- 1 Department of Surface Waters – Research and Management, Eawag: Swiss Federal Institute of Aquatic Science and Technology, Kastanienbaum, Switzerland
- 2 Department of Environmental Microbiology, UFZ, Helmholtz Centre for Environmental Research, Leipzig, Germany
At present, very little is known about the fate and persistence of multiresistant bacteria (MRB) and their resistance genes in natural aquatic environments. Treated, but partly also untreated sewage of the city of Lausanne, Switzerland is discharged into Vidy Bay (Lake Geneva) resulting in high levels of contamination in this part of the lake. In the present work we have studied the prevalence of MRB and resistance genes in the wastewater stream of Lausanne. Samples from hospital and municipal raw sewage, treated effluent from Lausanne’s wastewater treatment plant (WTP) as well as lake water and sediment samples obtained close to the WTP outlet pipe and a remote site close to a drinking water pump were evaluated for the prevalence of MRB. Selected isolates were identified (16S rRNA gene fragment sequencing) and characterized with regards to further resistances, resistance genes, and plasmids. Mostly, studies investigating this issue have relied on cultivation-based approaches. However, the limitations of these tools are well known, in particular for environmental microbial communities, and cultivation-independent molecular tools should be applied in parallel in order to take non-culturable organisms into account. Here we directly quantified the sulfonamide resistance genes sul1 and sul2 from environmental DNA extracts using TaqMan real-time quantitative PCR. Hospital sewage contained the highest load of MRB and antibiotic resistance genes (ARGs). Wastewater treatment reduced the total bacterial load up to 78% but evidence for selection of extremely multiresistant strains and accumulation of resistance genes was observed. Our data clearly indicated pollution of sediments with ARGs in the vicinity of the WTP outlet. The potential of lakes as reservoirs of MRB and potential risks are discussed.
Introduction
Antibiotic resistance has been the focus of research primarily in clinical settings (human and veterinary medicine), whereas the impact of environmental microbes as reservoirs of resistance factors and the impact of releasing antibiotic resistant bacteria (ARB) into the environment was not considered until quite recently. However, it is now known, that ARB and antibiotic resistance genes (ARGs) are ubiquitous in nature (Alonso et al., 2001; Martinez, 2008; Aminov, 2009) and can occur in high concentrations in clinical, industrial, and communal wastewater (Schwartz et al., 2003; Kümmerer, 2004; Volkmann et al., 2004) as well as in animal husbandry (Lanz et al., 2003; Perreten and Boerlin, 2003; Heuer et al., 2008). These environments frequently also contain elevated levels of antibiotics and other pharmaceuticals. Consequently they are considered to select for antibiotic resistance and to be important hot spots for horizontal gene transfer (HGT) of resistance genes, and thus sites of resistance evolution (Baquero et al., 2008; Kemper, 2008; Kümmerer, 2008). In addition, liquid and solids discharged from these environments have been described as major sources of ARB in surface waters (Young, 1993; Kümmerer, 2004; Blasco et al., 2009; Martinez, 2009) and soils (Sengelov et al., 2003; Heuer and Smalla, 2007; Heuer et al., 2008). There are two key concerns related to this continuous introduction of ARB and their resistance genes into the environment. The possible persistence and further dissemination of ARB in natural aquatic environments may contribute to the increase in infections with resistant pathogens. Secondly the dissemination of ARB and ARG may lead to an increase of the ARG pool in environmental bacteria – thus facilitating transfer of resistance into current and emerging pathogens.
Multiresistant bacteria (MRB) are assumed to emerge primarily from hospitals and other environments where high amounts of antibiotics are frequently used. Thus, an elevated occurrence of MRB in the environment is assumed to be a more appropriate incidence for anthropogenic pollution compared to strains carrying only a single resistance. In the culture-based part of the present study we directly selected for multiresistant strains by applying combinations of clinically relevant antibiotics representing seven different antibiotic classes. The antibiotics incorporated in our approach were chosen in order to include representatives of different modes of action (e.g., inhibition of folic acid synthesis as for sulfamethoxazole and trimethoprim, protein biosynthesis as for clarithromycin, tetracycline, and streptomycin, cell wall synthesis as for ceftazidime, and DNA replication as for norfloxacin), different time spans of clinical deployment (e.g., sulfamethoxazole representing an “old” and norfloxacin, ceftazidime representing “newer” antibiotic classes) and different origins (either natural, e.g., streptomycin, semi- or fully synthetic, e.g., clarithromycin and norfloxacin, respectively). Further criteria were the global but also local clinical relevance of the substances as well as their recent detection as micropollutants at our study sites (Blanc, 2010; Bonvin et al., 2011). Preference was given to broad-spectrum antibiotics. The three antibiotic combinations applied in the present study represent “old” (sulfamethoxazole, trimethoprim, and streptomycin), “mixed-old and new” (clarithromycin, tetracycline), and “new” (norfloxacin, ceftazidime) classes. Furthermore, sulfamethoxazole and trimethoprim are mostly applied together in clinics, which led us to apply them in combination also in our study.
Switzerland is a country with comparatively low antibiotic consumption (lowest defined daily dose per 1000 inhabitants among European countries; Filippini et al., 2006). Monitoring programs for antibiotic resistance in clinical settings have been established only recently (NRP49, 2007). Several studies on the release of antibiotics and other micropollutants and their occurrence in sewage and natural environments have been conducted in Switzerland (Alder et al., 2001; Giger et al., 2003; McArdell et al., 2003; Escher et al., 2011). However, the exposure of Swiss lakes to ARB and ARG has so far not received much attention, which is also the case for lakes in general. Most of the studies conducted on antibiotic resistance in surface waters focused on rivers (Iwane et al., 2001; Pei et al., 2006; Li et al., 2009). However, the significance of lakes as environments providing drinking water, food, and recreational activities marks them as highly relevant study objects. Lakes might not only be exposed to pollution with ARB and ARG but might also function as natural resistance reservoirs: Jones et al. (1986) found elevated levels of ARB in Lake Windermere and even higher resistance prevalence in two upland tarns. Picao et al. (2008) detected plasmid-mediated quinolone resistance genes in Aeromonas species isolated from Lake Lugano, Switzerland. However, Auerbach et al. (2007) found only 1 out of 10 tetracycline resistance genes in lake samples. The low number of studies and somewhat conflicting information they provide shows that there is still a considerable knowledge gap on the function of lakes as resistance reservoirs, which needs to be addressed.
Lake Geneva is the largest fresh water reservoir of Western Europe but at the same time receives wastewaters from the surrounding cities. The largest wastewater treatment plant (WTP) belongs to the city of Lausanne. The discharge of sewage from this plant has led to heavy pollution of its receiving water, the Vidy Bay, which is considered the most contaminated part of the lake. Several studies have reported on pollution of the sediment in the bay with heavy metals (Loizeau et al., 2004), micropollutants (Bonvin et al., 2011), and fecal indicator bacteria (Haller et al., 2009b; Poté et al., 2009a) as well as changes to the sediment conditions such as increased organic matter content (Poté et al., 2008; Bravo et al., 2011). To the best of our knowledge no previous studies have been conducted on the input of ARB and ARG into Vidy Bay/Lake Geneva.
In the present study, we aimed to evaluate the occurrence of ARB and ARGs in the wastewater stream of Lausanne and the role of Lausanne’s WTP. In particular, we wanted to evaluate whether the WTP acts as a barrier for MRB in the wastewater stream, or conversely, provides an environment for selection of MRB and horizontal transfer of resistance factors. Finally, the role of the lake as a potential reservoir of ARB and ARG is discussed. We combined application of culture-based and culture-independent methods to allow both: the identification of important key species that might carry and further disseminate antibiotic resistance into the aquatic environment, as well as a culture-independent description of the dynamics of ARGs in the various compartments of the Lausanne wastewater system and Lake Geneva.
Materials and Methods
Sampling Campaigns and Study Site
Lake Geneva is located in the South Western part of Switzerland (Figure 1) and has a volume of 89 km3, a surface area of 580 km2 and a maximum depth of 310 m. The Vidy Bay, which accounts for 0.3% of the lake’s total volume (Chèvre et al., 2011), is located at the northern shore of the lake, next to the city of Lausanne. Lausanne’s WTP treats sewage from 214000 inhabitants, including wastewater from several health care centers. The largest one is the Centre Hospitalier Universitaire Vaudois (CHUV). The most important building in terms of capacity and antibiotic consumption is the main building which accounts for 71% of the CHUV sewage output. On average 410 m3 day−1 of raw sewage are released from this building to the Lausanne municipal sewer system (Blanc, 2010). There is no pharmaceutical industry located in Lausanne, and intensive animal production is not prevalent in the vicinity. Hence it is assumed that the healthcare facilities are likely the main source of ARB. The WTP receives on average 107734 m3 day−1 of wastewater and discharges 86631 m3 day−1 of treated sewage (usually 1–3 m3 s−1 up to 5–6 m3 s−1) directly into Vidy Bay (Vioget et al., 2011). The discharge point is situated 700 m offshore at 30 m depth. During heavy rain events the capacity of the WTP is exceeded and untreated wastewaters are then discharged into the Bay. This has led to heavy pollution (Loizeau et al., 2004; Poté et al., 2009b). Only 3.2 km southwest from the WTP discharge point, Lausanne pumps lake water for its drinking water supply (St. Sulpice). Situated 1 km offshore and at 50 m depth the pump withdraws 100000 m3 of fresh water per day and drinking water is prepared via sand filtration and chlorination.
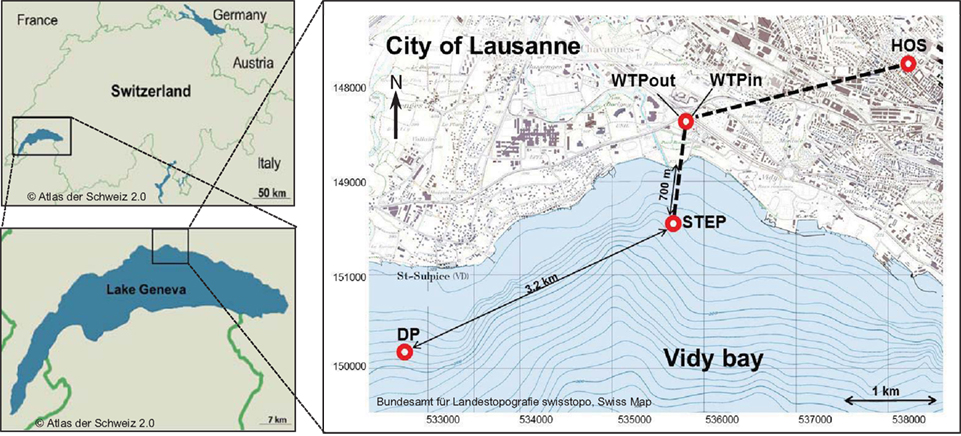
Figure 1. Map of the study area and sampling sites. Effluents of Lausanne’s wastewater treatment plant (WTP) are discharged via pipe 700 m off shore at 30 m depth (STEP, Station d’épuration, Swiss coordinates: 534672/ 151540). 3.2 km to the southwest, lake water is withdrawn for Lausanne’s drinking water supply (DP, Swiss coordinates: 531748/150195). Water was sampled at all points, sediment was sampled at STEP and DP.
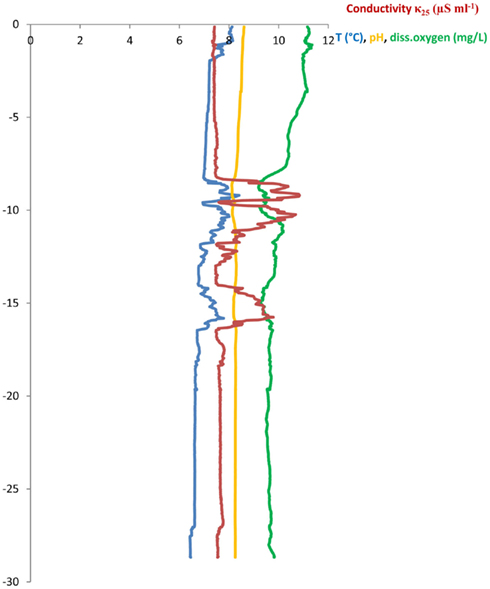
Three sampling campaigns were conducted between February and April 2010 in order to sample all of the following sites once. (a) Wastewater samples were taken from the sewage pipe exiting the main building of the CHUV (March 8, ∼09:00 a.m., sample code: HOS) and (b) at the in- and outlet of the WTP of Lausanne (Feb 22, ∼12:00 a.m., WTPin/WTPout). Additionally, (c) lake water and sediment cores were sampled at two points in the Vidy Bay: directly at the outlet of the wastewater discharge pipe (April 7, 12:00 a.m., sample code: STEP, “Station d’épuration”) and close to the end of the pipe supplying the drinking water pump (April 7, 04:15 p.m., DP) of St. Sulpice (Figure 1). All water samples were filled into ethanol-washed canisters after rinsing twice with sample water. One liter of wastewater was sampled using a repeatedly rinsed scoop and filled into the canisters. Lake water (5 l) and sediment were sampled from the R/V La Licorne (Institute Forel, Geneva), equipped with a crane to which either a sediment corer (Uwitec, Austria) or a rosette sampling device (1018 Rosette Sampling System, General Oceanics Inc., FL, USA), consisting of eleven 1.7-l Niskin bottles, were attached. The rosette was coupled to a CTD device (OCEAN SEVEN 316 Plus CTD, IDRONAUT S.r.l., General Oceanics), which allowed for online monitoring of dissolved oxygen, temperature, pH, and conductivity. Lake water was sampled at 20 m depth (below the thermocline) subsequent to recording a CTD profile (Figure A1 in Appendix). The maximum water depth at STEP and DP is 30 and 56 m, respectively.
Immediately after sampling, 45 ml aliquots of sample water were transferred to 50-ml polypropylene centrifuge tubes (Becton, Dickinson and Company, Sparks, NJ, USA) containing 5 ml of a 20% paraformaldehyde solution (final concentration 2%) in order to preserve cells for flow-cytometric counts of total bacteria (Troussellier et al., 1995). Samples were transported to the laboratory and stored at 4°C in the dark. Plating and filtration of water samples was carried out within 3–6 h of sampling. Sediment cores were stored for up to 1 week at 4°C in the dark before processing. Fixed samples for flow-cytometric counts were stored at −80°C and flow-cytometric analysis was carried out within 2–7 days.
Isolation and Quantification of Multiple Antibiotic Resistant Bacteria
For total and resistant viable cell counts (colony forming units – CFU), samples were processed as follows: (a) Lake water close to the DP: 4 l were filtered through sterile 0.2-μm-pore-sized polycarbonate Isopore™ membrane filters (147 mm in diameter, Millipore, Billerica, MA, USA). Filters were cut into small pieces and placed into 5–10 ml of sterile 0.05 M pyrophosphate (pp) solution (pH 8.5). Cells were suspended in pp by vigorous vortexing and saline solution was added to give a final volume of 40 ml and a 100-fold concentrated microbial suspension relative to the raw sample. (b) Sediment cores: In order to detach bacteria from particles of the sediment surface layer (0–3 cm depth) 3 g sediment were added to 30 ml pp and vortexed for 2 h (Lindahl, 1996; Amalfitano and Fazi, 2008). Particles were then allowed to settle for 18 min.
The concentrated DP microbial suspension from procedure (a) was applied directly as well as diluted 1–10. The suspensions of procedure (b) as well as raw HOS, WTPin, WTPout, and STEP samples were diluted from 1:10 up to 1:105 in sterile 0.9% saline solution. Hundred microliters of each dilution were plated in triplicate on the following agar media cast in 9 cm diameter Petri dishes: nutrient agar (NAg) for copiotrophic bacterial counts, R2A for heterotrophs, phenylethyl alcohol (PEA) agar to select for Gram-positive bacteria, Pseudomonas isolation (PIA) agar to select for pseudomonads, and eosin methylene blue (EMB) agar to select for Gram-negative enterobacteria. All media (Difco™, Becton, Dickinson and Company, Sparks, NJ, USA) were supplemented with three combinations of antibiotics (Sigma-Aldrich, MO, USA) at inhibitory concentrations: (a) “old” Sul/Trm/Str: sulfamethoxazole/trimethoprim (Sul/Trm, 64 μg ml−1, 8:1) and streptomycin (Str, 32 μg ml−1), (b) “new” Nor/Cef: norfloxacin (Nor, 2 μg ml−1)/ceftazidime (Cef, 32 μg ml−1), and (c) “mixed” Cla/Tet: clarithromycin (Cla, 4 μg ml−1)/tetracycline (Tet, 8 μg ml−1). Combination (c) was not tested with EMB and PIA media that enrich for Gram-negative organisms, as clarithromycin mainly selects against Gram-positive bacteria. The concentration of each substance was applied according to DIN norms [German Institute for Standardization (DIN) which develops official standards for susceptibility testing of antimicrobials in Germany and Europe in close collaboration with European Committee on Antimicrobial Susceptibility Testing (EUCAST) similar to CLSI guidelines in the US; Deutsches Institut für Normung e. V. [DIN], 2004]. Three plates of each medium without antibiotics served as positive controls and for determining total viable counts and one plate of each medium not receiving sample was incubated as a sterile control. In order to favor growth of bacteria which are adapted to lower temperatures, NAg and R2A plates were incubated at 25°C and evaluated for formation of colonies after 24, 48, and 72 h. PEA, PIA, and EMB plates were incubated at 37°C and evaluated after 24 and 48 h. All plates were incubated under aerobic conditions.
Total Bacterial Counts from Lake and Wastewater Samples
Total bacterial counts of all water samples were determined once per sample by flow-cytometry (FC) as follows: 1 ml of the fixed sample was diluted 100–1000 times (to give 1000–10000 events per second) and stained with 10 μl of diluted SYBR® Green I solution (Invitrogen, Basel, Switzerland, 1:100 dilution in anhydrous dimethyl sulfoxide). After 15 min of incubation at 20°C in the dark, samples were analyzed on an PARTEC-PAS-III CyFlow Space flow cytometer (Partec GmbH, Münster, Germany) according to Berney et al. (2008).
Identification of Multiple Resistant Bacterial Isolates from Wastewater by 16S RDNA Gene Fragment Sequencing
From R2A and EMB plates supplemented with Sul/Trm/Str approximately 25 colonies were picked from each of the following environments: HOS, WTPin, and WTPout. Antibiotic R2A and EMB plates inoculated with lake samples contained less than 25 colonies. For this reason, colonies from all media supplemented with Sul/Trm/Str were picked. For identification, we performed colony PCR amplification of bacterial 16S rDNA gene fragments. The final reaction volume of 100 μl contained 20 μl of 5× Colorless GoTaq® Flexi buffer (Promega, Madison, WI, USA), 3 mM MgCl2, 0.2 μM of each of the general bacterial primers 27f and 1492r (Table 1), 0.2 μM dNTPs, 1 mg ml−1 bovine serum albumin, 0.025 u μl−1 of GoTaq® Flexi DNA polymerase (Promega) and material from a single colony picked with sterile inoculation needles. 10 ng of DNA from E. coli served as a positive control and 1 μl of nuclease-free water (Qiagen, Germany) served as negative control in all PCRs carried out in this study. PCR reactions were run on a Touchgene Gradient Thermal Cycler (Techne, Cambridge, UK) with the following temperature program: initial denaturation for 5 min at 94°C followed by 35 cycles consisting of 30 s at 94°C, 1 min at 55°C and 1.5 min at 72°C, final extension for 5 min at 72°C.
PCR products were digested overnight at 37°C with HhaI (Promega) for analysis of restriction fragment length polymorphism (RFLP) typing. The final reaction volume of 20 μl contained 2 μl of 10× multicore buffer, 0.2 μl of enzyme (2 u), 7.8 μl water and 10 μl of PCR product. Lambda phage DNA (Promega) served as positive control. RFLP digests were visualized by gel electrophoresis on 3% NuSieve Agarose gels (Cambrex Bio Science Rockland, Inc., ME, USA) and stained with GelRed™ (Biotium, Inc., Hayward, CA, USA). Isolates were grouped into ribo-types according to identical restriction patterns and one representative of each group was selected for sequencing. The respective PCR products were cleaned using the Wizard® SV Gel and PCR Clean-Up System (Promega) and quantified using a NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific Inc., Wilmington, USA). Unidirectional sequencing was carried out by Microsynth AG (Balgach, Switzerland) using the 27f primer. Sequences were analyzed via the BLASTn program (Altschul et al., 1990) and the Ribosomal Database Project (release 10) Classifier tool (Wang et al., 2007). All identified bacterial isolates from the wastewater compartments underwent analysis of minimal inhibition concentrations (MICs; provided subculturing succeeded).
Determination of Minimal Inhibition Concentrations
Identified wastewater bacteria (HOS, WTPin, WTPout) isolated on R2A and EMB plates in the presence of Sul/Trm/Str were investigated for the MICs of various antibiotics using the Sensititre® broth dilution technique. As for the lake samples less than 25 colonies had formed on antibiotic supplemented plates, these were not included neither in MIC testing nor in plasmid and resistance gene detection (see below). Strains were grown on R2A and EMB agar plates. However, for several bacterial isolates subculturing on these media resulted in poor or even no growth for which reason also other media, including NAg, EMB, Columbia Agar with 5% sheep blood (LaboLife Sàrl, Pully, Switzerland) or Tryptic Soy Agar (Difco™) plates were used, according to the growth requirements of the different identified genera. All agar media were supplemented with Sul/Trm/Str as described above and strains were allowed to grow for 72 h at the temperatures which were used during isolation (25 or 37°C), and at 30°C for identified pseudomonads. One to four colonies per strain were picked (dependent on quantity of biomass formed) and transferred to 1 ml of sterile millipore water. After vortexing, 10 μl of the suspension were transferred to 11 ml of a liquid medium based on the agar medium used for growth [R2A broth, Mueller Hinton broth (MHB), nutrient broth (NB), brain heart infusion broth (BHB), tryptic soy broth (TSB); replacing solid media listed above in this order]. All media were purchased from Dicfo™, except R2A broth which was prepared according to Reasoner and Geldreich (1985).
Subsequently, Sensititre® 96 well susceptibility plates (TREK Diagnostic Systems, West Sussex, UK; plate formats applied in this study: EUMVS for Gram-negatives and NLM4 for Gram-positives) were inoculated with 50 μl culture medium per slot. A negative control was inoculated with sterile medium and a slot without antibiotic served as positive control. Plates were incubated at the appropriate temperature for each strain and evaluated for bacterial growth after 24 and 48 h. Plates were checked for biomass formation in each slot by measuring absorption at 570 nm using an Synergy™ HT Multi Detection Microplate Reader (BioTek, Winooski, VT, USA) and by eye. Strains were classified as resistant to the tested substances based on the observed minimum inhibitory concentration according to Deutsches Institut für Normung e. V. [DIN] (2004) norms and European Committee on Antimicrobial Susceptibility Testing [EUCAST] (2011). As for florfenicol no resistance breakpoints are available, strains were classified as resistant according to breakpoints for the closely related chloramphenicol (CHL).
Extraction and Replicon Typing of Plasmids from Isolated Strains
All Gram-negative wastewater bacteria referred to in the previous paragraphs were grown in 5 ml R2A broth, nutrient bouillon, LB, TSB, or BHB supplemented with 64 μg ml−1 Sul/Trm (8:1) and 32 μg ml−1 Str. The liquid cultures were shaken in 15-ml polypropylene tubes at 180 rpm and 25, 30, or 37°C for 24–48 h, depending on growth-preferences and growth rates of strains. Cells were harvested by centrifugation at 5000 rpm for 10 min at room temperature using an Eppendorf 5804R centrifuge with a swinging bucket rotor for 15-ml centrifuge tubes (Eppendorf, Hamburg, Germany). The supernatant was discarded and the pellets frozen at −20°C until plasmid extraction. Plasmid DNA was purified from frozen cell pellets using the PureYield™ Plasmid Miniprep System (Promega) for Gram-negative bacteria according to the manufacturers’ protocol. As positive controls we used E. coli J53 harboring the 33-kb-sized plasmid R388, which inherits the sul1 gene for sulfonamide resistance. Liquid cultures of this strain were prepared in LB with 20 μg ml−1 trimethoprim. For Gram-positive strains we applied the PureYield™ Plasmid Midiprep System (Promega) adapted according to Wegmüller and Schmid (2009) with a few modifications: in brief, cells were grown in 50 ml BHB for 20 h at 37°C and 180 rpm. Bacillus subtilis ssp. subtilis BD170 (DSMZ, Germany) carrying the 4.5-kb-sized plasmid pUB110 served as a positive control during extraction. Cell pellets were resuspended in 3 ml of the cell suspension solution supplied with the kit and incubated for 10 min at 37°C with 1 mg ml−1 lysozyme added (B. subtilis cells), or for 30 min at 37°C and 10 mg ml−1 of lysozyme and 1 ml of a 75% (w/v) saccharose solution (environmental isolates). Subsequently, cell lysis and neutralization was carried out according to the manufacturers guidelines, taking into account the recommended time to allow the formation of white flocculent before centrifugation of the lysate (Wegmüller and Schmid, 2009). Plasmid DNA was further purified following the Promega protocol and finally eluted in 400 μl of nuclease-free water.
Visualization of plasmid DNA was done by gel electrophoresis in 0.8% agarose gels at 50 V. Plasmid extracts showing several sharp bands were screened for the following Inc groups: IncP1α,β, IncP7, IncN, IncW, and IncQ, using the primers and PCR conditions as described previously (Table 1). PCR products were screened by gel electrophoresis. Plasmid extracts were evaluated as positive for the tested Inc groups if the PCR product exhibited a band of the correct size.
Detection of Sulfonamide Resistance Genes in Selected Isolates
All plasmid extracts were screened for the presence of the three sulfonamide resistance genes sul1, sul2, and sul3 using a multiplex PCR assay as described previously (Table 1). The presence of sul genes was determined from detection of a PCR amplicon band of the expected size following agarose gel electrophoresis. A post-PCR identification of which sul type was present was not performed. As positive controls for sul1, sul2, and sul3 served plasmids R388 (E. coli J53), RSF1010 (E. coli DH5α), and pVP440 (E. coli K12), respectively. The formation of PCR products of the correct size was evaluated via gel electrophoresis in 2% agarose gels.
DNA Extraction from Water Filters and Sediments
Water samples were filtered using 5- and 0.2-μm pore-size polycarbonate membrane filters (Isopore™, Ø 147-mm, Millipore). Filters were cut into quarters, folded, packed into clean self-seal bags, and shock-frozen in liquid nitrogen for subsequent storage at −80°C until DNA extraction (3–5 months). One quarter of the filter from each sampled location was extracted as follows: lysis of bacterial cells was carried out by a two-step procedure: (a) bead-beating in GOS buffer (Hönerlage et al., 1995) for 40 s at 4 ms−1 with a FastPrep®-24 Instrument (Biomedicals Europe, Illkirch, France). For this step we used 0.2 g of a 1:1 mixture of 106 μm and 150–212 μm glass beads (Sigma-Aldrich). (b) Freezing in liquid nitrogen for 1 min and thawing at room temperature (rt). Cell debris was removed by centrifugation for 30 min at maximum speed (13000 rpm, Eppendorf Centrifuge 5415R) at rt. The supernatant was treated for 30 min at 37°C with 10 mg ml−1 RNase A. DNA was extracted with 1/2 volume of phenol (pH 8, Sigma-Aldrich) and 1/2 volume of chloroform/isoamyl alcohol (24:1, CIA, both Sigma-Aldrich) followed by a second extraction with 1 volume of CIA. DNA was precipitated with 1 volume of isopropanol. After centrifugation at 4°C and maximum speed for 30 min the DNA pellet was rinsed with 70% cold ethanol and dissolved in 20–30 μl of sterile Tris–EDTA buffer.
Out of each sediment core, 0.5 g of surface layer material were transferred to sterile 2 ml screw-cap tubes (BRAND GmbH + Co KG, Wertheim, Germany) and stored at −80°C in 1.5 ml GOS buffer until DNA extraction. For cell lysis, a bead mixture as described above was added and after vortexing two times for 40 s each at 3000 rpm (maximum vortex velocity, Vortex Genie 2, Scientific Industries, Inc., NY, USA) the supernatant was extracted with 500 μl phenol (pH 8, Sigma-Aldrich), followed by extraction with 200 μl CIA and finally with 400 μl chloroform. Precipitation of DNA and subsequent steps were as described for the DNA extraction from filters. For both water filters and sediments one blank extract was prepared, containing only beads and chemicals, respectively one quarter of a clean filter.
Quantification of Sulfonamide Resistance Genes and Bacterial 16S RRNA Gene Fragments in Environmental DNA Extracts
The sulfonamide resistance genes sul1 and sul2 and bacterial 16S rRNA gene fragments were quantified from environmental DNA extracts using quantitative real-time Taqman®-PCR. Reactions were run in volumes of 20 μl (sul1 and sul2) and 30 μl (16S rRNA), containing 1× TaqMan® Fast Universal PCR-Master Mix (Applied Biosystems, Foster City, CA, USA) and 5 μl of DNA (dilutions of quantification standards or diluted (1:10 to 1:105) DNA extracts of samples). Extraction blanks (see above) as well as PCR blanks containing 5 μl of nuclease-free water were run as negative controls. To check for the presence of PCR inhibitors, standard PCR amplification of bacterial 16S rRNA gene fragments was carried out as described above, using 1 μl of template at various dilutions. Dilutions revealing successful amplification according to agarose gel electrophoresis were then used for real-time PCR. Primers and probes for sul1 and sul2 genes (Table 1), as well as their corresponding concentrations for PCR were as previously described (Heuer and Smalla, 2007; Heuer et al., 2008). PCR conditions for sul1 and sul2 were as follows: 2 min activation at 50°C, initial denaturation for 10 min at 94°C and 40 cycles of denaturation at 95°C for 15 s and annealing at 60°C for 3 min. For quantification of 16S rRNA genes we used universal primers and probes described by Suzuki et al. (2000). Primer and probe concentrations as well as PCR conditions were applied according to Newcombe et al. (2005). All samples and standards were run in triplicate in 96 well plates on a 7500 Fast Real-Time PCR System (Applied Biosystems) and analyzed using the automated settings of the 7500 Fast System SDS Software (Applied Biosystems). Standard curves were prepared from serial dilutions of the plasmids serving as positive controls for sul1 (R388) and sul2 (RSF1010) and a pGEM®-T Easy plasmid containing the target fragment of the 16S rRNA gene of E. coli. (position 1369–1492 of the E. coli reference genome, 123 bp). Dilution series were prepared as recommended by the Applied Biosystems Tutorial “Creating Standard Curves with Genomic DNA or Plasmid DNA Templates for Use in Quantitative PCR.” Efficiency (E) of each qPCR run was calculated from the slope of the standard curves according to the formula E = 10(−1/slope) − 1. The limit of quantification (LOQ) was determined by the most diluted standard in the standard curve that exhibited a standard deviation of Ct values of the triplicates smaller than 0.5 (Life Technologies Corporation, 2012).
Statistical Analysis
In order to determine significant differences of measurements in excess of the error of measurement, we performed pairwise t-tests in R, based on the triplicate measurements of either qPCR or plate counts. Correlation analyses for comparison of the outcome of different methods were performed using the data analysis tool in Microsoft Excel.
Results
Total Bacterial Load along the Wastewater Flow Line
We determined the total bacterial load in samples along the wastewater flow of Lausanne into the Vidy Bay, Lake Geneva using a culture-based (plate counts) and two culture-independent methods (total cell counts by FC and direct quantification of 16s rRNA gene fragments using quantitative real-time PCR). For 16S rRNA genes the PCR efficiency was calculated to be 109.94% and the LOQ was determined at 300 copies of the target per 5 μl.
All three methods show a decrease in total bacteria along the wastewater flow path from the urban catchment to the lake (Figures 2 and 3A). The two culture-independent methods, agreed very well in all analyzed water samples (correlation coefficient r2 = 0.94, Figure 3A). On all of the five tested media, and for all tested samples, viable bacterial numbers determined by plate counts were at least two orders of magnitude lower than the bacterial numbers determined with the culture-independent approaches. The highest numbers were detected on the two general media R2A and NAg.
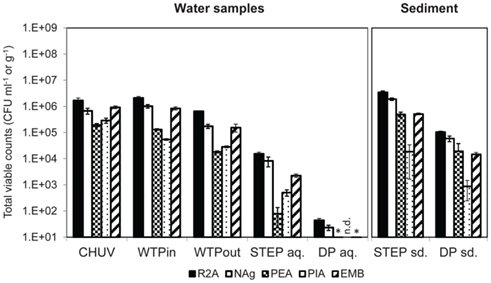
Figure 2. Total viable counts, normalized to sample volume or mass, on agar media without antibiotics. An asterisk (*) indicates less than 10 CFU ml−1. n.d., no colonies detected. Error bars indicate standard deviation of triplicate platings of the same sample.
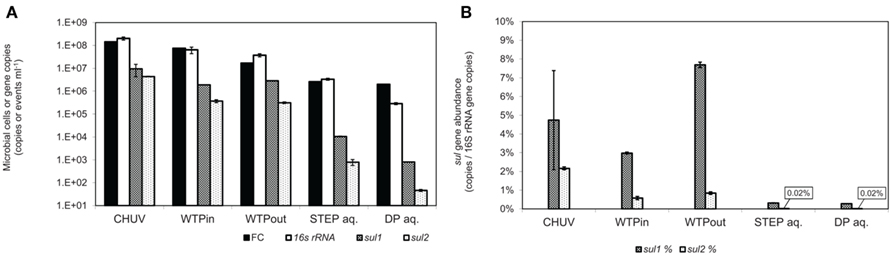
Figure 3. (A) Total gene copy numbers and flow-cytometric cell counts, normalized to sample volume. (B) Relative abundance of sulfonamide resistance gene copy numbers normalized to 16S rRNA gene copies. Error bars indicate standard deviation from triplicate real-time PCR amplification of the same sample.
The highest bacterial load was determined in the HOS sample based on FC and 16S rRNA gene copy numbers (p ≤ 0.01). Plate counts on the three selective media PIA (p ≤ 0.001), PEA (p ≤ 0.01), and EMB also revealed highest viable bacterial numbers in the HOS sample, although for EMB the difference was within the error of measurement. Both the general media R2A and NAg revealed slightly higher viable bacterial concentrations in the WTPin compared to the HOS sample but the difference was not significant (p ≥ 0.05).
The discrepancy between untreated and treated wastewater samples (WTPin and WTPout, respectively) was about 0.5–1 orders of magnitude for all three methods but for 16S rRNA gene copy numbers the difference was not significant. According to this data wastewater treatment reduced the total bacterial load is by 73% based on total viable counts (averaged over all five media), 78% according to FC and 42% according to 16S rRNA gene copy numbers, respectively.
Total viable counts in STEP lake water ranged from 101 to 104 CFU ml−1 but they were considerably lower (by 1–3 orders of magnitude) at the DP site (Figure 2). The trend of CFU-Total and FC counts along the flow path was mostly coherent, irrespective of the fact that the latter were generally 2–3 orders of magnitude higher. However, we observed an increasing divergence of the two methods in the lake samples: in contrast to the drop of bacterial plate counts from STEP to DP (p ≤ 0.05 for all media, except for PEA), FC counts revealed a similar bacterial load in both lake water samples of 106 events ml−1 (compare Figures 2 and 3A). 16S rRNA gene copy numbers were even 1 order of magnitude lower for the STEP water sample compared to the DP sample by p ≤ 0.01.
In sediment samples taken close to STEP total viable counts were approximately 10 times higher compared to DP sediments (Figure 2). Note that sediment viable counts per gram dry weight were at least 10 times higher compared to plate counts per milliliter of the overlying water columns at both lake sites. No qPCR quantification results for 16S rRNA genes are presented for sediments as we observed considerable inhibition for the method with these samples.
Multiresistant Bacteria and Resistance Genes along the Wastewater Flow Line
As for the total bacterial load we also detected a decreasing trend for MRB determined by plate counts along the wastewater flow path from the urban catchment to the lake (HOS > WTPin > WTPout > STEP > DP, Figures 4A–C). Plate counts for “new” antibiotics (Nor/Cef) were comparatively low along the whole wastewater flow line on four out of five of the tested media. Solely on PEA plates which select for Gram-positive bacteria, Nor/Cef resistance occurred in the same range as the numbers determined for the remaining antibiotics at the respective sampling sites (Figure 4C).
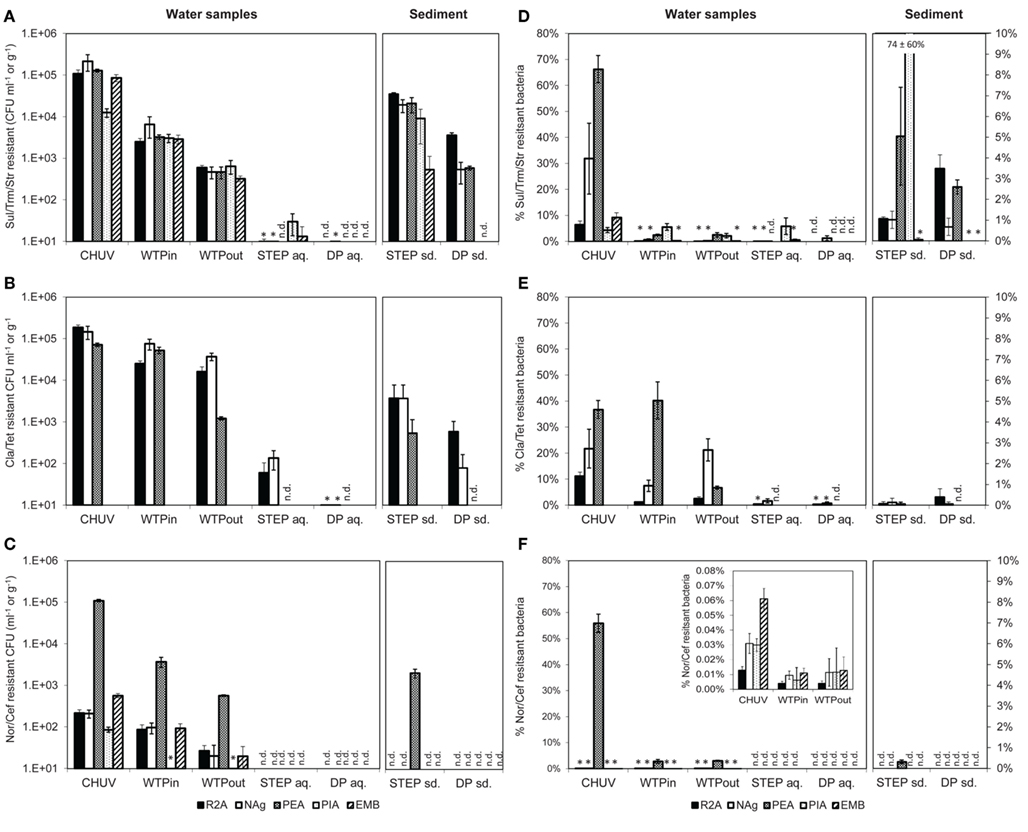
Figure 4. (A–C) Total viable counts, normalized to sample volume or mass, in the presence of (A) Sul/Trm/Str, (B) Cla/Tet, and (C) Nor/Cef. (D–F) Abundance, relative to total viable counts without antibiotics, in the presence of (D) Sul/Trm/Str, (E) Cla/Tet, and (F) Nor/Cef. The insert in (F) shows abundance of selected samples at an adjusted scale. An asterisk (*) indicates less than 10 CFU ml−1 in (A–C) and less than 1% relative abundance in (D–F). n.d., no colonies detected. Error bars indicate standard deviation of triplicate platings of the same sample.
The highest concentration of viable potentially MRB along the flow path was found in the HOS sample, regardless of the antibiotic combination or growth medium (Figures 4A–C). However, in case of Nag plates supplemented with Sul/Trm/Str and Cla/Tet counts on PEA plates the differences between HOS and WTPin samples were within the error of measurement (p = 0.08 for both, respectively).
Relative abundances of potentially MRB (i.e., MRB plate counts normalized to the total viable bacterial counts on the same medium) were mostly, but not exclusively, observed to be highest in HOS as well, ranging from 0.01 to 66% (Figures 4D–F). Besides the high values in HOS we also observed high relative abundance of Cla/Tet resistance on PEA plates in the WTPin sample (40%) and NAg plates showed high relative abundances for this antibiotic combination in the WTPout sample (21%). Both values were in the same range as the relative abundances determined for the HOS sample (36 and 21% for PEA and Nag, respectively; Figure 4E). For Nor/Cef resistance slightly elevated relative abundances were observed in WTPin and WTPout, but only for Gram-positives (PEA medium, 2.8 and 3.1%, respectively) whereas their abundance was low for the remaining media (less than 1%). Abundances of Sul/Trm/Str MRB were quite homogenous for all media and ranged from 0.01 to 5.6%.
Viable counts of potentially MRB were 0.5–1 orders of magnitude higher in the untreated versus the treated wastewater sample (WTPin and WTPout) which is similar to the trend observed for total viable counts. Calculated removal of MRB during wastewater treatment (averaged over all five media) was 62 ± 26, 78 ± 6, and 84 ± 6% for Cla/Tet, Nor/Cef (disregarding the much lower PIA counts), and Sul/Trm/Str MRB, respectively.
To some extent we noticed variations in removal efficiency among the different media and antibiotics. For instance, Cla/Tet resistant Gram-positives (PEA medium) seem to be more drastically reduced during wastewater treatment compared to other Cla/Tet resistant bacteria growing on R2A and NAg (Figure 4B). Nor/Cef resistant Gram-positives (PEA medium) were found at 10-fold higher number compared to viable bacteria on the remaining media, and appeared to retain this elevated number even after passing the WTP (Figure 4C). Although relative abundances of potentially MRB usually decreased from WTPin to WTPout, we also observed similar proportions in both samples, e.g., for Nor/Cef resistant Gram-positives (2.8 and 3.1%) and even increasing numbers, e.g., from 7.5 to 22% on NAg plates supplemented with Cla/Tet, respectively (Figure 4E).
The lake water generally exhibited low amounts of resistant bacteria. From STEP lake water samples we were able to isolate bacteria in the presence of Sul/Trm/Str (100 to 102 CFU ml−1) and Cla/Tet (101 to 102 CFU ml−1) but not in the presence of Nor/Cef. At the DP site, hardly any bacteria were cultured from lake water in the presence of antibiotics, despite the additional sample concentration step (Figures 4A–C).
In sediment samples Nor/Cef resistant colonies were formed solely on PEA plates from STEP, while none were obtained from DP sediments. Generally, STEP sediments contained higher numbers of MRB than DP sediments (Figure 4C). Compared to the overlying water column, the concentration of MRB in sediment were 1–3 orders of magnitude higher. Relative abundances of MRB in both sediment and lake samples were mostly below 1% except for Sul/Trm/Str plates which, on different selected media, reached up to 5% of the respective total viable counts (Figure 4D).
Real-time PCR quantification of sulfonamide resistance genes sul1 and sul2 in wastewater and lake water DNA-extracts revealed the highest load of both traits in the HOS sample and a general decrease along the flow path (Figure 3A). This trend is in good agreement with the outcome of the culture-based approach. sul1 gene copy numbers correlated with Sul/Trm/Str plate counts on R2A and NAg plates (r2 = 0.91, for both media), and the correlation was even closer for sul2 (r2 = 0.99, for both media). However, copy numbers per milliliter of water or gram sediment (dwt) of both ARGs were higher by 1–4 orders of magnitude compared to the respective viable counts in all analyzed samples (compare Figures 3A and 4A).
In contrast to the culture-based results, qPCR results revealed a twofold increased level of sul1 (p ≤ 0.001) and only a 15% lower level of sul2 (p = 0.3) in the WTPout sample compared to the WTPin sample.
Normalized to the 16S rRNA gene, the sul genes occurred at abundances of 0.28 to 7.7 and 0.02 to 2.1% for sul1 and sul2, respectively. The highest sul1 abundance was observed in the WTPout sample followed by HOS (Figure 3B).
The abundance of sul1 was found to be higher than sul2 in all samples with increasing discrepancy from wastewater to the lake samples. In lake water, the relative abundance of the two resistance genes was considerably lower (<1%) than in wastewater samples.
qPCR efficiencies derived by the slopes of the standard curves for both sul genes were 93% for sul1 and 102% for sul2. The LOQ for both standards was at 300 copies per 5 μl. All diluted samples amplified within the range of the standard curve, above the LOQ.
Important Genera Identified in Wastewater and Lake Samples
In total, 163 isolates from HOS, WTPin, and WTPout obtained on Sul/Trm/Str supplemented R2A and EMB plates were identified to the genus level by RFLP typing and sequencing of the rRNA gene. Sequences that passed quality control and the chimera check (using Bellerophon, version 3; DeSantis et al., 2006) were submitted to GenBank and are available under accession numbers: JQ595461–JQ595555 (the FASTA file provided as supplementary material also includes the lower quality sequences). We identified 27 different genera (Table 2), 16 in HOS, 14 in WTPin, and 13 in WTPout. The majority were Gram-negative bacteria, but we also identified a few Gram-positives in HOS (n = 6) and WTPin (n = 5), mostly Enterococci. Pseudomonads were found to be most abundant among isolates from all three sites. Enterococci and Brevundimonas were quite common in HOS, but the latter was also detected in WTPout. Escherichia/Shigella and Acidovorax species dominated the WTPin isolates. Escherichia/Shigella was also isolated frequently from WTPout, as were Sphingobacteria (accounting for ∼25% of the WTPout isolates). Furthermore, 17 genera were identified among the lake water bacteria (both STEP and DP) grown in the presence of Sul/Trm/Str (Table 2). In sediment from STEP and DP, Bacillus and Solibacillus, respectively, were the most frequently isolated strains. A further dominant genus in STEP sediment was again Brevundimonas.
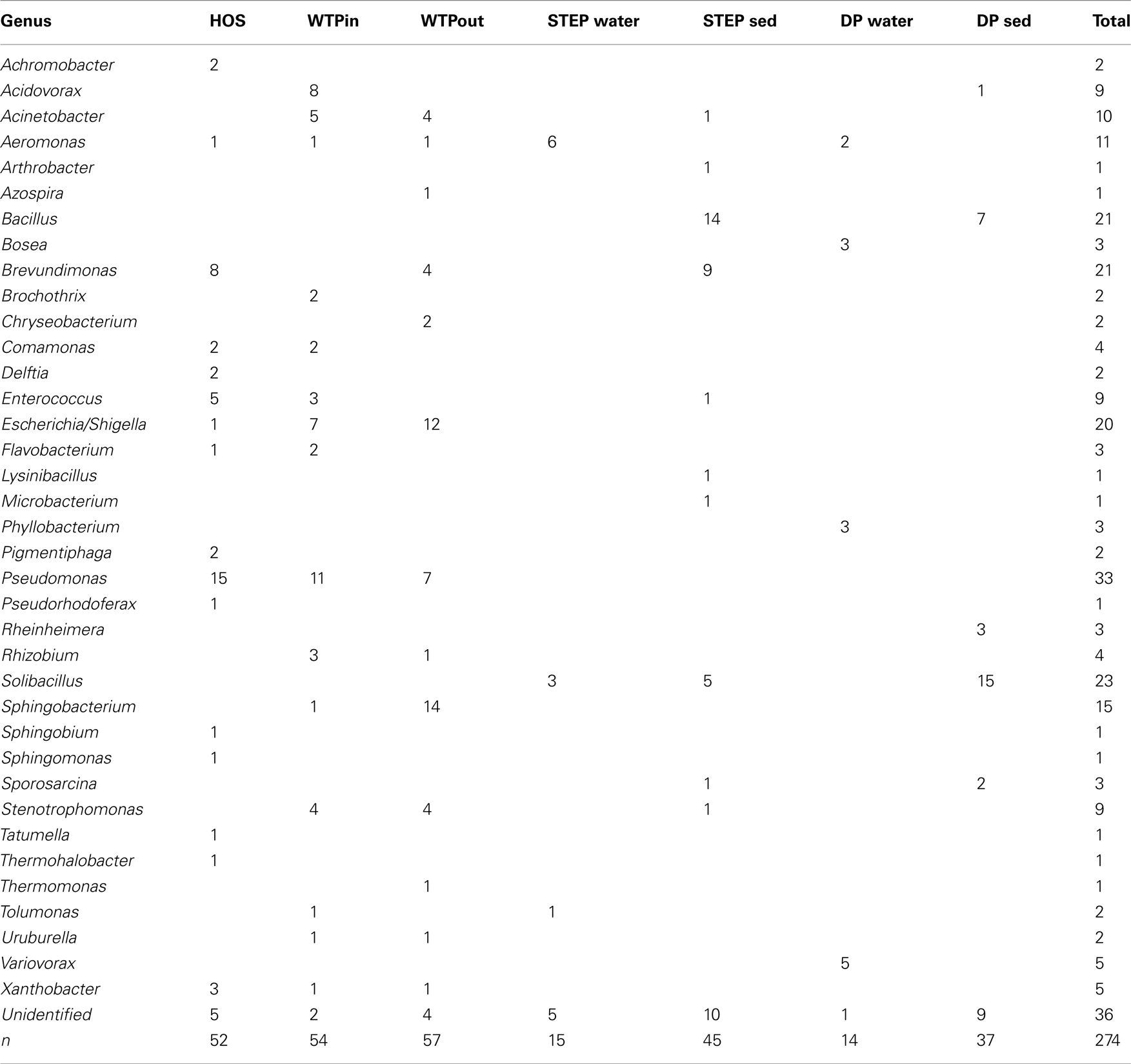
Table 2. Genera of Sul/Trm/Str resistant bacteria isolated from HOS, WTPin, and WTPout according to partial 16S rRNA gene sequences and their numerical distribution in the different wastewater compartments.
Prevalence of Highly and Extremely Multiresistant Bacteria along the Wastewater Flow Path
Resistance of identified multiresistant wastewater isolates obtained from Sul/Trm/Str plates was confirmed by determining the MICs of 14 antibiotics. The results of this experiment are summarized in Figure 5. Resistance toward the antibiotics applied in the isolation plates (Sul/Trm/Str) was confirmed. This resistance pattern usually coincided with a high tolerance or resistance to ampicillin, which was present in 76 and 77% of HOS and WTPin isolates (n = 42 and 22, respectively) and 87% of WTPout isolates (n = 45). Resistance patterns toward the 11 remaining substances tested varied among the different wastewater environments: the number of antibiotics to which more than 50% of the tested strains were resistant was 8 for the HOS sample. Among WTPin isolates only resistances against CHL and nalidixic acid (NAL) were detected frequently (in 55 and 67% of the isolates, respectively). When comparing the proportions of bacteria resistant against specific antibiotics in untreated (WTPin) and treated (WTPout) wastewater, similar or higher proportions were found in WTPout for 9 out of the 11 antibiotics not used in isolation. In the cases of gentamicin (GEN) and colistin (COL) the ratio was more than twice as high for WTPout. Only the resistances to fluoroquinolones (NAL and ciprofloxacin, CIP) occurred more frequently in WTPin isolates.
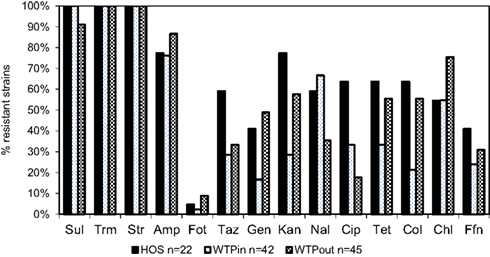
Figure 5. Percentage of bacterial isolates from HOS, WTPin, and WTPout that were classified as resistant to the listed antibiotics.
We have furthermore evaluated the number of resistances per strain. Strains with resistance against more than six or eight antibiotics were labeled as highly and extremely multiresistant, respectively. When comparing the occurrence of highly and extremely MRB at the three sites (Figure 6), highest proportions were again detected in the HOS sample (86 and 77%, respectively) and an increased level in WTPout (82 and 60%, respectively) compared to the WTPin sample (71 and 33%, respectively). Thus, we observed an 11 and 27% higher abundance of highly and extremely multiresistant strains, respectively, in the WTPout sample compared to WTPin. The most frequently isolated genera of highly multiresistant species were Pseudomonas and Escherichia/Shigella which were present at all three locations. Enterococcus and Brevundimonas were present in HOS. While the former was otherwise only detected in WTPin, the latter was only found in WTPout. Acinetobacter was prevalent in both WTPin and WTPout and a very dominant and highly multiresistant genus in the WTPout sample was Sphingobacterium.
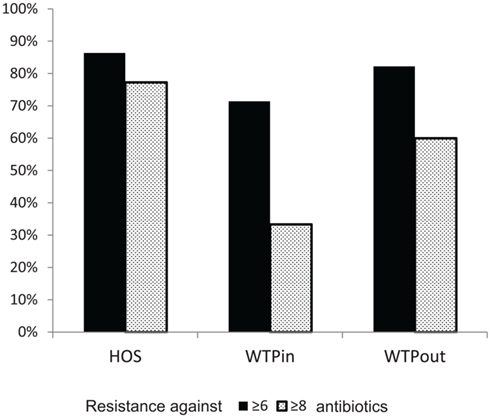
Figure 6. Comparison of percentages of bacteria inheriting more than six (“highly multiresistant”) and more than eight resistances (“extremely multiresistant”) at the three sampling sites HOS, WTPin, and WTPout.
Prevalence of Plasmids and of Sul Genes in Selected Wastewater Isolates
The extraction of plasmid DNA from 129 of the 169 selected wastewater isolates showed that 58, 77, and 46% of the strains isolated from HOS, WTPin, and WTPout were harboring plasmids. For many of the tested strains agarose gel electrophoresis of the plasmid DNA extract revealed multiple bands in the low kilobit size range, indicating multiple plasmids per strain. Replicon typing of 78 plasmid positive extracts revealed only a low number of specific amplifications for IncP1α,β (n = 3), IncP7 (n = 8), IncQ (n = 17), IncW (n = 2), and no amplicons were detected for IncN. Though the yield of positives for the tested Inc groups was quite low, the results nevertheless gave further indications for the presence of multiple plasmids (of replicon types IncQ and IncP7) in three of the tested strains. All plasmid extracts, independent of their band pattern, were screened for presence or absence of sulfonamide resistance genes sul1, sul2, and sul3. Sul genes were amplified in 76 out of 129 plasmid extracts, accounting for 77, 40, and 52% of the investigated bacterial isolates from HOS, WTPin, and WTPout, respectively.
Discussion
Our study focuses on a wastewater flow path in Lausanne, Switzerland. Switzerland is a country with comparatively low antibiotic consumption (Filippini et al., 2006). However, the impact of hospital effluents and wastewater treatment on the dissemination of resistant bacteria and resistance genes in the receiving (aquatic) environments is of global relevance. Several studies have documented high incidence of antibiotic resistance in such wastewaters all over the world (Reinthaler et al., 2003; Kümmerer, 2004; Baquero et al., 2008; Martinez, 2009). Our results confirmed our expectation that compared to the other tested water sources hospital effluents are most strongly contaminated with MRB and ARG. These findings are in agreement with the assumption that MRB are mainly selected in clinical environments (Kümmerer, 2009). Nevertheless, the overall impact of discharging untreated hospital effluents into the sewer system is still under debate. On the one hand, it has been demonstrated that significantly higher proportions of resistant bacteria were isolated from influents of WTPs receiving hospital wastewaters than from influents only treating municipal wastewater (Reinthaler et al., 2003; Garcia et al., 2007). On the other hand, Guardabassi et al. (2002) could not detect significantly elevated levels of resistant Acinetobacter species in wastewater downstream of a hospital. Taking into account that, typically, hospital effluent accounts for less than 1% of the total sewage arriving at the WTP (Kümmerer, 2008), its impact on the receiving environment remains open. In Lausanne, hospital wastewater contributes to approximately 0.4% of the total sewage volume processed at the WTP (calculated from release data of the main building, which accounts for 71% of the “CHUV” (Blanc, 2010). The reduced level of resistance at the WTPin site compared to HOS, is therefore expected. However the high levels of MRB and ARGs observed in the WTP inflow in this study were still high, indicating that municipal wastewater is probably the bigger net source of resistances entering the WTP. Nonetheless, a separate treatment of hospital sewage with the goal of removing MRB (as well as the pharmaceutical load, see below) to prevent their mixing with communal effluents and the environment is desirable.
Our observations indicate that the WTP of Lausanne strongly reduces the total number of MRB. This could, however, be primarily an effect of the reduction of the total bacterial load in the WTP effluent as we did not universally observe a strong reduction of the relative abundance of MRB according to the plate count analysis. In one case (Cla/Tet) we even observed an increase. The culture-independent approach also indicated a less efficient removal, respectively increase, of sul resistance genes during passage of the WTP. This data would suggest that there may be some selection for resistance factors during passage of the WTP. Finally, our results showed increased proportions of highly and extremely MRB among the isolated Sul/Trm/Str resistant strains in the sample of treated wastewater compared to the WTP inlet sample. As the respective bacterial genera of these strains were partly also detected in raw sewage but partly belong to genera only found at the outlet, we assume that there is both a poor removal of certain extremely multiresistant strains (pseudomonads and E. coli) during treatment, as well as a positive selection on multiresistance in other taxa, e.g., Brevundimonas (only detected in hospital sewage and WTPout).
As Sphingobacteria were quite abundant among the isolated highly MRB in the WTP it would be interesting to further investigate their potential as key players in the accumulation and subsequent transfer of highly multiresistant genotypes into the lake. Sphingobacteria are members of the Cytophaga/Flavobacteria/Bacteroidetes (CFB) group which are well known as regular members of freshwater aquatic microbial communities (Zwart et al., 2002). Close relatives of Sphingobacteria have been found in freshwater lakes (Grossart et al., 2008). The high degree of resistance to antimicrobial agents of the group is well known (Holmes, 2006). Another important genus we identified that carried multiple resistance to the lake is Brevundimonas which is infrequently isolated from clinical settings (Han and Andrade, 2005) but is typically established in fresh water communities (Zwart et al., 2002) and drinking water (World Health Organization, 2003), which would indicate a potential of these MRB to become established in the receiving water. These organisms might thus play a role in preserving and disseminating clinical multiresistant determinants in lakes and even promote their return to clinical settings via drinking water.
The ambivalent role of WTPs has been discussed in several reviews (Kim and Aga, 2007; Jury et al., 2010, 2011) and previous studies. The evidence presented so far is however in the majority based on using culture-dependent approaches. Some of these studies observed a general decrease in the prevalence of resistance against antimicrobial agents (Iwane et al., 2001; Guardabassi et al., 2002; Vilanova et al., 2002; Garcia et al., 2007) but still stated that the removal efficiency was unsatisfactory (Reinthaler et al., 2003; Duong et al., 2008; Prado et al., 2008) which is in line with our own findings. Auerbach et al. (2007) quantified various tetracycline resistance genes in influent and effluent of several WTPs and observed a general decrease in tet gene prevalence after sewage treatment. However, others observed a significant increase in antibiotic resistance prevalence, and concluded a selective effect for resistance genes during wastewater treatment (Da Silva et al., 2006; Silva et al., 2006; Zhang et al., 2009). In summary, the picture that emerges is that at least some WTPs select for certain resistance genes and certain multiresistant species, whereas for other species, or under different conditions, a significant removal may be possible. The factors governing the efficiency of removal remain to be determined.
An important factor fostering the selective effect of WTPs is the occurrence of HGT, which can be mediated via mobile genetic elements such as plasmids (conjugation), bacteriophages (transduction), or by uptake of free DNA from the surrounding environment (transformation). In the extreme case these processes might transfer resistance factors from multiresistant pathogens to bacteria which are further disseminated into the receiving aquatic environment. If these organisms subsequently become established in this environment, they will increase the resistance pool in the environment in the long term, which may allow other pathogens to recruit new resistances more easily. It has often been suggested that WTPs might be hot spots for HGT due to the favorable conditions which prevail in this environment, such as high bacterial densities, high oxygen, and high nutrient concentrations (Kim and Aga, 2007). We found a high prevalence of plasmids, partly identified as broad-host range plasmids such as IncP, IncQ, and IncW, in the studied multiresistant wastewater isolates. Detection of sul genes in the plasmid fraction indicated that plasmids carried transferrable antibiotic resistance determinants. This provides some evidence that the occurrence of HGT events and associated spread of ARGs is likely in the WTP environment.
Resistance to important new antibiotic classes such as fluoroquinolones and new generation cephalosporins were in general less prevalent at all our test sites compared to resistances against older antibiotics, such as sulfonamides. A similar outcome of a screening we conducted in 2009 (data not shown) supports this conclusion. Only Gram-positive bacteria showed some degree of tolerance against these substances. However, it remains unclear whether this is due to intrinsic resistance or whether Gram-positives have developed resistance to fluoroquinolones and cephalosporins more successfully. Moreover, whereas for most of the tested substances we found an increased tolerance among selected bacterial isolates at the outlet of the WTP, we could not confirm this pattern for fluoroquinolones. Fluoroquinolone resistance can be mediated by chromosomal mutations that are thought to be transferred vertically and the plasmid-borne qnr genes, which have been described to occur in Gram-negative bacteria (Martínez-Martínez et al., 1998; Cattoir et al., 2007, 2008). We assume that plasmid-mediated resistance is still a less prevalent mechanism at our site compared to chromosomal mutations. Hence, resistance to fluoroquinolones might not be as easily transferable to other bacteria, even in presence of high CIP concentrations in the effluents (Table 3). For instance, among the extremely multiresistant and highly prevalent strains of Sphingobactaria isolated at the outlet, none showed elevated tolerance to fluoroquinolones. It will be interesting to observe whether these resistances become more prevalent in wastewater and environmental communities over the next decades.
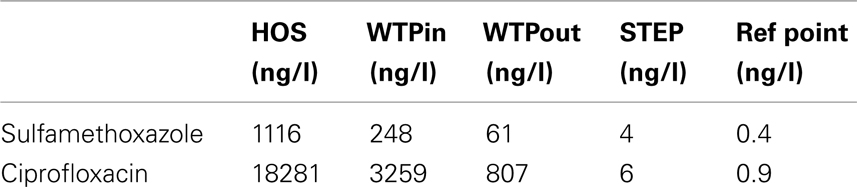
Table 3. Concentrations of ciprofloxacin and sulfamethoxazole determined in raw hospital sewage released from the main building of the CHUV (HOS), at sampling points WTPin and WTPout of Lausanne’s WTP (Blanc, 2010) as well as lake water above the outlet of the WTP discharge pipe (STEP) and a reference point (Bonvin et al., 2011 ), which is located southwest of STEP, 1.5 km from the DP (Swiss coordinates: 533048/150920).
The role of lakes as potential reservoirs of antibiotic resistance has been addressed only infrequently (Jones et al., 1986; Auerbach et al., 2007). Most comparable studies have focused on rivers (Castiglioni et al., 2008; Cummings et al., 2010). One goal of our study was to gain first insights on the impact of discharged wastewaters on the prevalence of MRB and ARGs in the receiving water body, the Vidy Bay of Lake Geneva. In this study we evaluated two sites, one directly at the WTP discharge point and a second site a few kilometers away, close to the drinking water pump of Lausanne. Our data indicate pollution of water and sediments with ARB and resistance genes close to the outlet of the WTP compared to the remote site. Both sites have been analyzed in preliminary experiments in 2009 (data not shown) which revealed a similar outcome as for the data presented here.
However, at both sites the sediments exhibited higher numbers of MRB, than the overlying water column. In the extreme case no MRB were isolated from the water column but isolates were always obtained from the sediments, as, e.g., for Nor/Cef resistant bacteria. As expected, the total bacterial counts were also higher in sediments. Thus, lake sediments appeared as sites where resistance traits persisted and accumulated. The potential of the heavily polluted (e.g., with heavy metals) sediments in Vidy Bay to preserve fecal indicator bacteria has been demonstrated in previous studies (Haller et al., 2009a,b; Poté et al., 2009b) and the impact of the WTP on contamination of the Vidy Bay and its bacterial community is a well-established fact (Pardos et al., 2004; Poté et al., 2008; Bravo et al., 2011; Haller et al., 2011; Thevenon et al., 2011). It is likely that selection and persistence of elevated levels of ARB and resistance genes in close proximity to the WTP outlet are favored, e.g., due to co-selection of antibiotic and heavy metal resistance (Wireman et al., 1997; Stepanauskas et al., 2006). The spatial distribution of resistances in the sediment is studied in more detail in ongoing research.
The question remains open whether the level of ARGs at the point close to the drinking water pump is also influenced by anthropogenic pollution (discharge of wastewaters) or represents typical natural levels of these genes in fresh water environments. Compared to rivers (Iwane et al., 2001; Vilanova et al., 2002; Li et al., 2009), addressing this question in lakes is not an easy task as transport of contaminated water masses and progress of dilution depends on constantly changing wind and temperature regimes, as well as bathymetric characteristics that determine currents and mixing. Tracer experiments releasing bacteriophages from the WTP outlet revealed their transport to within 1.5 km of the drinking water pump within 5–6 h during holomixing in winter time (Goldscheider et al., 2007). A similar tracer study carried out in 1997 even revealed bacteriophages released from the STEP in lake water in the immediate vicinity of the drinking water pump (Wildi and Rossi, 1997) within 48 h. Studies on the transport and fate of antibiotics and other pharmaceuticals in Vidy Bay, which can be ascribed to the WTP discharge with some certainty, were also detected at 1.5 km distance from the DP (reference point, compare Table 3) in direction of the drinking water pump at concentrations in the low nanogram per liter range (Bonvin et al., 2011). Thus, it cannot be ruled out that MRB and ARG released from the WTP might reach the DP and that the detected resistance levels in the present study might result or at least partly result from the impact of anthropogenic pollution and/or selection of resistance due to the presence of low antibiotic concentrations (Gullberg et al., 2011). In addition to direct contamination there is the question whether the chronic release of elevated levels of resistant bacteria, and transfer of resistance vectors into natural microbial populations can alter the natural resistance background in the long term. Studies in soil have indicated that such long-term trends may exist (Knapp et al., 2009). Currently however, we lack data on the natural resistance background and an understanding of the impact of elevated antibiotic resistance in natural environments, which leaves us unable to assess the risks associated with the levels observed here. This is clearly an important goal for research in the future.
In the present study we have analyzed a sample set of different aquatic compartments along a wastewater flow using different methodologies, which can be mainly differentiated into culture-based and culture-independent tools. The culture-based approach nicely allowed us to link antibiotic resistance to active (viable) bacterial cells present in the different environments and identify the bacterial taxa carrying resistances. Further, we were able to characterize detected strains with respect to multiple resistances and mobile genetic elements, such as plasmids and the genetic base for the observed resistance. The great limitation of this method is that only a small proportion of the natural bacterial community is accessible to culturing. It is well known that natural aquatic environments contain large proportions of non-culturable but viable bacteria (Oliver, 2005). Less than 1% of environmental bacteria are culturable, and large parts of the bacterial diversity have so far not been cultured at all (Amann et al., 1995). Even lab strains fail to be cultured when living under harsh conditions such as in fresh water environments (Arana et al., 1997). Large differences (2–4 log units) between numbers obtained from heterotrophic plate counts and FC have been described, e.g., for raw water samples by Hoefel et al. (2003).
The advantages of culture-independent approaches are evident particularly when dealing with oligotrophic environments such as fresh water and other natural ecosystems. Clearly, comparing plate counts to bacterial 16S rRNA gene copy numbers and flow-cytometric cell counts, results obtained from plate counts alone would have underestimated the actual prevalence of antibiotic resistance in the wastewater and especially at the two investigated sites in Lake Geneva. Of course biases for these methods exist as well, e.g., with respect to the method used for DNA extraction, PCR efficiencies, presence of PCR inhibitors or the gating parameters in the flow cytometer. Application of the culture-independent methods, FC, and qPCR, alone would have given no information on biological activity of the quantified bacterial traits (though for FC there exist procedures for-live-dead staining of cells) and no linkage of resistance genes and bacterial taxonomic identity. Determination of multiresistances and detection of the presence of mobile genetic elements within intact cells would likewise not have been possible. As such it has to be taken into account that for the high numbers (compared to the plate counts) of resistance genes and 16S RNA gene copy numbers or flow-cytometric counts, a certain fraction might result from disrupted or dead cells. The presence of multicopy plasmids carrying resistance genes, or the presence of abundant but non-culturable environmental strains may further contribute to the observed disparity between the two approaches. Taking the advantages and short-comings of the discussed methods into account it is perhaps surprising that, studies applying both culture-based and molecular tools in parallel are still limited in number although they can provide substantial information on the actual situation and related processes of antibiotic resistance in different compartments. Most previous studies discussing, e.g., the role of hospital effluents on potential contamination of nearby water sheds, focused on culture-based approaches (Esiobu et al., 2002; Novais et al., 2005; Schwartz et al., 2006; Duong et al., 2008; Manaia et al., 2010). Volkmann et al. (2004) could, e.g., confirm the contamination of different wastewater samples with genes conferring resistance to β-lactam antibiotics and vancomycin, whereas the staphylococci-specific methicillin resistance gene was found in significantly lower amounts. The result supported previous culture-based approaches in which methicillin resistant staphylococci were only isolated from clinical wastewaters (Schwartz et al., 2003).
Due to logistical constraints, particularly the high workload associated with culturing, it was not feasible to replicate the sampling, nor to sample all water sources at the same time, which of course implies that one should be careful in generalizing from the results presented here. Similar limitations are frequently encountered in comparable studies (Schwartz et al., 2006; Auerbach et al., 2007; Szczepanowski et al., 2009). Care was taken to sample under comparable circumstances (i.e., avoiding rainfall) and time of day, particularly in the actual sewage system, to obtain comparable samples. Temporal variation in total bacterial load and resistance counts might of course influence the general trends observed in our study. However, in two comparable studies where temporal variations were taken into account this seemed not to be the case. Da Silva et al. (2006) sampled a WTP in Chile during four separate sampling campaigns, representing spring, summer, autumn, and winter. They found higher fractions of resistant organisms in the warmer periods compared to the winter months, but the numbers usually varied within the same order of magnitude for all sampling events. The effect of treatment was consistent over time, i.e., treated effluents always exhibited higher resistance fractions compared to raw sewage. In contrast, Zhang et al. (2009) reported higher levels of resistance among Acinetobacter isolates from a WTP and its receiving river in the US during a cold and low flow sampling event compared to a warm and high flow sampling event. Still the trend of increased resistance from influent to effluent was observed during both campaigns. Finally, another study by Li et al. (2009) determined total cell counts (DAPI staining) and culturable bacteria (on TSA) of 3.3 ± 1.9 × 107 cells ml−1 and of 4.5 ± 2.1 × 104 CFU ml−1, respectively (averaged over three sampling campaigns carried out in December, April, and August) in the effluent of a WTP treating wastewater from an oxytetracycline production facility in China, indicating a seasonal variability of around 50%. We have carried out a similar campaign to that compared in the present study in 2009 which resulted in comparable trends, further supporting the validity of the data presented here (Nadine Czekalski, unpublished data).
The molecular methods proved valuable and generally the trends observed were in good agreement with the cultivation-based data. Therefore, a more thorough time-resolved and replicated study based on this methodology would be a good approach to overcome the noted limitations.
For future studies aiming to provide deeper insights into resistance background of natural aquatic environments as well as their pollution with resistant bacteria from anthropogenic sources, the use of advanced culture-independent tools, such as metagenomics and high throughput sequencing will be of great value. With respect to linking activity, mobility, and functional traits to bacterial taxa single cell sequencing approaches might probably become a helpful tool too, potentially replacing and even improving on current culture-based approaches.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Francisco Vazquez for substantial support of our lab work, Holger Heuer for kindly providing E. coli strains carrying sul resistance genes, Frederik Hammes for the opportunity to conduct and his support during flow-cytometric measurements, Janine Halder for providing CTD profiles, Alois Zwyssig, Michael Schurter, Philippe Arpagaus for assistance during field campaigns, and Luca Rossi and Nathalie Chèvre for organizing access to the CHUV and WTP sampling sites and providing background data. We would like to thank all members of the ProDoc project for helpful discussions. Laetitia Stoykowski, Pascal Kistler, and Sandro Brunner for assisting with the plate count experiment. Funding was provided by the Swiss National Science Foundation grant PFMP2-123048 for research module 4 of the ProDoc project “Scientific Concepts for the Sustainable Management of Mid-sized Lakes in the 21st Century.”
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/Antimicrobials,_Resistance_and_Chemotherapy/10.3389/fmicb.2012.00106/abstract
References
Alder, A. C., McArdell, C. S., Golet, E. M., Ibric, S., Molnar, E., Nipales Norriel, S., and Giger, W. (2001). “Occurrence and fate of fluoroquinolone, macrolide, and sulfonamide antibiotics during wastewater treatment and in ambient waters in Switzerland,” in Pharmaceuticals and Care Products in the Environment: Scientific and Regulatory Issues, ACS Symposium Series, Vol. 791, C. G. Daughton and T. L. Jones-Lepp (Washington, DC: American Chemical Society), 56–69.
Alonso, A., Sánchez, P., and Martínez, J. L. (2001). Environmental selection of antibiotic resistance genes. Environ. Microbiol. 3, 1–9.
Altschul, S., Gish, W., Miller, W., Myers, E., and Lipman, D. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410.
Amalfitano, S., and Fazi, S. (2008). Recovery and quantification of bacterial cells associated with streambed sediments. J. Microbiol. Methods 75, 237–243.
Amann, R. I., Ludwig, W., and Schleifer, K. H. (1995). Phylogenetic identification and in-situ detection of individual microbial-cells without cultivation. Microbiol. Rev. 59, 143–169.
Aminov, R. I. (2009). The role of antibiotics and antibiotic resistance in nature. Environ. Microbiol. 11, 2970–2988.
Arana, I., Pocino, M., Muela, A., Fernandez-Astorga, A., and Barcina, I. (1997). Detection and enumeration of viable but non-culturable transconjugants of Escherichia coli during the survival of recipient cells in river water. J. Appl. Microbiol. 83, 340–346.
Auerbach, E. A., Seyfried, E. E., and Mcmahon, K. D. (2007). Tetracycline resistance genes in activated sludge wastewater treatment plants. Water Res. 41, 1143–1151.
Baquero, F., Martínez, J.-L., and Cantón, R. (2008). Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 19, 260–265.
Berney, M., Vital, M., Hülshoff, I., Weilenmann, H.-U., Egli, T., and Hammes, F. (2008). Rapid, cultivation-independent assessment of microbial viability in drinking water. Water Res. 42, 4010–4018.
Blanc, L. (2010). Characterisation of Hospital Wastewater: The Case of the CHUV. MSc thesis, Cranfield University, Cranfield.
Blasco, M. D., Esteve, C., and Alcaide, E. (2009). Multiresistant waterborne pathogens isolated from water reservoirs and cooling systems. J. Appl. Microbiol. 105, 469–475.
Bonvin, F., Rutler, R., Che‘Vre, N., Halder, J., and Kohn, T. (2011). Spatial and temporal presence of a wastewater-derived micropollutant plume in Lake Geneva. Environ. Sci. Technol. 45, 4702–4709.
Bravo, A. G., Bouchet, S., Amouroux, D., Pote, J., and Dominik, J. (2011). Distribution of mercury and organic matter in particle-size classes in sediments contaminated by a waste water treatment plant: Vidy Bay, Lake Geneva, Switzerland. J. Environ. Monit. 13, 974–982.
Carattoli, A., Bertini, A., Villa, L., Falbo, V., Hopkins, K. L., and Threlfall, E. J. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63, 219–228.
Castiglioni, S., Pomati, F., Miller, K., Burns, B. P., Zuccato, E., Calamari, D., and Neilan, B. A. (2008). Novel homologs of the multiple resistance regulator marA in antibiotic-contaminated environments. Water Res. 42, 4271–4280.
Cattoir, V., Poirel, L., Aubert, C., Soussy, C.-J., and Nordmann, P. (2008). Unexpected occurrence of plasmid-mediated quinolone resistance determinants in environmental Aeromonas spp. Emerg. Infect. Dis. 14, 231–237.
Cattoir, V., Poirel, L., Rotimi, V., Soussy, C.-J., and Nordmann, P. (2007). Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J. Antimicrob. Chemother. 60, 394–397.
Chèvre, N., Guignard, C., Rossi, L., Pfeiffer, H.-R., Bader, H.-P., and Scheidegger, R. (2011). Substance flow analysis as a tool for urban water management. Water Sci. Technol. 63, 8.
Cummings, D. E., Archer, K. F., Arriola, D. J., Baker, P. A., Faucett, K. G., Laroya, J. B., Pfeil, K. L., Ryan, C. R., Ryan, K. R. U., and Zuill, D. E. (2010). Broad dissemination of plasmid-mediated quinolone resistance genes in sediments of two urban coastal wetlands. Environ. Sci. Technol. 45, 447–454.
Da Silva, M. F., Tiago, I., Veríssimo, A., Boaventura, R. A., Nunes, O. C., and Manaia, C. M. (2006). Antibiotic resistance of enterococci and related bacteria in an urban wastewater treatment plant. FEMS Microbiol. Ecol. 55, 322–329.
DeSantis, T. Z., Hugenholtz, P., Larsen, N., Rojas, M., Brodie, E. L., Keller, K., Huber, T., Dalevi, D., Hu, P., and Andersen, G. L. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072.
Deutsches Institut Für Normung e. V. (DIN). (2004). Medizinische Mikrobiologie-Empfindlichkeit-sprüfung von Mikrobiellen Kran-kheitserregern gegen Chemothera-peutika-Teil 4: Bewertungsstufen für die minimale Hemmkonzentration; MHK-Grenzwerte von antibakteriellen Wirkstoffen; DIN 58940-4, Bbl, 1:2004-2. Berlin: Beuth Verlag.
Duong, H. A., Pham, N. H., Nguyen, H. T., Hoang, T. T., Pham, H. V., Pham, V. C., Berg, M., Giger, W., and Alder, A. C. (2008). Occurrence, fate and antibiotic resistance of fluoroquinolone antibacterials in hospital wastewaters in Hanoi, Vietnam. Chemosphere 72, 968–973.
Escher, B. I., Baumgartner, R., Koller, M., Treyer, K., Lienert, J., and McArdell, C. S. (2011). Environmental toxicology and risk assessment of pharmaceuticals from hospital wastewater. Water Res. 45, 75–92.
Esiobu, N., Armenta, L., and Ike, J. (2002). Antibiotic resistance in soil and water environments. Int. J. Environ. Health Res. 12, 133–144.
European Committee on Antimicrobial Susceptibility Testing (EUCAST). (2011). Data from the EUCAST MIC and zone diameter distribution website. Available at: http://www.eucast.org [accessed January 20, 2011].
Filippini, M., Masiero, G., and Moschetti, K. (2006). Socioeconomic determinants of regional differences in outpatient antibiotic consumption: evidence from Switzerland. Health Policy (New York) 78, 77–92.
Garcia, S., Wade, B., Bauer, C., Craig, C., Nakaoka, K., and Lorowitz, W. (2007). The effect of wastewater treatment on antibiotic resistance in Escherichia coli and Enterococcus sp. Water Environ. Res. 79, 2387–2395.
Giger, W., Alder, A. C., Golet, E. M., Kohler, H. P. E., McArdell, C. S., Molnar, E., Siegrist, H., and Suter, M. J. F. (2003). Occurrence and fate of antibiotics as trace contaminants in wastewaters, sewage sludges, and surface waters. Chimia (Aarau) 57, 485–491.
Goldscheider, N., Haller, L., Pote, J., Wildi, W., and Zopfi, J. (2007). Characterizing water circulation and contaminant transport in Lake Geneva using bacteriophage tracer experiments and limnological methods. Environ. Sci. Technol. 41, 5252–5258.
Gotz, A., Pukall, R., Smit, E., Tietze, E., Prager, R., Tschape, H., Van Elsas, J., and Smalla, K. (1996). Detection and characterization of broad-host-range plasmids in environmental bacteria by PCR. Appl. Environ. Microbiol. 62, 2621–2628.
Grossart, H.-P., Jezbera, J., Hornák, K., Hutalle, K. M. L., Buck, U., and Šimek, K. (2008). Top-down and bottom-up induced shifts in bacterial abundance, production and community composition in an experimentally divided humic lake. Environ. Microbiol. 10, 635–652.
Guardabassi, L., Lo Fo Wong, D. M. A., and Dalsgaard, A. (2002). The effects of tertiary wastewater treatment on the prevalence of antimicrobial resistant bacteria. Water Res. 36, 1955–1964.
Gullberg, E., Cao, S., Berg, O. G., Ilbäck, C., Sandegren, L., Hughes, D., and Andersson, D. I. (2011). Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog. 7, e1002158. doi:10.1371/journal.ppat.1002158
Haller, L., Amedegnato, E., Poté, J., and Wildi, W. (2009a). Influence of freshwater sediment characteristics on persistence of fecal indicator bacteria. Water Air Soil Pollut. 203, 217–227.
Haller, L., Pote, J., Loizeau, J. L., and Wildi, W. (2009b). Distribution and survival of faecal indicator bacteria in the sediments of the Bay of Vidy, Lake Geneva, Switzerland. Ecol. Indic. 9, 540–547.
Haller, L., Tonolla, M., Zopfi, J., Peduzzi, R., Wildi, W., and Poté, J. (2011). Composition of bacterial and archaeal communities in freshwater sediments with different contamination levels (Lake Geneva, Switzerland). Water Res. 45, 1213–1228.
Han, X. Y., and Andrade, R. A. (2005). Brevundimonas diminuta infections and its resistance to fluoroquinolones. J. Antimicrob. Chemother. 55, 853–859.
Heuer, H., Focks, A., Lamshöft, M., Smalla, K., Matthies, M., and Spiteller, M. (2008). Fate of sulfadiazine administered to pigs and its quantitative effect on the dynamics of bacterial resistance genes in manure and manured soil. Soil Biol. Biochem. 40, 1892–1900.
Heuer, H., and Smalla, K. (2007). Manure and sulfadiazine synergistically increased bacterial antibiotic resistance in soil over at least two months. Environ. Microbiol. 9, 657–666.
Hoefel, D., Grooby, W. L., Monis, P. T., Andrews, S., and Saint, C. P. (2003). Enumeration of water-borne bacteria using viability assays and flow cytometry: a comparison to culture-based techniques. J. Microbiol. Methods 55, 585–597.
Holmes, B. (2006). “The genera Flavobacterium, Sphingobacterium and Weeksella,” in The Prokaryotes, 3rd Edn, eds. M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (New York: Springer), 539–548.>
Hönerlage, W., Hahn, D., and Zeyer, J. (1995). Detection of mRNA of nRPM in Bacillus megaterium ATTC 14581 grown in soil by whole-cell hybridization. Arch. Microbiol. 163, 235–241.
Iwane, T., Urase, T., and Yamamoto, K. (2001). Possible impact of treated wastewater discharge on incidence of antibiotic resistant bacteria in river water. Water Sci. Technol. 43, 91–99.
Jones, J. G., Gardener, S., Simon, B. M., and Pickup, R. W. (1986). Antibiotic resistant bacteria in Windermere and two remote upland tarns in the English Lake District. J. Appl. Microbiol. 60, 443–453.
Jury, K. L., Khan, S. J., Vancov, T., Stuetz, R. M., and Ashbolt, N. J. (2011). Are sewage treatment plants promoting antibiotic resistance? Crit. Rev. Environ. Sci. Technol. 41, 243–270.
Jury, K. L., Vancov, T., Stuetz, R. M., and Khan, S. J. (2010). “Antibiotic resistance dissemination and sewage treatment plants,” in Current Research, Technology and Education Topics in Applied Microbiology and Microbial Biotechnology, ed. A. Mendez-Vilas (Badajoz: Formatex Research Center), 509–519.
Kemper, N. (2008). Veterinary antibiotics in the aquatic and terrestrial environment. Ecol. Indic. 8, 1–13.
Kim, S., and Aga, D. S. (2007). Potential ecological and human health impacts of antibiotics and antibiotic-resistant bacteria from wastewater treatment plants. J. Toxicol. Environ. Health B Crit. Rev. 10, 559–573.
Knapp, C. W., Dolfing, J., Ehlert, P. A. I., and Graham, D. W. (2009). Evidence of increasing antibiotic resistance gene abundances in archived soils since 1940. Environ. Sci. Technol. 44, 580–587.
Kümmerer, K. (2008). Pharmaceuticals in the Environment. Sources, Effects and Risk. Berlin/Heidelberg: Springer.
Kümmerer, K. (2009). Antibiotics in the aquatic environment – a review – part II. Chemosphere 75, 435–441.
Lane, D. J. (1991). “16S/23S rRNA sequencing,” in Nucleic Acid Techniques in Bacterial Systematics, eds E. Stackebrandt and M. Goodfellow (New York, NY: John Wiley and Sons), 115–175.
Lanz, R., Kuhnert, P., and Boerlin, P. (2003). Antimicrobial resistance and resistance gene determinants in clinical Escherichia coli from different animal species in Switzerland. Vet. Microbiol. 91, 73–84.
Levchuk, A., Bulyga, I., Izmalkova, T., Sevast’ianovich, I., Kosheleva, I., Thomas, C., and Titok, M. (2006). Nah-plasmids of IncP-9 group from natural strains of Pseudomonas. Mol. Biol. 40, 835–843.
Li, D., Yu, T., Zhang, Y., Yang, M., Li, Z., Liu, M., and Qi, R. (2009). Antibiotic resistance characteristics of environmental bacteria from an oxytetracycline production wastewater treatment plant and the receiving river. Appl. Environ. Microbiol. 76, 3444–3451.
Life Technologies Corporation. (2012). Available at: http://www.invitrogen.com/site/us/en/home/Products-and-Services/Applications/PCR/real-time-pcr/qpcr-education/real-time-pcr-troubleshooting-tool/gene-expression-quantitation-troubleshooting/poor-pcr-efficiency.html [accessed January 30, 2012].
Lindahl, V. (1996). Improved soil dispersion procedures for total bacterial counts, extraction of indigenous bacteria and cell survival. J. Microbiol. Methods 25, 279–286.
Loizeau, J.-L., Pardos, M., Monna, F., Peytremann, C., Haller, L., and Dominik, J. (2004). The impact of a sewage treatment plant’s effluent on sediment quality in a small bay in Lake Geneva (Switzerland–France). Part 2: Temporal evolution of heavy metals. Lakes Reserv. Res. Manag. 9, 53–63.
Manaia, C., Novo, A., Coelho, B., and Nunes, O. (2010). Ciprofloxacin resistance in domestic wastewater treatment plants. Water Air Soil Pollut. 208, 335–343.
Martinez, J. L. (2008). Antibiotics and antibiotic resistance genes in natural environments. Science 321, 365–367.
Martinez, J. L. (2009). Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ. Pollut. 157, 2893–2902.
Martínez-Martínez, L., Pascual, A., and Jacoby, G. A. (1998). Quinolone resistance from a transferable plasmid. Lancet 351, 797–799.
McArdell, C. S., Molnar, E., Suter, M. J. F., and Giger, W. (2003). Occurrence and fate of macrolide antibiotics in wastewater treatment plants and in the Glatt Valley watershed, Switzerland. Environ. Sci. Technol. 37, 5479–5486.
Newcombe, D., Stuecker, T., La Duc, M., and Venkateswaran, K. (2005). “Q-PCR based bioburden assessment of drinking water throughout treatment and delivery to the International Space Station,” in 35th International Conference on Environmental Systems (ICES); 8th European Symposium on Space Environmental Control Systems (ESSECS) (Rome, Italy/Pasadena, CA: Jet Propulsion Laboratory, National Aeronautics and Space Administration).
Novais, C., Coque, T. M., Ferreira, H., Sousa, J. C., and Peixe, L. (2005). Environmental contamination with vancomycin-resistant enterococci from hospital sewage in Portugal. Appl. Environ. Microbiol. 71, 3364–3368.
NRP49. (2007). National Research Programme NRP 49. Antibiotic Resistance. Final Report, eds F. Baggi, J. C. Piffaretti, and F. Lurati (Bern: Schweizerischer Nationalfonds zur Förderung der wissenschaftlichen Forschung. Abteilung IV, Orientierte Forschung, Nationale Forschungsprogramme).
Pardos, M., Benninghoff, C., De Alencastro, L. F., and Wildi, W. (2004). The impact of a sewage treatment plant’s effluent on sediment quality in a small bay in Lake Geneva (Switzerland–France). Part 1: Spatial distribution of contaminants and the potential for biological impacts. Lakes Reserv. Res. Manag. 9, 41–52.
Pei, R., Kim, S.-C., Carlson, K. H., and Pruden, A. (2006). Effect of river landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Water Res. 40, 2427–2435.
Perreten, V., and Boerlin, P. (2003). A new sulfonamide resistance gene (sul3) in Escherichia coli is widespread in the pig population of Switzerland. Antimicrob. Agents Chemother. 47, 1169–1172.
Picao, R. C., Poirel, L., Demarta, A., Silva, C. S. F., Corvaglia, A. R., Petrini, O., and Nordmann, P. (2008). Plasmid-mediated quinolone resistance in Aeromonas allosaccharophila recovered from a Swiss lake. J. Antimicrob. Chemother. 62, 948–950.
Poté, J., Goldscheider, N., Haller, L., Zopfi, J., Khajehnouri, F., and Wildi, W. (2009a). Origin and spatial–temporal distribution of faecal bacteria in a bay of Lake Geneva, Switzerland. Environ. Monit. Assess. 154, 337–348.
Poté, J., Haller, L., Kottelat, R., Sastre, V., Arpagaus, P., and Wildi, W. (2009b). Persistence and growth of faecal culturable bacterial indicators in water column and sediments of Vidy Bay, Lake Geneva, Switzerland. J. Environ. Sci. (China) 21, 62–69.
Poté, J., Haller, L., Loizeau, J.-L., Garcia Bravo, A., Sastre, V., and Wildi, W. (2008). Effects of a sewage treatment plant outlet pipe extension on the distribution of contaminants in the sediments of the Bay of Vidy, Lake Geneva, Switzerland. Bioresour. Technol. 99, 7122–7131.
Prado, T., Pereira, W. C., Silva, D. M., Seki, L. M., Carvalho, A. P. D. A., and Asensi, M. D. (2008). Detection of extended-spectrum β-lactamase-producing Klebsiella pneumoniae in effluents and sludge of a hospital sewage treatment plant. Lett. Appl. Microbiol. 46, 136–141.
Reasoner, D. J., and Geldreich, E. E. (1985). A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 49, 1–7.
Reinthaler, F. F., Posch, J., Feierl, G., Wüst, G., Haas, D., Ruckenbauer, G., Mascher, F., and Marth, E. (2003). Antibiotic resistance of E. coli in sewage and sludge. Water Res. 37, 1685–1690.
Schwartz, T., Kohnen, W., Jansen, B., and Obst, U. (2003). Detection of antibiotic-resistant bacteria and their resistance genes in wastewater, surface water, and drinking water biofilms. FEMS Microbiol. Ecol. 43, 325–335.
Schwartz, T., Volkmann, H., Kirchen, S., Kohnen, W., Schon-Holz, K., Jansen, B., and Obst, U. (2006). Real-time PCR detection of Pseudomonas aeruginosa in clinical and municipal wastewater and genotyping of the ciprofloxacin-resistant isolates. FEMS Microbiol. Ecol. 57, 158–167.
Sengelov, G., Agerso, Y., Halling-Sorensen, B., Baloda, S. B., Andersen, J. S., and Jensen, L. B. (2003). Bacterial antibiotic resistance levels in Danish farmland as a result of treatment with pig manure slurry. Environ. Int. 28, 587–595.
Silva, J., Castillo, G., Callejas, L., López, H., and Olmos, J. (2006). Frequency of transferable multiple antibiotic resistance amongst coliform bacteria isolated from a treated sewage effluent in Antofagasta, Chile. Electron. J. Biotechnol. 9, 533–540.
Stepanauskas, R., Glenn, T. C., Jagoe, C. H., Tuckfield, R. C., Lindell, A. H., King, C. J., and Mcarthur, J. V. (2006). Coselection for microbial resistance to metals and antibiotics in freshwater microcosms. Environ. Microbiol. 8, 1510–1514.
Suzuki, M. T., Taylor, L. T., and Delong, E. F. (2000). Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 66, 4605–4614.
Szczepanowski, R., Linke, B., Krahn, I., Gartemann, K.-H., Gützkow, T., Eichler, W., Pühler, A., and Schlüter, A. (2009). Detection of 140 clinically relevant antibiotic-resistance genes in the plasmid metagenome of wastewater treatment plant bacteria showing reduced susceptibility to selected antibiotics. Microbiology 155, 2306–2319.
Thevenon, F., Graham, N. D., Herbez, A., Wildi, W., and Poté, J. (2011). Spatio-temporal distribution of organic and inorganic pollutants from Lake Geneva (Switzerland) reveals strong interacting effects of sewage treatment plant and eutrophication on microbial abundance. Chemosphere 84, 609–617.
Troussellier, M., Courties, C., and Zettelmaier, S. (1995). Flow cytometric analysis of coastal lagoon bacterioplankton and picophytoplankton: fixation and storage effects. Estuar. Coast. Shelf Sci. 40, 621–633.
Vilanova, X., Manero, A., Cerdà-Cuéllar, M., and Blanch, A. R. (2002). The effect of a sewage treatment plant effluent on the faecal coliforms and enterococci populations of the reception river waters. J. Appl. Microbiol. 92, 210–214.
Vioget, P., Jaquerod, C.-A., Ruegg, E., and Poget, E. (2011). Bilans 2010 de l’épuration vaudoise. Orbe, Savigny, Switzerland: Service des eaux, sols et assainissement, Canton de Vaud.
Volkmann, H., Schwartz, T., Bischoff, P., Kirchen, S., and Obst, U. (2004). Detection of clinically relevant antibiotic-resistance genes in municipal wastewater using real-time PCR (TaqMan). J. Microbiol. Methods 56, 277–286.
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267.
Wegmüller, S., and Schmid, S. (2009). Plasmid DNA isolation from the Gram-positive Bacterium Pediococcus damnosus Using a Modified PureYield™ Plasmid Midiprep System Protocol Promega. Available at: http://www.promega.com/resources/articles/pubhub/enotes/plasmid-dna-isolation-from-the-gram-positive-bacterium-pediococcus-damnosus/ (retrieved April 7, 2012).
Wildi, W., and Rossi, P. (1997). Assainissement de la baie de Vidy: tarcage par bactériophages 19-25 février 1997 6-11 mai 1997 (Lausanne: Ville de Lausanne, Direction de la sécurité cociale et de lénvironnement, Service d’assainissement, Station d’épuration de Vidy, Université de Genève, Institut F.A. Forel, Section des sciences de la terre, Université Neuchâtel, Laboratoire de Microbiologie).
Wireman, J., Liebert, C., Smith, T., and Summers, A. (1997). Association of mercury resistance with antibiotic resistance in the Gram-negative fecal bacteria of primates. Appl. Environ. Microbiol. 63, 4494–4503.
World Health Organization. (2003). “Relationships between common water bacteria and pathogens in drinking-water,” in Heterotrophic Plate Counts and Drinking-water Safety, eds. J. Bertram, J. Cotruvo, M. Exner, C. Fricker, and A. Glasmacher (London: IWA Publishing), 80–118.
Young, H.-K. (1993). Antimicrobial resistance spread in aquatic environments. J. Antimicrob. Chemother. 31, 627–635.
Zhang, Y., Marrs, C. F., Simon, C., and Xi, C. (2009). Wastewater treatment contributes to selective increase of antibiotic resistance among Acinetobacter spp. Sci. Total Environ. 407, 3702–3706.
Zwart, G., Crump, B. C., Agterveld, M. P. K.-V., Hagen, F., and Han, S.-K. (2002). Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat. Microb. Ecol. 28, 141–155.
Appendix
Keywords: aquatic, environment, sewage, pollution, sediment, qPCR, antibiotics
Citation: Czekalski N, Berthold T, Caucci S, Egli A and Bürgmann H (2012) Increased levels of multiresistant bacteria and resistance genes after wastewater treatment and their dissemination into Lake Geneva, Switzerland. Front. Microbio. 3:106. doi: 10.3389/fmicb.2012.00106
Received: 07 November 2011; Accepted: 02 March 2012;
Published online: 22 March 2012.
Edited by:
Sylvie Nazaret, Centre National de la Recherche Scientifique, FranceReviewed by:
Chris Rensing, RTI International, USAJulie Zilles, University of Illinois Urbana-Champaign, USA
Copyright: © 2012 Czekalski, Berthold, Caucci, Egli and Bürgmann. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Nadine Czekalski, Department of Surface Waters – Research and Management, Eawag: Swiss Federal Institute of Aquatic Science and Technology, Seestrasse 79, 6047 Kastanienbaum, Switzerland. e-mail:bmFkaW5lLmN6ZWthbHNraUBlYXdhZy5jaA==