
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 05 January 2016
Sec. Evolutionary and Genomic Microbiology
Volume 6 - 2015 | https://doi.org/10.3389/fmicb.2015.01462
Microsatellites or short sequence repeats are widespread genetic markers which are hypermutable 1–6 bp long short nucleotide motifs. Significantly, their applications in genetics are extensive due to their ceaseless mutational degree, widespread length variations and hypermutability skills. These features make them useful in determining the driving forces of evolution by using powerful molecular techniques. Consequently, revealing important questions, for example, what is the significance of these abundant sequences in DNA, what are their roles in genomic evolution? The answers of these important questions are hidden in the ways these short motifs contributed in altering the microbial genomes since the origin of life. Even though their size ranges from 1 –to- 6 bases, these repeats are becoming one of the most popular genetic probes in determining their associations and phylogenetic relationships in closely related genomes. Currently, they have been widely used in molecular genetics, biotechnology and evolutionary biology. However, due to limited knowledge; there is a significant gap in research and lack of information concerning hypermutational mechanisms. These mechanisms play a key role in microsatellite loci point mutations and phase variations. This review will extend the understandings of impacts and contributions of microsatellite in genomic evolution and their universal applications in microbiology.
Microsatellites or simple sequence repeats (SSRs) are short tandem repeats (STRs) of DNA sequence motifs predominantly abundant in various genomes and have been widely used for genetic studies and molecular markers (Han et al., 2015). The term “microsatellites” was first coined in by Litt and Luty (1989) during their work on (TG)n in gene of cardiac actin. These repeats were developed for the study of neurological diseases in human and afterward their applications made them significant in various molecular fields.
They are also known as single nucleotide polymorphisms (SNPs) (Batley et al., 2007), are associated and located at telomeres, centromeres, genic regions, intergenic regions and even at interspersed sites within a genome (Kim et al., 2015). SSRs are named as the most versatile molecular markers used to identify a certain molecular sequence in a pool of unknown DNA; they have applications in various fields of molecular biology, biotechnology and evolutionary biology. Tandem repeats are ubiquitous and widely used in genetic studies of microbes (Grover and Sharma, 2012; Poczai et al., 2013; Abdul-Muneer, 2014; Zhao et al., 2015). These markers are principal tools of determining hyper mutational genetic diversity by recently developed advanced sequence techniques in genetics (McCouch et al., 2002).
DNA is a polymorphic molecule, extremely stable in hostile environments and accountable for the inheritance of traits through generations by conserving genetic code of the host organism (Gyllensten et al., 1991; McKusick, 1998; Shitara et al., 1998; Birky, 2001). It has been demonstrated that SSR markers are repeated frequently in a conserved DNA sequence and suitable for studying genetic diversity among species, populations, and individuals. Various techniques have been established to evaluate DNA polymorphism by measuring genetic diversity in situ. Consequently, it is easy to trace the fingerprints of all the organisms by examining molecular markers of DNA involved in determining the inherited characters and evolutionary history in a phyletic lineage (McCouch et al., 1997, 2002).
The difference in number of repeats of SSR motifs in different species shows polymorphism (Xu et al., 2012). Low DNA amounts are needed for the amplification of genomic DNA; therefore, SSRs are polymerase chain reaction (PCR-based) markers and mostly co-dominant, multi-allelic, reproducible, and highly polymorphic (Birren and Lai, 1987; Powell et al., 1996b; Feingold et al., 2005). Generally, they have been used in genic linkage mapping, genetic characterization of germplasmic resource investigation, phylogenetic analysis, DNA fingerprinting and other genetic studies (Liu et al., 1996; Pérez et al., 2005; Weng et al., 2007). Satellite DNAs are generally related with centromeric heterochromatin and are being progressively employed as a useful tool for genome analysis, mapping and for understanding chromosomal organization (Megan et al., 2014; Shen et al., 2015). They are used for genome mapping, population studies, and specie identification and continued to be the genetic marker of choice in most non-human systems and form an important genomic component (Amos et al., 2015).
Microsatellites are characterized by tandemly repeated short motifs with length 1 –to- 6 bp long core sequences. Their hypervariability is based on changes in the repeats of core sequences several times at a given locus (Tautz, 1993; Varshney et al., 2005; Zulini et al., 2005; Wheeler et al., 2014). They can be traced in both coding and non-coding regions (Ellegren, 2004). Generally, there are three classes of biological markers: (i) nucleic acid hybridization, e.g., restriction fragment length polymorphisms (RFLPs) (ii) PCR-based on DNA amplification, e.g., random amplification of polymorphic DNAs (RAPD), amplified fragment length polymorphisms (AFLP), SSRs and (iii), SNPs (Priyono and Putranto, 2014; Vijay et al., 2015).
Molecular biology became progressive and innovative with the invention of PCR technology in mid 1980s (Saiki et al., 1985; Mullis and Faloona, 1987), this revolutionary technique facilitated in various biological fields, i.e., diagnostics, breeding programs, forensics, microbiology etc. Consequently, microsatellite maker systems are widely used in evolutionary biology due to their hypervariability and hypermutability (Dallas, 1992; Weber and Wong, 1993; Di Rienzo et al., 1994; Ellegren, 1995). Microsatellites are tandem repeated motifs of variable lengths found throughout cellular nuclear genomes (Jarne and Lagoda, 1996). They also appear in organelle genomes, e.g., chloroplast (Powell et al., 1995, 1996a) and mitochondria which were predominantly widespread in primitive microbial world (Soranzo et al., 1999). It is convenient to genotype microsatellites instead of their polymorphic variability nature, because, they are densely populated throughout genomes. Therefore, they are useful genetic markers in high resolution genetic mapping (Dib et al., 1996; Dietrich et al., 1996; Schuler et al., 1996; Knapik et al., 1998; Cooper et al., 1999).
In 1986, the role of microsatellites in microbial DNA was identified in Neisseria gonorrhoeae; a bacterium which is responsible for infamous sexually transmitted disease (STD) gonorrhea. This bacterium possesses family of 12 outer-membrane proteins which are encoded by Opas genes. These proteins upon expression help bacterium to adhere invading epithelial cells. The Opas genes retain multiple copies of microsatellites comprising of 5 based motif CTCTT (Moxon et al., 1994; Hung and Christodoulides, 2013). Several SSRs have been identified with their physiological and morphological functions in microbial genomes as shown in Table 1 (Peak et al., 1996; Burch et al., 1997; Inzana et al., 1997; Karlin et al., 1997; Grimwood et al., 2001; Rocha and Blanchard, 2002).
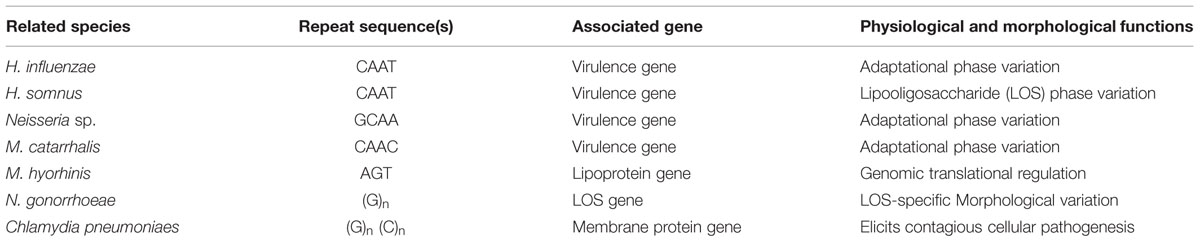
TABLE 1. Microbial coding regions containing simple sequence repeat (SSRs), physiological and morphological effects in various species.
Microsatellites are the most useful molecular markers with an advantage of easy and low-cost detection by PCR due to high mutation rates and new sequencing technologies. Therefore, their applications in microbiology are widespread for the determination of genomic evolution (Paglia and Morgante, 1998). As compared to RAPD and AFLP, which can detect the location of locus in a genome, microsatellites have an advantage, because, they can detect all the physiological parameters of a genome (Maughan et al., 1996; Powell et al., 1996b; Beismann et al., 1997; Gaiotto et al., 1997; Russell et al., 1997; Witsenboer et al., 1997; Semblat et al., 1998; Nybom et al., 2014). The present study aimed to investigate the roles of microsatellites in shaping the genomes over time and to develop better understandings of their characteristic hypermutability and hypervariability by employing advanced molecular techniques. This will help extend substantial knowledge about their significant importance in genome evolution.
The origin of microsatellites in microbial genomes is non-random, with various differences among the mechanisms which stimulated for SSRs genes, these mechanisms consisted of insertions, deletions, recombination and repair, transpositions, horizontal gene transfer and replication slippage (Hancock and Santibáñez-Koref, 1998; Primmer and Ellegren, 1998; Alba et al., 1999, 2001; Chambers and MacAvoy, 2000; Hartenstine et al., 2000; Jakupciak and Wells, 2000; Schlotterer, 2000; Zhu et al., 2000; Bhargava and Fuentes, 2010; Holder et al., 2015).
Currently, there are two non-mutually special hypotheses to describe the source of microsatellites: (i) De novo theory suggests that the microsatellites originated from a proto-microsatellite in microbes, a small region of as few as three or four repetitive elements within simple sequences, which are distinct as a struggle of repetitive motifs deficient in clear tandem organization (Messier et al., 1996; Buschiazzo and Gemmell, 2006; Wang et al., 2015). Consequently after formation, the conservation and proliferation was selected by strand slippage through replication and subjected to the repeat motif, it had a capacity to form unusual DNA conformations and contributed in recombination and transposition events. The number of repeat units runs parallel with the variability of microsatellite, but the least repeat number which is significant for strand slippage and other mutations is uncertain (Jentzsch et al., 2008). Slippage mutations occur repeatedly at runs of 3–4 bases in prokaryotic genomes (Foster and Trimarchi, 1994; Rosenberg et al., 1994; Sébastien et al., 2010).
(ii) Adopted microsatellites theory suggests their beginning from other genomic sections via transposable elements. The transposable elements consisted of one or more locations susceptible to microsatellite development and favored the distribution of microsatellites in genomes. This advocates a mutual association in which microsatellites acted as “retroposition navigator sequences,” while retrotransposons produced more microsatellites during their scattering in genomes. An example of a retrotransposon-mediated microsatellite birth is the origin of A/T rich microsatellites with motifs extending from 1 to 6 bases in length from Alu elements (Wilder and Hollocher, 2001; Jentzsch et al., 2008; Sand et al., 2015).
Microsatellites are DNA sequences of 1–6 bp units repeated in tandem and widely dispersed in the microbial genome (Powell et al., 1996a). Numerous repetitive sequences including microsatellites are found in up to 5% of the prokaryotic DNA (Ussery et al., 2004; Wheeler et al., 2014). The frequency and spreading of SSR is centered on species and motif specificity (Karlin et al., 1996, 1997; Bachtrog et al., 1999; Butcher et al., 2000; Crollius et al., 2000; Metzgar et al., 2000; Tóth et al., 2000; Gentles and Karlin, 2001; Morgante et al., 2002; Katoh et al., 2015). SSRs with 1–6 bp were used for phase variation in bacterial adaptations (Holder et al., 2015).
Microsatellites can be amplified with the help of PCR in rigid conditions with the amplification of single loci (Bravo et al., 2006; Buschiazzo and Gemmell, 2006). They are broadly distributed in various genomes and highly polymorphic in nature. Therefore, establishes the foundation of their success in wide range of biological fields (Chistiakov et al., 2006).
Simple sequence repeats in various organisms are also noticeable from the diverse genome regions, e.g., 3′-UTRs, 5′-UTRs, exons and introns (Rajendrakumar et al., 2007). Their localization can be altered by different aspects of DNA structures (Chistiakov et al., 2006). The transposable elements help in the formation and dispersion of microsatellite throughout the genome (Bhargava and Fuentes, 2010). Kashi et al. (1997) described the length of SSRs which influences the transcriptional activity in promoter regions.
The effect of length variations in the mononucleotide repeats and polymorphisms within these regions of chloroplast genome are used to study both intraspecific and interspecific variability (Powell et al., 1995). Length variation at a mitochondrial SSR locus was first reported by Soranzo et al. (1999). The descriptive analysis of microsatellite content in genome sequences reflects their roles in genome organization, recombination, gene regulation, quantitative genetic variation and gene evolution (Katti et al., 2001).
Classification of SSRs is based on their isolation and sequencing. They have variable length of repeat motifs from just a single base to thousands of bases; microsatellites can be classified on the number of bases, i.e., short repeats (10–30 bases) known as minisatellites and with longer repeats (between 10 and 100 bases) are called macrosatellites, satellites with even shorter repeat motifs, called microsatellites (Figure 1). Based on the length of the repeat units, SSRs are categorized into three groups (Class I>20 bp, Class II=between 11 and 20 bp, and Class III<11 bp), Scattered repetitive elements are determined at the flanking sites of the SSRs. (Temnykh et al., 2001; Varshney et al., 2002).
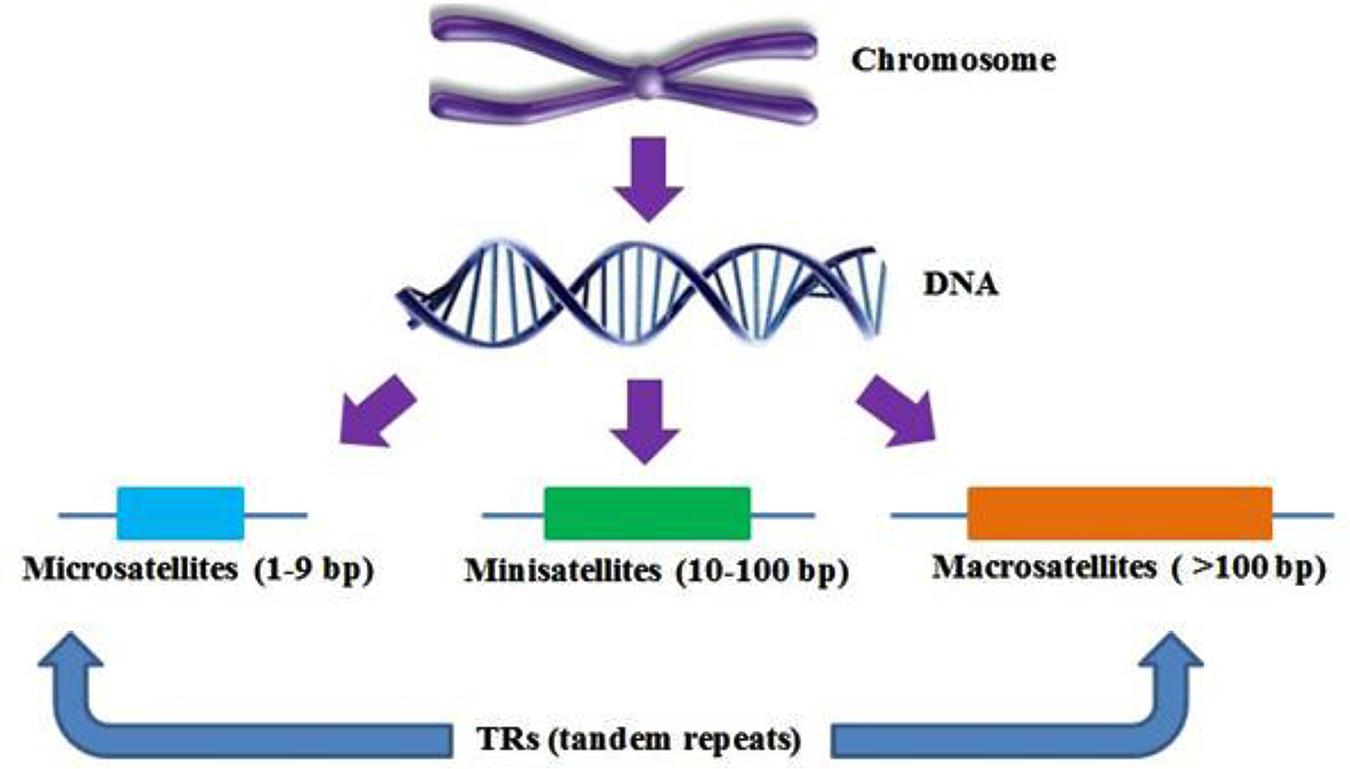
FIGURE 1. Diagram illustrating the different types of tandem repeats (TRs). The width of boxes has been shown to develop visual precision of the figure.
Abundance and length variations in microsatellites motifs are evaluated on mono, di, tri, tetra, penta and hexanucleotide. (Rabello et al., 2005; Merritt et al., 2015). They are also classified according to the type of repeated sequence presented: (i) perfect repeats, with perfect repetitions, e.g., (AT)20, have sequences of ten or more mononucleotide repeats, six or more dinucleotide repeats, tri, tetra and pentanucleotide repeats. (ii) Imperfect repeats, with interruption by different nucleotides which are usually not repeated, e.g., (AT)12 GC(AT)8, and (iii) composite, with two or more different motifs in tandem, e.g., (AT)7(GC)6. FORESTs database showed complementary sequences belonging to the same class (e.g., AC, CA, TG, GT). (Temnykh et al., 2001; Selkoe and Toonen, 2006). Compound microsatellites are present in the same expressed sequence tag (EST) at a distance by a maximum of 100 bp. A repeat having more than 50 bp distance from the 3′ end of sequences is not considered as microsatellite (Rabello et al., 2005; Vogiatzi et al., 2011; Wang et al., 2015).
Lactobacillus species revealed a wealth of compound imperfect microsatellites clustered in the coding regions of genomes. They were consisted of variant motifs with maximum distance (dMAX) increments of 10–50. The variations analyzed in compound microsatellite of Escherichia coli, and lactobacilli suggested diverse genomic features and evolutionary traces of compound microsatellites between these organisms (Basharat and Yasmin, 2015).
Simple sequence repeats like microsatellites are found abundantly in prokaryotic genomes, these repeats are extremely important molecular markers for the investigation of population genetics of genomes on the bases of excessive polymorphism, reproductivity, and codominance (Field and Wills, 1998; Schlotterer, 2000). 1,117 microsatellite patterns in about 3.8 Mb of unique sequences (0.47% of the total DNA used in the analysis) were identified in Paracoccidioides brasiliensis. 87.5% microsatellites were found in non-coding sequences (Nascimento et al., 2004). The applications of SSRs in genomic studies on molecular basis (Jarne and Lagoda, 1996), their evaluation of population dissemination and evolutionary relationships (Queller et al., 1993), have been used frequently in the study of parentage investigation, phylogenetic studies (Bowcock et al., 1994), studies on population diversity (Paetkau, 1999), determination of inbreeding (Coltman et al., 1998; Coulson et al., 1998), genetic recombinations, population genetic assembly, genomic mapping, and phylogeography (Sunnucks, 2000; Zhang and Hewitt, 2003).
Microsatellites are significant in evaluation of the ability of individual migrations, resemblances in vast extent of the organisms, ranging from mammals and higher chordates to lower microbes such as fungi and even prokaryotes and viruses (Ashley and Dow, 1994; Dib et al., 1996; Selkoe and Toonen, 2006; Breurec et al., 2011).
Debatable hypothesis had been confronted by the genetic evidences came from microsatellites like famous hypothesis put forth by Bass-Becking, “Everything is everywhere, the environment selects” (Baas Becking, 1934). These repeats are vital to differentiate morphologically different species on molecular basis. (Katz et al., 2005). Hatcher et al. (2015) reported that the poxvirus genomes consisted of 24% microsatellites nucleotide sequences. They exhibit hypervariations in poxvirus proteins, gene truncation, and reductive evolution. They are also widely used in the fields of genomic mapping, sex determination, environmental resource and genetics, evolutionary lineages of microbial strains and analysis of phylogenetic relationships in closely related species (Jarne and Lagoda, 1996; Hennequin et al., 2001; Luikart et al., 2003; Lim et al., 2004).
To study the microbial evolution and phylogenetic relationships, Escherichia coli (E. coli) reference strains are significant and most often used in determining the evolutionary relationships among microbes (Ochman and Selander, 1984), Several E. coli strains have been classified into six phylogenetic groups (A, B1, B2, C, D, and E) on the bases of multilocus enzyme electrophoresis (MLEE) method (Goullet and Picard, 1989), most importantly, these strains do not make assemblies within distinct phylogeny on the bases of rep-PCR DNA fingerprinting arrays (Johnson and O’Bryan, 2000). Metzgar et al. (2001) also reported similar applications for utilization of microsatellites at a greater extent in evolutionary analyses to characterize microbial strains.
Microsatellites are hypermutable in every generation, tetranucleotide repeats lose and gain units at a rate of 1 × 10-4 (De Bolle et al., 2000) suggesting that this high decline rate in prevalence reveals evolution by natural selection. Excessive rate of loci mutations results into harmful fitness effects rather than beneficial. SSRs are found abundantly in some host-adapted bacteria as compared to other genomes (Mrázek et al., 2007). It is shown that long tracts of tetranucleotide repeat sequences are abundantly found in the Hi strain Rd KW20 genome; these repeats have an association with the genes which control commensal and virulence behavior (Hood et al., 1996).
To study microsatellites, several approaches have been established with the recent development of advanced molecular techniques. These protocols can be grouped into three types: (i) the standard method, where a library is screened (ii) the automated method, sequences are searched in sequence databases and (iii) the sequencing method, whole or parts of the genome are sequenced. These methods are modified and optimized on the bases of species and conditions (Zane et al., 2002; Weising et al., 2005).
In the 1960s, simple repeats were identified in density gradient centrifugations of randomly sheared genomic DNAs by way of a ‘satellite peak’ and found dispersed throughout various genomes (Park et al., 2009). Different techniques have been introduced to identify microsatellites (Dutech et al., 2007). The most common methods in used for the identification of repeats are the target enrichment of DNAs (Hamilton et al., 1999; Zane et al., 2002). One method being employed is known as inter simple sequence repeat PCR (ISSR-PCR), in which ISSR primers containing microsatellites motifs along with three anchored nucleotides at 5′ terminal end are used for amplifying microsatellite sequence regions which are known to be abundant in genomes, the PCR products are then cloned and sequenced for determination of microsatellites (Zietkiewicz et al., 1994; Van Der Nest et al., 2000).
With recent development in molecular biology, modifications in DNA enrichment strategies are made, linking hybridization with probes to identify and compare a vast range of microsatellite sequences to genomic DNA fragments (Zane et al., 2002). One of the current approaches being used is called fast isolation by AFLPs of sequences containing repeats (FIASCO), which follows amplified fragment length polymorphism (AFLP) (Vos et al., 1995) Both ISSR-PCR and FIASCO methods are routinely applied in studies related to the identification and characterization of SSRs and they have been used to isolate microsatellites from various microbial species (Luque et al., 2002; Squirrell et al., 2003; Pfunder and Frey, 2006; Barnes et al., 2008; Santana et al., 2009).
In advent of recent development in identification strategies of microsatellites, various methods have been devised for characterization of microsatellites.
One method is the development of a library with the help of various protocols to create and screen a cDNA or PCR fragment library, in this method the DNA is fragmented by sonication or enzymatic digestion, then fragments are ligated into a vector and transformed into E. coli, following clones are analyzed by southern blot for SSR and finally the positive clones are sequenced (Weising et al., 2005).
The positive clone obtained ranges from 0.04 to 12% (Zane et al., 2002). The plasmids of fragment library can be screened by the use of biotinylated oligonucleotides (Ito et al., 1992). In another method, the genomic library was amplified using biotinylated oligonucleotides complementary to SSRs, as primers (Paetkau, 1999). A high enrichment efficiency of almost 90% for CA repeats was generated by using two rounds of amplification and hybridization with biotin/streptavidin (Kandpal et al., 1994).
Microsatellite identification and development can be done by using public DNA databases, such as BLASTN (Altschul et al., 1990; Dhillon et al., 2014). Various programs and reference lists are available in the database (Mittal and Dubey, 2009). Numerous studies have been used to search for more conserved and gene related microsatellites by using EST-SSRs (Varshney et al., 2005).
Expressed or whole genome sequencing can be made by new high-tech sequencing techniques (Abdelkrim et al., 2009; Mikheyev et al., 2010). With the use of inconsistent PCR amplification, approximately half of all microsatellite loci are lost (Arthofer et al., 2011)
Microsatellite markers from microbial genomes of model and non-model organisms are being isolated by use of next generation sequencing (NGS) like Roche 454 GS-FLX Titanium pyrosequencing platform, this technique has a potential for the isolation of microsatellite markers from the genome of both model and non-model species with no former reference genome existing (Margulies et al., 2005; Malausa et al., 2011). Four hundred and fifty-four pyrosequencing has many proficient advantages over customary enrichment techniques in isolating microsatellite markers because of high throughput, cost effective, rapid and low labor supplies (Rothberg and Leamon, 2008).
Currently, a new technique Comparative genomic hybridization (CGH-style) array manufactured by Nimblgen/Roche has been used to rapidly measure the complete microsatellite content of a genome. CGH-microarray measures DNA samples labeled with different fluorescent dye from a reference genome and a test genome, and hybridizes them competitively to develop a micro-assay based array comprised of immobilized DNA fragments from sequence of the reference individual (Hazen and Kay, 2003; Hardiman, 2004; Dorrell et al., 2005; Fan et al., 2006).
This technique sums the contributions for a specific repeated motif from number of sites in which that particular motif exists across the whole genome. CGH-array has the ability to assess 1 -to- 6 mer repeats. This method provides significant information about genetic distances for entire genes between pairs of entities in one assay and has made CGH array an attractive tool for phylogenetic analysis. Numerous research approaches applied this microarray to compare the evolutionary relationships of bacterial species (Israel et al., 2001; Chan et al., 2003; Wolfgang et al., 2003; Rasmussen et al., 2008; Igboin et al., 2009; Dorrell et al., 2011).
Guidot et al. (2007), Wan et al. (2007), and Dagerhamn et al. (2008) reported applications of CGH array to recover clusters of bacteria from large clone libraries; it is parallel with formerly described MLSA phylogenies. Solheim et al. (2009) described comparison of MLST phylogeny with CGH array used for Enteroccocus species to define lineage-specific genes in entire reference genome.
Recently, NGS technologies is the most powerful method available to generate cost effective DNA markers including SSRs and SNPs. NGS technologies are integrated with tools like association mapping studies. The NGS method is far more powerful than any existing in generating DNA markers and dramatically increased the yield of potential microsatellite primer pairs, generating 1000s of individual reads (Ekblom and Galindo, 2011; Hoffman and Nichols, 2011; Castoe et al., 2012; Smulders et al., 2012; Yang et al., 2012; Lance et al., 2013; Andersen and Mills, 2014; Vukosavljev et al., 2015), the development of molecular markers is based on short-length sequences from genomic DNA sequences or cDNA (RNA-seq) (Yang and Smith, 2013).
The analysis of loci is determined by the number of repeated motifs and on polymorphic level with specificity in population (Weising et al., 2005). Several statistical analysis based on genetic distances can be utilized along with the use of similarity index and band sharing data (Labate, 2000; Weising et al., 2005; Excoffier and Heckel, 2006). Excoffier and Heckel (2006) accredited two conversion programs for formatting input data files: Convert (Glaubitz, 2004) and Formatomatic (Manoukis, 2007).
Microsatellites are extremely hypermutable as associated with point mutations in coding, non-coding genes and mutation rates which range from 10-6 to 10-2 events per locus per microbial generation. These rates are greatly affected by numerous features, which affect both the likelihood of mutational generations and the restoration proficiency of these mutations (Jarne and Lagoda, 1996).
Evolution has operated on bacterial microsatellite loci at mutation rates up to 1 × 10-3 per division in combination with trans-acting factors; this mutability in bacterial pathogens is known as localized hypermutation. The mechanisms involved site-specific recombination, homologous recombination of tandem duplications of DNA sequences, SSR and G-quartet-mediated gene conversion in pilin sub-unit of Neisseria. This gave rise to specific phenotypes by presumptive, high frequency, reversible switches of associated gene expression. These switches are also responsible for phase variations observed in various bacterial genomes (Bidmos and Bayliss, 2014).
Mutation mechanisms, DNA healing, organizational features of microsatellite, genomic specific framework and selective biological impacts are important factors which relate and control the evolutionary dynamics of microsatellites. In prokaryotes, resilient progressive selective pressures are related with extremely mutable microsatellite loci stretches in genomes that regulate pathogenicity. The average mutation rate of a bacterial gene is 1 × 10-9 mutations/division, but mutation rates of microsatellites are significantly higher than this average. (Moxon et al., 1994; Bidmos and Bayliss, 2014). Large numbers of SSRs are supposed to evolve neutrally; the most extensively considered exclusions are the increasing number of triplet-repeat loci which are the source of genetic diseases (Sutherland and Richards, 1995). It is clear that the investigation of the evolutionary associations of tandem repeat sequences in microbial genomes with respect to genome volatility and utility is significantly supported by rapid emergence of many newly sequenced genomes (Strauss and Falkow, 1997).
Microsatellites are easily transferable to the related genomes which have high proportion of similar conserved transcribed domains (Cordeiro et al., 2001; Decroocq et al., 2003; Hempel and Peakall, 2003; Varshney et al., 2005). The detection of fractional polymorphism with these repeats showed high rates of portability within genomic regions (Cho et al., 2000; Scott et al., 2000; Eujayl et al., 2002), this ability is also associated with differences in gene expression rooted in various microbial species (Gao et al., 2004). Pandian et al. (2000) examined transferability of SSRs in many genomes and revealed a high level of sequence conservation.
The prevalence of flanking regions among microsatellites allows cross-species amplification (Rico et al., 1996; Peakall et al., 1998). Around 20 microsatellite markers are used for characterizing transferability and polymorphism by EST databases (Faria et al., 2010). Pépin et al. (1995) showed 40% of microsatellites are useful to study genomes of important loci. Dawson et al. (2010) developed primer sets from 33 polymorphic loci. The capacity of transferability can be determined by the extent of genomic sequence matching and by the use of interspecies sequence markers (Gupta et al., 2013).
Microsatellites constructed for specific species can be applied to other species closely related to each other. But if the genetic distance increases, the percentage of successful amplification of loci decreases (Jarne and Lagoda, 1996). “Null alleles” are formed with the occurrence of primer annealing point mutations and microsatellites fail to amplify the PCR product (Jarne and Lagoda, 1996; Dakin and Avise, 2004).
Microsatellites are tandemly repeated number of times. They are predominant genetic markers in molecular biology with DNA sequences of 1–6 bp in length. Essentially, the repeat-motifs containing more than mono-nucleotide are selected to develop molecular markers. To pursue a SSR, different parameters such as repeat sequence length, coding position, repeat category (mono- hexa), and sequence motifs are employed (Dikhit et al., 2014).
The molecular processes which expose DNA individual strands result in sequence repeat length mutations comprising of replication, recombination, DNA damage repair and rest of DNA metabolic processes (Wells et al., 2005; Lopez Castel et al., 2010). Microsatellite is prone to length mutations because of intrinsic features of repeat sequences such as unit length, number of repeats, and its structural purity (Fondon et al., 1998; Legendre et al., 2007). Mutation rates due to replication slippage at microsatellite loci are hypervariable extending from unnoticeable level to roughly about 8 × 10-3 (Mahtani and Willard, 1993; Weber and Wong, 1993; Strand et al., 1994; Tautz and Schlötterer, 1994).
Length changes in SSRs are occurred due to the replication slippage and loops because of mismatched DNA strands during replication, excluding Helicobacter pylori which has remarkably extended mono- dinucleotide repeats since they are physiologically functional (Tomb et al., 1997) or in case its genome lacks mismatch DNA repair (Eisen et al., 1997). Upon denaturation of daughter strand in replication, it will pair with wrong sequence complementary to the template strand and will result in sequence deletion or insertion. This type of microsatellite mutation occurs roughly once per 1,000 generations and are more prevalent than the point mutations in other genomic sites (Weber and Wong, 1993; Tautz and Schlötterer, 1994; Jarne and Lagoda, 1996).
SSRs are susceptible to replication mispairing slippage. Slippage involves a region of non-pairing (shown as a loop) containing backward or forward slippage loop repeats of nascent daughter strand or of the parental strand, results in an insertion or a deletion on both strands respectively (Figures 2 and 3). Subsequently, it is possible that slipped strand mispairing can also cause insertions/deletions in non-replicating DNA. In such cases, non-pairing is occurred in two regions of repeats positioned on both complementary DNA strands (Levinson and Gutman, 1987). The replication slippage predicts persistent deletions, duplications and insertions at infinite degree between non-contiguous repeats; this type of slippage is a leading cause of genomic evolution (Dover, 1995).
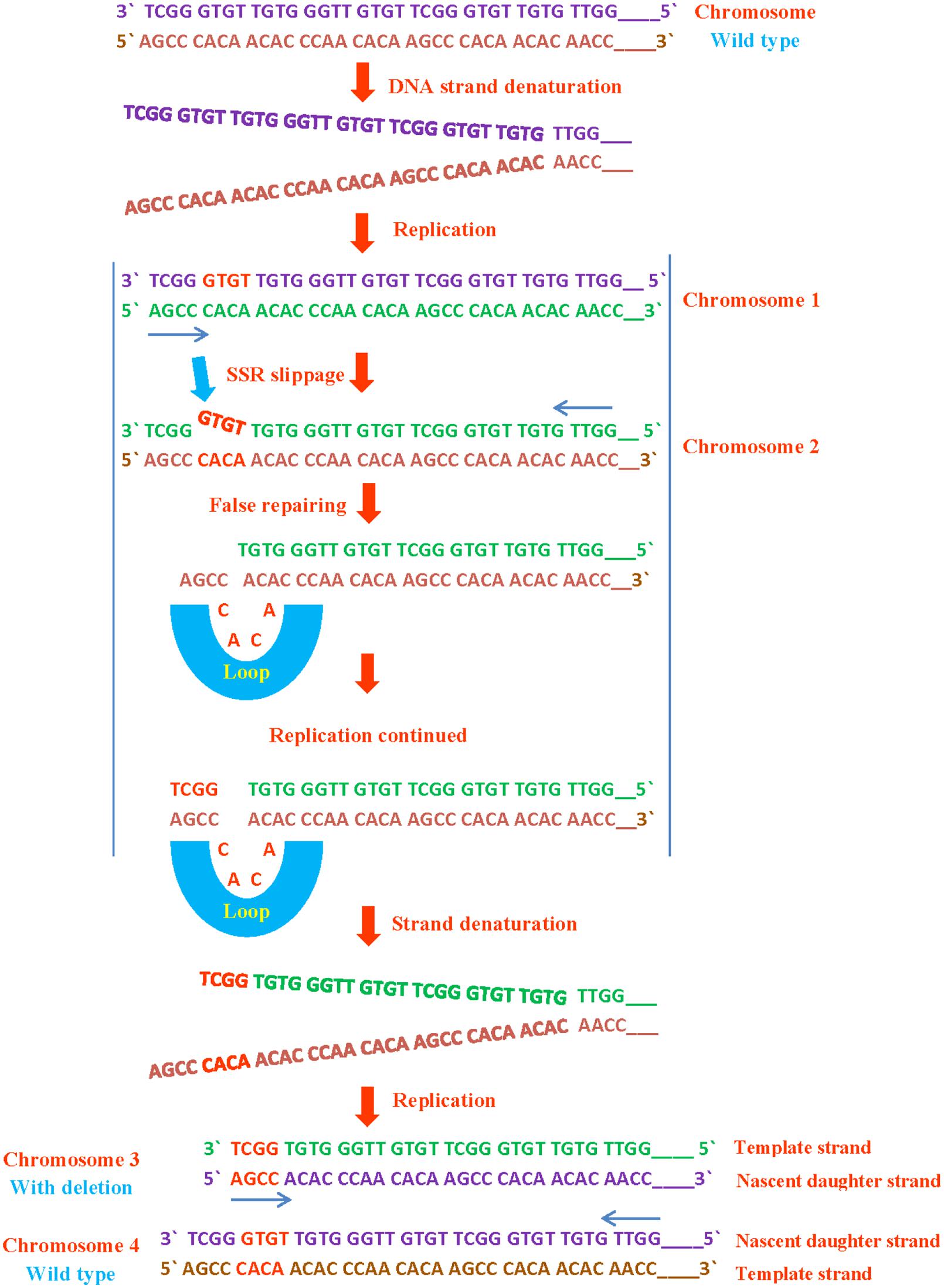
FIGURE 2. SSR deletion during DNA replication. If a SSR slips or loops out from template strand, it results in deletion. These mutations cause detrimental effects on normal protein function due to replacement of amino acid as has been seen in various microbes following genomic evolution.
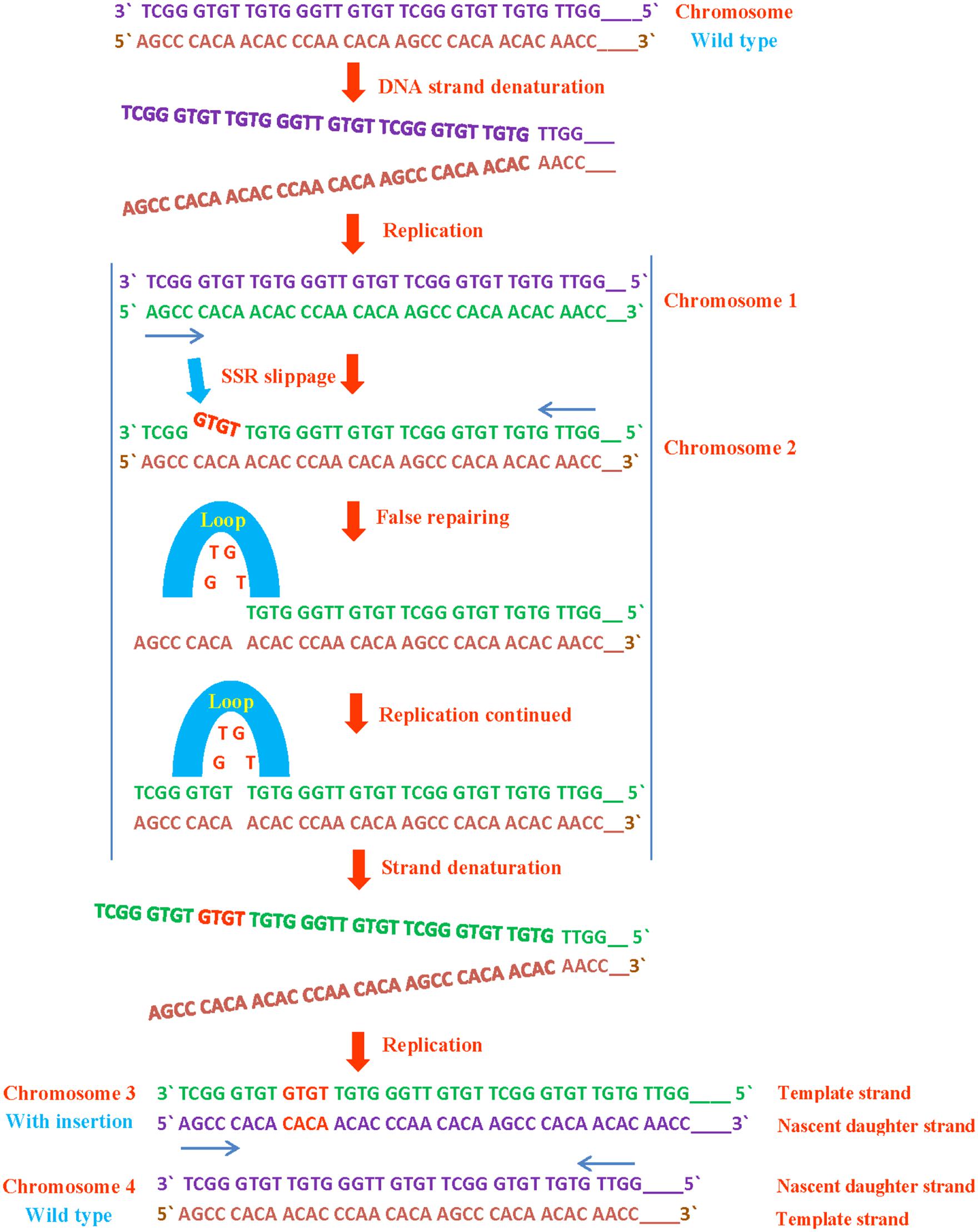
FIGURE 3. SSR insertion during DNA replication. If a SSR slips or loops out backward from template strand, it rearranges and inserted in form of duplication or at other site in template strand which mutates the normal sequence leading to the translational and translational mutations.
Microsatellites have been produced a vast number of amino acid repeat sequences in roughly 20 to 40% proteins found in various genomes (Marcotte et al., 1998). These repeats occur at protein coding sites in a genome and consist of trinucleotides (Sutherland and Richards, 1995). In yeast, these sequences are transcribed repeatedly in same amino acids such as glutamine, glutamic acid, asparagine, aspartic acid and serine affecting physical and chemical properties of the proteins. Such variations gradually modify the normal protein functions (Hancock and Santibáñez-Koref, 1998). The mutation rate measured for average microsatellite loci was 2.97 × 10-4 observed in yeast Aspergillus fumigatus. Yeast genome contains large number of microsatellites to offer targets for direct investigation (Strand et al., 1994, 1995; Séré et al., 2014).
Length mutations in FLO1 gene regulate the adhesive properties in bacterial membrane. These sequence mutations provide evolutionary modifications to the membrane surface proteins. Consequently, varying the adhesive features which assist pathogenic microbes to resist immunological changes in the hosts (Moxon et al., 1994; Verstrepen et al., 2005). Michael et al. (2008) reported length variations in microsatellites in fungus (Neurospora crassa); these variations control the time length of circadian clock cycle. Unwanted evolution induced by microsatellite deletions and indels can rapidly decline the performance of genetically engineered circuits and metabolic pathways in microbes (Jack et al., 2015).
Fimbriae formation in Haemophilus influenza is stimulated by unit mutations in microsatellite sequences by the modification of promoter spacing (Moxon et al., 1994). Microsatellites cause mutation instability in colorectal cancer infected with viruses (Rooney et al., 2014) by altering splicing or gene regulation. This includes nucleotide variations projected to cause missense swaps, small in-frame insertions/deletions or intragenic/intergenic sequence (Thompson et al., 2014; Thompson and Spurdle, 2015). SSRs have been accountable for the phase changes with the support of variation in promoter activity and gene transcription (Van Ham et al., 1993; Dawid et al., 1999; Martin et al., 2005). Oligopurine/oligopyrimidine with long tracts was discovered in bacterial genomes near regulatory regions (Holder et al., 2015).
Currently, molecular markers are very expensive for most wide-ranging applications; they have weaknesses in sequence determination, sequence information, unsuitable across species, numerous bands per reaction and misinterpretation in terms of loci and alleles (Miah et al., 2013). Due to the limited availability of genomic sequences of prokaryotic species at various genomic databases, it is not easy to analyze microsatellite sequences in vast reaches of DNA. Sometimes, microsatellite loci are not accommodating in determining the evolutionary relationships in distantly related species (Barbará et al., 2007), so in order to evaluate the occurrence of repeats for their identification and de novo characterization in individual genomes, massive degree of time duration and expensive research work is needed. The key drawback is that, microsatellites are isolated de novo from the species studied first time (Zane et al., 2002). Because of two main facts: (i) They are located in non-coding regions with higher rate of nucleotide substitution compared to the coding regions. Therefore, it is problematic to design universal primers corresponding conserved sequences. (ii) When engaging the identical primer pair, nucleotide switches within the repeats are observed between species (Clisson et al., 2000).
Therefore, study and construct of unknown microsatellites clone libraries depends on the occurrence of particular SSRs in genomes of interest (Zane et al., 2002; Selkoe and Toonen, 2006). The occurrence of microsatellites reported in various microbes employed in molecular studies is significantly different (Tóth et al., 2000). Sometimes, it has been documented that it is extremely difficult to obtain microsatellites and other repeat sequences from a particular DNA sequence (Dutech et al., 2007).
Microsatellites and their significance in determination and understanding microbial genome evolution have been established in present study. Microsatellites are important evolutionary markers which are useful in tracking SSRs length variations such as point mutations, duplications, DNA repair, and replication slippage in phyletic lineages stretched across the entire genomes. Additionally, novel SSR analysis techniques and sequencing methods are discussed in this study, which are useful for the determination of potent evolutionary markers for previously deserted microbial genomes. Microsatellite repetitions can be traced by pursuing these advanced sequences techniques and more refined research databases. This review will highlight new insights into these biologically active and significant marker tools for studying genomic evolutions in future research and will also extend further investigations on microsatellites and other sequence repeats in the field of microbiology.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
I would like to thank Prof. SW for providing ideas and encouragement that contributed to this study.
Abdelkrim, J., Robertson, B. C., Stanton, J. A. L., and Gemmell, N. J. (2009). Fast, cost-effective development of species-specific microsatellite markers by genomic sequencing. BioTechniques 46, 185–192. doi: 10.2144/000113084
Abdul-Muneer, P. M. (2014). Application of microsatellite markers in conservation genetics and fisheries management: recent advances in population structure analysis and conservation strategies. Genet. Res. Int. 2014:11. doi: 10.1155/2014/691759
Alba, M. M., Santibanez-Koref, M. F., and Hancock, J. M. (1999). Amino acid reiterations in yeast are overrepresented in particular classes of proteins and show evidence of a slippage-like mutational process. J. Mol. Evol. 49, 789–797. doi: 10.1007/PL00006601
Alba, M. M., Santibanez-Koref, M. F., and Hancock, J. M. (2001). The comparative genomics of polyglutamine repeats: extreme difference in the codon organization of repeat-encoding regions between mammals and Drosophila. J. Mol. Evol. 52, 249–259.
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Amos, W., Kosanovic, D., and Eriksson, A. (2015). Inter-allelic interactions play a major role in microsatellite evolution. Proc. R. Soc. B 282:20152125. doi: 10.1098/rspb.2015.2125
Andersen, J. C., and Mills, N. J. (2014). iMSAT: a novel approach to the development of microsatellite loci using barcoded Illumina libraries. BMC Genomics 15:858. doi: 10.1186/1471-2164-15-858
Arthofer, W., Steiner, F. M., and Schlick-Steiner, B. C. (2011). Rapid and cost-effective screening of newly identified microsatellite loci by high-resolution melting analysis. Mol. Genet. Genomics 286, 225–235. doi: 10.1007/s00438-011-0641-0
Ashley, M., and Dow, B. (1994). The use of microsatellite analysis in population biology: background, methods and potential applications. EXS 69, 185–201.
Baas Becking, L. (1934). Geobiologie of Inleiding tot de Milieukunde. Den Haag: W.P. Van Stockum & Zoon.
Bachtrog, D., Weiss, S., Zangerl, B., Brem, G., and Schlötterer, C. (1999). Distribution of dinucleotide microsatellites in the Drosophila melanogaster genome. Mol. Biol. Evol. 16, 602–610. doi: 10.1093/oxfordjournals.molbev.a026142
Barbará, T., Palma-Silva, C., Paggi, G. M., Bered, F., Fay, M. F., Lexer, C., et al. (2007). Cross species transfer of nuclear microsatellite markers: potential and limitations. Mol. Ecol. 16, 3759–3767. doi: 10.1111/j.1365-294X.2007.03439.x
Barnes, I., Cortinas, M. N., Wingfield, M. J., and Wingfield, B. D. (2008). Microsatellite markers for the red band needle blight pathogen, Dothistroma septosporum. Mol. Ecol. Resour. 8, 1026–1029. doi: 10.1111/j.1755-0998.2008.02142.x
Basharat, Z., and Yasmin, A. (2015). Survey of compound microsatellites in multiple Lactobacillus genomes. Can. J. Microbiol. 61, 898–902. doi: 10.1139/cjm-2015-0136
Batley, J., Jewell, E., and Edwards, D. (2007). Automated discovery of single nucleotide polymorphism and simple sequence repeat molecular genetic markers. Methods Mol. Biol. 406, 473–494.
Beismann, H., Barker, J. H. A., Karp, A., Speck, T., et al. (1997). AFLP analysis sheds light on distribution of two Salix species and their hybrid along a natural gradient. Mol. Ecol. 6, 989–993. doi: 10.1046/j.1365-294X.1997.00273.x
Bhargava, A., and Fuentes, F. F. (2010). Mutational dynamics of microsatellites. Mol. Biotechnol. 44, 250–266. doi: 10.1007/s12033-009-9230-4
Bidmos, F., and Bayliss, C. (2014). Genomic and global approaches to unravelling how hypermutable sequences Influence bacterial pathogenesis. Pathogens 3, 164–184. doi: 10.3390/pathogens3010164
Birky, C. W. Jr. (2001). The inheritance of genes in mitochondria and chloroplasts: laws, mechanisms, and models. Annu. Rev. Genet. 35, 125–148. doi: 10.1146/annurev.genet.35.102401.090231
Birren, B., and Lai, E. (1987). Nonmammalian Genomic Analysis A Practical Guide. San Diego, CA: Academic Press, 75–134.
Bowcock, A. M., Ruiz-Linares, A., Tomfohrde, J., Minch, E., Kidd, J. R., and Cavalli-Sforza, L. L. (1994). High resolution of human evolutionary trees with polymorphic microsatellites. Nature 368, 455–457. doi: 10.1038/368455a0
Bravo, J. P., Hoshino, A. A., Angelici, C. M. L. C. D., Lopes, C. R., and Gimenes, M. A. (2006). Transferability and use of microsatellite markers for the genetic analysis of the germplasm of some Arachis section species of the genus Arachis. Genet. Mol. Biol. 29, 516–524. doi: 10.1590/S1415-47572006000300021
Breurec, S., Guillard, B., Hem, S., Brisse, S., Dieye, F. B., Huerre, M., et al. (2011). Evolutionary history of Helicobacter pylori sequences reflect past human migrations in Southeast Asia. PLoS ONE 6:e22058. doi: 10.1371/journal.pone.0022058
Burch, C. L., Danaher, R. J., and Stein, D. C. (1997). Antigenic variation in Neisseria gonorrhoeae: production of multiple lipooligosaccharides. J. Bacteriol. 179, 982–986.
Buschiazzo, E., and Gemmell, N. J. (2006). The rise, fall and renaissance of microsatellites in eukaryotic genomes. Bioessays 28, 1040–1050. doi: 10.1002/bies.20470
Butcher, R. D., Hubbard, S. F., and Whitfield, W. G. (2000). Microsatellite frequency and size variation in the parthenogenetic parasitic wasp Venturia canescens (Gravenhorst) (Hymenoptera: Ichneumonidae). Insect Mol. Biol. 9, 375–384. doi: 10.1046/j.1365-2583.2000.00199.x
Castoe, T. A., Poole, A. W., de Koning, A. P., Jones K. L., Tomback, D. F., Oyler-McCance S. J., et al. (2012). Rapid microsatellite identification from Illumina paired-end genomic sequencing in two birds and a snake. PLoS ONE 7:e30953. doi: 10.1371/journal.pone.0030953
Chambers, K. G., and MacAvoy, E. S. (2000). Microsatellites: consensus and controversy. Comp. Biochem. Physiol. (Part B). 126, 455–476. doi: 10.1016/S0305-0491(00)00233-9
Chan, K., Baker, S., Kim, C. C., Detweiler, C. S., Dougan, G., and Falkow, S. (2003). Genomic comparison of Salmonella enterica serovars and Salmonella bongori by use of an S. enterica serovar typhimurium DNA Microarray. J. Bacteriol. 185, 553–563. doi: 10.1128/JB.185.2.553-563.2003
Chistiakov, D. A., Helleman, B., and Volckaert, F. A. M. (2006). Microsatellites and their genomic distribution, evolution, function and applications: a review with special reference to fish genetics. Aquaculture 255, 1–29. doi: 10.1016/j.aquaculture.2005.11.031
Cho, Y. G., Ishii, T., Temnykh, S., Chen, X., Lipovich, L., McCouch, S. R., et al. (2000). Diversity of microsatellites derived from genomic libraries and GenBank sequences in rice (Oriza sativa L.). Theor. Appl. Genet. 100, 713–722. doi: 10.1007/s001220051343
Clisson, I., Lathuilliere, M., and Crouau-Roy, B. (2000). Conservation and evolution of microsatellite loci in primate taxa. Am. J. Primatol. 50, 205–214. doi: 10.1002/(SICI)1098-2345(200003)50:3<205::AID-AJP3>3.0.CO;2-Y
Coltman, D. W., Bowen, W. D., and Wright, J. M. (1998). Birth weight and neonatal survival of harbour seal pups are positively correlated with genetic variation measured by microsatellites. Proc. R. Soc. Lon. Ser. B Biol. Sci. 265, 803–809. doi: 10.1098/rspb.1998.0363
Cooper, G., Amos, W., Bellamy, R., Siddiqui, M. R., Frodsham, A., Hill, A. V. S., et al. (1999). An empirical exploration of the (delta mu)2 genetic distance for 213 human microsatellite markers. Am. J. Hum. Genet. 65, 1125–1133. doi: 10.1086/302574
Cordeiro, G. M., Casu, R., McIntyre, C. L., Manners, J. M., Henry, R. J., et al. (2001). Microsatellite markers from sugarcane (Sacharum spp.) ESTs cross transferable to erianthus and sorghum. Plant Sci. 160, 1125–1133. doi: 10.1016/S0168-9452(01)00365-X
Coulson, T. N., Pemberton, J. M., Albon, S. D., Beaumont, M., Marshall, T. C., Slate, J., et al. (1998). Microsatellites reveal heterosis in red deer. Proc. R. Soc. Lon. Ser. B Biol. Sci. 267, 489–495.
Crollius, R. H., Jaillon, O., Dasilva, C., Ozouf-Costaz, C., Fischer, C., Bouneau, L., et al. (2000). Characterization and repeat analysis of the compact genome of the fresh water Pufferfish Tetraodon nigroviridis. Genome Res. 10, 939–949. doi: 10.1101/gr.10.7.939
Dagerhamn, J., Blomberg, C., Browall, S., Sjostrom, K., Morfeldt, E., and Henriques-Normark, B. (2008). Determination of accessory gene patterns predicts the same relatedness among strains of Streptococcus pneumoniae as sequencing of housekeeping genes does and represents a novel approach in molecular epidemiology. J. Clin. Microbiol. 46, 863–868. doi: 10.1128/JCM.01438-07
Dakin, E. E., and Avise, J. C. (2004). Microsatellite null alleles in parentage analysis. Heredity (Edinb.) 93, 504–509. doi: 10.1038/sj.hdy.6800545
Dallas, J. F. (1992). Estimation of microsatellite mutation rates in recombinant inbred strains of mouse. Mamm. Genome 5, 32–38.
Dawid, S., Barenkamp, S. J., and St. Geme, J. W. (1999). Variation in expression of the Haemophilus influenzae HMW adhesins: a prokaryotic system reminiscent of eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 96, 1077–1082. doi: 10.1073/pnas.96.3.1077
Dawson, D. A., Horsburgh, G. J., Küpper, C., Stewart, I. R., Ball, A. D., Durrant, K. L., et al. (2010). New methods to identify conserved microsatellite loci and develop primer sets of high cross-species utility as demonstrated for birds. Mol. Ecol. Resour. 10, 475–494. doi: 10.1111/j.1755-0998.2009.02775.x
De Bolle, X., Bayliss, C. D., Field, D., van de Ven, T., Saunders, N. J., Hood, D. W., et al. (2000). The length of a tetranucleotide repeat tract in Haemophilus influenzae determines the phase variation rate of a gene with homology to type III DNA methyltransferases. Mol. Microbiol. 35, 211–222. doi: 10.1046/j.1365-2958.2000.01701.x
Decroocq, V., Favé, M. G., Hagen, L., Bordenave, L., and Decroocq, S. (2003). Development and transferability of apricot and grape EST microsatellite markers across taxa. Theor. Appl. Genet. 106, 912–922.
Dhillon, B., Gill, N., Hamelin, R. C., and Goodwin, S. B. (2014). The landscape of transposable elements in the finished genome of the fungal wheat pathogen Mycosphaerella graminicola. BMC Genomics 15:1132. doi: 10.1186/1471-2164-15-1132
Dietrich, W. F., Miller, J., Steen, R., Merchant, M. A., Damron-Boles, D., Husain, Z., et al. (1996). A comprehensive genetic map of the mouse genome. Nature 14, 149–152. doi: 10.1038/380149a0
Dikhit, M. R., Moharana, K. C., Sahoo, B. R., Sahoo, G. C., and Das, P. (2014). LeishMicrosatDB: open source database of repeat sequences detected in six fully sequenced Leishmania genomes. Database 2014, pii: bau078. doi: 10.1093/database/bau078
Di Rienzo, A., Peterson, A. C., Garza, J. C., Valdes, A. M., Slatkin, M., and Freimer, N. B. (1994). Mutational processes of simple-sequence repeat loci in human populations. Proc. Natl. Acad. Sci. U.S.A. 91, 3166–3170. doi: 10.1073/pnas.91.8.3166
Dib, C., Faure, S., Fizames, C., Samson, D., Drouot, N., Vignal, A., et al. (1996). A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380, 152–154. doi: 10.1038/380152a0
Dorrell, N., Hinchliffe, S. J., and Wren, B. W. (2005). Comparative phylogenomics of pathogenic bacteria by microarray analysis. Curr. Opin. Microbiol. 8, 620–626. doi: 10.1016/j.mib.2005.08.012
Dorrell, N., Mangan, J. A., Laing, K. G., Hinds, J., Linton, D., Al-Ghusein, H., et al. (2011). Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 11, 1706–1715.
Dover, G. A. (1995). Slippery DNA runs on and on and on. Nat. Genet. 10, 254–256. doi: 10.1038/ng0795-254
Dutech, C., Enjalbert, J., Fournier, E., Delmotte, F., Barrès, B., Carlier, J., et al. (2007). Challenges of microsatellite isolation in fungi. Fungal Genet. Biol. 44, 933–949. doi: 10.1016/j.fgb.2007.05.003
Eisen, J. A., Kaiser, D., and Myers, R. M. (1997). Gastrogenomic delights: a movable feast. Nat. Med. 3, 1076–1078. doi: 10.1038/nm1097-1076
Ekblom, R., and Galindo, J. (2011). Applications of next generation sequencing in molecular ecology of non-model organisms. Heredity (Edinb.) 107, 15. doi: 10.1038/hdy.2010.152
Ellegren, H. (1995). Mutation rates at porcine microsatellite loci. Mamm. Genome 6, 376–377. doi: 10.1007/BF00364807
Ellegren, H. (2004). Microsatellites: simple sequences with complex evolution. Nat. Rev. Genet. 5, 435–445. doi: 10.1038/nrg1348
Eujayl, I., Sorrells, M. E., Baum, M., Wolters, P., and Powell, W. (2002). Isolation of EST-derived microsatellite markers for phenotyping the A and B genomes of wheat. Theor. Appl. Genet. 104, 399–407. doi: 10.1007/s001220100738
Excoffier, L., and Heckel, G. (2006). Computer programs for population genetics data analysis: a survival guide. Nat. Rev. Genet. 7, 745–758. doi: 10.1038/nrg1904
Fan, J. B., Chee, M. S., and Gunderson, K. L. (2006). Highly parallel genomic assays. Nat. Rev. Genet. 7, 632–644. doi: 10.1038/nrg1901
Faria, A. D., Mamani, E. M. C., Pappas, M. R., Pappas, G. J. Jr., and Gratapaglia, D. (2010). A Selected Set of EST-Derived microsatellites, polymorphic and transferable across 6 species of eucalyptus. J. Heredity 101, 512–520. doi: 10.1093/jhered/esq024
Feingold, S., Lloyd, J., Norero, N., Bonierbale, M., and Lorenzen, J. (2005). Mapping and characterization of new EST-derived microsatellites for potato (Solanum tuberosum L.). Theor. Appl. Genet. 111, 456–466. doi: 10.1007/s00122-005-2028-2
Field, D., and Wills, C. (1998). Abundant microsatellite polymorphism in Saccharomyces cerevisiae, and the different distributions of microsatellites in eight prokaryotes and S. cerevisiae, result from strong mutation pressures and a variety of selective forces. Proc. Natl. Acad. Sci. U.S.A. 95, 1647–1652. doi: 10.1073/pnas.95.4.1647
Fondon, J. W., Mele, G. M., Brezinschek, R. I., Cummings, D., Pande, A., Wren, J., et al. (1998). Computerized polymorphic marker identification: experimental validation and a predicted human polymorphism catalog. Proc. Natl. Acad. Sci. U.S.A. 95, 7514–7519. doi: 10.1073/pnas.95.13.7514
Foster, P. L., and Trimarchi, J. M. (1994). Adaptive reversion of a frameshift mutation in Escherichia coli by simple base deletions in homopolymeric runs. Science 265, 407–409. doi: 10.1126/science.8023164
Gaiotto, F. A., Bramucci, M., and Grattapaglia, D. (1997). Estimation of outcrossing rate in a breeding population of Eucalyptus urophylla with dominant RAPD and AFLP markers. Theor. Appl. Genet. 95, 842–849. doi: 10.1007/s001220050634
Gao, L. F., Jing, R. L., Huo, N. X., Li, Y., Li, X. P., Zhou, R. H., et al. (2004). One hundred and one new microsatellite loci derived from ESTs (EST-SSRs) in bread wheat. Theor. Appl. Genet. 108, 1392–1400. doi: 10.1007/s00122-003-1554-z
Gentles, A. J., and Karlin, S. (2001). Genome-scale compositional comparisons in eukaryotes. Genome Res. 11, 540–546. doi: 10.1101/gr.163101
Glaubitz, J. C. (2004). CONVERT: a user-friendly program to reformat diploid genotypic data for commonly used population genetic software packages. Mol. Ecol. Notes 4, 309–310. doi: 10.1111/j.1471-8286.2004.00597.x
Goullet, P., and Picard, B. (1989). Comparative electro- phoretic polymorphism of esterases and other enzymes in Escherichia coli. J. Gen. Microbiol. 135, 135–143.
Grimwood, J., Olinger, L., and Stephens, R. S. (2001). Expression of Chlamydia pneumoniae polymorphic membrane protein family genes. Infect. Immun. 69, 2383–2389. doi: 10.1128/IAI.69.4.2383-2389.2001
Grover, A., and Sharma, P. (2012). Tandem repetitions in transcriptomes of some Solanaceae species. Am. J. Mol. Biol. 2, 140–152. doi: 10.4236/ajmb.2012.22016
Guidot, A., Prior, P., Schoenfeld, J., Carrere, S., Genin, S., and Boucher, C. (2007). Genomic structure and phylogeny of the plant pathogen ralstonia solanacearum inferred from gene distribution analysis. J. Bacteriol. 189, 377–387. doi: 10.1128/JB.00999-06
Gupta, S., Gupta, D. S., Anjum, K. T., Pratap, A., and Kumar, J. (2013). Transferability of simple sequence repeat markers in blackgram (Vigna mungo L. Hepper). Aus. J. Crop Sci. 7, 345–353.
Gyllensten, U., Wharton, D., Josefsson, A., and Wilson, A. C. (1991). Paternal inheritance of mitochondrial DNA in mice. Nature 352, 255–257. doi: 10.1038/352255a0
Hamilton, M. B., Pincus, E. L., Di Fiore, A., and Fleischer, R. C. (1999). Universal linker and ligation procedures for construction of genomic DNA libraries enriched for microsatellites. BioTechniques 27, 500–507.
Han, B., Wang, C., Tang, Z., Ren, Y., Li, Y., Zhang, D., et al. (2015). Genome-wide analysis of microsatellite markers based on sequenced database in chinese spring wheat (Triticum aestivum L.). PLoS ONE 10:e0141540. doi: 10.1371/journal.pone.0141540
Hancock, J. M., and Santibáñez-Koref, M. F. (1998). Trinucleotide expansion diseases in the context of micro- and minisatellite evolution. EMBO J. 17, 5521–5524. doi: 10.1093/emboj/17.19.5521
Hardiman, G. (2004). Microarray platforms comparisons and contrasts. Pharmacogenomics 5, 487–502. doi: 10.1517/14622416.5.5.487
Hartenstine, M. J., Goodman, M. F., and Petruska, J. (2000). Base stacking and even/odd behavior of hairpin loops in DNA triplet repeat slippage and expansion with DNA polymerase. J. Biol. Chem. 275, 18382–18390. doi: 10.1074/jbc.275.24.18382
Hatcher, L. E., Wang, C., and Lefkowitz, J. E. (2015). Genome variability and gene content in chordopoxviruses: dependence on microsatellites. Viruses 7, 2126–2146. doi: 10.3390/v7042126
Hazen, S. P., and Kay, S. A. (2003). Gene arrays are not just for measuring gene expression. Trends Plant Sci. 8, 413–416. doi: 10.1016/S1360-1385(03)00186-9
Hempel, K., and Peakall, R. (2003). Cross-species amplification from crop soybean Glycine max provides informative microsatellite markers for the study of inbreeding wild relatives. Genome 46, 382–393. doi: 10.1139/g03-013
Hennequin, C., Thierry, A., Richard, G. F., Lecointre, G., Nguyen, H. V., Gaillardin, C., et al. (2001). Microsatellite typing as a new tool for identification of Saccharomyces cerevisiae strains. J. Clin. Microbiol. 39, 551–559. doi: 10.1128/JCM.39.2.551-559.2001
Hoffman, J. I., and Nichols, H. J. (2011). A novel approach for mining polymorphic microsatellite markers in silico. PLoS ONE 6:e23283. doi: 10.1371/journal.pone.0023283
Holder, I. T., Wagner, S., Xiong, P., Sinn, M., Frickey, T., Meyer, A., et al. (2015). Intrastrand triplex DNA repeats in bacteria: a source of genomic instability. Nucleic Acids Res. 43, 10126–10142. doi: 10.1093/nar/gkv1017
Hood, D. W., Deadman, M. E., Jennings, M. P., Bisercic, M., Fleischmann, R. D., Venter, J. C., et al. (1996). DNA repeats identify novel virulence genes in Haemophilus influenzae. Proc. Natl. Acad. Sci. U.S.A. 93, 11121–11125. doi: 10.1073/pnas.93.20.11121
Hung, M. C., and Christodoulides, M. (2013). The biology of neisseria, Adhesins. Biol. 2, 1054–1109. doi: 10.3390/biology2031054
Igboin, C. O., Griffen, A. L., and Leys, E. J. (2009). Porphyromonas gingivalis strain diversity. J. Clin. Microbiol. 47, 3073–3081. doi: 10.1128/JCM.00569-09
Inzana, T. J., Hensley, J., McQuiston, J., Lesse, A. J., Campagnari, A. A., Boyle, S. M., et al. (1997). Phase variation and conservation of lipooligosaccharide epitopes in Haemophilus somnus. Infect. Immun. 65, 4675–4681.
Israel, D. A., Salama, N., Krishna, U., Rieger, U. M., Atherton, J. C., Falkow, S., et al. (2001). Helicobacter pylori genetic diversity within the gastric niche of a single human host. Proc. Natl. Acad. Sci. U.S.A. 98, 14625–14630. doi: 10.1073/pnas.251551698
Ito, T., Smith, C. L., and Cantor, C. R. (1992). Sequence-specific DNA purification by triplex affinity capture. Proc. Natl. Acad. Sci. U.S.A. 89, 495–498. doi: 10.1073/pnas.89.2.495
Jack, B. R., Leonard, S. P., Mishler, D. M., Renda, B. A., Leon, D., Suárez, G. A., et al. (2015). Predicting the genetic stability of engineered DNA sequences with the EFM calculator. ACS Synth. Biol. 4, 939–943. doi: 10.1021/acssynbio.5b00068
Jakupciak, J. P., and Wells, R. D. (2000). Gene conversion (recombination) mediates expansions of CTG⋅CAG repeats. J. Biol. Chem. 275, 40003–40013. doi: 10.1074/jbc.M007153200
Jarne, P., and Lagoda, P. J. L. (1996). Microsatellites, from molecules to populations and back. Trends Ecol. Evolut. 11, 424–429. doi: 10.1016/0169-5347(96)10049-5
Jentzsch, V. I. M., Bagshaw, A., Buschiazzo, E., Merkel, A., and Gemmell, J. E. (2008). Evolution of Microsatellite DNA. In: Encyclopedia of Life Sciences (ELS), Chichester: John Wiley & Sons Ltd, 1–12.
Johnson, J. R., and O’Bryan, T. T. (2000). Improved repetitive- element PCR fingerprinting for resolving pathogenic and nonpathogenic phylogenetic groups within Escherichia coli. Clin. Vaccine Immunol. 7, 265–273. doi: 10.1128/CDLI.7.2.265-273.2000
Kandpal, R. P., Kandpal, G., and Weissman, S. M. (1994). Construction of libraries enriched for sequence repeats and jumping clones, and hybridization selection for region- specific markers. Proc. Natl. Acad. Sci. U.S.A. 91, 88–92. doi: 10.1073/pnas.91.1.88
Karlin, S., Mrazek, J., and Campbell, A. M. (1996). Frequent oligonucleotides and peptides of the Hemophilus influenzae genome. Nucleic Acids Res. 21, 4263–4272. doi: 10.1093/nar/24.21.4263
Karlin, S., Mrazek, J., and Campbell, A. M. (1997). Compositional biases of bacterial genomes and evolutionary implications. J. Bacteriol. 179, 3899–3913.
Kashi, Y., King, D., and Soller, M. (1997). Simple sequence repeats as a source of quantitative genetic variation. Trends Genet. 13, 74–78. doi: 10.1016/S0168-9525(97)01008-1
Katoh, H., Inoue, H., and Iwanami, T. (2015). Changes in variable number of tandem repeats in ‘Candidatus Liberibacter asiaticus’ through insect transmission. PLoS ONE 10:e0138699. doi: 10.1371/journal.pone.0138699
Katti, M. V., Ranjekar, P. K., and Gupta, V. S. (2001). Differential distribution of simple sequence repeats in eukaryotic genome sequences. Mol. Biol. Evol. 18, 1161–1167. doi: 10.1093/oxfordjournals.molbev.a003903
Katz, L., McManus, G., Snoeyenbos-West, O., Griffin, A., Pirog, K., Costas, B., et al. (2005). Reframing the ‘everything is everywhere’ debate: evidence for high gene flow and diversity in ciliate morphospecies. Aquat. Microb. Ecol. 41, 55–65. doi: 10.3354/ame041055
Kim, C. K., Lee, G. S., Mo, J. S., Bae, S. H., and Lee, T. H. (2015). Molecular marker database for efficient use in agricultural breeding programs. Bioinformation 11, 444–446. doi: 10.6026/97320630011444
Knapik, E. W., Goodman, A., Ekker, M., Chevrette, M., Delgado, J., Neuhauss, S., et al. (1998). A microsatellite genetic linkage map for zebrafish (Danio rerio). Nat. Genet. 18, 338–343. doi: 10.1038/ng0498-338
Labate, J. A. (2000). Software for population genetic analyses of molecular marker data. Crop Sci. 40, 1521–1528. doi: 10.2135/cropsci2000.4061521x
Lance, S. L., Love, C. N., Nunziata, S. O., O’Bryhim, J. R., Scott, D. E., Flynn, R. W., et al. (2013). 32 species validation of a new Illumina paired-end approach for the development of microsatellites. PLoS ONE 8:e81853. doi: 10.1371/journal.pone.0081853
Legendre, M., Pochet, N., Pak, T., and Verstrepen, K. J. (2007). Sequence-based estimation of minisatellite and microsatellite repeat variability. Genome Res. 17, 1787–1796. doi: 10.1101/gr.6554007
Levinson, G., and Gutman, G. A. (1987). Slipped strand mispairing: a major mechanism for DNA sequence evolution. Mol. Biol. Evol. 4, 203–221.
Lim, S., Notley-McRobb, L., Lim, M., and Carter, D. A. (2004). A comparison of the nature and abundance of microsatellites in 14 fungal genomes. Fungal Genet. Biol. 41, 1025–1036. doi: 10.1016/j.fgb.2004.08.004
Litt, M., and Luty, J. A. (1989). A hypervariable microsatellite revealed by in vitro amplification of a dinucleotide repeat within the cardiac muscle actin gene. Am. J. Hum. Genet. 44, 397–401.
Liu, Z. W., Biyashev, R. M., and Maroof, M. A. S. (1996). Development of simple sequence repeat DNA markers and their integration into a barley linkage map. Theor. Appl. Genet. 93, 869–876. doi: 10.1007/BF00224088
Lopez Castel, A., Cleary, J. D., and Pearson, C. E. (2010). Repeat instability as the basis for human diseases and as a potential target for therapy. Nat. Rev. Mol. Cell Biol. 11, 165–170. doi: 10.1038/nrm2854
Luikart, G., England, P. R., Tallmon, D., Jordan, S., and Taberlet, P. (2003). The power and promise of population genomics: from genotyping to genome typing. Nat. Rev. Genet. 4, 981–994. doi: 10.1038/nrg1226
Luque, C., Legal, L., Heidi, S., Gers, C., and Wink, M. (2002). ISSR (inter simple sequence repeats) as genetic markers in Noctuids (Lepidoptera). Hereditas 136, 251–253. doi: 10.1034/j.1601-5223.2002.1360312.x
Mahtani, M. M., and Willard, H. F. A. (1993). Polymorphic X-linked tetranucleotide repeat locus displaying a high rate of new mutation: implications for mechanisms of mutation at short tandem repeat loci. Hum. Molec. Genet. 2, 431–437. doi: 10.1093/hmg/2.4.431
Malausa, T., Gilles, A., Meglecz, E., Blanquart, H., Duthoy, S., Costedoat, C., et al. (2011). High-throughput microsatellite isolation through 454 GS-FLX titanium pyrosequencing of enriched DNA libraries. Mol. Ecol. Resour. 11, 638–644. doi: 10.1111/j.1755-0998.2011.02992.x
Manoukis, N. C. (2007). Formatomatic: a program for converting diploid allelic data between common formats for population genetic analysis. Mol. Ecol. Notes 7, 592–593. doi: 10.1111/j.1471-8286.2007.01784.x
Marcotte, E. M., Pellegrini, M., Yeates, T. O., and Eisenberg, D. (1998). A census of protein repeats. J. Mol. Biol. 293, 151–160. doi: 10.1006/jmbi.1999.3136
Margulies, M., Egholm, M., Altman, W. E., Attiya, S., Bader, J. S., Bemben, L. A., et al. (2005). Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380.
Martin, P., Makepeace, K., Hill, S., Hood, D., and Moxon, E. (2005). Microsatellite instability regulates transcription factor binding and gene expression. Proc. Natl. Acad. Sci. U.S.A. 102, 3800–3804. doi: 10.1073/pnas.0406805102
Maughan, P. J., Saghai Maroof M. A., Buss, G. R., and Huestis, G. M. (1996). Amplified fragment length polymorphism (AFLP) in soybean: species diversity, inheritance, and near-isogenic line analysis. Theor. Appl. Genet. 93, 392–401. doi: 10.1007/BF00223181
McCouch, S. R., Chen, X., Panaud, O., Temnykh, S., Xu, Y., Cho, Y. G., et al. (1997). Microsatellite marker development, mapping and applications in rice genetics and breeding. Plant Mol. Biol. 35, 89–99. doi: 10.1023/A:1005711431474
McCouch, S. R., Teytelman, L., Xu, Y., Lobos, K. B., Clare, K., Walton, M., et al. (2002). Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res. 9, 199–207. doi: 10.1093/dnares/9.6.199
McKusick, V. A. (1998). Mendelian Inheritance in Man. A Catalog of Human Genes and Genetic Disorders, 12th Edn. Baltimore, MD: Johns Hopkins University Press.
Megan, E., MacDonald, A., and Sullivan, A. B. (2014). The past, present, and future of human centromere genomics. Genes (Basel) 33, 50.
Merritt, B. J., Culley, T. M., Avanesyan, A., Stokes, R., and Brzyski, J. (2015). An empirical review: characteristics of plant microsatellite markers that confer higher levels of genetic variation. Appl. Plant Sci. 3:1500025. doi: 10.3732/apps.1500025
Messier, M., Li, S. H., and Stewart, C. B. (1996). The birth of microsatellites. Nature 381:483. doi: 10.1038/381483a0
Metzgar, D., Bytof, J., and Wills, C. (2000). Selection against frameshift mutations limits microsatellite expansion in coding DNA. Genome Res. 10, 7–80.
Metzgar, D., Thomas, E., Davis, C., Field, D., and Wills, C. (2001). The microsatellites of Escherichia coli: rapidly evolving repetitive DNAs in a non-pathogenic prokaryote. Mol. Microbiol. 39, 183–190. doi: 10.1046/j.1365-2958.2001.02245.x
Miah, G., Rafii, M. Y., Ismail, M. R., Puteh, A. B., Rahim, H. A., Latif, M. A., et al. (2013). A review of microsatellite markers and their applications in rice breeding programs to improve blast disease resistance. Int. J. Mol. Sci. 14, 22499–22528. doi: 10.3390/ijms141122499
Michael, T. P., Park, S., Kim, T. S., Booth, J., Byer, A., Sun, Q., et al. (2008). Redfield, Rosemary, ed. “Simple sequence repeats provide a substrate for phenotypic variation in the Neurospora crassa circadian clock”. PLoS ONE 2:e795. doi: 10.1371/journal.pone.0000795
Mikheyev, A. S., Vo, T., Wee, B., Singer, M. C., and Parmesan, C. (2010). Rapid Microsatellite isolation from a butterfly by de novo transcriptome sequencing: performance and a comparison with AFLP-derived distances. PLoS ONE 5:e11212. doi: 10.1371/journal.pone.0011212
Mittal, N., and Dubey, A. K. (2009). Microsatellite markers - A new practice of DNA based markers in molecular genetics. Pharmacogn. Rev. 3, 235–246.
Morgante, M., Hanafey, M., and Powell, W. (2002). Microsatellites are preferentially associated with nonrepetitive DNA in plant genome. Nat. Genet. 30, 194–200. doi: 10.1038/ng822
Moxon, E., Rainey, P., Nowak, M., and Lenski, R. (1994). Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr. Biol. 4, 24–33. doi: 10.1016/S0960-9822(00)00005-1
Mrázek, J., Guo, X., and Shah, A. (2007). Simple sequence repeats in prokaryotic genomes. Proc. Natl. Acad. Sci. U.S.A. 104, 8472–8477. doi: 10.1073/pnas.0702412104
Mullis, K., and Faloona, F. (1987). Specific synthesis of DNA in vitro via a polymerase catalyzed chain reaction. Methods Enzymol. 155, 335–350. doi: 10.1016/0076-6879(87)55023-6
Nascimento,É, Martinez, R., and Rodrigues Lopes, A. (2004). Detection and Selection of microsatellites in the genome of Paracoccidioides brasiliensis as molecular markers for clinical and epidemiological studies. J. Clin. Microbiol. 42, 5007–5014. doi: 10.1128/JCM.42.11.5007-5014.2004
Nybom, H., Weising, K., and Rotter, B. (2014). DNA fingerprinting in botany: past, present, future. Investig. Genet. 5:1. doi: 10.1186/2041-2223-5-1
Ochman, H., and Selander, R. K. (1984). Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157, 690–693.
Paetkau, D. (1999). Microsatellites obtained using strand extension: an enriched protocol. Biotechniques 26, 690–697.
Paglia, G., and Morgante, M. (1998). PCR-based multiplex DNA fingerprinting techniques for the analysis of conifer genomes. Mol. Breed. 4, 173–177. doi: 10.1023/A:1009637608702
Pandian, A., Ford, R., and Taylor, P. W. J. (2000). Transferability of sequence tagged microsatellite site (STMS) primers across four major pulses. Plant Mol. Biol. Reptr. 18, 395. doi: 10.1007/BF02825069
Park, Y. J., Lee, J. K., and Kim, N. S. (2009). Simple sequence repeat polymorphisms (SSRPs) for evaluation of molecular diversity and germplasm classification of minor crops. Molecules 14, 4546–4569. doi: 10.3390/molecules14114546
Peak, I. R. A., Jennings, M. P., Hood, D. W., Bisercic, M., and Moxon, E. R. (1996). Tetrameric repeat units associated with virulence factor phase variation in Hemophilus also occur in Neiserria spp. and Moraxella catarrhalis. FEMS Microbiol. Lett. 137, 109–114. doi: 10.1111/j.1574-6968.1996.tb08091.x
Peakall, R., Gilmore, S., Keys, W., Morgante, M., and Rafalski, A. (1998). Cross-species amplification of soybean (Glycine max) simple sequence repeats (SSRs) within the genus and other legume genera: implications for the transferability of SSRs in plants. Mol. Biol. Evol. 15, 1275–1287. doi: 10.1093/oxfordjournals.molbev.a025856
Pépin, L., Amigues, Y., Lépingle, A., Berthier, J. L., Bensaid, A., and Vaiman, D. (1995). Sequence conservation of microsatellites between Bos taurus (cattle), Capra hircus (goat) and related species. Examples of use in parentage testing and phylogeny analysis. Heredity 53:61.
Pérez, F., Ortiz, J., Zhinaula, M., Gonzabay, C., Calderón, J., and Volckaert, F. (2005). Development of EST-SSR markers by data mining in three species of shrimp: Litopenaeus vannamei, Litopenaeus stylirostris, and Trachypenaeus birdy. Mar. Biotechnol. 7, 554–569. doi: 10.1007/s10126-004-5099-1
Pfunder, M., and Frey, J. E. (2006). Isolation of microsatellite markers for Contarinia nasturtii, a European pest invading the New World. Mol. Ecol. Notes 6, 191–193. doi: 10.1111/j.1471-8286.2005.01189.x
Poczai, P., Varga, I., Laos, M., Cseh, A., Bell, N., Valkonen, J. P., et al. (2013). Advances in plant gene-targeted and functional markers: a review. Plant Methods 9:6. doi: 10.1186/1746-4811-9-6
Powell, W., Machray, G. C., and Provan, J. (1996a). Polymorphism revealed by simple sequence repeats. Trends Plant Sci. 1, 215–222. doi: 10.1016/S1360-1385(96)86898-0
Powell, W., Michele, M., Chaz, A., Michael, H., Julie, V., Scott, T., et al. (1996b). The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol. Breed. 2, 225–238. doi: 10.1007/BF00564200
Powell, W., Morgante, M., Andre, C., McNicol, J. W., Machray, G. C., Doyle, J. J., et al. (1995). Hypervariable microsatellites provide a general source of polymorphic DNA markers for the chloroplast genome. Curr. Biol. 5, 1023–1029. doi: 10.1016/S0960-9822(95)00206-5
Primmer, C. R., and Ellegren, H. (1998). Patterns of molecular evolution in avian microsatellites. Mol. Biol. Evol. 15, 997–1008. doi: 10.1093/oxfordjournals.molbev.a026015
Priyono, and Putranto, R. A. (2014). Molecular markers and their application for DNA fingerprinting and genetic diversity studies in Coffea species. Menara Perkebunan 82, 39–50.
Queller, D. C., Strassmann, J. E., and Hughes, C. R. (1993). Microsatellites and kinship. Trends Ecol. Evol. 8, 285–288. doi: 10.1016/0169-5347(93)90256-O
Rabello, E., de Souza, A. N., Adriane, N. D. S., Daniel, T., and Siu, M. (2005). In silico characterization of microsatellites in Eucalyptus spp.: abundance, length variation and transposon associations. Genet. Mol. Biol. 28, 582–588. doi: 10.1590/S1415-47572005000400013
Rajendrakumar, P., Biswal, A. K., Balachandran, S. M., Srinivasarao, K., and Sundaram, R. M. (2007). Simple sequence repeats in organellar genomes of rice: frequency and distribution in genic and intergenic regions. Bioinformatics 23, 1–4. doi: 10.1093/bioinformatics/btl547
Rasmussen, T. B., Danielsen, M., Valina, O., Garrigues, C., Johansen, E., and Pedersen, M. B. (2008). Streptococcus thermophilus core genome: comparative genome hybridization study of 47 Strains. Appl. Environ. Microbiol. 74, 4703–4710. doi: 10.1128/AEM.00132-08
Rico, C., Rico, I., and Hewitt, G. (1996). 470 million years of conservation of microsatellite loci among fish species. Proc. Biol. Sci. 263, 549–557. doi: 10.1098/rspb.1996.0083
Rocha, E. P. C., and Blanchard, A. (2002). Genomic repeats, genome plasticity and the dynamics of Mycoplasma evolution. Nucleic Acids Res. 30, 2031–2042. doi: 10.1093/nar/30.9.2031
Rooney, S. M., Sachet, S. A., Wu, C. J., Getz, G., Hacohen, N., et al. (2014). Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160, 48–61. doi: 10.1016/j.cell.2014.12.033
Rosenberg, S. M., Longerich, S., Gee, P., and Harris, R. S. (1994). Adaptive mutation by deletions in small mononucleotide repeats. Science 265, 405–407. doi: 10.1126/science.8023163
Rothberg, J. M., and Leamon, J. (2008). The development and impact of 454 sequencing. Nat. Biotechnol. 26, 1117–1124. doi: 10.1038/nbt1485
Russell, J. R., Fuller, J. D., Macaulay, M., Hatz, B. G., Jahoor, A., Powell, W., et al. (1997). Direct comparison of levels of genetic variation among barley accessions detected by RFLPs, AFLPs, SSRs and RAPDs. Theor. Appl. Genet. 95, 714–722. doi: 10.1007/s001220050617
Saiki, R. K., Scharf, S. J., Fallona, F., Mullis, K. B., Horn, G. T., Erlich, H. A., et al. (1985). Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 230, 1350–1354. doi: 10.1126/science.2999980
Sand, L. G. L., Szuhai, K., and Hogendoorn, P. C. W. (2015). Sequencing Overview of ewing sarcoma: a journey across genomic, epigenomic and transcriptomic landscapes. Int. J. Mol. Sci. 16, 16176–16215. doi: 10.3390/ijms160716176
Santana, C. Q., Coetzee, A. P. M., Steenkamp, T. E., Mlonyeni, X. O., Hammond, A. N. G., Wingfield, J. M., et al. (2009). Microsatellite discovery by deep sequencing of enriched genomic libraries. Biotechniques 46, 217–223. doi: 10.2144/000113085
Schlotterer, C. (2000). Evolutionary dynamics of microsatellite DNA. Chromosoma 109, 365–371. doi: 10.1007/s004120000089
Schuler, G. D., Boguski, M. S., Stewart, E. A., Stein, L. D., Gyapay, G., Rice, K., et al. (1996). A gene map of the human genome. Science 274, 540–546. doi: 10.1126/science.274.5287.540
Scott, K., Eggler, P., Seaton, G., Rossetto, M., Ablet, E., Lee, L., et al. (2000). Analysis of SSRs derived from grape ESTs. Theor. Appl. Genet. 100, 723–726. doi: 10.1007/s001220051344
Sébastien, L., Rivals, E., and Philippe, J. (2010). DNA slippage occurs at microsatellite loci without minimal threshold length in humans: a comparative genomic approach. Genome Biol. Evol. 2, 325–335. doi: 10.1093/gbe/evq023
Selkoe, K. A., and Toonen, R. J. (2006). Microsatellites for ecologists: a practical guide to using and evaluating microsatellite markers. Ecol. Lett. 9, 615–629. doi: 10.1111/j.1461-0248.2006.00889.x
Semblat, J. P., Wajnberg, E., Dalmasso, A., Abad, P., and Castagnone-Sereno, P. (1998). High-resolution DNA fingerprinting of parthenogenetic root-knot nematodes using AFLP analysis. Mol. Ecol. 7, 119–125. doi: 10.1046/j.1365-294x.1998.00326.x
Séré, M., Kaboré, J., Jamonneau, V., Marie G., Belem, A., Ayala, J. F., et al. (2014). Null allele, allelic dropouts or rare sex detection in clonal organisms: simulations and application to real data sets of pathogenic microbes. Parasites Vect. 7:331. doi: 10.1016/j.meegid.2013.08.006
Shen, X., Liu, Z. Q., Mocoeur, A., Xia, Y., and Jing, H. C. (2015). PAV markers in Sorghum bicolour: genome pattern, affected genes and pathways, and genetic linkage map construction. TAG. Theor. Appl. Genet. 128, 623–637. doi: 10.1007/s00122-015-2458-4
Shitara, H., Hayashi, J. I., Takahama, S., Kaneda, H., and Yonekawa, H. (1998). Maternal inheritance of mouse mtDNA in interspecific hybrids: segregation of leaked paternal mtDNA followed by the prevention of subsequent paternal leakage. Genetics 148:857.
Smulders, M. J. M., Vukosavljev, M., Shahin, A., van de Weg, W. E., and Arens, P. (2012). High throughput marker development and application in horticultural crops. Acta Hortic. (ISHS) 961, 547–551.
Solheim, M., Aakra, A., Snipen, L., Brede, D., and Nes, I. (2009). Comparative genomics of Enterococcus faecalis from healthy Norwegian infants. BMC Genomics 10:194. doi: 10.1186/1471-2164-10-194
Soranzo, N., Provan, J., and Powell, W. (1999). An example of microsatellite length variation in the mitochondrial genome of conifers. Genome 42, 158–161. doi: 10.1139/g98-111
Squirrell, J., Hollingsworth, P. M., Woodhead, M., Russell, J., Lowe, A. J., Gibby, M., et al. (2003). How much effort is required to isolate nuclear microsatellites from plants? Mol. Ecol. 12:1348. doi: 10.1046/j.1365-294X.2003.01825.x
Strand, M., Earley, M. C., Crouse, G. F., and Petes, T. D. (1995). Mutations in the MSH3 gene preferentially lead to deletions within tracts of simple repetitive DNA in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 92, 10418–10421. doi: 10.1073/pnas.92.22.10418
Strand, M., Prolla, T., Liskay, R., and Petes, T. (1994). Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature (London) 365, 274–276. doi: 10.1038/365274a0
Strauss, S. J., and Falkow, S. (1997). Microbial pathogenesis: genomics and beyond. Science 276, 707–711. doi: 10.1126/science.276.5313.707
Sunnucks, P. (2000). Efficient genetic markers for population biology. Trends Ecol. Evol. 15, 199–203. doi: 10.1016/S0169-5347(00)01825-5
Sutherland, G. R., and Richards, R. I. (1995). Simple tandem DNA repeats and human genetic disease. Proc. Natl. Acad. Sci. U.S.A. 92, 3636–3641. doi: 10.1073/pnas.92.9.3636
Tautz, D. (1993). “Notes on the definition and nomenclature of tandemly repetitive DNA sequences,” in DNA Fingerprinting: State of the Science, eds S. D. J. Pena, R. Chakraborty, J. T. Epplen, and A. J. Jeffreys (Basel: Birkhaiiser Verlag), 21–28.
Tautz, D., and Schlötterer, C. (1994). Simple sequences. Curr. Opin. Genet. Dev. 4, 832–837. doi: 10.1016/0959-437X(94)90067-1
Temnykh, S., DeClerck, G., Lukashova, A., Lipovich, L., Cartinhour, S., and McCouch, S. (2001). Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res. 11, 1441–1452. doi: 10.1101/gr.184001
Thompson, B. A., and Spurdle, A. B. (2015). Microsatellite Instability use in mismatch repair gene sequence variant classification. Genes (Basel) 6, 150–162. doi: 10.3390/genes6020150
Thompson, B. A., Spurdle, A. B., Plazzer, J. P., Greenblatt, M. S., Akagi, K., Al-Mulla, F., et al. (2014). Application of a 5-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants in the InSiGHT locus-specific database. Nat. Genet. 46, 107–115. doi: 10.1038/ng.2854
Tomb, J., White, O., Kerlavage, A. R., Clayton, R. A., and Sutton, G. G. (1997). The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature (London) 388, 539–547. doi: 10.1038/41483
Tóth, G., Gáspari, Z., and Jurka, J. (2000). Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res. 10, 967–981. doi: 10.1101/gr.10.7.967
Ussery, D. W., Binnewies, T. T., Gouveia-Oliveira, R., Jarmer, H., and Hallin, P. F. (2004). Genome update: DNA repeats in bacterial genomes. Microbiology 150, 3519–3521. doi: 10.1099/mic.0.27628-0
Van Der Nest, M. A., Emma, S., Brenda, D., and Wingfield, M. J. (2000). Development of simple sequence repeat (SSR) markers in Eucalyptus from amplified inter simple sequence repeats (ISSR). Plant Breed. 119, 433–436. doi: 10.1046/j.1439-0523.2000.00515.x
Van Ham, S., Van Alphen, L., Mooi, F., and Van Putten, J. (1993). Phase variation of H. influenzae fimbriae: transcriptional control of two divergent genes through a variable combined promoter region. Cell 73, 1187–1196. doi: 10.1016/0092-8674(93)90647-9
Varshney, R. K., Graner, A., and Sorrells, M. E. (2005). Genic microsatellite markers in plants: features and applications. Trends Biotechnol. 23, 48–55. doi: 10.1016/j.tibtech.2004.11.005
Varshney, R. K., Thiel, T., Stein, N., Langridge, P., and Graner, A. (2002). In silico analysis on frequency and distribution of microsatellites in ESTs of some cereal species. Cell Mol. Biol. Lett. 7, 537–546.
Verstrepen, K. J., Jansen, A., Lewitter, F., and Fink, G. R. (2005). Intragenic tandem repeats generate functional variability. Nat. Gen. 37, 986–990. doi: 10.1038/ng1618
Vijay, L., Ghori, P. I. S., Harshul, D., Parikh, M. G., Upadhyay, P. D., and Vaghela, D. (2015). Genetic markers in plant: conceptions, types and its medicinal and breeding application. Int. J. Pharm. Res. Bio-Sci. 4, 111–128.
Vogiatzi, E., Lagnel, J., Pakaki, V., Louro, B., Canario, A. V., Reinhardt, R., et al. (2011). In silico mining and characterization of simple sequence repeats from gilthead sea bream (Sparus aurata) expressed sequence tags (EST-SSRs); PCR amplification, polymorphism evaluation and multiplexing and cross-species assays. Mar. Genomics 4, 83–91. doi: 10.1016/j.margen.2011.01.003
Vos, P., Hogers, R., Bleeker, M., Reijans, M., van de Lee, T., Hornes, M., et al. (1995). A FLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23, 4407–4414. doi: 10.1093/nar/23.21.4407
Vukosavljev, M., Esselink, G. D., Van’t Westende, W. P., Cox, P., Visser, R. G., Arens, P., et al. (2015). Efficient development of highly polymorphic microsatellite markers based on polymorphic repeats in transcriptome sequences of multiple individuals. Mol. Ecol. Resour. 15, 17–27. doi: 10.1111/1755-0998.12289
Wan, Y., Broschat, S. L., and Call, D. R. (2007). Validation of mixed-genome microarrays as a method for genetic discrimination. Appl. Environ. Microbiol. 73, 1425–1432. doi: 10.1128/AEM.01647-06
Wang, L., Wang, Z., Chen, J., Liu, C., Zhu, W., Wang, L., et al. (2015). De Novo transcriptome assembly and development of novel microsatellite markers for the traditional chinese medicinal herb, Veratrilla baillonii Franch (Gentianaceae). Evol. Bioinform. 11, 39–45. doi: 10.4137/EBO.S20942
Weber, J. L., and Wong, C. (1993). Mutation of human short tandem repeats. Hum. Mol. Genet. 2, 1123–1128.
Weising, K., Nybom, H., Wolff, K., and Kahl, G. (2005). DNA Fingerprinting in Plants: Principles, Methods and Applications. Boca Raton: CRC Press.
Wells, R. D., Dere, R., Hebert, M. L., Napierala, M., and Son, L. S. (2005). Advances in mechanisms of genetic instability related to hereditary neurological diseases. Nucleic Acids Res. 33, 3785–3798. doi: 10.1093/nar/gki697
Weng, Y., Azhaguvel, P., Michels, G. J., and Rudd, J. C. (2007). Cross-species transferability of microsatellite markers from six aphid (Hemiptera: Aphididae) species and their use for evaluating biotypic diversity in two cereal aphids. Insect Mol. Biol. 16, 613–622.
Wheeler, G. L., Dorman, H. E., Buchanan, A., Challagundla, L., and Wallace, L. E. (2014). A review of the prevalence, utility, and caveats of using chloroplast simple sequence repeats for studies of plant biology. Appl. Plant Sci. 2:1400059. doi: 10.3732/apps.1400059
Wilder, J., and Hollocher, H. (2001). Mobile elements and the genesis of microsatellites in dipterans. Mol. Biol. Evol. 18, 384–392. doi: 10.1093/oxfordjournals.molbev.a003814
Witsenboer, H., Vogel, J., and Michelmore, R. W. (1997). Identification, genetic localization, and allelic diversity of selectively amplified microsatellite polymorphic loci in lettuce and wild relatives (Lactuca spp.). Genome 40, 923–936. doi: 10.1139/g97-119
Wolfgang, M. C., Kulasekara, B. R., Liang, X., Boyd, D., Wu, K., Yang, Q., et al. (2003). Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 100, 8484–8489. doi: 10.1073/pnas.0832438100
Xu, J. S., Wu, Y. T., Ye, S. J., Wang, L., and Feng, Y. Z. (2012). SSR primer screening and assessment on pear germplasm resources. J. Central South Univ. Forest. Technol. 32, 80–85.
Yang, H., Tao, Y., Zheng, Z., Li, C., Sweetingham, M. W., and Howieson, J. G. (2012). Application of next-generation sequencing for rapid marker development in molecular plant breeding: a case study on anthracnose disease resistance in Lupinus angustifolius L. BMC Genomics 13:318. doi: 10.1186/1471-2164-13-318
Yang, Y., and Smith, S. A. (2013). Optimizing de novo assembly of short-read RNA-seq data for phylogenomics. BMC Genomics 14:328. doi: 10.1186/1471-2164-14-328
Zane, L., Bargelloni, L., and Patarnello, T. (2002). Strategies for microsatellite isolation: a review. Mol. Ecol. 11, 1–16.
Zhang, D. X., and Hewitt, G. M. (2003). Nuclear DNA analyses in genetic studies of population: practice, problems, and prospects. Mol. Ecol. 12, 563–584. doi: 10.1046/j.1365-294X.2003.01773.x
Zhao, H., Yang, L., Peng, Z., Sun, H., Yue, X., Lou, Y., et al. (2015). Developing genome-wide microsatellite markers of bamboo and their applications on molecular marker assisted taxonomy for accessions in the genus Phyllostachys. Sci. Rep. 5:8018. doi: 10.1038/srep08018
Zhu, Y., Strassmann, J. E., and Queller, D. (2000). Insertions substitutions and origin of microsatellites. Genome Res. 76, 227–236.
Zietkiewicz, E., Rafalski, A., and Labuda, D. (1994). Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 20, 176–183. doi: 10.1006/geno.1994.1151
Keywords: microsatellites, short sequence repeats, DNA sequences, genetics, evolution, molecular, microbiology
Citation: Saeed AF, Wang R and Wang S (2016) Microsatellites in Pursuit of Microbial Genome Evolution. Front. Microbiol. 6:1462. doi: 10.3389/fmicb.2015.01462
Received: 11 April 2015; Accepted: 07 December 2015;
Published: 05 January 2016.
Edited by:
John R. Battista, Louisiana State University and A & M College, USAReviewed by:
Abd El-Latif Hesham, Assiut University, EgyptCopyright © 2016 Saeed, Wang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Abdullah F. Saeed, YWJkdWxsYWhmYXJoYW5AaG90bWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.