- 1Department of Sustainable Agro-Ecosystems and Bioresources, Research and Innovation Centre, Fondazione Edmund Mach, San Michele all'Adige, Italy
- 2Department of Agricultural and Environmental Science (DISA), PhD School of Agricultural Science and Biotechnology, University of Udine, Udine, Italy
- 3Department of Computational Biology, Research and Innovation Centre, Fondazione Edmund Mach, San Michele all'Adige, Italy
Lysobacter capsici AZ78 has considerable potential for biocontrol of phytopathogenic microorganisms. However, lack of information about genetic cues regarding its biological characteristics may slow down its exploitation as a biofungicide. In order to obtain a comprehensive overview of genetic features, the L. capsici AZ78 genome was sequenced, annotated and compared with the phylogenetically related pathogens Stenotrophomonas malthophilia K729a and Xanthomonas campestris pv. campestris ATCC 33913. Whole genome comparison, supported by functional analysis, indicated that L. capsici AZ78 has a larger number of genes responsible for interaction with phytopathogens and environmental stress than S. malthophilia K729a and X. c. pv. campestris ATCC 33913. Genes involved in the production of antibiotics, lytic enzymes and siderophores were specific for L. capsici AZ78, as well as genes involved in resistance to antibiotics, environmental stressors, fungicides and heavy metals. The L. capsici AZ78 genome did not encompass genes involved in infection of humans and plants included in the S. malthophilia K729a and X. c. pv. campestris ATCC 33913 genomes, respectively. The L. capsici AZ78 genome provides a genetic framework for detailed analysis of other L. capsici members and the development of novel biofungicides based on this bacterial strain.
Introduction
Genome sequencing represents an excellent tool for biological characterization of bacterial species; especially in the case of species that have been largely underexplored. In the family Xanthomonadaceae (Saddler and Bradbury, 2005), attention has been paid particularly to members that are pathogenic to humans (Stenotrophomonas) and plants (Xanthomonas and Xylella; Simpson et al., 2000; da Silva et al., 2002; Crossman et al., 2008). In contrast, other bacterial genera have been underexplored, as in the case of the genus Lysobacter, which was established in 1978 (Christensen and Cook, 1978). Since most Lysobacter spp. were wrongly classified as Myxobacteriales and Cythophagales, and several Lysobacter strains were wrongly assigned to Stenotrophomonas and Xanthomonas spp. (Christensen and Cook, 1978; Giesler and Yuen, 1998; Sakka et al., 1998; Nakayama et al., 1999), the importance of this genus was underestimated for a long time. The increasing number of 16S rDNA gene sequences available in public databases and the polyphasic approach to the identification of bacterial strains has led to an increase in the identification of new Lysobacter species. So far the genus has expanded to include 37 species (Singh et al., 2015) from the initial four: L. antibioticus, L. brunescens, Lysobacter enzymogenes, and L. gummosus (Christensen and Cook, 1978).
Some bacterial strains of the Lysobacter species act as biological control agents (BCAs) of plant diseases (Kobayashi and Yuen, 2007; Hayward et al., 2010). To date, most of the BCAs characterized have belonged to L. enzymogenes (Folman et al., 2003; Sullivan et al., 2003; Qian et al., 2009). Antagonistic mechanisms have received most attention in recent years, with the production of antibiotics and lytic enzymes by L. enzymogenes 3.1T8, C3, and OH11 and the related regulatory mechanisms being studied in some of these bacterial strains (Folman et al., 2004; Kobayashi et al., 2005; Palumbo et al., 2005; Yu et al., 2007; Zhang et al., 2011; Qian et al., 2012, 2013). Similarly to L. enzymogenes, L. capsici strains possess characteristics exploitable for the control of phytopathogenic microorganisms (Park et al., 2008). For example, the type strain L. capsici YC5194 produces secondary metabolites that inhibit the growth of phytopathogenic fungi (Park et al., 2008) and the L. capsici strain PG4 controls tomato foot and root rot caused by Fusarium oxysporum f. sp. radicis-lycopersici (Puopolo et al., 2010). Some L. capsici strains have been isolated from soils suppressive to Rhizoctonia solani and have been shown to be involved in the control of other phytopathogenic fungi (Postma et al., 2010). Other L. capsici strains can control nematodes, as in the case of L. capsici YS1275, used against Meloidogyne incognita (Lee et al., 2014), or oomycetes, as in the case of L. capsici AZ78 (Lc AZ78) used to control Phytophthora (P.) infestans and Plasmopara (Pl.) viticola (Puopolo et al., 2014a,b), indicating their high potential as broad spectrum BCAs. Lc AZ78's resistance to copper is an additional positive feature for a BCA, because it can be integrated within plant protection strategies including the use of copper fungicides (Puopolo and Pertot, 2014).
In comparison to L. enzymogenes, much less is known about the biological features of L. capsici (Puopolo et al., 2015). As understanding the biological characteristics of a microorganism is crucial for its development as a biopesticide, we sequenced the Lc AZ78 genome using PacBio technology and carried out functional experiments to assess the biological properties predicted by the genome analysis. To obtain a comprehensive overview of the genetic cues of L. capsici-specific biological characteristics, we compared the Lc AZ78 genome with the genome of two phylogenetically similar bacteria (Kobayashi and Yuen, 2007; Hayward et al., 2010): the opportunistic human pathogen S. malthophilia K729a (Sm K729a) and the phytopathogen X. campestris pv. campestris ATCC 33913 (Xcc ATCC 33913).
Materials and Methods
Microorganisms
L. capsici AZ78 was stored at length in glycerol 40% at −80°C and routinely grown on Luria-Bertani Agar at 27°C (LBA, Sigma-Aldrich, USA). In all the experiments Lc AZ78 cell suspensions were prepared by flooding LBA dishes with 5 ml of sterile saline solution (0.85% NaCl) after 72 h growth at 27°C. L. capsici AZ78 cells were then scraped from the medium surface using sterile spatulas and collected in sterile 15 ml tubes. The resulting Lc AZ78 cell suspensions were centrifuged (11,200 g, 5 min) and the pelleted cells were suspended in sterile distilled water to a final absorbance of 0.1 at 600 nm, corresponding to 1 × 108 Colony Forming Units (CFU)/ml. L. capsici AZ78 was used at this concentration in all experiments, except when otherwise indicated.
The phytopathogenic bacteria and fungi used in this work (Table S1) were grown respectively on Nutrient Agar (NA, Oxoid, United Kingdom) at 28°C and Potato Dextrose Agar (PDA, Oxoid) at 25°C. Bacterial strains were stored at length in glycerol 40% at −80°C, while fungal strains were stored on PDA slants at room temperature. P. infestans isolate was maintained on Pea Agar Medium (PAM, 12.5% frozen peas and 1.2% agar in distilled water) at 17°C and stored at length in glycerol 20% at −80°C.
DNA Extraction, Genome Sequencing, and Assembly
L. capsici AZ78 genomic DNA was extracted with a PureLink Genomic DNA Mini Kit (Thermo Fisher Scientific, Invitrogen, USA) according to the manufacturer's instructions. Once extracted, DNA integrity and the absence of RNA contamination was checked on a 1% agarose gel. Subsequently, the whole genomic DNA of Lc AZ78 was sequenced using PacBio technology at Baseclear B.V. (Leiden, Netherlands). A 10-kb PacBio single-molecule real-time (SMRT) cell was employed (Chin et al., 2013). The generated subreads were de novo assembled using the RS hierarchical genome assembly process (HGAP) protocol version 3.0, as available in SMRT Portal v2.0 (https://github.com/PacificBiosciences/Bioinformatics-Training/wiki/HGAP-in-SMRT-Analysis). The SMRT Portal was configured and used with a public machine image that Pacific Biosciences maintains and upgrades on Amazon Cloud (https://github.com/PacificBiosciences/Bioinformatics-Training/wiki/%22Installing%22-SMRT-Portal-the-easy-way—Launching-A-SMRT-Portal-AMI).
This Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession no. JAJA00000000. The version described in this paper is version JAJA02000000.
Genome Annotation and Comparative Analysis
The genome of Lc AZ78 was annotated with the online platform Rapid Annotation using Subsystem Technology (RAST) version 2.0 (Aziz et al., 2008). For genome comparison, the genomes of Sm K729a (AM743169) and Xcc ATCC 33913 (AE008922) were submitted to the same online platform to eliminate bias deriving from the different annotation systems employed. Genetic content comparison between genomes was conducted at nucleotide and amino acid level using BLASTN and BLASTP, respectively. Both minimum length > 70 and identity ≥ 70% at amino acid level were used as threshold parameters.
Proteolytic Activity in the Interaction between Lysobacter Capsici AZ78 and Phytophthora infestans
Plugs (5 mm) were cut from the edge of 7-day-old P. infestans colonies and transferred onto cellophane film overlying PAM dishes and incubated at 20°C for 7 days. After the incubation period, P. infestans macrocolonies originating from the plugs were transferred into 2 ml sterile tubes containing 500 μl of Phosphate Buffer Solution (PBS, 0.8% NaCl; 0.02% KCl; 0.145% NaH2PO4; 0.025% KH2PO4). Subsequently, the tubes were inoculated with 50 μl of a Lc AZ78 cell suspension (1 × 109 CFU/ml). Tubes containing PBS, PBS with Lc AZ78 and PBS with P. infestans were used as controls.
All the tubes were incubated at 25°C for 48 h and were processed at 6, 24, and 48 h to determine proteolytic activity. Briefly, tubes were centrifuged (16,100 g, 5 min) and 225 μl of supernatants were mixed with 150 μl of 1% Casein stock solution (50 mM Tris-HCL, pH 8.8) in new sterile 2 ml tubes. Subsequently, the tubes were incubated at 37°C for 1 h and undigested substrates were precipitated by adding 375 μl of 5% Trichloroacetic acid. The tubes were then centrifuged at 16,100 g for 3 min. The resulting supernatants were transferred into new sterile 2 ml tubes containing 400 μl of 1 M NaOH, and absorbance at 405 nm (AOD405 nm) was assessed using a spectrophotometer. At each time point, three 2 ml tubes (replicates) for each treatment were used, and the experiment was repeated.
Production of Lytic Enzymes and Siderophores
L. capsici AZ78 was evaluated in terms of its ability to degrade cellulose, chitin, laminarin and proteins using classic methods (Cowan, 1974; Sambrook and Russell, 2001). The occurrence of a clear halo surrounding Lc AZ78 colonies was checked after 48 h incubation at 27°C.
To determine siderophore production, LBA dishes were overlaid with CAS agar medium (Schwyn and Neilands, 1987). The final medium looked dark blue. Five microliter of Lc AZ78 cell suspension were spot inoculated onto these dishes. Siderophore production associated with the change in the color of CAS agar medium (Schwyn and Neilands, 1987) was assessed after 72 h incubation at 27°C.
In vitro Antifungal and Antibacterial Activity
The antifungal activity of Lc AZ78 against 23 phytopathogenic fungi (Table S1) was evaluated by using the classic dual-culture method. Briefly, 50 μl of Lc AZ78 cell suspension were spotted on two opposite edges of a PDA plate. After 24 h incubation at 27°C, plugs of mycelium (5 mm) were cut from the edge of young fungal colonies grown on PDA and placed at the center of the plates containing the Lc AZ78 macrocolonies. PDA plates seeded only with mycelium plugs were used as controls. After 4 days incubation at 25°C, inhibition of mycelial growth was evaluated by scoring the diameters of fungal colonies. Each test was performed in triplicate and the experiment was repeated.
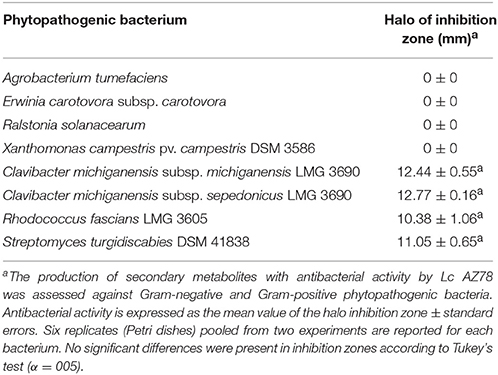
In vitro antibacterial activity of Lc AZ78 against eight phytopathogenic bacteria (Table S1) was evaluated. NA dishes were spot inoculated with 50 μl of Lc AZ78 cell suspension and incubated for 72 h at 27°C. L. capsici AZ78 cells were then killed by exposure to chloroform vapor for 60 min. The plates were subsequently aerated under the laminar flow for 60 min. Dishes were overlaid with 8 ml of 0.4% agar PBS, mixed with 2 ml of a suspension containing 1 × 108 CFU/ml of the test bacterial strains. NA dishes not seeded with Lc AZ78 and NA dishes overlaid with 0.4% agar PBS only were used as controls. Each test was performed in triplicate and the experiment was repeated. The diameters of inhibition haloes were scored after 48 h incubation at 28°C.
Determination of Lysobacter capsici AZ78 Resistance to Cobalt and Zinc
To determine the ability of Lc AZ78 to resist heavy metals, Lc AZ78 cells were grown on LBA amended with CoCl2 and ZnSO4 (Sigma-Aldrich). Briefly, filter-sterilized CoCl2 and ZnSO4 solutions were added to LBA to obtain the final concentrations of 0.5 and 1 mM, respectively. Subsequently, 100 μl of a serial dilution, from 10−1 to 10−7, of Lc AZ78 cell suspension were spread onto LBA and LBA amended with CoCl2 and ZnSO4 using sterile spatulas. L. capsici AZ78 CFU were counted after 4 days of incubation at 27°C. Three replicates (Petri dishes) of each combination (dilution and heavy metal concentration) were prepared and the experiments were repeated. Bacillus amyloliquefaciens FZB42 was used as control. The number of Lc AZ78 and B. amyloliquefaciens FZB42 CFU were log10 transformed before statistical analysis.
Resistance of Lysobacter capsici AZ78 to Chemical Fungicides and Insecticides
Thirty-two plant protection products commonly applied for the chemical control of grapevine plant diseases were used in this experiment (Table S2). Each plant protection product was dissolved in distilled water, filter-sterilized and added to LBA to achieve the maximum concentration commonly applied in the field (Table S2). A volume of 100 μl of Lc AZ78 cell suspension (1 × 103 CFU/ml) was spread onto LBA and LBA amended with plant protection products with sterile spatulas. L. capsici AZ78 CFUs were counted after 72 h of incubation at 27°C. Viability reduction was calculated according to this formula:
(CFU grown on LBA – CFU grown on LBA amended with the plant protection product) / (CFU grown on LBA)
Three Petri dishes (replicates) were used for each plant protection product, and the experiments were repeated.
Sensitivity of Lysobacter capsici AZ78 to Antibiotics
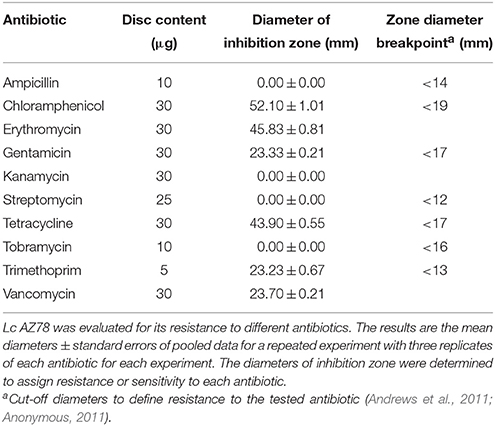
The standardized disc susceptibility testing method (Andrews et al., 2011) was used to determine the sensitivity of Lc AZ78 to ampicillin, chloramphenicol, erythromycin, gentamicin, kanamycin, streptomycin, tetracycline, tobramycin, trimethoprim, and vancomycin. Briefly, Mueller Hinton Agar (MHA, Sigma-Aldrich) was poured into sterile Petri dishes to reach a depth of 4 mm (Atlas, 2004). A volume of 100 μl of Lc AZ78 cell suspension (2 × 108 CFU/ml) was spread over the entire surface of MHA dishes using sterile spatulas. Subsequently, three discs of the same antibiotic (Oxoid) were placed on each inoculated Petri dish. After 48 h incubation at 27°C, the diameters of the inhibition zones were measured with a caliper. Three Petri dishes (replicates) were used for each antibiotic and the experiments were repeated. Resistance or sensitivity to antibiotics was assigned on the basis of the inhibition zone diameters (Andrews et al., 2011; Anonymous, 2011).
Statistical Analysis
The data obtained in the experiments aimed at assessing (i) proteolytic activity; (ii) antifungal and antibacterial activity; (iii) resistance to cobalt and zinc, were subjected to two-way ANOVA. The data on experimental repetitions were pooled when no significant differences were found according to the F-test (α > 0.05). In the case of proteolytic activity, antibacterial activity, and resistance to cobalt and zinc, the data were subsequently analyzed using one-way ANOVA, and mean comparisons were performed with Tukey's test (α = 0.05). Student's T-test (α = 0.05) was used as a post hoc test for mean comparison in the case of antifungal activity. All these statistical tests were carried out using Statistica 7.0 (StatSoft, USA).
Results
Genome Assembly and Sequence Comparison
The PacBio SMRT cell yielded output data with average genome coverage of ~44x to generate a de novo assembly of the complete genome sequence of Lc AZ78. The PacBio RSII sequencing system generated 298,751,307 base pairs (bp) through 67,899 reads (N50 read length 6848 and mean read length 4399). The genome of Lc AZ78 (Figure 1) consists of 6,272,844 bp assembled into three contigs, and the G+C content is 66.4% (Table 1), similarly to the L. capsici type strain (65.4%; Park et al., 2008). The Lc AZ78 genome contains 5292 predicted coding sequences and 93 predicted non-coding RNAs, including one transfer-messenger RNA (tmRNA), seven rRNAs, and 85 tRNAs (Table 1).
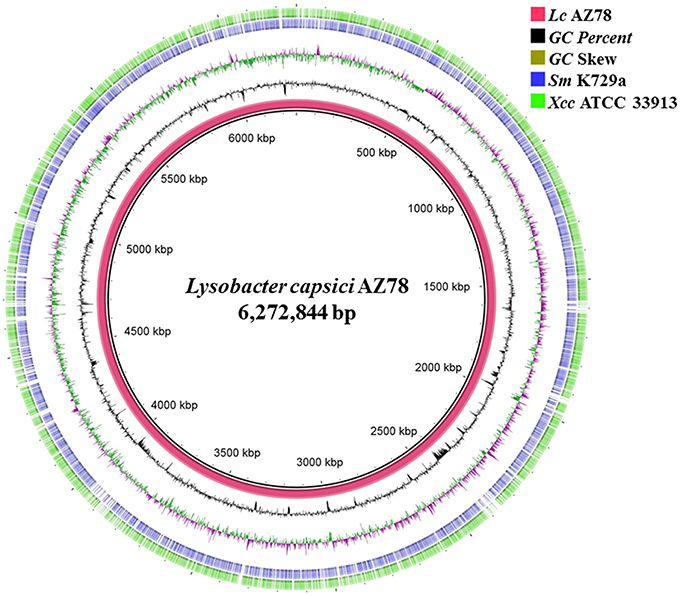
Figure 1. Graphic representation of the Lysobacter capsici AZ78 genome. Circles show (from the inside): (1) Lc AZ78 genome; (2) GC percent; (3) GC skew (4,5); Blast comparison of the Lc AZ78 genome with Sm K729a and Xcc ATCC 33913.
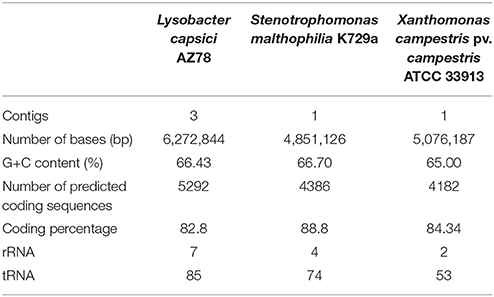
Table 1. Summary of the genomic characteristics of Lysobacter capsici AZ78, Stenotrophomonas malthophilia K279a and Xanthomonas campestris pv. campestris ATCC 33913.
The Lc AZ78 genome was compared with the genomes of the opportunistic human pathogen Sm K729a and the phytopathogen Xcc ATCC 33913 (da Silva et al., 2002; Crossman et al., 2008). Genome characteristics such as chromosome size, G+C content and the number of predicted coding sequences differed in the three bacterial strains (Table 1). They share a core genome of 2910 orthologs, mostly involved in primary metabolism (Figure 2). Genes that are associated with the capacity of Sm K729a to infect humans, such as the smlt0598, smlt3048, smlt4452, and wbpv genes (Crossman et al., 2008), are absent in the genome of Lc AZ78 and Xcc ATCC 33913. Similarly, the avrBs1, avrBs1.1, avrBs2, avrXccA1, avrXccA2, avrXccB and avrXccC genes involved in the pathogenic interaction (da Silva et al., 2002; Wang et al., 2007) are specific for the Xcc ATCC 33913. The Lc AZ78 genome contains several unique genes involved in interaction processes with other microorganisms and environmental factors (Table 2).
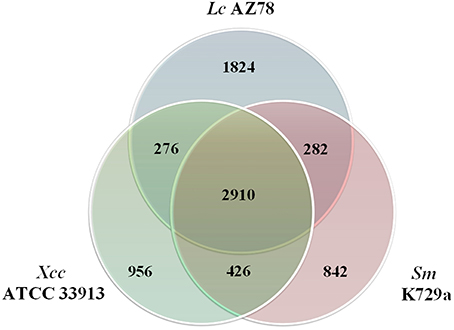
Figure 2. Comparison between the genomes of Lysobacter capsici AZ78, Stenotrophomonas malthophilia K729a and Xanthomonas campestris pv. campestris ATCC 33913. The Venn Diagram shows the number of shared and genome-specific genes in Lc AZ78, Sm K729a and Xcc ATCC 33913.
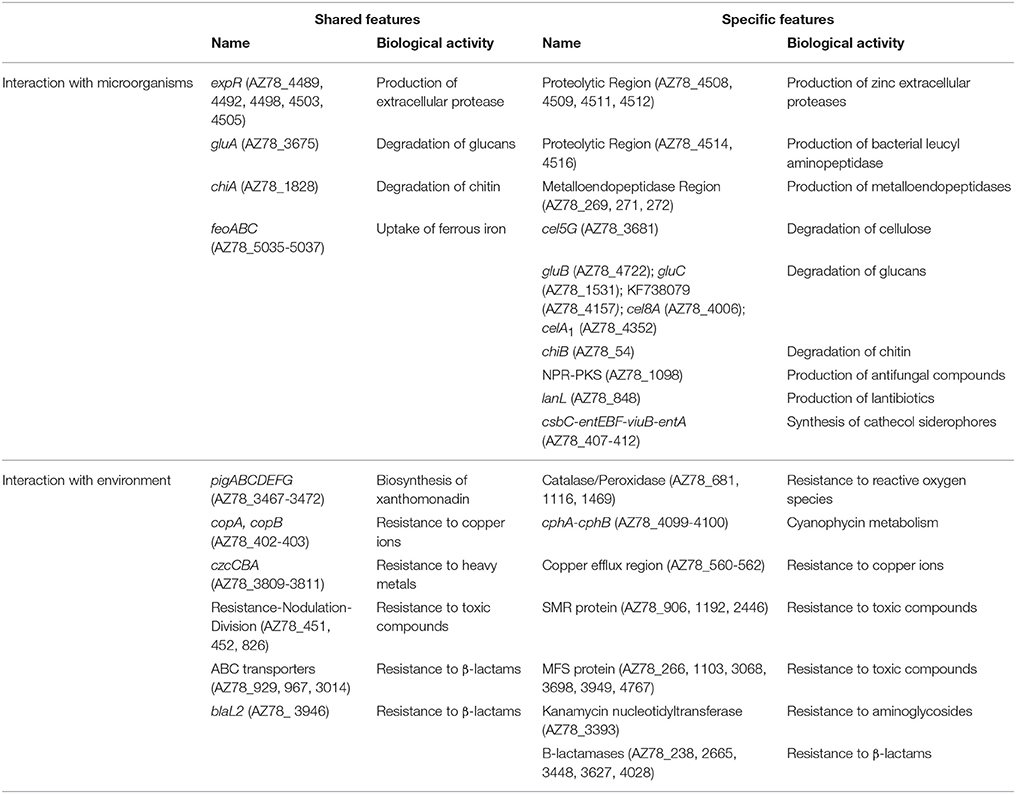
Table 2. List of genes specific to Lysobacter capsici AZ78 or shared with Stenotrophomonas malthophilia K279a and Xanthomonas campestris pv. campestris ATCC 33913.
Interaction with Microorganisms: Production of Lytic Enzymes
The presence in Lc AZ78 genome of 79 genes encoding proteolytic enzymes represents a substantial difference with Sm K729a and Xcc ATCC 33913 genomes. A region of 40,829 bp in length is specific to the Lc AZ78 genome and is missing from the genome of the other two bacterial species (Proteolytic Region, Table 2). Although this region contains five genes (AZ78_4489, 4492, 4498, 4503, and 4505) encoding extracellular proteases homologous to an extracellular protease present in the genome of Sm K729a (expR, Smlt0861; Table 2) and Xcc ATCC 33913 (XCC0851), it has other Lc AZ78-specific genes. In particular, four genes (AZ78_4508, 4509, 4511, and 4512) encoding extracellular zinc proteases (EC 3.4.24.26), and two genes (AZ78_4514 and 4516) encoding bacterial leucyl aminopeptidase (EC 3.4.11.10) have no orthologs in the Sm K729a and Xcc ATCC 33913 genomes. Another Lc AZ78-specific region (Metalloendopeptidase Region, Table 2) contains three Lc AZ78-specific genes (AZ78_269, 271 and 272) encoding metalloendopeptidases that do not share homology with any gene of Sm K729a and Xcc ATCC 33913. However, peptidase genes that have already been characterized in other Lysobacter strains were found in Lc AZ78, such as three genes encoding endopeptidases and a peptidyl Asp-metalloendopeptidase homologous to the lepA (AB045676) and lepB (AB094439) genes of Lysobacter sp. IB-9374, respectively (Chohnan et al., 2002, 2004).
Other genes located in these two regions share homology with protease genes (KF738078, KF738082, KF738069, and KF738070) of L. gummosus UASM 402 (Gökçen et al., 2014). The high number of proteases found in Lc AZ78 genome, led to investigate whether proteolytic activity could be involved in the interaction between Lc AZ78 and the phytopathogen P. infestans. A significant increase in proteolytic activity occurred when the two microorganisms were co-cultured at 25°C for 24 h (Figure 3).
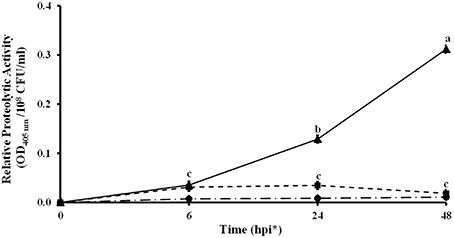
Figure 3. Proteolytic activity in the in vitro interaction between Lysobacter capsici AZ78 and Phytophthora infestans. Proteolytic activity was monitored 6, 24, and 48 h after the following treatments: Lc AZ78 (●), P. infestans (■) and Lc AZ78 + P. infestans (▲). Mean and standard error values for six replicates (2 ml-tubes) pooled from two experiments are reported for each condition. Different letters indicate significant differences according to Tukey's test (α = 0.05).
Other lytic enzymes of Lc AZ78 may be involved in the degradation of other components of P. infestans cell wall such as cellulose that was degraded in vitro by Lc AZ78 (Figure 4A). Cellulose degradation is related to the presence in Lc AZ78 genome of the AZ78_3681 gene encoding a cellulase belonging to glycosyl hydrolase family 5 that has no homology with cellulases included Sm K729a and Xcc ATCC 33913 genomes (Table 2). However, cellulase from Lc AZ78 shows homology at amino acid level with the Cel5G, a putative cellulase found in the genome of Cellvibrio japonicus Ueda107 (CP000934; DeBoy et al., 2008).
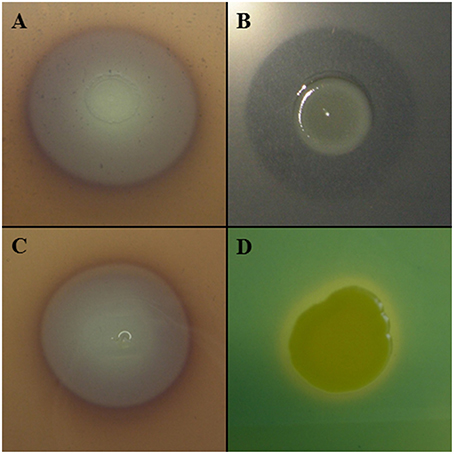
Figure 4. Characterization of Lysobacter capsici AZ78. Lc AZ78 produces (A) cellulases; (B) β-glucanases; (C) chitinases, and (D) siderophores.
L. capsici AZ78 was able to degrade laminarin in vitro (Figure 4B) and this lytic activity was confirmed by the presence in its genome of three genes encoding two enzymes (GluA, GluC) belonging to glycosyl hydrolase family 16, and one enzyme (GluB) belonging to glycosyl hydrolase family 64. Specifically, three genes (AZ78_3675, 4722, 1531) are orthologs of gluA, gluB and gluC previously characterized in L. enzymogenes C3 (AY667477; AY667478; AY667479) and N4-7 (AY157838; AY157839; AY157840; Palumbo et al., 2003, 2005). The Sm K729a genome lacks genes encoding endo β-1,3 glucanases, while Xcc ATCC 33913 has a gene (XCC1188) encoding an endo β-1,3 glucanase homologous to gluA of Lc AZ78 (Table 2). Unlike L. enzymogenes C3, N4-7 and Xcc ATCC 33913, Lc AZ78 has a second gene (AZ78_406) encoding an endo β-1,3 glucanase homologous to GluA, a gene (AZ78_4157) encoding an enzyme belonging to glycosyl hydrolase family 16 showing considerable similarity with the KF738079 gene identified in L. gummosus UASM 402 (Gökçen et al., 2014), and another gene (AZ78_4006; Table 2) which is homologous to cel8A of Lysobacter sp. IB-9374 (AB244037) encoding an enzyme belonging to glycosyl hydrolase family 8 with β-1,4 glucanase and chitosanase activity (Ogura et al., 2006). L. capsici AZ78 also has a gene (AZ78_4352) that encodes a β-1,4 endoglucanase belonging to glycosyl hydrolase family 6, which has never been characterized to date in Lysobacter members. This is missing in the genome of Sm K729a and Xcc ATCC 33913 (Table 2), although it is homologous to celA1 (Z12157) described for Streptomyces halstedii JM8 (Fernández-Abalos et al., 1992).
As regards phytopathogenic fungi, Lc AZ78 degraded chitin in vitro (Figure 4C), and this activity is related to the presence in its genome of the AZ78_1828 gene encoding a chitinase A present in the Sm K729a genome (Smlt0682, Table 2) and not in the Xcc ATCC 33913 genome. Unlike Sm K729a, the Lc AZ78 genome contains another gene (AZ78_3859) encoding a chitinase A that shares high similarity with the chiA gene (AB014770) described in Xanthomonas sp. AK (Sakka et al., 1998). L. capsici AZ78 genome also has a gene (AZ78_54) encoding a chitinase B (chiB, Table 2) sharing homology with chiB genes identified in the two strains Burkholderia gladioli CHB101 (AB038998) and BSR3 (CP002600; Shimosaka et al., 2001; Seo et al., 2011).
Interaction with Microorganisms: Production of Antibiotics
L. capsici AZ78 released secondary metabolites with antifungal activity in vitro and reduced the in vitro mycelial growth of 22 phytopathogenic fungi, with the sole exception of Pyrenochaeta (Py.) lycopersici (Table 3). L. capsici AZ78 genome was mined for genes potentially involved in the production of antibiotics and a genomic region of 9489 bp missing in Sm K729a and Xcc ATCC 33913 was identified. Specifically, the AZ78_1098 gene (Table 2) has homology with a gene encoding a hybrid polyketide synthase and a non-ribosomal peptide synthetase (NPR-PKS) involved in the biosynthesis of Heat Stable Antifungal Factor (HSAF) in L. enzymogenes C3 (EF028635; Yu et al., 2007).
In our antibacterial activity tests, Lc AZ78 released compounds that are toxic to the Gram-positive phytopathogenic bacteria Clavibacter michiganensis subsp. michiganensis LMG 7333, C. michiganensis subsp. sepedonicus LMG 2889, Rhodococcus fascians LMG 3605 and Streptomyces turgidiscabies DSM 41997, while no toxic activity was shown against the tested Gram-negative phytopathogenic bacteria (Table 4). L. capsici AZ78 genome hosts genes involved in the biosynthesis of ribosomally encoded antibacterial peptides named lantibiotics (Chatterjee et al., 2005), and these genes are missing in the genomes of Sm K729a and Xcc ATCC 33913 (Table 2). L. capsici AZ78 contains a gene (AZ78_848) homologous to the venL gene (WP_015031826) identified in Streptomyces venezuelae ATCC 10712 (Goto et al., 2010). As for venL in S. venezuelae ATCC 10712, the AZ78_848 gene is followed by two genes (AZ78_847 and AZ78_846) encoding an ATP-binding protein and an efflux transporter, indicating synteny of this region in the two bacterial species.
Interaction with Microorganisms: Production of Siderophores
L. capsici AZ78 produced siderophores on CAS agar plates (Figure 4D) and has genes involved in uptake and transport of iron ions that are homologous to genes present in the genome of Sm K729a and Xcc ATCC 33913. For example, the Lc AZ78 genome includes the feoABC operon (AZ78_5035-5037, Table 2) involved in the uptake of ferrous iron, which shares high homology with the feoABC operon present in Sm K729a (Smlt2211-2213), Xcc ATCC 33913 (XCC1834-1836). L. capsici AZ78 contains an additional gene cluster (AZ78_407-AZ78_412, Table 2) whose genes show homology with the entAFBE-csbC (CP001157) genes responsible for the production of catechol siderophores in Azotobacter vinelandii strains DJ and ATCC 12837 (Setubal et al., 2009; Yoneyama et al., 2011). The corresponding region in Lc AZ78 contains one gene (AZ78_411) encoding a protein similar to ViuB (CP001235), involved in the utilization of exogenous ferric vibriobactin complex in Vibrio cholerae 0395 (Butterton and Calderwood, 1994).
Interaction with the Environment: Tolerance to Environmental Stressors
The genetic information needed for resistance to environmental stressors is shared by the Lc AZ78, Sm K729a and Xcc ATCC 33913 genomes. For instance, Lc AZ78 genome has genes involved in the biosynthesis of xanthomonadin (AZ78_ 3467-3472, Table 2), a pigment responsible for protection against UV light irradiation in Xanthomonas spp. (Rajagopal et al., 1997). Moreover, Lc AZ78 includes several genes related to reactive oxygen species (ROS) resistance and shared with Sm K729a and Xcc ATCC 33913 and genes encoding a cytochrome c551 peroxidase (EC 1.11.1.5) (AZ78_681), a catalase (EC 1.11.1.6)/Peroxidase (EC 1.11.1.7) (AZ78_1116) and a superoxide dismutase (EC 1.15.1.1) (AZ78_1469) with no orthologs in the Sm K729a and Xcc ATCC 33913 genomes (Table 2).
Another specificity of Lc AZ78 genome is represented by a two-gene cluster (AZ78_4099-4100) responsible for the biosynthesis and degradation of cyanophycin with no orthologs in Sm K729a and Xcc ATCC 33913 (Table 2). The synthesis and degradation of cyanophycin are catalyzed respectively by cyanophycin synthetase and cyanophycinase encoded by the two-gene cluster cphA-cphB (Li et al., 2001; Krehenbrink et al., 2002). L. capsici AZ78 genome hosts a cyanophycinase (AZ78_4099), followed immediately by a cyanophycyn synthetase (AZ78_4100).
Interaction with the Environment: Tolerance to Heavy Metals
The Lc AZ78, Sm K729a, and Xcc ATCC 33913 genomes have the copA and copB genes, encoding a multicopper oxidase and the copper resistance protein B respectively (AZ78_402-403, Table 2). Unlike the other two Xandomonadaceae members, Lc AZ78 has a genomic region encoding proteins involved in the efflux of copper ions (Table 2). This region contains two Cu2+ exporting ATPases (EC 3.6.3.4; AZ78_560-561), homologous with CP002600 of B. gladioli BSR3 (Seo et al., 2011). The product of the gene AZ78_562, located downstream of the AZ78_560-561 genes, belongs to the MerR superfamily of transcriptional activators (Hobman and Brown, 1997) and is homologous to a Cu(I)-responsive transcriptional activator (cueR) of the copper efflux system in γ-proteobacteria (Stoyanov et al., 2001).
The Lc AZ78 genome has a 4396 bp region containing three genes (AZ78_3809-3811) that are homologous with the czcCBA operon involved in resistance to cadmium, cobalt and zinc (Nies, 2003). This operon (Table 2) is also present in the genomes of Sm K729a (Smlt2456-2458) and Xcc ATCC 33913 (XCC4036-4038). The Lc AZ78 genome also hosts a gene (AZ78_3808) encoding an ortholog of the CzcD protein involved in the expression regulation of czcCBA in Ralstonia sp. CH34 (Anton et al., 1999). The presence of these genes was associated with the ability of Lc AZ78 to grow on LBA amended with different concentrations of cobalt and zinc (Figure 5). In this in vitro experiments, B. amyloliquefaciens FZB42 was used as control since its genome is missing of czcCBA operon (CP000560; Chen et al., 2007). The viability of Lc AZ78 was not negatively affected by ZnSO4 at the concentrations tested while B. amyloliquefaciens FZB42 viability was reduced of an order of magnitude at both the concentrations tested (Figure 5A). CoCl2 resulted more toxic against Lc AZ78 cells and a decrease of two orders of magnitude was registered on LBA amended with 1 mM CoCl2 (Figure 5B). However, a more drastic reduction in B. amyloliquefaciens FZB42 was observed at the same conditions (Figure 5B).
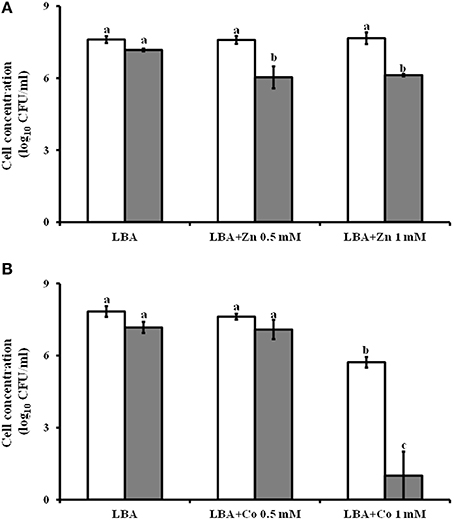
Figure 5. Assessment of Lysobacter capsici AZ78 resistance to heavy metals. Lc AZ78 (white columns) was evaluated to assess its ability to resist zinc (A) and cobalt (B) at two concentrations (0.5 and 1 mM). B. amyloliquefaciens FZB42 (gray columns) was used as control. Mean and standard error values (columns) for six replicates (Petri dishes) pooled from two experiments are reported for each heavy metal concentration. Different letters indicate significant differences according to Tukey's test (α = 0.05).
Interaction with the Environment: Tolerance to Fungicides and Antibiotics
L. capsici AZ78 was resistant in vitro to several fungicides and insecticides commonly applied in viticulture (Figure 6). This resistance may rely on the presence of efflux systems such as ABC transporters, Resistance-Nodulation-Division (RND), Small Multidrug Resistance protein (SMR), and proteins belonging to the Major Facilitator Super-family (MFS) and the Multidrug Toxic compound Extrusion family (MTE; Poole, 2001), which Lc AZ78 shares with Sm K729a and Xcc ATCC 33913 (Table 2). Moreover, there are some Lc AZ78-specific genes related to fungicide and antibiotic resistance, such as three genes (AZ78_906, 1192, and 2446) encoding putative SMR proteins and six genes (AZ78_266, 1103, 3068, 3688, 3949, and 4767) encoding putative MFS proteins (Table 2).
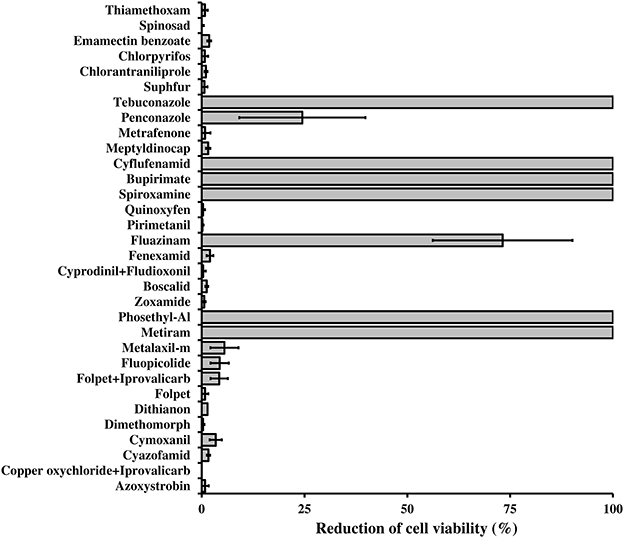
Figure 6. Resistance of Lysobacter capsici AZ78 to plant protection products. The survival of Lc AZ78 cells in the presence of fungicides and insecticides was assessed by growing the bacterium on LBA amended with plant protection products at concentrations commonly applied in the field. The reduction in cell viability was calculated as the ratio between the CFU difference for Lc AZ78 grown on LBA and Lc AZ78 grown on LBA amended with plant protection products, over Lc AZ78 CFU grown on LBA. Mean and standard error values (columns) for six replicates (Petri dishes) pooled from two experiments are reported for each plant protection product.
Since all these efflux systems can also contribute to resistance to antibiotics, we evaluated the sensitivity of Lc AZ78 to several antibiotics in vitro (Table 5). L. capsici AZ78 is sensitive to chloramphenicol, erythromycin, gentamicin, tetracycline, trimethoprim and vancomycin whereas it is resistant to ampicillin, kanamycin, streptomycin, and tobramycin. The resistance to kanamycin and streptomycin may depend on inactivation mediated by aminoglycoside phosphotransferase or adenylyltransferase enzymes (Shaw et al., 1993). The Lc AZ78 genome contains a gene (AZ78_3072) encoding putative streptomycin 3″-kinase (EC 2.7.1.87) that does not show homology with aph(3′′) and aph(3′)-IIc involved in streptomycin and kanamycin resistance in Sm K729a (Okazaki and Avison, 2007; Crossman et al., 2008). Moreover, the Lc AZ78 genome includes another gene (AZ78_3393) encoding a putative kanamycin nucleotidyltransferase (Table 2). Other putative antibiotic resistance genes in Lc AZ78 genome are responsible for resistance to β-lactams such as ampicillin: 12 genes showed homology with the β-lactamases of Sm K729a and Xcc ATCC 33913 and five β-lactamases genes (AZ78_238, 2665, 3488, 3627, and 4028) were unique to Lc AZ78 (Table 2).
Discussion
Development of new biofungicides based on BCAs are becoming an important task to reduce the use of chemical fungicides for the control of phytopathogenic microorganisms. Recently, Lc AZ78 was shown to effectively control two important phytopathogenic oomycetes and a first formulation prototype was designed for its application in vineyards for the control of Pl. viticola (Puopolo et al., 2014a,b; Segarra et al., 2015a,b). However, information regarding the biological characteristics of Lc AZ78 are needed for the registration process and to foster the development of this bacterial strain as the main ingredient of novel biofungicides. Thus, the genome of Lc AZ78 was sequenced by PacBio sequencing system. This technology was chosen because the advantage of longer reads during assembly ensures a higher quality final result (Roberts et al., 2013). The drawback of higher error rate for PacBio sequences can be overcome with a higher genome coverage, a choice that already proved to be the best strategy nowadays for bacterial sequencing projects (Booher et al., 2015; Cameron et al., 2015; Lee et al., 2015). Accordingly, PacBio RSII sequencing system resulted more effective than the Illumina GAIIx system previously used for obtaining a first draft genome of Lc AZ78 (Puopolo et al., 2014c) and number of contigs was reduced from 142 achieved with Illumina to three achieved with PacBio in this work.
Since comparison of the genomes highlighted important similarities and differences between microorganisms (Studholme et al., 2009; Straub et al., 2013), the Lc AZ78 genome was compared with the genomes of the opportunistic human pathogen Sm K729a and the phytopathogen Xcc ATCC 33913 originally sequenced using Sanger sequencing technology (da Silva et al., 2002; Crossman et al., 2008). This technology is no longer used for whole genome assembly projects, however the quality of Sanger sequencing technology is comparable to the quality achieved with PacBio technology (Eid et al., 2009). The comparative analysis of the three genomes revealed high diversity between the three bacterial strains and showed that Lc AZ78 does not have the genetic basis for pathogenic interaction with humans and plants. Particularly, the comparison highlights the lack in Lc AZ78 genome of genes responsible for the instauration of human diseases present in Sm K729a genome. Similarly, the lack of these genes represents one of the factors differentiating the opportunistic human pathogen Sm K729a from the closely related beneficial bacterium Stenotrophomonas rhizophila DSM14405T (Alavi et al., 2014). This comparative analysis also allowed to identify several Lc AZ78-specific genes that are related to bacterial responses to other microorganisms and environmental factors.
A vast number of Lc AZ78-specific genes are involved in competition with other microorganisms highlighting the biocontrol properties of Lc AZ78 associated with the release of extracellular enzymes that can lyse the cell wall of both fungi and oomycetes. Members of the genus Lysobacter are known to produce a plethora of extracellular enzymes with lytic activity capable of degrading the cell wall components of several phytopathogenic microorganisms (Kobayashi and Yuen, 2007; Hayward et al., 2010). Accordingly, the Lc AZ78 genome includes a repertoire of genes encoding lytic enzymes capable of degrading cellulose, chitin, laminarin and proteins in vitro.
One of the main differences between Lc AZ78, Sm K729a and Xcc ATCC 33913 genomes, relies on the presence of a large number of genes encoding extracellular proteases in the Lc AZ78 genome that may display specific proteolytic activities. Particularly, Lc AZ78 genome has four gene sharing homology with proteases characterized in L. gummosus UASM 402, which are involved in the digestion of biofilm produced by Staphylococcus epidermidis (Gökçen et al., 2014). The presence of these genes in the genome of Lc AZ78 highlights the importance of members of the L. capsici species as potential new sources of enzymes exploitable for the control of important human pathogens through degradation of their extracellular matrix. Proteolitic activity of Lc AZ78 increased by in vitro incubation with P. infestans, and this may represent a key process in Lc AZ78′s biocontrol activity as shown for L. enzymogenes 3.1T8 (Folman et al., 2003, 2004). Furthermore, Lc AZ78 degraded in vitro β-glucans (laminarin) and cellulose other components of the oomycete cell wall. The degradation of these polymers is associated with the presence in the Lc AZ78 genome of genes encoding cellulases and a vast array of β-glucanases that clearly indicates that phytopathogenic oomycetes represent an optimal target of this BCA.
The Lc AZ78 genome also includes unique genes encoding chitinases that are commonly involved in the control of phytopathogenic fungi and they play a significant role in the biocontrol activity of L. enzymogenes C3 against Bipolaris sorokiniana (Zhang and Yuen, 2000). The Lc AZ78 genome has a gene encoding for a ChiA enzyme already characterized in the L. enzymogenes C3 (AY667480), N4-7 (AY667481) and OH11 (DQ888611) strains (Zhang et al., 2001; Sullivan et al., 2003; Qian et al., 2009). Moreover, the analysis of genes encoding chitinases in Lc AZ78 genome revealed the presence of a second gene with a high level of identity with the chiA gene previously characterized in Xanthomonas sp. AK (Sakka et al., 1998). This homology, and the 16S rDNA analysis carried out by Folman et al. (2003), represents strong evidence for misidentification in the case of Xanthomonas sp. AK, which in our opinion should be considered a Lysobacter sp. Analysis of the Lc AZ78 genome highlighted the presence of a gene encoding a chitinase B, a novelty for the genus Lysobacter and Xanthomonadaceae. Indeed, enzymes such as ChiB belong to glycosyl hydrolase family 19, which includes chitinases mostly identified in actinomycetes (Watanabe et al., 1999). Within Proteobacteria, chiB genes were only identified in the two strains B. gladioli CHB101 and BSR3 (Shimosaka et al., 2001; Seo et al., 2011). The presence of both ChiA and ChiB chitinases in the Lc AZ78 genome supports the potential of this bacterial strain to attack and degrade the cell wall of phytopathogenic fungi.
The genomic and functional information provided in this work demonstrates that Lc AZ78 has unique genes responsible for the synthesis of macrocyclic lactams toxic against phytopathogenic microorganisms. L. enzymogenes C3 produces HSAF a compound consisting of dihydromaltophilin and related macrocyclic lactams, toxic for several phytopathogenic fungi and oomycetes (Yu et al., 2007; Li et al., 2008). Lysobacter sp. SB-K88 synthesizes the macrocyclic lactams Xanthobaccin A, B and C, which are highly active in vitro against Aphanomyces cochlioides and Pythium ultimum (Nakayama et al., 1999). The Lc AZ78 genome includes regions involved in the production of macrocyclic lactams and the in vitro experiments let to hypothesize that the putative toxic compounds released by Lc AZ78 have probable similarities with xanthobaccins. Indeed, Nakayama et al. (1999) reported that Py. lycopersici was not sensitive to xanthobaccins produced in vitro by Lysobacter sp. SB SB-K88. Similarly, this phytopathogenic fungus was not sensitive to the toxic compounds released in vitro by Lc AZ78 in our experiments. Future work will be aimed at determining the chemical structure of the secondary metabolites with antifungal activity produced by Lc AZ78 to study the involvement of this class of antibiotics in the biological control of phytopathogenic fungi and oomycetes.
L. capsici AZ78 also released secondary metabolites toxic to four phytopathogenic Gram-positive bacteria and the presence of genes involved in their biosynthesis differed in Lc AZ78, Sm K729a and Xcc ATCC 33913 strains. Particularly, Lc AZ78 genome contains genes involved in the production of lantibiotics, compounds toxic to Gram-positive bacteria (Chatterjee et al., 2005) that are missing in the genome of the other two bacterial strains. Little is known about lantibiotic production in Lysobacter members, whereas it was reported that L. enzymogenes OH11 produces the cyclic lipodepsipeptide WAP-8294A2 active against the human pathogenic Gram-positive bacterium Staphylococcus aureus (Zhang et al., 2011). Therefore, we cannot rule out the possibility that the antibacterial activity of Lc AZ78 could be associated with the production of other toxic secondary metabolites, and chemical analysis is needed to further investigate this topic.
The comparison of the three genomes higlighted genetic informations regarding the ability of Lc AZ78 to scavenge ferrous ions from the environment through the production of siderophores (Neilands, 1995; Chu et al., 2010). Altough the production of these secondary metabolites is known to be important for human pathogenic, plant pathogenic and plant beneficial bacteria (Hamdan et al., 1991; Pandey and Sonti, 2010; Skaar, 2010), few information are available about siderophore production in Lysobacter spp. Differently from Sm K729a and Xcc ATCC 33913, the genome of Lc AZ78 is provided with the entAFBE-csbC operon responsible for the production of cathecol siderophores indicating that Lc AZ78 may also compete with other microorganisms for iron ions in the environment.
Our results also highlight key genes involved in the resistance of Lc AZ78 to UV-light irradiation and starvation. Previously, we have shown that Lc AZ78 resisted to starvation stress for 15 days and can be stored at 4°C in distilled water for a year (Puopolo et al., 2014a; Segarra et al., 2015a). This ability may be associated with the presence of the cphA-cphB operon responsible for the production and degradation of cyanophycin, that is missing in the genome of Sm K729a and Xcc ATCC 33913. This compound is a branched non-ribosomally synthesized polypeptide that accumulates in cyanobacteria and proteobacteria (Allen et al., 1980; Krehenbrink et al., 2002) and acts as a temporary nitrogen and carbon reserve (Li et al., 2001; Krehenbrink et al., 2002). Therefore, L. capsici members could have the genetic basis to promptly adapt to environments lacking in nutrients. L. capsici AZ78 is able to resist UV-light irradiation (Puopolo et al., 2014a) and this ability may be associated with the presence of genes involved in the biosynthesis of xanthomonadin, in agreement with the production of a xanthomonadin-like aryl polyene group reported for L. enzymogenes OH11 (Wang et al., 2013). L. capsici AZ78 has a pronounced resistance to copper ions, which renders this BCA a prime candidate for combination with copper-based fungicides for more efficient control of Pl. viticola on grapevine plants (Puopolo and Pertot, 2014). Resistance to copper frequently arises in phytopathogenic xanthomonads (Stall et al., 1986; Behlau et al., 2011), and resistant Stenotrophomonas strains have been isolated from copper-polluted soils (Altimira et al., 2012). Some of the genes involved in the resistance to copper are shared among the Lc AZ78, Sm K729a and Xcc ATCC 33913 genomes, whereas genes encoding copper exporting ATPases are specific for Lc AZ78 genomes. Based on these differences, L. capsici members seem to have the genome makeup necessary to guarantee better survival in an environment with greater concentrations of copper. L. capsici AZ78 is also resistant to other heavy metals such as cobalt and zinc and this phenotype is associated with the presence of the czcCBA operon and several genes involved in the efflux of cadmium, cobalt and zinc. Most of these genes encode RND proteins known as key multidrug efflux transporters for resistance to antibiotics, dyes, fungicides and solvents in Gram-negative (Kumar and Schweizer, 2005; Bazzini et al., 2011; Yamaguchi et al., 2015). Accordingly, the presence of a high number of RND proteins and other efflux systems (SMR, MFS, and MTE) is also associated with the Lc AZ78 resistance to fungicides and insecticides in vitro. Regarding resistance to antibiotics, Lc AZ78 was sensitive to chloramphenicol and erythromycin although several genes involved in antibiotic resistance are shared with Sm K729a. Indeed, this opportunistic human pathogen is resistant to these antibiotics, making it very risky for treating infections in immunocompromised patients (Crossman et al., 2008; Ryan et al., 2009). Thus, it is conceivable that Sm K729a genome has genes involved in chloramphenicol and erythromycin resistance with no orthologs in Lc AZ78 genome.
In conclusion, the sequence and annotation of the Lc AZ78 genome provide a genetic framework for detailed analysis of potential biocontrol mechanisms against phytopathogens. In particular, the comparison of Lc AZ78, Sm K729a, and Xcc ATCC 33913 genomes allows to state that Lc AZ78 is missing of the genetic information needed to establish pathogenic interaction with humans and plants, an aspect that is crucial for the registration of new BCAs. This comparative approach highlights the genetic basis determining the Lc AZ78 aptitude to compete with phytopathogenic microorganisms through the release of (i) extracellular lytic enzymes; (ii) secondary metabolites with antibacterial and antifungal activity; and (iii) catechol siderophores. Furthermore, the Lc AZ78 genome contains a vast number of genes involved in resistance to environmental stress, antibiotics, heavy metals and plant protection products. Analysis of the Lc AZ78 genome will help to provide more accurate characterization of bacterial strains belonging to the Lysobacter genus and lead to important advances in the further development of Lc AZ78 as an active ingredient in new biofungicides.
Author Contributions
GP conceived the work, designed the experiments, carried out annotation of the genome and the experiments, analyzed the data, and wrote and edited the manuscript. ST carried out annotation of the genome and the experiments, analyzed the data, and wrote and edited the manuscript. PS, MM, and KE assembled the genome, wrote and edited the manuscript. MP and IP contributed to the conception of the work, designed the experiments and edited the manuscript. All the authors have read the manuscript and agree with its content.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This project has received funding from the European Union's Seventh Framework Programme for research, technological development and demonstration under grant agreement no. 289497 (project CO-FREE, theme KBBE.2011.1.2-06).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00096
References
Alavi, P., Starcher, M. R., Thallinger, G. G., Zachow, C., Müller, H., and Berg, G. (2014). Stenotrophomonas comparative genomics reveals genes and functions that differentiate beneficial and pathogenic bacteria. BMC Genomics 15:482. doi: 10.1186/1471-2164-15-482
Allen, M. M., Hutchison, F., and Weathers, P. J. (1980). Cyanophycin granule polypeptide formation and degradation in the cyanobacterium Aphanocapsa 6308. J. Bacteriol. 141, 687–693.
Altimira, F. A., Yáñez, C., Bravo, G., González, M., Rojaset, L. A., and Seeger, M. (2012). Characterization of copper-resistant bacteria and bacterial communities from copper-polluted agricultural soils of central Chile. BMC Microbiol. 12:193. doi: 10.1186/1471-2180-12-193
Andrews, J. M., Howe, R. A., and BSAC Working Party on Susceptibility (2011). Testing BSAC standardized disc susceptibility testing method (version 10). J. Antimicrob. Chemother. 66, 2726–2757. doi: 10.1093/jac/dkr359
Anonymous (2011). Etude de la Sensibilité aux Antimicrobiens. 2010-2011. Documentation Technique Extraite des Notices Techniques Commerciales et de Différentes Publications. Lycée des Métiers du Tertiaire, de la Santé et du Social - Louise Michel - Grenoble. Available online at: http://www.ac-grenoble.fr/disciplines/sti-biotechnologies/file/Microbiologie/Doc_tech_atb_1011.pdf
Anton, A., Grosse, C., Reissmann, J., Pribyl, T., and Nies, D. H. (1999). CzcD is a heavy metal ion transporter involved in regulation of heavy metal resistance in Ralstonia sp. strain CH34. J. Bacteriol. 181, 6876–6881.
Aziz, R. K., Bartels, D., Best, A. A., De Jongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Bazzini, S., Udine, C., Sass, A., Pasca, M. R., Longo, F., Emiliani, G., et al. (2011). Deciphering the role of RND efflux transporters in Burkholderia cenocepacia. PLoS ONE 6:e18902. doi: 10.1371/journal.pone.0018902
Behlau, F., Canteros, B. I., Minsavage, G. V., Jones, J. B., and Graham, J. H. (2011). Molecular characterization of copper resistance genes from Xanthomonas citri subsp. citri and Xanthomonas alfalfae subsp. citrumelonis. Appl. Environ. Microbiol. 77, 4089–4096. doi: 10.1128/AEM.03043-10
Booher, N. J., Carpenter, S. C. D., Sebra, R. P., Wang, L., Salzberg, S. L., Leach, J. E., et al. (2015). Single molecule real-time sequencing of Xanthomonas oryzae genomes reveals a dynamic structure and complex TAL (transcription activator-like) effector gene relationships. MGen. doi: 10.1099/mgen.0.000032. [Epub ahead of print].
Butterton, J. R., and Calderwood, S. B. (1994). Identification, cloning, and sequencing of a gene required for ferric vibriobactin utilization by Vibrio cholerae. J. Bacteriol. 176, 5631–5638.
Cameron, D. R., Jiang, J. H., Hassan, K. A., Elbourne, L. D., Tuck, K. L., Paulsen, I. T., et al. (2015). Insights on virulence from the complete genome of Staphylococcus capitis. Front. Microbiol. 6:980. doi: 10.3389/fmicb.2015.00980
Chatterjee, C., Paul, M., Xie, L., and van der Donk, W. A. (2005). Biosynthesis and mode of action of lantibiotics. Chem. Rev. 105, 633–684. doi: 10.1021/cr030105v
Chen, X. H., Koumoutsi, A., Scholz, R., Eisenreich, A., Schneider, K., Heinemeyer, I., et al. (2007). Comparative analysis of the complete genome sequence of the plant growth-promoting bacterium Bacillus amyloliquefaciens FZB42. Nat. Biotechnol. 25, 1007–1014. doi: 10.1038/nbt1325
Chin, C. S., Alexander, D. H., Marks, P., Klammer, A. A., Drake, J., Heiner, C., et al. (2013). Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569. doi: 10.1038/nmeth.2474
Chohnan, S., Nonaka, J., Teramoto, K., Taniguchi, K., Kameda, Y., Tamura, H., et al. (2002). Lysobacter strain with high lysyl endopeptidase production. FEMS Microbiol. Lett. 213, 13–20. doi: 10.1111/j.1574-6968.2002.tb11279.x
Chohnan, S., Shiraki, K., Yokota, K., Ohshima, M., Kuroiwa, N., Ahmed, K., et al. (2004). A second lysine-specific serine protease from Lysobacter sp. strain IB-9374. J. Bacteriol. 186, 5093–5100. doi: 10.1128/JB.186.15.5093-5100.2004
Christensen, P., and Cook, F. D. (1978). Lysobacter, a new genus of nonfruiting, gliding bacteria with a high base ratio. Int. J. Syst. Bacteriol. 28, 367–393. doi: 10.1099/00207713-28-3-367
Chu, B. C., Garcia-Herrero, A., Johanson, T. H., Krewulak, K. D., Lau, C. K., Peacock, R. S., et al. (2010). Siderophore uptake in bacteria and the battle for iron with the host; a bird's eye view. Biometals 23, 601–611. doi: 10.1007/s10534-010-9361-x
Cowan, S. T. (1974). Cowan and Steel's Manual for the Identification of Medical Bacteria. London; New York, NY: Cambridge University Press.
Crossman, L. C., Gould, V. C., Dow, J. M., Vernikos, G. S., Okazaki, A., Sebaihia, M., et al. (2008). The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol. 9, R74. doi: 10.1186/gb-2008-9-4-r74
da Silva, A. C., Ferro, J. A., Reinach, F. C., Farah, C. S., Furlan, L. R., Quaggio, R. B., et al. (2002). Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417, 459–463. doi: 10.1038/417459a
DeBoy, R. T., Mongodin, E. F., Fouts, D. E., Tailford, L. E., Khouri, H., Emerson, J. B., et al. (2008). Insights into plant cell wall degradation from the genome sequence of the soil bacterium Cellvibrio japonicus. J. Bacteriol. 190, 5455–5463. doi: 10.1128/JB.01701-07
Eid, J., Fehr, A., Gray, J., Luong, K., Lyle, J., Otto, G., et al. (2009). Real-Time DNA sequencing from single polymerase molecules. Science 323, 133–138. doi: 10.1126/science.1162986
Fernández-Abalos, J. M., Sanchez, P., Coll, P. M., Villanueva, J. R., Pérez, P., and Santamaria, R. I. (1992). Cloning and nucleotide sequence of celA1, an endo-β-1,4-glucanase-encoding gene from Streptomyces halstedii JM8. J. Bacteriol. 174, 6368–6376.
Folman, L. B., De Klein, M. J. E. M., Postma, J., and van Veen, J. A. (2004). Production of antifungal compounds by Lysobacter enzymogenes isolate 3.1T8 under different conditions in relation to its efficacy as a biocontrol agent of Pythium aphanidermatum in cucumber. Biol. Control 31, 145–154. doi: 10.1016/j.biocontrol.2004.03.008
Folman, L. B., Postma, J., and van Veen, J. A. (2003). Characterisation of Lysobacter enzymogenes (Christensen and Cook 1978) strain 3.1T8, a powerful antagonist of fungal diseases of cucumber. Microbiol. Res. 158, 107–115. doi: 10.1078/0944-5013-00185
Giesler, L. J., and Yuen, G. Y. (1998). Evaluation of Stenotrophomonas maltophilia C3 for biocontrol of brown patch disease. Crop Prot. 17, 509–513. doi: 10.1016/S0261-2194(98)00049-0
Gökçen, A., Vilcinskas, A., and Wiesner, J. (2014). Biofilm-degrading enzymes from Lysobacter gummosus. Virulence 5, 378–387. doi: 10.4161/viru.27919
Goto, Y., Li, B., Claesen, J., Shi, Y., Bibb, M. J., and van der Donk, W. A. (2010). Discovery of unique lanthionine synthetases reveals new mechanistic and evolutionary insights. PLoS Biol. 8:e1000339. doi: 10.1371/journal.pbio.1000339
Hamdan, H., Weller, D. M., and Thomashow, L. S. (1991). Relative importance of fluorescent siderophores and other factors in biological control of Gaeumannomyces graminis var. tritici by Pseudomonas fluorescens 2-79 and M4-80R. Appl. Environ. Microbiol. 57, 3270–3277.
Hayward, A. C., Fegan, N., Fegan, M., and Stirling, G. R. (2010). Stenotrophomonas and Lysobacter: ubiquitous plant-associated gamma-proteobacteria of developing significance in applied microbiology. J. Appl. Microbiol. 108, 756–770. doi: 10.1111/j.1365-2672.2009.04471.x
Hobman, J. L., and Brown, N. L. (1997). “Bacterial mercury-resistance genes,” in Metal Ions in Biological Systems, Vol. 34, eds A. Sigel and H. Sigel (New York, NY: Marcel Dekker), 527–568.
Kobayashi, D. Y., Reedy, R. M., Palumbo, J. D., Zhou, J. M., and Yuen, G. Y. (2005). A clp gene homologue belonging to the Crp gene family globally regulates lytic enzyme production, antimicrobial activity, and biological control activity expressed by Lysobacter enzymogenes strain C3. Appl. Environ. Microbiol. 71, 261–269. doi: 10.1128/AEM.71.1.261-269.2005
Kobayashi, D. Y., and Yuen, G. Y. (2007). The potential of Lysobacter spp. for biological control of plant diseases. CAB Rev. Persp. Agr. Vet. Sci. Nutr. Nat. Res. 2, 11. doi: 10.1079/PAVSNNR20072007
Krehenbrink, M., Oppermann-Sanio, F. B., and Steinbüchel, A. (2002). Evaluation of non-cyanobacterial genome sequences for occurrence of genes encoding proteins homologous to cyanophycin synthetase and cloning of an active cyanophycin synthetase from Acinetobacter sp. strain DSM 587. Arch. Microbiol. 177, 371–380. doi: 10.1007/s00203-001-0396-9
Kumar, A., and Schweizer, H. P. (2005). Bacterial resistance to antibiotics: active efflux and reduced uptake. Adv. Drug. Deliv. Rev. 57, 1486–1513. doi: 10.1016/j.addr.2005.04.004
Lee, W. C., Anton, B. P., Wang, S., Baybayan, P., Singh, S., Ashby, M., et al. (2015). The complete methylome of Helicobacter pylori UM032. BMC Genomics 16:424. doi: 10.1186/s12864-015-1585-2
Lee, Y. S., Naning, K. W., Nguyen, X. H., Kim, S. B., Moon, J. H., and Kim, K. Y. (2014). Ovicidal activity of lactic acid produced by Lysobacter capsici YS1215 on eggs of root-knot nematode, Meloidogyne incognita. J. Microbiol. Biotechnol. 24, 1510–1515. doi: 10.4014/jmb.1405.05014
Li, H., Sherman, D. M., Bao, S., and Sherman, L. A. (2001). Pattern of cyanophycin accumulation in nitrogen-fixing and non-nitrogen-fixing cyanobacteria. Arch. Microbiol. 176, 9–18. doi: 10.1007/s002030100281
Li, S., Jochum, C. C., Yu, F., Zaleta-Rivera, K., Du, L., Harris, S. D., et al. (2008). An antibiotic complex from Lysobacter enzymogenes strain C3: antimicrobial activity and role in plant disease control. Phytopathology 98, 695–701. doi: 10.1094/PHYTO-98-6-0695
Nakayama, T., Homma, Y., Hashidoko, Y., Mizutani, J., and Tahara, S. (1999). Possible role of xanthobaccins produced by Stenotrophomonas sp. strain SB-K88 in suppression of sugar beet damping-off disease. Appl. Environ. Microbiol. 65, 4334–4339.
Neilands, J. B. (1995). Siderophores: structure and function of microbial iron transport compounds. J. Biol. Chem. 270, 26723–26726. doi: 10.1074/jbc.270.45.26723
Nies, D. H. (2003). Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 27, 313–339. doi: 10.1016/S0168-6445(03)00048-2
Ogura, J., Toyoda, A., Kurosawa, T., Chong, A. L., Chohnan, S., and Masaki, T. (2006). Purification, characterization, and gene analysis of cellulase (Cel8A) from Lysobacter sp. IB-9374. Biosci. Biotechnol. Biochem. 70, 2420–2428. doi: 10.1271/bbb.60157
Okazaki, A., and Avison, M. B. (2007). Aph(3')-IIc, an aminoglycoside resistance determinant from Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 51, 359–360. doi: 10.1128/AAC.00795-06
Palumbo, J. D., Sullivan, R. F., and Kobayashi, D. Y. (2003). Molecular characterization and expression in Escherichia coli of three beta-1,3-glucanase genes from Lysobacter enzymogenes strain N4-7. J. Bacteriol. 185, 4362–4370. doi: 10.1128/JB.185.15.4362-4370.2003
Palumbo, J. D., Yuen, G. Y., Jochum, C. C., Tatum, K., and Kobayashi, D. Y. (2005). Mutagenesis of β-1,3-glucanase genes in Lysobacter enzymogenes strain C3 results in reduced biological control activity toward Bipolaris leaf spot of tall fescue and Pythium damping-off of sugar beet. Phytopathology 95, 701–707. doi: 10.1094/PHYTO-95-0701
Pandey, A., and Sonti, R. V. (2010). Role of the FeoB protein and siderophore in promoting virulence of Xanthomonas oryzae pv. oryzae on rice. J. Bacteriol. 192, 3187–3203. doi: 10.1128/JB.01558-09
Park, J. H., Kim, R., Aslam, Z., Jeon, C. O., and Chung, Y. R. (2008). Lysobacter capsici sp. nov., with antimicrobial activity, isolated from the rhizosphere of pepper, and emended description of the genus Lysobacter. Int. J. Syst. Evol. Microbiol. 58, 387–392. doi: 10.1099/ijs.0.65290-0
Poole, K. (2001). Multidrug resistance in Gram-negative bacteria. Curr. Opin. Microbiol. 4, 500–508. doi: 10.1016/S1369-5274(00)00242-3
Postma, J., Nijhuis, E. H., and Yassin, A. F. (2010). Genotypic and phenotypic variation among Lysobacter capsici strains isolated from Rhizoctonia suppressive soils. Syst. Appl. Microbiol. 33, 232–235. doi: 10.1016/j.syapm.2010.03.002
Puopolo, G., Cimmino, A., Palmieri, M. C., Giovannini, O., Evidente, A., and Pertot, I. (2014b). Lysobacter capsici AZ78 produces cyclo(L-Pro- L-Tyr), a 2,5-diketopiperazine with toxic activity against sporangia of Phytophthora infestans and Plasmopara viticola. J. Appl. Microbiol. 117, 1168–1180. doi: 10.1111/jam.12611
Puopolo, G., Giovannini, O., and Pertot, I. (2014a). Lysobacter capsici AZ78 can be combined with copper to effectively control Plasmopara viticola on grapevine. Microbiol. Res. 169, 633–642. doi: 10.1016/j.micres.2013.09.013
Puopolo, G., Palmieri, M. C., Giovannini, O., and Pertot, I. (2015). Impact of temperature on the survival and the biocontrol efficacy of Lysobacter capsici AZ78 against Phytophthora infestans. BioControl 60, 681–689. doi: 10.1007/s10526-015-9672-5
Puopolo, G., and Pertot, I. (2014). A New Bacterial Lysobacter capsici Strain and Uses Thereof. International Application No.: PCT/EP2014/058151. Pub. No.: WO/2014/173906.
Puopolo, G., Raio, A., and Zoina, A. (2010). Identification and characterization of Lysobacter capsici strain PG4: a new plant health-promoting rhizobacterium. J. Plant Pathol. 92, 157–164. doi: 10.4454/jpp.v92i1.25
Puopolo, G., Sonego, P., Engelen, K., and Pertot, I. (2014c). Draft genome sequence of Lysobacter capsici AZ78, a bacterium antagonistic to plant-pathogenic oomycetes. Genome Announc. 2:e00325-14. doi: 10.1128/genomeA.00325-14
Qian, G., Hu, B., Jiang, Y., and Liu, F. (2009). Identification and characterization of Lysobacter enzymogenes as a biological control agent against some fungal pathogens. Agric. Sci. China 8, 68–75. doi: 10.1016/S1671-2927(09)60010-9
Qian, G., Wang, Y., Liu, Y., Xu, F., He, Y. W., Du, L., et al. (2013). Lysobacter enzymogenes uses two distinct cell-cell signaling systems for differential regulation of secondary-metabolite biosynthesis and colony morphology. Appl. Environ. Microbiol. 79, 6604–6616. doi: 10.1128/AEM.01841-13
Qian, G., Wang, Y., Qian, D., Fan, J., Hu, B., and Liu, F. (2012). Selection of available suicide vectors for gene mutagenesis using chiA (a chitinase encoding gene) as a new reporter and primary functional analysis of chiA in Lysobacter enzymogenes strain OH11. World J. Microbiol. Biotechnol. 28, 549–557. doi: 10.1007/s11274-011-0846-8
Rajagopal, L., Sundari, C. S., Balasubramanian, D., and Sonti, R. V. (1997). The bacterial pigment xanthomonadin offers protection against photodamage. FEBS Lett. 415, 125–128. doi: 10.1016/S0014-5793(97)01109-5
Roberts, R. J., Carneiro, M. O., and Schatz, M. C. (2013). The advantages of SMRT sequencing. Genome Biol. 14, 405. doi: 10.1186/gb-2013-14-6-405
Ryan, R. P., Monchy, S., Cardinale, M., Taghavi, S., Crossman, L., Avison, M. B., et al. (2009). The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat. Rev. Microbiol. 7, 514–525. doi: 10.1038/nrmicro2163
Saddler, G. S., and Bradbury, J. F. (2005). “Family I. Xanthomonadaceae fam. nov.” in Bergeys Manual of Systematic Bacteriology, 2nd Edn., Vol. 2 (The Proteobacteria), part B (The Gammaproteobacteria), eds D. J. Brenner, N. R. Krieg, J. T. Staley, and G. M. Garrity (New York, NY: Springer), 63.
Sakka, K., Kusaka, R., Kawano, A., Karita, S., Sukhumavasi, J., and Kimura, T. (1998). Cloning and sequencing of the gene encoding chitinase ChiA from Xanthomonas sp. strain AK and some properties of ChiA. J. Ferment. Bioeng. 86, 527–533. doi: 10.1016/S0922-338X(99)80001-4
Sambrook, J., and Russell, D. W. (2001). Molecular Cloning: a Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
Schwyn, B., and Neilands, J. B. (1987). Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160, 47–56. doi: 10.1016/0003-2697(87)90612-9
Segarra, G., Puopolo, G., Giovannini, O., and Pertot, I. (2015a). Stepwise flow diagram for the development of formulations of non spore-forming bacteria against foliar pathogens: the case of Lysobacter capsici AZ78. J. Biotechnol. 216, 56–64. doi: 10.1016/j.jbiotec.2015.10.004
Segarra, G., Puopolo, G., Porcel-Rodríguez, E., Giovannini, O., and Pertot, I. (2015b). Monitoring Lysobacter capsici AZ78 using strain specific qPCR reveals the importance of the formulation for its survival in vineyards. FEMS Microbiol. Lett. 363:fnv243. doi: 10.1093/femsle/fnv243
Seo, Y.-S., Lim, J., Choi, B.-S., Kim, H., Goo, E., Lee, B., et al. (2011). Complete Genome Sequence of Burkholderia gladioli BSR3. J. Bacteriol. 193, 3149. doi: 10.1128/JB.00420-11
Setubal, J. C., dos Santos, P., Goldman, B. S., Ertesvåg, H., Espin, G., Rubio, L. M., et al. (2009). Genome sequence of Azotobacter vinelandii, an obligate aerobe specialized to support diverse anaerobic metabolic processes. J. Bacteriol. 191, 4534–4545. doi: 10.1128/JB.00504-09
Shaw, K. J., Rather, P. N., Hare, R. S., and Miller, G. H. (1993). Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 57, 138–163.
Shimosaka, M., Fukumori, Y., Narita, T., Zhang, X., Kodaira, R., Nogawa, M., et al. (2001). The bacterium Burkholderia gladioli strain CHB101 produces two different kinds of chitinases belonging to families 18 and 19 of the glycosyl hydrolases. J. Biosci. Bioeng. 91, 103–105. doi: 10.1016/S1389-1723(01)80123-7
Skaar, E. P. (2010). The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog. 6:e1000949. doi: 10.1371/journal.ppat.1000949
Simpson, A. J., Reinach, F. C., Arruda, P., Abreu, F. A., Acencio, M., Alvarenga, R., et al. (2000). The genome sequence of the plant pathogen Xylella fastidiosa. Nature 406, 151–159. doi: 10.1038/35018003
Singh, H., Du, J., Won, K.-H., Yang, J.-E., Akter, S., Kim, K.-Y., et al. (2015). Lysobacter novalis sp. nov. isolated from fallow farmland soil, South Korea. Int. J. Syst. Evol. Microbiol. 65, 3131–3136. doi: 10.1099/ijsem.0.000389
Stall, R. E., Loschke, D. C., and Jones, J. B. (1986). Linkage of copper resistance and avirulence loci on a self-transmissible plasmid in Xanthomonas campestris pv. vesicatoria. Phytopathology 76, 240–243. doi: 10.1094/Phyto-76-240
Stoyanov, J. V., Hobman, J. L., and Brown, N. L. (2001). CueR (YbbI) of Escherichia coli is a MerR family regulator controlling expression of the copper exporter CopA. Mol. Microbiol. 39, 502–511. doi: 10.1046/j.1365-2958.2001.02264.x
Straub, D., Rothballer, M., Hartmann, A., and Ludewig, U. (2013). The genome of the endophytic bacterium H. frisingense GSF30(T) identifies diverse strategies in the Herbaspirillum genus to interact with plants. Front. Microbiol. 4:168. doi: 10.3389/fmicb.2013.00168
Studholme, D. J., Ibanez, S. G., MacLean, D., Dangl, J. L., Chang, J. H., and Rathjen, J. P. (2009). A draft genome sequence and functional screen reveals the repertoire of type III secreted proteins of Pseudomonas syringae pathovar tabaci 11528. BMC Genomics 10:395. doi: 10.1186/1471-2164-10-395
Sullivan, R. F., Holtman, M. A., Zylstra, G. J., White, J. F., and Kobayashi, D. Y. (2003). Taxonomic positioning of two biological control agents for plant diseases as Lysobacter enzymogenes based on phylogenetic analysis of 16S rDNA, fatty acid composition and phenotypic characteristics. J. Appl. Microbiol. 94, 1079–1086. doi: 10.1046/j.1365-2672.2003.01932.x
Wang, L., Tang, X., and He, C. (2007). The bifunctional effector AvrXccC of Xanthomonas campestris pv. campestris requires plasma membrane-anchoring for host recognition. Mol. Plant Pathol. l 8, 491–501. doi: 10.1111/j.1364-3703.2007.00409.x
Wang, Y., Qian, G., Li, Y., Wang, Y., Wang, Y., Wright, S., et al. (2013). Biosynthetic mechanism for sunscreens of the biocontrol agent Lysobacter enzymogenes. PLoS ONE 8:e66633. doi: 10.1371/journal.pone.0066633
Watanabe, T., Kanai, R., Kawase, T., Tanabe, T., Mitsutomi, M., Sakuda, S., et al. (1999). Family 19 chitinases of Streptomyces species: characterization and distribution. Microbiology 145, 3353–3363. doi: 10.1099/00221287-145-12-3353
Yamaguchi, A., Nakashima, R., and Sakurai, K. (2015). Structural basis of RND-type multidrug exporters. Front. Microbiol. 6:327. doi: 10.3389/fmicb.2015.00327
Yoneyama, F., Yamamoto, M., Hashimoto, W., and Murata, K. (2011). Azotobacter vinelandii gene clusters for two types of peptidic and catechol siderophores produced in response to molybdenum. J. Appl. Microbiol. 111, 932–938. doi: 10.1111/j.1365-2672.2011.05109.x
Yu, F., Zaleta-Rivera, K., Zhu, X., Huffman, J., Millet, J. C., Harris, S. D., et al. (2007). Structure and biosynthesis of Heat-Stable Antifungal Factor (HSAF), a broad-spectrum antimycotic with a novel mode of action. Antimicrob. Agents Chemother. 51, 64–72. doi: 10.1128/AAC.00931-06
Zhang, W., Li, Y., Qian, G., Wang, Y., Chen, H., Li, Y. Z., et al. (2011). Identification and characterization of the anti-methicillin-resistant Staphylococcus aureus WAP-8294A2 biosynthetic gene cluster from Lysobacter enzymogenes OH11. Antimicrob. Agents Chemother. 55, 5581–5589. doi: 10.1128/AAC.05370-11
Zhang, Z., and Yuen, G. Y. (2000). The role of chitinase production by Stenotrophomonas maltophilia strain C3 in biological control of Bipolaris sorokiniana. Phytopathology 90, 384–389. doi: 10.1094/PHYTO.2000.90.4.384
Keywords: Lysobacter, biological control, lytic enzymes, siderophores, environmental stress
Citation: Puopolo G, Tomada S, Sonego P, Moretto M, Engelen K, Perazzolli M and Pertot I (2016) The Lysobacter capsici AZ78 Genome Has a Gene Pool Enabling it to Interact Successfully with Phytopathogenic Microorganisms and Environmental Factors. Front. Microbiol. 7:96. doi: 10.3389/fmicb.2016.00096
Received: 16 October 2015; Accepted: 18 January 2016;
Published: 05 February 2016.
Edited by:
Stéphane Hacquard, Max Planck Institute for Plant Breeding Research, GermanyReviewed by:
Zakira Naureen, University of Nizwa, OmanYang Bai, Max Planck Institute for Plant Breading Research, China
Copyright © 2016 Puopolo, Tomada, Sonego, Moretto, Engelen, Perazzolli and Pertot. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gerardo Puopolo, Z2VyYXJkby5wdW9wb2xvQGZtYWNoLml0
 Gerardo Puopolo
Gerardo Puopolo Selena Tomada
Selena Tomada Paolo Sonego
Paolo Sonego Marco Moretto
Marco Moretto Kristof Engelen
Kristof Engelen Michele Perazzolli
Michele Perazzolli Ilaria Pertot
Ilaria Pertot