- 1Department of Microbiology, Obafemi Awolowo University, Ile-Ife, Nigeria
- 2Institute of Medical Microbiology and Hygiene, Saarland University, Homburg, Germany
- 3Department of Biological Sciences, College of Science, Engineering and Technology, Osun State University, Osogbo, Nigeria
- 4Department of Medical Laboratory Services, Federal Medical Centre, Makurdi, Nigeria
- 5Department of Microbiology and Immunology, Alfaisal University, Riyadh, Saudi Arabia
- 6Institute of Medical Microbiology, University Hospital Münster, Münster, Germany
Staphylococcus aureus is an important human pathogen causing nosocomial and community-acquired infections worldwide. In the characterization of this opportunistic pathogen, DNA microarray hybridization technique is used as an alternative to sequence based genotyping to obtain a comprehensive assessment on the virulence, resistance determinants, and population structure. The objective of this study was to characterize a defined collection of S. aureus isolates from Nigeria using the microarray technique, and to assess the extent that it correlates with sequence-based genotyping methods. The clonal diversity and genomic content of 52 methicillin-susceptible Staphylococcus aureus (MSSA) were investigated by spa typing, MLST and DNA microarray hybridization. More than half (55.8%) of these isolates were associated with clonal complexes (CCs) typically associated with methicillin-resistant S. aureus (MRSA) clones i.e., CC1, CC5, CC8, CC30, and CC45. Certain genes linked with virulence (hlgA and clfA) and adherence (ebpS, fnbA, sspA, sspB, and sspP) were detected in all isolates. A number of genes or gene clusters were associated with distinct clonal types. The enterotoxin gene cluster (egc) was linked with CC5, CC25, CC30, CC45, and CC121, enterotoxin H gene (seh) with CC1, exfoliative toxin D gene (etd) with CC25 and CC80, and the epidermal cell differentiation inhibitor B gene (edinB) with CC25, CC80, and CC152. The excellent agreement between data from DNA microarray and MLST in the delineation of Nigerian MSSA isolates indicates that the microarray technique is a useful tool to provide information on antibiotic resistance, clonal diversity and virulence factors associated with infection and disease.
Introduction
Staphylococcus aureus is implicated in a variety of human infections with high rates of morbidity and mortality (Lowy, 1998; Corey, 2009). In infection, S. aureus exhibits a coordinated and regulated expression for a wide variety of cell and surface-associated virulence factors (Foster and Höök, 1998; Novick, 2006). These factors mediate adherence to host cells and damaged tissue, facilitate tissue destruction and spreading, promote iron uptake and evasion of host immune system, as well as tissue damage (Skaar and Schneewind, 2004; Grumann et al., 2014). Recent studies in Cameroon (Kihla et al., 2014), Egypt (Ahmed et al., 2014), Gabon (Alabi et al., 2013), Nigeria (Jido and Garba, 2012; Oladeinde et al., 2013), South Africa (Groome et al., 2012; Naidoo et al., 2013), and Tanzania (Kayange et al., 2010; Mhada et al., 2012) have identified S. aureus as the main etiological agent for various infections in Africa. Moreover, this species has been recognized as one main cause of community-acquired neonatal sepsis in Africa (Waters et al., 2011). These studies clearly establish the important role of this major human pathogen in tropical Africa.
In many health care institutions in sub-Saharan Africa, the lack of skilled laboratory manpower and resources is a major constraint in the identification of bacterial pathogens from clinical samples. If such analysis can be provided at all, identification of S. aureus typically relies on phenotypic methods precluding in-depth strain characterization. Molecular analysis of clonal attribution and presence of single genes contained in S. aureus isolates have emerged in pilot studies from select African centers, areas and populations (Ateba Ngoa et al., 2012; Shittu et al., 2012; Seni et al., 2013; Aiken et al., 2014; Egyir et al., 2014; Oosthuysen et al., 2014; Conceição et al., 2015; De Boeck et al., 2015; Kraef et al., 2015; Schaumburg et al., 2015). Nevertheless, in view of the impact of S. aureus disease in sub-Saharan Africa, the clonal characterization in concert with a comprehensive analysis of the hitherto ill-described virulence factor armamentarium of S. aureus isolates from this region is urgently warranted. Such analyses should target a broad spectrum of variable staphylococcal factors such as genes or gene clusters conferring antibiotic resistance, toxins, virulence, adhesion or immune evasion factors. These analyses have not been performed on a collection of S. aureus isolates in Nigeria, and reports from African countries are limited and only addressed a limited and select analytical spectrum (Raji et al., 2013; Aiken et al., 2014; Rovira et al., 2015).
The DNA microarray used for this analysis is a unique and comprehensive genotyping technique based on the analysis of 334 target sequences corresponding to approximately 170 distinct genes and their allelic variants. It enables the simultaneous identification of various gene classes including species markers, genes encoding resistance and virulence properties, exotoxin and adhesion factors, accessory gene regulator (agr), capsule, and SCCmec types (Monecke et al., 2011). Based on the observation of a high level of genetic diversity from previous investigations on methicillin-susceptible S. aureus (MSSA) in Nigeria (Shittu et al., 2011, 2012; Kolawole et al., 2013), we studied MSSA isolates obtained from various clinical sources in Nigeria using this comprehensive, array-based approach to provide an insight on the major factors associated with infection and disease.
Materials and Methods
Identification and Antibiotic Susceptibility Testing of S. aureus Isolates
The isolates (n = 52) were obtained from samples processed as part of surveillance activities in the microbiology laboratories of six health care institutions located in Ado-Ekiti, Ile-Ife, Osogbo, Lagos, and Ibadan in South-West Nigeria, and Maiduguri in North-East Nigeria. The duration of collection of isolates was from March 2009 to April, 2010. Only the isolates were analyzed in this study. Preliminary verification as S. aureus was based on colony characteristics on blood agar, positive results for catalase, coagulase and DNase tests. Twelve isolates from a previous study (Shittu et al., 2011) were also included in this investigation. Identification was confirmed by Matrix-Assisted Laser Desorption/Ionization-Time Of Flight analysis (MALDI-TOF). Susceptibility testing to penicillin (10 units), cefoxitin (30 μg), doxycycline (30 μg), erythromycin (15 μg), clindamycin (2 μg), gentamicin (10 μg), chloramphenicol (30 μg), and trimethoprim-sulfamethoxazole (1.25/23.75 μg) were determined using the disk diffusion method according to the Clinical Laboratory Standards Institute guidelines (Clinical and Laboratory Standards Institute (CLSI), 2009).
DNA Extraction
S. aureus genomic DNA was extracted from an 18–24 h old culture on sheep blood agar using lysis buffer and lysis enhancer (StaphyType Kit, Alere Technologies GmbH, Jena, Germany) and processed using a DNeasy tissue kit (Qiagen, Hilden, Germany).
Molecular Typing of the Isolates
Typing of S. aureus was based on sequencing of the hypervariable region of the protein A gene (spa). The spa types were determined using the Ridom StaphType software (Ridom GmbH, Würzburg, Germany, version 2.1.1) (Harmsen et al., 2003). Multilocus sequence typing (MLST) was performed for one isolate of each spa type (Enright et al., 2000), as a spa type usually belongs to one sequence type (ST) with few exceptions due to homoplasies (Basset et al., 2009, 2012). The allelic profiles and STs were assigned using the MLST S. aureus database (www.mlst.net), and the sequence types of the remaining isolates were inferred from the derived MLST data.
DNA Microarray Hybridization
The DNA microarray of the StaphyType™ kit (Alere Technologies GmbH, Jena, Germany) was used in this study according to previously established protocols (Monecke et al., 2008). The isolates were grouped with various clonal complexes (CCs) by the imaging software Iconoclust based on comparison of hybridization profiles to a collection of reference strains previously characterized by MLST.
Splits Graph Construction
The SplitsTree algorithm (Huson and Bryant, 2006) and software was used to analyze the similarities between hybridization patterns, and network tree construction was performed using SplitsTree 4.10 on default settings (characters transformation, uncorrected P; distance transformation, Neighbor-Net; and variance, ordinary least squares).
Results
Identification of S. aureus Isolates
A total of 52 MSSA (3 and 49 isolates from nasal and clinical sources, respectively) were analyzed (Table 1). The clinical isolates were obtained from wounds and associated infections (n = 29; 59.2%), urinary tract infections (n = 6; 12.2%), semen/infertility diagnosis (n = 4; 8.2%), ocular infections (n = 3; 6.1%), and pneumonia (n = 2; 4.1%). One isolate each was from otitis media, and blood related infections, while information on three isolates was not available. The clinical isolates were obtained from health care institutions located in Ile-Ife (n = 26; 53.1%), Osogbo (n = 11; 22.4%), Maiduguri (n = 5; 10.2%), Lagos (n = 4; 8.2%), Ibadan, and Ado-Ekiti (n = 2 isolates each: 4.1%).
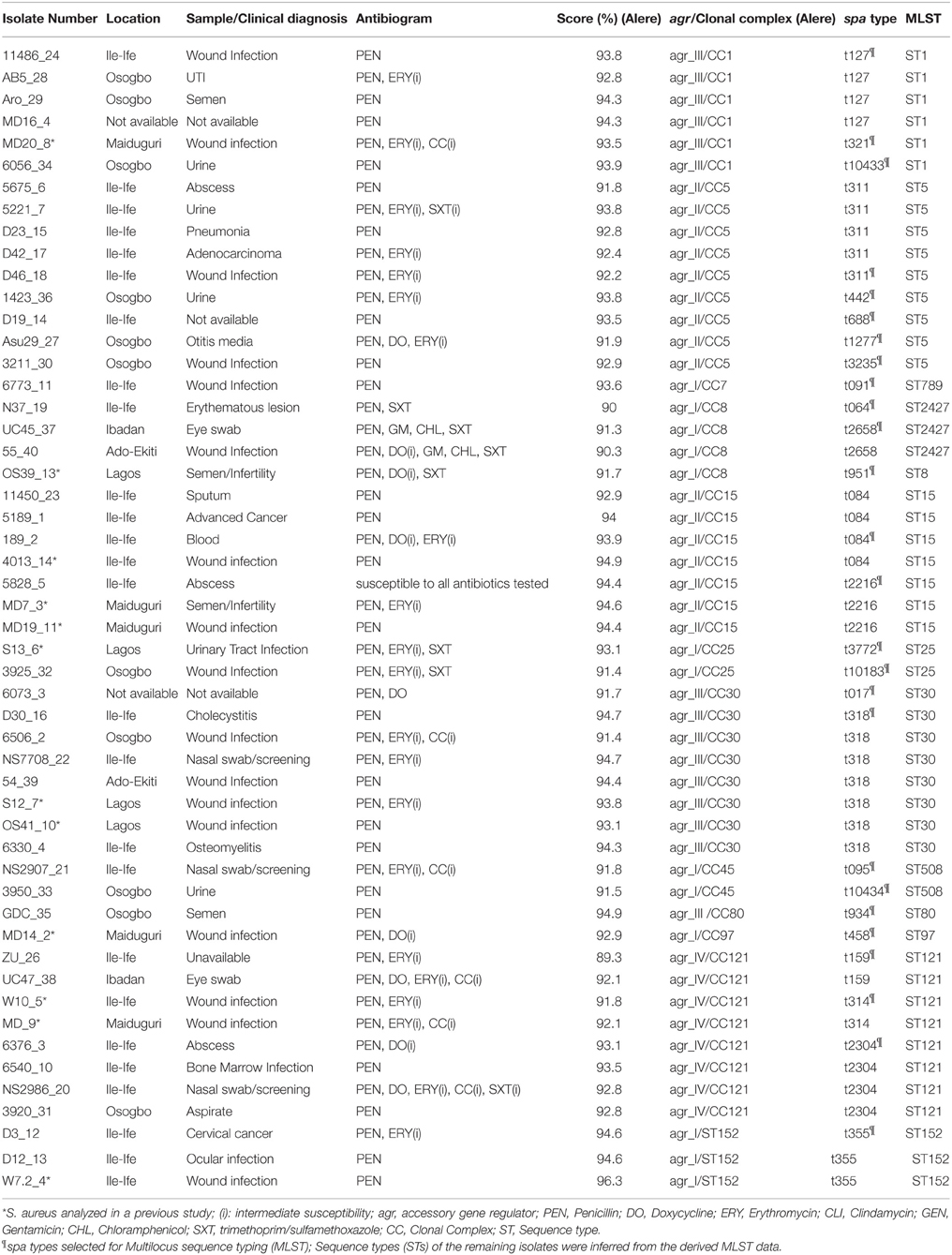
Table 1. Characterization of the methicillin-susceptible S. aureus (MSSA) from Nigeria based on antibiotyping, microarray analysis, spa typing, and MLST.
Antibiotic Susceptibility Testing
All the isolates were susceptible to cefoxitin and 98.1% (n = 51) were resistant to penicillin. Only two isolates each exhibited resistance to chloramphenicol and gentamicin, and four to doxycycline. Intermediate susceptibility to clindamycin and erythromycin were identified in six and 21 isolates, respectively. The predominant antibiotype was resistance only to penicillin (n = 23; 44.2%), and resistance to penicillin with intermediate susceptibility to erythromycin (n = 10; 19.2%) (Table 1).
Sequence based Typing (spa and MLST)
A total of 26 spa types were identified among the 52 MSSA isolates and the most common were t318 (n = 7), t311 (n = 5), t084, t127, and t2304 (n = 4 each). Based on MLST, the MSSA were classified into 13 sequence types (STs) (Table 1).
DNA Microarray Analysis
The assay confirmed the identity of the isolates (S. aureus) by positive results for specific markers including rndD1 (domain 1 of 23S rRNA), protein A (spa), glyceraldehyde 3-phosphate dehydrogenase (gapA), catalase A (katA), thermostable nuclease (nuc), and staphylococcal accessory regulator A (sarA) (Supplementary Materials 1, 2). The hybridization profiles revealed that the 52 MSSA isolates clustered in 12 different CCs. More than half (55.8%) of the CCs were associated with the genetic background common to the major methicillin-resistant S. aureus (MRSA) clones i.e., CC1 (n = 6 isolates), CC5 (n = 9), CC8 (n = 4), CC30 (n = 8), and CC45 (n = 2). The rest were assigned with CC7 (n = 1), CC15 (n = 7), CC25 (n = 2), CC80 (n = 1), CC97 (n = 1), CC121 (n = 8), and CC152 (n = 3).
Antibiotic Resistance Genes
A total of 69.2% (n = 36) of the isolates yielded a hybridization signal for the beta-lactamase gene (blaZ) and only 10 and three isolates were positive for the tetracycline resistance genes (tetK and tetM), respectively. The two MSSA in CC8 which exhibited phenotypic resistance to chloramphenicol and gentamicin possessed the corresponding resistance genes (cat and aacA-aphD). In addition, the single CC80 isolate was positive for the lincosamide resistance gene (lnuA).
Accessory Gene Regulator and Capsular Typing
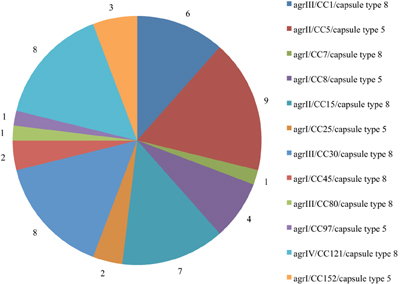
The distribution of agr/CCs/capsule types for the MSSA is indicated in Figure 1. Overall, 13 (25%) isolates assigned to different clonal lineages (CC7, CC8, CC25, CC45, CC97, and CC152) were associated with agr group I, 16 (30.2%; CC5 and CC15) with group II, and 15 (28.8%; CC1, CC30 and CC80) with group III. CC121 was the only representative for agr group IV (n = 8; 15.4%) (Table 1). The capsule type 8 was the most frequent and detected in 33 (63.5%) isolates affiliated with CC1, CC7, CC15, CC30, CC45, CC80, and CC121. The remaining isolates (20; 38.5%) belonged to capsule type 5 (assigned with CC5, CC8, CC25, CC97, and CC152).
Enterotoxin Genes
PVL-positive isolates (n = 27) belonged to CC1, CC5, CC15, CC30, CC80, CC121, and CC152 (Supplementary Material 1). Moreover, the lukF gene (haemolysin gamma; component B) was universally detected in all the CCs and the lukE genes was a common feature except with MSSA isolates in CC30, CC45, and CC152 (Supplementary Material 3). With respect to the carriage of superantigen genes, only three MSSA (one isolate in CC1 and two in CC45) tested positive for the toxic shock syndrome toxin gene (tst-1) (Supplementary Material 1). All the isolates in this study lacked a hybridization signal for the enterotoxin E gene (see Supplementary Material 2) and the enterotoxin genes were not detected in MSSA assigned with CC80, CC97, and CC152. In the haemolysin gene family, almost all (98.1%) the isolates in the various CCs possessed the haemolysin alpha and delta genes (hla, hld), while the haemolysin beta gene (hlb) was identified in the various CCs except in CC15, CC45, and CC152.
Microbial Surface Components Recognizing Adhesive Matrix Molecule (MSCRAMM) Genes
All the isolates were negative for the surface protein involved in biofilm production (bap), but possessed the genes for the inter-cellular adhesion protein (icaA/C/D) (CC152 isolates were icaC negative). The genes for clumping factor A (clfA), cell surface elastin binding protein (ebpS), fibronectin-binding protein A (fnbA) and proteases (sspA, sspB, and sspP) were detected in all the isolates (Supplementary Materials 1, 3).
Splits Tree Analysis
The analysis identified four main clusters (CC5/CC25; CC8/CC97; CC1/CC7/CC80, and CC30/CC45) indicating the phylogenetic relationship between the isolates (Figure 2).
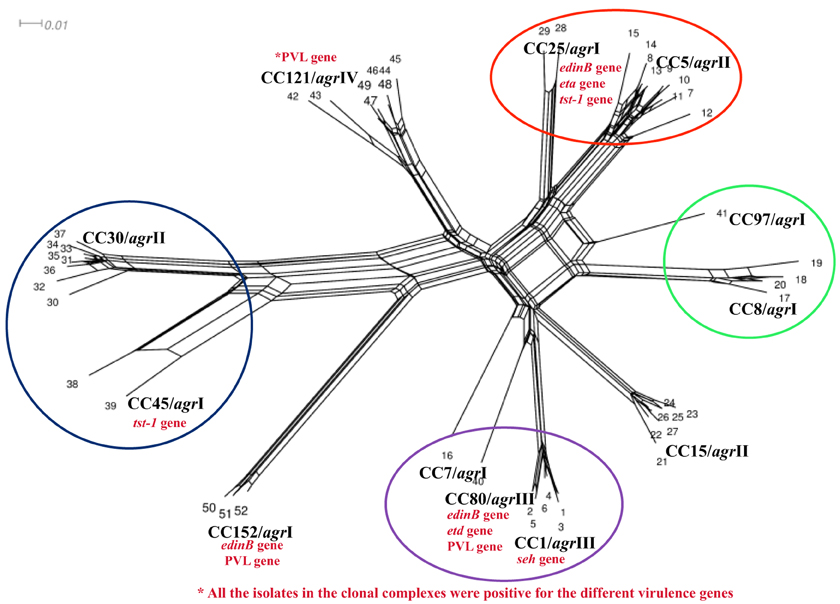
Figure 2. Splits tree graph based on hybridization profile of the MSSA isolates. The results of all array hybridization experiments were arranged in a matrix. Columns represent the target genes and the rows represent the number of experiments; 1, positive; 0, negative; −, ambiguous. Converted to “sequences”: 1, c; 0, g; −, c.
Discussion
We observed a complete agreement between DNA microarray analysis and MLST in the delineation of the isolates (Table 1), showing that the hybridization profile could be used to predict the lineages. Furthermore, the heterogeneous and divergent nature of the isolates observed in this study provided evidence on the overall higher diversity of MSSA compared with MRSA (Deurenberg and Stobberingh, 2008; Goering et al., 2008; Ghasemzadeh-Moghaddam et al., 2011; Ruffing et al., 2012; Blomfeldt et al., 2013; Rasmussen et al., 2013, 2014). In Nigeria, many diagnostic microbiology laboratories rely on the disc diffusion technique for antibiotic susceptibility testing, but this protocol does not provide information on the nature of resistance genes. The antibiotic susceptibility results observed in this study were in accordance with the corresponding resistance gene profiles by DNA microarray. MSSA isolates that exhibited full resistance to trimethoprim-sulfamethoxazole clustered with CC8 and CC25, but were dfrS1 negative indicating that a different mechanism could be attributed to resistance. A recent study (Nurjadi et al., 2014) has provided strong evidence that the dfrG gene is the predominant trimethoprim resistance determinant on S. aureus in Africa. Overall, resistant determinants for antibiotics, heavy metal and quaternary ammonium compounds were observed more often in CC8 than other CCs (Supplementary Materials 1, 3).
The accessory gene regulator (agr) and capsule typing methods are useful front-line tools for the characterization of S. aureus (Goerke et al., 2005). Hybridization signals for agr type I and IV were observed for one, three, and four isolates grouped with CC25, CC152, and CC121, respectively (Supplementary Materials 1, 2). This could be attributed to possible cross-hybridization as the alleles for the two agr types are closely related (Monecke et al., 2010). Our observations on CCs and agr groups were similar to previous reports on MSSA in five major African towns (Breurec et al., 2010), Gabon (Ateba Ngoa et al., 2012), and Nigeria (Ghebremedhin et al., 2009; Kolawole et al., 2013). In addition, our study also support the view (Wright et al., 2005; Holtfreter et al., 2007; Rasmussen et al., 2014) that an agr type may be detected in isolates which are assigned to genetically diverse CCs, whereas, it is also associated with specific CCs. The dominance of capsule type 8 in MSSA is consistent with data from Gabon (Schaumburg et al., 2011), Norway (Blomfeldt et al., 2013), and Sweden (Rasmussen et al., 2013, 2014).
Staphylococcal enterotoxins are typically encoded by genes located on mobile genetic elements (Baba et al., 2002). The egc cluster (seg+sei+sem+sen+seo+seu) is located on the genomic island vSAβ and reported to be associated with specific clonal types regardless of the geographical strain distribution (Lindsay and Holden, 2006). In this investigation, the egc-enterotoxin gene cluster was a unique feature for CC5, CC25, CC30, CC45, and CC121. Previous studies have indicated that the cluster is predominantly present in MSSA assigned with CC5, CC25, CC30, and CC45 (Van Trijp et al., 2010; Rasmussen et al., 2013). The seh gene is linked to the staphylococcal cassette chromosome mec (SCCmec elements) and reported to be restricted to the CC1 genomic background (Baba et al., 2002). Moreover, the seh gene has also been reported mainly in MSSA-CC30 (Blomfeldt et al., 2013). Nevertheless, our observation on seh-positive MSSA-CC1 is in agreement with previous reports (Chen et al., 2013; Rasmussen et al., 2013).
The genes associated with staphylococcal complement inhibitor (scn) and staphylokinase (sak) were also widely distributed across the CCs but CC15 isolates were sak gene negative. Virulence associated with the exfoliative toxins has been identified to cause epidermal cleavage in staphylococcal scalded skin syndrome (SSSS) and bullous impetigo (Ladhani et al., 1999). The exfoliative toxin D (ETD) is a 27-kDa protein which causes epidermal blisters in newborn mice (Yamasaki et al., 2006). The epidermal cell differentiation factors (EDIN) target and inhibit the small host protein RhoA, a master regulator of the host cell actin cytoskeleton (Inoue et al., 1991; Jaffe and Hall, 2005; Aktories, 2011). Furthermore, the edin-isoform (edinB) and etd genes are located in tandem in a S. aureus etd pathogenicity island in a chromosome of etd-positive S. aureus strains (Yamaguchi et al., 2002). A strong association of the etd gene with invasive CC25 S. aureus isolates has also been reported. In this study, all the isolates assigned with CC25 and CC80 were etd-positive, which is in agreement with a previous study in Nigeria (Shittu et al., 2011). Moreover, MSSA grouped with CC25, CC80, and CC152 were edinB positive but CC152 isolates were etd negative. Our observations were similar to a study on the distribution of the edin gene in S. aureus from diabetic foot ulcers (Messad et al., 2013). A study in MSSA bacteremia isolates in Sweden showed that the collagen binding protein (Cna) was detected in CC1, CC30, and CC45. Our report identified the gene in isolates assigned with CC1, CC30, CC45, CC121, and CC152.
Our study has a number of limitations. Although all isolates were of human origin, and the large majority was obtained from clinical samples, a clear distinction between commensal and clinical strains could not be made based on the available information. An association of isolates within the context of endemicity i.e., nosocomial vs. community associated infections, is also not clear. Furthermore, whereas the microarray analytical database is exhaustive, well-characterized, and validated with isolates from all continents, the attribution of CCs is based on the hybridization reactions and resulting microarray profile rather than gene sequencing, and a positive signal does not necessarily imply the presence of gene product (e.g., protein). In addition, the microarray method was unable to separate ST8 from ST2427. This might be due to the close phylogenetic relation of both STs as they are single locus variants (ST8: 3−3−1−1−4−4−3 and ST2427: 3−3−297−1−4−4−3). Finally, with a collection of 52 isolates studied, and a large number of genes and genetic profile ascertained by microarray, the potential for individual statistical comparisons is limited. Yet, with this comprehensive genetic-analytical approach performed on a clinical isolate collection obtained from patients of various medical institutions in a sub-Saharan African country, Nigeria, a number of important observations could be made which clearly characterize and demarcate the clonal distribution as well as the virulence gene equipment.
More than one half (55.8%; n = 29) of these MSSA isolates were associated with a genetic background which is attributable to classic methicillin-resistant S. aureus (MRSA) clones. PVL-positive isolates were identified in seven of the 12 CCs. Moreover, toxin genes were observed to be distributed mainly with certain clonal types, and in agreement with previous investigations (Holtfreter et al., 2007; Monecke et al., 2008). Antibiotic resistance gene profiles of the isolates by the DNA microarray demonstrated concordant results with data on antibiotic susceptibility testing. The array-based, comprehensive approach has been shown to yield such diverse CC and gene specific results on an isolate collection from sub-Saharan Africa. Overall, microarray analysis proved to be a useful tool to provide useful information on antibiotic resistance, population structure and various virulence factor profiles associated with infection and disease. It is assumed that these findings might be useful for a better understanding of clinical staphylococcal disease presentation, patient care and for assistance in outbreak investigation in health care institutions in a country such as Nigeria. Moreover, our study also underlines the need for further trials employing well-controlled, prospectively collected clinical isolates to delineate the genetic pathogen profile in conjunction with the clinical disease presentation in sub-Saharan Africa.
Author Contributions
AS, UR, GP, FS, LM, and MH conceived the study, OO, KO, AR conducted the sample collection and preliminary identification of the isolates. AS performed the microarray technique, AS and UR analyzed the microarray data, and AS wrote the manuscript (with input from all authors). All authors read and approved the final version of the manuscript.
Funding
The stay of AS at the Institute of Medical Microbiology and Hygiene, Institute of Medical Microbiology, Saarland University Medical Centre, Homburg/Saar and the University Hospital Münster, Münster, Germany was supported by the Third World Academy of Science and Deutsche Forschungsgemeinschaft (TWAS-DFG award) and the Deutsche Forschungsgemeinschaft (PAK296, Ei 247/8, He 1850/9−1, and He 1850/11−1).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the management and laboratory staff of the hospitals for their support in isolate collection, and Eniola Dolapo for her assistance in the preparation of this manuscript. We gratefully acknowledge the technical assistance of the staff at the Institute of Medical Microbiology, Saarland University Medical Centre, Homburg/Saar, the Institute of Medical Microbiology, University Hospital Münster and the staff at the Sequencing Unit at the Institute of Hygiene, University Hospital Münster, Germany. We thank Professor Iruka Okeke for the critical review of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.01160
Abbreviations
Agr, accessory gene regulator; CC, Clonal complex; CLSI, Clinical Laboratory Standards Institute; MSSA, Methicillin susceptible Staphylococcus aureus; MLST, Multilocus sequence typing; PVL, Panton-Valentine Leukocidin; S.aureus, Staphylococcus aureus; SCCmec, Staphylococcal chromosome cassette mec; spa, Staphylococcus aureus protein A; ST, Sequence Type.
References
Ahmed, E. F., Gad, G. F., Abdalla, A. M., Hasaneen, A. M., and Abdelwahab, S. F. (2014). Prevalence of methicillin resistant Staphylococcus aureus among Egyptian patients after surgical interventions. Surg. Infect. (Larchmt). 15, 404–411. doi: 10.1089/sur.2013.212
Aiken, A. M., Mutuku, I. M., Sabat, A. J., Akkerboom, V., Mwangi, J., Scott, J. A., et al. (2014). Carriage of Staphylococcus aureus in Thika Level 5 Hospital, Kenya: a cross-sectional study. Antimicrob. Resist. Infect. Control 3:22. doi: 10.1186/2047-2994-3-22
Aktories, K. (2011). Bacterial protein toxins that modify host regulatory GTPases. Nat. Rev. Microbiol. 9, 487–498. doi: 10.1038/nrmicro2592
Alabi, A. S., Frielinghaus, L., Kaba, H., Kösters, K., Huson, M. A., Kahl, B. C., et al. (2013). Retrospective analysis of antimicrobial resistance and bacterial spectrum of infection in Gabon, Central Africa. BMC Infect. Dis. 13:455. doi: 10.1186/1471-2334-13-455
Ateba Ngoa, U., Schaumburg, F., Adegnika, A. A., Kosters, K., Moller, T., Fernandes, J. F., et al. (2012). Epidemiology and population structure of Staphylococcus aureus in various population groups from a rural and semi urban area in Gabon, Central Africa. Acta Trop. 124, 42–47. doi: 10.1016/j.actatropica.2012.06.005
Baba, T., Takeuchi, F., Kuroda, M., Yuzawa, H., Aoki, K., Oguchi, A., et al. (2002). Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359, 1819–1827. doi: 10.1016/S0140-6736(02)08713-5
Basset, P., Hammer, N. B., Kuhn, G., Vogel, V., Sakwinska, O., and Blanc, D. S. (2009). Staphylococcus aureus clfB and spa alleles of the repeat regions are segregated into major phylogenetic lineages. Infect. Genet. Evol. 9, 941–947. doi: 10.1016/j.meegid.2009.06.015
Basset, P., Nübel, U., Witte, W., and Blanc, D. S. (2012). Evaluation of adding a second marker to overcome Staphylococcus aureus spa typing homoplasies. J. Clin. Microbiol. 50, 1475–1477. doi: 10.1128/JCM.00664-11
Blomfeldt, A., Aamot, H. V., Eskesen, A. N., Müller, F., and Monecke, S. (2013). Molecular characterization of methicillin-sensitive Staphylococcus aureus isolates from bacteremic patients in a Norwegian University Hospital. J. Clin. Microbiol. 51, 345–347. doi: 10.1128/JCM.02571-12
Breurec, S., Fall, C., Pouillot, R., Boisier, P., Brisse, S., Diene-Sarr, F., et al. (2010). Epidemiology of methicillin-susceptible Staphylococcus aureus lineages in five major African towns: high prevalence of Panton-Valentine leukocidin genes. Clin. Microbiol. Infect. 17, 633–639. doi: 10.1111/j.1469-0691.2010.03320.x
Chen, X., Wang, W.-K., Han, L.-Z., Liu, Y., Zhang, H., Tang, J., et al. (2013). Epidemiological and genetic diversity of Staphylococcus aureus causing bloodstream infection in Shanghai, 2009-2011. PLoS ONE 8:e72811. doi: 10.1371/journal.pone.0072811
Clinical Laboratory Standards Institute (CLSI). (2009). Performance Standards for Antimicrobial Disk Susceptibility Tests. Approved Standard, 10th Edn. Wayne, Penn.
Conceição, T., Coelho, C., Santos Silva, I., de Lencastre, H., and Aires-de-Sousa, M. (2015). Staphylococcus aureus in former Portuguese colonies from Africa and the far east: missing data to help fill the world map. Clin. Microbiol. Infect. 21, 842.e1–842.e10. doi: 10.1016/j.cmi.2015.05.010
Corey, G. R. (2009). Staphylococcus aureus bloodstream infections: definitions and treatment. Clin. Infect. Dis. 48, S254–S259. doi: 10.1086/598186
De Boeck, H., Vandendriessche, S., Hallin, M., Batoko, B., Alworonga, J. P., Mapendo, B., et al. (2015). Staphylococcus aureus nasal carriage among health care workers in Kisangani, the democratic republic of Congo. Eur. J. Clin. Microbiol. Infect. Dis. 34, 1567–1572. doi: 10.1007/s10096-015-2387-9
Deurenberg, R. H., and Stobberingh, E. E. (2008). The evolution of Staphylococcus aureus. Infect. Genet. Evol. 8, 747–763. doi: 10.1016/j.meegid.2008.07.007
Egyir, B., Guardabassi, L., Sørum, M., Nielsen, S. S., Kolekang, A., Frimpong, E., et al. (2014). Molecular epidemiology and antimicrobial susceptibility of clinical Staphylococcus aureus from healthcare institutions in Ghana. PLoS ONE 9:e89716. doi: 10.1371/journal.pone.0089716
Enright, M. C., Day, N. P., Davies, C. E., Peacock, S. J., and Spratt, B. G. (2000). Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38, 1008–1015.
Foster, T. J., and Höök, M. (1998). Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 6, 484–488. doi: 10.1016/S0966-842X(98)01400-0
Ghasemzadeh-Moghaddam, H., Ghaznavi-Rad, E., Sekawi, Z., Yun-Khoon, L., Aziz, M. N., Hamat, R. A., et al. (2011). Methicillin susceptible Staphylococcus aureus from clinical and community sources are genetically diverse. Int. J. Med. Microbiol. 301, 347–353. doi: 10.1016/j.ijmm.2010.10.004
Ghebremedhin, B., Olugbosi, M. O., Raji, A. M., Layer, F., Bakare, R. A., König, B., et al. (2009). Emergence of a community-associated methicillin-resistant Staphylococcus aureus strain with a unique resistance profile in Southwest Nigeria. J. Clin. Microbiol. 47, 2975–2980. doi: 10.1128/JCM.00648-09
Goering, R. V., Shawar, R. M., Scangarella, N. E., O'Hara, F. P., Amrine-Madsen, H., West, J. M., et al. (2008). Molecular epidemiology of methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates from global clinical trials. J. Clin. Microbiol. 46, 2842–2847. doi: 10.1128/JCM.00521-08
Goerke, C., Esser, S., Kümmel, M., and Wolz, C. (2005). Staphylococcus aureus strain designation by agr and cap polymorphism typing and delineation of agr diversification by sequence analysis. Int. J. Med. Microbiol. 295, 67–75. doi: 10.1016/j.ijmm.2005.01.004
Groome, M. J., Albrich, W. C., Wadula, J., Khoosal, M., and Madhi, S. A. (2012). Community-onset Staphylococcus aureus bacteraemia in hospitalized African children: high incidence in HIV-infected children and high prevalence of multidrug resistance. Paediatr. Int. Child Health. 32, 140–146. doi: 10.1179/1465328111Y.0000000044
Grumann, D., Nübel, U., and Bröker, B. M. (2014). Staphylococcus aureus toxins – their functions and genetics. Infect. Genet. Evol. 21, 583–592. doi: 10.1016/j.meegid.2013.03.013
Harmsen, D., Claus, H., Witte, W., Rothgänger, J., Claus, H., Turnwald, D., et al. (2003). Typing of methicillin-resistant Staphylococcus aureus in a university setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41, 5442–5448. doi: 10.1128/JCM.41.12.5442-5448.2003
Holtfreter, S., Grumann, D., Schmudde, M., Nguyen, H. T. T., Eichler, P., Strommenger, B., et al. (2007). Clonal distribution of superantigen genes in clinical Staphylococcus aureus isolates. J. Clin. Microbiol. 45, 2669–2680. doi: 10.1128/JCM.00204-07
Huson, D. H., and Bryant, D. (2006). Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267. doi: 10.1093/molbev/msj030
Inoue, S., Sugai, M., Murooka, Y., Paik, S. Y., Hong, Y. M., Ohgai, H., et al. (1991). Molecular cloning and sequencing of the epidermal cell differentiation inhibitor gene from Staphylococcus aureus. Biochem. Biophys. Res. Commun. 174, 459–464. doi: 10.1016/0006-291X(91)91438-I
Jaffe, A. B., and Hall, A. (2005). RHO GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 21, 247–269. doi: 10.1146/annurev.cellbio.21.020604.150721
Jido, T., and Garba, I. (2012). Surgical-site infection following cesarean section in Kano, Nigeria. Ann. Med. Health Sci. Res. 2, 33–36. doi: 10.4103/2141-9248.96934
Kayange, N., Kamugisha, E., Mwizamholya, D. L., Jeremiah, S., and Mshana, S. E. (2010). Predictors of positive blood culture and deaths among neonates with suspected neonatal sepsis in a tertiary hospital, Mwanza-Tanzania. BMC Pediatr. 10:39. doi: 10.1186/1471-2431-10-39
Kihla, A. J., Ngunde, P. J., Evelyn, M. S., Gerard, N., and Ndip, R. N. (2014). Risk factors for wound infection in health care facilities in Buea, Cameroon: aerobic bacterial pathogens and antibiogram of isolates. Pan. Afr. Med. J. 18, 6. doi: 10.11604/pamj.2014.18.6.2304
Kolawole, D. O., Adeyanju, A., Schaumburg, F., Akinyoola, A. L., Lawal, O. O., Amusa, Y. B., et al. (2013). Characterization of colonizing Staphylococcus aureus isolated from surgical wards' patients in a Nigerian university hospital. PLoS ONE 8:e68721. doi: 10.1371/journal.pone.0068721
Kraef, C., Alabi, A. S., Peters, G., Becker, K., Kremsner, P. G., Rossatanga, E. G., et al. (2015). Co-detection of Panton-Valentine leukocidin encoding genes and cotrimoxazole resistance in Staphylococcus aureus in Gabon: implications for HIV-patients' care. Front. Microbiol. 6:60. doi: 10.3389/fmicb.2015.00060
Ladhani, S., Joannou, C. L., Lochrie, D. P., Evans, R. W., and Poston, S. M. (1999). Clinical, microbial and biochemical aspects of the exfoilative toxins causing staphylococcal scalded-skin syndrome. Clin. Microbiol. Rev. 12, 224–242.
Lindsay, J. A., and Holden, M. T. (2006). Understanding the rise of the superbug: investigation of the evolution and genomic variation of Staphylococcus aureus. Funct. Integr. Genomics 6, 186–201. doi: 10.1007/s10142-005-0019-7
Lowy, F. D. (1998). Staphylococcus aureus infections. N. Engl. J. Med. 339, 520–532. doi: 10.1056/NEJM199808203390806
Messad, N., Landraud, L., Canivet, B., Lina, G., Richard, J.-L., and Sotto, A. (2013). Distribution of edin in Staphylococcus aureus isolated from diabetic foot ulcers. Clin. Microbiol. Infect. 19, 875–880. doi: 10.1111/1469-0691.12084
Mhada, T. V., Fredrick, F., Matee, M. I., and Massawe, A. (2012). Neonatal sepsis at Muhimbili National Hospital, Dar es Salaam, Tanzania; aetiology, antimicrobial sensitivity pattern and clinical outcome. BMC Public Health 12:904. doi: 10.1186/1471-2458-12-904
Monecke, S., Coombs, G., Shore, A. C., Coleman, D. C., Akpaka, P., Borg, M., et al. (2011). A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS ONE 6:e17936. doi: 10.1371/journal.pone.0017936
Monecke, S., Kanig, H., Rudolph, W., Müller, E., Coombs, G., Hotzel, H., et al. (2010). Characterisation of Australian MRSA strains ST75- and ST883-MRSA-IV and analysis of their accessory gene regulator locus. PLoS ONE 5:e14025. doi: 10.1371/journal.pone.0014025
Monecke, S., Slickers, P., and Ehricht, R. (2008). Assignment of Staphylococcus aureus isolates to clonal complexes based on microarray analysis and pattern recognition. FEMS Immunol. Med. Microbiol. 53, 237–251. doi: 10.1111/j.1574-695X.2008.00426.x
Naidoo, R., Nuttali, J., Whitelaw, A., and Eley, B. (2013). Epidemiology of Staphylococcus aureus Bacteraemia at a Tertiary Children's Hospital in Cape Town, South Africa. PLoS ONE 8:e78396. doi: 10.1371/journal.pone.0078396
Novick, R. P. (2006). “Staphylococcal pathogenesis and pathogenicity factors: genetics and regulation,” in Gram-Positive Pathogens, eds V. Fischetti, R. Novick, J. Ferretti, D. Portnoy, and J. Rood (Washington, DC: ASM Press), 496–516.
Nurjadi, D., Olalekan, A. O., Layer, F., Shittu, A. O., Alabi, A., Ghebremedhin, B., et al. (2014). Emergence of trimethoprim resistance gene dfrG in Staphylococcus aureus causing human infection and colonization in sub-Saharan Africa and its import to Europe. J. Antimicrob. Chemother. 69, 2361–2368. doi: 10.1093/jac/dku174
Oladeinde, B. H., Omoregie, R., Olley, M., Anunibe, J. A., and Onifade, A. A. (2013). A 5-year surveillance of wound infections at a rural tertiary hospital in Nigeria. Afr. Health Sci. 13, 351–356. doi: 10.4314/ahs.v13i2.22
Oosthuysen, W. F., Orth, H., Lombard, C. J., Sinha, B., and Wasserman, E. (2014). Population structure analyses of Staphylococcus aureus at Tygerberg Hospital, South Africa, reveals a diverse population, high prevalence of Panton-Valentine leukocidin genes and unique local MRSA clones. Clin. Microbiol. Infect. 20, 652–659. doi: 10.1111/1469-0691.12452
Raji, A., Ojemhen, O., Umejiburu, U., Ogunleye, A., Blanc, D. S., and Basset, P. (2013). High genetic diversity of Staphylococcus aureus in a tertiary care hospital in South west Nigeria. Diagn. Microbiol. Infect. Dis. 77, 367–369. doi: 10.1016/j.diagmicrobio.2013.08.030
Rasmussen, G., Monecke, S., Brus, O., Ehricht, R., and Soderquist, B. (2014). Long term molecular epidemiology of methicillin-susceptible Staphylococcus aureus bacteremia isolates in Sweden. PLoS ONE 9:e114276. doi: 10.1371/journal.pone.0114276
Rasmussen, G., Monecke, S., Ehricht, R., and Soderquist, B. (2013). Prevalence of clonal complexes and virulence genes among commensal and invasive Staphylococcus aureus isolates in Sweden. PLoS ONE 8:e77477. doi: 10.1371/journal.pone.0077477
Rovira, G., Cooke, E. F., Mucavele, H., Sitoe, A., Madrid, L., Nhampossa, T., et al. (2015). The challenge of diagnosing and treating Staphylococcus aureus invasive infections in a resource-limited sub-Saharan Africa setting: a case report. J. Trop. Pediatr. 61, 397–402. doi: 10.1093/tropej/fmv045
Ruffing, U., Akulenko, R., Bischoff, M., Helms, V., Herrmann, M., and von Müller, L. (2012). Matched-Cohort DNA microarray diversity analysis of methicillin sensitive and methicillin-resistant Staphylococcus aureus isolates from hospital admission patients. PLoS ONE 7:e52487. doi: 10.1371/journal.pone.0052487
Schaumburg, F., Ngoa, U. A., Kösters, K., Köck, R., Adegnika, A. A., Kremsner, P. G., et al. (2011). Virulence factors and genotypes of Staphylococcus aureus from infection and carriage in Gabon. Clin. Microbiol. Infect. 17, 1507–1513. doi: 10.1111/j.1469-0691.2011.03534.x
Schaumburg, F., Pauly, M., Anoh, E., Mossoun, A., Wiersma, L., Schubert, G., et al. (2015). Staphylococcus aureus complex from animals and humans in three remote African regions. Clin. Microbiol. Infect. 21, e1–e8. doi: 10.1016/j.cmi.2014.12.001
Seni, J., Bwanga, F., Najjuka, C. F., Makobore, P., Okee, M., and Mshana, S. E. (2013). Molecular characterization of Staphylococcus aureus from patients with surgical site infections at Mulago Hospital in Kampala, Uganda. PLoS ONE 8:e66153. doi: 10.1371/journal.pone.0066153
Shittu, A. O., Okon, K., Adesida, S., Oyedara, O., Witte, W., Strommenger, B., et al. (2011). Antibiotic resistance and molecular epidemiology of Staphylococcus aureus in Nigeria. BMC Microbiol. 11:92. doi: 10.1186/1471-2180-11-92
Shittu, A., Oyedara, O., Abegunrin, F., Okon, K., Raji, A., Taiwo, S., et al. (2012). Characterization of methicillin-susceptible and -resistant staphylococci in the clinical setting: a multicentre study in Nigeria. BMC Infect. Dis. 12:286. doi: 10.1186/1471-2334-12-286
Skaar, E. P., and Schneewind, O. (2004). Iron-regulated surface determinants (lsd) of Staphylococcus aureus: stealing iron from heme. Microbes Infect. 6, 390–397. doi: 10.1016/j.micinf.2003.12.008
Van Trijp, M. J. C. A., Melles, D. C., Snijders, S. V., Wertheim, H. F. L., Verbrugh, H. A., van Belkum, A., et al. (2010). Genotypes, superantigen gene profiles, and presence of exfoliative toxin genes in clinical methicillin-susceptible Staphylococcus aureus isolates. Diagn. Microbiol. Infect. Dis. 66, 222–224. doi: 10.1016/j.diagmicrobio.2009.08.021
Waters, D., Jawad, I., Ahmad, A., Luksic, I., Nair, H., Zgaga, L., et al. (2011). Aetiology of community-acquired neonatal sepsis in low- and middle-income countries. J. Glob. Health 1, 154–170.
Wright, J. S., Traber, K. E., Corrigan, R., Benson, S. A., Musser, J. M., and Novick, R. P. (2005). The agr radiation: an early event in the evolution of staphylococci. J. Bacteriol. 187, 5585–5594. doi: 10.1128/JB.187.16.5585-5594.2005
Yamaguchi, T., Nishifuji, K., Sasaki, M., Fudaba, Y., Aepfelbacher, M., and Takata, T. (2002). Identification of the Staphylococcus aureus etd pathogenicity island which encodes a novel exfoliative toxin, ETD, and EDIN-B. Infect. Immun. 70, 5835–5845. doi: 10.1128/IAI.70.10.5835-5845.2002
Keywords: Staphylococcus aureus, microarray, MLST, genotyping, Nigeria
Citation: Shittu AO, Oyedara O, Okon K, Raji A, Peters G, von Müller L, Schaumburg F, Herrmann M and Ruffing U (2015) An assessment on DNA microarray and sequence-based methods for the characterization of methicillin-susceptible Staphylococcus aureus from Nigeria. Front. Microbiol. 6:1160. doi: 10.3389/fmicb.2015.01160
Received: 12 August 2015; Accepted: 05 October 2015;
Published: 20 October 2015.
Edited by:
Miklos Fuzi, Semmelweis University, HungaryReviewed by:
Dmitri Debabov, NovaBay Pharmaceuticals, USADinesh Sriramulu, Shres Consultancy (Life Sciences), India
Karl Hassan, Macquarie University, Australia
Copyright © 2015 Shittu, Oyedara, Okon, Raji, Peters, von Müller, Schaumburg, Herrmann and Ruffing. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Adebayo O. Shittu, YmF5b19zaGl0dHVAeWFob28uY29t
†Present Address: Lutz von Müller, Institute of Laboratory Medicine, Microbiology and Hygiene (LMH), Christophorus-Kliniken GmbH, Coesfeld, Germany
 Adebayo O. Shittu
Adebayo O. Shittu Omotayo Oyedara
Omotayo Oyedara Kenneth Okon
Kenneth Okon Adeola Raji
Adeola Raji Georg Peters
Georg Peters Lutz von Müller
Lutz von Müller Frieder Schaumburg
Frieder Schaumburg Mathias Herrmann
Mathias Herrmann Ulla Ruffing
Ulla Ruffing