- Department of Applied Microbiology, Institute of Microbiology, Faculty of Biology, University of Warsaw, Warsaw, Poland
The plasmids of the incompatibility (Inc) group IncP-1, also called IncP, as extrachromosomal genetic elements can transfer and replicate virtually in all Gram-negative bacteria. They are composed of backbone genes that encode a variety of essential functions and accessory genes that have implications for human health and environmental bioremediation. Broad-host-range IncP plasmids are known to spread genes between distinct phylogenetic groups of bacteria. These genes often code for resistances to a broad spectrum of antibiotics, heavy metals, and quaternary ammonium compounds used as disinfectants. The backbone of these plasmids carries modules that enable them to effectively replicate, move to a new host via conjugative transfer and to be stably maintained in bacterial cells. The adaptive, resistance, and virulence genes are mainly located on mobile genetic elements integrated between the functional plasmid backbone modules. Environmental studies have demonstrated the wide distribution of IncP-like replicons in manure, soils and wastewater treatment plants. They also are present in strains of pathogenic or opportunistic bacteria, which can be a cause for concern, because they may encode multiresistance. Their broad distribution suggests that IncP plasmids play a crucial role in bacterial adaptation by utilizing horizontal gene transfer. This review summarizes the variety of genetic information and physiological functions carried by IncP plasmids, which can contribute to the spread of antibiotic and heavy metal resistance while also mediating the process of bioremediation of pollutants. Due to the location of the resistance genes on plasmids with a broad-host-range and the presence of transposons carrying these genes it seems that the spread of these genes would be possible and quite hazardous in infection control. Future studies are required to determine the level of risk of the spread of resistance genes located on these plasmids.
Characteristics of IncP-1 Plasmids
A characteristic feature of all plasmids, including IncP-1 plasmids, is that they belong to specific incompatibility (Inc) groups. Plasmids are classified into Inc groups based on their replication and partitioning systems: two plasmids belonging to the same Inc group are unable to coexist in a single bacterial cell (Shintani et al., 2010b).
Members of the Enterobacteriaceae family and the genus Pseudomonas carry plasmids belonging to more than 30 Inc groups. Plasmids from four of these groups (IncP, W, N, and Q) can transfer between and maintain themselves in both enteric bacteria and strains of Pseudomonas (Dröge et al., 2000). In particular, IncP plasmids are widely distributed in Gram-negative bacteria, e.g., in Escherichia coli, Pseudomonas spp., Klebsiella aerogenes, and Sphingomonas (Thomas, 2000; Shintani et al., 2010a). It should be added that IncP-1 in the Pseudomonas plasmid classification corresponds to IncP in the E. coli plasmid classification (Thomas and Haines, 2004).
The plasmids of the subgroup IncP-1 are of considerable interest to both molecular biologists and environmentalists due to their highly efficient conjugative transfer and ability to replicate in a broad range of hosts (Shintani et al., 2010a). These plasmids demonstrate great diversity – a phylogenetic study has shown the presence of 45 backbone genes on IncP-1 plasmids, but only 33 are shared by all the plasmids that have been completely sequenced to date.
A detailed phylogenetic analysis and study of the amino acid sequence of protein TrfA, which initiates plasmid replication, have allowed the classification of group IncP-1 into six subgroups: -α, -β, -γ, -ε, -δ (Bahl, 2009), and -ζ (Norberg et al., 2011) and also into an unnamed subgroup (Pachulec and van der Does, 2010). Unpublished data also indicate the existence of a η subgroup (Sen et al., 2012). IncP-1 plasmids have been identified in clinical and environmental, phylogenetically very distant, host bacterial species from around the world. Bacteria carrying these plasmids have been isolated from soil in areas contaminated by industry, pig manure, wastewater of industrial origin, river sediment, and fresh water (Bahl, 2009; Norberg et al., 2011; Sen et al., 2012). Comparative analysis of the nucleotide sequences of plasmids belonging to the IncP-1 subgroups α and β demonstrated the high similarity of their gene organization and also led to the identification of adaptive genes called “load genes.” The backbone sequences of these plasmids are responsible for the regulation of replication, stable maintenance in a bacterial cell and efficient conjugative transfer. In plasmids of the IncP-1β subgroup, the Tra1 and Tra2 regions of the backbone, that mediate conjugative transfer, are always separated by regions of clustered restriction sites, which may represent hotspots for the integration of various determinants. These regions can be a site for the insertion of mobile genetic elements (MGE) carrying catabolic genes, and antibiotic and heavy metal resistance determinants. The capacity for MGE integration is highly variable among IncP-1 plasmids. Plasmids pJP4, pSS60, and pSS50, belonging to the subgroup IncP-1β, contain one site for the integration of catabolic genes in their plasmid backbone, which is located between the oriV and trfA system replication sequences. The number of insertions in the backbone varies among IncP-1 plasmids (Trefault et al., 2004). pB4 is the only plasmid known to have three insertion sites: catabolic genes grouped into an operon are located near the oriV sequence, and antibiotic resistance determinants are located at both ends of region Tra2 (Adamczyk and Jagura-Burdzy, 2003).
There are currently only three known representatives of the subgroup IncP-1δ, namely plasmids pEST4011, pIJB1, and pAKD4. Plasmid pAKD4 is regarded as the prototype of this subgroup because it is the only one to possess the complete backbone sequence. Like other IncP-1 plasmids, those of subgroup δ carry specific genes that are located on transposons integrated within the plasmid backbone (Sen et al., 2010, 2012).
More than 50 plasmids belonging to the subgroup IncP-1ε have recently been identified and characterized. These plasmids confer resistance to a broad spectrum of antibiotics. This is due to the presence of a class 1 integron that carries the gene sul1 responsible for sulphonamide resistance at its 5 ′ end, plus gene cassettes carrying several determinants of resistance to other antibiotics (Heuer et al., 2012). Plasmids belonging to subgroup IncP-1ε exhibit great diversity (Sen et al., 2011).
Antibiotic Resistance Genes Present on IncP Plasmids
The extensive use of antibiotics for the treatment of humans and animals, and to stimulate the growth of livestock, has led to the rapid emergence and spread of resistance determinants to a broad spectrum of antibiotic substances (Allen et al., 2010; Aminov, 2010; Popowska et al., 2010, 2012; Tello et al., 2012). These determinants are located mainly on MGE such as plasmids and conjugative transposons, which ensures their spread by horizontal gene transfer. Horizontal gene transfer involving the acquisition of foreign DNA by transduction, transformation, or conjugation, is quite common among bacteria and plasmids play a key role in this process (Aminov, 2011; Martinez, 2009; Stokes and Gillings, 2011). Among the plasmids conferring resistance to antibiotics, representatives of the incompatibility groups P, Q, N, and W have been identified. These are characterized by their wide host range, including pathogenic bacteria, and so pose a serious threat to human health (Dröge et al., 2000). Numerous studies have demonstrated that the prevalence of such plasmids in different environments is very high, e.g., in soil, surface waters, wastewaters, and natural fertilizers (Götz et al., 1996; Haines et al., 2006; Binh et al., 2008; Sen et al., 2011; Heuer et al., 2012).
There are four basic mechanisms of plasmid-encoded antibiotic resistance: (i) enzymatic hydrolysis of the antibiotic molecule, which results in inactivation of the compound, (ii) enzymatic modification of the antibiotic to prevent interaction with the cellular target, (iii) efflux, which removes the antibiotic from the interior of the cell via specific cell membrane pumps, and (iv) a reaction called “bypass,” which is the formation of an alternative metabolic pathway that allows bypassing of the blocked stage and modification of the antibiotic target (Alekshun and Levy, 2007). The genes encoding the proteins responsible for these mechanisms are found on plasmids present in both clinical and environmental bacterial isolates (Aminov and Mackie, 2007; Allen et al., 2010; Wright, 2010).
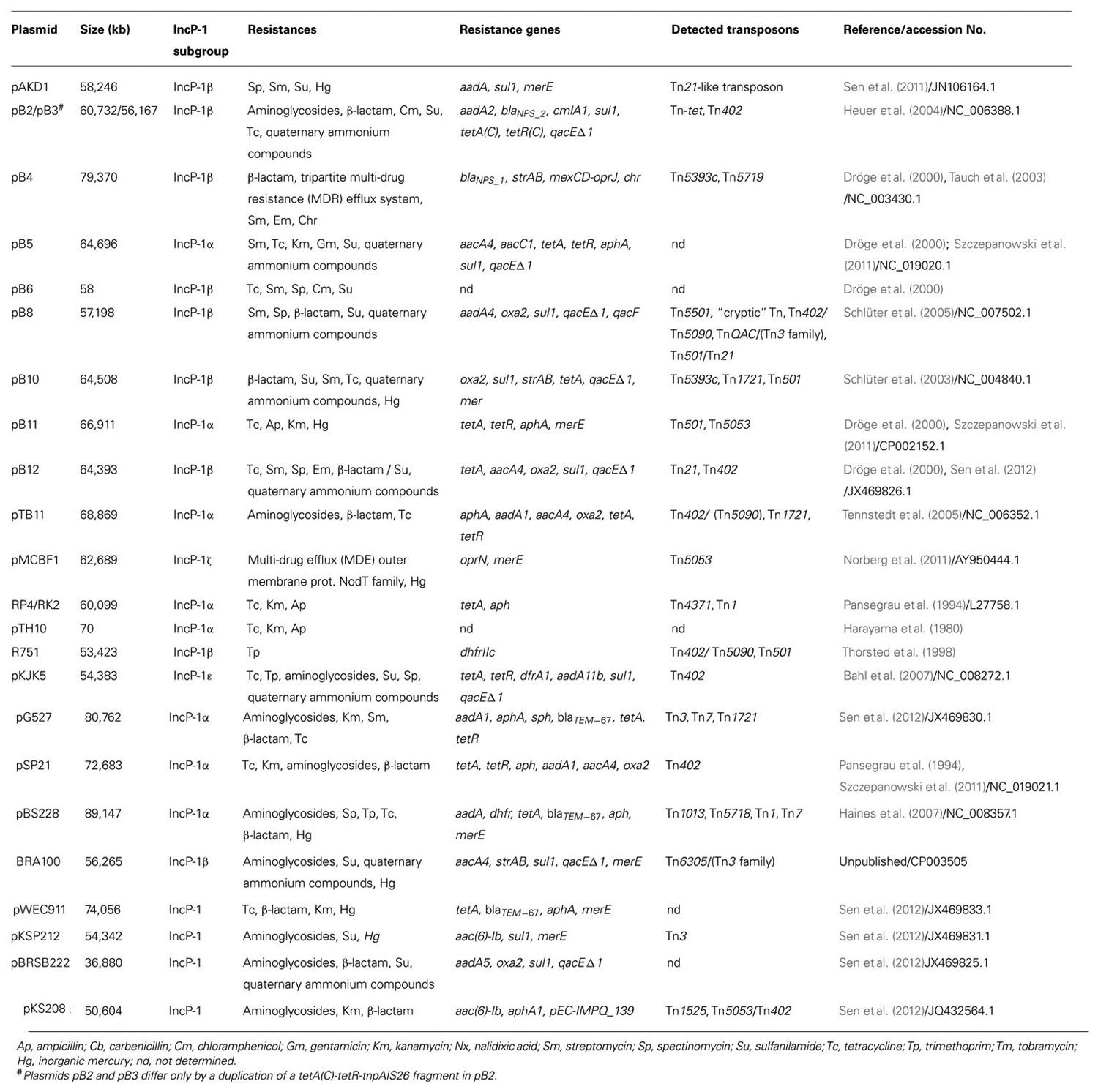
Table 1 shows plasmids from IncP-1 group, well characterized in terms of phenotype and molecular mechanisms of observed resistance to antibiotics and heavy metals. Resistance plasmids classified within subgroup IncP-1α were first identified in hospital isolates of Pseudomonas aeruginosa and K. aerogenes. These were found to encode multi-drug resistance (MDR) against antibiotics including penicillin, kanamycin, and tetracycline (Adamczyk and Jagura-Burdzy, 2003). Using the exogenous plasmid isolation method Dröge et al. (2000) found resistance plasmids, including those belonging to the subgroup IncP-1α, in bacterial communities from activated sludge derived from wastewater treatment plant (WWTP). Twelve plasmids were identified carrying an antibiotic and heavy metal resistance determinant, and ten of these could be classified to group IncP. These studies confirmed the wide distribution of this Inc group in these environments.
To date, the complete nucleotide sequences of six resistance plasmids belonging to subgroup IncP-1α have been determined. These plasmids, RP4/RK2, pTB11, pBS228, pB5, pB11, and pSP21, show a high degree of similarity with respect to elements of their plasmid backbone, while they are distinguished by the nature of the genes located within their adaptive modules (e.g., ISTB11, ISSP21, Tn402 derivatives, Tn1, Tn7). Plasmid pB5 contains an integron of class 1 carrying a resistance cassette with aacC1 and aacA4 genes, which encode enzymes that provide resistance to kanamycin, gentamicin, and tobramycin (Szczepanowski et al., 2011). The IncP-1α plasmids contain different variants of the integron aadA gene cassette (aadA1, aadA2, aadA4, and aadA5), which encode streptomycin/spectinomycin-300-O-adenylyltransferases conferring resistance to the aminoglycosides streptomycin and spectinomycin (Tennstedt et al., 2003, 2005). The aadA2 gene cassette is widely distributed, having been identified in the integrons of many bacteria belonging to at least 11 genera. Numerous studies have demonstrated that different environmental and pathogenic bacteria share a common pool of integron-specific resistance gene cassettes (Tennstedt et al., 2003, 2005).
In the case of plasmids pTB11 and pSP21, a class 1 integron carrying genes that determine bacterial resistance to aminoglycosides is located within transposon Tn402. In addition, these plasmids possess a region containing genes for tetracycline resistance (tetA and tetR; Tennstedt et al., 2005; Szczepanowski et al., 2011).
The IncP-1α plasmid pTB11 carries the aphA-orf3 module, encoding aminoglycoside-30-phosphotransferase responsible for aminoglycoside resistance, that shares the highest degree of similarity with the corresponding gene of plasmid RP4, originating from a clinical isolate. Another aminoglycoside resistance gene cassette located on pTB11 and pSp32 encodes an aminoglycoside-60-N-acetyltransferase mediating resistance to different aminoglycosides such as tobramycin, amikacin, and gentamicin. Two other aminoglycoside resistance gene cassettes, aadB and aacA29b, which respectively, code for an aminoglycoside-200-O-adenylyltransferase and an aminoglycoside-60-N-acetyltransferase, have also been identified on IncP-1α plasmids. The aadB gene was found on a plasmid from a clinical isolate of Acinetobacter baumannii, while an aacA29b gene cassette, present on a plasmid, represents a new variant of aacA29a previously found on In59 in a clinical P. aeruginosa isolate (Siarkou et al., 2009).
Resistance plasmids are also represented in subgroup IncP-1β. These include the fully sequenced plasmids pB2/pB3, PB4, PB8, and PB10. These plasmids carry a class 1 integron or, in the case of PB4, a fragment of Tn402. Besides sul1 that confers resistance to sulfonamides, these plasmids may also contain genes for resistance to other antibiotics and chemotherapeutics, e.g., oxa (PB8, PB10, and pB11) and bla (PB4 and PB3), encoding β-lactamase enzymes responsible for resistance to β-lactam antibiotics. Parts of the oxa1 gene cassette in plasmid pSp33 and blaNPS1 on the Tn402 remnant in this plasmid are nearly identical to corresponding regions identified in different Korean clinical isolates of Salmonella enterica ssp. enterica (Lee et al., 2003, 2004) and P. aeruginosa (Jacoby and Matthew, 1979; Pai and Jacoby, 2001), respectively. However, some bla genes identified in this subgroup of plasmids have novel sequences. All of the aforementioned IncP-1β plasmids also carry genes encoding resistance to streptomycin, a member of the aminoglycoside antibiotic family. Plasmids PB4 and PB10 contain transposon Tn5393c, which includes the genes strA and strB, encoding aminoglycoside-3′′-O-phosphotransferase and aminoglycoside-6-O-phosphotransferase, respectively, that modify the streptomycin molecule, thus causing its inactivation. Identical or almost identical copies of Tn5393 were recently identified in different human, fish, and plant pathogens. Plasmid pB10 also possesses the tetracycline resistance transposon Tn1721, that was first described in E. coli (Allmeier et al., 1992). A very similar transposon has been found in conjugative plasmids from the fish pathogen Aeromonas salmonicida (Sørum et al., 2003), the human pathogen Salmonella enterica ssp. enterica (Chiu et al., 2005) and a clinical isolate of E. coli (Boyd et al., 2004). With the use of plasmid pB10, carrying amoxicillin, streptomycin, sulfonamide, and tetracycline resistance genes, is has been shown that this broad-host-range IncP-1 plasmid can be transferred to foodborne pathogens (Salmonella spp. and E. coli O157:H7) under laboratory conditions (Van Meervenne et al., 2012). These results show that the antibiotic resistance genes encoded on plasmid pB10 can be expressed in the new hosts (pathogenic organisms) making these pathogens resistant to multiple antibiotics. These studies have shown that there is a serious risk to human health in cases of such transfer. The plasmid PB3 carries the gene clmA1 conferring resistance to chloramphenicol. The product of the clmA1 gene is a protein that acts as an efflux pump, which removes the drug from the bacterial cell (Schlüter et al., 2007). The genetic modules on these IncP-1β plasmids consist mainly of MGE that carry diverse determinants conferring resistance to different aminoglycosides, β-lactams, macrolides, sulphonamides, tetracycline, trimethoprim, disinfectants, and mercury ions. The greatest MGE diversity has been observed in plasmids of this subgroup isolated from bacteria living in WWTP (Szczepanowski et al., 2004). WWTP bacteria represent a reservoir for β-lactamase genes that have yet to be isolated from clinical strains. Mobile integron gene cassettes seem to function in the dissemination of β-lactamase genes in sewage habitats.
An important role in the spread of resistance genes in agricultural systems is played by representatives of subgroup IncP-1ε. The study of Heuer et al. (2012) showed that fifty plasmids belonging to this subgroup, isolated from samples of soil and manure, contain integrons of class 1 with gene cassettes plus the gene sul1 (sulphonamide resistance). The nucleotide sequences of four of these plasmids have been determined (pKS77, pHH3414, pHH128, and pHH3408) and they differ mainly in the organization of the elements in transposon Tn402. These elements are a class 1 integron IS1326 and, in the case of plasmids pHH3414, pKS77, and pHH128, transposon Tn1721 carrying the tetracycline resistance genes tetA and tetR. The resistance integrons contain the gene cassettes aadA, (pHH3414), aadB (pKS77), aadA1b, dfrA1b, and two copies of catB (pHH128; Heuer et al., 2012). The different aad gene variants encode adenyltransferases that modify the streptomycin and spectinomycin molecules, resulting in their inactivation. A similar mechanism is employed by the chloramphenicol acetyltransferase encoded by the cat genes to confer resistance to chloramphenicol. The dfr gene permits bacterial cells to survive in an environment with high concentrations of trimethoprim (Schlüter et al., 2007).
Interestingly, many studies have demonstrated a close relationship between the occurrence of MDR and resistance/tolerance to heavy metal ions (Lazar et al., 2002). Bacterial strains isolated from agricultural soil receiving wastewater can exhibit high levels of both metal and antibiotic resistance. Shafiani and Malik (2003) isolated 64 bacteria belonging to the genera Pseudomonas, Azotobacter, and Rhizobium from wastewater-irrigated soil. All the Pseudomonas isolates were resistant to heavy metals and cloxacillin, and more than half were also resistant to methicillin. Among these strains were MDR isolates. Ansari et al. (2008) found that 40 strains isolated from agricultural soils irrigated with wastewater showed resistance to most of the tested heavy metals (Ni, Cu, Zn, Pb). Moreover, around 75% of these strains were resistant to tetracycline, 57.5% to doxycycline, and 50% to ampicillin and nalidixic acid. The plasmids isolated from these resistant strains all belonged to the IncP group. Out of 12 pAKD plasmids (IncP-1 group) isolated from Norwegian soils only one, pAKD1, encoded antibiotic resistance in addition to mercury resistance. This plasmid carries a MDR Tn21-like transposon with a class 1 integron (Sen et al., 2011). These data suggest heavy metals treatment might have co-selected for MDR genes. This is consistent with previous and present observations that pollution with heavy metals can cause a rise in the abundance of drug resistance genes (Baker-Austin et al., 2006; Seiler and Berendonk, 2012).
Conclusion
The increasing use of antibiotics in human and veterinary medicine and agricultural production systems has caused the increasing development of high levels of and novel antibiotic resistances and the rapid global spread of antibiotic resistance genes (Aminov, 2009, 2010; Martinez, 2009; Schlüter et al., 2007; Sen et al., 2011). A consequence of the horizontal transfer of the genes located on MGE, especially on Broad-host-range plasmids is the rising detection and levels of resistant bacteria, particularly of MDR bacteria, both in clinical and natural environments (Dröge et al., 2000; Aminov and Mackie, 2007).
IncP-1 plasmids are characterized by their wide distribution in the environment and ability to efficiently replicate in the cells of diverse hosts. In addition, the presence of transposons carrying both antibiotic and heavy metal resistance genes on these plasmids permits the host bacteria to survive and multiply in environments contaminated with these compounds. The broad-host-range IncP-1 plasmids carry determinants for resistance to at least one heavy metal (Ni, Cd, Co, Cu, Hg, Pb, Zn), and antibiotics of different groups, i.e., tetracyclines, quinolones, aminoglycosides, sulfonamides, β-lactams, and chemotherapeutics. This fact is not surprising in light of recent data showing that heavy metals present in the environment might have co-selected for MDR genes (Seiler and Berendonk, 2012). The two main mechanisms of antibiotic resistance encoded on IncP-1 plasmids are mediated by efflux pumps and enzymatic modification or hydrolysis of the antibiotic molecule. IncP-1 plasmid-containing bacteria are also key players in the horizontal transfer of antibiotic resistance in agricultural systems. It has been suggested that the spread of multi-resistant bacteria in soil, water, and WWTPs is mainly due to their possession of IncP-1 plasmids (Schlüter et al., 2003, 2005; Bahl et al., 2007; Ansari et al., 2008; Norberg et al., 2011; Sen et al., 2011).
Based on their DNA sequences, it can be concluded that the vast majority of these resistance genes are located on MGE such as transposons and/or integrons (Tn402-like transposon with a class 1 integron), embedded within these transposons. In addition, the presence of different combinations of MGE and integrons in IncP plasmids as well as of characteristic hot spots for the integration of accessory elements creates unlimited possibilities for rearrangements of the genetic material of these plasmids. It therefore appears that the spread of resistance genes would be possible and quite hazardous in infection control. Literature data show that many representatives of this group of plasmids carrying resistance genes have been identified in pathogenic bacteria of the genus Escherichia, Salmonella, Shigella, Klebsiella, Aeromonas, and Pseudomonas. Of course, in order to determine the level of risk of the spread of resistance genes located on these plasmids extensive experimental studies are required. However, the demonstrated ability to transfer multiresistance pB10 plasmid, isolated from a WWTP, to the foodborne pathogens Salmonella spp. or E. coli O157: H7 with expression of most of the transferred resistance genes in these strains poses a real threat to the life and health of people (Van Meervenne et al., 2012).
Detailed analysis of IncP-1 plasmid genomes can shed light on the adaptive mechanisms underlying bacterial evolution and this information may be used to develop effective methods of bioremediation of contaminated land with xenobiotic and antibiotics and heavy metals. Intensive research on the biodegradation of compounds that are a serious threat to the environment has led to the isolation and characterization of bacterial strains that can use these substances as a source of carbon and energy. In many cases, the adaptation of these microorganisms is due to plasmids (mainly of Inc group P) that carry genes encoding enzymes involved in the degradation of xenobiotics. It is known, that the degradative plasmids (IncP-1, P-7, and P-9) contain catabolic genes, often arranged as operons, encoding enzymes required for the degradation and utilization of many toxic compounds including naphthalene, toluene, chlorobenzene, p-toluenesulfonate, 2,4-D, haloacetate, and atrazine (Williams, 2004; Shintani et al., 2010a,b). The majority of bacterial genes encoding enzymes of catabolic pathways are located on MGE such as plasmids, insertion sequences (e.g., as a form of composite transposons), transposons, integrons, genomic islands, and phage genomes (Top et al., 2002; Top and Springael, 2003; Nojiri et al., 2004; Springael and Top, 2004).
Theoretically, genetic modules present in this type of element (both in catabolic and resistance plasmids) may be used in the construction of genetically modified strains that are optimized for use in bioremediation and biotechnology (Vidali, 2001; Urgun-Demirtas et al., 2006; Singh et al., 2011). This is of great importance due to the fact that these antimicrobial agents are powerful inhibitors of xenobiotic biodegradation activities, mainly due to their bacteriostatic or bactericidal activities. However, before the broader implementation of such microorganisms, further research is required to thoroughly characterize them in order to limit the spread of antibiotic resistance genes.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Adamczyk, M., and Jagura-Burdzy, G. (2003). Spread and survival of promiscuous IncP-1 plasmids. Acta Biochim. Pol. 50, 425–453.
Alekshun, M. N., and Levy, S. B. (2007). Molecular mechanisms of antibacterial multidrug resistance. Cell 128, 1037–1050.
Allen, H. K., Donato, J., Wang, H. H., Cloud-Hansen, K. A., Davies, J., and Handelsman, J. (2010). Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 8, 251–259.
Allmeier, H., Cresnar, B., Greck, M., and Schmitt, R. (1992). Complete nucleotide sequence of Tn1721: gene organization and a novel gene product with features of a chemotaxis protein. Gene 111, 11–20.
Aminov, R. I. (2009). The role of antibiotics and antibiotic resistance in nature. Environ. Microbiol. 11, 2970–2988.
Aminov, R. I. (2010). A brief history of the antibiotic era: lessons learned and challenges for the future. Front. Microbiol. 1:134. doi: 10.3389/fmicb.2010.00134
Aminov, R. I. (2011). Horizontal gene exchange in environmental microbiota. Front. Microbiol. 2:158. doi: 10.3389/fmicb.2011.00158
Aminov, R., and Mackie, R. I. (2007). Evolution and ecology of antibiotic resistance genes. FEMS Microbiol. Lett. 271, 147–161.
Ansari, M. I., Grohmann, E., and Malik, A. (2008). Conjugative plasmids in multi-resistant bacterial isolates from Indian soil. J. Appl. Microbiol. 104, 1774–1781.
Bahl, M. I. (2009). All IncP-1 plasmid subgroups, including the novel e subgroup, are prevalent in the influent of a Danish wastewater treatment plant. Plasmid 62, 134–139.
Bahl, M. I., Hansen, L. H., Goesmann, A., and Sørensen, S. J. (2007). The multiple antibiotic resistance IncP-1 plasmid pKJK5 isolated from a soil environment is phylogenetically divergent from members of the previously established alpha, beta and delta sub-groups. Plasmid 58, 31–43.
Baker-Austin, C., Wright, M. S., Stepanauskas, R., and McArthur, J. V. (2006). Co-selection of antibiotic and metal resistance. Trends Microbiol. 14, 176–182.
Binh, C. T., Heuer, H., Kaupenjohann, M., and Smalla, K. (2008). Piggery manure used for soil fertilization is a reservoir for transferable antibiotic resistance plasmids. FEMS Microbiol. Ecol. 66, 25–37.
Boyd, D. A., Tyler, S., Christianson, S., McGeer, A., Muller, M. P., Willey, B. M., et al. (2004). Complete nucleotide sequence of a 92-kilobase plasmid harboring the CTX-M-15 extended-spectrum beta-lactamase involved in an outbreak in long-term-care facilities in Toronto, Canada. Antimicrob. Agents Chemother. 48, 3758–3764.
Chiu, C. H., Tang, P., Chu, C., Hu, S., Bao, Q., Yu, J., et al. (2005). The genome sequence of Salmonella enterica serovar Choleraesuis, a highly invasive and resistant zoonotic pathogen. Nucleic Acids Res. 33, 1690–1698.
Dröge, M., Puhler, A., and Selbitschka, W. (2000). Phenotypic and molecular characterization of conjugative antibiotic resistance plasmids isolated from bacterial communities of activated sludge. Mol. Gen. Genet. 263, 471–482.
Götz, A., Pukall, R., Smit, E., Tietze, E., Prager, R., Tschäpe, H., et al. (1996). Detection and characterization of broad-host-range plasmids in environmental bacteria by PCR. Appl. Environ. Microbiol. 62, 2621–2628.
Haines, A. S., Akhtar, P., Stephens, E. R., Jones, K., Thomas, C. M., Perkins, C. D., et al. (2006). Plasmids from freshwater environments capable of IncQ retrotransfer are diverse and include pQKH54, a new IncP-1 subgroup archetype. Microbiology 152, 2689–2701.
Haines, A. S., Jones, K., Batt, S. M., Kosheleva, I. A., and Thomas, C. M. (2007). Sequence of plasmid pBS228 and reconstruction of the IncP-1α phylogeny. Plasmid 58, 76–83.
Harayama, S., Tsuda, M., and Iino, T. (1980). High frequency mobilization of the chromosome of Escherichia coli by a mutant of plasmid RP4 temperature-sensitive for maintenance. Mol. Gen. Genet. 180, 47–56.
Heuer, H., Binh, C. T., Jechalke, S., Kopmann, C., Zimmerling, U., Krögerrecklenfort, E., et al. (2012). IncP-1ε plasmids are important vectors of antibiotic resistance genes in agricultural systems: diversification driven by class 1 integron gene cassettes. Front. Microbiol. 3:2. doi: 10.3389/fmicb.2012.00002
Heuer, H., Szczepanowski, R., Schneiker, S., Pühler, A., Top, E. M., and Schlüter, A. (2004). The complete sequences of plasmids pB2 and pB3 provide evidence for a recent ancestor of the IncP-1β group without any accessory genes. Microbiology 150, 3591–3599.
Jacoby, G. A., and Matthew, M. (1979). The distribution of β-lactamase genes on plasmids found in Pseudomonas. Plasmid 2, 41–47.
Lazar, V., Cernat, R., Balotsen, C., Cotar, A., Coipan, E., and Cojocaru, C. (2002). Correlation between multiple antibiotic resistance and heavy metal tolerance among some E. coli strains isolated from polluted waters. Bacteriol. Virusol. Parazitol. Epidemiol. 47, 155–160.
Lee, K., Yong, D., Yum, J. H., Kim, H. H., and Chong, Y. (2003). Diversity of TEM-52 extended-spectrum beta-lactamase-producing nontyphoidal Salmonella isolates in Korea. J. Antimicrob. Chemother. 52, 493–496.
Lee, K., Yong, D., Yum, J. H., Lim, Y. S., Kim, H. S., Lee, B. K., et al. (2004). Emergence of multidrug-resistant Salmonella enterica serovar typhi in Korea. Antimicrob. Agents Chemother. 48, 4130–4135.
Martinez, J. L. (2009). Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ. Pollut. 157, 2893–2902.
Nojiri, H., Shintani, M., and Omori, T. (2004). Divergence of mobile genetic elements involved in the distribution of xenobiotic-catabolic capacity. Appl. Microbiol. Biotechnol. 64, 154–174.
Norberg, P., Bergstrom, M., Jethava, V., Dubhashi, D., and Hermansson, M. (2011). The IncP-1 plasmid backbone adapts to different host bacterial species and evolves through homologous recombination. Nat. Commun. 2, 1–11.
Pachulec, E., and van der Does, C. (2010). Conjugative plasmids of Neisseria gonorrhoeae. PLoS ONE 5:e9962. doi: 10.1371/journal.pone.0009962
Pai, H., and Jacoby, G. A. (2001). Sequences of the NPS-1 and TLE-1 blactamase genes. Antimicrob. Agents Chemother. 45, 2947–2948.
Pansegrau, W., Lanka, E., Barth, P. T., Figurski, D. H., Guiney, D. G., Haas, D., et al. (1994). Complete nucleotide sequence of Birmingham IncP a plasmids. Compilation and comparative analysis. J. Mol. Biol. 239, 623–663.
Popowska, M., Miernik, A., Rzeczycka, M., and Łopaciuk, A. (2010). The impact of environmental contamination with antibiotics on levels of resistance in soil bacteria. J. Environ. Qual. 39, 1679–1687.
Popowska, M., Rzeczycka, M., Miernik, A., Krawczyk-Balska, A., Walsh, F., and Duffy, B. (2012). Influence of soil use on prevalence of tetracycline, streptomycin, and erythromycin resistance and associated resistance genes. Antimicrob. Agents Chemother. 56, 1434–1443.
Schlüter, A., Heuer, H., Szczepanowski, R., Forney, L. J., Thomas, C. M., Puhler, A., et al. (2003). The 64508 bp IncP-1β antibiotic multiresistance plasmid pB10 isolated from a waste-water treatment plant provides evidence for recombination between members of different branches of the IncP-1β group. Microbiology 149, 3139–3153.
Schlüter, A., Heuer, H., Szczepanowski, R., Poler, S. M., Schneiker, S., Pühler, A., et al. (2005). Plasmid pB8 is closely related to the prototype IncP-1β plasmid R751 but transfers poorly to Escherichia coli and carries a new transposon encoding a small multidrug resistance (SMR) efflux protein. Plasmid 54, 135–148.
Schlüter, A., Szczepanowski, R., Pühler, A., and Top, E. M. (2007). Genomics of IncP-1 antibiotic resistance plasmids isolated from wastewater treatment plants provides evidence for a widely accessible drug resistance gene pool. FEMS Microbiol. Rev. 31, 449–477.
Seiler, C., and Berendonk, T. U. (2012). Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front. Microbiol. 3:399. doi: 10.3389/fmicb.2012.00399
Sen, D., Brown, C. J., Top, E. M., and Sullivan, J. (2012). Inferring the evolutionary history of IncP-1 plasmids despite incongruence among backbone gene trees. Mol. Biol. Evol. 30, 154–166.
Sen, D., Van Der Auwera, G., Rogers, L., Thomas, C. M., Brown, C. J., and Top, E. M. (2011). Broad-host-range plasmids from agricultural soils have IncP-1 backbones with diverse accessory genes. Appl. Environ. Microbiol. 77, 7975–7983.
Sen, D., Yano, H., Suzuki, H., Krol, J. E., Rogers, L., Brown, C. J., et al. (2010). Comparative genomics of pAKD4, the prototype IncP-1δ plasmid with a complete backbone. Plasmid 63, 98–107.
Shafiani, S., and Malik, A. (2003). Tolerance of pesticides and antibiotic resistance in bacteria isolated from wastewater-irrigated soil. World J. Microbiol. Biotechnol. 19, 897–901.
Shintani, M., Takahashi, Y., Yamane, H., and Nojiri, H. (2010a). The behavior and significance of degradative plasmids belonging to Inc groups in Pseudomonas within natural environments and microcosms. Microbes Environ. 25, 253–265.
Shintani, M., Yamane, H., and Nojiri, H. (2010b). Behaviour of various hosts of the IncP-7 carbazole-degradative plasmid pCAR1 in artificial microcosms. Biosci. Biotechnol. Biochem. 74, 343–349.
Siarkou, V. I., Vitti, D., Protonotariou, E., Ikonomidis, A., and Sofianou, D. (2009). Molecular epidemiology of outbreak-related Pseudomonas aeruginosa strains carrying the novel variant blaVIM-17 metallo-β-lactamase gene. Antimicrob. Agents Chemother. 53, 1325–1330.
Singh, J. S., Abhilash, P. C., Singh, H. B., Singh, R. P., and Singh, D. P. (2011). Genetically engineered bacteria: an emerging tool for environmental remediation and future research perspectives. Gene 480, 1–9.
Sørum, H., L’Abe’e-Lund, T. M., Solberg, A., and Wold, A. (2003). Integron-containing IncU R plasmids pRAS1 and pAr-32 from the fish pathogen Aeromonas salmonicida. Antimicrob. Agents Chemother. 47, 1285–1290.
Springael, D., and Top, E. M. (2004). Horizontal gene transfer and microbial adaptation to xenobiotics: new types of mobile genetic elements and lessons from ecological studies. Trends Microbiol. 12, 53–58.
Stokes, H. W., and Gillings, M. R. (2011). Gene flow, mobile genetic elements and the recruitment of antibiotic resistance genes into Gram negative pathogens. FEMS Microbiol. Rev. 35, 790–819.
Szczepanowski, R., Eikmeyera, F., Harfmanna, J., Blomb, J., Rogersc, L. M., Top, E. M., et al. (2011). Sequencing and comparative analysis of IncP-1 antibiotic resistance plasmids reveal a highly conserved backbone and differences within accessory regions. J. Biotechnol. 155, 95–103.
Szczepanowski, R., Krahn, I., Linke, B., Goesmann, A., Pühler, A., and Schlüter, A. (2004). Antibiotic multiresistance plasmid pRSB101 isolated from a wastewater treatment plant is related to plasmids residing in phytopathogenic bacteria and carries eight different resistance determinants including a multidrug transport system. Microbiology 150, 3613–3630.
Tauch, A., Schlüter, A., Bischoff, N., Goesmann, A., Meyer, F., and Pühler, A. (2003). The 79,370-bp conjugative plasmid pB4 consists of an IncP-1β backbone loaded with a chromate resistance transposon, the strA-strB streptomycin resistance gene pair, the oxacillinase gene blaNPS-1, and a tripartite antibiotic efflux system of the resistance-nodulation-division family. Mol. Gen. Genomics 268, 570–584.
Tello, A., Austin, B., and Telfer, T. C. (2012). Selective pressure of antibiotic pollution on bacteria of importance to public health. Environ. Health Perspect. 120, 1100–1106.
Tennstedt, T., Szczepanowski, R., Braun, S., Pühler, A., and Schlüter, A. (2003). Occurrence of integron-associated resistance gene cassettes located on antibiotic resistance plasmids isolated from a wastewater treatment plant. FEMS Microbiol. Ecol. 45, 239–252.
Tennstedt, T., Szczepanowski, R., Krahn, I., Pühler, A., and Schlüter, A. (2005). Sequence of the 68,869 bp IncP-1α plasmid pTB11 from a waste-water treatment plant reveals a highly conserved backbone, a Tn402-like integron and other transposable elements. Plasmid 53, 218–238.
Thomas, C. M. (2000). The Horizontal Gene Pool: Bacterial Plasmids and Gene Spread. Amsterdam: Harwood Academic Publishers.
Thomas, C. M., and Haines, A. S. (2004). “Plasmids of the genus Pseudomonas,” in Pseudomonas, ed. J.-L. Ramos (London: Kluwer/Plenum), 197–231.
Thorsted, P. B., Macartney, D. P., Akhtar, P., Haines, A. S., Ali, N., Davidson, P., et al. (1998). Complete sequence of the IncP-1β plasmid R751: implications for evolution and organisation of the IncP backbone. J. Mol. Biol. 282, 969–990.
Top, E., Springael, D., and Boon, N. (2002). Catabolic mobile genetic elements and their potential use in bioaugmentation of polluted soils and waters. FEMS Microbiol. Ecol. 42, 199–208.
Top, E. M., and Springael, D. (2003). The role of mobile genetic elements in bacterial adaptation to xenobiotic organic compounds. Curr. Opin. Biotechnol. 14, 262–269.
Trefault, N., De la Iglesia, R., Molina, A. M., Manzano, M., Ledger, T., Pérez-Pantoja, D., et al. (2004). Genetic organization of the catabolic plasmid pJP4 from Ralstonia eutropha JMP134 (pJP4) reveals mechanisms of adaptation to chloroaromatic pollutants and evolution of specialized chloroaromatic degradation pathways. Environ. Microbiol. 6, 655–668.
Urgun-Demirtas, M., Stark, B., and Pagilla, K. (2006). Use of genetically engineered microorganisms (GEMs) for the bioremediation of contaminants. Crit. Rev. Biotechnol. 26, 145–164.
Van Meervenne, E., Van Coillie, E., Kerckhof, F.-M., Devlieghere, F., Herman, L., De Gelder, L. S. P., et al. (2012). Strain-specific transfer of antibiotic resistance from an environmental plasmid to foodborne pathogens. J. Biomed. Biotechnol. 2012, 834598.
Williams, P. A. (2004). Catabolic plasmids: fast-track bacterial evolution to combat pollution. Microbiol. Today 31, 168–170.
Keywords: IncP plasmid, antibiotic resistance, heavy metals, xenobiotic, catabolism, horizontal gene transfer, bioremediation
Citation: Popowska M and Krawczyk-Balska A (2013) Broad-host-range IncP-1 plasmids and their resistance potential. Front. Microbiol. 4:44. doi: 10.3389/fmicb.2013.00044
Received: 15 November 2012; Accepted: 19 February 2013;
Published online: 07 March 2013.
Edited by:
Fiona Walsh, Agroscope Changins Wädenswil, SwitzerlandReviewed by:
Holger Heuer, Julius Kühn-Institut, Federal Research Centre for Cultivated Plants, GermanyYoshimi Matsumoto, Osaka University, Japan
Copyright: © 2013 Popowska and Krawczyk-Balska. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Magdalena Popowska, Department of Applied Microbiology, Institute of Microbiology, Faculty of Biology, University of Warsaw, Ilji Miecznikowa 1, 02-096 Warsaw, Poland. e-mail:bWFnZGFwb3BAYmlvbC51dy5lZHUucGw=