- 1Ecology and Evolutionary Biology, University of Arizona, Tucson, AZ, USA
- 2Environmental Science, Policy and Management, University of California Berkeley, Berkeley, CA, USA
Adaptive radiations are characterized by an increased rate of speciation and expanded range of habitats and ecological niches exploited by those species. The Hawaiian Drosophilidae is a classic adaptive radiation; a single ancestral species colonized Hawaii approximately 25 million years ago and gave rise to two monophyletic lineages, the Hawaiian Drosophila and the genus Scaptomyza. The Hawaiian Drosophila are largely saprophagous and rely on approximately 40 endemic plant families and their associated microbes to complete development. Scaptomyza are even more diverse in host breadth. While many species of Scaptomyza utilize decomposing plant substrates, some species have evolved to become herbivores, parasites on spider egg masses, and exploit microbes on living plant tissue. Understanding the origin of the ecological diversity encompassed by these nearly 700 described species has been a challenge. The central role of microbes in drosophilid ecology suggests bacterial and fungal associates may have played a role in the diversification of the Hawaiian Drosophilidae. Here we synthesize recent ecological and microbial community data from the Hawaiian Drosophilidae to examine the forces that may have led to this adaptive radiation. We propose that the evolutionary success of the Hawaiian Drosophilidae is due to a combination of factors, including adaptation to novel ecological niches facilitated by microbes.
Introduction
Symbioses are broadly defined as persistent interactions between two or more species. While one view of symbioses is restricted to mutualistic relationships, most biologists now consider any type of long-standing interaction between species (e.g., commensalism, mutualism, parasitism) as a symbiotic relationship (Bradford and Schwab, 2013). Many insects have developed intimate evolutionary interactions with microbes that enhance nutrient acquisition or reproduction (Moran and Baumann, 2000; Currie et al., 2003, 2006; Mikheyev et al., 2007; Werren et al., 2008; Hansen and Moran, 2014), or defense against natural enemies (Oliver et al., 2009; Jaenike et al., 2010). A different type of relationship is seen in saprophagous insects, such as many fly species in the family Drosophilidae, which require microbes to break down plant material and make nutrients available for uptake.
Yeasts and bacteria associated with drosophilid flies can influence mating behavior, oviposition behavior, larval feeding choice, and food processing, and these ecological roles can have important evolutionary consequences for insects. Here we propose the Hawaiian Drosophilidae as a model system for studying the role of microbial associations in insect diversification. We focus on two systems: (1) the fungal associates of the largely saprophagous Hawaiian Drosophila lineage and (2) the bacterial species encountered by drosophilids, especially herbivorous members of the genus Scaptomyza.
Hawaiian Drosophilidae
The Hawaiian Drosophilidae is one of the best-characterized examples of an adaptive radiation (Carson and Kaneshiro, 1976). This group includes 687 described species (Magnacca and Price, 2012) and 200–300 more taxa that await description (O’Grady et al., 2011). Hawaiian Drosophilidae have adapted to a diverse array of niches and plant substrates (Kambysellis and Craddock, 1997), and their interactions with microbes are a central part of Drosophila ecology. Microbes have been implicated in providing direct and indirect nutrition sources (Northrop, 1918; Starmer and Aberdeen, 1990), generating chemosensory signals (Dobzhansky et al., 1956; Melcher and Pankratz, 2005), and extensively colonizing larvae and adults (Gilbert, 1980; Ganter, 1988; Coluccio et al., 2008). Although microbes can influence insect ecology (Feldhaar, 2011), promote speciation (Brucker and Bordenstein, 2012, 2013; Joy, 2013), and promote niche differentiation (Janson et al., 2008; Joy, 2013), the potential role of microbes in the diversification of Hawaiian Drosophilidae has not been explored in depth.
Drosophilidae is the oldest known lineage of endemic Hawaiian plants or insects (Price and Clague, 2002). A single colonizing species is estimated to have arrived in the Hawaiian Islands ∼25 million years ago (Thomas and Hunt, 1993; Russo et al., 1995), although recent estimates suggest a slightly older age (Tamura et al., 2004; Obbard et al., 2012). The Hawaiian Drosophilidae has since radiated into two species-rich lineages, the endemic Hawaiian Drosophila and the cosmopolitan genus Scaptomyza. Although the inclusion of Scaptomyza within a larger Drosophila group is confusing taxonomically, this is due to the large-scale polyphyly of the genus Drosophila (O’Grady and DeSalle, 2008; O’Grady et al., 2008a,b; O’Grady and Markow, 2009; O’Grady, 2010). The Hawaiian Drosophilidae (Hawaiian Drosophila+ Scaptomyza) is strongly supported as monophyletic in every rigorous phylogenetic study (Throckmorton, 1975; Thomas and Hunt, 1991, 1993; Baker and DeSalle, 1997; Remsen and DeSalle, 1998; Bonacum, 2001; Remsen and O’Grady, 2002; O’Grady and DeSalle, 2008; O’Grady et al., 2011). Most members of the genus Drosophila, including those endemic to Hawaii, are saprophagous and have adapted to a diverse array of substrates for oviposition, larval development and adult nutrition (Markow and O’Grady, 2005, 2008). While many Scaptomyza species are saprophagous on a variety of larval substrates, including plant leaves and flowers, some now specialize on spider egg sacs or land snails (Magnacca et al., 2008), and herbivory has evolved at least once within this lineage (Lapoint et al., 2013).
Hawaiian Drosophila and Associated Yeasts
The Hawaiian Drosophilidae (Figure 1A) utilize nearly 40% of the native Hawaiian plant families and an array of substrate types (leaves, bark, fruits, sap flux, fungus; Magnacca et al., 2008). Hawaiian Drosophila adults use volatile compounds as cues to identify host plants and stimulate mating and oviposition, although the identity and origin of these cues are unknown (Ohta, 1978). Among the closely related species in the cactophilic Drosophila repleta group, such cues can include byproducts of microbial metabolism (Fogleman and Foster, 1989), raising the possibility that host finding in the Hawaiian Drosophilidae may also be microbially mediated.
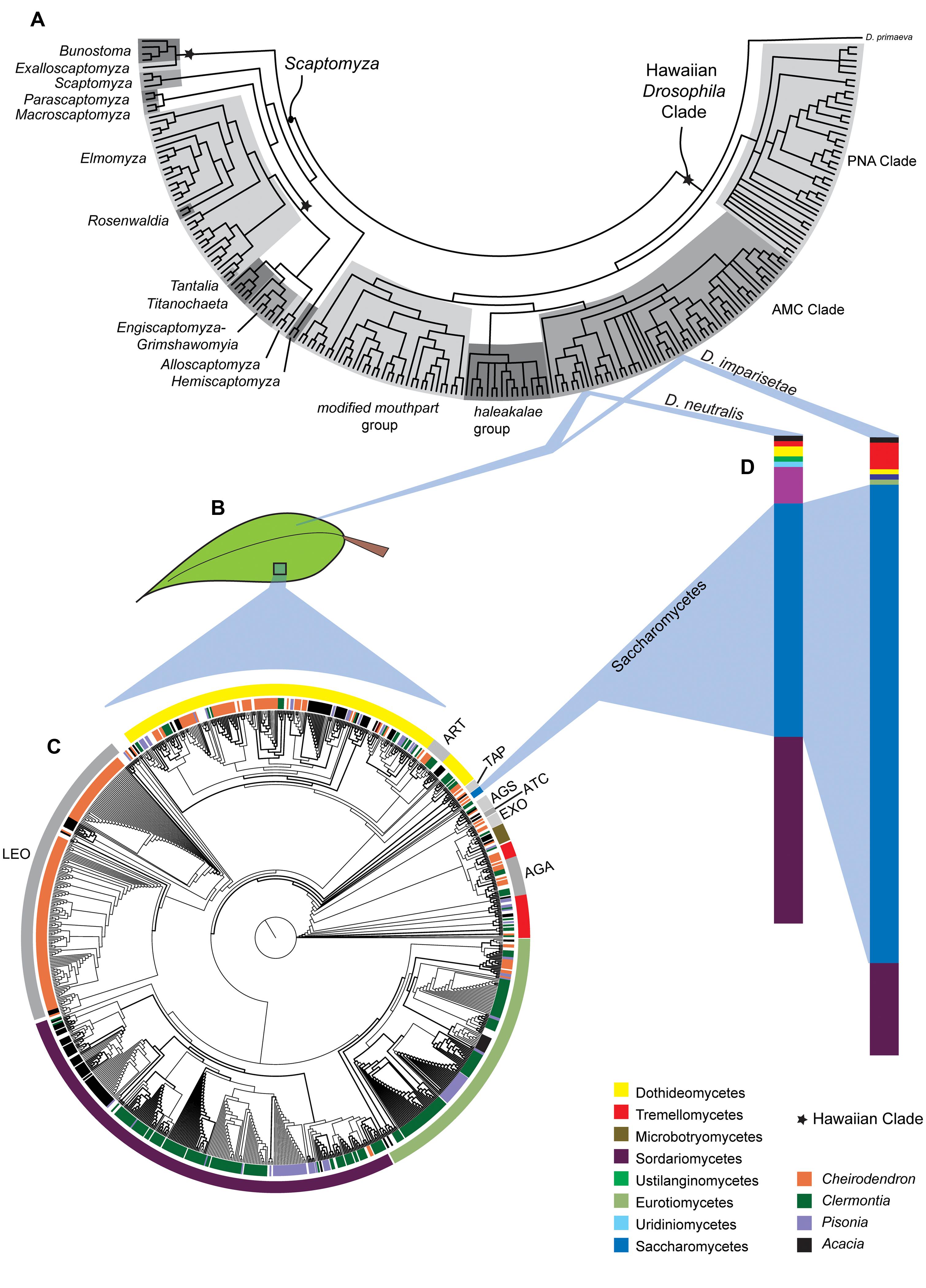
FIGURE 1. Ecological and evolutionary relationships between Hawaiian Drosophilidae, their endemic host plants and fungal decomposers. (A) Phylogenetic relationships among the Hawaiian Drosophila and Scaptomyza. (B) Two species in the AMC Clade, Drosophila imparisetae and Drosophila neutralis, both oviposit in rotting leaves of Araliaceae (Cheirodendron). (C) The phylogeny of the fungal taxa present in decomposing leaves from Cheirodendron (and other Hawaiian plant taxa) shows a diverse fungal community. The inner circle indicates the plant of origin, while the outer circle indicates fungal taxonomy. (D) The relative dearth of Saccharomycetes representatives in leaf communities is interesting given the prevalence, this fungal class in and on the bodies of the adult Drosophila species sampled. Aspects of this figure have been modified with permission from O’Grady et al. (2011),Ort et al. (2012), and Lapoint et al. (2013).
Ort et al. (2012) surveyed four endemic Hawaiian plants (Figure 1B) to determine whether microbial communities played a role in host plant specificity in Hawaiian Drosophila. Over 160 OTUs, representing 113 genera and 13 fungal classes, were discovered (Figure 1C). Ort et al. (2012) found little sharing of fungal taxa between different substrates, and fungal communities differed significantly between substrate type (e.g., leaves vs. stems) and among plant genera. It is clear that different substrates support correspondingly distinct fungal communities, which may provide unique oviposition cues or nutrition to flies that use those substrates.
Relative to their host plants, the fungal communities of the two Drosophila species examined, Drosophila imparisetae and Drosophila neutralis, were relatively simple: only seven or eight fungal lineages were present (Figure 1D). This suggests that Drosophila vector a limited number of fungal species from plant to plant. Interestingly, the most abundant fungal class associated with Drosophila adults, Saccharomycetes, was only modestly represented in the Cheirodendron leaf samples (Figure 1C), suggesting that Hawaiian flies select and vector their own yeasts from rotting plant to rotting plant, as in the cactophilic Drosophila (Barker and Starmer, 1982).
Drosophila-associated microbes may contribute to reproductive isolation of closely related species, which is critical to sustaining adaptive radiations. In the cactophilic species Drosophila buzzatii, heritable variation in oviposition behavior is mediated by attraction to different yeasts (Barker et al., 1994), which might contribute to assortative mating among genotypes. Isolation among races and species of cactophilic Drosophila species may have evolved due to chemical variation in larval substrates that are a combination of necrotic host plant tissues and microbial communities (Etges and de Oliveira, 2014). Combined with evidence that bacterial communities play a direct role in mating preference in Drosophila melanogaster (Sharon et al., 2010), this suggests that microbes can directly or indirectly influence speciation of drosophilid flies through mechanisms that are dependent on larval feeding substrates. Ohta (1980) provides strong evidence that post-mating barriers involving large chromosomal inversion in Hawaiian Drosophila can be explained by variation in host plant use. Thus, the combined effects of host plant phenotype and microbial community phenotypes have likely played an important role in driving the diversification of Hawaiian Drosophilidae.
Scaptomyza and Associated Bacteria
Most drosophilid flies likely feed on microbes associated with rotting vegetation or on the fruiting bodies of fungi. However, in a few lineages (including Scaptomyza), feeding on living plant tissues as a primary source of nutrition (herbivory) has evolved. This transition to herbivory is a remarkable feature of the Hawaiian Drosophilidae radiation, the challenges of which are underscored by the paucity of insect orders with herbivorous members (Mitter et al., 1988). Given the change in the nature of the relationship between Scaptomyza and its food source, the nature of its relationship to its microbial communities is expected to also change. It is likely that herbivorous drosophilid lineages encounter distinct groups of plant-associated microbes that may influence fly adaptation to these larval substrates.
Leaf-mining Scaptomyza larvae are extensively colonized by plant-associated bacteria during feeding. Gut bacterial composition of larval Scaptomyza flava collected from leaves of wild Barbarea vulgaris (Brassicaceae) resembled that of their host plant more than any other drosophilid: 99.7% of gut bacterial sequences matched OTUs found in Barbarea vulgaris (Figure 2A). Pseudomonadaceae predominated in Scaptomyza flava guts and Barbarea vulgaris leaves (Figure 2B). Also found in both samples were Enterobacter cloacae, which includes some strains that degrade isothiocyanates (Tang et al., 1972), a group of potent foliar toxins in the Brassicaceae that are released upon plant wounding. Although untested, Enterobacter cloacae or other bacteria may supplement the endogenous isothiocyanate detoxification abilities of Scaptomyza, which involve modification of ancient evolutionarily conserved detoxification pathways (Gloss et al., 2014).
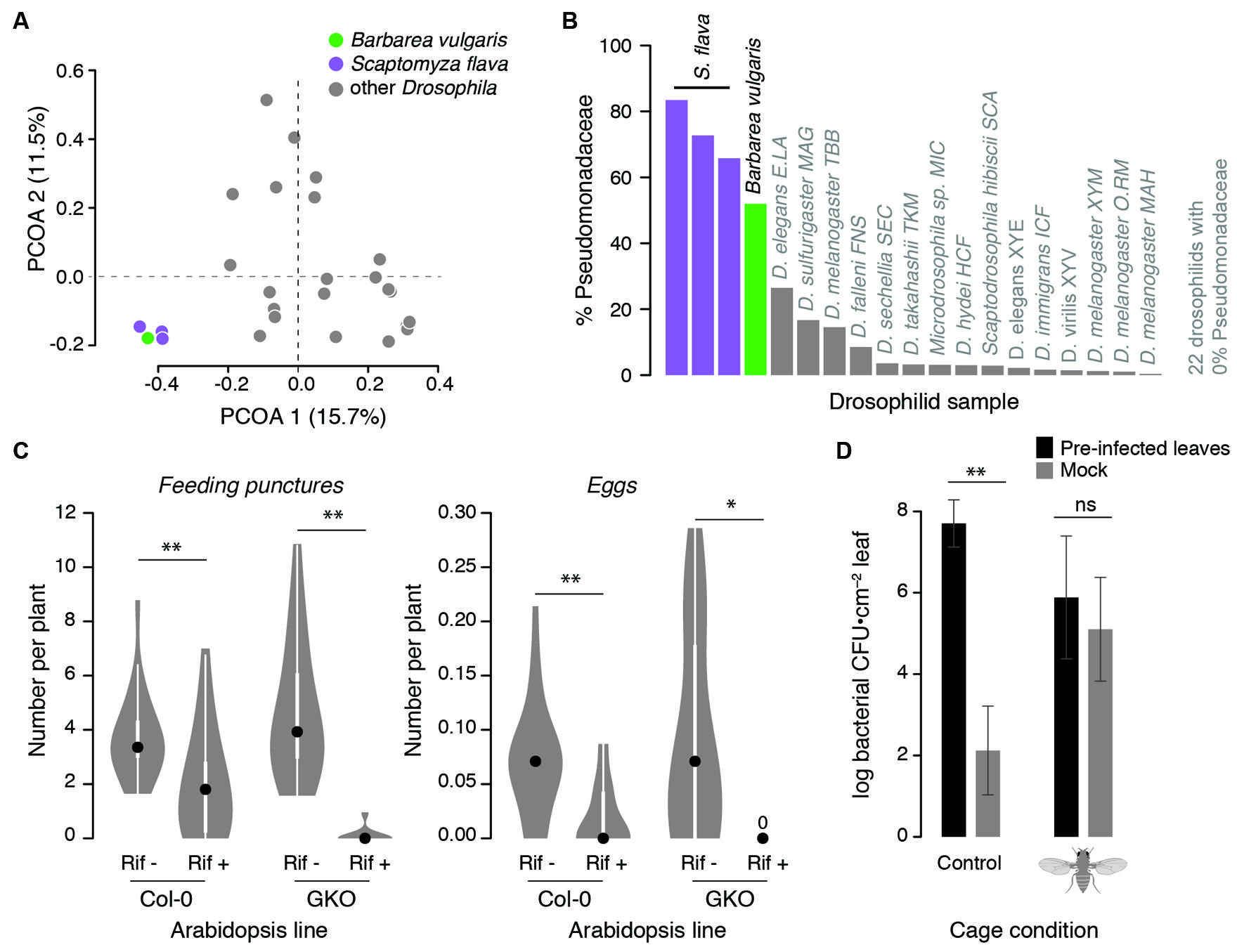
FIGURE 2. Observational and experimental evidence linking Scaptomyza ecology with microbes. (A,B) The gut bacterial community of Scaptomyza flava larvae more closely resembles that of a host plant (Barbarea vulgaris) than that of other drosophilids. Field-collected Scaptomyza flava and Barbarea vulgaris microbial communities were characterized with Illumina sequencing (according to Caporaso et al., 2012) and compared to other Drosophila communities sequenced by Chandler et al. (2011). (A) Principal coordinate analysis (PCOA) of unweighted UniFrac distances summarizing differences in gut bacterial community composition among species. Gut bacteria of Scaptomyza flava are significantly different from those of other drosophilids (PERMANOVA, P < 0.001). (B) The relative abundance of bacteria classified to Pseudomonadaceae is greater in Scaptomyza flava than other drosophilids and comparable to levels found in Barbarea vulgaris. (C) Pre-treating Scaptomyza flava with antibiotics reduces feeding and fecundity on Arabidopsis thaliana. Lab-reared flies were fed for 4 days on 5% sucrose with (Rif+) or without (Rif–) 50 μg/mL rifampicin (Sigma). Treatments were delivered into feeding chambers with 5 μL microcapillary tubes that were refreshed daily. After 4 days, all surviving flies were randomized to cages with plants of either line (wild-type Col-0 or glucosinolate knock-out GKO) and were allowed to feed and oviposit for 24 h, after which feeding puncture and eggs on each plant were counted. The number of feeding punctures and eggs per plant were normalized by number of flies released into each cage (each Rif– condition had 14 females; each Rif+ condition had 23 females). Data are presented as boxplots with 50% quantiles around the medians (dots) and symmetrical marginal frequency distributions. **P < 0.01, *P < 0.05 (Mann–Whitney U-test, two-tailed). (D) Scaptomyza flava females enhance transmission of Pseudomonas syringae between A. thaliana leaves in the laboratory; Pseudomonas syringae grew in un-treated leaves only when flies were present. Three lower leaves of 5 weeks old A. thaliana Col-0 were infected with 105/ml Pseudomonas syrinage pv. maculicola str. 4326. Four days later, leaves were removed and petioles were inserted into 60 mm petri dishes containing 1% Phytagel to maintain leaf hydration. Equal numbers of infected or un-infected leaves were randomized into one of two mesh cages. Into one of these cages, we released 20 adult female Scaptomyza flava for 2 days after which feeding punctures were counted on all leaves. Leaf discs were taken and homogenized in 10 mM MgSO4 and dilution-plated onto King’s B medium, and fluorescent colonies were counted 4 days later. Error bars indicate standard errors; **P < 0.01, ‘ns’ non-significant, unpaired t-tests on log10-transformed CFU counts.
One way herbivorous Scaptomyza may have adapted to feeding on living plant tissue is by acquiring novel microbial symbionts that aid in substrate utilization, for instance by catabolizing polysaccharides or plant secondary compounds. Bacteria that can metabolize plant-derived molecules in ways that contribute to insect fitness are most likely to be found already associated with the novel substrate (reviewed in Janson et al., 2008; Mason et al., 2014). Indeed, recent work suggests Scaptomyza flava depends upon their gut bacteria for fitness within plants. Laboratory-reared Scaptomyza flava treated with the antibiotic rifampicin showed reduced feeding rates and lower fecundity on Arabidopsis thaliana (Arabidopsis) compared to control flies (Figure 2C). This effect does not appear to be due to direct antibiotic toxicity, because treatment and control flies survived at similar rates, nor interactions with plant defensive compounds. This is because results were consistent when flies were reared on wild-type Arabidopsis as well as a mutant line deficient in the production of two defensive compounds. Although particular bacteria have not been implicated, this experiment indicates that gut bacterial communities may be important for degrading or processing plant tissues.
Diffuse interactions between insects, bacteria, and plants may also be involved in the evolution and maintenance of herbivory. For instance, insects can vector plant-associated bacteria between plants, including common pathogens, to make those plants more suitable hosts. Drosophila melanogaster can serve as both host and vector to the plant pathogen Erwinia carotovora (Basset et al., 2000, 2003), and several insects vector Pseudomonas syringae between plants (Snyder et al., 1998; Stavrinides et al., 2009).
Scaptomyza flava adult female flies can enhance the transmission of a model pathogenic strain of Pseudomonas syringae (Figure 2D); Pseudomonas syringae moved from pre-inoculated Arabidopsis leaves to un-treated control leaves in cages with adult female flies added, while control leaves only showed background levels of non-Pseudomonas syringae bacteria in cages without flies (Figure 2D). Not only are Pseudomonas spp. widespread within tissues of Scaptomyza flava and its host plant, but Pseudomonas syringae are overrepresented within plant leaves damaged by a close relative, Scaptomyza nigrita, in the wild (Humphrey et al., 2014). Furthermore, experimental plant infection with Pseudomonas species can enhance feeding by adults of the specialist Scaptomyza nigrita in the laboratory, indicating that plant exposure to certain Pseudomonas spp. can induce susceptibility to this herbivore (Humphrey et al., 2014). Thus, the potential exists for drosophilid herbivores to frequently encounter and transmit Pseudomonas species between host plants in ways that enhance insect fitness. Frequent exposure to, and transmission of, defense-altering microbes can ultimately lead to novel and potentially mutualistic interactions between microbes and insects (Luan et al., 2012).
Inoculating plants with bacteria may allow Scaptomyza species to subvert anti-herbivore defense by exploiting mutual antagonism between plant defense pathways, including the canonical plant defense hormones salicylic acid (SA) and jasmonic acid (JA). The SA pathway, typically triggered by bacterial infections, represses the JA pathway, which is typically triggered by chewing herbivores (Thaler et al., 2012). Bacterial infection can thus alter plant chemistry in ways that also affect herbivores, including via mechanisms independent of SA–JA antagonism (Cui et al., 2002, 2005; Groen et al., 2013). The Colorado potato beetle was recently shown to locally disable plant defenses by secreting bacteria—including Pseudomonas syringae—into plant tissues while feeding (Chung et al., 2013), likely via SA–JA antagonism. Actively suppressing host plant defenses with bacteria may be a common behavior among herbivorous insects, including Scaptomyza.
Plant interactions with leaf-colonizing bacteria are ubiquitous, and the phenotypic impacts of bacteria on plants likely have always been a part of the context in which insect herbivory evolves. Indirect interactions are numerous in diverse ecological communities, and can impact selection on focal herbivore traits such as feeding preference both within (Utsumi et al., 2012) or between plant host individuals (Tack et al., 2012). Specifically, herbivores within the Hawaiian Drosophilidae hold great potential to shed light on the role of plant-mediated indirect effects in the ecology and evolution of herbivore traits given that the focal players are themselves—or are related to—genetic model organisms (Whiteman et al., 2011).
Host Shifts and Symbioses
Plant defenses, especially secondary compounds, are key obstacles to host plant switching for herbivorous insects (Ehrlich and Raven, 1964). The tendency of herbivorous insect lineages to specialize upon plants with similar secondary chemistry suggests that mechanisms to overcome these defenses may be difficult for already-specialized insects to evolve (Berenbaum et al., 1989). However, many microbes are known to degrade plant secondary compounds and structurally similar chemicals (Winkelmann, 1992). Freeland and Janzen (1974) hypothesized that mammalian herbivores might rely on gut bacteria to detoxify secondary compounds of novel plants during host shifts, and a similar bacterial role has been suggested for insects (Broderick et al., 2004; Dillon and Dillon, 2004; Janson et al., 2008). External microbial associates, including yeasts, also have great detoxifying potential for saprophagous species that must also contend with plant secondary chemistry. Because associations with symbiotic microbes are likely to be more evolutionarily labile than endogenous detoxification mechanisms, symbiosis might facilitate colonization of and adaptation to novel plant substrates.
The detoxification abilities of Drosophila-associated yeasts have been extensively demonstrated in the cactophilic Drosophila. Diplodocus and Pichia species found on Stenocereus thurberi cacti hydrolyze plant lipids that inhibit both larval fly and yeast growth (Starmer, 1982) while Candida and Cryptococcus species consume byproducts of cactus fermentation (2-propanol and acetone), which are toxic to Drosophila mojavensis (Starmer et al., 1986). Fungi associated with other insects degrade a wide variety of common plant secondary compounds, including tannins, terpenes, chlorinated hydrocarbons, and phenolics, among others (Dowd, 1992). The Hawaiian Drosophila utilize a chemically diverse collection of host plants and may similarly benefit from the detoxifying activity of yeasts.
Examples of detoxification by insect gut bacteria are limited, but several recent reports suggest this phenomenon may be more widespread than is currently appreciated. Terpene-degrading bacteria are associated with pine bark beetles (Dendoctronus ponderosae) that mine galleries in terpene-rich subcortical tissues of pines (Adams et al., 2013; Boone et al., 2013), and gypsy moths (Lymantria dispar) rely upon plant-derived bacteria to supplement endogenous detoxification of phenolic glycosides (Mason et al., 2014). These results suggest that environmentally acquired bacteria can be important contributors to insect fitness by directly detoxifying plant compounds, which may facilitate invasion of novel niches.
Plant-associated bacteria may also contribute to insect detoxification capacities via horizontal gene transfer from ingested bacteria to gut residents. Such a transfer has been described in humans, where the bacterium Bacteroides plebeius from Japanese populations apparently acquired genes from marine Bacteroidetes that degrade seaweed polysaccharides (Hehemann et al., 2010). Like human guts, insect guts are hotspots of horizontal gene transfer (reviewed in Dillon and Dillon, 2004). Conjugative plasmids are shared promiscuously within the guts of silkworm larvae (Watanabe et al., 1998), although gut conditions may not be conducive to natural transformation (Ray et al., 2007). Some pathways for plant secondary compound detoxification are encoded in small genomic regions that might facilitate their transfer, such as genes in the pyrrolidine pathway responsible for nicotine catabolism in plant-associated Pseudomonas putida (Tang et al., 2013). Whether gene transfer from environmental bacteria to Drosophila gut bacteria is common is not yet known.
Concluding Remarks
Understanding the factors that generated and maintain the staggering diversity of the Hawaiian Drosophilidae has informed general hypotheses of how other organisms diversify. The diversity of these flies appears to be due to many different factors including geography, mating behaviors, and ecology. We propose that the interaction between the Hawaiian Drosophilidae, their host plants, and host-associated microbes is another important aspect driving the diversification of the Hawaiian Drosophilidae and/or maintaining this diversity. The role of microbial associates in nearly every aspect of drosophilid ecology—development, nutrition, host finding, and reproduction—presents many opportunities for those microbes to influence diversification. Experimental data from other Drosophila species suggest that bacteria, yeast and other fungi may allow host shifts and even trophic shifts, as well as instigate major changes in mating behaviors that subdivide populations in the Hawaiian Drosophilidae. Because yeasts are critical to host finding and substrate processing, Hawaiian Drosophila may not be radiating on host plants directly, but instead on fungal diversity. Additional studies are poised to expose the importance of these interactions.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Support for this research was provided by the National Science Foundation (DEB-662 1256758 to Noah K. Whiteman; DDIG DEB-1309493 to Parris T. Humphrey; GRF and IGERT to Timothy K. O’Connor), the John Templeton Foundation (Grant 664 ID #41855 to Noah K. Whiteman), National Institutes of Health (K12-631 GM000708 PERT to Richard T. Lapoint) and the National Geographic Society (9097-12 to Noah K. Whiteman).
References
Adams, A. S., Aylward, F. O., Adams, S. M., Erbilgin, N., Aukema, B. H., Currie, C. R.,et al. (2013). Mountain pine beetles colonizing historical and naive host trees are associated with a bacterial community highly enriched in genes contributing to terpene metabolism. Appl. Environ. Microbiol. 79, 3468–3475. doi: 10.1128/AEM.00068-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Baker, R. H., and DeSalle, R. (1997). Multiple sources of character information and the phylogeny of Hawaiian drosophilids. Syst. Biol. 46, 654–673. doi: 10.1093/sysbio/46.4.654
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Barker, J. S. F., and Starmer, W. T. (1982). Ecological Genetics and Evolution: the Cactus-Yeast-Drosophila Model System. New York: Academic Press.
Barker, J., Starmer, W. T., and Fogleman, J. C. (1994). Genotype-specific habitat selection for oviposition sites in the cactophilic species Drosophila buzzatii. Heredity (Edinb.) 72, 384–395. doi: 10.1038/hdy.1994.55
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Basset, A., Khush, R. S., Braun, A., Gardan, L., Boccard, F., Hoffmann, J. A.,et al. (2000). The phytopathogenic bacteria Erwinia carotovora infects Drosophila and activates an immune response. Proc. Natl. Acad. Sci. U.S.A. 97, 3376–3381. doi: 10.1073/pnas.97.7.3376
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Basset, A., Tzou, P., Lemaitre, B., and Boccard, F. (2003). A single gene that promotes interaction of a phytopathogenic bacterium with its insect vector, Drosophila melanogaster. EMBO Rep. 4, 205–209. doi: 10.1038/sj.embor.embor730
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Berenbaum, M. R., Zangerl, A. R., and Lee, K. (1989). Chemical barriers to adaptation by a specialist herbivore. Oecologia 80, 501–506. doi: 10.1007/BF00380073
Bonacum, J. (2001). Molecular Systematics of the Hawaiian Drosophilidae. Ph.D. thesis, Yale University, New Haven, CT.
Boone, C. K., Keefover-Ring, K., Mapes, A. C., Adams, A. S., Bohlmann, J., and Raffa, K. F. (2013). Bacteria associated with a tree-killing insect reduce concentrations of plant defense compounds. J. Chem. Ecol. 39, 1003–1006. doi: 10.1007/s10886-013-0313-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bradford, M. D., and Schwab, E. (2013). Current usage of symbiosis and associated terminology. Int. J. Biol. 5, 32–45. doi: 10.5539/ijb.v5n1p32
Broderick, N. A., Raffa, K. F., Goodman, R. M., and Handelsman, J. (2004). Census of the bacterial community of the gypsy moth larval midgut by using culturing and culture-independent methods. Appl. Environ. Microbiol. 70, 293–300. doi: 10.1128/AEM.70.1.293-300.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brucker, R. M., and Bordenstein, S. R. (2012). The roles of host evolutionary relationships (genus: Nasonia) and development in structuring microbial communities. Evolution 66, 349–362. doi: 10.1111/j.1558-5646.2011.01454.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brucker, R. M., and Bordenstein, S. R. (2013). The hologenomic basis of speciation: gut bacteria cause hybrid lethality in the genus Nasonia. Science 341, 667–669. doi: 10.1126/science.1240659
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Huntley, J., Fierer, N.,et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. doi: 10.1038/ismej.2012.8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Carson, H. L., and Kaneshiro, K. Y. (1976). Drosophila of Hawaii: systematics and ecological genetics. Annu. Rev. Ecol. Syst. 7, 311–345. doi: 10.1146/annurev.es.07.110176.001523
Chandler, J. A., Lang, J. M., Bhatnagar, S., Eisen, J. A., and Kopp, A. (2011). Bacterial communities of diverse Drosophila species: ecological context of a host–microbe model system. PLoS Genet. 7:e1002272. doi: 10.1371/journal.pgen.1002272
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chung, S. H., Rosa, C., Scully, E. D., Peiffer, M., Tooker, J. F., Hoover, K.,et al. (2013). Herbivore exploits orally secreted bacteria to suppress plant defenses. Proc. Natl. Acad. Sci. U.S.A. 110, 15728–15733. doi: 10.1073/pnas.1308867110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Coluccio, A. E., Rodriguez, R. K., Kernan, M. J., and Neiman, A. M. (2008). The yeast spore wall enables spores to survive passage through the digestive tract of Drosophila. PLoS ONE 3:e2873. doi: 10.1371/journal.pone.0002873
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cui, J., Bahrami, A. K., Pringle, E. G., Hernandez-Guzman, G., Bender, C. L., Pierce, N. E.,et al. (2005). Pseudomonas syringae manipulates systemic plant defenses against pathogens and herbivores. Proc. Natl. Acad. Sci. U.S.A. 102, 1791–1796. doi: 10.1073/pnas.0409450102
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cui, J., Jander, G., Racki, L. R., Kim, P. D., Pierce, N. E., and Ausubel, F. M. (2002). Signals involved in Arabidopsis resistance to Trichoplusia ni caterpillars induced by virulent and avirulent strains of the phytopathogen Pseudomonas syringae. Plant Physiol. 129, 551–564. doi: 10.1104/pp.010815
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Currie, C. R., Bot, A. N., and Boomsma, J. J. (2003). Experimental evidence of a tripartite mutualism: bacteria protect ant fungus gardens from specialized parasites. Oikos 101, 91–102. doi: 10.1034/j.1600-0706.2003.12036.x
Currie, C. R., Poulsen, M., Mendenhall, J., Boomsma, J. J., and Billen, J. (2006). Coevolved crypts and exocrine glands support mutualistic bacteria in fungus-growing ants. Science 311, 81–83. doi: 10.1126/science.1119744
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dillon, R., and Dillon, V. (2004). The gut bacteria of insects: nonpathogenic interactions. Annu. Rev. Entomol. 49, 71–92. doi: 10.1146/annurev.ento.49.061802.123416
Dobzhansky, T., Cooper, D., Phaff, H., Knapp, E., and Carson, H. (1956). Differential attraction of species of Drosophila to different species of yeasts. Ecology 37, 544–550. doi: 10.2307/1930178
Dowd, P. F. (1992). Insect fungal symbionts: a promising source of detoxifying enzymes. J. Ind. Microbiol. 9, 149–161. doi: 10.1007/BF01569619
Ehrlich, P., and Raven, P. (1964). Butterflies and plants – a study in coevolution. Evolution 18, 586–608. doi: 10.2307/2406212
Etges, W. J., and de Oliveira, C. C. (2014). Premating isolation is determined by larval rearing substrates in cactophilic Drosophila mojavensis. X. Age-specific dynamics of adult epicuticular hydrocarbon expression in response to different host plants. Ecol. Evol. 4, 2033–2045. doi: 10.1002/ece3.1088
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Feldhaar, H. (2011). Bacterial symbionts as mediators of ecologically important traits of insect hosts. Ecol. Entomol. 36, 533–543. doi: 10.1111/j.1365-2311.2011.01318.x
Fogleman, J. C., and Foster, J. L. (1989). Microbial colonization of injured cactus tissue (Stenocereus gummosus) and its relationship to the ecology of cactophilic Drosophila mojavensis. Appl. Environ. Microbiol. 55, 100–105.
Freeland, W. J., and Janzen, D. H. (1974). Strategies in herbivory by mammals: the role of plant secondary compounds. Am. Nat. 108, 269–289. doi: 10.1086/282907
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ganter, P. F. (1988). The vectoring of cactophilic yeasts by Drosophila. Oecologia 75, 400–404. doi: 10.1007/BF00376943
Gilbert, D. G. (1980). Dispersal of yeasts and bacteria by Drosophila in a temperate forest. Oecologia 46, 135–137. doi: 10.1007/BF00346979
Gloss, A. D., Giddings-Vassao, D., Schramm, K., Reichelt, M., Hailey, A., Rast, T.,et al. (2014). Evolution in an ancient detoxification pathway is coupled with a transition to herbivory in the Drosophilidae. Mol. Biol. Evol. 31, 2441–2456. doi: 10.1093/molbev/msu201
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Groen, S. C., Whiteman, N. K., Bahrami, A. K., Wilczek, A. M., Cui, J., Russell, J. A.,et al. (2013). Pathogen-triggered ethylene signaling mediates systemic-induced susceptibility to herbivory in Arabidopsis. Plant Cell 25, 4755–4766. doi: 10.1105/tpc.113.113415
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hansen, A. K., and Moran, N. A. (2014). The impact of microbial symbionts on host plant utilization by herbivorous insects. Mol. Ecol. 23, 1473–1496. doi: 10.1111/mec.12421
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hehemann, J. H., Correc, G., Barbeyron, T., Helbert, W., Czjzek, M., and Michel, G. (2010). Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature 464, 908–912. doi: 10.1038/nature08937
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Humphrey, P. T., Nguyen, T. T., Villalobos, M. M., and Whiteman, N. K. (2014). Diversity and abundance of phyllosphere bacteria are linked to insect herbivory. Mol. Ecol. 23, 1497–1515. doi: 10.1111/mec.12657
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jaenike, J., Unckless, R., Cockburn, S. N., Boelio, L. M., and Perlman, S. J. (2010). Adaptation via symbiosis: recent spread of a Drosophila defensive symbiont. Science 329, 212–215. doi: 10.1126/science.1188235
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Janson, E. M., Stireman, J. O., Singer, M. S., and Abbot, P. (2008). Phytophagous insect–microbe mutualisms and adaptive evolutionary diversification. Evolution 62, 997–1012. doi: 10.1111/j.1558-5646.2008.00348.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Joy, J. B. (2013). Symbiosis catalyses niche expansion and diversification. Proc. Bial. Sci. 280, 20122820. doi: 10.1098/rspb.2012.2820
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kambysellis, M. P., and Craddock, E. M. (1997). “Ecological and reproductive shifts in the diversification of the endemic Hawaiian Drosophila,” in Molecular Evolution and Adaptive Radiation, eds T. J. Givnish and K. J. Sytsma (Cambridge: Cambridge University Press), 475–509.
Lapoint, R. T., O’Grady, P. M., and Whiteman, N. K. (2013). Diversification and dispersal of the Hawaiian Drosophilidae: the evolution of Scaptomyza. Mol. Phylogenet. Evol. 69, 95–108. doi: 10.1016/j.ympev.2013.04.032.
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Luan, J.-B., Yao, D.-M., Zhang, T., Walling, L. L., Yang, M., Wang, Y.-J.,et al. (2012). Suppression of terpenoid synthesis in plants by a virus promotes its mutualism with vectors. Ecol. Lett. 16, 390–398. doi: 10.1111/ele.12055
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Magnacca, K. N., Foote, D., and O’Grady, P. M. (2008). A review of the endemic Hawaiian drosophilidae and their host plants. Zootaxa 1728, 1–58.
Magnacca, K. N., and Price, D. K. (2012). New species of Hawaiian picture wing Drosophila (Diptera: Drosophilidae), with a key to species. Zootaxa 3188, 1–30.
Markow, T. A., and O’Grady, P. (2005). Drosophila: A Guide to Species Identification and Use. Amsterdam: Academic Press.
Markow, T. A., and O’Grady, P. (2008). Reproductive ecology of Drosophila. Funct. Ecol. 22, 747–759. doi: 10.1111/j.1365-2435.2008.01457.x
Mason, C. J., Couture, J. J., and Raffa, K. F. (2014). Plant-associated bacteria degrade defense chemicals and reduce their adverse effects on an insect defoliator. Oecologia 175, 901–910. doi: 10.1007/s00442-014-2950-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Melcher, C., and Pankratz, M. J. (2005). Candidate gustatory interneurons modulating feeding behavior in the Drosophila brain. PLoS Biol. 3:e305. doi: 10.1371/journal.pbio.0030305
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mikheyev, A., Mueller, U. G., and Boomsma, J. J. (2007). Population genetic signatures of diffuse co-evolution between leaf-cutting ants and their cultivar fungi. Mol. Ecol. 16, 209–216. doi: 10.1111/j.1365-294X.2006.03134.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mitter, C., Farrell, B. D., and Wiegmann, B. (1988). The phylogenetic study of adaptive zones: has phytophagy promoted insect diversification? Am. Nat. 132, 107–128. doi: 10.1086/284840
Moran, N. A., and Baumann, P. (2000). Bacterial endosymbionts in animals. Curr. Opin. Microbiol. 3, 270–275. doi: 10.1016/S1369-5274(00)00088-6
Northrop, H. (1918). The role of yeast in the nutrition of an insect (Drosophila). Studies from the Rockefeller Institute for Medical Research. Reprints 28:281.
Obbard, D. J., Maclennan, J., Kim, K., Rambaut, A., O’Grady, P. M., and Jiggins, F. M. (2012). Estimating divergence dates and substitution rates in the Drosophila phylogeny. Mol. Biol. Evol. 20, 3459–3473. doi: 10.1093/molbev/mss150
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
O’Grady, P. M. (2010). Whither Drosophila? Genetics 185, 703–705. doi: 10.1534/genetics.110.118232
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
O’Grady, P. M., Ashburner, M., Castrezana, S., DeSalle, R., Kaneshiro, K. Y., Lapoint, R. T.,et al. (2008a). Comment on the proposed conservation of usage of Drosophila fallen, 1823 (Insecta, Diptera). Bull. Zool. Nomencl. 65, 141–144.
O’Grady, P. M., Lapoint, R. T., and Bennett, G. M. (2008b). The potential and peril of the supertree approach: a response to van der Linde and Houle. Insect Syst. Evol. 39, 269–280. doi: 10.1163/187631208788784219
O’Grady, P. M., and DeSalle, R. (2008). Out of Hawaii: the biogeographic history of the genus Scaptomyza (Diptera: Drosophilidae). Biol. Lett. 4, 195–199. doi: 10.1098/rsbl.2007.0575
O’Grady, P. M., Lapoint, R. T., Bonacum, J., Lasola, J., Owen, E., Wu, Y.,et al. (2011). Phylogenetic and ecological relationships of the Hawaiian Drosophila inferred by mitochondrial DNA analysis. Mol. Phylogenet. Evol. 58, 244–256. doi: 10.1016/j.ympev.2010.11.022
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
O’Grady, P. M., and Markow, T. A. (2009). Phylogenetic taxonomy in Drosophila: problems and prospects. Fly 3, 1–5. doi: 10.4161/fly.3.1.7748
Ohta, A. T. (1978). Ethological isolation and phylogeny in the grimshawi species complex of Hawaiian Drosophila. Evolution 32, 485–492. doi: 10.2307/2407716
Ohta, A. T. (1980). Coadaptive gene complexes in incipient species of Hawaiian Drosophila. Am. Nat. 115, 121–132. doi: 10.1086/283549
Oliver, K. M., Degnan, P. H., Hunter, M. S., and Moran, N. A. (2009). Bacteriophages encode factors required for protection in a symbiotic mutualism. Science 325, 992–994. doi: 10.1126/science.1174463
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ort, B. S., Bantay, R. M., Pantoja, N. A., and O’Grady, P. M. (2012). Fungal diversity associated with Hawaiian Drosophila host plants. PLoS ONE 7:e40550. doi: 10.1371/journal.pone.0040550
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Price, J. P., and Clague, D. A. (2002). How old is the Hawaiian biota? Geology and phylogeny suggest recent divergence. Proc. R. Soc. B 269, 2429–2435. doi: 10.1098/rspb.2002.2175
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ray, J. L., Anderson, H. K., Young, S., Nielsen, K. M., and O’Callaghan, M. (2007). An assessment of the potential of herbivorous insect gut bacteria to develop competence for natural transformation. Environ. Biosafety Res. 6, 135–147. doi: 10.1111/j.1439-0418.2009.01458.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Remsen, J., and DeSalle, R. (1998). Character congruence of multiple data partitions and the origin of the Hawaiian Drosophilidae. Mol. Phylogenet. Evol. 9, 225–235. doi: 10.1006/mpev.1997.0484
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Remsen, J., and O’Grady, P. M. (2002). Phylogeny of Drosophilidae (Diptera), with comments on combined analysis and character support. Mol. Phylogenet. Evol. 24, 248–263. doi: 10.1016/S1055-7903(02)00226-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Russo, C. A. M., Takezaki, N., and Nei, M. (1995). Molecular phylogeny and divergence times of drosophilid species. Mol. Biol. Evol. 12, 391–404.
Sharon, G., Segal, D., Ringo, J. M., Hefetz, A., Zilber-Rosenberg, I., and Rosenberg, E. (2010). Commensal bacteria play a role in mating preference of Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 107, 20051–20056. doi: 10.1073/pnas.1009906107
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Snyder, W. E., Tonkyn, D. W., and Kluepfel, D. A. (1998). Insect-mediated dispersal of the rhizobacterium Pseudomonas chlororaphis. Phytopathology 88, 1248–1254. doi: 10.1094/PHYTO.1998.88.12.1248
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Starmer, W. T. (1982). “Associations and interactions among yeasts, Drosophila, and their habitats,” in Ecological Genetics and Evolution: The Cactus–Yeast–Drosophila Model System, eds J. S. F. Barker and W. T. Starmer (Sydney: Academic Press), 159–174.
Starmer, W. T., Barker, J. S. F., Phaff, H. J., and Fogleman, J. C. (1986). Adaptations of Drosophila and yeasts: their interactions with the volatile 2-propanol in the cactus-microorganism-Drosophila model system. Aust. J. Biol. Sci. 39, 69–77.
Starmer, W. T., and Aberdeen, V. (1990). “The nutritional importance of pure and mixed cultures of yeasts in the development of Drosophila mulleri larvae in opuntia tissues and its relationship to host plant shifts,” in Ecological and Evolutionary Genetics of Drosophila, eds J. S. F. Barker, W. T. Starmer, and R. J. MacIntyre (New York: Springer), 145–160.
Stavrinides, J., McCloskey, J. K., and Ochman, H. (2009). Pea aphid as both host and vector for the phytopathogenic bacterium Pseudomonas syringae. Appl. Environ. Microbiol. 75, 2230–2235. doi: 10.1128/AEM.02860-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tack, A. J. M., Gripenberg, S., and Roslin, T. (2012). Cross-kingdom interactions matter: fungal mediated interactions structure an insect community on oak. Ecol. Lett. 15, 177–185. doi: 10.1111/j.1461-0248.2011.01724.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tamura, K., Subramanian, S., and Kumar, S. (2004). Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol. Biol. Evol. 21, 36–44. doi: 10.1093/molbev/msg236
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tang, C., Bhothipaksa, K., and Frank, H. A. (1972). Bacterial degradation of benzyl isothiocyanate. Appl. Microbiol. 23, 1145–1148.
Tang, H., Wang, L., Wang, W., Yu, H., Zhang, K., Yao, Y.,et al. (2013). Systematic unraveling of the unsolved pathway of nicotine degradation in Pseudomonas. PLoS Genet. 9:e1003923. doi: 10.1371/journal.pgen.1003923
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Thaler, J. S., Humphrey, P. T., and Whiteman, N. K. (2012). Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci. 17, 260–270. doi: 10.1016/j.tplants.2012.02.010
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Thomas, R. H., and Hunt, J. A. (1991). The molecular evolution of the alcohol dehydrogenase locus and the phylogeny of Hawaiian Drosophila. Mol. Biol. Evol. 8, 687–702.
Thomas, R. H., and Hunt, J. A. (1993). Phylogenetic relationships in Drosophila: a conflict between molecular and morphological data. Mol. Biol. Evol. 10, 362–374.
Throckmorton, L. H. (1975). “The phylogeny, ecology, and geography of Drosophila,” in Handbook of Genetics, Vol. 3, ed. R. C. King (New York: Plenum), 421–469.
Utsumi, S., Ando, Y., Roininen, H., Takahashi, J.-I., and Ohgushi, T. (2012). Herbivore community promotes trait evolution in a leaf beetle via induced plant response. Ecol. Lett. 16, 362–370. doi: 10.1111/ele.12051
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Watanabe, K., Hara, W., and Sato, M. (1998). Evidence for growth of strains of the plant epiphytic bacterium Erwinia herbicola and transconjugation among the bacterial strains in guts of the silkworm Bombyx mori. J. Invert. Path. 72, 104–111. doi: 10.1006/jipa.1998.4764
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Werren, J. H., Baldo, L., and Clark, M. E. (2008). Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6, 741–751. doi: 10.1038/nrmicro1969
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Whiteman, N. K., Groen, S. C., Chevasco, D., Bear, A., Beckwith, N., Gregory, T. R.,et al. (2011). Mining the plant-herbivore interface with a leafmining Drosophila of Arabidopsis. Mol. Ecol. 20, 995–1014. doi: 10.1111/j.1365-294X.2010.04901.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: Hawaiian Drosophila, Scaptomyza, symbiosis, fungi, Pseudomonas, herbivory, adaptive radiation
Citation: O’Connor TK, Humphrey PT, Lapoint RT, Whiteman NK and O’Grady PM (2014) Microbial interactions and the ecology and evolution of Hawaiian Drosophilidae. Front. Microbiol. 5:616. doi: 10.3389/fmicb.2014.00616
Received: 17 May 2014; Accepted: 29 October 2014;
Published online: 18 December 2014.
Edited by:
M. Pilar Francino, Center for Public Health Research, SpainReviewed by:
Rob DeSalle, American Museum of Natural History, USAJens Walter, University of Nebraska, USA
Copyright © 2014 O’Connor, Humphrey, Lapoint, Whiteman and O’Grady. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Patrick M. O’Grady, Environmental Science, Policy and Management, University of California Berkeley (University, Not-for-profit), 137 Hilgard Hall, Berkeley, CA 94720, USA e-mail:b2dyYWR5QGJlcmtlbGV5LmVkdQ==
† Timothy K. O’Connor and Parris T. Humphrey have contributed equally to this work.
 Timothy K. O’Connor
Timothy K. O’Connor Parris T. Humphrey
Parris T. Humphrey Richard T. Lapoint
Richard T. Lapoint Noah K. Whiteman
Noah K. Whiteman Patrick M. O’Grady
Patrick M. O’Grady