- Tularemia Pathogenesis Section, Laboratory of Intracellular Parasites, Rocky Mountain Laboratories, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Hamilton, MT, USA
The tularemia-causing bacterium Francisella tularensis is a facultative intracellular organism with a complex intracellular lifecycle that ensures its survival and proliferation in a variety of mammalian cell types, including professional phagocytes. Because this cycle is essential to Francisella pathogenesis and virulence, much research has focused on deciphering the mechanisms of its intracellular survival and replication and characterizing both bacterial and host determinants of the bacterium’s intracellular cycle. Studies of various strains and host cell models have led to the consensual paradigm of Francisella as a cytosolic pathogen, but also to some controversy about its intracellular cycle. In this review, we will detail major findings that have advanced our knowledge of Francisella intracellular survival strategies and also attempt to reconcile discrepancies that exist in our molecular understanding of the Francisella–phagocyte interactions.
Introduction
Many pathogenic microorganisms are characterized by an intracellular lifestyle and their ability to enter, survive, proliferate, and/or persist within cells of the infected host, therefore avoiding recognition and elimination by extracellular immune surveillance mechanisms. To achieve this, these pathogens have evolved sophisticated strategies to circumvent intracellular recognition and microbicidal processes, ultimately ensuring their survival and proliferation within the host. The Gram-negative bacterium Francisella tularensis is the causative agent of tularemia, a vector-borne zoonosis of the Northern Hemisphere that can affect humans and cause severe morbidity and mortality if untreated or misdiagnosed. Human tularemia is a fulminant disease that can be contracted by exposure to as few as 10 bacteria, the pneumonic form of which is the most severe (Oyston et al., 2004). Three subspecies of F. tularensis, F. tularensis subsp. tularensis (Type A), F. tularensis subsp. holarctica (Type B), and F. tularensis subsp. mediasiatica are recognized, among which strains from subspecies tularensis and holarctica can cause tularemia in humans (Oyston et al., 2004). Additionally, F. novicida, an avirulent Francisella species in humans that retains high virulence in rodents, has been used extensively as a surrogate model for F. tularensis due to lower biocontainment requirements (Baron and Nano, 1998; Lauriano et al., 2004; Santic et al., 2005a,b, 2007, 2008; Brotcke et al., 2006; de Bruin et al., 2007; Mohapatra et al., 2007a,b, 2008, 2010; Weiss et al., 2007; Brotcke and Monack, 2008; Barker et al., 2009a,b; Schmerk et al., 2009a,b; Al-Khodor and Abu Kwaik, 2010; Asare and Abu Kwaik, 2010; Asare et al., 2010). Essential to the development of tularemia is the bacterium’s ability to infect and proliferate within mononuclear phagocytes, such as macrophages and dendritic cells, although this bacterium can also infect polymorphonuclear neutrophils, hepatocytes, epithelial, and endothelial cells (Oyston et al., 2004).
Because of the importance of its intracellular lifestyle, the interactions of F. tularensis with host cells have been studied thoroughly, with a particular emphasis on those with macrophages. Using a variety of Francisella strains, such as F. novicida, the attenuated holarctica vaccine strain LVS, or virulent strains such as SchuS4 or clinical tularensis isolates, and host cell models, including murine or human primary macrophages and macrophage-like cell lines, several laboratories have defined the intracellular cycle of Francisella (Figures 1 and 2). Following phagocytic uptake, Francisella initially resides within a phagosome called the Francisella-containing phagosome (FCP) that interacts with early and late compartments of the endocytic pathway (Clemens et al., 2004; Santic et al., 2005a; Checroun et al., 2006; Chong et al., 2008), prior to phagosomal membrane disruption that allows bacterial release in the macrophage cytosol (Golovliov et al., 2003; Clemens et al., 2004; Santic et al., 2005a; Checroun et al., 2006; Chong et al., 2008; Wehrly et al., 2009). Once cytosolic, bacteria undergo extensive replication that culminates in the apoptotic and pyroptotic deaths of the infected cells (Lai et al., 2001; Lai and Sjostedt, 2003; Mariathasan et al., 2005; Santic et al., 2010) and/or reentry into the endocytic compartments in Francisella-containing vacuoles (FCV) in murine primary macrophages via an autophagy-mediated process (Checroun et al., 2006; Wehrly et al., 2009). Our current knowledge of the Francisella intracellular cycle indicates that this bacterium can be considered a cytosolic pathogen, yet cycles through a variety of intracellular compartments with distinct environments to ensure survival, proliferation, and eventual release. For this purpose, it has likely evolved a battery of mechanisms to subvert various host cell processes. In this review, we will focus on Francisella–macrophage interactions and discuss recent findings about both bacterial and host factors that contribute to the intracellular pathogenesis of this bacterium.
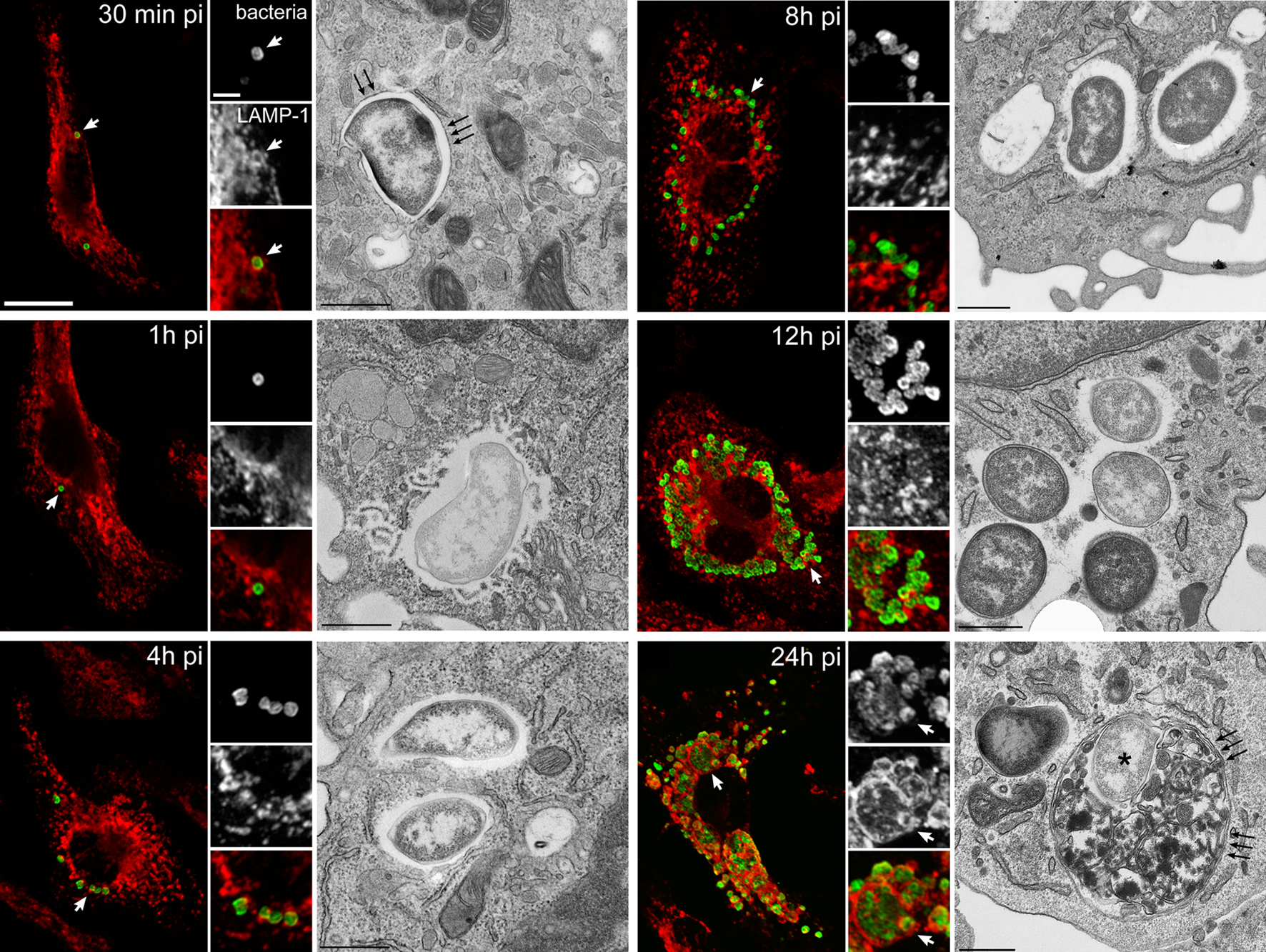
Figure 1. Infection cycle of F. tularensis Schu S4 within C57BL/6J murine bone marrow-derived macrophages (BMMs). BMMs were infected with Schu S4 and processed at various times post infection (pi) for immunofluorescence or transmission electron microscopy. Representative confocal and electron (TEM) micrographs of intracellular SchuS4 at 30 min, 1, 4, 8, 12, and 24 h pi, showing the early phagosomal stage, phagosomal disruption, bacterial release in the cytosol, cytosolic replication, and FCV formation. Bacteria appear in green and endosomal, LAMP-1-positive membranes appear in red. White arrowheads indicate either regions of interest in whole images or bacteria enclosed within a LAMP-1-positive compartment in insets. Black arrows in TEM micrographs indicate single or double membranes surrounding intracellular bacteria. Scale bars, 10 and 2 μm (confocal images) or 0.5 μm (TEM images). Reproduced with permission from John Wiley and Sons Publishing.
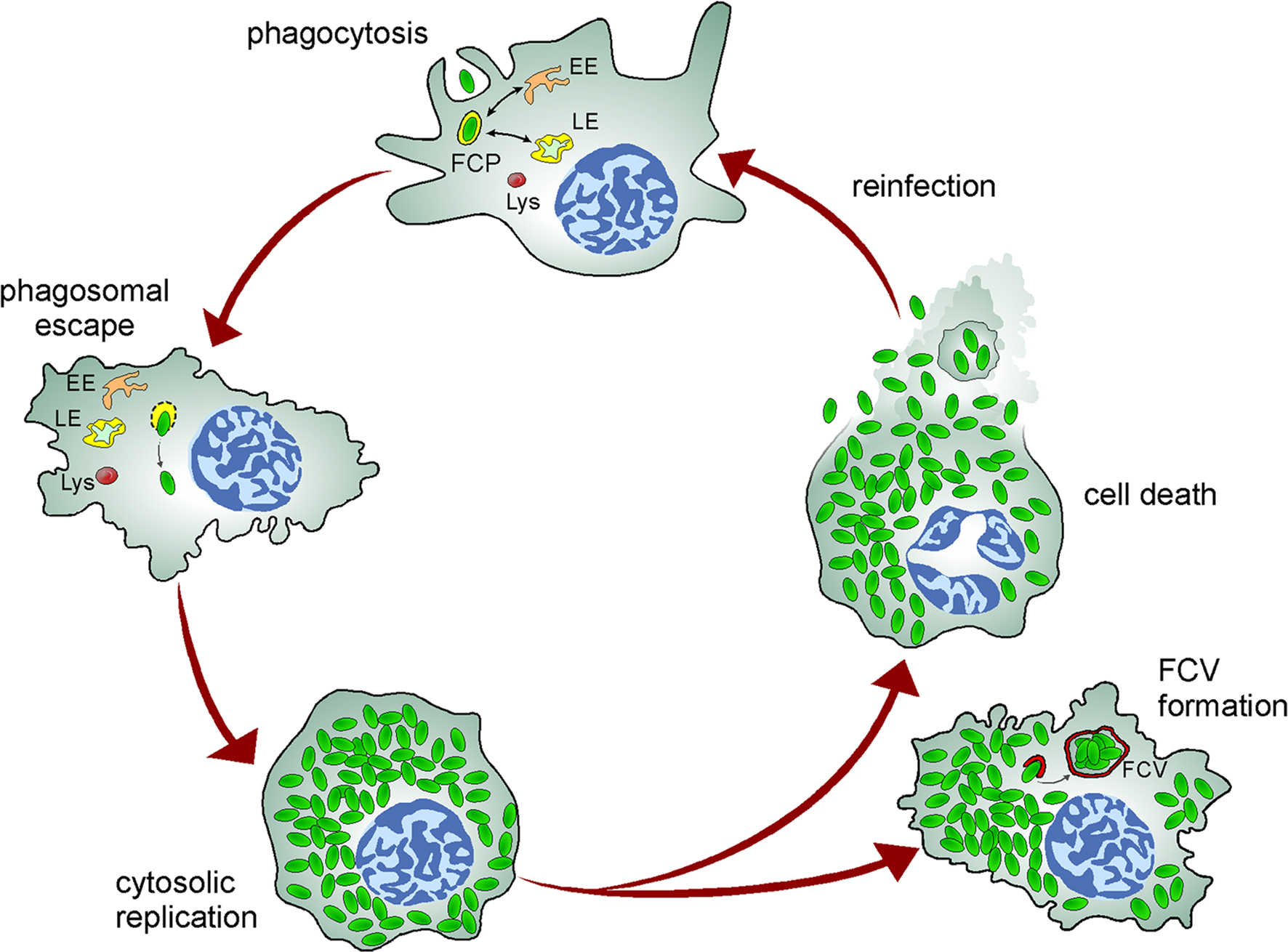
Figure 2. Model of the Francisella intracellular cycle in macrophages. Upon phagocytosis, bacteria reside in an early phagosome (FCP) that interacts with early (EE) and late (LE) endocytic compartments but not lysosomes (Lys). Bacteria rapidly disrupt the FCP membrane and reach the cytosol where they undergo extensive replication, a process followed by cell death and bacterial release or reentry of cytosolic bacteria within Francisella-containing vacuoles (FCV) via autophagy in murine macrophages.
Entry into Mammalian Cells
An essential step in the lifestyle of intracellular pathogens is binding and entry into host cells where they can undergo survival and proliferation. Such interactions require engagement of receptors that triggers specific signaling cascades, phagosome maturation events, and host microbicidal responses devoted to the destruction of the microorganism (Underhill and Ozinsky, 2002). Thus, the mode of uptake likely impacts the intracellular fate of a given pathogen.
The underlying mechanisms that are important for the phagocytosis of Francisella by macrophages are beginning to be defined. Uptake of Francisella into macrophages is markedly enhanced by serum opsonization (Clemens et al., 2004, 2005; Balagopal et al., 2006; Schulert and Allen, 2006). In an ultrastructural analysis of the entry process of a clinical F. tularensis subspecies tularensis isolate and the attenuated Francisella live vaccine strain (LVS) into human monocyte derived macrophages (MDMs) or THP-1 cells, Clemens et al. (2005)observed bacteria engulfed within asymmetric pseudopod loops, a process that was dependent on filamentous actin, serum with intact complement factor C3, and complement receptors. Several studies have also described the contribution of complement and the complement receptor CR3 in the phagocytosis of serum-opsonized F. novicida and LVS by human and murine macrophages, as well as human neutrophils and dendritic cells (Balagopal et al., 2006; Ben Nasr et al., 2006; Pierini, 2006; Schulert and Allen, 2006; Barker et al., 2009a). Fcγ receptors, scavenger receptor class A (SR-A), nucleolin, and the lung surfactant protein A (SP-A) are also involved in the uptake of serum-opsonized F. novicida and LVS into human and murine macrophages (Balagopal et al., 2006; Pierini, 2006; Barel et al., 2008), yet the predominant entry pathway appears to be mediated by complement receptor CR3 (Table 2). Although it remains to be determined whether serum opsonization of Francisella is important during pathogenesis in vivo, CR3-mediated phagocytosis may contribute to the bacterium’s intracellular survival, since this route of uptake does not induce the macrophage oxidative burst (Underhill and Ozinsky, 2002) and could therefore limit antimicrobial activities by the infected cells. Compared to opsonized bacteria, phagocytosis of non-opsonized F. novicida and LVS by human MDMs is mediated, in part, by the mannose receptor (MR; Balagopal et al., 2006; Schulert and Allen, 2006). The contribution of this receptor was further confirmed by the enhanced entry of LVS into J774A.1 macrophage-like cells expressing the MR and human MDMs with upregulated expression of the MR (Schulert and Allen, 2006).
A variety of pathogens target lipid rafts for internalization into host cells to avoid intracellular degradation (Zaas et al., 2005). Francisella appears to exploit this portal of entry, since cholesterol and caveolin-1, two components of lipid rafts, are incorporated into the newly formed FCP and cholesterol-rich lipid domains are required for uptake of LVS into the murine macrophage-like cell lines J774A.1 and RAW264.7 (Tamilselvam and Daefler, 2008). Francisella may target lipid rafts via the engagement of receptors that preferentially reside within these membrane signaling platforms, such as CR3 and Fcγ receptors (Peyron et al., 2000; Beekman et al., 2008). Consistently, depletion of lipid raft-associated GPI-anchored proteins abolished LVS entry and intracellular survival, suggesting a requirement for signaling events mediated through these proteins. Intracellular signals that accompany phagocytosis of Francisella involve the tyrosine kinase Syk and the MAP kinase ERK pathway (Parsa et al., 2008). Syk was activated upon infection of RAW264.7 cells with non-opsonized F. novicida (Parsa et al., 2008). Inhibition of Syk blocked uptake of F. novicida whereas overexpression enhanced uptake, demonstrating the involvement of this kinase in promoting bacterial entry (Parsa et al., 2008). Further analysis revealed that Syk regulates the activation of ERK and PI3K/Akt pathways. Inhibition of ERK, but not the PI3K/Akt pathway, decreased phagocytosis of F. novicida by macrophages (Parsa et al., 2008). Thus, the ERK pathway appears to be important for the phagocytosis of Francisella, but neither the precise mechanism of ERK activation nor the phagocytic pathways initiating this signaling have been determined.
Francisella Intracellular Survival Strategies
Regardless of their replication niche, most intracellular pathogens initiate their intracellular cycle within the membrane-enclosed environment of the nascent phagosome. Newly formed phagosomes typically undergo a maturation process involving sequential interactions with the endosomal–lysosomal network that progressively modify their composition into a degradative compartment (Haas, 2007). To survive the destructive nature of a mature phagolysosome, intracellular pathogens must either evade or withstand this hostile environment (Haas, 2007). Although Francisella disrupts its phagosome to proliferate in the macrophage cytosol, the bacterium is likely outfitted to combat the microbicidal arsenal of this compartment during its interim phagosomal stage.
Intracellular Survival: Inhibition of Nadph Oxidase Activity
Among the microbicidal agents encountered by newly ingested pathogens are the reactive oxygen species (ROS) generated by NADPH oxidase complexes assembled at the nascent phagosomal membrane. NADPH oxidase assembly involves phosphorylation of cytosolic p47phox, which is complexed with p67phox and p40phox, and the entire heterotrimer translocates to cell membrane sites where the gp91phox and p22phox subunits accumulate. Participation of a small G-protein, either Rac1 or Rac2, is also required for NADPH oxidase function (Babior, 2004). Francisella avoids this early killing mechanism by blocking NADPH oxidase activation on the newly formed FCP in human neutrophils, as well as human and murine macrophages (McCaffrey and Allen, 2006; Buchan et al., 2009; Schulert et al., 2009; McCaffrey et al., 2010; Mohapatra et al., 2010). Whereas virulent strains of the tularensis (strain Schu S4) and holarctica (strain 1547-57) subspecies and the attenuated holarctica LVS strain exclude gp91phox/gp22phox heterodimers from FCPs and limit p47phox phosphorylation (McCaffrey and Allen, 2006; McCaffrey et al., 2010), F. novicida appears to mainly depend upon dephosphorylation of Phox components to disrupt NADPH oxidase function (Mohapatra et al., 2010). Furthermore, SchuS4 and the virulent holarctica strain FSC200 are more resistant to killing by ROS in vitro than the avirulent LVS, via mechanisms independent of the catalase KatG (Lindgren et al., 2007). These data highlight the subtle differences in intracellular survival mechanisms used by different Francisella species. Additionally, Francisella impairs ROS production by subsequent heterologous stimulation (McCaffrey and Allen, 2006; Edwards et al., 2010; McCaffrey et al., 2010; Mohapatra et al., 2010). McCaffrey et al. (2010)determined that LVS does so by retaining dysfunctional NADPH oxidase complexes on the phagosomal membranes. Altogether, these studies clearly document that Francisella affects pre- and post-NADPH oxidase assembly mechanisms to inhibit the phagocyte respiratory burst (McCaffrey et al., 2010), a process that likely contributes to its survival within the FCP.
More controversial, species-specific findings have been made on the role of other Francisella proteins in inhibiting antimicrobial processes. An early study showed that the acid phosphatase AcpA, purified from an attenuated F. tularensis type A strain (ATCC6223), inhibits the oxidative burst of activated porcine neutrophils (Reilly et al., 1996). Consistent with this report, Mohapatra et al. (2010) found that loss of AcpA function in F. novicida resulted in ROS production and impaired intracellular survival in human neutrophils and MDMs. Moreover, infection with a quadruple acid phosphatase deletion mutant (ΔacpAΔacpBΔacpCΔhap) triggered even higher levels of oxidant generation and increased bacterial killing than infection with a ΔacpA mutant, illustrating the cumulative contribution of acid phosphatases to evasion of oxidative killing (Mohapatra et al., 2010). Contrary to the observations regarding AcpA in F. novicida, studies in SchuS4 revealed that AcpA activity is dispensable for inhibition of the phagocyte oxidative defense, indicating that observations made using F. novicida do not always apply to F. tularensis subspecies strains (McCaffrey et al., 2010). Other bacterial factors have also been implicated in impairing oxidase activation. Pyrimidine biosynthesis genes (carA, carB, and pyrB) were identified in LVS by a random transposon mutagenesis screen for factors affecting neutrophil NADPH oxidase activation (Schulert et al., 2009). These uracil auxotroph mutants triggered an oxidative burst in neutrophils and were killed in phagosomes containing superoxide (Schulert et al., 2009). Additionally, these mutant strains were unable to block subsequent neutrophil activation by heterologous stimuli; however, the effects on neutrophil activation may be due indirectly to the diminished fitness of these mutants (Schulert et al., 2009). Interestingly, these auxotrophic mutants did not elicit a respiratory burst in human monocytes or MDMs but were, nonetheless, unable to survive within these phagocytes (Schulert et al., 2009). Together, these data suggest that Francisella evades oxidative killing by different mechanisms in monocytes and macrophages versus neutrophils: in the former, by utilizing uptake receptors (e.g., CR3) that do not trigger an oxidative burst, and in the latter, by actively inhibiting NADPH oxidase assembly and activation (Schulert et al., 2009). Two Francisella virulence gene regulators, fevR and migR, were also shown to be required by LVS for regulating NADPH oxidase activity in neutrophils (Buchan et al., 2009; McCaffrey et al., 2010), suggesting they regulate expression of specific effector proteins. Studies in SchuS4 confirmed that fevR is required to block NADPH oxidase activity, but two proteins encoded by the Francisella pathogenicity island (FPI) affecting phagosomal escape, iglI and iglJ (see below), are not. These observations demonstrate that distinct genes within the fevR regulon are required for inhibiting oxidative burst and disrupting the FCP (McCaffrey et al., 2010). Altogether, these data indicate that Francisella applies a multi-factorial, multi-pronged strategy to circumvent phagocyte oxidative defenses.
Intracellular Survival: Bacterial Escape from the Francisella-Containing Phagosome
Although an early study suggested that Francisella inhabit atypical phagosomes within rodent macrophages (Anthony et al., 1991), recent findings by several laboratories have established that various Francisella strains escape from their initial phagosome into the cytosol of human and murine primary macrophages (Golovliov et al., 2003; Clemens et al., 2004; Santic et al., 2005a; Checroun et al., 2006), yet with conflicting kinetics since phagosomal escape has been shown to occur within one to several hours post infection (pi). In several ultrastructural studies, bacteria were observed in intact phagosomes until 2–4 h pi, from which point the phagosome membrane was gradually degraded and the bacteria eventually became cytosolic (Golovliov et al., 2003; Clemens et al., 2004; Lindgren et al., 2004; Santic et al., 2005a; McCaffrey and Allen, 2006; Schulert et al., 2009). Other studies, using ultrastructural analysis and phagosome integrity assays based upon the delivery of anti-Francisella antibodies to the cytosol of infected cells, have reported more rapid escape, where the majority of bacteria are cytosolic by 1 h pi (Checroun et al., 2006; Santic et al., 2007, 2008; Chong et al., 2008; Wehrly et al., 2009). While such variations in phagosomal escape kinetics may be a reflection of strain differences, as illustrated by side-by-side comparison of phagosomal escape kinetics of various Francisella strains (Chong et al., 2008), technical differences and differential sensitivity in assays and criteria used to assess phagosomal disruption may have also led to these different conclusions (Clemens et al., 2004, 2009; Santic et al., 2005a; Checroun et al., 2006; Chong et al., 2008). In particular, different opsonic and non-opsonic infection models used by various laboratories may have contributed to these discrepancies in the kinetics of Francisella phagosomal escape. A systematic analysis of how the mode of uptake affects phagosomal escape should address this question.
Prior to phagosomal escape, Francisella initially resides within a phagosome that interacts with early and late endocytic compartments (Clemens et al., 2004; Checroun et al., 2006; Chong et al., 2008; Santic et al., 2008; Wehrly et al., 2009). While the kinetics of acquisition of endosomal markers differ depending on the infection models used, confocal and electron microscopy studies have shown that nascent phagosomes containing F. tularensis, LVS, or F. novicida transiently acquire the early endosomal antigen 1 (EEA1) followed by acquisition of late endosomal markers CD63, LAMP1, LAMP2, and the Rab7 GTPase, the eventual loss of which correlates with phagosomal disruption (Golovliov et al., 2003; Clemens et al., 2004; Santic et al., 2005a, 2008; Checroun et al., 2006; Bonquist et al., 2008; Chong et al., 2008). These early trafficking events resemble those of a normal maturation process, although the FCP does not mature into a phagolysosome as it fails to acquire the acid hydrolase cathepsin D or lysosomal tracers (Anthony et al., 1991; Clemens et al., 2004; Santic et al., 2005a; Bonquist et al., 2008). Whether this results from Francisella actively inhibiting fusion with terminal lysosomes or from the loss of phagosomal integrity during bacterial escape still remains debatable. Santic et al. (2005b) have proposed that the process of phagosomal escape prevents fusion with lysosomes, since a phagosomal escape-deficient, F. novicida mutant in the FPI gene iglC (see below) is delivered to a Cathepsin D-positive, lysosomal compartment. However, Bonquist et al. (2008) have observed limited fusion of vacuoles containing various phagosomal escape-deficient mutants of LVS, including mutants in iglC and iglD, arguing that some inhibition of fusion with terminal lysosomes may still occur despite a lack of phagosomal escape. It is possible that phagosomes containing mutants deficient in phagosomal escape still undergo some residual phagosomal membrane alteration that, although not disruptive enough to ensure bacterial release, affects FCP maturation and lysosomal fusion. While these discrepancies could be due to differential phagosomal escape defects between the mutant strains examined, mutants in the same genes (iglC, iglD) were used in these studies, making these findings difficult to reconcile at this stage.
Phagosomal acidification, a process dependent upon the activity of the vacuolar ATPase (vATPase) pump, is critical for the fission and fusion of membranes that promote maturation and for the optimal activity of a number of microbicidal enzymes; thus, acidification is both a requirement and a consequence of phagosomal maturation (Huynh and Grinstein, 2007). The extent of FCP acidification and whether acidification influences Francisella phagosomal escape are contentious topics. In some studies FCPs harboring LVS, SchuS4, or a clinical isolate of F. tularensis subspecies tularensis in human macrophages and J774A.1 cells resisted acidification and acquired only limited amounts of the vATPase pump, suggesting that Francisella impairs phagosome maturation (Clemens et al., 2004, 2009; Bonquist et al., 2008). Moreover, the efficiency of Francisella phagosomal escape was unaffected in the presence of a vATPase pump inhibitor (Clemens et al., 2009). Other studies in human and murine macrophages reported that FCPs harboring F. novicida or SchuS4 became acidified and acquired the vATPase pump (Chong et al., 2008; Santic et al., 2008) prior to phagosomal disruption. Inhibition of phagosome acidification significantly delayed, but did not block, phagosomal disruption, indicating that FCP maturation is important for optimal bacterial escape to the cytosol (Chong et al., 2008; Santic et al., 2008). These studies suggest that the FCP acidic pH may provide conditions that induce and/or activate factors that promote the phagosomal disruption process, some of which may be related to pH-dependent, phagosomal iron availability (Fortier et al., 1995), although this needs to be formally demonstrated. Interestingly, studies reporting a lack of FCP acidification were performed using serum-opsonized bacteria, while those reporting transient FCP acidification were performed using non-opsonic conditions, regardless of the bacterial strains used, suggesting that the mode of uptake may affect FCP maturation.
Determinants of Intracellular Survival – the Francisella Pathogenicity Island
Phagosomal escape is an essential aspect of the intracellular survival strategy of Francisella, since it is a process conserved among all Francisella strains studied to date and is a pre-requisite for cytosolic replication. Mutants impaired in phagosomal escape are defective for intracellular growth (Lindgren et al., 2004; Santic et al., 2005b; Bonquist et al., 2008; Schmerk et al., 2009a), and delaying translocation to the cytosol impedes the onset of replication (Chong et al., 2008). Thus, considerable effort has focused on determining the factors that contribute to phagosomal disruption. Several genes encoded within the FPI, an approximately 30 kb locus of 17 genes with similarity to components of the recently described type VI secretion system of Vibrio cholera and Pseudomonas aeruginosa (Mougous et al., 2006; Pukatzki et al., 2006), are involved in the phagosomal escape and intracellular survival of Francisella (Nano et al., 2004) (Table 1). Two identical copies of the FPI are present in the genomes of F. tularensis subsp. tularensis and subsp. holarctica and a single copy is present in F. novicida (Nano et al., 2004), although whether FPI copy number is a factor in the virulence differences between Francisella species remains unclear. The FPI-encoded gene iglC encodes a 23-kDa hypothetical protein that is prominently induced during the intracellular growth of LVS in J774A.1 cells (Golovliov et al., 1997) and SchuS4 in primary bone marrow-derived macrophages (BMMs; Wehrly et al., 2009). Disruption of iglC in LVS and F. novicida impairs phagosomal escape and intracellular growth (Lindgren et al., 2004; Santic et al., 2005b; Bonquist et al., 2008; Chong et al., 2008). In LVS, this defect was partially reversed by in trans complementation. In F. novicida, phagosomal escape was restored only with the expression of both IglC and IglD in the iglC mutant due to the polar effect of the mutation on IglD expression (Chong et al., 2008). Therefore, observations drawn from the use of this F. novicida mutant regarding the role of IglC (Santic et al., 2005b) need to be re-evaluated in the absence of genetic complementation. In addition, disruption of IglCD functions significantly reduced, but did not completely block, FCP disruption, indicating the contribution of additional factors in the phagosomal escape of Francisella (Chong et al., 2008). In J774A.1 cells, the iglD mutant of LVS demonstrated an increased and persistent propensity for colocalization with the late endosomal marker LAMP-1 which was reversed by complementation with iglD, indicating that IglD function is required for escape from the FCP (Bonquist et al., 2008). Contradictory to this observation, Santic et al. (2007) reported that the iglD mutant of F. novicida rapidly escaped into the cytosol but was unable to replicate, suggesting that IglD does not play a role in phagosomal disruption but is required for cytosolic proliferation. The distinct iglD mutant trafficking phenotypes reported may be a result of methodological and strain differences. It is however difficult to envision how highly conserved genes within a major virulence locus such as the FPI can have different functions between Francisella species. Additional mutagenesis studies in F. novicida have identified other FPI proteins required for intracellular survival: PdpA (Schmerk et al., 2009a), VgrG (Barker et al., 2009b), and IglI (Barker et al., 2009b) are required for phagosomal escape and intramacrophage growth; iglA (de Bruin et al., 2007), iglB (Gray et al., 2002), pdpB (Brotcke et al., 2006; Tempel et al., 2006), and cds2 (Brotcke et al., 2006) are also required for intramacrophage growth, although their contributions to intracellular trafficking have not been examined. Despite the lack of information regarding the precise functions of the FPI genes, these data highlight the importance of the FPI-encoded proteins in the intracellular pathogenesis of Francisella.
Other Bacterial Determinants of Intracellular Survival
While the FPI is to date the most prominent virulence locus required for phagosomal escape, intracellular survival and growth of Francisella, other proteins encoded outside the FPI have also been implicated in the proper translocation of Francisella into the cytosol and subsequent replication. Intracellular trafficking and survival defects of mutants in the transcriptional regulators mglA (Baron and Nano, 1998; Santic et al., 2005b; Bonquist et al., 2008), fevR (Brotcke and Monack, 2008; Buchan et al., 2009; Wehrly et al., 2009), and migR (Buchan et al., 2009) have been documented. These are likely a result of the dysregulation of effector proteins that contribute to phagosomal escape and cytosolic proliferation, rather than a direct participation, since these transcription factors regulate virulence genes both within and outside of the FPI (Brotcke et al., 2006; Brotcke and Monack, 2008). Other loci, such as FTT1103 (Qin et al., 2009) and FTT1676 (Wehrly et al., 2009), both encoding hypothetical lipoproteins, are required for the phagosomal escape of SchuS4 in J774A.1 cells and murine BMMs, respectively. Three genes involved in pyrimidine biosynthesis, carA, carB, and pyrB, were identified in LVS to also play a role in phagosome disruption in human MDMs (Schulert et al., 2009), although their mechanistic contribution to phagosomal escape is elusive and may be indirect.
The contribution of acid phosphatases to the intracellular pathogenesis of Francisella has been a subject of much investigation and remains controversial. Francisella spp. contain at least six acid phosphatase-encoding genes, the conservation of which varies depending on the Francisella species (Child et al., 2010). While the most conserved genes are acpA, acpB, and acpC, others have been disrupted through genome rearrangements in virulent Type A and Type B strains, raising the question of their importance for Francisella virulence (Child et al., 2010). An acpA truncation mutant in F. novicida, lacking phosphatase activity but retaining phospholipase activity, was found to exhibit wild-type level replication in J774A.1 cells (Baron et al., 1999), indicating it is not essential to intracellular growth. Citing differences in mutant constructs, Mohapatra et al. (2007a) found that deletion of the entire acpA gene in F. novicida resulted in bacteria that were impaired for phagosomal escape and intracellular growth in human MDM and THP-1 macrophages. These defects were more pronounced with the accumulation of acp deletions (acpA, acpB, acpC, and hapA) indicating a cumulative role for these proteins (Mohapatra et al., 2008). In F. tularensis subspecies tularensis, however, neither the deletion of acpA nor the combined deletion of acpA, acpB, and acpC had an effect on the phagosomal escape or cytosolic proliferation in murine BMMs or human MDMs (Child et al., 2010). These contradictory findings between F. novicida and F. tularensis may illustrate species-specific differences. Yet, one would expect that proteins involved in an essential virulence process such as phagosomal escape to be highly conserved and not subjected to evolutionary genome reduction. While these functional differences require future clarification, they emphasize again that observations made from less virulent strains do not necessarily reflect those from virulent F. tularensis subspecies.
Our current knowledge of bacterial factors required for phagosomal escape indicates that it is a multi-factorial process. Consistently, a recent genetic screen performed in human macrophages using a F. novicida transposon mutant library identified 91 genes, encoding a variety of functions, as required for phagosomal escape (Asare and Abu Kwaik, 2010). Despite progress in identifying the factors involved in this crucial stage of Francisella trafficking, the actual mechanism of phagosome membrane disruption remains elusive and effector proteins of phagosomal disruption have yet to be characterized at the functional level.
Intracellular Replication of Francisella
Besides the importance of phagosomal escape, another critical aspect of Francisella intracellular pathogenesis is its ability to proliferate extensively within the cytosol of host cells (Oyston et al., 2004). Because phagosomal escape is pre-requisite to cytosolic proliferation, the identification of factors specifically required for cytosolic proliferation using mutagenesis approaches has been challenging. Since phagosomal escape-deficient mutants are also affected in intracellular growth, assigning a specific role in cytosolic replication to a gene requires characterization of the intracellular trafficking of the corresponding mutant. A vast number of genes affecting the intracellular survival and proliferation of Francisella has indeed been identified through mutagenesis studies (Brotcke et al., 2006; Qin and Mann, 2006; Tempel et al., 2006; Su et al., 2007; Weiss et al., 2007; Alkhuder et al., 2009; Schulert et al., 2009; Wehrly et al., 2009; Asare and Abu Kwaik, 2010). Yet, few have been confirmed by targeted mutagenesis, and even fewer have been characterized and specifically implicated in cytosolic replication. Alternative approaches, such as transcriptional profiling of intracellular Francisella, have also revealed genetic determinants of intracellular survival based on their upregulation during the intracellular cycle (Wehrly et al., 2009). What has emerged from this wealth of data suggests that Francisella uses specific mechanisms to successfully adapt to its intracellular lifestyle, because many candidate virulence determinants encode hypothetical proteins of unknown function. Highlighted here are the determinants for which a contribution to intracellular replication has been confirmed via targeted mutagenesis, or specifically demonstrated.
Bacterial factors implicated in the intramacrophage growth of Francisella that have been confirmed by targeted mutagenesis studies include pmrA (Mohapatra et al., 2007b; Sammons-Jackson et al., 2008), FTT0989 (Brotcke et al., 2006), htpG (Tempel et al., 2006; Weiss et al., 2007), dsbB (Maier et al., 2006; Tempel et al., 2006; Weiss et al., 2007; Qin et al., 2008), iglD (Santic et al., 2007), iglA (de Bruin et al., 2007), iglB (Gray et al., 2002), pdpB (Brotcke et al., 2006; Tempel et al., 2006), cds2 (Brotcke et al., 2006), and mglB (Baron and Nano, 1998)(Table 1). While the role of FPI genes, such as iglD, iglA, iglB, pdpB, and cds2, has been discussed above with regard to phagosomal escape, the specific roles in cytosolic replication of the transcriptional regulators PmrA (Mohapatra et al., 2007b; Sammons-Jackson et al., 2008) and MglB (Baron and Nano, 1998), the stress response protein HtpG (Tempel et al., 2006; Weiss et al., 2007), the inner membrane disulfide bond forming protein DsbB (Maier et al., 2006; Tempel et al., 2006; Weiss et al., 2007; Qin et al., 2008) and the putative transglutaminase FTT0989 (Brotcke et al., 2006) remain speculative until intracellular trafficking studies of the mutants are performed. The roles of these genes in intracellular growth may indeed be an indirect or non-specific consequence of their contribution to other aspects of the bacterium’s intracellular adaptation.
More extensive studies have nonetheless identified genes required for cytosolic proliferation of Francisella. Characterization of purine auxotroph mutants (ΔpurMCD) in LVS and SchuS4 revealed no defects in phagosomal escape, but bacteria failed to replicate in the cytosol of J774A.1 or primary murine macrophages (Pechous et al., 2006, 2008), indicating that Francisella cannot scavenge nucleotides from the macrophage and must synthesize them. Another metabolism-related gene, ggt, encoding a γ-glutamyl transpeptidase, was identified using an in vitro negative screen of a library of LVS transposon mutants (Alkhuder et al., 2009). Disruption of ggt in LVS impaired cytosolic replication in murine J774A.1, RAW264.7, BMMs, and human THP-1 macrophages. Although the efficiency of escape was not thoroughly investigated, ggt mutant bacteria were observed in the cytoplasm of infected RAW264.7 cells at 24 h pi by ultrastructural analysis, suggesting that mutant bacteria were able to breach the FCP. Further characterization showed that ggt is required for bacterial acquisition of cysteine from glutathione (the most abundant source of this amino acid in the cytosol) and γ-glutamyl peptides to promote intracellular growth in murine macrophages (Alkhuder et al., 2009). Consistently, genes involved in the catabolism of amino acids and encoding small peptide transporters are upregulated intracellularly in SchuS4, further indicating nutrient requirements and source for cytosolic Francisella (Wehrly et al., 2009). The contribution of ripA, which encodes an inner membrane protein, to the intracellular growth of LVS was identified through random mutagenesis and confirmed by characterization of a targeted deletion mutant (Fuller et al., 2008). ΔripA mutants escaped from their phagosomes with similar efficiency to wild-type LVS, but were unable to replicate in murine lung epithelial cells, TC-1, or J774A.1 cells (Fuller et al., 2008). The expression of ripA was upregulated during the early cytosolic replication stage during infection of J774A.1 macrophages, corroborating its role at this stage of the Francisella intracellular cycle (Fuller et al., 2009). Lacking homology to any previously characterized genes, the function of this Francisella specific factor remains to be uncovered. The FTT0369c locus, encoding a hypothetical protein with Sel1-family tetratricopeptide (TPR) repeat domains, was identified as transcriptionally upregulated during the cytosolic stage of SchuS4 infection of murine BMMs (Wehrly et al., 2009). Upon deletion of FTT0369c, SchuS4 bacteria were not able to replicate in the cytosol of murine BMMs, but retained phagosomal escape kinetics similar to wild-type SchuS4 (Wehrly et al., 2009), indicating a specific role of this locus in cytosolic proliferation.
As accentuated by the short list of determinants discussed above, characterization of the exact functions of bacterial factors specifically required for cytosolic proliferation has only just begun.
Host Factors Affecting Francisella Intracellular Survival
Understanding Francisella–macrophage interactions not only requires the characterization of the bacterial determinants but also that of host factors that either contribute to, or interfere with, the bacterium’s intracellular cycle (Table 2). Despite its ability to survive and proficiently replicate within quiescent macrophages, Francisella is unable to do so within interferon (IFN)-γ-activated macrophages (Anthony et al., 1992; Fortier et al., 1992; Polsinelli et al., 1994), indicating that host innate immune responses are capable of antagonizing Francisella intracellular proliferation. However, the mechanisms induced by this cytokine to control infection by Francisella are unclear. Several studies have ascribed the killing of LVS in IFN-γ-activated murine peritoneal exudates cells (PECs) to the actions of the inducible nitric oxide (NO) synthase (iNOS), which generates reactive nitrogen species (RNS; Anthony et al., 1992; Fortier et al., 1992; Lindgren et al., 2005, 2007). In IFN-γ-activated murine alveolar macrophages, NO was determined to play a minimal role in inhibiting the intracellular growth of LVS (Polsinelli et al., 1994). Addition of superoxide dismutase, catalase, excess iron or L-tryptophan did not reverse the growth inhibition of LVS observed in IFN-γ-activated alveolar macrophages, indicating that the antibacterial activity was not mediated by ROS or limiting concentrations of L-tryptophan or iron (Polsinelli et al., 1994). Similar to LVS, control of SchuS4 replication in IFN-γ-activated human and murine macrophages did not involve ROS, L-tryptophan sequestration or iron limitation, nor did it seem to involve autophagy (Edwards et al., 2010). In studies using SchuS4, the restriction on bacterial growth was reported to be iNOS-dependent in murine PECs (Lindgren et al., 2007), but was iNOS-independent in primary human and murine macrophages (Edwards et al., 2010).
In examining the interactions of LVS with IFN-γ-activated PECs, Lindgren et al. (2004) found that IFN-γ treatment reduced, but did not completely block, LVS escape into the macrophage cytosol. Phagosomal escape of LVS in IFN-γ-activated J774A.1 cells was however not affected, but its intracellular growth remained restricted (Bonquist et al., 2008). Consistent with these findings, IFN-γ treatment did not hinder phagosomal escape of SchuS4 in murine BMMs and human MDMs (Edwards et al., 2010). In contrast, Santic et al. (2005a) found that F. novicida is unable to escape from the FCP and is killed within phagolysosomes in IFN-γ-activated human MDMs. Altogether, these results indicate that inhibition of intracellular growth by IFN-γ is exerted at the cytosolic stage of the F. tularensis intracellular cycle, whereas growth restriction is determined during the phagosomal stage for F. novicida. Thus, the IFN-γ-induced mechanisms inhibiting Francisella intracellular replication seem to be dependent on macrophage type, as well as the infecting strain, although one cannot exclude that variations in the experimental designs of these studies have yielded contrasting conclusions. Regardless, the actual effector mechanisms induced by IFN-γ remain elusive.
Alternative activation of macrophages by mast cells has been shown to effectively control the intramacrophage growth of LVS (Ketavarapu et al., 2008). Both contact-dependent and secreted factors from mast cells contribute to this inhibitory effect since entry and replication of LVS within murine BMMs were diminished when co-cultured with infected murine bone marrow-derived mast cells or in the presence of infected mast cell-spent media. Among the factors secreted by mast cells, IL-4 was determined to be an important component mediating bacterial growth inhibition in BMMs. An earlier report demonstrated that IL-4 mediated upregulation of MR enhanced Francisella uptake by macrophages (Schulert and Allen, 2006); this mechanism of entry may ultimately be detrimental to the intracellular survival of Francisella as Rodriguez et al. (2010) found that restriction of intramacrophage replication by IL-4 was associated with increased ATP production, upregulated MR expression and prolonged FCP acidification, but how IL-4 affects Francisella intracellular fate remains to be directly demonstrated. Taken together, however, these data reflect the idea that mode of entry affects the intracellular fate of the invading pathogen.
Interactions of Francisella with the Autophagic Pathway
The cytosolic location of Francisella during its intracellular proliferation makes this pathogen a target for innate immune recognition and delivery to lysosomes via the autophagic pathway, a process known to recover and eliminate various cytosolic pathogens (Deretic, 2006). Yet, surprisingly, this bacterium can proliferate within the cytosol without any evidence for an autophagic response, suggesting it is either capable of preventing its recognition by the autophagic machinery or of inhibiting the autophagic process. Nonetheless, it was shown that a subset of cytosolic bacteria reentered the endocytic compartment in an autophagy-dependent process following replication in murine BMMs (Checroun et al., 2006), arguing for a belated autophagic response from the macrophage. While it remains unclear whether FCVs play a role in the intracellular cycle of Francisella or if they are a macrophage response to contain bacterial proliferation, their discovery indicated an involvement of autophagy in Francisella–macrophage interactions. Interestingly, FCV formation does not occur in human MDMs (Akimana et al., 2010; Edwards et al., 2010). While this suggests that their formation may require murine-specific signaling, one cannot exclude that higher and earlier Francisella-induced cytotoxicity observed in human MDMs may precede, hence prevent, FCV formation (Edwards et al., 2010). Additional evidence that autophagy may be manipulated by Francisella is that several autophagy-related genes, including beclin1, ATG5, ATG12, ATG16L, ATG7, and ATG4a, were down regulated during infection of human monocytes with SchuS4 or F. novicida, suggesting that Francisella suppresses the host autophagy response at the gene expression level (Butchar et al., 2008; Cremer et al., 2009). It was therefore proposed that by delaying autophagy, Francisella gains time in the macrophage cytosol to allow for replication and/or development of resistance to the acidic environment of the FCV (Cremer et al., 2009). However, whether autophagy is still functional in infected macrophages was not tested in these studies, so it remains to be demonstrated whether Francisella can actually inhibit the autophagic pathway during its intracellular cycle.
Signaling Pathways in Francisella Intracellular Pathogenesis
While innate immune processes that control Francisella intracellular growth have been extensively studied, additional host factors that influence the intracellular fate of Francisella have recently come to light from studies using F. novicida as a model system (Table 2). The PI3-Kinase/Akt signaling pathway is activated early during F. novicida infection (Parsa et al., 2006; Rajaram et al., 2009). Although this pathway was irrelevant for the phagocytosis of F. novicida (Parsa et al., 2008), continuous activation of Akt or deletion of the inositol phosphatase SHIP, which down-regulates the PI3K/Akt pathway, inhibited phagosomal disruption, and increased FCP fusion with lysosomes in murine BMMs and RAW264.7 cells (Rajaram et al., 2009), suggesting that activation of this pathway is restrictive to Francisella. Indeed, numerous gene members of the PI3K/Akt pathway were substantially downregulated in SchuS4 and F. novicida-infected human monocytes (Butchar et al., 2008), suggesting that Francisella may down modulate the PI3-K/Akt pathway.
Another signaling pathway initiated by F. novicida involves the small G-protein Ras. Intracellular F. novicida transiently activated Ras within 5 min of infection of human embryonic kidney cell line HEK293T through a signaling cascade involving SOS2, Grb2, PKCα, and PKCβ1 (Al-Khodor and Abu Kwaik, 2010). Knockdown of these components eliminated Ras activation, and impaired F. novicida intracellular replication without affecting the efficiency of phagosomal escape (Al-Khodor and Abu Kwaik, 2010). Furthermore, silencing of these signaling molecules had no impact on the phosphorylation of Akt or ERK, indicating that Ras activation does not trigger signaling through these pathways (Al-Khodor and Abu Kwaik, 2010). A recent genome-wide RNAi screen was used to identify 186 host factors that influence the intracellular replication of F. novicida in Drosophila melanogaster-derived S2R+ cells (Akimana et al., 2010). Among these, three were confirmed by knockdown of their corresponding mammalian homologues to impact the intracellular survival of F. novicida. Knockdown of the ubiquitin ligase CDC27 diminished the capacity of F. novicida to disrupt its phagosome and to replicate in the host cytosol, while silencing of the type III PI4-kinase α subunit PI4KC and the ubiquitin specific peptidase USP22 resulted in a cytosolic replication specific defect in HEK293T cells (Akimana et al., 2010). These data indicate that F. novicida exploits multiple host signaling pathways, as well as the host ubiquitin system, to ensure its intracellular survival. Future work should examine whether these signaling pathways and host machineries are also relevant to the intracellular pathogenesis of virulent Francisella species.
Concluding Remarks
Because the ability of Francisella spp. to survive and proliferate within mammalian cells is essential to their virulence, much research has focused over the last few years on understanding the intracellular cycle of these pathogens. While there is a consensus on the overall survival strategy of this pathogen, much controversy remains on more specific aspects of its intracellular cycle, such as the maturation of the FCP and the bacterial factors of phagosomal escape. Additionally, recent genomic and functional evidence indicates that caution should be exerted when assuming that conclusions drawn from studies using species of low virulence apply to virulent species of Francisella. A plethora of genetic screens for bacterial and host factors required for intracellular pathogenesis of various Francisella species has generated a wealth of information and identified key determinants of the Francisella–host cell interaction. Despite all these efforts, the molecular mechanisms governing the various stages of the Francisella intracellular cycle remain elusive. Future research on Francisella intracellular pathogenesis must now focus on characterizing the molecular functions of the bacterial and host factors identified, in order to gain a much-needed understanding of this bacterium’s pathogenesis at the molecular level.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We wish to thank Leigh Knodler for critical reading of the manuscript and Anita Mora for assistance with visual arts. This work was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Allergy, and Infectious Diseases.
References
Akimana, C., Al-Khodor, S., and Abu Kwaik, Y. (2010). Host factors required for modulation of phagosome biogenesis and proliferation of Francisella tularensis within the cytosol. PLoS ONE 5, e11025. doi: 10.1371/journal.pone.0011025.
Al-Khodor, S., and Abu Kwaik, Y. (2010). Triggering Ras signalling by intracellular Francisella tularensis through recruitment of PKCalpha and betaI to the SOS2/GrB2 complex is essential for bacterial proliferation in the cytosol. Cell Microbiol. 12, 1604–1621.
Alkhuder, K., Meibom, K. L., Dubail, I., Dupuis, M., and Charbit, A. (2009). Glutathione provides a source of cysteine essential for intracellular multiplication of Francisella tularensis. PLoS Pathog. 5, e1000284. doi: 10.1371/journal.ppat.1000284.
Anthony, L. D., Burke, R. D., and Nano, F. E. (1991). Growth of Francisella spp. in rodent macrophages. Infect. Immun. 59, 3291–3296.
Anthony, L. S., Morrissey, P. J., and Nano, F. E. (1992). Growth inhibition of Francisella tularensis live vaccine strain by IFN-gamma-activated macrophages is mediated by reactive nitrogen intermediates derived from L-arginine metabolism. J. Immunol. 148, 1829–1834.
Asare, R., and Abu Kwaik, Y. (2010). Molecular complexity orchestrates modulation of phagosome biogenesis and escape to the cytosol of macrophages by Francisella tularensis. Environ. Microbiol. 12, 2559–2586.
Asare, R., Akimana, C., Jones, S., and Abu Kwaik, Y. (2010). Molecular bases of proliferation of Francisella tularensis in arthropod vectors. Environ. Microbiol. 12, 2587–2612.
Balagopal, A., MacFarlane, A. S., Mohapatra, N., Soni, S., Gunn, J. S., and Schlesinger, L. S. (2006). Characterization of the receptor-ligand pathways important for entry and survival of Francisella tularensis in human macrophages. Infect. Immun. 74, 5114–5125.
Barel, M., Hovanessian, A. G., Meibom, K., Briand, J. P., Dupuis, M., and Charbit, A. (2008). A novel receptor – ligand pathway for entry of Francisella tularensis in monocyte-like THP-1 cells: interaction between surface nucleolin and bacterial elongation factor Tu. BMC Microbiol. 8, 145. doi:10.1186/1471-2180-8-145.
Barker, J. H., McCaffrey, R. L., Baman, N. K., Allen, L. A., Weiss, J. P., and Nauseef, W. M. (2009a). The role of complement opsonization in interactions between F. tularensis subsp. novicida and human neutrophils. Microbes Infect. 11, 762–769.
Barker, J. R., Chong, A., Wehrly, T. D., Yu, J. J., Rodriguez, S. A., Liu, J., Celli, J., Arulanandam, B. P., and Klose, K. E. (2009b). The Francisella tularensis pathogenicity island encodes a secretion system that is required for phagosome escape and virulence. Mol. Microbiol. 74, 1459–1470.
Baron, G. S., and Nano, F. E. (1998). MglA and MglB are required for the intramacrophage growth of Francisella novicida. Mol. Microbiol. 29, 247–259.
Baron, G. S., Reilly, T. J., and Nano, F. E. (1999). The respiratory burst-inhibiting acid phosphatase AcpA is not essential for the intramacrophage growth or virulence of Francisella novicida. FEMS Microbiol. Lett. 176, 85–90.
Beekman, J. M., van der Linden, J. A., van de Winkel, J. G., and Leusen, J. H. (2008). FcgammaRI (CD64) resides constitutively in lipid rafts. Immunol. Lett. 116, 149–155.
Bell, B. L., Mohapatra, N. P., and Gunn, J. S. (2010). Regulation of virulence gene transcripts by the Francisella novicida orphan response regulator PmrA: role of phosphorylation and evidence of MglA/SspA interaction. Infect. Immun. 78, 2189–2198.
Ben Nasr, A., Haithcoat, J., Masterson, J. E., Gunn, J. S., Eaves-Pyles, T., and Klimpel, G. R. (2006). Critical role for serum opsonins and complement receptors CR3 (CD11b/CD18) and CR4 (CD11c/CD18) in phagocytosis of Francisella tularensis by human dendritic cells (DC): uptake of Francisella leads to activation of immature DC and intracellular survival of the bacteria. J. Leukoc. Biol. 80, 774–786.
Bonquist, L., Lindgren, H., Golovliov, I., Guina, T., and Sjostedt, A. (2008). MglA and Igl proteins contribute to the modulation of Francisella tularensis live vaccine strain-containing phagosomes in murine macrophages. Infect. Immun. 76, 3502–3510.
Brotcke, A., and Monack, D. M. (2008). Identification of fevR, a novel regulator of virulence gene expression in Francisella novicida. Infect. Immun. 76, 3473–3480.
Brotcke, A., Weiss, D. S., Kim, C. C., Chain, P., Malfatti, S., Garcia, E., and Monack, D. M. (2006). Identification of MglA-regulated genes reveals novel virulence factors in Francisella tularensis. Infect. Immun. 74, 6642–6655.
Buchan, B. W., McCaffrey, R. L., Lindemann, S. R., Allen, L. A., and Jones, B. D. (2009). Identification of migR, a regulatory element of the Francisella tularensis live vaccine strain iglABCD virulence operon required for normal replication and trafficking in macrophages. Infect. Immun. 77, 2517–2529.
Butchar, J. P., Cremer, T. J., Clay, C. D., Gavrilin, M. A., Wewers, M. D., Marsh, C. B., Schlesinger, L. S., and Tridandapani, S. (2008). Microarray analysis of human monocytes infected with Francisella tularensis identifies new targets of host response subversion. PLoS ONE 3, e2924. doi: 10.1371/journal.pone.0002924.
Checroun, C., Wehrly, T. D., Fischer, E. R., Hayes, S. F., and Celli, J. (2006). Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc. Natl. Acad. Sci. U.S.A. 103, 14578–14583.
Child, R., Wehrly, T. D., Rockx-Brouwer, D., Dorward, D. W., and Celli, J. (2010). Acid phosphatases do not contribute to the pathogenesis of type A Francisella tularensis. Infect. Immun. 78, 59–67.
Chong, A., Wehrly, T. D., Nair, V., Fischer, E. R., Barker, J. R., Klose, K. E., and Celli, J. (2008). The early phagosomal stage of Francisella tularensis determines optimal phagosomal escape and Francisella pathogenicity island protein expression. Infect. Immun. 76, 5488–5499.
Clemens, D. L., and Horwitz, M. A. (2007). Uptake and intracellular fate of Francisella tularensis in human macrophages. Ann. N. Y. Acad. Sci. 1105, 160–186.
Clemens, D. L., Lee, B. Y., and Horwitz, M. A. (2004). Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect. Immun. 72, 3204–3217.
Clemens, D. L., Lee, B. Y., and Horwitz, M. A. (2005). Francisella tularensis enters macrophages via a novel process involving pseudopod loops. Infect. Immun. 73, 5892–5902.
Clemens, D. L., Lee, B. Y., and Horwitz, M. A. (2009). Francisella tularensis phagosomal escape does not require acidification of the phagosome. Infect. Immun. 77, 1757–1773.
Cremer, T. J., Amer, A., Tridandapani, S., and Butchar, J. P. (2009). Francisella tularensis regulates autophagy-related host cell signaling pathways. Autophagy 5, 125–128.
de Bruin, O. M., Ludu, J. S., and Nano, F. E. (2007). The Francisella pathogenicity island protein IglA localizes to the bacterial cytoplasm and is needed for intracellular growth. BMC Microbiol. 7, 1. doi: 10.1186/1471-2180-7-1.
Edwards, J. A., Rockx-Brouwer, D., Nair, V., and Celli, J. (2010). Restricted cytosolic growth of Francisella tularensis subsp. tularensis by IFN-gamma activation of macrophages. Microbiology 156, 327–339.
Fortier, A. H., Leiby, D. A., Narayanan, R. B., Asafoadjei, E., Crawford, R. M., Nacy, C. A., and Meltzer, M. S. (1995). Growth of Francisella tularensis LVS in macrophages: the acidic intracellular compartment provides essential iron required for growth. Infect. Immun. 63, 1478–1483.
Fortier, A. H., Polsinelli, T., Green, S. J., and Nacy, C. A. (1992). Activation of macrophages for destruction of Francisella tularensis: identification of cytokines, effector cells, and effector molecules. Infect. Immun. 60, 817–825.
Fuller, J. R., Craven, R. R., Hall, J. D., Kijek, T. M., Taft-Benz, S., and Kawula, T. H. (2008). RipA, a cytoplasmic membrane protein conserved among Francisella species, is required for intracellular survival. Infect. Immun. 76, 4934–4943.
Fuller, J. R., Kijek, T. M., Taft-Benz, S., and Kawula, T. H. (2009). Environmental and intracellular regulation of Francisella tularensis ripA. BMC Microbiol. 9, 216. doi: 10.1186/1471-2180-9-216.
Golovliov, I., Baranov, V., Krocova, Z., Kovarova, H., and Sjostedt, A. (2003). An attenuated strain of the facultative intracellular bacterium Francisella tularensis can escape the phagosome of monocytic cells. Infect. Immun. 71, 5940–5950.
Golovliov, I., Ericsson, M., Sandstrom, G., Tarnvik, A., and Sjostedt, A. (1997). Identification of proteins of Francisella tularensis induced during growth in macrophages and cloning of the gene encoding a prominently induced 23-kilodalton protein. Infect. Immun. 65, 2183–2189.
Gray, C. G., Cowley, S. C., Cheung, K. K., and Nano, F. E. (2002). The identification of five genetic loci of Francisella novicida associated with intracellular growth. FEMS Microbiol. Lett. 215, 53–56.
Huynh, K. K., and Grinstein, S. (2007). Regulation of vacuolar pH and its modulation by some microbial species. Microbiol. Mol. Biol. Rev. 71, 452–462.
Ketavarapu, J. M., Rodriguez, A. R., Yu, J. J., Cong, Y., Murthy, A. K., Forsthuber, T. G., Guentzel, M. N., Klose, K. E., Berton, M. T., and Arulanandam, B. P. (2008). Mast cells inhibit intramacrophage Francisella tularensis replication via contact and secreted products including IL-4. Proc. Natl. Acad. Sci. U.S.A. 105, 9313–9318.
Lai, X. H., Golovliov, I., and Sjostedt, A. (2001). Francisella tularensis induces cytopathogenicity and apoptosis in murine macrophages via a mechanism that requires intracellular bacterial multiplication. Infect. Immun. 69, 4691–4694.
Lai, X. H., and Sjostedt, A. (2003). Delineation of the molecular mechanisms of Francisella tularensis-induced apoptosis in murine macrophages. Infect. Immun. 71, 4642–4646.
Lauriano, C. M., Barker, J. R., Yoon, S. S., Nano, F. E., Arulanandam, B. P., Hassett, D. J., and Klose, K. E. (2004). MglA regulates transcription of virulence factors necessary for Francisella tularensis intraamoebae and intramacrophage survival. Proc. Natl. Acad. Sci. U.S.A. 101, 4246–4249.
Lindgren, H., Golovliov, I., Baranov, V., Ernst, R. K., Telepnev, M., and Sjostedt, A. (2004). Factors affecting the escape of Francisella tularensis from the phagolysosome. J. Med. Microbiol. 53, 953–958.
Lindgren, H., Shen, H., Zingmark, C., Golovliov, I., Conlan, W., and Sjostedt, A. (2007). Resistance of Francisella tularensis strains against reactive nitrogen and oxygen species with special reference to the role of KatG. Infect. Immun. 75, 1303–1309.
Lindgren, H., Stenman, L., Tarnvik, A., and Sjostedt, A. (2005). The contribution of reactive nitrogen and oxygen species to the killing of Francisella tularensis LVS by murine macrophages. Microbes Infect. 7, 467–475.
Maier, T. M., Pechous, R., Casey, M., Zahrt, T. C., and Frank, D. W. (2006). In vivo Himar1-based transposon mutagenesis of Francisella tularensis. Appl. Environ. Microbiol. 72, 1878–1885.
Mariathasan, S., Weiss, D. S., Dixit, V. M., and Monack, D. M. (2005). Innate immunity against Francisella tularensis is dependent on the ASC/caspase-1 axis. J. Exp. Med. 202, 1043–1049.
McCaffrey, R. L., and Allen, L. A. (2006). Francisella tularensis LVS evades killing by human neutrophils via inhibition of the respiratory burst and phagosome escape. J. Leukoc. Biol. 80, 1224–1230.
McCaffrey, R. L., Schwartz, J. T., Lindemann, S. R., Moreland, J. G., Buchan, B. W., Jones, B. D., and Allen, L. A. (2010). Multiple mechanisms of NADPH oxidase inhibition by type A and type B Francisella tularensis. J. Leukoc. Biol. 88, 791–805.
Mohapatra, N. P., Balagopal, A., Soni, S., Schlesinger, L. S., and Gunn, J. S. (2007a). AcpA is a Francisella acid phosphatase that affects intramacrophage survival and virulence. Infect. Immun. 75, 390–396.
Mohapatra, N. P., Soni, S., Bell, B. L., Warren, R., Ernst, R. K., Muszynski, A., Carlson, R. W., and Gunn, J. S. (2007b). Identification of an orphan response regulator required for the virulence of Francisella spp. and transcription of pathogenicity island genes. Infect. Immun. 75, 3305–3314.
Mohapatra, N. P., Soni, S., Rajaram, M. V., Dang, P. M., Reilly, T. J., El-Benna, J., Clay, C. D., Schlesinger, L. S., and Gunn, J. S. (2010). Francisella acid phosphatases inactivate the NADPH oxidase in human phagocytes. J. Immunol. 184, 5141–5150.
Mohapatra, N. P., Soni, S., Reilly, T. J., Liu, J., Klose, K. E., and Gunn, J. S. (2008). Combined deletion of four Francisella novicida acid phosphatases attenuates virulence and macrophage vacuolar escape. Infect. Immun. 76, 3690–3699.
Mougous, J. D., Cuff, M. E., Raunser, S., Shen, A., Zhou, M., Gifford, C. A., Goodman, A. L., Joachimiak, G., Ordonez, C. L., Lory, S., Walz, T., Joachimiak, A., and Mekalanos, J. J. (2006). A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312, 1526–1530.
Nano, F. E., Zhang, N., Cowley, S. C., Klose, K. E., Cheung, K. K., Roberts, M. J., Ludu, J. S., Letendre, G. W., Meierovics, A. I., Stephens, G., and Elkins, K. L. (2004). A Francisella tularensis pathogenicity island required for intramacrophage growth. J. Bacteriol. 186, 6430–6436.
Oyston, P. C., Sjostedt, A., and Titball, R. W. (2004). Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat. Rev. Microbiol. 2, 967–978.
Parsa, K. V., Butchar, J. P., Rajaram, M. V., Cremer, T. J., and Tridandapani, S. (2008). The tyrosine kinase Syk promotes phagocytosis of Francisella through the activation of Erk. Mol. Immunol. 45, 3012–3021.
Parsa, K. V., Ganesan, L. P., Rajaram, M. V., Gavrilin, M. A., Balagopal, A., Mohapatra, N. P., Wewers, M. D., Schlesinger, L. S., Gunn, J. S., and Tridandapani, S. (2006). Macrophage pro-inflammatory response to Francisella novicida infection is regulated by SHIP. PLoS Pathog. 2, e71. doi: 10.1371/journal.ppat.0020071.
Pechous, R., Celli, J., Penoske, R., Hayes, S. F., Frank, D. W., and Zahrt, T. C. (2006). Construction and characterization of an attenuated purine auxotroph in a Francisella tularensis live vaccine strain. Infect. Immun. 74, 4452–4461.
Pechous, R. D., McCarthy, T. R., Mohapatra, N. P., Soni, S., Penoske, R. M., Salzman, N. H., Frank, D. W., Gunn, J. S., and Zahrt, T. C. (2008). A Francisella tularensis Schu S4 purine auxotroph is highly attenuated in mice but offers limited protection against homologous intranasal challenge. PLoS ONE 3, e2487. doi: 10.1371/journal.pone.0002487.
Peyron, P., Bordier, C., N’Diaye, E. N., and Maridonneau-Parini, I. (2000). Nonopsonic phagocytosis of Mycobacterium kansasii by human neutrophils depends on cholesterol and is mediated by CR3 associated with glycosylphosphatidylinositol-anchored proteins. J. Immunol. 165, 5186–5191.
Pierini, L. M. (2006). Uptake of serum-opsonized Francisella tularensis by macrophages can be mediated by class A scavenger receptors. Cell Microbiol. 8, 1361–1370.
Polsinelli, T., Meltzer, M. S., and Fortier, A. H. (1994). Nitric oxide-independent killing of Francisella tularensis by IFN-gamma-stimulated murine alveolar macrophages. J. Immunol. 153, 1238–1245.
Pukatzki, S., Ma, A. T., Sturtevant, D., Krastins, B., Sarracino, D., Nelson, W. C., Heidelberg, J. F., and Mekalanos, J. J. (2006). Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U.S.A. 103, 1528–1533.
Qin, A., and Mann, B. J. (2006). Identification of transposon insertion mutants of Francisella tularensis tularensis strain Schu S4 deficient in intracellular replication in the hepatic cell line HepG2. BMC Microbiol. 6, 69. doi: 10.1186/1471-2180-6-69.
Qin, A., Scott, D. W., and Mann, B. J. (2008). Francisella tularensis subsp. tularensis Schu S4 disulfide bond formation protein B, but not an RND type efflux pump, is required for virulence. Infect. Immun. 76, 3086–3092.
Qin, A., Scott, D. W., Thompson, J. A., and Mann, B. J. (2009). Identification of an essential Francisella tularensis subsp. tularensis virulence factor. Infect. Immun. 77, 152–161.
Rajaram, M. V., Butchar, J. P., Parsa, K. V., Cremer, T. J., Amer, A., Schlesinger, L. S., and Tridandapani, S. (2009). Akt and SHIP modulate Francisella escape from the phagosome and induction of the Fas-mediated death pathway. PLoS ONE 4, e7919. doi: 10.1371/journal.pone.0007919.
Reilly, T. J., Baron, G. S., Nano, F. E., and Kuhlenschmidt, M. S. (1996). Characterization and sequencing of a respiratory burst-inhibiting acid phosphatase from Francisella tularensis. J. Biol. Chem. 271, 10973–10983.
Rodriguez, A. R., Yu, J. J., Murthy, A. K., Guentzel, M. N., Klose, K. E., Forsthuber, T. G., Chambers, J. P., Berton, M. T., and Arulanandam, B. P. (2010). Mast cell/IL-4 control of Francisella tularensis replication and host cell death is associated with increased ATP production and phagosomal acidification. Mucosal. Immunol. doi: 10.1038/mi.2010.59. [Epub ahead of print].
Sammons-Jackson, W. L., McClelland, K., Manch-Citron, J. N., Metzger, D. W., Bakshi, C. S., Garcia, E., Rasley, A., and Anderson, B. E. (2008). Generation and characterization of an attenuated mutant in a response regulator gene of Francisella tularensis live vaccine strain (LVS). DNA Cell Biol. 27, 387–403.
Santic, M., Asare, R., Skrobonja, I., Jones, S., and Abu Kwaik, Y. (2008). Acquisition of the vacuolar ATPase proton pump and phagosome acidification are essential for escape of Francisella tularensis into the macrophage cytosol. Infect. Immun. 76, 2671–2677.
Santic, M., Molmeret, M., and Abu Kwaik, Y. (2005a). Modulation of biogenesis of the Francisella tularensis subsp. novicida-containing phagosome in quiescent human macrophages and its maturation into a phagolysosome upon activation by IFN-gamma. Cell Microbiol. 7, 957–967.
Santic, M., Molmeret, M., Klose, K. E., Jones, S., and Kwaik, Y. A. (2005b). The Francisella tularensis pathogenicity island protein IglC and its regulator MglA are essential for modulating phagosome biogenesis and subsequent bacterial escape into the cytoplasm. Cell Microbiol. 7, 969–979.
Santic, M., Molmeret, M., Barker, J. R., Klose, K. E., Dekanic, A., Doric, M., and Abu Kwaik, Y. (2007). A Francisella tularensis pathogenicity island protein essential for bacterial proliferation within the host cell cytosol. Cell Microbiol. 9, 2391–2403.
Santic, M., Pavokovic, G., Jones, S., Asare, R., and Kwaik, Y. A. (2010). Regulation of apoptosis and anti-apoptosis signalling by Francisella tularensis. Microbes Infect. 12, 126–134.
Schmerk, C. L., Duplantis, B. N., Howard, P. L., and Nano, F. E. (2009a). A Francisella novicida pdpA mutant exhibits limited intracellular replication and remains associated with the lysosomal marker LAMP-1. Microbiology 155, 1498–1504.
Schmerk, C. L., Duplantis, B. N., Wang, D., Burke, R. D., Chou, A. Y., Elkins, K. L., Ludu, J. S., and Nano, F. E. (2009b). Characterization of the pathogenicity island protein PdpA and its role in the virulence of Francisella novicida. Microbiology 155, 1489–1497.
Schulert, G. S., and Allen, L. A. (2006). Differential infection of mononuclear phagocytes by Francisella tularensis: role of the macrophage mannose receptor. J. Leukoc. Biol. 80, 563–571.
Schulert, G. S., McCaffrey, R. L., Buchan, B. W., Lindemann, S. R., Hollenback, C., Jones, B. D., and Allen, L. A. (2009). Francisella tularensis genes required for inhibition of the neutrophil respiratory burst and intramacrophage growth identified by random transposon mutagenesis of strain LVS. Infect. Immun. 77, 1324–1336.
Su, J., Yang, J., Zhao, D., Kawula, T. H., Banas, J. A., and Zhang, J. R. (2007). Genome-wide identification of Francisella tularensis virulence determinants. Infect. Immun. 75, 3089–3101.
Tamilselvam, B., and Daefler, S. (2008). Francisella targets cholesterol-rich host cell membrane domains for entry into macrophages. J. Immunol. 180, 8262–8271.
Tempel, R., Lai, X. H., Crosa, L., Kozlowicz, B., and Heffron, F. (2006). Attenuated Francisella novicida transposon mutants protect mice against wild-type challenge. Infect. Immun. 74, 5095–5105.
Underhill, D. M., and Ozinsky, A. (2002). Phagocytosis of microbes: complexity in action. Annu. Rev. Immunol. 20, 825–852.
Wehrly, T. D., Chong, A., Virtaneva, K., Sturdevant, D. E., Child, R., Edwards, J. A., Brouwer, D., Nair, V., Fischer, E. R., Wicke, L., Curda, A. J., Kupko, J. J. 3rd, Martens, C., Crane, D. D., Bosio, C. M., Porcella, S. F., and Celli, J. (2009). Intracellular biology and virulence determinants of Francisella tularensis revealed by transcriptional profiling inside macrophages. Cell Microbiol. 11, 1128–1150.
Weiss, D. S., Brotcke, A., Henry, T., Margolis, J. J., Chan, K., and Monack, D. M. (2007). In vivo negative selection screen identifies genes required for Francisella virulence. Proc. Natl. Acad. Sci. U.S.A. 104, 6037–6042.
Keywords: Francisella, macrophage, phagosome, pathogenesis
Citation: Chong A and Celli J (2010) The Francisella intracellular life cycle: toward molecular mechanisms of intracellular survival and proliferation. Front. Microbio. 1:138. doi: 10.3389/fmicb.2010.00138
Received: 02 November 2010;
Accepted: 05 December 2010;
Published online: 28 December 2010.
Edited by:
Anders Sjostedt, Umeå University, SwedenCopyright: © 2010 Chong and Celli. This is an open-access article subject to an exclusive license agreement between the authors and the Frontiers Research Foundation, which permits unrestricted use, distribution, and reproduction in any medium, provided the original authors and source are credited.
*Correspondence: Jean Celli, Laboratory of Intracellular Parasites, Rocky Mountain Laboratories, National Institute of Allergy and Infectious Diseases, National Institutes of Health, 903 South 4th Street, Hamilton, MT 59840, USA. e-mail:amNlbGxpQG5pYWlkLm5paC5nb3Y=