- 1Unit of Microbiology, Department of Biomedical Sciences and Public Health, School of Medicine, Polytechnic University of Marche, Ancona, Italy
- 2Unit of Microbiology, Department of Life and Environmental Sciences, Polytechnic University of Marche, Ancona, Italy
In streptococci mef(I) and catQ, two relatively uncommon macrolide and chloramphenicol resistance genes, respectively, are typically linked in a genetic module designated IQ module. Though variable, the module consistently encompasses, and is sometimes reduced to, a conserved ∼5.8-kb mef(I)-catQ fragment. The prototype IQ module was described in Streptococcus pneumoniae. IQ-like modules have subsequently been detected in Streptococcus pyogenes and in different species of viridans group streptococci, where mef(E) may be found instead of mef(I). Three genetic elements, one carrying the prototype IQ module from S. pneumoniae and two carrying different, defective IQ modules from S. pyogenes, have recently been characterized. All are integrative and conjugative elements (ICEs) belonging to the Tn5253 family, and have been designated ICESpn529IQ, ICESpy029IQ and ICESpy005IQ, respectively. ICESpy029IQ and ICESpy005IQ were the first Tn5253 family ICEs to be described in S. pyogenes. A wealth of new information has been obtained by comparing their genetic organization, chromosomal integration, and transferability. The origin of the IQ module is unknown. The mechanism by which it spreads in streptococci is discussed.
The macrolide resistance gene mef(I) and the chloramphenicol resistance gene catQ are relatively uncommon genetic determinants accounting respectively for macrolide and chloramphenicol resistance. In streptococci, however, they are typically associated and genetically linked, since neither has yet been detected without the other. This review illustrates current data on the association and discusses its genetic basis.
mef Genes
In streptococci, mef-class genes encode efflux-mediated macrolide resistance. Compared to erm-class genes encoding methylase-mediated target site modification, mef genes usually produce lower-level resistance, only affecting 14- and 15-membered macrolides (M phenotype; Sutcliffe et al., 1996). In streptococci, mef genes include a number of subclasses, of which mef(A) and mef(E) are the most significant. mef(A), the first subclass to be discovered (Clancy et al., 1996), is widespread in Streptococcus pyogenes but is also common in Streptococcus pneumoniae. mef(E), discovered in S. pneumoniae (Tait-Kamradt et al., 1997), is frequently found in this and other Streptococcus species, but is uncommon in S. pyogenes (Klaassen and Mouton, 2005). mef(I), discovered in S. pneumoniae (Cochetti et al., 2005; accession no. AJ971089) and subsequently detected in S. pyogenes (Blackman Northwood et al., 2009) and viridans group streptococci (VGS; Brenciani et al., 2014), is the most widely investigated among the other mef subclasses; these include mef(O), mef(B), and mef(G), detected respectively in S. pyogenes (Sangvik et al., 2005), Streptococcus agalactiae (Cai et al., 2007), and group G streptococci (Amezaga and McKenzie, 2006; Cai et al., 2007). An msr-class gene with homology to msr(A), involved in macrolide efflux in Staphylococcus aureus (Ross et al., 1990), is located immediately downstream of the mef gene: it is usually designated msr(D), although a number of variants are associated with different mef genes. Cotranscription of mef(E) and msr(D) in S. pneumoniae suggested that the products of the two genes may act as a dual efflux system (Gay and Stephens, 2001).
cat Genes
The first and still predominant mechanism of bacterial resistance to chloramphenicol is enzymatic inactivation of the drug by different chloramphenicol acetyltransferases (CATs) encoded by cat genes. Two types of CATs have been recognized, A and B, each including a number of structurally and phylogenetically diverse groups (Schwarz et al., 2004). The catQ gene, encoding a type-A CAT assigned to a distinct group (Schwarz et al., 2004), was originally described (Rood et al., 1989) and then sequenced (Bannam and Rood, 1991; accession no. M55620) in Clostridium perfringens CW531. However, no further catQ gene from this species has been described or is available in GenBank, even though genes with 77–92% DNA identities to catQ from C. perfringens CW531 can be found in several other Clostridium species by in silico analysis. Different type-A CATs encoded by a number of cat genes have been described in streptococci (Trieu-Cuot et al., 1993; Schwarz et al., 2004). With regard to catQ, an early study reported a catQ-like gene in a S. agalactiae strain (Trieu-Cuot et al., 1993). More recently, Mingoia et al. (2007) described catQ in two S. pneumoniae isolates and found that the catQ gene from S. pneumoniae Spn529 (the reference isolate) displayed 96.2% DNA identity (accession no. AJ971089) to the original catQ from C. perfringens CW531.
The IQ Module
When catQ was first detected in S. pneumoniae, it was found to be linked to mef(I) in an ∼15.1-kb DNA fragment, containing two identical tnp1 transposase genes at either end, that was designated ‘IQ element’ (Mingoia et al., 2007). Here, we prefer to call it ‘IQ module,’ since the term ‘element’ might be confusing. Two defective IQ modules were subsequently described in S. pyogenes: one was characterized by a shorter region upstream of mef(I), formed only by tnp1 and a truncated recombinase gene rec2; the other consisted of the sole mef(I)-catQ fragment (Del Grosso et al., 2011). Very recently, a number of variously defective IQ-like modules have been detected in different VGS species (Brenciani et al., 2014). An interesting characteristic of VGS modules is that they most often bear mef(E) instead of mef(I). Moreover, all IQ-like modules detected in VGS lack the right tnp1, i.e., the transposase gene downstream of catQ that is found in the IQ module of S. pneumoniae. Despite variability upstream of the mef gene and downstream of catQ, all reported IQ modules appear to share a conserved mef-catQ fragment [six open reading frames (ORFs), ∼5.8 kb; Figure 1].
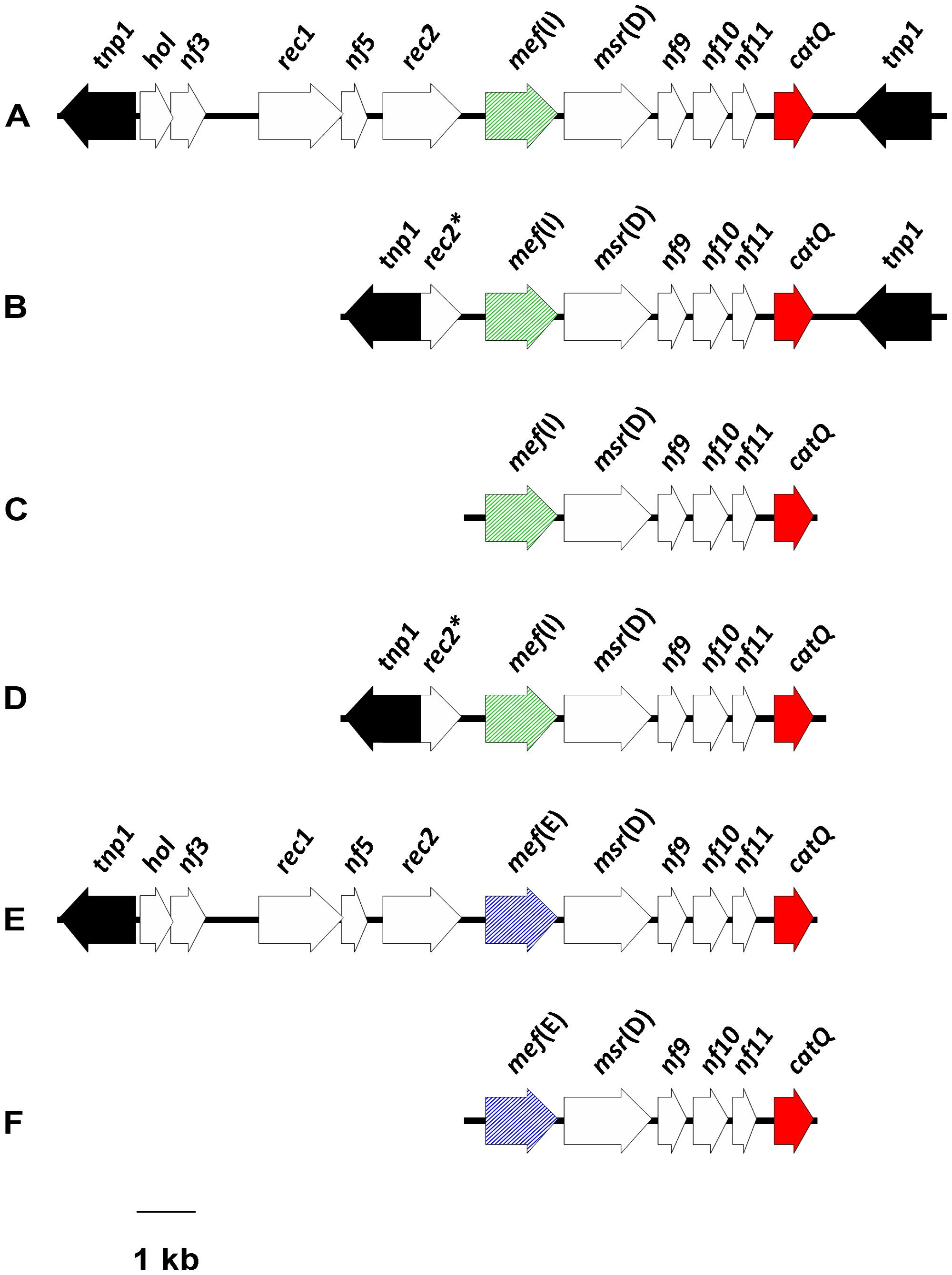
FIGURE 1. Schematic illustration of the IQ modules detected so far in Streptococcus species. (A) S. pneumoniae; prototype IQ module (Mingoia et al., 2007); (B) S. pyogenes (Del Grosso et al., 2011); (C) S. pyogenes (Del Grosso et al., 2011); (D) S. mitis (Brenciani et al., 2014); (E) S. oralis (Brenciani et al., 2014); (F) S. mitis, S. sanguinis, S. parasanguinis (Brenciani et al., 2014). The ORFs are depicted as arrows pointing in the direction of transcription: the mef gene is diagonally striped (green, mef(I); blue, mef(E)); catQ is red; the transposase gene tnp1 is black; and the other ORFs are white.
The origin of the IQ module is unknown. The prototype IQ module of pneumococcal origin has been proved to be capable of undergoing excision in circular form, but was unable to be transferred to a recipient (Mingoia et al., 2014).
From the 5216IQ Complex to a Tn5253 Family Ice
The first attempt to clarify the genetic basis of the mef(I)-catQ association dates back to 2007, when Mingoia et al. (2007) identified in S. pneumoniae a composite structure, that they designated 5216IQ complex (∼30.5 kb); it consisted of two moieties, one represented by the IQ module and the other formed by fragments of transposons Tn5252 and Tn916. The Tn5252 fragment corresponds to the conjugal transfer-related (CTR) functional module of Tn5252 (Alarcon-Chaidez et al., 1997); the Tn916 fragment contains a silent tet(M), unexpressed because it lacks the promoter, the ribosome-binding site, and part of the leader peptide. As realized later (Mingoia et al., 2014) also thanks to the new notion of integrative and conjugative element (ICE; Seth-Smith and Croucher, 2009; Wozniak and Waldor, 2010), the so-called 5216IQ complex is not in fact a genetic element (Varaldo et al., 2009), but rather part of a larger element, a Tn5253 family ICE.
Genetic Elements Carrying IQ Modules
The genetic location of the IQ module has recently been investigated in three strains: one S. pneumoniae (Spn529, serotype 11A [Mingoia 07]) and two S. pyogenes (Spy029 and Spy005, respectively emm types 25.0 and 12.42 [Del Grosso 11]). All three IQ modules were found to be inserted in Tn5253 family ICEs, which were respectively designated ICESpn529IQ, ICESpy029IQ, and ICESpy005IQ (Mingoia et al., 2014). It is worth noting that this was the first time that Tn5253 family ICEs were described in natural isolates of S. pyogenes. The family prototype, Tn5253, is a long-established conjugative composite element resulting from the insertion of Tn5251 (virtually identical to Tn916) into Tn5252 (Ayoubi et al., 1991). Several studies have recently addressed the heterogeneity of Tn5253-like composite elements (Croucher et al., 2009, 2011; Ding et al., 2009; Henderson-Begg et al., 2009; Mingoia et al., 2011; Wyres et al., 2013), whose integrase genes may belong to one of two types: int5252 (the gene of the Tn5253 prototype) or intSp23FST81 [first described in ICESp23FST81 (Croucher et al., 2009)]. The complete sequence of Tn5253 is available (accession no. EU351020) and has been extensively analyzed (Iannelli et al., 2014).
ICESpn529IQ has been completely sequenced (accession no. HG965092): it is 59,466 bp long; its G+C content is 36%; and sequence analysis disclosed 66 ORFs. The IQ module spans from orf22 to orf36 (which correspond to the two identical tnp1 transposase genes located at either end of the module); catQ is orf23 and mef(I) is orf28. ICESpy029IQ and ICESpy005IQ have been characterized based on PCR mapping, partial DNA sequencing, and restriction analysis. The three ICEs are shown and compared in Figure 2.
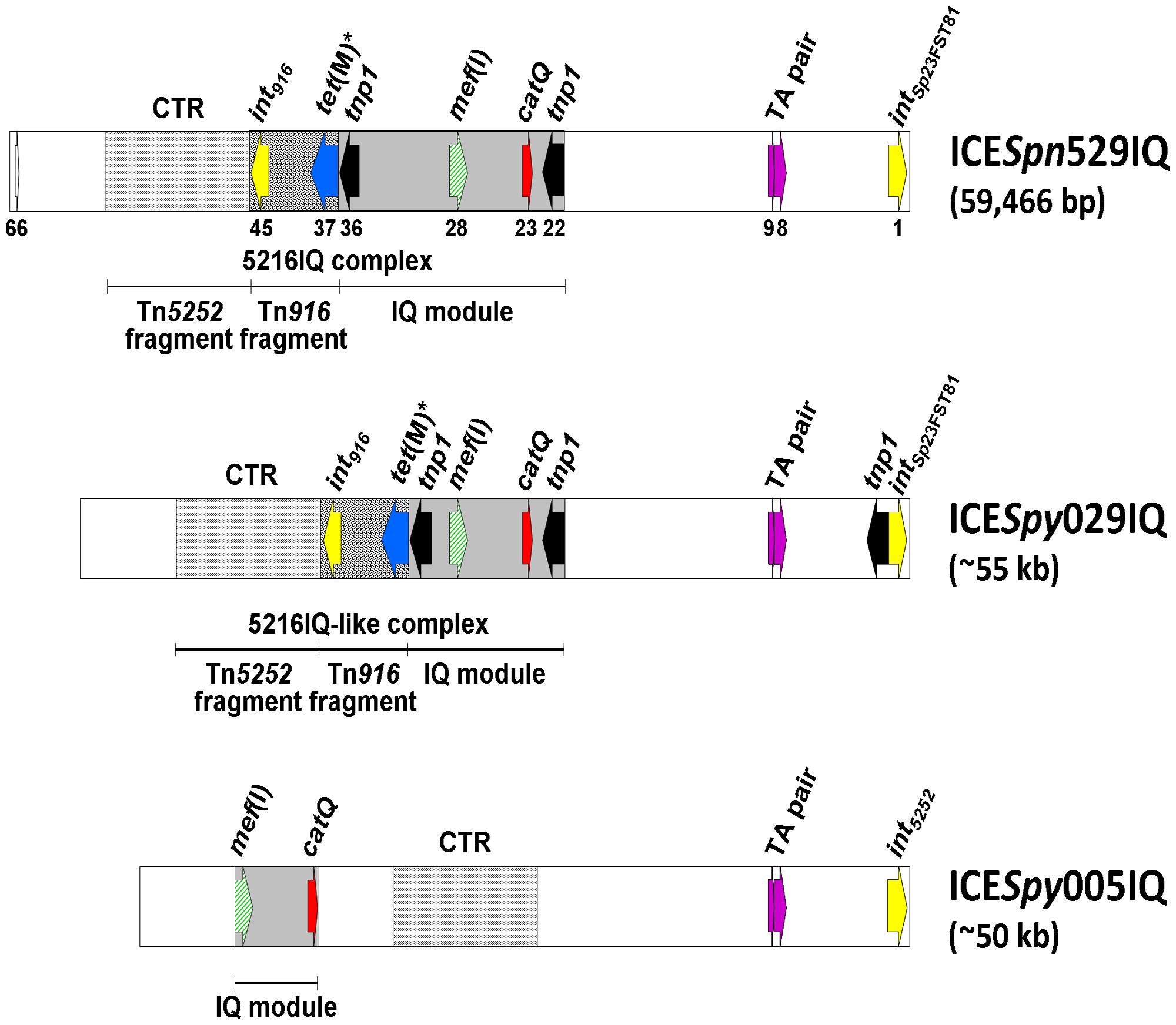
FIGURE 2. Schematic representation of ICESpn529IQ, completely sequenced, and of ICESpy029IQ and ICESpy005IQ, characterized by PCR mapping, partial DNA sequencing and restriction analysis. The ICE genomes are represented as white rectangles with some ORFs and some modules or fragments in evidence. The IQ module is depicted as a gray rectangle, the Tn916 fragment as a spotted rectangle, and the CTR module as a dotted rectangle. The ORFs are depicted as arrows pointing in the direction of transcription, including mef(I) (diagonally striped green), catQ (red), tnp1 (black), integrase genes (yellow), TA gene pair (purple), and silent tet(M) (blue). orf66 of ICESpn529IQ is white. The progressive number is reported only below selected ones of the 66 ORFs of ICESpn529IQ.
ICESpn529IQ and ICESpy029IQ appear to share a very similar organization. Both carry the integrase gene intSp23FST81 and an identical Tn916 fragment. Aside from the different size of their IQ modules, which nevertheless share the same position and insertion site in the respective ICE, the only noticeable difference between the two ICEs is a transposase gene tnp1 adjacent to intSp23FST81; as a result ICESpy029IQ carries a third tnp1 in addition to those found at the ends of the IQ module. In contrast, ICESpy005IQ has the integrase gene int5252 and no Tn916 fragment, and its defective IQ module is found in a different position compared to the IQ modules of ICESpn529IQ and ICESpy029IQ.
All three ICEs lack the linearized plasmid pC194, which contains the chloramphenicol resistance determinant catpC194 and is usually a distinctive cargo of the Tn5252-like moiety of Tn5253 family ICEs. This might be a fitness-related characteristic: as if the ICE no longer needed the plasmid since chloramphenicol resistance is already assured by catQ. It is well known since early studies that the DNA region containing the linearized pC194 is flanked by direct repeats whose recombination may lead to spontaneous curing of the region (Ayoubi et al., 1991; Kiliç et al., 1994). Recent analysis of the sequence of Tn5253 (Iannelli et al., 2014) disclosed a 7,627-bp Ωcat(pC194) flanked by two 1,169-bp direct repeats, which contain a toxin-antitoxin gene pair (Palmieri et al., 2013) that has previously been detected in the pathogenicity island PPI-1 of S. pneumoniae strain TIGR4 (Khoo et al., 2007). In all three ICEs the lack of the integrated plasmid pC194 is associated with the presence in the corresponding region of a single copy of this toxin-antitoxin operon (TA pair), which likely represents the recombination site of Ωcat(pC194) and might be the outcome of repair after plasmid excision. Of the three ICEs, only ICESpy005IQ has been detected as a free circular form, a critical condition for an ICE to undergo conjugal transfer (Wozniak and Waldor, 2010).
Transferability of mef(I) and catQ
The transferability of mef(I) and catQ varies with the transfer mechanism (conjugation or transformation) and the strain harboring the IQ module. Using S. pneumoniae donors, transfer of mef(I) and catQ has never been obtained to any recipient by conjugation, whereas mef(I)- and catQ-positive transformants have been obtained by transformation (Nielsen et al., 2010; Mingoia et al., 2014). It is worth noting that, in experiments using S. pneumoniae Spn529 as the donor, transformants were obtained at a very low frequency (around 10-9) and only using a crude lysate as the transforming DNA: this suggests that DNA may undergo changes during the extraction process, which may account for the early negative results (Mingoia et al., 2007). Conversely, using S. pyogenes donors, conjugation assays yielded no transconjugants from Spy029, whereas transconjugants harboring the entire ICESpy005IQ were obtained at high frequencies from Spy005 (Del Grosso et al., 2011; Mingoia et al., 2014). Transformants were obtained with both S. pyogenes donors, but at a high frequency from strain Spy005 and at a very low frequency from strain Spy029.
Spread of mef(I) and catQ in Streptococci
It has been argued by analogy with other mef gene subclasses (especially in pneumococcal populations) that the spread of mef(I) — and of the IQ module, where it is consistently found — could largely depend on transformation mechanisms (Mingoia et al., 2014). In S. pneumoniae, the two major mef determinants, mef(E) and mef(A), are carried respectively by the mega element (Gay and Stephens, 2001) and Tn1207.1 (Santagati et al., 2000), whose relationship with the relevant mef gene is roughly comparable to that of the IQ module with mef(I). Similar to the IQ module, the mega element and Tn1207.1 are transferable by transformation rather than by conjugation. While the mega element has a variety of insertion sites in the pneumococcal chromosome and its dissemination is erratic, Tn1207.1 is integrated into the chromosome at a specific site and its dissemination is mainly clonal (Del Grosso et al., 2002). Non-clonal dissemination of the IQ module is suggested by the wide range of serotypes of the S. pneumoniae isolates that have so far been found to be mef(I)-positive (Cochetti et al., 2005; Reinert et al., 2008; Nielsen et al., 2010); moreover S. pyogenes strains Spy029 and Spy005 also belong to different emm types (Del Grosso et al., 2011). Altogether, these data suggest that the behavior of mef(I) is likely to be more similar to that of mef(E) than of mef(A).
Concluding Remarks
In streptococci, the macrolide resistance determinant mef(I) and the chloramphenicol resistance determinant catQ, both relatively uncommon within the respective class of resistance genes, are typically linked in a genetic module, designated IQ module. Although the IQ module is uncommon in streptococci, it is likely to be unique to them: it has not been reported outside the genus Streptococcus, and its origin is unknown. Though variable, the IQ module consistently encompasses, and may be reduced to, a conserved ∼5.8-kb DNA fragment spanning from mef(I) (or mef(E) in some VGS) to catQ. After early report in S. pneumoniae (Mingoia et al., 2007), IQ-like modules have been detected in S. pyogenes (Del Grosso et al., 2011) and VGS (Brenciani et al., 2014); moreover, Mingoia et al. (2014) have recently reported on ongoing studies of isolates of other Streptococcus species (S. agalactiae) harboring linked mef(I) and catQ genes.
In recent years, neither mef(I) nor catQ have been described in the documented absence of the other in streptococci. However, the association is unlikely to have been the rule in earlier investigations. Indeed, the S. agalactiae strain where a catQ-like gene was first described in a streptococcus was erythromycin susceptible (Trieu-Cuot et al., 1993). The same is probably true of clostridia: C. perfringens CW531, where catQ was originally detected and sequenced even though its genetic environment was not investigated, was reported to be resistant, besides chloramphenicol, to tetracycline, but not to erythromycin (Rood et al., 1989); and there is no evidence of associated mef genes in the sequenced clostridial genomes or contigs containing catQ-related genes.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Alarcon-Chaidez, F., Sampath, J., Srinivas, P., and Vijayakumar, M. N. (1997). Tn5252: a model for complex Streptococcal conjugative transposons. Adv. Exp. Med. Biol. 418, 1029–1032. doi: 10.1007/978-1-4899-1825-3_242
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Amezaga, M. R., and McKenzie, H. (2006). Molecular epidemiology of macrolide resistance in β-haemolytic streptococci of Lancefield groups A, B, C and G and evidence for a new mef element in group G streptococci that carries allelic variants of mef and msr(D). J. Antimicrob. Chemother. 57, 443–449. doi: 10.1093/jac/dki490
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ayoubi, P., Kiliç, A. O., and Vijayakumar, M. N. (1991). Tn5253, the pneumococcal omega (cat tet) BM6001 element, is a composite structure of two conjugative transposons, Tn5251 and Tn5252. J. Bacteriol. 173, 1617–1622.
Bannam, T. L., and Rood, J. I. (1991). Relationship between the Clostridium perfringens catQ gene product and chloramphenicol acetyltransferases from other bacteria. Antimicrob. Agents Chemother. 35, 471–476. doi: 10.1128/AAC.35.3.471
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Blackman Northwood, J., Del Grosso, M., Cossins, L. R., Coley, M. D., Creti, R., Pantosti, A.,et al. (2009). Characterization of macrolide efflux pump mef subclasses detected in clinical isolates of Streptococcus pyogenes isolated between 1999 and 2005. Antimicrob. Agents Chemother. 53, 1921–1925. doi: 10.1128/AAC.01065-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brenciani, A., Tiberi, E., Tili, E., Mingoia, M., Palmieri, C., Varaldo, P. E.,et al. (2014). Genetic determinants and elements associated with antibiotic resistance in viridans group streptococci. J. Antimicrob. Chemother. 69, 1197–1204. doi: 10.1093/jac/dkt495
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cai, Y., Kong, F., and Gilbert, G. L. (2007). Three new macrolide efflux (mef) gene variants in Streptococcus agalactiae. J. Clin. Microbiol. 45, 2754–2755. doi: 10.1128/JCM.00579-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Clancy, J., Petitpas, J., Dib-Hajj, F., Yuan, W., Cronan, M., Kamath, A. V.,et al. (1996). Molecular cloning and functional analysis of a novel macrolide resistance determinant, mefA, from Streptococcus pyogenes. Mol. Microbiol. 22, 867–879. doi: 10.1046/j.1365-2958.1996.01521.x
Cochetti, I., Vecchi, M., Mingoia, M., Tili, E., Catania, M. R., Manzin, A.,et al. (2005). Molecular characterization of pneumococci with efflux-mediated erythromycin resistance and identification of a novel mef gene subclass, mef(I). Antimicrob. Agents Chemother. 49, 4999–5006. doi: 10.1128/AAC.49.12.4999-5006.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Croucher, N. J., Harris, S. R., Fraser, C., Quail, M. A., Burton, J., van der Linden, M.,et al. (2011). Rapid pneumococcal evolution in response to clinical interventions. Science 331, 430–434. doi: 10.1126/science.1198545
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Croucher, N. J., Walker, D., Romero, P., Lennard, N., Paterson, G. K., Bason, N. C.,et al. (2009). Role of conjugative elements in the evolution of the multidrug-resistant pandemic clone Streptococcus pneumoniae Spain23F ST81. J. Bacteriol. 191, 1480–1489. doi: 10.1128/JB.01343-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Del Grosso, M., Camilli, R., Barbabella, G., Blackman Northwood, J., Farrell, D. J., and Pantosti, A. (2011). Genetic resistance elements carrying mef subclasses other than mef(A) in Streptococcus pyogenes. Antimicrob. Agents Chemother. 55, 3226–3230. doi: 10.1128/AAC.01713-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Del Grosso, M., Iannelli, F., Messina, C., Santagati, M., Petrosillo, N., Stefani, S.,et al. (2002). Macrolide efflux genes mef(A) and mef(E) are carried by different genetic elements in Streptococcus pneumoniae. J. Clin. Microbiol. 40, 774–778. doi: 10.1128/JCM.40.3.774-778.2002
Ding, F., Tang, T., Hsu, M. H., Cui, P., Hu, S., Ju, J.,et al. (2009). Genome evolution driven by host adaptations results in a more virulent and antimicrobial-resistant Streptococcus pneumoniae serotype 14. BMC Genomics 10:158. doi: 10.1186/1471-2164-10-158
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gay, K., and Stephens, D. S. (2001). Structure and dissemination of a chromosomal insertion element encoding macrolide efflux in Streptococcus pneumoniae. J. Infect. Dis. 184, 56–65. doi: 10.1086/321001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Henderson-Begg, S. K., Roberts, A. P., and Hall, L. M. C. (2009). Diversity of putative Tn5253-like elements in Streptococcus pneumoniae. Int. J. Antimicrob. Agents 33, 364–367. doi: 10.1016/j.ijantimicag.2008.10.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Iannelli, F., Santoro, F., Oggioni, M. R., and Pozzi, G. (2014). Nucleotide sequence analysis of integrative conjugative element Tn5253 of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 58, 1235–1239. doi: 10.1128/AAC.01764-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Khoo, S. K., Loll, B., Chan, W. T., Shoeman, R. L., Ngoo, L., Yeo, C. C.,et al. (2007). Molecular and structural characterization of the PezAT chromosomal toxin-antitoxin system of the human pathogen Streptococcus pneumoniae. J. Biol. Chem. 282, 19606–19618. doi: 10.1074/jbc.M701703200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kiliç, A. O., Vijayakumar, M. N., and Al-Khaldi, S. F. (1994). Identification and nucleotide sequence analysis of a transfer-related region in the streptococcal conjugative transposon Tn5252. J. Bacteriol. 176, 5145–5150.
Klaassen, C. H., and Mouton, J. W. (2005). Molecular detection of the macrolide efflux genes: to discriminate or not between mef(A) and mef(E). Antimicrob. Agents Chemother. 49, 1271–1278. doi: 10.1128/AAC.49.4.1271-1278.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mingoia, M., Morici, E., Morroni, G., Giovanetti, E., Del Grosso, M., Pantosti, A.,et al. (2014). Tn5253 family integrative and conjugative elements carrying mef(I) and catQ determinants in Streptococcus pneumoniae and Streptococcus pyogenes. Antimicrob. Agents Chemother. 58, 5886–5893. doi: 10.1128/AAC.03638-14
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mingoia, M., Tili, E., Manso, E., Varaldo, P. E., and Montanari, M. P. (2011). Heterogeneity of Tn5253-like composite elements in clinical Streptococcus pneumoniae isolates. Antimicrob. Agents Chemother. 55, 1453–1459. doi: 10.1128/AAC.01087-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mingoia, M., Vecchi, M., Cochetti, I., Tili, E., Vitali, L. A., Manzin, A.,et al. (2007). Composite structure of Streptococcus pneumoniae containing the erythromycin efflux resistance gene mef(I) and the chloramphenicol resistance gene catQ. Antimicrob. Agents Chemother. 51, 3983–3987. doi: 10.1128/AAC.00790-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nielsen, K. L., Hammerum, A. M., Lambertsen, L. M., Lester, C. H., Arpi, M., Knudsen, J. D.,et al. (2010). Characterization and transfer studies of macrolide resistance genes in Streptococcus pneumoniae from Denmark. Scand. J. Infect. Dis. 42, 586–593. doi: 10.3109/00365541003754451
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Palmieri, C., Mingoia, M., and Varaldo, P. E. (2013). Unconventional circularizable bacterial genetic structures carrying antibiotic resistance determinants. Antimicrob. Agents Chemother. 57, 2440–2441. doi: 10.1128/AAC.02548-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Reinert, R. R., Filimonova, O. Y., Al-Lahham, A., Grudinina, S. A., Ilina, E. N., Weigel, L. M.,et al. (2008). Mechanisms of macrolide resistance among Streptococcus pneumoniae isolates from Russia. Antimicrob. Agents Chemother. 52, 2260–2262. doi: 10.1128/AAC.01270-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rood, J. I., Jefferson, S., Bannam, T. L., Wilkie, J. M., Mullany, P., and Wren, B. W. (1989). Hybridization analysis of three chloramphenicol resistance determinants from Clostridium perfringens and Clostridium difficile. Antimicrob. Agents Chemother. 33, 1569–1574. doi: 10.1128/AAC.33.9.1569
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ross, J. I., Eady, E. A., Cove, J. H., Cunliffe, W. J., Baumberg, S., and Wootton, J. C. (1990). Inducible erythromycin resistance in staphylococci is encoded by a member of the ATP-binding transport super-gene family. Mol. Microbiol. 4, 1207–1214. doi: 10.1111/j.1365-2958.1990.tb00696.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sangvik, M., Littauer, P., Simonsen, G. S., Sundsfjord, A., and Dahl, K. H. (2005). mef(A), mef(E) and a new mef allele in macrolide-resistant Streptococcus spp. isolates from Norway. J. Antimicrob. Chemother. 56, 841–846. doi: 10.1093/jac/dki327
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Santagati, M., Iannelli, F., Oggioni, M. R., Stefani, S., and Pozzi, G. (2000). Characterization of a genetic element carrying the macrolide efflux gene mef(A) in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44, 2585–2587. doi: 10.1128/AAC.44.9.2585-2587.2000
Schwarz, S., Kehrenberg, C., Doublet, B., and Cloeckaert, A. (2004). Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol. Rev. 28, 519–542. doi: 10.1016/j.femsre.2004.04.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Seth-Smith, H., and Croucher, N. J. (2009). Genome watch: breaking the ICE. Nat. Rev. Microbiol. 8, 328–329. doi: 10.1038/nrmicro2137
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sutcliffe, J., Tait-Kamradt, A., and Wondrack, L. (1996). Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob. Agents Chemother. 40, 1817–1824.
Tait-Kamradt, A., Clancy, J., Cronan, M., Dib-Hajj, F., Wondrack, L., Yuan, W.,et al. (1997). mefE is necessary for erythromycin-resistant M phenotype in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 41, 2251–2255.
Trieu-Cuot, P.,de Cespédès, G., Bentorcha, F., Delbos, F., Gaspar, E., and Horaud, T. (1993). Study of heterogeneity of chloramphenicol acetyltransferase (CAT) genes in streptococci and enterococci by polymerase chain reaction: characterization of a new CAT determinant. Antimicrob. Agents Chemother. 37, 2593–2598. doi: 10.1128/AAC.37.12.2593
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Varaldo, P. E., Montanari, M. P., and Giovanetti, E. (2009). Genetic elements responsible for erythromycin resistance in streptococci. Antimicrob. Agents Chemother. 53, 343–353. doi: 10.1128/AAC.00781-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wozniak, R. A., and Waldor, M. K. (2010). Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 8, 552–563. doi: 10.1038/nrmicro2382
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wyres, K. L., van Tonder, A., Lambertsen, L. M., Hakenbeck, R., Parkhill, J., Bentley, S. D.,et al. (2013). Evidence of antimicrobial resistance-conferring genetic elements among pneumococci isolated prior to 1974. BMC Genomics 14:500. doi: 10.1186/1471-2164-14-500
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: mef(I), catQ, macrolide resistance, chloramphenicol resistance, streptococci, IQ module, integrative and conjugative elements (ICEs)
Citation: Mingoia M, Morici E, Brenciani A, Giovanetti E and Varaldo PE (2015) Genetic basis of the association of resistance genes mef(I) (macrolides) and catQ (chloramphenicol) in streptococci. Front. Microbiol. 5:747. doi: 10.3389/fmicb.2014.00747
Received: 19 September 2014; Paper pending published: 16 October 2014;
Accepted: 09 December 2014; Published online: 06 January 2015.
Edited by:
Adam Paul Roberts, University College London, UKReviewed by:
Laura Pagani, University of Pavia, ItalyRoberta Creti, Istituto Superiore di Sanità, Italy
Copyright © 2015 Mingoia, Morici, Brenciani, Giovanetti and Varaldo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marina Mingoia, Unit of Microbiology, Department of Biomedical Sciences and Public Health, School of Medicine, Polytechnic University of Marche, Via Tronto 10/A, 60126 Ancona, Italy e-mail:bS5taW5nb2lhQHVuaXZwbS5pdA==
 Marina Mingoia
Marina Mingoia Eleonora Morici
Eleonora Morici Andrea Brenciani
Andrea Brenciani Eleonora Giovanetti
Eleonora Giovanetti Pietro E. Varaldo
Pietro E. Varaldo