- 1Foodborne and Waterborne Diseases Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 2Department of Medical Parasitology & Mycology, School of Public Health, Tehran University of Medical Sciences, Tehran, Iran
- 3Department of Biology, Science and Research Branch, Islamic Azad University, Tehran, Iran
- 4Gastroenterology and Liver Diseases Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 5Basic and Molecular Epidemiology of Gastrointestinal Disorder Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran
Objective: Irritable bowel syndrome (IBS) and inflammatory bowel disease (IBD) are gastrointestinal disorders, which can be triggered by gut microbiota dysbiosis. The development of IBS-like symptoms has been linked to the overgrowth of Candida spp. In addition, the critical role of fungi has been highlighted in the pathogenesis of IBD. This study investigated the association between Blastocystis and selected yeasts in IBS and IBD patients.
Methods: This investigation is a cross-sectional study from 2022 to 2024, performed on 91 participants, including 20 healthy individuals, 27 patients with IBS, and 44 IBD patients [39 with ulcerative colitis (UC; 88.63%) and 5 (11.37%) Crohn’s disease (CD)], who were also categorized based on the presence of Blastocystis. Total DNA was extracted from stool samples, and the presence and quantity of yeasts including C. albicans, C. tropicalis, C. glabrata, C. parapsilosis, C. krusei, Geotrichum candidum, Rhodotorula spp., Cryptococcus neoformans, and Saccharomyces cerevisiae were evaluated by real-time PCR. Statistical tests were used to assess significant associations between variables.
Results: Saccharomyces cerevisiae and C. albicans were the most prevalent yeasts in all groups. Candida tropicalis and C. neoformans were identified in neither patients nor healthy subjects. The presence/absence of C. albicans was not significantly different between patients with IBD, IBS, and the control groups. This was similar for G. candidum. However, there was a difference in the presence of S. cerevisiae among patients, although it was insignificant (p-value = 0.077). There was a significant difference in the quantity of C. albicans between IBD (880.421 ± 2140.504), IBS (10.307 ± 15.206), and controls (2875.888 ± 8383.889) (p-value = 0.020). Specifically, the source of difference was seen between IBD patients and the control group (p-value = 0.005). In addition, considering the presence of Blastocystis, a statistically significant association was seen between the number of C. albicans and the sample groups (p-value = 0.013). The quantity of C. albicans was significantly different between IBS and IBD patients.
Conclusion: Regarding the presence of Blastocystis, the quantity of C. albicans and S. cerevisiae was increased and decreased in the studied groups, respectively. This is a preliminary study, and eukaryote–eukaryote association in IBS and IBD patients should be considered in further studies.
1 Introduction
Irritable bowel syndrome (IBS) is one of the most frequently reported conditions among gut–brain interaction (DGBI) disorders (1). The prevalence of IBS varies regarding the diagnostic criteria, from 1.1% for Rome III in Iran to 45% for Rome II in Pakistan (1, 2). According to the latest estimation, the prevalence of IBS in developed countries such as the United States (US) ranges from 4.7 to 5.3% according to the Rome IV criteria (3). In addition, the prevalence of IBS in women seems to be higher than men (2, 4).
The main reason for IBS has not been determined; however, visceral hypersensitivity, gut microbiota alteration, immunological disorders, and increased gut permeability are suggested to be potential agents (5–7). Gut microbiota dysbiosis is thought to trigger visceral hypersensitivity, gut permeability, and immunological disorders in IBS (8, 9). Although metagenomics studies have illustrated that eukaryotes and viruses comprise a small portion of the gut microbiota core, the role of eukaryotes in maintaining the gut’s homeostasis seems to be determinative (10, 11).
Inflammatory bowel disease (IBD) is a well-known gastrointestinal (GI) tract disorder that is described by chronic inflammation. Ulcerative colitis (UC) and Crohn’s disease (CD) are two main conditions in IBD patients, which are characterized based on the regions of involvement through the GI tract and the clinical manifestations (12, 13). Inflammatory bowel disease is thought to be related to Western diets and industrialization. During recent decades, the increased incidence rate of IBD in developing countries infers a drift in the distribution of the disease (14, 15).
Inflammatory bowel disease results from a disturbance in immune responses through the gut. However, the leading hypothesis is an interplay between the gut microbes and the immune system in genetically susceptible patients under special conditions (16). Recent studies have highlighted the crucial role of gut microbiota in the development and amelioration/deterioration of clinical manifestations in patients with IBD (17–19). It was documented that the microbiota significantly contributes to the homeostasis and healthy conditions of the gut (20, 21). Therefore, disturbance in the gut microbiota may provide a proper condition for the development of GI tract disorders such as IBD (21). Fecal calprotectin is a protein, mainly released from neutrophils, which plays a crucial role in regulating innate immunity and responses to intestinal microbes. Measurement of fecal calprotectin indicates mucosal inflammation and inflammatory activity in IBD patients (22).
The role of the gut mycobiome in human health via interaction with bacteria has been recently highlighted (23, 24). It was hypothesized that the composition of the mycobiome and its metabolites may induce dysfunctions in mucosal immunity, leading to gut inflammation (10, 25–27). For example, β-glucan, as the major component of the cell wall of fungi, can modulate immune responses and induce inflammation (25). Many studies have evaluated the correlation between fungi and bacteria in healthy and disease conditions (28–30). Compared to the bacteria, in which the cell number per gram of stool may reach 1011, the number of fungal cells may vary from 105 to 106 (31, 32). It was demonstrated that a high carbohydrate diet has been weakly associated with overgrowth of yeasts, particularly Candida spp. (33), and probably “unexplained clinical symptoms,” which are linked to IBS (11, 34). The promising results of fungicide therapy in mice suffering from visceral hypersensitivity support the critical role of the gut mycobiome in the clinical manifestations of IBS (35). Moreover, metagenomics studies have demonstrated alteration, mainly changes in gut mycobiome species (intra-species or β-diversity) in IBS patients compared to healthy controls (23, 36, 37).
Apart from fungi, other eukaryotes, such as parasitic protozoa, seem to be associated with a healthy gut. Blastocystis is a hyper-prevalent protist of the human gut, and its prevalence in IBS patients is significantly higher than healthy controls (38, 39). Blastocystis is usually reported with lower prevalence in IBD patients compared to healthy subjects, although controversial findings disconfirm these results. Blastocystis has been linked to gut microbiota alteration in a broad spectrum of GI disorders (27). Although controversial (40), it was documented that Blastocystis is mainly linked to enriched beneficial bacteria of the gut (41, 42). However, there are no data describing intra-kingdom interplay between fungi and parasites in the healthy or unhealthy gut. During recent years, many studies have investigated the bilateral interaction of fungi or Blastocystis with the gut bacterial composition in IBS and IBD patients; still, there is no evidence exploring the association between fungi and Blastocystis in these patients. The current study aimed to depict a probable association between Blastocystis and selected yeasts in IBS and IBD patients.
2 Materials and methods
2.1 Ethics approval and consent to participate
All experimental protocols followed the ethical principles and the national norms and standards for conducting Medical Research in Iran. The study was performed according to the relevant guidelines and declaration. The current study was also approved by the Research Institute for Gastroenterology and Liver, Shahid Beheshti University of Medical Sciences (SBMU) (IR.SBMU.RIGLD.REC.1403.014 and IR.SBMU.RIGLD.REC.1400.020). Informed consent was also obtained from all participants or their legal guardian(s) before the study.
2.2 Sample collection
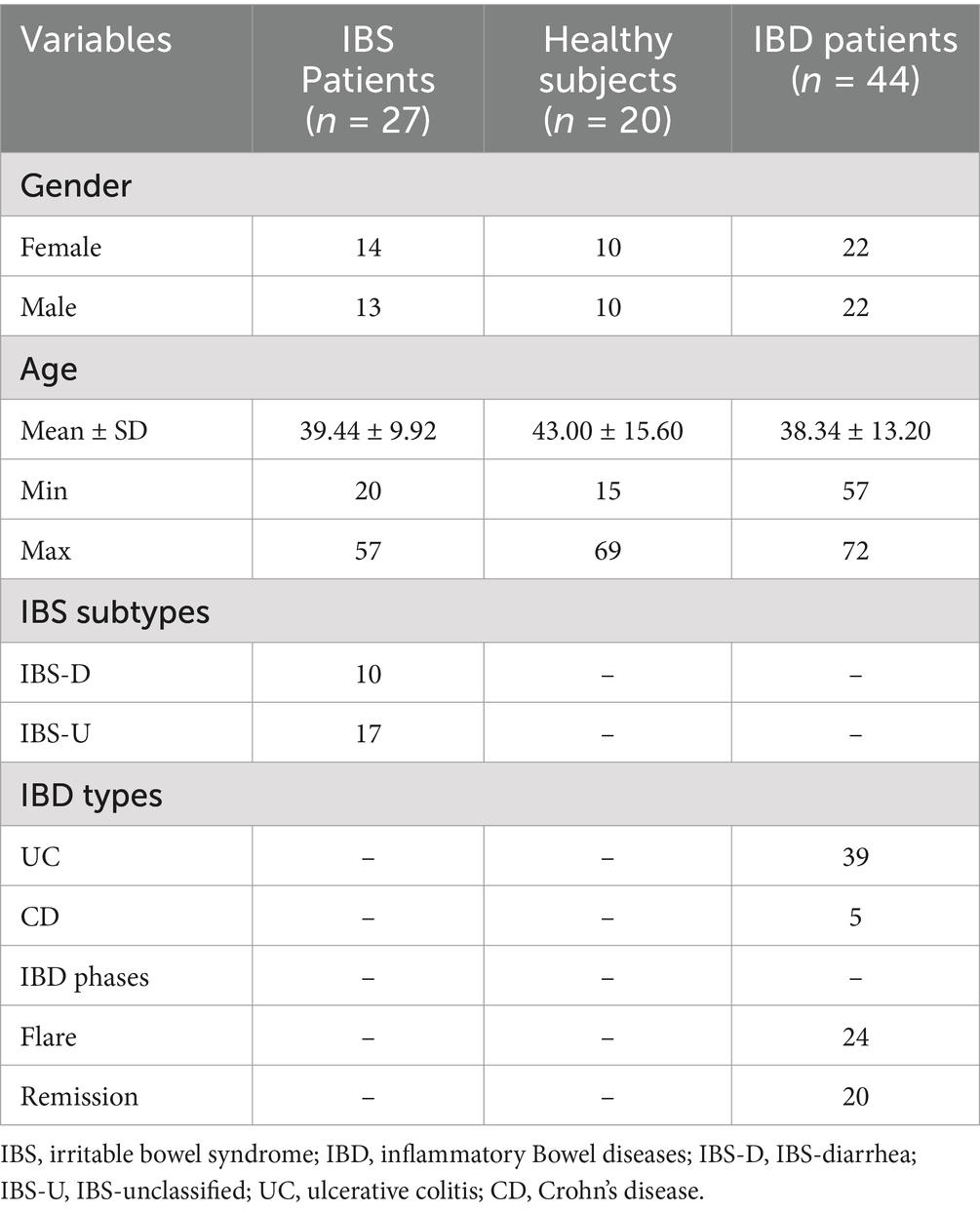
This cross-sectional study was performed on stool samples of 91 subjects consisting of 44, 27, and 20 IBD, IBS, and healthy individuals, respectively, collected from February 2022 to July 2024 (Figure 1). The IBD patients had been confirmed by colonoscopy, and the participants were from patients: (1) newly diagnosed as IBD, (2) patients in remission phase who were referred to the clinic for checkup, and (3) symptomatic patients. Samples were collected from all enrolled patients before consumption of any IBD drugs. Demographic data, drug consumption, stool appearance at the time of sampling, and symptoms during sampling were recorded. Consumption of any antibiotic and systemic antifungal and antiparasitic agents 1 month before sampling was considered exclusion criteria.
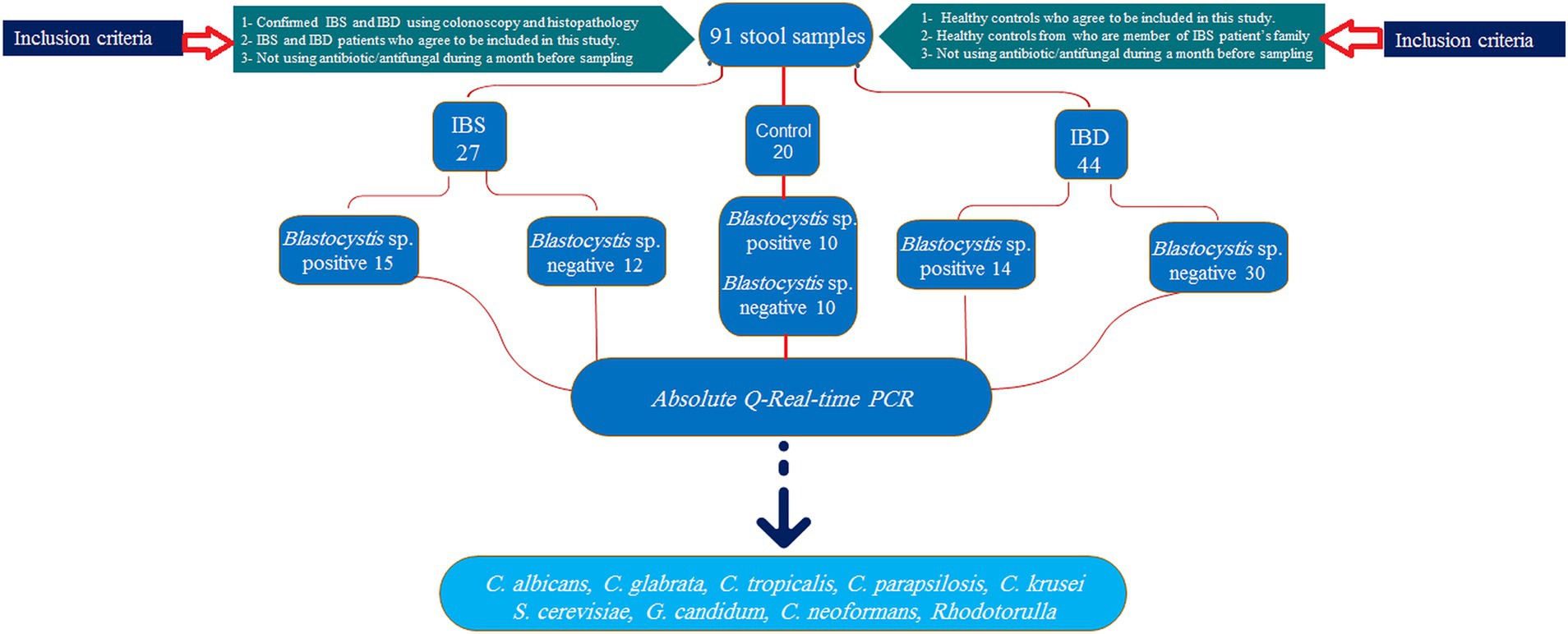
Figure 1. Sampling flowchart of study groups, allocated samples, inclusion criteria, and selected yeasts.
Regarding the presence of Blastocystis, groups were divided into healthy-Blastocystis (10), healthy (10), IBS-Blastocystis (15), IBS (12), IBD-Blastocystis (14), and IBD (30). Stool samples were immediately transported to the Parasitology lab in the Research Institute for Gastroenterology and Liver Diseases for further analysis. The calprotectin values were considered to distinguish flare and remission in IBD patients. Accordingly, to the clinical consultants, values more than 300 μg/g were considered activity indicators for flare conditions.
2.3 Fecal calprotectin
Fecal calprotectin was measured for all IBD samples using an ELISA Calprotectin kit (BÜHLMANN fCAL, Germany) with a working range of 10–3,600 μg/g, as recommended by the instrument. The values of calprotectin were categorized as 1–4, indicating >299, 300–999, 1,000–1999, and >2000 μg/g, respectively.
2.4 DNA extraction and Blastocystis identification
Total DNA was isolated from all samples using a commercial stool DNA extraction kit (Yekta Tajhiz, Tehran, Iran). In brief, 200 mg of stool samples were washed three times with sterile phosphate buffer saline (PBS; pH = 7.5). After the final washing step, the supernatant was discarded, and the DNA extraction kit treated the pellet. After 30 min incubation of samples at 60°C together with 40 μg of proteinase k and 250 μL of lysis buffer, DNA was isolated and purified according to the instructions. Purified DNA was stored at −20°C until use.
To identify Blastocystis in stool samples, the “barcoding region” of the small subunit ribosomal RNA (SSU rRNA) gene was targeted using primers mentioned elsewhere (43) and according to PCR conditions in our previous study (44).
2.5 Quantitative real-time PCR for fungi
To detect the presence and quantify the number of fungi in each sample, specific primers were designed from identical regions of the internal transcribed spacer (ITS) of selected yeasts (Table 1). Real-time PCR was carried out in a 15 μL total volume by a Rotor-Gene Q (QIAGEN, Germany) real-time instrument with the following conditions: 7.5 μL of 2X real-time PCR Master Mix (BioFACT™, Korea), 0.5 μL of each primer (5 ρM), 3.5 μL of distilled water, and 3 μL of template DNA. Targeted fragments were amplified by cycling program: 95°C for 10 min followed by 40 cycles: 95°C for 25 s, 59°C for 30 s, and 72°C for 20 s and ramping from 70°C to 95°C at 1°Cs−1. An appropriate positive control and sterile distilled water as negative control were tested in each run. The melting profiles were also analyzed using Rotor-Gene Q software to exclude non-specific amplifications and primer dimers. In addition, a cycle of threshold (Ct) value of more than 35 or no amplification curve was considered negative.
Absolute quantification real-time PCR was used to quantify the number of fungi in each sample. Positive controls were selected from the collection bank of Tehran Medical Mycology Laboratory (TMML) located in the Dept. of Parasitology and Mycology, Tehran University of Medical Sciences, with collection numbers: TMML-1227, TMML-1226, TMML-1223, TMML-1218, TMML-1322, TMML-621, TMML-695, TMML-2651, and TMML-181 for C. albicans, C. tropicalis, C. glabrata, C. parapsilosis, C. krusei, G. candidum, Rhodotorula spp., Cryptococcus neoformans, and S. cerevisiae, respectively. For this purpose, 1 × 106 were counted as follows: A loop full of a colony of each yeast was suspended in sterile PBS (pH = 7.5), and the number of yeasts was counted using spectrophotometry with a wavelength of 600. An optical density (OD) between 0.95 and 1.05 was considered 1 × 106. Afterward, DNA was extracted from each yeast, and six serial dilutions with log −10 were prepared from extracted DNAs.
Serial dilutions for each yeast were included in each real-time PCR experiment together with samples. A standard curve was drawn using serial dilutions to quantify the number of yeasts in each run. An R higher than 0.985 was considered as perfect.
2.6 Statistical analysis
A statistical analysis using the Chi-square test was conducted to compare the presence or absence of various fungi. The number of different fungi was compared using either the Mann–Whitney U-test or Kruskal–Wallis test. A logistic regression model was utilized to identify significant predictors for the existence of fungal species. In addition, a Spearman correlation coefficient was calculated to determine relationships. The R software, version 4.3.3, was used for all statistical analyses, and a p-value of less than 0.05 was considered statistically significant.
3 Results
In total, 44 IBD patients, 20 healthy controls, and 27 IBS patients were included in our study, of which 45 and 46 were males and females, respectively, with an age range from 15 to 72 years. The mean age ± standard deviation (SD) of participants was 39.44 ± 9.91, 38.34 ± 13.2, and 43.00 ± 15.60 for IBS, IBD, and healthy subjects, respectively. According to the patient reports and observations, 17 and 10 IBS patients were classified as IBS-U (IBS-unclassified) and IBS-D (IBS-diarrhea), respectively. From 44 IBD patients, 39 (88.63%) and 5 (11.37%) suffered from UC and CD, respectively. Among IBD cases, 23 (52.27%) and 21 (47.73%) subjects were recorded as flare and remission phases, respectively (Table 2). According to the IBD activity scores extracted from the calprotectin values, 20, 7, 5, and 10 for patients were classified into 4 groups with calprotectin values >299, 300–999, 1,000–1999, and >2000 μg/g, respectively. The flare phase was determined based on the calprotectin scores recommended by the clinical consultant. The stool appeared formed and watery samples in 35 (79.55%) and 9 (20.45%) patients, respectively. All watery stool samples were seen in patients suffering from flare IBD. The presence of Blastocystis was identified among 15 (55.55%), 10 (50%), and 14 (31.82%) for IBS patients, healthy controls, and IBD patients, respectively. Accordingly, the mean ± SD of calprotectin in patients with flare and remission phases was 1787.24 ± 788.56 and 124.8 ± 119.07, respectively. As a fact, in IBD, there was a significant difference in the levels of fecal calprotectin between IBD patients in the flare phase and those in remission (p-value <0.001) (Figure 2). Interestingly, there was also no significant difference in fecal calprotectin levels in IBD patients with and without Blastocystis (p-value = 0.099) (Figure 2). The association between Blastocystis and activity scores released from fecal calprotectin values was also assessed, which was near-significant (p-value = 0.051). There was a significant association between the stool appearance and the presence of Blastocystis (p-value = 0.019).
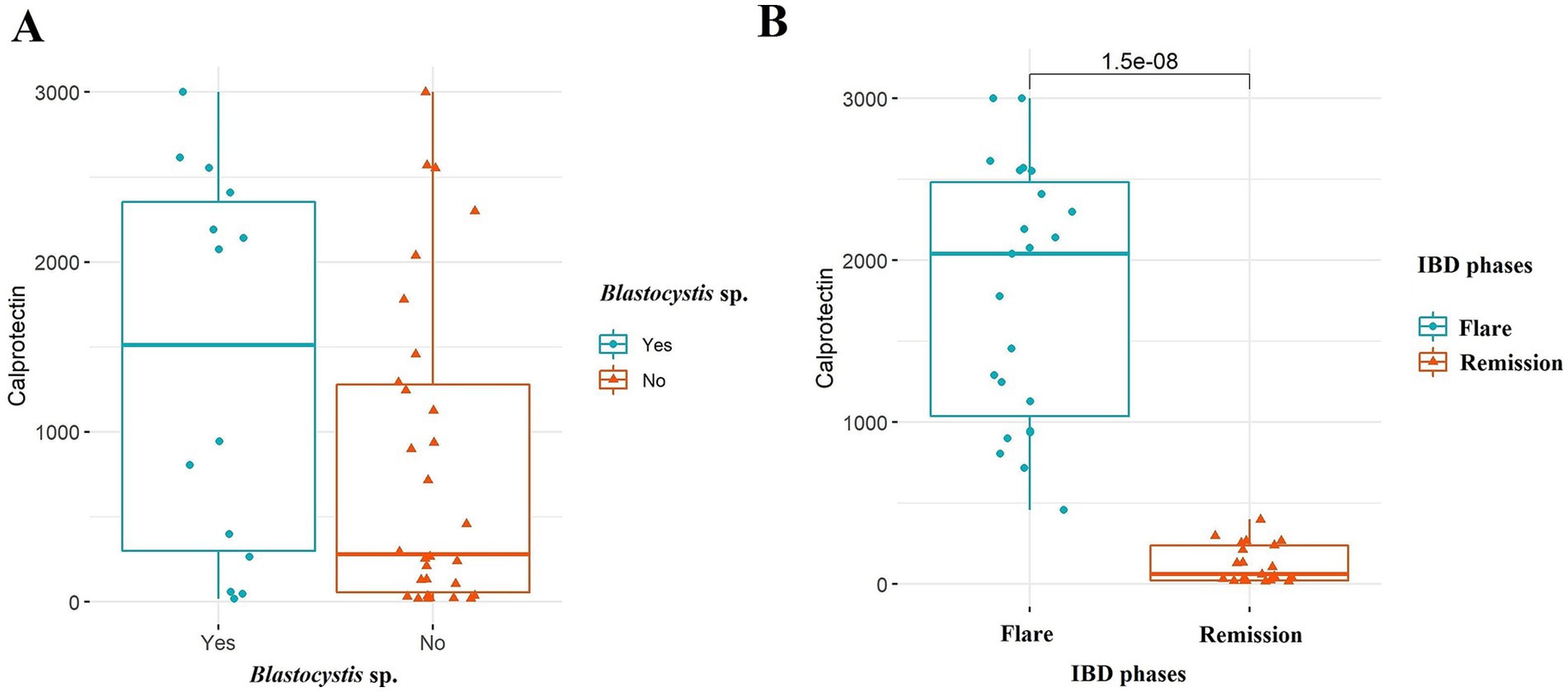
Figure 2. Comparison of fecal calprotectin levels among the IBD patients in (A) Blastocystis sp. and (B) IBD phases. Mann–Whitney U-test was used to analyze probable associations.
3.1 Yeast distribution
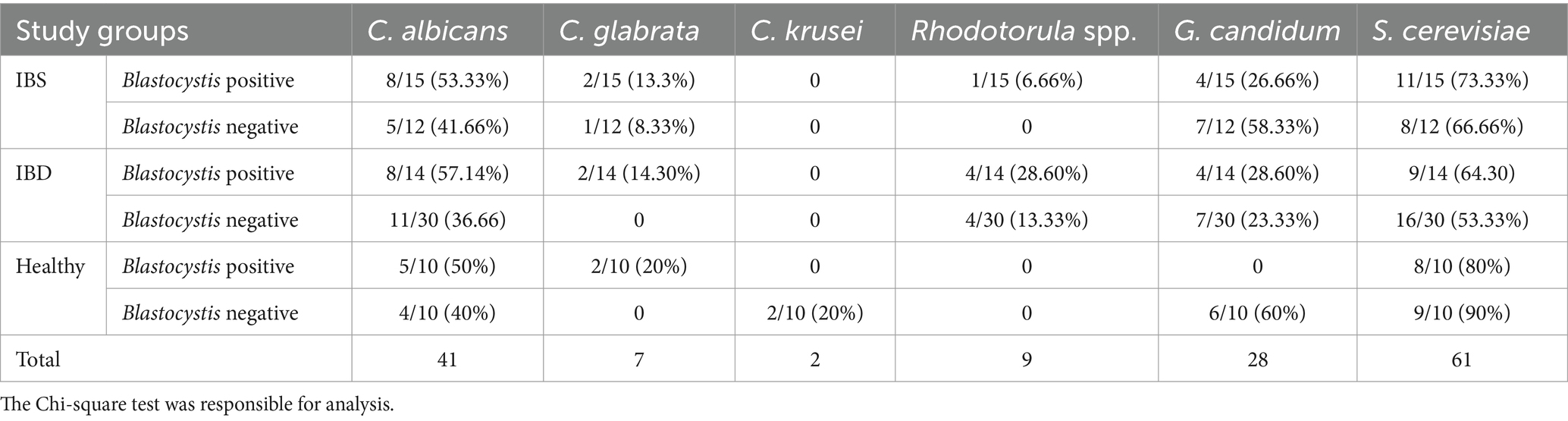
Real-time PCR was used to identify yeasts in samples. Our results showed the presence of C. albicans, C. glabrata, C. krusei, C. parapsilosis, S. cerevisiae, G. candidum, and Rhodotorula spp. in samples. Accordingly, S. cerevisiae was the most prevalent yeast in IBS, IBD, and healthy subjects: 19/27 (70.37%), 25/44 (56.82%), and 17/20 (85%), respectively. In IBS patients, the detected yeasts were C. albicans (13/27; 48.14%), G. candidum (12/27; 44.44%), C. glabrata (3/27; 11.11%), and Rhodotorula spp. (1/27; 3.7%). Saccharomyces cerevisiae was the dominant yeast in IBD (25/44; 56.82%), followed by C. albicans (19/44; 43.18%), G. candidum (11/44; 25%), Rhodotorula spp. (7/44; 15.91%), C. glabrata (2/44; 4.54%), and C. parapsilosis (1/44; 2.27%). In healthy controls, S. cerevisiae (17/20; 85%), C. albicans (9/20; 45%), G. candidum (6/20; 30%), C. glabrata (2/20; 10%), and C. krusei (2/20; 10%) were detected. Candida krusei in IBD and IBS patients and Rhodotorula spp. and C. parapsilosis in healthy subjects were not detected. Cryptococcus neoformans and C. tropicalis were identified in neither IBD nor IBS and healthy subjects. In addition, 12, 4, and 3 of IBD, IBS, and healthy subjects were negative for all tested yeasts, respectively (Table 3). There was no significant association between the activity scores and the presence of each yeast. In addition, except for C. glabrata (p-value = 0.038), the association between the presence of yeasts and stool forms was not statistically significant. The probable association between fecal calprotectin score and the presence of yeasts was also investigated which was not statistically significant.
Based on the presence of Blastocystis, S. cerevisiae (11/15; 73.33%), C. albicans (8/15; 53.33%), G. candidum (4/15; 26.66%), C. glabrata (2/15; 13.33%), and Rhodotorula spp. (1/15; 6.66%) were detected in IBS patients. Except for G. candidum (7/12; 58.33%), the prevalence of all tested yeasts in Blastocystis-negative IBS patients was lower than in Blastocystis-positive IBS patients. A significant association between the prevalence of yeasts and the presence of Blastocystis was not seen in IBS patients (Table 3).
In IBD patients, the presence of S. cerevisiae, C. albicans, G. candidum, Rhodotorula, and C. glabrata in Blastocystis-positive samples was (9/14; 64.28%), (8/14; 57.14%), (4/14; 28.57%), (4/14; 28.57%), and (2/14; 14.28%), respectively. In IBD group without Blastocystis, the presence of S. cerevisiae, C. albicans, G. candidum, and Rhodotorula was confirmed in (16/30; 53.33%), (11/30; 36.66%), (8/30; 26.66%), and (4/30; 13.33%) patients, respectively (Table 3).
In healthy subjects, the prevalence of S. cerevisiae, C. albicans, and C. glabrata in Blastocystis-positive samples was (8/10; 80%), (5/10; 50%), and (2/10; 20%), respectively, while in Blastocystis-negative samples, S. cerevisiae, C. albicans, G. candidum, and C. krusei were detected in (9/10; 90%), (4/10; 40%), (6/10; 60%), and (2/10; 20%), respectively. In addition, the Chi-square test showed that the correlation between the prevalence of yeasts and the presence of Blastocystis was not significant in healthy subjects (Table 3). The presence/absence of C. albicans and G. candidum was not significantly different between patients with IBD, IBS, and the control groups. However, there was no significant difference in the presence of S. cerevisiae among patients (p-value = 0.077) (Table 4).
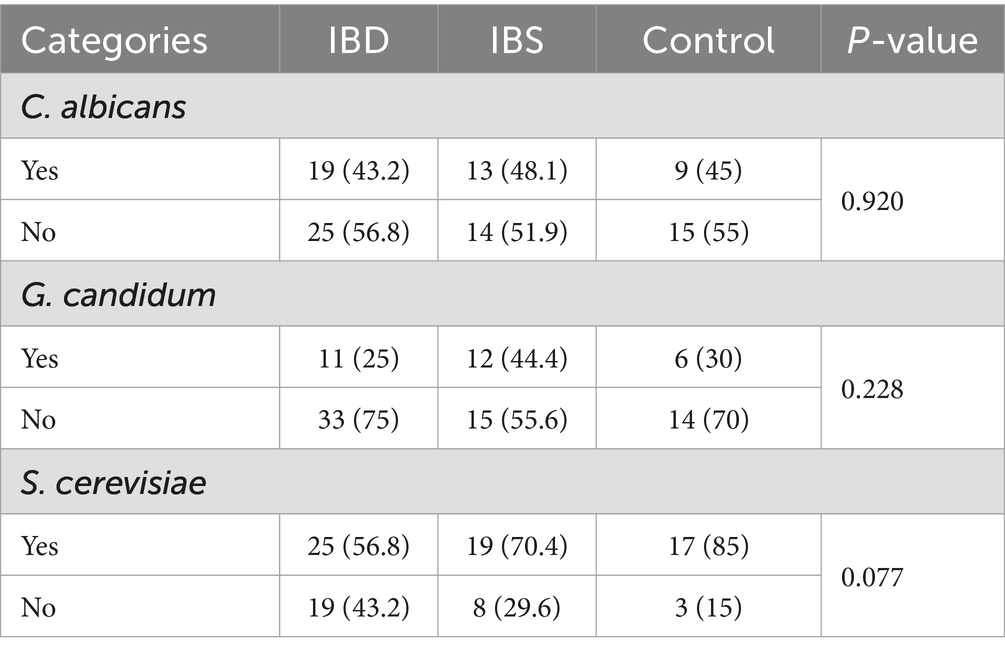
Table 4. Comparison of fungal distribution between IBD patients, IBS patients, and healthy controls using the chi-square test.
A logistic regression analysis was conducted to determine the presence of yeasts among patients (Tables 5, 6). The results revealed that IBD patients had a higher chance of having S. cerevisiae compared to the control group (OR (95% CI) = 4.15 (1.05, 16.37); p-value =0.042). The presence of Blastocystis also showed a near-significant relationship with G. candidum (OR (95% CI) = 2.69 (0.99, 7.30); p-value =0.053).
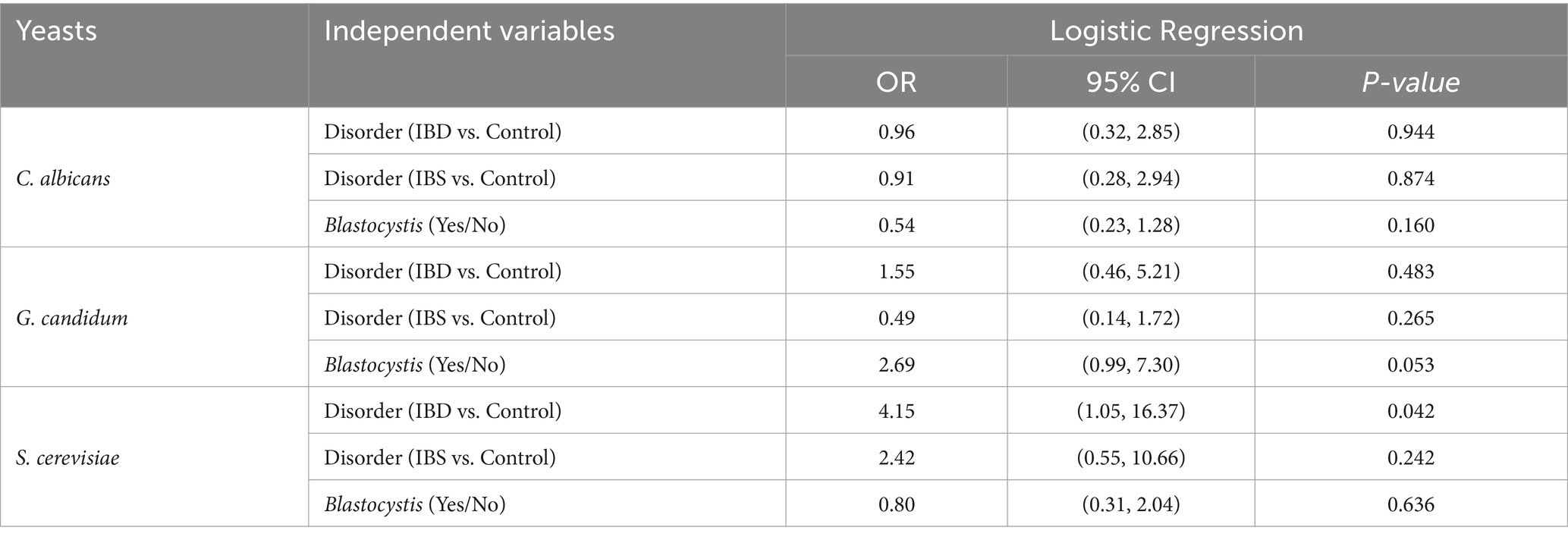
Table 5. Logistic regression analysis of the association between fungi and Blastocystis in patients.
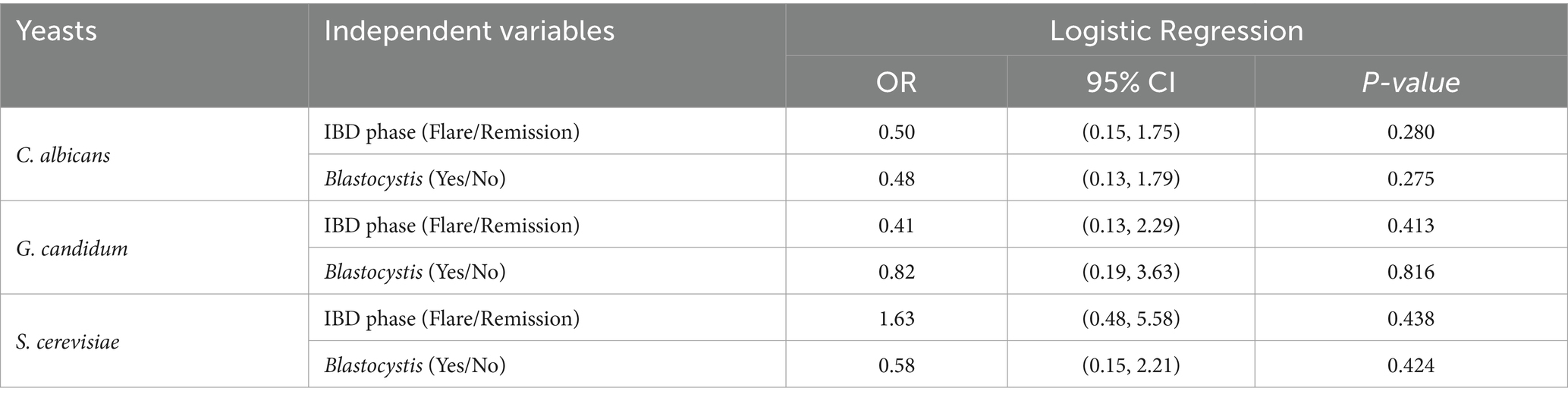
Table 6. Logistic regression analysis of the association between fungi species among IBD patients with different phases and Blastocystis.
We did not find any significant associations between the presence of different yeasts and the IBD phases or Blastocystis among IBD patients. In addition, the coefficient correlation between C. albicans and calprotectin was near-significant (r = 0.433, p-value =0.064) (Figure 3).
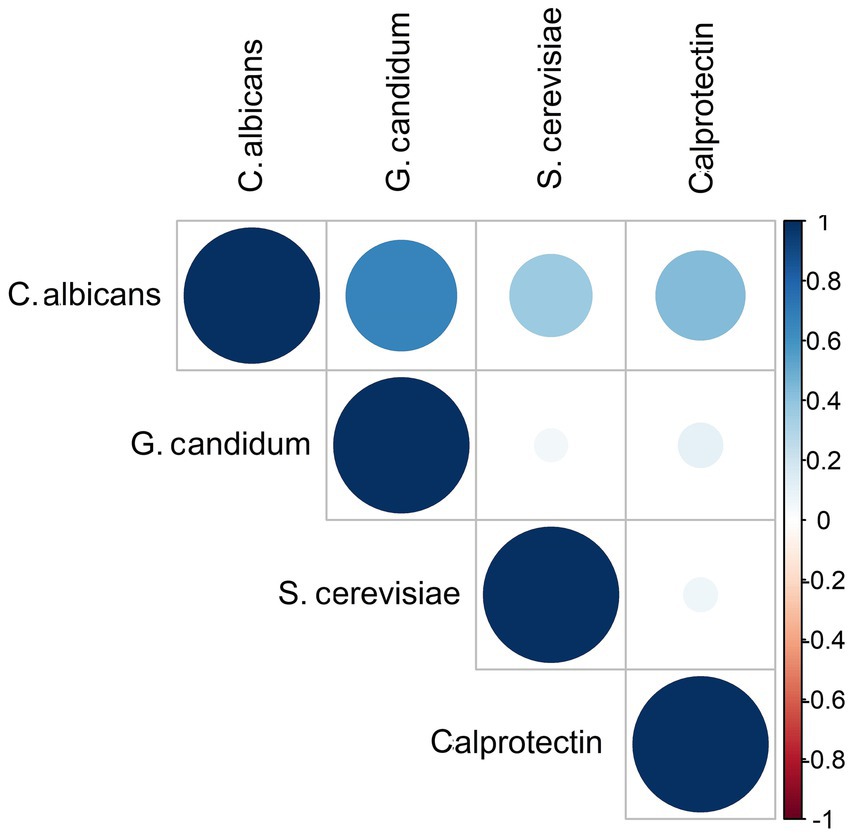
Figure 3. Correlation matrix diagram for the strength of the relationships between fungi species and fecal calprotectin among IBD patients. The size and the color intensity of the dark blue show an association between variables.
3.2 Yeast quantification
When looking at the number of C. albicans, there was a significant difference among patients (p-value = 0.020), in which the source of difference was seen between IBD and IBS patients (p-value = 0.005). There were no statistically significant differences in the quantity of G. candidum and S. cerevisiae across the different disorders of the patients (Figure 4).
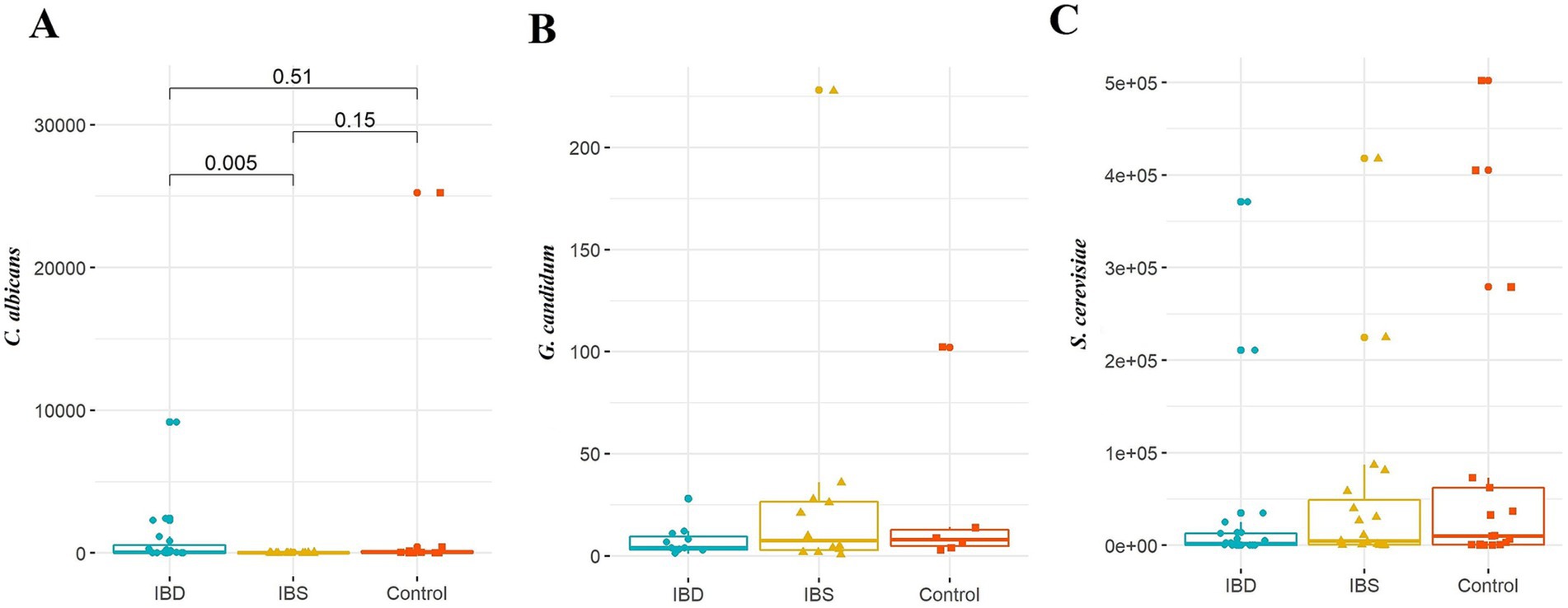
Figure 4. Comparison of fungal species: (A) C. albicans, (B) G. candidum, and (C) S. cerevisiae between IBD patients, IBS patients, and healthy controls. Kruskal–Wallis was used to evaluate the probable association between the quantity of selected yeasts and disorders. A post-hoc test was carried out to identify differences between groups.
Regarding the presence of Blastocystis, a statistically significant association was only seen between the number of C. albicans and the sample groups. Accordingly, the number of C. albicans in IBS patients who carried Blastocystis was significantly lower than two other groups (p-value = 0.013) (Table 7).
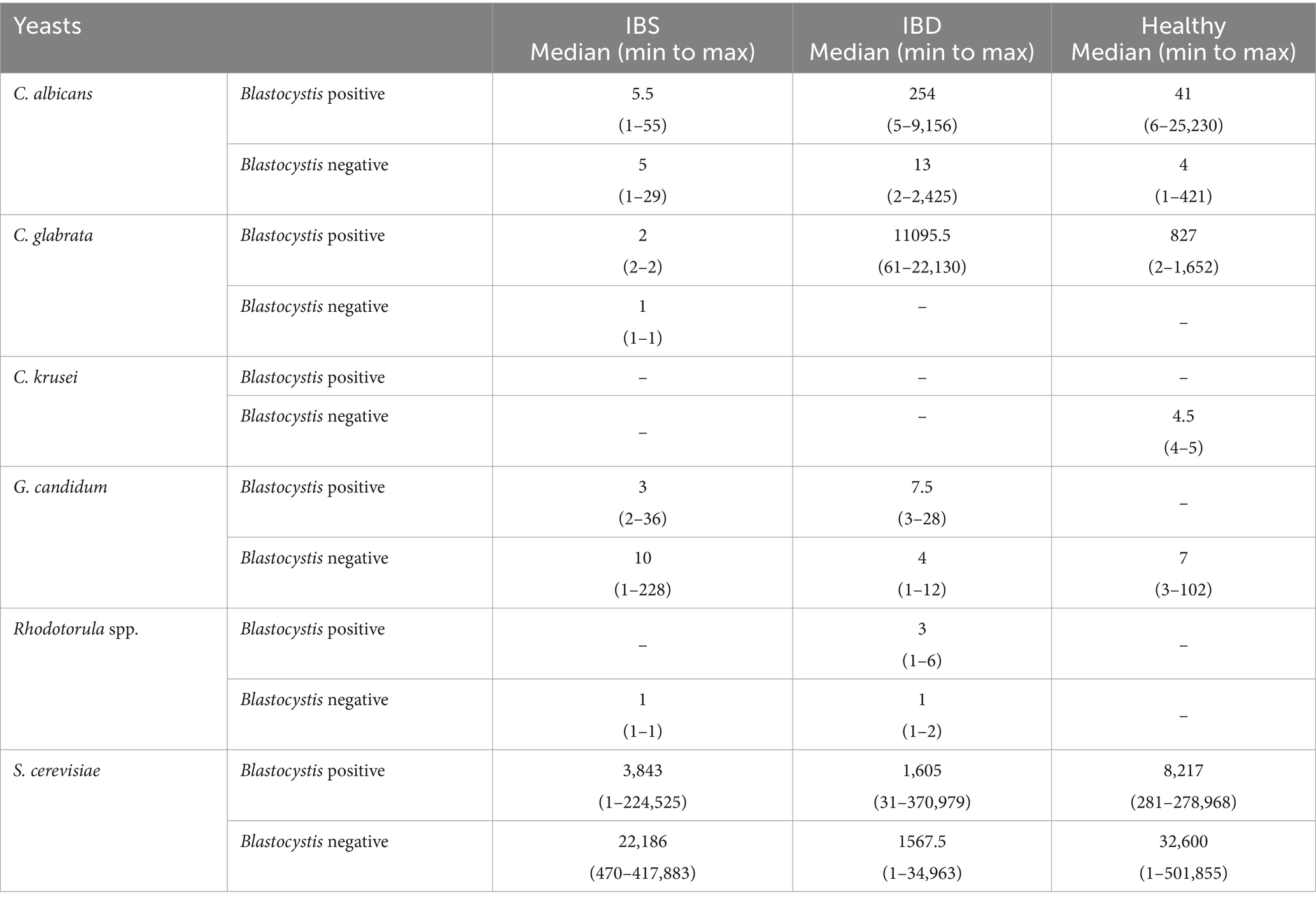
Table 7. Median (min to max) of the quantity of tested yeasts based on the absolute quantitative real-time PCR.
4 Discussion
The gut microbiota is a complex population of bacteria, viruses, protozoa, and fungi, whose number is approximately 10 times higher than the number of cells of host origin (45). It was estimated that only 0.1% of the gut microbiota are of eukaryotic origin, including protozoa and fungi (46, 47).
The role of mycobiome in GI disorders has significantly been highlighted in recent years. It has been suggested that although fungi and protozoa cover few portions of the gut microbiota, they are suggested to significantly shape the gut microbiota (10). A bacteria–fungi interaction was documented in GI disorders such as IBS (23) and IBD (16). It was demonstrated that gut microbiota dysbiosis may alter the fungal composition of the gut; however, this inter-kingdom correlation has been controversially depicted. For example, Hong et al. (23) proposed more susceptibility of gut mycobiome core compared to the gut bacteriome core in IBS-D patients, in which gut mycobiome changes could be considered a diagnostic signature. This finding confirmed the determinative role of gut mycobiome in presenting symptoms in IBS-like rats that suffered from visceral sensitivity (35). Such a co-variation between mycobiome and bacteriome was reported by Das et al. (36), who reported a co-variation between the gut mycobiome and bacteriome in IBS patients, which was suggested to be a potential clinical diagnostic value.
In IBS patients, Blastocystis is one of the most prevalent detected protozoa (27); therefore, investigating the interaction between this protist and prevalent yeasts in this group of patients could be interesting. In the current study, we failed to significantly correlate the prevalence of S. cerevisiae and C. albicans, two landmark yeasts of the gut, with the presence of Blastocystis in IBS patients, although our findings showed co-variation between these yeasts and Blastocystis, in which the prevalence of S. cerevisiae and C. albicans was increased from 41.6 and 66.66% in Blastocystis-negative to 53.3 and 73.3% in Blastocystis-positive subjects, respectively. In contrast, the co-variation between the presence of Blastocystis and the prevalence of S. cerevisiae and C. albicans was not observed in healthy subjects. Therefore, in addition to the available data indicating an increase in the presence of S. cerevisiae and C. albicans in IBS-like animal models (48), the gut condition and gut microbial disturbance in IBS subjects may increase eukaryotic–eukaryotic interactions. Blastocystis colonization seems to provide a favorable niche for the colonization of S. cerevisiae and C. albicans in IBS condition. However, the presence of Blastocystis increased and decreased the quantity of C. albicans and S. cerevisiae, respectively, regardless of the presence of IBS.
In IBD patients, a lack of co-variation was reported in CD patients between bacteria and fungi (16), while Imai et al. (49) showed mycobiome–bacteriome co-variation in CD patients compared to healthy and UC subjects. As a finding, although a statistically significant association between the presence of selected fungi and IBD was not seen, the quantity of C. albicans was correlated with the presence of IBD. In addition, the association between fecal calprotectin and the presence of C. albicans in IBD patients was close but not significant. Such a close association between C. albicans and IBD was documented by Yan et al. (50), who suggested a correlation between elevated levels of T helper 17 (Th17)-mediated immune responses and pathogenesis of IBD as well as the presence of C. albicans. Nevertheless, the lack of significant association between the presence of yeasts and the fecal calprotectin scores suggests that, most probably, there are more variables affecting the colonization of yeasts in the GI than just the increased number of Th17 cells. Interestingly, similar to C. albicans, the presence of Blastocystis in IBD patients was associated with elevated fecal calprotectin levels, despite it was insignificance.
On the other hand, Blastocystis has been associated with gut microbiota variation in healthy subjects and patients who suffer from GI disorders (40, 51–53). Owing to this, there is no knowledge of intra-kingdom interaction between fungi and eukaryotic microorganisms such as Blastocystis. Due to available studies, mining the correlation of Candida overgrowth with IBS-like symptoms (36, 54) and the high prevalence of Blastocystis in IBS patients (38, 39), the eukaryote–eukaryote interaction and synergistic effects of Blastocystis on the growth of C. albicans and the presentation of IBS symptoms should be considered. Furthermore, the quantity of S. cerevisiae, experienced as a potential probiotic in IBS patients to ameliorate the symptoms (55, 56), was reduced in the Blastocystis-positive group. This effect of Blastocystis on the richness of C. albicans and S. cerevisiae may rebut the term “healthy indicator” for this protozoan (41, 57, 58) not only in IBS patients but also in healthy subjects and IBD patients, in which the presence of Blastocystis increased and decreased the richness of C. albicans and S. cerevisiae, respectively, in healthy and IBD subjects.
This study is one of the first investigations evaluating the probable association between yeasts and Blastocystis in two significant GI disorders, IBS and IBD. Although this study is a pilot investigation, released results can provide interesting clue about intra-kingdom associations for further studies. However, the critical limitation of this study is the lack of a comprehensive molecular method for investigating a broad range of microorganisms. In addition, working on a higher number of samples can provide more accurate results. Another weakness of this study may be attributed to the lack of endoscopic scores that could help to extract more accurate data about the associations between clinical variables and yeast distribution in IBD patients.
5 Conclusion
In this study, regardless of the presence of Blastocystis, only the quantity of C. albicans was significantly different between IBS and IBD patients. Our findings showed a co-variation between the amount of C. albicans and the presence of Blastocystis. In contrast, the association between the number of S. cerevisiae and the presence of Blastocystis was reversed in all groups. The presence of Blastocystis was significantly associated with the stool forms, as well. Studies of mycobiome composition and intra-kingdom interaction between fungi and protozoa are limited. Although eukaryotes comprise a small portion of the gut microbiome, they are supposed to play a significant role in the development and symptoms of GI disorders. This investigation is a preliminary study mining eukaryote–eukaryote correlation in IBS and IBD patients. However, considering the probable determinative role of eukaryotes in shaping the gut microbiota composition, investigations of bilateral correlation between potential indicator microorganisms, such as Blastocystis, C. albicans, and S. cerevisiae, may provide interesting data.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving humans were approved by Research Institute for Gastroenterology and Liver, Shahid Beheshti University of Medical Sciences (SBMU) (IR.SBMU.RIGLD.REC.1403.014 and IR.SBMU.RIGLD.REC.1400.020). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
Author contributions
ZK: Data curation, Formal analysis, Methodology, Visualization, Writing – original draft, Writing – review & editing. SK: Methodology, Resources, Supervision, Writing – original draft. AO: Investigation, Methodology, Writing – original draft. AS: Investigation, Methodology, Resources, Writing – review & editing. SN: Investigation, Software, Validation, Writing – review & editing. ShS: Investigation, Resources, Writing – review & editing. SM: Investigation, Methodology, Resources, Writing – review & editing. SaS: Formal analysis, Software, Visualization, Writing – review & editing. HMR: Investigation, Writing – review & editing. HM: Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Validation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was financially supported by Shahid Beheshti University of Medical Sciences, Tehran, Iran (Grant no.: 43009717).
Acknowledgments
The authors thank all members of the Foodborne and Waterborne Diseases Research Center for their support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Sperber, AD. Epidemiology and burden of irritable bowel syndrome: an international perspective. Gastroenterol Clin North Am. (2021) 50:489–503. doi: 10.1016/j.gtc.2021.04.001
2. Sperber, AD, Dumitrascu, D, Fukudo, S, Gerson, C, Ghoshal, UC, Gwee, KA, et al. The global prevalence of IBS in adults remains elusive due to the heterogeneity of studies: a Rome foundation working team literature review. Gut. (2017) 66:1075–82. doi: 10.1136/gutjnl-2015-311240
3. Almario, CV, Sharabi, E, Chey, WD, Lauzon, M, Higgins, CS, and Spiegel, BMR. Prevalence and burden of illness of Rome IV irritable bowel syndrome in the United States: results from a nationwide cross-sectional study. Gastroenterology. (2023) 165:1475–87. doi: 10.1053/j.gastro.2023.08.010
4. Lovell, RM, and Ford, AC. Global prevalence of and risk factors for irritable bowel syndrome: a meta-analysis. Clin Gastroenterol Hepatol. (2012) 10:712–721.e4. doi: 10.1016/j.cgh.2012.02.029
5. Hanning, N, Edwinson, AL, Ceuleers, H, Peters, SA, De Man, JG, Hassett, LC, et al. Intestinal barrier dysfunction in irritable bowel syndrome: a systematic review. Ther Adv Gastroenterol. (2021) 14:1756284821993586. doi: 10.1177/1756284821993586
6. Mamieva, Z, Poluektova, E, Svistushkin, V, Sobolev, V, Shifrin, O, Guarner, F, et al. Antibiotics, gut microbiota, and irritable bowel syndrome: what are the relations? World J Gastroenterol. (2022) 28:1204–19. doi: 10.3748/wjg.v28.i12.1204
7. Moshiree, B, Heidelbaugh, JJ, and Sayuk, GS. A narrative review of irritable bowel syndrome with diarrhea: a primer for primary care providers. Adv Therap. (2022) 39:4003–20. doi: 10.1007/s12325-022-02224-z
8. Quigley, EMM. The gut-brain axis and the microbiome: clues to pathophysiology and opportunities for novel management strategies in irritable bowel syndrome (IBS). J Clin Med. (2018) 7:6. doi: 10.3390/jcm7010006
9. Holtmann, GJ, Ford, AC, and Talley, NJ. Pathophysiology of irritable bowel syndrome. Lancet Gastroenterol Hepatol. (2016) 1:133–46. doi: 10.1016/s2468-1253(16)30023-1
10. Beheshti-Maal, A, Shahrokh, S, Ansari, S, Mirsamadi, ES, Yadegar, A, Mirjalali, H, et al. Gut mycobiome: the probable determinative role of fungi in IBD patients. Mycoses. (2021) 64:468–76. doi: 10.1111/myc.13238
11. Santelmann, H, and Howard, JM. Yeast metabolic products, yeast antigens and yeasts as possible triggers for irritable bowel syndrome. Europ J Gastroenterol Hepatol. (2005) 17:21–6. doi: 10.1097/00042737-200501000-00005
12. Mulder, DJ, Noble, AJ, Justinich, CJ, and Duffin, JM. A tale of two diseases: the history of inflammatory bowel disease. J Crohns Colitis. (2014) 8:341–8. doi: 10.1016/j.crohns.2013.09.009
13. Kaser, A, Zeissig, S, and Blumberg, RS. Inflammatory bowel disease. Ann Rev Immunol. (2010) 28:573–621. doi: 10.1146/annurev-immunol-030409-101225
14. Benchimol, EI, Kaplan, GG, Otley, AR, Nguyen, GC, Underwood, FE, Guttmann, A, et al. Rural and urban residence during early life is associated with risk of inflammatory bowel disease: a population-based inception and birth cohort study. Am J Gastroenterol. (2017) 112:1412–22. doi: 10.1038/ajg.2017.208
15. Kaplan, GG. The global burden of IBD: from 2015 to 2025. Nat Rev Gastroenterol Hepatol. (2015) 12:720–7. doi: 10.1038/nrgastro.2015.150
16. Sokol, H, Leducq, V, Aschard, H, Pham, HP, Jegou, S, Landman, C, et al. Fungal microbiota dysbiosis in IBD. Gut. (2017) 66:1039–48. doi: 10.1136/gutjnl-2015-310746
17. Catalán-Serra, I, Thorsvik, S, Beisvag, V, Bruland, T, Underhill, D, Sandvik, AK, et al. Fungal microbiota composition in inflammatory bowel disease patients: characterization in different phenotypes and correlation with clinical activity and disease course. Inflamm Bowel Dis. (2024) 30:1164–77. doi: 10.1093/ibd/izad289
18. Jangi, S, Hsia, K, Zhao, N, Kumamoto, CA, Friedman, S, Singh, S, et al. Dynamics of the gut mycobiome in patients with ulcerative colitis. Clin Gastroenterol Hepatol. (2024) 22:821–830.e7. doi: 10.1016/j.cgh.2023.09.023
19. Yoon, H, Park, S, Jun, YK, Choi, Y, Shin, CM, Park, YS, et al. Evaluation of bacterial and fungal biomarkers for differentiation and prognosis of patients with inflammatory bowel disease. Microorganisms. (2023) 11:2882. doi: 10.3390/microorganisms11122882
20. Nishida, A, Inoue, R, Inatomi, O, Bamba, S, Naito, Y, and Andoh, A. Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin J Gastroenterol. (2018) 11:1–10. doi: 10.1007/s12328-017-0813-5
21. Sartor, RB, and Wu, GD. Roles for intestinal bacteria, viruses, and fungi in pathogenesis of inflammatory bowel diseases and therapeutic approaches. Gastroenterology. (2017) 152:327–339.e4. doi: 10.1053/j.gastro.2016.10.012
22. Kapel, N, Ouni, H, Benahmed, NA, and Barbot-Trystram, L. Fecal calprotectin for the diagnosis and management of inflammatory bowel diseases. Clin Transl Gastroenterol. (2023) 14:e00617. doi: 10.14309/ctg.0000000000000617
23. Hong, G, Li, Y, Yang, M, Li, G, Qian, W, Xiong, H, et al. Gut fungal dysbiosis and altered bacterial-fungal interaction in patients with diarrhea-predominant irritable bowel syndrome: An explorative study. Neurogastroenterol Motil. (2020) 32:e13891. doi: 10.1111/nmo.13891
24. Sam, QH, Chang, MW, and Chai, LY. The fungal mycobiome and its interaction with gut bacteria in the host. Int J Mol Sci. (2017) 18:330. doi: 10.3390/ijms18020330
25. Romani, L. Immunity to fungal infections. Nat Rev Immunol. (2011) 11:275–88. doi: 10.1038/nri2939
26. Peleg, AY, Hogan, DA, and Mylonakis, E. Medically important bacterial-fungal interactions. Nat Rev Microbiol. (2010) 8:340–9. doi: 10.1038/nrmicro2313
27. Olyaiee, A, Sadeghi, A, Yadegar, A, Mirsamadi, ES, and Mirjalali, H. Gut microbiota shifting in irritable bowel syndrome: the mysterious role of Blastocystis sp. Front Med. (2022) 9:890127. doi: 10.3389/fmed.2022.890127
28. Li, Q, Wang, C, Tang, C, He, Q, Li, N, and Li, J. Dysbiosis of gut fungal microbiota is associated with mucosal inflammation in Crohn's disease. J Clin Gastroenterol. (2014) 48:513–23. doi: 10.1097/mcg.0000000000000035
29. Hoarau, G, Mukherjee, PK, Gower-Rousseau, C, Hager, C, Chandra, J, Retuerto, MA, et al. Bacteriome and mycobiome interactions underscore microbial dysbiosis in familial Crohn’s disease. MBio. (2016) 7:e01250. doi: 10.1128/mBio.01250-16
30. Scanu, M, Toto, F, Petito, V, Masi, L, Fidaleo, M, Puca, P, et al. An integrative multi-omic analysis defines gut microbiota, mycobiota, and metabolic fingerprints in ulcerative colitis patients. Front Cell Infect Microbiol. (2024) 14:1366192. doi: 10.3389/fcimb.2024.1366192
31. Sender, R, Fuchs, S, and Milo, R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. (2016) 14:e1002533. doi: 10.1371/journal.pbio.1002533
32. Matijašić, M, Meštrović, T, Paljetak, H, Perić, M, Barešić, A, and Verbanac, D. Gut microbiota beyond bacteria-mycobiome, virome, archaeome, and eukaryotic parasites in IBD. Int J Mol Sci. (2020) 21:2668. doi: 10.3390/ijms21082668
33. Weig, M, Werner, E, Frosch, M, and Kasper, H. Limited effect of refined carbohydrate dietary supplementation on colonization of the gastrointestinal tract of healthy subjects by Candida albicans. Am J Clin Nutr. (1999) 69:1170–3. doi: 10.1093/ajcn/69.6.1170
34. Gu, Y, Zhou, G, Qin, X, Huang, S, Wang, B, and Cao, H. The potential role of gut mycobiome in irritable bowel syndrome. Front Microbiol. (2019) 10:1894. doi: 10.3389/fmicb.2019.01894
35. Botschuijver, S, Roeselers, G, Levin, E, Jonkers, DM, Welting, O, Heinsbroek, SEM, et al. Intestinal fungal dysbiosis is associated with visceral hypersensitivity in patients with irritable bowel syndrome and rats. Gastroenterology. (2017) 153:1026–39. doi: 10.1053/j.gastro.2017.06.004
36. Das, A, O'Herlihy, E, Shanahan, F, O'Toole, PW, and Jeffery, IB. The fecal mycobiome in patients with irritable bowel syndrome. Sci Rep. (2021) 11:124. doi: 10.1038/s41598-020-79478-6
37. Sciavilla, P, Strati, F, Di Paola, M, Modesto, M, Vitali, F, Cavalieri, D, et al. Gut microbiota profiles and characterization of cultivable fungal isolates in IBS patients. Appl Microbiol Biotechnol. (2021) 105:3277–88. doi: 10.1007/s00253-021-11264-4
38. Das, R, Khalil, S, Mirdha, BR, Makharia, GK, Dattagupta, S, and Chaudhry, R. Molecular characterization and subtyping of Blastocystis species in irritable bowel syndrome patients from North India. PLoS One. (2016) 11:e0147055. doi: 10.1371/journal.pone.0147055
39. Yakoob, J, Jafri, W, Jafri, N, Khan, R, Islam, M, Beg, MA, et al. Irritable bowel syndrome: in search of an etiology: role of Blastocystis hominis. Am J Trop Med Hyg. (2004) 70:383–5. doi: 10.4269/ajtmh.2004.70.383
40. Nourrisson, C, Scanzi, J, Pereira, B, NkoudMongo, C, Wawrzyniak, I, Cian, A, et al. Blastocystis is associated with decrease of fecal microbiota protective bacteria: comparative analysis between patients with irritable bowel syndrome and control subjects. PLoS One. (2014) 9:e111868. doi: 10.1371/journal.pone.0111868
41. Audebert, C, Even, G, Cian, A, Loywick, A, Merlin, S, Viscogliosi, E, et al. Colonization with the enteric protozoa Blastocystis is associated with increased diversity of human gut bacterial microbiota. Sci Rep. (2016) 6:25255. doi: 10.1038/srep25255
42. Yañez, CM, Hernández, AM, Sandoval, AM, Domínguez, MAM, Muñiz, SAZ, and Gómez, JOG. Prevalence of Blastocystis and its association with Firmicutes/Bacteroidetes ratio in clinically healthy and metabolically ill subjects. BMC Microbiol. (2021) 21:339. doi: 10.1186/s12866-021-02402-z
43. Scicluna, SM, Tawari, B, and Clark, CG. DNA barcoding of Blastocystis. Protist. (2006) 157:77–85. doi: 10.1016/j.protis.2005.12.001
44. Norouzi, M, Pirestani, M, Arefian, E, Dalimi, A, Sadraei, J, and Mirjalali, H. Exosomes secreted by Blastocystis subtypes affect the expression of proinflammatory and anti-inflammatory cytokines (TNFα, IL-6, IL-10, IL-4). Front Med. (2022) 9:940332. doi: 10.3389/fmed.2022.940332
45. Turnbaugh, PJ, Ley, RE, Hamady, M, Fraser-Liggett, CM, Knight, R, and Gordon, JI. The human microbiome project. Nature. (2007) 449:804–10. doi: 10.1038/nature06244
46. Qin, J, Li, R, Raes, J, Arumugam, M, Burgdorf, KS, Manichanh, C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. (2010) 464:59–65. doi: 10.1038/nature08821
47. Underhill, DM, and Iliev, ID. The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol. (2014) 14:405–16. doi: 10.1038/nri3684
48. van Thiel, IAM, Stavrou, AA, de Jong, A, Theelen, B, Davids, M, Hakvoort, TBM, et al. Genetic and phenotypic diversity of fecal Candida albicans strains in irritable bowel syndrome. Sci Rep. (2022) 12:5391. doi: 10.1038/s41598-022-09436-x
49. Imai, T, Inoue, R, Kawada, Y, Morita, Y, Inatomi, O, Nishida, A, et al. Characterization of fungal dysbiosis in Japanese patients with inflammatory bowel disease. J Gastroenterol. (2019) 54:149–59. doi: 10.1007/s00535-018-1530-7
50. Yan, Q, Li, S, Yan, Q, Huo, X, Wang, C, Wang, X, et al. A genomic compendium of cultivated human gut fungi characterizes the gut mycobiome and its relevance to common diseases. Cell. (2024) 187:2969–2989.e24. doi: 10.1016/j.cell.2024.04.043
51. Deng, L, Wojciech, L, Png, CW, Koh, EY, Aung, TT, Kioh, DYQ, et al. Experimental colonization with Blastocystis ST4 is associated with protective immune responses and modulation of gut microbiome in a DSS-induced colitis mouse model. Cell Mol Life Sci. (2022) 79:245. doi: 10.1007/s00018-022-04271-9
52. Kim, MJ, Lee, YJ, Kim, TJ, and Won, EJ. Gut microbiome profiles in colonizations with the enteric protozoa Blastocystis in Korean populations. Microorganisms. (2021) 10:34. doi: 10.3390/microorganisms10010034
53. Nourrisson, C, Scanzi, J, Brunet, J, Delbac, F, Dapoigny, M, and Poirier, P. Prokaryotic and eukaryotic fecal microbiota in irritable bowel syndrome patients and healthy individuals colonized with Blastocystis. Front Microbiol. (2021) 12:713347. doi: 10.3389/fmicb.2021.713347
54. Wang, ZK, Yang, YS, Stefka, AT, Sun, G, and Peng, LH. Review article: fungal microbiota and digestive diseases. Aliment Pharmacol Therap. (2014) 39:751–66. doi: 10.1111/apt.12665
55. Cayzeele-Decherf, A, Pélerin, F, Leuillet, S, Douillard, B, Housez, B, Cazaubiel, M, et al. Saccharomyces cerevisiae CNCM I-3856 in irritable bowel syndrome: An individual subject meta-analysis. World J Gastroenterol. (2017) 23:336–44. doi: 10.3748/wjg.v23.i2.336
56. Mourey, F, Decherf, A, Jeanne, JF, Clément-Ziza, M, Grisoni, ML, Machuron, F, et al. Saccharomyces cerevisiae I-3856 in irritable bowel syndrome with predominant constipation. World J Gastroenterol. (2022) 28:2509–22. doi: 10.3748/wjg.v28.i22.2509
57. Andersen, LO, and Stensvold, CR. Blastocystis in health and disease: are we moving from a clinical to a public health perspective? J Clin Microbiol. (2016) 54:524–8. doi: 10.1128/jcm.02520-15
58. Mirjalali, H, Abbasi, MR, Naderi, N, Hasani, Z, Mirsamadi, ES, Stensvold, CR, et al. Distribution and phylogenetic analysis of Blastocystis sp. subtypes isolated from IBD patients and healthy individuals in Iran. Europ J Clin Microbiol infect dis: official publication of the European Society of Clinical. Microbiology. (2017) 36:2335–42. doi: 10.1007/s10096-017-3065-x
Keywords: irritable bowel syndrome, inflammatory bowel diseases, calprotectin, mycobiome, Blastocystis , Candida albicans , Saccharomyces cerevisiae
Citation: Khosravany Z, Khodavaisy S, Olyaiee A, Sadeghi A, Nemati S, Shahrokh S, Mohammad Ali Gol S, Shojaei S, Mohammad Rahimi H and Mirjalali H (2025) A preliminary study of the association between Blastocystis and quantification of selected yeasts in IBD and IBS patients. Front. Med. 12:1514587. doi: 10.3389/fmed.2025.1514587
Edited by:
Ponsiano Ocama, Makerere University, UgandaReviewed by:
Daniela Elena Serban, University of Medicine and Pharmacy Iuliu Hatieganu, RomaniaJulia María Alatorre Cruz, Meritorious Autonomous University of Puebla, Mexico
Copyright © 2025 Khosravany, Khodavaisy, Olyaiee, Sadeghi, Nemati, Shahrokh, Mohammad Ali Gol, Shojaei, Mohammad Rahimi and Mirjalali. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hamed Mirjalali, aGFtZWRfbWlyamFsYWxpQGhvdG1haWwuY29t; aGFtZWRtaXJqYWxhbGlAc2JtdS5hYy5pcg==
 Zohre Khosravany
Zohre Khosravany Sadegh Khodavaisy
Sadegh Khodavaisy Alireza Olyaiee
Alireza Olyaiee Amir Sadeghi
Amir Sadeghi Sara Nemati
Sara Nemati Shabnam Shahrokh
Shabnam Shahrokh Sara Mohammad Ali Gol5
Sara Mohammad Ali Gol5 Sajad Shojaei
Sajad Shojaei Hamed Mirjalali
Hamed Mirjalali