
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 29 September 2015
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 6 - 2015 | https://doi.org/10.3389/fmicb.2015.00977
This article is part of the Research Topic Antibiotic resistance in aquatic systems View all 10 articles
 Aziz Ghaderpour1,2
Aziz Ghaderpour1,2 Wing Sze Ho1
Wing Sze Ho1 Li-Lee Chew1,2
Li-Lee Chew1,2 Chui Wei Bong1,2
Chui Wei Bong1,2 Ving Ching Chong1,2
Ving Ching Chong1,2 Kwai-Lin Thong1,2
Kwai-Lin Thong1,2 Lay Ching Chai1,2*
Lay Ching Chai1,2*E.coli, an important vector distributing antimicrobial resistance in the environment, was found to be multi-drug resistant, abundant, and genetically diverse in the Matang mangrove estuaries, Malaysia. One-third (34%) of the estuarine E. coli was multi-drug resistant. The highest antibiotic resistance prevalence was observed for aminoglycosides (83%) and beta-lactams (37%). Phylogenetic groups A and B1, being the most predominant E. coli, demonstrated the highest antibiotic resistant level and prevalence of integrons (integron I, 21%; integron II, 3%). Detection of phylogenetic group B23 downstream of fishing villages indicates human fecal contamination as a source of E. coli pollution. Enteroaggregative E. coli (1%) were also detected immediately downstream of the fishing village. The results indicated multi-drug resistance among E. coli circulating in Matang estuaries, which could be reflective of anthropogenic activities and aggravated by bacterial and antibiotic discharges from village lack of a sewerage system, aquaculture farms and upstream animal husbandry.
Escherichia coli (E. coli), a component of the common intestinal microbiota in humans and warm-blooded animals, is an important indicator of fecal contamination in aquatic environments and food. Certain strains of E. coli are able to cause gastrointestinal and extraintestinal infections in humans. The pathogenic E. coli that cause gastrointestinal infections include enterohemorrhagic E. coli (EHEC), also referred to as Shiga toxin-producing E. coli (STEC) or verocytotoxic E. coli (VTEC), enteropathogenic E. coli (EPEC), enteroaggregative E. coli (EAEC), enterotoxigenic E. coli (ETEC), enteroinvasive E. coli (EIEC), and diffusely adherent E. coli (DAEC) (Galvin et al., 2010; Koczura et al., 2013). These pathogenic E. coli, together with other virulent strains of E. coli could cause extraintestinal infections in humans such as pyelonephritis, urinary tract infection, cystitis, neonatal meningitis, and bacteremia (Johnson and Stell, 2000; Galvin et al., 2010; Koczura et al., 2013).
E. coli is divided into four main phylogenetic groups A, B1, B2, and D, based on the presence and absence of, chuA, a gene that is responsible for heme transport in enterohemorrhagic O157:H7 E. coli; yjaA, an unknown functional gene which is identified in the recent complete genome sequence of E. coli K-12; and TSPE4.C2, an anonymously designated DNA fragment which is the non-coding region in E. coli strains (Clermont et al., 2000). These four phylogenetic groups of E. coli appear to have distinctive genetic and phenotypic characteristics that are associated with different ecological niches (Bergthorsson and Ochman, 1998; Johnson et al., 2001; Gordon and Cowling, 2003; Gordon, 2004; Walk et al., 2007). It is interesting to note that most of the E. coli strains that are able to endure environmental stresses in the aquatic environment belong to the B1 phylogenetic group (Walk et al., 2007). In addition, E. coli strains that belong to the phylogenetic groups B2 and D harbor more virulent genes than those belonging to A and B1 (Johnson et al., 2001). The extraintestinal pathogenic E. coli strains are mostly phylogenetic group B2 and to a lesser extent of group D; while the commensal E. coli strains are commonly categorized into phylogenetic group A and B1 (Maynard et al., 2004; Vignaroli et al., 2012; Koczura et al., 2013; Pereira et al., 2013). Escobar-Páramo et al. (2004) found that most ETEC, EHEC, and EIEC belong to the phylogenetic groups A and B1; while EPEC, EAEC, and DAEC are usually not linked to any phylogenetic grouping.
The occurrence of E. coli in the environment, particularly in aquatic systems, is a global public health concern, especially when the water is used for human activities. The problem is recently exacerbated by the emergence of antimicrobial resistant E. coli strains in aquatic environments worldwide (Koczura et al., 2013; Pereira et al., 2013). Antibiotic resistant E. coli and other enteric bacteria, that survived the extensive antibiotic treatments in the gut of humans or animals, can enter aquatic systems through discharge from poultry and livestock production, and hospital and municipal wastewaters (Pruden et al., 2006; Pereira et al., 2013). Therefore, the rivers that are used for recreational activities, irrigation, and other purposes, can be efficient vehicles that disseminate the antibiotic resistant bacteria (Pruden et al., 2006; Su et al., 2012; Pereira et al., 2013).
Antibiotic resistance is mostly transferred among bacteria by mobile genetic elements such as integrons, plasmids and transposons. Integrons are well-organized gene expression systems that can integrate one or more non-functional gene cassettes and convert them into expressed genes (Hall and Collis, 1995; Recchia and Hall, 1995; Su et al., 2012). Integrons can easily spread the antibiotic resistance among bacterial species due to their association with plasmids. There are three types of integrons, class 1, 2, and 3 integrons and their classification is according to the integrase gene (intI) (Carattoli, 2003; Cambray et al., 2010; Su et al., 2012).
In addition to their use in human medicine, antibiotics are also widely used as therapeutics, prophylactics and metaphylactics in animals and aquaculture farms (Jaime et al., 2012). Twenty different antibiotics (e.g., erythromycin, tetracycline, oxalinic acid, oxytetracycline, amplicillin, norfloxacine, trimethoprim, sulfudimethoxine) and chemicals (e.g., calcium carbonate, potassium permanganate, formalin, and thiodane) are commonly used in aquaculture farms in Malaysia (Keh, 2003). Generally, antibiotics are mixed with feeds, or dissolved directly in water. However, fish are not able to metabolize the antibiotics, which are largely returned to the environment via faces that pose a potential threat to public health (Burridge et al., 2010; Jaime et al., 2012).
Several estuaries within Matang Mangrove Forest Reserve (MMFR), Perak were selected as study sites in this study. The MMFR bears the distinctive features of being one of the best managed mangrove forests in the world, an important nursery area sustaining the country's largest fisheries landings and site to the state's largest brackish water aquaculture farms (Ariffin and Nik Mustafa, 2013). The MMFR exemplifies significant economic benefits and ecological services that could be obtained through sustained forest management. However, the Matang area also demonstrates the increasing dilemma of multiple-use conflicts of land and water resources (Chong et al., 2010). Twenty eight of the 34 permanent settlements in the MMFR are fishing villages sited along the mangrove estuaries with a total population of 31,800 people (Lim and Mohamad Parit, 2001).
A recent study shows that the Matang mangrove estuaries may be facing a great challenge of anthropogenic pollution due to contamination by various potentially pathogenic bacteria including high counts of E. coli in both water and sediment (Ghaderpour et al., 2014). This kind of anthropogenic pollution could become a more significant public health issue if these bacteria are resistant to antibiotics. Therefore, in this study, we aimed to (i) investigate the genetic diversity of E. coli in Matang estuaries, (ii) determine the virulence factors of the estuarine E. coli isolates, and (iii) detect antibiotic resistant E. coli and their resistance mechanisms in Matang mangrove estuaries.
The MMFR is located in the northwest coast of peninsular Malaysia (Figure 1). It is a silviculture production forest of 41,000 ha managed on a sustainable basis since 1902. Within the sheltered waterways of the forest reserve, nearly 8000 floating fish cages are present, while 385 shrimp and fish ponds cover a total area of 223 ha on the landward margin of the mangrove forests (Ariffin and Nik Mustafa, 2013). Cockle culture covers an extensive area of 4726 ha within and outside the Matang estuaries.
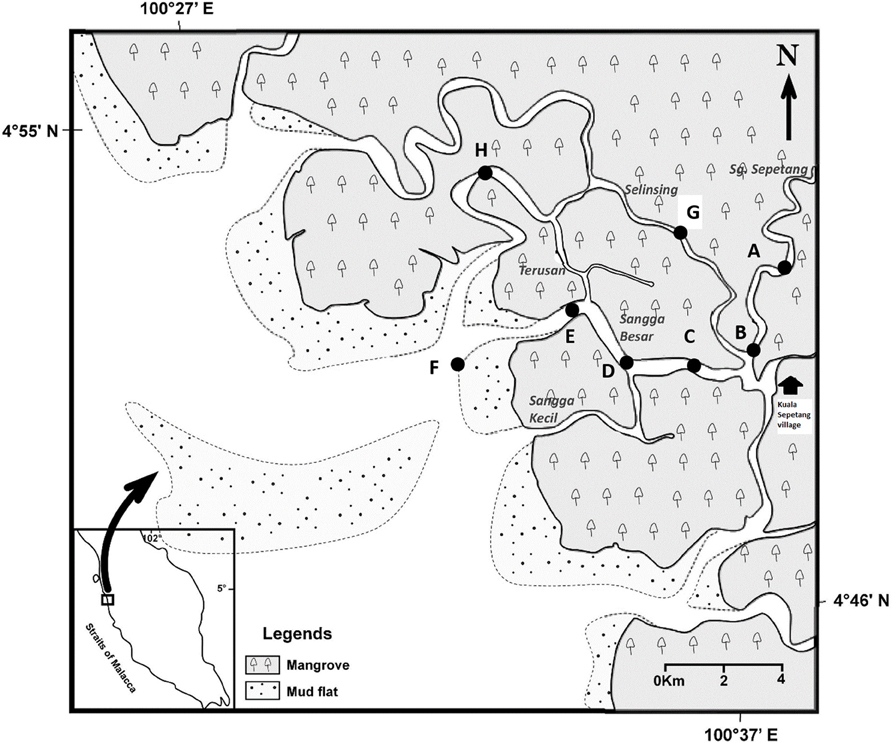
Figure 1. Locations of eight sampling stations (A–H) along Sepetang, Sangga Besar and Selinsing rivers in Matang Mangrove Forest Reserve, Perak, Malaysia.
Water and sediment samplings were carried out in the various estuaries, particularly the Sangga Besar, Sepetang and Selinsing Rivers. Sangga Besar River is the main waterway (8 km) traversed by fishing boats between the fishing village at Kuala Sepetang and the coastal fishing grounds. Unlike the Sangga Besar, the Selinsing River is a long waterway (18 km) relatively less used by boats as well as having few floating fish farms at its river mouth. Eight sampling sites (A–H) were set up, from upstream to downstream of the estuary (Figure 1). Station A is situated at the upper estuary of the Sepetang River, while stations B to F are located along the Sangga Besar River. Two further sampling sites, G and H were set up at the upstream and downstream of the Selinsing River, respectively. Station A is located upstream of the Kuala Sepetang village, while Station B, C, and G are situated 1–7 km downstream of the village, receiving the untreated sewage and other anthropogenic pollution from the village. Station D and E are located near to cockle culture beds and fish cages, respectively. Station F is situated at the river mouth downstream of the fish cages close to other cockle culture areas on the coastal mudflat. Station H is located 15 km downstream from the Kuala Sepetang village on Selinsing River, with very little human activity (Figure 1).
Four water and sediment samples were collected at each sampling station in October 2011 (wet season) and May 2012 (dry season). Water samples (0.5 m depths) were collected in acid washed bottles. A 15 cm × 15 cm Ekman grab (Wildco, USA) was used to collect bottom sediment. The top 5 cm surface of the collected sediment samples was sampled using a spatula and kept in sterile bottles. All samples were stored in ice, and immediately transferred into the laboratory freezer kept at 4°C until the analyses were carried out within 24 h of sampling.
Membrane filtration technique was used to isolate E. coli. Water (5 ml) and sediment (1 g in 9 ml of sterile 0.85% saline) samples were filtered through 0.45 mm nitrocellulose filters (47 mm diameter). The filters were then transferred on CHROMagar ECC (CHROMagar Inc., Paris, France) and incubated at 42 ± 0.5°C for 24 h. Blue colonies as presumptive E. coli were randomly picked and purified on Luria Bertani medium. After purification, presumptive E. coli isolates were kept in stab and in glycerol forms (Luria Bertani Broth with 50% glycerol) at 37°C and −20°C, respectively for the future analysis. The identity of the presumptive E. coli isolates was confirmed by PCR targeting the phoA gene, an E. coli housekeeping gene (Yu and Thong, 2009).
Two different multiplex PCR assays were carried out for all confirmed E. coli to identify the pathogenic E. coli (EHEC, EPEC, EAEC, ETEC, EIEC and DAEC). For both multiplex PCR, reaction mixture was carried out in 25 μL volume consisting of 1X green buffer (5X green GoTaq reaction buffer, pH 8.5), 0.5U of Taq DNA polymerase (Promega, Madison,Wis, USA), 1.65 mM MgCl2, 0.3μM of the selected primers, 220 μM of each deoxynucleoside triphosphate (dNTP), and 5 μL of DNA template. PCR amplification and primers used in this study were previously described by Gómez-Duarte et al. (2009). Primers sequences are listed in Supplementary Table 1. Six positive control strains (E. coli 2060–004, E2348/69, JM221, E9034A, C1845, and EC-12) were used in this study and sterile water was included as negative control. The six positive strains were provided by Oscar G. Gomez-Duarte, International Enteric Vaccines Research Program [IEVRP], University of Iowa Children's Hospital, Iowa City, IA (Gómez-Duarte et al., 2009).
A triplex PCR assay was used to determine the phylogenetic groups (A, B1, B2, and D) of E. coli. PCR conditions for presence or absence of chuA, yjaA, and TSPE4-C2 were used as described by Clermont et al. (2000) and Carlos et al. (2010) (Table 1). Each reaction was performed in 25 μL volume containing 5 μL (appr 10 ng) DNA template, 120 μM each dNTP, 1X PCR buffer, 0.5 U Taq polymerase, 1.5 mM MgCl2, 0.4 μM of yjaA primers, 0.24 μM each chuA, and TSPE4-C2 primers (Supplementary Table 1). Combination of presence or absence of chuA, yjaA, and TSPE4-C2 was used for identification of phylogenetic grouping as follows: phylogenetic group A (A0, −∕−∕−; A1, −∕+∕−), phylogenetic group B (B1, −∕−∕+), phylogenetic group B2 (B22, +∕+∕−; B23, +∕+∕+) and phylogenetic group D (D1, +∕−∕−; D2, +∕−∕+) (Carlos et al., 2010).
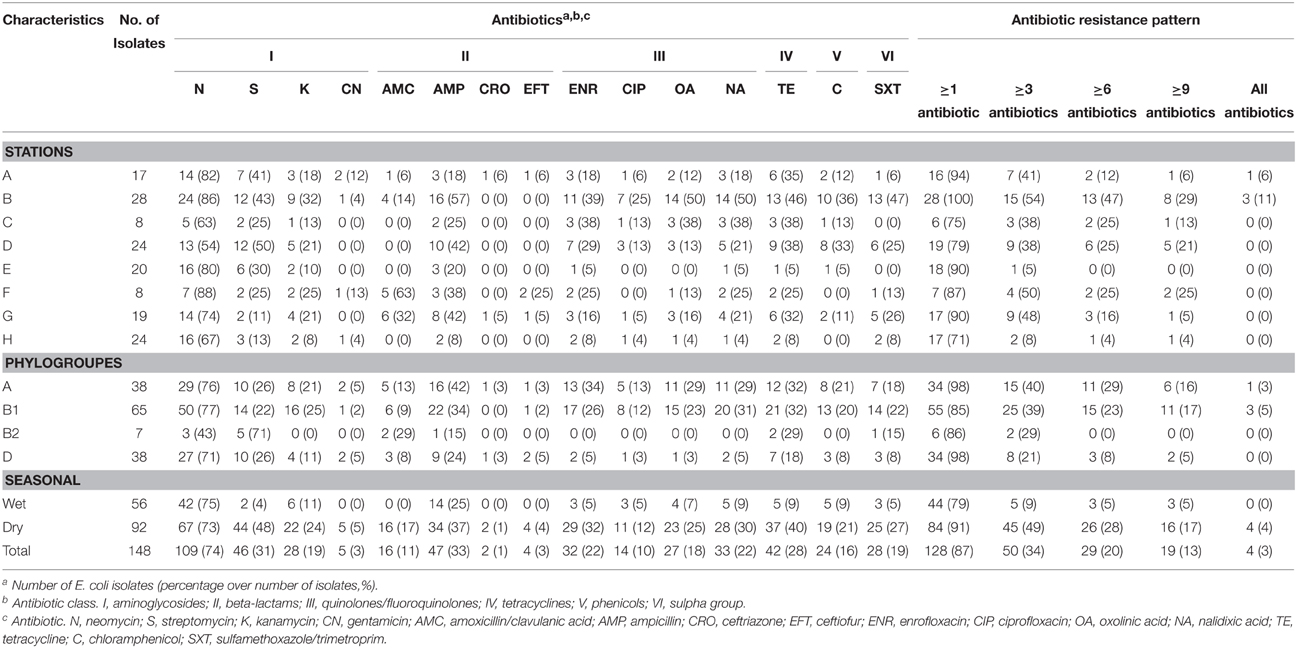
Table 1. Antibiotic susceptibility characterization of 148 isolates of E. coli isolated from estuarine water and surface sediments of Matang mangrove estuary, Perak, Malaysia.
Agar disc diffusion method on Mueller-Hinton Agar (Oxoid, Basingstoke, UK) was used to determine the antibiotic resistance patterns among E. coli isolates. A total of 15 antibiotics belonging to 7 classes were tested, including aminoglycosidase: gentamicin (CN, 10 μg), kanamycin (K, 30 μg), streptomycin (S, 25 μg), and neomycin (N, 30 μg); β-lactams: amoxicillin/clavulanic acid (AMC, 30μg), ampicillin (AMP, 10 μg), ceftriaxone (CRO, 30 μg), ceftiofur (EFT, 30 μg); quinolones: nalidixic acid (NA, 30 μg), oxolinic acid (OA, μg); fluoroquinolone: enrofloxacin (ENR, 5 μg), ciprofloxacin (CIP, 5 μg); tetracycline: tetracycline (TE, 30 μg); phenicols: chloramphenicol (C, 30 μg), and sulfamethoxazole/trimethoprim (SXT, 25 μg) (Oxoid Ltd.,Basingtoke, Hampshire, England). The bacteria were designated as resistant, intermediate and sensitive according to the Clinical Laboratory Standards Institute (CLSI)'s recommendations (CLSI, 2012). Bacteria strains that were resistant and intermediate were categorized as non-susceptible while the sensitive strains were categorized as susceptible.
All E. coli isolates were screened for integrase genes. Three different sets of primers were used to detect class 1, 2, and 3 integrase genes (Supplementary Table 1). Primers IntI1-F, IntI1-R and 5′CS and 3′CS were used to detect intI1 gene and class 1 integron gene cassettes, respectively. For class two integrons, IntI2-F, IntI2-R, and attI2-F/orfX-R primers were used for amplifying intI2 and class 2 integron gene cassettes, respectively. The primer IntI3-F/IntI3-R was used to amplify intI3 gene.
The reaction mixture for all PCR assays was performed in 25 μL volume which consisted of 1X PCR green buffer, 1.4 mM MgCl2, 120 μM of each dNTP, 0.06 μM of each primers, and 0.5 U of Taq DNA polymerase (Promega, Madison, Wisconsin, USA). The multiplex PCR amplification of integrase type 1 and 2, and class 1 integron gene cassettes was performed as previously described by Lim et al. (2009).
PCR products were purified and submitted to a commercial company (First BASE Laboratories, Malaysia) for sequencing using BigDye® Terminator v3.1 cycle sequencing kit chemistry. Homologs search against the GenBank databases of nr/nt using BLASTN (http://blast.ncbi.nlm.nih.gov/blast) were carried out for all the DNA sequences obtained.
Genetic diversity of 148 E. coli isolates was analyzed by Repetitive Extragenic Palindromic-PCR (REP-PCR) using REP primer (Supplementary Table 1). Bacterial isolates were grown in 1 ml of sterile Luria Bertani broth overnight at 37°C. Bacterial suspension in 1.5 ml microfuge tubes were centrifuged and washed with sterile Phosphate buffered saline (PBS) and 1X TE (Tris-EDTA) buffer. Bacterial pellets were suspended in sterile water and were then heated at 99°C for 10 min and chilled on ice for 20 min. The extracted crude DNA was used for amplification with REP oligonucleotides (Operon Biotechnologies GmBH, Germany) as previously reported (Lim et al., 2009). REP-PCR assay was carried out in 25 μL volume which included of 1X buffer (5X buffer GoTaq reaction buffer, pH 8.5), 1U of Taq DNA polymerase (Promega, Madison, Wisconsin, USA), 2.5 mM MgCl2, 200 μM of each dNTP 0.5 μM of the primer and 4 μL of DNA template. Temperature program for this assay consisted of initial denaturing at 94°C for 7 min, followed by 30 cycles of 30 s at 94°C, 1 min at 44°C, 8 min at 72°C and a final extension step 16 min at 65°C. PCR products were loaded in 1.5% agarose gels and stained in Gel red nucleic acid stain (Biotium Inc, USA). Banding patterns were analyzed by GelCompar II, version 2.5 (Applied Maths, Kortrijk, Belgium).
Prevalence of the antibiotic resistance was determined using x/y 100% equation, where x is defined as number of resistant E. coli isolates to antibiotics, and y is total isolated number of E. coli from each sample. Principle component analysis (PCA) was used to depict the distribution of E. coli with respect to phylogenetic groups, integron genes, and antibiotics resistance among eight sampling stations in Matang mangrove estuary. To perform the PCA procedure, the number of isolates of E. coli containing the phylogenetic groups, integron genes and antibiotics resistance was square root transformed to meet the parametric assumptions. PCA was performed using CANOCO 4.5 software (Ter Braak and Smilauer, 2002). Pearson's chi-squared goodness-of-fit test was applied to evaluate the significant difference among phylogenetic groups based on the frequency data. Also, comparisons of associations between phylogroups and antibiotic resistance were done using the chi- square test when no more than 20% of the expected cell frequencies were < 5 and all individual expected counts were ≥1, otherwise Fisher's exact test (two-tailed) was used. The degree of association between the presence of antibiotic resistance and presence of integron genes were tested using the correlation (Rn) of Ives and Gibbons for dichotomous normal-scale data in Zar (2010). Significance was tested using the chi-square contingency test. All tests were performed at the 5% level of significance.
A total of 175 presumptive E. coli bacteria were isolated from 64 water and sediment samples, during dry and wet seasons. Out of 175 presumptive E. coli isolates, 148 (85) were confirmed as E. coli, of which 17 were from station A, 28 from station B, 8 from station C, 24 from station D, 20 from station E, 8 from station F, 19 from station G and 24 from station H.
Among the 148 E. coli isolates, phylogenetic group B1 was the most prevalent (n = 65, 44%) followed by phylogenetic group A and D which had similar prevalence (n = 38; 26%). The phylogenetic group B2 was the least prevalent (5%) in Matang mangrove estuaries, and was only detected during the dry season. Phylogenetic groups B1 and D were present at all the stations located along Sangga Besar and Selinsing Rivers. Phylogenetic group B1 was the most frequent isolate from all stations except station E and H, where phylogenetic groups A and D predominated, respectively (Figure 2). Phylogenetic group D was more frequently isolated in the wet season (32%) than in the dry season (22%). The prevalence of group A was significantly higher at station E, located in Sangga Besar River (χ2 = 16.526, df = 6, p = 0.011); while station E recorded a significantly lower occurrence of E. coli subset D. Station H located downstream of Selinsing River had significantly higher occurrence of E. coli subset D (χ2 = 20.105, df = 6, p = 0.005) (Figures 1, 2).
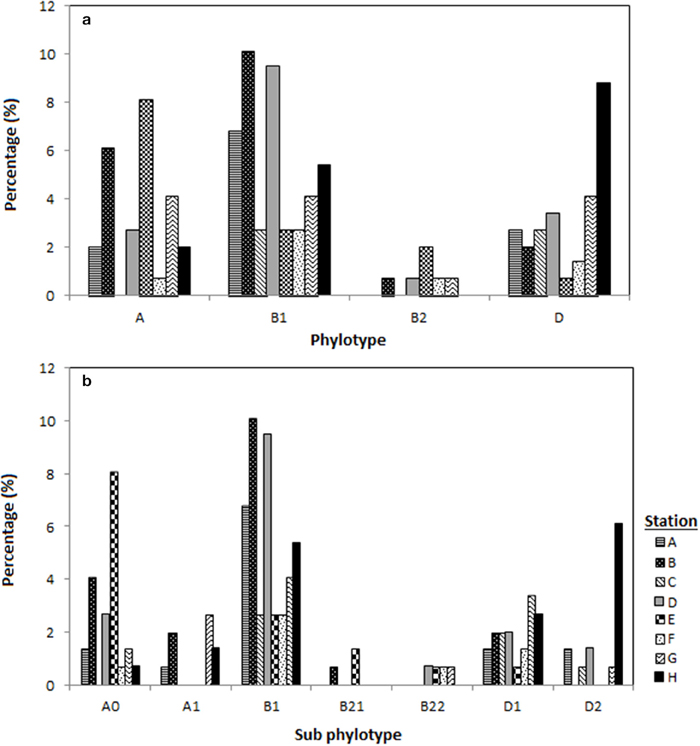
Figure 2. Distribution of the E. coli phylogenetic groups in eight different stations located in Sepetang River (A), Sangga Besar River (B–E) and Selinsing River (G and H) and coastal waters (F), (a) distribution of E. coli according to the respective phylogenetic groups (A, B1, B2, D) and (b) according to phylogroup subtyping (A0, A1, B1, B21, B22, D1, D2).
The genotypes of each phylogenetic group (A0/A1, B22/B23, and D1/D2) were not distributed homogenously among all stations. Genotype A0 comprised almost two thirds of phylogenetic group A and could be detected in almost all stations, while genotype A1 was only present in the upstream of Sangga Besar River (station A and B) and Selinsing River (station G and H). Genotype B23 was only detected from station D, E, F, and G which are all located downstream of a fishing village (Figure 2b).
None of the 148 isolates was EHEC, EPEC, ETEC, DAEC, or EIEC. However, two isolates originated from station D located upstream of Sangga Besar River and station G located upstream of Selinsing River were identified to be EAEC.
Among the 148 E. coli isolates from Matang mangrove estuaries, 128 (87%) were non-susceptible to at least one of 15 antibiotics tested (Table 2). Only 20 (14%) E. coli isolates were sensitive to all the 15 tested antibiotics. The antibiotic resistance percentages were: N (74%), S (31%), K (19%), CN (3%), AMC (11%), AMP (33%), CRO (1%), EFT (3%), ENR (22%), CIP (10%), OA (18%), NA (22%), TE (28%), C (16%), and SXT (19%) (Table 1 and Supplementary Table 2).
In comparisons among stations, Principle Component Analysis (PCA) showed that high antibiotic resistance among the E. coli isolates was detected at the stations (B and D) near to the fishing village (positive direction of PCA1, Figure 3). Most E. coli isolated from these stations were phylogenetic group B1 and carried class 1 integron. Moreover, high antibiotic resistances especially to aminoglycosides and beta-lactams were also detected in E. coli isolates from station F located on the cockle mud flat. On the other hand, low antibiotic resistance of E. coli isolates was detected at the stations with less anthropogenic influence (station H) and close to the fish cages (station E) (Table 1 and Figure 3). Overall, E. coli isolates from Matang mangrove estuaries demonstrated high prevalence of multi-drug resistance (MDR). One third of the tested E. coli isolates (34%) were found to be non-susceptible to three or more antibiotics (3R); and about one in every five isolates (20%) were non-susceptible to six or more antibiotics (6R) tested in this study (Table 1 and Supplementary Table 2). Further, four out of 148 isolates (3%) were non-susceptible to all of the 15 antibiotics examined in this work (Table 1 and Supplementary Table 2). Fifty out of 148 (34%) E. coli samples isolated from Matang mangrove estuaries demonstrated resistance to at least three antibiotic classes and 10% were non-susceptible to all the seven antibiotic classes tested (Table 1 and Supplementary Table 2).
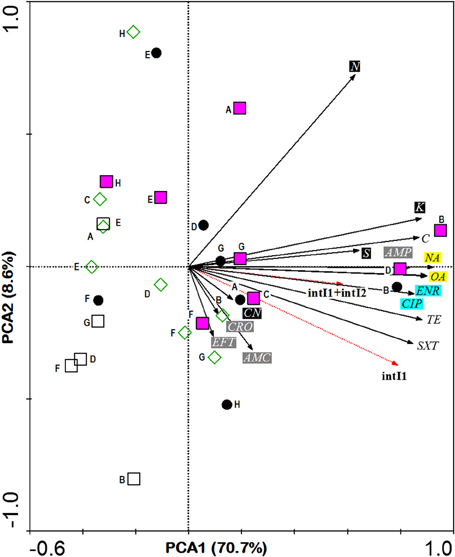
Figure 3. PCA ordination biplot showing the distribution of E. coli phylogenetic groups by sampling stations with respect to their resistance to various antibiotics and the integras genes they carried. The first (PCA1) and second (PCA2) principal components respectively explained 70.7 and 8.6% of the total variance of the explanatory variables. Phylogroups are denoted by various symbols: black circle A, pink square B1, empty square B2 and empty diamond D. Bold letter beside symbol denotes sampling stations located in Sepetang River (A), Sangga Besar River (B–E), Selinsing River (G and H) and adjacent coastal water (F). Black solid arrows indicate isolates resistant to 15 antibiotics: Class I (black box), CN, aminoglycosidase: gentamicin; K, kanamycin; S, streptomycin; N, neomycin; Class II (gray box); AMC, β-lactams: amoxicillin/clavulanic acid; AMP, ampicillin; CRO, ceftriaxone; EFT, ceftiofur; Class III (blue box); ENR, fluoroquinolone: enrofloxacin; CIP, ciprofloxacin; Class IV (yellow box); NA, quinolones: nalidixic acid; OA, oxolinic acid; Class V; TE, tetracycline: tetracycline; Class VI; C, phenicols: chloramphenicol and Class VII; SXT, sulfamethoxazole/trimethoprim. Red dotted arrows indicate E. coli integron gene (s).
The analysis of the phylogenetic groups with respect to their antibiotic resistance indicated that all groups showed some level of resistance to antibiotics. However, E. coli isolates in A and B1 phylogenetic groups (positive direction of PCA1) generally shows increasing resistance to most antibiotics as compared to the B2 and D phylogenetic groups (negative direction of PCA1). The phylogenetic group A and B1 demonstrated higher non-susceptibility level toward beta-lactams, (fluoro) quinolones, tetracycline, phenicols, and sulfamethoxazole/trimethoprim. Additionally, multi-drug resistance phenotype was more frequently identified in groups A and B1 (Table 1 and Supplementary Table 2). Chi-square independence test found only significant association between phylogroups with enrofloxacin (ENR; χ2 = 12.27; df = 3; p = 0.007), oxolinic acid (OA; χ2 = 11.71; df = 3; p = 0.008) and nalidixic acid (NA; χ2 = 12.04; df = 3; p = 0.001), which are all (fluoro) quinolones.
All 148 E. coli isolates were screened for class I, II and III integrons. Class I and II integrons were detected in 31 (21%) and 4 (3%) isolates, respectively (Table 2). Class III integron was absent in the E. coli isolates in Matang mangrove estuaries. Four isolates (3%) harbored both class I and II integrons. Of the 31 intI1-positive isolates, only 7 (23%) harbored gene cassettes; while none of the intI2 positive isolates carried any gene cassette. The size of the gene cassette in class 1 integron ranged from 700 to 1200 bp. Only one out of 148 isolates was found to carry more than one gene cassette, and this particular isolate was obtained from downstream of the Selinsing River (station H). The DNA sequences of all class I integrons have been deposited in GenBank under accession nos. KR952338-KR952345.
Five types of the resistance gene cassettes were detected in this study (Table 2). Three types of dfrA (dfrA 1, dfrA 5, and dfrA 17) were identified, which encoded the dihydrofolate reductase enzyme, mediating resistance to trimethoprim. Additionally, two types of the aadA (aadA 1 and aadA 22) were detected in class 1 integron that confers resistance to spectinomycin and streptomycin. The aadA and dfrA genes can be present alone or in combination with each other and other resistance genes as only one cassette combination (dfrA1 + aadA1) was recovered in this study (Table 2). All E. coli isolates that carried the aadA and dfrA genes were resistant to aminoglycosides and SXT, respectively (Table 2).
Half of the intI positive E.coli belonged to group B1 (n = 15, 48%) followed by nine isolates (29%) in group A and six isolates (19%) in group D. Only one isolate in group B2 was detected as intI1 positive. This integron gene was closely associated with most of the antibiotic resistance. In the PCA biplot, the gradients of increasing number of IntI positive isolates were in the same direction as the gradients of antibiotic resistance (Figure 3). Resistance to all antibiotics (except N, AMC, CN, and CRO), i.e., 73.3% of the 15 tested antibiotics, was significantly correlated to intI1-positive isolates (p < 0.001), with correlation (Rn) values that ranged from 0.50 (S) to 0.81 (SXT) (Supplementary Table 3). Of the eleven antibiotic resistant isolates that were intI1-positive, seven were also significantly correlated to intI2-positive isolates (p < 0.03), with Rn values that ranged from 0.49 (TE) to 0.73 (C). Among four intI2 positive isolates, three and one isolates belonged to phylogenetic group A and B1, respectively. Moreover, four out of seven isolates (57%) harbored gene cassette class 1 belonged to phylogenetic group D (Table 2).
All the confirmed 148 E. coli isolates that were recovered in October 2011 and May 2012 were subjected to REP-PCR. REP-PCR was carried out to determine the diversity of E. coli using UPGMA (unweighted pair-group method using arithmetic averages). REP-PCR subtyped the 148 E. coli isolates into 118 different REP profiles comprising of 200 to 2000 bp DNA fragments. At 80% similarity, the coefficient of similarity (F-value) ranged from 0.69 to 1.0 and the 148 isolates were clustered into seven groups (Supplementary Table 4). Cluster I and V included 76 and 27 E. coli isolates from different stations, respectively. Another 45 E. coli isolates were grouped into five clusters with each comprising of two to 18 isolates. Most of resistant isolates including MDR phenotypes were grouped into cluster, I, V, and III of the REP-PCR profile (Supplementary Table 4).
In the present study, the Matang mangrove estuaries harbor a high prevalence of antibiotic resistant E. coli in surface waters and sediments., the results indicated that four in every five E. coli isolates from Matang estuaries were non-susceptible to one or more antibiotics tested (1R); and one out of four 1R Isolates was non-susceptible to six or more antibiotics tested (Table 1). About one-third of the E. coli isolated from Matang estuaries were MDR and this figure is relatively higher compared to Tagus estuary in Portugal, in which 19% of the E. coli isolates were MDR (Pereira et al., 2013). However, the antibiotic resistant level was lower compared to the 88% found in Dongjiang River, China (Su et al., 2012). Laroche et al. (2009) also reported a relatively high level of MDR E. coli (39%) in Seine River, France. The high level of MDR E. coli observed in all of these studies from different parts of the world, including the current work highlighted the potential risk of dissemination of antibiotic resistance traits via the aquatic system.
PCA analysis showed that the prevalence of antibiotic resistance in E. coli was related to the sampling stations, in which antibiotic resistant E. coli was more frequently isolated from the upstream of both Sangga Besar and Selinsing Rivers. Station B was one of the stations with high numbers of MDR E. coli isolates recovered. This is likely due to its proximity to Kuala Sepetang fishing village where there is open sewage disposal and poor sanitation (Ghaderpour et al., 2014). Majority of E. coli isolates that demonstrated 100% non-susceptibility to all 15 antibiotics tested were isolated from station B. Also, E. coli isolates that showed non-susceptibility toward aminoglycosides and beta-lactams were more frequently isolated from stations located downstream of Kuala Sepetang or between the latter and Sangga Besar fishing village at the river mouth (Table 1). The findings suggested anthropogenic sources as the major contributor to the presence of antibiotic resistant E. coli in Matang mangrove estuaries. As the results show, beta-lactams resistant E. coli (i.e., non-susceptible to amocixillin-clavulanic acid, ampicillin, ceftriazone, or ceftiofur) was least frequently isolated from station H, but more frequently isolated from station F (75%) and station B (57%) located downstream of the villages. The highest antibiotic resistance prevalence was detected for neomycin (74%), ampicillin (33%), streptomycin (31%), and tetracycline (28%). The major antibiotic resistance found among E. coli isolates was in agreement with previous studies in aquatic environments (Hamelin et al., 2007; Laroche et al., 2009; Pereira et al., 2013). This resistance may reflect the abuse of antibiotics in human and veterinary medicine and aquaculture farms in the study area. The direct discharge of untreated sewage from the villages into the estuarine system, active fish and shrimp farming, as well as the presence of upstream swine farms are the main contributing factors to the high prevalence of MDR E. coli in Matang estuaries. Aquaculture could be a significant contributor to antibiotic resistance since the use of antibiotics, pesticides, and other chemotherapeutic agents in feed additives and treatment baths is as yet unregulated in Malaysia (Majusha et al., 2005; Ibrahim et al., 2010). Sulfonamides, tetracyclines, oxytetracycline, ampicillin, chloramphenicol, and quinoline are common antibiotics used in aquaculture farms in Perak as well as in the country (Mohamed et al., 2000; Keh, 2003). Nevertheless, reservoir sources of E.coli may include shorebirds and other feral animals such as monkeys, flying foxes and bats, as well as domestic animals, especially dogs (Frenzel and Couvillion, 2002).
It is interesting to note that even though the E. coli count at station F was low as reported by Ghaderpour et al. (2014), which resulted in lower isolation rate, a high level of antibiotic resistance was found in E. coli isolates from station F (Table 1 and Figure 2). A high bacteria count is expected at the river mouth due to bacterial contamination from the upstream, but due to the fact that E. coli is sensitive to high salinity (Gao et al., 2013), at station F, most of the E. coli would have been inactivated except antibiotic resistant strains that are also tolerant to high salinity. On the other hand, low antibiotic resistance was found in station H, downstream of Selinsing River, despite the high count of E. coli reported previously (Ghaderpour et al., 2014). Since station H is far away from obvious anthropogenic sources, its high count of E. coli could originate from wildlife. It has been shown in other studies that the prevalence of antibiotic resistance was low in wild animals compared to human and agricultural sources (Sayah et al., 2005; Edge and Hill, 2007).
Integron is an important factor in developing MDR resistance phenotypes among E. coli and other bacterial species since they are able to transfer different resistance genes simultaneously (Martinez, 2009). In the present study, 21% of E. coli isolates were integron positive. It was higher than those values reported in previous studies, 11% in France and Portugal (Laroche et al., 2009; Pereira et al., 2013), and 15% in Czech Republic (Dolejská et al., 2009). The frequency of the class 2 integron (3%) among E. coli isolates in Matang estuaries concurred with earlier studies in estuary (Laroche et al., 2009), in animal and human E. coli isolates (Kang et al., 2005; Skurnik et al., 2005; Cocchi et al., 2007). No class 3 integron was found in this study since integron class 3 is rare even among the human and animal E. coli. This finding is consistent with other reports (Laroche et al., 2009; Su et al., 2012; Pereira et al., 2013).
Class Integron 1 can be considered as a marker for antibiotic resistance even without the presence of the gene cassette as they are usually located on the transposons or plasmids that carry other resistance genes (Leverstein-van Hall et al., 2003; Mokracka et al., 2011). This might explain why 90% of the integron class 1 positive isolates were found as multi-drug resistance (Tables 1, 2).
Among the 31 integron positive E. coli isolates, only seven (22%) were detected carrying gene cassettes. All the gene cassettes were detected in class 1 integron positive E. coli isolates. Five different gene cassettes were found in this study (Table 2), and these have been previously reported among clinical E. coli (Lim et al., 2009; Ho et al., 2012; Su et al., 2012; Koczura et al., 2013). It is interesting to note that the gene cassettes found in class 1 integron among E. coli isolates in Matang estuaries were also reported in E. coli isolates from swine (Lapierre et al., 2010), poultry (Soufi et al., 2009), and humans (Lim et al., 2009; Ho et al., 2012).
The E. coli phylogenetic group B1 was the most prevalent (44%) in Matang mangrove estuaries, followed by phylogenetic groups A and D. Phylogenetic groups A and B1 comprised more than half (70%) of the total E. coli isolated from Matang mangrove estuaries. MLST genotyping has grouped phylogenetic groups A and B1 together, implying groups A and B1 to be sister groups (Gordon et al., 2008). Commensal E. coli was found to be predominantly in phylogenetic groups A and B1; whilst extraintestinal E. coli strains usually belong to groups B2 and D (Bingen et al., 1998; Picard et al., 1999; Johnson and Stell, 2000). Other studies have also reported that groups A and B1 were usually less pathogenic compared to groups B2 and D (Bingen et al., 1998; Picard et al., 1999; Johnson et al., 2001). Strains of A and B1 groups were recovered more frequently from aquatic environments than group B2 and D strains (Figueira et al., 2011). Walk et al. (2007) demonstrated that the majority of the E. coli strains that are able to persist in the environment belong to the B1 phylogenetic group. The findings from the present study are in agreement with previous studies in that A and B1 groups are predominant in the aquatic system, although phylogenetic group B1 comprised the most frequently isolated group in the present study. However, the distribution of these common phylogenetic groups did not hold true at station H, where phylogenetic group D was predomiant. Although, many studies have demonstrated groups B2 and D are usually more virulent (Bingen et al., 1998; Picard et al., 1999; Johnson et al., 2001), E. coli O157:H7 and other EHEC are in group D (Bidet et al., 2005), where cattle and birds (Escobar-Paìramo et al., 2006) are the main reservoirs. The findings obtained from the present study are realistic since station H was less influenced by human activity compared to the other stations where human activity was the main factor affecting the quality of the aquatic environment. Station H was least impacted by human activity where wild animals are thought to be the main source of E. coli at station H. However, due to the fact that pathogenic strains usually belong to group D, fecal contamination by feral animals can present a potential risk of pathogenic E. coli. Therefore, the identification of this kind of fecal contamination is necessary for appropriate management practices and remediation strategies to improve sustainable water resources.
Low prevalence of phylogenetic group B2 was found in our study which is in agreement with previous studies (Pereira et al., 2013). These isolates were detected in stations that were influenced more by human activities. It may show that this group is more associated with anthropogenic contamination as has been previously reported (Carlos et al., 2010). However, the percentage of E. coli in each group differs according to the sites of infection, geographical location, and level of antibiotic resistance (Duriez et al., 2001; Bukh et al., 2009). Carlos et al. (2010) reported that genotype B23 was found only in human feces and could be a good indicator for human fecal contamination in water. In the present study, genotype B23 was only detected from stations D, E, F, and G which were all located downstream of the fishing villages.
Many studies have reported the association of E. coli phylogenetic groups to their virulence, antibiotic resistance, and antibiotic resistant genes (Skurnik et al., 2005; Rijavec et al., 2006; Bukh et al., 2009; Garcia-Aljaro et al., 2009; Mataseje et al., 2009; Pereira et al., 2013; Mosquito et al., 2015). However, variable observations were reported from different studies. For instance, Mosquito et al. (2015) and Bukh et al. (2009) both reported that strains belonging to phylogenetic group D presented significantly higher percentages of multidrug resistance than the rest of the groups; while Skurnik et al. (2005), Rijavec et al. (2006), Garcia-Aljaro et al. (2009), Mataseje et al. (2009) and Pereira et al. (2013) had all found antibiotic resistance to be lower in E. coli phylogenetic group D and B2. In this study, we observed a lower percentage of antibiotic resistance in phylogenetic groups B2 (0%) and D (8%) as compared to group A (29%) and B1 (23%) (Table 1). However, the variation in observations from different studies could be due to the different origin of E. coli. Both studies (Bukh et al., 2009; Mosquito et al., 2015) reported an association of multidrug resistance with E. coli phylogenetic group D was of clinical origins but not environmental. Therefore, in our study, the higher percentage of multidrug resistance observed in group A and B1 was plausible. However, both E. coli phylogenetic group B2 and D demonstrated higher percentage of non-susceptibility to aminoglycosides, but not quinolones/fluoroquinolones. On the other hand, group A and B1 appear to be associated with resistance toward aminoglycosides and (fluoro) quinolones (Table 1 and Figure 3). The low non-susceptibility percentage in group D is out of our expectation as most studies have reported on its high resistance toward (fluoro) quinolones (Sabaté et al., 2008; Bukh et al., 2009; Mosquito et al., 2015). Probably, a study into the origins of E. coli phylogenetic group D isolated in this study will yield an explanation for this variation.
Our present finding also indicated that two isolates (1%) were pathogenic and identified as EAEC. Both EAEC isolates (SD4 and WGO3) belonged to phylogenetic group D and they were grouped in different clusters (Supplementary Table 4). Low percentage of the pathotypes in aquatic systems was also reported in a previous study (Hamelin et al., 2007). EAEC is known as an emergent pathogen causing diarrhea in developing countries and gastroenteritis outbreaks in industrialized countries, especially among children (Keskimäki et al., 2000; Villaseca et al., 2005). In Japan in 1993, a massive outbreak due to EAEC infection occurred among children (Keskimäki et al., 2000).
The source of EAEC remains unknown. The isolated EAEC were from stations D and G, and likely came from municipal wastewater as a potential source of pathogenic E. coli. The contaminated water may contaminate aquaculture products, while it is conceivable that their consumption could cause foodborne illness in humans. Water and food have been reported as the possible sources of EAEC infection in developing counties (Villaseca et al., 2005). This risk could become more significant if such bacteria are resistant to antibiotics.
Both EAEC isolates, SD4 and WGO3, contains class 1 integron with gene cassettes aadA1 (1000 bp) and dfrA5 (700 bp), respectively. The same gene cassettes with same length have been reported among clinical isolates of E. coli in the stools of infected persons in Malaysia (Lim et al., 2009; Ho et al., 2012). In addition, both EAEC isolates were MDR but were sensitive to fluoroquinolones. Fluoroquinolones has been found to be negatively associated with the presence of the virulence factors among the clinical E. coli strain (Johnson et al., 2003; Kawamura-Sato et al., 2010), but has not been observed among isolates from animals (Johnson et al., 2003; Kawamura-Sato et al., 2010). This may indicate that E. coli population in Matang estuaries is contributed by different sources of contamination.
REP-PCR is known as a fast and discriminatory technique to characterize the genomic diversity of E. coli isolates (Rademaker et al., 2000). Although the present results indicate a high diversity of E. coli isolates, the cluster analysis showed that the 148 isolates could be grouped into seven clusters at 80% similarity.
Overall, 76 (51%) and 27 (18%) E. coli isolates from eight different sampling sites were grouped into cluster I and V, respectively. The close similarity between some of the E. coli isolates over large spatial scale reflects both the hydrodynamics and interconnected waterways that characterize the Matang estuarine system. However, the diverse and variable nature of the E. coli population could be due to their survival rate, growth and inputs from different sources in the aquatic environment and, which differ according to the strain characteristic (Anderson et al., 2006; Kon et al., 2007, 2009). This explains why eight E. coli isolates in station F were grouped into four different clusters in the present study, while diversity was less in station C (n = 8) with two clusters, and station D (n = 24) with three clusters.
Although Selinsing River (stations G and H) was less used for aquaculture production compared to Sangga Besar River, the diversity of E. coli isolates was slightly higher. Besides the prolonged persistence and adaptation of E. coli in the water, heterogeneity among isolates could be due to erosion of the river banks and the presence of more wildlife that act as vectors of E. coli transmission (Solo-Gabriele et al., 2000; Kon et al., 2009). The high diversity was also found at stations A, B, and E as these stations were located next to human settlements and fish cage farms (E). However, there is no a single source that contributes to E.coli diversity and multiple sources are more likely (Kon et al., 2009).
Several studies reported different sources of E. coli isolates such as bird droppings (Edge and Hill, 2005), wildlife (Somarelli et al., 2007), and humans (Ram et al., 2004). However, it was not the main aim of this study to trace the sources of E. coli in the estuarine system. Perhaps, future work should compare the REP-PCR of E. coli isolates with those from humans and known animals in order to determine the sources of E. coli isolates in the Matang area.
Our study shows a high prevalence of the MDR (34%) among E. coli isolated from Matang estuaries. Phylogenetic group B1 has high resistance to antibiotics, thus playing a major role in the transmission of antimicrobial resistance via the aquatic environment. In particular, a high level of antibiotic resistance was observed at the upstream of the estuaries, which could be attributable to the adjacent human settlement and fish farms. Resistance to aminoglycosides, beta-lactams and tetracycline were most frequently detected in this study. Presence of integrons (21%) is another interesting result reflecting the contaminated MMFR estuaries that could act as a reservoir or source of MDR E. coli for the transmission of antibiotic resistance to the environment.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This research was financially supported by University of Malaya High Impact Research Grant UM.C/625/1/HIR/MOHE/SC/20 (UM.S/P/HIR/ MOHE/24) and IOES HiCoE Research Grant (IOES-2014E) from Ministry of Education Malaysia. Six strains (2060-004, E2348/69, JM221, E9034A, C1845, EC-12) were included as positive control for E. coli pathotyping, a gift from Oscar G. Gomez-Duarte, International Enteric Vaccines Research Program [IEVRP], University of Iowa Children's Hospital, Iowa City, IA.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00977
Anderson, M. A., Whitlock, J. E., and Harwood, V. J. (2006). Diversity and distribution of Escherichia coli genotypes and antibiotic resistance phenotypes in feces of humans, cattle, and horses. Appl. Environ. Microbiol. 27, 6914–6922. doi: 10.1128/AEM.01029-06
Ariffin, R., and Nik Mustafa, N. M. S. (eds.). (2013). A Working Plan for the Matang Mangrove Forest Reserve, Perak. 6th Revision. Ipoh: Perak State Forestry Department.
Bergthorsson, U., and Ochman, H. (1998). Distribution of chromosome length variation in natural isolates of Escherichia coli. Mol. Biol. Evol. 15, 6–16. doi: 10.1093/oxfordjournals.molbev.a025847
Bidet, P., Mariani-Kurkdjian, P., Grimont, F., Brahimi, N., Courroux, C., Grimont, P., et al. (2005). Characterization of Escherichia coli O157: H7 isolates causing haemolytic uraemic syndrome in France. J. Med. Microbiol. 54, 71–75. doi: 10.1099/jmm.0.45841-0
Bingen, E., Picard, B., Brahimi, N., Mathy, S., Desjardins, P., Elion, J., et al. (1998). Phylogenetic analysis of Escherichia coli strains causing neonatal meningitis suggests horizontal genetransfer from a predominant pool of highly virulent B2 group strain. J. Infect. Dis. 177, 642–650. doi: 10.1086/514217
Bukh, A. S., Schønheyder, H. C., Emmersen, J. M. G., Søgaard, M., Bastholm, S., and Roslev, P. (2009). Escherichia coli phylogenetic groups are associated with site of infection and level of antibiotic resistance in community-acquired bacteraemia: a 10 year population-based study in Denmark. J. Antimicrob. Chemother. 64, 163–168. doi: 10.1093/jac/dkp156
Burridge, L., Judith, S. W., Felipe, C., Jaime, P., and Katherine, B. (2010). Chemical use in salmon aquaculture: a review of current practices and possible environmental effects. Aquaculture 306, 7–23. doi: 10.1016/j.aquaculture.2010.05.020
Cambray, G., Guerout, A., and Mazel, D. (2010). Integrons. Annu. Rev. Genet. 44, 141–166. doi: 10.1146/annurev-genet-102209-163504
Carattoli, A. (2003). Plasmid-mediated antimicrobial resistance in Salmonella enterica. Curr. Issues. Mol. Biol. 5, 113–122. Available online at: http://www.horizonpress.com/cimb/v/v5/10.pdf
Carlos, C., Pires, M. M., Stoppe, N. C., Hachich, E. M, Sato, M. I., Gomes, T. A. T., et al. (2010). Escherichia coli phylogenetic group determination and its application in the identification of the major animal source of fecal contamination. BMC Microbiol. 10:161. doi: 10.1186/1471-2180-10-161
Chong, V. C., Lee, P. K. Y., and Lau, C. M. (2010). Diversity, extinction risk and conservation of Malaysian fishes. J. Fish. Biol. 76, 2009–2066. doi: 10.1111/j.1095-8649.2010.02685.x
Clermont, O., Bonacorsi, S., and Bingen, E. (2000). Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66, 4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000
CLSI. (2012). Performance Standards for Antimicrobial Susceptibility Testing. CLSI Document M100-S22. Wayne, PA: Clinical and Laboratory Standards Institute.
Cocchi, S., Grasselli, E., Gutacker, M., Benagli, C., Convert, M., and Piffaretti, J. C. (2007). Distribution and characterization of integrons in Escherichia coli strains of animal and human origin. FEMS. Immunol. Med. Microbiol. 50, 126–132. doi: 10.1111/j.1574-695X.2007.00242.x
Dolejská, M., Bierosová, B., Kohoutová, L., Literak, I., and Cízek, A. (2009). Antibiotic resistant Salmonella and Escherichia coli isolates with integrons and extended spectrum beta-lactamases in surface water and sympatric black-headed gulls. J. Appl. Microbiol. 106, 1941–1950. doi: 10.1111/j.1365-2672.2009.04155.x
Duriez, P., Clermont, O., Bonacorsi, S., Bingen, E., Chaventre, A., Elion, J., et al. (2001). Commensal Escherichia coli isolates are phylogenetically distributed among geographically distincthuman populations. Microbiology 147, 1671–1676. doi: 10.1099/00221287-147-6-1671
Edge, T. A., and Hill, S. (2005). Occurrence of antibiotic resistance in Escherichia coli from surface waters and fecal pollution sources near Hamilton, Ontario. Can. J. Microbiol 51, 501–505. doi: 10.1139/w05-028
Edge, T. A., and Hill, S. (2007). Multiple lines of evidence to identify the source of fecal pollution at a fresh water beach in Hamilton Harbour, Lake Ontario. Water Res. 41, 3585–3594. doi: 10.1016/j.watres.2007.05.012
Escobar-Páramo, P., Clermont, O., Blanc-Potard, A. B., Bui, H., Le Bouguénec, C., and Denamur, E. (2004). A specific genetic back ground is required for acquisition and expression of virulence factors in Escherichia coli. Mol. Biol. Evol. 21, 1085–1094. doi: 10.1093/molbev/msh118
Escobar-Paìramo, P., Le Menac'h, A., Le Gall, T., Amorin, C., Gouriou, S., Picard, B., et al. (2006). Identification of forces shaping the commensal Escherichia coli genetic structure by comparing animal and human isolates. Environ. Microbiol. 8, 1975–1984. doi: 10.1111/j.1462-2920.2006.01077.x
Figueira, V., Serra, E., and Manaia, C. M. (2011). Differential patterns of antimicrobial resistance in population subsets of Escherichia coli isolated from waste and surface waters. Sci. Total. Environ. 409, 1017–1023. doi: 10.1016/j.scitotenv.2010.12.011
Frenzel, S. A., and Couvillion, C. S. (2002). Fecal-indicator bacteria in streams along a gradient ofresidential development. J. Am. Water. Resource. Ass. 38, 265–273. doi: 10.1111/j.1752-1688.2002.tb01550.x
Galvin, S., Boyle, F., Hickey, P., Vellinga, A., Morris, D., and Cormican, M. (2010). Enumeration and characterization of antimicrobial-resistant Escherichia coli bacteria in effluent from municipal, hospital, and secondary treatment facility sources. Appl. Environ. Microbiol. 76, 4772–4779. doi: 10.1128/AEM.02898-09
Gao, G., Falconer, R. A., and Lin, B. (2013). Modelling importance of sediment effects on fate and transport of enterococci in the Severn Estuary, UK. Mar. Pollut. Bull. 67, 45–54. doi: 10.1016/j.marpolbul.2012.12.002
Garcia-Aljaro, C., Moreno, E., Andreu, A., Prats, G., and Blanch, A. R. (2009). Phylogroups, virulence determinants and antimicrobial resistance in stx2 gene-carrying Escherichia coli isolated from aquatic environments. Res. Microbiol. 160, 585–591. doi: 10.1016/j.resmic.2009.08.004
Ghaderpour, A., KhairulNazrin, M. N., Chew, L. L., Chong, V. C., Thong, K. L., and Chai, L. C. (2014). Detection of multiple potentially pathogenic bacteria in Matang mangrove estuaries, Malaysia. Mar. Pollut. Bull. 8, 324–330. doi: 10.1016/j.marpolbul.2014.04.029
Gómez-Duarte, O. G., Bai, J., and Newel, E. (2009). Detection of E. coli, Salmonella spp., Shigella spp., Yersinia enterocolitica, Vibrio cholerae, and Campylobacter spp. enteropathogens by Three-reaction Multiplex PCR. Diagn. Microbiol. Infect. Dis. 63, 1–9. doi: 10.1016/j.diagmicrobio.2008.09.006
Gordon, D. M. (2004). “The influence of ecological factors on the distribution and genetic structure of Escherichia coli,” in Escherichia coli and Salmonella Typhimurium: Cellular and Molecular Biology, ed F. Neidhardt (Washington, DC: American Society for Microbiology).
Gordon, D. M., Clermont, O., Tolley, H., and Denamur, E. (2008). Assigning Escherichia coli strains to phylogenetic groups: multi-locus sequence typing versus the PCR triplex method. Environ. Microbiol. 10, 2484–2496. doi: 10.1111/j.1462-2920.2008.01669.x
Gordon, D. M., and Cowling, A. (2003). The distribution and genetic structure of Escherichia coli in Australian vertebrates: host and geographic effects. Microbiology 149, 3575–3586. doi: 10.1099/mic.0.26486-0
Hall, R. M., and Collis, C. M. (1995). Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol. Microbiol. 15, 593–600. doi: 10.1111/j.1365-2958.1995.tb02368.x
Hamelin, K., Bruant, G., El-Shaarawi, A., Hill, S., Edge, T. A., Fairbrother, J., et al. (2007). Occurrence of virulence and antimicrobial resistance genes in Escherichia coli isolates from different aquatic ecosystems within the St. Clair River and Detroit river areas. Appl. Environ. Microbiol. 73, 477–484. doi: 10.1128/AEM.01445-06
Ho, W. S., Balan, G., Puthucheary, S., Kong, B. H., Lim, K. T., Tan, L. K., et al. (2012). Prevalence and characterization of multidrug-resistant and extended-spectrum beta-lactamase-producing Escherichia coli from pediatric wards of a Malaysian hospital. Microb. Drug. Resist. 18, 408–416. doi: 10.1089/mdr.2011.0222
Ibrahim, A. B., Mohd khan, A., Ayob, M. Y., and Norrakiah, A. S. (2010). Pesticide and antibiotic residues in freshwater aquaculture fish: Chemical risk assessment from farm to table. Asian J. Food Agro Indus. 3, 328–334.
Jaime, R., Carmen, G. F., and Paola, N. (2012). “Antibiotics in aquaculture use, abuse and alternatives,” in Health and Environment in Aquaculture, ed E. Carvalho, ISBN: 978-953-51-0497-1 (InTech). Available online at: http://www.intechopen.com/books/health-and-environment-in-aquaculture/antibiotics-in-aquaculture-use-abuse-and-alternatives
Johnson, J. R., Delavari, P., Kuskowski, M., and Stell, A. L. (2001). Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J. Infect. Dis. 183, 78–88. doi: 10.1086/317656
Johnson, J. R., Kuskowski, M. A., Owens, K., Gajewski, A., and Winokur, P. L. (2003). Phylogenetic origin and virulence genotype inrelation to resistance to fluoroquinolones and/or extended spectrum cephalosporins and cephamycins among Escherichia coli isolates from animals and humans. J. Infect. Dis. 188, 759–768. doi: 10.1086/377455
Johnson, J. R., and Stell, A. L. (2000). Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181, 261–272. doi: 10.1086/315217
Kang, H. Y., Jeong, Y. S., Oh, J. Y., Tae, S. H., Choi, C. H., Moon, D. C., et al. (2005). Characterization of antimicrobial resistance and class 1 integrons found in Escherichia coli isolates from humans and animals in Korea. J. Antimicrob. Chemother. 55, 639–644. doi: 10.1093/jac/dki076
Kawamura-Sato, K., Yoshida, R., Shibayama, K., and Ohta, M. (2010). Virulence genes, quinolone and fluoroquinolone resistance, and phylogenetic background of uropathogenic Escherichia coli strains isolated in Japan. Jpn. J. Infect. Dis. 63, 113–115. Available online at: http://www.nih.go.jp/niid/JJID/63/113.pdf
Keh, C. L. (2003). Survey of the Use of Drugs and Chemicals in Aquaculture in Malaysia. M.Sc. thesis, University of Malaya. 71.
Keskimäki, M., Mattila, L., Peltola, H., and Siitonen, A. (2000). Prevalence of diarrheagenic Escherichia coli in Finns with or without diarrhea during a round-the-world trip. J. Clin. Microbiol. 38, 4425–4429. Available online at: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC87616/
Koczura, R., Mokracka, J., Barczak, A., Krysiak, N., and Kaznowski, A. (2013). Association between the presence of class 1 integrons, virulence genes, and phylogenetic groups of Escherichia coli isolates from river water. Microb. Ecol. 65, 84–90. doi: 10.1007/s00248-012-0101-3
Kon, T., Weir, S. C., Howell, T. E., Lee, H., and Trevors, T. (2009). Repetitive element (REP)-polymerase chain reaction (PCR) analysis of Escherichia coli isolates from recreational waters of southeastern Lake Huron, Can. J. Microbiol. 55, 269–276. doi: 10.1139/W08-123
Kon, T., Weir, S. C., Howell, E. T., Lee, H., and Trevor, J. T. (2007). Genetic Relatedness of Escherichia coli isolates in interstitial water from Lake Huron (Canada) Beach. Environ. Microbiol. 73, 1961–1967. doi: 10.1128/AEM.02437-06
Lapierre, L., Martín, B. S., Araya-Jordán, C., and Borie, C. (2010). Comparison of integron linked antibiotic resistance genes in strains of Salmonella spp. isolated from swine in Chile in 2005 and 2008. Can. J. Microbiol. 56, 515–521. doi: 10.1139/W10-033
Laroche, E., Pawlak, B., Berthe, T., Skurnik, D., and Petit, F. (2009). Occurrence of antibiotic resistance andclass 1, 2 and 3 integrons in Escherichia coli isolated from a densely populated estuary (Seine, France). FEMS. Microbiol. Ecol. 68, 118–130. doi: 10.1111/j.1574-6941.2009.00655.x
Leverstein-van Hall, M. A., Blok, H. E. M., Rogier, A., Donders, T., Paauw, A., Fluit, A. C., et al. (2003). Multidrug resistance among Enterobacteriaceae is strongly associated with the presence of integronsand is independent of species or isolate origin. J. Infect. Dis. 187, 251–259. doi: 10.1086/345880
Lim, H. F., and Mohamad Parit, M. (2001). Sustainable Development of the Matang Mangroves and Its Impacts on Local Socio-economic Livelihood. Intensification of Research in Priority Areas Report, Government of Malaysia.
Lim, K. T., Yasin, R., Yeo, C. C., Puthucheary, S., and Thong, K. L. (2009). Characterization of multidrug resistant ESBL-producing Escherichia coli isolates from hospitals in Malaysia. J. Biomed. Biotechnol. 2009:165637. doi: 10.1155/2009/165637
Majusha, S., Sarita, G. B., Elyas, K. K., and Chandrasekaran, M. (2005). Multiple antibiotic resistances of vibrio isolates from coastal and brackish water areas. Am. J. Biochem. Biotechnol. 1, 201–206. doi: 10.3844/ajbbsp.2005.201.206
Martinez, J. L. (2009). Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ. Pollut. 157, 2893–2902. doi: 10.1016/j.envpol.2009.05.051
Mataseje, L. F., Neumann, N., Crago, B., Baudry, P., Zhanel, G. G., Louie, M., et al. (2009). ARO water study group characterization of cefoxitin-resistant Escherichia coli isolates fromrecreational beaches and private drinking water in Canada between 2004 and 2006. Antimicrob. Agents. Ch. 53, 3126–3130. doi: 10.1128/AAC.01353-08
Maynard, C., Bekal, S., Sanschagrin, F., Levesque, R. C., Brousseau, R., Masson, L., et al. (2004). Heterogeneity among virulence and antimicrobial resistance gene profiles of extraintestinal Escherichia coli isolates of animal and human origin. J. Clin. Microbiol. 42, 5444–5452. doi: 10.1128/JCM.42.12.5444-5452.2004
Mohamed, S., Nagaraj, G., Chua, F. H. C., and Wang, Y. G. (2000). “The use of chemicals in aquaculture in Malaysia and Singapore,” in Use of Chemicals in Aquaculture in Asia: Proceedings of the Meeting on the Use of Chemicals in Aquaculture in Asia 20-22 May 1996, Tigbauan, Iloilo, Philippines, eds J. R. Arthur, C. R. Lavilla-Pitogo, and R. P. Subasinghe (Tigbauan: Aquaculture Department, Southeast Asian Fisheries Development Center), 127–140. Available online at: http://hdl.handle.net/10862/601
Mokracka, J., Koczura, R., Pawłowski, K., and Kaznowski, A. (2011). Resistance patterns and integron cassette arrays of Enterobacter cloacae complex strains of human origin. J. Med. Microbiol. 60, 737–743. doi: 10.1099/jmm.0.027979-0
Mosquito, S., Pons, M. J., Riveros, M., Ruis, J., and Ochoa, T. J. (2015). Diarrheagenic Escherichia coli phylogroups are associated with antibiotic resistance and duration of diarrheal episode. Sci. World J. 2015:610403. doi: 10.1155/2015/610403
Pereira, A., Santos, A., Tacão, M., Alves, A., Henriques, I., and Correia, A. (2013). Genetic diversity and antimicrobial resistance of Escherichia coli from Tagus estuary (Portugal). Sci. Total. Environ. 461, 65–71. doi: 10.1016/j.scitotenv.2013.04.067
Picard, B., Garcia, J. S., Gouriou, S., Duriez, P., Brahimi, N., Bingen, E., et al. (1999). The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect. Immun. 67, 546–553.
Pruden, A., Pei, R., Storteboom, H., and Carlson, K. H. (2006). Antibiotic resistance genesas emerging contaminants: studies in northern Colorado. Environ. Sci. Technol. 40, 7445–7450. doi: 10.1021/es060413l
Rademaker, J. L. W., Hoste, B., Louws, F. J., Kersters, K., Swings, J., Vauterin, L., et al. (2000). Comparison of AFLPand REP-PCR genomic fingerprinting with DNA-DNA homologystudies: Xanthomonas as a model system. Int. J. Sys. Evol. Micr. 50, 665–677. doi: 10.1099/00207713-50-2-665
Ram, J. L., Ritchie, R. P., Fang, J., Gonzales, F. S., and Selegean, J. P. (2004). Sequence based source tracking of E. coli based on genetic diversity of beta glucuronidase. J. Environ. Q. 33, 1024–1032. doi: 10.2134/jeq2004.1024
Recchia, G. D., and Hall, R. M. (1995). Gene cassettes: a new class of mobile element. Microbiol-UK 141, 3015–3027. doi: 10.1099/13500872-141-12-3015
Rijavec, M., Starcic-Erjavec, M., Ambrozic-Avgustin, J., Reissbrodt, R., Fruth, A., Krizan-Hergouth, V., et al. (2006). High prevalence of multidrug resistance and random distribution of mobile genetic elements among uropathogenic Escherichia coli (UPEC) of the four major phylogenetic groups. Curr. Microbiol. 53, 158–162. doi: 10.1007/s00284-005-0501-4
Sabaté, M., Prats, G., Moreno, E., Ballest,é, E., Blanch, A. R., and Andreu, A. (2008). Virulence and antimicrobial resistance profiles among Escherichia coli strains isolated from human and animal wastewater. Res. Microbiol. 159, 288–293. doi: 10.1016/j.resmic.2008.02.001
Sayah, R. S., Kaneene, J. B., Johnson, Y., and Miller, R. (2005). Patterns ofantimicrobial resistance observed in Escherichia coli isolates obtained from domestic- and wild-animal fecal samples, human septage, and surface water. Appl. Environ. Microbiol. 71, 1394–1404. doi: 10.1128/AEM.71.3.1394-1404.2005
Skurnik, D., Le Menac'h, A., Zurakowski, D., Mazel, D., Courvalin, P., Denamur, E., et al. (2005). Integron associated antibiotic resistance and phylogenetic grouping of Escherichia coli isolates from healthy subjects free of recent antibiotic exposure. Antimicrob. Agents. Ch. 49, 3062–3065. doi: 10.1128/AAC.49.7.3062-3065.2005
Solo-Gabriele, H. M., Wolfert, M. A., Desmarais, T. R., and Palmer, C. J. (2000). Sources of Escherichia coli in a coastal subtropical environment. Appl. Environ. Microbiol. 66, 230–237. doi: 10.1128/AEM.66.1.230-237.2000
Somarelli, J. A., Makarewicz, J. C., Sia, R., and Simon, R. (2007). Wildlife identified as major source of E. coli in agriculturally dominated watersheds by BOX- AIR-derived genetic fingerprints. J. Environ. Manage. 82, 60–65. doi: 10.1016/j.jenvman.2005.12.013
Soufi, L., Abbassi, M. S., Saenz, Y., Vinue, L., Somalo, S., Zarazaga, M., et al. (2009). Torres C. Prevalence and diversity of integrons and associated resistance genes in Escherichia coli isolates from poultry meat in Tunisia. Foodborne. Pathoge. Dis. 6, 1067–1073. doi: 10.1089/fpd.2009.0284
Su, H. C., Ying, G. G., Tao, R., Zhang, Q. R., Zhao, L. J., and Liu, S. Y. (2012). Class 1 and 2 integrons, sul resistance genes and antibiotic resistance in Escherichia coli isolated from Dongjiang River, South China. Environ. Pollut. 169, 42–49. doi: 10.1016/j.envpol.2012.05.007
Ter Braak, C. J. F., and Smilauer, P. (2002). CANOCO Reference Manual and CanoDraw for Windows User's Guide: Software for Canonical Community Ordination (Version 4.5). Ithaca, NY: Microcomputer Power. 500.
Vignaroli, C., Luna, G. M., Rinaldi, C., Di Cesare, A., Danovaroand, R., and Biavasco, F. (2012). New sequence types and multidrug resistance among pathogenic escherichia coli isolates from coastal marine sediments. Appl. Environ. Microbiol. 78, 3916–3922. doi: 10.1128/AEM.07820-11
Villaseca, J. M., Hernández, U., Sainz-Espuñes, T. R., Rosario, C., and Eslava, C. (2005). Enteroaggregative Escherichia coli an emergent pathogen with different virulence properties. Rev. Latinoam. Microbiol. 47, 140–159. Available online at: http://new.medigraphic.com/cgi-bin/resumen.cgi?IDREVISTA=23&IDARTICULO=5182IDPUBLICACION=642
Walk, S. T., Alm, E. W., Calhoun, L. M., Mladonicky, J. M., and Whittam, T. S. (2007). Genetic diversity and population structure of Escherichia coli isolated from freshwater beaches. Environ. Microbiol. 9, 2274–2288. doi: 10.1111/j.1462-2920.2007.01341.x
Yu, X. K., and Thong, K. L. (2009). Multiplex PCR for simultaneous detection of virulence genes in Escherichia coli. Malays. J. Med. Sci. 28, 1–14. Available online at: http://eprints.um.edu.my/5657/1/Multiplex_PCR_for_simultaneous_detection_of_virulence_Genes_in_Escherichia_coli.pdf
Keywords: E. coli, Matang mangrove estuaries, antibiotic resistance, phylogenetic groups
Citation: Ghaderpour A, Ho WS, Chew L-L, Bong CW, Chong VC, Thong K-L and Chai LC (2015) Diverse and abundant multi-drug resistant E. coli in Matang mangrove estuaries, Malaysia. Front. Microbiol. 6:977. doi: 10.3389/fmicb.2015.00977
Received: 10 April 2015; Accepted: 02 September 2015;
Published: 29 September 2015.
Edited by:
Rustam Aminov, Technical University of Denmark, DenmarkReviewed by:
Veljo Kisand, University of Tartu, EstoniaCopyright © 2015 Ghaderpour, Ho, Chew, Bong, Chong, Thong and Chai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lay Ching Chai, Faculty of Science, Institute of Biological Sciences, University of Malaya, 50603 Kuala Lumpur, Malaysia,bGNjaGFpQHVtLmVkdS5teQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.