- 1Department of Biological Sciences, Faculty of Agriculture, Science and Technology, North-West University, Mmabatho, South Africa
- 2Food Security and Safety Niche Area, Faculty of Agriculture, Science and Technology, North-West University, Mmabatho, South Africa
Macergens are bacteria capable of releasing pectic enzymes (pectolytic bacteria). These enzymatic actions result in the separation of plant tissues leading to total plant destruction. This can be attributed to soft rot diseases in vegetables. These macergens primarily belong to the genus Erwinia and to a range of opportunistic pathogens namely: the Xanthomonas spp., Pseudomonas spp., Clostridium spp., Cytophaga spp., and Bacillus spp. They consist of taxa that displayed considerable heterogeneity and intermingled with members of other genera belonging to the Enterobacteriaceae. They have been classified based on phenotypic, chemotaxonomic and genotypic which obviously not necessary in the taxonomy of all bacterial genera for defining bacterial species and describing new ones These taxonomic markers have been used traditionally as a simple technique for identification of bacterial isolates. The most important fields of taxonomy are supposed to be based on clear, reliable and worldwide applicable criteria. Hence, this review clarifies the taxonomy of the macergens to the species level and revealed that their taxonomy is beyond complete. For discovery of additional species, further research with the use modern molecular methods like phylogenomics need to be done. This can precisely define classification of macergens resulting in occasional, but significant changes in previous taxonomic schemes of these macergens.
Introduction
Marcergens are soft rot causing bacteria, responsible for plant tissue maceration resulting in total tissue collapse (Beattie, 2006; Bhai et al., 2012). Soft rot diseases of vegetables are the most characteristic symptom of tissue maceration in a plant. These begin as small water soaked lesion, expands and intensifies until the tissue turns soft and watery (Reddy, 2015). Apparently, the outer surface of the diseased plant might stay unbroken, while tanning and depressed, or enclosed in an exuding bacterial mucus layer (Heyman et al., 2013). Foul smells are common owing to the discharge of explosive complexes through tissue degradation. Best bacterial growth follows plant cell lysis in these diseases (Rich, 2013). Soft-rotting bacteria are distinguished for the speed at which they stimulate soft rot. Warehoused crop may turn to liquid in only a few hours (Reddy, 2015). These pathogens usually enter through wound spots or natural openings such as lenticels and persist in the intercellular spaces and vascular tissues till the environmental conditions become fit for disease development. Parenchymatous tissues are macerated by massive quantities of pectic exoenzymes exudates produced during this period. These enzymes comprise of cellulolytic enzymes, pectate lyases, and pectin methylesterases, which are responsible for the total tissue destruction (Parthiban et al., 2012).
Soft rot can be found worldwide, anywhere ample storage tissues of vegetables and ornamentals are found (Golkhandan et al., 2013; Elbanna et al., 2014). Potatoes, carrots, and onions are among the most affected vegetables, along with tomato and cucumber (Mir et al., 2010) (Figure 1). Soft rot of fleshy vegetables and ornamental plants can be caused by more than six genera of pectolytic bacteria comprising; Erwinia, Pseudomonas, Clostridium, Bacillus, Cytophaga, and Xanthomonas (Elbanna et al., 2014). The estimated rate of infection of macergens on harvested crop ranges from 15 to 30%. Erwinia are the major macergens causing tissue degradation in vegetables (Choi and Kim, 2013; Waleron et al., 2014). Although, Erwinia are the primary macergens, it is not a single taxon. It is reclassified into genera such as Pectobacterium and Dickeya (Brady et al., 2012; Nabhan et al., 2012; Czajkowski et al., 2013). Macergens comprise of multiple groups ranging from the very complex Pseudomonas, a gamma-proteobacteria to as diverse as Bacillus and Clostridium which are firmicutes. Bacillus spp., Clostridium spp., Pseudomonas marginalis, and Pantoea agglomerans only cause soft rot when conditions are favorable to do so, thus are secondary invader called opportunistic pathogens (da Silva, 2013). Among all these pectolytic bacteria, soft rot Erwinias are the most important primary macergens that can macerate both the growing and harvested crop (Baz et al., 2012). All other bacteria are referred to as secondary because they can only destroy the parenchymatous tissues of plant under extreme environmental conditions or secondary invaders after Erwinias or other pathogens have infected the plant.
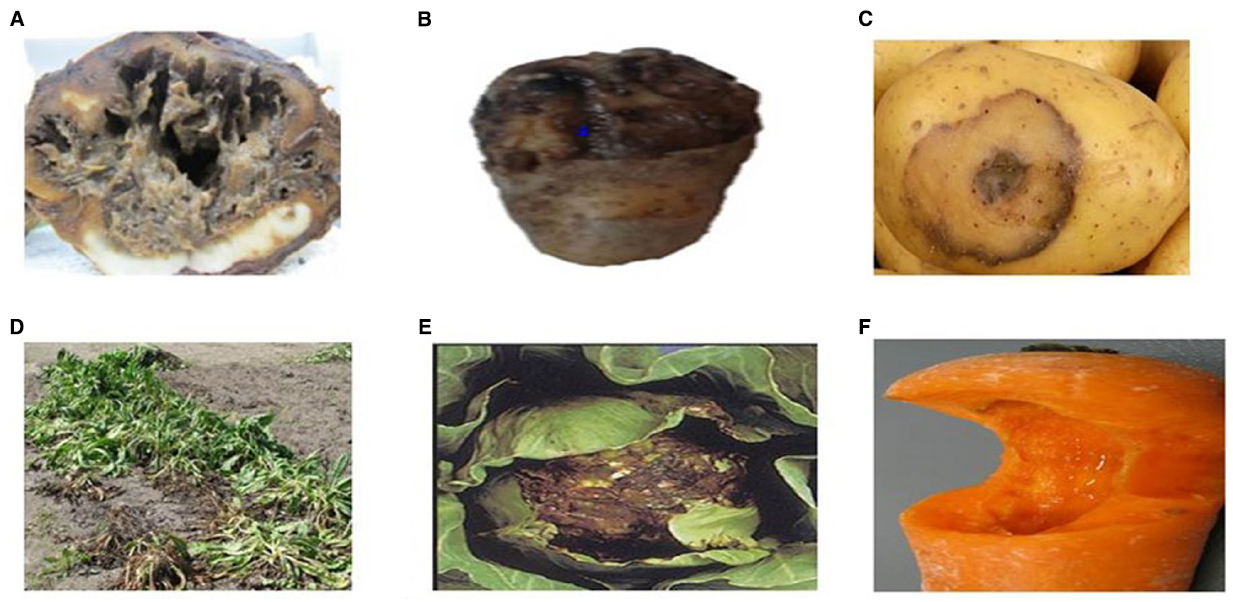
Figure 1. Unmarketable Vegetable as a Result of Macergen Infestation (A). Chicory root affected by soft rot diseases, (B,C). Potato with soft rot diseases, (D). Chicory leaves with soft rot disease, (E). Cabbage with soft rot disease, (F). Carrot with soft rot disease. Adapted from Lindsey du Toit, Washington State University, North Carolina Cooperative Extension Sevice (Lan et al., 2013).
These macergens infect and destroy plant tissues both pre- and post-harvest and this species causes the greatest damage to harvested vegetables (Lee et al., 2012). There is need to ensure a continuous cold chain from immediately after post-harvest, to retail for successful management of these ubiquitous spoilage bacteria that only thrive well at temperatures of 20°C and above (Tournas, 2005). The fluorescent Pseudomonads (P. fluorescens and P. viridiflava) can macerate plant parenchymatous tissues at a temperature below 4°C. This cause higher occurrence of these bacteria on decayed vegetables both at wholesale and retail markets. These soft-rotting fluorescent Pseudomonads and Erwinia therefore become the major threat to commercial fresh product operations and fresh vegetables precisely, from the farm to retail and wholesale outlets (Saranraj et al., 2012). There are currently no commercial agents available specifically for controlling soft rot (Yaganza et al., 2014).
Despite advances in vegetable production and disease management, many challenges face growers of vegetables, out of which the major one is the damage caused by macergens (Wu et al., 2012). Macergens damage the tissues of vegetable thereby reducing the quality, yield, shelf-life and consumer satisfaction of these plants (Akhtar, 2015). They usually cause great economic losses due to their ability to infect and macerate vegetable tissues at any point in time, be it, the field, transit, storage or marketing period (Lee et al., 2012). In the nature of today’s worldwide market, there are extremely high expectations for growers to provide ample supplies of high-quality, disease-free produce that have extended shelf-life (Kewa, 2012; Cheverton, 2015). The traditional methods to identify these macergens are extremely slow, more complex and obsolete (Hawks, 2005). Also, resistance genes active against macergens have been found in multiple host species, but their sequences and mechanisms remain unknown (Nykyri et al., 2012). Hence, means of quick identification of these bacteria are very essential. But the understanding of the taxonomy of these macergens will go a long way in shedding light to understand their biology and ultimately to the best method of controlling them. At present, there is very few knowledge available on the biology, ecology and epidemiology of macergens affecting vegetables in lowland and highland tropics. In order to increase crop production an assessment of biology, ecology and epidemiology of these bacteria need to be successfully implemented. Thus, this review focuses on the classification and taxonomy of the macergens to the species level. This is very important for more exploration in biotechnology.
Types of Microorganisms on Vegetables
The majority of Gram negative rods identified from raw vegetables were fluorescent Pseudomonads spp., Klebsiella spp., Serratia spp., Flavobacterium spp., Xanthomonads spp., Chronobacterium spp., and Alcaligenes (Elbanna et al., 2014). In vegetables like broccoli, cabbage, mungbean sprouts and carrot, Gram positive rods were predominantly isolated. Coryneform bacteria and catalase negative cocci were also predominantly isolated from broccoli, raw peas and raw sweet corn. In India, the mesophilic microflora of potatoes mainly comprised Gram positive bacteria, Bacillus spp., and Micrococcus spp. as fluorescent Pseudomonads, Cytophaga spp., Flavobacterium spp., Xanthomonas. spp., and Erwinia spp. Leuconostoc mesenteroides was the most common and abundant species found in vegetables amongst lactic acid bacteria (Andrews and Harris, 2000).
Taxonomy of Macergens
Genus Erwinia
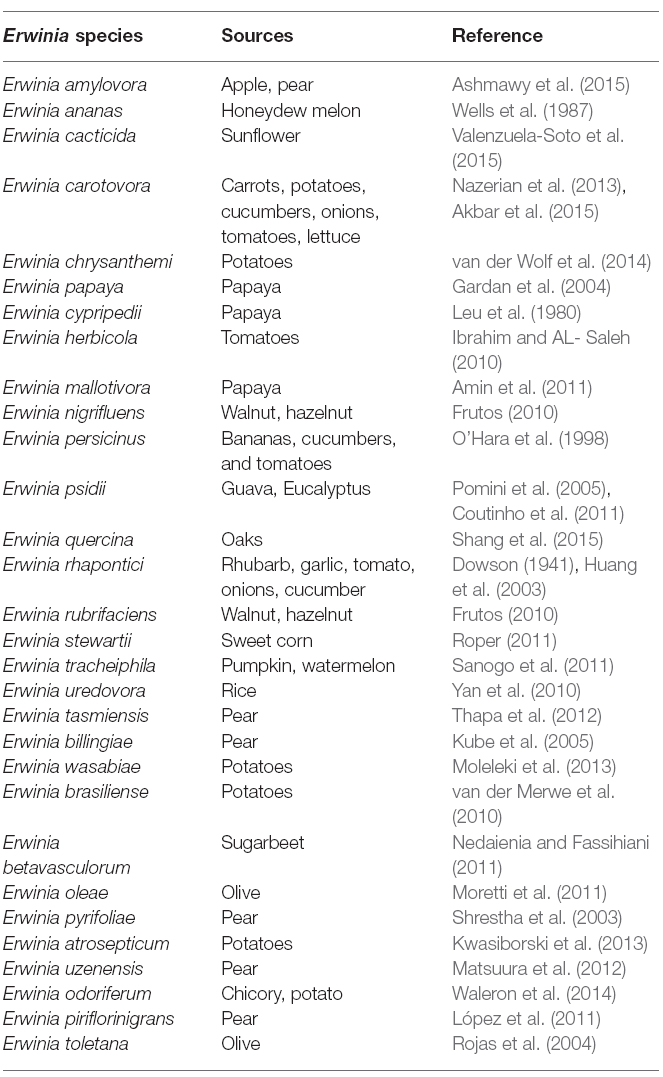
Erwinia belongs to the phylum Proteobacteria, class Gammaproteobacteria, order Enterobacteriales and family Enterobacteriaceae. For the past several decades, Enterobacteria that macerate and decay plants tissues, often referred to as the pectolytic Erwinias, were placed in genus Erwinia. Named after the eminent plant pathologist, Erwinin F. Smith. They are non-spore forming, facultative Gram negative rod-shaped anaerobes of approximately 0.7 × 1.5 μm in size with peritrichous flagella. This genus contains diverse set of group of organisms represented in Table 1. Since its establishment many new genera have been generated from Erwinia.
Nomenclature of Erwinia
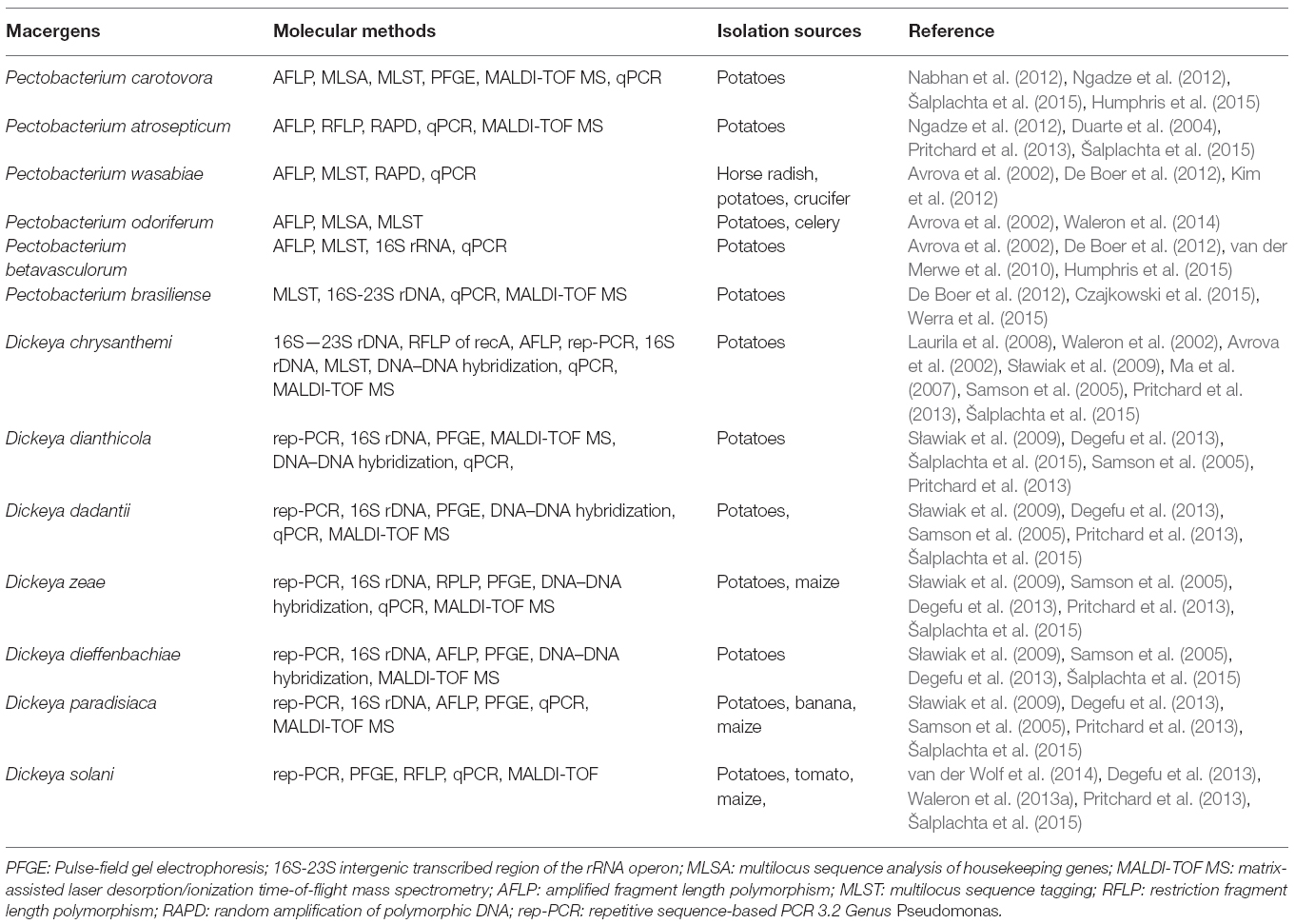
Traditionally two species (Erwinia carotovora and Erwinia chrysanthemi) are circumscribed as the important plant pathogenic strains, but has been reclassification into a new genus, Pectobacterium, with multiple species being proposed (Gardan et al., 2003). Pectobacterium spp. (Waldee, 1945; formerly Erwinia carotovora) and Dickeya spp. (formerly Erwinia chrysanthemi) species are related to soft rot Enterobacteria pathogens with broad host ranges. These species formerly were known as the soft rot Erwinia spp., but several studies have shown that the soft rot Enterobacteria and E. amylovora, the type strain of the Erwinia genus, are too divergent to be included in one clade. Therefore, the soft rot Erwinia spp. were moved to two new genera as Pectobacterium and Dickeya (Nabhan et al., 2013). Pectobacterium and Dickeya spp. are considered broad-host range pathogens in part because, they have been isolated from so many plant species and in part because, single strains are pathogens of numerous plant species under experimental conditions. Within the genus Pectobacterium, there are five major clades designated I, II, III, IV, and V, which differs from previous studies. These comprise five subspecies or species-level clades of Pectobacterium namely; Pectobacterium carotovorum subsp. carotovorum (syn. Erwinia carotovorum subsp. carotovorum), Pectobacterium atrosepticum (syn. Erwinia carotovorum subsp. atrosepticum) Pectobacterium wasabiae (syn. Erwinia carotovorum subsp. wasabiae), Pectobacterium betavasculorum (syn. Erwinia carotovorum subsp. betavasculorum), and Pectobacterium carotovorum subsp. brasiliense (Hauben et al., 2005; Nabhan et al., 2012). The reconstructed phylogenies agree that P. atrosepticum, P. betavasculorum, and P. wasabiae do form individual clades and place the brasiliensis strains in an individual clade.
Previous suggestions to separate the pectolytic Enterobacteria into the genus Pectobacterium has not found favor among phytobacteriologists. Initially the suggestion was made by Waldee (1945), who recommended the segregation on the basis of the unique pectolytic activity of the bacteria. Consequently, Hauben et al. (1998) revived the suggestion to support the proposal by adding evidence from the 16S ribosomal DNA sequence analysis of various plant-associated members of the Enterobacteriaceae. Although the phenotypic characterization and analysis of a single DNA fragment might have been considered insufficient for the subdivision at the generic level, the DNA-DNA hybridization study conducted by Gardan et al. (2003) provides further stimulation to change in favor of the new nomenclature. Samson et al. (2005), have proposed several new species from new genus, Dickeya for E. chrysanthemi, comprising of six genomic species namely: Dickeya dianthicola, D. dadantii, D. zeae, D. chrysanthemi, D. dieffenbachiae, D. paradisiaca.
A recently initiated multi-locus sequencing project, as well as DNA hybridization data from the 1970s, supports the transfer of E. carotovora and E. chrysanthemi to two separate genera as well as the elevation of some soft rot Erwinia subgroups to the species level (Brady et al., 2012).
All the phylogenetic analyses completed to date have suffered from the small number of strains available for some Enterobacteria species, which makes it difficult to determine the relatedness of these taxa. Unfortunately, the naming and re-naming of species has caused considerable confusion in the literature, resulting in manuscripts being published with names that were used for only a few years. Since Erwinia has remained the preferred name used in the literature, the comprehensive phylogenetic study of the entire group of soft rot Enterobacteria remains uncompleted (Charkowski, 2006; Elbanna et al., 2014). The pectolytic Erwinia are ubiquitous in environments that support plant growth, and because they may be found in association with asymptomatic plants, they have been viewed as opportunistic pathogens analogous to medical bacteria that infect only immunologically compromised individuals. Pectobacterium carotovorum, in the family Enterobacteriaceae, is a highly diverse species consisting of at least two valid names, P. carotovorum subsp. carotovorum and P. carotovorum subsp. odoriferum and a suggested third taxon, P. carotovorum subsp. brasiliense (De Boer et al., 2012). Despite the lack of valid carotovorum publication, the P. carotovorum subsp. brasiliense name has been used in more than 10 publications since first published in 2004 as Erwinia carotovora subsp. brasiliense (Ma et al., 2007). Assigning strains to this taxon was based mainly on the genetic information of the 16S-23S intergenic spacer region of the rRNA operon, partial sequence of 16S rRNA gene and multilocus sequence analysis (MLSA) of housekeeping genes and MALDI-TOF characterization (Wensing et al., 2012). Table 2 depicts the molecular method employed in the characterization of Pectobacterium and Dickeya species. Pectobacterium carotovorum subsp. brasiliense was first described as causing blackleg disease on potatoes (Solanum tuberosum L.) in Brazil and has since been described as also causing soft rot in Capsicum annum L., Ornithogalum spp., and Daucus carota subsp. Sativus. Strains of this taxon were isolated in the USA, Canada, South Africa, Peru, Germany, Japan, Israel, and Syria (Ngadze et al., 2012; Moleleki et al., 2013).
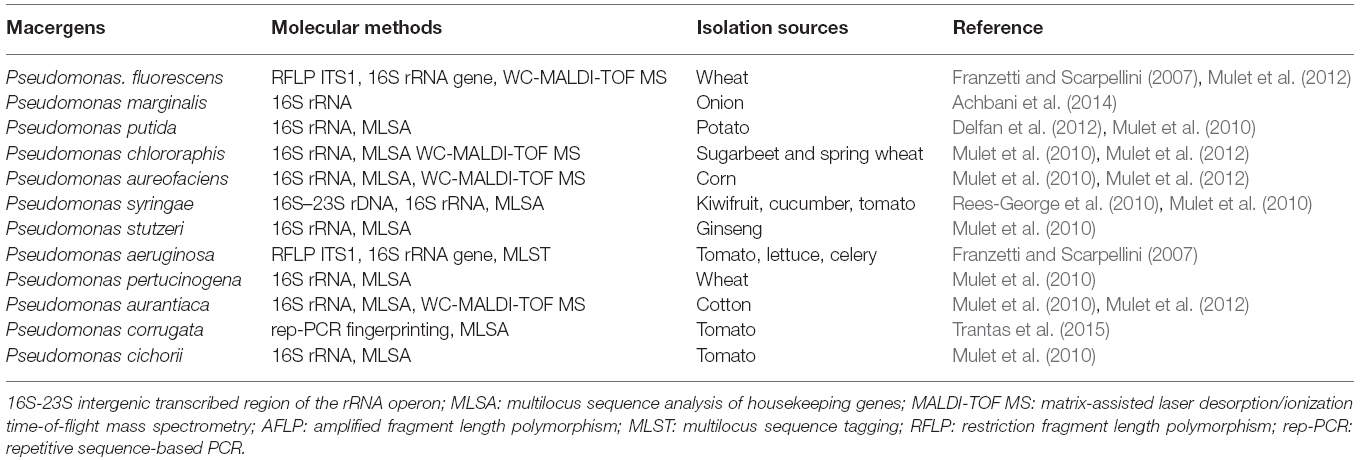
Genus Pseudomonas was first described in 1894 as one of the most diverse and ubiquitous bacterial genera whose species have been isolated worldwide from soil, decayed plant materials and rhizopheric region, quite a numerous plant species (Migula, 1894). They comprise a heterogeneous group of species which were grouped into five groups based on RNA homology (Saranraj et al., 2012). The RNA-homology group I belong to the fluorescent group because of their ability of producing pyoverdines. Pectolytic Pseudomonas belongs to this rRNA group I organism of gamma Proteobacteria. They are non-sporulating, Gram-negative, strict aerobic, rod-shape with polar flagella (Özen and Ussery, 2012). The strains of these bacteria called P. marginalis or P. fluorescens can be attributed to soft rot diseases in vegetables. The very complex groups of fluorescent, oxidase positive soft rot Pseudomonas are opportunistic macergens. Table 3 represents the molecular methods for the description of Pseudomonas species belonging to macergens.
Nomenclature of Pseudomonas
The nomenclature of bacteria in the genus Pseudomonas has changed considerably during the last decennia. P. marginalis or P. fluorescens are pectinolytic that cause strains soft rot on a wide range of hosts. The taxonomic and phytopathogenic status of P. marginalis is not well known. However, these are biochemically and phenotypically indistinguishable from saprophytic strains of P. fluorescens biovars II, P. putida, and P. chlororaphis (now includes P. aureofaciens). Based on their ability to degrade pectin and macerate the plant parenchymatous tissues they are referred to as P. marginalis. Recently, based on 16S rRNA analysis Anzai et al. (2000) came up with 57 strains of Pseudomonas sensu stricto with seven subclusters: P. syringae group, P. chlororaphis group, P. fluorescens group, P. putida group, P. stutzeri group, P. aeruginosa group, and P. pertucinogena group (Novik et al., 2015). Also, in the same genus Pseudomonas, some species have been found to be misclassified for instance P. aureofaciens and P. aurantiaca, which were reclassified into P. chlororaphis (Peix et al., 2007).
Ever since the discovery of genus Pseudomonas, it has undergone several taxonomic changes not only as far as the number of species included, but also as far as the criteria used for their definition and delineation. In Bergey’s Manual of Systematic Bacteriology’s current edition, an extensive list of methods used in Pseudomonas taxonomy was integrated (Palleroni, 2005). These methods, which consist of cell morphology and structure, cell wall composition, pigment types, nutritional and metabolic characteristics, susceptibility to different compounds, antibiotic production, pathogenicity of other organisms, antigenic structure and genetic and ecological studies revealed the efforts for characterizing Pseudomonas species. The phenotypic taxonomic markers comprise a set of tests, namely: cell shape, flagella type, consumption of carbon sources such as organic acids, polyalcohols and amino acids, ability to grow in different culture conditions, antibiotic resistance, production of antibiotic substances and exocellular enzymes (Palleroni, 2005).
In Pseudomonas taxonomy, the effectiveness of chemotaxonomic studies has been confirmed, such as quinone systems, fatty acid, protein, polar lipid or polyamine profiles, which are usually useful in the taxonomy of most bacterial groups. Generally, Pseudomonas species were reclassified by chemotaxonomic markers into other genera such as P. mephitica into Janthinobacterium lividum (Kämpfer et al., 2008). Janse et al. (1992), used whole fatty acid analysis in the study of a broad collection opportunistic phytopathogenic to clarify the taxonomic position of some P. marginalis strains included in the P. fluorescens group. Also, Janse et al. (1992) reported that other bacteria (P. putida, P. aureofaciens, and P. tolaasii) within the fluorescent oxidase positive pseudomonads group also exhibit pectinolytic ability. Hence, they are referred to as P. fluorescens supercluster. The study of polyamine composition in Proteobacteria revealed putrescine as the main polyamine present in the P. fluorescens complex, thus help in the delineation of species from this group. Recently, the polar lipid patterns of representative species of genus Pseudomonas were analyzed which showed the presence of phosphatidylglycerol, diphosphatidylglycerol and phosphatidylethanolamine as major polar lipids (Cámara et al., 2007).
Siderotyping an interesting taxonomic tool was used in characterizing fluorescent and then to non-fluorescent based on the isoelectrophoretic. Characterization of the major siderophores and pyoverdines and determination of strains pyoverdine mediated iron uptake specificity led to characterization of several Pseudomonas strains at species level, through species-specific pyoverdines (Novik et al., 2015). Mass spectrometry for the determination of molecular mass of pyoverdines has helped recently to improve siderotyping resolution power and accuracy (Meyer et al., 2008).
Currently fluorescent spectroscopy fingerprinting, the most modern techniques for biomolecules analysis are being applied to Pseudomonas taxonomy, by emission spectra of three intrinsic fluorophores (NADH, tryptophan, and the complex of aromatic amino acids and nucleic acid), which have been able to differentiate Pseudomonas at genus level from Burkholderia, Xanthomonas, or Stenotrophomonas with very high sensitivity, and moreover at species level P. chlororaphis, P. lundensis, P. fragi, P. taetrolens and P. stutzeri grouped separately from P. putida, P. pseudoalcaligenes, and P. fluorescens, which correlate with the phylogenetic clusters earlier obtained by Anzai et al. (2000); Peix et al. (2007), and Tourkya et al. (2009).
Hence, other gene sequences like housekeeping genes have been used in the last decade as phylogenetic molecular markers in taxonomic studies such as the recA, atpD, carA, gyrB, and rpoB, whose effectiveness has been demonstrated in genus Pseudomonas for species differentiation (Hilario et al., 2004). For instance, the effectiveness of rpoB has been reported in discriminating closely related Pseudomonas, with a phylogenetic resolution of the rpoB tree roughly three times higher than that of the 16S rRNA gene tree (Tayeb et al., 2005). These genes also enhanced differentiation of subspecies within P. chlororaphis (Hilario et al., 2004; Peix et al., 2007). Nevertheless, the analysis of housekeeping genes is not frequently used so far in Pseudomonas species description, but only gyrB, rpoB and rpoD have been integrated in the current description of P. xiamenensis (Lai and Shao, 2008).
16S-23S rRNA intergenic spacer is another phylogenetic marker used increasingly in taxonomic studies for discrimination of very closely related bacteria, at species and intraspecific levels, even at the strain level because of its high variability both in size and sequence (Sakamoto et al., 2001). This region can be amplified by using universal primers, and specific protocols (Locatelli et al., 2002). The efficacy of this phylogenetic marker has been reported in the differentiation of Pseudomonas species (Guasp et al., 2000). The selection of the minimal principles necessary for species delineation and description is selected for each bacterial genus by a committee created by experts in the given genus. The methods used in the taxonomy of the genus Pseudomonas and its related genera have been standardized by the subcommittee on the taxonomy. However, the minimal standards for genus Pseudomonas species description are yet to be cleared after the 2002 meeting of this subcommittee (De Vos and Yabuuchi, 2002). Hence, the new species description of this genus must be based on the general minimal standards for bacterial species characterization (Stackebrandt et al., 2002). These general minimal standards needed for the classification of new species and/or subspecies must comprise 16S rRNA sequencing, DNA-DNA hybridization, fatty acid analysis and phenotypic classification.
Genus Xanthomonas
The genus Xanthomonas belong to the family Xanthomonadaceae. This family composed of 10 genera that dwell in an extreme environment. The genus Xanthomonas belongs to the gamma proteolytic subdivision (Mbega et al., 2014). They are Gram-negative, aerobic, rod-shape, motile, non-spore forming with a single polar flagellum, comprises of 27 species infecting more than 400 dicots and monocots plant species (Rodriguez et al., 2012).
Nomenclature of Xanthomonas
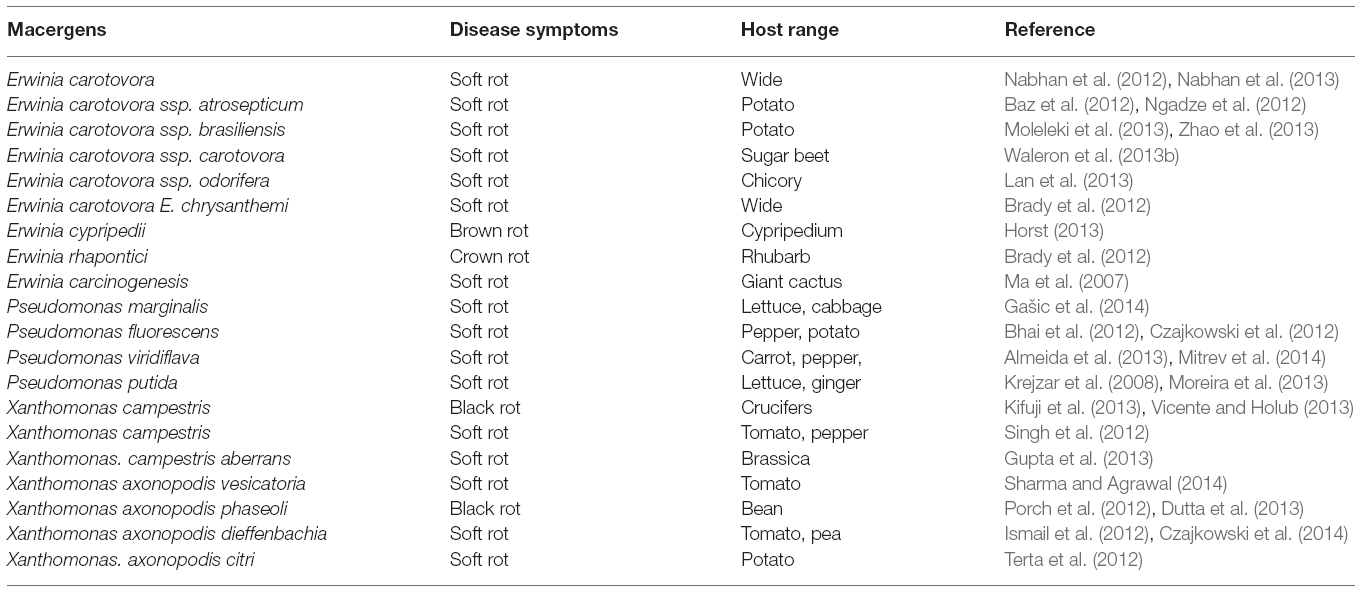
Traditionally, genus Xanthomonas is referred to as a taxon of pathogenic plant bacteria (Dye et al., 1974; Bradbury, 1984). Xanthomonas usually produce some extracellular polysaccharide namely: xanthan and xanthomonadin, a membrane-bound, brominated, aryl-polyene, yellow pigment (Adriko et al., 2014). This yellow pigment is responsible for their pathogenicity and virulence (Subramoni et al., 2006). However, the yellow-pigmented X. spp. (X. campestris) are the only one associated with tissue maceration of the post-harvest vegetables and fruits (Liao and Wells, 1987). They are opportunistic macergens because they are entering through natural openings or after infection of the plant by Erwinia spp.
Genetically, it can be differentiated into over 141 pathovars (pv.) based on specificity range (Swings and Civerolo, 1993). But Xanthomonas classification of X. campestris pathovar was based on the host pathogenicity system (Table 4)
Initially, this genus undergone diverse taxonomic and phylogenetic studies based on their phenotype and host specificity. Until Vauterin et al. (1995) revised the reclassification of Xanthomonas by DNA-DNA hybridization into 20 species based on their genomic relatedness. Phenotypic fingerprinting techniques such as 50S-polyacrylamide gel electrophoresis (50S-PAGE) of cellular proteins and gas chromatographic analysis of fatty acid methyl esters (FAME) reasonable supported these genomic groups to an extent. Hence, both techniques are useful tools in specific and interspecific differentiation of Xanthomonas levels (Rademaker et al., 2000).
Other analyses like Multi-Locus Sequence Analysis (MLSA), Amplified Fragment Length Polymorphism (AFLP) were also used in characterisation of this genus, revealing the complexity and diversity of the genus previously described by DNA-DNA hybridization (Ferreira-Tonin et al., 2012; Hamza et al., 2012). Not quite long, the phylogeny of species representing the principal lineages of the genus Xanthomonas were reported based on their genome (Rodriguez et al., 2012). The 16S ribosomal DNA sequences and MLSA classified Xanthomonas species into two major groups (Vicente and Holub, 2013). Group I comprising: X. albilineans, X. hyacinth, X. theicola, X. sacchari and X. translucens, and Group II made up of X. arboricola, X. axonopodis, X. bromi, X. campestris, X. cassavae, X. codiaei, X. cucurbitae, X. fragariae, X. hortorum, X. melonis, X. oryzae, X. pisi, X. populi, X. vasicola, and X. vesicatoria (Rodriguez et al., 2012). Thus, taxonomy of this genus are still subjected to debate since the last decade (Rodriguez et al., 2012; Vandroemme et al., 2013; Lamichhane, 2014).
Conclusion
The taxonomy of all these macergens is far from being complete because of the controversial issues arising from their classification which were based on host pathogenicity (Table 1). This may be affected by the sudden change in the ecosystem. This classification is not based on scientific research perspective for defined taxa and the consequences brought about by these marcergens may become difficult to understand. It is majorly based on symptoms that is similar in all the macergens, and this is unreliable according to (Sławiak et al., 2013). Although, some scientific method like MLSA were used for the classification they have limitation of single locus analysis. Thus, a proper classification is imperative, in order to reflect an understanding of their existing natural diversity and relationships among them. This will help plant breeders, farmers, and legislators to ensure quick and effective disease diagnosis and management, in order to avoid unnecessary destruction of economically valuable crops. The knowledge of genomic diversity within the macergens pathovars is necessary for host resistance disease based management strategies for the plant breeders.
As a concluding comment, we would like to stress that we applaud further developments in molecular methods of analyzing macergens for a better classification of these macergens. It is our belief, however, that any future progress in taxonomy as a scientific discipline will depend only on the availability of new experimental data that will broaden and refined the view on bacterial diversity.
Author Contributions
BR involved in data collection from internet, drafting of the manuscript or revising it critically for important intellectual content; have given final approval of the version to be published; and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. OO involved in collection of data, drafting of the manuscript, revising it critically, responsible for any aspect of the article and also help in the general supervision of the article.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank the North West University and National Research Foundation, South Africa for funds (Grant no. UID81192 OO Babalola) that have supported research in their laboratory.
Abbreviation
DNA: Deoxy Ribonucleic Acid; ITS: Internal transcribed spacer; MLSA: Multilocus Sequence Analysis; NADH: Nicotinamide Adenine Dinucleotide (Reduced); RNA: Ribonucleic Acid; rDNA: Ribosomal Deoxy Ribonucleic Acid; rRNA: Ribosomal Ribonucleic Acid
References
Achbani, E., Sadik, S., El Kahkahi, R., Benbouazza, A., and Mazouz, H. (2014). First report on Pseudomonas marginalis bacterium causing soft rot of onion in morocco. Atlas J. Biol. 3, 218–223. doi: 10.5147/ajb.2014.0136
Adriko, J., Mbega, E. R., Mortensen, C. N., Wulff, E. G., Tushemereirwe, W. K., Kubiriba, J., et al. (2014). Improved PCR for identification of members of the genus Xanthomonas. Eur. J. Plant Pathol. 138, 293–306. doi: 10.1007/s10658-013-0329-x
Akbar, A., Ahmad, M., Khan, S. Z., and Ahmad, Z. (2015). Characterization of the causal organism of soft rot of tomatoes and other vegetables and evaluation of its most aggressive isolates. Am. J. Plant Sci. 6, 511. doi: 10.4236/ajps.2015.64055
Akhtar, S. (2015). “Advances in conventional breeding approaches for postharvest quality improvement in vegetables,” in Postharvest Biology and Technology of Horticultural Crops: Principles and Practices for Quality Maintenance, ed. M. W. Siddiqui (New Jersey, USA: Apple Academic Press Inc.), 141–176.
Almeida, I., Maciel, K., Neto, J. R., and Beriam, L. (2013). Pseudomonas viridiflava in imported carrot seeds. Australas. Plant Dis. Notes 8, 17–19. doi: 10.1007/s13314-012-0086-2
Amin, N. M., Bunawan, H., Redzuan, R. A., and Jaganath, I. B. S. (2011). Erwinia mallotivora sp., a new pathogen of papaya (Carica papaya) in Peninsular Malaysia. Int. J. Mol. Sci. 12, 39–45. doi: 10.3390/ijms12010039
Andrews, J. H., and Harris, R. F. (2000). The ecology and biogeography of microorganisms on plant surface. Annu. Rev. Phytopathol. 38, 145–180. doi: 10.1146/annurev.phyto.38.1.145
Anzai, Y., Kim, H., Park, J. Y., Wakabayashi, H., and Oyaizu, H. (2000). Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. Int. J. Syst. Evol. Microbiol. 50, 1563–1589. doi: 10.1099/00207713-50-4-1563
Ashmawy, N. A., Zaghloul, T. I., and El-Sabagh, M. A. (2015). Isolation and molecular characterization of the fire blight pathogen, Erwinia amylovora, isolated from apple and pear orchards in Egypt. Plant Pathol. J. 14, 142. doi: 10.3923/ppj.2015.142.147
Avrova, A. O., Hyman, L. J., Toth, R. L., and Toth, I. K. (2002). Application of amplified fragment length polymorphism fingerprinting for taxonomy and identification of the soft rot bacteria Erwinia carotovora and Erwinia chrysanthemi. Appl. Environ. Microbiol. 68, 1499–1508. doi: 10.1128/AEM.68.4.1499-1508.2002
Baz, M., Lahbabi, D., Samri, S., Val, F., Hamelin, G., Madore, I., et al. (2012). Control of potato soft rot caused by Pectobacterium carotovorum and Pectobacterium atrosepticum by Moroccan actinobacteria isolates. World J. Microb. Biotechnol. 28, 303–311. doi: 10.1007/s11274-011-0820-5
Beattie, G. A. (2006). “Plant-associated bacteria: survey, molecular phylogeny, genomics and recent advances,” in Plant-associated Bacteria, ed. S. S. Gnanamanickam (Netherlands: Springer), 1–56. doi: 10.1007/978-1-4020-4538-7_1
Bhai, R., Kishore, V., Kumar, A., Anandaraj, M., and Eapen, S. (2012). Screening of rhizobacterial isolates against soft rot disease of ginger (Zingiber officinale Rosc.). J. Spices Aromat. Crops 14, 130–136.
Bradbury, J. F. (1984). “Genus II. Xanthomonas dowson 1939, 187,” in Bergey’s Manual of Systematic Bacteriology, eds N. R. Krieg and J. G. Holt (Baltimore: Williams & Wilkins), 199–210.
Brady, C. L., Cleenwerck, I., Denman, S., Venter, S. N., Rodríguez-Palenzuela, P., Coutinho, T. A., et al. (2012). Proposal to reclassify Brenneria quercina (Hildebrand and Schroth, 1967) Hauben et al. 1999 into a new genus, Lonsdalea gen. nov., as Lonsdalea quercina comb. nov., descriptions of Lonsdalea quercina subsp. quercina comb. nov., Lonsdalea quercina subsp. iberica subsp. nov., and Lonsdalea quercina subsp. britannica subsp. nov., emendation of the description of the genus Brenneria, reclassification of Dickeya dieffenbachiae as Dickeya dadantii subsp. dieffenbachiae comb. nov., and emendation of the description of Dickeya dadantii. Int. J. Syst. Evol. Microbiol. 62, 1592–1602. doi: 10.1099/ijs.0.035055-0
Cámara, B., Strömpl, C., Verbarg, S., Spröer, C., Pieper, D. H., and Tindall, B. J. (2007). Pseudomonas reinekei sp. nov., Pseudomonas moorei sp. nov. and Pseudomonas mohnii sp. nov., novel species capable of degrading chlorosalicylates or isopimaric acid. Int. J. Syst. Evol. Microb. 57, 923–931. doi: 10.1099/ijs.0.64703-0
Charkowski, A. O. (2006). “The soft rot Erwinia,” in Plant-Associated Bacteria, ed. S. S. Gnanamanickam (Heidelberg, NL: Springer), 423–505. doi: 10.1007/978-1-4020-4538-7_13
Cheverton, P. (2015). Key Account Management: Tools and Techniques for Achieving Profitable Key Supplier Status. Great Britain: Kogan Page Publishers.
Choi, O., and Kim, J. (2013). Pectobacterium carotovorum subsp. brasiliense causing soft rot on paprika in Korea. J. Phytopathol. 161, 125–127. doi: 10.1111/jph.12022
Coutinho, T. A., Brady, C. L., Van Der Vaart, M., Venter, S. N., Telechea, N., Rolfo, M., et al. (2011). A new shoot and stem disease of Eucalyptus species caused by Erwinia psidii. Australas. Plant Pathol. 40, 55–60. doi: 10.1007/s13313-010-0013-y
Czajkowski, R., De Boer, W., Van Der Zouwen, P., Kastelein, P., Jafra, S., De Haan, E., et al. (2013). Virulence of ‘Dickeya solani’ and Dickeya dianthicola biovar-1 and-7 strains on potato (Solanum tuberosum). Plant Pathol. 62, 597–610. doi: 10.1111/j.1365-3059.2012.02664.x
Czajkowski, R., De Boer, W., Van Veen, J., and Van Der Wolf, J. (2012). Characterization of bacterial isolates from rotting potato tuber tissue showing antagonism to Dickeya sp. biovar 3 in vitro and in planta. Plant Pathol. 61, 169–182. doi: 10.1111/j.1365-3059.2011.02486.x
Czajkowski, R., Ozymko, Z., and Lojkowska, E. (2014). Isolation and characterization of novel soilborne lytic bacteriophages infecting Dickeya spp. biovar 3 (‘D. solani’). Plant Pathol. 63, 758–772. doi: 10.1111/ppa.12157
Czajkowski, R., Pérombelon, M., Jafra, S., Lojkowska, E., Potrykus, M., Van Der Wolf, J., et al. (2015). Detection, identification and differentiation of Pectobacterium and Dickeya species causing potato blackleg and tuber soft rot: a review. Ann. Appl. Biol. 166, 18–38. doi: 10.1111/aab.12166
da Silva, W. L. (2013). Sweetpotato Storage Root Rots: Flooding-Associated Bacterial Soft Rot Caused by Clostridium spp. and Infection by Fungal End Rot Pathogens Prior to Harvest. Ph.D. thesis, Louisiana State University, Louisiana, USA.
De Boer, S. H., Li, X., and Ward, L. J. (2012). Pectobacterium spp. associated with bacterial stem rot syndrome of potato in Canada. Phytopathology 102, 937–947. doi: 10.1094/PHYTO-04-12-0083-R
De Vos, P., and Yabuuchi, E. (2002). International committee on systematic bacteriology: subcommittee on the taxonomy of Pseudomonas and related organisms international committee on systematic bacteriology: subcommittee on the taxonomy of Pseudomonas and related organisms. Int. J. Syst. Evol. Microbiol. 52, 2329. doi: 10.1099/ijs.0.02575-0
Degefu, Y., Potrykus, M., Golanowska, M., Virtanen, E., and Lojkowska, E. (2013). A new clade of Dickeya spp. plays a major role in potato blackleg outbreaks in North Finland. Ann. Appl. Biol. 162, 231–241.
Delfan, A., Etemadifar, Z., Bouzari, M., and Emtiazi, G. (2012). Screening of novel bacteriophage infection in Pseudomonas putida isolated from potato disease. Jundishapur J. Microbiol. 5, 550–554. doi: 10.5812/jjm.3786
Dowson, W. (1941). The identification of the bacteria commonly causing soft rot in plants. Ann. Appl. Biol. 28, 102–106. doi: 10.1111/j.1744-7348.1941.tb07543.x
Duarte, V., De Boer, S., Ward, L., and Oliveira, A. (2004). Characterization of atypical Erwinia carotovora strains causing blackleg of potato in Brazil. J. Appl. Microbiol. 96, 535–545. doi: 10.1111/j.1365-2672.2004.02173.x
Dutta, B., Block, C., Stevenson, K., Sanders, F. H., Walcott, R., and Gitaitis, R. (2013). Distribution of phytopathogenic bacteria in infested seeds. Seed Sci. Technol. 41, 383–397. doi: 10.15258/sst.2013.41.3.06
Dye, D., Lelliott, R., Buchanan, R., and Gibbons, N. (1974). “Genus II. Xanthomonas Dowson 1939, 187,” in Bergey’s Manual of Determinative Bacteriology, eds R. E. Buchanan, and N. E. Gibbons (Baltimore: Williams & Wilkins Company), 243–249.
Elbanna, K., Elnaggar, S., and Bakeer, A. (2014). Characterization of Bacillus altitudinis as a new causative agent of bacterial soft rot. J. Phytopathol. 162, 712–722. doi: 10.1111/jph.12250
Ferreira-Tonin, M., Rodrigues-Neto, J., Harakava, R., and Destéfano, S. A. L. (2012). Phylogenetic analysis of Xanthomonas based on partial rpoB gene sequences and species differentiation by PCR-RFLP. Int. J. Syst. Evol. Microbiol. 62, 1419–1424. doi: 10.1099/ijs.0.028977-0
Franzetti, L., and Scarpellini, M. (2007). Characterisation of Pseudomonas spp. isolated from foods. Ann. Microbiol. 57, 39–47. doi: 10.1007/BF03175048
Frutos, D. (2010). Bacterial diseases of walnut and hazelnut and genetic resources. J. Plant Pathol. 92, S79–S85.
Gardan, L., Christen, R., Achouak, W., and Prior, P. (2004). Erwinia papayae sp. nov., a pathogen of papaya (Carica papaya). Int. J. Syst. Evol. Microbiol. 54, 107–113. doi: 10.1099/ijs.0.02718-0
Gardan, L., Gouy, C., Christen, R., and Samson, R. (2003). Elevation of three subspecies of Pectobacterium carotovorum to species level: Pectobacterium atrosepticum sp. nov., Pectobacterium betavasculorum sp. nov. and Pectobacterium wasabiae sp. nov. Int. J. Syst. Evol. Microbiol. 53, 381–391. doi: 10.1099/ijs.0.02423-0
Gašić, K., Gavrilović, V., Dolovac, N., Trkulja, N., Živković, S., Ristić, D., et al. (2014). Pectobacterium carotovorum subsp. carotovorum-the causal agent of broccoli soft rot in Serbia. Pesticidi Fitomedicina 29, 249–255. doi: 10.2298/PIF1404249G
Golkhandan, E., Kamaruzaman, S., Sariah, M., Zainal Abidin, M., and Nasehi, A. (2013). Characterisation of Pectobacterium carotovorum causing soft rot on Kalanchoe gastonis-bonnierii in Malaysia. Archives Phytopathol. Plant Prot. 46, 1809–1815. doi: 10.1080/03235408.2013.778452
Guasp, C., Moore, E. R., Lalucat, J., and Bennasar, A. (2000). Utility of internally transcribed 16S-23S rDNA spacer regions for the definition of Pseudomonas stutzeri genomovars and other Pseudomonas species. Int. J. Syst. Evol. Microbiol. 50, 1629–1639. doi: 10.1099/00207713-50-4-1629
Gupta, M., Vikram, A., and Bharat, N. (2013). Black rot-A devastating disease of crucifers: a review. Agric. Rev. 34, 269–278. doi: 10.5958/j.0976-0741.34.4.012
Hamza, A., Robene-Soustrade, I., Jouen, E., Lefeuvre, P., Chiroleu, F., Fisher-Le Saux, M., et al. (2012). MultiLocus sequence analysis-and amplified fragment length polymorphism-based characterization of xanthomonads associated with bacterial spot of tomato and pepper and their relatedness to Xanthomonas species. Syst. Appl. Microbiol. 35, 183–190. doi: 10.1016/j.syapm.2011.12.005
Hauben, L. M., Edward, R. B., Vauterin, L., Steenackers, M., Mergaert, J., Verdonck, L., et al. (1998). Phylogenetic position of phytopathogens within the Enterobacteriaceae. Syst. Appl. Microbiol. 21, 384–397. doi: 10.1016/S0723-2020(98)80048-9
Hauben, L., Van Gijsegem, F., and Swings, J. (2005). “Genus XXIV. Pectobacterium Waldee 1945, 469AL emend,” in Bergey’s Manual of Systematic Bacteriology, 2nd Edn, eds L. Hauben, E. R. B. Moore, I. Vauterin, M. Steenakcers, J. Mergaert, J. Verdonck et al. (New York: Springer), 721–730.
Hawks, B. (2005). Agricultural bioterrorism protection act of 2002: possession, use, and transfer of biological agents and toxins; final rule. Federal Regist. 70, 13241–13292.
Heyman, F., Blair, J., Persson, L., and Wikström, M. (2013). Root rot of pea and faba bean in southern Sweden caused by Phytophthora pisi sp. nov. Plant Dis. 97, 461–471. doi: 10.1094/PDIS-09-12-0823-RE
Hilario, E., Buckley, T. R., and Young, J. M. (2004). Improved resolution on the phylogenetic relationships among Pseudomonas by the combined analysis of atpD, carA, recA, and 16S rDNA. Antonie Van Leeuwenhoek 86, 51–64. doi: 10.1023/B:ANTO.0000024910.57117.16
Horst, R. K. (ed) (2013). “Bacterial diseases,” in Westcott’s Plant Disease Handbook, (New York, USA: Springer), 69–90.
Huang, H., Hsieh, T., Erickson, R., and Erickson, R. (2003). Biology and epidemiology of Erwinia rhapontici, causal agent of pink seed and crown rot of plants. Plant Pathol. Bull. 12, 69–76.
Humphris, S. N., Cahill, G., Elphinstone, J. G., Kelly, R., Parkinson, N. M., Pritchard, L., et al. (2015). Detection of the bacterial potato pathogens Pectobacterium and Dickeya spp. using conventional and real-time PCR. Plant Pathol. Techniques Protocols 1302, 1–16. doi: 10.1007/978-1-4939-2620-6_1
Ibrahim, Y. E., and AL- Saleh, M. A. (2010). Isolation and characterization of Erwinia herbicola associated with internal discoloration of tomato fruits (Lycopersicon esculentum Mill) in Saudi Arabia. Emirates J. Food Agric. 22, 475–482. doi: 10.9755/ejfa.v22i6.4665
Ismail, M. E., Abdel-Monaim, M. F., and Mostafa, Y. M. (2012). Identification and pathogenicity of phytopathogenic bacteria associated with soft rot disease of girasole tuber. Not. Sci. Biol. 4, 75–81. doi: 10.5897/jbr11.015
Janse, J. D., Derks, J. H. J., Spit, B. E., and Van Der Tuin, W. R. (1992). Classification of fluorescent soft rot Pseudomonas Bacteria, Including P. marginalis Strains, using whole cell fatty acid analysis. Syst. Appl. Microbiol. 15, 538–553. doi: 10.1016/S0723-2020(11)80114-1
Kämpfer, P., Falsen, E., and Busse, H. (2008). Reclassification of Pseudomonas mephitica Claydon and Hammer 1939 as a later heterotypic synonym of Janthinobacterium lividum (Eisenberg 1891) De Ley et al. 1978. Int. J. Syst. Evol. Microbiol. 58, 136–138. doi: 10.1099/ijs.0.65450-0
Kewa, J. L. (2012). Supplying Customer Requirements in the Fresh Produce Chain in the Highlands of Papua New Guinea. Ph.D. thesis, Lincoln University, Lincoln.
Kifuji, Y., Hanzawa, H., Terasawa, Y., and Nishio, T. (2013). QTL analysis of black rot resistance in cabbage using newly developed EST-SNP markers. Euphytica 190, 289–295. doi: 10.1007/s10681-012-0847-1
Kim, M. H., Cho, M. S., Kim, B. K., Choi, H. J., Hahn, J. H., Kim, C., et al. (2012). Quantitative real-time polymerase chain reaction assay for detection of Pectobacterium wasabiae using YD repeat protein gene-based primers. Plant Dis. 96, 253–257. doi: 10.1094/PDIS-06-11-0511
Krejzar, V., Mertelík, J., Pánková, I., Kloudová, K., and Kudela, V. (2008). Pseudomonas marginalis associated with soft rot of Zantedeschia spp. Plant Prot. Sci. 44, 85–90.
Kube, M., Beck, A., Zinder, S., Kuhl, H., Reinhardt, R., and Adrian, L. (2005). Genome sequence of the chlorinated compound-respiring bacterium Dehalococcoides species strain CBDB1. Nat. Biotechnol. 23, 1269–1273. doi: 10.1038/nbt1131
Kwasiborski, A., Mondy, S., Beury-Cirou, A., and Faure, D. (2013). Genome sequence of the Pectobacterium atrosepticum strain CFBP6276, causing blackleg and soft rot diseases on potato plants and tubers. Genome Announc. 1, e00374-00313. doi: 10.1128/genomeA.00374-13
Lai, Q., and Shao, Z. (2008). Pseudomonas xiamenensis sp. nov., a denitrifying bacterium isolated from activated sludge. Int. J. Syst. Evol. Microbiol. 58, 1911–1915. doi: 10.1099/ijs.0.65459-0
Lamichhane, J. R. (2014). Xanthomonas arboricola diseases of stone fruit, almond, and walnut trees: progress toward understanding and management. Plant Dis. 98, 1600–1610. doi: 10.1094/PDIS-08-14-0831-FE
Lan, W. W., Nishiwaki, Y., Akino, S., and Kondo, N. (2013). Soft rot of root chicory in Hokkaido and its causal bacteria. J. General Plant Pathol. 79, 182–193. doi: 10.1007/s10327-013-0440-z
Laurila, J., Ahola, V., Lehtinen, A., Joutsjoki, T., Hannukkala, A., Rahkonen, A., et al. (2008). Characterization of Dickeya strains isolated from potato and river water samples in Finland. Eur. J. Plant Pathol. 122, 213–225. doi: 10.1007/s10658-008-9274-5
Lee, J.-H., Shin, H., Ji, S., Malhotra, S., Kumar, M., Ryu, S., et al. (2012). Complete genome sequence of phytopathogenic Pectobacterium carotovorum subsp. carotovorum bacteriophage PP1. J. Virol. 86, 8899–8900. doi: 10.1128/JVI.01283-12
Leu, L., Lee, C., and Huang, T. (1980). Papaya black rot caused by Erwinia cypripedii. Plant Prot. Bull. Taiwan 22, 377–384.
Liao, C. H., and Wells, J. M. (1987). Diversity of pectolytic, fluorescent pseudomonads causing soft rots of fresh vegetables at produce markets. Phytopathology 77, 673–677. doi: 10.1094/Phyto-77-673
Locatelli, L., Tarnawski, S., Hamelin, J., Rossi, P., Aragno, M., and Fromin, N. (2002). Specific PCR Amplification for the Genus Pseudomonas Targeting the 3′ Half of 16S rDNA and the Whole 16S–23S rDNA Spacer. Syst. Appl. Microbiol. 25, 220–227. doi: 10.1078/0723-2020-00110
López, M. M., Roselló, M., Llop, P., Ferrer, S., Christen, R., and Gardan, L. (2011). Erwinia piriflorinigrans sp. nov., a novel pathogen that causes necrosis of pear blossoms. Int. J. Syst. Evol. Microbiol. 61, 561–567. doi: 10.1099/ijs.0.020479-0
Ma, B., Hibbing, M. E., Kim, H. S., Reedy, R. M., Yedidia, I., Breuer, J., et al. (2007). Host range and molecular phylogenies of the soft rot enterobacterial genera Pectobacterium and Dickeya. Phytopathology 97, 1150–1163. doi: 10.1094/PHYTO-97-9-1150
Matsuura, T., Mizuno, A., Tsukamoto, T., Shimizu, Y., Saito, N., Sato, S., et al. (2012). Erwinia uzenensis sp. nov., a novel pathogen that affects European pear trees (Pyrus communis L.). Int. J. Syst. Evol. Microbiol. 62, 1799–1803. doi: 10.1099/ijs.0.032011-0
Mbega, E. R., Mabagala, R., Adriko, J., Lund, O. S., Wulff, E. G., and Mortensen, C. N. (2014). Five species of xanthomonads associated with bacterial leaf spot symptoms in tomato from Tanzania. Eur. J. Plant Pathol. 138, 293–306. doi: 10.1094/PDIS-01-12-0105-PDN
Meyer, J. M., Gruffaz, C., Raharinosy, V., Bezverbnaya, I., Schäfer, M., and Budzikiewicz, H. (2008). Siderotyping of fluorescent Pseudomonas: molecular mass determination by mass spectrometry as a powerful pyoverdine siderotyping method. Biometals 21, 259–271. doi: 10.1007/s10534-007-9115-6
Migula, N. (1894). Arbeiten aus dem Bakteriologischen. Inst. Technischen Hochschule Karlsruhe 1, 235–238.
Mir, S. A., Zargar, M. Y., Sheikh, P. A., Bhat, K. A., Bhat, N. A., and Masoodi, S. D. (2010). Studies on status and host range of soft rot disease of cabbage (Brassica oleracea var Capitata) Kashmir Valley. J. Phytol. 2, 55–59.
Mitrev, S., Karov, I., Kovacevik, B., and Kostadinovska, E. (2014). Pseudomonas population causing tomato pith necrosis in the Republic of Macedonia. J. Plant Pathol. 96, 589–592. doi: 10.4454/JPP.V96I3.002
Moleleki, L. N., Onkendi, E. M., Mongae, A., and Kubheka, G. C. (2013). Characterisation of Pectobacterium wasabiae causing blackleg and soft rot diseases in South Africa. Eur. J. Plant Pathol. 135, 279–288. doi: 10.1007/s10658-012-0084-4
Moreira, S. I., Dutra, D. D. C., Rodrigues, A. C., Oliveira, J. R. D., Dhingra, O. D., and Pereira, O. L. (2013). Fungi and bacteria associated with post-harvest rot of ginger rhizomes in Espírito Santo, Brazil. Trop. Plant Pathol. 38, 218–226.
Moretti, C., Hosni, T., Vandemeulebroecke, K., Brady, C., De Vos, P., Buonaurio, R., et al. (2011). Erwinia oleae sp. nov., isolated from olive knots caused by Pseudomonas savastanoi pv. savastanoi. Int. J. Syst. Evol. Microbiol. 61, 2745–2752. doi: 10.1099/ijs.0.026336-0
Mulet, M., Gomila, M., Scotta, C., Sánchez, D., Lalucat, J., and García-Valdés, E. (2012). Concordance between whole-cell matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry and multilocus sequence analysis approaches in species discrimination within the genus Pseudomonas. Syst. Appl. Microbiol. 35, 455–464. doi: 10.1016/j.syapm.2012.08.007
Mulet, M., Lalucat, J., and García-Valdés, E. (2010). DNA sequence-based analysis of the Pseudomonas species. Environ. Microbiol. 12, 1513–1530. doi: 10.1111/j.1462-2920.2010.02181.x
Nabhan, S., De Boer, S. H., Maiss, E., and Wydra, K. (2013). Pectobacterium aroidearum sp. nov., a soft rot pathogen with preference for monocotyledonous plants. Int. J. Syst. Evol. Microbiol. 63, 2520–2525. doi: 10.1099/ijs.0.046011-0
Nabhan, S., Wydra, K., Linde, M., and Debener, T. (2012). The use of two complementary DNA assays, AFLP and MLSA, for epidemic and phylogenetic studies of pectolytic enterobacterial strains with focus on the heterogeneous species Pectobacterium carotovorum. Plant Pathol. 61, 498–508. doi: 10.1111/j.1365-3059.2011.02546.x
Nazerian, E., Sijam, K., Ahmad, Z. A. M., and Vadamalai, G. (2013). Characterization of Pectobacterium carotovorum subsp. carotovorum as a new disease on Lettuce in Malaysia. Australas. Plant Dis. Notes 8, 105–107. doi: 10.1007/s13314-013-0107-9
Nedaienia, R., and Fassihiani, A. (2011). Host range and distribution of Pectobacterium betavasculorum the causal agent of bacterial vascular necrosis and root rot of sugarbeet in fars province. Iran. J. Plant Pathol. 47, 47–48.
Ngadze, E., Brady, C. L., Coutinho, T. A., and Van Der Waals, J. E. (2012). Pectinolytic bacteria associated with potato soft rot and blackleg in South Africa and Zimbabwe. Eur. J. Plant Pathol. 134, 533–549. doi: 10.1007/s10658-012-0036-z
Novik, G., Savich, V., and Kiseleva, E. (2015). “An insight into beneficial Pseudomonas bacteria,” in Microbiology in Agriculture and Human Health, (Europe: InTech), 73–105. doi: 10.5772/60502
Nykyri, J., Niemi, O., Koskinen, P., Nokso-Koivisto, J., Pasanen, M., Broberg, M., et al. (2012). Revised phylogeny and novel horizontally acquired virulence determinants of the model soft rot phytopathogen Pectobacterium wasabiae SCC3193. PLOS Pathog. 8:e1003013. doi: 10.1371/journal.ppat.1003013
O’Hara, C. M., Steigerwalt, A. G., Hill, B. C., Miller, J. M., and Brenner, D. J. (1998). First report of a human isolate of Erwinia persicinus. J. Clin. Microbiol. 36, 248–250.
Özen, A. I., and Ussery, D. W. (2012). Defining the Pseudomonas genus: where do we draw the line with Azotobacter? Microb. Ecol. 63, 239–248. doi: 10.1007/s00248-011-9914-8
Palleroni, N. J. (2005). Genus IX. Stenotrophomonas Palleroni and Bradbury 1993, 608VP. Bergey’s Manual Syst. Bacteriol. 2, 107–115.
Parthiban, V., Prakasam, V., and Prabakar, K. (2012). Enzymatic changes in carrot roots induced by Erwinia carotovora var. carotovora. Int. J. Pharma Biosci. 3, 246–252.
Peix, A., Valverde, A., Rivas, R., Igual, J. M., Ramírez-Bahena, M. H., Mateos, P. F., et al. (2007). Reclassification of Pseudomonas aurantiaca as a synonym of Pseudomonas chlororaphis and proposal of three subspecies, P. chlororaphis subsp. chlororaphis subsp. nov., P. chlororaphis subsp. aureofaciens subsp. nov., comb. nov. and P. chlororaphis subsp. aurantiaca subsp. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 57, 1286–1290. doi: 10.1099/ijs.0.64621-0
Pomini, A. M., Manfio, G. P., Araújo, W. L., and Marsaioli, A. J. (2005). Acyl-homoserine lactones from Erwinia psidii R. IBSBF 435T, a guava phytopathogen (Psidium guajava L.). J. Agric. Food Chem. 53, 6262–6265. doi: 10.1021/jf050586e
Porch, T. G., Urrea, C. A., Beaver, J. S., Valentin, S., Peña, P. A., and Smith, J. R. (2012). Registration of TARS-MST1 and SB-DT1 multiple-stress-tolerant black bean germplasm. J. Plant Regist. 6, 75–80. doi: 10.3198/jpr2010.08.0501crg
Pritchard, L., Humphris, S., Saddler, G., Parkinson, N., Bertrand, V., Elphinstone, J., et al. (2013). Detection of phytopathogens of the genus Dickeya using a PCR primer prediction pipeline for draft bacterial genome sequences. Plant Pathol. 62, 587–596. doi: 10.1111/j.1365-3059.2012.02678.x
Rademaker, J. L., Hoste, B., Louws, F. J., Kersters, K., Swings, J., Vauterin, L., et al. (2000). Comparison of AFLP and rep-PCR genomic fingerprinting with DNA-DNA homology studies: Xanthomonas as a model system. Int. J. Syst. Evol. Microbiol. 50, 665–677. doi: 10.1099/00207713-50-2-665
Reddy, P. P. (2015). Plant Protection in Tropical Root and Tuber Crops. (New Delhi, IND: Springer). doi: 10.1007/978-81-322-2389-4
Rees-George, J., Vanneste, J. L., Cornish, D. A., Pushparajah, I. P. S., Yu, J., Templeton, M. D., et al. (2010). Detection of Pseudomonas syringae pv. actinidiae using polymerase chain reaction (PCR) primers based on the 16S–23S rDNA intertranscribed spacer region and comparison with PCR primers based on other gene regions. Plant Pathol. 59, 453–464. doi: 10.1111/j.1365-3059.2010.02259.x
Rodriguez, R., Luis, M., Grajales, A., Arrieta-Ortiz, M., L., Salazar, C., et al. (2012). Genomes-based phylogeny of the genus Xanthomonas. BMC Microbiol. 12:43. doi: 10.1186/1471-2180-12-43
Rojas, A. M., Rios, J. E., Fischer-Le Saux, M., Jimenez, P., Reche, P., Bonneau, S., et al. (2004). Erwinia toletana sp. nov., associated with Pseudomonas savastanoi-induced tree knots. Int. J. Syst. Evol. Microbiol. 54, 2217–2222. doi: 10.1099/ijs.0.02924-0
Roper, M. C. (2011). Pantoea stewartii subsp. stewartii: lessons learned from a xylem-dwelling pathogen of sweet corn. Mol. Plant Pathol. 12, 628–637. doi: 10.1111/j.1364-3703.2010.00698.x
Sakamoto, M., Takeuchi, Y., Umeda, M., Ishikawa, I., and Benno, Y. (2001). Rapid detection and quantification of five periodontopathic bacteria by real-time PCR. Microbiol. Immunol. 45, 39. doi: 10.1111/j.1348-0421.2001.tb01272.x
Šalplachta, J., Kubesová, A., Horký, J., Matoušková, H., Tesařová, M., and Horká, M. (2015). Characterization of Dickeya and Pectobacterium species by capillary electrophoretic techniques and MALDI-TOF MS. Anal. Bioanal. Chem. 407, 7625–7635. doi: 10.1007/s00216-015-8920-y
Samson, R., Legendre, J., Christen, R., Fischer-Le Saux, Achouak, W., and Gardan, L. (2005). Transfer of Pectobacterium chrysanthemi (Burkholder et al. 1953) Brenner et al. 1973 and Brenneria paradisiaca to the genus Dickeya gen. nov. as Dickeya chrysanthemi comb. nov. and Dickeya paradisiaca comb. nov. and delineation of four novel species, Dickeya dadantii sp. nov., Dickeya dianthicola sp. nov. and Dickeya zeae sp. nov. Int. J. Syst. Evol. Microbiol. 55, 1415–1427. doi: 10.1099/ijs.0.02791-0
Sanogo, S., Etarock, B., and Clary, M. (2011). First report of bacterial wilt caused by Erwinia tracheiphila on pumpkin and watermelon in New Mexico. Plant Dis. 95, 1583–1583. doi: 10.1094/PDIS-06-11-0507
Saranraj, P., Stella, D., and Reetha, D. (2012). Microbial spoilage of vegetables and its control measures: a review. Int. J. Nat. Prod. Sci. 2, 1–12.
Shang, J., Liu, B., and He, W. (2015). A new method to detect Lonsdalea quercina in infected plant tissues by real-time PCR. Forest Pathol. 45, 28–35. doi: 10.1111/efp.12125
Sharma, D., and Agrawal, K. (2014). Incidence and histopathological study of Xanthomonas axonopodis. J. Agric. Technol. 10, 233–242.
Shrestha, R., Koo, J., Park, D., Hwang, I., Hur, J., and Lim, C. (2003). Erwinia pyrifoliae, a causal endemic pathogen of shoot blight of Asian pear tree in Korea. Plant Pathol. J. 19, 294–300. doi: 10.5423/PPJ.2003.19.6.294
Singh, U., Singh, R. P., and Kohmoto, K. (2012). “Pathogenesis and host specificity in plant pathogenic bacteria,” in Prokaryotes, ed. G. Meurant (Great Britain: Elsevier Science Ltd), 19–23.
Sławiak, M., Van Beckhoven, J. R., Speksnijder, A. G., Czajkowski, R., Grabe, G., and Van Der Wolf, J. M. (2009). Biochemical and genetical analysis reveal a new clade of biovar 3 Dickeya spp. strains isolated from potato in Europe. Eur. J. Plant Pathol. 125, 245–261. doi: 10.1007/s10658-009-9479-2
Sławiak, M., Van Doorn, R., Szemes, M., Speksnijder, A., Waleron, M., Van Der Wolf, J., et al. (2013). Multiplex detection and identification of bacterial pathogens causing potato blackleg and soft rot in Europe, using padlock probes. Ann. Appl. Biol. 163, 378–393. doi: 10.1111/aab.12075
Stackebrandt, E., Frederiksen, W., Garrity, G. M., Grimont, P. A. D., Kämpfer, P., Maiden, M. C. J., et al. (2002). Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 52, 1043–1047. doi: 10.1099/00207713-52-3-1043
Subramoni, S., Jha, G., and Sonti, R. (2006). “Virulence functions of xanthomonads,” in Plant-Associated Bacteria, ed. S. Gnanamanickam (Heidelberg, NL: Springer), 535–571. doi: 10.1007/978-1-4020-4538-7_15
Tayeb, A. L., Ageron, E., Grimont, F., and Grimont, P. (2005). Molecular phylogeny of the genus Pseudomonas based on rpoB sequences and application for the identification of isolates. Res. Microbiol. 156, 763–773. doi: 10.1016/j.resmic.2005.02.009
Terta, M., Azelmat, S., M’hand, R. A., Achbani, E. H., Barakate, M., Bouteau, F., et al. (2012). Molecular typing of Pectobacterium carotovorum isolated from potato tuber soft rot in Morocco. Ann. Microbiol. 62, 1411–1417. doi: 10.1007/s13213-011-0391-6
Thapa, S. P., Park, D. H., Kim, W. S., Choi, B. S., Lim, J. S., Choi, I. Y., et al. (2012). Comparative genomics of Japanese Erwinia pyrifoliae strain Ejp617 with closely related Erwinias. Genome 56, 83–90. doi: 10.1139/gen-2012-0094
Tourkya, B., Boubellouta, T., Dufour, E., and Leriche, F. (2009). Fluorescence spectroscopy as a promising tool for a polyphasic approach to pseudomonad taxonomy. Curr. Microbiol. 58, 39–46. doi: 10.1007/s00284-008-9263-0
Tournas, V. H. (2005). Spoilage of vegetable crops by bacteria and fungi and related health hazards. Crit. Rev. Microbiol 31, 33–44. doi: 10.1080/10408410590886024
Trantas, E., Sarris, P., Pentari, M., Mpalantinaki, E., Ververidis, F., and Goumas, D. (2015). Diversity among Pseudomonas corrugata and Pseudomonas mediterranea isolated from tomato and pepper showing symptoms of pith necrosis in Greece. Plant Pathol. 64, 307–318. doi: 10.1111/ppa.12261
Valenzuela-Soto, J., Maldonado-Bonilla, L., Hernández-Guzmán, G., Rincón-Enríquez, G., Martínez-Gallardo, N., Ramírez-Chávez, E., et al. (2015). Infection by a coronatine-producing strain of Pectobacterium cacticidum isolated from sunflower plants in Mexico is characterized by soft rot and chlorosis. J. Gen. Plant Pathol. 81, 368–381. doi: 10.1007/s10327-015-0606-y
van der Merwe, J. J., Coutinho, T. A., Korsten, L., and Van Der Waals, J. E. (2010). Pectobacterium carotovorum subsp. brasiliensis causing blackleg on potatoes in South Africa. Eur. J. Plant Pathol. 126, 175–185. doi: 10.1007/s10658-009-9531-2
van der Wolf, J. M., Nijhuis, E. H., Kowalewska, M. J., Saddler, G. S., Parkinson, N., Elphinstone, J. G., et al. (2014). Dickeya solani sp. nov., a pectinolytic plant-pathogenic bacterium isolated from potato (Solanum tuberosum). Int. J. Syst. Evol. Microbiol. 64, 768–774. doi: 10.1099/ijs.0.052944-0
Vandroemme, J., Cottyn, B., Baeyen, S., De Vos, P., and Maes, M. (2013). Draft genome sequence of Xanthomonas fragariae reveals reductive evolution and distinct virulence-related gene content. BMC Genom. 14:829. doi: 10.1186/1471-2164-14-829
Vauterin, L., Hoste, B., Kersters, K., and Swings, J. (1995). Reclassification of Xanthomonas. Int. J. Syst. Bacteriol. 45, 472–489. doi: 10.1099/00207713-45-3-472
Vicente, J. G., and Holub, E. B. (2013). Xanthomonas campestris pv. campestris (cause of black rot of crucifers) in the genomic era is still a worldwide threat to brassica crops. Mol. Plant Pathol. 14, 2–18. doi: 10.1111/j.1364-3703.2012.00833.x
Waldee, E. L. (1945). Comparative studies of some peritrichous phytopathogenic bacteria. Iowa State College J. Sci. 19, 435–484.
Waleron, M., Czajkowski, R., Waleron, K., and Lojkowska, E. (2013a). Restriction fragment length polymorphism-based identification of ‘Dickeya Solani’, A new genetic clade of Dickeya spp. J. Plant Pathol. 95, 609–613.
Waleron, M., Waleron, K., and Lojkowska, E. (2013b). Occurrence of Pectobacterium wasabiae in potato field samples. Eur. J. Plant Pathol. 137, 149–158. doi: 10.1007/s10658-013-0227-2
Waleron, M., Waleron, K., and Lojkowska, E. (2014). Characterization of Pectobacterium carotovorum subsp. odoriferum causing soft rot of stored vegetables. Eur. J. Plant Pathol. 139, 457–469. doi: 10.1007/s10658-014-0403-z
Waleron, M., Waleron, K., Podhajska, A. J., and Łojkowska, E. (2002). Genotyping of bacteria belonging to the former Erwinia genus by PCR-RFLP analysis of a recA gene fragment. Microbiology 148, 583–595. doi: 10.1099/00221287-148-2-583
Wells, J., Sheng, W., Ceponis, M., and Chen, T. (1987). Isolation and characterization of strains of Erwinia ananas from honeydew melons. Phytopathology 77, 511–514. doi: 10.1094/Phyto-77-511
Wensing, A., Gernold, M., and Geider, K. (2012). Detection of Erwinia species from the apple and pear flora by mass spectroscopy of whole cells and with novel PCR primers. J. Appl. Microbiol. 112, 147–158. doi: 10.1111/j.1365-2672.2011.05165.x
Werra, P. D., Bussereau, F., Keiser, A., and Ziegler, D. (2015). First report of potato blackleg caused by Pectobacterium carotovorum subsp. brasiliense in Switzerland. Plant Dis. 99, 551–552. doi: 10.1094/PDIS-07-14-0742-PDN
Wu, J., Liu, X., Diao, Y., Ding, Z., and Hu, Z. (2012). Authentication and characterization of a candidate antagonistic bacterium against soft rot of Amorphophallus konjac. Crop. Prot. 34, 83–87. doi: 10.1016/j.cropro.2011.12.008
Yaganza, E.-S., Tweddell, R., and Arul, J. (2014). Postharvest application of organic and inorganic salts to control potato (Solanum tuberosum L.) Storage softrot: plant tissue–salt physicochemical interactions. J. Agric. Food Chem. 62, 9223–9231. doi: 10.1021/jf5017863
Yan, H., Yu, S., Xie, G., Fang, W., Su, T., and Li, B. (2010). Grain discoloration of rice caused by Pantoea ananatis (synonym Erwinia uredovora) in China. Plant Dis. 94, 482–482. doi: 10.1094/PDIS-94-4-0482B
Keywords: classification, macergens, pectolytic, proteolytic, taxonomy, species
Citation: Aremu BR and Babalola OO (2015) Classification and Taxonomy of Vegetable Macergens. Front. Microbiol. 6:1361. doi: 10.3389/fmicb.2015.01361
Received: 21 August 2015; Accepted: 16 November 2015;
Published: 27 November 2015.
Edited by:
Giovanna Suzzi, Università degli Studi di Teramo, ItalyReviewed by:
Ivonne Delgadillo, University of Aveiro, PortugalRosanna Tofalo, Università degli Studi di Teramo, Italy
Copyright © 2015 Aremu and Babalola. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Olubukola O. Babalola, b2x1YnVrb2xhLmJhYmFsb2xhQG53dS5hYy56YQ==
 Bukola R. Aremu
Bukola R. Aremu