
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 11 August 2014
Sec. Plant Pathogen Interactions
Volume 5 - 2014 | https://doi.org/10.3389/fmicb.2014.00411
This article is part of the Research Topic Plants as alternative hosts for human and animal pathogens View all 11 articles
Certain human bacterial pathogens such as the enterohemorrhagic Escherichia coli and Salmonella enterica are not proven to be plant pathogens yet. Nonetheless, under certain conditions they can survive on, penetrate into, and colonize internal plant tissues causing serious food borne disease outbreaks. In this review, we highlight current understanding on the molecular mechanisms of plant responses against human bacterial pathogens and discuss salient common and contrasting themes of plant interactions with phytopathogens or human pathogens.
Bagged greens in the market are often labeled “pre-washed,” “triple-washed,” or “ready-to-eat,” and look shiny and clean. But are they really “clean” of harmful microbes? We cannot be so sure. Food safety has been threatened by contamination with human pathogens including bacteria, viruses, and parasites. Between 2000 and 2008, norovirus and Salmonella spp. contributed to 58 and 11% of forborne illnesses, respectively in the United States (Scallan et al., 2011). In those same years, non-typhoidal Salmonella alone was ranked as the topmost bacterial pathogen contributing to hospitalizations (35%) and deaths (28%) (Scallan et al., 2011). In 2007, 235 outbreaks were associated with a single food commodity; out of which 17% was associated with poultry, 16% with beef, and 14% with leafy vegetables that also accounted for the most episodes of illnesses (CDC, 2010).
Apart from the direct effects on human health, enormous economic losses are incurred by contaminated food products recalls. The 8-day recall of spinach in 2006 cost $350 million to the US economy (Hussain and Dawson, 2013). It should be realized that this is not the loss of one individual, but several growers, workers, and distributors. This is a common scenario for any multistate foodborne outbreak. Additionally, the skepticism of the general public toward consumption of a particular food product can lead to deficiencies of an important food source from the diet. Less demand would in turn lead to losses for the food industry. Economic analysis shows that money spent on prevention of foodborne outbreak by producers is much less than the cost incurred after the outbreak (Ribera et al., 2012).
Contamination of plants can occur at any step of food chain while the food travels from farm to table. Both pre-harvest and post-harvest steps are prone to contamination. Contaminated irrigation water, farm workers with limited means of proper sanitation, and fecal contamination in the farm by animals can expose plants to human pathogens before harvest of the edible parts (Lynch et al., 2009; Barak and Schroeder, 2012). After harvest, contamination can occur during unclean modes of transportation, processing, and bagging (Lynch et al., 2009). Mechanical damage during transport can dramatically increase the population of human pathogens surviving on the surface of edible plants (Aruscavage et al., 2008). Control measures to decrease pathogen load on plant surfaces have been defined by the Food Safety Modernization Act (US Food and Drug Administration) and Hazard Analysis and Critical Control Point system (HACCP). Using chlorine for post-harvest crop handling has been approved by US Department of Agriculture (USDA) under the National Organic Program. However, some studies indicated that internalized human pathogens escape sanitization (Seo and Frank, 1999; Saldaña et al., 2011). Thus, understanding the biology of human pathogen-plant interactions is now crucial to prevent pathogen colonization of and survival in/on plants, and to incorporate additional, complementing measures to control food borne outbreaks.
We reasoned that as plants are recognized vectors for human pathogens, enhancing the plant immune system against them creates a unique opportunity to disrupt the pathogen cycle. In this cross-kingdom interaction, the physiology of both partners contribute to the outcome of the interactions (i.e., colonization of plants or not). Bacterial factors important for interaction with plants have been discussed in recent, comprehensive reviews (Tyler and Triplett, 2008; Teplitski et al., 2009; Berger et al., 2010; Barak and Schroeder, 2012; Brandl et al., 2013). Plant factors contributing to bacterial contamination (or lack of) is much less studied and discussed. In this review, we highlight current knowledge on plants as vectors for human pathogens, the molecular mechanisms of plant responses to human bacterial pathogens, and discuss common themes of plant defenses induced by phytopathogens and human pathogens. We have focused on human bacterial pathogens that are not recognized plant pathogens such as Salmonella enterica and Escherichia coli (Barak and Schroeder, 2012; Meng et al., 2013), but yet are major threats to food safety and human health.
The leaf environment has long been considered to be a hostile environment for bacteria. The leaf surface is exposed to rapidly fluctuating temperature and relative humidity, UV radiation, fluctuating availability of moisture in the form of rain or dew, lack of nutrients, and hydrophobicity (Lindow and Brandl, 2003). Such extreme fluctuations, for example within a single day, are certainly not experienced by pathogens in animal and human gut. Thus, it is tempting to speculate that animal pathogens may not even be able to survive and grow in an environment as dynamic as the leaf surface. However, the high incidence of human pathogens such as S. enterica and E. coli O157:H7 on fresh produce, sprouts, vegetables, leading to foodborne illness outbreaks indicate a certain level of human pathogen fitness in/on the leaf.
The plant surface presents a barrier to bacterial invaders by the presence of wax, cuticle, cell wall, trichomes, and stomata. All except stomata, present a passive defense system to prevent internalization of bacteria. Nonetheless, several bacteria are able to survive on and penetrate within the plant interior. The surface of just one leaf is a very large habitat for any bacteria. The architecture of the leaf by itself is not uniform and provides areas of different environmental conditions. There are bulges and troughs formed by veins, leaf hair or trichomes, stomata, and hydathodes that form microsites for bacterial survival with increased water and nutrient availability, as well as temperature and UV radiation protection (Leveau and Lindow, 2001; Miller et al., 2001; Brandl and Amundson, 2008; Kroupitski et al., 2009; Barak et al., 2011). Indeed, distinct microcolonies or aggregates of S. enterica were found on cilantro leaf surfaces in the vein region (Brandl and Mandrell, 2002) In addition, preference to the abaxial side of lettuce leaf by S. enterica may be is an important strategy for UV avoidance (Kroupitski et al., 2011). Conversion of cells to viable but non-culturable (VNBC) state in E. coli O157:H7 on lettuce leaves may also be a strategy to escape harsh environmental conditions (Dinu and Bach, 2011). Hence, localization to favorable microsites, avoidance of harsh environments, and survival by aggregation or conversion to non-culturable state may allow these human pathogens to survive and at times multiply to great extent on the leaf surface.
As stomata are abundant natural pores in the plant epidermis which serve as entrance points for bacteria to colonize the leaf interior (intercellular space, xylem, and phloem), several studies addressed the question as to whether human bacterial pathogens could internalize leaves through stomata. Populations of E. coli O157:H7 and S. enterica SL1344 in the Arabidopsis leaf apoplast can be as large as four logs per cm2 of leaf after surface-inoculation under 60% relative humidity (Roy et al., 2013) suggesting that these bacteria can and access the apoplast of intact leaves. Several microscopy studies indicated association of pathogens on or near guard cells. For instance, S. enterica serovar Typhimurium SL1344 was shown to internalize arugula and iceberg lettuce through stomata and bacterial cells were located in the sub-stomatal space (Golberg et al., 2011). However, no internalization of SL1344 was observed into parsley where most cells were found on the leaf surface even though stomata were partially open (Golberg et al., 2011). Cells of S. enterica serovar Typhimurium MAE110 (Gu et al., 2011), enteroaggregative E. coli (Berger et al., 2009b), and E. coli O157:H7 (Saldaña et al., 2011) were found to be associated with stomata in tomato, arugula leaves, and baby spinach leaves, respectively. In the stem E. coli O157:H7 and Salmonella serovar Typhimurium were found to be associated with the hypocotyl and the stem tissues including epidermis, cortex, vascular bundles, and pith when seedlings were germinated from contaminated seeds (Deering et al., 2011a,b).
The plant rhizosphere is also a complex habitat for microorganisms with different life styles including plant beneficial symbionts and human pathogens. Nutritionally rich root exudate has been documented to attract S. enterica to lettuce roots (Klerks et al., 2007a). Although bacteria cannot directly penetrate through root cells, sites at the lateral root emergence and root cracks provide ports of entry for S. enterica and E. coli O157:H7 into root tissues (Cooley et al., 2003; Dong et al., 2003; Klerks et al., 2007b; Tyler and Triplett, 2008), and in some instances between the epidermal cells (Klerks et al., 2007b). High colonization of S. enterica has been observed in the root-shoot transition area (Klerks et al., 2007b). Once internalized both bacterial pathogens have been found in the intercellular space of the root outer cortex of Medicago truncatula (Jayaraman et al., 2014). Salmonella enterica was found in the parenchyma, endodermis, pericycle, and vascular system of lettuce roots (Klerks et al., 2007b) and in the inner root cortex of barley (Kutter et al., 2006). A detailed study on the localization of E. coli O157:H7 in live root tissue demonstrated that this bacterium can colonize the plant cell wall, apoplast, and cytoplasm (Wright et al., 2013). Intracellular localization of E. coli O157:H7 seems to be a rare event as most of the microscopy-based studies show bacterial cells in the intercellular space only. Bacterial translocation from roots to the phyllosphere may be by migration on the plant surface in a flagellum-dependent manner (Cooley et al., 2003) or presumably through the vasculature (Itoh et al., 1998; Solomon et al., 2002). The mechanism for internal movement of enteric bacterial cells from the root cortex to the root vasculature through the endodermis and casparian strips and movement from the roots to the phyllosphere through the vascular system is yet to be demonstrated.
Several outbreaks of S. enterica have also been associated with fruits, especially tomatoes. Salmonella enterica is unlikely to survive on surface of intact fruits (Wei et al., 1995) raising the question: what are the routes for human pathogenic bacteria penetration into fruits? It has been suggested that S. enterica can move from inoculated leaves (Barak et al., 2011), stems, and flowers (Guo et al., 2001) to tomato fruits. However, the rate of internal contamination of fruits was low (1.8%) when leaves were surface-infected with S. enterica (Gu et al., 2011). The phloem has been suggested as the route of movement of bacteria to non-inoculated parts of the plant as bacterial cells were detected in this tissue by microscopy (Gu et al., 2011). Figure 1 depicts the observed phyllosphere and rhizosphere niches colonized by bacteria in/on intact plants and probable sources of contamination.
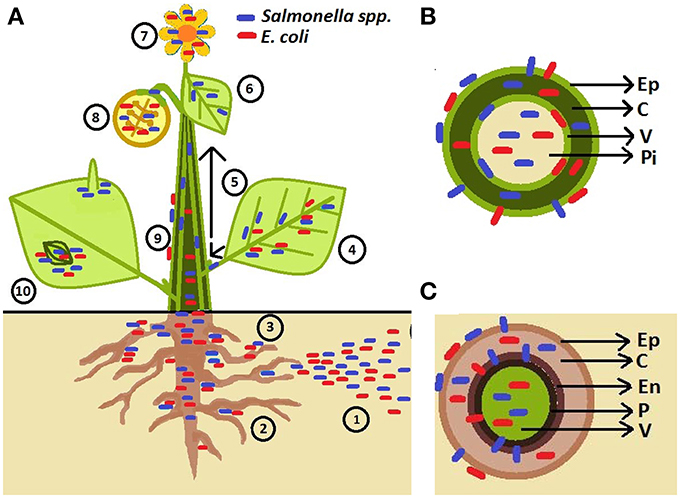
Figure 1. Schematic representation of human pathogen (HP) association with plants. (A) Pathogens are introduced to soil through contaminated irrigation water, fertilizers, manure, and pesticides (1). HPs are attracted to rhizosphere (2; Klerks et al., 2007a) and penetrate root tissues at the sites of lateral root emergence, root cracks as well as root-shoot transition area (3; Cooley et al., 2003; Dong et al., 2003; Klerks et al., 2007b; Tyler and Triplett, 2008). HPs were found to live on the leaf surface near veins (Brandl and Mandrell, 2002), in the leaf apoplast (intercellular space) (Brandl and Mandrell, 2002; Solomon et al., 2002; Niemira, 2007; Kroupitski et al., 2009; Barak et al., 2011; Dinu and Bach, 2011; Gu et al., 2011; Roy et al., 2013), and sometimes with affinity for abaxial side of leaf (e.g., S. enterica; (Kroupitski et al., 2011) (4). Salmonella enterica Typhimurium can enter tomato plants via leaves and move through vascular bundles (petioles and stems) (5) into non-inoculated leaves (6) and fruits (8) (Gu et al., 2011). HPs are also found to be associated with flower (7; Guo et al., 2001; Cooley et al., 2003). Salmonella could travel from infected leaves (4), stems (5), and flowers (7) to colonize the fruit interior (the diagram represents a cross-section of a fruit) and fruit calyx (8) Guo et al., 2001; Janes et al., 2005; Barak et al., 2011. Escherichia coli O157:H7 has also been observed in the internal parts of the apple and the seeds following contamination of the flower (8) (Burnett et al., 2000). Movement on the plant surface has also been observed (9; Cooley et al., 2003). Epiphytic Salmonella and E. coli O157:H7 can aggregate near stomata and sub-stomatal space (10; Shaw et al., 2008; Berger et al., 2009a,b; Golberg et al., 2011; Gu et al., 2011; Saldaña et al., 2011), reach the sub-stomatal cavity and survive/colonize in the spongy mesophyll (Solomon et al., 2002; Wachtel et al., 2002; Warriner et al., 2003; Jablasone et al., 2005; Franz et al., 2007). Salmonella cells were observed near trichomes (10; Barak et al., 2011; Gu et al., 2011). (B) Stem cross-section showing bacteria located in different tissues (Ep, epidermis; C, cortex; V, vascular tissue; Pi, pith) (Deering et al., 2011a,b). (C) Root cross-section showing bacteria on the root surface, internalizing between the epidermal cells, and colonizing root outer and inner cortex, endodermis (En), pericycle (P) and vascular system (Kutter et al., 2006; Klerks et al., 2007a,b; Jayaraman et al., 2014).
Plants possess a complex innate immune system to ward off microbial invaders (Jones and Dangl, 2006). Plants are able to mount a generalized step-one response that is triggered by modified/degraded plant products or conserved pathogen molecules. These molecules are known as damage or pathogen associated molecular patterns (DAMP/PAMP). In many cases, conserved PAMPs are components of cell walls and surface structures such as flagellin, lipopolysaccharides, and chitin (Zeng et al., 2010). Examples of intracellular PAMPs exist such as the elongation factor EF-Tu (Kunze et al., 2004). PAMPs are recognized by a diverse set of plant extracellular receptors called pattern-recognition receptors (PRRs) that pass intracellular signals launching an army of defense molecules to stop the invasion of the pathogens. This branch of the immune system known as pathogen-triggered immunity (PTI) is the first line of active defense against infection.
Human pathogen on plants (HPOP) is an emerging field that only recently has caught the attention of plant biologists and phytopathologists. A few studies have been reported in the last 5–10 years, which focused on the most well studied PAMPs, flagellin and lipopolysaccharide (LPS), in the interaction of human pathogens with plants. Table 1 lists the plants, bacterial strains, and method details for such studies.
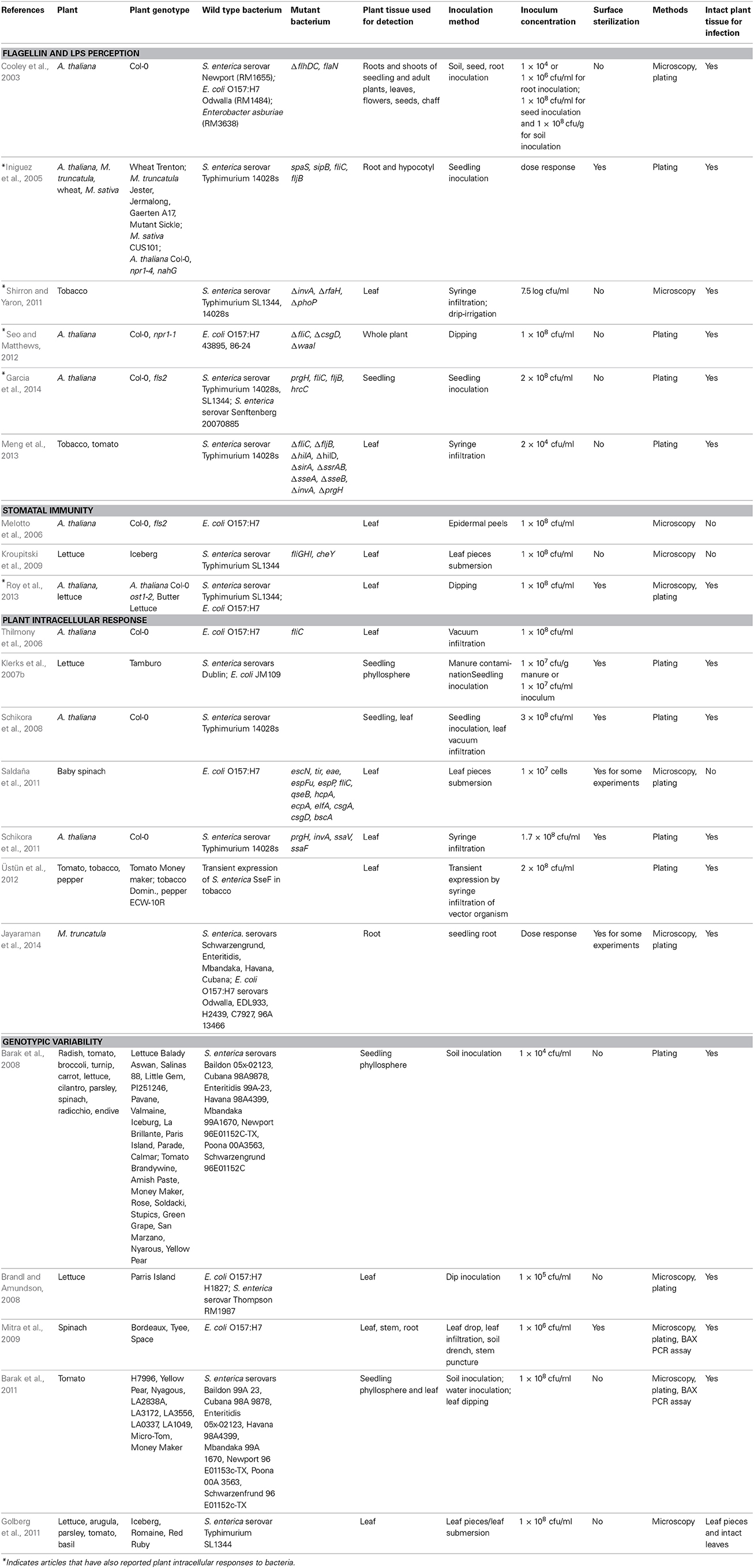
Table 1. Experimental conditions used in the studies reporting plant response to pathogenic Salmonella and E. coli.
Flagellin, the structural component of flagellum in bacteria, is involved in bacterial attachment and motility on the plant (Cooley et al., 2003), is recognized by plant through the FLS2 receptor (Garcia et al., 2014), and induces plant defenses (Meng et al., 2013; Garcia et al., 2014). Similar to the well-studied PTI elicitor flg22 (Felix et al., 1999), the flg22 epitope of S. enterica serovar Typhimurium 14028 is also an effective PAMP and elicitor of downstream immune responses in Arabidopsis (Garcia et al., 2014), tobacco, and tomato plants (Meng et al., 2013). Flagellum-deficient mutants of S. enterica serovar Typhimurium 14028 are better colonizers of wheat, alfalfa, and Arabidopsis roots as compared to the wild type bacterium (Iniguez et al., 2005) further suggesting that the Salmonella flagellum induces plant defenses that may restrict bacterial colonization of several plant organs. However, the Salmonella flg22 peptide is not the only PAMP for elicitation of plant immune response as fls2 mutant of Arabidopsis still shows a low level of PTI activation in response to this PAMP (Garcia et al., 2014).
Purified flagellin or derived epitopes of E. coli O157:H7 has not been used to induce plant defenses. However, flagellum-deficient mutant of this strain does not activate the SA-dependent BGL2 gene promoter as much as the wild type strain and shows larger population in Arabidopsis than the wild type strain (Seo and Matthews, 2012) further suggesting that surface structures in the bacterial cell are perceived by plants.
The differences in responses observed could be attributed to the presence of other microbial signatures eliciting plant defense. Variations in plant response to S. enterica flagellin could be owed to host-strain specificity as well. Although flagellin sequences from S. enterica strains and other bacteria are highly conserved, even a minor change of five amino acids in the flg22 epitope leads to reduced activation of PTI in Arabidopsis, tobacco, and tomato plants (Garcia et al., 2014). Adding to the specificity, it has also been shown that Brassicaceae and Solanocecae plants recognize specific flagellin (Robatzek et al., 2007; Clarke et al., 2013). Hence, evolving variations in flagellin sequences could be a strategy employed by the pathogens to avoid plant recognition, which in turn leads to the development of pathogen-specific immune responses in the plant.
Flagella also play an important role in bacterial behavior on the plant. Several studies have pointed out to the usefulness of flagella for attachment to leaf surfaces and movement on plant surfaces (Berger et al., 2009a,b; Xicohtencatl-Cortes et al., 2009; Saldaña et al., 2011; Shaw et al., 2011).
Lipopolysaccharide (LPS) is a component of the cell wall of Gram-negative bacterial pathogens of animals and plants. In the animal host, LPS is a well-characterized PAMP that is recognized by host Toll-like receptor 4 (de Jong et al., 2012). In plants however, receptors for LPS have not been discovered yet. Nonetheless, current evidence suggests that human pathogen-derived LPS can be perceived by plants resulting in PTI activation. For instance, on the leaf surface, purified LPS from Pseudomonas aeruginosa, S. Minnesota R595, and E. coli O55:B5 induces strong stomatal closure in Arabidopsis (Melotto et al., 2006). Purified LPS from Salmonella triggers of ROS production and extracellular alkalinization in tobacco cell suspension (Shirron and Yaron, 2011) but not on tomato leaves (Meng et al., 2013) suggesting that LPS recognition may be either dependent on experimental conditions or variable among plant species.
Genetic evidence suggests that the high activity of SA-dependent BGL2 gene promoter in Arabidopsis is dependent on the presence of LPS in E. coli O157:H7 as higher activity of this promoter was observed in the wild type bacterial as compared to its LPS mutant (Seo and Matthews, 2012). However, LPS-dependent responses seem not to be sufficient to restrict bacterial survival on plants as the population titer of E. coli O157:H7 LPS mutant or wild type in plant is essentially the same (Seo and Matthews, 2012). Additionally, live S. Typhimurium cells do not induce ROS in epidermal tissue of tobacco (Shirron and Yaron, 2011) suggesting that, at least Salmonella, can suppress LPS-induced ROS and extracellular alkalinization.
Similar to flagellin, the O-antigen moiety of LPS is not only important for plant perception of bacterial cells, but also for bacterial attachment, fitness, and survival on plants (Barak et al., 2007; Berger et al., 2011; Marvasi et al., 2013).
One of the earliest PTI responses in plants is stomatal closure that greatly decreases the rate of pathogen entry into plant's internal tissues. This response requires molecular components of PTI including such as flagellin and LPS perception and hormone perception and signaling (Melotto et al., 2006, 2008; Zeng and He, 2010; Sawinski et al., 2013). Stomatal immunity is also triggered by the presence of human pathogens S. enterica serovar Typhimurium SL1344 and E. coli O157:H7 (Melotto et al., 2006; Kroupitski et al., 2009; Roy et al., 2013), albeit at various levels. For instance, E. coli O157:H7 induces a strong stomatal immunity and Salmonella SL1344 elicits only a transient stomatal closure in both Arabidopsis (Melotto et al., 2006; Roy et al., 2013) and lettuce (Kroupitski et al., 2009; Roy et al., 2013) suggesting that the bacterial strain SL1344 can either induce weaker or subvert stomata-based defense. Active suppression of stomatal closure by SL1344 may be unlikely because it cannot re-open dark-closed stomata (Roy et al., 2013). However, it is possible that signaling pathways underlying bacterium-triggered and dark-induced stomatal closure are not entirely overlapping and SL1344 acts on immunity-specific signaling to subvert stomatal closure.
Recognition of PAMPs by PRRs leads to several hallmark cellular defense responses that are categorized based on the timing of response. Zipfel and Robatzek (2010) have discussed that early responses occur within seconds to minutes of recognition including ion fluxes, extracellular alkalinization, and oxidative burst. Intermediate responses occur within minutes to hours including stomatal closure, ethylene production, mitogen-activated protein kinase (MAPK) signaling, and transcriptional reprogramming. Late responses occur from hours to days and involve callose deposition, salicylic acid accumulation, and defense gene transcription.
These hallmark plant cellular defenses have also been tested for both E. coli and S. enterica (Figure 2). In particular, S. enterica infection results in the induction of MPK3/MPK6 kinase activity and plant defense-associated genes PDF1.2, PR1, and PR2 in Arabidopsis leaves (Schikora et al., 2008) as well as PR1, PR4, and PR5 in lettuce (Klerks et al., 2007b). MPK6 activation in Arabidopsis is independent of FLS2 (Schikora et al., 2008), indicating that flagellin is not the only active PAMP of Salmonella and plant response to other PAMPs may converge at MAPK signaling. Direct comparison of the PR1 gene expression in Arabidopsis indicated that both E. coli O157:H7 and Salmonella SL1344 are able to induce this defense marker gene, however at difference levels (Roy et al., 2013). The PR1 gene induction is low in SL1344-infected plants indicating that immune responses are either weaker or are suppressed by Salmonella.
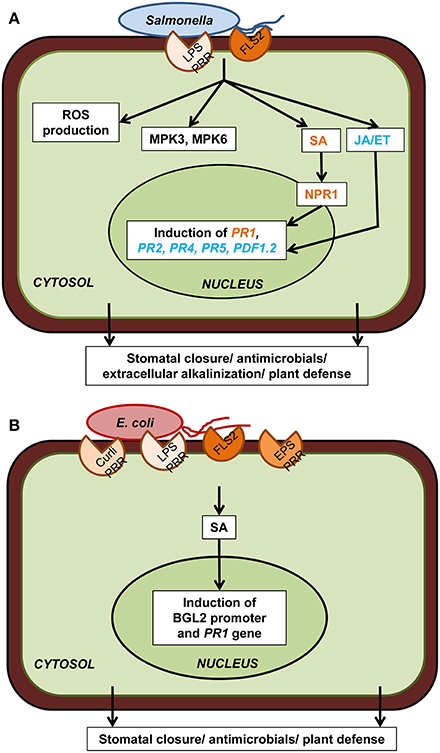
Figure 2. Plant cellular defense responses against human pathogens. (A) Upon reception of PAMP (flagellin, LPS) through PRR (FLS2 and putatively others), Salmonella spp. trigger downstream plant defense responses which include ROS production, MPK3/6, salicylic acid (SA) signaling through NPR1, jasmonic acid (JA) and ethylene (ET) signaling, defense-associated gene induction, and extracellular alkalinization. All these cellular events ultimately lead to stomatal closure, antimicrobial activity, and plant defense. (B) Escherichia coli PAMPs (curli, LPS, flagellin, EPS) are also perceived by PRRs (FLS2 and putatively others) present on plant cell surface which triggers the induction of the SA-dependent BGL2 promoter activity and PR1 gene expression. Only components that have been directly demonstrated experimentally are included in the diagram. Plant defense responses in case of both these human pathogens are strain specific as well as plant cultivar specific.
A few studies (Table 1) have addressed the role of plant hormones in response to endophytic colonization of human bacterial pathogens:
The ethylene-insensitive mutant of Arabidopsis, ein2, supports higher Salmonella 14028 inside whole seedlings as compared to the wild type Col-0 plants (Schikora et al., 2008). Furthermore, addition of a specific inhibitor of ethylene mediated signaling, 1-methylcyclopropene (1-MCP), to the growth medium resulted in increased S. enterica 14028 endophytic colonization of Medicago truncatula, but not M. sativum, roots and hypocotyls (Iniguez et al., 2005) suggesting that the role of endogenous ethylene signaling maybe be specific to each plant-bacterium interaction. However, ethylene signaling may play a contrasting role during fruit contamination. Tomato mutants (rin and nor) with defects in ethylene synthesis, perception, and signal transduction show significantly reduced Salmonella proliferation within their fruits as compared to the wild type control (Marvasi et al., 2014).
Similar to the ein2 mutant, the coronatine-insensitive mutant of Arabidopsis, coi1-16, also supports high Salmonella 14028 inside whole seedlings (Schikora et al., 2008). Along with the induction of the jasmonate-responsive gene PDF1.2 addressed in the same study and mentioned above, it seems that jasmonate signaling is also an important component to restrict Salmonella infection in, at least, Arabidopsis. These results are surprising as coi1 mutants are well known to have increased resistant to various bacterial pathogen of plants, such as P. syringae, but not to fungal or viral pathogens (Feys et al., 1994; Kloek et al., 2001).
Two genetic lines of Arabidopsis has been extensively used to determine the role of salicylic acid (SA) in plant defenses against phytopathogens, the transgenic nahG plant that cannot accumulate SA (Friedrich et al., 1995) and the null mutant npr1 that is disrupted in both SA-dependent and -independent defense responses (Ton et al., 2002). Both of these plant lines support higher populations of Salmonella 14028 inside their roots (Iniguez et al., 2005) and seedlings (Schikora et al., 2008) as compared to the wild type plant. NPR1-dependent signaling is important reduce the population of the curli-negative strain of E. coli O157:H7 43895 but not for the curli-positive strain 86-24 in Arabidopsis leaves (Seo and Matthews, 2012). Although only a few strains of Salmonella and E. coli have been used, there is an emerging patterns suggesting that SA itself and activation of SA-signaling can potentially restrict HPOP.
In attempts to understand the overall cellular transcriptional response to human bacterial pathogens, global transcriptomic analyses have been used. Thilmony et al. (2006) showed that E. coli O157:H7 regulates PTI-associated genes in Arabidopsis leaves, albeit in a flagellin-independent manner. A similar transcriptomic analysis with medium-grown Arabidopsis seedlings 2h after inoculation with S. enterica serovar Typhimurium 14028, E. coli K-12, and P. syringae pv. tomato DC3000 showed a strong overlap among genes responsive to each bacterial infection suggesting a common mechanism of plant basal response toward bacteria (Schikora et al., 2011). Gene expression analysis of Medicago truncatula seedlings root-inoculated with only two bacterial cells per plant indicated that 83 gene probes (30–40% of each data set) were commonly regulated in response to S. enterica and E. coli O157:H7 (Jayaraman et al., 2014). All together, these studies indicate that each human pathogenic bacterium can modulate specific plant genes beyond a basal defense response; however the mechanisms for plant-bacterium specificity are largely unknown.
Successful virulent pathogens of plants are able to defeat this army plant defense by employing its own set of artillery (such as the type three secretion system effectors and phytotoxins) and cause disease in the host plant (Melotto and Kunkel, 2013; Xin and He, 2013). In incompatible interactions (i.e., low bacterial colonization and no disease on leaves), the host plant already has pre-evolved molecules (R proteins) that recognize these effectors and cause a specific defense response to this pathogen. This specific response is called effector-triggered immunity (ETI). Because the type 3-secretion system (T3SS) is important for the virulence of both animal and plant pathogenic bacteria on their natural hosts as evidenced by the use of bacterial mutants, it is reasonable to expect that T3SS would be important for HPOP as well. However, animal and plant cell surfaces are structurally different; the plant cells wall seems to be impenetrable by the secretion needle of the extracellular animal pathogens (Salmonella and E. coli) as discussed by He et al. (2004) raising the question of how these effectors can reach the plant cytoplasm and interfere with plant defenses. To date, there is no evidence for the ability of human pathogens to inject T3SS effectors inside plant cells. It is possible that the T3SS is still active on the plant cell surface and the effectors are secreted into the plant apoplast. If that is the case, however, plant membrane receptors would be necessary to recognize the effectors and trigger plant cellular responses. Nevertheless, it has been observed that the T3SS mutant of E. coli O157:H7, escN, has reduced ability to attach to and colonize baby spinach leaves similar to the fliC mutant (Saldaña et al., 2011). Furthermore, apoplastic population of T3SS structural mutants of S. enterica serovar Typhimurium 14028 (invA, prgH, ssaV, and ssaJ) is smaller than that of the wild type bacterium in Arabidopsis leaves (Schikora et al., 2011) and plant defense-associated genes are up-regulated for longer time by the prgH mutant than wild type Salmonella in Arabidopsis seedlings (Garcia et al., 2014). Contrary to these findings, Iniguez et al. (2005) reported that two Salmonella 14028 T3SS-SPI1, the structural mutant spaS and the effector mutant sipB, hypercolonize roots and hypocotyls of M. sativum and fail to induce SA-dependent PR1 promoter in Arabidopsis leaves. More studies need to be conducted to conclude whether T3SS of Salmonella acts as “recognizable” surface structure similar to flagellum and/or as a conduit to deliver effectors in plant tissues and trigger ETI. It is worth mentioning that T3SS and effectors of the phytopathogen P. syringae pv. syringae have functions on ETI as well as bacterial fitness on plant surface (Lee et al., 2012) and the filamentous T3SS protein EspA is required for E. coli O157:H7 attachment to arugula leaves (Shaw et al., 2008).
The invA structural mutant, that is defective in all T3SS-1 system-associated phenotypes, induces high ROS and extracellular alkalinizing in tobacco BY-2 cell suspension and hypersensitive reaction (HR) in tobacco leaves as compared to the wild type strain (Shirron and Yaron, 2011) suggesting that T3SS is important for this suppression of immunity. However, Shirron and Yaron (2011) also reported that plant response to the regulatory mutant phoP that modulates the expression of many effector proteins and membrane components (Dalebroux and Miller, 2014), is no different to that of the wild type bacterium. These findings raised the question whether the phenotypes observed are due to the T3SS structure itself or due to the translocated effectors. A recent report shows that transient expression of the type three effector of Salmonella 14028 SseF in tobacco plants elicits HR, and this response is dependent on the SGT1 protein (Üstün et al., 2012). This study suggests that SseF can induce resistant-like response in plants and requires resistance (R) protein signaling components. Üstün et al. (2012) and Shirron and Yaron (2011) also showed that Salmonella 14028, which is able to deliver the SseF effector, cannot induce HR or any disease-like symptoms in tobacco leaves. Thus, it remains to be determined what would be the biological relevance of ETI in the Salmonella and other human pathogenic bacteria in their interaction with plants in nature.
Although S. enterica and E. coli O157:H7 have not been traditionally known to be closely associated with plants and modulate plant's physiology, the evidence tells us otherwise. An arms-race evolution in both the human pathogen and the plant is therefore, expected. A few studies (methodology details described in Table 1) have addressed whether genetic variability among plant species or within the same plant species (i.e., cultivars, varieties, and ecotypes) can be correlated with differential bacterial behavior and/or colonization of plants. Barak et al. (2011) described that different tomato cultivars can harbor different levels of S. enterica population after inoculation via water (sprinkler imitation) indicating plant factors may control the ability of bacterial to colonize the phyllosphere. However, they also found that the cultivar with the smallest S. enterica population also had the lowest number of speck lesions when infected with the tomato pathogen Pst DC3000 (Barak et al., 2011), suggesting that strong basal defense in this cultivar may account for low bacterial colonization. On a comparative study of S. enterica contamination of several crop species, Barak et al. (2008) reported that seedlings from Brassicaceae family have higher contamination than carrot, tomato, and lettuce when grown on contaminated soil. Seedling contamination correlated with the Salmonella population in the phyllosphere of all crop species, except tomato.
Golberg et al. (2011) reported variations in internalization of Salmonella SL1344 in different leafy vegetables and fresh herbs using confocal microscopy. Internalization incidence (% of microscopic fields containing bacterial cells) was high in iceberg lettuce and arugula, moderate in romaine lettuce, red lettuce, basil, and low in parsley and tomato. Attraction to stomata was seen in iceberg lettuce and basil, not in arugula, parsley, and tomato. Brandl and Amundson (2008) reported that the age of romaine lettuce leaves is correlated with population size of E. coli O157:H7 and S. enterica Thompson on leaves. Young leaves (inner) harbor greater number of cells than middle aged leaves. These authors also observed that exudates on the surface of younger leaves have higher nitrogen content than that of older leaves, which may contribute to determining the bacterial population size on the leaf. Thus, it is tempting to speculate that the genetic variability existent among plant genotypes regarding the chemical composition of their organ exudates may be a determinant for human pathogen behavior (such as chemotaxis and tropism toward stomata and roots) and ability to colonize plants.
Finally, Mitra et al. (2009) studied the effect of different methods of inoculation on internalization and survival of E. coli O157:H7 in three cultivars of spinach. Among the organs studied, the spinach phylloplane and the stem provided the most and least suitable niche for this bacterium colonization, respectively. Although the leaf surface was the best “territory” for E. coli, the leaf morphologies of each cultivar affected the ability of this bacterium to survive.
Collectively, all these studies point out that the plant genotype, age, leaf morphology, chemical composition of exudates, and the primarily infected organ affect the outcome of bacterial colonization of plants and the process may not be a generalized phenomenon, consequently shaping specific human pathogen and plant interactions.
The fundamental understanding of plant association with human bacterial pathogens that do not cause visual or macroscopic symptom in the plant, but yet are major food contaminants, are in its infancy. Both plant and bacterial factors are critical for these cross-kingdom interactions and emerging evidence suggests an overlap between plant molecular responses to human pathogens and phytopathogens. The future challenge will be to determine how these interactions differ. As this field of research is relatively new, we see differences in conclusions from different laboratories regarding multiplication vs. decline in bacterial populations overtime and disease-like symptoms vs. HR on inoculated plants. These differences are mainly associated with differences in methods of inoculation, bacterial strains, inoculum concentration, plant age, and plant cultivation methods (e.g., growth on medium, soil, or hydroponic solutions). Standard procedures for model systems, consensus, and collaborations must be developed among food scientists, microbiologists, plant pathologists, and molecular biologists to elucidate the specificity of each plant-bacterium interaction and avoid discrepancies in making general conclusions. A major point to be resolved is whether the observed plant defenses against Salmonella and its PAMPs are due to low recognition and/or active suppression. If Salmonella suppression of the plant immunity is a cause of weak defense responses, the major question becomes what is the responsible factor? This line of research might lead to a whole new paradigm that otherwise could not be revealed by only studying plant associations with its own natural pathogens.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Work in our laboratory is supported by funding from the US National Institute of Allergy and Infectious Disease (R01AI068718).
Aruscavage, D., Miller, S. A., Lewis Ivey, M. L., Lee, K., and LeJeune, J. T. (2008). Survival and dissemination of Escherichia coli O157:H7 on physically and biologically damaged lettuce plants. J. Food Prot. 71, 2384–2388. doi: 10.1111/j.1750-3841.2006.00157.x
Barak, J. D., Jahn, C. E., Gibson, D. L., and Charkowski, A. O. (2007). The role of cellulose and O-antigen capsule in the colonization of plants by Salmonella enterica. Mol. Plant Microbe Interact. 20, 1083–1091. doi: 10.1094/MPMI-20-9-1083
Barak, J. D., Kramer, L. C., and Hao, L. (2011). Colonization of tomato plants by Salmonella enterica is cultivar dependent, and type 1 trichomes are preferred colonization sites. Appl. Environ. Microbiol. 77, 498–504. doi: 10.1128/AEM.01661-10
Barak, J. D., Liang, A., and Narm, K.-E. (2008). Differential attachment and subsequent contamination of agricultural crops by Salmonella enterica. Appl. Environ. Microbiol. 74, 5568–5570. doi: 10.1128/AEM.01077-08
Barak, J. D., and Schroeder, B. K. (2012). Interrelationships of food safety and plant pathology: the life cycle of human pathogens on plants. Annu. Rev. Phytopathol. 50, 241–266. doi: 10.1146/annurev-phyto-081211-172936
Berger, C. N., Brown, D. J., Shaw, R. K., Minuzzi, F., Feys, B., and Frankel, G. (2011). Salmonella enterica strains belonging to O serogroup 1, 3, 9 induce chlorosis and wilting of Arabidopsis thaliana leaves. Environ. Microbiol. 13, 1299–1308. doi: 10.1111/j.1462-2920.2011.02429.x
Berger, C. N., Shaw, R. K., Brown, D. J., Mather, H., Clare, S., Dougan, G., et al. (2009a). Interaction of Salmonella enterica with basil and other salad leaves. ISME J. 3, 261–265. doi: 10.1038/ismej.2008.95
Berger, C. N., Shaw, R. K., Ruiz-Perez, F., Nataro, J. P., Henderson, I. R., Pallen, M. J., et al. (2009b). Interaction of enteroaggregative Escherichia coli with salad leaves. Environ. Microbiol. Rep. 1, 234–239. doi: 10.1111/j.1758-2229.2009.00037.x
Berger, C. N., Sodha, S. V., Shaw, R. K., Griffin, P. M., Pink, D., Hand, P., et al. (2010). Fresh fruit and vegetable as vehicles for the transmission of human pathogens. Environ. Microbiol. 12, 2385–2397. doi: 10.1111/j.1462-2920.2010.02297.x
Brandl, M. T., and Amundson, R. (2008). Leaf age as a risk factor in contamination of lettuce with Escherichia coli O157:H7 and Salmonella enterica. Appl. Environ. Microbiol. 74, 2298–2306. doi: 10.1128/AEM.02459-07
Brandl, M. T., Cox, C. E., and Teplitski, M. (2013). Salmonella interactions with plants and their associated microbiota. Phytopathology 103, 316–325. doi: 10.1094/PHYTO-11-12-0295-RVW
Brandl, M. T., and Mandrell, R. E. (2002). Fitness of Salmonella enterica serovar Thompson in the cilantro phyllosphere. Appl. Environ. Microbiol. 68, 3614–3621. doi: 10.1128/AEM.68.7.3614-3621.2002
Burnett, S. L., Chen, J., and Beuchat, L. R. (2000). Attachment of Escherichia coli O157:H7 to the surfaces and internal structures of apples as detected by confocal scanning laser microscopy. Appl. Environ. Microbiol. 66, 4679–4687. doi: 10.1128/AEM.66.11.4679-4687.2000
CDC. (2010). Surveillance for foodborne disease outbreaks: United States, 2007. Morb. Mortal. Wkly. Rep. 59, 973–1008. Available online at: http://www.cdc.gov/mmwr/preview/mmwrhtml/mm5931a1.htm?s_cid=mm5931a1_w
Clarke, C. R., Chinchilla, D., Hind, S. R., Taguchi, F., Miki, R., Ichinose, Y., et al. (2013). Allelic variation in two distinct Pseudomonas syringae flagellin epitopes modulates the strength of plant immune responses but not bacterial motility. New Phytol. 200, 847–680. doi: 10.1111/nph.12408
Cooley, M. B., Miller, W. G., and Mandrell, R. E. (2003). Colonization of Arabidopsis thaliana with Salmonella enterica and enterohemorrhagic Escherichia coli O157:H7 and competition by Enterobacter asburiae. Appl. Environ. Microbiol. 69, 4915–4926. doi: 10.1128/AEM.69.8.4915-4926.2003
Dalebroux, Z. D., and Miller, S. I. (2014). Salmonellae PhoPQ regulation of the outer membrane to resist innate immunity. Curr. Opin. Microbiol. 17, 106–113. doi: 10.1016/j.mib.2013.12.005
Deering, A. J., Pruitt, R. E., Mauer, L. J., and Reuhs, B. L. (2011a). Identification of the cellular location of internalized Escherichia coli O157:H7 in mung bean, Vigna radiata, using immunocytochemical techniques. J. Food Prot. 74, 1224–1230. doi: 10.4315/0362-028X.JFP-11-015
Deering, A. J., Pruitt, R. E., Mauer, L. J., and Reuhs, B. L. (2011b). Examination of the internalization of Salmonella serovar Typhimurium in peanut, Arachis hypogaea, using immunocytochemical techniques. Food Res. Int. 45, 1037–1043. doi: 10.1016/j.foodres.2011.01.061
de Jong, H. K., Parry, C. M., van der Poll, T., and Wiersinga, W. J. (2012). Host-pathogen interaction in invasive Salmonellosis. PLoS Pathog. 8:e1002933. doi: 10.1371/journal.ppat.1002933
Dinu, L. D., and Bach, S. (2011). Induction of viable but nonculturable Escherichia coli O157:H7 in the phyllosphere of lettuce: a food safety risk factor. Appl. Environ. Microbiol. 77, 8295–8302. doi: 10.1128/AEM.05020-11
Dong, Y., Iniguez, A. L., Ahmer, B. M., and Triplett, E. W. (2003). Kinetics and strain specificity of rhizosphere and endophytic colonization by enteric bacteria on seedlings of Medicago sativa and Medicago truncatula. Appl. Environ. Microbiol. 69, 1783–1790. doi: 10.1128/AEM.69.3.1783-1790.2003
Felix, G., Duran, J. D., Volko, S., and Boller, T. (1999). Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18, 265–276. doi: 10.1046/j.1365-313X.1999.00265.x
Feys, B. J. F., Benedetti, C. E., Penfold, C. N., and Turner, J. G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6, 751–759. doi: 10.1105/tpc.6.5.751
Franz, E., Visser, A. A., Van Diepeningen, A. D., Klerks, M. M., Termorshuizen, A. J., and van Bruggen, A. H. (2007). Quantification of contamination of lettuce by GFP-expressing Escherichia coli O157:H7 and Salmonella enterica serovar Typhimurium. Food Microbiol. 24, 106–112. doi: 10.1016/j.fm.2006.03.002
Friedrich, L., Vernooij, B., Gaffney, T., Morse, A., and Ryals, J. (1995). Characterization of tobacco plants expressing a bacterial salicylate hydroxylase gene. Plant Mol. Biol. 29, 959–968. doi: 10.1007/BF00014969
Garcia, A. V., Charrier, A., Schikora, A., Bigeard, J., Pateyron, S., de Tauzia-Moreau, M., et al. (2014). Salmonella enterica flagellin is recognized via FLS2 and activates PAMP-triggered immunity in Arabidopsis thaliana. Mol. Plant 7, 657–674. doi: 10.1093/mp/sst145
Golberg, D., Kroupitski, Y., Belausov, E., Pinto, R., and Sela, S. (2011). Salmonella Typhimurium internalization is variable in leafy vegetables and fresh herbs. Int. J. Food Microbiol. 145, 250–257. doi: 10.1016/j.ijfoodmicro.2010.12.031
Gu, G., Hu, J., Cevallos-Cevallos, J. M., Richardson, S. M., Bartz, J. A., and van Bruggen, A. H. C. (2011). Internal colonization of Salmonella enterica serovar Typhimurium in tomato plants. PLoS ONE 6:e27340. doi: 10.1371/journal.pone.0027340
Guo, X., Chen, J., Brackett, R. E., and Beuchat, L. R. (2001). Survival of Salmonellae on and in tomato plants from the time of inoculation at flowering and early stages of fruit development through fruit ripening. Appl. Environ. Microbiol. 67, 4760–4764. doi: 10.1128/AEM.67.10.4760-4764.2001
He, S. Y., Nomura, K., and Whittam, T. S. (2004). Type III protein secretion mechanism in mammalian and plant pathogens. Biochim. Biophys. Acta 1694, 181–206. doi: 10.1016/j.bbamcr.2004.03.011
Hussain, M. A., and Dawson, C. O. (2013). Economic impact of food safety outbreaks on food businesses. Foods 2, 585–589. doi: 10.3390/foods2040585
Iniguez, A. L., Dong, Y., Carter, H. D., Ahmer, B. M. M., Stone, J. M., and Triplett, E. W. (2005). Regulation of enteric endophytic bacterial colonization by plant defenses. Mol. Plant-Microbe Interact. 18, 169–178. doi: 10.1094/MPMI-18-0169
Itoh, Y., Sugita-Konishi, Y., Kasuga, F., Iwaki, M., Hara-Kudo, Y., Saito, N., et al. (1998). Enterohemorrhagic Escherichia coli O157:H7 present in radish sprouts. Appl. Environ. Microbiol. 64, 1532–1535.
Jablasone, J., Warriner, K., and Griffiths, M. (2005). Interactions of Escherichia coli O157:H7, Salmonella typhimurium and Listeria monocytogenes plants cultivated in agnotobiotic system. Int. J. Food Microbiol. 99, 7–18. doi: 10.1016/j.ijfoodmicro.2004.06.011
Janes, M. E., Kim, K. S., and Johnson, M. G. (2005). Transmission electron microscopy study of enterohemorrhagic Escherichia coli O157:H7 in apple tissue. J. Food Prot. 68, 216–224.
Jayaraman, D., Valdés-López, O., Kaspar, C. W., and Ané, J.-M. (2014). Response of Medicago truncatula Seedlings to Colonization by Salmonella enterica and Escherichia coli O157:H7. PLoS ONE 9:e87970. doi: 10.1371/journal.pone.0087970
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Klerks, M. M., Franz, E., vanGent-Pelzer, M., Zijlstra, C., and van Bruggen, A. H. C. (2007a). Differential interaction of Salmonella enterica serovars with lettuce cultivars and plant-microbe factors influencing the colonization efficiency. ISME J. 1, 620–631. doi: 10.1038/ismej.2007.82
Klerks, M. M., vanGent-Pelzer, M., Franz, E., Zijlstra, C., and van Bruggen, A. H. C. (2007b). Physiological and molecular responses of Lactuca sativa to colonization by Salmonella enterica serovar Dublin. Appl. Environ. Microbiol. 73, 4905–4914. doi: 10.1128/AEM.02522-06
Kloek, A. P., Verbsky, M. L., Sharma, S. B., Schoelz, J. E., Vogel, J., Klessig, D. F., et al. (2001). Resistance to Pseudomonas syringae conferred by an Arabidopsis thaliana coronatine-insensitive (coi1) mutation occurs through two distinct mechanisms. Plant J. 26, 509–522. doi: 10.1046/j.1365-313x.2001.01050.x
Kroupitski, Y., Golberg, D., Belausov, E., Pinto, R., Swartzberg, D., Granot, D., et al. (2009). Internalization of Salmonella enterica in leaves is induced by light and involves chemotaxis and penetration through open stomata. Appl. Environ. Microbiol. 75, 6076–6086. doi: 10.1128/AEM.01084-09
Kroupitski, Y., Pinto, R., Belausov, E., and Sela, S. (2011). Distribution of Salmonella typhimurium in romaine lettuce leaves. Food Microbiol. 28, 990–997. doi: 10.1016/j.fm.2011.01.007
Kunze, G., Zipfel, C., Robatzek, S., Niehaus, K., Boller, T., and Felix, G. (2004). The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16, 3496–3507. doi: 10.1105/tpc.104.026765
Kutter, S., Hartmann, A., and Schmid, M. (2006). Colonization of barley (Hordeum vulgare) with Salmonella enterica and Listeria spp. FEMS Microbiol. Ecol. 56, 262–271. doi: 10.1111/j.1574-6941.2005.00053.x
Lee, J., Teitzel, G. M., Munkvold, K., del Pozo, O., Martin, G. B., Michelmore, R. W., et al. (2012). Type III secretion and effectors shape the survival and growth pattern on Pseudomonas syringae on leaf surfaces. Plant Physiol. 158, 1803–1818. doi: 10.1104/pp.111.190686
Leveau, J. H., and Lindow, S. E. (2001). Appetite of an epiphyte: quantitative monitoring of bacterial sugar consumption in the phyllosphere. Proc. Natl. Acad. Sci. U.S.A. 98, 3446–3453. doi: 10.1073/pnas.061629598
Lindow, S. E., and Brandl, M. T. (2003). Microbiology of the phyllosphere. Appl. Environ. Microbiol. 69, 1875–1883. doi: 10.1128/AEM.69.4.1875-1883.2003
Lynch, M. F., Tauxe, R. V., and Hedberg, C. W. (2009). The growing burden of foodborne outbreaks due to contaminated fresh produce: risks and opportunities. Epidemiol. Infect. 137, 307–315. doi: 10.1017/S0950268808001969
Marvasi, M., Cox, C. E., Xu, Y., Noel, J. T., Giovannoni, J. J., and Teplitski, M. (2013). Differential regulation of Salmonella typhimurium genes involved in O-antigen capsule production and their role in persistence within tomato fruit. Mol. Plant-Microbe Interact. 26, 793–800. doi: 10.1094/MPMI-09-12-0208-R
Marvasi, M., Noel, J. T., George, A. S., Farias, M. A., Jenkins, K. T., Hochmuth, G., et al. (2014). Ethylene signaling affects susceptibility of tomatoes to Salmonella. Microb. Biotechnol. doi: 10.1111/1751-7915.12130. [Epub ahead of print].
Melotto, M., and Kunkel, B. N. (2013). “Virulence strategies of plant pathogenic bacteria,” in The Prokaryotes—Prokaryotic Physiology and Biochemistry, 4th Edn., eds E. Rosenberg, E. Stackebrand, E. F. DeLong, F. Thompson, and S. Lory (Berlin; Heidelberg: Springer-Verlag), 61–82.
Melotto, M., Underwood, W., and He, S. Y. (2008). Role of stomata in plant innate immunity and foliar bacterial diseases. Ann. Rev. Phytopathol. 46, 101–122. doi: 10.1146/annurev.phyto.121107.104959
Melotto, M., Underwood, W., Koczan, J., Nomura, K., and He, S. Y. (2006). Plant stomata function in innate immunity against bacterial invasion. Cell 126, 969–980. doi: 10.1016/j.cell.2006.06.054
Meng, F., Altier, C., and Martin, G. B. (2013). Salmonella colonization activates the plant immune system and benefits from association with plant pathogenic bacteria. Environ. Microbiol. 9, 2418–2430. doi: 10.1111/1462-2920.12113
Miller, W. G., Brandl, M. T., Quiñones, B., and Lindow, S. E. (2001). Biological sensor for sucrose availability: relative sensitivities of various reporter genes. Appl. Environ. Microbiol. 67, 1308–1317. doi: 10.1128/AEM.67.3.1308-1317.2001
Mitra, R., Cuesta-Alonso, E., Wayadande, A., Talley, J., Gilliland, S., and Fletcher, J. (2009). Effect of route of introduction and host cultivar on the colonization, internalization, and movement of the human pathogen Escherichia coli O157:H7 in spinach. J. Food Prot. 72, 1521–1530.
Niemira, B. A. (2007). Relative efficacy of sodium hypochlorite wash versus irradiation to inactivate Escherichia coli O157:H7 internalized in leaves of romaine lettuce and baby spinach. J. Food Prot. 70, 2526–2532.
Ribera, L. A., Palma, M. A., Paggi, M., Knutson, R., Masabni, J. G., and Anciso, J. (2012). Economic analysis of food safety compliance costs and foodborne illness outbreaks in the United States. Hort Technol. 22, 150–156.
Robatzek, S., Bittel, P., Chinchilla, D., Köchner, P., Felix, G., Shiu, S.-H., et al. (2007). Molecular identification and characterization of the tomato flagellin receptor LeFLS2, an orthologue of Arabidopsis FLS2 exhibiting characteristically different perception specificities. Plant Mol. Biol. 64, 539–547. doi: 10.1007/s11103-007-9173-8
Roy, D., Panchal, S., and Melotto, M. (2013). Escherichia coli O157:H7 induces stronger plant immunity than Salmonella enterica Typhimurium SL1344. Phytopathology 103, 326–332. doi: 10.1094/PHYTO-09-12-0230-FI
Saldaña, Z., Sánchez, E., Xicohtencatl-Cortes, J., Puente, J. L., and Girón, J. A. (2011). Surface structures involved in plant stomata and leaf colonization by shiga-toxigenic Escherichia coli O157:H7. Front. Microbiol. 2:119. doi: 10.3389/fmicb.2011.00119
Sawinski, K., Mersmann, S., Robatzek, S., and Böhmer, M. (2013). Guarding the green: pathways to stomatal immunity. Mol. Plant-Microbe Interact. 26, 626–632. doi: 10.1094/MPMI-12-12-0288-CR
Scallan, E., Hoekstra, R. M., Angulo, F. J., Tauxe, R. V., Widdowson, M. A., Roy, S. L., et al. (2011). Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17, 7–15. doi: 10.3201/eid1701.P11101
Schikora, A., Carreri, A., Charpentier, E., and Hirt, H. (2008). The dark side of the salad: Salmonella Typhimurium overcomes the innate immune response of Arabidopsis thaliana and shows an endopathogenic lifestyle. PLoS ONE 3:e2279. doi: 10.1371/journal.pone.0002279
Schikora, A., Virlogeux-Payant, I., Bueso, E., Garcia, A. V., Nilau, T., Charrier, A., et al. (2011). Conservation of Salmonella infection mechanisms in plants and animals. PLoS ONE 6:e24112. doi: 10.1371/journal.pone.0024112
Seo, K. H., and Frank, J. F. (1999). Attachment of Escherichia coli O157:H7 to lettuce leaf surface and bacterial viability in response to chlorine treatment as demonstrated by using confocal scanning laser microscopy. J. Food Prot. 62, 3–9.
Seo, S., and Matthews, K. R. (2012). Influence of the plant defense response to Escherichia coli O157:H7 cell surface structures on survival of that enteric pathogen on plant surfaces. Appl. Environ. Microbiol. 78, 5882–5889. doi: 10.1128/AEM.01095-12
Shaw, R. K., Berger, C. N., Feys, B., Knutton, S., Pallen, M. J., and Frankel, G. (2008). Enterohemorrhagic Escherichia coli exploits EspA filaments for attachment to salad leaves. Appl. Environ. Microbiol. 74, 2908–2914. doi: 10.1128/AEM.02704-07
Shaw, R. K., Berger, C. N., Pallen, M. J., Sjöling, A., and Frankel, G. (2011). Flagella mediate attachment of enterotoxigenic Escherichia coli to fresh salad leaves. Environ. Microbiol. Rep. 3, 112–117. doi: 10.1111/j.1758-2229.2010.00195.x
Shirron, N., and Yaron, S. (2011). Active suppression of early immune response in tobacco by the human pathogen Salmonella Typhimurium. PLoS ONE 6:e18855. doi: 10.1371/journal.pone.0018855
Solomon, E. B., Yaron, S., and Matthews, K. R. (2002). Transmission of Escherichia coli O157:H7 from contaminated manure and irrigation water to lettuce plant tissue and its subsequent internalization. Appl. Environ. Microbiol. 68, 397–400. doi: 10.1128/AEM.68.1.397-400.2002
Üstün, S., Müller, P., Palmisano, R., Hensel, M., and Börnke, F. (2012). SseF, a type III effector protein from the mammalian pathogen Salmonella enterica, requires resistance-gene-mediated signaling to activate cell death in the model plant Nicotiana benthamiana. New Phytol. 194, 1046–1060. doi: 10.1111/j.1469-8137.2012.04124.x
Teplitski, M., Barak, J. D., and Schneider, K. R. (2009). Human enteric pathogens in produce: un-answered ecological questions with direct implications for food safety. Curr. Opin. Biotechnol. 20, 166–171. doi: 10.1016/j.copbio.2009.03.002
Thilmony, R., Underwood, W., and He, S. Y. (2006). Genome-wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringae pv. tomato DC3000 and the human pathogen Escherichia coli O157:H7. Plant J. 46, 34–53. doi: 10.1111/j.1365-313X.2006.02725.x
Ton, J., De Vos, M., Robben, C., Buchala, A., Métraux, J. P., Van Loon, L. C., et al. (2002). Characterization of Arabidopsis enhanced disease susceptibility mutants that are affected in systemically induced resistance. Plant J. 29, 11–21. doi: 10.1046/j.1365-313x.2002.01190.x
Tyler, H. L., and Triplett, E. W. (2008). Plants as a habitat for beneficial and/or human pathogenic bacteria. Annu. Rev. Phytopathol. 46, 53–73. doi: 10.1146/annurev.phyto.011708.103102
Wachtel, M. R., Whitehand, L. C., and Mandrell, R. E. (2002). Association of Escherichia coli O157:H7 with preharvest leaf lettuce upon exposure to contaminated irrigation water. J. Food Prot. 65, 18–25.
Warriner, K., Ibrahim, F., Dickinson, M., Wright, C., and Waites, W. M. (2003). Interaction of Escherichia coli with growing salad spinach plants. J. Food Prot. 66, 1790–1797.
Wei, C. I., Huang, J. M., Lin, W. F., Tamplin, M. L., and Bartz, J. A. (1995). Growth and survival of Salmonella Montevideo on tomatoes and disinfection with chlorinated water. J. Food Prot. 8, 829–836.
Wright, K. M., Chapman, S., McGeachy, K., Humphrism, S., Campbell, E., Toth, I. K., et al. (2013). The endophytic lifestyle of Escherichia coli O157:H7: quantification and internal localization in roots. Phytopathology 103, 333–340. doi: 10.1094/PHYTO-08-12-0209-FI
Xicohtencatl-Cortes, J., Sánchez Chacón, E., Saldaña, Z., Freer, E., and Girón, J. A. (2009). Interaction of Escherichia coli O157:H7 with leafy green produce. J. Food Prot. 72, 1531–1537.
Xin, X.-F., and He, S. Y. (2013). Pseudomonas syringae pv. tomato DC3000: a model pathogen for probing disease susceptibility and hormone signaling in plants. Annu. Rev. Phytopathol. 51, 473–498. doi: 10.1146/annurev-phyto-082712-102321
Zeng, W., and He, S. Y. (2010). A prominent role of the flagellin receptor FLAGELLIN-SENSING2 in mediating stomatal response to Pseudomonas syringae pv. tomato DC3000 in Arabidopsis. Plant Physiol. 153, 1188–1198. doi: 10.1104/pp.110.157016
Zeng, W., Melotto, M., and He, S. Y. (2010). Plant stomata: a checkpoint of host immunity and pathogen virulence. Curr. Opin. Biotech. 21, 599–603. doi: 10.1016/j.copbio.2010.05.006
Keywords: leafy vegetables, fresh produce, Salmonella enterica, Escherichia coli O157:H7, plant defense
Citation: Melotto M, Panchal S and Roy D (2014) Plant innate immunity against human bacterial pathogens. Front. Microbiol. 5:411. doi: 10.3389/fmicb.2014.00411
Received: 31 March 2014; Accepted: 21 July 2014;
Published online: 11 August 2014.
Edited by:
Adam Schikora, JLU Giessen, GermanyCopyright © 2014 Melotto, Panchal and Roy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maeli Melotto, Department of Plant Sciences, University of California, Davis, One Shields Avenue, Davis, CA 95616, USA e-mail:bWVsb3R0b0B1Y2RhdmlzLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.