- Department of Traditional and Western Medical Hepatology, Hebei Medical University Third Hospital, the Key Laboratory of Hepatic Fibrosis Mechanisms of Chronic Liver Diseases in Hebei Province, Hebei International Science and Technology Cooperation Base – Hebei International Joint Research Center for Molecular Diagnosis of Liver Cancer, Shijiazhuang, China
Background and aims: This research aimed to examine the association between hepatitis B virus (HBV) covalently closed circular DNA (cccDNA) and liver inflammation in chronic hepatitis B (CHB) infection patients.
Methods: From August 2013 to June 2022, CHB patients at Hebei Medical University Third Hospital (Hebei, China) were recruited. Intrahepatic cccDNA was quantified and its association with liver inflammation was analyzed. Liver inflammation was assessed using the Ishak-modified histological activity index (HAI). Biochemical and viral indicators as well as hepatic inflammation biomarkers were monitored.
Results: In total, 55 CHB patients were enrolled. The average HBV-cccDNA level was markedly elevated in HBeAg+ patients compared to HBeAg patients. Intrahepatic cccDNA levels differed significantly in different liver inflammation groups and showed a positive correlation with the HAI score for hepatic inflammation.
Conclusion: HBV-cccDNA level was associated with liver inflammation.
1 Introduction
Hepatitis B virus (HBV) is a significant contributor to viral hepatitis, which affects over 257–291 million individuals globally. The interplay between HBV viral propagation and the immune system’s reaction gives rise to various health consequences, from liver inflammation to hepatocellular carcinoma (HCC) and cirrhosis (1). HBV-cccDNA formation is critical for the virus’s life cycle and is crucial in the persistence and recurrence of infection. Hence, intrahepatic HBV-cccDNA acts as an essential biomarker of disease progression and is widely utilized to assess the effectiveness of antiviral treatments and establish treatment goals (2–6).
The host anti-HBV-specific immune response can control infection by clearing infected hepatocytes. However, the virus can weaken and deplete the host’s specific antiviral cellular immune system by continuously expressing high levels of hepatitis-B-e-antigen (HBeAg) and hepatitis-B-surface-antigen (HBsAg), making it ineffective in clearing infected hepatocytes. In clinical practice, the negative conversion of HBeAg commonly signifies partial immune control over chronic hepatitis B (CHB) infection. Therefore, patients are assigned to HBeAg+ chronic HBV group, HBeAg+ CHB group, HBeAg chronic HBV infection group, and HBeAg CHB group according to their HBeAg status and severity of liver inflammation. The progression of persistent HBV relies on the interplay between the virus and host immunity. However, due to the lack of simple and effective host-specific immune evaluation indicators for HBV, the dynamic changes of host immune factors in persistent HBV and their role in disease progression remain largely unknown. Previous studies have shown that host immune activation against HBV can effectively suppress viral replication by targeting infected liver cells, but this response is typically seen in the early HBV stage in HBeAg+ patients with high levels HBV-DNA levels (7). HBV can undergo spontaneous reactivation or reactivate in response to immunosuppressive or anti-inflammatory treatments (8). The existing nucleos(t)ide analogs (NAs) inhibit the replication of cytoplasmic HBV; however, they do not address cccDNA and are not curative treatments (9). Magri et al. (10) revealed a gene signature related to immune responses involved in HBV-cccDNA transcription. In addition to aspartate transaminase (AST) and ALT, serum HBsAg level was found to correlate with the grade of inflammation in HBeAg+ CHB patients prior to NA treatment. Additionally, combining HBsAg with AST demonstrated outstanding diagnostic performance for predicting severe inflammation (11).
Therefore, this research aimed to evaluate the changes in HBV-cccDNA levels of CHB patients receiving liver biopsy based on their HBeAg status, liver function, liver tissue inflammation activity, viral load, and cccDNA levels.
2 Materials and methods
2.1 Objects
From August 2013 to June 2022, 55 patients with CHB at Hebei Medical University Third Hospital (Hebei, China) were recruited in this study. Persistent HBV patients receiving liver biopsy and screening for serum biochemical and viral indicators were also included. The following criteria were used for inclusion: (1) age 18–65 years; (2) the serum level of HBsAg was detected in the last 6 months; (3) the patient provided consent for a liver biopsy and testing of serum biochemical and viral indicators; and (4) informed consent in writing was provided for treatment and participation in this research. The following criteria were used for exclusion: (1) the patient had previously undergone antiviral therapy for HBV; (2) co-infection with other viruses, including HCV, HDV, and HIV; (3) diagnosed with compensated/decompensated cirrhosis or HCC; (4) autoimmune hepatitis, primary biliary cirrhosis, alcoholic/non-alcoholic liver diseases; and (5) coexisting conditions, or severe metabolic imbalance or psychiatric disorders. This research was granted approval from the Ethics Committee of the same institute.
2.2 Data collection
Demographic and laboratory parameters including age, gender, red blood cell (RBC) count, white blood cell (WBC) count, hemoglobin (HGB) level, platelet (PLT) count, and the levels of alanine aminotransferase (ALT), AST, alkaline phosphatase (ALP), albumin (ALB), total bilirubin (TBIL), glutamyl transpeptidase (GGT), HBsAb, HBsAg, HBcAb, HBeAg, HBeAb, and HBV-DNA were obtained retrospectively from the electronic medical records system.
2.3 Assessment of liver inflammation
Liver biopsies were conducted through standard procedures. Ultrasound-guided liver biopsy was conducted based on the standard procedure (12). At least 2 pieces with 2.0 cm length were collected to ensure the presence of 11 portal tracts for histological evaluation. Two pathologists independently evaluated the biopsy specimens, blinded to both the biopsy timing and clinical information. In cases of inconsistency, both pathologists reexamined the samples to reach a consensus. Liver inflammation was assessed using the Ishak-modified histological activity index (HAI) (13, 14).
2.4 Detection of intrahepatic HBV DNA and cccDNA levels
About 30 μm formalin fixed paraffin-embedded liver biopsy tissue in sections of 6 μm each was used for DNA extraction. To prevent contamination, disposable tweezers, brush, and interleaver were used and sectioning blades were carefully cleaned with 70% ethanol after every sampling. Genomic DNA and intracellular-free microchromosomal DNA were isolated using the QIAamp-DNA-Mini-Kit in accordance with the manufacturer’s guidelines (QIAGEN, Germany). PSAD was utilized for digestion of HBV double-stranded DNA, relaxed circular DNA, and single-stranded DNA (Epicentre, USA). Subsequently, cccDNA-selective amplification was carried out using the rolling-circle-amplification (RCA) technique with Phi29 (New England Biolabs, USA). RCA products were further amplified and quantified via TaqMan real-time PCR, employing probes that target the unfilled portions of the viral genome along with cccDNA-specific primer pairs. Notably, Phi29 and PSAD were unnecessary for detecting total HBV-DNA. The cell count was determined using probes and primers as a standard control, specifically DNA fragments of human β-actin. The LLOQ was 0.01 copies per cell.
2.5 Statistical analysis
All statistical tests were conducted using GraphPad Prism v8.0, MedCalc v15.0, and SPSS v26.0. Continuous data are presented as mean ± SD or median (IQR). They were compared using the Kruskal–Wallis test. Categorical data were examined using the χ2 test. They are presented as numbers (percentages). The correlations between two parameters were determined using Pearson’s correlation. Two-tailed tests of significance were conducted, and p < 0.05 was regarded as statistically significant.
3 Results
3.1 Baseline features
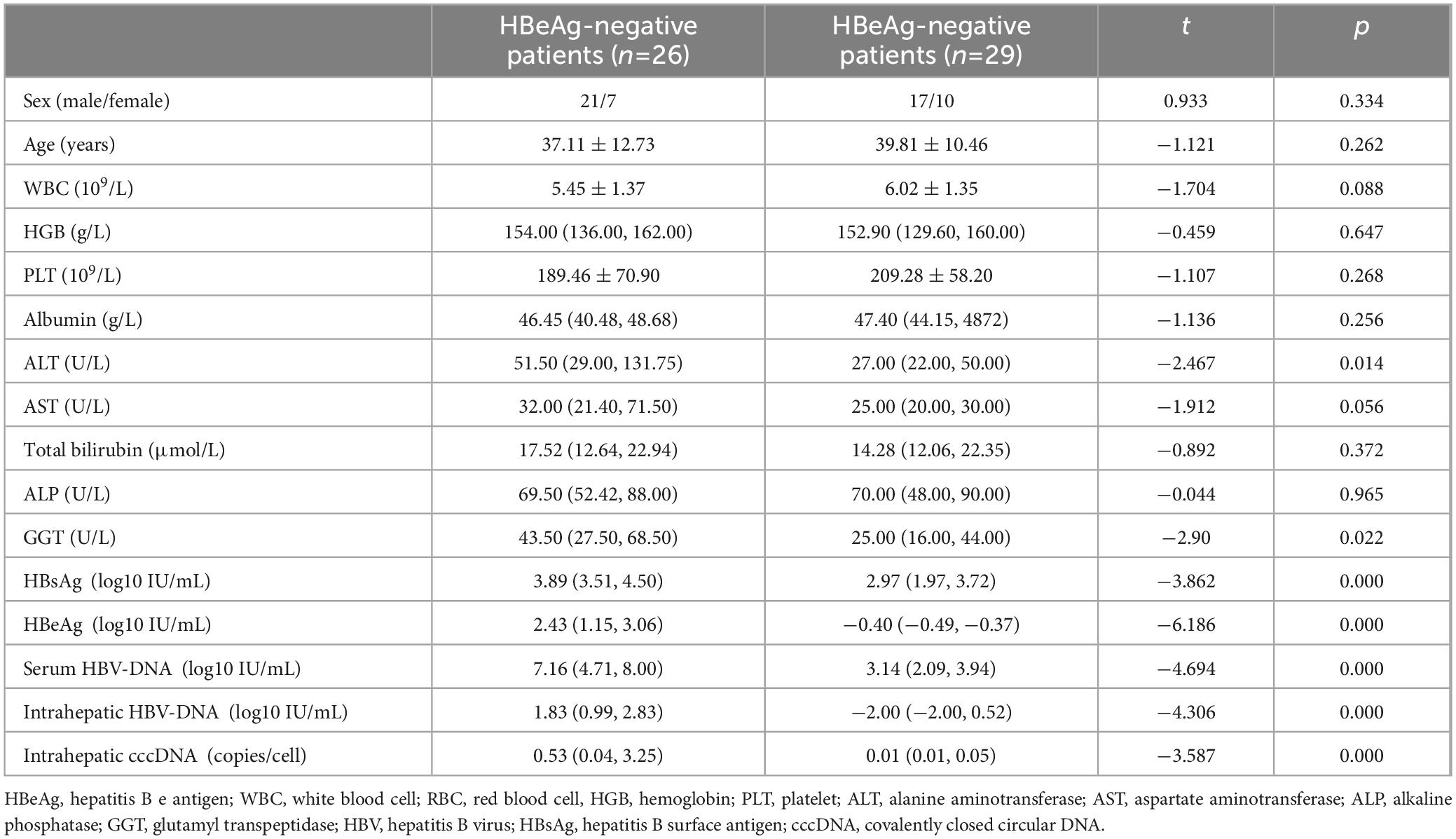
In total, 55 patients were included between August 2013 and June 2022. These patients were categorized into two groups according to their HBeAg status. No remarkable differences were observed between the two groups regarding sex, age, AST, total bilirubin, and albumin between the HBeAg and HBeAg+ groups. However, obvious differences were noted between the two groups regarding AST, GGT, serum HBV-DNA, serum HBsAg, intrahepatic HBV-DNA, and intrahepatic HBV-cccDNA levels (Table 1).
3.2 Variation in HBV-cccDNA and serum-based viral indicators
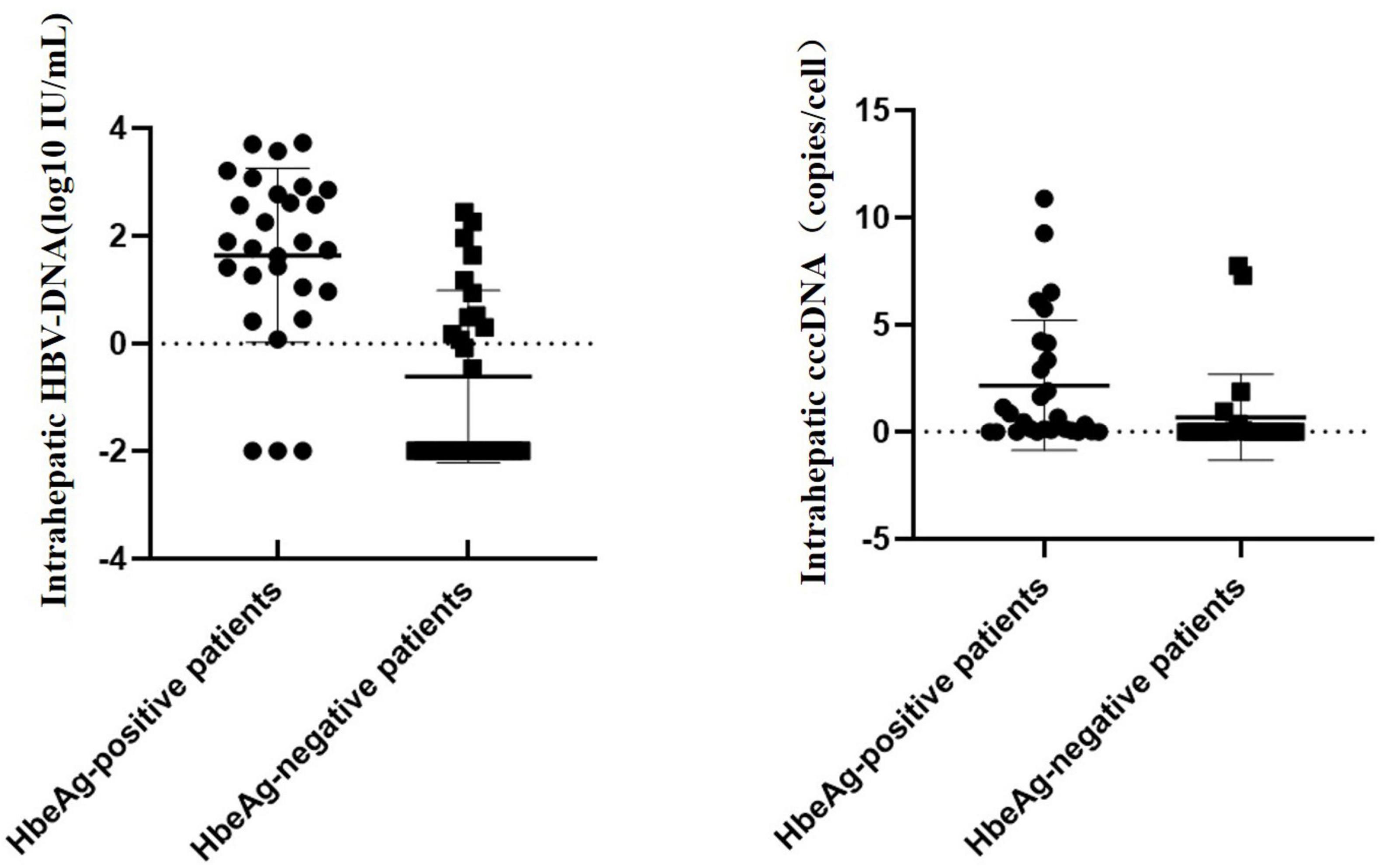
When stratifying patients by the HBeAg status, the mean HBV-cccDNA and intrahepatic HBV-DNA levels of patients with HBeAg+ were remarkably increased compared to those with HBeAg (0.53 vs. 0.01 log10 copies/cell, 1.83 vs. 2 log10 IU/mL, p < 0.001) (Figure 1).
3.3 HBV-cccDNA and hepatic inflammation
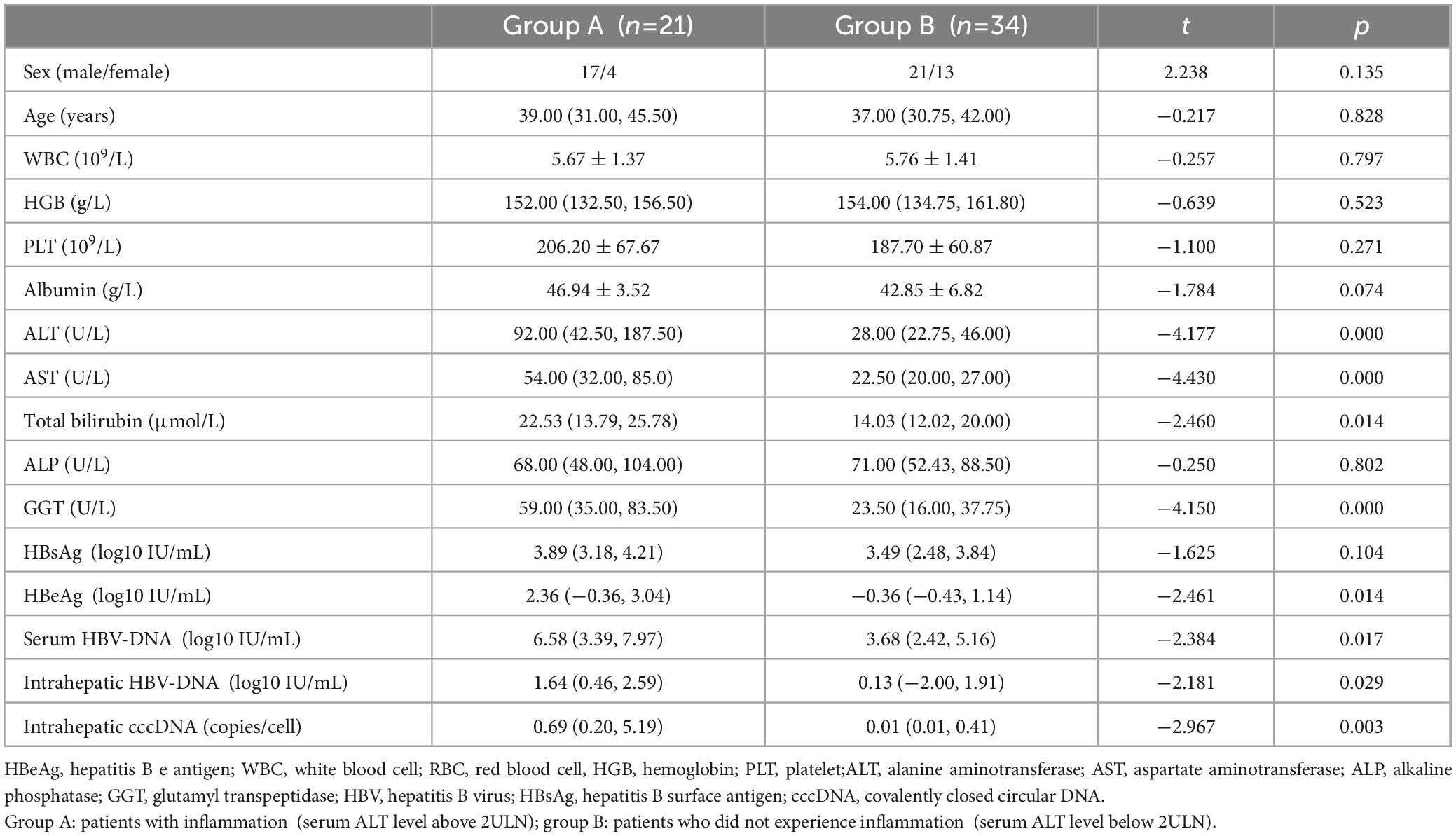
Fifty-five patients with ALT < 2 ULN were categorized into two groups according to the severity of hepatic inflammation. Twenty-one patients with liver inflammation (ALT more than 2 ULN) were allocated into group A. Thirty-four patients without liver inflammation (ALT less than 2 ULN) were allocated into group B. Age, sex, and ALB did not markedly vary between the two groups. There were obvious differences in AST, ALT, GGT, total bilirubin, serum HBsAg, serum HBeAg, intrahepatic HBV-DNA levels, serum HBV-DNA, and intrahepatic HBV-cccDNA levels between the two groups (Table 2). Besides, logistic regression analysis revealed positive correlations between HBV-cccDNA levels and liver inflammation HAI scores (Figure 2).
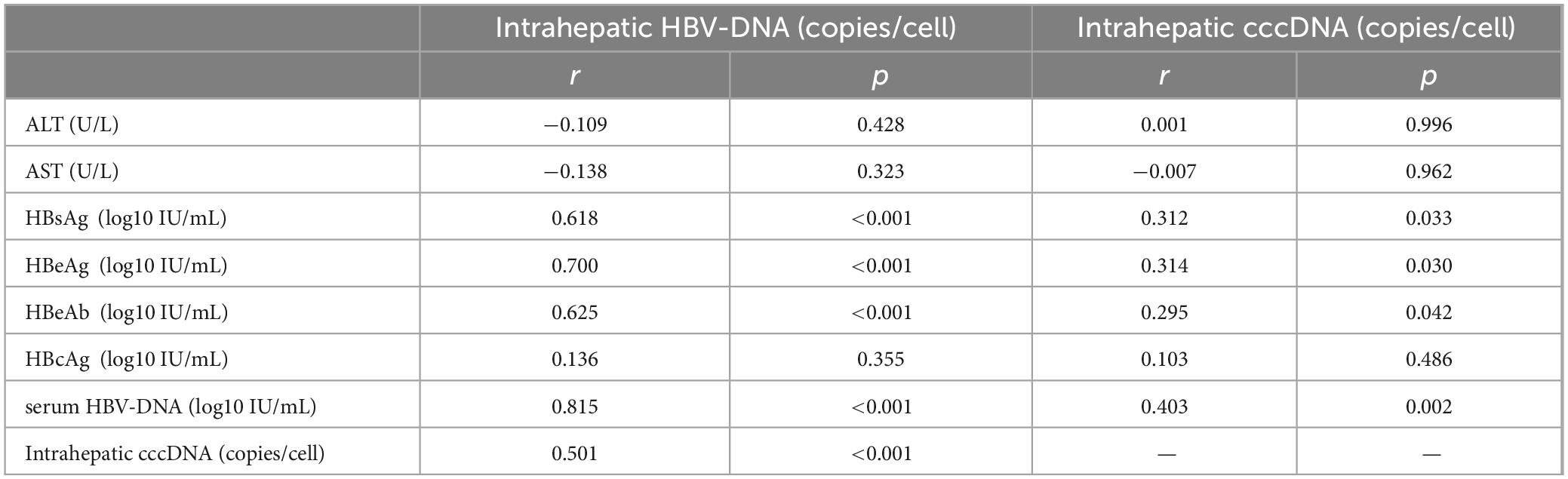
3.4 Correlations between HBV-cccDNA and viral indicators
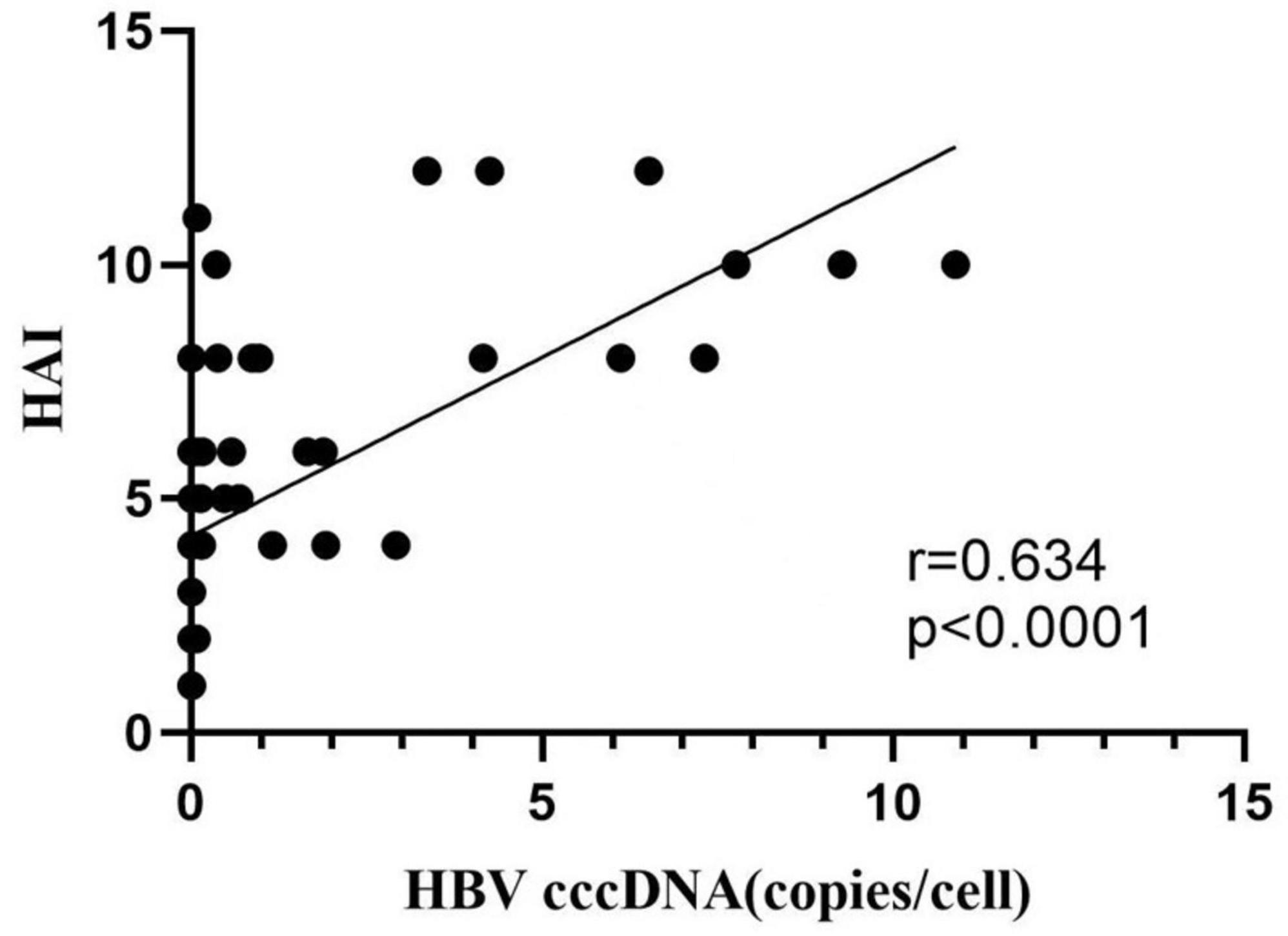
Pearson correlation analysis indicated that intrahepatic cccDNA level was positively correlated with HBsAg, HBeAg, HBeAb, and serum HBV-DNA levels (r = 0.312, 0.314, 0.295, 0.403, p < 0.05). Intrahepatic HBV-DNA level was positively correlated with HBsAg, HBeAg, HBeAb, serum HBV-DNA, and intrahepatic HBV-cccDNA levels (r = 0.618, 0.700, 0.625, 0.815, and 0.501, p < 0.001) (Figure 3 and Table 3).
4 Discussion
Our findings indicated that the HBV-cccDNA level was markedly higher in HBeAg+ patients than in HBeAg patients. The reduced viral load in serum during the HBeAg phase seems to be associated with mechanisms other than merely the repression of intracellular HBV-cccDNA, and the replicative effectiveness of HBV is also involved in this phenomenon (15). Furthermore, the reduced levels of intracellular pregenomic RNA also indicate the suppression of serum HBV-DNA in HBeAg patients. Additionally, the serum HBV-DNA concentration is affected by the amount of intracellular HBV-cccDNA and the virus’s replication efficiency (16).
Our results indicated that serum viral markers positively related to intrahepatic HBV-cccDNA levels in HBsAg, HBeAg, HBeAb, and HBV-DNA, which are in agreement with prior research (17–19). For instance, Thompson et al. (18) found that numerous hepatocytes stained positive for HBsAg in HBeAg+ patients with suppressed viral propagation, suggesting that the link between HBV replication and HBsAg production weakens during the HBeAg stage. This is likely due to HBsAg being produced from sources other than intracellular HBV-cccDNA. Additionally, prior research has reported a correlation between serum HBsAg levels and cccDNA copy number (20).
In our study, intrahepatic HBV-cccDNA levels, along with HBV-DNA viral load, HBeAg, and HBsAg titers, were substantially higher in HBeAg+ patients than in HBeAg patients, likely due to increased viral protein synthesis and release in individuals with elevated intracellular HBV-cccDNA levels. Such immune activation may worsen inflammation, resulting in elevated serum ALT concentrations. Intrahepatic HBV-cccDNA level was evaluated through liver biopsies and histological assessment. This is usually accompanied by hepatocyte damage, hepatic flares, and histologic changes. HBV-cccDNA transcribes mRNA encoding HBV-specific proteins and can recruit and contribute to inflammatory cells that cause liver injury (21). The findings indicated that elevated intrahepatic HBV-cccDNA level could elevate the likelihood of hepatic inflammation. More research is needed to confirm HBV-cccDNA as a viral indicator for patients potentially requiring further outpatient support. Currently, there is controversy regarding the association between intrahepatic HBV-cccDNA and histopathological liver inflammation (22–25). Some investigators have found no significant association between HBV-cccDNA levels and hepatic inflammation grade in HBeAg patients, whereas other groups report that HBeAg patients with significant hepatic inflammation show elevated intrahepatic HBV-cccDNA levels. Magri et al. (10) showed that transcripts derived from cccDNA are linked to biomarkers of hepatic inflammation. The inflammatory hepatic transcriptome analysis indicated that 24 genes are remarkably correlated with cccDNA transcriptional activity (10). The study identified a gene signature related to the immune response associated with HBV-cccDNA transcription.
Herein, we prospectively examined the association between intracellular HBV-cccDNA content and hepatic inflammation in patients. HBV-cccDNA can be used as a biomarker for assessing liver disease severity in clinical practice. However, further research is warranted to delineate the immune pathways controlling viral replication.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by the Ethics Committee of Hebei Medical University Third Hospital. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
SZ: Methodology, Writing – original draft, Data curation, Formal analysis, Project administration, Supervision, Writing – review & editing. CD: Methodology, Project administration, Writing – original draft. CC: Methodology, Writing – original draft. JZ: Methodology, Writing – review & editing. JL: Data curation, Methodology, Writing – review & editing. XZ: Data curation, Methodology, Writing – original draft. WR: Investigation, Methodology, Writing – original draft. YZ: Investigation, Methodology, Writing – original draft. YN: Methodology, Writing – original draft, Formal Analysis, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by Hebei Province Medical Applicable Technology Tracking Project (G2019T009), the Introduction of Foreign Intelligence Program in Hebei Province in 2024, and Fourth High-end Talents in Hebei Province.
Acknowledgments
We would like to thank all donators for donating research blood samples and liver biopsies. We are grateful to Yan Liu and Lei Li for their professional technical support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Liu J, Liang W, Jing W, Liu M. Countdown to 2030:eliminating hepatitis B disease, China. Bull World Health Organ. (2019) 97:230–8.
2. Mason WS, Aldrich C, Summers J, Taylor JM. Asymmetric replication of duck hepatitis B virus DNA in liver cells: Free minus strand DNA. Proc Natl Acad Sci U S A. (1982) 79:3997–4001.
3. Wu TT, Coates L, Aldrich CE, Summers J, Mason WS. In hepatocytes infected with duck hepatitis B virus, the template for viral RNA synthesis is amplified by an intracellular pathway. Virology. (1990) 175:255–61.
4. Zoulim F. New insight on hepatitis B virus persistence from the study of intrahepatic viral cccDNA. J Hepatol. (2005) 42:302–8.
5. Dandri M, Burda MR, Will H, Petersen J. Increased hepatocyte turnover and inhibition of woodchuck hepatitis B virus replication by adefovir in vitro do not lead to reduction of the closed circular DNA. Hepatology. (2000) 32:139–46.
6. Moraleda G, Saputelli J, Aldrich CE, Averett D, Condreay L, Mason WS. Lack of effect of antiviral therapy in nondividing hepatocyte cultures on the closed circular DNA of woodchuck hepatitis virus. J Virol. (1997) 71:9392–9.
7. Xing TJ, Zhao KY, Li WT, Wang LJ, Lu FM. [Association between HBV viral load and severity of liver inflammation in patients with chronic hepatitis B virus infection]. Zhonghua Gan Zang Bing Za Zhi. (2023) 31:954–60. doi: 10.3760/cma.j.cn501113-20230820-00061
8. Loomba R, Liang TJ. Hepatitis B reactivation associated with immune suppressive and biological modifier therapies: Current concepts, management strategies, and future directions. Gastroenterology. (2017) 152:1297–309.
9. Revill PA, Chisari FV, Block JM, Dandri M, Gehring AJ, Guo H, et al. A global scientific strategy to cure hepatitis B. Lancet Gastroenterol Hepatol. (2019) 4:545–58.
10. Magri A, D’Arienzo V, Minisini R, Jühling F, Wing PAC, Rapetti R, et al. Inflammatory gene expression associates with hepatitis B virus cccDNA- but not integrant-derived transcripts in HBeAg negative disease. Viruses. (2022) 14:1070. doi: 10.3390/v14051070
11. Zhao J, Bian D, Liao H, Wang Y, Ren Y, Jiang Y, et al. Serum HBsAg and HBcrAg is associated with inflammation in HBeAg-positive chronic hepatitis B patients. Front Cell Infect Microbiol. (2023) 13:1083912. doi: 10.3389/fcimb.2023.1083912
12. Rockey DC, Caldwell SH, Goodman ZD, Nelson RC, Smith AD, American Association for the Study of Liver Diseases, et al. Liver biopsy. Hepatology. (2009) 49:1017–44.
13. Ishak K, Baptista A, Bianchi L, Callea F, De Groote J, Gudat F, et al. Histological grading and staging of chronic hepatitis. J Hepatol. (1995) 22:696–9.
14. Knodell RG, Ishak KG, Black WC, Chen TS, Craig R, Kaplowitz N, et al. Formulation and application of a numerical scoring system for assessing histological activity in asymptomatic chronic active hepatitis. Hepatology. (1981) 1:431–5.
15. Volz T, Lutgehetmann M, Wachtler P, Jacob A, Quaas A, Murray JM, et al. Impaired intrahepatic hepatitis B virus productivity contributes to low viremia in most HBeAg-negative patients. Gastroenterology. (2007) 133:843–52.
16. Levrero M, Pollicino T, Petersen J, Belloni L, Raimondo G, Dandri M. Control of cccDNA function in hepatitis B virus infection. J Hepatol. (2009) 51:581–92.
17. Gao Y, Li Y, Meng Q, Zhang Z, Zhao P, Shang Q, et al. Serum hepatitis B virus DNA, RNA, and HBsAg: Which correlated better with intrahepatic covalently closed circular DNA before and after Nucleos(t)ide Analogue Treatment? J Clin Microbiol. (2017) 55, 2972–82.
18. Thompson AJ, Nguyen T, Iser D, Ayres A, Jackson K, Littlejohn M, et al. Serum hepatitis B surface antigen and hepatitis B e antigen titers: Disease phase influences correlation with viral load and intrahepatic hepatitis B virus markers. Hepatology. (2010) 51:1933–44.
19. Li W, Zhao J, Zou Z, Liu Y, Li B, Sun Y, et al. Analysis of hepatitis B virus intrahepatic covalently closed circular DNA and serum viral markers in treatment-naive patients with acute and chronic HBV infection. PLoS One. (2014) 9:e89046. doi: 10.1371/journal.pone.0089046
20. Wang Q, Luan W, Warren L, Fiel MI, Blank S, Kadri H, et al. Serum hepatitis B surface antigen correlates with tissue covalently closed circular DNA in patients with hepatitis -associated hepatocellular carcinoma. J Med Virol. (2016) 88:244–51.
21. Cheng PN, Liu WC, Tsai HW, Wu IC, Chang TT, Young KC, et al. Association of intrahepatic cccDNA reduction with the improvement of liver histology in chronic hepatitis B patients receiving oral antiviral agents. J Med Virol. (2011) 83:602–7.
22. Lin LY, Wong VW, Zhou HJ, Chan HY, Gui HL, Guo SM, et al. Relationship between serum hepatitis B virus DNA and surface antigen with covalently closed circular DNA in HBeAg-negative patients. J Med Virol (2010) 82:1494–500.
23. Guner R, Karahocagil M, Buyukberber M, Kandemir O, Ural O, Usluer G, et al. Correlation between intrahepatic hepatitis B virus cccDNA levels and other activity markers in patients with HBeAg-negative chronic hepatitis B infection. Eur J Gastroenterol Hepatol. (2011) 23:1185–91.
24. Takkenberg RB, Zaaijer HL, Menting S, Weegink CJ, Terpstra V, Cornelissen M, et al. Detection of hepatitis B virus covalently closed circular DNA in paraffin embedded and cryo-preserved liver biopsies of chronic hepatitis B patients. Eur J Gastroenterol Hepatol. (2010) 22:952–60.
Keywords: covalently closed circular DNA (cccDNA), inflammation, liver, HBV, chronic hepatitis B
Citation: Zhao S, Dong C, Chang C, Zhang J, Li J, Zhang X, Ren W, Zhang Y and Nan Y (2025) Association between intrahepatic cccDNA and the severity of liver inflammation in chronic hepatitis B virus infection patients. Front. Med. 11:1519686. doi: 10.3389/fmed.2024.1519686
Received: 30 October 2024; Accepted: 16 December 2024;
Published: 24 January 2025.
Edited by:
Krzysztof Tomasiewicz, Medical University of Lublin, PolandReviewed by:
Tuo Shao, Massachusetts General Hospital and Harvard Medical School, United StatesGiuseppina Maria Elena Colomba, Azienda Ospedaliera Ospedali Riuniti Villa Sofia Cervello, Italy
Copyright © 2025 Zhao, Dong, Chang, Zhang, Li, Zhang, Ren, Zhang and Nan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuemin Nan, bmFueXVlbWluQDE2My5jb20=
 Suxian Zhao
Suxian Zhao Chen Dong
Chen Dong Yuemin Nan
Yuemin Nan