
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 13 January 2016
Sec. Terrestrial Microbiology
Volume 6 - 2015 | https://doi.org/10.3389/fmicb.2015.01537
The microbial community in the rhizosphere environment is critical for the health of land plants and the processing of soil organic matter. The objective of this study was to determine the extent to which rice plants shape the microbial community in rice field soil over the course of a growing season. Rice (Oryza sativa) was cultivated under greenhouse conditions in rice field soil from Vercelli, Italy and the microbial community in the rhizosphere of planted soil microcosms was characterized at four plant growth stages using quantitative PCR and 16S rRNA gene pyrotag analysis and compared to that of unplanted bulk soil. The abundances of 16S rRNA genes in the rice rhizosphere were on average twice that of unplanted bulk soil, indicating a stimulation of microbial growth in the rhizosphere. Soil environment type (i.e., rhizosphere versus bulk soil) had a greater effect on the community structure than did time (e.g., plant growth stage). Numerous phyla were affected by the presence of rice plants, but the strongest effects were observed for Gemmatimonadetes, Proteobacteria, and Verrucomicrobia. With respect to functional groups of microorganisms, potential iron reducers (e.g., Geobacter, Anaeromyxobacter) and fermenters (e.g., Clostridiaceae, Opitutaceae) were notably enriched in the rhizosphere environment. A Herbaspirillum species was always more abundant in the rhizosphere than bulk soil and was enriched in the rhizosphere during the early stage of plant growth.
Plants influence the spatial structure in soil by the growth of their roots (Angers and Caron, 1998). Further, they shape the chemical composition of the rhizosphere and provide microbial growth substrates by rhizodeposition (Lynch and Whipps, 1990; Hirsch et al., 2013; Philippot et al., 2013). Depending on the plant species, between 20 and 50% of plant photosynthate is transported belowground (reviewed by Kuzyakov and Domanski, 2000), and an average of 17% is released to the soil environment (reviewed by Jones et al., 2009). In return for growth substrates, rhizosphere microorganisms benefit plants by providing nutrients, phytohormones, suppressing phytopathogens or increasing resilience to abiotic stress such as heat, high salt, or drought (Welbaum et al., 2004; van der Heijden et al., 2008; Yang et al., 2009; Mendes et al., 2011; Nihorimbere et al., 2011; Haney et al., 2015). The microbial community is also largely responsible for the decomposition of organic matter in soil (Kuzyakov, 2002), which has consequences for biogeochemical cycling, soil formation, soil fertility and atmospheric trace gas formation (Conrad, 1996; Hinsinger et al., 2009).
Various factors shape the microbial communities associated with plants. A detailed study of Arabidopsis thaliana indicated that the endophytic microbial community has relatively low diversity compared with the bulk soil and is similar in plants cultivated in geochemically distinct soils (Lundberg et al., 2012). This indicates that the plant selects specific microorganisms to colonize its tissues. The A. thaliana rhizosphere microbial community also differed from the bulk soil, but soil geochemistry played a larger role in shaping the community. The same was true for maize; the microbial community of the rhizosphere was distinct from the bulk soil, but more similar than the rhizosphere communities of plants grown in different soils (Peiffer et al., 2013). Whereas soil geochemistry appears to be the primary determinant of the rhizosphere community structure, plant species (Marschner et al., 2004), cultivar (Micallef et al., 2009b; Inceoğlu et al., 2010; Edwards et al., 2015) and growth stage (Micallef et al., 2009a; Chaparro et al., 2014; Sugiyama et al., 2014; Sun et al., 2014) have all been shown to have significant additional effects.
Rice differs from most crops in that it is typically cultivated in flooded soil, resulting in oxic and anoxic zones within the rice rhizosphere that select for specific physiological groups of microorganisms with either aerobic, anaerobic, or facultivative metabolism (Brune et al., 2000). Methanogenesis in the rhizosphere and bulk soil of rice fields results in high methane (CH4) production, with rice agriculture currently contributing ∼10% of the global CH4 budget (Conrad, 2009). The primary substrates for methanogens are acetate or H2 + CO2 produced from the breakdown of complex carbon by the microbial community, including fermenters and acetogens (McInerney et al., 2008). Approximately 60% of CH4 produced in rice fields originates from root exudates or decaying root material (Watanabe et al., 1999).
The microbial communities inhabiting the rice field ecosystem have been described previously. For instance, the microbes within the rice root interior, the rhizoplane and the rhizosphere have been analyzed (Edwards et al., 2015). In addition, the microbial communities in various zones, such as rhizosphere, anoxic bulk soil, and oxic surface soil have been reported (Großkopf et al., 1998; Lüdemann et al., 2000; Lu et al., 2004; Asakawa and Kimura, 2008; Breidenbach and Conrad, 2015; Lee et al., 2015). Further studies have investigated the rice phyllosphere microbial community by 16S rRNA pyrotag sequencing (Ren et al., 2014) as well as endophytic and rhizospheric communities with metagenomic and metaproteomic approaches (Knief et al., 2012). Experiments have identified the bacteria (Lu et al., 2006; Hernández et al., 2015) and archaea (Lu and Conrad, 2005; Zhu et al., 2014) that consume plant-derived carbon in the rhizosphere. Also, specific functional groups of microorganisms, such as methanogens (Ramakrishnan et al., 2001; Lee et al., 2014) and methanotrophs (Henckel et al., 1999; Eller and Frenzel, 2001; Ho et al., 2011; Lee et al., 2014), have been extensively analyzed in rice systems. However, none of these studies focused on the impact of the rice plant on the total microbial community and over several growth stages of rice.
The objective of this study was to determine the extent to which rice plants influence the microbial community in a rice field soil. Secondly, we aimed to determine if plant growth stage had an effect on the microbial community composition in the rhizosphere. The experiment was performed under greenhouse conditions to minimize confounding factors, and using a soil from Vercelli (Italy) with a long history of rice cultivation to avoid changes that could result in a soil that was not adapted to flooding or the growth of rice plants. Similarly, it was necessary to use the rice variety cultivated in the Vercelli rice fields, otherwise there could have been changes to the microbial community as a result of adaptation to an unfamiliar plant genotype (Edwards et al., 2015). The first sampling was past the initial dynamic phase after flooding in order to focus on the influence of the plant and not the effects of flooding. The analysis provides information on the difference between the rice rhizosphere and bulk soil microbial community across different plant growth stages showing how the rice plant shapes the microbial community composition in a mature rice field soil.
Soil was sampled from rice fields at the Italian Rice Research Institute in Vercelli, air-dried and stored at room temperature until the start of the experiment. A detailed analysis of the physiochemical properties of the soil has been published previously (Pump and Conrad, 2014). Immediately prior to the establishment of microcosms, soil was sieved through a stainless steel screen (0.2 mm mesh) and 2.5 kg was added to opaque plastic pots (16 cm height, 17.5 cm diameter). The pots were flooded with deionized water 1 week before planting. Fertilizers included urea (CH4N2O, 45 g l-1) as nitrogen source, phosphorus (Na2HPO4⋅2H2O, 17 g l-1), potassium (KCl, 50 g l-1), and magnesium (MgSO4⋅7H2O, 2 g l-1). The phosphorus, potassium, and magnesium solutions were added at a ratio of 10 ml kg-1 soil 1 day before planting, whereas 5 ml kg-1 of the urea was added twice, 1 day before planting and after 14 days of plant growth. Rice seeds (Oryza sativa var. Koral) were also obtained from the Rice Research Institute in Vercelli, Italy. The rice seeds were treated with the fungicide Aatiram and germinated at 25°C and 75% humidity in a greenhouse. Three germinated rice seedlings were planted each in a total of 20 pots. A further five pots were left unplanted. The pots were incubated in a greenhouse at 25°C and 75% humidity with a 12 h light/dark cycle. Pots were watered daily to maintain approximately 3 cm water overlying the soil. Plant heights and tiller number were recorded weekly. Five planted pots were sacrificed after 34, 52, 62, and 90 days after planting (stages 1–4) and rhizosphere soil (planted pots), bulk soil (unplanted pots) and pore water were collected aseptically using sterilized equipment. Plants were extracted from the pots and shaken to remove large soil aggregates and adhering soil. The soil remaining attached on the roots was considered to be rhizosphere soil and was sampled using a sterile spatula. Samples were immediately frozen in liquid nitrogen and stored at -80°C until further analysis. The water content of each soil was determined gravimetrically by drying subsamples at 60°C until they reached a constant weight.
Soil samples (100 g) were centrifuged for 10 min at 20,000 g and 4°C in 50-ml centrifuge tubes. The supernatant was filter sterilized using 0.2 μm acetate-free filters (GE Healthcare Life Science, Freiburg, Germany) and stored at -20°C until analysis. Organic acids were analyzed with high performance liquid chromatography [HPLC; pump S1000, oven S4110 (Sykam, Germany), sampler Jasco 851-AS Intelligent Sampler (JapanSpectroscopy Co. Ltd., Japan) with an Aminex HPX-87 H organic acid column (Bio-Rad, Germany)]. Technical specifications were as follows: 65°C oven temperature, 2 mM H2SO4 as eluent, 0.5 ml/min flow rate and a UV detector at 40°C. Inorganic ions including chloride, nitrate, nitrite, phosphate, and sulfate were detected using ion chromatography [IC; pump S1121, sampler S5200 (Sykam); Bak et al., 1991]. Concentrations of organic acids and inorganic ions between the treatments were compared using analysis of variance (ANOVA) followed by Tukey’s post hoc test in R version 3.02 (R Core Team, 2013).
Soil DNA was extracted using the NucleoSpin® Soil Kit (Macherey-Nagel, Düren, Germany) following the manufacturer’s instructions. DNA concentration and purity were determined spectrophotometrically (NanoDrop Technologies, USA). All DNA samples had absorbance ratios of A260:A230 > 1.7 and A260:A280 > 1.8.
Quantitative PCR (qPCR) was used to quantify bacterial and archaeal 16S rRNA gene copies using primers Ba519f/Ba907r (Stubner, 2002) and Ar364f/Ar934br (Burggraf et al., 1997; Großkopf et al., 1998) respectively. All reactions were set-up on ice with minimal light exposure. Each reaction included the following: 4 mM MgCl2, 0.01 μM fluorescein calibration dye (Bio-Rad), 0.625 U Jump StartTM TaqReadyMixTM, 0.60 μM of each primer, 1 μL template DNA. Standards containing known numbers of DNA copies of the target gene were serially diluted to serve as calibration curves. The qPCRs were performed on an iCycler thermocycler equipped with a MyiQ detection system (Bio-Rad, Munich, Germany). The following program was used for amplification of archaeal 16S rRNA gene copies: 94°C for 6 min, followed by 40 cycles of 94°C for 35 s, 66°C for 30 s and 72°C for 45 s. Fluorescence was measured after each cycle at a temperature of 86.5°C. A melting curve was performed from 75 to 95°C. The program for amplification of bacterial 16S rRNA gene copies was as follows: 94°C for 8 min, followed by 50 cycles of 94°C for 20 s, 50°C for 20 s, and 72°C for 50 s. A melting curve was performed from 75 to 95°C. Data analysis was performed using Bio-Rad IQ5 2.0 Standard Edition Optical System Software (Bio-Rad). A Kolmogoroff–Smirnoff test indicated that the qPCR data was normally distributed. Means of copy numbers between the treatments were compared using ANOVA followed by Tukey’s post hoc test in R version 3.0.2 (R Core Team, 2013).
A total of 24 samples were chosen for amplicon pyrosequencing. Samples corresponded to three replicate microcosms chosen at random from each of the two treatments (rhizosphere, bulk soil) and the four sampling time points. The 16S rRNA genes of bacteria and archaea were targeted with primers F515 and R806 described previously (Bates et al., 2011). The forward primer was tagged with 6-base barcodes. Sequencing of the PCR products was performed at the Max Planck Genome Centre in Cologne using a Roche 454 Genome Sequencer GS FLX+. Data are available in the NCBI sequence read archive (SRA) with the accession number SRP057189.
Sequences were quality filtered and grouped into OTUs (97% identity) with the UPARSE pipeline (Edgar, 2013). A maximum expected error threshold of 0.5 was used and sequences were trimmed to 200 bp. Sequences were excluded if they contained mismatches to the forward primer, were shorter than 200 bp or contained ambiguities. Chimeras were removed using the UCHIME de novo algorithm. Representative sequences of OTUs were classified using the silva taxonomy and the Wang (naïve Bayesian classifier) method implemented in mothur version 1.31.2 (Schloss et al., 2009). Relative abundances of OTUs between samples were analyzed using the vegan package version 2.2-1 (Oksanen et al., 2013) in R version 3.0.2 (R Core Team, 2013).
Statistical analyses were performed using R version 3.0.2 (R Core Team, 2013). ANOVA and principal components analysis (PCA) were performed using package stats (R Core Team, 2013). Data was tested for normality before performing ANOVA. Linear regression models were calculated using the function lm within stats. Constrained correspondence analysis (CCA) was performed using the function cca in the vegan package (Oksanen et al., 2013). PCA was performed using the prcomp function of Hellinger distances (i.e., Euclidean distances of Hellinger transformed data) and the 50 OTUs contributing the largest absolute loadings in the first dimension were obtained from the rotation output file. A heatmap was prepared using the gplots package (Warnes et al., 2015); the samples were clustered with the hclust function using the ‘Ward’ method based on various distances/dissimilarities (Euclidean, Manhattan, Bray–Curtis) calculated with the vegdist function in the vegan package. Diversity analyses (Shannon index, coverage) were performed using the diversity calculators within the vegan package; the dataset was first randomly subsampled to 3699 sequences per sample using the rrarefy function in vegan. Differences in population structure were tested by ANOSIM (Clarke, 1993) based on Bray–Curtis dissimilarities within the vegan package. Differences between sites (rhizosphere versus bulk soil) or differences based on sampling time were examined.
The determination of the growth stage was conducted by monitoring rice plant height and tiller number (Supplementary Figure S1). Four sampling time points were selected corresponding to stage 1 (day 34, early vegetative), stage 2 (day 52, late vegetative), stage 3 (day 62, reproductive), and stage 4 (day 90, maturity). Soil pore water analyses indicated that lactate, formate, acetate, chloride, and propionate concentrations were higher in the planted pots, but these differences were only statistically significant for chloride and propionate (Table 1). The concentrations of malate, nitrate, and sulfate were similar between rhizosphere (planted pots) and bulk soil (unplanted pots).
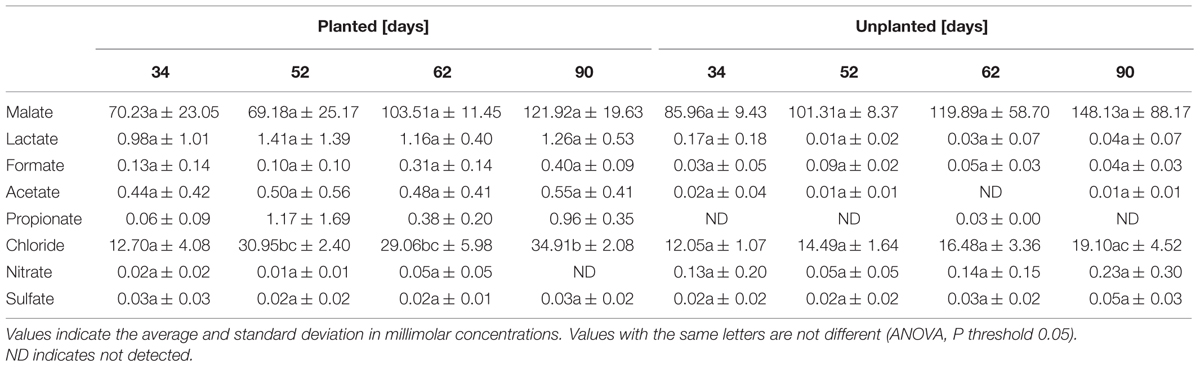
TABLE 1. Concentrations of organic acids and inorganic ions detected in the pore water of rhizosphere (planted) and bulk soil (unplanted) at stages 1–4, corresponding to days 34, 52, 62, and 90.
The influence of the rice plant on the microbial community size was determined by qPCR assays targeting the bacterial and archaeal 16S rRNA gene. Both bacterial and archaeal 16S rRNA abundances were about twofold higher in rhizosphere (planted pots) than in soil from bulk soil (unplanted pots; Figure 1). Temporal changes were not detected in the bulk soil. The abundance of bacterial and archaeal 16S rRNA gene copies decreased from stages 1 to 4, but this was not statistically significant (ANOVA, P > 0.05). The 16S rRNA gene copy numbers of bacteria were approximately 20 times larger than those of archaea.
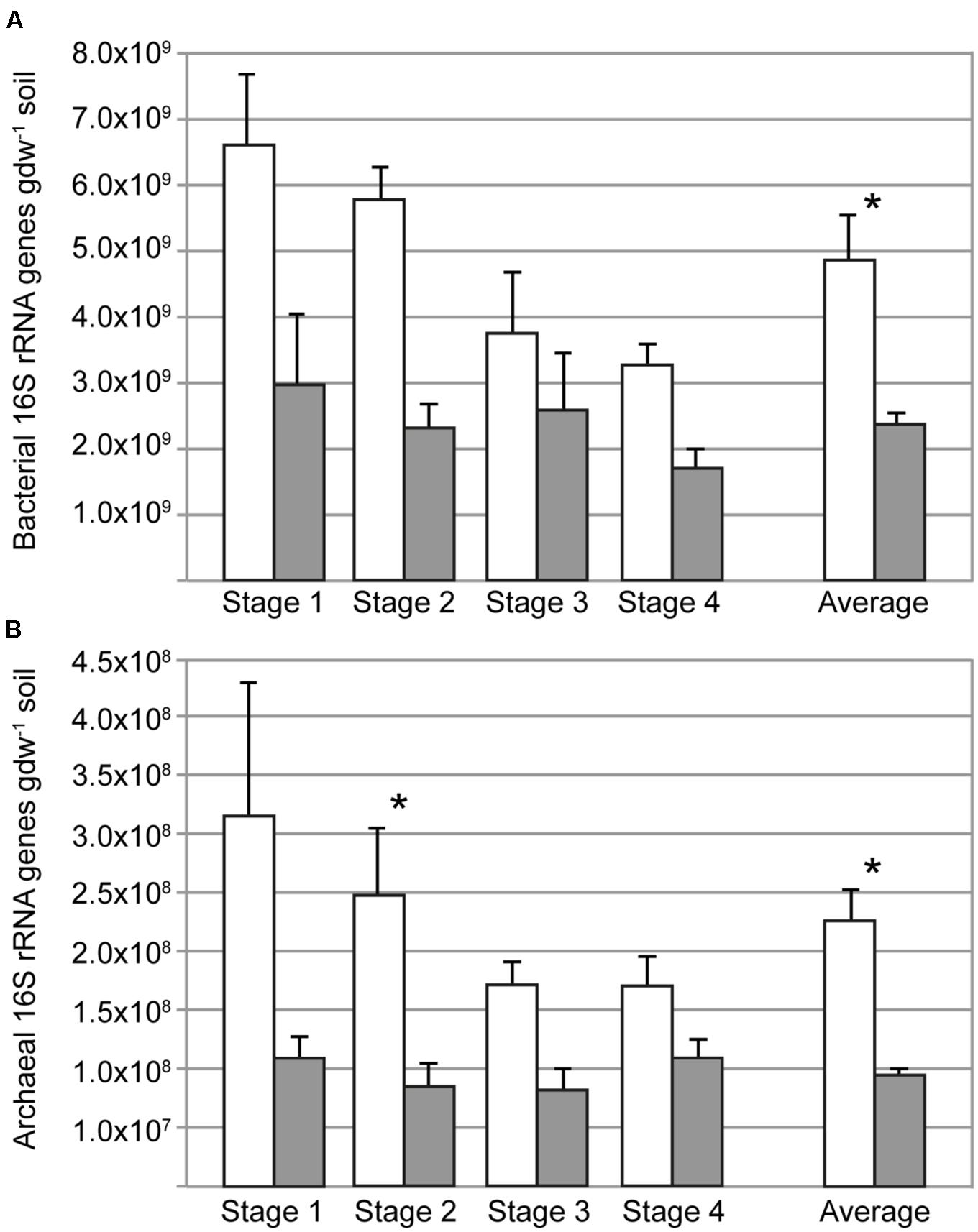
FIGURE 1. Copy numbers of bacterial (A) and archaeal (B) 16S rRNA genes in rhizosphere (open bars) and bulk soil (filled bars) at different sampling times (days 34, 52, 62, 90); the average of the four sampling times is also shown. Asterisks indicate when the abundance in rhizosphere and bulk soil was different (ANOVA, P < 0.05). Error bars correspond to standard errors of means from replicate soil samples (n = 5).
Pyrosequencing of the bacterial 16S rRNA gene was performed to characterize the microbial communities. After processing and chimera removal, an average of 7300 sequences (±2500) were obtained for each sample. The sequences grouped into 8685 OTUs at 97% identity. Coverage was between 91 and 95% for each of the eight sample types (bulk soil and rhizosphere at four sampling times). Diversity indices were neither different between rhizosphere and bulk soils nor between sampling times.
The microbial communities separated by rhizosphere and bulk soil based on PCA of OTUs (97% 16S rRNA identity), indicating that there was a difference in the community composition (ANOSIM, R = 0.39, P = 0.001; Figure 2). Despite this clear separation, the first axis only explained 8.6% of the variance in the data, indicating that the relative abundance of most OTUs were similar between rhizosphere and bulk soils. There was no apparent clustering of rhizosphere samples based on plant growth stage or the corresponding sampling time in unplanted (bulk) soil microcosms. Sequences were classified and summarized by bacterial phylum, which did not show differences (ANOVA, P > 0.05) between rhizosphere and bulk soil samples (Supplementary Figure S2), indicating the overall composition of the community at low phylogenetic resolution did not change between these environments. CCA was performed to test the extent to which pore water data (Table 1) explained the community composition (Supplementary Figure S3). Only chloride was significant.
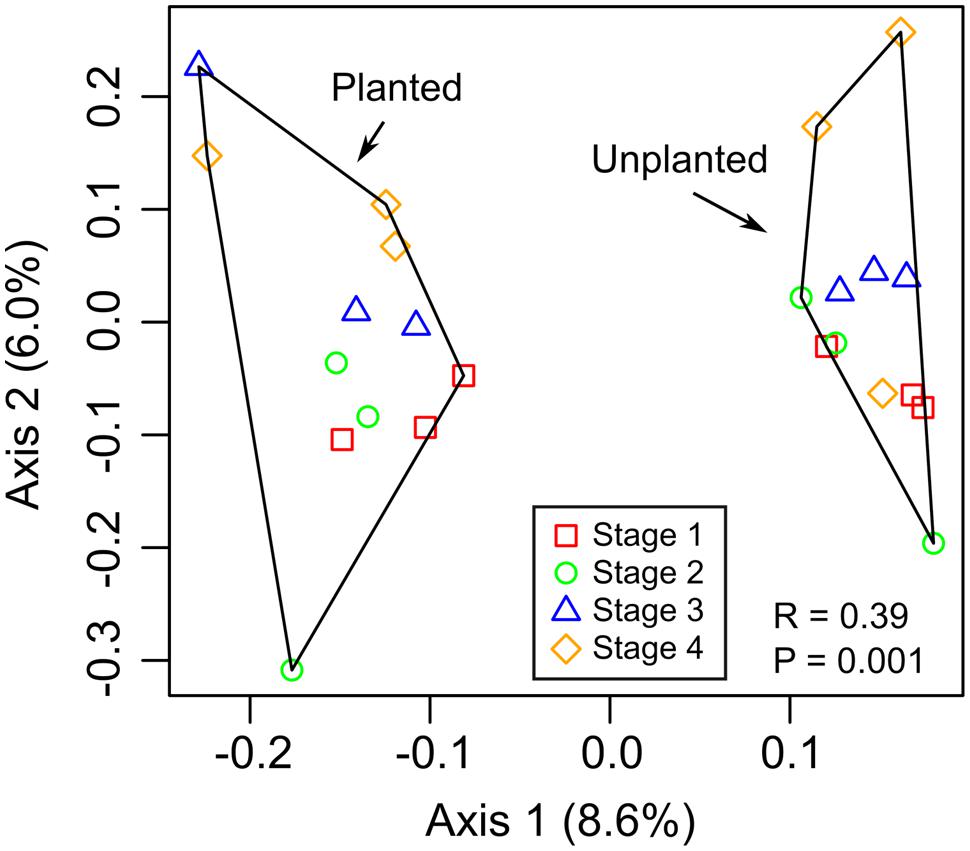
FIGURE 2. Principal component analysis (PCA) based on relative abundance of 16S rRNA gene OTUs. The ANOSIM statistic was applied to the ordinations to test for differences between rhizosphere (planted) and bulk (unplanted) soil communities, with corresponding R- and P-values shown.
A heatmap was constructed to depict the relative abundance of OTUs that best represent the dissimilarity between the microbial communities in rhizosphere versus bulk soil by selecting 50 with the largest loadings of the PCA. Among these 50 OTUs, 34 could be classified to family level or lower and are depicted in a heatmap (Figure 3); data for all 50 OTUs are provided in the supplement. The rhizosphere and bulk soil samples clustered independently with each of the distance/dissimilarity indices tested. ANOVA was used to test if their relative abundance between rhizosphere and bulk soil was significantly different. Most of the OTUs were significant, with some exceptions including several Cyanobacteria that were abundant in one rhizosphere sample in stage 2 (Figure 3). The rhizosphere soil contained a higher abundance of rice mitochondria and plastid 16S rRNA sequences. This was anticipated since root cells are sloughed-off into the rhizosphere, and therefore provides a confirmation of the ability of the analysis to detect differences between the rhizosphere and bulk soil communities. The Shannon diversity index for these 50 OTUs in rhizosphere was 3.08 ± 0.14 and 2.55 ± 0.25 for the bulk soil. Among these 50 OTUs, 42 had higher average relative abundance in rhizosphere than bulk soil and eight had higher abundance in bulk than rhizosphere soil (Supplementary data); given that the number of 16S rRNA genes was twofold larger in the rhizosphere than bulk soil, it is possible that even more than 42 of the OTUs were higher in rhizosphere soil based on absolute abundance.
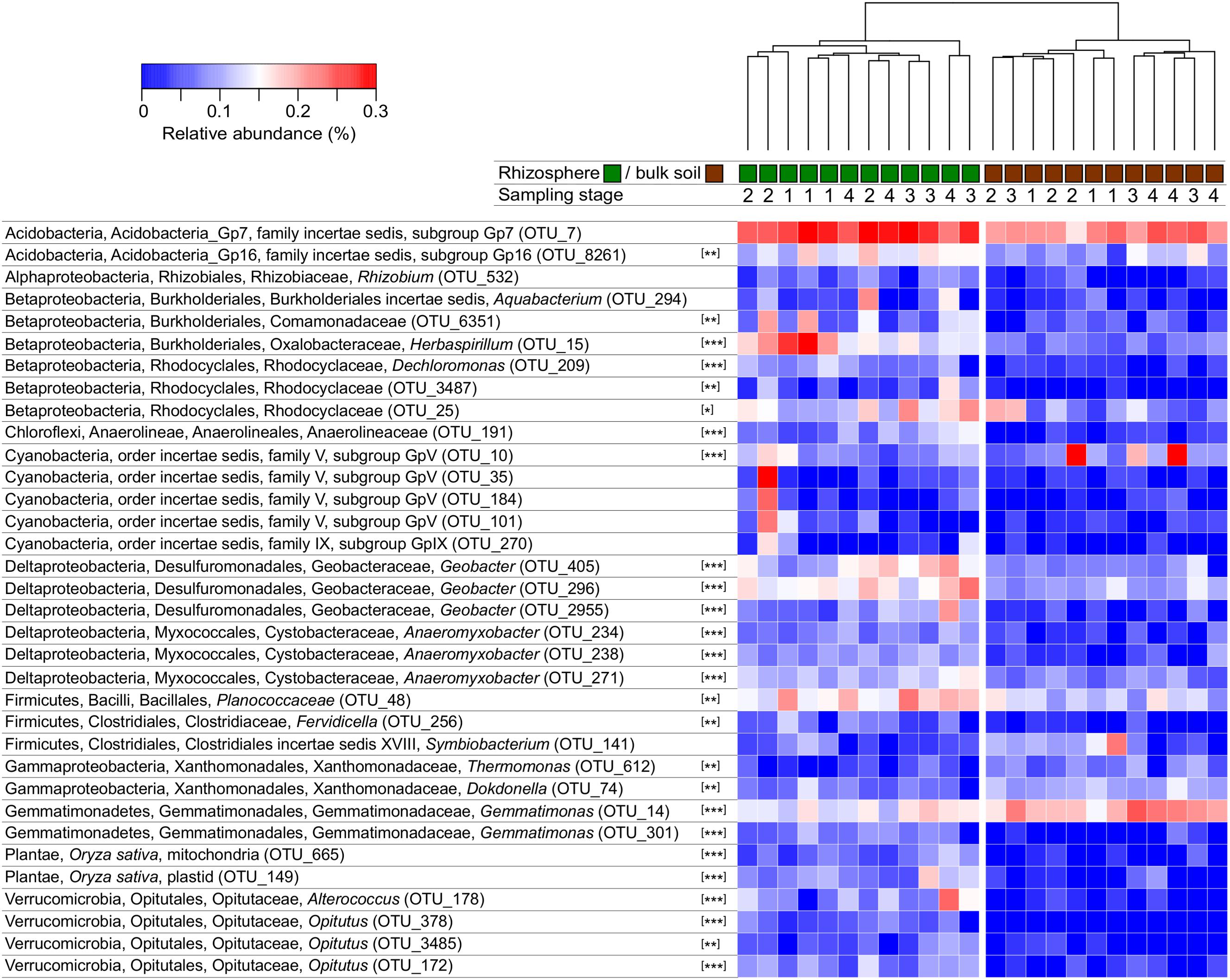
FIGURE 3. Heatmap depicting the relative abundance of OTUs with greatest dissimilarity between rhizosphere and bulk soils. Clustering of samples is shown based on Manhattan distances and was similar using Bray–Curtis dissimilarity. OTUs were classified and only those that could be classified to order or lower were included. The sampling times (1–4) correspond to 34, 52, 62, and 90 days after planting. Asterisks indicate when the abundance of an OTU was different in rhizosphere and bulk soil (ANOVA, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001).
Among the 42 OTUs that were enriched in rhizosphere soil, we aimed to determine if some were selected at a specific plant growth stage. The five OTUs with higher relative abundance in one stage compared with any other stage (ANOVA, P < 0.05) are depicted in a PCA analysis (Figure 4). OTU_15, corresponding to an uncultivated Herbaspirillum species, was the only OTU enriched in stage 1. There was a difference in relative abundance for OTU_15 between stages 1 and 4 (Tukey’s post hoc test). The other four OTUs were more abundant in stages 3 or 4 compared with stage 1. These corresponded to two Geobacter species (OTU_405, OTU_2955), one Anaerolineaceae (OTU_191) and one Alterococcus (OTU_178).
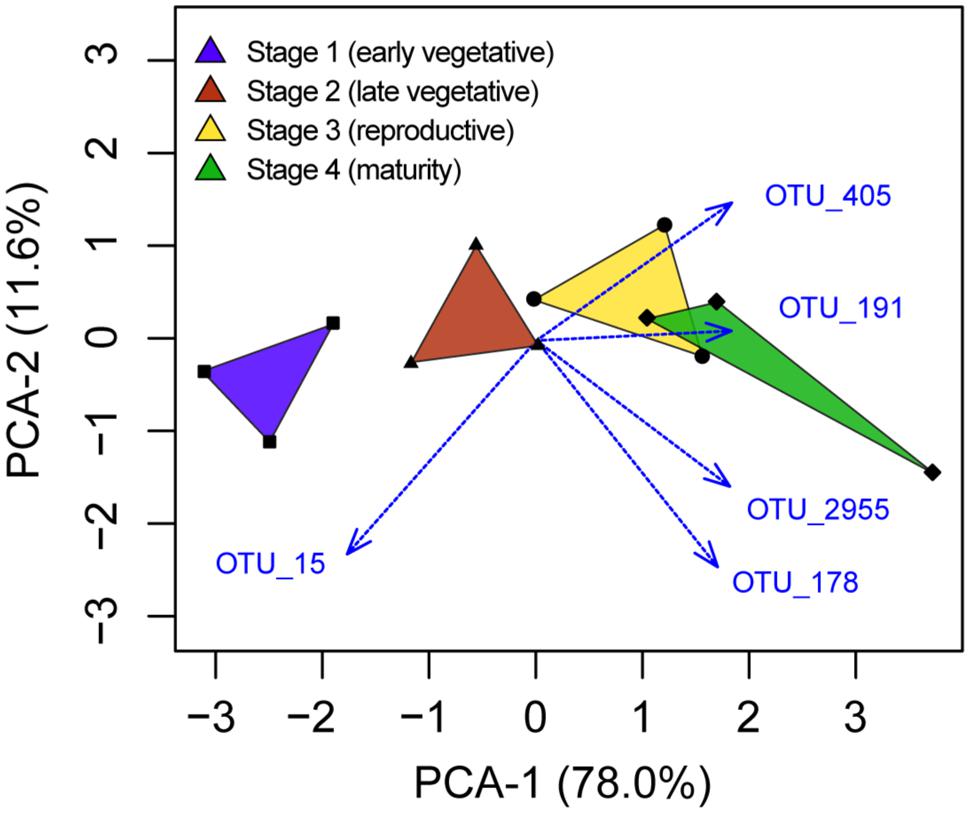
FIGURE 4. Principal component analysis depiction of the five OTUs identified to have different relative abundances between plant growth stages.
Although the PCA/heatmap provided hints as to which OTUs were differentially abundant between rhizosphere and bulk soil microcosms, we wanted to determine if this pattern could be seen for individual phyla alone. To determine this, OTUs were classified and separated into phylum-level groups, which were then analyzed individually by principal coordinates analysis (PCoA; Figure 5). When testing for differences between rhizosphere and bulk soil communities, the ANOSIM statistic was largest (R ≥ 0.5) for Gemmatimonadetes, Proteobacteria and Verrucomicrobia, indicating clear differences in community structure for these taxa. No difference was detected (R = 0) for the candidate phylum BRC1, or differences were not significant (P > 0.05) for Planctomycetes and Archaea. Differences in population structures based on growth stages were only significant for Firmicutes and Archaea, but in both cases there was overlap between the stages (Figure 5). Separate analyses of Bacillales and Clostridiales within the Firmicutes showed a difference in communities between bulk and rhizosphere soils for both classes, but the temporal pattern was only significant for Clostridiales (R = 0.18, P = 0.001). For Archaea, one cause of the temporal pattern was a higher relative abudance of Methanosarcina and Methanosaeta in stage 4 compared with the earlier sampling times, although this difference was not statistically significant. Linear models were used to test for correlations between Methanosarcina and Methanosaeta relative abundances with pore water data (Table 1). Among these, the only significant relationship was a positive correlation between Methanosarcina and formate concentration.
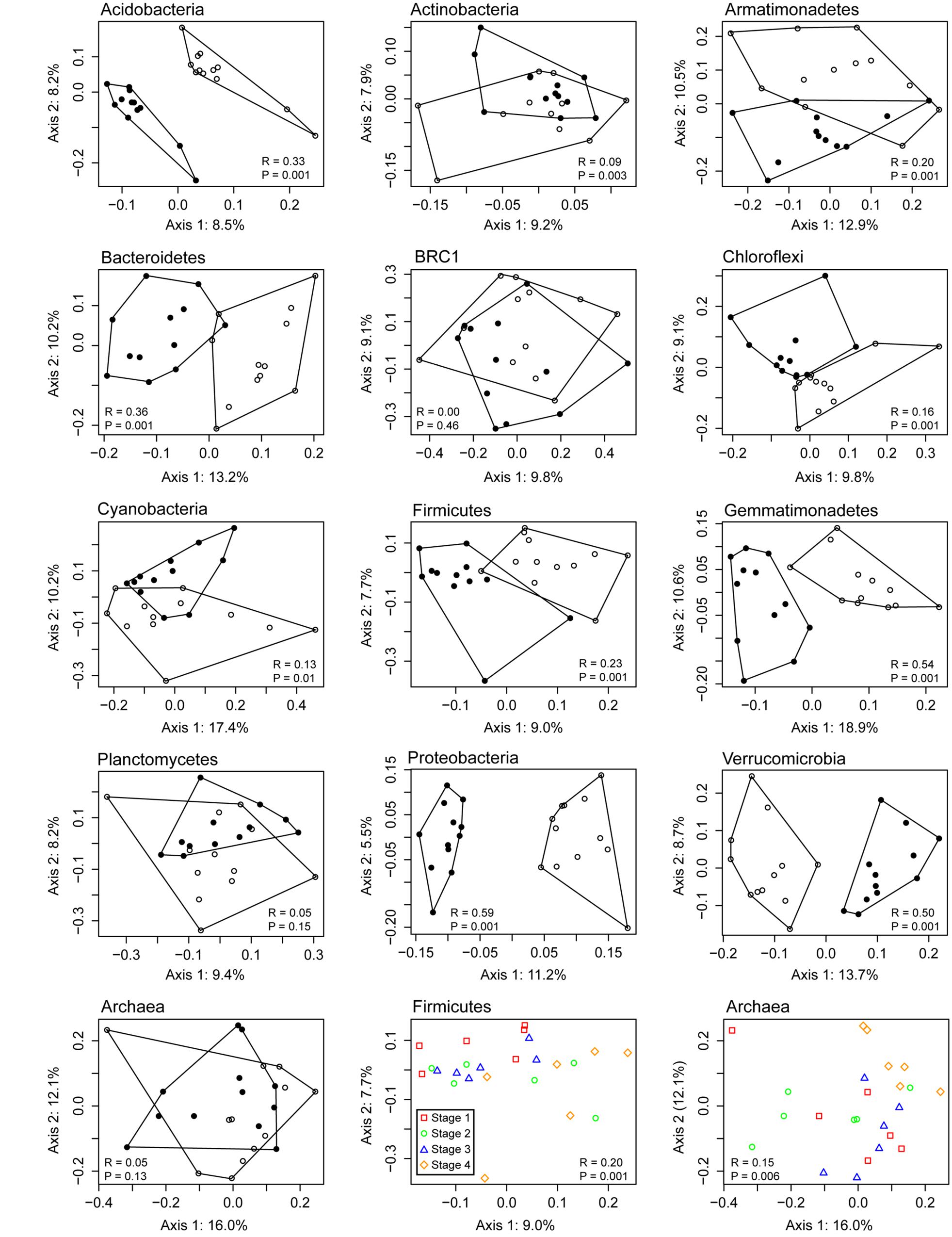
FIGURE 5. Principal coordinates analysis (PCoA) of 16S rRNA gene OTUs assigned to major microbial phyla detected in Vercelli rice field soil. Bulk soil samples are depicted with filled symbols and rhizosphere samples with open symbols. The ANOSIM statistic was applied to the ordinations to test for differences between rhizosphere and bulk soil communities, with corresponding R- and P-values shown; the last two plots show the R- and P-values based on differences between sampling times for Firmicutes and Archaea respectively.
The rhizosphere is known as a compartment in the soil influenced by the plant and where organic matter is introduced via rhizodeposition and sloughed-off cells. Rhizodeposition by rice plants has been described (Aulakh et al., 2001; Wu et al., 2009) and shown to enhance microbial activity in the rhizosphere compared with bulk soil (Butler et al., 2003). Oxygen is also released from rice roots (Frenzel et al., 1992; Colmer and Pedersen, 2008) and serves as an electron acceptor for aerobic microorganisms and is likely to influence the microbial populations and the chemical transformations that occur. The volume and composition of organic molecules released from roots changes somewhat with rice plant growth stage (Aulakh et al., 2001), which in theory could also cause temporal shifts in the rhizosphere microbial community. Although not statistically significant, our data (Table 1) supports other studies that show the combined influence of the rice plant and the microorganisms in the rhizosphere act to modify the chemical composition of the soil environment. Most importantly, in this study we show the extent to which these changes occur in the rice rhizosphere and the degree to which growth stage influences the microbial community.
There were relatively few temporal changes in the microbial community either in bulk or rhizosphere soil. This is consistent with a previous study (Noll et al., 2005) of Vercelli rice field soil where changes in the bacterial community were observed only during the first 21 days after flooding. This initial phase is dominated by a succession of r-strategists that presumably consume readily available growth substrates, energy sources and high-redox potential electron acceptors (Noll et al., 2005; Shrestha et al., 2007). Interestingly in that study, a difference between the surface and subsurface soil was also not observed beyond day 21 (Noll et al., 2005). Another study also examined changes in a time-series within the first 2 weeks after planting (Edwards et al., 2015). Our first sampling time was 34 days after planting, equivalent to 41 days after flooding, and therefore well-beyond this initial dynamic phase. This was important for our experimental design as we could focus on the influence of the plant and not the effects of flooding.
Previous reports have shown that the difference in microbial communities between the bulk and rhizosphere soil of land plants is less than the difference between soils with different locations or geochemistry (Lundberg et al., 2012; Peiffer et al., 2013; Edwards et al., 2015). A study of rice field showed that the microbial communities clustered by soil depths, which affects oxygenation and redox potential, rather than by bulk and rhizosphere soil (Lee et al., 2015). We sampled only the soil adhered to the rice root or unplanted bulk soil below the oxygen diffusion zone, therefore, making the comparison of rhizosphere and bulk soil communities more straightforward. Although there was a doubling of the bacterial community in the rhizosphere as determined by qPCR targeting bacteria 16S rRNA genes, there was no significant difference in the relative abundance at the phylum level. This indicates that the changes in relative abundance was not restricted to a small number of taxa within a limited number of phyla, otherwise the relative abundance of the stimulated phyla would have increased and unresponsive phyla would have decreased relative to the bulk soil. This was confirmed by the separate analysis of phylum-level microbial populations, which showed that with the exception of candidate division BRC1, Planctomycetes and Archaea, all the community structures of individual phyla were distinct between rhizosphere and bulk soil. Therefore, the rice plant restructured nearly all the phylum-level microbial groups, which can explain the doubling of bacterial abundance in the rhizosphere soil.
The absolute abundance of Archaea, like Bacteria, was on average more than twofold greater in rhizosphere than bulk soil, but the archaeal community composition based on relative abundance of OTUs was not significantly shaped by these locations. As discussed above, an increase in the absolute abundance of a group without a change in the relative abundance of the individual OTUs would indicate an equal stimulation of all taxa, which appeared to be the case of Archaea in our experimental system. Studies have shown that archaeal communities are relatively stable once established and are resistant to environmental perturbation, such as drainage (Krüger et al., 2005; Watanabe et al., 2006; Scavino Fernandez et al., 2013; Breidenbach and Conrad, 2015), or oxygenation in the case of strict anaerobes (Yuan et al., 2009). Methanogens such as Methanosarcinaceae and Methanocellaceae possess various genes encoding for oxygen detoxifying enzymes (Erkel et al., 2006; Angel et al., 2011) thus probably allowing them to survive exposure to oxygen in the rhizosphere. Another study has proposed Methanosaetaceae to tolerate oxygen better than previously believed (Brandt et al., 2015). The lower abundance of Archaea in the bulk soil compared with rhizosphere might be a reflection of energy or carbon limitation in the absence of rice plants. Our current hypothesis is that the relative abundance of Archaea in Vercelli rice field soil is shaped by their growth in the rhizosphere, and that archaeal OTUs respond proportionally to carbon or energy limitation. Whereas the community is relatively stable, their activity is likely to fluctuate. For example, studies have shown that mcrA gene expression in methanogens varies during rice plant development (Lee et al., 2014), after exposure to oxygen (Yuan et al., 2009) or at different times of the day in the rice rhizosphere (Xu et al., 2012), but under most of these conditions there was no change in methanogen numbers.
We also identified specific OTUs that had different relative abundance between rhizosphere and bulk soil. Three OTUs corresponding to each of Geobacter and Anaeromyxobacter were significantly more abundant in the rhizosphere. Both these genera have been shown to be important iron reducers in Vercelli soil (Hori et al., 2010). Hori et al. (2010) specifically showed that they use acetate as a carbon source and oxidized iron, in the form of either ferrihydrite or goethite, as an electron acceptor. They also identified uncultivated Rhodocyclaceae that grew in the presence of goethite, which might also explain the higher abundance of Rhodocyclaceae in rhizosphere soil in our experiment. Iron reduction can be sustained in the rice rhizosphere by the exudation of acetate by roots and the chemical or biological oxidation of reduced iron (Conrad and Frenzel, 2002). The rhizosphere also contained higher relative abundances of Opitutus and Clostridia OTUs. These are well known as anaerobes with fermentative metabolism (Andreesen and Schaupp, 1973; Chin et al., 2001) and OTUs belonging to both these genera were identified consumers of plant-derived carbon in the rice rhizosphere of Vercelli soil (Hernández et al., 2015).
Compared with the difference between bulk and rhizosphere soil, there was only a relatively minor effect of plant growth stage on the microbial community composition. The ANOSIM analysis showed significant temporal patterns for Firmicutes and Archaea only, and both demonstrated high overlap between the sampling stages. This temporal pattern of Firmicutes was restricted to Clostridiales, which are known fermenters of plant material in flooded soil (Rui et al., 2009). Previous studies in rice field soil has shown them to be among the first to proliferate after flooding (Noll et al., 2005; Rui et al., 2009). The difference in the composition of Firmicutes between bulk and rhizosphere soil was still slightly stronger than the temporal differences. In contrast, we observed a difference in archaeal community composition only with time and not between bulk and rhizosphere soils. In other words, although the rice plants caused a doubling of the Archaea in the rhizosphere compared with the bulk soil, their community composition was influenced more by time after flooding than by the rice plant. Previous studies of methanogens, which are the dominant Archaea in Vercelli soil, have shown their activity to change with time after flooding (Yao and Conrad, 1999; Yao et al., 1999). This temporal effect is controlled or influenced by a variety of factors, including the presence of high potential inorganic electron acceptors and the availability of degradable organic substrates (Conrad and Frenzel, 2002).
We were interested in identifying OTUs enriched in the rhizosphere at one of the plant growth stages. OTU_15, corresponding to a Herbaspirillum species, was the only OTU identified as enriched in stage 1. Many Herbaspirillum have been identified as endophytic plant growth promoting bacteria (Elbeltagy et al., 2001; Rothballer et al., 2008). Baldani et al. (2000) identified Herbaspirillum seropedicae strains that could provide up to 54% of rice seedling nitrogen. Another study showed that phosphate-solubilizing Herbaspirillum strains could provide further rice plant growth enhancement under certain conditions (Estrada et al., 2013) and may have other plant growth promoting mechanisms. More research is needed to determine how beneficial strains of Herbaspirillum could be enriched within the rhizosphere and endosphere early during plant development and how this might be used to decrease fertilization requirements.
Finally, there were four OTUs identified as being significantly enriched in the late growth stage 4. Two of the OTUs corresponded to Geobacter. As discussed above, Geobacter are known iron reducers that consume plant carbon in the rice rhizosphere (Hori et al., 2010; Hernández et al., 2015). Their activity is of particular importance since it consumes organic substrates that could otherwise be converted to CH4, and studies have shown that the addition of oxidized iron decreases methanogenesis in rice field soil (Achtnich et al., 1995; Chidthaisong and Conrad, 2000; Jäckel et al., 2005). CH4 released to the atmosphere from rice fields is a global concern (Bodelier, 2015), and understanding how the abundance and activity of Geobacter can be enhanced in the rice rhizosphere might lead to strategies for decreasing methanogenesis.
This study showed that rice plants have a extensive effect on the microbial community composition, which was detectable across all bacterial phyla except BRC-1 and Planctomycetes. The majority of the OTUs with a significant response had higher relative abundance in the rhizosphere compared with bulk soil. Although the differences in relative anbundance of OTUs between bulk and rhizosphere were moderate (maximum ∼5-fold), it shows how the rice plant shapes and maintains the microbial community composition in rice field soil. The archaeal community composition did not change between bulk soil and rhizosphere despite an increased abundance in the latter, suggesting that their community composition is relatively stable in Vercelli rice field soil. Future studies should focus on understanding how specific microbial species, such as Herbaspirillum and Geobacter, are enriched in the rhizosphere at different growth stages and whether this involves plant signaling mechanisms (Hassan and Mathesius, 2012). Strategies to enhance the activity of these microorganisms might decrease fertilization requirements or lead to a mitigation in greenhouse gas formation, both of which are priorities for rice agriculture.
BB designed and performed experiments, analyzed the data, and wrote the manuscript. JP provided reagents and designed and performed experiments. MD designed the experiments, analyzed the data, and wrote the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Elisabetta Lupotto (CRA-RIS, Vercelli, Italy) for providing rice seeds and soil. BB was financially supported the German Research Foundation (DFG) for funding (FOR 1701, “Introducing Non-Flooded Crops in Rice-Dominated Landscapes: Impacts on Carbon, Nitrogen and Water Cycles [ICON].” JP was financially supported by the Research Center for Synthetic Microbiology (‘Synmikro’) of the Landes-Offensive zur Entwicklungwissenschaftlich-ökonomischer Exzellenz (LOEWE).
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.01537
Achtnich, C., Bak, F., and Conrad, R. (1995). Competition for electron donors among nitrate reducers, ferric iron reducers, sulfate reducers, and methanogens in anoxic paddy soil. Biol. Fertil. Soils 19, 65–72. doi: 10.1007/BF00336349
Andreesen, J. R., and Schaupp, A. (1973). Fermentation of glucose, fructose, and xylose by Clostridium thermoaceticum: effect of metals on growth yield, enzymes, and the synthesis of acetate from CO2. J. Bacteriol. 114, 743–751.
Angel, R., Matthies, D., and Conrad, R. (2011). Activation of methanogenesis in arid biological soil crusts despite the presence of oxygen. PLoS ONE 6:e20453. doi: 10.1371/journal.pone.0020453
Angers, D., and Caron, J. (1998). Plant induced changes in soil structure: processes and feedbacks. Biogeochemistry 42, 55–72. doi: 10.1023/A:1005944025343
Asakawa, S., and Kimura, M. (2008). Comparison of bacterial community structures at main habitats in paddy field ecosystem based on DGGE analysis. Soil Biol. Biochem. 40, 1322–1329. doi: 10.1016/j.soilbio.2007.09.024
Aulakh, M. S., Wassmann, R., Bueno, C., Kreuzwieser, J., and Rennenberg, H. (2001). Characterization of root exudates at different growth stages of ten rice (Oryza sativa L.) cultivars. Plant Biol. 3, 139–148. doi: 10.1055/s-2001-15205
Bak, F., Scheff, G., and Jansen, K. H. (1991). A rapid and sensitive ion chromatographic technique for the determination of sulfate and sulfate reduction rates in freshwater lake sediments. FEMS Microbiol. Ecol. 85, 23–30. doi: 10.1111/j.1574-6968.1991.tb04694.x
Baldani, V. L. D., Baldani, J. I., and Döbereiner, J. (2000). Inoculation of rice plants with the endophytic diazotrophs Herbaspirillum seropedicae and Burkholderia spp. Biol. Fertil. Soils 30, 485–491. doi: 10.1007/s003740050027
Bates, S. T., Cropsey, G. W. G., Caporaso, J. G., Knight, R., and Fierer, N. (2011). Bacterial communities associated with the lichen symbiosis. Appl. Environ. Microbiol. 77, 1309–1314. doi: 10.1128/AEM.02257-10
Bodelier, P. L. E. (2015). Sustainability: bypassing the methane cycle. Nature 523, 534–535. doi: 10.1038/nature14633
Brandt, F. B., Martinson, G. O., Pommerenke, B., Pump, J., and Conrad, R. (2015). Drying effects on archaeal community composition and methanogenesis in bromeliad tanks. FEMS Microbiol. Ecol. 91, 1–10. doi: 10.1093/femsec/fiu021
Breidenbach, B., and Conrad, R. (2015). Seasonal dynamics of bacterial and archaeal methanogenic communities in flooded rice fields and effect of drainage. Front. Microbiol. 5:752. doi: 10.3389/fmicb.2014.00752
Brune, A., Frenzel, P., and Cypionka, H. (2000). Life at the oxic-anoxic interface: microbial activities and adaptations. FEMS Microbiol. Rev. 24, 691–710. doi: 10.1016/S0168-6445(00)00054-1
Burggraf, S., Huber, H., and Stetter, K. O. (1997). Reclassification of the crenarchaeal orders and families in accordance with 16S rRNA sequence data. Int. J. Syst. Evol. Microbiol. 47, 657–660.
Butler, J. L., Williams, M. A., Bottomley, P. J., and Myrold, D. D. (2003). Microbial community dynamics associated with rhizosphere carbon flow. Appl. Environ. Microbiol. 69, 6793–6800. doi: 10.1128/AEM.69.11.6793-6800.2003
Chaparro, J. M., Badri, D. V., and Vivanco, J. M. (2014). Rhizosphere microbiome assemblage is affected by plant development. ISME J. 8, 790–803. doi: 10.1038/ismej.2013.196
Chidthaisong, A., and Conrad, R. (2000). Turnover of glucose and acetate coupled to reduction of nitrate, ferric iron and sulfate and to methanogenesis in anoxic rice field soil. FEMS Microbiol. Ecol. 31, 73–86. doi: 10.1111/j.1574-6941.2000.tb00673.x
Chin, K. J., Liesack, W., and Janssen, P. H. (2001). Opitutus terrae gen. nov., sp. nov., to accommodate novel strains of the division “Verrucomicrobia” isolated from rice paddy soil. Int. J. Syst. Evol. Microbiol. 51, 1965–1968. doi: 10.1099/00207713-51-6-1965
Clarke, K. R. (1993). Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 18, 117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x
Colmer, T. D., and Pedersen, O. (2008). Oxygen dynamics in submerged rice (Oryza sativa). New Phytol. 178, 326–334. doi: 10.1111/j.1469-8137.2007.02364.x
Conrad, R. (1996). Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO. Microbiol. Rev. 60, 609–640.
Conrad, R. (2009). The global methane cycle: recent advances in understanding the microbial processes involved. Environ. Microbiol. Rep. 1, 285–292. doi: 10.1111/j.1758-2229.2009.00038.x
Conrad, R., and Frenzel, P. (2002). “Flooded soils,” in Encylopedia of Environmental Microbiology, ed. G. Britton (New York, NY: John Wiley & Sons), 1316–1333.
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–1000. doi: 10.1038/nmeth.2604
Edwards, J., Johnson, C., Santos-Medellín, C., Lurie, E., Podishetty, N. K., Bhatnagar, S., et al. (2015). Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. U.S.A. 112, 911–920. doi: 10.1073/pnas.1414592112
Elbeltagy, A., Nishioka, K., Sato, T., Suzuki, H., Ye, B., Hamada, T., et al. (2001). Endophytic colonization and in planta nitrogen fixation by a Herbaspirillum sp. isolated from wild rice species. Appl. Environ. Microbiol. 67, 5285–5293. doi: 10.1128/AEM.67.11.5285-5293.2001
Eller, G., and Frenzel, P. (2001). Changes in activity and community structure of methane oxidising bacteria over the growth period of rice. Appl. Environ. Microbiol. 67, 2395–2403. doi: 10.1128/AEM.67.6.2395-2403.2001
Erkel, C., Kube, M., Reinhardt, R., and Liesack, W. (2006). Genome of Rice Cluster I archaea–the key methane producers in the rice rhizosphere. Science 313, 370–372. doi: 10.1126/science.1127062
Estrada, G. A., Baldani, V. L. D., de Oliveira, D. M., Urquiaga, S., and Baldani, J. I. (2013). Selection of phosphate-solubilizing diazotrophic Herbaspirillum and Burkholderia strains and their effect on rice crop yield and nutrient uptake. Plant Soil 369, 115–129. doi: 10.1007/s11104-012-1550-7
Frenzel, P., Rothfuss, F., and Conrad, R. (1992). Oxygen profiles and methane turnover in a flooded rice microcosm. Biol. Fertil. Soils 14, 84–89. doi: 10.1007/BF00336255
Großkopf, R., Janssen, P. H., and Liesack, W. (1998). Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl. Environ. Microbiol. 64, 960–969.
Haney, C. H., Samuel, B. S., Bush, J., and Ausubel, F. M. (2015). Associations with rhizosphere bacteria can confer an adaptive advantage to plants. Nat. Plants 1, 15051. doi: 10.1038/nplants.2015.51
Hassan, S., and Mathesius, U. (2012). The role of flavonoids in root-rhizosphere signalling: opportunities and challenges for improving plant-microbe interactions. J. Exp. Bot. 63, 3429–3444. doi: 10.1093/jxb/err430
Henckel, T., Friedrich, M., and Conrad, R. (1999). Molecular analyses of the methane-oxidizing microbial community in rice field soil by targeting the genes of the 16S rRNA, particulate methane monooxygenase, and methanol dehydrogenase. Appl. Environ. Microbiol. 65, 1980–1990.
Hernández, M., Dumont, M. G., Yuan, Q., and Conrad, R. (2015). Different bacterial populations associated with the roots and rhizosphere of rice incorporate plant-derived carbon. Appl. Environ. Microbiol. 81, 2244–2253. doi: 10.1128/AEM.03209-14
Hinsinger, P., Bengough, A. G., Vetterlein, D., and Young, I. M. (2009). Rhizosphere: biophysics, biogeochemistry and ecological relevance. Plant Soil 321, 117–152. doi: 10.1007/s11104-008-9885-9
Hirsch, P. R., Miller, A. J., and Dennis, P. G. (2013). “Do root exudates exert more influence on rhizosphere bacterial community structure than other rhizodeposits?,” in Molecular Microbial Ecology of the Rhizosphere, ed. F. J. de Bruijn (Hoboken, NJ: John Wiley & Sons), 229–242.
Ho, A., Lüke, C., Cao, Z., and Frenzel, P. (2011). Ageing well: methane oxidation and methane oxidizing bacteria along a chronosequence of 2000 years. Environ. Microbiol. Rep. 3, 738–743. doi: 10.1111/j.1758-2229.2011.00292.x
Hori, T., Müller, A., Igarashi, Y., Conrad, R., and Friedrich, M. W. (2010). Identification of iron-reducing microorganisms in anoxic rice paddy soil by 13C-acetate probing. ISME J. 4, 267–278. doi: 10.1038/ismej.2009.100
Inceoğlu, O., Salles, J. F., van Overbeek, L., and van Elsas, J. D. (2010). Effects of plant genotype and growth stage on the betaproteobacterial communities associated with different potato cultivars in two fields. Appl. Environ. Microbiol. 76, 3675–3684. doi: 10.1128/AEM.00040-10
Jäckel, U., Russo, S., and Schnell, S. (2005). Enhanced iron reduction by iron supplement: a strategy to reduce methane emission from paddies. Soil Biol. Biochem. 37, 2150–2154. doi: 10.1016/j.soilbio.2005.03.003
Jones, D. L., Nguyen, C., and Finlay, R. D. (2009). Carbon flow in the rhizosphere: carbon trading at the soil–root interface. Plant Soil 321, 5–33. doi: 10.1007/s11104-009-9925-0
Knief, C., Delmotte, N., Chaffron, S., Stark, M., Innerebner, G., Wassmann, R., et al. (2012). Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J. 6, 1378–1390. doi: 10.1038/ismej.2011.192
Krüger, M., Frenzel, P., Kemnitz, D., and Conrad, R. (2005). Activity, structure and dynamics of the methanogenic archaeal community in a flooded Italian rice field. FEMS Microbiol. Ecol. 51, 323–331. doi: 10.1016/j.femsec.2004.09.004
Kuzyakov, Y. (2002). Review: factors affecting rhizosphere priming effects. J. Plant Nutr. Soil Sci. 165, 382–396. doi: 10.1002/1522-2624(200208)165:4<382::AID-JPLN382>3.0.CO;2-#
Kuzyakov, Y., and Domanski, G. (2000). Carbon input by plants into the soil. J. Plant Nutr. Soil Sci. 163, 421–431. doi: 10.1002/1522-2624(200008)163:4<421::AID-JPLN421>3.0.CO;2-R
Lee, H. J., Jeong, S. E., Kim, P. J., Madsen, E. L., and Jeon, C. O. (2015). High resolution depth distribution of Bacteria, Archaea, methanotrophs, and methanogens in the bulk and rhizosphere soils of a flooded rice paddy. Front. Microbiol. 6:639. doi: 10.3389/fmicb.2015.00639
Lee, H. J., Kim, S. Y., Kim, P. J., Madsen, E. L., and Jeon, C. O. (2014). Methane emission and dynamics of methanotrophic and methanogenic communities in a flooded rice field ecosystem. FEMS Microbiol. Ecol. 88, 195–212. doi: 10.1111/1574-6941.12282
Lu, Y., and Conrad, R. (2005). In situ stable isotope probing of methanogenic archaea in the rice rhizosphere. Science 309, 1088–1090. doi: 10.1126/science.1113435
Lu, Y., Murase, J., Watanabe, A., Sugimoto, A., and Kimura, M. (2004). Linking microbial community dynamics to rhizosphere carbon flow in a wetland rice soil. FEMS Microbiol. Ecol. 48, 179–186. doi: 10.1016/j.femsec.2004.01.004
Lu, Y., Rosencrantz, D., Liesack, W., and Conrad, R. (2006). Structure and activity of bacterial community inhabiting rice roots and the rhizosphere. Environ. Microbiol. 8, 1351–1360. doi: 10.1111/j.1462-2920.2006.01028.x
Lüdemann, H., Arth, I., and Liesack, W. (2000). Spatial changes in the bacterial community structure along a vertical oxygen gradient in flooded paddy soil cores. Appl. Environ. Microbiol. 66, 754–762. doi: 10.1128/AEM.66.2.754-762.2000
Lundberg, D. S., Lebeis, S. L., Paredes, S. H., Yourstone, S., Gehring, J., Malfatti, S., et al. (2012). Defining the core Arabidopsis thaliana root microbiome. Nature 488, 86–90. doi: 10.1038/nature11237
Lynch, J. M., and Whipps, J. M. (1990). Substrate flow in the rhizosphere. Plant Soil 129, 1–10. doi: 10.1007/BF00011685
Marschner, P., Crowley, D., and Yang, C. H. (2004). Development of specific rhizosphere bacterial communities in relation to plant species, nutrition and soil type. Plant Soil 261, 199–208. doi: 10.1023/B:PLSO.0000035569.80747.c5
McInerney, M. J., Struchtemeyer, C. G., Sieber, J., Mouttaki, H., Stams, A. J. M., Schink, B., et al. (2008). Physiology, ecology, phylogeny, and genomics of microorganisms capable of syntrophic metabolism. Ann. N. Y. Acad. Sci. 1125, 58–72. doi: 10.1196/annals.1419.005
Mendes, R., Kruijt, M., de Bruijn, I., Dekkers, E., van der Voort, M., Schneider, J. H. M., et al. (2011). Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332, 1097–1100. doi: 10.1126/science.1203980
Micallef, S. A., Channer, S., Shiaris, M. P., and Colón-Carmona, A. (2009a). Plant age and genotype impact the progression of bacterial community succession in the Arabidopsis rhizosphere. Plant Signal. Behav. 4, 777–780. doi: 10.1093/jxb/erp053
Micallef, S. A., Shiaris, M. P., and Colón-Carmona, A. (2009b). Influence of Arabidopsis thaliana accessions on rhizobacterial communities and natural variation in root exudates. J. Exp. Bot. 60, 1729–1742. doi: 10.1093/jxb/erp053
Nihorimbere, V., Ongena, M., Smargiassi, M., and Thonart, P. (2011). Beneficial effect of the rhizosphere microbial community for plant growth and health. Biotechnol. Agron. Soc. Environ. 15, 327–337.
Noll, M., Matthies, D., Frenzel, P., Derakshani, M., and Liesack, W. (2005). Succession of bacterial community structure and diversity in a paddy soil oxygen gradient. Environ. Microbiol. 7, 382–395. doi: 10.1111/j.1462-2920.2005.00700.x
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O’Hara, R. B., et al. (2013) Vegan: Community Ecology Package. R Package Version 2.0-10. Available at: http://cran.r-project.org/package=vegan
Peiffer, J. A., Spor, A., Koren, O., Jin, Z., Green, S., Dangl, J. L., et al. (2013). Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. U.S.A. 110, 6548–6553. doi: 10.1073/pnas.1302837110
Philippot, L., Raaijmakers, J. M., Lemanceau, P., and van der Putten, W. H. (2013). Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11, 789–799. doi: 10.1038/nrmicro3109
Pump, J., and Conrad, R. (2014). Rice biomass production and carbon cycling in 13CO2 pulse-labeled microcosms with different soils under submerged conditions. Plant Soil 384, 213–229. doi: 10.1007/s11104-014-2201-y
R Core Team (2013) R: A Language and Environment for Statistical Computing. Available at: http://www.R-project.org/
Ramakrishnan, B., Lueders, T., Dun, P. F., Conrad, R., and Friedrich, M. W. (2001). Archaeal community structures in rice soils from different geographical regions before and after initiation of methane production. FEMS Microbiol. Ecol. 37, 175–186. doi: 10.1111/j.1574-6941.2001.tb00865.x
Ren, G., Zhang, H., Lin, X., Zhu, J., and Jia, Z. (2014). Response of phyllosphere bacterial communities to elevated CO2 during rice growing season. Appl. Microbiol. Biotechnol. 98, 9459–9471. doi: 10.1007/s00253-014-5915-0
Rothballer, M., Eckert, B., Schmid, M., Fekete, A., Schloter, M., Lehner, A., et al. (2008). Endophytic root colonization of gramineous plants by Herbaspirillum frisingense. FEMS Microbiol. Ecol. 66, 85–95. doi: 10.1111/j.1574-6941.2008.00582.x
Rui, J., Peng, J., and Lu, Y. (2009). Succession of bacterial populations during plant residue decomposition in rice field soil. Appl. Environ. Microbiol. 75, 4879–4886. doi: 10.1128/AEM.00702-09
Scavino Fernandez, A., Ji, Y., Pump, J., Klose, M., Claus, P., and Conrad, R. (2013). Structure and function of the methanogenic microbial communities in Uruguayan soils shifted between pasture and irrigated rice fields. Environ. Microbiol. 15, 2588–2602. doi: 10.1111/1462-2920.12161
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Shrestha, P. M., Noll, M., and Liesack, W. (2007). Phylogenetic identity, growth-response time and rRNA operon copy number of soil bacteria indicate different stages of community succession. Environ. Microbiol. 9, 2464–2474. doi: 10.1111/j.1462-2920.2007.01364.x
Stubner, S. (2002). Enumeration of 16S rDNA of Desulfotomaculum lineage 1 in rice field soil by real-time PCR with SybrGreenTM detection. J. Microbiol. Methods 50, 155–164. doi: 10.1016/S0167-7012(02)00024-6
Sugiyama, A., Ueda, Y., Zushi, T., Takase, H., and Yazaki, K. (2014). Changes in the bacterial community of soybean rhizospheres during growth in the field. PLoS ONE 9:e100709. doi: 10.1371/journal.pone.0100709
Sun, J., Zhang, Q., Zhou, J., and Wei, Q. (2014). Illumina amplicon sequencing of 16S rRNA tag reveals bacterial community development in the rhizosphere of apple nurseries at a replant disease site and a new planting site. PLoS ONE 9:e111744. doi: 10.1371/journal.pone.0111744
van der Heijden, M. G. A., Bardgett, R. D., and van Straalen, N. M. (2008). The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 11, 296–310. doi: 10.1111/j.1461-0248.2007.01139.x
Warnes, A. G. R., Bolker, B., Bonebakker, L., Huber, W., Liaw, A., Lumley, T., et al. (2015) Package Gplots: Various R Programming Tools for Plotting Data. Available at: http://cran.r-project.org/
Watanabe, A., Takeda, T., and Kimura, M. (1999). Evaluation of origins of CH4 carbon emitted from rice paddies. J. Geophys. Res. 104, 23623–23629. doi: 10.1029/1999JD900467
Watanabe, T., Kimura, M., and Asakawa, S. (2006). Community structure of methanogenic archaea in paddy field soil under double cropping (rice–wheat). Soil Biol. Biochem. 38, 1264–1274. doi: 10.1016/j.soilbio.2005.09.020
Welbaum, G. E., Sturz, A. V., Dong, Z., and Nowak, J. (2004). Managing soil microorganisms to improve productivity of agro-ecosystems. CRC. Crit. Rev. Plant Sci. 23, 175–193. doi: 10.1080/07352680490433295
Wu, W. X., Liu, W., Lu, H. H., Chen, Y. X., Medha, D., and Janice, T. (2009). Use of 13C labeling to assess carbon partitioning in transgenic and nontransgenic (parental) rice and their rhizosphere soil microbial communities. FEMS Microbiol. Ecol. 67, 93–102. doi: 10.1111/j.1574-6941.2008.00599.x
Xu, Y., Ma, K., Huang, S., Liu, L., and Lu, Y. (2012). Diel cycle of methanogen mcrA transcripts in rice rhizosphere. Environ. Microbiol. Rep. 4, 655–663. doi: 10.1111/j.1758-2229.2012.00392.x
Yang, J., Kloepper, J. W., and Ryu, C.-M. (2009). Rhizosphere bacteria help plants tolerate abiotic stress. Trends Plant Sci. 14, 1–4. doi: 10.1016/j.tplants.2008.10.004
Yao, H., and Conrad, R. (1999). Thermodynamics of methane production in different rice paddy soils from China, the Philippines and Italy. Soil Biol. Biochem. 31, 463–473. doi: 10.1016/S0038-0717(98)00152-7
Yao, H., Conrad, R., Wassmann, R., and Neue, H. U. (1999). Effect of soil characteristics on sequential reduction and methane production in sixteen rice paddy soils from China, the Philippines, and Italy. Biogeochemistry 47, 269–295. doi: 10.1023/A:1006186420644
Yuan, Y., Conrad, R., and Lu, Y. (2009). Responses of methanogenic archaeal community to oxygen exposure in rice field soil. Environ. Microbiol. Rep. 1, 347–354. doi: 10.1111/j.1758-2229.2009.00036.x
Keywords: Oryza sativa, rhizosphere, community ecology, bacteria, Archaea, Herbaspirillum, plant growth stage
Citation: Breidenbach B, Pump J and Dumont MG (2016) Microbial Community Structure in the Rhizosphere of Rice Plants. Front. Microbiol. 6:1537. doi: 10.3389/fmicb.2015.01537
Received: 28 September 2015; Accepted: 21 December 2015;
Published: 13 January 2016.
Edited by:
Paul Bodelier, Netherlands Institute of Ecology, NetherlandsReviewed by:
Zhongjun Jia, Institute of Soil Science, Chinese Academy of Sciences, ChinaCopyright © 2016 Breidenbach, Pump and Dumont. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marc G. Dumont, bS5nLmR1bW9udEBzb3Rvbi5hYy51aw==
†Present address: Marc G. Dumont, Centre for Biological Sciences, University of Southampton, Southampton SO17 1BJ, UK
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.