- 1Division of Genetics and Molecular Biology, Faculty of Science, Institute of Biological Sciences, University of Malaya, Kuala Lumpur, Malaysia
- 2Department of Medical Microbiology, Faculty of Medicine, University of Malaya, Kuala Lumpur, Malaysia
Background
The genus of Pandoraea was first proposed in 2000 following the isolation from the sputum of cystic fibrosis patients (Coenye et al., 2000). Five species were initially assigned to the novel genus namely Pandoraea apista, Pandoraea pulmonicola, Pandoraea pnomenusa, Pandoraea sputorum, and Pandoraea norimbergensis but the description of four new species and another four genomospecies in the subsequent years led to a total of nine species and four genomospecies within the genus of Pandoraea (Daneshvar et al., 2001; Anandham et al., 2010; Sahin et al., 2011). The isolation of Pandoraea spp. from various environmental samples such as water, sludge, and soils have been reported, but to date, only P. pnomenusa, P. apista, P. pulmonicola, and P. sputorum were isolated from clinical specimens such as blood, sputum and bronchial fluid of patients with cystic fibrosis or chronic lung diseases (Coenye et al., 2000; Daneshvar et al., 2001; Stryjewski et al., 2003; Han-Jen et al., 2013). Members of Pandoraea tend to exhibit broad resistance to ampicillin, extended-spectrum cephalosporins, aztreonam, aminoglycosides, and meropenem but they are sensitive to imipenem (Daneshvar et al., 2001; Stryjewski et al., 2003). However, the clinical significance and prevalence of these multi-drug resistant bacteria among patients with cystic fibrosis or respiratory diseases remained unknown since Pandoraea spp. are usually misidentified as Burkholderia cepacia complex, Ralstonia pickettii, or Ralstonia paucula (Segonds et al., 2003). Ambiguity in differentiating between B. cepacia complex, Ralstonia spp. and Pandoraea spp. can be resolved by 16S ribosomal DNA-PCR (Coenye et al., 2001) and gyrB gene restriction fragment length polymorphism (Coenye and LiPuma, 2002) but the limited use of molecular typing methods in routine clinical microbiological laboratory has resulted in the underreporting of Pandoraea spp. in clinical cases.
The first case of mortality due to P. pnomenusa was documented in 2003 whereby the patient developed bacteremia and subsequently died from multiple organ failure after undergoing a bilateral lung transplant operation (Stryjewski et al., 2003). Isolation of P. pnomenusa from purulent secretions collected by bronchoalveolar lavage on postoperative day 10 indicated that the lungs could be the source of infection and the cause of fatality was subsequently established through the repeated isolation of P. pnomenusa from blood cultures on postoperative day 8 and at the time of death (Stryjewski et al., 2003). In a study conducted by Costello et al. (2011), P. pnomenusa was found to be invasive in lung cells expressing cystic fibrosis transmembrane regulator and the species exhibited the greatest level of transepithelial translocation when compared to P. pulmonicola, P. apista, and B. cenocepacia. Reports of Pandoraea -associated bacteraemia and the subsequent isolation of P. pnomenusa from blood culture suggest this species may be highly pathogenic (Daneshvar et al., 2001; Stryjewski et al., 2003; Costello et al., 2011). Furthermore, P. pnomenusa could trigger the accumulation of pro-inflammatory cytokines, particularly interleukins 8 and 6, which could lead to lung tissue damage. Chronic lung inflammation is one of the main causes of fatality in individuals with cystic fibrosis and hence, the ability of P. pnomenusa to elicit the secretion of interleukins 8 and 6 as a part of the host response may serve as one of the mechanisms of pathogenesis for this species (Caraher et al., 2008).
In the last decade, only a few studies have been done to investigate the underlying pathogenic mechanisms of P. pnomenusa. In order to gain a better understanding of this species, we studied the complete genome sequence of P. pnomenusa type strain DSM 16536T which was isolated from the sputum of a cystic fibrosis patient originating from Edinburgh, United Kingdom. Availability of this genome sequence of P. pnomenusa DSM 16536T will serve as a basis for further in-depth analysis of the virulence, pathogenesis and genetics of P. pnomenusa.
Materials and Methods
DNA Purification
P. pnomenusa DSM 16536T, a clinical strain isolated from a cystic fibrosis patient in United Kingdom, was acquired from the German Collection of Microorganisms and Cell cultures (DSMZ). This bacterium was grown aerobically in LB broth at 37°C. Isolation and purification of bacterial genomic DNA was performed with MasterPure DNA Purification Kit (EpiCenter, CA, USA) according to the manufacturer's instruction. The purity, quality and quantity of purified genomic DNA were assessed using NanoDrop 2000 UV-Vis spectrophotometer (Thermo Scientific, MA, USA) and Qubit 2.0 fluorometer (Life Technologies, MA, USA), respectively.
Genome Sequencing and Assembly
The sheared genomic DNA of P. pnomenusa DSM 16536T was prepared following the “Procedure a Checklist-20kb Template Preparation using BluePippinTM Size Selection” protocol, in which size selection of the constructed SMRTbell templates was performed with a cutoff length of 7kb. Purified and size-selected SMRTbell library was sequenced on four SMRT cells using P5C3 chemistry on Pacific Biosciences (PacBio) Single Molecule, Real-Time (SMRT) RS II instrument. Sub-reads which were generated from the raw sequencing reads after adapter removal were further filtered and mapped prior to de novo assembly using Hierarchical Genome Assembly Process (HGAP) version 3. The polished assembly generated was examined for circularity based on the presence of overlapping sequences at both ends of the contig. Location of the overlapping sequence were determined using Gepard dotplot program (Krumsiek et al., 2007). Subsequently, circularization of the contig was performed by removing one of the overlapping ends to generate a blunt-ended circular assembly yielding a complete genome map.
Genome Annotation
Gene prediction and annotation were performed using Rapid Annotation Search Tool (RAST) (Aziz et al., 2008), Rapid Prokaryotic Genome Annotation (Prokka) (Seemann, 2014), and NCBI Prokaryotic Genome Annotation Pipeline (PGAP) based on the Best-placed reference protein set and GeneMarkS+. Additional gene identification was done using KEGG database (Kanehisa et al., 2015), Pathosystem Resource Integration Center (PATRIC) (Wattam et al., 2014), and IMG ER (Markowitz et al., 2009). Both IMG ER and NCBI PGAP were used for the identification of tRNA and rRNA genes. Supplementary annotation of antimicrobial resistance genes was analyzed using ResFinder (Carattoli et al., 2014).
Results
Genome Characteristics
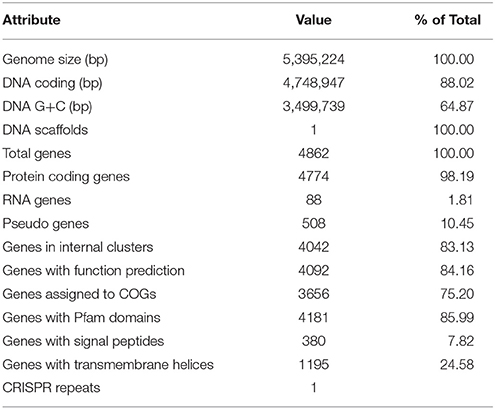
The genome of P. pnomenusa DSM 16536T consists of a single circular chromosome of 5,389,285bp with a mean genome coverage of 244.62-fold and an average GC content of 64.87% (Table 1). No plasmid was found in the genome sequence of this bacterium. A total of 4811 genes was predicted of which 4586 genes were identified as protein coding genes. There are 88 RNA genes consisting of 12 rRNA (4 5S rRNA, 4 16S rRNA, and 4 23S rRNA) and 65 tRNA genes. Besides GenBank (accession number: CP009553.2; http://www.ncbi.nlm.nih.gov/nuccore/CP009553.2), in which the genome sequences data are available in FASTA, annotated GenBank flat file, graphical and ASN.1 formats, functional annotation results of this genome are also accessible through KEGG resources.
The overview of this genome record can be attained from the complete genome directory of KEGG ORGANISMS database (http://www.genome.jp/kegg/catalog/org_list.html) with the organism prefix of ppnm. Further, through the ppnm hyperlink, cross-reference information in the form of protein, and small-molecules interaction network maps (http://www.genome.jp/kegg-bin/show_organism?menu_type=pathway_maps&org=ppnm), BRITE biological systems hierarchical classifications (http://www.genome.jp/kegg-bin/show_organism?menu_type=gene_catalogs&org=ppnm), KEGG modules (http://www.genome.jp/kegg-bin/show_organism?menu_type=pathway_modules&jp/kegg-bin/show_organism?menu_type=pathway_modules&org=ppnm), and whole genome map which can be visualized using genome map browser (http://www.genome.jp/kegg-bin/show_genomemap_top?org_id=ppnm) can be accessed through the subdirectory panel. Further insights to the potential pathogenicity of this clinical strain can also be gained from its genomic information through KEGG Pathogen resource (http://www.genome.jp/kegg/disease/pathogen.html).
Virulence Genes
From the complete genome sequences, various genes which could potentially contributing to pathogenicity were identified using specialty gene annotations in PATRIC server via a combination of few curated virulence factors databases including Virulence Factors Database (VFDB), Victors and PATRIC curated virulence database (PATRIC_VF). A total of 16 virulence factors were identified, namely: Phosphoribosylformylglycinamidine cyclo-ligase (LV28_09200), RNA polymerase sigma factor RpoE (LV28_09850), phosphoenolpyruvate synthase (LV28_01060), RecA protein (LV28_10630), aminase component of anthranilate synthase (LV28_11620), imidazole glycerol phosphate synthase cyclase subunit (LV28_12510), translation elongation factor Tu (LV28_12945; LV28_13105), argininosuccinate synthase (LV28_16050), endonuclease III (LV28_23310), 3-isopropylmalate dehydrogenase (LV28_23930), 3-isopropylmalate dehydrogenase (LV28_23930), RNA-binding protein Hfq (LV28_24345), chorismate synthase (LV28_04475), acetolactate synthase large and small subunit (LV28_04625 and LV28_04620), and chemotaxis protein CheY (LV28_14025). These identified virulence factors are well-characterized virulence determinants in the Burkholderia pseudomallei (close phylogenetic neighbor of P. pnomenusa) and other pathogens such as Listeria monocytogens, Actinobacillus pleuropneumoniae, Neisseria meningitis, and Shigella flexneri (Sun et al., 2000; Sheehan et al., 2003; Dons et al., 2004; Pilatz et al., 2006; Sharma and Payne, 2006; Thongboonkerd et al., 2007). Hence, these virulence factors identified could serve as promising gene candidates in studying the underlying pathogenic mechanism of persistent P. pnomenusa infection. Furthermore, previous studies also demonstrated targeting against these genes resulted in attenuation suggesting that these predicted virulence factors are potential targets for new therapeutic strategies and vaccines development (Cuccui et al., 2007; Breitbach et al., 2008; Srilunchang et al., 2009).
Antimicrobial Resistance Genes
From RAST analysis, a total of 44 genes responsible for antibiotic resistance were identified in the genome. Majority of these genes were previously characterized in various pathogens, including Pseudomonas aeruginosa and Escherichia coli (Morita et al., 2012; Weatherspoon-Griffin et al., 2014). Full details of these antibiotic resistance genes, including efflux pumps-encoding genes and beta-lactamases-encoding genes (Okusu et al., 1996; Piddock, 2006; Poole, 2011; Poirel et al., 2012; Morita et al., 2014, 2015; Weatherspoon-Griffin et al., 2014; Podnecky et al., 2015) are available in Supplementary Table 1. Furthermore, analysis of antimicrobial resistance gene with ResFinder also revealed the presence of OXA-62, a carbapenem-hydrolyzing oxacillinase, further affirming the finding of Schneider et al. (2006) which demonstrated that OXA-62 is distributed specifically within the P. pnomenusa species and producing the imipenem-resistant phenotype.
Data Access
The assembled and annotated genome of P. pnomenusa DSM 16536T described in this paper has been deposited in GenBank (accession number of CP009553.2); KEGG database (entry number of T03411) and JGI portal with GOLD ID of Gp0107448 and IMG taxon ID of 2606217238.
Author Contributions
YL, RE, DY, CY, GA, WY perform the experiments and collected the data. KC conceived the idea, obtained the funding and WY managed the finance of the project. YL, RE, DY, CY, GA, WY, and KT prepared the draft, and KC proofread the final draft. All authors approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This project was financially funded by University of Malaya-Ministry of Higher Education High Impact Research Grants (UM.C/625/1/HIR/MOHE/CHAN/01, Grant No. A-000001-50001) and UM-MOHE HIR Grant (UM.C/625/1/HIR/MOHE/CHAN/14/1, no. H-50001-A000027) which was awarded to KC.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00109
References
Anandham, R., Indiragandhi, P., Kwon, S. W., Sa, T. M., Jeon, C. O., Kim, Y. K., et al. (2010). Pandoraea thiooxydans sp. nov., a facultatively chemolithotrophic, thiosulfate-oxidizing bacterium isolated from rhizosphere soils of sesame (Sesamum indicum L.). Int. J. Syst. Evol. Microbiol. 60, 21–26. doi: 10.1099/ijs.0.012823-0
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Breitbach, K., Köhler, J., and Steinmetz, I. (2008). Induction of protective immunity against Burkholderia pseudomallei using attenuated mutants with defects in the intracellular life cycle. Trans. R. Soc. Trop. Med. Hyg. 102(Suppl. 1), S89–S94. doi: 10.1016/S0035-9203(08)70022-1
Caraher, E., Collins, J., Herbert, G., Murphy, P. G., Gallagher, C. G., Crowe, M. J., et al. (2008). Evaluation of in vitro virulence characteristics of the genus Pandoraea in lung epithelial cells. J. Med. Microbiol. 57, 15–20. doi: 10.1099/jmm.0.47544-0
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Coenye, T., Falsen, E., Hoste, B., Ohlen, M., Goris, J., Govan, J. R., et al. (2000). Description of Pandoraea gen. nov. with Pandoraea apista sp. nov., Pandoraea pulmonicola sp. nov., Pandoraea pnomenusa sp. nov., Pandoraea sputorum sp. nov. and Pandoraea norimbergensis comb. nov. Int. J. Syst. Evol. Microbiol. 50, 887–899. doi: 10.1099/00207713-50-2-887
Coenye, T., and LiPuma, J. J. (2002). Use of the gyrB gene for the identification of Pandoraea species. FEMS Microbiol. Lett. 208, 15–19. doi: 10.1111/j.1574-6968.2002.tb11053.x
Coenye, T., Liu, L., Vandamme, P., and LiPuma, J. J. (2001). Identification of Pandoraea species by 16S ribosomal DNA-based PCR assays. J. Clin. Microbiol. 39, 4452–4455. doi: 10.1128/JCM.39.12.4452-4455.2001
Costello, A., Herbert, G., Fabunmi, L., Schaffer, K., Kavanagh, K. A., Caraher, E. M., et al. (2011). Virulence of an emerging respiratory pathogen, genus Pandoraea, in vivo and its interactions with lung epithelial cells. J. Med. Microbiol. 60, 289–299. doi: 10.1099/jmm.0.022657-0
Cuccui, J., Easton, A., Chu, K. K., Bancroft, G. J., Oyston, P. C., Titball, R. W., et al. (2007). Development of signature-tagged mutagenesis in Burkholderia pseudomallei to identify genes important in survival and pathogenesis. Infect. Immun. 75, 1186–1195. doi: 10.1128/IAI.01240-06
Daneshvar, M. I., Hollis, D. G., Steigerwalt, A. G., Whitney, A. M., Spangler, L., Douglas, M. P., et al. (2001). Assignment of CDC weak oxidizer group 2 (WO-2) to the genus Pandoraea and characterization of three new Pandoraea genomospecies. J. Clin. Microbiol. 39, 1819–1826. doi: 10.1128/JCM.39.5.1819-1826.2001
Dons, L., Eriksson, E., Jin, Y., Rottenberg, M. E., Kristensson, K., Larsen, C. N., et al. (2004). Role of flagellin and the two-component CheA/CheY system of Listeria monocytogenes in host cell invasion and virulence. Infect. Immun. 72, 3237–3244. doi: 10.1128/IAI.72.6.3237-3244.2004
Han-Jen, R. E., Wai-Fong, Y., and Kok-Gan, C. (2013). Pandoraea sp. RB-44, a novel quorum sensing soil bacterium. Sensors 13, 14121–14132. doi: 10.3390/s131014121
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M., and Tanabe, M. (2015). KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462. doi: 10.1093/nar/gkv1070
Krumsiek, J., Arnold, R., and Rattei, T. (2007). Gepard: a rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics 23, 1026–1028. doi: 10.1093/bioinformatics/btm039
Markowitz, V. M., Mavromatis, K., Ivanova, N. N., Chen, I. M., Chu, K., and Kyrpides, N. C. (2009). IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 25, 2271–2278. doi: 10.1093/bioinformatics/btp393
Morita, Y., Tomida, J., and Kawamura, Y. (2012). MexXY multidrug efflux system of Pseudomonas aeruginosa. Front. Microbiol. 3:408. doi: 10.3389/fmicb.2012.00408
Morita, Y., Tomida, J., and Kawamura, Y. (2014). Responses of Pseudomonas aeruginosa to antimicrobials. Front. Microbiol. 4:422. doi: 10.3389/fmicb.2013.00422
Morita, Y., Tomida, J., and Kawamura, Y. (2015). Efflux-mediated fluoroquinolone resistance in the multidrug-resistant Pseudomonas aeruginosa clinical isolate PA7: identification of a novel MexS variant involved in upregulation of the mexEF-oprN multidrug efflux operon. Front. Microbiol. 6:8. doi: 10.3389/fmicb.2015.00008
Okusu, H., Ma, D., and Nikaido, H. (1996). AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J. Bacteriol. 178, 306–308.
Piddock, L. J. V. (2006). Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin. Microbiol. Rev. 19, 382–402. doi: 10.1128/CMR.19.2.382-402.2006
Pilatz, S., Breitbach, K., Hein, N., Fehlhaber, B., Schulze, J., Brenneke, B., et al. (2006). Identification of Burkholderia pseudomallei genes required for the intracellular life cycle and in vivo virulence. Infect. Immun. 74, 3576–3586. doi: 10.1128/IAI.01262-05
Podnecky, N. L., Rhodes, K. A., and Schweizer, H. P. (2015). Efflux pump-mediated drug resistance in Burkholderia. Front. Microbiol. 6:305. doi: 10.3389/fmicb.2015.00305
Poirel, L., Cattoir, V., and Nordmann, P. (2012). Plasmid-mediated quinolone resistance; interactions between human, animal, and environmental ecologies. Front. Microbiol. 3:24. doi: 10.3389/fmicb.2012.00024
Poole, K. (2011). Pseudomonas aeruginosa: resistance to the max. Front. Microbiol. 2:65. doi: 10.3389/fmicb.2011.00065
Sahin, N., Tani, A., Kotan, R., Sedlacek, I., Kimbara, K., and Tamer, A. U. (2011). Pandoraea oxalativorans sp. nov., Pandoraea faecigallinarum sp. nov. and Pandoraea vervacti sp. nov., isolated from oxalate-enriched culture. Int. J. Syst. Evol. Microbiol. 61, 2247–2253. doi: 10.1099/ijs.0.026138-0
Schneider, I., Queenan, A. M., and Bauernfeind, A. (2006). Novel carbapenem-hydrolyzing oxacillinase OXA-62 from Pandoraea pnomenusa. Antimicrob. Agents Chemother. 50, 1330–1335. doi: 10.1128/AAC.50.4.1330-1335.2006
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Segonds, C., Paute, S., and Chabanon, G. (2003). Use of amplified ribosomal DNA restriction analysis for identification of Ralstonia and Pandoraea species: interest in determination of the respiratory bacterial flora in patients with cystic fibrosis. J. Clin. Microbiol. 41, 3415–3418. doi: 10.1128/JCM.41.7.3415-3418.2003
Sharma, A. K., and Payne, S. M. (2006). Induction of expression of hfq by DksA is essential for Shigella flexneri virulence. Mol. Microbiol. 62, 469–479. doi: 10.1111/j.1365-2958.2006.05376.x
Sheehan, B. J., Bossé, J. T., Beddek, A. J., Rycroft, A. N., Kroll, J. S., and Langford, P. R. (2003). Identification of Actinobacillus pleuropneumoniae genes important for survival during infection in its natural host. Infect. Immun. 71, 3960–3970. doi: 10.1128/IAI.71.7.3960-3970.2003
Srilunchang, T., Proungvitaya, T., Wongratanacheewin, S., Strugnell, R., and Homchampa, P. (2009). Construction and characterization of an unmarked aroC deletion mutant of Burkholderia pseudomallei strain A2. Southeast Asian J. Trop. Med. Public Health 40, 123–130.
Stryjewski, M. E., LiPuma, J. J., Messier, R. H. Jr., Reller, L. B., and Alexander, B. D. (2003). Sepsis, multiple organ failure, and death due to Pandoraea pnomenusa infection after lung transplantation. J. Clin. Microbiol. 41, 2255–2257. doi: 10.1128/JCM.41.5.2255-2257.2003
Sun, Y. H., Bakshi, S., Chalmers, R., and Tang, C. M. (2000). Functional genomics of Neisseria meningitidis pathogenesis. Nat. Med. 6, 1269–1273. doi: 10.1038/81380
Thongboonkerd, V., Vanaporn, M., Songtawee, N., Kanlaya, R., Sinchaikul, S., Chen, S. T., et al. (2007). Altered proteome in Burkholderia pseudomallei rpoE operon knockout mutant: insights into mechanisms of rpoE operon in stress tolerance, survival, and virulence. J. Proteome Res. 6, 1334–1341. doi: 10.1021/pr060457t
Wattam, A. R., Abraham, D., Dalay, O., Disz, T. L., Driscoll, T., Gabbard, J. L., et al. (2014). PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 42, D581–D591. doi: 10.1093/nar/gkt1099
Weatherspoon-Griffin, N., Yang, D., Kong, W., Hua, Z., and Shi, Y. (2014). The CpxR/CpxA two-component regulatory system up-regulates the multidrug resistance cascade to facilitate Escherichia coli resistance to a model antimicrobial peptide. J. Biol. Chem. 289, 32571–32582. doi: 10.1074/jbc.M114.565762
Keywords: Pandoraea pnomenusa, cystic fibrosis, opportunistic pathogen, complete genome, Single Molecule Real-Time (SMRT) sequencing
Citation: Lim Y-L, Ee R, Yong D, Yu C-Y, Ang G-Y, Tee K-K, Yin W-F and Chan K-G (2016) Complete Genome Sequence Analysis of Pandoraea pnomenusa Type Strain DSM 16536T Isolated from a Cystic Fibrosis Patient. Front. Microbiol. 7:109. doi: 10.3389/fmicb.2016.00109
Received: 06 December 2015; Accepted: 21 January 2016;
Published: 08 February 2016.
Edited by:
Yuji Morita, Aichi Gakuin University, JapanReviewed by:
Dinesh Sriramulu, Shres Consultancy (Life Sciences), IndiaDerek Sarovich, Menzies School of Health Research, Australia
Copyright © 2016 Lim, Ee, Yong, Yu, Ang, Tee, Yin and Chan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kok-Gan Chan, a29rZ2FuQHVtLmVkdS5teQ==
 Yan-Lue Lim
Yan-Lue Lim Robson Ee
Robson Ee Delicia Yong
Delicia Yong Choo-Yee Yu
Choo-Yee Yu Geik-Yong Ang1
Geik-Yong Ang1 Kok-Gan Chan
Kok-Gan Chan