- 1Department of Molecular and Biomedical Sciences, University of Maine, Orono, ME, USA
- 2Department of Molecular and Cell Biology, University of Connecticut, Storrs, CT, USA
Plants in terrestrial systems have evolved in direct association with microbes functioning as both agonists and antagonists of plant fitness and adaptability. As such, investigations that segregate plants and microbes provide only a limited scope of the biotic interactions that dictate plant community structure and composition in natural systems. Invasive plants provide an excellent working model to compare and contrast the effects of microbial communities associated with natural plant populations on plant fitness, adaptation, and fecundity. The last decade of DNA sequencing technology advancements opened the door to microbial community analysis, which has led to an increased awareness of the importance of an organism’s microbiome and the disease states associated with microbiome shifts. Employing microbiome analysis to study the symbiotic networks associated with invasive plants will help us to understand what microorganisms contribute to plant fitness in natural systems, how different soil microbial communities impact plant fitness and adaptability, specificity of host–microbe interactions in natural plant populations, and the selective pressures that dictate the structure of above-ground and below-ground biotic communities. This review discusses recent advances in invasive plant biology that have resulted from microbiome analyses as well as the microbial factors that direct plant fitness and adaptability in natural systems.
Introduction
Symbiotic relationships shaped the origin, organization, and evolution of all life on Earth. Originally defined as “the living together of unlike named organisms” (de Bary, 1878), the term symbiosis has traditionally been applied to associations like mutualism, commensalism, and even parasitism (Parniske, 2008). More recent symbiosis research is expanding this definition to encompass a role of microbial symbiotic relationships in far-reaching themes of biology such as speciation, evolution, and coadaptation (Margulis, 1993; Klepzig et al., 2009; Carrapiço, 2010; Lankau, 2012). The association and close relationships of organisms that cohabitate are vital for the growth and development of all eukaryotic organisms (Carrapiço, 2010; McFall-Ngai et al., 2013). These associations (=symbiotic networks of microorganisms) shape natural landscapes and directly influence the evolutionary trajectory of individual species and entire ecosystems (Gilbert, 2002; Klepzig et al., 2009).
Plant invasions are a global concern because they pose a direct threat to biodiversity and natural resource management, especially in protected areas (i.e., public lands, refuges, conservations, etc.; Foxcroft et al., 2013). For a plant to be considered invasive (and not just naturalized) it must be non-native to the ecosystem in question and it must cause environmental damage (i.e., detrimental effects on native flora and fauna) or harm humans (Invasive Species Advisory Committee [ISAC], 2006). Invasive plant science represents a crossroads of diverse opinions derived from many economic, ecological and societal interest groups, and this has lead to disputes regarding the correct approach to invasive plant issues (Simberloff et al., 2013). To further complicate the issue, plant classification as “invasive” or “weedy” is often based more on human perceptions and opinions than on actual data regarding the economic, societal, or environmental impact of the plant taxon (Hayes and Barry, 2008). However, the environmental consensus supports severe ecological damage by plants deemed invasive in protected areas and significant reductions in the biodiversity of native species resulting from plant invasions. Comprehensive reviews of invasive plant impacts have covered the ecological effects of invaders (Pyšek et al., 2012), nutrient cycling modifications (Ehrenfeld, 2003; Liao et al., 2007), mechanisms of plant invasion (Levine et al., 2003), hybridization, and competition (Vila et al., 2004). Synthesizing accurate predictions of the invasive potential of specific plant taxa has proven difficult and there is no universal trait that can be collectively applied to predict invasiveness (Rejmanek and Richardson, 1996; Richardson and Pysek, 2006; Hayes and Barry, 2008; Thompson and Davis, 2011; Morin et al., 2013). A standard approach is needed for accurate impact assessment and the development of a new global database suitable to make future predictions of problem taxa (Morin et al., 2013).
The rhizosphere microbiome comprises the greatest diversity of microorganisms directly interacting with a given plant; therefore, it has a tremendous capacity to impact plant fitness and adaptation. Bacterial and fungal communities in the rhizosphere affect plant immunity (van Wees et al., 2008; Ronald and Shirasu, 2012), pathogen abundance (Berendsen et al., 2012), nutrient acquisition (Jones et al., 2009; Richardson et al., 2009), and stress tolerance (Doubkova et al., 2012; Marasco et al., 2012). Traditional hypotheses for plant invasion, such as enemy release hypothesis (ERH; Klironomonos, 2002; Mitchell and Power, 2003; Blumenthal, 2006; Liu and Stiling, 2006; Reinhart and Callaway, 2006; Blumenthal et al., 2009; Eschtruth and Battles, 2009), accumulation of local pathogens (ALP; Eppinga et al., 2006), enhanced mutualist hypothesis (EMH; Marler et al., 1999; Reinhart and Callaway, 2004; Parker et al., 2006), and plant–soil feedbacks (Ehrenfeld, 2003; Ehrenfeld et al., 2005; Bever et al., 2012), all point directly to the rhizosphere microbiome, in its entirety, as the primary mediator of plant establishment and success.
The study of soil microbial communities once relied on laboratory culture techniques, phospholipid fatty acid analysis (PFLA), denaturing gel gradient electrophoresis (DGGE), and terminal restriction fragment length polymorphism (TRFLP; Zhang and Xu, 2008; van Elsas and Boersma, 2011). Early on, culture-based approaches revealed “the great plate count anomaly” wherein only about 1% of visible microscopic cells can be cultured using conventional techniques (Staley and Konopka, 1985; Zhang and Xu, 2008; Stein and Nicol, 2011). The DNA technologies available today use genetic information to model the structure and composition of a microbial community (Venter et al., 2004; Tringe and Rubin, 2005; Hugenholtz and Tyson, 2008; Kunin et al., 2008; Vakhlu et al., 2008; Marguerat and Bähler, 2009; Metzker, 2010; Wooley et al., 2010; Simon and Daniel, 2011; Sun et al., 2011; van Elsas and Boersma, 2011; Thomas et al., 2012; Yousuf et al., 2012; Bibby, 2013; Mathieu et al., 2013). Capable of generating millions of base pairs in a matter of hours for only a few thousand dollars, the primary limitation to next-gen sequencing technologies is handling the expansive datasets and applying appropriate statistical analyses to address the biological questions at hand (Metzker, 2010).
The link between the rhizosphere microbial community and invasive plant success has been studied for many years (Van der Putten et al., 2007; Pringle et al., 2009; Berendsen et al., 2012; Bakker et al., 2013). Invasive plants provide a unique perspective to study the effects of the rhizosphere microbiome on plant fitness, the role evolutionary interactions play in structuring the plant ecology observed at present, and the potential for directed control and management of invasive plants. The aim of this review was to focus on recent insights into plant–microbe interactions in the rhizosphere of invasive plants. We were interested in studies that used a sequencing based approach to investigate the rhizosphere microbiome of invasive plants. Surprisingly, we found that few invasive plant scientists have moved beyond traditional methods of soil community analysis (i.e., DGGE) regardless of the increasing availability of next-gen sequencing platforms. We discuss the current microbiome data for invasive plants with regard to popular mechanisms of plant invasion (i.e., enemy release, novel symbiont, etc.). Particular attention has been given to rhizosphere microbiome analysis and what this methodology reveals about microbial symbiotic networks in the soil as contributing factors to the development and progression of plant invasions in terrestrial ecosystems.
Rhizosphere Microbiota are a Key Component of Plant Fitness
Over 400 million years ago, during the Paleozoic era, the evolution of land plants was made possible by a symbiosis between mycorrhizal fungi and the common ancestor of land plants (Wang and Qiu, 2006; Humphreys et al., 2010). This association resulted in a fitness advantage and enhanced stress tolerance that was critical for the establishment of terrestrial plants (i.e., increased access to water and mineral nutrients). Evidence of microbial symbiosis is apparent in the oldest lineages of land plants, the liverworts. The arbuscular mycorrhizal (AM) symbioses of liverworts significantly promote photosynthetic C uptake, acquisition of P and N from the soil, growth, and asexual reproduction (Humphreys et al., 2010). Mycorrhizal symbioses undoubtedly demonstrate the importance of symbiotic relationships in terrestrial ecosystems and have been credited for stimulating the diversification of both plant hosts and fungal symbionts (Wang and Qiu, 2006).
The soil microbial community constitutes a major portion of a plant’s symbiotic network. Soil is the greatest reservoir of microbes that affect plant growth, fitness, fecundity, and stress tolerance (reviewed by Buée et al., 2009; Faure et al., 2009; Lambers et al., 2009; Lugtenberg and Kamilova, 2009; Chaparro et al., 2012; Doornbos et al., 2012; Bakker et al., 2013). All plants maintain a direct interaction with soil microbes in the rhizosphere, which is the soil compartment immediately surrounding the root wherein plant root exudates directly influence the structure and function of the soil microbial community (Figure 1; Hiltner, 1904; Hartmann et al., 2008). The sugars, amino acids, flavonoids, proteins, and fatty acids secreted by plant roots help to structure the associated soil microbiome (Badri et al., 2009; Dennis et al., 2010; Doornbos et al., 2012) and these exudates vary among plant species and between genotypes (Rovira, 1969; Micallef et al., 2009). The quantity and composition of root exudate fluctuates with plant developmental stage and the proximity to neighboring species (Chaparro et al., 2012). Microbes growing in the nutrient rich rhizosphere produce molecular signals that promote plant fitness and growth (i.e., hormones) and can disrupt inter-plant communication in natural systems (Faure et al., 2009; Sanon et al., 2009).
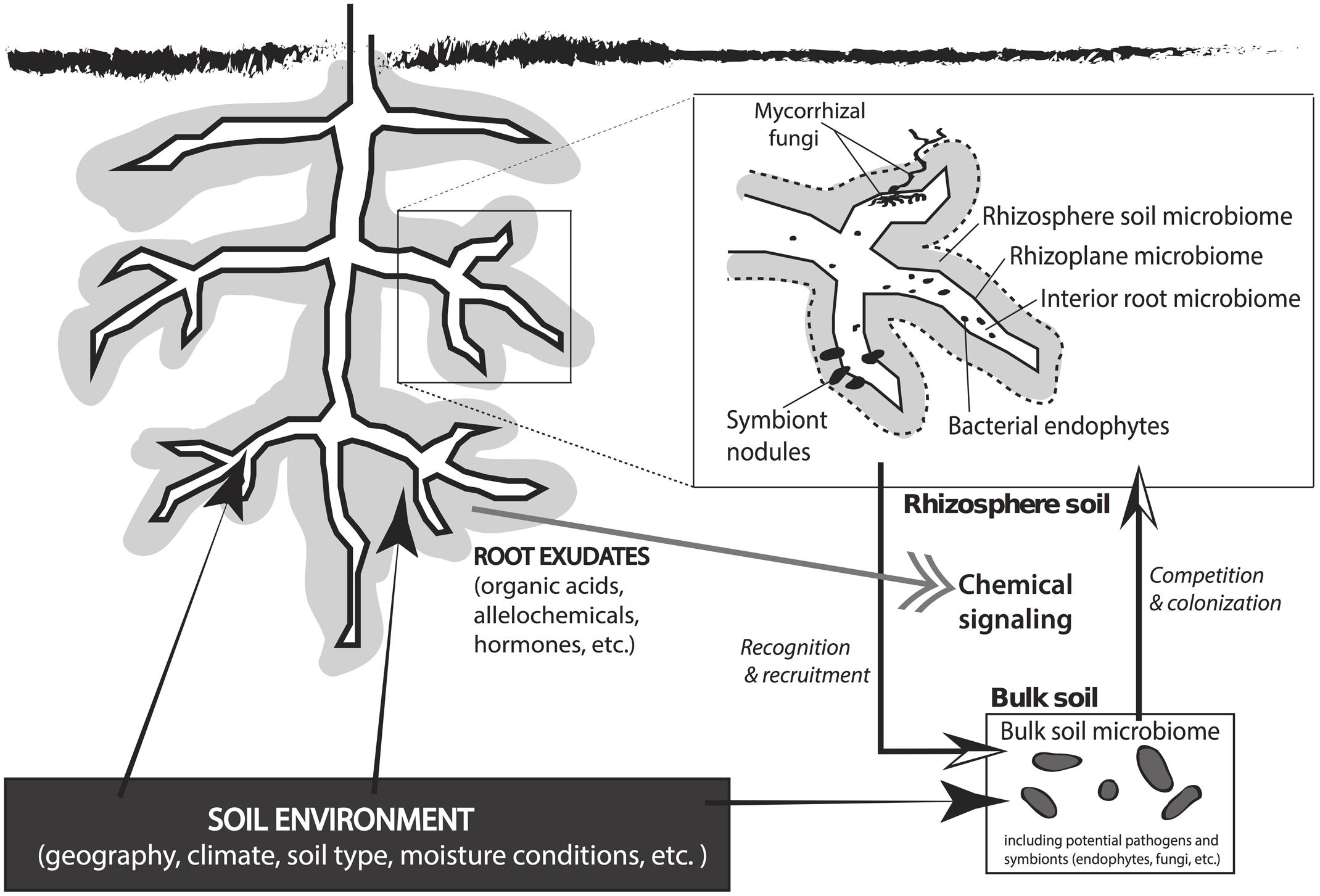
FIGURE 1. An overview of plant–microbe interactions that occur in rhizosphere and bulk soils beneath a plant. The soil environment has a direct effect on the plant, the rhizosphere microclimate, and the microbial community in the bulk soil. Root exudates from the plant direct chemical signaling between the plant and the microbial symbiotic network in the soil matrix. Rhizobiota recognize root exudate signals and are recruited to the rhizoplane or root interior. Bulk soil microbes compete for space to colonize the rhizosphere, which results in a rhizosphere microbial community that is derived from the total microbial population in the bulk soil. The microenvironment in the rhizosphere includes the rhizosphere microbiome (<3–5 mm of the root), rhizoplane microbiome (at root–soil interface), and the interior root microbiome. Common symbiotic interactions in the root zone include mycorrhizal fungi, bacterial endophytes, and symbiont nodules.
Microbes in the rhizosphere can provide a direct access to limiting nutrients (e.g., N2 fixing symbiont) or increase the total surface area of the root system (e.g., mycorrhizal fungi). Many reviews have already covered the positive effects of beneficial root symbionts in the rhizosphere (Buée et al., 2009; Bakker et al., 2013), factors affecting rhizosphere microbial communities (Philippot et al., 2013), and the microbial effects on plant health (Berendsen et al., 2012; Berlec, 2012; Bever et al., 2012) and stress tolerance (Rodriguez et al., 2008).
Antagonistic interactions derived from microbial pathogens play critical roles in determining the genetic structure and spatiotemporal abundance of a plant (Gilbert, 2002; Blumenthal et al., 2009). Pathogenic microbes impose selective pressures on a plant population that favor a specific genetic structure within the host plant community and this stimulates evolutionary change over time (Gilbert, 2002). In natural systems, pathogens mediate plant competition and affect spatiotemporal distribution of individuals within the plant community by creating inhabitable and uninhabitable areas within the ecosystem (Gilbert, 2002). The Janzen-Connell hypothesis postulated that pathogen and host densities are responsible for the observed distribution of a plant species by affecting the establishment success of seedlings (Packer and Clay, 2000). A high density of Pythium sp. in the soil beneath parental Prunus serotina trees was observed to prohibit the establishment of seedlings in the immediate vicinity (0–5 m), but not seedlings growing at greater distances (25–30 m; Packer and Clay, 2000). Thus, pathogen accumulation beneath parent plants functions to promote seedling distribution and reduce competition between the parent plant and its offspring.
Invasive Plants Disrupt Native Symbiotic Networks
The introduction of non-native plants can disrupt native symbiotic networks in the soil and change local grazing patterns for insects and fauna (Elias et al., 2006; Klepzig et al., 2009). Introduced plants alter patterns of nutrient cycling (Laungani and Knops, 2009) and cause chemical changes in the soil environment (i.e., allelopathy; Cipollini et al., 2012). Often these non-native invaders bring novel traits to the environment that put native plants at a disadvantage (Van der Putten et al., 2007; Laungani and Knops, 2009; Perkins et al., 2011). Plant–microbe interactions may assist invasive plants with outcompeting native flora using mechanisms that include allelopathy-mediated suppression of native rhizosphere microbes and beneficial symbionts (Stinson et al., 2006; Callaway et al., 2008), the accumulation of native plant pathogens in the invaded soils (Mangla et al., 2008), and changes in nutrient cycling dynamics that favor the exotic plant (Ehrenfeld et al., 2001; Ehrenfeld, 2003; Laungani and Knops, 2009). Increased availability or access to vital nutrients provides a competitive advantage to invasive plants and facilitates significant biomass accumulation (Blumenthal, 2006; Blumenthal et al., 2009).
Allelopathic plants are among the most aggressive invaders of non-native ecosystems because non-native plants with the ability to synthesize toxic chemicals are often at a competitive advantage (Lankau, 2012). Allaria petiolata (garlic mustard) produces allelopathic chemicals that target beneficial microbes like AM symbionts of native plants (Stinson et al., 2006; Callaway and Vivanco, 2007; Callaway et al., 2008). A. petiolata also demonstrated an increased production of toxic chemicals when growing in non-native regions that contain a greater competitive interspecific density, implicating the allelopathic effects as the primary invasive characteristic (Lankau, 2012). The introduction of novel allelochemicals into an environment affects the structure of the soil microbial community and the microbial biodiversity, especially if these chemicals have antimicrobial activity or function as metal chelators (Inderjit et al., 2011). Soil microbes are the first line of defense toward novel chemicals in a native ecosystem. They mediate much of the allelopathic effect in ways as simple as the ability to degrade or detoxify compounds before they accumulate in the soil and inhibit native plant growth (Cipollini et al., 2012).
Invasive plants outcompete native plants by accumulating large concentrations of native plant pathogens in the soil (Eppinga et al., 2006; Mangla et al., 2008). A release from microbial pathogens, insect pests, and herbivores of the native range is one mechanism behind the success of invasive plants (Klironomonos, 2002; Mitchell and Power, 2003; Reinhart and Callaway, 2006; Blumenthal et al., 2009), but the distribution of pathogens in the invasive range is just as important for defining competition with native flora. Root exudates of Chromolaena odorata, a severely destructive tropical weed, concentrate Fusarium sp. spores to a level 25-times greater than that observed in the root zone of native plants (Mangla et al., 2008). Thus, these plants exacerbate and exploit the native biotic interactions and gain a competitive advantage.
Many, but not all, invasive plants alter patterns of nutrient cycling in the invasive range (Perkins et al., 2011). Changes in the N cycling dynamics in the soil are a frequent consequence of invasive plant introduction (Ehrenfeld, 2003; Mack and D’Antonio, 2003; Laungani and Knops, 2009; Perkins et al., 2011). Non-native species can change the quality and quantity of leaf litter (Ehrenfeld et al., 2001), modify local decomposition rates (Kourtev et al., 2002a; Elgersma et al., 2012), and disrupt local feedback mechanisms in the soil system (Ehrenfeld et al., 2005). For example, Pinus strobus is an invader of N-poor grasslands that demonstrates a higher N residence time in the plant tissues than native species (Laungani and Knops, 2009). This increased residence time facilitates the accumulation of twice as much N in plant tissues and up to four times as much N in the photosynthetic tissues, relative to native grasses (Laungani and Knops, 2009). The differences in N utilization between non-native and native plants create a positive feedback in the soil that significantly increases N availability and results in increased total C gains, both of which allow P. strobus to gain a competitive advantage (Laungani and Knops, 2009).
Microbial Impacts on Plant Establishment and Proliferation
Not all microbes are found ubiquitously throughout soils around the world, and thus, soil microbes are not exempt from fundamental evolutionary processes of geographic isolation and natural selection (Rout and Callaway, 2012). Plant–microbe interactions in the rhizosphere (beneficial, pathogen, etc.) can dictate whether the plant is capable of naturalization and the possibility of an invasive growth habit. Pringle et al. (2009) proposed three criteria to model how mycorrhizal symbioses influence the outcome of a plant invasion: (1) the type of plant–fungi relationship (obligate or facultative) from the plant perspective; (2) if the relationship was specific or flexible, meaning the plant associates with one mycorrhizal fungus versus many; and (3) whether these microbial symbionts were found in the introduced range (Pringle et al., 2009). According to this model, obligate symbionts prevent the growth of non-native plants if the microbial symbiont is not already present in the introduced region, nor is it co-introduced with the host plant. Facultative symbioses are often less restrictive because the plants may form novel beneficial symbioses with suitable replacement microbes in the non-native range, or survive without the symbiont. Consequently, the symbiotic flexibility in facultative symbioses enhances the likelihood of favorable plant adaptations and the development of invasive populations in the introduced region (Pringle et al., 2009).
In the introduced region, the soil microbial community mediates plant abundance and disturbance of the soil can influence the progression of a plant invasion. A removal of the above-ground plant community coupled with little or no physical disruption of the soil is classified as Type I soil disturbance. A Type II soil disturbance includes physical disruption of the soil matrix in addition to removal of the above-ground plant biomass (Fukano et al., 2013). Type I disturbances leave the soil microbial community intact, whereas Type II disturbances completely disrupt the structure of the microbial community. Interestingly, the growth of non-native species is enhanced when they are rare in the ecosystems subjected to Type I disturbance (Fukano et al., 2013). In contrast, type II disturbances give native species an advantage and require non-native invaders to maintain a higher competitive ability. Thus, a physical disturbance that alters the composition of the soil microbial community favors native plants, yet the opposite result occurs (enhanced fitness of non-native plants) if the soil microbial community remains intact.
The Rhizosphere Microbiota of Invasive Plants
The rhizosphere microbiota of non-cultivated plant systems provide a better platform to study the critical plant–microbe interactions that affect plant fitness and adaptability because they are under less anthropogenic control than agricultural systems (Philippot et al., 2013). Figure 2 depicts seven biotic and abiotic factors that together determine the presence or absence of specific microbiota in the soil microbiome of natural systems. Factors such as soil disturbance, local flora and fauna, and allelopathic effects from the plant each impose a selective pressure on the soil microbial community. The cumulative effect of these selective pressures is what determines the frequency and abundance of microbes in the soil, and thus, what microbes the plant is able to recruit into the rhizosphere.
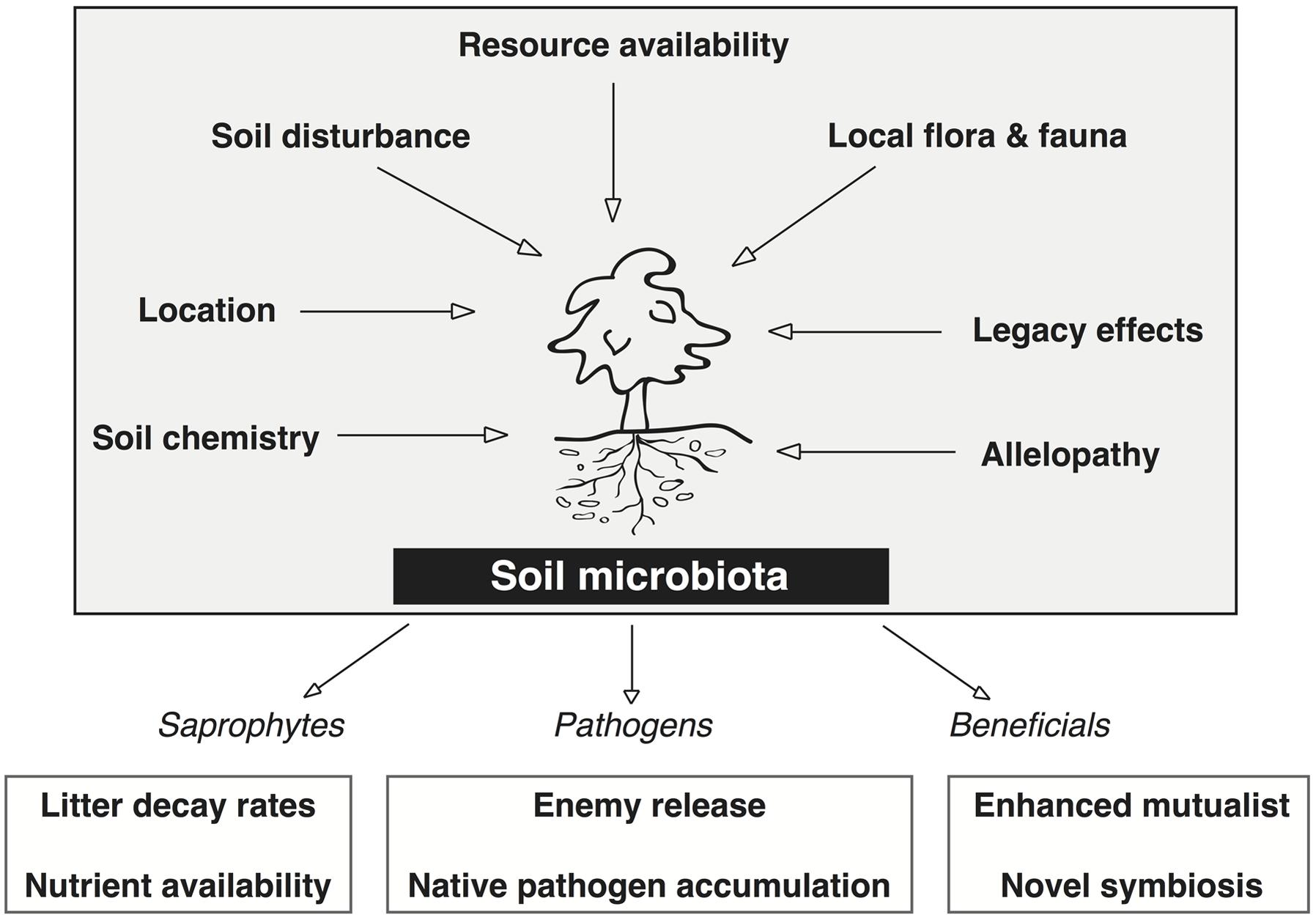
FIGURE 2. Factors that directly affect the soil microbiota associated with invasive plants and the positive feedbacks on the plant invasion derived from each major group (saprophytes, pathogens, and beneficials). Each of the seven factors directly affect the microbial community structure and function in the soil by imposing some degree of selective pressure wherein certain microbes are not capable of surviving. These seven factors dictate the relative abundance of saprophytes, pathogens, and beneficials that are able to associate with the plant. Two mechanisms of plant invasion that lead to positive feedbacks from these plant–microbe interactions are shown for each group of soil microbes.
Microbiome analysis of rhizosphere microbiota associated with invasive Berberis thunbergii in Maine showed that environmental factors alone cannot explain the structure of the rhizosphere microbial community associated with this plant in the invasive range. Coats et al. (2014) used amplicon pyrosequencing to assess effects of environmental factors on the bacterial and fungal communities in the rhizosphere of B. thunbergii (Japanese barberry) from invasive stands in coastal Maine, USA. The effects of soil chemistry, location, and surrounding plant canopy cover were investigated and a high degree of spatial variation in the rhizosphere microbial communities of B. thunbergii was reported. Bulk soil chemistry had more of an effect on the bacterial community structure than the fungal community. An effect of location was detected in the rhizosphere microbial community, but it was less significant than the effect of surrounding plant canopy cover. The significant effects of these environmental factors on the structure of the rhizosphere microbial community associated with B. thunbergii suggests some soils and/or plant communities are more prone to plant invasions based on the soil microbial communities they foster.
The microbial diversity in the rhizosphere includes many species of bacteria, archaea, fungi, oomycetes, viruses, and various microfauna (nematodes, protozoa, etc.; reviewed by Buée et al., 2009; Bever et al., 2012; Philippot et al., 2013). The rhizosphere microbiome differs from the bulk soil and between plant species. Using a metatranscriptomic approach, Turner et al. (2013) identified kingdom level differences in the rhizosphere bacterial communities of wheat, oat, and pea plants. The fungal diversity in the rhizosphere also varied significantly between these crop plants. Investigations that have focused on the interactive effects between major microbial groups in the rhizosphere have revealed a joint effect of fungal endophytes and AM fungi that promotes plant growth (Larimer et al., 2010). Bacterial endophytes have been observed to enhance competition by invasive plants through providing the plant with increased access to nutrients (Fe and P) and by producing plant growth promoting hormones (IAA; Rout et al., 2013). When comparing native and non-native plants with DGGE, Xiao et al. (2014) found that the soil fungal communities were more affected by the invasive plant than the native plant and the modifications to the fungal community promoted invasive plant growth. Differences in the rhizosphere pathogen communities of related Phragmites australis haplotype populations (a native and non-native) have also demonstrated that non-native species cultivate different soil pathogen communities than native plants regardless of the genetic similarity of the host plant (Nelson and Karp, 2013).
Rhizosphere Microbiome in Native and Invasive Range Soils
Recent investigations that have contrasted plant–microbe interactions in the native and invasive range have focused on the net effect of soil biota on plant growth, plant allelopathic responses, and the rhizosphere microbiome. The rhizosphere microbiota (saprophytes, pathogens, and beneficials) each have positive effects on invasive plant growth (lower boxes of Figure 2). Stimulating saprophyte growth creates a positive feedback in the soil of invasive plants by increasing litter decay rates and nutrient availability (Van der Putten et al., 2007; Bever et al., 2012). The mutualistic associations and/or novel symbioses in the introduced range can enhance plant fitness by promoting plant growth, nutrient acquisition, and disease suppression (Van der Putten et al., 2007; Pringle et al., 2009; Berendsen et al., 2012; Bakker et al., 2013). The empirical evidence obtained from studies that compare plant–microbe interactions in each range support current microbe based theories of plant invasions and provide evidence for microbe enhanced plant fitness in the invasive range.
Triadica sebifera (Chinese tallow) is native in China and invasive in the US. Yang et al. (2013) studied the net effect of native and invasive range soil microbiota on the growth of T. sebifera and four co-occurring plant genera (Liquidambar, Ulmus, Celtis, and Platanus). Native range soils had no effect, or a negative effect, on T. sebifera performance yet there was always a positive effect of invasive range soil on plant survival and biomass production. A greater biomass was observed for the invasive plants grown in active soil mix than in sterilized or fungicide-treated soils. Higher mycorrhizal colonization of T. sebifera was found on plants growing in the invasive range soil. Interestingly, there was no effect of native or invasive range soil on the other four genera examined, and native plants maintained higher mycorrhizal colonization rates in native soil than invasive range soil. These results not only support Enhanced Mutualist and Pathogen Release Hypotheses, they also indicate a significant specificity in the plant–microbe interactions for some plant species that contribute to invasive plant growth.
The allelopathic response of invasive plants can differ between native and invasive ranges with greater allelopathic effects observed in the invasive range. Yuan et al. (2013) observed increased allelochemical content (total phenolics, total flavones, and total saponins) for Solidago canadensis, a native of the US that has developed invasive populations in China. The increased production of allelopathic chemicals by S. canadensis in the invasive range also coincided with a greater inhibition of native plant seedlings. Whether the increase in allelochemical production is solely a result of the plant–microbe interactions remains unclear, although it would seem to be a beneficial plant response to the development of novel interactions with foreign soil microbiota.
The most comprehensive investigation of a rhizosphere microbiome associated with an invasive plant was conducted on B. thunbergii, a native of central Japan that is invasive in the US. The microbial community (Bacteria, Archaea, and Eukaryota) structure was modeled using amplicon pyrosequencing to compare rhizosphere communities of native B. thunbergii from central Japan (n = 8) with those from an invasive stand in the US (n = 5; Coats, 2013). A total of 432 genera were identified from all three domains in Japan and US rhizosphere soils combined, although only Eukaryotes from the lineage Fungi were included in this analysis. B. thunbergii rhizosphere soils from Japan and the US shared 171 genera, most of which were Proteobacteria (Bacteria) and Ascomycota (Fungi). Rhizosphere soil from Japan contained 71 unique genera and the US soils harbored 190 unique genera. A high degree of phylogenetic redundancy was observed within the microbial community at the phyla level, although the community structure was significantly different between samples from each region (Coats, 2013).
The apparent difference in the rhizosphere microbiota of B. thunbergii in native and invasive (non-native) soil supports our hypothesis that soil microbial communities are the primary mediators of invasive plant growth in non-native habitats. The data showed a significant effect of geographic location with less species diversity and increased abundance of pathogenic species observed in rhizosphere soils from the native range compared to the invasive range (Coats, 2013). Therefore, the microbial community shifts observed between the rhizosphere soil in the native and non-native ranges support Enemy Release and Enhanced Mutualist Hypotheses, as well as an increased access to nutrients via saprophyte stimulation and/or novel symbiont acquisition. Interestingly, Bacteria communities were more significantly different between rhizosphere samples from the two ranges than the Archaea or the Eukaryota communities (Coats, 2013).
Pathogen release, wherein exotic plants are not subjected to the heavy pathogen loads characteristic of native range soils in the non-native range, has been implicated as a common mechanism for plant invasions, especially when coupled with increased access to nutrients (Blumenthal, 2006; Blumenthal et al., 2009). The impacts of enemy release on a plant invasion are determined from two opposing factors: (1) plants’ “escape” from heavy pathogen loads in the native range and (2) the rate of accumulating pathogens in the introduced range (release = escape - accumulation; Mitchell and Power, 2003). Many genera that were found strictly in B. thunbergii rhizosphere soils from Japan are common plant pathogens, including Clostridium, Enterobacter (Pantoea), and Serratia (Schaad et al., 2001; Grimont and Grimont, 2006), and these putatively pathogenic microbes occurred in greater abundance in the native soils. For instance, two pathogenic Serratia species (S. proteamaculans and S. marcescens) constituted 1.8% of the total reads in some rhizosphere samples from Japan and as much as 52% of the total for other Japan rhizosphere samples (Grimont and Grimont, 2006; Coats, 2013). Buttiauxella was detected in every rhizosphere sample from Japan (compared to three US samples) and it comprised 8.5–70.1% of the total reads, although the average was approximately 30–35% per sample. Stenotrophomonas, another putative Berberis pathogen, comprised approximately 1–9% of the total reads in the native Japan soils but contributed very little (∼0.1% of the total reads) to the microbial community in the rhizosphere soil from the US (Coats, 2013).
The rhizosphere microbial communities associated with B. thunbergii also implicate a role for enhanced mutualism as one factor in the development of invasive populations (Coats, 2013). Some genera that are likely to be putative beneficial symbionts, such as Glomus (mycorrhizal fungi) and Frankia (N2-fixing actinomycete), were detected solely in rhizosphere communities of the invasive range. Other genera that also contain putative beneficials were detected in both regions, although their abundance was greater in the rhizosphere soil from the invasive range. Some of these genera are capable of symbiotic or free-living (diazotrophic) N fixation (e.g., Bradyrhizobium, Rhizobium, Azospira, etc.), whereas others are likely to function more like plant growth promoting rhizobacteria (e.g., Bacillus and Pseudomonas) that promote plant fitness by producing growth simulating phytohormones (Faure et al., 2009; Effmert et al., 2012), enhancing stress tolerance (Dimkpa et al., 2009; Kang et al., 2010; Pineda et al., 2010), or antagonizing pathogenic microbes that inhabit the root zone (Berendsen et al., 2012).
Alterations to N cycling dynamics are a commonly reported feature of B. thunbergii invasions in North American soils, which suggests saprophyte stimulation (via increased litter decay rates) and/or novel symbiont acquisition are responsible for the observed changes in the invasive range (Coats, 2013). Relative to native Vaccinium shrubs, B. thunbergii plants produce large quantities of N-rich biomass, N-rich leaf litter, and N-rich secondary metabolites (Ehrenfeld et al., 2001; Elgersma et al., 2012) and they harbor higher levels of extractable nitrate in the soil (Ehrenfeld, 1999). B. thunbergii preferentially uses nitrate (Ehrenfeld et al., 2001), a trait that facilitates out-competing ammonium utilizing plants (Gilliam, 2006), and these exotic plants have increased rates of nitrification in the soil rather than high N availability from mineralization (Kourtev et al., 2002b, 2003; Elgersma et al., 2011). The rhizosphere soil from B. thunbergii showed an increased abundance of nitrifying bacteria such as Nitrospirales (0.0–2.4%) and Nitrosomonadales (0.4–1.6%) in the invasive range soils relative to rhizosphere soils from the native range (0.0–0.3% and 0.0–0.2% for Nitrospirales and Nitrosomonadales, respectively; Coats, 2013). The data acquired by microbiome analysis show that differences in the microbial community structure between the two ranges corroborate previous investigations of soil N cycling beneath B. thunbergii in the invasive range. This metagenomic approach also identifies specific organisms that are likely to be the culprits behind changes in the N cycling patterns in the invasive range soil and that can be targeted during future investigation of the microbial function in the rhizosphere.
Future Research
Given the recent advances in high-throughput DNA sequencing and the availability of cost-effective microbiome analysis, it is time invasive plant biologists begin to focus on a full characterization of soil microbial communities in an effort to understand how changes or shifts in the rhizosphere microbiome are affecting the above-ground ecology. Metagenomics and metatranscriptomics provide a rapid means to investigate the genomics and gene expression that mediate plant–microbe interactions in the rhizosphere as well as provide much needed information regarding the metabolic capacity and ecological function of rhizosphere microbes. These plant–microbe interactions not only contribute to invasive plant growth and fitness, they also define the range of suitable habitats and areas of competitive advantage. Obtaining high quality predictions for the most susceptible habitats is the best way to prevent invasive plant introduction and subsequent damage. Microbiome profiling of soil, by programs such as the Earth Microbiome Project (http://www.earthmicrobiome.org/; Gilbert et al., 2010), will undoubtedly enhance prediction algorithms and help identify microbial components in regions of high or low susceptibility. However, the information gained from rhizosphere microbiome analysis is not limited to predictions and promoting a better understanding of plant–microbe interactions in natural ecosystems. Microbiome-based investigations will greatly assist in the development of microbial probiotics and/or targeted approaches to reclaiming habitats that have become heavily invaded (Berlec, 2012). Such an approach would continue to build on current methods of reducing cost and environmental damage caused by terrestrial invaders and focus efforts on prohibiting the initial establishment.
Conclusion
The introduction and prevalence of invasive plants, and the threat of increasing invasion rates, substantiates the need to understand the mechanisms underlying the success of plants that become invasive. Symbiotic networks of microorganisms in the soil undoubtedly affect the naturalization of non-native plants in the introduced region and the ability of these plants to outcompete native species. Plant–microbe interactions in the rhizosphere directly contribute to plant fitness, nutrient acquisition, and stress tolerance. Therefore, the rhizosphere microbiome of a plant harbors a tremendous capacity to promote or inhibit invasive growth characteristics. Invasion mechanisms employed by some plants involve rhizosphere microbiome shifts between the native and invasive ranges. These microbial community shifts provide evidence in support of the Enemy Release and Enhanced Mutualist Hypotheses as well as corroborating plant–microbe feedbacks that lead to an enhanced resource acquisition beyond the limits of native flora.
Author Contributions
The manuscript was drafted by Vanessa C. Coats with editorial remarks from Mary E. Rumpho.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by U.S. Department of Agriculture CSREES grants no. 2009-38914-19786 and 2010-38914-20996 to Mary E. Rumpho, and a 2010 LL Bean Scientific Research Fellowship in Acadia National Park from the Friends of Acadia National Park (http://www.friendsofacadia.org/) granted to Vanessa C. Coats. This is Maine Agricultural and Forest Experiment Station Publication Number 3368, Hatch Project no. ME08362-07H.
References
Badri, D. V., Weir, T. L., Van der Lelie, D., and Vivanco, J. M. (2009). Rhizosphere chemical dialogues: plant–microbe interactions. Curr. Opin. Biotechnol. 20, 642–650. doi: 10.1016/j.copbio.2009.09.014
Bakker, P., Berendsen, R. L., and Doornbos, R. F. (2013). The rhizosphere revisited: root microbiomics. Front. Plant Sci. 4:165. doi: 10.3389/fpls.2013.00165
Berendsen R. L., Pieterse, C. M. J., and Bakker, P. A. H. M. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Berlec, A. (2012). Novel techniques and findings in the study of plant microbiota: search for plant probiotics. Plant Sci. 193–194, 96–102. doi: 10.1016/j.plantsci.2012.05.010
Bever, J. D., Platt, T. G., and Morton, E. R. (2012). Microbial population and community dynamics on plant roots and their feedbacks on plant communities. Annu. Rev. Microbiol. 66, 265–283. doi: 10.1146/annurev-micro-092611-150107
Bibby, K. (2013). Metagenomic identification of viral pathogens. Trends Biotechnol. 31, 275–279. doi: 10.1016/j.tibtech.2013.01.016
Blumenthal, D. (2006). Interactions between resource availability and enemy release in plant invasion. Ecol. Lett. 9, 887–895. doi: 10.1111/j.1461-0248.2006.00934.x
Blumenthal, D., Mitchell, C. E., Pyšek, P., and Jarošík, V. (2009). Synergy between pathogen release and resource availability in plant invasion. Proc. Natl. Acad. Sci. U.S.A. 106, 7899–7904. doi: 10.1073/pnas.0812607106
Buée, M., Boer, W., Martin, F., Overbeek, L., and Jurkevitch, E. (2009). The rhizosphere zoo: an overview of plant-associated communities of microorganisms, including phages, bacteria, archaea, and fungi, and of some of their structuring factors. Plant Soil 321, 189–212. doi: 10.1007/s11104-009-9991-3
Callaway, R. M., Cipollini, D., Barto, K., Thelen, G. C., Hallett, S. G., Prati, D.,et al. (2008). Novel weapons: invasive plant suppresses fungal mutualists in America but not in its native Europe. Ecology 89, 1043–1055. doi: 10.1890/07-0370.1
Callaway, R. M., and Vivanco, J. M. (2007). Invasion of plants into native communities using the underground information superhighway. Allelopathy J. 19, 143–151.
Carrapiço, F. (2010). How symbiogenic is evolution? Theor. Biosci. 129, 135–139. doi: 10.1007/s12064-010-0100-1
Chaparro, J. M., Sheflin, A. M., Manter, D. K., and Vivanco, J. M. (2012). Manipulating the soil microbiome to increase soil health and plant fertility. Biol. Fertil. Soils 48, 489–499. doi: 10.1007/s00374-012-0691-4
Cipollini, D., Rigsby, C. M., and Barto, E. K. (2012). Microbes as targets and mediators of allelopathy in plants. J. Chem. Ecol. 38, 714–727. doi: 10.1007/s10886-012-0133-7
Coats, V. C. (2013, August). Microbial Associates of Berberis thunbergii (Japanese barberry). Ph.D. dissertation, University of Maine. Ann Arbor: ProQuest/UMI.
Coats, V. C., Pelletreau, K. N., and Rumpho, M. E. (2014). Amplicon pyrosequencing reveals the soil microbial diversity associated with invasive Japanese barberry (Berberis thunbergii DC.). Mol. Ecol.
de Bary, A. (1878). Ueber Symbiose-Tageblatt 51 Versamml. Cassel: Deutscher Naturforscher u. Aerzte.
Dennis, P. G., Miller, A. J., and Hirsch, P. R. (2010). Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol. Ecol.
Dimkpa, C., Weinand, T., and Asch, F. (2009). Plant–rhizobacteria interactions alleviate abiotic stress conditions. Plant Cell Environ.
Doornbos, R. F., Loon, L. C., and Bakker, P. A. H. M. (2012). Impact of root exudates and plant defense signaling on bacterial communities in the rhizosphere. Agron. Sustain. Dev.
Doubkova, P., Suda, J., and Sudova, R. (2012). The symbiosis with arbuscular mycorrhizal fungi contributes to plant tolerance to serpentine edaphic stress. Soil Biol. Biochem.
Effmert, U., Kalderás, J., Warnke, R., and Piechulla, B. (2012). Volatile mediated interactions between bacteria and fungi in the soil. J. Chem. Ecol.
Ehrenfeld, J. G. (1999). Structure and dynamics of populations of Japanese barberry (Berberis thunbergii DC.). Biol. Invasions
Ehrenfeld, J. G. (2003). Effects of exotic plant invasions on soil nutrient cycling processes. Ecosystems
Ehrenfeld, J. G., Kourtev, P., and Huang, W. (2001). Changes in soil functions following invasions of exotic understory plants in deciduous forests. Ecol. Appl. 11, 1287–1300. doi: 10.1890/1051-0761(2001)011[1287:CISFFI]2.0.CO;2
Ehrenfeld, J. G., Ravit, B., and Elgersma, K. (2005). Feedback in the plant–soil system. Annu. Rev. Environ. Resour. 30, 75–115. doi: 10.1146/annurev.energy.30.050504.144212
Elgersma, K. J., Ehrenfeld, J. G., Yu, S., and Vor, T. (2011). Legacy effects overwhelm the short-term effects of exotic plant invasion and restoration on soil microbial community structure, enzyme activities, and nitrogen cycling. Oecologia 167, 733–745. doi: 10.1007/s00442-011-2022-0
Elgersma, K. J., Yu, S., Vor, T., and Ehrenfeld, J. G. (2012). Microbial-mediated feedbacks of leaf litter on invasive plant growth and interspecific competition. Plant Soil 356, 341–355. doi: 10.1007/s11104-011-1117-z
Elias, S. P., Lubelczyk, C. B., Rand, P. W., Lacombe, E. H., Holman, M. S., and Smith, R. P. (2006). Deer browse resistant exotic-invasive understory: an indicator of elevated human risk of exposure to Ixodes scapularis (Acari:Ixodidae) in southern coastal Maine woodlands. J. Med. Entomol. 43, 1142–1152. doi: 10.1603/0022-2585(2006)43[1142:DBREUA]2.0.CO;2
Eppinga, M. B., Rietkerk, M., Dekker, S. C., De Ruiter, P. C., and Van der Putten, W. H. (2006). Accumulation of local pathogens: a new hypothesis to explain exotic plant invasions. Oikos 114, 168–176. doi: 10.1111/j.2006.0030-1299.14625.x
Eschtruth, A. K., and Battles, J. J. (2009). Acceleration of exotic plant invasion in a forested ecosystem by a generalist herbivore. Conserv. Biol. 23, 388–399. doi: 10.1111/j.1523-1739.2008.01122.x
Faure, D., Vereecke, D., and Leveau, J. H. J. (2009). Molecular communication in the rhizosphere. Plant Soil 321, 279–303. doi: 10.1007/s11104-008-9839-2
Foxcroft, L. C., Pysek, P., Richardson, D. M., Pergi, J., and Hulme, P. E., eds. (2013). “Ch. 2: The bottom line: impacts of alien plant invasions in protected areas,” in Plant Invasions in Protected Areas: Patterns, Problems, and Challenges, Invading Nature. Springer Series in Invasion Ecology 7 (Dordrecht: Springer Science+Business Media).
Fukano, Y., Tachiki, Y., Yahara, T., and Iwasa, Y. (2013). Soil disturbances can suppress the invasion of alien plants under plant–soil feedback. Ecol. Model. 260, 42–49. doi: 10.1016/j.ecolmodel.2013.03.022
Gilbert, G. S. (2002). Evolutionary ecology of plant diseases in natural ecosystems. Annu. Rev. Phytopathol. 40, 13–43. doi: 10.1146/annurev.phyto.40.021202.110417
Gilbert, J. A., Meyer, F., Jansson, J., and Gordon, J. (2010). The Earth Microbiome Project: meeting report of the “1st EMP meeting on sample selection and acquisition” at Argonne National Laboratory October 6th 2010. Stand. Genomic Sci. 3, 249–253. doi: 10.4056/aigs.1443528
Gilliam, F. S. (2006). Response of the herbaceous layer of forest ecosystems to excess nitrogen deposition. J. Ecol. 94, 1176–1191. doi: 10.1111/j.1365-2745.2006.01155.x
Grimont, F., and Grimont, P. A. (2006). The genus Serratia. Prokaryotes 6, 219–244. doi: 10.1007/0-387-30746-X_11
Hartmann, A., Rothballer, M., and Schmid, M. (2008). Lorenz Hiltner, a pioneer in rhizosphere microbial ecology and soil bacteriology research. Plant Soil 312, 7–14. doi: 10.1007/s11104-007-9514-z
Hayes, K. R., and Barry S. C. (2008). Are there any consistent predictions of invasion success? Biol. Invasions 10, 483–506. doi: 10.1007/s10530-007-9146-5
Hiltner, L. (1904). Uber neuere Erfahrungen und Probleme auf dem Gebiete der Bodenbakteriologie unter besonderden berucksichtigung und Brache. Arb. Dtsch. Landwirtsch. Gesellschaft. 98, 59–78.
Hugenholtz, P., and Tyson, G. W. (2008). Microbiology: metagenomics. Nature 455, 481–483. doi: 10.1038/455481a
Humphreys, C. P., Franks, P. J., Rees, M., Bidartondo, M. I., Leake, J. R., and Beerling, D. J. (2010). Mutualistic mycorrhiza-like symbiosis in the most ancient group of land plants. Nat. Commun. 1, 1–7. doi: 10.1038/ncomms1105
Inderjit, Wardle, D. A., Karban, R., and Callaway, R. M. (2011). The ecosystem and evolutionary contexts of allelopathy. Trends Ecol. Evol. 26, 655–662. doi: 10.1016/j.tree.2011.08.003
Invasive Species Advisory Committee [ISAC]. (2006). Invasive Species Definition Clarification and Guidance White Paper. The National Invasive Species Council, Submitted by the Definitions Subcommittee of the Invasive Species Advisory Committee. Available at: http://www.invasivespecies.gov/global/ISAC/ISAC_documents/ISAC%20Definititions%20White%20Paper
Jones, D. L., Nguyen, C., and Finlay, R. D. (2009). Carbon flow in the rhizosphere: carbon trading at the soil–root interface. Plant Soil 321, 5–33. doi: 10.1007/s11104-009-9925-0
Kang, B. G., Kim, W. T., Yun, H. S., and Chang, S. C. (2010). Use of plant growth-promoting rhizobacteria to control stress responses of plant roots. Plant Biotechnol. Rep. 4, 179–183. doi: 10.1007/s11816-010-0136-1
Klepzig, K. D., Adams, A. S., Handelsman, J., and Raffa, K. F. (2009). Symbioses: a key driver of insect physiological processes, ecological interactions, evolutionary diversification, and impacts on humans. Environ. Entomol. 38, 67–77. doi: 10.1603/022.038.0109
Klironomonos, J. N. (2002). Feedback with soil biota contributes to plant rarity and invasiveness in communities. Nature 417, 67–70. doi: 10.1038/417067a
Kourtev, P., Ehrenfeld, J., and Huang, W. (2002a). Enzyme activities during litter decomposition of two exotic and two native plant species in hardwood forests of New Jersey. Soil Biol. Biochem. 34, 1207–1218. doi: 10.1016/S0038-0717(02)00057-3
Kourtev, P. S., Ehrenfeld, J. G., and Häggblom, M. (2002b). Exotic plant species alter the microbial community structure and function in the soil. Ecology 83, 3152–3166. doi: 10.1890/0012-9658(2002)083[3152:EPSATM]2.0.CO;2
Kourtev, P. S., Ehrenfeld, J. G., and Häggblom, M. (2003). Experimental analysis of the effect of exotic and native plant species on the structure and function of soil microbial communities. Soil Biol. Biochem. 35, 895–905. doi: 10.1016/S0038-0717(03)00120-2
Kunin, V., Copeland, A., Lapidus, A., Mavromatis, K., and Hugenholtz, P. (2008). A bioinformatician’s guide to metagenomics. Microbiol. Mol. Biol. Rev. 72, 557–578. doi: 10.1128/MMBR.00009-08
Lambers, H., Mougel, C., Jaillard, B., and Hinsinger, P. (2009). Plant–microbe–soil interactions in the rhizosphere: an evolutionary perspective. Plant Soil 321, 83–115. doi: 10.1007/s11104-009-0042-x
Lankau, R. A. (2012). Coevolution between invasive and native plants driven by chemical competition and soil biota. Proc. Natl. Acad. Sci. U.S.A. 109, 11240–11245. doi: 10.1073/pnas.1201343109
Larimer, A. L., Bever, J. D., and Clay, K. (2010). The interactive effects of plant microbial symbionts: a review and meta-analysis. Symbiosis 51, 139–148. doi: 10.1007/s13199-010-0083-1
Laungani, R., and Knops, J. M. H. (2009). Species-driven changes in nitrogen cycling can provide a mechanism for plant invasions. Proc. Natl. Acad. Sci. U.S.A. 106, 12400–12405. doi: 10.1073/pnas.0900921106
Levine, J. M., Vila, M., D’Antonio, C. M., Dukes, J. S., Grigulis, K., and Lavorel, S. (2003). Mechanisms underlying the impacts of exotic plant invasions. Proc. Biol. Sci. 270, 775–781. doi: 10.1098/rspb.2003.2327
Liao, C., Peng, R., Luo, Y., Zhou, X., Wu, X., Fang, C.,et al. (2007). Altered ecosystem carbon and nitrogen cycles by plant invasion: a meta-analysis. New Phytol. 177, 706–714. doi: 10.1111/j.1469-8137.2007.02290.x
Liu, H., and Stiling, P. (2006). Testing the enemy release hypothesis: a review and meta-analysis. Biol. Invasions 8, 1535–1545. doi: 10.1007/s10530-005-5845-y
Lugtenberg, B., and Kamilova, F. (2009). Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63, 541–556. doi: 10.1146/annurev.micro.62.081307.162918
Mack, M. C., and D’Antonio, C. M. (2003). Exotic grasses alter controls over soil nitrogen dynamics in a Hawaiian woodland. Ecol. Appl. 13, 154–166. doi: 10.1890/1051-0761(2003)013[0154:EGACOS]2.0.CO;2
Mangla, S., Inderjit, and Callaway, R. M. (2008). Exotic invasive plant accumulates native soil pathogens which inhibit native plants. J. Ecol. 96, 58–67.
Marasco, R., Rolli, E., Ettoumi, B., Vigani, G., Mapelli, F., Borin, S.,et al. (2012). A drought resistance-promoting microbiome is selected by root system under desert farming. PLoS ONE 7:e48479. doi: 10.1371/journal.pone.0048479
Marguerat, S., and Bähler, J. (2009). RNA-seq: from technology to biology. Cell. Mol. Life Sci. 67, 569–579. doi: 10.1007/s00018-009-0180-6
Margulis, L. (1993). Origins of species: acquired genomes and individuality. Biosystems 31, 121–125. doi: 10.1016/0303-2647(93)90039-F
Marler, M. J., Zabinski, C. A., and Callaway, R. N. (1999). Mycorrhizae indirectly enhance competitive effects of an invasive forb on a native bunchgrass. Ecology 80, 1180–1186. doi: 10.1890/0012-9658(1999)080[1180:MIECEO]2.0.CO;2
Mathieu, A., Delmont, T. O., Vogel, T. M., Robe, P., Nalin, R., and Simonet, P. (2013). Life on human surfaces: skin metagenomics. PLoS ONE 8:e65288. doi: 10.1371/journal.pone.0065288
McFall-Ngai, M., Hadfield, M. G., Bosch, T. C., Carey, H. V., Domazet-Lošo, T., Douglas, A. E.,et al. (2013). Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. U.S.A. 110, 3229–3236. doi: 10.1073/pnas.1218525110
Metzker, M. L. (2010). Sequencing technologies – the next generation. Nat. Rev. Genet. 11, 31–46. doi: 10.1038/nrg2626
Micallef, S. A., Channer, S., Shiaris, M. P., and Colón-Carmona, A. (2009). Plant age and genotype impact the progression of bacterial community succession in the Arabidopsis rhizosphere. Plant Signal. Behav. 4, 777–780. doi: 10.4161/psb.4.8.9229
Mitchell, C. E., and Power, A. G. (2003). Release of invasive plants from fungal and viral pathogens. Nature 421, 625–627. doi: 10.1038/nature01317
Morin, L., Paini, D. R., and Randall, R. P. (2013). Can global weed assemblages be used to predict future weeds? PLoS ONE 8:e55547. doi: 10.1371/journal.pone.0055547
Nelson, E. B., and Karp, M. A. (2013). Soil pathogen communities associated with native and non-native Phragmites australis populations in freshwater wetlands. Ecol. Evol. 3, 5254–5267. doi: 10.1002/ece3.900
Packer, A., and Clay, K. (2000). Soil pathogens and spatial patterns of seedling mortality in a temperate tree. Nature 404, 278–281. doi: 10.1038/35005072
Parniske, M. (2008). Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat. Rev. Microbiol. 6, 763–775. doi: 10.1038/nrmicro1987
Parker, M. A., Malek, W., and Parker, I. M. (2006). Growth of an invasive legume is symbiont limited in newly occupied habitats. Divers. Distrib. 12, 563–571. doi: 10.1111/j.1366-9516.2006.00255.x
Perkins, L. B., Johnson, D. W., and Nowak, R. S. (2011). Plant-induced changes in soil nutrient dynamics by native and invasive grass species. Plant Soil 345, 365–374. doi: 10.1007/s11104-011-0788-9
Philippot, L., Raaijmakers, J. M., Lemanceau, P., and van der Putten, W. H. (2013). Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11, 789–799. doi: 10.1038/nrmicro3109
Pineda, A., Zheng, S. J., van Loon, J. J. A., Pieterse, C. M. J., and Dicke, M. (2010). Helping plants to deal with insects: the role of beneficial soil-borne microbes. Trends Plant Sci. 15, 507–514. doi: 10.1016/j.tplants.2010.05.007
Pringle, A., Bever, J. D., Gardes, M., Parrent, J. L., Rillig, M. C., and Klironomos, J. N. (2009). Mycorrhizal symbioses and plant invasions. Annu. Rev. Ecol. Evol. Syst. 40, 699–715. doi: 10.1146/annurev.ecolsys.39.110707.173454
Pyšek, P., Jarošík, V., Hulme, P. E., Pergl, J., Hejda, M., Schaffner, U.,et al. (2012). A global assessment of invasive plant impacts on resident species, communities and ecosystems: the interaction of impact measures, invading species’ traits and environment. Glob. Change Biol. 18, 1725–1737. doi: 10.1111/j.1365-2486.2011.02636.x
Reinhart, K. O., and Callaway, R. M. (2004). Soil biota facilitate exotic Acer invasions in Europe and North America. Ecol. Appl. 14, 1737–1745. doi: 10.1890/03-5204
Reinhart, K. O., and Callaway, R. M. (2006). Soil biota and invasive plants. New Phytol. 170, 445–457. doi: 10.1111/j.1469-8137.2006.01715.x
Rejmanek, M., and Richardson D. M. (1996). What attributes make some plant species more invasive? Ecology 77, 1655–1661. doi: 10.2307/2265768
Richardson, A. E., Barea, J. M., McNeill, A. M., and Prigent-Combaret, C. (2009). Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant Soil 321, 305–339. doi: 10.1007/s11104-009-9895-2
Richardson, D. M., and Pysek, P. (2006). Plant invasions: merging the concepts of species invasiveness and community invasibility. Prog. Phys. Geogr. 30, 409–431. doi: 10.1191/0309133306pp490pr
Rodriguez, R. J., Henson, J., Van Volkenburgh, E., Hoy, M., Wright, L., Beckwith, F.,et al. (2008). Stress tolerance in plants via habitat-adapted symbiosis. ISME J. 2, 404–416. doi: 10.1038/ismej.2007.106
Ronald, P. C., and Shirasu, K. (2012). Front-runners in plant–microbe interactions. Curr. Opin. Plant Biol. 15, 345–348. doi: 10.1016/j.pbi.2012.06.001
Rout, M. E., and Callaway, R. M. (2012). Interactions between exotic invasive plants and soil microbes in the rhizosphere suggest that “everything is not everywhere.” Microb. Ecol. 110, 460–470. doi: 10.1093/aob/mcs061
Rout, M. E., Chrzanowski, T. H., Westlie, T. K., DeLuca, T. H., Callaway, R. M., and Holben, W. E. (2013). Bacterial endophytes enhance competition by invasive plants. Am. J. Bot. 100, 1726–1737. doi: 10.3732/ajb.1200577
Sanon, A., Andrianjaka, Z. N., Prin, Y., Bally, R., Thioulouse, J., Comte, G.,et al. (2009). Rhizosphere microbiota interfers with plant–plant interactions. Plant Soil 321, 259–278. doi: 10.1007/s11104-009-0010-5
Schaad, N. W., Jones, J. B., and Chun, W., eds. (2001). Laboratory Guide for Identification of Plant Pathogenic Bacteria, Vol. 373. Minnesota: APS Press.
Simberloff, D., Martin, J.-L., Genovesi, P., Maris, V., Wardle, D. A., Aronson, J.,et al. (2013). Impacts of biological invasions: what’s what and the way forward. Trends Ecol. Evol. 28, 58–66. doi: 10.1016/j.tree.2012.07.013
Simon, C., and Daniel, R. (2011). Metagenomic analyses: past and future trends. Appl. Environ. Microb. 77, 1153–1161. doi: 10.1128/AEM.02345-10
Staley, J. T., and Konopka, A. (1985). Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 39, 321–346. doi: 10.1146/annurev.mi.39.100185.001541
Stein, L. Y., and Nicol, G. W. (2011). Grand challenges in terrestrial microbiology. Front. Microbiol. 2:6. doi: 10.3389/fmicb.2011.00006
Stinson, K. A., Campbell, S. A., Powell, J. R., Wolfe, B. E., Callaway, R. M., Thelen, G. C.,et al. (2006). Invasive plant suppresses the growth of native tree seedlings by disrupting belowground mutualisms. PLoS Biol. 4:e140. doi: 10.1371/journal.pbio.0040140
Sun, Y., Wolcott, R. D., and Dowd, S. E. (2011). “Tag-encoded FLX-amplicon pyrosequencing for the elucidation of microbial and function gene diversity in any environment,” in Methods in Molecular Biology, Vol. 733, eds Y. M. Kwon and S. C. Ricke (Totowa, NJ: Humana Press), 129–141.
Thomas, T., Gilbert, J., and Meyer, F. (2012). Metagenomics – a guide from sampling to data analysis. Microb. Inform. Exp. 2:3. doi: 10.1186/2042-5783-2-3
Thompson, K., and Davis, M. A. (2011). Why research on traits of invasive plants tells us very little. Trends Ecol. Evol. 26, 155–156. doi: 10.1016/j.tree.2011.01.007
Tringe, S. G., and Rubin, E. M. (2005). Metagenomics: DNA sequencing of environmental samples. Nat. Rev. Genet. 6, 805–814. doi: 10.1038/nrg1709
Turner, T. R., Ramakrishnan, K., Walshaw, J., Heavens, D., Alston, M., Swarbeck, D.,et al. (2013). Comparative metatranscriptomics reveals kingdom level changes in the rhizosphere microbiome of plants. ISME J. 7, 2248–2258. doi: 10.1038/ismej.2013.119
Vakhlu, J., Sudan, A. K., and Johri, B. (2008). Metagenomics: future of microbial gene mining. Indian J. Microbiol. 48, 202–215. doi: 10.1007/s12088-008-0033-2
Van der Putten, W. H., Klironomos, J. N., and Wardle, D. A. (2007). Microbial ecology of biological invasions. ISME J. 1, 28–37. doi: 10.1038/ismej.2007.9
van Elsas, J. D., and Boersma, F. G. H. (2011). A review of molecular methods to study the microbiota of soil and the mycosphere. Eur. J. Soil Biol. 47, 77–87. doi: 10.1016/j.ejsobi.2010.11.010
van Wees, S. C., Van der Ent, S., and Pieterse, C. M. (2008). Plant immune responses triggered by beneficial microbes. Curr. Opin. Plant Biol. 11, 443–448. doi: 10.1016/j.pbi.2008.05.005
Venter, J. C., Remington, K., Heidelberg, J. F., and Halpern, A. L. (2004). Environmental genome shotgun sequencing of the Sargasso Sea. Science 304, 66–74. doi: 10.1126/science.1093857
Vila, M., Williamson, M., and Lonsdale, M. (2004). Competition experiments on alien weeds with crops: lessons for measuring plant invasion impact? Biol. Invasions 6, 59–69. doi: 10.1023/B:BINV.0000010122.77024.8a
Wang, B., and Qiu, Y. L. (2006). Phylogenetic distribution and evolution of mycorrhizas in land plants. Mycorrhiza 16, 299–363. doi: 10.1007/s00572-005-0033-6
Wooley, J. C., Godzik, A., and Friedberg, I. (2010). A primer on metagenomics. PLoS Comput. Biol. 6:e1000667. doi: 10.1371/journal.pcbi.1000667
Xiao, H. F., Feng, Y. L., Schaefer, D. A., and Yang, X. D. (2014). Soil fungi rather than bacteria were modified by invasive plants, and that benefited invasive plant growth. Plant Soil 378, 253–264. doi: 10.1007/s11104-014-2040-x
Yang, Q., Carrillo, J., Jin, H., Shang, L., Hovick, S. M., Nijjer, S.,et al. (2013). Plant–soil biota interactions of an invasive species in its native and introduced ranges: Implications for invasion success. Soil Biol. Biochem. 65, 78–85. doi: 10.1016/j.soilbio.2013.05.004
Yousuf, B., Keshri, J., Mishra, A., and Jha, B. (2012). Application of targeted metagenomics to explore abundance and diversity of CO2-fixing bacterial community using cbbL gene from the rhizosphere of Arachis hypogaea. Gene 506, 18–24. doi: 10.1016/j.gene.2012.06.083
Yuan, Y., Wang, B., Zhang, S., Tang, J., Tu, C., Hu, S.,et al. (2013). Enhanced allelopathy and competitive ability of invasive plant Solidago canadensis in its introduced range. J. Plant Ecol. 6, 253–263. doi: 10.1093/jpe/rts033
Keywords: rhizosphere, microbiome, plant–microbe interactions, invasive plant, soil
Citation: Coats VC and Rumpho ME (2014) The rhizosphere microbiota of plant invaders: an overview of recent advances in the microbiomics of invasive plants. Front. Microbiol. 5:368. doi: 10.3389/fmicb.2014.00368
Received: 29 April 2014; Paper pending published: 27 May 2014;
Accepted: 01 July 2014; Published online: 23 July 2014.
Edited by:
Monica Medina, Pennsylvania State University, USAReviewed by:
Scott Clingenpeel, DOE Joint Genome Institute, USADetmer Sipkema, Wageningen University, Netherlands
Copyright © 2014 Coats and Rumpho. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mary E. Rumpho, Department of Molecular and Cell Biology, University of Connecticut, 91 North Eagleville Road, Unit 3125, Storrs, CT 06269, USA e-mail:rumpho@uconn.edu
 Vanessa C. Coats
Vanessa C. Coats