
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 30 October 2015
Sec. Food Microbiology
Volume 6 - 2015 | https://doi.org/10.3389/fmicb.2015.01199
This article is part of the Research Topic Regulation of gene expression in enteropathogenic bacteria View all 42 articles
 Tilman Schultze1
Tilman Schultze1 Rolf Hilker1,2
Rolf Hilker1,2 Gopala K. Mannala1
Gopala K. Mannala1 Katrin Gentil1
Katrin Gentil1 Markus Weigel1
Markus Weigel1 Neda Farmani1
Neda Farmani1 Anita C. Windhorst3
Anita C. Windhorst3 Alexander Goesmann2
Alexander Goesmann2 Trinad Chakraborty1
Trinad Chakraborty1 Torsten Hain1*
Torsten Hain1*Listeria monocytogenes is a bacterial pathogen and causative agent for the foodborne infection listeriosis, which is mainly a threat for pregnant, elderly, or immunocompromised individuals. Due to its ability to invade and colonize diverse eukaryotic cell types including cells from invertebrates, L. monocytogenes has become a well-established model organism for intracellular growth. Almost 10 years ago, we and others presented the first whole-genome microarray-based intracellular transcriptome of L. monocytogenes. With the advent of newer technologies addressing transcriptomes in greater detail, we revisit this work, and analyze the intracellular transcriptome of L. monocytogenes during growth in murine macrophages using a deep sequencing based approach. We detected 656 differentially expressed genes of which 367 were upregulated during intracellular growth in macrophages compared to extracellular growth in Brain Heart Infusion broth. This study confirmed ∼64% of all regulated genes previously identified by microarray analysis. Many of the regulated genes that were detected in the current study involve transporters for various metals, ions as well as complex sugars such as mannose. We also report changes in antisense transcription, especially upregulations during intracellular bacterial survival. A notable finding was the detection of regulatory changes for a subset of temperate A118-like prophage genes, thereby shedding light on the transcriptional profile of this bacteriophage during intracellular growth. In total, our study provides an updated genome-wide view of the transcriptional landscape of L. monocytogenes during intracellular growth and represents a rich resource for future detailed analysis.
Listeria monocytogenes is a ubiquitously distributed pathogen. It grows under a variety of adverse conditions like high salt concentration or low temperatures thereby resisting many common food preservation techniques. L. monocytogenes contaminated food is a serious health threat and mainly immunocompromised individuals, elderly persons and pregnant women suffer from severe complications (Hamon et al., 2006; Allerberger and Wagner, 2010; Cossart and Lebreton, 2014; Elinav et al., 2014). Upon ingestion of contaminated food, L. monocytogenes reaches the gastrointestinal tract where it invades epithelial cells. Via infection of macrophages and escape from their phagosome, L. monocytogenes ultimately reaches liver and spleen. Due to its versatile ability of infecting mammalian and invertebrate cells, L. monocytogenes has become a popular model organism for intracellular pathogenicity (Hamon et al., 2006; Lecuit, 2007; Cossart and Toledo-Arana, 2008; Cossart, 2011).
Recently, non-coding RNA elements were frequently studied in L. monocytogenes (Mellin and Cossart, 2012). Non-coding RNA elements are widely implicated in adaptation to different environmental conditions, metabolism, and virulence. The role of regulatory RNA elements (Johansson et al., 2002; Mansjö and Johansson, 2011), small RNA (Christiansen et al., 2006; Mandin et al., 2007; Nielsen et al., 2008; Oliver et al., 2009; Toledo-Arana et al., 2009; Mraheil et al., 2011; Nielsen et al., 2011; Sievers et al., 2014, 2015) and antisense RNA (Toledo-Arana et al., 2009; Wurtzel et al., 2012; Behrens et al., 2014; Wehner et al., 2014) was investigated. A new class of regulatory elements, the excludon was defined as long transcript that function as both sense transcript for one gene (or set of genes) as well as antisense transcript for a divergently oriented neighboring gene (or set of genes; Wurtzel et al., 2012).
Intracellular growth of L. monocytogenes was included as experimental condition in several genome-wide transcriptional profiling studies, however, few publications have focused on the changes in the mRNA transcriptome. These studies were conducted under very diverse conditions, making comparisons between studies difficult. The mRNA transcriptional profile of intracellular L. monocytogenes 6 h after infection of Caco-2-ephithelial cells was assessed by DNA microarray analysis (Joseph et al., 2006). In this study – as in every other that followed – the transcriptome of L. monocytogenes grown in rich media Brain Heart Infusion (BHI) served as reference. A similar approach was used to examine the transcriptome of L. monocytogenes when grown in murine macrophages P388D1 (Chatterjee et al., 2006). Here, the transcriptome of bacteria after 4 and 8 h post-infection as well as during intravacuolar growth was analyzed. Camejo et al. (2009) introduced in vivo work that shed light on the transcriptional profile of L. monocytogenes in murine spleens at 24, 48, and 72 h post-intravenous infection.
Despite the introduction of next generation sequencing (NGS) technologies, no study has addressed transcriptional changes in L. monocytogenes in detail. We now revisit the earlier microarray-based work (Chatterjee et al., 2006) complementing it with recent RNA-seq data (Wehner et al., 2014) which exclusively investigated non-coding RNAs of L. monocytogenes under intra- and extracellular growth conditions.
Here, we report the transcriptional changes in L. monocytogenes during intracellular growth in murine macrophages using an approach that also detects alteration of non-coding RNA elements. Furthermore, we construct a framework for further analysis targeting intracellular survival of L. monocytogenes. Finally, we outline common transcriptional responses of L. monocytogenes during intracellular growth among all the diverse growth conditions investigated to date.
All bacterial strains and plasmids used in this study are listed in Table 1. Wild type L. monocytogenes EGD-e serotype 1/2a (Glaser et al., 2001) and its isogenic deletion mutants Δlmo2316 or Δlmo1119 were cultivated under aerobic conditions in BHI broth (Difco, Becton Dickinson, Franklin Lakes, NJ, USA) or on BHI agar plates at 37°C. For RNA-seq experiments, L. monocytogenes EGD-e was grown in BHI broth overnight at 37°C with shaking at 180 rpm. Bacterial cultures were supplemented with fresh medium and grown until OD600 reached 1.0.
Escherichia coli strain (DH10β) was grown in Luria-Bertani (LB) broth or on LB agar plates at 37°C. Cultures were supplemented with 300 μg/ml erythromycin (Sigma-Aldrich, St. Louis, MO, USA) for E. coli and 5 or 10 μg/ml for erythromycin for L. monocytogenes or 50 μg/ml ampicillin (Sigma-Aldrich, St. Louis, MO, USA) for E. coli and 200 μg/ml ampicillin for L. monocytogenes.
Generation of lmo2316 or lmo1119 in frame deletion mutants was performed as previously described (Seifart Gomes et al., 2011). A detailed description is provided as Supplemental Text 1.
Bacterial RNA isolation was carried out as described previously (Wehner et al., 2014). The quality of total RNA was assessed using Bioanalyzer 2100 Total RNA nano chip (Agilent, Santa Clara, CA, USA) and RNA concentration was measured using Qubit 2.0 fluorimeter (Invitrogen/Life Technologies, Carlsbad, CA, USA). rRNA was depleted from 1 μg of total RNA using Ribo-Zero Magnetic Kit (Bacteria; Epicentre, Madison, WI, USA). Depleted RNA was then treated with tobacco acid pyrophosphatase (Epicentre, Madison, WI, USA) and cleaned up with the RiboMinus concentration module (Life Technologies, Carlsbad, CA, USA).
For fragmentation and further library preparation TruSeq Stranded Total RNA Seq Kit (Illumina, San Diego, CA, USA) was used according to manufacturer’s instruction. In this process a barcoded-approach was chosen to facilitate multiplexed sequencing. For reverse transcription Superscript II polymerase (Life Technologies, Carlsbad, CA, USA) was used and libraries were purified with AMPure XP Reagent (Beckman Coulter, Pasadena, CA, USA). The yield and size distribution of the amplified cDNA were assessed with DNA High sensitivity kit (Agilent, Santa Clara, CA, USA). Libraries were then diluted to 4 nM, pooled, denatured and further diluted to 10 pM. Sequencing was carried out on the MiSeq using v2 chemistry (Illumina, San Diego, CA, USA).
FastQ-files of sequencing runs performed with Ion Torrent PGM (Wehner et al., 2014) were downloaded from EBI (accession number PRJEB6949).
FastQ-files for MiSeq runs were obtained after image processing, basecalling and demultiplexing of reads performed by Illumina MiSeq-Control Software version 2.1.13 using RTA-version 1.17.22. For online accessibility, fastQ-files of the MiSeq runs were stored at the European Nucleotide Archive (accession number PRJEB10393).
Sequencing reads were mapped against L. monocytogenes EGD-e (NCBI RefSeq NC_003210.1). Reads were aligned with bowtie2 version 2.1 (Langmead and Salzberg, 2012). Read type was set to single-end for all runs. SAM-files of all PGM runs were used as input for read counting using HTSeq count version 0.6.1 (Anders et al., 2014). Strand-specificity was taken into account and rRNA (lmor), tRNA (lmot) as well as small RNA (lmos or former rli) were excluded from counting. The default counting mode ‘union’ was selected. All differential expression analyses were performed in R version 3.1.1 R Core Team (2014) using the package DESeq2 (Love et al., 2014). p-values were adjusted by the default method in DESeq2 and genes were considered differentially expressed when adjusted p-values below 0.05 were obtained.
While only the PGM runs were used for the differential expression model, all visualizations of read mappings and coverage plots were generated from the additional experiments run on the Illumina MiSeq. 3,556,623 reads aligned to the reference exactly one time for the extracellular condition and 6,503,703 reads aligned for the intracellular condition. The difference was caused by variances in sequencing output, as mapping rates were alike for both experiments (75 and 79%). Visualizations were then created using ReadXplorer version 2.0.1. (Hilker et al., 2014).
mRNA levels of lmo1119 and lmo2316 from both extra- and intracellularly grown bacteria were assessed using quantitative real-time-PCR (qRT-PCR) as previously described (Wehner et al., 2014). Each qRT-PCR was performed in triplicate (locus tag _5′ and locus tag _3′, Supplementary Table S1; 7900 HT Fast Real Time System, Applied Biosystems/Thermo Fisher Scientific, Waltham, MA, USA). Gene expression was normalized to 16S rRNA and relative expression of lmo1119 and lmo2316 was calculated using the mathematical method published by Pfaffl and Pfaffl (2001).
P388D1 murine macrophage were infected with L. monocytogenes EGD-e and its isogenic deletion mutants as described previously (Mraheil et al., 2011).
All experimental work was repeated for a minimum of three times. Statistical analyses for qRT-PCR and survival assays were performed using R version 3.1.3. Data was log-transformed. Differential expression detected by qRT-PCR was tested by Student’s one-sample t-test. Differences in survival were analyzed using a linear regression model (Chambers and Hastie, 1992), where the amount of colony forming units (CFUs) was corrected by the inoculation amount as covariable. Pairwise comparisons were performed using the R package lsmeans version 2.171 and multcomp (Hothorn et al., 2008). p < 0.05 was considered significant.
We based our work on a recent study which included six sequencing runs (three runs for extracellular and three runs for intracellular growth conditions) performed with IonTorrent semiconductor sequencing technology (Wehner et al., 2014). This study investigated regulatory RNA, changes of mRNA expression were beyond the scope of the work.
To extend and further back up this data set additional experiments were included using the reversible dye-terminator based sequencing approach of the Illumina MiSeq. These additional runs also facilitate detailed analysis of read mappings due to a higher amount of mapped reads and thus higher overall depth of coverage. In addition, it enables the evaluation whether transcriptomic changes occur platform-independently. Overall, pairwise comparison of normalized read counts provided no indication that variances in read counts between Illumina and Ion torrent runs exceeded variances of biological repeats among the Ion torrent runs (data not shown).
Comparing extracellular to intracellular growth conditions, our results show differential regulation for 656 genes (nearly 23% of all genes).The genes with the most dramatic changes in expression are listed in Table 2.
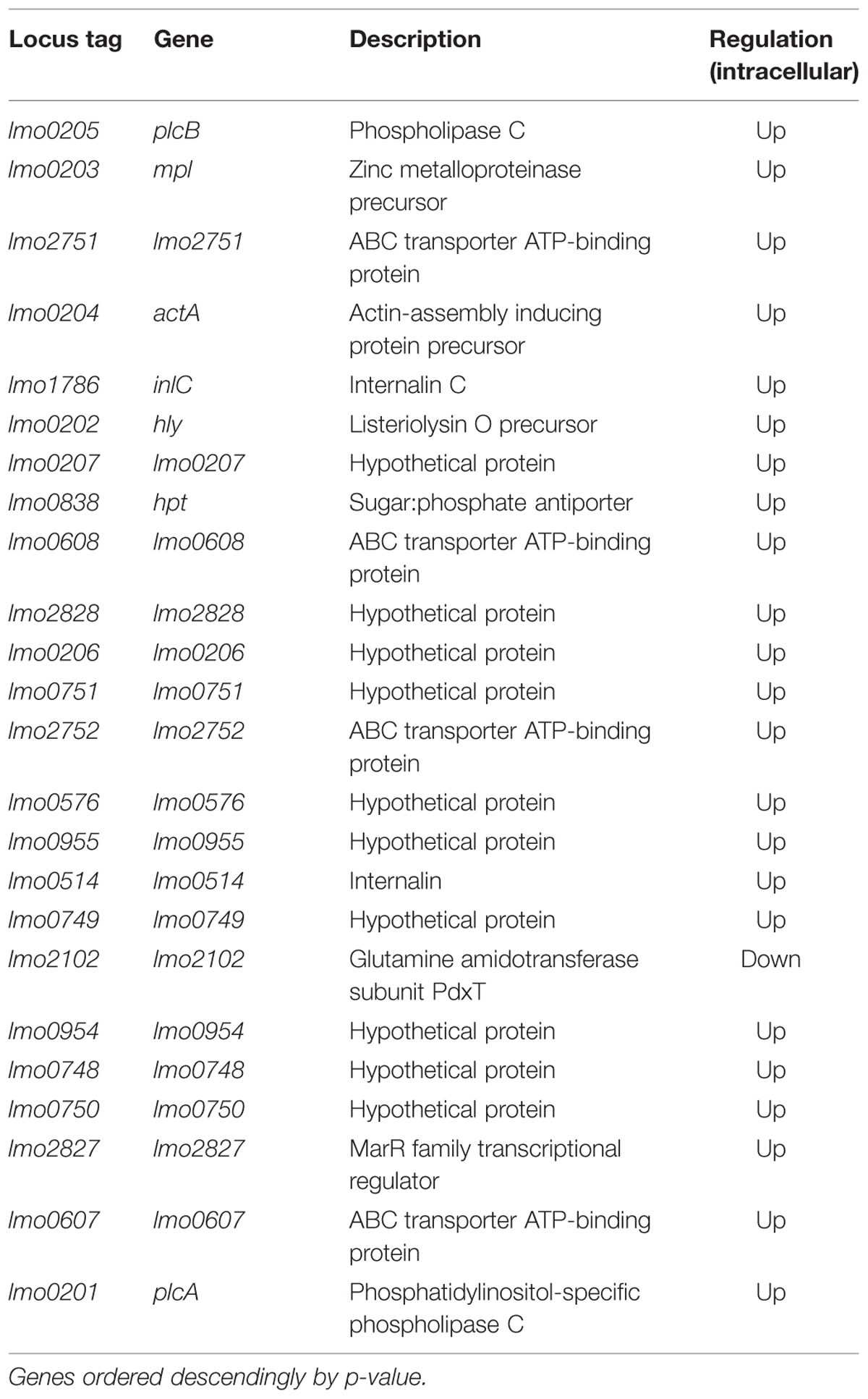
TABLE 2. Except of differentially expressed genes under intracellular growth conditions compared to extracellular growth.
Moreover, Supplementary Table S2 provides a complete list of all genes that were differentially expressed under intracellular growth conditions. Of those 656 genes, 367 were predicted to be up- and 289 to be downregulated when grown in murine macrophages. These genes were further characterized using the previously introduced operon model (Toledo-Arana et al., 2009). Among the 656 differentially regulated genes, 386 are organized in operon structures while 270 are transcribed monocistronically. Further analysis of the differential expression pattern of these operons is given in Supplemental Text 2 as well as Supplementary Figure S1.
When comparing our current results to the results of the previous microarray approach, we confirmed 55% of all differentially expressed genes found by the microarray-based approach. In both cases, an arbitrary threshold (p-value below 0.05) was applied to distinguish between differentially and not differentially expressed genes. A threshold of p < 0.1 in the current analysis increases the number to 64% of the previously found genes.
Five differentially regulated genes show opposing directions of regulation in both studies. These genes, one hypothetical gene as well as the metal transport operons lmo1423–1424 and lmo1848–1849, suggest that there have been minor experimental differences between both experimental conditions.
We then compared how the differentially expressed genes translate into alterations of functional pathways by applying the Clusters of Orthologous Groups of proteins (COG)2 classification. The number of differentially expressed genes is higher in the current RNA-seq study, however, the effects on the regulation of functional pathways is very similar (Figure 1).
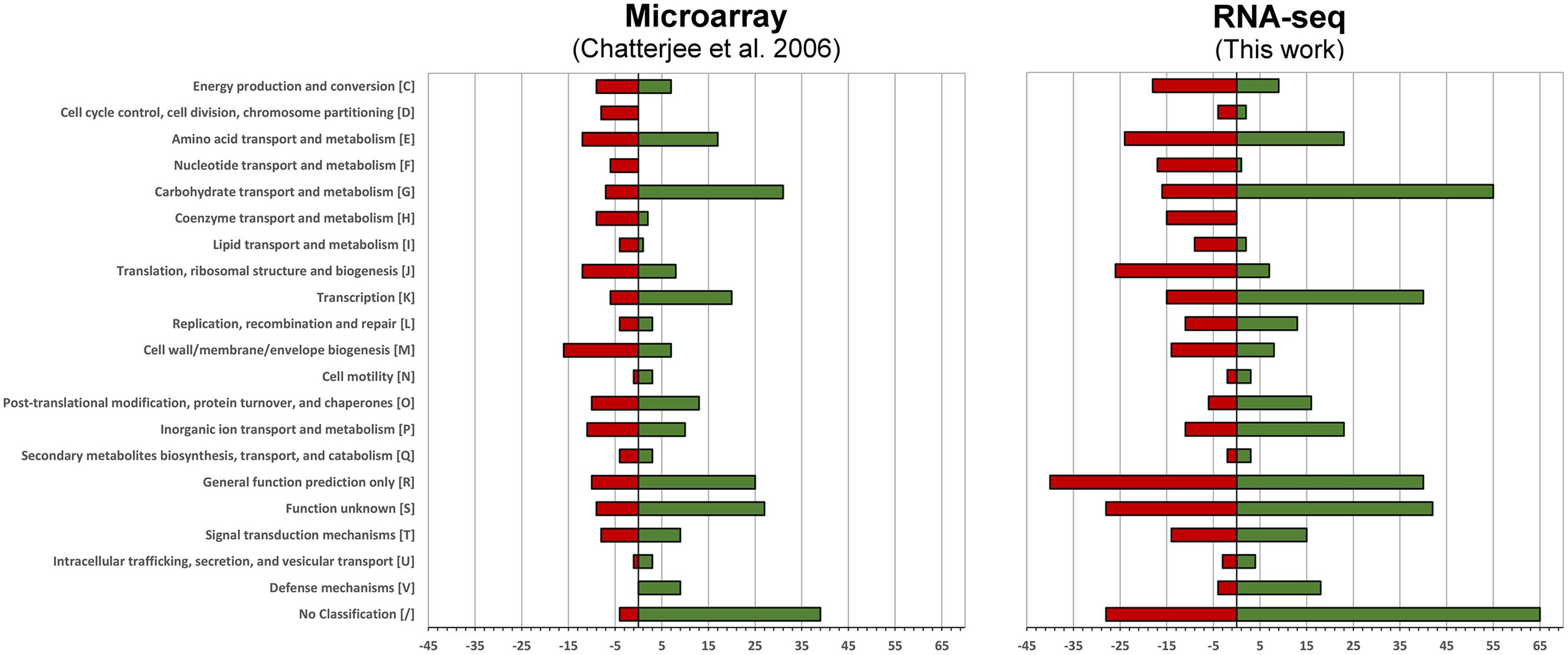
FIGURE 1. Functional changes in response to intracellular growth conditions based on Cluster of Orthologous Groups of proteins (COG) classification. Shown are the functional changes for the current RNA-seq approach (right) and the earlier microarray-based work (left). For each class the number of differentially regulated genes among this class is visualized by a bar. Red bars represent the downregulated and green bars the upregulated genes when grown in murine macrophages.
The investigated intracellular growth conditions vary widely among all published studies (Chatterjee et al., 2006; Joseph et al., 2006; Camejo et al., 2009; Toledo-Arana et al., 2009). We identified common transcriptional responses among these studies and our current work that are described in detail.
Regardless whether epithelial cells, macrophages, blood, intestine, or spleens were infected, upregulation of the gene cluster lmo0201–0205 was reported under intracellular growth conditions. Genes encoded by this virulence gene cluster are actA (intracellular motility), metalloprotease (mpl), two phospholipases (plcA, plcB), and the pore forming toxin listeriolysin (hly). The virulence gene cluster is controlled by the positive regulatory factor PrfA. prfA was consistently upregulated during intracellular growth in all studies. In addition to the virulence cluster, PrfA-regulated expression has been reported for lmo0207 (orfZ), lmo0796, lmo0434 (inlB), lmo1786 (inlC), lmo2219 (prsA2), and lmo0838 (hpt). lmo0207 and lmo0206, which were significantly upregulated in all studies except one (Camejo et al., 2009), are genes of poorly understood function adjacent to the virulence cluster. lmo0434 (inlB) and lmo1786 (inlC) encoding internalins needed for invasion as well as lmo2219 (prsA2) encoding a predicted peptidyl-prolyl isomerase. The PrfA-regulated gene lmo0838 is involved in the transport of phosphorylated sugars. Apart from PrfA-regulated genes, the studies by Camejo, Chatterjee as well as our current study report upregulation of four VirR-regulated genes. Two of these genes are encoding hypothetical proteins (lmo0604 and lmo2177) and two are encoding ABC transporter (lmo2114 and lmo2115).
Upregulation of genes involved in stress response represents a second principal finding. This mechanism was found for L. monocytogenes grown in epithelial cells, macrophages, splenocytes, and bacteria grown in the intestine (Chatterjee et al., 2006; Joseph et al., 2006; Camejo et al., 2009; Toledo-Arana et al., 2009) Specifically, the upregulation of lmo1473 (dnaK), lmo1474 (grpE), and lmo2068 (groEL) belonging to heat shock molecular chaperons (class I) as well as lmo2206 (clpB) which is an ATP-dependent protease (class III), was found in all studies. In addition to the induced transcription of lmo1473 and lmo1474, upregulation of lmo1471 (prmA), lmo1472 (dnaJ) and lmo1475 (hrcA), being part of the same operon, was found by our study as well as by the previous microarray-approach. Furthermore, our study suggests a broader transcriptional upregulation for class III heat shock genes. Transcripts of the whole major heat shock class III operon consisting of lmo0229 to lmo0232 were significantly increased. Especially, lmo0232 (clpC) was significantly upregulated in all studies except the one using epithelial cells as host. Similarly, the caseinolytic protease proteins lmo0997 (clpE) was upregulated in splenocytes and macrophages.
The last common finding among all studies concerns the intracellular transcriptional upregulation for lmo1538. lmo1538 encodes a glycerol kinase that is required for intracellular growth. Regulation of glycolysis under intracellular growth conditions was also addressed by the previous studies. Central genes of glycolysis were upregulated when L. monocytogenes was grown in splenocytes (Camejo et al., 2009), the studies in epithelial cells and macrophages found downregulation (Chatterjee et al., 2006; Joseph et al., 2006). For instance, it was observed that the operon lmo2455–2460, encoding five essential enzymes for the terminal glycolysis steps, has been downregulated.
Additional metabolic changes extending previous findings involve the transport of sugar and sugar phosphates. Our results provide evidence for the downregulation of mannose transporter subunits lmo0096–0098 as well as lmo1389–1390, genes involved in sugar transport. In contrast, upregulation was observed for lmo0736–0739, contributing to sugar transport and processing of phosphorylated ribulose, and an entire operon (lmo2733–2736), encoding fructose transporter, sugar hydrolase and sucrose phosphorylase. Remarkably, the operon lmo2670–2672 was also uniformly upregulated during intracellular growth, including lmo2671, a hypothetical protein that has similarity to a lactoylglutathione lyase. Genes required for phosphate transport were also differentially expressed. One of those loci consists of the genes lmo2494–2499, encoding proteins for phosphate uptake and transport. In addition, operon lmo2248 (a putative phosphate transport regulator)/lmo2249 (a low-affinity inorganic phosphate transporter) was downregulated during intracellular growth. Finally, several genes involved in DNA repair, e.g., recA (lmo1398) and the excinucleases lmo2050, lmo2488 (uvrA)/lmo2489 (uvrB) were differentially expressed during intracellular growth conditions.
A benefit of strand-specific RNA-seq studies is the ability to detect antisense transcripts as well as sense transcripts. We investigated changes in antisense expression between intra- and extracellular growth conditions. These analyses were not possible with previous microarray based approaches.
For many of the genes showing dramatic differential antisense expression during intracellular growth, adjacent divergent genes displayed drastic changes in sense expression. Examples are antisense transcripts against lmo0208–0209 and lmo0198–0199 both flanking the virulence gene cluster. Similarly, antisense transcripts of lmo0839 (neighboring the divergently oriented hpt) and of the sugar transferase gene lmo0497 (adjacent a highly upregulated SOS response gene lmo0496), were highly induced.
We also detect strong antisense upregulation for lmo1705 that was previously reported only as marginal note in the supplement (Wehner et al., 2014). With the additional sequencing runs that provided a higher read coverage, we obtained a distinct picture. Sense transcripts are slightly downregulated during intracellular growth while antisense transcripts are upregulated. When analyzing the read pattern (Figure 2) hardly any read was spanning the intergenic region between lmo1705 and lmo1706. Hence, the increase of antisense reads could not be attributed to an erroneous lack of transcription termination of the adjacent lmo1706. Moreover, a high coverage was found for the intergenic region between lmo1705 and lmo1704. Operon lmo1702–1704 appears to have an additional 5′ UTR transcription start site, which overlaps lmo1705. The complete list with genes for which putative changes in antisense expression were found is provided in Supplementary Table S3.
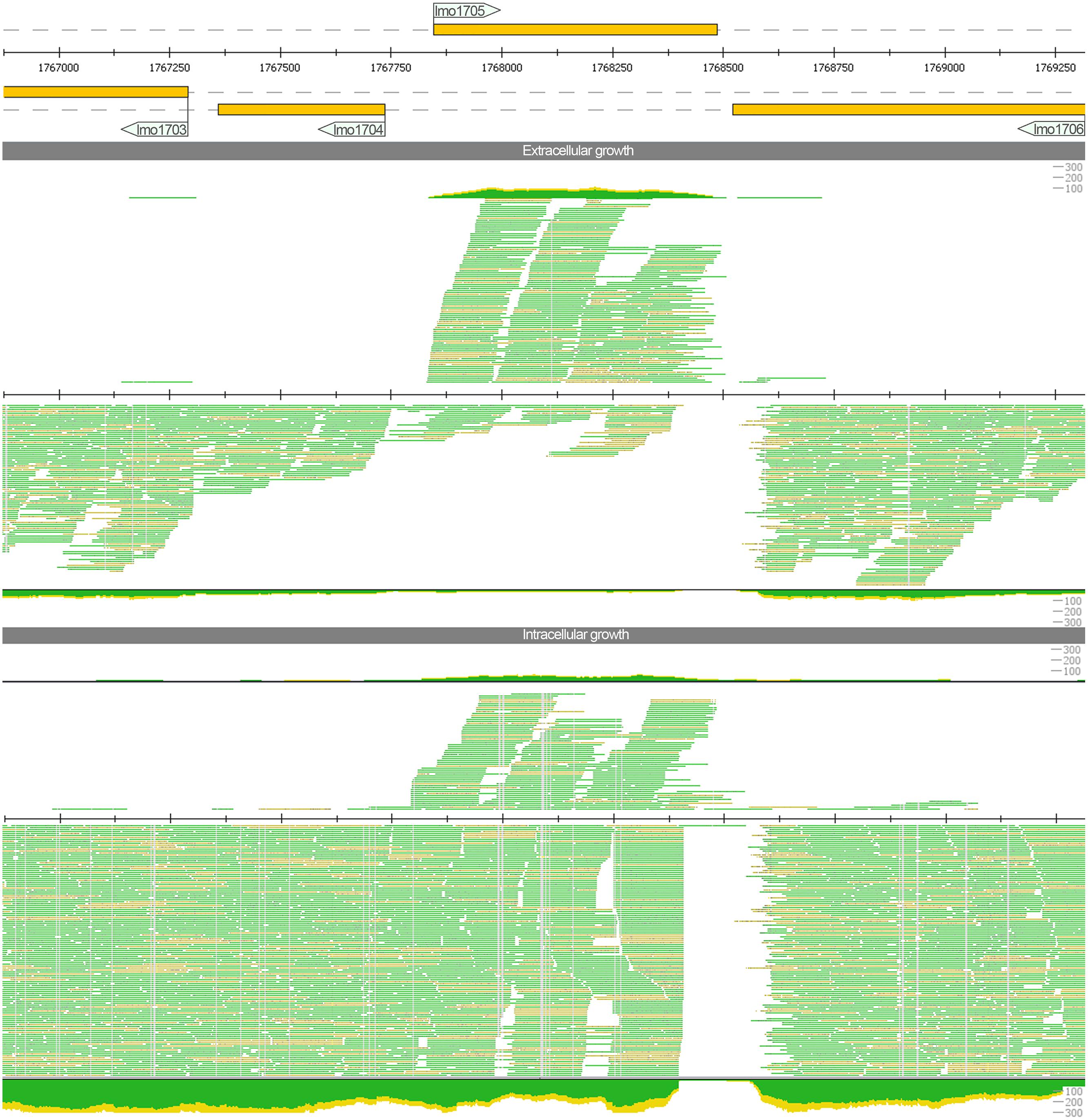
FIGURE 2. Read mapping for the locus lmo1705 under extracellular and intracellular growth conditions generated with ReadXplorer (Hilker et al., 2014). The three tracks represent genome annotation of this locus, read mapping for extra- and for intracellular conditions (in top–down order). For each track, genes on the plus strand (respectively their transcripts) are depicted above the central line, while minus strand genes (or their transcripts) are shown below. For the two read mapping tracks (extra- and intracellular growth condition), a detailed alignment up to a depth of 35 reads and a coverage plot is provided, too. The color thereby indicates whether reads are uniquely mapped and match the reference perfectly (green) or whether they map uniquely, but with mismatches (yellow). The read mapping indicates a strong increase in antisense transcripts for lmo1705 during intracellular growth.
In our study, we found increased expression of a cassette of A118-like prophage genes (lmo2271–2332) under intracellular growth conditions. In contrast, only few genes were predicted to be upregulated by the previous studies.
In analogy to the bacteriophage A118 described previously (Loessner et al., 2000), three organizational clusters, namely ‘late genes,’ ‘early genes,’ and ‘lysogeny control,’ can be distinguished in the phage genome. The expression among these clusters differs widely (Figure 3).
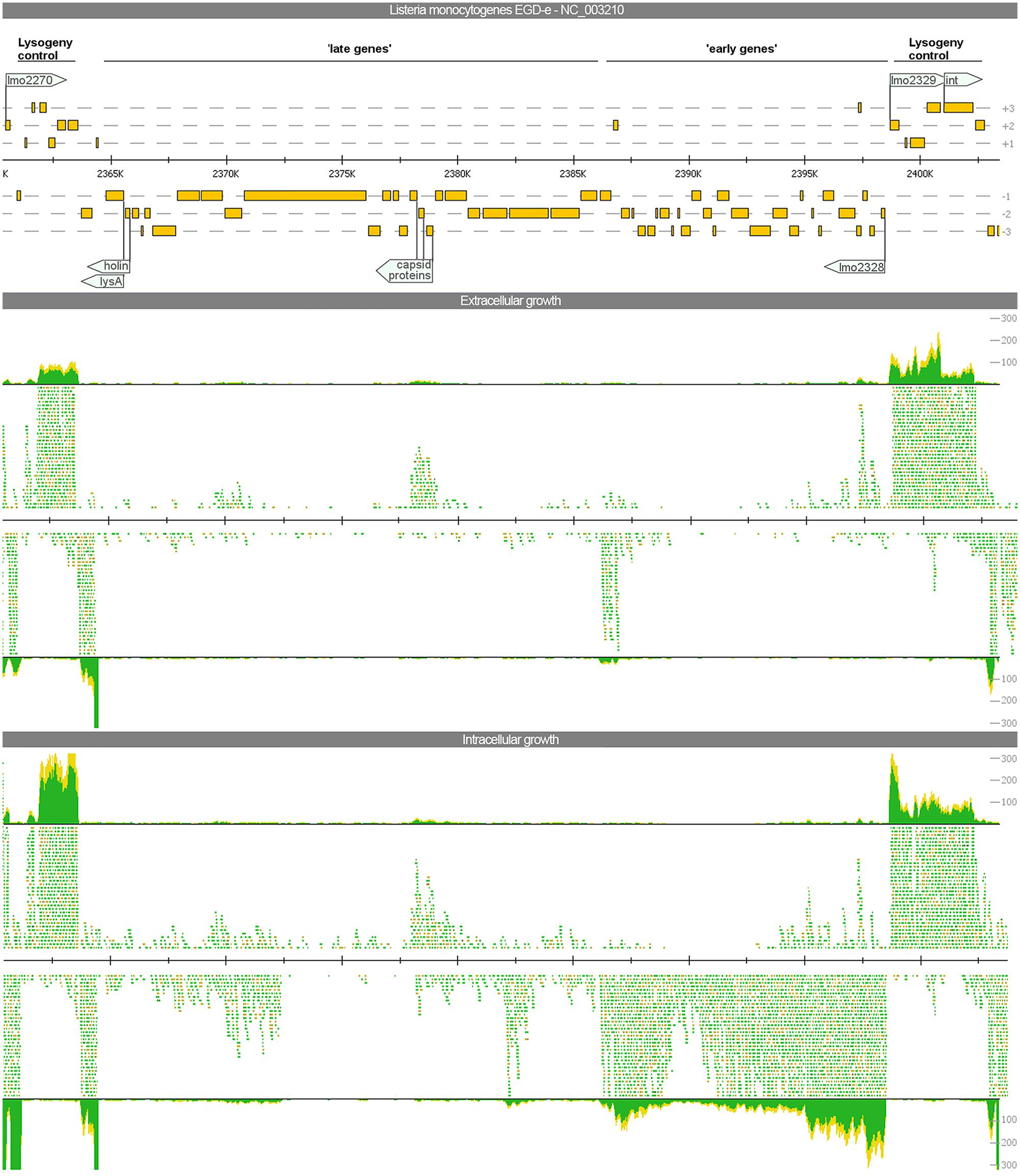
FIGURE 3. Visualization of the gene expression profiles of the A118-like prophage locus (lmo2271–2332) under extra- and intracellular growth condition. The read mapping illustrates a strong increase of transcripts for the ‘early genes’ whereas ‘late genes’ are poorly expressed under both conditions.
The late genes (lmo2278–2301) comprise genes coding for proteins required for host cell lysis, structural and assembly proteins as well as proteins for DNA packaging. All these genes are poorly expressed under any growth condition. Evidence for upregulation under intracellular growth condition was only found for lmo2299 most likely encoding a portal protein. Beside a generally low expression, antisense transcripts were detected for all genes of this locus expressed under both investigated conditions. For the lysis factors holin and lysA, antisense transcripts were almost as frequent as sense transcripts and for three minor capsid proteins (lmo2292–2294) antisense transcripts even exceeded the amount of sense transcripts.
The early genes (lmo2302–2328), encoding for products of replication, recombination, and modification of phage DNA, showed an identical expression pattern compared to the late genes during extracellular growth. During intracellular growth, gene expression of early genes was strongly induced with 23 genes significantly upregulated.
The third cluster involved in lysogeny control (lmo2271–2277 and lmo2329–2332) flanks both insertion sites and contains several putative transcriptional factors as well as the integrase. On a transcriptional level, all of these genes were well-expressed under both intra- and extracellular growth Of particular interest are the genes lmo2328 and lmo2329. Both of these adjacent and divergently oriented genes possess a helix-turn-helix motif common for transcriptional regulators. This constellation is a component of several bacteriophage genomes. In E. coli phage aaa, homologs of these genes are involved in the switch between lysogeny and lytic behavior (Oppenheim et al., 2005). In our study, lmo2329 is highly expressed under both conditions, but lmo2328 is exclusively expressed under intracellular growth conditions.
To assess whether phage excision takes place thereby restoring functionality of the previously split competence activator comK (lmo2270 and lmo2333), we mapped the sequencing reads against L. monocytogenes EGD (downloaded from NCBI NC_022568) using relaxed mapping conditions. L. monocytogenes EGD is highly related to L. monocytogenes EGD-e but does not have the A118-like prophage insertion. The read mapping (Figure 4) highlights that not even a single read is spanning the phage insertion site under intracellular growth conditions. Therefore, no evidence for excision restoring comK was detected. In addition, no significant difference was found between intra- and extracellular growth for the gene expression of competence cluster comF and comG as well as the genes comEA and comEB. Only expression of comEC was increased under intracellular conditions (Supplementary Figure S2).
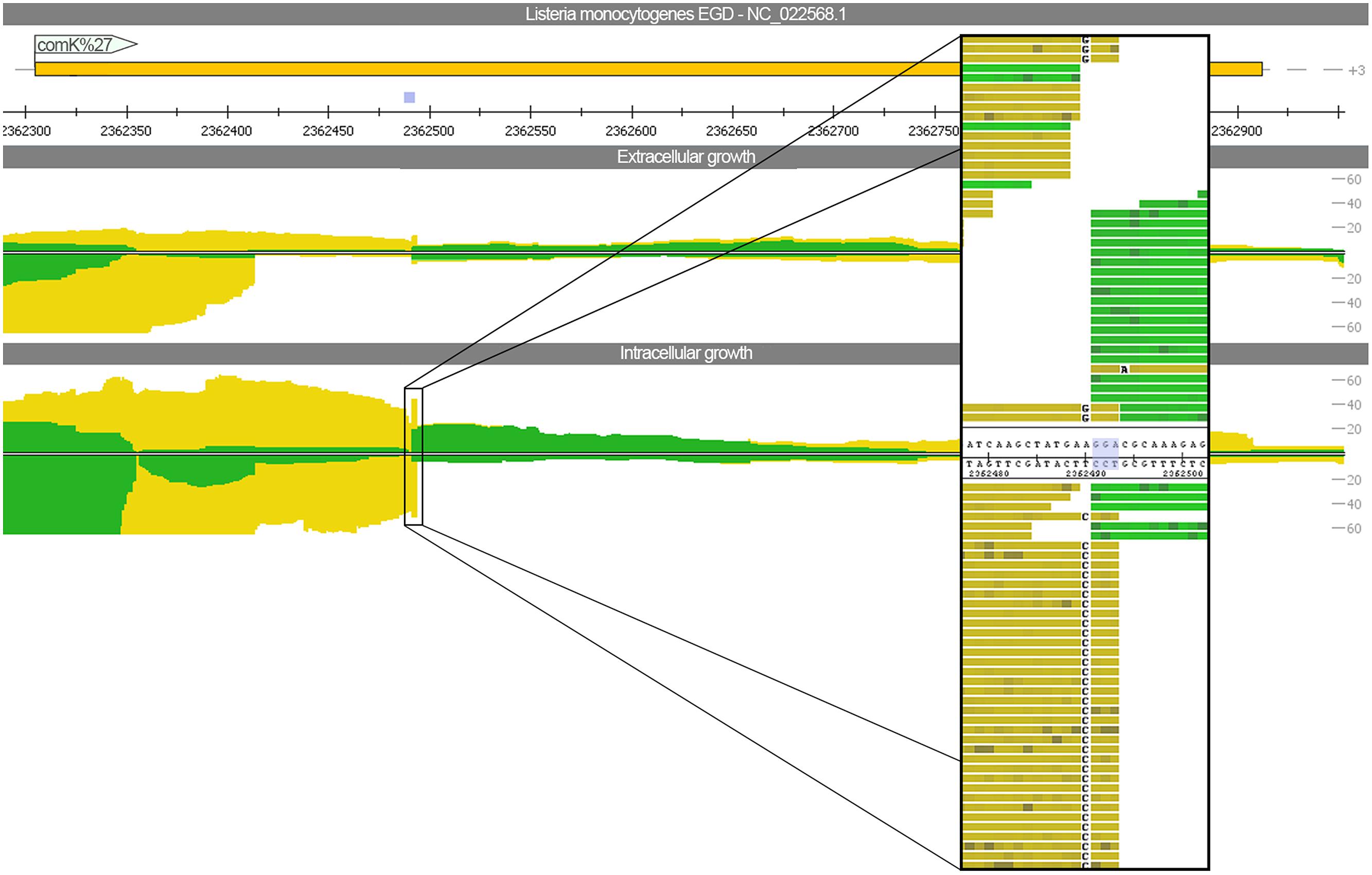
FIGURE 4. Read mapping for intra- and extracellular growth against Listeria monocytogenes EGD. In contrast to L. monocytogenes EGD-e, the strain EGD has no A118-like prophage insertion. The view depicts the comK gene, which is split and thereby inactivated by the bacteriophage insertion for the strain EGD-e. A close-up picture highlights that no reads are spanning the three nucleotide insertion site.
Within the phage region, lmo2316 encoding a putative site-specific adenine methylase (based on KEGG database prediction) was highly upregulated during intracellular growth. Methylases are known to contribute to pathogenicity and intracellular survival. lmo2316 was further characterized and the bacterial methyltransferase lmo1119, which is located elsewhere in the bacterial genome and is predicted to have similar functions, was included in the analysis as phage-independent control. Of note, the gene locus of lmo1119 indicates significant difference in its GC content compared to the whole genome sequence (Supplementary Figure S3).
While expression of the phage gene lmo2316 was strongly induced during intracellular growth, no difference in lmo1119 expression was detected under intracellular growth conditions. However, lmo1119 was well-expressed under both growth conditions whereas lmo2316 is hardly expressed at all under extracellular growth conditions.
We used qRT-PCR analysis to confirm the transcriptional changes of lmo1119 and lmo2316 indicated by RNA-seq. As predicted, lmo2316 was significantly upregulated (p = 0.0014) and lmo1119 showed a trend toward downregulation during intracellular growth (p = 0.1091; Figure 5A).
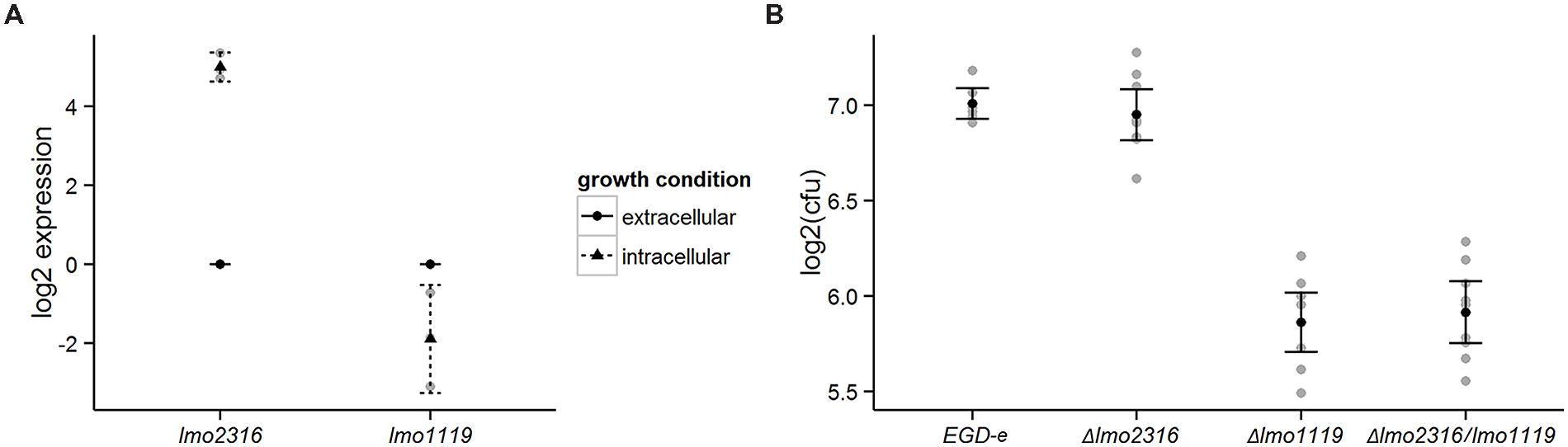
FIGURE 5. (A) Validation of RNA-seq data with qRT-PCR for the genes lmo1119 and lmo2316. Transcripts of each gene were assessed during extracellular growth and compared to intracellular expression. Values represent means of at least three independent experiments ± 2 SE. (B) Survival of Δlmo1119, Δlmo2316 and the double mutant Δlmo1119/lmo2316 in P388D1 murine macrophages. Macrophages were infected with wild type L. monocytogenes and its isogenic deletion mutants and bacterial CFU counts were measured after 4 h post-infection. Linear models to estimate intracellular survival of infection, log2-ratios of individual experiments are shown in gray. Supporting statistical information is given in Supplementary Table S4 or for a repetition of this experiment including a prfA deletion mutant strain as negative control in Supplementary Table S5, respectively.
In order to assess the effect of the phage methylase on intracellular growth properties of L. monocytogenes, we generated a knockout mutant and investigated its ability to survive in P388D1 murine macrophages compared with the wild type strain (L. monocytogenes EGD-e). lmo1119 single and lmo1119/lmo2316 double mutants were included as controls in this experiment. Δlmo1119 and Δlmo1119/lmo2316 were significantly impaired in their intracellular survival compared to L. monocytogenes EGD-e. In contrast, Δlmo2316 survived similar to L. monocytogenes EGD-e. Similarly, no additive effect of Δlmo1119/lmo2316 was observed in comparison to Δlmo1119 single mutant (Figure 5B, Supplementary Table S4). Growth in BHI was not altered in the knockout strains in comparison to wild type L. monocytogenes EGD-e (data not shown).
In conclusion, even though lmo2316 was significantly induced during intracellular growth, no contribution to intracellular survival of L. monocytogenes grown in murine macrophages was found.
This study reveals significant differences in the transcriptional response of L. monocytogenes for 23% of the genes when grown in murine macrophages compared to growth in BHI. This exceeds the microarray findings where 17% were differentially regulated (Chatterjee et al., 2006). Given the enormous differences between growth in rich media such as BHI and the cytosolic growth conditions including induction of stress responses, 23% seems reasonable if not rather low. The direct comparison between results of both studies show a high degree of similarity particularly regarding methodological differences.
This work reveals several transcriptional adaptations to intracellular growth conditions that appear to be universal, as they have been reported in previous studies after infection of epithelial cells, macrophages, and splenocytes (Chatterjee et al., 2006; Joseph et al., 2006; Camejo et al., 2009). Transcriptional upregulation of prfA and PrfA-dependent genes is one of those strategies. The majority of these upregulated genes are organized in the virulence gene cluster (lmo0201–0205). The phospholipases plcA, plcB, and listeriolysin are crucial for phagosomal escape. The metalloprotease Mpl is involved in maturation of plcB and the product of actA facilitates actin assembly beneficial for cell-to-cell spread of Listeria (Cossart, 2011). Apart from this locus, two internalin genes, prsA2 and hpt were upregulated. While Internalin B (inlB) is required for cell entry (Gaillard et al., 1991), internalin C (inlC) is involved in cell-to-cell (Engelbrecht et al., 1996; Rajabian et al., 2009) spread and interferes with the innate immune response of the host (Gouin et al., 2010). The chaperonin lmo2219 (prsA2) is thought to contribute to virulence, e.g., by promoting the stability of listeriolysin (Alonzo et al., 2009; Cahoon and Freitag, 2015). Hpt is involved in the transport of sugar-phosphates and is required for efficient intracellular proliferation (Chico-Calero et al., 2002). Thus, a predominant carbon source other than glucose has been suggested during intracellular growth (Joseph et al., 2006). Our sequencing results show differential regulation of several operons involved in sugar transport. Among those, the mannose/glucose transport operon (lmo0096–0098) was downregulated. Expression of this operon has recently been demonstrated to play a role in glucose mediated virulence gene repression (Zébré et al., 2015).
Of note, lmo1538, encoding a glycerol kinase, was found to be upregulated after infection of epithelial cells, macrophages and splenocytes (Chatterjee et al., 2006; Joseph et al., 2006; Camejo et al., 2009). Therefore, glycerol represents an alternative carbon source (Joseph et al., 2008; Grubmüller et al., 2014). In this context, the downregulation of central genes for glycolysis was discussed. While this downregulation occurred for L. monocytogenes infecting epithelial cells and macrophages (Chatterjee et al., 2006; Joseph et al., 2006; Grubmüller et al., 2014), L. monocytogenes grown in spleens showed an opposite trend (Camejo et al., 2009). Our study confirmed this downregulation of several glycolysis genes and showed upregulation of an operon including a predicted glyoxolase (lmo2671). Thus, it is interesting to figure out whether the glyoxolase pathway, also known to function as an offshoot to glycolysis, is involved in the adaptation to intracellular growth conditions.
Another universal transcriptional response is the upregulation of genes involved in stress response. Maintenance of proper protein folding during a stress response is achieved by chaperons and ATP-dependent proteases that prevent the accumulation of misfolded proteins. Our results showed upregulation of the dnaK-operon and caseinolytic protease genes (clpB, clpE, and clpC). The importance for intracellular survival of all these caseinolytic proteases has been demonstrated (Rouquette et al., 1996; Nair et al., 1999; Chastanet et al., 2004).
In our study, we address both intracellular sense and antisense regulation in L. monocytogenes for the first time. We provide evidence for regulation of several genes under intracellular growth conditions. Antisense regulation in L. monocytogenes has not been investigated thoroughly although it might be of utmost importance for translation (Mellin and Cossart, 2012; Behrens et al., 2014; Schultze et al., 2014; Wehner et al., 2014). We found antisense regulation targeting lmo1705, a deoxyguanosine kinase/deoxyadenosine kinase. Read mappings of this region prompt a linkage to the transcription of the adjacent divergent operon lmo1702–1704. Therefore, this locus might represent a putative excludon activated under intracellular growth conditions.
In addition to the bacterial transcriptional profile, we also provide the transcriptional response of a bacteriophage to intracellular growth conditions. During the last few years, bacteriophages and their response to environmental changes were increasingly investigated as they represent a mean to counteract food-borne pathogens (Denes and Wiedmann, 2014).
The greatest transcriptional changes in the prophage region occurred in the ‘early genes.’ The function of ‘early genes’ includes DNA replication, modification, recombination, and gene expression modification. Among the upregulated genes, we focused on a site-specific DNA methylase lmo2316. It is well-known that DNA methylation is crucial for epigenetic control of gene expression in bacteria. Phenomena such as timing of DNA replication, DNA repair, protection against foreign bacteriophage DNA, timing of transposition, pilus expression and conjugal plasmid transfer are sensitive to the methylation states of specific DNA regions (Sánchez-Romero et al., 2015). DNA methylases are also involved in virulence (Heusipp et al., 2007). Yet, intracellular survival in macrophages was not affected by the deletion of lmo2316. Recent studies outline the importance of methylases in the evolutionary arms race between bacteria and bacteriophages (Labrie et al., 2010; Goldfarb et al., 2015). Given that, it is more likely that this particular DNA methylase modifies newly synthesized phage DNA to evade bacterial restriction modification (R–M) systems. Several R–M systems have been previously reported for L. monocytogenes epidemic clone I and II (Sau3AI-like, LmoH7, LmoJ2, and LmoJ3) which were involved in temperature dependent phage resistance (Yildirim et al., 2010; Kim et al., 2012; Lee et al., 2012). Impaired growth of Δlmo1119 in macrophages might be explained by the role of lmo1119 in maintenance of the bacteria-specific DNA methylation pattern. Thus, a knockout of this gene can be detrimental due to its own R-M system. Of note, both gene loci (lmo2316 and lmo1119) indicate an unusually low GC content. lmo2316 is located in the well-known A118-like prophage locus whereas lmo1119 is a downstream neighbor of TN916. The latter chromosomal locus was previously designated as “hypervariable hotspot 8” where LIPI-3 is inserted in lineage I strains L. monocytogenes (Glaser et al., 2001; Cotter et al., 2008; Kuenne et al., 2013). This suggests that both gene regions originated by horizontal gene transfer via transduction and/or transposition.
Whether the temperate phage remains in lysogenic state or switches into the lytic cycle and thereby eventually lyses its host, is of crucial importance for survival of the bacteria. For the E. coli phage aaa, the two adjacent and divergently oriented transcriptional regulators cI and cro are crucial for the ‘lysis-lysogeny decision.’ Gene products of cI repress the ‘early phage’ genes including cro. Gene products of cro in turn repress transcription of cI directly as well as indirectly by decreasing transcription of an activator of cI. Currently, the indirect route is thought to have the major effect (Dodd et al., 2005; Oppenheim et al., 2005).
Homologs of cI and cro have been described for Lactobacillus phage (Kodaira et al., 1997) or the L. monocytogenes A118 phage (Loessner et al., 2000). Similar to A118 phage, the A118-like bacteriophage in L. monocytogenes EGD-e contains the genes lmo2328 and lmo2329 exhibiting similarity to cI and cro, respectively.
However, RNA-seq data contradicts the proposed mechanism of mutual repression of lmo2328 and lmo2329 for the A118-like prophage. During intracellular growth, the homologs to both cro and cI were highly expressed. Hence, the mechanism of switching between lysogeny and lytic state known from E. coli phage aaa is probably not completely applicable in L. monocytogenes EGD-e.
Yet, upregulation of lmo1398 (recA) during intracellular growth might explain the expression of the ‘early genes’ including lmo2328 (homolog to cro) despite the presence of lmo2329 (homolog to the inhibitor of early genes cI) transcripts. In E. coli, RecA supports cleavage of cI and thereby derepresses the ‘early genes.’
Of note, the ‘late genes’ are only poorly expressed under both growth conditions. The discovery of antisense transcripts in this region indicates another inhibitory mechanism.
Our study also provided further insights concerning the excision of the A118-like prophage of L. monocytogenes EGD-e when grown in macrophages. In L. monocytogenes strain 10403S a comparable phage insertion was investigated in murine bone-marrow derived macrophages under intracellular growth conditions (Rabinovich et al., 2012). This study demonstrated that prophage excision takes place under intracellular growth conditions thereby restoring functionality of the previously split competence activator comK (Rabinovich et al., 2012). We found no evidence for such an excision of the prophage genome. This is corroborated by unaltered transcript expression of the phage integrase under both growth conditions.
Moreover, the increased gene expression of all three competence gene clusters (comE, comG, and comK) in L. monocytogenes 10403S due to restoration of comK (Rabinovich et al., 2012) was not confirmed for L. monocytogenes EGD-e. In contrast, our transcriptional profile showed no intracellular upregulation of the clusters comG and comK as well as no changes in comEA and comEB expression. Only comEC expression was intracellularly upregulated in our study. Interestingly, only comG and comEC were shown to be beneficial for intracellular survival probably by alleviating the phagolysosomal escape (Rabinovich et al., 2012). We propose that in L. monocytogenes EGD-e, comEC can be alternatively activated bypassing the global competence activator comK.
Here, we present the first intracellular mRNA transcriptome for L. monocytogenes obtained using an NGS approach. RNA-seq-based studies have the power to study both coding and non-coding RNA phenomena. Our data is supported by previous microarray studies. In summary, we provide a base for future work targeting intracellular survival of L. monocytogenes covering both sense and antisense transcript changes. By comparing mRNA transcriptome data from different studies and different stages of listerial infection (epithelial cells, macrophages, and spleen as target organ), we identified a common set of genes always upregulated during intracellular growth. Furthermore, we provide the transcriptional profile of a temperate A118-like phage thereby offering novel insights in phage control and the intertwined coexisting between bacteriophage and bacteria. Taken together, this work expands the view on the transcriptional changes arising from intracellular growth conditions.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Alexandra Amend for excellent technical assistance. This work was supported by LOEWE Medical RNomics and the German Centre for Infection Research, Justus-Liebig University Giessen to TH and TC.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.01199
FIGURE S1 |Coverage plot of all operons for which differential expression patterns were in discord with the predicted operon model (Toledo-Arana et al., 2009). SigB operon (lmo0887-0896) shown in (A), lmo1416-1420 in (B) and lmo2463-2465 given in (C).
FIGURE S2 |Read mappings of all competence clusters. The compentence cluster comG and comF are poorly expressed. Only comEC (lmo1482) was significantly upregulated during intracellular growth.
FIGURE S3 |Visualization of the chromosomal regions of lmo1119 (A) and lmo2316 (B) using GECO (Kuenne et al., 2007). The line below the illustrated genes indicates the deviation from the average GC content. Values greater than zero are highlighted by brown colour while those below zero are blue. (C) Comparison of the A118-like prophage region between the Listeria monocytogenes strains EGD-e, 10403S and EGD. Genes are colored according to similarity on protein level. Core genes are indicated in black.
Allerberger, F., and Wagner, M. (2010). Listeriosis: a resurgent foodborne infection. Clin. Microbiol. Infect. 16, 16–23. doi: 10.1111/j.1469-0691.2009.03109.x
Alonzo, F., Port, G. C., Cao, M., and Freitag, N. E. (2009). The posttranslocation chaperone PrsA2 contributes to multiple facets of Listeria monocytogenes pathogenesis. Infect. Immun. 77, 2612–2623. doi: 10.1128/IAI.00280-09
Anders, S., Pyl, P. T., and Huber, W. (2014). HTSeq A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. doi: 10.1101/002824
Behrens, S., Widder, S., Mannala, G. K., Qing, X., Madhugiri, R., Kefer, N., et al. (2014). Ultra deep sequencing of Listeria monocytogenes sRNA transcriptome revealed new antisense RNAs. PLoS ONE 9:e83979. doi: 10.1371/journal.pone.0083979
Cahoon, L. A., and Freitag, N. E. (2015). Identification of conserved and species-specific functions of the Listeria monocytogenes PrsA2 secretion chaperone. Infect. Immun. 83, 4028–4041. doi: 10.1128/IAI.00504-15
Camejo, A., Buchrieser, C., Couvé, E., Carvalho, F., Reis, O., Ferreira, P., et al. (2009). In vivo transcriptional profiling of Listeria monocytogenes and mutagenesis identify new virulence factors involved in infection. PLoS Pathog. 5:449. doi: 10.1371/journal.ppat.1000449
Chambers, J. M., and Hastie, T. J. (1992). “Linear models,” in Statistical Models, Chap. 4, eds J. M. Chambers and T. J. Hast (Pacific Grove, CA: Wadsworth Brooks Chambers JM).
Chastanet, A., Derre, I., Nair, S., and Msadek, T. (2004). ClpB, a Novel Member of the Listeria monocytogenes CtsR Regulon. Is involved in virulence but not in general stress tolerance. J. Bacteriol. 186, 1165–1174. doi: 10.1128/JB.186.4.1165
Chatterjee, S. S., Hossain, H., Otten, S., Kuenne, C., Kuchmina, K., Machata, S., et al. (2006). Intracellular gene expression profile of Listeria monocytogenes. Infect. Immun. 74, 1323–1338. doi: 10.1128/IAI.74.2.1323
Chico-Calero, I., Suárez, M., González-Zorn, B., Scortti, M., Slaghuis, J., Goebel, W., et al. (2002). Hpt, a bacterial homolog of the microsomal glucose- 6-phosphate translocase, mediates rapid intracellular proliferation in Listeria. Proc. Natl. Acad. Sci. U.S.A. 99, 431–436. doi: 10.1073/pnas.012363899
Christiansen, J. K., Nielsen, J. S., Ebersbach, T., Valentin-Hansen, P., Søgaard-Andersen, L., and Kallipolitis, B. H. (2006). Identification of small Hfq-binding RNAs in Listeria monocytogenes. RNA 12, 1383–1396. doi: 10.1261/rna.49706
Cossart, P. (2011). Illuminating the landscape of host-pathogen interactions with the bacterium Listeria monocytogenes. Proc. Natl. Acad. Sci. U.S.A. 108, 19484–19491. doi: 10.1073/pnas.1112371108
Cossart, P., and Lebreton, A. (2014). A trip in the “new microbiology” with the bacterial pathogen Listeria monocytogenes. FEBS Lett. 588, 2437–2445. doi: 10.1016/j.febslet.2014.05.051
Cossart, P., and Toledo-Arana, A. (2008). Listeria monocytogenes, a unique model in infection biology: an overview. Microbes Infect. 10, 1041–1050. doi: 10.1016/j.micinf.2008.07.043
Cotter, P. D., Draper, L. A., Lawton, E. M., Daly, K. M., Groeger, D. S., Casey, P. G., et al. (2008). Listeriolysin S, a novel peptide haemolysin associated with a subset of lineage I Listeria monocytogenes. PLoS Pathog. 4:e1000144. doi: 10.1371/journal.ppat.1000144
Denes, T., and Wiedmann, M. (2014). Environmental responses and phage susceptibility in foodborne pathogens: implications for improving applications in food safety. Curr. Opin. Biotechnol. 26, 45–49. doi: 10.1016/j.copbio.2013.09.001
Dodd, I. B., Shearwin, K. E., and Egan, J. B. (2005). Revisited gene regulation in bacteriophage aaa. Curr. Opin. Genet. Dev. 15, 145–152. doi: 10.1016/j.gde.2005.02.001
Elinav, H., Hershko-Klement, A., Valinsky, L., Jaffe, J., Wiseman, A., Shimon, H., et al. (2014). Pregnancy-associated listeriosis: clinical characteristics and geospatial analysis of a 10-year period in israel. Clin. Infect. Dis. 59, 953–961. doi: 10.1093/cid/ciu504
Engelbrecht, F., Chun, S. K., Ochs, C., Hess, J., Lottspeich, F., Goebel, W., et al. (1996). A new PrfA-regulated gene of Listeria monocytogenes encoding a small, secreted protein which belongs to the family of internalins. Mol. Microbiol. 21, 823–837.
Gaillard, J. L., Berche, P., Frehel, C., Gouin, E., and Cossart, P. (1991). Entry of L. monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from gram-positive cocci. Cell 65, 1127–1141. doi: 10.1016/0092-8674(91)90009-N
Glaser, P., Frangeul, L., Buchrieser, C., Rusniok, C., Amend, A., Baquero, F., et al. (2001). Comparative genomics of listeria species. Science 294, 849–852. doi: 10.1126/science.1063447
Goldfarb, T., Sberro, H., Weinstock, E., Cohen, O., Doron, S., Charpak-amikam, Y., et al. (2015). BREX is a novel phage resistance system widespread in microbial genomes. EMBO J. 34, 169–184.
Gouin, E., Adib-Conquy, M., Balestrino, D., Nahori, M.-A., Villiers, V., Colland, F., et al. (2010). The Listeria monocytogenes InlC protein interferes with innate immune responses by targeting the IκB kinase subunit IKKα. Proc. Natl. Acad. Sci. U.S.A. 107, 17333–17338. doi: 10.1073/pnas.1007765107
Grubmüller, S., Schauer, K., Goebel, W., Fuchs, T. M., and Eisenreich, W. (2014). Analysis of carbon substrates used by Listeria monocytogenes during growth in J774A.1 macrophages suggests a bipartite intracellular metabolism. Front. Cell. Infect. Microbiol. 4:156. doi: 10.3389/fcimb.2014.00156
Hamon, M., Bierne, H., and Cossart, P. (2006). Listeria monocytogenes: a multifaceted model. Nat. Rev. Microbiol. 4, 423–434. doi: 10.1038/nrmicro1413
Heusipp, G., Fälker, S., and Alexander Schmidt, M. (2007). DNA adenine methylation and bacterial pathogenesis. Int. J. Med. Microbiol. 297, 1–7. doi: 10.1016/j.ijmm.2006.10.002
Hilker, R., Stadermann, K. B., Doppmeier, D., Kalinowski, J., Stoye, J., Straube, J., et al. (2014). ReadXplorer - visualization and analysis of mapped sequences. Bioinformatics 30, 2247–2254. doi: 10.1093/bioinformatics/btu205
Hothorn, T., Bretz, F., and Westfall, P. (2008). Simultaneous inference in general parametric models. Biom. J. 50, 346–363. doi: 10.1002/bimj.200810425
Johansson, J., Mandin, P., Renzoni, A., Chiaruttini, C., Springer, M., and Cossart, P. (2002). An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 110, 551–561. doi: 10.1016/S0092-8674(02)00905-4
Joseph, B., Joseph, B., Schauer, K., Schauer, K., Fuchs, T. M., Fuchs, T. M., et al. (2006). Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. Society 188, 556–568. doi: 10.1128/JB.188.2.556
Joseph, B., Mertins, S., Stoll, R., Schar, J., Umesha, K. R., Luo, Q., et al. (2008). Glycerol metabolism and PrfA activity in Listeria monocytogenes. J. Bacteriol. 190, 5412–5430. doi: 10.1128/JB.00259-08
Kim, J. W., Dutta, V., Elhanafi, D., Lee, S., Osborne, J. A., and Kathariou, S. (2012). A novel restriction-modification system is responsible for temperature-dependent phage resistance in Listeria monocytogenes ECII. Appl. Environ. Microbiol. 78, 1995–2004. doi: 10.1128/AEM.07086-11
Kodaira, K., Oki, M., Kakikawa, M., Watanabe, N., Hirakawa, M., Yamada, K., et al. (1997). Genome structure of the Lactobacillus temperate phage φg1e: the whole genome sequence and the putative promoter/repressor system. Gene 187, 45–53. doi: 10.1016/S0378-1119(96)00687-7
Kuenne, C., Billion, A., Mraheil, M. A., Strittmatter, A., Daniel, R., Goesmann, A., et al. (2013). Reassessment of the Listeria monocytogenes pan-genome reveals dynamic integration hotspots and mobile genetic elements as major components of the accessory genome. BMC Genomics 14:47. doi: 10.1186/1471-2164-14-47
Kuenne, C. T., Ghai, R., Chakraborty, T., and Hain, T. (2007). GECO - Linear visualization for comparative genomics. Bioinformatics 23, 125–126. doi: 10.1093/bioinformatics/btl556
Labrie, S. J., Samson, J. E., and Moineau, S. (2010). Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8, 317–327. doi: 10.1038/nrmicro2315
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Lecuit, M. (2007). Human listeriosis and animal models. Microbes Infect. 9, 1216–1225. doi: 10.1016/j.micinf.2007.05.009
Lee, S., Ward, T. J., Siletzky, R. M., and Kathariou, S. (2012). Two novel type II restriction-modification systems occupying genomically equivalent locations on the chromosomes of Listeria monocytogenes strains. Appl. Environ. Microbiol. 78, 2623–2630. doi: 10.1128/AEM.07203-11
Loessner, M. J., Inman, R. B., and Lauer, P. (2000). Complete nucleotide sequence, molecular analysis and genome structure of bacteriophage A118 of Listeria monocytogenes: implications for phage evolution. Mol. Microbiol. 35, 324–340. doi: 10.1046/j.1365-2958.2000.01720.x
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21. doi: 10.1186/s13059-014-0550-8
Mandin, P., Geissmann, T., Cossart, P., Repoila, F., and Vergassola, M. (2007). Identification of new noncoding RNAs in Listeria monocytogenes and prediction of mRNA targets. Nucleic Acids Res. 35, 962–974. doi: 10.1093/nar/gkl1096
Mansjö, M., and Johansson, J. (2011). The riboflavin analog roseoflavin targets an FMN-riboswitch and blocks Listeria monocytogenes growth, but also stimulates virulence gene-expression and infection. RNA Biol. 8, 674–680. doi: 10.4161/rna.8.4.15586
Mellin, J. R., and Cossart, P. (2012). The non-coding RNA world of the bacterial pathogen Listeria monocytogenes. RNA Biol. 9, 372–378. doi: 10.4161/rna.19235
Mraheil, M. A., Billion, A., Mohamed, W., Mukherjee, K., Kuenne, C., Pischimarov, J., et al. (2011). The intracellular sRNA transcriptome of Listeria monocytogenes during growth in macrophages. Nucleic Acids Res. 39, 4235–4248. doi: 10.1093/nar/gkr033
Nair, S., Frehel, C., Nguyen, L., Escuyer, V., and Berche, P. (1999). ClpE, a novel member of the HSP100 family, is involved in cell division and virulence of Listeria monocytogenes. Mol. Microbiol. 31, 185–196. doi: 10.1046/j.1365-2958.1999.01159.x
Nielsen, J. S., Larsen, M. H., Lillebaek, E. M. S., Bergholz, T. M., Christiansen, M. H. G., Boor, K. J., et al. (2011). A small RNA controls expression of the chitinase ChiA in Listeria monocytogenes. PLoS ONE 6:e19019. doi: 10.1371/journal.pone.0019019
Nielsen, J. S., Olsen, A. S., Bonde, M., Valentin-Hansen, P., and Kallipolitis, B. H. (2008). Identification of a sigma B-dependent small noncoding RNA in Listeria monocytogenes. J. Bacteriol. 190, 6264–6270. doi: 10.1128/JB.00740-08
Oliver, H. F., Orsi, R. H., Ponnala, L., Keich, U., Wang, W., Sun, Q., et al. (2009). Deep RNA sequencing of L. monocytogenes reveals overlapping and extensive stationary phase and sigma B-dependent transcriptomes, including multiple highly transcribed noncoding RNAs. BMC Genomics 10:641. doi: 10.1186/1471-2164-10-641
Oppenheim, A. B., Kobiler, O., Stavans, J., Court, D. L., and Adhya, S. (2005). Switches in bacteriophage lambda development. Annu. Rev. Genet. 39, 409–429. doi: 10.1146/annurev.genet.39.073003.113656
Pfaffl, M. W., and Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Rabinovich, L., Sigal, N., Borovok, I., Nir-Paz, R., and Herskovits, A. A. (2012). Prophage excision activates listeria competence genes that promote phagosomal escape and virulence. Cell 150, 792–802. doi: 10.1016/j.cell.2012.06.036
Rajabian, T., Gavicherla, B., Heisig, M., Muller-Altrock, S., Goebel, W., Gray-Owen, S. D., et al. (2009). The bacterial virulence factor InlC perturbs apical cell junctions and promotes cell-to-cell spread of Listeria. Nat. Cell Biol. 11, 1212–1218. doi: 10.1038/ncb1964
R Core Team (2014). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. Available at: http://www.R-project.org/
Rouquette, C., Ripio, M. T., Pellegrini, E., Bolla, J. M., Tascon, R. I., Vázquez-Boland, J. A., et al. (1996). Identification of a ClpC ATPase required for stress tolerance and in vivo survival of Listeria monocytogenes. Mol. Microbiol. 21, 977–987. doi: 10.1046/j.1365-2958.1996.641432.x
Sánchez-Romero, M. A., Cota, I., and Casadesús, J. (2015). DNA methylation in bacteria: from the methyl group to the methylome. Curr. Opin. Microbiol. 25, 9–16. doi: 10.1016/j.mib.2015.03.004
Schäferkordt, S., and Chakraborty, T. (1995). Vector plasmid for insertional mutagenesis and directional cloning in Listeria spp. Biotechniques 19, 720–722, 724–725.
Schultze, T., Izar, B., Qing, X., Mannala, G. K., and Hain, T. (2014). Current status of antisense RNA-mediated gene regulation in Listeria monocytogenes. Front. Cell. Infect. Microbiol. 4:135. doi: 10.3389/fcimb.2014.00135
Seifart Gomes, C., Izar, B., Pazan, F., Mohamed, W., Mraheil, M. A., Mukherjee, K., et al. (2011). Universal stress proteins are important for oxidative and acid stress resistance and growth of Listeria monocytogenes EGD-e in vitro and in vivo. PLoS ONE 6:e24965. doi: 10.1371/journal.pone.0024965
Sievers, S., Lund, A., Menendez-Gil, P., Nielsen, A., Storm Mollerup, M., Lambert Nielsen, S., et al. (2015). The multicopy sRNA LhrC controls expression of the oligopeptide-binding protein OppA in Listeria monocytogenes. RNA Biol. 12, 985–997. doi: 10.1080/15476286.2015.1071011
Sievers, S., Sternkopf Lillebaek, E. M., Jacobsen, K., Lund, A., Mollerup, M. S., Nielsen, P. K., et al. (2014). A multicopy sRNA of Listeria monocytogenes regulates expression of the virulence adhesin LapB. Nucleic Acids Res. 42, 9383–9398. doi: 10.1093/nar/gku630
Toledo-Arana, A., Dussurget, O., Nikitas, G., Sesto, N., Guet-Revillet, H., Balestrino, D., et al. (2009). The Listeria transcriptional landscape from saprophytism to virulence. Nature 459, 950–956. doi: 10.1038/nature08080
Wehner, S., Mannala, G. K., Qing, X., Madhugiri, R., Chakraborty, T., Mraheil, M. A., et al. (2014). Detection of very long antisense transcripts by whole transcriptome RNA-seq analysis of Listeria monocytogenes by semiconductor sequencing technology. PLoS ONE 9:e108639. doi: 10.1371/journal.pone.0108639
Wurtzel, O., Sesto, N., Mellin, J. R., Karunker, I., Edelheit, S., Bécavin, C., et al. (2012). Comparative transcriptomics of pathogenic and non-pathogenic Listeria species. Mol. Syst. Biol. 8, 1–14. doi: 10.1038/msb.2012.11
Yildirim, S., Elhanafi, D., Lin, W., Hitchins, A. D., Siletzky, R. M., and Kathariou, S. (2010). Conservation of genomic localization and sequence content of Sau3AI-like restriction-modification gene cassettes among Listeria monocytogenes epidemic clone I and selected strains of serotype 1/2a. Appl. Environ. Microbiol. 76, 5577–5584. doi: 10.1128/AEM.00648-10
Zébré, A. C., Aké, F. M., Ventroux, M., Koffi-Nevry, R., Noirot-Gros, M.-F., Deutscher, J., et al. (2015). Interaction with enzyme IIBMpo (EIIBMpo) and phosphorylation by phosphorylated EIIBMpo exert antagonistic effects on the transcriptional activator ManR of Listeria monocytogenes. J. Bacteriol. 197, 1559–1572. doi: 10.1128/JB.02522-14
Keywords: Listeria monocytogenes, intracellular, mRNA transcriptome analysis, RNA-seq, human pathogenic bacteria
Citation: Schultze T, Hilker R, Mannala GK, Gentil K, Weigel M, Farmani N, Windhorst AC, Goesmann A, Chakraborty T and Hain T (2015) A detailed view of the intracellular transcriptome of Listeria monocytogenes in murine macrophages using RNA-seq. Front. Microbiol. 6:1199. doi: 10.3389/fmicb.2015.01199
Received: 29 May 2015; Accepted: 15 October 2015;
Published: 30 October 2015.
Edited by:
Dongsheng Zhou, Beijing Institute of Microbiology and Epidemiology, ChinaReviewed by:
Luca Cocolin, University of Turin, ItalyCopyright © 2015 Schultze, Hilker, Mannala, Gentil, Weigel, Farmani, Windhorst, Goesmann, Chakraborty and Hain. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Torsten Hain, dG9yc3Rlbi5oYWluQG1pa3JvYmlvLm1lZC51bmktZ2llc3Nlbi5kZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.