
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 08 May 2015
Sec. Terrestrial Microbiology
Volume 6 - 2015 | https://doi.org/10.3389/fmicb.2015.00379
This article is part of the Research Topic Microbial Responses to Environmental Changes View all 20 articles
 Xiaohong Wu1,2
Xiaohong Wu1,2 Tida Ge1,2*
Tida Ge1,2* Wei Wang1
Wei Wang1 Hongzhao Yuan1
Hongzhao Yuan1 Carl-Eric Wegner3
Carl-Eric Wegner3 Zhenke Zhu1,2
Zhenke Zhu1,2 Andrew S. Whiteley2,4
Andrew S. Whiteley2,4 Jinshui Wu1,2
Jinshui Wu1,2The effect of different cropping systems on CO2 fixation by soil microorganisms was studied by comparing soils from three exemplary cropping systems after 10 years of agricultural practice. Studied cropping systems included: continuous cropping of paddy rice (rice-rice), rotation of paddy rice and rapeseed (rice-rapeseed), and rotated cropping of rapeseed and corn (rapeseed-corn). Soils from different cropping systems were incubated with continuous 14C-CO2 labeling for 110 days. The CO2-fixing bacterial communities were investigated by analyzing the cbbL gene encoding ribulose-1,5-bisphosphate carboxylase oxygenase (RubisCO). Abundance, diversity and activity of cbbL-carrying bacteria were analyzed by quantitative PCR, cbbL clone libraries and enzyme assays. After 110 days incubation, substantial amounts of 14C-CO2 were incorporated into soil organic carbon (14C-SOC) and microbial biomass carbon (14C-MBC). Rice-rice rotated soil showed stronger incorporation rates when looking at 14C-SOC and 14C-MBC contents. These differences in incorporation rates were also reflected by determined RubisCO activities. 14C-MBC, cbbL gene abundances and RubisCO activity were found to correlate significantly with 14C-SOC, indicating cbbL-carrying bacteria to be key players for CO2 fixation in these soils. The analysis of clone libraries revealed distinct cbbL-carrying bacterial communities for the individual soils analyzed. Most of the identified operational taxonomic units (OTU) were related to Nitrobacter hamburgensis, Methylibium petroleiphilum, Rhodoblastus acidophilus, Bradyrhizobium, Cupriavidus metallidurans, Rubrivivax, Burkholderia, Stappia, and Thiobacillus thiophilus. OTUs related to Rubrivivax gelatinosus were specific for rice-rice soil. OTUs linked to Methylibium petroleiphilum were exclusively found in rice-rapeseed soil. Observed differences could be linked to differences in soil parameters such as SOC. We conclude that the long-term application of cropping systems alters underlying soil parameters, which in turn selects for distinct autotrophic communities.
Autotrophic bacteria with the capacity to fix CO2 are widespread in extreme terrestrial ecosystems (Giri et al., 2004; Nanba et al., 2004; Nakai et al., 2012). Recently, an isotope incubation experiment revealed high CO2 assimilation rates by autotrophic bacteria in agricultural soils, which represented a potential carbon sequestration of 0.6–4.9 Pg C year−1 (Yuan et al., 2012a; Ge et al., 2013). Autotrophic bacteria evolved six pathways for CO2 fixation: (1) the Calvin-Benson-Bassham cycle, (2) the reductive tricarboxylic acid cycle, (3) the reductive acetyl-CoA pathway, (4) the 3-hydroxypropionate cycle, (5) the 3-hydroxypropionate/4-hydroxybutyrate pathway, and (6) the dicarboxylate/4-hydroxybutyrate cycle (Fuchs, 2011). The predominant pathway for autotrophic bacteria to assimilate CO2 is the Calvin-Benson-Bassham cycle (CBB) (Selesi et al., 2005). Ribulose-1,5-bisphosphate carboxylase oxygenase (RuBisCO), the enzyme which catalyzes the rate-limiting step in the CBB cycle, exists in four distinct holoenzyme forms (I, II, III, and IV). These forms differ in structure, catalytic activity, and O2 sensitivity (Tabita, 1999). Form I RubisCO, composed of eight large subunits and eight small subunits, is the most abundant among the four forms (Tabita et al., 2008). Four clades of the cbbL gene that encodes the large subunits of form I RubisCO are known, namely IA to ID (Tabita, 1999). The presence of the cbbL gene has been documented in diverse phylogenetic groups from obligate autotrophic bacteria (form IA) to facultative autotrophic bacteria (form IC) (Kusian and Bowien, 1997; Kong et al., 2012).
In recent years, the cbbL gene has been widely used as a functional marker to analyze the diversity of autotrophic bacteria in diverse environments. Based on cbbL gene analysis, an unexpected level of cbbL diversity has been reported in agricultural soils (Selesi et al., 2005; Tolli and King, 2005; Yuan et al., 2012b; Xiao et al., 2014a). Phylogenetic analysis showed that Azospirillum lipoferum, Rhodopseudomonas palustris, Bradyrhizobium japonicum, Ralstonia eutropha are the dominant autotrophic bacteria in these soils (Yuan et al., 2012a). Different management practices including fertilizer treatments, land use alterations and different plant covers showed effects on the diversity and abundance of autotrophic bacterial communities in soils (Selesi et al., 2005; Tolli and King, 2005; Yuan et al., 2012b; Xiao et al., 2014a). Moreover, a link between microbial autotrophy and edaphic factors such as soil organic carbon (SOC), pH and clay content was identified (Selesi et al., 2007; Yuan et al., 2012a,b; Xiao et al., 2014a,b). A recent study revealed that differences in community composition, abundance and activity of autotrophic bacteria affect microbial carbon fixation across soil depth (Wu et al., 2014). A large proportion of the fixed C was restricted to surface soil (0–1 cm), and the assimilated 14C was mainly aliphatically stabilized in the humin fraction of agricultural soils (Hart et al., 2013a,b; Jian et al., 2014). Relating to agricultural soils, previous studies focused on the process of CO2 fixation by autotrophic bacteria in continuous cropping systems. However, information on autotrophic bacteria involved in CO2 fixation in rotated cropping systems is limited.
The rotation of paddy rice and upland crop is a common agricultural practice in the subtropical area of China (Zhu et al., 2010). Field studies indicated that paddy-upland rotated soils are characterized by different physical and chemical properties in comparison to paddy and upland soils (Nishida et al., 2013; Liu et al., 2014). The different soil condition in paddy-upland rotated soil was shown to affect soil microbial communities, especially functional guilds like purple phototrophic bacteria and methanogens (Feng et al., 2011; Bernard et al., 2012). The effect of paddy-upland rotation on autotrophic bacteria remains unclear. Therefore, three cropping systems: (i) double cropping of paddy rice, (ii) rotation of paddy rice and rapeseed, and (iii) double cropping of rapeseed and corn in an experimental field with a known cultivation record of continuous paddy rice farming were examined to study how different cropping systems affect CO2 fixation by autotrophic bacteria. Continuous 14C-CO2 labeling was applied to quantify the incorporation of microbial fixed C to the soil organic matter pool (14C-SOC) and soil microbial biomass (14C-MBC) at different depths (0–1, 1–5, 5–17 cm). Based on cbbL gene analysis, the abundance, diversity and composition of autotrophic bacterial communities in different cropping systems were investigated.
The sampling was conducted in a long term agricultural management experiment site at Pantang in subtropical China (29° 10′−29° 18′ N, 111° 18′−111° 33′ E). The experimental site was characterized by a typical subtropical climate with an annual mean precipitation of approximately 1400 mm and an average annual temperature of 16.8°C. Three cropping systems, namely double cropping of paddy rice (rice-rice), rotation of paddy rice and rapeseed (rice-rapeseed), and double cropping of rapeseed and corn (rapeseed-corn) were established in 2000. Four replicate plots of each cropping system were randomly arranged in the fields. Soils at the field site were developed from quaternary red earth and were used for rice farming for decades prior to the implementation of the cropping systems. For rice-rice, the field was permanently flooded during the spring and autumn rice growing seasons. For rice-rapeseed, the field was flooded in the spring rice growing season while it was drained in the rapeseed season. For rapeseed-corn, crops were planted in rapeseed-corn sequence in a drained paddy field. These treatments were maintained for more than 10 years when we conducted this study. After the harvest of the late crop, one soil core was retrieved from each field by inserting a PVC column (10 cm diameter, 20 cm height) to 17 cm depth at a random location within each 33 m2 plot. Visible crops or grass at the surface were removed. Basic geochemical parameters for all soils are given in Table 1.
All PVC columns were incubated in a growth chamber (80 × 250 cm, height 120 cm) for 110 days with continuous 14C-CO2 labeling as described previously (Ge et al., 2012; Yuan et al., 2012a; Wu et al., 2014). The 14C-CO2 was generated by forcing a Na142CO3 solution (1.0M, a radioactivity of 1.68 × 104 Bqμg−1 C) into an acid bath (HCl, 2M) and the gas concentration (14C-CO2) was maintained at approximately 350 μL L−1. During the incubation period, all soils were illuminated by a parabolic aluminum reflector lamp with an intensity of about 500 mmol photons m−2 s−1 for 12 h each day (8:00 a.m.–8:00 p.m.). The day/night air temperature inside the chamber was maintained at 31 ± 1°C/24 ± 1°C and the relative humidity was kept at 80–90%. Soils from rice-rice plots remained flooded with a 1–2 cm water layer while those from rapeseed-corn plots were drained. Rice-rapeseed soils were also incubated under the waterlogged condition, due to rice being the crop plant for the following growing season in this cropping system. At the end of the 110 day incubation, the flooded water was removed and soils from 0 to 1, 1 to 5, and 5 to 17 cm depth intervals were sampled. The sectioned soil layers, each with four replicates, were divided into two sub-samples. One sub-sample was stored at 4°C for biochemical analysis while the other was kept at −70°C for molecular analysis. For each sectioned soil sample, the soil moisture content was measured immediately after sampling.
Soil pH was determined using a pH meter (Delta 320, Mettler-Toledo Instruments Ltd., China) at a 1:2.5 (w:v) soil-to-H2O ratio. Soil organic carbon (SOC) and total nitrogen (TN) contents were measured by dry combustion with a macro elemental analyzer (Vario MAX C/N, Elementar Analyse Systeme, Germany). Total phosphorus (TP) was determined using the Mo-Sb colorimeteric method (Lu, 2000). Clay content was measured using the pipette method and cation exchange capacity (CEC) was determined by titration (Rhoades, 1982; Müller and Höper, 2004). 14C-SOC (mg kg−1) was determined according to Wu and O'Donnell (1997), and14C-MBC (mg kg−1) was analyzed using the fumigation-extraction method (Wu et al., 1990). The amounts of 14C-SOC and 14C-MBC were calculated using the following formulas:
where F1 and F2 represent the factors to convert the counting volume (1 ml from 40 ml plus soil water volume in ml for F1 and 1 ml from 80 ml plus soil water volume in ml for F2); Rs and Rp, radioactivity (Bq ml−1; blank counts omitted) for the trap solution and that for Na142CO3 (Bq mg−1 C l−1) used to produce 14C-CO2 in the growth chamber; Rf and Ruf, radioactivity (Bq l−1; blank counts subtracted for the extractants of the fumigated soil and unfumigated soil, respectively); W, the weight (kg) of digested soil on an oven-dry basis; Kc, the factor (0.45) converting measured 14C into biomass 14C (Wu et al., 1990).
DNA was extracted in triplicate from 500 mg (fresh weight) soil from each independent replicate, using the FastDNA Spin Kit (BIO101, Qbiogene Inc., Carlsbad, CA) according to the manufacturer's protocol. The integrity and quantity of the extracted DNA were evaluated by standard agarose gel electrophoresis and a spectrophotometer (Nanodrop ND-1000, PeqLab, Germany). The cbbL gene fragments from one randomly chosen replicate of rice-rice and rapeseed-corn cropping systems were amplified using the same thermal profile as previously described by Wu et al. (2014). PCR reactions were set up as follows: 12.5 μl 2 × PCR MasterMix (Tiangen, China), approximately 50 ng soil DNA, and 0.1 μM of each cbbL primer, modified by Tolli and King (2005) per reaction. In order to show the reproducibility of our approach, the cbbL gene fragments from two replicate samples originating from the rice-rapeseed rotation were generated separately. Subsequently, PCR products were purified with an agarose gel DNA purification kit (Tiangen, China) and ligated into the pGEM-T Easy Vector System (Promega, Mannheim, Germany), and then transformed into E. coli DH5α-competent cells. Positive clones were sequenced at the Beijing Genome Institute (Beijing, China).
Clone sequences were grouped into operational taxonomic units (OTUs) based on 95% nucleotide sequence similarity using Mothur (Schloss et al., 2009). The OTUs primarily responsible for the differences in cbbL-carrying bacterial community among samples were identified based on the similarity percentage analysis (SIMPER) using PAST (Hammer et al., 2001). The representative nucleotide sequences of these OTUs were subsequently translated into amino acid sequences and aligned with closely related known sequences in GenBank using Clustal W (http://www.ebi.ac.uk/clustalw). If necessary, alignments were manually refined. The resulting alignment was used to construct a neighbor-joining tree using MEGA 5.0 (Tamura et al., 2011). Bootstrap analysis of 1000 replicates was conducted to estimate the robustness of the tree topologies.
Rarefaction curves were generated by the Analytic Rarefaction program (http://strata.uga.edu/software/Software.html) to assess the sampling effort. Shannon indices were computed using Mothur to compare the diversity of the cbbL-bearing bacterial communities in three cropping systems (Schloss and Handelsman, 2008). The coverage rate was computed as C = [1− (n/N)] × 100, where n represents the number of OTUs containing one individual sequence and N is the total number of sequences.
The cbbL gene abundance was quantified using an ABI 7900 real-time PCR system (ABI 7900, Foster City, CA, USA) using SYBR Green I based assays. Quantitative PCR was performed in 10 μL reaction mixtures containing 5 μL 1 × SYBR Premix Extaq (Takara Bio Inc., Shiga, Japan), 5 ng of template DNA, 0.1 μM of primers with the following thermal profile: 30 s at 95°C, followed by 5 cycles of 5 s at 95°C, 45 s annealing temperature decreased from 66 to 62°C and an extension at 72°C for 30 s. In addition, another 35 cycles at 95°C for 5 s, 62°C for 45 s, and 72°C for 30 s. A final melting curve was generated to evaluate the amplification specificity. Ten-fold serial dilutions of plasmid DNA extracted from positive clones were used to establish a standard curve. The real-time PCR assays were performed in triplicate for each replicate sample. The copy number of the cbbL gene was calculated directly using SDS 2.3 software.
RubisCO enzyme activity was assayed according to Yuan et al. (2012a). Briefly, 2 g soil (four replicates) were thoroughly homogenized by an ultrasonic cell mixer (JY92-II Scientz, China) in an extraction buffer containing Tris-HCl (100 mM, pH 7.8) and Dithioreitol (DTT, 1 mM). The supernatant was collected by centrifugation and was precipitated with solid ammonium sulfate to reach 80% saturation. The resulting pellets were collected and dissolved in Tris-HCl/DTT. RubisCO activity was measured at 30°C using spectrophotometry (UV-2450, Shimadzu, Japan) and calculated according to Takai et al. (2005).
Canonical correspondence analysis (CCA) was performed using CANOCO 5.0 for Windows (Microcomputer Power, Ithaca, NY, USA) to characterize the effect of measured soil properties on the composition of bacteria communities carrying cbbL gene. Significant differences in community composition were tested by permutational Two-Way analysis of variance or multivariate analysis of variance (MANOVA) implemented in PAST (Hammer et al., 2001). PERMANOVA is a distance-based non-parametric MANOVA that allows the analysis of multivariate (or univariate) data in response to treatments in an experimental design. Statistical significant differences between data sets based on metadata (soil parameters, cbbL copy numbers, RubisCO activities) were identified by two-way analysis of variance (ANOVA) and differences were considered significant at P < 0.05. A multiple regression model was built by stepwise regression with significance being defined as P < 0.05. ANOVA and multiple regression analyses were carried out using SPSS (version 16.0, SPSS Inc., USA). The reproducibility of the carried out clone library analysis was tested using the aforementioned replicated clone libraries originating from the rice-rapeseed rotation. The robustness of clone library analysis was assessed based on calculated unweighted UniFrac distances (Lozupone and Knight, 2005).
Nucleotide sequences were deposited in the EMBL European Nucleotide Database (http://www.ebi.ac.uk/ena/data/view/) under accession numbers HG940678–HG941631.
The 14C-SOC and 14C-MBC concentrations were significantly different in the three cropping systems according to Two-Way ANOVA analysis. In 0–1 cm, the maximum 14C-SOC and 14C-MBC concentrations were detected in rice-rice rotated soil, reaching 935 mg kg−1 and 375 mg kg−1 respectively. The 14C-SOC and 14C-MBC concentrations in corresponding depths of rice-rapeseed rotated soil were 1.6 and 4.0 times lower than those in rice-rice rotated soil (Table 2). In 1–5 cm, the radioactivity, in terms of SOC and MBC was higher in rice-rice rotated soil than in rice-rapeseed rotated soil (Table 2). The amount of 14C incorporated in rapeseed-corn rotated soil was much lower compared to all other soils when investigating the 0–1 and 1–5 cm soil layers (Table 2). In addition, the 14C-SOC and 14C-MBC contents decreased with increasing depth, irrespective of the cropping systems. No radioactivity was detected in the 5–17 cm layer with the exception of rice-rapeseed rotated soil (Table 2). A statistically significant correlation was observed between 14C-SOC and 14C-MBC concentration (P < 0.05). The cbbL gene abundance and RubisCO activity were significantly related to 14C-SOC concentration (P < 0.05).
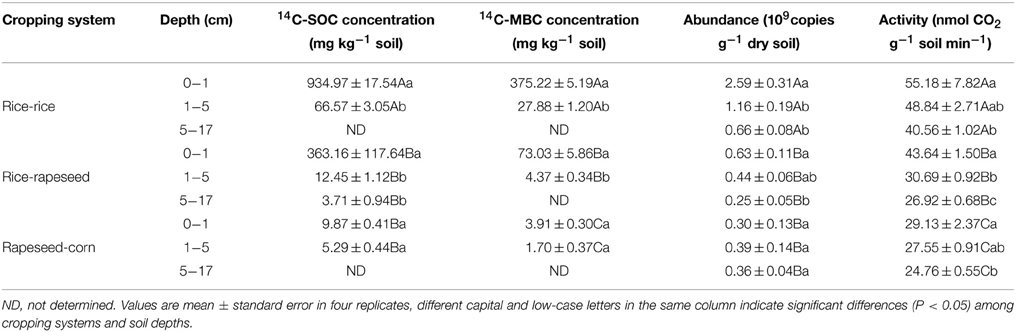
Table 2. Amounts of 14C-SOC, 14C-MBC, abundance and activity of cbbL-carrying bacteria in three cropping systems.
Nine clone libraries of 106 clones each were obtained from different depth intervals for the three cropping systems. Overall, sequences were grouped into 148, 155, and 111 OTUs for rice-rice, rice-rapeseed and rapeseed-corn rotated soils, respectively (Table 3). High levels of diversity were observed based on the number of OTUs as well as calculated Shannon indices. Differences in diversity between cropping systems were small (Table 3). Rarefaction analyses suggested that additional sequencing effort is needed to cover the full diversity of cbbL sequences in our systems (Figure 1). The underestimated diversity was also reflected by the coverage of libraries, which ranged from 52 to 71% (Table 3). Determined Pearson coefficients revealed no significant correlations between diversity and monitored soil properties (P > 0.05).
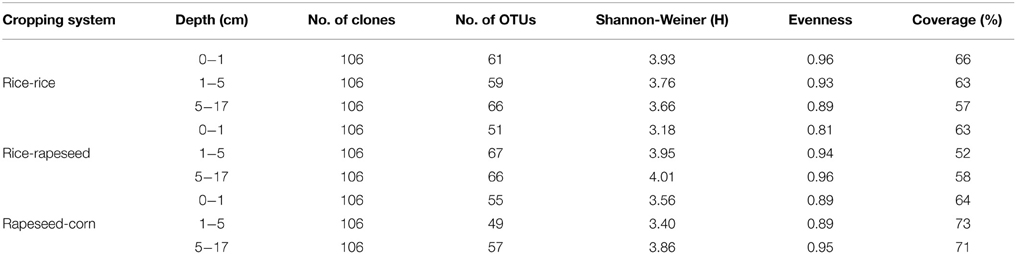
Table 3. Diversity of cbbL-containing bacterial community in sectioned soil depths from different cropping systems.
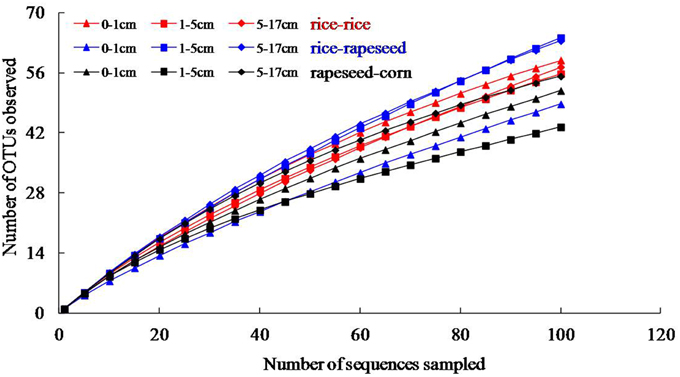
Figure 1. Rarefaction analysis of cbbL clone libraries based on 95% nucleotide sequences similarity.
The comparison of cbbL-bearing bacterial communities in two rice-rapeseed rotated soils revealed no statistically significant differences in community structures for replicated samples (Table S1), suggesting that our sampling approach is robust enough to draw reliable conclusions. The cbbL-carrying bacterial communities within the three cropping systems clustered into different groups, as revealed by the CCA analysis (Figure 2). Samples from different depth layers of the same cropping system formed relatively tight clusters (Figure 2). PERMANOVA analysis showed that the individual cropping system was a statistically significant determinant of community composition (P < 0.05), whereas the community structure did not change markedly at different sampling depths (P > 0.05). CCA analysis revealed that the SOC content (P < 0.05) was the main environmental driver for changes in the cbbL-bearing bacterial communities.
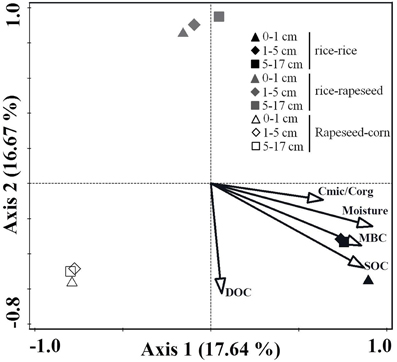
Figure 2. Changes in the cbbL-containing bacterial communities in response to different cropping systems. Cmic/Corg describes the ratio of microbial biomass carbon to total organic carbon.
The cbbL gene abundances in rice-rice rotated soil were significantly higher than those determined in rice-rapeseed and rapeseed-corn rotated soils (Table 2). Gene copy numbers were four times higher in the 0–1 cm layer, when compared to the 5–17 cm layer in rice-rice rotated soil (Table 2). Similar vertical trends were found for rice-rapeseed rotated soil, where copy numbers decreased by 63% in the 5–17 cm layer compared to the 0–1 cm layer (Table 2). cbbL copy numbers changed only slightly with depth in rapeseed-corn soil (Table 2). The abundance of cbbL-carrying bacteria was significantly correlated with DOC and MBC (P < 0.05).
RubisCO activity differed in the individual cropping systems. Highest activities were seen in rice-rice rotated soil (Table 2). Activities were generally found to decrease with soil depth. Multiple regression analysis based on stepwise showed that MBC was the main factor affecting RubisCO activity (P < 0.05).
A total of 57 OTUs were identified as main phylotypes responsible for observed differences in community structure, cumulatively contributing 50% of the community variation (Figure 3). Sequences from these OTUs were dominated by facultative cbbL-carrying bacterial communities (Form IC), and they varied in their relative abundances in relation to cropping system and soil depth. Sequences from rapeseed-corn soil mainly formed four clades, with two clusters relating to Nitrobacter hamburgensis and Nocardia asteroides respectively and two novel clades without known representatives (Figure 4). Sequences from rice-rapeseed rotated soil were phylogenetically diverse, but closely related to sequences from Methylibium petroleiphilum, Rhodoblastus acidophilus, Bradyrhizobium, and Cupriavidus metallidurans (Figure 4). Sequences retrieved from rice-rice soil were widely distributed, grouping with various facultative and obligate autotrophic groups such as Rubrivivax, Burkholderia, Bradyrhizobium, Stappia, and Thiobacillus thiophilus (Figure 4).

Figure 3. Heatmap of the 57 OTUs primarily responsible for community differences. Respective cumulative contributions to community dissimilarities are given in addition.
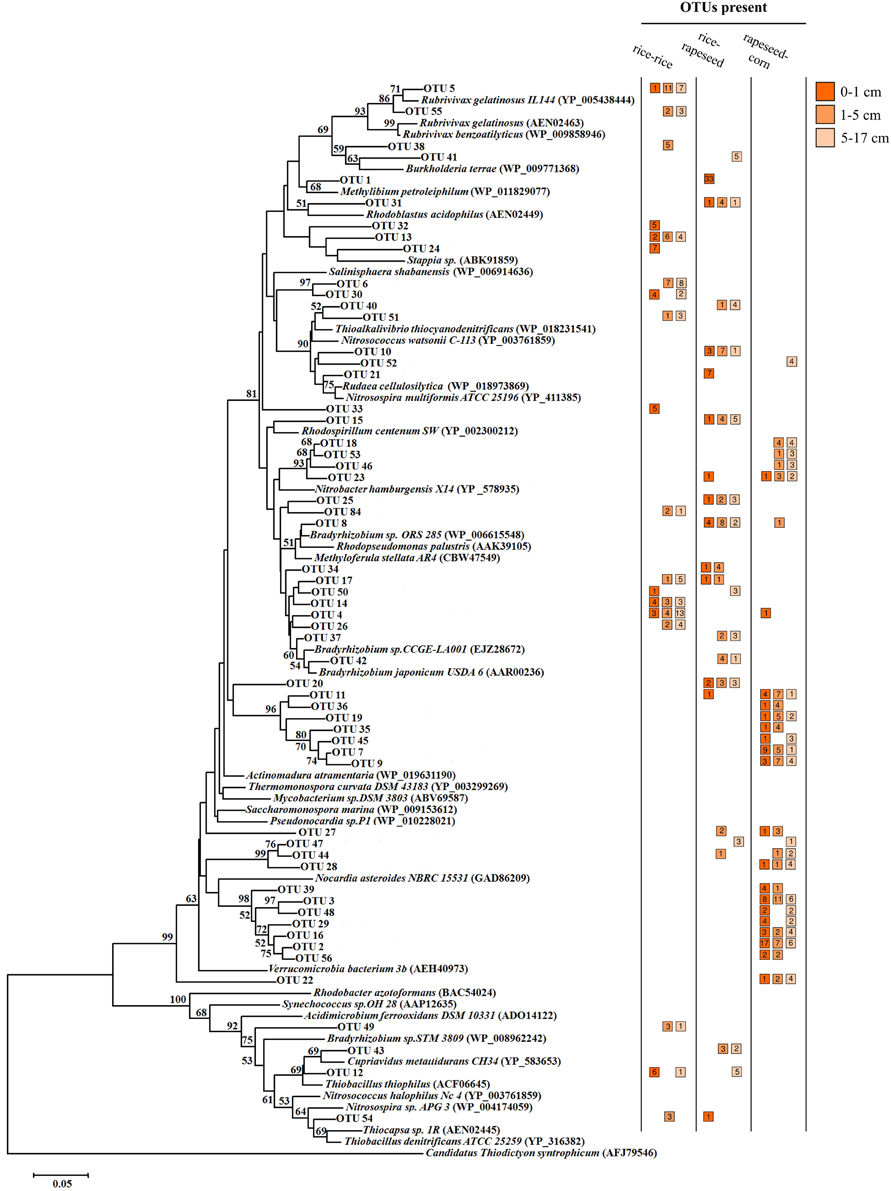
Figure 4. Neighbor-joining tree illustrating the phylogeny of the OTUs primarily responsible for the differences in cbbL-carrying bacterial communities. The tree was constructed using 165 deduced amino acids of corresponding nucleic acid cbbL clone sequences and 36 reference sequences from public databases. The cbbM gene from Candidatus “Thiodictyon syntrophicum” strain Cad 16 (accession number AFJ79546) was used as outgroup. The number of sequences retrieved from different samples is displayed in the colored squares next to the OTUs. Bootstrap values above 50% are indicated at the branch nodes. The scale bar represents 0.05 substitutions per amino acid based on a p-distance matrix analysis.
According to our previous microcosm experiments, autotrophic bacteria contribute significantly to CO2 fixation in agricultural soils (Yuan et al., 2012a; Ge et al., 2013; Wu et al., 2014). We previously ascertained that the 14C incorporation, as a measure for the autotrophic soil carbon sink mediated by autotrophic bacteria, was 2–13-folds larger in continuous paddy rice soils than upland crop soils (Yuan et al., 2012a; Ge et al., 2013). Here we extended previous work to gain insights into the effect of different cropping systems on microbial CO2 fixation processes, using three different cropping systems, including rice-rice, rice-rapeseed and rapeseed-corn rotated soils. The significant linear correlation between 14C-SOC and 14C-MBC concentrations indicated that the fixed 14C in three cropping systems was derived from microbial fixation (Yuan et al., 2012a; Ge et al., 2013; Wu et al., 2014). Autotrophic bacteria, as revealed by the positive relationships between cbbL gene abundance, RubisCO activity and 14C-SOC concentration, were the major microbial players behind 14CO2 incorporation into SOC. The cbbL-carrying bacteria recovered in these soils were dominated by sequences related to facultative autotrophs like phototrophic, nitrogen fixing, nitrifying and CO and hydrogen oxidizing bacteria. However, some members of the cbbL-carrying bacterial communities were exclusive in one cropping system. For example, clone sequences in OTUs specific to rice-rice soil were closely related to Rubrivivax gelatinosus, which is a phototrophic bacterium with two cbbL gene copies surviving in aquatic ecosystems and moist soils (Kuske et al., 1997; Badger and Bek, 2008). While sequences closely affiliated to methylotrophic bacterium Methylibium petroleiphilum PM1 were exclusive to rice-rapeseed soil, whose presence has been documented in aquatic systems previously (Chen et al., 2009). Although facultative chemoautotrophy has been identified as an alternative metabolism in the methylotrophic bacterium Beijerinckia mobilis, a potential autotrophic metabolism of Methylibium petroleiphilum PM1 has not yet been demonstrated (Dedysh et al., 2005; Kane et al., 2007).
The diversity of cbbL-carrying bacterial communities suggested the presence of metabolically versatile autotrophic bacteria in the three cropping systems under study here. Much lower diversity patterns were reported in previous work regarding different managed agricultural soils using T-RFLP analysis (Selesi et al., 2005; Yuan et al., 2013; Xiao et al., 2014a). Applying clone library analyses improved the resolution of cbbL sequence analysis in comparison to previous studies (Marsh, 1999). Observed high diversities are presumably a consequence of changing underlying soil properties due to the applied cropping systems. Numerous studies have established a link between the cbbL diversity and soil properties (Nanba et al., 2004; Selesi et al., 2005; Yuan et al., 2012a,b, 2013; Xiao et al., 2014a). In our study, all the tested soils were developed from the quaternary red earth, which is characterized by a high clay content. The high amount of available nutrients in soil clay fractions were reported to favor the development of diverse cbbL-carrying bacterial communities (Paul et al., 2001; Selesi et al., 2007). This likely explains higher diversities in our soils in comparison to those observed in coastal barren saline soils based on clone library analysis (Yousuf et al., 2012).
The contributions of autotrophic bacteria to CO2 fixation in the three cropping systems were different, with the highest 14C incorporation rate observed in rice-rice, followed by rice-rapeseed, with the lowest value detected in rapeseed-corn rotated soil (Table 1). Paddy-upland rotation cropping systems differ from normal paddy rice and upland crop systems, where various water regimes are practiced in different crop growth seasons (Nishimura et al., 2008; Liu et al., 2010). During the submerged period of paddy rice cultivation, the anoxic conditions restrict the mineralization processes in soil, whereas the drainage of paddy fields for upland crop cultivation resulted in an oxic condition which enhances the decomposition processes within the soil (Chang Chien et al., 2006; Iqbal et al., 2009). As a result, physicochemical properties (e.g., SOC) are significantly altered in paddy-upland rotated systems when compared to paddy and upland soil traditional cultivation methods (Wang and Yang, 2003; Liu et al., 2010; Zhu et al., 2010). In this study, all soils were collected from the same climatic condition, and have the same origin and cultivation history. Soil properties such as SOC and TN were changing in response to the three cropping systems (Table 1), due to the differences in field management and crop regime over the 10 year period. The variations in soil properties (SOC, TN, MBC, and DOC) caused by the applied cropping systems affected the abundance, activity and composition of cbbL-carrying bacteria (Selesi et al., 2005; Xiao et al., 2014a), and thus resulted in the differences in 14C incorporation rates in soils. We observed an almost two orders of magnitude difference in 14C assimilation rates between rotation systems but only two- to four-fold differences in cbbL gene abundance and RubisCO activity. Considering that only cbbL copy numbers have been determined, differences on the level of transcription and translation cannot be ruled out. It appears reasonable to assume that 14C was at least partially assimilated by alternative CO2 fixation pathways, such as the reductive acetyl-CoA cycle that is known to be commonly active under anoxic conditions (Campbell and Cary, 2004; Nakagawa et al., 2005).
Labeling experiments as the one presented here are easily affected by the availability of unlabeled substrate. Dependent on the availability of unlabeled CO2, determined incorporation rates are eventually influenced due to a dilution effect. The availability of CO2 in soil pore space is strongly correlated with ongoing respiration processes, which are influenced by present organic substrates (Van Hees et al., 2005; Iqbal et al., 2009). Against this background our results appeared robust against the outlined dilution effect. The SOC content was highest in rice-rice soil, which presumably stimulated respiration. As a result CO2 availability would have been rather high, potentially diluting added 14C-CO2. Nevertheless, 14C incorporation was the highest in these soils.
In line with previous work, fixed 14C significantly decreased with soil depth in three cropping systems, indicating that a large proportion of microbially fixed 14C was restricted to the surface soil (Wu et al., 2014). Nishimura et al. (2008) reported that land use change from paddy rice to upland crop not only affected soil properties within the surface soil layer, but also caused changes within the deeper soil layers. Therefore, the differences in soil properties across soil depths may affect the availability of substrate (14CO2) and electron donors to autotrophic bacteria, resulting in changes in 14C fixation rates with soil depth (Jeffery et al., 2009; Kellermann et al., 2012; Wu et al., 2014). In addition, 14CO2 reduced to methane in the flooded surface soil could be accessed by methane oxidizing bacteria, which might in part explain the large assimilation in the surface of rice-rice and rice-rapeseed soils as well. It cannot be ruled out that incorporated 14CO2 was at least partially derived from CO2 fixation by soil algae, which eventually released metabolites that could have been processed by heterotrophic organisms. Nevertheless, soil algae were presumably playing a minor role. From previous work it is known that their abundance is at least one order of magnitude lower compared to autotrophic bacteria (Yuan et al., 2012a). Future isotope based work could help to gain an insights regarding the active community fraction consuming CO2 among cbbL-carrying bacteria and algae as a whole, since our community structure analyses revealed only minor changes for cbbL–carrying bacterial communities within different depths.
The present study showed variations in CO2 fixation by autotrophic bacteria in response to different cropping systems. Statistical analysis revealed higher CO2 assimilation rates in rice-rice than rice-rapeseed and rapeseed-corn rotated soils. Observed differences in soil parameters caused by the applied cropping systems lead to changes in cbbL abundance, activity and bacterial community structure, and thus resulted in differences in 14C incorporation rates in the three cropping systems. These results broaden our knowledge about the importance of autotrophic bacteria involved in the soil carbon sink. However, questions still remain, including the true extent of cbbL diversity. Here, next generation technologies such as high throughout sequencing appear represent promising follow up approaches, because the resolution of the analysis would substantially increase. Identifying active autotrophs involved in CO2 fixation by studying community gene expression would provide a better understanding about organisms playing major roles under different soil management conditions and how active organisms eventually interact with each other.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This study was supported financially by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB15020401), the National Natural Science Foundation of China (41271279; 41430860) and the Chinese Academy of Sciences Visiting Professorship for Senior International Scientists awarded to Prof. AW (2013T2S0013). We also thank the editor and the reviewers for their helpful advice, which led to improvements in this paper.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00379/abstract
Badger, M. R., and Bek, E. J. (2008). Multiple Rubisco forms in proteobacteria: their functional significance in relation to CO2 acquisition by the CBB cycle. J. Exp. Bot. 59, 1525–1541. doi: 10.1093/jxb/erm297
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bernard, E., Larkin, R. P., Tavantzis, S., Erich, M. S., Alyokhin, A., Sewell, G., et al. (2012). Compost, rapeseed rotation, and biocontrol agents significantly impact soil microbial communities in organic and conventional potato production systems. Appl. Soil Ecol. 52, 29–41. doi: 10.1016/j.apsoil.2011.10.002
Campbell, B. J., and Cary, S. C. (2004). Abundance of reverse tricarboxylic acid cycle genes in free-living microorganisms at deep-sea hydrothermal vent. Appl. Environ. Microbiol. 70, 6282–6289. doi: 10.1128/AEM.70.10.6282-6289.2004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chang Chien, S. W., Wang, M. C., Hsu, J. H., and Seshaiah, K. (2006). Influence of fertilizers applied to a paddy-upland rotation on characteristics of soil organic carbon and humic acids. J. Agric. Food Chem. 54, 6790–6799. doi: 10.1021/jf061275m
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chen, Y., Wu, L. Q., Boden, R., Hillebrand, A., Kumaresan, D., Moussard, H., et al. (2009). Life without light: microbial diversity and evidence of sulfur- and ammonium-based chemolithotrophy in Movile Cave. ISME J. 3, 1093–1104. doi: 10.1038/ismej.2009.57
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dedysh, S. N., Smirnova, K. V., Khmelenina, V. N., Suzina, N. E., Liesack, W., and Trotsenko, Y. A. (2005). Methylotrophic autotrophy in Beijerinckia mobilis. J. Bacteriol. 187, 3884–3888. doi: 10.1128/JB.187.11.3884-3888.2005
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Feng, Y., Lin, X., Zhang, J., Mao, T., and Zhu, J. (2011). Soil purple phototrophic bacterial diversity under double cropping (rice-wheat) with free-air CO2 enrichment (FACE). Eur. J. Soil Sci. 62, 533–540. doi: 10.1111/j.1365-2389.2011.01357.x
Fuchs, G. (2011). Alternative pathways of carbon dioxide fixation: insights into the early evolution of life? Annu. Rev. Microbiol. 65, 631–658. doi: 10.1146/annurev-micro-090110-102801
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ge, T. D., Wu, X. H., Chen, X. J., Yuan, H. Z., Zou, Z. Y., Li, B. Z., et al. (2013). Microbial phototrophic fixation of atmospheric CO2 in China subtropical upland and paddy soils. Geochim. Cosmochim. AC 113, 70–78. doi: 10.1016/j.gca.2013.03.020
Ge, T. D., Yuan, H. Z., Zhu, H. H., Wu, X. H., Nie, S. A., Liu, C., et al. (2012). Biological carbon assimilation and dynamics in a flooded rice-soil system. Soil Biol. Biochem. 48, 39–49. doi: 10.1016/j.soilbio.2012.01.009
Giri, B. J., Bano, N., and Hollibaugh, J. T. (2004). Distribution of rubisCO genotypes along a redox gradient in Mono Lake, California. Appl. Environ. Microb. 70, 3443–3448. doi: 10.1128/AEM.70.6.3443-3448.2004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hammer, Ø., Harper, D. A. T., and Ryan, P. D. (2001). PAST: paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4, 1–9.
Hart, K. M., Kulakova, A. N., Allen, C. C. R., Simpson, A., Oppenheimer, S. F., Masoom, H., et al. (2013a). Tracking the fate of microbially sequestered carbon dioxide in soil organic matter. Environ. Sci. Technol. 47, 5128–5137. doi: 10.1021/es3050696
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hart, K. M., Oppenheimer, S. F., Moran, B. W., Allen, C. C. R., Kouloumbos, V., Simpson, A. J., et al. (2013b). CO2 uptake by a soil microcosm. Soil Biol. Biochem. 57, 615–624. doi: 10.1016/j.soilbio.2012.10.036
Iqbal, J., Hu, R. G., Lin, S., Ahamadou, B., and Feng, M. L. (2009). Carbon dioxide emissions from Ultisol under different land uses in mid-subtropical China. Geoderma 152, 63–73. doi: 10.1016/j.geoderma.2009.05.011
Jeffery, S., Harris, J. A., Rickson, R. J., and Ritz, K. (2009). The spectral quality of light influences the temporal development of the microbial phenotype at the arable soil surface. Soil Biol. Biochem. 41, 553–560. doi: 10.1016/j.soilbio.2008.12.014
Jian, Y., Zeng, G. J., Zhou, P., Yuan, H. Z., Ge, T. D., Zou, D. S., et al. (2014). Input and distribution of autotrophic microbe-assimilated carbon in humus and aggregate fractions of soils (in Chinese). Int. J. Environ. Res. 27, 1499–1504. doi: 10.13198/j.issn.1001-6929.2014.12.16
Kane, S. R., Chakicherla, A. Y., Chain, P. S. G., Schmidt, R. S., Shin, M. W., Legler, T. C., et al. (2007). Whole-genome analysis of the Methyl tert-Butyl Ether-Degrading Beta-Proteobacterium Methylibium petroleiphilum PM1. J. Bacteriol. 189, 1931–1945. doi: 10.1128/JB.01259-06
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kellermann, C., Selesi, D., Lee, N., Hügler, M., Esperschütz, J., Hartmann, A., et al. (2012). Microbial CO2 fixation potential in a tar-oil-contaiminated porous aquifer. FEMS Microbiol. Ecol. 81, 172–187. doi: 10.1111/j.1574-6941.2012.01359.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kong, W. D., Ream, D. C., Priscu, J. C., and Morgan-Kiss, R. M. (2012). Diversity and expression of RubisCO genes in a perennially ice-covered Antarctic lake during the polar night transition. Appl. Environ. Microb. 78, 4538–4366. doi: 10.1128/AEM.00029-12
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kusian, B., and Bowien, B. (1997). Organization and regulation of cbb CO2 assimilation genes in autotrophic bacteria. FEMS Microbiol. Rev. 21, 135–155. doi: 10.1111/j.1574-6976.1997.tb00348.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kuske, C. R., Barns, S. M., and Busch, J. D. (1997). Diverse uncultivated bacterial groups from soils of the arid southwestern united states that are present in many geographic regions. Appl. Environ. Microb. 63, 3614–3621.
Liu, S. L., Huang, D. Y., Chen, A. L., Wei, W. X., Brookes, P. C., Li, Y., et al. (2014). Differential responses of crop yields and soil organic carbon stock to fertilization and rice straw incorporation in three cropping systems in the subtropics. Agr. Ecosyst. Environ. 184, 51–58. doi: 10.1016/j.agee.2013.11.019
Liu, S. L., Li, Y., Wu, J. S., Huang, D. Y., Su, Y. R., and Wei, W. X. (2010). Spatial variability of soil microbial biomass carbon, nitrogen and phosphorus in a hilly red soil landscape in subtropical China. Soil Sci. Plant Nutr. 56, 693–704. doi: 10.1111/j.1747-0765.2010.00510.x
Lozupone, C., and Knight, R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microb. 71, 8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lu, R. K. (2000). Analytical Methods of Soil Agrochemistry (in Chinese). Beijing: Chinese Agricultural Science and Technology Press. 638.
Marsh, T. L. (1999). Terminal restriction fragment length polymorphism (T-RFLP): an emerging method for characterizing diversity among homologous populations of amplification products. Curr. Opin. Microbiol. 2, 323–327. doi: 10.1016/S1369-5274(99)80056-3
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Müller, T., and Höper, H. (2004). Soil organic matter turnover as a function of the soil clay content: consequences for model applications. Soil Biol. Biochem. 36, 877–888. doi: 10.1016/j.soilbio.2003.12.015
Nakagawa, S., Takai, K., Inagaki, F., Chiba, H., Ishibashi, J., Kataoka, S., et al. (2005). Variability in microbial community and venting chemistry in a sediment-hosted backarc hydro-thermal system: impacts of subseafloor phase-separation. FEMS Microbiol. Ecol. 54, 141–155. doi: 10.1016/j.femsec.2005.03.007
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Nakai, R., Abe, T., Baba, T., Imura, S., Kagoshima, H., Kanda, H., et al. (2012). Diversity of RuBisCO gene responsible for CO2 fixation in an Antarctic moss pillar. Polar Biol. 35, 1641–1650. doi: 10.1007/s00300-012-1204-5
Nanba, K., King, G. M., and Dunfield, K. (2004). Analysis of facultative lithotroph distribution and diversity on volcanic deposits by use of the large subunit of ribulose 1,5-bisphosphate carboxylase/oxygenase. Appl. Environ. Microb. 71, 2245–2253. doi: 10.1128/AEM.70.4.2245-2253.2004
Nishimura, S., Yonemura, S., Sawamoto, T., Shirato, Y., Akiyama, H., Sudo, S., et al. (2008). Effect of land use change from paddy rice cultivation to upland crop cultivation on soil budget of a cropland in Japan. Agr. Ecosyst. Environ. 125, 9–20. doi: 10.1016/j.agee.2007.11.003
Nishida, M., Sekiya, H., and Yoshida, K. (2013). Status of paddy soils as affected by paddy rice and upland soybean rotation in northeat Japan, with special reference to nitrogen fertility. Soil Sci. Plant Nutr. 59, 208–217. doi: 10.1080/00380768.2012.762588
Paul, E. A., Collins, H. P., and Leavitt, S. W. (2001). Dynamics of resistant soil carbon of Midwestern agricultural soils measured by naturally occurring 14C abundance. Geoderma. 104, 239–256. doi: 10.1016/S0016-7061(01)00083-0
Rhoades, J. D. (1982). “Cation exchangeable capacity,” in Methods of Soil Analysis, Part 2: Chemical and Microbiological Properties, 2nd Edn., eds A. L. Pace, R. H. Miller, and D. R. Keeney (Madison, WI: American Society of Agronomy, Inc., and Soil Science Society of America, Inc.), 149–165.
Schloss, P. D., and Handelsman, J. (2008). A statistical toolbox for metagenomics: assessing functional diversity in microbial communities. BMC Bioinformatics 9:34. doi: 10.1186/1471-2105-9-34
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microb. 75, 7537–7541. doi: 10.1128/AEM.01541-09
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Selesi, D., Pattis, I., Schmid, M., Kandeler, E., and Hartmann, A. (2007). Quantification of bacterial RubisCO genes in soils by cbbL targeted real-time PCR. J. Microbiol. Meth. 69, 497–503. doi: 10.1016/j.mimet.2007.03.002
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Selesi, D., Schmid, M., and Hartmann, A. (2005). Diversity of green–like and red–like ribulose–1,5–bisphosphate carboxylase/oxygenase large–subunit genes (cbbL) in differently managed agricultural soils. Appl. Environ. Microb. 71, 175–184. doi: 10.1128/AEM.71.1.175-184.2005
Tabita, F. R. (1999). Microbial ribulose-1,5-bisphosphatecarboxylase/oxygenase:a different perspective. Photosynth. Res. 60, 1–28. doi: 10.1023/A:1006211417981
Tabita, F. R., Satagopan, S., Hanson, T. E., Kreel, N. E., and Scott, S.S. (2008). Distinct form I, II, III, and IV Rubisco proteins from the three kingdoms of life provide clues about Rubisco evolution and structure/function relationships. J. Exp. Bot. 59, 1515–1524. doi: 10.1093/jxb/erm361
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Takai, K., Campbell, B. J., Cary, S. C., Suzuki, M., Oida, H., Nunoura, T., et al. (2005). Enzymatic and genetic characterization of carbon and energy metabolisms by deep-sea hydrothermal chemolithoautotrophic isolates of Epsilonproteobacteria. Appl. Environ. Microb. 71, 7310–7320. doi: 10.1128/AEM.71.11.7310-7320.2005
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using Maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tolli, J., and King, G. M. (2005). Diversity and structure of bacterial chemolithotrophic communities in pine forest and agroecosystem soils. Appl. Environ. Microbiol. 71, 8411–8418. doi: 10.1128/AEM.71.12.8411-8418.2005
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Van Hees, P. A. W., Jones, D. L., Finlay, R., Godbold, D. L., and Lundström, U. S. (2005). The carbon we do not see-the impact of low molecular weight compounds on carbon dynamics and respiration in forest soils: a review. Soil Biol. Biochem. 37, 1–13. doi: 10.1016/j.soilbio.2004.06.010
Wang, M. C., and Yang, C. H. (2003). Type of fertilizer applied to a paddy-upland rotation affects selected soil quality attributes. Geoderma 114, 93–108. doi: 10.1016/S0016-7061(02)00356-7
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wu, J., Joergensen, R. G., Pommerening, B., Chaussod, R., and Brookes, P. C. (1990). Measurement of soil microbial biomass C by fumigation-extraction-an automated procedure. Soil Biol. Biochem. 22, 1167–1169. doi: 10.1016/0038-0717(90)90046-3
Wu, J., and O'Donnell, A. G. (1997). Procedure for the simultaneous analysis of total and radioactive carbon in soil and plant materials. Soil Biol. Biochem. 29, 199–202.
Wu, X. H., Ge, T. D., Yuan, H. Z., Li, B. Z., Zhu, H. H., Zhou, P., et al. (2014). Changes in bacteria CO2 fixation with depth in agricultural soils. Appl. Microbiol. Biot. 98, 2309–2319. doi: 10.1007/s00253-013-5179-0
Xiao, K. Q., Bao, P., Bao, Q. L., Jia, Y., Huang, F. Y., Su, J. Q., et al. (2014a). Quantitative analysis of ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO) large-subunit genes (cbbL) in typical paddy soils. FEMS Microbiol. Ecol. 87, 89–101. doi: 10.1111/1574-6941.12193
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Xiao, K. Q., Nie, S. A., Bao, P., Wang, F. H., Bao, Q. L., and Zhu, Y. G. (2014b). Rhizosphere effect has no effect on marker genes related to autotrophic CO2 fixation in paddy soils? J. Soil Sediment 14, 1082–1087. doi: 10.1007/s11368-014-0864-x
Yousuf, B., Sanadhya, P., Keshri, J., and Jha, B. (2012). Comparative molecular analysis of chemolithoautotrophic bacterial diversity and community structure from coastal saline soils, Gujarat, India. BMC Microbiol. 12, 150. doi: 10.1186/1471-2180-12-150
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yuan, H. Z., Ge, T. D., Chen, C. Y., O'Donnel, A. G., and Wu, J. S. (2012a). Significant role for microbial autotrophy in the sequestration of soil carbon. Appl. Environ. Microb. 78, 2328–2336. doi: 10.1128/AEM.06881-11
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yuan, H. Z., Ge, T. D., Wu, X. H., Liu, S. L., Tong, C. L., Qin, H. L., et al. (2012b). Long–term field fertilization alters the diversity of autotrophic bacteria based on the ribulose–1,5–biphosphate carboxylase/oxygenase (RubisCO) large–subunit genes in paddy soil. Appl. Microbiol. Biot. 95, 1061–1071. doi: 10.1007/s00253-011-3760-y
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yuan, H. Z., Ge, T. D., Wu, X. H., Liu, S. L., Zhou, P., Chen, X. J., et al. (2013). Effect of land use on the abundance and diversity of autotrophic bacteria as measured by ribulose-1,5-biphosphate carboxylase/oxygenase (RubisCO) large subunit gene abundance in soils. Biol. Fert. Soils 49, 609–616. doi: 10.1007/s00374-012-0750-x
Keywords: cropping systems, autotrophic bacteria CO2 fixation, RubisCO, cbbL genes, 14C continuous labeling, 14C-SOC, soil depth
Citation: Wu X, Ge T, Wang W, Yuan H, Wegner C-E, Zhu Z, Whiteley AS and Wu J (2015) Cropping systems modulate the rate and magnitude of soil microbial autotrophic CO2 fixation in soil. Front. Microbiol. 6:379. doi: 10.3389/fmicb.2015.00379
Received: 03 August 2014; Accepted: 13 April 2015;
Published: 08 May 2015.
Edited by:
Jérôme Comte, Laval University, CanadaReviewed by:
Nathan Basiliko, Laurentian University, CanadaCopyright © 2015 Wu, Ge, Wang, Yuan, Wegner, Zhu, Whiteley and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tida Ge, Institute of Subtropical Agriculture, Chinese Academy of Sciences, No.644, Yuanda'er Road, Changsha, Hunan 410125, China,Z3RkQGlzYS5hYy5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.