
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 10 April 2012
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 3 - 2012 | https://doi.org/10.3389/fmicb.2012.00139
This article is part of the Research Topic Role and prevalence of antibiosis and the related resistance genes in the environment View all 12 articles
Elevated levels of antibiotic resistance genes (ARGs) in soil and water have been linked to livestock farms and in some cases feed antibiotics may select for antibiotic resistant gut microbiota. The purpose of this study was to examine the establishment of ARGs in the feces of calves receiving milk replacer containing no antibiotics versus subtherapeutic or therapeutic doses of tetracycline and neomycin. The effect of antibiotics on calf health was also of interest. Twenty-eight male and female dairy calves were assigned to one of the three antibiotic treatment groups at birth and fecal samples were collected at weeks 6, 7 (prior to weaning), and 12 (5 weeks after weaning). ARGs corresponding to the tetracycline (tetC, tetG, tetO, tetW, and tetX), macrolide (ermB, ermF), and sulfonamide (sul1, sul2) classes of antibiotics along with the class I integron gene, intI1, were monitored by quantitative polymerase chain reaction as potential indicators of direct selection, co-selection, or horizontal gene transfer of ARGs. Surprisingly, there was no significant effect of antibiotic treatment on the absolute abundance (gene copies per gram wet manure) of any of the ARGs except ermF, which was lower in the antibiotic-treated calf manure, presumably because a significant portion of host bacterial cells carrying ermF were not resistant to tetracycline or neomycin. However, relative abundance (gene copies normalized to 16S rRNA genes) of tetO was higher in calves fed the highest dose of antibiotic than in the other treatments. All genes, except tetC and intI1, were detectable in feces from 6 weeks onward, and tetW and tetG significantly increased (P < 0.10), even in control calves. Overall, the results provide new insight into the colonization of calf gut flora with ARGs in the early weeks. Although feed antibiotics exerted little effect on the ARGs monitored in this study, the fact that they also provided no health benefit suggests that the greater than conventional nutritional intake applied in this study overrides previously reported health benefits of antibiotics. The results suggest potential benefit of broader management strategies, and that cost and risk may be avoided by minimizing incorporation of antibiotics in milk replacer.
The extensive use of antibiotics in animal agriculture and the development of antibiotic resistant bacteria have been cause for increasing concern. Livestock operations are often cited as a reservoir for resistant bacteria and antibiotic resistance genes (ARGs; Chee-Sanford et al., 2001; Smith et al., 2004; Sawant et al., 2007; McKinney et al., 2010); and antibiotic use has implications for both animal and human health. Antibiotics are fed to livestock at subtherapeutic levels for both growth promotion and disease prevention, because their use can reduce morbidity and mortality (e.g., Wileman et al., 2009). However, the unintentional selection of bacteria that are resistant to antibiotics could have important human health consequences, with several studies noting identical resistance elements in both humans and food animals (e.g., Boerlin et al., 2001; Lauderdale et al., 2002; Ho et al., 2010).
Based on concerns about human health effects, subtherapeutic use of antibiotics was banned in the European Union in the late 1990s (Boerlin et al., 2001). A major trigger for earlier bans in Denmark and other Nordic countries, was linkage of avoparcin use in broiler chickens and swine to vancomycin resistant Enterococcal infections (VRE) in humans (Bager et al., 1997; Bates, 1997). In the United States (U.S.), the Food and Drug Administration (FDA) recently issued new rules limiting antibiotic use in milk replacers for calves (21CFR § 520.1484; 21CFR § 520.1660d). The precise effects of antibiotics in milk replacer is currently unknown, and the general consequences of antibiotic amendment in livestock feed are complex (Marshall and Levy, 2011). In the 3-years following the European ban, a slight decrease in antibiotic resistance was observed in rectal swabs collected from food animals at slaughter, and a documentable decrease in acquired fecal enterococci resistance in humans was also observed (Casewell et al., 2003). Following the Taiwan avoparcin ban in chickens in 2000, the incidence of VRE decreased significantly (Lauderdale et al., 2007). However, VRE did still persist following the ban and resistance to other classes of antibiotics, including tetracyclines and macrolides, stayed the same or even increased. In support of this observation, vancomycin and macrolide ARGs have been noted to be present together on the same genetic element from isolates obtained from Danish pig herds (Aarestrup, 2000). Thus, it is critical to consider co-selection of resistance across antibiotic classes in evaluating impacts of feed antibiotics.
Effects of subtherapeutic antibiotic use on animal health have been difficult to assess. Based on medicated feed prescription rates, decreased rates of respiratory infections were observed in Swiss piglets and fattening pigs in the 3-years following the antibiotic ban relative to the 3-years prior, but gastrointestinal tract infections appeared to go up (Arnold et al., 2004). A marked increase in necrotic enteritis was noted in broiler chickens in Norway following the avoparcin ban, but this seemed to be ameliorated by the subsequent approval of narisin as a feed additive (Grave et al., 2004). Also, while therapeutic antibiotic use rates did indeed decrease in Switzerland (Arnold et al., 2004), Norway, and Sweden (Grave et al., 2006) following the E.U. ban, therapeutic use increased in weaning piglets in Denmark (Grave et al., 2006). Generally, a comprehensive examination of banning subtherapeutic antibiotic use in piglets, beef cattle, and poultry in Sweden, indicated that although temporary health problems and increases in antibiotic uses were noted, strategic management practices make possible competitive animal production and reduced antibiotic use (Wierup, 2001). Few controlled studies have examined the effects of antibiotic use on health (Berge et al., 2009a) or antibiotic resistance (Langford et al., 2003; Berge et al., 2006a; Kaneene et al., 2008; Pereira et al., 2011) in pre-weaned calves, and none have incorporated culture-independent techniques to quantify the establishment of ARGs in calf manure.
The objective of this study was to compare the establishment of a cross-section of ARGs in the feces of calves receiving milk replacer dosed with subtherapeutic and therapeutic antibiotics and their effect on overall health. In particular, the effect of oxytetracycline and neomycin in the milk replacer and weaning to starter grain containing monensin were of interest as typical practices in the U.S. The inclusion of oxytetracycline and neomycin in milk replacer for young calves is one of the most common uses of subtherapeutic antibiotics in the U.S. dairy industry. Neomycin is absorbed only very slightly by the animal (Aschbacher and Feil, 1994) and thus remains in the digestive tract where it controls growth of pathogenic organisms. Oxytetracycline, in contrast, is highly absorbed, so is considered useful for prevention and treatment of respiratory illnesses. One intentional contrast to conventional dairy practice incorporated in this study was feeding calves >2 times more protein than in traditional feeding programs, and >1.5 times more fat. This is because while conventional U.S. milk replacer feeding rates generally provide sufficient nutrients only for maintenance and modest growth [National Research Council (NRC), 2001], which could bias the evaluation of the health benefits of antibiotics.
In this study, 28 male and female dairy calves were assigned to control, subtherapeutic, or therapeutic antibiotic treatment groups at birth and fecal samples were collected at weeks 6, 7 (prior to weaning), and 12 (5 weeks after weaning). The abundance of a representative range of tetracycline (tetO, tetW, tetC, tetG, and tetX), sulfonamide (sul1 and sul2), and macrolide (ermB and ermF) ARGs as well as a class 1 transferable genetic element (intI1) was quantified in the calf feces as a culture-independent assessment of resistance. The specific ARGs examined were selected as indicators of direct antibiotic selection (i.e., tet ARGs) as well as potential co-selection of resistance to clinically relevant antibiotics (i.e., sul and erm ARGs) and overall horizontal gene transfer potential (i.e., intI1). Quantitative polymerase chain reaction (qPCR) was incorporated to provide a culture-independent assessment of the overall resistance potential incurred by these ARGs. Identifying factors that affect fecal excretion of ARGs by dairy calves will help assess the contribution of dairy farms to the overall loading of anthropogenic sources of antibiotic resistance to the environment and provide insight into effective management strategies for limiting the spread of antibiotic resistance.
Forty-one newborn calves were blocked according to breed (Holstein or crossbred), birth order (two blocks), and gender. Crossbred calves were either Holstein- or Jersey-sired and out of Jersey × Holstein, Jersey × Holstein × Brown Swiss, or Jersey × Holstein × Swedish Red dams. Within each block, calves were randomly assigned to treatments at birth, resulting in the assignments summarized in Table A1 in Appendix. Treatments were control (containing no antibiotics in the milk replacer), subtherapeutic (neomycin sulfate and oxytetracycline hydrochloride each fed at 10 mg/calf/day provided from day 1 until weaning), and therapeutic (no antibiotics in the milk replacer until day 36, then neomycin sulfate and oxytetracycline hydrochloride each fed at 1000 mg/calf/day for 14 day). All protocols and procedures were approved by the Virginia Tech Institutional Animal Care and Use Committee.
Within 2 h of birth, calves were moved from the calving pen to individual stalls in an enclosed barn. Calf navels were dipped in a 7% tincture of iodine, and calves were vaccinated intranasally with Nasalgen (Merck Animal Health, Millsboro, DE). Birth date, weight, and identity of the dam were recorded. Calves received 1.89 l of high quality thawed colostrum as soon as possible after birth, usually within 2 h. A second feeding of 1.89 l of colostrum was administered 6 h after the first feeding and calf navels were dipped a second time.
Calves were enrolled and treatments were imposed at 1 day of age. Calves were fed milk replacer (Cow’s Match, Land O’Lakes Animal Products Co., Arden Hills, MN, USA) twice daily, at 6:00 AM and 6:00 PM. Milk replacer contained 28% crude protein and 20% fat, with whey protein as the protein source and human grade edible lard as the fat source. Milk replacer powder was reconstituted with water to contain 17.6% solids and fed individually to calves via nipple buckets. Amount of liquid fed provided 1.1–0.68 kg of dry matter per calf, depending upon birth weight, resulting in feeding 2.1–3.4 times more protein and 1.5–2.4 times more fat than traditional feeding programs. This feeding rate has been termed “intensive feeding” is higher than conventional calf feeding programs, which deliver only 0.454 kg of milk replacer dry matter containing 20% fat and 20% protein. The intensive feeding program thus more closely approaches amounts calves would consume if allowed free choice to liquid diets. Antibiotic treatments were added directly to the nipple buckets, which were used exclusively for a specific treatment to avoid cross-contamination. Calves were supervised during feeding, and 15 min was allowed for consumption. After feeding, all equipment was thoroughly cleaned. Calves were fed starter grain (Intensity −22% crude protein, 10.13 mg/kg of monensin, Southern States Cooperative, Richmond, VA, USA) beginning at 1 day of age.
Calves were housed at the Virginia Tech Dairy Cattle Center in individual fiberglass or plastic hutches (1.83 m × 1.37 m) with metal hog panels to create a fenced area (∼2.5 m2). Bull calves were housed in hutches until 39 day, when they were moved into metabolism crates to facilitate total feces and urine collection (described below). After the total collection period was complete, bull calves were moved back to their original hutches until weaned.
Weaning was initiated at day 50 by reducing milk replacer offered by 50% and feeding only once daily at 6:00 AM. Calves were initially fed 0.22 kg of starter grain, and the amount offered increased in 0.22 kg increments as consumption increased. When starter grain consumption reached 1.81 kg daily, calves were fully weaned (59 ± 2 day of age). Post-weaning, calves were housed in the calf hutches for 7 days then were housed in small groups in a 3-sided barn with access to a drylot. Weaned calves were group-fed first cutting alfalfa hay and 22% crude protein starter grain ad lib. Starter grain contained monensin at a concentration of 10.13 mg/kg.
Following the 6:00 AM feeding, body temperatures, fecal scores, and respiratory scores (Larson et al., 1977) were recorded daily. Calves were weighed at birth and at weaning to calculate average daily gain.
Bull calves were housed in metabolism crates (1.2 m × 0.6 m) to facilitate total collection of feces beginning at 39 day of age, for 3 day of adaptation to the stall followed by 7 day of total collection. Trays were placed under the metabolism crates to collect excreted feces. Feces were collected daily, weighed, and subsampled. Because heifer calves are anatomically ill-suited for total collection of feces uncontaminated by urine, grab samples of feces were collected from heifer calves at 6:00 AM, 2:00 PM, 8:00 PM, and 12:00 AM for 7 day beginning at 6 weeks of age. Fecal samples were collected from the gravel surface if a fresh sample was available at collection time. If no fresh sample was available, calves were rectally stimulated to produce a fresh sample. After weaning, samples were collected weekly from calves (now group-housed) via rectal palpation.
A total of 13 calves were removed from the study (3, 7, and 3 were on control, subtherapeutic, and therapeutic treatments, respectively), because of health problems requiring clinical antibiotic treatment.
Feces samples were frozen immediately at −20°C after collection. Samples collected from heifer calves were thawed and pooled by date collected on an equal wet weight basis. Pooling was not necessary for bull calf feces samples because one sample was collected of each day’s total excretion. DNA was extracted from 500 mg of wet feces per calf from week 6, 7, and 12. The FastDNA® Spin Kit for soil was applied across all samples. The FastPrep® instrument (MP Biomedicals, Santa Ana, CA, USA) was employed with homogenization for 40 s at a speed of 6.0. The DNA was suspended in purified water and aliquots were stored in 0.5 ml cryovials at −80°C.
Previously reported qPCR protocols were implemented to quantify the following ARGs: ermB, ermF (Chen et al., 2007), sul1, sul2 (Pei et al., 2006), tetC, tetG (Aminov et al., 2002), tetO, tetW (Aminov et al., 2001), and tetX (Ng et al., 2001) as well as intI1 (Hardwick et al., 2008) and 16S rRNA (Suzuki et al., 2000) genes. Primer sequences are provided in Table A2 in Appendix. The 9-μl per reaction master mix consisted of 2.8 μl of sterile water, 0.6 μl (5 M) of forward primer, 0.6 μl (5 M) of reverse primer, and 5 μl of SsoFastEvaGreen (Bio-Rad, Hercules, CA, USA) with 1 μl of diluted DNA extract added. A dilution series qPCR was conducted for each gene and each sample matrix, through which it was determined that a 1:70 dilution provided the most consistent results across all samples. All qPCR were conducted in triplicate on a CFX-96 (Bio-Rad). Calibration curves were constructed from serial dilutions of positive controls over seven orders of magnitude. Positive controls were obtained from cloned PCR products of target genes verified by DNA sequencing. Gene copies for any samples with numbers of gene copies below the limit of quantification were recorded as 0 for the purposes of statistical analysis.
Fecal scores, respiratory scores, and body temperatures were analyzed using the Mixed procedure of SAS (9.2, 2008) with the model:
where:
μ = overall mean;
D = effect of day (i = 1–65);
T = effect of treatment (j = control, subtherapeutic, therapeutic);
B = effect of breed (k = crossbred, Holstein);
G = effect of gender (l = bull, heifer); and
e = error (interaction of day, treatment, breed, and gender).
Average daily gains were analyzed using the Mixed procedure of SAS (9.2, 2008) with the model:
where:
μ = overall mean;
T = effect of treatment (i = control, subtherapeutic, therapeutic);
B = effect of breed (j = crossbred, Holstein);
G = effect of gender (k = bull, heifer); and
e = error (interaction of treatment, breed, and gender).
Because they were not normally distributed, ARG data were log-transformed prior to statistical analysis to determine significance of main effects and interactions. Log-transformed values were used to calculate LSM. All gene data were analyzed using the GLIMMIX procedure of SAS (9.2, 2008) with the model:
where:
μ = overall mean;
T = effect of treatment (i = control, subtherapeutic, therapeutic);
B = effect of breed (j = crossbred, Holstein);
G = effect of gender (k = bull, heifer);
W = effect of week (l = 6, 7, 12); and
e = error (interaction of treatment, breed, gender, and week).
After the log-transformed data set was used to determine significance, contrasts were performed to separate means for significant effects and interactions. All data are reported as LSM ± SE. Significance was determined at P < 0.10.
No effects of antibiotic treatment, breed or gender were observed on average daily gain (1.30 ± 0.13 kg/day), fecal scores (2.5 ± 0.17), respiratory scores (1.2 ± 0.04), or body temperature (102.2° ± 0.95).
Two of the analyzed genes (tetC and intI1) were not detected in fecal samples of any calves. Four genes encoding resistance to tetracyclines were detected in feces (tetG, tetO, tetW, tetX; Figure 1) as were two genes encoding resistance to sulfonamides (sul1, sul2) and two genes encoding resistance to erythromycin (ermB, ermF). All calves harbored at least one of the tet genes in the feces by week 12, and multiple ARGs related to tetracyclines were present in nearly all of the fecal samples analyzed. ARGs corresponding to antibiotics not fed to calves were less abundant across all three treatments (Figure 2). About one third of the calves did not have sul1 present in their feces at any individual sampling point.
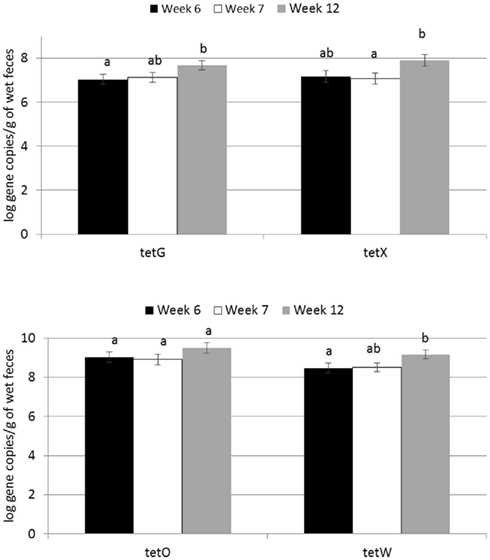
Figure 1. Effect of time on abundance of tetracycline ARGs in calf feces. Bars represent the pooled averages across calves fed medicated or non-medicated milk replacers. Letters above the bars indicate significantly different LSM groupings (P < 0.10).
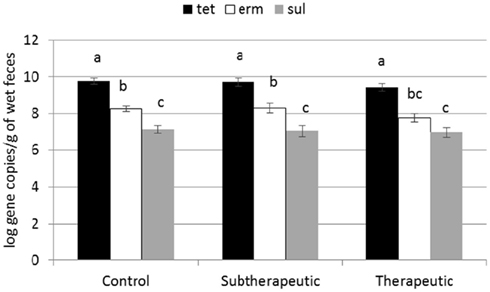
Figure 2. Effect of antibiotics in milk replacer on abundance of tetracycline (tet), macrolide (erm), and sulfonamide (sul) classes of ARGs. Bars represent the pooled averages of the total ARGs quantified within each class. Letters above the bars indicate significantly different LSM groupings (P < 0.10).
Feeding milk replacer with subtherapeutic or therapeutic doses of neomycin and oxytetracycline had no effect on the absolute abundance (log copy per gram wet weight) of tetG, tetO, tetW, tetX, sul1, sul2, or ermB in manure, but calves fed medicated milk replacers yielded reduced abundance of ermF as compared to control calves (Table 1). The relative abundance (ARG per 16S rRNA; normalized) of ermF was not different between antibiotic fed and non-antibiotic control groups (Table 2). Calves fed the higher (therapeutic) dose of antibiotics resulted in increased relative abundance of tetO as compared to subtherapeutic and control calves (Table 2).
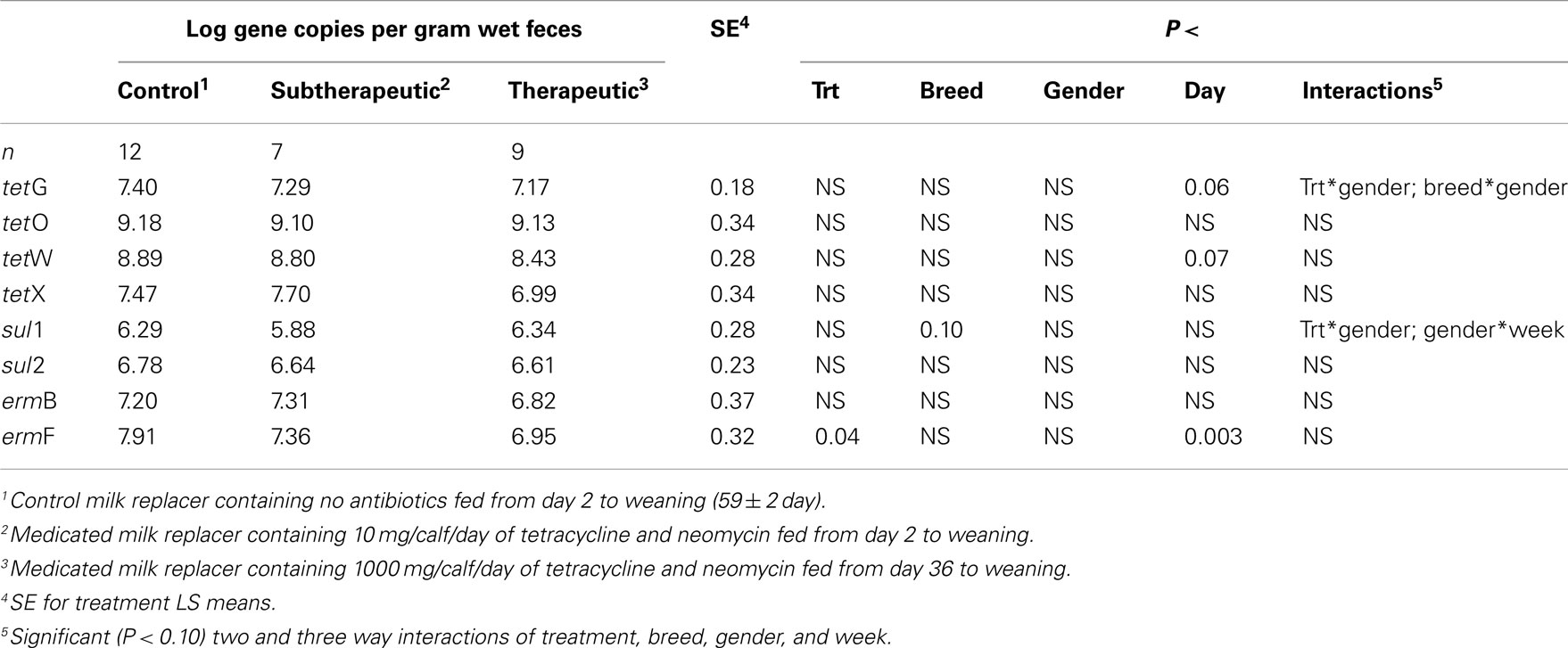
Table 1. Effect of milk replacer medication, breed, gender, week, and the interaction of milk replacer with these on abundance of selected antibiotic resistance genes in the feces of dairy calves.
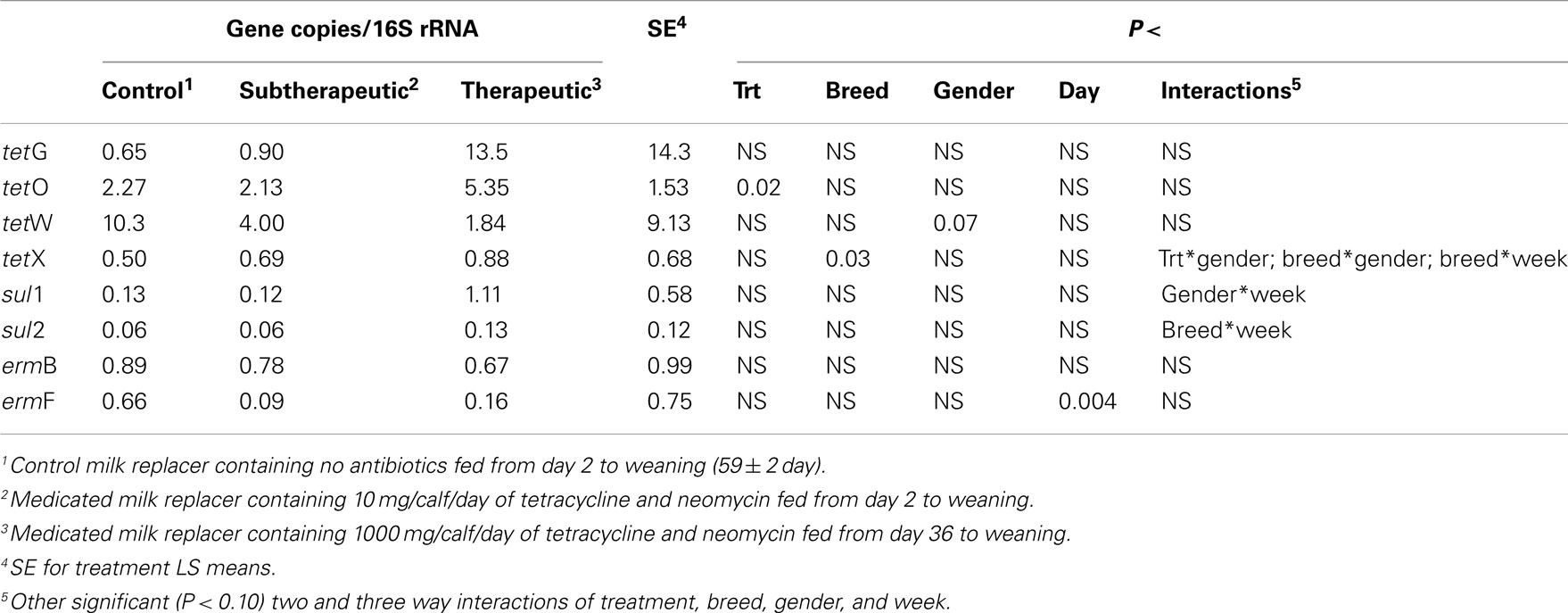
Table 2. Effect of milk replacer medication, breed, gender, week, and the interaction of milk replacer with these on selected antibiotic resistance genes in the feces of dairy calves.
Abundance of the ARGs analyzed was not different between genders or between breeds, but abundance of all tet ARGs except tetO increased with time in all treatments (Figure 1), including the no antibiotic control. For tetG and tetX, gene abundance increased between weeks 6 and 12. For tetW, the increase in abundance occurred during the period following weaning, and was detected at the week 12 sampling point. Effects of time on tet ARGs were no longer observed when the data were expressed per unit of 16S rRNA gene (Table 2). However, relative abundance of ermF was observed to increase with time across treatments, only when expressed per unit of 16S rRNA gene (Figure 3).
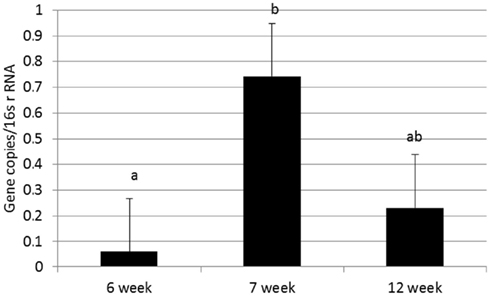
Figure 3. Effect of time on relative abundance of ermF ARGs in calf feces. Bars represent the pooled averages across calves fed medicated or non-medicated milk replacers. Letters above the bars indicate significantly different LSM groupings (P < 0.10).
For the majority of the ARGs, breed, gender, and week did not have any effect except that crossbred and bull calves harbored reduced relative abundance of tetX and tetW, respectively (Table 2). Interaction of treatment with breed, gender, and week of treatment had no significant effects on gene abundance for the majority of the analyzed genes. The interactions of treatment and gender were significant for tetG and sul1 (Table 1). Among control calves, heifers harbored higher abundance of tetG more than did bulls; that was reversed in the subtherapeutic group but the difference was not significant (Figure 4). There was no effect of gender on abundance of sul1 among calves fed the subtherapeutic milk replacer, but among control calves, heifers had far more copies of sul1 in their feces than did bulls (Figure 5). The interaction of week and gender was significant for sul1 (Table 1). Heifers had higher abundance of sul1 at week 6 than did bulls, (Figure 6). The interaction of breed with gender was significant for tetG (Table 1). Crossbred bulls had the higher abundance of tetG than crossbred heifers; there was no effect of gender among Holstein calves (Figure 7).
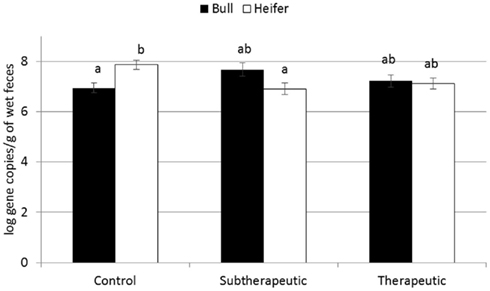
Figure 4. Effect of the gender and antibiotic treatment interaction on abundance of tetG ARGs in calf feces. Bars represent the pooled averages across the three sampling events. Letters above the bars indicate significantly different LSM groupings (P < 0.10).
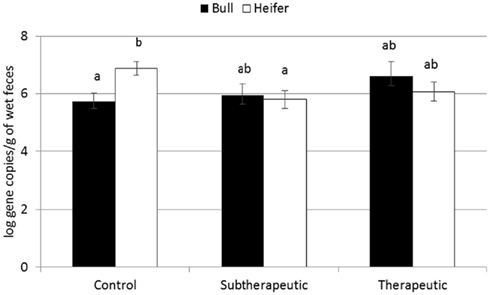
Figure 5. Effect of the gender and antibiotic treatment interaction on abundance of sul1 ARGs in calf feces. Bars represent the pooled averages across the three sampling events. Letters above the bars indicate significantly different LSM groupings (P < 0.10).
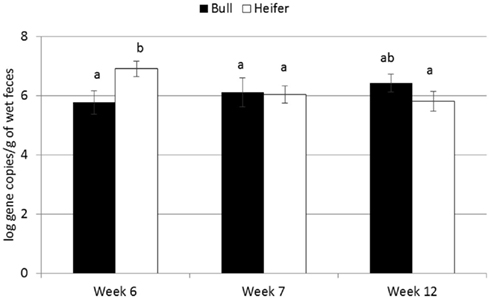
Figure 6. Effect of the gender by time interaction on abundance of sul1 ARGs in calf feces. Bars represent the pooled averages across calves fed medicated or non-medicated milk replacers. Letters above the bars indicate significantly different LSM groupings (P < 0.10).
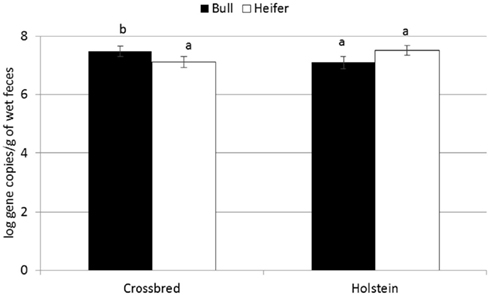
Figure 7. Effect of gender by breed interaction on abundance of tetG ARGs in calf feces. Bars represent the pooled averages across calves fed medicated or non-medicated milk replacers and over the three sampling events. Letters above the bars indicate significantly different LSM groupings (P < 0.10).
The lack of effect of milk replacer medication on growth and health is in contrast to some observations (Quigley and Drew, 2000; Berge et al., 2005; Stanton et al., 2010), probably because calves in the current study were fed a more nutrient-dense diet with higher intake than in most early research. Consequently, calves completing the course of the study were relatively healthy and grew quickly. Notably, of the 13 calves that were removed due to health problems, 7 were in the subtherapeutic treatment group. Health problems that were observed in calves in the current study were largely due to protozoan and viral microorganisms, which are not susceptible to antibiotics. Langford et al. (2003) observed no effect of penicillin content of milk in calves fed ad libitum; they also attributed the lack of effect of antibiotic to the nutritional benefits to the calf of offering an unlimited supply of milk. Interestingly, Berge et al. (2009a) actually observed antibiotic addition to pasteurized waste milk to be associated with a higher percentage of diarrhea days in calves compared to no antibiotic addition, while respiratory scores were equivalent between the two treatments. They partially attributed this effect to adequate passage of immunity via colostrum, as advocated in their parallel studies (Berge et al., 2009b). Their study also highlights potentially negative effects of antibiotics on gastrointestinal flora. Kaneene et al. (2008) similarly observed no health benefits of adding oxytetracycline and neomycin to milk replacer and also noted substantial colostrum provisions to the calves. Thus, nutritional status and overall health may override benefits provided by subtherapeutic antibiotic treatment.
The present study represents the first controlled, culture-independent evaluation of the effect of antibiotic addition in milk replacer on antibiotic resistance in calf feces through weaning. The results provide an indication of the effects of subtherapeutic and therapeutic tetracycline and neomycin treatments typical in the U.S. Among the eight ARGs examined across three classes, tetO was the only one to exhibit an increase in response to antibiotic treatment, but only when normalized to 16S rRNA genes. The results were somewhat unexpected, given reports of feed antibiotics increasing resistance in manure of mature cattle (Alexander et al., 2009) and other livestock (Marshall and Levy, 2011). In a prior study of calves fed “waste milk” containing penicillin residues, resistance of fecal bacteria to penicillin increased with increasing dose fed (Langford et al., 2003). One possible cause for discrepancy is that of the 25 known tetracycline ARGs (Aminov et al., 2001, 2002; Yang et al., 2004), only five were quantified in this study. Although these represented the three main classes of resistance (efflux, ribosomal protection, and degradation) and have previously been reported to be found in cattle waste (Storteboom et al., 2007; Alexander et al., 2009; Dahshan et al., 2010; McKinney et al., 2010), it is quite possible that key tetracycline ARGs responding to antibiotic treatment in this study were overlooked. Interestingly, though tetX was previously reported to be confined within the genus Bacteroides (Yang et al., 2004), its abundance was comparable with that of tetG, with recent reports suggesting it is more widespread than previously thought (Ghosh et al., 2009). Regardless, the results of this study suggest that removal of chlortetracycline and neomycin from milk replacer will not eliminate shedding of ARGs into the environment. McKinney et al. (2010) similarly observed only a very modest reduction of tet and sul ARG abundance in waste lagoons on organic dairy farms relative to those of conventional dairies.
The reduced abundance of ermF observed in feces of calves fed antibiotics as compared to control calves was intriguing. Macrolide and sulfonamide ARGs were examined in this study as potential indicators of co-selection of resistance to antibiotics other than the oxytetracycline and neomycin administered in the milk replacer. In particular, sul1 is carried within the 3′ conserved region of class 1 integrons (Mazel, 2006), while tetracycline resistance has been noted to be prevalent in macrolide-resistant Streptococcus pyogenes (Nielsen et al., 2004). In a study by Berge et al. (2006a) it was found that milk replacer containing neomycin sulfate and tetracycline HCl selected for E. coli resistant to classes of antimicrobials not used, including: aminoglycosides, chloramphenicol, and sulfonamides. The reduced abundance of ermF in the current study may be explained by effects of medicated milk replacer on specific populations of excreted bacteria. For example, Berge et al. (2006b) observed that medicated milk replacer decreased fecal shedding of Salmonella enterica by dairy calves. Decreased fecal shedding of certain bacteria could explain the decreased numbers of gene copies of ermF with antibiotic feeding; the species of bacteria carrying erythromycin resistance being shed in the feces was unknown in this study. It seems probable that one of the two antibiotics fed in the milk replacer were effective against fecal bacteria carrying the ermF gene. Interestingly, the effect of antibiotic treatment was no longer observed when ermF was normalized to 16S rRNA genes (Table 2). Normalization to the 16S rRNA gene, present in all bacteria, provides an indication of the proportion of the bacterial community carrying the ARG of interest and also aids in accounting for minor variations in sample processing. The similarity in relative abundance (ARGs per 16S rRNA; normalized) of ermF between antibiotic fed and non-antibiotic control groups suggests that there was a decrease in overall fecal bacterial shedding in antibiotic-treated groups. Reduced 16S rRNA genes in the manure of antibiotic-treated calves would also be consistent with the tetO increase only being significant when normalized to 16S rRNA genes. However, the range of 16S rRNA gene concentrations encountered among the fecal samples was wide, and there was no significant effect found of antibiotic treatment on overall 16S rRNA gene abundance.
The increased absolute abundance of tet and relative ermF ARGs with time in all calves in the current study (including calves not fed antibiotics) suggests that the calf gut environment itself was amenable to the establishment of resistance regardless of antibiotic content of the feed. Berge et al. (2006a) similarly reported an age-related increase in antibiotic resistance of E. coli (measured phenotypically) even in non-treated calves. On the other hand, no effect of time on tet ARGs was observed when data were expressed per unit of 16S rRNA. This suggests that the overall bacterial population density increased in corresponding samples with time and that the bacteria carrying tet ARGs were neither negatively nor positively impacted. However, the range of 16S rRNA gene concentrations was again wide and a statistically significant increase with time was not identified as a general phenomenon.
Few studies have specifically characterized the establishment of antibiotic resistance in calf manure with time. Similar to the present study, increased overall resistance to 12 antimicrobials was noted among fecal E. coli isolates over the first 4 months of life in calves not receiving antibiotics (Berge et al., 2006a). A recent study by Pereira et al. (2011) examined the antibiotic resistance patterns of E. coli isolates on day 2 and day 6 of calves from two farms that did not administer antibiotics in the feed milk as compared to a third farm that administered both chlortetracycline and sulfamethazine. Odds of resistance to most antibiotics observed for the farm administering antibiotics was higher compared to control farms. The inclusion of sulfonamide, rather than neomycin, could have been the driving factor in the effects observed relative to the present study. While neomycin was included in this study because of its widespread implementation in the U.S., neo ARGs (i.e., aminoglycoside phosphotransferases) were not monitored. Nonetheless, as neomycin is rarely used in humans, the present study did provide insight into potential effects of common practice on resistance to three classes of antibiotics of high clinical relevance, in addition to the clinically relevant integron, intI1 (Hall and Collis, 1998). Also important to consider, in comparison to the Pereira et al. (2011) study, is that in a study design in which the antibiotic treatments are geographically isolated, the actual established microbiota of the farm, rather than the antibiotic present in the milk, could be an overarching factor. The present study thus has value in its focus of the effects of the three controlled treatments on a single farm.
Of note, as is routine practice in the U.S., monensin was included in the grain fed to all calves. While very little grain was consumed during the early weeks, the monensin-containing grain became the sole source of nutrition post-weaning (7 weeks). Tet ARGs did generally increase with time, tetG and tetW prior to weaning and tetX 5 weeks post-weaning. It is not possible to determine from this study whether monensin was related to the observed increases of tet ARGs; however, all significant increases with time were lost when normalized to 16S rRNA genes. Therefore, it can be concluded that monensin did not generally suppress the total bacterial density to produce the observed effect. Co-selection of bacteria carrying tet ARGs by monensin is one possible explanation for their increase with time; however, acquired resistance of bacteria to monensin has not previously been observed (Butaye et al., 2001). Furthermore, bacteria that are tolerant of monensin have been observed to be no more or less sensitive to medically important antibiotics than monensin-susceptible bacteria (Houlihan and Russell, 2003).
The effects of interactions of time, treatment, gender, and/or breed are not easily explained. They may be attributed to the small number of observations associated with each interaction, although each individual group was comprised of n ranging from six to eight. If the effects observed across the gender and breed interactions are indeed real, they highlight the complexity of factors governing the microbial ecology of the bovine gut and the establishment of antibiotic resistance.
To our knowledge, this study provides the first insight into the establishment of antibiotic resistance in calf feces in the early weeks using qPCR to directly quantify ARGs. This approach is advantageous in that it circumvents biases associated with culturing and thus enables broad quantification of resistance elements, including those harbored by unculturable strains. However, molecular techniques possess their own limitations, which must be considered in interpreting the findings of this study. Firstly, although 10 genes were quantified by qPCR, these likely only scratch the surface of the full array of resistance elements present in calf feces. Indeed, recent examination of the human gut microbiome has revealed a vast array of resistance elements, most of which not previously described (Sommer et al., 2009). Thus, it is quite possible that ARGs not monitored or detectable in this study did in fact respond strongly to antibiotic treatment. Secondly, qPCR is limited by the quality of the DNA extraction. While no ideal DNA extraction method exists, we viewed the FastDNA® Spin Kit for soil to be the best option. While kits specifically formulated to manure are available, head to head comparison has actually indicated downstream detection of a broader range of bacteria when the soil kit was applied to extract DNA from feces (Ariefdjohan et al., 2010). Further, the FastPrep® instrument applied in this study was noted to result in the least DNA damage, relative to other cell lysis techniques. The FastDNA® Spin Kit for soil was successfully applied for qPCR analysis of a range of manures by Layton et al. (2006), although they employed a 1:10 dilution prior to extraction, while our study did not. A potential consequence of undiluted samples is overloading of the extraction column and thus lower overall DNA yield. Normalization to 16S rRNA genes should help account for such variation. Also important to note is that our methods were able to detect significant absolute difference in ARGs in several instances, which would not be expected to be discernible if extraction column overloading was the dominant factor driving the results.
Up to 80% of E. coli isolates obtained from mature heifers have been observed to be multi-drug resistant (resistant to three to six drugs), typically displaying resistance to tetracycline, ampicillin, ceftiofur, florfenicol, chloramphenicol, spectinomycin, and streptomycin (Sawant et al., 2007). However, the effect of antimicrobial use on multiple antimicrobial resistance has only recently been studied in calves. Pereira et al. (2011) noted that 81% of the E. coli isolates obtained from both medicated and non-medicated calves were resistant to three or more antibiotics already within the first 6 days of life. Berge et al. (2006a) observed that tetracycline and neomycin in the feed were associated with higher levels of multiple antimicrobial resistant fecal E. coli in calves monitored up to 4 weeks of age. The same research group (Berge et al., 2010) further noted that fecal E. coli were more likely to be resistant to multiple antibiotics in older calves (14- or 28-2) than in newborn calves if calves were fed antimicrobials in milk. In the current study, 8 of the 10 target ARGs were detected in the feces of the experimental calves. Fecal samples from all calves carried multiple types of ARGs within the same sample. Also, feces of all calves carried at least one gene encoding resistance to each of the three classes of antibiotics of interest (tetracyclines, sulfonamides, erythromycins), but class 1 integrons (intI1) were not detected in any of the fecal samples. In contrast, bacterial strains carrying class 1 integrons were isolated from dairy manure-impacted soil, but not from a corresponding unimpacted soil (Srinivasan et al., 2008). Class 1 integrons were also detected in 10% of samples from scouring calves (Ahmed et al., 2009); calves in the current study were not diarrheic during sample collections. Wu et al. (2011) observed that sulfonamide use was associated with higher occurrence of intI1 genes among tetracycline resistant E. coli from beef cattle. That intI1 was not detectable but that multiple ARGs were present in feces of all calves is intriguing. It is plausible that the ARGs identified were carried by different bacterial species in the feces sample, but also the analyzed genes may be carried by the same bacterial species, without linkage to a transferable element such as intI1. Thus, conclusive evidence of multiple drug resistance in the calf manure was not obtained in this study. Nonetheless, multiple drug resistance has been observed to be significantly higher in dairy manure-impacted environments, such as soils (Srinivasan et al., 2008), thus the evolution of antibiotic resistance from calf to mature cow to the environment remains an important issue.
Improved calf health and growth commonly observed with feeding of medicated milk replacers were not observed, likely because the calves in this experiment were maintained on a high plane of nutrition. In situations where management or nutrition is not optimal, health differences may be more apparent. Genes related to tetracycline resistance increased over time regardless of treatment, thus the calf gut itself appears to be an environment conducive to the proliferation of certain ARGs. Genes encoding resistance to antibiotics that were not administered were less abundant, thus co-selection was not observed in this study. However, macrolide and sulfonamide ARGs were still present and persisted in the feces of these experimental calves. Other ARGs not examined were also likely present and may have responded to antibiotic treatment. The results have important management implications, indicating that if nutritional requirements are appropriately managed, the cost and risk of subtherapeutic antibiotic use may not be necessary. Nonetheless, ARGs across three classes persisted and increased with time, even in the feces of calves not administered antibiotics. Thus, comprehensive management strategies that limit cross-dissemination between animals and control transport of manure from farms, in addition to prudent use of antibiotics, are called for.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Financial support was provided by the Virginia Agricultural Council. The authors wish to thank Chad McKinney for technical assistance.
Aarestrup, F. M. (2000). Characterization of glycopeptide-resistant Enterococcus faecium (GRE) from broilers and pigs in Denmark: genetic evidence that persistence of GRE in pig herds is associated with coselection by resistance to macrolides. J. Clin. Microbiol. 38, 2774–2777.
Ahmed, A. M., Younis, E. E. A., Osman, S. A., Ishida, Y., El-khodery, S. A., and Shimamoto, T. (2009). Genetic analysis of antimicrobial resistance in Escherichia coli isolated from diarrheic neonatal calves. Vet. Microbiol. 136, 397–402.
Alexander, T. W., Reuter, T., Sharma, R., Yanke, L. J., Topp, E., and McAllister, T. A. (2009). Longitudinal characterization of resistant Escherichia coli in fecal deposits from cattle fed subtherapeutic levels of antimicrobials. Appl. Environ. Microbiol. 75, 7125–7134.
Aminov, R. I., Chee-Sanford, J. C., Garrigues, N., Teferedegne, B., Krapac, I. J., White, B. A., and Mackie, R. I. (2002). Development, validation and application of PCR primers for detection of tetracycline efflux genes of Gram-negative bacteria. Appl. Environ. Microbiol. 68, 1786–1793.
Aminov, R. I., Garrigues-Jeanjean, N., and Mackie, R. I. (2001). Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl. Environ. Microbiol. 67, 22–32.
Ariefdjohan, M. W., Savaiano, D. A., and Nakatsu, C. H. (2010). Comparison of DNA extraction kits for PCR-DGGE analysis of human intestinal microbial communities from fecal specimens. Nutr. J. 9, 23.
Arnold, S., Gassner, B., Giger, T., and wahlen, R. Z. (2004). Banning antimicrobial growth promoters in feedstuffs does not result in increased therapeutic use of antibiotics in medicated feed in pig farming. Pharmacoepidemiol. Drug Saf. 13, 323–331.
Aschbacher, P. W., and Feil, V. J. (1994). Neomycin metabolism in calves. J. Anim. Sci. 72, 683–689.
Bager, F., Madsen, M., Christensen, J., and Aarestrup, F. M. (1997). Avoparcin used as a growth promoter is associated with the occurrence of vancomycin-resistant Enterococcus faecium on Danish poultry and pig farms. Prevent. Vet. Med. 31, 95–112.
Bates, J. (1997). Epidemiology of vancomycin-resistant enterococci in the community and the relevance of farm animals to human infection. J. Hosp. Infect. 37, 89–101.
Berge, A. C., Epperson, W. B., and Pritchard, R. H. (2005). Assessing the effect of a single dose florfenicol treatment in feedlot cattle on the antimicrobial resistance patterns in fecal Escherichia coli. Vet. Res. 36, 723–734.
Berge, A. C., Hancock, D. D., Sischo, W. M., and Besser, T. E. (2010). Geographic, farm and animal factors associated with multiple antimicrobial resistance in fecal Escherichia coli isolates from cattle in the Western United States. J. Am. Vet. Med. Assoc. 236, 1338–1344.
Berge, A. C., Moore, D. A., Besser, T. E., and Sischo, W. M. (2009a). Targeting therapy to minimize antimicrobial use in preweaned calves: effects on health, growth, and treatment costs. J. Dairy Sci. 92, 4704–4714.
Berge, A. C., Besser, T. E., Moore, D. A., and Sischo, W. M. (2009b). Evaluation of the effects of oral colostrum supplementation during the first fourteen days on the health and performance of preweaned calves. J. Dairy Sci. 92, 286–295.
Berge, A. C., Moore, D. A., and Sischo, W. M. (2006a). Field trial evaluating the influence of prophylactic and therapeutic antimicrobial administration on antimicrobial resistance of fecal Escherichia coli in dairy calves. Appl. Environ. Microbiol. 72, 3872–3878.
Berge, A. C., Moore, D. A., and Sischo, W. M. (2006b). Prevalence and antimicrobial resistance patterns of Salmonella enterica in preweaned calves from dairies and calf ranches. Am. J. Vet. Res. 67, 1580–1588.
Boerlin, P., Wissing, A., Aarestrup, F. M., Frey, J., and Nicolet, J. (2001). Antimicrobial growth promoter ban and resistance to macrolides and vancomycin in enterococci from pigs. J. Clin. Microbiol. 39, 4193–4195.
Butaye, P., Devriese, L. A., and Haesebrouck, F. (2001). Differences in antibiotic resistance patterns of Enterococcus faecalis and Enterococcus faecium strains isolated from farm and pet animals. Antimicrob. Agents Chemother. 45, 1374–1378.
Casewell, M., Friis, C., Marco, E., McMullin, P., and Phillips, I. (2003). The European ban on growth-promoting antibiotics and emerging consequences for human and animal health. J. Antimicrob. Chemother. 52, 159–161.
Chee-Sanford, J. C., Aminov, R. I., Krapac, I. J., Garrigues-Jeanjean, N., and Mackie, R. I. (2001). Occurrence and diversity of tetracycline resistance genes in lagoons and groundwater underlying two swine production facilities. Appl. Environ. Microbiol. 67, 1494–1502.
Chen, J., Yu, Z., Michel, F. C., Wittum, T., and Morrison, M. (2007). Development and application of real-time PCR assays for quantification of erm genes conferring resistance to macrolides-lincosamides-streptogramin B in livestock manure and manure management systems. Appl. Environ. Microbiol. 73, 4407–4416.
Dahshan, H., Shahada, F., Chuma, T., Moriki, H., and Okamoto, K. (2010). Genetic analysis of multidrug-resistant Salmonella enterica serovars Stanley and Typhimurium from cattle. Vet. Microbiol. 145, 76–83.
Ghosh, S., Sadowsky, M. J., Roberts, M. C., Gralnick, J. A., and LaPara, T. M. (2009). Sphingobacterium sp strain PM2-P1-29 harbours a functional tet(X) gene encoding for the degradation of tetracycline. J. Appl. Microbiol. 106, 1336–:1342.
Grave, K., Jensen, V. F., Odensvik, K., Wierup, M., and Bangen, M. (2006). Usage of veterinary therapeutic antimicrobials in Denmark, Norway and Sweden following termination of antimicrobial growth promoter use. Prevent. Vet. Med. 75, 123–132.
Grave, K., Kaldhusdal, M., Krused, H., Fevang Harre, L. M., and Flatlandsmo, K. (2004). What has happened in Norway after the ban of avoparcin? Consumption of antimicrobials by poultry. Prevent. Vet. Med. 62, 59–72.
Hall, R., and Collis, C. (1998). Antibiotic resistance in Gram-negative bacteria: the role of gene cassettes and integrons. Drug Resist. Updat. 1, 109–119.
Hardwick, S. A., Stokes, H. W., Findlay, S., Taylor, M., and Gillings, M. R. (2008). Quantification of class 1 integron abundance in natural environments using real-time quantitative PCR. FEMS Microbiol. Lett. 278, 207–212.
Ho, P. L., Wong, R. C., Lo, S. W., Chow, K. H., Wong, S. S., and Que, T. L. (2010). Genetic identity of aminoglycoside-resistance genes in Escherichia coli isolates from human and animal sources. J. Med. Microbiol. 59, 702–707.
Houlihan, A. J., and Russell, J. B. (2003). The susceptibility of ionophore-resistant Clostridium aminophilum F to other antibiotics. J. Antimicrob. Chemother. 52, 623–628.
Kaneene, J. B., Warnick, L. D., Bolin, C. A., Erskine, R. J., May, K., and Miller, R. A. (2008). Changes in tetracycline susceptibility of enteric bacteria following switching to nonmedicated milk replacer for dairy calves. J. Clin. Microbiol. 46, 1968–1977.
Langford, F. M., Weary, D. M., and Fisher, L. (2003). Antibiotic resistance in gut bacteria from dairy calves: a dose response to the level of antibiotics fed in milk. J. Dairy Sci. 86, 3963–3966.
Larson, L. L., Owen, F. G., Albright, J. L., Appleman, R. D., Lamb, R. C., and Muller, L. D. (1977). Guidelines toward more uniformity in measuring and reporting calf experimental data. J. Dairy Sci. 60, 989–991.
Lauderdale, T. L., McDonald, L. C., Shiau, Y. R., Chen, P. C., Wang, H. Y., Lai, J. F., and Ho, M. (2002). Vancomycin-resistant enterococci from humans and retail chickens in Taiwan with unique VanB phenotype-vanA genotype incongruence. Antimicrob. Agents Chemother. 46, 525–527.
Lauderdale, T. L., Shiau, Y. R., Wang, H. Y., Lai, J. F., Huang, I. W., Chen, P. C., Chen, H. Y., Lai, S. S., Liu, Y. F., and Ho, M. (2007). Effect of banning vancomycin analogue avoparcin on vancomycin-resistant enterococci in chicken farms in Taiwan. Environ. Microbiol. 9, 819–823.
Layton, A., McKay, L., Williams, D., Garrett, V., Gentry, R., and Sayler, G. (2006). Development of bacteroides 16S rRNA gene TaqMan-based real-time PCR assays for estimation of total, human, and bovine fecal pollution in water. Appl. Environ. Microbiol. 72, 4214–4224.
Marshall, B. M., and Levy, S. B. (2011). Food animals and antimicrobials: impacts on human health. Clin. Microbiol. Rev. 24, 718.
McKinney, C. W., Loftin, K. A., Meyer, M. T., Davis, J. G., and Pruden, A. (2010). tet and sul antibiotic resistance genes in livestock lagoons of various operation type, configuration, and antibiotic occurrence. Environ. Sci. Technol. 44, 6102–6109.
National Research Council (NRC). (2001). Nutrient Requirements of Dairy Cattle, 7th Edn. Washington, DC: The National Academies Press.
Ng, L. K., Martin, I., Alfa, M., and Mulvey, M. (2001). Multiplex PCR for the detection of tetracycline resistant genes. Mol. Cell. Probes 15, 209–215.
Nielsen, H. U. K., Hammerum, A. M., Ekelund, K., Bang, D., Pallesen, L. V., and Frimodt-Moller, N. (2004). Tetracycline and macrolide co-resistance in Streptococcus pyogenes: co-selection as a reason for increase in macrolide-resistant S-pyogenes? Microb. Drug Resist. 10, 231–238.
Pei, R., Kim, S. C., Carlson, K. H., and Pruden, A. (2006). Effect of river landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Water Res. 40, 2427–2435.
Pereira, R. V. V., Santos, T. M. A., Bicalho, M. L., Caixeta, L. S., Machado, V. S., and Bicalho, R. C. (2011). Antimicrobial resistance and prevalence of virulence factor genes in fecal Escherichia coli of Holstein calves fed milk with and without antimicrobials. J. Dairy Sci. 94, 4556–4565.
Quigley, J. D., and Drew, M. D. (2000). Effects of oral antibiotics or bovine plasma on survival, health and growth in dairy calves challenged with Escherichia coli. Food Agric. Immunol. 12, 311–318.
Sawant, A. A., Hegde, N. V., Straley, B. A., Donaldson, S. C., Love, B. C., Knabel, S. J., and Jayarao, B. M. (2007). Antimicrobial-resistant enteric bacteria from dairy cattle. Appl. Environ. Microbiol. 73, 156–163.
Smith, M. S., Yang, R. K., Knapp, C. W., Niu, Y., Peak, N., Hanfelt, M. M., Galland, J. C., and Graham, D. W. (2004). Quantification of tetracycline resistance genes in feedlot lagoons by real-time PCR. Appl. Environ. Microbiol. 70, 7372–7377.
Sommer, M. O., Danatas, A. G., and Church, G. M. (2009). Functional characterization of the antibiotic resistance reservoir in the human microflora. Science 325, 1128–1131.
Srinivasan, V., Nam, H.-M., Sawant, A. A., Headrick, S. I., Nguyen, L. I., and Oliver, S. P. (2008). Distribution of tetracycline and streptomycin resistance genes and class 1 integrons in Enterobacteriaceae isolated from dairy and nondairy farm soils. Microb. Ecol. 55, 184–193.
Stanton, A. L., Kelton, D. F., LeBlanc, S. J., Millman, S. T., Wormuth, J., Dingwell, R. T., and Leslie, K. E. (2010). The effect of treatment with long-acting antibiotic at postweaning movement on respiratory disease and on growth in commercial dairy calves. J. Dairy Sci. 93, 574–581.
Storteboom, H. N., Kim, S. C., Doesken, K., Davis, J., Carlson, K. H., and Pruden, A. (2007). Response of antibiotics and resistance genes to high-intensity and low-intensity manure management. J. Environ. Qual. 36, 1695–1703.
Suzuki, M. T., Taylor, L. T., and DeLong, E. F. (2000). Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 66, 4605–4614.
Wierup, M. (2001). The Swedish experience of the 1986 year ban of antimicrobial growth promoters, with special reference to animal health, disease prevention, productivity, and usage of antimicrobials. Microb. Drug Resist. 7, 183–190.
Wileman, B. W., Thomson, D. U., Reinhardt, C. D., and Renter, D. G. (2009). Analysis of modern technologies commonly used in beef cattle production: conventional beef production versus nonconventional production using meta-analysis. J. Anim. Sci. 87, 3418–3426.
Wu, R. B., Alexander, T. W., Li, J. Q., Munns, K., Sharma, R., and McAllister, T. A. (2011). Prevalence and diversity of class 1 integrons and resistance genes in antimicrobial-resistant Escherichia coli originating from beef cattle administered subtherapeutic antimicrobials. J. Appl. Microbiol. 111, 511–523.
Yang, W. R., Moore, I. F., Koteva, K. P., Bareich, D. C., Hughes, D. W., and Wright, D. G. (2004). TetX is a flavin-dependent monooxygenase conferring resistance to tetracycline antibiotics. J. Biol. Chem. 279, 52346–52352.
Keywords: antibiotic resistance genes, manure, dairy calves, milk replacer
Citation: Thames CH, Pruden A, James RE, Ray PP and Knowlton KF (2012) Excretion of antibiotic resistance genes by dairy calves fed milk replacers with varying doses of antibiotics. Front. Microbio. 3:139. doi: 10.3389/fmicb.2012.00139
Received: 02 November 2011; Accepted: 23 March 2012;
Published online: 10 April 2012.
Edited by:
Rustam I. Aminov, University of Aberdeen, UKReviewed by:
Jun Lin, The University of Tennessee, USACopyright: © 2012 Thames, Pruden, James, Ray and Knowlton. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Amy Pruden, Department of Civil and Environmental Engineering, Virginia Tech, 403 Durham Hall (0246), Blacksburg, VA 24061, USA. e-mail:YXBydWRlbkB2dC5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.