
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 16 March 2015
Sec. Microbial Physiology and Metabolism
Volume 6 - 2015 | https://doi.org/10.3389/fmicb.2015.00187
This article is part of the Research Topic Regulatory potential of post-translational modifications in bacteria View all 21 articles
Low molecular weight (LMW) thiols function as thiol-redox buffers to maintain the reduced state of the cytoplasm. The best studied LMW thiol is the tripeptide glutathione (GSH) present in all eukaryotes and Gram-negative bacteria. Firmicutes bacteria, including Bacillus and Staphylococcus species utilize the redox buffer bacillithiol (BSH) while Actinomycetes produce the related redox buffer mycothiol (MSH). In eukaryotes, proteins are post-translationally modified to S-glutathionylated proteins under conditions of oxidative stress. S-glutathionylation has emerged as major redox-regulatory mechanism in eukaryotes and protects active site cysteine residues against overoxidation to sulfonic acids. First studies identified S-glutathionylated proteins also in Gram-negative bacteria. Advances in mass spectrometry have further facilitated the identification of protein S-bacillithiolations and S-mycothiolation as BSH- and MSH-mixed protein disulfides formed under oxidative stress in Firmicutes and Actinomycetes, respectively. In Bacillus subtilis, protein S-bacillithiolation controls the activities of the redox-sensing OhrR repressor and the methionine synthase MetE in vivo. In Corynebacterium glutamicum, protein S-mycothiolation was more widespread and affected the functions of the maltodextrin phosphorylase MalP and thiol peroxidase (Tpx). In addition, novel bacilliredoxins (Brx) and mycoredoxins (Mrx1) were shown to function similar to glutaredoxins in the reduction of BSH- and MSH-mixed protein disulfides. Here we review the current knowledge about the functions of the bacterial thiol-redox buffers glutathione, bacillithiol, and mycothiol and the role of protein S-thiolation in redox regulation and thiol protection in model and pathogenic bacteria.
The cytoplasm is a reducing environment and protein thiols are maintained in their reduced state by low molecular weight (LMW) thiol-redox buffers and enzymatic thiol-disulfide oxidoreductases, including the thioredoxin and glutaredoxin systems (Fahey, 2013; Van Laer et al., 2013). In their natural environment or during infections, bacteria encounter different reactive species, such as reactive oxygen, nitrogen, chlorine, and electrophilic species (ROS, RNS, RCS, RES) (Antelmann and Helmann, 2011; Gray et al., 2013a). These reactive species cause different post-translational thiol-modifications in proteins and activate or inactivate specific transcription factors resulting in expression of detoxification pathways. LMW thiol-redox buffers function in detoxification of different reactive species and are often present in millimolar concentrations in the cytoplasm.
The best studied LMW thiol is glutathione (GSH) present in eukaryotes and Gram-negative bacteria (Fahey, 2013). Most Gram-positive bacteria do not produce GSH. Instead, the Actinomycetes utilize mycothiol (MSH) as thiol-redox buffer (Jothivasan and Hamilton, 2008; Newton et al., 2008). In Bacillus megaterium, Bacillus cereus, and Staphylococcus aureus, coenzyme A (CoASH) serves as an abundant LMW thiol (Newton et al., 1996). Many Firmicutes bacteria, including Bacillus and Staphylococcus species utilize bacillithiol (BSH) and cysteine as major thiol-redox buffers (Newton et al., 2009). Alternative LMW thiols include also the betaine-histidine derivative ergothioneine that compensates for the absence of MSH in Mycobacterium smegmatis mshA mutants (Ta et al., 2011). Cysteine is used for alternative S-thiolations in the absence of BSH and MSH in Bacillus subtilis and Corynebacterium glutamicum since S-cysteinylated proteins were identified in bsh and msh mutants (Chi et al., 2011, 2014).
The protozoa Leishmania and Trypanosoma produce the glutathione-derivative trypanothione (bis-glutathionyl-spermidine or TSH2). In Escherichia coli, glutathionylspermidine (GSP) was detected during the stationary phase (Fahey, 2013). Some microaerophilic γ-proteobacteria utilize glutathione amide (GASH) which forms a persulfide (GASSH) during photoautotrophic growth on high concentrations of sulfide (Bartsch et al., 1996).
Under conditions of oxidative stress, LMW thiols form mixed disulfides with protein thiols which is termed protein S-thiolation. In eukaryotes, protein S-glutathionylation has emerged as major redox-regulatory mechanism that controls the activity of redox sensing transcription factors and protects active site Cys residues against irreversible oxidation to sulfonic acids (Dalle-Donne et al., 2009). S-glutathionylation controls numerous physiological processes, such as cellular growth and differentiation, cell cycle progression, transcriptional activity, cytoskeletal function, cellular metabolism, and apoptosis (Klatt and Lamas, 2000; Ghezzi, 2005, 2013; Dalle-Donne et al., 2007, 2009). S-glutathionylation must meet several criteria to function as redox-control mechanism: (1) reversibility, (2) specificity to active site Cys, (3) change in protein function/activity, and (4) induction by ROS or RNS. S-glutathionylation serves as a form of GSH storage to prevent the export of GSSG under oxidative stress (Dalle-Donne et al., 2009). Many eukaryotic proteins, like α-ketoglutarate dehydrogenase, glyceraldehyde 3-phosphate dehydrogenase, ornithine δ-aminotransferase, pyruvate kinase, heat specific chaperones, and regulatory proteins (c-Jun, NF-κB) are reversibly inactivated or activated by S-glutathionylation (Dalle-Donne et al., 2009; Kehr et al., 2011). However, the regulatory role of protein S-thiolation for bacterial physiology has only recently been investigated. Here we review the current knowledge about the functions of the bacterial redox buffers GSH, MSH, and BSH and their roles for protein S-thiolations in GSH-, MSH- and BSH-producing bacteria.
Bacteria encounter ROS during respiration or by the oxidative burst of activated neutrophils during infections (Imlay, 2003, 2008, 2013). The incomplete stepwise reduction of molecular oxygen (O2) leads to generation of superoxide anions (O2•−), hydrogen peroxide (H2O2) and the highly reactive hydroxyl radical (OH•) (Figure 1). Superoxide anion and H2O2 are also produced by autoxidation of flavoenzymes (Mishra and Imlay, 2012; Imlay, 2013). Superoxide dismutases (SOD) convert O2•− to H2O2. Several peroxide scavenging enzymes, such as catalases and peroxidases catalyze the detoxification of H2O2. H2O2 reacts with ferrous iron (Fe2+) in the Fenton reaction generating the highly toxic hydroxyl radical (OH•) which can damage all cellular macromolecules (Imlay, 2003, 2008). H2O2 destroys the Fe-S-cluster of dehydratases and inactivates single ferrous iron-centers of redox enzymes (Mishra and Imlay, 2012; Imlay, 2013).
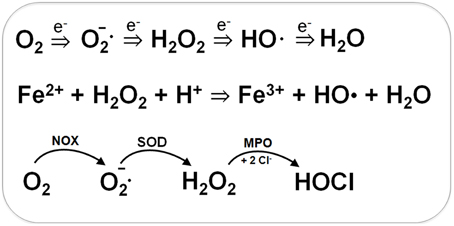
Figure 1. Generation of Reactive Oxygen Species (ROS) during respiration and HOCl production by activated neutrophils during infections. ROS are generated in bacteria during respiration by stepwise one-electron transfer to O2 producing superoxide anion, hydrogen peroxide and hydroxyl radical. The highly reactive hydroxyl radical is also produced from H2O2 and Fe2+ in the Fenton reaction. During infections, activated neutrophils generate superoxide anion by the NADPH oxidase (NOX) that is converted to H2O2 by the superoxide dismutase (SOD). Myeloperoxidase (MPO) is released upon degranulocytosis producing the highly reactive hypochlorous acids (HOCl) from H2O2 and Cl− as potent killing agent for pathogenic bacteria.
During the oxidative burst, activated neutrophils release O2•−, H2O2, nitric oxide (NO), and hypochlorous acid (HOCl) with the aim to kill invading pathogenic bacteria (Forman and Torres, 2001; Winterbourn and Kettle, 2013). The neutrophil NADPH oxidase (NOX) shuttles electrons from NADPH to O2 in the phagosomal lumen and generates around 20 μM superoxide anion. Myeloperoxidase (MPO) is released upon degranulation in millimolar concentrations. MPO catalyzes the dismutation of O2•− to H2O2 and subsequent conversion of H2O2 and chloride to HOCl (Figure 1) (Winterbourn and Kettle, 2013). NO is generated in neutrophils by the inducible nitric oxide synthase (iNOS) catalyzing the oxidation of L-arginine to L-citrulline. The reaction of NO with O2•− leads to formation of peroxynitrite (ONOO−). Thus, neutrophils release ROS, RNS, and the highly reactive HOCl as antimicrobial defense mechanism.
Reactive electrophilic species (RES) have electron-deficient centers and can react with the nucleophilic Cys thiol group via the thiol-S-alkylation chemistry (Figure 2) (Antelmann and Helmann, 2011). RES include quinones, aldehydes, epoxides, diamide and α,β-unsaturated dicarbonyl compounds. RES are often generated as secondary reactive intermediates from oxidation products of amino acids, lipids or carbohydrates (Marnett et al., 2003; Rudolph and Freeman, 2009). Quinones are lipid-electron carriers of the respiratory chain, including ubiquinone and menaquinone (Farrand and Taber, 1974). Soil bacteria encounter quinones as redox active components of humic substances and in dissolved organic matter (Ratasuk and Nanny, 2007). The toxic dicarbonyl compound methylglyoxal is produced in all organisms from triose-phosphate intermediates as byproduct of the glycolysis and can be generated also from amino acids metabolism (Ferguson et al., 1998; Booth et al., 2003; Kalapos, 2008). Bacteria also have to cope with the carbonyl compound formaldehyde. Formaldehyde is an intermediate in the C1-metabolism of methanotrophic and methylotrophic bacteria and is ubiquitously distributed in the environment. Thus, bacteria have evolved conserved pathways for detoxification of formaldehyde and methylglyoxal that involve LMW thiols.
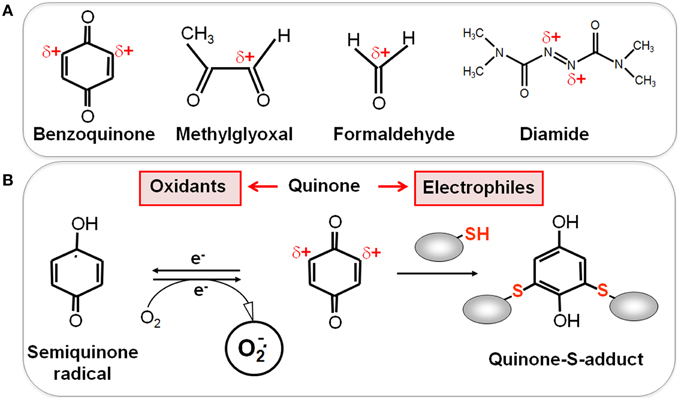
Figure 2. Reactive Electrophilic Species (RES) with partial positive charges (δ+) (A) and the reaction of quinones with thiols via the S-alkylation and oxidation chemistry (B). (A) In quinones and aldehydes the electrons are drawn to carbonyl oxygen leaving the partial positive charges at neighboring carbon atoms that become electrophilic. Diamide is an electrophilic azocompound that causes disulfide stress. (B) Quinones can react as electrophiles with the nucleophilic thiol group of Cys residues via thiol-S-alkylation leading to irreversible thiol-S-adduct formation. In the oxidative mode, quinones are incompletely reduced to semiquinone radicals that generate superoxide anions and can oxidize protein thiols to disulfides.
In eukaryotic cells, RES are implicated in many pathophysiological processes and modulate signaling pathways (Mackay and Knock, 2015). Eukaryotic cells produce lipid-derived RES, such as malondialdehyde (MDA) and 4-hydroxy-2-nonenal (HNE) (Rudolph and Freeman, 2009). HNE is generated from polyunsaturated fatty acids in biological membranes by a radical-based peroxidation chain reaction (Jacobs and Marnett, 2010). Furthermore, 15-deoxy-Δ12,14-prostaglandin J2 (15d-PGJ2) is generated from arachidonic acid during inflammation and 2-trans-hexadecenal (2-HD) is produced during sphingolipid metabolism which promotes apoptosis (Wang et al., 2014). Bacterial membrane lipids also contain unsaturated fatty acids which are synthesized at higher levels during adaptation to cold shock to maintain the fluidity of the membrane (De Mendoza, 2014). These unsaturated fatty acids in bacterial membrane lipids could be the target for ROS leading to lipid peroxidation products in bacteria. Lipid hydroperoxides, such as linoleic acid hydroperoxide are sensed by the redox-sensing MarR-type repressor OhrR. OhrR regulates the peroxiredoxin OhrA that functions in detoxification of organic hydroperoxides (Atichartpongkul et al., 2001; Fuangthong et al., 2001). However, the fatty acid-derived peroxidation product which is sensed by OhrR in vivo remains to be identified.
ROS, RES, and RCS can damage all cellular macromolecules including proteins, nucleic acids or carbohydrates (Imlay, 2008, 2013; Jacobs and Marnett, 2010; Gray et al., 2013a). However, in eukaryotes low levels of ROS and RES act also as second messengers to modulate signal transduction pathways (Rudolph and Freeman, 2009; Mackay and Knock, 2015). Bacterial transcription factors often use redox-sensitive Cys residues for sensing of ROS, RES, and RCS to control the expression of specific detoxification pathways (Antelmann and Helmann, 2011; Gray et al., 2013a). The thiol group of cysteine is subject to reversible and irreversible post-translational thiol-modifications that lead to inactivation or activation of the transcription factor. Protein thiols can be reversibly oxidized to protein disulfides and irreversibly overoxidized to sulfinic or sulfonic acids by ROS (Antelmann and Helmann, 2011). ROS lead first to oxidation of protein thiols to Cys sulfenic acids as unstable intermediates (R-SOH) (Figure 3). Cys sulfenic acid rapidly reacts further with other thiols to form intramolecular and intermolecular protein disulfides or mixed disulfides with LMW thiols, collectively termed as S-thiolations (e.g., S-cysteinylations, S-glutathionylations, S-mycothiolations, and S-bacillithiolations). Protein S-thiolations protect the thiol groups against the irreversible overoxidation to Cys sulfinic (R-SO2H) and sulfonic acid (R-SO3H). This is particularly important for essential and abundant proteins whose overoxidation would lead to loss of cell viability and requires new protein synthesis to replace inactivated proteins. However, eukaryotic sulfiredoxins are able to reduce sulfinic acids in 2-Cys peroxiredoxins, but sulfiredoxins are not present in bacteria (Lowther and Haynes, 2011).
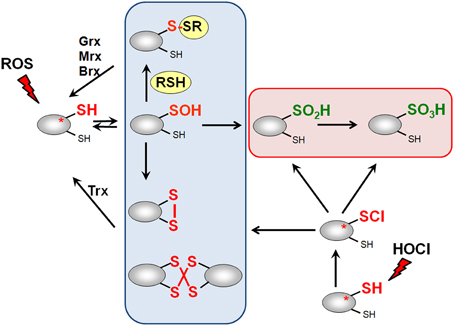
Figure 3. Thiol-chemistry of ROS and HOCl with thiol-containing proteins. The Cys thiol group is oxidized by ROS to an unstable Cys sulfenic acid intermediate (Cys-SOH) that reacts further with proximal thiols to form intramolecular and intermolecular disulfides or mixed disulfides with LMW thiols (RSH), such as glutathione, bacillithiol or cysteine, termed as S-thiolations. HOCl leads to chlorination of protein thiols to sulfenylchloride intermediates (Cys-SCl) that react further to form disulfides. In the absence of proximal thiols, the chlorinated Cys is overoxidized to Cys sulfinic and sulfonic acids. Disulfides function as redox switches to control protein activity and protect thiol groups against overoxidation to Cys sulfinic and sulfonic acids.
Hypochloric acid (HOCl) is a strong two-electron oxidant and chlorinating agent with a high redox potential [E0′(HOCl/Cl−) = 1.28 mV] (Davies, 2011). HOCl targets most strongly the sulfur-containing amino acids cysteine and methionine with the second-order rate constant of k = 3 × 107 M−1 s−1 (Hawkins et al., 2003). HOCl first chlorinates the thiol group to form the unstable sulfenylchloride intermediate that reacts further with another thiol group to disulfides. In the absence of another thiol, the chlorinated thiol group is overoxidized very rapidly to sulfinic or sulfonic acids (Hawkins et al., 2003) (Figure 3). We confirmed that strong disulfide stress responses are caused by HOCl in different Gram-positive bacteria in vivo and detected mixed protein disulfides with Cys, BSH, and MSH as major oxidation products (Chi et al., 2011, 2013, 2014).
RNS cause reversible thiol-modifications: nitric oxide (NO) leads to S-nitrosothiol formation (RS-NO) and peroxinitrite (ONOO−) causes S-nitrothiol (RS-NO2) formation. Alternatively, S-nitrosothiol (e.g., GSNO or MSNO) can be formed by direct reaction of NO with LMW thiols (Antelmann and Helmann, 2011).
RES can react via the thiol-S-alkylation chemistry with Cys thiols. However, quinones have two modes of action, an oxidation and an alkylation mode. In the oxidation mode, the one-electron reduction of quinones generates the highly reactive semiquinone radical leading to generation of superoxide anions (Figure 2). The electrophilic reaction of quinones involves the 1,4-reductive Michael-type addition of thiols to quinones (Marnett et al., 2003). Quinones lead to irreversible thiol-S-alkylation and protein aggregation to deplete protein thiols in the proteome in vivo (Liebeke et al., 2008). However, non-toxic quinone concentrations resulted in reversible thiol-disulfide switches in RES-sensing redox regulators (e.g., YodB, CatR, QsrR, NemR) to activate the expression of specific quinone detoxification pathways (Antelmann and Helmann, 2011; Gray et al., 2013a; Lee et al., 2013). Methylglyoxal reacts with nucleophilic centers of the DNA and with the amino acids arginine, lysine and cysteine causing advanced glycation end-products (Bourajjaj et al., 2003). The lipid-derived electrophiles MDA and HNE were shown to alkylate DNA bases and protein thiols leading to DNA and membrane damages in eukaryotes (Rudolph and Freeman, 2009).
The tripeptide glutathione (γ-glutamylcysteinyl-glycine; GSH) is utilized as major LMW thiol-redox buffer in Gram-negative bacteria and in some Gram-positive Firmicutes bacteria, including Streptococcus agalactiae, Listeria monocytogenes, and Clostridium acetobutylicum (Figure 4). In E. coli, GSH biosynthesis occurs in two steps: The γ-glutamate cysteine ligase (GshA) catalyzes the formation of γ-glutamylcysteine (γ-Glu-Cys) from glutamate and cysteine. In the second step, ligation of glycine to γ-Glu-Cys is catalyzed by glutathione synthase (GshB) (Meister, 1995; Anderson, 1998). In S. agalactiae and L. monocytogenes, a bifunctional fusion protein GshF is present that exhibits both GshA and GshB activities (Gopal et al., 2005; Janowiak and Griffith, 2005). Interestingly, Lactococcus lactis, Streptococcus pneumoniae and Haemophilus influenzae do not synthesize GSH, but encode GSH-uptake mechanisms. In S. pneumoniae, the GSH-uptake from the host is mediated by the ABC transporter binding protein GshT (Potter et al., 2012; Vergauwen et al., 2013). In addition, the cystine importer TcyBC was shown to be primed for GSH import by GshT. In H. influenzae, GSH import is mediated by the ABC-transporter DppBCDF and requires the periplasmic GSH-binding protein GbpA (Vergauwen et al., 2010). Strikingly, these pathogens utilize host-derived GSH as protection mechanism against the host immune defense.
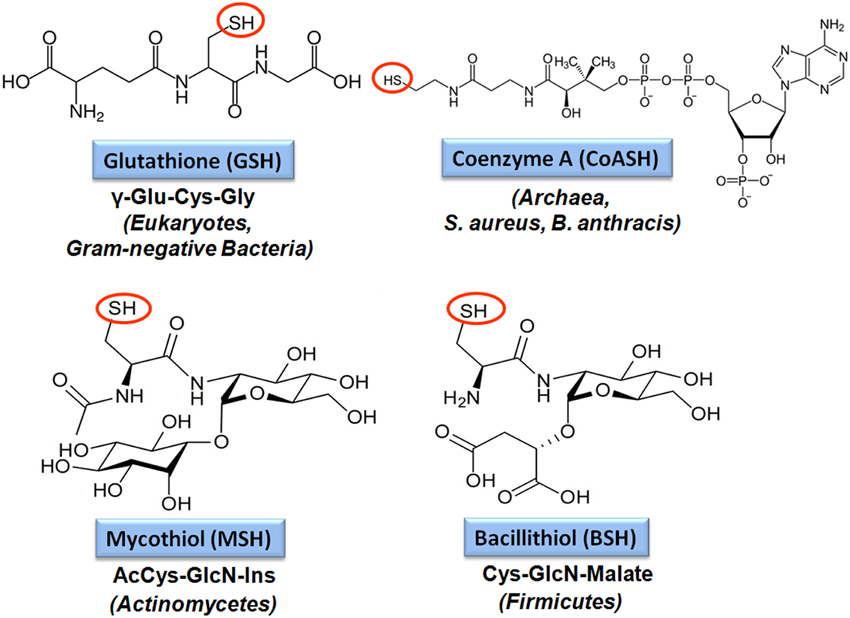
Figure 4. Structures of major bacterial low molecular weight (LMW) thiols. Major LMW thiols are glutathione (GSH) in eukaryotes and Gram-negative bacteria, mycothiol (MSH) in Actinomycetes and bacillithiol (BSH) in Firmicutes. Coenzyme A (CoASH) also serves as a LMW thiol-redox buffer in some bacteria, like in S. aureus and B. anthracis.
GSH is present in millimolar concentrations in the cytoplasm of E. coli (Masip et al., 2006; Fahey, 2013). GSH maintains protein thiols in the reduced state and serves as a storage form of cysteine. In contrast to cysteine, GSH is resistant to metal-catalyzed autooxidation because of its bound Cys amino and carboxyl groups that prevent ligation of heavy metal ions (Fahey, 2013). During the bacterial growth and under oxidative stress, GSH is oxidized to glutathione disulfide (GSSG). The NADPH-dependent glutathione reductase (Gor) keeps GSH in its reduced state to maintain a high GSH/GSSG ratio ranging from 30:1 to 100:1 in the cytoplasm. The standard thiol-disulfide redox potential of the GSH redox couple was calculated as E0′(GSSG/GSH) = −240 mV at physiological pH values (Hwang et al., 1995; Van Laer et al., 2013).
The various detoxification functions of GSH have been extensively studied in E. coli gsh mutants. In E. coli, GSH functions in detoxification of ROS, RES, RCS, RNS, xenobiotics, antibiotics, toxic metals, and metalloids (Masip et al., 2006) (Table 1). Detoxification of xenobiotics, electrophiles and antibiotics by GSH occurs either spontaneously by S-conjugation or by the catalytic activity of GSH-S-transferases (Fahey, 2013). The GSH-S-conjugates are usually excreted from the cell as non-toxic mercapturic acid derivatives. GSH was shown to function as a cofactor in methylglyoxal detoxification in E. coli. The major pathway for methylglyoxal detoxification in E. coli is the GSH-dependent glyoxalase pathway. The glyoxalase-I (GloA) catalyzes formation of S-lactoyl-GSH from GSH-hemithioacetal and glyoxalase-II (GloB) converts S-lactoyl-GSH to D-lactate (Ferguson et al., 1998; Booth et al., 2003). In addition, glyoxalase-III operates GSH-independently to convert methylglyoxal to lactate. The glyoxalase-I encoding gloA gene and the nemRA operon are redox-regulated by the NemR repressor under methylglyoxal, quinone and HOCl stress (Gray et al., 2013b; Lee et al., 2013; Ozyamak et al., 2013). The glyoxalase GloA and the oxidoreductase NemA are important for methylglyoxal survival and confer resistance to methylglyoxal in E. coli (Ozyamak et al., 2013). The resistance to methylglyoxal is also linked to the activation of potassium efflux by the S-lactoyl-GSH intermediate leading to cytoplasmic acidification (Ferguson et al., 1998; Booth et al., 2003). The cytoplasmic acidification limits the interaction of methylglyoxal with DNA bases. Thus, potassium efflux and detoxification by GloA protect against methylglyoxal toxicity in E. coli.
Interestingly, expression of the E. coli gshAB genes in the industrial important Clostridium acetobutylicum enhances robustness and alcohol production as a promising strategy for engineering industrial production strains. Thus, GSH protects also against large-scale ethanol and butanol production in C. acetobutylicum during fermentation (Hou et al., 2013).
GSH has many detoxification functions to maintain the redox balance of the cytoplasm, but only recently the role of GSH for the control of virulence functions has been explored in the pathogenic bacteria S. pneumoniae, L. monocytogenes, and Salmonella Typhimurium (Potter et al., 2012; Song et al., 2013; Reniere et al., 2015) (Table 2). In S. pneumoniae, the glutathione reductase Gor and the GSH-importer GshT were required for oxidative stress protection and metal ion resistance. Moreover, the gshT mutant was attenuated in colonization and invasion in a mouse model of pneumococcal infection (Potter et al., 2012). Thus, GSH protects against the host immune defense and contributes to fitness of S. pneumoniae.
The intracellular pathogen L. monocytogenes is able to synthesize GSH via the gshF fusion protein, but GSH can be also imported from the host (Reniere et al., 2015). The L. monocytogenes gshF mutant was two-fold less virulent compared to the wild type in a mice model. In addition, the gshF mutant was sensitive to oxidative stress, contains lower levels of ActA and formed small plaques in tissue culture assays that measure cell-to-cell spread (Reniere et al., 2015). The Actin assembly-inducing protein ActA is controlled by the virulence regulator PrfA and used by L. monocytogenes to move through the host cells (Freitag et al., 2009). It was shown that the virulence phenotype of the gshF mutant is caused by the lack of PrfA activation by bacterial and host-derived GSH (Reniere et al., 2015). Interestingly, activation of PrfA is mediated by an allosteric binding of GSH to PrfA, but not by S-glutathionylation. Thus, GSH plays a role as signaling molecule to activate virulence gene expression in an intracellular pathogen.
In S. Typhimurium, GSH antagonizes the bacteriostatic effects of RNS in vivo and gshA mutants were sensitive to ROS and RNS (Song et al., 2013). Thus, GSH presents a first line defense against ROS and RNS produced by the host immune system. This was shown in an acute model of salmonellosis in mice expressing the wild-type NRAMP1R allele (natural resistance-associated macrophage protein 1) which is linked to high NO production of the macrophages. The gshA and gshB mutants were attenuated in this acute model of salmonellosis. It was further shown that GSH protects against ROS and RNS produced by the NADPH phagocyte oxidase and inducible nitric oxide synthase (iNOS) in this mice model (Song et al., 2013). These recent studies highlight the important roles of GSH in the control of virulence functions, expression of virulence factors and pathogen fitness under infection conditions in S. pneumoniae, L. monocytogenes, and S. Typhimurium. As shown for L. monocytogenes, GSH might play similar roles to activate virulence gene expression by redox control of virulence gene regulators in other pathogens which remains to be elucidated.
Advances in probe design and mass spectrometry-based thiol-trapping approaches have facilitated the detection of specific reversible thiol-modifications, including sulfenylation, nitrosylation, glutathionylation, and sulfhydrations of proteins at a global scale (Leonard and Carroll, 2011; Pan and Carroll, 2013; Paulsen and Carroll, 2013; Gupta and Carroll, 2014; Zhang et al., 2014). Different methods have been applied for specific detection of S-glutathionylations in eukaryotic cells, including the use of GSH-specific antibodies and the labeling of S-thiolations with 35S-cysteine followed by 2D gel electrophoresis and phosphoimaging (Fratelli et al., 2002, 2003, 2004). However, the specificity of the GSH antibodies is questionable and the gel-based detection of S-thiolations using 35S-cysteine does not distinguish between S-cysteinylations and S-glutathionylations or other forms of S-thiolations. Hence, more sensitive mass spectrometry-based redox proteomics methods have been developed, including the glutaredoxin-coupled NEM-biotin switch assay and the treatment of cell extracts with N,N-biotinyl glutathione disulfide (BioGSSG) (Lind et al., 2002; Brennan et al., 2006; Kehr et al., 2011; Zaffagnini et al., 2012a) (Figure 5). Both methods make use of the specific streptavidin enrichment of biotinylated peptides that improve the identification of S-glutathionylated peptides using mass spectrometry. The NEM biotin-switch assay was successfully applied to detect protein S-glutathionylation in eukaryotic endothelial cells and malaria parasites which applies glutaredoxin for reduction of protein-SSG followed by NEM-biotin alkylation and enrichment using streptavidin columns (Lind et al., 2002; Kehr et al., 2011). The biotin-GSSG approach has been applied to identify S-glutathionylated proteins in the green algae Chlamydomonas reinhardtii (Zaffagnini et al., 2012a) and in the photosynthetic cyanobacterium Synechocystis sp. PCC6803 (Chardonnet et al., 2015). In total, 383 S-glutathionylated proteins were identified in Synechocystis sp. PCC6803 and 125 glutathionylation sites were mapped by mass spectrometry. In addition, the peroxiredoxin PrxII (Sll1621) and the 3-phosphoglycerate dehydrogenase PGDH (Sll1908) could be S-glutathionylated by BioGSSG in vitro (Chardonnet et al., 2015).
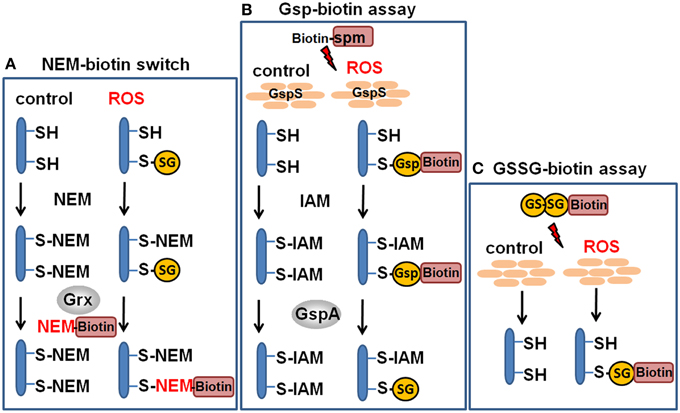
Figure 5. Redox proteomics methods to study protein S-glutathionylation at a global scale. Mass spectrometry-based methods for identification of S-glutathionylations include the glutaredoxin-coupled NEM-biotin switch assay (A), the biotin-Gsp assay, (B) or the N,N-biotinyl glutathione disulfide (BioGSSG) assay, (C) (Lind et al., 2002; Brennan et al., 2006; Kehr et al., 2011; Zaffagnini et al., 2012a). In the biotin-Gsp assay, E. coli GspS is expressed in mammalian cells and converts GSH and biotinyl-spermine (biotine-spm) to biotin-glutathionylspermidine (biotin-Gsp). Proteins in ROS-treated cells are modified by biotin-Gsp-S-thiolation (Chiang et al., 2012; Lin et al., 2015). The biotin-spm is removed from the enriched biotin-Gsp-S-thiolated peptides by GspA and the GSS-peptides are identified by mass spectrometry.
In another approach, biotinyl-spermine (biotine-spm) and in vivo expressed E. coli GspS have been applied for mammalian cells to convert GSH to biotin-glutathionylspermidine (biotin-Gsp) which subsequently modified proteins by Biotin-Gsp-S-thiolation (Chiang et al., 2012; Lin et al., 2015). The biotine-spm is removed enzymatically by GspA from the enriched biotin-Gsp-S-thiolated peptides and the GSS-peptides are identified by mass spectrometry. This approach allows the identification of S-glutathionylation sites without the biotin-tag and enhances the coverage for S-glutathionylated proteins. In mammalian cells, 1409 S-glutathionylated cysteines in 913 proteins were identified using the Gsp-biotin approach (Chiang et al., 2012; Lin et al., 2015). This makes the application of this chemoenzymatic approach using the GspS enzyme attractive for global and specific studies of S-glutathionylated proteins.
In S. Typhimurium, protein S-glutathionylation has been studied at a global scale by top-down and bottom-up proteomic approaches (Ansong et al., 2013). Top-down proteomics uses whole proteins for separation and fragmentation directly in the mass spectrometer. In bottom-up proteomics approaches proteins are digested by a protease and the peptide mixtures are analyzed by mass spectrometry to identify proteins at the peptide level. The top-down proteomic approach identified 563 proteins with 1665 post-translational modifications in S. Typhimurium. The authors identify 25 S-thiolated proteins in cells grown in complete LB medium including 16 S-glutathionylated proteins and nine S-cysteinylated proteins. Interestingly, a subset of nine S-glutathionylated are modified by S-cysteinylation in infection-like minimal LPM medium (Table 3). This could indicate a shift from S-glutathionylation to S-cysteinylation under infection-like conditions in S. Typhimurium. These S-thiolated proteins include phosphoglycerate kinase (Pgk), elongation factor (Tuf) and enolase (Eno) that are also targets for S-glutathionylations in endothelial cells (Fratelli et al., 2002). The top-down proteomics results were verified by bottom-up proteomics approaches to identify the specific S-thiolated Cys peptides (Ansong et al., 2013). Structural analysis revealed that S-glutathionylation occurred mostly at buried Cys residues and not at surface-exposed Cys. S-glutathionylation on buried Cys was also shown for the enolase whose activity is known to be modified by S-thiolation in human cells (Fratelli et al., 2002). It is postulated that S. Typhimurium switches from S-glutathionylation to S-cysteinylation during infection conditions as novel redox-control mechanism. In agreement with the proteome data, transcriptome results point to an up-regulation of Cys biosynthesis and down-regulation of GSH biosynthesis under infection-like conditions. However, the physiological role of this S-thiolation switch for redox control of the identified protein targets under ROS stress remains to be elucidated. Furthermore, no blocking of reduced thiols with NEM or IAM was performed to avoid artificial disulfide formation. Thus, it remains to be verified that the observed S-thiolations are not caused by artificial thiol-disulfide exchange. Previous studies have also shown that L. monocytogenes is able to both import and synthesize GSH (Reniere et al., 2015). Furthermore, the non-GSH-utilizing S. aureus was shown to import GSH during growth in LB medium (Pöther et al., 2013). Thus, the possible uptake of GSH in S. Typhimurium from LB-medium could contribute to the observed S-glutathionylations which needs to be investigated.
The role of protein S-glutathionylation for redox control has been studied in few Gram-negative bacteria, including E. coli, S. Typhimurium, Neisseria meningitidis, Pseudoalteromonas haloplanktis, and Synechocystis sp. PCC6803 (Table 3). In E. coli, the peroxide-sensing regulator OxyR is activated by S-glutathionylation at its redox-sensing Cys199 in vitro (Kim et al., 2002). In addition, the activities of glyceraldehyde-3-phosphate dehydrogenase, methionine synthase and the PAPS reductase are inhibited by S-glutathionylation in E. coli (Lillig et al., 2003; Hondorp and Matthews, 2004; Brandes et al., 2009). A recent study provides a model for the S-glutathionylation of the conserved active site Cys in GapDH and explains the reactivity of the active site toward H2O2 (Peralta et al., 2015). Reaction of the active site Cys with H2O2 is catalyzed by a mechanism which stabilizes the transition state and promotes leaving group departure by providing a proton relay. This model suggests the conserved redox-regulation of GapDH by S-thiolation of its active site Cys across all domains of life.
In Neisseria meningitidis, an esterase EstD acts together with the GSH-dependent alcohol dehydrogenase AdhC in formaldehyde detoxification. EstD is inactivated via S-glutathionylation at its conserved Cys54 by its substrate S-formyl-GSH during formaldehyde detoxification in vitro (Chen et al., 2013). In the psychrophilic bacterium Pseudoalteromonas haloplanktis, S-glutathionylation of the iron-superoxide dismutase PhSOD at the single Cys57 protected the enzyme from tyrosine nitration and peroxynitrite inactivation in vitro and in vivo (Castellano et al., 2008).
In Synechocystis sp. PCC6803, a MerA-like enzyme that functions in mercury and uranium reduction was shown to be redox-controlled by S-glutathionylation (Marteyn et al., 2013). MerA was S-glutathionylated at Cys78 that is required for mercury reduction resulting in inhibition of MerA activity. MerA redox regulation and reactivation required reduction by glutaredoxin-1 (Grx1). The active site Cys31 and Cys86 of Grx-1 operate in MerA interactions and both Cys are required for MerA reactivation. Furthermore, S-glutathionylation was shown to control the activity of the transcription factor AbrB2 in Synechocystis sp. PCC6803 (Cassier-Chauvat et al., 2014). AbrB2 is a repressor of the hydrogenase-encoding hoxEFUYH operon and also down-regulates antioxidant genes, such as cydAB encoding the cytochrome bd-quinol oxidase and norB encoding the nitric oxide reductase. The production of hydrogen is thought to be an antioxidant mechanism to eliminate electrons for oxygen reduction and ROS generation. AbrB2 contains a conserved single cysteine that is essential for redox-regulation and oligomerisation of AbrB2 as shown in C34S mutants. S-glutathionylation of Cys34 affected the binding of AbrB2 to the hox promoter and the stability of AbrB2 in vitro. In conclusion, S-glutathionylation has been shown to function in the redox-control of two transcriptional regulators, OxyR and AbrB2 in Gram-negative bacteria in vitro. However, compared to the many targets for S-glutathionylation that have been studied in eukaryotic organisms, there is much to be discovered about the regulatory potential of S-glutathionylation in bacteria.
Protein S-glutathionylation is a reversible redox switch mechanism. The glutaredoxin (Grx)/GSH/GSH reductase (Gor) system catalyzes specific de-glutathionylation of S-glutathionylated proteins (Fernandes and Holmgren, 2004; Inaba, 2009). Grx were first discovered in E. coli (Holmgren, 1976) where they have important functions as electron donors for ribonucleotide reductase (RNR), adenosine-5′-phosphosulfate (APS) reductase, 3′-phosphoadenosine-5′-phosphosulfate (PAPS) reductase and arsenate reductases (Holmgren, 1981; Aslund et al., 1994). Grx are structurally classified into the classical di-thiol Grxs with a CPTC redox active site and the monothiol Grx containing a CGPS redox active site (Lillig et al., 2008). In E. coli, three di-thiol Grx proteins (Grx1, Grx2, and Grx3) and one monothiol protein (Grx4) have been characterized.
The de-glutathionylation by Grx enzymes involves thiol-disulfide exchange reactions with GSH via nucleophilic double displacement (ping–pong) mechanisms and occurs via mono- or di-thiol mechanisms. Most di-thiol Grx use monothiol mechanisms that take place in two steps: In the first step, the nucleophilic thiolate anion attacks the S-glutathionylated substrate protein, resulting in reduction of the mixed disulfide and the S-glutathionylated Grx (Grx-SSG) intermediate. This Grx-SSG intermediate is regenerated by GSH and Gor at expense of NADPH (Allen and Mieyal, 2012). The di-thiol mechanism involves a second active site Cys that forms an intramolecular disulfide to resolve the Grx-SSG intermediate that has been shown for some plant Grx enzymes (Zaffagnini et al., 2012b). However, this di-thiol mechanism of Grx is less efficient for protein de-glutathionylation and more involved in the reduction of intermolecular protein disulfides (Lillig et al., 2008; Allen and Mieyal, 2012). Thus far, the knowledge about Grx functions and substrates in most GSH-producing bacteria is scarce and remains an important subject for future studies.
Bacillithiol (BSH) is composed of Cys-GlcN-malate and serves as major LMW thiol in many Firmicutes bacteria, including Bacillus and Staphylococcus species, Deinococcus radiodurans, and Streptococcus agalactiae (Newton et al., 2009) (Figure 4). The BSH biosynthesis pathway was first identified in B. subtilis. In the first step, the glycosyltransferase BshA couples UDP-GlcNAc to L-malic acid for generation GlcNAc-Mal (Ruane et al., 2008; Gaballa et al., 2010; Parsonage et al., 2010). The deacetylase BshB1 catalyzes deacetylation of GlcNAc-Mal to GlcN-Mal. The last step involves the putative cysteine ligase YllA (BshC) that presumably adds Cys to GlcN-Mal (Gaballa et al., 2010). BshB1 has a paralog BshB2 and both enzymes have deacetylase activity. The functional redundancy of BshB1 and BshB2 in B. subtilis suggests that BshB2 might function as BSH-S-conjugate amidase Bca in detoxification of RES similar to the MSH-S-conjugate amidase Mca (Parsonage et al., 2010). The functions of the BshB1/2 homologs of B. anthracis (BA1557 and BA3888) and B. cereus (BC1534 and BC3461) in the deacetylation of GlcNAc-Mal have been demonstrated in vitro. In addition, BA3888 was shown to function as BSH-S-conjugate amidase (Bca) (Fang et al., 2013). In contrast to BshA and BshB, the activity of the putative cysteine ligase BshC has never been demonstrated biochemically in vitro. The structure of BshC was resolved revealing a core Rossmann fold with connecting peptide motifs (CP1 and CP2) and an α-helical coiled-coil domain required for dimerization (Vanduinen et al., 2015). BshC was crystallized with citrate and glycerol in the canonical active site and ADP bound in a second binding pocket that is different from the ADP-binding pocket in the related MshC structure. The active sites are solvent exposed and open for possible interactions with a protein, substrate or cofactor that remain to be elucidated to understand the catalytic mechanism of BshC (Vanduinen et al., 2015).
The regulation of the BSH biosynthesis genes has been studied in B. subtilis. The bshA and bshB1 genes belong to a large operon of seven genes including mgsA which encodes a methylglyoxal synthase. The bshB2 and bshC genes are encoded by two different operons. The bshA, bshB, and bshC genes are induced under conditions of disulfide stress provoked by diamide or NaOCl and positively controlled by the disulfide stress regulator Spx (Chi et al., 2011; Rochat et al., 2012; Gaballa et al., 2013). Consistent with the Spx-dependent control of the BSH biosynthesis genes, lower BSH levels were detected in the spx mutant using thiol-metabolomics (Chi et al., 2011; Rochat et al., 2012; Gaballa et al., 2013). It is interesting to note, that the Trx pathway and BSH biosynthesis genes are both regulated by the major disulfide stress regulator Spx in B. subtilis (Zuber, 2004, 2009; Chi et al., 2011; Rochat et al., 2012; Gaballa et al., 2013).
BSH is predominantly present in its reduced form in the cytoplasm with BSH/BSSB ratios ranging from 100:1 to 400:1 in B. subtilis indicating the presence of an efficient bacillithiol disulfide reductase (Sharma et al., 2013). The FAD-dependent pyridine nucleotide disulfide oxidoreductase YpdA (IPR023856) was suggested to function as BSSB reductase because of its phylogenetic relationship to the BSH biosynthesis enzymes as revealed by a STRING search (Gaballa et al., 2010). However, the function of YpdA has not yet been demonstrated.
The standard thiol-redox potential of BSH was calculated as E0′(BSSB/BSH) = −221 mV which is higher than the GSH redox potential [E0′(GSSG/GSH) = −240 mV] (Sharma et al., 2013). The microscopic pKa values of the thiol group of BSH were determined as pKa = 7.97 when the amino group of the Cys is protonated and as pKa = 9.55 in the presence of the deprotonated amino group of Cys (Sharma et al., 2013). Thus, the thiol group in BSH is more acidic compared to the thiol group in Cys suggesting an enhanced level and reactivity of the BSH thiolate anions to detoxify reactive species. The BSH concentrations in B. subtilis vary during the growth in LB medium and increase strongly during the stationary phase to 3.5–5.2 mM. In contrast, the cellular Cys concentration is kept at a relatively low level (0.13–0.28 mM). Thus, BSH concentrations are ~17-fold higher compared to the level of Cys (Sharma et al., 2013). Similar concentrations of BSH (2 mM) were measured in Bacillus pumilus during growth. In B. pumilus, BSH levels increased under peroxide stress to 6 mM which is caused by an increased bshB expression (Handtke et al., 2014). BSH levels are also two-fold increased under diamide and NaOCl stress in B. subtilis due to Spx-dependent induction of bshA, bshB, and bshC (Chi et al., 2013; Gaballa et al., 2013). In S. aureus, the BSH levels are lower (0.3–1 mM) in the different clinical isolates (COL, USA300, Mu50, or N315) and BSH levels are not up-regulated during the stationary phase (Posada et al., 2014).
The physiological functions of BSH were studied in bsh mutants of B. subtilis and S. aureus (Table 1). Phenotype analyses showed increased sensitivities of bsh mutants toward hypochlorite, diamide, methylglyoxal, ROS (paraquat, H2O2), osmotic, and acidic stress, alkylating agents and fosfomycin in B. subtilis (Gaballa et al., 2010; Chi et al., 2011). The fosfomycin-sensitive phenotype of bsh mutants depends on the epoxide hydrolase FosB that requires BSH as a cofactor to open the ring structure for fosfomycin detoxification (Lamers et al., 2012; Roberts et al., 2013; Thompson et al., 2013) (Figure 6). FosB shows a preference for BSH as thiol cofactor and does only work poorly with Cys. The biochemical activity has been demonstrated for various Bacillus and Staphylococcus FosB homologs (Lamers et al., 2012; Roberts et al., 2013; Thompson et al., 2013). In B. subtilis and S. aureus, both FosB and BSH confer resistance to fosfomycin treatment in survival assays in vivo (Gaballa et al., 2010; Thompson et al., 2014). Co-crystallization of S. aureus FosB with L-Cys or BSH revealed a mixed disulfide at the active site Cys9 of FosB which is unique in FosB from S. aureus (Thompson et al., 2014).
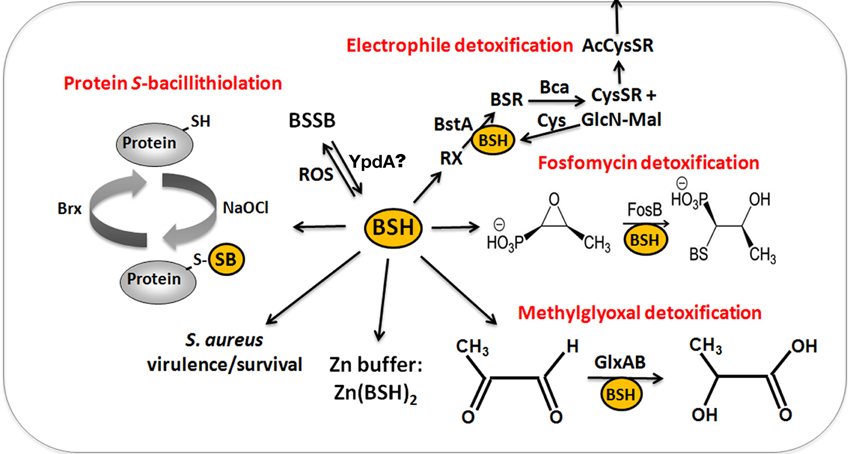
Figure 6. The functions of bacillithiol (BSH) in B. subtilis and S. aureus. Bacillithiol functions in detoxification of ROS, RES, HOCl, and antibiotics (fosfomycin, rifampicin) in B. subtilis and S. aureus. BSH is oxidized by ROS to bacillithiol disulfide (BSSB). Electrophiles (RX) are conjugated to BSH by the BSH S-transferase BstA to form BS-electrophiles (BSR) which are cleaved by the BSH S-conjugate amidase Bca to CysSR and mercapturic acids (AcCySR) that are exported from the cell. BSH serves as a cofactor for the epoxide hydrolase FosB which adds BSH to fosfomycin to open the ring structure for its detoxification. BSH functions in methylglyoxal detoxification as a cofactor for the glyoxalases I/II (GlxA and GlxB) in B. subtilis. GlxA converts BSH-hemithioacetal to S-lactoyl-BSH that is further converted by GlxB to D-lactate. BSH serves as Zn buffer under conditions of Zn excess in B. subtilis. In S. aureus, BSH is important under infection-related conditions and increased the survival of S. aureus in phagocytosis assays using murine macrophages. Under conditions of NaOCl stress, proteins are oxidized to mixed disulfides with BSH, termed as S-bacillithiolations which is reversed by bacilliredoxins.
Reactive electrophiles, such as monobromobimane are detoxified by direct conjugation to BSH or by conjugation reactions catalyzed by BSH S-transferases. BSH functions as a cofactor for DinB-family S-transferases that are widely distributed among GSH-, BSH-, and MSH-producing bacteria (Newton et al., 2011; Perera et al., 2014). The B. subtilis DinB-family YfiT protein was active as S-transferase with BSH to conjugate monochlorobimane, but inactive with MSH or GSH (Newton et al., 2011). The yfiT gene is flanked by yfiS and yfiU encoding putative efflux transporters for mercapturic acids produced during electrophile detoxification. The YfiT-homolog of S. aureus BstA catalyzed the conjugation of BSH to monochlorobimane, 1-chloro-2,4-dinitrobenzene and cerulenin, while rifampicin was BstA-independently conjugated to BSH (Perera et al., 2014).
BSH is involved in methylglyoxal detoxification and functions as a cofactor for BSH-dependent glyoxalases in B. subtilis (Chandrangsu et al., 2014). Methylglyoxal rapidly depletes BSH leading to BSH-hemithioacetal formation that is converted to S-lactoyl BSH by the glyoxalase-I (GlxA). The glyoxalase-II (GlxB) catalyzes conversion of S-lactoyl-BSH to lactate (Figure 6). Phenotype studies further indicated that BSH can detoxify heavy metal ions, such as tellurite and selenite in B. subtilis (Helmann, 2011). In addition, BSH functions as Zn buffer in metal ion homeostasis (Ma et al., 2014). The Cys thiol and carboxylate moieties of BSH can bind and store Zn(II) as BSH2:Zn complex under conditions of Zn(II) stress (Ma et al., 2014). BSH binding to Zn(II) occurred at much higher affinity compared to GSH. Mutants lacking BSH are more sensitive to Zn(II) stress and induced the Zn efflux CadA system at lower Zn levels compared to the wild type. BSH also protected against Zn(II) toxicity in cells lacking Zn efflux pumps. In addition, Zn efflux is elevated under conditions of diamide stress when the pool of reduced BSH is depleted. These results establish a new role of BSH as buffer for the labile Zn pool that are likely important for related pathogens under infection conditions.
In conclusion, functional analyses of bsh mutants established important roles of BSH as GSH surrogate in Firmicutes bacteria, including similar detoxification functions and BSH-dependent enzymes, such as DinB-family S-transferases and glyoxalases that are widely conserved across bacteria. However, the conserved role of FosB as BSH-dependent fosfomycin hydrolase and the function of BSH as Zn buffer have been described only in BSH-producing bacteria.
Phenotype analyses of S. aureus bsh mutants were conducted for different clinical isolates of methicillin-resistant S. aureus strains (MRSA) that revealed a role of BSH for stress resistance and under infection conditions (Pöther et al., 2013; Posada et al., 2014) (Table 2). In survival assays, S. aureus USA300 LAC transposon bsh mutants were more sensitive to alkylating agents (iodoacetamide and CDNB), methylglyoxal, peroxide and superoxide stress, diamide, fosfomycin, cerulenin, rifamycin and metals ions, like copper and cadmium (Rajkarnikar et al., 2013). In S. aureus COL and USA300 backgrounds, bshA and fosB mutants with clean deletions showed increased sensitivities to fosfomycin, diamide and H2O2 and the levels of NADPH and BSH were decreased in fosB mutants suggesting a function of FosB as S-transferase in the oxidative stress resistance (Posada et al., 2014). The S. aureus COL and USA300 bshA mutants showed a decreased survival in human whole-blood survival assays (Posada et al., 2014). Microarray analyses of the bshA mutant further revealed that staphyloxanthin biosynthetic genes are induced while the level of staphyloxanthin was strongly decreased in the S. aureus bshA mutant. Interestingly, the widely used strains of the S. aureus NCTC8325 lineage including SH1000 harbor natural yllA (bshC) null mutations that are caused by a 8 bp duplication in the bshC gene and these strains do not produce BSH (Gaballa et al., 2010; Newton et al., 2012; Posada et al., 2014). In contrast, S. aureus Newman encodes a functional bshC gene and produces BSH as revealed by thiol metabolomics (Newton et al., 2012; Pöther et al., 2013). BSH biosynthesis in S. aureus SH1000 could be restored by plasmid-encoded expression of the bshC gene (Pöther et al., 2013; Posada et al., 2014). In phagocytosis assays using murine macrophages and human epithelial cell lines the survival of the SH1000 strain was decreased compared to the bshC complemented S. aureus strain (Pöther et al., 2013; Posada et al., 2014). Thus, BSH is involved in the defense against the host-immune system and contributes to pathogen fitness in S. aureus clinical MRSA isolates under infection-related conditions. It will be exciting to unravel the regulatory mechanisms that contribute to virulence control by BSH in S. aureus.
Protein S-bacillithiolation was recently discovered as a widespread thiol protection and redox-regulatory mechanism in different Firmicutes bacteria (Chi et al., 2011, 2013) (Figure 7). S-bacillithiolation functions as a redox-switch mechanism to control the activity of redox-sensing transcription factors and metabolic enzymes, including OhrR and MetE (Lee et al., 2007; Chi et al., 2011) (Table 3). S-bacillithiolation of the OhrR repressor occurs at its lone Cys15 residue leading to inactivation of OhrR and expression of the thiol-dependent OhrA peroxiredoxin for detoxification of organic hydroperoxides and NaOCl (Fuangthong et al., 2001; Chi et al., 2011). S-bacillithiolation is also widespread among other Firmicutes with eight common and 29 unique S-bacillithiolated proteins identified in B. subtilis, Bacillus amyloliquefaciens, Bacillus pumilus, B. megaterium, and Staphylococcus carnosus (Chi et al., 2011, 2013). The S-bacillithiolome contains mainly biosynthetic enzymes for amino acids (methionine, cysteine, branched chain and aromatic amino acids), cofactors (thiamine), nucleotides (GTP), as well as translation factors, chaperones, redox, and antioxidant proteins. Among the most conserved protein-SSB were abundant and essential proteins like TufA, MetE, GuaB that are targets for S-thiolation also in MSH-producing bacteria (Chi et al., 2014).
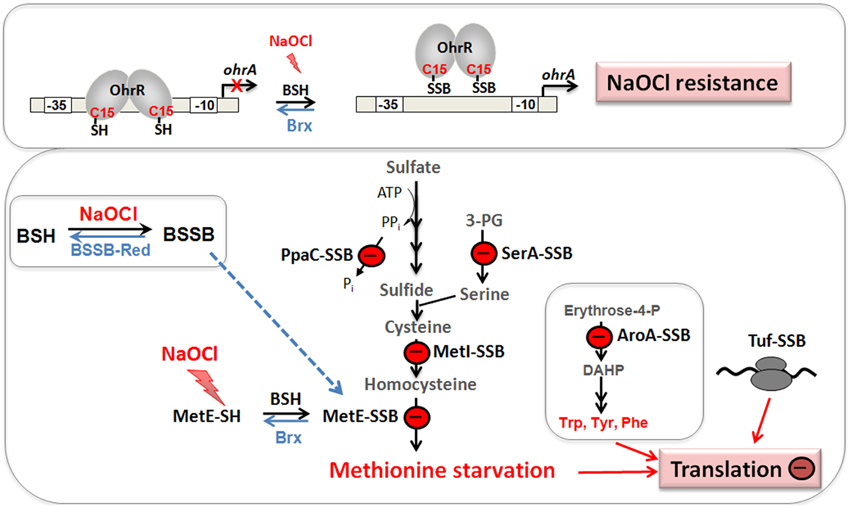
Figure 7. Physiological roles of S-bacillithiolations in B. subtilis and other Firmicutes. NaOCl leads to S-bacillithiolation of OhrR, MetE, YxjG, PpaC, SerA, AroA, GuaB, YumC, TufA, and YphP in B. subtilis (Chi et al., 2011). S-bacillithiolation of OhrR inactivates the repressor and causes induction of the OhrA peroxiredoxin that confers NaOCl resistance. S-bacillithiolation of the methionine synthase MetE at its active site Cys730 and other enzymes of the Cys and Met biosynthesis pathway (YxjG, PpaC, SerA, MetI) leads to methionine auxotrophy (Chi et al., 2011, 2013). In addition, other amino acids biosynthesis enzymes, translation factors and ribosomal proteins are S-bacillithiolated in Firmicutes bacteria. Thus, we hypothesize that S-bacillithiolation leads to a transient translation stop during the time of NaOCl detoxification to prevent further protein damage. NaOCl stress causes oxidation of BSH to BSSB and a two-fold decreased BSH/BSSB redox ratio that possibly contributes to S-bacillithiolation. The reduction of MetE-SSB and OhrR-SSB is catalyzed by bacilliredoxins (BrxA/B) in B. subtilis.
The methionine synthase MetE is the most abundant S-bacillithiolated protein in Bacillus species after NaOCl exposure. S-bacillithiolation of MetE occurs at its Zn-binding active site Cys730 and at the non-essential surface-exposed Cys719, leading to methionine starvation in NaOCl-treated cells (Chi et al., 2011). Similarly, methionine auxotrophy is caused by S-glutathionylation of MetE in E. coli after diamide stress (Hondorp and Matthews, 2004). The active site Zn center of MetE is also S-mycothiolated in C. glutamicum (Chi et al., 2014). Since formyl methionine is required for initiation of translation, MetE inactivation could stop translation during the time of hypochlorite detoxification. This translation arrest caused by S-bacillithiolation is supported by the strong repression of the stringent response RelA regulon under NaOCl stress, which includes genes for ribosomal proteins and translation factors (Chi et al., 2011).
Our studies revealed that S-bacillithiolations were observed under diamide and NaOCl stress, but not under control conditions. This confirms previous results about the mechanisms of S-glutathionylations which requires activation of protein thiols by ROS. S-glutathionylation can be caused via thiol-disulfide exchange with GSSG and by activation of thiols to sulfenic acid, sulfenylamides, thiyl radicals, thiosulfinate or S-nitrosyl intermediates (Gallogly and Mieyal, 2007; Mieyal et al., 2008; Allen and Mieyal, 2012; Mieyal and Chock, 2012). Hypochlorite leads to chlorination of the thiol group to form sulfenylchloride that is unstable and rapidly reacts further to form mixed BSH protein disulfides (Hawkins et al., 2003; Davies, 2011). The increased BSSB level under NaOCl-stress might also contribute to S-bacillithiolation via thiol-disulfide exchange.
Among the S-bacillithiolated proteins, the thioredoxin-like proteins YtxJ, YphP, and YqiW were identified in B. subtilis and Staphylococcus that occur only in BSH-producing bacteria (Chi et al., 2013). These Trx-like enzymes were suggested to function as bacilliredoxins (Brx) in the de-bacillithiolation process. YtxJ could functions as monothiol Brx and contains a single Cys in the conserved TCPIS motif. YphP (BrxA) and YqiW (BrxB) are paralogs of the uncharacterized DUF1094 family (53% identity) with unusual CGC active sites (Gaballa et al., 2010). YphP has also weak thiol-disulfide isomerase activity and a relatively high standard redox potential of E0′ = −130 mV (Derewenda et al., 2009). It was demonstrated that BrxA and BrxB function in the reduction of the S-bacillithiolated substrates MetE and OhrR in vitro (Gaballa et al., 2014) (Figure 8). The BrxBCxA resolving Cys mutant protein was able to reduce S-bacillithiolated OhrR to restore the DNA-binding activity of OhrR. However, the BrxBCxA mutant was unable to reduce S-cysteinylated OhrR. These results provide first evidence for the function of glutaredoxin-like enzymes in BSH-producing bacteria. However, phenotype analyses revealed that both, BrxA and BrxB are not essential and rather dispensable for oxidative stress resistance under conditions of S-bacillithiolations in B. subtilis (Gaballa et al., 2014). Thus, the bacilliredoxin pathway is redundant with other thiol-disulfide oxidoreductases or the thioredoxin pathway in vivo for reduction of BSH mixed disulfides. In conclusion, the redox regulation of enzymes and transcription regulators by S-bacillithiolation and bacilliredoxins has been studied in detail in the model bacterium B. subtilis. Future studies should be directed to elucidate if S-bacillithiolation and bacilliredoxins control virulence functions and pathogen fitness in the major pathogen S. aureus.
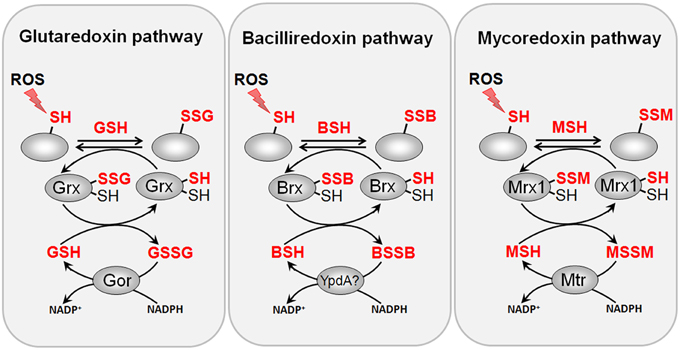
Figure 8. Reduction of protein S-glutathionylations, S-bacillithiolations and S-mycothiolations by glutaredoxin, bacilliredoxin and mycoredoxin pathways. The S-glutathionylated proteins are reduced by glutaredoxins (Grx) leading to a Grx-SSG intermediate that is reduced by GSH leading to GSSG which is recycled back to GSH by the NADPH-dependent GSSG reductase (Gor). Analogous bacilliredoxin and mycoredoxin pathways have been characterized in BSH- and MSH-utilizing Gram-positive bacteria. The S-bacillithiolated proteins are reduced by bacilliredoxins (Brx) leading to Brx-SSB formation. Brx-SSB is reduced by BSH with the generation of BSSB that likely requires the NADPH-dependent BSSB reductase YpdA for regeneration of BSH. In Actinomycetes, mycoredoxin1 catalyzes reduction of S-mycothiolated proteins leading to Mrx1-SSM generation that is recycled by MSH and the NADPH-dependent MSSM reductase Mtr.
Mycothiol (MSH) is composed of N-Acetyl-Cys-GlcN-myoinositol (Figure 4) and is present in high-GC Gram-positive Actinomycetes, such as Streptomycetes, Mycobacteria and Corynebacteria (Jothivasan and Hamilton, 2008; Newton et al., 2008). The biosynthesis of MSH proceeds from myo-inositol-1-phosphate, UDP-GlcNAc and cysteine and occurs in five steps (Jothivasan and Hamilton, 2008; Newton et al., 2008). The glycosyltransferase MshA conjugates myo-inositol-1-P to UDP-GlcNAc and produces GlcNAc-Ins-P. Dephosphorylation of GlcNAc-Ins-P by the phosphatase MshA2 generates GlcNAc-Ins which is the substrate for the deacetylase MshB. The MshB enzyme is homologous to the MSH S-conjugate amidase (Mca), and has both deacetylase and amidase activities. The cysteine ligase MshC adds Cys to GlcN-Ins to generate Cys-GlcN-Ins. The final acetylation of the Cys is catalyzed by the acetyltransferase MshD to produce MSH (Jothivasan and Hamilton, 2008; Newton et al., 2008). The structure of MSH is similar to that of BSH and the glycosyltransferase BshA and deacetylase BshB of B. subtilis are homologs of the MshA and MshB enzymes of Mycobacteria.
MSH biosynthesis enzymes in Streptomycetes are redox-controlled under diamide stress by the disulfide stress specific σR ECF sigma factor/RsrA anti sigma factor system (Kim et al., 2012). σR is sequestered by its redox-sensitive anti sigma factor RsrA in non-stressed cells. RsrA is oxidized at redox-sensing Cys residues in the Zn-binding site under disulfide stress that leads to relief of σR. Free σR transcribes genes required to maintain the thiol-redox homeostasis, including the genes for TrxAB and MSH biosynthesis, such as mshA, mshB, mshC, mshD, mca (Bae et al., 2004; Newton and Fahey, 2008; Park and Roe, 2008). In C. glutamicum, the homologous ECF sigma factor σH/RshA system controls the disulfide stress response genes for the Trx/TrxR system (trxB, trxB1, trxC) and for MSH biosynthesis and recycling (mshC, mca, mtr) (Ehira et al., 2009; Busche et al., 2012). The regulation of the Trx and MSH pathways by σR/RsrA or σH/RshA is conserved among Actinomycetes (Park and Roe, 2008; Antelmann and Helmann, 2011; Kim et al., 2012). Thus, it is common in Gram-positive bacteria that the genes for BSH and MSH biosynthesis pathways are under redox-control of the major disulfide stress regulators, Spx in Firmicutes bacteria and RsrA/RshA in Actinomycetes, respectively.
MSH serves as the major thiol-redox buffer in Actinomycetes. MSH is oxidized to MSH disulfide (MSSM) under oxidative stress conditions. The mycothiol disulfide reductase Mtr maintains MSH in its reduced state at the expense of NADPH. MSH is involved in protection against oxidative and electrophile stress, alkylating agents, toxins, antibiotics (erythromycin, vancomycin, rifampin, azithromycin), heavy metal stress, aromatic compounds, ethanol and glyphosate in Streptomycetes, Mycobacteria and Corynebacteria (Buchmeier et al., 2003, 2006; Rawat et al., 2007; Newton et al., 2008; Liu et al., 2013) (Table 1). MSH is used as a cofactor for MSH-dependent enzymes during detoxification of toxins, electrophiles and antibiotics in Actinomycetes (Figure 9, Table 1). MSH forms conjugates with xenobiotics and antibiotics either spontaneously or by the DinB-family MSH S-transferases (Newton et al., 2011). The MSH S-transferase Mst of M. smegmatis was shown to catalyze the conjugation of monochlorobimane and DTNB to MSH but its natural substrate is not known (Newton et al., 2011). MSH-S-conjugates are rapidly cleaved by the MSH-S-conjugate amidase (Mca) to glucoseamine-myo-inositol (GlcN-Ins) and mercapturic acid derivatives (AcCysSR) that are excreted from the cell. Mca is the major detoxification enzyme for MSH S-conjugates with antibiotics, including cerulenin and rifamycin in Mycobacteria (Newton et al., 2008, 2011). Interestingly, MSH and the Mca-homologs LmbT, LmbV and LmbE play also a direct role in the biosynthesis of the sulfur-containing lincosamide antibiotic lincomycin in Streptomyces lincolnensis (Zhao et al., 2015). MSH functions as the sulfur donor for incorporation of the methylmercapto group into lincomycin after thiol exchange. In addition, ergothioneine (EGT), that is utilized as another thiol by Actinomycetes, acts as a carrier for the assembly of the N-methylated 4-propyl-L-proline (PPL) and lincosamide moieties to form lincomycin. EGT and MSH were shown to function in lincomycin biosynthesis through unusual S-glycosylations documenting a first biochemical role of LMW thiols in bacteria. Since the biosynthetic pathways for many sulfur-containing natural compounds include Mca homologs, the involvement of LMW thiols in natural product biosynthesis might be a common mechanism (Zhao et al., 2015).
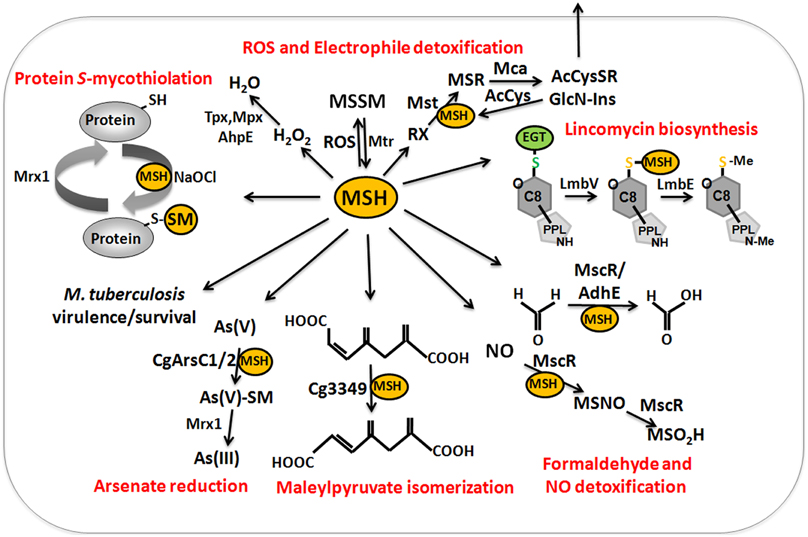
Figure 9. The functions of mycothiol (MSH) in Mycobacteria and Corynebacteria. Mycothiol (MSH) is oxidized by ROS to mycothiol disulfide (MSSM). MSSM is reduced back to MSH by the mycothiol disulfide reductase Mtr on expense of NADPH. MSH-dependent peroxidases, such as Mpx, Tpx, and AhpE function in peroxide detoxification. Electrophiles (RX) are conjugated to MSH by the MSH S-transferase Mst to form MS-electrophiles (MSR) which are cleaved by the MSH S-conjugate amidase Mca to mercapturic acids (AcCySR) that are exported from the cell. The Mca-homologs LmbT, LmbV, and LmbE function also in the assembly and biosynthesis of the sulfur-containing lincosamide antibiotic lincomycin in Streptomyces lincolnensis (Zhao et al., 2015). MSH serves as a cofactor for the alcohol dehydrogenase AdhE/MscR in Mycobacteria and Corynebacteria for detoxification of formaldehyde to formate and MSNO to MSO2H. MSH functions in detoxification of maleylpyruvate as a cofactor for maleylpyruvate isomerase in C. glutamicum. Arsenate reductases CgArsC1 and CgArsC2 conjugate MSH and arsenate As(V) to form As(V)-SM that is reduced to As(III) by Mrx1. In M. tuberculosis, MSH is important under infection conditions and for growth and survival. Under conditions of NaOCl stress, proteins are oxidized to mixed disulfides with MSH, termed as S-mycothiolations which is reversed by mycoredoxins.
MSH functions as a cofactor for many redox enzymes that are involved in the detoxification of peroxides, electrophiles (formaldehyde), NO, aromatic compounds (maleylpyruvate) and arsenate (Fahey, 2013) (Table 1). There is evidence for a MSH-peroxidase Mpx involved in peroxide detoxification that was identified as S-mycothiolated Gpx-homolog under oxidative stress in C. glutamicum (Chi et al., 2014). The MSH-dependent alcohol dehydrogenase MscR (MSNO reductase/formaldehyde dehydrogenase) catalyzes the detoxification of formaldehyde and S-nitrosyl-mycothiol (MSNO) (Newton et al., 2008). MSH reacts with formaldehyde to MS-CH2OH that is converted to formate by MscR. MscR also converts MSNO to MSH sulfinamide (MSONH2). In C. glutamicum, a similar MSH-dependent pathway for formaldehyde oxidation by the MSH-dependent formaldehyde dehydrogenase AdhE/FadH has been characterized (Lessmeier et al., 2013; Witthoff et al., 2013). In C. glutamicum, MSH is further involved in degradation of aromatic compounds, including gentisate, 3-hydroxybenzoate, maleylpyruvate, resorcinol, and naphthalene and msh mutants were unable to grow on these substrates (Liu et al., 2013). MSH functions as a cofactor for the maleylpyruvate isomerase in the gentisate ring-cleavage pathway to catalyze the isomerization of maleylpyruvate to fumaryl pyruvate in C. glutamicum (Feng et al., 2006). Similarly, MSH was suggested as a cofactor for enzymes of the naphthalene and resorcinol degradation pathway (Liu et al., 2013).
MSH confers resistance to metal ions, such as Cr(VI), Zn(II), Cd(II), Co(II), and Mn(II) in C. glutamicum (Liu et al., 2013). The detoxification of arsenate [As-(V)] to arsenite [As(III)] depends on the MSH-dependent arsenate reductases ArsC1/C2 (Ordonez et al., 2009). ArsC1/C2 function similar to S-transferases in arsenate detoxification by formation of an arseno-MSH conjugate that requires the mycoredoxin-1/MSH/Mtr electron pathway for reduction. In contrast, another arsenate reductase Cg_ArsC1′ detoxifies arsenate with electrons from the Trx pathway (Villadangos et al., 2011).
MSH enhanced also the robustness of C. glutamicum during industrial production of glutamate and L-lysine (Liu et al., 2014). The overexpression of mshA resulted in increased MSH biosynthesis and higher resistance of C. glutamicum to peroxides, methylglyoxal, antibiotics (erythromycin and streptomycin), metal ions, organic acids, furfural and ethanol (Liu et al., 2014). Thus, the increased biosynthesis of LMW thiol redox buffers, as shown for GSH in C. acetobutylicum and MSH in C. glutamicum, might be a promising strategy to engineer robust industrial production strains.
In Mycobacterium tuberculosis, MSH is essential for growth and survival of M. tuberculosis under infection conditions (Sareen et al., 2003; Sassetti and Rubin, 2003). In addition, MSH is required to activate the antituberculosis prodrug isoniazid and hence M. tuberculosis mshA mutants are resistant to isoniazid (Buchmeier et al., 2003). Tuberculosis (TB) causes still nearly 2 million death each year and multiple and extensive drug resistant strains occur that require new targets for antituberculosis drugs. Thus, inhibitors of MSH biosynthesis enzymes are promising candidates for antituberculosis drug developments. Several MSH biosynthesis inhibitors have been applied that target the MSH-S-conjugate amidase Mca, the deacetylase MshB, the cysteine ligase MshC and the MSSM reductase Mtr that are attractive antituberculosis drug targets (Nilewar and Kathiravan, 2014).
Protein S-mycothiolation was first studied in C. glutamicum and 25 S-mycothiolated proteins could be identified under NaOCl stress by mass spectrometry (Chi et al., 2014) (Table 3). The thiol-peroxidase Tpx and the putative MSH peroxidase Mpx were S-mycothiolated under control and NaOCl stress conditions at their active site Cys residues. The fragment ion spectra of the S-mycothiolated Cys-peptides are characterized by diagnostic myoinositol-loss precursor ions (−180 Da) that serve as markers for identification. The 25 S-mycothiolated proteins overlap with 16 NaOCl-sensitive proteins identified in the fluorescent-label thiol-redox proteome. These include Tuf, GuaB1, GuaB2, SerA, and MetE as conserved abundant targets for S-thiolations across Gram-positive bacteria (Chi et al., 2013). The S-mycothiolated proteins are involved in the metabolism of carbohydrates, such as glycolysis (Fba, Pta, XylB), glycogen and maltodextrin degradation (MalP) and several biosynthesis pathways for serine, cysteine, methionine (SerA, Hom, MetE), nucleotides and thiamine (GuaB1, GuaB2, PurL, NadC, ThiD1, and ThiD2) and myo-inositol-1-phosphate (Ino-1 or Cg3323) (Figure 10). Further protein-SSM function in peroxide detoxification (Tpx, Gpx), methionine sulfoxide reduction (MsrA), heme degradation for iron mobilization (HmuO) and protein translation (RpsF, RpsC, RpsM, RplM, Tuf). The glycogen phosphorylase MalP is one of the most abundantly S-mycothiolated proteins in NaOCl-treated cells (Chi et al., 2014). S-mycothiolation of MalP is important for oxidative stress resistance in C. glutamicum since the malP deletion mutant is NaOCl-sensitive in growth assays. MalP functions in glycogen degradation during the stationary phase. S-mycothiolation of MalP may prevent glycogen degradation under NaOCl stress since the glycogen content remained stable despite a strongly decreased glucose uptake rate.
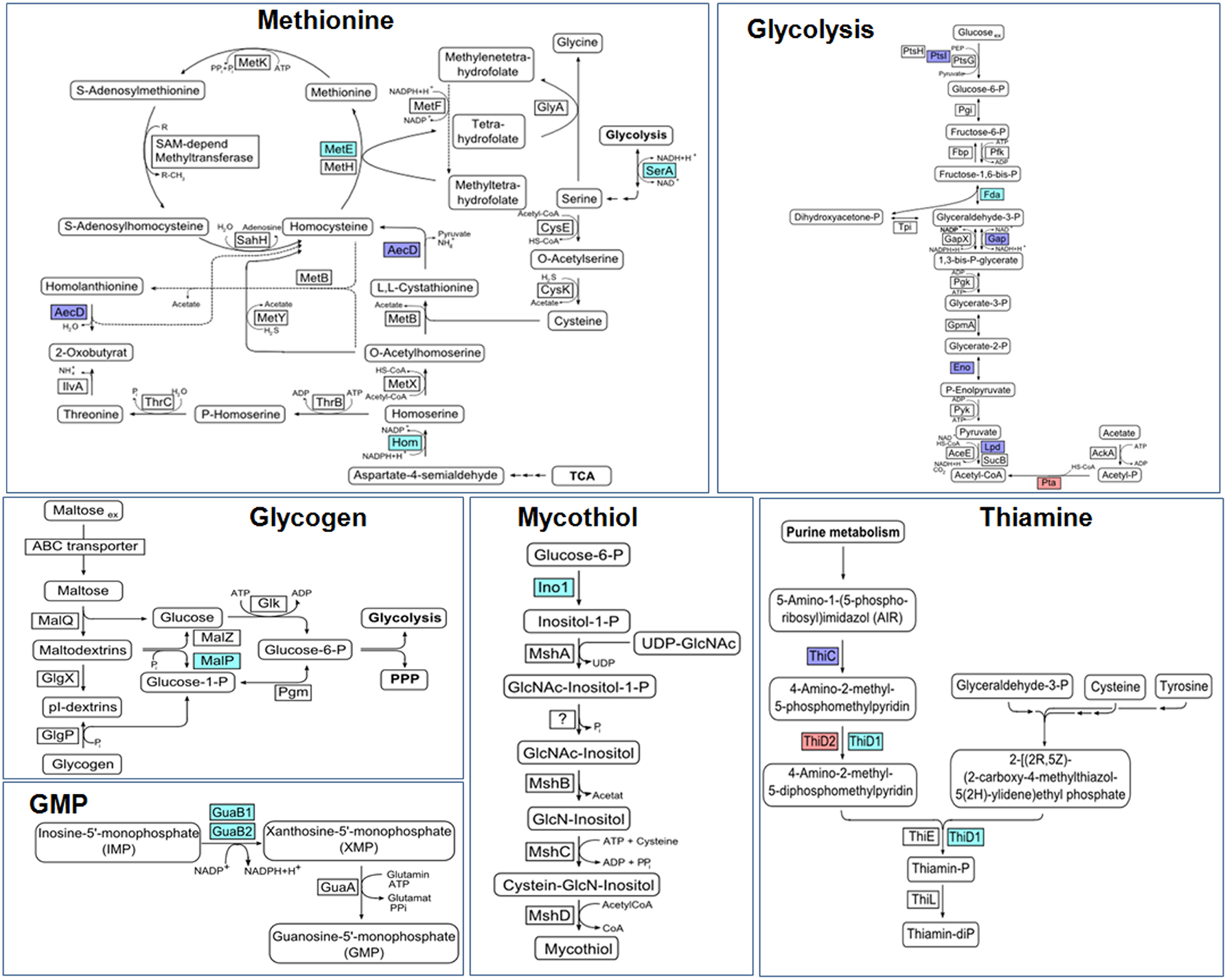
Figure 10. Physiological roles of S-mycothiolations in Corynebacterium glutamicum. The metabolic pathways for glycolysis, biosynthesis of methionine, thiamine, GMP, MSH, and glycogen metabolism are shown including identified S-mycothiolated proteins. The identified S-mycothiolated or oxidized proteins are labeled with colors (S-mycothiolated proteins are red; reversibly oxidized proteins are magenta; both reversibly oxidized and S-mycothiolated are blue). The selected S-mycothiolated metabolic enzymes include MetE, SerA, Hom (Met biosynthesis); Fba, Pta (glycolysis); MalP (glycogen utilization); Ino-1 (MSH biosynthesis); ThiD1, ThiD2 (thiamine biosynthesis); GuaB1, GuaB2 (GMP biosynthesis). Further proteins with Cys-SSM sites are involved in translation (Tuf, PheT, RpsC, RpsF, RpsM, RplM) and antioxidant functions (Tpx, Bcp, MsrA) that are not shown here. The figure is adapted from (Chi et al., 2014).
The mycoredoxin-1 (Mrx1) has been characterized as glutaredoxin-homolog of Actinomycetes in reduction of MSH mixed disulfides (Van Laer et al., 2012) (Figure 8). Mrx-1 has a typical Trx-like fold with a CGYC motif and a cis-Pro57 in a groove that presumable binds MSH. The redox potential of Mrx-1 was calculated as E0′ = −218 mV and the pKa of the active site Cys17 was 5.1–5.6. Mrx-1 catalyzed de-mycothiolation in a hydroxyethyl disulfide (HED) assay and is coupled to the MSH/Mtr/NADPH pathway. Mrx-1 operates via a monothiol reaction mechanism in the de-mycothiolation reaction analogous to most glutaredoxins that are involved in de-glutathionylation. The first Mrx1 substrate was identified as the thiol-peroxidase Tpx that was S-mycothiolated at its active site Cys60 and resolving site Cys94 in C. glutamicum in vivo under hypochlorite stress (Chi et al., 2014). Tpx showed NADPH-linked peroxidase activity and reduced H2O2 in a Trx/TrxR-coupled electron assay. S-mycothiolation of Tpx inhibits the peroxidase activity which was restored after reduction by the Mrx1/MSH/Mtr pathway. Thus, S-mycothiolation controls Tpx activity and protects the peroxidatic Cys against overoxidation. In M. tuberculosis, Mrx1 has been shown to reduce the one-Cys peroxiredoxin AhpE (Hugo et al., 2014). AhpE is a membrane-associated peroxidase that detoxifies peroxinitrite and fatty acid hydroperoxides as preferred substrates (Hugo et al., 2009; Reyes et al., 2011). AhpE is oxidized by peroxides to form a sulfenic acid intermediate (AhpE-SOH) that can be reduced directly by Mrx1. Alternatively, AhpE-SOH can react with MSH to S-mycothiolated AhpE-SSM which is reduced by the Mrx1/MSH/Mtr electron pathway (Hugo et al., 2014). The direct AhpE-SOH reduction may occur in the membrane when MSH is not available and the formation of AhpE-SSM and subsequent Mrx1-reduction was suggested to predominate in the cytosol. Interestingly, the reducing mechanism of AhpE-SSM is similar to the detoxification of arsenate by CgArsC1 and CgArsC2. Arsenate reacts with MSH to an arseno-(V)-MSH complex that is reduced by Mrx1 releasing As(III) and Mrx1-SSM that is recycled by the MSH/Mtr/NADPH electron pathway (Ordonez et al., 2009; Villadangos et al., 2011). It remains to be shown if AhpE is mycothiolated under oxidative stress in M. tuberculosis cells in vivo. These results show that Mrx1 functions as glutaredoxin homolog in C. glutamicum and M. tuberculosis in the reduction of S-mycothiolated peroxiredoxins (Tpx and AhpE), when coupled to the MSH/Mtr/NADPH electron pathway and as electron donor for arsenate reductase in arsenate detoxification.
Recently, Mrx1 has been coupled to redox sensitive GFP (roGFP2) to construct a new genetically encoded biosensor for dynamic measurements of the MSH redox potential in different M. tuberculosis strains (Bhaskar et al., 2014). This study revealed phenotypic redox heterogeneity of E0′(MSSM/MSH) within Mycobacteria inside infected macrophages that are caused by sub-vacuolar compartments. Those sub-populations with higher E0′(MSSM/MSH) were more susceptible to clinical relevant antibiotics whereas populations with lower MSH redox potentials were resistant to antibiotics. The results further show that several anti-TB drugs induce oxidative stress in M. tuberculosis during infections. In conclusion, this Mrx1-roGFP2 biosensor is a promising tool to study MSH redox potential changes of M. tuberculosis under infections and antibiotic treatments. This is the first example for a genetically encoded redox biosensor that measures dynamic changes of the mycothiol redox potential in bacteria. Future studies should be directed to apply similar biosensors in other pathogenic bacteria to study the dynamics of redox potential changes during infections.
In this review, we provide an overview about the biosynthesis pathways and functions of the bacterial redox buffers glutathione, bacillithiol and mycothiol and their regulatory roles for protein S-thiolations. Bacterial redox buffers maintain the reduced state of the cytoplasm and function as cofactors of conserved enzymes for detoxification of ROS, RES, chlorines, antibiotics and xenobiotics. These thiol-dependent enzymes include NADPH-dependent disulfide reductases (Gor, Mtr, YpdA) and related glutaredoxin-like enzymes (Grx, Mrx, Brx), DinB-family S-transferases (Gst, Mst, BstA), S-conjugate amidases (Mca, Bca) and glyoxalases (GloAB, GlxAB). However, some detoxification enzymes still need to be characterized in BSH-utilizing bacteria, including the BSH-dependent formaldehyde reductase (AdhA), the putative BSH peroxidase (Bpx) or thiol-dependent dioxygenases (MhqA, MhqE and MhqO) (Antelmann et al., 2008). The discovery of the biochemical functions of MSH, EGT and S-transferases in the lincomycin antibiotic biosynthesis opens perspectives to characterize the roles of thiol-redox buffers in the biosynthesis of sulfur-containing co-factors, natural compounds and antibiotics in other bacteria.
The structures of BSH and MSH are similar and the BSH biosynthesis enzymes BshA, BshB and BshC are homologous to the MSH biosynthesis enzymes MshA, MshB, and MshC. However, the crystal structure of BshC has revealed significant differences compared to MshC which requires further studies to understand the still unknown cysteine ligation mechanism of BshC (Vanduinen et al., 2015). It is further interesting, that the levels of BSH and MSH vary strongly between Firmicutes and Actinomycetes and also during growth and stress conditions. While Mycobacteria produce up to 20 mM MSH, the levels of BSH are much lower reaching 1–6 mM in Firmicutes bacteria. The differences in BSH and MSH levels during growth and under stress can be explained by the redox control of the BSH and MSH biosynthesis enzymes by the major thiol-based redox sensors (Spx and RsrA/RshA), presumably to enhance the redox buffer capacity under certain conditions to keep the reduced state of the cytoplasm. In contrast, redox regulation of GSH biosynthesis genes has not been shown. However, the pathogen L. monocytogenes is able to synthesize GSH and to import host-derived GSH as adaptation strategy under infection conditions (Reniere et al., 2015). Importantly, synthesized and host-derived GSH both contribute to virulence factor regulation in L. monocytogenes, while GSH-import was required for full virulence in S. pneumoniae (Potter et al., 2012; Reniere et al., 2015). Overall, the roles of GSH, BSH and MSH for virulence and pathogen fitness have been shown for many important human pathogens, including L. monocytogenes, S. pneumoniae, S. Typhimurium, S. aureus, and M. tuberculosis. Future studies in the field of infection biology should be directed to understand the molecular mechanisms of virulence factor regulation by thiol-redox buffers that might involve also protein S-thiolation mechanisms. The GSH, BSH and MSH biosynthesis enzymes, GSH uptake systems as well as S-thiolated proteins could be promising drug targets for the development of novel anti-infectives against emerging drug resistant strains of S. pneumoniae, S. aureus and M. tuberculosis. Thus, the large scale identification and quantification of S-thiolated proteins in pathogens is an important topic for future research.
Advances in mass spectrometry and chemical probe design have facilitated the development of more sensitive redox proteomics methods, such as the NEM-biotin switch assay or the Gsp-biotin assay to study targets for protein S-glutathionylation at a global scale (Lind et al., 2002; Kehr et al., 2011; Lin et al., 2015). In addition, numerous BSH- and MSH-mixed protein disulfides have been identified recently under disulfide stress conditions, such as NaOCl and diamide. However, more quantitative MS-based redox proteomics approaches are required to determine the level of mixed BSH- and MSH-protein disulfides by combining the direct shotgun approach with OxICAT or the NEM-biotin switch assay coupled to Brx or Mrx1 (Leichert et al., 2008; Kehr et al., 2011). In addition, the regulatory roles for only few S-bacillithiolated and S-mycothiolated proteins have been studied thus far, including the redox regulator OhrR and the methionine synthase MetE (Lee et al., 2007; Chi et al., 2011). However, many interesting metabolic enzymes, redox-sensing transcription factors or virulence factors might be controlled by protein S-thiolations in the pathogenic bacteria S. aureus and M. tuberculosis that remain to be elucidated in future research. Thus, it is an exciting field for new frontiers of science to unravel the regulatory potential of emerging protein S-thiolations in bacteria.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG) AN746/3-1 and AN746/4-1 within the DFG priority program SPP1710, by the DFG Research Training Group GRK1947, project [C1] and by an ERC Consolidator Grant (GA 615585) MYCOTHIOLOME to H.A.
Allen, E. M., and Mieyal, J. J. (2012). Protein-thiol oxidation and cell death: regulatory role of glutaredoxins. Antioxid. Redox Signal. 17, 1748–1763. doi: 10.1089/ars.2012.4644
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Anderson, M. E. (1998). Glutathione: an overview of biosynthesis and modulation. Chem. Biol. Interact. 111–112, 1–14. doi: 10.1016/S0009-2797(97)00146-4
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ansong, C., Wu, S., Meng, D., Liu, X., Brewer, H. M., Deatherage Kaiser, B. L., et al. (2013). Top-down proteomics reveals a unique protein S-thiolation switch in Salmonella Typhimurium in response to infection-like conditions. Proc. Natl. Acad. Sci. U.S.A. 110, 10153–10158. doi: 10.1073/pnas.1221210110
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Antelmann, H., Hecker, M., and Zuber, P. (2008). Proteomic signatures uncover thiol-specific electrophile resistance mechanisms in Bacillus subtilis. Expert Rev. Proteomics 5, 77–90. doi: 10.1586/14789450.5.1.77
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Antelmann, H., and Helmann, J. D. (2011). Thiol-based redox switches and gene regulation. Antioxid. Redox Signal. 14, 1049–1063. doi: 10.1089/ars.2010.3400
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Aslund, F., Ehn, B., Miranda-Vizuete, A., Pueyo, C., and Holmgren, A. (1994). Two additional glutaredoxins exist in Escherichia coli: glutaredoxin 3 is a hydrogen donor for ribonucleotide reductase in a thioredoxin/glutaredoxin 1 double mutant. Proc. Natl. Acad. Sci. U.S.A. 91, 9813–9817. doi: 10.1073/pnas.91.21.9813
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Atichartpongkul, S., Loprasert, S., Vattanaviboon, P., Whangsuk, W., Helmann, J. D., and Mongkolsuk, S. (2001). Bacterial Ohr and OsmC paralogues define two protein families with distinct functions and patterns of expression. Microbiology 147, 1775–1782.
Bae, J. B., Park, J. H., Hahn, M. Y., Kim, M. S., and Roe, J. H. (2004). Redox-dependent changes in RsrA, an anti-sigma factor in Streptomyces coelicolor: zinc release and disulfide bond formation. J. Mol. Biol. 335, 425–435. doi: 10.1016/j.jmb.2003.10.065
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bartsch, R. G., Newton, G. L., Sherrill, C., and Fahey, R. C. (1996). Glutathione amide and its perthiol in anaerobic sulfur bacteria. J. Bacteriol. 178, 4742–4746.
Bhaskar, A., Chawla, M., Mehta, M., Parikh, P., Chandra, P., Bhave, D., et al. (2014). Reengineering redox sensitive GFP to measure mycothiol redox potential of Mycobacterium tuberculosis during infection. PLoS Pathog. 10:e1003902. doi: 10.1371/journal.ppat.1003902
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Booth, I. R., Ferguson, G. P., Miller, S., Li, C., Gunasekera, B., and Kinghorn, S. (2003). Bacterial production of methylglyoxal: a survival strategy or death by misadventure? Biochem. Soc. Trans. 31, 1406–1408.
Bourajjaj, M., Stehouwer, C. D., Van Hinsbergh, V. W., and Schalkwijk, C. G. (2003). Role of methylglyoxal adducts in the development of vascular complications in diabetes mellitus. Biochem. Soc. Trans. 31, 1400–1402. doi: 10.1042/BST0311400
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Brandes, N., Schmitt, S., and Jakob, U. (2009). Thiol-based redox switches in eukaryotic proteins. Antioxid. Redox Signal. 11, 997–1014. doi: 10.1089/ars.2008.2285
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Brennan, J. P., Miller, J. I., Fuller, W., Wait, R., Begum, S., Dunn, M. J., et al. (2006). The utility of N,N-biotinyl glutathione disulfide in the study of protein S-glutathiolation. Mol. Cell. Proteomics 5, 215–225. doi: 10.1074/mcp.M500212-MCP200
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Buchmeier, N. A., Newton, G. L., and Fahey, R. C. (2006). A mycothiol synthase mutant of Mycobacterium tuberculosis has an altered thiol-disulfide content and limited tolerance to stress. J. Bacteriol. 188, 6245–6252. doi: 10.1128/JB.00393-06
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Buchmeier, N. A., Newton, G. L., Koledin, T., and Fahey, R. C. (2003). Association of mycothiol with protection of Mycobacterium tuberculosis from toxic oxidants and antibiotics. Mol. Microbiol. 47, 1723–1732. doi: 10.1046/j.1365-2958.2003.03416.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Busche, T., Silar, R., Picmanova, M., Patek, M., and Kalinowski, J. (2012). Transcriptional regulation of the operon encoding stress-responsive ECF sigma factor SigH and its anti-sigma factor RshA, and control of its regulatory network in Corynebacterium glutamicum. BMC Genomics 13:445. doi: 10.1186/1471-2164-13-445
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Cassier-Chauvat, C., Veaudor, T., and Chauvat, F. (2014). Advances in the function and regulation of hydrogenase in the cyanobacterium Synechocystis PCC6803. Int. J. Mol. Sci. 15, 19938–19951. doi: 10.3390/ijms151119938
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Castellano, I., Ruocco, M. R., Cecere, F., Di Maro, A., Chambery, A., Michniewicz, A., et al. (2008). Glutathionylation of the iron superoxide dismutase from the psychrophilic eubacterium Pseudoalteromonas haloplanktis. Biochim. Biophys. Acta 1784, 816–826. doi: 10.1016/j.bbapap.2008.02.003
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chandrangsu, P., Dusi, R., Hamilton, C. J., and Helmann, J. D. (2014). Methylglyoxal resistance in Bacillus subtilis: contributions of bacillithiol-dependent and independent pathways. Mol. Microbiol. 91, 706–715. doi: 10.1111/mmi.12489
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chardonnet, S., Sakr, S., Cassier-Chauvat, C., Le Marechal, P., Chauvat, F., Lemaire, S. D., et al. (2015). First proteomic study of S-glutathionylation in cyanobacteria. J. Proteome Res. 14, 59–71. doi: 10.1021/pr500625a
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chen, N. H., Counago, R. M., Djoko, K. Y., Jennings, M. P., Apicella, M. A., Kobe, B., et al. (2013). A glutathione-dependent detoxification system is required for formaldehyde resistance and optimal survival of Neisseria meningitidis in biofilms. Antioxid. Redox Signal. 18, 743–755. doi: 10.1089/ars.2012.4749
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chi, B. K., Busche, T., Van Laer, K., Basell, K., Becher, D., Clermont, L., et al. (2014). Protein S-mycothiolation functions as redox-switch and thiol protection mechanism in Corynebacterium glutamicum under hypochlorite stress. Antioxid. Redox Signal. 20, 589–605. doi: 10.1089/ars.2013.5423
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chi, B. K., Gronau, K., Mader, U., Hessling, B., Becher, D., and Antelmann, H. (2011). S-bacillithiolation protects against hypochlorite stress in Bacillus subtilis as revealed by transcriptomics and redox proteomics. Mol. Cell. Proteomics 10, M111.009506. doi: 10.1074/mcp.M111.009506
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chi, B. K., Roberts, A. A., Huyen, T. T., Basell, K., Becher, D., Albrecht, D., et al. (2013). S-bacillithiolation protects conserved and essential proteins against hypochlorite stress in firmicutes bacteria. Antioxid. Redox Signal. 18, 1273–1295. doi: 10.1089/ars.2012.4686
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chiang, B. Y., Chou, C. C., Hsieh, F. T., Gao, S., Lin, J. C., Lin, S. H., et al. (2012). In vivo tagging and characterization of S-glutathionylated proteins by a chemoenzymatic method. Angew. Chem. Int. Ed Engl. 51, 5871–5875. doi: 10.1002/anie.201200321
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dalle-Donne, I., Rossi, R., Colombo, G., Giustarini, D., and Milzani, A. (2009). Protein S-glutathionylation: a regulatory device from bacteria to humans. Trends Biochem. Sci. 34, 85–96. doi: 10.1016/j.tibs.2008.11.002
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dalle-Donne, I., Rossi, R., Giustarini, D., Colombo, R., and Milzani, A. (2007). S-glutathionylation in protein redox regulation. Free Radic. Biol. Med. 43, 883–898. doi: 10.1016/j.freeradbiomed.2007.06.014
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Davies, M. J. (2011). Myeloperoxidase-derived oxidation: mechanisms of biological damage and its prevention. J. Clin. Biochem. Nutr. 48, 8–19. doi: 10.3164/jcbn.11-006FR
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
De Mendoza, D. (2014). Temperature sensing by membranes. Annu. Rev. Microbiol. 68, 101–116. doi: 10.1146/annurev-micro-091313-103612
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Derewenda, U., Boczek, T., Gorres, K. L., Yu, M., Hung, L. W., Cooper, D., et al. (2009). Structure and function of Bacillus subtilis YphP, a prokaryotic disulfide isomerase with a CXC catalytic motif. Biochemistry 48, 8664–8671. doi: 10.1021/bi900437z
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ehira, S., Teramoto, H., Inui, M., and Yukawa, H. (2009). Regulation of Corynebacterium glutamicum heat shock response by the extracytoplasmic-function sigma factor SigH and transcriptional regulators HspR and HrcA. J. Bacteriol. 191, 2964–2972. doi: 10.1128/JB.00112-09
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fahey, R. C. (2013). Glutathione analogs in prokaryotes. Biochim. Biophys. Acta 1830, 3182–3198. doi: 10.1016/j.bbagen.2012.10.006
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fang, Z., Roberts, A. A., Weidman, K., Sharma, S. V., Claiborne, A., Hamilton, C. J., et al. (2013). Cross functionalities of Bacillus deacetylases involved in bacillithiol biosynthesis and bacillithiol-S-conjugate detoxification pathways. Biochem. J. doi: 10.1042/BJ20130415
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Farrand, S. K., and Taber, H. W. (1974). Changes in menaquinone concentration during growth and early sporulation in Bacillus subtilis. J. Bacteriol. 117, 324–326.
Feng, J., Che, Y., Milse, J., Yin, Y. J., Liu, L., Ruckert, C., et al. (2006). The gene ncgl2918 encodes a novel maleylpyruvate isomerase that needs mycothiol as cofactor and links mycothiol biosynthesis and gentisate assimilation in Corynebacterium glutamicum. J. Biol. Chem. 281, 10778–10785. doi: 10.1074/jbc.M513192200
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ferguson, G. P., Totemeyer, S., Maclean, M. J., and Booth, I. R. (1998). Methylglyoxal production in bacteria: suicide or survival? Arch. Microbiol. 170, 209–218.
Fernandes, A. P., and Holmgren, A. (2004). Glutaredoxins: glutathione-dependent redox enzymes with functions far beyond a simple thioredoxin backup system. Antioxid. Redox Signal. 6, 63–74. doi: 10.1089/152308604771978354
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Forman, H. J., and Torres, M. (2001). Redox signaling in macrophages. Mol. Aspects Med. 22, 189–216. doi: 10.1016/S0098-2997(01)00010-3
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fratelli, M., Demol, H., Puype, M., Casagrande, S., Eberini, I., Salmona, M., et al. (2002). Identification by redox proteomics of glutathionylated proteins in oxidatively stressed human T lymphocytes. Proc. Natl. Acad. Sci. U.S.A. 99, 3505–3510. doi: 10.1073/pnas.052592699
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fratelli, M., Demol, H., Puype, M., Casagrande, S., Villa, P., Eberini, I., et al. (2003). Identification of proteins undergoing glutathionylation in oxidatively stressed hepatocytes and hepatoma cells. Proteomics 3, 1154–1161. doi: 10.1002/pmic.200300436
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fratelli, M., Gianazza, E., and Ghezzi, P. (2004). Redox proteomics: identification and functional role of glutathionylated proteins. Expert Rev. Proteomics 1, 365–376. doi: 10.1586/14789450.1.3.365
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Freitag, N. E., Port, G. C., and Miner, M. D. (2009). Listeria monocytogenes – from saprophyte to intracellular pathogen. Nat. Rev. Microbiol. 7, 623–628. doi: 10.1038/nrmicro2171
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fuangthong, M., Atichartpongkul, S., Mongkolsuk, S., and Helmann, J. D. (2001). OhrR is a repressor of ohrA, a key organic hydroperoxide resistance determinant in Bacillus subtilis. J. Bacteriol. 183, 4134–4141. doi: 10.1128/JB.183.14.4134-4141.2001
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gaballa, A., Antelmann, H., Hamilton, C. J., and Helmann, J. D. (2013). Regulation of Bacillus subtilis bacillithiol biosynthesis operons by Spx. Microbiology 159, 2025–2035. doi: 10.1099/mic.0.070482-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gaballa, A., Chi, B. K., Roberts, A. A., Becher, D., Hamilton, C. J., Antelmann, H., et al. (2014). Redox regulation in Bacillus subtilis: the bacilliredoxins BrxA(YphP) and BrxB(YqiW) function in de-bacillithiolation of S-bacillithiolated OhrR and MetE. Antioxid. Redox Signal. 21, 357–367. doi: 10.1089/ars.2013.5327
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gaballa, A., Newton, G. L., Antelmann, H., Parsonage, D., Upton, H., Rawat, M., et al. (2010). Biosynthesis and functions of bacillithiol, a major low-molecular-weight thiol in Bacilli. Proc. Natl. Acad. Sci. U.S.A. 107, 6482–6486. doi: 10.1073/pnas.1000928107
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gallogly, M. M., and Mieyal, J. J. (2007). Mechanisms of reversible protein glutathionylation in redox signaling and oxidative stress. Curr. Opin. Pharmacol. 7, 381–391. doi: 10.1016/j.coph.2007.06.003
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ghezzi, P. (2005). Regulation of protein function by glutathionylation. Free Radic. Res. 39, 573–580. doi: 10.1080/10715760500072172
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ghezzi, P. (2013). Protein glutathionylation in health and disease. Biochim. Biophys. Acta 1830, 3165–3172. doi: 10.1016/j.bbagen.2013.02.009
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gopal, S., Borovok, I., Ofer, A., Yanku, M., Cohen, G., Goebel, W., et al. (2005). A multidomain fusion protein in Listeria monocytogenes catalyzes the two primary activities for glutathione biosynthesis. J. Bacteriol. 187, 3839–3847. doi: 10.1128/JB.187.11.3839-3847.2005
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gray, M. J., Wholey, W. Y., and Jakob, U. (2013a). Bacterial responses to reactive chlorine species. Annu. Rev. Microbiol. 67, 141–160. doi: 10.1146/annurev-micro-102912-142520
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gray, M. J., Wholey, W. Y., Parker, B. W., Kim, M., and Jakob, U. (2013b). NemR is a bleach-sensing transcription factor. J. Biol. Chem. 288, 13789–13798. doi: 10.1074/jbc.M113.454421
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gupta, V., and Carroll, K. S. (2014). Sulfenic acid chemistry, detection and cellular lifetime. Biochim. Biophys. Acta 1840, 847–875. doi: 10.1016/j.bbagen.2013.05.040
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Handtke, S., Schroeter, R., Jurgen, B., Methling, K., Schluter, R., Albrecht, D., et al. (2014). Bacillus pumilus reveals a remarkably high resistance to hydrogen peroxide provoked oxidative stress. PLoS ONE 9:e85625. doi: 10.1371/journal.pone.0085625
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hawkins, C. L., Pattison, D. I., and Davies, M. J. (2003). Hypochlorite-induced oxidation of amino acids, peptides and proteins. Amino Acids 25, 259–274. doi: 10.1007/s00726-003-0016-x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Helmann, J. D. (2011). Bacillithiol, a new player in bacterial redox homeostasis. Antioxid. Redox Signal. 15, 123–133. doi: 10.1089/ars.2010.3562
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Holmgren, A. (1976). Hydrogen donor system for Escherichia coli ribonucleoside-diphosphate reductase dependent upon glutathione. Proc. Natl. Acad. Sci. U.S.A. 73, 2275–2279. doi: 10.1073/pnas.73.7.2275
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Holmgren, A. (1981). Regulation of ribonucleotide reductase. Curr. Top. Cell. Regul. 19, 47–76. doi: 10.1016/B978-0-12-152819-5.50019-1
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hondorp, E. R., and Matthews, R. G. (2004). Oxidative stress inactivates cobalamin-independent methionine synthase (MetE) in Escherichia coli. PLoS Biol. 2:e336. doi: 10.1371/journal.pbio.0020336
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hou, X., Peng, W., Xiong, L., Huang, C., Chen, X., and Zhang, W. (2013). Engineering Clostridium acetobutylicum for alcohol production. J. Biotechnol. 166, 25–33. doi: 10.1016/j.jbiotec.2013.04.013
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hugo, M., Turell, L., Manta, B., Botti, H., Monteiro, G., Netto, L. E., et al. (2009). Thiol and sulfenic acid oxidation of AhpE, the one-cysteine peroxiredoxin from Mycobacterium tuberculosis: kinetics, acidity constants, and conformational dynamics. Biochemistry 48, 9416–9426. doi: 10.1021/bi901221s
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hugo, M., Van Laer, K., Reyes, A. M., Vertommen, D., Messens, J., Radi, R., et al. (2014). Mycothiol/mycoredoxin 1-dependent reduction of the peroxiredoxin AhpE from Mycobacterium tuberculosis. J. Biol. Chem. 289, 5228–5239. doi: 10.1074/jbc.M113.510248
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hwang, C., Lodish, H. F., and Sinskey, A. J. (1995). Measurement of glutathione redox state in cytosol and secretory pathway of cultured cells. Meth. Enzymol. 251, 212–221. doi: 10.1016/0076-6879(95)51123-7
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Imlay, J. A. (2003). Pathways of oxidative damage. Annu. Rev. Microbiol. 57, 395–418. doi: 10.1146/annurev.micro.57.030502.090938
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Imlay, J. A. (2008). Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 77, 755–776. doi: 10.1146/annurev.biochem.77.061606.161055
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Imlay, J. A. (2013). The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat. Rev. Microbiol. 11, 443–454. doi: 10.1038/nrmicro3032
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Inaba, K. (2009). Disulfide bond formation system in Escherichia coli. J. Biochem. 146, 591–597. doi: 10.1093/jb/mvp102
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jacobs, A. T., and Marnett, L. J. (2010). Systems analysis of protein modification and cellular responses induced by electrophile stress. Acc. Chem. Res. 43, 673–683. doi: 10.1021/ar900286y
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Janowiak, B. E., and Griffith, O. W. (2005). Glutathione synthesis in Streptococcus agalactiae. One protein accounts for gamma-glutamylcysteine synthetase and glutathione synthetase activities. J. Biol. Chem. 280, 11829–11839. doi: 10.1074/jbc.M414326200
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jothivasan, V. K., and Hamilton, C. J. (2008). Mycothiol: synthesis, biosynthesis and biological functions of the major low molecular weight thiol in actinomycetes. Nat. Prod. Rep. 25, 1091–1117. doi: 10.1039/b616489g
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kalapos, M. P. (2008). The tandem of free radicals and methylglyoxal. Chem. Biol. Interact. 171, 251–271. doi: 10.1016/j.cbi.2007.11.009
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kehr, S., Jortzik, E., Delahunty, C., Yates, J. R. III, Rahlfs, S., and Becker, K. (2011). Protein S-glutathionylation in malaria parasites. Antioxid. Redox Signal. 15, 2855–2865. doi: 10.1089/ars.2011.4029
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kim, M. S., Dufour, Y. S., Yoo, J. S., Cho, Y. B., Park, J. H., Nam, G. B., et al. (2012). Conservation of thiol-oxidative stress responses regulated by SigR orthologues in actinomycetes. Mol. Microbiol. 85, 326–344. doi: 10.1111/j.1365-2958.2012.08115.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kim, S. O., Merchant, K., Nudelman, R., Beyer, W. F. Jr., Keng, T., Deangelo, J., et al. (2002). OxyR: a molecular code for redox-related signaling. Cell 109, 383–396. doi: 10.1016/S0092-8674(02)00723-7
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Klatt, P., and Lamas, S. (2000). Regulation of protein function by S-glutathiolation in response to oxidative and nitrosative stress. Eur. J. Biochem. 267, 4928–4944. doi: 10.1046/j.1432-1327.2000.01601.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lamers, A. P., Keithly, M. E., Kim, K., Cook, P. D., Stec, D. F., Hines, K. M., et al. (2012). Synthesis of bacillithiol and the catalytic selectivity of FosB-type fosfomycin resistance proteins. Org. Lett. 14, 5207–5209. doi: 10.1021/ol302327t
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lee, C., Shin, J., and Park, C. (2013). Novel regulatory system nemRA-gloA for electrophile reduction in Escherichia coli K-12. Mol. Microbiol. 88, 395–412. doi: 10.1111/mmi.12192
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lee, J. W., Soonsanga, S., and Helmann, J. D. (2007). A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc. Natl. Acad. Sci. U.S.A. 104, 8743–8748. doi: 10.1073/pnas.0702081104
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Leichert, L. I., Gehrke, F., Gudiseva, H. V., Blackwell, T., Ilbert, M., Walker, A. K., et al. (2008). Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc. Natl. Acad. Sci. U.S.A. 105, 8197–8202. doi: 10.1073/pnas.0707723105
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Leonard, S. E., and Carroll, K. S. (2011). Chemical ‘omics’ approaches for understanding protein cysteine oxidation in biology. Curr. Opin. Chem. Biol. 15, 88–102. doi: 10.1016/j.cbpa.2010.11.012
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lessmeier, L., Hoefener, M., and Wendisch, V. F. (2013). Formaldehyde degradation in Corynebacterium glutamicum involves acetaldehyde dehydrogenase and mycothiol-dependent formaldehyde dehydrogenase. Microbiology 159, 2651–2662. doi: 10.1099/mic.0.072413-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Liebeke, M., Pother, D. C., Van Duy, N., Albrecht, D., Becher, D., Hochgrafe, F., et al. (2008). Depletion of thiol-containing proteins in response to quinones in Bacillus subtilis. Mol. Microbiol. 69, 1513–1529. doi: 10.1111/j.1365-2958.2008.06382.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lillig, C. H., Berndt, C., and Holmgren, A. (2008). Glutaredoxin systems. Biochim. Biophys. Acta 1780, 1304–1317. doi: 10.1016/j.bbagen.2008.06.003
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lillig, C. H., Potamitou, A., Schwenn, J. D., Vlamis-Gardikas, A., and Holmgren, A. (2003). Redox regulation of 3′-phosphoadenylylsulfate reductase from Escherichia coli by glutathione and glutaredoxins. J. Biol. Chem. 278, 22325–22330. doi: 10.1074/jbc.M302304200
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lin, J. C., Chiang, B. Y., Chou, C. C., Chen, T. C., Chen, Y. J., and Lin, C. H. (2015). Glutathionylspermidine in the modification of protein SH groups: the enzymology and its application to study protein glutathionylation. Molecules 20, 1452–1474. doi: 10.3390/molecules20011452
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lind, C., Gerdes, R., Hamnell, Y., Schuppe-Koistinen, I., Von Lowenhielm, H. B., Holmgren, A., et al. (2002). Identification of S-glutathionylated cellular proteins during oxidative stress and constitutive metabolism by affinity purification and proteomic analysis. Arch. Biochem. Biophys. 406, 229–240. doi: 10.1016/S0003-9861(02)00468-X
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Liu, Y. B., Chen, C., Chaudhry, M. T., Si, M. R., Zhang, L., Wang, Y., et al. (2014). Enhancing Corynebacterium glutamicum robustness by over-expressing a gene, mshA, for mycothiol glycosyltransferase. Biotechnol. Lett. 36, 1453–1459. doi: 10.1007/s10529-014-1501-x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Liu, Y. B., Long, M. X., Yin, Y. J., Si, M. R., Zhang, L., Lu, Z. Q., et al. (2013). Physiological roles of mycothiol in detoxification and tolerance to multiple poisonous chemicals in Corynebacterium glutamicum. Arch. Microbiol. 195, 419–429. doi: 10.1007/s00203-013-0889-3
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lowther, W. T., and Haynes, A. C. (2011). Reduction of cysteine sulfinic acid in eukaryotic, typical 2-Cys peroxiredoxins by sulfiredoxin. Antioxid. Redox Signal. 15, 99–109. doi: 10.1089/ars.2010.3564
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ma, Z., Chandrangsu, P., Helmann, T. C., Romsang, A., Gaballa, A., and Helmann, J. D. (2014). Bacillithiol is a major buffer of the labile zinc pool in Bacillus subtilis. Mol. Microbiol. 94, 756–770. doi: 10.1111/mmi.12794
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mackay, C. E., and Knock, G. A. (2015). Control of vascular smooth muscle function by Src-family kinases and reactive oxygen species in health and disease. J. Physiol. (Lond). doi: 10.1113/jphysiol.2014.285304
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Marnett, L. J., Riggins, J. N., and West, J. D. (2003). Endogenous generation of reactive oxidants and electrophiles and their reactions with DNA and protein. J. Clin. Invest. 111, 583–593. doi: 10.1172/JCI200318022
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Marteyn, B., Sakr, S., Farci, S., Bedhomme, M., Chardonnet, S., Decottignies, P., et al. (2013). The Synechocystis PCC6803 MerA-like enzyme operates in the reduction of both mercury and uranium under the control of the glutaredoxin 1 enzyme. J. Bacteriol. 195, 4138–4145. doi: 10.1128/JB.00272-13
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Masip, L., Veeravalli, K., and Georgiou, G. (2006). The many faces of glutathione in bacteria. Antioxid. Redox Signal. 8, 753–762. doi: 10.1089/ars.2006.8.753
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Meister, A. (1995). Glutathione biosynthesis and its inhibition. Meth. Enzymol. 252, 26–30. doi: 10.1016/0076-6879(95)52005-8
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mieyal, J. J., and Chock, P. B. (2012). Posttranslational modification of cysteine in redox signaling and oxidative stress: focus on s-glutathionylation. Antioxid. Redox Signal. 16, 471–475. doi: 10.1089/ars.2011.4454
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mieyal, J. J., Gallogly, M. M., Qanungo, S., Sabens, E. A., and Shelton, M. D. (2008). Molecular mechanisms and clinical implications of reversible protein S-glutathionylation. Antioxid. Redox Signal. 10, 1941–1988. doi: 10.1089/ars.2008.2089
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mishra, S., and Imlay, J. (2012). Why do bacteria use so many enzymes to scavenge hydrogen peroxide? Arch. Biochem. Biophys. 525, 145–160. doi: 10.1016/j.abb.2012.04.014
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Newton, G. L., Arnold, K., Price, M. S., Sherrill, C., Delcardayre, S. B., Aharonowitz, Y., et al. (1996). Distribution of thiols in microorganisms: mycothiol is a major thiol in most actinomycetes. J. Bacteriol. 178, 1990–1995.
Newton, G. L., Buchmeier, N., and Fahey, R. C. (2008). Biosynthesis and functions of mycothiol, the unique protective thiol of Actinobacteria. Microbiol. Mol. Biol. Rev. 72, 471–494. doi: 10.1128/MMBR.00008-08
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Newton, G. L., and Fahey, R. C. (2008). Regulation of mycothiol metabolism by sigma(R) and the thiol redox sensor anti-sigma factor RsrA. Mol. Microbiol. 68, 805–809. doi: 10.1111/j.1365-2958.2008.06222.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Newton, G. L., Fahey, R. C., and Rawat, M. (2012). Detoxification of toxins by bacillithiol in Staphylococcus aureus. Microbiology 158, 1117–1126. doi: 10.1099/mic.0.055715-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Newton, G. L., Leung, S. S., Wakabayashi, J. I., Rawat, M., and Fahey, R. C. (2011). The DinB superfamily includes novel mycothiol, bacillithiol, and glutathione S-transferases. Biochemistry 50, 10751–10760. doi: 10.1021/bi201460j
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Newton, G. L., Rawat, M., La Clair, J. J., Jothivasan, V. K., Budiarto, T., Hamilton, C. J., et al. (2009). Bacillithiol is an antioxidant thiol produced in Bacilli. Nat. Chem. Biol. 5, 625–627. doi: 10.1038/nchembio.189
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Nilewar, S. S., and Kathiravan, M. K. (2014). Mycothiol: a promising antitubercular target. Bioorg. Chem. 52, 62–68. doi: 10.1016/j.bioorg.2013.11.004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ordonez, E., Van Belle, K., Roos, G., De Galan, S., Letek, M., Gil, J. A., et al. (2009). Arsenate reductase, mycothiol, and mycoredoxin concert thiol/disulfide exchange. J. Biol. Chem. 284, 15107–15116. doi: 10.1074/jbc.M900877200
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ozyamak, E., De Almeida, C., De Moura, A. P., Miller, S., and Booth, I. R. (2013). Integrated stress response of Escherichia coli to methylglyoxal: transcriptional readthrough from the nemRA operon enhances protection through increased expression of glyoxalase I. Mol. Microbiol. 88, 936–950. doi: 10.1111/mmi.12234
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pan, J., and Carroll, K. S. (2013). Persulfide reactivity in the detection of protein s-sulfhydration. ACS Chem. Biol. 8, 1110–1116. doi: 10.1021/cb4001052
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Park, J. H., and Roe, J. H. (2008). Mycothiol regulates and is regulated by a thiol-specific antisigma factor RsrA and sigma(R) in Streptomyces coelicolor. Mol. Microbiol. 68, 861–870. doi: 10.1111/j.1365-2958.2008.06191.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Parsonage, D., Newton, G. L., Holder, R. C., Wallace, B. D., Paige, C., Hamilton, C. J., et al. (2010). Characterization of the N-acetyl-alpha-D-glucosaminyl l-malate synthase and deacetylase functions for bacillithiol biosynthesis in Bacillus anthracis. Biochemistry 49, 8398–8414. doi: 10.1021/bi100698n
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Paulsen, C. E., and Carroll, K. S. (2013). Cysteine-mediated redox signaling: chemistry, biology, and tools for discovery. Chem. Rev. 113, 4633–4679. doi: 10.1021/cr300163e
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Peralta, D., Bronowska, A. K., Morgan, B., Doka, E., Van Laer, K., Nagy, P., et al. (2015). A proton relay enhances H2O2 sensitivity of GAPDH to facilitate metabolic adaptation. Nat. Chem. Biol. 11, 156–163. doi: 10.1038/nchembio.1720
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Perera, V. R., Newton, G. L., Parnell, J. M., Komives, E. A., and Pogliano, K. (2014). Purification and characterization of the Staphylococcus aureus bacillithiol transferase BstA. Biochim. Biophys. Acta 1840, 2851–2861. doi: 10.1016/j.bbagen.2014.05.001
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Posada, A. C., Kolar, S. L., Dusi, R. G., Francois, P., Roberts, A. A., Hamilton, C. J., et al. (2014). Importance of bacillithiol in the oxidative stress response of Staphylococcus aureus. Infect. Immun. 82, 316–332. doi: 10.1128/IAI.01074-13
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pöther, D. C., Gierok, P., Harms, M., Mostertz, J., Hochgrafe, F., Antelmann, H., et al. (2013). Distribution and infection-related functions of bacillithiol in Staphylococcus aureus. Int. J. Med. Microbiol. 303, 114–123. doi: 10.1016/j.ijmm.2013.01.003
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Potter, A. J., Trappetti, C., and Paton, J. C. (2012). Streptococcus pneumoniae uses glutathione to defend against oxidative stress and metal ion toxicity. J. Bacteriol. 194, 6248–6254. doi: 10.1128/JB.01393-12
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rajkarnikar, A., Strankman, A., Duran, S., Vargas, D., Roberts, A. A., Barretto, K., et al. (2013). Analysis of mutants disrupted in bacillithiol metabolism in Staphylococcus aureus. Biochem. Biophys. Res. Commun. 436, 128–133. doi: 10.1016/j.bbrc.2013.04.027
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ratasuk, N., and Nanny, M. A. (2007). Characterization and quantification of reversible redox sites in humic substances. Environ. Sci. Technol. 41, 7844–7850. doi: 10.1021/es071389u
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rawat, M., Johnson, C., Cadiz, V., and Av-Gay, Y. (2007). Comparative analysis of mutants in the mycothiol biosynthesis pathway in Mycobacterium smegmatis. Biochem. Biophys. Res. Commun. 363, 71–76. doi: 10.1016/j.bbrc.2007.08.142
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Reniere, M. L., Whiteley, A. T., Hamilton, K. L., John, S. M., Lauer, P., Brennan, R. G., et al. (2015). Glutathione activates virulence gene expression of an intracellular pathogen. Nature 517, 170–173. doi: 10.1038/nature14029
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Reyes, A. M., Hugo, M., Trostchansky, A., Capece, L., Radi, R., and Trujillo, M. (2011). Oxidizing substrate specificity of Mycobacterium tuberculosis alkyl hydroperoxide reductase E: kinetics and mechanisms of oxidation and overoxidation. Free Radic. Biol. Med. 51, 464–473. doi: 10.1016/j.freeradbiomed.2011.04.023
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Roberts, A. A., Sharma, S. V., Strankman, A. W., Duran, S. R., Rawat, M., and Hamilton, C. J. (2013). Mechanistic studies of FosB: a divalent-metal-dependent bacillithiol-S-transferase that mediates fosfomycin resistance in Staphylococcus aureus. Biochem. J. 451, 69–79. doi: 10.1042/BJ20121541
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rochat, T., Nicolas, P., Delumeau, O., Rabatinova, A., Korelusova, J., Leduc, A., et al. (2012). Genome-wide identification of genes directly regulated by the pleiotropic transcription factor Spx in Bacillus subtilis. Nucleic Acids Res. 40, 9571–9583. doi: 10.1093/nar/gks755
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ruane, K. M., Davies, G. J., and Martinez-Fleites, C. (2008). Crystal structure of a family GT4 glycosyltransferase from Bacillus anthracis ORF BA1558. Proteins 73, 784–787. doi: 10.1002/prot.22171
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rudolph, T. K., and Freeman, B. A. (2009). Transduction of redox signaling by electrophile-protein reactions. Sci Signal 2:re7. doi: 10.1126/scisignal.290re7
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sareen, D., Newton, G. L., Fahey, R. C., and Buchmeier, N. A. (2003). Mycothiol is essential for growth of Mycobacterium tuberculosis Erdman. J. Bacteriol. 185, 6736–6740. doi: 10.1128/JB.185.22.6736-6740.2003
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sassetti, C. M., and Rubin, E. J. (2003). Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. U.S.A. 100, 12989–12994. doi: 10.1073/pnas.2134250100
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sharma, S. V., Arbach, M., Roberts, A. A., Macdonald, C. J., Groom, M., and Hamilton, C. J. (2013). Biophysical features of bacillithiol, the glutathione surrogate of Bacillus subtilis and other firmicutes. Chembiochem 14, 2160–2168. doi: 10.1002/cbic.201300404
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Song, M., Husain, M., Jones-Carson, J., Liu, L., Henard, C. A., and Vazquez-Torres, A. (2013). Low-molecular-weight thiol-dependent antioxidant and antinitrosative defences in Salmonella pathogenesis. Mol. Microbiol. 87, 609–622. doi: 10.1111/mmi.12119
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ta, P., Buchmeier, N., Newton, G. L., Rawat, M., and Fahey, R. C. (2011). Organic hydroperoxide resistance protein and ergothioneine compensate for loss of mycothiol in Mycobacterium smegmatis mutants. J. Bacteriol. 193, 1981–1990. doi: 10.1128/JB.01402-10
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Thompson, M. K., Keithly, M. E., Goodman, M. C., Hammer, N. D., Cook, P. D., Jagessar, K. L., et al. (2014). Structure and function of the genomically encoded fosfomycin resistance enzyme, FosB, from Staphylococcus aureus. Biochemistry 53, 755–765. doi: 10.1021/bi4015852
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Thompson, M. K., Keithly, M. E., Harp, J., Cook, P. D., Jagessar, K. L., Sulikowski, G. A., et al. (2013). Structural and chemical aspects of resistance to the antibiotic fosfomycin conferred by FosB from Bacillus cereus. Biochemistry 52, 7350–7362. doi: 10.1021/bi4009648
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Van Laer, K., Buts, L., Foloppe, N., Vertommen, D., Van Belle, K., Wahni, K., et al. (2012). Mycoredoxin-1 is one of the missing links in the oxidative stress defence mechanism of Mycobacteria. Mol. Microbiol. 86, 787–804. doi: 10.1111/mmi.12030
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Van Laer, K., Hamilton, C. J., and Messens, J. (2013). Low-molecular-weight thiols in thiol-disulfide exchange. Antioxid. Redox Signal. 18, 1642–1653. doi: 10.1089/ars.2012.4964
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Vanduinen, A. J., Winchell, K. R., Keithly, M. E., and Cook, P. D. (2015). X-ray crystallographic structure of BshC, a unique enzyme involved in bacillithiol biosynthesis. Biochemistry 54, 100–103. doi: 10.1021/bi501394q
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Vergauwen, B., Elegheert, J., Dansercoer, A., Devreese, B., and Savvides, S. N. (2010). Glutathione import in Haemophilus influenzae Rd is primed by the periplasmic heme-binding protein HbpA. Proc. Natl. Acad. Sci. U.S.A. 107, 13270–13275. doi: 10.1073/pnas.1005198107
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Vergauwen, B., Verstraete, K., Senadheera, D. B., Dansercoer, A., Cvitkovitch, D. G., Guedon, E., et al. (2013). Molecular and structural basis of glutathione import in Gram-positive bacteria via GshT and the cystine ABC importer TcyBC of Streptococcus mutans. Mol. Microbiol. 89, 288–303. doi: 10.1111/mmi.12274
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Villadangos, A. F., Van Belle, K., Wahni, K., Dufe, V. T., Freitas, S., Nur, H., et al. (2011). Corynebacterium glutamicum survives arsenic stress with arsenate reductases coupled to two distinct redox mechanisms. Mol. Microbiol. 82, 998–1014. doi: 10.1111/j.1365-2958.2011.07882.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wang, C., Weerapana, E., Blewett, M. M., and Cravatt, B. F. (2014). A chemoproteomic platform to quantitatively map targets of lipid-derived electrophiles. Nat. Methods 11, 79–85. doi: 10.1038/nmeth.2759
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Winterbourn, C. C., and Kettle, A. J. (2013). Redox reactions and microbial killing in the neutrophil phagosome. Antioxid. Redox Signal. 18, 642–660. doi: 10.1089/ars.2012.4827
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Witthoff, S., Muhlroth, A., Marienhagen, J., and Bott, M. (2013). C1 metabolism in Corynebacterium glutamicum: an endogenous pathway for oxidation of methanol to carbon dioxide. Appl. Environ. Microbiol. 79, 6974–6983. doi: 10.1128/AEM.02705-13
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zaffagnini, M., Bedhomme, M., Groni, H., Marchand, C. H., Puppo, C., Gontero, B., et al. (2012a). Glutathionylation in the photosynthetic model organism Chlamydomonas reinhardtii: a proteomic survey. Mol. Cell. Proteomics 11:M111.014142. doi: 10.1074/mcp.M111.014142
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zaffagnini, M., Bedhomme, M., Lemaire, S. D., and Trost, P. (2012b). The emerging roles of protein glutathionylation in chloroplasts. Plant Sci. 185–186, 86–96. doi: 10.1016/j.plantsci.2012.01.005
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhang, D., Macinkovic, I., Devarie-Baez, N. O., Pan, J., Park, C. M., Carroll, K. S., et al. (2014). Detection of protein S-sulfhydration by a tag-switch technique. Angew. Chem. Int. Ed Engl. 53, 575–581. doi: 10.1002/anie.201305876
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhao, Q., Wang, M., Xu, D., Zhang, Q., and Liu, W. (2015). Metabolic coupling of two small-molecule thiols programs the biosynthesis of lincomycin A. Nature 518, 115–119. doi: 10.1038/nature14137
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zuber, P. (2004). Spx-RNA polymerase interaction and global transcriptional control during oxidative stress. J. Bacteriol. 186, 1911–1918. doi: 10.1128/JB.186.7.1911-1918.2004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zuber, P. (2009). Management of oxidative stress in Bacillus. Annu. Rev. Microbiol. 63, 575–597. doi: 10.1146/annurev.micro.091208.073241
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keywords: oxidative stress, protein S-thiolation, thiol-redox buffer, glutathione, bacillithiol, mycothiol
Citation: Loi VV, Rossius M and Antelmann H (2015) Redox regulation by reversible protein S-thiolation in bacteria. Front. Microbiol. 6:187. doi: 10.3389/fmicb.2015.00187
Received: 25 November 2014; Accepted: 20 February 2015;
Published: 16 March 2015.
Edited by:
Jörg Stülke, Georg-August-Universität Göttingen, GermanyReviewed by:
Ulrike Kappler, University of Queensland, AustraliaCopyright © 2015 Loi, Rossius and Antelmann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haike Antelmann, Institute of Microbiology, Ernst-Moritz-Arndt-University of Greifswald, Friedrich-Ludwig-Jahnstr. 15, Greifswald, GermanyYW50ZWxtYW5AdW5pLWdyZWlmc3dhbGQuZGU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.