
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Med. , 10 March 2025
Sec. Hepatobiliary Diseases
Volume 12 - 2025 | https://doi.org/10.3389/fmed.2025.1513233
This article is part of the Research Topic Genomic Dissection and Therapy Development in Liver Cancer View all articles
Background: Hepatocellular carcinoma (HCC) is the third most prevalent cause of cancer-related mortality globally and the sixth most common cancer overall. It is critical to investigate new biomarkers and prognostic variables because there are currently no early diagnostic indicators. Actin-related proteins (ARPs) are involved in transcriptional regulation, chromatin remodeling, and DNA repair—all processes that have been connected to the development of cancer. However, it’s still unclear how ARPs and HCC are related.
Methods: Through the examination of databases like The Cancer Genome Atlas (TCGA) and The International Cancer Genome Consortium (ICGC), we examined the variations in the expression of ARPs between the transcriptomes of normal tissue and HCC. Furthermore, univariate and multivariate Cox analysis were used to assess the prognostic effects of ARPs. The investigation of immune cell infiltration and possible functional enrichment followed. Additionally, tissue chips containing regional liver cancer specimens were used to confirm ACTR6 expression and the clinical impact of prognosis using an immunohistochemistry (IHC) test. Finally, to investigate the expression and function of ACTR6 in liver cancer cells, real-time qPCR (RT-qPCR) assays, CCK-8, clone creation, cell cycle, and transwell migration and invasion experiments were carried out.
Results: We found that, in addition to ACTR3C, 17 ARPs were significantly overexpressed in HCC compared with normal tissues. In both univariate and multivariate Cox models, ACTR6 and ACTL6A were identified as potential independent risk factors for the prognosis of HCC, with ACTR6 having the lowest p-value. Clinical samples also confirmed this conclusion. Furthermore, ACTR6 overexpression showed a strong connection with immune cell infiltration levels and clinical and pathological factors linked to a poor prognosis. Functionally, knocking down ACTR6 inhibited cell migration and proliferation, produced a G1 cell cycle arrest, and decreased the viability of liver cancer cells.
Conclusion: These findings demonstrate that ACTR6 is highly expressed in HCC and is associated with poor prognosis. In addition, ACTR6 may induce immune cell infiltration and promote hepatocarcinogenesis by regulating the cell cycle.
In 2020, hepatocellular carcinoma (HCC) accounted for approximately 906,000 new cases and 830,000 deaths worldwide, making it the sixth most prevalent cancer and the third highest cause of cancer-related deaths (1). Due to inadequate therapy, the 5-year overall survival of HCC patients is less than 20% (2, 3). In developing countries, the prevalence of HCC and associated cancer deaths is rising (4). More than a million individuals will pass away from HCC in 2030, according to a World Health Organization report (5). The pathophysiology of hepatocellular carcinoma (HCC) is complex, including fibrosis, liver inflammation, and abnormal hepatocyte regeneration. At the moment, surgery, chemotherapy, radiation therapy, and targeted therapy are the primary clinical therapies for liver cancer (6). Surgery is the most effective treatment for people with early-stage liver cancer when compared to other options. However, the majority of patients were detected with advanced liver cancer, losing the best opportunity for therapy since they lack diagnostic markers and show no apparent symptoms in the early stages (7). Thus, finding precise biomarkers for early diagnosis and efficacious treatment approaches is of crucial clinical importance.
Actin is widely recognized for its capacity to control membrane development, shape the cytoplasm, and guarantee the cytoskeleton’s dynamic behavior (8). Its activity is accomplished in collaboration with actin-related proteins (ARPs), a class of proteins that have structural similarities with actin. The actin superfamily, which is made up of both conventional actin and ARPs, is a highly conserved and ancient collection of proteins. Reuniting and expanding the identification and categorization of ARPs, Muller et al. (9) used a comparative genomic study of around 700 protein sequences. The ARP subfamily was split into Arp1 ~ 11, with Arp1 being the most closely linked to actin and Arp10 and Arp11 being the least (9), based on sequence consistency and similarity with typical actin. With the continuous update of understanding, ARPs family species are divided into more and more detailed. We found 18 members of the ARPs family, including ACTR1A/ACTR1B/ACTR2/ACTR3/ACTR3B/ACTR3C/ACTR5/ACTR6/ACTR8/ACTR10/ARPC1A/ARPC2/ARPC3/ARPC4/ARPC5/ARPC5L/ACTL6A/ACTL8.
Arps and actin exhibit significant sequence identity and similarity, indicating that the two proteins may share a tertiary structure that is based on a highly conserved ATPase domain—an ATP/ADP-binding pocket called the “actin fold” (9). They play a role in DNA repair, transcription control, and chromatin modification. The significance of ARPs in the development of cancer has progressively come to light recently. ACTL6A has been shown to be essential in controlling the glutathione (GSH) metabolic pathway because it increases the production of gamma-glutamylcysteine ligase catalytic subunits (GCLC). This reduces reactive oxygen species (ROS) levels and prevents iron mortality in gastric cancer cells (10). Furthermore, ACTR2 induced Wnt signaling in diffuse large B-cell lymphoma (DLBCL) and used Wnt signaling to cause DLBCL to proliferate both in vitro and in vivo (11). In addition, as early as 2020, a study on non-small cell lung cancer found that ACTR6 affects the progression of the disease by influencing the biology of tumor-associated macrophages (TAM), and served as a potential prognostic marker for lung cancer (12). In 2022, a study showed that ACTR6 is also one of the prognostic risk model genes for HER2+ breast cancer (13).
However, we do not yet understand how ARPs affect the development of liver cancer. In this study, the ARP gene family was used as a breakthrough point to discuss ACTR6’s transcriptome, genomics, immune infiltration, and prognosis in liver cancer. Immunohistochemistry (IHC) was applied to validate the expression of ACTR6 in liver cancer. The physiologic roles of ACTR6 in liver cancer cells were investigated using real-time qPCR (RT-qPCR) assays, CCK-8, clone creation, cell cycle, and transwell migration and invasion tests. Our findings will provide additional information on the significance of ARPs, especially ACTR6, in liver cancer progression.
The International Cancer Genome Consortium (ICGC)1 and TCGA2 provided the HCC expression and clinical prognostic data for the HCC tissue samples. There were 371 HCC tissue samples and 50 normal liver tissue samples in the TCGA dataset. Additionally, the ICGC provided information on 202 normal tissues and 243 HCC tissues.
OS was chosen as the survival outcome since the majority of HCC patients had a poor prognosis. Using a univariate Cox analysis, we eliminated ARPs that were connected to survival (p < 0.05). Furthermore, multivariate Cox analysis was used for analyzing the ARPs significantly related to OS and its relationship with clinicopathological factors. Besides, OS and progression-free survival (PFS) time were calculated through Kaplan–Meier plotter.3
We initially analyzed 22 different immune cells in each normal and HCC sample using the CIBERSORT algorithm in R language. Next, we used TIMER, a platform accessible at https://cistrome.shinyapps.io/timer/, to evaluate ACTR6 expression in HCC, as well as its correlation with immune cell numbers and its impact on immune cell markers. The CIBERSORT and TIMER algorithms were used to assess the variations in immune cell infiltration or immunological responses between the ACTR6 high- and low-expression groups. Heatmaps with various methods showed the variations in immune cell infiltration.
The LinkedOmics database4 provided a total of 8,599 co-expression genes. Then we were able to access the target gene ACTR6’s correlation gene table and correlation volcano map on the LinkFinder plate. Heatmaps of the top 50 genes that are positively and negatively correlated with ACTR6 were also acquired.
A characteristic gene list of the cell cluster, or the most important genes of ACTR6, was uploaded to the Database for Annotation, Visualization, and Integrated Discovery (DAVID, v6.8). Homo sapiens was chosen as the species and the official gene symbol as an identification. Ultimately, enrichment findings were acquired using Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis and Gene Ontology (GO) analysis. In this study, the top six outcomes were shown in increasing order of p-value (p < 0.05).
Genes associated with the cell cycle were compiled using the AmiGO 2 portal.5 Every HCC sample’s functional enrichment score was determined using the provided package (R environment) and default settings. Using the pheatmap package (R environment), a heatmap of the enrichment results was created. Pearson correlation analysis was used to ascertain the relationship between ACTR6 and cell cycle.
Huh7, HepG2, and Hep3B were human HCC cell lines obtained from the Chinese Academy of Sciences Cell Bank. These cell lines were maintained in DMEM (Invitrogen, # 11965-118; CA) with 1% penicillin–streptomycin and 10% FBS added. We acquired the human normal liver cell line Thle-2 from Keycell Biotechnology (Wuhan, China). The Thle-2 cells were kept alive in a unique growth medium that Keycell Biotechnology supplied. The cells were cultivated at 37°C with 5% CO2 in a humidified incubator. Every 3 months, the absence of mycoplasma infection was confirmed in all cell lines. Shanghai Outdo Biotech Co., Ltd. provided the formalin-fixed paraffin-embedded HCC cancer tissue microarrays (HLivH180Su30).
The rabbit polyclonal antibody against ACTR6 (Catalog # PA5-58453) was from Thermo Fisher Scientific (Waltham, MA). ImmPRESS Universal Polymer Reagent (Horse Anti-Mouse/Rabbit IgG) was from Vector Laboratories (S.F, CA).
SiRNAs were transfected using the lipofectamine 3000 Transfection Kit (ThermoFisher Scientific, #L3000-015) after being obtained from Hippobio in Huzhou, China. CCGAGAUAAUCCUUCCGAAUUTT (ACTR6 siRNA1), GCCUGACUUCAGUACAAUUAATT (ACTR6 siRNA2), and GCACAUAGGUAUUUCCGAGAUTT (ACTR6 siRNA3) were the siRNA sequences aimed against ACTR6.
Following the manufacturer’s instructions, total RNA was isolated from cultivated cells using the FastPure® Cell/Tissue Total RNA Isolation Kit V2 (Vazyme, # RC112; Nanjing, China). Using a StepOnePlusTM Real-Time PCR System (ThermoFisher Scientific), 15 ng of cDNA that had been generated from 900 ng of total RNA using the PrimScriptTM RT reagent Kit with gDNA Eraser (TaKaRa, #RR047A; Dalian, China) was exposed to qPCR. The relative gene expression levels were normalized against β-actin using the 2−ΔΔCT technique. Supplementary Table S1 has a listing of primer sequences.
Five thousand cells per well of 96-well plates were used to plate Huh7 cells transfected with NC or siACTR6. At the 24-, 48-, and 72-h mark, CCK-8 (DOJINDO, # CK04, Japan) was added, and the mixture was incubated for a further 2 h. Then, the plates were assayed by testing the absorbance at 450 nm.
One thousand transfected cells were cultivated for 10–14 days at 37°C with 5% CO2 after being seeded onto 6-well plates. After discarding all liquids, the samples were fixed for 30 min at room temperature in 4% paraformaldehyde (Servicebio, #G1101; Wuhan, China), and then stained with crystal violet (Beyotime, #C0121; Beijing, China). Bio-Rad’s Quantity One® software was used to take counts and photos.
Serum-free media was used to sustain the cells after they were planted into the top chambers of 24-well transwell inserts (8 μm, Corning, #3422, 5 × 104 cells/well). The medium was added to the bottom chambers along with 10% FBS. Following a 16-h incubation period, the cells grown in inserts were fixed for 30 min using 4% paraformaldehyde (Servicebio, #G1101; Wuhan) and stained for 8 min with crystal violet dye solution (Beyotime, #C0121; Beijing, China). Using a cotton-tipped swab, the non-migrated cells on the membranes’ top surface were scrubbed away. Before being quantified using the ImageJ program, cells that moved to the membranes’ bottom surface were further photographed using an Olympus IX73 Fluorescence Microscope System.
Using inserts that had been pre-coated with a suitable quantity of Matrigel (BD Biosciences, # 356234; Franklin Lakes, NJ) combined with precooled media at a ratio of 1:8, cell invasion was measured. The cells were cultured in serum-free media and seeded into the top chambers of 24-well transwell inserts (8 μm, Corning, #3422, 1 × 105 cells/well). The next experiment followed a similar protocol to the transwell migration test previously reported.
The anti-ACTR6 antibody (dilution ratio, 1:200) was incubated for 10 h at 4°C on HCC and nearby tissues. After that, the tissues were treated for 30 min at 37°C with Horse Anti-Mouse/Rabbit IgG. The samples were then treated with DAB Peroxidase Substrate Kit for the reaction, and hematoxylin was used as a counterstain. Two different, skilled pathologists evaluated each IHC staining; they were unaware of the patient’s clinical status or diagnosis beforehand. ACTR6 staining score, including staining density and intensity. The scoring methods were as follows: (1) The percentage of staining positive cells was divided into 5 grades, namely <1%, 1–25%, 26–50%, 51–75%, and 76–100%, which were defined as 0, 1, 2, 3, and 4 points respectively; (2) The staining intensity was divided into 4 grades: negative staining (no staining, 0 points), weak staining intensity (light brown, 1 point), medium staining intensity (brown, 2 points), and strong staining intensity (dark brown, 3 points). (3) Immunoreactive score (IRS) = percentage of positive cells × staining intensity score, with a total score of 0 ~ 12, in which 0 ~ 5 is defined as low expression, and 6 ~ 12 is defined as high expression.
For all statistical analyses, SPSS 25.0 (SPSS Software, Chicago, United States), GraphPad Prism 9and R v 4.3.26 were used. For statistical analysis, the Student’s t test, Mann–Whitney test, one-way analysis of variance (ANOVA) test, or Pearson correlation coefficient were used. In addition, we investigated the connection between genes and illness prognosis using COX analysis.
ARPC3/ARPC5L/ARPC5/ARPC2/ARPC4/ARPC1A/ACTL8/ACTL6A/ACTR6/ACTR5/ACTR8/ACTR1B/ACTR3/ACTR10/ACTR3B/ACTR2/ACTR1A were significantly overexpressed in HCC tissues more than in normal liver tissues, according to a combination of the analysis results of The Cancer Genome Atlas (TCGA) and International Cancer Genome Consortium (ICGC) databases (Figure 1; Supplementary Figure S1).
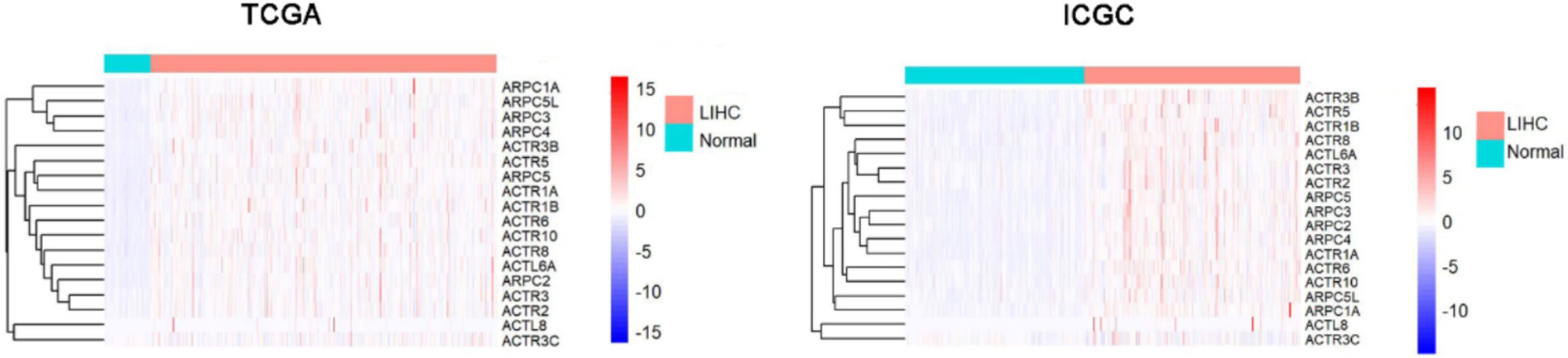
Figure 1. ARPs family transcriptional levels in TCGA and ICGC comparing HCC to normal tissues. HCC, hepatocellular carcinoma; ICGC, The International Cancer Genome Consortium; ARP, actin-related proteins; TCGA, The Cancer Genome Atlas.
Further investigation into the relationship between ARPs expression and patient OS in the TCGA and ICGC databases revealed that, according to Univariate analysis, shorter OS in HCC was correlated with higher levels of ARPC2/ARPC3/ARPC4/ARPC5L/ACTR6/ACTL6A mRNA expression (Figures 2A,B), as well as with T stage, pathological stage, and tumor status (Figure 2C). Further multivariate analysis revealed that longer OS in HCC patients was correlated with higher ACTL6A/ACTR6 mRNA expression (Supplementary Figure S2). This suggests that ACTL6A/ACTR6 may be independent risk factors for predicting the prognosis of HCC patients, with ACTR6 showing the lowest p-value of 0.001 in multivariate Cox analysis (Figure 2D). Additionally, we investigated the survival data for ACTR6 expression in HCC using a Kaplan–Meier plotter (Figures 2E,F).
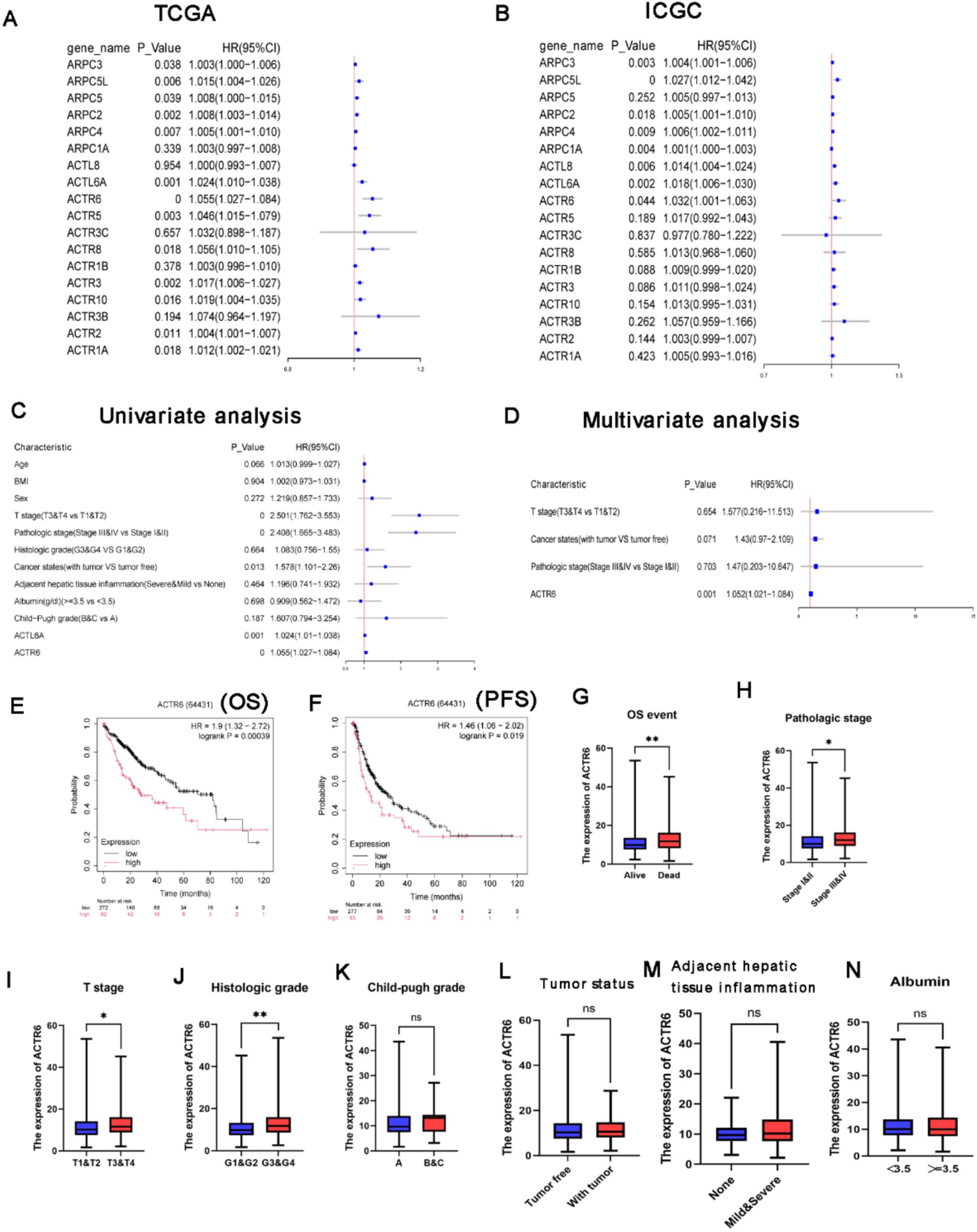
Figure 2. Value of distinct ARPs mRNA expressions for prognosis in hepatocellular cancer. (A,B) In HCC patients, there was a correlation between the mRNA expression level of ARPs and OS. ACTR6 and patient OS were analyzed using univariate (C) and multivariate (D) Cox regression. The association between ACTR6 and several clinicopathological variables as determined by the Student’s t test. In patients with HCC, shorter overall (E) and progression-free survival (F) times were associated with ACTR6 mRNA expression. Events related to OS (G), pathologic stage (H), T stage (I), histologic grade (J), child-pugh grade (K), tumor states (L), inflammation of the surrounding hepatic tissue (M), and albumin (N).
Based on the research mentioned above, it was shown that ACTR6 may be used as an independent prognostic factor to predict the prognosis of patients with HCC. After examining the relationship between ACTR6 expression and clinicopathological features of HCC patients, we found that ACTR6 expression was higher in deceased patients (Figure 2G) and higher in pathological stages (Figure 2H), T stages (Figure 2I) and histological grades (Figure 2J). Nevertheless, increased child-pugh grade (Figure 2K), tumor status (Figure 2L), surrounding hepatic tissue inflammation (Figure 2M), and albumin (Figure 2N) were not linked to increased expression of ACTR6.
We selected 8,599 DCGs from the LinkedOmics database, including 5,969 positively correlated and 2,630 negatively correlated genes, in order to investigate the possible mechanism by which ACTR6 operates in HCC (Figure 3A). The top 50 DCGs that are correlated with ACTR6 were shown on heatmaps (Figures 3B,C). To investigate the biological activities associated with ACTR6, the TCGA databases’ most closely related genes were eliminated by Pearson correlation analysis (|R| > 0.4, p < 0.05). In order to examine the important biological processes and functions, these linked genes were selected for GO and KEGG enrichment analysis. The GO analysis found that the DCGs were mostly involved in the cell division, mRNA splicing, and ribosomal small subunit biogenesis at the biological process level (Figure 3D). The cellular component enrichment study indicates that the most enriched category is nucleoplasm (Figure 3E). The most enriched categories at the molecular function level were RNA binding and protein binding (Figure 3F). The KEGG enrichment analysis showed that the cell cycle pathway was most significantly regulated by DCGs (Figure 3G). These findings suggested that ACTR6 on HCC probably plays an essential role in cell cycle.
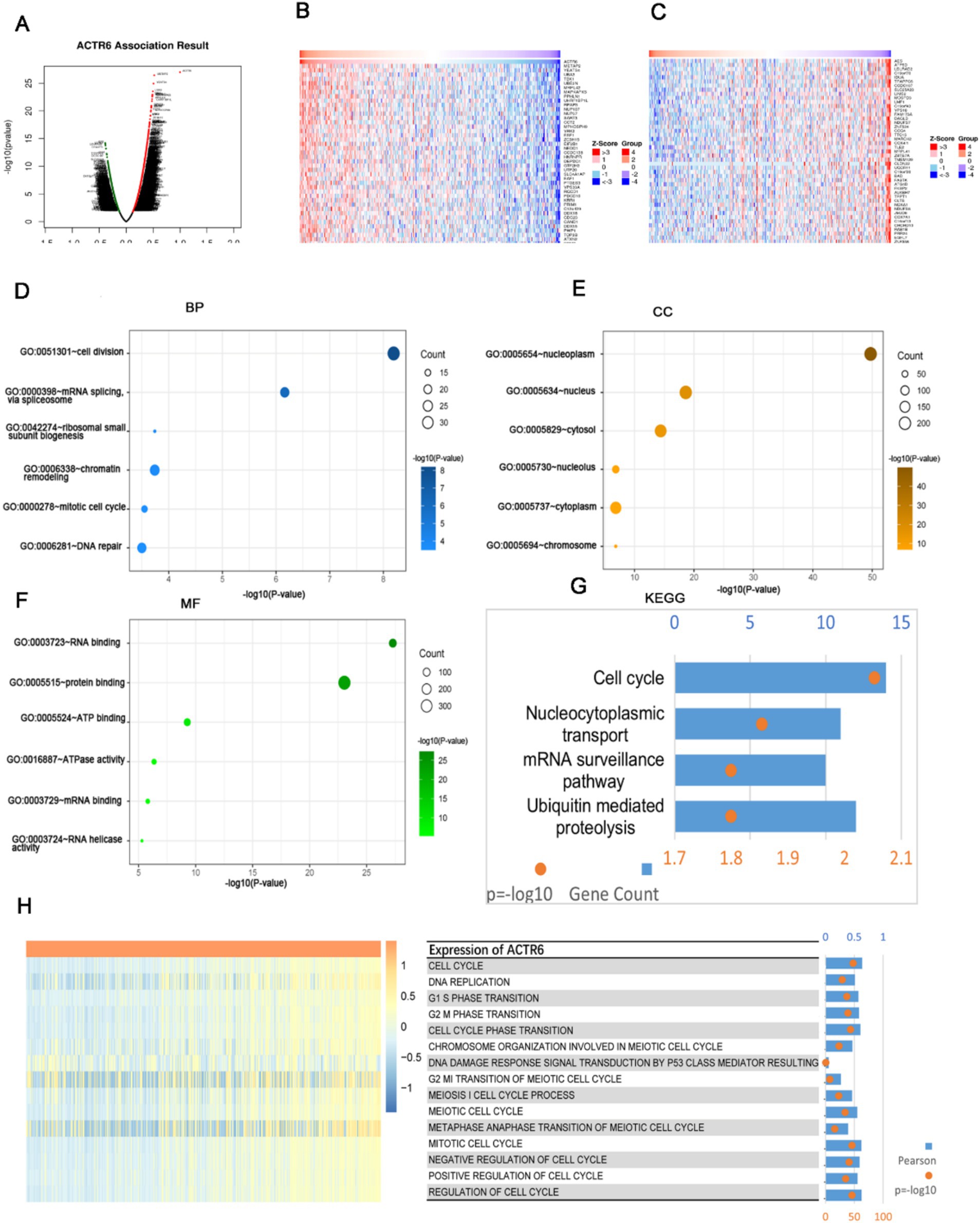
Figure 3. Analysis of functional enrichment of ACTR6 genes that are differently co-expressed in HCC. (A) Volcano plots of ACTR6 genes that are differently co-expressed. The top 50 were shown both negatively (C) and positively (B) on the heatmap. In HCC, ACTR6 is intimately related to the cell cycle. (D–F) The TCGA database links ACTR6 to biological processes (BP), cellular components (CC), and molecular functions (MF). (G) ACTR6 pathway analysis from the Kyoto Encyclopedia of Genes and Genomes (KEGG) in the TCGC database. (H) Pearson connection between cell cycle (GSVA) and ACTR6. The R-value was expressed by the band’s width.
The TCGA databases’ gene set variation analysis was utilized to calculate the cell cycle’s enrichment score. The enrichment score and ACTR6 expression were connected, and the results indicated that ACTR6 expression was positively correlated with most cell cycle events, with the exception of damage response signal transduction via p53 class mediator (Figure 3H). These findings revealed a connection between cell cycle and ACTR6.
The development and spread of HCC are significantly influenced by immune infiltrations (14). We assessed the degrees of immune cell infiltration in both normal and HCC patients using the CIBERSORT and TIMER algorithms. The findings demonstrated that HCC patients had a greater degree of immune cell infiltration than did healthy individuals (Figure 4A). Furthermore, among patients with HCC, the group exhibiting high expression of ACTR6 had a greater amount of immune cell infiltration (Figure 4B). ACTR6 was positively correlated with Eosinophils, Macrophages M0, T cells CD4 memory activated, etc., and negatively correlated with T cells CD4 naive, NK cells activated, etc. (Figure 4C). Additionally, we employed TIMER online analysis to examine the correlation between ACTR6 expression and six types of immunological infiltrations (Figure 4D). To better understand the relationship between ACTR6 expression and immune infiltration, we also used the TIMER database to investigate the relationship between ACTR6 and immune cell markers. Following purity-based correction, we found that most immune cell indicators positively correlated with ACTR6 expression (Table 1).
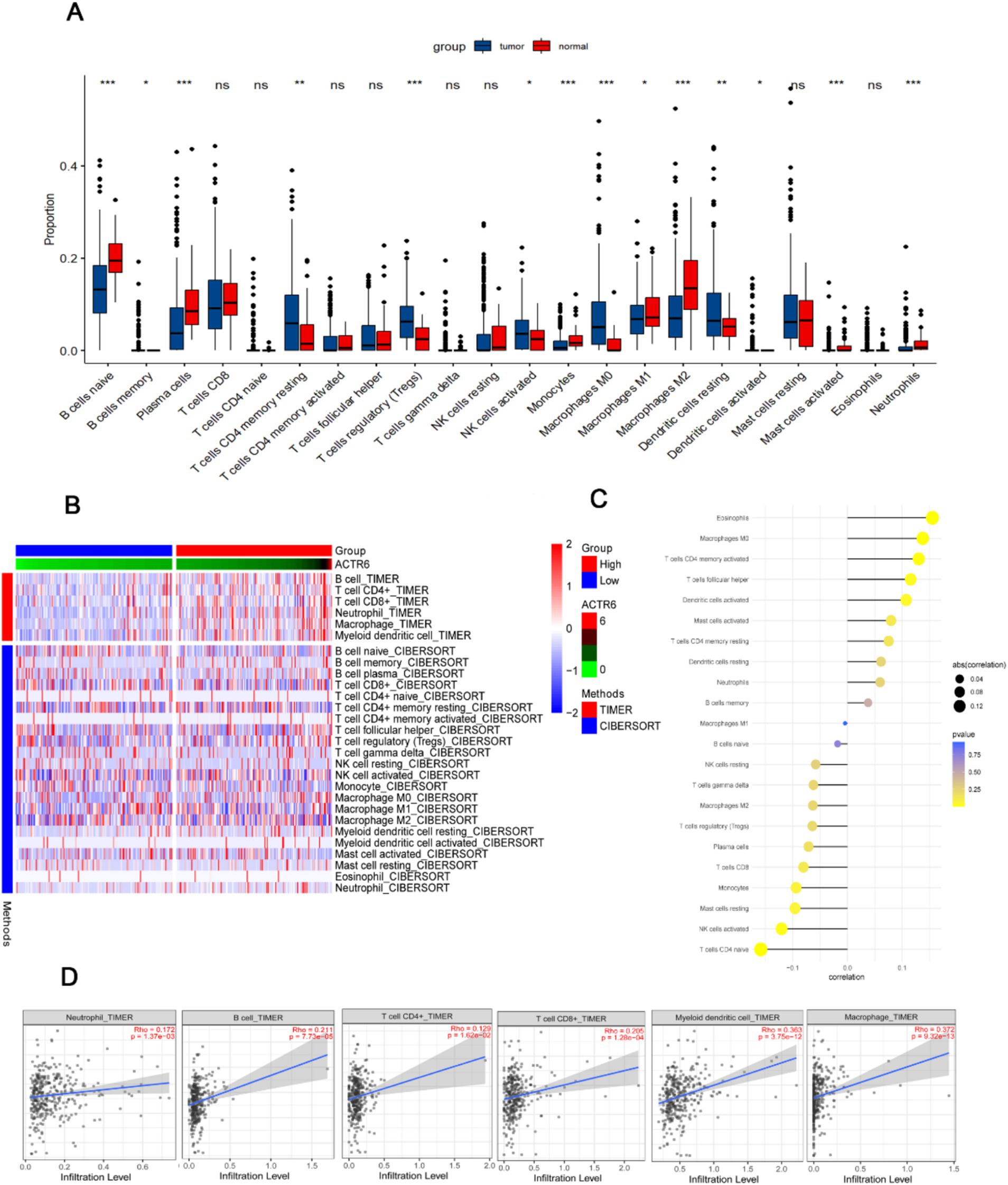
Figure 4. Relationship between immune infiltration in HCC and ACTR6 mRNA expression. (A) The comparison of estimated fractions of 22 immune cells between the normal and HCC patients. (B) The heat map illustrates how immune cells infiltrate differently in HCC samples with high and low ACTR6 expression. (C) The Lollipop graph illustrates the relationship between immune cells and ACTR6 expression. (D) The TIMER online analysis revealed a favorable correlation between immune cell expression and ACTR6 expression levels.
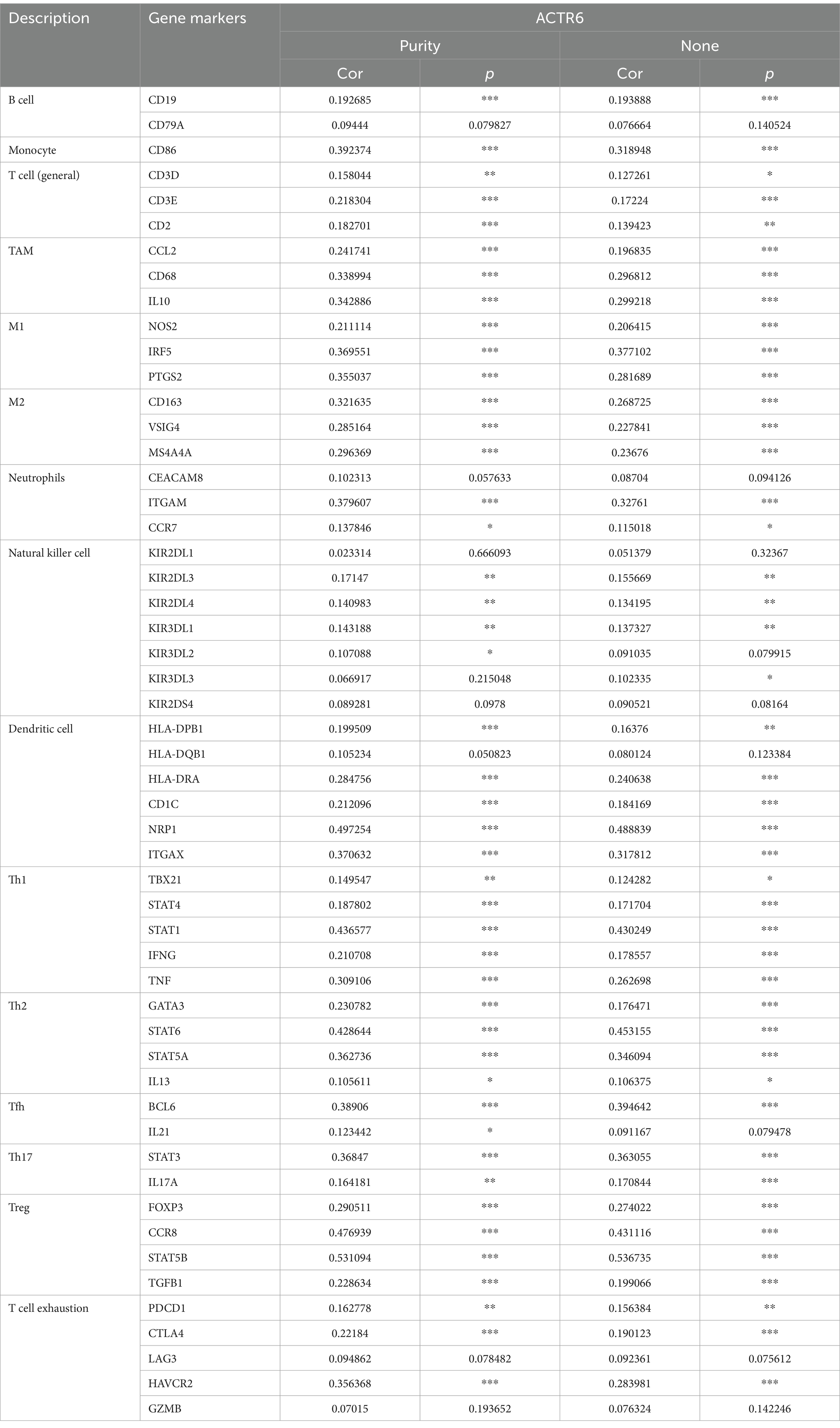
Table 1. Analysis of the correlation between ACTR6 and related immune cell genes and markers in TIMER.
Through RT-qPCR we found that the expression level of ACTR6 in the three HCC cell lines was higher than that in normal liver cells (Figure 5A) Furthermore, to confirm the expression of ACTR6 in hepatocellular carcinoma and its prognostic effect, we used immunohistochemical methods to detect the protein expression of ACTR6 in 90 pairs (six pairs of which were lost) of formalin-fixed paraffin-embedded (FFPE) liver tissue samples (Figure 5B). It is determined that hepatocellular carcinoma has greater levels of ACTR6 protein expression (Figure 5C). However, the high expression of ACTR6 does not appear to be related to various clinicopathological factors (Table 2), which may be due to insufficient sample size. Further survival analysis showed that the higher the expression of ACTR6 in liver cancer tissues, the shorter the patients’ OS (Figure 5D). In addition, multivariate Cox regression analysis also showed that high expression of ACTR6 was correlated with OS (Table 3). Taken together, these results suggest that ACTR6 expression is up-regulated in HCC, and its high expression is associated with poor prognosis in HCC patients.
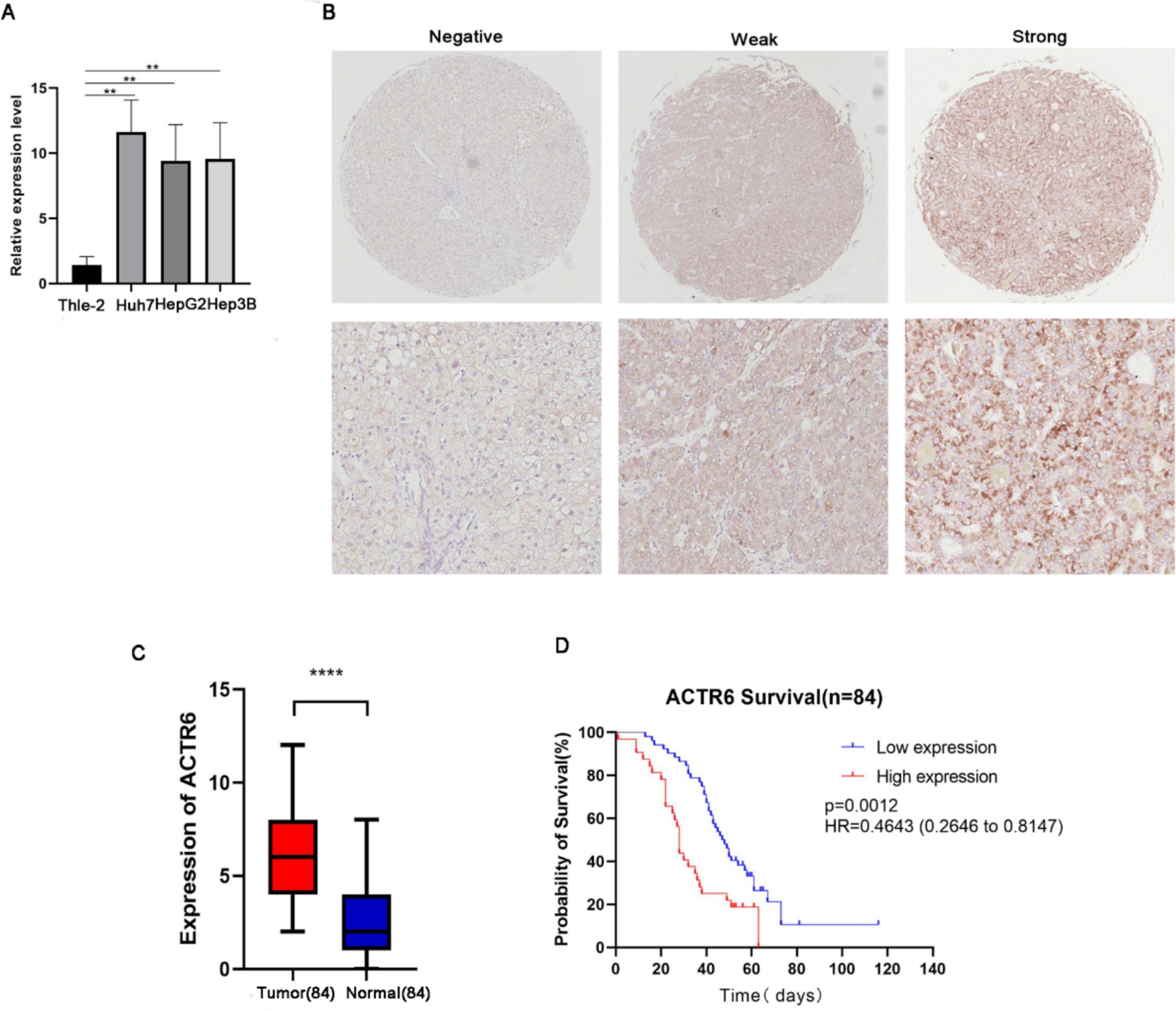
Figure 5. The expression of ACTR6 in normal liver cell line and liver cancer cell line Thle-2 was detected by RT-PCR (A). A comparison of paired neighboring normal tissues and ACTR6 expression in formalin-fixed paraffin-embedded (FFPE) hepatocellular carcinoma (n = 84 physiologically independent samples) using representative microphotographs (B) and a student’s t test analysis (C). (D) Kaplan–Meier analysis of the overall survival probability of 84 HCC patients, stratified by ACTR6 expression.
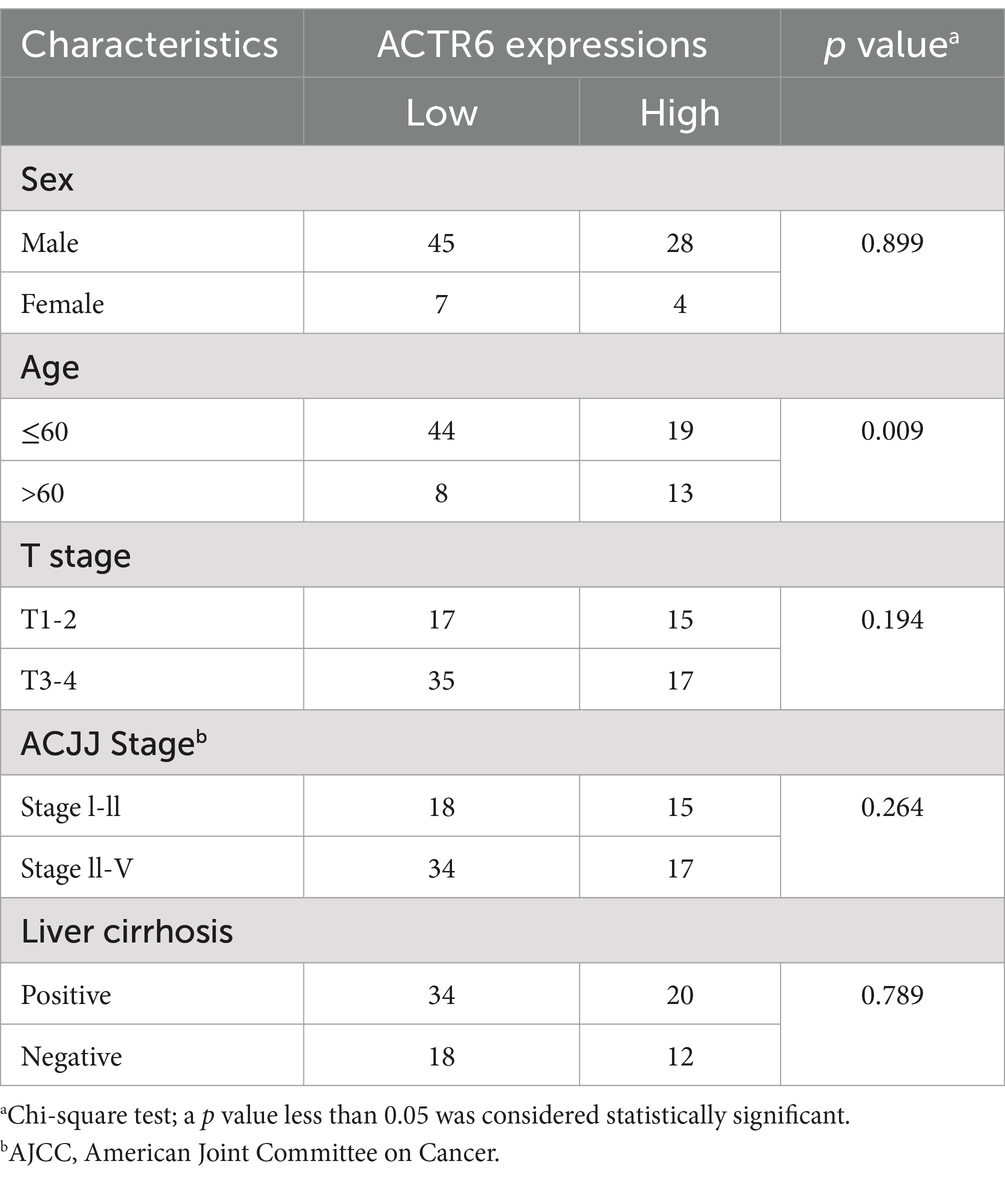
Table 2. Relationship between ACTR6 expression and clinicopathological features of hepatocellular carcinoma patients.
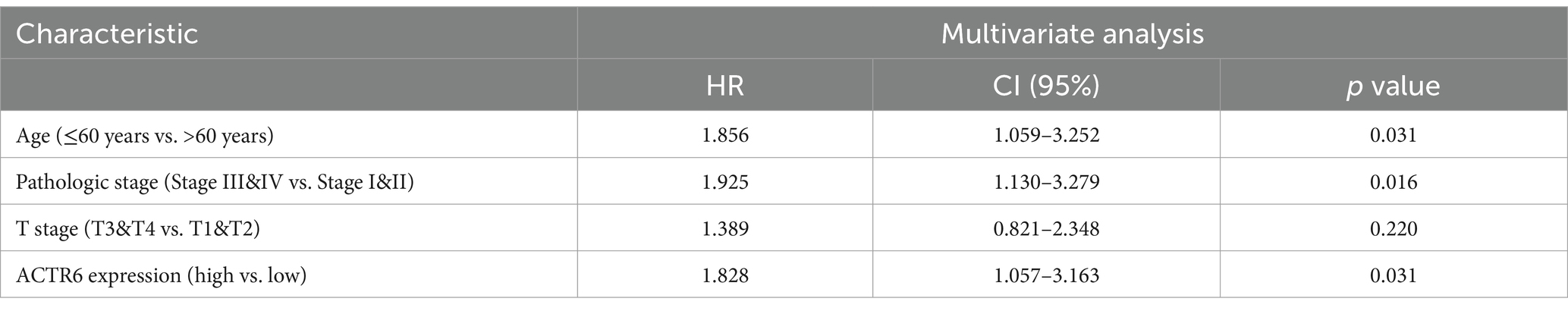
Table 3. Multifactorial analysis affecting the overall survival of patients with hepatocellular carcinoma.
Two significant indicators of the development of cancer are the modification of the extracellular matrix and dysregulated cell cycle (14). We tried to ascertain ACTR6’s function in controlling cell migration and proliferation based on the GSVA results. We used three separate siRNAs to knock down ACTR6 in Huh7 and HepG2 cells in order to functionally investigate the function of this gene in controlling the development of hepatocellular carcinoma (Figures 6A,B). We discovered that ACTR6 knockdown markedly reduced cell proliferation through investigations on clonal formation and cell proliferation (Figures 6C–F). Furthermore, transwell experiment demonstrated a substantial reduction in cell migration following ACTR6 siRNA transfection (Figures 6G,H). Ultimately, we demonstrated by flow cytometry that ACTR6 knockdown resulted in G1 cell cycle arrest (Figure 6I).
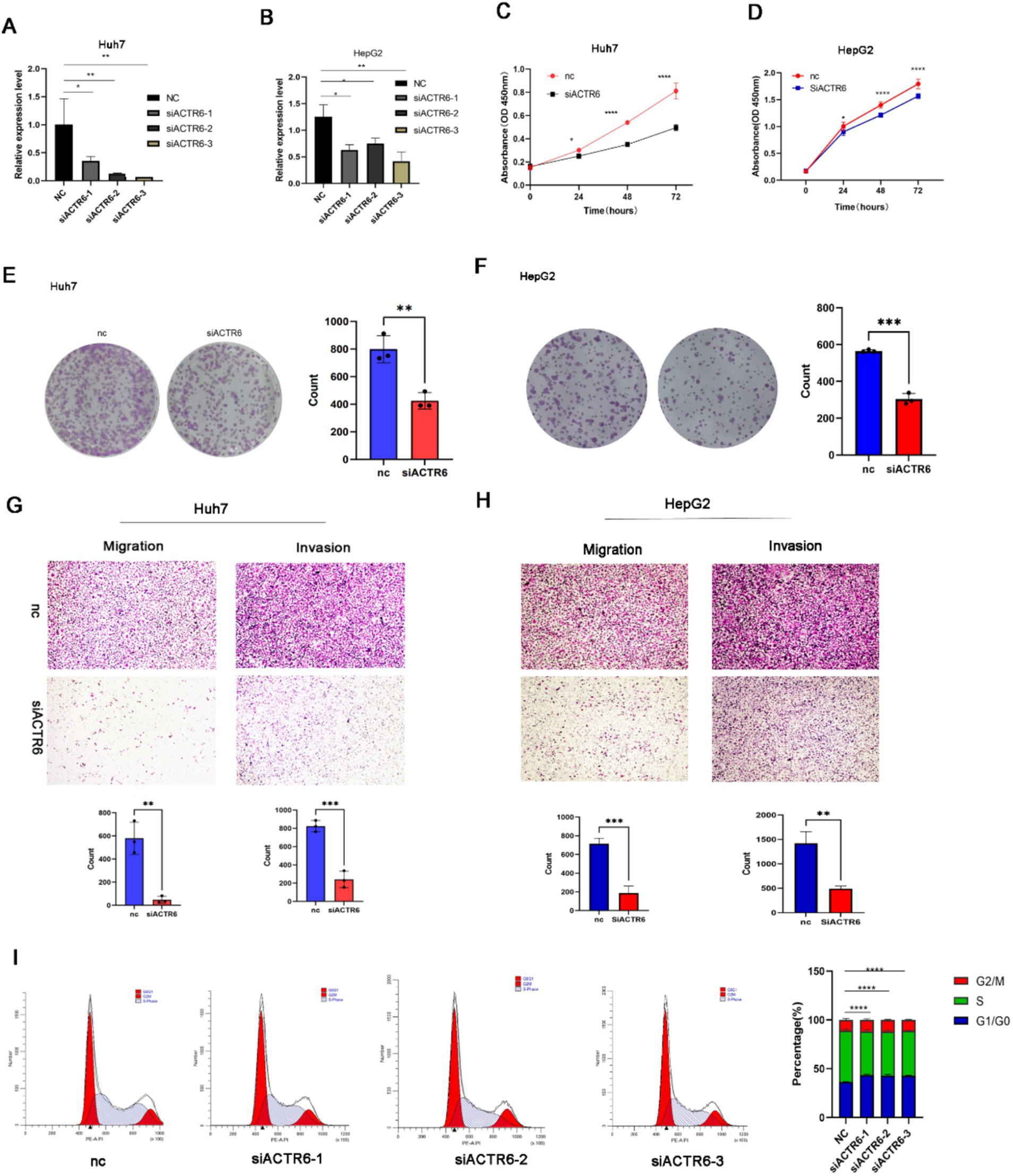
Figure 6. ACTR6 knockdown inhibited cell migration and proliferation. SiRNA suppression of ACTR6 (A,B) reduced proliferation (C,D) and clonal formation (E,F) as well as migration and invasion (G,H) of Huh7 and HepG2 cells. Scale bars, 200 μm (cell migration and invasion assay). Using the ImageJ program, the numbers of clonal formation, migratory, and invasive cells were quantified. After 24 h, Huh7 cells had a G1 cell cycle arrest due to ACTR6 knockdown (I).
Many factors, such as obesity, type 2 diabetes, metabolic syndrome, and nonalcoholic fatty liver disease (NAFLD), contribute to the development of HCC. It is still among the most deadly malignant tumors in the world (15). Abnormal malignant cell activities such as proliferation, migration, invasion, and autophagy are associated with the formation and progression of HCC (16, 17).
ARPs are necessary for the function of chromatin remodeling complexes (18). Previous studies have shown that Arp4–Arp9 are found in chromatin remodeling and histone modification complexes (19). They play a role in DNA repair, transcription control, and chromatin modification (20). For instance, ACTL6A, also referred to as Arp4, is involved in both the activation and repression of transcription from genes (21–24). ACTR5 and ACTR8, also referred to as Arp5 and Arp8, are particular INO80 complex components that are preserved. Both of them are mainly found within the INO80 complex in yeast. They are essential to the enzymatic activity of INO80 (25). Unlike other nuclear Arps, which are primarily associated with transcriptional activation, ACTR6, also known as Arp6, is required to maintain gene silence in heterochromatin. Research has demonstrated that human and chicken ACTR6 directly interact with heterochromatin protein 1 (HP1) in vitro (26, 27). Furthermore, the vertebrate ACTR6 is necessary for the SRCAP complex to function (18). The histone variation H2A.Z is incorporated into nucleosomes at promoter areas by the SRCAP complex. Via transcriptional control and chromosomal architecture, H2A.Z is linked to several physiological processes, such as stem cell maintenance, cellular proliferation, and stress response (27–29). It also plays significant roles in epigenetic regulation and chromosome architecture.
However, the involvement of ARP family proteins in HCC has not been completely investigated. We are in charge of this investigation of the expression, mutation, and prognostic significance of different ARP family members in HCC. After analyzing the TCGA and ICGC datasets, we discovered that patients with HCC had highly overexpressed versions of 17 ARPs. A favorable correlation was found between short OS and overexpression of ARPC2/ARPC3/ARPC4/ARPC5L/ACTR6/ACTL6A. Additional multivariate analysis showed that ACTR6/ACTL6A may function as separate risk variables for the prognosis of patients with colorectal cancer. We confirmed the significant expression of ACTR6 in liver cancer cells and liver tumor tissues, as well as its impact on the prognosis of liver cancer patients, using cell assays and immunohistochemistry analysis. Next, we looked at ACTR6’s involvement in liver cancer in more detail. On the one hand, aberrant proliferation of malignant tumors is primarily caused by dysregulation of the cell cycle. In order to estimate ACTR6’s biological role, we used GO, KEGG, and GSVA analysis. This revealed that ACTR6 may control the cell cycle, which in turn controls cell migration and proliferation. The next functional tests confirmed that ACTR6 knockdown inhibited cell proliferation by causing a G1/S cell cycle arrest as well as limiting cell migration. Cancer is characterized by persistent proliferative signaling, which promotes unending and excessive cycles of cell division. It has recently become clear that, rather than causing unchecked cell cycle advancement, these mutations that impede cell cycle exit and prevent apoptosis are what propel this uncontrollably dividing cell. These include mutations in the signaling pathways that trigger cell cycle exit or encourage entrance into the S phase (30), although they are significantly less common in the pathways that obstruct mitotic entry (31–34) and exit (35–39). On the other hand, it is thought that a crucial factor contributing to the development of HCC is the chronic inflammatory response (40–42). Our research revealed a significant association between ACTR6 and immune cells, particularly in macrophage and dendritic cells. Through the upregulation of proinflammatory cytokines and immunological checkpoints, immune cells aid in the development of tumors by immune escape (43).
Due to the limitations of our work, additional research is required to explore the possible molecular mechanism behind the carcinogenesis of ARP family proteins. However, we present the first information on the ARPprotein family’s differential expression in HCC, specifically in relation to ACTR6, as well as its possible diagnostic and prognostic significance. The expression of ACTR6 is strongly correlated with a bad prognosis in HCC. It can be explored as a potential target for HCC treatment and prognostic biomarkers. Preliminary mechanistic investigations have indicated that it increases HCC formation and progression via regulating cell cycle and immune cell infiltration. In the future, more in vivo experiments should be done to verify the expression of ACTR6 and further explore the specific mechanism of ACTR6 promoting the cell cycle conversion of liver cancer.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
The studies involving humans were approved by the Ethics Committee of Shanghai Xinchao Biotechnology Co. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
JW: Conceptualization, Data curation, Methodology, Writing – original draft. MS: Conceptualization, Data curation, Formal analysis, Methodology, Writing – original draft. JT: Conceptualization, Methodology, Writing – review & editing. HY: Methodology, Visualization, Writing – review & editing. XG: Data curation, Methodology, Writing – review & editing. ZC: Methodology, Writing – review & editing. XS: Methodology, Writing – review & editing. MC: Conceptualization, Methodology, Supervision, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This project was funded by the Henan Provincial Science and Technology Tackling Key Project (242102310230).
We thank all the volunteers who participated in this study and the Cancer Genome Atlas project.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declare that no Gen AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2025.1513233/full#supplementary-material
2. ^https://cancergenome.nih.gov/
3. ^http://kmplot.com/analysis/
4. ^https://www.linkedomics.org/login.php
1. Sung, H, Ferlay, J, Siegel, RL, Laversanne, M, Soerjomataram, I, Jemal, A, et al. Global Cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
2. Zeng, H, Chen, W, Zheng, R, Zhang, S, Ji, JS, Zou, X, et al. Changing cancer survival in China during 2003-15: a pooled analysis of 17 population-based cancer registries. Lancet Glob Health. (2018) 6:e555–67. doi: 10.1016/S2214-109X(18)30127-X
3. Siegel, RL, Miller, KD, Fuchs, HE, and Jemal, A. Cancer statistics, 2021. CA Cancer J Clin. (2021) 71:7–33. doi: 10.3322/caac.21654
4. Chen, W. Cancer statistics: updated cancer burden in China. Chin J Cancer Res. (2015) 27:1. doi: 10.3978/j.issn.1000-9604.2015.02.07
5. Villanueva, A. Hepatocellular Carcinoma. N Engl J Med. (2019) 380:1450–62. doi: 10.1056/NEJMra1713263
6. Chen, S, Cao, Q, Wen, W, and Wang, H. Targeted therapy for hepatocellular carcinoma: challenges and opportunities. Cancer Lett. (2019) 460:1–9. doi: 10.1016/j.canlet.2019.114428
7. Vogel, A, Cervantes, A, Chau, I, Daniele, B, Llovet, JM, Meyer, T, et al. Correction to: “hepatocellular carcinoma: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. (2019) 30:871–3. doi: 10.1093/annonc/mdy510
8. Pollard, TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. (2007) 36:451–77. doi: 10.1146/annurev.biophys.35.040405.101936
9. Muller, J, Oma, Y, Vallar, L, Friederich, E, Poch, O, and Winsor, B. Sequence and comparative genomic analysis of actin-related proteins. Mol Biol Cell. (2005) 16:5736–48. doi: 10.1091/mbc.e05-06-0508
10. Yang, Z, Zou, S, Zhang, Y, Zhang, J, Zhang, P, Xiao, L, et al. ACTL6A protects gastric cancer cells against ferroptosis through induction of glutathione synthesis. Nat Commun. (2023) 14:4193. doi: 10.1038/s41467-023-39901-8
11. Chen, D, and Jiang, L. Upregulation of actin-related protein 2 (ACTR2) exacerbated the malignancy of diffuse large B-cell lymphoma through activating Wnt signaling. Comput Math Methods Med. (2022) 2022:1–11. doi: 10.1155/2022/9351921
12. Zheng, X, Weigert, A, Reu, S, Guenther, S, Mansouri, S, Bassaly, B, et al. Spatial density and distribution of tumor-associated macrophages predict survival in non-small cell lung carcinoma. Cancer Res. (2020) 80:4414–25. doi: 10.1158/0008-5472.CAN-20-0069
13. Xu, J, Qin, S, Yi, Y, Gao, H, Liu, X, Ma, F, et al. Delving into the heterogeneity of different breast Cancer subtypes and the prognostic models utilizing scRNA-Seq and bulk RNA-Seq. Int J Mol Sci. (2022) 23:9936. doi: 10.3390/ijms23179936
14. Li, W, and Liu, J. The prognostic and immunotherapeutic significance of AHSA1 in Pan-Cancer, and its relationship with the proliferation and metastasis of hepatocellular carcinoma. Front Immunol. (2022) 13:845585. doi: 10.3389/fimmu.2022.845585
15. Kulik, L, and El-Serag, HB. Epidemiology and Management of Hepatocellular Carcinoma. Gastroenterology. (2019) 156:477–91.e1. doi: 10.1053/j.gastro.2018.08.065
16. Jiang, Y, Han, Q, Zhao, H, and Zhang, J. The mechanisms of HBV-induced hepatocellular carcinoma. J Hepatocell Carcinoma. (2021) 8:435–50. doi: 10.2147/JHC.S307962
17. Chidambaranathan-Reghupaty, S, Fisher, PB, and Sarkar, D. Hepatocellular carcinoma (HCC): epidemiology, etiology and molecular classification. Adv Cancer Res. (2021) 149:1–61. doi: 10.1016/bs.acr.2020.10.001
18. Schneider, R, and Grosschedl, R. Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes Dev. (2007) 21:3027–43. doi: 10.1101/gad.1604607
19. Oma, Y, and Harata, M. Actin-related proteins localized in the nucleus: from discovery to novel roles in nuclear organization. Nucleus (Austin, Tex). (2011) 2:38–46. doi: 10.4161/nucl.2.1.14510
20. Miralles, F, and Visa, N. Actin in transcription and transcription regulation. Curr Opin Cell Biol. (2006) 18:261–6. doi: 10.1016/j.ceb.2006.04.009
21. Harata, M, Zhang, Y, Stillman, DJ, Matsui, D, Oma, Y, Nishimori, K, et al. Correlation between chromatin association and transcriptional regulation for the Act3p/Arp4 nuclear actin-related protein of Saccharomyces cerevisiae. Nucleic Acids Res. (2002) 30:1743–50. doi: 10.1093/nar/30.8.1743
22. Görzer, I, Schüller, C, Heidenreich, E, Krupanska, L, Kuchler, K, and Wintersberger, U. The nuclear actin-related protein Act3p/Arp4p of Saccharomyces cerevisiae is involved in transcription regulation of stress genes. Mol Microbiol. (2003) 50:1155–71. doi: 10.1046/j.1365-2958.2003.03759.x
23. Steinboeck, F, Krupanska, L, Bogusch, A, Kaufmann, A, and Heidenreich, E. Novel regulatory properties of Saccharomyces cerevisiae Arp4. J Biochem. (2006) 139:741–51. doi: 10.1093/jb/mvj080
24. Lindstrom, KC, Vary, JC Jr, Parthun, MR, Delrow, J, and Tsukiyama, T. Isw1 functions in parallel with the NuA4 and Swr1 complexes in stress-induced gene repression. Mol Cell Biol. (2006) 26:6117–29. doi: 10.1128/MCB.00642-06
25. Shen, X, Ranallo, R, Choi, E, and Wu, C. Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol Cell. (2003) 12:147–55. doi: 10.1016/S1097-2765(03)00264-8
26. Frankel, S, Sigel, EA, Craig, C, Elgin, SC, Mooseker, MS, and Artavanis-Tsakonas, S. An actin-related protein in Drosophila colocalizes with heterochromatin protein 1 in pericentric heterochromatin. J Cell Sci. (1997) 110:1999–2012. doi: 10.1242/jcs.110.17.1999
27. Ohfuchi, E, Kato, M, Sasaki, M, Sugimoto, K, Oma, Y, and Harata, M. Vertebrate Arp6, a novel nuclear actin-related protein, interacts with heterochromatin protein 1. Eur J Cell Biol. (2006) 85:411–21. doi: 10.1016/j.ejcb.2005.12.006
28. Brickner, JH, and Walter, P. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. (2004) 2:e342. doi: 10.1371/journal.pbio.0020342
29. Schmid, M, Arib, G, Laemmli, C, Nishikawa, J, Durussel, T, and Laemmli, UK. Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol Cell. (2006) 21:379–91. doi: 10.1016/j.molcel.2005.12.012
30. Sanchez-Vega, F, Mina, M, Armenia, J, Chatila, WK, Luna, A, La, KC, et al. Oncogenic signaling pathways in the Cancer genome atlas. Cell. (2018) 173:321–37.e10. doi: 10.1016/j.cell.2018.03.035
31. Lecona, E, and Fernandez-Capetillo, O. Targeting ATR in cancer. Nat Rev Cancer. (2018) 18:586–95. doi: 10.1038/s41568-018-0034-3
32. Luserna, G, di Rorà, A, Cerchione, C, Martinelli, G, and Simonetti, G. A WEE1 family business: regulation of mitosis, cancer progression, and therapeutic target. J Hematol Oncol. (2020) 13:126. doi: 10.1186/s13045-020-00959-2
33. Peyressatre, M, Prével, C, Pellerano, M, and Morris, MC. Targeting cyclin-dependent kinases in human cancers: from small molecules to peptide inhibitors. Cancers. (2015) 7:179–237. doi: 10.3390/cancers7010179
34. Liu, K, Zheng, M, Lu, R, Du, J, Zhao, Q, Li, Z, et al. The role of CDC25C in cell cycle regulation and clinical cancer therapy: a systematic review. Cancer Cell Int. (2020) 20:213. doi: 10.1186/s12935-020-01304-w
35. Pérez de Castro, I, de Cárcer, G, and Malumbres, M. A census of mitotic cancer genes: new insights into tumor cell biology and cancer therapy. Carcinogenesis. (2007) 28:899–912. doi: 10.1093/carcin/bgm019
36. Bates, M, Furlong, F, Gallagher, MF, Spillane, CD, McCann, A, O'Toole, S, et al. Too MAD or not MAD enough: the duplicitous role of the spindle assembly checkpoint protein MAD2 in cancer. Cancer Lett. (2020) 469:11–21. doi: 10.1016/j.canlet.2019.10.005
37. Wang, L, Zhang, J, Wan, L, Zhou, X, Wang, Z, and Wei, W. Targeting Cdc20 as a novel cancer therapeutic strategy. Pharmacol Ther. (2015) 151:141–51. doi: 10.1016/j.pharmthera.2015.04.002
38. Xie, Y, Wang, A, Lin, J, Wu, L, Zhang, H, Yang, X, et al. Mps1/TTK: a novel target and biomarker for cancer. J Drug Target. (2017) 25:112–8. doi: 10.1080/1061186X.2016.1258568
39. Borah, NA, and Reddy, MM. Aurora kinase B inhibition: a potential therapeutic strategy for Cancer. Molecules (Basel, Switzerland). (2021) 26:1981. doi: 10.3390/molecules26071981
40. Leonardi, GC, Candido, S, Cervello, M, Nicolosi, D, Raiti, F, Travali, S, et al. The tumor microenvironment in hepatocellular carcinoma (review). Int J Oncol. (2012) 40:1733–47. doi: 10.3892/ijo.2012.1408
41. Mossanen, JC, and Tacke, F. Role of lymphocytes in liver cancer. Onco Targets Ther. (2013) 2:e26468. doi: 10.4161/onci.26468
42. Kim, SY, Kyaw, YY, and Cheong, J. Functional interaction of endoplasmic reticulum stress and hepatitis B virus in the pathogenesis of liver diseases. World J Gastroenterol. (2017) 23:7657–65. doi: 10.3748/wjg.v23.i43.7657
Keywords: hepatocellular carcinoma, actin-related protein family, ACTR6, cell cycle, immune infiltration
Citation: Wang J, Song M, Tang J, Yue H, Guo X, Chen Z, Shen X and Cao M (2025) Expression, prognosis and preliminary investigation of the mechanism of action of ACTR6, a member of the ARPs gene family, in hepatocellular carcinoma. Front. Med. 12:1513233. doi: 10.3389/fmed.2025.1513233
Received: 18 October 2024; Accepted: 20 February 2025;
Published: 10 March 2025.
Edited by:
Guangyan Zhangyuan, Shanghai Jiao Tong University, ChinaReviewed by:
Jin Wu, Roswell Park Comprehensive Cancer Center, United StatesCopyright © 2025 Wang, Song, Tang, Yue, Guo, Chen, Shen and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingbo Cao, bWluZ2JvY2FvQDEyNi5jb20=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.