
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 20 April 2011
Sec. Cellular and Infection Microbiology - closed section
volume 2 - 2011 | https://doi.org/10.3389/fmicb.2011.00084
This article is part of the Research Topic Salmonella host-pathogen interactions View all 10 articles
Nitric oxide (NO) and its congeners contribute to the innate immune response to Salmonella. This enteric pathogen is exposed to reactive nitrogen species (RNS) in the environment and at different anatomical locations during its infectious cycle in vertebrate hosts. Chemical generation of RNS enhances the gastric barrier to enteropathogenic bacteria, while products of the Salmonella pathogenicity island 1 type III secretion system and Salmonella-associated molecular patterns stimulate transcription of inducible NO synthase (iNOS) by cells of the mononuclear phagocytic cell lineage. The resulting NO, or products that arise from its interactions with oxygen (O2) or iron and low-molecular weight thiols, are preferentially bacteriostatic against Salmonella, while reaction of NO and superoxide (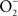 ) generates the bactericidal compound peroxynitrite (ONOO−). The anti-Salmonella activity of RNS emanates from the modification of redox active thiols and metal prosthetic groups of key molecular targets of the electron transport chain, central metabolic enzymes, transcription factors, and DNA and DNA-associated proteins. In turn, Salmonella display a plethora of defenses that modulate the delivery of iNOS-containing vesicles to phagosomes, scavenge and detoxify RNS, and repair biomolecules damaged by these toxic species. Traditionally, RNS have been recognized as important mediators of host defense against Salmonella. However, exciting new findings indicate that Salmonella can exploit the RNS produced during the infection to foster virulence. More knowledge of the primary RNS produced in response to Salmonella infection, the bacterial processes affected by these toxic species, and the adaptive bacterial responses that protect Salmonella from nitrosative and oxidative stress associated with NO will increase our understanding of Salmonella pathogenesis. This information may assist in the development of novel therapeutics against this common enteropathogen.
) generates the bactericidal compound peroxynitrite (ONOO−). The anti-Salmonella activity of RNS emanates from the modification of redox active thiols and metal prosthetic groups of key molecular targets of the electron transport chain, central metabolic enzymes, transcription factors, and DNA and DNA-associated proteins. In turn, Salmonella display a plethora of defenses that modulate the delivery of iNOS-containing vesicles to phagosomes, scavenge and detoxify RNS, and repair biomolecules damaged by these toxic species. Traditionally, RNS have been recognized as important mediators of host defense against Salmonella. However, exciting new findings indicate that Salmonella can exploit the RNS produced during the infection to foster virulence. More knowledge of the primary RNS produced in response to Salmonella infection, the bacterial processes affected by these toxic species, and the adaptive bacterial responses that protect Salmonella from nitrosative and oxidative stress associated with NO will increase our understanding of Salmonella pathogenesis. This information may assist in the development of novel therapeutics against this common enteropathogen.
Gram-negative bacilli of the species Salmonella enterica are frequent causes of disease in humans as well as domestic and wild animals. Invasive salmonellosis is commonly associated with a variety of syndromes that range from gastroenteritis to severe systemic infections. Although non-typhoidal Salmonella such as S. Typhimurium or S. Enteritidis are common causes of gastroenteritis in healthy individuals, these bacteria can cause life-threatening disseminated infections in immunocompromised hosts. Salmonella infections are primarily acquired through the ingestion of contaminated food or water. As food-borne pathogens capable of disseminating extraintestinally, Salmonella are exposed to a variety of innate defenses in the stomach, intestinal lumen, gastrointestinal mucosa, and the intracellular environment of phagocytes. The availability of immunocompromised strains of mice has revealed that reactive nitrogen species (RNS) produced by the enzymatic activity of the inducible NO synthase (iNOS) hemoprotein are integral components of the host armamentarium against Salmonella. As it has been shown for many phylogenetically diverse organisms, the contribution of RNS in resistance to Salmonella does not appear to be limited to direct cytotoxicity, but also involves host and pathogen signaling cascades that indirectly affect the outcome of the infection. Many of the mechanisms by which RNS contribute to Salmonella pathogenesis have been elucidated in murine models of infection using S. enterica serovar Typhimurium, although exciting evidence indicates that these reactive species are also used by diverse animal and human cells in their defense against multiple serovars of S. enterica. This review will discuss what we know about the contribution of RNS to Salmonella pathogenesis, paying particular attention to our current understanding of the mechanisms by which nitric oxide (NO) helps control Salmonella infections and the strategies used by this facultative intracellular pathogen to lessen the cytotoxicity of NO and its nitrosative and oxidative derivatives.
As is the case for other enteropathogenic bacteria, Salmonella are exposed to NO at different stages during the infectious cycle. Salmonella encounter RNS in transit through the environment, the gastrointestinal lumen and mucosa, and phagosomes of mononuclear cells populating gut-associated lymphoid tissues or systemic sites.
Salmonella outbreaks are often associated with contaminated produce. This suggests that Salmonella must establish relationships with plants to ensure persistence in the environment while in transit between animal hosts. The plant surface is for the most part a hostile environment in which bacteria can suffer the adverse effects of ultraviolet light and desiccation. Stomata (small openings in leaves involved in H2O and gas exchange) provide a favorable niche for the survival of enteric bacteria in the plant host. Symmetrical guard cells are responsible for closing the pore of stomata, thereby limiting gas diffusion. Work with E. coli has shown that lipopolysaccharide (LPS) in the cell envelope triggers the closing of stomata, thus eliciting a protective response that limits bacterial colonization (Melotto et al., 2006). Elegant microscopic and pharmacological evidence has demonstrated that NO is the signal that triggers closing of stomata in response to E. coli. Given the similarities in the structure of LPS from E. coli and Salmonella, it is likely that the NO-dependent innate response that regulates the closing of stomata is not limited to E. coli, but represents a general strategy used by plants against Salmonella as well.
Whether associated with tainted vegetables, animal products, or water, Salmonella are encountered in the gastrointestinal tract by most vertebrate hosts. The acidic environment of the stomach is an insurmountable barrier for most microorganisms. Salmonella and many other enteropathogenic bacteria can resist the extreme acidity of the stomach for brief periods of time. In addition, Salmonella, either in response to moderate acidic environmental conditions or upon brief contact to mildly acidic pockets within the gastric content, express a genetic program known as the acid tolerance response (ATR) that enhances resistance to low pH and promotes oral virulence (Foster and Hall, 1990; Bourret et al., 2008). In addition to the accepted role of acidity as a non-specific host defense of the gastric lumen, extensive work by Lundberg et al. (2004) indicates that nitrogen oxides represent another component of the antimicrobial arsenal of the stomach. Although the gastric mucosa can express iNOS in response to infection (Jones-Carson et al., 1995), the bulk of RNS in the gastric lumen originate via nitrate(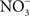 ).
).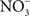 accumulates in blood from both endogenous inflammatory and physiological processes, and exogenous dietary sources. This anion is actively secreted in saliva from the enterosalivary circulation. Upon delivery in the mouth, salivary
accumulates in blood from both endogenous inflammatory and physiological processes, and exogenous dietary sources. This anion is actively secreted in saliva from the enterosalivary circulation. Upon delivery in the mouth, salivary 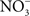 is reduced to nitrite (
is reduced to nitrite (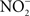 ) by resident microbiota populating the posterior, anaerobic regions of the tongue (reaction 1; Duncan et al., 1995). In the low pH of the stomach,
) by resident microbiota populating the posterior, anaerobic regions of the tongue (reaction 1; Duncan et al., 1995). In the low pH of the stomach, 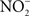 is protonated to its nitrous acid (HNO2) conjugate, which serves as a precursor for various RNS such as NO, nitrogen dioxide (NO2), and dinitrogen trioxide (N2O3; reactions 2–4).
is protonated to its nitrous acid (HNO2) conjugate, which serves as a precursor for various RNS such as NO, nitrogen dioxide (NO2), and dinitrogen trioxide (N2O3; reactions 2–4).




The concentration of NO in the gastric space of human volunteers fluctuates from 100 μM under resting conditions to 500 μM after intake of the daily 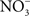 dietary consumption (McKnight et al., 1997). It is becoming increasingly clear that the battery of nitrogen oxides produced in the stomach adds to antimicrobial defense against diverse enteric pathogens. At pH 2.0, RNS are directly bactericidal against Salmonella, whereas at a more moderate pH 4.0, RNS exert indirect antimicrobial activity (Bourret et al., 2008). RNS encountered at pH 4.0 interfere with the ATR of rapidly growing Salmonella by selectively inhibiting the PhoPQ two-component regulatory signaling cascade. Similar to the phenotypes seen in bacteria lacking a functional PhoQ sensor kinase, the NO-dependent inhibition of the ATR prevents the adaptation of Salmonella to pH 3.0, and reduces both oral virulence and fecal shedding (Bourret et al., 2008). Despite the harsh host defenses present in the gastric lumen, some salmonellae must survive the rigors imposed by low pH and RNS, since these enteric bacteria are common causes of gastroenteritis in healthy individuals.
dietary consumption (McKnight et al., 1997). It is becoming increasingly clear that the battery of nitrogen oxides produced in the stomach adds to antimicrobial defense against diverse enteric pathogens. At pH 2.0, RNS are directly bactericidal against Salmonella, whereas at a more moderate pH 4.0, RNS exert indirect antimicrobial activity (Bourret et al., 2008). RNS encountered at pH 4.0 interfere with the ATR of rapidly growing Salmonella by selectively inhibiting the PhoPQ two-component regulatory signaling cascade. Similar to the phenotypes seen in bacteria lacking a functional PhoQ sensor kinase, the NO-dependent inhibition of the ATR prevents the adaptation of Salmonella to pH 3.0, and reduces both oral virulence and fecal shedding (Bourret et al., 2008). Despite the harsh host defenses present in the gastric lumen, some salmonellae must survive the rigors imposed by low pH and RNS, since these enteric bacteria are common causes of gastroenteritis in healthy individuals.
Salmonella utilize the type III secretion system encoded within Salmonella pathogenicity island 1 (SPI1) to gain access to the mucosa of the small intestine. Secretion of SPI1 effector proteins into enterocytes and M cells of Peyer’s patches of the ileum induces actin rearrangements that promote bacterial engulfment. In the lamina propria, Salmonella infect gut-associated macrophages, which undergo a proinflammatory cell death named pyroptosis (Bergsbaken et al., 2009). Molecular characterization of this event has shown that the SipB and SopE SPI1 effectors stimulate the formation of inflammasomes, multicellular complexes containing caspase-1, and the NOD-LRR family of proteins (Hersh et al., 1999; Muller et al., 2009). The proteolytic activity of caspase-1 activates the secretion of interleukin (IL)-1β and IL-18, stimulating the innate immune response against Salmonella (Raupach et al., 2006). Of importance to this review, the inflammatory response of the gut mucosa also induces the expression of iNOS and the consequent generation of NO. Perhaps unexpectedly, caspase-1, but not the proinflammatory cytokine IL-1β, induces the transcription of iNOS (Buzzo et al., 2010). The induction of iNOS appears to be a highly coveted event in the interaction of Salmonella with the gastrointestinal mucosa, because inflammasomes can be independently triggered through the injection of flagellin into the host cell cytosol (Gewirtz et al., 2001). Interestingly, flagellin is delivered into host cells via the SPI1 type III secretion system (Sun et al., 2007). The recognition of flagellin by the cytosolic NOD proteins Nirk4 and Naip5 induces the expression of iNOS (Sun et al., 2007; Buzzo et al., 2010). Therefore, it appears that several Salmonella effectors can independently trigger iNOS transcription.
The NO generated in response to Salmonella appears to play two seemingly contradictory roles in the gastrointestinal phase of the infection. On one hand, NO exerts important antimicrobial activity that helps limit the bacterial burden in an acute murine model of salmonellosis. Accordingly, the absence of iNOS increases both the numbers of Salmonella in Peyer’s patches and the translocation of the bacteria into the underlying gut mucosa (Ackermann et al., 2008; Alam et al., 2008). On the other hand, NO and the ensuing proinflammatory cascade can be of benefit to Salmonella, because inflammation can paradoxically promote intestinal colonization and the systemic spread of Salmonella (Stecher et al., 2007). This observation appears to be in conflict with the more conventional view that intestinal inflammation protects against Salmonella. The effects of reactive species on the normal microbiota may explain this apparent paradox. Because the majority of the intestinal flora consists of anaerobes, it is expected that reactive oxygen and nitrogen species generated in response to Salmonella are detrimental for the abundant microbiota that colonize the gastrointestinal tract. Following this line of reasoning, the inflammation caused by Salmonella eliminates a significant proportion of the gut microbiota, allowing Salmonella to more freely colonize the gastrointestinal mucosa (Stecher et al., 2007; Ackermann et al., 2008). Disrupting the microbial balance in the gastrointestinal tract is a SPI1-dependent process, because mutants in this type III secretion system are at a disadvantage for colonization and induction of inflammation (Barman et al., 2008).
The beneficial effects of NO may not be limited to the elimination of the competing microbiota. NO produced in the inflammatory process could potentiate Salmonella growth by supplying alternative electron acceptors. 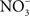 can arise in the gut as either an auto-oxidative product or through enzymatic detoxification of NO or nitrification reactions (Tannenbaum et al., 1978).
can arise in the gut as either an auto-oxidative product or through enzymatic detoxification of NO or nitrification reactions (Tannenbaum et al., 1978). 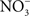 could be used as alternative electron acceptor to maintain electron transport chain function in the O2-limited environment of the gut (Jones et al., 2007). Failure of Salmonella to use O2 as a terminal electron acceptor in the gut is likely compounded by the NO-dependent inhibition of the enzymatic activity of terminal quinol cytochrome oxidases in the electron transport chain (Husain et al., 2008). Under nitrosative stress in the O2-depleted environment of the gastrointestinal lumen, Salmonella likely utilizes available alternative electron acceptors. In favor of this idea, the Salmonella
could be used as alternative electron acceptor to maintain electron transport chain function in the O2-limited environment of the gut (Jones et al., 2007). Failure of Salmonella to use O2 as a terminal electron acceptor in the gut is likely compounded by the NO-dependent inhibition of the enzymatic activity of terminal quinol cytochrome oxidases in the electron transport chain (Husain et al., 2008). Under nitrosative stress in the O2-depleted environment of the gastrointestinal lumen, Salmonella likely utilizes available alternative electron acceptors. In favor of this idea, the Salmonella
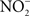 transporter NirC contributes to Salmonella oral virulence (Das et al., 2009). In addition, the Salmonella genome encodes the membrane-bound narGHJI and narZYWV and periplasmic nap nitrate reductases that could use
transporter NirC contributes to Salmonella oral virulence (Das et al., 2009). In addition, the Salmonella genome encodes the membrane-bound narGHJI and narZYWV and periplasmic nap nitrate reductases that could use 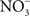 as a respiratory substrate (Prior et al., 2009). However, the role of these nitrate reductases to Salmonella pathogenesis awaits investigation. In contrast to
as a respiratory substrate (Prior et al., 2009). However, the role of these nitrate reductases to Salmonella pathogenesis awaits investigation. In contrast to 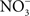 , other metabolites generated through reaction with reactive oxygen species have been demonstrated to serve as alternative terminal electron acceptors. For instance, oxyradicals synthesized by the NADPH phagocyte oxidase react with sulfur compounds in the gut lumen, generating the alternative electron acceptor tetrathionate (Winter et al., 2010).
, other metabolites generated through reaction with reactive oxygen species have been demonstrated to serve as alternative terminal electron acceptors. For instance, oxyradicals synthesized by the NADPH phagocyte oxidase react with sulfur compounds in the gut lumen, generating the alternative electron acceptor tetrathionate (Winter et al., 2010).
In conclusion, the growth advantage of Salmonella in the midst of an inflamed mucosa could be attributed to the ability of this enteric pathogen to utilize tetrathionate and 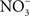 as electron acceptors, while anaerobic residents are eradicated by the ensuing inflammation. In addition to disrupting the normal balance in the microbiota, RNS may contribute more directly to the gastroenteritis and diarrheal syndrome associated with Salmonella infection. Coexpression of iNOS and soluble guanylate cyclase (Closs et al., 1998), whose heme cofactor is allosterically stimulated by NO, could synergize with effectors of the SPI1 type III secretion system for the stimulation of secretion of fluid into the intestinal lumen (Tsolis et al., 1999).
as electron acceptors, while anaerobic residents are eradicated by the ensuing inflammation. In addition to disrupting the normal balance in the microbiota, RNS may contribute more directly to the gastroenteritis and diarrheal syndrome associated with Salmonella infection. Coexpression of iNOS and soluble guanylate cyclase (Closs et al., 1998), whose heme cofactor is allosterically stimulated by NO, could synergize with effectors of the SPI1 type III secretion system for the stimulation of secretion of fluid into the intestinal lumen (Tsolis et al., 1999).
The ability of Salmonella to survive within professional phagocytes is a hallmark of the pathogenesis of this enteric bacterium (Fields et al., 1986). The interaction of Salmonella with professional phagocytes occurs shortly after infection, as Salmonella can be found within the confines of lamina propria CD18+ phagocytic cells just minutes after challenge (Vazquez-Torres et al., 1999). In addition, Salmonella manipulate CD18+ phagocytic cells as Trojan horses for their extraintestinal dissemination to systemic viscera (Vazquez-Torres et al., 1999; Worley et al., 2006). Phagocytes can provide a safe place for the intracellular replication of Salmonella (Fields et al., 1986; Jantsch et al., 2003; Das et al., 2009). However, cells of the mononuclear phagocytic cell lineage also serve as bottlenecks that eliminate a substantial number of intracellular Salmonella (Vazquez-Torres et al., 2000). Several effectors, of which the enzymatic production of reactive oxygen and nitrogen species are probably the best characterized, mediate the anti-Salmonella activity of macrophages. Importantly, both human and rodent macrophages have been shown to express iNOS in response to Salmonella (Witthoft et al., 1998; Eriksson et al., 2000; Khan et al., 2001; Stevanin et al., 2002; Giacomodonato et al., 2003; Bourret et al., 2008; Azenabor et al., 2009). The expression of iNOS takes place in vivo 3 days after intraperitoneal challenge (Umezawa et al., 1997). As discussed in more detail in the next section, the paucity in the expression of iNOS is indicative of the inducible nature of the response. The RNS generated by the enzymatic activity of iNOS are not simple markers of infection, but crucial host defenses that limit the replication of Salmonella in the spleen and liver (Mastroeni et al., 2000).
Investigations using a murine system have defined in detail the contribution of iNOS to the anti-Salmonella activity of professional phagocytes (Vazquez-Torres et al., 2000). The main anti-Salmonella activity of iNOS is expressed at later stages of the infection and manifests itself as an inhibition of Salmonella replication. Although more limited in scope, the NO congener ONOO−, which is the product of the rate-limited reaction of 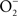 and NO, also contributes to the early killing of Salmonella by IFNγ-primed macrophages.
and NO, also contributes to the early killing of Salmonella by IFNγ-primed macrophages.
In addition to exerting direct anti-Salmonella activity, NO congeners regulate the ensuing innate immune response. For instance, NO produced in response to Salmonella prevents apoptosis (Cerquetti et al., 2002). Accordingly, Salmonella-infected, iNOS-deficient mice harbor abnormally high numbers of apoptotic cells in the liver and Peyer’s patches (Alam et al., 2002, 2008). In addition, these immunodeficient mice suffer from enhanced septicemia, suggesting that the anti-apoptotic role associated with NO limits the extraintestinal dissemination of Salmonella.
The expression of iNOS is controlled at the transcriptional level. Both innate and IFNγ-dependent signaling cascades upregulate the expression of iNOS mRNA and protein in the course of Salmonella infection. The pattern recognition receptor Toll-like receptor 4 (TLR4) binds to lipid A acyl chains of LPS located in the outer leaflet of the outer membrane of Gram-negative bacteria. Binding of LPS to TLR4 activates MyD88, TRIF, and NFκB signaling that stimulates IFNβ production and STAT1 phosphorylation (Xie et al., 1994; Toshchakov et al., 2002; Talbot et al., 2009). On the other hand, erythropoietin antagonizes the activation of NFκB signaling in response to Salmonella (Nairz et al., 2011). The IFN/JAK/STAT-dependent activation of the IRF1 transcription factor, in turn, activates iNOS expression in response to Salmonella LPS (Kamijo et al., 1994). The importance that the TLR4-lipid A signaling cascade plays in Salmonella pathogenesis is demonstrated by the fact that C3H/HeJ mice bearing a defective TLR4 allele are extraordinarily sensitive to Salmonella infection (Vazquez-Torres et al., 2004). The contribution of TLR4 signaling to host defense against Salmonella is partially mediated through the regulation of iNOS expression, because macrophages lacking TLR4 are not only low producers of NO, but are also less capable of controlling intracellular Salmonella (Vazquez-Torres et al., 2004). Salmonella have an invested interest in controlling TLR4 signaling as demonstrated by the fact that 3′-O-deacylation of Salmonella lipid A reduces both NO generation and antimicrobial activity of macrophages (Kawano et al., 2010).
The Nramp1 (Slc11a1) divalent metal transporter associated with phagosomal membranes also optimizes the innate expression of iNOS in response to Salmonella (Nairz et al., 2009). The mechanisms by which Nramp1 induces iNOS are, however, not completely understood, and may reflect the pleiotropic effects associated with disturbances in cytosolic metal concentrations associated with the Nramp1-dependent efflux of metals from the phagosome (reviewed in Cellier et al., 2007). The expression of this metal transporter can activate IRF1 (Fritsche et al., 2003), which, as seen above, is a positive signal of iNOS transcription. Nramp1 can also work synergistically with TNFα for the induction of iNOS expression (Ables et al., 2001). However, Salmonella can induce iNOS enzymatic activity in the absence of signaling through the TNFα p55 receptor (Vazquez-Torres et al., 2001). Finally, the increased NO synthesis seen in Nramp1+ macrophages has been associated with decreases in IL-10 (Fritsche et al., 2008). Of biological importance, mice lacking Nramp1 are hyper-susceptible to Salmonella infection (Govoni and Gros, 1998). Possibly the most dramatic evidence linking Nramp1 to the NO-dependent host defense against Salmonella comes from the fact that attenuation of certain Salmonella mutants deficient in antinitrosative defenses is uniquely exposed in an iNOS-dependent manner in Nramp1+ models of salmonellosis (Bang et al., 2006; Richardson et al., 2009; Husain et al., 2010).
Salmonella induce the production of both IL-12 by macrophages, and IFNγ by natural killer and T helper cells (Schwacha and Eisenstein, 1997). As is the case for other activation markers such as MHC class II, the expression of iNOS seen in response to IFNγ is optimal in the presence of a triggering signal. Findings in macrophages deficient in TLR4 indicate that the expression of iNOS seen in IFNγ-primed macrophages does not require LPS as the triggering signal (Vazquez-Torres et al., 2004). Given the direct activation of iNOS transcription by LPS, these findings might seem surprising. However, it is quite possible that in a hyper-activated state, Salmonella fimbriae, porins, and other surface structures may synergize with IFNγ in the activation of iNOS transcription (Vitiello et al., 2008). The elevated NO fluxes of IFNγ-primed macrophages are associated with enhanced anti-Salmonella activity (Vazquez-Torres et al., 2000).
Human mononuclear cells, which similar to their murine counterparts activate iNOS expression in response to Salmonella LPS, use NO in their anti-Salmonella activity (Stevanin et al., 2002; Azenabor et al., 2009; Gomes et al., 2010). Despite these exciting findings generated with in vitro cell cultures, there still is a lack of clinical evidence correlating defects in iNOS with the predisposition of humans to Salmonella infection. Having said this, individuals bearing mutations in the IFNγ or IL-12 receptors, or IL-12 p40 are extremely susceptible to Salmonella (de Jong et al., 1998). It remains to be investigated whether and to what extent the IFNγ-signaling pathway contributes to the resistance of humans to salmonellosis through the upregulation of iNOS expression and high NO output.
Despite being a radical, NO is remarkably unreactive. The biological chemistry of NO is derived from both direct and indirect effects of this radical with molecular targets (Figure 1). NO can react directly with a limited number of metalloproteins and organic radicals. Moreover, the auto-oxidation, nitrosative, and oxidative products of the reaction of NO with O2, 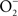 , or iron and low-molecular weight thiols can diversify the biological chemistry of this diatomic radical. As presented above, most of the anti-Salmonella activity exerted by RNS is expressed in the form of bacteriostasis. The chemistries and the Salmonella molecular targets underlying the NO-mediated bacteriostasis are being elucidated.
, or iron and low-molecular weight thiols can diversify the biological chemistry of this diatomic radical. As presented above, most of the anti-Salmonella activity exerted by RNS is expressed in the form of bacteriostasis. The chemistries and the Salmonella molecular targets underlying the NO-mediated bacteriostasis are being elucidated.
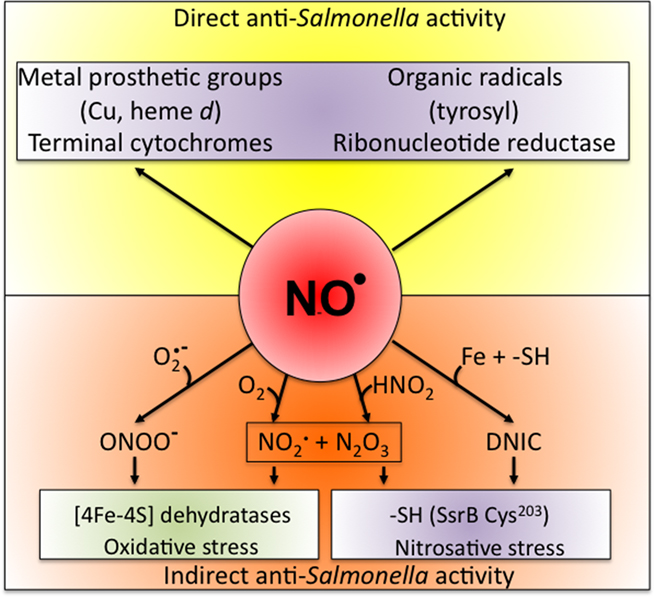
Figure 1. Biological chemistry of NO and Salmonella targets. NO can react directly (yellow box) with metal prosthetic groups or other radicals. Binuclear centers containing copper (Cu) or iron heme d in terminal quinol cytochrome oxidases of the electron transport chain are directly nitrosylated by NO. The covalent attachment of NO to molecular targets causes nitrosative stress (purple boxes). Investigations in E. coli and mitochondria have shown that a tyrosyl radical in the active site of ribonucleotide reductase can be modified directly by NO. NO can also exert biological functions indirectly (orange box) by forming RNS through its interactions with superoxide anion (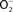 ), molecular oxygen (O2), or iron and low-molecular weight thiols (-SH). Nitrogen dioxide (NO2) and dinitrogen trioxide (N2O3) can independently arise from the condensation of acidified nitrite (HNO2) or the auto-oxidation of NO in the presence of O2. Peroxynitrite (ONOO−), NO2, and N2O3 are strong oxidants. [4Fe–4S] clusters of dehydratases are frequent targets of ONOO− (green box). N2O3 and dinitrosyl–iron complexes (DNIC), which frequently target thiol-containing proteins, are common sources of nitrosative stress. The redox active thiol of Cys203 in the dimerization domain of the SsrB response regulator that controls SPI2 gene transcription is a bona fide sensor of nitrosative stress in Salmonella.
), molecular oxygen (O2), or iron and low-molecular weight thiols (-SH). Nitrogen dioxide (NO2) and dinitrogen trioxide (N2O3) can independently arise from the condensation of acidified nitrite (HNO2) or the auto-oxidation of NO in the presence of O2. Peroxynitrite (ONOO−), NO2, and N2O3 are strong oxidants. [4Fe–4S] clusters of dehydratases are frequent targets of ONOO− (green box). N2O3 and dinitrosyl–iron complexes (DNIC), which frequently target thiol-containing proteins, are common sources of nitrosative stress. The redox active thiol of Cys203 in the dimerization domain of the SsrB response regulator that controls SPI2 gene transcription is a bona fide sensor of nitrosative stress in Salmonella.
NO can react directly with metal prosthetic groups or with other radicals. Direct binding of NO to redox active iron cofactors is well characterized. For example, the high affinity of NO for terminal quinol cytochrome oxidases of the electron transport chain can inhibit the ability of Salmonella and E. coli to reduce O2 to H2O (Butler et al., 1997; Husain et al., 2008). Salmonella has four terminal cytochrome oxidases, of which bd and bo are the best characterized for their reactivities with NO and O2. Work done in E. coli indicates that NO binds with higher affinity to cytochrome bd than cytochrome bo (Mason et al., 2009). So, it is expected that cytochrome bd will preferentially be inhibited at low NO rates, thereby protecting the proton-translocating and O2-reducing capacities of cytochrome bo. Accordingly, cytochrome bd has been shown to be nitrosylated at heme d in Salmonella exposed to NO (Husain et al., 2008). At high levels of NO, such as those produced in the inflammatory response to Salmonella, both cytochromes would be expected to form metal nitrosyl compounds, and thus the overall respiratory activity ought to be repressed. Accordingly, Salmonella exposed to high levels of NO stop respiring (Husain et al., 2008). The NO-mediated repression of respiration could have dramatic consequences in the pathogenesis of Salmonella. First, carbon utilization in Salmonella experiencing intense nitrosative stress would be expected to flux toward fermentative pathways, which yield lower ATP per hexose molecule consumed. Second, reduced respiratory activity is likely to affect some signaling pathways that rely on the activity of the electron transport chain. For example, the ArcB sensor kinase in E. coli responds to the reduced quinone pool (Georgellis et al., 2001). Third, the NO-mediated inhibition of respiration may limit the transport of nutrients and other molecules across the membrane, which relies on the proton motive force. Fourth, the repression of respiration and a drop in proton motive force may, however, be of benefit to Salmonella in some settings, as shown by the fact that the NO-dependent repression of respiration increases resistance of Salmonella to aminoglycosides (McCollister et al., 2011). And, fifth, the NO-dependent repression of respiration can boost the antioxidant defenses of Salmonella (Husain et al., 2008).
In E. coli, NO forms iron–nitrosyl complexes with the ferric uptake regulatory protein Fur, thereby derepressing iron-regulated gene transcription (D’Autreaux et al., 2002). In addition to affecting iron metabolism, the nitrosylation of Fur could have broad implications in Salmonella gene expression. For example, Fur negatively regulates transcription of HN-S in Salmonella (Troxell et al., 2010). Because H-NS binds to AT-rich DNA and represses transcription through topological constraint (Lucchini et al., 2006; Navarre et al., 2006), the nitrosylation of Fur could influence the regulation of horizontally acquired Salmonella SPI1 and SPI2 genes. This interesting possibility awaits investigation.
Bearing an unpaired electron, NO reacts with high affinity and specificity with organic radicals. For example, NO directly reacts with the tyrosyl radical in the active site of ribonucleotide reductase (Lepoivre et al., 1991). The generation of the tyrosyl radical is a crucial step in the catalytic transfer of electrons to ribonucleotides for the reduction of the 2′ carbon of ribose-5-phosphate and formation of the deoxy derivative. Therefore, nitrosylation of ribonucleotide reductase disrupts the formation of deoxyribonucleotides needed for repair and synthesis of DNA. The concerted inhibition of respiratory activity and ribonucleotide reductase very likely contributes to the bacteriostatic effects of NO against Salmonella.
The indirect effects of NO on biological targets are mediated through the RNS generated from the reaction of NO with other molecules. NO can generate biologically relevant RNS through its interactions with molecular O2 or 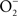 . Because of kinetic and temporal constraints, both of these chemistries appear to be limited to highly activated macrophages. IFNγ-activated macrophages synthesize about three- to fourfold higher amounts of NO than non-activated controls (Vazquez-Torres et al., 2000; McCollister et al., 2007). The increased NO fluxes of IFNγ-activated macrophages allow the following chemistries:
. Because of kinetic and temporal constraints, both of these chemistries appear to be limited to highly activated macrophages. IFNγ-activated macrophages synthesize about three- to fourfold higher amounts of NO than non-activated controls (Vazquez-Torres et al., 2000; McCollister et al., 2007). The increased NO fluxes of IFNγ-activated macrophages allow the following chemistries:


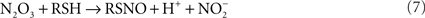
Since reaction (5) is second order for NO, it follows that N2O3 is only detected in the highly activated macrophages sustaining high NO fluxes (reaction 6; McCollister et al., 2007). In addition to being a potent oxidizing agent, N2O3 is a powerful S- and N-nitrosating species that promotes the formation of S-nitrosothiols (reaction 7) and N-nitrosamines. Similar to its formation in the gastric lumen, N2O3 can independently be generated in the phagosomal lumen through the condensation of HNO2. In fact, about a third of the N2O3 generated in IFNγ-primed macrophages appears to form via HNO2 (McCollister et al., 2007). Dinitrosyl–iron complexes (DNICs) generated from the reaction of NO with iron and low-molecular weight thiols are alternative nitrosating agents to N2O3. Lancaster and colleagues have argued that transnitrosation from DNICs is the primary means of S-nitrosothiol formation in vivo (Bosworth et al., 2009).
The reaction of NO with 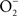 is also limited to IFNγ-activated macrophages. The temporal dissociation between the activities of the NADPH oxidase and iNOS hemoproteins appears to be the main reason for the lack of ONOO− synthesis by non-activated macrophages. Treatment of macrophages with IFNγ stimulates iNOS expression during early phases of the infection, thereby allowing the simultaneous production of
is also limited to IFNγ-activated macrophages. The temporal dissociation between the activities of the NADPH oxidase and iNOS hemoproteins appears to be the main reason for the lack of ONOO− synthesis by non-activated macrophages. Treatment of macrophages with IFNγ stimulates iNOS expression during early phases of the infection, thereby allowing the simultaneous production of 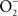 and NO (Vazquez-Torres et al., 2000). A lack of ONOO− production in the absence of iNOS or NADPH oxidase demonstrates that the generation of ONOO− in IFNγ-primed macrophages results from the host response. Detection of nitrotyrosine formation in systemic sites suggests that the highly reactive oxidant ONOO− is indeed produced in the course of Salmonella infection (Alam et al., 2002). As seen in macrophages, the nitrotyrosine signature found in Salmonella-infected mice could reflect ONOO− generated from the interaction of NADPH oxidase and iNOS enzymatic activities. Nonetheless, it is also possible that the ONOO− detected in vivo could have been generated from NO produced by macrophages and
and NO (Vazquez-Torres et al., 2000). A lack of ONOO− production in the absence of iNOS or NADPH oxidase demonstrates that the generation of ONOO− in IFNγ-primed macrophages results from the host response. Detection of nitrotyrosine formation in systemic sites suggests that the highly reactive oxidant ONOO− is indeed produced in the course of Salmonella infection (Alam et al., 2002). As seen in macrophages, the nitrotyrosine signature found in Salmonella-infected mice could reflect ONOO− generated from the interaction of NADPH oxidase and iNOS enzymatic activities. Nonetheless, it is also possible that the ONOO− detected in vivo could have been generated from NO produced by macrophages and 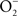 generated adventitiously from the reduction of O2 by electrons in NADH dehydrogenases of the electron transport chain. Conditions that reduce the flow of electrons diminish terminal cytochrome activity and lead to the stasis of electrons upstream in the electron transport chain, a situation that can stimulate
generated adventitiously from the reduction of O2 by electrons in NADH dehydrogenases of the electron transport chain. Conditions that reduce the flow of electrons diminish terminal cytochrome activity and lead to the stasis of electrons upstream in the electron transport chain, a situation that can stimulate 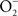 formation (Boveris and Chance, 1973). Moreover, nitrotyrosine could be a signature of peroxidase enzymatic activity using
formation (Boveris and Chance, 1973). Moreover, nitrotyrosine could be a signature of peroxidase enzymatic activity using 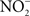 as a substrate (Eiserich et al., 1998). These interesting possibilities need to be investigated. It is also possible that reactive oxygen species such as H2O2 may synergize with NO for antimicrobial activity (Pacelli et al., 1995).
as a substrate (Eiserich et al., 1998). These interesting possibilities need to be investigated. It is also possible that reactive oxygen species such as H2O2 may synergize with NO for antimicrobial activity (Pacelli et al., 1995).
ONOO− generated by the reaction of NO with 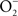 preferentially targets [4Fe–4S] clusters of dehydratases (Castro et al., 1994; Hausladen and Fridovich, 1994; Keyer and Imlay, 1997). Several enzymes of intermediary metabolism containing [4Fe–4S] prosthetic groups in their catalytic cores are prime targets of ONOO−. Aconitase and fumarase A of the citric acid cycle, dihydroxyacid dehydratase involved in branch chain amino acid synthesis, and phosphogluconate dehydratase of the Entner–Doudoroff pathway can all be inhibited with low concentrations of ONOO− (Keyer and Imlay, 1997; Hyduke et al., 2007). Alternatively, nitrosative species such as N2O3, S-nitrosoglutathione, and DNICs stimulate S-nitrosothiol formation. Some enzymes of intermediary metabolism utilize redox active cysteine residues for catalysis. For instance, redox active thiols in glyceraldehyde-3-phosphate dehydrogenase of glycolysis, the dihydrolipoamide acetyltransferase subunit of pyruvate dehydrogenase, ketol-acid reductoisomerase, and the small subunit of glutamate synthase responsible for amino acid synthesis are primary targets of nitrosative stress (Keyer and Imlay, 1997; Brandes et al., 2007). Collectively, the inhibition of redox active enzymes in central metabolism by oxidative and nitrosative stress likely induces global changes in bacterial physiology. Salmonella undergoing nitrosative stress downregulate translational machinery (Bourret et al., 2008), which not only is a key signature of the stringent response, but may also represent a physiological adaptation to RNS. Although the mechanism by which RNS activate the stringent response is not known, it is possible that the inhibition of amino acid synthesis by RNS could be the signal (Brandes et al., 2007; Hyduke et al., 2007).
preferentially targets [4Fe–4S] clusters of dehydratases (Castro et al., 1994; Hausladen and Fridovich, 1994; Keyer and Imlay, 1997). Several enzymes of intermediary metabolism containing [4Fe–4S] prosthetic groups in their catalytic cores are prime targets of ONOO−. Aconitase and fumarase A of the citric acid cycle, dihydroxyacid dehydratase involved in branch chain amino acid synthesis, and phosphogluconate dehydratase of the Entner–Doudoroff pathway can all be inhibited with low concentrations of ONOO− (Keyer and Imlay, 1997; Hyduke et al., 2007). Alternatively, nitrosative species such as N2O3, S-nitrosoglutathione, and DNICs stimulate S-nitrosothiol formation. Some enzymes of intermediary metabolism utilize redox active cysteine residues for catalysis. For instance, redox active thiols in glyceraldehyde-3-phosphate dehydrogenase of glycolysis, the dihydrolipoamide acetyltransferase subunit of pyruvate dehydrogenase, ketol-acid reductoisomerase, and the small subunit of glutamate synthase responsible for amino acid synthesis are primary targets of nitrosative stress (Keyer and Imlay, 1997; Brandes et al., 2007). Collectively, the inhibition of redox active enzymes in central metabolism by oxidative and nitrosative stress likely induces global changes in bacterial physiology. Salmonella undergoing nitrosative stress downregulate translational machinery (Bourret et al., 2008), which not only is a key signature of the stringent response, but may also represent a physiological adaptation to RNS. Although the mechanism by which RNS activate the stringent response is not known, it is possible that the inhibition of amino acid synthesis by RNS could be the signal (Brandes et al., 2007; Hyduke et al., 2007).
Reactive nitrogen species can also disassemble zinc-fingers as suggested by the observation that NO-treated Salmonella accumulate chelatable zinc in their cytoplasms (Schapiro et al., 2003). It has been proposed that NO inactivates the zinc-finger-containing proteins PriA, DnaG, and DnaJ (Schapiro et al., 2003). However, biochemical evidence indicating that NO modifies the zinc-finger motifs in these DNA-binding proteins has not been experimentally demonstrated. Together with repression of the electron transport chain, inhibition of ribonucleotide reductase, and inhibition of enzymes in central metabolism, disruption of zinc-fingers in proteins associated with DNA replication could contribute to the bacteriostasis and cell filamentation of NO-treated Salmonella (De Groote et al., 1995; Vazquez-Torres et al., 2000; Schapiro et al., 2003).
Redox active thiols are preferred targets of the RNS generated during the auto-oxidation of NO. The thiol of Cys203 in the dimerization domain of the SPI2 master regulator SsrB is a bona fide example of a Salmonella target of S-nitrosylation (Husain et al., 2010). SsrB was recently recognized as a redox active protein that senses nitrosative stress. The relevance of the NO-sensing activity of SsrB in Salmonella pathogenesis is manifested by the fact that a strain of Salmonella expressing a redox resistant SsrB C203S variant is attenuated in an Nramp1 model of oral salmonellosis, and that Cys203 is conserved in SsrB of both typhoidal and non-typhoidal strains of Salmonella. The only exception is a strain of S. enterica sv. St. Paul, which instead has a tyrosine at position 203. This substitution is quite interesting since tyrosines and cysteines are preferred targets of RNS. It is still unknown how the sensing of NO congeners by SsrB Cys203 enhances Salmonella fitness. It is possible that, by decreasing either Salmonella-induced apoptosis or reducing recognition by T and B lymphocytes, the tight control of SPI2 expression exerted by a redox active SsrB may increase intracellular survival or limit the specific immune response to SPI2 effectors. The idea that the downregulation of SPI2 by the NO-sensing activity of SsrB is key to some aspect of Salmonella pathogenesis is in keeping with previous observations that showed that the repression of SPI2 is as important for Salmonella virulence as its positive regulation (Coombes et al., 2005).
Many pathogenic bacteria have developed or adapted mechanisms to counteract RNS encountered in the host (Figure 2). As an intracellular pathogen, Salmonella possesses several strategies that avoid contact with iNOS-containing vesicles, detoxify NO, or repair lesions incurred by RNS.
Figure 2. Antinitrosative defenses of Salmonella. Effectors secreted by the type III secretion system encoded within SPI2 actively avoid NADPH oxidase- and iNOS-containing vesicles. Superoxide ( ) produced by the enzymatic activity of NADPH oxidase enters the periplasmic space of Salmonella through porins, while NO can freely diffuse across membranes. Overlapping NADPH phagocyte oxidase and iNOS activities can generate the potent oxidant peroxynitrite (ONOO−) through the reaction of NO with
) produced by the enzymatic activity of NADPH oxidase enters the periplasmic space of Salmonella through porins, while NO can freely diffuse across membranes. Overlapping NADPH phagocyte oxidase and iNOS activities can generate the potent oxidant peroxynitrite (ONOO−) through the reaction of NO with  . The negative effects of ONOO− are alleviated directly by the enzymatic activity of alkyl hydroperoxidase (AhpC) or indirectly by periplasmic SodCI, which detoxifies
. The negative effects of ONOO− are alleviated directly by the enzymatic activity of alkyl hydroperoxidase (AhpC) or indirectly by periplasmic SodCI, which detoxifies 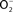 and thereby limits ONOO− formation. NO can be directly reduced by the denitrosylase activity of the flavohemoprotein Hmp. Transcription of hmpA is de-repressed by the inactivation of the [2Fe–2S] redox active prosthetic group of the NsrR transcriptional repressor by NO. In hypoxic or anaerobic conditions, both the flavorubredoxin (NorV) and the cytochrome c nitrite reductase (NrfA) can reduce NO to nitrous oxide (N2O). NO interacts with iron (Fe) and small-molecular weight thiols (SMWT) to generate dinitrosyl–iron complexes (DNICs), while its reaction with O2 gives rise to dinitrogen trioxide (N2O3). Iron storage proteins (ISP) such as ferritins can limit the generation of DNICs by restricting available Fe. Homocysteine or SMWT antagonize the S-nitrosylation associated with N2O3 or DNIC. The SPI2 regulator SsrB is an example of a Salmonella protein that gets S-nitrosylated. A variety of RNS can oxidize purines and pyrimidines in the chromosome of Salmonella. The combined action of base excision repair (BER) glycosylases and Xth/Nfo endonucleases can repair these lesions.
and thereby limits ONOO− formation. NO can be directly reduced by the denitrosylase activity of the flavohemoprotein Hmp. Transcription of hmpA is de-repressed by the inactivation of the [2Fe–2S] redox active prosthetic group of the NsrR transcriptional repressor by NO. In hypoxic or anaerobic conditions, both the flavorubredoxin (NorV) and the cytochrome c nitrite reductase (NrfA) can reduce NO to nitrous oxide (N2O). NO interacts with iron (Fe) and small-molecular weight thiols (SMWT) to generate dinitrosyl–iron complexes (DNICs), while its reaction with O2 gives rise to dinitrogen trioxide (N2O3). Iron storage proteins (ISP) such as ferritins can limit the generation of DNICs by restricting available Fe. Homocysteine or SMWT antagonize the S-nitrosylation associated with N2O3 or DNIC. The SPI2 regulator SsrB is an example of a Salmonella protein that gets S-nitrosylated. A variety of RNS can oxidize purines and pyrimidines in the chromosome of Salmonella. The combined action of base excision repair (BER) glycosylases and Xth/Nfo endonucleases can repair these lesions.
Salmonella actively avoid iNOS-containing vesicles of professional phagocytes. SPI2 and iNOS are optimally expressed after 8 h of the innate response of macrophages to Salmonella (Eriksson et al., 2003). Effectors secreted through the SPI2 type III secretion system minimize trafficking of iNOS-containing vesicles to the proximity of phagosomes (Chakravortty et al., 2002). It might seem unclear why segregation of iNOS from the Salmonella phagosome might be advantageous, since NO diffuses freely through membranes. Moreover, others have noticed that iNOS, associated with cortical actin, vesicles, or cytosol, is not significantly mobilized in response to Salmonella infection (Webb et al., 2001; McCollister et al., 2007). Avoidance of iNOS-containing vesicles might be advantageous in the context of limiting exposure to ONOO− (Chakravortty et al., 2002), which being an anion does not cross freely through membranes. IFNγ-activated macrophages can, nonetheless, downregulate the intracellular expression of SPI2 (McCollister et al., 2005; Bourret et al., 2009). Biochemical and genetic lines of evidence indicate that NO congeners produced by the enzymatic activity of iNOS mediate repression of SPI2 in IFNγ-primed macrophages. This agrees with the fact that the chemical generation of NO represses SPI2 transcription (Bourret et al., 2009). Although it is not known how SPI2 transcription gets downregulated by RNS of activated macrophages, this process is independent of the repression of PhoPQ signaling (Bourret et al., 2009). It is also unclear if downregulation of SPI2 is a strategy sought by the bacteria or a target of host defense. The fact that NO congeners produced by IFNγ-activated macrophages promote the maturation of the Salmonella phagosome along the degradative pathway would argue to the latter. In this context, the NO-dependent repression of SPI2 transcription accounts for a great part of the enhanced killing of Salmonella by IFNγ-primed macrophages (McCollister et al., 2005).
Thiol-based scavenging systems serve as a means of directly removing RNS. Homocysteine, an intermediate in the methionine biosynthetic pathway, has been shown to enhance the resistance of Salmonella to S-nitrosothiols (De Groote et al., 1996). Furthermore, homocysteine adds to the antinitrosative defenses of Salmonella in a murine model of salmonellosis (De Groote et al., 1996). Other thiol-based scavengers, including cysteine and the tripeptide glutathione, could serve similar roles. Nonetheless, the contribution of these low-molecular weight thiols to the antinitrosative defenses of Salmonella awaits investigation.
The flavohemoprotein Hmp is the primary means of NO detoxification in Salmonella (Bang et al., 2006). This flavohemoprotein, which is expressed by Salmonella within the intracellular environment of professional phagocytes (Eriksson et al., 2003), contributes to the inducible antinitrosative response of Salmonella and many other organisms by denitrosylating NO to 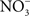 , utilizing in the process O2, NADH, and FAD (Crawford and Goldberg, 1998; Hausladen et al., 2001). Hmp limits the accumulation of low-molecular weight nitrosothiols in Salmonella-infected macrophages, and protects Salmonella against authentic NO while minimizing the anti-Salmonella activity of RNS generated by murine and human macrophages (Stevanin et al., 2002; Bang et al., 2006; Gilberthorpe et al., 2007; Laver et al., 2010). Moreover, Hmp contributes to the antinitrosative defenses of Salmonella in an Nramp1+ murine model of salmonellosis (Bang et al., 2006). The constitutive expression of hmpA, nonetheless, leads to a loss of Salmonella fitness through its
, utilizing in the process O2, NADH, and FAD (Crawford and Goldberg, 1998; Hausladen et al., 2001). Hmp limits the accumulation of low-molecular weight nitrosothiols in Salmonella-infected macrophages, and protects Salmonella against authentic NO while minimizing the anti-Salmonella activity of RNS generated by murine and human macrophages (Stevanin et al., 2002; Bang et al., 2006; Gilberthorpe et al., 2007; Laver et al., 2010). Moreover, Hmp contributes to the antinitrosative defenses of Salmonella in an Nramp1+ murine model of salmonellosis (Bang et al., 2006). The constitutive expression of hmpA, nonetheless, leads to a loss of Salmonella fitness through its 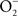 -producing capacity (McLean et al., 2010). Transcription of hmpA is greatly increased in response to NO (Crawford and Goldberg, 1998). Expression of hmpA is under the control of the redox active repressor NsrR (Bang et al., 2006), whose [Fe–S] cluster is reversibly inactivated by NO. In addition, hmpA expression is upregulated under iron-limiting conditions through NsrR, but independently of Fur (Bang et al., 2006).
-producing capacity (McLean et al., 2010). Transcription of hmpA is greatly increased in response to NO (Crawford and Goldberg, 1998). Expression of hmpA is under the control of the redox active repressor NsrR (Bang et al., 2006), whose [Fe–S] cluster is reversibly inactivated by NO. In addition, hmpA expression is upregulated under iron-limiting conditions through NsrR, but independently of Fur (Bang et al., 2006).
Since significant antinitrosative activity of Hmp requires the consumption of O2, the role of this flavohemoprotein might be limited in hypoxic or anoxic environments. The flavorubredoxin (NorV) and cytochrome c nitrite reductase (NrfA) not only reduce NO to nitrous oxide (N2O), but are also important for the resistance of Salmonella to RNS under anaerobic conditions (Mills et al., 2005, 2008). However, these enzymes appear to contribute minimally to the antinitrosative defenses of Salmonella in vivo, because mutants lacking either norV or nrfA are virulent when inoculated intraperitoneally (Bang et al., 2006). It remains possible that NorV and NrfA may be important for resistance to nitrosative stress in the anoxic environment of the intestine. Salmonella also possess pathways for the denitrification of NO. The role of nitrogen metabolism in Salmonella has not been evaluated in the context of pathogenesis. However, similar to E. coli O157 (Jones et al., 2007), the terminal electron acceptor 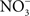 might also be important for colonization of the gastrointestinal tract by Salmonella. O2 is thought to be limiting in the gut and in the intracellular environment of phagocytes and, therefore, alternative electron acceptors must be used instead. A limited respiratory activity can be aggravated under nitrosative stress that inhibits terminal cytochromes of the electron transport chain. Thus, the nitrate reductase complex NarGHIJ and the global regulator Fnr could help in maintaining NO homeostasis and resistance to RNS during some phases of the infection (Gilberthorpe and Poole, 2008).
might also be important for colonization of the gastrointestinal tract by Salmonella. O2 is thought to be limiting in the gut and in the intracellular environment of phagocytes and, therefore, alternative electron acceptors must be used instead. A limited respiratory activity can be aggravated under nitrosative stress that inhibits terminal cytochromes of the electron transport chain. Thus, the nitrate reductase complex NarGHIJ and the global regulator Fnr could help in maintaining NO homeostasis and resistance to RNS during some phases of the infection (Gilberthorpe and Poole, 2008).
ONOO− is formed through the reaction of NO with 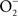 (Koppenol et al., 1992). ONOO− is a strong oxidant capable of modifying lipids, amino acids, DNA, and redox active metal centers of dehydratases (Radi, 2004). In analogy to the extensive functional overlap in the enzymatic detoxification of reactive oxygen species (Hebrard et al., 2009), Salmonella can antagonize ONOO− through both indirect and direct mechanisms. By consuming the
(Koppenol et al., 1992). ONOO− is a strong oxidant capable of modifying lipids, amino acids, DNA, and redox active metal centers of dehydratases (Radi, 2004). In analogy to the extensive functional overlap in the enzymatic detoxification of reactive oxygen species (Hebrard et al., 2009), Salmonella can antagonize ONOO− through both indirect and direct mechanisms. By consuming the 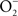 precursor, Salmonella periplasmic Cu/Zn superoxide dismutase SodCI prevents ONOO− formation (De Groote et al., 1997). SodCI has been shown to be crucial for Salmonella resistance to the synergistic cytotoxicity of
precursor, Salmonella periplasmic Cu/Zn superoxide dismutase SodCI prevents ONOO− formation (De Groote et al., 1997). SodCI has been shown to be crucial for Salmonella resistance to the synergistic cytotoxicity of 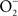 and NO produced by NADPH oxidase and iNOS hemoproteins (De Groote et al., 1997; Sansone et al., 2002). Alternatively, ONOO− can be detoxified to
and NO produced by NADPH oxidase and iNOS hemoproteins (De Groote et al., 1997; Sansone et al., 2002). Alternatively, ONOO− can be detoxified to 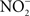 by the peroxiredoxin-alkyl hydroperoxide reductase AhpC (Chen et al., 1998). In contrast to sodCI, ahpC is not essential for Salmonella pathogenesis (Taylor et al., 1998), raising the possibility that SodCI is the primary means of protection against ONOO−in vivo.
by the peroxiredoxin-alkyl hydroperoxide reductase AhpC (Chen et al., 1998). In contrast to sodCI, ahpC is not essential for Salmonella pathogenesis (Taylor et al., 1998), raising the possibility that SodCI is the primary means of protection against ONOO−in vivo.
Given that NO can inhibit several enzymes of central metabolic pathways (Castro et al., 1994; Keyer and Imlay, 1997; Brandes et al., 2007; Hyduke et al., 2007), Salmonella likely coordinate a metabolic response to this diatomic radical. In accordance with this idea, glucose-6-phosphate dehydrogenase (Zwf) of the pentose phosphate pathway is important for resistance to RNS in vitro and in vivo (Lundberg et al., 1999). The gene encoding Zwf is part of the SoxR regulon, an [Fe–S] cluster-containing transcription factor activated by NO in E. coli (Ding and Demple, 2000). Zwf shuffles the flow of carbon through the pentose phosphate pathway, producing NADPH reducing equivalents in the process. NADPH could fuel glutathione oxidoreductase or thioredoxin reductase to repair damage caused by RNS. Moreover, the downstream non-oxidative branch of the pentose phosphate pathway generates precursors for the biosynthesis of nucleotides, which are needed for repair of RNS-mediated DNA damage.
Reactive nitrogen species produced in response to Salmonella can damage DNA by oxidizing purines and pyrimidines. The concerted actions of base excision repair glycosylases and apurinic/apyrimidinic Xth/Nfo endonucleases protect Salmonella against products of iNOS in macrophages and contribute to Salmonella virulence in an acute model of infection (Suvarnapunya et al., 2003; Richardson et al., 2009). Because the repair of damaged DNA requires nucleotides, Salmonella must use NO-resistant pathways for the biosynthesis of deoxyribonucleotides. As described above, NO can inhibit ribonucleotide reductase by reacting with the tyrosyl radical. Interestingly, Salmonella encodes other ribonucleotide reductases that may be resistant to the inhibitory effects of NO (Panosa et al., 2010).
Bacterial pathogens must adapt to changing environmental conditions to survive and cause disease. Salmonella experiences the stress imposed by RNS generated during the course of infection. NO produced by iNOS in response to Salmonella infection is involved in a broad range of pathophysiological processes, acting both as a signaling molecule and a potent antimicrobial mediator. RNS inhibit assorted bacterial targets involved in a variety of cellular processes. Given this strong selective pressure, Salmonella have developed mechanisms to counteract the cytotoxicity of RNS. Even more unexpectedly, recent investigations have shown that Salmonella can take advantage of the RNS to bolster growth in infected tissues.
By actively invading the gastrointestinal epithelia, Salmonella induce inflammation that promotes its colonization and spread. The NO produced as part of the inflammatory process may generate alternative electron acceptors that can be utilized by Salmonella in the O2-depleted environment of the gut. The ability of Salmonella to utilize 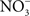 as an alternative electron acceptor may be especially important under nitrosative stress conditions that repress respiratory activity. Unexpectedly, independent lines of recent evidence indicate the RNS increase the fitness of Salmonella and allow the bacteria to outcompete intestinal microbiota (Stecher et al., 2007; Ackermann et al., 2008). Outcompeting the normal flora not only promotes colonization of the intestine and spread to systemic locations, but also promotes diarrhea that serves to disseminate this enteropathogen in the environment. NO sensing and regulation of virulence expression is a fascinating aspect of the emerging view that Salmonella do in fact co-opt RNS generated in the host response. The SPI2 regulator SsrB is the first example of a Salmonella regulatory protein required for fine-tuning virulence in the context of RNS generated in the host response. The redox active Cys203 in SsrB serves as a molecular switch to tightly control gene expression during the course of salmonellosis. It is highly likely that Salmonella possesses a variety of sensors, such as Fur, devoted to coordinating responses to NO.
as an alternative electron acceptor may be especially important under nitrosative stress conditions that repress respiratory activity. Unexpectedly, independent lines of recent evidence indicate the RNS increase the fitness of Salmonella and allow the bacteria to outcompete intestinal microbiota (Stecher et al., 2007; Ackermann et al., 2008). Outcompeting the normal flora not only promotes colonization of the intestine and spread to systemic locations, but also promotes diarrhea that serves to disseminate this enteropathogen in the environment. NO sensing and regulation of virulence expression is a fascinating aspect of the emerging view that Salmonella do in fact co-opt RNS generated in the host response. The SPI2 regulator SsrB is the first example of a Salmonella regulatory protein required for fine-tuning virulence in the context of RNS generated in the host response. The redox active Cys203 in SsrB serves as a molecular switch to tightly control gene expression during the course of salmonellosis. It is highly likely that Salmonella possesses a variety of sensors, such as Fur, devoted to coordinating responses to NO.
Recent investigations have uncovered novel mechanisms by which Salmonella circumvent the detrimental effects of RNS produced during the host response to this facultative intracellular pathogen. Understanding the molecular mechanisms that coordinate Salmonella virulence in response to NO will advance our understanding of host–pathogen interactions taking place in the course of salmonellosis. This information can, in turn, illuminate novel therapeutic strategies to decrease the health burden that Salmonella infections inflict across the globe.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This review was supported by grants from the Burroughs Welcome Fund, NIH project AI54959, and the Institutional Training grant T32 AI052066. We would like to thank Dr. Jessica Jones-Carson for discussions.
Ables, G. P., Takamatsu, D., Noma, H., El-Shazly, S., Jin, H. K., Taniguchi, T., Sekikawa, K., and Watanabe, T. (2001). The roles of Nramp1 and Tnfα genes in nitric oxide production and their effect on the growth of Salmonella typhimurium in macrophages from Nramp1 congenic and tumor necrosis factor α−/− mice. J. Interferon Cytokine Res. 21, 53–62.
Ackermann, M., Stecher, B., Freed, N. E., Songhet, P., Hardt, W. D., and Doebeli, M. (2008). Self-destructive cooperation mediated by phenotypic noise. Nature 454, 987–990.
Alam, M. S., Akaike, T., Okamoto, S., Kubota, T., Yoshitake, J., Sawa, T., Miyamoto, Y., Tamura, F., and Maeda, H. (2002). Role of nitric oxide in host defense in murine salmonellosis as a function of its antibacterial and antiapoptotic activities. Infect. Immun. 70, 3130–3142.
Alam, M. S., Zaki, M. H., Sawa, T., Islam, S., Ahmed, K. A., Fujii, S., Okamoto, T., and Akaike, T. (2008). Nitric oxide produced in Peyer’s patches exhibits antiapoptotic activity contributing to an antimicrobial effect in murine salmonellosis. Microbiol. Immunol. 52, 197–208.
Azenabor, A. A., Kennedy, P., and York, J. (2009). Free intracellular Ca2+ regulates bacterial lipopolysaccharide induction of iNOS in human macrophages. Immunobiology 214, 143–152.
Bang, I. S., Liu, L., Vazquez-Torres, A., Crouch, M. L., Stamler, J. S., and Fang, F. C. (2006). Maintenance of nitric oxide and redox homeostasis by the Salmonella flavohemoglobin hmp. J. Biol. Chem. 281, 28039–28047.
Barman, M., Unold, D., Shifley, K., Amir, E., Hung, K., Bos, N., and Salzman, N. (2008). Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect. Immun. 76, 907–915.
Bergsbaken, T., Fink, S. L., and Cookson, B. T. (2009). Pyroptosis: host cell death and inflammation. Nat. Rev. Microbiol. 7, 99–109.
Bosworth, C. A., Toledo, J. C. Jr., Zmijewski, J. W., Li, Q., and Lancaster, J. R. Jr. (2009). Dinitrosyliron complexes and the mechanism(s) of cellular protein nitrosothiol formation from nitric oxide. Proc. Natl. Acad. Sci. U.S.A. 106, 4671–4676.
Bourret, T. J., Porwollik, S., McClelland, M., Zhao, R., Greco, T., Ischiropoulos, H., and Vazquez-Torres, A. (2008). Nitric oxide antagonizes the acid tolerance response that protects Salmonella against innate gastric defenses. PLoS ONE 3, e1833. doi: 10.1371/journal.pone.0001833
Bourret, T. J., Song, M., and Vazquez-Torres, A. (2009). Codependent and independent effects of nitric oxide-mediated suppression of PhoPQ and Salmonella pathogenicity island 2 on intracellular Salmonella enterica serovar typhimurium survival. Infect. Immun. 77, 5107–5115.
Boveris, A., and Chance, B. (1973). The mitochondrial generation of hydrogen peroxide. General properties and effect of hyperbaric oxygen. Biochem. J. 134, 707–716.
Brandes, N., Rinck, A., Leichert, L. I., and Jakob, U. (2007). Nitrosative stress treatment of E. coli targets distinct set of thiol-containing proteins. Mol. Microbiol. 66, 901–914.
Butler, C. S., Seward, H. E., Greenwood, C., and Thomson, A. J. (1997). Fast cytochrome bo from Escherichia coli binds two molecules of nitric oxide at CuB. Biochemistry 36, 16259–16266.
Buzzo, C. L., Campopiano, J. C., Massis, L. M., Lage, S. L., Cassado, A. A., Leme-Souza, R., Cunha, L. D., Russo, M., Zamboni, D. S., Amarante-Mendes, G. P., and Bortoluci, K. R. (2010). A novel pathway for inducible nitric oxide synthase activation through inflammasomes. J. Biol. Chem. 285, 32087–32095.
Castro, L., Rodriguez, M., and Radi, R. (1994). Aconitase is readily inactivated by peroxynitrite, but not by its precursor, nitric oxide. J. Biol. Chem. 269, 29409–29415.
Cellier, M. F., Courville, P., and Campion, C. (2007). Nramp1 phagocyte intracellular metal withdrawal defense. Microbes Infect. 9, 1662–1670.
Cerquetti, M. C., Goren, N. B., Ropolo, A. J., Grasso, D., Giacomodonato, M. N., and Vaccaro, M. I. (2002). Nitric oxide and apoptosis induced in Peyer’s patches by attenuated strains of Salmonella enterica serovar Enteritidis. Infect. Immun. 70, 964–969.
Chakravortty, D., Hansen-Wester, I., and Hensel, M. (2002). Salmonella pathogenicity island 2 mediates protection of intracellular Salmonella from reactive nitrogen intermediates. J. Exp. Med. 195, 1155–1166.
Chen, L., Xie, Q. W., and Nathan, C. (1998). Alkyl hydroperoxide reductase subunit C (AhpC) protects bacterial and human cells against reactive nitrogen intermediates. Mol. Cell 1, 795–805.
Closs, E. I., Enseleit, F., Koesling, D., Pfeilschifter, J. M., Schwarz, P. M., and Forstermann, U. (1998). Coexpression of inducible NO synthase and soluble guanylyl cyclase in colonic enterocytes: a pathophysiologic signaling pathway for the initiation of diarrhea by gram-negative bacteria? FASEB J. 12, 1643–1649.
Coombes, B. K., Wickham, M. E., Lowden, M. J., Brown, N. F., and Finlay, B. B. (2005). Negative regulation of Salmonella pathogenicity island 2 is required for contextual control of virulence during typhoid. Proc. Natl. Acad. Sci. U.S.A. 102, 17460–17465.
Crawford, M. J., and Goldberg, D. E. (1998). Role for the Salmonella flavohemoglobin in protection from nitric oxide. J. Biol. Chem. 273, 12543–12547.
Das, P., Lahiri, A., and Chakravortty, D. (2009). Novel role of the nitrite transporter NirC in Salmonellapathogenesis: SPI2-dependent suppression of inducible nitric oxide synthase in activated macrophages. Microbiology 155, 2476–2489.
D’Autreaux, B., Touati, D., Bersch, B., Latour, J. M., and Michaud-Soret, I. (2002). Direct inhibition by nitric oxide of the transcriptional ferric uptake regulation protein via nitrosylation of the iron. Proc. Natl. Acad. Sci. U.S.A. 99, 16619–16624.
De Groote, M. A., Granger, D., Xu, Y., Campbell, G., Prince, R., and Fang, F. C. (1995). Genetic and redox determinants of nitric oxide cytotoxicity in a Salmonella typhimurium model. Proc. Natl. Acad. Sci. U.S.A. 92, 6399–6403.
De Groote, M. A., Ochsner, U. A., Shiloh, M. U., Nathan, C., McCord, J. M., Dinauer, M. C., Libby, S. J., Vazquez-Torres, A., Xu, Y., and Fang, F. C. (1997). Periplasmic superoxide dismutase protects Salmonella from products of phagocyte NADPH oxidase and nitric oxide synthase. Proc. Natl. Acad. Sci. U.S.A. 94, 13997–14001.
De Groote, M. A., Testerman, T., Xu, Y., Stauffer, G., and Fang, F. C. (1996). Homocysteine antagonism of nitric oxide-related cytostasis in Salmonella typhimurium. Science 272, 414–417.
de Jong, R., Altare, F., Haagen, I. A., Elferink, D. G., Boer, T., van Breda Vriesman, P. J., Kabel, P. J., Draaisma, J. M., van Dissel, J. T., Kroon, F. P., Casanova, J. L., and Ottenhoff, T. H. (1998). Severe mycobacterial and Salmonella infections in interleukin-12 receptor-deficient patients. Science 280, 1435–1438.
Ding, H., and Demple, B. (2000). Direct nitric oxide signal transduction via nitrosylation of iron–sulfur centers in the SoxR transcription activator. Proc. Natl. Acad. Sci. U.S.A. 97, 5146–5150.
Duncan, C., Dougall, H., Johnston, P., Green, S., Brogan, R., Leifert, C., Smith, L., Golden, M., and Benjamin, N. (1995). Chemical generation of nitric oxide in the mouth from the enterosalivary circulation of dietary nitrate. Nat. Med. 1, 546–551.
Eiserich, J. P., Hristova, M., Cross, C. E., Jones, A. D., Freeman, B. A., Halliwell, B., and van der Vliet, A. (1998). Formation of nitric oxide-derived inflammatory oxidants by myeloperoxidase in neutrophils. Nature 391, 393–397.
Eriksson, S., Bjorkman, J., Borg, S., Syk, A., Pettersson, S., Andersson, D. I., and Rhen, M. (2000). Salmonella typhimurium mutants that downregulate phagocyte nitric oxide production. Cell. Microbiol. 2, 239–250.
Eriksson, S., Lucchini, S., Thompson, A., Rhen, M., and Hinton, J. C. (2003). Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47, 103–118.
Fields, P. I., Swanson, R. V., Haidaris, C. G., and Heffron, F. (1986). Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl. Acad. Sci. U.S.A. 83, 5189–5193.
Foster, J. W., and Hall, H. K. (1990). Adaptive acidification tolerance response of Salmonella typhimurium. J. Bacteriol. 172, 771–778.
Fritsche, G., Dlaska, M., Barton, H., Theurl, I., Garimorth, K., and Weiss, G. (2003). Nramp1 functionality increases inducible nitric oxide synthase transcription via stimulation of IFN regulatory factor 1 expression. J. Immunol. 171, 1994–1998.
Fritsche, G., Nairz, M., Werner, E. R., Barton, H. C., and Weiss, G. (2008). Nramp1-functionality increases iNOS expression via repression of IL-10 formation. Eur. J. Immunol. 38, 3060–3067.
Georgellis, D., Kwon, O., and Lin, E. C. (2001). Quinones as the redox signal for the arc two-component system of bacteria. Science 292, 2314–2316.
Gewirtz, A. T., Simon, P. O. Jr., Schmitt, C. K., Taylor, L. J., Hagedorn, C. H., O’Brien, A. D., Neish, A. S., and Madara, J. L. (2001). Salmonella typhimurium translocates flagellin across intestinal epithelia, inducing a proinflammatory response. J. Clin. Invest. 107, 99–109.
Giacomodonato, M. N., Goren, N. B., Sordelli, D. O., Vaccaro, M. I., Grasso, D. H., Ropolo, A. J., and Cerquetti, M. C. (2003). Involvement of intestinal inducible nitric oxide synthase (iNOS) in the early stages of murine salmonellosis. FEMS Microbiol. Lett. 223, 231–238.
Gilberthorpe, N. J., Lee, M. E., Stevanin, T. M., Read, R. C., and Poole, R. K. (2007). NsrR: a key regulator circumventing Salmonella enterica serovar Typhimurium oxidative and nitrosative stress in vitro and in IFNγ-stimulated J774.2 macrophages. Microbiology 153, 1756–1771.
Gilberthorpe, N. J., and Poole, R. K. (2008). Nitric oxide homeostasis in Salmonella typhimurium: roles of respiratory nitrate reductase and flavohemoglobin. J. Biol. Chem. 283, 11146–11154.
Gomes, N. E., Brunialti, M. K., Mendes, M. E., Freudenberg, M., Galanos, C., and Salomao, R. (2010). Lipopolysaccharide-induced expression of cell surface receptors and cell activation of neutrophils and monocytes in whole human blood. Braz. J. Med. Biol. Res. 43, 853–858.
Govoni, G., and Gros, P. (1998). Macrophage NRAMP1 and its role in resistance to microbial infections. Inflamm. Res. 47, 277–284.
Hausladen, A., and Fridovich, I. (1994). Superoxide and peroxynitrite inactivate aconitases, but nitric oxide does not. J. Biol. Chem. 269, 29405–29408.
Hausladen, A., Gow, A., and Stamler, J. S. (2001). Flavohemoglobin denitrosylase catalyzes the reaction of a nitroxyl equivalent with molecular oxygen. Proc. Natl. Acad. Sci. U.S.A. 98, 10108–10112.
Hebrard, M., Viala, J. P., Meresse, S., Barras, F., and Aussel, L. (2009). Redundant hydrogen peroxide scavengers contribute to Salmonella virulence and oxidative stress resistance. J. Bacteriol. 191, 4605–4614.
Hersh, D., Monack, D. M., Smith, M. R., Ghori, N., Falkow, S., and Zychlinsky, A. (1999). The Salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1. Proc. Natl. Acad. Sci. U.S.A. 96, 2396–2401.
Husain, M., Bourret, T. J., McCollister, B. D., Jones-Carson, J., Laughlin, J., and Vazquez-Torres, A. (2008). Nitric oxide evokes an adaptive response to oxidative stress by arresting respiration. J. Biol. Chem. 283, 7682–7689.
Husain, M., Jones-Carson, J., Song, M., McCollister, B. D., Bourret, T. J., and Vazquez-Torres, A. (2010). Redox sensor SsrB Cys203 enhances Salmonella fitness against nitric oxide generated in the host immune response to oral infection. Proc. Natl. Acad. Sci. U.S.A. 107, 14396–14401.
Hyduke, D. R., Jarboe, L. R., Tran, L. M., Chou, K. J., and Liao, J. C. (2007). Integrated network analysis identifies nitric oxide response networks and dihydroxyacid dehydratase as a crucial target in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 104, 8484–8489.
Jantsch, J., Cheminay, C., Chakravortty, D., Lindig, T., Hein, J., and Hensel, M. (2003). Intracellular activities of Salmonella enterica in murine dendritic cells. Cell. Microbiol. 5, 933–945.
Jones, S. A., Chowdhury, F. Z., Fabich, A. J., Anderson, A., Schreiner, D. M., House, A. L., Autieri, S. M., Leatham, M. P., Lins, J. J., Jorgensen, M., Cohen, P. S., and Conway, T. (2007). Respiration of Escherichia coli in the mouse intestine. Infect. Immun. 75, 4891–4899.
Jones-Carson, J., Vazquez-Torres, A., van der Heyde, H. C., Warner, T., Wagner, R. D., and Balish, E. (1995). γδ T cell-induced nitric oxide production enhances resistance to mucosal candidiasis. Nat. Med. 1, 552–557.
Kamijo, R., Harada, H., Matsuyama, T., Bosland, M., Gerecitano, J., Shapiro, D., Le, J., Koh, S. I., Kimura, T., Green, S. J., Mak, T. W., Taniguchi, T., and Vilcek, J. (1994). Requirement for transcription factor IRF-1 in NO synthase induction in macrophages. Science 263, 1612–1615.
Kawano, M., Manabe, T., and Kawasaki, K. (2010). Salmonella enterica serovar Typhimurium lipopolysaccharide deacylation enhances its intracellular growth within macrophages. FEBS Lett. 584, 207–212.
Keyer, K., and Imlay, J. A. (1997). Inactivation of dehydratase [4Fe–4S] clusters and disruption of iron homeostasis upon cell exposure to peroxynitrite. J. Biol. Chem. 272, 27652–27659.
Khan, S. A., Strijbos, P. J., Everest, P., Moss, D., Stratford, R., Mastroeni, P., Allen, J., Servos, S., Charles, I. G., Dougan, G., and Maskell, D. J. (2001). Early responses to Salmonella typhimurium infection in mice occur at focal lesions in infected organs. Microb. Pathog. 30, 29–38.
Koppenol, W. H., Moreno, J. J., Pryor, W. A., Ischiropoulos, H., and Beckman, J. S. (1992). Peroxynitrite, a cloaked oxidant formed by nitric oxide and superoxide. Chem. Res. Toxicol. 5, 834–842.
Laver, J. R., Stevanin, T. M., Messenger, S. L., Lunn, A. D., Lee, M. E., Moir, J. W., Poole, R. K., and Read, R. C. (2010). Bacterial nitric oxide detoxification prevents host cell S-nitrosothiol formation: a novel mechanism of bacterial pathogenesis. FASEB J. 24, 286–295.
Lepoivre, M., Fieschi, F., Coves, J., Thelander, L., and Fontecave, M. (1991). Inactivation of ribonucleotide reductase by nitric oxide. Biochem. Biophys. Res. Commun. 179, 442–448.
Lucchini, S., Rowley, G., Goldberg, M. D., Hurd, D., Harrison, M., and Hinton, J. C. (2006). H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2, e81. doi: 10.1371/journal.ppat.0020081
Lundberg, B. E., Wolf, R. E. Jr., Dinauer, M. C., Xu, Y., and Fang, F. C. (1999). Glucose 6-phosphate dehydrogenase is required for Salmonella typhimurium virulence and resistance to reactive oxygen and nitrogen intermediates. Infect. Immun. 67, 436–438.
Lundberg, J. O., Weitzberg, E., Cole, J. A., and Benjamin, N. (2004). Nitrate, bacteria and human health. Nat. Rev. Microbiol. 2, 593–602.
Mason, M. G., Shepherd, M., Nicholls, P., Dobbin, P. S., Dodsworth, K. S., Poole, R. K., and Cooper, C. E. (2009). Cytochrome bd confers nitric oxide resistance to Escherichia coli. Nat. Chem. Biol. 5, 94–96.
Mastroeni, P., Vazquez-Torres, A., Fang, F. C., Xu, Y., Khan, S., Hormaeche, C. E., and Dougan, G. (2000). Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. I. Effects on microbial proliferation and host survival in vivo. J. Exp. Med. 192, 237–248.
McCollister, B. D., Bourret, T. J., Gill, R., Jones-Carson, J., and Vazquez-Torres, A. (2005). Repression of SPI2 transcription by nitric oxide-producing, IFNγ-activated macrophages promotes maturation of Salmonella phagosomes. J. Exp. Med. 202, 625–635.
McCollister, B. D., Hoffman, M., Husain, M., and Vazquez-Torres, A. (2011). Nitric oxide protects bacteria from aminoglycosides by blocking the energy-dependent phases of drug uptake. Antimicrob. Agents Chemother. doi: 10.1128/AAC.01203-10. [Epub ahead of print].
McCollister, B. D., Myers, J. T., Jones-Carson, J., Husain, M., Bourret, T. J., and Vazquez-Torres, A. (2007). N2O3 enhances the nitrosative potential of IFNγ-primed macrophages in response to Salmonella. Immunobiology 212, 759–769.
McKnight, G. M., Smith, L. M., Drummond, R. S., Duncan, C. W., Golden, M., and Benjamin, N. (1997). Chemical synthesis of nitric oxide in the stomach from dietary nitrate in humans. Gut 40, 211–214.
McLean, S., Bowman, L. A., and Poole, R. K. (2010). Peroxynitrite stress is exacerbated by flavohaemoglobin-derived oxidative stress in Salmonella Typhimurium and relieved by NO. Microbiology 156, 3556–3565.
Melotto, M., Underwood, W., Koczan, J., Nomura, K., and He, S. Y. (2006). Plant stomata function in innate immunity against bacterial invasion. Cell 126, 969–980.
Mills, P. C., Richardson, D. J., Hinton, J. C., and Spiro, S. (2005). Detoxification of nitric oxide by the flavorubredoxin of Salmonella enterica serovar Typhimurium. Biochem. Soc. Trans. 33, 198–199.
Mills, P. C., Rowley, G., Spiro, S., Hinton, J. C., and Richardson, D. J. (2008). A combination of cytochrome c nitrite reductase (NrfA) and flavorubredoxin (NorV) protects Salmonella enterica serovar Typhimurium against killing by NO in anoxic environments. Microbiology 154, 1218–1228.
Muller, A. J., Hoffmann, C., Galle, M., Van Den Broeke, A., Heikenwalder, M., Falter, L., Misselwitz, B., Kremer, M., Beyaert, R., and Hardt, W. D. (2009). The Typhimurium, S. effector SopE induces caspase-1 activation in stromal cells to initiate gut inflammation. Cell Host Microbe 6, 125–136.
Nairz, M., Fritsche, G., Crouch, M. L., Barton, H. C., Fang, F. C., and Weiss, G. (2009). Slc11a1 limits intracellular growth of Salmonella enterica sv. Typhimurium by promoting macrophage immune effector functions and impairing bacterial iron acquisition. Cell. Microbiol. 11, 1365–1381.
Nairz, M., Schroll, A., Moschen, A. R., Sonnweber, T., Theurl, M., Theurl, I., Taub, N., Jamnig, C., Neurauter, D., Huber, L. A., Tilg, H., Moser, P. L., and Weiss, G. (2011). Erythropoietin contrastingly affects bacterial infection and experimental colitis by inhibiting nuclear factor-κB-inducible immune pathways. Immunity 31, 61–74.
Navarre, W. W., Porwollik, S., Wang, Y., McClelland, M., Rosen, H., Libby, S. J., and Fang, F. C. (2006). Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313, 236–238.
Pacelli, R., Wink, D. A., Cook, J. A., Krishna, M. C., DeGraff, W., Friedman, N., Tsokos, M., Samuni, A., and Mitchell, J. B. (1995). Nitric oxide potentiates hydrogen peroxide-induced killing of Escherichia coli. J. Exp. Med. 182, 1469–1479.
Panosa, A., Roca, I., and Gibert, I. (2010). Ribonucleotide reductases of Salmonella typhimurium: transcriptional regulation and differential role in pathogenesis. PLoS ONE 5, e11328. doi: 10.1371/journal.pone.0011328
Prior, K., Hautefort, I., Hinton, J. C., Richardson, D. J., and Rowley, G. (2009). All stressed out. Salmonella pathogenesis and reactive nitrogen species. Adv. Microb. Physiol. 56, 1–28.
Radi, R. (2004). Nitric oxide, oxidants, and protein tyrosine nitration. Proc. Natl. Acad. Sci. U.S.A. 101, 4003–4008.
Raupach, B., Peuschel, S. K., Monack, D. M., and Zychlinsky, A. (2006). Caspase-1-mediated activation of interleukin-1β (IL-1β) and IL-18 contributes to innate immune defenses against Salmonella enterica serovar Typhimurium infection. Infect. Immun. 74, 4922–4926.
Richardson, A. R., Soliven, K. C., Castor, M. E., Barnes, P. D., Libby, S. J., and Fang, F. C. (2009). The base excision repair system of Salmonella enterica serovar Typhimurium counteracts DNA damage by host nitric oxide. PLoS Pathog. 5, e1000451. doi: 10.1371/journal.ppat.1000451
Sansone, A., Watson, P. R., Wallis, T. S., Langford, P. R., and Kroll, J. S. (2002). The role of two periplasmic copper- and zinc-cofactored superoxide dismutases in the virulence of Salmonella choleraesuis. Microbiology 148, 719–726.
Schapiro, J. M., Libby, S. J., and Fang, F. C. (2003). Inhibition of bacterial DNA replication by zinc mobilization during nitrosative stress. Proc. Natl. Acad. Sci. U.S.A. 100, 8496–8501.
Schwacha, M. G., and Eisenstein, T. K. (1997). Interleukin-12 is critical for induction of nitric oxide-mediated immunosuppression following vaccination of mice with attenuated Salmonella typhimurium. Infect. Immun. 65, 4897–4903.
Stecher, B., Robbiani, R., Walker, A. W., Westendorf, A. M., Barthel, M., Kremer, M., Chaffron, S., Macpherson, A. J., Buer, J., Parkhill, J., Dougan, G., von Mering, C., and Hardt, W. D. (2007). Salmonella enterica serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 5, 2177–2189. doi: 10.1371/journal.pbio.0050244
Stevanin, T. M., Poole, R. K., Demoncheaux, E. A., and Read, R. C. (2002). Flavohemoglobin Hmp protects Salmonella enterica serovar Typhimurium from nitric oxide-related killing by human macrophages. Infect. Immun. 70, 4399–4405.
Sun, Y. H., Rolan, H. G., and Tsolis, R. M. (2007). Injection of flagellin into the host cell cytosol by Salmonella enterica serotype Typhimurium. J. Biol. Chem. 282, 33897–33901.
Suvarnapunya, A. E., Lagasse, H. A., and Stein, M. A. (2003). The role of DNA base excision repair in the pathogenesis of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 48, 549–559.
Talbot, S., Totemeyer, S., Yamamoto, M., Akira, S., Hughes, K., Gray, D., Barr, T., Mastroeni, P., Maskell, D. J., and Bryant, C. E. (2009). Toll-like receptor 4 signalling through MyD88 is essential to control Salmonella enterica serovar Typhimurium infection, but not for the initiation of bacterial clearance. Immunology 128, 472–483.
Tannenbaum, S. R., Fett, D., Young, V. R., Land, P. D., and Bruce, W. R. (1978). Nitrite and nitrate are formed by endogenous synthesis in the human intestine. Science 200, 1487–1489.
Taylor, P. D., Inchley, C. J., and Gallagher, M. P. (1998). The Salmonella typhimurium AhpC polypeptide is not essential for virulence in BALB/c mice but is recognized as an antigen during infection. Infect. Immun. 66, 3208–3217.
Toshchakov, V., Jones, B. W., Perera, P. Y., Thomas, K., Cody, M. J., Zhang, S., Williams, B. R., Major, J., Hamilton, T. A., Fenton, M. J., and Vogel, S. N. (2002). TLR4, but not TLR2, mediates IFNβ-induced STAT1α/β-dependent gene expression in macrophages. Nat. Immunol. 3, 392–398.
Troxell, B., Sikes, M. L., Fink, R. C., Vazquez-Torres, A., Jones-Carson, J., and Hassan, H. M. (2010). Fur negatively regulates hns and is required for the zxpression of HilA and virulence in Salmonella enterica serovar Typhimurium. J. Bacteriol. 193, 497–505.
Tsolis, R. M., Adams, L. G., Ficht, T. A., and Baumler, A. J. (1999). Contribution of Salmonella typhimurium virulence factors to diarrheal disease in calves. Infect. Immun. 67, 4879–4885.
Umezawa, K., Akaike, T., Fujii, S., Suga, M., Setoguchi, K., Ozawa, A., and Maeda, H. (1997). Induction of nitric oxide synthesis and xanthine oxidase and their roles in the antimicrobial mechanism against Salmonella typhimurium infection in mice. Infect. Immun. 65, 2932–2940.
Vazquez-Torres, A., Fantuzzi, G., Edwards, C. K. III, Dinarello, C. A., and Fang, F. C. (2001). Defective localization of the NADPH phagocyte oxidase to Salmonella-containing phagosomes in tumor necrosis factor p55 receptor-deficient macrophages. Proc. Natl. Acad. Sci. U.S.A. 98, 2561–2565.
Vazquez-Torres, A., Jones-Carson, J., Baumler, A. J., Falkow, S., Valdivia, R., Brown, W., Le, M., Berggren, R., Parks, W. T., and Fang, F. C. (1999). Extraintestinal dissemination of Salmonella by CD18-expressing phagocytes. Nature 401, 804–808.
Vazquez-Torres, A., Jones-Carson, J., Mastroeni, P., Ischiropoulos, H., and Fang, F. C. (2000). Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. Effects, I. on microbial killing by activated peritoneal macrophages in vitro. J. Exp. Med. 192, 227–236.
Vazquez-Torres, A., Vallance, B. A., Bergman, M. A., Finlay, B. B., Cookson, B. T., Jones-Carson, J., and Fang, F. C. (2004). Toll-like receptor 4 dependence of innate and adaptive immunity to Salmonella: importance of the Kupffer cell network. J. Immunol. 172, 6202–6208.
Vitiello, M., D’Isanto, M., Finamore, E., Ciarcia, R., Kampanaraki, A., and Galdiero, M. (2008). Role of mitogen-activated protein kinases in the iNOS production and cytokine secretion by Salmonella enterica serovar Typhimurium porins. Cytokine 41, 279–285.
Webb, J. L., Harvey, M. W., Holden, D. W., and Evans, T. J. (2001). Macrophage nitric oxide synthase associates with cortical actin but is not recruited to phagosomes. Infect. Immun. 69, 6391–6400.
Winter, S. E., Thiennimitr, P., Winter, M. G., Butler, B. P., Huseby, D. L., Crawford, R. W., Russell, J. M., Bevins, C. L., Adams, L. G., Tsolis, R. M., Roth, J. R., and Baumler, A. J. (2010). Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467, 426–429.
Witthoft, T., Eckmann, L., Kim, J. M., and Kagnoff, M. F. (1998). Enteroinvasive bacteria directly activate expression of iNOS and NO production in human colon epithelial cells. Am. J. Physiol. 275, G564–G571.
Worley, M. J., Nieman, G. S., Geddes, K., and Heffron, F. (2006). Salmonella typhimurium disseminates within its host by manipulating the motility of infected cells. Proc. Natl. Acad. Sci. U.S.A. 103, 17915–17920.
Keywords: inducible nitric oxide synthase, macrophages, reactive nitrogen species, Salmonella, redox chemistry, virulence, intracellular, enteric bacteria
Citation: Henard CA and Vázquez-Torres A (2011) Nitric oxide and Salmonella pathogenesis. Front. Microbio. 2:84. doi: 10.3389/fmicb.2011.00084
Received: 02 February 2011; Accepted: 08 April 2011;
Published online: 20 April 2011.
Edited by:
John S. Gunn, The Ohio State University, USAReviewed by:
Miguel A. Valvano, University of Western Ontario, CanadaCopyright: © 2011 Henard and Vázquez-Torres. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Andrés Vázquez-Torres, Department of Microbiology, University of Colorado School of Medicine, Mail Box 8333, P.O. Box 6511, Room P18-9131, Aurora, CO 80045, USA. e-mail:YW5kcmVzLnZhenF1ZXotdG9ycmVzQHVjZGVudmVyLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.