
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol. , 10 May 2012
Sec. Fungi and Their Interactions
Volume 3 - 2012 | https://doi.org/10.3389/fmicb.2012.00176
This article is part of the Research Topic Vaccines and immunotherapy against fungi: the new frontier View all 15 articles
 Agostinho Carvalho1,2,3*
Agostinho Carvalho1,2,3* Cristina Cunha1
Cristina Cunha1 Rossana G. Iannitti1
Rossana G. Iannitti1 Andrea Casagrande1,4
Andrea Casagrande1,4 Francesco Bistoni1
Francesco Bistoni1 Franco Aversa5
Franco Aversa5 Luigina Romani1
Luigina Romani1
Fungal vaccines have long been a goal in the fields of immunology and microbiology to counter the high mortality and morbidity rates owing to fungal diseases, particularly in immunocompromised patients. However, the design of effective vaccination formulations for durable protection to the different fungi has lagged behind due to the important differences among fungi and their biology and our limited understanding of the complex host–pathogen interactions and immune responses. Overcoming these challenges is expected to contribute to improved vaccination strategies aimed at personalized efficacy across distinct target patient populations. This likely requires the integration of multifaceted approaches encompassing advanced immunology, systems biology, immunogenetics, and bioinformatics in the fields of fungal and host biology and their reciprocal interactions.
Fungal diseases are epidemiological hallmarks of distinct settings of at-risk patients, not only in terms of their underlying condition but in the spectrum of diseases they develop (Segal, 2009). Although fungi are responsible for pulmonary manifestations and cutaneous lesions in apparently immunocompetent individuals, their impact is most relevant in patients with severe immune dysfunction, in which they can cause severe, life-threatening forms of infection. As an increasing number of immunocompromised individuals resulting from intensive chemotherapy regimens, bone marrow or solid organ transplantation, and autoimmune diseases has been witnessed in the last decades, so has the incidence of fungal diseases (Segal, 2009). Therefore, fungal vaccination has been regarded as a particularly promising strategy in these groups of highly susceptible individuals. Indeed, the fact that a number of well-defined risk factors manifest before the onset of infection affords a window of opportunity to vaccinate. However, many challenges confront the development of fungal vaccines for humans. Among them, the insufficient understanding of the critical immune defects that predispose to pathogen-specific vulnerability in primary or secondary immunodeficient patients and the historical assumption that the immune system of these patients would not respond properly to strategies relying on immunological memory. However, it is noteworthy that the immunogenicity and efficacy of vaccines has been confirmed even in patients with profound lymphocyte defects, such as the case of human immunodeficiency virus (HIV)-infected patients (Klugman et al., 2003). However, a further degree of complexity has been recently provided by the acknowledgment that immune responses critically rely on individual genetic profiles (Carvalho et al., 2010). Hence, and despite the obvious advantages of “universal vaccine” strategies to address protection from fungi (Cassone and Rappuoli, 2010), immunogenetic-based approaches have also revealed the significant contribution of the host’s genetic background to efficient vaccine responses (Carvalho et al., 2012b), thereby suggesting that a more personalized approach would ultimately be of additional interest. The purpose of this review is therefore to present an update of concepts relevant for the design of ideal antifungal vaccines and the challenges faced in delivering them to specific target populations.
Although the global incidence of fungal diseases is currently rivaling those of many of the best known bacterial diseases, humans have coevolved with ubiquitous or commensal fungi in host–fungus relationships that for the most part are positive or neutral (Romani, 2011). This is illustrated by a number of cases, including that of Candida albicans, Pneumocystis jiroveci, and Malassezia spp., that live as benign commensals in one or more body locations in a majority of healthy individuals. As opportunistic pathogens, they are poised to overgrow cavities and penetrate tissue in response to alterations in host physiology that presumably compromises the complex mechanisms of immune adaptation that normally suppress their growth. Most fungi, however, such as Aspergillus fumigatus, Cryptococcus neoformans, and the thermally dimorphic fungi (Histoplasma capsulatum, Blastomyces dermatitidis, Paracoccidioides brasiliensis, Coccidioides immitis, Penicillium marneffei, and Sporothrix schenckii) are found ubiquitously in nature and can cause a wide spectrum of diseases ranging from acute pulmonary manifestations and cutaneous lesions in immunocompetent individuals to allergic syndromes and severe life-threatening infections in patients with primary or secondary immune dysfunction.
It is now clear that the clinical manifestations of a given fungal disease depend, to a great extent, on the immune ability of the host (Casadevall and Pirofski, 2003; Romani, 2011). Indeed, the paradoxical association of fungal diseases with either deficient or hyper-reactive states of immune activation is closely related with the two types of defense mechanisms a host can evolve to increase its fitness when challenged with a pathogen: resistance and tolerance (Schneider and Ayres, 2008). Mechanisms of resistance delineate the host’s ability to limit the fungal growth by directly countering pathogens through recognition and elimination systems. Mechanisms of tolerance, however, regulate the self-harm that can be caused by an overactive immune response and other mechanisms not directly related to immune resistance. Given the different pathological and epidemiological effects these mechanisms may prompt, a further detailed understanding of the wide spectrum of host–pathogen interactions and immune responses will ultimately be paramount for the design of effective vaccination formulations affording comprehensive and durable protection to different fungi (Figure 1). The design of fungal vaccines is however not only constrained by the nature of the target populations, which may be genetically and immunologically different – not necessarily immunocompromised – but also by the dynamics of fungal diversity. Indeed, and even considering the premise that a fungal vaccine would be feasible even in patients with severe immune dysfunction (Spellberg, 2011), no examples can be cited up until today. Attempts at fungal vaccination have been restricted to pre-clinical research essentially because of safety concerns, as complex and ill-defined antigenic mixtures do not cope with present day safety restrictions. However, whole genome sequencing and proteomic approaches have made available most – if not all – fungal proteins, thereby allowing the selection of a discrete number of fungal antigens to test for protection. This has directed interest in subunit antigens, which however lack the natural adjuvant properties of whole-cell or live vaccines, and consequently optimal immunogenicity properties. In addition, the human microbiota and their role in programming human metabolism is currently emerging as a key component required for the definition of immune responses to fungi, in particular adaptive immunity (Littman and Pamer, 2011). Thus, the symbiotic relationship of the microbial species with the host requires a tuned response that prevents host damage while tolerating the presence of potentially beneficial microbes, meaning that the host and the fungus exert control over each other in a way that fungal commensalism ultimately benefits the host (Bonifazi et al., 2009). As a corollary, the shaping of intestinal and lung immune responses by microbiota to achieve protection to vaccines will likely become an area of intense research.
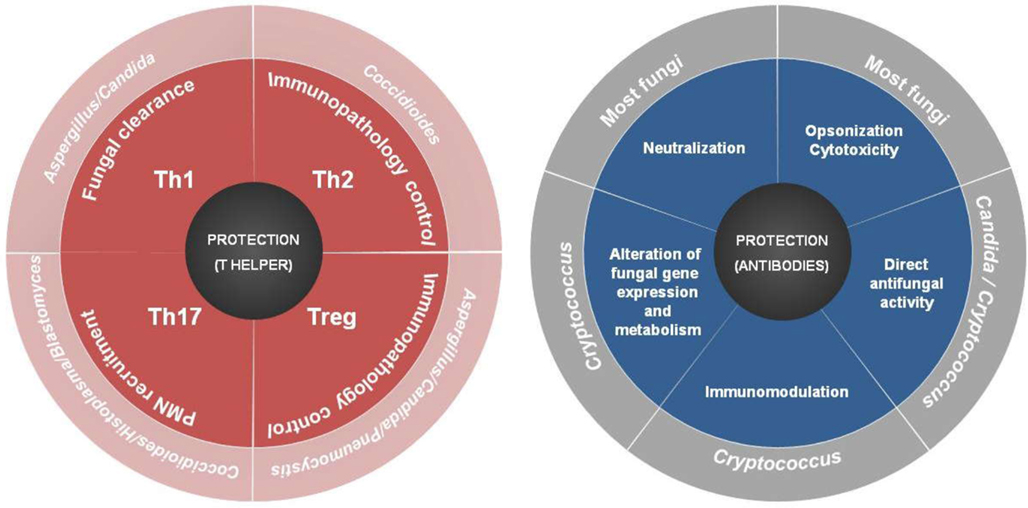
Figure 1. Varieties of T helper (Th) and antibody cell responses to fungi and their correlates of protection.
The growing understanding that fungal pathogens may thrive in regulatory environments has to be integrated within the protective immune responses developed in a context of vaccination. Although protective immunity may be accomplished by means of preventing regulatory T (Treg) cell induction or function – as Treg cells can indeed control the intensity of secondary responses to fungal infections (Cavassani et al., 2006; Deepe and Gibbons, 2008; Loures et al., 2009) – their presence upon secondary antigen exposure may prevent immunopathology in the context of vaccination and favor long-term memory (Romani and Puccetti, 2006; Bozza et al., 2009). This notion is crucially exemplified in infections spawned by the reactivation of latent commensal organisms, in which a vaccine candidate is expected to elicit protective memory responses in a Treg-rich environment – that is, long-lasting sterilizing immunity through the generation of effector T cells is not needed here. This can be achieved by concurrently focusing on effector mechanisms of resistance as well as on manipulation of tolerance to restrain immunopathology. However straightforward this approach may sound, these mechanisms lie on a precarious balance that may differ with each fungal pathogen and even sites of infection. This demands for a fine prediction and definition of fungal antigens and adjuvants that trigger the most appropriate classes of resistance and tolerance mechanisms as well as the selection of sites for vaccination where their contribution to protective memory could be properly and most significantly achieved.
A successful vaccination relies on the eliciting of pathogen-specific immune responses and consequent immunological memory that mediates long-term protection from infection or disease. A plethora of chemical and antigenic formulations has already been considered for active vaccination against all major fungal pathogens in pre-clinical models of infection (Table 1; Cassone, 2008) and it is well accepted nowadays that the immunogenic potential of fungal stimuli critically relies on their innate immune recognition, particularly by pattern recognition receptors (PRRs). The most well-known PRRs for fungi include Toll-like receptors (TLRs), C-type lectin receptors and the nucleotide binding domain leucine-rich repeat containing receptors which detect a vast array of fungal molecules or danger signals (Romani, 2011). In this regard, systems biology analyses of naïve to effector to memory transition has revealed changes in expression of innate immune receptors to be one major early molecular signature upon vaccination (Pulendran et al., 2010).
Given the array of fungal ligands present at the cell surface, as well as those that become available to immune sensing upon processing of the fungus by phagocytic cells, it is now clear that vaccine-induced protection to attenuated fungal strains occurs through distinct PRRs and downstream signaling adapters (Wuthrich et al., 2011; De Luca et al., 2012). For instance, T helper (Th)17-induced acquisition of vaccine immunity to live attenuated strains of B. dermatitidis, H. capsulatum, and C. posadasii was found to require myeloid differentiation primary response gene 88 (MyD88) signaling (Wuthrich et al., 2011), whereas Th1-induced protection to A. fumigatus relied on TIR-domain-containing adapter inducing interferon-β (De Luca et al., 2012). Of interest, vaccination with purified A. fumigatus antigens was found to be dependent on the MyD88 pathway in the presence of the appropriate adjuvant (Carvalho et al., 2012b; De Luca et al., 2012), a finding pointing to the crucial role of adjuvants in promoting T cell differentiation along specific effector pathways. Thus, fungal innate sensing is one critical step in mounting immune responses eventually defining appropriate effector responses to maximize protection (Levitz and Golenbock, 2012). Moreover, given the intricacies of the complex innate immune signaling networks activated in response to fungal antigens (Romani, 2011), the use of individual PRRs or in combinations will have to be weighed in order to achieve the best vaccine-specific responses appropriate for each fungal pathogen.
The thorough dissection of mechanisms regulating the magnitude, quality, and persistence of vaccine-induced humoral and T cell dependent immunity will add to a more rational design of potentially useful vaccines (Pulendran and Ahmed, 2011). Examples of such approaches include the development of a novel vaccine platform consisting of hollow yeast-derived β-glucan particles that combine adjuvancity and high load antigen delivery to induce strong humoral and Th1- and Th17-biased T cell responses (Huang et al., 2010) and the glycoconjugate vaccines which elicit B-cell responses of increased potency by provision of immunogenic epitopes to CD4+ T cells (Torosantucci et al., 2005; Rachini et al., 2007; Xin et al., 2008; Bromuro et al., 2010). T cells are critical for protective immunity, as they monitor host cells for infection and mobilize appropriate effector functions, either by inducing cytokines and effector cytolytic molecules or by attracting professional phagocytes to the site of microbial deposition, where they activate their antimicrobial capacities. Although CD4+ Th1 cells have been historically considered the cornerstone of cell-mediated defense against intracellular fungi, CD8+ T cells have also been found to perform effector functions against these pathogens (Cutler et al., 2007). Indeed, in a mouse model of vaccination against blastomycosis, both the numbers and function of protective antifungal memory CD8+ T cells were maintained even in the absence of CD4+ T cell help (Nanjappa et al., 2012). In any case, Th1-mediated protection has been reported across nearly all clinically relevant fungal infections. For example, crude antigen preparations from A. fumigatus or recombinant fungal antigens alone (Diaz-Arevalo et al., 2011) or in conjunction with CpG oligonucleotides as adjuvants (Cenci et al., 2000; Ito et al., 2006; Bozza et al., 2009; Stuehler et al., 2011), mannosylated cryptococcal antigens (Lam et al., 2005), B. dermatitidis adhesin antigen (Wuthrich et al., 2003), heat shock protein 60 from P. brasiliensis (de Bastos Ascenco Soares et al., 2008) and H. capsulatum (Deepe and Gibbons, 2002) and the multivalent vaccines, comprised of complexes of protein antigens of Coccidioides spp., administered in combinations with adjuvants (Shubitz et al., 2006; Tarcha et al., 2006) have been associated with induction of strong Th1 responses.
The persistence of immunological memory and how it pertains to vaccination strategies is also a question of central importance. Memory T cells are derived from normal T cells that have learned how to overcome a pathogen by “remembering” the strategy used to defeat previous infections (Sallusto et al., 2010). In addition to central memory T cells present in secondary lymphoid organs which scrutinize the presence of remote pathogens via dendritic cells (DCs), effector memory T cells reside in peripheral non-lymphoid tissues such as the skin and mucosa. The latter are heterogeneous in terms of homing receptor expression and effector function and comprise the Th1, Th2, Th17, and Th22 cell subsets, as well as Treg cells and cytotoxic T lymphocytes. Although Th1 and Th17 cells mediate vaccine-induced protection from fungal infection through a variety of antifungal effector mechanisms, Th22 cells are instead required for antifungal resistance at mucosal surfaces (De Luca et al., 2010). Memory CD8+ cytotoxic T cells are also induced in fungal infections (Nanjappa et al., 2012) and exhibit a pleiotropic activity by mediating protection via production of IFN-γ and cytolytic activity against fungus-laden cells or the fungus itself (Carvalho et al., 2012b; De Luca et al., 2012). As such, CD8+ T cells, especially if long-lasting, are regarded as ideal candidates for expansion at mucosal surfaces by vaccination strategies. The recent evidence proposing a role for metabolism (Pearce et al., 2009) and bioenergetic stability (van der Windt et al., 2012) in harnessing T cell memory opens up new perspectives on how epigenetic and environmental mechanisms modulate memory differentiation and quality, thus opening new avenues for vaccine development. Finally, additional subsets of T cells may also become important targets for new vaccines, such as the newly described invariant natural killer T cells that activate antifungal responses through the recognition of fungal cell wall β-1,3 glucans (Cohen et al., 2011).
Fungal vaccines are active immunotherapies in the sense they boost the immune system to specifically attack fungi, honing in on one or more specific fungal antigens (Carvalho et al., 2012a). Alternatively, passive immunotherapy strategies are comprised of laboratory-synthesized antibodies or other immune system components that are administered to patients. Thus, passive immunotherapies do not stimulate the immune system to “actively” respond to infection in the way a vaccine does. In this regard, a number of monoclonal human recombinant antibodies and their fragments have already been tested in experimental fungal infection (Table 1). Antifungal vaccines are known to exploit the redundancy in the immune system to afford protection through a multiplicity of mechanisms (Cutler et al., 2007; Cassone, 2008). Indeed, antibody responses are induced by most antifungal vaccines and antibody titer threshold may therefore predict vaccine efficacy and may serve as a vaccine surrogate marker even when the mechanism of protection is cell-mediated (Spellberg et al., 2008). Indeed, and even though protection against intracellular pathogens might be prevalently provided by CD4+ and CD8+ T cells, antibodies are now known to participate in all aspects of the immune response, globally contributing to the optimal function of T cell-mediated immunity (Casadevall and Pirofski, 2012). This also suggests that administering antibodies together with a vaccine may be potentially exploited to further enhance or modulate the immune response. Given that passive antibody administration has been deemed effective against fungal infection, it is now accepted that vaccine-mediated protection not only relies on the production and maintenance of specific antibodies, but also on their direct activity (Cutler et al., 2007; Cassone, 2008). This is the case of anti-β-glucan antibodies generated by immunization with laminarin, a β-glucan from algae, conjugated with a genetically detoxified diphtheria toxin (Bromuro et al., 2010) or antibodies generated through idiotypic vaccination (Magliani et al., 2005) that proved to be protective in passive vaccination experiments in different fungal infection settings by acting directly on fungal cells. Because of quantity restrictions, high cost, and the limited effectiveness inherent to a pure antibody approach, it is difficult to envisage antibody therapy against fungal infections in a near future. Indeed, the development of efungumab (Mycograb), a monoclonal recombinant antibody fragment against fungal HSP90 (Matthews et al., 2003) has recently been discontinued. This may have been related with concerns regarding specificity, affinity, and even isotype. For instance, different immunoglobulin G (IgG) subclasses with identical variable regions but different capacities to bind Fc receptors displayed distinct efficacy in terms of protection from cryptococcosis (Beenhouwer et al., 2007). In addition, given that immunocompromised patients may lack efficient effector functions, the use of antibodies that inhibit fungal growth or viability should be favored in these patients.
The deciphering of the complexity of immune responses to vaccines demands for the integration of advanced immunology approaches, systems biology, immunogenetic profiling, and bioinformatics in the areas of pathogen biology, host biology, and the interaction between the two. Vaccinomics is an emergent term in the field of vaccinology that encompasses the use of immunogenetics to the appreciation of mechanisms of heterogeneity in immune responses to vaccines (Poland and Oberg, 2010). A number of genetic variants in immune genes has already been disclosed as major determinants of the immune response to fungi (Carvalho et al., 2010) and are regarded as promising targets to exploit toward improved diagnosis and therapy of fungal diseases, particularly in immunocompromised patients (Cunha and Carvalho, 2012). A systematic evaluation of the functional impact of genetic variability in the immune system will pave the way to the discovery and interpretation of immunogenetic signatures and immune profiles that may be used to discriminate response efficiencies to antifungal vaccines. The recent finding that genetic deficiency of TLR3 was associated with susceptibility to invasive aspergillosis and concomitant failure to activate memory protective CD8+ T cells in allogeneic stem cell transplanted patients is one first example (Carvalho et al., 2012b). Given the high degree of complexity in human immune responses, overcoming the many challenges currently restraining accurate prediction of vaccination efficiencies has been recently proposed to rely on five state-of-the-art approaches (Kennedy and Poland, 2011). By using whole genome immunogenetics, next generation sequencing, cutting-edge “omics” techniques, advanced bioinformatics, and systems biology applied to immune profiling and vaccine responses, it may be possible to identify the best predictors of vaccine efficacy or adverse responses – predictive vaccinology – in each target population, thereby improving the management of these severe, often fatal diseases.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Cristina Massi Benedetti for editorial assistance. The studies were supported by the Specific Targeted Research Project “ALLFUN” (FP7-HEALTH-2009-260338) and the Fondazione per la Ricerca sulla Fibrosi Cistica (FFC#21/2010, with the contribution of Francesca Guadagnin, Coca Cola Light® Tribute to Fashion and Delegazione FFC di Belluno). Agostinho Carvalho and Cristina Cunha were financially supported by fellowships from Fundação para a Ciência e Tecnologia, Portugal (contracts SFRH/BPD/46292/2008 and SFRH/BD/65962/2009, respectively).
Beenhouwer, D. O., Yoo, E. M., Lai, C. W., Rocha, M. A., and Morrison, S. L. (2007). Human immunoglobulin G2 (IgG2) and IgG4, but not IgG1 or IgG3, protect mice against Cryptococcus neoformans infection. Infect. Immun. 75, 1424–1435.
Bonifazi, P., Zelante, T., D’Angelo, C., De Luca, A., Moretti, S., Bozza, S., Perruccio, K., Iannitti, R. G., Giovannini, G., Volpi, C., Fallarino, F., Puccetti, P., and Romani, L. (2009). Balancing inflammation and tolerance in vivo through dendritic cells by the commensal Candida albicans. Mucosal Immunol. 2, 362–374.
Bozza, S., Clavaud, C., Giovannini, G., Fontaine, T., Beauvais, A., Sarfati, J., D’Angelo, C., Perruccio, K., Bonifazi, P., Zagarella, S., Moretti, S., Bistoni, F., Latge, J. P., and Romani, L. (2009). Immune sensing of Aspergillus fumigatus proteins, glycolipids, and polysaccharides and the impact on Th immunity and vaccination. J. Immunol. 183, 2407–2414.
Bromuro, C., Romano, M., Chiani, P., Berti, F., Tontini, M., Proietti, D., Mori, E., Torosantucci, A., Costantino, P., Rappuoli, R., and Cassone, A. (2010). Beta-glucan-CRM197 conjugates as candidates antifungal vaccines. Vaccine 28, 2615–2623.
Carvalho, A., Cunha, C., Bistoni, F., and Romani, L. (2012a). Immunotherapy of aspergillosis. Clin. Microbiol. Infect. 18, 120–125.
Carvalho, A., De Luca, A., Bozza, S., Cunha, C., D’Angelo, C., Moretti, S., Perruccio, K., Iannitti, R. G., Fallarino, F., Pierini, A., Latge, J. P., Velardi, A., Aversa, F., and Romani, L. (2012b). TLR3 essentially promotes protective class I-restricted memory CD8 T-cell responses to Aspergillus fumigatus in hematopoietic transplanted patients. Blood 119, 967–977.
Carvalho, A., Cunha, C., Pasqualotto, A. C., Pitzurra, L., Denning, D. W., and Romani, L. (2010). Genetic variability of innate immunity impacts human susceptibility to fungal diseases. Int. J. Infect. Dis. 14, e460–e468.
Casadevall, A., and Pirofski, L. A. (2003). The damage-response framework of microbial pathogenesis. Nat. Rev. Microbiol. 1, 17–24.
Casadevall, A., and Pirofski, L. A. (2012). A new synthesis for antibody-mediated immunity. Nat. Immunol. 13, 21–28.
Cassone, A. (2008). Fungal vaccines: real progress from real challenges. Lancet Infect. Dis. 8, 114–124.
Cassone, A., and Rappuoli, R. (2010). Universal vaccines: shifting to one for many. MBio 1, e00042–10.
Cavassani, K. A., Campanelli, A. P., Moreira, A. P., Vancim, J. O., Vitali, L. H., Mamede, R. C., Martinez, R., and Silva, J. S. (2006). Systemic and local characterization of regulatory T cells in a chronic fungal infection in humans. J. Immunol. 177, 5811–5818.
Cenci, E., Mencacci, A., Bacci, A., Bistoni, F., Kurup, V. P., and Romani, L. (2000). T cell vaccination in mice with invasive pulmonary aspergillosis. J. Immunol. 165, 381–388.
Cohen, N. R., Tatituri, R. V., Rivera, A., Watts, G. F., Kim, E. Y., Chiba, A., Fuchs, B. B., Mylonakis, E., Besra, G. S., Levitz, S. M., Brigl, M., and Brenner, M. B. (2011). Innate recognition of cell wall beta-glucans drives invariant natural killer T cell responses against fungi. Cell Host Microbe 10, 437–450.
Cunha, C., and Carvalho, A. (2012). Host genetics and invasive fungal diseases: towards improved diagnosis and therapy? Expert Rev. Anti. Infect. Ther. 10, 257–259.
Cutler, J. E., Deepe, G. S. Jr., and Klein, B. S. (2007). Advances in combating fungal diseases: vaccines on the threshold. Nat. Rev. Microbiol. 5, 13–28.
de Bastos Ascenco Soares, R., Gomez, F. J., De Almeida Soares, C. M., and Deepe, G. S. Jr. (2008). Vaccination with heat shock protein 60 induces a protective immune response against experimental Paracoccidioides brasiliensis pulmonary infection. Infect. Immun. 76, 4214–4221.
De Luca, A., Iannitti, R. G., Bozza, S., Beau, R., Casagrande, A., D’Angelo, C., Moretti, S., Cunha, C., Giovannini, G., Massi-Benedetti, C., Carvalho, A., Boon, L., Latge, J. P., and Romani, L. (2012). CD4 T cell vaccination overcomes defective cross-presentation of fungal antigens in murine chronic granulomatous disease. J. Clin. Invest. doi:10.1172/JCI60862. [Epub ahead of print].
De Luca, A., Zelante, T., D’Angelo, C., Zagarella, S., Fallarino, F., Spreca, A., Iannitti, R. G., Bonifazi, P., Renauld, J. C., Bistoni, F., Puccetti, P., and Romani, L. (2010). IL-22 defines a novel immune pathway of antifungal resistance. Mucosal Immunol. 3, 361–373.
Deepe, G. S. Jr., and Gibbons, R. S. (2002). Cellular and molecular regulation of vaccination with heat shock protein 60 from Histoplasma capsulatum. Infect. Immun. 70, 3759–3767.
Deepe, G. S. Jr., and Gibbons, R. S. (2008). TNF-alpha antagonism generates a population of antigen-specific CD4+ CD25+ T cells that inhibit protective immunity in murine histoplasmosis. J. Immunol. 180, 1088–1097.
Diaz-Arevalo, D., Bagramyan, K., Hong, T. B., Ito, J. I., and Kalkum, M. (2011). CD4+ T cells mediate the protective effect of the recombinant Asp f3-based anti-aspergillosis vaccine. Infect. Immun. 79, 2257–2266.
Huang, H., Ostroff, G. R., Lee, C. K., Specht, C. A., and Levitz, S. M. (2010). Robust stimulation of humoral and cellular immune responses following vaccination with antigen-loaded beta-glucan particles. MBio 1, e00164–10.
Ito, J. I., Lyons, J. M., Hong, T. B., Tamae, D., Liu, Y. K., Wilczynski, S. P., and Kalkum, M. (2006). Vaccinations with recombinant variants of Aspergillus fumigatus allergen Asp f 3 protect mice against invasive aspergillosis. Infect. Immun. 74, 5075–5084.
Kennedy, R. B., and Poland, G. A. (2011). The top five “game changers” in vaccinology: toward rational and directed vaccine development. OMICS 15, 533–537.
Klugman, K. P., Madhi, S. A., Huebner, R. E., Kohberger, R., Mbelle, N., and Pierce, N. (2003). A trial of a 9-valent pneumococcal conjugate vaccine in children with and those without HIV infection. N. Engl. J. Med. 349, 1341–1348.
Lam, J. S., Mansour, M. K., Specht, C. A., and Levitz, S. M. (2005). A model vaccine exploiting fungal mannosylation to increase antigen immunogenicity. J. Immunol. 175, 7496–7503.
Levitz, S. M., and Golenbock, D. T. (2012). Beyond empiricism: informing vaccine development through innate immunity research. Cell 148, 1284–1292.
Littman, D. R., and Pamer, E. G. (2011). Role of the commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe 10, 311–323.
Loures, F. V., Pina, A., Felonato, M., and Calich, V. L. (2009). TLR2 is a negative regulator of Th17 cells and tissue pathology in a pulmonary model of fungal infection. J. Immunol. 183, 1279–1290.
Magliani, W., Conti, S., Frazzi, R., Ravanetti, L., Maffei, D. L., and Polonelli, L. (2005). Protective antifungal yeast killer toxin-like antibodies. Curr. Mol. Med. 5, 443–452.
Matthews, R. C., Rigg, G., Hodgetts, S., Carter, T., Chapman, C., Gregory, C., Illidge, C., and Burnie, J. (2003). Preclinical assessment of the efficacy of mycograb, a human recombinant antibody against fungal HSP90. Antimicrob. Agents Chemother. 47, 2208–2216.
Nanjappa, S. G., Heninger, E., Wuthrich, M., Sullivan, T., and Klein, B. (2012). Protective antifungal memory CD8+ T cells are maintained in the absence of CD4+ T cell help and cognate antigen in mice. J. Clin. Invest. 122, 987–999.
Pearce, E. L., Walsh, M. C., Cejas, P. J., Harms, G. M., Shen, H., Wang, L. S., Jones, R. G., and Choi, Y. (2009). Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature 460, 103–107.
Poland, G. A., and Oberg, A. L. (2010). Vaccinomics and bioinformatics: accelerants for the next golden age of vaccinology. Vaccine 28, 3509–3510.
Pulendran, B., and Ahmed, R. (2011). Immunological mechanisms of vaccination. Nat. Immunol. 12, 509–517.
Rachini, A., Pietrella, D., Lupo, P., Torosantucci, A., Chiani, P., Bromuro, C., Proietti, C., Bistoni, F., Cassone, A., and Vecchiarelli, A. (2007). An anti-beta-glucan monoclonal antibody inhibits growth and capsule formation of Cryptococcus neoformans in vitro and exerts therapeutic, anticryptococcal activity in vivo. Infect. Immun. 75, 5085–5094.
Romani, L., and Puccetti, P. (2006). Protective tolerance to fungi: the role of IL-10 and tryptophan catabolism. Trends Microbiol. 14, 183–189.
Sallusto, F., Lanzavecchia, A., Araki, K., and Ahmed, R. (2010). From vaccines to memory and back. Immunity 33, 451–463.
Schneider, D. S., and Ayres, J. S. (2008). Two ways to survive infection: what resistance and tolerance can teach us about treating infectious diseases. Nat. Rev. Immunol. 8, 889–895.
Shubitz, L. F., Yu, J. J., Hung, C. Y., Kirkland, T. N., Peng, T., Perrill, R., Simons, J., Xue, J., Herr, R. A., Cole, G. T., and Galgiani, J. N. (2006). Improved protection of mice against lethal respiratory infection with Coccidioides posadasii using two recombinant antigens expressed as a single protein. Vaccine 24, 5904–5911.
Spellberg, B., Ibrahim, A. S., Lin, L., Avanesian, V., Fu, Y., Lipke, P., Otoo, H., Ho, T., and Edwards, J. E. Jr. (2008). Antibody titer threshold predicts anti-candidal vaccine efficacy even though the mechanism of protection is induction of cell-mediated immunity. J. Infect. Dis. 197, 967–971.
Stuehler, C., Khanna, N., Bozza, S., Zelante, T., Moretti, S., Kruhm, M., Lurati, S., Conrad, B., Worschech, E., Stevanovic, S., Krappmann, S., Einsele, H., Latge, J. P., Loeffler, J., Romani, L., and Topp, M. S. (2011). Cross-protective TH1 immunity against Aspergillus fumigatus and Candida albicans. Blood 117, 5881–5891.
Tarcha, E. J., Basrur, V., Hung, C. Y., Gardner, M. J., and Cole, G. T. (2006). A recombinant aspartyl protease of Coccidioides posadasii induces protection against pulmonary coccidioidomycosis in mice. Infect. Immun. 74, 516–527.
Torosantucci, A., Bromuro, C., Chiani, P., De Bernardis, F., Berti, F., Galli, C., Norelli, F., Bellucci, C., Polonelli, L., Costantino, P., Rappuoli, R., and Cassone, A. (2005). A novel glyco-conjugate vaccine against fungal pathogens. J. Exp. Med. 202, 597–606.
van der Windt, G. J., Everts, B., Chang, C. H., Curtis, J. D., Freitas, T. C., Amiel, E., Pearce, E. J., and Pearce, E. L. (2012). Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity 36, 68–78.
Wuthrich, M., Filutowicz, H. I., Warner, T., Deepe, G. S. Jr., and Klein, B. S. (2003). Vaccine immunity to pathogenic fungi overcomes the requirement for CD4 help in exogenous antigen presentation to CD8+ T cells: implications for vaccine development in immune-deficient hosts. J. Exp. Med. 197, 1405–1416.
Wuthrich, M., Gern, B., Hung, C. Y., Ersland, K., Rocco, N., Pick-Jacobs, J., Galles, K., Filutowicz, H., Warner, T., Evans, M., Cole, G., and Klein, B. (2011). Vaccine-induced protection against 3 systemic mycoses endemic to North America requires Th17 cells in mice. J. Clin. Invest. 121, 554–568.
Keywords: fungi, immune responses, vaccines, immunotherapy, vaccinomics
Citation: Carvalho A, Cunha C, Iannitti RG, Casagrande A, Bistoni F, Aversa F and Romani L (2012) Host defense pathways against fungi: the basis for vaccines and immunotherapy. Front. Microbio. 3:176. doi: 10.3389/fmicb.2012.00176
Received: 03 April 2012; Accepted: 21 April 2012;
Published online: 10 May 2012.
Edited by:
Carlos Pelleschi Taborda, University of São Paulo, BrazilReviewed by:
Carlos Pelleschi Taborda, University of São Paulo, BrazilCopyright: © 2012 Carvalho, Cunha, Iannitti, Casagrande, Bistoni, Aversa and Romani. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Agostinho Carvalho, Department of Experimental Medicine and Biochemical Sciences, University of Perugia, Via del Giochetto, 06122 Perugia, Italy. e-mail:YWFjYXJ2YWxobzIwMDhAZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.