- 1 Department of Microbiology, University of Georgia, Athens, GA, USA
- 2 Department of Marine Sciences, University of Georgia, Athens, GA, USA
Dimethylsulfoniopropionate (DMSP) is a metabolite produced primarily by marine phytoplankton and is the main precursor to the climatically important gas dimethylsulfide (DMS). DMS is released upon bacterial catabolism of DMSP, but it is not the only possible fate of DMSP sulfur. An alternative demethylation/demethiolation pathway results in the eventual release of methanethiol, a highly reactive volatile sulfur compound that contributes little to the atmospheric sulfur flux. The activity of these pathways control the natural flux of sulfur released to the atmosphere. Although these biochemical pathways and the factors that regulate them are of great interest, they are poorly understood. Only recently have some of the genes and pathways responsible for DMSP catabolism been elucidated. Thus far, six different enzymes have been identified that catalyze the cleavage of DMSP, resulting in the release of DMS. In addition, five of these enzymes appear to produce acrylate, while one produces 3-hydroxypropionate. In contrast, only one enzyme, designated DmdA, has been identified that catalyzes the demethylation reaction producing methylmercaptopropionate (MMPA). The metabolism of MMPA is performed by a series of three coenzyme-A mediated reactions catalyzed by DmdB, DmdC, and DmdD. Interestingly, Candidatus Pelagibacter ubique, a member of the SAR11 clade of Alphaproteobacteria that is highly abundant in marine surface waters, possessed functional DmdA, DmdB, and DmdC enzymes. Microbially mediated transformations of both DMS and methanethiol are also possible, although many of the biochemical and molecular genetic details are still unknown. This review will focus on the recent discoveries in the biochemical pathways that mineralize and assimilate DMSP carbon and sulfur, as well as the areas for which a comprehensive understanding is still lacking.
Introduction
Dimethylsulfoniopropionate (DMSP) is ubiquitous in marine surface waters, ranging in concentration from less than 1 nM in the open oceans to several micromolar in phytoplankton blooms (Van Duyl et al., 1998). The primary sources of DMSP in marine surface waters are micro and macro-algae (Yoch, 2002), although some halophytic plants also produce DMSP (Otte et al., 2004). DMSP is released from phytoplankton upon cellular lysis caused by zooplankton grazing (Wolfe and Steinke, 1997), senescence (Stefels and Van Boeckel, 1993), and viral infection (Hill et al., 1998). DMSP is produced by marine phytoplankton where it has been shown to possess a variety of functions, although its osmotic potential to regulate cell volume is the most widely recognized (Kirst, 1990). In some organisms it may function as an antioxidant (Sunda et al., 2002), predator deterrent (Wolfe and Steinke, 1997), and cryoprotectant (Karsten et al., 1996). These functions are common properties of other well-studied organic osmolytes (Yancey, 2005), thus DMSP may have different roles in different organisms. Consistent with its function as an organic osmolyte, DMSP accumulates to very high and osmotically significant concentrations in some marine phytoplankton, ranging from 0.1 to 1 M (reviewed in Stefels, 2000; Yoch, 2002).
The importance of DMSP lies not only in its availability as a source of reduced sulfur and carbon for marine microbes, but also because DMSP is the precursor for the climatically active gas dimethylsulfide (DMS; Lovelock et al., 1972). DMS is the primary natural source of sulfur to the atmosphere, where it is oxidized to sulfate, sulfur dioxide, methanesulfonic acid, and other products that act as cloud condensation nuclei (Hatakeyama et al., 1982). Although the total flux of DMS is less than half that of anthropogenic sulfur dioxide emissions, the longer residence time of DMS oxidation products in the atmosphere and the global distribution of DMS release result in a greater contribution of DMS to the atmospheric sulfur burden (Chin and Jacob, 1996). The relationship between solar radiation and DMS concentration is known as the CLAW hypothesis, an acronym from the first letter of the author’s surnames (Charlson et al., 1987), which states that increased levels of solar radiation and the resulting higher temperatures encourage growth of DMSP-producing marine phytoplankton and increased total DMSP production. The resulting increase in the amount of DMS released into the atmosphere then causes an increase in the abundance of cloud condensation nuclei, which then causes a decrease in solar radiation, slower growth of marine phytoplankton, and decreased DMSP production. These coupled processes then form a negative feedback loop. James Lovelock, an author on the original CLAW hypothesis, later proposed the “anti-CLAW” hypothesis, which described a positive feedback between global temperature and DMS production. Increasing global temperatures and resulting surface water temperatures may cause increased stratification of the oceans. Stratification would then decrease the flux of nutrients from deeper waters to the surface, resulting in decreased phytoplankton growth, thereby decreasing DMSP and DMS production (Lovelock, 2006).
In support of the CLAW hypothesis, strong correlations between DMS concentration and the dose of solar radiation have been reported (Vallina and Simo, 2007). However, the factors governing the production and atmospheric release of DMS are complicated. Marine bacteria were only identified as the primary mediators of DMSP catabolism after the publication of the CLAW hypothesis. It was also discovered that marine bacteria consume DMSP through an alternative pathway that does not produce DMS. Instead, it produces the more highly reactive volatile sulfur species methanethiol (MeSH) that contributes little to the atmospheric sulfur flux (Kiene and Taylor, 1988b; Taylor and Gilchrist, 1991). This phenomenon led to the proposal of a “bacterial switch,” in which marine bacteria shift between producing more or less DMS and MeSH (Simo, 2001). The gene responsible for the initial demethylation of DMSP leading to MeSH production, whose expression and activity contribute to control for the bacterial switch, was identified in 2006 (Howard et al., 2006). This was first of several genes identified that encode enzymes that directly consume DMSP, the details of which are discussed below. Many of these studies were performed in cultured representatives of the well-studied roseobacters, a phylogenetically coherent clade of clade of Alphaproteobacteria that are mostly marine in origin (Buchan et al., 2005; Wagner-Dobler and Biebl, 2006).
Background
Studies involving the enzymatic reactions of important sulfur transformations have been ongoing for years. Despite this, there are large gaps in our understanding of the specific transformations of DMSP and its degradation products. While a number of enzymes were previously purified and characterized as discussed below, the genes were not identified at the time. The lack of identified genes was unfortunate as the explosion in metagenomic and metatranscriptomic data provided an opportunity to further our understanding of their distribution and expression in the environment. However, in the last few years the identification of gene products responsible for the direct transformations of DMSP through the cleavage or demethylation pathways, as well as down-stream metabolic pathways, have made such studies possible (Howard et al., 2008; Raina et al., 2010; Varaljay et al., 2010; Vila-Costa et al., 2010; Reisch et al., 2011).
DMSP Synthesis
Three pathways have been described for the biosynthesis of DMSP: in the beach sunflower [Wollastonia biflora (L.) DC; Rhodes et al., 1997], the smooth cordgrass (Spartina alterniflora Loisel; Kocsis and Hanson, 2000), and sea lettuce (Ulva lactuca L.; Gage and Rhodes, 1997). However, the genes that encode most of the enzymes are not known. Each of these pathways share methionine as the starting compound but differ in the subsequent steps. Thus, the sulfur from DMSP and methionine share the same origin. Most evidence suggests that this sulfur is assimilated from sulfate using the adenosine 5′-phosphosulfate/3′-phosphoadenosine 5′-phosphosulfate (APS/PAPS) system that produces sulfite (Stefels, 2000). Sulfite is then reduced to sulfide in a reaction catalyzed by sulfite reductase and requiring six electrons. Sulfide is incorporated into cysteine, the precursor of methionine biosynthesis.
Environmental Fate of DMSP
There are three separate fates of DMSP sulfur: (1) production of volatile species and evolution to the atmosphere, (2) assimilation by marine microorganisms, (3) oxidation followed by release or re-assimilation. These three fates were demonstrated in a study using a 35S DMSP tracer to track the partitioning of DMSP sulfur to various products in oceanic and coastal seawater (Kiene and Linn, 2000). About 15% of added DMSP was taken up by bacterial cells but not further metabolized even after 24 h of incubation, suggesting an intracellular accumulation of DMSP. This phenomenon was also observed in studies of chemostat-grown Ruegeria pomeroyi, a model organism for the roseobacter clade of marine Alphaproteobacterium (Reisch et al., 2008). In the Kiene and Linn (2000) study most of the DMSP was incorporated into protein or transformed to dissolved non-volatile products (DNVS). The DNVS was probably formed by oxidation to DMSO and sulfate. There was also a large difference in the amount of DMSP routed through the demethylation pathway and partitioned as protein or DNVS between coastal and open ocean waters. In coastal samples, 60% of DMSP was assimilated. In ocean samples, only 16% was assimilated, and the remainder was found as DNVS. The reason for this difference is thought to be related to the sulfur demand of the cells from different marine environments. Cells within the coastal samples are likely to have higher growth rates and therefore an increased sulfur demand, causing more sulfur to be assimilated, and less oxidized. In both coastal and oceanic samples, only a small portion of the total DMSP, an average of 10%, was routed through the DMSP-cleavage pathway and DMS production.
Phytoplankton Cleavage
Marine phytoplankton are the primary synthesizers of DMSP, while marine bacteria are the primary degraders. However, field studies have suggested that dinoflagellates may contribute significantly to the release of DMS in phytoplankton blooms (Steinke et al., 2002), and some marine phytoplankton also have the capacity to degrade DMSP through the cleavage pathway. Four out of five cultured strains of the dinoflagellate Symbiodinium microadriaticum possessed DMSP lyase activity, although the rates varied significantly between strains (Yost and Mitchelmore, 2009). In addition, while DMSP production in coccolithophores is ubiquitous, DMSP lyase activity is not. A study of 10 strains of coccolithophores found that only those closely related to Emiliania huxleyi (Lohm.) Hay and Mohler and Gephyrocapsa oceanica Kamptner were capable of DMS production (Franklin et al., 2010). E. huxleyi is a well-studied model organism that is highly abundant in marine surface waters where it is often, but not always, numerically dominant. The distribution of DMSP lyase activity across the phylogenetic range of the coccolithophores is not yet known, complicating our understanding of the coccolithophore contribution to DMS production (Franklin et al., 2010). Studies of E. huxleyi showed that significant amounts of DMS are only produced upon cell damage. This evidence suggests that the DMSP lyase is physically separated from the cell’s cytoplasm, where intracellular DMSP is stored and may act as a signaling molecule (Wolfe and Steinke, 1996).
Dimethylsulfoniopropionate lyase enzymes have been purified from the green macroalgae [Ulva curvata (Kütz.) De Toni; De Souza and Yoch, 1995a] and the red macroalgae (Polysiphonia paniculata Montagne; Nishiguchi and Goff, 1995). The identities of genes encoding these enzymes remain unknown, and it is not known whether or not the two enzymes are related.
While marine bacteria are the primary mediators of DMSP degradation, there is evidence that phytoplankton may be responsible for a large part of the DMSP-cleavage reaction and DMS release in the ocean. A number of modeling studies have recently attributed increased importance to the phytoplankton contribution to DMS production (Toole and Siegel, 2004; Toole et al., 2006, 2008). One study found that the solar radiation-induced release of DMS from phytoplankton cells was necessary to produce realistic DMS predictions and reproduce the summer DMS accumulation found in surface waters (Vallina et al., 2008). Models that omit DMS release from phytoplankton underestimate the surface-to-air flux by 25%, indicating a significant phytoplankton contribution to the DMSP-cleavage pathway (Van Den Berg et al., 1996).
Bacterial Cleavage
Dimethylsulfoniopropionate in marine waters undergoes a non-enzymatic hydrolysis that releases DMS and acrylate. In the absence of biotic processes, the half-life of DMSP in seawater is about 8 years, a rate of hydrolysis that is far too low to account for the observed turnover of DMSP in natural waters (Dacey and Blough, 1987). This realization and the identification of DMS-producing bacteria suggested that bacteria were the primary mediators of DMSP degradation. A bacterial DMSP lyase (E.C. 4.4.1.3) was first purified and characterized in 1995 from a marine isolate, Alcaligenes faecalis M3A (De Souza and Yoch, 1995b). The enzyme had a Km for DMSP of 1.41 mM and a Vmax of 402 μmol min−1 mg of protein−1. However, the protein-encoding gene was not identified until about 15 years later.
DMSP-Cleavage Enzyme DddY
A recent re-examination of A. facecalis M3A identified the gene responsible for encoding the DMSP lyase (Curson et al., 2011). This protein, designated DddY, possessed no known functional domains, and unlike the other DMSP-cleavage enzymes identified was located in the bacterial periplasmic space, as originally found in 1995 (De Souza and Yoch, 1995b). The dddY gene was located on the chromosome near genes that conferred the ability to metabolize acrylate, much like the pathway discussed below from Halomonas HTNK1. Interestingly, this gene was not present in the global ocean survey (GOS) marine metagenomic database (Rusch et al., 2007), which suggests that it does not play a major role in DMSP processing in marine surface waters. However, it may be abundant in anoxic areas of marine sediment, where it may have some ecological significance (Curson et al., 2011).
DMSP-Cleavage Enzyme DddD
Most early predictions regarding the pathways of DMSP catabolism assumed that the DMSP-cleavage pathway would split DMSP into DMS and acrylate. Identification of genes catalyzing the cleavage reaction have mostly proven these early hypotheses correct, with the exception of the first cleavage enzyme identified, DddD. This enzyme turned out to produce 3-hydroxypropionate instead of acrylate (Todd et al., 2007, 2009b). The gene was identified in a bacterial isolate cultured from the rhizosphere of the salt marsh grass Spartina anglica C.E.Hubb. Based upon 16S rRNA gene sequencing, the bacterium was closely related to a member of the genus Marinomonas, and was designated strain MWYL1. A dddD mutation in this organism completely abolished DMS production. However, in R. pomeroyi, dddD does not appear to encode a major pathway of DMSP-cleavage. Inactivation of the dddD gene in R. pomeroyi had no effect on DMSP metabolism under the conditions tested, but this organism contains three additional DMSP-cleavage enzymes, as discussed below (Todd et al., 2010). The dddD gene possessed similarity to acyl-CoA transferases, which was unexpected for a lyase. Because of this annotation, the enzyme was originally hypothesized to catalyze the formation of a DMSP-coenzyme-A-thioester, which would spontaneously hydrolyze to DMS and 3-hydroxypropionyl-CoA. Although the DddD enzyme has not been purified and characterized biochemically, the activity of the recombinant dddD from Halomonas HTNK1 was investigated (Todd et al., 2009b). When dddD alone was expressed and the culture was provided with [1-13C] or [1-14C] DMSP, only 3-hydroxypropionate was detected after overnight incubation. The authors conclude that 3-hydroxypropionate, not a DMSP-CoA thioester as initially proposed, was the product of the DddD catalyzed reaction. However, these experiments used millimolar concentrations of DMSP, and it is unlikely that an equimolar buildup of a coenzyme-A thioester would occur. Thus, the actual product of DddD may be a coenzyme-A thioester, such as acryloyl-CoA or 3-hydroxypropionyl-CoA, which E. coli then metabolizes releasing 3-hydroxypropionate. The authors note that, for unknown reasons, cell extracts with DddD possess no activity, thus precluding in vitro biochemical characterization.
DMSP-Cleavage Enzyme DddL
Identification of the first gene encoding an authentic DMSP lyase was reported in 2008 and designated dddL (Curson et al., 2008). The gene was identified by expression of a cosmid library of the Sulfitobacter sp. EE-36 genome in the Rhizobium leguminosarum strain J391. This strain was used for expression because it was an Alphaproteobacterium, like Sulfitobacter sp. EE-36, making expression of recombinant proteins more likely. The clone that possessed dddL was able to produce low levels of DMS and consequently a dddL deletion mutant in Sulfitobacter EE-36 was unable to produce DMS from DMSP. The amino acid sequence of DddL lacked similarity to any proteins of known function, and homologous genes in both the cultured and metagenomic databases were rare. However, two strains of Rhodobacter sphaeroides, a well-studied organism not previously known to consume DMSP, possessed dddL, while a third strain did not. Accordingly, the two strains with dddL produced DMS from DMSP but not the third. Although DddL was not purified and characterized in vitro, recombinant E. coli expressing DddL released large amounts of acrylate into the medium when provided DMSP, suggesting that DddL was a DMSP lyase.
DMSP-Cleavage Enzyme DddP
In 2009 a third enzyme, designated DddP, was identified using a cosmid library from Roseovarius nubinhibens ISM (Todd et al., 2009a). Mutation of the dddP gene in both R. nubinhibens and R. pomeroyi significantly decreased but did not abolish the production of DMS from DMSP, suggesting that these organisms possessed a second DMSP-cleaving enzyme (Todd et al., 2009a, 2010). Upon purification and characterization of the enzyme, 13C, and 14C-DMSP isotope studies showed that the enzyme was a true DMSP lyase, releasing DMS, and acrylate (Kirkwood et al., 2010). The gene was originally annotated as an M24 metallopeptidase, but the subsequent characterization showed that the DddP was neither a metalloenzyme nor a peptidase. This observation was unusual, but not unprecedented. Creatinase from Paracoccus sp. WB1 was also annotated as a metallopeptidase but also did not contain metals (Wang et al., 2006). The enzyme had a Km of 13.8 mM and Vmax of 0.3 μmol min−1 mg of protein−1 with DMSP as the substrate. Compared to the kinetic values described above for DddY, both the affinity and maximum rate of catalysis are low. Low affinities for DMSP were previously reported for a DMSP-cleavage enzyme (De Souza and Yoch, 1995b) and the DMSP demethylase discussed below. Unfortunately, substrate specificity of the enzyme was not examined, and given the low Vmax it is possible that DMSP is not the only physiological substrate. Thus, the enzyme may have a broad substrate specificity and catalyze multiple reactions in the cell. While the enzymes specificity may have physiological implications, it would nonetheless catalyze the DMSP-cleavage reaction. Close homologs to the dddP gene were found in several roseobacters and, surprisingly, in a few fungal species. It was confirmed that fungal species with the dddP gene produced DMS, while those without the gene were not, suggesting that horizontal gene transfer was responsible for this unusual gene distribution (Todd et al., 2009a).
DMSP-Cleavage Enzyme DddQ
The fourth enzyme identified was encoded by a gene designated dddQ (Todd et al., 2010). As stated above, the dddP gene knockout in Roseovarius nubinhibens did not abolish the production of DMS. Therefore, the cosmid library was searched for a second gene capable of conferring DMSP-cleavage activity. Two adjacent genes in the middle of a 10 gene cluster were identified. When cloned and expressed in E. coli, each gene conferred the ability to produce DMS from DMSP, and they were designated dddQ. Assays with cell extracts from the recombinant E. coli with [1-13C] and [1-14C] DMSP showed that the three carbon moiety produced in the reaction was acrylate. A few roseobacters possessed dddQ homologs, including R. pomeroyi. Unlike R. nubinhibens, which has two adjacent copies, R. pomeroyi possesses only one copy. R. pomeroyi with a mutation in the dddQ gene was still capable of DMS production, though the rate was diminished by 95%, which is consistent with the presence of additional DMSP-cleavage enzymes.
DMSP-Cleavage Enzyme DddW
The sixth enzyme identified that catalyzes DMSP-cleavage was encoded by a gene designated dddW (Todd et al., 2011). This gene was identified in R. pomeroyi, where microarray experiments showed that the gene was significantly induced in the presence of DMSP. Cloning and expressing the gene in E. coli conferred the ability to form DMS from DMSP. Cell-free extracts possessed activity that cleaved DMSP into acrylate and DMS. This gene possessed no sequence similarity to genes with known function, but the polypeptide sequence possessed a predicted cupin-binding fold, like that of DddP and DddQ.
Distribution of DMSP-Cleavage Enzymes
The distribution and abundance of the DMSP-catabolizing genes in the metagenomic database have recently been reported (Howard et al., 2008; Raina et al., 2010; Moran et al., 2012). While numbers alone are unlikely to fully decipher the ecological role or significance of DMSP catabolism in the environment, they do reveal which genes are likely to be important on a global scale. As discussed below, the DMSP demethylase gene, dmdA, is the most abundant gene found in the GOS that acts directly on DMSP. Of the DMSP-cleavage enzymes identified thus far, dddP is by far the most abundant, found in 6% of bacteria examined in the GOS (Moran et al., 2012). This abundance is due to the genes presence in many roseobacters, as well as Candidatus Puniceispirillum marinum, the cultured representative of the SAR116 clade of Alphaproteobacteria. The abundance is consistent with the ribotype abundance identified in the GOS, where the SAR116 cluster represented 2.7% and the roseobacters 2.6% of sequences (Biers et al., 2009). In contrast, the SAR11 clade of the Alphaproteobacteria constitutes 31% of sequenced ribotypes in the GOS (Biers et al., 2009), which is consistent with the dmdA abundance of 27% (Moran et al., 2012).
The diversity of the DMSP-cleavage enzymes identified thus far is surprising. While there are numerous examples of non-homologous isofunctional enzymes (Omelchenko et al., 2010), six examples in closely related bacteria is unusual. This diversity suggests that there may be more, yet-unidentified enzymes catalyzing the cleavage reaction. For example, dddW, the most recently identified DMSP lyase gene, has only one highly similar homolog in the entire genomic database (Todd et al., 2011). This gene was identified in the well-studied bacterium R. pomeroyi, and it is likely that if other bacteria were screened with similar depth, by either whole-genome transcriptional analysis or whole-genome cloning, more cleavage enzymes would be found. The contribution of these low-abundance DMSP-cleavage enzymes to the flux of DMSP in the environment is probably minimal on an individual basis. But if there are in fact more of these novel DMSP lyases harbored by less understood bacteria, they may contribute significantly to the total flux.
DMSP Demethylation
The gene catalyzing the initial demethylation of DMSP was identified in 2006 in R. pomeroyi using a transposon mutant library. The gene, designated dmdA, was originally annotated as a glycine cleavage T-protein (GcvT), one of four proteins in the glycine degradation system (Okamuraikeda et al., 1982). Like GcvT, DmdA requires tetrahydrofolate (THF) to accept the methyl group from DMSP. DmdA from both Candidatus P. ubique and R. pomeroyi were purified and characterized, and both possessed similar kinetic properties (Reisch et al., 2008). The enzymes had low affinities for DMSP, with Kms of 13.2 and 5.4 mM for the enzymes from Candidatus P. ubique and R. pomeroyi, respectively. During growth on DMSP, R. pomeroyi cultures maintained an intracellular concentration of DMSP of ∼70 mM, which would allow for near maximal activity of DmdA in vivo. Such a high concentration of DMSP is osmotically significant and suggests that R. pomeroyi accumulates DMSP as an organic osmolyte. It is notable that the enzyme had a strict substrate specificity, indicating that the enzyme evolved to function with DMSP and was not a promiscuous enzyme that catalyzed multiple reactions.
Subsequent analysis of dmdA in marine metagenomic data showed that this gene is particularly abundant in ocean surface waters, with estimates placing the number of cells that possess this gene ranged at 27% (Moran et al., 2012). One reason for this high abundance was confirmed recently. In laboratory experiments the SAR11 clade bacterium Candidatus P. ubique required an exogenous source of reduced sulfur, such as DMSP or methionine, to reach high cell density in culture (Tripp et al., 2008). Despite very high concentrations of sulfate in most of the surface ocean, Candidatus P. ubique is incapable of utilizing this potential sulfur source as it does not possess the genes required for assimilatory sulfate reduction (Tripp et al., 2008). Instead, the bacterium uses only reduced sources of sulfur, and they are even selective in their use of reduced species. For instance, cysteine did not increase growth yields to the same extent as DMSP and methionine. Presumably, the inability to assimilate oxidized sulfur compounds results from the energetic costs associated with sulfate reduction. For these extreme oligotrophs, electron donors may be very scarce.
First Demethylation Carbon
There are two products of the DMSP demethylation reaction catalyzed by DmdA, MMPA, and 5-methyl-THF, which carries the methyl group removed from DMSP (Figure 1). The fate of 5-methyl-THF has not been directly studied, but the possibilities are numerous as it is a major donor of single carbon units in bacterial cells (Figure 2). Many organisms oxidize 5-methyl-THF to 5,10-methylene-THF by 5,10-methylene-THF reductase (MetF, 1.5.1.20) and subsequently to 5-formyl-THF by methylene-THF dehydrogenase (FolD, 1.5.1.5). 5-methyl-THF is also the methyl donor for methionine and S-adenosyl-methionine synthesis, while 5-formyl-THF is the source of two carbon atoms in purine nucleoside biosynthesis. 5,10-methylene-THF serves as a carbon donor for the conversion of glycine to serine, a reaction that is also part of the serine assimilation pathway for C-1 assimilation.
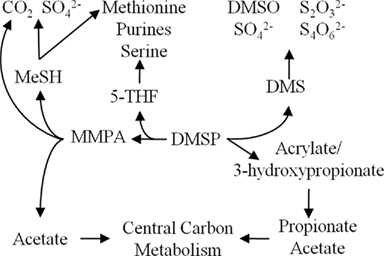
Figure 1. Overview of DMSP catabolic pathways in marine bacteria and the fates of carbon and sulfur.
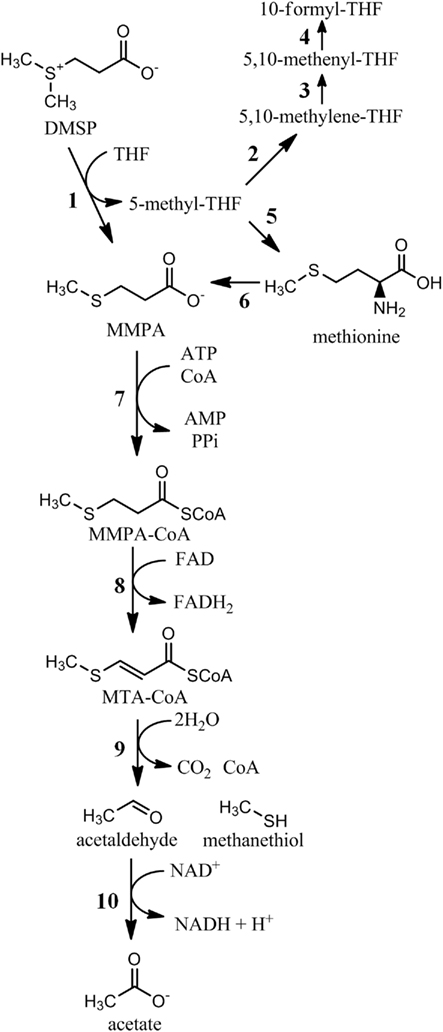
Figure 2. Biochemical pathways of DMSP demethylation. 1, DMSP demethylase (DmdA); 2, 5,10-methylene-THF reductase (MetF, E.C. 1.5.1.20); 3, methylene-THF dehydrogenase (FolD, E.C. 1.5.1.5); 4, methenyl-THF-cyclohydrolase (E.C. 3.5.4.9); 5, methionine synthase (MetH, E.C. 2.1.1.13); 6, methionine salvage pathway (multiple enzymes); 7, MMPA-CoA ligase (DmdB); 8, MMPA-CoA dehydrogenase (DmdC); 9, methylthioacryloyl-CoA hydratase (DmdD); 10, acetaldehyde dehydrogenase (E.C. 1.2.1.10).
MMPA Demethiolation
Most MMPA produced during DMSP degradation is further processed through a demethiolation pathway that releases the volatile sulfur compound MeSH (Figure 3). This transformation was long thought be the result of a cleavage or a reductive cleavage reaction, producing the three carbon intermediate propionate or acrylate (Kiene and Taylor, 1988b; Taylor and Gilchrist, 1991). However, an alternative hypothesis suggested that MMPA may be catabolized in a fatty acid β-oxidation-like pathway (Taylor and Visscher, 1996; Bentley and Chasteen, 2004). Recently, it was confirmed that the latter pathway is present in R. pomeroyi and required for MeSH production (Reisch et al., 2011). In this pathway, a series of three coenzyme-A mediated reactions catalyze the demethiolation of MMPA. First, a MMPA-CoA thioester is formed in an ATP-dependent reaction catalyzed by an enzyme designated as DmdB, encoded by a gene originally annotated as a medium chain fatty-acid-CoA ligase. Next, the MMPA moiety of MMPA-CoA is dehydrogenated between its α and β carbons, creating a double bond, transferring two electrons to FAD, and forming methylthioacryloyl-CoA (MTA-CoA). The third step, which mediates the actual demethiolation, is a unique reaction catalyzed by a crotonase-type enzyme. Hydration of MTA-CoA leads to its decomposition into MeSH, acetaldehyde, CO2, and free CoA. Like dmdA, the genes that encode dmdB and dmdC are abundant in the marine metagenomic database. In contrast, dmdD is rare, although an Ruegeria lacuscaerulensis, which did not possess dmdD, was also capable of the DmdD-catalyzed reaction, suggesting a non-orthologous gene had replaced dmdD in at least some bacteria. Unlike dmdA though, the MMPA-CoA pathway was shown to be present in a wide variety of bacteria, including many that are not associated with DMSP catabolism.
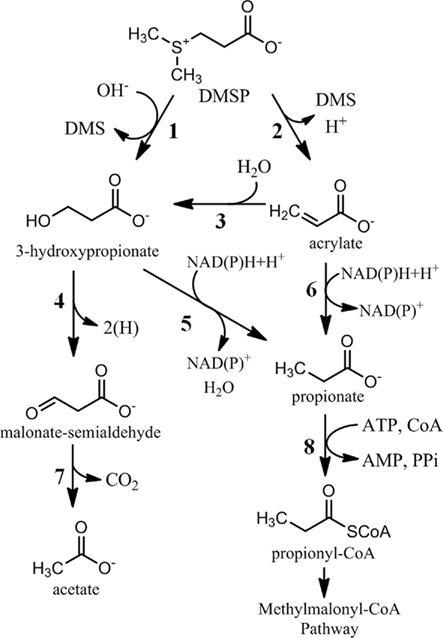
Figure 3. Dimethylsulfoniopropionate cleavage pathways leading to central carbon metabolism. Reactions 5 and 6 may be coenzyme-A mediated and would therefore bypass reaction 8. 1, DMSP-cleavage enzyme (DddD); 2, DMSP lyase (DddL, DddP, DddQ, DddY, DddW, E.C. 4.4.1.3); 3, acrylate hydratase; 4, 3-hydroxypropionate dehydrogenase; 5, 3-hydroxypropionate reductase; 6, acrylate reductase (1.3.99.3); 7, malonate semialdehyde dehydrogenase/decarboxylase (E.C. 1.2.1.18); 8, propionate-CoA ligase (PrpE, E.C. 6.2.1.17).
Methionine as a Source of MMPA
The distribution of the key genes of the MMPA-CoA pathway (dmdB and dmdC) is much broader than that of dmdA. Moreover, recombinant proteins from many terrestrial bacteria possess these activities and whole cells of several distantly related bacteria release MeSH from MMPA (Reisch et al., 2011). Conservation of this pathway amongst such a diverse group of bacteria suggests physiological importance but raises the question of how cells that are incapable of demethylating DMSP obtain MMPA.
A possible source of MMPA may be the methionine salvage pathway, which is found in many types of organisms and has a primary function of recycling the reduced thiomethyl moiety from methionine (reviewed in Albers, 2009). The condensation of methionine and ATP results in production of the one-carbon donor S-adenosyl-methionine (SAM). In polyamine synthesis, the aminopropyl group of decarboxylated SAM is transferred to putrescine, yielding spermine, and methylthioadenosine (MTA). MTA, which contains the methylthio moiety of methionine, is the start of the methionine salvage pathway. After several steps there is a branch point where an aci-reductone dioxygenase can catalyze one of two reactions, depending upon the presence of either Fe2+ or Ni2+. When bound to Fe2+, the enzyme catalyzes the production of formate and 4-methylthio-2-oxobutyrate, which can be aminated to methionine in a single step (Heilbronn et al., 1999). However, when bound to Ni2+, the enzyme produces MMPA, carbon monoxide, and formate in an “off-pathway” reaction (Dai et al., 1999). The physiological significance of this off-pathway transformation is unknown and it has not been demonstrated that it is an alternative source of MMPA.
MeSH Assimilation
The product of MMPA degradation, MeSH, is a source of cellular sulfur for marine bacteria. A report in 1999 examined the fate of DMSP sulfur in pure cultures of several marine Alphaproteobacteria (Gonzalez et al., 1999). Using 35S-labeled DMSP, this work showed that nearly all the sulfur from DMSP was converted into TCA-insoluble material, most of which was protein. Interestingly, the percentage of 35S that was found in TCA-insoluble material decreased significantly as the concentration of DMSP increased. The authors hypothesized that once the bacterial sulfur demand was fulfilled; most DMSP was routed through the cleavage pathway and released as DMS. A more extensive study describing the fate of the sulfur and methyl carbons of DMSP was performed using both pure cultures and natural populations in marine surface waters (Kiene et al., 1999). These experiments also showed that much of the DMSP-derived sulfur was found in TCA-insoluble material. Pure cultures of an organism incapable of producing MeSH from DMSP were nonetheless capable of assimilating sulfur from 35S-MeSH, which is consistent with the widespread distribution of the MMPA-CoA pathway. Furthermore, with [3H-methyl] MeSH, the methyl group was incorporated into the methyl group of methionine. Similar trends and rates of assimilation were observed when using 35S- and 3H-MeSH, suggesting that both the S and methyl groups are directly incorporated into methionine, possibly by the enzyme cystathionine γ-synthetase (Kanzaki et al., 1987; Kiene et al., 1999). This hypothesis is supported by the observation that two inhibitors of the enzyme, vinylglycine and propargylglycine, caused a significant decrease in the incorporation of 35S into TCA-insoluble material.
While the direct incorporation of MeSH into sulfur-containing amino acids is likely, it is also possible that the sulfur and methyl moieties are incorporated independently. The initial demethylation of DMSP transfers one methyl group to tetrahydrofolate, producing 5-methyl-tetrahydrofolate (Reisch et al., 2008). The traditional route of methionine biosynthesis through methionine synthase transfers a methyl group from 5-methyl-THF to homocysteine. Therefore, organisms rapidly consuming DMSP will likely have an abundance of 5-methyl-THF that can be used in methylating reactions, including methionine synthesis. It is also likely that different organisms possess different metabolic capabilities, leaving open the possibility that both the direct incorporation of MeSH and the separate incorporation of C and S are possible in different organisms. Biochemical confirmation of these hypothesized pathways and elucidation of the genes catalyzing these reactions will allow for further investigation of the cellular assimilation of MeSH.
MeSH Oxidation
While one major fate of MeSH in marine surface waters is the incorporation into sulfur-containing amino acids, much of the MeSH is completely oxidized (Figure 4). This MeSH oxidation pathway is expected to be initiated by a MeSH-oxidase (E.C. 1.8.3.4), which produces formaldehyde, hydrogen sulfide, and hydrogen peroxide. This enzyme has been purified from Thiobacillus thioparus (Gould and Kanagawa, 1992), Hyphomicrobium EG (Suylen et al., 1987), and Rhodococcus rhodochrous (Kim et al., 2000), but the gene encoding this enzyme has not been reported. None of the enzymes require exogenous cofactors, but the reported molecular weights are different, making it unclear as to whether these enzymes are related. Given the high turnover reported for MeSH in marine surface waters (Kiene, 1996; Kiene et al., 1999) and for cultured marine bacteria (Gonzalez et al., 1999), identification of this enzyme would be a significant step in understanding of the fate of MeSH in marine systems.
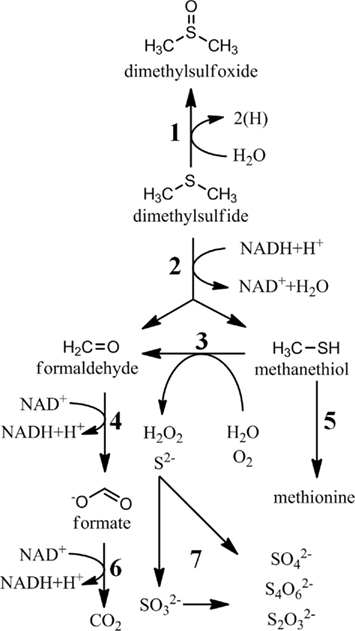
Figure 4. Bioconversions of dimethylsulfide and methanethiol. 1, DMS dehydrogenase; 2, DMS monooxygenase; 3, methanethiol oxidase (E.C. 1.8.3.4); 4, formaldehyde oxidation (various enzymes); 5, cystathionine γ-synthetase (E.C. 2.5.1.48); 6, formate dehydrogenase (E.C. 1.2.1.2); 7, sulfide oxidation (many enzymes).
DMS Consumption
The fate of DMS in marine surface waters is of great interest due to its contribution to cloud condensation nuclei in the atmosphere. Due to biotic and abiotic consumption of DMS in the ocean mixed layer, only 2–10% of the DMS produced is released to the atmosphere (Kiene and Bates, 1990; Archer et al., 2002; Zubkov et al., 2002). The biological degradation of DMS in seawater has long been recognized, but these transformations are still poorly understood. Unlike MeSH, very little sulfur from DMS is assimilated into cells in natural populations, suggesting that only specialized methylotrophs are capable of its assimilation (Vila-Costa et al., 2006). Nonetheless, numerous bacteria have been isolated by their ability to grow on DMS, although many were not isolated from marine sources (reviewed in Schafer et al., 2010). The biological fates of DMS are oxidation to DMSO, sulfate, thiosulfate, and tetrathionate (De Zwart et al., 1997; Vila-Costa et al., 2006; Del Valle et al., 2007; Boden et al., 2010; Figures 1 and 4). In marine surface water, most data indicates that DMSO and sulfate are the primary products of DMS oxidation (Kiene and Linn, 2000; Vila-Costa et al., 2006; Del Valle et al., 2007). However, it is unknown if these studies attempted to quantify thiosulfate and tetrathionate concentrations, as they were not generally believed to be products of DMS degradation. Another possibility is that much of the flux is routed through thiosulfate or tetrathionate, which are then rapidly consumed to produce sulfate.
A dimethylsulfide dehydrogenase that catalyzes the oxidation of DMS to DMSO was identified in Rhodovulum sulfidophilum, a purple non-sulfur member of the Alphaproteobacteria (Mcdevitt et al., 2002). The protein belonged to the DMSO reductase family of molybdoproteins, and showed high similarity to nitrate reductase. Few highly similar homologs are found in the genomic or metagenomic databases, thus the ecological significance of this reaction is unclear. However, the biological production of DMSO from DMS in marine surface has been demonstrated, so other DMSO dehydrogenase reactions must occur (Del Valle et al., 2007).
A DMS monooxygenase, which oxidizes DMS to MeSH and formaldehyde, was characterized from Hyphomicrobium sulfonivorans, an Alphaproteobacterium belonging to the order Rhizobiales that was originally isolated from garden soil (Boden et al., 2011). Homologs to the gene encoding the monooxygenase gene were abundant in the genomic database, but the organisms harboring the genes were mostly terrestrial in origin. Only one member of the roseobacters possessed close homologs, Citreicella sp. SE45. Likewise, GOS metagenomic database contained only a few homologs. Thus, it is unlikely that this DMS monooxygenase plays a significant role in marine surface waters. Nevertheless it is possible that an unrelated enzyme catalyzes a similar reaction in the marine systems.
A member of the Gammaproteobacteria, Methylophaga thiooxydans possessed a novel pathway of DMS oxidation that produced tetrathionate with thiosulfate as an intermediate (Boden et al., 2010). In the environment, tetrathionate would probably be rapidly oxidized because it is rich in electrons. The genes involved in the production of tetrathionate through this pathway and, therefore, its distribution and abundance in the environment are unknown. Similarly, Methylophaga sulfidovorans was able to transform DMS into thiosulfate (De Zwart et al., 1997). Biochemical and molecular specifics of this conversion were not reported, and it is unclear if this is a common fate of DMS in marine systems.
Sulfur Oxidation
A major fate of DMSP-derived sulfur in environmental studies is the complete oxidation to sulfate (Kiene and Linn, 2000), but there is remarkably little information on the enzymes and pathways utilized by bacteria that inhabit marine surface waters. Overall, there is a large diversity of bacterial sulfur oxidation systems (reviewed in Ghosh and Dam, 2009), but many of these systems were identified in extremophilic microbes or phototrophic bacteria that are unlikely to inhabit marine surface waters where most DMSP cycling occurs. Thus, the distribution of these sulfur oxidation systems in marine surface waters remains unclear.
It is assumed that most, if not all, of the sulfur transformed into sulfate is routed through the demethylation/demethiolation pathway of DMSP degradation. Since the sulfur moiety from the demethiolation pathway is in the form MeSH, complete oxidation of the sulfur and carbon would yield 14 electrons. If methanethiol oxidase initiates the oxidation of MeSH in marine bacteria, the sulfur moiety is transformed to sulfide. Two enzymes are known that oxidize sulfide to elemental sulfur; flavocytochrome c-sulfide dehydrogenase and sulfide: quinone reductase (Dolata et al., 1993; Schutz et al., 1997). The oxidation of elemental sulfur is not well understood and different mechanisms exist in different bacteria. Some meso-acidophilic bacteria were shown to use a reduced thiol, such as glutathione, to activate elemental sulfur in a reaction that yields glutathione persulfide, which was then oxidized to sulfite (Rohwerder and Sand, 2003). The green sulfur bacteria possess a reverse-acting dissimilatory sulfite reduction pathway that also oxidizes a persulfide to yield sulfite (Pott and Dahl, 1998). Other bacteria use the Kelly–Friedrich pathway, discussed below, to oxidize elemental sulfur to sulfate (Mukhopadhyaya et al., 2000).
The oxidation of sulfite to sulfate may proceed through at least two mechanisms. One possibility is that a sulfite dehydrogenase (E.C. 1.8.2.1) directly oxidizes sulfite while transferring two electrons to a cytochrome (Kappler, 2011). The second mechanism uses an AMP-dependent pathway that has reverse activities of APS reductase (E.C. 1.8.99.2) and ATP sulfhydrolase (E.C. 2.7.7.4), resulting in the production of ATP (reviewed in Kappler and Dahl, 2001). Interestingly, Candidatus P. ubique possesses gene homologs for the AMP-dependent pathway, but does not possess genes enabling oxidation of inorganic sulfur to sulfite (Kuever and Meyer, 2007). Thus, the sulfite may be derived from the sulfur of DMSP, but the pathways producing sulfite are unknown.
In the Alphaproteobacteria, it is likely that sulfide oxidation proceeds through the well-studied Kelly–Friedrich pathway (Lu et al., 1985; Friedrich et al., 2001). This system completely oxidizes sulfide to sulfate without the formation of sulfite. Interestingly, the Alphaproteobacterium Starkeya novella possesses a both the sulfite dehydrogenase described above as well as parts of the Kelly–Friedrich pathway (Kappler et al., 2001). The Kelly–Friedrich pathway is widely distributed in the marine roseobacters, as 23 of the 32 sequenced genomes have the sox genes that encode for this pathway (Newton et al., 2010). Those roseobacters that possess the Kelly–Friedrich pathway are able to gain energy from the oxidation of sulfide derived from MeSH or possibly DMS degradation. However, a number of roseobacters that possess DMSP degradation genes do not possess the sox genes. In these bacteria the fate of the DMSP derived sulfur is unclear. It is possible that some of these roseobacters that have been cultured and sequenced, but not physiologically characterized, may possess sulfur oxidation pathways similar to those discussed above, whose molecular characteristics are still unknown.
Oxidation of thiosulfate, produced in inorganic sulfur oxidation or DMS consumption, proceeds through a pathway in which tetrathionate is formed. Despite extensive investigations, the molecular specifics of this pathway are mostly unknown, and the biochemical specifics may differ between groups of bacteria (Ghosh and Dam, 2009). A thiosulfate dehydrogenase first forms tetrathionate from two molecules of thiosulfate. A hydrolase then releases sulfite from tetrathionate, before being oxidized to sulfate by one of the mechanisms discussed above.
Double Demethylation of DMSP
One possible fate of DMSP that has remained largely unstudied involves a second demethylation of DMSP and the conversion of MMPA to mercaptopropionate (MPA). The production of MPA from DMSP was first reported in 1988 in anoxic surface sediments (Kiene and Taylor, 1988a), but the importance of MPA in aerobic surface waters remains unclear. An aerobic bacterium showed a stoichiometric production of MPA from DMSP or MMPA (Visscher and Taylor, 1994), suggesting it may be a dead-end product of limited physiological significance. Other reports have indicated that pure cultures of anaerobic bacteria metabolized MPA to H2S and acrylate (Taylor and Visscher, 1996). Methane production from MMPA has also been demonstrated in a limited number of strains of the strictly anaerobic methanogens of the genus Methanosarcina, where it was hypothesized that a transmethylation of coenzyme-M from MMPA was the likely mechanism (Vandermaarel et al., 1995). The biological implications of this reaction may be significant in anoxic areas, but most evidence suggests that double demethylation is not a major fate of DMSP in marine surface waters. However, until progress is made in understanding the biochemical reactions and gene products catalyzing these reactions, the possibility of a major role cannot be ruled out.
Acrylate and 3-Hydroxypropionate Assimilation
In addition to the uncertainty in the initial steps of DMSP metabolism, the fate of the three carbon moiety of DMSP is not well understood. As detailed above, five of the DMSP-cleavage enzymes identified thus far likely result in the production of acrylate, while the other results in 3-hydroxypropionate (Figure 3). In Halomonas HTNK1, acrylate is believed to be metabolized to 3-hydroxypropionate, indicating that DMSP and acrylate degradation share a common intermediate (Todd et al., 2009b). 3-hydroxypropionate is further metabolized by an alcohol dehydrogenase, encoded by dddA, which produces an intermediate hypothesized to be malonate semialdehyde (Figure 3). The gene product of dddC, which encodes for an enzyme annotated as a methylmalonate-semialdehyde dehydrogenase, may transform malonate-semialdehyde to acetyl-CoA (Stines-Chaumeil et al., 2006; Todd et al., 2009b). In E. coli expressing dddC and provided [1-14C] DMSP, the 14C is released as carbon dioxide, presumably by decarboxylation of malonate-semialdehyde. The distribution of this first pathway identified for bacterial DMSP-cleavage and its ecological significance remain unclear, as homologs for dddA and dddC in the metagenomic databases are rare.
Another possibility is that acrylate is assimilated as a three carbon moiety in a pathway similar to that of propionate. In the methylmalonyl-CoA pathway, propionyl-CoA is carboxylated to the four carbon intermediate methylmalonyl-CoA, which is then re-arranged to the TCA cycle intermediate succinyl-CoA (Flavin and Ochoa, 1957). The genes for this pathway are widespread throughout members of roseobacter clade. The transformation of acrylate to propionyl-CoA could occur through at least two different mechanisms. One possibility is the direct reduction of acrylate or acryl-CoA to propionate or propionyl-CoA, respectively. The CoA-dependent reduction of acrylate to propionate was observed in the Crenarchaeota and in Clostridium propionicum (Hetzel et al., 2003; Teufel et al., 2009). Given the distant taxonomic relationship between these prokaryotes and Proteobacteria, it is difficult to identify an ortholog in the roseobacter genomes solely on sequence similarity. An alternative route for forming propionyl-CoA would involve an initial hydration of acrylate or acryloyl-CoA to 3-hydroxypropionate or 3-hydroxypropionyl-CoA, as discussed above for Halomonas HTNK1. A reduction to propionate or propionyl-CoA is then possible. A trifunctional enzyme that catalyzes the complete conversion of 3-hydroxypropionate to propionyl-CoA was identified in Chloroflexus atlanticus (Alber and Fuchs, 2002). Again, the distant taxonomic relationship between Chloroflexus and Proteobacteria make it difficult to identify this enzyme in bacteria in marine surface waters.
Acetate Assimilation
As described above, DMSP catabolism frequently yields acetate. The simplest form of acetate assimilation, the glyoxylate shunt, is absent from most roseobacters but present in the cultured representatives of the SAR11 clade. The glyoxylate shunt requires only two enzymes, isocitrate lyase (E.C. 4.1.3.1) and malate synthase (E.C. 2.3.3.9) in addition to the enzymes of the TCA cycle. The glyoxylate shunt bypasses the two decarboxylation steps of the TCA cyle, resulting in the net assimilation of acetate. The pathway for acetate assimilation in isocitrate lyase-negative organisms has been the subject of investigation for decades, but recently a complete pathway was described and designated the ethylmalonyl-CoA pathway (Erb et al., 2007). All of the enzymes in the pathway were subsequently identified (Erb et al., 2009), and homologous genes for most of the proteins in this pathway are present in R. pomeroyi and other roseobacters. However, further investigations are needed to confirm the physiological significance of this pathway for the assimilation of DMSP.
Conclusion
Several major discoveries regarding the molecular biology and enzymology of DMSP metabolism have occurred during the last several years. These discoveries have enabled the use of molecular approaches to dissect DMSP biogeochemistry and interrogate the environmental significance of DMSP transformations. While these breakthroughs have significantly enhanced our understanding of DMSP and sulfur transformation in the environment, many of the microbial sulfur transformations remain “black boxes.” Elucidation of the carbon and sulfur transformations within these black boxes on both the molecular and biochemical level is critical to our understanding of the marine microbial food web and global sulfur cycle.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Financial support was provided by a dissertation completion grant from the University of Georgia, the National Science Foundation (MCB-07021258 and OCE-0724017), and the Gordon and Betty Moore Foundation.
References
Alber, B. E., and Fuchs, G. (2002). Propionyl-coenzyme A synthase from Chloroflexus aurantiacus, a key enzyme of the 3-hydroxypropionate cycle for autotrophic CO2 fixation. J. Biol. Chem. 277, 12137–12143.
Albers, E. (2009). Metabolic characteristics and importance of the universal methionine salvage pathway recycling methionine from 5′-methylthioadenosine. IUBMB Life 61, 1132–1142.
Archer, S. D., Gilbert, F. J., Nightingale, P. D., Zubkov, M. V., Taylor, A. H., Smith, G. C., and Burkill, P. H. (2002). Transformation of dimethylsulphoniopropionate to dimethyl sulphide during summer in the North Sea with an examination of key processes via a modelling approach. Deep Sea Res. 49, 3067–3101.
Bentley, R., and Chasteen, T. G. (2004). Environmental VOSCs–formation and degradation of dimethyl sulfide, methanethiol and related materials. Chemosphere 55, 291–317.
Biers, E. J., Sun, S. L., and Howard, E. C. (2009). Prokaryotic genomes and diversity in surface ocean waters: interrogating the global ocean sampling metagenome. Appl. Environ. Microbiol. 75, 2221–2229.
Boden, R., Borodina, E., Wood, A. P., Kelly, D. P., Murrell, J. C., and Schafer, H. (2011). Purification and characterization of dimethylsulfide monooxygenase from Hyphomicrobium sulfonivorans. J. Bacteriol. 193, 1250–1258.
Boden, R., Kelly, D. P., Murrell, J. C., and Schafer, H. (2010). Oxidation of dimethylsulfide to tetrathionate by Methylophaga thiooxidans sp. nov.: a new link in the sulfur cycle. Environ. Microbiol. 12, 2688–2699.
Buchan, A., Gonzalez, J. M., and Moran, M. A. (2005). Overview of the marine Roseobacter lineage. Appl. Environ. Microbiol. 71, 5665–5677.
Charlson, R. J., Lovelock, J. E., Andreae, M. O., and Warren, S. G. (1987). Oceanic phytoplankton, atmospheric sulfur, cloud albedo and climate. Nature 326, 655–661.
Chin, M., and Jacob, D. J. (1996). Anthropogenic and natural contributions to tropospheric sulfate: a global model analysis. J. Geophys. Res. 101, 18691–18699.
Curson, A. R., Sullivan, M. J., Todd, J. D., and Johnston, A. W. (2011). DddY, a periplasmic dimethylsulfoniopropionate lyase found in taxonomically diverse species of Proteobacteria. ISME J. 5, 1191–1200.
Curson, A. R. J., Rogers, R., Todd, J. D., Brearley, C. A., and Johnston, A. W. B. (2008). Molecular genetic analysis of a dimethylsulfoniopropionate lyase that liberates the climate-changing gas dimethylsulfide in several marine alpha-proteobacteria and Rhodobacter sphaeroides. Environ. Microbiol. 10, 1099–1099.
Dacey, J. W. H., and Blough, N. V. (1987). Hydroxide decomposition of dimethylsulfoniopropionate to form dimethylsulfide. Geophys. Res. Lett. 14, 1246–1249.
Dai, Y., Wensink, P. C., and Abeles, R. H. (1999). One protein, two enzymes. J. Biol. Chem. 274, 1193–1195.
De Souza, M. P., and Yoch, D. C. (1995a). Comparative physiology of dimethyl sulfide production by dimethylsulfoniopropionate lyase in Pseudomonas doudoroffii and Alcaligenes sp. strain M3A. Appl. Environ. Microbiol. 61, 3986–3991.
De Souza, M. P., and Yoch, D. C. (1995b). Purification and characterization of dimethylsulfoniopropionate lyase from an Alcaligenes-like dimethyl sulfide-producing marine isolate. Appl. Environ. Microbiol. 61, 21–26.
De Zwart, J., Sluis, J., and Kuenen, J. G. (1997). Competition for dimethyl sulfide and hydrogen sulfide by Methylophaga sulfidovorans and Thiobacillus thioparus T5 in continuous cultures. Appl. Environ. Microbiol. 63, 3318–3322.
Del Valle, D. A., Kieber, D. J., and Kiene, R. P. (2007). Depth-dependent fate of biologically-consumed dimethylsulfide in the Sargasso Sea. Mar. Chem. 103, 197–208.
Dolata, M. M., Vanbeeumen, J. J., Ambler, R. P., Meyer, T. E., and Cusanovich, M. A. (1993). Nucleotide-sequence of the heme subunit of flavocytochrome c from the purple phototrophic bacterium, chromatium-vinosum – a 2.6-kilobase pair dna fragment contains 2 multiheme cytochromes, a flavoprotein, and a homolog of human ankyrin. J. Biol. Chem. 268, 14426–14431.
Erb, T. J., Berg, I. A., Brecht, V., Muller, M., Fuchs, G., and Alber, B. E. (2007). Synthesis of C-5-dicarboxylic acids from C-2-units involving crotonyl-CoA carboxylase/reductase: the ethylmalonyl-CoA pathway. Proc. Natl. Acad. Sci. U.S.A. 104, 10631–10636.
Erb, T. J., Fuchs, G., and Alber, B. E. (2009). (2S)-Methylsuccinyl-CoA dehydrogenase closes the ethylmalonyl-CoA pathway for acetyl-CoA assimilation. Mol. Microbiol. 73, 992–1008.
Flavin, M., and Ochoa, S. (1957). Metabolism of propionic acid in animal tissues. 1. Enzymatic conversion of propionate to succinate. J. Biol. Chem. 229, 965–979.
Franklin, D. J., Steinke, M., Young, J., Probert, I., and Malin, G. (2010). Dimethylsulphoniopropionate (DMSP), DMSP-lyase activity (DLA) and dimethylsulphide (DMS) in 10 species of coccolithophore. Mar. Ecol. Prog. Ser. 410, 13–23.
Friedrich, C. G., Rother, D., Bardischewsky, F., Quentmeier, A., and Fischer, J. (2001). Oxidation of reduced inorganic sulfur compounds by bacteria: emergence of a common mechanism? Appl. Environ. Microbiol. 67, 2873–2882.
Gage, D. A., and Rhodes, D. (1997). A new route for synthesis of dimethylsulphoniopropionate in marine algae. Nature 387, 891.
Ghosh, W., and Dam, B. (2009). Biochemistry and molecular biology of lithotrophic sulfur oxidation by taxonomically and ecologically diverse bacteria and archaea. FEMS Microbiol. Rev. 33, 999–1043.
Gonzalez, J. M., Kiene, R. P., and Moran, M. A. (1999). Transformation of sulfur compounds by an abundant lineage of marine bacteria in the alpha-subclass of the class Proteobacteria. Appl. Environ. Microbiol. 65, 3810–3819.
Gould, W. D., and Kanagawa, T. (1992). Purification and properties of methyl mercaptan oxidase from Thiobacillus thioparus TK-m. J. Gen. Microbiol. 138, 217–221.
Hatakeyama, S., Okuda, M., and Akimoto, H. (1982). Formation of sulfur-dioxide and methanesulfonic-acid in the photo-oxidation of dimethyl sulfide in the air. Geophys. Res. Lett. 9, 583–586.
Heilbronn, J., Wilson, J., and Berger, B. J. (1999). Tyrosine aminotransferase catalyzes the final step of methionine recycling in Klebsiella pneumoniae. J. Bacteriol. 181, 1739–1747.
Hetzel, M., Brock, M., Selmer, T., Pierik, A. J., Golding, B. T., and Buckel, W. (2003). Acryloyl-CoA reductase from Clostridium propionicum. An enzyme complex of propionyl-CoA dehydrogenase and electron-transferring flavoprotein. Eur. J. Biochem. 270, 902–910.
Hill, R. W., White, B. A., Cottrell, M. T., and Dacey, J. W. H. (1998). Virus-mediated total release of dimethylsulfoniopropionate from marine phytoplankton: a potential climate process. Aquat. Microb. Ecol. 14, 1–6.
Howard, E. C., Henriksen, J. R., Buchan, A., Reisch, C. R., Burgmann, H., Welsh, R., Ye, W., Gonzalez, J. M., Mace, K., Joye, S. B., Kiene, R. P., Whitman, W. B., and Moran, M. A. (2006). Bacterial taxa that limit sulfur flux from the ocean. Science 314, 649–652.
Howard, E. C., Sun, S. L., Biers, E. J., and Moran, M. A. (2008). Abundant and diverse bacteria involved in DMSP degradation in marine surface waters. Environ. Microbiol. 10, 2397–2410.
Kanzaki, H., Kobayashi, M., Nagasawa, T., and Yamada, H. (1987). Purification and characterization of cystathionine gamma-synthase type-II from Bacillus sphaericus. Eur. J. Biochem. 163, 105–112.
Kappler, U., and Dahl, C. (2001). Enzymology and molecular biology of prokaryotic sulfite oxidation. FEMS Microbiol. Lett. 203, 1–9.
Kappler, U., Friedrich, C. G., Truper, H. G., and Dahl, C. (2001). Evidence for two pathways of thiosulfate oxidation in Starkeya novella (formerly Thiobacillus novellus). Arch. Microbiol. 175, 102–111.
Karsten, U., Kuck, K., Vogt, C., and Kirst, G.O. (1996). “Dimethylsulphoniopropionate production in phototrophic organisms and its physiological function as a cryoprotectant,” in Biological and Environmental Chemistry of DMSP and Related Sulfonium Compounds, eds R. P. Kiene P. T. Visscher M. D. Keller, and G. O. Kirst (New York: Plenum Press), 143–153.
Kiene, R. P. (1996). Production of methanethiol from dimethylsulfoniopropionate in marine surface waters. Mar. Chem. 54, 69–83.
Kiene, R. P., and Bates, T. S. (1990). Biological removal of dimethyl sulfide from sea-water. Nature 345, 702–705.
Kiene, R. P., and Linn, L. J. (2000). The fate of dissolved dimethylsulfoniopropionate (DMSP) in seawater: tracer studies using S-35-DMSP. Geochim. Cosmochim. Acta 64, 2797–2810.
Kiene, R. P., Linn, L. J., Gonzalez, J., Moran, M. A., and Bruton, J. A. (1999). Dimethylsulfoniopropionate and methanethiol are important precursors of methionine and protein-sulfur in marine bacterioplankton. Appl. Environ. Microbiol. 65, 4549–4558.
Kiene, R. P., and Taylor, B. F. (1988a). Biotransformations of organosulfur compounds in sediments via 3-mercaptopropionate. Nature 332, 148–150.
Kiene, R. P., and Taylor, B. F. (1988b). Demethylation of dimethylsulfoniopropionate and production of thiols in anoxic marine sediments. Appl. Environ. Microbiol. 54, 2208–2212.
Kim, S. J., Shin, H. J., Kim, Y. C., Lee, D. S., and Yang, J. W. (2000). Isolation and purification of methyl mercaptan oxidase from Rhodococcus rhodochrous for mercaptan detection. Bioprocess Biosyst. Eng. 5, 465–468.
Kirkwood, M., Le Brun, N. E., Todd, J. D., and Johnston, A. W. B. (2010). The dddP gene of Roseovarius nubinhibens encodes a novel lyase that cleaves dimethylsulfoniopropionate into acrylate plus dimethyl sulfide. Microbiology 156, 1900–1906.
Kirst, G. O. (1990). Salinity tolerance of eukaryotic marine algae. Annu. Rev. Plant Physiol. 41, 21–53.
Kocsis, M. G., and Hanson, A. D. (2000). Biochemical evidence for two novel enzymes in the biosynthesis of 3-dimethylsulfoniopropionate in Spartina alterniflora. Plant Physiol. 123, 1153–1161.
Kuever, J., and Meyer, B. (2007). Phylogeny of the alpha and beta subunits of the dissimilatory adenosine-5′-phosphosulfate (APS) reductase from sulfate-reducing prokaryotes – origin and evolution of the dissimilatory sulfate-reduction pathway. Microbiology 153, 2026–2044.
Lovelock, J. (2006). The Revenge of Gaia: Earth’s Climate in Crisis and the Fate of Humanity. New York: Basic Books.
Lovelock, J. E., Maggs, R. J., and Rasmussen, R. A. (1972). Atmospheric dimethyl sulfide and natural sulfur cycle. Nature 237, 452–453.
Lu, W. P., Swoboda, B. E. P., and Kelly, D. P. (1985). Properties of the thiosulfate-oxidizing multi-enzyme system from Thiobacillus versutus. Biochim. Biophys. Acta 828, 116–122.
Mcdevitt, C. A., Hugenholtz, P., Hanson, G. R., and Mcewan, A. G. (2002). Molecular analysis of dimethyl sulphide dehydrogenase from Rhodovulum sulfidophilum: its place in the dimethyl sulphoxide reductase family of microbial molybdopterin-containing enzymes. Mol. Microbiol. 44, 1575–1587.
Moran, M. A., Reisch, C. R., Kiene, R. P., and Whitman, W. B. (2012). Genomic insights into bacterial DMSP transformations. Annu. Rev. Mar. Sci. 4. doi: 10.1146/annurevmarine-120710-100827
Mukhopadhyaya, P. N., Deb, C., Lahiri, C., and Roy, P. (2000). A soxA gene, encoding a diheme cytochrome c, and a sox locus, essential for sulfur oxidation in a new sulfur lithotrophic bacterium. J. Bacteriol. 182, 4278–4287.
Newton, R. J., Griffin, L. E., Bowles, K. M., Meile, C., Gifford, S., Givens, C. E., Howard, E. C., King, E., Oakley, C. A., Reisch, C. R., Rinta-Kanto, J. M., Sharma, S., Sun, S. L., Varaljay, V., Vila-Costa, M., Westrich, J. R., and Moran, M. A. (2010). Genome characteristics of a generalist marine bacterial lineage. ISME J. 4, 784–798.
Nishiguchi, M. K., and Goff, L. J. (1995). Isolation, purification, and characterization of DMSP lyase [dimethylpropiothetin dethiomethylase (4.4.1.3)] from the red alga Polysiphonia paniculata. J. Phycol. 31, 567–574.
Okamuraikeda, K., Fujiwara, K., and Motokawa, Y. (1982). Purification and characterization of chicken liver T-protein, a component of the glycine cleavage system. J. Biol. Chem. 257, 135–139.
Omelchenko, M. V., Galperin, M. Y., Wolf, Y. I., and Koonin, E. V. (2010). Non-homologous isofunctional enzymes: A systematic analysis of alternative solutions in enzyme evolution. Biol. Direct 5, 1–20.
Otte, M. L., Wilson, G., Morris, J. T., and Moran, B. M. (2004). Dimethylsulphoniopropionate (DMSP) and related compounds in higher plants. J. Exp. Bot. 55, 1919–1925.
Pott, A. S., and Dahl, C. (1998). Sirohaem sulfite reductase and other proteins encoded by genes at the dsr locus of Chromatium vinosum are involved in the oxidation of intracellular sulfur. Microbiology 144, 1881–1894.
Raina, J. B., Dinsdale, E. A., Willis, B. L., and Bourne, D. G. (2010). Do the organic sulfur compounds DMSP and DMS drive coral microbial associations? Trends Microbiol. 18, 101–108.
Reisch, C. R., Moran, M. A., and Whitman, W. B. (2008). Dimethylsulfoniopropionate-dependent demethylase (DmdA) from Pelagibacter ubique and Silicibacter pomeroyi. J. Bacteriol. 190, 8018–8024.
Reisch, C. R., Stoudemayer, M. J., Varaljay, V. A., Amster, I. J., Moran, M. A., and Whitman, W. B. (2011). Novel pathway for assimilation of dimethylsulphoniopropionate widespread in marine bacteria. Nature 473, 208–211.
Rhodes, D., Gage, D. A., Cooper, A., and Hanson, A. D. (1997). S-Methylmethionine conversion to dimethylsulfoniopropionate: evidence for an unusual transamination reaction. Plant Physiol. 115, 1541–1548.
Rohwerder, T., and Sand, W. (2003). The sulfane sulfur of persulfides is the actual substrate of the sulfur-oxidizing enzymes from Acidithiobacillus and Acidiphilium spp. Microbiology 149, 1699–1709.
Rusch, D. B., Halpern, A. L., Sutton, G., Heidelberg, K. B., Williamson, S., Yooseph, S., Wu, D. Y., Eisen, J. A., Hoffman, J. M., Remington, K., Beeson, K., Tran, B., Smith, H., Baden-Tillson, H., Stewart, C., Thorpe, J., Freeman, J., Andrews-Pfannkoch, C., Venter, J. E., Li, K., Kravitz, S., Heidelberg, J. F., Utterback, T., Rogers, Y. H., Falcon, L. I., Souza, V., Bonilla-Rosso, G., Eguiarte, L. E., Karl, D. M., Sathyendranath, S., Platt, T., Bermingham, E., Gallardo, V., Tamayo-Castillo, G., Ferrari, M. R., Strausberg, R. L., Nealson, K., Friedman, R., Frazier, M., and Venter, J. C. (2007). The sorcerer ii global ocean sampling expedition: Northwest Atlantic through eastern tropical pacific. PLoS Biol. 5, e77. doi: 10.1371/journal.pbio.0050077
Schafer, H., Myronova, N., and Boden, R. (2010). Microbial degradation of dimethylsulphide and related C-1-sulphur compounds: organisms and pathways controlling fluxes of sulphur in the biosphere. J. Exp. Bot. 61, 315–334.
Schutz, M., Shahak, Y., Padan, E., and Hauska, G. (1997). Sulfide-quinone reductase from Rhodobacter capsulatus – purification, cloning, and expression. J. Biol. Chem. 272, 9890–9894.
Simo, R. (2001). Production of atmospheric sulfur by oceanic plankton: biogeochemical, ecological and evolutionary links. Trends Ecol. Evol. (Amst.) 16, 287–294.
Stefels, J. (2000). Physiological aspects of the production and conversion of DMSP in marine algae and higher plants. J. Sea Res. 43, 183–197.
Stefels, J., and Van Boeckel, W. (1993). Production of DMS from dissolved DMSP in axenic cultures of the marine phytoplankton species Phaeocystis sp. Mar. Ecol. Prog. Ser. 97, 11–18.
Steinke, M., Malin, G., Archer, S. D., Burkill, P. H., and Liss, P. S. (2002). DMS production in a coccolithophorid bloom: evidence for the importance of dinoflagellate DMSP lyases. Aquat. Microb. Ecol. 26, 259–270.
Stines-Chaumeil, C., Talfournier, F., and Branlant, G. (2006). Mechanistic characterization of the MSDH (methylmalonate semialdehyde dehydrogenase) from Bacillus subtilis. Biochem. J. 395, 107–115.
Sunda, W., Kieber, D. J., Kiene, R. P., and Huntsman, S. (2002). An antioxidant function for DMSP and DMS in marine algae. Nature 418, 317–320.
Suylen, G. M. H., Large, P. J., Vandijken, J. P., and Kuenen, J. G. (1987). Methyl mercaptan oxidase, a key enzyme in the metabolism of methylated sulfur-compounds by Hyphomicrobium EG. J. Gen. Microbiol. 133, 2989–2997.
Taylor, B. F., and Gilchrist, D. C. (1991). New routes for aerobic biodegradation of dimethylsulfoniopropionate. Appl. Environ. Microbiol. 57, 3581–3584.
Taylor, B. F., and Visscher, P. T. (1996). “Metabolic pathways involved in DMSP degradation,” in Biological and Environmental Chemistry of DMSP and Related Sulfonium Compounds, eds R. P. Kiene P. T. Visscher, M. D. Keller and G. O. Kirst (New York: Plenum Press), 265–276.
Teufel, R., Kung, J. W., Kockelkorn, D., Alber, B. E., and Fuchs, G. (2009). 3-hydroxypropionyl-coenzyme A dehydratase and acryloyl-coenzyme A reductase, enzymes of the autotrophic 3-hydroxypropionate/4-hydroxybutyrate cycle in the Sulfolobales. J. Bacteriol. 191, 4572–4581.
Todd, J. D., Curson, A. R., Dupont, C. L., Nicholson, P., and Johnston, A. W. (2009a). The dddP gene, encoding a novel enzyme that converts dimethylsulfoniopropionate into dimethyl sulfide, is widespread in ocean metagenomes and marine bacteria and also occurs in some ascomycete fungi. Environ. Microbiol. 11, 1376–1385.
Todd, J. D., Curson, A. R., Nikolaidou-Katsaraidou, N., Brearley, C. A., Watmough, N. J., Chan, Y., Page, P. C., Sun, L., and Johnston, A. W. (2009b). Molecular dissection of bacterial acrylate catabolism – unexpected links with dimethylsulfoniopropionate catabolism and dimethyl sulfide production. Environ. Microbiol. 12, 327–343.
Todd, J. D., Curson, A. R., Kirkwood, M., Sullivan, M. J., Green, R. T., and Johnston, A. W. (2010). DddQ, a novel, cupin-containing, dimethylsulfoniopropionate lyase in marine roseobacters and in uncultured marine bacteria. Environ. Microbiol. 13, 427–438.
Todd, J. D., Kirkwood, M., Newton-Payne, S., and Johnston, A. W. (2011). DddW, a third DMSP lyase in a model Roseobacter marine bacterium, Ruegeria pomeroyi DSS-3. ISME J. doi: 10.1038/ismej.2011.79
Todd, J. D., Rogers, R., Li, Y. G., Wexler, M., Bond, P. L., Sun, L., Curson, A. R., Malin, G., Steinke, M., and Johnston, A. W. (2007). Structural and regulatory genes required to make the gas dimethyl sulfide in bacteria. Science 315, 666–669.
Toole, D. A., and Siegel, D. A. (2004). Light-driven cycling of dimethylsulfide (DMS) in the Sargasso Sea: closing the loop. Geophys. Res. Lett. 31, 1–4.
Toole, D. A., Siegel, D. A., and Doney, S. C. (2008). A light-driven, one-dimensional dimethylsulfide biogeochemical cycling model for the Sargasso Sea. J. Geophys. Res. 113, 1–20.
Toole, D. A., Slezak, D., Kiene, R. P., Kieber, D. J., and Siegel, D. A. (2006). Effects of solar radiation on dimethylsulfide cycling in the western Atlantic Ocean. Deep Sea Res. 53, 136–153.
Tripp, H. J., Kitner, J. B., Schwalbach, M. S., Dacey, J. W. H., Wilhelm, L. J., and Giovannoni, S. J. (2008). SAR11 marine bacteria require exogenous reduced sulphur for growth. Nature 452, 741–744.
Vallina, S. M., and Simo, R. (2007). Strong relationship between DMS and the solar radiation dose over the global surface ocean. Science 315, 506–508.
Vallina, S. M., Simo, R., Anderson, T. R., Gabric, A., Cropp, R., and Pacheco, J. M. (2008). A dynamic model of oceanic sulfur (DMOS) applied to the Sargasso Sea: simulating the dimethylsulfide (DMS) summer paradox. J. Geophys. Res. 113, 1–23.
Van Den Berg, A. J., Turner, S. M., Vanduyl, F. C., and Ruardij, P. (1996). Model structure and analysis of dimethylsulphide (DMS) production in the southern North Sea, considering phytoplankton dimethylsulphoniopropionate- (DMSP) lyase and eutrophication effects. Mar. Ecol. Prog. Ser. 145, 233–244.
Van Duyl, F. C., Gieskes, W. W. C., Kop, A. J., and Lewis, W. E. (1998). Biological control of short-term variations in the concentration of DMSP and DMS during a Phaeocystis spring bloom. J. Sea Res. 40, 221–231.
Vandermaarel, M. J. E. C., Jansen, M., and Hansen, T. A. (1995). Methanogenic conversion of 3-S-methylmercaptopropionate to 3-mercaptopropionate. Appl. Environ. Microbiol. 61, 48–51.
Varaljay, V. A., Howard, E. C., Sun, S., and Moran, M. A. (2010). Deep sequencing of a dimethylsulfoniopropionate-degrad ing gene (dmdA) by using PCR primer pairs designed on the basis of marine metagenomic data. Appl. Environ. Microbiol. 76, 609–617.
Vila-Costa, M., Del Valle, D. A., Gonzalez, J. M., Slezak, D., Kiene, R. P., Sanchez, O., and Simo, R. (2006). Phylogenetic identification and metabolism of marine dimethylsulfide-consuming bacteria. Environ. Microbiol. 8, 2189–2200.
Vila-Costa, M., Rinta-Kanto, J. M., Sun, S., Sharma, S., Poretsky, R., and Moran, M. A. (2010). Transcriptomic analysis of a marine bacterial community enriched with dimethylsulfoniopropionate. ISME J. 4, 1410–1420.
Visscher, P. T., and Taylor, B. F. (1994). Demethylation of dimethylsulfoniopropionate to 3-mercaptopropionate by an aerobic marine bacterium. Appl. Environ. Microbiol. 60, 4617–4619.
Wagner-Dobler, I., and Biebl, H. (2006). Environmental biology of the marine Roseobacter lineage. Annu. Rev. Microbiol. 60, 255–280.
Wang, Y. Y., Ma, X. H., Zhao, W. F., Jia, X. M., Kai, L., and Xu, X. H. (2006). Study on the creatinase from Paracoccus sp strain WB1. Process Biochem. 41, 2072–2077.
Wolfe, G. V., and Steinke, M. (1996). Grazing-activated production of dimethyl sulfide (DMS) by two clones of Emiliania huxleyi. Limnol. Oceanogr. 41, 1151–1160.
Wolfe, G. V., and Steinke, M. (1997). Grazing-activated chemical defence in a unicellular marine alga. Nature 387, 894–897.
Yancey, P. H. (2005). Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. J. Exp. Biol. 208, 2819–2830.
Yoch, D. C. (2002). Dimethylsulfoniopropionate: its sources, role in the marine food web, and biological degradation to dimethylsulfide. Appl. Environ. Microbiol. 68, 5804–5815.
Yost, D. M., and Mitchelmore, C. L. (2009). Dimethylsulfoniopropionate (DMSP) lyase activity in different strains of the symbiotic alga Symbiodinium microadriaticum. Mar. Ecol. Prog. Ser. 386, 61–70.
Keywords: roseobacters, dimethylsulfoniopropionate, Pelagibacter ubique (SAR11), methyl mercaptopropionate, acetate, acrylate, dimethylsulfide, methanethiol
Citation: Reisch CR, Moran MA and Whitman WB (2011) Bacterial catabolism of dimethylsulfoniopropionate (DMSP). Front. Microbio. 2:172. doi: 10.3389/fmicb.2011.00172
Received: 29 April 2011; Accepted: 28 July 2011;
Published online: 12 August 2011.
Edited by:
Thomas E. Hanson, University of Delaware, USAReviewed by:
Andrew W. B. Johnston, University of East Anglia, UKRich Boden, University of Warwick, UK
Copyright: © 2011 Reisch, Moran and Whitman. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: William B. Whitman, Department of Microbiology, University of Georgia, Athens, GA 30602, USA. e-mail:d2hpdG1hbkB1Z2EuZWR1

 Mary Ann Moran2
Mary Ann Moran2