- 1Hepatobiliary Department, The Affiliated Traditional Chinese Medicine Hospital, Southwest Medical University, Luzhou, China
- 2Sichuan Clinical Research Center for Traditional Chinese Medicine of Liver Diseases, The Affiliated Traditional Chinese Medicine Hospital, Southwest Medical University, Luzhou, China
- 3Department of Biomedical Science, Faculty of Medicine and Health Sciences, Universiti Putra Malaysia, Serdang, Selangor, Malaysia
- 4UPM-MAKNA Cancer Research Laboratory, Institute of Bioscience, Universiti Putra Malaysia, Serdang, Selangor, Malaysia
Introduction: Non-alcoholic fatty liver disease (NAFLD) has become the leading cause of chronic liver disease worldwide. Hepatocellular carcinoma (HCC) is the fifth most common malignancy worldwide, with high morbidity and mortality. The rapidly increasing incidence of NAFLD is becoming an essential precursor of HCC globally. MicroRNAs (miRNAs) are involved in the progression of NAFLD and HCC.
Method: Potential miRNAs associated with NAFLD in HCC tumorigenesis were identified through a systematic review, and their roles were evaluated by data mining analysis. The biological function of the potential miRNA and its target genes in NAFLD and HCC were evaluated by bioinformatic analysis.
Result: MIR122 was identified as the potential miRNA associated with NAFLD and HCC. Then, MIR122 expression was significantly lower in HCC patients, and higher MIR122 levels were associated with significantly better overall survival. Next, the biological functions of MIR122 and target genes were predicted to be involved in inflammation, fibrosis, cell proliferation, invasion, metastasis, and apoptosis. In particular, the FOXO signaling pathway may regulate the above biological functions.
Conclusion: MIR122 was suggested to be involved in progressing from NAFLD to HCC through the PI3K/AKT/FOXO pathway.
Systematic review registration: PROSPERO, identifier: CRD 42024517940.
1 Introduction
Nonalcoholic fatty liver disease (NAFLD) is characterized by the histological presence of steatosis in more than 5% of hepatocytes without alcohol abuse (1). Metabolic syndrome is the primary risk factor for NAFLD, which is characterized by obesity, hyperglycemia, dyslipidemia, and systemic hypertension (HTN) (2). The spectrum of NAFLD encompasses nonalcoholic fatty liver (NAFL), nonalcoholic steatohepatitis (NASH), and their related fibrosis, cirrhosis, and hepatocellular carcinoma (HCC) (3). The global prevalence of NAFLD is estimated to be around 25.2%, comprising 27.4% in Asia, with the highest being in China at over 33%, and its incidence and prevalence are escalating worldwide (4, 5).
More than 80% of cases of Primary Liver cancer (PLC) are HCC, which stands as the fifth most common cancer globally and is the second most mortality malignancy (6). Risk factors for HCC include hepatitis B and C, exposure to dietary toxins, alcohol abuse, and metabolic liver diseases (7). The annual incidence of HCC in NASH patients with cirrhosis is estimated to range from 0.5 to 2.6%, and the rate in non-cirrhotic patients is approximately 0.01% to 0.13% (5). During the obesity epidemic, the incidence of NAFLD-related HCC is experiencing a pronounced and exponential increase (8). NAFLD is becoming an essential precursor of HCC worldwide (9). However, the molecular mechanism between NAFLD and HCC has not been thoroughly studied until now.
MicroRNAs (miRNAs) are a subtype of small, endogenous, noncoding RNAs with lengths typically ranging from 19 to 25 nucleotides. MiRNAs exert their regulatory influence by inversely modulating gene expression through direct induction of messenger RNA (mRNA) degradation or suppressing translation via base pairing with complementary sites in target mRNAs' 3′-untranslated regions (3′-UTRs) (10, 11). Due to this feature, miRNAs have been identified to play critical post-transcription roles in many diseases, including NAFLD and HCC (12). As reported, miRNAs regulate tumor genesis and progression in HCC, such as proliferation, invasion, recurrence, and metastasis (13). Meanwhile, miRNAs also have crucial roles in regulating NAFLD, including the pathological process of steatosis and inflammation in liver cells (13). However, the co-regulated roles of miRNAs between NAFLD and HCC remain unclear.
In this study, we aimed to evaluate the roles of miRNAs associated with NAFLD in HCC tumorigenesis through systematic review and data mining analysis, and the mechanism was evaluated by bioinformatic analysis.
2 Materials and methods
2.1 System review
A systematic review proceeded for the research objective by following the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) statement. This system review has been officially registered in the Prospective Registry of International Systematic Reviews (PROSPERO) database (CRD 42024517940).
2.2 Search strategy
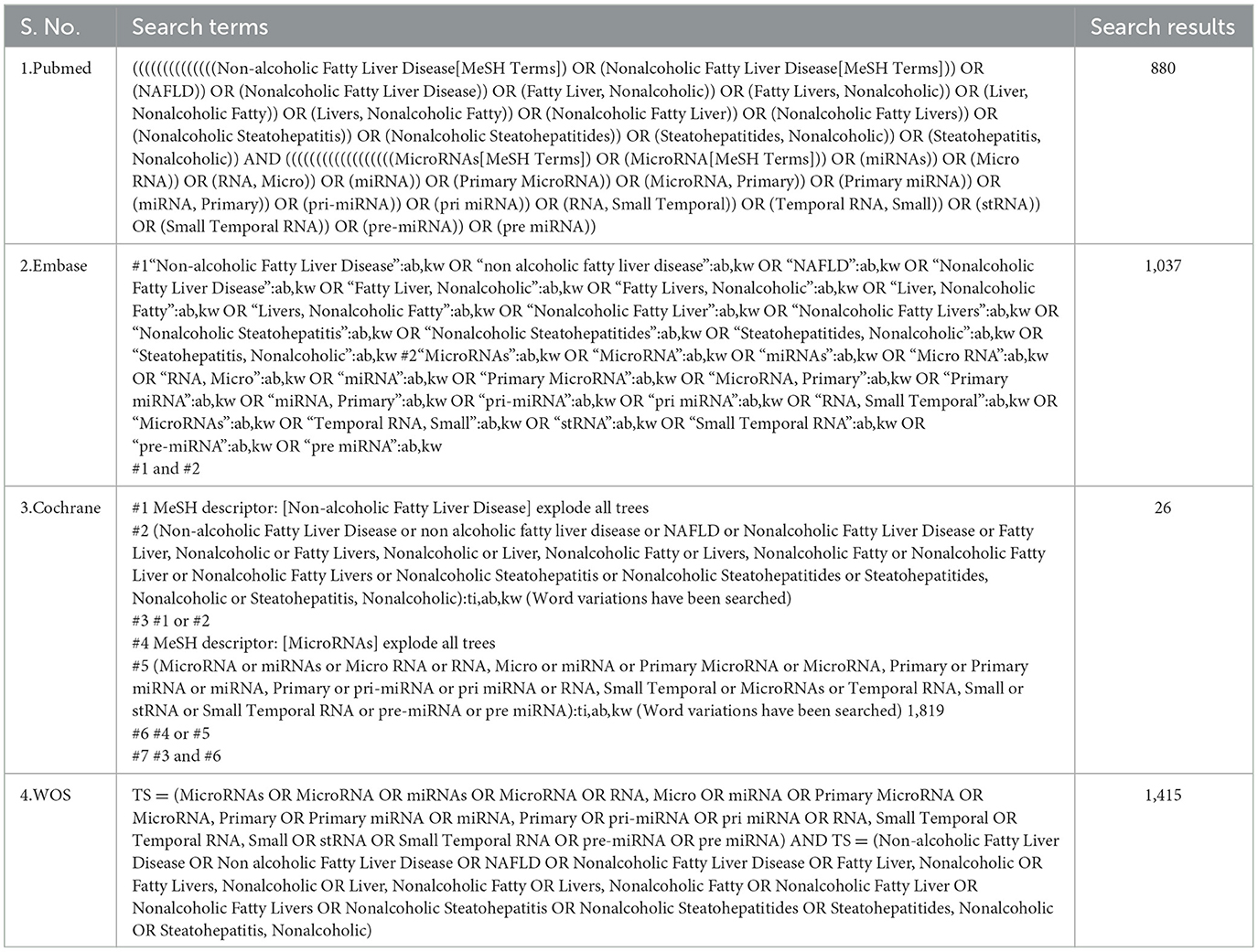
The researchers searched four public databases (PubMed, EMBASE, Cochrane, and Web of Science) from their creation to January 2024. The search strategy was constructed around the PICOS tool: (P) Population: Patients with NAFLD, (I) Intervention: miRNA, (C) Comparison: Significant high or low expression of miRNAs, (O) Outcomes: Progression of steatosis or fibrosis in population with NAFLD, (S) Study type: Comparative research. The complete detailed search strategy is shown in Table 1.
2.3 Eligibility criteria
The inclusion criteria were as follows: (1) Experimental group with altered miRNA as an intervention for people with NAFLD, (2) Control group with significantly high or low expression of miRNAs, (3) Cohort studies, (4) Outcome included the progression of steatosis, or fibrosis in population with NAFLD.
The exclusion criteria were as follows: (1) non-human studies, (2) non-comparative studies, (3) reviews and systematic evaluations meta-analyses, and (4) case reports, reviews, letters, and editorials.
2.4 Study selection
The articles underwent screening and exclusion using the Rayyan website. Firstly, two researchers (Y. Jiang and H Peng) screened the literature titles to identify duplicates, non-randomized controlled trial studies, reviews, systematic evaluations, meta-analyses, case reports, letters, and editorials. Then, the same two researchers (Y. Jiang and H Peng) reviewed the literature's abstracts to determine inclusion and exclusion criteria. Finally, both researchers (Y. Jiang and H Peng) independently read the full articles of the selected ones and further refined the inclusion criteria. Throughout this process, the researchers worked independently, and disputes were resolved by the third author (X Zhu).
2.5 Data mining and bioinformatic analysis
To identify the potential miRNAs associated with HCC tumorigenesis, we employed the COREMINE database (https://www.coremine.com/), which utilizes a text-mining algorithm. Searching keywords, including “Liver neoplasms” and “Cell Transformation, Neoplastic,” were selected in COREMINE to compile a list of microRNAs linked to HCC tumorigenesis.
The difference in expression and prognosis efficacy of miRNA in HCC patients was investigated in the ENCORI database (https://rnasysu.com/encori/) and Kaplan-Meier plotter (https://kmplot.com/analysis/). The miRNA targets and their biological roles were identified using the ONCO.IO database (https://onco.io/). Next, a regulatory network model was constructed to understand miRNA-target interactions better. The Enrichr database (https://amp.pharm.mssm.edu/Enrichr/) enriched the significant pathways related to the miRNA target genes.
2.6 Statistical analysis
Statistical analysis followed the specific rules of the COREMINE database, Onco.io database, Kaplan–Meier plotter, and Enrichr database online tools. In the Onco.io and Enrichr databases, P < 0.05 was considered statistically significant. In the COREMINE database, Extracted Associations were set as co-mentioned with all selected nodes, and P < 0.05.
3 Results
3.1 Study characteristics
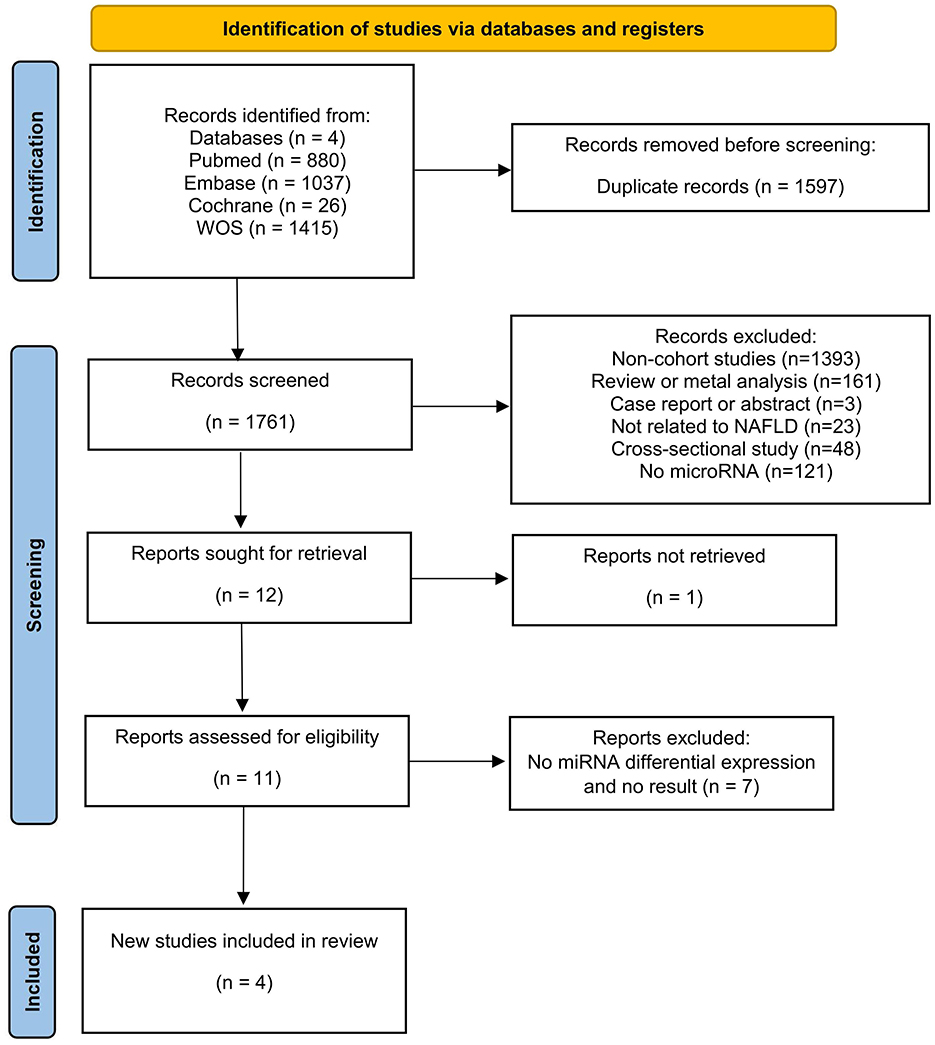
The literature search yielded 3,358 articles (PubMed = 880, Web of Science = 1,415, Embase = 1,037, Cochrane = 26), with 1,597 identified as duplicates and subsequently removed. Among the remaining 1,761 articles, 1,749 abstracts were reviewed and removed based on the inclusion and exclusion criteria. Then, one of the remaining 12 articles was excluded because the study was not retrieved. After that, 11 articles underwent thorough full-text reading, excluding eight due to a lack of differentially expressed miRNAs and non-compliance with outcome metrics. Four articles met the criteria and were included in the current review (Figure 1).
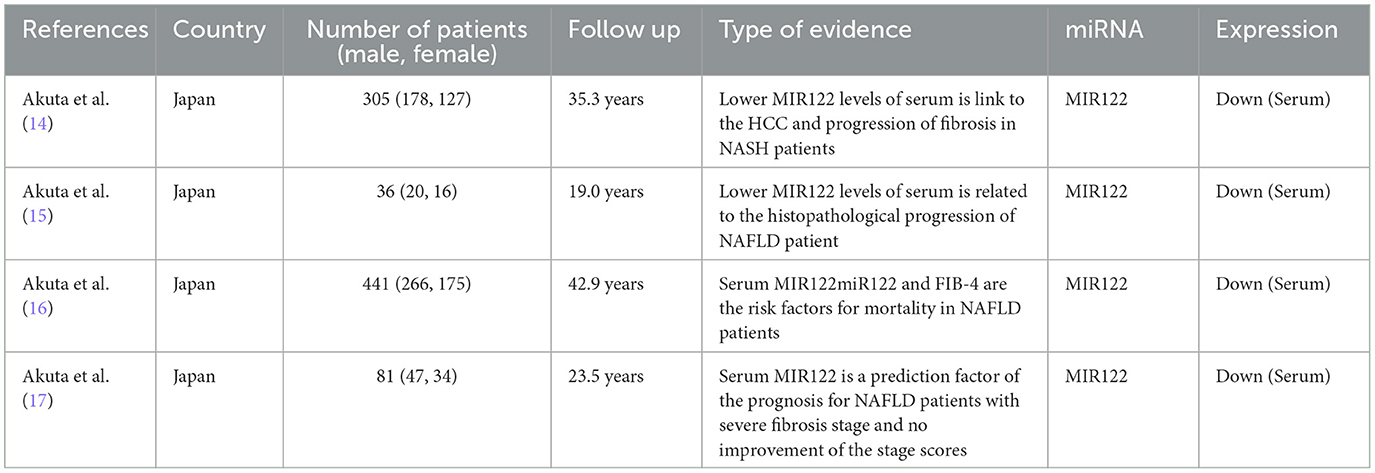
The four studies included 863 patients who were diagnosed with NAFLD by liver biopsy (14–17). Among them, 21 patients died during the follow-up period, and 13 developed HCC. Serum microRNA 122 (MIR122) levels tend to decrease before progression to the fibrotic stage, and reduced levels may reflect an increased risk of hepatocellular carcinoma. Serum MIR122 and a history of hepatocellular carcinoma are independent poor prognostic factors. Detailed characteristics of the included studies are shown (Table 2).
3.2 MIR122 is the potential miRNA associated with NAFLD and tumor genesis of HCC
One hundred fourteen miRNAs related to liver tumors and tumor cell transformation were identified from the COREMINE database (P < 0.05). Subsequently, MIR122 was detected as involved in the NAFLD progression and HCC tumorigenesis (Figure 2A). The expression of MIR122 decreased in HCC patients, and the 5-year overall survival rate in the group with high MIR122 expression was significantly higher than in the group with lower expression (P = 0.014; Figures 2B–D).
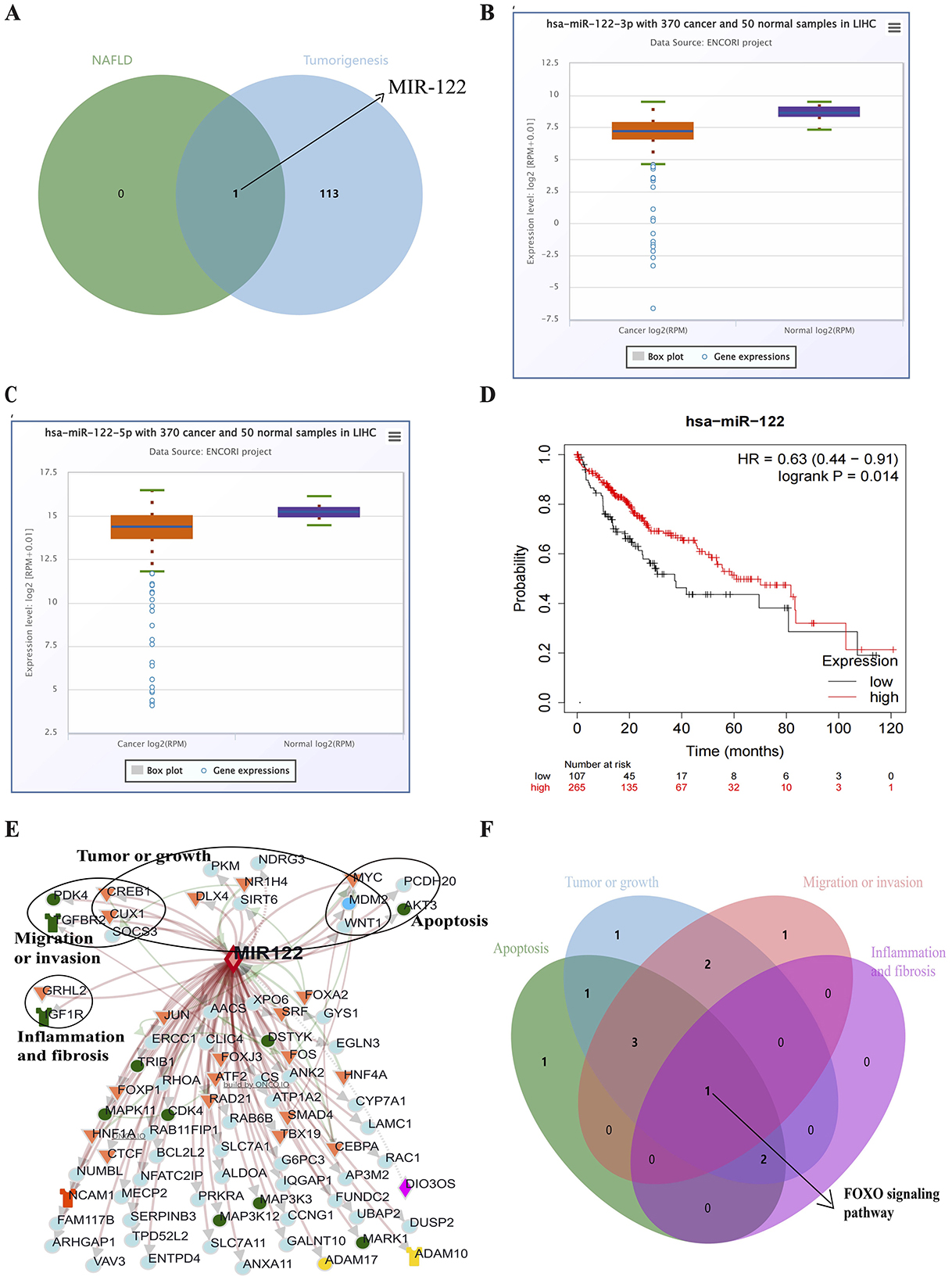
Figure 2. (A) Venn diagram showing all common microRNAs in both pathways. (B) The Starbase online website analyzes MIR122-3p's relative expression in HCC. (C) The Starbase online website analyzes MIR122-5p's relative expression in HCC. (D) The 10-year survival rate of the MIR122 low-expression group decreased dramatically. (E) A network of MIR122 and its target genes. (F) Target signaling pathways associated with biological processes.
3.3 The potential biological roles of the target genes of MIR122 in cancer
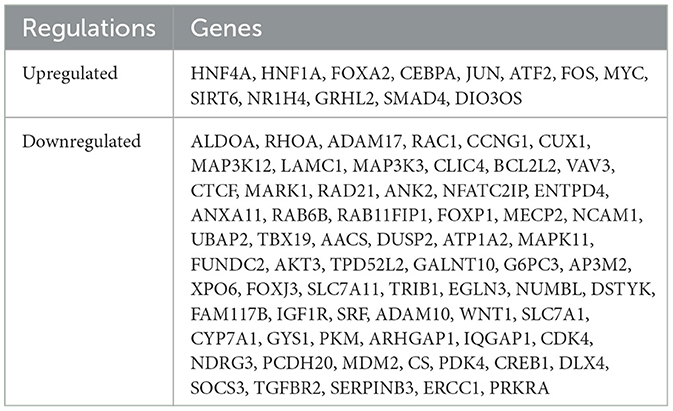
The ONCO.IO database was used to obtain the regulatory network of MIR122 and its target genes in cancer. There are 80 target genes, including 13 upregulated and 67 downregulated genes (Table 3). The results showed that MIR122 interacted significantly and formed a target gene network (Figure 2E). These target genes significantly regulate essential biological processes such as inflammation, fibrosis, tumor growth, migration, and apoptosis. Some targets, such as DLX4, NDRG3, WNT1, MYC, CREB1, and SIRT6, are involved in tumor or cell proliferation. Some other targets are involved in inflammation and fibrosis, such as TGFBR2 and GRHL2. In addition, some targets co-regulate the same biological processes. For example, IGF1R, PDK4, and SOCS3 are involved in tumor proliferation, migration, and invasion. MYC and MDM2 are involved in inflammation and fibrosis and are associated with tumor proliferation. The roles of MIR122 target genes in regulating these biological processes are listed in Table 4.
The potential pathways of target genes were further investigated using the Enrichr database. These biological processes involve common signaling pathways, such as the PI3K-Akt signaling pathway, FoxO signaling pathway, TGF-beta signaling pathway, MAPK signaling pathway, and JAK-STAT signaling pathway. The FoxO signaling pathway involves all biological processes (Figure 2F).
4 Discussion
MiRNAs are post-transcriptional regulators that play essential roles in various processes, and their dysregulation has been associated with NAFLD and HCC (18, 19). For example, miRNAs can target multiple genes involved in developing NAFLD, including cholesterol biosynthesis, de novo lipogenesis, hepatic energy metabolism, inflammation, cell regeneration, and fibrotic signaling (13, 20). Adipokines secreted by adipose tissue tend to induce systemic chronic inflammation, which increases the risk of various types of carcinogenesis (21). MiRNAs play an essential role in the progression from NAFLD to HCC by regulating the expression of key signaling pathways and targets (21). Thus, dysregulated miRNAs serve as a bridge linking NAFLD and HCC, such as MIR122. Meanwhile, chronic liver inflammation of different etiologies, including NAFLD, is associated with reduced MIR122 expression in hepatocytes, and MIR122 expression levels are relatively low in NAFLD-related HCC compared to healthy tissues (22–24).
The involvement of MIR122 has been reported to be a tumor suppressor in inflammation, fibrosis, cancer cell proliferation, invasion, metastasis, and apoptosis (25). We detected that the FOXO signaling pathway collectively participates in these biological processes, and MIR122 target genes are mainly concentrated in the FOXO signaling pathway (26, 27). FOXO proteins are growth and stress-regulated transcription factors, which play essential roles in glucose and lipid metabolism and are involved in various cellular functions (28–32). FOXO family members, FOXO1, FOXO3a, and FOXO4, can be phosphorylated by AKT, leading to loss of FOXO DNA-binding activity. Inactivation of FOXO1 and FOXO3 also results in hypertriglyceridemia and hypercholesterolemia due to increased hepatic lipid secretion and mild steatosis, indicating that FOXO1 and FOXO3 work together to inhibit critical pathways in adipogenesis as well as enhance gluconeogenic genes (33). In addition, it has been shown that FOXO1 increases MIR122 promoter activity by binding to the MIR122 promoter, which can be inhibited by leptin-induced phosphorylation of FOXO1 by the PI3K/Akt signaling pathway, and it was demonstrated that FOXO1 attenuates leptin-associated HSC activation and hepatic fibrosis in the ob/ob mouse model (34). MIR122 also directly inhibits FOXO3 to promote the development of NAFLD (35).
FOXO1 is the primary member of the FOXO family that can promote HCC tumorigenesis and progression (36). FOXO family members can be regulated by the PI3K/Akt pathway and act as a vital target of the insulin/insulin-like growth factor (IGF)-1 signaling pathway to regulate cellular functions (37–39). Studies have shown that FOXO isoforms exhibit differential expression in HCC's primary tumor tissues and cells (36, 40, 41).
In HCC patients, increased FOXO1 expression was found to predict a favorable prognosis for HCC patients and was negatively correlated with vascular infiltration (41). FOXO1 is essential in multiple signaling pathways, especially the insulin/PI3K/AKT signaling pathway, which negatively regulates FOXO1 (42, 43). Activation of this pathway increases cell survival, promotes cell proliferation, and induces cancer development (44, 45). In HCC, overactive Akt signaling inhibits the transcriptional activity of FOXO1, weakening the defense against oxidative stress. FOXO1 typically inhibits the expression of epithelial-mesenchymal transition (EMT)-inducing transcription factors and transforming growth factor-beta (TGF-β), leading to a subsequent increase in EMT and HCC cell migration and invasion (46, 47).
This study has some limitations. First, considering the long period of progression from NAFLD to HCC and acknowledging that steatosis and fibrosis are crucial factors for NAFLD progression to HCC, we set the search strategy of the potential miRNAs as one that can significantly change during the progression of steatosis or fibrosis in a population with NAFLD. Except for MIR122, other relevant miRNAs with significant expression differences were not identified in this systematic review after searching multiple databases due to a lack of cohort data in NAFLD studies. The second limitation is that the population of the included studies was limited to Japanese patients. On the one hand, this is one of the few groups that have conducted long-term follow-up cohort studies on NAFLD patients. On the other hand, other studies found that MIR122 levels are significantly lower in NASH patients in Western populations and that MIR122 expression decreases significantly with the progression of hepatic steatosis (23, 48). Additionally, MIR122 levels were found to be downregulated in the serum of HCC patients from Egypt (49). These results supported evidence that MIR122 may be the potential bridging factor between NAFLD and HCC. However, the value of the MIR122's biomarker and treatment target must still be determined through the long-term cohort study and experiment. Nevertheless, low expression of MIR122 is associated with a poor prognosis in patients with NAFLD-associated HCC, providing a new view for discovering new biomarkers and their associated targets and pathways through systematic review and data mining approaches.
5 Conclusions
In summary, MIR122 may be a potential regulator correlated to NAFLD and HCC and involved in the process of NAFLD to HCC through the PI3K/AKT/FOXO pathway. However, the specific mechanism needs further experimental verification.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found at: COREMINE database (https://www.coremine.com/) ENCORI database (https://rnasysu.com/encori/) Kaplan–Meier plotter (https://kmplot.com/analysis/) ONCO.IO database (https://onco.io/) Enricher database (https://amp.pharm.mssm.edu/Enrichr/).
Author contributions
QP: Methodology, Writing – original draft. XZ: Writing – original draft, Writing – review & editing. YJ: Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. MP: Data curation, Methodology, Writing – review & editing. DZ: Data curation, Formal analysis, Writing – review & editing. XW: Data curation, Formal analysis, Project administration, Writing – review & editing. YC: Methodology, Supervision, Writing – review & editing. JW: Funding acquisition, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Natural Science Foundation of Sichuan Province No. 2022NSFSC1536, Sichuan Science and Technology Program (2022YFS0619), the Ministry of Chinese Medical of Sichuan Province (2021MS158 and 2021MS140), The project of the affiliated traditional Chinese medicine hospital of Southwest medical university (No. 2023ZYYJ08), the National Traditional Chinese Medicine Clinical Research Base Construction Unit of the Affiliated Traditional Chinese Medicine Hospital of Southwest Medical University, Luzhou Science and Technology Innovation Team, and Science and Technology Innovation Team of Affiliated Traditional Chinese Medicine Hospital of Southwest Medical University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2024.1462513/full#supplementary-material
References
1. Meroni M, Longo M, Rustichelli A, Dongiovanni P. Nutrition and genetics in NAFLD: the perfect binomium. Int J Mol Sci. (2020) 21:2986. doi: 10.3390/ijms21082986
2. Friedman SL, Neuschwander-Tetri BA, Rinella M, Sanyal AJ. Mechanisms of NAFLD development and therapeutic strategies. Nat Med. (2018) 24:908–22. doi: 10.1038/s41591-018-0104-9
3. Wong RJ, Aguilar M, Cheung R, Perumpail RB, Harrison SA, Younossi ZM, et al. Nonalcoholic steatohepatitis is the second leading etiology of liver disease among adults awaiting liver transplantation in the United States. Gastroenterology. (2015) 148:547–55. doi: 10.1053/j.gastro.2014.11.039
4. Fang J, Yu C-H, Li X-J, Yao J-M, Fang Z-Y, Yoon S-H, et al. Gut dysbiosis in nonalcoholic fatty liver disease: pathogenesis, diagnosis, and therapeutic implications. Front Cell Infect Microbiol. (2022) 12:997018. doi: 10.3389/fcimb.2022.997018
5. Huang DQ, El-Serag HB, Loomba R. Global epidemiology of NAFLD-related HCC: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol. (2021) 18:223–38. doi: 10.1038/s41575-020-00381-6
6. Chen K, Li Y, Wang B, Yan X, Tao Y, Song W, et al. Patient-derived models facilitate precision medicine in liver cancer by remodeling cell-matrix interaction. Front Immunol. (2023) 14:1101324. doi: 10.3389/fimmu.2023.1101324
7. Li Y, Zhang R, Xu Z, Wang Z. Advances in nanoliposomes for the diagnosis and treatment of liver cancer. Int J Nanomedicine. (2022) 17:909–25. doi: 10.2147/IJN.S349426
8. Shah PA, Patil R, Harrison SA. NAFLD-related hepatocellular carcinoma: the growing challenge. Hepatology. (2023) 77:323–38. doi: 10.1002/hep.32542
9. Ioannou GN. Epidemiology and risk-stratification of NAFLD-associated HCC. J Hepatol. (2021) 75:1476–84. doi: 10.1016/j.jhep.2021.08.012
10. Liu W, Zheng L, Zhang R, Hou P, Wang J, Wu L, et al. Circ-ZEB1 promotes PIK3CA expression by silencing miR-199a-3p and affects the proliferation and apoptosis of hepatocellular carcinoma. Mol Cancer. (2022) 21:72. doi: 10.1186/s12943-022-01529-5
11. Lou W, Liu J, Gao Y, Zhong G, Ding B, Xu L, et al. MicroRNA regulation of liver cancer stem cells. Am J Cancer Res. (2018) 8:1126−41.
12. Greene CM, Varley RB, Lawless MW. MicroRNAs and liver cancer associated with iron overload: therapeutic targets unravelled. World J Gastroenterol. (2013) 19:5212–26. doi: 10.3748/wjg.v19.i32.5212
13. Michelotti GA, Machado MV, Diehl AM. NAFLD, NASH and liver cancer. Nat Rev Gastroenterol Hepatol. (2013) 10:656–65. doi: 10.1038/nrgastro.2013.183
14. Akuta N, Kawamura Y, Suzuki F, Saitoh S, Arase Y, Fujiyama S, et al. Analysis of association between circulating miR-122 and histopathological features of nonalcoholic fatty liver disease in patients free of hepatocellular carcinoma. BMC Gastroenterol. (2016) 16:1–9. doi: 10.1186/s12876-016-0557-6
15. Akuta N, Kawamura Y, Suzuki F, Saitoh S, Arase Y, Kunimoto H, et al. Impact of circulating miR-122 for histological features and hepatocellular carcinoma of nonalcoholic fatty liver disease in Japan. Hepatol Int. (2016) 10:647–56. doi: 10.1007/s12072-016-9729-2
16. Akuta N, Kawamura Y, Arase Y, Saitoh S, Fujiyama S, Sezaki H, et al. Circulating microRNA-122 and fibrosis stage predict mortality of Japanese patients with histopathologically confirmed NAFLD. Hepatol Commun. (2020) 4:66–76. doi: 10.1002/hep4.1445
17. Akuta N, Kawamura Y, Suzuki F, Saitoh S, Arase Y, Muraishi N, et al. Dynamics of circulating miR-122 predict liver cancer and mortality in Japanese patients with histopathologically confirmed NAFLD and severe fibrosis stage. Oncology. (2022) 100:31–8. doi: 10.1159/000519995
18. O'Brien J, Hayder H, Zayed Y, Peng C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol. (2018) 9:402. doi: 10.3389/fendo.2018.00402
19. Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Target Ther. (2016) 1:15004. doi: 10.1038/sigtrans.2015.4
20. Su Q, Kumar V, Sud N, Mahato RI. MicroRNAs in the pathogenesis and treatment of progressive liver injury in NAFLD and liver fibrosis. Adv Drug Deliv Rev. (2018) 129:54–63. doi: 10.1016/j.addr.2018.01.009
21. Morishita A, Oura K, Tadokoro T, Fujita K, Tani J, Kobara H, et al. MicroRNAs and nonalcoholic steatohepatitis: a review. Int J Mol Sci. (2023) 24:14482. doi: 10.3390/ijms241914482
22. Ambade A, Satishchandran A, Szabo G. Alcoholic hepatitis accelerates early hepatobiliary cancer by increasing stemness and miR-122-mediated HIF-1α activation. Sci Rep. (2016) 6:21340. doi: 10.1038/srep21340
23. Cheung O, Puri P, Eicken C, Contos MJ, Mirshahi F, Maher JW, et al. Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology. (2008) 48:1810–20. doi: 10.1002/hep.22569
24. Rusu I, Pirlog R, Chiroi P, Nutu A, Puia VR, Fetti AC, et al. The implications of noncoding RNAs in the evolution and progression of nonalcoholic fatty liver disease (NAFLD)-related HCC. Int J Mol Sci. (2022) 23:12370. doi: 10.3390/ijms232012370
25. Pelizzaro F, Cardin R, Sartori A, Imondi A, Penzo B, Aliberti C, et al. Circulating microRNA-21 and microRNA-122 as prognostic biomarkers in hepatocellular carcinoma patients treated with transarterial chemoembolization. Biomedicines. (2021) 9:890. doi: 10.3390/biomedicines9080890
26. Fang Y, Yan D, Wang L, Zhang J, He Q. Circulating microRNAs (miR-16, miR-22, miR-122) expression and early diagnosis of hepatocellular carcinoma. J Clin Lab Anal. (2022) 36:e24541. doi: 10.1002/jcla.24541
27. Zhang X, Wang X, Wu J, Peng J, Deng X, Shen Y, et al. The diagnostic values of circulating miRNAs for hypertension and bioinformatics analysis. Biosci Rep. (2018) 38:BSR20180525. doi: 10.1042/BSR20180525
28. Farhan M, Wang H, Gaur U, Little PJ, Xu J, Zheng W, et al. FOXO signaling pathways as therapeutic targets in cancer. Int J Biol Sci. (2017) 13:815–27. doi: 10.7150/ijbs.20052
29. Gui T, Burgering BMT. FOXOs: masters of the equilibrium. FEBS J. (2022) 289:7918–39. doi: 10.1111/febs.16221
30. Pan X, Zhang Y, Kim H-G, Liangpunsakul S, Dong XC. FOXO transcription factors protect against the diet-induced fatty liver disease. Sci Rep. (2017) 7:44597. doi: 10.1038/srep44597
31. Webb AE, Brunet A. FOXO transcription factors: key regulators of cellular quality control. Trends Biochem Sci. (2014) 39:159–69. doi: 10.1016/j.tibs.2014.02.003
32. Zhang W, Bu SY, Mashek MT, O-Sullivan I, Sibai Z, Khan SA, et al. Integrated regulation of hepatic lipid and glucose metabolism by adipose triacylglycerol lipase and FoxO proteins. Cell Rep. (2016) 15:349–59. doi: 10.1016/j.celrep.2016.03.021
33. Zhang K, Li L, Qi Y, Zhu X, Gan B, DePinho RA, et al. Hepatic suppression of foxo1 and Foxo3 causes hypoglycemia and hyperlipidemia in mice. Endocrinology. (2012) 153:631–46. doi: 10.1210/en.2011-1527
34. Cao Q, Zhu X, Zhai X, Ji L, Cheng F, Zhu Y, et al. Leptin suppresses microRNA-122 promoter activity by phosphorylation of foxO1 in hepatic stellate cell contributing to leptin promotion of mouse liver fibrosis. Toxicol Appl Pharmacol. (2018) 339:143–50. doi: 10.1016/j.taap.2017.12.007
35. Hu Y, Peng X, Du G, Zhang Z, Zhai Y, Xiong X, et al. MicroRNA-122-5p inhibition improves inflammation and oxidative stress damage in dietary-induced non-alcoholic fatty liver disease through targeting FOXO3. Front Physiol. (2022) 13:803445. doi: 10.3389/fphys.2022.803445
36. Tu S, Qiu Y. Molecular subtypes and scoring tools related to Foxo signaling pathway for assessing hepatocellular carcinoma prognosis and treatment responsiveness. Front Pharmacol. (2023) 14:1213506. doi: 10.3389/fphar.2023.1213506
37. Jiang J, Liu Z, Ge C, Chen C, Zhao F, Li H, et al. NK3 homeobox 1 (NKX3.1) up-regulates forkhead box O1 expression in hepatocellular carcinoma and thereby suppresses tumor proliferation and invasion. J Biol Chem. (2017) 292:19146–59. doi: 10.1074/jbc.M117.793760
38. Qin M, Ren X, Zhang M, Chen Z, Shen J. Molecular mechanism of microRNA-mediated hypoglycemic effect of whole grain highland barley. Gene. (2024) 895:148021. doi: 10.1016/j.gene.2023.148021
39. Tan C, Liu S, Tan S, Zeng X, Yu H, Li A, et al. Polymorphisms in microRNA target sites of forkhead box O genes are associated with hepatocellular carcinoma. PLoS ONE. (2015) 10:e0119210. doi: 10.1371/journal.pone.0119210
40. Compagnoni C, Capelli R, Zelli V, Corrente A, Vecchiotti D, Flati I, et al. MiR-182-5p is upregulated in hepatic tissues from a diet-induced NAFLD/NASH/HCC C57BL/6J mouse model and modulates Cyld and Foxo1 expression. Int J Mol Sci. (2023) 24:9239. doi: 10.3390/ijms24119239
41. Cui X, Zhao H, Wei S, Du Q, Dong K, Yan Y, et al. Hepatocellular carcinoma-derived FOXO1 inhibits tumor progression by suppressing IL-6 secretion from macrophages. Neoplasia. (2023) 40:100900. doi: 10.1016/j.neo.2023.100900
42. Guo W, Yao X, Lan S, Zhang C, Li H, Chen Z, et al. Metabolomics and integrated network pharmacology analysis reveal SNKAF decoction suppresses cell proliferation and induced cell apoptisis in hepatocellular carcinoma via PI3K/Akt/P53/FoxO signaling axis. Chin Med. (2022) 17:76. doi: 10.1186/s13020-022-00628-1
43. Wang Q, Yu W-N, Chen X, Peng X-D, Jeon S-M, Birnbaum MJ, et al. Spontaneous hepatocellular carcinoma after the combined deletion of Akt isoforms. Cancer Cell. (2016) 29:523–35. doi: 10.1016/j.ccell.2016.02.008
44. Carbajo-Pescador S, Mauriz JL, García-Palomo A, González-Gallego J. FoxO proteins: Regulation and molecular targets in liver cancer. Curr Med Chem. (2014) 21:1231–46. doi: 10.2174/0929867321666131228205703
45. Sun EJ, Wankell M, Palamuthusingam P, McFarlane C, Hebbard L. Targeting the PI3K/Akt/mTOR pathway in hepatocellular carcinoma. Biomedicines. (2021) 9:1639. doi: 10.3390/biomedicines9111639
46. Buczek ME, Miles AK, Green W, Johnson C, Boocock DJ, Pockley AG, et al. Cytoplasmic PML promotes TGF-β-associated epithelial-mesenchymal transition and invasion in prostate cancer. Oncogene. (2016) 35:3465–75. doi: 10.1038/onc.2015.409
47. Yang J, Li T, Gao C, Lv X, Liu K, Song H, et al. FOXO1 3'UTR functions as a ceRNA in repressing the metastases of breast cancer cells via regulating miRNA activity. FEBS Lett. (2014) 588:3218–24. doi: 10.1016/j.febslet.2014.07.003
48. Celikbilek M, Baskol M, Taheri S, Deniz K, Dogan S, Zararsiz G, et al. Circulating microRNAs in patients with non-alcoholic fatty liver disease. World J Hepatol. (2014) 6:613–20. doi: 10.4254/wjh.v6.i8.613
Keywords: nonalcoholic fatty liver disease, hepatocellular carcinoma, microRNAs, biological functions, signaling pathway, systematic review
Citation: Peng Q, Zhu X, Jiang Y, Peng M, Zheng D, Wang X, Cheah YK and Wang J (2024) Evaluating the roles of microRNAs associated with nonalcoholic fatty liver disease in hepatocellular carcinoma tumorigenesis: a systematic review and network analysis. Front. Med. 11:1462513. doi: 10.3389/fmed.2024.1462513
Received: 11 July 2024; Accepted: 28 October 2024;
Published: 15 November 2024.
Edited by:
Ana Sandoval-Rodriguez, University of Guadalajara, MexicoReviewed by:
Rebeca Escutia Gutiérrez, University of Guadalajara, MexicoRebeca Rosas Campos, University of Guadalajara, Mexico
Copyright © 2024 Peng, Zhu, Jiang, Peng, Zheng, Wang, Cheah and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Wang, bHl3ajY4QDEyNi5jb20=; Yoke Kqueen Cheah, eWtjaGVhaEB1cG0uZWR1Lm15
†These authors have contributed equally to this work
 Qinghua Peng
Qinghua Peng Xiaoning Zhu
Xiaoning Zhu Yuanyuan Jiang1,2
Yuanyuan Jiang1,2 Xiaodong Wang
Xiaodong Wang Yoke Kqueen Cheah
Yoke Kqueen Cheah Jing Wang
Jing Wang