- 1Faulty of Science, Institute of Biological Sciences, Universiti Malaya, Kuala Lumpur, Malaysia
- 2Faculty of Medicine, Biomedical Research Centre, Universiti Sultan Zainal Abidin, Kuala Terengganu, Malaysia
- 3Faculty of Bioresources and Food Industry, Universiti Sultan Zainal Abidin, Kuala Terengganu, Malaysia
Acinetobacter baumannii is a Gram-negative nosocomial pathogen of importance due to its uncanny ability to acquire resistance to most antimicrobials. These include carbapenems, which are the drugs of choice for treating A. baumannii infections, and polymyxins, the drugs of last resort. Whole genome sequencing was performed on two clinical carbapenem-resistant A. baumannii AC29 and AC30 strains which had an indistinguishable ApaI pulsotype but different susceptibilities to polymyxin. Both genomes consisted of an approximately 3.8 Mbp circular chromosome each and several plasmids. AC29 (susceptible to polymyxin) and AC30 (resistant to polymyxin) belonged to the ST195 lineage and are phylogenetically clustered under the International Clone II (IC-II) group. An AbaR4-type resistance island (RI) interrupted the comM gene in the chromosomes of both strains and contained the blaOXA−23 carbapenemase gene and determinants for tetracycline and streptomycin resistance. AC29 harbored another copy of blaOXA−23 in a large (~74 kb) conjugative plasmid, pAC29b, but this gene was absent in a similar plasmid (pAC30c) found in AC30. A 7 kb Tn1548::armA RI which encodes determinants for aminoglycoside and macrolide resistance, is chromosomally-located in AC29 but found in a 16 kb plasmid in AC30, pAC30b. Analysis of known determinants for polymyxin resistance in AC30 showed mutations in the pmrA gene encoding the response regulator of the two-component pmrAB signal transduction system as well as in the lpxD, lpxC, and lpsB genes that encode enzymes involved in the biosynthesis of lipopolysaccharide (LPS). Experimental evidence indicated that impairment of LPS along with overexpression of pmrAB may have contributed to the development of polymyxin resistance in AC30. Cloning of a novel variant of the blaAmpC gene from AC29 and AC30, and its subsequent expression in E. coli also indicated its likely function as an extended-spectrum cephalosporinase.
Introduction
Acinetobacter baumannii is a significant nosocomial pathogen that infects immunocompromised patients, including patients with underlying diseases, especially those who are warded in intensive care units (ICU; Bergogne-Bérézin and Towner, 1996; Nordmann and Poirel, 2008; Camp and Tatum, 2010). These nosocomial infections include ventilator-associated pneumonia, secondary meningitis, endocarditis, urinary tract infections, surgical site infections, and blood stream infections (Camp and Tatum, 2010; Huang et al., 2012). A. baumannii is intrinsically resistant to commonly used antibiotics such as aminopenicillins, first- and second-generation cephalosporins and chloramphenicol (Dijkshoorn et al., 2007). The notoriety of this pathogen stems from its ability to develop and acquire resistance to almost all antimicrobial drugs as well as its tolerance to desiccation and ability to survive on inanimate surfaces for prolonged periods of time (Camp and Tatum, 2010; Roca et al., 2012; Doi et al., 2015). In depth analyses of the resistance mechanisms in A. baumannii revealed that its multidrug resistance phenotype is mediated by all the major resistance mechanisms that are known to occur in bacteria, including modification of target sites, enzymatic inactivation, active efflux, and decreased influx of drugs (Dijkshoorn et al., 2007).
Carbapenem resistance in A. baumannii is usually acquired with the most significant mechanism being the production of carbapenemases (Poirel and Norman, 2006). A. baumannii naturally produces the OXA-51-group carbapenemase at a low level. The transposition of an insertion sequence (usually ISAba1 or ISAba9) upstream of the blaOXA−51 gene provides a strong promoter for expression of the carbapenemase leading to elevation of carbapenem MICs and thus, resistance (Higgins et al., 2010). A. baumannii also readily acquires several OXA-group β-lactamases usually through transposons and plasmids with OXA-23 being the most prevalent (Poirel and Norman, 2006; Roca et al., 2012).
A. baumannii naturally produces the AmpC-type β-lactamase and overexpression of the blaAmpC gene has been associated with the presence of ISAba1 providing a strong promoter leading to cephalosporin resistance (Segal et al., 2005; Tian et al., 2011). These Acinetobacter-derived cephalosporinases (ADCs) typically hydrolyze penicillins, narrow- and extended-spectrum cephalosporins but not cefepime or carbapenems (Rodríguez-Martínez et al., 2010). Recently, several ADC variants have been identified that are capable of hydrolyzing cefepime and these are termed as extended-spectrum AmpCs (ESACs) (Rodríguez-Martínez et al., 2010; Tian et al., 2011).
It is of great concern when carbapenems, which are the drugs of choice for treatment, are increasingly compromised. As the efficacy of these drugs decreases, polymyxins were re-introduced as the drug of “last resort” (Landman et al., 2008). The renewed interest in polymyxins (polymyxin B and colistin, or polymyxin E) as therapeutic agents was due to the pathogen's outer membrane, polyanionic lipopolysaccharide (LPS) having high affinity toward the cationic polymyxin molecules (Landman et al., 2008). Binding of the LPS and polymyxin molecules resulted in a detergent-like effect that disrupts the outer membrane thereby causing cytoplasm leakage of the pathogen (Landman et al., 2008; Arroyo et al., 2011). Although polymyxins were once discarded due to their neurotoxicity and nephrotoxicity, polymyxins have a more effective antibacterial activity against most Gram negative pathogens, including A. baumannii (Landman et al., 2008). Previous studies from Garnacho-Montero et al. (2003) and Moffatt et al. (2010) showed that intravenous polymyxins were safe to use as an effective treatment to Acinetobacter infections. However, uncontrolled use or overuse of polymyxins in the hospital environment may lead to the development of polymyxin-resistance in A. baumannii (Arroyo et al., 2011).
Polymyxin resistance in A. baumannii appeared to develop intrinsically as a result of exposure to this class of drugs. Two major mechanisms of polymyxin resistance have been described for A. baumannii. The first is the modification of the lipid A moiety of LPS with phosphoethanolamine as a result of mutations in the pmrA/pmrB two-component signal transduction system which leads to the up-regulated expression of the pmrCAB operon. Overexpression of pmrC which encodes the enzyme responsible for phosphoethanolamine addition to lipid A, impairs the binding of polymyxin to the outer membrane thereby leading to resistance (Adams et al., 2009; Arroyo et al., 2011; Beceiro et al., 2011; Park et al., 2011). The second mechanism is the complete loss of the LPS caused either by mutations or the insertional inactivation of the lipid A biosynthesis genes, namely lpxA, lpxC, and lpxD (Moffatt et al., 2010, 2011). Mutations in the lpsB gene that encodes a glycosyltransferase involved in the biosynthesis of the LPS core have also been implicated in polymyxin resistance (Hood et al., 2013).
We have previously characterized 54 A. baumannii strains obtained from a tertiary hospital in Terengganu, Malaysia (Lean et al., 2014). Out of these, 39 were carbapenem- and multidrug-resistant (MDR). Among the 39 carbapenem resistant strains, 14 were also resistant to polymyxin B and categorized as extensive-drug resistant (XDR). Two strains, A. baumannii AC29 and AC30 were isolated from the wounds of different patients, and shared an identical ApaI pulsotype. However, AC29 is susceptible to polymyxin B whereas AC30 was resistant with an MIC value of 128 μg/ml. Here, we report the comparative genomic analyses of these two A. baumannii strains, AC29 and AC30, to show that despite sharing an identical pulsotype, there are significant changes in the genome structure particularly in the resistance islands and the plasmid content of the two isolates. We also present experimental evidence to elucidate possible mechanisms for the development of polymyxin resistance in A. baumannii AC30 and the likely implication of a novel blaAmpC cephalosporinase gene in resistance to extended-spectrum cephalosporins as well as imipenem in both AC29 and AC30.
Materials and Methods
Whole Genome Sequencing
Strains Selection, Antibiotic Resistance Profiles, and DNA Extraction
Two strains of A. baumannii AC29 and AC30 from a tertiary hospital in Terengganu, Malaysia were selected for this study (Lean et al., 2014). Both strains were obtained from the wounds of different patients using standard microbiology procedures. A. baumannii AC29 and AC30 shared an identical ApaI pulsotype and showed resistance to gentamycin, tobramycin, amikacin, ciprofloxacin, levofloxacin, piperacillin-tazobactam, cefotaxime, ceftazidime, cefepime, ampicillin-sulbactam, tetracycline, and doxycycline. Both isolates were also resistant to carbapenem with MIC values of >32 μg/ml for both imipenem and meropenem (Clinical and Laboratory Standards Institute, 2013). However, AC29 was susceptible to polymyxin B whereas AC30 was resistant with an MIC value of 128 μg/ml (Magiorakos et al., 2011; Lean et al., 2014).
Genomic DNA of AC29 and AC30 were extracted using Wizard Genomic DNA Purification Kit (Promega, USA) according to the manufacturer's instructions. Extracted DNA was quantified using the spectrophotometer at OD260 and the purity was determined by OD260/OD280 ratio (Ausubel et al., 2002).
Genomic Sequencing, Assembly, and Annotations
Whole genome sequencing of A. baumannii AC29 and AC30 was carried out by a commercial vendor using the Illumina Genome Analyzer IIx platform. CLC Bio software package was used to assemble the genome sequence data. Open reading frame (ORF) prediction and gene functional assignments were done using Prodigal 2.60 (Hyatt et al., 2010), RNAmmer 1.2 (Lagesen et al., 2007), and tRNAscan-SE (Lowe and Eddy, 1997). Functional annotation of the genome was performed using Blast2Go and the Rapid Annotation using Subsystem Technology (RAST) server (Aziz et al., 2008).
MLST and Phylogenetic Analysis
To determine the sequence types, multilocus sequence typing (MLST) of AC29 and AC30 was performed according to the Bartual or Oxford scheme using the seven housekeeping genes cpn60, gdhB, gltA, gpi, gyrB, recA, and rpoD (Bartual et al., 2005) and the Pasteur scheme using the genes cpn60, fusA, gltA, pyrG, recA, rplB, and rpoB (Diancourt et al., 2010). The gene sequences were compared to the PubMLST database for A. baumannii (http://pubmlst.org/abaumannii/) and assigned to the appropriate sequence types.
The phylogenetic relationship of AC29 and AC30 to sequenced A. baumannii strains was inferred using composition vector tree (CVTree) version 2 (Xu and Hao, 2009) based on the concatenated nucleotide sequences of the seven reference genes used in the Bartual MLST scheme. The CVTree web-based server (http://tlife.fudan.edu.cn/cvtree/) generates phylogenetic trees based on k-tuple values (Xu and Hao, 2009). The completed A. baumannii genomes used in the analysis were downloaded via the NCBI FTP site and the reference gene sequences extracted for phylogenetic analysis.
Comparative Genomics Analyses
Whole genome sequences of these two A. baumannii AC29 and AC30 strains were compared to our previously reported A. baumannii AC12 genome (Lean et al., 2015) and the completed A. baumannii genomes available from NCBI FTP site using Mauve (Darling et al., 2004). Circular chromosome and plasmid map of these strains were constructed using CGView Server developed by the Stothart Research Group (Grant and Stothard, 2008).
Accession Numbers
Genome sequences of A. baumannii strains AC29 and AC30 were deposited under the accession number CP007535 and CP007577, respectively. Plasmid sequences of pAC29a, pAC29b, pAC30a, pAC30b, and pAC30c were deposited under the accession number CP008850, CP008851, CP007578, CP007579, and CP007580, respectively. Accession numbers for our previously reported A. baumannii strain AC12 (CP007549) and pAC12 (CP007550) were as in (Lean et al., 2015).
Quantitative Real-Time PCR Of pmrAB
One of the gene loci that have been implicated in polymyxin resistance is the pmrCAB operon with pmrAB encoding the two-component signal transduction system and pmrC encoding the enzyme that catalyzes the addition of phosphoethanolamine to the lipid A moiety of lipopolysaccharide (LPS; Arroyo et al., 2011; Park et al., 2011; Hood et al., 2013). To determine the transcript levels of pmrAB from the polymyxin-susceptible AC29, and the polymyxin-resistant isolates AC30 as well as AC12 (Lean et al., 2015), quantitative real-time PCR (qRT-PCR) was employed. A. baumannii ATCC19606 was used as the polymyxin-susceptible control. Total RNA was extracted from A. baumannii strains grown in LB broth using Qiagen RNeasy Mini Kit (Qiagen).
The extracted total RNA was subjected to reverse transcription using the Quantitect Reverse Transcription Kit (Qiagen) in a T-Gradient PCR machine from Biometra/Applied Biosystems. The resulting cDNA produced was used as template for quantitative real-time PCR using the SYBR Green PCR Kit (Qiagen) performed in a Rotor-Gene 6000 Real-Time PCR Machine (Corbett Life Science/Qiagen). Prior to the actual amplification cycling conditions, the mixture was subjected to initial heat activation at 95°C for 5 min followed by 35 cycles of the two-step cycling condition of denaturation at 95°C for 10 s and annealing at 60°C for 30 s. The melting curve profile for each amplification reaction and the relevant CT value were automatically generated using the software provided (Corbett Life Science/Qiagen). The rpoB gene was used as the housekeeping gene for normalization (Park et al., 2011). Relative quantification using the ΔΔCT method (Pfaffl, 2001) was then applied to quantify the expression of pmrAB.
Lipopolysaccharide Analysis
Lipopolysaccharide (LPS) analysis was carried out for the polymyxin resistant strains AC30 and AC12 (Lean et al., 2015), the polymyxin susceptible strain AC29 along with A. baumannii ATCC19606 as the polymyxin-susceptible control. LPS extraction was carried out according to Hood et al. (2013). Briefly, bacteria were swabbed from LB agar plates into 154 mM NaCl, and the cell density was adjusted to an OD600 of 1.5, pelleted and resuspended in lysis buffer (2% SDS, 4% 2-mercaptoethanol, 10% glycerol, 0.1 M Tris-HCl, pH 6.8). Samples were then boiled for 10 min, cooled to 60°C and treated with proteinase K for 1 h. These samples were electrophoresed through a 15% polyacrylamide gel using standard methods and stained with Pro-Q Emerald 300 LPS stain according to the manufacturer's recommendations (Invitrogen). Molecular weight marker used in the experiment was the Precision Plus Protein Dual Xtra Standards (BioRad). The stained minigels were then viewed and photographed under UV light using a UV Gel Imager (Alpha-Innotech).
Cloning and Expression of the Novel Variant blaAmpC Gene
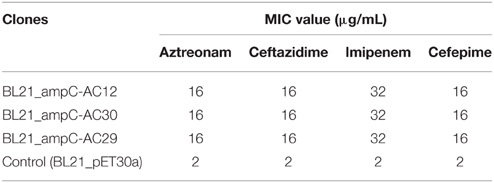
The novel variant blaAmpC gene was PCR-amplified from the genomes of A. baumannii AC29 and AC30 using specific primers ampC_BamHI: 5′-GGATCCATGGCTGTGGGTGT TATTCAA-3′ and ampC_HindIII: 5′-AAGCTTTTATTTCTTT ATTGCATTCAGCAC-3′ with the following conditions—initial hold at 95°C for 3 min, followed by 40 cycles of 95°C for 50 s, 55°C for 50 s, 72°C for 1 min, and final extension at 72°C for 90 s. Purified PCR products were then cloned into pGEM-T Easy (Promega, USA) according to the manufacturer's instructions and transformed into E. coli JM109. Transformants were selected on LB agar supplemented with 100 mM ampicillin with 100 mM IPTG and 50 mg/ml X-gal. Plasmids were prepared from white colonies and screened by digestion with BamHI and HindIII which cut at the restriction sites incorporated into the primers used for amplification. Plasmids with the expected restriction fragments were validated by conventional Sanger dideoxy sequencing prior to subcloning into the pET30a expression vector. The resulting pET30a-ampC recombinant plasmids were then transformed into E. coli BL21 (DE3/pLysS) and expression of the cloned blaAmpC gene carried out by inducing the cells with 0.1 mM IPTG. After that, the E. coli BL21 recombinant clones were tested for their MIC values for extended-spectrum cephalosporins (i.e., ceftazidime and cefepime) as well as aztreonam and imipenem at concentrations of 2, 4, 8, 16, and 32 μg/ml using the agar dilution method.
Results
Strain Characteristics and Genome Analyses
Basic Genome Features and Sequence Types
Analyses of the whole genome sequencing indicated that the A. baumannii AC29 and AC30 genomes were 3,935,134 and 3,925,274 bp, with GC content of 38.84 and 38.98%, respectively. Predicted ORFs from the genomes of A. baumannii AC29 and AC30 were 3728 and 3646, respectively. Both strains contained only one chromosome with varying number of plasmids, as featured in Table 1. The Bartual or Oxford scheme for MLST utilizes seven housekeeping genes cpn60, gdhB, gltA, gpi, gyrB, recA, and rpoD to determine the sequence type (ST) of A. baumannii (Bartual et al., 2005). Both AC29 and AC30 were designated ST195 (derived from cpn60-2, gdhB-3, gltA-1, gpi-96, gyrB-3, recA-2, and rpoD-3) whereas using the Pasteur MLST scheme (Diancourt et al., 2010), both A. baumannii strains were designated ST2 (cpn60-2, fusA-2, gltA-2, pyrG-2, recA-2, rplB-2, and rpoB-2). Thus, AC29 and AC30 were categorized as belonging to the International Clone-II (or IC-II, also referred to as Global Clonal 2, or GC2) group of strains.
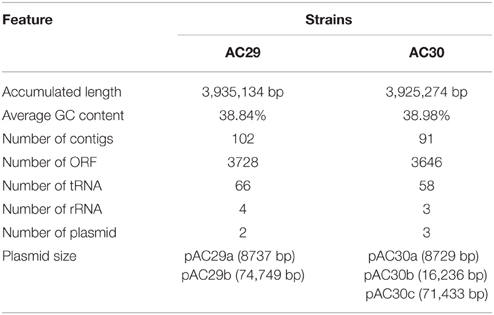
Table 1. General genomic features of the whole genome sequences of A. baumannii strains AC29 and AC30
Phylogenetic analysis of the A. baumannii AC29 and AC30 genomes were performed with other available A. baumannii genomes from the NCBI's FTP site and using A. baumannii SDF as an outgroup to root the phylogenetic tree based on the concatenated nucleotide sequences of the seven reference genes utilized in the Bartual MLST scheme (Farrugia et al., 2013). Based on the phylogenetic tree (Figure 1), A. baumannii AC29 and AC30 were closely related to A. baumannii AC12 which was also isolated from the same hospital in Terengganu and was also typed as ST195 (Lean et al., 2015) along with A. baumannii M1 which was also isolated from Malaysia (accession no. LAIL0100000). These strains were clustered together with other A. baumannii isolates of the IC-II group and this was also reflected in their genome grouping based on genomic BLAST (as indicated at http://www.ncbi.nlm.nih.gov/genome/genomegroups/403? accessed on 19 August 2015).
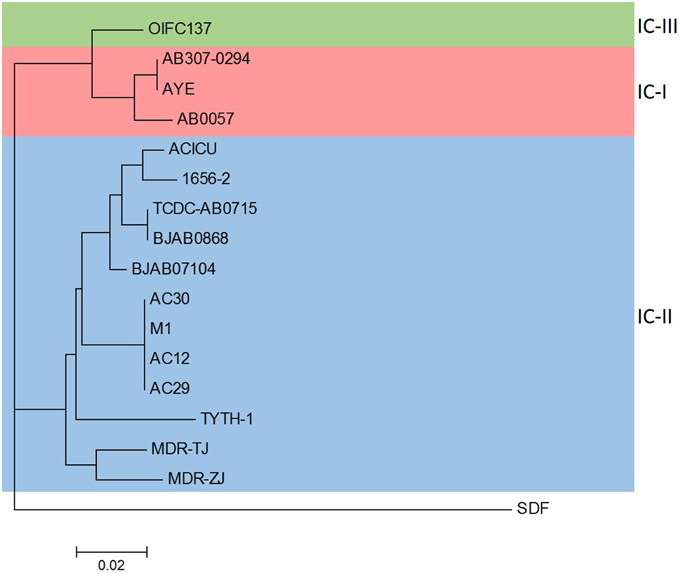
Figure 1. Phylogenetic analysis of A. baumannii AC12, AC29, AC30, and completed A. baumannii genomes obtained from NCBI. The phylogenetic tree was constructed by composition vector tree version 2 (CVTree 2) using concatenated nucleotide sequences of the seven reference genes based on the Bartual/Oxford MLST scheme (cpn60, gdhB, gltA, gpi, gyrB, recA, and rpoD). Strains belonging to the international clonal groups IC-I, IC-II, and IC-III are indicated. The accession nos. of the strains used in the analysis are as follows: AB307-0294 (CP001172.1), AYE (CU459141.1), AB0057 (CP001182.1), ACICU (CP000863.1), 1656-2 (CP001921.1), TCDC-AB0715 (CP002522.2), BJAB0868 (CP003849.1), BJAB07104 (CP003846.1), M1 (LAIL01000001.1), TYTH-1 (CP003856.1), MDR-TJ (CP003500.1), MDR-ZJ (CP001937.1), OIFC137 (NZ_AFDK01000002.1), SDF (NC_010400.1).
The AbaR4-Type Resistance Island
The genomes of A. baumannii AC29 and AC30 contained an AbaR4-type resistance island (RI) which interrupts the comM gene (Nigro and Hall, 2012). The island found in AC29 is 23 kb and designated AC29-RI1 whereas in AC30, it is 26 kb and designated AC30-RI1. Tn6022 and/or ΔTn6022 is part of the important features for this type of RI; in AC29-RI1, only the entire tniC gene is deleted in ΔTn6022 whereas AC30-RI1 carries a complete copy of Tn6022 (Figure 2). In AC29-RI1, the ΔTn6022 is missing the tniC gene on its far left end, but full-length tniD and tniE genes are present in the transposon. AC30-RI1 contains the full length Tn6022. In both AC29-RI1 and AC30-RI1 islands, only one copy of either ΔTn6022 or Tn6022 is found.
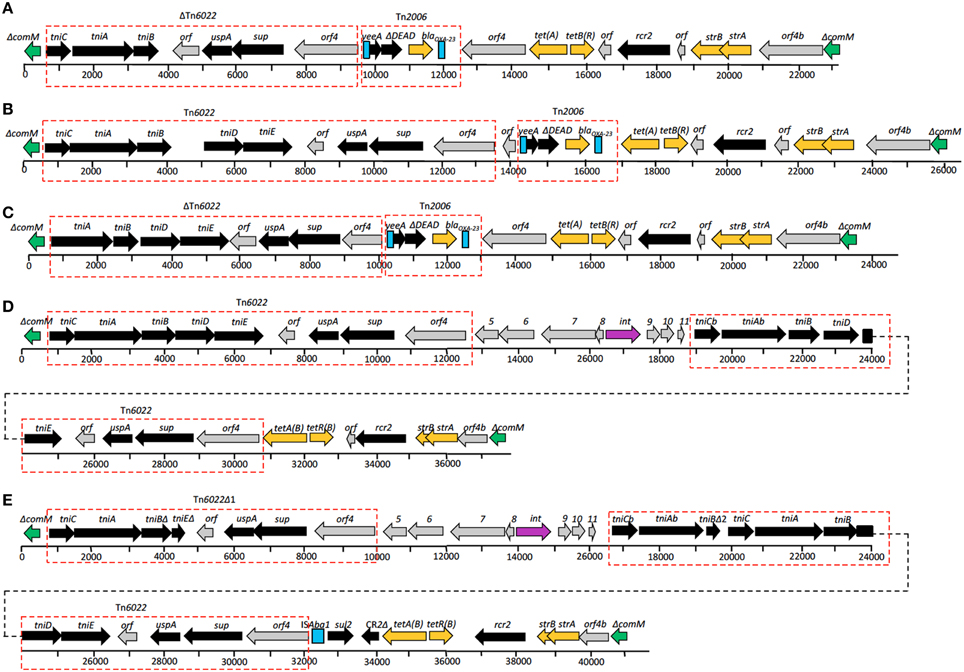
Figure 2. Structures of resistance islands present in A. baumannii AC12, AC30, and AC29 as compared to similar islands in A. baumannii MDR-ZJ and MDR-TJ. (A) Structure of resistance island ACRI12-1 present in A. baumannii AC12. ΔTn6022, Tn6022, and Tn2006 are indicated in the red dotted-line box. Directions of genes and ORFs are indicated by arrows above the central thick line and the names are as given above. Green boxes represent truncated ΔcomM, turquoise blue boxes represent insertion element, ISAba1, and orange boxes represent genes conferring antibiotic resistance. Numbered boxes in gray represent genes encoding for hypothetical proteins. Comparison between structures of (A) AC12-RI1, (B) AC30-RI1, (C) AC29-RI1, (D) AbaR22, and (E) RIMDR−TJ, revealed similarities in the islands, as indicated by the same colors and red dotted-line box.
Drug resistance genes found in the AC29-RI1 and ACRI30-1 islands are sulI (conferring sulphonamide resistance), blaOXA−23 (conferring carbapenem resistance), tetA and tetB (conferring tetracycline resistance), and strA with strB (conferring streptomycin resistance). The blaOXA−23 gene is flanked by two copies of the ISAba1 insertion element in a composite transposon structure similar to Tn2006 (Figure 2). Tn2006 comprises of the blaOXA−23 carbapenemase-encoding gene, a DEAD/DEAH box helicase-like gene and an ATPase gene (yeeA) flanked by two copies of ISAba1.
Plasmids of A. baumannii AC29 and AC30
A Small 8.7 kb Plasmid Found in Both AC29 and AC30
Sequence analysis revealed the presence of a small 8.7 kb plasmid in both the genomes of A. baumannii AC29 and AC30, designated pAC29a and pAC30a, respectively (Table 1). These plasmids contained a total of nine coding sequences (CDS) (Figure 3). No antibiotic resistance gene was found on these plasmids. Two plasmid replication genes, designated repA and repB, were found on these plasmids along with an iteron sequence made up of four direct repeats of 5′-ATA TGT CCA CGT TTA CCT TGC A-3′ located 53 nucleotides upstream of the repB gene. Other features on this cryptic plasmid are a Sel1 repeat protein-encoding gene (sel1) which is flanked by XerC/XerD-like recombination sites in an inverted repeat formation, an outer membrane TonB-dependent receptor gene, a gene encoding for putative septicolysin and two hypothetical protein-encoding genes. A toxin-antitoxin (TA) system designated AbkB/AbkA (Mosqueda et al., 2014) was also encoded on these 8.7 kb plasmids.
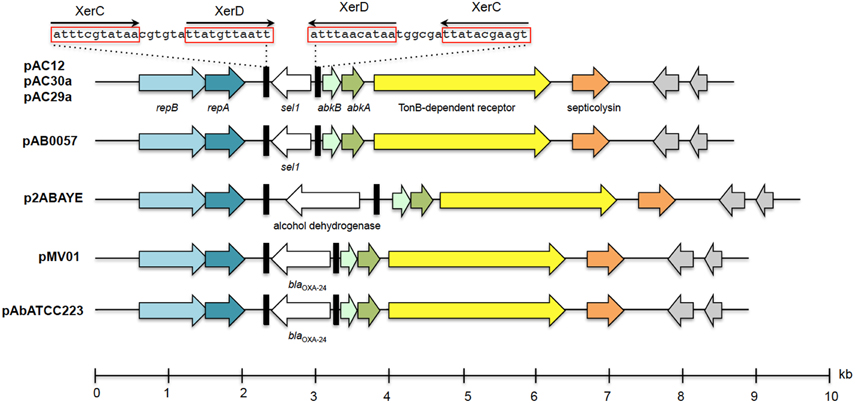
Figure 3. Linear map showing the genetic organization of plasmids pAC29a, pAC30a, and pAC12 in comparison with other similar A. baumanii plasmids. The XerC/XerD-like recombination sites are indicated in black rectangular boxes with the nucleotide sequences as indicated above the boxes. The two plasmid replication genes, repB and repA, are indicated in blue whereas the putative abkA/abkB toxin-antitoxin genes are indicated in green arrows. Hypothetical ORFs are shaded gray. The TonB-dependent receptor gene is indicated in yellow whereas the putative septicolysin gene is in orange.
PAC30b, a 16.2 kb Resistance Plasmid Found in A. baumannii AC30
Plasmid pAC30b is 16,236 bp in size and is found only in A. baumannii AC30. pAC30b contained 11 CDS and is a resistance plasmid, as indicated by the presence of the 16S rRNA methylase gene (armA) and aminoglycoside 3′-phosphotransferase gene (aphA1) which confer resistance to aminoglycosides, along with macrolide 2′-phosphotransferase (mph2) and macrolide efflux protein-coding (mel) genes which confer resistance to macrolides (Figure 4; Zhou et al., 2011). The armA, mph2, and mel genes along with adjacent putative transposase genes tnpD and tnpU make up a small 7 kb RI designated Tn1548::armA which have been reported in several other plasmids from Enterobacteriaceae (Dolejska et al., 2013).
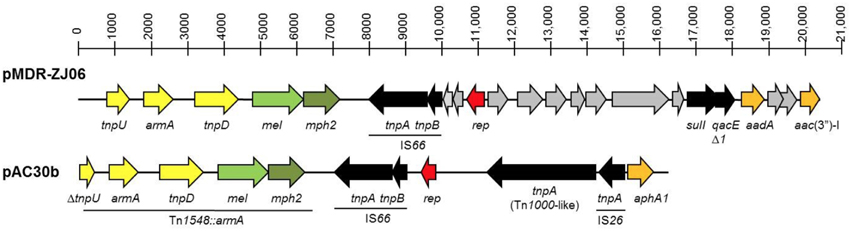
Figure 4. Linear map comparison of the Tn1548::armA region in plasmid pAC30b from A. baumannii AC30 and pMDR-ZJ06 from A. baumannii MDR-ZJ06 (NC_017172). The Tn1548::armA region and insertion elements (IS) were underlined and indicated in the diagram.
The tnpU putative transposase gene located at the 3′-end of Tn1548::armA is truncated in pAC30b whereby only the last 480 bp of the 836-bp gene could be found and was thus designated ΔtnpU. The pAC30b plasmid encodes a number of putative transposases: ΔtnpU and tnpD from Tn1548::armA, tnpA, and tnpB transposases from IS66, a 2888-bp tnpA from the Tn3 family of transposases (or Tn1000-like) and immediately adjacent to that, another smaller 704-bp tnpA transposase belonging to IS26. The aminoglycoside resistance gene aphA1 is found downstream from IS26.
A Large ca. 70 kb Conjugative Plasmid in the Genomes of A. baumannii AC29 and AC30
Both A. baumannii AC29 and AC30 were found to harbor similar conjugative plasmids of ca. 70 kb in size designated pAC29b and pAC30c, respectively. There are a total of 101 CDS in the 74,749 bp pAC29b whereas pAC30b is 71,433 bp and contains a total of 96 CDS. Both pAC29b and pAC30c encode a complete tra locus (Figure 5) similar to the tra locus found in pACICU2 (Iacono et al., 2008), p2ABTCDC0715 (Chen et al., 2011), pAb-G7-2 (Hamidian and Hall, 2014), and pA85-3 (Hamidian et al., 2014). Genes for the T4SS in pAC30c and pAC29b are clustered in two separate regions: the main tra region responsible for mating pair formation spans approximately 20 kb from traL to traG, and a second smaller region containing traD and traI (or trwB and trwC) responsible for plasmid mobilization. In pAC30c, the space between these two regions contains several hypothetical ORFs and a solitary relE toxin gene without the corresponding relB antitoxin gene (Figure 5).
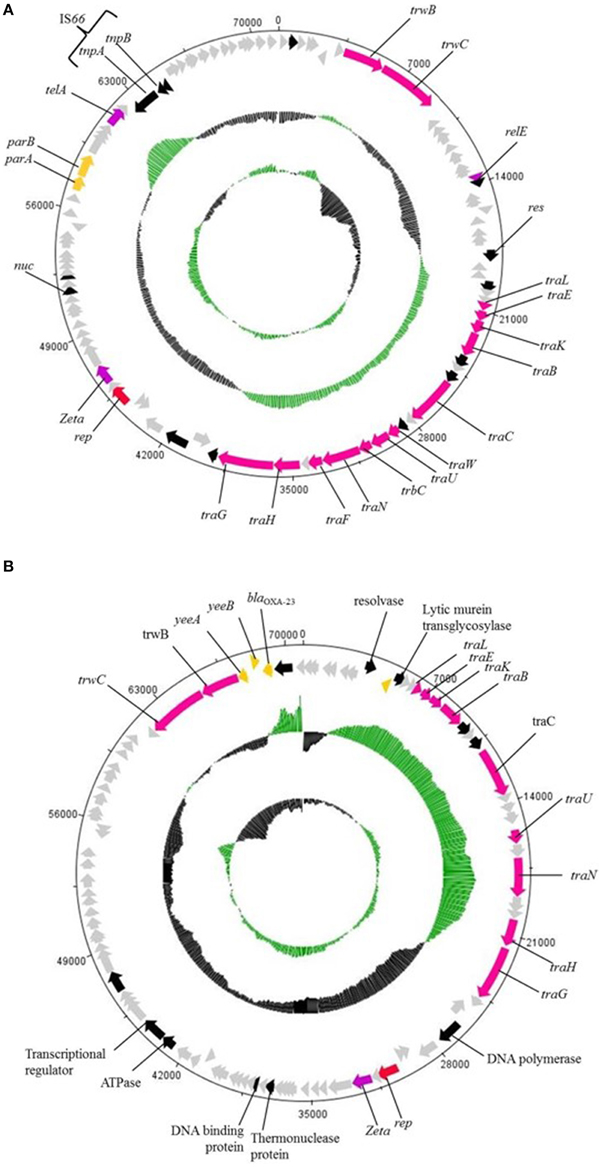
Figure 5. Circular map of the ~70 kb plasmids pAC30c (A) and pAC29b (B). Outer circle of the map represents ORFs found in the plasmid, gray ORFs are genes encoding for hypothetical proteins. The two inner circles represent GC plot and GC skew, whereby the green circle stands for above average and black circle stands for below average G+C content. Red colored arrow represents plasmid replication gene, rep; orange colored arrow represents resistant determinants; pink colored arrow represents tra genes; black colored arrow represents genes encoding proteins with known functional homologs; purple colored arrow represents putative toxin gene homologs of toxin-antitoxin systems; and gray colored arrow represents genes encoding hypothetical proteins.
Plasmid pAC29b is larger by 3316 bp when compared to pAC30c. However, there is a 14,374 bp fragment in pAC30c which has no homology with pAC29b. The additional fragment found in pAC29b included the β-lactamase gene blaOXA−23 and two transcriptional regulators, tetR and asnC. Other identifiable genes within this pAC29b-unique fragment include yeeA (encoding DNA methyltransferase), yeeB (encoding ATP-dependent helicase), yhbS (encoding N-acetyltransfease), mscM (encoding mini-conductance mechanosensitive channel), iutA (encoding ferric aerobactin receptor), lrp (encoding leucine-responsive regulatory protein), and aroQ (encoding monofunctional chorismate mutase). Thus, AC29 harbors two copies of blaOXA−23—one within the AbaR4-like AC29RI-1 island in the chromosome, and another encoded on the pAC29b plasmid. However, plasmid pAC30c does not encode for any known resistance determinant.
Investigations into the Possible Contributors of Polymyxin Resistance in A. baumannii AC30
To investigate if differential expression of pmrAB occurred in the polymyxin-resistant AC30 as well as AC12 strains, relative quantification of the pmrAB transcript levels were determined by qRT-PCR with A. baumannii ATCC19606 as the polymyxin-susceptible control. When compared to ATCC19606, the relative expression levels of pmrA in AC30 was surprisingly lower at about 0.2-folds but levels of pmrB were higher at 4.8-folds (Figure 6). In contrast, the other polymyxin-resistant strain, AC12 displayed about 8.5-folds higher pmrA expression levels and about two-folds higher pmrB levels in comparison with ATCC19606. The polymyxin-susceptible AC29 showed dramatically lower pmrAB expression, at about 0.05- and 0.03-folds, respectively.
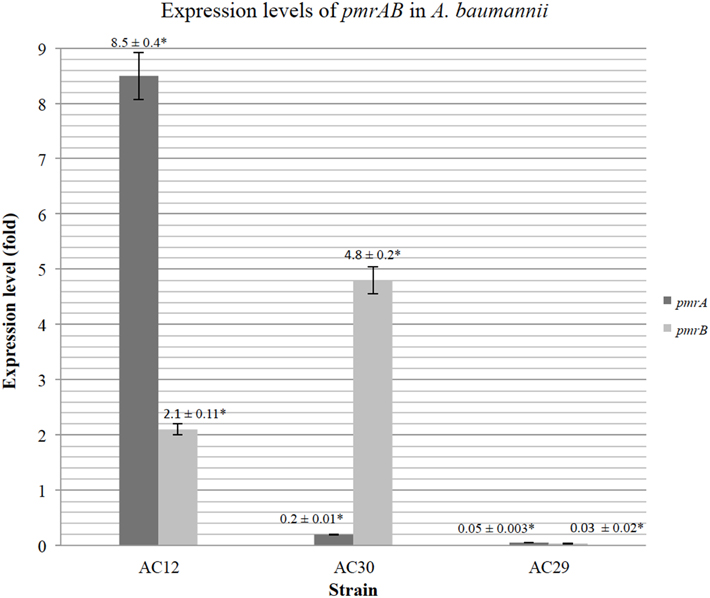
Figure 6. Relative expression levels of pmrAB of the polymyxin-resistant A. baumannii AC12 and AC30 as compared to the polymyxin-susceptible A. baumannii AC29 as determined by Quantitative Real-Time reverse transcriptase-PCR (qRT-PCR). The data represents the mean fold change ± standard deviation (SD; indicated as error bars in the graph) taken from three replicates performed for each qRT-PCR reaction. Asterisk (*) indicate statistical significance, as determined by using two-tailed, unpaired Student's t-test with p < 0.05.
LPS from the two polymyxin-resistant strains, AC12 and AC30, along with two polymxyin-susceptible strains AC29 and ATCC19606 were extracted and analyzed on 15% SDS-polyacrylamide gels (Figure 7). SDS-PAGE of the extracted LPS yielded a band of ~10 kD, which was within the expected molecular weight (between 6 and 10 kDa) for LPS. Results indicated that the LPS in the polymyxin resistant strains AC12 and AC30 were not totally absent as had been previously reported in other polymyxin-resistant strains (Moffatt et al., 2010, 2011; Hood et al., 2013) but the intensity of the LPS band was considerably less when compared with the LPS band of the polymyxin-susceptible strains.
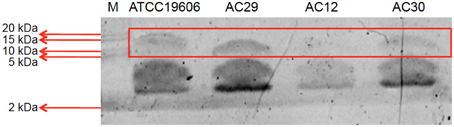
Figure 7. SDS-PAGE analysis of the extracted lipopolysaccharide (LPS) layer of the A. baumannii ATCC19606 (control; polymyxin susceptible strain), AC12 (polymyxin resistant strain), AC29 (polymyxin susceptible strain), and AC30 (polymyxin resistant strain). M stands for the protein marker Precision Plus Protein Dual Xtra Standards (BioRad) with the sizes as indicated in kDa.
AmpC-Mediated Resistance to Extended-Spectrum Cephalosporins
The AmpC cephalosporinase encoded by A. baumannii AC29 and AC30 displayed identical non-synonymous mutations, R80S and G246S (with reference to ADC-7, the reference A. baumannii-encoded AmpC; Hujer et al., 2005; Rodríguez-Martínez et al., 2010) to that found in A. baumannii AC12 (Lean et al., 2015) but the effects of these mutations on cephalosporin resistance have not been reported. To investigate if the mutations in the blaAmpC gene harbored by A. baumannii AC29 and AC30 has any effect on resistance against β-lactams especially cephalosporins, these genes were cloned and expressed in E. coli BL21 through the IPTG-inducible T7 promoter of the pET30a vector. Recombinant E. coli BL21 carrying the blaAmpC from AC29 and AC30 displayed resistance to ceftazidime, cefepime, aztreonam, and even imipenem (Table 2). Thus, the blaAmpC encoded by AC29 and AC30 is likely an extended-spectrum Acinetobacter-derived AmpC (ESAC).
Other Resistance Determinants and Efflux Pumps
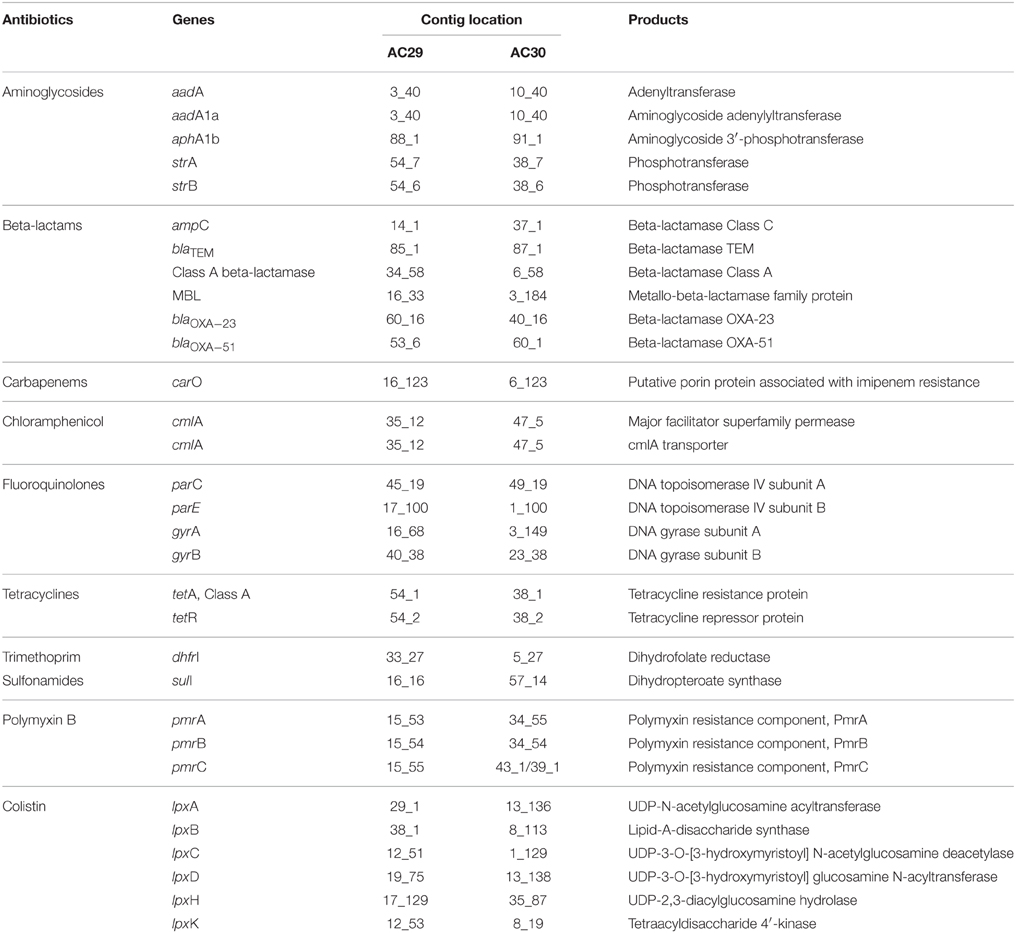
Besides the blaAmpC-encoded cephalosporinase (belonging to Ambler Class C β-lactamases) and the Class D carbapenemases encoded by blaOXA−23 and blaOXA−51 that were described in the previous sections, two other β-lactamase genes were found in the genomes of AC29 and AC30 (Table 3). These are the blaTEM gene which encodes class A extended-spectrum β-lactamase and a gene identified as belonging to the metalo-β-lactamase family (Ambler Class B). The contributions of these genes to the β-lactam resistance phenotype of AC12 and AC30 are currently unknown.
One of the main mechanisms of fluoroquinolone resistance is mutations that alter the drug targets. The targets of fluoroquinolone action are the bacterial enzymes DNA gyrase (encoded by gyrA and gyrB) and DNA topoisomerase IV (encoded by parC and parE), both of which work together in replication, transcription, recombination and repair of DNA (Jacoby, 2009). The AC29 and AC30-encoded gyrA and parC showed the Ser → Leu amino acid substitutions at positions 83 and 80, respectively, which have been implicated in fluoroquinolone resistance (Wisplinghoff et al., 2003; Fournier et al., 2006; Maragakis and Perl, 2008). However, four additional novel point mutations (G145D, S118G, L644P, and T872A) were observed in the AC29 and AC30-encoded gyrA. Whether these mutations contribute to fluoroquinolone resistance in A. baumannii AC29 and AC30 would require further investigations.
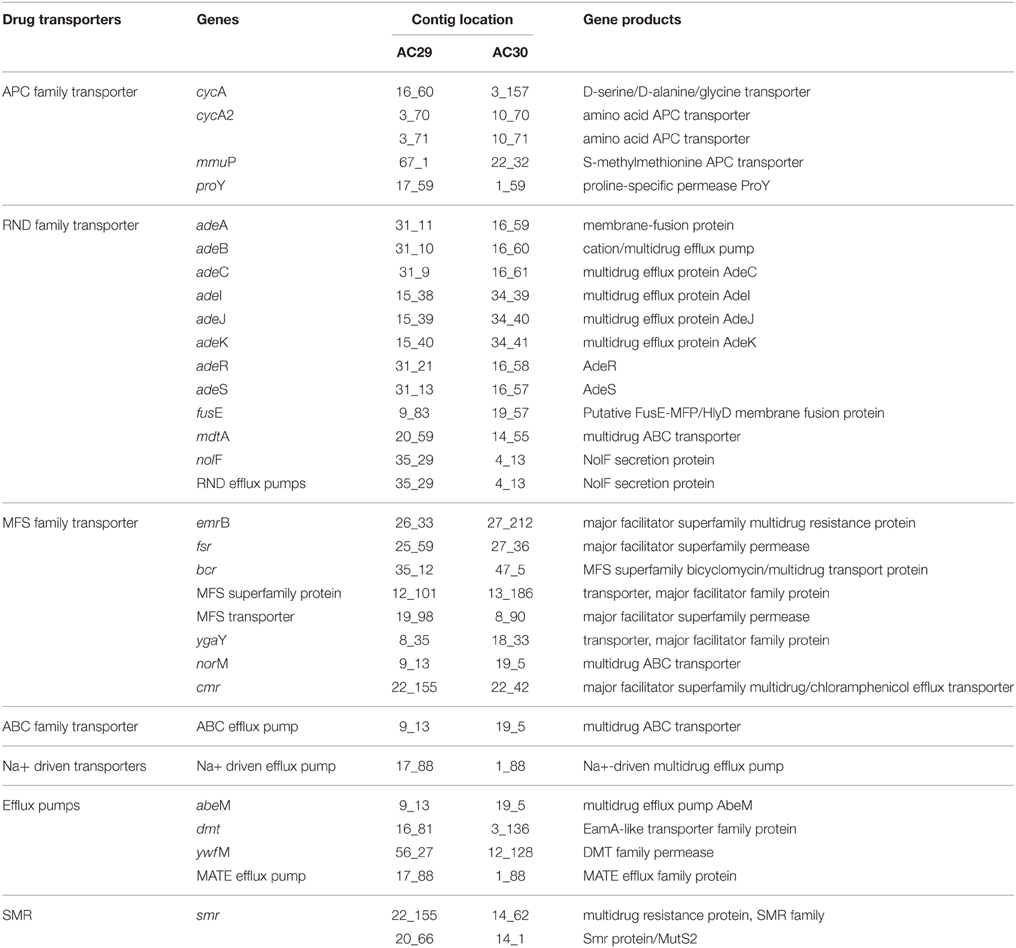
Multidrug efflux pumps and porins also play important roles in A. baumannii antimicrobial resistance (Vila et al., 2007). Among the five major families of bacterial efflux pumps (i.e., RND, MFS, APC, ABC, and MATE; Table 4), the RND family is widely disseminated in Gram-negative bacteria (Poole, 2004; Bonomo and Szabo, 2006; Nordmann and Poirel, 2008; Wieczorek et al., 2008). The genome analysis of A. baumannii AC29 and AC30 showed the presence of the complete adeABC genes along with its two-component regulatory genes, adeRS. Another member of the RND family encoded by adeIJK was also present in AC29 and AC30. However, a third RND efflux pump encoded by the adeFGH operon was absent in both isolates. Overexpression of adeABC and to a certain extent, adeIJK, has also been associated with the multidrug resistance phenotype in A. baumannii (Vila et al., 2007) and it would be of interest to investigate if this is likewise in A. baumannii AC29 and AC30.
Discussions
Whole Genome Sequencing and Sequence Types
In this study, we applied comparative genomics on two A. baumannii strains that are indistinguishable by PFGE but have different antibiograms to better understand their mechanism of resistance. Although these two strains only differed by their susceptibility toward polymyxin B (AC29 was susceptible whereas AC30 was resistant), detailed genomic analysis indicated a number of differences in the RI structures and the plasmid contents. Both A. baumannii AC29 and AC30 were typed as ST195 and, not surprisingly, their closest neighbor was A. baumannii AC12 which was also another ST195 strain as previously reported from (Lean et al., 2015). Another close neighbor was A. baumannii M1, an isolate from Malaysia that was recently deposited and from its genome sequence, was deduced to be also ST195. All these strains were clearly clustered in the IC-II group of global A. baumannii clones (Figure 1). Strains that were typed as ST195 under the Bartual MLST scheme are grouped into the clonal complex 92 (CC92) which are characterized as under IC-II (Kim et al., 2013; Zarrilli et al., 2013).
The AbaR4-Type Resistance Island
One of the known hotspots for the insertion of resistance islands in A. baumannii is the comM gene (Post et al., 2010; Zhou et al., 2011; Huang et al., 2012; Kim et al., 2013). The genomes of A. baumannii AC29 and AC30 also contained a similar island within comM but with slight differences. Previous analysis of the A. baumannii AC12 genome showed the presence of a 23 kb AbaR4-type island designated AC12-RI1 within comM (Lean et al., 2015) which consisted of a backbone of Tn6167 and a truncated version of Tn6022 designated ΔTn6022. In all three islands (i.e., AC12RI-1, AC29RI-1, and AC30RI-1), only one copy of either ΔTn6022 or Tn6022 is found whereas other similar resistance islands usually contained two copies of ΔTn6022 and/or Tn6022. For instance, in RIMDR−TJ and AbaR22, another copy of the complete Tn6022 was located next to the tetA(B) gene (Figure 2). Besides that, the blaOXA−23 gene in AC12-RI1, AC29-RI1, and AC30-RI1 was found flanked by two copies of the ISAba1 insertion element in a composite transposon structure similar to Tn2006 (Figure 2). The presence of ISAba1 upstream of blaOXA−23 may contribute to carbapenem resistance by increasing the expression level of the gene by virtue of its outward-directed promoter (Zhu et al., 2013). The Tn2006-like structure was inserted immediately downstream from ΔTn6022 or Tn6022, and interestingly in AC12-RI1 and AC29-RI1, the orf4 gene found at the right-most end of ΔTn6022 was duplicated, resulting in the ΔTn2006-like structure being flanked by orf4 on either end (Figure 2). Such an arrangement has not been reported in similar resistance islands before. Besides that, comparison between the Tn2006 present in the three RIs and Tn6167 showed some differences. Tn6167 harbors Tn2006, but at a different location whereas both RIMDR−TJ and AbaR22 do not contain Tn2006. However, the right end of the islands were identical with Tn6167, RIMDR−TJ and AbaR22 and comprises of tetA(B)-tetR(B), the small mobile element CR2, strA-strB and orf4b, a hypothetical ORF related to orf4 of Tn6022 and Tn6022Δ1 (Figure 2).
The structures of AC29-RI1 and AC30-RI1 indicated that they are novel variants of the AbaR4-type RIs that were recently reported in a survey of RIs found in A. baumannii strains throughout Asia (Kim et al., 2013). Kim et al. (2013) also reported that AbaR4-type RIs were commonly found among carbapenem-resistant CC92 strains which included the ST195 lineage.
Plasmids of A. baumannii AC29 and AC30
A Small 8.7 kb Plasmid Found in Both AC29 and AC30
Plasmid pAC29a and pAC30a were almost identical to pAC12 (Lean et al., 2015), pAB0057 (Adams et al., 2008), p1ABTCDC0715 (Chen et al., 2011), and pPKAB07 (Saranathan et al., 2014). A similar A. baumannii plasmid, pABVA01 (8963 bp), was found to harbor the blaOXA−24 carbapenese gene flanked by XerC/XerD-like recombination sites (D'Andrea et al., 2009) and this arrangement was also subsequently reported in the 8771 bp plasmid pMMCU1 (Merino et al., 2010). The XerC/XerD-like recombination sites were present in pAC29a/pAC30a but with the selI gene instead of blaOXA−24 within the potential recombination region. The pAC12 plasmid from A. baumannii AC12 (Lean et al., 2015) and pAB0057 from A. baumannii AB0057 (Adams et al., 2008) also contained the sel1 gene in between the XerC/XerD sites. The function of the sel1 gene is currently unknown. Other similar Acinetobacter plasmids harbored different sized fragments between the XerC/XerD recombination sites with p2ABAYE from A. baumannii AYE (Fournier et al., 2006) for example, harboring a putative alcohol dehydrogenase gene (Figure 3). The blaOXA−24 gene is so far reported only in strains from Italy and Spain (D'Andrea et al., 2009; Merino et al., 2010). This was corroborated in a recent multicenter study which showed that the blaOXA−24/blaOXA40−like genes in A. baumannii strains isolated from Spain was predominantly carried by small 8–12 kb plasmids with two of the sequenced plasmids, pAbATCC223 and pAbATCC329, harboring the blaOXA−24 gene in between the XerC/XerD sites (Mosqueda et al., 2013). Xer recombination is a site-specific recombination mechanism involved in events such as the integration of phage CTX-Φ at the dif1 site in the Vibrio cholerae chromosome (D'Andrea et al., 2009). Moreover, the proteins required for Xer recombination, such as the XerC and XerD recombinases and PepA are reportedly encoded in the chromosome of several A. baumannii strains (Merino et al., 2010). Thus, the XerC/XerD-like sites on these 8.7 kb plasmids could act as site-specific recombination targets responsible for mobilization of discrete gene modules such as blaOXA−24 and sel1 within Acinetobacter plasmids (D'Andrea et al., 2009; Merino et al., 2010).
Other highlights of this small plasmid are the presence of a toxin-antitoxin (TA) system designated AbkB/AbkA (Mosqueda et al., 2014) and a gene encoding a possible TonB-dependent receptor protein. Most TA systems are usually characterized by two co-transcribed genes with the antitoxin gene preceding the toxin gene (Chan et al., 2012; Hayes and Kêdzierska, 2014). AbkB/AbkA seems to differ from canonical TA systems as the abkB toxin gene precedes the abkA antitoxin gene. To date, only three characterized TA loci have been reported to display this unusual genetic arrangement: the mqsRA (Brown et al., 2009), the higBA (Tian et al., 1996), and hicAB (Jørgensen et al., 2009) modules. AbkB/AbkA was previously identified as one of the four functional TA systems in A. baumannii (it was designated SplT/SplA or DUF497/COG3914) whereby overexpression of the toxin was shown to inhibit growth in E. coli and this was overcame by co-expression of the cognate antitoxin (Jurenaite et al., 2013). The AbkB (or SplT) toxin was shown to inhibit translation when overexpressed in E. coli with cleavage of lpp mRNA and transfer-messenger RNA (tmRNA) demonstrated, thus indicating that the AbkB toxin likely functions as an endoribonuclease or RNA interferase. The AbkB/AbkA locus was found to be highly prevalent in small plasmids of A. baumannii clinical strains (88.6% prevalence among 476 clinical isolates from Lithuania; Jurenaite et al., 2013). The presence of a TA system on these plasmids would explain their stability in the absence of any apparent selection pressure, particularly for the small plasmids without the blaOXA−24/blaOXA40−like gene such as pAC30a and pAC29a.
Some TonB-dependent receptors, in particular BauA, play important roles in the acquisition of iron in A. baumannii (Dorsey et al., 2004; Mihara et al., 2004) with recent transcriptomic and proteomic analyses indicating approximately 20 TonB-dependent receptors in A. baumannii, some of which are regulated by iron (Antunes et al., 2011; Eijkelkamp et al., 2011; Nwugo et al., 2011). TonB-dependent receptors are usually found in the outer membrane where they interact with TonB and associated inner membrane proteins (ExbB and ExbD) that provide energy needed to transport host iron-carrier and iron-siderophore complexes into the periplasm once these complexes are bound to the TonB-dependent receptors (Zimbler et al., 2013). Thus, along with TonB, the TonB-depended receptors play an important role in the virulence of A. baumannii. Whether the TonB-dependent receptor protein encoded by these small plasmids plays a similar role in iron acquisition and hence, virulence, awaits further experimentation. Another possible virulence-associated gene encoded in these small plasmids is found downstream of the gene encoding the TonB-dependent receptor. This gene is predicted to encode a 152-amino acids-residue protein homologous to septicolysin, a putative virulence factor (Mosqueda et al., 2014). Septicolysin is a member of thiol-activated cytolysins which have been implicated in the pathogenesis of infections by several Gram-positive pathogens such as Clostridium perfringens, Listeria monocytogenes, and Streptococcus pneumoniae and are characterized by their cytolytic activity for eukaryotic cells (Billington et al., 2000). With two putative virulence factors encoded on these small plasmids, it would therefore be of interest to investigate if these plasmids play a role in the virulence and pathogenesis of A. baumannii infections, especially in view of their prevalence among clinical A. baumannii isolates.
pAC30b, a 16.2 kb Resistance Plasmid Found in A. baumannii AC30
Both pAC30b and pMDR-ZJ06 are resistance plasmids, as indicated by the presence of genes which confer resistance to aminoglycosides and macrolides within the 7 kb Tn1548::armA resistance island (Zhou et al., 2011). Interestingly, Tn1548::armA is located in the chromosome of A. baumannii AC12 (Lean et al., 2015) as well as in A. baumannii AC29. In both cases, Tn1548::armA was found downstream of a cluster of five genes encoding proteins annotated as paraquat-inducible protein A and protein B. The pMDR-ZJ06 plasmid harbors a class 1 integron encoding the aminoglycoside acetyltransferase (aacC1) and adenyltransferase (aadA1) along with sulI that confers sulphonamide resistance (Zhou et al., 2011) but this integron was absent in pAC30b.
Besides pMDR-ZJ06, pAC30b also shared sequence identity with p3BJAB0868 from A. baumannii BJAB0868 and p2BJAB07104 from A. baumannii BJAB07104 (Zhu et al., 2013) and this is mainly an approximately 10 kb fragment that spanned Tn1548::armA, IS66 and the rep gene. Two other parts of pAC30b that were identical with pMDR-ZJ06, p3BJAB0868, and p2BJAB07104 are IS26 and the aphA1 gene, both of which were in different locations in pAC30b when compared with pMDR-ZJ06 (Figure 4), p3BJAB0868, and p2BJAB07104. The aphA1 gene is flanked by two copies of IS26 in a composite transposon-like structure designated Tn6210 in p3BJAB0868 and p2BJAB07104 (Zhu et al., 2013) as well as pMDR-ZJ06 but in pAC30b, only one copy of IS26 is found adjacent to aphA1. It is possible that a deletion had occurred in pAC30b that took out the other copy of IS26 as well as part of the tnpU gene of Tn1548::armA which is located adjacent to the IS26-aphA1 structure. All three plasmids pMDR-ZJ06, p3BJAB0868, and p2BJAB07104 which were isolated from A. baumannii strains in China, harbored a class 1 integron but this was absent in pAC30b.
It is noteworthy that in contrast to pAC30a and pAC29a, pAC30b has very few similar plasmids in other A. baumannii isolates besides pMDR-ZJ06, p3BJAB0868, and p2BJAB07104, suggesting that these plasmids are not so prevalent. What the four A. baumannii plasmids (namely, pAC30b, pMDR-ZJ06, p3BJAB0868, and p2BJAB07104) have in common are (1) a number of transposases encoded by IS elements and transposons, and (2) a lack of any plasmid stability genes such as toxin-antitoxin systems. Thus, there is a likelihood that these plasmids are much less stable as compared to the smaller pAC29a/pAC30a plasmid which is endowed with the AbkB/AbkA toxin-antitoxin system. Hence, in A. baumannii AC29 (and AC12), portions of pAC30b are chromosomally-located and there is no widespread occurrence of pAC30b-like plasmids in other A. baumannii strains.
A Large ca. 70 kb Conjugative Plasmid in the Genomes of A. baumannii AC29 and AC30
The tra locus in pAC29b and pAC30c encodes for a type IV secretion system (T4SS) which is related to the bacterial conjugation machinery and mediates horizontal gene transfer (Juhas et al., 2008). Plasmids pAb-G7-2, pA85-3, and even pACICU2 have been demonstrated to be transmissible by conjugation to other A. baumannii strains (Hamidian and Hall, 2014; Hamidian et al., 2014). It is thus very likely that pAC29b and pCA30c are also conjugative as the tra locus in these plasmids is almost identical. In pAb-G7-2, the space between these two regions contained the aminoglycoside resistance transposon, TnaphA6 (Hamidian and Hall, 2014) whereas in pA85-3, the AbaR4 resistance island is located in the same area (Hamidian et al., 2014). In pAC30c, this region contained several hypothetical ORFs and a solitary relE toxin gene without the corresponding relB antitoxin gene. However, the two genes flanking relE could perhaps function as the antitoxin as they encode for hypothetical proteins of about the same size as the putative RelE toxin. Toxin-antitoxin pairs are usually about the same size with a few exceptions such as the Zeta toxin which is much larger (~270 amino acids) as compared to their cognate Epsilon antitoxin (~90 amino acids; Chan et al., 2012; Jurenaite et al., 2013). Although toxins usually interact with their cognate antitoxin pair, sometimes mixing and matching between different toxin and antitoxin families do occur (Chan et al., 2012; Hayes and Kêdzierska, 2014). Nevertheless, the functionality of the plasmid pAC30c and pAC29b-encoded relE toxin gene needs to be ascertained.
Both pAC30c and pAC29b carry the rep gene designated repAci6, similar to pACICU2, pAb-G7-2, and pA85-3. Another solitary toxin, this time from the Zeta family, is located downstream from the repAci6 gene in both plasmids. Experimental evidence had suggested that its overexpression is non-toxic to E. coli and thus, may not function as a typical toxin (Jurenaite et al., 2013). Its putative antitoxin partner, located upstream of its reading frame, does not bear any homology to the Epsilon antitoxin. These solitary Zeta-like toxins have been observed in several other plasmids in diverse bacterial species and their function is currently unknown (Chan et al., 2012; Jurenaite et al., 2013). In the absence of typical canonical toxin-antitoxin systems, the only other identifiable genes that could contribute to plasmid stability in pAC30c and pAC29b are the parAB genes which likely encode proteins that are involved in plasmid partitioning.
Plasmid pAC29b harbors a blaOXA−23 gene but unlike the A. baumannii A85 plasmid pA85-3 (Hamidian et al., 2014), the gene is not located within an AbaR4 island. Both pAb-G7-2 and pACICU2 harbor the aminoglycoside resistance gene aphA6 within a composite transposon designated TnaphA6 (Hamidian and Hall, 2014; Hamidian et al., 2014). Plasmid pAC30c however, does not encode any known resistance determinant.
Investigations into the Possible Contributors of Polymyxin Resistance in A. baumannii AC30
The mechanism for polymyxin resistance in A. baumannii has only recently been elucidated and the main mechanism appeared to be either covalent modification of the lipid A portion of LPS (Arroyo et al., 2011; Beceiro et al., 2011) or disruption of LPS biosynthesis (Moffatt et al., 2010; Park et al., 2011). By modification and/or mutations in the amino acid sequences, negative charges on the outer membrane can be reduced, leading to reduction in the affinity of the positively-charged polymyxin component, hence giving rise to polymyxin resistance (Adams et al., 2009; Arroyo et al., 2011; Beceiro et al., 2011). Overexpression of the two-component signal transduction system pmrAB and mutations within these genes, especially pmrB, were reported to contribute to polymyxin resistance (Adams et al., 2009; Arroyo et al., 2011; Park et al., 2011). These two genes are part of an operon along with pmrC, which encodes the enzyme responsible for the covalent addition of phosphoethanolamine to lipid A. Sequence analysis of pmrCAB from the polymyxin-resistant AC30 in comparison with the polymyxin-susceptible AC29 and other susceptible strains in the database (including ATCC19606 and ATCC17978) showed a P102H mutation within pmrA. Identical mutations were found within pmrA of AC12 (Lean et al., 2015) and other polymyxin-resistant isolates from Terengganu (Lean et al., 2014). Polymyxin-resistant isolate AC12 displayed higher pmrA expression levels (8.5-folds) and also higher pmrB levels (about two-folds) in comparison with ATCC19606. In contrast, the polymyxin-susceptible AC29 isolate showed dramatically lower pmrAB expression (0.05- and 0.03 folds). It should be noted that in this case, no isogenic polymyxin-susceptible strains for the polymyxin-resistant strains were available for comparison. Thus, the expression levels for AC29, AC30, and AC12 were compared with the non-isogenic reference strain ATCC19606 and therefore may not yield an accurate picture for the pmrAB levels. Nevertheless, the results do indicate upregulation particularly for pmrB in both the polymyxin-resistant isolates AC30 and AC12.
The lpxA, lpxC, and lpxD genes encode the first three enzymes in the lipid A biosynthesis pathway (Moffatt et al., 2010, 2011). No mutation was found in lpxA. In contrast, lpxD showed three amino acid mutations (S102T, V141I, R173G) in AC30 while lpxC had a K141R substitution. The lpsB gene encodes a glycosyltransferase is involved in the biosynthesis of the LPS core and was recently implicated in A. baumannii colistin resistance (Hood et al., 2013). Comparison of lpsB sequences indicated a H181Y substitution in AC30. Identical mutations in lpxD, lpxC, and lpsB were reported in the polymyxin-resistant A. baumannii AC12 (Lean et al., 2015); likewise, these and other mutations had been previously reported in the polymyxin-resistant A. baumannii strains from Terengganu (Lean et al., 2014). Results from the LPS analysis indicated that the LPS in the polymyxin resistant strains AC12 and AC30 were considerably less when compared with the LPS band of the polymyxin-susceptible strains, which has been previously reported in other polymyxin-resistant strains (Moffatt et al., 2010, 2011; Hood et al., 2013). This suggested that the mutations found in the lpxD, lpxC, and lpsB genes in the two polymyxin-resistant strains may have led to impairment but not a total loss of the LPS. It is likely that in the case of AC12 and AC30, polymyxin resistance could be the result of a combination of increased pmrAB expression leading to covalent modification of the lipid A moiety of LPS and possibly impaired LPS synthesis as well. Nevertheless, it should be noted that in a recent study (Hood et al., 2013), screening of transposon mutant libraries led to the identification of more than 20 genes that may be involved in inducible colistin resistance in A. baumannii. Most of these genes converged on pathways involved in osmotolerance, cell envelope biosynthesis along with protein folding (Hood et al., 2013). The role that these factors may play in the development of polymyxin resistance in the Terengganu A. baumannii strains would also need to be investigated.
AmpC-Mediated Resistance to Extended-Spectrum Cephalosporins
Resistance to β-lactam antibiotics via synthesis of β-lactamase encoded by the chromosome and/or plasmids is the most common resistance mechanism observed in A. baumannii (Bou and Martínez-Beltrán, 2000). Resistance to broad-spectrum cephalosporins in A. baumannii are usually related to the over production of extended spectrum β-lactamases (ESBL), especially AmpC-type β-lactamases designated ADCs (Acinetobacter-derived cephalosporinases) (Rodríguez-Martínez et al., 2010). ADCs typically hydrolyze penicillins, narrow- and extended-spectrum cephalosporins but not zwitterionic cephalosporins such as cefepime or carbapenems (Rodríguez-Martínez et al., 2010). However, extended-spectrum AmpCs (ESACs) have been reported in A. baumannii that confer reduced susceptibility to all cephalosporins and this includes ADC-33 (Rodríguez-Martínez et al., 2010) and ADC-56 (Tian et al., 2011).
Since the recombinant E. coli BL21 carrying the blaAmpC from AC29 and AC30 displayed resistance to ceftazidime, cefepime, aztreonam, and even imipenem, this strongly suggests that the AmpC from AC29 and AC30 is an ESAC cephalosphorinase. ADC-33 possessed a P210R substitution together with a duplication of the Ala residue at position 215 within the Ω loop, both of which are required for extended spectrum activity (Rodríguez-Martínez et al., 2010). ADC-56 possesed an R148Q mutation also within the Ω loop, that enabled the enzyme to hydrolyze cefepime (Tian et al., 2011). Thus, the G246S mutation within the Ω loop of the AmpC from AC29 and AC30 could be responsible for the extended spectrum activity. This could be examined and verified by site-directed mutagenesis of the pET30a recombinant clones. Likewise, the contribution of the R80S mutation toward extended spectrum activity should be investigated even though it is located in a non-active site.
Conclusions
In this study, we presented the comparative genome analyses of two Malaysian A. baumannii strains AC29 and AC30 that belonged to the ST195 lineage and had identical ApaI pulsotype but different susceptibilities to polymyxin. Their MLST profiles and phylogenetic clustering based on their concatenated MLST sequences clearly showed these strains belonging to the International Clone II (IC-II) group. Novel resistance island (RI) variants and plasmids were discovered from the genome sequence of these strains. Both strains shared a similar AbaR4-type RI of approximately 22 kb interrupting the comM gene insertional hotspot and which contains the carbapenem resistance gene, blaOXA−23 within a composite transposon, Tn2006. The island also encodes genes conferring resistance to tetracyclines, sulphonomides and streptomycin. Both A. baumannii strains harbored a small ~8 kb cryptic plasmid which encode putative virulence determinants (TonB-dependent receptor and septicolysin) as well as a XerC/XerD recombination site. Plasmid pAC30b is found only in AC30 but not AC29 and contained the Tn1548::armA island that confers resistance to aminoglycosides and macrolides. Interestingly, this island was found in the chromosome of AC29. Both AC30 and AC29 harbored a ~70 kb conjugative plasmid designated pAC30c and pAC29b with pAC29b containing a copy of blaOXA−23. Thus, genomic islands and, to a lesser extent, conjugative plasmids, appeared to play an important role in the dissemination and acquisition of antibiotic resistance determinants in the Terengganu A. baumannii strains. Experimental evidence also indicated that polymyxin resistance in AC30 may have developed through a combination of pmrAB upregulation and partial impairment of the lipopolysaccharide layer. The blaAmpC variant encoded by both AC29 and AC30 was also shown to be likely an extended-spectrum Acinetobacter-derived AmpC (ESAC) conferring resistance to cefepime as well as imipenem. Whole genome sequencing of the two Terengganu A. baumannii clinical strains and subsequent experiments enabled a detailed characterization of their genetic repertoire of resistance, thereby giving us an insight into the genetic blueprint of Malaysian isolates of this increasingly important and deadly nosocomial pathogen.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank University of Malaya and Universiti Sultan Zainal Abidin for facilities and support. This project was supported by High Impact Research Grant UM.C/625/HIR/MOHE/02 to KLT and UniSZA research grant UDM/09/BR(006) to CCY and ZS. SSL is supported by a University of Malaya Postgraduate Fellowship.
References
Adams, M. D., Goglin, K., Molyneaux, N., Hujer, K. M., Lavender, H., Jamison, J. J., et al. (2008). Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J. Bacteriol. 190, 8053–8064. doi: 10.1128/JB.00834-08
Adams, M. D., Nickel, G. C., Bajaksouzian, S., Lavender, H., Murthy, A. R., Jacobs, M. R., et al. (2009). Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob. Agents Chemother. 53, 3628–3634. doi: 10.1128/AAC.00284-09
Antunes, L. C. S., Imperi, F., Towner, K. J., and Visca, P. (2011). Genome-assisted identification of putative iron-utilization genes in Acinetobacter baumannii and their distribution among a genotypically diverse collection of clinical isolates. Res. Microbiol. 162, 279–284. doi: 10.1016/j.resmic.2010.10.010
Arroyo, L. A., Herrera, C. M., Fernandez, L., Hankins, J. V., Trent, M. S., and Hancock, R. E. W. (2011). The pmrCAB operon mediates polymyxin resistance in Acinetobacter baumannii ATCC 17978 and clinical isolates through phosphoethanolamine modification of lipid A. Antimicrob. Agents Chemother. 55, 3743–3751. doi: 10.1128/AAC.00256-11
Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. D., Smith, J. A., et al. (2002). Short Protocols in Molecular Biology (5th Edn). John Wiley & Sons.
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Bartual, S. G., Seifert, H., Hippler, C., Rodríguez-valera, F., and Domı, M. A. (2005). Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 43, 4382–4390. doi: 10.1128/JCM.43.9.4382-4390.2005
Beceiro, A., Llobet, E., Aranda, J., Bengoechea, J. A., Doumith, M., Hornsey, M., et al. (2011). Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob. Agents Chemother. 55, 3370–3379. doi: 10.1128/AAC.00079-11
Bergogne-Bérézin, E., and Towner, K. J. (1996). Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 9, 148–165.
Billington, S. J., Jost, B. H., and Songer, J. G. (2000). Thiol-activated cytolysins: structure, function and role in pathogenesis. FEMS Microbiol. Lett. 182, 197–205 doi: 10.1111/j.1574-6968.2000.tb08895.x
Bonomo, R. a., and Szabo, D. (2006). Mechanisms of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clin. Infecti. Dis. 43, 49–56. doi: 10.1086/504477
Bou, G., and Martínez-Beltrán, J. (2000). Cloning, nucleotide sequencing, and analysis of the gene encoding an AmpC beta-lactamase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 44, 428–432 doi: 10.1128/AAC.44.2.428-432.2000
Brown, B. L., Grigoriu, S., Kim, Y., Arruda, J. M., Davenport, A., Wood, T. K., et al. (2009). Three dimensional structure of the MqsR: MqsA complex: a novel TA pair comprised of a toxin homologous to RelE and an antitoxin with unique properties. PLoS Pathog. 5:e1000706. doi: 10.1371/journal.ppat.1000706
Camp, C., and Tatum, O. L. (2010). A review of Acinetobacter baumannii as a highly successful pathogen in times of war. Lab. Med. 41, 649–657. doi: 10.1309/LM90IJNDDDWRI3RE
Chan, W. T., Moreno-Córdoba, I., Yeo, C. C., and Espinosa, M. (2012). Toxin-antitoxin genes of the Gram-positive pathogen Streptococcus pneumoniae: so few and yet so many. Microbiol. Mol. Bio. Rev. 76, 773–791. doi: 10.1128/MMBR.00030-12
Chen, C.-C., Lin, Y.-C., Sheng, W.-H., Chen, Y.-C., Chang, S.-C., Hsia, K.-C., et al. (2011). Genome sequence of a dominant, multidrug-resistant Acinetobacter baumannii strain, TCDC-AB0715. J. Bacteriol. 193, 2361–2372. doi: 10.1128/JB.00244-11
Clinical Laboratory Standards Institute (2013). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Third Informational Supplement. Wayne, PA: CLSI (Seventeen Informational Supplement M100–S23).
D'Andrea, M. M., Giani, T., D'Arezzo, S., Capone, A., Petrosillo, N., Visca, P., et al. (2009). Characterization of pABVA01, a plasmid encoding the OXA-24 carbapenemase from Italian isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 53, 3528–3533. doi: 10.1128/AAC.00178-09
Darling, A. C. E., Mau, B., Blattner, F. R., and Perna, N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. doi: 10.1101/gr.2289704
Diancourt, L., Passet, V., Nemec, A., Dijkshoorn, L., and Brisse, S. (2010). The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 5:e10034. doi: 10.1371/journal.pone.0010034
Dijkshoorn, L., Nemec, A., and Seifert, H. (2007). An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 5, 939–951. doi: 10.1038/nrmicro1789
Doi, Y., Murray, G. L., and Peleg, A. Y. (2015). Acinetobacter baumannii: evolution of antimicrobial resistance – treatment options. Sem. Respir. Crit. Care Med. 36, 95–98. doi: 10.1055/s-0034-1398388
Dolejska, M., Villa, L., Poirel, L., Nordmann, P., and Carattoli, A. (2013). Complete sequencing of an IncHI1 plasmid encoding the carbapenemase NDM-1, the ArmA 16S RNA methylase and a resistance-nodulation-cell division/multidrug efflux pump. J. Antimicrob. Chemother. 68, 3657–3667 doi: 10.1093/jac/dks357
Dorsey, C. W., Tomaras, A. P., Connerly, P. L., Tolmasky, M. E., Crosa, J. H., and Actis, L. A. (2004). The siderophore-mediated iron acquisition systems of Acinetobacter baumannii ATCC 19606 and Vibrio anguillarum 775 are structurally and functionally related. Microbology 150, 3657–3667. doi: 10.1099/mic.0.27371-0
Eijkelkamp, B. A., Hassan, K. A., Paulsen, I. T., and Brown, M. H. (2011). Investigation of the human pathogen Acinetobacter baumannii under iron limiting conditions. BMC Genomics 12:126 doi: 10.1186/1471-2164-12-126
Farrugia, D. N., Elbourne, L. D. H., Hassan, K. A., Eijkelkamp, B. A., Tetu, S. G., Brown, M. H., et al. (2013). The complete genome and phenome of a community-acquired Acinetobacter baumannii. PLoS ONE 8:e58628. doi: 10.1371/journal.pone.0058628
Fournier, P.-E., Vallenet, D., Barbe, V., Audic, S., Ogata, H., Poirel, L., et al. (2006). Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet. 2:e7 doi: 10.1371/journal.pgen.0020007
Garnacho-Montero, J., Ortiz-Leyba, C., Jimenez-Jimenez, F. J., Barrero-Almodovar, A. E., Garcia-Garmendia, J. L., Bernabeu-Wittell, M., et al. (2003). Treatment of multidrug-resistant Acinetobacter baumannii ventilator-associated pneumonia (VAP) with intravenous colistin: a comparison with imipenem-susceptible VAP. Clin. Infect. Dis. 36, 1111–1118. doi: 10.1086/374337
Grant, J. R., and Stothard, P. (2008). The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 36, 181–194. doi: 10.1093/nar/gkn179
Hamidian, M., and Hall, R. M. (2014). pACICU2 is a conjugative plasmid of Acinetobacter carrying the aminoglycoside resistance transposon TnaphA6. J. Antimicrob. Chemother. 69, 1146–1158. doi: 10.1093/jac/dkt488
Hamidian, M., Kenyon, J. J., Holt, K. E., Pickard, D., and Hall, R. M. (2014). A conjugative plasmid carrying the carbapenem resistance gene blaOXA−23 in AbaR4 in an extensively resistant GC1 Acinetobacter baumannii isolate. J. Antimicrob. Chemother. 69, 2625–2628. doi: 10.1093/jac/dku188
Hayes, F., and Kêdzierska, B. (2014). Regulating toxin-antitoxin expression: controlled detonation of intracellular molecular timebombs. Toxins 6, 337–358 doi: 10.3390/toxins6010337
Higgins, P. G., Dammhayn, C., Hackel, M., and Seifert, H. (2010). Global spread of carbapenem-resistant Acinetobacter baumannii. J. Antimicrob. Chemother. 65, 233–238. doi: 10.1093/jac/dkp428
Hood, M. I., Becker, K. W., Roux, C. M., Dunman, P. M., and Skaar, E. P. (2013). Genetic determinants of intrinsic colistin tolerance in Acinetobacter baumannii. Infect. Immun. 81, 542–551. doi: 10.1128/IAI.00704-12
Huang, H., Yang, Z.-L., Wu, X.-M., Wang, Y., Liu, Y.-J., Luo, H., et al. (2012). Complete genome sequence of Acinetobacter baumannii MDR-TJ and insights into its mechanism of antibiotic resistance. J. Antimicrob. Chemother. 67, 2825–2832. doi: 10.1093/jac/dks327
Hujer, K. M., Hamza, N. S., Hujer, A. M., Perez, F., Helfand, M. S., Bethel, C. R., et al. (2005). Identification of a new allelic variant of the Acinetobacter baumannii cephalosporinase, ADC-7 beta-lactamase: defining a unique family of class C enzymes. Antimicrob. Agents Chemother. 49, 2941–2948. doi: 10.1128/AAC.49.7.2941-2948.2005
Hyatt, D., Chen, G. L., Locascio, P. F., Land, M. L., Larimer, F. W., and Hauser, L. J. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119
Iacono, M., Villa, L., Fortini, D., Bordoni, R., Imperi, F., Bonnal, R. J. P., et al. (2008). Whole-genome pyrosequencing of an epidemic multidrug-resistant Acinetobacter baumannii strain belonging to the European clone II group. Antimicrob. Agents Chemother. 52, 2616–2625 doi: 10.1128/AAC.01643-07
Jacoby, G. A. (2009). AmpC beta-lactamases. Clin. Microbiol. Rev. 22, 161–182. doi: 10.1128/CMR.00036-08
Jørgensen, M. G., Pandey, D. P., Jaskolska, M., and Gerdes, K. (2009). HicA of Escherichia coli defines a novel family of translation-independent mRNA interferases in bacteria and archaea. J. Bacteriol. 191, 1191–1199. doi: 10.1128/JB.01013-08
Juhas, M., Crook, D. W., and Hood, D. W. (2008). Type IV secretion systems: tools of bacterial horizontal gene transfer and virulence. Cell. Microbiol. 10, 2377–2386 doi: 10.1111/j.1462-5822.2008.01187.x
Jurenaite, M., Markuckas, A., and Suziedeliene, E. (2013). Identification and characterization of type II toxin-antitoxin systems in the opportunistic pathogen Acinetobacter baumannii. J. Bacteriol. 14, 3165–3172. doi: 10.1128/jb.00237-13
Kim, D. H., Choi, J.-Y., Kim, H. W., Kim, S. H., Chung, D. R., Peck, K. R., et al. (2013). Spread of carbapenem-resistant Acinetobacter baumannii global clone 2 in Asia and AbaR-type resistance islands. Antimicrob. Agents Chemother. 57, 5239–5246. doi: 10.1128/AAC.00633-13
Lagesen, K., Hallin, P., Rødland, E. A., Staerfeldt, H.-H., Rognes, T., and Ussery, D. W. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108. doi: 10.1093/nar/gkm160
Landman, D., Georgescu, C., Martin, D. A., and Quale, J. (2008). Polymyxins revisited. Clin. Microbiol. Rev. 21, 449–465. doi: 10.1128/CMR.00006-08
Lean, S.-S., Suhaili, Z., Salwani, I., Nor Iza, A. R., Norlela, O., Fatimah, H. A., et al. (2014). Prevalence and genetic characterization of polymyxin-resistant Acinetobacter baumannii isolated from a tertiary hospital in Terengganu, Malaysia. ISRN Microbiol. 2014, 1–28 doi: 10.1155/2014/953417
Lean, S.-S., Yeo, C. C., Suhaili, Z., and Thong, K.-L. (2015). Whole-genome analysis of an extensively drug-resistant clinical isolate of Acinetobacter baumannii AC12: insights into the mechanisms of resistance of an ST195 clone from Malaysia. Int. J. Antimicrob. Agents 45, 178–182. doi: 10.1016/j.ijantimicag.2014.10.015
Lowe, T. M., and Eddy, S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964 doi: 10.1093/nar/25.5.0955
Magiorakos, A., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2011). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. Dis. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Maragakis, L. L., and Perl, T. M. (2008). Acinetobacter baumannii: epidemiology, antimicrobial resistance, and treatment options. Clin. Infect. Dis. 46, 1254–1263 doi: 10.1086/529198
Merino, M., Acosta, J., Poza, M., Sanz, F., Beceiro, A., Chaves, F., et al. (2010). OXA-24 carbapenemase gene flanked by XerC/XerD-like recombination sites in different plasmids from different Acinetobacter species isolated during a nosocomial outbreak. Antimicrob. Agents Chemother. 54, 2724–2727. doi: 10.1128/AAC.01674-09
Mihara, K., Tanabe, T., Yamakawa, Y., Funahashi, T., Nakao, H., Narimatsu, S., et al. (2004). Identification and transcriptional organization of a gene cluster involved in biosynthesis and transport of acinetobactin, a siderophore produced by Acinetobacter baumannii ATCC 19606. Microbiology 50, 2587–2597. doi: 10.1099/mic.0.27141-0
Moffatt, J. H., Harper, M., Adler, B., Nation, R. L., Li, J., and Boyce, J. D. (2011). Insertion sequence ISAba11 is involved in colistin resistance and loss of lipopolysaccharide in Acinetobacter baumannii. Antimicrob. Agents Chemother. 55, 3022–3024. doi: 10.1128/AAC.01732-10
Moffatt, J. H., Harper, M., Harrison, P., Hale, J. D. F., Vinogradov, E., Seemann, T., et al. (2010). Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob. Agents Chemother. 54, 4971–4977. doi: 10.1128/AAC.00834-10
Mosqueda, N., Espinal, P., Cosgaya, C., Viota, S., Plasensia, V., Lerma, F. Á., et al. (2013). Globally expanding carbapenemase finally bursts in Spain: nosocomial outbreak of Acinetobacter baumannii producing a plasmid-encoded OXA-23 in Barcelona, Spain. Antimicrob. Agents Chemother. 57, 5155–5157. doi: 10.1128/AAC.01486-13
Mosqueda, N., Gato, E., Roca, I., López, M., de Alegría, C. R., Fernández-Cuenca, F., et al. (2014). Characterization of plasmids carrying the blaOXA-24/40 carbapenemase gene and the genes encoding the AbkA/AbkB proteins of a toxin/antitoxin system. J. Antimicrob. Chemother. 69, 2629–2633. doi: 10.1093/jac/dku179
Nigro, S. J., and Hall, R. M. (2012). Tn6167, an antibiotic resistance island in an Australian carbapenem-resistant Acinetobacter baumannii GC2, ST92 isolate. J. Antimicrob. Chemother. 67, 1342–1346. doi: 10.1093/jac/dks037
Nordmann, P., and Poirel, L. (2008). Acinetobacter baumannii – Basic and Emerging Mechanisms of Resistance. Euro. Infect. Dis. 2, 94–97.
Nwugo, C. C., Gaddy, J. A., Zimbler, D. L., and Actis, L. A. (2011). Deciphering the iron response in Acinetobacter baumannii: a proteomics approach. J. Proteomics. 74, 44–58 doi: 10.1016/j.jprot.2010.07.010
Park, Y. K., Choi, J. Y., Shin, D., and Ko, K. S. (2011). Correlation between overexpression and amino acid substitution of the PmrAB locus and colistin resistance in Acinetobacter baumannii. Int. J. Antimicrob. Agents. 37, 525–530. doi: 10.1016/j.ijantimicag.2011.02.008
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT–PCR. Nucl. Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Poirel, L., and Norman, P. (2006). Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin. Micro. Infect. 12, 826–836. doi: 10.1111/j.1469-0691.2006.01456.x
Poole, K. (2004). Resistance to beta-lactam antibiotics. Cell. Mol. Life Sci. 61, 2200–2223. doi: 10.1007/s00018-004-4060-9
Post, V., White, P. A., and Hall, R. M. (2010). Evolution of AbaR-type genomic resistance islands in multiply antibiotic-resistant Acinetobacter baumannii. J. Antimicrob. Chemother. 65, 1162–1170. doi: 10.1093/jac/dkq095
Roca, I., Espinal, P., Villa-Faréz, X., and Villa, J. (2012). The Acinetobacter baumannii oxymoron: commensal hospital dweller turned pan-drug-resistant menace. Front. Microbiol. 3:148 doi: 10.3389/fmicb.2012.00148
Rodríguez-Martínez, J.-M., Poirel, L., and Nordmann, P. (2010). Genetic and functional variability of AmpC-type β-lactamases from Acinetobacter baumannii. Antimicrob. Agents Chemother. 54, 4930–4933. doi: 10.1128/AAC.00427-10
Saranathan, R., Tomar, A., Sudhakar, P., Arunkumar, K. P., and Prashanth, K. (2014). Draft genome sequence of a multidrug-resistant Acinetobacter baumannii PKAB07 clinical strain from India belonging to sequence type 195. Genome Announ. 2, e00184–e00114. doi: 10.1128/genomeA.00184-14
Segal, H., Garnym, S. M., and Elisha, B. G. (2005). Is ISABA-1 customized for Acinetobacter? FEMS Microbiol. Lett. 243, 425–429. doi: 10.1016/j.femsle.2005.01.005
Tian, G.-B., Adams-Haduch, J. M., Taracila, M., Bonomo, R. A., Wang, H.-N., and Doi, Y. (2011). Extended-spectrum AmpC cephalosporinase in Acinetobacter baumannii: ADC-56 confers resistance to cefepime. Antimicrob. Agents Chemother. 55, 4922–4925. doi: 10.1128/AAC.00704-11
Tian, Q. B., Ohnishi, M., Tabuchi, A., and Terawaki, Y. (1996). A new plasmid-encoded proteic killer gene system: cloning, sequencing, and analyzing hig locus of plasmid Rts1. Biochem. Biophys. Res. Commun. 220, 280–284 doi: 10.1006/bbrc.1996.0396
Vila, J., Martí, S., and Sánchez-Céspedes, J. (2007). Porins, efflux pumps and multidrug resistance in Acinetobacter baumannii. J. Antimicrob. Chemother. 59, 1210–1215. doi: 10.1093/jac/dkl509
Wieczorek, P., Sacha, P., Hauschild, T., Zórawski, M., Krawczyk, M., and Tryniszewska, E. (2008). Multidrug resistant Acinetobacter baumannii–the role of AdeABC (RND family) efflux pump in resistance to antibiotics. Folia Histochem. Cytobiol. 46, 257–267. doi: 10.2478/v10042-008-0056-x
Wisplinghoff, H., Decker, M., Haefs, C., Krut, O., Plum, G., and Seifert, H. (2003). Mutations in gyrA and parC associated with resistance to fluoroquinolones in epidemiologically defined clinical strains of Acinetobacter baumannii. J. Antimicrob. Chemother. 51, 177–180. doi: 10.1128/AAC.39.5.1201
Xu, Z., and Hao, B. (2009). CVTree update: a newly designed phylogenetic study platform using composition vectors and whole genomes. Nucleic Acids Res. 37, 174–178. doi: 10.1093/nar/gkp278
Zarrilli, R., Pournaras, S., Giannouli, M., and Tsakris, A. (2013). Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int. J. Antimicrob. Agents 41, 11 doi: 10.1016/j.ijantimicag.2012.09.008
Zhou, H., Zhang, T., Yu, D., Pi, B., Yang, Q., Zhou, J., et al. (2011). Genomic analysis of the multidrug-resistant Acinetobacter baumannii Strain MDR-ZJ06 widely spread in China. Antimicrob. Agents Chemother. 55, 4506–4512. doi: 10.1128/AAC.01134-10
Zhu, L., Yan, Z., Zhang, Z., Zhou, Q., Zhou, J., Wakeland, E. K., et al. (2013). Complete genome analysis of three Acinetobacter baumannii clinical isolates in China for insight into the diversification of drug resistance elements. PLoS ONE 8:e66584. doi: 10.1371/journal.pone.0066584
Keywords: Acinetobacter baumannii, carbapenem-resistant, polymyxin, whole genome sequencing, resistance island, resistance mechanisms
Citation: Lean S-S, Yeo CC, Suhaili Z and Thong K-L (2016) Comparative Genomics of Two ST 195 Carbapenem-Resistant Acinetobacter baumannii with Different Susceptibility to Polymyxin Revealed Underlying Resistance Mechanism. Front. Microbiol. 6:1445. doi: 10.3389/fmicb.2015.01445
Received: 26 June 2015; Accepted: 03 December 2015;
Published: 05 January 2016.
Edited by:
Margaret Ip, The Chinese University of Hong Kong, ChinaReviewed by:
Benjamin Andrew Evans, University of Edinburgh, UKRaffaele Zarrilli, University of Naples Federico II, Italy
Copyright © 2016 Lean, Yeo, Suhaili and Thong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kwai-Lin Thong, dGhvbmdrbEB1bS5lZHUubXk=
 Soo-Sum Lean
Soo-Sum Lean Chew Chieng Yeo
Chew Chieng Yeo Zarizal Suhaili
Zarizal Suhaili Kwai-Lin Thong
Kwai-Lin Thong