
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 28 February 2012
Sec. Terrestrial Microbiology
Volume 3 - 2012 | https://doi.org/10.3389/fmicb.2012.00072
This article is part of the Research TopicMicrobiology of wetlandsView all 18 articles
Freshwater wetlands are a major source of the greenhouse gas methane but at the same time can function as carbon sink. Their response to global warming and environmental pollution is one of the largest unknowns in the upcoming decades to centuries. In this review, we highlight the role of sulfate-reducing microorganisms (SRM) in the intertwined element cycles of wetlands. Although regarded primarily as methanogenic environments, biogeochemical studies have revealed a previously hidden sulfur cycle in wetlands that can sustain rapid renewal of the small standing pools of sulfate. Thus, dissimilatory sulfate reduction, which frequently occurs at rates comparable to marine surface sediments, can contribute up to 36–50% to anaerobic carbon mineralization in these ecosystems. Since sulfate reduction is thermodynamically favored relative to fermentative processes and methanogenesis, it effectively decreases gross methane production thereby mitigating the flux of methane to the atmosphere. However, very little is known about wetland SRM. Molecular analyses using dsrAB [encoding subunit A and B of the dissimilatory (bi)sulfite reductase] as marker genes demonstrated that members of novel phylogenetic lineages, which are unrelated to recognized SRM, dominate dsrAB richness and, if tested, are also abundant among the dsrAB-containing wetland microbiota. These discoveries point toward the existence of so far unknown SRM that are an important part of the autochthonous wetland microbiota. In addition to these numerically dominant microorganisms, a recent stable isotope probing study of SRM in a German peatland indicated that rare biosphere members might be highly active in situ and have a considerable stake in wetland sulfate reduction. The hidden sulfur cycle in wetlands and the fact that wetland SRM are not well represented by described SRM species explains their so far neglected role as important actors in carbon cycling and climate change.
Freshwater wetlands (from here on referred to as wetlands) comprise diverse habitats ranging from peatlands with and without permafrost (e.g., ombrotrophic bogs and minerotrophic fens), freshwater marshes (e.g., the Everglades or the Okavango delta), freshwater swamps including riparian zones, constructed wetlands for waste water treatment, and agricultural wetlands (Mitsch et al., 2009; Kögel-Knabner et al., 2010; Vymazal, 2010). All wetlands have in common that they are periodically or permanently water-saturated soil environments with a characteristic vegetation and a water table at or close to the soil surface (Mitsch and Gosselink, 2007). As a consequence, they are characterized by steep gradients in soil redox conditions that sustain a complex pattern of biogeochemical cycling of elements (Limpens et al., 2008; Kögel-Knabner et al., 2010). Natural wetlands can act as a net sink of carbon depending on wetland type, age, and location as well as the prevailing climate and environmental conditions (Blodau, 2002; Bridgham et al., 2006; Kayranli et al., 2010). Over the past millennia, they accumulated up to one third of the terrestrial organic carbon just considering peatlands (Limpens et al., 2008), which represents half of the carbon that is in the atmosphere as carbon dioxide (Dise, 2009). At the same time, anthropogenic wetlands such as rice paddies are maintained for agriculture to provide food for more than 50% of the world’s population (http://beta.irri.org).
Organic carbon degradation in wetlands is catalyzed by different functional guilds of aerobic and anaerobic microorganisms, whose competition for electron donors or syntrophic cooperation determines how much of the carbon loss from wetlands proceeds through the emission of carbon dioxide or the more potent greenhouse gas methane (Clymo, 1984; Yao et al., 1999; Dedysh et al., 2001; Frolking et al., 2001; Conrad, 2002; Horn et al., 2003; Loy et al., 2004; Beer et al., 2008; Hamberger et al., 2008; Küsel et al., 2008; Wüst et al., 2009). Although natural and anthropogenic wetlands together cover just 7% of Earth’s land surface (Batzer and Sharitz, 2006; Limpens et al., 2008; Kögel-Knabner et al., 2010), they are made responsible for 30–40% of the global emission of methane (Houweling et al., 1999; Wuebbles and Hayhoe, 2002). It is currently not clear how the carbon balance of wetlands will change in the upcoming decades to centuries due to global warming, which is connected to rising atmospheric carbon dioxide levels and changes in precipitation amount and frequency (IPCC, 2007; Dise, 2009). Such variations of environmental conditions on short- and long-term scales have important implications for the biogeochemical cycling of elements in wetlands and govern transitions between synergistic and antagonistic trophic interactions among wetland microorganisms, which ultimately also determine the extent of carbon mineralization that is channeled through methanogenesis.
Sulfur cycling in wetlands has been studied since the 1980s, revealing that sulfate reduction operates at rates that are comparable to marine surface sediments, where this process is the most important anaerobic degradation pathway for organic matter (Jørgensen, 1982 and references in Table 1). Nevertheless, the importance of sulfate reduction for wetland biogeochemistry has remained underestimated because standing pools of sulfate are typically in the lower micromolar-range and were thus generally interpreted to be too low to sustain sulfate reduction over longer periods of time. This perception was perpetuated by the fact that known taxa of sulfate-reducing microorganisms (SRM) were usually not detected at all or constituted only a very minor fraction of the wetland microbiota (Costello and Schmidt, 2006; Dedysh et al., 2006; Kraigher et al., 2006). However, a series of studies provided cumulative evidence for a hidden sulfur cycle that contributes to rapid recycling of sulfide to sulfate in low-sulfate environments and thus sustains the observed high sulfate reduction rates (SRR; Wieder and Lang, 1988; Jørgensen, 1990; Elsgaard and Jørgensen, 1992; Wind and Conrad, 1997; Mandernack et al., 2000; Heitmann and Blodau, 2006; Blodau et al., 2007; Heitmann et al., 2007; Knorr and Blodau, 2009; Knorr et al., 2009).
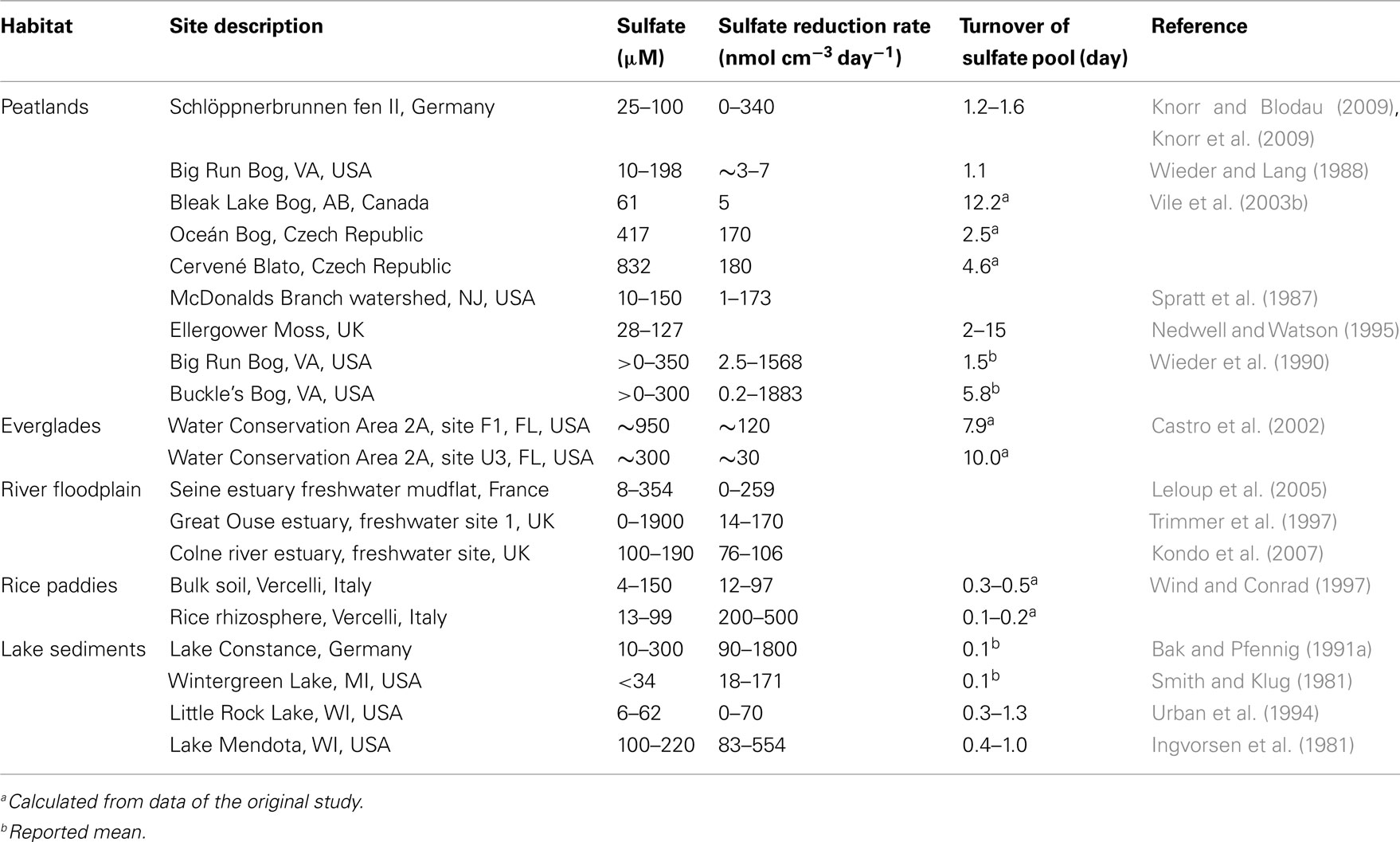
Table 1. Summary of sulfate reduction rates in environments with low-sulfate concentrations (μM-range) as determined with 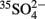 -radiotracer methods.
-radiotracer methods.
Because SRM couple sulfate dissimilation with heterotrophic carbon degradation or carbon dioxide fixation, sulfate reduction directly influences the carbon cycle in wetlands. SRM are known to be metabolically versatile and are able to utilize a great variety of substrates ranging from hydrogen, short-chained fatty acids, and other degradation intermediates like ethanol and lactate up to monosaccharides, amino acids, aromatic compounds, alkanes, and alkenes (Rabus et al., 2006; Muyzer and Stams, 2008). In contrast, organic polymers such as cellulose, proteins, or DNA and RNA are typically not degraded by known SRM (Muyzer and Stams, 2008). Of importance is that SRM are energetically favored in the competition for substrates with microorganisms involved in the methanogenic degradation pathways, resulting in a considerable diversion of the carbon flow from methane to carbon dioxide (Gauci et al., 2004). This mitigating effect of sulfate reduction on the methane flux from wetlands is expected to become even more pronounced in the near future (Gauci et al., 2004). Despite successful efforts to reduce aerial sulfur pollution in developed countries, global SO2 emission is predicted to increase in the next decades due to increasing untreated combustion of coal and other fossil fuels in developing countries situated mainly in Asia (Smith et al., 2005; Ward, 2009). Subsequent sulfuric acid deposition on wetlands by acid rain is predicted to stimulate sulfate reduction and thus to cause a suppression of global wetland methane emission by up to 15% (Gauci et al., 2004).
Here, we provide an overview on the process of sulfate reduction in wetlands, with a focus on ombrotrophic and minerotrophic peatlands, river floodplains, and rice paddies. The first part of this review highlights the biogeochemistry of sulfate reduction in such wetlands, with insights into possible mechanisms that could sustain rapid sulfur cycling. The second part summarizes current knowledge on the identity and activity of SRM in wetlands, including a meta-analysis of all sequences of the marker genes dsrAB that were recovered from wetland microorganisms.
Sulfate concentrations in natural wetlands and rice paddy soils are typically in the micromolar-range and are thus often considered to be too low to sustain long-term sulfate reduction. However, δ34S values and the fact that wetlands can function as sulfate sink despite their often low anion exchange capacity indicate that sulfate reduction is occurring over long time periods in these low-sulfate environments (Spratt et al., 1987; Alewell and Gehre, 1999; Groscheova et al., 2000; Alewell and Novak, 2001; Jacks and Norrström, 2004; Novak et al., 2005; Paul et al., 2006; Alewell et al., 2008). Even more striking, measured SRR can vary dramatically over time and space in freshwater wetlands, ranging from non-detectable to >100 nmol cm−3 day−1 and in extreme cases reaching even values of >1.000 nmol cm−3 day−1 (Table 1). Often these rates are in the same order of magnitude as observed SRR in marine surface sediments, which are characterized by much higher sulfate concentrations (mM-range) and where sulfate reduction was shown to be the most important anaerobic degradation process (Jørgensen, 1982; Howarth and Jørgensen, 1984; Skyring, 1987). These high SRR in freshwater wetlands initially raised concerns about the reliability of such measurements, because these numbers would indicate a very rapid turnover of the complete sulfate pools within hours to days and necessitate a rapid recycling of sulfate. In many studies, SRR in low-sulfate environments were measured by the 35S-radiotracer assay, where 35S-labeled sulfate is added to either intact soil or sediment cores or to slurries thereof (Jørgensen, 1978; Chapman and Davidson, 2001; King, 2001). Thereafter, the turnover of labeled sulfate to sulfide is analyzed after a given time interval (e.g., 90 min). The produced sulfide either stays in solution as hydrogen sulfide or rapidly reacts to various forms of metal mono- or disulfides (mostly in combination with iron) or to elemental sulfur. Therefore, the 35S-label is recovered from the sum of these inorganic reduced sulfur species and used in combination with the initial amount of 35S-sulfate tracer, the overall sulfate pool, and the incubation time to calculate SRR (for details see Kallmeyer et al., 2004). Although regarded as the most reliable method to determine SRR, there are indeed a number of caveats (Jørgensen, 1978; Moeslund et al., 1994; Kallmeyer et al., 2004; Canfield et al., 2005). For example, an overestimation of sulfate concentrations, 35S-isotope exchange between sulfate and more reduced sulfur species (S2−, 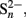 S0), and a stimulation of SRM by the added 35S-sulfate could lead to an overestimation of the measured SRR. However, sulfate concentrations can be reliably measured down to 0.15 μM (Tarpgaard et al., 2011), 35S-isotope exchange between sulfate and the more reduced sulfur species was shown not to occur within 144 h (Fossing and Jørgensen, 1990), which is much longer than the typical incubations times of 30–90 min (Wind and Conrad, 1997; Knorr and Blodau, 2009; Knorr et al., 2009) or the sometimes extended incubations times of up to 48 h (e.g., Vile et al., 2003b), and the added tracer (1 nmol per injection of ca. 50 kBq
S0), and a stimulation of SRM by the added 35S-sulfate could lead to an overestimation of the measured SRR. However, sulfate concentrations can be reliably measured down to 0.15 μM (Tarpgaard et al., 2011), 35S-isotope exchange between sulfate and the more reduced sulfur species was shown not to occur within 144 h (Fossing and Jørgensen, 1990), which is much longer than the typical incubations times of 30–90 min (Wind and Conrad, 1997; Knorr and Blodau, 2009; Knorr et al., 2009) or the sometimes extended incubations times of up to 48 h (e.g., Vile et al., 2003b), and the added tracer (1 nmol per injection of ca. 50 kBq 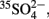 , Kallmeyer et al., 2004) represents a negligible fraction of the total sulfate pool, even at wetland sulfate concentrations of only 1 μM. Also impurities in commercially supplied
, Kallmeyer et al., 2004) represents a negligible fraction of the total sulfate pool, even at wetland sulfate concentrations of only 1 μM. Also impurities in commercially supplied 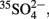 , which can constitute up to 0.05% of the total radioactivity and co-extract with reduced sulfur compounds during turnover measurements (Kallmeyer et al., 2004), would not explain the high SRR measured in wetland samples. On the contrary, an underestimation of SRR in freshwater wetlands is often more likely, especially when prolonged incubations times are used (several hours to days), because of (i) re-oxidation of reduced sulfur compounds to sulfate under anoxic conditions (Jørgensen, 1990; Elsgaard and Jørgensen, 1992; Fossing, 1995), (ii) 35S-tracer back flux from sulfide to sulfate due to the reversibility of enzyme reactions within the sulfate reduction pathway (Holler et al., 2011), and (iii) the possible incorporation of reduced sulfur species and thus of tracer into organic matter (Wieder and Lang, 1988; Wind and Conrad, 1997; Chapman and Davidson, 2001), which would not be detected by the standard 35S-radiotracer assay (Canfield et al., 1986; Fossing and Jørgensen, 1989; Kallmeyer et al., 2004).
, which can constitute up to 0.05% of the total radioactivity and co-extract with reduced sulfur compounds during turnover measurements (Kallmeyer et al., 2004), would not explain the high SRR measured in wetland samples. On the contrary, an underestimation of SRR in freshwater wetlands is often more likely, especially when prolonged incubations times are used (several hours to days), because of (i) re-oxidation of reduced sulfur compounds to sulfate under anoxic conditions (Jørgensen, 1990; Elsgaard and Jørgensen, 1992; Fossing, 1995), (ii) 35S-tracer back flux from sulfide to sulfate due to the reversibility of enzyme reactions within the sulfate reduction pathway (Holler et al., 2011), and (iii) the possible incorporation of reduced sulfur species and thus of tracer into organic matter (Wieder and Lang, 1988; Wind and Conrad, 1997; Chapman and Davidson, 2001), which would not be detected by the standard 35S-radiotracer assay (Canfield et al., 1986; Fossing and Jørgensen, 1989; Kallmeyer et al., 2004).
A number of peatland studies reported excess carbon dioxide over methane production under anoxic conditions; results that could not be explained by methanogenesis as the sole terminal degradation process (Segers and Kengen, 1998; Vile et al., 2003a; Yavitt and Seidmann-Zager, 2006; Blodau et al., 2007; Keller and Bridgham, 2007; Wüst et al., 2009). It was hypothesized that the excess carbon dioxide may stem from fermentation (Vile et al., 2003a,b), but the typically lacking accumulation of fermentation products (such as hydrogen or short chain fatty acids) lead to the hypothesis that this carbon dioxide may stem from alternative anaerobic degradation processes such as sulfate reduction, iron reduction, organic matter reduction, or a combination thereof (Wieder et al., 1990; Segers and Kengen, 1998; Blodau et al., 2007; Küsel et al., 2008; Limpens et al., 2008). This was supported by the fact that high SRR prevailed even under prolonged anoxic periods (16–228 days) and under depletion of the sulfate pools below a detection limit of about 0.5 μM in large peat soil mesocosms in the laboratory as well as in field plots (Figures 1A–D; Knorr and Blodau, 2009; Knorr et al., 2009). The actual contribution of sulfate reduction to anaerobic carbon mineralization in wetlands has not yet been conclusively elucidated and so far mostly relies on incubation assays rather than on in situ measurements. Nevertheless, for peatlands current estimates range from 0 to 50% using inhibition assays of SRM by the addition of molybdate, which functions as an intracellular competitor to sulfate (Blodau et al., 2007; Keller and Bridgham, 2007), and from 1 to 36% using the 35S-radiotracer assay (Vile et al., 2003b).
Figure 1. Compilation of sulfate reduction rates from large peat soil mesocosms [60 cm in diameter and 60 cm in depth; left column (A,C,E); Knorr and Blodau, 2009] and from field samples [right column (B,D,F); Knorr et al., 2009] of the German peatland Schlöppnerbrunnen II. Both, data from mesocosm and field samples demonstrate decreasing maximum sulfate reduction rates with increasing exposure to water logging, which represents anoxic conditions (A,B). Maximum sulfate reduction rates depended on prevailing sulfate concentrations (C,D) and a similar wide span in sulfate reduction rates was observed above and below the water table with the water table representing the transition zone between oxic to anoxic conditions (E,F). Time spans in the legend represent the incubation time (days) of soils below the water table (WT). Negative and positive values on the abscissa (x-axis) in subfigure (E) and (F) represent relative positions above and below the water table, respectively.
From a thermodynamic point of view, SRM typically outcompete microorganisms involved in the methanogenic degradation pathways such as primary and secondary fermenters, homoacetogens, and methanogens for substrates (Muyzer and Stams, 2008), thus diverting anaerobic carbon mineralization away from methanogenesis to carbon dioxide production. However, SRM are also known to grow in syntrophic association with hydrogen-scavenging methanogens or can themselves constitute the hydrogen-scavenging partner in syntrophic associations (Rabus et al., 2006). Especially under sulfate-limiting conditions, it has been proposed that such syntrophic associations play an important role in organic matter degradation, with hydrogen-utilizing methanogens being replaced by hydrogen-utilizing SRM (Muyzer and Stams, 2008). Indeed, SRM were shown to effectively compete with methanogens for hydrogen and acetate at the naturally occurring low sulfate and substrate concentrations in different freshwater habitats ranging from peat and rice paddy soil to lake sediments (lower micromolar-range of sulfate and acetate and nanomolar-range for hydrogen; Lovley and Klug, 1983; Achtnich et al., 1995; Watson and Nedwell, 1998). At the same time, molybdate inhibition assays of different peat soils revealed that SRM and methanogens do not always compete but can also form syntrophic associations as revealed by co-inhibition of sulfate reduction and methanogenesis upon molybdate addition (Watson and Nedwell, 1998; Blodau et al., 2007), with the inhibition of methanogenesis being reversible when additional hydrogen was supplied (Watson and Nedwell, 1998). If anaerobic oxidation of methane (AOM) coupled to sulfate reduction, an important process in anoxic marine environments that is catalyzed by syntrophic consortia of archaeal methanotrophs and SRM (Knittel and Boetius, 2009), takes also place in freshwater wetlands is not completely clear. From a thermodynamic point of view, AOM coupled to sulfate reduction would yield a ΔG of −14 to −19 kJ/reaction under the substrate concentrations occurring in wetlands (Smemo and Yavitt, 2011), which overlaps with energy yields of AOM observed in marine sediments (ΔG = −18 to −35 kJ/reaction, Thauer, 2011). There are a few studies that found hints for AOM in wetlands, however, with no clear connection to sulfate reduction (Smemo and Yavitt, 2011). Adding to these intertwined relationships between SRM and methanogens in freshwater wetlands, seasonal shifts in the dominance of sulfate reduction over methanogenesis and vice versa were observed for two peatlands in the UK (Nedwell and Watson, 1995; Watson and Nedwell, 1998). When combining the carbon flow through sulfate reduction and methanogenesis (but not taking other anaerobic degradation processes into account), the contribution of sulfate reduction ranged from 6–96%. However, it was not clear whether the dominance of sulfate reduction was an effect of higher natural sulfate concentrations (up to 700 μM), lower temperatures during winter time, or a combination of both (Nedwell and Watson, 1995; Watson and Nedwell, 1998).
On the scale of field manipulation studies, deposition or experimental addition of sulfate has been shown to reduce net methane emissions from natural wetlands (Dise and Verry, 2001; Gauci et al., 2002, 2004) and rice paddies (Lindau et al., 1993; Yagi et al., 1997; van der Gon et al., 2001) for prolonged time periods spanning from several weeks to 2 years. In this respect, it should be noted that not only stimulated sulfate reduction might have led to a suppression of methanogenesis but also increased plant growth by a fertilization effect of added sulfur in sulfur-deficient soils such as nitrogen/phosphorous-fertilized rice paddies or ombrotrophic peatlands. For rice plants, it was suggested that this fertilization effect would lead to a redirection of photosynthate to grain production, accompanied by reduced rhizospheric carbon leakage and thus reduced root-derived substrate supply to methanogenesis (Gauci et al., 2008). Also in ombrotrophic peatlands, a large fraction of sulfur deposited by rain was repeatedly observed to be scavenged by the living plant layer (Urban et al., 1989; Steinmann and Shotyk, 1997; Blodau et al., 2007; Bottrell et al., 2010). However, if this directly influences peatland methane gross production or emission has not been investigated in detail so far.
Ongoing sulfate reduction in wetlands, which can be observed over long time periods despite small pool sizes, needs either an effective recycling mechanism for sulfate to prevent rapid depletion of the sulfate pool and/or a constant external input of sulfate, e.g., through atmospheric deposition, groundwater flow, and/or the overlaying water. Reported turnover times of sulfate pools were often as short as hours to days (Table 1) and only a fraction thereof could be explained by externally supplied sulfate (Wieder and Lang, 1988; Urban et al., 1994; Nedwell and Watson, 1995; Wind and Conrad, 1997). This has led to the suggestions that recycling of reduced sulfur compounds to sulfate is the main mechanisms that drives sulfate reduction in wetlands (Wieder and Lang, 1988; Jørgensen, 1990; Wieder et al., 1990; Urban et al., 1994; Nedwell and Watson, 1995; Wind and Conrad, 1997; Blodau et al., 2007).
Recycling of reduced sulfur compounds to sulfate can be complex and may occur at different spatiotemporal scales as well as under oxic and anoxic conditions. On large spatial scale, modeling studies suggest that water movement and mixing in the capillary fringe (i.e., the soil layer in which groundwater seeps up from the water table to fill soil pores by capillary action) and shallow groundwater (Reeve et al., 2006) as well as different flow paths of water through wetland soils (Fleckenstein et al., 2011) may be important for creating redox gradients and for the provision and recycling of electron acceptors. Also a temporal decoupling of sulfate reduction and re-oxidation of reduced sulfur compounds, such as during short-term (Deppe et al., 2009; Knorr and Blodau, 2009) or seasonal water table fluctuations (Alewell and Novak, 2001), which influences the transition zone from oxic to anoxic conditions, is possible. This is supported by experimental evidence, where recycling of sulfur compounds along an elevational transect through a North American peatland (Mandernack et al., 2000) and heterogeneous distribution of sulfate reduction and sulfide re-oxidation on the range of tens to hundreds of meters in a small catchment in Central Europe (Alewell et al., 2006) was observed over the course of different seasons by δ34S signatures and 35S-sulfate incubations.
Recycling of reduced sulfur compounds does also occur on a smaller scale of millimeters and a number of mechanisms have been identified to chemically or microbiologically re-oxidize sulfide to sulfate, thiosulfate, or elemental sulfur (Kelly et al., 1997; Wind and Conrad, 1997; Canfield et al., 2005; Friedrich et al., 2005; Gregersen et al., 2011). A common feature of sedges and graminoids in wetlands and also of rice plants is the formation of aerenchymatic tissues to provide oxygen to their rooting system (Wind and Conrad, 1997; Brune et al., 2000; Pezeshki, 2001). Therefore, root surfaces of aerenchymatic plants have been shown to provide highly reactive interfaces of oxic and anoxic conditions that allow for rapid recycling of electron acceptors, such as reduced iron and sulfides (Roden and Wetzel, 1996; Wind and Conrad, 1997; Segers and Leffelaar, 2001). In addition, electric currents in marine sediments, which were proposed to be mediated by bacterial pili (nanowires), were shown to span from the sulfidic zone over a 12-mm distance to the oxic surface layers, thereby fueling sulfide re-oxidation at depth and effectively extending the zone, in which electron acceptor resupply can fuel ongoing respiratory activity (Nielsen et al., 2010). This surprising finding has so far not been described for freshwater wetlands but there is no reason why this phenomenon should be limited to marine surface sediments.
Recycling of sulfides to oxidized sulfur species may also occur in the absence of oxygen, which is often not evident at first sight. However, high SRR that proceed over long time periods (>100–200 days) under anoxic conditions as observed, e.g., in peat monoliths and in field studies of the minerotrophic peatland Schlöppnerbrunnen II in Central Europe (Knorr and Blodau, 2009; Knorr et al., 2009; Figures 1A,B,E,F), can only be explained by an anoxic sulfur cycle. Although there is no final proof yet how this anoxic recycling proceeds, there is cumulating evidence that iron minerals and redox-active organic matter might play a role in the first step of sulfide oxidation to either elemental sulfur/polysulfide or thiosulfate (Figure 2). In marine sediments, it has been demonstrated that sulfides react with iron oxides to form Fe(II) (Canfield, 1989). The sulfides in this reaction are oxidized mainly to elemental sulfur and to a smaller extend to thiosulfate as inferred from incubations mimicking environmental conditions but free of microorganisms (Pyzik and Sommer, 1981). This process is strongly dependent on the reactivity of the present iron oxides and their surface properties (Canfield et al., 1992; Peiffer and Gade, 2007) and subsequent formation of iron sulfides, Fe(II) sorption, and associated surface passivation of iron minerals could limit this process over time (Elsgaard and Jørgensen, 1992; Roden and Urrutia, 2002). In rice paddy soil and iron-rich peatlands, formation of Fe(II) under anoxic conditions was repeatedly observed and often attributed solely to microbial iron reduction (Yao et al., 1999; Küsel et al., 2008; Knorr et al., 2009; Hori et al., 2010). However, Fe(II) formation could, at least partially, also be attributed to sulfide re-oxidation on iron oxides since this is a fast process taking place in the order of minutes to days (Peiffer and Gade, 2007). Therefore, it could be implied that not only in marine sediments but also in iron-rich wetland soils there is a competition between microbial Fe(III) reduction and chemical reduction of Fe(III) mediated by sulfide oxidation. The interaction of iron and sulfur has so far not been adequately investigated in wetlands and further investigations about the complex geochemistry of iron and sulfur in organic rich soils are needed.
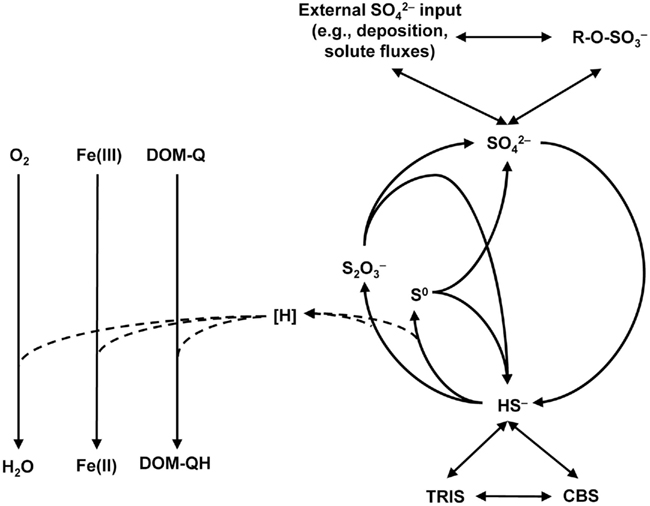
Figure 2. Schematic overview of the proposed sulfur cycle in freshwater wetlands. Abbreviations: DOM-Q, quinone moieties of dissolved organic matter,  organic sulfate esters, TRIS, total reactive inorganic sulfur, CBS, carbon bonded sulfur.
organic sulfate esters, TRIS, total reactive inorganic sulfur, CBS, carbon bonded sulfur.
From diagenetic studies it is known that sulfides may react with organic matter and form organic forms of sulfur also during early stages of diagenesis (Brown, 1986; Ferdelman et al., 1991; Bottrell et al., 2010), e.g., by addition of sulfur to quinone moieties, polysulfide bonds, formation of thiol-groups (S-H), or sulfate esters (Vairavamurthy et al., 1992; Ghani et al., 1993; Kertesz, 2000; Perlinger et al., 2002). Sulfate esters of small molecular size have been demonstrated to be utilized as alternative terminal electron acceptor by some SRM (Lie et al., 1996, 1998; Visscher et al., 1999), thus also supporting SRM activity in absence of inorganic sulfate. More recently, it has been demonstrated that humic substances can also mediate re-oxidation of sulfides and thus fuel an anoxic sulfur cycle (Heitmann and Blodau, 2006; Heitmann et al., 2007). Although the electron transfer capacity of dissolved organic matter toward sulfides was relatively low in the study of Heitmann et al. (2007), advances in methodological approaches such as electrochemical determination of electron accepting capacities of dissolved organic matter (Aeschbacher et al., 2010) or including the solid phase material (Roden et al., 2010) may reveal a greater contribution of organic matter to total electron accepting capacities. Therefore, natural organic matter may be a key to an anoxic cycling of sulfur (Blodau et al., 2007). However, it needs to be kept in mind that the addition of sulfur to organic matter also provides a sink for sulfur and thus reduces the amount of sulfur to be recycled. One may thus hypothesize that during longer exposure to anoxic conditions, SRR tend to decrease possibly also because of a loss of sulfur into the organic fraction (e.g., Figure 1A). Indeed, a large part of sulfur in peatlands and rice paddy soils (25 to >80%) is bound to the organic fraction (Wieder and Lang, 1988; Urban et al., 1989; Steinmann and Shotyk, 1997; Wind and Conrad, 1997; Mandernack et al., 2000) and investigations of stable sulfur isotopes support that a large fraction of non-humic organosulfur compounds (e.g., S-containing fulvic acids) are formed by reactions of microbiologically produced sulfides and organic matter, while humic organosulfur may preferentially originate from plant material (Bottrell et al., 2010). If non-humic organosulfur compounds are transformed to humic acid-bound sulfur during diagenesis or if this sulfur is mainly released again as inorganic sulfur available for re-oxidation, as observed in estuarine sediments (Brüchert, 1998), is yet poorly understood.
The known products of anoxic sulfide re-oxidation include elemental sulfur, thiosulfate, or polysulfides (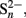 Jørgensen, 1990; Canfield et al., 1992; Thamdrup et al., 1993; Heitmann and Blodau, 2006; Peiffer and Gade, 2007). Thiosulfate and elemental sulfur are known to be used for energy generation by anaerobic microorganisms including SRM. These sulfur compounds can be either reduced back to sulfide, disproportionated to sulfate and sulfide, or in the case of elemental sulfur and in the presence of Mn(IV) be oxidized completely to sulfate (Bak and Cypionka, 1987; Thamdrup et al., 1993; Lovley and Phillips, 1994; Rabus et al., 2006). For freshwater sediments at neutral pH, 35S-radiotracer assays showed that of all the sulfur that is anoxically recycled through thiosulfate 6% gets completely oxidized to sulfate, 50% gets completely reduced to sulfide, and 44% gets disproportionated to sulfate and sulfide (Jørgensen, 1990). Due to this large shunt of thiosulfate directly back to sulfide, only 28% of the recycled sulfide ends up again in the sulfate pool, which would only partially explain how the high SRR are maintained in wetlands under anoxic conditions. Comparable flux measurements do not exist for elemental sulfur because of rapid exchange of isotopes between the pools of S0,
Jørgensen, 1990; Canfield et al., 1992; Thamdrup et al., 1993; Heitmann and Blodau, 2006; Peiffer and Gade, 2007). Thiosulfate and elemental sulfur are known to be used for energy generation by anaerobic microorganisms including SRM. These sulfur compounds can be either reduced back to sulfide, disproportionated to sulfate and sulfide, or in the case of elemental sulfur and in the presence of Mn(IV) be oxidized completely to sulfate (Bak and Cypionka, 1987; Thamdrup et al., 1993; Lovley and Phillips, 1994; Rabus et al., 2006). For freshwater sediments at neutral pH, 35S-radiotracer assays showed that of all the sulfur that is anoxically recycled through thiosulfate 6% gets completely oxidized to sulfate, 50% gets completely reduced to sulfide, and 44% gets disproportionated to sulfate and sulfide (Jørgensen, 1990). Due to this large shunt of thiosulfate directly back to sulfide, only 28% of the recycled sulfide ends up again in the sulfate pool, which would only partially explain how the high SRR are maintained in wetlands under anoxic conditions. Comparable flux measurements do not exist for elemental sulfur because of rapid exchange of isotopes between the pools of S0, 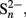 H2S, and FeS (Fossing and Jørgensen, 1990). However, microbially mediated disproportionation of elemental sulfur would lead to a 1:3 stoichiometry of produced sulfate to sulfide and would also not explain sufficient replenishment of the sulfate pool.
H2S, and FeS (Fossing and Jørgensen, 1990). However, microbially mediated disproportionation of elemental sulfur would lead to a 1:3 stoichiometry of produced sulfate to sulfide and would also not explain sufficient replenishment of the sulfate pool.
An alternative explanation that could support a major flux from sulfide to sulfate is through the intermediate elemental sulfur with its complete oxidation being mediated by unknown oxidants (e.g., redox-active organic matter). That such a reaction is in principle possible is known from the complete oxidation of elemental sulfur mediated by Mn(IV) as observed in enrichment cultures (Lovley and Phillips, 1994) or by the stimulation of sulfate production from sulfide by addition of nitrate to a bioreactor inoculated with wetland soil and operating under anoxic, freshwater conditions or to marine surface sediments (Elsgaard and Jørgensen, 1992; Haaijer et al., 2007). However, manganese and nitrate pools are generally too small in wetland types such as peatlands and rice paddies (Murase and Kimura, 1997; Yao et al., 1999; Alewell et al., 2006; Koretsky et al., 2007) to sustain massive sulfur oxidation over prolonged time periods. Minding the redox potential of the MnO2/Mn2+ couple 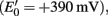 manganese could theoretically function as a redox shuttle between reduced sulfur compounds and electron acceptors with a higher redox potential than manganese (e.g., oxygen). The insolubility of MnO2 in water may nonetheless hamper the activity range of such a MnO2/Mn2+ redox shuttle. Also the idea of an
manganese could theoretically function as a redox shuttle between reduced sulfur compounds and electron acceptors with a higher redox potential than manganese (e.g., oxygen). The insolubility of MnO2 in water may nonetheless hamper the activity range of such a MnO2/Mn2+ redox shuttle. Also the idea of an 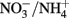 redox shuttle seems unlikely because this would necessitate a close coupling of oxic ammonia oxidation and anoxic nitrate ammonification under the exclusion of competing processes such as denitrification of nitrate to N2 and ammonia/nitrate uptake by plants, which both would further deplete the inorganic N pool in wetland soils. On the other hand, redox potentials and associated electron transfer capacities of humic acids, and probably also of other dissolved and solid organic matter, span a wide range (> +150 to < −350 mV; Aeschbacher et al., 2010) and may therefore especially in carbon rich wetlands provide a significant electron accepting capacity.
redox shuttle seems unlikely because this would necessitate a close coupling of oxic ammonia oxidation and anoxic nitrate ammonification under the exclusion of competing processes such as denitrification of nitrate to N2 and ammonia/nitrate uptake by plants, which both would further deplete the inorganic N pool in wetland soils. On the other hand, redox potentials and associated electron transfer capacities of humic acids, and probably also of other dissolved and solid organic matter, span a wide range (> +150 to < −350 mV; Aeschbacher et al., 2010) and may therefore especially in carbon rich wetlands provide a significant electron accepting capacity.
Adding to the complexity of anoxic sulfate recycling, Schippers and Jørgensen (2001) could show for marine sediments that pyrite (FeS2), a major sulfur compound in sediments that forms from the reaction of H2S or  with iron minerals such as FeS (Schippers and Jørgensen, 2001; Holmkvist et al., 2011a and references therein), is not a stable end product of sediment sulfur transformations but can also be completely re-oxidized to sulfate in the presence of MnO2 as a terminal electron acceptor with Fe(III)/Fe(II) functioning as a redox shuttle. Also the addition of nitrate to pyrite-amended groundwater (Jørgensen et al., 2009) and pure cultures of Thiobacillus denitrificans (Bosch et al., 2012) triggered oxidation of pyrite to sulfate, further highlighting the potential role of pyrite as an intermediate in sulfur cycling. Currently, it is not clear if similar processes also occur in freshwater wetlands, possibly involving organic matter as alternative electron acceptor to MnO2 and nitrate. In summary, several concepts have been put forward how the observed high SRR can be maintained over long periods of time under anoxic conditions. However, each of these concepts needs further experimental validation.
with iron minerals such as FeS (Schippers and Jørgensen, 2001; Holmkvist et al., 2011a and references therein), is not a stable end product of sediment sulfur transformations but can also be completely re-oxidized to sulfate in the presence of MnO2 as a terminal electron acceptor with Fe(III)/Fe(II) functioning as a redox shuttle. Also the addition of nitrate to pyrite-amended groundwater (Jørgensen et al., 2009) and pure cultures of Thiobacillus denitrificans (Bosch et al., 2012) triggered oxidation of pyrite to sulfate, further highlighting the potential role of pyrite as an intermediate in sulfur cycling. Currently, it is not clear if similar processes also occur in freshwater wetlands, possibly involving organic matter as alternative electron acceptor to MnO2 and nitrate. In summary, several concepts have been put forward how the observed high SRR can be maintained over long periods of time under anoxic conditions. However, each of these concepts needs further experimental validation.
Anoxic conditions, high sulfate concentrations (up to 28 mM), and the large extent of marine surface sediments render sulfate reduction a main driver of anaerobic carbon mineralization in the oceans. Marine surface sediments are thus considered the prime habitats for SRM. Consequently, numerous studies have determined the microbial community structures in these habitats through isolation and taxonomic description of sulfate-reducing strains and complementary molecular diversity surveys. We thus know today that a dominant fraction (up to 20–40%) of the microbial community in marine sediments consists of microorganisms that are related to validly described SRM species (e.g., Ravenschlag et al., 1999; Musat et al., 2006; Uthicke and McGuire, 2007; Orcutt et al., 2011; Teske et al., 2011). This is in stark contrast to freshwater wetlands, where taxa including known SRM are rarely detected or constitute only a minor fraction (<1%) of the microbial community, if general molecular diversity surveys based on bacterial or archaeal 16S rRNA gene clone libraries are performed (e.g., Costello and Schmidt, 2006; Dedysh et al., 2006; Kraigher et al., 2006; Lu et al., 2006; Jackson et al., 2009; Wilhelm et al., 2011). Only the application of cultivation or targeted molecular approaches revealed that members of recognized SRM taxa are actually present in freshwater wetlands. First evidence for the existence of SRM that thrive in low-sulfate environments were provided by enrichments from lake sediments (Bak and Pfennig, 1991b; Ramamoorthy et al., 2006), rice paddy fields (Dalsgaard and Bak, 1994; Wind and Conrad, 1995; Wind et al., 1999), permafrost soil (Vatsurina et al., 2008), and constructed wetlands (Lee et al., 2009) and included Desulfovibrio, Desulfobulbus, Desulfotomaculum, and Desulfosporosinus spp. In parallel, taxa including known SRM were also detected by targeted molecular analyses such as (quantitative) PCR assays with taxa-specific 16S rRNA gene-targeted primers (Scheid and Stubner, 2001; Stubner, 2002, 2004; Stahl et al., 2007) and/or with the DSR1F–DSR4R primer variants targeting the functional marker genes dsrAB (Wagner et al., 2005; Steger et al., 2011; Table 2). The latter genes encode the alpha- and beta-subunits of dissimilatory (bi)sulfite reductase that is used by SRM for energy conservation. Although a few horizontal gene transfer events have slightly worsened the usefulness of dsrAB as phylogenetic marker genes (Zverlov et al., 2005), the foundation of these molecular 16S rRNA gene- or dsrAB-based diversity studies is that an unknown environmental sequence can be phylogenetically assigned with high likelihood to a described taxon, if the sequence branches unambiguously within this taxon. Using this rationale, members of the families Desulfobacteraceae and Syntrophobacteraceae and bacteria related to Desulfobacca acetoxidans were identified in more than half of the wetlands listed in Table 2. Members of other SRM families/family level lineages in the Deltaproteobacteria and the Firmicutes can also be regularly detected using these targeted approaches. Given their thermophilic lifestyle, it is not surprising that bacterial SRM of the phylum Thermodesulfobacteria and the genus Thermodesulfovibrio or archaeal SRM of the genera Archaeoglobus and Caldivirga have so far not been found in freshwater wetlands.
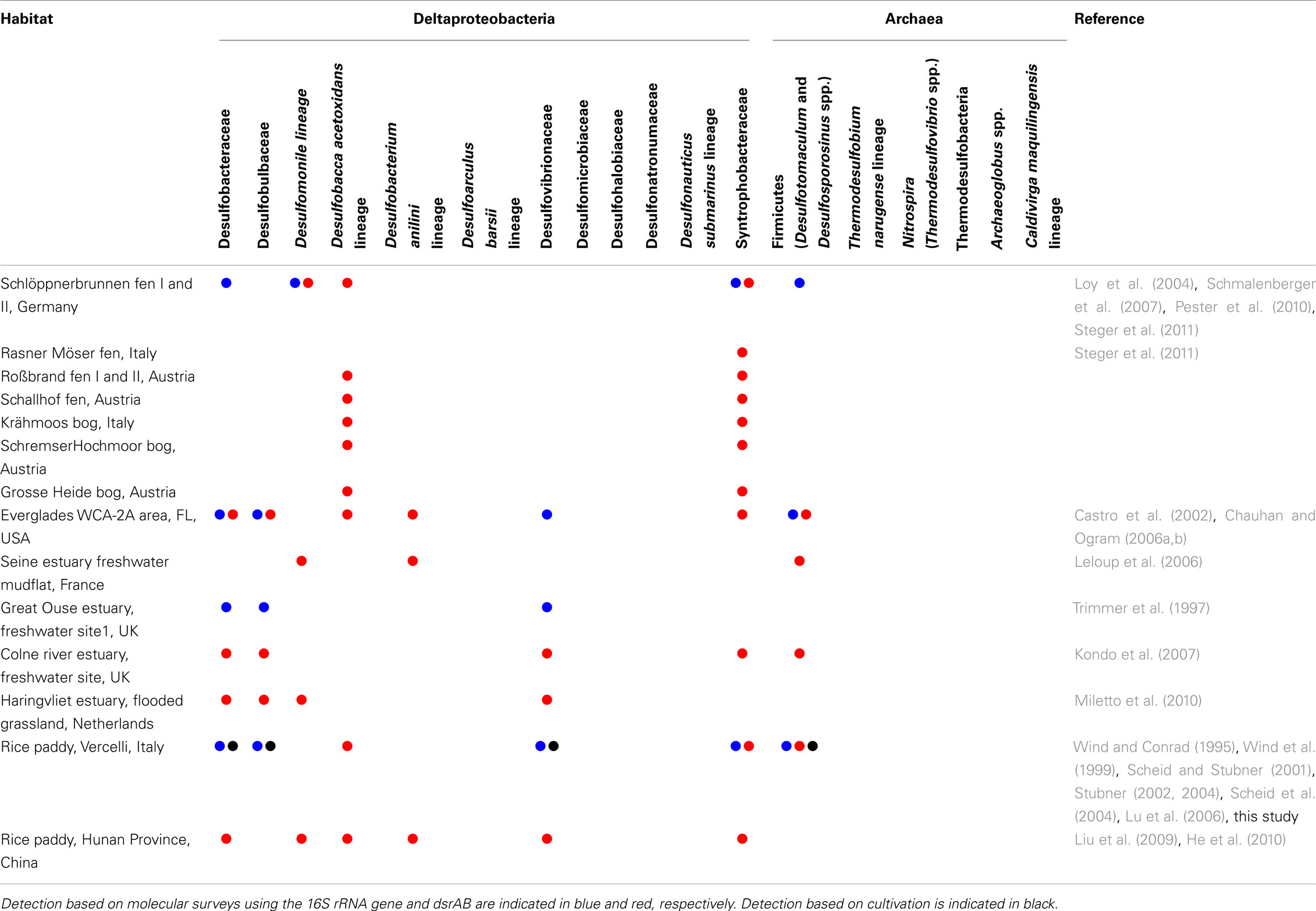
Table 2. Occurrence of members of known SRM families (i.e., described families or approximately family level lineages that harbor known SRM isolates) in freshwater wetlands as reported in the analyzed studies.
However, inferring ecophysiological properties of the detected SRM taxa in wetlands is notoriously difficult because even main metabolic properties of SRM, such as complete versus incomplete organic substrate mineralization, substrate utilization preferences, and alternative options for energy conservation such as fermentation or syntrophy with methanogens cannot be clearly inferred from phylogenetic analyses of 16S rRNA genes or dsrAB (Stahl et al., 2007; Loy et al., 2008). For many of the wetland microorganisms that belong to known SRM taxa and were detected using cultivation-independent approaches, it even remains unknown if they are truly capable of dissimilatory sulfate reduction. This is due to exceptions of the generally close correlation of sulfate-reducing metabolism and 16S rRNA and/or dsrAB phylogeny. For example, the butyrate-utilizing marine syntroph Algorimarina butyrica is not capable of reducing sulfate but, based on 16S rRNA phylogeny, is a member of the deltaproteobacterial family Desulfobacteraceae (Kendall et al., 2006), which has been long thought to exclusively consist of SRM. Furthermore, some syntrophs related to sulfate-reducing Desulfotomaculum spp., such as members of the genera Pelotomaculum and Sporotomaculum, are not able to anaerobically grow with sulfate as electron acceptors but harbor dsrAB (Zverlov et al., 2005; Imachi et al., 2006).
Pure snapshot analyses of dsrAB diversity in the environment have low informative value regarding the ecophysiology of the microorganisms carrying these genes, especially if contextual information such as biogeochemical process data is missing. Nevertheless, the dsrAB approach has allowed important insights into the natural diversity of microorganisms carrying these genes and has revealed, for various environments, the presence of many dsrAB sequences that are not closely related to dsrAB from known SRM (Figure 3). We have performed a comprehensive phylogenetic re-analysis of all phylogenetically novel dsrAB sequences retrieved from different freshwater wetland habitats (Table 3) in combination with representative sequences from other environments (see legend of Figure 3 for methodological details). The novel dsrAB sequences from yet uncultivated microorganisms formed 10 monophyletic lineages (which we designated “uncultured dsrAB lineage 1 to 10”; Figure 3), each corresponding to a new family in the consensus tree and emphasizing the large diversity of elusive microorganisms harboring these genes. Besides uncultured dsrAB lineages 5, 6, and 7 that formed independent branches close to the root, a further level of monophyletic clustering was apparent in the consensus DsrAB tree that we followed to tentatively designate three phylogenetically stable “superclusters,” each having high bootstrap support and comprising at least two uncultured dsrAB family level lineages and/or known SRM families. Two of the superclusters harbor cultured representatives and were thus named accordingly. The “Deltaproteobacteria supercluster” includes all dsrAB sequences from deltaproteobacterial species (please note that the deep-branching D. acetoxidans lineage is tentatively considered part of this supercluster despite weak bootstrap support), laterally acquired, Deltaproteobacteria-like dsrAB from Firmicutes and Thermodesulfobacteria (Zverlov et al., 2005), uncultured dsrAB lineage 1, and other environmental dsrAB. The Thermodesulfovibrio supercluster includes dsrAB of Thermodesulfovibrio spp., uncultured dsrAB lineage 10, and other environmental dsrAB. The third supercluster consists exclusively of environmental dsrAB sequences from yet uncultivated microorganisms, including uncultured dsrAB lineages 8 and 9, and was therefore named “environmental supercluster 1”. The consensus tree provides some indication for another supercluster, including dsrAB from recognized bacteria in the Firmicutes, uncultured dsrAB lineages 2, 3, and 4, and other environmental dsrAB. However, the monophyletic origin of this supercluster is not supported by bootstrap analysis and is also not evident in other previously published DsrAB trees (Pester et al., 2010; Steger et al., 2011). We have thus not named this putative supercluster because its members and their relative branching orders currently remain unresolved (Figure 3).
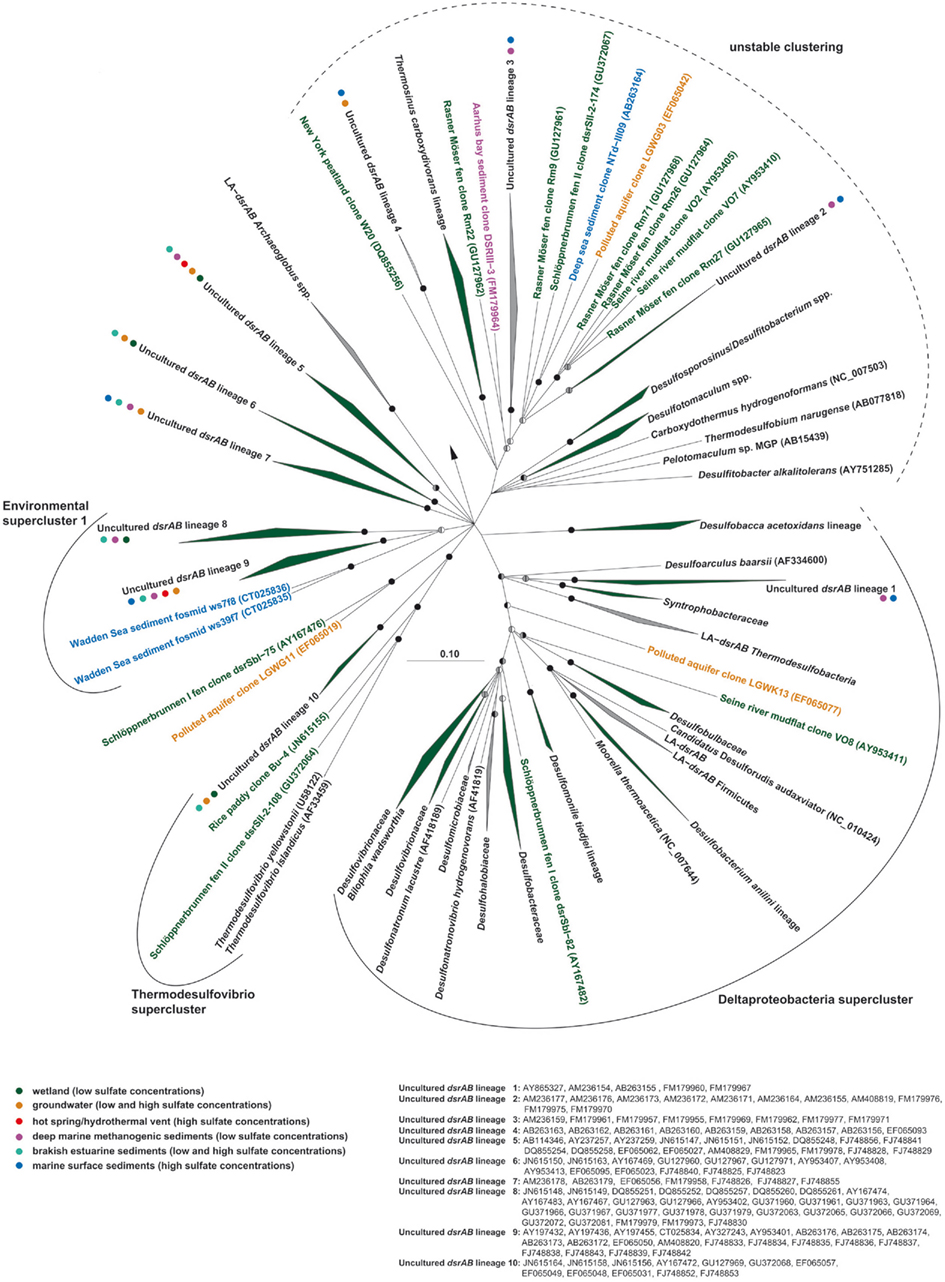
Figure 3. DsrAB consensus tree showing the affiliation of dsrAB sequences from freshwater wetlands (sequences and lineages marked in green). Environmental DsrAB sequences not affiliated with sequences from cultured microorganisms were grouped into an “uncultured dsrAB lineage” on the approximate family level if at least two sequences with ≥64% amino acid identity formed a monophyletic cluster and contained no sequence that was ≥64% identical to a sequence outside this lineage. The conservative 64% limit was inferred from DsrAB of cultured representatives belonging to 10 known families with a minimum intra-family amino acid sequence identity of 64–89%. Bootstrap support for identified clusters is shown by split circles (right: maximum likelihood, 1000 re-samplings; left: maximum parsimony, 100 re-samplings) at the respective branches with black indicating ≥90% support, gray indicating ≥70% support, and white/absence of circles indicating <70% support. Family level DsrAB lineages were summarized to superclusters if their monophyletic origin was supported by bootstrap values of ≥70%. The color code of environmental DsrAB sequences or of dots behind uncultured family level DsrAB lineages indicates the habitat where the respective sequences were retrieved from (this data is not provided for recognized families). For phylogenetic inference of deduced DsrAB amino acid sequences, insertions and deletions were removed from the data set by using an alignment mask (indel filter), which resulted in 502 amino acid positions for comparative analyses. Distance matrix (Neighbor Joining with PAM as amino acid replacement model), maximum likelihood (RAxML with PAM as amino acid replacement model), and maximum-parsimony algorithms were used as provided in the ARB software package (Ludwig et al., 2004) to determine the phylogenetic relatedness of the analyzed DsrAB sequences. Reverse DsrAB of sulfur-oxidizing bacteria were used as outgroup (Loy et al., 2009). A strict consensus tree was constructed from the individual trees obtained with the different algorithms using the Phylip (Felsenstein, 1989) and ARB (Ludwig et al., 2004) software packages. Branch lengths of the consensus tree were inferred by the Fitch algorithm using a Jukes–Cantor derived distance matrix (Phylip), the scale bar represents 10% estimated sequence divergence. Affiliation of short DsrA or DsrB sequences (<542 amino acids) retrieved from freshwater wetlands was inferred using the consensus tree and the quick-add-parsimony tool within ARB. Uncultured family level DsrAB lineages that comprise such short DsrA and DsrB sequences but no near full-length DsrAB sequences from freshwater wetlands are colored green but are not marked with a green dot.
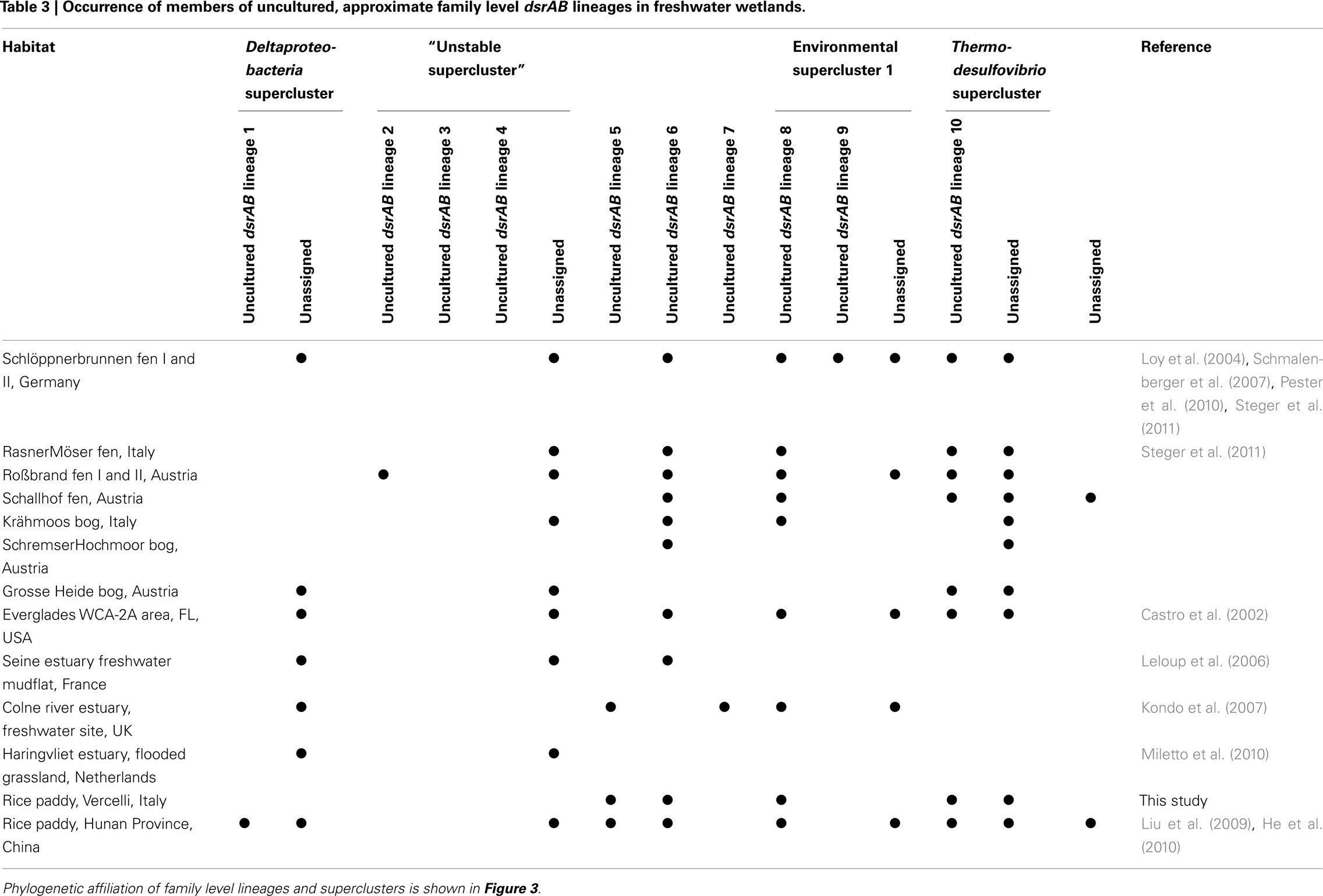
Table 3. Occurrence of members of uncultured, approximate family level dsrAB lineages in freshwater wetlands.
With the exception of uncultured dsrAB lineages 3 and 4, dsrAB sequences from freshwater wetlands are present in eight of the ten uncultured family level lineages. There is additional evidence of dsrAB novelty in wetlands through individual (or groups) of wetland dsrAB sequences, but these did not meet our criteria to designate them as a novel “uncultured dsrAB lineage” (Figure 3). Overall, dsrAB belonging to lineage 6 (branching off close to the root), lineage 8 (environmental supercluster 1), and lineage 10 (Thermodesulfovibrio supercluster) were most often detected in freshwater wetlands (Table 3). In addition, dsrAB sequences not affiliated to any of the uncultured family level lineages but belonging to the Deltaproteobacteria supercluster, the Thermodesulfovibrio supercluster, and the putative, phylogenetically unstable supercluster also contributed to the dsrAB diversity in most analyzed freshwater wetlands.
The dsrAB diversity in wetlands is dominated by sequences from lineages without cultivated microorganisms (Figure 3). Key questions are (i) if each of these phylogenetically novel dsrAB sequence variants is representative of an individual microorganism and (ii) if these genes potentially encode functional enzymes. Functional marker genes can be present as different copies within the same microorganism as is known, e.g., for the alpha subunit of the particulate methane monooxygenase within type II methanotrophs (Tchawa Yimga et al., 2003; Baani and Liesack, 2008) and the novel dsrAB types could likewise represent phylogenetically different versions of dissimilatory (bi)sulfite reductases. However, SRM that possess multiple dsr operons with significantly different sequences have not been reported so far. The only exception to this observation is the homoacetogen Moorella thermoacetica, which is not known to perform dissimilatory sulfate reduction but possesses two very distinct dsrAB copies (Loy et al., 2009). In principle, the novel dsrAB types found in wetlands could also be pseudogenes. However, no internal stop codons were found in these sequences and a previous analysis revealed sequence conservation of the functional site in both, subunit A and B of the enzyme, as well as a low rate of non-synonymous to synonymous substitutions (Loy et al., 2004), indicating that these genes could be expressed into functionally active proteins.
Although our dsrAB meta-analysis included data from geographically and biogeochemically distinct wetlands (comprising minerotrophic and ombrotrophic peatlands, river floodplains, freshwater marshes within the Florida Everglades, and rice paddy fields), clear distribution patterns of major dsrAB lineages or superclusters with geography or environmental parameters like pH were not evident at this coarse level of phylogenetic resolution. However, when the dsrAB diversity of various Central European wetlands was analyzed at higher phylogenetic resolution by combined dsrB-fingerprinting and sequencing, signatures of biogeographical distribution became apparent and were best explained by soil pH and wetland type (Steger et al., 2011).
Thriving in freshwater wetland habitats seems to be a trait that is widely distributed in the different cultivated and uncultivated family level lineages of the DsrAB tree. Furthermore, all of the uncultured dsrAB lineages that contained dsrAB from freshwater wetlands also contained dsrAB from many other environments (Figure 3). These environments include deep marine methanogenic sediments (Leloup et al., 2007, 2009), which are also characterized by low-sulfate concentrations and ongoing, albeit very slow, dissimilatory sulfate reduction (Holmkvist et al., 2011a,b). In addition to environments with low-sulfate concentrations, all of the uncultured family level dsrAB lineages that contained dsrAB from freshwater wetlands also contained dsrAB from habitats influenced by both low and high sulfate concentrations such as polluted coastal aquifers (Wu et al., 2009) and estuarine sediments (Jiang et al., 2009) or habitats with permanently high sulfate concentrations comprising marine surface sediments (Leloup et al., 2007, 2009), hydrothermal vents (Dhillon et al., 2003; Mussmann et al., 2005), and hot springs (Fishbain et al., 2003). This observation emphasizes the ubiquitous distribution of members of these uncultured dsrAB lineages, suggesting that adaptation to different sulfate concentrations is not a trait that leads to ecologically coherent dsrAB sequence groups at the approximate taxonomic/phylogenetic rank of families.
Peatlands in the forested Lehstenbach catchment (Bavaria, Germany) represent attractive model ecosystems for wetland SRM studies due to the extensive characterization of their biogeochemistry and microbiota and the deposition of sulfur by acid rain during the second half of the last century (Alewell and Novak, 2001; Matzner, 2004; Novak et al., 2005; Alewell et al., 2006; Schmalenberger et al., 2007; Küsel et al., 2008; Knorr and Blodau, 2009; Knorr et al., 2009). They are currently also the best studied wetlands with respect to the composition and environmental dynamics of dsrAB-containing microorganisms (Loy et al., 2004; Schmalenberger et al., 2007; Pester et al., 2010; Steger et al., 2011). In particular, the fen Schlöppnerbrunnen II comprises at least 53 different operational taxonomic units (OTUs) of dsrAB at the approximate species level (Loy et al., 2004; Schmalenberger et al., 2007; Pester et al., 2010; Steger et al., 2011). More than half of these dsrAB OTUs (28 out of 53) belong to the uncultured lineage 8 within the “environmental supercluster 1”. Based on a dsrAB microarray analysis over a 6-year period, two species level OTUs of lineage 8 (OTU 2 and 14) and two OTUs loosely affiliated to D. acetoxidans (OTU 1 and 4) were shown to dominate the dsrAB diversity in the Schlöppnerbrunnen fen II. A parallel quantitative PCR analysis of OTU 1 and OTU 2 revealed that microorganisms carrying the respective dsrAB each represent up to about 1–2% of the microbial community (Steger et al., 2011; Figure 4). Given this relatively high abundance for a single microbial species in soils, these microorganisms represent “core” members of the autochthonous microbial community in this wetland and certainly have an important, albeit unknown function in biogeochemical cycling (Pedrós-Alió, 2012). It would be tempting to speculate that these unknown microorganisms represent novel SRM that are actively involved in sulfate reduction in situ. However, dsrAB is present not only in SRM but also in microorganisms capable of sulfite (e.g., Desulfitobacterium spp., Spring and Rosenzweig, 2006) or organosulfonate reduction (e.g., Bilophila wadsworthia, Laue et al., 2001). Furthermore, some microorganisms harboring dsrAB can switch between a sulfate reducing and syntrophic lifestyle or, in the extreme case, are not capable of sulfate/sulfite reduction at all. Members of the deltaproteobacterial genus Syntrophobacter are prime examples for microorganisms that can perform dissimilatory sulfate reduction for energy generation but prefer to release generated reducing equivalents to syntrophically associated methanogens or other SRM (Wallrabenstein et al., 1994, 1995; Harmsen et al., 1998). Even more intriguing examples are Pelotomaculum spp. within the Desulfotomaculum cluster I. These syntrophic microorganisms, which typically thrive in anoxic, methanogenic environments, possibly lost the capability for sulfate reduction due to the need to adapt to low-sulfate conditions during the course of evolution. In accordance with their physiological properties, dsrAB could not be detected by PCR assays in most Pelotomaculum spp. – with one exception. The type strain Pelotomaculum propionicicum MGP possesses dsrAB and even transcribes these genes in the presence and, most astonishingly, also in the absence of sulfate (Imachi et al., 2006). Since P. propionicicum is an obligate syntroph (Imachi et al., 2007), this exemplifies that the mere presence or even transcription of dsrAB does not necessarily mean that the corresponding microorganisms perform dissimilatory sulfate or sulfite reduction. Interestingly, syntrophic dsrAB-containing non-sulfite/sulfate reducers, syntrophic SRM, and “bona fide” sulfite/sulfate reducers can be very closely related, indicating an evolutionary association between the ability to respire sulfite/sulfate and the syntrophic lifestyle.
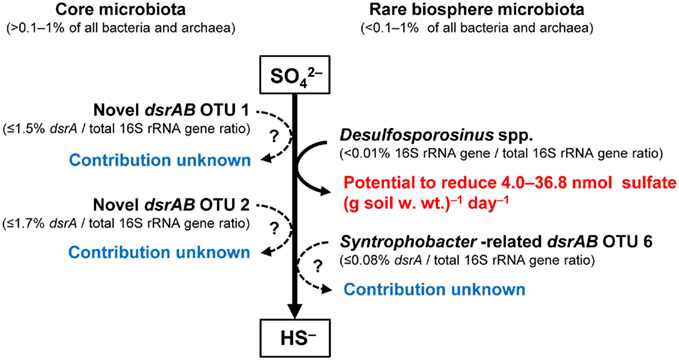
Figure 4. Relative abundances (as inferred from quantitative dsrA- or 16S rRNA gene-targeted PCR analyses) of recognized and putative SRM in the model peatland Schlöppnerbrunnen II. This figure is based on data from Loy et al. (2004); Schmalenberger et al. (2007); Pester et al. (2010); Steger et al. (2011). The abundance of the large number of other dsrAB OTUs (including dsrAB related to Desulfomonile and Desulfobacca spp.), which were also detected at Schlöppnerbrunnen II, and their contribution to sulfate reduction is currently unknown (Loy et al., 2004; Schmalenberger et al., 2007; Pester et al., 2010; Steger et al., 2011).
Linking phylogenetic information obtained from a molecular marker gene such as 16S rRNA or dsrAB with the physiological capability for dissimilatory sulfate reduction is not straightforward. However, great progress has been made through development of methods that combine molecular and isotope analysis to identify microorganisms metabolizing a supplemented isotope-labeled substrate under defined experimental conditions (Loy and Pester, 2010). A recent DNA stable isotope probing study of soil from Schlöppnerbrunnen fen II, using in situ concentrations of typical 13C-labeled carbon degradation intermediates (mixture of lactate, acetate, formate, and propionate) found no differential labeling of novel dsrAB under sulfate-reducing versus methanogenic conditions, leaving it unexplained whether the corresponding microorganisms were active under none or both of the provided conditions (Pester et al., 2010). Therefore, it still remains unresolved whether wetland microorganisms with phylogenetically novel dsrAB types are SRM, syntrophs, or capable of switching between both lifestyles.
Microbial communities in the environment are typically composed of abundant or “core” taxa, which are considered to carry out most ecosystem functions, and a very large number of highly diverse but low abundance taxa (less than 0.1–1% of all bacteria and archaea), which are referred to as the “rare biosphere” (Pedrós-Alió, 2006; Sogin et al., 2006; Fuhrman, 2009). Other than an inventory of their 16S rRNA genes very little is known about the ecological role of the “rare biosphere,” which is also true for freshwater wetlands. Experimental evidence is gathering that rare microorganisms result from historical ecological change with the potential to become dominant in response to shifts in environmental conditions – the “microbial seed bank” concept (Dethlefsen et al., 2008; Brazelton et al., 2010; Lennon and Jones, 2011) or that they simply result from random dispersal (Hubert et al., 2009). In contrast to the majority of these dormant populations, it was recently found that “rare biosphere” members could also actively contribute to ecosystem functions and in this particular case to peatland sulfate reduction and carbon cycling (Figure 4). Comparative DNA stable isotope probing (Loy and Pester, 2010) of Schlöppnerbrunnen fen II soil under sulfate-reducing and methanogenic conditions showed that a Desulfosporosinus spp., which constitutes only a minor proportion of the archaeal and bacterial community (0.006% relative 16S rRNA gene abundance), could be an important SRM in this particular peatland. For the identified Desulfosporosinus population, a high cell-specific SRR of up to 341 fmol 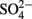 cell−1 day−1 was estimated (Pester et al., 2010), which was comparable to the highest cell-specific rates reported for SRM in pure culture (Detmers et al., 2001). Quantifying the Desulfosporosinus populations at different soil depths and relating the observed abundances to the estimated cell-specific rates revealed that they had the potential to reduce sulfate in situ at a rate of up to 4.0–36.8 nmol (g soil w. wt.)−1 day−1 (Pester et al., 2010). This could explain a considerable fraction of the radiotracer-measured SRRs of the same peatland, which ranged from 0 to ca. 340 nmol (g soil w. wt.)−1 day−1 over a depth profile of 0–30 cm and a 300-days period (Knorr and Blodau, 2009; Knorr et al., 2009). Modeling of sulfate diffusion to such highly active cells revealed sufficient sulfate supply even at bulk concentrations as low as 10 μM (Pester et al., 2010). In addition, apparent sulfate half-saturation concentrations, Km, for sulfate reducers in pure culture (Pallud and Van Cappellen, 2006 and references therein) and in sediment slurries (Tarpgaard et al., 2011) have been reported to be as low as 2–5 μM indicating no kinetic limitation from the electron acceptor side as well. These findings indicated that the identified peatland Desulfosporosinus sp. has the potential to contribute considerably to peatland sulfate reduction despite its very low abundance.
cell−1 day−1 was estimated (Pester et al., 2010), which was comparable to the highest cell-specific rates reported for SRM in pure culture (Detmers et al., 2001). Quantifying the Desulfosporosinus populations at different soil depths and relating the observed abundances to the estimated cell-specific rates revealed that they had the potential to reduce sulfate in situ at a rate of up to 4.0–36.8 nmol (g soil w. wt.)−1 day−1 (Pester et al., 2010). This could explain a considerable fraction of the radiotracer-measured SRRs of the same peatland, which ranged from 0 to ca. 340 nmol (g soil w. wt.)−1 day−1 over a depth profile of 0–30 cm and a 300-days period (Knorr and Blodau, 2009; Knorr et al., 2009). Modeling of sulfate diffusion to such highly active cells revealed sufficient sulfate supply even at bulk concentrations as low as 10 μM (Pester et al., 2010). In addition, apparent sulfate half-saturation concentrations, Km, for sulfate reducers in pure culture (Pallud and Van Cappellen, 2006 and references therein) and in sediment slurries (Tarpgaard et al., 2011) have been reported to be as low as 2–5 μM indicating no kinetic limitation from the electron acceptor side as well. These findings indicated that the identified peatland Desulfosporosinus sp. has the potential to contribute considerably to peatland sulfate reduction despite its very low abundance.
In the Schlöppnerbrunnen fen II, currently only one additional putative SRM, the Syntrophobacter-related dsrAB OTU 6, has been shown by quantitative PCR to be a autochthonous member of the “rare biosphere” (Figure 4; Steger et al., 2011). However, based on microarray analysis, a large fraction of the analyzed Schlöppnerbrunnen dsrAB OTUs were of low relative abundance or below the detection limit (Steger et al., 2011) and it can be assumed that a large part of the detected dsrAB diversity belongs to microorganisms that are part of the “rare biosphere” as well. However, in contrast to the rare peatland Desulfosporosinus spp., their contribution to peatland sulfate reduction is still unclear.
It has long been known that dissimilatory sulfate reduction occurs in freshwater wetlands, with pioneering studies dating back to the 1980s (Table 1 and references therein). Although the measured rates were often comparable to or partly exceeded SRR in marine environments (Jørgensen, 1982; Howarth and Jørgensen, 1984; Skyring, 1987), the importance of sulfate reduction to anaerobic carbon mineralization in wetlands is still often neglected. This can be mainly attributed to the current inability to quantitatively explain how the small sulfate pools are rapidly replenished to maintain the high SRR that can be repeatedly measured in different wetland types and over prolonged periods of time. Several concepts for this apparently hidden sulfur cycle, which is not only occurring at oxic–anoxic interfaces but also in anoxic soil layers, have been put forward but need further experimental confirmation, especially in the context of the intertwined redox-cycles of sulfur and iron compounds as well as of redox-active organic matter (Figure 2). In particular, there is a need to systematically quantify the varying contribution of sulfate reduction to overall anaerobic carbon degradation and to determine fluxes of sulfur recycling through the proposed intermediates elemental sulfur and thiosulfate in the oxic–anoxic transition zone and under completely anoxic conditions.
The identification of microorganisms responsible for sulfate reduction in freshwater wetlands has just begun. Molecular surveys have revealed that many members of the autochthonous wetland microbiota harbor diverse and novel variants of the marker genes dsrAB. However, at the moment it is not known whether these novel dsrAB-containing microorganisms perform dissimilatory sulfate reduction, are capable of alternative energy generation through syntrophy with methanogens, or switch between these different lifestyles depending on the prevailing environmental conditions. Targeted cultivation and substrate-mediated isotope labeling approaches thus need to be employed more systematically to detect “true” SRM in wetlands. The identification of a peatland Desulfosporosinus spp. by stable isotope probing as an active “rare biosphere” member with potentially high cell-specific SRR (Pester et al., 2010) and the cultivation of SRM from rice paddy soil (Wind and Conrad, 1995; Wind et al., 1999) provide examples for the success of such approaches. Comparative genomics of SRM strains isolated from contrasting environments that differ in biogeochemical parameters, such as sulfate concentration, availability of nutrients and/or pH, provide useful information on the different genomic and metabolic features that SRM have evolved to become a ubiquitously distributed microbial guild (Plugge et al., 2011; Zhou et al., 2011). Extending genome analyses to single cells (Podar et al., 2007; Woyke et al., 2009, 2010) retrieved directly from wetland soils could lay the foundation to identify microorganisms that harbor the great diversity of novel dsrAB found in wetlands and to get insights into their evolution and potential metabolic capabilities. Along with high-throughput amplicon sequencing of dsrA/B- or 16S rRNA genes and their transcripts in context with measurements of biogeochemical parameters and processes, these approaches will be crucial to identify SRM and understand their community dynamics and various life strategies in such heterogeneous environments such as wetlands.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are grateful to Kirsten Küsel, Harold Drake, and Christian Blodau for introducing us to the ecology of SRM and to the biogeochemical cycles at the Schlöppnerbrunnen fens. We heartily thank Kasper U. Kjeldsen for his continuous contributions to the dsrAB database. Bianca Pommerenke and Albert Müller are acknowledged for sequencing of dsrAB from rice paddy soil and for dsrAB database update and maintenance, respectively. This research was funded by the Austrian Science Fund (FWF): P23117-B17 to Michael Pester; P18836-B17 and P20185-B17 to Alexander Loy.
Achtnich, C., Bak, F., and Conrad, R. (1995). Competition for electron donors among nitrate reducers, ferric iron reducers, sulfate reducers, and methanogens in anoxic paddy soil. Biol. Fertil. Soils 19, 65–72.
Aeschbacher, M., Sander, M., and Schwarzenbach, R. P. (2010). Novel electrochemical approach to assess the redox properties of humic substances. Environ. Sci. Technol. 44, 87–93.
Alewell, C., and Gehre, M. (1999). Patterns of stable S isotopes in a forested catchment as indicators for biological S turnover. Biogeochemistry 47, 319–333.
Alewell, C., and Novak, M. (2001). Spotting zones of dissimilatory sulfate reduction in a forested catchment: the 34S-35S approach. Environ. Pollut. 112, 369–377.
Alewell, C., Paul, S., Lischeid, G., and Storck, F. R. (2008). Co-regulation of redox processes in freshwater wetlands as a function of organic matter availability? Sci. Total Environ. 404, 335–342.
Alewell, C., Paul, S., Lischeld, G., Kuesel, K., and Gehre, M. (2006). Characterizing the redox status in three different forested wetlands with geochemical data. Environ. Sci. Technol. 40, 7609–7615.
Baani, M., and Liesack, W. (2008). Two isozymes of particulate methane monooxygenase with different methane oxidation kinetics are found in Methylocystis sp. strain SC2. Proc. Natl. Acad. Sci. U.S.A. 105, 10203–10208.
Bak, F., and Cypionka, H. (1987). A novel type of energy metabolism involving fermentation of inorganic sulphur compounds. Nature 326, 891–892.
Bak, F., and Pfennig, N. (1991a). Microbial sulfate reduction in littoral sediment of Lake Constance. FEMS Microbiol. Ecol. 85, 31–42.
Bak, F., and Pfennig, N. (1991b). Sulfate-reducing bacteria in littoral sediment of Lake Constance. FEMS Microbiol. Ecol. 85, 43–52.
Batzer, D. P., and Sharitz, R. R. (2006). “Ecology of freshwater and estuarine wetlands. An introduction,” in Ecology of Freshwater and Estuarine Wetlands, eds D. P. Batzer, and R. R. Sharitz (Berkeley: University of California Press), 1–6.
Beer, J., Lee, K., Whiticar, M., and Blodau, C. (2008). Geochemical controls on anaerobic organic matter decomposition in a northern peatland. Limnol. Oceanogr. 53, 1393–1407.
Blodau, C. (2002). Carbon cycling in peatlands – a review of processes and controls. Environ. Rev. 10, 111–134.
Blodau, C., Mayer, B., Peiffer, S., and Moore, T. R. (2007). Support for an anaerobic sulfur cycle in two Canadian peatland soils. J. Geophys. Res. 112, 1–10.
Bosch, J., Lee, K.-Y., Jordan, G., Kim, K.-W., and Meckenstock, R. U. (2012). Anaerobic, nitrate-dependent oxidation of pyrite nanoparticles by Thiobacillus denitrificans. Environ. Sci. Technol. doi: 10.1021/es2022329
Bottrell, S. H., Hatfield, D., Bartlett, R., Spence, M. J., Bartle, K. D., and Mortimer, R. J. G. (2010). Concentrations, sulfur isotopic compositions and origin of organosulfur compounds in pore waters of a highly polluted raised peatland. Org. Geochem. 41, 55–62.
Brazelton, W. J., Ludwig, K. A., Sogin, M. L., Andreishcheva, E. N., Kelley, D. S., Shen, C.-C., Edwards, R. L., and Baross, J. A. (2010). Archaea and bacteria with surprising microdiversity show shifts in dominance over 1,000-year time scales in hydrothermal chimneys. Proc. Natl. Acad. Sci. U.S.A. 107, 1612–1617.
Bridgham, S. D., Megonigal, J. P., Keller, J. K., Bliss, N. B., and Trettin, C. (2006). The carbon balance of North American wetlands. Wetlands 26, 889–916.
Brown, K. A. (1986). Formation of organic sulfur in anaerobic peat. Soil Biol. Biochem. 18, 131–140.
Brüchert, V. (1998). Early diagenesis of sulfur in estuarine sediments: the role of sedimentary humic and fulvic acids. Geochim. Cosmochim. Acta 62, 1567–1586.
Brune, A., Frenzel, P., and Cypionka, H. (2000). Life at the oxic-anoxic interface: microbial activities and adaptations. FEMS Microbiol. Rev. 24, 691–710.
Canfield, D. E., Kristensen, E., and Thamdrup, B. (2005). “The sulfur cycle,” in Aquatic Geomicrobiology, eds D. E. Canfield, E. Kristensen, and B. Thamdrup (London: Academic Press), 313–381.
Canfield, D. E., Raiswell, R., and Bottrell, S. (1992). The reactivity of sedimentary iron minerals toward sulfide. Am. J. Sci. 292, 659–683.
Canfield, D. E., Raiswell, R., Westrich, J. T., Reaves, C. M., and Berner, R. A. (1986). The use of chromium reduction in the analysis of reduced inorganic sulfur in sediments and shales. Chem. Geol. 54, 149–155.
Castro, H., Reddy, K. R., and Ogram, A. (2002). Composition and function of sulfate-reducing prokaryotes in eutrophic and pristine areas of the Florida Everglades. Appl. Environ. Microbiol. 68, 6129–6137.
Chapman, S. J., and Davidson, M. S. (2001). 35S-sulphate reduction and transformation in peat. Soil Biol. Biochem. 33, 593–602.
Chauhan, A., and Ogram, A. (2006a). Fatty acid-oxidizing consortia along a nutrient gradient in the Florida Everglades. Appl. Environ. Microbiol. 72, 2400–2406.
Chauhan, A., and Ogram, A. (2006b). Phylogeny of acetate-utilizing microorganisms in soils along a nutrient gradient in the Florida Everglades. Appl. Environ. Microbiol. 72, 6837–6840.
Clymo, R. S. (1984). The limits to peat bog growth. Philos. Trans. R. Soc. Lond. B Biol. Sci. 303, 605–654.
Conrad, R. (2002). Control of microbial methane production in wetland rice fields. Nutr. Cycl. Agroecosyst. 64, 59–69.
Costello, E. K., and Schmidt, S. K. (2006). Microbial diversity in alpine tundra wet meadow soil: novel Chloroflexi from a cold, water-saturated environment. Environ. Microbiol. 8, 1471–1486.
Dalsgaard, T., and Bak, F. (1994). Nitrate reduction in a sulfate-reducing bacterium, Desulfovibrio desulfuricans, isolated from rice paddy soil: sulfide inhibition, kinetics, and regulation. Appl. Environ. Microbiol. 60, 291–297.
Dedysh, S. N., Derakshani, M., and Liesack, W. (2001). Detection and enumeration of methanotrophs in acidic Sphagnum peat by 16S rRNA fluorescence in situ hybridization, including the use of newly developed oligonucleotide probes for Methylocella palustris. Appl. Environ. Microbiol. 67, 4850–4857.
Dedysh, S. N., Pankratov, T. A., Belova, S. E., Kulichevskaya, I. S., and Liesack, W. (2006). Phylogenetic analysis and in situ identification of bacteria community composition in an acidic Sphagnum peat bog. Appl. Environ. Microbiol. 72, 2110–2117.
Deppe, M., McKnight, D. M., and Blodau, C. (2009). Effects of short-term drying and irrigation on electron flow in mesocosms of a northern bog and an alpine fen. Environ. Sci. Technol. 44, 80–86.
Dethlefsen, L., Huse, S., Sogin, M. L., and Relman, D. A. (2008). The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 6, e280. doi: 10.1371/journal.pbio.0060280
Detmers, J., Bruchert, V., Habicht, K. S., and Kuever, J. (2001). Diversity of sulfur isotope fractionations by sulfate-reducing prokaryotes. Appl. Environ. Microbiol. 67, 888–894.
Dhillon, A., Teske, A., Dillon, J., Stahl, D. A., and Sogin, M. L. (2003). Molecular characterization of sulfate reducing bacteria in the Guaymas Basin. Appl. Environ. Microbiol. 69, 2765–2772.
Dise, N., and Verry, E. (2001). Suppression of peatland methane emission by cumulative sulfate deposition in simulated acid rain. Biogeochemistry 53, 143–160.
Elsgaard, L., and Jørgensen, B. B. (1992). Anoxic transformations of radiolabeled hydrogen-sulfide in marine and fresh-water sediments. Geochim. Cosmochim. Acta 56, 2425–2435.
Ferdelman, T. G., Church, T. M., and Luther Iii, G. W. (1991). Sulfur enrichment of humic substances in a Delaware salt marsh sediment core. Geochim. Cosmochim. Acta 55, 979–988.
Fishbain, S., Dillon, J. G., Gough, H. L., and Stahl, D. A. (2003). Linkage of high rates of sulfate reduction in Yellowstone hot springs to unique sequence types in the dissimilatory sulfate respiration pathway. Appl. Environ. Microbiol. 69, 3663–3667.
Fleckenstein, J. H., Frei, S., and Knorr, K. H. (2011). “Modelling interactions between hydrologic dynamics and biogeochemical processes in a riparian wetland of a low-order stream,” in IUGG 2011, eds C. Abesser, G. Nützmann, M. C. Hill, G. Blöschl, and E. Lakshmanan (Melbourne: IAHS Red Books), 87–92.
Fossing, H. (1995). “S-35-radiolabeling to probe biogeochemical cycling of sulfur,” in Geochemical Transformations of Sedimentary Sulfur, eds M. A. Vairavamurthy, and M. A. A. Schoonen (Washington: American Chemical Society), 348–364.
Fossing, H., and Jørgensen, B. B. (1989). Measurement of bacterial sulfate reduction in sediments – evaluation of a single-step chromium reduction method. Biogeochemistry 8, 205–222.
Fossing, H., and Jørgensen, B. B. (1990). Isotope exchange reactions with radiolabeled sulfur compounds in anoxic seawater. Biogeochemistry 9, 223–245.
Friedrich, C. G., Bardischewsky, F., Rother, D., Quentmeier, A., and Fischer, J. (2005). Prokaryotic sulfur oxidation. Curr. Opin. Microbiol. 8, 253–259.
Frolking, S., Roulet, N. T., Moore, T. R., Richard, P. J. H., Lavoie, M., and Muller, S. D. (2001). Modeling northern peatland decomposition and peat accumulation. Ecosystems 4, 479–498.
Fuhrman, J. A. (2009). Microbial community structure and its functional implications. Nature 459, 193–199.
Gauci, V., Dise, N., and Fowler, D. (2002). Controls on suppression of methane flux from a peat bog subjected to simulated acid rain sulfate deposition. Global Biogeochem. Cycles 16, doi: 10.1029/2000gb001370
Gauci, V., Dise, N. B., Howell, G., and Jenkins, M. E. (2008). Suppression of rice methane emission by sulfate deposition in simulated acid rain. J. Geophys. Res. 113. doi: 10.1029/2007JG000501
Gauci, V., Matthews, E., Dise, N., Walter, B., Koch, D., Granberg, G., and Vile, M. (2004). Sulfur pollution suppression of the wetland methane source in the 20th and 21st centuries. Proc. Natl. Acad. Sci. U.S.A. 101, 12583–12587.
Ghani, A., McLaren, R. G., and Swift, R. S. (1993). Mobilization of recently formed soil organic sulfur. Soil Biol. Biochem. 25, 1739–1744.
Gregersen, L. H., Bryant, D. A., and Frigaard, N.-U. (2011). Mechanisms and evolution of oxidative sulfur metabolism in green sulfur bacteria. Front. Microbio. 2:116. doi: 10.3389/fmicb.2011.00116
Groscheova, H., Novak, M., and Alewell, C. (2000). Changes in the delta S-34 ratio of pore-water sulfate in incubated Sphagnum peat. Wetlands 20, 62–69.
Haaijer, S. C. M., Lamers, L. P. M., Smolders, A. J. P., Jetten, M. S. M., and Den Camp, H. (2007). Iron sulfide and pyrite as potential electron donors for microbial nitrate reduction in freshwater wetlands. Geomicrobiol. J. 24, 391–401.
Hamberger, A., Horn, M. A., Dumont, M. G., Murrell, J. C., and Drake, H. L. (2008). Anaerobic consumers of monosaccharides in a moderately acidic fen. Appl. Environ. Microbiol. 74, 3112–3120.
Harmsen, H. J. M., Van Kuijk, B. L. M., Plugge, C. M., Akkermans, A. D. L., De Vos, W. M., and Stams, A. J. M. (1998). Syntrophobacter fumaroxidans sp. nov., a syntrophic propionate-degrading sulfate-reducing bacterium. Int. J. Syst. Bacteriol. 48, 1383–1387.
He, J. Z., Liu, X. Z., Zheng, Y., Shen, J. P., and Zhang, L. M. (2010). Dynamics of sulfate reduction and sulfate-reducing prokaryotes in anaerobic paddy soil amended with rice straw. Biol. Fertil. Soils 46, 283–291.
Heitmann, T., and Blodau, C. (2006). Oxidation and incorporation of hydrogen sulfide by dissolved organic matter. Chem. Geol. 235, 12–20.
Heitmann, T., Goldhammer, T., Beer, J., and Blodau, C. (2007). Electron transfer of dissolved organic matter and its potential significance for anaerobic respiration in a northern bog. Glob. Chang. Biol. 13, 1771–1785.
Holler, T., Wegener, G., Niemann, H., Deusner, C., Ferdelman, T. G., Boetius, A., Brunner, B., and Widdel, F. (2011). Carbon and sulfur back flux during anaerobic microbial oxidation of methane and coupled sulfate reduction. Proc. Natl. Acad. Sci. U.S.A. 108, E1484–E1490.
Holmkvist, L., Ferdelman, T. G., and Jørgensen, B. B. (2011a). A cryptic sulfur cycle driven by iron in the methane zone of marine sediment (Aarhus Bay, Denmark). Geochim. Cosmochim. Acta 75, 3581–3599.
Holmkvist, L., Kamyshny, A. Jr., Vogt, C., Vamvakopoulos, K., Ferdelman, T. G., and Jørgensen, B. B. (2011b). Sulfate reduction below the sulfate-methane transition in Black Sea sediments. Deep Sea Res. Part I Oceanogr. Res. Pap. 58, 493–504.
Hori, T., Muller, A., Igarashi, Y., Conrad, R., and Friedrich, M. W. (2010). Identification of iron-reducing microorganisms in anoxic rice paddy soil by C-13-acetate probing. ISME J. 4, 267–278.
Horn, M. A., Matthies, C., Kuesel, K., Schramm, A., and Drake, H. L. (2003). Hydrogenotrophic methanogenesis by moderately acid-tolerant methanogens of a methane-emitting acidic peat. Appl. Environ. Microbiol. 69, 74–83.
Houweling, S., Kaminski, T., Dentener, F., Lelieveld, J., and Heimann, M. (1999). Inverse modeling of methane sources and sinks using the adjoint of a global transport model. J. Geophys. Res. 104, 26137–26160.
Howarth, R. W., and Jørgensen, B. B. (1984). Formation of 35S-labelled elemental sulfur and pyrite in coastal marine sediments (Limfjorden and Kysing Fjord, Denmark) during short-term 35SO42-reduction measurements. Geochim. Cosmochim. Acta 48, 1807–1818.
Hubert, C., Loy, A., Nickel, M., Arnosti, C., Baranyi, C., Bruchert, V., Ferdelman, T., Finster, K., Christensen, F. M., Rosa De Rezende, J., Vandieken, V., and Jørgensen, B. B. (2009). A constant flux of diverse thermophilic bacteria into the cold arctic seabed. Science 325, 1541–1544.
Imachi, H., Sakai, S., Ohashi, A., Harada, H., Hanada, S., Kamagata, Y., and Sekiguchi, Y. (2007). Pelotomaculum propionicicum sp. nov., an anaerobic, mesophilic, obligately syntrophic propionate-oxidizing bacterium. Int. J. Syst. Evol. Microbiol. 57, 1487–1492.
Imachi, H., Sekiguchi, Y., Kamagata, Y., Loy, A., Qiu, Y. L., Hugenholtz, P., Kimura, N., Wagner, M., Ohashi, A., and Harada, H. (2006). Non-sulfate-reducing, syntrophic bacteria affiliated with Desulfotomaculum cluster I are widely distributed in methanogenic environments. Appl. Environ. Microbiol. 72, 2080–2091.
Ingvorsen, K., Zeikus, J. G., and Brock, T. D. (1981). Dynamics of bacterial sulfate reduction in a eutrophic lake. Appl. Environ. Microbiol. 42, 1029–1036.
IPCC. (2007). Contribution of Working Groups I, II and III to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge: Cambridge University Press.
Jacks, G., and Norrström, A. C. (2004). Hydrochemistry and hydrology of forest riparian wetlands. For. Ecol. Manage. 196, 187–197.
Jackson, C. R., Liew, K. C., and Yule, C. M. (2009). Structural and functional changes with depth in microbial communities in a tropical Malaysian peat swamp forest. Microb. Ecol. 57, 402–412.
Jiang, L. J., Zheng, Y. P., Peng, X. T., Zhou, H. Y., Zhang, C. L., Xiao, X., and Wang, F. P. (2009). Vertical distribution and diversity of sulfate-reducing prokaryotes in the Pearl River estuarine sediments, Southern China. FEMS Microbiol. Ecol. 70, 249–262.
Jørgensen, B. B. (1978). Comparison of methods for the quantification of bacterial sulfate reduction in coastal marine sediments. 1. Measurements with radiotracer techniques. Geomicrobiol. J. 1, 11–27.
Jørgensen, B. B. (1982). Mineralization of organic matter in the sea bed – the role of sulphate reduction. Nature 296, 643–645.
Jørgensen, B. B. (1990). The sulfur cycle of fresh-water sediments – role of thiosulfate. Limnol. Oceanogr. 35, 1329–1342.
Jørgensen, C. J., Jacobsen, O. S., Elberling, B., and Aamand, J. (2009). Microbial oxidation of pyrite coupled to nitrate reduction in anoxic groundwater sediment. Environ. Sci. Technol. 43, 4851–4857.
Kallmeyer, J., Ferdelman, T. G., Weber, A., Fossing, H., and Jørgensen, B. B. (2004). A cold chromium distillation procedure for radiolabeled sulfide applied to sulfate reduction measurements. Limnol. Oceanogr. Methods 2, 171–180.
Kayranli, B., Scholz, M., Mustafa, A., and Hedmark, A. (2010). Carbon storage and fluxes within freshwater wetlands: a critical review. Wetlands 30, 111–124.
Keller, J. K., and Bridgham, S. D. (2007). Pathways of anaerobic carbon cycling across an ombrotrophic-minerotrophic peatland gradient. Limnol. Oceanogr. 52, 96–107.
Kelly, D. P., Shergill, J. K., Lu, W. P., and Wood, A. P. (1997). Oxidative metabolism of inorganic sulfur compounds by bacteria. Antonie Van Leeuwenhoek 71, 95–107.
Kendall, M. M., Liu, Y., and Boone, D. R. (2006). Butyrate- and propionate-degrading syntrophs from permanently cold marine sediments in Skan Bay, Alaska, and description of Algorimarina butyrica gen. nov., sp. nov. FEMS Microbiol. Lett. 262, 107–114.
Kertesz, M. A. (2000). Riding the sulfur cycle – metabolism of sulfonates and sulfate esters in Gram-negative bacteria. FEMS Microbiol. Rev. 24, 135–175.
King, G. M. (2001). “Radiotracer assays (S-35) of sulfate reduction rates in marine and freshwater sediments,” in Methods in Microbiology, ed. J. H. Paul (San Diego: Academic Press Inc.), 489–500.
Knittel, K., and Boetius, A. (2009). Anaerobic oxidation of methane: progress with an unknown process. Annu. Rev. Microbiol. 63, 311–334.
Knorr, K.-H., and Blodau, C. (2009). Impact of experimental drought and rewetting on redox transformations and methanogenesis in mesocosms of a northern fen soil. Soil Biol. Biochem. 41, 1187–1198.
Knorr, K. H., Lischeid, G., and Blodau, C. (2009). Dynamics of redox processes in a minerotrophic fen exposed to a water table manipulation. Geoderma 153, 379–392.
Kögel-Knabner, I., Amelung, W., Cao, Z., Fiedler, S., Frenzel, P., Jahn, R., Kalbitz, K., Koelbl, A., and Schloter, M. (2010). Biogeochemistry of paddy soils. Geoderma 157, 1–14.
Kondo, R., Purdy, K. J., Silva, S. D. Q., and Nedwell, D. B. (2007). Spatial dynamics of sulphate-reducing bacterial compositions in sediment along a salinity gradient in a UK estuary. Microbes Environ. 22, 11–19.
Koretsky, C. M., Haveman, M., Beuving, L., Cuellar, A., Shattuck, T., and Wagner, M. (2007). Spatial variation of redox and trace metal geochemistry in a minerotrophic fen. Biogeochemistry 86, 33–62.
Kraigher, B., Stres, B., Hacin, J., Ausec, L., Mahne, I., Van Elsas, J. D., and Mandic-Mulec, I. (2006). Microbial activity and community structure in two drained fen soils in the Ljubljana Marsh. Soil Biol. Biochem. 38, 2762–2771.
Küsel, K., Blothe, M., Schulz, D., Reiche, M., and Drake, H. L. (2008). Microbial reduction of iron and porewater biogeochemistry in acidic peatlands. Biogeosciences 5, 1537–1549.
Laue, H., Friedrich, M., Ruff, J., and Cook, A. M. (2001). Dissimilatory sulfite reductase (desulfoviridin) of the taurine-degrading, non-sulfate-reducing bacterium Bilophila wadsworthia RZATAU contains a fused DsrB-DsrD subunit. J. Bacteriol. 183, 1727–1733.
Lee, Y.-J., Romanek, C. S., and Wiegel, J. (2009). Desulfosporosinus youngiae sp. nov., a spore-forming, sulfate-reducing bacterium isolated from a constructed wetland treating acid mine drainage. Int. J. Syst. Evol. Microbiol. 59, 2743–2746.
Leloup, J., Fossing, H., Kohls, K., Holmkvist, L., Borowski, C., and Jørgensen, B. B. (2009). Sulfate-reducing bacteria in marine sediment (Aarhus Bay, Denmark): abundance and diversity related to geochemical zonation. Environ. Microbiol. 11, 1278–1291.
Leloup, J., Loy, A., Knab, N. J., Borowski, C., Wagner, M., and Jørgensen, B. B. (2007). Diversity and abundance of sulfate-reducing microorganisms in the sulfate and methane zones of a marine sediment, Black Sea. Environ. Microbiol. 9, 131–142.
Leloup, J., Petit, F., Boust, D., Deloffre, J., Bally, G., Clarisse, O., and Quillet, L. (2005). Dynamics of sulfate-reducing microorganisms (dsrAB genes) in two contrasting mudflats of the Seine estuary (France). Microb. Ecol. 50, 307–314.
Leloup, J., Quillet, L., Berthe, T., and Petit, F. (2006). Diversity of the dsrAB (dissimilatory sulfite reductase) gene sequences retrieved from two contrasting mudflats of the Seine estuary, France. FEMS Microbiol. Ecol. 55, 230–238.
Lennon, J. T., and Jones, S. E. (2011). Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat. Rev. Microbiol. 9, 119–130.
Lie, T. J., Leadbetter, J. R., and Leadbetter, E. R. (1998). Metabolism of sulfonic acids and other organosulfur compounds by sulfate-reducing bacteria. Geomicrobiol. J. 15, 135–149.
Lie, T. J., Pitta, T., Leadbetter, E. R., Godchaux, W., and Leadbetter, J. R. (1996). Sulfonates: novel electron acceptors in anaerobic respiration. Arch. Microbiol. 166, 204–210.
Limpens, J., Berendse, F., Blodau, C., Canadell, J. G., Freeman, C., Holden, J., Roulet, N., Rydin, H., and Schaepman-Strub, G. (2008). Peatlands and the carbon cycle: from local processes to global implications a synthesis. Biogeosciences 5, 1475–1491.
Lindau, C. W., Bollich, P. K., Delaune, R. D., Mosier, A. R., and Bronson, K. F. (1993). Methane mitigation in flooded Louisiana rice fields. Biol. Fertil. Soils 15, 174–178.
Liu, X. Z., Zhang, L. M., Prosser, J. I., and He, J. Z. (2009). Abundance and community structure of sulfate reducing prokaryotes in a paddy soil of southern China under different fertilization regimes. Soil Biol. Biochem. 41, 687–694.
Lovley, D. R., and Klug, M. J. (1983). Sulfate reducers can outcompete methanogens at freshwater sulfate concentrations. Appl. Environ. Microbiol. 45, 187–192.
Lovley, D. R., and Phillips, E. J. P. (1994). Novel processes for anaerobic sulfate production from elemental sulfur by sulfate-reducing bacteria. Appl. Environ. Microbiol. 60, 2394–2399.
Loy, A., Duller, S., Baranyi, C., Mußmann, M., Ott, J., Sharon, I., Béjà, O., Le Paslier, D., Dahl, C., and Wagner, M. (2009). Reverse dissimilatory sulfite reductase as phylogenetic marker for a subgroup of sulfur-oxidizing prokaryotes. Environ. Microbiol. 11, 289–299.
Loy, A., Duller, S., and Wagner, M. (2008). “Evolution and ecology of microbes dissimilating sulfur compounds: insights from siroheme sulfite reductases,” in Microbial Sulfur Metabolism, eds C. Dahl, and C. G. Friedrich (Berlin: Springer), 46–59.
Loy, A., Küsel, K., Lehner, A., Drake, H. L., and Wagner, M. (2004). Microarray and functional gene analyses of sulfate-reducing prokaryotes in low-sulfate, acidic fens reveal cooccurrence of recognized genera and novel lineages. Appl. Environ. Microbiol. 70, 6998–7009.
Loy, A., and Pester, M. (2010). “Probing identity and physiology of uncultured microorganisms with isotopic labeling techniques,” in Geomicrobiology: Molecular and Environmental Perspective, eds L. Barton, M. Mandl, and A. Loy (Heidelberg: Springer), 127–146.
Lu, Y. H., Rosencrantz, D., Liesack, W., and Conrad, R. (2006). Structure and activity of bacterial community inhabiting rice roots and the rhizosphere. Environ. Microbiol. 8, 1351–1360.
Ludwig, W., Strunk, O., Westram, R., Richter, L., Meier, H., Yadhukumar, Buchner, A., Lai, T., Steppi, S., Jobb, G., Forster, W., Brettske, I., Gerber, S., Ginhart, A. W., Gross, O., Grumann, S., Hermann, S., Jost, R., Konig, A., Liss, T., Lussmann, R., May, M., Nonhoff, B., Reichel, B., Strehlow, R., Stamatakis, A., Stuckmann, N., Vilbig, A., Lenke, M., Ludwig, T., Bode, A., and Schleifer, K.-H. (2004). ARB: a software environment for sequence data. Nucleic Acids Res. 32, 1363–1371.
Mandernack, K. W., Lynch, L., Krouse, H. R., and Morgan, M. D. (2000). Sulfur cycling in wetland peat of the New Jersey Pinelands and its effect on stream water chemistry. Geochim. Cosmochim. Acta 64, 3949–3964.
Matzner, E. (2004). Biogeochemistry of Forested Catchments in a Changing Environment – A German Case Study. Berlin: Springer Verlag.
Miletto, M., Loeb, R., Antheunisse, A. M., Bodelier, P. L. E., and Laanbroek, H. J. (2010). Response of the sulfate-reducing community to the re-establishment of estuarine conditions in two contrasting soils: a mesocosm approach. Microb. Ecol. 59, 109–120.
Mitsch, W. J., Gosselink, J. G., Anderson, C. J., and Zhang, L. (2009). Wetland Ecosystems. New York: John Wiley & Sons, Inc.
Moeslund, L., Thamdrup, B., and Jørgensen, B. B. (1994). Sulfur and iron cycling in a coastal sediment – radiotracer studies and seasonal dynamics. Biogeochemistry 27, 129–152.
Murase, J., and Kimura, M. (1997). Anaerobic reoxidation of Mn2+, Fe2+, S0 and S2- in submerged paddy soils. Biol. Fertil. Soils 25, 302–306.
Musat, N., Werner, U., Knittel, K., Kolb, S., Dodenhof, T., Van Beusekom, J. E. E., De Beer, D., Dubilier, N., and Amann, R. (2006). Microbial community structure of sandy intertidal sediments in the North Sea, Sylt-Rømø Basin, Wadden Sea. Syst. Appl. Microbiol. 29, 333–348.
Mussmann, M., Richter, M., Lombardot, T., Meyerdierks, A., Kuever, J., Kube, M., Glockner, F. O., and Amann, R. (2005). Clustered genes related to sulfate respiration in uncultured prokaryotes support the theory of their concomitant horizontal transfer. J. Bacteriol. 187, 7126–7137.
Muyzer, G., and Stams, A. J. M. (2008). The ecology and biotechnology of sulphate-reducing bacteria. Nat. Rev. Microbiol. 6, 441–454.
Nedwell, D. B., and Watson, A. (1995). CH4 production, oxidation and emission in a UK ombrotrophic peat bog – influence of SO42– from acid rain. Soil Biol. Biochem. 27, 893–903.
Nielsen, L. P., Risgaard-Petersen, N., Fossing, H., Christensen, P. B., and Sayama, M. (2010). Electric currents couple spatially separated biogeochemical processes in marine sediment. Nature 463, 1071–1074.
Novak, M., Vile, M. A., Bottrell, S. H., Stepanova, M., Jackova, I., Buzek, F., Prechova, E., and Newton, R. J. (2005). Isotope systematics of sulfate-oxygen and sulfate-sulfur in six European peatlands. Biogeochemistry 76, 187–213.
Orcutt, B. N., Sylvan, J. B., Knab, N. J., and Edwards, K. J. (2011). Microbial ecology of the dark ocean above, at, and below the seafloor. Microbiol. Mol. Biol. Rev. 75, 361–422.
Pallud, C., and Van Cappellen, P. (2006). Kinetics of microbial sulfate reduction in estuarine sediments. Geochim. Cosmochim. Acta 70, 1148–1162.
Paul, S., Kuesel, K., and Alewell, C. (2006). Reduction processes in forest wetlands: tracking down heterogeneity of source/sink functions with a combination of methods. Soil Biol. Biochem. 38, 1028–1039.
Pedrós-Alió, C. (2006). Marine microbial diversity: can it be determined? Trends Microbiol. 14, 257–263.
Peiffer, S., and Gade, W. (2007). Reactivity of ferric oxides toward H2S at low pH. Environ. Sci. Technol. 41, 3159–3164.
Perlinger, J. A., Kalluri, V. M., Venkatapathy, R., and Angst, W. (2002). Addition of hydrogen sulfide to juglone. Environ. Sci. Technol. 36, 2663–2669.
Pester, M., Bittner, N., Pinsurang, D., Wagner, M., and Loy, A. (2010). A “rare biosphere” microorganism contributes to sulfate reduction in a peatland. ISME J. 4, 1591–1602.
Plugge, C. M., Zhang, W., Scholten, J. C. M., and Stams, A. J. M. (2011). Metabolic flexibility of sulfate reducing bacteria. Front. Microbiol. 2:81. doi: 10.3389/fmicb.2011.00081
Podar, M., Abulencia, C. B., Walcher, M., Hutchison, D., Zengler, K., Garcia, J. A., Holland, T., Cotton, D., Hauser, L., and Keller, M. (2007). Targeted access to the genomes of low-abundance organisms in complex microbial communities. Appl. Environ. Microbiol. 73, 3205–3214.
Pyzik, A. J., and Sommer, S. E. (1981). Sedimentary iron monosulfides: kinetics and mechanism of formation. Geochim. Cosmochim. Acta 45, 687–698.
Rabus, R., Hansen, T. A., and Widdel, F. (2006). “Dissimilatory sulfate- and sulfur-reducing prokaryotes,” in The Prokaryotes: A Handbook on the Biology of Bacteria, 3rd Edn, eds M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (Singapore: Springer), 659–768.
Ramamoorthy, S., Sass, H., Langner, H., Schumann, P., Kroppenstedt, R. M., Spring, S., Overmann, J., and Rosenzweig, R. F. (2006). Desulfosporosinus lacus sp nov., a sulfate-reducing bacterium isolated from pristine freshwater lake sediments. Int. J. Syst. Evol. Microbiol. 56, 2729–2736.
Ravenschlag, K., Sahm, K., Pernthaler, J., and Amann, R. (1999). High bacterial diversity in permanently cold marine sediments. Appl. Environ. Microbiol. 65, 3982–3989.
Reeve, A. S., Evensen, R., Glaser, P. H., Siegel, D. I., and Rosenberry, D. (2006). Flow path oscillations in transient ground-water simulations of large peatland systems. J. Hydrol. 316, 313–324.
Roden, E. E., Kappler, A., Bauer, I., Jiang, J., Paul, A., Stoesser, R., Konishi, H., and Xu, H. F. (2010). Extracellular electron transfer through microbial reduction of solid-phase humic substances. Nat. Geosci. 3, 417–421.
Roden, E. E., and Urrutia, M. M. (2002). Influence of biogenic Fe(II) on bacterial crystalline Fe(III) oxide reduction. Geomicrobiol. J. 19, 209–251.
Roden, E. E., and Wetzel, R. G. (1996). Organic carbon oxidation and suppression of methane production by microbial Fe(III) oxide reduction in vegetated and unvegetated freshwater wetland sediments. Limnol. Oceanogr. 41, 1733–1748.
Scheid, D., and Stubner, S. (2001). Structure and diversity of Gram-negative sulfate-reducing bacteria on rice roots. FEMS Microbiol. Ecol. 36, 175–183.
Scheid, D., Stubner, S., and Conrad, R. (2004). Identification of rice root associated nitrate, sulfate and ferric iron reducing bacteria during root decomposition. FEMS Microbiol. Ecol. 50, 101–110.
Schippers, A., and Jørgensen, B. B. (2001). Oxidation of pyrite and iron sulfide by manganese dioxide in marine sediments. Geochim. Cosmochim. Acta 65, 915–922.
Schmalenberger, A., Drake, H. L., and Küsel, K. (2007). High unique diversity of sulfate-reducing prokaryotes characterized in a depth gradient in an acidic fen. Environ. Microbiol. 9, 1317–1328.
Segers, R., and Kengen, S. W. M. (1998). Methane production as a function of anaerobic carbon mineralization: a process model. Soil Biol. Biochem. 30, 1107–1117.
Segers, R., and Leffelaar, P. A. (2001). Modeling methane fluxes in wetlands with gas-transporting plants 1. Single-root scale. J. Geophys. Res. 106, 3511–3528.
Smemo, K. A., and Yavitt, J. B. (2011). Anaerobic oxidation of methane: an underappreciated aspect of methane cycling in peatland ecosystems? Biogeosciences 8, 779–793.
Smith, R. L., and Klug, M. J. (1981). Reduction of sulfur-compounds in the sediments of a eutrophic lake basin. Appl. Environ. Microbiol. 41, 1230–1237.
Smith, S. J., Pitcher, H., and Wigley, T. M. L. (2005). Future sulfur dioxide emissions. Clim. Change 73, 267–318.
Sogin, M. L., Morrison, H. G., Huber, J. A., Mark Welch, D., Huse, S. M., Neal, P. R., Arrieta, J. M., and Herndl, G. J. (2006). Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Natl. Acad. Sci. U.S.A. 103, 12115–12120.
Spratt, H. G., Morgan, M. D., and Good, R. E. (1987). Sulfate reduction in peat from a New Jersey pinelands cedar swamp. Appl. Environ. Microbiol. 53, 1406–1411.
Spring, S., and Rosenzweig, F. (2006). “The genera Desulfitobacterium and Desulfosporosinus: taxonomy,” in The Prokaryotes: A Handbook on the Biology of Bacteria, 3rd Edn, eds M. Dworkin, S. Falkow, E. Rosenberg, K. H. Schleifer, and E. Stackebrandt (Singapore: Springer), 771–786.
Stahl, D. A., Loy, A., and Wagner, M. (2007). “Molecular strategies for studies of natural populations of sulphate-reducing microorganisms,” in Sulphate-Reducing Bacteria, eds L. L. Barton, and W. A. Hamilton (Cambridge: Cambridge University Press), 39–115.
Steger, D., Wentrup, C., Braunegger, C., Deevong, P., Hofer, M., Pester, M., Wagner, M., and Loy, A. (2011). Microorganisms with novel dissimilatory (bi)sulfite reductase genes are widespread and core microbiota in low-sulfate peatlands. Appl. Environ. Microbiol. 77, 1231–1242.
Steinmann, P., and Shotyk, W. (1997). Chemical composition, pH, and redox state of sulfur and iron in complete vertical porewater profiles from two Sphagnum peat bogs, Jura Mountains, Switzerland. Geochim. Cosmochim. Acta 61, 1143–1163.
Stubner, S. (2002). Enumeration of 16S rDNA of Desulfotomaculum lineage 1 in rice field soil by real-time PCR with SybrGreenTM detection. J. Microbiol. Methods 50, 155–164.
Stubner, S. (2004). Quantification of Gram-negative sulphate-reducing bacteria in rice field soil by 16S rRNA gene-targeted real-time PCR. J. Microbiol. Methods 57, 219–230.
Tarpgaard, I. H., Røy, H., and Jørgensen, B. B. (2011). Concurrent low- and high-affinity sulfate reduction kinetics in marine sediment. Geochim. Cosmochim. Acta 75, 2997–3010.
Tchawa Yimga, M., Dunfield, P. F., Ricke, P., Heyer, J., and Liesack, W. (2003). Wide distribution of a novel pmoA-like gene copy among type II methanotrophs, and its expression in Methylocystis strain SC2. Appl. Environ. Microbiol. 69, 5593–5602.
Teske, A., Durbin, A., Ziervogel, K., Cox, C., and Arnosti, C. (2011). Microbial community composition and function in permanently cold seawater and sediments from an arctic fjord of Svalbard. Appl. Environ. Microbiol. 77, 2008–2018.
Thamdrup, B., Finster, K., Hansen, J. W., and Bak, F. (1993). Bacterial disproportionation of elemental sulfur coupled to chemical reduction of iron or manganese. Appl. Environ. Microbiol. 59, 101–108.
Thauer, R. K. (2011). Anaerobic oxidation of methane with sulfate: on the reversibility of the reactions that are catalyzed by enzymes also involved in methanogenesis from CO2. Curr. Opin. Microbiol. 14, 292–299.
Trimmer, M., Purdy, K. J., and Nedwell, D. B. (1997). Process measurement and phylogenetic analysis of the sulfate reducing bacterial communities of two contrasting benthic sites in the upper estuary of the Great Ouse, Norfolk, UK. FEMS Microbiol. Ecol. 24, 333–342.
Urban, N. R., Brezonik, P. L., Baker, L. A., and Sherman, L. A. (1994). Sulfate reduction and diffusion in sediments of Little-Rock Lake, Wisconsin. Limnol. Oceanogr. 39, 797–815.
Urban, N. R., Eisenreich, S. J., and Grigal, D. F. (1989). Sulfur cycling in a forested Sphagnum bog in Northern Minnesota. Biogeochemistry 7, 81–109.
Uthicke, S., and McGuire, K. (2007). Bacterial communities in Great Barrier Reef calcareous sediments: contrasting 16S rDNA libraries from nearshore and outer shelf reefs. Estuar. Coast. Shelf Sci. 72, 188–200.
Vairavamurthy, A., Mopper, K., and Taylor, B. F. (1992). Occurance of particle-bound polysulfides and significance of their reaction with organic matters in marine sediments. Geophys. Res. Lett. 19, 2043–2046.
van der Gon, D. H., Van Bodegom, P., Wassmann, R., Lantin, R., and Metra-Corton, T. (2001). Sulfate-containing amendments to reduce methane emissions from rice fields: mechanisms, effectiveness and costs. Mitigation Adapt. Strateg. Glob. Change 6, 71–89.
Vatsurina, A., Badrutdinova, D., Schumann, P., Spring, S., and Vainshtein, M. (2008). Desulfosporosinus hippei sp. nov., a mesophilic sulfate-reducing bacterium isolated from permafrost. Int. J. Syst. Evol. Microbiol. 58, 1228–1232.
Vile, M. A., Bridgham, S. D., and Wieder, R. K. (2003a). Response of anaerobic carbon mineralization rates to sulfate amendments in a boreal peatland. Ecol. Appl. 13, 720–734.
Vile, M. A., Bridgham, S. D., Wieder, R. K., and Novak, M. (2003b). Atmospheric sulfur deposition alters pathways of gaseous carbon production in peatlands. Global Biogeochem. Cycles 17, 1058.
Visscher, P. T., Gritzer, R. F., and Leadbetter, E. R. (1999). Low-molecular-weight sulfonates, a major substrate for sulfate reducers in marine microbial mats. Appl. Environ. Microbiol. 65, 3272–3278.
Vymazal, J. (2010). Constructed wetlands for wastewater treatment: five decades of experience. Environ. Sci. Technol. 45, 61–69.
Wagner, M., Loy, A., Klein, M., Lee, N., Ramsing, N. B., Stahl, D. A., and Friedrich, M. W. (2005). Functional marker genes for identification of sulphate-reducing prokaryotes. Methods Enzymol. 397, 469–489.
Wallrabenstein, C., Hauschild, E., and Schink, B. (1994). Pure culture and cytological properties of Syntrophobacter wolinii. FEMS Microbiol. Lett. 123, 249–254.
Wallrabenstein, C., Hauschild, E., and Schink, B. (1995). Syntrophobacter pfennigii sp nov, new syntrophically propionate-oxidizing anaerobe growing in pure culture with propionate and sulfate. Arch. Microbiol. 164, 346–352.
Ward, P. L. (2009). Sulfur dioxide initiates global climate change in four ways. Thin Solid Films 517, 3188–3203.
Watson, A., and Nedwell, D. B. (1998). Methane production and emission from peat: the influence of anions (sulphate, nitrate) from acid rain. Atmos. Environ. 32, 3239–3245.
Wieder, R. K., and Lang, G. E. (1988). Cycling of inorganic and organic sulfur in peat from Big Run Bog, West Virginia. Biogeochemistry 5, 221–242.
Wieder, R. K., Yavitt, J. B., and Lang, G. E. (1990). Methane production and sulfate reduction in 2 Appalachian peatlands. Biogeochemistry 10, 81–104.
Wilhelm, R. C., Niederberger, T. D., Greer, C., and Whyte, L. G. (2011). Microbial diversity of active layer and permafrost in an acidic wetland from the Canadian High Arctic. Can. J. Microbiol. 57, 303–315.
Wind, T., and Conrad, R. (1995). Sulfur compounds, potential turnover of sulfate and thiosulfate, and numbers of sulfate-reducing bacteria in planted and unplanted paddy soil. FEMS Microbiol. Ecol. 18, 257–266.
Wind, T., and Conrad, R. (1997). Localization of sulfate reduction in planted and unplanted rice field soil. Biogeochemistry 37, 253–278.
Wind, T., Stubner, S., and Conrad, R. (1999). Sulfate-reducing bacteria in rice field soil and on rice roots. Syst. Appl. Microbiol. 22, 269–279.
Woyke, T., Tighe, D., Mavromatis, K., Clum, A., Copeland, A., Schackwitz, W., Lapidus, A., Wu, D., McCutcheon, J. P., McDonald, B. R., Moran, N. A., Bristow, J., and Cheng, J.-F. (2010). One bacterial cell, one complete genome. PLoS ONE 5, e10314. doi: 10.1371/journal.pone.0010314
Woyke, T., Xie, G., Copeland, A., Gonzalez, J. M., Han, C., Kiss, H., Saw, J. H., Senin, P., Yang, C., Chatterji, S., Cheng, J.-F., Eisen, J. A., Sieracki, M. E., and Stepanauskas, R. (2009). Assembling the Marine metagenome, one cell at a time. PLoS ONE 4, e5299. doi: 10.1371/journal.pone.0005299
Wu, X.-J., Pan, J.-L., Liu, X.-L., Tan, J., Li, D.-T., and Yang, H. (2009). Sulfate-reducing bacteria in leachate-polluted aquifers along the shore of the East China Sea. Can. J. Microbiol. 55, 818–828.
Wuebbles, D. J., and Hayhoe, K. (2002). Atmospheric methane and global change. Earth Sci. Rev. 57, 177–210.
Wüst, P. K., Horn, M. A., and Drake, H. L. (2009). Trophic links between fermenters and methanogens in a moderately acidic fen soil. Environ. Microbiol. 11, 1395–1409.
Yagi, K., Tsuruta, H., and Minami, K. (1997). Possible options for mitigating methane emission from rice cultivation. Nutr. Cycl. Agroecosyst. 49, 213–220.
Yao, H., Conrad, R., Wassmann, R., and Neue, H. U. (1999). Effect of soil characteristics on sequential reduction and methane production in sixteen rice paddy soils from China, the Philippines, and Italy. Biogeochemistry 47, 269–295.
Yavitt, J. B., and Seidmann-Zager, M. (2006). Methanogenic conditions in northern peat soils. Geomicrobiol. J. 23, 119–127.
Zhou, J., He, Q., Hemme, C. L., Mukhopadhyay, A., Hillesland, K., Zhou, A., He, Z., Van Nostrand, J. D., Hazen, T. C., Stahl, D. A., Wall, J. D., and Arkin, A. P. (2011). How sulphate-reducing microorganisms cope with stress: lessons from systems biology. Nat. Rev. Microbiol. 9, 452–466.
Keywords: dsrAB, dissimilatory (bi)sulfite reductase, rare biosphere, sulfur cycle, peatland, rice paddy, sulfate-reducing microorganisms, sulfur pollution
Citation: Pester M, Knorr K-H, Friedrich MW, Wagner M and Loy A (2012) Sulfate-reducing microorganisms in wetlands – fameless actors in carbon cycling and climate change. Front. Microbio. 3:72. doi: 10.3389/fmicb.2012.00072
Received: 25 November 2011; Paper pending published: 05 January 2012;
Accepted: 11 February 2012; Published online: 28 February 2012.
Edited by:
Paul Bodelier, Netherlands Institute of Ecology, NetherlandsReviewed by:
Timothy Ferdelman, Max Planck Institute for Marine Microbiology, GermanyCopyright: © 2012 Pester, Knorr, Friedrich, Wagner and Loy. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Alexander Loy, Department for Microbial Ecology, University of Vienna, Althanstrasse 14, A-1090 Vienna, Austria. e-mail:bG95QG1pY3JvYmlhbC1lY29sb2d5Lm5ldA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.