
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 15 July 2014
Sec. Terrestrial Microbiology
Volume 5 - 2014 | https://doi.org/10.3389/fmicb.2014.00346
This article is part of the Research Topic The Impact of Microorganisms on Consumption of Atmospheric Trace GasesView all 15 articles
Isoprene and monoterpenes constitute a significant fraction of new plant biomass. Emission rates into the atmosphere alone are estimated to be over 500 Tg per year. These natural hydrocarbons are mineralized annually in similar quantities. In the atmosphere, abiotic photochemical processes cause lifetimes of minutes to hours. Microorganisms encounter isoprene, monoterpenes, and other volatiles of plant origin while living in and on plants, in the soil and in aquatic habitats. Below toxic concentrations, the compounds can serve as carbon and energy source for aerobic and anaerobic microorganisms. Besides these catabolic reactions, transformations may occur as part of detoxification processes. Initial transformations of monoterpenes involve the introduction of functional groups, oxidation reactions, and molecular rearrangements catalyzed by various enzymes. Pseudomonas and Rhodococcus strains and members of the genera Castellaniella and Thauera have become model organisms for the elucidation of biochemical pathways. We review here the enzymes and their genes together with microorganisms known for a monoterpene metabolism, with a strong focus on microorganisms that are taxonomically validly described and currently available from culture collections. Metagenomes of microbiomes with a monoterpene-rich diet confirmed the ecological relevance of monoterpene metabolism and raised concerns on the quality of our insights based on the limited biochemical knowledge.
Annually about 120 Pg of carbon dioxide are assimilated by plants. A part is transformed into chemically complex molecules and released into the environment by emission or excretion (Ghirardo et al., 2011). Volatile organic compounds (VOCs) comprise a large number of molecules, including various hydrocarbons, single carbon compounds (e.g. methane), isoprene and terpenes (e.g. mono- and sesquiterpenes). The atmosphere is loaded with an estimated VOC emission rate of about 1150 Tg C yr−1 (Stotzky and Schenck, 1976; Guenther et al., 1995; Atkinson and Arey, 2003). These estimates included only non-methane VOCs of biogenic origin (BVOCs); a second source are anthropogenic VOCs. Among the BVOCs, isoprene and monoterpenes dominate with estimated emission rates of about 500 Tg C yr−1 and 127 Tg C yr−1, respectively (Guenther et al., 1995). Monoterpenes (C10H16) consist of two linked isoprene (C5H8) units and include in the strict sense only hydrocarbons. Often the term monoterpene is applied including monoterpenoids which are characterized by oxygen-containing functional groups. Structural isomers—acyclic, mono-, and bicyclic monoterpenes—, stereoisomers as well as a variety of substitutions result in a large diversity of molecules. Today, more than 55,000 different isoprenoids are known (Ajikumar et al., 2008). Monoterpenes are not only emitted as cooling substances (Sharkey et al., 2008), but can also be stored intracellularly serving mainly as deterrent or infochemical (Dudareva et al., 2013). Wood plants mainly accumulate pinene and other pure hydrocarbon monoterpenes as constituents of their resins, whereas citrus plants are the major source of limonene. Flowers, however, produce and emit a variety of oxygenated monoterpenes (e.g. linalool) (Kesselmeier and Staudt, 1999 and references therein, Sharkey and Yeh, 2001; Bicas et al., 2009).
In the atmosphere, monoterpenes are transformed in purely chemical reactions within hours. Photolysis and reactions with molecular oxygen, ozone, hydroxyl radicals, NOx species, and chlorine atoms result in carbonyls, alcohols, esters, halogenated hydrocarbons, and peroxynitrates. These products condense and lead to the formation of secondary aerosols. Rain or precipitation transports them to soils (Atkinson and Arey, 2003; Fu et al., 2009; Ziemann and Atkinson, 2012). Monoterpenes reach the surface layers of soils by leaf fall and excreted resins. Also roots emit monoterpenes into the rhizosphere (Wilt et al., 1993; Kainulainen and Holopainen, 2002). Deeper soil layers do contain significant less monoterpenes than the surface soil layer. Emission into the atmosphere and biotransformations in the surface layer mainly by microorganisms are the major sinks. An alternative, abiotic photoreactions like in the atmosphere, is limited by light availability in soil (Kainulainen and Holopainen, 2002; Insam and Seewald, 2010).
Bacteria encountering monoterpenes have to deal with their toxic effects (reviewed by Bakkali et al., 2008). In order to prevent the accumulation of monoterpenes in the cell and cytoplasmatic membrane, bacteria modify their membrane lipids, transform monoterpenes and use active transport by efflux pumps (Papadopoulos et al., 2008; Martinez et al., 2009). Below toxic concentrations monoterpenes are used by microorganisms as sole carbon and energy source. The mineralization of the hydrocarbons requires the introduction of functional groups to access beta-oxidation like fragmentation reactions yielding central metabolites, e.g. acetyl-CoA. In many aerobic microorganisms molecular oxygen serves as reactive agent to functionalize the monoterpenes (Figure 1). Strains of Pseudomonas and Rhodococcus have become model organisms for the elucidation of pathways in aerobic bacteria. Nearly 40 years after the first reports on aerobic mineralization (Seubert, 1960; Seubert and Fass, 1964; Dhavalikar and Bhattacharyya, 1966; Dhavalikar et al., 1966), the mineralization of monoterpenes in denitrifying bacteria and methanogenic communities was discovered (Harder and Probian, 1995; Harder and Foss, 1999). Betaproteobacterial strains of the genera Castellaniella and Thauera are the study objects for the elucidation of anaerobic pathways. All these bacteria were obtained in single-fed batch enrichments with high substrate concentrations (mmol*L−1), in contrast to low concentrations in nature (μmol*L−1). Consequently, in batch enrichments isolated strains exhibit often a solvent tolerance; they grow in the presence of a pure monoterpene phase. Cultivation was rarely attempted by physical separation followed by single-fed batch cultivations. Such dilution-to-extinction series performed in replicates—also known as most-probable-number (MPN) method—revealed a frequent presence of the degradative capacities in natural populations: denitrifying communities in sewage sludge and forest soil yielded 106–107 monoterpene-utilizing cells ml−1, representing 0.7–100% of the total cultivable nitrate-reducing microorganisms (Harder et al., 2000). MPN cultivations for aerobic bacteria have not been reported so far, and for both cases the highly abundant bacteria with the capacity to grow on monoterpenes have not been identified.
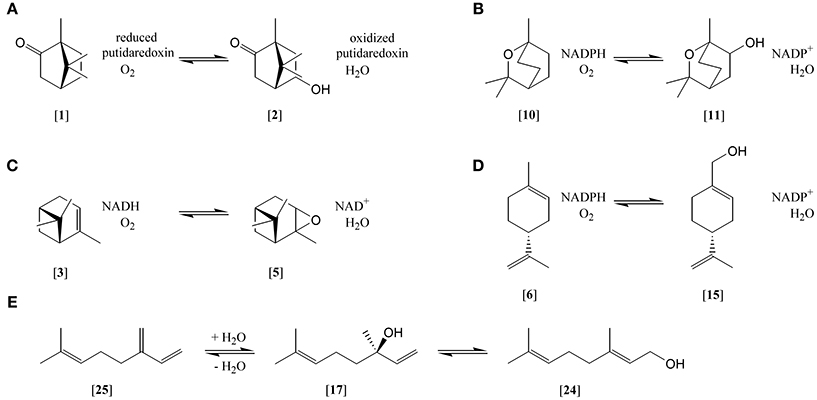
Figure 1. Selected monoterpene transformations. (A) (+)-camphor [1] hydroxlation to 5-hydroxycamphor [2]; (B) 1,8-cineole [10] hydroxylation to hydroxy-1,8-cineole [11]; (C) α-pinene [3] epoxidation to α-pinene oxide [5]; (D) (R)-limonene [6] hydroxylation to perillyl alcohol [15]; (E) myrcene [25] hydration to (S)-(+)-linalool [17] and isomerization to geraniol [24].
Over the last 50 years, many monoterpene transformations have been reported for microbial cultures, but the biochemical pathways were rarely disclosed. More important for the maintenance of our knowledge, only a small portion of the investigated strains were deposited in culture collections. Without detailed knowledge of genes or the availability of strains, the observations of biotransformation experiments are of limited value for future studies. Therefore, this review on the transformation of monoterpenes focusses on enzymes for which the gene and protein sequences are available in public databases as well as on microorganisms that at least have been deposited in a public culture collection and ideally are validly described (Table 1). A broad overview on microbial biotransformations is also provided by a number of older review articles (Trudgill, 1990, 1994; van der Werf et al., 1997; Hylemon and Harder, 1998; Duetz et al., 2003; Ishida, 2005; Li et al., 2006; Bicas et al., 2009; Li and Lan, 2011; Schewe et al., 2011; Tong, 2013). KEGG and MetaCyc, two widely used reference datasets of metabolic pathways (reviewed by Altman et al., 2013), include degradation pathways of limonene, pinene, geraniol, and citronellol. Single reactions of p-cymene and p-cumate degradation are covered. MetaCyc additionally covers the metabolism of myrcene, camphor, eucalyptol, and carveol.
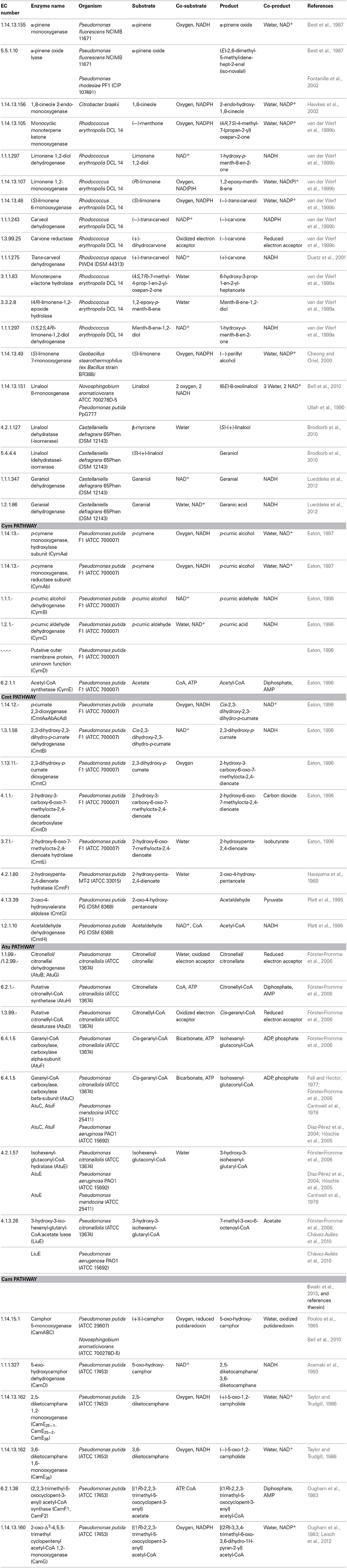
Table 1. Summary table of monoterpene transforming enzymes in validly described species of Bacteria.
(+)-Camphor [1, Figure 2] (C10H16O) is the substrate of one of the first and best described monoterpene transforming enzymes, a specific cytochrome P450 monooxygenase (camABC, P450cam, EC 1.14.15.1) from Pseudomonas putida (ATCC 17453). Initially, (+)-camphor is hydroxylated. The resulting 5-exo-hydroxycamphor [2] is oxidized by a NAD-reducing dehydrogenase (EC 1.1.1.327) which gene camD is part of the operon camDCAB. The diketone is oxidized in a Baeyer–Villiger like oxidation to a lactone, either by a 2,5-diketocamphane 1,2-monooxygenase or a 3,6-diketocamphane 1,6-monooxygenase (camE25−1E25−2 or camE36, EC 1.14.13.162). The lactone spontaneously hydrolyses to 2-oxo-Δ3-4,5,5-trimethylcyclopentenyl-acetic acid which is activated as coenzyme A thioester by a specific synthase (camF1,2, EC 6.2.1.38). This CoA-ester serves as substrate for another specific monooxygenase (camG, EC 1.14.13.160), which initiates the cleavage of the second ring by formation of a lactone. After hydrolysis of the lactone, the linear product is oxidized to isobutanoyl-CoA and three acetyl-CoA. All corresponding genes (camABCDEFG) have been identified on a linear plasmid (Ougham et al., 1983; Taylor and Trudgill, 1986; Aramaki et al., 1993; Kadow et al., 2012; Leisch et al., 2012; Iwaki et al., 2013).
The most abundant bicyclic monoterpene is pinene with the isomers α-pinene [3] and β-pinene [4] (C10H16), a main constituent of wood resins (e.g. conifers). Pseudomonas rhodesiae (CIP 107491) and P. fluorescens (NCIMB 11671) grew on α-pinene as sole carbon source. α-pinene is oxidized to α-pinene oxide [5] by a NADH-dependent α-pinene oxygenase (EC 1.14.12.155) and undergoes ring cleavage by action of a specific α-pinene oxide lyase (EC 5.5.1.10), forming apparently isonovalal as first product which is isomerized to novalal (Best et al., 1987; Bicas et al., 2008; Linares et al., 2009). The cleavage reaction of α-pinene oxide was also described for a Nocardia sp. strain P18.3 (Griffiths et al., 1987; Trudgill, 1990, 1994).
An alternative route for pinene degradation via a monocyclic p-menthene derivate has been described for Pseudomonas sp. strain PIN (Yoo and Day, 2002). Bacillus pallidus BR425 degrades α- and β-pinene apparently via limonene [6] and pinocarveol. While α-pinene is transformed into limonene and pinocarveol, β-pinene yields pinocarveol only. Both intermediates may be further transformed into carveol [7] and carvone. The activity of a specific monooxygenases has been suggested, but experimental evidence is lacking (Savithiry et al., 1998). Serratia marcescens uses α-pinene as sole carbon source. Trans-verbenol [8] was a detectable metabolite. In glucose and nitrogen supplemented medium, this strain formed α-terpineol [9]. The two oxidation products were considered to be dead-end products as they accumulated in cultures (Wright et al., 1986). A general precaution has to be mentioned here for many biotransformation studies: monoterpenes contain often impurities and oxidation products which may be utilized as substrates resulting in traces of monoterpene and monoterpenoid transformation products that are not further metabolized. Stoichiometric experiments have to show that the amount of metabolite is larger than the amount of impurity in the substrate. Only such careful stoichiometric experiments, mutants in functional genes or the characterization of enzymes in vitro can provide a proof of the presence of a biotransformation.
Eucalyptol, the bicyclic monoterpene 1,8-cineole [10] (C10H18O), is transformed in several pathways. Novosphingobium subterranea converts 1,8-cineole initially into 2-endo-hydroxycineole, 2,2-oxo-cineole, and 2-exo-hydroxycineole. Acidic products from ring cleavages have been identified in situ (Rasmussen et al., 2005). Hydroxy-cineole formation occurred in 1,8-cineole-grown cultures of Pseudomonas flava (Carman et al., 1986). A cytochrome P450 monooxygenase from Bacillus cereus UI-1477 catalyzes the hydroxylation of 1,8-cineole, yielding either 2R-endo- or 2R-exo-hydroxy-1,8-cineole [11] (Liu and Rosazza, 1990, 1993). Another 1,8-cineole-specific P450 monooxygenase (EC 1.14.13.156) has been purified and characterized from Citrobacter braakii, which yielded 2-endo-hydroxy-1,8-cineole only. Further oxidation and lactonization were followed by a spontaneous lactone ring hydrolysis (Hawkes et al., 2002). Biotransformation in Rhodococcus sp. C1 involves an initial hydroxylation to 6-endo-hydroxycineol [12] and further oxidation to 6-oxocineole by a 6-endo-hydroxycineol dehydrogenase (EC 1.1.1.241). A 6-oxocineole monooxygenase (EC 1.14.13.51) converts the ketone into an unstable lactone. Spontaneous decomposition results in (R)-5,5-dimethyl-4-(3′-oxobutyl)-4,5-dihydrofuran-2(3H)-one. An initial monooxygenase activity has not been detected in cell-free systems, while the dehydrogenase and oxygenase activities have been measured in crude cell extracts (Williams et al., 1989).
Limonene [6, Figure 3] (C10H16) is the most abundant monocyclic monoterpene, besides toluene the second most abundant VOC indoors (Brown et al., 1994). It represents the main component of essential oils from citrus plants, e.g. lemon and orange. Rhodococcus erythropolis DCL14 transforms (R/S)-limonene via limonene-1,2-epoxide into limonene-1,2-diol [13, Figure 5], applying a limonene-1,2 monooxygenase (EC 1.14.13.107) and a limonene-1,2-epoxide hydrolase (EC 3.3.2.8), respectively. A specific dehydrogenase (EC 1.1.1.297) forms the ketone, 1-hydroxy-2-oxolimonene, which is oxidized to a lactone by a 1-hydroxy-2-oxolimonene 1,2-monooxygenase (EC 1.14.13.105). Enzyme activities were only detected in limonene-induced cells, suggesting a tight regulation of the limonene degradation. R. erythropolis DCL14 harbors a second pathway for limonene degradation. Initially, (R)-limonene is hydroxylated by a NADPH-dependent limonene 6-monooxygenase (EC 1.14.13.48) to trans-carveol [7]. Subsequently, trans-carveol is oxidized to carvone and dihydrocarvone by a carveol dehydrogenase (EC 1.1.1.243) and carvone reductase (EC 1.3.99.25), respectively. A monocyclic monoterpene ketone monooxygenase (EC 1.14.13.105) inserts an oxygen atom, forming isopropenyl-7-methyl-2-oxo-oxepanone [14, Figure 6]. This lactone is cleaved by a specific ε-lactone hydrolase (EC 3.1.1.83) yielding hydroxyl-3-isopropenyl-heptanoate. Oxidation and activation as coenzyme A thioester enable a further degradation in accordance to the beta-oxidation (van der Werf et al., 1999b; van der Werf and Boot, 2000). R. opacus PWD4 uses (R)-limonene on the same pathway. Biomass from a glucose-toluene chemostat culture transformed limonene into trans-carveol, which was further oxidized to carvone by a trans-carveol dehydrogenase (EC 1.1.1.275) (Duetz et al., 2001).
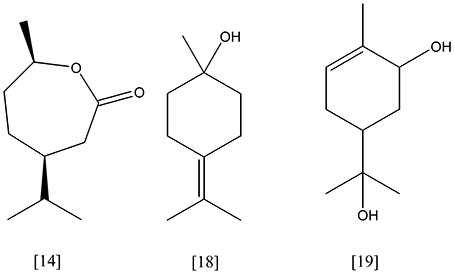
Figure 6. isopropenyl-7-methyl-2-oxo-oxepanone [14]; γ -terpineol [18]; p-menth-1-ene-6,8-diol [19].
Studies on the limonene metabolism in P. gladioli identified α-terpineol [9, Figure 4] and perillyl alcohol [15] as major metabolites. However, none of the involved enzymes has been purified or further characterized (Cadwallader et al., 1989). A α-terpineol dehydratase from P. gladioli was isolated and partially purified. The hydration reaction to the isopropenyl double bond of (4R)-(+)-limonene resulted in (4R)-(+)-α-terpineol as only product (Cadwallader et al., 1992). Geobacillus stearothermophilus (ex Bacillus) showed growth on limonene as sole carbon source. The main limonene transformation product was perillyl alcohol, while α-terpineol and perillyl aldehyde were found in minor concentrations. After heterologous expression of a putative limonene degradation pathway in E. coli, α-terpineol was identified as major product of the biotransformation. Other studies reported a limonene hydroxylation on the methyl group yielding perillyl alcohol, which underwent further oxidation to perillic acid (Chang and Oriel, 1994; Chang et al., 1995). Additional studies on the recombinant limonene hydroxylase confirmed the production of perillyl alcohol from limonene but revealed in addition the formation of carveol. The limonene hydroxylase showed dependency on molecular oxygen and NADH as cofactors and was suggested to belong to the (S)-limonene 7-monooxygenase family (EC 1.14.13.49) (Cheong and Oriel, 2000).
Enterobacter agglomerans 6L and Kosakonia cowanii 6L (ex Enterobacter cowanii) transformed (R)-limonene [6]. The main metabolites detected in ether extracts of E. agglomerans 6L cultures were γ-valerolactone and cryptone [16]. In assays using four recombinant expressed limonene-transforming enzymes from K. cowanii 6L, linalool [17, Figure 8] was identified as main product besides smaller amounts of dihydrolinalool. It was proposed that the potential limonene hydroxylase converts limonene into linalool, perillyl alcohol, α-terpineol and γ-terpineol [18] (Park et al., 2003; Yang et al., 2007).
Pseudomonas putida (MTCC 1072) converts limonene to p-menth-1-ene-6,8-diol [19] and perillyl alcohol (Chatterjee and Bhattacharyya, 2001). No sequence information was found in public databases. Two other strains of Pseudomonas putida (F1 and GS1) have been found to convert (+)-limonene to perillic acid in co-substrate fed-batch cultures (Speelmans et al., 1998). Experimental results indicated the participation of the p-cymene pathway (CYM) (Mars et al., 2001). Castellaniella defragrans grows anaerobically on cyclic monoterpenes as sole carbon and energy source under denitrifying conditions (Foss et al., 1998). Recent experiments suggested an oxygen-independent hydroxylation on the methyl group of limonene to perillyl alcohol as the initial activation step, followed by subsequent oxidation to perillic acid (Petasch et al., 2014).
P-cymene [20, Figure 7] (C10H14) is an aromatic monoterpene (p-isopropyl-toluene). Pseudomonas putida F1 (ATCC 700007) degrades p-cymene to p-cumate [21] via the CYM-pathway (cymBCAaAbDE). A two-component p-cymene monooxygenase (cymAaAb, EC 1.14.13.-) introduces a hydroxyl group on the methyl group of p-cymene. The resulting p-cumic alcohol is oxidized to the corresponding carboxylic acid by an alcohol and an aldehyde dehydrogenase (cymB and cymC, EC 1.1.1.- and EC 1.2.1.-). The genes cymD and cymE encode for a putative outer membrane protein and an acetyl coenzyme A synthetase, respectively. However, their role in the pathway remains unclear (Eaton, 1997). Upstream of the cym-operon, the genes for the further degradation of p-cumate are located. They are organized in another operon and comprise eight genes (cmtABCDEFGH). P. putida F1 has been shown to use p-cumate as sole carbon source. It is hydroxylated by a ferredoxin dependent p-cumate 2,3-dioxygenase. The genes cmtAaAd encode a ferredoxin reductase and a ferredoxin, and cmtAbAc encode the large and the small subunits of the dioxygenase (EC 1.14.12.-). The resulting cis-2,3-dihydroxy-2,3-dihydro-p-cumate is oxidized and ring cleavage occurs by introduction of another oxygen molecule. The responsible enzymes are a specific dehydrogenase (cmtB, EC 1.3.1.58) and a 2,3-dihydroxy-p-cumate dioxygenase (cmtC, EC 1.13.11.-), respectively. Further degradation is accomplished by a decarboxylation and elimination of an isobutyrate molecule, catalyzed by a 2-hydroxy-3-carboxy-6-oxo-7-methylocta-2,4-dienoate decarboxylase (cmtD, EC 4.1.1.-) and a 2-hydroxy-6-oxo-7-methylocta-2,4-dienoate hydrolase (cmtE, EC 3.7.1.-). The product, 2-hydroxypenta-2,4-dienoate, undergoes a water addition by a specific hydratase (cmtF, EC 4.2.1.80). Then, a carbon-carbon lyase reaction yields pyruvate and acetaldehyde, catalyzed by 2-oxo-4-hydroxyvalerate aldolase (cmtG, EC 4.1.3.39). Acetaldehyde is oxidized and enters as acetyl-CoA the citrate cycle (Eaton, 1996).
Thauera terpenica 21 Mol utilizes menthol [22] as sole carbon source. The proposed degradation mechanism involves two initial oxidation reactions leading to menth-2-enone, followed by a hydration and an additional oxidation step. Finally, ring cleavage may occur and the molecule is attached to coenzyme A to yield 3,7-dimethyl-5-oxo-octyl-CoA (Foss and Harder, 1998; Hylemon and Harder, 1998).
First studies on acyclic monoterpenoids in the early sixties by Seubert and colleagues described the degradation of citronellol [23], geraniol [24], and nerol via an oxidation of the alcohol to an acid, followed by the formation of a CoA-thioester and subsequent beta-oxidation in Pseudomonas citronellolis (ATCC 13674) (Seubert, 1960; Seubert and Remberger, 1963; Seubert et al., 1963; Seubert and Fass, 1964). This knowledge has been extended toward other Pseudomonas strains (Cantwell et al., 1978). The complete degradation pathway has been classified as the acyclic terpene utilization and leucine utilization (ATU/LIU) pathway involving the genes atuABCDEFGH and liuRABCDE. After the initial formation of cis-geranyl-CoA, a geranyl-coenzyme-A carboxylase (atuCF, EC 6.4.1.5) elongates the methylgroup. A hydroxyl group is introduced by an isohexenyl-glutaconyl-CoA hydratase (atuE, EC 4.2.1.57), followed by a water addition and elimination of an acetate molecule catalyzed by a 3-hydroxy-3-isohexenylglutaryl-CoA lyase (liuE, EC 4.1.3.26). The resulting 7-methyl-3-oxooct-6-enoyl-CoA is further degraded via two beta-oxidation like reactions to yield 3-methylcrotonyl-CoA, which enters the leucine degradation pathway (liuRABCDE) (Höschle et al., 2005; Aguilar et al., 2006; Förster-Fromme et al., 2006; Chávez-Avilés et al., 2010; Förster-Fromme and Jendrossek, 2010). Citronellol degradation is reported for many Pseudomonas strains, including P. aeruginosa PAO1 (ATCC 15692), P. mendocina (ATCC 25411), and P. delhiensis (DSM 18900) (Cantwell et al., 1978; Prakash et al., 2007; Förster-Fromme and Jendrossek, 2010). Among the few reactions described in detail is a molybdenum dependent dehydrogenase responsible for the geranial oxidation to geranylate in P. aeruginosa PAO1 (Höschle and Jendrossek, 2005).
The acyclic monoterpene β-myrcene [25] (C10H16) is transformed by Pseudomonas aeruginosa (PTCC 1074) into dihydrolinalool, 2,6-dimethyloctane and α-terpineol. Limonene has been proposed as possible intermediate in α-terpineol formation but was not detected in the culture broth (Esmaeili and Hashemi, 2011). Pseudomonas sp. M1 accomplishes degradation by hydroxylation on the C8 position to myrcene-8-ol, which is further oxidized, linked to coenzyme A and metabolized in a beta-oxidation like manner (Iurescia et al., 1999). The formation of geraniol from β-myrcene has been observed with resting cells of Rhodococcus erythropolis MLT1, regardless of the presence of a cytochrome P450 inhibitor. The reaction was dependent on aerobic conditions, however it remains unclear if a monooxygenase or lyase system is involved (Thompson et al., 2010).
The tertiary alcohol linalool is also transformed at the C8 position. A linalool monooxygenase (EC 1.14.13.151) has been described in P. putida PpG777 and Novosphingobium aromaticivorans (ATCC 700278D-5) (Ullah et al., 1990; Bell et al., 2010). In the absence of molecular oxygen, Castellaniella defragrans 65Phen has an unique enzyme for the linalool transformation, the linalool dehydratase-isomerase (Brodkorb et al., 2010). Castellaniella and Thauera strains were the first anaerobic microorganisms shown to anaerobically degrade and mineralize monoterpenes (Harder and Probian, 1995; Harder et al., 2000). The linalool dehydratase-isomerase (EC 4.2.1.127 and 5.4.4.4) of C. defragrans 65Phen catalyzes a regio- and stereo-specific hydration of β-myrcene yielding the tertiary alcohol (S)-(+)-linalool [17] and the isomerization to the primary alcohol geraniol (Brodkorb et al., 2010; Lueddeke and Harder, 2011). Geraniol and geranial dehydrogenases formed geranic acid (Heyen and Harder, 2000; Lueddeke et al., 2012). T. linaloolentis 47Lol grows on linalool as sole carbon and energy source. A similar isomerization of linalool to geraniol with subsequent oxidation of geraniol to geranial has been observed in cultures (Foss and Harder, 1997).
Fungi excrete laccases which are copper-containing oxidases. Utilizing molecular oxygen as a cosubstrate, an unspecific oxidation of organic molecules is initiated by these enzymes. Additionally, fungi express a variety of cytochrome P450 mono- and di-oxygenases. Thus, several fungi were described to transform monoterpenes during growth in rich medium (reviewed by Farooq et al., 2004). Species with a reported capacity to transform monoterpenes are Aspergillus niger, Botrytis cinerea, Diplodia gossypina, Mucor circinelloides, Penicillium italicum, Penicillium digitatum, Corynespora cassiicola, and Glomerella cingulata. For a long time, no species have been described to use monoterpenes as sole carbon and energy source for growth (Trudgill, 1994 and references therein). Recently, Grosmannia clavigera, a bark beetle-associated fungal pathogen of pine trees, was shown to grow on a mono- and diterpene mixture, containing α/β-pinene and 3-carene (Diguistini et al., 2011). ABC efflux transporter and cytochrome P450 enzymes confer a monoterpene resistance to the blue-stain fungi (Lah et al., 2013; Wang et al., 2013).
Habitats with a dense vegetation of wood and flowers are expected to contain larger populations of monoterpene transforming microorganisms. Whereas coniferous forests emit up to 6.7 g carbon*m−2*yr−1, broadleaf evergreen forest and grassland emit only 3.5 and 2.5 g carbon*m−2*yr−1, respectively (Tanaka et al., 2012). Monoterpene emission rates between 0.3 and 7 g carbon*m−2*yr−1 for the United States—mainly α- and β-pinene, limonene and β-myrcene (Geron et al., 2000)—can support the aerobic growth of 0.15–3.5 g bacteria*m−2*yr−1, assuming 50% of carbon incorporated into biomass. This is a significant potential, considering the presence of around 10 g microbial biomass in the top centimeter of soil per square meter.
In marine systems, isoprene and monoterpenes (mainly α-pinene) are produced by phytoplankton and algae and partially emitted into the atmosphere (reviewed by Yassaa et al., 2008; Shaw et al., 2010). Isoprene emission was estimated to 0.2–1.2 Tg carbon*yr−1 (Palmer and Shaw, 2005; Gantt et al., 2009; Shaw et al., 2010). For the ocean surface area this results in an emission rate of 0.0025 g carbon*m−2*yr−1. Current uncertainties in the size of emission based on shipborne measurements in comparison to satellite data (Luo and Yu, 2010) may be resolved by incorporating an export from the continental atmosphere to the oceanic atmosphere (Hu et al., 2013). Isoprene-amended samples from marine habitats were enriched in bacteria affiliating with Actinobacteria, Alphaproteobacteria, and Bacteroidetes and first strains were shown to degrade isoprene and aliphatic hydrocarbons (Acuña Alvarez et al., 2009).
In summary, these findings indicate a higher abundance of monoterpene transforming and mineralizing bacteria in soils than in the ocean. Indeed, most monoterpene transforming bacteria have been enriched or isolated from soil and freshwater samples in habitats with monoterpene emitting vegetation.
Databases are nowadays available for the analysis of enzymatic reactions and metabolic pathways in metagenomic and genomic sequence datasets. The most relevant are the Kyoto Encyclopedia of Genes and Genomes (KEGG), MetaCyc and the Biocatalysis/Biodegradation database of the University of Minnesota.
First studies used KEGG to identify monoterpene-related genes in metagenomes of microbiomes in insects and nematodes feeding on a monoterpene-rich diet. Pine beetles encounter the high terpenoid concentrations of conifers and may take advantage of detoxification processes catalyzed by their symbionts/microbiomes (Adams et al., 2013). The KEGG pathway for limonene and pinene degradation (ko00903) was used to identify genes encoding enzymes putatively involved in monoterpene degradation. Five enzymes were present and more abundant in the metagenomes than in a combined metagenomic set of plant biomass-degrading communities. These enzymes were an aldehyde dehydrogenase, an oxidoreductase, an enoyl-CoA hydratase and two hydratases/epimerases. Whether these genes are truly involved in monoterpene metabolism or the degradation of cyclic compounds, e.g. related aromatic lignin monomers, is an open question. Taxonomically, these genes affiliated with the genera Pseudomonas, Rahnella, Serratia, and Stenotrophomonas.
The pinewood nematode Bursaphlenchus xylophilus transcribes cytochrome P450 genes as main metabolic pathway for xenobiotics detoxification, but not all enzymes needed for terpenoid metabolism were detected by transcriptomic analysis. Metagenomic data of nematode bacterial symbionts included the complete α-pinene degradation pathway (Cheng et al., 2013). Annotation based on KEGG revealed that the degradation pathways for limonene and pinene (map00903) and for geraniol (map00281) accounted for 2.5% of mapped metagenes. The majority of these genes affiliated to Pseudomonas, Achromobacter, and Agrobacterium. Strains isolated from the nematode and capable of growth on α-pinene affiliated to Pseudomonas, Achromobacter, Agrobacterium, Cytophaga, Herbaspirillum, and Stenotrophomonas.
The synthesis and transformation of BVOCs, especially terpenoids, by plants is well studied (Kesselmeier and Staudt, 1999). Corresponding pathways have been elucidated and a variety of corresponding enzymes have been isolated and characterized (Mahmoud and Croteau, 2002; Yu and Utsumi, 2009). In contrast, the exploration of the microbial transformation and mineralization of monoterpenes has accumulated a small coverage of the field. Simply, over the last 50 years, research on bacterial monoterpene metabolism had only found the interest of very few principal investigators. Now, large sequence datasets of organisms and biological communities provide an unprecedented insight into the diversity of pathways and provide us with challenging hypotheses. However, the basis for the annotation is the biochemical characterization of enzymes which is only available for few monoterpenes. Only three pathways are completely known on the genetic and enzymatic level: the ones for camphor (CAM), p-cymene (CYM/CMT), and citronellol/geraniol (ATU/LIU). For pinene, the gene for a key enzyme, the α-pinene oxide lyase (EC 5.5.1.10), is still unknown. The lack of such a key enzyme sequence for a KEGG pathway (map00903) illustrates our uncertainty in the interpretation of metagenomic and genomic datasets. Progress in proteomic and metabolomic analyses in the last years support now biochemical and genetic experiments which will swiftly reveal the desired identification of key enzymes in the monoterpene metabolism.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acuña Alvarez, L., Exton, D. A., Timmis, K. N., Suggett, D. J., and McGenity, T. J. (2009). Characterization of marine isoprene-degrading communities. Environ. Microbiol. 11, 3280–3291. doi: 10.1111/j.1462-2920.2009.02069.x
Adams, A. S., Aylward, F. O., Adams, S. M., Erbilgin, N., Aukema, B. H., Currie, C. R., et al. (2013). Mountain pine beetles colonizing historical and native host trees are associated with a bacterial community highly enriched in genes contributing to terpene metabolism. Appl. Environ. Microbiol. 79, 3468–3475. doi: 10.1128/AEM.00068-13
Aguilar, J. A., Zavala, A. N., Díaz-Pérez, C., Cervantes, C., Díaz-Pérez, A. L., and Campos-García, J. (2006). The atu and liu clusters are involved in the catabolic pathways for acyclic monoterpenes and leucine in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 72, 2070–2079. doi: 10.1128/AEM.72.3.2070-2079.2006
Ajikumar, P. K., Tyo, K., Carlsen, S., Mucha, O., Phon, T. H., and Stephanopoulos, G. (2008). Terpenoids: opportunities for biosynthesis of natural product drugs using engineered microorganisms. Mol. Pharm. 5, 167–190. doi: 10.1021/mp700151b
Altman, T., Travers, M., Kothari, A., Caspi, R., and Karp, P. D. (2013). A systematic comparison of the MetaCyc and KEGG pathway databases. BMC Bioinformatics 14:112. doi: 10.1186/1471-2105-14-112
Aramaki, H., Koga, H., Sagara, Y., Hosoi, M., and Horiuchi, T. (1993). Complete nucleotide-sequence of the 5-exo-hydroxycamphor dehydrogenase gene on the CAM plasmid of Pseudomonas putida (ATCC-17453). Biochim. Biophys. Acta 1174, 91–94. doi: 10.1016/0167-4781(93)90098-X
Atkinson, R., and Arey, J. (2003). Atmospheric degradation of volatile organic compounds. Chem. Rev. 103, 4605–4638. doi: 10.1021/cr0206420
Bakkali, F., Averbeck, S., Averbeck, D., and Waomar, M. (2008). Biological effects of essential oils - a review. Food Chem. Toxicol. 46, 446–475. doi: 10.1016/j.fct.2007.09.106
Bell, S. G., Dale, A., Rees, N. H., and Wong, L. L. (2010). A cytochrome P450 class I electron transfer system from Novosphingobium aromaticivorans. Appl. Microbiol. Biotechnol. 86, 163–175. doi: 10.1007/s00253-009-2234-y
Best, D. J., Floyd, N. C., Magalhaes, A., Burfield, A., and Rhodes, P. M. (1987). Initial enzymatic steps in the degradation of alpha-pinene by Pseudomonas fluorescens Ncimb 11671. Biocatal. Biotransfor. 1, 147–159. doi: 10.3109/10242428709040139
Bicas, J. L., Dionisio, A. P., and Pastore, G. M. (2009). Bio-oxidation of terpenes: an approach for the flavor industry. Chem. Rev. 109, 4518–4531. doi: 10.1021/cr800190y
Bicas, J. L., Fontanille, P., Pastore, G. M., and Larroche, C. (2008). Characterization of monoterpene biotransformation in two pseudomonads. J. Appl. Microbiol. 105, 1991–2001. doi: 10.1111/j.1365-2672.2008.03923.x
Brodkorb, D., Gottschall, M., Marmulla, R., Lueddeke, F., and Harder, J. (2010). Linalool dehydratase-isomerase, a bifunctional enzyme in the anaerobic degradation of monoterpenes. J. Biol. Chem. 285, 30436–30442. doi: 10.1074/jbc.M109.084244
Brown, S. K., Sim, M. R., Abramson, M. J., and Gray, C. N. (1994). Concentrations of volatile organic compounds in indoor air - a review. Indoor Air 4, 123–134.
Cadwallader, K. R., Braddock, R. J., and Parish, M. E. (1992). Isolation of alpha-terpineol dehydratase from Pseudomonas gladioli. J. Food Sci. 57, 241. doi: 10.1111/j.1365-2621.1992.tb05464.x
Cadwallader, K. R., Braddock, R. J., Parish, M. E., and Higgins, D. P. (1989). Bioconversion of (+)-limonene by Pseudomonas gladioli. J. Food Sci. 54, 1241–1245. doi: 10.1111/j.1365-2621.1989.tb05964.x
Cantwell, S. G., Lau, E. P., Watt, D. S., and Fall, R. R. (1978). Biodegradation of acyclic isoprenoids by Pseudomonas species. J. Bacteriol. 135, 324–333.
Carman, R. M., Macrae, I. C., and Perkins, M. V. (1986). The oxidation of 1,8-cineole by Pseudomonas flava. Aust. J. Chem. 39, 1739–1746. doi: 10.1071/CH9861739
Chang, H. C., Gage, D. A., and Oriel, P. J. (1995). Cloning and expression of a limonene degradation pathway from Bacillus stearothermophilus in Escherichia coli. J. Food Sci. 60, 551–553. doi: 10.1111/j.1365-2621.1995.tb09824.x
Chang, H. C., and Oriel, P. (1994). Bioproduction of perillyl alcohol and related monoterpenes by isolates of Bacillus stearothermophilus. J. Food Sci. 59, 660. doi: 10.1111/j.1365-2621.1994.tb05588.x
Chatterjee, T., and Bhattacharyya, D. K. (2001). Biotransformation of limonene by Pseudomonas putida. Appl. Microbiol. Biotechnol. 55, 541–546. doi: 10.1007/s002530000538
Chávez-Avilés, M., Díaz-Pérez, A. L., and Campos-García, J. (2010). The bifunctional role of LiuE from Pseudomonas aeruginosa, displays additionally HIHG-CoA lyase enzymatic activity. Mol. Biol. Rep. 37, 1787–1791. doi: 10.1007/s11033-009-9611-6
Cheng, X. Y., Tian, X. L., Wang, Y. S., Lin, R. M., Mao, Z. C., Chen, N. S., et al. (2013). Metagenomic analysis of the pinewood nematode microbiome reveals a symbiotic relationship critical for xenobiotics degradation. Sci. Rep. 3:1869. doi: 10.1038/srep01869
Cheong, T. K., and Oriel, P. J. (2000). Cloning and expression of the limonene hydroxylase of Bacillus stearothermophilus BR388 and utilization in two-phase limonene conversions. Appl. Biochem. Biotechnol. 84–86, 903–915. doi: 10.1385/ABAB:84-86:1-9:903
Dhavalikar, R., and Bhattacharyya, P. (1966). Microbiological transformation of terpenes. 8. Fermentation of limonene by a soil Pseudomonad. Indian J. Biochem. 3, 144–157.
Dhavalikar, R., Rangachari, P., and Bhattacharyya, P. (1966). Microbiological transformations of terpenes. 9. Pathways of degradation of limonene in a soil Pseudomonad. Indian J. Biochem. 3, 158–164.
Díaz-Pérez, A. L., Zavala-Hernandez, A. N., Cervantes, C., and Campos-Garcia, J. (2004). The gnyRDBHAL cluster is involved in acyclic isoprenoid degradation in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 70, 5102–5110. doi: 10.1128/AEM.70.9.5102-5110.2004
Diguistini, S., Wang, Y., Liao, N. Y., Taylor, G., Tanguay, P., Feau, N., et al. (2011). Genome and transcriptome analyses of the mountain pine beetle-fungal symbiont Grosmannia clavigera, a lodgepole pine pathogen. Proc. Natl. Acad. Sci. U.S.A. 108, 2504–2509. doi: 10.1073/pnas.1011289108
Dudareva, N., Klempien, A., Muhlemann, J. K., and Kaplan, I. (2013). Biosynthesis, function and metabolic engineering of plant volatile organic compounds. New Phytol. 198, 16–32. doi: 10.1111/nph.12145
Duetz, W. A., Bouwmeester, H., Van Beilen, J. B., and Witholt, B. (2003). Biotransformation of limonene by bacteria, fungi, yeasts, and plants. Appl. Microbiol. Biotechnol. 61, 269–277. doi: 10.1007/s00253-003-1221-y
Duetz, W. A., Fjallman, A. H. M., Ren, S. Y., Jourdat, C., and Witholt, B. (2001). Biotransformation of D-limonene to (+) trans-carveol by toluene-grown Rhodococcus opacus PWD4 cells. Appl. Environ. Microbiol. 67, 2829–2832. doi: 10.1128/AEM.67.6.2829-2832.2001
Eaton, R. W. (1996). p-Cumate catabolic pathway in Pseudomonas putida F1: cloning and characterization of DNA carrying the cmt operon. J. Bacteriol. 178, 1351–1362.
Eaton, R. W. (1997). p-Cymene catabolic pathway in Pseudomonas putida F1: cloning and characterization of DNA encoding conversion of p-cymene to p-cumate. J. Bacteriol. 179, 3171–3180.
Esmaeili, A., and Hashemi, E. (2011). Biotransformation of myrcene by Pseudomonas aeruginosa. Chem. Cent. J. 5:26. doi: 10.1186/1752-153X-5-26
Fall, R. R., and Hector, M. L. (1977). Acyl-coenzyme-a carboxylases. Homologous 3-methylcrotonyl-coA and geranyl-coA carboxylases from Pseudomonas citronellolis. Biochemistry 16, 4000–4005.
Farooq, A., Atta-Ur-Rahman, and Choudhary, M. I. (2004). Fungal transformation of monoterpenes. Curr. Org. Chem. 8, 353–367. doi: 10.2174/1385272043485945
Fontanille, P., Le Flèche, A., and Larroche, C. (2002). Pseudomonas rhodesiae PF1: a new and efficient biocatalyst for production of isonovalal from α-pinene oxide. Biocatal. Biotransformation 20, 413–421. doi: 10.1080/1024242021000058702
Förster-Fromme, K., Höschle, B., Mack, C., Bott, M., Armbruster, W., and Jendrossek, D. (2006). Identification of genes and proteins necessary for catabolism of acyclic terpenes and leucine/isovalerate in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 72, 4819–4828. doi: 10.1128/AEM.00853-06
Förster-Fromme, K., and Jendrossek, D. (2010). Catabolism of citronellol and related acyclic terpenoids in pseudomonads. Appl. Microbiol. Biotechnol. 87, 859–869. doi: 10.1007/s00253-010-2644-x
Foss, S., and Harder, J. (1997). Microbial transformation of a tertiary allylalcohol: regioselective isomerisation of linalool to geraniol without nerol formation. FEMS Microbiol. Lett. 149, 71–75. doi: 10.1016/S0378-1097(97)00057-8
Foss, S., and Harder, J. (1998). Thauera linaloolentis sp. nov. and Thauera terpenica sp. nov., isolated on oxygen-containing monoterpenes (linalool, menthol, and eucalyptol) and nitrate. Syst. Appl. Microbiol. 21, 365–373. doi: 10.1016/S0723-2020(98)80046-5
Foss, S., Heyen, U., and Harder, J. (1998). Alcaligenes defragrans sp. nov., description of four strains isolated on alkenoic monoterpenes ((+)-menthene, alpha-pinene, 2-carene, and alpha-phellandrene) and nitrate. Syst. Appl. Microbiol. 21, 237–244. doi: 10.1016/S0723-2020(98)80028-3
Fu, P. Q., Kawamura, K., Chen, J., and Barrie, L. A. (2009). Isoprene, monoterpene, and sesquiterpene oxidation products in the high arctic aerosols during late winter to early summer. Environ. Sci. Technol. 43, 4022–4028. doi: 10.1021/es803669a
Gantt, B., Meskhidze, N., and Kamykowski, D. (2009). A new physically-based quantification of marine isoprene and primary organic aerosol emissions. Atmos. Chem. Phys. 9, 4915–4927. doi: 10.5194/acp-9-4915-2009
Geron, C., Rasmussen, R., Arnts, R. R., and Guenther, A. (2000). A review and synthesis of monoterpene speciation from forests in the United States. Atmos. Environ. 34, 1761–1781. doi: 10.1016/S1352-2310(99)00364-7
Ghirardo, A., Gutknecht, J., Zimmer, I., Bruggemann, N., and Schnitzler, J. P. (2011). Biogenic volatile organic compound and respiratory CO2 emissions after 13C-labeling: online tracing of C translocation dynamics in poplar plants. PLoS ONE 6:e17393. doi: 10.1371/journal.pone.0017393
Griffiths, E. T., Harries, P. C., Jeffcoat, R., and Trudgill, P. W. (1987). Purification and properties of alpha-pinene oxide lyase from Nocardia sp. strain P18.3. J. Bacteriol. 169, 4980–4983.
Guenther, A., Hewitt, C. N., Erickson, D., Fall, R., Geron, C., Graedel, T., et al. (1995). A global model of natural volatile organic compound emissions. J. Geophys. Res. Atmos. 100, 8873–8892. doi: 10.1029/94JD02950
Harayama, S., Rekik, M., Ngai, K. L., and Ornston, L. N. (1989). Physically associated enzymes produce and metabolize 2-hydroxy-2,4-dienoate, a chemically unstable intermediate formed in catechol metabolism via meta-cleavage in Pseudomonas putida. J. Bacteriol. 171, 6251–6258.
Harder, J., and Foss, S. (1999). Anaerobic formation of the aromatic hydrocarbon p-cymene from monoterpenes by methanogenic enrichment cultures. Geomicrobiol. J. 16, 295–305. doi: 10.1080/014904599270550
Harder, J., Heyen, U., Probian, C., and Foss, S. (2000). Anaerobic utilization of essential oils by denitrifying bacteria. Biodegradation 11, 55–63. doi: 10.1023/A:1026552724696
Harder, J., and Probian, C. (1995). Microbial degradation of monoterpenes in the absence of molecular oxygen. Appl. Environ. Microbiol. 61, 3804–3808.
Hawkes, D. B., Adams, G. W., Burlingame, A. L., De Montellano, P. R. O., and De Voss, J. J. (2002). Cytochrome P450cin (CYP176A), isolation, expression, and characterization. J. Biol. Chem. 277, 27725–27732. doi: 10.1074/jbc.M203382200
Heyen, U., and Harder, J. (2000). Geranic acid formation, an initial reaction of anaerobic monoterpene metabolism in denitrifying Alcaligenes defragrans. Appl. Environ. Microbiol. 66, 3004–3009. doi: 10.1128/AEM.66.7.3004-3009.2000
Höschle, B., Gnau, V., and Jendrossek, D. (2005). Methylcrotonyl-CoA and geranyl-CoA carboxylases are involved in leucine/isovalerate utilization (Liu) and acyclic terpene utilization (Atu), and are encoded by liuB/liuD and atuC/atuF, in Pseudomonas aeruginosa. Microbiology 151(pt 11), 3649–3656. doi: 10.1099/mic.0.28260-0
Höschle, B., and Jendrossek, D. (2005). Utilization of geraniol is dependent on molybdenum in Pseudomonas aeruginosa: evidence for different metabolic routes for oxidation of geraniol and citronellol. Microbiology 151, 2277–2283. doi: 10.1099/mic.0.27957-0
Hu, Q. H., Xie, Z. Q., Wang, X. M., Kang, H., He, Q. F., and Zhang, P. F. (2013). Secondary organic aerosols over oceans via oxidation of isoprene and monoterpenes from Arctic to Antarctic. Sci. Rep. 3:2280. doi: 10.1038/srep02280
Hylemon, P. B., and Harder, J. (1998). Biotransformation of monoterpenes, bile acids, and other isoprenoids in anaerobic ecosystems. FEMS Microbiol. Rev. 22, 475–488. doi: 10.1111/j.1574-6976.1998.tb00382.x
Insam, H., and Seewald, M. S. A. (2010). Volatile organic compounds (VOCs) in soils. Biol. Fertil. Soils 46, 199–213. doi: 10.1007/s00374-010-0442-3
Ishida, T. (2005). Biotransformation of terpenoids by mammals, microorganisms, and plant-cultured cells. Chem. Biodivers. 2, 569–590. doi: 10.1002/cbdv.200590038
Iurescia, S., Marconi, A. M., Tofani, D., Gambacorta, A., Paterno, A., Devirgiliis, C., et al. (1999). Identification and sequencing of beta-myrcene catabolism genes from Pseudomonas sp. strain M1. Appl. Environ. Microbiol. 65, 2871–2876.
Iwaki, H., Grosse, S., Bergeron, H., Leisch, H., Morley, K., Hasegawa, Y., et al. (2013). Camphor pathway redux: functional recombinant expression of 2,5- and 3,6-diketocamphane monooxygenases of Pseudomonas putida ATCC 17453 with their cognate flavin reductase catalyzing Baeyer-Villiger reactions. Appl. Environ. Microbiol. 79, 3282–3293. doi: 10.1128/AEM.03958-12
Kadow, M., Loschinski, K., Sass, S., Schmidt, M., and Bornscheuer, U. T. (2012). Completing the series of BVMOs involved in camphor metabolism of Pseudomonas putida NCIMB 10007 by identification of the two missing genes, their functional expression in E. coli, and biochemical characterization. Appl. Microbiol. Biotechnol. 96, 419–429. doi: 10.1007/s00253-011-3859-1
Kainulainen, P., and Holopainen, J. K. (2002). Concentrations of secondary compounds in scots pine needles at different stages of decomposition. Soil Biol. Biochem. 34, 37–42. doi: 10.1016/S0038-0717(01)00147-X
Kesselmeier, J., and Staudt, M. (1999). Biogenic volatile organic compounds (VOC): an overview on emission, physiology and ecology. J. Atmos. Chem. 33, 23–88. doi: 10.1023/A:1006127516791
Lah, L., Haridas, S., Bohlmann, J., and Breuil, C. (2013). The cytochromes P450 of Grosmannia clavigera: genome organization, phylogeny, and expression in response to pine host chemicals. Fungal Genet. Biol. 50, 72–81. doi: 10.1016/j.fgb.2012.10.002
Leisch, H., Shi, R., Grosse, S., Morley, K., Bergeron, H., Cygler, M., et al. (2012). Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-Δ3-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453. Appl. Environ. Microbiol. 78, 2200–2212. doi: 10.1128/AEM.07694-11
Li, H. J., and Lan, W. J. (2011). Biotransformation of limonene by microorganisms. Prog. Chem. 23, 2318–2325.
Li, H. J., Lan, W. J., Cai, C. H., Zhou, Y. P., and Lin, Y. C. (2006). Biotransformation of limonene by marine bacteria. Chin. J. Anal. Chem. 34, 946–950. doi: 10.1016/S1872-2040(06)60046-7
Linares, D., Fontanille, P., and Larroche, C. (2009). Exploration of A-pinene degradation pathway of Pseudomonas rhodesiae cip 107491. Application to novalic acid production in a bioreactor. Food Res. Int. 42, 461–469. doi: 10.1016/j.foodres.2008.12.001
Liu, W. G., and Rosazza, J. P. N. (1990). Stereospecific hydroxylation of 1,8-cineole using a microbial biocatalyst. Tetrahedron Lett. 31, 2833–2836. doi: 10.1016/0040-4039(90)80160-N
Liu, W. G., and Rosazza, J. P. N. (1993). A soluble Bacillus cereus cytochrome-P-450cin system catalyzes 1,4-cineole hydroxylations. Appl. Environ. Microbiol. 59, 3889–3893.
Lueddeke, F., and Harder, J. (2011). Enantiospecific (S)-(+)-linalool formation from beta-myrcene by linalool dehydratase-isomerase. Z. Naturforsch. C 66, 409–412. doi: 10.5560/ZNC.2011.66c0409
Lueddeke, F., Wuelfing, A., Timke, M., Germer, F., Weber, J., Dikfidan, A., et al. (2012). Geraniol and geranial dehydrogenases induced in anaerobic monoterpene degradation by Castellaniella defragrans. Appl. Environ. Microbiol. 78, 2128–2136. doi: 10.1128/AEM.07226-11
Luo, G., and Yu, F. (2010). A numerical evaluation of global oceanic emissions of α-pinene and isoprene. Atmos. Chem. Phys. 10, 2007–2015. doi: 10.5194/acp-10-2007-2010
Mahmoud, S. S., and Croteau, R. B. (2002). Strategies for transgenic manipulation of monoterpene biosynthesis in plants. Trends Plant Sci. 7, 366–373. doi: 10.1016/S1360-1385(02)02303-8
Mars, A. E., Gorissen, J. P. L., Van Den Beld, I., and Eggink, G. (2001). Bioconversion of limonene to increased concentrations of perillic acid by Pseudomonas putida GS1 in a fed-batch reactor. Appl. Microbiol. Biotechnol. 56, 101–107. doi: 10.1007/s002530100625
Martinez, J. L., Sánchez, M. B., Martínez-Solano, L., Hernandez, A., Garmendia, L., Fajardo, A., et al. (2009). Functional role of bacterial multidrug efflux pumps in microbial natural ecosystems. FEMS Microbiol. Rev. 33, 430–449. doi: 10.1111/j.1574-6976.2008.00157.x
Ougham, H. J., Taylor, D. G., and Trudgill, P. W. (1983). Camphor revisited - Involvement of a unique monooxygenase in metabolism of 2-oxo-Δ3-4,5,5-trimethylcyclopentenylacetic acid by Pseudomonas putida. J. Bacteriol. 153, 140–152.
Palmer, P. I., and Shaw, S. L. (2005). Quantifying global marine isoprene fluxes using modis chlorophyll observations. Geophys. Res. Lett. 32, L09805. doi: 10.1029/2005GL022592
Papadopoulos, C. J., Carson, C. F., Chang, B. J., and Riley, T. V. (2008). Role of the mexab-oprm efflux pump of Pseudomonas aeruginosa in tolerance to tea tree (Melaleuca altenifolia) oil and its monoterpene components terpinen-4-ol, 1,8-cineole, and alpha-terpineol. Appl. Environ. Microbiol. 74, 1932–1935. doi: 10.1128/AEM.02334-07
Park, Y. J., Kim, I. C., and Chang, H. C. (2003). Microbial conversion of (+)-limonene by an Enterobacter agglomerans isolate. J. Microbiol. Biotechnol. 13, 636–639.
Petasch, J., Disch, E.-M., Markert, S., Becher, D., Schweder, T., Hüttel, B., et al. (2014). The oxygen-independent metabolism of cyclic monoterpenes in Castellaniella defragrans 65Phen. BMC Microbiol. 14:164. doi: 10.1186/1471-2180-14-164
Platt, A., Shingler, V., Taylor, S. C., and Williams, P. A. (1995). The 4-hydroxy-2-oxovalerate aldolase and acetaldehyde dehydrogenase (acylating) encoded by the nahM and nahO genes of the naphthalene catabolic plasmid pWW60-22 provide further evidence of conservation of meta-cleavage pathway gene sequences. Microbiology 141, 2223–2233.
Poulos, T. L., Finzel, B. C., Gunsalus, I. C., Wagner, G. C., and Kraut, J. (1985). The 2.6-A crystal structure of Pseudomonas putida cytochrome-P-450. J. Biol. Chem. 260, 6122–6130.
Prakash, O., Kumari, K., and Lal, R. (2007). Pseudomonas delhiensis sp. nov., from a fly ash dumping site of a thermal power plant. Int. J. Syst. Evol. Microbiol. 57, 527–531. doi: 10.1099/ijs.0.64456-0
Rasmussen, J. A. M., Henderson, K. A., Straffon, M. J., Dumsday, G. J., Coulton, J., and Zachariou, M. (2005). Two new biocatalysts for improved biological oxidation of 1,8-cineole. Aust. J. Chem. 58, 912–916. doi: 10.1071/CH05204
Savithiry, N., Gage, D., Fu, W. J., and Oriel, P. (1998). Degradation of pinene by Bacillus pallidus BR425. Biodegradation 9, 337–341. doi: 10.1023/A:1008304603734
Schewe, H., Mirata, M. A., Holtmann, D., and Schrader, J. (2011). Biooxidation of monoterpenes with bacterial monooxygenases. Process Biochem. 46, 1885–1899. doi: 10.1016/j.procbio.2011.06.010
Seubert, W. (1960). Degradation of isoprenoid compounds by microorganisms. i. isolation and characterization of an isoprenoid-degrading bacterium, Pseudomonas citronellolis n. sp. J. Bacteriol. 79, 426–434.
Seubert, W., and Fass, E. (1964). Untersuchungen über den bakteriellen Abbau von Isoprenoiden. 5. Der Mechanismus des Isoprenabbaus. Biochem. Z. 341, 35–44.
Seubert, W., Fass, E., and Remberger, U. (1963). Untersuchungen über den bakteriellen Abbau von Isoprenoiden. 3. Reinigung und Eigenschaften der Geranylcarboxylase. Biochem. Z. 338, 265–275.
Seubert, W., and Remberger, U. (1963). Untersuchungen über den bakteriellen Abbau von Isoprenoiden. 2. Die Rolle der Kohlensäure. Biochem. Z. 338, 245–264.
Sharkey, T. D., Wiberley, A. E., and Donohue, A. R. (2008). Isoprene emission from plants: why and how. Ann. Bot. 101, 5–18. doi: 10.1093/aob/mcm240
Sharkey, T. D., and Yeh, S. S. (2001). Isoprene emission from plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 407–436. doi: 10.1146/annurev.arplant.52.1.407
Shaw, S. L., Gantt, B., and Meskhidze, N. (2010). Production and emissions of marine isoprene and monoterpenes: a review. Adv. Meteorol. 2010:408696. doi: 10.1155/2010/408696
Speelmans, G., Bijlsma, A., and Eggink, G. (1998). Limonene bioconversion to high concentrations of a single and stable product, perillic acid, by a solvent-resistant Pseudomonas putida strain. Appl. Microbiol. Biotechnol. 50, 538–544. doi: 10.1007/s002530051331
Stotzky, G., and Schenck, S. (1976). Volatile organic compounds and microorganisms. Crit. Rev. Microbiol. 4, 333–382. doi: 10.3109/10408417609102303
Tanaka, K., Kim, H. J., Saito, K., Takahashi, H. G., Watanabe, M., Yokohata, T., et al. (2012). How have both cultivation and warming influenced annual global isoprene and monoterpene emissions since the preindustrial era? Atmos. Chem. Phys. 12, 9703–9718. doi: 10.5194/acp-12-9703-2012
Taylor, D. G., and Trudgill, P. W. (1986). Camphor revisited: studies of 2,5-diketocamphane 1,2-monooxygenase from Pseudomonas putida ATCC 17453. J. Bacteriol. 165, 489–497.
Thompson, M. L., Marriott, R., Dowle, A., and Grogan, G. (2010). Biotransformation of beta-myrcene to geraniol by a strain of Rhodococcus erythropolis isolated by selective enrichment from hop plants. Appl. Microbiol. Biotechnol. 85, 721–730. doi: 10.1007/s00253-009-2182-6
Tong, W.-Y. (2013). “Biotransformation of terpenoids and steroids,” in Natural Products, eds K. G. Ramawat and J.-M. Mérillon (Berlin; Heidelberg: Springer), 2733–2759. doi: 10.1007/978-3-642-22144-6_122
Trudgill, P. W. (1990). Microbial metabolism of monoterpenes - recent developments. Biodegradation 1, 93–105. doi: 10.1007/BF00058829
Trudgill, P. W. (1994). Microbial Metabolism And Transformation Of Selected Monoterpenes. Dordrecht; Norwell: Kluwer Academic Publishers.
Ullah, A. J. H., Murray, R. I., Bhattacharyya, P. K., Wagner, G. C., and Gunsalus, I. C. (1990). Protein components of a cytochrome P-450 linalool 8-methyl hydroxylase. J. Biol. Chem. 265, 1345–1351.
van der Werf, M., De Bont, J. A. M., and Leak, D. J. (1997). “Opportunities in microbial biotransformation of monoterpenes,” in Advances In Biochemical Engineering Biotechnology; Biotechnology Of Aroma Compounds, ed R. G. Berger (Berlin; New York, NY: Springer-Verlag), 147–177.
van der Werf, M. J., and Boot, A. M. (2000). Metabolism of carveol and dihydrocarveol in Rhodococcus erythropolis DCL14. Microbiology 146, 1129–1141.
van der Werf, M. J., Orru, R. V. A., Overkamp, K. M., Swarts, H. J., Osprian, I., Steinreiber, A., et al. (1999a). Substrate specificity and stereospecificity of limonene-1,2-epoxide hydrolase from Rhodococcus erythropolis DCL14; an enzyme showing sequential and enantioconvergent substrate conversion. Appl. Microbiol. Biotechnol. 52, 380–385.
van der Werf, M. J., Swarts, H. J., and De Bont, J. A. M. (1999b). Rhodococcus erythropolis DCL14 contains a novel degradation pathway for limonene. Appl. Environ. Microbiol. 65, 2092–2102.
Wang, Y., Lim, L., Diguistini, S., Robertson, G., Bohlmann, J., and Breuil, C. (2013). A specialized ABC efflux transporter GcABC-G1 confers monoterpene resistance to Grosmannia clavigera, a bark beetle-associated fungal pathogen of pine trees. New Phytol. 197, 886–898. doi: 10.1111/nph.12063
Williams, D. R., Trudgill, P. W., and Taylor, D. G. (1989). Metabolism of 1,8-cineole by a Rhodococcus species: ring cleavage reactions. J. Gen. Microbiol. 135, 1957–1967.
Wilt, F. M., Miller, G. C., Everett, R. L., and Hackett, M. (1993). Monoterpene concentrations in fresh, senescent, and decaying foliage of single-leaf pinyon (Pinus monophylla Torr And Frem: Pinaceae) from the Western Great-Basin. J. Chem. Ecol. 19, 185–194. doi: 10.1007/BF00993688
Wright, S. J., Caunt, P., Carter, D., and Baker, P. B. (1986). Microbial oxidation of α-pinene by Serratia marcescens. Appl. Microbiol. Biotechnol. 23, 224–227.
Yang, J. E., Park, Y. J., and Chang, H. C. (2007). Cloning of four genes involved in limonene hydroxylation from Enterobacter cowanii 6L. J. Microbiol. Biotechnol. 17, 1169–1176.
Yassaa, N., Peeken, I., Zollner, E., Bluhm, K., Arnold, S., Spracklen, D., et al. (2008). Evidence for marine production of monoterpenes. Environ. Chem. 5, 391–401. doi: 10.1071/EN08047
Yoo, S. K., and Day, D. F. (2002). Bacterial metabolism of alpha- and beta-pinene and related monoterpenes by Pseudomonas sp. strain PIN. Process Biochem. 37, 739–745. doi: 10.1016/S0032-9592(01)00262-X
Yu, F. N. A., and Utsumi, R. (2009). Diversity, regulation, and genetic manipulation of plant mono- and sesquiterpenoid biosynthesis. Cell. Mol. Life Sci. 66, 3043–3052. doi: 10.1007/s00018-009-0066-7
Keywords: isoprenoids, acyclic monoterpene utilization, camphor, pinene, limonene, linalool, myrcene, eucalyptol
Citation: Marmulla R and Harder J (2014) Microbial monoterpene transformations—a review. Front. Microbiol. 5:346. doi: 10.3389/fmicb.2014.00346
Received: 30 April 2014; Accepted: 21 June 2014;
Published online: 15 July 2014.
Edited by:
Colin Murrell, University of East Anglia, UKReviewed by:
Terry John McGenity, University of Essex, UKCopyright © 2014 Marmulla and Harder. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jens Harder, Max Planck Institute for Marine Microbiology, Celsiusstr. 1, Bremen 28359, Germany e-mail:amhhcmRlckBtcGktYnJlbWVuLmRl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.