
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 25 March 2011
Sec. Microbial Physiology and Metabolism
volume 2 - 2011 | https://doi.org/10.3389/fmicb.2011.00055
This article is part of the Research Topic The microbial sulfur cycle View all 21 articles
Marine micro-oxic to sulfidic environments are sites of intensive biogeochemical cycling and elemental sequestration, where prokaryotes are major driving forces mediating carbon, nitrogen, sulfur, phosphorus, and metal cycles, important from both biogeochemical and evolutionary perspectives. Associations between single-celled eukaryotes and bacteria and/or archaea are common in such habitats. Here we describe a ciliate common in the micro-oxic to anoxic, typically sulfidic, sediments of Santa Barbara Basin (CA, USA). The ciliate is 95% similar to Parduzcia orbis (18S rRNA). Transmission electron micrographs reveal clusters of at least three different endobiont types organized within membrane-bound sub-cellular regions. Catalyzed reporter deposition–fluorescent in situ hybridization and 16S rRNA clone libraries confirm the symbionts include up to two sulfate reducers (Desulfobulbaceae, Desulfobacteraceae), a methanogen (Methanobacteriales), and possibly a Bacteroidete (Cytophaga) and a Type I methanotroph, suggesting synergistic metabolisms in this environment. This case study is discussed in terms of implications to biogeochemistry, and benthic ecology.
Anoxic habitats are common, including certain marine and freshwater sediments and water columns, water-logged soils, sewage, the gastro-intestinal tracts of animals, the carcasses of dead mammals on the sea-floor, and the interiors of some suspended organic aggregates. Many of these habitats not only lack oxygen, but have considerable, and sometimes physiologically significant (e.g., Bagarinao, 1992) concentrations of hydrogen sulfide, which inhibits aerobic respiration. The abundant bacteria and archaea found in anoxic and potentially sulfidic environments are grazed upon by anaerobic protists (Fenchel and Finlay, 1990). Through grazing on prokaryotic and other eukaryotic prey, protists modify or re-mineralize organic matter, and regenerate nutrients (Taylor, 1982; Jumars et al., 1989; Sherr and Sherr, 2002). Protist grazing in marine environments also can affect the quantity, activity and physiological state of their prey, and hence, through these direct and indirect effects, may help determine the metabolic potential of prey communities (Madsen et al., 1991; Sherr and Sherr, 2002; Frias-Lopez et al., 2009). Anaerobic protists, including many groups of eukaryotes that lack mitochondria, such as anaerobic ciliates, were first documented by Fenchel et al. (1977). Many anaerobic eukaryotes, including the anaerobic ciliates are currently considered to be secondarily amitochondriate, and more recently adapted to an anaerobic lifestyle (see discussion by Embley and Finlay, 1994). Fenchel et al. (1977) first showed that several ciliate genera, including Caenomorpha, Metopus, Parablepharisma, Plagiopyla, Saprodinium, and Sonderia lack mitochondria and cytochrome oxidase. Like many other anaerobic eukaryotes however, they have specialized redox organelles called hydrogenosomes (possibly of mitochondrial origin, see Müller, 1988 and recent discussion in Barbera et al., 2010). Hydrogenosomes carry out an H2-evolving fermentation thought to play an integral role in the symbiosis between anaerobic ciliates and certain bacteria and archaea including methanogens (commonly members of Methanobacteriales and Methanomicrobiales; Embley and Finlay, 1994).
Endosymbiotic associations are common within protists (for recent review see Nowack and Melkonin, 2010). The acquisition of endosymbionts has been proposed to represent a general evolutionary strategy in protists to acquire novel biochemical functions (Nowack and Melkonin, 2010) and, in the case of micro-oxic (up to 0.1 ml/L; Bernhard and Sen Gupta, 1999) or anoxic, and potentially sulfidic marine environments, symbiosis may represent a strategy for exploiting these otherwise inhospitable habitats. Examples of symbiotic relationships between bacteria, archaea, and eukaryotes in deep-sea oxygen-depleted marine environments have been well documented for some groups, starting with the discovery of associations between metazoa and bacteria at hydrothermal vents (Cavanaugh et al., 1981; Cavanaugh, 1994), cold seeps (Barry et al., 1996), and the edges of silled basins (e.g., Distel and Felbeck, 1988). Chemosynthetic autotrophy supports many of these associations and often involves the oxidation of hydrogen sulfide or methane by endosymbiotic bacteria within the animal hosts.
The symbionts of some marine ciliates have been described phylogenetically, including some from oxygenated habitats (Amman et al., 1991; Springer et al., 1993; Beier et al., 2002; Vannini et al., 2004) and some from anoxic habitats (Embley et al., 1992; Embley and Finlay, 1994). Much information about ciliate endosymbionts has come from morphological and functional approaches (e.g., Fenchel and Finlay, 1991b; Fenchel and Bernard, 1993; and for review see Görtz, 2002). Multiple endosymbionts have been described in individuals of certain ciliate species. For example, in the parasitic ciliate Ichthyophthirius multifiliis, a common pathogen of freshwater fish, 16S rRNA gene sequencing and fluorescent in situ hybridization (FISH) demonstrated that three classes of bacteria were present, including Alphaproteobacteria (Rickettsiales), Sphingobacteria (in the cytoplasm), and Flavobacterium columnare (associated with cilia; Sun et al., 2009). In another freshwater ciliate, Cyclidium porcatum, an internal structure is present that consists of hydrogenosomes interspersed with methanogens and unidentified bacteria. This association is stable and persistent, indicative of an anaerobic symbiotic consortium of three functional partners (Esteban et al., 1993).
Methanogens are common endosymbionts in free-living anaerobic ciliates (e.g., Fenchel and Finlay, 1991a; van Hoek et al., 2000). In the anaerobic free-living ciliates Metopus contortus, M. striatus, M. palaeformis, Trimyema sp., and C. porcatum, methanogens from three different genera were identified on the basis of rRNA gene sequencing and microscopy (Embley and Finlay, 1993, 1994). These authors determined that such endosymbioses have formed repeatedly and independently, and that most likely, some were recent events. There have been different levels of association observed between hydrogenosomes in anaerobic ciliates and their endosymbiotic methanogens (Embley and Finlay, 1994). The endosymbiotic methanogens may use the H2 produced by hydrogenosomes to reduce CO2 and generate energy (Van Bruggen et al., 1983; Jones et al., 1987), and in the process may release dissolved organics utilizable by the host (Fenchel and Finlay, 1991a). Some ciliates clearly benefit from this association because the growth rate of these ciliates is much lower if their methanogens are inhibited (Fenchel and Finlay, 1991b; Finlay and Fenchel, 1992).
Ciliates are perhaps the most studied protist group in terms of taxonomy. A unifying feature of ciliates is that they all contain two types of nuclei, a germ or micronucleus and a polyploid somatic macronucleus, the latter of which is most actively transcribed (Raikov, 1985). The karyorelictid ciliates have been hypothesized to be primitive within the phylum Ciliophora because of their simple form of nuclear dualism whereby the macronuclei are nearly diploid and do not divide once they differentiate from a micronucleus (Corliss, 1979; Hirt et al., 1995). Here we describe a karyorelictid ciliate and its bacterial and archaeal associates recovered from the oxygen-depleted sediments of Santa Barbara Basin, CA, USA.
The Santa Barbara Basin, which is located off California (USA), has a maximum depth of ∼600 m and sill depth of ∼475 m. The samples used for this study were collected using a Soutar box corer or an MC800 multicorer from sea-floor sediments (580–592 m depth) collected between September 2007 and June 2009 using the RV Robert Gordon Sproul. Initial examination of sediments revealed a relatively abundant brown ciliate. Samples bearing the targeted ciliate were collected along a north-south trending transect along 120°02′W, from 34°17.6′N to 34°13.0′N. Our samples were collected from box cores that exhibited a surface covering of sulfide-oxidizing bacteria (either Thioploca or Beggiatoa, both of which require sulfide and little or no oxygen; e.g., McHatton et al., 1996). Surface ∼0–2 cm sediments were transferred to 100–250 ml high-density polyethylene (HDPE) bottles, completely filled with bottom water, and stored at ∼7°C. Sample bottles were kept tightly closed until aliquots were removed for ciliate harvest. Once a bottle was opened, the redox chemistry changed sufficiently so that within about a week most of the target ciliates died. We did not amend bottles with substrates or food. This approach has been used successfully by Bernhard for many studies (e.g., Bernhard et al., 2000, 2006; Edgcomb et al., 2010).
For specimen counts, aliquots of archived quantitative surface 1-cm3 samples were counted for ciliate abundances. Quantitative samples were taken from boxcores using 60-cc syringe cores. Concurrent with sediment sampling, oxygen concentrations were obtained for bottom waters using the microwinkler method (Browenkow and Cline, 1969).
For light microscope observation, cells were observed under the dissecting microscope in ice-chilled Petri dishes containing sample water and sediment. Cells were selected using micropipettes. Live ciliates were studied at magnifications of 40–1000× with brightfield, phase contrast, and differential interference contrast (DIC) using a Zeiss Axioskop 2 plus microscope. A minimum of 50 living target ciliates were observed and photographed. Cells lysed within minutes of illumination under a cover glass making in vivo observations quite difficult. Consistent with the experience of others studying marine karyorelictids, we had great difficulty with fixation and silver impregnation of these ciliates (Foissner and Dragesco, 1996; Dragesco, 1999; Foissner and Al-Rasheid, 1999). We employed the techniques described by Foissner and Dragesco (1996) and Foissner and Al-Rasheid (1999), including many of our own modifications, without success. Consequently protargol impregnations were of suboptimal quality. Although some temporary preparations done according to the method of Foissner and Dragesco (1996) were adequate for determining general morphologic features, many details were often obscured by densely impregnating cortical granules. Microphotographs and measurements of fixed cells were made using a Zeiss AxioCam and Axiovision imaging software or a Flex digital camera and Spot imaging software (Diagnostic Instruments Inc., Sterling Heights, MI, USA). Microphotographs of uncompressed cells were used for in vivo measurements. Terminology is according to Dragesco (1999), Foissner and Xu (2007), and Lynn (2008). Classification is according to Lynn (2008).
Aboard the ship, aliquots of bulk sediments were fixed in 3% glutaraldehyde (final concentration) buffered with 0.1 M Na-cacodylate (pH 7.2), from which specimens were isolated from the coarse residue after sieving with buffer over a 63-μm screen. Specimens were processed for transmission electron microscopy (TEM) following our standard procedures (Bernhard et al., 2000). Sections from approximately 10 individuals were examined with a Zeiss 10CA transmission electron microscope.
Single cells of the brown-pigmented ciliate were picked from sediments using a dissecting microscope. In order to greatly minimize contamination, cells were rinsed three times in sterile seawater (0.2 μm filtered) before being placed into 2.0 ml microfuge tubes and frozen at −20°C for DNA extraction. Surface sterilization with antibiotic such as streptomycin was not possible because these ciliates lysed within ∼10 min of being removed from their bottles for hand picking. Exposure to an antibiotic solution as per methods of Vannini et al. (2004) caused immediate lysis of this ciliate. Individuals were divided into two groups: single individuals/PCR tube for direct PCR amplification or pools of ∼25–50 individuals for DNA extraction. DNA was extracted from pooled samples using the DNAeasy kit (Qiagen, USA) following the manufacturer’s recommendations.
Bacteria-, eukarya-, and archaea-specific primers were tested for positive PCR amplification. Bacterial primers were Bact8F (Amman et al., 1995) paired with U1492R (Longnecker and Reysenbach, 2001). Archaeal primers were ARC4F (Jolivet et al., 2003) paired with 1492R (Lane, 1991), followed by ARC21F (DeLong, 1992) paired with ARC915R (Stahl and Amann, 1991). Eukaryotic primers were 82F (Dawson and Pace, 2002), 360F (Medlin et al., 1988), and 528F (Elwood et al., 1985) paired with U1492R. PCR amplification conditions for the bacterial and eukaryotic primer pairs were: 95°C for 5 min, followed by 40 cycles of 95°C for 1 min, 45°C for 1 min, 72°C for 1.5 min, and a final cycle of 72°C for 7 min. For archaea, a nested amplification was used with SpeedSTAR HS DNA Polymerase (TaKaRa) as described by the manufacturer: 98°C for 2 min followed by 30 cycles of 98°C for 5 s, 55°C for 15 s, and 72°C for 20 s, and a final cycle of 72°C for 2 min. This was followed by another round of 98°C for 2 min followed by 30 cycles of 98°C for 5 s, 58°C for 15 s, and 72°C for 20 s with a final cycle of 72°C for 2 min. We also amplified one of the key genes of sulfate reduction, dsrAB using the primer mixes DSR1F and DSR4R according to the protocols in Loy et al. (2004). Cycling conditions were: 95°C for 5 min followed by 35 cycles of 94°C for 40 s, 48°C for 40 s, and 72°C for 90 s with a final 72°C incubation for 7 min.
Amplified DNA was checked for quality by agarose gel electrophoresis, bands of the expected sizes were gel purified using the Zymoclean Gel DNA Recovery Kit (Zymo Research) and positive reactions were cloned into the vector pCR4-TOPO using the TOPO TA Cloning Kit (Invitrogen) following the manufacturer’s instructions. Plasmid DNA from 16 clones (12 clones for dsrAB gene sequences) per primer pair was prepared using a MWG Biotech RoboPrep2500, and inserts were sequenced bi-directionally using the universal M13F and M13R primers and an Applied Biosystems 3730XL capillary sequencer at the Keck Facility at the Josephine Bay Paul Center at the Marine Biological Laboratory (MBL), Woods Hole, MA, USA. Processing of sequence data used PHRED and PHRAP (Ewing and Green, 1998; Ewing et al., 1998) and a pipeline script. The sequences were checked for chimeras using the Bellerophon Chimera Check and the Check_Chimera utilities (Ribosomal Database Project; Cole et al., 2003).
For phylogenetic analyses, we aligned the clone sequences from archaeal and bacterial amplifications to 16S rRNA sequences available in the ARB package (Ludwig et al., 2004; http://www.arb-home.de). The rRNA alignment was corrected manually according to secondary structure information. Only unambiguously aligned positions (961 bp for the host ciliate alignment, 844 for the archaeal data set, and 982 for the bacterial data set) were used to construct phylogenetic trees. To these alignments, we added the closest relatives of our original sequences retrieved from GenBank using BLASTn. Alignments were subjected to Bayesian and Maximum Likelihood inference using RAxML (Stamatakis et al., 2008) and MrBayes (Ronquist and Huelsenbeck, 2003). All phylogenetic analyses were performed on the CIPRES portal (Miller et al., 2009) under the GTR + Gamma model using 1000 bootstrap replicates and estimation of the proportion of invariable sites. Bayesian analyses consisted of two independent runs with 4 × 106 generations. Trees were sampled every 1000 generations with 25% discarded as burn-in. Topologies of ML and Bayesian trees were compared and the tree with the best likelihood was chosen for presentation.
Catalyzed reporter deposition (CARD)–FISH was performed with only minor modifications to the methods of Pernthaler et al. (2002). A minimum of 80 cells were hand picked and rinsed in sterile seawater and fixed in Bouin’s solution for 2 h (Stoeck et al., 2003), then rinsed three times with 5 ml sterile phosphate buffered saline (PBS) by filtration onto a 0.2-μm pore size, 25 mm Isopore GTTP filter (Millipore, USA). Air-drying caused the cells to collapse and release their contents, and so in most cases we did not permit the filters to dry completely before overlaying them with 37°C 0.2% (w/v) Metaphor agarose and drying at 50°C. To inactivate endogenous peroxidases, filter sections were incubated in 10 ml of 0.01 M HCl for 10 min at room temperature. Filters were washed in 50 ml 1× PBS, then in 50 ml of distilled, deionized water (ddH20). The host and endobiont cells were permeabilized by incubating the individual filter pieces in 2.0 ml Eppendorf microfuge tubes for 60 min at 37°C in a lysozyme solution (0.05 M EDTA, pH 8.0; 0.1 M Tris HCl, pH 8.0; 10 mg/ml lysozyme). The filters were washed in 1× PBS for 2 min, followed by treatment for 20 min in a solution of proteinase K (50 μl of 1064 U/μl in 10 ml Tris EDTA buffer) at room temperature. Proteinase K was inactivated in a solution of 0.01 M HCL for 20 min at room temperature, and filters were washed in 50 ml 1× PBS for 1 min and then 50 ml ddH20 for 2 min, followed by 50 ml of absolute ethanol (96%) and air-dried or partially air-dried prior to embedding in Metaphor agarose. For visualization of archaeal endobionts, we tested 0.5–5.0 U of pseudomurein endoisopeptidase (PeiW) per filter according to the methods of Nakamura et al. (2006). Hybridization buffer and probe were mixed 300:1 in 2.0 ml Eppendorf tubes (probe at 50 ng/μl). For 50 ml of hybridization buffer we mixed 3.6 ml 5 M NaCl, 0.4 ml 1 M Tris HCl and ddH20 depending on formamide concentration for each probe used (see Table 1). The remainder of the CARD–FISH procedure followed the protocol of Edgcomb et al. (2010). The Alexa488-labeled probes used include EUB338 I-III (Daims et al., 2001), NON338 (Wallner et al., 1993), Arch915 (Stahl and Amann, 1991), Alf968 (Neef, 1997), Gam42a (Manz et al., 1992) and Gam42a competitor (Yeates et al., 2003), BET42a (Manz et al., 1992) and BET42a competitor (Yeates et al., 2003), DELTA495a, b, and c and the corresponding competitor probes for each, cDELTA495a, b, and c (Lucker et al., 2007), EPS549 (Lin et al., 2006), DSBAC357 (Lucker et al., 2007), DSB706 (Loy et al., 2002), DSB658 (Manz et al., 1998), and EPS549 (Lin et al., 2006). Formamide and NaCl concentrations used in wash buffers are noted in Table 1. CARD–FISH images were taken on a Zeiss Axioplan 2 microscope equipped with a Zeiss AxioCam camera. Confocal microscope images were taken with an Olympus Fluoview 300 Laser Scanning Confocal Microscope equipped with an Argon laser for FITC/Alexa488.
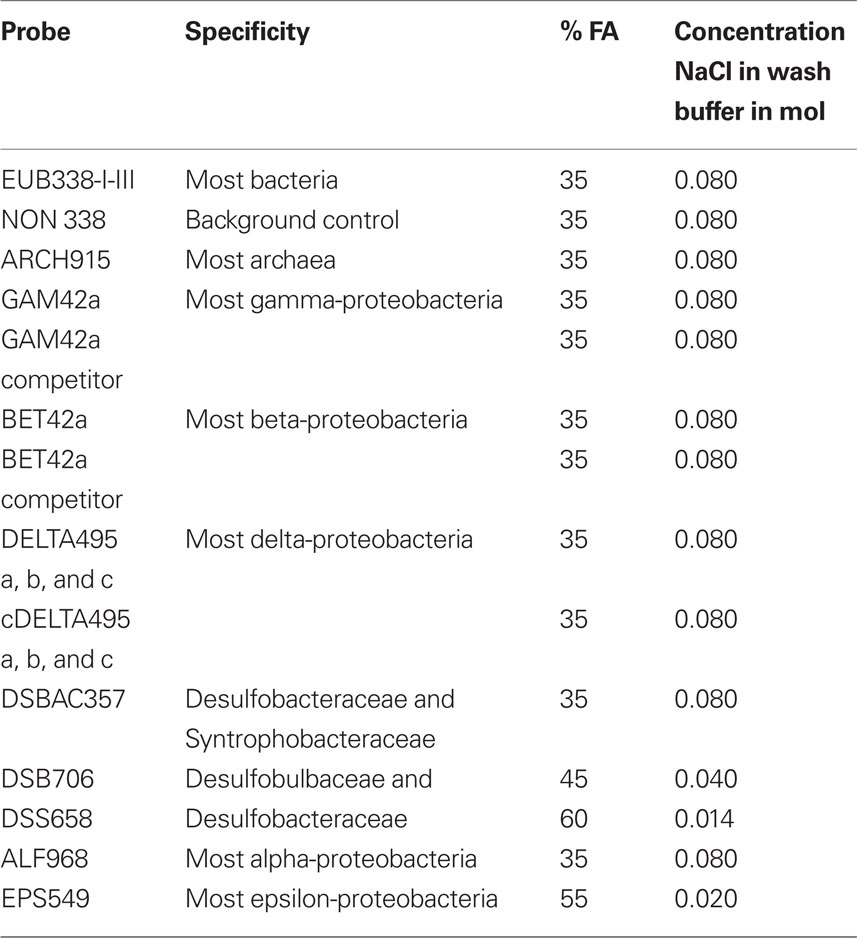
Table 1. Percent formamide concentrations and concentration of NaCl used in wash buffers for probes used in this study.
Dissolved oxygen concentrations in bottom waters overlying sediments used for recovery of the brown-pigmented ciliate analyzed for CARD–FISH ranged from 0.2 to 0.5 μM. Concentrations <1 μM are typical for these sites (Bernhard et al., 2000, 2003). It should be noted that bottom-water oxygen and sulfide concentrations vary considerably in Santa Barbara Basin depending on timing of flushing events (e.g., Reimers et al., 1990, 1996; Kuwabara et al., 1999; Bernhard et al., 2003).
Concentrations of the brown-pigmented ciliate ranged from an average of 19.1 cells/cm3 when bottom waters were 10.8 μM O2 to 71.6 cells/cm3 when bottom waters were 0.1 μM O2 (Table 2), indicating a clear preference for low oxygen sedimentary environments.
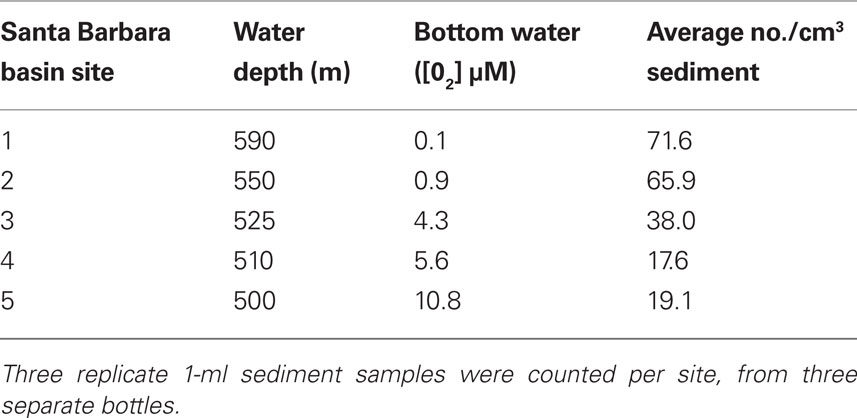
Table 2. Average counts of the Geleiid karyorelictean ciliate from different sites in Santa Barbara Basin with varying bottom-water oxygen concentrations.
The size of the host ciliate in vivo was ∼280–365 × 120–150 μm (n = 20). The body shape is obpyriform, roughly elliptical in cross-section and slightly laterally compressed. The anterior part forms a broad leftward-curving neck, terminating in a bluntly rounded rostrum. The anterior is lanceolate in more strongly contracted individuals (Figures 1A,B). The posterior is bluntly tapered. This ciliate moves slowly, is sluggishly contractile, writhing through detritus, and does not swim. The cell shape is broadly ovate after fixation, ∼325 μm × 250 μm. The buccal overture is subapical. The overhanging rostrum forms a raised lip around the anterior margin of the buccal cavity (Figure 1C). The nuclear apparatus is located centrally, obscured by cortical granules in vivo, and consists of two adjacent but unattached spherical macronuclear nodules ∼20–25 μm across in impregnated specimens. A single ∼ 8 μm diameter spherical micronucleus is present between but not attached to the macronuclei. The macronuclei contain ∼20 densely impregnating nucleoli ∼3–7 μm across after protargol (Figure 2B, inset). There was no evidence of a contractile vacuole. The cortex is flexible.
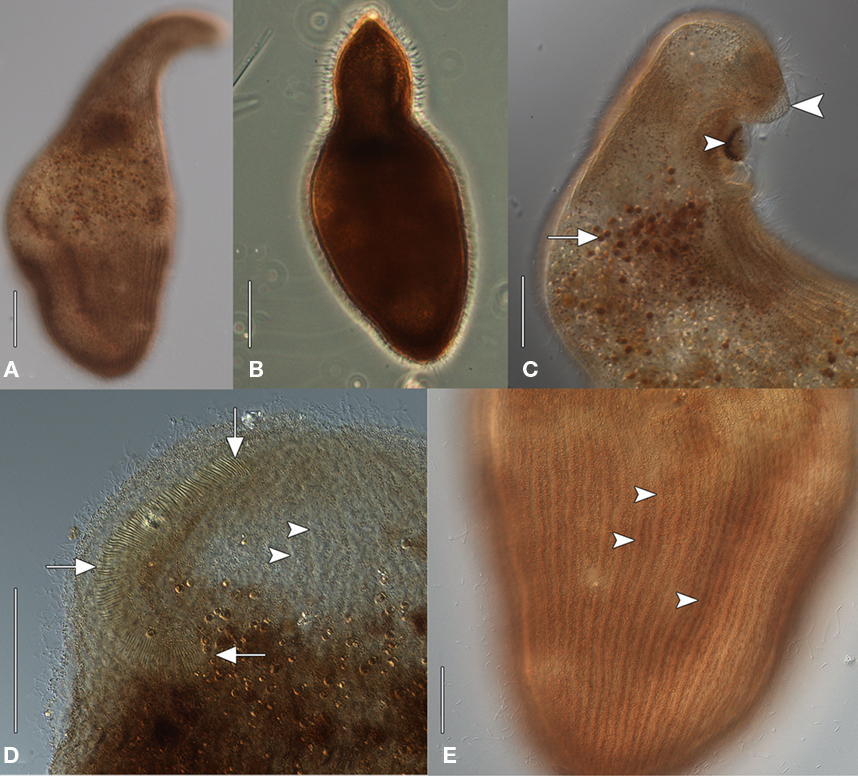
Figure 1. Geleiid karyorelictean ciliate recovered from sulfidic sediments, in vivo [(A, C–D), differential interference contrast, DIC; (B) phase contrast]. (A) Cell shape, dorsal view. (B) Strongly contracted cell with more acutely tapered anterior end. (C) Anterior end, lateral view. A mass of pigmentocysts is located at the cytostome (small arrowhead). The blunt rostrum forms a lip that overhangs the anterior margin of buccal overture (large arrowhead). Pigmentocysts are scattered throughout the cytoplasm (arrow). (D) Detail of anterior end, ventral view. The right paroral polykinety (arrows) extends for a short distance onto the left side of the buccal overture. Arrowheads mark probable longitudinal interkinetidal myonemes. (E) Detail of cortex, ventral view. Densely packed cortical pigment granules are located within somatic kineties (arrowheads). Scale bars: (A–D) = 50 μm, (E) = 25 μm.
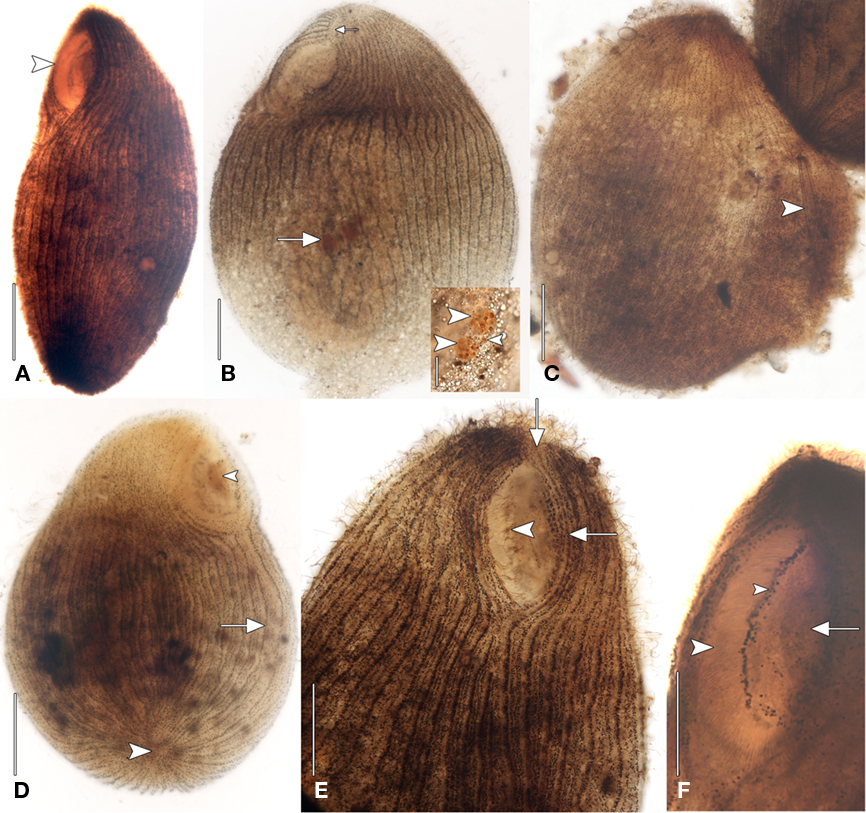
Figure 2. Geleiid karyorelictean ciliate recovered from sulfidic sediments after protargol. (A) Left anterolateral view showing position of cytostome (arrowhead). (B) Left anterolateral view of infraciliature. Right and left somatic kineties originate at short preoral suture (small arrow). The nuclear apparatus consisting of two macronuclear nodules and a single nearby micronucleus is located centrally (large arrow). Inset, detail of nuclear apparatus. The macronuclear nodules (large arrowheads) contain densely impregnating nucleoli. The single micronucleus is located between the macronuclei (small arrowhead). (C) Dorsal view of infraciliature. The cytoplasm, containing an ingested diatom (arrowhead), has ruptured through the cortex. (D) Posteroventral view of infraciliature. Somatic kineties converge at the posterior pole (large arrowhead). Some somatic kineties are incomplete (arrow). A mass of impregnated pigmentocysts is seen at the cytostome (small arrowhead). (E) Anteroventral view. Long, fine cilia arise from the right paroral polykinety (arrowhead). Kinety n probably becomes the first adoral kinety and continues into the preoral suture (arrows). (F) Left anteroventral view. A line of granules marks the course of a paroral intrabuccal kinety. Scale bars: (A–E) = 50 μm, Inset (B,F)= 25 μm.
A disorganized concentration of ∼0.6 μm diameter brownish-pink pigment granules is located within each kinety with two rows of more loosely spaced granules between kineties (Figures 1E and 2D,E). Cortical granules and larger cytoplasmic pigmentocysts give the cells a deep brownish-pink color under DIC and phase contrast illumination (Figures 1A–C,E). Wavy fibrillar structures, likely contracted myonemes, are located between kineties (Figure 1D) and the cytoplasm contains numerous shiny brownish-pink pigmentocysts, ∼3–5 μm in diameter (Figure 1C). Food vacuoles were observed rarely, occasionally containing pennate diatoms (Figure 2C). The somatic infraciliature is comprised of ∼40–50 meridional kineties, ∼9 μm apart in impregnated specimens, and consisting of ciliated dikinetids. Kineties originate anteriorly at a short preoral suture, and converge at the posterior pole (Figures 2B,D,E). There are occasional incomplete somatic kineties but no true postoral kineties (Figures 2D,E). There is no postoral suture. The somatic cilia are ∼13 μm long. Kinety 1 and kinety n diverge at the posterior end of the buccal overture to pass right and left of it, respectively (Figure 2E). The elliptical, cup-shaped oral apparatus is subapical, and extends ∼20–25% of the cell length. An aggregation of pigmentocysts is present at the centerof the cytostome in vivo and in protargol impregnated specimens(Figures 1C and 2A,D,F). The right paroral polykinety is ∼12 μm wide, and consists of ∼100 densely spaced files of basal bodies. The paroral polykinety occupies the entire right margin of the buccal overture, curving around it posteriorly onto the left margin for a short distance (Figures 1D and 2F). Paroral polykinety cilia are longer, ∼17–20 μm, and finer than somatic cilia (Figure 2E). A densely impregnating line of granules medial to the right oral polykinety probably represents a “paraoral intrabuccal kinety” described in Parduczia and other Geleiides (Dragesco, 1999, Figure 180 in that text). Better-quality silver impregnations are necessary for a complete characterization of the oral infraciliature and statistically meaningful morphometric data.
Transmission electron microscopy analysis revealed a consistent dorso-centrally located region of double membrane-bound vesicles arranged in a kidney bean-shape (shape description based on FISH images only), each containing numerous prokaryotic cells (Figure 3A). Cells inside the membrane-bound structures exhibiting satisfactory preservation usually showed a consistent distribution within each vesicle (Figures 3A–C). A minimum of three morphotypes of endobionts were observed, which may or may not each represent unique species. One prominent electron-dense rod-shaped endobiont often lined the inner surface of the membrane-bound vesicle. This rod ranged in size from ∼2 to 7 μm in length and ∼1 to 2 μm in diameter (Figures 3B,C). Many of these showed internal membranes typical of Type I methanotrophs (Garrity et al., 2005; Figures 3C,D). A second morphotype was represented by smaller rod-shaped cells (∼0.5–1.0 μm diameter and ∼1–2 μm in length), many with apparent internal granules reminiscent of some sulfate-reducing bacteria within the Desulfobacteraceae and Desulfobulbaceae (Widdel and Bak, 1992; Figures 3B,C). The third morphotype is a curved rod approximately 1–3 μm in length, and ∼0.2–0.5 μm in diameter (Figure 3E). Outside of each membrane-bound vesicle in the kidney bean-shaped cluster of vesicles in the host ciliate, there are pear-shaped double membrane-bound objects reminiscent of hydrogenosomes (Figures 3B,C). We observed these to be typically nestled between the vesicles and in contact with the membrane of at least one vesicle.
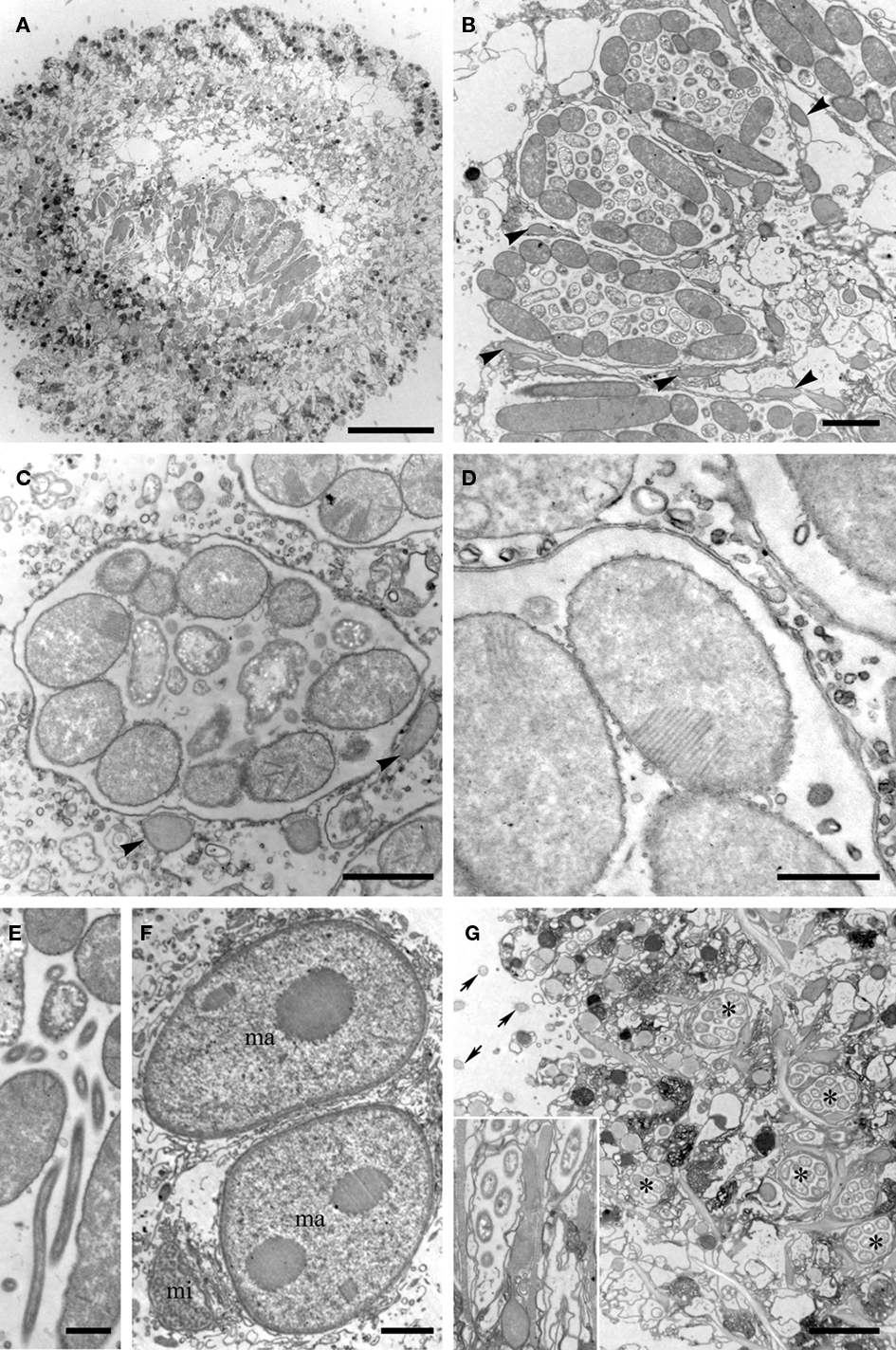
Figure 3. Transmission electron micrographs of the Santa Barbara Geleiid karyorelictean ciliate. (A) View of cross-section through ciliate showing portion of the kidney-shaped membrane-bound vesicle region. (B) Cross-section view through three double membrane-bound vesicles. Note the structured orientations of the long rod-shaped endobionts vs. shorter forms. Arrowheads = putative hydrogenosomes. (C) Close up of endobionts in a vesicle, showing vesicle double membrane. Note stacked membranes in many of the endobionts. Arrowheads = putative hydrogenosomes. (D) Close up showing stacked membranes in endobiont and double membrane of vesicle. (E) Close up of suspected Cytophaga. (F) View of two macronuclei (ma) and micronucleus (mi). (G) Ciliate periphery showing additional vesicles (*) with single endobiont morphotype. Arrows = Cilia. Inset shows close up including rod-shaped endobiont. Scale bars: (A) = 10 μm; (B,F,G) = 2 μm; (C–E) = 0.5 μm.
The nuclei were consistent in appearance with observations made by light microscopy (Figure 3F). It should also be noted that other membrane-bound vesicles containing what appears to be a single prokaryote morphotype were consistently located along the host cell periphery (Figure 3G). Epibiotic organisms were not observed with either light microscopy or TEM.
The eukaryotic primer sets 82F/U1492R, 528F/U1492R, and 360F/U1492R all provided positive amplication. BLASTn analysis of the small subunit ribosomal RNA (SSU rRNA) gene indicated that this sequence was most closely related to P. orbis (sequences are 95% similar). A phylogenetic analysis including all major groups of ciliates confirmed the affiliation with P. orbis within the Karyorelictea with 100% bootstrap support under maximum likelihood and a Bayesian posterior probability of 1.0 (Figure 4). Tree topologies under Bayesian and maximum likelihood analyses were congruent.
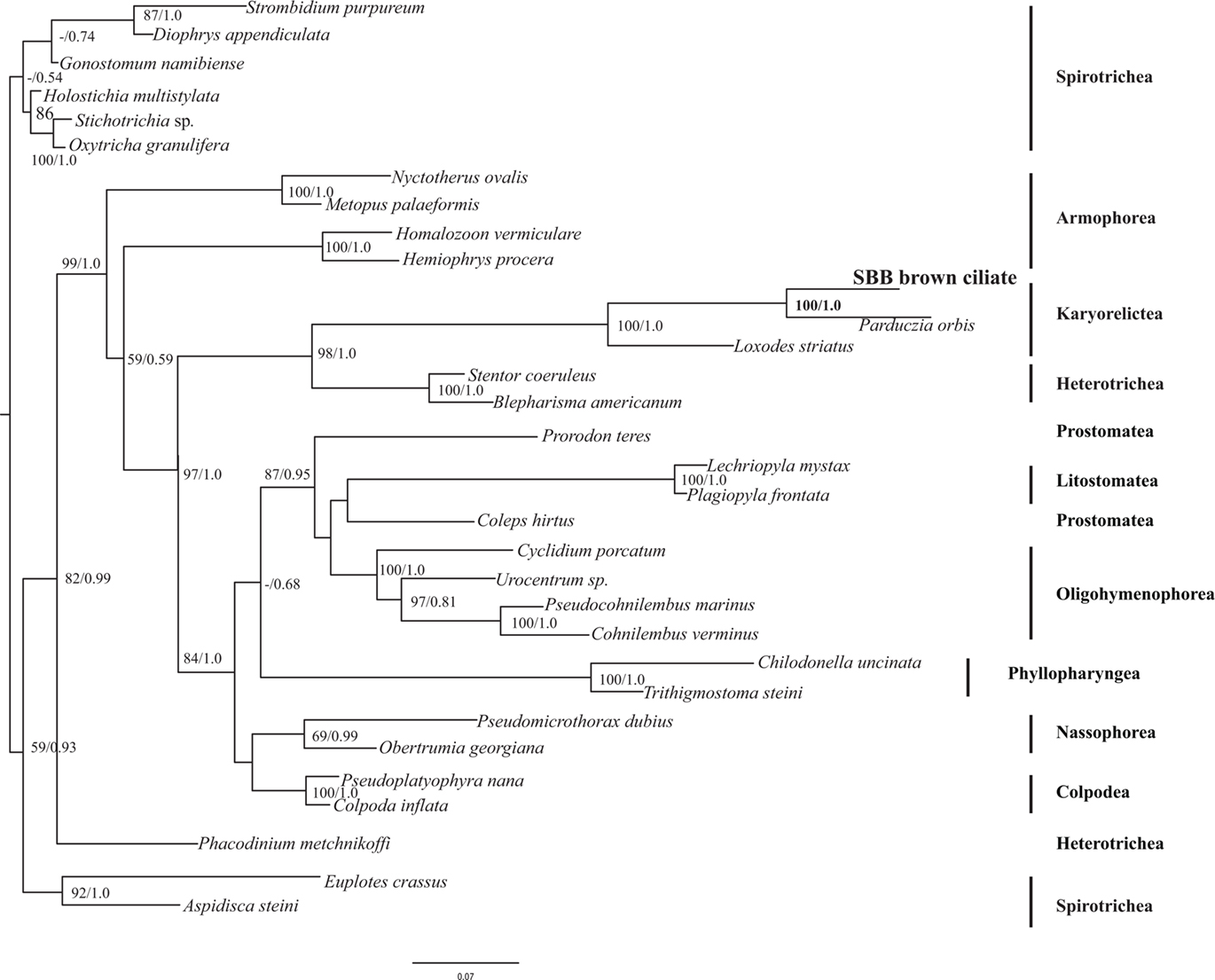
Figure 4. Phylogenetic analysis of 18S rRNA gene from the Santa Barbara Basin Geleiid karyorelictean ciliate. Tree is based on an alignment of 961 nucleotides. Bootstrapping and determination of the best estimate of the ML tree topology for this data set was conducted with the Rapid Bootstrapping algorithm of RAxML version 7.0 under the GTR + Gamma model. Maximum likelihood and Bayesian analyses were run on the CIPRES portal (www.phylo.org). Numbers at nodes present maximum likelihood bootstrap support above 50% and Bayesian posterior probabilities above 0.5, respectively.
Positive PCR amplification was obtained with bacterial primers, and we cloned and sequenced 12 clones. The results of phylogenetic analyses indicate the presence of two types of sulfate-reducing bacteria, one affiliating with the Desulfobacteraceae with 100% bootstrap support under maximum likelihood, and one affiliating with the Desulfobulbaceae with 97% bootstrap support under maximum likelihood (Figure 5). A positive PCR amplification was also obtained with the nested archaeal primer sets ARC4F/1492R and ARC21F/ARC915R. Phylogenetic analysis indicates this sequence is most closely related to members of the Methanobacteria with 100% bootstrap support under maximum likelihood (Figure 6). Successful amplification was also achieved using primers to one of the functional genes of sulfate reduction dsrAB. Seven of the 12 dsrAB clones we sequenced BLASTed as “uncultured sulfate-reducing bacterium dsr A/B” (EF065041). Two of the dsrAB clones were then sequenced with internal primers DSR1F1 and DSR1R1 (Bahr et al., 2005) resulting in nearly full-length sequences of 1964 bp. These two sequences were 99.6% identical. The sequence belonging to the closest cultured relative was a sequence from a Desulfobacteraceae. SSU rRNA gene (for the ciliate and the endobionts) and dsrAB sequences have been deposited in GenBank under the accession numbers JF327423-JF327426 and JF439663-JF439664.
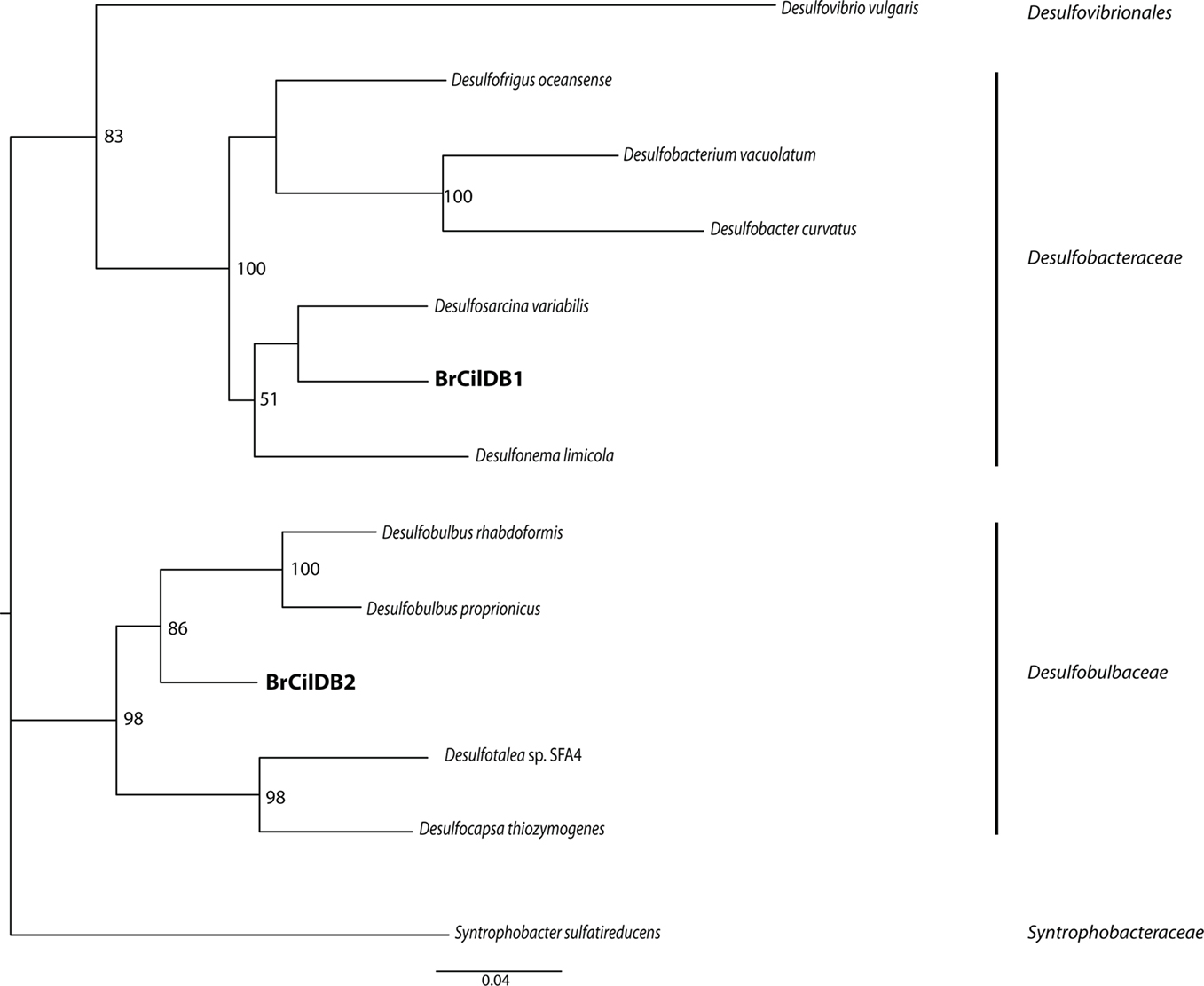
Figure 5. Phylogenetic analysis of 16S rRNA gene sequences from bacterial endobionts of the Santa Barbara Basin Geleiid karyorelictean ciliate. Tree is based on an alignment of 982 nucleotides. Bootstrapping and determination of the best estimate of the ML tree topology for this data set was conducted with the Rapid Bootstrapping algorithm of RAxML version 7.0 under the GTR + Gamma model, run on the CIPRES portal (www.phylo.org). Numbers at nodes present maximum likelihood bootstrap supportvalues above 50%.
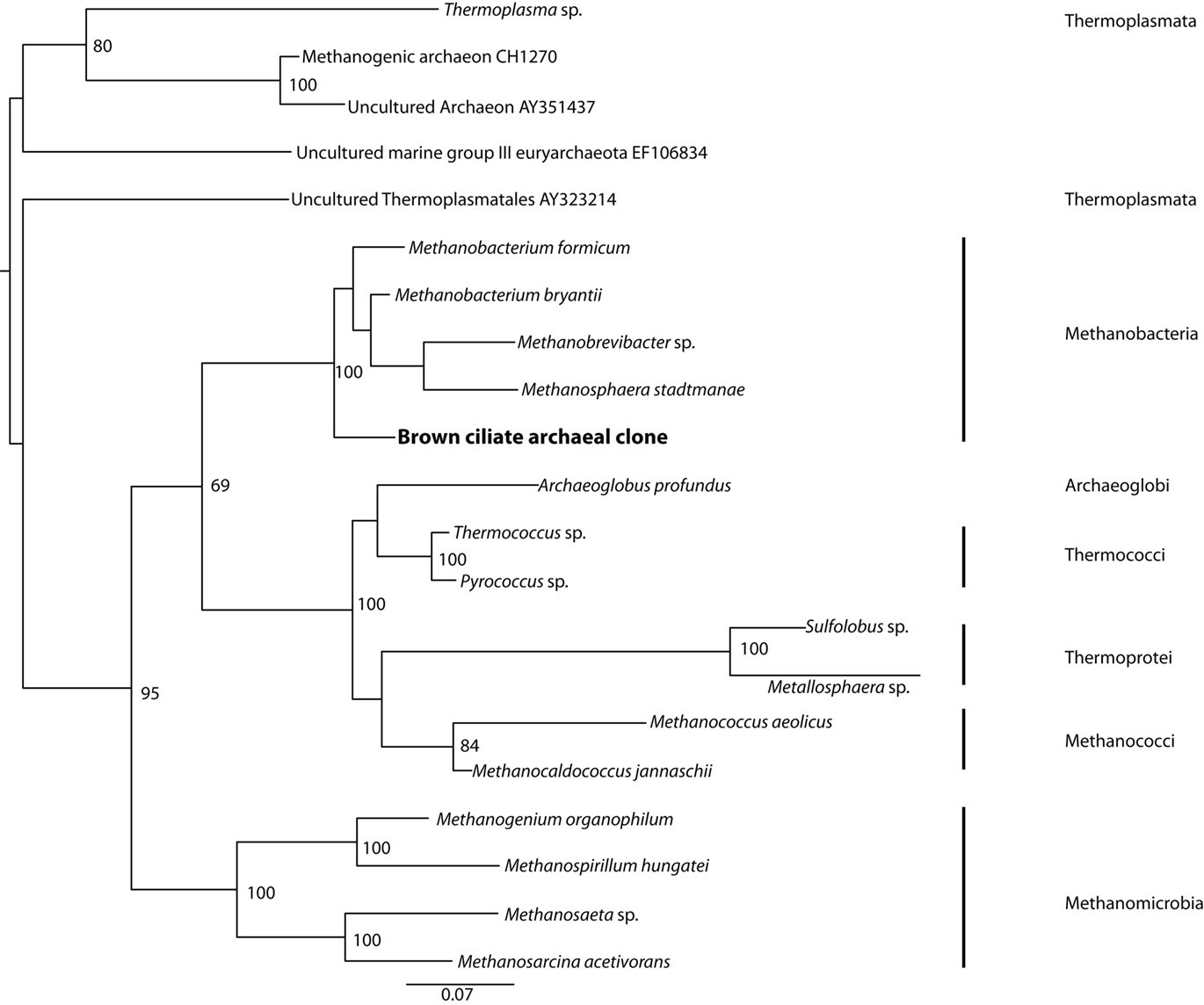
Figure 6. Phylogenetic analysis of 16S rRNA gene sequences from archaeal endobionts of the Santa Barbara Basin Geleiid karyorelictean ciliate. Tree is based on an alignment of 844 nucleotides. Bootstrapping and determination of the best estimate of the ML tree topology for this data set was conducted with the Rapid Bootstrapping algorithm of RAxML version7.0 under the GTR + Gamma model, run on the CIPRES portal (www.phylo.org). Numbers at nodes present maximum likelihood bootstrap support values above 50%.
Fixation did not always permit the visualization of original vesicle orientation/content structure, as cells that were agar-overlaid after drying collapsed and the cell membrane was often compromised. Permeabilization of intact host ciliates was challenging, as treatment with lysozyme, proteinase K, or PeiW often caused lysis, but visualization of CARD–FISH probe hybridization to the endobionts was not successful without these treatments. Both positive (general eubacterial probe) and negative (no probe and NON probe) control filters were included to be sure that permeabilization was sufficient in any individual experiment to interpret a negative result correctly. DAPI staining was included in all experiments, and this consistently revealed a ∼30- to 50-μm diameter and ∼200- to 250-μm long kidney bean-shaped area within the ciliate endoplasm where a dense region of endobiont cells was located (Figure 7A insert, Figures 7E,G). A representative image of the NON probe hybridization is shown in Figure 7B. The general eubacterial probe produces strong hybridization signal in this same region, showing clearly the ∼10- to 12-μm long vesicles containing the endobionts (Figures 7C,D). Hybridization signal with the gammaproteobacterial probe was positive, with signal also coming from the central region of the double membrane-bound vesicles (Figure 7F). Hybridization with the general sulfate-reducing bacteria (SRB) probe also was detected in the region of the densely-packed vesicles (Figure 7H), and also could be seen in the periphery of the cell (data not shown). The probe specific to the Desulfobacteraceae produced strong signal from within the cluster of membrane-bound vesicles (Figure 7I) and laser scanning confocal microscopy shows these positive signals to be coming from cells no larger than ∼1–1.5 μm (Figure 7J), consistent with the size and morphology of some of the cells located on the interior of the vesicle-bound consortium as described above. Signal from the second group-specific SRB probe to the Desulfobulbaceae was generally more sparsely distributed within the region of the densely-packed vesicles (Figure 7K). Positive signal emanated from the region of the vesicles with the general archaeal probe, showing ∼5 μm long cells (Figures 7L,M). Coenzyme F420-based blue-green autofluorescence typical of many methanogens was also observed (Figure 7N). The host nucleus was usually (but not always) visible with DAPI, depending on the cell’s orientation and degree of flattening (Figure 7G). Hybridizations with the alpha-, beta-, and epsilon-proteobacterial probes were all negative (data not shown).
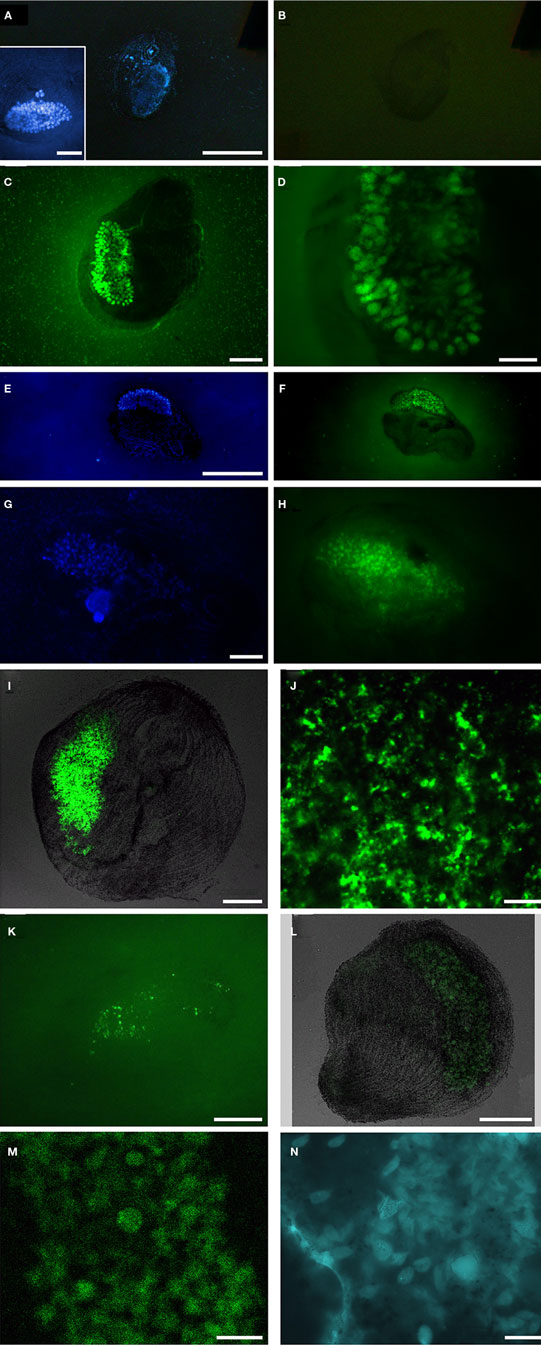
Figure 7. Catalyzed reporter deposition–fluorescent in situ hybridization analyses of Santa Barbara Geleiid karyorelictean ciliate. (A,B) DAPI and Alexa488-NON338 (negative control) images, insert presents DAPI of cell showing host nuclei, (C,D) Alexa488-EUB338I-III (general eubacteria) probe, (E,F) DAPI and Alexa488-GAM42a (gammaproteobacteria) probe, (G,H) DAPI and Alexa488-DELTA495a, b, and c (general deltaproteobacteria) probe, (I,J) Alexa488-DSBAC357 (Desulfobacteraceae) probe, (K) Alexa488-DSS658 (Desulfobulbaceae) probe, (L,M) Alexa488-ARCH915 (general archaea) probe, (N) autofluorescence in region of membrane-bound vesicles. Scale bars: (A–C,E–I,L), inset (A) = 50 μm, (D,K,N) = 20 μm; (J,M) = 10 μm.)
Abundance data indicate that this conspicuous ciliate species is prevalent in micro-oxic to anoxic and sulfidic Santa Barbara Basin sediments, yet is rare in adjacent well oxygenated sediments. During periodic oxygenation events in the basin, we presume that the ciliate migrates deeper into the sediments to avoid oxygen exposure. The vertical migration of benthic Santa Barbara Basin benthos in response to changing environmental conditions was documented by Bernhard et al. (2003).
Phylogenetic analysis of the SSU rRNA gene from this brown-pigmented ciliate confirms that it is a karyorelictid, as implied by light microscopy, and together with BLASTn analysis, that the most closely related organism in public databases is another karyorelictid, P. orbis (95% sequence similarity), a member of the Geleiidae. Morphological observations suggest a close relationship to Parduczia or Avelia. It has oral pigmentocysts like Avelia sp. (Figures 2E,F), however unlike Avelia sp., and more like Parduczia sp., this ciliate has a subapical position of the oral apparatus, and a beak or rostrum at the anterior end. Setting this ciliate apart from other members of Geleiidae are (1) the markedly smaller size (∼10–20% that of other geleiids) and (2) the amphoriform/obpyriform, rather than a very slender, filiform shape (Figure 8). As noted, due to difficulties encountered during fixation and silver impregnation, there are not yet sufficient taxonomic details to formally describe this as a new species (or genus).
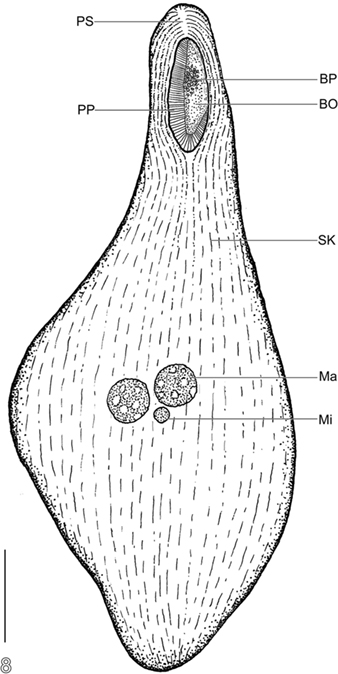
Figure 8. Semi-schematic body plan of Santa Barabara Geleiid karyorelictean ciliate. BO, buccal overture; BP, buccal pigmentocysts; Ma, macronuclear nodule; Mi, micronucleus; PP, right paroral polykinety; PS, preoral suture; SK, somatic kinety. Scale bar = 50 μm.
Most protist taxa observed in the micro-oxic and sulfidic sediments of Santa Barbara Basin harbor bacterial epibionts and/or endobionts (e.g., Bernhard et al., 2000, 2006), so it is not a surprise that this brown-pigmented, Parduczia-like ciliate also harbors endobionts. Within this Geleiid karyorelictean ciliate the internal double membrane-bound vesicles containing endobionts were consistently observed in the same dorso-central region of cells in every specimen examined. These structures were persistent in ciliates maintained in sample bottles containing sediments and bottom water in our cold room for more than 6 months after collection. The consistent observation of a highly specific ultrastructural affinity between this karyorelictid and its bacterial and archaeal endobionts, as well as the phylogenetic identity of some of the endobionts, leads us to infer symbiotic relationships that are likely mutualistic.
Phylogenetic and CARD–FISH analyses indicate that the endobionts include at least one archaeon within the Methanobacteria, and two types of sulfate-respiring bacteria. This ensemble of endobionts is surrounded by a double membrane, which may suggest a secondary endosymbiosis series of events. CARD–FISH and cellular ultrastructure also suggest the possibility of a methane-oxidizing gammaproteobacterium in the consortium. Although demonstrating metabolic exchange between the host and endobionts was outside the scope of this project, there are four reasons to allow inferences about the putative roles and metabolic interactions of these varied microbes. These factors include (1) the array of morphotypes observed in our TEM analyses, (2) probable physiotypes deduced from positive FISH hybridization results, (3) the positive recovery of one of the key genes of sulfate reduction, and (4) phylogenetic analysis of bacterial and archaeal small subunit ribosomal RNA genes.
Examples of these potential physiological contributions include sulfate reduction, methane oxidation, methanogenesis, and hydrogen and acetate generation (Figure 9). Evidence for the presence of two types of sulfate reducers comes from the recovery of one SSU rRNA gene signature affiliated with the Desulfobulbaceae, and another affiliated with the Desulfobacteraceae, as well as positive CARD–FISH hybridization to cells within the membrane-bound vesicles using probes to these two different groups of sulfate-reducing bacteria. As noted, we also recovered sequences affiliating most closely with the dsrAB gene from an uncultured sulfate-reducing bacterium. This gene is one of the key genes involved in sulfate reduction (Wagner et al., 1998; Klein et al., 2001). Evidence for the presence of a methanogen comes from the recovery of an archaeal SSU rRNA gene sequence affiliating with the Methanobacteria, positive CARD–FISH hybridization with a general archaeal probe, and from the presence of autofluorescence typical of most methanogens within the kidney bean-shaped area inside the host ciliate. Evidence for the presence of a gammaproteobacterial methanotroph is not as strong. We surmised the presence of this organism within the internal vesicle-associated consortia on the basis of positive CARD–FISH hybridization within the described region of membrane-bound vesicles using a probe specific to gammaproteobacteria, and cell ultrastructure reminiscent of Type I methanotrophs based on internal membrane structure. We did not recover a SSU rRNA gene sequence of a gammaproteobacteria, but we attribute this to the relatively low sequencing effort in this study (only 12 clones). Confirming the presence of a methanotroph with deeper sequencing, as well as demonstrating the metabolic activities associated with sulfate reduction and methanogenesis via other avenues of investigation are areas of future research in our laboratories.
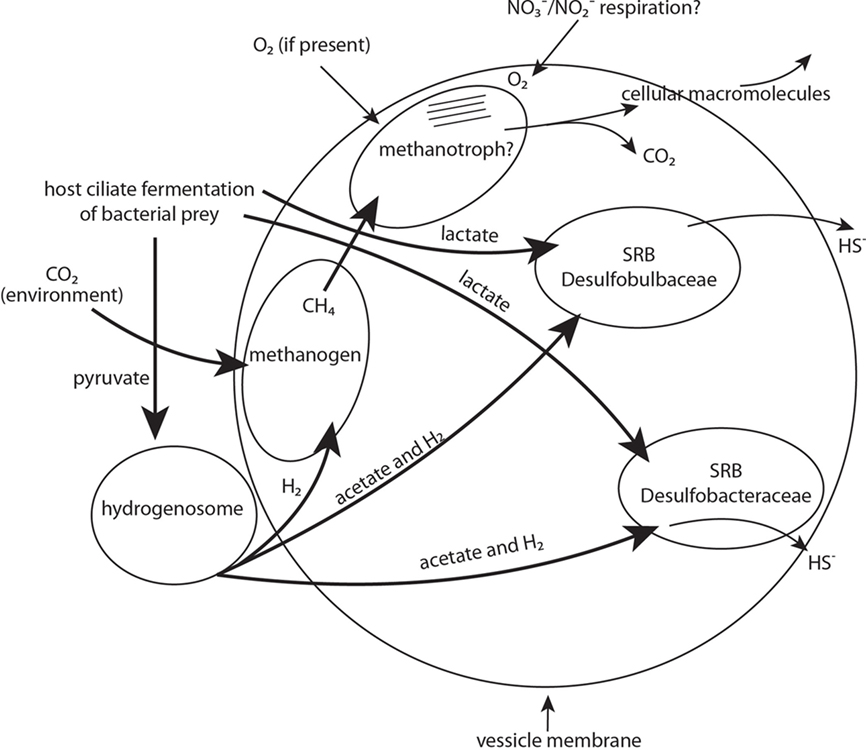
Figure 9. Schematic representation of proposed diverse endobiont and host cell metabolisms and their potential interactions. Major physiotypes include sulfate respiration, methanogenesis, methanotrophy, and hydrogenosome activity. Other activities might include nitrate and nitrite respiration. Bold arrows indicate carbon source/electron donor.
Elucidating the benefits of putative symbioses such as these for both the host and endobionts is not always straightforward. Although food vacuoles were only occasionally observed with light microscopy, this ciliate likely grazes on bacteria present in the sediments, and the fermentation of this food may produce a range of low molecular weight metabolites (including lactate and pyruvate and nitrogen compounds). Any lactate available from ciliate fermentation processes could be used for the growth of the sulfate-reducing bacteria (Figure 9). Although not directly demonstrated, the small double membrane-bound objects nestled along the outside of each vesicle are likely to be hydrogenosomes, as previously surmised. This is a logical explanation considering that many anaerobic ciliates described to date contain hydrogenosomes (Embley et al., 1995; Fenchel and Finlay, 1995). Hydrogenosomes could utilize available pyruvate resulting from the host fermentation processes as a carbon source, and would release molecular hydrogen and acetate (Figure 9). Both products, plus any available pyruvate, could be utilized as an electron donor and carbon source for growth and sulfate reduction by the sulfate-reducing bacteria. Methanogens, which are common endobionts of anaerobic ciliates (e.g., Finlay and Fenchel, 1989, and see recent synopsis by Hackstein, 2011) likely utilize some of the hydrogen produced by the hydrogenosomes along with CO2 diffused either from the ciliate’s metabolism or from the environment to produce methane (Figure 9). This methane could possibly be oxidized anaerobically via the coupling of this oxidation with the reduction of sulfate by sulfate-reducing bacteria (e.g., Hinrichs et al., 1999; Boetius et al., 2000; Orphan et al., 2001) but this coupling is usually between a sulfate-reducing bacterium and an archaeon.
Alternatively, we consider the possibility that enough oxygen may be present for aerobic methane oxidation by the bacterial methanotroph. Oxygen could be delivered via diffusion into the ciliate at periodic intervals when the ciliate migrates to micro-oxic zones, or via a mechanism similar to the recently described process of nitrite-driven anaerobic methane oxidation (Ettwig et al., 2010). Ettwig et al. (2010) report an anaerobic denitrifying bacterium that bypasses the denitrification intermediate nitrous oxide by the conversion of two nitric oxide molecules to dinitrogen and oxygen, which is used to oxidize methane by an unknown mechanism. Since this ciliate inhabits anoxic to micro-oxic sediments, it is likely that any methane oxidizer present must be able to conduct this process anaerobically. However, if the host ciliate were to migrate to sediments where limited oxygen is present, this gammaproteobacterium might switch to aerobic oxidation of methane since this is energetically more favorable (Caldwell et al., 2008). A methanotroph would likely evolve CO2 and cellular materials (e.g., polysaccharides, lipids, and proteins). It should be noted that even aerobic metabolism of Escherichia coli can function at unusually low oxygen concentrations (Stolper et al., 2010). An avenue for future investigation will be to obtain a small subunit ribosomal RNA signature from a gammaproteobacterium and to perhaps trace the movement of isotopically labeled compounds through this consortium. Although we obtained a SSU rRNA sequence from the Cytophaga/Flexibacter group, and a thin (∼0.2 μm by 1–3 μm) curved endobiont was observed with TEM within the membrane-bound vesicles, we did not perform FISH analyses to confirm that the observed curved cell morphology is a member of that group. Members of this group are often decomposers of polysaccharides (Dworkin et al., 1999).
In sum, the endobionts of this ciliate likely benefit by obtaining a constant supply of nutrients via the host digestive processes. They also live in a protected environment, and are delivered by the ciliate to the appropriate redox gradient in the sediments for their cellular metabolisms. The benefits to the host are less clear, although the endobionts through their respective metabolisms likely provide the host with a wide range of conveniently internalized nutrients. The TEM images of the membrane-bound vesicles containing endobionts are consistent with speculations about their function: the highly organized arrangement of different endobiont morphologies within the vesicles was found consistently in this region of the cell, and was not reminiscent of food vacuoles. The prokaryote cells inside these vesicles appear morphologically intact, unlike those typical of food vacuoles. We cannot rule out the possibility however that under certain circumstances, the ciliate host may turn to these functional consortia of endobionts as a food source. This ciliate is host to a complex consortium of prokaryotic endobionts that may function to elegantly and intricately meld the sulfur cycle with cycling of carbon and nitrogen. We look forward to learning more about the metabolic interactions between these endobionts and to elucidating additional protist – prokaryote symbioses from such habitats. These avenues of investigation will expand our knowledge of the diversity of life on our planet and of the potential impacts of these microbial populations on major biogeochemical cycles.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the captain and crew of the RV Robert Gordon Sproul and Richard Sperduto who helped with sampling, Louis Kerr for TEM sectioning, Dagmar Woebkin for helpful discussions about CARD–FISH methods, Barbara MacGregor for help with deltaproteobacterial probes, Hilary Morrison and Rich Fox at MBL for the use of their pipeline script, David Patterson for assistance with initial ciliate identification and photo-documentation, and Margot Wilstermann and Marius Karolinski for performing counts of ciliates in our quantitative samples. We also thank Colleen Cavanaugh, Tom Fenchel, and Andreas Teske for helpful discussions about the endobionts. This research was supported by grants from NSF (MCB-0604084 to Virginia P. Edgcomb and Joan M. Bernhard and MCB-0702491 to Joan M. Bernhard, Virginia P. Edgcomb, and K. L. Casciotti).
Amman, R., Springer, N., Ludwig, W., Görtz, H.-D., and Schleifer, K.-H. (1991). Identification in situ and phylogeny of uncultured bacterial endosymbionts. Nature 351, 161–164.
Amman, R. I., Ludwig, W., and Schleifer, K.-H. (1995). Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59, 143–169.
Bagarinao, T. (1992). Sulfide as an environmental factor and toxicant – tolerance and adaptations in aquatic organisms. Aquat. Toxicol. 24, 21–62.
Bahr, M., Crump, B. C., Klepac-Ceraj, V., Teske, A., Sogin, M. L., and Hobbie, J. E. (2005). Molecular characterization of sulfate-reducing bacteria in a New England salt marsh. Environ. Microbiol. 7, 1175–1185.
Barbera, M. J., Ruiz-Trillo, I., Tufts, J. Y. A., Bery, A., Silberman, J. D., and Roger, A. J. (2010). Sawyeria marylandensis (Heterolobosea) has a hydrogenosome with novel metabolic properties. Eukaryot. Cell 9, 1913–1924.
Barry, J. P., Greene, H. G., Orange, D. L., Baxter, C. H., Robinson, B. H., Kochevar, R. E., Nybakken, J. W., Reed, D. L., and McHugh, C. M. (1996). Biologic and geologic characteristics of cold seeps in Monterey Bay, California. Deep Sea Res. 43, 1739–1762.
Beier, C. L., Horn, M., Michel, R., Schweikert, M., Görtz, H.-D., and Wagner, M. (2002). The genus Caedibacter comprises endosymbionts of Paramecium spp. related to the Rickettsiales (Alphaproteobacteria) and to Francisella tularensis (Gammaproteobacteria). Appl. Environ. Microbiol. 68, 6043–6050.
Bernhard, J. M., Buck, K. R., Farmer, M. A., and Bowser, S. S. (2000). The Santa Barbara Basin is a symbiosis oasis. Nature 403, 77–80.
Bernhard, J. M., Habura, A., and Bowser, S. S. (2006). An endobiont-bearing allogromiid from the Santa Barbara Basin: implications for the early diversitfication of foraminifera. J. Geophys. Res. 111, G03002.
Bernhard, J. M., and Sen Gupta, B. K. (1999). “Foraminifera of oxygen-depleted environments,” in Modern Foraminifera, ed. B. K. Sen Gupta (Dordrecht: Kluwer Academic Publishers), 201–216.
Bernhard, J. M., Visscher, P. T., and Bowser, S. S. (2003). Submillimeter life positions of bacteria, protists, and metazoans in laminated sediments of the Santa Barbara Basin. Limnol. Oceanogr. 45, 813–828.
Boetius, A., Ravenschlag, K., Shubert, C. J., Rickert, D., Widdel, F., Gieseke, A., Amann, R., Jørgensen, B.B., Witte, U., and Pfannkuche, O. (2000). Microscopic identification of a microbial consortium apparently mediating anaerobic methane oxidation above marine gas hydrates. Nature 407, 623–626.
Browenkow, W. W., and Cline, J. D. (1969). Colorimetric determination of dissolved oxygen at low concentrations. Limnol. Oceanogr. 14, 450–454.
Caldwell, S. L., Laidler, J. R., Brewer, E. A., Eberly, J. O., Sandborgh, S. C., and Colwell, F. S. (2008). Anaerobic oxidation of methane: mechanisms, bioenergetics, and the ecology of associated microorganisms. Environ. Sci. Technol. 42, 6791–6799.
Cavanaugh, C. M. (1994). Microbial symbiosis: patterns of diversity in the marine environment. Am. Zool. 34, 79–89.
Cavanaugh, C. M., Gardiner, S. L., Jones, M. L., Jannasch, H. W., and Waterbury, J. B. (1981). Prokaryotic cells in the hydrothermal vent tube worm Riftia pachyptila Jones: possible chemoautotrophic symbionts. Science 213, 340–341.
Cole, J. R., Chai, B., Marsh, T. L., Farris, R. J., Wang, Q., Kulam, S. A., Chandra, S., McGarrell, D. M., Schmidt, T. M., Garrity, G. M., and Tiedje, J. M. (2003). The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31, 442–443.
Daims, H., Ramsing, N. B., Schleifer, K.-H., and Wagner, M. (2001). Cultivation-independent, semiautomatic determination of absolute bacterial cell numbers in environmental samples by fluorescence in situ hybridization. Appl. Environ. Microbiol. 67, 5810–5818.
Dawson, S. C., and Pace, N. R. (2002). Novel kingdom-level eukaryotic diversity in anoxic environments. Proc. Natl. Acad. Sci. U.S.A. 99, 8324–8329.
DeLong, E. F. (1992). Archaea in coastal marine environments. Proc. Natl. Acad. Sci. U.S.A. 89, 5685–5689.
Distel, D. L., and Felbeck, H. (1988). Pathways of inorganic carbon fixation in the endosymbiont-bearing lucinid clam Lucinoma aequizonata. I. Purification and characterization of endosymbiotic bacteria. J. Exp. Zool. 247, 1–10.
Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., and Stackebrandt, E. (eds). (1999). The Prokaryotes: A Handbook on the Biology of Bacteria, 3rd Edn. Singapore: Springer.
Edgcomb, V., Breglia, S. A., Yubuki, N., Beaudoin, D., Patterson, D. J., Leander, B. S., and Bernhard, J. M. (2010). Identity of epibiotic bacteria on symbiontid euglenozoans in O2-depleted marine sediments: evidence for symbiont and host co-evolution. ISME J. 5, 1–13.
Elwood, H. J., Olsen, G. J., and Sogin, M. L. (1985). The small-subunit ribosomal RNA gene sequences from the hypotrichous ciliates Oxytricha nova and Stylonychia pustulata. Mol. Biol. Evol. 2, 399–410.
Embley, T. M., and Finlay, B. J. (1993). Systematic and morphological diversity of endosymbiotic methanogens in anaerobic ciliates. Antonie Van Leeuwenhoek 64, 261–271.
Embley, T. M., and Finlay, B. J. (1994). The use of small subunit rRNA sequences to unravel the relationships between anaerobic ciliates and their methanogen endosymbionts. Microbiology 140, 225–235.
Embley, T. M., Finlay, B. J., Dyal, P., Hirt, R. P., Wilkinson, M., and Williams, A. G. (1995). Multiple origins of anaerobic ciliates with hydrogenosomes within the radiation of aerobic ciliates. Proc. R. Soc. Lond. B Biol. Sci. 262, 87–93.
Embley, T. M., Finlay, B. J., Thomas, R. H., and Dyal, P. L. (1992). The use of rRNA sequences and fluorescent probes to investigate the phylogenetic positions of the anaerobic ciliate Metopus palaeformis and its archaeobacterial endosymbiont. J. Gen. Microiol. 138, 1479–1487.
Esteban, G., Guhl, B., Clarke, K., Embley, T., and Finlay, B. (1993). Cyclidium porcatum n. sp.: a free-living anaerobic scuticociliate containing a stable complex of hydrogenosomes, eubacteria and archaeobacteria. Eur. J. Protistol. 29, 262–270.
Ettwig, K. F., Butler, M. K., Le Paslier, D., Pelletier, E., Mangenot, S., Kuypers, M. M., Schreiber, F., Dutih, B. E., Zedelius, J., de Beer, D., Gloerich, J., Wessels, H. J., van Alen, T., Luesken, F., Wu, M. L., van de Pas-Schoonen, K. T., Op den Camp, H. J., Janssen-Megens, E. M., Francoijs, K. J., Stunnenberg, H., Weissenbach, J., Jetten, M. S., and Strous, M. (2010). Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464, 543–548.
Ewing, B., and Green, P. (1998). Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8, 186–194.
Ewing, B., Hillier, L., Wendl, M. C., and Green, P. (1998). Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8, 175–185.
Fenchel, T., and Bernard, C. (1993). Endosymbiotic purple non-sulphur bacteria in an anaerobic ciliated protozoon. FEMS Microbiol. Lett. 110, 21–25.
Fenchel, T., and Finlay, B. J. (1990). Anaerobic free-living protozoa: growth efficiencies and the structure of anaerobic communities. FEMS Microbiol. Ecol. 74, 269–276.
Fenchel, T., and Finlay, B. J. (1991a). The biology of free-living anaerobic ciliates. Eur. J. Protistol. 26, 201–215.
Fenchel, T., and Finlay, B. J. (1991b). Endosymbiotic methanogenic bacteria in anaerobic ciliates: significance for the growth efficiency of the host. J. Protozool. 38, 18–22.
Fenchel, T., and Finlay, B. J. (1995). Ecology and Evolution in Anoxic Worlds. Oxford: Oxford University Press.
Fenchel, T., Perry, T., and Thane, A. (1977). Anaerobiosis and symbiosis with bacteria in free-living ciliates. J. Protozool. 24, 154–163.
Finlay, B. J., and Fenchel, T. (1989). Hydrogenosomes in some anaerobic protozoa resemble mitochondria. FEMS Microbiol. Ecol. 65, 311–314.
Finlay, B. J., and Fenchel, T. (1992). Methanogens and other bacteria as symbionts of free-living anaerobic ciliates. Symbiosis 14, 375–390.
Foissner, W., and Al-Rasheid, K. A. S. (1999). Updating the Tracheleocercids (Ciliophora, Karyorelictea). VI. A detailed description of Sultanophrys arabica nov. gen., nov. spec. (Sultanophryidae nov. fam.). Eur. J. Protistol. 35, 146–160.
Foissner, W., and Dragesco, J. (1996). Updating the Trachelocercids (Ciliophora, Karyorelictea). I. A detailed description of the infraciliature of Trachelolophos giga N.G., N. sp. and T. filum (Dragesco and Dragesco-Kernelis, 1986) N. Comb. J. Eukaryot. Microbiol. 43, 12–25.
Foissner, W., and Xu, K. (2007). Monograph of the Spathidiida (Ciliophora, Haptoria). Vol. I: Protospathidiidae, Arcuospathidiidae, Apertospathulidae. Monogr. Biol. 81, 1–485.
Frias-Lopez, J., Thompson, A., Waldbauer, J., and Chisholm, S. W. (2009). Use of stable isotope-labelled cells to identify active grazers of picocyanobacteria in ocean surface waters. Environ. Microbiol. 11, 512–525.
Garrity, G. M., Brenner, D. J., Krieg, N. R., and Staley, J. R. (eds). (2005). Bergey’s Manual of Systematic Bacteriology, Vol. 2: The Proteobacteria. Baltimore, MD: Williams and Wilkins.
Görtz, H.-D. (2002). “Symbiotic associations between ciliates and prokaryotes,” in The Prokaryotes: an Evolving Electronic Resource for the Microbiological Community, 3rd Edn. Available at http://141.150.157.117:8080/prokPUB/chaprender/jsp/showchap.jsp?chapnum=355
Hackstein, J. H. P. (2011). Anaerobic ciliates and their methanogenic endosymbionts. Microbiol. Monogr. 19, 12–23.
Hinrichs, K. U., Hayes, J. M., Sylva, S. P., Brewster, P. G., and DeLong, E. F. (1999). Methane-consuming archaebacteria in marine sediments. Nature 398, 802–805.
Hirt, R. P., Dyal, P. L., Wilkinson, M., Finlay, B. J., Roberts, D. M., and Embley, T. M. (1995). Phylogenetic relationships among Karyorelictids and Heterotrichs inferred from small subunit rRNA sequences: resolution at the base of the ciliate tree. Mol. Phylogenet. Evol. 4, 77–87.
Jolivet, E., L’Haridon, S., Corre, E., Forterre, P., and Prieur, D. (2003). Thermococcus gammatolerans sp. nov., a hyperthermophilic archaeon from a deep-sea hydrothermal vent that resists ionizing radiation. Int. J. Syst. Evol. Microbiol. 53, 847–851.
Jones, W. J., Nagle, D. P., and Whitman, W. B. (1987). Methanogens and the diversity of archaebacteria. Microbiol. Rev. 51, 135–177.
Jumars, P. A., Penry, D. L., Baross, J. A., Perry, M. J., and Frost, B. W. (1989). Closing the the microbial loop: dissolved carbon pathway to heterotrophic bacteria from incomplete ingestion, digestion and absorption in animals. Deep Sea Res. 36, 483–495.
Klein, M., Friedrich, M., Roger, A. J., Fishbain, S., Hugenholtz, P., Abicht, A., Blackall, L. L., Stahl, D. A., and Wagner, M. (2001). Multiple lateral transfer events of dissimilatory sulfite-reductase genes between major lineages of bacteria. J. Bacteriol. 183, 6028–6034.
Kuwabara, J. S., van Geen, A., McCorkle, D. C., and Bernhard, J. M. (1999). Dissolved sulfide distributions in the water column and sediment pore waters of the Santa Barbara basin. Geochim. Cosmochim. Acta 63, 2199–2209.
Lane, D. J. (1991). “16S/23S rRNA sequencing,” in Nucleic Acid Techniques in Bacterial Systematics, eds E. Stackebrandt and M. Goodfellow (New York: Wiley), 115–148.
Lin, X., Wakeham, S. G., Putnam, I. F., Astor, Y. M., Scranton, M. I., Chistoserdov, A. Y., and Taylor, G. T. (2006). Comparison of vertical distributions of prokaryotic assemblages in the anoxic Cariaco Basin and Black Sea by use of fluorescence in situ hybridization. Appl. Environ. Microbiol. 72, 2679–2690.
Longnecker, K., and Reysenbach, A. L. (2001). Expansion of the geographic distribution of a novel lineage of epsilon-Proteobacteria to a hydrothermal vent site on the Southern East Pacific Rise. FEMS Microbiol. Ecol. 35, 287–293.
Loy, A., Kusel, K., Lehner, A., Drake, H. L., and Wagner, M. (2004). Microarray and functional gene analysis of sulfatereducing prokaryotes in low sulfate, acidic fens reveal co-occurrence of recognized genera and novel lineages. Appl. Environ. Microbiol. 70, 6998–7009.
Loy, A., Lehner, A., Lee, N., Adamczyk, J., Meier, H., Ernst, J., Schleifer, K. H., and Wagner, M. (2002). Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl. Environ. Microbiol. 68, 5064–5081.
Lucker, S., Steger, D., Kjeldsen, K. U., MacGregor, B. J., Wagner, M., and Loy, A. (2007). Improved 16S rRNA-targeted probe set for analysis of sulfate-reducing bacteria by fluorescence in situ hybridization. J. Microbiol. Methods 69, 523–528.
Ludwig, W., Strunk, O., Westram, R., Richter, L., Meier, H., Yadhukumar, Buchner, A., Lai, T., Steppi, S., Jobb, G., Förster, W., Brettske, I., Gerber, S., Ginhart, A. W., Gross, O., Grumann, S., Hermann, S., Jost, R., König, A., Liss, T., Lüssmann, R., May, M., Nonhoff, B., Reichel, B., Strehlow, R., Stamatakis, A., Stuckmann, N., Vilbig, A., Lenke, M., Ludwig, T., Bode, A., and Schleifer, K. H. (2004). ARB: a software environment for sequence data. Nucleic Acids Res. 32, 1363–1371.
Lynn, D. H. (2008). The Ciliated Protozoa: Characterization, Classification and Guide to the Literature. Dordrecht: Springer.
Madsen, E. L., Sinclair, J. L., and Ghiorse, W. C. (1991). In situ biodegradation: microbiological patterns in a contaminated aquifer. Science 252, 830–833.
Manz, W., Amann, R., Ludwig, W., Wagner, M., and Schleifer, K.-H. (1992). Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria – problems and solutions. Syst. Appl. Microbiol. 15, 539–600.
Manz, W., Eisenbrecher, M., Neu, T. R., and Szewzyk, U. (1998). Abundance and spatial organization of Gram-negative sulfate-reducing bacteria in activated sludge investigated by in situ probing with specific 16S rRNA targeted oligonucleotides. FEMS Microbiol. Ecol. 25, 43–61.
McHatton, S. C., Barry, J. P., Jannasch, H. W., and Nelson, D. C. (1996). High nitrate concentrations in vacuolate, autotrophic marine Beggiatoa spp. Appl. Environ. Microbiol. 62, 954–958.
Medlin, L., Elwood, H. J., Stickel, S., and Sogin, M. L. (1988). The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71, 491–499.
Miller, M. A., Holder, M. T., Vos, R., Midford, P. E., Liebowitz, T., Chan, L., Hoover, P., and Warnow, T. (2009). The CIPRES Portals. Available at: http://www.webcitation.org/5imQUeQa
Müller, M. (1988). Energy metabolism of protozoa without mitochondria. Annu. Rev. Microbiol. 42, 465–488.
Nakamura, K., Terada, T., Sekiguchi, Y., Shinzato, N., Meng, X.-Y., Enoki, M., and Kamagata, Y. (2006). Application of pseudomurein endoisopeptidase to fluorescence in situ hybridization of methanogens within the family Methanobacteriaceae. Appl. Environ. Microbiol. 72, 6907–6913.
Neef, A. (1997). Anwendung der in situ-Einzelzell-Identifizierung von Bakterien zur Populationsanalyse in komplexen mikrobiellen Biozonose. Munchen: TU.
Nowack, E. C., and Melkonin, M. (2010). Endosymbiotic associations within protists. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 699–712.
Orphan, V. J., Hinrichs, K. U., Ussler, I. W., Paull, C. K., Taylor, L. T., Sylva, S. P., Hayes, J. M., and Delong, E. F. (2001). Comparative analysis of methane-oxidizing archaea and sulfate-reducing bacteria in anoxic marine sediments. Appl. Environ. Microbiol. 67, 1922–1934.
Pernthaler, A., Pernthaler, J., and Amann, R. (2002). Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria, Appl. Environ. Microbiol. 68, 3094–3101.
Raikov, I. B. (1985). Primitive never-dividing macronuclei of some lower ciliates. Int. Rev. Cytol. 95, 267–325.
Reimers, C. E., Lange, C. B., Tabak, M. and Bernhard, J. M. (1990). Seasonal spillover and varve formation in the Santa Barbara Basin, California. Limnol. Oceanogr. 35, 1577–1585.
Reimers, C. E., Ruttenberg, K. C., Canfield, D. E., Christiansen, M. B., and Martin, J. B. (1996). Porewater pH and authigenic phases formed in the uppermost sediments of the Santa Barbara Basin. Geochim. Cosmochim. Acta 60, 4037–4057.
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574.
Sherr, E. B., and Sherr, B. F. (2002). Significance of predation by protists in aquatic microbial food webs. Antonie Van Leeuwenhoek 81, 293–308.
Springer, N., Ludwig, W., Amman, R., Schmidt, H. J., Görtz, H.-D., and Schleifer, K.-H. (1993). Occurrence of fragmented 16S rRNA in an obligate bacterial endosymbiont of Paramecium caudatum. Proc. Natl. Acad. Sci. U.S.A. 90, 9892–9895.
Stahl, D. A., and Amann, R. (eds). (1991). Development and Application of Nucleic Acid Probes in Bacterial Systematics. Chichester: Wiley & Sons Ltd.
Stamatakis, A., Hoover, P., and Rougemont, J. (2008). A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 57, 758–771.
Stoeck, T., Fowle, W. H., and Epstein, S. S. (2003). Methodology of protistan discovery: from rRNA detection to quality scanning electron microscope images. Appl. Environ. Microbiol. 69, 6856–6863.
Stolper, D. A., Revsbech, N. P., and Canfield, D. E. (2010). Aerobic growth at nanomolar oxygen concentrations. Proc. Natl. Acad. Sci. U.S.A. 107, 18755–18760.
Sun, H. Y., Noe, J., Barber, J., Coyne, R. S., Cassidy-Hanley, D., Clark, T. G., Findly, R. C., and Dickerson, H. W. (2009). Endosymbiotic bacteria in the parasitic ciliate Ichthyophthirius multifillis. Appl. Environ. Microbiol. 75, 7445–7452.
Taylor, G. T. (1982). The role of pelagic heterotrophic protozoa in nutrient cycling: a review. Ann. Inst. Oceanogr. (Suppl.), Paris 58, 227–241.
Van Bruggen, J. J. A., Stumm, C. K., and Vogels, G. D. (1983). Symbiosis of methanogenic bacteria and sapropelic protozoa. Arch. Microbiol. 136, 89–96.
van Hoek, A. H., van Alen, T. A., Sprakel, V. S., Leunissen, J. A., Brigge, T., Vogels, G. D., and Hackstein, J. H. (2000). Multiple acquisition of methanogenic archaeal symbionts by anaerobic ciliates. Mol. Biol. Evol. 17, 251–258.
Vannini, C., Rosati, G., Verni, F., and Petroni, G. (2004). Identification of the bacterial endosymbionts of the marine ciliate Euplotes magnicirratus (Ciliophora, Hypotrichia) and proposal of ‘Candidatus Dovosia euplotis’. Int. J. Syst. Evol. Microbiol. 54, 1151–1156.
Wagner, M., Roger, A. J., Flax, J. L., Brusseau, G. A., and Stahl, D. A. (1998). Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate reduction. J. Bacteriol. 180, 2975–2982.
Wallner, G., Amman, R. I., and Beisker, W. (1993). Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14, 136–143.
Widdel, F., and Bak, F. (eds). (1992). Gram-Negative Mesophilic Sulfate-Reducing Bacteria. New York, NY: Springer-Verlag.
Keywords: ciliate, anoxia, symbiosis, TEM, SSU rRNA, FISH
Citation: Edgcomb VP, Leadbetter ER, Bourland W, Beaudoin D and Bernhard JM (2011) Structured multiple endosymbiosis of bacteria and archaea in a ciliate from marine sulfidic sediments: a survival mechanism in low oxygen, sulfidic sediments? Front. Microbio. 2:55. doi: 10.3389/fmicb.2011.00055
Received: 27 January 2011;
Paper pending published: 14 February 2011;
Accepted: 10 March 2011;
Published online: 25 March 2011
Edited by:
Martin G. Klotz, University of Louisville, USAReviewed by:
Thorsten Stoeck, University of Kaiserslautern, GermanyCopyright: © 2011 Edgcomb, Leadbetter, Bourland, Beaudoin and Bernhard. This is an open-access article subject to an exclusive license agreement between the authors and Frontiers Media SA, which permits unrestricted use, distribution, and reproduction in any medium, provided the original authors and source are credited.
*Correspondence: Virginia P. Edgcomb, Geology and Geophysics Department, Woods Hole Oceanographic Institution, Woods Hole, MA 02543-1050, USA. e-mail:dmVkZ2NvbWJAd2hvaS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.