in the United States (Smith and Coast, 2013). Currently, more than 70% of nosocomial pathogens are resistant to at least one of the antimicrobials commonly used to treat infections caused by them (Mishra et al., 2012). The rapid emergence of ARB compounded with the decline in new antimicrobial discoveries further complicates the problem. For example, two new antimicrobials that have been clinically approved in the past decade are linezolid and daptomycin. However, bacteria resistant to these antimicrobials were already reported shortly after their introduction (Meka and Gold, 2004; Hayden et al., 2005). Therefore, there is a dire need to discover new antimicrobials to combat ARB related infections.
One strategy to look for novel bioactive compounds is bioprospecting in extreme, understudied ecosystems. The discovery of abyssomycin produced by Verrucosispora spp. isolated from South China Sea sediment is an example of successful bioprospecting (Bister et al., 2004). Bacteria produce a wide array of compounds to obtain a competitive advantage by suppressing their competitors and to colonize new habitats. Bacteria capable of thriving in such extreme conditions are very likely to evolve special adaptations and produce unique molecules or compounds that may have beneficial uses (Kohama et al., 1993; Pettit, 2011).
Indo-Malaysian tropical peat swamp forests are an example of a unique ecosystem, characterized by the formation of deep layers of peat in an acidic, waterlogged, nutrient poor environment. Despite such a challenging habitat, there is an enormous microbial diversity in tropical peat swamp forests. Kanokratana et al. (2011) performed a metagenomic study on Thai tropical peat soil and showed that 80% of the bacterial community could be potentially novel species or strains. The North Selangor tropical peat swamp forest in Malaysia was therefore chosen to be the site of study and during the course of isolating antimicrobial producing bacteria, a novel bacteria strain MSt1T was isolated (Aw et al., 2014). Strain MSt1T exerts broad spectrum antimicrobial activity against many clinically relevant pathogens such as methicillin resistant Staphylococcus aureus (MRSA), vancomycin resistant Enterococcus (VRE), Escherichia coli, Pseudomonas aeruginosa, Salmonella Typhimurium, Candida albicans and Cryptococcus neoformans to name a few. Further analysis revealed that strain MSt1T belonged to the genus Paenibacillus.
The genus Paenibacillus was proposed by Ash et al. (1993) to distinguish members of the “16S rRNA group 3″ bacilli from other members in the genus Bacillus. Currently, there are 165 species in the genus Paenibacillus and the type species is Paenibacillus polymyxa. Members of Paenibacillus are known to be Gram variable, rod shaped, aerobic or facultatively anaerobic, endospore forming and motile via peritrichous flagella. DNA G+C content of Paenibacillus species is between 39 and 54 mol%, the major cellular fatty acid is anteiso-C15:0, the cell wall peptidoglycan diamino acid is meso-diaminopimelic acid and the major menaquinone is menaquinone-7 (MK-7; Shida et al., 1997). Members belonging to the genus Paenibacillus are well known to produce a wide range of antimicrobial compounds e.g., P. ehimensis that produces a cyclic lipopeptide (Aktuganov et al., 2008; Huang et al., 2013), P. koreensis that produces an iturin-like antifungal (Chung et al., 2000), P. elgii that produces pelgipeptins (Kim et al., 2004; Wu et al., 2010; Ding et al., 2011) and P. polymyxa that produces polymyxin B (Paulus and Gray, 1964; Shaheen et al., 2011).
In this study, Paenibacillus sp. strain MSt1T was subjected to a polyphasic taxonomic approach as described by Logan et al. (2009) with respect to the minimal standards for description of aerobic, endospore-forming bacteria and results indicated that strain MSt1T represented a novel species within the genus Paenibacillus, for which the name Paenibacillus tyrfis sp. nov. is proposed. The antimicrobial activity of the extract produced by strain MSt1T was characterized and chemical analysis was performed to determine the constituents present in the extract.
Strain MSt1T was isolated as described in Aw et al. (2014). Briefly, strain MSt1T was isolated from North Selangor tropical peat swamp soil (3° 39′ 30.8″ N; 101° 19′ 18.4″ E) from surface peat soil. Strain MSt1T was selected for further study due to its wide spectrum antimicrobial activity against these indicator pathogens. Pure cultures of strain MSt1T were routinely maintained on TSA at 30°C and long term storage was performed via cryopreservation with 25% (v/v) glycerol at −80°C.
Genomic DNA was extracted from a 3 day old culture of strain MSt1T on TSA using GF-1 DNA extraction kit (Vivantis, Malaysia) according to manufacturer's instructions. The almost complete 16S rRNA gene sequence of strain MSt1T (1492 bp; DDBJ/EMBL/GenBank accession number KT216503) was amplified according to Lee et al. (2014) using the primer pairs of 27f and 1492r. Briefly, 16S rRNA gene sequence was amplified using universal primer pair 27f (5′ AGAGTTTGATCMTGGCTCAG 3′) and 1492r (5′ TACGGYTACCTTGTTACGACTT 3′). PCR reaction mixture contained 5 × MyTaq Red Buffer, 1.25 U MyTaq DNA polymerase (Bioline, United Kingdom), 0.5 μM of 27f forward primer, 0.5 μM of 1492r reverse primer and 3 μL of DNA template with the final volume of 50 μL. The thermal cycling profile of initial denaturation at 95°C for 5 min followed by 30 cycles of denaturation at 95°C for 1 min, annealing at 55°C for 1 min and elongation at 72°C for 1 min were used.
The resulting 16S rRNA gene sequence was compared to the sequences in the EzTaxon database (http://eztaxon-e.ezbiocloud.net/; Kim et al., 2012). The 16S rRNA gene sequence of strain MSt1T was aligned with sequences of closely related type species that had been retrieved from the GenBank/EMBL/DDBJ databases using CLUSTAL-X software (Thompson et al., 1997). The alignment was manually verified and adjusted prior to the construction of the phylogenetic tree using the neighbor-joining algorithm (Saitou and Nei, 1987) with the MEGA version 6 software (Tamura et al., 2013). The EzTaxon-e server (http://eztaxon-e.ezbiocloud.net/; Kim et al., 2012) was used for calculations of sequence similarity level. The stability of the resultant tree topologies were evaluated by using the bootstrap resampling method (1000 resampling) of Felsenstein (1985). The evolutionary distances were computed using Kimura's two-parameter model (Kimura, 1980). The 16S rRNA gene sequence of strain MSt1T was deposited at DDBJ/EMBL/GenBank under the accession no. KT216503.
The draft genome sequence of strain MSt1T was obtained as described in Aw et al. (2014). The draft genome of strain MSt1T was deposited at DDBJ/EMBL/GenBank under the accession no. JNVM00000000.
As the 16S rRNA gene sequence of strain MSt1T showed more than 97% similarity when compared with its closely related type species, analyses that compare between genomes of closely related species such as ANI and DDH using the draft genome sequence of strain MSt1T were performed. ANI determination was performed at EzGenome (http://www.ezbiocloud.net/ezgenome/ani) as described by Goris et al. (2007) by comparing the draft genome of strain MSt1T to its closely related isolates as determined from the phylogenetic analysis of the 16S rRNA sequences. Estimation of in silico pairwise DDH was performed with genome to genome distance calculator (GGDC 2.0; ; Meier-Kolthoff et al., 2013).
Phenotypic analysis was performed using growths on TSA at 30°C for 3 days unless otherwise mentioned. Colony morphology was determined using growths on TSA at 30°C for 3 days. Cellular morphology (vegetative cells, sporulating cells, and flagellation) was observed using a light microscope (Olympus EX43) and scanning electron microscopy (Hitachi SU8010) following growth in tryptone soy broth (TSB) at 30°C for 5 days (Figure S1). Sporulation was promoted by growing the strain on TSB supplemented with 10 μg/mL MnSO4 at 30°C for 5 days and stained with Schaeffer-Fulton stain (Schaeffer and Fulton, 1933). Gram staining was performed using standard Gram staining procedure and Gram reaction was confirmed using the KOH lysis test (Cerny, 1978; Moaledj, 1986). Flagellation was determined using Ryu's flagella stain (Kodaka et al., 1982). Motility was determined using semi-solid agar as described by Tittsler and Sandholzer (1936).
The following phenotypic tests were performed as described by Wu et al. (2011) and Glaeser et al. (2013). Optimal temperature study was performed using growth in TSB at 4, 15, 25, 30, 37, 42, and 50°C for 7 days. Optimal pH for growth was studied with pH between 3 and 10 (1 pH unit interval) and salt tolerance was determined with 0–5% (w/v) NaCl (1% intervals) using TSB at 30°C for 7 days. Catalase activity was tested by bubble production with 3% (v/v) hydrogen peroxide solution (Merck). Oxidase activity was tested with color change using 1% tetramethyl-p-phenylenediamine (BD). Acid production from a sole carbon source were determined using API 50CH (bioMérieux) according to the manufacturer's instructions. API ZYM and API 20E (bioMérieux) strips were used to determine enzyme activity of strain MSt1T.
For the chemotaxonomic approach, analyses of polar lipid, respiratory quinone, cellular fatty acids, and cell wall peptidoglycan were performed by Identification Service of DSMZ (Braunschweig, Germany). Biomass of strain MSt1T was grown in TSB at 30°C for 5 days. Polar lipids and major respiratory quinones were extracted and analyzed using thin layer chromatography (TLC) as described by Kates (1986). Fatty acid analysis was performed according to Miller (1982) and Kuykendall et al. (1988). Biomass of strain MSt1T and its reference species were grown in TSB at 30°C for 5 days. Determination of cell wall peptidoglycan was performed according to Rhuland et al. (1955) with the freeze-dried biomass of strain MSt1T, grown in TSB at 30°C for 5 days.
Partial purification of extract produced by strain MSt1T was performed according to Ding et al. (2011) with modification. Briefly, strain MSt1T was grown on TSA for 3–4 days at 30°C. Whole agar including the bacteria was placed into a glass jar and extraction was performed three times using 100% acetonitrile (Merck, Germany). The extract was concentrated under reduced pressure using a rotary evaporator and it was then freeze-dried. The dried crude extract was then packed into a C-18 (Merck) column and eluted with 50:50, 70:30, and 100:0% methanol:water. The active fraction (100% methanol) was collected and concentrated under reduced pressure using a rotary evaporator followed by freeze-drying. The active fraction was further fractionated using a preparative HPLC system [Agilent Infinity 1260 Quaternary LC system (Agilent, USA)] with a reverse phase column [Purospher STAR RP-18 endcapped (Merck), 4.6 μm, 250 × 20 mm]. Mobile phase A consisted of 100% Milli-Q water and mobile phase B consisted of 100% methanol. A linear gradient of 60–95% B in 15 min at a flow rate of 18 mL/min were used as elution and monitored at 254 nm. All isolated peaks were collected and the active fraction was evaporated and stored at -20°C for further analysis.
Antimicrobial activity of the active fraction from strain MSt1T was tested against E. coli ATCC 25922, P. aeruginosa ATCC 10145, methicillin sensitive S. aureus (MSSA) ATCC 29213, MRSA 700699, VRE ATCC 700802, and C. albicans from IMR (Institute for Medical Research, Malaysia) via the broth microdilution method as described by CLSI (2012). Briefly, pathogens were grown overnight in brain heart infusion broth (BHI) and adjusted to 0.5 McFarland standard (OD625 at 0.08–0.13). This adjusted culture was diluted 100 times before being used as the inoculum for broth microdilution. The active fraction was reconstituted in phosphate buffered saline (PBS; Oxoid, UK) at 50 mg/mL and serially diluted in the 96-well microtitre plate (SPL Life Science, Korea) using sterile BHI broth. Diluted inoculum (100 μL) was added to the microtiter plate. Positive control used against bacteria was 1.25 mg/mL chloramphenicol (Nacalai-Tesque, Japan) whereas positive control used against C. albicans was 5 mg/mL cycloheximide (Nacalai-Tesque). The microtitre plate was incubated at 37°C for 18–24 h. All MIC determination were performed in triplicates. MIC was determined by the lowest concentration of active fraction where no visible growth was observed in the well according to CLSI (2012).
The mode of action of the active fraction produced by strain MSt1T was studied by looking at the effect of the antimicrobial compound on the surface structure of the E. coli ATCC 25922. The cellular structure of E. coli after treatment with the active fraction was examined using field emission scanning electron microscopy (FE-SEM). Briefly, E. coli was grown in BHI at 37°C for 18 h. The culture was adjusted to McFarland 0.5 (OD625 at 0.08–0.13) and a final concentration of 0.1 mg/mL of active fraction was added and incubated at 37°C for 4 h. The culture was centrifuged at 5000 × g for 3 min and the supernatant was removed. The pellet was washed with PBS (Oxoid) and subjected to centrifugation at 5000 × g for 3 min. This washing process was performed three times. The washed pellet was reconstituted with minimal volume of PBS, placed onto a glass slide and allowed to air dry for 30 min.
The slide was fixed using 2.5% (v/v) gluteraldehyde (Sigma, USA) in PBS overnight and washed three times using PBS. The slide was then subjected to serial dehydration from 20% ethanol to 100% ethanol for 10 min on each step and kept in a desiccator overnight. The slide was spur-coated with platinum using Q150R Rotary-Pumped Sputter Coater before being observed using Hitachi SU8010 FE-SEM.
The 16S rRNA gene sequence of strain MSt1T was found to be similar to P. elgii SD17T (99.5%), P. ehimensis KCTC 3748T (98.8%), and P. tianmuensis B27T (98.2%), whereas sequences similarities of less than 97.9% were obtained to other Paenibacillus species. Phylogenetic trees were constructed to determine the phylogenetic position of this strain (Figure 1). The phylogenetic analysis revealed that strain MSt1T was closely related to the type strain P. elgii SD17T as they formed a distinct clade supported by a high bootstrap value (94%), indicating a high confidence level of this association (Figure 1). The closest related species, P. elgii SD17T, P. ehimensis KCTC 3748T, and P. tianmuensis B27T were used as reference strains for further polyphasic analysis.

Figure 1. Neighbor-joining tree based on 16S rRNA sequences showing relationships between strain MSt1T and representatives of other related taxa. Bootstrap values (>50%) based on 1000 resampled datasets are shown at branch nodes. Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446T was used as the outgroup. Bar, 2 substitutions per 100 nucleotide positions.
The draft genome sequence of strain MSt1T was reported in Aw et al. (2014). As the 16S rRNA gene sequence of strain MSt1T showed more than 97% similarity when compared with its closely related type species, alternative methods such as DDH should be performed as recommended by Tindall et al. (2010). With the advance in genomics and whole genome sequencing, in lieu of DDH, direct comparison between genome sequences between strains can be performed with methods such as ANI and in silico pairwise DDH. Using the draft genome of strain MSt1T and compared with P. elgii B69 and P. ehimensis A2, the ANI value were 93.45 and 90.42%, respectively. The threshold of 70% genomic relatedness with DDH is generally recommended for species delineation (Wayne et al., 1987) and has been found to correlate to 95–96% average nucleotide identity (ANI; Goris et al., 2007; Richter and Rosselló-Móra, 2009; Kim et al., 2014). As the ANI value obtained was below the 95–96% threshold, this supports the proposal that strain MSt1T belongs to a novel species.
Further analyses using the draft genome sequence were performed using in silico pairwise DDH determination. Using the draft genome of strain MSt1T and compared with P. elgii B69 and P. ehimensis A2, the in silico pairwise DDH value was estimated to be 55.40 ± 2.73% and 43.70 ± 2.54%, which was below the recommended threshold of 70% as recommended by Wayne et al. (1987). Therefore, the results of ANI and in silico DDH both agreed that strain MSt1T represents a new species at the genome level.
Strain MSt1T was found to be Gram variable rods, facultatively anaerobic, endospore-forming, and motile via peritrichous flagella. Colonies grown on TSA are milky white, round, sticky colonies. Colonies are approximately 5–8 mm in diameter after incubation for 3 days at 30°C on TSA. Cells are approximately 3–5 μm in length and 0.6–1.0 μm in width. Ellipsoidal endospores were found in a terminal position. The growth temperature ranges from 20 to 42°C with an optimal temperature of 30°C. Growth occurs between pH 3 and 10 (optimum at pH 6–7) and between 0 and 2% (w/v) NaCl (optimum growth at 1%). Strain MSt1T is both catalase, oxidase and haemolysis positive.
The physiological and biochemical characteristics of strain MSt1T were compared with its most closely related type strains (Table 1) and showed distinctive differences from these type strains. Strain MSt1T was found to show distinctive differences as compared to its closely related type strains in enzymatic reaction of α-glucosidase and β-glucosidase together with the inability to produce indole and ferment L-arabinose.
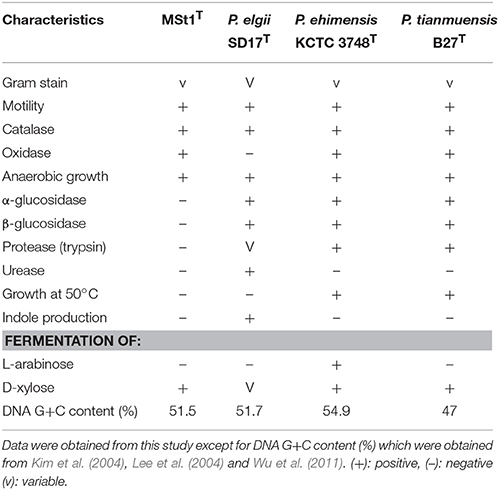
Table 1. Distinctive phenotypic characteristics of strain MSt1T when compared to its closely related type species of Paenibacillus.
The polar lipid profile of strain MSt1T consisted of phosphatidylglycerol (PG), phosphatidylethanolamine (PE), diphosphatidylglycerol (DPG), one unknown lipid, two unknown glycolipids and one unknown phospholipid (Figure S2). Members of the genus Paenibacillus are known to have DPG as the major polar lipid with some species containing PE and PG. The major respiratory quinones of strain MSt1T was found to be MK-7 which is characteristic of members of Paenibacillus (Shida et al., 1997).
The major fatty acid found in strain MSt1T was anteiso-C15:0 (48.2%) and C16:0 (29.0%; Table 2). The fatty acid profile of strain MSt1T was found to be similar to the reference strains where all three reference species also contained anteiso-C15:0 (39.9–50.0%) and C16:0 (22.0–37.0%) as their major fatty acid. The main difference between strain MSt1T and P. elgii SD17T was that strain MSt1T was nearly double that of anteiso-C15:0as compared to C16:0 whereas P. elgii SD17T contained almost equal amounts of both anteiso-C15:0 and C16:0. It was also found that the cell wall peptidoglycan of strain MSt1T was meso-diaminopimelic acid which was consistent with other species of the genus Paenibacillus (Shida et al., 1997).
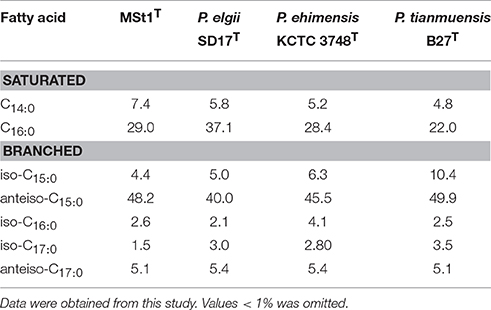
Table 2. Cellular fatty acid profile of strain MSt1T when compared to its closely related type species of Paenibacillus.
In view of morphological, physiological, chemotaxonomic, phylogenetic and genomic results obtained for strain MSt1T, it is evident that strain MSt1T belongs to the genus Paenibacillus. Distinct differences in physiological, chemotaxonomic and genomic data between strain MSt1T and its closely related type species make it evident that strain MSt1T represent a novel species for which the name P. tyrfis sp. nov. is proposed.
The active fraction from strain MSt1T showed strong antimicrobial effects against all six pathogens tested (Table 3). The active fraction was found to be more effective against Gram negative bacteria such as E. coli (MIC = 1.5 μg/mL) and yeasts such as C. albicans (MIC = 12.5 μg/mL) as compared to Gram positive bacteria such as E. faecalis (MIC = 50 μg/mL).
As the active fraction produced by strain MSt1T was found to be most effective against E. coli, SEM imaging was used to study the antimicrobial effect on the surface morphology of E. coli. E. coli treated with 0.1 mg/mL of the active fraction resulted in cell rupture as compared to the control that was being treated with PBS (Figure 2). Cell rupture can be caused by several reasons such as disruption of cell membrane or weakening of cell wall (Thimon et al., 1995; Domenech et al., 2009; Velkov et al., 2014). Several classes of antimicrobial compounds are well known to target those sites such as glycopeptide antibiotics that disrupts the D-ala-D-ala sequence of lipid II, resulting in the disruption of transpeptidation and transglycosylation of peptidoglycan synthesis (Higgins et al., 2005; Domenech et al., 2009). This causes the weakening of the cell wall that leads to cell lysis. Another class of antimicrobial known as lipopeptide acts as a detergent that disrupts the stability of cell membrane, also leading to cell lysis (Domenech et al., 2009).

Figure 2. Scanning electron microscopy images on the antimicrobial effect of active fraction produced by strain MSt1T against E. coli ATCC 25922 at 5000x magnification. (A) Negative control (treatment with PBS); (B) Treatment with 0.1 mg/mL of active fraction produced by strain MSt1T. Arrows points at the cell rupture of E. coli after treatment.
Members of Paenibacillus are well known to produce a wide array of antimicrobials, in particular antimicrobials belonging to the class of lipopeptides eg. polymyxin B produced by P. polymyxa (He et al., 2007), battacin by Paenibacillus tianmuensis (Qian et al., 2012) and pelgipeptin by Paenibacillus elgii (Wu et al., 2010; Ding et al., 2011). Preliminary chemical analysis of the active fraction using mass spectrometry revealed possible presence of lipopeptide class of antimicrobial compounds (data not shown). However, further purification of the antimicrobial is required for accurate identification of the antimicrobial compounds.
This study highlighted several key points. Although tropical peat swamp forests are known to be challenging environments for the survival of microorganisms, this study showed that tropical peat swamp forests harbor novel bacteria with the isolation and characterization of strain MSt1T as P. tyrfis. Furthermore, strain MSt1T also exhibited strong antimicrobial activity against a wide spectrum of pathogens tested; particularly the induction of cell rupture of E. coli when treated with the active fraction produced by strain MSt1T. Preliminary characterization of the active fraction highly suggests that the active fraction contains a lipopeptide-like compound. Further investigation in terms of identification and the exact mode of action of the antimicrobial compound produced by strain MSt1T will be carried out.
P. tyrfis (ty.r'fis G. gen. n. tyrfi of peat, referring to the tropical peat swamp soil, the geographical origin of the type strain).
Cells are stained Gram variable rods, facultatively anaerobic, endospore-forming, and motile via peritrichous flagella. Colonies grown on TSA are milky white, round, sticky colonies. Colonies are approximately 5–8 mm in diameter after incubation for 3 days at 30°C on TSA. Cells are approximately 3–5 μm in length and 0.6–1.0 μm in width. Ellipsoidal endospores were found in a terminal position. The growth temperature ranges from 20 to 42°C with an optimal temperature of 30°C. Growth occurs between pH 3 and 10 (optimum at pH 6 and 7) and between 0 and 2% (w/v) NaCl (optimum growth at 1%). Strain MSt1T is both catalase, oxidase and haemolysis positive. Based on API ZYM test strip results, strain MSt1T is positive for alkaline phosphatase, esterase (C4), esterase lipase (C8), lipase (C4), leucine arylamidase, acid phosphatse, naphthol-AS-bi-phosphohydrolase, β-glucuronidase, but negative for valine arylamidase, cysteine arylamidase, trypsin, α-chymotrypsin, α-galactosidase, β-galactosidase, α-glucosidase, β-glucosidase, n-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase. API 20E showed that strain MSt1T is positive for arginine dihydrolase, deaminase, nitrate reduction, and gelatinase, but negative for lysine decarboxylase, ornithine decarboxylase, citrate utilization, H2S production, urea hydrolysis, indole production and Voges-Proskauer. With API 50CH, acid is produced when strain MSt1T ferments glycerol, ribose, D-xylose, D-galactose, D-glucose, D-fructose, D-mannose, D-mannitol, D-sorbitol, methyl-αD-mannopyranoside, methyl-αD-glucopyranoside, amygdaline, arbutine, esculine, salicine, D-cellobiose, D-maltose, D-lactose, D-melibiose, D-saccharose, D-trehalose, D-raffinose, D-turanose, and D-arabitol. Acid is not produced erythritol, D-arabinose, L-arabinose, L-xylose, adonitol, Methyl-βD-xylopyranoside, L-sorbose, L-rhamnose, dulcitol, inositol, N-acetylglucosamine, inuline, D-melezitose, amidon, glycogen, xylitol, gentiobiose, D-lyxose, D-tagatose, D-fucose, L-fucose, L-arabitol, potassium gluconate, potassium 2-ketogluconate, and potassium 5-ketogluconate. Cell-wall peptidoglycan contains meso-diaminopimelic acid. The predominant menaquinone is MK-7. The major cellular fatty acids are anteiso-C15:0 and C16:0. The polar lipid profile consists of phosphatidylglycerol, phosphatidylethanolamine, diphosphatidylglycerol, one unknown lipid, two unknown glycolipids and one unknown phospholipid. The DNA G+C content of strain MSt1T is 51.5 mol%.
The type strain, MSt1T (= DSM 100708T = MCCC 1K01247T) was isolated from tropical peat swamp soil in North Selangor peat swamp, Malaysia. The 16S rRNA gene sequence of type strain MSt1T has been deposited in DDBJ/EMBL/GenBank under the accession number KT216503.
AYK performed the laboratory experiments, analyzed the data, and written up the manuscript. LSM supervised the entire study. LLH and CYL co-supervised the study. LSM, AYK, and OKS contributed to the experimental designs. LLH and AYK contributed to the polyphasic taxonomy. CYL and AYK contributed to the chemical analysis. CY, LSM, and AYK conceived the idea of bioprospecting in tropical peat swamp forest. All authors proofread and reviewed the manuscript.
The authors express gratitude to External Industry Grant from Biotek Abadi (GBA-808138 & GBA-808813) awarded to Dr. LL for paying the article publication charges of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors express gratitude to Monash University Malaysia, Tropical Medicine and Biology Multidisciplinary Platform and School of Science, Monash University Malaysia for funding this research.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00219
Aktuganov, G., Melentjev, A., Galimzianova, N., Khalikova, E., Korpela, T., and Susi, P. (2008). Wide-range antifungal antagonism of Paenibacillus ehimensis IB-X-b and its dependence on chitinase and β-1,3-glucanase production. Can. J. Microbiol. 54, 577–587. doi: 10.1139/W08-043
Ash, C., Priest, F., and Collins, M. D. (1993). Molecular identification of rRNA group 3 bacilli (Ash, Farrow, Wallbanks and Collins) using a PCR probe test. Antonie van Leeuwenhoek 64, 253–260. doi: 10.1007/BF00873085
Aw, Y. K., Ong, K. S., Yule, C. M., Gan, H. M., and Lee, S. M. (2014). Draft genome sequence of Paenibacillus sp. strain MSt1 with broad antimicrobial activity, isolated from Malaysian tropical peat swamp soil. Genome Announc. 2:e01024-14. doi: 10.1128/genomea.01024-14
Bister, B., Bischoff, D., Ströbele, M., Riedlinger, J., Reicke, A., Wolter, F., et al. (2004). Abyssomicin C—a polycyclic antibiotic from a marine Verrucosispora strain as an inhibitor of the p-aminobenzoic acid/tetrahydrofolate biosynthesis pathway. Angew. Chem. Int. Ed. Engl. 43, 2574–2576. doi: 10.1002/anie.200353160
Boucher, H. W., Talbot, G. H., Bradley, J. S., Edwards, J. E., Gilbert, D., Rice, L. B., et al. (2009). Bad bugs, no drugs: No ESKAPE! An update from the infectious diseases society of America. Clin. Infect. Dis. 48, 1–12. doi: 10.1086/595011
Cerny, G. (1978). Studies on the aminopeptidase test for the distinction of gram-negative from gram-positive bacteria. Eur. J. Appl. Microbiol. Biotechnol. 5, 113–122. doi: 10.1007/BF00498805
Chung, Y. R., Kim, C. H., Hwang, I., and Chun, J. (2000). Paenibacillus koreensis sp. nov., a new species that produces an iturin-like antifungal compound. Int. J. Syst. Evol. Microbiol. 50, 1495–1500. doi: 10.1099/00207713-50-4-1495
CLSI (2012). Performance Standard for Antimicrobial Susceptibility Testing; Twenty-Second Informational Supplement M100-A9. Pennsylvania, PA: CLSI.
Davies, J., and Davies, D. (2010). Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 74, 417–433. doi: 10.1128/MMBR.00016-10
Ding, R., Wu, X.-C., Qian, C.-D., Teng, Y., Li, O., Zhan, Z.-J., et al. (2011). Isolation and identification of lipopeptide antibiotics from Paenibacillus elgii B69 with inhibitory activity against methicillin-resistant Staphylococcus aureus. J. Microbiol. 49, 942–949. doi: 10.1007/s12275-011-1153-7
Domenech, O., Francius, G., Tulkens, P. M., Van Bambeke, F., Dufrêne, Y., and Mingeot-Leclercq, M.-P. (2009). Interactions of oritavancin, a new lipoglycopeptide derived from vancomycin, with phospholipid bilayers: effect on membrane permeability and nanoscale lipid membrane organization. Biochim. Biophys. Acta 1788, 1832–1840. doi: 10.1016/j.bbamem.2009.05.003
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.2307/2408678
Glaeser, S. P., Falsen, E., Busse, H.-J., and Kämpfer, P. (2013). Paenibacillus vulneris sp. nov., isolated from a necrotic wound. Int. J. Syst. Evol. Microbiol. 63, 777–782. doi: 10.1099/ijs.0.041210-0
Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., Vandamme, P., and Tiedje, J. M. (2007). DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91. doi: 10.1099/ijs.0.64483-0
Hayden, M. K., Rezai, K., Hayes, R. A., Lolans, K., Quinn, J. P., and Weinstein, R. A. (2005). Development of daptomycin resistance in vivo in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43, 5285–5287. doi: 10.1128/JCM.43.10.5285-5287.2005
He, Z., Kisla, D., Zhang, L., Yuan, C., Green-Church, K. B., and Yousef, A. E. (2007). Isolation and identification of a Paenibacillus polymyxa strain that coproduces a novel lantibiotic and polymyxin. Appl. Environ. Microbiol. 73, 168–178. doi: 10.1128/AEM.02023-06
Higgins, D. L., Chang, R., Debabov, D. V., Leung, J., Wu, T., Krause, K. M., et al. (2005). Telavancin, a multifunctional lipoglycopeptide, disrupts both cell wall synthesis and cell membrane Integrity in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 49, 1127–1134. doi: 10.1128/AAC.49.3.1127-1134.2005
Huang, Z., Hu, Y., Shou, L., and Song, M. (2013). Isolation and partial characterization of cyclic lipopeptide antibiotics produced by Paenibacillus ehimensis B7. BMC Microbiol. 13:87. doi: 10.1186/1471-2180-13-87
Kanokratana, P., Uengwetwanit, T., Rattanachomsri, U., Bunterngsook, B., Nimchua, T., Tangphatsornruang, S., et al. (2011). Insights into the phylogeny and metabolic potential of a primary tropical peat swamp forest microbial community by metagenomic analysis. Microb. Ecol. 61, 518–528. doi: 10.1007/s00248-010-9766-7
Kates, M. (1986). “Lipid extraction procedures,” in Techniques of Lipidology, eds T. S. Work and E. Work (Amsterdam: Elsevier), 100–111.
Kim, D.-S., Bae, C.-Y., Jeon, J.-J., Chun, S.-J., Oh, H. W., Hong, S. G., et al. (2004). Paenibacillus elgii sp. nov., with broad antimicrobial activity. Int. J. Syst. Evol. Microbiol. 54, 2031–2035. doi: 10.1099/ijs.0.02414-0
Kim, M., Oh, H.-S., Park, S.-C., and Chun, J. (2014). Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 64, 346–351. doi: 10.1099/ijs.0.059774-0
Kim, O.-S., Cho, Y.-J., Lee, K., Yoon, S.-H., Kim, M., Na, H., et al. (2012). Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 62, 716–721. doi: 10.1099/ijs.0.038075-0
Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Kodaka, H., Armfield, A. Y., Lombard, G. L., and Dowell, V. R. (1982). Practical procedure for demonstrating bacterial flagella. J. Clin. Microbiol. 16, 948–952.
Kohama, Y., Iida, K., Tanaka, K., Semba, T., Itoh, M., Teramoto, T., et al. (1993). Studies on thermophile products. VI. activation of mouse peritoneal macrophages by bis(2-hydroxyethyl) trisulfide. Biol. Pharm. Bull. 16, 973–977. doi: 10.1248/bpb.16.973
Kuykendall, L. D., Roy, M. A., Neill, J. J., and Devine, T. E. (1988). Fatty acids, antibiotic resistance, and deoxyribonucleic acid homology groups of Bradyrhizobium japonicum. Int. J. Syst. Evol. Microbiol. 38, 358–361. doi: 10.1099/00207713-38-4-358
Lee, J.-S., Pyun, Y.-R., and Bae, K. S. (2004). Transfer of Bacillus ehimensis and Bacillus chitinolyticus to the genus Paenibacillus with emended descriptions of Paenibacillus ehimensis comb. nov. and Paenibacillus chitinolyticus comb. nov. Int. J. Syst. Evol. Microbiol. 54, 929–933. doi: 10.1099/ijs.0.02765-0
Lee, L. H., Zainal, N., Azman, A. S., Eng, S. K., Ab Mutalib, N. S., Yin, W. F., et al. (2014). Streptomyces pluripotens sp. nov., a bacteriocin-producing streptomycete that inhibits meticillin-resistant Staphylococcus aureus. Int. J. Syst. Evol. Microbiol. 64, 3297–3306. doi: 10.1099/ijs.0.065045-0
Logan, N. A., Berge, O., Bishop, A. H., Busse, H.-J., De Vos, P., Fritze, D., et al. (2009). Proposed minimal standards for describing new taxa of aerobic, endospore-forming bacteria. Int. J. Syst. Evol. Microbiol. 59, 2114–2121. doi: 10.1099/ijs.0.013649-0
Meier-Kolthoff, J., Auch, A., Klenk, H.-P., and Göker, M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. doi: 10.1186/1471-2105-14-60
Meka, V. G., and Gold, H. S. (2004). Antimicrobial resistance to linezolid. Clin. Infect. Dis. 39, 1010–1015. doi: 10.1086/423841
Miller, L. T. (1982). Single derivatization method for routine analysis of bacterial whole-cell fatty acid methyl esters, including hydroxy acids. J. Clin. Microbiol. 16, 584–586.
Mishra, R. P. N., Oviedo-Orta, E., Prachi, P., Rappuoli, R., and Bagnoli, F. (2012). Vaccines and antibiotic resistance. Curr. Opin. Microbiol. 15, 596–602. doi: 10.1016/j.mib.2012.08.002
Moaledj, K. (1986). Comparison of Gram-staining and alternate methods, KOH test and aminopeptidase activity in aquatic bacteria: their application to numerical taxonomy. J. Microbiol. Methods 5, 303–310. doi: 10.1016/0167-7012(86)90056-4
Paulus, H., and Gray, E. (1964). The biosynthesis of polymyxin B by growing cultures of Bacillus polymyxa. J. Biol. Chem. 239, 865–871.
Pettit, R. K. (2011). Culturability and secondary metabolite diversity of extreme microbes: expanding contribution of deep sea and deep-sea vent microbes to natural product discovery. Mar. Biotechnol. 13, 1–11. doi: 10.1007/s10126-010-9294-y
Qian, C.-D., Wu, X.-C., Teng, Y., Zhao, W.-P., Li, O., Fang, S.-G., et al. (2012). Battacin (Octapeptin B5), a new cyclic lipopeptide antibiotic from Paenibacillus tianmuensis active against multidrug-resistant Gram-negative bacteria. Antimicrob. Agents Chemother. 56, 1458–1465. doi: 10.1128/AAC.05580-11
Rhuland, L. E., Work, E., Denman, R., and Hoare, D. (1955). The behavior of the isomers of α, ε-diaminopimelic acid on paper chromatograms. J. Am. Chem. Soc. 77, 4844–4846. doi: 10.1021/ja01623a047
Richter, M., and Rosselló-Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 106, 19126–19131. doi: 10.1073/pnas.0906412106
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Schaeffer, A. B., and Fulton, M. D. (1933). A simplified method of staining endospores. Science 77, 194–194. doi: 10.1126/science.77.1990.194
Shaheen, M., Li, J., Ross, A. C., Vederas, J. C., and Jensen, S. E. (2011). Paenibacillus polymyxa PKB1 produces variants of polymyxin B-type antibiotics. Chem. Biol. 18, 1640–1648. doi: 10.1016/j.chembiol.2011.09.017
Shida, O., Takagi, H., Kadowaki, K., Nakamura, L. K., and Komagata, K. (1997). Transfer of Bacillus alginolyticus, Bacillus chondroitinus, Bacillus curdlanolyticus, Bacillus glucanolyticus, Bacillus kobensis, and Bacillus thiaminolyticus to the genus Paenibacillus and emended description of the genus Paenibacillus. Int. J. Syst. Bacteriol. 47, 289–298. doi: 10.1099/00207713-47-2-289
Smith, R., and Coast, J. (2013). The true cost of antimicrobial resistance. BMJ 346:f1493. doi: 10.1136/bmj.f1493
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Thimon, L., Peypoux, F., Wallach, J., and Michel, G. (1995). Effect of the lipopeptide antibiotic, iturin A, on morphology and membrane ultrastructure of yeast cells. FEMS Microbiol. Lett. 128, 101–106. doi: 10.1111/j.1574-6968.1995.tb07507.x
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Tindall, B. J., Rosselló-Mora, R., Busse, H.-J., Ludwig, W., and Kämpfer, P. (2010). Notes on the characterization of prokaryote strains for taxonomic purposes. Int. J. Syst. Evol. Microbiol. 60, 249–266. doi: 10.1099/ijs.0.016949-0
Tittsler, R. P., and Sandholzer, L. A. (1936). The use of semi-solid agar for the detection of bacterial motility. J. Bacteriol. 31, 575–580.
Velkov, T., Roberts, K. D., Nation, R. L., Wang, J., Thompson, P. E., and Li, J. (2014). Teaching ‘old’ polymyxins new tricks: new-generation lipopeptides targeting gram-negative ‘superbugs’. ACS Chem. Biol. 9, 1172–1177. doi: 10.1021/cb500080r
Wayne, L., Brenner, D., Colwell, R., Grimont, P., Kandler, O., Krichevsky, M., et al. (1987). Report of the Ad Hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37, 463–464. doi: 10.1099/00207713-37-4-463
Wu, X., Fang, H., Qian, C., Wen, Y., Shen, X., Li, O., et al. (2011). Paenibacillus tianmuensis sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 61, 1133–1137. doi: 10.1099/ijs.0.024109-0
Keywords: Paenibacillus, antimicrobial, peat swamp, Malaysia, lipopeptide
Citation: Aw Y-K, Ong K-S, Lee L-H, Cheow Y-L, Yule CM and Lee S-M (2016) Newly Isolated Paenibacillus tyrfis sp. nov., from Malaysian Tropical Peat Swamp Soil with Broad Spectrum Antimicrobial Activity. Front. Microbiol. 7:219. doi: 10.3389/fmicb.2016.00219
Received: 26 October 2015; Accepted: 10 February 2016;
Published: 01 March 2016.
Edited by:
Tzi Bun Ng, The Chinese University of Hong Kong, ChinaReviewed by:
Sylvie Lautru, Université Paris Sud, FranceCopyright © 2016 Aw, Ong, Lee, Cheow, Yule and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sui-Mae Lee, bGVlLnN1aS5tYWVAbW9uYXNoLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.