
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 10 February 2014
Sec. Aquatic Microbiology
Volume 5 - 2014 | https://doi.org/10.3389/fmicb.2014.00045
This article is part of the Research TopicVibrio ecology, pathogenesis and evolution.View all 26 articles
 Bradd J. Haley1
Bradd J. Haley1 Tamar Kokashvili2
Tamar Kokashvili2 Ana Tskshvediani2
Ana Tskshvediani2 Nino Janelidze2
Nino Janelidze2 Nino Mitaishvili2
Nino Mitaishvili2 Christopher J. Grim1,3†
Christopher J. Grim1,3† Guillaume Constantin de Magny4
Guillaume Constantin de Magny4 Arlene J. Chen1
Arlene J. Chen1 Elisa Taviani1†
Elisa Taviani1† Tamar Eliashvili2
Tamar Eliashvili2 Marina Tediashvili2
Marina Tediashvili2 Chris A. Whitehouse5
Chris A. Whitehouse5 Rita R. Colwell1,3,6,7
Rita R. Colwell1,3,6,7 Anwar Huq1,8*
Anwar Huq1,8*Vibrio parahaemolyticus is a leading cause of seafood-related gastroenteritis and is also an autochthonous member of marine and estuarine environments worldwide. One-hundred seventy strains of V. parahaemolyticus were isolated from water and plankton samples collected along the Georgian coast of the Black Sea during 28 months of sample collection. All isolated strains were tested for presence of tlh, trh, and tdh. A subset of strains were serotyped and tested for additional factors and markers of pandemicity. Twenty-six serotypes, five of which are clinically relevant, were identified. Although all 170 isolates were negative for tdh, trh, and the Kanagawa Phenomenon, 7 possessed the GS-PCR sequence and 27 the 850 bp sequence of V. parahaemolyticus pandemic strains. The V. parahaemolyticus population in the Black Sea was estimated to be genomically heterogeneous by rep-PCR and the serodiversity observed did not correlate with rep-PCR genomic diversity. Statistical modeling was used to predict presence of V. parahaemolyticus as a function of water temperature, with strongest concordance observed for Green Cape site samples (Percent of total variance = 70, P < 0.001). Results demonstrate a diverse population of V. parahaemolyticus in the Black Sea, some of which carry pandemic markers, with increased water temperature correlated to an increase in abundance of V. parahaemolyticus.
Vibrio parahaemolyticus, a halophilic bacterium, is a causative agent of seafood-related gastroenteritis, wound infections, and septicemia and is known to occur in marine, estuarine, and brackish water environments globally with sporadic occurrence in fresh water (Sarkar et al., 1985; DePaola et al., 2000; Wong et al., 2000; Alam et al., 2009). In addition to notoriety as a causative agent of human infection, the organism is autochthonous to marine and brackish water ecosystems and, similar to other Vibrio spp., degrades chitin (Kaneko and Colwell, 1974; Kadokura et al., 2007). One of its main virulence factors, the type three secretion system-2 (TTSS2), plays an important role in preventing predation of its host by higher organisms, suggesting the virulence factors have evolved via environmental selection (Matz et al., 2011). Little work has been done on non-anthropocentric roles of this organism, but its ubiquity and association with animals demonstrate that its ecology extends beyond the human body.
The majority of clinical strains encode the thermostable direct hemolysin (TDH), within the V. parahaemolyticus pathogenicity island (Vp-PAI), one of the virulence factors responsible for enterotoxicity (Honda, 1993; Guang-Qing et al., 1995). However, some clinical isolates do not encode TDH, but other hemolysins instead, such as the TDH-related hemolysin (TRH), while all encode the thermolabile hemolysin (TLH). It has also been reported that two type three secretion systems (TTSS1 and TTSS2) are involved in V. parahaemolyticus pathogenicity (Bhattacharjee et al., 2006; Ono et al., 2006; Kodama et al., 2007; Matlawska-Wasowska et al., 2010). The TTSS1 found in all V. parahaemolyticus strains examined to date has been shown to translocate an effector protein (VP1686) into the cytosol of macrophages and induce DNA fragmentation and another effector protein (VP1680) has been shown to play a role in cytotoxicity in eukaryotic cells (Bhattacharjee et al., 2006; Ono et al., 2006). Interestingly, V. parahaemolyticus strains lacking TDH, TRH, and TTSS2 have frequently been isolated from patients not colonized by TDH-, TRH-, and TTSS2-positive strains, suggesting TTSS1 is also responsible for illness in humans (Suthienkul et al., 1995; Okuda et al., 1997; Vuddhakul et al., 2000; Laohaprertthisan et al., 2003; Cabanillas-Beltrán et al., 2006; Bhoopong et al., 2007; Meador et al., 2007; Serichantalergs et al., 2007; Chao et al., 2009, 2010; García et al., 2009; Harth et al., 2009).
V. parahaemolyticus has been frequently isolated from water samples collected from the Black Sea and sporadic cases of gastroenteritis caused by this bacterium and related vibrios have historically been reported in the Sea of Azov region (Libinzon et al., 1974, 1980, 1981; Shikulov et al., 1980; Clark et al., 1998; WHO, 2011). Further, human pathogenic vibrios are known to be endemic to the greater Caucasus (Narkevich et al., 1993; Gurbanov et al., 2011; Rashid et al., 2013) but the ecologies of these organisms are not well-elucidated in this region. The increasing global incidence of V. parahaemolyticus infections suggests it is important to fully understand the ecology of these regions in multiple locations so that public health assessments can be made more accurately (Baker-Austin et al., 2010). Members of the Vibrionaceae are known to have an intimate association with planktonic organisms and many studies have demonstrated the role of environmental conditions (namely water temperature and salinity) on the density of these organisms in water bodies. Generally, an increase in temperature of a water body is associated with an increase in Vibrio density (Turner et al., 2009; Oberbeckmann et al., 2012). To further understand the ecology of V. parahaemolyticus along the Georgian coast of the Black Sea we evaluated the presence of these organisms in water and plankton fractions over a 28 month period (June 2006 to October 2008) and modeled their presence in relation to environmental conditions (salinity, water temperature, pH, and dissolved oxygen). We further evaluated the molecular diversity and presence of virulence factors in a subset of V. parahaemolyticus isolates collected during this study.
Water samples were collected monthly, except July to September when water was collected biweekly, from five stations on the coast of the Black Sea (Figure 1). One hundred liters of water were filtered through 200- and 64-μm plankton nets, to separate size fractions of plankton. Water temperature, salinity, pH, and dissolved oxygen were recorded at the time of sampling. The water fraction (100 ml) was filtered using a 0.45 μm nitrocellulose membrane, which was incubated in alkaline peptone water (APW) at 37°C for 24 h. An aliquot (1- to 5-ml) of each plankton fraction (64- and 200-μm) was also inoculated in APW and incubated at 37°C for 24 h. A 10 microliter loop of the enrichment cultures were streaked onto thiosulfate citrate bile salts (TCBS) agar plates, which were incubated overnight at 37°C. All colonies that appeared yellow to green at 24 h were considered presumptive Vibrio spp., picked with a sterile toothpick, and streaked to isolate colonies on Luria–Bertani (LB) agar. Presumptive V. parahaemolyticus colonies were confirmed by streaking onto CHROMagar™ Vibrio (mauve colonies) the latter were confirmed by PCR (presence of tlh, and V. parahaemolyticus-specific collagenase).
For molecular analyses, the following PCR primers were used; collagenase (Di Pinto et al., 2005), tdh, trh, and tlh (Bej et al., 1999), GS-PCR (Matsumoto et al., 2000), ORF8 (Nasu et al., 2000), Mtase (Wang et al., 2006), histone-like DNA-binding protein (HU-α ORF) (Williams et al., 2004), the 850 bp pandemic strain sequence (VPF2/VPR2) (Khan et al., 2002), VP1346 (yop) and VP1339 (escC) of TTSS2 (Chao et al., 2010), VP1680 (Whitaker et al., 2012) and VP1686 of TTSS1 (This study). Primer sequences for VP1686 were VP1686-F: TGCTTTTGTGATCGCTTTTG and VP1686-R: TGAAGGCAAACTCAGCATTG (Ta = 56°C; amplicon size = 169 bp) and were designed in silico using V. parahaemolyticus RIMD2210633 (NC_004603.1/NC_004605.1). DNA (25.0 ng) was mixed with 2.5 mM of dNTP, 15 mM of PCR buffer, and 5 U μL−1 of Taq DNA polymerase, using 20 μm of appropriate primer for each analysis. Amplicons were visualized on 1.5% agarose gel stained with ethidium bromide and examined under a UV transilluminator.
To approximate the molecular diversity of the V. parahaemolyticus isolates, rep-PCR was executed on a randomly selected subset of strains following the methods of Chokesajjawatee et al. (2008). PCR products were separated on a 1% agarose gel in TAE buffer. The resulting fingerprint patterns were documented using the GelDoc-It™ Imaging System (Ultra-Violet Products, Upland, CA). Banding patterns were identified by visual observation and dendrograms were calculated by the unweighted pair-group method using average linkages (UPGMA). Serotyping was performed as follows. Strains were streaked on LB agar with 3% NaCl and incubated overnight at 37°C. One 10 μl loopful of growth was homogenized in 1 mL of saline solution (0.9% NaCl). This solution was divided into two 500 μl tubes, one of which was boiled for 2 h. Ten microliters of the boiled cell solution was then mixed with 10 μl of each O-antisera and 10 μl of the cell suspension that had not been boiled was mixed with 10 μl of K-antisera on a glass slide and agglutination visually determined (Denka Seiken Co., Niigata-ken, Japan). Distilled water was used as a negative control for serotyping assays. V. parahaemolyticus strain RIMD2210633 (KP positive; serotype O3:K6) for assays.
Predictive models of V. parahaemolyticus detection were determined by examining the relationship between presence/absence (response variable) and recorded environmental parameters (explanatory variables) at the time of sample collection. Environmental parameters were also evaluated as explanatory variables by determining the distance from optimality for each data point. This was performed by subtracting the median values of all parameters for those samples in which V. parahaemolyticus had been detected (optimal parameters) from all data points following the methods of Jacobs et al. (2010) and Banakar et al. (2011). The absolute values of differences were used as explanatory variables in binary logistic regression analysis. For all measures of association, p-values ≤ 0.05 were considered significant. Statistical analyses were conducted on R (http://www.r-project.org/) and SAS softwares (Cary, NC, USA).
In total, 170 isolates of V. parahaemolyticus were recovered from Black Sea water and plankton samples collected along the Georgian coast, of which 101 were from water, 30 from the 64 μm fraction, and 39 from the 200 μm fraction of plankton (Figure 2).
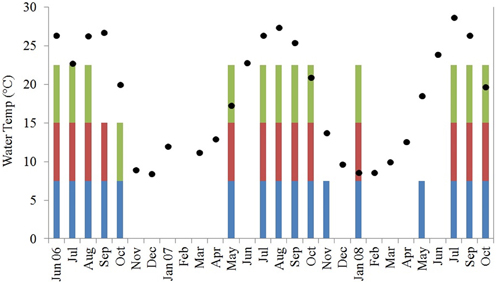
Figure 2. Water temperature in degrees C (black diamonds, Y-axis) and V. parahaemolyticus detection in water (blue bars) and plankton [200 μm (green bars) and 64 μm (magenta bars)]. Water temperature is averaged across all sites for each sampling month and colored bars demonstrate at least 1 positive sample for that fraction across all sites for each sampling month.
Vibrio parahaemolyticus was isolated from 40 of a total of 106 water samples collected and 19 of 106 and 26 of 106 of 64- and 200-μm plankton fractions, respectively. Based on Cochran's Q-test, water samples yielded V. parahaemolyticus significantly more frequently than either of the plankton fractions. The difference in V. parahaemolyticus isolation frequency was not significantly different between the two plankton fractions. When these distributions were binned to water temperature quartiles (11, 19.8, and 25.8°C), water samples with temperature between 11 and 19.8°C were significantly more likely to yield V. parahaemolyticus isolates than plankton.
Median water temperatures and salinities for all fractions positive for V. parahaemolyticus were higher than those that were negative for V. parahaemolyticus, while the opposite was observed for dissolved oxygen (Table 1). Median pH levels were slightly lower for all fractions positive for V. parahaemolyticus than those that were negative, excluding the P64 fraction (Table 1).
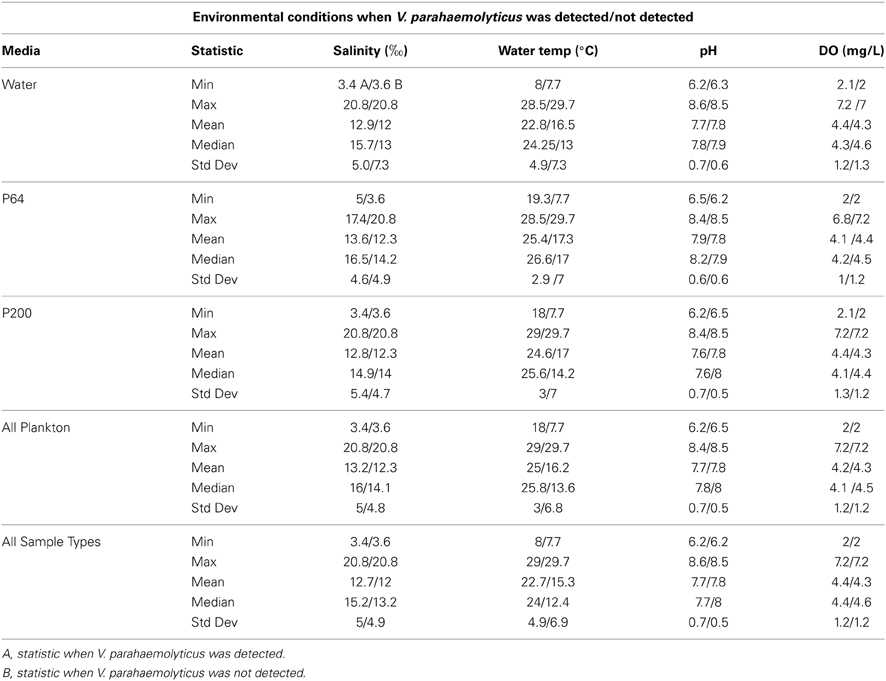
Table 1. Recorded environmental parameters when V. parahaemolyticus was/was not detected for each sample type.
Twenty-seven serotypes of V. parahaemolyticus were detected the majority of which were O2:K28 (7 isolates), O3:K31 (7), O3:KUT (7), O4:KUT (7), and untypable (24) (Table 2). Vibrio parahaemolyticus O3 O-antigenic type was the most common, comprising 35% of the isolates. Untypable strains may represent strains with novel serology for which V. parahaemolyticus anti-sera has not yet been developed, or strains in which antigenic expression is altered or repressed.
None of the V. parahaemolyticus isolates carried the genes for thermostable direct hemolysin (tdh) thermostable-related hemolysin (trh), TTSS-2, or MTase; all were both, Kanagawa phenomenon and urease negative (Table 2). Nineteen isolates resulted in PCR amplicons for the pandemic GS-PCR marker (toxRS sequence of pandemic strains), but only seven were 651 bp and 12 were ca. 750 bp. Twenty seven isolates carried the 850-bp pandemic sequence (VPF2/VPR2). Three of the 651 bp, GS-PCR-positive strains were positive for the 850 bp pandemic sequence, whereas six of the 750 bp, GS-PCR-positive isolates encoded this region. Each of the 651 bp, GS-PCR-positive isolates were different serotypes and were typed as O1:KUT, O3:KUT, O3:K31, O3:K33 O3:K65, OUT:K33, and UT, the most notable was the O1:KUT, related to pandemicity. This isolate was also positive for the 850 bp pandemic sequence but lacked all other markers of virulence except TTSS1. Rep-PCR was performed on 45 of the strains (Figure 3). A dendrogram of banding patterns revealed a high level of diversity suggesting a non-clonal population of V. parahaemolyticus in this environment.
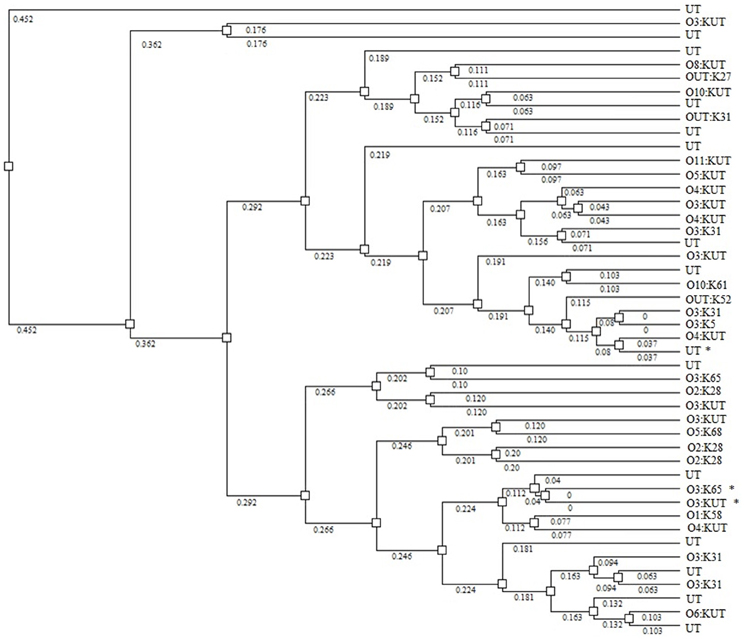
Figure 3. Dendrogram showing relatedness of V. parahaemolyticus strains by rep-PCR. Asterisks identify strains that are GS-PCR-positive. Numbers on branches indicate degree of divergence between isolates.
Among four explanatory variables in a logistic regression used to model presence/absence of V. parahaemolyticus as the response variable, water temperature was the only significant predictor (Table 3). When data from all sites were combined, water temperature explained 37.3% of variance in isolation of V. parahaemolyticus, suggesting the dynamics of the population are driven by multiple factors. In the Chorokhi and Supsa estuaries, the proportion of variance in V. parahaemolyticus isolation explained by water temperature was 22 and 32.1%, respectively, but higher for Batumi Bulvard and Green Cape sites, 43.2 and 70.1%, respectively (Table 3).
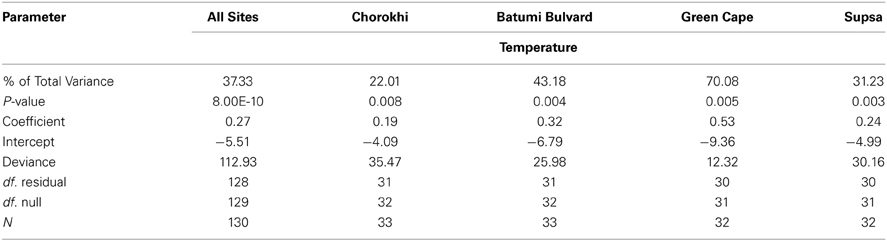
Table 3. Results of binary logistic regression analysis between V. parahaemolyticus water temperature.
Although commonly isolated from brackish waters, presence of V. parahaemolyticus suggests a public health concern to those utilizing these water sources or consuming products harvested from these waters. This risk is appreciable regardless of pathogenicity island presence in the genomes of circulating V. parahaemolyticus, since some infections are caused by isolates lacking tdh, trh, and TTSS2 (Suthienkul et al., 1995; Okuda et al., 1997; Vuddhakul et al., 2000; Laohaprertthisan et al., 2003; Cabanillas-Beltrán et al., 2006; Bhoopong et al., 2007; Meador et al., 2007; Serichantalergs et al., 2007; Chao et al., 2009, 2010; García et al., 2009; Harth et al., 2009). Isolates recovered in this study lacked the major virulence factors associated with the majority of clinical cases. However, these results are not surprising since typically <1% of environmental isolates encode these elements (McLaughlin et al., 2005). The historical reporting of V. parahaemolyticus infections in this region suggests that either infections have been caused by strains lacking major virulence factors, resident strains encoding these virulence factors were not detected using the methods employed by this study, or both.
Results of this study demonstrated a high level of diversity among isolates as measured by serotype distribution, presence/absence of pandemic markers, and rep-PCR banding patterns. Strains isolated in this study represented 9 O-antigens and 27 K-antigens, as well as untypable strains, a measure of antigenic diversity of natural isolates in this region. Mutations within antigen coding regions of the genome are common, as well as lateral transfer, allowing strains to adapt to microenvironments of the environment or evade predation by grazing protozoa (Lerouge et al., 2001; Woo et al., 2001; Wildschutte et al., 2004). Molecular divergence was noted by the heterogeneity observed among O3:K31 and O2:K28 strains by rep-PCR analysis suggesting that serology does not necessarily correlate with genome architecture. This genomic heterogeneity indicates the necessity of classifying strains by methods other than serology. The high degree of divergence among environmental V. parahaemolyticus strains in the Black Sea is corroborated by reports of similar findings in geographically distant regions (Wong et al., 1999; Matsumoto et al., 2000; Alam et al., 2009; Yu et al., 2002; Ellis et al., 2012; Paranjpye et al., 2012).
V. parahaemolyticus was detected across a broad range of salinities (3.4–20.8‰) (Table 2). However, it was not significantly associated with V. parahaemolyticus presence in our model. This is most likely due to the relative stability of salinity readings at each site over the course of the study (data not shown). V. parahaemolyticus is a known member of estuarine and marine environments and salinity values detected during this study were typical of brackish waters (0.5 > 30‰) suggesting a suitable salinity regime for V. parahaemolyticus presence at most sampling points. V. parahaemolyticus seasonality was observed at all sites, with a clear trend of increasing numbers as water temperatures increased from May to September. The organism was isolated from water samples at temperatures as low as 8°C, but more frequently (ca. 93% of strains) at temperatures greater than 17°C (Table 1). The highest percentage of total variance in detection, related to temperature, was at Green Cape (percent of total variance = 70, P < 0.05). At each site, the total variance in V. parahaemolyticus detection was significantly related to an increase in water temperature. However, these associations were not as strong for the Batumi Bulvard (43.18), Chorokhi estuary (22.01), and Supsa estuary (31.23) sites (Table 3). Interestingly, the associations between water temperature and V. parahaemolyticus detection were weaker for the two estuarine sites. Salinities at these two sites were much lower than the non-estuarine sites (Batumi and Green Cape) suggesting that either salinity played a role in V. parahaemolyticus presence, even though it did not show up as significant in our model, or that an unmonitored parameter common to both estuarine environments influenced V. parahaemolyticus presence. This trend is indicative of the patchiness of V. parahaemolyticus distribution in water bodies suggesting that environmental conditions are noticeably different at different locations within the same water body and that these differences contribute to V. parahaemolyticus presence.
In summary, an antigenically diverse population of V. parahaemolyticus inhabits the Georgian coast of the Black Sea. Although none of the strains collected during this study were Kanagawa phenomena-positive or tdh and trh-positive, the TTSS1 effector proteins and TLH were present in some isolates, which included a possible serovariant of the V. parahaemolyticus O3:K6 pandemic clone. These results, together with epidemiological data demonstrating strains lacking pathogenicity islands can cause disease, suggest there is a risk associated with occurrence of V. parahaemolyticus in Black Sea coastal waters. Warmer temperatures in the spring and summer lead to increased densities of V. parahaemolyticus. Recent clinical data on isolation of TDH-, TRH-, and TTSS2-negative V. parahaemolyticus suggests these strains represent underreported etiological agents of diarrhea, similar to V. cholerae non-O1/non-O139 strains lacking major virulence factors (Safrin et al., 1988; Ko et al., 1998; Lukinmaa et al., 2006; Shannon and Kimbrough, 2006; Chatterjee et al., 2009; Hasan et al., 2012; Marin et al., 2013). The high frequency of detection of V. parahaemolyticus lacking major virulence factors but associated with severe infection, suggests recreational water and shellfish harvesting areas in Georgia should be monitored, especially when water temperatures are seasonally high.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The Associate Editor declares that despite being affiliated to the same institution as the authors Bradd J. Haley, Christopher J. Grim, Arlene J. Chen, Elisa Taviani, Rita R. Colwell and Anwar Huq, the review process was handled objectively and no conflict of interest exists.
The research described in this report was made possible by financial support provided by the Biological Threat Reduction Program of the U.S. Defense Threat Reduction Agency (DTRA) (Project # GG-13) through Bechtel National Inc., sponsor account number 24914416HC4W00000006. Christopher J. Grim was supported by an IC Postdoctoral Research Fellowship (NGA Grant #HM15820612010). Partial funding for this study was provided by NIH Grant No. 2RO1A1039129-11A2 and NSF Grant No. 0813066. Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the U.S. Army.
Alam, M., Chowdhury, W. B., Bhuiyan, N. A., Islam, A., Hasan, N. A., Nair, G. B., et al. (2009). Serogroup, virulence, and genetic traits of Vibrio parahaemolyticus in the estuarine ecosystem of Bangladesh. Appl. Environ. Microbiol. 75, 6268–6274. doi: 10.1128/AEM.00266-09
Baker-Austin, C., Stockley, L., Rangdale, R., and Martinez-Urtaza, J. (2010). Environmental occurrence and clinical impact of Vibrio vulnificus and Vibrio parahaemolyticus: a European perspective. Environ. Microbiol. Rep. 2, 7–18. doi: 10.1111/j.1758-2229.2009.00096.x
Banakar, V., Constantin de Magny, G., Jacobs, J., Murtugudde, R., Huq, A., Wood, R. J., et al. (2011). Temporal and spatial variability in the distribution of Vibrio vulnificus in the Chesapeake Bay: a hindcast study. Ecohealth 8, 456–467. doi: 10.1007/s10393-011-0736-4
Bej, A. K., Patterson, D. P., Brasher, C. W., Vickery, M. C., Jones, D. D., and Kaysner, C. A. (1999). Detection of total and hemolysin-producing Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tl, tdh and trh. J. Microbiol. Methods 36, 215–225. doi: 10.1016/S0167-7012(99)00037-8
Bhattacharjee, R. N., Park, K. S., Kumagai, Y., Okada, K., Yamamoto, M., Uematsu, S., et al. (2006). VP1686, a Vibrio type III secretion protein, induces toll-like receptor-independent apoptosis in macrophage through NF-kappaB inhibition. J. Biol. Chem. 281, 36897–36904. doi: 10.1074/jbc.M605493200
Bhoopong, P., Palittapongarnpim, P., Pomwised, R., Kiatkittipong, A., Kamruzzaman, M., Nakaguchi, Y., et al. (2007). Variability of properties of Vibrio parahaemolyticus strains isolated from individual patients. J. Clin. Microbiol. 45, 1544–1550. doi: 10.1128/JCM.02371-06
Cabanillas-Beltrán, H., LLausás-Magaña, E., Romero, R., Espinoza, A., García-Gasca, A., Nishibuchi, M., et al. (2006). Outbreak of gastroenteritis caused by the pandemic Vibrio parahaemolyticus O3:K6 in Mexico. FEMS Microbiol. Lett. 265, 76–80. doi: 10.1111/j.1574-6968.2006.00475.x
Chao, G., Jiao, X., Zhou, X., Wang, F., Yang, Z., Huang, J., et al. (2010). Distribution of genes encoding four pathogenicity islands (VPaIs), T6SS, biofilm, and type I pilus in food and clinical strains of Vibrio parahaemolyticus in China. Foodborne Pathog. Dis. 7, 649–658. doi: 10.1089/fpd.2009.0441
Chao, G., Jiao, X., Zhou, X., Yang, Z., Huan, J., Pan, Z., et al. (2009). Serodiversity, pandemic O3, K6 clone, molecular typing, and antibiotic susceptibility of foodborne and clinical Vibrio parahaemolyticus isolates in Jiangsu, China. Foodborne Pathog. Dis. 6, 1021–1028. doi: 10.1089/fpd.2009.0295
Chatterjee, S., Ghosh, K., Raychoudhuri, A., Chowdhury, G., Bhattacharya, M. K., Mukhopadhyay, A. K., et al. (2009). Incidence, virulence factors, and clonality among clinical strains of non-O1, non-O139 Vibrio cholerae isolates from hospitalized diarrheal patients in Kolkata, India. J. Clin. Microbiol. 47, 1087–1095. doi: 10.1128/JCM.02026-08
Chokesajjawatee, N., Zo, Y. G., and Colwell, R. R. (2008). Determination of clonality and relatedness of Vibrio cholerae isolates by genomic fingerprinting, using long-range repetitive element sequence-based PCR. Appl. Environ. Microbiol. 74, 5392–5401. doi: 10.1128/AEM.00151-08
Clark, C., Kravetz, A., Dendy, C., Wang, G., Tyler, K., and Johnson, W. (1998). Investigation of the 1994–5 Ukrainian Vibrio cholerae epidemic using molecular methods. Epidemiol. Infect. 121, 15–29. doi: 10.1017/S0950268898008814
DePaola, A., Kaysner, C. A., Bowers, J., and Cook, D. W. (2000). Environmental investigations of Vibrio parahaemolyticus in oysters after outbreaks in Washington, Texas, and New York (1997 and 1998). Appl. Environ. Microbiol. 66, 4649–4654. doi: 10.1128/AEM.66.11.4649-4654.2000
Di Pinto, A., Ciccarese, G., Tantillo, G., Catalano, D., and Forte, V. T. (2005). A collagenase-targeted multiplex PCR assay for identification of Vibrio alginolyticus, Vibrio cholerae, and Vibrio parahaemolyticus. J. Food Prot. 68, 150–153.
Ellis, C. N., Schuster, B. M., Striplin, M. J., Jones, S. H., Whistler, C. A., and Cooper, V. S. (2012). Influence of seasonality on the genetic diversity of Vibrio parahaemolyticus in New Hampshire shellfish waters as determined by multilocus sequence analysis. Appl. Environ. Microbiol. 78, 3778–3782. doi: 10.1128/AEM.07794-11
García, K., Torres, R., Uribe, P., Hernández, C., Rioseco, M., Romero, J., et al. (2009). Dynamics of clinical and environmental Vibrio parahaemolyticus strains during seafood-related summer diarrhea outbreaks in Southern Chile. Appl. Environ. Microbiol. 75, 7482–7487. doi: 10.1128/AEM.01662-09
Guang-Qing, T., Tetsuya, L., Koichiro, Y., and Takeshi, H. (1995). Ca2+ independent cytotoxicity of Vibrio parahaemolyticus thermostable direct hemolysin (TDH) on Intestine 407, a cell line derived from human embryonic intestine. FEMS Microbiol. Lett. 134, 233–238. doi: 10.1111/j.1574-6968.1995.tb07943.x
Gurbanov, S., Akhmadov, R., Shamkhalova, G., Akhmadova, S., Haley, B., Colwell, R., et al. (2011). Occurrence of Vibrio cholerae in municipal and natural waters and incidence of cholera in Azerbaijan. Ecohealth 8, 468–477. doi: 10.1007/s10393-012-0756-8
Harth, E., Matsuda, L., Hernández, C., Rioseco, M., Romero, J., González-Escalona, N., et al. (2009). Epidemiology of Vibrio parahaemolyticus outbreaks, Southern Chile. Emerg. Infect. Dis. 15, 163–168. doi: 10.3201/eid1502.071269
Hasan, N. A., Choi, S. Y., Eppinger, M., Clark, P. W., Chen, A., Alam, M., et al. (2012). Genomic diversity of 2010 Haitian cholera outbreak strains. Proc. Natl. Acad. Sci. U.S.A. 109, E2010–E2017. doi: 10.1073/pnas.1207359109
Honda, T. (1993). The pathogenicity of Vibrio parahaemolyticus and the role of the thermostable direct haemolysin and related haemolysins. Rev. Med. Microbiol. 4, 106–113. doi: 10.1097/00013542-199304000-00006
Jacobs, J. M., Rhodes, M., Brown, C. W., Hood, R. R., Leigh, A., Long, W., et al. (2010). “Predicting the distribution of Vibrio vulnificus in Chesapeake Bay,” in NOAA Technical Memorandum NOS NCCOS 112. NOAA National Centers for Coastal Ocean Science, Center for Coastal Environmental Health and Biomolecular Research, Cooperative Oxford Laboratory (Oxford, MD).
Kadokura, K., Rokutani, A., Yamamoto, M., Ikegami, T., Sugita, H., Itoi, S., et al. (2007). Purification and characterization of Vibrio parahaemolyticus extracellular chitinase and chitin oligosaccharide deacetylase involved in the production of heterodisaccharide from chitin. Appl. Microbiol. Biotechnol. 75, 357–365. doi: 10.1007/s00253-006-0831-6
Kaneko, T., and Colwell, R. (1974). Distribution of Vibrio parahaemolyticus and related organisms in the atlantic ocean off South Carolina and Georgia. Appl. Environ. Microbiol. 28 1009–1017.
Khan, A. A., McCarthy, S., Wang, R. F., and Cerniglia, C. E. (2002). Characterization of United States outbreak isolates of Vibrio parahaemolyticus using enterobacterial repetitive intergenic consensus (ERIC) PCR and development of a rapid PCR method for detection of O3, K6 isolates. FEMS Microbiol. Lett. 206, 209–214. doi: 10.1016/S0378-1097(01)00545-6
Ko, W. C., Chuang, Y. C., Huang, G. C., and Hsu, S. Y. (1998). Infections due to non-O1 Vibrio cholerae in southern Taiwan: predominance in cirrhotic patients. Clin. Infect. Dis. 27, 774–780. doi: 10.1086/514947
Kodama, T., Rokuda, M., Park, K. S., Cantarelli, V. V., Matsuda, S., Iida, T., et al. (2007). Identification and characterization of VopT, a novel ADP-ribosyltransferase effector protein secreted via the Vibrio parahaemolyticus type III secretion system 2. Cell. Microbiol. 9, 2598–2609. doi: 10.1111/j.1462-5822.2007.00980.x
Laohaprertthisan, V., Chowdhury, A., Kongmuang, U., Kalnauwakul, S., Ishibashi, M., Matsumoto, C., et al. (2003). Prevalence and serodiversity of the pandemic clone among the clinical strains of Vibrio parahaemolyticus isolated in Southern Thailand. Epidemiol. Infect. 130, 395–406. doi: 10.1017/S095026880300845
Lerouge, I., Laeremans, T., Verreth, C., Vanderleyden, J., Van Soom, C., Tobin, A., et al. (2001). Identification of an ATP-binding cassette transporter for export of the O-antigen across the inner membrane in Rhizobium etli based on the genetic, functional, and structural analysis of an lps mutant deficient in O-antigen. J. Biol. Chem. 276, 17190–17198. doi: 10.1074/jbc.M101129200
Libinzon, A., Brudnyi, R., Nagornaia, A., Demina, A., and Krasnova, N. (1981). Halophilic Black Sea vibrios and their role in human pathology. Zh. Mikrobiol. Epidemiol. Immunobiol. 2, 97–101.
Libinzon, A. E., Domaradskii, I. V., Us, Z. I., Demina, A. I., and Nagornaia, A. F. (1974). Parahemolytic vibrios and related halophilic microorganisms of the Black Sea. Zh. Mikrobiol. Epidemiol. Immunobiol. 00, 80–84.
Libinzon, A. E., Luneva, E. I., Kashirova, A. K., Velichko, S. A., and Sakhno, E. I. (1980). Halophilic vibrios in human excreta and seawater. Gig. Sanit. 12, 73–74.
Lukinmaa, S., Mattila, K., Lehtinen, V., Hakkinen, M., Koskela, M., and Siitonen, A. (2006). Territorial waters of the Baltic Sea as a source of infections caused by Vibrio cholerae non-O1, non-O139, report of 3 hospitalized cases. Diagn. Microbiol. Infect. Dis. 54, 1–6. doi: 10.1016/j.diagmicrobio.2005.06.020
Marin, M. A., Thompson, C. C., Freitas, F. S., Fonseca, E. L., Aboderin, A. O., Zailani, S. B., et al. (2013). Cholera outbreaks in Nigeria are associated with multidrug resistant atypical El Tor and non-O1/non-O139 Vibrio cholerae. PLoS Negl. Trop. Dis. 7:e2049. doi: 10.1371/journal.pntd.0002049
Matlawska-Wasowska, K., Finn, R., Mustel, A., O'Byrne, C. P., Baird, A. W., Coffey, E. T., et al. (2010). The Vibrio parahaemolyticus type III secretion systems manipulate host cell MAPK for critical steps in pathogenesis. BMC Microbiol. 10:329. doi: 10.1186/1471-2180-10-329
Matsumoto, C., Okuda, J., Ishibashi, M., Iwanaga, M., Garg, P., Rammamurthy, T., et al. (2000). Pandemic spread of an O3, K6 clone of Vibrio parahaemolyticus and emergence of related strains evidenced by arbitrarily primed PCR and toxRS sequence analyses. J. Clin. Microbiol. 38, 578–585.
Matz, C., Nouri, B., McCarter, L., and Martinez-Urtaza, J. (2011). Acquired type III secretion system determines environmental fitness of epidemic Vibrio parahaemolyticus in the interaction with bacterivorous protists. PLoS ONE 6:e20275. doi: 10.1371/journal.pone.0020275
McLaughlin, J. B., DePaola, A., Bopp, C. A., Martinek, K. A., Napolilli, N. P., Allison, C. G., et al. (2005). Outbreak of Vibrio parahaemolyticus gastroenteritis associated with Alaskan oysters. N. Engl. J. Med. 353, 1463–1470. doi: 10.1056/NEJMoa051594
Meador, C., Parsons, M., Bopp, C., Gerner-Smidt, P., Painter, J., and Vora, G. (2007). Virulence Gene- and pandemic group-specific marker profiling of clinical Vibrio parahaemolyticus isolates. J. Clin. Microbiol. 45, 1133–1139. doi: 10.1128/JCM.00042-07
Narkevich, M. I., Onischenko, G. G., Lomov, J. M., Moskvitina, E. A., Podosinnikova, L. S., and Medinsky, G. M. (1993). The seventh pandemic of cholera in the USSR, 1961-1989. Bull. World Health Organ. 71, 189–196.
Nasu, H., Iida, T., Sugahara, T., Yamaichi, Y., Park, K. S., Yokoyama, K., et al. (2000). A filamentous phage associated with recent pandemic Vibrio parahaemolyticus O3, K6 strains. J. Clin. Microbiol. 38, 2156–2161.
Oberbeckmann, S., Fuchs, B. M., Meiners, M., Wichels, A., Wiltshire, K. H., and Gerdts, G. (2012). Seasonal dynamics and modeling of a Vibrio community in coastal waters of the North Sea. Microb. Ecol. 63, 543–551. doi: 10.1007/s00248-011-9990-9
Okuda, J., Ishibashi, M., Hayakawa, E., Nishino, T., Takeda, Y., Mukhopadhyay, A. K., et al. (1997). Emergence of a unique O3, K6 clone of Vibrio parahaemolyticus in Calcutta, India, and isolation of strains from the same clonal group from Southeast Asian travelers arriving in Japan. J. Clin. Microbiol. 35, 3150–3155.
Ono, T., Park, K. S., Ueta, M., Iida, T., and Honda, T. (2006). Identification of proteins secreted via Vibrio parahaemolyticus type III secretion system 1. Infect. Immun. 74, 1032–1042. doi: 10.1128/IAI.74.2.1032-1042.2006
Paranjpye, R., Hamel, O. S., Stojanovski, A., and Liermann, M. (2012). Genetic diversity of clinical and environmental Vibrio parahaemolyticus strains from the Pacific Northwest. Appl. Environ. Microbiol. 78, 8631–8638. doi: 10.1128/AEM.01531-12
Rashid, A., Haley, B. J., Rajabov, M., Ahmadova, S., Gurbanov, S., Colwell, R. R., et al. (2013). Detection of Vibrio cholerae in environmental waters including drinking water reservoirs of Azerbaijan. Environ. Microbiol. Rep. 5, 30–38. doi: 10.1111/j.1758-2229.2012.00369.x
Safrin, S., Morris, J. G. Jr., Adams, M., Pons, V., Jacobs, R., and Conte, J. E. Jr. (1988). Non-O:1 Vibrio cholerae bacteremia: case report and review. Rev. Infect. Dis. 10, 1012–1017. doi: 10.1093/clinids/10.5.1012
Sarkar, B. L., Nair, G. B., Banerjee, A. K., and Pal, S. C. (1985). Seasonal distribution of Vibrio parahaemolyticus in freshwater environs and in association with freshwater fishes in Calcutta. Appl. Environ. Microbiol. 49, 132–136.
Serichantalergs, O., Bhuiyan, N. A., Nair, G. B., Chivaratanond, O., Srijan, A., Bodhidatta, L., et al. (2007). The dominance of pandemic serovars of Vibrio parahaemolyticus in expatriates and sporadic cases of diarrhoea in Thailand, and a new emergent serovar (O3: K46) with pandemic traits. J. Med. Microbiol. 56, 608–613. doi: 10.1099/jmm.0.47006-0
Shannon, J. D., and Kimbrough, R. C. (2006). Pulmonary cholera due to infection with a non-O1 Vibrio cholerae strain. J. Clin. Microbiol. 44, 3459–3460. doi: 10.1128/JCM.02343-05
Shikulov, V., Khaitovich, A., and Bogatyreva, L. (1980). Isolation of halophilic vibrios from humans and the environment. Zh. Mikrobiol. Epidemiol. Immunobiol. 6, 38–40.
Suthienkul, O., Ishibashi, M., Iida, T., Nettip, N., Supavej, S., Eampokalap, B., et al. (1995). Urease production correlates with possession of the trh gene in Vibrio parahaemolyticus strains isolated in Thailand. J. Infect. Dis. 172, 1405–1408. doi: 10.1093/infdis/172.5.1405
Turner, J. W., Good, B., Cole, D., and Lipp, E. K. (2009). Plankton composition and environmental factors contribute to Vibrio seasonality. ISME J. 3, 1082–1092. doi: 10.1038/ismej.2009.50
Vuddhakul, V., Chowdhury, A., Laohaprertthisan, V., Pungrasamee1, P., Patararungrong, N., Thianmontri, P., et al. (2000). Isolation of a pandemic O3, K6 clone of a Vibrio parahaemolyticus strain from environmental and clinical sources in Thailand. Appl. Environ. Microbiol. 66, 2685–2689. doi: 10.1128/AEM.66.6.2685-2689.2000
Wang, H. Z., Wong, M. M. L., O'Toole, D., Mak, M. M. H., Wu, R. S. S., and Kong, R. Y. C. (2006). Identification of a DNA methyltransferase gene carried on a pathogenicity island-like element (VPAI) in Vibrio parahaemolyticus and its prevalence among clinical and environmental isolates. Appl. Environ. Microb. 72, 4455–4460. doi: 10.1128/AEM.02095-05
Whitaker, W. B., Parent, M. A., Boyd, A., Richards, G. P., and Boyd, E. F. (2012). The Vibrio parahaemolyticus ToxRS regulator is required for stress tolerance and colonization in a novel orogastric streptomycin-induced adult murine model. Infect. Immun. 80, 1834–1845. doi: 10.1128/IAI.06284-11
WHO. (2011). Available online at: http://www.euro.who.int/en/what-we-do/health-topics/emergencies/international-health-regulations/news/news/2011/06/ukraine-reports-14-cholera-cases (Accessed March 20, 2012).
Wildschutte, H., Wolfe, D. M., Tamewitz, A., and Lawrence, J. G. (2004). Protozoan predation, diversifying selection, and the evolution of antigenic diversity in Salmonella. Proc. Natl. Acad. Sci. U.S.A. 101, 10644–10649. doi: 10.1073/pnas.0404028101
Williams, T. L., Musser, S. M., Nordstrom, J. L., DePaola, A., and Monday, S. R. (2004). Identification of a protein biomarker unique to the pandemic O3, K6 clone of Vibrio parahaemolyticus. J. Clin. Microbiol. 42, 1657–1665. doi: 10.1128/JCM.42.4.1657-1665.2004
Wong, H. C., Chen, M. C., Liu, S. H., and Liu, D. P. (1999). Incidence of highly genetically diversified Vibrio parahaemolyticus in seafood imported from Asian countries. Int. J. Food Microbiol. 52, 181–188. doi: 10.1016/S0168-1605(99)00143-9
Wong, H. C., Liu, S. H., Ku, L. W., Lee, I. Y., Wang, T. K., Lee, Y. S., et al. (2000). Characterization of Vibrio parahaemolyticus isolates obtained from foodborne illness outbreaks during 1992 through 1995 in Taiwan. J. Food Prot. 63, 900–906.
Woo, P. C. Y., Fung, A. M. Y., Wong, S. S. Y., Tsoi, H.-W., and Yuen, K.-Y. (2001). Isolation and characterization of a Salmonella enterica serotype typhi variant and its clinical and public health implications. J. Clin. Microbiol. 39, 1190–1194. doi: 10.1128/JCM.39.3.1190-1194.2001
Keywords: Vibrio parahaemolyticus, predictive modeling, Vibrionaceae, Black Sea, aquatic microbiology
Citation: Haley BJ, Kokashvili T, Tskshvediani A, Janelidze N, Mitaishvili N, Grim CJ, Constantin de Magny G, Chen AJ, Taviani E, Eliashvili T, Tediashvili M, Whitehouse CA, Colwell RR and Huq A (2014) Molecular diversity and predictability of Vibrio parahaemolyticus along the Georgian coastal zone of the Black Sea. Front. Microbiol. 5:45. doi: 10.3389/fmicb.2014.00045
Received: 26 November 2013; Paper pending published: 13 December 2013;
Accepted: 21 January 2014; Published online: 10 February 2014.
Edited by:
Daniela Ceccarelli, University of Maryland, USAReviewed by:
Christopher Staley, University of Minnesota, USACopyright © 2014 Haley, Kokashvili, Tskshvediani, Janelidze, Mitaishvili, Grim, Constantin de Magny, Chen, Taviani, Eliashvili, Tediashvili, Whitehouse, Colwell and Huq. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anwar Huq, Maryland Pathogen Research Institute, University of Maryland, 3102 Biosciences Research Building, MD 20742, USA e-mail:aHVxQHVtZC5lZHU=
†Present address: Christopher J. Grim, US Food and Drug Administration, Laurel, MD, USA
Elisa Taviani, Biotechnology Center University Eduardo Mondlane, Mozambique
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.