- 1Department of Biology and Biotechnology “Charles Darwin”, Sapienza University of Rome, Rome, Italy
- 2Department of Sciences, Universita degli Studi Roma Tre, Rome, Italy
- 3Research Core Unit Metabolomics, Institute of Pharmacology, Hannover Medical School, Hannover, Germany
- 4Pasteur Institute – Cenci Bolognetti Foundation, Sapienza University of Rome, Rome, Italy
In Pseudomonas aeruginosa the Gac signaling system and the second messenger cyclic diguanylate (c-di-GMP) participate in the control of the switch between planktonic and biofilm lifestyles, by regulating the production of the two exopolysaccharides Pel and Psl. The Gac and c-di-GMP regulatory networks also coordinately promote the production of the pyoverdine siderophore, and the extracellular polysaccharides Pel and Psl have recently been found to mediate c-di-GMP-dependent regulation of pyoverdine genes. Here we demonstrate that Pel and Psl are also essential for Gac–mediated activation of pyoverdine production. A pel psl double mutant produces very low levels of pyoverdine and shows a marked reduction in the expression of the pyoverdine-dependent virulence factors exotoxin A and PrpL protease. While the exopolysaccharide-proficient parent strain forms multicellular planktonic aggregates in liquid cultures, the Pel and Psl-deficient mutant mainly grows as dispersed cells. Notably, artificially induced cell aggregation is able to restore pyoverdine-dependent gene expression in the pel psl mutant, in a way that appears to be independent of iron diffusion or siderophore signaling, as well as of recently described contact-dependent mechanosensitive systems. This study demonstrates that cell aggregation represents an important cue triggering the expression of pyoverdine-related genes in P. aeruginosa, suggesting a novel link between virulence gene expression, cell–cell interaction and the multicellular community lifestyle.
Introduction
Pseudomonas aeruginosa is a metabolically versatile Gram-negative bacterium and an opportunistic pathogen in cystic fibrosis (CF) and otherwise critical patients, causing both chronic and acute infections (Driscoll et al., 2007). The ability of P. aeruginosa to rapidly adapt to diverse ecological niches and to switch from acute to chronic infections is related to the tightly regulated expression of specific sub-sets of genes in response to environmental cues (Stover et al., 2000). Characteristic traits of P. aeruginosa chronic infection are the microcolony and biofilm mode of growth (Parsek and Singh, 2003). Microcolonies are small aggregates of cells that open the way to the communal organization of biofilms, which are surface-associated communities of bacteria encased in a self-generated polymeric matrix (Zhao et al., 2013). Extracellular polysaccharides represent a key component of the P. aeruginosa biofilm matrix, and are involved in surface attachment and cell–cell interaction (Ma et al., 2009).
Pseudomonas aeruginosa strains can produce three main exopolysaccharides, namely alginate, Pel, and Psl (Colvin et al., 2012). Alginate confers the typical mucoid phenotype to producing strains (Sherbrock-Cox et al., 1984), and plays a crucial role in CF lung colonization. Psl and Pel are normally produced by non-mucoid strains, and failure to produce Pel and/or Psl exopolysaccharides impairs biofilm formation in vitro (Colvin et al., 2012). The exopolysaccharide Psl consists of repeating pentamers of D-mannose, L-rhamnose, and D-glucose (Ma et al., 2007). The helical distribution of Psl on the cell surface promotes cell–cell and cell–surface interactions in microcolonies (Zhao et al., 2013). Psl also plays an essential role in the maintenance of the mature biofilm structure (Jackson et al., 2004; Ma et al., 2009). The exopolysaccharide Pel was originally identified by a transposon mutagenesis screening for the loss of surface pellicle formation in P. aeruginosa PA14 (Friedman and Kolter, 2004). Pel structure has not yet been determined, although carbohydrate analysis suggested a glucose-rich composition (Friedman and Kolter, 2004).
The production of the Pel and Psl exopolysaccharides is controlled by many regulatory networks at the level of transcription, translation, and biosynthesis (Goodman et al., 2004; Ventre et al., 2006; Lee et al., 2007; Sakuragi and Kolter, 2007; Gilbert et al., 2009). The Gac system and the signaling molecule cyclic diguanylate (c-di-GMP) are the best characterized regulatory networks involved in the regulation of pel and psl gene expression and, consequently, in the switch from the planktonic to the biofilm lifestyle. The Gac system relies on the sensor kinase GacS that, in response to a still unknown signal, activates the transcriptional regulator GacA, which in turn promotes the transcription of two small non-coding RNAs (sRNAs), RsmZ and RsmY (Brencic et al., 2009). These sRNAs bind to and sequester the mRNA binding protein RsmA, thereby inhibiting its activity as translational repressor (Heeb et al., 2006). While psl gene expression is directly repressed by RsmA at the translational level (Irie et al., 2010), there is no evidence of a direct effect of RsmA on the pel genes, although transcriptomic analysis showed that the active state of the Gac network (i.e., rsmA mutation) also promotes transcription of the pel operon (Brencic and Lory, 2009). The Gac system has recently been shown to positively affect the intracellular levels of the signaling molecule c-di-GMP, which also induces exopolysaccharides production (Moscoso et al., 2011, 2014; Irie et al., 2012; Frangipani et al., 2014). This second messenger regulates Pel production both at the transcriptional and post-transcriptional level, by inhibiting the activity of the transcriptional repressor FleQ (Hickman and Harwood, 2008), and activating the Pel biosynthesis protein PelD (Lee et al., 2007). Although the molecular mechanism has not been elucidated yet, high intracellular levels of c-di-GMP also increase the expression of psl genes (Hickman et al., 2005).
Besides exopolysaccharides production and biofilm formation, Gac and c-di-GMP also act in a concerted way to promote the expression of pyoverdine genes (Frangipani et al., 2014), and it has recently been reported that the exopolysaccharides Pel and Psl are important for c-di-GMP regulation of pyoverdine production (Chen et al., 2015). Pyoverdine is a green fluorescent siderophore which plays a prominent role in P. aeruginosa pathogenicity (Visca et al., 2007; Cornelis and Dingemans, 2013). Pyoverdine acts not only as a high-affinity iron scavenger, but also serves as a signal molecule to promote the expression of important P. aeruginosa virulence factors, via a cell–surface signaling cascade which involves the other membrane ferri-pyoverdine receptor FpvA and the cytoplasmic membrane-spanning antisigma factor FpvR, ultimately leading to the activation of the extracytoplasmic function (ECF) sigma factor PvdS (Lamont et al., 2002; Llamas et al., 2014). PvdS directs the transcription of almost thirty P. aeruginosa genes, including those involved in pyoverdine biosynthesis and transport, the gene for the extracellular protease PrpL and, indirectly, the exotoxin A gene toxA (Ochsner et al., 2002; Llamas et al., 2014). The dual function of pyoverdine in iron uptake and virulence renders this siderophore essential for P. aeruginosa infectivity, as demonstrated in different mouse models (Meyer et al., 1996; Takase et al., 2000; Imperi et al., 2013). As for any iron-uptake system, pyoverdine production needs to be promptly shut down when intracellular iron levels are sufficiently high. This iron-mediated control occurs through the ferric uptake regulator Fur, an iron-sensing transcriptional repressor which binds to its co-repressor Fe2+ and inhibits transcription of the sigma factor gene pvdS (Ochsner et al., 2002). Besides Fur-Fe2+ mediated repression, pyoverdine production was found to be influenced by a number of environmental signals and regulatory pathways, including oxygen and nutrient availability, cellular communication and oxidative stress (reviewed in Llamas et al., 2014).
In the present study we demonstrate that the extracellular polysaccharides Pel and Psl are also essential for Gac-mediated regulation of pyoverdine production, which appears strongly repressed in a pel psl double mutant. We also provide evidence that artificially induced cell aggregation is able to restore pyoverdine-dependent gene expression in the Pel and Psl-deficient mutant. Our findings support the hypothesis that cell aggregation, rather than polysaccharide production per se, is an important cue triggering production of pyoverdine and pyoverdine-controlled virulence factors in P. aeruginosa.
Materials and Methods
Bacterial Strains, Growth Conditions, and Plasmids
Bacterial strains and plasmids used in this study are listed in Supplementary Table S1. The iron-depleted complex medium TSBD (Ohman et al., 1980) or the M9 minimal medium supplemented with 20 mM sodium succinate (Sambrook et al., 1989) were used as iron-poor media, to which FeCl3 was added at the indicated concentrations when required. Virulence factor production and gene expression assays were performed on bacteria grown at 37°C in 96-well microtiter plates (250 μl of medium in each well) under static conditions, unless otherwise stated. When specified, TSBD medium was supplemented with L-arabinose, agar, phytagel, or sucrose (Sigma–Aldrich) at the indicated final concentrations. Polystyrene beads (Polybead® Microspheres 3.00 μm, Polysciences) were washed several times with sterile water and then resuspended in TSBD at the desired concentrations.
Generation of Deletion and Conditional Mutants
All the primers and restriction sites used for PCR and cloning are listed in Supplementary Table S2. Deletion mutants in pelABCD, pslABCD, fpvR, pilY1, or pilA were generated using specific derivatives of the pDM4 suicide plasmid (Supplementary Table S1). The fur conditional mutant was generated using a recently described strategy (Lo Sciuto et al., 2014), based on the mini-CTX1-mediated insertion of the fur coding sequence under the control of an arabinose-dependent promoter into a neutral site of the P. aeruginosa genome, followed by the in-frame deletion of the endogenous fur gene using the pDM4Δfur suicide plasmid (Supplementary Table S1), under permissive conditions (i.e., arabinose-containing media). Gene replacements and insertions were verified by PCR and DNA sequencing.
Growth and Pyoverdine Measurements
Growth was measured as the OD600 of appropriate dilutions of bacterial cultures in sterile growth medium. Pyoverdine production was measured as the OD405 of culture supernatants appropriately diluted in 0.1 M Tris-HCl (pH 8), and normalized to the OD600 of the corresponding cultures (Imperi et al., 2009). When indicated, pyoverdine production was normalized to the number of colony forming units/ml.
β-Galactosidase and c-di-GMP Assays
The β-galactosidase activity from P. aeruginosa cells carrying reporter plasmids (Supplementary Table S1) was determined spectrophotometrically using o-nitrophenyl-β-D-galactopyranoside as the substrate, normalized to the OD600 of the bacterial culture and expressed in Miller (1972) units. Intracellular c-di-GMP levels were determined by liquid chromatography coupled with tandem mass spectrometry as described (Spangler et al., 2010), and normalized to the corresponding cellular protein content determined using the DC protein assay kit (Bio-Rad) and bovine serum albumin as the standard.
Western Blot Analysis of ToxA and PrpL Enzymatic Assay
For detection of ToxA, 100-μl aliquots of culture supernatants were supplemented with 20 μl of 6× SDS-PAGE loading dye [375 mM Tris-HCl (pH 6.8), 9% SDS, 50% Glycerol, 0.03% Bromophenol blue]. To normalize the amount of secreted proteins to bacterial growth, the volume of supernatants loaded into SDS-PAGE gels was calculated according to the formula: loading volume (μl) = 10/OD600 of the corresponding bacterial culture. Proteins resolved by SDS-PAGE were electrotransferred onto a nitrocellulose filter (Hybond-C extra, Amersham), and probed for ToxA using a rabbit polyclonal anti-ToxA antibody (Sigma–Aldrich). Filters were developed with 5-bromo-4-chloro-3-indoyl-phosphate and nitro blue tetrazolium chloride reagents for colorimetric alkaline phosphatase detection (Promega).
PrpL activity was determined as previously described (Imperi et al., 2010), using the chromogenic substrate Chromozym PL (tosyl-Gly-Pro-Lys-p-nitroanilide; Sigma–Aldrich) which is specific for PrpL (protease IV) and is not cleaved by other P. aeruginosa proteases (Caballero et al., 2001). Briefly, 10 μl of culture supernatants were mixed with 10 μl of 2 mg/ml Chromozym PL and 180 μl of phosphate buffer (pH 7.0) in 96-well microtiter plates. The OD410 was read at 2 min intervals for 30 min in a Victor3V plate reader (Perkin-Elmer), and the change in optical density (ΔOD410) per minute was determined. PrpL activity units were determined as: (ΔOD410 × total assay volume/ml)/(sample volume × E410 × light path); where total assay volume was 200 μl, sample volume was 10 μl, the extinction coefficient (E) of the product (p-nitroaniline) at 410 nm was 9.75, and the light path was 0.35 cm. PrpL activity was then normalized to the OD600 of the corresponding bacterial culture.
Quantitative Real-Time Reverse-Transcription PCR (qRT-PCR)
Total RNA was purified by using RNeasy minicolumns, treated with DNase and re-purified with the RNeasy MinElute cleanup kit (Qiagen). cDNA was reverse transcribed from 0.5 μg of total RNA with PrimeScript RT reagent Kit (TaKaRa). cDNA was then used as the template for qRT-PCR in a 7300 Real-Time PCR System (Applied Biosystems) using SYBR green with the ROX detection system (Bio-Rad). The primers used for qRT-PCR are listed in Supplementary Table S2. At least three wells were run for each cDNA sample. Relative gene expression with respect to the housekeeping gene rpsL was calculated using the 2-ΔΔCt method (Livak and Schmittgen, 2001).
Confocal Microscopy
Thirty microliter of P. aeruginosa cultures in TSBD or M9 were spotted on a glass slide that was freshly coated with 0.5% agarose in water, and covered with a cover slip. Images were acquired with a Leica TCS SP5 inverted confocal microscope equipped with a HCX PLAPO lamba blue 40X/1.25 OIL objective (Zeiss). Images were recorded using specific sets for GFP (excitation at 488 nm, emission window from 500 to 600 nm).
Laser Diffraction Analysis (LDA)
Laser diffraction analysis was performed in a Mastersizer 2000 (Malvern Instrument, UK) as previously described (Schleheck et al., 2009) with minor modifications. In particular, the following settings were used: stirring speed 1000 rpm, laser intensity 79%, 5,000 size scan, Frauenhofer count mode. The amount of bacterial suspension was adjusted to result in an LDA-obscurity of 2–6%. Size distribution scans are quantified by LDA and expressed as volume percentage, corresponding to the volumetric contribution of each particle size class to the total volume of all particle size classes (Schleheck et al., 2009).
Statistical Analysis
Statistical analysis was performed with the software GraphPad Instat, using One-Way Analysis of Variance (ANOVA), followed by Tukey–Kramer multiple comparison tests.
Results
Exopolysaccharide Production is Essential for Gac-Mediated Regulation of Pyoverdine Production
It has recently been demonstrated that the presence of the Pel and/or Psl polysaccharides is required for the ability of the second messenger c-di-GMP to positively regulate the production of the P. aeruginosa siderophore pyoverdine (Chen et al., 2015).
In order to verify whether the Pel and Psl exopolysaccharides are also involved in the Gac-mediated control of pyoverdine gene expression, we generated single and double deletion mutants in pel and psl genes in P. aeruginosa PAO1 (wild-type) and in isogenic mutants in which the Gac signaling is inactive (rsmY rsmZ mutant) or constitutively active (rsmA mutant; Supplementary Table S1). In agreement with previous observations (Frangipani et al., 2014), pyoverdine production in the iron-poor TSBD medium was higher in the rsmA mutant and strongly reduced in the rsmY rsmZ mutant compared with the wild type (WT; Figure 1A). This pyoverdine production profile was maintained in strains lacking either Pel or Psl, while pyoverdine production was drastically reduced in the pel psl double mutant irrespective of the activation state of the Gac system (Figure 1A). This response appeared to be independent of the previously reported effect of the Gac system on c-di-GMP production (Moscoso et al., 2011; Frangipani et al., 2014), since mass spectrometry analysis revealed that the Gac system is still able to control intracellular c-di-GMP levels in a Pel/Psl deficient background (Figure 1B). Thus, both the Gac system (Figure 1A) and the c-di-GMP second messenger (Chen et al., 2015) require at least one of two exopolysaccharides Pel and Psl to exert their control on pyoverdine biosynthesis, indicating that these exopolysaccharides, or an exopolysaccharide-dependent phenotype, are implicated in pyoverdine gene regulation. Indeed, we observed that the expression of the pyoverdine biosynthetic gene pvdD was greatly reduced in the pel psl mutant relative to the WT, as well as that of the pyoverdine-dependent virulence genes toxA and prpL (Figure 1C). Accordingly, the levels of the corresponding virulence factors in culture supernatants were significantly lower in the pel psl double mutant as compared to the WT strain (Figures 1D,E), as also observed for pyoverdine (Figure 1A). Therefore, lack of the Pel and Psl exopolysaccharides in P. aeruginosa causes a significant reduction not only in the production of the siderophore pyoverdine but also of pyoverdine-dependent virulence factors.
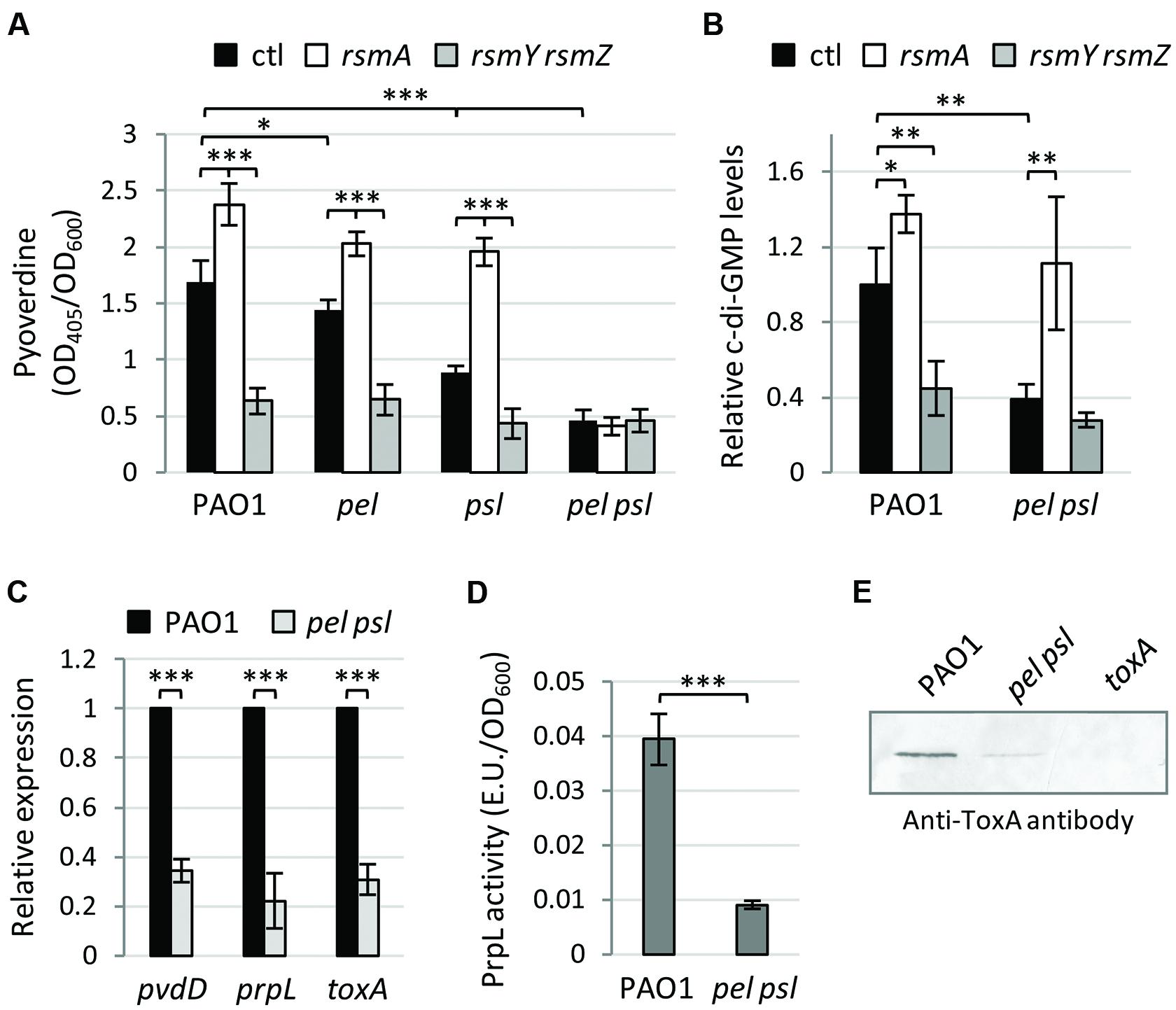
FIGURE 1. The exopolysaccharides Pel and Psl are essential for Gac-mediated control on pyoverdine production and positively regulate pyoverdine-dependent virulence factors. (A) Pyoverdine production by Pseudomonas aeruginosa PAO1, pel, psl, and pel psl mutants, and their derivatives deleted in the rsmA or in the rsmY and rsmZ genes. (B) Intracellular levels of c-di-GMP (relative to wild type PAO1) in P. aeruginosa PAO1, the pel psl mutant, and their derivatives deleted in the rsmA or the rsmY and rsmZ genes. (C) Relative mRNA levels of pvdD, toxA, and prpL as determined by qRT-PCR, and (D) PrpL enzymatic activity in culture supernatants of P. aeruginosa PAO1 and the pel psl mutant. (E) Western-blot showing ToxA levels in culture supernatants of P. aeruginosa PAO1, the pel psl mutant, and the toxA mutant (used as negative control). Bacteria were grown in TSBD at 37°C under static conditions for 14 h. Values in (A–D) are the mean (±SD) of at least three independent assays. Asterisks indicate statistically significant differences with respect to the corresponding parental strain (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001). The image in panel (E) is representative of four independent experiments giving similar results.
Exopolysaccharides Affect Pyoverdine Production Independently of Fur and Pyoverdine Signaling
The presence/absence of extracellular polysaccharides on the bacterial cell surface could influence the transport of nutrients and/or signal molecules. Since pyoverdine gene regulation is directly or indirectly controlled by the intracellular levels of iron, through the iron sensor Fur, and by the availability of iron-loaded pyoverdine on the cell surface, through the pyoverdine signaling cascade, we investigated the role of Fur and pyoverdine signaling in the Pel and Psl control of pyoverdine-dependent gene expression.
Since Fur is considered an essential protein in P. aeruginosa due to the inability to obtain fur null mutants in this species (Barton et al., 1996; Cornelis et al., 2009), to verify the involvement of Fur in exopolysaccharide-mediated control of pyoverdine production we generated a fur conditional mutants in WT and pel psl backgrounds, by replacing the native fur gene with an arabinose-inducible allele. The PAO1 fur conditional mutant grew poorly in the iron-poor TSBD medium, unless arabinose was added to the medium (Supplementary Figure S1). Compared with the WT strain, pyoverdine levels were significantly higher in the fur conditional mutant grown without arabinose, and were not shut down by the addition of iron to the growth medium (Supplementary Figure S1). This evidence confirmed the suitability of our conditional mutagenesis strategy to obtain Fur-depleted P. aeruginosa cells. Notably, the differences in pyoverdine production between fur and pel psl fur conditional mutants in the absence of arabinose were comparable to those observed for the WT and the pel psl mutant (Figure 2A), clearly indicating that Pel and Psl control pyoverdine gene expression in a way that is independent of Fur. Although not strictly instrumental in the present study, the fur conditional mutant generated in this work will represent a valuable tool for better understanding iron uptake regulation and metabolism in P. aeruginosa.
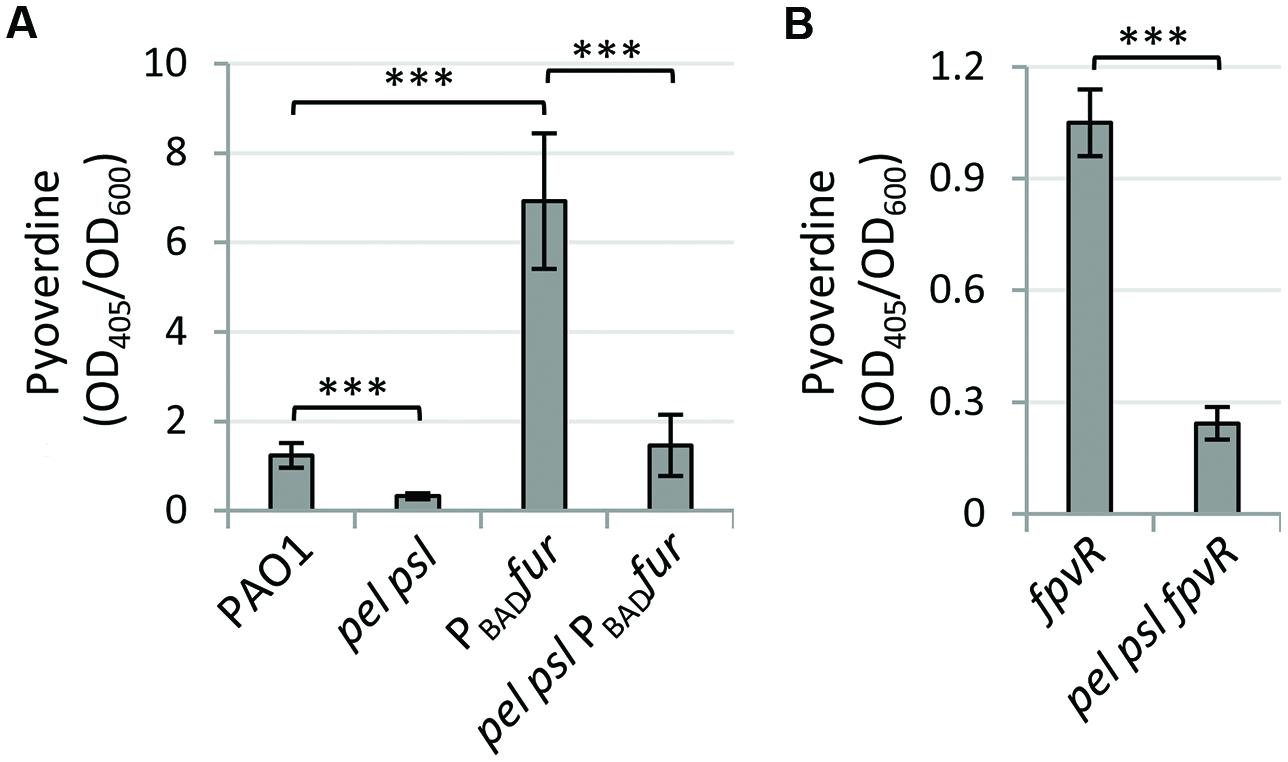
FIGURE 2. Fur and pyoverdine signaling are not involved in exopolysaccharide-mediated pyoverdine regulation. Pyoverdine production in (A) P. aeruginosa PAO1, the pel psl mutant, and their derivatives in which the native fur gene was replaced by an arabinose-inducible allele (PBADfur), and (B) P. aeruginosa PAO1 fpvR and pel psl fpvR mutants. Bacteria were cultured in TSBD without arabinose at 37°C under static conditions for 14 h, and values are the mean (±SD) of at least three independent assays. Asterisks indicate statistically significant differences with respect to the corresponding parental strain (∗∗∗p < 0.001).
To test the effect of exopolysaccharides on pyoverdine signaling, we deleted the fpvR gene in both the WT and Pel- and Psl-negative backgrounds, in order to obtain strains in which PvdS activity cannot be influenced by pyoverdine signaling through the anti-sigma factor FpvR (Lamont et al., 2002; Tiburzi et al., 2008). Again, deletion of fpvR did not restore pyoverdine production in the pel psl mutant (Figure 2B), ruling out the involvement of FpvR and, thus, of the pyoverdine signaling cascade on the exopolysaccharides-mediated control of pyoverdine production.
Artificially Induced Cell Aggregation Restores Production of Virulence Factors in Exopolysaccharide-Defective Cells
Our findings argue for a prominent role of extracellular polysaccharides in triggering the expression of pyoverdine-dependent virulence genes. As shown in Figure 1A, each single exopolysaccharide has the ability to promote pyoverdine production, suggesting that a phenotype that depends on the presence of exopolysaccharide(s), rather than a specific exopolysaccharide molecule, could be important for pyoverdine-related gene expression. It is well known that extracellular polysaccharides have a role in surface attachment and biofilm formation (Colvin et al., 2012). However, exopolysaccharides have also been proposed to mediate planktonic aggregation (Klebensberger et al., 2007; Alhede et al., 2011), as confirmed by our observation that Pel and Psl-deficient cells prevalently grow in TSBD medium as dispersed cells, while Pel and Psl-proficient WT cells grow as large aggregates including hundreds of cells (Figure 3A). These bacterial aggregates appear quite loose, and can be partially dispersed by vigorous pipetting (data not shown). This qualitative evidence was confirmed by determining the size and distribution of planktonic cells and cell aggregates by LDA. This analysis revealed that cell aggregates with a size ranging from 30 to 600 μm represent more than 85% of the bacterial population in planktonic cultures of the WT strain. In contrast, cell aggregates were not detectable in planktonic cultures of the pel psl mutant, which only contained single cells (Figure 3B). Such planktonic aggregates were also observed in cultures of the WT PAO1 grown in M9 minimal medium, while they were not detected in pel psl cultures (Supplementary Figure S2). Notably, also in this medium pyoverdine production was strongly reduced in the pel psl mutant with respect to the WT strain (Supplementary Figure S2).
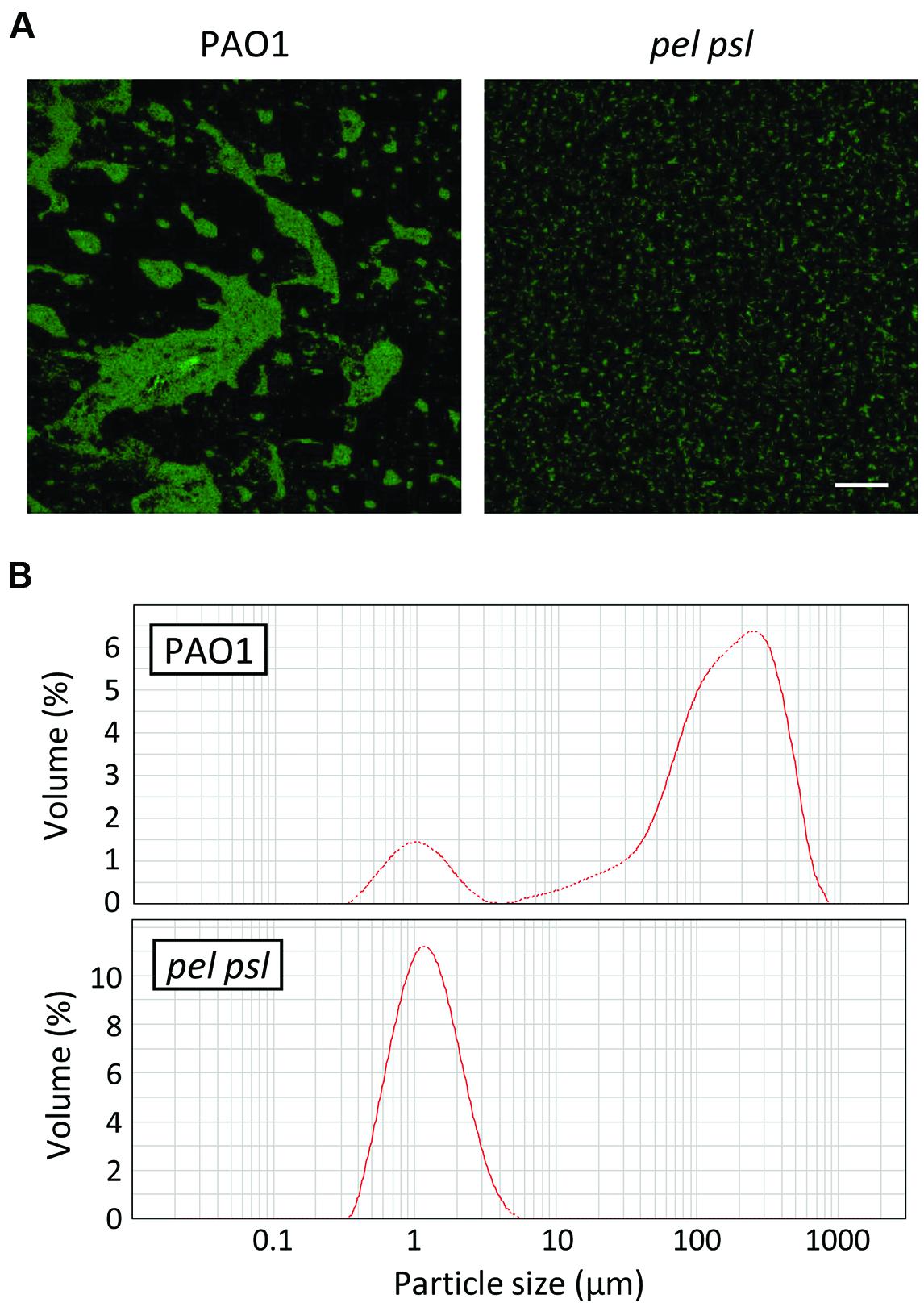
FIGURE 3. Role of Pel and Psl exopolysaccharides in planktonic aggregation of P. aeruginosa cells. (A) Confocal microscopy images of P. aeruginosa PAO1 and pel psl cells harboring the GFP-expressing vector pMMG cultured in TSBD at 37°C under static conditions for 14 h. The images are representative of several micrographs from five independent experiments. Bar: 50 μm. (B) Representative laser diffraction analysis (LDA) particle-size scans of P. aeruginosa PAO1 and pel psl liquid cultures in TSBD after 14 h of growth at 37°C.
We thus hypothesized that cell-to-cell contacts and/or cell aggregation, instead of exopolysaccharides by themselves, could represent the signal triggering pyoverdine-dependent gene expression. This hypothesis was first tested by growing WT and pel psl mutant cells as colonies on the surface of TSBD medium solidified with different gelling substances, such as polyacrylamide and the polysaccharides agar and phytagel, and qualitatively assessing pyoverdine production by comparing fluorescence emission under UV light (Visca et al., 2007). While the fluorescence of the pel psl mutant growing as single cells in liquid medium was much lower than that of the aggregated WT and comparable to that of the pyoverdine-deficient mutant PAO1 pvdA (Imperi et al., 2008), the Pel and Psl-deficient mutant showed fluorescence levels that were indistinguishable from those of the WT, and much higher than those of the pvdA mutant, during aggregated (colony) growth on solid surfaces (Figure 4A). Although this assay clearly showed that growth on solid surfaces, where cells can interact with each other irrespective of aggregative polysaccharides, enhanced pyoverdine production in the pel psl mutant, the qualitative nature of the assay did not allow a quantitative comparison of pyoverdine levels between strains. To overcome this limitation, strains were cultured in TSBD medium supplemented with different sub-gelling concentrations of agar (0.0125–0.2%, Figure 4B) or phytagel (0.08–0.125%, Supplementary Figure S3), thus allowing to quantitatively assess pyoverdine levels in culture supernatants. Pyoverdine levels in the supernatants of pel psl mutant cultures were increased by sub-gelling concentrations of agar in a concentration-dependent manner, while agar had negligible effects on pyoverdine production by the WT strain (Figure 4B). Substantially similar results were obtained with phytagel (Supplementary Figure S3). LDA and microscopic analyses confirmed that the presence of sub-gelling concentrations of agar in the growth medium forced the pel psl mutant to grow as large clusters of cells comparable to those observed for the WT (Figure 4C), consistent with the conclusion that cell aggregation stimulates pyoverdine production.
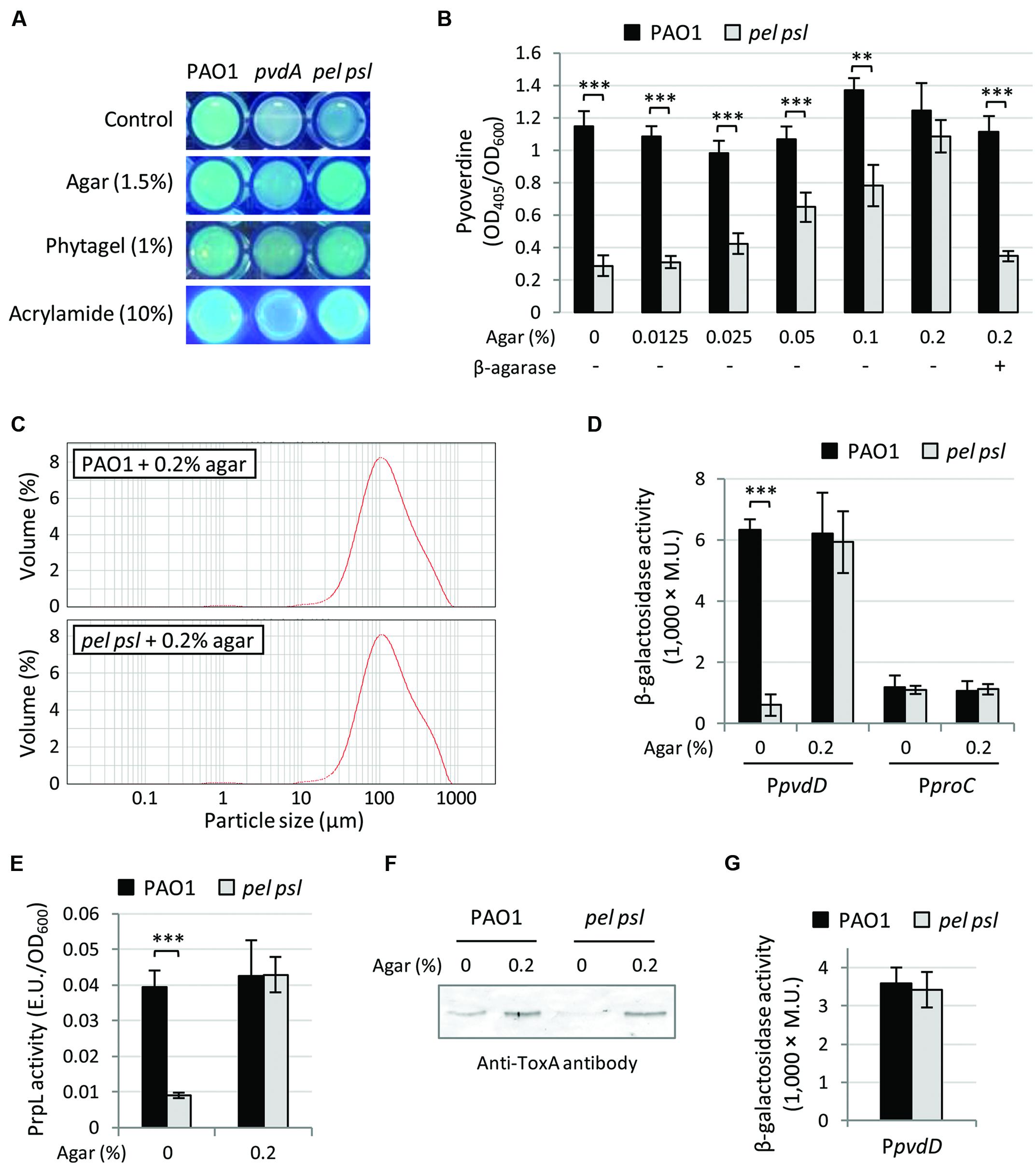
FIGURE 4. Cell aggregation is involved in Pel and Psl-mediated control of pyoverdine-dependent virulence factors. (A) Fluorescent phenotype upon UV light exposure of P. aeruginosa PAO1, the pel psl mutant and the pvdA mutant (used as pyoverdine-deficient negative control) grown in liquid TSBD medium (Control) or on TSBD solidified with 1.5% agar, 1% phytagel, or 10% polyacrylamide. (B) Pyoverdine production by P. aeruginosa PAO1 and the pel psl mutant grown in TSBD supplemented with increasing concentrations of agar (0–0.2%) and/or β-agarase I (3.3 units/ml). (C) Representative LDA particle-size scans of P. aeruginosa PAO1 and pel psl cultures in TSBD supplemented with 0.2% agar after 14 h of growth at 37°C. (D) Activity of the PpvdD::lacZ and PproC’-‘lacZ reporter fusions, (E) PrpL enzymatic activity and (F) ToxA levels in culture supernatants from P. aeruginosa PAO1 and pel psl cultures in TSBD supplemented or not with 0.2% agar. Bacteria were cultured in TSBD at 37°C under static conditions for 14 h. (G) Activity of the PpvdD::lacZ transcriptional fusion in P. aeruginosa PAO1 and pel psl cells cultured on TSBD solidified with 10% polyacrylamide. Values in (B,D,E,G) are the mean (±SD) of at least three independent assays. Asterisks indicate statistically significant differences with respect to the wild type strain (PAO1) grown under the same culture conditions (∗∗p < 0.01, ∗∗∗p < 0.001). Images in (A,F) are representative of two independent experiments giving similar results.
Notably, pyoverdine production by the pel psl mutant in the presence of agar was abrogated by agar degradation with β-agarase I, which however, had no effect on pyoverdine production by WT PAO1 (Figure 4B). This result indicates that the effect of agar on the pel psl mutant is not related to any specific constituent of the polysaccharide matrix or to a general increase in the osmolarity of the growth medium. In fact, differently from agar and phytagel, high concentrations of sucrose (up to 10%) did not stimulate pyoverdine production by the exopolysaccharides-null mutant (Supplementary Figure S3). These evidences are in line with the finding that gelling agents only promoted pyoverdine production in the pel psl mutant but not in the WT (Figure 4 and Supplementary Figure S3), indicating that this effect is specific to Pel and Psl-deficient cells. Coherent with the pyoverdine production profile, addition of 0.2% agar stimulated pvdD promoter activity (Figure 4D), as well as PrpL and ToxA production by the pel psl mutant (Figures 4E,F), while it had no effect on the expression of the housekeeping gene proC (Figure 4D). Notably, pvdD promoter activity in the pel psl mutant was also restored to WT levels by growing cells on the surface of TSBD solidified with polyacrylamide (Figure 4G), indicating that bacterial aggregation and/or cell contacts can trigger pyoverdine gene expression in the absence of any endogenous or exogenously added exopolysaccharide.
It appears therefore that artificially induced cell aggregation, obtained by growing cells either as colonies on solid surface or in liquid cultures in the presence of aggregating agents, is able to restore pyoverdine-dependent phenotypes in the Pel and Psl-deficient mutant, supporting the idea that cell-to-cell contacts and/or growth as aggregates are by themselves cues that stimulate pyoverdine production and virulence in P. aeruginosa.
Contacts with Abiotic Surface and the Mechanosensors PilY1 and Type IV Pili are not involved in Aggregation-Mediated Activation of Pyoverdine Production
In order to verify whether the stimulation of pyoverdine production by cell aggregation was due to specific cell-to-cell interactions or to an increase in surface contacts during growth as cellular aggregates, the pel psl mutant was cultured in the presence of 5 × 107 or 5 × 108 polystyrene beads per ml (3-μm size, Polysciences), under vigorous shaking in order to maintain beads in suspension. We calculated that the addition of 5 × 108 beads/ml with a 3-μm diameter in the well of a 96-wells microtiter plate (containing 250 μl of medium) leads to >20-fold increase in the plastic surface available for cell contacts, relative to cultures without beads. While the WT strain formed large aggregates of bacterial cells around the beads, the pel psl mutant only attached to the beads without developing cell aggregates (Figure 5A), indicating that the presence of the beads actually increases the number of contacts between bacterial cells and the inert plastic material. However, polystyrene beads had no effect on pyoverdine production by the pel psl mutant or the WT used as control (Figure 5B), suggesting that cell aggregation, rather than non-specific surface contacts, represents the cue which triggers pyoverdine gene expression in P. aeruginosa.
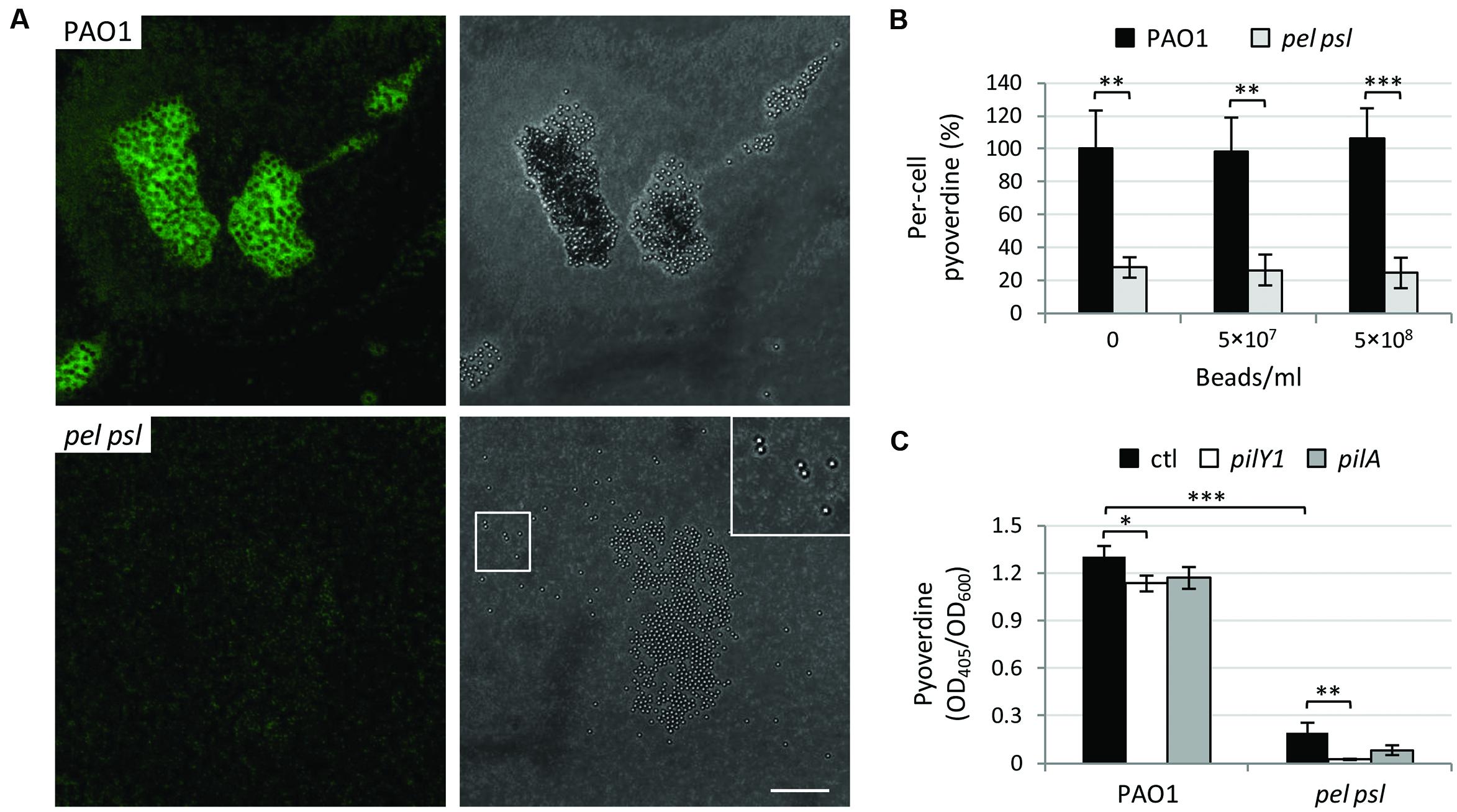
FIGURE 5. Aggregation does not stimulate pyoverdine production thorough non-specific physical contacts or the mechanosensors PilY1 and type IV pili. (A) Confocal microscopy images of P. aeruginosa PAO1 and pel psl cells harboring the GFP-expressing vector pMMG cultured in TSBD in the presence of 5 × 108 polystyrene beads/ml. Images are representative of several micrographs showing similar results. Bar: 30 μm. The inset is a 2× magnification of the highlighted area. (B) Pyoverdine production by P. aeruginosa PAO1 and pel psl in the presence or in the absence of 3-μm size polystyrene beads (5 × 107 or 5 × 108 beads/ml), normalized to the number of colony forming units/ml and expressed as percentage with respect to the untreated wild type. Bacteria were grown in TSBD at 37°C for 14 h under vigorous shaking (220 rpm). (C) Pyoverdine production by P. aeruginosa PAO1, pel psl and their corresponding pilY1 or pilA deletion mutants after 14 h of growth in TSBD at 37°C under static conditions. Values are the mean (±SD) of at least three independent assays. Asterisks indicate statistically significant differences with respect to the corresponding parental strain grown under the same culture conditions (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
To further verify this hypothesis, we investigated the possible involvement in aggregation-mediated induction of pyoverdine production of two P. aeruginosa mechanosensors, namely PilY1 and type IV pili, which have recently been implicated in surface contact-dependent activation of virulence gene expression (Siryaporn et al., 2014; Persat et al., 2015). Should PilY1 or type IV pili also be involved in pyoverdine activation in response to planktonic aggregation, the deletion of pilY1 or pilA (encoding the major pilin subunit of type IV pili) should either reduce pyoverdine production in the WT or increase pyoverdine production in the pel psl mutant, in case these mechanosensors act as positive or negative aggregation-dependent regulators, respectively. However, deletion of pilY1 or pilA caused only minor reduction (about 10%) of pyoverdine levels in the WT background, while it was not able to restore pyoverdine production in the pel psl mutant background (Figure 5C). This result indicates that formerly characterized mechanosensors, such as PilY1 and type IV pili, are not the effectors of aggregation-dependent activation of pyoverdine genes.
Discussion
In the last decades, the old vision of bacteria as strictly unicellular organisms living in a planktonic single-cell status was swept away by the finding that bacterial cells in natural, industrial and many clinical settings predominantly exist as biofilms, i.e., structured microbial communities attached to a surface and encased in an extracellular matrix (Mann and Wozniak, 2012). Also during planktonic growth in liquid cultures bacteria can assemble into aggregates of densely packed cells, and it is believed that planktonic aggregation can play a role in resistance to stresses and antibiotics (Schleheck et al., 2009; Blom et al., 2010; Haaber et al., 2012), as well as in microbe–host cell interaction (Lepanto et al., 2011). Although hundreds of studies have investigated the physiology of biofilm-living bacterial cells, very little is known about the effects of cell aggregation during planktonic growth.
Here we provide evidence that growth as planktonic aggregates promotes production of three major virulence factors in the opportunistic pathogen P. aeruginosa, namely pyoverdine, extracellular protease PrpL and exotoxin A. Indeed, we observed that a Pel and Psl-null mutant unable to aggregate in liquid cultures is also defective in the expression of virulence factor genes, and that this effect can be rescued by artificially induced cell aggregation (Figure 4 and Supplementary Figure S3).
Although in this study, we did not elucidate the mechanism by which cell aggregation is perceived by bacterial cells and translated into a transcriptional response which affects pyoverdine-related genes, two hypotheses can reasonably be made to explain the observed effect of planktonic aggregation on gene expression. First, a sensory machinery could transduce a contact signal deriving from the cell envelope into a cytoplasmic response. Our experiments lead to exclude the involvement of physical changes in the cell envelope due to contacts with abiotic surfaces, as well as any role of the two mechanosensors PilY and type IV pili (Figure 5), which have recently been found to induce P. aeruginosa virulence in response to surface contacts (Siryaporn et al., 2014; Persat et al., 2015). Also the P. aeruginosa Wsp system, a chemotaxis-like signal transduction complex which promotes c-di-GMP production by activating the diguanylate cyclase WspR, is stimulated by growth on surfaces (Güvener and Harwood, 2007). However, it has recently been shown that exopolysaccharides depletion in a c-di-GMP overproducing P. aeruginosa strain abrogated the ability of c-di-GMP to promote pyoverdine production (Chen et al., 2015). Accordingly, we found that the overexpression of a constitutively active WspR variant, which results in almost 100-fold increase in intracellular c-di-GMP levels, is unable to induce pyoverdine production in Pel and Psl-deficient cells (Supplementary Figure S4), reasonably excluding also the involvement of the Wsp system. However, other mechanosensitive factors or contact-dependent systems, which likely respond to specific cell-to-cell interactions, could exist in P. aeruginosa. For instance, the P. aeruginosa PAO1 genome is predicted to encode 26 methyl accepting chemotaxis proteins (Whitchurch et al., 2004), 13 cell–surface signaling systems (Llamas et al., 2014), up to 50 canonical two-component systems and more than 10 orphan sensor kinases (Gooderham and Hancock, 2009), many of them being implicated in virulence gene regulation.
The alternative hypothesis is that growth as aggregates determines changes in the cell surrounding environment which could influence virulence gene expression. Studies on the physiology of planktonic aggregates are still at an early stage, but it has recently been reported that localized oxygen depletion occurs in aggregates of P. aeruginosa cells grown in a gelatin-based microtrap (Wessel et al., 2014). Notably, oxygen is a well known inducer of pyoverdine gene expression (Ochsner et al., 1996; Llamas et al., 2014); thus oxygen limitation cannot explain the observed induction of pyoverdine production in bacterial aggregates. On the other hand, since exotoxin A expression is induced under microaerobic conditions through a PvdS-independent mechanism (Gaines et al., 2007), we cannot exclude that the increased exotoxin A expression levels in planktonic aggregates (Figures 1 and 4) could be related, at least in part, to oxygen depletion. It has also been reported that pyoverdine concentration is heterogeneous in P. aeruginosa microcolonies, with a maximum at the colony center (Julou et al., 2013). Although this heterogeneity could influence the efficiency of pyoverdine signaling and/or iron uptake in cell aggregates, we have demonstrated that the aggregation-mediated effect on pyoverdine production is independent of pyoverdine signaling and the iron sensor Fur (Figure 2), ruling out that increased virulence gene expression in aggregated cells may be due to altered pyoverdine signal transduction or dysregulated iron homeostasis. Thus, it is plausible that still uncharacterized chemical and/or physiological changes occurring in densely packed bacterial cells are responsible for the observed activation of pyoverdine-related virulence genes in P. aeruginosa planktonic aggregates.
In summary, our work provides the first evidence that formation of planktonic aggregates stimulates the production of pyoverdine-dependent virulence factors in P. aeruginosa. Further studies are necessary to clarify the mechanistic link between cell aggregation and activation of virulence gene expression. Such a kind of cell contact- or aggregation-dependent induction of virulence could represent a further strategy to modulate bacterial pathogenicity in response to population density, additional or complementary to chemical signaling via quorum sensing. Cellular aggregation also represents the first committed step of biofilm formation. Although some recent transcriptomics and proteomics studies highlighted an overall attenuation of virulence gene expression in mature P. aeruginosa biofilms (Li et al., 2014; Park et al., 2014), which include many slowly growing or quiescent cells, our finding could imply that the virulence potential of P. aeruginosa is increased during the first stages of biofilm formation, when siderophores, extracellular enzymes, and toxins would provide cells with essential nutrients for the energy-demanding process of biofilm development. This hypothesis is in line with the evidence that the gene expression profile of developing P. aeruginosa biofilms is more similar to that of exponential phase cultures rather than mature biofilms (Waite et al., 2006). Finally, since planktonic aggregation seems to be widespread among bacteria (Fazli et al., 2009; Schleheck et al., 2009; Blom et al., 2010; Haaber et al., 2012), it would be interesting to verify whether the correlation between cell aggregation and virulence is conserved in other bacterial pathogens.
Author Contributions
Conceived and designed experiments: DV and FI. Performed the experiments: DV, MP, CB, and FI. Analyzed the data: DV, CB, VK, PV, and FI. Contributed reagents/materials/analysis tools: VK, PV, and FI. Wrote the paper: DV, PV, and FI. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Annette Garbe for technical assistance in c-di-GMP measurements. We also thank Livia Leoni for providing pel and psl mutagenesis plasmids, Miguel Camara for the pMMG plasmid, Alain Filloux for plasmid pBBR1MCS-4-wspRR242A, and Francesco Basile for assistance with LDA analysis. This work was supported by the Italian Cystic Fibrosis Research Foundation (grant FFC#10/2013 to FI), the Sapienza University of Rome (grant Ateneo 2013 to FI) and the Italian Ministry of University and Research-PRIN 2012 (prot. 2012WJSX8K to PV).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00902
References
Alhede, M., Kragh, K. N., Qvortrup, K., Allesen-Holm, M., van Gennip, M., Christensen, L. D., et al. (2011). Phenotypes of non-attached Pseudomonas aeruginosa aggregates resemble surface attached biofilm. PLoS ONE 6:e27943. doi: 10.1371/journal.pone.0027943
Barton, H. A., Johnson, Z., Cox, C. D., Vasil, A. I., and Vasil, M. L. (1996). Ferric uptake regulator mutants of Pseudomonas aeruginosa with distinct alterations in the iron-dependent repression of exotoxin A and siderophores in aerobic and microaerobic environments. Mol. Microbiol. 21, 1001–1017. doi: 10.1046/j.1365-2958.1996.381426.x
Blom, J. F., Horňák, K., Simek, K., and Pernthaler, J. (2010). Aggregate formation in a freshwater bacterial strain induced by growth state and conspecific chemical cues. Environ. Microbiol. 12, 2486–2495. doi: 10.1111/j.1462-2920.2010.02222.x
Brencic, A., and Lory, S. (2009). Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol. Microbiol. 72, 612–632. doi: 10.1111/j.1365-2958.2009.06670.x
Brencic, A., McFarland, K. A., McManus, H. R., Castang, S., Mogno, I., Dove, S. L., et al. (2009). The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol. Microbiol. 73, 434–445. doi: 10.1111/j.1365-2958.2009.06782.x
Caballero, A. R., Moreau, J. M., Engel, L. S., Marquart, M. E., Hill, J. M., and O’Callaghan, R. J. (2001). Pseudomonas aeruginosa protease IV enzyme assays and comparison to other Pseudomonas proteases. Anal. Biochem. 290, 330–337. doi: 10.1006/abio.2001.4999
Chen, Y., Yuan, M., Mohanty, A., Yam, J. K., Liu, Y., Chua, S. L., et al. (2015). Multiple-Diguanylate Cyclase-Coordinated regulation of pyoverdine synthesis in Pseudomonas aeruginosa. Environ. Microbiol. Rep. 7, 498–507. doi: 10.1111/1758-2229.12278
Colvin, K. M., Irie, Y., Tart, C. S., Urbano, R., Whitney, J. C., Ryder, C., et al. (2012). The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ. Microbiol. 14, 1913–1928. doi: 10.1111/j.1462-2920.2011.02657.x
Cornelis, P., and Dingemans, J. (2013). Pseudomonas aeruginosa adapts its iron uptake strategies in function of the type of infections. Front. Cell. Infect. Microbiol. 3:75. doi: 10.3389/fcimb.2013.00075
Cornelis, P., Matthijs, S., and Van Oeffelen, L. (2009). Iron uptake regulation in Pseudomonas aeruginosa. Biometals 22, 15–22. doi: 10.1007/s10534-008-9193-0
Driscoll, J. A., Brody, S. L., and Kollef, M. H. (2007). The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs 67, 351–368. doi: 10.2165/00003495-200767030-00003
Fazli, M., Bjarnsholt, T., Kirketerp-Møller, K., Jørgensen, B., Andersen, A. S., Krogfelt, K. A., et al. (2009). Nonrandom distribution of Pseudomonas aeruginosa and Staphylococcus aureus in chronic wounds. J. Clin. Microbiol. 47, 4084–4089. doi: 10.1128/JCM.01395-09
Frangipani, E., Visaggio, D., Heeb, S., Kaever, V., Cámara, M., Visca, P., et al. (2014). The Gac/Rsm and cyclic-di-GMP signalling networks coordinately regulate iron uptake in Pseudomonas aeruginosa. Environ. Microbiol. 16, 676–688. doi: 10.1111/1462-2920.12164
Friedman, L., and Kolter, R. (2004). Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51, 675–690. doi: 10.1046/j.1365-2958.2003.03877.x
Gaines, J. M., Carty, N. L., Tiburzi, F., Davinic, M., Visca, P., Colmer-Hamood, J. A., et al. (2007). Regulation of the Pseudomonas aeruginosa toxA, regA and ptxR genes by the iron-starvation sigma factor PvdS under reduced levels of oxygen. Microbiology 153, 4219–4233. doi: 10.1099/mic.0.2007/011338-0
Gilbert, K. B., Kim, T. H., Gupta, R., Greenberg, E. P., and Schuster, M. (2009). Global position analysis of the Pseudomonas aeruginosa quorum-sensing transcription factor LasR. Mol. Microbiol. 73, 1072–1085. doi: 10.1111/j.1365-2958.2009.06832.x
Gooderham, W. J., and Hancock, R. E. (2009). Regulation of virulence and antibiotic resistance by two-component regulatory systems in Pseudomonas aeruginosa. FEMS Microbiol. Rev. 33, 279–294. doi: 10.1111/j.1574-6976.2008.00135.x
Goodman, A. L., Kulasekara, B., Rietsch, A., Boyd, D., Smith, R. S., and Lory, S. (2004). A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell 7, 745–754. doi: 10.1016/j.devcel.2004.08.020
Güvener, Z. T., and Harwood, C. S. (2007). Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol. Microbiol. 66, 1459–1473. doi: 10.1111/j.1365-2958.2007.06008.x
Haaber, J., Cohn, M. T., Frees, D., Andersen, T. J., and Ingmer, H. (2012). Planktonic aggregates of Staphylococcus aureus protect against common antibiotics. PLoS ONE 7:e41075. doi: 10.1371/journal.pone.0041075
Heeb, S., Kuehne, S. A., Bycroft, M., Crivii, S., Allen, M. D., Haas, D., et al. (2006). Functional analysis of the post-transcriptional regulator RsmA reveals a novel RNA-binding site. J. Mol. Biol. 355, 1026–1036. doi: 10.1016/j.imb.2005.11.045
Hickman, J. W., and Harwood, C. S. (2008). Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol. Microbiol. 69, 376–389. doi: 10.1111/j.1365-2958.2008.06281.x
Hickman, J. W., Tifrea, D. F., and Harwood, C. S. (2005). A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. U.S.A. 102, 14422–14427. doi: 10.1073/pnas.0507170102
Imperi, F., Massai, F., Facchini, M., Frangipani, E., Visaggio, D., Leoni, L., et al. (2013). Repurposing the antimycotic drug flucytosine for suppression of Pseudomonas aeruginosa pathogenicity. Proc. Natl. Acad. Sci. U.S.A. 110, 7458–7463. doi: 10.1073/pnas.1222706110
Imperi, F., Putignani, L., Tiburzi, F., Ambrosi, C., Cipollone, R., Ascenzi, P., et al. (2008). Membrane-association determinants of the omega-amino acid monooxygenase PvdA, a pyoverdine biosynthetic enzyme from Pseudomonas aeruginosa. Microbiology 154, 2804–2813. doi: 10.1099/mic.0.2008/018804-18800
Imperi, F., Tiburzi, F., Fimia, G. M., and Visca, P. (2010). Transcriptional control of the pvdS iron starvation sigma factor gene by the master regulator of sulfur metabolism CysB in Pseudomonas aeruginosa. Environ. Microbiol. 12, 1630–1642. doi: 10.1111/j.1462-2920.2010.02210.x
Imperi, F., Tiburzi, F., and Visca, P. (2009). Molecular basis of pyoverdine siderophore recycling in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 106, 20440–20445. doi: 10.1073/pnas.0908760106
Irie, Y., Borlee, B. R., O’Connor, J. R., Hill, P. J., Harwood, C. S., Wozniak, D. J., et al. (2012). Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 109, 20632–20636. doi: 10.1073/pnas.1217993109
Irie, Y., Starkey, M., Edwards, A. N., Wozniak, D. J., Romeo, T., and Parsek, M. R. (2010). Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol. Microbiol. 78, 158–172. doi: 10.1111/j.1365-2958.2010.07320.x
Jackson, K. D., Starkey, M., Kremer, S., Parsek, M. R., and Wozniak, D. J. (2004). Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J. Bacteriol. 186, 4466–4475. doi: 10.1128/JB.186.14.4466-4475.2004
Julou, T., Mora, T., Guillon, L., Croquette, V., Schalk, I. J., Bensimon, D., et al. (2013). Cell-cell contacts confine public goods diffusion inside Pseudomonas aeruginosa clonal microcolonies. Proc. Natl. Acad. Sci. U.S.A. 110, 12577–12582. doi: 10.1073/pnas.1301428110
Klebensberger, J., Lautenschlager, K., Bressler, D., Wingender, J., and Philipp, B. (2007). Detergent-induced cell aggregation in subpopulations of Pseudomonas aeruginosa as a preadaptive survival strategy. Environ. Microbiol. 9, 2247–2259. doi: 10.1128/JB.186.14.4466-4475.2004
Lamont, I. L., Beare, P. A., Ochsner, U., Vasil, A. I., and Vasil, M. L. (2002). Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 99, 7072–7077. doi: 10.1073/pnas.092016999
Lee, V. T., Matewish, J. M., Kessler, J. L., Hyodo, M., Hayakawa, Y., and Lory, S. A. (2007). cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol. Microbiol. 65, 1474–1484. doi: 10.1111/j.1365-2958.2007.05879.x
Lepanto, P., Bryant, D. M., Rossello, J., Datta, A., Mostov, K. E., and Kierbel, A. (2011). Pseudomonas aeruginosa interacts with epithelial cells rapidly forming aggregates that are internalized by a Lyn-dependent mechanism. Cell. Microbiol. 13, 1212–1222. doi: 10.1111/j.1462-5822.2011.01611.x
Li, Y., Petrova, O. E., Su, S., Lau, G. W., Panmanee, W., Na, R., et al. (2014). BdlA, DipA and induced dispersion contribute to acute virulence and chronic persistence of Pseudomonas aeruginosa. PLoS Pathog. 10:e1004168. doi: 10.1371/journal.ppat.1004168
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Llamas, M. A., Imperi, F., Visca, P., and Lamont, I. L. (2014). Cell-surface signaling in Pseudomonas: stress responses, iron transport, and pathogenicity. FEMS Microbiol. Rev. 38, 569–597. doi: 10.1111/1574-6976.12078
Lo Sciuto, A., Fernández-Piñar, R., Bertuccini, L., Iosi, F., Superti, F., and Imperi, F. (2014). The periplasmic protein TolB as a potential drug target in Pseudomonas aeruginosa. PLoS ONE 9:e103784. doi: 10.1371/journal.pone.0103784
Ma, L., Conover, M., Lu, H., Parsek, M. R., Bayles, K., and Wozniak, D. J. (2009). Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog. 5:e1000354. doi: 10.1371/journal.ppat.1000354
Ma, L., Lu, H., Sprinkle, A., Parsek, M. R., and Wozniak, D. J. (2007). Pseudomonas aeruginosa Psl is a galactose- and mannose-rich exopolysaccharide. J. Bacteriol. 189, 8353–8356. doi: 10.1128/JB.00620-07
Mann, E. E., and Wozniak, D. J. (2012). Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol. Rev. 36, 893–916. doi: 10.1111/j.1574-6976.2011.00322.x
Meyer, J. M., Neely, A., Stintzi, A., Georges, C., and Holder, I. A. (1996). Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect. Immun. 64, 518–523.
Miller, J. H. (1972). Experiments in Molecular Genetics. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory), 252–255.
Moscoso, J. A., Jaeger, T., Valentini, M., Hui, K., Jenal, U., and Filloux, A. (2014). The diguanylate cyclase SadC is a central player of the Gac/Rsm-mediated biofilm formation in Pseudomonas aeruginosa. J. Bacteriol. 196, 4081–4088. doi: 10.1128/JB.01850-1814
Moscoso, J. A., Mikkelsen, H., Heeb, S., Williams, P., and Filloux, A. (2011). The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environ. Microbiol. 13, 3128–3138. doi: 10.1111/j.1462-2920.2011.02595.x
Ochsner, U. A., Johnson, Z., Lamont, I. L., Cunliffe, H. E., and Vasil, M. L. (1996). Exotoxin a production in Pseudomonas aeruginosa requires the iron-regulated pvdS gene encoding an alternative sigma factor. Mol. Microbiol. 21, 1019–1028. doi: 10.1046/j.1365-2958.1996.481425.x
Ochsner, U. A., Wilderman, P. J., Vasil, A. I., and Vasil, M. L. (2002). Gene Chip expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol. Microbiol. 45, 1277–1287. doi: 10.1046/j.1365-2958.2002.03084.x
Ohman, D. E., Sadoff, J. C., and Iglewski, B. H. (1980). Toxin A-deficient mutants of Pseudomonas aeruginosa PA103: isolation and characterization. Infect. Immun. 28, 899–908.
Park, A. J., Murphy, K., Krieger, J. R., Brewer, D., Taylor, P., Habash, M., et al. (2014). A temporal examination of the planktonic and biofilm proteome of whole cell Pseudomonas aeruginosa PAO1 using quantitative mass spectrometry. Mol. Cell. Proteomics 13, 1095–1105. doi: 10.1074/mcp.M113.033985
Parsek, M. R., and Singh, P. K. (2003). Bacterial biofilms: an emerging link to disease pathogenesis. Annu. Rev. Microbiol. 57, 677–701. doi: 10.1146/annurev.micro.57.030502.090720
Persat, A., Inclan, Y. F., Engel, J. N., Stone, H. A., and Gitai, Z. (2015). Type IV pili mechanochemically regulate virulence factors in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 112, 7563–7568. doi: 10.1073/pnas.1502025112
Sakuragi, Y., and Kolter, R. (2007). Quorum-sensing regulation of the biofilm matrix genes (pel) of Pseudomonas aeruginosa. J. Bacteriol. 189, 5383–5386. doi: 10.1128/JB.00137-07
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
Schleheck, D., Barraud, N., Klebensberger, J., Webb, J. S., McDougald, D., Rice, S. A., et al. (2009). Pseudomonas aeruginosa PAO1 preferentially grows as aggregates in liquid batch cultures and disperses upon starvation. PLoS ONE 4:e5513. doi: 10.1371/journal.pone.0005513
Sherbrock-Cox, V., Russell, N. J., and Gacesa, P. (1984). The purification and chemical characterisation of the alginate present in extracellular material produced by mucoid strains of Pseudomonas aeruginosa. Carbohydr. Res. 135, 147–154. doi: 10.1016/0008-6215(84)85012-0
Siryaporn, A., Kuchma, S. L., O’Toole, G. A., and Gitai, Z. (2014). Surface attachment induces Pseudomonas aeruginosa virulence. Proc. Natl. Acad. Sci. U.S.A. 111, 16860–16865. doi: 10.1073/pnas.1415712111
Spangler, C., Böhm, A., Jenal, U., Seifert, R., and Kaever, V. (2010). A liquid chromatography-coupled tandem mass spectrometry method for quantitation of cyclic di-guanosine monophosphate. J. Microbiol. Methods 81, 226–231. doi: 10.1016/j.mimet.2010.03.020
Stover, C. K., Pham, X. Q., Erwin, A. L., Mizoguchi, S. D., Warrener, P., Hickey, M. J., et al. (2000). Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406, 959–964. doi: 10.1038/35023079
Takase, H., Nitanai, H., Hoshino, K., and Otani, T. (2000). Impact of siderophore production on Pseudomonas aeruginosa infections in immunosuppressed mice. Infect. Immun. 68, 1834–1839. doi: 10.1128/IAI.68.4.1834-1839.2000
Tiburzi, F., Imperi, F., and Visca, P. (2008). Intracellular levels and activity of PvdS, the major iron starvation sigma factor of Pseudomonas aeruginosa. Mol. Microbiol. 67, 213–227. doi: 10.1111/j.1365-2958.2007.06051.x
Ventre, I., Goodman, A. L., Vallet-Gely, I., Vasseur, P., Soscia, C., Molin, S., et al. (2006). Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc. Natl. Acad. Sci. U.S.A. 103, 171–176. doi: 10.1073/pnas.0507407103
Visca, P., Imperi, F., and Lamont, I. L. (2007). Pyoverdine siderophores: from biogenesis to biosignificance. Trends Microbiol. 15, 22–30. doi: 10.1016/j.tim.2006.11.004
Waite, R. D., Paccanaro, A., Papakonstantinopoulou, A., Hurst, J. M., Saqi, M., Littler, E., et al. (2006). Clustering of Pseudomonas aeruginosa transcriptomes from planktonic cultures, developing and mature biofilms reveals distinct expression profiles. BMC Genomics 7:162. doi: 10.1186/1471-2164-7-162
Wessel, A. K., Arshad, T. A., Fitzpatrick, M., Connell, J. L., Bonnecaze, R. T., Shear, J. B., et al. (2014). Oxygen limitation within a bacterial aggregate. MBio 5:e00992. doi: 10.1128/mBio.00992-14
Whitchurch, C. B., Leech, A. J., Young, M. D., Kennedy, D., Sargent, J. L., Bertrand, J. J., et al. (2004). Characterization of a complex chemosensory signal transduction system which controls twitching motility in Pseudomonas aeruginosa. Mol. Microbiol. 52, 873–893. doi: 10.1111/j.1365-2958.2004.04026.x
Keywords: cell aggregates, extracellular polysaccharide, gene regulation, iron uptake, mechanosensor, Pseudomonas aeruginosa, siderophore, virulence
Citation: Visaggio D, Pasqua M, Bonchi C, Kaever V, Visca P and Imperi F (2015) Cell aggregation promotes pyoverdine-dependent iron uptake and virulence in Pseudomonas aeruginosa. Front. Microbiol. 6:902. doi: 10.3389/fmicb.2015.00902
Received: 02 July 2015; Accepted: 17 August 2015;
Published: 28 August 2015.
Edited by:
Dongsheng Zhou, Beijing Institute of Microbiology and Epidemiology, ChinaReviewed by:
Pierre Cornelis, Vrije Universiteit Brussel, BelgiumLuyan Zulie Ma, Institute of Microbiology Chinese Academy of Sciences, China
Copyright © 2015 Visaggio, Pasqua, Bonchi, Kaever, Visca and Imperi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Francesco Imperi, Department of Biology and Biotechnology “Charles Darwin”, Sapienza University of Rome, Via dei Sardi 70, Rome 00185, Italy,ZnJhbmNlc2NvLmltcGVyaUB1bmlyb21hMS5pdA==
 Daniela Visaggio
Daniela Visaggio Martina Pasqua1
Martina Pasqua1 Volkhard Kaever
Volkhard Kaever Paolo Visca
Paolo Visca Francesco Imperi
Francesco Imperi