- 1Medical School, Huanghe Science and Technology College, Zhengzhou, China
- 2Jice Medical Institute, Xi’an, Shaanxi, China
Background: Many studies have shown that cytokines play an important role in the pathogenesis of asthma, but their biological effects on asthma remain unclear. The Mendelian randomization (MR) method was used to evaluate the causal relationship between various cytokines [such as interleukins (ILs), interferons (IFNs), tumor necrosis factors (TNFs), colony-stimulating factors (CSFs), transforming growth factor (TGF), etc.,] and asthma.
Methods: In this study, inverse variance weighting was used to evaluate the causal relationship between asthma and cytokines. In addition, the reliability of the results is ensured by multiple methods such as MR-Egger, weighted median, MR-Raps, MR-Presso, and RadialMR, as well as sensitivity analysis.
Results: The results showed that none of the 11 cytokines was associated with the risk of asthma. In contrast, asthma can increase levels of IL-5 [odds ratio (OR) = 1.112, 95% confidence interval (CI): 1.009–1.224, P = 0.032] and IL-9 (OR = 1.111, 95% CI: 1.013–1.219, P = 0.025).
Conclusion: Genetically predicted asthma was positively associated with elevated levels of IL-5 and IL-9, indicating the downstream effects of IL-5 and IL-9 on asthma. Medical treatments can thus be designed to target IL-5 and IL-9 to prevent asthma exacerbations.
1 Introduction
Asthma is a chronic lower respiratory disease that is difficult to cure, with prevalence rates reaching 15% in children and 10% in adults, and cases continue to rise for unknown reasons (1, 2). The clinical symptoms of asthma include recurrent coughing, wheezing, chest tightness, and difficulty breathing, etc., which seriously reduces the patient’s quality of daily life and increases social medical expenses and medical burden of social services (3, 4). Therefore, there is a need for a deeper understanding of the complex molecular mechanisms of asthma to develop personalized treatments that target specific immune mechanisms.
The main features of asthma are airway inflammation and airway hyperresponsiveness. Interventional therapy targeting airway inflammation is one of the main solutions to control asthma. Several cytokines such as interleukins (ILs), interferons (IFNs), tumor necrosis factors (TNFs), colony-stimulating factors (CSFs), and transforming growth factors (TGFs) are associated with the development of airway inflammation (5, 6). Both IL-4 and IL-13 play inflammatory roles in the development of asthma and their expression is regulated. Kotsimbos et al. found that IL-13 and IL-4 were co-expressed and significantly upregulated in the bronchial mucosa of 9 asthma patients and 10 healthy controls (7). IL-4 can exacerbate asthma by inducing B-cell autophagy, promoting immunoglobulin E (IgE) production, and receptor expression (3, 7, 8). Injections of TNF-α into the airways of normal volunteers lead to increased bronchial reactivity, and prospective studies have shown that increased maternal weight leads to increased concentrations of TNF-α in the blood of infants, thereby enhancing the risk of asthma in infants (9). Sun et al. conducted a meta-analysis of 50 studies and found that TNF-α-238G/A, -308G/A, and -857C/T polymorphisms were significantly correlated with the risk of asthma in different ethnic groups (10). IFNs have immunomodulatory properties, and studies have shown that during the onset of asthma, interferon levels in the respiratory tract are significantly changed, and reduced IFN-γ production was associated with the severity of asthma (11–13).
Although studies have demonstrated a close relationship between cytokines and asthma, the causal relationship between the two has not been fully elucidated. In this paper, the causal relationship between asthma and various cytokines (interleukin, interferon, tumor necrosis factor, etc.) has been analyzed by Mendelian randomization (MR), a method of causality assessment by genetic tools, which can provide a better understanding of the mechanism of cytokine action associated with inflammation in asthma. The results lay the theoretical foundation for proposing more targeted and effective therapeutic regimens for better control of patients’ ast.
2 Materials and methods
2.1 Data sources
Genome-wide association study (GWAS) summary datasets for asthma were obtained from the IEU Open GWAS Database.1 We selected for analysis a variety of cytokines that are altered in the onset of asthma, and dominated by ILs, with IL-2, IL-4, IL-5, IL-9, IL-13, IL-17, IL-27, TNF-α, TNF-β, IFN-γ, G-CSF, M-CSF, and TGF-β. Among them, IL-27 and TGF-β were obtained from different GWAS of inflammatory cytokines levels (14, 15). The rest of the cytokines GWAS data were obtained from a multicenter study in Finland. Subjects aged 25–74 years were randomly selected from five regions in Finland. Cytokine data of the subjects were analyzed, and peripheral blood was extracted for expression quantitative trait loci (eQTL) analysis to quantify their gene expression profiles (16). See Supplementary Table 1 for more details.
2.2 Quality control and identifying genetic instruments
To confirm the independence of single nucleotide polymorphism (SNPs), we selected SNPs that exceeded the genome-wide association threshold (p < 5 × 10–6) as independent variables (IVs), because the small number of SNPs met the threshold for significance (p < 5 × 10–8); and SNPs with a physical distance less than 5000 kb and r2< 0.01 were removed to avoid linkage disequilibrium (LD). For SNPs not found in the outcome dataset, proxy-SNPs were used based on the LD-threshold r2 > 0.8, and SNPs that failed to find alternative sites were eliminated. Data were collated so that the effect and other alleles are indeed the same between exposure and outcome datasets.
2.3 MR analysis
This study was a bidirectional MR analysis based on a two-sample MR analysis. The inverse-variance weighted MR method (IVW-MR) was used as the main analysis to estimate the causal effects between cytokines levels and asthma. The IVW-MR method specifically requires that SNPs affect the outcome only through exposure in the study. Although many known confounding SNPs were excluded as much as possible in this study, there are still many unknown confounding factors that may lead to gene pleiotropy and bias for the estimation of effect values. Therefore, we added other methods, include MR-Egger, weighted median, MR-Raps, MR-Presso, and RadialMR, as supplements to test the reliability and stability of the results.
We performed a series of sensitivity tests to ensure that our results were robust. Cochran’s Q test was used to account for possible heterogeneity, with p < 0.05 deeming significantly heterogeneous, and a random-effect IVW model was applied. The horizontal pleiotropy effect was estimated by MR-Eggers. The statistical tests for MR analysis were performed using the “Two-Sample-MR” package (version 0.5.4) in the R environment (R version 4.2). The statistically significant association is defined as p < 0.05.
3 Results
3.1 Influence of cytokines on asthma
All selected cytokines were analyzed using IVW as the main analytical method, and the results are shown in Figure 1 and Supplementary Table 2. Results of the MR analysis showed that genetically determined higher levels of cytokines did not affect the risk of asthma, and the results of IVW were consistent with those of other analysis methods. For the MR-Raps analysis method, some cytokines do not have a P-value, because only five or fewer SNPs are within the screening conditions, making it impossible to perform MR-Raps analysis. In the sensitivity analysis, the MR-Egger intercept test suggested no evidence of pleiotropy (Supplementary Table 4). There was significant heterogeneity in the Cochran’s Q test of IL-2 and IL-27 (Supplementary Table 4), the random-effect model were performed for their IVW analyses, and it supports the existing results.
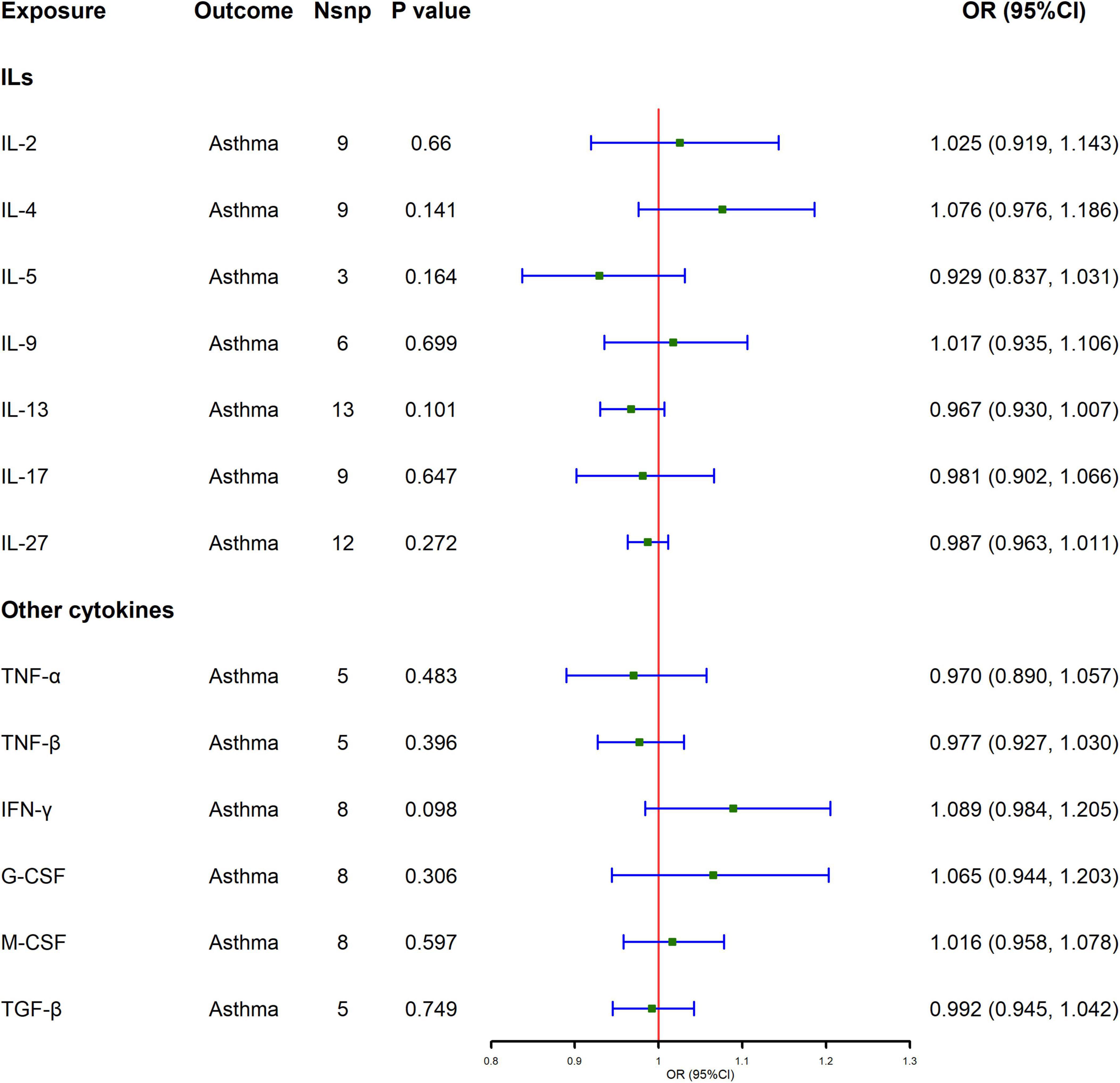
Figure 1. Causal effect of cytokines levels on asthma using Mendelian randomization. Odds ratio (OR) and 95% confidence interval (CI) represent the change in the odds ratio of asthma per 1-SD increase in cytokine level.
3.2 Influence of asthma on cytokines
Figure 2 and Supplementary Table 3 show the MR analysis results of the impact of asthma on the levels of various cytokines. The results show that asthma is significantly correlated with elevated levels of IL-5 and IL-9 [IL-5, odds ratio (OR) = 1.112, 95% confidence interval (CI): 1.009–1.224, P = 0.032; IL-9, OR = 1.111, 95% CI: 1.013–1.219, P = 0.025], and the results of IVW are also supported by Radial-MR. In sensitivity analysis, no evidence of directional pleiotropy was found in the MR-Egger analysis. The test for heterogeneity confirmed that there is no evidence of heterogeneity (Supplementary Table 4).
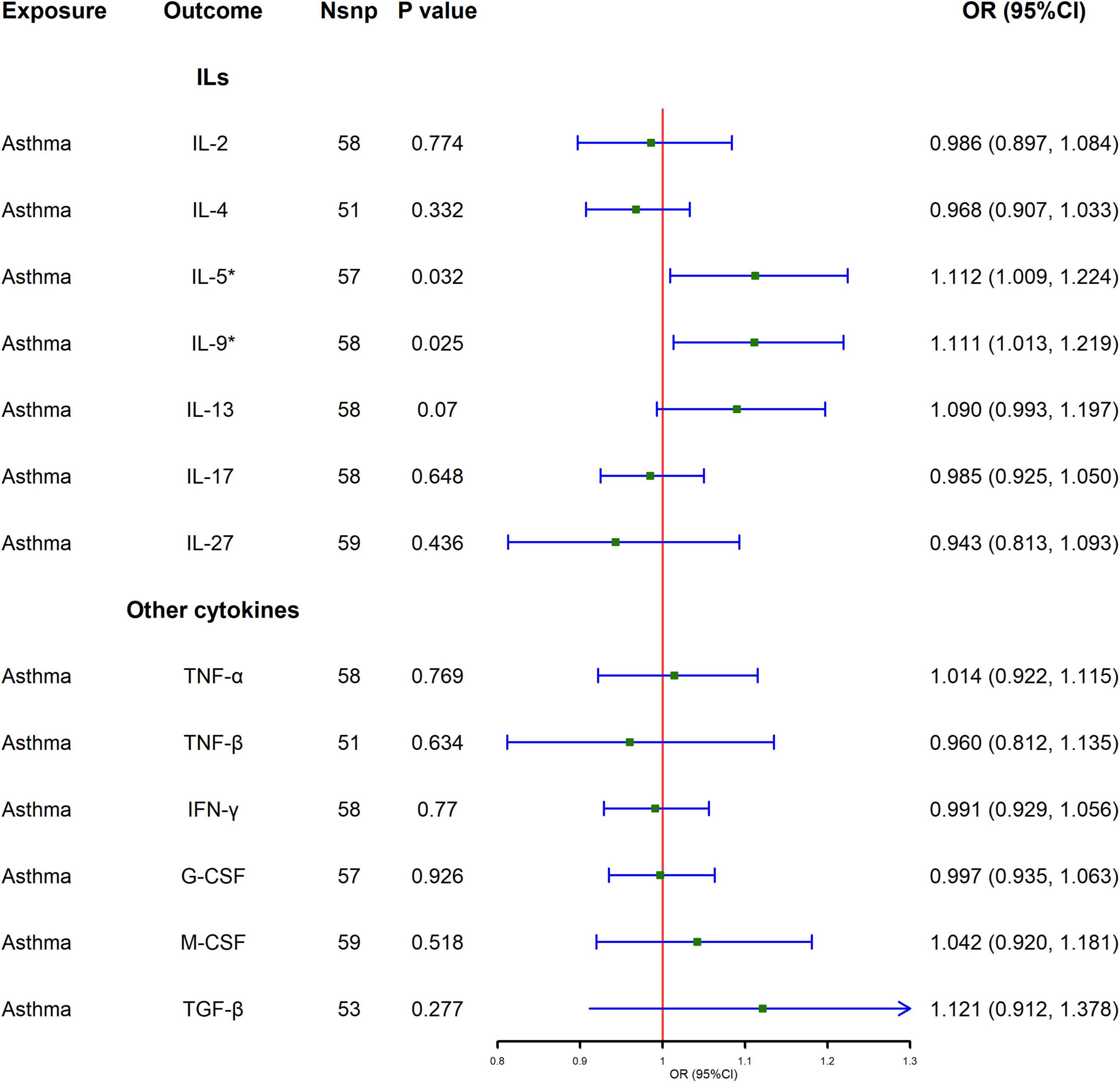
Figure 2. Causal effect of asthma on cytokines levels using Mendelian randomization. Odds ratio (OR) and 95% confidence interval (CI) represent the change in the odds ratio of cytokines level per 1-SD increase in the risk of asthma.
4 Discussion
Since asthma and cytokines are closely related, we investigated the causality between asthma and cytokines through a comprehensive bidirectional MR analysis. The results of the MR analysis showed that asthma increased the levels of IL-5 and IL-9, but did not affect levels of other cytokines. There was no evidence that these cytokines increase or decrease the risk of asthma.
Several observational studies have also demonstrated changes in IL-5 levels during the onset of asthma. Compared to controls, bronchial biopsy results in both early and advanced asthma patients showed increased IL-5 mRNA expression (17, 18). Higher concentrations of IL-5 were detected in the induced sputum of patients with acute exacerbations of asthma, and IL-5 mRNA expression was associated with clinical severity of asthma (17, 19). IL-5 is produced by a variety of immune cells, such as regulating T helper 2 (Th2) cells, Class 2 innate lymphocytes (ILC2), mast cells, natural killer T (NKT) cells, and eosinophils (20). IL-5 levels increase when eosinophilic counts also increase during asthma development. The results of our MR analysis do not support the genetic prediction that IL-5 increases the risk of asthma. The discrepancy may be due to the effect of IL-5 on asthma risk through a complex immune cascade. IL-5 is considered to be the main regulator of eosinophilic increase and activation. IL-5 specifically binds to the receptor subunit IL-5Rα to regulate the differentiation, proliferation, migration, and activation of eosinophils. Upon activation, eosinophils release cytotoxins that induce functional damage to surrounding cells and tissues (21, 22). IL-5 has been identified as a therapeutic target for eosinophilic diseases and several monoclonal antibodies have been approved for clinical use in the treatment of severe asthma, for example, mepolizumab and reslizumab targeting IL-5, and benralizumab targeting the IL-5 receptor (23, 24).
The results of the MR analysis did not support that increased IL-9 levels increased asthma risk. A case-control study of 70 asthmatic patients and 77 healthy control adults aged 18–60 years. The patients’ asthma and its severity were confirmed by medical diagnosis and the pulmonary function test. Serum levels of IL-9 and the SNP of the IL-9 promoter rs2069882 were detected by molecular assay. The results showed that IL-9 serum levels were not significantly associated with asthma severity or atopic type, and the SNP of the IL-9 promoter rs2069882 was not significantly associated with asthma susceptibility (25). Similarly, Waldman and Robinson (26) meta-analysis of linkage studies between asthma and the IL-9 gene showed that the IL-9 gene has little correlation with the pathogenesis of asthma (26). Consistent with MR results, another observational study noted segmental allergen excitation in patients with atopic asthma led to increased expression of IL-9 in lymphocytes in bronchoalveolar lavage fluid (27). Elevated levels of IL-9 have also been detected in asthmatic patients’ lungs, sputum, and serum (28). In addition, elevated mRNA expression levels of IL-9 and IL-9 receptor (IL-9R) were detected in patients with asthma (29). IL-9 is derived primarily from T helper cells, a subgroup of T cells that specialize in producing IL-9, called Th9 cells. IL-9 can also be produced by different immune cells such as mast cells, macrophages, eosinophils, and neutrophils (28–30). The expression of IL-9 can promote the production of IgG and IgE in B-cell lymphocytes, promote the survival and maturation of eosinophils synergistically with IL-5, and activate airway epithelial cells by stimulating the expression of various proteases, chemokines and selective mucins (31, 32).
For other cytokines, there are some differences in our results compared to previous studies showing a strong relationship with asthma. In patients with eosinophilic asthma, the level of CSF in the respiratory tract is significantly increased, causing allergenic sensitization in the lungs, promoting neutrophils proliferation in the bone marrow, and ultimately leading to asthma exacerbation (33). Prophylactic injection of IL-27 reduced the concentration of Th2 cytokines and increased the number of type 1 regulatory T cells in mice, thereby reducing the lung inflammatory environment and improving asthma (34). In this study, there was no evidence that asthma development could increase or decrease the levels of these cytokines, and whether the changes in the levels of these cytokines had an effect on the risk of asthma. This suggests that previous observational findings may be due to environmental influences, complex regulatory mechanisms in the body, or some other unknown mechanism.
In the study, we selected a variety of cytokines whose expression levels are altered in the onset of asthma, as well as data with the large sample sizes used in most of the analyses we conducted. It provides some reference for further study. Secondly, MR analysis can reduce the influence of confounding factors and reverse causality to a certain extent. Both cytokines and asthma-related genetic IVs are derived from European population studies, which can greatly reduce the bias caused by population stratification.
However, there are some limitations to this MR study. Firstly, the samples selected for this paper were patients who were clearly diagnosed with asthma after various tests in hospital visits, there was no sub-type categorization of asthma to allow for in-depth study of a specific asthma group, and the pooled data of multiple phenotypes made it possible for some small bias in the results. Secondly, regulatory mechanisms or cascading responses during growth and development may attenuate the effects of genetic tools. MR analyses may reflect the effects of exposure to elevated cytokines on asthma throughout the human lifespan. However, high levels of cytokines presented only at a certain time of life (e.g., middle age and old age) may contribute to the risk of asthma development. Finally, the study originated from a European population, which may be partially different from other populations and cannot be applied to other Asian populations, African populations, and others. The main reason for selecting a European population is that the sample sizes of relevant GWAS studies in European populations are relatively larger and the data is more comprehensive, making the results more reliable.
5 Conclusion
The MR analysis showed that asthma increased the levels of IL-5 and IL-9, and there was no evidence that cytokines increased or decreased the risk of asthma. Future work could further investigate whether altered levels of IL-5 and IL-9 can be used as biomarkers of asthma development, and further explore the potential mechanism of action of IL-5 and IL-9 in asthma, and evaluate their potential for clinical prevention and treatment of asthma.
Data availability statement
The original contributions presented in this study are included in this article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
YZ: Conceptualization, Project administration, Software, Writing – original draft, Writing – review and editing. QC: Conceptualization, Formal analysis, Investigation, Writing – review and editing. XS: Conceptualization, Data curation, Validation, Writing – review and editing. LL: Data curation, Validation, Visualization, Writing – review and editing. DW: Conceptualization, Resources, Writing – original draft, Writing – review and editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
We thanks for the contributions of the IEU Open GWAS Database for providing high quality GWAS data for researchers.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2024.1447673/full#supplementary-material
Footnotes
References
1. Desai D, Brightling C. Cytokine and anti-cytokine therapy in asthma: Ready for the clinic? Clin Exp Immunol. (2009) 158:10–9. doi: 10.1111/j.1365-2249.2009.03998.x
2. Fang L, Sun Q, Roth M. Immunologic and non-immunologic mechanisms leading to airway remodeling in asthma. Int J Mol Sci. (2020) 21:757. doi: 10.3390/ijms21030757
3. Nur Husna SM, Md Shukri N, Mohd Ashari NS, Wong KK. IL-4/IL-13 axis as therapeutic targets in allergic rhinitis and asthma. PeerJ. (2022) 10:e13444. doi: 10.7717/peerj.13444
4. Izuhara K. The role of interleukin-4 and interleukin-13 in the non-immunologic aspects of asthma pathogenesis. Clin Chem Lab Med. (2003) 41:860–4. doi: 10.1515/CCLM.2003.130
5. Dong H, Hao Y, Li W, Yang W, Gao P. IL-36 cytokines: Their roles in asthma and potential as a therapeutic. Front Immuno.l (2022) 13:921275. doi: 10.3389/fimmu.2022.921275
6. Hudey SN, Ledford DK, Cardet JC. Mechanisms of non-type 2 asthma. Curr Opin Immunol. (2020) 66:123–8. doi: 10.1016/j.coi.2020.10.002
7. Kotsimbos TC, Ernst P, Hamid QA. Interleukin-13 and interleukin-4 are coexpressed in atopic asthma. Proc Assoc Am Physicians. (1996) 108:368–73.
8. Xia F, Deng C, Jiang Y, Qu Y, Deng J, Cai Z, et al. IL4 (interleukin 4) induces autophagy in B cells leading to exacerbated asthma. Autophagy. (2018) 14:450–64. doi: 10.1080/15548627.2017.1421884
9. Halonen M, Lohman IC, Stern DA, Ellis WL, Rothers J, Wright AL. Perinatal tumor necrosis factor-alpha production, influenced by maternal pregnancy weight gain, predicts childhood asthma. Am J Respir Crit Care Med. (2013) 188:35–41. doi: 10.1164/rccm.201207-1265OC
10. Xu L, Liu C, Zheng Y, Huang Y, Zhong Y, Zhao Z, et al. Association of TNF-alpha-308G/A, -238G/A, -863C/A, -1031T/C, -857C/T polymorphisms with periodontitis susceptibility: Evidence from a meta-analysis of 52 studies. Medicine (Baltimore). (2020) 99:e21851. doi: 10.1097/MD.0000000000021851
11. Krammer S, Gutu CS, Grund JC, Chiriac MT, Zirlik S, Finotto S. Regulation and function of interferon-lambda (IFNlambda) and its receptor in asthma. Front Immunol. (2021) 12:731807. doi: 10.3389/fimmu.2021.731807
12. Hansel TT, Tunstall T, Trujillo-Torralbo M-B, Shamji B, Del-Rosario A, Dhariwal J, et al. A comprehensive evaluation of nasal and bronchial cytokines and chemokines following experimental rhinovirus infection in allergic asthma: Increased interferons (IFN-gamma and IFN-lambda) and type 2 inflammation (IL-5 and IL-13). EBioMedicine. (2017) 19:128–38. doi: 10.1016/j.ebiom.2017.03.033
13. Brooks GD, Buchta KA, Swenson CA, Gern JE, Busse WW. Rhinovirus-induced interferon-gamma and airway responsiveness in asthma. Am J Respir Crit Care Med. (2003) 168:1091–4. doi: 10.1164/rccm.200306-737OC
14. Gilly A, Park Y-C, Png G, Barysenka Ai, Fischer I, Bjørnland T, et al. Whole-genome sequencing analysis of the cardiometabolic proteome. Nat Commun. (2020) 11:6336. doi: 10.1038/s41467-020-20079-2
15. Hillary RF, Trejo-Banos D, Kousathanas A, McCartney DL, Harris SE, Stevenson AJ, et al. Multi-method genome- and epigenome-wide studies of inflammatory protein levels in healthy older adults. Genome Med. (2020) 12:60. doi: 10.1186/s13073-020-00754-1
16. Ahola-Olli AV, Würtz P, Havulinna AS, Aalto K, Pitkänen N, Lehtimäki T, et al. Genome-wide association study identifies 27 loci influencing concentrations of circulating cytokines and growth factors. Am J Hum Genet. (2017) 100:40–50. doi: 10.1016/j.ajhg.2016.11.007
17. Principe S, Porsbjerg C, Ditlev SB, Klein DK, Golebski K, Dyhre-Petersen N, et al. Treating severe asthma: Targeting the IL-5 pathway. Clin Exp Allergy. (2021) 51:992–1005. doi: 10.1111/cea.13885
18. Wood LJ, Sehmi R, Dorman S, Hamid Q, Tulic MK, Watson RM, et al. Allergen-induced increases in bone marrow T lymphocytes and interleukin-5 expression in subjects with asthma. Am J Respir Crit Care Med. (2002) 166:883–9. doi: 10.1164/rccm.2108015
19. Nakagome K, Nagata M. Involvement and possible role of eosinophils in asthma exacerbation. Front Immunol. (2018) 9:2220. doi: 10.3389/fimmu.2018.02220
20. Moldaver DM, Larche M, Rudulier CD. An update on lymphocyte subtypes in asthma and airway disease. Chest. (2017) 151:1122–30. doi: 10.1016/j.chest.2016.10.038
21. Kay AB, Klion AD. Anti-interleukin-5 therapy for asthma and hypereosinophilic syndrome. Immunol Allergy Clin North Am. (2004) 2:645–66. doi: 10.1016/j.iac.2004.06.007
22. Stein ML, Munitz A. Targeting interleukin (IL) 5 for asthma and hypereosinophilic diseases. Recent Pat Inflamm Allergy Drug Discov. (2010) 4:201–9. doi: 10.2174/187221310793564290
23. Lombardi C, Comberiati P, Ridolo E, Cottini M, Yacoub MR, Casagrande S, et al. Anti-IL-5 pathway agents in eosinophilic-associated disorders across the lifespan. Drugs. (2024) 84:661–84. doi: 10.1007/s40265-024-02037-0
24. Pelaia C, Crimi C, Vatrella A, Tinello C, Terracciano R, Pelaia G. Molecular targets for biological therapies of severe asthma. Front Immunol. (2020) 11:603312. doi: 10.3389/fimmu.2020.603312
25. Mahdaviani SA, Eskian M, Khorasanizadeh M, Bashardoost B, Nejad ST, Jamaati HR, et al. Interleukin 9 serum level and single nucleotide polymorphism in patients with asthma. Acta Biomed. (2021) 92:e2021206. doi: 10.23750/abm.v92i3.10544
26. Waldman ID, Robinson BF. Meta-analysis of sib pair linkage studies of asthma and the interleukin-9 gene (IL9). Genet Epidemiol. (2001) 21:S109–14. doi: 10.1002/gepi.2001.21.s1.s109
27. Erpenbeck VJ, Hohlfeld JM, Volkmann B, Hagenberg A, Geldmacher H, Braun A, et al. Segmental allergen challenge in patients with atopic asthma leads to increased IL-9 expression in bronchoalveolar lavage fluid lymphocytes. J Allergy Clin Immunol. (2003) 111:1319–27. doi: 10.1067/mai.2003.1485
28. Gong F, Pan YH, Huang X, Zhu HY, Jiang DL. From bench to bedside: Therapeutic potential of interleukin-9 in the treatment of asthma. Exp Ther Med. (2017) 13:389–94. doi: 10.3892/etm.2017.4024
29. Shimbara A, Christodoulopoulos P, Soussi-Gounni A, Olivenstein R, Nakamura Y, Levitt RC, et al. IL-9 and its receptor in allergic and nonallergic lung disease: Increased expression in asthma. J Allergy Clin Immunol. (2000) 105:108–15. doi: 10.1016/s0091-6749(00)90185-4
30. Neurath MF, Finotto S. IL-9 signaling as key driver of chronic inflammation in mucosal immunity. Cytokine Growth Factor Rev. (2016) 29:93–9. doi: 10.1016/j.cytogfr.2016.02.002
31. Louahed J, Toda M, Jen J, Hamid Q, Renauld JC, Levitt RC, et al. Interleukin-9 upregulates mucus expression in the airways. Am J Respir Cell Mol Biol. (2000) 22:649–56. doi: 10.1165/ajrcmb.22.6.3927
32. Gounni AS, Gregory B, Nutku E, Aris F, Latifa K, Minshall E, et al. Interleukin-9 enhances interleukin-5 receptor expression, differentiation, and survival of human eosinophils. Blood. (2000) 96:2163–71.
33. Kim YM, Kim H, Lee S, Kim S, Lee J-U, Choi Y, et al. Airway G-CSF identifies neutrophilic inflammation and contributes to asthma progression. Eur Respir J. (2020) 55:2019. doi: 10.1183/13993003.00827-2019
Keywords: asthma, interleukins, interferons, tumor necrosis factors, Mendelian randomization
Citation: Zheng Y, Chen Q, Shi X, Lei L and Wang D (2024) Causality between various cytokines and asthma: a bidirectional two-sample Mendelian randomization analysis. Front. Med. 11:1447673. doi: 10.3389/fmed.2024.1447673
Received: 12 June 2024; Accepted: 29 July 2024;
Published: 08 August 2024.
Edited by:
Kelly Domvri, Aristotle University of Thessaloniki, GreeceReviewed by:
Antonio Ferrante, South Australia Pathology, AustraliaMaria Paparoupa, University Medical Center Hamburg-Eppendorf, Germany
Copyright © 2024 Zheng, Chen, Shi, Lei and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Donglin Wang, ZGxfd2FuZ0AxNjMuY29t
†These authors have contributed equally to this work
 Yansen Zheng
Yansen Zheng Qi Chen
Qi Chen Xiaqing Shi2
Xiaqing Shi2 Donglin Wang
Donglin Wang