- Evolution, Ecology and Behavior Program, Department of Biology, Indiana University, Bloomington, IN, USA
The field of microbiome research is arguably one of the fastest growing in biology. Bacteria feature prominently in studies on animal health, but fungi appear to be the more prominent functional symbionts for plants. Despite the similarities in the ecological organization and evolutionary importance of animal-bacterial and plant–fungal microbiomes, there is a general failure across disciplines to integrate the advances made in each system. Researchers studying bacterial symbionts in animals benefit from greater access to efficient sequencing pipelines and taxonomic reference databases, perhaps due to high medical and veterinary interest. However, researchers studying plant–fungal symbionts benefit from the relative tractability of fungi under laboratory conditions and ease of cultivation. Thus each system has strengths to offer, but both suffer from the lack of a common conceptual framework. We argue that community ecology best illuminates complex species interactions across space and time. In this synthesis we compare and contrast the animal-bacterial and plant–fungal microbiomes using six core theories in community ecology (i.e., succession, community assembly, metacommunities, multi-trophic interactions, disturbance, restoration). The examples and questions raised are meant to spark discussion amongst biologists and lead to the integration of these two systems, as well as more informative, manipulatory experiments on microbiomes research.
Introduction
Communities, or species assemblages, are a fundamental unit of ecological organization, just as cell or tissue types are fundamental units of study for molecular biology and physiology. Originally developed for macroorganismal systems (Clements, 1916), many community ecology theories attempt to elucidate complex species interactions across space and time. Increasingly, these concepts are being applied to the study of some of our planet’s most complex and intimate communities—host-associated microbiomes. Both plants and animals are colonized by an astonishing number of symbiotic microbes (Raaijmakers et al., 2008; Zilber-Rosenberg and Rosenberg, 2008; Rodriguez et al., 2009; Lecuit and Eloit, 2013), and recent advances in sequencing technologies and data processing are finally affording researchers the opportunity to uncover the cryptic diversity and functions of these microbiomes (Zimmerman et al., 2014).
The microbiome can be made up of myriad prokaryotic and eukaryotic organisms, including bacteria, archaea, viruses, fungi, and protozoans. These groups of organisms have garnered attention due to their collective functional role in controlling host nutrition, metabolism, physiology, and immunology (Ottman et al., 2012). The importance of these different taxa can differ depending on the macroorganism with which they are associated. In animals, the bacterial microbiome significantly outnumbers and exerts more control over its host’s health and well-being than the fungal microbiome (Huffnagle and Noverr, 2013; Box 1). However, the opposite appears true for plants. Fungi, including leaf- and shoot-inhabiting fungi and root-associated mycorrhizae, appear to be the functionally prominent symbionts (Rodriguez et al., 2009; Porras-Alfaro and Bayman, 2011), despite being numerically less abundant than their bacterial counterparts (Lundberg et al., 2012; Bulgarelli et al., 2013, 2012). In particular, leaf- and shoot-inhabiting fungi (i.e., endophytes) are increasingly recognized for their impact on plant host health and utility in the study of community-level processes. Located in one of the most critical organs for energy processing (i.e., photosynthetically active leaves and shoots), these horizontally-transmitted endophytic fungi are readily culturable under laboratory settings and known colonizers of all plant species studied to date, including both wild plant species and model organisms such as Arabidopsis. Because endophytic fungi are easily cultured, they lend themselves well to studying the ecology of natural systems under controlled conditions in the laboratory. Although there are strong similarities in the ecological organization and functional significance of both animal-bacterial and aboveground plant-endophyte symbioses, the two systems historically have not been critically evaluated in conjunction with one another, potentially neglecting fruitful avenues for theoretical and experimental comparison. In this synthesis we highlight the commonalities of these two distinct research areas, which have had little communication between them, and outline how they may be unified by a common theoretical foundation.
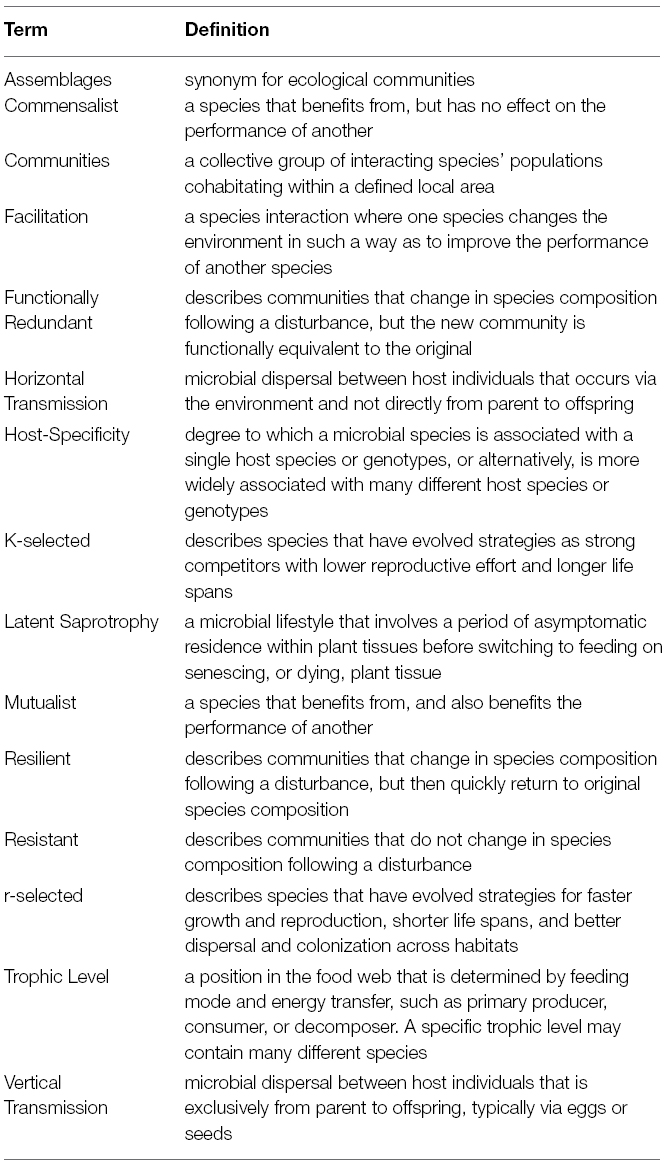
The specific goals of this synthesis are twofold; first, to highlight the ecological and functional similarities between animal-bacterial and aboveground plant-endophyte microbiomes using six core theories in community ecology, specifically: successional theory, seed banks in community assembly theory, metacommunity theory, multi-trophic interactions, disturbance ecology, and restoration ecology (for a glossary of useful terms in community ecology, please refer to Table 1). These represent major theories in ecology that have been used independently and in conjunction for decades as a way to describe and predict macroorganismal abundance and distribution patterns. Our second goal is to show how these theories may be applied to animal and plant microbiomes and to further outline experiments that will spark research and discussion among a wide range of biologists (e.g., ecologists, animal scientists, agronomists, medical scientists, microbiologists). We do not intend this to be an exhaustive review of the current state of microbiome research nor that of community ecology, but rather to offer a novel perspective on the study of both using two exemplar systems.
Defining Communities
The numeric diversity of microbial symbionts is astounding. Within the human-bacterial microbiome, up to 90–240 bacterial genera alone are associated with the skin (The Human Microbiome Project Consortium, 2012b), and at least 5 million non-redundant bacterial and phage genes are encompassed by the microbial community of the human gut (The Human Microbiome Project Consortium, 2012a). These communities are a product of at least 500 million years of coevolution between animals and their bacterial symbionts (Ley et al., 2008; McFall-Ngai et al., 2013). Similarly, evidence suggests that plants have played host to suites of aboveground fungal symbionts since their initial colonization of land 450 million years ago (Krings et al., 2007). Collectively known as “endophytes,” these fungal symbionts are increasingly recognized for their diversity and impact on host functioning (e.g., host physiology and immunology; Schulz and Boyle, 2005; Rodriguez et al., 2009). For the purposes of our paper, we define endophytic fungi (hereafter, “EF”) as those fungal symbionts that reside cryptically within healthy aboveground plant tissues such as leaves and shoots, form localized infections, and are horizontally transmitted amongst hosts (Rodriguez et al., 2009). This definition serves to distinguish EF from the vertically-transmitted systemic fungi typical of many grasses, as well as from bacterial endophytes, transient surface-dwelling microbes, and belowground fungal symbionts such as mycorrhizae, which form partially external symbioses with multiple host plants at the same time, and dark-septate root fungi that are generally restricted in their transmission from host-to-host by a structured soil medium (Box 2). The root-associated microbiome, known collectively as the rhizosphere, has been the subject of extensive research (Lundberg et al., 2012; Bulgarelli et al., 2013, 2012; Philippot et al., 2013; Box 1). However, increasingly researchers are using leaf -and shoot-associated fungal microbiomes (i.e., the “phyllosphere”) to move beyond characterization studies and into more manipulative exploration of the collective form and function of microbial communities in hosts.
Leaf and shoot EF have been isolated from all plant species sampled to date, including aquatic and basal plant lineages (Bayman, 2006; Higgins et al., 2007; U’Ren et al., 2012; Sandberg et al., 2014). They are considered to be the most speciose and phylogenetically diverse members of the fungal kingdom (Arnold et al., 2000). Tens to hundreds of different fungal species may coexist within the foliage of a single host (Gamboa et al., 2002), where they may constitute up to 2.5% of photosynthetic biomass (Davey et al., 2009). Unlike most bacteria, which switch frequently between leaf surfaces and internal tissue, fungi maintain a more stable and intimate relationship with their plant hosts (Beattie and Lindow, 1995; Hallmann et al., 1997).
Roles of the Microbiome Community
Just as free-living organisms provide extensive ecosystem services (e.g., pollination, nutrient cycling, water purification), microbial symbionts can significantly impact their surrounding host ecosystems. Although important defensive and nutritive roles are well-studied in the vertically-transmitted bacterial symbionts of insects and other invertebrates (Box 2), horizontally-transmitted bacterial symbionts of humans also manifest a variety of functional roles in their hosts and are now even considered analogous to an “organ” in and of itself (Lepage et al., 2013). The gut microbiome assists in the breakdown of dietary products and production of essential nutrients, such as vitamins B and D (Ley et al., 2008; Qin et al., 2010). Beyond their nutritional role, bacterial symbionts of vertebrates actively shape the mucosal layer of the small intestine and colon during development (Sommer and Bäckhed, 2013), which is later used as a selective barrier to reject pathogenic species (Hooper et al., 2012). Some gut bacteria (i.e., bifidobacteria) also take on a direct non-host immunity role by fermenting macronutrients into short-chain fatty acids as an energy source for host T-cells fighting off pathogenic bacterial blooms (Fukuda et al., 2011). Many other animal organs play host to bacterial symbionts (Box 3), including the skin (Chen and Tsao, 2013). In one study, mice grown without skin bacteria exhibited abnormal cytokine production and their T-cell populations were unable to mount an adequate immune response against the skin parasite Leishmania major (Naik et al., 2012). It is becoming increasingly clear that many human diseases are associated with an imbalance in the numerical composition or nutritive and immunological function of the microbiome, termed “dysbiosis.” The medical community now even recognizes the potential to use these shifts in bacterial abundance as a diagnostic tool to document and quantify disease severity (Hollister et al., 2014). A disrupted human microbiome has been linked to diverse pathologies, including kwashiorkor, a severe form of acute malnutrition (Smith et al., 2013); psoriasis (Statnikov et al., 2013); sexually-transmitted diseases (Brotman et al., 2012); and inflammatory bowel disease (Frank et al., 2007). A key role of the bacterial microbiota in carcinogenesis has also been proposed (Schwabe and Jobin, 2013).
Similarly, numerous studies have documented that EF may confer pathogen resistance to their plant hosts (Arnold et al., 2003; Ganley et al., 2008; Lee et al., 2009). Mechanistically, this is thought to occur via direct secretion of antimicrobial substances, competitively “out-crowding” fungal pathogens for plant tissue habitat (Rodriguez Estrada et al., 2011), or priming the host plant’s immune system against future pathogen invasion (Alabouvette et al., 2009; Hartley et al., 2015). For example, EF render the palatability of leaves unpredictable to herbivores by increasing the spatial heterogeneity of the chemical landscape within host-plant tissues (Carroll, 1988; Herre et al., 2007). Several studies of temperate trees have found negative effects of EF on leafmining or galling insects (Wilson and Carroll, 1997; Lawson et al., 2014), including evidence suggesting that leafmining insects are more likely to lay eggs on leaves with lower endophyte densities (Wilson and Faeth, 2001). Moreover, endophytes may also strengthen plants’ innate pathogen and herbivore defenses; colonization by a single, common endophyte (i.e., Colletotrichum tropicale) induced the upregulation of over 100 different host genes in Theobroma cacao, including many related to chemical defense and the hardening of cell walls (Mejia et al., 2014). The anti-microbials secreted by EF are being investigated for their potential as medicinal therapies for human diseases and some forms of agricultural pest control, a field known as bioprospecting (Strobel and Daisy, 2003; Porras-Alfaro and Bayman, 2011). Beyond host defense, endophyte colonization has been implicated in conferring abiotic stress tolerance to hosts, often by altering plant physiology (e.g., hormonal manipulation, water consumption; Rodriguez et al., 2009, 2008). Inoculation by a single endophyte has been shown to reduce wilting in cacao under drought conditions (Bae et al., 2009), although other studies using the same host have shown that infection by a suite of endophytes increases water loss in host plants (Arnold and Engelbrecht, 2007). This suggests that some EF may be conditionally mutualistic, or that crowding, while beneficial for pathogen defense, can more quickly drain hosts of necessary resources. Under what conditions compositionally distinct microbiomes can either prevent disease or induce stress in their hosts are parallel questions for future research in both animal and plant hosts.
Applying Community Ecology Theories to Microbiomes
The field of community ecology has its roots in studies of plant and animal communities from the 1920s and 1930s (Clements, 1916; Gleason, 1926; Elton, 1927). In its infancy, single and isolated theories were often used to delineate complex species interactions and make predictions about the abundance and distributions of species across space and time (Mittelbach, 2012). By the 1980s, however, a paradigm shift toward a more pluralistic and integrative approach began to take hold. This modern view of community ecology considers the multitude of processes that can regulate species diversity and abundance at the local as well as regional scales. The integration of theories has catapulted research of macroorganismal communities in the last 30 years and we believe the time is ripe to begin advancing the study of microorganismal communities, using a balance of theory and technology, in order to gain a fuller appreciation for the ecology of these unseen systems.
At its core, the field of community ecology seeks to understand temporal and spatial dynamics of communities, interactions between a community’s component members, and the emergent properties of communities in response to change. We focus on six major theories in community ecology that are particularly relevant to testing for patterns of association and interaction within microbial communities: succession, community assembly, metacommunity dynamics, multi-trophic interactions, disturbance, and restoration (See Figure 1). While these six theories are not exhaustive, they are widely considered as cornerstone theories in studies of ecological communities (Begon et al., 2006; Mittelbach, 2012; Molles Jr, 2013) and have important application in the agricultural, medicinal, and environmental science fields. We conclude with a brief discussion of how these theories may be applied to the study of host-associated microbial communities versus the macroorganismal communities for which they were originally developed.
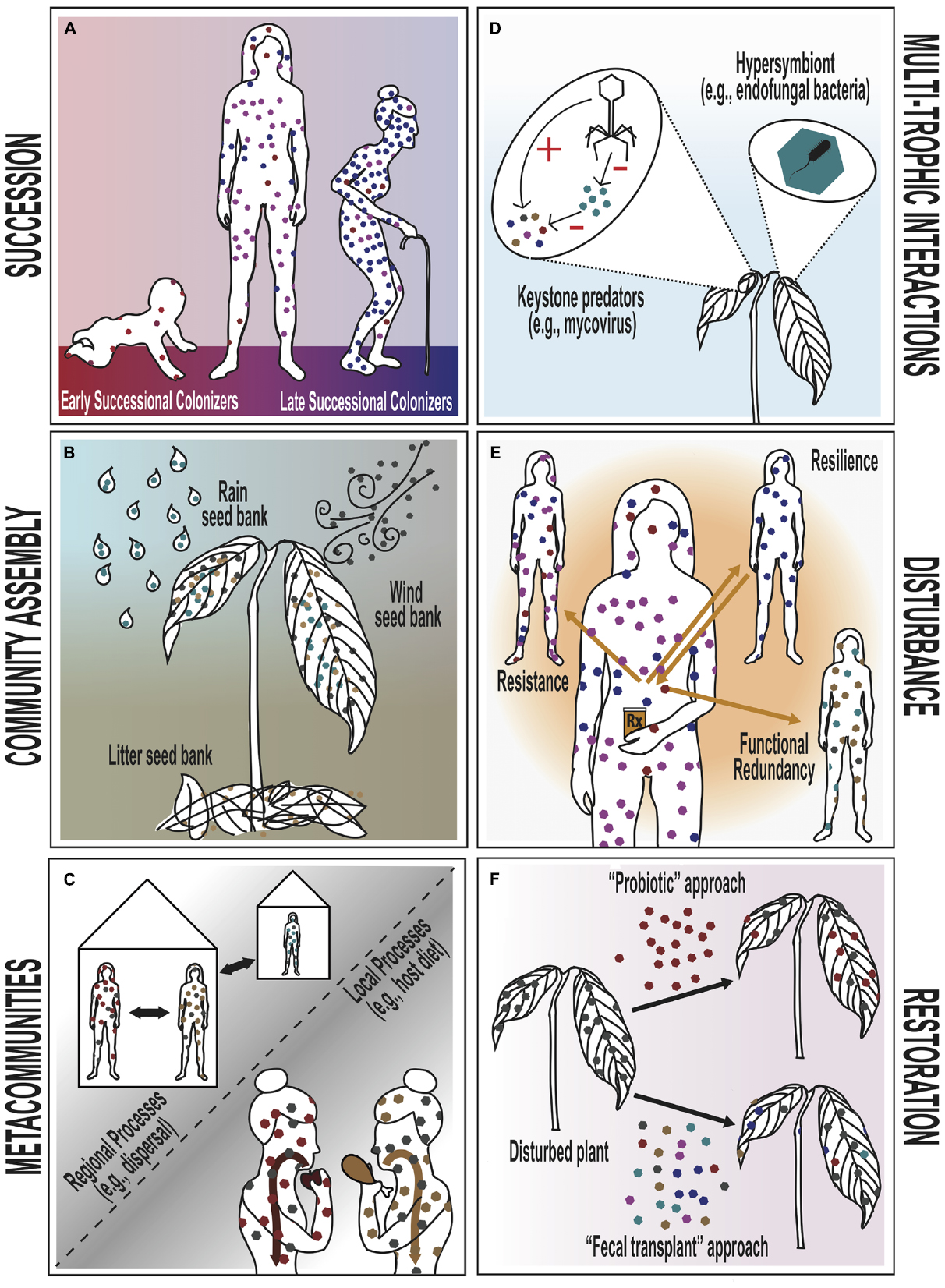
Figure 1. Conceptual diagrams of six classical theories in community ecology, applied to the animal/human and plant microbiomes. Each panel represents a separate theory. Hexagons are used throughout to represent the bacterial and fungal constituents of the human and plant microbiome, respectively. (A) Successional theory. In humans, late successional colonizers replace early successional colonizers. As succession proceeds, the density of the bacterial microbiome increases steadily, but diversity only increases into adulthood and declines thereafter in old age. (B) Community assembly theory. The primary sources of endophytic fungal colonizers for the plant microbiome are rain, wind and plant litter (e.g., leaves, twigs, bark). These spore sources can be considered akin to “seed banks” in community assembly theory. (C) Metacommunity theory. Human microbiomes can be influenced by both local processes, such as species interactions and habitat suitability (e.g., an individual’s diet), and regional processes (e.g., dispersal and extinction among or between households). The “local” and “regional” scales of microbiome communities may be defined flexibly (e.g., as organs, individuals, or households/populations). (D) Multi-trophic interactions. Within plants, endophytic fungi interact with many other organisms. Keystone predators and their analogs (e.g., mycoviruses) may suppress a dominant EF and indirectly promote a more diverse microbiome. Bacteria that reside intra-cellularly within fungal cells are known as “hypersymbionts” and may alter fungal behavior and have cascading health effects on the ultimate plant host. (E) Disturbance theory. Antibiotics represent an example of disturbance to the microbiome in humans. Four distinct types of microbiome community response to such a disturbance are theoretically possible: (1) microbiome composition remains unchanged (resistance), (2) microbiome composition changes but returns quickly to its original state (resilience), (3) microbiome composition changes but the new microbial constituents maintain the same function as the original community (functional redundancy), (4) or microbiome composition changes and does not retain original community function (not pictured). (F) Restoration ecology. A disturbed plant microbiome may be restored to its original composition or function through different approaches. In the “probiotic” approach, a plant is seeded with a single, presumably beneficial, EF species in order to restore the microbiome community, while the “fecal transplant” approach relies on inoculation by an entire healthy microbiome community in order to restore the target microbiome community.
Successional Theory
The theory of successional and temporal change in communities (i.e., predictable changes in species composition over time) has a long history in ecology (Connell and Slatyer, 1977). There is increasing evidence that the archetypal concept of successional transitions could inform changes in community composition and function for microbiomes (Figure 1A; Costello et al., 2012; Lozupone et al., 2012a). In human infants, as well as other mammals, primary succession of the microbiome begins prior to birth (Ottman et al., 2012). For example, in humans this consists of early successional microbial communities that are dominated by bifidobacteria (Hinde and Lewis, 2015). As we age, the microbiome community transitions into one with both higher numerical abundance and greater species diversity. Species diversity peaks in adulthood, with a microbiome dominated by Bacteroidetes and Firmicutes, but then declines (Koenig et al., 2011). By contrast, numerical abundance of the bacterial microbiome only increases with age (Koenig et al., 2011). Events such as disease onset, antibiotic treatments, and changes in diet can then cause non-random shifts in the composition of the microbiome (Koenig et al., 2011). Unlike higher animals, however, plants grow in size and develop new leaves and other organs continuously throughout their lives (Barthélémy and Caraglio, 2007; Klimesová and Klimeš, 2007; Box 3). This continual production of new habitat is denoted by increasing EF species diversity in individual leaves as they age (Arnold and Herre, 2003; Voříšková and Baldrian, 2013). Furthermore, leaf aging is thought to be associated with functional shifts in the roles of EF species from more mutualistic and commensalistic in mid-aged leaves to “latent saprotrophy” and decomposition acceleration in older leaves (Osorio and Stephan, 1991). The presence of such intra-host variation in age structure of habitats and communities within the plant–fungal microbiome presents itself as a unique opportunity to study temporal patterns of community assembly as the habitat (i.e., the leaf) ages and the host ages. To our knowledge no corollary exists for this concept in mammalian organs or tissues, including for skin microbiomes, making plants an excellent system in which to study microbiome succession while controlling for individual host variation and genetics.
One mechanism of succession occurs when early colonizers of a habitat facilitate the success of later colonizers (Connell and Slatyer, 1977). A transition in constituency from facultative to obligate anaerobes over the course of development is a common pattern seen in the fecal microbiomes of human infants (Sharon et al., 2013; Sommer and Bäckhed, 2013), as well as in bovine rumen microbiome (Jami et al., 2013). Facultative anaerobes are better able to colonize and tolerate these highly oxygenated and dynamic “virgin habitats,” but over time they engineer their environment to contain less oxygen and facilitate colonization by more stable obligate anaerobic communities. It is also well known that early colonizers of the human tooth microbiome, such as Streptococcus oralis, Streptococcus gordonii, and Actinomyces oris, perform the ecological role of adherence to the smooth and barren surface of the new teeth, subsequently acting as “bridges” and facilitators for secondary bacterial colonizers such as Veillonella parvula and Porphyromonas gingivalis as plaque formation proceeds (Kuramitsu et al., 2007; Kolenbrander, 2011). Successional transitions in microbiome constituency have also been documented in plant–fungal microbiomes, although there are far fewer examples of this ecological theory for EF (but see Martinson et al., 2012). In one study on two more pathogenic cousins of asymptomatic endophytes, primary residency by Fusarium graminearum increased the establishment success of Fusarium verticillioides in their mutual maize host (Picot et al., 2012). Another study documented facilitation or inhibition models of succession, which are contingent upon the order of arrival of pathogens and endophytes to a naïve environment. Secondary inoculation of EF can facilitate disease spread of Pseudomonas spp. in wild lima-bean hosts, but inhibit the disease-causing agent if inoculated first (Adame-Álvarez et al., 2014). This inhibition model, as well as other models of succession (i.e., “Tolerance”) are less explored than facilitation with regards to microbiomes.
Many experiments incorporating both traditional in vitro assays and advanced sequencing technologies, such as metagenomic and metatranscriptomic comparisons, could and should be used to test the components of successional theory as it applies to microbiomes in animals and plants. Late colonizing species may be stronger competitors (K-selected) in one-on-one in vitro assays or demonstrate higher population growth rates and tolerance in the face of external stress (e.g., antibiotics, fungicides, acidity, drought). Studies of microbial gene expression could reveal if early colonizers are faster growing (r-selected) or more tolerant of stressful conditions (e.g., oxidative stress; UV radiation; Lozupone et al., 2012b). Additionally, the question of whether it is possible to alter the trajectory of succession from healthy and stable to degraded and chaotic, or vice versa, has important implications for applied scientists studying long-term human health and plant fitness. If different successional trajectories can be engineered in animal and plant microbiomes, then medicinal and agricultural practices that induce stability should be promoted, while those practices that cause dysbiosis should be dissuaded (Lemon et al., 2012).
“Seed Banks” and Community Assembly
Community assembly theory is used to understand the processes that shape the structure and function of communities and has been important in identifying patterns of macroorganismal communities across islands and continental landscapes. Given the horizontal mode of transmission of mammal-associated bacteria and leaf- and shoot-associated EF communities, community assembly theory may be particularly helpful in informing how microbiomes come together when their component members originate from different sources. In some special cases hosts directly facilitate the colonization of microbes from the environment. These cases have been studied in the squid-Vibrio model, where the host excretes substances to encourage Vibrio fisherii to colonize its light organs (McFall-Ngai et al., 2013) and leguminous plants that excrete substances to encourage colonization by rhizobia (Kiers et al., 2003; Nelson and Sadowsky, 2015; Box 1). However, for most other animals and plants, environmental transmission of microbial symbionts is much less directed. It is important to know the source populations, or “seed banks,” for horizontally-transmitted microbial species in the microbiome in order to make predictions about host health and function for animals and plants (Figure 1B; Harrison and Cornell, 2008). For example, it is known that the microbiomes of infants born via cesarean section (C-section) are fundamentally different from those born vaginally (Penders et al., 2006; Dominguez-Bello et al., 2010). Specifically, naturally-delivered infants are primarily colonized by the Lactobacillus-dominated vaginal microbiome, while C-section infants are more broadly colonized by non-lactobacillus-dominated bacteria from the surrounding environment, in particular the skin microbiome of attending physicians and the child’s mother. These differences in seed bank sources for C-section newborns have functional implications, and have been connected to susceptibility to methicillin-resistant Staphylococcus aureus (MRSA) skin infections (Dominguez-Bello et al., 2010). After birth, important source populations for the bacterial microbiome are related to life events, such as breastfeeding and the onset of a solid food diet (Matamoros et al., 2013; Bergström et al., 2014). Environmental surroundings such as the presence of a family dog (Azad et al., 2013) and types of fruits and vegetables consumed (Leff and Fierer, 2013), can also shape the assembly of a child’s microbiome community. One study has even demonstrated up to 14 bacterial types in common between children and the house dust from the home in which they were raised (Konya et al., 2014).
For EF communities, horizontal transmission from the environment is thought to be the predominant mode of dispersal (Rodriguez et al., 2009, but see Hodgson et al., 2014). Seedlings propagated in sterile environments (i.e., in growth chambers or greenhouses) have been shown to be virtually free of EF (Arnold and Herre, 2003), and studies artificially excluding air- and rain-borne spores from plants in the wild show reduced colonization by EF (Kaneko and Kaneko, 2004). In addition, insects are thought to ferry spores from plant to plant during feeding or pollination (Herrera et al., 2009), and neighborhood leaf litter is known to be an important source of fungal colonizers for aboveground leaf tissue (Kaneko and Kakishima, 2001; Herre et al., 2007). Some EF, such as Coccomyces sinensis, go on living within the leaf tissue even upon leaf abscission and decay (Koukol et al., 2011; Hirose et al., 2013) and thus are able to sporulate and re-colonize living tissues of their host or host’s offspring. Alternatively, leaf litter from one plant species could serve as a seed source for other plant species in the neighborhood, depending on the degree of host-specificity of the microbial symbiont. The functional and host-health consequences of these divergent sources of colonization for the plant microbiome could be important if, for example, the colonizers from litter are more often beneficial to their original host plant or host plant offspring. Alternatively, fungal seed banks from the air could represent cosmopolitan, or weedy, species with little benefit to the host they colonize.
By manipulating the relative contribution of different modes and sources of transmission for microbiome communities we can better understand how these communities differentially affect host functions. While this is a relatively straightforward pursuit in plant systems, it is ethically problematic to manipulate “seed” sources in humans. However, model organisms such as mice and flies could provide fruitful insights to human bacterial community assembly. It will be important in the future to alter and compare the effects of factors such as diet, kin, birthing mode, breastfeeding, and local environment in order to better understand inter-individual variation in microbiome community assembly (Costello et al., 2012), as well as host’s ability to facilitate colonization by specific beneficial microbes.
Metacommunity Theory
Metacommunity theory is a relatively recent development in community ecology (Holyoak et al., 2005). The basic tenets of the theory posit that small-scale, local communities are interconnected at a regional scale via the processes of dispersal and extinction (Leibold et al., 2004). The specific species composition of the local communities is determined both by these two regional processes and the local processes of species interactions and habitat suitability (Holyoak et al., 2005; Leibold and McPeek, 2006; Mihaljevic, 2012). Metacommunity theory has been used to characterize communities of free-living macroorganisms, such as mosquito communities inhabiting water-filled tree cavities (Ellis et al., 2006), but researchers are increasingly advocating a role for metacommunity theory in studies of symbiotic microbes as well (Figure 1C; Costello et al., 2012; Mihaljevic, 2012). Symbiotic communities may even provide an advantage over the study of free-living communities in that “local” and “regional” scales can be defined flexibly, within nested levels of organization (Mihaljevic, 2012; O’Dwyer et al., 2012). For example, local microbiome communities can be defined at the organ level (e.g., gut, skin, leaf, stem), at the host level (e.g., individual person, plant), or even at the host population level (e.g., one family household, one forest). Locally, bacterial and fungal symbionts may compete for resources or avoid common enemies, which can influence community composition. But regionally, dispersal and extinction among these organs, hosts, or host populations can also influence microbiome communities across larger spatial scales. The flexible definition of local microbiome communities and nested organization of organs within hosts within host populations could be used as a tool to compare and contrast the importance of local and regional processes in a way not possible for free-living communities.
Despite this potential utility, few direct examples exist for the application of metacommunity theory to microbiome communities in either animals or plants. Indirectly, many studies have considered individual components of the theory, yet never under a unified framework. For example, the diet and societal role of the human host (e.g., hunters, gatherers, farmers) can be thought of as local processes that determine the quality and suitability of the gut habitat for different bacterial species in the gut microbiome. For instance, one study of traditional hunter-gatherer communities found a marked absence of Bifidobacterium relative to westernized urban controls, as well as strong differences in bacterial composition between the sexes, likely reflecting the sexual division of labor in this society (Schnorr et al., 2014). Similarly, EF colonization success and subsequent reproduction in plants has been shown to differ between shaded and full-sun leaf habitats, with increased prevalence of in C. cladosporioides and A. alternata for full-sun leaves in the same Japanese beech tree (Osono and Mori, 2003). Again, these patterns reflect the role of local habitat suitability processes in shaping the microbiome. Local species interactions have also been shown to shape the microbiome. In maize, interspecific interactions between EF species have frequently been detected. However, the outcome of these interactions, whether mutually beneficial or antagonistic, was dependent on the definition of “local community” used by the researchers (i.e., individual plant organs or entire plants, respectively; Pan and May, 2009). The regional process of dispersal is consistent with the higher degree of microbiome similarity among cohabiting family members (or plant individuals) than between families (or plant populations). For instance, research shows that the highest degree of MB similarity among family members exists for the skin microbiome (Song et al., 2013), suggesting that the regional process of dispersal is of greater relative importance in shaping the skin organ microbiome than the local metacommunity processes of species interactions or habitat suitability.
Future studies are needed to unite and compare the local and regional scale processes that shape the microbiome. Competition and dispersal assays could be performed in vitro for culturable symbionts, such as many EF, as a way to explain the absence, or differential abundance, of microbial species across hosts and host habitats. Efficacy of colonization success achieved by artificial inoculations could reveal how important the processes of host and organ specificity are to local community formation. Additionally, techniques such as molecular genotyping can be used to track specific microbial genotypes across space and time and quantify the patterns of dispersal and extinction for different microbiome species at a regional scale (Cockburn et al., 2013; McCormack et al., 2013).
Multi-trophic Interactions
A major goal in ecology is to understand how multi-trophic level interactions (i.e., interactions amongst different groups and types of species) influence the diversity and abundance of species in communities. The ecology and evolution of symbiotic microbes is not just the simple byproduct of pairwise interactions between host and symbiont, but rather, is a function of complex multi-trophic interactions with predatory species, competing parasites, and other symbionts within the host, to name a few (Figure 1D; Agrawal et al., 2007). For instance, the population dynamics of many bacterial species and their specialist predatory phages cycle in classic predator-prey fashion over the course of human development (Sharon et al., 2013). In this case, viruses function analogously to the keystone predators in many food web networks because they control the population growth of otherwise dominant bacteria and maintain species diversity (Paine, 1966; Rodriguez-Valera et al., 2009). “Myco”-viruses have also been isolated from many fungi existing in plant species, including grapevines (Al Rwahnih et al., 2011), and chestnut trees (Springer et al., 2013), as well as corn and wheat (Chu et al., 2002). Mycoviruses have been implicated in reducing the virulence of fungal pathogens from elm and chestnut trees (Heiniger and Rigling, 1994; Buck and Brasier, 2001). However, whether mycoviruses control the population dynamics of more commensal or mutualistic fungal symbionts, such as EF, is a needed area for future research.
Other types of multi-trophic interactions are also important regulators of the microbiome in humans and plants. For example, recent work has uncovered the presence of endohyphal bacteria in the EF, Pestalotiopsis spp. This additional layer of symbiosis (i.e., hypersymbiosis) between the bacteria and its fungal host has been implicated in regulating hormonal transfer to the ultimate coniferous host, Platycladus orientalis (Hoffman et al., 2013). In mammals, studies have demonstrated that a high diversity of eukaryotic protists and fungi interact with bacteria during their residence within the gut (Parfrey et al., 2014) and that their direct contact with the bacterial microbiome can influence host health (Box 1). Intriguingly many of the factors thought to influence the abundance and distribution of the gut microbiome (e.g., host diet, age, environment) may also influence helminthic parasite communities. Helminth infection in pigs was followed by significant decreases in bacterial genes related to carbohydrate metabolism in the gut, but infection by another helminth species in mice models was correlated with increases in commensal bacterial species abundance (reviewed in Glendinning et al., 2014). Factorial experiments manipulating host diet (animals) or environmental resources (plants), different community members of the microbiome, and other trophic players could yield insights into the defensive or parasitic roles of various symbionts for their host. Such experiments could furthermore facilitate the design of therapeutic treatments of parasites using microbiome manipulation.
Disturbance Ecology
Disturbance can be defined as a single disruptive event, or select set of events, that significantly changes community structure and function (Connell, 1978; Pickett, 2012). Ecological communities can respond in one of four ways to disturbance. Resistant communities experience no change in composition following the disturbance. Resilient assemblages change initially, but then return to their original state. Functionally redundant communities experience a change in species composition that has no impact on the overall community function. Finally, some communities are fundamentally altered by disturbance in both species composition and function (Allison and Martiny, 2008). Disturbances that disrupt the microbiome vary widely in magnitude and type (Figure 1E). Antibiotics, though largely considered one of modern medicines’ greatest advances, are being increasingly cited as an unprecedented type of disturbance to the human microbiome. Broad-spectrum antibiotics indiscriminately target bacterial wall components, thus eliminating many non-target commensalist and mutualistic microbes in addition to undesired bacterial pathogens (i.e., low community resistance; Lozupone et al., 2012a). Similarly, increasing the frequency of this form of antibiotic disturbance can lead to long-term shifts in the microbiome composition of healthy human adults (i.e., low community resilience; Dethlefsen and Relman, 2011). Gorilla populations from central Africa that face a higher degree of anthropogenic disturbance and habituation have compositionally distinct gut microbiomes from those populations that face less anthropogenic disturbance. Although the causal mechanism for these changes is unknown, the structural differences in gut microbiome profiles are correlated with functional changes in short-chain fatty acid and metabolite concentrations (Gomez et al., 2015).
Clearly the integration of disturbance theory in the animal microbiome remains incomplete. However, there is even less evidence documenting the effects of EF disturbance for plant health and performance. Preliminary evidence suggests that EF community composition exhibits little resistance to disturbance. Physical disturbances, such as hail storms, have been shown to decrease the diversity of foliar EF communities in the Brazilian plant Coccoloba cereifera (Fernandes et al., 2011). Other recent research shows startlingly long-term effects of physical disturbance for plant EF communities following the 2010 Deepwater Horizon Oil spill in the Gulf of Mexico. Characterization of these EF communities in smooth cordgrass showed near total loss of leaf EF, even 3 years after the original oil spill occurred (Kandalepas et al., 2015). In both of these situations, however, the functional consequences of reduced microbiome diversity remain unknown.
Future research is needed to characterize the response of host-associated microbiomes to diverse types of disturbance. This could be done by first manipulating disturbance frequency and/or intensity across a gradient and then measuring the subsequent changes in microbiome community composition and function. In humans and other animal systems, such efforts could improve our understanding of how certain pathologies are initiated in disturbed hosts. Likewise, empirical explorations of EF response to disturbance will be needed in order to make predictions about the effects of shifting anthropogenic activity and climate on plant stress tolerance and performance (Allison and Martiny, 2008; Porras-Alfaro and Bayman, 2011). Alternatively, understanding such functional consequences of microbiome disturbance could be used as a management technique for reducing the fitness of noxious or invasive plants or animals. For example, the EF microbiome of the invasive plant Phragmites australis (common reed) is currently being investigated with the ultimate intention of using disturbance as a method to perturb the microbiome and reduce this plant’s competitive ability (Kowalski et al., 2015).
Restoration Ecology
Restoration of a disturbed ecological community to its former healthy state can only be undertaken with a deep understanding of both the biology of the species to be restored and the physical nature of the habitat itself (Bradshaw, 1996). As we continue to gain a better understanding of the factors that disrupt the composition and functioning of the microbiome in animals and plants, we are increasingly left with the problem of how to effectively restore microbiome function for the host following degradation or dysbiosis. For the human microbiome, this has led to the recent development of two methodologies in particular: fecal transplantation and over-the-counter probiotics (Figure 1F; Lemon et al., 2012). Although both restoration methods involve seeding presumed beneficial microbes into the existing host microbiome, they operate in fundamentally different ways. Fecal transplant, by literal definition, is a form of restoration in which the entirety of the gastrointestinal microbiota from a healthy host is seeded into an unhealthy individual experiencing dysbiosis (Kassam et al., 2013). The procedure has gained much public interest due to its high success rate at curing Clostridium difficile infections (CDI; Jorup-Rönström et al., 2012; Kassam et al., 2013). There are some indications that fecal transplants may also benefit patients suffering from a variety of other gastrointestinal and non-gastrointestinal conditions (reviewed in Aroniadis and Brandt, 2013), but these treatments are still in the early stages of testing (Ravel et al., 2014). In contrast, the probiotic approach typically involves seeding a single strain (e.g., Lactobacillus spp.), or limited number of strains, into the degraded microbiome of an unhealthy host. The goal of probiotic ingestion or application is for the symbiont(s) to either directly target pathogenic invaders using antagonistic secretions (Buffie et al., 2014), or indirectly facilitate the transition to more desirable microbiome metabolic functioning (Lemon et al., 2012). In one successful example, the use of a probiotic, Lactobacillus delbrueckii, to treat bacterial vaginosis syndrome in women was significantly more effective at long-term restoration toward a healthy vaginal microbiome than the traditional antibiotic treatment (Ling et al., 2013). Looking forward, we suggest that the narrow definitions of fecal transplants and probiotics in medicinal therapy could be applied to other forms of dysbiosis, representing methods of community restoration using high or low species diversity, respectively.
As an example of the probiotic restoration approach in the plant–fungal system, inoculation with the EF species Trichoderma hamatum promoted seedling growth and reduced wilt in cacao plants under drought conditions (Bae et al., 2009). Similarly, inoculation with a generalist foliar EF (Alternaria spp.) was shown to alter chemical secretions in its host forb, indirectly enhancing host competitive ability (Aschehoug et al., 2014, 2012). Preliminary evidence also suggests that inoculation with certain EF species can alter biomass production in forage crops (Kleczewski et al., 2012). Whole community seeding (the fecal transplant approach) is a less-explored concept in aboveground plant microbiomes. One example, however, showed that plant leaves artificially inoculated with a cocktail of seven EF were more resistant to pathogen infection than endophyte-free leaves (Arnold et al., 2003).
An important question to the future of both medicinal and agricultural therapies is which general method of restoration is the most appropriate for different states of dysbiosis, and will require careful consideration of the causes of dysbiosis and their pathologic outcomes. Inoculation studies with various levels of microbiome diversity (e.g., none, single species/low diversity, whole community/high diversity) could qualify whether and how host or microbiome community function is restored following pathogen invasion or other sources of disturbance. Furthermore, in order to assess whether these restoration techniques are more effective than traditional antibiotic or fungicide administration, a greater integration of molecular toolsets such as transcriptomics, metagenomics, and metabolomics will be needed to assign roles to the various states of “restored” microbiomes.
Community Ecology Theory: Microbial vs. Macroorganismal Systems
Each of the six theories outlined here has provided important insights and directions for the study of macroorganismal communities. For example, successional theory predicts changes in forest communities through time (Halpern, 1989; Peterson and Pickett, 1995), while food web and multi-trophic interaction theories predict feeding relationships and patterns of energy transfer in aquatic communities (Wallace et al., 1997). However, community ecology theory has been less commonly used to understand the structure and function of microbial communities. By definition, host-associated microbiomes represent integrated communities occurring inside of a living host habitat. Therefore, the community ecology perspective may be especially relevant to microbiomes compared to other free-living microbial communities. The question remains, however, if such theories are more or less applicable to microbiomes than they are to macroorganismal communities. For example, the rapid generation times of microorganisms would likely accelerate the rate of community succession in microbiomes in comparison with macroorganismal communities. For similar reasons, microbiomes may be more resilient to external disturbances than macroorganismal communities, for which the impact of disturbance can persist for much longer. The local community assembly and metacommunity dynamics of both microbiomes and macroorganismal communities depend on the distribution, dispersal, dormancy, and extinction capabilities of their constituent species, as well as the local interactions amongst species and with their environment. More research is needed in order to determine whether microbial and macroorganismal communities differ fundamentally in the rate and magnitude of these processes. In contrast, the theory of multi-trophic interactions may be more pertinent to animal and plant macroorganismal systems due to their more complex food webs and patterns of energy transfer. In general, we do not know the relative applicability of each ecological theory to microbial and macroorganismal systems, or to human-bacterial and plant-EF systems specifically, but incorporating an ecological perspective into microbiome research will certainly help to answer this question.
Future Directions in Microbiome Ecology
For upward of 30 years, a pluralistic approach to community ecology has helped explain complex species interactions of macroorganisms. The last decade has revolutionized our characterization of formerly “unseen” microbial communities, due to significant advancements in the quality and affordability of sequencing technologies and data processing. Paradoxically, although our datasets for microbial symbionts continue to expand, we are increasingly unable to interpret the findings, primarily due to a lack of basic ecological information for individual community members (Peay, 2014). Thus, despite technological improvements, and perhaps because of them, simple characterization studies are far more common than functional assays or manipulative experiments in animal and plant microbiome studies. Moreover, many regions of the microbial tree of life remain poorly described (Arnold et al., 2000; Kyrpides et al., 2014) and recognition of high genetic and functional diversity within and among microbial isolates is often ignored in the effort to assign workable taxonomic identities (Boon et al., 2014; Shapiro and Polz, 2014). In order to move the field of Microbiome Ecology forward, we need a conceptual shift that places value not only on describing what community members are present in the microbiome, but also on understanding what the ecological roles of these community members are. We must consider microbiome studies across multiple spatial, temporal, and trophic scales in order to better understand and predict community change. Likewise, identifying sources of degradation in these symbiotic communities, and implementing changes to restore them will be crucial if we are to make use of the knowledge gained from studying our microbial partners to improve human and animal health, agricultural productivity, and maintenance of healthy ecosystems. Essentially, we face the same challenges as animal, plant, and marine ecologists of the preceding century, except that the tools to measure and describe the “unseen majority” have only become available more recently. The unification of technological advances with community ecology theory could lead to both an increased breadth of described bacterial and fungal species, as well as more consistent predictions and understanding of their functional roles in nature.
In this synthesis we have defined and described lines of inquiry for core ecological theories established over the course of the previous century, as they may be used to delineate the functional significance of symbiotic microbes. Notably, the six community ecology theories described here are not mutually exclusive, nor exhaustive. Researchers should integrate and test different theories based on their system and experimental agendas. For instance, a conservation biologist could purposefully disrupt the microbiome of an invasive species using the principles of disturbance ecology, while simultaneously working to restore the microbiomes of native species and using the metacommunity framework to track microbial dispersal across habitats. On the other hand, a medical researcher might place greater emphasis on community assembly and successional theories as methods to predict which lifestyle factors alter microbial colonization in humans. Or similarly, how bacterial communities in the gut respond to multi-trophic interactions with viruses or other eukaryotes. In this way, disease onset as a result of microbial dysbiosis could be predicted or prevented. We believe that microbiome studies are at a critical turning point, moving from a simply descriptive phase into one that uses ecological principals and experimental manipulation to achieve better understanding and application. In order to move this emerging field forward, microbiome researchers and applied scientists alike must collaborate and communicate the theories and results of their respective fields. Community ecology should then be seen as fulfilling an important niche amongst a broad array of disciplines: a science developed to understand complex species interactions and make cross-system comparisons.
Author Contributions
Both NC and BW equally drafted and edited this manuscript, with substantial editorial remarks from KC.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Irene G. Newton, Jay Lennon, Evie Rynkiewicz, and several members of the Clay Lab, in particular Luke P. Henry, Zackery Shearin, and Chai Qing for their feedback and comments on the manuscript. BW was supported by an NIH Genetics, Cellular and Molecular Sciences (GCMS) Training Grant during the time of writing and NC was supported by an NSF Graduate Research Fellowship. Additionally, we thank the reviewers for their constructive comments. This publication was financially supported through USGS Grant G13AC00285 and NSF Grant DEB-03268742.
References
Adame-Álvarez, R.-M. M., Mendiola-Soto, J., and Heil, M. (2014). Order of arrival shifts endophyte-pathogen interactions in bean from resistance induction to disease facilitation. FEMS Microbiol. Lett. 355, 100–107. doi: 10.1111/1574-6968.12454
Agrawal, A. A., Ackerly, D. D., Adler, F., Arnold, A. E., Caceres, C., Doak, D. F., et al. (2007). Filling key gaps in population and community ecology. Front. Ecol. Environ. 5:145–152. doi: 10.1890/1540-9295(2007)5[145:FKGIPA]2.0.CO;2
Alabouvette, C., Olivain, C., Migheli, Q., and Steinberg, C. (2009). Microbiological control of soil-borne phytopathogenic fungi with special emphasis on wilt-inducing Fusarium oxysporum. New Phytol. 184, 529–544. doi: 10.1111/j.1469-8137.2009.03014.x
Allison, S. D., and Martiny, J. B. (2008). Resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. U.S.A. 105, 11512–11519. doi: 10.1073/pnas.0801925105
Al Rwahnih, M., Daubert, S., Urbez-Torres, J. R., Cordero, F., and Rowhani, A. (2011). Deep sequencing evidence from single grapevine plants reveals a virome dominated by mycoviruses. Arch. Virol. 156, 397–403. doi: 10.1007/s00705-010-0869-8
Arnold, A. E., and Engelbrecht, B. M. J. (2007). Fungal endophytes nearly double minimum leaf conductance in seedlings of a neotropical tree species. J. Trop. Ecol. 23, 369–372. doi: 10.1017/S0266467407004038
Arnold, A. E., Maynard, Z., Gilbert, G. S., Coley, P. D., and Kursar, T. A. (2000). Are tropical fungal endophytes hyperdiverse? Ecol. Lett. 3, 267–274. doi: 10.1046/j.1461-0248.2000.00159.x
Arnold, A. E., Mejía, L. C., Kyllo, D., Rojas, E. I., Maynard, Z., Robbins, N., et al. (2003). Fungal endophytes limit pathogen damage in a tropical tree. Proc. Natl. Acad. Sci. U.S.A. 100, 15649–15654. doi: 10.1073/pnas.2533483100
Arnold, A., and Herre, E. (2003). Canopy cover and leaf age affect colonization by tropical fungal endophytes: ecological pattern and process in Theobroma cacao (Malvaceae). Mycologia 95, 388–398. doi: 10.2307/3761880
Aroniadis, O. C., and Brandt, L. J. (2013). Fecal microbiota transplantation: past, present and future. Curr. Opin. Gastroenterol. 29, 79–84. doi: 10.1097/MOG.0b013e32835a4b3e
Aschehoug, E., Metlen, K., Callaway, R., and Newcombe, G. (2012). Fungal endophytes directly increase the competitive effects of an invasive forb. Ecology 93, 3–8. doi: 10.1890/11-1347.1
Aschehoug, E. T., Callaway, R. M., Newcombe, G., Tharayil, N., and Chen, S. (2014). Fungal endophyte increases the allelopathic effects of an invasive forb. Oecologia 175, 285–291. doi: 10.1007/s00442-014-2891-0
Azad, M. B., Konya, T., Maughan, H., Guttman, D. S., Field, C. J., Sears, M. R., et al. (2013). Infant gut microbiota and the hygiene hypothesis of allergic disease: impact of household pets and siblings on microbiota composition and diversity. Allergy Asthma Clin. Immunol. 9, 15. doi: 10.1186/1710-1492-9-15
Bae, H., Sicher, R. C., Kim, M. S., Kim, S.-H., Strem, M. D., Melnick, R. L., et al. (2009). The beneficial endophyte Trichoderma hamatum isolate DIS 219b promotes growth and delays the onset of the drought response in Theobroma cacao. J. Exp. Bot. 60, 3279–3295. doi: 10.1093/jxb/erp165
Barthélémy, D., and Caraglio, Y. (2007). Plant architecture: a dynamic, multilevel and comprehensive approach to plant form, structure and ontogeny. Ann. Bot. 99, 375–407. doi: 10.1093/aob/mcl260
Bayman, P. (2006). “Diversity, scale and variation of endophytic fungi in leaves of tropical plants,” in Microbial Ecology of Aerial Plant Surfaces, eds M. J. Bailey, A. K. Lilley, and T. M. Timms-Wilson (Cambridge: CABI International), 37–50.
Beattie, G. A., and Lindow, S. E. (1995). The secret life of foliar bacterial pathogens on leaves. Annu. Rev. Phytopathol. 33, 145–172. doi: 10.1146/annurev.py.33.090195.001045
Begon, M., Townsend, C. R., and Harper, J. L. (2006). Ecology: From Individuals to Ecosystems. 4th Edn. Malden, MA: Blackwell Publishing, Ltd.
Bergström, A., Skov, T. H., Bahl, M. I., Roager, H. M., Christensen, L. B., Ejlerskov, K. T., et al. (2014). Establishment of intestinal microbiota during early life: a longitudinal, explorative study of a large cohort of Danish infants. Appl. Environ. Microbiol. 80, 2889–2900. doi: 10.1128/AEM.00342-14
Boon, E., Meehan, C. J., Whidden, C., Wong, D. H.-J., Langille, M. G. I., and Beiko, R. G. (2014). Interactions in the microbiome: communities of organisms and communities of genes. FEMS Microbiol. Rev. 38, 90–118. doi: 10.1111/1574-6976.12035
Bradshaw, A. (1996). Underlying principles of restoration. Can. J. Fish. Aquat. Sci. 53, 3–9. doi: 10.1139/f95-265
Brotman, R. M., Bradford, L. L., Conrad, M., Gajer, P., Ault, K., Peralta, L., et al. (2012). Association between Trichomonas vaginalis and vaginal bacterial community composition among reproductive-age women. Sex Transm. Dis. 39, 807–812. doi: 10.1097/OLQ.0b013e3182631c79
Buck, K. W., and Brasier, C. M. (2001). “Viruses of the Dutch Elm disease fungi,” in dsRNA Genetic Elements: Concepts and Applications in Agriculture, Forestry, and Medicine, ed. S. M. Tavantzis (Boca Raton, FL: CRC Press), 165–190.
Buffie, C. G., Bucci, V., Stein, R. R., McKenney, P. T., Ling, L., Gobourne, A., et al. (2014). Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208. doi: 10.1038/nature13828
Bulgarelli, D., Rott, M., Schlaeppi, K., Ver Loren van Themaat, E., Ahmadinejad, N., Assenza, F., et al. (2012). Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91–95. doi: 10.1038/nature11336
Bulgarelli, D., Schlaeppi, K., Spaepen, S., Ver Loren van Themaat, E., and Schulze-Lefert, P. (2013). Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 64, 807–838. doi: 10.1146/annurev-arplant-050312-120106
Carroll, G. (1988). Fungal endophytes in stems and leaves: from latent pathogen to mutualistic symbiont. Ecology 69, 2–9.
Chen, Y. E., and Tsao, H. (2013). The skin microbiome: current perspectives and future challenges. J. Am. Acad. Dermatol. 69, 143–155. doi: 10.1016/j.jaad.2013.01.016
Chu, Y.-M., Jeon, J.-J., Yea, S.-J., Kim, Y.-H., Yun, S.-H., Lee, Y.-W., et al. (2002). Double-stranded RNA mycovirus from Fusarium graminearum. Appl. Environ. Microbiol. 68, 2529–2534. doi: 10.1128/AEM.68.5.2529-2534.2002
Clay, K. (1988). Fungal endophytes of grasses: a defensive mutualism between plants and fungi. Ecology 69, 10–16.
Clay, K., and Schardl, C. (2002). Evolutionary origins and ecological consequences of endophyte symbiosis with grasses. Am. Nat. 160, S99–S127. doi: 10.1086/342161
Clements, F. E. (1916). Plant succession, an analysis of the development of vegetation. Carnegie Inst. Washingt. 242, 1–512.
Cockburn, S. N., Haselkorn, T. S., Hamilton, P. T., Landzberg, E., Jaenike, J., and Perlman, S. J. (2013). Dynamics of the continent-wide spread of a Drosophila defensive symbiont. Ecol. Lett. 16, 609–616. doi: 10.1111/ele.12087
Connell, J. H., and Slatyer, R. O. (1977). Mechanisms of succession in natural communities and their role in community stability and organization. Am. Nat. 111, 1119–1144.
Costello, E. K., Lauber, C. L., Hamady, M., Fierer, N., Gordon, J. I., and Knight, R. (2009). Bacterial community variation in human body habitats across space and time. Science 326, 1694–1697. doi: 10.1126/science.1177486
Costello, E. K., Stagaman, K., Dethlefsen, L., Bohannan, B. J. M., and Relman, D. A. (2012). The application of ecological theory toward an understanding of the human microbiome. Science 336, 1255–1262. doi: 10.1126/science.1224203
Davey, M. L., Nybakken, L., Kauserud, H., and Ohlson, M. (2009). Fungal biomass associated with the phyllosphere of bryophytes and vascular plants. Mycol. Res. 113, 1254–1260. doi: 10.1016/j.mycres.2009.08.001
Dethlefsen, L., and Relman, D. A. (2011). Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl. Acad. Sci. U.S.A. 108, 4554–4561. doi: 10.1073/pnas.1000087107
Dominguez-Bello, M. G., Costello, E. K., Contreras, M., Magris, M., Hidalgo, G., Fierer, N., et al. (2010). Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. U.S.A. 107, 11971–11975. doi: 10.1073/pnas.1002601107
Ellis, A. M. A., Lounibos, L. P., and Holyoak, M. (2006). Evaluating the long-term metacommunity dynamics of tree hole mosquitoes. Ecology 87, 2582–2590. doi: 10.1890/0012-9658(2006)87[2582:ETLMDO]2.0.CO;2
Fernandes, G. W., Oki, Y., Sanchez-Azofeifa, A., Faccion, G., and Amaro-Arruda, H. C. (2011). Hail impact on leaves and endophytes of the endemic threatened Coccoloba cereifera (Polygonaceae). Plant Ecol. 212, 1687–1697. doi: 10.1007/s11258-011-9941-z
Frank, D. N., St. Amand, A. L., Feldman, R. A., Boedeker, E. C., Harpaz, N., and Pace, N. R. (2007). Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U.S.A. 104, 13780–13785. doi: 10.1073/pnas.0706625104
Fukuda, S., Toh, H., Hase, K., Oshima, K., Nakanishi, Y., Yoshimura, K., et al. (2011). Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469, 543–547. doi: 10.1038/nature09646
Gamboa, M. A., Laureano, S., and Bayman, P. (2002). Measuring diversity of endophytic fungi in leaf fragments: does size matter? Mycopathologia 156, 41–45. doi: 10.1023/A:1021362217723
Ganley, R. J., Sniezko, R. A., and Newcombe, G. (2008). Endophyte-mediated resistance against white pine blister rust in Pinus monticola. For. Ecol. Manage. 255, 2751–2760. doi: 10.1016/j.foreco.2008.01.052
Gans, J., Wolinsky, M., and Dunbar, J. (2005). Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science 309, 1387–1390. doi: 10.1126/science.1126853
Gleason, H. (1926). The individualistic concept of the plant association. Bull. Torrey Bot. Club 53, 7–26.
Glendinning, L., Nausch, N., Free, A., Taylor, D. W., and Mutapi, F. (2014). The microbiota and helminths: sharing the same niche in the human host. Parasitology 141, 1255–1271. doi: 10.1017/S0031182014000699
Gomez, A., Petrzelkova, K., Yeoman, C. J., Vlckova, K., Koppova, I., Carbonero, F., et al. (2015). Gut microbiome composition and metabolomic profiles of wild western lowland gorillas (Gorilla gorilla gorilla) reflect host ecology. Mol. Ecol. 24, 2551–2565. doi: 10.1111/mec.13181
Grice, E. A., Kong, H. H., Conlan, S., Deming, C. B., Davis, J., Young, A. C., et al. (2009). Topographical and temporal diversity of the human skin microbiome. Science 324, 1190–1192. doi: 10.1126/science.1171700
Grice, E. A., and Segre, J. A. (2011). The skin microbiome. Nat. Rev. Microbiol. 9, 244–253. doi: 10.1038/nrmicro2537
Hallmann, J., Quadt-Hallmann, A., Mahaffee, W. F., and Kloepper, J. W. (1997). Bacterial endophytes in agricultural crops. Can. J. Microbiol. 43, 895–914. doi: 10.1139/m97-131
Halpern, C. B. (1989). Early successional patterns of forest species: interactions of life history traits and disturbance. Ecology 70, 704–720.
Harrison, S., and Cornell, H. (2008). Toward a better understanding of the regional causes of local community richness. Ecol. Lett. 11, 969–979. doi: 10.1111/j.1461-0248.2008.01210.x
Hartley, S. E., Eschen, R., Horwood, J. M., Gange, A. C., and Hill, E. M. (2015). Infection by a foliar endophyte elicits novel arabidopside-based plant defence reactions in its host, Cirsium arvense. New Phytol. 205, 816–827. doi: 10.1111/nph.13067
Heiniger, U., and Rigling, D. (1994). Biological control of chestnut blight. Annu. Rev. Phytopathol. 32, 581–599. doi: 10.1126/science.215.4532.466
Herre, E. A., Mejía, L. C., Kyllo, D. A., Rojas, E., Maynard, Z., Butler, A., et al. (2007). Ecological implications of anti-pathogen effects of tropical fungal endophytes and mycorrhizae. Ecology 88, 550–558. doi: 10.1890/05-1606
Herrera, C. M., de Vega, C., Canto, A., and Pozo, M. I. (2009). Yeasts in floral nectar: a quantitative survey. Ann. Bot. 103, 1415–1423. doi: 10.1093/aob/mcp026
Higgins, K. L., Arnold, A. E., Miadlikowska, J., Sarvate, S. D., and Lutzoni, F. (2007). Phylogenetic relationships, host affinity, and geographic structure of boreal and arctic endophytes from three major plant lineages. Mol. Phylogenet. Evol. 42, 543–555. doi: 10.1016/j.ympev.2006.07.012
Hinde, K., and Lewis, Z. T. (2015). Mother’s littlest helpers. Science 348, 1427–1428. doi: 10.1126/science.aac7436
Hirose, D., Matsuoka, S., and Osono, T. (2013). Assessment of the fungal diversity and succession of ligninolytic endophytes in Camellia japonica leaves using clone library analysis. Mycologia 105, 837–843. doi: 10.3852/12-385
Hodgson, S., de Cates, C., Hodgson, J., Morley, N. J., Sutton, B. C., and Gange, A. C. (2014). Vertical transmission of fungal endophytes is widespread in forbs. Ecol. Evol. 4, 1199–1208. doi: 10.1002/ece3.953
Hoffman, M. T., Gunatilaka, M. K., Wijeratne, K., Gunatilaka, L., and Arnold, A. E. (2013). Endohyphal bacterium enhances production of indole-3-acetic acid by a foliar fungal endophyte. PLoS ONE 8:e73132. doi: 10.1371/journal.pone.0073132
Hollister, E. B., Gao, C., and Versalovic, J. (2014). Compositional and functional features of the gastrointestinal microbiome and their effects on human health. Gastroenterology 146, 1449–1458. doi: 10.1053/j.gastro.2014.01.052
Holyoak, M., Leibold, M. A., Mouquet, N., Holt, R. D., and Hoopes, M. F. (2005). “Metacommunities: a framework for large-scale community ecology,” in Metacommunities: Spatial Dynamics and Ecological Communities, eds M. Holyoak, M. A. Leibold, N. Mouquet, and R. D. Holt (Chicago, IL: University of Chicago Press), 1–31.
Hooper, L. V., Littman, D. R., and Macpherson, A. J. (2012). Interactions between the microbiota and the immune system. Science 336, 1268–1273. doi: 10.1126/science.1223490
Huffnagle, G. B., and Noverr, M. C. (2013). The emerging world of the fungal microbiome. Trends Microbiol. 21, 334–341. doi: 10.1016/j.tim.2013.04.002
Jami, E., Israel, A., Kotser, A., and Mizrahi, I. (2013). Exploring the bovine rumen bacterial community from birth to adulthood. ISME J. 7, 1069–1079. doi: 10.1038/ismej.2013.2
Jorup-Rönström, C., Håkanson, A., Sandell, S., Edvinsson, O., Midtvedt, T., Persson, A.-K., et al. (2012). Fecal transplant against relapsing Clostridium difficile-associated diarrhea in 32 patients. Scand. J. Gastroenterol. 47, 548–552. doi: 10.3109/00365521.2012.672587
Kandalepas, D., Blum, M. J., and Van Bael, S. A. (2015). Shifts in symbiotic endophyte communities of a foundational salt marsh grass following oil exposure from the Deepwater Horizon oil spill. PLoS ONE 10:e0122378. doi: 10.1371/journal.pone.0122378
Kaneko, R., and Kakishima, M. (2001). Mycosphaerella buna sp. nov. with a Pseudocercospora anamorph isolated from the leaves of Japanese beech. Mycoscience 42, 59–66. doi: 10.1007/BF02463976
Kaneko, R., and Kaneko, S. (2004). The effect of bagging branches on levels of endophytic fungal infection in Japanese beech leaves. For. Pathol. 34, 65–78. doi: 10.1111/j.1439-0329.2004.00350.x
Kassam, Z., Lee, C. H., Yuan, Y., and Hunt, R. H. (2013). Fecal microbiota transplantation for Clostridium difficile infection: systematic review and meta-analysis. Am. J. Gastroenterol. 108, 500–508. doi: 10.1038/ajg.2013.59
Kiers, E. T., Rousseau, R. A., West, S. A., and Denison, R. F. (2003). Host sanctions and the legume-rhizobium mutualism. Nature 425, 78–81. doi: 10.1038/nature01931
Kleczewski, N. M., Bauer, J. T., Bever, J. D., Clay, K., and Reynolds, H. L. (2012). A survey of endophytic fungi of switchgrass (Panicum virgatum) in the Midwest, and their putative roles in plant growth. Fungal Ecol. 5, 521–529. doi: 10.1016/j.funeco.2011.12.006
Klimesová, J., and Klimeš, L. (2007). Bud banks and their role in vegetative regeneration—A literature review and proposal for simple classification and assessment. Perspect. Plant Ecol. Evol. Syst. 8, 115–129. doi: 10.1016/j.ppees.2006.10.002
Koenig, J. E., Spor, A., Scalfone, N., Fricker, A. D., Stombaugh, J., Knight, R., et al. (2011). Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. U.S.A. 108, 4578–4585. doi: 10.1073/pnas.1000081107
Kolenbrander, P. E. (2011). Multispecies communities: interspecies interactions influence growth on saliva as sole nutritional source. Int. J. Oral Sci. 3, 49–54. doi: 10.4248/IJOS11025
Konya, T., Koster, B., Maughan, H., Escobar, M., Azad, M. B., Guttman, D. S., et al. (2014). Associations between bacterial communities of house dust and infant gut. Environ. Res. 131, 25–30. doi: 10.1016/j.envres.2014.02.005
Koukol, O., Kolařík, M., Kolářová, Z., and Baldrian, P. (2011). Diversity of foliar endophytes in wind-fallen Picea abies trees. Fungal Divers. 54, 69–77. doi: 10.1007/s13225-011-0112-2
Kowalski, K. P., Bacon, C., Bickford, W., Braun, H., Clay, K., Leduc-Lapierre, M., et al. (2015). Advancing the science of microbial symbiosis to support invasive species management: a case study on Phragmites in the Great Lakes. Front. Microbiol. 6:95. doi: 10.3389/fmicb.2015.00095
Krings, M., Taylor, T. N., Hass, H., Kerp, H., Dotzler, N., and Hermsen, E. J. (2007). Fungal endophytes in a 400-million-yr-old land plant: infection pathways, spatial distribution, and host responses. New Phytol. 174, 648–657. doi: 10.1111/j.1469-8137.2007.02008.x
Krom, B. P., Kidwai, S., and ten Cate, J. M. (2014). Candida and other fungal species: forgotten players of healthy oral microbiota. J. Dent. Res. 93, 445–451. doi: 10.1177/0022034514521814
Kuramitsu, H. K., He, X., Lux, R., Anderson, M. H., and Shi, W. (2007). Interspecies interactions within oral microbial communities. Microbiol. Mol. Biol. Rev. 71, 653–670. doi: 10.1128/MMBR.00024-07
Kyrpides, N. C., Hugenholtz, P., Eisen, J. A., Woyke, T., Göker, M., Parker, C. T., et al. (2014). Genomic encyclopedia of bacteria and archaea: sequencing a myriad of type strains. PLoS Biol. 12:e1001920. doi: 10.1371/journal.pbio.1001920
Lawson, S. P., Christian, N., and Abbot, P. (2014). Comparative analysis of the biodiversity of fungal endophytes in insect-induced galls and surrounding foliar tissue. Fungal Divers. 66, 86–97. doi: 10.1007/s13225-013-0268-z
Lecuit, M., and Eloit, M. (2013). The human virome: new tools and concepts. Trends Microbiol. 21, 510–515. doi: 10.1016/j.tim.2013.07.001
Lee, K., Pan, J. J., and May, G. (2009). Endophytic Fusarium verticillioides reduces disease severity caused by Ustilago maydis on maize. FEMS Microbiol. Lett. 299, 31–37. doi: 10.1111/j.1574-6968.2009.01719.x
Leff, J. W., Del Tredici, P., Friedman, W. E., and Fierer, N. (2015). Spatial structuring of bacterial communities within individual Ginkgo biloba trees. Environ. Microbiol. V17, 2352–2361. doi: 10.1111/1462-2920.12695
Leff, J. W., and Fierer, N. (2013). Bacterial communities associated with the surfaces of fresh fruits and vegetables. PLoS ONE 8:e59310. doi: 10.1371/journal.pone.0059310
Leibold, M. A., Holyoak, M., Mouquet, N., Amarasekare, P., Chase, J. M., Hoopes, M. F., et al. (2004). The metacommunity concept: a framework for multi-scale community ecology. Ecol. Lett. 7, 601–613. doi: 10.1111/j.1461-0248.2004.00608.x
Leibold, M. A., and McPeek, M. A. (2006). Coexistence of the niche and neutral perspectives in community ecology. Ecology 87, 1399–1410. doi: 10.1890/0012-9658(2006)87[1399:COTNAN]2.0.CO;2
Lemon, K. P., Armitage, G. C., Relman, D. A., and Fischbach, M. A. (2012). Microbiota-targeted therapies: an ecological perspective. Sci. Transl. Med. 4, 137rv5. doi: 10.1126/scitranslmed.3004183
Lepage, P., Leclerc, M. C., Joossens, M., Mondot, S., Blottière, H. M., Raes, J., et al. (2013). A metagenomic insight into our gut’s microbiome. Gut 62, 146–158. doi: 10.1136/gutjnl-2011-301805
Ley, R., Lozupone, C., Hamady, M., Knight, R., and Gordon, J. I. (2008). Worlds within worlds: evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 6, 776–788. doi: 10.1038/nrmicro1978
Ling, Z., Liu, X., Chen, W., Luo, Y., Yuan, L., Xia, Y., et al. (2013). The restoration of the vaginal microbiota after treatment for bacterial vaginosis with metronidazole or probiotics. Microb. Ecol. 65, 773–780. doi: 10.1007/s00248-012-0154-3
Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K., and Knight, R. (2012a). Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230. doi: 10.1038/nature11550
Lozupone, C., Faust, K., Raes, J., Faith, J. J., Frank, D. N., Zaneveld, J., et al. (2012b). Identifying genomic and metabolic features that can underlie early successional and opportunistic lifestyles of human gut symbionts. Genome Res. 22, 1974–1984. doi: 10.1101/gr.138198.112
Lundberg, D. S., Lebeis, S. L., Paredes, S. H., Yourstone, S., Gehring, J., Malfatti, S., et al. (2012). Defining the core Arabidopsis thaliana root microbiome. Nature 488, 86–90. doi: 10.1038/nature11237
Martín, J. A., Witzell, J., Blumenstein, K., Rozpedowska, E., Helander, M., Sieber, T. N., et al. (2013). Resistance to Dutch elm disease reduces presence of xylem endophytic fungi in Elms (Ulmus spp.). PLoS ONE 8:e56987. doi: 10.1371/journal.pone.0056987
Martinson, E. O., Herre, E. A., Machado, C. A., and Arnold, A. E. (2012). Culture-free survey reveals diverse and distinctive fungal communities associated with developing figs (Ficus spp.) in Panama. Microb. Ecol. 64, 1073–1084. doi: 10.1007/s00248-012-0079-x
Matamoros, S., Gras-Leguen, C., Le Vacon, F., Potel, G., and de La Cochetiere, M.-F. (2013). Development of intestinal microbiota in infants and its impact on health. Trends Microbiol. 21, 167–173. doi: 10.1016/j.tim.2012.12.001
McCormack, J. E., Hird, S. M., Zellmer, A. J., Carstens, B. C., and Brumfield, R. T. (2013). Applications of next-generation sequencing to phylogeography and phylogenetics. Mol. Phylogenet. Evol. 66, 526–538. doi: 10.1016/j.ympev.2011.12.007
Mcfall-Ngai, M. (2007). Adaptive immunity: care for the community. Nature 445, 153–153. doi: 10.1038/445153a
McFall-Ngai, M., Hadfield, M. G., Bosch, T. C. G., Carey, H. V., Domazet-Lošo, T., Douglas, A. E., et al. (2013). Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. U.S.A. 110, 3229–3236. doi: 10.1073/pnas.1218525110
Mejia, L. C., Herre, E. A., Sparks, J. P., Winter, K., Garcia, M. N., Van Bael, S. A., et al. (2014). Pervasive effects of a dominant foliar endophytic fungus on host genetic and phenotypic expression in a tropical tree. Front. Microbiol. 5:479. doi: 10.3389/fmicb.2014.00479
Mihaljevic, J. R. (2012). Linking metacommunity theory and symbiont evolutionary ecology. Trends Ecol. Evol. 27, 323–329. doi: 10.1016/j.tree.2012.01.011
Mishra, A., Gond, S. K., Kumar, A., Sharma, V. K., Verma, S. K., Kharwar, R. N., et al. (2012). Season and tissue type affect fungal endophyte communities of the Indian medicinal plant Tinospora cordifolia more strongly than geographic location. Microb. Ecol. 64, 388–398. doi: 10.1007/s00248-012-0029-7
Molles Jr., M. C. (2013). Ecology: Concepts and Applications. 6th Edn. Boston, MA: McGraw-Hill Publishing Company.
Naik, S., Bouladoux, N., Wilhelm, C., Molloy, M. J., Salcedo, R., Kastenmuller, W., et al. (2012). Compartmentalized control of skin immunity by resident commensals. Science 337, 1115–1119. doi: 10.1126/science.1225152
Nelson, M. S., and Sadowsky, M. J. (2015). Secretion systems and signal exchange between nitrogen-fixing rhizobia and legumes. Front. Plant Sci. 6:491. doi: 10.3389/fpls.2015.00491
O’Dwyer, J. P., Kembel, S. W., and Green, J. L. (2012). Phylogenetic diversity theory sheds light on the structure of microbial communities. PLoS Comput. Biol. 8:e1002832. doi: 10.1371/journal.pcbi.1002832
Oliver, K. M., Smith, A. H., and Russell, J. A. (2014). Defensive symbiosis in the real world—advancing ecological studies of heritable, protective bacteria in aphids and beyond. Funct. Ecol. 28, 341–355. doi: 10.1111/1365-2435.12133
Osono, T., and Mori, A. (2003). Colonization of Japanese beech leaves by phyllosphere fungi. Mycoscience 44, 437–441. doi: 10.1007/s10267-003-0135-y
Osorio, M., and Stephan, B. R. (1991). Life cycle of Lophodermium piceae in Norway spruce needles. Eur. J. Plant Pathol. 21, 152–163.
Ottman, N., Smidt, H., Vos De, W. M., and Belzer, C. (2012). The function of our microbiota: who is out there and what do they do? Front. Cell. Infect. Microbiol. 2:104. doi: 10.3389/fcimb.2012.00104
Panaccione, D. G., Beaulieu, W. T., and Cook, D. (2014). Bioactive alkaloids in vertically transmitted fungal endophytes. Funct. Ecol. 28, 299–314. doi: 10.1111/1365-2435.12076
Pan, J. J., and May, G. (2009). Fungal–fungal associations affect the assembly of endophyte communities in maize (Zea mays). Microb. Ecol. 58, 668–678. doi: 10.1007/s00248-009-9543-7
Parfrey, L. W., Walters, W. A., Lauber, C. L., Clemente, J. C., Berg-Lyons, D., Teiling, C., et al. (2014). Communities of microbial eukaryotes in the mammalian gut within the context of environmental eukaryotic diversity. Front. Microbiol. 5:298. doi: 10.3389/fmicb.2014.00298
Peay, K. G. (2014). Back to the future: natural history and the way forward in modern fungal ecology. Fungal Ecol. 12, 4–9. doi: 10.1016/j.funeco.2014.06.001
Peiffer, J. A., Spor, A., Koren, O., Jin, Z., Tringe, S. G., Dangl, J. L., et al. (2013). Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. 110, 6548–6553. doi: 10.1073/pnas.1302837110
Penders, J., Thijs, C., Vink, C., Stelma, F. F., Snijders, B., Kummeling, I., et al. (2006). Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 118, 511–521. doi: 10.1542/peds.2005-2824
Peterson, C. J., and Pickett, S. T. A. (1995). Forest reorganization: a case study in an old-growth forest catastrophic blowdown. Ecology 76, 763–774.
Philippot, L., Raaijmakers, J. M., Lemanceau, P., and van der Putten, W. H. (2013). Going back to the roots: the microbial ecology of the rhizosphere. Nature 11, 789–799. doi: 10.1038/nrmicro3109
Pickett, S. T. (2012). The Ecology of Natural Disturbance and Patch Dynamics. New York: Academic Press.
Picot, A., Hourcade-Marcolla, D., Barreau, C., Pinson-Gadais, L., Caron, D., Richard-Forget, F., et al. (2012). Interactions between Fusarium verticillioides and Fusarium graminearum in maize ears and consequences for fungal development and mycotoxin accumulation. Plant Pathol. 61, 140–151. doi: 10.1111/j.1365-3059.2011.02503.x
Porras-Alfaro, A., and Bayman, P. (2011). Hidden fungi, emergent properties: endophytes and microbiomes. Annu. Rev. Phytopathol. 49, 291–315. doi: 10.1146/annurev-phyto-080508-081831
Qin, J., Li, R., Raes, J., Arumugam, M., Burgdorf, K. S., Manichanh, C., et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65. doi: 10.1038/nature08821
Raaijmakers, J. M., Paulitz, T. C., Steinberg, C., Alabouvette, C., and Moënne-Loccoz, Y. (2008). The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 321, 341–361. doi: 10.1007/s11104-008-9568-6
Ravel, J., Blaser, M. J., Braun, J., Brown, E., Bushman, F. D., Chang, E. B. et al. (2014). Human Microbiome Science: Vision for the Future. Bethesda, MD: Microbiome. doi: 10.1186/2049-2618-2-16
Rodriguez Estrada, A. E., Hegeman, A., Kistler, H. C., and May, G. (2011). In vitro interactions between Fusarium verticillioides and Ustilago maydis through real-time PCR and metabolic profiling. Fungal Genet. Biol. 48, 874–885. doi: 10.1016/j.fgb.2011.06.006
Rodriguez, R. J., Henson, J., Van Volkenburgh, E., Hoy, M., Wright, L., Beckwith, F., et al. (2008). Stress tolerance in plants via habitat-adapted symbiosis. ISME J. 2, 404–416. doi: 10.1038/ismej.2007.106
Rodriguez, R. J., White, J. F. Jr., Arnold, A. E., and Redman, R. S. (2009). Fungal endophytes: diversity and functional roles. New Phytol. 182, 314–330. doi: 10.1111/j.1469-8137.2009.02773.x
Rodriguez-Valera, F., Martin-Cuadrado, A.-B., Rodriguez-Brito, B., Pasić, L., Thingstad, T. F., Rohwer, F., et al. (2009). Explaining microbial population genomics through phage predation. Nat. Rev. Microbiol. 7, 828–836. doi: 10.1038/nrmicro2235
Rosenblueth, M., and Martínez-Romero, E. (2006). Bacterial endophytes and their interactions with hosts. Mol. Plant Microbe Interact. 19, 827–837. doi: 10.1094/MPMI-19-0827
Ryan, R. P., Germaine, K., Franks, A., Ryan, D. J., and Dowling, D. N. (2008). Bacterial endophytes: recent developments and applications. FEMS Microbiol. Lett. 278, 1–9. doi: 10.1111/j.1574-6968.2007.00918.x
Sandberg, D. C., Battista, L. J., and Arnold, A. E. (2014). Fungal endophytes of aquatic macrophytes: diverse host-generalists characterized by tissue preferences and geographic structure. Microb. Ecol. 67, 735–747. doi: 10.1007/s00248-013-0324-y
Santamaria, O., and Diez, J. J. (2005). Fungi in leaves, twigs and stem bark of Populus tremula from northern Spain. For. Pathol. 35, 95–104. doi: 10.1111/j.1439-0329.2004.00389.x
Schnorr, S. L., Candela, M., Rampelli, S., Centanni, M., Consolandi, C., Basaglia, G., et al. (2014). Gut microbiome of the Hadza hunter-gatherers. Nat. Commun. 5:3654. doi: 10.1038/ncomms4654
Schulz, B., and Boyle, C. (2005). The endophytic continuum. Mycol. Res. 109, 661–686. doi: 10.1017/S095375620500273X
Schwabe, R. F., and Jobin, C. (2013). The microbiome and cancer. Nat. Rev. Cancer 13, 800–812. doi: 10.1038/nrc3610
Shade, A., McManus, P. S., and Handelsman, J. (2013). Unexpected diversity during community succession in the apple flower microbiome. MBio 4:e00602–12. doi: 10.1128/mBio.00602-12
Shapiro, B. J., and Polz, M. F. (2014). Ordering microbial diversity into ecologically and genetically cohesive units. Trends Microbiol. 22, 235–247. doi: 10.1016/j.tim.2014.02.006
Sharon, I., Morowitz, M. J., Thomas, B. C., Costello, E. K., Relman, D. A., and Banfield, J. F. (2013). Time series community genomics analysis reveals rapid shifts in bacterial species, strains, and phage during infant gut colonization. Genome Res. 23, 111–120. doi: 10.1101/gr.142315.112
Smith, M. I., Yatsunenko, T., Manary, M. J., Trehan, I., Mkakosya, R., Cheng, J., et al. (2013). Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science 339, 548–554. doi: 10.1126/science.1229000
Sommer, F., and Bäckhed, F. (2013). The gut microbiota–masters of host development and physiology. Nat. Rev. Microbiol. 11, 227–238. doi: 10.1038/nrmicro2974
Song, S. J., Lauber, C., Costello, E. K., Lozupone, C. A., Humphrey, G., Berg-Lyons, D., et al. (2013). Cohabiting family members share microbiota with one another and with their dogs. Elife 2, e00458. doi: 10.7554/eLife.00458
Springer, J. C., Davelos Baines, A. L., Fulbright, D. W., Chansler, M. T., and Jarosz, A. M. (2013). Hyperparasites influence population structure of the chestnut blight pathogen, Cryphonectria parasitica. Phytopathology 103, 1280–1286. doi: 10.1094/PHYTO-10-12-0273-R
Statnikov, A., Alekseyenko, A. V, Li, Z., Henaff, M., Perez-Perez, G. I., Blaser, M. J., and Aliferis, C. F. (2013). Microbiomic signatures of psoriasis: feasibility and methodology comparison. Sci. Rep. 3, 2620. doi: 10.1038/srep02620
Stearns, J. C., Lynch, M. D. J., Senadheera, D. B., Tenenbaum, H. C., Goldberg, M. B., Cvitkovitch, D. G., et al. (2011). Bacterial biogeography of the human digestive tract. Sci. Rep. 1, 170. doi: 10.1038/srep00170
Strobel, G., and Daisy, B. (2003). Bioprospecting for microbial endophytes and their natural products. Microbiol. Mol. Biol. Rev. 67, 491–502.
The Human Microbiome Project Consortium. (2012a). A framework for human microbiome research. Nature 486, 215–221. doi: 10.1038/nature11209
The Human Microbiome Project Consortium. (2012b). Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. doi: 10.1038/nature11234
U’Ren, J. M., Lutzoni, F., Miadlikowska, J., Laetsch, A. D., and Arnold, A. E. (2012). Host and geographic structure of endophytic and endolichenic fungi at a continental scale. Am. J. Bot. 99, 898–914. doi: 10.3732/ajb.1100459
Voříšková, J., and Baldrian, P. (2013). Fungal community on decomposing leaf litter undergoes rapid successional changes. ISME J. 7, 477–486. doi: 10.1038/ismej.2012.116
Wallace, J. B., Eggert, S. L., Meyer, J. L., and Webster, J. R. (1997). Multiple trophic levels of a forest stream linked to terrestrial litter inputs. Science 277, 102–104.
Wearn, J. A., Sutton, B. C., Morley, N. J., and Gange, A. C. (2012). Species and organ specificity of fungal endophytes in herbaceous grassland plants. J. Ecol. 100, 1085–1092. doi: 10.1111/j.1365-2745.2012.01997.x
Wilson, D., and Carroll, G. (1997). Avoidance of high-endophyte space by gall-forming insects. Ecology 78, 2153–2163
Wilson, D., and Faeth, S. H. (2001). Do fungal endophytes result in selection for leafminer ovipositional preference? Ecology 82, 1097–1111. doi: 10.2307/2679906
Zilber-Rosenberg, I., and Rosenberg, E. (2008). Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol. Rev. 32, 723–735. doi: 10.1111/j.1574-6976.2008.00123.x
Keywords: community ecology, microbiome, bacteria, fungal endophyte, symbiosis, functional similarity
Citation: Christian N, Whitaker BK and Clay K (2015) Microbiomes: unifying animal and plant systems through the lens of community ecology theory. Front. Microbiol. 6:869. doi: 10.3389/fmicb.2015.00869
Received: 01 June 2015; Accepted: 10 August 2015;
Published: 07 September 2015.
Edited by:
Mike Taylor, The University of Auckland, New ZealandReviewed by:
Devin Coleman-Derr, Department of Energy Joint Genome Institute, USAGavin Lear, The University of Auckland, New Zealand
Copyright © 2015 Christian, Whitaker and Clay. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Natalie Christian and Briana K. Whitaker, Evolution, Ecology and Behavior Program, Department of Biology, Indiana University, Jordan Hall, 1001 East 3rd Street, Bloomington, IN 47405, USA,bnNjaHJpc3RAaW5kaWFuYS5lZHU=;YnJrYXdoaXRAaW5kaWFuYS5lZHU=
†These authors have contributed equally to this work.
 Natalie Christian
Natalie Christian