- 1Department of Infectious Diseases, People’s Hospital of Xiushan County, Xiushan, China
- 2Department of Tuberculosis Diseases, Chongqing Public Health Medical Center, Chongqing, China
- 3Centers for Disease Control and Prevention of Xiushan County, Xiushan, China
- 4Department of Clinical Laboratory, People’s Hospital of Xiushan County, Xiushan, China
The gram-negative bacterium Vibrio cholerae (VC) is divided into multiple serogroups, with groups O1 and O139 responsible for cholera. Conversely, Vibrio cholerae belonging to the non-O1/non-O139 group (NOVC) does not produce cholera-causing toxins. Insufficient understanding of the frequency of NOVC causes fear during the early detection phase. Acute gastroenteritis is often caused by NOVC, while extra gastrointestinal infections are less common. In the case described here, the patient had a postoperative choledochoduodenal fistula due to prior choledochotomy. In August 2023, he was hospitalized with fever and diarrhea. The gram-negative bacilli Vibrio cholerae was isolated from a blood specimen using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). The strain was identified as non-O1/O139 by serum agglutination tests. Subsequent whole-genome sequencing and database analysis revealed that the strain possessed resistance genes such as CRP, varG, almG, and QnrVC4, as well as various virulence factors such as RTX, hlyA, VAS, and T3SS. The phylogenetic tree analysis indicated that CQ23-0008VC had close relationship with cholerae strains isolated from aquatic environments. The patient was treated promptly and discharged after being admitted with severe symptoms. However, Bioinformatics analysis indicated that the virulence factors that were identified in the bacteria were significant; thus, these virulence factors can indicate to medical professionals that a patient could have a septicemia caused by NOVC.
1 Introduction
The severe infectious diarrheal illness known as cholera is caused by the O1 and O139 strains of Vibrio cholerae (1). Although NOVC primarily leads to intestinal infections, it can also result in parenteral infections, particularly septicemias, in individuals with compromised immune systems. Vibrio cholerae can induce diarrhea resembling cholera. Although the exact etiology is unknown, several case reports indicate that individuals with neoplasia, diabetes mellitus, and chronic liver disease have higher rates of NOVC septicemias (2–5). Thus, highlighting the high fatality rate associated with these parenteral septicemias is crucial, as they frequently lead to severe complications such as infectious shock and multiorgan failure (6).
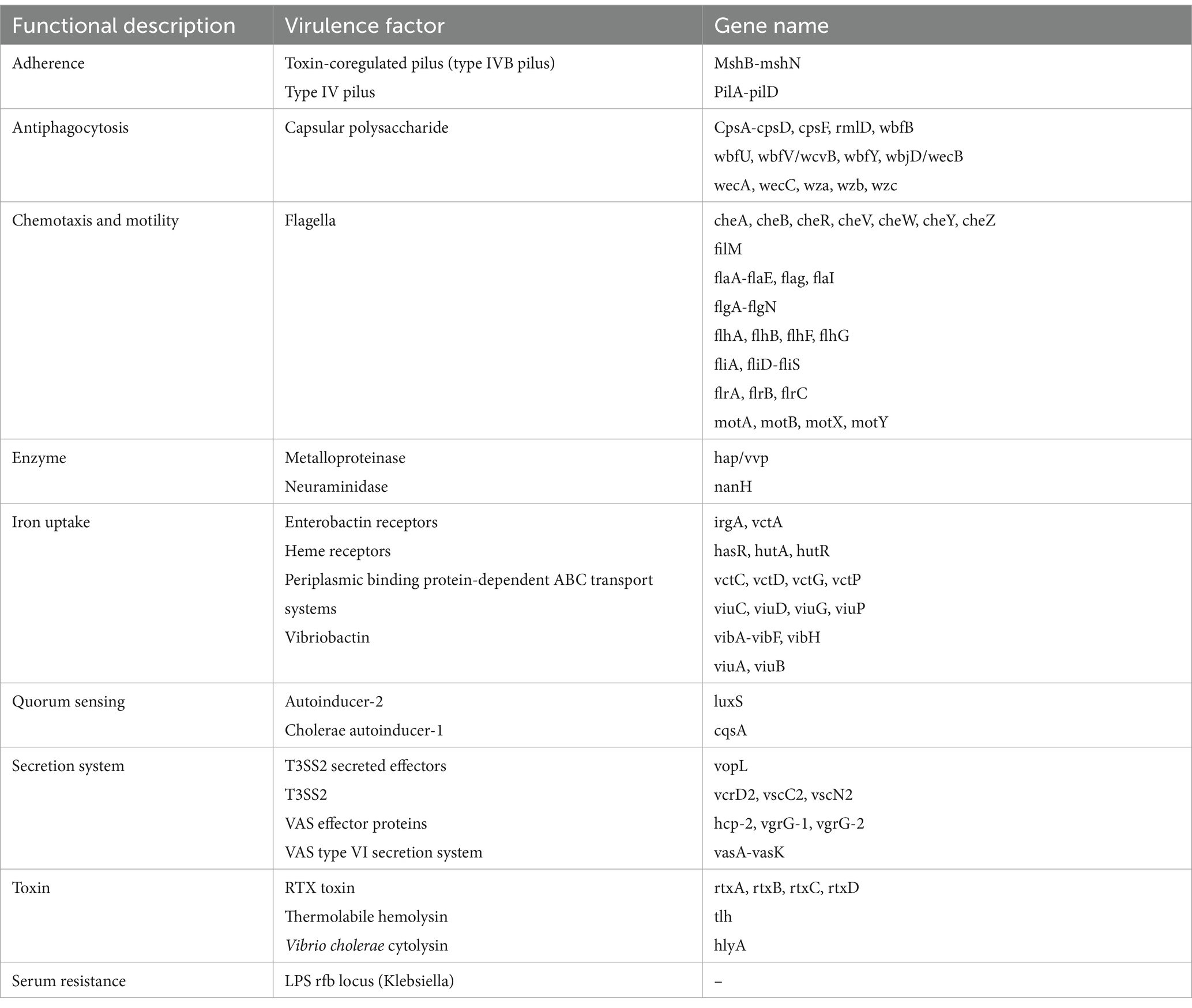
The etiology of NOVC remains unknown; however, virulence gene analysis in published cases revealed significant virulence factors, including gene clusters for hlyA, toxR, hapA, rtxA/rtxC, nanH, zot, stn, ompU, luxS, the type VI secretion system (T6SS), and the type III secretion system (T3SS) (7–9). Variations in the distribution of these virulence factors are directly linked to the pathogenicity of NOVC and its various clinical manifestations (10). In addition, sepsis is relatively rare in patients with Choledochoduodenal fistula. The present study describes the whole sequence of an isolate of NOVC from a choledochoduodenal fistula patient with septicemia.
2 Materials and methods
2.1 Strain isolation and identification
Blood samples from a patient with a choledochoduodenal fistula were collected for strain identification. The strain was subjected to gram staining and examined under a microscope. Subsequently, the strain was transferred to a blood plate and incubated for 16 h. Individual colonies were isolated for serological and biochemical identification.
2.2 Antibiotic susceptibility test
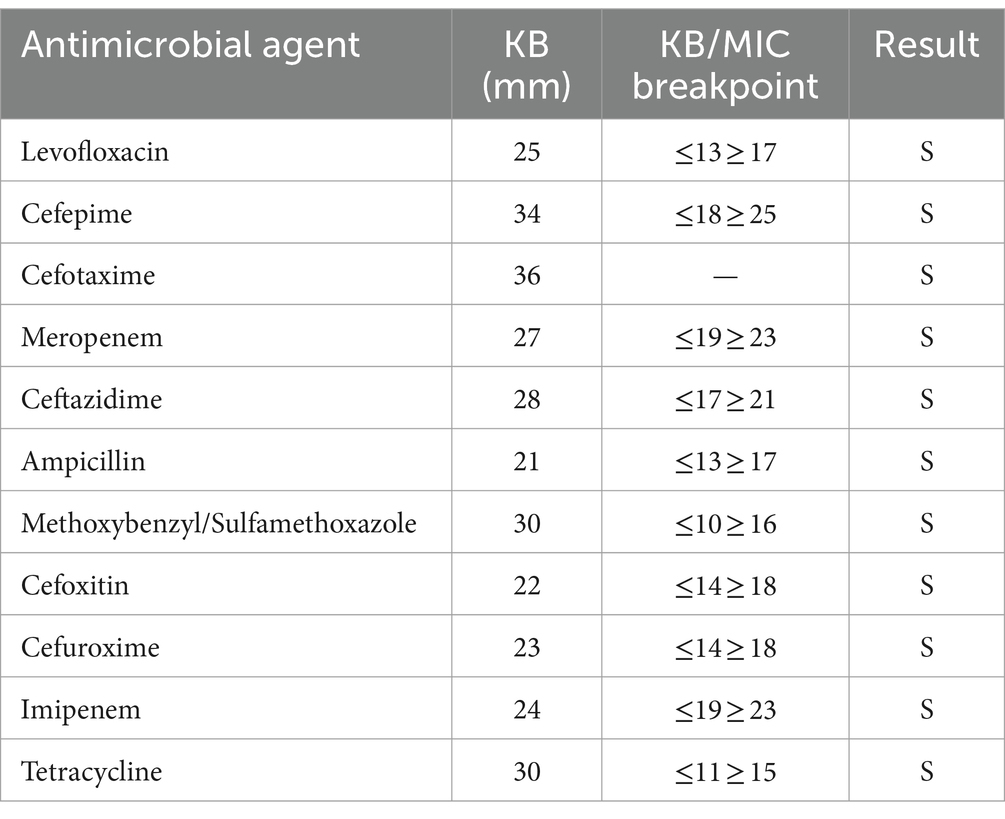
The drug susceptibility test was performed using the Kirby-Bauer (K-B) method recommended by the National Clinical Laboratory Procedures (China). The 2016 edition of the American Committee for Standardization of Clinical Laboratories’ Drug Susceptibility Test Standards was used to evaluate the results (11). The findings indicated sensitivity to the following drugs: levofloxacin, tetracycline, cefoxitin, cefuroxime, imipenem, meropenem, ampicillin, ceftazidime, cefotaxime, cefepime and cotrimoxazole.
2.3 Whole-genome sequencing and gene function analysis
Sequencing was performed using the MGISEQ200RS (Beijing Genomics Institute, China) platform with Illumina PE100 technology for bipartite sequencing. The MGAP (Beijing Genomics Institute, China) platform was used for sequence assembly and splicing. Average nucleotide identity (ANI) analysis and strain identification were conducted on the assembled sequences using the pubMLST database (https://pubmlst.org/species-id) and the ANI calculator (Environmental Microbial Genomics Laboratory gatech.edu). Gene sequences were compared to the KEGG, COG, CAZyme, VFDB, and CARD databases using Prokka software (version 1.14.6) to obtain genome annotation information. The final assembled complete genome was uploaded to the NCBI database (www.ncbi.nlm.nih.gov) under accession number JBCQWS000000000.
2.4 Phylogenetic tree construction
To understand the possible origin of CQ23-0008VC bacteria about the genetic evolution of different vibrio, a phylogenetic tree was constructed through the Mega software (version 11) according to the. V. cholerae O1 biovar EI Tor str. L-3226 (access number: GCA_000600255.1) served as the reference strain, the genome of Vibrio was retrieved from the NCBI database (https://www.ncbi.nlm. nih.gov).
3 Results
3.1 Case presentation
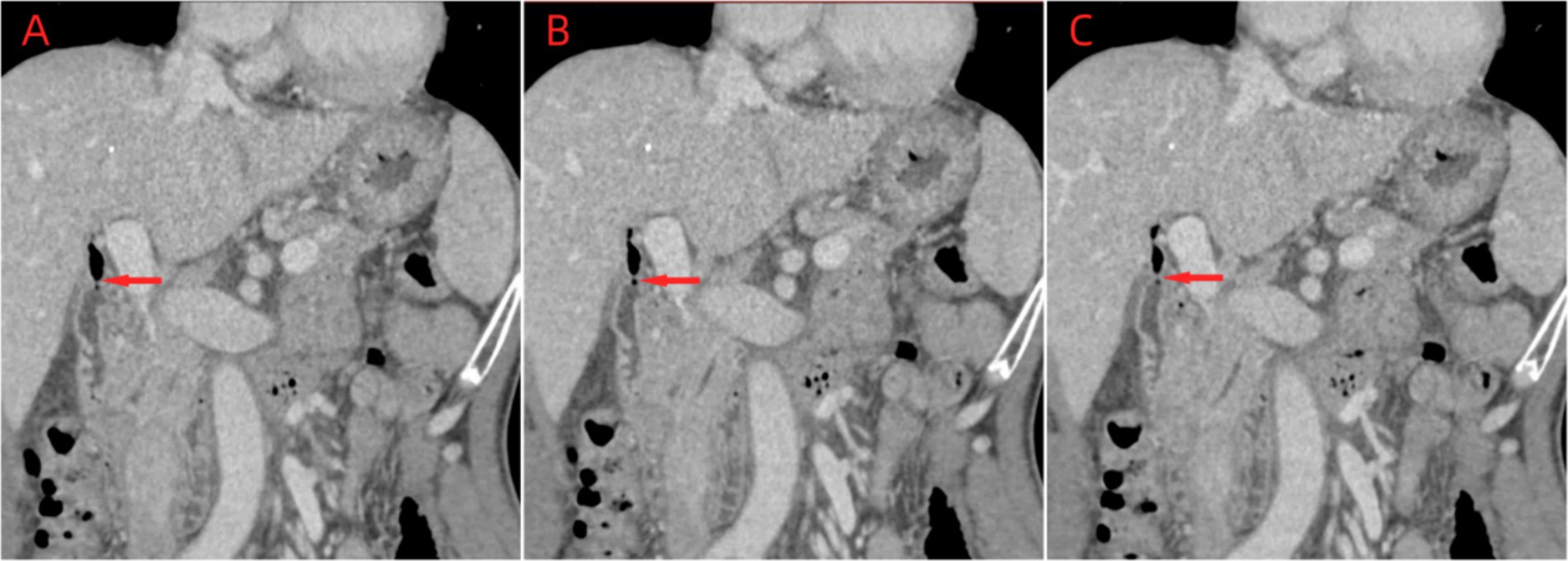
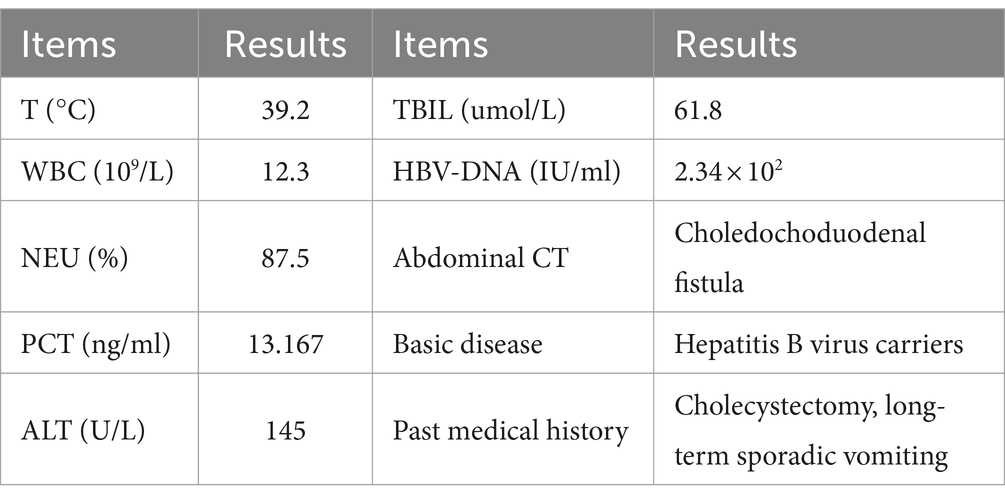
In August 2023, a 64-year-old male patient who experienced chills, fever, nausea, vomiting, diarrhea, and altered stomach contents. He had undergone a cholecystectomy 30 years prior, suffered from long-term sporadic vomiting, and maintained a regular diet. Enhanced computed tomography (CT) of the abdomen revealed Choledochoduodenal fistula (Figure 1). On the night of admission, the patient presents with chills and fever. A blood culture was obtained, the patient was treated with ceftazidime (2 g every 8 h) for anti-infection. On August 7, blood culture revealed the growth of Gram negative bacteria, which were identified as Vibrio cholerae and transferred to blood agar plates. The strain was identified as V. cholerae of the non-O1/non-O139 group, which did not produce the virulence factors ctxAB and tcpA. On August 8, the strain was sent to the Center for Disease Control and Prevention (CDC) of Chongqing for gene sequencing. Following an 8-day course of ceftazidime anti-infection therapy, the patient was cured and discharged from the hospital. Prior to illness, the patient had no contact with other patients with diarrhea or with sea or river water. The main clinical features and laboratory tests during the hospitalization of this patient are presented in Table 1.
3.2 Strain isolation and identification
After sixteen hours, blood cultures were positive for gram-negative bacteria upon microscopic examination (Figure 2). Within a day, β-hemolytic colonies appeared on the transgenic blood agar plate. MALDI-TOF MS confirmed the identification of this strain as V. cholerae. The local CDC identified the bacterium as a non-O1, non-O139 V. cholerae strain, designated CQ23-0008VC, after O1 and O139 serum agglutination tests returned negative results.
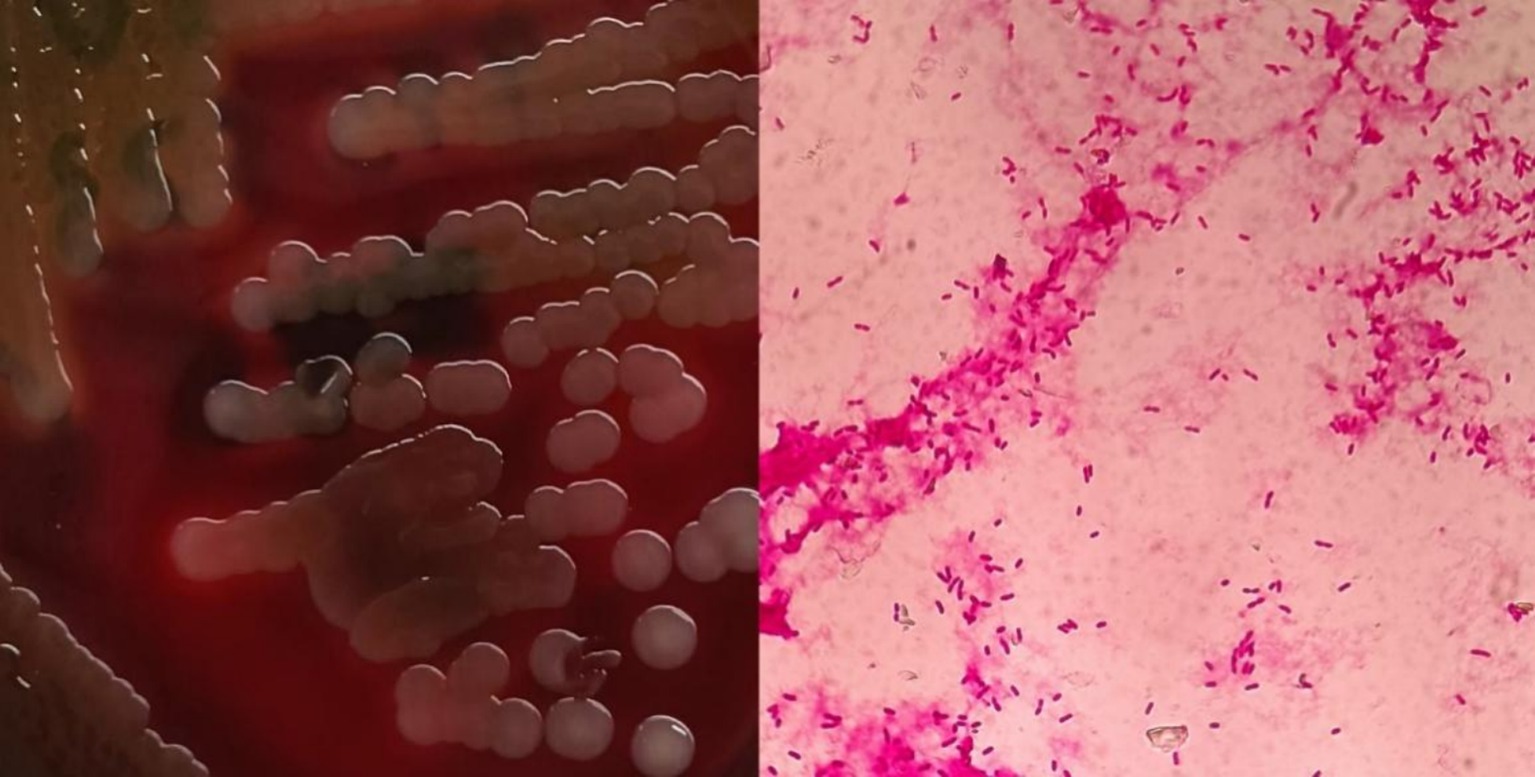
Figure 2. Blood agar plate and morphological characteristics were observed under microscopy after gram staining.
3.3 Genome assembly and genome annotation information analysis
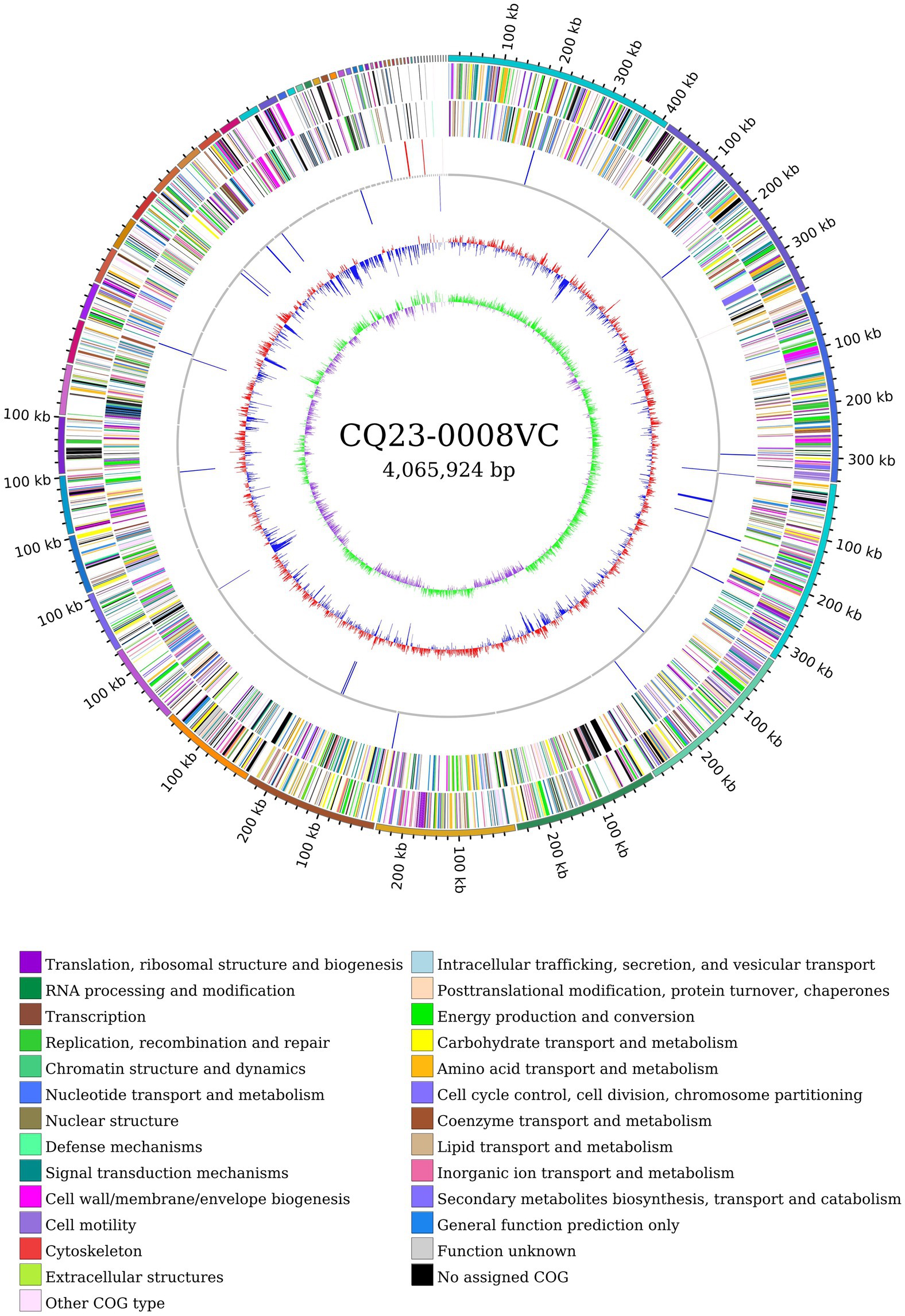
Following sequencing, the original CQ23-0008VC sequence was subjected to quality control, assessment, assembly, and circular genome mapping using Circos software (Figure 3). The CQ23-0008VC genome was estimated to comprise 4,065,924 bp, encoding 3,664 genes with a 47.48% GC content. The genome included 76 tRNAs, 7 rRNAs, and 1 tmRNA among the noncoding RNAs. The sequences were posted to the PubMLST website’s rMLST database, where they matched 100% to V. cholerae ST680. The identification results revealed alleles encoding adk 2, gyrB 23, mdh 15, metE 9, pntA 59, purM 1, and pyrC 35. Using the ANI calculator, 98.37% of the sequences were identified as ANIbs. A total of 3,580 (97. 71%) genes were annotated through comparison with other databases: 2,814 genes were annotated by the COG database, 2,323 genes by the KEGG database, and 186 genes by the VFDB database.
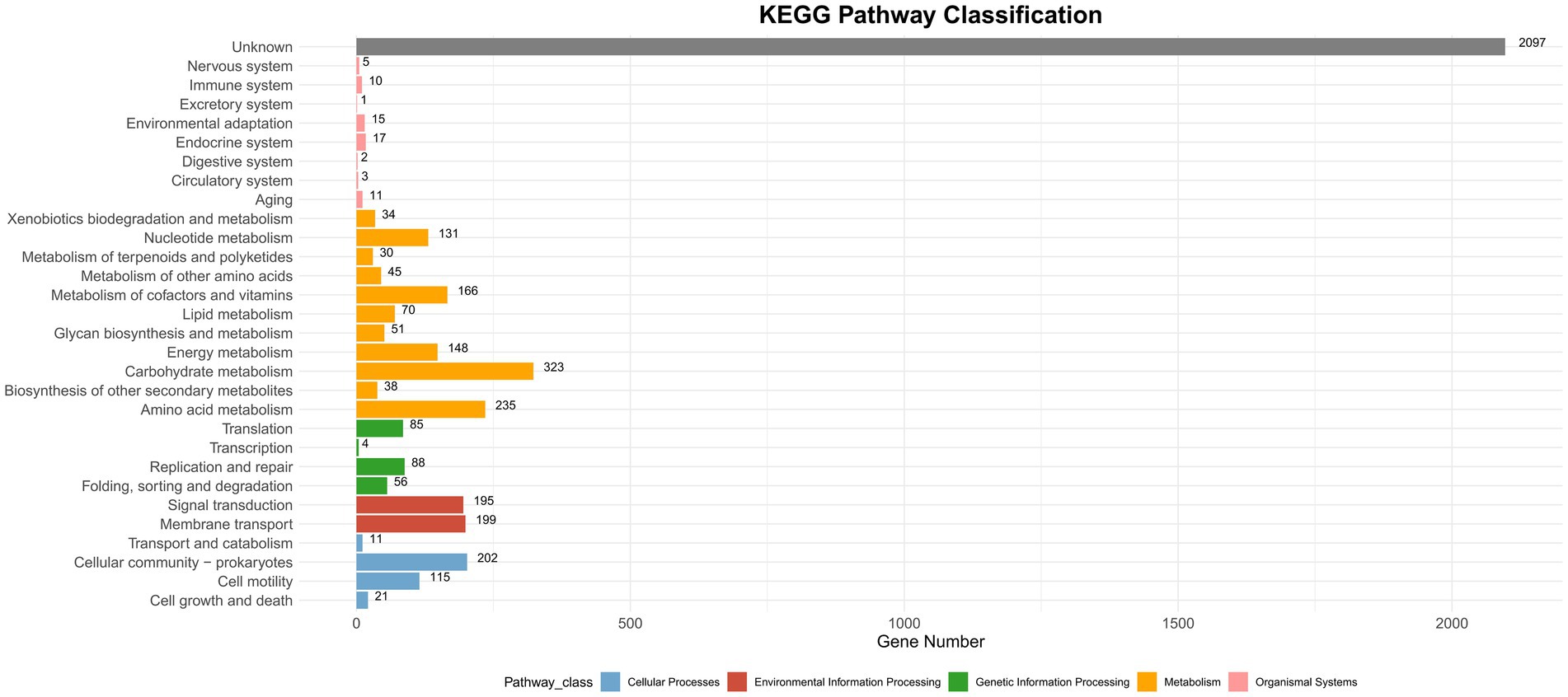
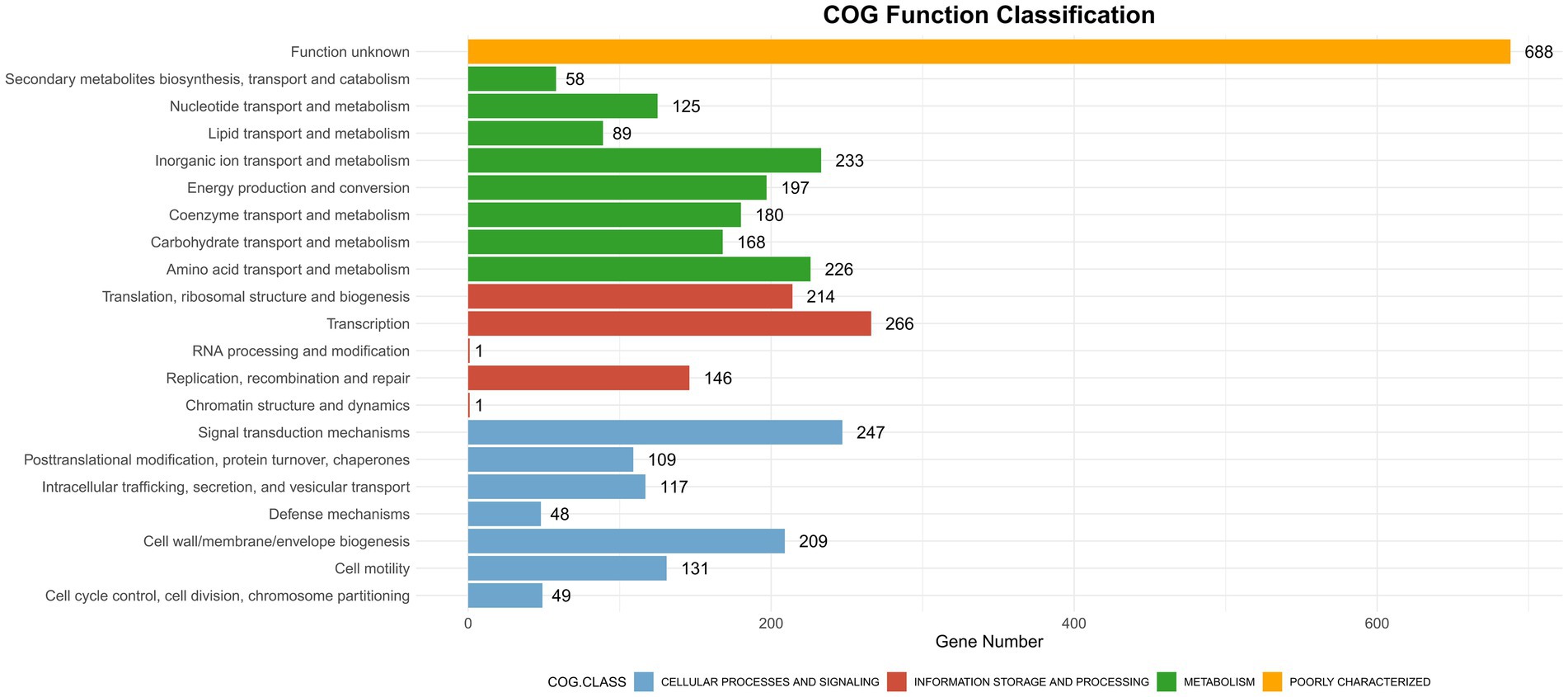
CAZyme database analysis was performed to identify six major categories of carbohydrate-active enzymes in the CQ23-0008VC genome, suggesting that the bacterium had diverse functional capabilities and adaptability. The CQ23-0008VC genome contained 2,323 genes linked to metabolic pathways, which were identified by analysis via the KEGG database. These genes were classified into four categories: metabolism, genetic information processing, cellular motility, and organic systems (Figure 4). Genes were classified into three main functional types and assigned to twenty categories in the COG database. These categories included metabolism, information processing and storage, cellular activities, and signaling. The metabolic pathways of 688 genes remain unknown (Figure 5). Using the VFDB database, genes in the CQ23-0008VC genome that may encode virulence factors were investigated. The genome contained 160 other virulence-related factors, such as rtxA and hlyA, but lacked the CTX virulence factor (Table 2).
3.4 Resistance phenotypes and resistance genes
The strain was found to be sensitive to ampicillin, methotrexate/sulfamethoxazole, cefoxitin, cefuroxime, imipenem, tetracycline, levofloxacin, cefepime, cefotaxime, meropenem, and ceftazidime (Table 3). The CQ23-0008VC genome harbored four resistance genes: varG, almG, QnrVC4, and CRP (Table 4).
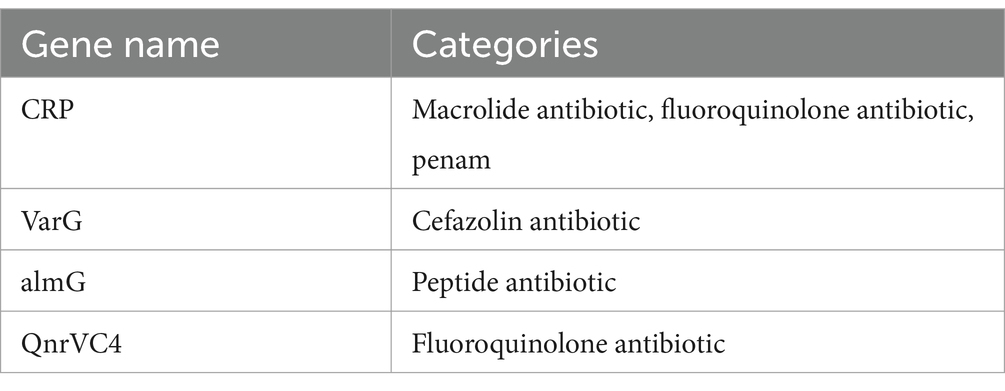
Table 4. The antibiotic resistance genes of the Vibrio cholerae Strain CQ23-0008VC annotated in CARD.
3.5 Phylogenetic tree analysis of Vibrio cholerae CQ23-0008VC
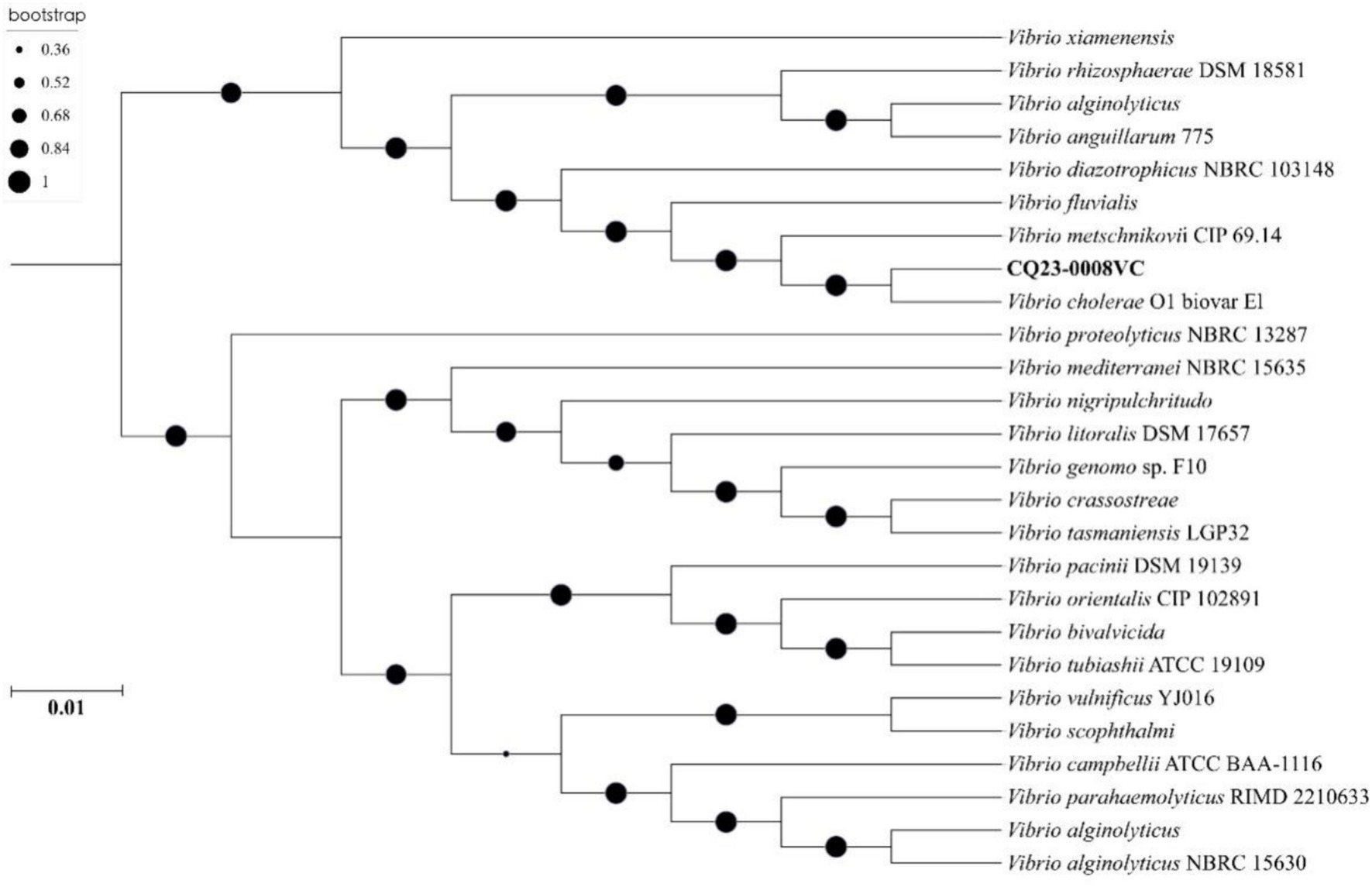
Phylogenetic tree was constructed using nucleotide sequences of these 25 isolates strains from GenBank. The results indicated that CQ23-0008VC was the closest in genetic relationship to a V. cholerae strain O1 biovar EI. Besides, CQ23-0008VC belonged to the same genetic evolutionary lineage as Vibrio, which originates from marine, soil, and river environments (Figure 6).
4 Discussion
Clinicians have started to take NOVC more seriously due to documented cases of extraintestinal infections, despite its typical reputation as a weakly pathogenic or asymptomatic colonizer (12). These infections increasingly include skin infections, meningitis, septicemias, lung infections, and plasma infections. In some instances, these infections can escalate into infectious shock and potentially fatal multiorgan failure (13–16). Given the relatively large hepatopathic population in China, paying sufficient attention to NOVC is crucial as it is more likely to infect immunocompromised patients with chronic liver disease, diabetes mellitus, malignant tumors, and other conditions, often resulting in serious septicemias (17–19).
In 2018, a retrospective analysis of three disseminated cases of NOVC bacteremia by Chinese scholars found that extraintestinal infections of nontoxigenic non-O1/ non-O139 V. cholerae are highlighted in hepatitis B cirrhosis patients (7). Multiple literature reviews have also confirmed that liver diseases, such as hepatic cirrhosis, liver cancer are a risk factor for NOVC infection (10, 20, 21). In this case study, the patient had a choledochoduodenal fistula, which could be the pathological pathway of NOVC invasion, and was also an HBsAg carrier, but the patient did not know that he carried hepatitis B virus before. In addition, this patient had been diagnosed with NOVC infection in August, which is consistent with the time reported in the literature reviews (10). Epidemiological analysis revealed that the patient’s history of pathogenic exposure was unclear. In several instances, there was no obvious source of exposure, and NOVC was not found in other family members or water supplies.
Comparison of the strain’s sequencing data to the KEGG, COG, and CAZyme databases indicated that the strain’s growth mode and metabolic pathways were similar to those of other NOVCs (7, 22). Although the patient’s diarrhea symptoms were not as severe as those caused by cholera, strain CQ23-0008VC carried more virulence factors than did other strains in the VFDB database. This finding could be related to the stimulation of other virulence factors or to dysbiosis of the intestinal flora resulting from septicemia. The clinical presentation and virulence factors reported in this case align more closely with those observed in previous instances of septicemia caused by NOVC (23, 24).
The drug sensitivity results demonstrated that the CQ23-0008VC strain was sensitive to other antimicrobial agents, including quinolones, cephalosporins, and sulfonamides. This strain did not develop resistance to ampicillin or imipenem, which may be related to its origin. Bacterial resistance is closely linked to patient prognosis. According to the CARD database, the resistance genes detected in strain CQ23-0008VC were CRP, varG, almG, and QnrVC4, There were fewer resistance genes in this strain than those found in other reported NOVC strains, which may explain its sensitivity to most antimicrobial agents and suggests variability in resistance rates among different NOVC strains. This result was further supported by a meta-analysis (25). Despite being susceptible to antibiotics such as quinolone, penicillin, aminoglycoside, and macrolides, strain CQ23-0008VC possessed related resistance genes. This phenomenon may be related to the expression levels of resistance genes, their silencing or inactivation, and the genetic backgrounds of the bacteria (26). Therefore, clinicians should consider their assessments on the phenotype characteristics of the strains’ in vito antimicrobial susceptibility, rather than relying solely on evaluations from databases (27, 28). Conversely, when resistance genes are detected in the genome but the isolates show susceptibility in phenotype tests, a critical approach should be taken to interpret the antimicrobial spectrum accurately. Therefore, in vitro drug sensitivity testing should be combined with resistance gene monitoring to choose an antibiotic regimen more precisely and prevent the development of drug-resistant bacteria.
Hemolysin A (HlyA), which is extensively distributed in Vibrio cholerae and has a distinctive structural domain that attaches to target cells, may significantly contribute to the ability of NOVC to infiltrate the bloodstream (29, 30). The distributions of the T3SS and the T6SS differed between strains; comparison with the VFDB database revealed that the strain in this case had a T3SS rather than a T6SS. Determining the difference in bacterial pathogenicity caused by a T3SS or a T6SS will require further investigation. The T3SS/T6SS might increase the virulence of NOVC strains lacking CT and TCP genes, potentially explaining the severity of some septicemias (31–33). Numerous virulence factors work together during parenteral infection with NOVC, thereby necessitating further in-depth molecular biology research to fully comprehend the pathogenic mechanisms involved in parenteral infection caused by NOVC.
5 Conclusion
This study shows that NOVC carries multiple virulence factors and resistance genes and is potentially pathological in humans. Although there is an inconsistency between drug-resistant phenotypes and drug-resistant genes, the monitoring of NOVC resistance genes is helpful for the rational use of antimicrobial drugs. Although NOVC infection is rarely, clinicians should be aware of the possibility of infection with this pathogen in patients with hepatic and biliary tract diseases. Therefore, parenteral infections caused by NOVC should be taken more seriously.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, JBCQWS000000000.
Ethics statement
The studies involving humans were approved by the Ethics Committee of People’s Hospital of Xiushan County. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. Written informed consent was obtained from the participant/patient(s) for the publication of this case report.
Author contributions
WY: Project administration, Writing – original draft, Conceptualization, Formal analysis, Resources. PL: Formal analysis, Supervision, Writing – review & editing. SoL: Investigation, Methodology, Writing – original draft. YY: Conceptualization, Data curation, Visualization, Writing – original draft. ShL: Methodology, Software, Validation, Visualization, Writing – original draft. CY: Conceptualization, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
The authors would like to thank Peibo Li for technical assistance during the experiments.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Clemens, JD, Nair, GB, Ahmed, T, Qadri, F, and Holmgren, J. Cholera. Lancet. (2017) 390:1539–49. doi: 10.1016/S0140-6736(17)30559-7
2. Lu, B, Zhou, H, Li, D, Li, F, Zhu, F, Cui, Y, et al. The first case of bacteraemia due to non-O1/non-O139 Vibrio cholerae in a type 2 diabetes mellitus patient in mainland China. Int J Infect Dis. (2014) 25:116–8. doi: 10.1016/j.ijid.2014.04.015
3. Yang, C-J, Wang, C-S, Lu, P-L, Chen, T-C, Chen, Y-H, Huang, M-S, et al. Bullous cellulitis in cirrhotic patients – a rare but life-threatening infection caused by non-O1, non-O139 Vibrio cholerae bacteraemia. J Med Microbiol. (2011) 60:861–2. doi: 10.1099/jmm.0.024497-0
4. Marinello, S, Marini, G, Parisi, G, Gottardello, L, Rossi, L, Besutti, V, et al. Vibrio cholerae non-O1, non-O139 bacteraemia associated with pneumonia, Italy 2016. Infection. (2017) 45:237–40. doi: 10.1007/s15010-016-0961-4
5. Deris, ZZ, Leow, VM, Hassan, WMNW, Lah, NAZN, Lee, SY, Hawa, HS, et al. Non-O1, non-O139 Vibrio cholerae bacteraemia in splenectomised thalassaemic patient from Malaysia. Trop Biomed. (2009) 26:320–5. Available at: https://www.msptm.org/files/320_-_325_Deris_Z_Z.pdf
6. Rodríguez, JY, Duarte, C, Rodríguez, GJ, Montaño, LA, Benítez-Peñuela, MA, Díaz, P, et al. Bacteremia by non-O1/non-O139 Vibrio cholerae: case description and literature review. Biomedica. (2023) 43:323–9. doi: 10.7705/biomedica.6716
7. Jiang, F, Bi, R, Deng, L, Kang, H, Gu, B, and Ma, P. Virulence-associated genes and molecular characteristics of non-O1/non-O139 Vibrio cholerae isolated from hepatitis B cirrhosis patients in China. Int J Infect Dis. (2018) 74:117–22. doi: 10.1016/j.ijid.2018.06.021
8. Li, F, Du, P, Li, B, Ke, C, Chen, A, Chen, J, et al. Distribution of virulence-associated genes and genetic relationships in non-O1/O139 Vibrio cholerae aquatic isolates from China. Appl Environ Microbiol. (2014) 80:4987–92. doi: 10.1128/AEM.01021-14
9. Zhang, Q, Alter, T, Strauch, E, Eichhorn, I, Borowiak, M, Deneke, C, et al. German coasts harbor non-O1/non-O139 Vibrio cholerae with clinical virulence gene profiles. Infect Genet Evol. (2024) 120:105587. doi: 10.1016/j.meegid.2024.105587
10. Li, X, Wu, Y, Sun, X, Ma, J, Li, X, Liu, C, et al. Non-O1/non-O139 Vibrio cholerae bacteraemia in mainland China from 2005 to 2019: clinical, epidemiological and genetic characteristics. Epidemiol Infect. (2020) 148:e186. doi: 10.1017/S0950268820001545
11. Humphries, R, Bobenchik, AM, Hindler, JA, and Schuetz, AN. Overview of changes to the clinical and laboratory standards institute performance standards for antimicrobial susceptibility testing, M100, 31st edition. J Clin Microbiol. (2021) 59:e0021321. doi: 10.1128/JCM.00213-21
12. Wu, M, Zhou, L, and Cao, L. Acute hemorrhagic necrotizing enterocolitis caused by non-O1/non-O139 Vibrio cholerae infection: a case report. Medicine. (2021) 100:e26460. doi: 10.1097/MD.0000000000026460
13. Chen, J, Huang, J, Huang, M, Chen, Z, Chen, A, Yang, J, et al. Two cases of septic shock with different outcomes caused by non-O1/non-O139 Vibrio cholerae isolates. J Int Med Res. (2020) 48:030006052093345. doi: 10.1177/0300060520933459
14. Hao, Y, Wang, Y, Bi, Z, Sun, B, Jin, Y, Bai, Y, et al. A case of non-O1/non-O139 Vibrio cholerae septicemia and meningitis in a neonate. Int J Infect Dis. (2015) 35:117–9. doi: 10.1016/j.ijid.2015.05.004
15. Maraki, S, Christidou, A, Anastasaki, M, and Scoulica, E. Non-O1, non-O139 Vibrio cholerae bacteremic skin and soft tissue infections. Infect Dis. (2016) 48:171–6. doi: 10.3109/23744235.2015.1104720
16. Lin, P-H, Lin, Y-H, Chen, C-C, and Lin, H-C. An uncommon case of necrotizing fasciitis caused by non-O1, non-O139 vibrio cholerae. Asian J Surg. (2023) 46:3829–30. doi: 10.1016/j.asjsur.2023.03.126
17. Shanley, J, Kanj, A, El Zein, S, Tabaja, H, Trzcinski, B, Horman, J, et al. Non-O1, non-O139 Vibrio cholerae bacteremia in an urban academic medical center in the United States. IDCases. (2019) 15:e00527. doi: 10.1016/j.idcr.2019.e00527
18. Kadkhoda, K, Adam, H, Gilmour, MW, and Hammond, GW. Nontoxigenic Vibrio cholerae septicemia in an immunocompromised patient. Case Rep Infect Dis. (2012) 2012:698746. doi: 10.1155/2012/698746
19. Sachu, A, Johnson, D, Thomas, S, and Mathew, R. Non O1/ O139 Vibrio Cholerae septicemia in a patient with hepatocellular carcinoma. Ethiop J Health Sci. (2021) 31:1303–6. doi: 10.4314/ejhs.v31i6.27
20. Deshayes, S, Daurel, C, Cattoir, V, Parienti, J-J, Quilici, M-L, and De La Blanchardière, A. Non-O1, non-O139 Vibrio cholerae bacteraemia: case report and literature review. Springerplus. (2015) 4:575. doi: 10.1186/s40064-015-1346-3
21. Zhang, X, Lu, Y, Qian, H, Liu, G, Mei, Y, Jin, F, et al. Non-O1, non-O139 Vibrio cholerae (NOVC) bacteremia: case report and literature review, 2015–2019. IDR. (2020) 13:1009–16. doi: 10.2147/IDR.S245806
22. Tang, J, Li, S, Zhang, M, Li, F, Tang, Y, and Yang, F. Whole genome analysis of a non-O1, non-O139 Vibrio cholerae detected from human blood in China. IDR. (2023) 16:5453–61. doi: 10.2147/IDR.S420095
23. Yan, D, Sun, M, Xiao, L, Chen, H, Zhang, Q, Zhi, Y, et al. Interspecies interactions of non-O1/O139 Vibrio cholerae and Salmonella typhimurium: a rare coinfection case report. Diagn Microbiol Infect Dis. (2023) 107:116049. doi: 10.1016/j.diagmicrobio.2023.116049
24. Zhao, Y, He, T, Tu, B, Mao, X, Jiang, J, Jiang, X, et al. Death in a farmer with underlying diseases carrying Vibrio cholerae non-O1/non-O139 producing zonula occludens toxin. Int J Infect Dis. (2022) 120:83–7. doi: 10.1016/j.ijid.2022.04.020
25. Wu, Q, Vaziri, AZ, Omidi, N, Hassan Kaviar, V, Maleki, A, Khadivar, P, et al. Antimicrobial resistance among clinical Vibrio cholerae non-O1/non-O139 isolates: systematic review and meta-analysis. Pathogens Global Health. (2023) 117:235–44. doi: 10.1080/20477724.2022.2114620
26. Das, B, Verma, J, Kumar, P, Ghosh, A, and Ramamurthy, T. Antibiotic resistance in Vibrio cholerae: understanding the ecology of resistance genes and mechanisms. Vaccine. (2020) 38 Suppl 1:A83–92. doi: 10.1016/j.vaccine.2019.06.031
27. Nahar, N, and Rashid, RB. In silico assessment of the genotypic distribution of virulence and antibiotic resistance genes in Vibrio cholerae. Dhaka Univ J Pharm Sci. (2017) 16:77–85. doi: 10.3329/dujps.v16i1.33385
28. Lepuschitz, S, Baron, S, Larvor, E, Granier, SA, Pretzer, C, Mach, RL, et al. Phenotypic and genotypic antimicrobial resistance traits of Vibrio cholerae non-O1/non-O139 isolated from a large Austrian Lake frequently associated with cases of human infection. Front Microbiol. (2019) 10:2600. doi: 10.3389/fmicb.2019.02600
29. Al-Tawfiq, JA, Chopra, H, Dhama, K, Sah, R, Schlagenhauf, P, and Memish, ZA. The cholera challenge: how should the world respond? New Microbes New Infect. (2023) 51:101077. doi: 10.1016/j.nmni.2022.101077
30. Ruenchit, P, Reamtong, O, Siripanichgon, K, Chaicumpa, W, and Diraphat, P. New facet of non-O1/non-O139 Vibrio cholerae hemolysin a: a competitive factor in the ecological niche. FEMS Microbiol Ecol. (2017) 94:1–12. doi: 10.1093/femsec/fix113
31. Chaand, M, Miller, KA, Sofia, MK, Schlesener, C, Weaver, JWA, Sood, V, et al. Type 3 secretion System Island encoded proteins required for colonization by non-O1/non-O139 serogroup Vibrio cholerae. Infect Immun. (2015) 83:2862–9. doi: 10.1128/IAI.03020-14
32. Santoriello, F, and Pukatzki, S. Type VI secretion systems: environmental and intra-host competition of Vibrio cholerae. Adv Exp Med Biol. (2023) 1404:41–63. doi: 10.1007/978-3-031-22997-8_3
Keywords: Vibrio cholerae, bioinformatics analysis, choledochoduodenal fistula, virulence factor, septicemia
Citation: Yang W, Li P, Lei S, Yu Y, Liu S and You C (2024) Case report: Characterization and bioinformatics analysis of non-O1/O139 Vibrio cholerae strain isolated from a choledochoduodenal fistula patient with septicemia. Front. Med. 11:1458143. doi: 10.3389/fmed.2024.1458143
Edited by:
Ansel Hsiao, University of California, Riverside, United StatesReviewed by:
Arkaprabha Banerjee, University of Pennsylvania, United StatesAlejandro A. Hidalgo, Andres Bello University, Chile
Copyright © 2024 Yang, Li, Lei, Yu, Liu and You. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chengdong You, dW5pcXVleWNkQGhvdG1haWwuY29t
 Wei Yang1
Wei Yang1 Chengdong You
Chengdong You