- 1Department of Molecular Genetics, Groningen Biomolecular Sciences and Biotechnology Institute, University of Groningen, Groningen, Netherlands
- 2Department of Bioinformatics and Biotechnology, Government College University, Faisalabad, Pakistan
- 3Department of Microbiology, Tumor and Cell Biology, Karolinska Institutet, Stockholm, Sweden
In this study, we have explored the impact of ascorbic acid on the transcriptome of Streptococcus pneumoniae D39. The expression of several genes and operons, including the ula operon (which has been previously shown to be involved in ascorbic acid utilization), the AdcR regulon (which has been previously shown to be involved in zinc transport and virulence) and a PTS operon (which we denote here as ula2 operon) were altered in the presence of ascorbic acid. The ula2 operon consists of five genes, including the transcriptional activator ulaR2. Our β-galactosidase assay data and transcriptome comparison of the ulaR2 mutant with the wild-type demonstrated that the transcriptional activator UlaR2 in the presence of ascorbic acid activates the expression of the ula2 operon. We further predict a 16-bp regulatory site (5′-ATATTGTGCTCAAATA-3′) for UlaR2 in the Pula2. Furthermore, we have explored the effect of ascorbic acid on the expression of the AdcR regulon. Our ICP-MS analysis showed that addition of ascorbic acid to the medium causes zinc starvation in the cell which leads to the activation of the AdcR regulon.
Introduction
Streptococcus pneumoniae is an opportunistic Gram-positive human nasopharyngeal pathogen that causes millions of deaths each year worldwide due to pneumococcal infections like pneumonia, meningitis, bacteremia, otitis media (Mitchell, 2003; Kadioglu et al., 2008; O'Brien et al., 2009). It relies on different carbohydrate sources for its survival in the host, although there is a limited availability of free carbohydrate sources in its initial colonization niche, the nasopharynx (Ispahani et al., 2004; Iyer et al., 2005; Kadioglu et al., 2008; Bidossi et al., 2012; Buckwalter and King, 2012). However, the pneumococcus can make use of the complex glycoconjugates available in the host environment to obtain energy required for colonization (King et al., 2006; Burnaugh et al., 2008; Marion et al., 2011).
Like many other bacteria, glucose is the preferred carbon source for S. pneumoniae, but due to the presence of different sugar-specific PTSs (phosphotransferase systems), it can make use of various other carbon sources including ascorbic acid (Cochu et al., 2003; Bidossi et al., 2012). Many bacteria have been shown to ferment ascorbic acid and use it as a carbon source (Yew and Gerlt, 2002; Bidossi et al., 2012; Mehmeti et al., 2013). Ascorbic acid enters the cell through the ascorbate-specific PTS (UlaABC) that putatively carries out the assimilation of ascorbic acid in many bacteria including Escherichia coli (Yew and Gerlt, 2002; Campos et al., 2004) and Klebsiella pneumoniae (Campos et al., 2008). In our previous study, we showed that S. pneumoniae also harbors the ula operon, consisting of nine genes, that shares high homology with the ula gene cluster responsible for the assimilation of ascorbic acid in E. coli and K. pneumoniae (Afzal et al., 2015). However, there are a number of variations in the organization and regulation of the ula locus in S. pneumoniae compared to E. coli and K. pneumoniae. One major difference is the mode of regulation of the ula operon. In S. pneumoniae, the ula operon is activated by transcriptional regulator UlaR in the presence of ascorbic acid, whereas in E. coli and K. pneumoniae the ula gene cluster is repressed by UlaR in the absence of ascorbic acid (Campos et al., 2004, 2008; Afzal et al., 2015).
K. pneumoniae and E. coli are reported to have two systems (ulaABCDEFG and yiaKLX1X2PQRS) for the transport and utilization of ascorbic acid (Campos et al., 2007, 2008). This may also suggest the presence of another ascorbic acid-specific PTS in S. pneumoniae. To explore this, we investigated the effect of ascorbic acid on the whole transcriptome of S. pneumoniae. We did find another PTS operon (denoted as ula2 operon) that was upregulated in the presence of ascorbic acid. The ula2 operon consists of five genes, including one coding for the transcriptional regulator UlaR2 and has high sequence identity with the ula operon of S. pneumoniae D39. We further demonstrated with the help of microarray analysis and β-galactosidase assays that the transcriptional regulator UlaR2 acts as a transcriptional activator of the ula2 operon in the presence of ascorbic acid. We have also predicted the putative regulatory site (5′-ATATTGTGCTCAAATA-3′) in the promoter region of the ula2 operon for the transcriptional activator UlaR2. Furthermore, we have also shown through β-galactosidase assays and ICP-MS (Inductively Coupled Plasma Mass Spectrometry) analysis that ascorbic acid in the medium leads to zinc starvation in the cell, which results in the upregulation of the AdcR regulon in the presence of ascorbic acid.
Materials and Methods
DNA Techniques, Bacterial Strains, Growth Conditions, and β-Galactosidase Assays
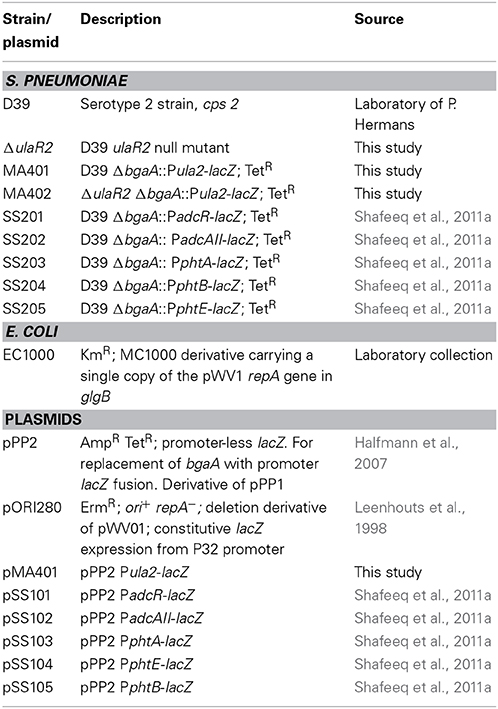
All DNA manipulation techniques, growth conditions and media were the same as described previously (Afzal et al., 2014), unless indicated otherwise. β-Galactosidase assays were performed as described before (Israelsen et al., 1995), using cells that were grown in M17 medium with appropriate carbon sources as mentioned in the Results section (Terzaghi and Sandine, 1975). For β-galactosidase assays, derivatives of S. pneumoniae D39 were grown in M17 medium supplemented with different sugars (arabinose, ascorbic acid, cellobiose, dextrose, fructose, fucose, galactose, glucose, lactose, maltose, mannose, melibiose, sorbitol, sucrose, trehalose, and xylose) with concentrations mentioned in the Results section and harvested in their respective mid-exponential phase of growth. For selection on antibiotics, the medium was supplemented with the following concentrations of antibiotics: tetracycline, 2.5 μg/ml for S. pneumoniae; and ampicillin, 100 μg/ml for E. coli. All bacterial strains used in this study were stored in 10% (v/v) glycerol at −80°C. Bacterial strains and plasmids used in this study are listed in Table 1. Primers used in this study are based on the DNA sequence of the D39 genome (Lanie et al., 2007) and listed in Table 2.
Construction of the ulaR2 Mutant
A markerless ulaR2 mutant (ΔulaR2) was constructed in the S. pneumoniae D39 strain using the pORI280 plasmid, as described before (Kloosterman et al., 2006). Primer pairs ulaR2-1/ulaR2-2 and ulaR2-3/ulaR2-4 were used to generate PCR fragments of the left and right flanking regions of ulaR2, respectively. Mutants were further examined for the presence of the ulaR2 deletion by PCR and DNA sequencing.
Construction of Transcriptional lacZ-fusions
An ectopic lacZ-fusion to the ulaR2 (spd_1961) promoter was constructed in the integrated plasmid pPP2 (Halfmann et al., 2007) with the primer pairs mentioned in Table 2, yielding pMA401. This construct was subsequently introduced into the wild-type and ΔulaR2 strains (via double crossover in the bgaA locus), resulting in strains MA401 and MA402, respectively. All plasmid constructs were checked by PCR and DNA sequencing.
Inductively Coupled Plasma Mass Spectrometry (ICP-MS) Analysis
To determine the intracellular concentration of metal ions by ICP-MS analysis, S. pneumoniae D39 was grown in 15 ml of M17 with and without 10 mM ascorbic acid. The cells were harvested at the mid-exponential growth phase. Cultures were centrifuged and washed (at 4°C) twice with M17 medium, and twice with phosphate-buffered saline (PBS). The cell pellets were dried overnight in a vacuum concentrator at room temperature. The ICP-MS analysis was performed on the cells as described before (Jacobsen et al., 2011; Shafeeq et al., 2011a). Results were expressed as μg of metal ions per g dry weight of cells.
Microarray Analysis and Reverse Transcription (RT)-PCR
For microarray analysis of the effect of ascorbic acid on the transcriptome of S. pneumoniae, wild-type D39 was grown in three independent replicates in M17 and in AM17 (10 mM ascorbic acid + M17) medium, and harvested at their respective mid-exponential growth phase.
To analyze the effect of the ulaR2 deletion on the transcriptome of S. pneumoniae, D39 wild type and ΔulaR2 strains were grown in three independent replicates in AM17 (10 mM ascorbic acid + M17) medium and were harvested at their respective mid-exponential growth phase. All other procedures regarding the DNA microarray experiments were performed essentially as described previously (Shafeeq et al., 2011b, 2012).
To confirm the polycistronic nature of the ula2 operon, RT-PCR (reverse transcription PCR) reactions were performed as described before (Afzal et al., 2014) on the total RNA isolated from wild-type S. pneumoniae D39 grown in AM17 (10 mM ascorbic acid + M17) medium. For the fair comparison of PCR products, 100 ng of RNA and 20 ng of DNA were used.
Microarray Data Analysis
DNA microarray data were analyzed as previously described (Shafeeq et al., 2011a,b). Briefly, microarray slide images were scanned using GenPix Pro 6.1 (MSD analytical technologies). Processing and normalization (LOWESS spotpin-based) of slides were done with the in-house developed MicroPrep software. DNA microarray data were obtained from independent biological replicates hybridized to glass slides, of which one was a dye swap. Expression ratios were calculated from the measurements of at least five spots. Differential expression tests were performed on expression ratios with a local copy of the Cyber-T implementation of a variant of the t-test. False discovery rates were calculated as described (Van Hijum et al., 2005). A gene was considered differentially expressed when p-value was <0.001 and false discovery rate was <0.05 and when at least five measurements were available. For the identification of differentially expressed genes, only genes with a fold change >2 were selected. Microarray data have been submitted to GEO (Gene Expression Omnibus) under the accession number GSE64107.
Results
Ascorbic Acid-Dependent Gene Regulation in S. pneumoniae
To elucidate the effect of ascorbic acid on the transcriptional responses of S. pneumoniae D39 strain, we compared the transcriptome of D39 wild-type grown in AM17 (10 mM ascorbic acid + M17) to that grown in M17 medium (no added sugar). The expression of several genes and operons with diverse functions were altered in the presence of ascorbic acid (Table 3). As expected, the expression of the ula operon was highly upregulated in the presence of ascorbic acid. In a previous study, we have shown that the expression of the ula operon is activated by the transcriptional regulator UlaR in the presence of ascorbic acid (Afzal et al., 2015). Therefore, the altered expression of the ula operon indicates that the conditions used for our transcriptome studies are suitable to explore the ascorbic acid-specific gene expression. The AdcR regulon was also upregulated in our microarray experiment. The AdcR regulon consists of zinc-dependent ABC transporters (AdcCBA), the zinc-dependent transcriptional repressor (AdcR), adhesion lipoprotein (LmB/AdcAII) and pneumococcal histidine triad (Pht) family proteins (PhtA, PhtB, PhtD, and PhtE). This regulon is shown to be involved in zinc transport and the transcriptional repressor AdcR represses its expression in the presence of zinc (Shafeeq et al., 2011a). The upregulation of the AdcR regulon in the presence of ascorbic acid suggests that the presence of ascorbic acid in the medium may cause zinc starvation.
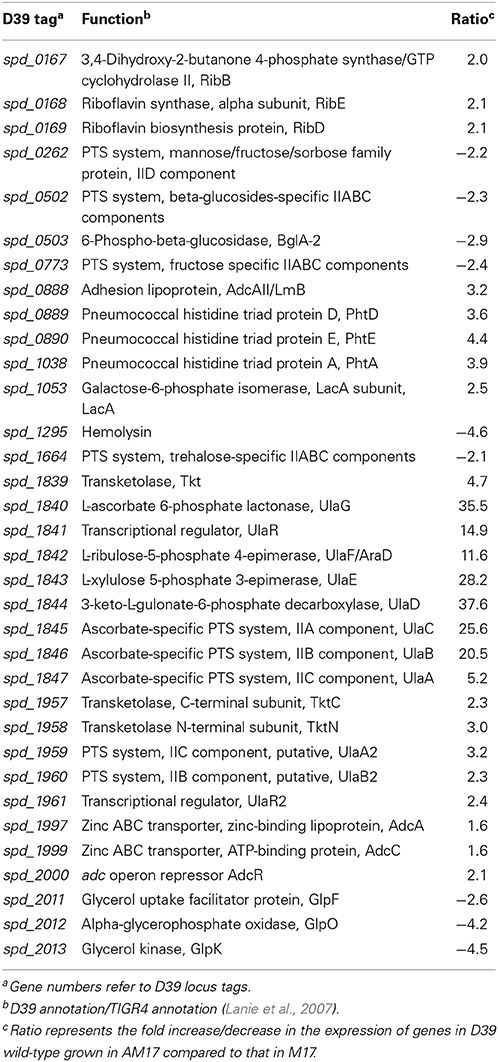
Table 3. Summary of transcriptome comparison of S. pneumoniae strain D39 wild-type grown in AM17 (10 mM ascorbic acid + M17) and in M17.
The expression of the ribDEBH (spd_0166-69) operon was also upregulated in our microarray analysis. This operon is putatively involved in riboflavin metabolism and utilization of ribulose 5-phospate produced from L-ascorbate through the pentose phosphate pathway. Expression of the genes putatively involved in mannose, fructose, sorbose, β-glucosides, and trehalose uptake and utilization was also altered in our ascorbic acid-dependent microarray. An operon (spd_2011-13) consisting of glycerol uptake and utilization genes was downregulated in the presence of ascorbic acid. It has been shown that ascorbic acid causes inhibition of transport of these sugars in E. coli (Loewen and Richter, 1983). Hemolysin (spd-1295) was also downregulated about five times in the presence of ascorbic acid. A PTS operon (spd_1957-61) was upregulated in our microarray analysis. This operon has sequence homology with the ula operon of S. pneumoniae. Based on its sequence homology with the ula operon, we denoted this operon here as the ula2 operon Upregulation of the ula2 operon in the presence of ascorbic acid and its sequence homology with the ula operon may suggest its putative involvement in ascorbic acid utilization and metabolism. Therefore, we decided to further explore the organization and regulation of this operon in the presence of ascorbic acid.
Organization of the ula2 Operon in S. pneumoniae D39
The ula2 operon consists of five genes in S. pneumoniae D39 (Figure 1A), which encode an ascorbic acid-specific PTS (ulaB2A2: spd_1959-60), a transketolase N-terminal subunit (tktN: spd_1958) and a transketolase C-terminal subunit (tktC: spd_1957). ulaB2 has 37% sequence identity to ulaB and ulaA2 has 29% sequence identity to ulaA of the ula operon. Similarly, tktC and tktN have 24 and 35% identity to tkt of the ula operon.
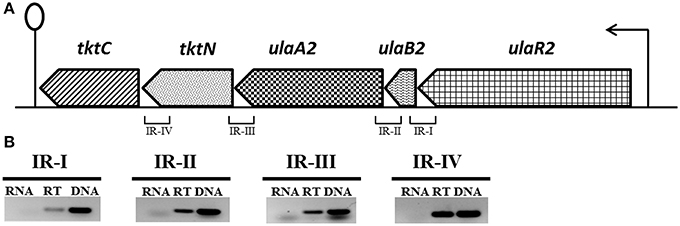
Figure 1. (A) Organization of the ula2 operon in S. pneumoniae D39. Lollipop structure represents the putative transcriptional terminator while black arrows indicate the promoter regions. See text for further details. We take 1 kb = 2.5 cm here for the representation of gene size. (B) Reverse transcriptase (RT) PCR analysis to confirm the polycistronic nature of the ula2 operon in S. pneumoniae D39. RT-PCR was performed on the total RNA isolated from D39 wild-type grown in AM17 (10 mM ascorbic acid + M17) medium with (RT) and without (RNA) reverse transcriptase treatment using the IR-I, IR-II, IR-III, and IR-IV intergenic region primer pairs. DNA was used as a positive control.
RT-PCR using the primer pairs mentioned in Table 2 was performed to confirm that the ula2 operon is transcribed as a single transcriptional unit. RT-PCR data showed that the ula2 operon is transcribed as one transcriptional unit (Figure 1B).
Ascorbic Acid Induces the ula2 Operon, While Glucose, like Other Tested Sugars, Diminishes the Expression of the ula2 Operon
To verify that the altered expression of the ula2 operon in our microarray analysis is due to the specific effect of ascorbic acid in the medium, we made a transcriptional lacZ-fusion to the promoter region of the ula2 operon (Pula2) and transformed it into S. pneumoniae D39 wild type. The wild-type strain containing Pula2-lacZ was grown in M17 medium supplemented with 10 mM of different carbon sources (arabinose, ascorbic acid, cellobiose, dextrose, fructose, fucose, galactose, glucose, lactose, maltose, mannose, melibiose, sorbitol, sucrose, trehalose, and xylose) and β-galactosidase assays were performed. The expression of Pula2-lacZ was highest in the presence of ascorbic acid and lowest in the presence of all other sugars including glucose (Figure 2B). This data confirms our microarray results and also shows that ascorbic acid activates the expression of the ula2 operon, while other tested carbon sources do not play a role in the activation of the ula2 operon.
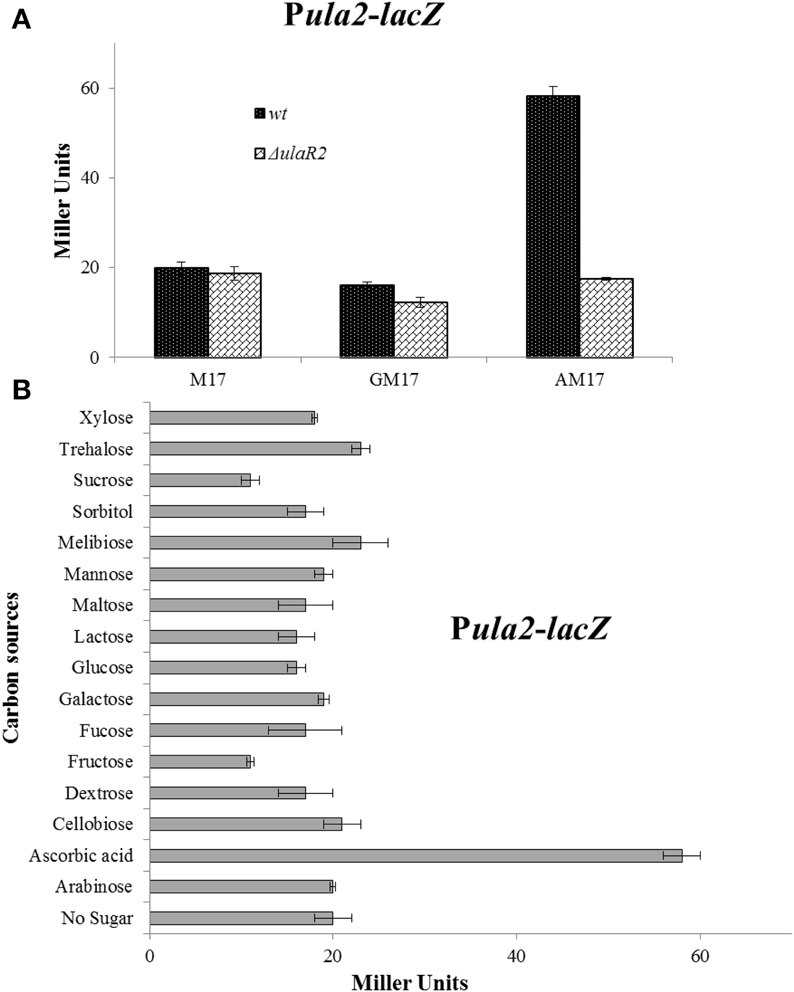
Figure 2. (A) Expression levels (in Miller units) of Pula2-lacZ in D39 wild-type and ΔulaR2 grown in M17 (without any sugar), GM17 (10 mM Glucose + M17) and AM17 (10 mM ascorbic acid + M17) medium. Standard deviation of three independent experiments is indicated in bars. (B) Expression levels (in Miller units) of Pula2-lacZ in D39 wild-type grown in M17 medium with different added sugars (10 mM). Standard deviation of three independent experiments is indicated in bars. The results of One-Way ANOVA are provided in the supplementary data (S1).
UlaR2 Acts as a Transcriptional Activator of the ula2 Operon in the Presence of Ascorbic Acid in S. pneumoniae
The gene for the DNA-binding transcriptional regulator UlaR2 is present in the ula2 operon. The presence of ulaR2 in the ula2 operon might be an indication of its involvement in the regulation of the ula2 operon. To demonstrate the role of UlaR2 in the regulation of the ula2 operon, we created a markerless knockout of ulaR2 in S. pneumoniae D39 and introduced Pula2-lacZ in the ΔulaR2 strain to perform the β-galactosidase assay. Our β-galactosidase assay data show that expression of Pula2-lacZ was abolished in the ΔulaR2 strain compared to the wild-type strain even in the presence of ascorbic acid (Figure 2A). This data suggests the role of ulaR2 as a transcriptional activator of the ula2 operon in the presence of ascorbic acid.
DNA Microarray Analysis of the ΔulaR2 Strain
To study the impact of the ulaR2 deletion on the transcriptome of S. pneumoniae, the transcriptome of wild-type D39 was compared to that of D39 ΔulaR2 grown in AM17 (10 mM ascorbic acid + M17) medium. AM17 medium was used to perform transcriptome analysis, as our β-galactosidase assay with Pula2-lacZ in the ΔulaR2 strain suggests that UlaR2 activates the expression of its targets in the presence of ascorbic acid. The transcriptomic changes incurred in S. pneumoniae D39 due to the deletion of ulaR2 in the presence of ascorbic acid are summarized in Table 4. ulaR2 was nearly six-fold downregulated in ΔulaR2, confirming the deletion of ulaR2 in the ΔulaR2 strain. The ula2 operon was the only operon that was significantly downregulated in the ΔulaR2 strain after applying the criteria of >2.0-fold difference as the threshold change and a P-value < 0.001 was chosen. Downregulation of the ula2 operon in the ΔulaR2 strain confirms the role of UlaR2 as a transcriptional activator of the ula2 operon in the presence of ascorbic acid and also suggests that the ula2 operon is the only target of UlaR2. Moreover, this data is also in accordance with our β-galactosidase data mentioned above. We further decided to explore the regulatory site of UlaR2 in Pula2.
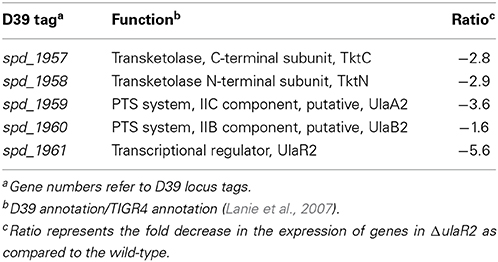
Table 4. Summary of transcriptome comparison of S. pneumoniae ΔulaR2 strain with D39 wild-type grown in AM17 (10 mM ascorbic acid + M17).
Prediction of an UlaR2 Regulatory Site in Pula2
A 16-bp DNA sequence (5′-ATATTGTGCTCAAATA-3′) located upstream of the ula2 operon of S. pneumoniae D39 was predicted by using a MEME motif sampler search (Bailey and Elkan, 1994) which might act as an UlaR2 regulatory site. This predicted UlaR2 regulatory site is found conserved in other sequenced strains of S. pneumoniae available in the KEGG database. Streptococcus agalactiae, Streptococcus equi, Streptococcus mitis, Streptococcus pyogenes, Streptococcus suis, and Streptococcus uberis also encode a putative ula2 operon with a similar gene composition as in S. pneumoniae (Figure 3A). To find the conservation of the UlaR2 regulatory site in these streptococci, we analyzed the ula2 promoter region in these species for the presence of an UlaR2 regulatory site. The analysis revealed that the UlaR2 regulatory site is highly conserved in these streptococci. We further generated a weight matrix of a 16-bp wide UlaR2 regulatory site (5′-AACACARRMCTGTGTK-3′) with the help of Genome 2D Software (Baerends et al., 2004) by aligning the predicted UlaR2 regulatory sequences in the Pula2 of S. agalactiae, S. equi, S. mitis, S. pyogenes, S. suis, and S. uberis with that of S. pneumoniae (Figure 3B). To find more targets of UlaR2 in the D39 genome, we performed a genome-wide search with the pneumococcal UlaR2 regulatory site by using Genome 2D software (Baerends et al., 2004). We could not find any other stretch of DNA similar to the UlaR2 regulatory site in the D39 genome, suggesting that the ula2 operon is the only target of UlaR2. This observation is also consistent with our transcriptome analysis with ΔulaR2.

Figure 3. Identification of the UlaR2 regulatory site in different streptococci. (A) Weight matrix of the identified UlaR2 regulatory site in the Pula2 of different streptococci. (B) Position of the UlaR2 regulatory site in the Pula2 of different streptococci. PN, S. pneumoniae; ST, S. mitis; SA, S. agalactiae; SE, S. equi; SP, S. pyogenes; SS, S. suis; SU, S. uberis. Putative −10 sequences are underlined, translational start sites and putative UlaR2 regulatory sites are bold.
Ascorbic Acid Activates/Derepresses the Expression of the AdcR Regulon by Decreasing the Zinc Concentration
In our microarray analysis in the presence of ascorbic acid, expression of the AdcR regulon was upregulated. This might suggest the role of ascorbic acid in the regulation of the AdcR regulon. AdcR is a zinc-dependent transcriptional regulator that represses the expression of the AdcR regulon (which consists of genes that encode zinc transport and Pht-family proteins) in the presence of zinc (Shafeeq et al., 2011a, 2013). To study the role of ascorbic acid in the regulation of the AdcR regulon, we tested promoter-lacZ fusions of the genes that were upregulated in our microarray experiment in the presence of ascorbic acid (adcAII-phtD, phtA, phtB, phtE, and adcRCBA). The expression of these promoters was significantly higher in AM17 (10 mM ascorbic acid + M17) medium compared to M17 (no added sugar) and GM17 (10 mM glucose + M17) medium, confirming our microarray results (Figure 4). We further studied the role of zinc in the regulation of the AdcR regulon in the presence of ascorbic acid. We added various concentrations of zinc (0.05, 0.1, and 0.2 mM zinc) in AM17 medium (10 mM ascorbic acid + M17). Addition of zinc to the medium in the presence of ascorbic acid led to the repression of the following genes (adcAII-phtD, phtA, phtB, phtE, and adcRCBA) to the expression level without ascorbic acid (Figure 4). This data suggests that the presence of ascorbic acid in the medium causes zinc limitation in the cell.
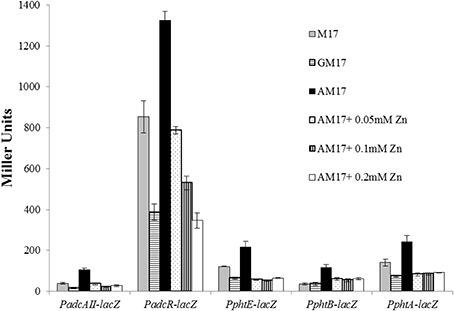
Figure 4. Expression levels (in Miller units) of PadcAII-lacZ, PphtB-lacZ, PphtE-lacZ, PphtA-lacZ, and PadcR-lacZ in D39 wild-type grown in M17 (without any sugar), GM17 (10 mM Glucose + M17) and AM17 (10 mM ascorbic acid + M17) with the addition of increasing concentrations of zinc. Standard deviation of three independent experiments is indicated in bars.
To further investigate the role of ascorbic acid in zinc limitation, ICP-MS analysis was performed on the cells grown with and without 10 mM ascorbic acid in the medium (Table 5). Our ICP-MS analysis showed that the intracellular concentration of zinc decreases to less than half in the presence of ascorbic acid in the medium (Table 5). The concentration of zinc was 12 μg/g dry mass of cells grown in AM17, whereas this concentration was 28 μg/g dry mass of cells grown in M17 (without any added sugar). This data confirms that the altered expression of the AdcR regulon is due to zinc limitation in the medium caused by ascorbic acid. The levels of cobalt and nickel were below the detection level. Similarly, the manganese concentration was four times lower and the iron concentration was about three times lower in the presence of ascorbic acid. We could not observe any significant change in the expression of manganese- and iron-responsive genes in our microarray in the presence of ascorbic acid. This suggests that the concentrations of manganese and iron in the medium are already high enough to repress the expression of manganese-/iron-dependent genes. We also checked the concentration of metal ions in AM17, M17, and GM17 media by ICP-MS analysis. No difference in the concentration of metal ions, including zinc, was observed among these media (data not shown).
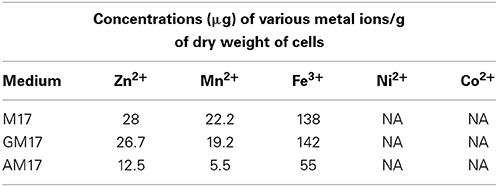
Table 5. ICP-MS analysis of the cells grown in M17, GM17 (10 mM Glucose + M17) and AM17 (10 mM Ascorbic acid + M17) medium.
Discussion
The concentration of ascorbic acid in human blood plasma is about 15 mg/L whereas this concentration may increase up to 50–70 times in lungs, brain, lymph nodes, liver, spleen, pituitary gland, and retina. During infection, this ascorbic acid can be a very useful carbon source for S. pneumoniae (Oreopoulos et al., 1993; Hediger, 2002). Ascorbic acid-dependent expression of the ula operon in S. pneumoniae was explored in our previous study that highlights the presence of an ascorbic acid-specific ula system positively regulated by a transcriptional regulator UlaR (Afzal et al., 2015). Deletion of the ascorbic acid-specific PTS (ulaA) encoded by the ula operon, did not lead to the abolition of growth of S. pneumoniae in the presence of ascorbic acid (Bidossi et al., 2012). This might suggest the presence of another ascorbic acid PTS in S. pneumoniae. The current study is aimed to explore the global gene expression of S. pneumoniae in the presence of ascorbic acid to identify the second putative ascorbic acid system. The ula operon expression was highly upregulated in our microarray analysis in the presence of ascorbic acid, consistent with the results presented in our previous study (Afzal et al., 2015). Furthermore, we found another PTS operon (the ula2 operon) consisting of five genes (spd_1957-61) that is upregulated in the presence of ascorbic acid. We further explored the regulation of this operon in the presence of ascorbic acid and demonstrated that transcriptional regulator UlaR2 acts as a transcriptional activator of the ula2 operon.
Our bioinformatic analysis shows that the ula2 operon shares sequence homology with the ascorbate system in K. pneumoniae and E. coli, which has been shown to be involved in the transport and metabolism of ascorbic acid (Campos et al., 2008). However, there are some differences between them. The number of genes in the second ascorbic acid system varies among these bacteria. The ula2 operon is composed of five genes in S. pneumoniae and regulated by a transcriptional activator, UlaR2, whereas yiaK-S consists of eight genes (yiaKLX1X2PQRS) that are organized in one operon in K. pneumoniae and regulated by a transcriptional repressor YiaJ. Similarly, E. coli also possesses the yiaK-S operon consisting of nine genes (yiaKLMNOPQRS) that is also regulated by a transcriptional repressor YiaJ and this second system has been shown to be involved in the ascorbic acid transport and metabolism (Campos et al., 2007). yiaMNO encodes the tripartite ATP-independent periplasmic transporter and has been reported to recognize L-xylulose as its substrate in E. coli (Plantinga et al., 2004). Another study using different techniques demonstrated that 2,3-diketogulonate, the breakdown product of L-ascorbate, is the substrate of yiaMNO in E. coli (Thomas et al., 2006). yiaX2 encodes a secondary transport protein and yiaX1 codes for a chemotaxis-like protein in K. pneumoniae. Similarly, ula2 operon contains ulaB2A2 as the transport genes in S. pneumoniae. The alternative presence of ulaB2A2, yiaMNO, and yiaX2 with different substrate specificities and the presence of yiaX1 suggest the extent of variation this system can exhibit and this diversification may suggest that this system is beneficial only under certain conditions.
In our transcriptome analysis, expression of several other sugar-specific genes was also altered in the presence of ascorbic acid. Ascorbic acid has been shown to inhibit the uptake of other sugars by various PTSs and this inhibition ranges from 0% (in case of mannitol PTS) to 90% (for glucose PTS) in E. coli (Loewen and Richter, 1983). This sugar uptake inhibition was not limited to the PTSs but also to the facilitated-diffusion uptake of glycerol. Other sugar transport systems were also inhibited to varying degrees (Loewen and Richter, 1983). The riboflavin metabolism operon ribDEBH (spd_0166-69) was also upregulated in ascorbic acid metabolism. RibH (spd_0166) is a 6,7-dimethyl-8-ribityllumazine synthase and RibB (spd_0167) is a 3,4-dihydroxy-2-butanone 4-phosphate synthase/GTP cyclohydrolase II that converts ribulose 5-phosphate into 3,4-dihydroxy-2-butanone 4-phosphate. RibE (spd_0168) is the alpha subunit of riboflavin synthase, whereas RibD (spd_0169) is the riboflavin biosynthesis protein carrying out the next steps of riboflavin metabolism in S. pneumoniae. Three transketolases (Tkt, TktN, and TktC), encoded by genes present in the ula and ula2 operons, are also part of the pentose phosphate pathway and are involved in the subsequent reactions on the intermediates produced in this pathway. The upregulation of this operon in the presence of ascorbic acid suggests that ribulose 5-phosphate produced from ascorbic acid is utilized by S. pneumoniae for the biosynthesis and metabolism of riboflavin.
The presence of ascorbic acid in the medium also resulted in the upregulation of the AdcR regulon, consisting of zinc-responsive genes (adcRCBA adcAII, phtA, phtB, phtD, and phtE) that are involved in zinc transport and virulence (Reyes-Caballero et al., 2010). These genes are negatively regulated by the transcriptional repressor AdcR in the presence of zinc (Shafeeq et al., 2011a). Our ICP-MS analysis showed that the intracellular concentration of zinc decreased about twofold in the presence of ascorbic acid, suggesting that the altered expression of the AdcR regulon in the presence of ascorbic acid is due to zinc starvation in the cell caused by ascorbic acid. Ascorbic acid has already been shown to chelate iron and lead (Lynch and Cook, 1980; Simon and Hudes, 1999). The zinc starvation in the cell caused by ascorbic acid may be due to its metal chelating ability. The intracellular concentration of iron and manganese was also decreased three and four times, respectively, in the presence of ascorbic acid in the medium. However, we could not observe an effect on the expression of manganese- and iron-dependent genes in our ascorbic acid microarray, suggesting that cells still have a high enough concentration of these metal ions for the unchanged expression of the manganese- and iron-dependent genes. A more sophisticated study (involving chemically-defined medium) might provide more insight into the mechanisms by which ascorbic acid causes the starvation of the metal ions inside the cell.
Our bioinformatic analysis showed that the UlaR2 in S. pneumoniae has PTS regulation domain (PRD). The PRD is the phosphorylatable regulatory domain present in bacterial transcriptional activators that usually activates the expression of genes and operons involved in the carbohydrate metabolism (van Tilbeurgh and Declerck, 2001). These transcriptional regulators contain two PRDs with highly conserved histidine residues (van Tilbeurgh and Declerck, 2001). The histidine residues in these PRDs are the targets of the PTS-catalyzed phosphorylation events (Rothe et al., 2012). In addition to PRD, UlaR2 also contains an HTH (Helix Turn Helix) domain, a PTS_EIIA-like and a PTS_EIIB-like domain in S. pneumoniae. Domain organization of UlaR2 is similar to transcriptional regulator UlaR which has two PRDs, an HTH domain and a PTS_EIIB domain (Afzal et al., 2015) and CelR in S. pneumoniae which also has an HTH, two PRDs in addition to an EIIA and an EIIB domain (Shafeeq et al., 2011c). The presence of PRD in UlaR2 suggests that UlaR2 in S. pneumoniae is a PRD-regulated transcriptional regulator belonging to the PRD-containing transcriptional regulators family.
CcpA (Carbon catabolite protein A) is a master transcriptional regulator that represses the expression of genes other than those involved in the transport and metabolism of a preferred carbon source i.e., glucose, through a mechanism called CCR (Carbon Catabolite Repression) (Lulko et al., 2007; Zomer et al., 2007). In this study, we have studied the role of different sugars on the expression of Pula2-lacZ and observed that the expression of Pula2-lacZ was highest in the presence of ascorbic acid compared to other tested sugars. To determine the role of CcpA in the repression of the ula2 operon caused by glucose, we analyzed the promoter region of ulaR2 for a putative cre (catabolite repression element) box. We could not find any cre box in the promoter region of the ula2 operon. The absence of a cre box in Pula2 suggests that CcpA may not be involved in the regulation of the ula2 operon. These findings are consistent with the observations mentioned in a previous study (Carvalho et al., 2011) where no effect on the expression of ula2 operon was observed in a ΔccpA strain in the presence of galactose and glucose.
The two ascorbic acid systems (ula and ula2) are highly conserved in other sequenced strains of S. pneumoniae available in the KEGG database. These ula and ula2 operons are present in a similar organization as in the D39 strain in these pneumococcal strains, except for the G54 strain that has an additional PTS subunit IIA present in the ula2 operon. The conservation and similar organization of the ula and ula2 operons in these pneumococcal strains indicate a similar pattern of regulation of the ula operon by UlaR and the ula2 operon by UlaR2 in the presence of ascorbic acid. Moreover, conservation of these two systems in other pneumococcal strains suggests the important physiological function of these two systems in the life style of S. pneumoniae. Further studies involving animal models will highlight the importance of these two systems in the pathogenicity of S. pneumoniae.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Muhammad Afzal is supported by the G C University, Faisalabad, Pakistan under the faculty development program of HEC Pakistan. We also thank Anne de Jong for his help in bioinformatic analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmicb.2015.00072/abstract
References
Afzal, M., Shafeeq, S., Henriques-Normark, B., and Kuipers, O. P. (2015). UlaR activates expression of the ula operon in Streptococcus pneumoniae in the presence of ascorbic acid. Microbiol. Read. Engl. 161, 41–49. doi: 10.1099/mic.0.083899-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Afzal, M., Shafeeq, S., and Kuipers, O. P. (2014). LacR is a repressor of lacABCD and LacT is an activator of lacTFEG, constituting the lac gene cluster in Streptococcus pneumoniae. Appl. Environ. Microbiol. 80, 5349–5358. doi: 10.1128/AEM.01370-14
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Baerends, R. J. S., Smits, W. K., de Jong, A., Hamoen, L. W., Kok, J., and Kuipers, O. P. (2004). Genome2D: a visualization tool for the rapid analysis of bacterial transcriptome data. Genome Biol. 5:R37. doi: 10.1186/gb-2004-5-5-r37
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bailey, T. L., and Elkan, C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36.
Bidossi, A., Mulas, L., Decorosi, F., Colomba, L., Ricci, S., Pozzi, G., et al. (2012). A functional genomics approach to establish the complement of carbohydrate transporters in Streptococcus pneumoniae. PLoS ONE 7:e33320. doi: 10.1371/journal.pone.0033320
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Buckwalter, C. M., and King, S. J. (2012). Pneumococcal carbohydrate transport: food for thought. Trends Microbiol. 20, 517–522. doi: 10.1016/j.tim.2012.08.008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Burnaugh, A. M., Frantz, L. J., and King, S. J. (2008). Growth of Streptococcus pneumoniae on human glycoconjugates is dependent upon the sequential activity of bacterial exoglycosidases. J. Bacteriol. 190, 221–230. doi: 10.1128/JB.01251-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Campos, E., Baldoma, L., Aguilar, J., and Badia, J. (2004). Regulation of expression of the divergent ulaG and ulaABCDEF operons involved in LaAscorbate dissimilation in Escherichia coli. J. Bacteriol. 186, 1720–1728. doi: 10.1128/JB.186.6.1720-1728.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Campos, E., de la Riva, L., Garces, F., Giménez, R., Aguilar, J., Baldoma, L., et al. (2008). The yiaKLX1X2PQRS and ulaABCDEFG gene systems are required for the aerobic utilization of L-ascorbate in Klebsiella pneumoniae strain 13882 with L-ascorbate-6-phosphate as the inducer. J. Bacteriol. 190, 6615–6624. doi: 10.1128/JB.00815-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Campos, E., Montella, C., Garces, F., Baldoma, L., Aguilar, J., and Badia, J. (2007). Aerobic L-ascorbate metabolism and associated oxidative stress in Escherichia coli. Microbiol. Read. Engl. 153, 3399–3408. doi: 10.1099/mic.0.2007/009613-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Carvalho, S. M., Kloosterman, T. G., Kuipers, O. P., and Neves, A. R. (2011). CcpA ensures optimal metabolic fitness of Streptococcus pneumoniae D39. PLoS ONE 6:e26707. doi: 10.1371/journal.pone.0026707
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cochu, A., Vadeboncoeur, C., Moineau, S., and Frenette, M. (2003). Genetic and biochemical characterization of the phosphoenolpyruvate:glucose/mannose phosphotransferase system of Streptococcus thermophilus. Appl. Environ. Microbiol. 69, 5423–5432. doi: 10.1128/AEM.69.9.5423-5432.2003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Halfmann, A., Hakenbeck, R., and Bruckner, R. (2007). A new integrative reporter plasmid for Streptococcus pneumoniae. FEMS Microbiol. Lett. 268, 217–224. doi: 10.1111/j.1574-6968.2006.00584.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hediger, M. A. (2002). New view at C. Nat. Med. 8, 445–446. doi: 10.1038/nm0502-445
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ispahani, P., Slack, R. C. B., Donald, F. E., Weston, V. C., and Rutter, N. (2004). Twenty year surveillance of invasive pneumococcal disease in Nottingham: serogroups responsible and implications for immunisation. Arch. Dis. Child. 89, 757–762. doi: 10.1136/adc.2003.036921
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Israelsen, H., Madsen, S. M., Vrang, A., Hansen, E. B., and Johansen, E. (1995). Cloning and partial characterization of regulated promoters from Lactococcus lactis Tn917-lacZ integrants with the new promoter probe vector, pAK80. Appl. Environ. Microbiol. 61, 2540–2547.
Iyer, R., Baliga, N. S., and Camilli, A. (2005). Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J. Bacteriol. 187, 8340–8349. doi: 10.1128/JB.187.24.8340-8349.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jacobsen, F. E., Kazmierczak, K. M., Lisher, J. P., Winkler, M. E., and Giedroc, D. P. (2011). Interplay between manganese and zinc homeostasis in the human pathogen Streptococcus pneumoniae. Metallomics 3, 38–41. doi: 10.1039/c0mt00050g
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kadioglu, A., Weiser, J. N., Paton, J. C., and Andrew, P. W. (2008). The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat.Rev.Microbiol. 6, 288–301. doi: 10.1038/nrmicro1871
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
King, S. J., Hippe, K. R., and Weiser, J. N. (2006). Deglycosylation of human glycoconjugates by the sequential activities of exoglycosidases expressed by Streptococcus pneumoniae. Mol. Microbiol. 59, 961–974. doi: 10.1111/j.1365-2958.2005.04984.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kloosterman, T. G., Bijlsma, J. J. E., Kok, J., and Kuipers, O. P. (2006). To have neighbour's fare: extending the molecular toolbox for Streptococcus pneumoniae. Microbiol. Read. Engl. 152, 351–359. doi: 10.1099/mic.0.28521-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lanie, J. A., Ng, W. L., Kazmierczak, K. M., Andrzejewski, T. M., Davidsen, T. M., Wayne, K. J., et al. (2007). Genome sequence of Avery's virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J. Bacteriol. 189, 38–51. doi: 10.1128/JB.01148-06
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Leenhouts, K., Venema, G., and Kok, J. (1998). A lactococcal pWV01 based integration toolbox for bacteria. Methods Cell Sci. 20, 35–50. doi: 10.1023/A:1009862119114
Loewen, P. C., and Richter, H. E. (1983). Inhibition of sugar uptake by ascorbic acid in Escherichia coli. Arch. Biochem. Biophys. 226, 657–665. doi: 10.1016/0003-9861(83)90335-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lulko, A. T., Buist, G., Kok, J., and Kuipers, O. P. (2007). Transcriptome analysis of temporal regulation of carbon metabolism by CcpA in Bacillus subtilis reveals additional target genes. J. Mol. Microbiol. Biotechnol. 12, 82–95. doi: 10.1159/000096463
Lynch, S. R., and Cook, J. D. (1980). Interaction of vitamin C and iron. Ann. N. Y. Acad. Sci. 355, 32–44. doi: 10.1111/j.1749-6632.1980.tb21325.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Marion, C., Aten, A. E., Woodiga, S. A., and King, S. J. (2011). Identification of an ATPase, MsmK, which energizes multiple carbohydrate ABC transporters in Streptococcus pneumoniae. Infect. Immun. 79, 4193–4200. doi: 10.1128/IAI.05290-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mehmeti, I., Solheim, M., Nes, I. F., and Holo, H. (2013). Enterococcus faecalis grows on ascorbic acid. Appl. Environ. Microbiol. 79, 4756–4758. doi: 10.1128/AEM.00228-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mitchell, T. J. (2003). The pathogenesis of streptococcal infections: from tooth decay to meningitis. Nat. Rev. Microbiol. 1, 219–230. doi: 10.1038/nrmicro771
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
O'Brien, K. L., Wolfson, L. J., Watt, J. P., Henkle, E., Deloria-Knoll, M., McCall, N., et al. (2009). Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374, 893–902. doi: 10.1016/S0140-6736(09)61204-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Oreopoulos, D. G., Lindeman, R. D., VanderJagt, D. J., Tzamaloukas, A. H., Bhagavan, H. N., and Garry, P. J. (1993). Renal excretion of ascorbic acid: effect of age and sex. J. Am. Coll. Nutr. 12, 537–542. doi: 10.1080/07315724.1993.10718349
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Plantinga, T. H., Van Der Does, C., Badia, J., Aguilar, J., Konings, W. N., and Driessen, A. J. M. (2004). Functional characterization of the Escherichia coli K-12 yiaMNO transport protein genes. Mol. Membr. Biol. 21, 51–57. doi: 10.1080/09687680310001607369
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Reyes-Caballero, H., Guerra, A. J., Jacobsen, F. E., Kazmierczak, K. M., Cowart, D., Koppolu, U. M. K., et al. (2010). The metalloregulatory zinc site in Streptococcus pneumoniae AdcR, a zinc-activated MarR family repressor. J. Mol. Biol. 403, 197–216. doi: 10.1016/j.jmb.2010.08.030
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rothe, F. M., Bahr, T., Stülke, J., Rak, B., and Görke, B. (2012). Activation of Escherichia coli antiterminator BglG requires its phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 109, 15906–15911. doi: 10.1073/pnas.1210443109
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shafeeq, S., Kloosterman, T. G., and Kuipers, O. P. (2011a). Transcriptional response of Streptococcus pneumoniae to Zn(2+) limitation and the repressor/activator function of AdcR. Metallomics. 3, 609–618. doi: 10.1039/c1mt00030f
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shafeeq, S., Kloosterman, T. G., and Kuipers, O. P. (2011c). CelR-mediated activation of the cellobiose-utilization gene cluster in Streptococcus pneumoniae. Microbiol. Read. Engl. 157, 2854–2861. doi: 10.1099/mic.0.051359-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shafeeq, S., Kloosterman, T. G., Rajendran, V., and Kuipers, O. P. (2012). Characterization of the ROK-family transcriptional regulator RokA of Streptococcus pneumoniae D39. Microbiol. Read. Engl. 158, 2917–2926. doi: 10.1099/mic.0.062919-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shafeeq, S., Kuipers, O. P., and Kloosterman, T. G. (2013). The role of zinc in the interplay between pathogenic streptococci and their hosts. Mol. Microbiol. 88, 1047–1057. doi: 10.1111/mmi.12256
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shafeeq, S., Yesilkaya, H., Kloosterman, T. G., Narayanan, G., Wandel, M., Andrew, P. W., et al. (2011b). The cop operon is required for copper homeostasis and contributes to virulence in Streptococcus pneumoniae. Mol. Microbiol. 81, 1255–1270. doi: 10.1111/j.1365-2958.2011.07758.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Simon, J. A., and Hudes, E. S. (1999). Relationship of ascorbic acid to blood lead levels. JAMA 281, 2289–2293. doi: 10.1001/jama.281.24.2289
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Terzaghi, B. E., and Sandine, W. E. (1975). Improved medium for lactic streptococci and their bacteriophages. Appl. Environ. Microbiol. 29, 807–813.
van Tilbeurgh, H., and Declerck, N. (2001). Structural insights into the regulation of bacterial signalling proteins containing PRDs. Curr. Opin. Struct. Biol. 11, 685–693. doi: 10.1016/S0959-440X(01)00267-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Thomas, G. H., Southworth, T., León-Kempis, M. R., Leech, A., and Kelly, D. J. (2006). Novel ligands for the extracellular solute receptors of two bacterial TRAP transporters. Microbiol. Read. Engl. 152, 187–198. doi: 10.1099/mic.0.28334-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Van Hijum, S. A. F. T., de Jong, A., Baerends, R. J. S., Karsens, H. A., Kramer, N. E., Larsen, R., et al. (2005). A generally applicable validation scheme for the assessment of factors involved in reproducibility and quality of DNA-microarray data. BMC Genomics 6:77. doi: 10.1186/1471-2164-6-77
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yew, W. S., and Gerlt, J. A. (2002). Utilization of L-ascorbate by Escherichia coli K-12: assignments of functions to products of the yjf-sga and yia-sgb operons. J. Bacteriol. 184, 302–306. doi: 10.1128/JB.184.1.302-306.2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zomer, A. L., Buist, G., Larsen, R., Kok, J., and Kuipers, O. P. (2007). Time-resolved determination of the CcpA regulon of Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 189, 1366–1381. doi: 10.1128/JB.01013-06
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: ascorbic acid, UlaR2, pneumococcus, ula2 operon, AdcR, Zinc
Citation: Afzal M, Shafeeq S and Kuipers OP (2015) Ascorbic acid-dependent gene expression in Streptococcus pneumoniae and the activator function of the transcriptional regulator UlaR2. Front. Microbiol. 6:72. doi: 10.3389/fmicb.2015.00072
Received: 28 November 2014; Accepted: 20 January 2015;
Published online: 11 February 2015.
Edited by:
Biswarup Mukhopadhyay, Virginia Tech, USAReviewed by:
Sven Hammerschmidt, Universität Greifswald, GermanyChristopher McDevitt, The University of Adelaide, Australia
Jens Kreth, University of Oklahoma Health Sciences Center, USA
Copyright © 2015 Afzal, Shafeeq and Kuipers. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Oscar P. Kuipers, Department of Molecular Genetics, Groningen Biomolecular Sciences and Biotechnology Institute, University of Groningen, Nijenborgh 7, 9747 AG Groningen, Netherlands e-mail:by5wLmt1aXBlcnNAcnVnLm5s
 Muhammad Afzal
Muhammad Afzal Sulman Shafeeq
Sulman Shafeeq Oscar P. Kuipers
Oscar P. Kuipers