
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 20 August 2014
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 5 - 2014 | https://doi.org/10.3389/fmicb.2014.00426
This article is part of the Research Topic Antibiotic resistance in aquatic systems View all 10 articles
The aim of this study was to examine antibiotic resistance (AR) dissemination in coastal water, considering the contribution of different sources of fecal contamination. Samples were collected in Berlenga, an uninhabited island classified as Natural Reserve and visited by tourists for aquatic recreational activities. To achieve our aim, AR in Escherichia coli isolates from coastal water was compared to AR in isolates from two sources of fecal contamination: human-derived sewage and seagull feces. Isolation of E. coli was done on Chromocult agar. Based on genetic typing 414 strains were established. Distribution of E. coli phylogenetic groups was similar among isolates of all sources. Resistances to streptomycin, tetracycline, cephalothin, and amoxicillin were the most frequent. Higher rates of AR were found among seawater and feces isolates, except for last-line antibiotics used in human medicine. Multi-resistance rates in isolates from sewage and seagull feces (29 and 32%) were lower than in isolates from seawater (39%). Seawater AR profiles were similar to those from seagull feces and differed significantly from sewage AR profiles. Nucleotide sequences matching resistance genes blaTEM, sul1, sul2, tet(A), and tet(B), were present in isolates of all sources. Genes conferring resistance to 3rd generation cephalosporins were detected in seawater (blaCTX-M-1 and blaSHV-12) and seagull feces (blaCMY-2). Plasmid-mediated determinants of resistance to quinolones were found: qnrS1 in all sources and qnrB19 in seawater and seagull feces. Our results show that seawater is a relevant reservoir of AR and that seagulls are an efficient vehicle to spread human-associated bacteria and resistance genes. The E. coli resistome recaptured from Berlenga coastal water was mainly modulated by seagulls-derived fecal pollution. The repertoire of resistance genes covers antibiotics critically important for humans, a potential risk for human health.
Fecal contamination in aquatic environments contributes to the spread of human pathogens along with the dissemination of antibiotic-resistant bacteria. Overexposure to antibiotics is leading to increasing levels of resistance in the human (and other animals) commensal microbiota (Austin et al., 1999). The resistomes of fecal bacteria, once in environmental settings, contribute with antibiotic resistance (AR) genes to non-resistant indigenous microorganisms (Aminov, 2011; Tacão et al., 2014). In aquatic systems the cycle may include subsequent transmission of AR to human-associated bacteria (Devirgiliis et al., 2011; Figueira et al., 2011). At the same time, enrichment in AR bacteria is promoted by the presence of antimicrobials or other contaminants in the environment. This facts reinforce the need to identify the sources of antibiotic-resistant bacteria in aquatic environments of human usage (Rosewarne et al., 2010; Gomez-Alvarez et al., 2012).
Escherichia coli is a natural member of the intestinal microbiota of human and other homeothermic animals and is a fecal indicator bacteria of election (see review by Harwood et al., 2014). Multi-resistant strains are found in environments such as coastal waters. Although E. coli is considered harmless and commensal, several pathotypes have been recognized as significant human pathogens, comprising strains implicated in gastro-intestinal, urinary, or respiratory infections (Kaper et al., 2004). E. coli strains can be assigned to four main phylogenetic groups: A, B1, B2 and D (Clermont et al., 2000). Extraintestinal pathogenic bacteria usually belong to groups B2 and D, precisely those that include strains with a larger diversity of virulence factors. The commensal strains belong to groups A and B1 (Picard et al., 1999; Johnson et al., 2001).
In recent years, E. coli has been recognized as a major player in the dissemination of AR (Henriques et al., 2006a; Zhao and Dang, 2012). Resistance genotypes so far characterized include genes for resistance to last-line antibiotics such as 3rd generation cephalosporins and fluoroquinolones (Alouache et al., 2012; Wellington et al., 2013; Tacão et al., 2014). To some extent, similar resistance phenotypes and genotypes are shared by strains exposed to similar environmental pressures (Wicki et al., 2011). Considering this fact, some animal hosts may favor the preferential development of certain genotypes.
Seagulls readily utilize food sources of human origin, especially garbage, which may contribute to the high levels of AR associated to their microbiota (Bonnedahl et al., 2009; Poirel et al., 2012). Multi-resistant phenotypes are found in E. coli strains isolated from seagull droppings in different geographic areas (Gionechetti et al., 2008; Bonnedahl et al., 2009; Dolejska et al., 2009; Poirel et al., 2012). Genes encoding ESBLs (extended spectrum beta-lactamases), such as blaCTX−M−1 and blaCTX−M−15 (Bonnedahl et al., 2009; Dolejska et al., 2009; Simões et al., 2010), and resistance to quinolones (like the plasmid-mediated qnrS) (Literak et al., 2010; Vredenburg et al., 2014) integrate the repertoire of seagull's resistome.
Freshwater environments are well-recognized reactors for the dissemination and evolution of AR (Baquero et al., 2008; Figueira et al., 2011; Tacão et al., 2012) but processes occurring in seawater are less understood. The presence of AR strains of E. coli in coastal water represents a health issue in areas that are used for recreational activities.
Berlenga is an uninhabited island, recently added to the World Network of Biosphere Reserves due to its ecological relevance. Episodes of fecal pollution have been recorded in its coastal water (Araújo et al., 2014) with seagulls confirmed as the main source of fecal contamination. Human-derived sewage (from visiting tourists) was also suggested as a secondary source (Araújo et al., 2014).
Because it is a well-delimited ecosystem, the island constitutes a privileged location to study pathways implicated in dissemination of AR in natural environments. Resistance to 3rd generation cephalosporins or fluoroquinolones, used as front-line drugs for treatment of Gram-negative infections (Cantón and Coque, 2006; Cattoir et al., 2007), deserved special attention.
In this study the focus was put in coastal water and in the specific contribution of the two previously identified sources of fecal pollution. To accomplish our aims, E. coli strains were characterized at the phenotypic level with respect to their resistances profiles; at the genotypic level, specific genes were used as indicators for the resistance to last-line use antibiotics. Phenotypic and genotypic data from coastal water strains was compared with data obtained from the strains of each of the two sources of fecal contamination. Main contributions to the water resistome are hypothesized and discussed.
Berlenga Island is the main island of the Berlengas archipelago, located in the Portuguese continental shelf, 5.7 miles northwest of Cape Carvoeiro. The island has a great diversity of habitats, marine species, and avifauna, including a large population of the yellow-legged gull (Larus [cachinnans] michahellis). Recently, Berlengas was added to the UNESCO'S World Biosphere Reserve. Although uninhabited, many tourists visit the island in the summer mainly for recreational activities such as swimming and diving (Araújo et al., 2014). Human-derived sewage is collected on a settlement tank and discharged near the coastline of the island.
Samples of seagull feces, human-derived sewage and seawater from the Berlenga beach were collected as described in Araújo et al. (2014). Sampling was performed every 2 weeks between May and September 2011. Briefly, samples of seagull feces, human-derived sewage and seawater were collected in each sampling date and consisted in: (i) 5 composite samples of seagull feces scattered on the beach; (ii) 250 mL of raw human-derived sewage taken from the effluent of the sanitary infrastructures; and (iii) 2 L of seawater collected about 30 cm below the surface. Samples were collected in sterile containers and kept on ice until processing.
A total of 939 E. coli isolates were retrieved in Chromocult Coliform Agar plates (Merck, Germany) from seagull feces (n = 427), sewage (n = 170) and seawater (n = 342) as described by Araújo et al. (2014). Genetic diversity of the E. coli isolates was inspected by BOX-PCR with primers and conditions previously described (Araújo et al., 2014). Fingerprinting analysis allowed the selection of 414 non-clonal isolates, which were included in the present study. From those, 179 isolates were from seagull feces, 69 isolates were from sewage and 166 were from seawater.
The triplex PCR developed by Clermont et al. (2000), with primers for genes chuA and yjaA and for the DNA fragment TSPE4-C2, was used to determine the phylogenetic groups of the E. coli strains. Template DNA was obtained by suspending 2 bacterial colonies in 100 μL of sterile water. For each PCR reaction, 1 μL of this suspension was used as template. Appropriate positive and negative controls were included in the assay.
The E. coli isolates were tested for susceptibility to 16 antibiotics by disk diffusion method on Mueller-Hinton Agar (Oxoid, Basingstoke, UK) according to the Clinical and Laboratory Standards Institute guidelines (CLSI, 2012). The following disks (Oxoid, UK) were used: ampicillin (10 μg), amoxicillin (10 μg), amoxicillin/clavulanic acid (20/10 μg), piperacillin (100 μg), piperacillin/tazobactam (100/10 μg), cephalothin (30 μg), ceftazidime (30 μg), cefotaxime (30 μg), gentamicin (10 μg), streptomycin (10 μg), imipenem (10 μg), nalidixic acid (30 μg), ciprofloxacin (5 μg), tetracycline (30 μg), chloramphenicol (30 μg) and trimethoprim/sulfamethoxazole (1.25/23.75 μg). E. coli ATCC 25922 was used as quality control. Isolates were classified as sensitive, intermediate or resistant according to the CLSI recommendations after 24 h incubation at 37°C. To determine the percentage of resistant strains to each antibiotic and to define resistance phenotypes, intermediate and resistant isolates were subsequently grouped in a same resistant class.
Escherichia coli strains displaying resistance and intermediate resistance phenotypes were screened by PCR to detect genes conferring resistance to β-lactams (blaTEM, blaSHV, blaCTX−M, blaIMP, blaVIM, blaKPC, blaOXA−48, blaGES, blaAmpC−like), tetracycline [tet(A), tet(B)], quinolones (qnrA, qnrB, qnrS) and sulfonamides (sul1, sul2). The genes tested were chosen among the most prevalent in clinical and environmental E. coli isolates. The PCR reactions were performed in a MyCycler Thermal cycler (Bio-Rad, USA). Primers and PCR conditions are presented in Table 1 and Table S1 (Supporting Information). The reaction mixtures (25 μL total volume) consisted of 6.25 μL NZYTaq 2x Green Master Mix (2.5 mM MgCl2; 200 μM dNTPs; 0.2 U/μL DNA polymerase) (NZYtech, Portugal), 16.25 μL of ultrapure water, 0.75 μL of each primer (reverse and forward), and 1 μL of cell suspension prepared as described above. The presence of blaAmpC−like genes was inspected using a multiplex PCR as described by Dallenne et al. (2010). Negative and positive controls were included in each PCR experiment. The negative control differed from other reaction mixtures by substituting the cell suspension for the same volume of dH2O; positive control strains are indicated in Table 1. PCR products were analyzed by electrophoresis on a 1.5% agarose gel and stained with ethidium bromide. All amplicons obtained using primers for blaSHV, blaCTX−M, blaAmpC−like, qnrA, qnrB, and qnrS and PCR products obtained using primers for blaTEM from 3rd generation cephalosporins-resistant isolates were sequenced (accession numbers KM094197 to KM094211). For this PCR products were purified with DNA Clean & Concentrator (Zymo Research, USA) following manufacturer's instructions, and used as template in the sequencing reactions. Online similarity searches were performed with the BLAST software at the National Center for Biotechnology Information website.
The genomic contexts of both genes were determined by PCR using previously described primers and conditions (Saladin et al., 2002; Eckert et al., 2006; Tacão et al., 2012). Primers targeted either blaCTX−M−1 or blaCMY−2 and genes previously found in the vicinity of these resistance genes (i.e., ISECP1 for blaCTX−M−1 and blaCMY−2; ORF477 for blaCTX−M−1) (Saladin et al., 2002; Eckert et al., 2006; Tacão et al., 2012). PCR fragments were sequenced as described above.
AR phenotypes for each isolate were converted into a numeric code for each antibiotic: 0 signifying susceptibility, 1 representing intermediate resistance and 2 representing resistance. Principal Component Analysis (PCA, using a covariance matrix model) was used to explore AR patterns, reducing the multidimensional data matrix to a bidimensional biplot fit for interpretation, as performed in suchlike studies (Parveen et al., 1997; Su et al., 2012; Pereira et al., 2013). One-way analyses of variance (ANOVA) were performed on the PCA sample scores (principal components 1 and 2—PC1 and PC2) to assess significant differences (p ≤ 0.05) in the AR profiles of E. coli isolates among sampling sources or phylogenetic groups. When applicable, post-hoc Tukey tests followed ANOVA tests. ANOVAs and PCA were conducted using Minitab and CANOCO for Windows version 4.5 (Scientia Ltd., UK), respectively.
The set of 414 E. coli isolates were assigned to the main phylogenetic groups: A, B1, B2, and D (Figure 1). Overall, group A was the most prevalent (45% of the total number of isolates) followed by group B1 (31%), group D (21%) and group B2 (3%). Considering the different sampling sources (e.g., seawater, seagull feces and human-derived sewage) distribution among phylogenetic groups was rather similar, except for group B2 isolates that were only detected in sewage (10% of the total number of isolates retrieved from this source) and in seagull feces (4%).
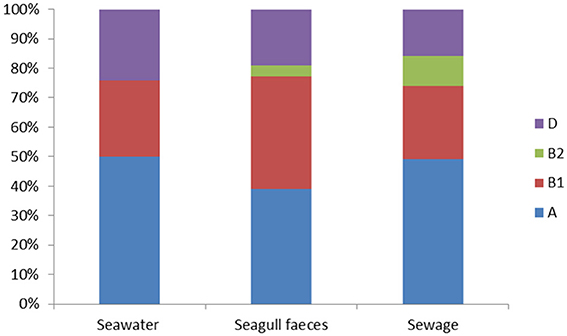
Figure 1. Distribution of the E. coli phylogenetic groups (A, B1, B2 and D) among seawater (n = 166), seagull feces (n = 179) and human-derived sewage (n = 69).
The susceptibility patterns of the isolates to 16 antibiotics are shown in Figure 2. Resistance (or intermediate resistance) was detected to all antibiotics tested and 94% of the isolates were resistant to at least one of the antibiotics. The most prevalent AR was toward streptomycin (83–100%), followed by tetracycline and cephalothin in seawater (48 and 42% respectively), and tetracycline and amoxicillin in the fecal sources (35 and 34% in seagull feces and 23% for both antibiotics in sewage). Isolates were more susceptible to imipenem, 3rd generation cephalosporins, gentamicin and the combination piperacillin/tazobactam.
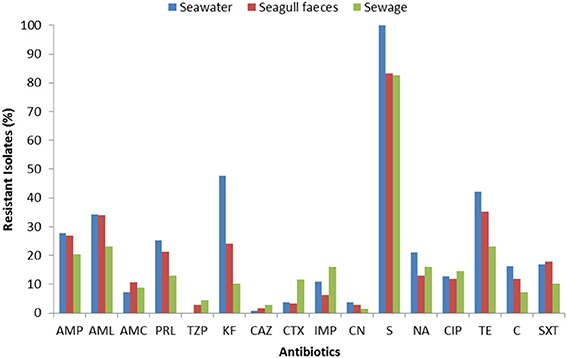
Figure 2. Total resistance frequencies to 16 antibiotics of E. coli strains isolated from seawater (n = 166), seagull feces (n = 179) and human-derived sewage (n = 69). AMP, ampicillin; AML, amoxicillin; AMC, amoxicillin/clavulanic acid; PRL, piperacillin; TZP, piperacillin/tazobactam; KF, cephalothin; CAZ, ceftazidime; CTX, cefotaxime; IMP, imipenem; CN, gentamicin; S, streptomycin; NA, nalidixic acid; CIP, ciprofloxacin; TE, tetracycline; C, chloramphenicol; SXT, sulfamethoxazole/trimethoprim.
For the majority of the antibiotics tested (e.g., penicillins, cephalothin, tetracycline), resistance rates were higher in isolates from seawater and seagull feces (Figure 2). Resistance to imipenem, 3rd generation cephalosporins and ciprofloxacin was most common in sewage. Multi-resistance levels (i.e., resistance to antibiotics of at least three different classes) were higher among isolates recovered from seawater (39%), followed by isolates from seagull feces (32%) and sewage (29%).
PCA analysis confirmed a clear AR gradient, with less resistant isolates (isolates resistant to a lower number of antibiotics) consistently displaying lower PC1 scores (i.e., being located at the left of the biplot) and more resistant isolates displaying higher PC1 scores (i.e., being located at the right of the biplot) (Figure 3). Although resistant and multi-resistant isolates were found in all sampling sources, significant differences in PC1 scores were found among sources [ANOVA: F(2, 411) = 3.7, p = 0.025]. This was mostly due to a significantly higher PC1 score in seawater isolates comparatively to sewage isolates (Tukey test, p < 0.05). No significant differences in terms of the AR profile were found across phylogenetic groups, either for PC1 [ANOVA: F(2, 397) = 1.59, p = 0.206] or PC2 [ANOVA: F(2, 397) = 0.43, p = 0.649]; group B2 was excluded from this analysis due to low representativeness.
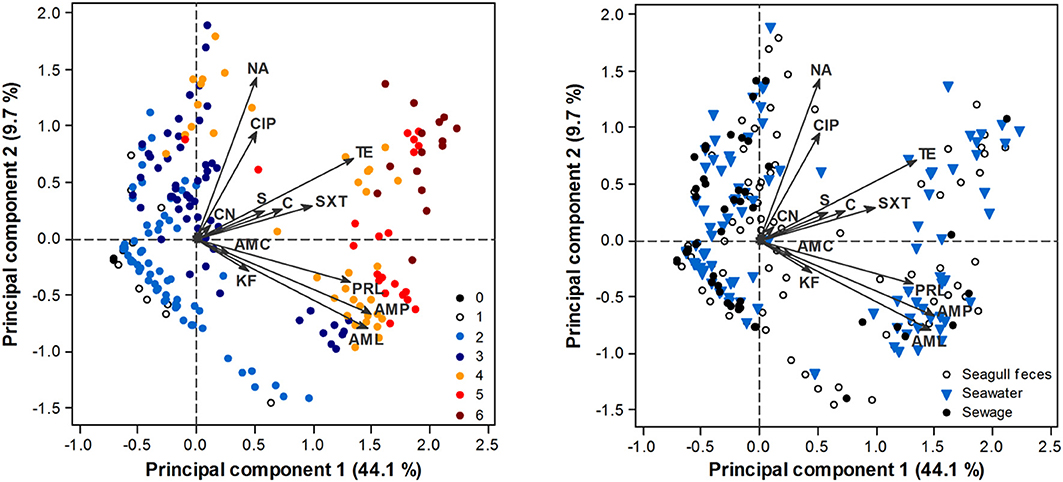
Figure 3. PCA biplot of E. coli isolate scores in terms of their AR profile. Arrows represent antibiotics: AMP, ampicillin; AML, amoxicillin; AMC, amoxicillin/clavulanic acid; PRL, piperacillin; TZP, piperacillin/tazobactam; KF, cephalothin; CAZ, ceftazidime; CTX, cefotaxime; IMP, imipenem; CN, gentamicin; S, streptomycin; NA, nalidixic acid; CIP, ciprofloxacin; TE, tetracycline; C, chloramphenicol; SXT, sulfamethoxazole/trimethoprim. TZP, CAZ, CTX, IMP have little influence in the ordination, thus respective arrows are not perceptible. Scores are depicted according to the number of antibiotic classes that isolates are resistant to (0–6; left panel) or source of origin (right panel).
A total of 126 different resistance (or intermediate resistance) phenotypes were observed but most (n = 86) were represented by only one isolate. Resistance to streptomycin was the most common phenotype, identified in 90 isolates, followed by resistance to streptomycin and tetracycline (30 isolates) and streptomycin and cephalothin (27 isolates). Several multi-resistance phenotypes were detected. Among these, 10 phenotypes included 8 antibiotics and 9 phenotypes included 9 antibiotics. Twenty-four phenotypes (Table 2) were common to seawater and to at least one of the putative pollution sources (i.e., sewage and seagull feces). From these, 45% (n = 11) were common to seawater and seagull feces but were absent from human-derived sewage, while only 8% (n = 2) of the phenotypes were exclusively detected in both seawater and sewage.
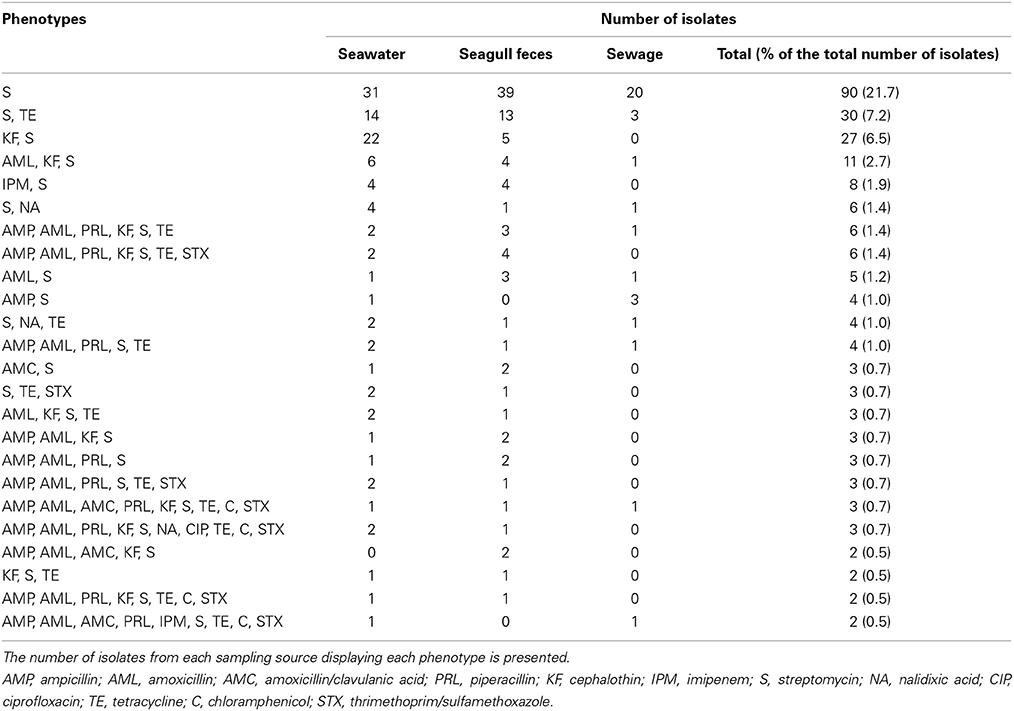
Table 2. AR phenotypes detected in seawater and in at least one of the putative pollution sources (i.e., seagull feces and sewage).
The resistance genes elected for this study were detected in 157 isolates out of 390 displaying a resistance phenotype (Table 3). The most commonly detected genes were blaTEM, sul1, sul2, tet(A), and tet(B), which were prevalent in 6 out of 8 sampling moments (data not shown). The blaTEM gene (blaTEM−1 according to sequence analysis of representative isolates) was more prevalent in isolates from seawater (69% of 68 isolates resistant to penicillins) than in isolates from seagull feces (38% of) or sewage (24%). Both tet(A) and sul2 were commonly detected in seawater (69 and 64%, respectively) and seagull feces (70 and 60%), but were less frequent in sewage (31 and 43%). Eighty six isolates displaying resistance to quinolones were tested for the presence of qnr determinants. Among these, qnrS (identified as qnrS1 by sequence analysis) was the most prevalent, being more frequent in seagull feces (17% of 30 resistant isolates). This gene was detected in 5 out of 8 sampling dates. Gene qnrB (qnrB19 according to sequence analysis) was detected in two sampling dates in isolates from seawater and seagull feces and qnrA was not detected. Gene blaSHV−12 was detected in one isolate from seawater. Genes blaCTX−M and blaAmpC−like were detected in one isolate each, from seawater and seagull feces respectively. Sequencing analysis revealed 100% identity with blaCTX−M−1 and blaCMY−2. The genomic contexts of both genes were determined and results revealed genomic contexts identical to the ones previously described in clinical isolates (Kang et al., 2006; Tacão et al., 2012): ISECP1 was detected upstream of both genes and ORF477 was identified downstream blaCTX−M−1. The genes encoding resistance to carbapenems (i.e., blaOXA−48, blaIMP, blaVIM, blaGES and blaKPC) were not detected in any of the E. coli isolates studied.
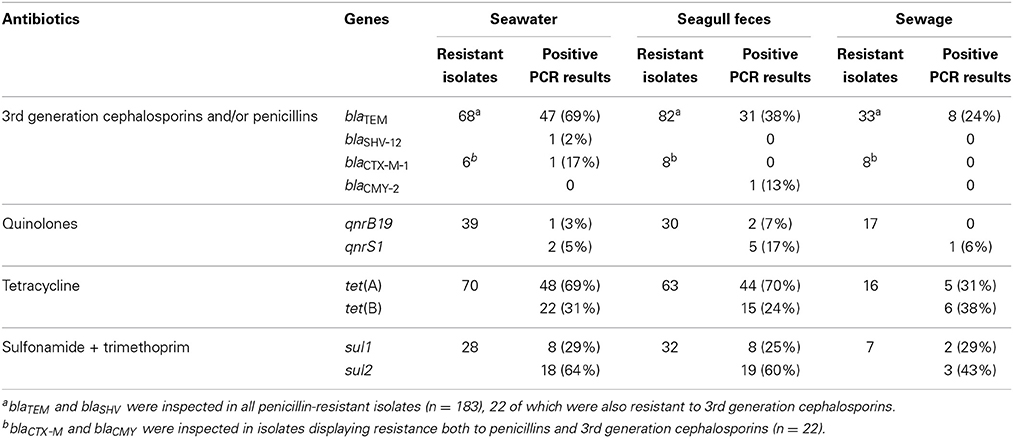
Table 3. Prevalence of resistance genes detected in the E. coli strains according to the isolation source.
A total of 27 different AR genotypes were observed. Among these, 7 included only 1 resistance gene, 7 included 2 genes and 8 included 3 genes. Four genotypes included 4 genes: blaTEM-tet(A)-sul1-sul2, detected in 4 isolates from seawater and 5 isolates from seagull feces; blaTEM-tet(B)-sul1-sul2, detected in 2 isolates from seawater and 2 isolates from sewage; blaTEM-blaSHV−12-tet(A)-sul2 and blaTEM-blaCTX−M−1-tet(A)-qnrS1, each detected in 1 seawater isolate. The genotype blaTEM-tet(B)-qnrS1-sul1-sul2 was detected in 1 isolate from seagull feces. Fourteen genotypes were shared between seawater and at least 1 of the putative pollution sources (Table 4). From these, 50% (n = 7) were shared between seawater and seagull feces, exclusively. Only 1 genotype was common to seawater and sewage, and absent from seagull feces.
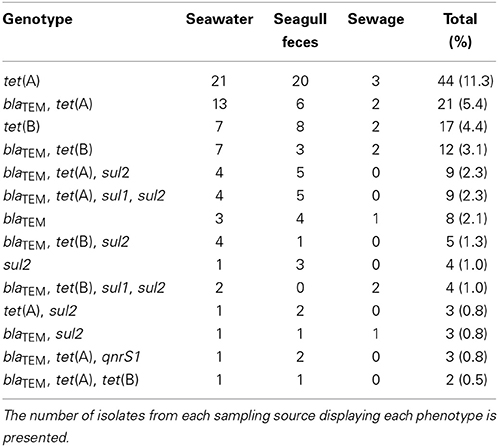
Table 4. AR genotypes detected in seawater and in at least one of the putative pollution sources (i.e., seagull feces and sewage).
The threats for human health represented by fecal pollution in aquatic environments depend on whether the source is human or non-human (Field and Samadpour, 2007). Dissemination of AR is certainly a risk factor. The particular characteristics of the Berlenga Island allowed evaluation of the contribution given by human-derived sewage and seagull feces, to the AR patterns found in E. coli strains that can be grown and isolated from coastal water samples.
High prevalence of resistance to streptomycin, tetracycline, cephalothin, and amoxicillin was detected in all sources. Results are in agreement with previous studies that demonstrated a wide dissemination of resistance to these antibiotics among E. coli isolates from Portuguese clinical settings (Freitas et al., 2014), animals (Radhouani et al., 2009) and aquatic systems (Pereira et al., 2013). For antibiotics mainly used in human medicine such as 3rd generation cephalosporins, ciprofloxacin and imipenem, levels of resistance were low. An uncommon phenotype combining resistance to imipenem and streptomycin and susceptibility to penicillins was detected in eight strains, six of which were isolated from sewage. Genes encoding carbapenemases were not detected in these strains, an indication that non-enzymatic mechanisms, such as the expression of efflux pumps or porin loss (Papp-Wallace et al., 2011), may contribute to the phenotype. Concerning 3rd generation cephalosporins, the resistance phenotype was explained only in three out of twenty-two resistant isolates. Novel or uncommon resistance genes may be present in the genomes of the remaining strains. On the other hand, mutations in the promoter and attenuator sequences of the chromosomal ampC gene may also contribute to reduced susceptibility to these antibiotics. These mutations have been described in E. coli isolates from different sources, including aquatic systems (Mataseje et al., 2009).
Levels of AR in coastal water isolates were expected to be low, considering the moderate human impact and the limits to survival imposed to E. coli by water salinity. However, experimental data revealed resistance levels similar to those previously reported for polluted freshwater (Ham et al., 2012; Pereira et al., 2013) or wastewater (Figueira et al., 2011). Multi-resistance levels were higher in coastal water (39%) than in the two sources of fecal contamination (32% for seagull feces and 29% found in human sewage).
Genes frequently hosted by clinical isolates and sharing similar genomic contexts (Saladin et al., 2002; Eckert et al., 2006), were also hosted by seawater isolates. Examples are ESBL-encoding genes such as blaCTX−M−1 and blaSHV−12 or plasmid-mediated quinolone resistance genes (e.g., qnrB19). It is to point that dissemination of these genes or related ones is a matter of serious concern in a scenario of lack of innovation in what concerns antimicrobial therapies. In fact, 3rd generation cephalosporins and fluoroquinolones are front-line therapeutics for the treatment of Gram-negative infections (Cantón and Coque, 2006; Cattoir et al., 2007).
AR features of seawater E. coli strains are mainly dependent on their animal source (Wicki et al., 2011). At the same time, E. coli strains are able to adapt to non-human environments, a process that implies changes in their genetic background (van Elsas et al., 2011). Adaptation to the more stressful saline conditions of coastal waters may favor strains with genotypes that are more efficient to deal with other stresses.
The results strongly suggest that the E. coli resistome found in coastal water was mainly impacted by fecal pollution from seagulls. The AR patterns found on seagull feces and seawater shared abundant common elements, while differing significantly from the resistance patterns found on sewage. Differences were not attributable to a different distribution in terms of phylogenetic groups. Based on these observations it is possible to hypothesize that the enteric environment of seagulls positively selects E. coli strains more prone to survive in marine environments. The same data allows proposing resistome analysis as a microbial source-tracking tool (see Harwood et al., 2014). Such approach has been previously tested, using almost exclusively data resulting from the analysis of resistance phenotypes (Parveen et al., 1997; Field and Samadpour, 2007; Carroll et al., 2009). Data on resistance genotypes add valuable information for the identification of the contamination source. The resistance gene pool in the analyzed sources was quite stable along the period of the study, except for the sporadic detection of infrequent genes (data not shown). Our recommendation is toward the use of genotypic data only in combination with other data. Our data further confirmed the conclusion of Araújo et al. (2014) establishing seagulls as the main source of fecal pollution on Berlenga Island.
This study highlights the risks associated to seagull-derived pollution. This is a critical aspect since wild life-derived pollution is frequently neglected due to the assumption that host-specificity of pathogens (Field and Samadpour, 2007) is a preventive barrier, that makes difficult if not impossible, the establishment on a new host. Indeed, the contribution of seagull fecal bacteria to AR dissemination constitutes an undeniable risk to humans, especially because of the close contact between these birds and human waste. Previous studies have confirmed seagulls as a reservoir of AR (Gionechetti et al., 2008; Bonnedahl et al., 2009; Dolejska et al., 2009). In our study, besides the high level of multi-resistance in seagull feces, we retrieved isolates carrying genes conferring resistance to antibiotics that are critically important for human medicine. For example, the prevalence of isolates carrying the qnrS1 gene was higher in seagull feces than in sewage. This plasmid-encoded gene was previously identified in isolates from seagull feces (Literak et al., 2010) and is frequently found in human pathogens (Cattoir et al., 2007). Likewise, we detected the presence of qnrB19 in seawater and seagull feces, a gene which was previously detected in animal and human isolates (Dolejska et al., 2013). Moreover, the plasmid-encoded gene blaCMY−2, conferring resistance to 3rd generation cephalosporins, was detected in seagull feces and in a genomic context identical to the one found in clinical isolates (Verdet et al., 2009). Poirel et al. (2012) reported a high proportion of CMY-2-positive strains among resistant E. coli isolated from seagull feces in Miami Beach. So, data obtained in different contexts indicate seagulls as important vectors of antibiotic resistant bacteria.
A previous study in the same Island (Radhouani et al., 2009), showed that all isolates were susceptible to carbapenems and 3rd generation cephalosporins. These antibiotics are mainly used in human medicine (Cantón and Coque, 2006; Papp-Wallace et al., 2011) and resistance to them is known to be still limited in unpolluted or moderately polluted environments (Henriques et al., 2012; Tacão et al., 2012). As reported here, resistance to these antibiotics was most common in human-derived sewage, but its presence on seagull feces isolates suggests an undesirable progress in terms of resistance dissemination. Seagulls based on the Island fly to urban areas on the coast for feeding. Like suggested for other geographic regions (Bonnedahl et al., 2009), human activities may considerably influence the AR patterns associated to seagulls in remote areas, such as Berlenga. In fact, it is a matter of concern that levels of resistance to 3rd generation cephalosporins have been consistently increasing in isolates from clinical settings in Portugal (ECDC, 2013). For example, gene blaCMY−2, which was here detected in one isolate from seagull feces, has been detected in E. coli from clinical samples in Portuguese health care settings (Freitas et al., 2014).
This study confirms seawater as a relevant reservoir of E. coli strains adapted to marine environments and carrying AR determinants that are common to clinical strains. It is evidenced that the seawater E. coli resistome in Berlenga is mainly modulated by seagulls-derived pollution. This fact confirms seagulls as a main spread vehicle of AR bacteria to coastal water. Occurrence of high levels of multi-resistance and of clinically relevant genes underlies the need to carefully consider the human health risks subjacent to seagull-derived pollution. The assessment of these risks is essential for an effective management of coastal water used for recreational activities. Lastly, we believe that this study—along with correlated data from previous studies—has succeeded in showing that AR could be used as a tool to aid in the identification of the fecal sources of contamination.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Portuguese Foundation for Science and Technology (FCT) supported this study through projects PTDC/AAC-AMB/109155/2008 and FCOMP-01-0124-FEDER-008640. FCT also financed the fellowships of A Pereira (SFRH/BPD/26685/2006). I Henriques was supported by the project MARES (Sustainable Use of Marine Resources; ref. SCT_2011_02_033_4904) funded by National Strategic reference Framework. Bruno B. Castro was supported by the Programme Ciência 2008, co-funded by the Human Potential Operational Programme (National Strategic Reference Framework 2007–2013) and European Social Fund (EU). This work was supported by European Funds through COMPETE and by National Funds through the Portuguese Science Foundation (FCT) within project PEst-C/MAR/LA0017/2013.
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmicb.2014.00426/abstract
Alouache, S., Kada, M., Messai, Y., Estepa, V., Torres, C., and Bakour, R. (2012). Antibiotic resistance and extended-spectrum beta-lactamases in isolated bacteria from seawater of Algiers beaches (Algeria). Microb. Environ. 27, 80–86. doi: 10.1264/jsme2.ME11266
Aminov, R. I. (2011). Horizontal gene exchange in environmental microbiota. Front. Microbiol. 2:158. doi: 10.3389/fmicb.2011.00158
Araújo, S., Henriques, I. S., Leandro, S. M., Alves, A., Pereira, A., and Correia, A. (2014). Gulls identified as major source of faecal pollution in coastal waters: a microbial source tracking study. Sci. Total Environ. 470–471, 84–91. doi: 10.1016/j.scitotenv.2013.09.075
Austin, D. J., Kristinsson, K. G., and Anderson, R. M. (1999). The relationship between the volume of antimicrobial consumption in human communities and the frequency of resistance. Proc. Natl. Acad. Sci. U.S.A. 96, 1152–1156. doi: 10.1073/pnas.96.3.1152
Baquero, F., Martínez, J. L., and Cantón, R. (2008). Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 19, 260–265. doi: 10.1016/j.copbio.2008.05.006
Bonnedahl, J., Drobni, M., Gauthier-Clerc, M., Hernandez, J., Granholm, S., Kayser, Y., et al. (2009). Dissemination of Escherichia coli with CTX-M type ESBL between humans and yellow-legged gulls in the south of France. PLoS ONE 4:e5958. doi: 10.1371/journal.pone.0005958
Cantón, R., and Coque, T. M. (2006). The CTX-M beta-lactamase pandemic. Curr. Opin. Microbiol. 9, 466–475. doi: 10.1016/j.mib.2006.08.011
Carroll, S. P., Dawes, L., Hargreaves, M., and Goonetilleke, A. (2009). Faecal pollution source identifcation in an urbanizing catchment using antibiotic resistance profling, discriminant analysis and partial least squares regression. Water Res. 43, 1237–1246. doi: 10.1016/j.watres.2008.12.017
Cattoir, V., Poirel, L., and Nordmann, P. (2007). Plasmid-mediated quinolone resistance determinant QnrB4 identified in France in an Enterobacter cloacae clinical isolate coexpressing a QnrS1 determinant. Antimicrob. Agents Chemother. 51, 2652–2653. doi: 10.1128/AAC.01616-06
Clermont, O., Bonacorsi, S., and Bingen, E. (2000). Rapid and simple determination of Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66, 4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000
CLSI (2012). Performance Standard for Antimicrobial Susceptibility Testing e Document Approved Standard M100-S22. Wayne, PA: CLSI.
Dallenne, C., Da Costa, A., Decré, D., Favier, C., and Arlet, G. (2010). Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65, 490–495. doi: 10.1093/jac/dkp498
Devirgiliis, C., Barile, S., and Perozzi, G. (2011). Antibiotic resistance determinants in the interplay between food and gut microbiota. Genes Nutr. 6, 275–284. doi: 10.1007/s12263-011-0226-x
Dolejska, M., Bierosova, B., Kohoutova, L., Literak, I., and Cizek, A. (2009). Antibiotic-resistant Salmonella and Escherichia coli isolates with integrons and extended-spectrum beta-lactamases in surface water and sympatric black-headed gulls. J. Appl. Microbiol. 106, 1941–1950. doi: 10.1111/j.1365-2672.2009.04155.x
Dolejska, M., Villa, L., Hasman, H., Hansen, L., and Carattoli, A. (2013). Characterization of IncN plasmids carrying blaCTX-M-1 and qnr genes in Escherichia coli and Salmonella from animals, the environment and humans. J. Antimicrob. Chemother. 68, 333–339. doi: 10.1093/jac/dks387
ECDC. (2013). European Centre for Disease Prevention and Control. Antimicrobial resistance surveillance in Europe 2012. Annual report of the European Antimicrobial Resistence Surveillance Network (EARS-Net). Available online at http://ecdc.europa.eu/ (Accessed 07 July, 2014).
Eckert, C., Gautier, V., and Alert, G. (2006). DNA sequence analysis of the genetic environment of various blaCTX-M genes. J. Antimicrob. Chemother. 57, 14–23. doi: 10.1093/jac/dki398
Field, K. G., and Samadpour, M. (2007). Faecal source tracking, the indicator paradigm, and managing water quality. Water Res. 41, 3517–3538. doi: 10.1016/j.watres.2007.06.056
Figueira, V., Serra, E., and Manaia, C. M. (2011). Differential patterns of antimicrobial resistance in population subsets of Escherichia coli isolated from waste- and surface waters. Sci. Total Environ. 409, 1017–1023. doi: 10.1016/j.scitotenv.2010.12.011
Freitas, F., Machado, E., Ribeiro, T. G., Novais, Â., and Peixe, L. (2014). Long-term dissemination of acquired AmpC β-lactamases among Klebsiella spp. and Escherichia coli in Portuguese clinical settings. Eur. J. Clin. Microbiol. Infect. Dis. 33, 551–558. doi: 10.1007/s10096-013-1983-9
Gionechetti, F., Zucca, P., Gombac, F., Monti-Bragadin, C., Lagatolla, C., Tonin, E., et al. (2008). Characterization of antimicrobial resistance and class 1 integrons in Enterobacteriaceae isolated from Mediterranean herring gulls (Larus cachinnans). Microb. Drug Resist. 14, 93–99. doi: 10.1089/mdr.2008.0803
Gomez-Alvarez, V., Revetta, R. P., and Santo Domingo, J. W. (2012). Metagenomic analysis of drinking water receiving different disinfection treatments. Appl. Environ. Microbiol. 78, 6095–6102. doi: 10.1128/AEM.01018-12
Guillard, T., Moret, H., Brasmea, L., Carliera, A., Vernet-Garniera, V., Cambau, E., et al. (2011). Rapid detection of qnr and qepA plasmid mediated quinolone resistance genes using real-time PCR. Diagn. Microbiol. Infect. Dis. 70, 253–259. doi: 10.1016/j.diagmicrobio.2011.01.004
Ham, Y. S., Kobori, H., Kang, J. H., Matsuzaki, T., Iino, M., and Nomura, H. (2012). Distribution of antibiotic resistance in urban watershed in Japan. Environ. Pollut. 162, 98–103. doi: 10.1016/j.envpol.2011.11.002
Harwood, V. J., Staley, C., Badgley, B. D., Borges, K., and Korajkic, A. (2014). Microbial source tracking markers for detection of faecal contamination in environmental waters: relationships between pathogens and human health outcomes. FEMS Microbiol. Rev. 38, 1–40. doi: 10.1111/1574-6976.12031
Henriques, I., Araújo, S., Azevedo, J. S., Alves, M. S., Chouchani, C., Pereira, A., et al. (2012). Prevalence and diversity of carbapenem-resistant bacteria in untreated drinking water in Portugal. Microb. Drug Resist. 18, 531–537. doi: 10.1089/mdr.2012.0029
Henriques, I., Fonseca, F., Alves, A., Saavedra, M. J., and Correia, A. (2006a). Occurrence and diversity of integrons and beta-lactamase genes among ampicillin-resistant isolates from estuarine waters. Res. Microbiol. 157, 938–947. doi: 10.1016/j.resmic.2006.09.003
Henriques, I., Moura, A., Alves, A., Saavedra, M. J., and Correia, A. (2006b). Analysing diversity among beta-lactamase encoding genes in aquatic environments. FEMS Microbiol. Ecol. 56, 418–429. doi: 10.1111/j.1574-6941.2006.00073.x
Heuer, H., and Smalla, K. (2007). Manure and sulfadiazine synergistically increased bacterial antibiotic resistance in soil over at least two months. Environ. Microbiol. 9, 657–666. doi: 10.1111/j.1462-2920.2006.01185.x
Johnson, J. R., Delavari, P., Kuskowski, M., and Stell, A. L. (2001). Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J. Infect. Dis. 183, 78–88. doi: 10.1086/317656
Kang, M. S., Besser, T. E., and Call, D. R. (2006). Variability in the region downstream of the blaCMY-2 beta-lactamase gene in Escherichia coli and Salmonella enterica plasmids. Antimicrob. Agents Chemother. 50, 1590–1593. doi: 10.1128/AAC.50.4.1590-1593.2006
Kaper, J. B., Nataro, J. P., and Mobley, H. L. (2004). Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2, 123–140. doi: 10.1038/nrmicro818
Literak, I., Dolejska, M., Janoszowska, D., Hrusakova, J., Meissner, W., Rzyska, H., et al. (2010). Antibiotic-resistant Escherichia coli bacteria, including strains with genes encoding the extended-spectrum beta-lactamase and QnrS, in waterbirds on the Baltic Sea Coast of Poland. Appl. Environ. Microbiol. 76, 8126–8134. doi: 10.1128/AEM.01446-10
Lu, S., Zhang, Y., Geng, S., Li, T., Ye, Z., Zhang, D., et al. (2010). High diversity of extended-spectrum beta- lactamase-producing bacteria in an urban river sediment habitat. Appl. Environ. Microbiol. 76, 5972–5976. doi: 10.1128/AEM.00711-10
Mataseje, L. F., Neumann, N., Crago, B., Baudry, P., Zhanel, G. G., Louie, M., et al. (2009). Characterization of cefoxitin-resistant Escherichia coli isolates from recreational beaches and private drinking water in Canada between 2004 and 2006. Antimicrob. Agents Chemother. 53, 3126–3130. doi: 10.1128/AAC.01353-08
Nawaz, M., Sung, K., Khan, S. A., Khan, A. A., and Steel, R. (2006). Biochemical and molecular characterization of tetracycline-resistant Aeromonas veronii isolates from catfish. Appl. Environ. Microbiol. 72, 6461–6466. doi: 10.1128/AEM.00271-06
Papp-Wallace, K. M., Endimiani, A., Taracila, M. A., and Bonomo, R. A. (2011). Carbapenems: past, present, and future. Antimicrob. Agents Chemother. 55, 4943–4960. doi: 10.1128/AAC.00296-11
Parveen, S., Murphree, R. L., Edmiston, L., Kaspar, C. W., Portier, K. M., and Tamplin, M. L. (1997). Association of multiple-antibiotic-resistance profiles with point and nonpoint sources of Escherichia coli in Apalachicola Bay. Appl. Environ. Microbiol. 63, 2607–2612. doi: 10.1128/AEM.00763-07
Pereira, A., Santos, A., Tacão, M., Alves, A., Henriques, I., and Correia, A. (2013). Genetic diversity and antimicrobial resistance of Escherichia coli from Tagus estuary (Portugal). Sci. Total Environ. 461–462, 65–71. doi: 10.1016/j.scitotenv.2013.04.067
Picard, B., Garcia, J. S., Gouriou, S., Duriez, P., Brahimi, N., Bingen, E., et al. (1999). The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect. Immu. 67, 546–553.
Poirel, L., Héritier, C., Tolün, V., and Nordmann, P. (2004). Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48, 15–22. doi: 10.1128/AEM.00271-06
Poirel, L., Potron, A., de La Cuesta, C., Cleary, T., Nordmann, P., and Munoz-Price, L. S. (2012). Wild coastline birds as reservoirs of broad-spectrum-beta-lactamase-producing Enterobacteriaceae in Miami Beach, Florida. Antimicrob. Agents Chemother. 56, 2756–2758. doi: 10.1128/AAC.05982-11
Radhouani, H., Poeta, P., Igrejas, G., Gonçalves, A., Vinu,é, L., and Torres, C. (2009). Antimicrobial resistance and phylogenetic groups in isolates of Escherichia coli from seagulls at the Berlengas nature reserve. Vet. Rec. 165, 138–142. doi: 10.1136/vr.165.5.138
Rosewarne, C. P., Pettigrove, V., Stokes, H. W., and Parsons, Y. M. (2010). Class 1 integrons in benthic bacterial communities: abundance, association with Tn402-like transposition modules and evidence for co-selection with heavy-metal resistance. FEMS Microbiol. Ecol. 72, 35–46. doi: 10.1111/j.1574-6941.2009.00823.x
Saladin, M., Cao, V. T., Lambert, T., Donay, J. L., Herrmann, J. L., Ould-Hocine, Z., et al. (2002). Diversity of CTX-M beta-lactamases and their promoter regions from Enterobacteriaceae isolated in three Parisian hospitals. FEMS Microbiol. Lett. 209, 161-168. doi: 10.1111/j.1574-6968.2002.tb11126.x
Simões, R. R., Poirel, L., Da Costa, P. M., and Nordmann, P. (2010). Seagulls and beaches as reservoirs for multidrug-resistant Escherichia coli. Emerg. Infect. Dis. 6, 110–112. doi: 10.3201/eid1601.090896
Speldooren, V., Heym, B., Labia, R., and Nicolas-Chanoine, M. H. (1998). Discriminatory detection of inhibitor-resistant beta-lactamases in Escherichia coli by single-strand conformation polymorphism-PCR. Antimicrob. Agents Chemother. 42, 879–884.
Su, H. C., Ying, G. G., Tao, R., Zhang, R. Q., Zhao, J. L., and Liu, Y. S. (2012). Class 1 and 2 integrons, sul resistance genes and antibiotic resistance in Escherichia coli isolated from Dongjiang River, South China. Environ. Pollut. 169, 42–49. doi: 10.1016/j.envpol.2012.05.007
Tacão, M., Correia, A., and Henriques, I. (2012). Resistance to broad-spectrum antibiotics in aquatic systems: anthropogenic activities modulate the dissemination of blaCTX-M-like genes. Appl. Environ. Microbiol. 78, 4134–4140. doi: 10.1128/AEM.00359-12
Tacão, M., Moura, A., Correia, A., and Henriques, I. (2014). Co-resistance to different classes of antibiotics among ESBL-producers from aquatic systems. Water Res. 48, 100–107. doi: 10.1016/j.watres.2013.09.021
van Elsas, J. D., Semenov, A. V., Costa, R., and Trevors, J. T. (2011). Survival of Escherichia coli in the environment: fundamental and public health aspects. ISME J. 5, 173–183. doi: 10.1038/ismej.2010.80
Verdet, C., Gautier, V., Chachaty, E., Ronco, E., Hidri, N., Decré, D., et al. (2009). Genetic context of plasmid-carried blaCMY-2-like genes in Enterobacteriaceae. Antimicrob. Agents Chemother. 53, 4002–4006. doi: 10.1128/AAC.00753-08
Vredenburg, J., Varela, A. R., Hasan, B., Bertilsson, S., Olsen, B., Narciso-da-Rocha, C., et al. (2014). Quinolone-resistant Escherichia coli isolated from birds of prey in Portugal are genetically distinct from those isolated from water environments and gulls in Portugal, Spain and Sweden. Environ. Microbiol. 16, 995–1004. doi: 10.1111/1462-2920.12231
Wellington, E. M. H., Boxall, A. B. A., Cross, P., Feil, E. J., Gaze, W. H., Hawkey, P. M., et al. (2013). The role of natural environment in the emergence of antibiotic resistance in Gram-negative bacteria. Lancet Infect. Dis. 13, 155–165. doi: 10.1016/S1473-3099(12)70317-1
Wicki, M., Karabulut, F., Auckenthaler, A., Felleisen, R., Tanner, M., and Baumgartner, A. (2011). Identification of faecal input sites in spring water by selection and genotyping of multiresistant Escherichia coli. Appl. Environ. Microbiol. 77, 8427–8433. doi: 10.1128/AEM.05651-11
Keywords: Escherichia coli, antibiotic resistance, fecal pollution, water quality, Microbial source tracking
Citation: Alves MS, Pereira A, Araújo SM, Castro BB, Correia ACM and Henriques I (2014) Seawater is a reservoir of multi-resistant Escherichia coli, including strains hosting plasmid-mediated quinolones resistance and extended-spectrum beta-lactamases genes. Front. Microbiol. 5:426. doi: 10.3389/fmicb.2014.00426
Received: 29 May 2014; Accepted: 29 July 2014;
Published online: 20 August 2014.
Edited by:
Satoru Suzuki, Ehime University, JapanReviewed by:
Daniela Ceccarelli, University of Maryland, USACopyright © 2014 Alves, Pereira, Araújo, Castro, Correia and Henriques. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Isabel Henriques, Department of Biology and CESAM, University of Aveiro, Campus Universitário de Santiago, Aveiro, 3810-193 Aveiro, Portugal e-mail:aWhlbnJpcXVlc0B1YS5wdA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.